Abstract
The origin and early evolution of photosynthesis are reviewed from an ecophysiological perspective. Earth's first ecosystems were chemotrophic, fueled by geological H2 at hydrothermal vents and, required flavin-based electron bifurcation to reduce ferredoxin for CO2 fixation. Chlorophyll-based phototrophy (chlorophototrophy) allowed autotrophs to generate reduced ferredoxin without electron bifurcation, providing them access to reductants other than H2. Because high-intensity, short-wavelength electromagnetic radiation at Earth's surface would have been damaging for the first chlorophyll (Chl)-containing cells, photosynthesis probably arose at hydrothermal vents under low-intensity, long-wavelength geothermal light. The first photochemically active pigments were possibly Zn-tetrapyrroles. We suggest that (i) after the evolution of red-absorbing Chl-like pigments, the first light-driven electron transport chains reduced ferredoxin via a type-1 reaction center (RC) progenitor with electrons from H2S; (ii) photothioautotrophy, first with one RC and then with two, was the bridge between H2-dependent chemolithoautotrophy and water-splitting photosynthesis; (iii) photothiotrophy sustained primary production in the photic zone of Archean oceans; (iv) photosynthesis arose in an anoxygenic cyanobacterial progenitor; (v) Chl a is the ancestral Chl; and (vi), anoxygenic chlorophototrophic lineages characterized so far acquired, by horizontal gene transfer, RCs and Chl biosynthesis with or without autotrophy, from the architects of chlorophototrophy—the cyanobacterial lineage.
Keywords: hydrothermal light, Zn-tetrapyrroles, photothiotrophy, cyanobacteria, lateral gene transfer, reaction center evolution
Questions of how and where chlorophyll-based photosynthesis (chlorophototrophy) arose and how the process subsequently spread among bacteria are typically investigated using phylogenetic trees, but in prokaryotes horizontal gene transfer decouples physiology from phylogeny; here we address the evolution of photosynthesis not from the perspective of gene or lineage phylogenies, but from the physiological perspective of chemical processes.
INTRODUCTION
Autotrophs have fueled primary production on Earth for at least 3.95 billion years (Tashiro et al.2017). The advent of photosynthesis—light-dependent CO2 fixation—was a pivotal event in microbial evolution, yet several key aspects of its origin remain unresolved. Photosynthesis encompasses two discrete physiological processes: chlorophototrophy, the use of chlorophyll (Chl) and light to generate ATP and/or reducing power, and the fixation of CO2 for biomass production and growth. Photosynthesis furthermore exists in two basic forms: anoxygenic photosynthesis involving one reaction center (RC) and oxygen-producing photosynthesis involving two. There is broad consensus that H2-based chemosynthesis predated chlorophototrophy in microbial evolution and that anoxygenic photosynthesis predated the oxygenic form. There is, however, no consensus concerning the physiological processes that mediated either transition. This review will focus on the two main transitions in photosynthesis evolution: (i) the origin of chlorophototropy, including the advent of Chl itself as well as key physiological constraints that may have helped the first Chl-bearing cells to access reductants other than H2 for primary production, and (ii) physiological intermediate states in the emergence of oxygenic photosynthesis from simpler anoxygenic versions.
Gene phylogenies offer limited insight into the matter because phylogenies are inherently error-prone (Williams et al.2013; Graur 2016) and because horizontal gene transfer has played a substantial role in generating the highly dispersed distribution of photosynthesis that is observed among modern bacterial lineages (Fischer, Hemp and Johnson 2016). Moreover, there is no consensus concerning either the numbers of transfers that took place during evolution or the directions in which those transfers occurred (Olson and Pierson 1987; Baymann et al.2001).
The evolution of photosynthesis has been amply reviewed in recent years (inter alia, Xiong and Bauer 2002a, 2002b; Allen 2005; Björn and Govindjee 2009; Hohmann-Mariott and Blankenship 2011; Williamson et al.2011; Fischer, Hemp and Johnson 2016), so there is no need for a new overview. Yet there is room to consider specific aspects of the physiological and ecological setting for the origin of photosynthesis, the subsequent evolution of reaction centers (RC), their co-evolution (or not) with chlorophyll (Chl) biosynthesis and CO2 fixation pathways, and the nature and role of low-potential electron donors. Phylogeny in the sense of prokaryotic lineage relationships (Fox et al.1980) is not our focus because photosynthesis arose and evolved in bacteria, and horizontal gene transfer in prokaryotes often decouples physiology from phylogeny (Wagner et al. 2017a).
THE PROBLEM
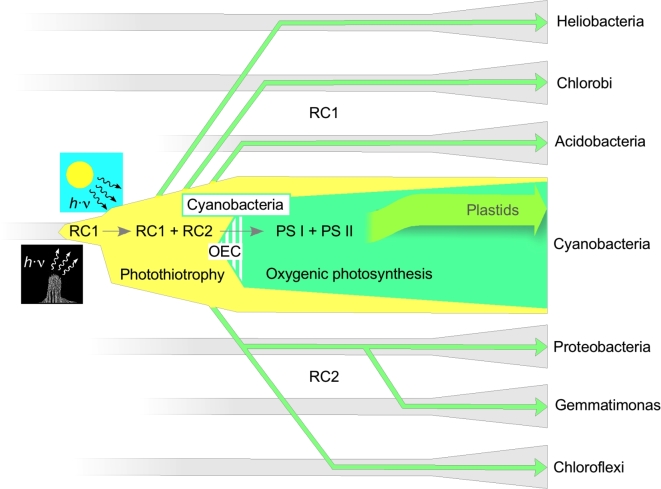
The basic puzzle concerning the evolution of photosynthesis on the basis of currently known Chl-harboring prokaryotic lineages is summarized in Table 1. The two types of RC, type-1 RC or RC1 and type-2 RC or RC2 (Golbeck 1993; Schubert et al.1998; Sadekar, Raymond and Blankenship 2006), may be subdivided further into four types of RC. There are homodimeric types of RC1 (in the green sulfur bacteria Chlorobia, Heliobacteriaceae and Acidobacteria) and heterodimeric types of RC1 (photosystem I, or PSI, in Cyanobacteria), and there are two types of RC2 (in Proteobacteria, Chloroflexi and Gemmatimonadetes; and photosystem II (PSII) of Cyanobacteria) that are both heterodimeric (Hohmann-Marriott and Blankenship 2011; Fischer, Hemp and Johnson 2016). The RCs are combined with three of the six known pathways of CO2 fixation (Fuchs 2011), and there are seven bacterial phyla within which Chl-based phototrophy occurs (Fischer, Hemp and Johnson 2016). Three of those lineages (Acidobacteria, Chlorobi and Chloroflexi) produce chlorosomes (Bryant and Liu 2013), antenna complexes containing self-assembling nanotubular, bacteriochlorophyll (BChl) suprastructures with a protein-stabilized, lipid monolayer envelope. Six of the phyla have members that grow aerobically (Acidobacteria, Chlorobi, Chloroflexi, Proteobacteria, Gemmatimonadetes and Cyanobacteria). Three lineages lack CO2 fixation pathways altogether (Firmicutes/heliobacteria, Acidobacteria and Gemmatimonadetes). Four lineages harbor photolithoautotrophic forms, whereas the remainder grow photoheterotrophically or photomixotrophically. The photolithoautotrophic Fe2+-oxidizing (photoferrotrophic) Rhodopseudomonas palustris strain TIE1 (Jiao et al.2005) is a notable exception among the Proteobacteria, which otherwise are notoriously versatile regarding their use of the Calvin-Benson-Bassham (CBB) cycle under aerobic or anaerobic conditions, in the light or in the dark, using H2, H2S or organic compounds as electron sources (Madigan and Gest 1979; McKinlay and Harwood 2010).
Table 1.
Some evolutionarily relevant physiological properties of chlorophyll-containing phototrophic bacteria.
| RC type | Taxon | PSa | Aerobicb heterotrophs | CO2 fixationc | Chlorosomesd | Photolithoautotrophy, e− donor |
|---|---|---|---|---|---|---|
| 1 | Firmicutes | Noe | ||||
| Heliobacterium | An | No | No | – | No | |
| 1 | Acidobacteria | Yesf | ||||
| Chloracidobacterium | μOx | Yes | No | + | No | |
| 1 | Chlorobi | Yesg | ||||
| Chlorobium | An | No | rTCA | + | H2S, S0, S2O32– | |
| C. ferrooxidans | An | No | rTCAh | + | Fe2+ | |
| 2 | Chloroflexi | Yesi | ||||
| Chloroflexus | An | Yes | 3HPBc | + | H2 or H2S | |
| Oscillochloris | An | ?j | CBB | + | H2 or H2S | |
| 2 | Proteobacteria | Yesk | ||||
| Purple sulfurl | An | Yesm | CBB | – | H2S, S0, S2O32– | |
| Purple non-sulfurn | An | Yeso | CBB | – | H2, H2S, S2O32– | |
| Rps. palustris TIE1p | An | Yes | CBB | – | Fe2+ | |
| Aerobic anoxygenicq | Ox | Yes | No | – | No | |
| 2 | Gemmatimonadetes | |||||
| Gemmatimonas | Ox | Yesr | No | – | No | |
| 1 + 2 | Cyanobacteria | Ox | Yes | CBBs | – | H2St or H2O |
| Oscillatoria | Anu | Yes | CBB | – | H2Sv or H2O | |
| Microcoleus | An | Yes | CBB | – | H2Sv or H2O |
Refers to O2 tolerance during phototrophic growth. An: anaerobic; Ox: aerobic; μOx: microoxic.
‘Yes’ indicates the ability for aerobic heterotrophic growth in the genus or the strain, or that the ability for aerobic heterotrophic growth is a widespread trait within the group. References in this column refer to the presence of terminal oxidase genes in the genome, or growth in the presence of O2.
CO2 fixation pathways that occur in combination with chlorophyll-based phototrophy. rTCA, reverse tricarboxylic acid (or Arnon-Buchanan) cycle; 3HPB, 3-hydroxypropionate bi-cycle; CBB, Calvin-Benson-Bassham cycle. The enzymes catalyzing the reductive steps in the rTCA cycle are ferredoxin-dependent, as in the case of the other two anaerobic pathways of CO2 fixation, the acetyl-CoA (or Wood-Ljungdahl) pathway and the dicarboxylate/4-hydroxybutyrate cycle (Fuchs 2011). The enzymes catalyzing the reductive steps in the 3HPB cycle and the CBB cycle are NADPH dependent (Fuchs 2011). We note that of the six pathways of CO2 fixation currently known: three have been named by the individuals who characterized them and the other three were characterized by Georg Fuchs.
The chlorosome protein CsmA, which binds BChl a and comprises the baseplate that connects the chlorosome to the FMO protein in GSB and Cab. thermophilum, is present in all lineages possessing chlorosomes studied so far (Bryant and Liu 2013) indicating a common ancestry of chlorosome antennae.
While there are aerobic Firmicutes, no phototrophic ones are aerobic. Heliobacteria are strictly anaerobic in phototrophic growth (Heinnickel and Golbeck 2007).
The acidobacteria were initially describes as aerobes (Kishomoti et al.1991; Bryant et al.2007). Chloracidobacterium thermophilum was initially described as an aerobe (Bryant et al.2007; Garcia Costas et al.2012). It has an absolute O2 requirement for growth, but O2 levels higher than ∼1% ambient O2 inhibit growth (Tank and Bryant 2015).
As shown in fig. 4.6 in Bryant and Liu (2013), O2-reducing terminal oxidases (bb3, bd) occur in most lineages of Chlorobi. Moreover, aa3, bb3 and bd are present in the non-photosynthetic ancestors of the photosynthetic Chlorobi. There are aerobic Chlorobi, but only ‘Candidatus Thermochlorobacter aerophilum’ is a chlorophototroph (Liu et al.2012; Tank et al.2017). All members of the family Chlorobiaceae are strictly anaerobic photolithoautotrophs.
The enzymes of the rTCA cycle are present in the draft genome of C. phaeoferrooxidans (Crowe et al.2017) and the bacterium grows autotrophically.
See Hanada and Pierson (2006). There is another phototroph in one of the deep branching families of the group, ‘Candidatus Roseilinea gracile’, that does not belong to the Chloroflexaceae. It has type 2 RCs, and BChl a but apparently lacks chlorosomes. Its terminal acceptors are not known but it grows as an anaerobe. See Tank et al. (2017) and Thiel et al. (2017).
We could not find reports for aerobic heterotrophic growth of Oscillochloris, but it branches in phylogenies after Chloroflexus and Roseiflexus, both of which grow aerobically in the dark (Hanada et al.2002)
Most Proteobacteria are capable of aerobic heterotrophic growth (Kersters et al.2006).
Purple sulfur bacteria can use H2S, S0 and other sulfur compounds during photoautotrophic growth (Dahl 2017), Chromatium vinosum is a well-studied example (Dahl 2017). There are also photoferrotrophic purple sulfur bacteria such as Thiodictyon (Hegler et al.2008; Camacho et al.2017).
Several photosynthetic members of the Chromatiaceae (purple sulfur) respire oxygen (Overmann and Pfennig 1992). Aerobic growth is present among both Chromatiaceae (Imhoff 2006b) and Ectothiorhodospiraceae (Imhoff 2006c).
Some Rhodobacter species are able to thrive as photolithoautotrophs, using reduced sulfur compounds (sulfide, thiosulfate) as electron donors (Pujalte et al.2014). Elemental sulfur (polysulfide) is the common end product of sulfide oxidation, although some species such as R. veldkampii can oxidize it to sulfate (Hansen and Imhoff 1985). Several Rhodopseudomonas species can use H2, H2S or S2O32– (de Souza et al.2014).
The photosynthetic purple non-sulfur bacteria are typically facultative anaerobes (McEwan 1994; Imhoff 2006a).
Rhodopseudomonas palustris strain TIE1 grows photoautotrophically with Fe2+ as the electron source (Bird et al.2011).
Aerobic anoxygenic phototrophs are found in α, β and γ subclasses of Proteobacteria and are very common in modern environments (Yurkov and Beatty 1998; Koblizeck 2015).
Zeng et al. (2014) reported Gemmatimonas phototrophica as growing semiaerobically in 10% O2 instead of 20% O2.
The Calvin cycle in different bacterial groups entails deeply divergent and in some cases unrelated enzymes, for example, classI/classII aldolase, classI/classII phosphoribulokinase or different forms of RubisCO (Martin and Schnarrenberger 1997).
See Allen (2005) and Oren and Padan (1978) for light-dependent anoxygenic H2S-dependent growth of O. limnetica to produce extracellular S±0; see De Wit and van Gemerden (1987) and Rabenstein et al. (1995) for evidence that cyanobacterial light-dependent H2S oxidation can generate thiosulfate (probably via sulfite).
See the text.
The chlorophototrophic lineages of prokaryotes are not closely related in any modern phylogenetic scheme (Fischer, Hemp and Johnson 2016), largely because chlorophototrophy has been spread among prokaryotes by horizontal gene transfer during evolution. There are ∼100 kb plasmids in some proteobacteria that contain all genes required for the RC, Chl and carotenoid biosynthesis in a cluster that is collinear with segments of proteobacterial chromosomes, and that is mobile among marine Roseobacter strains (Petersen et al.2012). Functional genes for PSI and PSII are mobile on phage genomes in the marine environment (Fridman, Flores-Uribe and Larom 2017). The most recently characterized chlorophototroph, a member of the phylum Gemmatimonadetes, clearly acquired its phototrophic gene cluster from a purple phototrophic bacterium (Zeng et al.2014). Many of the photosynthetic chlorophototrophic lineages can harness reduced sulfur species as an electron donor for photoautotrophic growth (Table 1), some lineages can use Fe2+, but only cyanobacteria can oxidize water. Notably, all of the photosynthetic lineages listed in Table 1 having members that can fix CO2 also include members that can use H2S as the electron donor during photolithoautotrophic growth. A number of cyanobacteria can grow photosynthetically with H2S as the electron source (Oren and Padan 1978; De wit and van Gemerden 1987; Rabenstein, Rethmeier and Fischer 1995; Grim and Dick 2016; Klatt et al.2015, 2016; Miller and Bebout 2004).
ITS ABOUT REDUCING CO2
Of the many things that the emergence of photosynthesis did for life (Judson 2017), perhaps the most important was to increase primary production. Before the origin of photosynthesis, there was only one significant source of electrons to fuel primary production on Earth: geochemical H2. Primary production via organic substances from space was not possible because of the paucity of fermentable substrates that have been found in such material and because of their structural heterogeneity, comprising a mixture of different isomers present at parts per billion concentrations each (Schönheit, Buckel and Martin 2016). CO2 was thus the starting material for organic biosynthesis, spurring the accumulation and diversification of the first forms of life and first ecosystems.
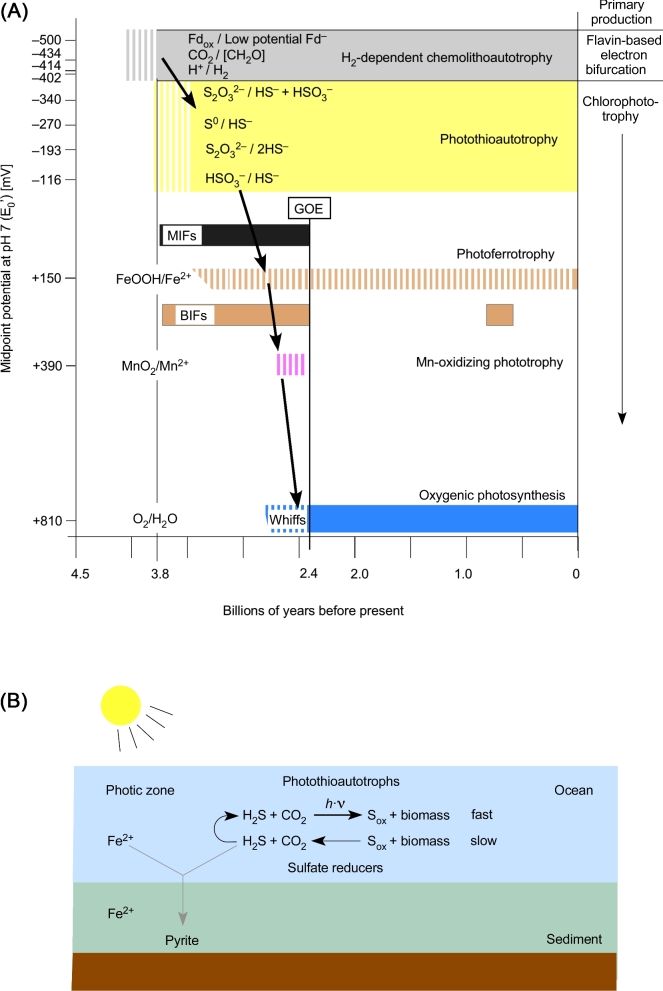
Among the electron donors that were widely available on the early Earth for CO2 reduction, only H2 has a sufficiently low midpoint potential to support CO2 reduction (Fig. 1; Table 2). The exhalation of H2 at hydrothermal vents stems from a spontaneous (exergonic) geochemical reaction called serpentinization (Sleep et al.2004; Martin et al.2008; Russell, Hall and Martin 2010; Sleep, Bird and Pope 2011). Serpentinization typically generates in the range of 10–20 mM H2 in the effluent of modern vents (Kelley, Baross and Delaney 2002; Schrenk, Brazelton and Lang 2013). During serpentinization, water circulating through hydrothermal systems comes into contact with Fe2+-bearing minerals in the crust that transfer electrons to water, producing H2 and leaving Fe3+ minerals such as magnetite (Fe3O4) behind (Bach et al.2006; Schrenk, Brazelton and Lang 2013). The serpentinization reaction can be written in a simplified form (Sleep, Bird and Pope 2011; McCollom and Seewald 2013) as
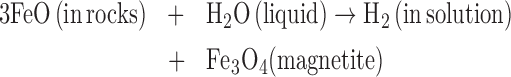 |
(1) |
with the relevant redox reaction being the oxidation of Fe2+ to Fe3+ to generate H2, which is released into the hydrothermal effluent (Sleep et al.2004; Bach et al.2006; Sleep, Bird and Pope 2011; Schrenk, Brazelton and Lang 2013).
Figure 1.
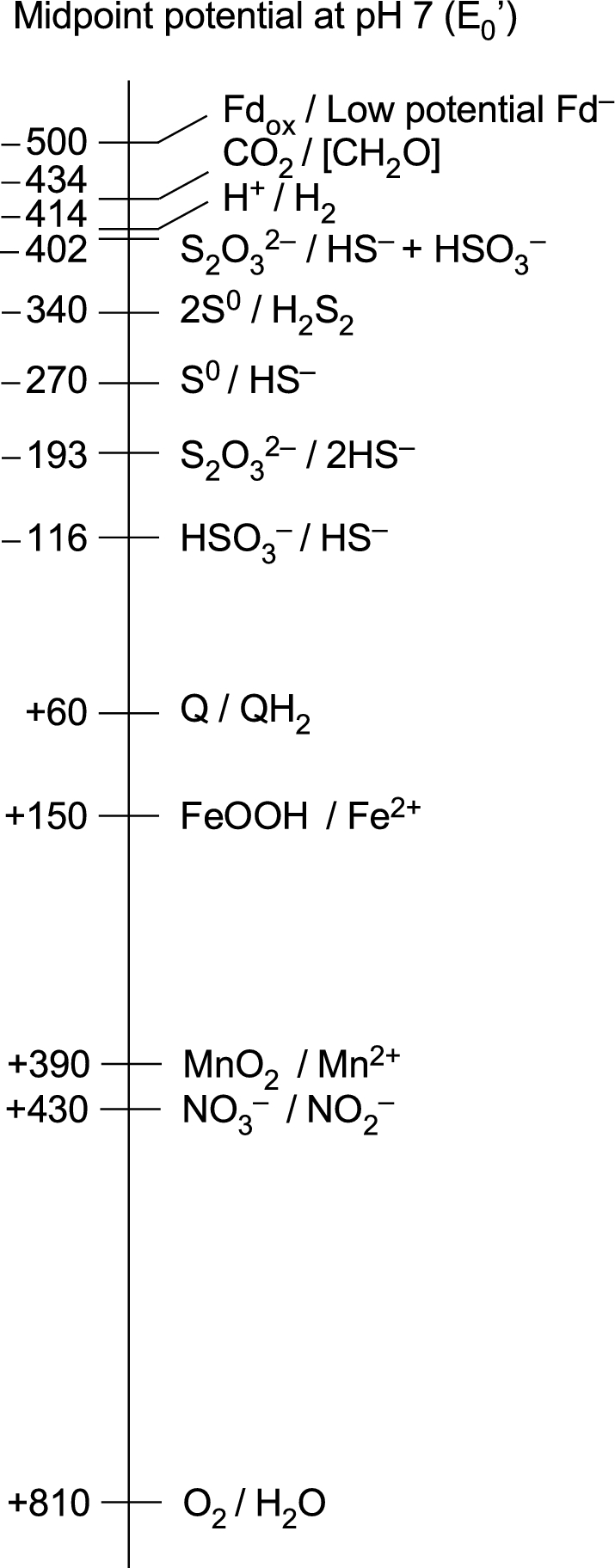
Midpoint potentials of some redox couples relevant to this paper. The relevant redox couples for chemolithoautotrophic primary production are the uppermost three. Values are from Thauer, Jungermann and Decker (1977), Brune (1989), Griffin, Schott and Schink (2007), Sharma et al. (2012) and Lengeler, Drews and Schlegel (1999). The H2AsO4/H3AsO3 couple (arsenate/arsenite, not shown) has a midpoint potential at pH 7 of +54 mV and is used by some chlorophototrophic proteobacteria (Budinoff and Hollibaugh 2008).
Table 2.
Key transitions in physiological evolution and primary production.
| Relative age | Tetrapyrrolea | Physiologya | Modern group | Electron sources |
|---|---|---|---|---|
| Modern | Chlorophyll | Chlorophototrophy with RCI and RCII | Cyanobacteria | H2S, H2O |
| Advanced | Chlorophyll | Chlorophototrophy with RCI | Cyanobacteria, chlorobia | H2Sb |
| Intermediate-2 | Porphyrins | Mixotrophy | None | H2S |
| Intermediate-1 | Heme | Anaerobic respiration | Sulfur reducersc, autotrophic ε-proteoc | Organicsd, H2 |
| Primordial | Cobalamine | Chemolithoautotrophy | Acetogens, methanogensf | H2 |
Heme before chlorophyll: Granick (1965); cobalamin before heme before chlorophyll: Decker et al. (1970). Chemolithoautotrophs before anaerobic respiration before anoyxgenic photosynthesis: Decker et al. (1970).
Before the advent of photosynthetic H2S oxidation, H2 was the sole reductant driving primary production. Chlorophotosynthetic use of the same reductant (H2) would not substantially increase primary production (see text) and would be restricted to environments where H2 from serpentinization was discharged in photic environments. Of course, H2 is also generated by anaerobic fermentations of reduced carbon compounds, but that does not permit a net increase in primary production.
Here, the term sulfur reducers designates organisms that gain energy by S, sulfite or sulfate reduction, a very broad definition (Rabus et al.2015). Many sulfate reducers grow chemolithoautotrophically using the acetyl-CoA pathway for carbon metabolism and sulfate reduction with H2 for energy metabolism, or chemoorganoheterotrophically on acetate or lactate (Rabus et al.2015). H2-dependent epsilonproteobacteria that use the rTCA cycle for autotrophy and anaerobic respiration via sulfur reduction, for example, would be another kind of early intermediate. See the text.
Of course, use of organics as electron donors does not increase primary production, but it increases metabolic flexibility and permits specialization of carbon and energy metabolism.
In acetogens and methanogens, the acetyl-CoA pathway is the central pathway of carbon assimilation and energy metabolism, whereas the ion gradient that drives ATP-synthase is generated during the process of CO2 reduction with electrons from H2 (Thauer et al.2008; Schuchmann and Müller 2014; Sousa and Martin. 2014).
The H2 so generated escapes from the crust into the ocean via the circulating hydrothermal vent effluent. Serpentinization has been going on for 4.2 billion years, since there was liquid water on Earth (Sleep, Bird and Pope 2011). In addition to reducing water to H2, it also reduces CO2 to simple organic compounds such as methane and acetate (McDermott et al.2015; McCollom 2016; Miller et al.2017). The serpentinization reactions involving CO2 reduction reveal strong similarities between a spontaneous geochemical process and the core physiology of primitive anaerobes that reduce CO2 with electrons from H2 as their main bioenergetic reaction (Sousa and Martin 2014). Geochemical evidence indicates that anoxygenic photosynthesis was probably in existence by 3.3 to 3.4 billion years ago (Tice and Lowe 2006; Westall et al.2006, 2011; Arndt and Nisbet 2012). It has been estimated that the transition from H2-based chemosynthesis to anoxygenic photosynthesis increased Earth's yearly primary production by a factor of ∼2000, and that the subsequent transition to oxygenic photosynthesis increased primary production by an additional factor of ∼30 (Raven 2009). Thus, prior to the origin of photosynthesis, primary production on Earth was very limited in magnitude and restricted to sites of H2 emission.
REDUCED FERREDOXIN BEFORE PHOTOSYNTHESIS
The origin of Chl is the starting point of photosynthetic evolution. Based on the order of biosynthetic precursors in modern pathways (Table 2), Chl arose in cells that could synthesize heme, and heme arose in cells that made cobalamin (Decker, Jungermann and Thauer 1970; Sousa et al.2013). Note that the primitive lineages of acetogens and methanogens synthesize cobalamin, but lack heme (cytochromes) and quinones (Thauer et al.2008; Schuchman and Müller 2014). We briefly consider the physiology of the cells within which photosynthesis might have arisen, keeping in mind that before the origin of photosynthesis, primary production was anaerobic and dependent on H2 generated by serpentinization in hydrothermal systems.
Primary production in strict anaerobes requires reduced ferredoxin, Fdred. The oxygen-sensitive (anaerobic) pathways of CO2 fixation—the acetyl-CoA pathway, the reductive (or reverse) TCA (rTCA) cycle and the dicarboxylate/4-hydroxybutyrate cycle—entail one or more Fd-dependent reduction steps and harbor one or more oxygen-sensitive enzymes (Fuchs 2011). The first cells were likely H2-dependent chemolithoautotrophs that used the linear, exergonic acetyl-CoA pathway for CO2 fixation (Fuchs 2011; Poehlein et al.2012; Takami et al.2012; Weiss et al.2016). The acetyl-CoA pathway, also called the Wood-Ljungdahl pathway, is unique among CO2 fixation pathways in that it is exergonic, allowing microbes with primitive redox physiology like acetogens and methanogens to generate protonmotive force for ATP synthesis from CO2 reduction (Thauer et al.2008; Poehlein et al.2012; Schuchmann and Müller 2014). All other CO2 fixation pathways require energetic input in the form of ATP (Fuchs 2011). The core CO2-fixing and bioenergetic reaction in acetogens and methanogens (that is, the exergonic synthesis of acetate or methane from H2 and CO2) is strikingly similar to spontaneous geochemical CO2 reduction during serpentinization (Sousa and Martin 2014; McDermott et al.2015; McCollom 2016; Miller et al.2017).
There is, however, a crucial mechanistic and energetic hurdle to CO2 fixation in acetogens and methanogens. Their CO2-reducing enzymes require Fdred with a very low midpoint potential, on the order of −500 mV (Fuchs 2011; Buckel and Thauer 2013). This must be generated using electrons from H2, with a midpoint potential of –414 mV. During the reduction of the FeS clusters in low-potential Fd by H2, electrons must flow energetically uphill (i.e. endergonically). To perform this energetic trick, cells employ a recently discovered mechanism called flavin-based electron bifurcation (Herrmann et al.2008; Li et al.2008; Buckel and Thauer 2013; Lubner et al.2017; Wagner et al. 2017), in which the electron pair in H2 is first transferred to FAD, a transducer of a two-electron to a one-electron transfer. One electron exits FADH energetically uphill to produce Fdred while the other goes to a sufficiently positive electron acceptor, such as NAD+ (in acetogens, Eo' = –320 mV) or the heterodisulfide CoM–S–S–CoB (in methanogens, Eo' = –140 mV), so that the overall energetics of the reaction are favorable, allowing it to proceed (Buckel and Thauer 2013).
How did the earliest organisms generate low-potential Fdred for CO2 reduction prior to evolutionary origin of electron bifurcation? Comparative genomic data indicate that the first cells had a physiology very similar to that of acetogens and methanogens and arose in a geochemical setting rich in FeS minerals (Weiss et al.2016), in which the Earth's spontaneous redox chemistry provided FeS minerals with the electron-donating and CO2-reducing function of Fdred, via serpentinization. In addition, zero valent transition metals such as Fe0 readily reduce CO2 to methanol (Guan et al.2003) and acetate (He et al.2010), they occur naturally in hydrothermal vents, for example as awaruite (Ni3Fe) which is a common constituent of serpentinizing systems (McCollom 2016), and they are electron sources for methanogenic growth (Daniels et al.1987). Early chemolithoautotrophs depended on interactions between rocks, water, metals, and H2.
H2-dependent primary production based on serpentinzation was stable on geological timescales, and it fueled Earth's first ecosystems. Some sulfate reducers also grow autotrophically using the acetyl-CoA pathway (Rabus et al.2015), and represent a slightly advanced state relative to acetogens and methanogens in which carbon and energy metabolism are both fueled by the reduction of CO2 by H2 (Buckel and Thauer 2013). In sulfate reducers that use the acetyl-CoA pathway autotrophically, CO2 is fixed while energy is obtained by the reduction of sulfur compounds with electrons from H2, or organic donors such as acetate or lactate (Rabus et al.2015).
With accumulating biomass on primordial Earth, the first heterotrophic metabolisms became possible (Schönheit, Buckel and Martin 2016). Bacterial cells consist of roughly 50%–60% protein, 20% RNA, 10% lipids and 5%–10% saccharides (Neidhardt, Ingraham and Schaechter 1990), and so fermentative breakdown of amino acids, purines and sugars became energetically favorable at low H2 partial pressures. These fermentations, along with organoheterotrophic and cytochrome-independent respiration using S±0 as the terminal acceptor, as found in heterotrophic Thermococcales (Schut, Bridger and Adams 2007; Schut et al.2013), likely were among the first heterotrophic metabolisms (Schönheit, Buckel and Martin 2016). Photosynthesis arose in ecosystems where primary production was chemolithoautotrophic and where heterotrophs lived by consuming the cell mass of the H2-dependent primary producers.
Coupling Fd reduction to CO2 reduction
Thoughts on the origin and early evolution of photoautotrophy as well as the occurrence of ancient light carbon isotopes in the geochemical record (Nisbet and Sleep 2001) have long been associated with ribulose-1,5-bisphosphate carboxylase/oxygenase, RuBisCO (Tabita 1999), the CO2-fixing enzyme of the Calvin cycle (also called the Calvin-Benson-Bassham or CBB cycle) and the quantitatively most significant entry point of CO2 into the modern carbon cycle (Raven 2009). More recent findings concerning the other five pathways of CO2 fixation indicate, however, that anaerobic CO2-fixing pathways, in particular the acetyl-CoA pathway, which is ferredoxin-dependent, and the reverse TCA cycle, which is both ferredoxin and NAD(P)H dependent, predate the Calvin cycle (Berg 2011; Fuchs 2011), which is fully NADPH dependent and has no Fd-dependent reactions. Consistent with that view, divergent and likely ancient forms of RuBisCO, called type IV RuBisCO or RuBisCO-like proteins (RLPs), have been shown to function in heterotrophic metabolism in diverse bacteria and archaea (Hanson and Tabita 2001; Tabita et al.2007; Sato and Atomi 2011; Tabita et al.2008; Aono et al.2012).
Such findings suggest that the ancestral function of RubisCO likely arose in a heterotrophic context, having later been co-opted into the CBB cycle for CO2 fixation. For example, in Bacillus species, RLP functions in a heterotrophic methionine salvage pathway (Ashida, Danchin and Yokota 2005; Ashida et al.2003) and the RLPs from methanogenic archaea, which populate the root in phylogenetic trees of RuBisCO sequences, incorporate ribulose bisphosphate-carboxylase activity into methanogen metabolism (Kono et al.2017), even though CO2 fixation in methanogens proceeds through the acetyl-CoA pathway (Thauer et al.2008; Fuchs 2011). Perhaps more revealing is the role of RuBisCO activity in the utilization of RNA as a substrate. The first heterotrophs likely lived on the cell mass of dead chemoautotrophs, with RNA as an important source of organic material.
Cells are roughly 20% RNA by weight (Neidhardt, Ingraham and Schaechter 1990). For the first heterotrophs, RNA was an excellent carbon and energy source (Schönheit, Buckel and Martin 2016). In the archaeon Thermococcus kodakarensis, RuBisCO participates in a short RNA degradation pathway (Aono et al.2012), the first enzyme of which phosphorolytically cleaves the base from ribonucleoside monophosphates (RNA breakdown products) to generate ribose-1,5-bisphosphate. The next step is catalyzed by an isomerase that generates ribulose-1,5-bisphosphate, which is cleaved by RuBisCO via carboxylation, generating two molecules of 3-phospho-D-glycerate (3PGA) for carbon and energy metabolism (Sato, Atomi and Imanaka 2007; Aono et al.2012; Schönheit, Buckel and Martin 2016). Although central carbon and energy metabolism in archaea is very different from that in bacteria (Reher et al.2010; Bräsen et al.2014), 3PGA is a universal metabolite among free-living cells, and the same RuBisCO like protein is also found in anaerobic heterotrophic bacteria that lack a CBB cycle (Wrighton et al.2012).
Because all forms of anoxygenic photosynthesis entail electron transport chains containing cytochromes and quinones, the cells that evolved photosynthesis must have been capable of some sort of respiration involving cytochromes and quinones (Decker, Jungermann and Thauer 1970; Xiong and Bauer 2002a, 2002b). This is consistent with the view that Chl biosynthesis arose subsequent to heme biosynthesis by pathway extension starting from late intermediates, in a specific evolutionary lineage (Granick 1965; Decker, Jungermann and Thauer 1970) (Table 2). The type of respiration is immaterial here, but it was before the advent of O2, and hence it cannot have involved high-potential acceptors like O2 (Fig. 1). SO2, a gas commonly emitted by volcanic activity, was present in the Earth's most ancient oceans, dissolved as sulfite SO32– (Halevy, Zuber and Schrag 2007), and could have been an early terminal electron acceptor. The energy-conserving segment of sulfate reduction starts from sulfite (Rabus et al.2015; Santos et al.2015); sulfite (sulfate) reduction and sulfur-based respirations, which generate H2S, were likely among the first to arise in metabolic evolution (Decker, Jungermann and Thauer 1970; Arndt and Nisbet 2012). Isotopic signatures from rocks 3.8 to 2.7 billion years of age indicate that the sulfur cycle was operating in a more or less modern form before the rise of oxygen (Grassineau et al.2006).
Sulfate reducers are replete with cytochromes and quinones (Rabus et al.2015). Many oxidize fermentation end products such as acetate and lactate for energy metabolism while using the acetyl-CoA pathway for carbon metabolism. Although we use the vernacular term ‘sulfate reducers’ here, in no passage does this wording necessitate the presence or involvement of sulfate. Sulfate reducers are thought to first activate sulfate to sulfite at the expense of ATP and a reductant (Santos et al.2015), whereas the subsequent six-electron reduction of sulfite to sulfide is exergonic if the electrons stem from H2 or organic compounds (Rabus et al.2015). From the physiological and energetic standpoints on ancient Earth, sulfate reduction is better seen as sulfite reduction. The heme- and cytochrome-containing anaerobes that evolved photosynthesis were most likely facultative chemoautotrophs/chemoheterotrophs. Perhaps they were similar in physiology to sulfate reducers that use the acetyl-CoA pathway (Rabus et al.2015), or to epsilonproteobacteria that reduce S0 using polysulfide reductase and fix CO2 by the rTCA cycle (Grote et al.2012).
The transition to phototrophic Fd reduction may have taken place in a cell that used the rTCA cycle. The rTCA cycle, also called the Arnon-Buchanan cycle, consumes ATP and thus must be supported by an independent energy metabolism generating ATP (Fuchs 2011), which is in contrast to a central role in generating ATP as in the acetyl-CoA pathway during chemoautotrophic growth. Separate pathways of carbon and energy metabolism would have been conducive to the onset of phototrophy. The incomplete 'horseshoe' (i.e. branched) TCA cycle manifest in acetogens and methanogens (Fuchs 1989; Simpson and Whitman 1993; Furdui and Ragsdale 2000) would be ancestral to the rTCA cycle (Martin and Russell 2007). The TCA cycle can function as a branched pathway in Escherichia coli and other chemotrophic bacteria during anaerobic fermentative growth (and largely during aerobic growth on glucose as well; Neidhardt, Ingraham and Schaechter 1990), whereas anaerobic phototrophs such as Rhodobacter can use light-driven, reverse electron transfer to catalyze the succinate dehydrogenase reaction and thereby run a complete, oxidative TCA cycle anaerobically (Beatty and Gest 1981). The mechanism of flavin-based electron bifurcation in epsilonproteobacteria that use the rTCA cycle has not been reported, but it could involve electron bifurcation at the heterotrimeric Fe-Fe hydrogenase, HydABC, which catalyzes the reversible reduction of NAD+ and Fd with H2 in Thermotoga spp. (Schut and Adams 2009) and acetogens (Schuchmann and Müller 2014). The evolution of phototrophic Fd reduction in a cell that used the rTCA cycle would have provided a net boost to autotrophic carbon metabolism without directly interfering in energy metabolism. It is worth noting that organisms utilizing the acetyl-CoA pathway and the rTCA cycle are common in hydrothermal vent environments (Chapelle et al.2002; Campbell and Cary 2004; Takai et al.2005; Lever et al.2010; Lever 2012).
ZN-TETRAPYRROLE TRIPLET AND FERREDOXIN REDUCTION
Primary productivity was limited by H2 production at vents until the origin of photosynthesis. Geochemical data indicate that photosynthesis-based primary production was in operation some 3.4 billion years ago (Tice and Lowe 2006; Westall et al.2006, 2011), long before the appearance of O2 roughly 2.4 billion years ago (Arndt and Nisbet 2012; Lyons, Reinhard and Planavsky 2014; Fischer, Hemp and Johnson 2016). It is widely agreed that anoxygenic photosynthesis preceded oxygenic photosynthesis, but how and where anoxygenic photosynthesis arose has been unresolved.
From our standpoint, the main initial benefit of anoxygenic photosynthesis was not the additional energy provided by cyclic electron flow using an RC2 to produce protonmotive force for ATP synthesis. Instead, the main benefit was that anoxygenic photosynthesis provided access to a new source of moderately low-potential electrons—i.e. from a donor other than H2—that could be used together with light energy to generate Fdred for the purpose of CO2 fixation. Therefore, the RC1, which provides linear electron flow to Fd, came first. Given that H2 was the lowest potential sustainable source of electrons on the early Earth, the only way to convert electrons from a higher potential donor to a much lower potential is, as far as we know, by harnessing light to generate electrons of sufficiently low potential to reduce Fd to Fdred. To rephrase, for emphasis: in a world where H2 was the electron donor with the most negative midpoint potential (Fig. 1), Chl-based phototrophy provided an alternative mechanism to flavin-based electron bifurcation as a means to generate low-potential Fdred for CO2 fixation.
As a short digression, and small caveat to the foregoing sentence, one might imagine that flavin-based electron bifurcation could, in principle, offer a mechanism to generate low-potential Fdred with electrons from donors with more positive midpoint potentials than H2, such as H2S or other reduced sulfur species, provided that very high potential acceptors (such as O2) were available in the environment prior to the origin of photosynthesis. However, there is no evidence for the existence on the early Earth of very high potential acceptors with midpoint potentials near or exceeding that of O2 (+810 mV) in amounts approaching those required to run an ecosystem. In that context, one might ask whether the levels of H2 generated by serpentinization are really sufficient to fuel chemolithoautotrophs. The answer is yes: the 10–20 mMol/kg levels of H2 commonly observed at vents of serpentinizing systems are orders of magnitude more than the H2 partial pressure of roughly 10 Pa that methanogens lacking cytochromes require for sustained growth (Thauer et al.2008). Digression aside, the capability to utilize alternative electron sources would have conferred on cells a powerful selective advantage in H2-limited environments.
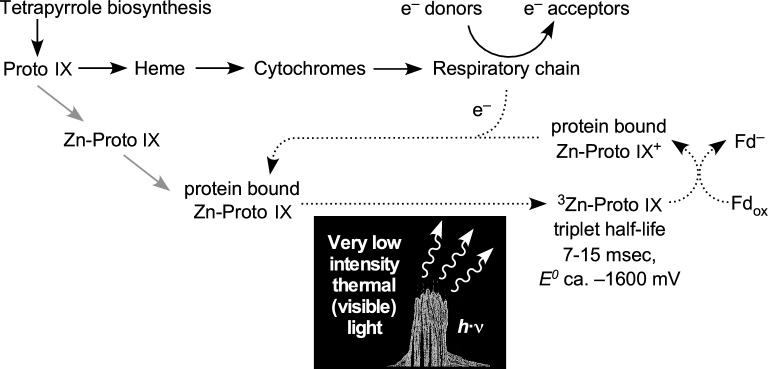
Zn-protoporphyrin IX: a functional bridge to Chl
There are two prerequisites for harnessing light energy. One is a source of light, with which we will deal in a subsequent section (focusing not on sunlight, however, but on light that is emitted from hydrothermal vents). The other prerequisite is a molecule with the photochemical properties of Chl, which we consider now. At the time of origin of the Chl biosynthetic pathway, the first enzymes involved were surely not finely tuned, and enzymes in related pathways could have promoted the accumulation of intermediates as side reactions. Preexisting enzymes could have furthermore participated in more than one pathway. Although insertion of Zn+2 into protoporphyrin IX (PPIX) occurs spontaneously (Taketani et al.2007; Becker et al.2012), the insertion of Zn+2 is catalyzed by ferrochelatase (Hunter, Sampson and Ferreira 2008; Chau et al.2011).
For example, in bchD mutants of Rhodobacter sphaeroides lacking Mg-chelatase, which catalyzes the first committed step of modern Chl/BChl synthesis, ferrochelatase from the heme biosynthetic pathway inserts Zn2+ into PPIX, leading to the production of Zn-PPIX monomethyl ester, Zn-divinyl-protochlorophyllide and Zn-BChl a (Jaschke et al.2011). As Williamson et al. (2011) have previously pointed out, and as we further develop in the following sections, Zn-tetrapyrroles have interesting properties in the context of Chl evolution, and we suggest that it is possible that Zn-tetrapyrroles predated Mg-derivatives.
What might cells have done with Zn-PPIX? Zinc typically exhibits five-coordinate geometry in porphyrins (Favereau et al.2015). Zinc in Zn-PPIX would not have been useful for electron transfer reactions as iron in heme (in cytochromes) because Zn has only one valence state, Zn+2, and therefore is biologically redox inert (Kręzèl and Maret 2016). However, if Zn-PPIX were inserted into a preexisting cytosolic heme-binding protein, such as a soluble apocytochrome b, that protein, although unable to catalyze cytochrome-type, one-electron transfer reactions, would have been able to absorb and store light energy by virtue of its Zn-PPIX chromophore. How so? Absorption of light (∼410, 540 and 580 nm) by Zn-PPIX (Jaschke et al.2011) produces a long-lived triplet excited state with a yield of ∼90% (Vanderkooi and Berger 1989) and—importantly—with a long lifetime of ∼7 to 15 ms (Dixit, Waring and Vanderkooi 1981).
Using light to reduce ferredoxin
The excited triplet state in Zn-PPIX has a half-life roughly 106-fold longer than Mg-Chl excited states, which typically are in the range of a few nanoseconds for the photochemically active singlet species (Björn et al.2009). A total of 7–15 ms is virtually an eternity for the purpose of catalyzing a light-driven electron transfer reaction, and brings possible activities into the time domain of diffusion rate-limited chemical reactions with other molecules in the cell. This is ample time for an exited state 3Zn-PPIX 'cytochrome' to find by diffusion a cytosolic electron acceptor, such as soluble Fd. Fd is not only a ubiquitous protein in anaerobes (Buckel and Thauer 2013), but is also extremely abundant—in a typical anaerobe, Fd has cytosolic concentrations on the order of 80–400 μM (Thamer et al.2003) (0.2–1 μmol Fd per gram of cytosolic protein, which is typically 400 mg/ml). The redox potential of a photoexcited Zn-PPIX triplet is about –1.6 V (Dixit, Moy and Vanderkooi 1984); for example, the potential of photo-excited triplet Zn-cytochrome c (3Zn-PPIX(cyt)) at 25°C and pH 7 is –1.7 V (Shen and Kostic 1996). Photoexcited 3Zn-PPIX(cyt) is a strong reductant (Shen and Kostic 1996) that readily reduces both plastocyanin and ferricytochrome b5 (Qin and Kostic 1994). The redox potentials from 3Zn(cyt) to Zn(cyt)+ (900 mV) and back to Zn(cyt) (800 mV) would fit with Fd reduction, and with reduction of ZnCyt+ by a respiratory chain (Shen and Kostic 1996). A protein-bound, photoexcited 3Zn-porphyrin, 3Zn-PPIX, or a 3Zn-PPIX-cytochrome should have more than sufficient driving potential to reduce soluble Fd. These features could constitute the core of a primordial phototrophic Fd reduction pathway (Fig. 2).
Figure 2.
A proposal for the origin of Chl-based phototrophy (see the text). In the primitive, ancestral pathway, a side activity of ferrochelatase produces Zn-PPIX as in modern cells although Zn2+ can also spontaneously insert into PPIX. Zn-PPIX could bind to an abundant soluble heme-binding protein, leading to a photoactive protein that could have reduced soluble Fd and replaced flavin-based electron bifurcation. Williamson et al. (2011) have discussed the possible role of Zn-tetrapryrroles as functional intermediates in Chl evolution.
A soluble heme-binding protein carrying Zn-PPIX could have become an alternative to H2-dependent, flavin-based electron bifurcation and furthermore paved the way to the advent of Chl biosynthesis. Both synthetic Zn-tetrapyrroles and engineered, cytochrome-bound Zn-tetrapyrroles perform light-dependent redox reactions and have been used in the study of artificial photosynthesis (Razeghifard and Wydrzynski 2003; Hay et al.2004). Synthetic protein-bound Zn-tetrapyroles continue to be of interest in that regard (Cohen-Ofri et al.2011). Furthermore, Zn-tetrapyrroles also function in nature. Zn-BChl a is used in the RC2 of Acidiphilium rubrum (Wakao et al.1996), and Zn-BChl a΄ apparently forms the special pair of the type-1 RC in Chloracidobacterium thermophilum (Tsukatani et al.2012). Thus, there is physiological relevance of Zn-tetrapyrroles in modern chlorophototrophy, lending weight to the possibility of their involvement in the origin of phototrophy in a heme-containing chemotroph.
FROM HEME TO CHL: RECRUITMENT FROM EXISTING PATHWAYS
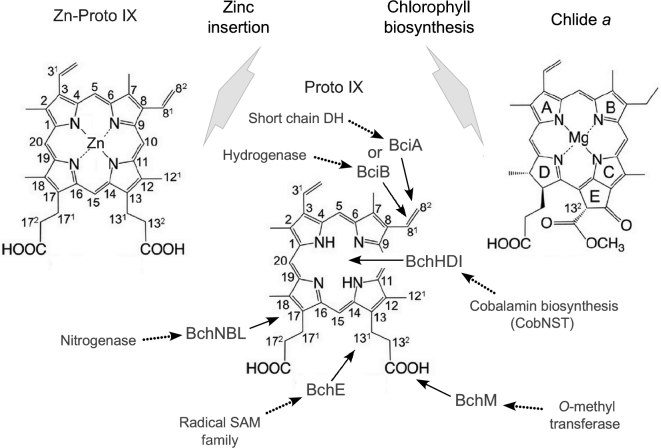
The next questions are how, and why, did the existence of Zn-PPIX lead to a biosynthetic pathway to produce chlorophyllide a and ultimately extend to Chl a? Zn-PPIX has very strong absorbance in the blue at about 423 nm but only weak absorbance at ∼550 and ∼600 nm. Because the light environment around vents (see the next section) is deficient in blue light, it would have been strongly advantageous, in terms of harnessing available light, to shift the absorbance of the Zn-PPIX into the red/near-IR region of the spectrum. Initially, Mg-chelatase may not have formed the first step in the Chl biosynthetic pathway. Substitution of Zn with Mg would have shifted the chemistry of the excited state from the triplet state for Zn-tetrapyrroles to the singlet state for Mg-tetrapyrroles. This would likely have required that a protein interacting with a chain of electron acceptors already existed to reduce the probability of the back reaction to reduce the oxidized pigment and increase the lifetime of the charge-separated state. However, it really makes little difference when the Mg chelation step evolved because enzymes of Chl biosynthesis tolerate, to differing degrees, variation with respect to the identity of the metal in the tetrapyrrole ring, Zn or Mg (Jaschke et al.2011). Fundamentally, these enzymes require that the tetrapyrrole coordinates a metal atom, the insertion of which is the function of the chelatase, the first enzyme in the pathway.
Thus, the next enzyme of concern would have been the enzyme magnesium-protoporphyrin IX methyltransferase (BchM/ChlM) to protect the carboxyl group of the propionate side chain at C-13 of the Chl precursor (Gomez Maqueo Chew and Bryant 2007a). This enzyme is a simple O-methyl transferase, a common enzyme activity that is widespread in ribosomal RNA modification and that was present in ancient cells (Weiss et al.2016). The next enzyme, BchE, which forms the isocyclic ring (ring E of (B)Chls), is an oxygen-independent oxidative ring cyclase. BchE is a radical-SAM enzyme, another very ancient family of proteins (Broderick et al.2014; Weiss et al.2016), and it furthermore bears some resemblance to coproporphyrinogen III oxidase, HemN, which removes the carboxyl groups from two propionate side chains of coproporphyrinogen III to form the two vinyl moieties of PPIX (Layer et al.2003). Protection of the carboxyl group to prevent decarboxylation from occurring would have promoted the ring closure reaction. Another radical SAM enzyme, BciD, probably arose later, but it also has a related activity (Thweatt et al.2017). BciD oxidizes the C-7 methyl group of BChl c to form the C-7 formyl group of BChl e, a 4-electron oxidation, by hydroxylating the methyl group twice to form a geminal-diol intermediate, which then apparently dehydrates spontaneously to form the formyl group. By analogy in a Zn-based phototrophic system, one component of the BchE reaction might involve radical-based, sequential hydroxylation of the C-131 position to form a geminal-diol that dehydrates spontaneously to form the keto group of isocyclic ring E. The product of this reaction, Zn-divinyl-protochlorophyllide, has absorption maxima at 438, 575 and 624 nm (Jaschke et al.2011), which would improve light absorbance relative to Zn-PPIX in the orange-red region of the visible light spectrum.
The reduction of the 8-vinyl group (BciA/BciB) may not have been required to produce a highly functional Chl-like molecule, and in any event, it leads to rather small differences in the absorbance spectra of Chls (Gomez Maqueo Chew and Bryant 2007b; Bjorn et al.2009). BciA and BciB are alternative enzymes and evolutionarily unrelated. As sketched in Figure 3, BciA is a member of the large NADPH dependent short chain dehydrogenase family while BciA is related to F420-reducing hydrogenases typical of methanogens (Sousa et al. 2013). Some cyanobacteria, such as Prochlorococcus spp., produce divinyl-Chl a, which functions equivalently to Chl a but has somewhat enhanced absorption of blue light (Chisholm et al.1988; Goericke and Repeta 1993). At least some cyanobacteria can grow when the gene encoding 8-vinyl-reductase is inactivated (Ito et al.2008). Finally, a key step in producing a red-absorbing pigment is the reduction of the D-ring double bond to produce the conjugation system of the chlorin ring. A multisubunit enzyme, BchNBL (ChlNBL), which is structurally related to Mo-nitrogenase, would have first catalyzed this reaction (Nomata et al.2006; Bröcker et al.2010; Muraki et al.2010). 8-Vinyl Zn-chlorophyllide a has very strong absorption in the red at 664 nm, like Chl a, which presumably would have been a desirable property for early light-driven processes (Tamiaki et al.2013).
Figure 3.
Structural changes and enzymes needed to convert PPIX into chlorophyllide a. Modified from Bryant and Liu (2013). PPIX with the changes that generate Zn-Proto IX (left) and chlorophyllide a (right). Zn-PPIX is photochemically active and accumulates in Mg-chelatase mutatants of R. sphaeroides through a side activity of ferrochelatase (Jaschke et al.2011). Chloracidobacterium thermophilum uses Zn-BChl a΄ in its RC (Tsukatani et al.2012). BciD (not shown in the figure) and BchE are both radical SAM enzymes. BciD catalyzes a 4-electron oxidation of the methyl group at C7 to generate a formyl group. A 4-electron oxidation also occurs during the second part of the BchE reaction, part one being the oxidative ring closure (which is possibly similar to coproporphyrinogen III oxidase), part two being the oxidation to produce the keto group of the isocyclic ring E. See the text.
Addition of a hydrophobic phytol tail by Chl synthase (ChlG) is all that would then have been required to transfer any phototrophic process(es) that could occur with soluble proteins into the membrane (Gomez Maqueo Chew and Bryant 2007a). This would have had important consequences, because this would have allowed the coupling of redox reactions to energy conservation via ion gradients using pre-existing cytochrome and quinone components. At some point, Zn2+ was replaced by Mg2+. This may have occurred comparatively late in the origin of phototrophy because the transition from triplet to singlet-based excited state dynamics would have meant that a protein with a series of electron acceptors must have existed to ensure efficient spatial charge separation could occur, and outcompete the rapid back reaction to the ground state that would otherwise have occurred. This may also be why some RCs still apparently employ Zn-BChl a΄ (Tsukatani et al.2012). Again, cells would probably have turned to preexisting enzymes. PPIX Mg-chelatase is related to the cobalt chelatase (CobNST) that functions in cobalamin biosynthesis (Debussche et al.1992), and a pathway to Mg-chelatase would have required only gene duplication and divergence. Thus, all of the enzymes needed to modify Zn-PPIX spontaneously produced from heme biosynthesis to form Chls were probably co-opted from pathways and enzymes already existing in cells (Fig. 3). The one component for which no obvious ancestor has yet been identified is the ancestor of the type-1 RC protein itself. Such a protein probably was a transmembrane, alpha-helical polypeptide, and it is possible that it could have bound a tetrapyrrole (e.g. heme) and/or a quinone. Xiong and Bauer (2002a, 2002b) have suggested that cytochrome b is a one such candidate.
ABYSSAL LIGHT, LOW LIGHT, GOOD LIGHT
When biologists (or geochemists) think of the origin of photosynthesis, they often think of harnessing sunlight at the Earth's surface. We are thinking of harvesting light initially from hydrothermal vents (Nisbet, Cann and Van Dover 1995) in the otherwise dark abyss of the ocean floor because, in addition to other reasons to be outlined in this section, that is the site of primary production that was required to support the growth of the heme-containing cells that made the transition to chlorophototrophy (Fig. 2). Hydrothermal vents emit thermal light and visible light.
Thermal light is black body radiation emitted from black smoker types of vents with very hot (>400°C) effluent (van Dover et al. 1996; White, Chave and Reynolds 2000; White et al. 2002a,b). At such temperatures the emitted light is mainly wavelengths >900 nm, but lesser amounts of light extend down to ∼750 nm, which could be absorbed by chlorosomes (if they existed). Black body light from a >400°C source typically has a vanishingly small component of what is commonly thought of as photosynthetically active radiation (400–700 nm). Because of its low flux in the Chl a absorption range, thermal light is not widely discussed as a source of photosynthetically relevant radiation. However, the idea has been alive and well in the literature for over 20 years, starting with the suggestion that the pathway to anoxygenic photosynthesis emerged as a heat- and light-sensing mechanism to guide motile prokaryotes to sources of chemical energy (Nisbet, Cann and Van Dover 1995).
Low-intensity visible light from hydrothermal vents
Ambient light at hydrothermal vents has a component with wavelengths in the visible range (van Dover et al.1996; White et al.2002a). The first hints for visible light at vents came from studies of a shrimp that inhabits the dark abyss near vents and possesses unusual photoreceptive organs (van Dover et al.1989). The mechanism(s) generating light in the visible spectrum at vents that exceeds the contribution from thermal light are still unknown; possible sources include sonoluminescence (the collapse of small bubbles) and triboluminescence (light emission from small photoactive crystals) (Tapley, Buettner and Shick 1999; White, Chave and Reynolds 2000). Ambient light flux at vents has been measured directly, and fluxes were reported to be too low to support photosynthetic life, on the order of 106 photons cm−2 s−1 in the 600–700 nm range (White et al.2002a), although higher photon fluxes were also reported, on the order of 109 cm−2 s−1 in the 700–800 nm range (White et al.2002b). Of course, the photon flux at vents decreases with the square of the distance from the source, and can be further reduced by turbidity; at a distance of 2 cm from flange pools at vents along the Juan de Fuca Ridge, photon fluxes on the order of 1011 cm−2 s−1 in the 600–1000 nm range were measured (White et al.2002b).
The isolation and cultivation of an obligately photoautotrophic, H2S-dependent green sulfur bacterium (GSB) from a hydrothermal vent sample raised the possibility that light emission at hydrothermal vents could indeed be sufficient to support photosynthesis (Beatty et al.2005). The photon fluxes at vents are on the same order as those observed near the chemocline of the Black Sea, where the flux was reported as 1.8 × 1011 photons cm−2 s−1 (∼3 nmol photons m−2 s−1). A stable population of a photosynthetic GSB, Prosthecochloris phaeobacteroides BS1, lives there at a depth of ∼100 to 110 m and has an estimated in situ doubling time of ≥2.8 years (Overmann and Pfennig 1992) or more (Manske et al.2005; Marschall et al.2010). The Black Sea GSB uses isorenieratene and BChl e to harvest light from the sun (Overmann and Pfennig 1992), which occurs in the blue/green region (ca. 450–550 nm) at that depth. The hydrothermal vent GSB uses chlorobactene and BChl c, which are better suited for using black body radiation because the in vivo BChl c absorbance peak is maximal at 750 nm, tailing off around 850 nm (Beatty et al.2005).
Photon fluxes at vents are about six orders of magnitude lower than surface irradiance for sunlight. A doubling time of 3 years or more might at first seem so slow that one can neglect it, but if we consider modern low-energy environments (Whitman, Coleman and Wiebe 1998), microbiologists are reporting turnover times (not doubling, just carbon turnover whether in the same cell or in a new one) on the order of 1000 years or more (Hoehler and Jorgensen 2013). Our point here is that the photon fluxes observed at vents in the relevant spectral range for chlorophototrophy are much lower than those at the surface, but they could support doubling times that are two to three orders of magnitude faster than those in modern microbial communities in low-energy environments. Thus, considering that microbes do not double in the time domain of hours in the wild, it is possible that the Chl-dependent, photosynthetic lifestyle arose using hydrothermal light. After all, with a doubling time of 3 years, it would take a 1 μm3 bacterium only 500 years to produce a cell mass that weighs more than the Earth, given enough substrate.
Better than sunlight for the origin of phototrophy
In our view, there are reasons to think that a low-light origin of photosynthesis is far more likely than an origin at the surface, where photons arrive a million times faster. The reasons are simple: photooxidation and UV light. Chl absorbs photons and is excited to a highly reducing species (which becomes highly oxidizing if an electron acceptor is nearby). Modern cells must direct those electrons in an orderly flow to acceptors, and obtain reductant at a rate that keeps light-activated Chl from causing harmful oxidations of the cell constituents (Krause and Weis 1991; Demmig-Adams and Adams 1992; Garcia-Medosa et al.2011). However, most modern Chl-related, high-light damage relates to interactions with O2 (Szabo, Bergantino and Giacometti 2005), whereas Chl synthesis arose in the absence of O2. Chl triplets can reduce yields, but they do little damage to cells when there is no O2 around, and as noted above, triplet states might have initially provided advantages to cells. However, UV light can easily produce the second excited state of Chl (or higher), and can lead to oxidized Chl, which is a very dangerous molecule, one of the strongest oxidants known in biology (Ishikita et al.2005). Modern cells that use Chl go to great lengths to protect themselves from its oxidative power. At the origin of Chl synthesis, cells would have possessed no mechanisms to protect themselves from oxidants the strength of oxidized Chl because they had never seen such a strong oxidant. Although it has recently been suggested that atmospheric gasses other than ozone might have absorbed some UV radiation during the Archean (Muller et al.2016), in the absence of an ozone layer, there was certainly a higher flux of photosynthetically active radiation at shallower depths of the water column than at vents on the ocean floor, and sunlight was accompanied by some level of UV radiation, which is not present in hydrothermal light.
Existence near the heavily irradiated, aquatic surface was thus a life-threatening situation for a primitive Chl-containing cell. Moreover, if early primary production was physically linked to H2 production via serpentinization at hydrothermal vents, then before the origin of photosynthesis there was no reductant at the surface from which cells could live; the UV-irradiated ocean surface harbored neither high local concentrations of a reductant (H2) for chemosynthetic primary production to support the growth of the first microbes that invented photosynthesis nor did it harbor high local concentrations of an alternative electron donor (H2S) that early photosynthesizers might have harnessed. One could argue that hydrothermal sites in shallow water could have provided the substrates required to support the origin of photosynthesis, but an origin of Chl-based photosynthesis from hydrothermal light seems more likely, because it offers the opportunity of harnessing light energy without the risk of photooxidative damage from UV radiation. In modern oceans, the deep penetration of UV light is attenuated by scattering and absorption due to the presence of living cells and associated chromophoric organic matter (Tedetti and Sempéré 2006). Before the origin of chlorophototrophy, such factors would not have attenuated UV light penetration. Therefore, from the standpoint of a bacterium in its environment, dim light emitted from hydrothermal vents—the only habitat where cells were stably growing on the early Earth—provides the most likely illumination for the origin of photosynthesis.
TYPE 1 RCs OR TYPE 2 RCs FIRST?
One of the classical questions in the evolution of photosynthesis is which kind of RC came first, type 1 or type 2. The two types of RC are related at the level of structure (Schubert et al.1998; Allen 2005; Sadekar, Raymond and Blankenship 2006; Blankenship 2010; Cardona, Murray and Rutherford 2015), but with undetectable homology in amino acid sequence alignments. In anoxygenic photosynthesis, type 2 RCs support cyclic electron flow. The electron acceptors of and donors to the RC (quinones and cytochromes and cytochrome-like electron carriers) are supplied by the cell, and an ion gradient is generated that is used for ATP synthesis or reverse electron transport to generate reductants. Anoxygenic type 1 RCs mostly support linear electron flow, and they typically generate Fdred from H2S or organic compounds.
If the type 2 RC arose first, then the initial function of chlorophototrophy in the context of increasing primary production was to support the synthesis of ATP and NAD(P)H via reverse electron transport. However, using organic electron donors such as succinate does not increase net primary production. In other words, if a type 2 RC arose first, then the synthesis of NAD(P)H via reverse electron transport, as in the case of photoferrotrophs like Rps. palustris TIE1 (Bird, Bonnefoy and Newman 2011), appeared in evolution before the RC1-dependent Fd reduction for CO2 fixation, as in the case of the GSB Chlorobaculum tepidum. That possibility seems unlikely because it would mean that autotrophs started off dependent on electron bifurcation to synthesize Fdred for CO2 fixation, took an evolutionary detour through NAD(P)H and then returned to Fdred with the origin of the type 1 RC.
If the type 1 RC arose first, the initial benefit of chlorophototrophy was to provide access to a new reductant—H2S—thereby providing a selective advantage and enabling increased net primary production. To us it seems more likely that photosynthesis started with RC1 than with RC2. Our logic here follows a traditional line of reasoning in photosynthesis evolution, but applies it to a different setting. That is, the traditional logic for the origin of oxygenic photosynthesis from anoxygenic photosynthesis has been that it afforded cells access to a new reductant: H2O. By the same reasoning, we suggest that the original function of photosynthesis was to provide access to a reductant other than H2, namely H2S. H2S, like H2, is abundant in hydrothermal effluents (Kelley, Baross and Delaney 2002). The initial physiological consequence of phototrophy (and the reason for its evolutionary success) was that it replaced flavin-based electron bifurcation as a mechanism to generate low-potential Fdred for CO2 fixation. The first chlorophototrophs made the step into an ecologically new world that was no longer dependent on H2. Chl-dependent photosynthesis represented light energy-supported access to a new reductant, H2S, in a highly reducing, low-potential redox environment that harbored three essential prerequisites for photosynthesis evolution:
a preexisting reductant (H2) to support the survival of cells while they were evolving Chl synthesis,
access to a new reductant (H2S) requiring light energy for oxidation but with a low midpoint potential (HS−/S° couple, Eo΄ = −270 mV), and
a continuous low light flux that was free of UV photons, providing the first cells that started to accumulate Chl an opportunity to adapt to its presence, rather than suffering damage from UV-induced oxidized Chl as the photobiological situation at shallow depths would have presented.
It is true that anoxygenic chlorophototrophic bacteria can use the type 2 RC for reverse electron transport and photoferrotrophic growth (Bird, Bonnefoy and Newman 2011), yet the main function associated with RC2 today is cyclic electron transport (except in conjunction with PSI as in cyanobacteria). The main function associated with RC1 is linear electron flow from H2S to produce Fdred. We suggest that it has always been that way, and that the origin of RC1 was thus the first decisive step in the origin of photosynthesis. We do not offer a suggestion for the precursor protein from which the RC1 proteins arose; it is possible that some Chl-lacking, heme-containing protein with structural similarity to RC1 might someday be found, conceivably a cytochrome b homolog (Xiong and Bauer 2002a, 2002b); however, the nature of the RC protein precursor is not essential here.
H2S FIRST OR Fe2+ FIRST?
Electron donors for anoxygenic photosynthesis include H2, H2S and Fe2+ (Table 1), and certainly all would have been present at a hydrothermal vent in the Archean. H2 is the least likely electron donor for the first chlorophototrophs. H2-dependent chemotrophs can grow at H2 partial pressures as low as 1 Pa (Thauer 2011) using one or more of the three kinds of hydrogenase known—[Fe-Ni], [Fe-Fe] and [Fe] (Shima et al.2008). Autotrophs that use H2 to reduce Fd via electron bifurcation would have had no benefit from photosynthesis. They would have remained dependent on H2 but with an additional dependence (on light). That is, they would have had to evolve and employ an energetically expensive machinery (Chl synthesis and RC biogenesis) that only added redundancy to a simpler, preexisting, highly tuned and fully functional chemolithoautotrophic metabolism, without eliminating H2 dependence or gaining any net benefit.
For that reason, the suggestion that H2 was perhaps the first photosynthetic electron donor (Tice and Lowe 2006) seems unlikely to us. Harnessing H2, which the first autotrophs could use without Chl anyway, would not have increased primary production, nor is light required for autotrophic growth on H2 (Thauer et al.2008; Buckel and Thauer 2013; Schuchmann and Müller 2014), which may explain why the acetyl-CoA pathway is not used in conjunction with phototrophy. Phrased another way, if H2 is the first electron donor for chlorophototrophs, it provides no benefit to the cell that evolved photosynthesis because the final benefit of the extensive evolutionary investment (Chl biosynthesis, RC origin, integration into the electron transport chain) is the ability to do what cells could do from the outset, namely access H2 as a reductant for CO2 fixation. In this regard, we note that an RC would affect neither the affinity of a preexisting hydrogenase for H2 nor that of a CO2 reducing enzyme for its substrates.
We suggest that the first electron donor for anoxygenic photosynthesis was H2S (HS−), not Fe2+, because the evolutionary and electrochemical leap in redox midpoint potential from H2 (–414 mV) to access the HS−/S° couple (–270 mV) is of far lesser magnitude than that needed to access Fe2+ (Eo' = ca. + 150 mV at pH 7). Furthermore, in photoferrotrophs characterized so far, Fe2+ oxidation entails cytochromes and high-potential iron-sulfur proteins (HiPIPs) with much more positive midpoint potentials (Bird, Bonnefoy and Newman 2011; Crowe et al.2017). For example, PioA from the phototrophic iron oxidation operon of Rps. palustris TIE1, which uses a type 2 RC, is a decaheme cytochrome, and PioC is a HiPIP (Bose and Newmann 2011); their midpoint potentials reside in the range of +385 to +450 mV (Bird, Bonnefoy and Newman 2011). FoxE in R. ferrooxidans SW2 is the iron-oxidizing protein and is a diheme cytochrome, which also has a high midpoint potential in the range +201 to +300 mV at pH 7 to 6 (Saraiva, Newman and Louro 2012). The Cyc2-type cytochromes that photoferrotrophs employ in iron oxidation typically have very high midpoint potentials in the range of +560 mV (Bird, Bonnefoy and Newman 2011), and appear to be the same type of protein that aerobic iron oxidizers, such as Acidithiobacillus ferrooxidans (Ishii et al.2015) or Mariprofundus ferrooxydans PV-1 (Barco et al.2015), use to oxidize Fe2+ to Fe3+, with O2 serving as the terminal acceptor. The same high-potential cytochrome, also called Cyc2PV-1 (Barco et al.2015), occurs in the iron-oxidizing photoferroautotrophic GSB, Chlorobium phaeoferrooxidans (Crowe et al.2017), which uses a type 1 RC. The electrons from Cyc2PV-1 in M. ferrooxydans PV-1 are thought to be transferred to a soluble periplasmic di-heme cytochrome, called Cyc1PV-1 (Barco et al.2015), which is thought to function similarly (electron transfer) to FoxE in Rhodobacter sp. SW2 and PioA in Rps. palustris TIE1 (Bird, Bonnefoy and Newman 2011). In the case of the photoferroautotrophic GSB electrons are transferred to a type-1 RC, whereas in photoferroautotrophic Rps. palustris TIE1 a type 2 RC performs cyclic electron transfer to energize reverse electron transport, but in both cases high-potential cytochromes are involved.
There is only one isolated report of photoferrotrophic growth of cyanobacteria (Cohen 1984). Siderophilic (iron-loving) cyanobacteria have been isolated from iron-rich environments (for example, Leptolyngbya sp. strain JSC-1, also known as Marsacia ferruginose; Brown et al.2010). This organism requires high concentrations of iron for growth and forms iron deposits inside and outside cells; however, convincing evidence for photoferrotrophy in this organism is still lacking. Because cyanobacteria oxidize water and produce oxygen, spontaneous oxidation of Fe2+ complicates analyses, and Fe3+ is toxic in the presence of oxygen. Given that water is 55 M and Fe2+ concentrations are generally in the micromolar range, there would seem to be little selection pressure for cyanobacteria to oxidize iron under most circumstances. Thus, if modern cyanobacteria do oxidize iron, one might expect this process to occur in environments that undergo alternating periods of oxic and anoxic conditions during the diel cycle (e.g. hot spring microbial mats (Jensen et al.2011)).
In RC2-containing photoferrotrophs (for example, Rhodobacter or Rhodopseudomonas spp.), CO2 fixation occurs via the CBB cycle, one of the most recent Fd-independent pathways of CO2 fixation to have arisen (Fuchs 2011). Of course, Rhodobacter spp. are capable of reducing Fd with the help of RC2 using Rnf (an NADPH:Fd oxidoreductase) that harnesses the photosynthetic transmembrane potential to produce Fdred for nitrogen fixation (Schmehl et al.1993; Biegel et al.2011) by reverse electron transport. However, the electrons for Fd reduction in Rhodobacter spp. typically come from substrates such as organic acids in RC2-based phototrophy and the process is driven by ATP hydrolysis (Hoffmann et al.2015).
Modern photoferrotrophy lacks ancient traits
One could argue that photoferrotrophy involving RC1, as in C. ferrooxidans, came before H2S oxidation involving RC1, but any strictly anaerobic cell that was in the process of synthesizing Chl in a manner that would not have been suicidal would have been far more likely to access the HS−/S° couple (Eo' = –270 mV), rather than the much more positive Fe2+ oxidation step (Eo' = ca. + 150 mV at pH 7), based on what we currently know about electron donors in anoxygenic RCs. Oxidation of either H2S or Fe2+ is not likely to have been an activity of the RC itself; it probably would have required an enzyme or intermediate cofactor (e.g. cytochrome). For the HS−/S° couple, preexisting pathways involved in S0 reduction could have been recruited to operate with the same substrate but in reverse. That would not have been an option in the case of the Fe2+/Fe3+ couple, for lack of environmental Fe3+ and hence Fe3+ reducers prior to the great oxidation event (GOE).
High-potential cytochromes involved in Fe2+ oxidation seem to be more typical of O2-respiring bacteria, and thus appear to be derived mechanisms of Fe2+ oxidation that might have come into combination with RCs later in evolution, possibly after the appearance of O2. We are confronted with the problem that not only is photosynthesis mobile across broad taxonomic boundaries, but also iron oxidation and O2 reduction (terminal oxidases) are probably mobile too, making it difficult to identify the directions and the timing of horizontal gene transfers (Castresana et al.1994; Soo et al.2017).
The existence of photoferrotrophy in GSB and proteobacteria does not preclude the possibility that other forms of photoferrotrophy might have existed before the appearance of O2. These would have entailed theoretical mechanisms of electron entry into the photosynthetic electron chain that are independent of high-potential cytochromes and HiPIPs. Such mechanisms of photoferrotrophy might exist but remain to be discovered. Importantly, we are not calling into question the idea that photoferrotrophy per se is a very ancient form of metabolism (Widdel et al.1993; Ehrenreich and Widdel 1994; Heising et al.1999). Neither are we questioning the possibility that photoferrotrophy may have been causal to the deposition of the banded iron formations (BIF) (Widdel et al.1993; Ehrenreich and Widdel 1994; Heising et al.1999; Kappler et al.2005). We do note, however, that Crowe et al. (2014) pointed out that the photoferrotrophy of GSB may have had a low contribution to BIF deposition under low light conditions, and that Grassineau et al. (2006) have pointed out that the sulfur cycle during the Archean was probably very similar to that of today based on sulfur isotope data.
We are simply stating that, from the standpoint of physiology, the nature of electron flow, and the cofactors involved in the forms of photoferrotrophy characterized so far, this metabolism does not look very ancient in any respect (far less ancient than H2S-dependent phototrophy in GSB, in particular). Rather, it appears to result from the acquisition of a couple of high-potential cytochromes (and HiPIPs) by horizontal gene transfer, which were incorporated into the metabolism of bacteria that were already able to grow photoautotrophically.
It is also possible that the photoferrotrophs characterized so far are different lineages than those supposedly involved in BIF deposition, even though the underlying chemical process (light-dependent Fe2+ oxidation) is the same. These same kinds of basic questions—namely, are the bacteria that perform these processes today direct descendants of the bacterial lineages that performed the processes 3 billion years ago, and are the processes even the same, pervade the literature on the evolution of photosynthesis. They also pervade the literature on Earth history because photosynthesis is so closely tied to geochemical evolution. Because horizontal gene transfer decouples physiology from phylogeny (Martin 2016; Wagner et al. 2017), it is important to make sense out of the evolution of photosynthesis in a manner that is not dependent on branching orders in trees (while not completely ignoring circumstances where trees might be relevant and provide insights).
Of course, photothiotrophy requires a sulfide-oxidizing enzyme to access the reductant. The enzymology of sulfide oxidation was probably not the limiting step, however, as there are at least three phylogenetically unrelated isoenzymes that oxidize H2S—sulfide:quinone oxidoreductase (SQR), flavocytochrome c/sulfide dehydrogenase (FccAB) and a rhodanese-like protein, SoxL (Dahl 2017)—in addition to several unrelated enzymes that can reduce S0 to H2S, including the NADPH-dependent sulfur reductase Nsr (Bridger et al.2011) and polysulfide reductase Psr (Jormakka et al.2008). The existence of such enzymatic diversity indicates that the evolution of H2S oxidation systems (or reversing electron flow through pre-existing S0-reducing enzymes) is not an evolutionary hurdle per se; the hurdle is the origin of Chl-based, light-dependent oxidation.
INTERMEZZO
To summarize so far, Chl biosynthesis (from the heme precursor PPIX) was the initial step of photosynthesis evolution. Zn-tetrapyrroles might have played a role as intermediates in Chl origin (Williamson et al.2011). Chl probably arose in an anaerobic bacterium that possessed cobalamin, cytochromes and quinones. That bacterium was furthermore a heterotroph or facultative heterotroph, and it probably lived near a hydrothermal vent, because that was where electron bifurcation-dependent primary production was occurring. This early evolutionary line could have made the first steps towards chlorophototrophy using hydrothermal light because at that time low-intensity, long-wavelength light presented an opportunity, whereas high-intensity UV light would have been deadly. Most lineages of photoautotrophic bacteria use H2S as an electron donor. The first RC probably functioned similarly to the type 1 RC of a GSB, coupling light-dependent H2S oxidation to the generation of Fdred, replacing the function of flavin-based electron bifurcation in H2-dependent chemolithoautotrophs as the fulcrum of primary production and affecting primary production in two significant ways: it allowed primary production to increase, and it released the physical constraint tying primary production to hydrothermal sources of H2. Obligately photoautotrophic GSB present at hydrothermal vents today might harbor an ancient kind of low-light phototrophy analogous to that existing at the onset of phototrophy, but need not represent the most ancient chlorophototrophic lineage, because of horizontal gene transfer.
With the ability to harness light in a linear electron transport chain leading from H2S to Fdred, light limitation would eventually become as important as reductant limitation in such an environment. This would have provided a selective advantage to improvements and modifications of Chl biosynthesis that generated functionally specialized pigments. Perhaps more immediately, it would have conferred advantages to cells that could most efficiently harvest low-intensity light: that is, to develop an antenna like the chlorosome, which is simple in design, unparalleled in light-harvesting efficiency and energetically inexpensive to produce compared to protein-based antenna systems (membrane-intrinsic antenna complexes, or phycobilisomes). Chlorosomes may resemble the ancestral state of light harvesting (Bryant and Liu 2013). Photoferrotrophy, as found in currently characterized proteobacteria and GSB (Table 1), is most plausibly interpreted as the recent horizontal acquisition of cytochromes that can oxidize Fe2+ (from bacteria that can oxidize Fe2+ using O2 as the terminal electron acceptor) by these chlorophototrophs.
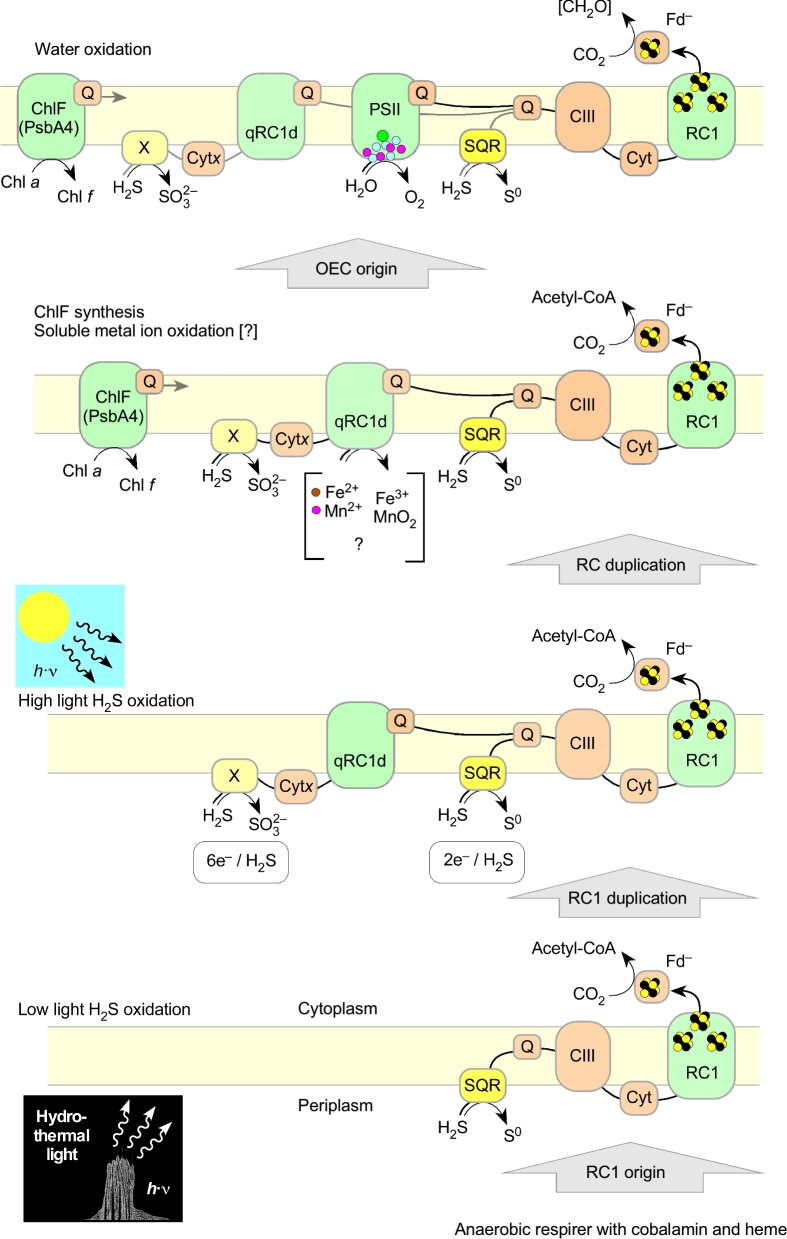
SULFIDE AND A POSSIBLE PATH TO WATER SPLITTING
A second photosystem, for what?
Evolution in prokaryotes does not proceed under direction, nor does it seek out new solutions; it proceeds via gene duplication, mutation, (re-)combination and horizontal transfer, and it is advanced by natural selection. Once cells had evolved the ability to access H2S and light using Chl, standard Darwinian trial-and-error tinkering would have begun to integrate photothiotrophy into the preexisting physiology and genetic composition of the cell (Bauer and Bird 1996; Allen 2005). Photosynthetic life at low light intensities would be a primitive trait in our scenario, and chlorosomes, exceedingly efficient antenna complexes requiring only a few conserved proteins (Bryant and Liu 2013), probably represent one of the earliest forms of light-harvesting antenna complexes. However, the limited and skewed phylogenetic distribution of chlorosomes, their occurrence in combination together with either RC1 or RC2 (Table 1) and the small number of proteins (beyond Chl biosynthesis) required for their biogenesis suggests that they, too, could be subject to horizontal transfer in evolution. Primary production based on the oxidation of H2S should have been a stable physiology.
There are two main questions about the origin of two types of RCs: how and why? The former is much easier to answer than the latter. The main contours of our proposal are summarized in Fig. 4.
Figure 4.
Photothiotrophy in the evolution of two photosystems. SQR: sulfide:quinone oxidoreductase. CIII: complex III-like cytochrome complex. Q: quinone. qRC1d, a quinone-reducing RC1 duplicate. X: Sulfite-generating protein. Cyt x: hypothetical carrier (probably a cytochrome). See the text.
There are two ideas for how two RCs came to be: gene duplication and functional diversification within one organism (Olson and Pierson 1987; Allen 2005), and vertical divergence by means of lineage-specific RC evolution prior to horizontal gene transfer to bring a second RC (RC1? RC2?) into a protocyanobacterium (Hohmann-Mariott and Blankenship 2011; Blankenship 2017). This question is readily answered on the basis of the Chl biosynthesis pathway. The RC proteins of RC1 and RC2 are structurally related but deeply divergent in terms of structure, subunits, pigment binding, amino acid sequences and function (Schubert et al.1998; Sadekar, Raymond and Blankenship 2006; Gisriel et al.2017). What they share in common, however, is that neither RC type can undergo any sustained evolution (that is, avoid pseudogenization and loss) without Chl (Sousa et al.2013). Thus, in the case of lineage diversification and vertical divergence, there should exist, and we should expect to observe, two deeply divergent pathways of Chl biosynthesis in nature: one associated with RC1 (at least sometimes) and one associated (at least sometimes) with RC2. In the case of gene duplication within a single lineage (RC1 yields RC2 or vice versa), Chl biosynthesis should be monomorphic—that is, not existing in two deeply divergent manifestations.
Although not widely acknowledged, this question has already been answered: there is only one pathway for Chl biosynthesis leading from Mg-protoporphyrin IX to chlorophyllide a in modern chlorophototrophs. This is strikingly different from the case of heme biosynthesis, for which pathway variations exist (Dailey et al.2017). Because the available evidence indicates a single pathway, it appears that the two RC types arose in the same genome via gene duplication (Sousa et al.2013). We note that there are alternative forms for the relatively recently evolved O2-dependent and more ancient O2-independent isoenzymes—sometimes found within the same genome (Chen, Canniffe and Hunter 2017), and so some extant Chl biosynthetic enzymes arose after the origin of O2. How RCs came to exist in the genomes where they are currently found, namely vertical inheritance vs horizontal transfer, is a different question that will be briefly addressed below.
Two RCs and two entry points for sulfide
We have outlined above reasons why RC1 should be the ancestral RC type. The original function of the product of gene duplication and divergence, RC2, is more difficult to ascertain. In a bacterium that was oxidizing H2S with the help of RC1 (and probably SQR), the function of the initial duplication of RC1, RC1d (meaning initially identical in sequence to RC1), would have been to do what it had been doing before the duplication; namely, the RC1d would help to oxidize H2S by transferring electrons from cytochromes to Fd. In anoxygenic bacteria, both RC1 and RC2 oxidize cytochromes. RC1 reduces Fd via three [4Fe4S] clusters at the cytosolic (acceptor) side of the protein complex whereas RC2 reduces quinones (Golbeck 1993). Both RC electron-transfer pathways involve the participation of a cytochrome c:quinol oxidoreductase, usually similar to Complex III, although Chloroflexi and Acidobacteria use alternative Complexes III (Bryant et al. 2012; Bryant and Liu 2013). We will not deal with possible modifications within the RC, but instead focus on substrate oxidation and product reduction.
Assuming RC1 to be the ancestral state, the evolutionary and functional modifications would occur in its duplicate gene. Loss of FeS clusters and their replacement by a quinone-binding pocket would have led to a quinone-reducing RC1 duplicate, labeled qRC1d in Fig. 4, which retained the ability to oxidize cytochrome, but probably a different cytochrome from that recognized by RC1 (hence labeled Cyt x in Fig. 4). This would have generated the capacity for combined linear and cyclic electron transport, and two types of RC that could be differentially expressed to serve ATP synthesis or Fd reduction, as needed by the cell (Allen 2005). This hypothetical intermediate state is almost identical to that suggested by Allen (2005), although our starting point is different. Allen (2005) suggested that from this intermediate state, which he termed a ‘heteronuclear anoxygenic phototroph’ (HAP), there was a loss of differential regulation from the ‘OR’ state to the ‘AND’ state, leading to expression of both RCs simultaneously. Evolutionary exploration led to an RC producing a very strong oxidant, initially oxidized Chl but eventually the tyrosyl radical of RC2 prior to the origin of a Mn-based water-splitting complex. Oxidation of soluble, environmental Fe2+ or Mn2+ may have represented an intermediate ancestral state from which the water-splitting complex arose. There is geochemical evidence to suggest that just prior to the GOE, soluble Mn2+ oxidation occurred in marine environments (Johnson et al.2013). The GOE is widely interpreted as marking the global appearance of O2 in the geochemical record (Fischer, Hemp and Johnson 2016).
Sulfide oxidation to sulfite and thiosulfate
We suggest that the initial physiological function of an RC1d module was sulfide oxidation to sulfite and thiosulfate, which is based on the following three observations: (i) many cyanobacteria oxidize H2S in a light-dependent manner to produce S2O32– (thiosulfate) rather than S±0, indicating that SQR is not the sulfide-oxidizing enzyme (de Wit and van Gemerden 1987; Rabenstein, Rethmeier and Fischer 1995); (ii) light-dependent H2S oxidation to thiosulfate in the cyanobacterium Microcoleus chthonoplastes was initially reported to be insensitive to DCMU (3-(3,4-dichlorophenyl)-1,1-dimethylurea)), an inhibitor of PSII-dependent quinone reduction (de Wit and van Gemerden 1987), but in a later study on the same organism, thiosulfate production was reported to be inhibited by DCMU (Rabenstein, Rethmeier and Fischer 1995); (iii) highly divergent copies of psbA, which encode products that lack the residues crucial for the binding of the Mn4CaO5 cluster in the oxygen-evolving complex (OEC), exist in many cyanobacterial genomes (Murray 2012). One of these was recently shown to perform a light-dependent oxidation in Chl biosynthesis to generate Chl f, and was hence renamed ChlF to reflect that function (Ho et al.2016). ChlF is a new type of homodimeric, DCMU-sensitive photo-oxidoreductase that uses light to perform a four-electron oxidation of Chl a or chlorophyllide a (Ho et al.2016; G. Shen and D. A. B., unpublished). In phylogenetic trees, ChlF is an early diverging member of the psbA superfamily that retains most of the functional features of the RC protein PsbA of PSII, except for the ability to bind the Mn4CaO5 cluster (Murray 2012; Cardona, Murray and Rutherford 2015; Ho et al.2016; Cardona et al.2017).
PSII itself is an unlikely candidate for H2S oxidation because it is highly specialized and not known to function with reductants other than H2O or H2O2. Furthermore, PSII activity is inhibited by sulfide, although some cyanobacterial strains show much greater tolerance to phototrophic sulfide inhibition than others (Miller and Bebout 2004). As depicted in Fig. 4, the main difference between our intermediate and Allen's HAP is that the new RC1-derived, sulfide-oxidizing module would allow cells to introduce electrons from H2S at different electron yields into a linear electron transport chain via two low-potential sites: qRC1d (H2S to S2O32–, Eo' = –193 mV, a six-electron reaction) and/or RC1 (H2S to S±0, Eo' = –270 mV, a two-electron reaction). This also constitutes a difference from the model of Klatt et al. (2015), who proposed a single site of sulfide entry to the electron transport chain via SQR. In a recent study of Geitlerinema sp. PCC 9228 (formerly Oscillatoria limnetica Solar Lake), Grim and Dick (2016) noted that several cyanobacteria, including Geitlerinema sp., possess an additional copy of SQR and that these are sometimes located in the genomic proximity of divergent psbA gene duplicates.
The benefit for a bacterium with the configuration we propose (i.e. two options for oxidizing the same substrate) is evident. The oxidation of H2S to SO32– via qRC1d would introduce six electrons per H2S into the electron transport chain (Brune 1989), whereas the oxidation of H2S to S0 via RC1 introduces only two electrons per H2S, providing a simple means via differential gene expression of RC1 and qRC1d to modulate the light-dependent reduction state of the quinone pool with respect to the physiological needs of the cell—either at constant light intensity and constant H2S concentrations, or in response to changing light and/or H2S availability. Obtaining six electrons from H2S for carbon reduction and ATP synthesis would obviously be advantageous over the SQR reaction in environments where H2S concentration decreased, or when more electrons were needed (for example, during N2 fixation). This aspect of our proposal (differential gene regulation to maintain redox balance in the bioenergetic membrane) is virtually identical to that of Allen (2005), but our substrates and gene expression regimens are different. Our dual sulfide-oxidizing intermediate (DSO) would be able to generate different quinone reduction states from the same low-potential substrate, H2S, by regulating RC gene expression, whereby the ultimate fate of the DSO and HAP (Allen 2005) hypothetical intermediates—further evolution resulting in water oxidation—is the same.
We offer no specific mechanistic proposal for how light-dependent H2S oxidation to sulfite or thiosulfate by protein X in Fig. 4 might occur—whether it entails one electron to two electron-transducing cofactors such as a flavin or quinone, or whether it might involve sulfur radicals. There are many thiyl radicals in biology (Buckel and Golding 2006), including thiyl to tyrosyl transfers as in the reaction mechanism of ribonucleotide reductase (Buckel and Golding 2012). However, the first step in H2S oxidation to sulfur or polysulfide is typically a two-electron reaction (Luther 2010; Luther et al.2011), suggesting that in both modern cyanobacteria and the phototrophic thiosulfate-generating reactions of ancient protocyanobacteria, X might catalyze a two-electron reaction. Nothing is yet known biochemically about how cyanobacteria oxidize sulfide to thiosulfate via sulfite as originally reported by de Wit and van Gemerden (1987) and confirmed by Rabenstein, Rethmeier and Fischer (1995). In particular, the sulfite/thiosulfate-generating enzyme(s) has not been identified. It is possible that thiosulfate results from a spontaneous or enzymatic reaction of a sulfite intermediate with the substrate, sulfide. Protein-bound trisulfides have recently been characterized as intermediates in bacterial sulfite reduction (Santos et al.2015). In Fig. 4, we designate the so far uncharacterized sulfite-producing enzyme as X and the corresponding intermediate carrier, probably a cytochrome, as Cyt x.
Donors more redox positive than sulfide
Further modification of the periplasmic, oxidizing side of qRC1d to accommodate oxidation of other substrates, such as soluble Mn2+, would have given rise to novel and different anoxygenic phototrophic activities as possible intermediates en route to water oxidation (Dismukes et al.2001; Allen and Martin 2007; Johnson et al.2013; Fischer, Hemp and Johnson 2016). Whether protocyanobacteria could have oxidized soluble Fe2+ has not been discussed. In light of the single isolated report of photoferrotrophic cyanobacterial growth (Cohen 1984), and given 2.4 billion years of global O2 and low concentrations of Fe2+, it is possible that cyanobacteria have lost this trait altogether, that they never had it or that it has not been studied in enough detail. However, with oxygenic photosynthesis in place and energy to burn, so to speak, the transition from rTCA-based autotrophy to the energetically expensive and inefficient CBB cycle became 'affordable’.
The origin of the OEC marks two important transitions. First, it would mark the end of reductant limitation for marine primary production (under the premise that photothiotrophy was quantitatively more significant than photoferrotrophy during the Archean). Second, it marks the endpoint of the evolutionary inference: an oxygenic photosynthetic bacterium possessing two photosystems linked in series, one capable of water oxidation (PSII) feeding electrons into a linear chain to RC1 (PSI) that generates reduced Fd (Fig. 4, top panel). In such an organism, the use of H2S as reductant becomes almost irrelevant, useful only under specialized environmental conditions, but essential under conditions of high H2S for reasons of PSII inhibition. Occasional essentiality (i.e. a conditionally advantageous capability) would explain why the ancestral trait (two kinds of sulfide oxidation differing in their electron yield) may have been retained in some modern generalist cyanobacterial lineages. By physiological definition (having PSI and PSII, and producing O2), such an organism would be a cyanobacterium (or a quasi-cyanobacterium), but it is not essential here whether the organism would branch basal to or within modern, chlorophototrophic cyanobacteria (sometimes also called Oxyphotobacteria), nor is it crucial when the bacterium evolved water oxidation capability, provided that it occurred prior to the GOE ca. 2.4 billion years ago and—a condition imposed by our present inference—provided that it occurred after the origin of photothiotrophy in the cyanobacterial lineage. The timing, ecological consequences and some possible geochemical consequences of a DSO are briefly considered in the next section.
PHOTOTHIOTROPHY: UNDERESTIMATED IN EARTH HISTORY
When we turn to the literature on the role of photosynthesis in Earth's geochemical history, three things stand out: BIFs (banded-iron formations), MIFs (mass-independent sulfur fractionations) and, with rare exceptions (for example, Allen 2005), the paucity of attention given to the possibility that photothiotrophy played a major role in Earth's geochemical history prior to the GOE. Johnston et al. (2009) proposed an important role for cyanobacterial photothiotrophy, but long after the GOE. Recalling that all modern phyla harboring chlorophotoautotrophs have members that oxidize sulfide (Table 1), and that Allen (2016) recently proposed an origin of Archean stromatolites involving the growth of protocyanobacteria that oxidized H2S at RC1—a suggestion that is fully compatible with our present considerations—our thoughts on the possible role of photothiotrophy in Earth's history, summarized in Fig. 5, are as follows.
Figure 5.
The possible role of photothiotrophy in Earth's history. (A) Redox potentials of reductants for primary production vs time. Midpoint potentials are taken from sources listed in the legend of Fig. 1. GOE: great oxidation event. MIFs: mass-independent sulfur fractionations (a proxy for O2 absence or presence under 10−5 present atmospheric levels in the atmosphere). Mass-dependent sulfur isotope effects are also pronounced prior to the GOE (not shown, see the text). Whiffs indicate the presence of oxidants in the water column or slight O2 presence prior to the GOE. BIFs: banded iron formations. The column ‘primary production’ at right underscores the point that before the origin of chlorophototrophy, primary production was H2-dependent and mechanistically dependent on flavin-based electron bifurcation (indicated by gray shading). The figure suggests that photothiotrophy preceded photoferrotrophy and water splitting in evolution. (B) A possible photothiotrophic sulfur cycle in Archean oceans before the GOE. See the text.
The salient Earth history observations as taken from three recent reviews (Arndt and Nisbet 2012; Lyons, Reinhard and Planavsky 2014; Fischer, Hemp and Johnson 2016) are fairly straightforward and consistent. Prior to the GOE 2.4 billion (Ga) years ago, there was basically no O2 in the atmosphere, as indicated by the presence of MIF. The MIFs disappear simultaneously with changes in other geochemical markers for the presence of oxygen at 2.4 Ga, indicating the presence of a strong oxidant, almost certainly O2, in the atmosphere. The BIFs are abundant before the GOE and rare after the GOE. The source of the BIFs is unresolved.
Photoferrotrophy has been widely discussed as a possible biological mechanism to generate BIFs (Camacho et al.2017), but the numbers observed for the activity of modern photoferrotrophs do not add up in a way that provides a compelling case for their role in BIF formation (Crowe et al.2014). Lyons, Reinhard and Planavsky (2014) went a step further and suggested that H2O was the source of electrons for primary production prior to the GOE, while Fischer, Hemp and Johnson (2016) pointed out that anoxygenic photosynthesis with inorganic electron donors other than H2O, and not excluding reduced sulfur species, could account for the same observations (organic-rich shales, high primary production) prior to the GOE.
We suggest the possibility of a quantitatively significant role for photothiotrophy in evolution long before the GOE. Our proposal could be integrated into an Earth history context as follows (see Fig. 5a). H2S-based photosynthesis is the ancestral form of anoxygenic photosynthesis, and hence would be ancient, consistent with earlier proposals (Allen 2005). At least some members of all modern photosynthetic phyla (i.e. those that fix CO2) are capable of using sulfide as an electron donor: Cyanobacteria, Chloroflexi, Proteobacteria and Chlorobi (Table 1). H2S would be the most likely source of high primary production pre-GOE. The duplication of RC1, leading to RC2, produced a highly specialized bacterium with a high affinity for H2S and other reduced sulfur species in ocean waters where light was not limiting. The respiratory action of heterotrophic sulfur, sulfite and sulfate reducers converted oxidized sulfur to the reduced state (H2S), but high light intensity and dense protocyanobacterial growth kept H2S concentrations in the photic zone low, and maintained sulfur in the oxidized state (Fig. 4b). This assumes that neither light nor CO2 was limiting for primary production, but that once again in Earth history the reductant for primary production—this time H2S instead of H2—was limiting. Oxidized S species from photothioautotrophic growth were reduced to H2S by sulfate reducers, but at a rate slower than photooxidation. S was cycled like an environmental cofactor, without H2S being removed from the photic zone as pyrite, FeS2, because H2S was oxidized by a light-driven system faster than it was produced by sulfate reducers (Fig. 5B).
Sulfur limitation (arising from variations in its environmental availability) in an Archean photothiotrophic ocean could have generated mass-dependent fractionation of sulfur isotopes prior to the GOE, which is observed (Johnston 2011). With the advent of water oxidation, MIFs would end for the conventional reason: because of the accumulation of atmospheric O2. With the replacement of H2S with H2O as the main reductant for primary production, S would no longer have been limiting for primary production, which should have influenced isotope fractionation effects for S species around the time of the GOE. Moreover, with the origin of water oxidation at the OEC, reductant would no longer have been limiting for primary production. That would have caused a substantial shift in ecosystem function and elemental cycling.
In the geochemical literature, photothiotrophy is sometimes considered to be a process that evolved late, after the origin of oxygenic photosynthesis (Johnston et al.2009; Johnston 2011) or that was quantitatively of insufficient significance to have produced geological effects (Lyons, Reinhard and Planavsky 2014). Alternatively, it is just not on the map at all, with the effects of MIFs and non-biological processes standing in the foreground (Ueno 2014). Other views mention H2S (Klatt et al. 2015; Fischer, Hemp and Johnson 2016) but devote little attention to the role of H2S as an electron donor in the evolution of photosynthesis. Like Allen (2005, 2016), we see an important role for photothiotrophy in the evolution of photosynthesis and in Earth's history.
Direct evidence for the workings of anoxygenic photosynthesis prior to the GOE is scarce. As Arndt and Nisbet (2012) point out, Frances Westall has reported the characterization of a 3.3 billion-year-old microbial biofilm from an air-exposed setting that was probably performing anoxygenic photosynthesis and may have contained sulfate reducers (Westall et al.2006, 2011). What kind of photosynthesis was supporting that biofilm? The simplest interpretation is that it was anoxygenic photosynthesis (Arndt and Nisbet 2012), as opposed to oxygenic photosynthesis (Lyons, Reinhard and Planavsky 2014), and a likely possibility is that it was photothiotrophy. Of the electron donors used by chlorophototrophs, H2S is the most common (Table 1), and it might also be the most ancient. The experiments on photothiotrophs showing relatively small S isotope fractionation effects in laboratory studies have not yet been applied to photothiotrophic cyanobacteria (Johnston 2011). In particular, it will be important to determine whether there is a significant isotopic signature difference between cyanobacteria that oxidize sulfide to sulfur (or polysulfide) and those that produce thiosulfate (see above).
Whiffs of oxidant
An Archean photothiotrophic ocean would have been stratified in terms of redox chemistry, not due to an absence of mixing but rather due to the bacterial utilization of sunlight. In the geochemical literature, there are phenomena called ‘whiffs’ (Anbar et al.2007; Knoll, Bergmann and Strauss 2016). Whiffs designate rock formations that were deposited before the GOE and that harbor evidence for the existence of oxidants in the marine water column (Lyons, Reinhard and Planavsky 2014; Fischer, Hemp and Johnson 2016). The causes of whiffs are debated. Some authors interpret them by as evidence for the existence of O2 pre-GOE, but at face value they simply indicate the existence of oxidants in the water column. If the Archean oceans were inhabited by bacteria that could perform light-dependent oxidation of sulfide to produce thiosulfate, such activities could be interpreted as evidence for a strong oxidant. Photooxidized Chls are indeed strong oxidants. In PSII, the oxidized special pair of Chls oxidizes tyrosine Z (TyrZ), generating a tyrosyl radical, which is also a very strong oxidant. TyrZ resides close enough to the Mn4CaO5 water-splitting, OEC to oxidize it (Umena et al.2011; Shen 2015; Barber 2016).
Prior to the origin of the OEC, the strong oxidant underpinning ‘whiffs’ prior to the GOE could have been a cellular protein-bound oxidant, a Chl cation (P+) or a TyrZ-like radical, but not O2 itself. In other words, ‘whiffs’ of oxygen prior to the GOE might reflect the existence of a prevalent biological oxidant (RC2) in the photic zone that possessed the midpoint potential to oxidize an OEC, and hence was capable of oxidizing dissolved metals such as Mn2+ (Dismukes et al.2001; Allen and Martin 2007; Johnson et al.2013; Fischer, Hemp and Johnson 2016), and possibly Fe2+, but operated before the OEC had fully evolved. Again, we do not call into question the existence of photoferrotrophy in BIF formation (Widdel et al.1993; Bird, Bonnefoy and Newman 2011; Camacho et al.2017) prior to the GOE, or the role of photosynthetic Mn+2 oxidation prior to the GOE (Johnson et al.2013). However, the question of which organisms might have been responsible is open, and we note that the strong oxidant in RC2 of a photothiotroph bearing two types of RCs (a proto-cyanobacterium) could have oxidized other substrates: Mn+2 for example, as does an engineered RC2 of R. sphaeroides (Allen et al.2012), and conceivably Fe2+. The observation that cyanobacteria oxidize Mn2+ in the process of OEC assembly is also relevant in this context (Tamura and Cheniae 1987).
Today, marine primary production is not limited by reductant but nutrients such as iron are limiting. Concerning geochemical models of Earth history, if photoferrotrophy was the predominant form of primary production prior to the GOE, reductant (Fe2+) would not have been limiting (although phototrophic biomass might have been), whereas if photothiotrophy was the predominant form of primary production prior to the GOE, reductant (H2S) would probably have been limiting until the origin of water splitting. Fitting of geochemical data points to mathematical models (Johnston et al.2009; Lyons, Reinhard and Planavsky 2014) together with measurements of cyanobacterial S isotope discrimination during photothiotrophic growth might help to discern between these alternatives.
PHYLOGENETIC DISTRIBUTION OF RCs
Finally, we return to the question of how RC1 and RC2 came to be distributed among modern Chl-producing bacteria. No modern treatment of RC evolution can realistically avoid the problem of horizontal gene transfer; thus, the questions descend to how much and in which direction. An important thing to keep in mind about RC evolution is not which lineages have RCs, but which lineages lack them. Sousa et al. (2013) provided one overview of that pattern, but the best rendering of phylogenetic photosystem distribution published to date (to our knowledge) is that in fig. 5 of Fischer, Hemp and Johnson (2016), to which the reader is referred rather than duplicating it here. Our proposal for the distribution of RCs among modern chlorophototrophic lineages has precedent based on bioinformatic surveys (Mulkidjanian et al.2006), yet differs from previous views in physiological context (photothiotrophy) and in implications: we suggest that RC1 and RC2 arose within the lineage leading to cyanobacteria (one can generically designate members of this lineage as protocyanobacteria), and that those RCs have been vertically inherited in that lineage since the advent of Chl biosynthesis. This suggestion carries a few corollaries, some novel, some not.
Origin of chlorophototrophy in cyanobacteria
As the first corollary, it would mean that the origin of chlorophototrophy occurred in cyanobacteria and after the separation of the ancestral protocyanobacterial lineage from other bacteria. Here, critics might point to new phylogenies based on ribosomal proteins that were interpreted as evidence for a 'merger hypothesis', that is, that two RCs were transferred into a non-photosynthetic cyanobacterial ancestor to give rise to the lineage (Mathis 1990; Xiong, Inoue and Bauer 1998; Shih et al.2017; Blankenship 2017; Soo et al.2017).
Looking at the issue openly, both before and after the publication by Soo et al. (2017), there are no bacterial lineages known that branch immediately ancestral to cyanobacteria and that possess only one RC or some other 'primitive' form of photosynthesis. In our view, designating new lineages such as Melainabacteria and Sericytochromatia and referring to them as ‘cyanobacteria’, is presently unsubstantiated. By their own analysis, Soo et al. (2017) concluded that extensive gene loss later occurred in respiring ancestors of gut-residing fermentative melainabacteria, and there is no evidence that loss of photosynthetic capability did not occur in the protocyanobacterial ancestors of these organisms prior to or at the time of the GOE. Hence, it is not evident how the tree of Soo et al. (2017), even if its depiction of ribosome lineage relationships holds up over the coming years, directly bears upon the origin and evolution of either Chl or RCs, both of which certainly evolve independently of the ribosome because of horizontal gene transfer. Soo et al. (2017) investigated the evolution of the ribosome as a proxy for evolution of the rest of the genome, whereas we are primarily concerned with the origin of the segments of the genome that specify Chl, RCs and autotrophy. The evolution of Chl and RCs is inextricably linked because RCs cannot undergo sustained evolution in the absence of Chl. In contrast, there is no functional linkage between the evolution of ribosomes on the one hand and Chl and RCs on the other. The evolution of ribosomes and chlorophototrophy need not coincide.
Chlorophototrophy did evolve somehow and somewhere, and we suggest that it occurred in the stem lineage of cyanobacteria. Furthermore, as Sousa et al. (2013) have pointed out, and as we stress again here, the 'merger' hypothesis for the presence of two RC types in cyanobacteria (Blankenship 2017; Soo et al.2017) is unlikely to be correct in any case because it predicts the existence of a deep and ancient dichotomy in Chl biosysnthetic pathways leading to the common precursor, chlorophyllide a. Such a dichotomy is not observed among chlorophototrophic lineages; Chl biosynthesis entails one ancestral pathway, combined in some genomes with the evolutionarily recent O2-dependent AcsF alternative to the radical SAM-dependent ring closure reaction catalyzed by BchE (Sousa et al.2013; Chen, Canniffe and Hunter 2017) (Fig. 3).
Note that nowhere in this discussion have we suggested a timeline for the origin of Chl. We simply suggest that it may have arisen from Zn-PPIX (Figs 2 and 3), that its origin was required for primary production that was not tied to geochemical H2 at hydrothermal vents, and that it arose before the GOE. In line with this, Bryant and Liu (2013) pointed out that trees based on (B)Chl biosynthesis suggest the divergence of three lineages that are nearly equally deep—Chlorobi-Chloroflexi, Heliobacteriaceae-Cyanobacteria and Acidobacteria-Proteobacteria—each of which contain members with either or both a type 1 and a type 2 RC. One could argue that the duplication leading to heterodimeric PSII, which occurred independently from that leading to PufLM-type 2 RCs, also occurred independently from that of a common RC1-type ancestor, in which (B)Chl biosynthesis already existed.
Chl a first because it arose that way
As the second corollary, an origin of chlorophototrophy in a cyanobacterial progenitor would mean that Chl a, which is prevalent in cyanobacteria and is the product of the shortest Chl biosynthetic route (Gomez Maqueo Chew and Bryant 2007a), is also the ancestral Chl, which fits exactly with the Granick hypothesis (Granick 1965) that the evolution of (at least Chl biosynthesis) pathways may recapitulate evolution.
Cyanobacteria are ancient (but not primordial)
Third, it would mean that the cyanobacteria descend from the most ancient Chl-bearing lineage, without placing a date on their age. This would mean in turn that cyanobacteria are a very ancient line whose origin greatly precedes the GOE, as some interpretations of the fossil record would suggest (Golubic and Lee 1999; Schopf 2012; Knoll, Bergmann and Strauss 2016). This is also very close to what Olson and Pierson (1987) suggested almost 30 years ago, although they had the Heliobacteriaceae (Firmicutes) lineage branching off before the duplication that gave rise to RCII. Twenty-nine years later, the 'lone ranger' status of Heliobacterium and related organisms as the sole chlorophototrophs in the clostridial lineage (Fischer, Hemp and Johnson 2016) might be better explained as horizontal acquisition instead of differential loss among all other Firmicutes. This would also fit with the absence of autotrophy in Heliobacteriaceae (Table 1), and would not contradict the ancestral features preserved in their RC structure (Gisriel et al.2017), provided that the transfer was ancient. From the first sections of this paper, it should be clear that we are not saying that the cyanobacterial lineage represents a primordial form of bacteria, we are simply suggesting that it represents the first lineage with chlorophototrophy.
The export hypothesis
Fourth, because none of the anoxygenic photosynthetic lineages branch from within the cyanobacteria, it would mean that all anoxygenic chlorophotosynthetic lineages acquired their RCs via horizontal gene transfer ultimately from a cyanobacterial progenitor, although sometimes the transfers might have occurred through intermediate lineages as in the case of Gemmatimonas phototrophica (Zeng et al.2014). This proposal, which we call the 'export' hypothesis (Fig. 6), is precisely the converse of the 'merger' hypothesis (Blankenship 2017); hence, it might not sit well with some proponents of the merger hypothesis, but it would explain a lot. It would directly explain why Chl synthesis undergoes variation and modification emanating from chlorophyllide a (requiring only esterification to produce Chl a) in anoxygenic photosynthetic lineages. It would also directly explain why RCs are found in combination with different CO2 fixation pathways: namely, via RC and (B)Chl operon transfer without cotransfer of the cbb (CBB cycle) operon; as well as RC + Chl transfer with cotransfer of the CBB cycle operon, but involving variation in the latter by replacement of aldolase, RubisCO, bisphosphatases, phosphoribulokinase and GAPDH types (Table 1).
Figure 6.
Export hypothesis for the distribution of RCs among oxygenic and anoxygenic chlorophototrophs. Ribosomal lineages are indicated with gray lines; transfer of RCs into recipient lineages are indicated with green arrows. OEC: oxygen evolving complex. The origin of plastids (Sanchez-Barraclado et al.2017), where cyanobacteria donated both RC1 and RC2 plus the genes for autotrophy to eukaryotes (Ku et al.2015), is symbolically indicated as a wide arrow branching within the cyanobacterial radiation. The figure makes no statement about the timing of export transfer events relative to the origin of the OEC (probably at or near the time of the GOE) or relative to other transfers. Assuming that a cyanobacterial progenitor invented chlorophototrophy, and that geochemical evidence for ancient phototrophy at around 3.2 to 3.4 Ga before present represents photothiotrophy, then photothiotrophy would trace to that age. The photothiotrophic cyanobacterial lineage existent prior to the origin of the OEC corresponds to protocyanobacteria in our terminology here. Note that we indicate no ribosomal lineage branching patterns at all (except plastids), including Melainabacteria or Sericytochromatia relative to cyanobacteria, because ancient lineage relationships and branching patterns inevitably change as new phylogenetic methods are employed and as new lineages become known (Williams et al.2013). Symbols for hydrothermal light at photosynthesis and RC1 origin and sunlight at RC2 origin are indicated. See the text.
Of course, RC transfer without Chl biosynthesis transfer cannot result in fixation of chlorophototrophic physiology, whereas RC + Chl transfer can give rise to photoheterotrophic lineages. It would also directly explain why so few bacterial lineages, among the countless lineages in nature, are chlorophototrophic, and those that are tend to occur as recent (tip) lineages in phylogenetic trees, rather than branching near the root in their respective groups (see fig. 5 of Fischer, Hemp and Johnson 2016). Acquisition of traits associated with oxygen utilization (a couple of Chl biosynthesis enzymes, terminal oxidases, iron oxidation) in some anoxygenic photosynthetic lineages can be attributed to horizontal gene transfers that occurred post-GOE (Table 1). Shih, Ward and Fischer (2017) suggested that Chloroflexus might have acquired its phototrophic ability via horizontal gene transfer, compatible with earlier transfer proposals (Mulkidijanian et al.2006) and with our present proposal, which, unlike these prior proposals, is not based on phylogenetic trees. Instead our proposal takes into account Roseobacter plasmids that carry phototrophic genes (Petersen et al.2012), the extremely sparse distribution of RCs across distantly related bacterial lineages (Sousa et al.2013; Fischer, Hemp and Johnson 2016), and the longstanding observation that chlorophototrophy occurs in combination with different CO2 fixation pathways—and sometimes in obligately aerobic heterotrophs (Table 1). Rather than having been imported into cyanobacteria, the export hypothesis has it that functional RCs were distributed among lineages as a result of modular export from the architects of chlorophototrophy—the cyanobacterial lineage.
Possible reductive RC evolution
Finally, were the origin of RCs in all current anoxygenic photosynthetic lineages due to horizontal gene transfer, it would also directly explain why loss of chlorophototrophy is rarely observed among the free-living bacterial lineages in which it has taken hold. Absence of chlorophototrophy among most but not all bacterial (and archaeal) lineages is not the result of rampant loss, it is the result of not having acquired the trait. As a case in point, if there are free-living secondarily non-photosynthetic cyanobacterial lineages in nature, they are likely to be very rare, if they exist at all, based on metagenomic analyses published so far. This evidently is because once the GOE event occurred—the greatest ecological niche expansion in Earth's history—and organisms evolved into committed chlorophototrophic or chemoheterotrophic lifestyles, loss of chlorophototrophy would mean that any newly created, non-phototrophic mutant would have to displace a more highly evolved heterotroph from an existing ecological niche, an improbable event. For example, the conditions that would provide a selective advantage for a cyanobacterium that had lost the capacity for photoautotrophic growth would be highly unusual and very rare; indeed, only two closely related examples have been identified to date. Zehr and coworkers have described two variants of ‘Candidatus Atelocyanobacterium thalassa’, which are photoheterotrophic cyanobacterial symbiont partners of prymnesiophyte algae that have lost PSII and RuBisCO, but which have retained nitrogenase and PSI (Thompson et al.2012; Bombar et al.2014). These are examples of reductive evolution of photosynthesis within the cyanobacterial lineage.
How far can reductive evolution of photosynthesis go? The traditional interpretation of the homodimeric RCs of GSB and heliobacteria is that they reflect an ancient state of RCs. If they are the result of export from cyanobacteria, they might well be relicts of a very ancient transfer (Gisriel et al.2017), and in that sense molecular fossils from the early evolution of chlorophototrophy. This seems more likely than the alternative scenario that the heliobacterial RC arose by reductive evolution from a cyanobacterial PSI. We note in passing, however, that the concept of reductive evolution, a recurrent theme in molecular and genomic evolution (Ku et al.2015; Albalat and Cañestro 2016), is not widely discussed in the context of the evolution of chlorophototrophy. Nevertheless, several examples exist, primarily among the metabolically flexible Proteobacteria (for example, Zheng et al.2012; Keppen et al.2013; Koblížek et al.2013; Kopetjka et al.2017).
CONCLUSIONS
From comparative physiology, we arrive at a list of main logical inferences about photosynthesis evolution as follows, all to be understood as conditional sentences, but written as declarative statements for readability. Life started off chemolithoautotrophically, with primary production at geological sources of H2 (hydrothermal vents). Flavin-based electron bifurcation was used by the first autotrophs for the reduction of low-potential Fd to produce Fdred, the essential electron donor in anaerobic pathways of CO2 fixation. Chl biosynthesis was the decisive invention for the origin of chlorophototrophy, which likely occurred in an anaerobe that had cobalamin, heme, cytochromes and quinones, and probably was capable of chemoheterotrophic growth. Chlorophototrophy evolved under low intensity hydrothermal-light conditions, in the midst of chemolithoautotrophic primary production, and where the photobiologically damaging effects of high-light flux and ultraviolet radiation were absent.
The first photochemically active pigments were possibly Zn-tetrapyrroles such as Zn-PPIX, the excited triplet of which (3Zn-PPIX) is strongly reducing (–1600 mV) with a long half-life (7–15 ms). The function (and physiological advantage) of the first chlorophototrophic electron transfer chain was Fd reduction via FeS clusters in RC1 with electrons from H2S, as occurs in modern-day GSB (for example, Chlorobium sp.). This freed primary production from H2 exhalation at vents. Photothioautotrophy, first with one RC then with two, was the physiological and evolutionary bridge between H2-dependent chemolithoautotrophy involving flavin-based electron biufurcation and chlorophototrophic oxidation of metal ions, both soluble and in the OEC. Photothiotrophy was a globally widespread (and the predominant) source of primary production in the photic zone of Archean oceans and was a trait of protocyanobacteria, with sulfate (sulfite) reducers regenerating H2S from photooxidized products (sulfur, sulfite and thiosulfate), giving rise to fluctuating mass-dependent sulfur isotope fractionation prior to the GOE. Whiffs of oxygen prior to the GOE reflect the existence of a strong oxidant in the oceans, which we suggest stems from an RC2 ancestor prior to the evolution of water oxidation.
Modern cyanobacterial photothiotrophy involves two different kinds of H2S oxidation that would permit, via gene regulation, and physiological modulation of the quinone pool redox state in the photosynthetic membrane: a two-electron reaction (to form S0) involving SQR and RCI, and a six-electron reaction (to form SO32–/S2O32–). Cyanobacterial ChlF is a relict from gene duplication that possibly gave rise to an ancestral homodimeric RC2. Photosynthesis arose in a cyanobacterial progenitor, and Chl a is the ancestral Chl. All anoxygenic chlorophotosynthetic lineages characterized so far are the result of acquisition via horizontal gene transfer of one RC and Chl biosynthesis, accompanied or not by a CO2 fixation pathway, via gene export from the cyanobacterial lineage, in some cases through intermediate lineages. Physiology speaks to the direction of those transfers but not to their geological timing. Prior to the origin of the OEC, reductants for CO2 fixation (H2 in cells without Chl and H2S in chlorophototrophs) were probably limiting for marine primary production.
Acknowledgements
We thank George Luther III, John F. Allen, Dianne Newman, Filipa Sousa, Christiane Dahl, Ines Periera, and Carlo Kleinermanns for discussions on aspects of the paper. We thank Verena Zimorski for help in preparing the manuscript. WFM thanks the ERC (666053 emicrobEvol) and the Volkswagen Foundation (93 046) for funding. DAB thanks the U. S. Dept. of Energy (DE-FG02-94ER20137), NASA Exobiology (NNX16AJ62G) and National Science Foundation (MCB-1613022) for support. JTB thanks Genome British Columbia (SOF153) and the Canadian Natural Sciences and Engineering Research Council (Discovery Grant 2796) for funding.
Conflict of interest. None declared.
REFERENCES
- Albalat R, Cañestro C. Evolution by gene loss. Nat Rev Genet 2016;17:379–91. [DOI] [PubMed] [Google Scholar]
- Allen JF. A redox switch hypothesis for the origin of two light reactions in photosynthesis. FEBS Lett 2005;579:963–8. [DOI] [PubMed] [Google Scholar]
- Allen JF. A proposal for formation of archaean stromatolites before the advent of oxygenic photosynthesis. Front Microbiol 2016;7:1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen JF, Martin W. Evolutionary biology: out of thin air. Nature 2007;445:610–2. [DOI] [PubMed] [Google Scholar]
- Allen JP, Olson TL, Oyala P et al. . Light-driven oxygen production from superoxide by Mn-binding bacterial reaction centers. P Natl Acad Sci USA 2012;109:2314–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anbar AD, Duan Y, Lyons TW et al. . A whiff of oxygen before the great oxidation event? Science 2007;317:1903–6. [DOI] [PubMed] [Google Scholar]
- Aono R, Sato T, Yano A et al. . Enzymatic characterization of AMP phosphorylase and ribose-1,5-bisphosphate isomerase functioning in an archaeal AMP metabolic pathway. J Bacteriol 2012;194:6847–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arndt NT, Nisbet EG. Processes on the young Earth and the habitats of early life. Annu Rev Earth Pl Sc 2012;40:521–49. [Google Scholar]
- Ashida H, Danchin A, Yokota A. Was photosynthetic RuBisCO recruited by acquisitive evolution from RuBisCO-like proteins involved in sulfur metabolism? Res Microbiol 2005;156:611–8. [DOI] [PubMed] [Google Scholar]
- Ashida H, Saito Y, Kojima C et al. . A functional link between RuBisCO-like protein of Bacillus and photosynthetic RuBisCO. Science 2003;302:286–90. [DOI] [PubMed] [Google Scholar]
- Bach W, Paulick H, Garrido CJ et al. . Unraveling the sequence of serpentinization reactions: petrography, mineral chemistry, and petrophysics of serpentinites from MAR 15°N (ODP Leg 209, Site 1274). Geophys Res Lett 2006;33:L13306. [Google Scholar]
- Barber J. Mn4Ca cluster of photosynthetic oxygen-evolving center: structure, function and evolution. Biochemistry 2016;55:5901–6. [DOI] [PubMed] [Google Scholar]
- Barco RA, Emerson D, Sylvan JB et al. . New insight into microbial iron oxidation as revealed by the proteomic profile of an obligate iron-oxidizing chemolithoautotroph. Appl Environ Microb 2015;81:5927–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer CE, Bird TH. Regulatory circuits controlling photosynthesis gene expression. Cell 1996;85:5–8. [DOI] [PubMed] [Google Scholar]
- Baymann F, Brugna M, Mühlenhoff U et al. . Daddy, where did (PS)I come from? Biochim Biophys Acta 2001;1507:291–310. [DOI] [PubMed] [Google Scholar]
- Beatty JT, Gest H. Biosynthetic and bioenergetic functions of citric acid cycle reactions in Rhodopseudomonas capsulata. J Bacteriol 1981;148:584–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beatty JT, Overmann J, Lince MT et al. . An obligately photosynthetic bacterial anaerobe from a deep-sea hydrothermal vent. P Natl Acad Sci USA 2005;102:9306–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker EM, Westermann S, Hansson M et al. . Parallel enzymatic and non-enzymatic formation of zinc protoporphyrin IX in pork. Food Chem 2012;130:832–40. [Google Scholar]
- Berg IA. Ecological aspects of the distribution of different autotrophic CO2 fixation pathways. Appl Environ Microb 2011;77:1925–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biegel E, Schmidt S, Gonzalez JM et al. . Biochemistry, evolution and physiological function of the Rnf complex, a novel ion-motive electron transport complex in prokaryotes. Cell Mol Life Sci 2011;68:613–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird LJ, Bonnefoy V, Newman DK. Bioenergetic challenges of microbial iron metabolisms. Trends Microbiol 2011;19:330–40. [DOI] [PubMed] [Google Scholar]
- Björn LO, Govindjee. The evolution of photosynthesis and chloroplasts. Curr Sci 2009;96:1466–74. [Google Scholar]
- Björn LO, Papageorgiou GC, Blankenship RE et al. . Why Chlorophyll a? Photosynth Res 2009;99:85–98. [DOI] [PubMed] [Google Scholar]
- Blankenship RE. Early evolution of photosynthesis. Plant Physiol 2010;154:434–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blankenship RE. How cyanobacteria went green. Science 2017;355:1372–3. [DOI] [PubMed] [Google Scholar]
- Bombar D, Heller P, Sanchez-Baracaldo P et al. . Comparative genomics reveals surprising divergence of two closely related strains of uncultured UCYN-A cyanobacteria. ISME J 2014;8:2530–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bose A, Newman DK. Regulation of the phototrophic iron oxidation (pio) genes in Rhodopseudomonas palustris TIE-1 is mediated by the global regulator, FixK. Mol Microbiol 2011;79:63–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bräsen C, Esser D, Rauch B et al. . Carbohydrate metabolism in archaea: current insights into unusual enzymes and pathways and their regulation. Microbiol Mol Biol R 2014;78:89–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridger SL, Clarkson SM, Stirrett K et al. . Deletion strains reveal metabolic roles for key elemental sulfur-responsive proteins in Pyrococcus furiosus. J Bacteriol 2011;193:6498–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bröcker MJ, Schomburg S, Heinz DW et al. . Crystal structure of the nitrogenase-like dark operative protochlorophyllide oxidoreductase catalytic complex (ChlN/ChlB)2. J Biol Chem 2010;285:27336–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broderick JB, Duffus BR, Duschene KS et al. . Radical S-adenosylmethionine enzymes. Chem Rev 2014;114:4229–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown II, Bryant DA, Casamatta D et al. . Polyphasic characterization of a thermotolerant, siderophilic filamentous cyanobacterium that produces intracellular and extracellular iron deposits. Appl Environ Microb 2010;76:6664–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brune DC. Sulfur oxidation by phototrophic bacteria. Biochim Biophys Acta 1989;975:189–221. [DOI] [PubMed] [Google Scholar]
- Bryant DA, Costas AM, Maresca JA et al. . Candidatus Chloracidobacterium thermophilum: an aerobic phototrophic acidobacterium. Science 2007;317:523–6. [DOI] [PubMed] [Google Scholar]
- Bryant DA, Liu Z. Green bacteria: insights into green bacterial evolution through genomic analyses. Adv Bot Res 2013;66:99–150. [Google Scholar]
- Bryant DA, Liu Z, Li T et al. . Comparartive and functional genomics of anoxygenic green bacteria from the taxa Chlorobi, Chloroflexi, and Acidobacteria. In: Burnap R, Vermass W (eds.). Functional Genomics and Evolution of Photosynthetic Systems. Dodrecht, Heidelberg, London, New York: Springer, 2012, 47–102. [Google Scholar]
- Buckel W, Golding BT. Radical enzymes in anaerobes. Annu Rev Microbiol 2006;60:27–49. [DOI] [PubMed] [Google Scholar]
- Buckel W, Golding BT. Radical enzymes. In: Chatgilialoglu C, Studer A (eds). Encyclopedia of Radicals in Chemistry, Biology and Materials. Chicester, West Sussex, Hoboken, NJ: John Wiley & Sons, Ltd., 2012, 1501–46. [Google Scholar]
- Buckel W, Thauer RK. Energy conservation via electron bifurcating ferredoxin reductase ans proton/NA+ translocating ferredoxin oxidation. Biochim Biophys Acta 2013;1827:94–113. [DOI] [PubMed] [Google Scholar]
- Budinoff CR, Hollibaugh JT. Arsenite-dependent photoautotrophy by an Ectothiorhodospira-dominated consortium. ISME J 2008;2:340–3. [DOI] [PubMed] [Google Scholar]
- Camacho A, Walter XA, Picazo A et al. . Photoferrotrophy: remains of an ancient photosynthesis in modern environments. Front Microbiol 2017;8:323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell BJ, Cary SC. Abundance of reverse tricarboxylic acid cycle genes in free-living microorganisms at deep-sea hydrothermal vents. Appl Environ Microb 2004;70:6282–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardona T, Murray JW, Rutherford AW. Origin and evolution of water oxidation before the last common ancestor of the Cyanobacteria. Mol Biol Evol 2015;32:1310–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardona T, Sanchez-Baracaldo P, Rutherford AW et al. . Molecular evidence for the early evolution of photosynthetic water oxidation. BioRxiv 2017, DOI: 10.1101/109447. [DOI] [Google Scholar]
- Castresana J, Lübben M, Saraste M et al. . Evolution of cytochrome oxidase, an enzyme older than atmospheric oxygen. EMBO J 1994;13:2516–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapelle FH, O’Neill K, Bradley PM et al. . A hydrogen-based subsurface microbial community dominated by methanogens. Nature 2002;415:312–5. [DOI] [PubMed] [Google Scholar]
- Chau TT, Ishigaki M, Kataoka T et al. . Ferrochelatase catalyzes the formation of Zn-protoporphyrin IX in dry-cured ham via the conversion reaction from heme. J Agr Food Chem 2011;59:12238–45. [DOI] [PubMed] [Google Scholar]
- Chen GE, Canniffe DP, Hunter CN. Three classes of oxygen-dependent cyclase involved in chlorophyll and bacteriochlorophyll biosynthesis. P Natl Acad Sci USA 2017;114:6280–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chisholm SW, Olson RJ, Zettler ER et al. . A novel free-living prochlorophyte abundant in the oceanic euphotic zone. Nature 1988;334:340–3. [Google Scholar]
- Cohen Y. Comparative N and S cycles. In: Klug MJ, Reddy CA (eds). Current Perspectives in Microbial Ecology. Washington, DC: American Society for Microbiology, 1984, 436–41. [Google Scholar]
- Cohen-Ofri I, van Gastel M, Grzyb J et al. . Zinc-bacteriochlorophyllide dimers in de novo designed four-helix bundle proteins. A model system for natural light energy harvesting and dissipation. J Am Chem Soc 2011;133:9526–35. [DOI] [PubMed] [Google Scholar]
- Crowe SA, Hahn AS, Morgan-Lang C et al. . Draft genome sequence of the pelagic photoferrotroph Chlorobium phaeoferrooxidans. Genome Announc 2017;5:e01584–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowe SA, Maresca JA, Jones C et al. . Deep-water anoxygenic photosythesis in a ferruginous chemocline. Geobiology 2014;12:322–39. [DOI] [PubMed] [Google Scholar]
- Dahl C. Sulfur metabolism in phototrophic bacteria. In: Hallenbeck PC. (ed.). Modern Topics in the Phototrophic Prokaryotes. Cham: Springer International, 2017, 27–66. [Google Scholar]
- Dailey HA, Dailey TA, Gerdes S et al. . Prokaryotic heme biosynthesis: multiple pathways to a common essential product. Microbiol Mol Biol Rev 2017;81:e00048–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels L, Belay N, Rajagopal BS et al. . Bacterial methanogenesis and growth from CO2 with elemental iron as the sole source of electrons. Science 1987;237:509–11. [DOI] [PubMed] [Google Scholar]
- de Souza JAM, Carrareto Alves LM, de Mello Varani A et al. . The family Bradyrhizobiaceae. In: Rosenberg E, DeLong EF, Lory S et al. (eds). The Prokaryotes – Alphaproteobacteria and Betaproteobacteria. Berlin, Heidelberg: Springer, 2014, 135–54. [Google Scholar]
- de Wit R, van Gemerden H. Oxidation of sulfide to thiosulfate by Microcoleus chtonoplastes. FEMS Microbiol Ecol 1987;45:7–13. [Google Scholar]
- Debussche L, Couder M, Thibaut D et al. . Assay, purification, and characterization of cobaltochelatase, a unique complex enzyme catalyzing cobalt insertion in hydrogenobyrinic acid a,c-diamide during coenzyme B12 biosynthesis in Pseudomonas denitrificans. J Bacteriol 1992;174:7445–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker K, Jungermann K, Thauer RK. Energy production in anaerobic organisms. Angew Chem Int Edit 1970;9:138–58. [DOI] [PubMed] [Google Scholar]
- Demmig-Adams B, Adams WW. Photoprotection and other responses of plants to high light stress. Annu Rev Plant Phys 1992;43:599–626. [Google Scholar]
- Dismukes GC, Klimov VV, Baranov SV et al. . The origin of atmospheric oxygen on Earth: the innovation of oxygenic photosynthesis. P Natl Acad Sci USA 2001;98:2170–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixit BPSN, Moy VT, Vanderkooi JM. Reactions of excited-state cytochrome c derivatives. Delayed fluorescence and phosphorescence of zinc, tin, and metal-free cytochrome c at room temperature. Biochemistry 1984;23:2103–7. [Google Scholar]
- Dixit SN, Waring AJ, Vanderkooi JM. Triplet absorption and phosphorence emission in zinc cytochrome c. FEBS Lett 1981;125:86–8. [DOI] [PubMed] [Google Scholar]
- Ehrenreich A, Widdel F. Anaerobic oxidation of ferrous iron by purple bacteria, a new type of phototrophic metabolism. Appl Environ Microb 1994;60:4517–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favereau L, Cnossen A, Kelber JB et al. . Six-coordinate zinc porphyrins for template-directed synthesis of spiro-fused nanorings. J Am Chem Soc 2015;137:14256−9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer WW, Hemp J, Johnson JE. Evolution of oxygenic photosynthesis. Ann Rev Earth Pl Sc 2016;44:647–83. [Google Scholar]
- Fox G, Stackebrandt E, Hespell R et al. . The phylogeny of prokaryotes. Science 1980;209:457–63. [DOI] [PubMed] [Google Scholar]
- Fridman S, Flores-Uribe J, Larom S. A myovirus encoding both photosystem I and II proteins enhances cyclic electron flow in infected Prochlorococcus cells. Nat Microbiol 2017; doi: 10.1038/s41564-017-0002-9. [DOI] [PubMed] [Google Scholar]
- Fuchs G. Alternative pathways of autotrophic CO2 fixation. In: Schlegel HG, Bowien B (eds). Autotrophic Bacteria. Madison, WI: Science Tech Publishers, 1989, 365–82. [Google Scholar]
- Fuchs G. Alternative pathways of carbon dioxide fixation: insights into the early evolution of life? Annu Rev Microbiol 2011;65:631–58. [DOI] [PubMed] [Google Scholar]
- Furdui C, Ragsdale SW. The role of pyruvate ferredoxin oxidoreductase in pyruvate synthesis during autotrophic growth by the Wood–Ljungdahl pathway. J Biol Chem 2000;275:28494–9. [DOI] [PubMed] [Google Scholar]
- Garcia Costas AM, Liu Z, Tomsho LP et al. . Complete genome of the Candidatus Chloracidobacterium thermophilum, a chlorophyll-based photoheterotroph belonging to the phylum acidobacteria. Environ Microbiol 2012;14:177–90. [DOI] [PubMed] [Google Scholar]
- Garcia-Mendoza E, Ocampo-Alvarez H, Govindjee. Photoprotection in the brown alga Macrocystis pyrifera: evolutionary implications. J Photoch Photobio B 2011;104:377–85. [DOI] [PubMed] [Google Scholar]
- Gisriel C, Sarrou I, Ferlez B et al. . Structure of a symmetric photosynthetic reaction center–photosystem. Science 2017. doi: 10.1126/science.aan5611. [DOI] [PubMed] [Google Scholar]
- Goericke R, Repeta DJ. Chlorophylls a and b and divinyl chlorophylls a and b in the open substropical North Atlantic Ocean. Mar Ecol Progr 1993;101:307–13. [Google Scholar]
- Golbeck JH. Shared thematic elements in photochemical reaction centers. P Natl Acad Sci USA 1993;90:1642–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golubic S, Lee S-J. Early cyanobacterial fossil record: preservation, palaeoenvironments and identification. Eur J Phycol 1999;34:339–48. [Google Scholar]
- Gomez Maqueo Chew A, Bryant DA. Chlorophyll biosynthesis in bacteria: the origins of structural and functional diversity. Annu Rev Microbiol 2007a;61:113–29. [DOI] [PubMed] [Google Scholar]
- Gomez Maqueo Chew A, Bryant DA. Characterization of a plant-like protochlorophyllide a divinyl reductase in green sulfur bacteria. J Biol Chem 2007b;282:2967–75. [DOI] [PubMed] [Google Scholar]
- Granick S. Evolution of heme and chlorophyll. In: Bryson G, Vogel HJ (eds.). Evolving Genes and Proteins. New York: Academic Press, 1965, 67–88. [DOI] [PubMed] [Google Scholar]
- Grassineau NV, Abell P, Appel PWU et al. . Early life signatures in sulphur and carbon isotopes from Isua, Barberton, Wabigoon (Steep Rock) and Belingwe greenstone belts (3.8 to 2.7Ga). In: Kesler SE, Ohmoto H (eds). Evolution of Early Earth's Atmosphere, Hydrosphere and Biosphere – Constraints from Ore Deposits. Geological Society of America. Special Publication; 2006;198:33–52. [Google Scholar]
- Graur D. Molecular and Genome Evolution. Sunderland, MA: Sinauer Associates, 2016. [Google Scholar]
- Griffin BM, Schott J, Schink B. Nitrite, an electron donor for anoxygenic photosynthesis. Science 2007;316:1870. [DOI] [PubMed] [Google Scholar]
- Grim SL, Dick GJ. Photosynthetic versatility in the genome of Geitlerinema sp. PCC 9228 (formerly Oscillatoria limnetica ‘Solar Lake’), a model anoxygenic photosynthetic cyanobacterium. Front Microbiol 2016;7:1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grote J, Schott T, Bruckner CG et al. . Genome and physiology of a model Epsilonproteobacterium responsible for sulfide detoxification in marine oxygen depletion zones. P Natl Acad Sci USA 2012;109:506–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan G, Kida T, Ma T et al. . Reduction of aqueous CO2 at ambient temperature using zero-valent iron-based composites. Green Chem 2003;5:630–34. [Google Scholar]
- Halevy I, Zuber MT, Schrag DP. A sulfur dioxide climate feedback on early Mars. Science 2007;318:1903–7. [DOI] [PubMed] [Google Scholar]
- Hanada S, Pierson BK. The family Chloroflexaceae. In: Dworkin M, Falkow S, Rosenberg E et al. (eds.). The Prokaryotes – A Handbook on the Biology of Bacteria. Volume 7: Proteobacteria: Delta and Epsilon Subclasses. Deeply Rooting Bacteria. New York: Springer, 2006, 815–42. [Google Scholar]
- Hanada S, Takaichi S, Matsuura K et al. . Roseiflexus castenholzii gen. nov., sp. nov., a thermophilic, filamentous, photosynthetic bacterium that lacks chlorosomes. Int J Syst Evol Micr 2002;52:187–93. [DOI] [PubMed] [Google Scholar]
- Hansen TA, Imhoff JF. Rhodobacter veldkampii, a new species of phototrophic purple nonsulfur bacteria. Int J Syst Bacteriol 1985;35:115–6. [Google Scholar]
- Hanson TE, Tabita FR. A ribulose-1,5-bisphosphate carboxylase/oxygenase (RubisCO)-like protein from Chlorobium tepidum that is involved with sulfur metabolism and the response to oxidative stress. P Natl Acad Sci USA 2001;98:4397–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay S, Wallace BB, Smith TA et al. . Protein engineering of cytochrome b562 for quinone binding and light-induced electron transfer. P Natl Acad Sci USA 2004;101:17675–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He C, Tian G, Liu Z et al. . A mild hydrothermal route to fix carbon dioxide to simple carboxylic acids. Org Lett 2010;12:649–51. [DOI] [PubMed] [Google Scholar]
- Hegler F, Posth NR, Jiang J et al. . Physiology of phototrophic iron(II)-oxidizing bacteria: implications for modern and ancient environments. FEMS Microbiol Ecol 2008;66:250–60. [DOI] [PubMed] [Google Scholar]
- Heinnickel M, Golbeck JH. Heliobacterial photosynthesis. Photosynth Res 2007;92:35–53. [DOI] [PubMed] [Google Scholar]
- Heising S, Richter L, Ludwig W et al. . Chlorobium ferrooxidans sp. nov., a phototrophic green sulfur bacterium that oxidizes ferrous iron in coculture with a “Geospirillum” sp. strain. Arch Microbiol 1999;172:116–24. [DOI] [PubMed] [Google Scholar]
- Herrmann G, Jayamani E, Mai G et al. . Energy conservation via electron-transferring flavoprotein in anaerobic bacteria. J Bacteriol 2008;190:784–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho M-Y, Shen G, Canniffe DP et al. . Light-dependent chlorophyll f synthase is a highly divergent paralog of PsbA of photosystem II. Science 2016;353:aaf9178. [DOI] [PubMed] [Google Scholar]
- Hoehler TM, Jörgensen BB. Microbial life under extreme energy limitation. Nat Rev Microbiol 2013;11:83–94. [DOI] [PubMed] [Google Scholar]
- Hoffmann MC, Wagner E, Langklotz S et al. . Proteome profiling of the Rhodobacter capsulatus molybdenum response reveals a role of IscN in nitrogen fixation by Fe-nitrogenase. J Bacteriol 2015;198:33–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohmann-Marriott MF, Blankenship RE. Evolution of photosynthesis. Annu Rev Plant Biol 2011;62:515–48. [DOI] [PubMed] [Google Scholar]
- Hunter GA, Sampson MP, Ferreira GC. Metal ion substrate inhibition of ferrochelatase. J Biol Chem 2008;283:23685–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imhoff JF. The phototrophic alpha proteobacteria. In: Dworkin M, Falkow S, Rosenberg E et al. (eds.). The Prokaryotes – A Handbook on the Biology of Bacteria. Volume 5: Proteobacteria: Alpha and Beta Subclasses. New York: Springer, 2006a, 41–64. [Google Scholar]
- Imhoff JF. The Chromatiaceae. In: Dworkin M, Falkow S, Rosenberg E et al. (eds.). The Prokaryotes – A Handbook on the Biology of Bacteria. Volume 6: Proteobacteria: Gamma Subclasses. New York: Springer, 2006b, 846–73. [Google Scholar]
- Imhoff JF. The family Ectothiorhodospiraceae. In: Dworkin M, Falkow S, Rosenberg E et al. (eds.). The Prokaryotes – A Handbook on the Biology of Bacteria. Volume 6: Proteobacteria: Gamma Subclasses. New York: Springer, 2006c, 874–86. [Google Scholar]
- Ishii T, Kawaichi S, Nakagawa H et al. . From chemolithoautotrophs to electrolithoautotrophs: CO2 fixation by Fe(II)-oxidizing bacteria coupled with direct uptake of electrons from solid electron sources. Front Microbiol 2015;6:994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikita H, Loll B, Biesiadka J et al. . Redox potentials of chlorophylls in the photosystem II reaction center. Biochemistry 2005;44:4118–24. [DOI] [PubMed] [Google Scholar]
- Ito H, Yokono M, Tanaka R et al. . Identification of a novel vinyl reductase gene essential for the biosynthesis of monovinyl chlorophyll in Synechocystis sp. PCC6803. J Biol Chem 2008;283:9002–11. [DOI] [PubMed] [Google Scholar]
- Jaschke PR, Hardjasa A, Digby EL et al. . A bchD (magnesium chelatase) mutant of Rhodobacter sphaeroides synthesizes zinc bacteriochlorophyll through novel zinc-containing intermediates. J Biol Chem 2011;286:20313–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen SI, Steunou A-S, Bhaya D et al. . In situ dynamics of O2, pH and cyanobacterial transcripts associated with CCM, photosynthesis and detoxification of ROS. ISME J 2011;5:317–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y, Kappler A, Croal LR et al. . Isolation and characterization of a genetically tractable photoautotrophic Fe(II)-oxidizing bacterium, Rhodopseudomonas palustris strain TIE-1. Appl Environ Microb 2005;71:4487–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JE, Webb SM, Thomas K et al. . Manganese-oxidizing photosynthesis before the rise of cyanobacteria. P Natl Acad Sci USA 2013;110:11238–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston DT. Multiple sulfur isotopes and the evolution of Earth's surface sulfur cycle. Earth-Sci Rev 2011;106:161–83. [Google Scholar]
- Johnston DT, Wolfe-Simon F, Pearson A et al. . Anoxygenic photosynthesis modulated Proterozoic oxygen and sustained Earth's middle age. P Natl Acad Sci USA 2009;106:16925–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jormakka M, Yokoyama K, Yano T et al. . Molecular mechanism of energy conservation in polysulfide respiration. Nat Struct Mol Biol 2008;15:730–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Judson OP. The energy expansions of evolution. Nat Ecol Evol 2017;1:0138. [DOI] [PubMed] [Google Scholar]
- Kappler A, Pasquero C, Konhauser KO et al. . Deposition of banded iron formations by anoxygenic phototrophic Fe(II)- oxidizing bacteria. Geology 2005;33:865–8. [Google Scholar]
- Kelley DS, Baross JA, Delaney JR. Volcanoes, fluids, and life at mid-ocean ridge spreading centers. Annu Rev Earth Pl Sc 2002;30:385–491. [Google Scholar]
- Keppen OI, Krasil’nikova EN, Lebedeva NV et al. . Comparative study of metabolism of the purple photosynthetic bacteria grown in the light and the dark under anaerobic and aerobic conditions. Microbiology 2013;82:547–53. [PubMed] [Google Scholar]
- Kersters K, De Vos P, Gillis M et al. . Introduction to the Proteobacteria. In: Dworkin M, Falkow S, Rosenberg E et al. (eds). The Prokaryotes – A Handbook on the Biology of Bacteria. Volume 5: Proteobacteria: Alpha and Beta Subclasses. New York: Springer, 2006, 3–37. [Google Scholar]
- Kishimoto N, Kosako Y, Tano T. Acidobacterium capsulatum, gen. nov., sp. nov.: an acidophilic chemoorganotrophic bacterium containing menaquinone from acidic mineral environment. Curr Microbiol 1991;22:1–7. [Google Scholar]
- Klatt JM, Al-Najjar MAA, Yilmaz P et al. . Anoxygenic photosynthesis controls oxygenic photosynthesis in a cyanobacterium from a sulfidic spring. Appl Environ Microb 2015;81:2025–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klatt JM, Meyer S, Häusler S et al. . Structure and function of natural sulphide-oxidizing microbial mats under dynamic input of light and chemical energy. ISME J 2016;10:921–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoll AH, Bergmann KD, Strauss JV. Life: the first two billion years. Philos T Roy Soc B 2016;371:20150493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koblizek M. Ecology of aerobic anoxygenic phototrophs in aquatic environments. FEMS Microbiol Rev 2015;39:854–70. [DOI] [PubMed] [Google Scholar]
- Koblížek M, Zeng Y, Horák A et al. . Regressive evolution of photosynthesis in the Roseobacter clade. Adv Bot Res 2013;66:385–405 [Google Scholar]
- Kopejtka K, Tomasch J, Zeng Y et al. . Genomic analysis of the evolution of phototrophy among haloalkaliphilic Rhodobacterales. Genome Biol Evol 2017;9:1950–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kono T, Mehrotra S, Endo C et al. . A RuBisCO-mediated carbon metabolic pathway in methanogenic archaea. Nat Commun 2017;8:14007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause GH, Weis E. Chlorophyll fluorescence and photosynthesis – the basics. Annu Rev Plant Phys 1991;42:313–49. [Google Scholar]
- Kręzèl A, Maret W. The biological inorganic chemistry of zinc ions. Arch Biochem Biophys 2016;611:3–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku C, Nelson-Sathi S, Roettger M et al. . Endosymbiotic origin and differential loss of eukaryotic genes. Nature 2015;524:427–32. [DOI] [PubMed] [Google Scholar]
- Layer G, Moser J, Heinz DW et al. . Crystal structure of coproporphyrinogen III oxidase reveals cofactor geometry of radical SAM enzymes. EMBO J 2003;22:6214–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengeler JW, Drews G, Schlegel HG. Biology of the Prokaryotes. Stuttgart: Thieme, 1999. [Google Scholar]
- Lever MA. Acetogenesis in the energy-starved deep biosphere – a paradox? Front Microbiol 2012;2:284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lever MA, Heuer VB, Morono Y et al. . Acetogenesis in deep subseafloor sediments of the Juan de Fuca ridge flank: a synthesis of geochemical, thermodynamic, and gene-based evidence. Geomicrobiol J 2010;27:183–211. [Google Scholar]
- Li F, Hinderberger J, Seedorf H et al. . Coupled ferredoxin and crotonyl coenzyme A (CoA) reduction with NADH catalyzed by the butyryl-CoA dehydrogenase/Etf complex from Clostridium kluyveri. J Bacteriol 2008;190:843–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Klatt CG, Ludwig M et al. . Candidatus Thermochlorobacter aerophilum: an aerobic chlorophotoheterotrophic member of the phylum Chlorobi defined by metagenomics and metatranscriptomics. ISME J 2012;6:1869–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubner CE, Jennings DP, Mulder DW et al. . Mechanistic insights into energy conservation by flavin-based electron bifurcation. Nat Chem Biol 2017;13:655–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luther GW., 3rd The role of one- and two-electron transfer reactions in forming thermodynamically unstable intermediates as barriers in multi-electron redox reactions. Aquat Geochem 2010;16:395–420. [Google Scholar]
- Luther GW 3rd, Findlay AJ, MacDonald DJ et al. . Thermodynamics and kinetics of sulfide oxidation by oxygen: a look at inorganically controlled reactions and biologically mediated processes in the environment. Front Microbiol 2011;2:62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons TW, Reinhard CT, Planavsky NJ. The rise of oxygen in Earth's early ocean and atmosphere. Nature 2014;506:307–15. [DOI] [PubMed] [Google Scholar]
- McCollom TM. Abiotic methane formation during experimental serpentinization of olivine. P Natl Acad Sci USA 2016;113:13965–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCollom TM, Seewald JS. Serpentinites, hydrogen, and life. Elements 2013;9:129–34. [Google Scholar]
- McDermott JM, Seewald JS, German CR et al. . Pathways for abiotic organic synthesis at submarine hydrothermal fields. P Natl Acad Sci USA 2015;112:7668–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEwan AG. Photosynthetic electron-transport and anaerobic metabolism in purple nonsulfur phototrophic bacteria. Anton Leeuw 1994;66:151–64. [DOI] [PubMed] [Google Scholar]
- McKinlay JB, Harwood CS. Carbon dioxide fixation as a central redox cofactor recycling mechanism in bacteria. P Natl Acad Sci USA 2010;107:11669–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madigan MT, Gest H. Growth of the photosynthetic bacterium Rhodopseudomonas capsulata chemoautotrophically in darkness with H2 as the energy source. J Bacteriol 1979;137:524–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manske AK, Glaeser J, Kuypers MAM et al. . Physiology and phylogeny of green sulfur bacteria forming a monospecific phototrophic assemblage at a depth of 100 meters in the Black Sea. Appl Environ Microb 2005;71:8049–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marschall E, Jogler M, Hessge U et al. . Large-scale distribution and activity patterns of an extremely low-light-adapted population of green sulfur bacteria in the Black Sea. Environ Microbiol 2010;12:1348–62. [DOI] [PubMed] [Google Scholar]
- Martin W, Baross J, Kelley D et al. . Hydrothermal vents and the origin of life. Nat Rev Microbiol 2008;6:805–14. [DOI] [PubMed] [Google Scholar]
- Martin W, Russell MJ. On the origin of biochemistry at an alkaline hydrothermal vent. Philos T Roy Soc B 2007;367:1887–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin W, Schnarrenberger C. The evolution of the Calvin cycle from prokaryotic to eukaryotic chromosomes: a case study of functional redundancy in ancient pathways through endosymbiosis. Curr Genet 1997;32:1–18. [DOI] [PubMed] [Google Scholar]
- Martin WF. Physiology, phylogeny and the energetic roots of life. Period Biol 2016;18:343–52. [Google Scholar]
- Mathis P. Compared structure of plant and bacterial photosynthetic reaction centers. Evolutionary implications. Biochim Biophys Acta 1990;1018:163–7. [Google Scholar]
- Miller H, Mayhew LE, Ellison ET et al. . Low temperature hydrogen production during experimental hydration of partially-serpentinized dunite. Geochim Cosmochim Acta 2017;209:161–83. [Google Scholar]
- Miller SR, Bebout BM. Variation in sulfide tolerance of photosystem II in phylogenetically diverse cyanobacteria from sulfidic habitats. Appl Environ Microb 2004;70:736–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulkidjanian AY, Koonin EV, Makrova KS et al. . The cyanobacterial genome core and the origins of photosynthesis. P Natl Acad Sci USA 2006;103:13126–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller E, Philippot P, Rollion-Bard C et al. . Multiple sulfur-isotope signatures in Archean sulfates and their implications for the chemistry and dynamics of the early atmosphere. P Natl Acad Sci USA 2016;113:7432–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muraki N, Nomata J, Ebata K et al. . X-ray crystal structure of the light-independent protochlorophyllide reductase. Nature 2010;465:110–4. [DOI] [PubMed] [Google Scholar]
- Murray JW. Sequence variation at the oxygen-evolving centre of photosystem II: a new class of ‘rogue’ cyanobacterial D1 proteins. Photosynth Res 2012;110:177–84. [DOI] [PubMed] [Google Scholar]
- Neidhardt FC, Ingraham JL, Schaechter M. Physiology of the Bacterial Cell. A Molecular Approch. Sunderland, MA: Sinauer Associates, 1990. [Google Scholar]
- Nisbet EG, Cann JR, Van Dover CL. Origins of photosynthesis. Nature 1995;373:479–80. [Google Scholar]
- Nisbet EG, Sleep NH. The habitat and nature of early life. Nature 2001;409:1083–91. [DOI] [PubMed] [Google Scholar]
- Nomata J, Mizoguchi T, Tamiaki H et al. . A second nitrogenase-like enzyme for bacteriochlorophyll biosynthesis. J Biol Chem 2006;281:15021–8. [DOI] [PubMed] [Google Scholar]
- Olson JM, Pierson BK. Evolution of reaction centers in photosynthetic prokaryotes. Int Rev Cytol 1987;108:209–48. [DOI] [PubMed] [Google Scholar]
- Oren A, Padan E. Induction of anaerobic, photoautotrophic growth in the cyanobacterium Oscillatoria limnetica. J Bacteriol 1978;133:558–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overmann J, Pfennig N. Continuous chemotrophic growth and respiration of Chromatiaceae species at low oxygen concentrations. Arch Microbiol 1992;158:59–67. [Google Scholar]
- Petersen J, Brinkmann H, Bunk B et al. . Think pink: photosynthesis, plasmids and the Roseobacter clade. Environ Microbiol 2012;14:2661–72. [DOI] [PubMed] [Google Scholar]
- Poehlein A, Schmidt S, Kaster AK et al. . An ancient pathway combining carbon dioxide fixation with the generation and utilization of a sodium ion gradient for ATP synthesis. PLoS One 2012;7:e33439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pujalte MJ, Lucena T, Ruvira MA et al. . The Family Rhodobacteraceae. In: Dworkin M, Falkow S, Rosenberg E et al. (eds.). The Prokaryotes – A Handbook on the Biology of Bacteria. Volume 5: Proteobacteria: Alpha and Beta Subclasses. Heidelberg, New York, Dodrecht, London: Springer, 2014, 439–512. [Google Scholar]
- Qin L, Kostic NM. Photoinduced electron transfer from the triplet state of zinc cytochrome c to ferricytochrome b5 is gated by configurational fluctuations of the diprotein complex. Biochemistry 1994;33:12592–9. [DOI] [PubMed] [Google Scholar]
- Rabenstein A, Rethmeier J, Fischer U. Sulphite as intermediate sulphur compound in anaerobic sulphide oxidation to thiosulphate by marine cyanobacteria. Z Naturforsch 1995;50c:769–74. [Google Scholar]
- Rabus R, Venceslau SS, Wöhlbrand L et al. . A post-genomic view of the ecophysiology, catabolism and biotechnological relevance of sulphate-reducing prokaryotes. Adv Microb Physiol 2015;66:55–321. [DOI] [PubMed] [Google Scholar]
- Raven JA. Contributions of anoxygenic and oxygenic phototrophy and chemolithotrophy to carbon and oxygen fluxes in aquatic environments. Aquat Microb Ecol 2009;56:177–92. [Google Scholar]
- Razeghifard AR, Wydrzynski T. Binding of Zn-chlorin to a synthetic four-helix bundle peptide through histidine ligation. Biochemistry 2003;42:1024–30. [DOI] [PubMed] [Google Scholar]
- Reher M, Fuhrer T, Bott M et al. . The nonphosphorylative Entner-Doudoroff pathway in the thermoacidophilic euryarchaeon Picrophilus torridus involves a novel 2-keto-3-deoxygluconate-specific aldolase. J Bacteriol 2010;192:964–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell MJ, Hall AJ, Martin W. Serpentinization as a source of energy at the origin of life. Geobiology 2010;8:355–71. [DOI] [PubMed] [Google Scholar]
- Sadekar S, Raymond J, Blankenship RE. Conservation of distantly related membrane proteins: photosynthetic reaction centers share a common structural core. Mol Biol Evol 2006;23:2001–7. [DOI] [PubMed] [Google Scholar]
- Sánchez-Baracaldo P, Raven JA, Pisani D et al. . Early photosynthetic eukaryotes inhabited low-salinity habitats. P Natl Acad Sci USA 2017;11:E7737–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos AA, Venceslau SS, Grein F et al. . A protein trisulfide couples dissimilatory sulfate reduction to energy conservation. Science 2015;350:1541–5. [DOI] [PubMed] [Google Scholar]
- Saraiva IH, Newman DK, Louro RO. Functional characterization of the FoxE iron oxidoreductase from the photoferrotroph Rhodobacter ferrooxidans SW2. J Biol Chem 2012;287:25541–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T, Atomi H. Novel metabolic pathways in archaea. Curr Opin Microbiol 2011;14:307–14. [DOI] [PubMed] [Google Scholar]
- Sato T, Atomi H, Imanaka T. Archaeal type III RuBisCOs function in a pathway for AMP metabolism. Science 2007;315:1003–6. [DOI] [PubMed] [Google Scholar]
- Schmehl M, Jahn A, Meyer zu Vilsendorf A et al. . Identification of a new class of nitrogen fixation genes in Rhodobacter capsulatus: a putative membrane complex involved in electron transport to nitrogenase. Mol Gen Genet 1993;241:602–15. [DOI] [PubMed] [Google Scholar]
- Schopf JF. The fossil record of cyanobacteria. In: Whitton BA. (ed.) Ecology of Cyanobacteria II: Their Diversity in Space and Time. Berlin: Springer, 2012, 15–36. [Google Scholar]
- Schönheit P, Buckel W, Martin WF. On the origin of heterotrophy. Trends Microbiol 2016;24:12–25. [DOI] [PubMed] [Google Scholar]
- Schrenk MO, Brazelton WJ, Lang SQ. Serpentinization, carbon, and deep life. Rev Mineral Geochem 2013;75:575–606. [Google Scholar]
- Schubert WD, Klukas O, Saenger W et al. . A common ancestor for oxygenic and anoxygenic photosynthetic systems: a comparison based on the structural model of photosystem I. J Mol Biol 1998;280:297–314. [DOI] [PubMed] [Google Scholar]
- Schuchmann K, Müller V. Autotrophy at the thermodynamic limit of life: a model for energy conservation in acetogenic bacteria. Nat Rev Microbiol 2014;12:809–21. [DOI] [PubMed] [Google Scholar]
- Schut GJ, Adams MWW. The iron-hydrogenase of Thermotoga maritima utilizes ferredoxin and NADH synergistically: a new perspective on anaerobic hydrogen production. J Bacteriol 2009;191:4451–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schut GJ, Bridger SL, Adams MWW. Insights into the metabolism of elemental sulfur by the hyperthermophilic archaeon Pyrococcus furiosus: characterization of a coenzyme A-dependent NAD(P)H sulfur oxidoreductase. J Bacteriol 2007;189:4431–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schut GJ, Boyd ES, Peters JW et al. . The modular respiratory complexes involved in hydrogen and sulfur metabolism by heterotrophic hyperthermophilic archaea and their evolutionary implications. FEMS Microbiol Rev 2013;37:182–203. [DOI] [PubMed] [Google Scholar]
- Sharma P, de Mattos MJT, Hellingwerf KJ et al. . On the function of the various quinone species in Escherichia coli. FEBS J 2012;279:3364–73. [DOI] [PubMed] [Google Scholar]
- Shen C, Kostic NM. Reductive quenching of the triplet state of zinc cytochrome c by the hexacyanoferrate(II) anion and by conjugate bases of ethylenediaminetetraacetic acid. Inorg Chem 1996;35:2780–4. [Google Scholar]
- Shen JR. The structure of photosystem II and the mechanism of water oxidation in photosynthesis. Annu Rev Plant Biol 2015;66:23–48. [DOI] [PubMed] [Google Scholar]
- Shih PM, Hemp J, Ward LM et al. . Crown group Oxyphotobacteria postdate the rise of oxygen. Geobiology 2017;15:19–29. [DOI] [PubMed] [Google Scholar]
- Shih PM, Ward LM, Fischer WW. Evolution of the 3-hydroxypropionate bicycle and recent transfer of anoxygenic photosynthesis into the Chloroflexi. P Natl Acad Sci USA 2017;114:10749–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shima S, Pilak O, Vogt S et al. . The crystal structure of [Fe]-hydrogenase reveals the geometry of the active site. Science 2008;321:572–5. [DOI] [PubMed] [Google Scholar]
- Simpson PG, Whitman WB. Anabolic pathways in methanogens. In: Ferry JG. (ed.) Methanogenesis: Ecology, Physiology, Biochemistry, and Genetics. New York, NY: Chapman and Hall, 1993, 445–72. [Google Scholar]
- Sleep NH, Bird DK, Pope EC. Serpentinite and the dawn of life. Philos T R Soc B 2011;366:2857–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sleep NH, Meibom A, Fridriksson T et al. . H2-rich fluids from serpentinization: geochemical and biotic implications. P Natl Acad Sci USA 2004;101:12818–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soo RM, Hemp J, Parks DH et al. . On the origins of oxygenic photosynthesis and aerobic respiration in cyanobacteria. Science 2017;355:1436–40. [DOI] [PubMed] [Google Scholar]
- Sousa FL, Martin WF. Biochemical fossils of the ancient transition from geoenergetics to bioenergetics in prokaryotic one carbon compound metabolism. Biochim Biophys Acta 2014;1837:964–81. [DOI] [PubMed] [Google Scholar]
- Sousa FL, Shavit-Grievink L, Allen JF et al. . Chlorophyll biosynthesis gene evolution indicates photosystem gene duplication, not photosystem merger, at the origin of oxygenic photosynthesis. Genome Biol Evol 2013;5:200–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo I, Bergantino E, Giacometti GM. Light and oxygenic photosynthesis: energy dissipation as a protection mechanism against photo-oxidation. EMBO Rep 2005;6:629–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabita FR. Microbial ribulose 1,5-bisphosphate carboxylase/oxygenase: a different perspective. Photosynth Res 1999;60:1–28. [Google Scholar]
- Tabita FR, Hanson TE, Satagopan S et al. . Phylogenetic and evolutionary relationships of RubisCO and the RubisCO-like proteins and the functional lessons provided by diverse molecular forms. Philos T R Soc B 2008;363:2629–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabita FR, Hanson TE, Li H et al. . Function, structure, and evolution of the RubisCO-like proteins and their RubisCO homologs. Microbiol Mol Biol Rev 2007;71:576–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takai K, Campbell BJ, Cary SC et al. . Enzymatic and genetic characterization of carbon and energy metabolisms by deep-sea hydrothermal chemolithoautotrophic isolates of Epsilonproteobacteria. Appl Environ Microb 2005;71:7310–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takami H, Noguchi H, Takaki Y et al. . A deeply branching thermophilic bacterium with an ancient acetyl-CoA pathway dominates a subsurface ecosystem. PLoS One 2012;7:e30559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taketani S, Ishigaki M, Mizutani A et al. . Heme synthase (ferrochelatase) catalyzes the removal of iron from heme and demetalation of metalloporphyrins. Biochemistry 2007;46:15054–61. [DOI] [PubMed] [Google Scholar]
- Tamiaki H, Xu M, Tanaka T et al. . Photoreduction of zinc 8-vinylated chlorophyll derivative to bacteriochlorophyll-b/g analog possessing an 8-ethylidene group. Bioorg Med Chem Lett 2013;23:2377–9. [DOI] [PubMed] [Google Scholar]
- Tamura N, Cheniae G. Photoactivation of the water-oxidizing complex in photosystem II membranes depleted of Mn and extrinsic proteins. I. Biochemical and kinetic characterization. Biochim Biophys Acta 1987;890:179–94. [Google Scholar]
- Tank M, Bryant DA. Nutrient requirements and growth physiology of the photoheterotrophic Acidobacterium, Chloracidobacterium thermophilum. Front Microbiol 2015;6:226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tank M, Thiel V, Ward DM et al. . A panoply of phototrophs: an overview of chlorophototrophs found in the microbial mats of alkaline siliceous hot springs in Yellowstone National Park, WY, USA. In: Hallenbeck PC. (ed.). Modern Topics in the Phototrophic Prokaryotes: Environmental and Applied Aspects. Berlin: Springer, 2017, 87–137. [Google Scholar]
- Tapley DW, Buettner GR, Shick JM. Free radicals and chemiluminescence as products of the spontaneous oxidation of sulfide in seawater, and their biological implications. Biol Bull 1999;196:52–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tashiro T, Ishida A, Hori M et al. . Early trace of life from 3.95 Ga sedimentary rocks in Labrador, Canada. Nature 2017;549:516–8. [DOI] [PubMed] [Google Scholar]
- Tedetti M, Sempéré R. Penetration of ultraviolet radiation in the marine environment. A review. Photochem Photobiol 2006;82:389–97. [DOI] [PubMed] [Google Scholar]
- Thamer W, Cirpus I, Hans M et al. . A two [4Fe-4S]-cluster-containing ferredoxin as an alternative electron donor for 2-hydroxyglutaryl-CoA dehydratase from Acidaminococcus fermentans. Arch Microbiol 2003;179:197–204. [DOI] [PubMed] [Google Scholar]
- Thauer RK. Hydrogenases and the global H2 cycle. Eur J Inorg Chem 2011;2011:919–21. [Google Scholar]
- Thauer RK, Jungermann KK, Decker K. Energy-conservation in chemotrophic anaerobic bacteria. Bacteriol Rev 1977;41:100–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thauer RK, Kaster AK, Seedorf H et al. . Methanogenic archaea: ecologically relevant differences in energy conservation. Nat Rev Microbiol 2008;6:579–91. [DOI] [PubMed] [Google Scholar]
- Thiel V, Hügler M, Ward DM et al. . The dark side of the mushroom spring microbial mat: life in the shadow of chlorophototrophs. II. Metabolic functions of abundant community members predicted from metagenomic analyses. Front Microbiol 2017;8:943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson AW, Foster RA, Krupke A et al. . Unicellular cyanobacterium symbiotic with a single-celled eukaryotic alga. Science 2012;337:1546–50. [DOI] [PubMed] [Google Scholar]
- Thweatt JL, Ferlez BH, Golbeck JH et al. . BciD is a radical-S-adenosyl-L-methionine (SAM) enzyme that completes bacteriochlorophyllide e biosynthesis by oxidizing a methyl group into a formyl group at C-7. J Biol Chem 2017;292:1361–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tice MM, Lowe DR. Hydrogen-based carbon fixation in the earliest known photosynthetic organisms. Geology 2006;34:37–40. [Google Scholar]
- Tsukatani Y, Romberger SP, Golbeck JH et al. . Isolation and characterization of homodimeric type-I reaction center complex from Candidatus Chloracidobacterium thermophilum, an aerobic chlorophototroph. J Biol Chem 2012;287:5720–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueno Y. Coping with low ocean sulfate. Science 2014;346:703–4. [DOI] [PubMed] [Google Scholar]
- Umena Y, Kawakami K, Shen J-R et al. . Crystal structure of oxygen-evolving photosystem II at a resolution of 1.9 Å. Nature 2011;473:55–60. [DOI] [PubMed] [Google Scholar]
- Van Dover CL, Reynolds GT, Chave AD et al. . Light at deep-sea hydrothermal vents. Geophys Res Lett 1996;23:2049–52. [Google Scholar]
- Van Dover CL, Szuts EZ, Chamberlain SC et al. . A novel eye in eyeless shrimp from hydrothermal vents of the mid-atlantic ridge. Nature 1989;337:458–60. [DOI] [PubMed] [Google Scholar]
- Vanderkooi JM, Berger JW. Excited triplet states used to study biological macromolecules at room temperature. Biochim Biophys Acta 1989;976:1–27. [DOI] [PubMed] [Google Scholar]
- Wagner A, Whitaker RJ, Krause DJ et al. . Mechanisms of gene flow in archaea. Nat Rev Microbiol 2017a;15:492–501. [DOI] [PubMed] [Google Scholar]
- Wagner T, Koch J, Ermler U et al. . Methanogenic heterodisulfide reductase (HdrABC-MvhAGD) uses two noncubane [4Fe-4S] clusters for reduction. Science 2017b;357:699–702. [DOI] [PubMed] [Google Scholar]
- Wakao N, Yokoi N, Isoyama N et al. . Discovery of natural photosynthesis using Zn-containing bacteriochlorophyll in an aerobic bacterium Acidiphilium rubrum. Plant Cell Physiol 1996;37:889–93. [Google Scholar]
- Weiss MC, Sousa FL, Mrnjavac N et al. . The physiology and habitat of the last universal common ancestor. Nat Microbiol 2016;1:16116. [DOI] [PubMed] [Google Scholar]
- Westall F, Cavalazzi B, Lemelle L et al. . Implications of in situ calcification for photosynthesis in a ∼3.3 Ga-old microbial biofilm from the Barberton greenstone belt, South Africa. Earth Planet Sci Lett 2011;310:468–79. [Google Scholar]
- Westall F, de Ronde CEJ, Southam G et al. . Implications of a 3.472–3.333 Gyr old subaerial microbial mat from the Barberton Greenstone Belt, South Africa for the UV environmental conditions on the early Earth. Philos T R Soc B 2006;361:1857–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White SN, Chave AD, Reynolds GT. Investigations of ambient light emission at dep sea hydrothermal vents. J Geophys Res 2000;107:EPM1–13. [Google Scholar]
- White SN, Chave AD, Reynolds GT et al. . Ambient light emission from hydrothermal vents on the Mid-Atlantic Ridge. Geophys Res Lett 2002a;29:34-1–4. [Google Scholar]
- White SN, Chave AD, Reynolds GT et al. . Variations in ambient light emissions from black smokers and flange pools on the Juan de Fuca Ridge. Geophys Res Lett 2002b;27:1151–4. [Google Scholar]
- Whitman WB, Coleman DC, Wiebe WJ. Prokaryotes: the unseen majority. P Natl Acad Sci USA 1998;95:6578–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widdel F, Schnell S, Heising S et al. . Ferrous iron oxidation by anoxygenic phototrophic bacteria. Nature 1993;362:834–6. [Google Scholar]
- Williams TA, Foster PG, Cox CJ. et al, An archaeal origin of eukaryotes supports only two primary domains of life. Nature 2013;504:231–6. [DOI] [PubMed] [Google Scholar]
- Williamson A, Conlan B, Hillier W et al. . The evolution of Photosystem II: insights into the past and future. Photosynth Res 2011;107:71–86. [DOI] [PubMed] [Google Scholar]
- Wrighton KC, Thomas BC, Sharon I et al. . Fermentation, hydrogen, and sulfur metabolism in multiple uncultivated bacterial phyla. Science 2012;337:1661–5. [DOI] [PubMed] [Google Scholar]
- Xiong J, Bauer CE. Complex evolution of photosynthesis. Annu Rev Plant Biol 2002a;53:503–21. [DOI] [PubMed] [Google Scholar]
- Xiong J, Bauer CE. A cytochrome b origin of photosynthetic reaction centers: an evolutionary link between respiration and photosynthesis. J Mol Biol 2002b;322:1025–37. [DOI] [PubMed] [Google Scholar]
- Xiong J, Inoue K, Bauer CE. Tracking molecular evolution of photosynthesis by characterization of a major photosynthesis gene cluster from Heliobacillus mobilis. P Natl Acad Sci USA 1998;95:14851–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yurkov VV, Beatty JT. Aerobic anoxygenic phototrophic bacteria. Microbiol Mol Biol R 1998;62:695–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Y, Feng F, Medová H et al. . Functional type 2 photosynthetic reaction centers found in the rare bacterial phylum Gemmatimonadetes. P Natl Acad Sci USA 2014;111:7795–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Q, Zhang R, Fogg PCM et al. Gain and loss of phototrophic genes revealed by comparison of two Citromicrobium bacterial genomes. PLoS One 2012;7:e35790. [DOI] [PMC free article] [PubMed] [Google Scholar]