Abstract
Coevolution between hosts and pathogens generates strong selection pressures to maintain resistance and infectivity, respectively. Genomes of plant pathogens often encode major effect loci for the ability to successfully infect specific host genotypes. Hence, spatial heterogeneity in host genotypes coupled with abiotic factors could lead to locally adapted pathogen populations. However, the genetic basis of local adaptation is poorly understood. Rhynchosporium commune, the pathogen causing barley scald disease, interacts at least partially in a gene-for-gene manner with its host. We analyzed global field populations of 125 R. commune isolates to identify candidate genes for local adaptation. Whole genome sequencing data showed that the pathogen is subdivided into three genetic clusters associated with distinct geographic and climatic regions. Using haplotype-based selection scans applied independently to each genetic cluster, we found strong evidence for selective sweeps throughout the genome. Comparisons of loci under selection among clusters revealed little overlap, suggesting that ecological differences associated with each cluster led to variable selection regimes. The strongest signals of selection were found predominantly in the two clusters composed of isolates from Central Europe and Ethiopia. The strongest selective sweep regions encoded protein functions related to biotic and abiotic stress responses. Selective sweep regions were enriched in genes encoding functions in cellular localization, protein transport activity, and DNA damage responses. In contrast to the prevailing view that a small number of gene-for-gene interactions govern plant pathogen evolution, our analyses suggest that the evolutionary trajectory is largely determined by spatially heterogeneous biotic and abiotic selection pressures.
Keywords: population genomics, genome-wide selection scans, plant pathogen, fungi
Introduction
Pathogens pose a major threat to human health and global food security. Hosts and pathogens are often locked into strong antagonistic coevolution, which is a major force driving biodiversity. During coevolution, adaptation in either of the partners generates selection pressure on the other partner, which in turn evolves counteradaptations (Brockhurst and Koskella 2013). Pathogens are considered to be at an advantage in the coevolutionary race as they often exhibit shorter generation times, higher mutation rates, and larger effective population sizes relative to their host (Hamilton et al. 1990; Kaltz and Shykoff 1998; Gandon 2002). Under reduced levels of gene flow, this asymmetry can lead to locally adapted pathogen populations, that is, situations where a pathogen population is better adapted to its sympatric host population than allopatric host populations (“local” vs “foreign” host) (Kaltz and Shykoff 1998). In a heterogeneous environment, a diverse set of host populations connected by variable levels of gene flow will generate complex selection pressures on pathogen populations over space and time and strong frequency-dependent selection can lead to local extinction and recolonization events (Kaltz and Shykoff 1998). Hence, the degree of local adaptation exhibited by hosts and their pathogens will be shaped by multiple factors, including the supply of genetic variation and their respective population sizes (Gandon et al. 1996).
Studies of local adaptation in pathogens have been sparse. Laine (2005) observed a geographical mosaic in the degrees of local adaptation in metapopulations of Podosphaera plantaginis, an obligate fungal pathogen causing mildew in the perennial Plantago lanceolata. The major factor shaping the extent of local adaptation was likely the rate of pathogen dispersal among host populations. A similar observation was reported earlier by Thrall and Burdon (2002) in a study of the flax rust pathogen Melampsora lini and its host wild flax (Linum marginale). The most resistant host populations harbored on average more virulent pathogen genotypes. A main driver for this correlation was likely a trade-off in spore production (i.e., reproduction) and virulence. The causal agent of common bean anthracnose, Colletotrichum lindemuthianum, has lower levels of population differentiation compared with its host plant, which likely favored local adaptation of pathogen populations (Sicard et al. 1997, 2007; Cattan-Toupance et al. 1998). Local adaptation is not only shaped by rates of gene flow, but also by heterogeneity in the abiotic environment such as differences in rainfall and temperature. Gorter et al. (2016) coevolved the bacterium Pseudomonas fluorescens and its viral parasite bacteriophage Φ2 at three different temperatures and found that the bacteria and phages were more resistant and infectious at the temperature where they previously coevolved. Similarly, the coevolution of P. lanceolata–P. plantaginis was found to be tightly linked to climatic factors (Laine 2008). However, what environmental factors are conducive to local adaptation and the relative effect of individual selective agents remain poorly understood.
The process of local adaptation in agricultural ecosystems may differ from the process that operates in natural host–pathogen systems (Croll and McDonald 2017). Agricultural ecosystems usually exhibit homogeneous environments with genetically uniform hosts planted over large areas. Though planting a single elite crop variety is thought to improve crop yields, the resulting massive monoculture in turn imposes strong directional selection for pathogens to specialize on a specific host genotype (Stukenbrock and McDonald 2008). Highly specialized agricultural pathogens cause significant damage on host plants and lead to large-scale losses. The Irish famine in the 19th century was caused by the attack of an oomycete pathogen specialized on potato clones that destroyed the food supply of one-third of the population (Scholthof 2007). Widespread planting of a single banana clone “Gros Michel” in Panama and Costa Rica led to the emergence of the specialized pathogen Fusarium oxysporum f. sp. cubense (Ploetz 2000). After the failure of “Gros Michel”, a new banana variety named Cavendish was more widely planted. However, this favored the emergence of a new pathogen strain genotype named tropical race 4 and a widespread breakdown of Cavendish resistance that was observed across Asia and Africa (Ordonez et al. 2015). More recently, a new race of the wheat stem rust pathogen Puccinia graminis f. sp. tritici surmounted the widely deployed stem rust resistance gene Sr31 (Pretorius et al. 2000).
With the exception of a few highly clonal crops such as bananas, most agroecosystems exhibit both complex environmental differences and diversified host genotypes at the scale of countries and continents (Bianchi et al. 2006). The resulting mosaic in host genotypes and ecological niches creates the opportunity for divergent selection among pathogen populations. Zhan and McDonald (2011) found evidence for local thermal adaptation across global populations of the wheat pathogen Zymoseptoria tritici. Temperature-dependent local adaptation was also observed among isolates of Puccinia striiformis f.sp. tritici (PST) collected in northern and southern France (Mboup et al. 2012). The northern PST population harbored all virulence genes necessary to infect wheat varieties deployed in the south, but it failed to invade the southern region due to a lack of adaptation to the warmer Mediterranean climate. In addition to climatic limitations, extensive use of fungicides can impose strong directional selection on pathogen populations and lead to local adaptation. Selection for fungicide tolerance likely led to locally adapted Z. tritici populations (Zhan et al. 2006). Although evidence for divergent selection can be found through careful assessment of phenotypes, direct identification of the genes underlying the process of local adaptation remains rare.
The genetic basis of pathogen local adaptation in agricultural ecosystems is generally not well understood, though many microbial pathogens should be highly tractable models for genomic analyses (Croll and McDonald 2017). Until recently, the identification of adaptive loci in plant pathogens was largely limited to population analyses of pathogenicity genes (Schürch et al. 2004; Stukenbrock and McDonald 2007). However, pathogens likely experience a multitude of biotic and abiotic selection pressures. Hence, many loci across the pathogen genome are likely to harbor genetic variation under selection. The analyses of loci under recent selection have been revolutionized by genome sequencing at the population level. A number of statistical tests were specifically designed to retrieve signatures of adaptive loci in large-scale data sets (Voight et al. 2006; Lao et al. 2007; Bigham et al. 2010; Qian et al. 2013; Liu et al. 2017). Many tests of selection focused on one of the following signatures of a positively selected allele: an increase in linkage disequilibrium (LD), shifts in allele frequency spectra, or higher than expected levels of population differentiation (Vitti et al. 2013). Tools implementing these statistics have been widely used to detect signatures of recent selection in various organisms including humans, flies, the malaria pathogen, and livestock animals (Duffy et al. 2015; Zhao et al. 2015; Garud et al. 2015; Martin et al. 2016). One of the widely used approaches is to calculate integrated haplotype homozygosity (iHS), which estimates the decay of the extended haplotype homozygosity (EHH) between an ancestral and derived allele at each SNP position (Sabeti et al. 2002; Voight et al. 2006). The rationale for this method is that the increase in frequency of a beneficial mutation will occur too quickly for recombination to homogenize its genetic background. Hence, a positively selected allele will be embedded in a less common and longer stretch of homogenous chromosomal sequence compared with neutral loci residing in more common and shorter haplotypes. Analyses of EHH are particularly suited to detection of soft (i.e., incomplete) selective sweeps (Voight et al. 2006). An extension to within-population EHH analyses is the cross-population EHH (XP-EHH), which is a more powerful method to detect nearly fixed selective sweeps because it compares haplotypes in a pair of populations (Sabeti et al. 2002). A combination of iHS and XP-EHH analyses should lead to a comprehensive set of candidate loci underlying recent local adaptation.
Rhynchosporium commune is an important fungal pathogen that causes barley scald disease globally with particularly high prevalence in temperate regions with cool and moist winters (Linde et al. 2003; Brunner et al. 2007; Aoki et al. 2011). R. commune is a member of the host-specialized Rhynchosporium species complex that split from a common ancestor 1,200–3,600 years ago (Zaffarano et al. 2008, 2011). The center of origin of R. commune is likely in Scandinavia, because this region is the diversity hotspot of the pathogen, unlike many other pathogens of crops that were domesticated in the Fertile Crescent (Zaffarano et al. 2006; Brunner et al. 2007). Individual field populations of this sexual pathogen are highly diverse with an average of 76% of the global genetic diversity found within a single barley field (Linde et al. 2003, 2009). The genetic basis of adaptation to its host is poorly understood with a few exceptions. Three genes encoding necrosis inducing peptides (NIP1−3) contribute to the development of necrosis (localized cell death promoting the infection) (Wevelsiep et al. 1991, 1993; Rohe et al. 1995; Schürch et al. 2004; Kirsten et al. 2012; Stefansson, Willi, et al. 2014) and a previously unknown pathogen-associated molecular pattern named Cell Death Inducing 1 (RcCDI1) is highly expressed during early infection of the host (Franco-Orozco et al. 2017). A gene-for-gene interaction of the NIP1 gene with the Rrs1 resistance locus in barley has been established in which the pathogen overcame NIP1 recognition through mutations and complete deletions of the NIP1 gene (Schürch et al. 2004). R. commune populations can rapidly adapt to overcome host resistance (Xi et al. 2000) and were shown to adapt to abiotic selection pressures including temperature variation and fungicide applications (Stefansson et al. 2013; Brunner et al. 2015; Mohd-Assaad et al. 2016). A genome-wide association study (GWAS) revealed that resistance to azole fungicides emerged from mutations in multiple loci including the gene encoding the protein targeted by azoles (Mohd-Assaad et al. 2016).
A genome-wide analysis of nine global populations of R. commune showed that the species is subdivided into three main genetic groups (Mohd-Assaad et al. 2016). Each of these main groups was primarily associated with distinct geographic and climatic regions including Scandinavia, Central Europe and Oceania, and Ethiopia. Here we aim to identify candidate regions of divergent selection in these three genetic groups of R. commune. For this, we analyzed 125 whole-genome sequences collected from isolates across the distribution range of the pathogen. We performed genome-wide analyses of selection first at the population level using iHS and then at the between-population level using XP-EHH.
Materials and Methods
Isolates Collection, DNA Preparation, and Full Genome Sequencing
A total of 125 strains of R. commune were collected from nine countries: New Zealand, Australia, Ethiopia, Switzerland, Spain, Norway, Finland, Iceland, and USA (fig. 1A). Fourteen genetically distinct haplotypes were chosen from each field population with the exception of the USA population (n = 13). All isolates were previously characterized using microsatellite markers for population genetics studies (Stefansson et al. 2013; Stefansson, McDonald, et al. 2014; Stefansson, Willi, et al. 2014). We added isolates from two closely related Rhynchosporium species as outgroups, including nine isolates of R. secalis from Switzerland, France, and Russia as well as eight isolates of R. agropyri collected from different locations in Switzerland. Details for all Rhynchosporium spp. isolates used in this study are summarized in supplementary table 1, Supplementary Material online. Genomic DNA was isolated from mycelium grown in Potato Dextrose Broth (PDB) using DNeasy Plant Mini Kits (Qiagen) according to the manufacturer’s protocol. Paired-end sequencing of 125 bp reads was performed on a HiSeq2000 Illumina sequencer with an insert size of approximately 500 bp. The Illumina sequencing was performed to an average depth of 37×.
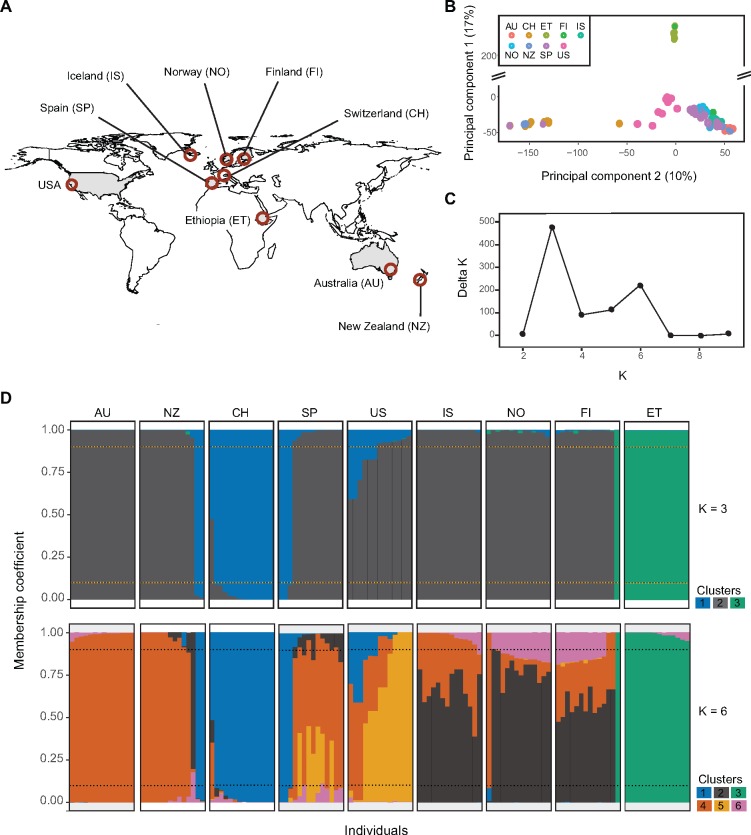
Fig. 1.
—Population structure analyses of 125 Rhynchosporium commune isolates from a worldwide collection based on genome-wide single nucleotide polymorphisms. (A) Sampling locations of R. commune included in this study. (B) Principal component analysis of genetic differentiation among isolates. The percentage of variance explained by the first two principal components is shown in parentheses. (C) Delta K plot of the STRUCTURE Bayesian clustering analyses for Ks of 2–9. (D) Genetic clustering using STRUCTURE for K = 3 (above) and K = 6 (below). Each colour represents one cluster and the vertical bar represents the proportion of cluster assignment for each genotype. Dotted lines are the minimum threshold value of membership coefficient for cluster assignment (90%).
Read Mapping, SNP Calling, and Quality Control
Summaries of the read mapping, SNP calling and quality control pipeline are shown in supplementary figure 1, Supplementary Material online. Low quality reads and traces of Illumina adapter sequences were trimmed using Trimmomatic version 0.32 (Bolger et al. 2014) with the following settings: trailing = 10, sliding-window = 4: 10, and minlen = 50. Resulting high-quality reads were mapped to the reference genome of R. commune (Penselin et al. 2016) using Bowtie2 version 2.2.3 (Langmead et al. 2009). Aligned reads were flagged for duplicates using the MarkDuplicates program in Picard tools version 1.119 (http://broadinstitute.github.io/picard/; last accessed August 2017).
Multi-sample single nucleotide polymorphism (SNP) calling was performed using two independent variant-calling software suites to obtain high confidence SNPs: the Genome Analysis Toolkit (GATK) version 3.3-0 (McKenna et al. 2010) and Freebayes version v1.0.2-29-g41c1313 (Garrison and Marth 2012). For GATK, SNPs were called using the tools HaplotypeCaller and GenotypeGVCF. The maximum number of alternative alleles was set to two with a minimum phred-scaled quality score of 500. In addition, SNPs were only retained if they matched the following criteria: QualByDepth > 20 (a measure of alternative allele quality independent of read depth), MQ > 30 (the minimum phred-scaled mapping quality of the reads supporting the alternative allele), FisherStrand < 10 (whether an alternative allele was predominantly supported by one read orientation only), −2 < BaseQualityRankSumTest < 2 (a test statistic to assess whether the base quality of reads supporting the alternative allele was significantly worse than reads supporting the reference allele), −2 < ReadPosRankSumTest < 2 (a test statistic to assess whether the base position in a read supporting the alternative allele was significantly different than the base position in a read supporting the reference allele), −2 < MappingQualityRankSumTest < 2 (a test statistic to assess whether the mapping quality of reads supporting the alternative allele was significantly worse than reads supporting the reference allele), RMSMappingQuality > 30 (an estimation of the overall mapping quality of reads supporting an alternative allele). For Freebayes, we genotyped only SNPs and ignored insertion/deletions (indels), multi-nucleotide polymorphisms (MNPs), and complex events. Reads were required to have a minimum mapping quality of 30. The maximum number of alternative alleles was set to the best two alleles ranked by the sum of supporting quality scores.
SNPs called by GATK and Freebayes were compared using vcftools version 0.1.12b (Danecek et al. 2011). We only retained SNPs that were detected by both SNP caller suites for subsequent analyses. A 92% of all SNPs retained from the GATK pipeline could be confirmed by FreeBayes (supplementary fig. 1, Supplementary Material online). The joint SNP data set was further filtered to retain only SNPs with a genotyping rate >90%. The SNP phred-scaled quality (QUAL) values and SNP allele frequencies called by GATK-HaplotypeCaller and FreeBayes were highly correlated (supplementary fig. 2A and B, Supplementary Material online). We found a strong positive correlation between QUAL and alternative allele frequencies at the SNP loci as expected. The QUAL of a SNP depends among other factors on the number of reads supporting the alternative (nonreference) allele. We identified a small number of SNPs in proximity to repeat-rich regions of the genome exhibiting low QUAL values regardless of the alternative allele frequency (supplementary fig. 2C and D, Supplementary Material online). The genotyping rate per isolate was >99% for 95% of all R. commune isolates. The remaining isolates had a genotyping rate >95%.
Isolates from the closely related sister species R. secalis (n = 9) and R. agropyri (n = 8) were sequenced to an average depth of 45× and 56×, respectively, and processed using the same procedure described for GATK above with the following exception: SNPs were retained if the genotyping rate was >50%. The relaxed genotyping threshold in these species stems from the fact that the sequencing reads of R. secalis and R. agropyri were mapped to the reference genome of R. commune. The multisample SNPs was performed with the inclusion of all R. commune isolates to ensure better annotation of SNPs identified in R. secalis and R. agropyri. The genotyping rate for R. secalis and R. agropyri isolates ranged from 94% to 96%. The lower genotyping rates are most likely due to the higher divergence between the R. commune reference genome used for mapping and the sister species. SNP genotypes retained for R. secalis and R. agropyri were used as outgroups to infer the ancestral state of SNPs in R. commune. An R. commune SNP allele was designated as ancestral if the allele was identified as fixed in both R. secalis and R. agropyri.
Prediction of Gene Functions
We predicted putative functions of all genes identified in the R. commune genome using InterProScan v.5.23-62.0 (Jones et al. 2014). The assignment of protein sequence motifs to protein families (Pfam) and gene ontology (GO) terms was performed based on hidden Markov models (HMM) implemented in InterProScan. Amino acid sequences were screened for evidence of secretion signals, and transmembrane, cytoplasmic and extracellular domains using a combination of SignalP v.4.1 (Petersen et al. 2011), Phobius v.1.01 (Käll et al. 2007), and TMHMM v.2.0 (Krogh et al. 2001). The R. commune secretome was defined as proteins predicted to include a secretion signal based on analyses using SignalP and Phobius. We removed any proteins with a predicted transmembrane domain based on Phobius and TMHMM analyses and a predicted cytoplasmic domain based on Phobius. We used the machine-learning approach implemented in EffectorP version 1.0 (Sperschneider et al. 2016) to identify the most likely effector proteins among the secreted proteins. We retained only predicted effectors with a posterior probability >0.8. All predicted secretomes were also screened for carbohydrate-active modules using the carbohydrate-active enzyme annotation (dbCAN) release 5.0 (Yin et al. 2012) for the identification of carbohydrate-active enzymes (CAZymes). We retained only protein hits with e-values <1e−17 and a coverage >45% following dbCAN recommendations (http://csbl.bmb.uga.edu/dbCAN/download/readme.txt; last accessed July 1, 2017). Repetitive elements in the reference genome of R. commune were annotated using RepeatModeler version 1.0.8 with default settings (Smit and Hubeley, RepeatModeler Open-1.0, 2008–2015; http://www.repeatmasker.org; last accessed August 1, 2017). RepeatModeler combines two de novo repeat finding algorithms (RECON and RepeatScout) to identify repetitive sequences in a genome. In the final step, the RepeatModeler annotation of repetitive sequences was matched against known repeat elements (Repbase version 20160629) using repeatmasker 4.0.6 (Smit, Hubley, and Green, RepeatMasker Open-4.0, 2013–2015; http://www.repeatmasker.org; last accessed August 1, 2017).
Analyses of Population Structure
Genetic structure among R. commune samples was analysed using a principal component analysis (PCA) implemented in TASSEL version 20150625 (Bradbury et al. 2007). We also performed an unsupervised model-based Bayesian clustering implemented in STRUCTURE version 2.34 (Pritchard et al. 2000) to assign individuals to subgroups. To reduce the computational load, the SNP data set used in STRUCTURE was reduced to one variant per 5 kb using the “–thin” option in vcftools, leaving 8,235 SNPs. The assignment of genotypes to clusters was run for total cluster numbers ranging from K = 1 to 10. We used the admixture model with correlated allele frequencies and no prior information about the demography. Each of the different Ks was replicated 10 times with a burn-in period of 50,000 samples followed by 100,000 Monte Carlo Markov chain replicates. Parameter convergence was inspected visually. The output of the STRUCTURE analysis was processed using STRUCTURE HARVESTER (Earl and vonHoldt 2012) and the optimal number of subgroups was determined using the ΔK method (Evanno et al. 2005).
Genome-Wide Scans for Selection
Scans for selective sweeps were performed using two haplotype-based methods: integrated haplotype scores (iHS) and cross-population EHH (XP-EHH) implemented in the rehh package version 2.0.2 (Gautier et al. 2017) in R. For both analyses, we restricted our data set to include only SNPs for which the ancestral allele was known (see above). We also removed all SNP loci which were not segregating variation in the cluster for which the selection scan was performed to avoid potential issues introduced by SNP window-based analyses. The iHS analyses were performed independently for each of the genetic clusters identified with STRUCTURE using only SNPs that were polymorphic within the genetic cluster. Significant SNPs were defined by selecting the top 0.05% of |iHS| for clusters 1 and 3, and top 0.01% of |iHS| for the large cluster 2. We used a more stringent outlier threshold for cluster 2 because this cluster was genetically (and geographically) more heterogeneous than clusters 1 and 3. For the XP-EHH analyses, we performed the test using all SNPs which were genotyped in at least 90% of isolates within each of the genetic clusters. SNPs were only retained if the ancestral allele was assigned (see above). The XP-EHH analyses were performed on pairwise comparisons of cluster 1 and cluster 3 against cluster 2 as the reference population unless stated otherwise. Significant SNPs were selected from the top 0.05% of |XP-EHH| for all clusters. We used default options for all analyses. However, we set the maxgap option to 20,000 whenever calc_ehh, calc_ehhs, or scan_hh was used. In addition to that, the threshold of missing data for haplotypes and SNPs was set to 90%.
Significant SNPs identified from either of the two selection scan methods and located on the same scaffold were grouped using a hierarchical clustering approach. If the distance between significant SNPs was below 50,000 bp, the SNPs were grouped into a single region under selection. Then, the extent of the region under selection was further refined by computing the EHH for each significant SNP within a region under selection. The extent of the EHH above 0.05 was used to define windows around each SNP contained within a region and overlapping windows were then merged.
We analyzed coding sequences for ratios of synonymous and nonsynymous polymorphisms. For this, we used SnpEff version 4.3p (Cingolani et al. 2012) to distinguish SNP variant effects. We also performed a McDonald–Kreitman (MK) test (McDonald and Kreitman 1991) on coding sequence SNPs between R. commune and R. secalis (as an outgroup). We used a perl script to extract fixed and polymorphic sites from a variant call format (vcf) file and perform the MK test. The script mkt.pl was written by Santiago Sánchez-Ramírez and is available from https://sites.google.com/site/santiagosnchezrmirez/home/software/perl; last accessed February 1, 2018.
Gene Function Enrichment Analyses
Enrichment analysis of GO categories was performed using the R package GSEABase and GOstats (Falcon and Gentleman 2007; Anders et al. 2015) with a false discovery rate set to 0.05. The minimum GO term size to be considered for enrichment analyses was set to at least five members in the reference genome. The enriched terms were then summarized by REVIGO (Supek et al. 2011) by removing redundant GO terms. We also analyzed evidence of enrichment in secreted proteins, effectors, cell wall degrading enzymes, and major facilitator superfamily (MFS) transporters using a hypergeometric test in R.
Results
Identification of High Quality Single SNPs with Known Ancestral State
We sequenced 125 R. commune isolates collected from single field populations in New Zealand, Australia, Ethiopia, Spain, Switzerland, Norway, Finland, Iceland, and the USA. The sample size per population was 13–14 isolates. We identified a total of 736,839 high quality SNPs using the GATK pipeline. We could confirm 92% of these SNPs by performing an independent SNP calling procedure using FreeBayes (supplementary fig. 1, Supplementary Material online). For all further analyses, we retained only SNPs identified by both SNP callers and genotyped in >90% of the isolates, resulting in a data set of 584,053 SNPs (supplementary fig. 1, Supplementary Material online). SNPs sharing a fixed allele between the two sister species were used to assign the ancestral state of SNPs identified in R. commune. Ancestral states were assigned for 481,424 SNPs (82.4% of all retained R. commune SNPs). These SNPs were unevenly distributed along scaffolds (fig. 2B) and regions of high repeat density were largely devoid of callable SNPs due to the strict filtering procedure (fig. 2C).
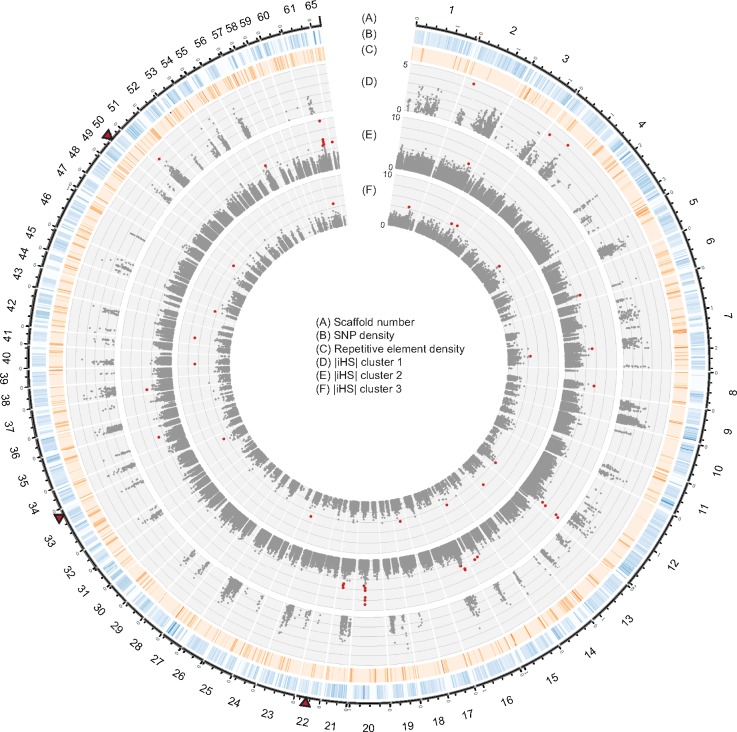
Fig. 2.
—Genome-wide selection scans using the standardized integrated haplotype |iHS| score in three genetic clusters of Rhynchosporium commune. (A) Scaffolds of the reference genome and positions in Mb. (B) Density of single nucleotide polymorphisms (SNPs) across the whole genome shown in 10 kb nonoverlapping windows (gradient shows coverage between 0% and 5%). The total number of single nucleotide polymorphisms (SNPs) with known ancestry was 481,424. (C) Coverage of repetitive elements across the whole genome shown in 10 kb nonoverlapping windows (gradient shows coverage between 0% and 100%). (D–F) Dots show |iHS| values for each SNP within the clusters 1, 2, and 3, respectively. The most significant SNPs (top 0.05% for clusters 1 and 3, and top 0.01% for cluster 2) were highlighted in red. Triangles near the outermost plot indicate the location of loci most significantly associated with resistance to the fungicide cyproconazole (Bonferroni threshold α = 0.05) in a genome-wide association study (Mohd-Assaad et al. 2016).
Population Structure Analyses Based on Genome-Wide SNPs
We performed principal component (PC) and Bayesian clustering to analyse evidence for population subdivisions. The first two PCs explained 17% and 10% of the total genetic variation, respectively, and grouped the isolate genotypes into three clusters. Most populations including Scandinavia, New Zealand, Australia, and Spain were clustered into a single large group, whereas Ethiopian and Swiss populations formed two individual clusters distinct from the main group (fig. 1B). The unsupervised Bayesian STRUCTURE analysis identified two probable genotype clustering scenarios at K = 3 and K = 6. K = 3 received the highest support consistent with the PCA (fig. 1B and C). For K = 3, cluster 2 was found to be the most dominant cluster (fig. 1C and D). All genotypes from the Ethiopian population were assigned to a distinct cluster (fig. 1D). Genotypes from the Swiss population were mostly assigned to cluster 1, however, several genotypes showed mixed ancestries from cluster 1 and 2 (fig. 1D). Interestingly, two and three genotypes from New Zealand and Spain, respectively, were assigned to the cluster containing mainly Swiss isolates. In addition, six genotypes from the US population showed mixed ancestry with significant contributions from clusters 1 and 2. One genotype from the Finland population clustered with Ethopian genotypes. Because a migration event from Ethiopia to Finland is highly unlikely, we suspected human error and excluded this isolate from further analyses. After discarding admixed isolates, we retained a total of 18, 85, and 14 isolates in cluster 1, cluster 2, and cluster 3, respectively, for the selection analyses.
Evidence for Recent Positive Selection Identified by iHS
We screened for loci that experienced recent positive selection using EHH-based statistics. These statistics contrast the frequency of a haplotype to its relative EHH (defined from a core ancestral or derived allele). For all analyses, we used only SNP markers, for which the ancestral state could be assigned. First, we calculated the iHS statistics that compare the area under the EHH curve of the ancestral and derived alleles from the core allele. Performing the analyses separately for each genetic cluster, we identified a total of 39 genomic regions with signatures of selection across 27 scaffolds (fig. 2; supplementary table 2, Supplementary Material online). The length of regions under selection ranged from 1,851 to 401,914 bp and contained between 2 and 108 genes (supplementary table 2, Supplementary Material online). The length and total number of genes located within the selective sweep regions identified by the iHS test for each of the cluster is summarized in supplementary figure 3, Supplementary Material online. We found that all regions under selection identified by iHS were each unique to an individual cluster. The highest percentage of identified loci was found in cluster 2 (46%), followed by cluster 3 (44%) and cluster 1 (10%). As the SNP density varied considerably along scaffolds, we tested whether SNP density had an impact on the detection of selection. We found that the SNP density of regions identified to be under selection did not vary substantially from the SNP density across the genome (supplementary fig. 4, Supplementary Material online). Exceptions were regions lacking any confidently identified SNPs, as we required a maximum distance between SNPs of 20 kb for selection scans. Regions devoid of confidently identified SNPs showed higher percentages of repetitive elements than the genome-wide average (supplementary fig. 4, Supplementary Material online). We also found that the length of selective sweep regions was not correlated with SNP density (supplementary fig. 5, Supplementary Material online). For each cluster, we analyzed the ratio of synonymous versus nonsynonymous polymorphisms segregating in genes located in selective sweep regions (supplementary fig. 6, Supplementary Material online). We found that the ratio was not meaningfully different compared with the genome-wide background. We performed MK tests on polymorphism detected in R. commune and R. secalis as an outgroup. We found only 21 genes with completely fixed differences between the species. No gene showed a significantly different ratio for synonymous versus nonsynonymous sites within and between species.
To investigate candidate genes likely to be under the strongest selection pressure, we focused on the three most significant SNPs identified by iHS in all clusters. We found a total of 15 candidate genes that were located within 5,000 bp of the most significant SNPs (table 1). For cluster 1, we found two predicted candidate genes on scaffold 2, including the gene RCO7_04421 encoding a glucose–methanol–choline (GMC) oxidoreductase involved in plant cell wall degradation and RCO7_04423 encoding a multi-antimicrobial extrusion (MATE) multidrug transporter involved in the secretion of a wide range of metabolic and xenobiotic substances. In addition, we identified the gene RCO7_06862 on scaffold 4, encoding a KES1 oxysterol-binding protein with a role in the fungal ergosterol biosynthesis pathway. The gene RCO7_03674 encodes a protein belonging to the acyltransferase (GNAT) family that catalyzes the transfer of an acyl group from acyl coenzyme A to an amino group of various substrates. In close proximity to the third most significant SNP on scaffold 51, we identified RCO7_03675 which encodes an alpha-mannosidase involved in the degradation of the plant cell wall. In cluster 2, we found the candidate gene RCO7_01309 on scaffold 61, which encodes an enzyme that catalyses the removal of an ammonia group from glutamine in a variety of substrates. The gene RCO7_02709 on scaffold 12 encodes a membrane-embedded UbiA prenyltransferase. In cluster 3, we found the candidate gene RC07_07301 on scaffold 42 encoding a concentrative nucleoside transporter, which mediates the uptake of nucleosides and nucleobases across the plasma membrane. We found no overlap between loci under selection and SNPs significantly associated with azole resistance in a previous GWAS analysis (Mohd-Assaad et al. 2016). As we found no effector candidate genes in the regions likely under the strongest selection pressure, we tested whether SNP density had an effect on the ability to detect selection on effector candidate genes. However, we found that effector candidate genes were on average in relatively SNP-dense regions (supplementary fig. 4, Supplementary Material online).
Table 1.
Genes in Selective Sweep Regions Identified Using Integrated Haplotype Score (|iHS|) Analyses in the Three Genetic Clusters of Rhynchosporium commune
| Cluster | Scaffold | SNP Position | |iHS| | Genes in Sweep Region |
|---|---|---|---|---|
| Cluster 1 | RCO7_scaffold002 | 262395 | 4.57691295 | RCO7_04421 (glucose–methanol–choline (GMC) oxidoreductase) |
| RCO7_04422 (related to DUF 202 domain protein) | ||||
| RCO7_04423 (multi-antimicrobial extrusion (MATE) multidrug transporter) | ||||
| Cluster 1 | RCO7_scaffold004 | 707629 | 4.655198545 | RCO7_06862 (KES1 oxysterol-binding protein involved in ergosterol biosynthesis) |
| Cluster 1 | RCO7_scaffold051 | 125465 | 3.966106351 | RCO7_03673 (uncharacterized protein) |
| RCO7_03674 (acyltransferase GNAT family) | ||||
| RCO7_03675 (glycosyl hydrolase family 47 alpha-mannosidase) | ||||
| Cluster 2 | RCO7_scaffold061 | 469958 | 9.75730438 | RCO7_01309 (class I glutamine amidotransferase (GAT1)-like protein) |
| Cluster 2 | RCO7_scaffold012 | 1864317 | 9.31713174 | RCO7_02708 (uncharacterized protein) |
| RCO7_02709 (UbiA prenyltransferase family) | ||||
| Cluster 2 | RCO7_scaffold020 | 646208 | 8.83413424 | RCO7_14617 (uncharacterized protein) |
| Cluster 3 | RCO7_scaffold042 | 286253 | 7.03521568 | RCO7_07300 (uncharacterized protein) |
| RCO7_07301 (concentrative nucleoside transporter (CNT)) | ||||
| Cluster 3 | RCO7_scaffold040 | 75641 | 6.70227808 | Intergenic region |
| Cluster 3 | RCO7_scaffold013 | 900028 | 5.4617104 | RCO7_00065 (uncharacterized protein with SKP1/BTB/POZ domain) |
| RCO7_00064 (uncharacterized protein) |
Note.—Genes were selected if they were within 5,000 bp from the most significant SNPs (top 0.05% in clusters 1 and 3, and 0.01% in cluster 2).
Positive Selection Identified by Cross-Population Extended Haplotype Homozygosity
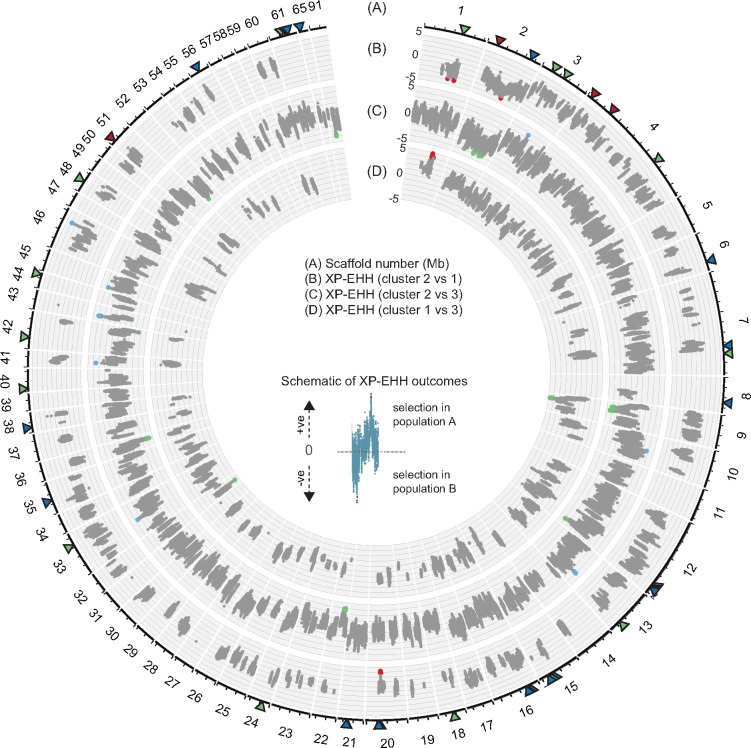
Unlike analyses of iHS comparing haplotype lengths within clusters, the XP-EHH test compares the profile of EHH between pairs of clusters at each focal SNP. We identified a total of 29 selective sweeps distributed across 19 scaffolds in all 3 pairwise cluster comparisons (fig. 3; supplementary table 2, Supplementary Material online). The length of regions under selection ranged from 1,851 to 721,862 bp and contained 2 to 224 genes among the pairwise XP-EHH analyses (supplementary table 2, Supplementary Material online). We summarized the length and total number of genes within the selective sweep regions in supplementary figure 3, Supplementary Material online. Fifty-two percent of these selective sweeps were identified in cluster 3 while the remaining sweeps were shared evenly between cluster 1 and cluster 2. Interestingly, the selection targets on scaffold 1 (616,499–1,059,480 bp) and scaffold nine (159,071–349,486 bp) were identified in two different pairwise comparisons (fig. 3; supplementary table 2, Supplementary Material online). In addition to that, a selective sweep on scaffold 2 (589,559–926,564 bp) was identified in cluster 1 and cluster 3 when XP-EHH was performed using cluster 2 as the reference cluster. Four of these selective sweeps (scaffold 1: 961,484–984,586 bp, scaffold 3: 623,832–625,683 bp, scaffold 12: 1,753,708–1,809,408 bp and scaffold 20: 533,694–710,548 bp) overlapped with selective sweeps identified using iHS. A total of 85 genes were found in overlapping selective sweep regions. However, each of these selective sweeps was detected in different genetic clusters than the sweeps identified by iHS. Using the same procedure as for selective sweeps detected by iHS, we tested whether SNP density had an effect on the detection of selection. We found no evidence that SNP density had a meaningful impact on the ability to detect selection or on the length of identified selective sweeps (supplementary figs. 4 and 5, Supplementary Material online). For each XP-EHH analysis of cluster pairs, we analyzed the ratio of synonymous versus nonsynonymous polymorphisms segregating in genes located in selective sweep regions (supplementary fig. 6, Supplementary Material online). We found that the ratio was not meaningfully different compared with the genome-wide background.
Fig. 3.
—Genome-wide scan of the cross-population extended haplotype homozygosity (XP-EHH) analyses. (A) Scaffolds of the reference genome and positions in Mb. XP-EHH analyses of (B) cluster 2 versus cluster 1, (C) cluster 2 versus cluster 3, (D) cluster 1 versus cluster 3. The positive and negative values in (B)–(D) indicate the direction of selection. Positive values indicate positive selection in the reference population (cluster 2 was used as the reference population in (B) and (C), cluster 1 in (D)). The most significant SNPs (top 0.05% of |XP-EHH|) are highlighted in red, blue, and green for signals of positive selection in cluster 1, 2, and 3, respectively. Triangles near the outermost plot show the most significant SNPs identified by the standardized integrated haplotype |iHS| analyses (red for cluster 1, blue for cluster 2 and green for cluster 3).
We focused on genes located in selective sweep regions identified by both XP-EHH and iHS analyses. We found a total of 10 genes that were located within 5,000 bp of the most significant SNPs identified by both XP-EHH and iHS analyses (table 2). The two genes identified on scaffold 1 were RCO7_01116, encoding a phthalate 4,5-dioxygenase oxygenase reductase subunit involved in the degradation of xenobiotics, and an unknown protein encoded by RCO7_01117. We identified an ubiquitin-conjugating enzyme E2 encoded by RCO7_02480 and a MON2 protein encoded by RCO7_02479 on scaffold 3. MON2 plays an important role in endomembrane trafficking and Golgi homeostasis. We identified the RCO7_02696 gene on scaffold 12 that encodes a mediator complex subunit 7 (MED7) protein involved in the transcriptional regulation of nearly all RNA polymerase II-dependent genes. In addition to that we identified RCO7_02697, which encodes a type 2 phosphatidic acid phosphatase (PAP2) gene important for lipid metabolism and signaling. We tested whether SNP density had an effect on the ability to detect selection on effector candidate genes in the XP-EHH analyses. However, effector candidate genes were on average in relatively SNP-dense regions as for the SNP sets used for iHS scans (supplementary fig. 4, Supplementary Material online).
Table 2.
Genes Identified in Overlapping Selective Sweep Regions
| Regions | Scaffold | Selection Analyses/Cluster Identities | SNP Position | |iHS|/XPEHH | Candidate Genes under Selection |
|---|---|---|---|---|---|
| 1 | RCO7_scaffold001 | (a) XP-EHH—cluster 1 versus Cluster 3 | 962864 | 4.785 | (a) RCO7_01115 (transcription factor Som1) |
| (b) iHS—cluster 3 | 963272 | 4.343 | RCO7_01116 (phthalate 4,5-dioxygenase oxygenase reductase subunit) | ||
| RCO7_01117 (uncharacterized protein) | |||||
| 2 | RCO7_scaffold003 | (a) XP-EHH—cluster 2 versus Cluster 3 | 624918 | 3.564 | RCO7_02480 (ubiquitin-conjugating enzyme E2) |
| (b) iHS—cluster 3 | 624918 | 4.6574 | RCO7_02479 (MON2) | ||
| 3 | RCO7_scaffold012 | (a) XP-EHH—cluster 2 versus cluster 3 | 1822306 | −3.977 | (a) RCO7_02695 (uncharacterized protein) |
| (b) iHS—cluster 2 | 1823054 | 8.694 | RCO7_02696 (mediator complex subunit 7 (MED7)) | ||
| 1823058 | 8.694 | RCO7_02697 (Type 2 phosphatidic acid phosphatase (PAP2)) | |||
| 4 | RCO7_scaffold020 | (a) XP-EHH—cluster 2 versus cluster 1 | 647275 | −4.251 | RCO7_14617 (uncharacterized protein) |
| (b) iHS—cluster 2 | 646208 | 8.834 | (a) RCO7_03084 (uncharacterized protein) | ||
| 5 | RCO7_scaffold001 | (a) XPEHH—cluster2 versus cluster1 | 1026013 | 3.914 | RCO7_01132 (G-protein alpha chain) |
| (b) XPEHH—cluster1 versus cluster3 | 1025700 | 4.130 | |||
| 6 | RCO7_scaffold009 | (a) XPEHH—cluster 2 versus cluster 3 | 205732 | −4.091 | (a) intergenic |
| (b) XPEHH—cluster 1 versus cluster 3 | 204366 | −4.684 | (b) RCO7_06601 (Cu/Zn superoxide dismutase) | ||
| 7 | RCO7_scaffold002 | (a) XPEHH—cluster 2 versus cluster 1 | 818548 | −3.749 | RCO7_04583 (transmembrane protein with MARVEL domain) |
| (b) XPEHH—cluster 2 versus cluster 3 | 818548 | −3.574 | RCO7_04584 (domain of unknown function DUF2012) |
Note.—Regions 1–4 show selective sweep regions identified by both integrated haplotype score (|iHS|) and cross-population extended haplotype homozygosity (XP-EHH) analyses. Regions 5–7 show selective sweep regions identified in two separate XP-EHH analyses. Genes were reported if they were within 5,000 bp of the most significant single nucleotide polymorphism (SNP) locus (top 0.05%). For each XP-EHH selection analyses, the genetic cluster in which selection was detected is shown in bold. Gene names shown in bold are genes found in the overlap of both selection analyses. Gene names labelled with either (a) or (b) refer to which selection analyses identified these genes (see third column).
In order to identify additional candidate genes for local adaptation, we focused on selective sweep regions identified by two sets of cross-population tests. We found the gene RCO7_01132 located on scaffold 1 under positive selection in cluster 1 (using the cluster pairs 2–1 and 1–3 for XP-EHH analyses). This gene codes for a guanine nucleotide binding protein (G-protein) alpha chain, which plays an important role in various signaling systems. However, the two most significant SNPs identified on scaffold 9 in cluster 3 were not in proximity to any known gene. Using cluster 2 as the reference cluster, the strongest signal of a selective sweep was found in a large intergenic region. Using cluster 1 as the reference cluster, we identified RCO7_06601 encoding a Cu/Zn superoxide dismutase located 1,758 bp away from the top significant SNP. Cu/Zn superoxide dismutases play a role in protecting cells from damage caused by oxygen-mediated free radicals. Finally, we found two candidate genes on scaffold 2 shared by clusters 1 and 3 (when compared against cluster 2). The first candidate gene was RCO7_04583, encoding a transmembrane gene with a MARVEL domain which plays a role in membrane apposition events. The function of the second gene (RCO7_04584) is unknown.
Gene Functions Overrepresented in Selective Sweep Regions
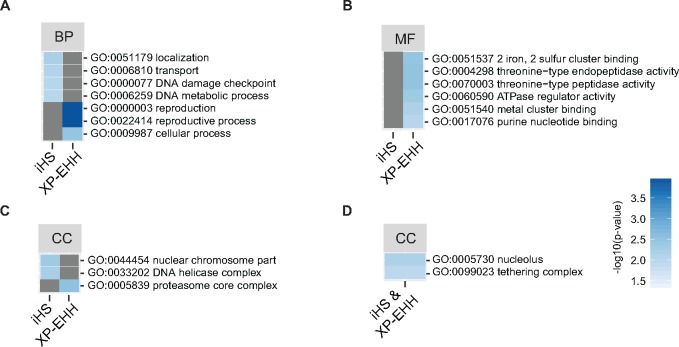
We found a total of 972 and 811 genes in selective sweep regions detected by iHS and XP-EHH analyses, respectively. We analyzed whether genes under selection were enriched for specific functions by performing a GO enrichment analysis. We found 16 terms that were significantly enriched (P < 0.01) for different functional categories (fig. 4; supplementary table 3, Supplementary Material online). Enriched GO terms did not overlap between gene sets identified by iHS and XP-EHH analyses, respectively. In iHS analyses, we found enrichment for localization (i.e., any process involved in the establishment and maintenance of cellular location) (GO: 0051179), transport (GO: 0006810), DNA damage checkpoint (GO: 0000077), and DNA metabolic process (GO: 0006259). Interestingly, we also found strong enrichment of the term reproduction (GO: 0000003, P = 0.0001) and reproductive processes (GO: 0022414, P = 0.0001) in regions under selection identified by XP-EHH. We investigated the genes encoding functions assigned to the GO term reproduction and related functions and found that these genes were related to meiosis and plasma membrane fusion, including a meiosis specific protein Spo22/ZIP4/TEX (RCO7_00959), a plasma membrane fusion protein PRM1 (RCO7_01086), and a DNA repair protein RAD51 (RCO7_01125). In addition, we investigated whether selective sweep regions identified by both iHS and XP-EHH were enriched for specific gene functions. We found enrichment for nucleus associated functions (GO: 0005730, P = 0.0055) and vesicle tethering (GO: 0099023, P = 0.0092; fig. 4; supplementary table 3, Supplementary Material online). We found no evidence for enrichment in genes encoding secreted proteins, effectors, plant cell wall degrading enzymes or MFS transporters in regions under selection (supplementary table 4, Supplementary Material online).
Fig. 4.
—Gene ontology (GO) enrichment analyses for genes within selective sweep regions. Enriched GO terms related to biological process (BP), molecular functions (MF) and cellular components (CC) are shown for analyses of the integrated haplotype score (iHS) and cross-population extended haplotype homozygosity (XP-EHH). Loci were combined for all individual analyses of the three genetic clusters. GO terms were simplified using REVIGO prior to enrichment analyses and only significantly enriched terms P < 0.01 are shown.
Discussion
We used population whole-genome sequencing to identify signatures of recent selection in populations of the barley scald pathogen. Using two different haplotype-based selection scans, we found widespread signals of divergent selection in three genetic clusters of R. commune. The strongest signatures of selection were predominantly found in two of the three clusters. Contrary to expectations for a host specialized pathogen, loci showing strong evidence for selection were largely unrelated to adaptation to the host plant. Instead, we found enrichment in genes encoding functions in cellular localization, protein transport activity, DNA damage response, and general metabolism. Genes related to reproductive processes were found to be significantly enriched in selective sweeps identified by cross-population analyses.
The global collection of the pathogen R. commune showed strong evidence for subdivision into three genetic clusters. A total of 85 genotypes were assigned to a predominantly northern European cluster. This cluster also comprised the genotypes collected near the center of origin of the pathogen (i.e., Scandinavia) (Zaffarano et al. 2006). Previous findings based on multilocus sequence data showed that R. commune likely expanded across the world from its center of origin (Brunner et al. 2007; Zaffarano et al. 2009). Nevertheless, contemporary gene flow was estimated to be low among continents. Our analyses of genome-wide SNPs confirmed the strong subdivision among continents. For example, the Swiss and Ethiopian populations showed striking genetic differentiation from the main cluster and each constituted largely independent clusters. However, multiple genotypes showed admixture between clusters suggesting ongoing genetic exchange. Interestingly, two and three genotypes of New Zealand and Spain, respectively, were assigned to the Swiss genetic cluster (>90% posterior probability), suggesting recent migration events and the absence of significant admixture. The US population showed frequent admixture from Swiss and Scandinavian populations showing that these two donor populations are playing an important role in the spread of R. commune to the North American continent. Previous studies suggested that gene flow from Europe was facilitated by the introduction of infected seeds during European colonization and may be ongoing to the present day (Brunner et al. 2007; Zaffarano et al. 2008; McDonald 2015). Despite evidence for migration events, gene flow was severely restricted among the genetic clusters. This should favor the emergence of local adaptation.
We performed genome-wide selection scans independently for each of the three genetic clusters and excluded admixed genotypes to avoid confounding effects of population structure. We found widespread evidence for selective sweeps across the genome, yet there was little overlap in loci under selection among clusters. The divergent selection pressures among clusters strongly suggest that differences in ecological factors imposed different selection regimes. For instance, in Ethiopia barley is grown over a broad climatic and edaphic range (barley is grown at altitudes from 1,400 to over 4,000 m above the sea level). Nevertheless, the pathogen is exposed to less seasonal temperature fluctuations than in Scandinavia (Asfaw 2000; Stefansson et al. 2013). Similarly, the climate in Oceania covers different seasonal fluctuations than Central European localities. Furthermore, our samples were isolated from genetically diverse barley landraces in Ethiopia (Hadado et al. 2010) while the Scandinavian and Swiss clusters comprised pathogen populations that were exposed to genetically homogeneous elite barley cultivars. Additional differences in environmental conditions include the early application of azole fungicides in Europe. Azole resistance was detected in many fungal pathogens in Europe, including R. commune, because the late 1990s (Cools et al. 2013). The Swiss population of R. commune already accumulated a number of azole resistance mutations as early as 1999 (Brunner et al. 2015; Mohd-Assaad et al. 2016). Taken together, the three genetic clusters comprised populations exposed to quite different environmental conditions, providing substantial opportunity for local adaptation.
Our scans for selective sweeps in each of the three genetic clusters using iHS revealed a large number of loci under recent selection. These loci were broadly distributed across the genome. Importantly, we found little overlap in loci among the major genetic clusters, strongly suggesting differences in selection pressures among these clusters. Because R. commune has a well assembled and annotated genome, we investigated genes in immediate proximity to each of the selective sweep regions. We focused on the strongest signals of selection in each cluster and retrieved a total of 15 genes adjacent to these sweep regions. Half of these genes coded for proteins with a conserved function, including two membrane-bound transporters. The first MATE family transporter is involved in various metabolic pathways (Moriyama et al. 2008; Kuroda and Tsuchiya 2009; Liu et al. 2016). In the rice pathogen Pyricularia oryzae, an MATE-family pump regulates glucose assimilation, sporulation, and pathogenicity (Fernandez et al. 2012). The second transporter mediates the uptake and release of nucleosides (Young et al. 2013). It is unclear what selection pressure would act on such a highly conserved transporter. Previous studies showed that nucleoside transporters can help attenuate nitrogen starvation, restore nucleotide pools and regenerate mycelium growth in nitrogen-free medium (Hamari et al. 2009; Dean et al. 2014; Daumann et al. 2016). Uptake of nucleoside and nitrogenous compounds are important for pathogenicity in Candida albicans, Xenorhabdus nemaptophila and obligate parasitic protozoan (de Koning et al. 2005; Orchard and Goodrich-Blair 2005; Chitty and Fraser 2017). We also identified a GNAT acyltransferase that plays a role in multiple biological processes including the regulation of growth, morphogenesis, cellulase expression, and stress–response pathways related to the host and abiotic environment (O’Meara et al. 2010; Xin et al. 2013; Xue-Franzén et al. 2013). Additional genes in immediate proximity to selective sweeps encoded a GMC oxidoreductase and an alpha-mannosidase contributing to the degradation of plant cell walls including lignin and mannose (Choi et al. 2013; Takahashi et al. 2015).
Fungicide resistance, in particular to azoles, evolved repeatedly in many plant pathogens (Cools et al. 2013). Several of the R. commune populations analyzed in this study were exposed to azoles and multiple loci were shown to confer increased resistance (Brunner et al. 2015; Mohd-Assaad et al. 2016). Azole resistance was found to be particularly high among isolates assigned to cluster 1. Interestingly, we identified a gene encoding the KES1 oxysterol-binding protein to be under selection in cluster 1. Mutations in the KES1 gene can result in hypersensitivity to fungicides such as azoles that target ergosterol biosynthesis (Fang et al. 2012). Nevertheless, we found no evidence for positive selection at the loci previously associated with azole resistance by GWAS (Mohd-Assaad et al. 2016). This may be due to a combination of factors. First, fungicides were applied only for the last 30–40 years and the number of generations that has elapsed may have been too short for a strong selection signal to arise. Second, previous studies showed that azole resistance is a polygenic trait with possibly dozens of loci contributing to the overall level of resistance (Mohd-Assaad et al. 2016) making the detection of selective sweeps at individual loci less likely. Third, many mutations that conferred higher levels of resistance were also found in sensitive populations, suggesting that azole resistance at least partially arose from standing genetic variation. Fourth, some resistance mutations had negative pleiotropic effects on growth rates (Mohd-Assaad et al. 2016). Negative pleiotropy would weaken the response to selection due to fungicide exposure. Furthermore, the pathogen is only exposed to significant levels of fungicides during the crop growing season. Hence, resistance mutations with negative pleiotropic effects are likely under fluctuating selection which is not expected to generate strong signatures of selection.
In addition to our scans of recent positive selection within each genetic cluster, we used XP-EHH among pairs of clusters to gain sensitivity in the detection and to identify differences in signals among clusters. We focused in particular on loci that were confirmed to be under selection in a specific cluster by analyzing both pairwise comparisons with the other two clusters. We found such cross-validated loci for clusters 1 and 3. A gene encoding a guanine nucleotide binding protein was under strong selection specific to cluster 1. This protein is a part of heterotrimeric complex that has wide ranging roles in signaling, including known effects on reproduction, developmental processes and virulence in fungi (Gao and Nuss 1996; Horwitz et al. 1999; Gronover et al. 2001; Yamagishi et al. 2006; Tan et al. 2008). In cluster 3, a cross-validated gene encodes a Cu/Zn superoxide dismutase. These proteins enable pathogens to withstand reactive oxygen species, which are among the primary defense responses of plants during attack by pathogens (Yao et al. 2016; Rolke et al. 2004). Given the signatures of selection unique to individual clusters, both genes are strong candidates for genes underlying local adaptation.
Detecting signatures of the most recent episodes of selection is challenging because neutral processes can generate signatures indistinguishable from selection (Vitti et al. 2013; Xiang-Yu et al. 2016). Loci across the genome can show shifts in allele frequency spectra due to recent population bottlenecks or expansions. Genome-wide selection scans can control for the effects of demographic changes to some extent because neutral processes are affecting all loci across the genome in contrast to the localized impact of selection (Biswas and Akey 2006). Haplotype block structure-based tests for selection such as iHS and XP-EHH used in this study are particularly powerful in identifying the specific alleles under recent selection by taking ancestral state information into account (Voight et al. 2006). iHS is robust to heterogeneity in recombination rates across the genome (Sabeti et al. 2002; Xiang-Yu et al. 2016). iHS is also fairly robust to SNP ascertainment bias affecting the allele frequency spectrum and demography (Voight et al. 2006; Xiang-Yu et al. 2016). However, false positive rates can be high if the populations experienced extreme bottlenecks. Historic population sizes of R. commune fluctuated over time, with a bottleneck estimated to have occurred 250–500 years before present ending a slow contraction over thousands of years (Zaffarano et al. 2008). The bottleneck experienced by R. commune was unlikely to be strong enough to explain a substantial portion of the observed haplotype block structure. We focused our analyses mainly on the strongest signals of selection. However, relatively weak signals of selection in the analyzed populations should be cautiously evaluated for their significance.
We performed cross-population tests of selection that aimed at identifying loci potentially involved in local adaptation. However, highly differentiated genomic regions among populations can also arise in the absence of local adaptation (Bierne et al. 2011). Underappreciated environment-independent factors such as pre- and post-zygotic isolation or epistasis can lead to strong reductions in gene flow at specific loci. If such factors coincide with ecological variation, local adaptation is expected to produce indistinguishable signatures at loci under selection. However, we analyzed complete genomes instead of a relying on marker techniques that generate only reduced marker sets. Hence, we were able to analyze putative functions within selective sweep regions and found that many loci were indeed likely to encode functions related to biotic or abiotic adaptation. Analyzing the contribution of individual loci to phenotypic traits will enable a clear distinction between loci underlying local adaptation and endogeneous barriers.
Genes identified in selective sweep loci were only weakly connected with the repertoire of genes usually associated with highly specific host–pathogen interactions. Genes underlying host specialization often encode small secreted proteins (i.e., effector candidates). Such proteins directly or indirectly interact with a host receptor and modulate the host physiology to facilitate infection (de Jonge et al. 2011). In the genomes of many filamentous pathogens, genes encoding small effector genes are predominantly located in proximity to repeat-rich regions (Raffaele and Kamoun 2012; Grandaubert et al. 2014; Plissonneau et al. 2016; Hartmann et al. 2017). Repetitive sequences are often devoid of high-quality callable SNPs. We tested whether genes encoding small secreted proteins (i.e., effector candidates) were located in regions without a sufficient density of high-quality SNPs. However, we found that regions containing genes encoding small secreted proteins showed similar levels of SNP density compared with the genomic background. Therefore, the lack of effector candidate genes in selective sweep regions is unlikely to be explained by callable SNP density variation.
A major gap in our understanding of host specialization is how many effectors play a role in modulating host immune responses to the pathogen's advantage. Bioinformatics screens of fungal pathogen genomes typically reveal hundreds of genes encoding putative effectors (Jones et al. 2018). However, population-level association mapping analyses suggested that a relatively small number of effectors could explain most variation in virulence (Fouché et al. 2018). Necrotrophic pathogens such as R. commune may not require a large arsenal of highly specialized effectors. We also found no evidence that genes encoding additional essential functions for plant pathogens such as cell wall degrading enzymes, proteases, lipases, and MFS transporters were over-represented among selective sweep loci. These proteins play critical roles in the degradation of cell walls (Esquerré-Tugayé et al. 2000; Kikot et al. 2009), the secretion of virulence factors or detoxification (Coleman and Mylonakis 2009). In contrast, we found that selective sweep loci were overrepresented in genes encoding functions in transport, cellular localization and DNA damage repair. Such functions are more likely to be associated with selection pressures exerted by the abiotic environment (e.g., tolerance to external stresses such as UV or extreme temperature). Genes in loci detected by XP-EHH were overrepresented in functions related to reproduction and meiosis. However, these genes were predominantly encoded on a single scaffold and hitchhiking selection may be an alternative explanation for this enrichment.
Similar to our study, Badouin (2017) found widespread selection across the genome of anther-smut fungi. Given the obligate association of anther-smut fungi with their hosts, selection pressure exerted by the host is likely to have a stronger impact on the pathogen. However, analyses of selective sweep loci revealed a multitude of possible targets not necessarily related to selection by the host. A number of studies focused on individual pathogenicity loci that encoded proteins that modulate the host immune system and may be detected by the host. Gene sequences encoding these proteins were found to show strong signatures of positive selection based on excessive nonsynonymous versus synonymous substitution rates (Schürch et al. 2004; Stukenbrock and McDonald 2007; Stukenbrock et al. 2010; Stukenbrock et al. 2011; Guyon et al. 2014). However, signatures of selection based on substitution rates arise over much longer time frames and reveal different selection regimes between species rather than selection pressure within species. Haplotype-based scans for selection across the genome generally detect much younger selection pressures (Sabeti et al. 2002; Vitti et al. 2013). We found no evidence for a shift in synonymous versus nonsynonymous polymorphism ratios in selective sweep regions compared with the genomic background. Extended haplotypes arise by rapid gains in beneficial allele frequencies coupled with insufficient recombination to breakdown LD between adjacent sites. Hence, no changes in synonymous versus nonsynonymous polymorphism ratios are expected for the most recent episodes of selection.
The emergence of plant pathogens in agricultural ecosystems is a primary concern to ensure food security. The evolutionary processes through which plant pathogens rapidly overcome resistance in host plants or become resistant to fungicides provide illuminating case studies for the rapid evolution of complex traits. Our study showed that selection pressures operating across the distribution range of a globally distributed pathogen are likely to vary extensively. We found that the loci under recent positive selection encoded a multitude of functions related to abiotic and biotic stress factors. We found no evidence that selection imposed by the barley host played a dominant role in shaping recent selection pressures. This is surprising given the fact that many pathogen populations are known to encode major effect loci for aggressiveness (de Sain and Rep 2015; Lo Presti et al. 2015). Instead, selection on R. commune led to selection on a large number of loci likely contributing to the rapid evolution of polygenic traits. Identifying the adaptive value of individual adaptive mutations and dissecting the genetic basis of complex traits will lead to major advances in understanding rapid evolutionary processes in populations with large standing genetic variation.
Authors’ Contributions
NMA and DC conceived and designed the study, NMA analyzed the data, NMA and DC wrote the manuscript. BAM provided funding, advice and edited the manuscript.
Data Accessibility
Raw sequencing data are available from the NCBI Short Read Archive under the BioProject accession number PRJNA327656.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
We carried out part of the research at the Quantitative Genomics Facility of the D-BSSE and Genetic Diversity Center (GDC) of ETH Zurich. N.M. was supported by the Ministry of Higher Education Malaysia (MOHE) and Universiti Kebangsaan Malaysia (UKM) under the SLAI scheme. D.C. is supported by the Swiss National Science Foundation (grant 31003A_173265). B.A.M. is supported by the Swiss National Science Foundation (grant 31003A_155955).
Literature Cited
- Anders S, Pyl PT, Huber W.. 2015. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 312:166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki E, Baba T, Yamaguchi O, Ito S, Moriwaki J.. 2011. Development of barley cultivars with resistance to scald (Rhynchosporium secalis (Oud.) Davis) in Japan. Jpn Agric Res Q. 454:349–357. [Google Scholar]
- Asfaw Z. 2000. The barleys of Ethiopia. Rome: International Plant Genetic Resources Institute. [Google Scholar]
- Badouin H. 2017. Widespread selective sweeps throughout the genome of model plant pathogenic fungi and identification of effector candidates. Mol Ecol 267:2041–2062. [DOI] [PubMed] [Google Scholar]
- Bianchi FJJA, Booij CJH, Tscharntke T.. 2006. Sustainable pest regulation in agricultural landscapes: a review on landscape composition, biodiversity and natural pest control. Proc Biol Sci. 2731595:1715–1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierne N, Welch J, Loire E, Bonhomme F, David P.. 2011. The coupling hypothesis: why genome scans may fail to map local adaptation genes. Mol Ecol. 2010:2044–2072. [DOI] [PubMed] [Google Scholar]
- Bigham A, et al. , . 2010. Identifying signatures of natural selection in Tibetan and Andean populations using dense genome scan data. PLoS Genet. 69:e1001116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas S, Akey JM.. 2006. Genomic insights into positive selection. Trends Genet. 228:437–446. [DOI] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B.. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 3015:2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradbury PJ, et al. , . 2007. TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics 2319:2633–2635. [DOI] [PubMed] [Google Scholar]
- Brockhurst MA, Koskella B.. 2013. Experimental coevolution of species interactions. Trends Ecol Evol. 286:367–375. [DOI] [PubMed] [Google Scholar]
- Brunner PC, Schürch S, McDonald BA.. 2007. The origin and colonization history of the barley scald pathogen Rhynchosporium secalis. J Evol Biol. 204:1311–1321. [DOI] [PubMed] [Google Scholar]
- Brunner PC, Stefansson TS, Fountaine J, Richina V, McDonald BA.. 2015. A global analysis of CYP51 diversity and azole sensitivity in Rhynchosporium commune. Phytopathology 1064:355–361. [DOI] [PubMed] [Google Scholar]
- Cattan-Toupance I, Michalakis Y, Neema C.. 1998. Genetic structure of wild bean populations in their South-Andean centre of origin. Theor Appl Genet. 96(6–7):844–851. [Google Scholar]
- Chitty JL, Fraser JA.. 2017. Purine acquisition and synthesis by human fungal pathogens. Microorganisms 52:33.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J, Kim K-T, Jeon J, Lee Y-H.. 2013. Fungal plant cell wall-degrading enzyme database: a platform for comparative and evolutionary genomics in fungi and Oomycetes. BMC Genomics 14(Suppl 5):S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cingolani P, et al. , . 2012. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: sNPs in the genome of Drosophila melanogaster strain w1118 ; iso-2; iso-3. Fly 62:80–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman JJ, Mylonakis E.. 2009. Efflux in fungi: la pièce de résistance. PLoS Pathog. 56:e1000486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cools HJ, Hawkins NJ, Fraaije BA.. 2013. Constraints on the evolution of azole resistance in plant pathogenic fungi. Plant Pathol. 62:36–42. [Google Scholar]
- Croll D, McDonald BA.. 2017. The genetic basis of local adaptation for pathogenic fungi in agricultural ecosystems. Mol Ecol. 267:2027–2040. [DOI] [PubMed] [Google Scholar]
- Danecek P, et al. , . 2011. The variant call format and VCFtools. Bioinformatics. 27:2156–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daumann M, Golfier P, Knüppel N, Hahn M, Möhlmann T.. 2016. Botrytis cinerea can import and utilize nucleosides in salvage and catabolism and BcENT functions as high affinity nucleoside transporter. Fungal Biol. 1208:904–916. [DOI] [PubMed] [Google Scholar]
- Dean P, Major P, Nakjang S, Hirt RP, Embley TM.. 2014. Transport proteins of parasitic protists and their role in nutrient salvage. Front. Plant Sci 5:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jonge R, Bolton MD, Thomma BPHJ. 2011. How filamentous pathogens co-opt plants: the ins and outs of fungal effectors. Curr Opin Plant Biol. 14:400–406. [DOI] [PubMed] [Google Scholar]
- de Koning HP, Bridges DJ, Burchmore RJS.. 2005. Purine and pyrimidine transport in pathogenic protozoa: from biology to therapy. FEMS Microbiol Rev 295:987–1020. [DOI] [PubMed] [Google Scholar]
- Duffy CW, et al. , . 2015. Comparison of genomic signatures of selection on Plasmodium falciparum between different regions of a country with high malaria endemicity. BMC Genomics. 16:527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earl DA, vonHoldt BM.. 2012. STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour. 42:359–361. [Google Scholar]
- Esquerré-Tugayé M-T, Boudart G, Dumas B.. 2000. Cell wall degrading enzymes, inhibitory proteins, and oligosaccharides participate in the molecular dialogue between plants and pathogens. Plant Physiol Biochem. 38(1–2):157–163. [Google Scholar]
- Evanno G, Regnaut S, Goudet J.. 2005. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol. 148:2611–2620. [DOI] [PubMed] [Google Scholar]
- Falcon S, Gentleman R.. 2007. Using GOstats to test gene lists for GO term association. Bioinformatics 232:257–258. [DOI] [PubMed] [Google Scholar]
- Fang Y, et al. , . 2012. A genomewide screen in Schizosaccharomyces pombe for genes affecting the sensitivity of antifungal drugs that target ergosterol biosynthesis. Antimicrob Agents Chemother. 564:1949–1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez J, et al. , . 2012. Principles of carbon catabolite repression in the rice blast fungus: tps1, Nmr1-3, and a MATE–family pump regulate glucose metabolism during infection. PLoS Genet. 85:e1002673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouché S, Plissonneau C, Croll D.. 2018. The birth and death of effectors in rapidly evolving filamentous pathogen genomes. Curr Opin Microbiol. 46:34–42. [DOI] [PubMed] [Google Scholar]
- Franco-Orozco B, et al. , . 2017. A new proteinaceous pathogen-associated molecular pattern (PAMP) identified in Ascomycete fungi induces cell death in Solanaceae. New Phytol. 2144:1657–1672. [DOI] [PubMed] [Google Scholar]
- Gandon A. 2002. Coevolution between parasite virulence and host life-history traits. Am Nat. 160:374, Michalakis. [DOI] [PubMed] [Google Scholar]
- Gandon S, Capowiez Y, Dubois Y, Michalakis Y, Olivieri I.. 1996. Local adaptation and gene-for-gene coevolution in a metapopulation model. Proc R Soc B Biol Sci. 2631373:1003–1009. [Google Scholar]
- Gao S, Nuss DL.. 1996. Distinct roles for two G protein α subunits in fungal virulence, morphology, and reproduction revealed by targeted gene disruption. Proc Natl Acad Sci USA. 9324:14122–14127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrison E, Marth G.. 2012. Haplotype-based variant detection from short-read sequencing. ArXiv12073907 Q-Bio. Available from: http://arxiv.org/abs/1207.3907 (accessed March 15, 2018).
- Garud NR, Messer PW, Buzbas EO, Petrov DA.. 2015. Recent selective sweeps in North American Drosophila melanogaster show signatures of soft sweeps. PLOS Genet. 112:e1005004.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautier M, Klassmann A, Vitalis R.. 2017. REHH 2.0: a reimplementation of the R package REHH to detect positive selection from haplotype structure. Mol Ecol Resour. 171:78–90. [DOI] [PubMed] [Google Scholar]
- Gorter FA, Scanlan PD, Buckling A.. 2016. Adaptation to abiotic conditions drives local adaptation in bacteria and viruses coevolving in heterogeneous environments. Biol Lett. 122:20150879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandaubert J, et al. , . 2014. Transposable element-assisted evolution and adaptation to host plant within the Leptosphaeria maculans-Leptosphaeria biglobosa species complex of fungal pathogens. BMC Genomics 151:891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gronover CS, Kasulke D, Tudzynski P, Tudzynski B.. 2001. The role of G protein Alpha subunits in the infection process of the gray mold fungus Botrytis cinerea. Mol Plant Microbe Interact. 1411:1293–1302. [DOI] [PubMed] [Google Scholar]
- Guyon K, Balagué C, Roby D, Raffaele S.. 2014. Secretome analysis reveals effector candidates associated with broad host range necrotrophy in the fungal plant pathogen Sclerotinia sclerotiorum. BMC Genomics 151:336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadado TT, Rau D, Bitocchi E, Papa R.. 2010. Adaptation and diversity along an altitudinal gradient in Ethiopian barley (Hordeum vulgare L.) landraces revealed by molecular analysis. BMC Plant Biol. 101:121–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamari Z, et al. , . 2009. Convergent evolution and orphan genes in the Fur4p-like family and characterization of a general nucleoside transporter in Aspergillus nidulans. Mol Microbiol. 731:43–57. [DOI] [PubMed] [Google Scholar]
- Hamilton WD, Axelrod R, Tanese R.. 1990. Sexual reproduction as an adaptation to resist parasites (a review). Proc Natl Acad Sci USA. 879:3566–3573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann FE, Sánchez-Vallet A, McDonald BA, Croll D.. 2017. A fungal wheat pathogen evolved host specialization by extensive chromosomal rearrangements. ISME J. 115:1189–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horwitz BA, et al. , . 1999. A G protein alpha subunit from Cochliobolus heterostrophus involved in mating and appressorium formation. Fungal Genet Biol. 261:19–32. [DOI] [PubMed] [Google Scholar]
- Jones DA, Bertazzoni S, Turo CJ, Syme RA, Hane JK.. 2018. Bioinformatic prediction of plant–pathogenicity effector proteins of fungi. Curr Opin Microbiol. 46:43–49. [DOI] [PubMed] [Google Scholar]
- Jones P, et al. , . 2014. InterProScan 5: genome-scale protein function classification. Bioinformatics 309:1236–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Käll L, Krogh A, Sonnhammer ELL.. 2007. Advantages of combined transmembrane topology and signal peptide prediction—the Phobius web server. Nucleic Acids Res 35(Web Server):W429–W432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaltz O, Shykoff JA.. 1998. Local adaptation in host–parasite systems. Heredity 814:361–370. [Google Scholar]
- Kikot GE, Hours RA, Alconada TM.. 2009. Contribution of cell wall degrading enzymes to pathogenesis of Fusarium graminearum: a review. J Basic Microbiol. 493:231–241. [DOI] [PubMed] [Google Scholar]
- Kirsten S, et al. , . 2012. Necrosis-inducing proteins of Rhynchosporium commune, effectors in quantitative disease resistance. Mol Plant-Microbe Interact. 2510:1314–1325. [DOI] [PubMed] [Google Scholar]
- Krogh A, Larsson B, von Heijne G, Sonnhammer EL.. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol 3053:567–580. [DOI] [PubMed] [Google Scholar]
- Kuroda T, Tsuchiya T.. 2009. Multidrug efflux transporters in the MATE family. Biochim Biophys Acta 17945:763–768. [DOI] [PubMed] [Google Scholar]
- Laine A-L. 2005. Spatial scale of local adaptation in a plant-pathogen metapopulation. J Evol Biol. 184:930–938. [DOI] [PubMed] [Google Scholar]
- Laine A-L. 2008. Temperature-mediated patterns of local adaptation in a natural plant-pathogen metapopulation. Ecol Lett. 114:327–337. [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL.. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 103:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lao O, de Gruijter JM, van Duijn K, Navarro A, Kayser M.. 2007. Signatures of positive selection in genes associated with human skin pigmentation as revealed from analyses of single nucleotide polymorphisms. Ann Hum Genet. 713:354–369. [DOI] [PubMed] [Google Scholar]
- Linde CC, Zala M, Ceccarelli S, McDonald BA.. 2003. Further evidence for sexual reproduction in Rhynchosporium secalis based on distribution and frequency of mating-type alleles. Fungal Genet Biol. 402:115–125. [DOI] [PubMed] [Google Scholar]
- Linde CC, Zala M, McDonald BA.. 2009. Molecular evidence for recent founder populations and human-mediated migration in the barley scald pathogen Rhynchosporium secalis. Mol Phylogenet Evol. 513:454–464. [DOI] [PubMed] [Google Scholar]
- Liu J, Li Y, Wang W, Gai J, Li Y.. 2016. Genome-wide analysis of MATE transporters and expression patterns of a subgroup of MATE genes in response to aluminum toxicity in soybean. BMC Genomics 171:223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, et al. , . 2017. Characterising private and shared signatures of positive selection in 37 Asian populations. Eur J Hum Genet 254:499–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo Presti L, et al. , . 2015. Fungal effectors and plant susceptibility. Annu Rev Plant Biol. 661:513–545. [DOI] [PubMed] [Google Scholar]
- Martin SH, et al. , . 2016. Natural selection and genetic diversity in the butterfly Heliconius melpomene. Genetics 2031:525–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mboup M, et al. , . 2012. Genetic structure and local adaptation of European wheat yellow rust populations: the role of temperature-specific adaptation. Evol Appl. 54:341–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald BA. 2015. How can research on pathogen population biology suggest disease management strategies? The example of barley scald (Rhynchosporium commune). Plant Pathol 645:1005–1013. [Google Scholar]
- McDonald JH, Kreitman M.. 1991. Adaptive protein evolution at the Adh locus in Drosophila. Nature 3516328:652–654. [DOI] [PubMed] [Google Scholar]
- McKenna A, et al. , . 2010. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 209:1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohd-Assaad N, McDonald BA, Croll D.. 2016. Multilocus resistance evolution to azole fungicides in fungal plant pathogen populations. Mol Ecol. 2524:6124–6142. [DOI] [PubMed] [Google Scholar]
- Moriyama Y, Hiasa M, Matsumoto T, Omote H.. 2008. Multidrug and toxic compound extrusion (MATE)-type proteins as anchor transporters for the excretion of metabolic waste products and xenobiotics. Xenobiotica 38(7–8):1107–1118. [DOI] [PubMed] [Google Scholar]
- O’Meara TR, Hay C, Price MS, Giles S, Alspaugh JA.. 2010. Cryptococcus neoformans histone acetyltransferase Gcn5 regulates fungal adaptation to the host. Eukaryot Cell 98:1193–1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orchard SS, Goodrich-Blair H.. 2005. Pyrimidine nucleoside salvage confers an advantage to Xenorhabdus nematophila in its host interactions. Appl Environ Microbiol 7110:6254–6259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ordonez N, et al. , . 2015. Worse comes to worst: bananas and Panama disease—when plant and pathogen clones meet. PLoS Pathog. 1111:e1005197.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penselin D, et al. , . 2016. Comparative genomics to explore phylogenetic relationship, cryptic sexual potential and host specificity of Rhynchosporium species on grasses. BMC Genomics 171:953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen TN, Brunak S, Heijne G, von Nielsen H.. 2011. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods 810:785–786. [DOI] [PubMed] [Google Scholar]
- Plissonneau C, Stürchler A, Croll D.. 2016. The evolution of orphan regions in genomes of a fungal pathogen of wheat. mBio 75:e01231-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ploetz RC. 2000. Panama Disease: a classic and destructive disease of banana. Plant Health Prog. doi: 10.1094/PHP-2000-1204-01-HM. [Google Scholar]
- Pretorius ZA, Singh RP, Wagoire WW, Payne TS.. 2000. Detection of virulence to wheat stem rust resistance gene Sr31 in Puccinia graminis. f. sp. tritici in Uganda. Plant Dis. 842:203–203. [DOI] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P.. 2000. Inference of population structure using multilocus genotype data. Genetics 1552:945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian W, Deng L, Lu D, Xu S.. 2013. Genome-wide landscapes of human local adaptation in Asia. PLoS ONE 81:e54224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raffaele S, Kamoun S.. 2012. Genome evolution in filamentous plant pathogens: why bigger can be better. Nat Rev Microbiol. 106:417–430. [DOI] [PubMed] [Google Scholar]
- Rohe M, et al. , . 1995. The race-specific elicitor, NIP1, from the barley pathogen, Rhynchosporium secalis, determines avirulence on host plants of the Rrs1 resistance genotype. EMBO J. 1417:4168–4177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolke Y, et al. , . 2004. Functional analysis of H(2)O(2)-generating systems in Botrytis cinerea: the major Cu-Zn-superoxide dismutase (BCSOD1) contributes to virulence on French bean, whereas a glucose oxidase (BCGOD1) is dispensable. Mol Plant Pathol. 5:17–27. [DOI] [PubMed] [Google Scholar]
- Sabeti PC, et al. , . 2002. Detecting recent positive selection in the human genome from haplotype structure. Nature 4196909:832–837. [DOI] [PubMed] [Google Scholar]
- de Sain M, Rep M.. 2015. The role of pathogen-secreted proteins in fungal vascular wilt diseases. Int J Mol Sci. 1610:23970–23993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholthof K-BG. 2007. The disease triangle: pathogens, the environment and society. Nat Rev Microbiol. 52:152–156. [DOI] [PubMed] [Google Scholar]
- Schürch S, Linde CC, Knogge W, Jackson LF, McDonald BA.. 2004. Molecular population genetic analysis differentiates two virulence mechanisms of the fungal avirulence gene NIP1. Mol Plant-Microbe Interact. 1710:1114–1125. [DOI] [PubMed] [Google Scholar]
- Sicard D, et al. , . 2007. Specialization and local adaptation of a fungal parasite on two host plant species as revealed by two fitness traits. Evol Int J Org Evol 611:27–41. [DOI] [PubMed] [Google Scholar]
- Sicard D, Buchet S, Michalakis Y, Neema C.. 1997. Genetic variability of Colletotrichum lindemuthianum in wild populations of common bean. Plant Pathol. 463:355–365. [Google Scholar]
- Sperschneider J, et al. , . 2016. EffectorP: predicting fungal effector proteins from secretomes using machine learning. New Phytol. 2102:743–761. [DOI] [PubMed] [Google Scholar]
- Stefansson TS, McDonald BA, Willi Y.. 2013. Local adaptation and evolutionary potential along a temperature gradient in the fungal pathogen Rhynchosporium commune. Evol Appl. 63:524–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefansson TS, McDonald BA, Willi Y.. 2014. The influence of genetic drift and selection on quantitative traits in a plant pathogenic fungus. PLoS ONE 911:e112523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefansson TS, Willi Y, Croll D, McDonald BA.. 2014. An assay for quantitative virulence in Rhynchosporium commune reveals an association between effector genotype and virulence. Plant Pathol. 632:405–414. [Google Scholar]
- Stukenbrock EH, et al. , . 2011. The making of a new pathogen: insights from comparative population genomics of the domesticated wheat pathogen Mycosphaerella graminicola and its wild sister species. Genome Res. 2112:2157–2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stukenbrock EH, et al. , . 2010. Whole-genome and chromosome evolution associated with host adaptation and speciation of the wheat pathogen Mycosphaerella graminicola. PLoS Genet. 612:e1001189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stukenbrock EH, McDonald BA.. 2007. Geographical variation and positive diversifying selection in the host-specific toxin SnToxA. Mol Plant Pathol. 83:321–332. [DOI] [PubMed] [Google Scholar]
- Stukenbrock EH, McDonald BA.. 2008. The origins of plant pathogens in agro-ecosystems. Annu Rev Phytopathol. 461:75–100. [DOI] [PubMed] [Google Scholar]
- Supek F, Bošnjak M, Škunca N, Šmuc T.. 2011. REVIGO summarizes and visualizes long lists of Gene Ontology terms. PLoS ONE 67:e21800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, et al. , . 2015. Membrane-associated glucose-methanol-choline oxidoreductase family enzymes PhcC and PhcD are essential for enantioselective catabolism of dehydrodiconiferyl alcohol. Appl Environ Microbiol. 8123:8022–8036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan K-C, et al. , . 2008. A signaling-regulated, short-chain dehydrogenase of Stagonospora nodorum regulates asexual development. Eukaryot Cell 711:1916–1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thrall PH, Burdon JJ.. 2002. The effect of spatial scale of host and pathogen dispersal. Plant Pathol. 512:169–184. [Google Scholar]
- Vitti JJ, Grossman SR, Sabeti PC.. 2013. Detecting natural selection in genomic data. Annu Rev Genet. 471:97–120. [DOI] [PubMed] [Google Scholar]
- Voight BF, Kudaravalli S, Wen X, Pritchard JK.. 2006. A map of recent positive selection in the human genome. PLoS Biol. 43:e72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wevelsiep L, Kogel K-H, Knogge W.. 1991. Purification and characterization of peptides from Rhynchosporium secalis inducing necrosis in barley. Physiol Mol Plant Pathol. 396:471–482. [Google Scholar]
- Wevelsiep L, Rupping E, Knogge W.. 1993. Stimulation of barley plasmalemma H+-ATPase by phytotoxic peptides from the fungal pathogen Rhynchosporium secalis. Plant Physiol. 1011:297–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi K, Xue AG, Burnett PA, Helm JH, Turkington TK.. 2000. Quantitative resistance of barley cultivars to Rhynchosporium secalis. Can J Plant Pathol. 223:217–223. [Google Scholar]
- Xiang-Yu J, Yang Z, Tang K, Li H.. 2016. Revisiting the false positive rate in detecting recent positive selection. Quant Biol. 43:207–216. [Google Scholar]
- Xin Q, Gong Y, Lv X, Chen G, Liu W.. 2013. Trichoderma reesei histone acetyltransferase Gcn5 regulates fungal growth, conidiation, and cellulase gene expression. Curr Microbiol. 675:580–589. [DOI] [PubMed] [Google Scholar]
- Xue-Franzén Y, Henriksson J, Bürglin TR, Wright APH.. 2013. Distinct roles of the Gcn5 histone acetyltransferase revealed during transient stress-induced reprogramming of the genome. BMC Genomics 141:479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagishi D, Otani H, Kodama M.. 2006. G protein signaling mediates developmental processes and pathogenesis of Alternaria alternata. Mol Plant-Microbe Interact. 1911:1280–1288. [DOI] [PubMed] [Google Scholar]
- Yao S-H, et al. . 2016. A cytoplasmic Cu-Zn superoxide dismutase SOD1 contributes to hyphal growth and virulence of Fusarium graminearum. Fungal Genet Biol. 91:32–42. [DOI] [PubMed] [Google Scholar]
- Yin Y, et al. , . 2012. dbCAN: a web resource for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 40(W1):W445–W451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young JD, Yao SYM, Baldwin JM, Cass CE, Baldwin SA.. 2013. The human concentrative and equilibrative nucleoside transporter families, SLC28 and SLC29. Mol Aspects Med. 34(2–3):529–547. [DOI] [PubMed] [Google Scholar]
- Zaffarano PL, Mcdonald BA, Linde CC.. 2009. Phylogeographical analyses reveal global migration patterns of the barley scald pathogen Rhynchosporium secalis. Mol Ecol. 182:279–293. [DOI] [PubMed] [Google Scholar]
- Zaffarano PL, McDonald BA, Linde CC.. 2008. Rapid speciation following recent host shifts in the plant pathogenic fungus Rhynchosporium. Evolution 626:1418–1436. [DOI] [PubMed] [Google Scholar]
- Zaffarano PL, McDonald BA, Linde CC.. 2011. Two new species of Rhynchosporium. Mycologia 1031:195–202. [DOI] [PubMed] [Google Scholar]
- Zaffarano PL, McDonald BA, Zala M, Linde CC.. 2006. Global hierarchical gene diversity analysis suggests the fertile crescent is not the center of origin of the barley scald pathogen Rhynchosporium secalis. Phytopathology 969:941–950. [DOI] [PubMed] [Google Scholar]
- Zhan J, McDonald BA.. 2011. Thermal adaptation in the fungal pathogen Mycosphaerella graminicola. Mol. Ecol 208:1689–1701. [DOI] [PubMed] [Google Scholar]
- Zhan J, Stefanato FL, Mcdonald BA.. 2006. Selection for increased cyproconazole tolerance in Mycosphaerella graminicola through local adaptation and in response to host resistance. Mol Plant Pathol. 74:259–268. [DOI] [PubMed] [Google Scholar]
- Zhao F, McParland S, Kearney F, Du L, Berry DP.. 2015. Detection of selection signatures in dairy and beef cattle using high-density genomic information. Genet Sel Evol. 471:49. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw sequencing data are available from the NCBI Short Read Archive under the BioProject accession number PRJNA327656.