Abstract
STUDY QUESTION
Are exosomal microRNAs (miRNAs) in seminal plasma (SP) useful as markers of the origin of azoospermia and the presence of sperm in the testis?
SUMMARY ANSWER
Our study demonstrated the potential of several miRNAs contained in small extracellular vesicles (sEVs) of seminal fluid as sensitive and specific biomarkers for selecting those azoospermic individuals with real chances of obtaining spermatozoa from the testicular biopsy.
WHAT IS KNOWN ALREADY
There are no precise non-invasive diagnostic methods for classifying the origin of the sperm defects in semen and the spermatogenic reserve of the testis in those infertile men with a total absence of sperm in the ejaculate (azoospermia). The diagnosis of such individuals is often based on the practice of biopsies. In this context it is reasonable to study the presence of organ-specific markers in human semen that contains fluid from the testis and the male reproductive glands, which could help in the diagnosis and prognosis of male infertility. Additionally, seminal fluid contains high concentrations of sEVs that are morphologically and molecularly consistent with exosomes, which originate from multiple cellular sources in the male reproductive tract.
STUDY DESIGN, SIZE, DURATION
A case and control prospective study was performed. This study compares the miRNA content of exosomes in semen samples obtained from nine normozoospermic fertile individuals (control group), 14 infertile men diagnosed with azoospermia due to spermatogenic failure, and 13 individuals with obstructive azoospermia and conserved spermatogenesis. Additionally, three severe oligozoospermic individuals (<5 × 106 sperm/ml) were included in the study.
PARTICIPANTS/MATERIALS, SETTING, METHODS
A differential high-throughput miRNA profiling analysis using miRNA quantitative PCR panels was performed in SP exosomes from azoospermic patients and fertile individuals.
MAIN RESULTS AND THE ROLE OF CHANCE
A total of 623 miRNAs were included in the miRNA profiling stage of the study. A total of 397 miRNAs (63.7%) were consistently detected in samples from all groups and statistically analysed, which revealed altered patterns of miRNA expression in infertile patients. We focused on the miRNAs that were differentially expressed between azoospermia as a result of an obstruction in the genital tract (i.e. having conserved spermatogenesis) and azoospermia caused by spermatogenic failure, and described, in a miRNA validation stage of the study, the expression values of one miRNA (miR-31-5p) in exosomes from semen as a predictive biomarker test for the origin of azoospermia with high sensitivity and specificity (>90%). The efficacy of the predictive test was even better when the blood FSH values were included in the analysis. Furthermore a model that included miR-539-5p and miR-941 expression values is also described as being useful for predicting the presence of residual spermatogenesis in individuals with severe spermatogenic disorders with diagnostic accuracy.
LIMITATIONS, REASONS FOR CAUTION
Further studies, with an independent second population involving a larger number of samples, are needed to confirm our findings.
WIDER IMPLICATIONS OF THE FINDINGS
Our findings contribute to the search for the most valuable genetic markers that are potentially useful as tools for predicting the presence of testicular sperm in azoospermic individuals.
STUDY FUNDING/COMPETING INTEREST(S)
This work was financially supported by grants from the Fondo de Investigaciones Sanitarias/Fondo Europeo de Desarrollo Regional “Una manera de hacer Europa” (FIS/FEDER) [Grant number PI15/00153], the Generalitat de Catalunya [Grant number 2014SGR5412]. S.L. is sponsored by the Researchers Stabilization Program (ISCIII/Generalitat de Catalunya) from the Spanish National Health System [CES09/020].
Keywords: azoospermia, miRNAs, small extracellular vesicles, seminal plasma, biomarker
Introduction
Approximately 4% of men worldwide suffer from infertility. In a high percentage of those men, the aetiology of infertility is closely related to alterations of the classic parameters of basic semen analysis, such as the concentration, motility and/or morphology of the spermatozoa (Guzick et al., 2001). Routine biochemical markers in semen, such as acid phosphatase, citric acid and zinc (prostate), fructose (seminal vesicles) and alpha-glucosidase (epididymis), are used to diagnose possible acquired and congenital obstructions or to identify functional abnormalities of the genital glands such as those that occur after infection or inflammation. Although still useful, these biochemical markers have limited clinical utility for classifying the origin of the sperm defects in semen, and the spermatogenic reserve of the testis in those men with a total absence of sperm in the ejaculate (azoospermia), a particularly challenging condition which accounts for more than 10% cases of male infertility (Tüttelmann and Nieschlag, 2010). There are no precise non-invasive diagnostic methods for determining such conditions, and in general the diagnosis is based on the practice of biopsies. In this context it is reasonable to study the presence of organ-specific markers in human fluids that could help in the diagnosis and prognosis of azoospermia. Semen, a complex fluid composed of cells and seminal plasma (SP), can be accessed with relative ease. The SP, which contains fluid from the testis and the male reproductive glands, is emerging as a source for potential biomarkers for reproductive diseases.
Several studies have shown that SP contains high concentrations of small extracellular vesicles (sEVs), that are morphologically and molecularly consistent with exosomes, which originate from multiple cellular sources in the male reproductive tract: namely prostate (prostasomes), epididymis (epididymosomes), seminal vesicles and the testis (Renneberg et al., 1997; Vojtech et al., 2014). Exosomes are cell-derived vesicles present in various biological fluids. It has been calculated that, in mammals, each ejaculate contains trillions of exosomes, characterized by a high content in cholesterol and sphingomyelin, and a very complex protein composition. Exosomes have immunosuppressive effects on the cells, which is relevant in the genital mucosa. The exosomes also contain coding and non-coding RNAs (Valadi et al., 2007) that can be transferred to recipient cells to modulate their function, thus mediating paracrine signalling (Valadi et al., 2007; Zomer et al., 2010). In particular, in male fertility the semen exosomes have been proposed as participating in the capacitation and acrosome reaction of sperm (Aalberts et al., 2013) and as functioning as a means of selectively transporting and delivering various regulatory molecules to the female reproductive system, thus contributing to fertilization (Vojtech et al., 2014).
Exosomes in semen contain a population of small non-coding RNAs (sncRNAs; from 20 to 100 nucleotides), including microRNAs (miRNAs) (21%), and several other RNAs such as Piwi-interacting RNAs (piRNAs), Y RNAs, rRNAs and tRNAs (Vojtech et al., 2014). Semen exosomes show a unique profile of miRNAs that greatly differs from that of other biological fluids (Vojtech et al., 2014). It seems that the content of RNA, and in particular of the miRNA, of the exosomes in semen varies according to the cell of origin of the sEVs, so it is able to reflect the pathophysiological conditions of the organ of origin. This fact is relevant when considering expression profiles of miRNAs in exosomes as potentially reliable biomarkers.
MiRNAs (19–22 nucleotides) negatively modulate gene expression at the post-transcriptional level, affecting mRNA stability and translation. Studies have shown that miRNAs play critical roles in a variety of biological processes such as cell proliferation, differentiation, apoptosis and carcinogenesis (Bartel, 2004; He and Hannon, 2004). miRNAs actively participate in diverse aspects of vertebrate differentiation and development (Lakshmipathy et al., 2007; Tang et al., 2007; Hammond, 2015), and, specifically, in testis differentiation in the embryo, male germline development and sperm production (Ro et al., 2007; Hayashi et al., 2008; Maatouk et al., 2008; Bouhallier et al., 2010). miRNAs are not only present in cells but also in extracellular milieu, especially in different biofluids including not only blood plasma (Arroyo et al., 2011), but also saliva, tears, urine, breast milk, colostrum, peritoneal fluid, cerebrospinal fluid, bronchial lavage and, strikingly, seminal fluid (Weber et al., 2010). Such extracellular miRNAs are presented either bound with protein complexes (Li et al., 2012) or contained in EVs such as the exosomes (Valadi et al., 2007). This attribute allows miRNAs to be remarkably resistant to the presence of high RNAse activity in the extracellular environment. Consequently, miRNAs in human fluids have been postulated as useful non-invasive biomarkers for human diseases.
The mRNAs and the miRNAs of SP that come from the testis or from the epididymis are hardly detectable in the blood, as they are contained in vesicles or bound to protein complexes and cannot cross the blood-testicular or hematoepididimary barriers. Studying them in SP would therefore seem to be a valuable approach but few studies have analysed the expression profile of miRNAs contained in the exosomes of the semen biofluid.
Our research group proposed the study of expression levels of exosome miRNAs in semen from men with severe spermatogenic disorders and vasectomized men to evaluate their use as non-invasive biomarkers that may contribute to the diagnosis of azoospermia. Additionally, it would help to describe the miRNAs contained in exosomes that come from testis and epididymis, as semen from vasectomized men does not contain secretions from these two organs. Furthermore, miRNAs derived from/associated with the presence of the germline would be identified when studying semen from men with deficient sperm production. The use of non-invasive biomarkers in azoospermia would help in determining the pathophysiological cause of the absence of sperm in semen and help with predicting the presence of sperm in testis to facilitate the optimal choice for sperm extraction.
Materials and Methods
Subjects of study
Patients and controls participating in the study were selected from men referred to the Andrology Service of the Fundació Puigvert. The study was approved by the Institutional Review Board of the Centre and all the participants signed a written informed consent.
Semen samples were obtained from nine normozoospermic (Nz) fertile individuals consulting for vasectomy (control group); 14 infertile men diagnosed with secretory azoospermia (SA) (no sperm in semen sample due to spermatogenic failure) or cryptozoospermia (<0.15 × 106 sperm/ml); and 13 individuals with obstructive azoospermia (OA) and conserved spermatogenesis including men successfully vasectomized (OA-V, n = 8) and individuals presenting pathological naturally occurring obstruction in the genital tract (OA-N, n = 5). Additionally, three severe oligozoospermic individuals (<5 × 106 sperm/ml) were included in the study (Table I).
Table I.
Clinical data of individuals included in the study of miRNA content of seminal plasma exosomes in azoospermia.
| Patient no. | Spermiogram | Subgroups | Male age (years)a | Testes volume (R, L) | FSH (IU/l) | Semen volume (ml) | pH (>7,2) | Sperm count (×106/ml) | Progressive motility (%) | Nomal morphology (%) | Fructose (umol/ejac) >13 | Citrate (umol/ejac) >52 | a-Glucosidase (mU/ejac) >20 | Naturally conceived children (n) | IVF conceived children (n, IVF cycles) | TESE value (million/ml) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Nz | Nz | 41 | 20, 20 | nd | 2.5 | 7.5 | 137 | 47 | 15 | nd | nd | nd | Yes (2) | – | – |
| 2 | Nz | Nz | 40 | 20, 20 | nd | 6.3 | 7.5 | 114 | 65 | 5 | nd | nd | nd | Yes (2) | – | – |
| 3 | Nz | Nz | 40 | 20, 20 | nd | 2.4 | 7.5 | 113 | 52 | 6 | nd | nd | nd | Yes (2) | – | – |
| 4 | Nz | Nz | 44 | 20, 20 | nd | 3.6 | 7.5 | 90 | 73 | 9 | nd | nd | nd | Yes (2) | – | – |
| 5 | Nz | Nz | 39 | 15, 15 | nd | 4.9 | 7.2 | 60 | 53 | 6 | nd | nd | nd | Yes (1) | – | – |
| 6 | Nz | Nz | 37 | 20, 20 | nd | 4.2 | 7.5 | 56 | 70 | 12 | nd | nd | nd | Yes (2) | – | – |
| 7 | Nz | Nz | 46 | 15, 15 | nd | 2.8 | 7.7 | 41 | 42 | 4 | nd | nd | nd | Yes (2) | – | – |
| 8 | Nz | Nz | 39 | 20, 20 | nd | 2.3 | 7.5 | 20 | 63 | 8 | nd | nd | nd | Yes (2) | – | – |
| 9 | Nz | Nz | 45 | 22, 22 | nd | 4.1 | 7.2 | 20 | 36 | 9 | nd | nd | nd | Yes (2) | – | – |
| 10 | AZO | SA (Sp+) | 39 | 6, 9 | 12.6 | 6.5 | 7.5 | 0.15 | 13 | 0 | 47 | 84 | 15 | No | – | nd |
| 11 | AZO | SA (Sp+) | 36 | 17, 0 | 15.3 | 4.0 | 7.2 | 0.1 | 4 | 0 | nd | nd | nd | No | No (1 IVF) | R: 0.029 |
| 12 | AZO | SA (Sp+) | 48 | 20, 10 | 17.2 | 2.6 | 7.2 | 0.01 | 0 | 0 | nd | nd | nd | No | Gest (2 IVF) | R: 0.02; L: 0.01 |
| 13 | AZO | SA* (Sp+) | 30 | 10, 9 | 0.4 | 3.8 | 7.7 | 0.001 | 0 | 0 | 51 | 48 | 18 | No | – | nd |
| 14 | AZO | SA* (Sp+) | 22 | 4, 4 | 0.5 | 1.7 | 7.7 | 0.001 | 0 | 0 | 50 | 56 | 18 | No | – | nd |
| 15 | AZO | SA (Sp+) | 51 | 8, 8 | 19.3 | 3.3 | 7.2 | 0-0.001 | 0 | 0 | nd | nd | nd | No | – | nd |
| 16 | AZO | SA (Sp+) | 30 | 10, 8 | 11.3 | 6.0 | 7.5 | 0 | – | – | 146 | 62 | 12 | No | Gest (1 IVF) | R + L: 0.001; μTESE L: 0.047 |
| 17 | AZO | SA (Sp+) | 45 | 15, 15 | 15.0 | 3.1 | 7.7 | 0 | – | – | 53 | 39 | 6 | No | No (1 IVF) | R: 0.07; L: 0.06 |
| 18 | AZO | SA (Sp−) | 38 | 10, 10 | 8.4 | 4.7 | 7.5 | 0 | – | – | 35 | 73 | 10 | No | – | R:0; L: 0 |
| 19 | AZO | SA (Sp−) | 45 | 10, 12 | 55.0 | 5.2 | 7.7 | 0 | – | – | nd | nd | nd | No | – | R: 0; L: 0.002 |
| 20 | AZO | SA (Sp−) | 30 | 9, 9 | 32.9 | 8.5 | 7.2 | 0 | – | – | 176 | 167 | 25 | No | – | R:0; L: 0 |
| 21 | AZO | SA (Sp−) | 30 | 13, 15 | 18.4 | 5.1 | 7.5 | 0 | – | – | 156 | 83 | 13 | No | – | R:0; L: 0 |
| 22 | AZO | SA | 34 | 12, 10 | 38.0 | 5.3 | 7.2 | 0 | – | – | nd | nd | nd | No | – | nd |
| 23 | AZO | SA | 32 | 8, 8 | 18.2 | 10.8 | 7.2 | 0 | – | – | 161 | 321 | 23 | No | – | nd |
| 24 | AZO | OA-N# | 35 | 20, 20 | 10.0 | 1.3 | 6.4 | 0 | – | – | nd | nd | nd | No | – | R: 0.3; L: 0.55 |
| 25 | AZO | OA-N# | 37 | 25, 25 | nd | 0.5 | 6.4 | 0 | – | – | nd | nd | nd | No | – | R: 0.42; L: 0.25 |
| 26 | AZO | OA-N | 44 | 0, 15 | 8.4 | 2.9 | 7.5 | 0 | – | – | 48 | 93 | 31 | No | No (2 IVF) | L: 0.5 |
| 27 | AZO | OA-N | 33 | 0, 20 | 6.6 | 4.5 | 7.2 | 0 | – | – | 57 | 97 | 3 | No | – | L: 0.182 |
| 28 | AZO | OA-N | 41 | 20, 20 | 1.7 | 2.1 | 7.5 | 0 | – | – | 86 | 69 | 12 | No | Gest (2 IVF) | R: 0.246 |
| 29 | AZO | OA-V | 41 | 15, 15 | – | 1.8 | 7.5 | 0 | – | – | nd | nd | nd | Yes (2) | – | nd |
| 30 | AZO | OA-V | 40 | 15, 15 | – | 2.7 | 7.7 | 0 | – | – | nd | nd | nd | Yes (2) | – | nd |
| 31 | AZO | OA -V | 40 | 25, 25 | – | 3.0 | 7.7 | 0 | – | – | nd | nd | nd | Yes (1) | – | nd |
| 32 | AZO | OA -V | 40 | 20, 20 | – | 2.7 | 7.5 | 0 | – | – | nd | nd | nd | Yes (2) | – | nd |
| 33 | AZO | OA -V | 40 | 20, 20 | – | 2.7 | 7.5 | 0 | – | – | nd | nd | nd | Yes (2) | – | nd |
| 34 | AZO | OA -V | 39 | 20, 20 | – | 4.2 | 7.2 | 0 | – | – | nd | nd | nd | Yes (2) | – | nd |
| 35 | AZO | OA -V | 36 | 25, 25 | – | 1.7 | 7.7 | 0 | – | – | nd | nd | nd | Yes (2) | – | nd |
| 36 | AZO | OA -V | 40 | 25, 25 | – | 1.9 | 7.5 | 0 | – | nd | nd | nd | Yes (2) | – | nd | |
| 37 | OLIGO | Oligozoospermia | 37 | 20, 20 | 4.8 | 3.8 | 7.7 | 2 | 54 | 10 | nd | nd | nd | No | No (1 IVF) | nd |
| 38 | OLIGO | Oligozoospermia | 35 | 15, 9 | 3.8–4.2 | 4.1 | 7.2 | 3 | 40 | 4 | nd | nd | nd | No | – | nd |
| 39 | OLIGO | Oligozoospermia | 21 | 15, 12 | 2.5 | 6.0 | 7.5 | 5 | 9 | 0 | nd | nd | nd | No (single) | – | nd |
Nz, normozoospermia; AZO, azoospermia and cryptozoospermia; OA-N, obstructive azoospermia due to pathological naturally occurring- obstruction in the genital tract; OA-V, obstructive azoospermia as a result of a vasectomy; SA, secretory azoospermia/cryptozoospermia; SA (Sp+), individuals with a positive TESE value > 0.01 × 106 sperm/ml; R, right testis; L, left testis; nd, not determined; TESE, testicular sperm extraction; Gest, gestation.
aAge at the time of clinical assessment.
*Hypogonadism. #CBAVD (congenital bilateral absence of the vas deferens).
Semen analysis was performed on all the individuals in accordance with World Health Organization guidelines (Cooper et al., 2010).
Men with spermatogenic disorders included in the study did not show clinical factors (varicocele, infection, immunologic factors, anatomic malformation or chemical insults) or genetic causes (chromosomal aberration or a Y-chromosome microdeletion) for their infertility.
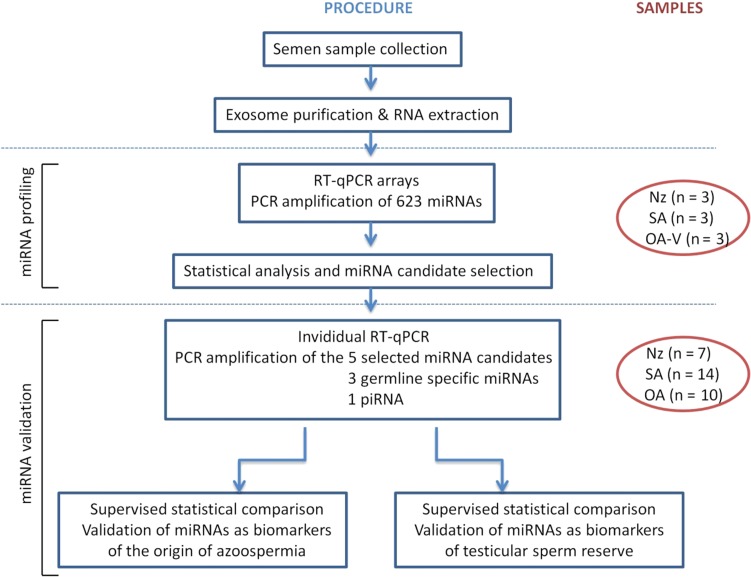
For the subsequent analysis of exosome miRNA levels in semen, samples were used into a two-stage (screening and validating stages) case–control designed study (Fig. 1).
Figure 1.
Flow chart outlining the two stages of miRNA profiling and validation performed in the study. The number of microRNAs (miRNAs) and individuals analysed at each stage are depicted. Nz, normozoospermia; SA, secretory azoospermia; OA-V, obstructive azoospermia from men successfully vasectomized; piRNA, Piwi-interacting RNA; RT-qPCR, quantitative RT-PCR.
Semen samples and exosome isolation
Semen samples were obtained by masturbation after 3–5 days of sexual abstinence. They were allowed to liquefy for 30 min at 37°C. Isolation of exosomal vesicles was performed by differential centrifugation steps including one microfiltration step and ultracentrifugation as described elsewhere (Li et al., 2012; Crescitelli et al., 2013). Briefly, cells and apoptotic bodies were pelleted by centrifugation at 1600 g for 10 min and microvesicles at 16 500 g for 10 min at 4°C to obtain SP (supernatant) which was carefully collected and immediately stored at −80°C until use. SP (200 μl) was filtered (0.22 μm pore size) to remove macromolecules and cell debris that they may have still contained. Subsequently, 9 ml of PBS was additionally filtered. The resulting filtrate was ultracentrifuged at 100 000 g for 2 h at 4°C to sediment the sEVs, which mainly contain exosomes. The pellet was resuspended in 100 μl PBS and frozen at −80°C. Nanoparticle tracking analysis was performed by NanoSight NS300 (Malvern Instruments Ltd, UK) showing an enrichment of particles with a size of <200 nm, accounting for 95% of the particles recovered (Supplementary Table SI). The vesicle distribution by size and the morphological parameters showed no significant differences among groups.
Small RNA-containing total RNA isolation
The exosome suspension was treated with RNAse A (Qiagen NV; Germany) (100 μg/ml final reaction concentration; 15 min at 37°C) to degrade the residual RNA outside the vesicles.
Total RNA was obtained from exosomes using the miRCURY RNA Isolation Kit-Cell and Plant (Exiqon; Denmark). RNA concentration was calculated by using the QUBIT fluorometer and the Quant-iT RNA Assay kit (Invitrogen; CA, USA). All RNA samples presented a OD 260/280 nm ratio ≥1.7 when using a Nanodrop UV–Vis spectrophotometer (Thermo Fisher Scientific; MA, USA).
Exosomal miRNA quantitative real-time PCR profiling
For the miRNA screening, RNA was reverse transcribed (RT) using the miRCURY LNA™ Universal RT miRNA PCR, Polyadenylation and cDNA synthesis kit (Exiqon; Denmark). cDNA was diluted 50× and assayed in 10 μl PCR reactions according to the protocol for miRCURY LNA™ Universal RT miRNA PCR. Each miRNA was assayed once by quantitative real-time PCR (qPCR) using ExiLENT SYBR Green mastermix on the miRNA Ready-to-Use PCR Human panels I and II that include 623 mature miRNAs of miRBase (www.mirbase.org/) in a LightCycler® 480 Instrument (Roche; Switzerland). Experiments were conducted at Exiqon Services. Details and conditions of analysis of the amplification curves and efficiencies are described elsewhere (Muñoz et al., 2015).
Next, to correct for potential overall differences in amount and quality between the samples, for each sample the raw data (Crossing points: Cp values) were normalized to the mean of the 50 most stable assays (mean 50) that were detected in all samples: dCp = mean 50 Cp – assay Cp. Those assays were previously selected as being the ones with the lowest coefficient of variation (CV < 0.018) of Cp values among samples in the study (Supplementary Table SII) as well as showing no statistical differences in absolute expression levels between groups, either individually or for the mean value.
The relative quantitative method of 2dCp was used to calculate the relative quantification (RQ) miRNA expression values.
Validation of miRNA candidates by RT-qPCR analysis
First-stranded cDNA specific for miRNA was obtained by RT of 50 ng of RNA in 10 μl, using the Universal cDNA synthesis kit II (Exiqon; Denmark). For qPCR analysis, cDNA was diluted (12×) and assayed in 10 μl PCR reactions containing ExiLENT SYBR Green mastermix (Exiqon; Denmark). Duplicate amplification reactions of individual assays (LNA™-enhanced miRNA qPCR primers; Supplementary Table SIII) were carried out on a Lightcycler® 96 Instrument (Roche; Switzerland). Target miRNA expression for exosomes in semen samples was calculated relative to the mean expression value of miR-30e-3p and miR-30d-5p, chosen from the exosomal miRNA qPCR profiling study as being among the most stable miRNAs (Supplementary Table SII). The RQ values were calculated using the 2dCp strategy. The same procedure was applied to determine tissue expression profiling of miRNA candidates.
Statistical analysis
The non-parametric Kruskal–Wallis test was used to analyse the differences in clinical data and absolute expression levels of reference genes. Unpaired two-tailed Student’s t test was used to analyse the differences in relative expression of miRNAs between groups in the miRNA profiling study and the Benjamini–Hochberg procedure for multiple testing correction, using a maximum discovery rate of 5%, was applied to calculate the False Discovery Rate (FDR) for each of the P-values obtained. The non-parametric Mann–Whitney U-test was used to evaluate differences in relative expression of selected miRNAs between groups. Pearson product-moment correlation coefficients (r) were calculated to determine the correlation between the miRNA RQ values and the various parameters in semen analysis or testicular biopsy in patient groups and controls. Multivariate binary logistic regression (backward stepwise, conditional, method) and receiver operating characteristic (ROC) curve analysis of the RQ values was used to distinguish the aetiology of azoospermia, as well as individuals with a positive testicular sperm extraction (TESE) value >0.01 × 106 sperm/ml (SA_Sp+). Accuracy was measured as the area under the ROC curve. All data analyses were performed using the SPSS software version 15.0 (SPSS Inc.; IBM; IL, USA). A P-value ≤0.05 was considered significant.
Results
Obstructive and secretory azoospermia show altered profiles of miRNA contained in SP exosomes
In order to identify global changes of the exosome miRNA levels in SP associated with azoospermia of different origin, we first analysed the level of expression of 623 human miRNAs in SP exosomes from three Nz individuals (Nz; sample nos. 1,3,4) who showed normal characteristics in a seminogram, and from azoospermic men resulting from vasectomy surgical procedure (OA-V; n = 3; sample nos. 29–31) or resulting from impaired sperm production (SA; n = 3; sample nos. 11,16,17).
No amplification values were obtained for 78 miRNAs, suggesting that the miRNA levels were beneath the detection threshold of the technique. Of the amplified miRNAs, 148 were excluded from further analysis owing to poor amplification efficiency across samples (missing expression values for six out of the nine samples). The remaining miRNAs (n = 397; 63.7%) were further statistically evaluated and the results are presented in Supplementary Table SIV.
The presence of 393 miRNAs (Cp value <38) was determined in the Nz samples (two of them were not detected in OA—suggesting that they are specifically expressed in somatic cells from epididymis or testis—and 16 miRNAs were not expressed in either OA or SA—suggesting a testicular specific expression), whereas four miRNAs were detected in azoospermic samples but not in Nz samples, which suggests that they are minor miRNAs coming from reproductive tissues other than the epididymis and testis.
We found 60 miRNAs in OA and/or SA that presented significant differences in expression when compared with Nz controls (Table II). In detail, regarding the OA-Nz comparison, nine miRNAs were significantly overexpressed (fold-change increase range: 1.47–3.70; >2-fold-increase n = 4) and 42 underexpressed (fold-change decrease range: 1.47–1000; >2-fold-decrease n = 41). Nine of these miRNAs (eight down-regulated and one up-regulated) passed the FDR correction (P-value ≤ 0.0006). Fifteen out of the 42 down-regulated miRNAs (36%) are located in chromosome X. Some of the down-regulated miRNAs map to miRNA clusters: chromosome 19 (a non-conserved cluster) (Bentwich et al., 2005), and chromosome X (four clusters) (www.mirbase.org) (Supplementary Tables SII and SIV). Additionally, in the SA samples, eight miRNAs were significantly overexpressed (fold-change increase range: 1.24–9.45; >2-fold-increase n = 2) and 26 were underexpressed (fold-change decrease range: 1.13–1000; >2-fold-decrease n = 25) (Table II) when compared with Nz controls. Four down-regulated miRNAs passed the FDR correction (P-value ≤ 0.0006).
Table II.
Seminal exosome-derived miRNAs differentially expressed in OA and/or SA compared with Nz individuals.
| Seminal plasma exosomal miRNA expression | ||||||
|---|---|---|---|---|---|---|
| miRNA | Location | Nz | OA | SA | Deduced miRNA origin from our work | Human miRNA expression profile in tissues |
| A. Underexpressed miRNAs | ||||||
| hsa-miR-202-3p | chr. 10 | 1 | 0.001** | 0.001** | Testis | T (Hu et al., 2014) |
| hsa-miR-514a-3p2cX | chr. X | 1 | 0.001** | 0.001** | Testis | T (Hu et al., 2014) |
| hsa-miR-202-5p | chr. 10 | 1 | 0.004** | 0.004** | Testis | T (Hu et al., 2014); Sertoli cell expressed |
| hsa-miR-509-3-5p1cX | chr. X | 1 | 0.004** | 0.016* | Testis | T (Hu et al., 2014) |
| hsa-miR-510-5p2cX | chr. X | 1 | 0.008** | 0.008** | Testis | |
| hsa-miR-513c-5p3cX | chr. X | 1 | 0.010** | 0.009** | Testis | |
| hsa-miR-518e-3p1c19 | chr. 19 | 1 | 0.029** | 0.080* | Testis | |
| hsa-miR-508-5p1cX | chr. X | 1 | 0.032** | 0.031** | Testis | |
| hsa-miR-520h2c19 | chr. 19 | 1 | 0.036** | 0.054* | Testis | |
| hsa-miR-9-3p | chr. 1, 5, 15 | 1 | 0.049** | 0.047* | Testis | |
| hsa-miR-506-3p1cX | chr. X | 1 | 0.050** | 0.047** | Testis | |
| hsa-miR-383-5p | chr. 8 | 1 | 0.059** | 0.057** | Testis | |
| hsa-miR-34c-5p | chr. 11 | 1 | 0.059** | 0.073* | Testis | T/EP (Hu et al., 2014) |
| hsa-miR-517c-3p2c19 | chr. 19 | 1 | 0.069** | 0.091* | Testis | |
| hsa-miR-873-5p | chr. 9 | 1 | 0.082** | 0.079** | Testis | |
| hsa-miR-34b-5p | chr. 11 | 1 | 0.007* | 0.167* | Testis | |
| hsa-miR-513a-3p1cX | chr. X | 1 | 0.036* | 0.034* | Testis | |
| hsa-miR-5211c19.2c19 | chr. 19 | 1 | 0.042* | 0.040* | Testis | |
| hsa-miR-452-5p | chr. X | 1 | 0.063* | 0.148* | Testis | |
| hsa-miR-122-5p | chr. 18 | 1 | 0.094* | 0.133* | Testis | |
| hsa-miR-449a | chr. 5 | 1 | 0.237* | 0.205** | Testis | |
| hsa-miR-499a-5p | chr. 20 | 1 | 0.343* | 0.315* | Testis | |
| hsa-miR-455-5p | chr. 9 | 1 | 0.382* | 0.297* | Testis | |
| hsa-miR-891b6cX | chr. X | 1 | 0.084* | 0.455 | Epididymis/Testis | EP (Li et al., 2010, 2012; Hu et al., 2014) |
| hsa-miR-8906cX | chr. X | 1 | 0.090* | 0.363 | Epididymis/Testis | EP (Li et al., 2010; Hu et al., 2014) |
| hsa-miR-34c-3p | chr. 11 | 1 | 0.093* | 0.382 | Epididymis/Testis | |
| hsa-miR-891a-5p | chr. X | 1 | 0.097* | 0.448 | Epididymis/Testis | EP (Li et al., 2010, 2012; Hu et al., 2014) |
| hsa-miR-888-5p6cX | chr. X | 1 | 0.098* | 0.391 | Epididymis/Testis | EP (Li et al., 2010; Hu et al., 2014) |
| hsa-miR-124-3p | chr. 8, 20 | 1 | 0.101** | 0.481 | Epididymis/Testis | |
| hsa-miR-892a6cX | chr. X | 1 | 0.107* | 0.544 | Epididymis/Testis | EP (Li et al., 2010; Hu et al., 2014) |
| hsa-miR-551b-3p | chr. 3 | 1 | 0.139** | 0.246 | Epididymis/Testis | |
| hsa-miR-424-5p | chr. X | 1 | 0.212* | 0.424 | Epididymis/Testis | T/EP (Hu et al., 2014) |
| hsa-miR-181b-5p | chr. 1, 9 | 1 | 0.246* | 0.362 | Epididymis/Testis | |
| hsa-miR-31-3p | chr. 9 | 1 | 0.250* | 0.367 | Epididymis/Testis | |
| hsa-miR-181a-5p | chr. 1, 9 | 1 | 0.292* | 0.459 | Epididymis/Testis | |
| hsa-miR-31-5p | chr. 9 | 1 | 0.302* | 0.548 | Epididymis/Testis | T/EP (Hu et al., 2014) |
| hsa-miR-10b-3p | chr. 2 | 1 | 0.313* | 0.923 | Epididymis/Testis | EP (Li et al., 2012) |
| hsa-miR-222-3p | chr. X | 1 | 0.323* | 0.570 | Epididymis/Testis | Wide expression (Hu et al., 2014) |
| hsa-miR-455-3p | chr. 9 | 1 | 0.364* | 0.439 | Epididymis/Testis | |
| hsa-miR-205-5p | chr. 1 | 1 | 0.367* | 0.734 | Epididymis/Testis | |
| hsa-miR-182-3p | chr. 7 | 1 | 0.399* | 1.454 | Epididymis/Testis | EP (Li et al., 2012) |
| hsa-miR-95-3p | chr. 4 | 1 | 0.678* | 1.398 | Epididymis/Testis | |
| hsa-miR-9-5p | chr. 1, 5, 15 | 1 | 0.342 | 0.328* | ||
| hsa-miR-132-5p | chr. 17 | 1 | 0.674 | 0.456* | ||
| hsa-miR-203a | chr. 14 | 1 | 0.995 | 0.880* | ||
| B. Overexpressed miRNAs | ||||||
| hsa-miR-363-3p5cX | chr. X | 1 | 1.474** | 1.241** | ||
| hsa-miR-365a-3p | chr. 16 | 1 | 1.689* | 1.394* | ||
| hsa-miR-29a-3p | chr. 7 | 1 | 1.614* | 1.288 | ||
| hsa-miR-296-5p | chr. 20 | 1 | 3.705* | 2.400 | ||
| hsa-miR-23b-5p | chr. 9 | 1 | 2.590* | 1.498 | ||
| hsa-miR-21-3p | chr. 17 | 1 | 2.202* | 1.485 | ||
| hsa-miR-193a-3p | chr. 17 | 1 | 2.879* | 1.783 | ||
| hsa-miR-29c-3p | chr. 1 | 1 | 1.687* | 1.415 | T/EP (Hu et al., 2014) | |
| hsa-miR-361-3p | chr. X | 1 | 1.741* | 1.575 | ||
| hsa-miR-550a-5p | chr. 7 | 1 | 2.465 | 9.455* | ||
| hsa-miR-423-5p | chr. 17 | 1 | 1.129 | 1.485* | T (Hu et al., 2014) | |
| hsa-let-7f-1-3p | chr. 9 | 1 | 1.093 | 1.808* | ||
| hsa-miR-153-3p | chr. 2, 7 | 1 | 1.667 | 2.251* | ||
| hsa-miR-196b-3p | chr. 7 | 1 | 1.123 | 1.451* | ||
| hsa-miR-96-5p | chr. 7 | 1 | 1.750 | 1.694* | ||
Statistically increased miRNA expression levels are depicted in green; statistically decreased miRNA expression levels are depicted in red
when compared with controls. Hsa, Homo sapiens.
*P < 0.05; ** P < 0.005.
1-6cX clusters in chromosome X.
1-2c19 clusters in chromosome 19.
T, preferential expression in the testis.
EP, preferentially expressed in epididymis.
Among the differentially expressed miRNAs, 25 miRNAs were shared among OA and SA groups, either down- (n = 23) or up-regulated (n = 2), when compared with Nz controls (Table II). Remarkably, among the shared down-regulated miRNAs in azoospermia, there was no expression value (Cp value > 38) for miR-202-3p, miR-202-5p, miR-383-5p, miR-506-3p, miR-508-5p, miR-510-5p, miR-513a-3p, miR-513c-5p, miR-514a-3p, miR-518e-3p, miR-520h, miR-521, miR-873-5p and miR-9-3p (Supplementary Table SIV). Those exosomal miRNAs that presented statistically lower levels in both OA and SA men were candidates for having preferentially been derived from the testicular cells associated with presence of the germline (n = 23), whereas the 19 miRNAs found to be down-regulated in OA but not in SA samples are suggested to be preferentially secreted by somatic cells of the testis and/or the epididymis (Table II). Interestingly, among the latter group of miRNAs we found several that were described to be epididymis-specific, such as the X-linked miR-888 cluster (Li et al., 2010; Belleannee et al., 2012).
Exosome miRNA levels in SP identify azoospermia with different origins
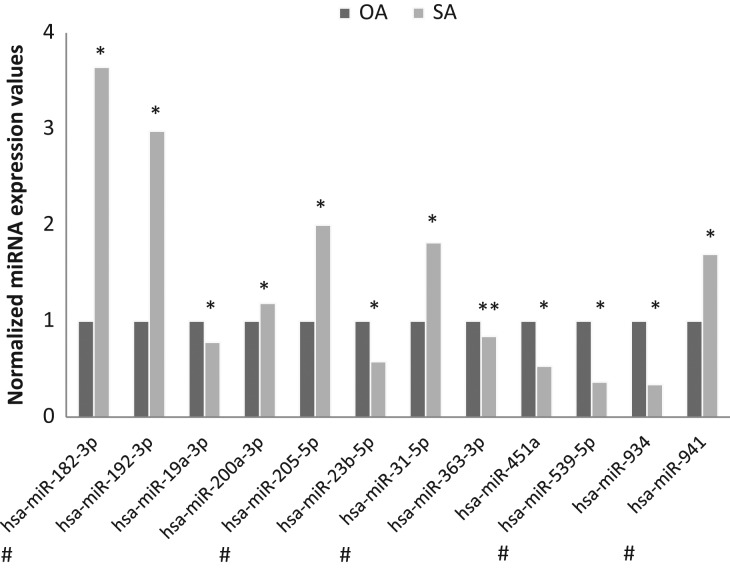
Subsequently, we compared the miRNA expression profile of SP exosomes between SA and OA-V samples with potential implications for diagnosis. We found that 12 miRNAs (Fig. 2) presented significant differences in expression between groups, although no miRNA passed the FDR correction. According to this profiling outcome, we proceeded to validate several miRNAs as candidate biomarkers of the origin of azoospermia.
Figure 2.
Differential abundance profile of exosomal miRNAs as assessed by quantitative RT-PCR arrays from OA-V and SA semen samples. Normalized expression levels relative to the mean of the 50 stable miRNAs are shown. Significant differences between groups are indicated: *P-value < 0.05; **P-value < 0.01 (Student’s t-test). Those miRNAs selected for validation are indicated by a # symbol.
Hsa miRNA, Homo sapiens miRNA
First, five miRNAs (miR-182-3p, miR-205-5p, miR-31-5p, miR-539-5p, miR-941) were selected for validation in a larger number of individuals based on the following criteria: we selected those miRNAs that presented ≥1.7-fold difference in expression between groups and a Cp value <34 in any of the azoospermic groups (Fig. 2). We included in the validation stage of the study three additional miRNAs, such as miR-122-5p, miR-34c-5p, miR-449a, that we found were differentially expressed in SP exosomes between azoospermic and Nz individuals (Table II) and were previously described as being preferentially produced from the meiotic and/or post-meiotic germ-cell stages of the testis (Yu et al., 2005; Bouhallier et al., 2010; Bao et al., 2012). Additionally, the cellular content of all these three miRNAs was found to be decreased in testicular developing germ-cells and in mature sperm from patients with deficient sperm production; and the combination of the expression values of these three miRNAs in testicular biopsies is able to predict the availability of sperm in the biopsy for ART (Muñoz et al., 2015). PiRNAs are sncRNAs predominantly expressed in the germline. Thus, the piR-58527 (piRNABank accession DQ591415) was additionally included in the validation study as it is specifically expressed in the postmeotic germ cells (Heyn et al., 2012).
In order to determine the expression level of each miRNA in the different organs of the reproductive tract, the expression of the eight miRNAs was first tested in testis, epididymis, prostate, SP and lymphocytes, the latter as external control cells (Supplementary Fig. S1). We could corroborate the preferential testicular expression of miR-122-5p, miR-34c-5p and miR-449a, whereas miR-205-5p is preferentially expressed in epididymis and prostate. The miR-182-3p, miR-31-5p, miR-539-5p and miR-941 are expressed in the three reproductive organs (testis, epididymis and prostate).
Therefore, these particular eight miRNAs and one piRNA were individually reanalysed in a subsequent set of semen samples (seven Nz samples, 14 SA samples, 10 OA samples including five OA-V samples and five OA-N samples, and three oligozoospermic samples) by RT-qPCR (Fig. 1). First, our results showed that the expression values of miR-122-5p (P < 0.0001), piR-58527 (P < 0.0001) and miR-539-5p (P = 0.034) were statistically different between azoospermic and Nz individuals (Supplementary Fig. S2). Interestingly, the miR-122-5p expression value was statistically correlated with the concentration of sperm in semen (r = 0.616, P < 0.0001).
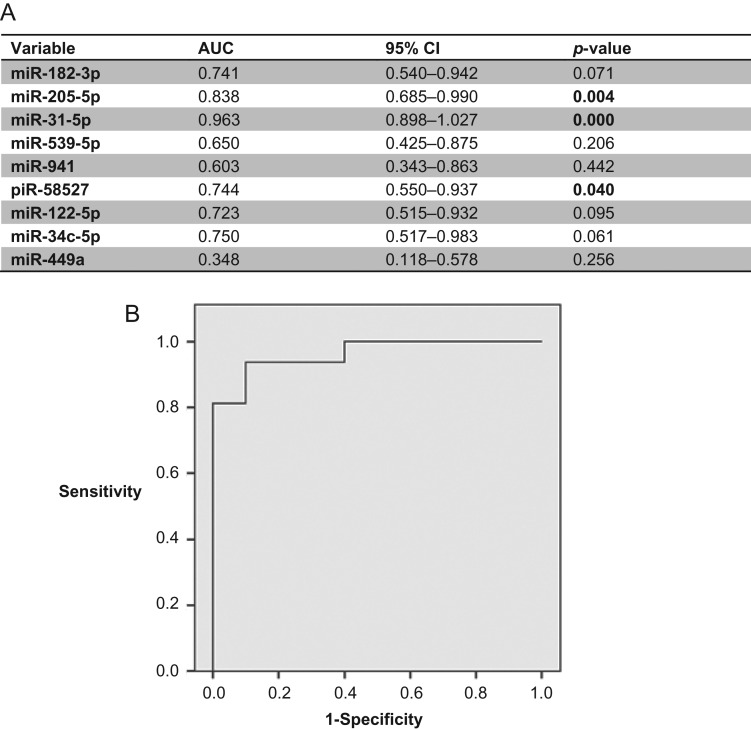
Furthermore, when the SA and OA samples were compared, our results show that the tendencies among the nine miRNAs/piRNA analysed were conserved among the two different approaches (miRNA qPCR array and RT-qPCR; r: 0.93), although only two of them (miR-205-5p, miR-31-5p; P < 0.004) as well as piR-58527 (P = 0.031) were found to be statistically different between both groups of azoospermia in the RT-qPCR validation analysis (Supplementary Fig. S2). The expression values of these three sncRNAs resulted in good predictive accuracy [miR-205-5p (AUC 0.843, P = 0.005); miR-31-5p (AUC: 0.957; P < 0.0001); piR-58527 (AUC: 0.764; P = 0.030)] (Fig. 3A), suggesting they have a potential use as indicators of the origin of the azoospermia. As a comparison, the ROC curve analysis of blood FSH levels was also determined (AUC 0.850, P = 0.004). To determine if a multiplex model could improve performance over single biomarkers for discriminating SA from OA samples, the three previously selected miRNAs were analysed in a multivariate logistic regression analysis. Interestingly, this analysis resulted in a model that only included the miR-31-5p expression values. The sensitivity and the specificity for predicting the origin of azoospermia were 92.9 and 90%, respectively (Fig. 3B). Strikingly, an increased value of sensitivity and specificity (100%) was obtained when FSH + miR-31-5p values were included in the model.
Figure 3.
Predictive efficiency of miRNA variables for distinguishing SA from OA samples. (A) Receiver operating characteristic (ROC) curve analysis showing the predictive efficiency of miRNA variables for distinguishing SA from OA samples at the validation stage. (B) ROC curve of miR-31-5p for predictive classification of azoospermic samples into SA and OA sub-phenotypes at the validation stage.
When the OA sample group was divided into OA-N and OA-V sub-phenotypes, three miRNAs [miR-205-5p (P = 0.010); miR-31-5p (P = 0.001); and miR-539-5p (P = 0.026)] were differentially expressed between OA-N and SA samples whereas miR-31-5p and miR-941 (P < 0.003) were down-regulated in OA-V compared with SA samples. This observation suggests that the profile of exosomal miRNA in semen is different in congenital obstruction of the genital tract from that obtained from the vasectomy procedure, and probably depends on the level at which the obstruction of the genital tract is produced. We then specifically focused on the differences between naturally occurring OA-N and SA samples. The expression values of the three differentially expressed miRNAs resulted in good predictive accuracy [miR-205-5p (AUC: 0.886, P = 0.012); miR-31-5p (AUC: 0.957; P = 0.003); miR-539-5p (AUC: 0.843; P = 0.026)], suggesting they have a potential use as indicators of the origin of the congenital azoospermia, as well as FSH (AUC: 0.843, P = 0.026). When the multivariate regression analysis was performed, it resulted in a model that, again, only included the miR-31-5p expression values. The sensitivity and the specificity for predicting the origin of naturally occurring azoospermia were 92.9 and 80%, respectively, confirming that exosomal miR-31-5p expression values in semen samples could be useful to predict the presence of a conserved spermatogenesis in the testis of azoospermic individuals. Once again, an increased value of sensitivity and specificity (100%) was obtained when (FSH + miR-31-5p) were included in the predictive model.
Exosome miRNA levels in SP and the presence of intratubular mature germ cells in spermatogenic disorders
In order to assess if SP miRNA expression and histological parameters from a SA biopsy were associated and to know the physiological relevance and/or the potential implications for reproductive treatments, we compared the miRNA profile of four SA individuals (nos. 18–21) with no spermatozoa in the testicular biopsy (Sp−) and eight SA samples (nos. 10–17) with presence of intra-testicular sperm (Sp+), the latter including the samples from cryptozoospermic individuals and those samples from individuals with a positive TESE value (>0.01 × 106 sperm/ml) (Table I). The TESE value is defined as the number of spermatozoa that was obtained directly after processing 100 mg of biopsy in 1 ml of medium.
No significant differences between SA−(Sp+) and SA−(Sp−) were found regarding the individual levels of the above eight miRNAs and the piRNA in SP exosomes. However, when the multivariate regression analysis was performed, it resulted in a model that included the miR-539-5p and the miR-941 expression values. Interestingly, the sensitivity and the specificity for predicting the presence of spermatozoa in a testicular biopsy were both 100% for the samples included in the study. When FSH was incorporated in the regression analysis, the model included miR-539-5p, miR-941 and FSH values resulting in true positive and negative rates for predicting residual spermatogenesis in the testes of again 100%.
Discussion
In the last decade multiple efforts have been made in order to develop specific biomarkers to contribute in the diagnosis and prognosis of male infertility and to increase the success of reproductive treatments. At present, assessment of azoospermic patients is based on medical history, physical findings, hormone analysis, karyotype and a limited number of genetic tests. However, in some cases the diagnosis remains elusive and testicular biopsy is necessary. Furthermore, even in suspected non-obstructive azoospermia, currently available markers, such as elevated levels of FSH in blood or diminished testicular volume among others, are unsatisfactory (Zitzmann et al., 2006; Cissen et al., 2016), and thus testis biopsy is also mandatory to identify men with some residual spermatogenesis. For these reasons, a non-invasive screening test that could identify those individuals with real chances of positive sperm recovery on the biopsy would be welcomed. Semen contains high concentrations of miRNAs, which can be identified and quantified, making them candidates as biomarkers for non-invasive diagnostic/prognostic purposes. Recently, a few studies have focused on cell-free miRNAs from semen and found an association between alterations of sperm quality and aberrant miRNA levels, both in whole SP (Wang et al., 2011; Wu et al., 2012, 2013) and in exosomes (Abu-Halima et al., 2016). In particular, miRNAs contained in exosomes can reflect the pathophysiological conditions of the organ of origin. These attributes support the idea that the study of exosomal miRNA content could be useful as a specific indicator of alterations of the organ/cells that released them.
First, in the present study we focused our attention on those miRNAs that were statistically down-regulated both in OA-V and SA individuals and identified 23 miRNAs associated with the presence of the germline in the testis (Table II). Interestingly, three of these miRNAs (miR-202-5p, miR-509-3-5p and miR-34c-5p) were previously described as being among the 10 most abundant miRNAs in human testis (Yang et al., 2013) and some of these miRNAs, such as miR-34c-5p, miR-449a and miR-122-5p, had been previously found to be underexpressed in testicular and sperm samples with deficient sperm production (Muñoz et al., 2015) suggesting they were reliable results. Additionally, from our results we come to the conclusion that 19 miRNAs have a preferential somatic origin from testis and/or epididymis (down-regulated in vasectomized but not in SA patients) (Table II). Among them, there are members of the X-linked primate-specific miR-888 cluster (i.e. miR-890, miR-891a, miR-891b, miR-892a, miR-888) previously described to be epididymis-specific and to play a determinant role in regulating physiological functions of this organ, such as sperm maturation (Li et al., 2010) supporting the veracity of our conclusion. About 37% of the azoospermic down-regulated SP exosomal miRNAs were located on the X-chromosome and most of them are placed in clusters (Table II). Many X-linked miRNAs are preferentially or exclusively expressed in spermatogenic cells (Ro et al., 2007). In line with this, other exosomal miRNAs located in clusters additionally have their expression altered in azoospermia, such as clusters of chromosome 19 and clusters miR-34b/c and miR-449, which were previously found to be significantly reduced in testis with severe maturation blockade of the germline (Muñoz et al., 2015).
There have been previous studies that compared the miRNA fingerprint, but in whole semen, from vasectomized and Nz individuals as well as secretory azoospermia and normozoospermia. A first study (Hu et al., 2014) identified 61 miRNAs that presented lower levels in semen from vasectomized men than in semen from Nz individuals. The altered miRNA profile is quite different from the one that we identified, and only 17 miRNAs were shared between the two studies (Table II). Other studies reported an altered miRNA fingerprint from SP in SA, although the profile is quite different from the one found in our miRNA study with SP exosomes. Specifically only miR-34c-5p, miR-122 were found markedly decreased in SA (Wang et al., 2011) in agreement with our data, but miR-146b-5p, miR-181a, miR-374b, miR-509-5p and miR-513a-5p (Wang et al., 2011), miR-19b and let-7a (Wu et al., 2012), and miR-141, miR-429 and miR-7-1-3p (Wu et al., 2013) were not found to be altered in our study. These data suggest that the miRNA profile in whole semen is quite different to the one contained in the seminal exosomes.
Specifically, we aimed to assess the potential of SP miRNAs contained in exosomes as sensitive and specific biomarkers for selecting those azoospermic individuals with real chances of obtaining spermatozoa from the testicular biopsy, either by themselves or combined in predictive models. To our knowledge, this is the first time that the miRNA expression profile from semen exosomes has been compared between azoospermic individuals of different origin: obstructive versus secretory azoospermia. Our results suggest that miRNAs in exosomes from SP are useful for establishing the origin of azoospermia and can predict the presence of sperm in testicular tissue. In fact, the levels of 12 miRNAs in exosomes differed significantly between obstructive as compared with secretory azoospermia when assessed by miRNA qPCR arrays, profiling 623 human miRNAs. These results were validated for nine miRNA/piRNA in a larger cohort of patients and two of them (miR-205-5p, miR-31-5p), as well as the germ-cell specific piR-58527, were confirmed to be significantly down-regulated in OA compared with SA. Interestingly, a positive correlation with a strong predictive accuracy between the individual expression values of these three sncRNAs and the selection of OA samples was determined for the first time, reaching very good diagnostic efficiency (AUC > 0.95) for the miR-31-5p. What is more, the sensitivity and specificity of miR-31-5p for predicting the origin of azoospermia were notable (92.9 and 90%, respectively, when comparing OA and SA samples; as well as 92.9 and 80%, respectively, when comparing congenital OA-N and SA samples); therefore, the use of miR-31-5p has potential implications for diagnosis. Additionally, the combination of the values of well-described blood biochemical markers, such as FSH, with those obtained from miR-31-5p in SP resulted in a better efficacy of the predictive model.
Our results additionally provide evidence that a logistic model combining the expression values of miR-539-5p and miR-941 was able to discriminate the presence or the absence of sperm in a testicular biopsy with a spermatogenic disorder with a high diagnostic accuracy. As a next step, further studies, with an independent second population involving a larger number of samples, are needed to confirm our findings.
In summary, our study identified several miRNAs altered in SP exosomes that potentially provide information about the origin of azoospermia and the spermatogenic reserve of the testis. We propose the analysis of miR-31-5p, miR-539-5p and miR-941 in exosomes from SP in patients with azoospermia. The first diagnostic step will consider the expression of miR-31-5p in exosomes in combination with FSH concentration in blood to distinguish those samples with an obstruction and conserved spermatogenesis from samples with a spermatogenic disorder. For the latter samples, the levels of miR-539-5p and miR-941 will be used to document the presence of residual sperm in the testis. Our findings contribute to the search for the most valuable genetic markers that are potentially useful as tools for predicting the presence of testicular sperm. We show that SP biomarkers for azoospermia represent a promising alternative or an addition to traditional biomarkers. Such information would be useful in the clinics, helping in the diagnosis of azoospermic patients but also providing realistic information about the chances of testicular sperm retrieval in order to avoid unnecessary biopsies.
Supplementary Material
Acknowledgements
We are indebted to the individuals who participated in the study. We thank the staff of the Seminology and Embryology Laboratory of Fundació Puigvert for providing seminal samples, Cristian Tebé for his advice on statistical analysis and Harvey Evans for the revision of the English text. The nanoparticle tracking analysis was performed by the Unit 6 of ICTS ‘NANBIOSIS’.
Authors’ roles
M.B. and S.L. performed the RNA experiments and analysed the data. A.M. provided samples and performed semen analyses. L.B. performed clinical and testis assessment, provided samples and critically reviewed the article. S.L. conceived and designed the experiments and wrote the article.
Funding
Grants from the Fondo de Investigaciones Sanitarias/Fondo Europeo de Desarrollo Regional ‘Una manera de hacer Europa’ (FIS/FEDER) [grant number PI15/00153], the Generalitat de Catalunya [grant number 2014SGR5412]. S.L. is sponsored by the Researchers Stabilization Program (ISCIII/Generalitat de Catalunya) from the Spanish National Health System [CES09/020].
Conflict of interest
S.L. and L.B. hold a patent entitled ‘Methods and markers for azoospermia characterization’.
References
- Aalberts M, Sostaric E, Wubbolts R, Wauben MW, Nolte-‘t Hoen EN, Gadella BM, Stout TA, Stoorvogel W. Spermatozoa recruit prostasomes in response to capacitation induction. Biochim Biophys Acta 2013;1834:2326–2335. [DOI] [PubMed] [Google Scholar]
- Abu-Halima M, Ludwig N, Hart M, Leidinger P, Backes C, Keller A, Hammadeh M, Meese E. Altered micro-ribonucleic acid expression profiles of extracellular microvesicles in the seminal plasma of patients with oligoasthenozoospermia. Fertil Steril 2016;106:1061–1069.e1063. [DOI] [PubMed] [Google Scholar]
- Arroyo JD, Chevillet JR, Kroh EM, Ruf IK, Pritchard CC, Gibson DF, Mitchell PS, Bennett CF, Pogosova-Agadjanyan EL, Stirewalt DL et al. . Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci U S A 2011;108:5003–5008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao J, Li D, Wang L, Wu J, Hu Y, Wang Z, Chen Y, Cao X, Jiang C, Yan W et al. . MicroRNA-449 and microRNA-34b/c function redundantly in murine testes by targeting E2F transcription factor-retinoblastoma protein (E2F-pRb) pathway. J Biol Chem 2012;287:21686–21698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004;116:281–297. [DOI] [PubMed] [Google Scholar]
- Belleannee C, Calvo E, Thimon V, Cyr DG, Legare C, Garneau L, Sullivan R. Role of microRNAs in controlling gene expression in different segments of the human epididymis. PLoS One 2012;7:e34996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentwich I, Avniel A, Karov Y, Aharonov R, Gilad S, Barad O, Barzilai A, Einat P, Einav U, Meiri E et al. . Identification of hundreds of conserved and nonconserved human microRNAs. Nat Genet 2005;37:766–770. [DOI] [PubMed] [Google Scholar]
- Bouhallier F, Allioli N, Lavial F, Chalmel F, Perrard MH, Durand P, Samarut J, Pain B, Rouault JP. Role of miR-34c microRNA in the late steps of spermatogenesis. RNA 2010;16:720–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cissen M, Meijerink AM, D’Hauwers KW, Meissner A, van der Weide N, Mochtar MH, de Melker AA, Ramos L, Repping S, Braat DD et al. . Prediction model for obtaining spermatozoa with testicular sperm extraction in men with non-obstructive azoospermia. Hum Reprod 2016;31:1934–1941. [DOI] [PubMed] [Google Scholar]
- Cooper TG, Noonan E, von Eckardstein S, Auger J, Baker HW, Behre HM, Haugen TB, Kruger T, Wang C, Mbizvo MT et al. . World Health Organization reference values for human semen characteristics. Hum Reprod Update 2010;16:231–245. [DOI] [PubMed] [Google Scholar]
- Crescitelli R, Lässer C, Szabó TG, Kittel A, Eldh M, Dianzani I, Buzás EI, Lötvall J. Distinct RNA profiles in subpopulations of extracellular vesicles: apoptotic bodies, microvesicles and exosomes. J Extracell Vesicles 2013;2 10.3402/jev.v2i0.20677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzick DS, Overstreet JW, Factor-Litvak P, Brazil CK, Nakajima ST, Coutifaris C, Carson SA, Cisneros P, Steinkampf MP, Hill JA et al. . Sperm morphology, motility, and concentration in fertile and infertile men. N Engl J Med 2001;345:1388–1393. [DOI] [PubMed] [Google Scholar]
- Hammond SM. An overview of microRNAs. Adv Drug Deliv Rev 2015;87:3–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi K, Chuva de Sousa Lopes SM, Kaneda M, Tang F, Hajkova P, Lao K, O’Carroll D, Das PP, Tarakhovsky A, Miska EA et al. . MicroRNA biogenesis is required for mouse primordial germ cell development and spermatogenesis. PLoS One 2008;3:e1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet 2004;5:522–531. [DOI] [PubMed] [Google Scholar]
- Heyn H, Ferreira HJ, Bassas L, Bonache S, Sayols S, Sandoval J, Esteller M, Larriba S. Epigenetic disruption of the PIWI pathway in human spermatogenic disorders. PLoS One 2012;7:e47892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu L, Wu C, Guo C, Li H, Xiong C. Identification of microRNAs predominately derived from testis and epididymis in human seminal plasma. Clin Biochem 2014;47:967–972. [DOI] [PubMed] [Google Scholar]
- Lakshmipathy U, Love B, Goff LA, Jornsten R, Graichen R, Hart RP, Chesnut JD. MicroRNA expression pattern of undifferentiated and differentiated human embryonic stem cells. Stem Cells Dev 2007;16:1003–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Huang S, Guo C, Guan H, Xiong C. Cell-free seminal mRNA and microRNA exist in different forms. PLoS One 2012;7:e34566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Liu Y, Dong D, Zhang Z. Evolution of an X-linked primate-specific micro RNA cluster. Mol Biol Evol 2010;27:671–683. [DOI] [PubMed] [Google Scholar]
- Maatouk DM, Loveland KL, McManus MT, Moore K, Harfe BD. Dicer1 is required for differentiation of the mouse male germline. Biol Reprod 2008;79:696–703. [DOI] [PubMed] [Google Scholar]
- Muñoz X, Mata A, Bassas L, Larriba S. Altered miRNA signature of developing germ-cells in infertile patients relates to the severity of spermatogenic failure and persists in spermatozoa. Sci Rep 2015;5:17991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renneberg H, Konrad L, Dammshauser I, Seitz J, Aumuller G. Immunohistochemistry of prostasomes from human semen. Prostate 1997;30:98–106. [DOI] [PubMed] [Google Scholar]
- Ro S, Park C, Sanders KM, McCarrey JR, Yan W. Cloning and expression profiling of testis-expressed microRNAs. Dev Biol 2007;311:592–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang F, Kaneda M, O’Carroll D, Hajkova P, Barton SC, Sun YA, Lee C, Tarakhovsky A, Lao K, Surani MA. Maternal microRNAs are essential for mouse zygotic development. Genes Dev 2007;21:644–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tüttelmann F, Nieschlag E. Classification of andrological disorders In: Nieschlag E, Behre HM, Nieschlag S (eds). Andrology: Male Reproductive Health and Dysfunction. Berlin, Germany: Springer, 2010, 87–92. [Google Scholar]
- Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol 2007;9:654–659. [DOI] [PubMed] [Google Scholar]
- Vojtech L, Woo S, Hughes S, Levy C, Ballweber L, Sauteraud RP, Strobl J, Westerberg K, Gottardo R, Tewari M et al. . Exosomes in human semen carry a distinctive repertoire of small non-coding RNAs with potential regulatory functions. Nucleic Acids Res 2014;42:7290–7304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Yang C, Chen X, Yao B, Yang C, Zhu C, Li L, Wang J, Li X, Shao Y et al. . Altered profile of seminal plasma microRNAs in the molecular diagnosis of male infertility. Clin Chem 2011;57:1722–1731. [DOI] [PubMed] [Google Scholar]
- Weber JA, Baxter DH, Zhang S, Huang DY, Huang KH, Lee MJ, Galas DJ, Wang K. The microRNA spectrum in 12 body fluids. Clin Chem 2010;56:1733–1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu W, Hu Z, Qin Y, Dong J, Dai J, Lu C, Zhang W, Shen H, Xia Y, Wang X. Seminal plasma microRNAs: potential biomarkers for spermatogenesis status. Mol Hum Reprod 2012;18:489–497. [DOI] [PubMed] [Google Scholar]
- Wu W, Qin Y, Li Z, Dong J, Dai J, Lu C, Guo X, Zhao Y, Zhu Y, Zhang W et al. . Genome-wide microRNA expression profiling in idiopathic non-obstructive azoospermia: significant up-regulation of miR-141, miR-429 and miR-7-1-3p. Hum Reprod 2013;28:1827–1836. [DOI] [PubMed] [Google Scholar]
- Yang Q, Hua J, Wang L, Xu B, Zhang H, Ye N, Zhang Z, Yu D, Cooke HJ, Zhang Y et al. . MicroRNA and piRNA profiles in normal human testis detected by next generation sequencing. PLoS One 2013;8:e66809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Z, Raabe T, Hecht NB. MicroRNA Mirn122a reduces expression of the posttranscriptionally regulated germ cell transition protein 2 (Tnp2) messenger RNA (mRNA) by mRNA cleavage. Biol Reprod 2005;73:427–433. [DOI] [PubMed] [Google Scholar]
- Zitzmann M, Nordhoff V, von Schonfeld V, Nordsiek-Mengede A, Kliesch S, Schuring AN, Luetjens CM, Kamischke A, Cooper T, Simoni M et al. . Elevated follicle-stimulating hormone levels and the chances for azoospermic men to become fathers after retrieval of elongated spermatids from cryopreserved testicular tissue. Fertil Steril 2006;86:339–347. [DOI] [PubMed] [Google Scholar]
- Zomer A, Vendrig T, Hopmans ES, van Eijndhoven M, Middeldorp JM, Pegtel DM. Exosomes: fit to deliver small RNA. Commun Integr Biol 2010;3:447–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.