Abstract
Summary
With the rapid development in next-generation sequencing, cost and time requirements for genomic sequencing are decreasing, enabling applications in many areas such as cancer research. Many tools have been developed to analyze genomic variation ranging from single nucleotide variants to whole chromosomal aberrations. As sequencing throughput increases, the number of variants called by such tools also grows. Often employed manual inspection of such calls is thus becoming a time-consuming procedure. We developed the Variant InsPector and Expert Rating tool (VIPER) to speed up this process by integrating the Integrative Genomics Viewer into a web application. Analysts can then quickly iterate through variants, apply filters and make decisions based on the generated images and variant metadata. VIPER was successfully employed in analyses with manual inspection of more than 10 000 calls.
Availability and implementation
VIPER is implemented in Java and Javascript and is freely available at https://github.com/MarWoes/viper.
Supplementary information
Supplementary data are available at Bioinformatics online.
1 Introduction
With the development of next-generation sequencing (NGS), genomic sequencing becomes applicable for many research areas, especially fields related to cancer research (Shen et al., 2015). Many tools have been developed to detect variation at different scales. For example, the GATK (McKenna et al., 2010) can be utilized to detect single nucleotide variants (SNVs) and small indels, while tools such as Pindel (Ye et al., 2009) aim at detecting larger structural variants (SVs). The output of variant callers should be handled with caution as there are many sources of errors such as sequencing errors or variants located in repetitive regions that lead to discordance between multiple callers (Hwang et al., 2016; Sandmann et al., 2017). To prevent erroneously called variants from distorting analysis results, expert based review and experimental validation of potential variants is often employed as a result, especially in clinical contexts. Reviewing variants usually includes visualizing the genomic regions surrounding the variant’s breakpoints with tools such as the Integrative Genomics Viewer (IGV) (Robinson et al., 2011; Thorvaldsdóttir et al., 2013).
However, with increasing throughput and decreasing cost of modern sequencing techniques, the rate at which genomes can be sequenced is also growing, leading to more sequenced genomes and more potential variants to be inspected. As a result, manually investigating and deciding which calls might be true positives becomes a cumbersome and time-consuming process. For example, using IGV investigation requires loading each sample, navigating to each breakpoint locus and documenting the decision for the variant call. This increases time requirements for expert review of variant calls and as a result hinders genomic analyses as well as development of novel detection algorithms that rely on annotated datasets with large numbers of calls.
2 Materials and methods
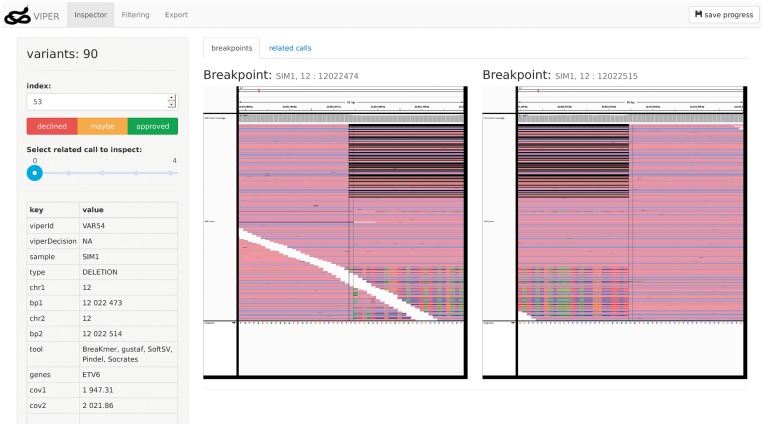
We present the Variant InsPector and Expert Rating tool (VIPER) to streamline the investigation and decision making process for variant calls. VIPER is a web application implemented in Java and Javascript that accepts variant calls as CSV or VCF files and alignment data as BAM files as input. It supports importing SNVs and small indels as well as larger SVs as described in detail in the Supplementary Material. Variant metadata such as coverage or quality information may be provided in the input. VIPER uses the IGV to generate images of breakpoint regions and displays these in the browser. The user can approve or decline variant calls based on these images, as shown in Figure 1. Additionally, calls can be filtered based on the metadata provided by the variant input file and the decisions made by the user. User decisions as filtering criteria enable complex incremental filtering without the need to specify a single complex filtering step.
Fig. 1.
VIPER’s Inspector tab. IGV images for regions containing the beginning and end of the currently inspected variant are presented on the right hand side. On the left side details about the variant are displayed. These contain breakpoint information and metadata such as coverage or quality metrics. Users can use the decision buttons to annotate the currently displayed variant
To reduce the number of calls to investigate, similar calls are optionally grouped together, treating them as single variants in the decision making process. This eases dealing with control-tumor pairs, as variants found in both control and tumor sample are grouped together. A detailed explanation of the filtering and the grouping can be found in the Supplementary Material.
After annotating the calls with decisions, the variant calls can be exported to CSV and XLSX files. VIPER is only dependent on Java (1.8) and a modern web browser and can be used on Windows, Linux and OS X systems. IGV is run in a headless environment on Linux systems.
3 Results and discussion
We applied VIPER to two explorative variant analyses for NGS datasets on a desktop machine with an Intel Core i5-4590 @ 3.30 GHz x 4 processor and 8 GB RAM running Ubuntu 16.04. We used ten tools to detect SVs on an amplicon-based dataset covering 111 patients diagnosed with myelodysplastic syndromes (MDS). The target region is ∼125 kbp in length with an average coverage of 3675×. The tools yielded 18 803 SV calls that were summarized to 8752 unique calls using VIPER. Since we were looking for variants with low variant allele frequency (VAF), we expected many false-positive calls. VIPER enabled discarding 8363 calls upon manual inspection and left only 389 candidate calls.
Another application consisted of 11 250 SNV and small indel calls for 491 control-tumor sample pairs. mtDNA (16 569 bp) was sequenced and analyzed for all samples with an average coverage of 4521×. Using VIPER it could be confirmed that all calls were correctly classified as true or false positives by the pipeline in use. Both analyses required ∼ 2 person-days each to manually inspect all calls. Despite the high number of calls, using VIPER enabled a fast inspection and decision making process.
Supplementary Material
Acknowledgements
We thank Dr. Aniek de Graaf, Radboud University Medical centre, Nijmegen, The Netherlands for providing the MDS dataset and Dr. Kornelius Kerl, Dr. Stefan Pfister and the DKFZ Heidelberg for providing the mtDNA dataset.
Funding
This work was supported by EU grants Triage-MDS (ERA-Net TRANSCAN grant no. BMBF 01KT1401), MDS-RIGHT (grant no. 634789) and Deutsche Krebshilfe (DKH grant 110495).
Conflict of Interest: none declared.
References
- Hwang S. et al. (2016) Systematic comparison of variant calling pipelines using gold standard personal exome variants. Sci. Rep., 5, 17875.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A. et al. (2010) The genome analysis toolkit: a mapreduce framework for analyzing next-generation dna sequencing data. Genome Res., 20, 1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson J.T. et al. (2011) Integrative genomics viewer. Nat. Biotechnol., 29, 24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandmann S. et al. (2017) Evaluating variant calling tools for non-matched next-generation sequencing data. Sci. Rep., 7, 43169.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen T. et al. (2015) Clinical applications of next generation sequencing in cancer: from panels, to exomes, to genomes. Front. Genet., 6, 215.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorvaldsdóttir H. et al. (2013) Integrative genomics viewer (IGV): high-performance genomics data visualization and exploration. Brief. Bioinf., 14, 178–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye K. et al. (2009) Pindel: a pattern growth approach to detect break points of large deletions and medium sized insertions from paired-end short reads. Bioinformatics, 25, 2865–2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.