ABSTRACT
One of the most important clinical obstacles in cystic fibrosis (CF) treatment is antibiotic treatment failure due to biofilms produced by Pseudomonas aeruginosa. The ability of this pathogen to survive eradication by tobramycin and pathoadapt into a hyperbiofilm state leading to chronic infections is key to its success. Retrospective studies have demonstrated that preventing this pathoadaptation by improving eradication is essential to extend the lives of CF patients. To identify adjuvants that enhance tobramycin eradication of P. aeruginosa, we performed a high-throughput screen of 6,080 compounds from four drug-repurposing libraries. We identified that the Food and Drug Administration (FDA)-approved compound triclosan, in combination with tobramycin, resulted in a 100-fold reduction of viable cells within biofilms at 6 h, but neither compound alone had significant antimicrobial activity against biofilms. This synergistic treatment significantly accelerated the killing of biofilms compared to that with tobramycin treatment alone, and the combination was effective against 6/7 CF clinical isolates compared to tobramycin treatment alone, including a tobramycin-resistant strain. Further, triclosan and tobramycin killed persister cells, causing a 100-fold reduction by 8 h and complete eradication by 24 h. Triclosan also enhances tobramycin killing of multiple Burkholderia cenocepacia and Staphylococcus aureus clinical isolates grown as biofilms. Additionally, triclosan showed synergy with other aminoglycosides, such as gentamicin or streptomycin. Triclosan is a well-tolerated aminoglycoside adjuvant shown to be safe for human use that could improve the treatment of biofilm-based infections.
KEYWORDS: Pseudomonas aeruginosa, biofilm, persister, tobramycin, triclosan
INTRODUCTION
Cystic fibrosis (CF) is the most common life-shortening genetic disease in Caucasians, affecting 70,000 people worldwide and over 30,000 people in the United States (1). Total hospital costs for the treatment of CF patients in the United States are approximately $1 billion per year, with hospitalizations due to bacterial infections accounting for the majority of these costs (2–4). CF is caused by a mutation in the cystic fibrosis transmembrane conductance regulator gene, resulting in either absent or altered chloride and bicarbonate transport in the lungs and throughout the body (5). In the lungs, this causes the airway mucus to become thick and dry, blocking the small airways and creating an environment that is prone to recurrent bacterial infections (6–8).
By adulthood, the most common pathogen found in the lungs of CF patients is Pseudomonas aeruginosa (1). Central to this pathogen's success is its ability to form biofilms, which are 10 to 1,000 times more resistant to antibiotics, due to a self-produced extracellular polysaccharide (EPS) matrix surrounding the cells (9, 10). In addition, biofilms have a greater number of dormant cells, termed “persisters,” which are metabolically inert, making them tolerant to antimicrobials that target metabolically active processes (11, 12). Finally, P. aeruginosa can pathoadapt in the lungs of CF patients, often producing a mucoid-type biofilm that produces a thicker EPS matrix. This matrix slows the diffusion of antibiotics and reduces their effectiveness (13, 14).
The current Pseudomonas eradication protocol used clinically is 300 mg of aerosolized tobramycin twice a day for 28 days in on/off cycles, reaching mean sputum concentrations of 737 μg/g of sputum (∼1,440 μM per dose) (15). Despite the routine use of Pseudomonas eradication therapies, by early adulthood, ∼80% of CF patients are chronically colonized with P. aeruginosa (16). Numerous retrospective studies have shown that eradication of transient infections by P. aeruginosa can extend the lives of CF patients (13, 17). Thus, there is a critical need to identify new agents that target cells within biofilms and avoid selecting for resistance. One possible strategy is to identify antiresistance compounds or adjuvants which are effective when combined with antimicrobials but are not effective on their own (18).
To this end, we performed a high-throughput screen (HTS) to identify adjuvants that are effective when combined with tobramycin against P. aeruginosa biofilms. From the HTS, we determined that the nonionic bisphenol triclosan in combination with tobramycin was significantly more effective at killing mature P. aeruginosa biofilms. Triclosan similarly showed synergy with the aminoglycosides gentamicin and streptomycin. We also found that the combination more effectively killed biofilms of Burkholderia cenocepacia and Staphylococcus aureus, two bacteria commonly isolated from CF patients, compared with killing by tobramycin alone. The combination of triclosan with aminoglycosides significantly enhanced the rate of biofilm killing and led to persister cell eradication.
Recently, after decades of overuse, the FDA has restricted the use of triclosan in consumer products due to concerns over bioaccumulation and cross-resistance, but this decision was driven primarily by a lack of demonstrated efficacy in consumer products rather than safety concerns. Numerous toxicity studies and the Scientific Committee on Consumer Safety published by the European Union have found that triclosan is safe when used appropriately (19–22). To further explore the clinical potential of triclosan in treating lung infections, we performed an acute and long-term triclosan toxicity study in rats and show that direct delivery of triclosan to the lungs did not elicit significant toxicity. Our results suggest that triclosan could provide a potential new aminoglycoside adjuvant for the treatment of P. aeruginosa biofilms for multiple indications, including CF, diabetic foot ulcers, and burn wounds (23–25).
RESULTS
Triclosan combined with aminoglycosides results in greater killing of P. aeruginosa biofilms.
We developed and carried out an HTS of 6,080 compounds from four drug-repurposing libraries at the University of Michigan (UM) Center for Chemical Genomics (CCG) to identify compounds that enhanced tobramycin-induced killing of P. aeruginosa strain PAO1 biofilms grown on the pegs of a 384-well disposable pin tool. A concentration of 250 μg/ml tobramycin (∼500 μM, 500× the planktonic MIC) was used, because we found that this concentration had only mild effects on biofilms, as seen below. Compounds were added at 10 μM, and biofilms were treated for 6 h before assaying for cell viability using the BacTiter-Glo assay. The average plate Z-factor was 0.6. One hundred eighteen compounds exhibiting >3 standard deviations from the control and 50% enhancement of biofilm killing were selected as initial hits. The initial hit rate for the screen was 1.9%, but this rate is likely inflated, as these libraries are enriched for biologically active molecules, including a number of antibiotics. Dose-response curves (DRC) for each of these 118 hits were generated in duplicate to determine the pAC50 (the inverse log10 of 50% activity) and the maximum killing percentage. This was performed both with and without 250 μg/ml tobramycin. Eighty-two compounds were considered active, with pAC50 values ranging from 8.4 to 3.4, showing a 71.3% confirmation rate. Thirty-one of these compounds were also active on their own and were given lower priority. As we are interested in targeting new pathways that are specific to antibiotic tolerance, we decided to focus first on compounds that are only effective in the presence of tobramycin. After removing compounds that contained problematic toxicity groups and exhibited promiscuous activity in prior HTSs, 26 compounds remained as promising antibiotic adjuvants for further investigation. Here, we describe our further analysis of one of these 26 compounds, triclosan.
Triclosan is a broad-spectrum antimicrobial that inhibits the enoyl-acyl carrier protein reductase FabI to prevent fatty acid synthesis in several bacterial species (Fig. 1) (26, 27). We chose to further characterize triclosan, as it showed significant synergy with tobramycin. Furthermore, it is known that P. aeruginosa is inherently resistant to triclosan, making it an intriguing candidate (28–30). Finally, triclosan has been widely used as an antimicrobial for decades and is still FDA approved to be used in toothpastes at millimolar concentrations (31).
FIG 1.
Molecular structure of triclosan and type II fatty acid synthesis. (A) Chemical structure of triclosan, 2,4,4′-trichloro-2′-hydroxydiphenyl ether. (B) Type II fatty acid synthesis (FASII) pathway. Triclosan is known to disrupt FASII by inhibiting FabI. However, FabV is more resistant to triclosan. ACP, acyl carrier protein; CoA, coenzyme A.
To determine the antibiofilm activities of triclosan and tobramycin, 24-h PAO1 biofilms were treated for 6 h with 100 μM triclosan and 500 μM tobramycin alone or in combination. Tobramycin alone resulted in an ∼2-fold reduction in viable cells compared to untreated biofilms, and triclosan alone exhibited no killing, but the combination resulted in an ∼100-fold reduction in viable cells (Fig. 2). These experiments were performed using the BacTiter-Glo assay to measure ATP in the biofilm, and in a control experiment, we demonstrated that the luminescence generated by this assay shows a direct linear relationship with CFUs (see Fig. S1 in the supplemental material).
FIG 2.
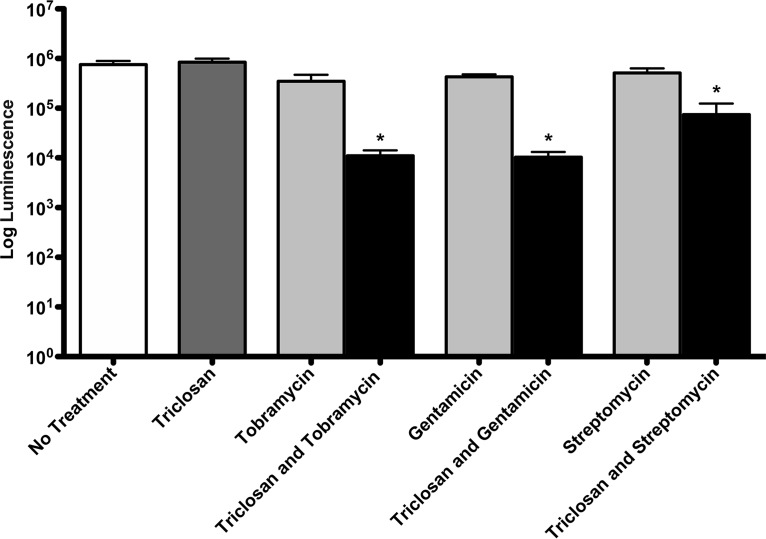
Triclosan enhances aminoglycoside killing of 24-h-old biofilms. Twenty-four-hour-old biofilms grown on minimum biofilm eradication concentration (MBEC) plates were treated for 6 h with 100 μM triclosan, 500 μM tobramycin, 100 μM gentamicin, or streptomycin, alone and in combination, and the number of viable cells within the biofilms was quantified using the BacTiter-Glo assay. The assay was performed at least three times in triplicate. The results represent the means ± the standard error of the mean (SEM). A one-way analysis of variance (ANOVA) followed by Bonferroni's multiple comparison post hoc test was used to determine statistical significance compared to each aminoglycoside alone (*, P < 0.05).
We also tested whether triclosan showed synergy with the antipseudomonal aminoglycosides gentamicin or streptomycin. Biofilms were treated with either 100 μM gentamicin or streptomycin alone or in combination with 100 μM triclosan. A concentration of 100 μM was chosen for gentamicin and streptomycin because this concentration results in a 50% reduction in viable cells. After 6 h of treatment, gentamicin reduced the number of cells by 2-fold, whereas triclosan and gentamicin reduced the number of cells by 100-fold (Fig. 2). Likewise, streptomycin alone had little effect on the cells within biofilms, whereas the combination of triclosan and streptomycin reduced the number of cells by 10-fold.
We also tested if triclosan could enhance several additional antipseudomonal antibiotics, including third- and fourth-generation carbapenems, which all exhibited poor activity on their own, and we found that triclosan only showed synergy with aminoglycosides (Fig. S2).
Triclosan demonstrates synergy with aminoglycosides at multiple concentrations.
DRC were performed to determine 50% effective concentrations (EC50s). Biofilms were treated with dilutions of triclosan and each aminoglycoside ranging from 0.390 to 400 μM in triplicate. Tobramycin alone showed modest activity at concentrations greater than 10 μM, killing approximately 50% of the cells after 6 h of treatment (Fig. 3). However, higher concentrations of tobramycin were less effective. This could be due to a phenomenon known as adaptive resistance, where aminoglycosides induce antibiotic resistance in P. aeruginosa (32, 33). In addition, the paradoxical effects of aminoglycosides in which higher concentrations of aminoglycosides are less effective than lower concentrations have previously been reported (34). Triclosan alone did not exhibit activity at any concentration tested. In contrast, the combination of triclosan and tobramycin increased biofilm killing at concentrations between 25 and 400 μM, with maximum activity at 400 μM, demonstrating 100-fold more activity than that of tobramycin alone. Like tobramycin, gentamicin or streptomycin treatment alone exhibited modest killing activity against biofilms that decreased at increasing concentrations (Fig. S3). Triclosan combined with streptomycin or gentamicin at higher concentrations led to an ∼100-fold reduction in cells. The EC50s for biofilm killing by these three combinations were between 20 and 30 μM (Table 1). The EC50 for tobramycin and gentamicin with triclosan showed wide variation, with 95% confidence intervals (95% CIs) of 7.65 to 83.78 μM and 12.08 to 44.02 μM, respectively, whereas streptomycin had a 95% CI of 24.5 to 34.31 μM. We attribute this greater variability in response to tobramycin and gentamicin with triclosan due to the paradoxical effect of these antibiotics exhibiting decreased activity at higher concentrations, which was not observed for streptomycin (Fig. 3 and S3) (34).
FIG 3.
Triclosan enhances tobramycin killing at multiple concentrations. Twenty-four-hour-old biofilms grown on MBEC plates were treated for 6 h with 2-fold dilutions of equal concentrations of triclosan, tobramycin, and triclosan combined with tobramycin, and the number of viable cells within the biofilms was quantified using the BacTiter-Glo assay. The assay was performed at least three times in triplicate. The results represent the means ± the SEM.
TABLE 1.
EC50 values for aminoglycoside combinationsa
| Antibiotic | Adjuvant | EC50 (μM) | 95% confidence interval (μM) |
|---|---|---|---|
| Tobramycin | Triclosan | 20.50 | 7.65–83.78 |
| Gentamicin | Triclosan | 23.06 | 12.08–44.02 |
| Streptomycin | Triclosan | 28.96 | 24.45–34.31 |
EC50 values were calculated using Prism version 5. Log10 (inhibitor) versus response–variable slope (four parameters) analyses were performed. EC50 values for aminoglycoside alone were not constructed, because no curve was established due to their ineffectiveness against biofilms.
Checkerboard dilution experiments were performed to determine the lowest possible combinations of tobramycin and triclosan that resulted in >1 log10 killing (Fig. 4). Biofilms were treated with dilutions of triclosan ranging from 12.5 to 100 μM in triplicate, and tobramycin ranging from 66 to 534 μM in triplicate. Triclosan at 50 μM with 66 to 534 μM tobramycin and 25 μM triclosan combined with 534 or 267 μM tobramycin resulted in significant killing of the biofilms (Fig. S4). Tobramycin held constant at a concentration of 66 μM combined with triclosan from 1.5 to 400 μM showed that triclosan demonstrated synergy with tobramycin at any concentration greater than 12.5 μM (Fig. S5).
FIG 4.

Triclosan enhances low concentrations of tobramycin. Twenty-four-hour-old biofilms grown on MBEC plates were treated for 6 h with checkerboard dilutions of triclosan combined with tobramycin. Numbers of viable cells within the biofilms were quantified using the BacTiter-Glo assay. The assay was performed at least three times in triplicate, and the mean is shown.
Planktonic MICs of tobramycin, gentamicin, or streptomycin are not changed when used in combination with triclosan.
We wondered if the synergy of triclosan with aminoglycosides was specific to biofilm-growing cells. We therefore determined the MICs of the combinations against planktonically grown cells. Surprisingly, we found that triclosan combined with tobramycin, gentamicin, or streptomycin did not impact the planktonic MIC of the aminoglycosides to kill P. aeruginosa. This suggests that triclosan and these aminoglycosides only function synergistically against biofilm-growing bacteria.
Triclosan combined with aminoglycosides accelerated killing of biofilm cells.
Time-kill assays were performed to determine the rate of killing of 100 μM triclosan, 500 μM tobramycin, or the combination of the two. Triclosan was ineffective alone, whereas tobramycin increased ATP concentrations and or cell numbers in the biofilm at 2 and 4 h (Fig. 5). This may be due to a stress response initiated by bacteria experiencing antimicrobial toxicity (35). Triclosan combined with tobramycin resulted in a shorter onset of action, with killing observed at 2 h compared to 6 h for tobramycin alone (Fig. 5). In addition, the combination exhibited killing at 2 h that was similar to that observed for tobramycin alone at 8 h and reduced the number of cells within the biofilm by over 100-fold at 4 h compared to tobramycin alone (Fig. 5). Analogous experiments were repeated with streptomycin and gentamicin, with similar outcomes (Fig. S6).
FIG 5.
Triclosan enhances the onset and maximum efficacy of tobramycin. Twenty-four-hour-old biofilms grown on MBEC plates were treated with triclosan (100 μM) tobramycin (500 μM), or a combination of the two. At 0, 2, 4, 6, and 8 h, the numbers of viable cells within the biofilms were determined using the BacTiter-Glo assay. The assay was performed at least three times in triplicate. The results represent means ± the SEM.
Triclosan and tobramycin effectively kill biofilms of CF clinical isolates.
We examined triclosan and tobramycin activities against seven P. aeruginosa clinical isolates that were collected from patients at the Michigan State University (MSU) or the University of Washington CF Clinic. Importantly, two clinical isolates, AMT0023_30 and AMT0023_34, were isolated from the same patient at 6 months or 8 years of age. In addition, clinical isolates CF110_N and CF110_O were isolated longitudinally from the same patient 3 months apart (Table 2). Using isolates collected from the same patient at different times allowed us to test the activities of triclosan and tobramycin against P. aeruginosa isolates unsuccessfully exposed to eradication therapies using tobramycin.
TABLE 2.
Bacterial strains used in this study
| Strain | Characteristicsa | Source or referenceb |
|---|---|---|
| PAO1 | Standard reference strain, isolated in 1954 (64) | Martha Mulks |
| Tn::fabI | ISlacZ/hah | 38 |
| AMT0023_30 | Early isolate, 6 mo | 65 |
| AMT0023_34 | Late isolate, 8 yr | 65 |
| CF_115_J | P. aeruginosa clinical CF isolate, Michigan | Martha Mulks |
| CF_110_N | P. aeruginosa clinical CF isolate, Michigan | Martha Mulks |
| CF_110_O | P. aeruginosa clinical CF isolate, Michigan | Martha Mulks |
| CF_131_M | P. aeruginosa clinical CF isolate, Michigan | Martha Mulks |
| CF_300_A | P. aeruginosa clinical CF isolate, Michigan | Martha Mulks |
| AU1054 | BCC clinical CF isolate, USA | J. J. LiPuma |
| PC184 | BCC clinical CF isolate, Cleveland Ohio | J. J. LiPuma |
| AU2289 | BCC clinical CF isolate, Michigan | J. J. LiPuma |
| H12424 | Soil, onion field, New York | J. J. LiPuma |
| J2315 | BCC clinical CF isolate, Edinburgh, UK | J. J. LiPuma |
| USA_300_JE2 | MRSA, wound, California | 61 |
| COL | MRSA, Colindale Hospital, England | 61 |
| Newman (25904) | MSSA, wound, endocarditis | Neal Hammer ATCC |
| Wichita (29213) | MSSA, better biofilm former | ATCC |
BCC, Burkholderia cenocepacia complex; MRSA, methicillin-resistant Staphylococcus aureus; MSSA, methicillin-sensitive Staphylococcus aureus; ATCC, American Tissue Type Collection.
J. J. LiPuma, U.S. Burkholderia cenocepacia Research Laboratory and Repository, UM, Ann Arbor, MI.
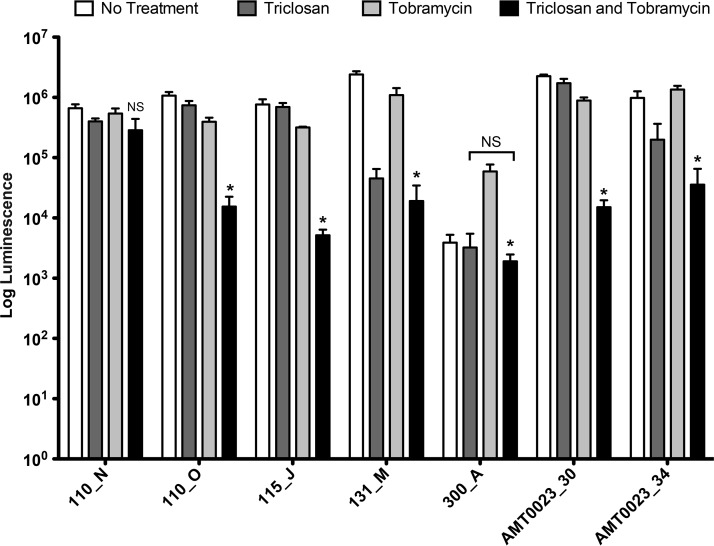
Triclosan at 100 μM combined with 500 μM tobramycin resulted in significantly greater killing of 6/7 P. aeruginosa clinical isolates compared to tobramycin treatment alone (Fig. 6). Strain 300_A without any treatment formed minimal biofilms that were barely above the limit of detection of our assay (103 log luminescence). However, treatment with tobramycin led to a 50-fold increase in the biofilm. The combination of tobramycin and triclosan reduced biofilm formation of this strain to background levels, indicating that the combination therapy is much more effective than tobramycin alone. Importantly, the combination of triclosan and tobramycin was effective against the tobramycin-resistant strain AMT023_34. This clinical isolate has mutations in the mexZ repressor causing increased expression of the MexXY-OpRM multidrug efflux pump that transports tobramycin and in the mutS gene, resulting in a hypermutator state. Moreover, this strain produces persister cells at an increased frequency (36).
FIG 6.
Tobramycin and triclosan are effective against P. aeruginosa CF isolates. Twenty-four-hour-old biofilms grown on MBEC plates were treated with triclosan (100 μM,) tobramycin (500 μM), or a combination of the two for 6 h. The numbers of viable cells within the biofilms were quantified using the BacTiter-Glo assay. The assay was performed at least three times in triplicate. The results represent means ± the SEM. A one-way ANOVA followed by Bonferroni's multiple comparison post hoc test was used to determine statistical significance compared to tobramycin alone (*, P < 0.05). NS, not significant.
As CF infections are polymicrobial, we assessed the activities of triclosan, tobramycin, and the combination of the two against other bacterial pathogens associated with CF. We first assessed the activity against five Burkholderia cenocepacia complex isolates, with four CF clinical isolates and one environmental isolate (Fig. S7). Two of the strains formed robust biofilms under our conditions, and the combination reduced viable cells ∼1,000-fold compared to tobramycin alone. In the other three isolates, which all poorly formed biofilms, the combination reduced viable cells in two of them, although this difference was not statistically significant, mainly due to the luminescence values being near the limit of detection for our assay. We also assessed activity against four strains of Staphylococcus aureus. Each of these strains increased biofilms in response to tobramycin, but all were sensitive to triclosan alone and the combination; however, most of the activity in the combination appeared to be driven primarily by triclosan (Fig. S8). This is expected, because S. aureus contains only fabI, which is known to be sensitive to triclosan (37).
Triclosan combined with aminoglycosides does not increase dispersal of P. aeruginosa biofilms.
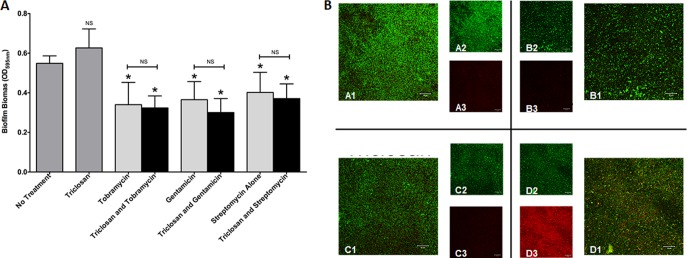
We examined whether triclosan combined with tobramycin, gentamicin, or streptomycin dispersed P. aeruginosa PAO1 biofilms grown under static conditions by staining biofilm biomass with crystal violet. Biofilms were treated for 6 h with 100 μM triclosan alone or 100 μM triclosan with gentamicin, streptomycin, or 500 μM tobramycin in combination. Triclosan treatment alone had no significant effect, while all three aminoglycosides caused significant biofilm dispersal. However, triclosan combined with the aminoglycosides did not significantly increase biofilm dispersal versus that with the aminoglycoside alone (Fig. 7A).
FIG 7.
Aminoglycosides combined with triclosan do not increase biofilm dispersal. (A) Twenty-four-hour-old biofilms grown on MBEC plates were treated with triclosan (100 μM), tobramycin (500 μM), gentamicin (100 μM), or streptomycin (100 μM), alone and in combination. The effect on biofilm biomass was quantified by staining with crystal violet. The experiment was performed at least five times in triplicate. The results represent the means ± SEM. A one-way ANOVA followed by Dunnett's multiple-comparison post hoc test was used to determine statistical significance compared to no treatment (*, P < 0.05). NS, not significant. (B) Twenty-four-hour-old biofilms grown in flow cells were treated with triclosan (100 μM), tobramycin (524 μM), or the combination for 6 h. Live cells are stained green, and dead cells are stained red. Representative images are shown: no treatment (top left quadrant), tobramycin alone (top right quadrant), triclosan alone (lower left quadrant), and a combination (lower right quadrant). Insets are shown for the live channel (A2, B2, C2, and D2) and for the dead channel (A3, B3, C3, and D3) for each condition.
To further evaluate the effects of tobramycin and triclosan on biofilm dispersal, we evaluated biofilms formed under flow conditions for 24 h treated for 6 h with 100 μM triclosan, 500 μM tobramycin, or a combination of the two. Following treatment, cells were stained with SYTO9 and propidium iodide to measure live (green) and dead (red) cells, respectively. We found that tobramycin or triclosan treatment alone did not promote cell killing. While the combination did not significantly disperse the biofilm, it did elicit increased cell death (Fig. 7B).
FabI inhibition by triclosan is not responsible for synergy.
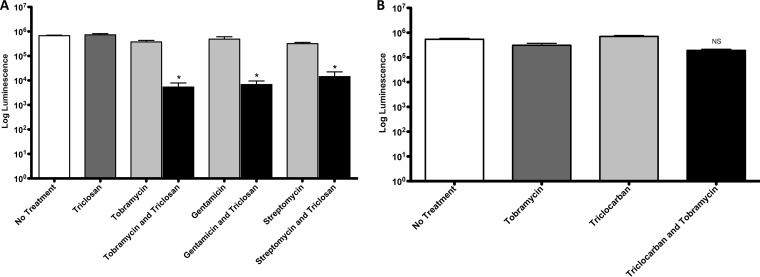
To determine if triclosan-induced inhibition of FabI accounts for the observed synergy, we measured the activities of triclosan and tobramycin against biofilms of a FabI-deficient strain (Tn::fabI mutant), which has a ISlacZ/hah transposon inserted in the FabI gene (38). If the synergistic activity of triclosan is solely due to FabI inhibition, we would expect the fabI transposon mutant to be sensitive to tobramycin alone in the absence of triclosan. Biofilms were treated with 100 μM triclosan alone or 100 μM triclosan with gentamicin, streptomycin, or 500 μM tobramycin in combination for 6 h. Contrary to our expectation, biofilms of the fabI transposon mutant did not exhibit increased sensitivity to aminoglycosides, and triclosan continued to significantly enhance tobramycin, gentamicin, and streptomycin killing of biofilms even when FabI was not present (Fig. 8A). In addition, the activity of triclocarban, which is a triclosan analog and is thought to also inhibit FabI (39, 40), did not enhance tobramycin activity against biofilms (Fig. 8B).
FIG 8.
FabI inhibition by triclosan is not responsible for the synergy. (A) Twenty-four-hour-old biofilms grown on MBEC plates by a FabI P. aeruginosa-deficient strain (Tn::fabI mutant) were treated for 6 h with triclosan (100 μM), tobramycin (500 μM), gentamicin (100 μM), or streptomycin (100 μM), alone and in combination. (B) Twenty-four-hour-old biofilms grown on MBEC plates by PAO1 were treated for 6 h with triclocarban (100 μM) or tobramycin (500 μM), alone or in combination. The number of viable cells within the biofilms were quantified using the BacTiter-Glo assay. The assay was performed at least three times in triplicate. The results represent means ± the SEM. A one-way ANOVA followed by Bonferroni's multiple-comparison post hoc test was used to determine statistical significance compared to tobramycin alone (*, P < 0.05). NS, not significant.
Triclosan and tobramycin kill persister cells.
We hypothesized that the combination therapy of triclosan and tobramycin may enhance killing by targeting persister cell populations within biofilms. Persister cells are dormant nongrowing cells that are recalcitrant to antimicrobial therapy (41). We examined the ability of the combination of triclosan and tobramycin to kill P. aeruginosa PAO1 persister cells by performing a time-kill assay on 20-h-old stationary cells, which are enriched for persister cells (42). We found that the combination of triclosan and tobramycin significantly enhanced persister cell killing compared to that with either antimicrobial alone (Fig. 9). By 8 h, the combination resulted in an ∼100-fold reduction in persister cells compared with that with tobramycin alone, and the classic persister biphasic killing pattern was not observed. At 24 h, the combination exhibited a 6-log10 increase in killing versus that with tobramycin alone, and viable cells could not be recovered (<10 CFU/ml).
FIG 9.
Tobramycin combined with triclosan kills persister cells. Twenty-hour-old stationary-phase cells were treated with triclosan (100 μM) or tobramycin (50 μM), alone and in combination, for 6 h. At 0, 2, 4, 6, 8, and 24 h, aliquots were taken for enumeration (CFU per milliliter). The experiment was performed three times in triplicate. The results represent means ± the SEM.
Intratracheal administration of triclosan to the lungs of rats exhibits mild clinical symptoms.
As there are limited data regarding the toxicity of triclosan delivered directly to the lungs, we performed 1-day acute-exposure and a 7-day repeated-exposure rat toxicity studies. In the 1-day acute-exposure study, triclosan concentrations of 10, 30, 100, 300, and 1,000 μg/kg of body weight were delivered by transoral intratracheal installation into the lungs of Sprague-Dawley rats, and 24 h following treatment, the animals were sacrificed for analysis. For reference, ∼10 μg/ml corresponds to the effective dose that we determined in vitro. Overall, both complete blood count (CBC) and serum chemistry analyses for triclosan-treated animals were within the reference ranges, with the exception of neutropenia observed at the highest triclosan dose of 1,000 μg/kg. The concentration of triclosan in plasma was assessed by liquid chromatography-tandem mass spectrometry (LC-MS/MS), and triclosan was not detected in any animal, suggesting limited to no systemic absorption. Histopathology assessment revealed mild perivascular edema within adventitia around pulmonary veins and mild type II pneumocyte hyperplasia. In general, pathological changes were mild and may not be sufficiently severe to present clinical signs.
In the repeated-exposure toxicity study, doses of 30, 100, and 300 μg/kg triclosan were administered by intratracheal instillation consecutively for 7 days. We found no significant difference in body weights of the treated rats compared to those of the controls. CBC and serum chemistry analyses for triclosan-treated animals were within reference ranges. At the conclusion of the study, blood was analyzed for triclosan by LC-MS/MS. Triclosan was below the limit of detection of our assay in each sample tested, again indicating limited to no systemic absorption despite repeated exposures. The histopathology assessment revealed perivascular edema, with severity correlating with increasing doses of triclosan. The overall histopathology assessment concluded that the observed triclosan-dependent changes were mild and of insufficient severity to present clinical signs.
DISCUSSION
The development of antibiotic adjuvants or antiresistance compounds that are effective when combined with antibiotics but have no effect on their own is an attractive strategy for new therapies to treat problematic bacterial infections (18). Further, drug repurposing or repositioning affords many benefits, including reduced costs and accelerated deployment (43–45). We identified triclosan as a promising lead to enhance aminoglycoside killing of P. aeruginosa biofilms. Triclosan exhibited no activity on its own, which should delay the development of resistance, while demonstrating activity with the greatest number of antipseudomonal aminoglycoside antibiotics, showing broad-spectrum augmentation. Together, these results demonstrate that triclosan is a viable lead for further research.
Triclosan has been used for the past 3 decades as a general antimicrobial and antifungal in toothpaste and plastics (31). Because of decades of overuse, the FDA has recently restricted the use of triclosan mainly due to concerns over bioaccumulation and the potential for induction of resistance to other antibiotics in bacteria. Importantly, in these rulings, the FDA declared that there was not enough evidence to consider triclosan a generally recognized as safe (GRAS) compound, but it did not otherwise address potential toxicity. However, numerous safety studies have concluded that triclosan has acceptable safety parameters when administered to humans. For example, a 4-year study on humans found that routine use of toothpaste containing 0.3% triclosan, as is found in Colgate Total (Colgate-Palmolive Company), had no adverse effects on the endocrine system (19). Importantly, this is 333× the concentration of triclosan that is effective at enhancing aminoglycoside activity. A second study evaluated the accumulation of triclosan in humans via exposure to consumer products and also found no adverse health outcomes (20). Furthermore, in a human safety study totaling 1,246 participants who used toothpaste and mouthwash containing up to 0.6% triclosan for up to 12 weeks, no adverse effects were found (21). Finally, the Scientific Committee of Consumer Products of the European Union recently released a comprehensive report that summarized hundreds of triclosan toxicity studies, including human oral dose toxicity studies (22). The toxicity level for the majority of the studies is >50 mg/kg of body weight, again orders of magnitude higher than the effective dose we report in our work. Together, these studies show that triclosan is safe when used appropriately.
Triclosan has been used routinely in consumer products, raising the specter of cross-resistance. Interestingly, cross-resistance studies have been inconclusive. Several studies have shown that even prolonged exposure to triclosan does not lead to cross-resistance (46). Furthermore, comprehensive environmental surveys have found no association between triclosan and resistance in the environment (31). Nevertheless, the potential for cross-resistance remains, and triclosan should only be used for appropriate applications.
We envision the use of triclosan at a low concentration, ∼30 μM, in combination with tobramycin or other aminoglycosides, as an inhaled aerosolized solution into the lungs of CF patients. This route of administration provides many benefits, including fewer side effects, due to reduced systemic absorption, and enhanced activity, due to direct delivery to the lungs (47, 48). To further assess triclosan toxicity when administered to the lungs, we performed both single and repeated intratracheal instillation toxicity studies on Sprague-Dawley rats using concentrations up to 1,000 μg/kg triclosan and found only mild clinical symptoms, with little significant change to lung histology or blood chemistry. Furthermore, we developed a sensitive LC-MS/MS assay for triclosan, and no triclosan was detected in the blood when administered to the lung, indicating little systemic absorption (limit of detection, 300 ng/ml). Although more toxicity studies are needed, the safety profile of triclosan suggests that it is a worthy candidate for further exploration.
P. aeruginosa is also responsible for a number of other infections, including nonhealing chronic wounds, such as diabetic foot ulcers and burns (23–25). These wounds are often multispecies infections consisting of both Gram-negative and Gram-positive bacteria. We have found that triclosan in combination with tobramycin and alone is effective at killing B. cenocepacia and S. aureus biofilms, indicating this combination could target both Gram-negative and Gram-positive pathogens. Both P. aeruginosa and S. aureus infect chronic nonhealing wounds; thus, this combination of triclosan with aminoglycosides might more effectively treat these infections (24).
Triclosan is an inhibitor of the enoyl-acyl carrier protein reductase FabI, corrupting fatty acid synthesis (Fig. 1) (26, 27). Intriguingly, P. aeruginosa is inherently resistant to triclosan due to three resistance mechanisms: (i) slowed diffusion through the outer membrane (OM), (ii) a triclosan-specific efflux pump, and (iii) a triclosan-resistant enoyl-acyl carrier, FabV (28–30). We hypothesize that aminoglycosides may first disrupt the OM creating fissures by “self-promoted uptake,” allowing triclosan to readily enter the cell bypassing the first two resistance mechanisms, potentially reaching concentrations that overcome FabV resistance (49, 50). Additional publications support this hypothesis, finding that triclosan is able to cause cell death in an efflux pump mutant, despite the presence of FabV (29). In fact, Neckles and colleagues found that FabV could be inhibited by micromolar concentrations of triclosan and diphenyl ethers (51). In addition, our data suggest that FabI is not the sole target of triclosan, given that the combination remains effective against a FabI-deficient strain (Fig. 8A). Moreover, our results suggests a biofilm-specific mechanism of action. This is a curious observation and highlights the genetic and phenotypic differences between planktonic cells and cells within biofilms (52). The mechanism of action of this combination is under investigation.
Persister cells represent a major tolerance mechanism in biofilms and are responsible for recurrent infections due to their ability to regrow and reform biofilms once antimicrobial levels drop or therapies are ended. Moreover, high-persister-cell mutants are often isolated from CF patients (36). For this reason, it is important to develop therapies that effectively target these dormant cells (53). There are currently few treatments that eradicate persister cells, and thus, the combination of triclosan with aminoglycosides is a significant advance. Understanding the mechanism by which this combination kills persister cells could identify new therapeutic targets.
We report that triclosan, combined with three commonly used antipseudomonal aminoglycosides, killed highly tolerant and tobramycin-resistant P. aeruginosa biofilms. Our results show that the combination increases both the rate and degree of killing of cells within P. aeruginosa biofilms, which has important clinical implications for the treatment of biofilm infections in CF patients. Moreover, killing activity against a variety of CF P. aeruginosa clinical isolates, including tobramycin-resistant isolates, further suggests clinical potential for the treatment of lung infections in CF patients. Importantly, we found that significantly lower concentrations of tobramycin can be used when combined with triclosan for maximum efficacy. This reduction in aminoglycosides would have significant benefits, as these antimicrobials are known to be nephrotoxic and ototoxic, especially in CF patients who receive high doses throughout their lives (54, 55).
The repositioning of drugs for use in humans has a history of past success. A notable example is ivermectin, originally used for livestock, as it has since been repurposed for head lice, scabies, and river blindness, among several other uses (56). The use of triclosan as an aminoglycoside adjuvant against biofilms could represent a viable option for treating biofilm-based infections.
MATERIALS AND METHODS
Bacterial strains, culture conditions, antibiotics, and compounds.
The strains used in this study are listed in Table 2. Unless stated, bacterial strains were grown in 8-ml glass test tubes (18 by 150 mm) at 35°C in cation-adjusted Mueller-Hinton broth II (MHB; Sigma-Aldrich) with agitation at 210 rpm. Biofilms were grown using the minimum biofilm eradication concentration (MBEC) assay (Innovotech), as previously described (57). For MBEC experiments, 1 ml of the culture was pelleted and washed three times in 10% MHB in Dulbecco's phosphate-buffered saline with magnesium and calcium (DPBS; Sigma-Aldrich) and diluted to an optical density at 600 nm (OD600) of 0.001. Ten percent MHB was used to slow bacterial growth and avoid the rapid exhaustion of nutrients that occurs when using more-nutrient-rich media. Diluted culture at a concentration of 160 μl/well was seeded into a 96-well MBEC plate and incubated for 24 h at 35°C in a humidified chamber with agitation at 150 rpm. Twenty-four-hour-old biofilms grown on the peg lid were then transferred to a 96-well plate filled with DPBS and washed for 5 min to remove nonadherent cells before being assayed. Bacteria were plated on Dey-Engley neutralizing agar plates (DEA), which neutralize the activities of disinfectants and antiseptics (Sigma-Aldrich), or on tryptic soy agar plates (TSA; Sigma-Aldrich). Antimicrobial activity was neutralized using Dey-Engley (D/E) medium before plating. All antibiotics were obtained from Sigma-Aldrich. Tobramycin sulfate, gentamicin sulfate, and streptomycin sulfate were dissolved in autoclaved deionized water and filter sterilized using 0.22-μm-pore-size filter membranes (Thomas Scientific). Triclosan (2,4,4′-trichloro-2′-hydroxydiphenyl ether) and triclocarban [1-(4-chlorophenyl)-3-(3,4-dichlorophenyl)urea] were dissolved in 100% ethanol. For this study, all susceptibility testing was performed in 1% MHB diluted in DPBS to prevent further growth.
BacTiter-Glo assay calibration curve.
The BacTiter-Glo microbial cell viability assay (Promega) is a bioluminescent assay that determines the number of viable cells present based on a quantification of ATP concentration, as previously described (58, 59). To confirm that the BacTiter-Glo assay can be used to reliably determine the number of cells, a calibration curve was performed. One milliliter of a 16-h overnight culture of P. aeruginosa PAO1 was washed three times and diluted 2-fold in a black 96-well ViewPlate (PerkinElmer) with 1% MHB. Aliquots were taken from each dilution series for CFU enumeration on TSA. The BacTiter-Glo assay was then performed according to the manufacturer's specifications to enumerate cell viability. The plate was incubated using the BacTiter-Glo assay for 5 min, and then the luminescence per well was measured using an EnVision multilabel plate reader (PerkinElmer, Waltham, MA). Data were plotted as the average luminescence for each dilution series in triplicate versus the average CFU per milliliter for each dilution series in triplicate. We derived a coefficient of determination (r2 = 0.9884) for luminescence versus CFU per milliliter using a linear regression (Fig. S1) and determined that the limit of detection for the BacTiter-Glo assay was 1,000 CFU/ml.
High-throughput screen.
We screened 6,080 compounds from the Prestwick (Prestwick Chemical), MS2400 (Spectrum Collection), LOPAC1280 (Sigma-Aldrich), and Focused Collections libraries at the University of Michigan (UM) Center for Chemical Genomics (CCG). An overnight culture was prepared as described above, 30 μl/well was seeded into a 384-well plate (Corning), and a 384-pin tool (Scinomix) was inserted. The plate-pin combination was incubated for 18 h at 37°C in a humidified chamber without agitation. The pin tool was then transferred to a plate filled with 40 μl/well DPBS to remove nonadherent cells and debris and then transferred to a 384-well plate filled with 40 μl/well compounds alone, compounds with tobramycin, or sterile medium. Compounds were used at a concentration of 10 μM in 1% MHB. Tobramycin was used at a concentration of 250 μg/ml (∼500 μM, 500× the MIC) in 1% MHB. A concentration of 250 μg/ml tobramycin was chosen because it leads to ∼50% killing of the biofilm, as described below. Compounds were dissolved in dimethyl sulfoxide (DMSO). Polymyxin B was used as a positive control at 10 μg/ml, and 1% DMSO was used as a negative control. After 6 h of static treatment at 37°C, the pin tool was washed in a 384-well plate filled with 40 μl/well DPBS for 5 min to reduce the carryover of various treatments. Then, the pin tool was transferred to a 384-white well plate (Greiner) to prevent luminescence cross talk, filled with 40 μl/well BacTiter-Glo (25% [vol/vol] in DPBS) for 5 min to enumerate cell viability, and the luminescence per peg was measured using a EnVision multilabel plate reader. Luminescence data were plotted as % reduction calculated as ([untreated luminescence − treated luminescence]/untreated luminescence) × 100.
Biofilm susceptibility testing using BacTiter-Glo assay.
Twenty-four-hour-old biofilms formed on the lid of an MBEC plate were transferred to the 96-well plate in which the dilutions had been made and incubated for 6 h at 35°C without agitation. The MBEC lid was washed for 5 min in DPBS to reduce carryover and transferred to a black 96-well ViewPlate filled with 160 μl/well 25% BacTiter-Glo to enumerate cell viability, as described above. Data were plotted as the average luminescence for each condition tested in triplicate. Dose-response curves (DRC), time-kill curves, and checkerboard experiments were performed similarly.
MIC.
MICs were determined using the broth microdilution technique (60). Microdilutions of each aminoglycoside and triclosan were made in a 96-well plate. Cells were added at a concentration of ∼1 × 106 CFU/ml. The plates were then incubated for 24 h at 35°C in a humidified chamber with agitation at 150 rpm. After the 24-h incubation, the absorbance at 595 nm was measured using a SpectraMax M5 microplate spectrophotometer system (Molecular Devices Sunnyvale, CA). MIC breakpoints were chosen as the minimum concentration in which no turbidity greater than background was measured.
Crystal violet staining.
To study biofilm dispersal under static conditions, crystal violet staining was performed as previously described (61). Twenty-four-hour-old biofilms formed on MBEC plates, as described above, were stained with 0.41% crystal violet solubilized in 12% ethanol in a 96-well plate following 6 h of treatment.
Flow cell assays.
To study biofilm dispersal under flow conditions, biofilms were grown in disposable flow cells (Stovall Life Science, Greensboro, NC), as previously described (62). Briefly, the inlet side of the flow cell was connected to a reservoir filled with 10% MHB, and the outlet side was connected to a waste reservoir. Each flow cell was injected with 0.5 ml of a 16-h overnight culture, and the chamber was incubated at 37°C for 1 h in 100% MHB. The flow was then resumed at a rate of 0.2 ml/min. After 24 h, biofilms were treated for 6 h with triclosan and tobramycin or with either compound alone. Biofilms were then stained with the LIVE/DEAD cell viability assay using SYTO9 and propidium iodide dyes (Thermo Fisher Scientific).
Persister enrichment and killing assays.
To determine the effects of triclosan combined with tobramycin on persister cells, planktonic stationary-phase cultures were used as previously described (63). Cultures were grown for 20 h and 100 μl/well cells were dispersed in a 96-well plate. Treatments were added, and the plate was incubated at 37°C without agitation. At 2, 4, 6, 8, and 24 h, 30-μl aliquots were serially diluted and plated on DEA, and the CFU were enumerated. The dilutions that contained 3 to 30 colonies per 10 μl were counted. Eradication was recorded if there were no colonies found in the drop plating in triplicate of a 1 × 10−10 dilution, and thus, the limit of detection for this assay is 10 CFU/ml.
Triclosan toxicity studies.
Both the 1-day acute-toxicity study and 7-day repeated-exposure toxicity studies were performed in collaboration with the Michigan State University In Vivo Pharmacology Facility.
(i) One-day acute study.
Sprague-Dawley rats were anesthetized with isoflurane and received treatments by transoral intratracheal instillation. A control group (n = 4) received 1% DMSO in sterile saline. Dosing groups received triclosan at 10, 30, 100, 300, or 1,000 μg/kg, at n = 3 per each dosing. Twenty-four hours after dosing, ≥2 ml blood was collected under isoflurane anesthesia, and the animals were subsequently euthanized. Blood samples were transferred to tubes containing dipotassium EDTA (K2-EDTA) to prevent clotting or to serum separator tubes for complete blood count (CBC), quantifying systemic triclosan absorption using liquid chromatography-tandem mass spectrometry (LC-MS/MS), and subjected to clinical chemistry evaluation. Lungs were collected en bloc, weighed, and inflation fixed in 10% formalin. The left lung was processed using standard hematoxylin and eosin (H&E) staining. Microscopic evaluations of the lungs underwent blind examination by the Department of Pathobiology and Diagnostic Investigation at MSU by a board-certified veterinary pathologist. Using GraphPad Prism 6.04, statistical analysis of body weight, organ weights, CBC, and clinical chemistry results were compared by a Kruskal-Wallis one-way analysis of variance, followed by post hoc testing using Dunn's multiple-comparison test.
(ii) Seven-day repeated-exposure study.
Sprague-Dawley rats were administered vehicle or triclosan at 30, 100, or 300 μg/kg via transoral intratracheal instillation daily for 7 consecutive days (n = 5). Rats were weighed prior to dosing on day 1 and on day 7. On day 7, ∼1.5 h after dosing, ≥2 ml blood was collected. Blood samples were processed as described above. Lungs were collected en bloc and processed, evaluated, and analyzed as described above.
Mass spectrometry quantification of triclosan in serum.
Serum samples were filtered through a 0.45-μm-pore-size syringe filter (Titan3 polyvinylidene difluoride [PVDF]; Thermo Scientific) using a 1-ml syringe (BD) into a glass insert (Agilent Technologies) of a 2-ml clear MS vial (Restek). Samples were analyzed for triclosan via LC-MS/MS on a Quattro Premier XE mass spectrometer (Waters) coupled with an Acquity ultraperformance LC (UPLC) system (Waters). Triclosan was detected using electrospray ionization using multiple-reaction monitoring in negative-ion mode at m/z 286.94→35.00. The MS parameters were as follows: capillary voltage, 3.5 kV; cone voltage, 35 V; collision energy, 25 V; source temperature, 120°C; desolvation temperature, 350°C; cone gas flow (nitrogen), 50 liters/h; desolvation gas flow (nitrogen), 800 liters/h; collision gas flow (nitrogen), 0.17 ml/min; and multiplier voltage, 650 V. Chromatography separation was reverse phase using an Acquity UPLC BEH C18 column (50 by 2.1 mm, 1.7 μM; Waters) at a temperature of 40°C, with a flow rate of 0.4 ml/min and the following gradient of solvent A (0.1% formic acid) to solvent B (acetonitrile in high-performance liquid chromatography water): t = 0 min, 95%A/5% B; t = 3 min, 5%A/95% B; t = 4.01 min, 95%A/5% B to 5 min.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants from the Hunt for a Cure and the NSF BEACON Center for Evolution in Action (grant DBI-0939454) to C.M.W. and M.H.M., NIH grants GM109259 and GM110444 to C.M.W., an MSU Strategic Partnership Grant to C.M.W., and the Cystic Fibrosis Foundation Traineeship to M.M.M. In addition, M.M.M. was supported by a Wentworth Fellowship from the MSU Microbial and Molecular Genetics department.
We thank all members of the Waters' laboratory for their helpful scientific discussions, including Geoff Severin, Nico Fernandez, and Meng-lun Hsieh. We thank Neal Hammer for generously providing the S. aureus strains USA_300_JE2 and Newman. We thank Martha Larsen and Tom McQuade at the Center for Chemical Genomics at the University of Michigan for performing the HTS. We also thank Marc Bailie, Halen Agnew, Matt Bernard, and Teresa Krieger-Burke at the MSU In Vivo Facility for performing the triclosan toxicity studies. We also thank the MSU Mass Spectrometry Core of the Research Technology Support Facility, specifically Lijun Chen, Tony Schilmiller, and Dan Jones.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.00146-18.
REFERENCES
- 1.O'Sullivan BP, Freedman SD. 2009. Cystic fibrosis. Lancet 373:1891–1904. doi: 10.1016/S0140-6736(09)60327-5. [DOI] [PubMed] [Google Scholar]
- 2.van Gool K, Norman R, Delatycki MB, Hall J, Massie J. 2013. Understanding the costs of care for cystic fibrosis: an analysis by age and health status. Value Health 16:345–355. doi: 10.1016/j.jval.2012.12.003. [DOI] [PubMed] [Google Scholar]
- 3.Ouyang L, Grosse SD, Amendah DD, Schechter MS. 2009. Healthcare expenditures for privately insured people with cystic fibrosis. Pediatr Pulmonol 44:989–996. doi: 10.1002/ppul.21090. [DOI] [PubMed] [Google Scholar]
- 4.Agrawal A, Agarwal A, Mehta D, Sikachi RR, Du D, Wang J. 2017. Nationwide trends of hospitalizations for cystic fibrosis in the United States from 2003 to 2013. Intractable Rare Dis Res 6:191–198. doi: 10.5582/irdr.2017.01043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cutting GR. 2015. Cystic fibrosis genetics: from molecular understanding to clinical application. Nat Rev Genet 16:45–56. doi: 10.1038/nrg3849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stoltz DA, Meyerholz DK, Welsh MJ. 2015. Origins of cystic fibrosis lung disease. N Engl J Med 372:1574–1575. doi: 10.1056/NEJMc1502191. [DOI] [PubMed] [Google Scholar]
- 7.Shah VS, Meyerholz DK, Tang XX, Reznikov L, Abou Alaiwa M, Ernst SE, Karp PH, Wohlford-Lenane CL, Heilmann KP, Leidinger MR, Allen PD, Zabner J, McCray PB, Ostedgaard LS, Stoltz DA, Randak CO, Welsh MJ. 2016. Airway acidification initiates host defense abnormalities in cystic fibrosis mice. Science 351:503–507. doi: 10.1126/science.aad5589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Borowitz D. 2015. CFTR, bicarbonate, and the pathophysiology of cystic fibrosis. Pediatr Pulmonol 50(Suppl 40):S24–S30. doi: 10.1002/ppul.23247. [DOI] [PubMed] [Google Scholar]
- 9.Lewis K. 2001. Riddle of biofilm resistance. Antimicrob Agents Chemother 45:999–1007. doi: 10.1128/AAC.45.4.999-1007.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Costerton JW, Lewandowski Z, Caldwell DE, Korber DR, Lappin-Scott HM. 1995. Microbial biofilms. Annu Rev Microbiol 49:711–745. doi: 10.1146/annurev.mi.49.100195.003431. [DOI] [PubMed] [Google Scholar]
- 11.Bigger J. 1944. Treatment of staphylococcal infections with penicillin by intermittent sterilisation. Lancet 244:497–500. doi: 10.1016/S0140-6736(00)74210-3. [DOI] [Google Scholar]
- 12.Keren I, Kaldalu N, Spoering A, Wang Y, Lewis K. 2004. Persister cells and tolerance to antimicrobials. FEMS Microbiol Lett 230:13–18. doi: 10.1016/S0378-1097(03)00856-5. [DOI] [PubMed] [Google Scholar]
- 13.Folkesson A, Jelsbak L, Yang L, Johansen HK, Ciofu O, Høiby N, Molin S. 2012. Adaptation of Pseudomonas aeruginosa to the cystic fibrosis airway: an evolutionary perspective. Nat Rev Microbiol 10:841–851. doi: 10.1038/nrmicro2907. [DOI] [PubMed] [Google Scholar]
- 14.Govan JR, Deretic V. 1996. Microbial pathogenesis in cystic fibrosis: mucoid Pseudomonas aeruginosa and Burkholderia cepacia. Microbiol Rev 60:539–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chmiel JF, Aksamit TR, Chotirmall SH, Dasenbrook EC, Elborn JS, LiPuma JJ, Ranganathan SC, Waters VJ, Ratjen FA. 2014. Antibiotic management of lung infections in cystic fibrosis. I. The microbiome, methicillin-resistant Staphylococcus aureus, Gram-negative bacteria, and multiple infections. Ann Am Thorac Soc 11:1120–1129. doi: 10.1513/AnnalsATS.201402-050AS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tingpej P, Smith L, Rose B, Zhu H, Conibear T, Nassafi Al K, Manos J, Elkins M, Bye P, Willcox M, Bell S, Wainwright C, Harbour C. 2007. Phenotypic characterization of clonal and nonclonal Pseudomonas aeruginosa strains isolated from lungs of adults with cystic fibrosis. J Clin Microbiol 45:1697–1704. doi: 10.1128/JCM.02364-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stuart B, Lin JH, Mogayzel PJ Jr. 2010. Early eradication of Pseudomonas aeruginosa in patients with cystic fibrosis. Paediatr Respir Rev 11:177–184. doi: 10.1016/j.prrv.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gill EE, Franco OL, Hancock REW. 2014. Antibiotic adjuvants: diverse strategies for controlling drug-resistant pathogens. Chem Biol Drug Des 85:56–78. doi: 10.1111/cbdd.12478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cullinan MP, Palmer JE, Carle AD, West MJ, Seymour GJ. 2012. Long term use of triclosan toothpaste and thyroid function. Sci Total Environ 416:75–79. doi: 10.1016/j.scitotenv.2011.11.063. [DOI] [PubMed] [Google Scholar]
- 20.Rodricks JV, Swenberg JA, Borzelleca JF, Maronpot RR, Shipp AM. 2010. Triclosan: a critical review of the experimental data and development of margins of safety for consumer products. Crit Rev Toxicol 40:422–484. doi: 10.3109/10408441003667514. [DOI] [PubMed] [Google Scholar]
- 21.Fang J-L, Stingley RL, Beland FA, Harrouk W, Lumpkins DL, Howard P. 2010. Occurrence, efficacy, metabolism, and toxicity of triclosan. J Environ Sci Health C Environ Carcinog Ecotoxicol Rev 28:147–171. doi: 10.1080/10590501.2010.504978. [DOI] [PubMed] [Google Scholar]
- 22.Scientific Committee on Consumer Safety (SCCS). 2010. Scientific Committee on Consumer Safety (SCCS): opinion on triclosan–antimicrobial resistance. Directorate-General for Health and Consumers, European Union, Brussels, Belgium: https://ec.europa.eu/health/scientific_committees/consumer_safety/docs/sccs_o_023.pdf. [Google Scholar]
- 23.Sivanmaliappan TS, Sevanan M. 2011. Antimicrobial susceptibility patterns of Pseudomonas aeruginosa from diabetes patients with foot ulcers. Int J Microbiol 2011:605195. doi: 10.1155/2011/605195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Omar A, Wright J, Schultz G, Burrell R, Nadworny P. 2017. Microbial biofilms and chronic wounds. Microorganisms 5:9–15. doi: 10.3390/microorganisms5010009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Altoparlak U, Erol S, Akcay MN, Celebi F, Kadanali A. 2004. The time-related changes of antimicrobial resistance patterns and predominant bacterial profiles of burn wounds and body flora of burned patients. Burns 30:660–664. doi: 10.1016/j.burns.2004.03.005. [DOI] [PubMed] [Google Scholar]
- 26.Heath RJ, Rubin JR, Holland DR, Zhang E, Snow ME, Rock CO. 1999. Mechanism of triclosan inhibition of bacterial fatty acid synthesis. J Biol Chem 274:11110–11114. doi: 10.1074/jbc.274.16.11110. [DOI] [PubMed] [Google Scholar]
- 27.McMurry LM, Oethinger M, Levy SB. 1998. Triclosan targets lipid synthesis. Nature 394:531–532. doi: 10.1038/28970. [DOI] [PubMed] [Google Scholar]
- 28.Zhu L, Lin J, Ma J, Cronan JE, Wang H. 2010. Triclosan resistance of Pseudomonas aeruginosa PAO1 is due to FabV, a triclosan-resistant enoyl-acyl carrier protein reductase. Antimicrob Agents Chemother 54:689–698. doi: 10.1128/AAC.01152-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chuanchuen R, Karkhoff-Schweizer RR, Schweizer HP. 2003. High-level triclosan resistance in Pseudomonas aeruginosa is solely a result of efflux. Am J Infection Control 31:124–127. doi: 10.1067/mic.2003.11. [DOI] [PubMed] [Google Scholar]
- 30.Champlin FR, Ellison ML, Bullard JW, Conrad RS. 2005. Effect of outer membrane permeabilisation on intrinsic resistance to low triclosan levels in Pseudomonas aeruginosa. Int J Antimicrob Agents 26:159–164. doi: 10.1016/j.ijantimicag.2005.04.020. [DOI] [PubMed] [Google Scholar]
- 31.Russell AD. 2004. Whither triclosan? J Antimicrob Chemother 53:693–695. doi: 10.1093/jac/dkh171. [DOI] [PubMed] [Google Scholar]
- 32.Barclay ML, Begg EJ. 2001. Aminoglycoside adaptive resistance: importance for effective dosage regimens. Drugs 61:713–721. doi: 10.2165/00003495-200161060-00001. [DOI] [PubMed] [Google Scholar]
- 33.Gilleland LB, Gilleland HE, Gibson JA, Champlin FR. 1989. Adaptive resistance to aminoglycoside antibiotics in Pseudomonas aeruginosa. J Med Microbiol 29:41–50. doi: 10.1099/00222615-29-1-41. [DOI] [PubMed] [Google Scholar]
- 34.Lorian V, Silletti RP, Biondo FX, De Freitas CC. 1979. Paradoxical effect of aminoglycoside antibiotics on the growth of Gram-negative bacilli. J Antimicrob Chemother 5:613–616. doi: 10.1093/jac/5.5.613. [DOI] [PubMed] [Google Scholar]
- 35.Kindrachuk KN, Fernández L, Bains M, Hancock REW. 2011. Involvement of an ATP-dependent protease, PA0779/AsrA, in inducing heat shock in response to tobramycin in Pseudomonas aeruginosa. Antimicrob Agents Chemother 55:1874–1882. doi: 10.1128/AAC.00935-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mulcahy LR, Burns JL, Lory S, Lewis K. 2010. Emergence of Pseudomonas aeruginosa strains producing high levels of persister cells in patients with cystic fibrosis. J Bacteriol 192:6191–6199. doi: 10.1128/JB.01651-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Heath RJ, Li J, Roland GE, Rock CO. 2000. Inhibition of the Staphylococcus aureus NADPH-dependent enoyl-acyl carrier protein reductase by triclosan and hexachlorophene. J Biol Chem 275:4654–4659. doi: 10.1074/jbc.275.7.4654. [DOI] [PubMed] [Google Scholar]
- 38.Jacobs MA, Alwood A, Thaipisuttikul I, Spencer D, Haugen E, Ernst S, Will O, Kaul R, Raymond C, Levy R, Chun-Rong L, Guenthner D, Bovee D, Olson MV, Manoil C. 2003. Comprehensive transposon mutant library of Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 100:14339–14344. doi: 10.1073/pnas.2036282100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Orsi M, Noro MG, Essex JW. 2011. Dual-resolution molecular dynamics simulation of antimicrobials in biomembranes. J R Soc Interface 8:826–841. doi: 10.1098/rsif.2010.0541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Walsh SE, Maillard J-Y, Russell AD, Catrenich CE, Charbonneau DL, Bartolo RG. 2003. Activity and mechanisms of action of selected biocidal agents on Gram-positive and -negative bacteria. J Appl Microbiol 94:240–247. doi: 10.1046/j.1365-2672.2003.01825.x. [DOI] [PubMed] [Google Scholar]
- 41.Lewis K. 2010. Persister cells. Annu Rev Microbiol 64:357–372. doi: 10.1146/annurev.micro.112408.134306. [DOI] [PubMed] [Google Scholar]
- 42.Lewis K. 2006. Persister cells, dormancy and infectious disease. Nat Rev Microbiol 5:48–56. [DOI] [PubMed] [Google Scholar]
- 43.Sharlow ER. 2016. Revisiting repurposing. Assay Drug Dev Technol 14:554–556. doi: 10.1089/adt.2016.766. [DOI] [PubMed] [Google Scholar]
- 44.Brown D. 2015. Antibiotic resistance breakers: can repurposed drugs fill the antibiotic discovery void? Nat Rev Drug Discov 14:821–832. doi: 10.1038/nrd4675. [DOI] [PubMed] [Google Scholar]
- 45.Brown ED, Wright GD. 2016. Antibacterial drug discovery in the resistance era. Nature 529:336–343. doi: 10.1038/nature17042. [DOI] [PubMed] [Google Scholar]
- 46.Saleh S, Haddadin RNS, Baillie S, Collier PJ. 2011. Triclosan–an update. Lett Appl Microbiol 52:87–95. doi: 10.1111/j.1472-765X.2010.02976.x. [DOI] [PubMed] [Google Scholar]
- 47.Quon BS, Goss CH, Ramsey BW. 2014. Inhaled antibiotics for lower airway infections. Ann Am Thorac Soc 11:425–434. doi: 10.1513/AnnalsATS.201311-395FR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Smith AL. 2002. Inhaled antibiotic therapy: what drug? What dose? What regimen? What formulation? J Cyst Fibros 1(Suppl 2):189–193. [DOI] [PubMed] [Google Scholar]
- 49.Montie T, Patamasucon P. 1995. Aminoglycosides: the complex problem of antibiotic mechanisms and clinical applications. Eur J Clin Microbiol Infect Dis 14:85–87. doi: 10.1007/BF02111863. [DOI] [PubMed] [Google Scholar]
- 50.Taber HW, Mueller JP, Miller PF, Arrow AS. 1987. Bacterial uptake of aminoglycoside antibiotics. Microbiol Rev 51:439–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Neckles C, Pschibul A, Lai C-T, Hirschbeck M, Kuper J, Davoodi S, Zou J, Liu N, Pan P, Shah S, Daryaee F, Bommineni GR, Lai C, Simmerling C, Kisker C, Tonge PJ. 2016. Selectivity of pyridone- and diphenyl ether-based inhibitors for the Yersinia pestis FabV enoyl-ACP reductase. Biochemistry 55:2992–3006. doi: 10.1021/acs.biochem.5b01301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Folsom JP, Richards L, Pitts B, Roe F, Ehrlich GD, Parker A, Mazurie A, Stewart PS. 2010. Physiology of Pseudomonas aeruginosa in biofilms as revealed by transcriptome analysis. BMC Microbiol 10:294. doi: 10.1186/1471-2180-10-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schmidt NW, Deshayes S, Hawker S, Blacker A, Kasko AM, Wong GCL. 2014. Engineering persister-specific antibiotics with synergistic antimicrobial functions. ACS Nano 8:8786–8793. doi: 10.1021/nn502201a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ratjen F, Brockhaus F, Angyalosi G. 2009. Aminoglycoside therapy against Pseudomonas aeruginosa in cystic fibrosis: a review. J Cyst Fibros 8:361–369. doi: 10.1016/j.jcf.2009.08.004. [DOI] [PubMed] [Google Scholar]
- 55.Pedersen SS, Jensen T, Osterhammel D, Osterhammel P. 1987. Cumulative and acute toxicity of repeated high-dose tobramycin treatment in cystic fibrosis. Antimicrob Agents Chemother 31:594–599. doi: 10.1128/AAC.31.4.594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Õmura S. 2008. Ivermectin: 25 years and still going strong. Int J Antimicrob Agents 31:91–98. doi: 10.1016/j.ijantimicag.2007.08.023. [DOI] [PubMed] [Google Scholar]
- 57.Ceri H, Olson ME, Stremick C, Read RR, Morck D, Buret A. 1999. The Calgary Biofilm Device: new technology for rapid determination of antibiotic susceptibilities of bacterial biofilms. J Clin Microbiol 37:1771–1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pothineni VR, Wagh D, Babar MM, Inayathullah M, Solow-Cordero D, Kim K-M, Samineni AV, Parekh MB, Tayebi L, Rajadas J. 2016. Identification of new drug candidates against Borrelia burgdorferi using high-throughput screening. Drug Des Devel Ther 10:1307–1322. doi: 10.2147/DDDT.S101486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ballell L, Bates RH, Young RJ, Alvarez-Gomez D, Alvarez-Ruiz E, Barroso V, Blanco D, Crespo B, Escribano J, González R, Lozano S, Huss S, Santos-Villarejo A, Martín-Plaza JJ, Mendoza A, Rebollo-Lopez MJ, Remuiñan-Blanco M, Lavandera JL, Pérez-Herran E, Gamo-Benito FJ, García-Bustos JF, Barros D, Castro JP, Cammack N. 2013. Fueling open-source drug discovery: 177 small-molecule leads against tuberculosis. ChemMedChem 8:313–321. doi: 10.1002/cmdc.201200428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Andrews JM. 2001. Determination of minimum inhibitory concentrations. J Antimicrob Chemother 48(Suppl 1):5–16. [DOI] [PubMed] [Google Scholar]
- 61.Sambanthamoorthy K, Gokhale AA, Lao W, Parashar V, Neiditch MB, Semmelhack MF, Lee I, Waters CM. 2011. Identification of a novel benzimidazole that inhibits bacterial biofilm formation in a broad-spectrum manner. Antimicrob Agents Chemother 55:4369–4378. doi: 10.1128/AAC.00583-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sambanthamoorthy K, Luo C, Pattabiraman N, Feng X, Koestler B, Waters CM, Palys TJ. 2014. Identification of small molecules inhibiting diguanylate cyclases to control bacterial biofilm development. Biofouling 30:17–28. doi: 10.1080/08927014.2013.832224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Spoering AL, Lewis K. 2001. Biofilms and planktonic cells of Pseudomonas aeruginosa have similar resistance to killing by antimicrobials. J Bacteriol 183:6746–6751. doi: 10.1128/JB.183.23.6746-6751.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Holloway BW. 1955. Genetic recombination in Pseudomonas aeruginosa. J Gen Microbiol 13:572–581. [DOI] [PubMed] [Google Scholar]
- 65.De Soyza A, Hall AJ, Mahenthiralingam E, Drevinek P, Kaca W, Drulis-Kawa Z, Stoitsova SR, Toth V, Coenye T, Zlosnik JEA, Burns JL, Sá-Correia I, De Vos D, Pirnay J-P, Kidd TJ, Reid D, Manos J, Klockgether J, Wiehlmann L, Tümmler B, McClean S, Winstanley C, EU FP7 funded COST Action BM1003 “Cell surface virulence determinants of cystic fibrosis pathogens.” 2013. Developing an international Pseudomonas aeruginosa reference panel. Microbiologyopen 2:1010–1023. doi: 10.1002/mbo3.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.