ABSTRACT
Clostridium difficile, recently renamed Clostridioides difficile, is the most common cause of antibiotic-associated nosocomial gastrointestinal infections worldwide. To differentiate endogenous infections and transmission events, highly discriminatory subtyping is necessary. Today, methods based on whole-genome sequencing data are increasingly used to subtype bacterial pathogens; however, frequently a standardized methodology and typing nomenclature are missing. Here we report a core genome multilocus sequence typing (cgMLST) approach developed for C. difficile. Initially, we determined the breadth of the C. difficile population based on all available MLST sequence types with Bayesian inference (BAPS). The resulting BAPS partitions were used in combination with C. difficile clade information to select representative isolates that were subsequently used to define cgMLST target genes. Finally, we evaluated the novel cgMLST scheme with genomes from 3,025 isolates. BAPS grouping (n = 6 groups) together with the clade information led to a total of 11 representative isolates that were included for cgMLST definition and resulted in 2,270 cgMLST genes that were present in all isolates. Overall, 2,184 to 2,268 cgMLST targets were detected in the genome sequences of 70 outbreak-associated and reference strains, and on average 99.3% cgMLST targets (1,116 to 2,270 targets) were present in 2,954 genomes downloaded from the NCBI database, underlining the representativeness of the cgMLST scheme. Moreover, reanalyzing different cluster scenarios with cgMLST were concordant to published single nucleotide variant analyses. In conclusion, the novel cgMLST is representative for the whole C. difficile population, is highly discriminatory in outbreak situations, and provides a unique nomenclature facilitating interlaboratory exchange.
KEYWORDS: Clostridium difficile, cgMLST, whole-genome sequencing, typing
INTRODUCTION
Clostridium difficile, recently renamed Clostridioides difficile, is an anaerobic, Gram-positive, endospore-forming rod-shaped bacterium and the most common cause of antibiotic-associated nosocomial gastrointestinal infections in Europe and the United States (1, 2). Over the last decades, severe C. difficile infections (CDI) have been increasingly detected in hospitals, making C. difficile an important nosocomial pathogen. CDI develop either from endogenous colonization under selecting conditions such as an antibiotic treatment or from an exogenous source, i.e., spores from the contaminated environment (3).
Several methods are described for C. difficile typing, of which PCR ribotyping is currently becoming a gold standard worldwide (1, 4). For an initial grouping of strains, multilocus sequence typing (MLST) (5, 6) and toxinotyping (7) are also widely used methods. For highly discriminatory subtyping of strains, which is necessary in the case of a suspected outbreak, these methods are sometimes complemented with pulsed-field gel electrophoresis (PFGE) or multilocus variable-number tandem-repeat (VNTR) analysis (MLVA) (4); both methods are able to differentiate among closely related isolates. Except for MLST, where a central database hosting the typing nomenclature is in place, the interlaboratory exchange of such typing data is hampered by the lack of a publicly available database ensuring a unique nomenclature and—in the case of PCR ribotyping and PFGE—by difficulties to standardize the interpretation of DNA banding patterns (4, 8).
Nowadays, sequence-based typing approaches using whole-genome sequence (WGS) data are overcoming these obstacles. Several studies on various bacterial species have already shown that WGS-based typing, based either on single nucleotide variants (SNVs) (9, 10) or on gene-by-gene allelic profiling of core genome genes, frequently named core genome MLST (cgMLST) (11–13), currently represents the ultimate tool for strain subtyping. Moreover, it was recently shown in an international ring trial that cgMLST is highly reproducible (14).
For C. difficile, initial studies also confirmed the general applicability of WGS-based typing (9, 15, 16). Nevertheless, the broad use of WGS-based typing of C. difficile is still hampered by the lack of standardized nomenclature (17); this has already been established for other pathogens (18–21) and would facilitate interlaboratory exchange of data.
Therefore, to obtain the basis of a standardized nomenclature for WGS-based C. difficile typing, we defined a novel C. difficile cgMLST scheme covering the genetic diversity within the C. difficile population based on well-characterized reference strains and subsequently challenged this scheme using a diverse set of strains from sporadic cases and outbreak investigations.
MATERIALS AND METHODS
C. difficile strains and genomes.
All strains and genome sequences used for the development of the novel C. difficile cgMLST scheme are listed in Table 1. The isolates were selected by covering the whole diversity of C. difficile organisms, i.e., representative isolates for each clade (downloaded from https://pubmlst.org/cdifficile/) and—based on a Bayesian analysis of the genetic population structure (BAPS; see below) using all available MLST sequence types (STs) as input data—randomly selected representative isolates for each BAPS partition were included (17, 22). The well-defined C. difficile strain 630 (23) was used as the reference sequence during cgMLST target definition. Moreover, the NCBI RefSeq sequences of C. difficile strains CD196 and M120 were used.
TABLE 1.
List of Clostridium difficile isolates and genomes used for cgMLST target definition
| Isolate | Clade | BAPS partition | MLST ST | PCR ribotype | Toxinotypea | NCBI RefSeq/ENA SRA accession no. (reference) |
|---|---|---|---|---|---|---|
| 630 (reference) | 1 | Cd06 | 54 | 012 | 0 | NC_009089 |
| 2402 | 1 | Cd06 | 199 | SLO 086 | XXXIII | ERS2050168 (this study) |
| CD196 (R12087) | 2 | Cd06 | 1 | 027 | IIIb | NC_013315 |
| 8785 | 2 | Cd06 | 196 | 109 | IXc | ERS2050173 (this study) |
| C00007686 | 3 | None | 5 | SAMEA2240504 (39) | ||
| 1470 | 4 | Cd05 | 37 | 017 | VIII | ERS2050166 (this study) |
| M120 | 5 | Cd03 | 11 | 078 | V | NC_017174 (40) |
| SUC36 | 5 | Cd03 | 195 | 078 | XVI | ERS2050188 (this study) |
| 173070 | C-IIb | Cd01 | 200 | 151 | XXXII | ERS2050167 (this study) |
| ZZV13-5576 | C-I | Cd02 | 297 | SLO 229 | Paloc negative | ERR2216002 (this study) |
| ZZV14-6045 | C-IIIb | Cd04 | 343 | SLO 205 | PaLoc negative | ERR2216003 (this study) |
For subsequent evaluation of the scheme, we used two different sets of isolates/genome sequences: first, a total of 70 well-defined C. difficile isolates (Table 2) were used comprising (i) the reference strains of all published toxinotypes (n = 38) to cover the diversity of toxigenic strains (7), (ii) isolates from two published clusters (n = 8) as examples to rule in or out nosocomial transmissions (9), and (iii) isolates detected during a surveillance study for infection control (n = 24) (15). As a second set for evaluation of the cgMLST scheme, we downloaded 268 assembled genome sequences from the NCBI database (ftp://ftp.ncbi.nih.gov/genomes/) and sequence reads (only data generated with any Illumina sequencing platform) from 3,482 C. difficile isolates from the NCBI Sequence Read Archive (SRA) that were available until 21 October 2015 and were assembled prior to use.
TABLE 2.
List of 70 C. difficile isolates and genomes (toxinotypes and cluster/outbreak isolates) for evaluation of the novel cgMLST scheme
| Isolate | Cladea | BAPS partition | MLST STa | PCR ribotypeb | Toxinotypec | % cgMLST targets | NCBI or ENA SRA accession no. (reference) |
|---|---|---|---|---|---|---|---|
| EX623 | 1 | Cd06 | 24 | 102 | I | 99.5 | ERS2039514 (this study) |
| IS 25 | 1 | Cd06 | 58 | 258 | XII | 99.6 | ERS2050180 (this study) |
| IS 58 | 5 | Cd03 | 11 | 033 | XIa | 97.7 | ERS2050181 (this study) |
| J9965 | 2 | Cd06 | 194 | SLO 032 | Xb | 98.7 | ERS2050182 (this study) |
| K095 | 1 | Cd06 | 2 | 014 | XVIII | 99.9 | ERS2039515 (this study) |
| KK2443/2006 | 1 | Cd06 | 19 | SLO 037 | XXVII | 98.9 | ERS2039516 (this study) |
| OCD 5/2 | 5 | Cd03 | 11 | 033 | XIc | 97.7 | ERS2039517 (this study) |
| R 10870 | 2 | None | 114 | 111 | XIVa | 99.1 | ERS2050183 (this study) |
| R 11402 | 5 | Cd03 | 11 | 288 (CE) | XIb | 97.6 | ERS2050184 (this study) |
| R 9385 | 2 | Cd06 | 116 | 122 | XIVb | 99.6 | ERS2050185 (this study) |
| R 9367 | 1 | Cd06 | 55 | 070 | XIII | 99.8 | ERS2039518 (this study) |
| SE 881 | 5 | Cd03 | 11 | 045 | V | 98.8 | ERS2050186 (this study) |
| SE 844 | 2 | None | 192 | 080 | IIIa | 99.6 | ERS2050187 (this study) |
| TFA/V14-10 | 2 | Cd06 | 231 | 153 (CE) | XId | 99.4 | ERS2039519 (this study) |
| TR13 | 1 | Cd06 | 17 | 018 | XIX | 99.4 | ERS2039541 (this study) |
| TR14 | 1 | Cd06 | 182 | SLO 005 | XX | 99.9 | ERS2039542 (this study) |
| 1732874 | 2 | Cd06 | 226 | SLO 228 | IXd | 99.6 | ERS2039496 (this study) |
| 3073 | 2 | Cd06 | 41 | SLO 042 | IIId | 99.6 | ERS2039506 (this study) |
| 51377 | 5 | Cd03 | 11 | 127 | VI | 98.8 | ERS2050169 (this study) |
| 51680 | 2 | Cd06 | 67 | 019 | IXa | 99.6 | ERS2050170 (this study) |
| 57267 | 5 | None | 193 | 063 | VII | 98.4 | ERS2050171 (this study) |
| 597B | None | 122 | 131 | 0/v | 99.3 | Reference 22 | |
| 55767 | 3 | None | 5 | 023 | IV | 98.7 | ERS2039507 (this study) |
| 7325 | 2 | Cd06 | 1 | 027 | XXV | 99.5 | ERS2050172 (this study) |
| 7459 | 1 | Cd06 | 16 | 050 (CE) | XXVI | 99.3 | ERS2039509 (this study) |
| 8864 | 2 | Cd06 | 62 | 591 (CE) | Xa | 99.7 | ERS2050174 (this study) |
| AC008 | 1 | Cd06 | 53 | 103 | II | 99.9 | ERS2039510 (this study) |
| AI 541 | 2 | Cd06 | 231 | 251 | IIIe | 99.6 | ERS2039511 (this study) |
| CD07-468 | 2 | Cd06 | 197 | 027 | XXII | 99.6 | ERS2050175 (this study) |
| CD07-140 | 1 | Cd06 | 3 | 001 | XXIX | 98.8 | ERS2039512 (this study) |
| CD08-070 | 5 | Cd03 | 11 | 126 | XXVIII | 99.1 | ERS2050176 (this study) |
| CD10-055 | Cd04 | 369 | SLO 201 | XXXIV | 96.2 | ERS2039513 (this study) | |
| CH6223 | 4 | None | 198 | SLO 035 | XXI | 98.5 | ERS2050177 (this study) |
| CH6230 | 2 | Cd06 | 123 | 251 | IIIc | 99.3 | ERS2050178 (this study) |
| ES 130 | 5 | Cd03 | 166 | SLO 101 | XXX | 98.7 | Reference 42 |
| WA 151 | 5 | Cd03 | 167 | SLO 098 | XXI | 98.4 | Reference 42 |
| VPI 10463 | 1 | Cd06 | 46 | 087 | 0 | 99.4 | ERS2039543 (this study) |
| TFA/V20-1 | 2 | Cd06 | 41 | 244 | IXb | 99.5 | ERS2039540 (this study) |
| C00006623 | 1 | Cd06 | 2 | 99.8 | ERX103559 (9) | ||
| C00006624 | 1 | Cd06 | 10 | 99.8 | ERX103560 (9) | ||
| C00006625 | 4 | Cd05 | 37 | 98.9 | ERX103561 (9) | ||
| C00006626 | 4 | Cd05 | 37 | 99.5 | ERX103562 (9) | ||
| C00006627 | 3 | None | 5 | 98.5 | ERX103563 (9) | ||
| C00006628 | 1 | Cd06 | 10 | 99.8 | ERX103564 (9) | ||
| C00006629 | 1 | Cd06 | 54 | 99.3 | ERX103565 (9) | ||
| C00006630 | 3 | None | 5 | 98.7 | ERX103566 (9) | ||
| M68 | 4 | Cd05 | 37 | 017 | VIII | 98.8 | NC_017175 (15, 40, 43) |
| R20291 | 2 | Cd06 | 1 | 027 | III | 99.6 | NC_013316 (15, 40, 43) |
| P1 | 2 | Cd06 | 1 | 99.6 | SRX821661 (15) | ||
| P2 | 2 | Cd06 | 1 | 99.7 | SRX821763 (15) | ||
| P3 | 2 | Cd06 | 1 | 99.7 | SRX821764 (15) | ||
| P4 | 2 | Cd06 | 1 | 99.7 | SRX821765 (15) | ||
| P5 | 2 | Cd06 | 1 | 99.7 | SRX821766 (15) | ||
| P6 | 2 | Cd06 | 1 | 99.7 | SRX821767 (15) | ||
| P7 | 2 | Cd06 | 1 | 99.7 | SRX821768 (15) | ||
| P8 | 1 | Cd06 | 2 | 99.8 | SRX821769 (15) | ||
| P9 | 4 | Cd05 | 37 | 99.6 | SRX821770 (15) | ||
| P10 | 4 | Cd05 | 37 | 99.6 | SRX821771 (15) | ||
| P11 | 1 | Cd06 | 2 | 99.9 | SRX821772 (15) | ||
| P12 | 4 | None | 81 | 99.5 | SRX821773 (15) | ||
| P13A | 2 | Cd06 | 1 | 99.7 | SRX821774 (15) | ||
| P13B | 2 | Cd06 | 1 | 99.7 | SRX821775 (15) | ||
| P13C | 2 | Cd06 | 1 | 99.7 | SRX821777 (15) | ||
| P14 | 1 | Cd06 | 8 | 99.7 | SRX821778 (15) | ||
| P15 | 1 | Cd06 | 8 | 99.7 | SRX821779 (15) | ||
| P16 | 2 | Cd06 | 1 | 99.7 | SRX821780 (15) | ||
| P17 | 2 | Cd06 | 1 | 99.7 | SRX821781 (15) | ||
| P18 | 2 | Cd06 | 1 | 99.7 | SRX821782 (15) | ||
| P19 | 4 | None | 81 | 99.5 | SRX821783 (15) | ||
| P20 | 4 | None | 81 | 99.5 | SRX821784 (15) |
MLST STs were in accordance to the C. difficile MLST database (https://pubmlst.org/cdifficile/), and clades were determined in this study (see Table S1).
Toxinotypes were given in accordance with the recent update on C. difficile toxinotyping (7).
BAPS.
To determine the overall C. difficile species variation, we used Bayesian Analysis of Population Structure (BAPS) version 6.0 (17, 24, 25). Sequences of all MLST STs available as of 31 March 2016 (n = 347 STs) were downloaded from the MLST website (https://pubmlst.org/cdifficile/) (6), and all allelic gene sequences per locus were multiply aligned using MUSCLE (26) and finally concatenated for each ST. BAPS was carried out using the clustering of linked molecular data functionality. Ten runs were performed, setting an upper limit of 30 partitions. Admixture analysis was performed using the following parameters: minimum population size considered, 1; iterations, 50; number of reference individuals simulated from each population, 50; and number of iterations for each reference individual, 10.
DNA extraction, whole-genome sequencing, and assembly.
Prior to sequencing, the isolates were cultured anaerobically for 48 h at 37°C on Columbia blood agar plates (Oxoid, Wesel, Germany) and DNA was extracted using a fast glass bead method (27). Sequencing libraries were prepared using Nextera XT chemistry (Illumina Inc., San Diego, CA) for a 250-bp paired-end sequencing run on an Illumina MiSeq sequencer. Samples were sequenced to aim for minimum coverage of 120-fold using Illumina's recommended standard protocols. The resulting FASTQ files were de novo assembled using the SPAdes assembler version 3.11 (28) integrated in Ridom SeqSphere+ software (29) (version 5.0 beta; Ridom GmbH, Münster, Germany) using the following SPAdes parameters: k, automatic selection based on read length and mismatch careful mode turned on.
cgMLST target gene definition.
To determine the cgMLST gene set, a genome-wide gene-by-gene comparison was performed using the cgMLST target definer (version 1.4) function of SeqSphere+ (Ridom GmbH) with relaxed parameters (≥80% gene sequence identity and 100% gene sequence overlap) reflecting the high diversity within C. difficile. These cgMLST target definer parameters comprised the following filters to exclude certain genes of the C. difficile strain 630 reference genome (GenBank accession number NC_009089.1) from the cgMLST scheme: a “minimum length filter that discards all genes that are shorter than 50 bases,” a “start codon filter that discards all genes that contain no start codon at the beginning of the gene,” a “stop codon filter that discards all genes that contain no stop codon, more than 1 stop codon or if the stop codon is not at the end of the gene,” a “homologous gene filter that discards all genes that have fragments that occur in multiple copies in reference genome (with identity of ≥90% and more than 100 bases overlap),” and a “gene overlap filter that discards the shorter gene from the cgMLST scheme if the two genes affected overlap more than 4 bases.” The remaining genes were then used in a pairwise comparison with BLAST version 2.2.12 (parameters used were word size 11, mismatch penalty −1, match reward 1, gap open costs 5, and gap extension costs 2) with the query C. difficile chromosomes. All genes of the reference genome that were common in all query genomes with a sequence identity of ≥80% and 100% overlap (with the default parameter stop codon percentage filter turned on; i.e., more than 80% of the query genomes do not contain internal stop codons) formed the final cgMLST scheme.
Evaluation of the cgMLST target gene set.
To evaluate the representativeness and the discriminatory power of the novel C. difficile cgMLST target gene set, we used the above-mentioned genomes (Table 2; see also Tables S3 and S4 in the supplemental material). To ensure the sequence quality of the downloaded genomes/reads prior to further analyses, only isolates with a coverage of ≥50 and a consensus base count that deviated at most ±10% from the median consensus base count were included. A well-defined cgMLST scheme should result, on average, in 97.5% extracted cgMLST target genes (30). To extract the cgMLST genes, the default parameters were used in the SeqSphere+ software: (i) for processing options, “Ignore contigs shorter than 200 bases”; (ii) for scanning options, “Matching scanning thresholds for creating targets from assembled genomes” with “required identity to reference sequence of 90%” and “required alignment to reference sequence with 99%”; and (iii) for BLAST options, word size 11, mismatch penalty −1, match reward 1, gap open costs 5, and gap extension costs 2. In addition, the target genes were assessed for quality, i.e., the absence of frameshifts and ambiguous nucleotides. A core genome gene was considered a “good target” only if all of the above-listed criteria were met, in which case the complete sequence was analyzed in comparison to the reference sequence. Alleles for each gene were assigned automatically by the SeqSphere+ software to ensure a unique nomenclature. The combination of all alleles in each strain formed an allelic profile that was used to generate minimum spanning trees (MST) using the parameter “pairwise ignore missing values” during distance calculation.
In order to maintain backwards compatibility with classical C. difficile MLST, the sequences of the seven genes comprising the allelic profile of the MLST scheme were extracted separately from the genome sequences and queried against the C. difficile MLST database in order to assign STs in silico using the SeqSphere+ software that queries the respective gene sequences, compares them with the allele library of each of the seven MLST target genes, and assigns alleles and STs.
Accession number(s).
All raw reads generated and/or contig sequences were submitted to the European Nucleotide Archive (http://www.ebi.ac.uk/ena/) under accession number PRJEB23450. The NCBI accession numbers for the sequences determined for this study are ERR2216002, ERR2216003, ERS2039496, ERS2039506, ERS2039507, ERS2039509 to ERS2039519, ERS2039540 to ERS2039543, ERS2050167 to ERS2050178, and ERS2050180 to ERS2050188 (see Tables 1 and 2).
RESULTS
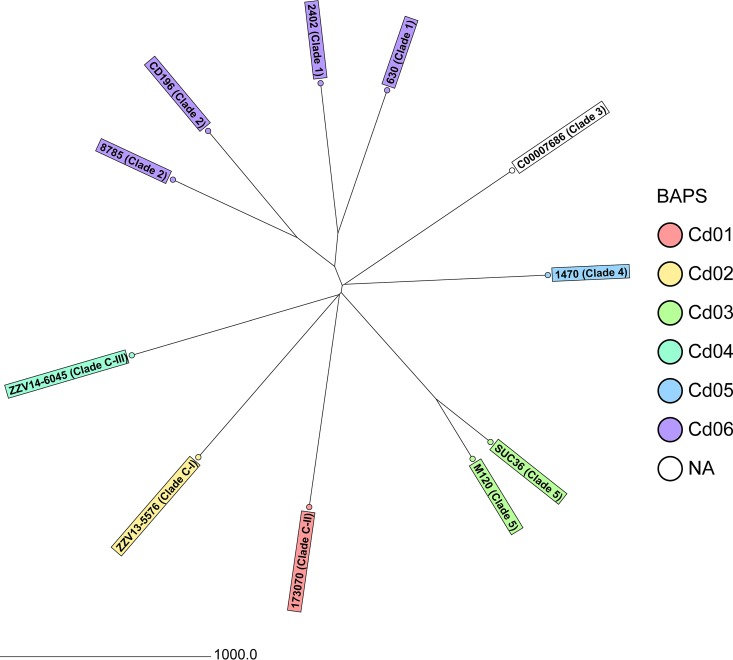
To develop a cgMLST scheme that sufficiently covers the diversity of the species C. difficile, we initially determined—besides the known partitioning into clades—the diversity using BAPS. This approach based on 347 STs resulted in six partitions comprising 306 STs; 41 STs were not assigned to any BAPS group (Table S1). Based on this grouping, 11 genome sequences, including that of C. difficile strain 630 (Table 1), were used to define the cgMLST scheme. Their comparison resulted in selection of 2,270 genes out of 3,756 genes present in strain 630 (50.4% of the 630 strain chromosome nucleotides) (Table S2). Figure 1 illustrates the diversity of the 11 isolates used for cgMLST target definition.
FIG 1.
Neighbor-joining tree of the 11 C. difficile isolates used for cgMLST target definition based on cgMLST target genes with pairwise ignore missing values. In addition to the sample name, the clade is given and the BAPS partitions are colored. The distance is given as the number of cgMLST genes.
This novel cgMLST scheme was then challenged with different sets of strains (Table 2; see also Table S3). Out of the genomes of the 38 reference strains of all published toxinotypes, 2,184 to 2,268 cgMLST targets (mean, 99.1%; median, 99.4%) could be extracted. Similarly, for the two published outbreaks, all isolates contained 2,237 to 2,267 cgMLST targets (mean, 99.5%; median, 99.7%), underlining the representativeness of the cgMLST scheme.
Moreover, we investigated the publicly available genome sequences from the NCBI (n = 268 assembled C. difficile genomes and reads from 3,482 isolates of the SRA). We first determined the median of the consensus base count (4,150,084 bp) and included only genomes of isolates in the analysis that exhibited ±10% of the median consensus base count. Furthermore, genomes of isolates with coverage <50-fold were excluded as well as NCBI assembled genomes, which also existed as SRA isolates. In total, we finally included 2,954 publicly available genomic data in our final analysis. Summarized from Table S4, on average 99.3% cgMLST targets were detected (median, 99.6%; 1,116 to 2,270 targets). Figure S1 illustrates the population structure and relationship to classical MLST STs.
To further ascertain the representativeness of our approach, especially using BAPS, we determined the ST distribution and percentage of isolates from the 2,954 isolates that were not grouped into any of the BAPS groups. After exclusion of 33 isolates with an unknown ST, only 9 of the 41 STs that were not assigned to any BAPS group were present in 123 (4.2%) isolates. Of these 123 isolates, however, all had, on average 98.8% cgMLST targets (Table S4).
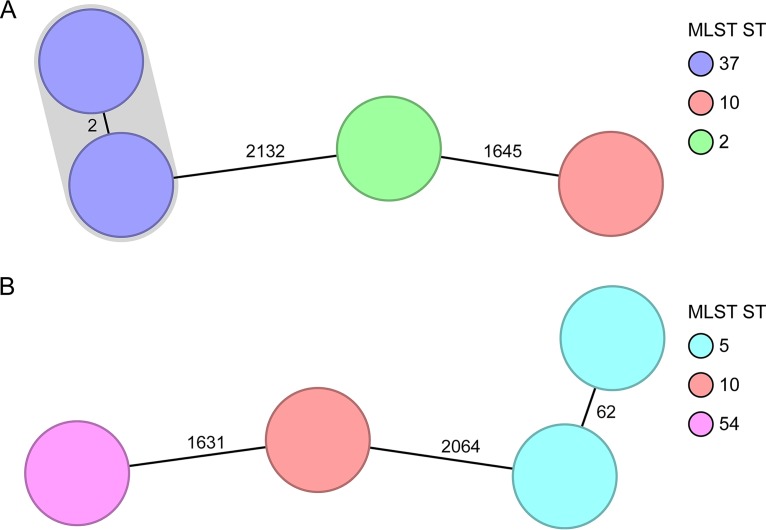
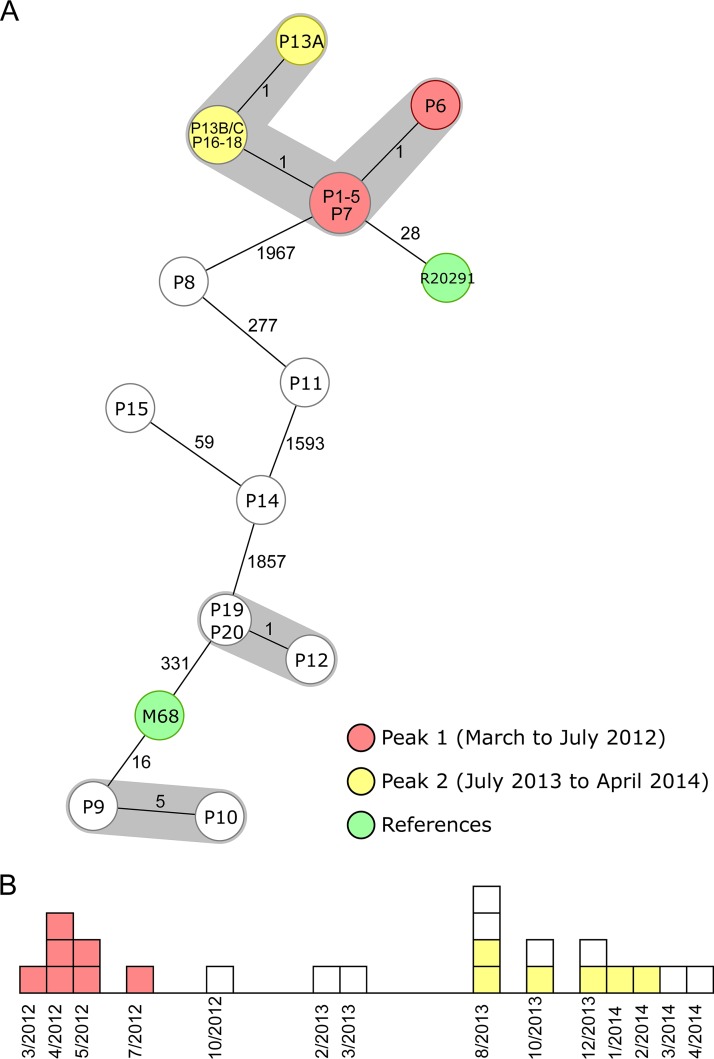
After confirmation of the representativeness of the novel cgMLST scheme, we analyzed the capability of the scheme to differentiate among closely related isolates from outbreak investigations. We reanalyzed different scenarios from the literature comprising short- and long-term scenarios (9, 15). In Fig. 2, two short-term spatiotemporal clusters spanning 17 to 22 days illustrate two typical clinical scenarios: while Fig. 2A shows that a clonal spread was detected among two isolates differing in only two cgMLST targets, Fig. 2B indicates that a transmission could be ruled out, as the suspected isolates belonged to the same ST type but differed in 62 cgMLST targets. These findings were in accordance with the published SNV analysis (9). Figure 3 shows the cgMLST typing results from a recent long-term outbreak investigation in a Chinese hospital from 2012 to 2014 (15). Two peaks (March to July 2012 and August 2013 to February 2014) of a clonal spread were recognized; again, our reanalysis using cgMLST corroborated the previous findings that these peaks were linked and belonged to the same outbreak clone. Based on these results, we finally defined the threshold, i.e., the maximum number of differing alleles for isolates that are likely to belong to the same clone, as ≤6 alleles. Isolates sharing genotypes within this threshold are then grouped within the same cluster type (CT) (21).
FIG 2.
Minimum-spanning tree of two spatiotemporal clusters (9). Each node represents a unique cgMLST allele profile. The numbers on connecting lines display the number of differing alleles between the genotypes (line length not to scale). The different nodes are colored by the MLST ST, and closely related genotypes (≤6 different cgMLST alleles) are shaded. (A) Short-term cluster of four cases, where one transmission event was epidemiologically confirmed (2,244 to 2,265 cgMLST target genes [mean, 99.5%] were analyzed). (B) Short-term cluster of four cases, where a clonal transmission was ruled out (2,237 to 2,265 cgMLST target genes [mean, 99.1%] were analyzed).
FIG 3.
Minimum-spanning tree and epidemiological curve illustrating a long-term spatiotemporal C. difficile cluster with two identified peaks (15). The 22 cluster isolates are colored according to their peaks, and the two reference strains are marked in green. (A) Minimum-spanning tree of the reanalyzed sequences based on cgMLST targets. Each node represents a unique allelic profile, and the size of the nodes represents the number of isolates. The numbers on connecting lines are the numbers of differing alleles between the genotypes (not to scale), and closely related genotypes (≤6 different cgMLST alleles) are shaded; 2,243 to 2,267 cgMLST target genes (mean of 99.6% of all cgMLST targets) were analyzed. All isolates of peaks 1 and 2 belonged to ST1. (B) Epidemiological curve. Each box represents one isolate, and boxes are colored according to their peak affiliation.
DISCUSSION
Here we describe the establishment of a novel cgMLST scheme for C. difficile, the most common cause of antibiotic-associated gastrointestinal infections worldwide. Based on a collection of isolates that represent the diversity of C. difficile organisms, we were able to construct a robust cgMLST scheme that contains 2,270 targets. This is in concordance with a previous estimate of the number of C. difficile core genes, where—depending on the number of strains and their characteristics—a range of 600 to 3,000 target genes were predicted (31).
Until today, SNV analysis was mainly reported for C. difficile WGS comparisons of circumscribed clinical or epidemiological settings (9, 15, 16). In this study, SNV results were calculated in comparison to a reference sequence among all strains included; any addition of strains would result in novel SNV results, which could lead to conflicting results, as SNV typing does not rely on a fixed nomenclature but is (re)calculated as soon as a novel strain is added. In contrast, cgMLST allows an easy curation of allelic data in a central database, which is a prerequisite for ensuring a universal typing nomenclature as already shown nearly 2 decades ago for “classical” MLST (32). The recently established database (http://www.cgmlst.org/) currently hosts (November 2017) the nomenclature for cgMLST schemes of nine different species (18–21, 33, 34), enabling a uniform typing nomenclature for thousands of genes using next-generation sequencing. The allele-based approach comprises another advantage in comparison to SNV-based approaches: it treats both a mutation that creates an SNV and a recombination that is likely to introduce multiple SNVs as a single evolutionary event (12, 19). Thereby, it compensates for recombination (32), which is helpful for a better definition of genetic relationships in bacteria with higher recombination rates, like C. difficile (35).
The subsequent evaluation of the novel cgMLST scheme employing a diverse collection of isolates as well as strains from clearly defined outbreak situations confirmed the representativeness of the novel scheme, i.e., the typeability (36) (99.3% of all cgMLST targets were successfully extracted from all 2,954 publicly available genomic data) and its ability to type all strains with sufficient discriminatory power to differentiate even among closely related isolates within nosocomial clusters. The high discriminatory power combined with a standardized typing nomenclature, which is crucial for outbreak investigations to facilitate comparison with historical data (12, 19, 37), enabled us to differentiate among epidemiologically related isolates detected during short- and long-term scenarios. Clonal transmissions as well as accidental spatiotemporal clusters could be exactly resolved (Fig. 2 and 3). Even isolates detected more than 1 year apart were still grouped together differing in ≤3 alleles (Fig. 3), which is in line with previous observations that expected 0 to 3 SNVs among transmitted samples within 1 year (16). Moreover, Eyre et al. suggested a threshold of ≥10 SNVs for genetically distinct isolates (16); analogously, we would suggest—adding a 2-fold-higher threshold as determined by our data as a precaution—a threshold of ≥7 alleles difference for isolates being unrelated and ≤6 alleles for isolates that are likely to belong to the same clone. Nonetheless, it has to be noted that typing efforts should always be evaluated in the context of the epidemiological situation.
When introducing novel typing approaches, backward comparability with previous typing methods and a high level of typeability, i.e., the representativeness of a method for any sample in a population, are always great demands. Backward comparability is possible for MLST, in which the ST can be easily extracted from the WGS data in silico, and clustering of cgMLST genotypes is concordant to MLST STs (Fig. S1). However, only limited backward comparability is possible with PCR ribotyping, currently the most widely used typing method for C. difficile. Due to the repetitive nature of the ribosomal operon (part of which is the internal transcribed spacer [ITS] region that is the target region for PCR ribotyping), PCR ribotypes cannot be extracted from draft genomes with any current methodology. To some extent, a correlation of PCR ribotypes and STs is known (38). To assign a PCR ribotype to a new cgMLST cluster, a representative strain would need to be PCR ribotyped. With respect to typeability, we have chosen the BAPS partitioning approach (17, 24, 25) to create an unbiased overview of the population diversity and subsequently randomly selected representative isolates for the cgMLST target definition and sequenced them to achieve highest sequence quality. Another way would be to analyze all available data from public databases; however, the quality is frequently unknown.
In summary, here we present the cgMLST typing scheme for C. difficile with a discriminatory power comparable to that of SNV analysis. The new scheme offers an excellent typing platform that enables local and international comparison of C. difficile isolates and could hence contribute to both better detection or clarification of outbreaks and a deeper understanding of the spread of C. difficile lineages.
Supplementary Material
ACKNOWLEDGMENTS
We thank Ursula Keckevoet and Isabell Höfig (Münster) for skillful technical assistance.
M.R. and S.J. were partially supported by the Slovenian Research Agency, grant J4-8224.
D.H. is one of the developers of the Ridom SeqSphere+ software mentioned in the article, which is a development of the company Ridom GmbH (Müenster, Germany) that is partially owned by him. The other authors have declared no conflict of interests.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JCM.01987-17.
REFERENCES
- 1.Martin JS, Monaghan TM, Wilcox MH. 2016. Clostridium difficile infection: epidemiology, diagnosis and understanding transmission. Nat Rev Gastroenterol Hepatol 13:206–216. doi: 10.1038/nrgastro.2016.25. [DOI] [PubMed] [Google Scholar]
- 2.Lessa FC, Mu Y, Bamberg WM, Beldavs ZG, Dumyati GK, Dunn JR, Farley MM, Holzbauer SM, Meek JI, Phipps EC, Wilson LE, Winston LG, Cohen JA, Limbago BM, Fridkin SK, Gerding DN, McDonald LC. 2015. Burden of Clostridium difficile infection in the United States. N Engl J Med 372:825–834. doi: 10.1056/NEJMoa1408913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ofosu A. 2016. Clostridium difficile infection: a review of current and emerging therapies. Ann Gastroenterol 29:147–154. doi: 10.20524/aog.2016.0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Knetsch CW, Lawley TD, Hensgens MP, Corver J, Wilcox MW, Kuijper EJ. 2013. Current application and future perspectives of molecular typing methods to study Clostridium difficile infections. Euro Surveill 18:20381. doi: 10.2807/ese.18.04.20381-en. [DOI] [PubMed] [Google Scholar]
- 5.Rupnik M, Braun V, Soehn F, Janc M, Hofstetter M, Laufenberg-Feldmann R, von Eichel-Streiber C. 1997. Characterization of polymorphisms in the toxin A and B genes of Clostridium difficile. FEMS Microbiol Lett 148:197–202. doi: 10.1111/j.1574-6968.1997.tb10288.x. [DOI] [PubMed] [Google Scholar]
- 6.Griffiths D, Fawley W, Kachrimanidou M, Bowden R, Crook DW, Fung R, Golubchik T, Harding RM, Jeffery KJ, Jolley KA, Kirton R, Peto TE, Rees G, Stoesser N, Vaughan A, Walker AS, Young BC, Wilcox M, Dingle KE. 2010. Multilocus sequence typing of Clostridium difficile. J Clin Microbiol 48:770–778. doi: 10.1128/JCM.01796-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rupnik M, Janezic S. 2016. An update on Clostridium difficile toxinotyping. J Clin Microbiol 54:13–18. doi: 10.1128/JCM.02083-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jenke C, Harmsen D, Weniger T, Rothganger J, Hyytia-Trees E, Bielaszewska M, Karch H, Mellmann A. 2010. Phylogenetic analysis of enterohemorrhagic Escherichia coli O157, Germany, 1987–2008. Emerg Infect Dis 16:610–616. doi: 10.3201/eid1604.091361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eyre DW, Golubchik T, Gordon NC, Bowden R, Piazza P, Batty EM, Ip CL, Wilson DJ, Didelot X, O'Connor L, Lay R, Buck D, Kearns AM, Shaw A, Paul J, Wilcox MH, Donnelly PJ, Peto TE, Walker AS, Crook DW. 2012. A pilot study of rapid benchtop sequencing of Staphylococcus aureus and Clostridium difficile for outbreak detection and surveillance. BMJ Open 2:e001124. doi: 10.1136/bmjopen-2012-001124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Turabelidze G, Lawrence SJ, Gao H, Sodergren E, Weinstock GM, Abubucker S, Wylie T, Mitreva M, Shaikh N, Gautom R, Tarr PI. 2013. Precise dissection of an Escherichia coli O157:H7 outbreak by single nucleotide polymorphism analysis. J Clin Microbiol 51:3950–3954. doi: 10.1128/JCM.01930-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mellmann A, Harmsen D, Cummings CA, Zentz EB, Leopold SR, Rico A, Prior K, Szczepanowski R, Ji Y, Zhang W, McLaughlin SF, Henkhaus JK, Leopold B, Bielaszewska M, Prager R, Brzoska PM, Moore RL, Guenther S, Rothberg JM, Karch H. 2011. Prospective genomic characterization of the German enterohemorrhagic Escherichia coli O104:H4 outbreak by rapid next generation sequencing technology. PLoS One 6:e22751. doi: 10.1371/journal.pone.0022751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maiden MC, Jansen van Rensburg MJ, Bray JE, Earle SG, Ford SA, Jolley KA, McCarthy ND. 2013. MLST revisited: the gene-by-gene approach to bacterial genomics. Nat Rev Microbiol 11:728–736. doi: 10.1038/nrmicro3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Willems S, Kampmeier S, Bletz S, Kossow A, Kock R, Kipp F, Mellmann A. 2016. Whole-genome sequencing elucidates epidemiology of nosocomial clusters of Acinetobacter baumannii. J Clin Microbiol 54:2391–2394. doi: 10.1128/JCM.00721-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mellmann A, Andersen PS, Bletz S, Friedrich AW, Kohl TA, Lilje B, Niemann S, Prior K, Rossen JW, Harmsen D. 2017. High interlaboratory reproducibility and accuracy of next-generation-sequencing-based bacterial genotyping in a ring trial. J Clin Microbiol 55:908–913. doi: 10.1128/JCM.02242-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jia H, Du P, Yang H, Zhang Y, Wang J, Zhang W, Han G, Han N, Yao Z, Wang H, Zhang J, Wang Z, Ding Q, Qiang Y, Barbut F, Gao GF, Cao Y, Cheng Y, Chen C. 2016. Nosocomial transmission of Clostridium difficile ribotype 027 in a Chinese hospital, 2012–2014, traced by whole genome sequencing. BMC Genomics 17:405. doi: 10.1186/s12864-016-2708-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eyre DW, Cule ML, Wilson DJ, Griffiths D, Vaughan A, O'Connor L, Ip CL, Golubchik T, Batty EM, Finney JM, Wyllie DH, Didelot X, Piazza P, Bowden R, Dingle KE, Harding RM, Crook DW, Wilcox MH, Peto TE, Walker AS. 2013. Diverse sources of C. difficile infection identified on whole-genome sequencing. N Engl J Med 369:1195–1205. doi: 10.1056/NEJMoa1216064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Corander J, Marttinen P, Siren J, Tang J. 2008. Enhanced Bayesian modelling in BAPS software for learning genetic structures of populations. BMC Bioinformatics 9:539. doi: 10.1186/1471-2105-9-539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ruppitsch W, Pietzka A, Prior K, Bletz S, Fernandez HL, Allerberger F, Harmsen D, Mellmann A. 2015. Defining and evaluating a core genome multilocus sequence typing scheme for whole-genome sequence-based typing of Listeria monocytogenes. J Clin Microbiol 53:2869–2876. doi: 10.1128/JCM.01193-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leopold SR, Goering RV, Witten A, Harmsen D, Mellmann A. 2014. Bacterial whole-genome sequencing revisited: portable, scalable, and standardized analysis for typing and detection of virulence and antibiotic resistance genes. J Clin Microbiol 52:2365–2370. doi: 10.1128/JCM.00262-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kohl TA, Diel R, Harmsen D, Rothganger J, Walter KM, Merker M, Weniger T, Niemann S. 2014. Whole-genome-based Mycobacterium tuberculosis surveillance: a standardized, portable, and expandable approach. J Clin Microbiol 52:2479–2486. doi: 10.1128/JCM.00567-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.de Been M, Pinholt M, Top J, Bletz S, Mellmann A, van Schaik W, Brouwer E, Rogers M, Kraat Y, Bonten M, Corander J, Westh H, Harmsen D, Willems RJ. 2015. Core genome multilocus sequence typing scheme for high-resolution typing of Enterococcus faecium. J Clin Microbiol 53:3788–3797. doi: 10.1128/JCM.01946-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Knetsch CW, Terveer EM, Lauber C, Gorbalenya AE, Harmanus C, Kuijper EJ, Corver J, van Leeuwen HC. 2012. Comparative analysis of an expanded Clostridium difficile reference strain collection reveals genetic diversity and evolution through six lineages. Infect Genet Evol 12:1577–1585. doi: 10.1016/j.meegid.2012.06.003. [DOI] [PubMed] [Google Scholar]
- 23.Riedel T, Bunk B, Thurmer A, Sproer C, Brzuszkiewicz E, Abt B, Gronow S, Liesegang H, Daniel R, Overmann J. 2015. Genome resequencing of the virulent and multidrug-resistant reference strain Clostridium difficile 630. Genome Announc 3(2):e00276-15. doi: 10.1128/genomeA.00276-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cheng L, Connor TR, Siren J, Aanensen DM, Corander J. 2013. Hierarchical and spatially explicit clustering of DNA sequences with BAPS software. Mol Biol Evol 30:1224–1228. doi: 10.1093/molbev/mst028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van Tonder AJ, Mistry S, Bray JE, Hill DM, Cody AJ, Farmer CL, Klugman KP, von Gottberg A, Bentley SD, Parkhill J, Jolley KA, Maiden MC, Brueggemann AB. 2014. Defining the estimated core genome of bacterial populations using a Bayesian decision model. PLoS Comput Biol 10:e1003788. doi: 10.1371/journal.pcbi.1003788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koser CU, Fraser LJ, Ioannou A, Becq J, Ellington MJ, Holden MT, Reuter S, Torok ME, Bentley SD, Parkhill J, Gormley NA, Smith GP, Peacock SJ. 2014. Rapid single-colony whole-genome sequencing of bacterial pathogens. J Antimicrob Chemother 69:1275–1281. doi: 10.1093/jac/dkt494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jünemann S, Sedlazeck FJ, Prior K, Albersmeier A, John U, Kalinowski J, Mellmann A, Goesmann A, von Haeseler A, Stoye J, Harmsen D. 2013. Updating benchtop sequencing performance comparison. Nat Biotechnol 31:294–296. doi: 10.1038/nbt.2522. [DOI] [PubMed] [Google Scholar]
- 30.Higgins PG, Prior K, Harmsen D, Seifert H. 2017. Development and evaluation of a core genome multilocus typing scheme for whole-genome sequence-based typing of Acinetobacter baumannii. PLoS One 12:e0179228. doi: 10.1371/journal.pone.0179228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Knight DR, Elliott B, Chang BJ, Perkins TT, Riley TV. 2015. Diversity and evolution in the genome of Clostridium difficile. Clin Microbiol Rev 28:721–741. doi: 10.1128/CMR.00127-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maiden MC, Bygraves JA, Feil E, Morelli G, Russell JE, Urwin R, Zhang Q, Zhou J, Zurth K, Caugant DA, Feavers IM, Achtman M, Spratt BG. 1998. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc Natl Acad Sci U S A 95:3140–3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Antwerpen MH, Prior K, Mellmann A, Hoppner S, Splettstoesser WD, Harmsen D. 2015. Rapid high resolution genotyping of Francisella tularensis by whole genome sequence comparison of annotated genes (“MLST+”). PLoS One 10:e0123298. doi: 10.1371/journal.pone.0123298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moran-Gilad J, Prior K, Yakunin E, Harrison TG, Underwood A, Lazarovitch T, Valinsky L, Luck C, Krux F, Agmon V, Grotto I, Harmsen D. 2015. Design and application of a core genome multilocus sequence typing scheme for investigation of Legionnaires' disease incidents. Euro Surveill 20:21186. doi: 10.2807/1560-7917.ES2015.20.28.21186. [DOI] [PubMed] [Google Scholar]
- 35.Scaria J, Ponnala L, Janvilisri T, Yan W, Mueller LA, Chang YF. 2010. Analysis of ultra low genome conservation in Clostridium difficile. PLoS One 5:e15147. doi: 10.1371/journal.pone.0015147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van Belkum A, Tassios PT, Dijkshoorn L, Haeggman S, Cookson B, Fry NK, Fussing V, Green J, Feil E, Gerner-Smidt P, Brisse S, Struelens M, European Society of Clinical Microbiology and Infectious Diseases (ESCMID) Study Group on Epidemiological Markers (ESGEM). 2007. Guidelines for the validation and application of typing methods for use in bacterial epidemiology. Clin Microbiol Infect 13(Suppl 3):S1–S46. doi: 10.1111/j.1469-0691.2007.01732.x. [DOI] [PubMed] [Google Scholar]
- 37.Schmid D, Allerberger F, Huhulescu S, Pietzka A, Amar C, Kleta S, Prager R, Preussel K, Aichinger E, Mellmann A. 2014. Whole genome sequencing as a tool to investigate a cluster of seven cases of listeriosis in Austria and Germany, 2011–2013. Clin Microbiol Infect 20:431–436. doi: 10.1111/1469-0691.12638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Janezic S, Rupnik M. 2015. Genomic diversity of Clostridium difficile strains. Res Microbiol 166:353–360. doi: 10.1016/j.resmic.2015.02.002. [DOI] [PubMed] [Google Scholar]
- 39.Eyre DW, Fawley WN, Best EL, Griffiths D, Stoesser NE, Crook DW, Peto TE, Walker AS, Wilcox MH. 2013. Comparison of multilocus variable-number tandem-repeat analysis and whole-genome sequencing for investigation of Clostridium difficile transmission. J Clin Microbiol 51:4141–4149. doi: 10.1128/JCM.01095-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kurka H, Ehrenreich A, Ludwig W, Monot M, Rupnik M, Barbut F, Indra A, Dupuy B, Liebl W. 2014. Sequence similarity of Clostridium difficile strains by analysis of conserved genes and genome content is reflected by their ribotype affiliation. PLoS One 9:e86535. doi: 10.1371/journal.pone.0086535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Janezic S, Potocnik M, Zidaric V, Rupnik M. 2016. Highly divergent Clostridium difficile strains isolated from the environment. PLoS One 11:e0167101. doi: 10.1371/journal.pone.0167101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Elliott B, Squire MM, Thean S, Chang BJ, Brazier JS, Rupnik M, Riley TV. 2011. New types of toxin A-negative, toxin B-positive strains among clinical isolates of Clostridium difficile in Australia. J Med Microbiol 60:1108–1111. doi: 10.1099/jmm.0.031062-0. [DOI] [PubMed] [Google Scholar]
- 43.He M, Sebaihia M, Lawley TD, Stabler RA, Dawson LF, Martin MJ, Holt KE, Seth-Smith HM, Quail MA, Rance R, Brooks K, Churcher C, Harris D, Bentley SD, Burrows C, Clark L, Corton C, Murray V, Rose G, Thurston S, van Tonder A, Walker D, Wren BW, Dougan G, Parkhill J. 2010. Evolutionary dynamics of Clostridium difficile over short and long time scales. Proc Natl Acad Sci U S A 107:7527–7532. doi: 10.1073/pnas.0914322107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.