Overexpression of rice OsPGIP2 in Brassica napus results in resistance to Sclerotinia sclerotiorum by increasing activation of defense mechanisms and inhibiting fungal SsPG3 and SsPG6 proteins.
Keywords: Brassica napus, defense pathways, OsPGIP2, damage/pathogen associated molecular patterns (DAMP/PAMP)-triggered immunity, polygalacturonase-inhibiting protein, polygalacturonase, resistance, Sclerotinia sclerotiorum
Abstract
Sclerotinia stem rot (SSR), caused by Sclerotinia sclerotiorum, is the most serious disease affecting the yield of the agriculturally and economically important crop Brassica napus (rapeseed). In this study, Oryza sativa polygalacturonase-inhibiting protein 2 (OsPGIP2) was found to effectively enhanced rapeseed immunity against S. sclerotiorum infection. Leaf extracts of B. napus plants overexpressing OsPGIP2 showed enhanced S. sclerotiorum resistance by delaying pathogen infection. The constitutive expression of OsPGIP2 in rapeseed plants provided a rapid and effective defense response, which included the production of reactive oxygen species, interactions with S. sclerotiorum polygalacturonases (SsPG3 and SsPG6), and effects on the expression of defense genes. RNA sequencing analysis revealed that the pathogen induced many differentially expressed genes associated with pathogen recognition, redox homeostasis, mitogen-activated protein kinase signaling cascades, hormone signaling pathways, pathogen-/defense-related genes, and cell wall-related genes. The overexpression of OsPGIP2 also led to constitutively increased cell wall cellulose and hemicellulose contents in stems without compromising seed quality. The results demonstrate that OsPGIP2 plays a major role in rapeseed defense mechanisms, and we propose a model for OsPGIP2-conferred resistance to S. sclerotiorum in these plants.
Introduction
Sclerotinia stem rot (SSR) caused by the fungus Sclerotinia sclerotiorum (Lib.) de Bary, has a broad host range (Boland and Hall, 1994) and its severe symptoms make it the most devastating disease of rapeseed (Brassica napus L.). The fungus attacks rapeseed plants at all growth stages, but most especially in the flowering period, and it can infect leaves, stems, and siliques. Depending on the production area and climatology, SSR can cause severe yield losses ranging from 10–80% and results in low oil quality (Alizadeh et al.,2006; Sharma et al., 2015). Despite the efforts of breeding programs, effective resistance sources against S. sclerotiorum have not yet been identified and the mechanisms underlying the interaction between rapeseed and S. sclerotiorum remain elusive. With the development of biotechnological tools, many candidate genes involved in B. napus defense against S. sclerotiorum have been identified (Zhao et al., 2007; Wei et al., 2016; Wu et al., 2016). Transgenic manipulation of genes associated with innate defense signaling pathways may offer alternative approaches for improving resistance to S. sclerotiorum in rapeseed varieties.
During the interaction between plants and pathogens, two main immune responses provide natural disease-resistance mechanisms. Pathogen-associated molecular patterns (PAMP)-triggered immunity (PTI) is the primary defense response that activates downstream gene expression and results in relatively weak but race-non-specific hypersensitive responses (HRs) (Boller and Felix, 2009; Baxter et al., 2014). Effector-triggered immunity (ETI) is induced by highly variable pathogen avirulence effectors that are recognized by host disease-resistance (R) proteins, and results in a rapid HR and long-lasting resistance (Boller and Felix, 2009). Both PTI and ETI can confer host-plant resistance to pathogens, but PTI overlapping with downstream immune responses is responsible for the responses against necrotrophs with a broad host range (Mengiste, 2012). The earliest defense reactions in PTI include the synthesis of reactive oxygen species (ROS), changes in ion fluxes across membranes, or increases in calcium (Ca2+) concentrations in the cellular space (Lecourieux et al., 2006; Torres, 2010; Meng and Zhang, 2013; Baxter et al., 2014; Seybold et al., 2014). Calcium then acts as a diffusible second messenger for several resistance responses, including the synthesis of pathogen-related (PR) proteins and phytoalexins, and programmed cell death in neighboring cells (Zurbriggen et al., 2009). Subsequent reactions include changes in plant endogenous hormone status, cell wall reinforcement by polysaccharide deposition and lignification, activation of downstream defense genes, and accumulation of secondary metabolites (Glawischnig, 2007; Peliter et al., 2009; Matern et al., 2011; Robert-Seilaniantz et al., 2011; Chowdhury et al., 2014).
During infection, pathogens secrete cell wall-degrading enzymes (CWDEs), such as pectinases, xylanases, polygalacturonases (PGs), and cellulases, in order to degrade cell wall polysaccharides and to penetrate and colonize host tissues (Bolton et al., 2006; Fróes et al., 2012). Concurrently, plants counteract these attacks with an array of inhibitor proteins (Juge, 2006). Interestingly, polygalacturonase inhibitor proteins (PGIPs) seem not only to recognize different surface motifs of functionally related structurally variable PGs but also to shift the breakdown process toward generating larger peptide fragments, which are damage-associated molecular pattern (DAMP)-active oligogalacturonides (OGs) (Federici et al., 2006; Maulik et al., 2012; Kalunke et al., 2015). Transgenic Arabidopsis thaliana plants expressing a particular PGIP–PG chimera exhibit accumulated OGs and enhanced resistance to a variety of pathogens, providing direct evidence for the function of OGs as in vivo elicitors of plant defense responses (Benedetti et al., 2015). Furthermore, treatment of plants with exogenous OGs promotes the production of ROS, up-regulates the expression of β-1,3-glucanase, increases the transcripts of phenylalanine ammonia-lyase (PAL) to accumulate phytoalexins and lignin (D’Ovidio et al., 2004; Ridley et al., 2001; Ferrari et al., 2013), and promotes changes in gene expression in the salicylic acid (SA), ethylene (ET), and jasmonic acid (JA) pathways (Dupont et al., 2006; Aziz et al., 2007; Osorio et al., 2008). Because the OG receptor WALL-ASSOCIATED KINASE 1 (WAK1) is capable of activating the kinase domain of the elongation factor Tu (EF-TU) receptor (EFR), OGs seem to be involved in PTI responses to necrotrophic pathogens (Mengiste, 2012).
The ‘PG–PGIP’ interaction mechanism provides a theoretical basis for plant disease resistance (Benedetti et al., 2011; Gutierrez, 2012). Recent studies have demonstrated that overexpressing exogenous PGIPs is associated with a reduction of symptoms caused by bacteria, fungal pathogens, and insects in wheat, grape, apple, Chinese cabbage, tomato, and mung bean (Szankowski et al., 2003; Agüero et al., 2005; Hwang et al., 2010; Schacht et al., 2011; Borras-Hidalgo, et al., 2012; Wang et al., 2015; Chotechung et al., 2016). In addition to PvPGIP2 (Phaseolus vulgaris) and BnPGIP2 (Akhgari et al., 2012; Huangfu et al., 2014), only a few other PGIP genes cause S. sclerotiorum resistance when overexpressed in B. napus. Although there are at least 16 PGIP genes that are highly induced by S. sclerotiorum infection in B. napus (Hegedus et al., 2008), few commercial rapeseed varieties exhibit partial resistance (Wu et al., 2016). Arabidopsis thaliana lines overexpressing BnPGIP2 exhibit smaller necrotic lesions than wild-type plants, but no long-term effect on S. sclerotiorum disease progression is observed (Bashi et al., 2013).
In Oryza sativa, the expression of OsPGIP2 is induced more immediately and strongly at 0–12 h after Rhizoctonia solani infection than OsFOR1, which is the first PGIP gene identified with inhibitory activity against the PGase from Aspergillus niger (Jang et al., 2003; Lu et al., 2012). Therefore, Lu et al. (2012) proposed that increasing the expression of OsPGIP2 by transgenic technology might correspondingly increase the resistance to pathogens in transgenic lines. In the present study, we transformed the exogenous OsPGIP2 gene and introduced it into two different rapeseed backgrounds, and then assessed the enhancement of S. sclerotiorum resistance in the transgenic rapeseed lines. Our results indicated that the ectopic expression of OsPGIP2 in rapeseed not only conferred resistance to S. sclerotiorum at both the seedling and adult stages, but also improved seed quality traits in the transgenic lines. Moreover, transient expression experiments showed that OsPGIP2 interacts with S. sclerotiorum SsPG3 and SsPG6 and, based on RNA sequencing analysis, we suggest that OsPGIP2 played an important role in the defense mechanisms in the transgenic lines. Overall, our results demonstrate that OsPGIP2 might be an effective gene for developing S. sclerotiorum-resistant rapeseed varieties.
Materials and methods
Plant transformation and growth conditions
The vector pCAMIBA1300-35S:OsPGIP2 (kindly provided by Dr Liaoxun Lu, Huazhong Agriculture University, China) was transformed into the partially resistant B. napus line 7-5 and the susceptible line T45 by Agrobacterium-mediated transformation (Jonoubi et al., 2005). Untransformed plants and transgenic lines were planted in an isolated nursery field at the Huazhong Agriculture University experimental farm in Wuhan, China.
PCR and Southern blot analyses
Total genomic DNA was extracted from fresh leaves of wild-type and transformed plants using the cetyltrimethylammonium bromide (CTAB) method (Doyle, 1990). Verifications of OsPGIP2 and the hygromycin resistance gene (HYG) in putative transgenic plants were performed by polymerase chain reaction (PCR) using the primers 333-L/R and 309-L/R. Each PCR was carried out in a 15-μl reaction that included 5 μM of each primer, 100 μM dNTP, 1× Taq buffer, 1.5 mM MgCl2, 2.0 units of Taq DNA polymerase (Thermo Scientific, Shanghai, China), and 50 ng of DNA template. PCR conditions consisted of 35 amplification cycles (94 °C for 30 s, 58 °C for 30 s, and 72 °C for 30 s) in total. PCR products were separated on a 1.0% (w/v) agarose gel and visualized after ethidium bromide staining. Southern blot analysis was carried out by the Towin biotechnology company (Wuhan, China) to confirm the integration of the introduced genes.
Detached leaf inoculation
The PCR-positive transformed plants and untransformed controls were grown in the field to the 9–10 leaf stage. The eighth leaf of each individual plant was removed for detached leaf inoculation with S. sclerotiorum in a greenhouse, as previously described (Wu et al., 2013). Ten biological replicates were performed for this experiment. At 48 h and 72 h post-inoculation (hpi), images were taken of inoculated leaves and lesion sizes were measured using ImageJ software (Nicholson, 2010).
Hydrogen peroxide detection on inoculated leaves
The accumulation of H2O2 in leaves of transgenic plants was observed using 3′-3-diaminobenzidine (DAB) staining. Leaves inoculated with S. sclerotiorum were cut to the same size and stained with DAB solution as previously described (Dong et al., 2008). Leaves were then washed in deionized water and treated with 96% ethanol to remove chlorophyll. Images were taken under a stereoscopic microscope (SZX16, Olympus, Japan).
Test of seedling resistance under laboratory conditions
To prepare the S. sclerotiorum inoculum, five hyphal agar discs (8 mm in diameter) were placed into 100 ml of PDB (potato-dextrose broth) liquid medium for 3 d at 22 ± 2 °C under continuous shaking (200 rpm). Hyphae were then disrupted in a blender to produce a 5% (v/v) S. sclerotiorum hyphal suspension, which was sprayed onto 6-week-old seedlings at 100 ml m–2. For each rapeseed line, approximately 20 seedlings were planted in 1-l pots, using three biological replicates. Lines 7-5WT and T45WT were used as untransformed controls; ZS11 (Zhongshuang11) and Westar were used as resistant and susceptible controls, respectively. Inoculated plants were incubated in a growth chamber at 22 ± 2 °C under 16 h light/8 h dark and 80% relative humidity. At 14 d post-inoculation (dpi), we counted the number of seedlings that were completely withered (dead), that were diseased but not wilting (susceptible), and that were without symptoms (healthy).
Design of field trial and stem inoculation at the flowering stage in the field
Six randomized-block replicates, consisting of 12 rows of approximately 120 plants per transgenic line, were used in field experiments. Two blocks were selected for stem inoculation of S. scleroterum, two were selected to measure artificial infection by the fungus, and the remaining two were not subject to any pathogen inoculation. Lines 7-5WT and T45WT were used as untransformed controls. Crop management followed the standard agronomic practice excluding pesticide application. Field stem inoculations were carried out using a method modified from Wu et al. (2013). Agar discs (8 mm in diameter) were excised from the edges of growing fungal colonies and up-ended into the lids of 1.5-ml or 2.0-ml centrifuge tubes, which were then affixed with plastic wrap onto rapeseed stems 50 cm from the ground. Disease severity was assessed by measuring lesion length per pathogen infection spot at 7 dpi. Inoculation, observation, and disease resistance were only determined for plants located in the center of blocks to avoid marginal effects.
Artificial infection using S. sclerotiorum suspension and evaluation of disease resistance
In the two randomized blocks selected to measure artificial infection, a 5% suspension of S. sclerotiorum hyphae was sprayed onto rapeseed plants at the full-flowering stage. Lines 7-5WT and T45WT were used as untransformed controls; ZS11 and Westar were used as resistant and susceptible varieties, respectively. At 7 d before harvest, disease severity was assessed and classified as follows: 0, no lesions; 1, superficial lesions or small branches affected; 2, large branches dead; 3, at least 50% of main stem circumference with lesions; 4, main stem girdled but plant produced good seeds; 5, main stem girdled and much reduced yield, or dead (Falak et al., 2011). The disease index (DI) was calculated as 100[∑(Nii)]/(kN), where i is the disease severity score, k is the highest score that was observed, Ni is the number of every line with each score, and N is total number of plants assessed per line (Liu et al., 2005). The relative resistance index (RRI) was calculated as ln[(100−DIck)/DIck]−ln[(100−DIm)/DIm] where DIck is the disease index of the control (variety ZS11), and DIm is that of the line being evaluated. The relative resistance was then evaluated as: highly resistant (HR), RRI≤–1.2; moderately resistant (MR), –1.2<RRI≤–0.7; lightly resistant (LR), –0.7<RRI≤0; lightly susceptible (LS), 0<RRI≤0.9; moderately susceptible (MS), 0.9<RRI≤2.0; and highly susceptible (HS), RRI>2.0 (Liu et al., 2005).
Analysis of seed quality traits
We harvested seed from fully open-pollinated rapeseed plants in the inoculated blocks. Nine plants were harvested from every line in the center of each plot. The 1000-seed weight from each line was calculated by averaging three measurements of 1000 well-filled seeds dried naturally for at least 3 weeks after harvest (Fan et al., 2010). Near-infrared reflectance spectroscopy (NIRS) (FOSS Analytical A/S, NIRS, USA) was used to measure the contents of protein, glucosinolate (GSL), oil, and erucic acid (Kumar et al., 2010).
Antifungal activity assay in vitro
Fresh leaves (5 g) were ground to powder in liquid nitrogen and then mixed with 30 ml extraction buffer (50 mM MES, 100 mM TrisHCl, 0.1 mM EDTA, 30 mM NaCl, pH=7.0) (Jiang et al., 2013). This mixture was then centrifuged at 10 000 g for 10 min at room temperature and the supernatant was collected as the crude leaf extract. A S. sclerotiorum mycelia plug was inoculated onto the center of a potato dextrose agar (PDA) medium, which was mixed with 50% (v/v) crude leaf extracts from transgenic lines or extraction buffer (control) in three biological replicates. These mixtures were incubated in the dark for 36 h before hyphae were visualized under a stereoscopic microscope (SZX16, Olympus, Japan).
Determination of cell wall monosaccharides in stems
The stems of rapeseed plants at the flowering and final flowering stages were cut 10–50 cm above ground and the leaves were removed. Three biological replicates were used in this assay. The stem tissue was completely dried at 60 °C, crushed in a pulverizer, and passed twice through a 60-mesh sieve (0.3 mm). Samples were first extracted with benzene-ethanol Soxhlet extractor to remove resin and pigments, and then hydrolyzed with 72% H2SO4 and 4% H2SO4 to obtain the lignocellulose components that are easy to quantify, including cellulose and hemicellulose. These were hydrolyzed to monosaccharides, which were quantified by high-performance liquid chromatography (HPLC, LC-20AT, Shimadzu, Japan); the content of acid-insoluble lignin was determined by the burning method and acid-soluble lignin was determined by UV-Visible spectrophotometry (UV-2550) (Sluiter et al., 2004).
RNA extraction and RNA- sequencing analysis of differentially expressed genes
At five time-points (0, 1, 3, 5, and 7 dpi), epidermal stem tissues extending 5 mm beyond the inoculation site and 1 mm deep were excised using a razor blade. Tissues were harvested from nine plants in the two stem-inoculated blocks in the field, and three harvested plants were pooled to form each sample, generating three biological replicates for quantitative real-time PCR (qPCR). Total RNA was extracted using an RNeasy Plant Mini Kit, (Promega, shanghai, China), according to the manufacturer’s instructions. For RNA sequencing (RNA-seq), equal RNA concentrations from three biological replicates at 3 dpi were pooled into a single sample, and samples of 7-5WT, 7-5D, T45WT, and T45B#2 were sequenced on an Illumina Hiseq 2000 platform (Novogene Bioinformatics Technology Co., Ltd, Beijing, China). The mRNA sequencing produced 39 654 044 (T45WT), 43 614 190 (T45B#2), 54 035 950 (7-5WT), and 47 130 844 (7-5D) clean raw reads. To identify genes corresponding to reads from each sample library, the reads were aligned to the B. napus reference genome (Chalhoub et al., 2014) and the S. sclerotiorum genome (Amselem et al., 2011) using TopHat (Trapnell et al., 2012). Gene expression levels were estimated based on fragments per kilobase of exon per million fragments mapped (FPKM), which were used to draw heatmaps for the differentially expressed genes (DEGs) identified with DEGseq based on the criteria q-value <0.005 and |log2 (FPKM-transgenic/FPKM-WT)| >1 between transgenic and wild-type lines (Wang et al., 2010). For gene ontology (GO) term annotations, all B. napus genes were searched against the National Center for Biotechnology Information (NCBI) non-redundant (Nr) protein database using GOSeq with corrected P-value <0.05 (Young et al., 2010). The transcriptome data has been deposited in NCBI Sequence Read Archive database (accession number, SRP145538).
Quantitative real-time polymerase chain reaction analysis
qPCR was performed on an ABI 7500 Real-Time PCR System using GO Taq® qPCR Master Mix (Promega) according to the manufacturer’s instructions. The primers used for amplifications are listed in Supplementary Table S4 at JXB online. Cycle threshold (Ct) values and relative abundance were calculated using the relative quantification method (Livak and Schmittgen, 2001). The genes BnACTIN7 (AF111812) and UBC21 (ubiquitin-conjugating enzyme 21) were used as references for the analysis of rapeseed gene expression. 28S ribosomal DNA (AF431951.1) and H3 (histone 3) were used as internal controls for S. sclerotiorum gene expression. Three biological replicates and three technical replicates were performed for each sample.
expression assay
The affinities between OsPGIP2–SsPGs were predicated using PPA-Pred2 (Protein–Protein Affinity Predictor; https://www.iitm.ac.in/bioinfo/PPA_Pred/prediction.html). The class of the protein–protein complex was set as ‘Enzyme-Inhibitor’. The smaller the dissociation constant, the more tightly bound the ligand is, or the higher the affinity between ligand and protein. Transient expression assays were performed in leaves of tobacco (Nicotiana benthamiana) as previously described (Li et al., 2013). The coding sequences of SsPG1 (AF501307), SsPG3 (AY312510), SsPG5 (AY496277), and SsPG6 (AF501308) were subcloned into the N-terminal luciferase-fusion vector JW771-NLUC. The coding sequence of OsPGIP2 (AM180653.1) was sub-cloned into the C-terminal luciferase-fusion vector JW772-CLUC. Relevant primer sequences are listed in Supplementary Table S4. For protein interaction analysis, the plasmids of two combinatory constructs were mobilized into Agrobacterium tumefaciens and transformed simultaneously into the leaves of 5-week-old tobacco plants using a needleless syringe (Sparkes et al., 2006). After infiltration, plants were incubated at 22 °C for 48 h before the leaf surface was sprayed with 100 mM Beetle luciferin potassium salt (Promega, Madison, WI, USA) and kept in darkness for 5 min. Luminescent images were captured using a Lumazone 1300B imaging apparatus with a cool charge-coupled device CCD (iXon; Andor Technology, Belfast, UK). Two biological replicates with two technical replicates were used in the assays.
Computational and statistical analyses
Quality traits of seeds, 1000-seed weight, cell wall monosaccharides of stems, lesion length, lesion area from the different transgenic and WT rapeseed lines, and correlations between qPCR and RNA-seq were analysed using SPSS version 22.0. Where the statistical test for significance was between two groups, one-way ANOVA was used, but when comparing between three or more groups, least-significant difference (LSD) multiple-comparison tests were used.
Results
Stable transgenic B. napus seedlings overexpressing OsPGIP2 are resistant to S. sclerotiorum
We constitutively expressed OsPGIP2 in the partially resistant B. napus line 7-5 and the susceptible line T45, and the transgenic plants did not show any major morphological or growth defects (see Supplementary Fig. S2A, B). The T4 homozygous OsPGIP2-expressing transgenic lines 7-5C, 7-5D, 7-5G, T45B#1, T45B#2, and T45C, which contained a single copy of OsPGIP2 as confirmed by Southern blotting (Supplementary Fig. S1A), were selected for further analyses. qPCR analysis of these lines revealed that the OsPGIP2 gene was expressed in all independent transgenic lines (Supplementary Fig. S1B); the PCR analysis showed that OsPGIP2 and HYG were stably inherited in these transgenic lines up to the T4 generation (Fig. S1C, D). To test the resistance to S. sclerotiorum, we used leaves detached from T4 seedlings at 48 and 72 hpi (Fig. 1A). In the 7-5 background, no significant differences in the area of necrotic lesions were detected between the transgenic lines and the wild-type at 48 hpi; however, at 72 hpi, the 7-5C and 7-5G lines had smaller necrotic lesions than the untransformed plants (P<0.05), and the 7-5D lines were significantly resistant to infection compared to inoculated untransformed leaves (P<0.01; Fig. 1B). In the T45 background, only T45B#2 was not significantly different from its control after 48 hpi; after 72 hpi, all the transgenic lines showed a significant reduction in lesion size compared to the inoculated wild-type (P<0.001; Fig. 1B). Moreover, after spraying the leaves of 6-week-old transgenic lines (7-5D and T45B#2) with hyphae suspended in PDB liquid medium, those from wild-type plants dropped and died while most of the lines overexpressing OsPGIP2 exhibited normal growth phenotypes (Fig. 1C). The transgenic line 7-5D showed a decreased death rate (13.64%) compared to 7-5WT (51.28%); T45B#2 lines had a 19.44% death rate while T45WT presented a death rate of 70.37% (Fig. 1D). Thus, between 48 and 72 hpi, the necrotic lesions in the transgenic lines expanded more slowly than in control plants, and the transgenic lines showed a higher survival rate than the wild-type lines when sprayed with hyphae. These results suggest that OsPGIP2 enhanced rapeseed resistance to S. sclerotiorum at the seedling stage under controlled disease stress conditions.
Fig. 1.
OsPGIP2 overexpression conferred resistance to S. sclerotiorum in transgenic rapeseed plants. (A) T4 homozygous lines (7-5D and T45B#2) and wild-types (WT) inoculated with 8-mm agar plugs of S. sclerotiorum hyphae at 22 °C. Images were taken at 48 and 72 h post-inoculation (hpi). Scale bars are 10 cm. (B) Lesion areas in transgenic and untransformed rapeseed plants at 48 and 72 hpi. Data mean values (±SE) (n=20), and significant differences were determined by one-way ANOVA: ***P<0.001, **P<0.01, *P<0.05. 7-5WT and T45WT are wild-type controls. (C) Seedlings of 6-week-old transgenic and control B. napus plants sprayed with hyphal solution. Infection phenotypes were assessed at 14 d post-inoculation. ZS11 was the resistant control and Westar was the susceptible control. 7-5WT and T45WT are wild-type controls. (D) Analysis of the survival of transgenic lines inoculated with S. sclerotiorum hyphae. ZS11 was the resistant control and Westar was the susceptible control. Data are means of three biological replicates (±SE). 7-5WT and T45WT are wild-type controls. (E) Stems of transgenic lines and wild-types 1 week after inoculation with S. sclerotiorum. Scale bars are 2 cm. (F) Stem lesion lengths in transgenic OsPGIP2-overexpressing lines and wild-types 1 week after inoculation with S. sclerotiorum. 7-5WT and T45WT are wild-type controls. Data are means (±SE) (n≥20). Significant differences were determined by one-way ANOVA: ***P<0.001, **P<0.01, *P<0.05.
Transgenic rapeseed lines respond to S. sclerotiorum infection in the field
At the flowering stage, the primary stems of transgenic rapeseed lines were inoculated with isolated S. sclerotiorum using hyphal agar plugs. After 7 d, all transgenic individuals in the 7-5 and T45 backgrounds showed smaller lesions on their stems compared with wild-type plants (Fig. 1E, F). Resistance under field conditions was assessed in three consecutive self-crossed generations (2013–2016) and remained relatively consistent throughout the years (see Supplementary Table S1). After plants were sprayed with S. sclerotiorum hyphae in the field, RRI screening revealed that transgenic lines 7-5D and T45B#1 were moderately resistant (MR) compared to their control lines; 7-5WT was moderately susceptible (MS), and T45WT was highly susceptible (HS) compared to the ZS11 resistant control (Table 1).
Table 1.
Resistance assessment of T4 homozygous transgenic rapeseed lines under controlled disease pressure in the field
| Variety | DIa | RRIb | Control | Categoryb |
|---|---|---|---|---|
| ZS11 | 0.09 | 0.00 | ZS11 | – |
| Westar | 0.89 | 2.31 | ZS11 | HS |
| 7-5C | 0.43 | –0.42 | 7-5WT | LR |
| 7-5D | 0.31 | –0.73 | 7-5WT | MR |
| 7-5G | 0.52 | –0.23 | 7-5WT | LR |
| 7-5WT | 0.65 | 1.99 | ZS11 | MS |
| T45B#1 | 0.42 | –0.74 | T45WT | MR |
| T45B#2 | 0.74 | –0.17 | T45WT | LR |
| T45C | 0.52 | –0.52 | T45WT | LR |
| T45WT | 0.87 | 2.29 | ZS11 | HS |
Data were collected at the harvest stage, after spraying plants (n>30) with S. sclerotiorum. WT indicates non-transformed rapeseed plants. ZS11 was the resistant cultivar control and Westar was the susceptible cultivar control.
a DI is the S. sclerotiorum disease incidence measured under controlled disease stress in field conditions, and is classified from 0 (low) to 5 (high).
b RRI is the relative disease resistance, and is the categories are classified as high (HR), medium (MR), or low (LR) resistance, or as low (LS), medium (MS), or high (HS) susceptibility.
Because the known detrimental effects of SSR on rapeseed, we assessed seed quality traits and determined the grain weight of transgenic and wild-type plants following stem infection and disease development. Although the T4 homozygous transgenic OsPGIP2 lines did not show any significant differences in seed protein or erucic acid contents compared with the wild-type plants after inoculation, the GSL content displayed a significant decrease in the transgenic lines (Table 2). In the 7-5 genetic background, the 1000-seed weight of the transgenic lines ranged from 2.99–3.25 g, which was higher than that of wild-type plants (1.62 g). The oil content was 38.38–40.05% higher than that of the wild-type lines (30.68%) (Table 2). In individual T45 transgenic plants, seed weight ranged from 2.75–3.29 g, which was greater than that of the wild-type plants (1.68 g). The oil content of the T45 transgenic lines was 38.72–40.42% while that of the untransformed line was 30.26% (Table 2). In summary, these measurements were consistent with the results of the previous S. Sclerotiorum hyphal treatment (Table 2, and see Supplementary Fig. S3), and indicated that transforming OsPGIP2 into rapeseed could maintain seed quality traits in plants exposed to S. sclerotiorum. In general, all the transgenic lines showed stable seed quality and improved resistance to S. Sclerotiorum infection in the adult stage under field conditions.
Table 2.
Quality traits of seeds of transgenic rapeseed lines evaluated after S. sclerotiorum inoculation in the field in 2016
| Variety | 1000-seed weight (g) | Oil (%) | Glucosinolate (µmol g–1) | Protein (%) | Erucic acid (%) |
|---|---|---|---|---|---|
| 7-5C | 2.99 ± 0.06a | 40.05 ± 1.87a | 15.31 ± 0.25c | 26.69 ± 0.28a | 0.77 ± 0.17a |
| 7-5D | 3.25 ± 0.15a | 39.74 ± 0.25a | 17.39 ± 0.83b | 26.45 ± 0.35a | 0.79 ± 0.06a |
| 7-5G | 3.07 ± 0.13a | 38.38 ± 0.24a | 13.85 ± 0.73d | 27.89 ± 0.15a | 0.75 ± 0.06a |
| 7-5WT | 1.62 ± 0.08b | 30.68 ± 0.45b | 22.46 ± 1.61a | 28.52 ± 0.26a | 0.70 ± 0.07a |
| T45B#1 | 2.75 ± 0.06b | 38.99 ± 0.47a | 16.16 ± 1.24d | 26.67 ± 0.26a | 0.75 ± 0.04a |
| T45B#2 | 3.29 ± 0.24a | 38.72 ± 0.69a | 23.61 ± 3.08b | 27.87 ± 0.39a | 0.91 ± 0.08a |
| T45C | 2.97 ± 0.04a | 40.42 ± 0.19a | 21.03 ± 1.11c | 26.34 ± 0.22a | 0.77 ± 0.08a |
| T45WT | 1.68 ± 0.11c | 30.26 ± 0.06b | 34.45 ± 0.12a | 27.41 ± 0.04a | 0.84 ± 0.04a |
The field assay in the T4 generation was performed in two completely randomized blocks and each replicate contained about 20 plants. WT indicates non-transformed rapeseed plants. Values are means of nine replicates ±SD. Seed quality traits were measured based on 1000 well-filled dry seeds. Multiple comparisons were used to test statistical significance. Different letters indicate significant differences at P<0.05.
Leaf extracts of OsPGIP2 transgenic lines delay S. sclerotiorum growth
To verify resistance in vitro, we cultivated S. sclerotiorum in uninfected transgenic OsPGIP2 rapeseed leaf extracts (Fig. 2A). We found that S. sclerotiorum grew significantly more on PDA without leaf extract than when cultured with 50% (v/v) leaf extract from transgenic lines and wild-type plants; however, the diameters of the S. sclerotiorum colonies in PDA with leaf extracts from transgenic rapeseed were significantly smaller than in PDA with control leaf extract (Fig. 2B). In addition, whilst the hyphal growth of S. sclerotiorum was evenly and sparsely distributed on the PDA plates with extraction buffer (control) (Fig. 2C), in the presence of leaf extracts from transgenic and wild-type lines growth was reduced, with dense distribution and entangled ends of the hyphae. The reduction in growth was more conspicuous on PDA plates containing leaf extracts from the transgenic lines (Fig. 2C). These experiments suggested that leaf extracts of transgenic lines might effectively delay the expansion of S. sclerotiorum.
Fig. 2.
Cultures of S. sclerotiorum on PDA plates with leaf extracts from OsPGIP2 transgenic and wild-type lines. (A) Images of S. sclerotiorum inoculated on PDA plates treated with either leaf extracts (50%) or a mock solution at 36 h post-infection (hpi). Scale bars are 4 cm. (B) Statistical analysis of the diffusion diameter of S. sclerotiorum on PDA plates treated with either leaf extracts (50%) or a mock solution at 36 hpi. Data are means from three biological replicates (±SD) (n=3). Significant differences were determined by one-way ANOVA: ***P<0.001, **P<0.01, *P<0.05. 7-5WT and T45WT are wild-type controls. (C) Hyphal growth of S. sclerotiorum on PDA plates treated with either OsPGIP2 transgenic leaf extracts (50%) or a mock solution at 36 hpi. Scale bars are 1 mm.
DEGs in transgenic plants infected with S. sclerotiorum
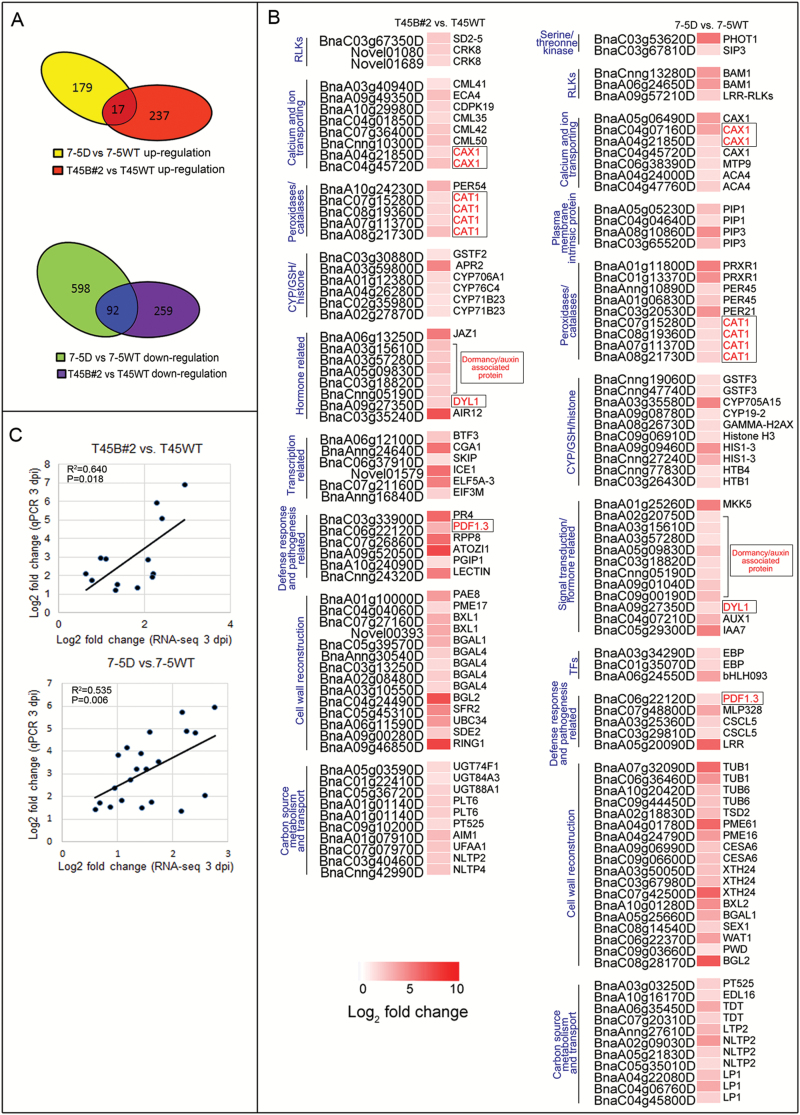
To elucidate the molecular network underlying OsPGIP2 disease resistance, we performed transcriptome profiling analysis on the stems of transgenic (7-5D and T45B#2) and wild-type (7-5WT and T45WT) lines at 3 dpi (Supplementary File S1). The proportions of reads mapped to the S. sclerotiorum genome were significantly higher in the wild-type than in the transgenic lines (see Supplementary Table S2), suggesting that S. sclerotiorum could infect and propagate more easily in the wild-type lines. To explore the biological functions of OsPGIP2 in 7-5D and T45B#2, reads were mapped to the reference genome of B. napus (Chalhoub et al., 2014). We found that 17 up-regulated genes were common to both genotypes, 179 genes were specific to the 7-5 background, and 237 were specific to the T45 background (Fig. 3A). There were 92 down-regulated genes that were common to both genotypes; 598 were specific to 7-5, and 259 were specific to T45. To validate the data obtained from the RNA-seq, 13 and 21 genes from the T45 and 7-5 transgenic lines, respectively, were selected for qPCR. For all 34 genes, the results obtained using both techniques were highly related at 3 dpi (T45B#2: R2=0.640, P<0.018; 7-5D: R2=0.535, P=0.006), confirming the reliability of RNA-seq data (Fig. 3C). In addition, 18 genes were also tested at 0–7 dpi in the T45 and 7-5 backgrounds, and this revealed their higher expression levels in transgenic than in non-transgenic lines after inoculation (Supplementary Figs S6, S7).
Fig. 3.
Differential gene expression between transgenic and wild-type B. napus lines in response to S. sclerotiorum infection. (A) Venn diagrams representing the up- and down-regulation of B. napus differentially expressed genes (DEGs) in response to S. sclerotiorum infection at 3 d post-inoculation (dpi). (B) Heatmap of a subset of significantly up-regulated DEGs related to S. sclerotiorum infection. DEGs were considered statistically significant if q-value <0.005 and |log2-fold change| >1. The log2-fold change is indicated according to the scale bar. The intensity of the colour indicates the expression of genes, with a darker colour meaning higher gene expression. The common up-regulated DEGs are highlighted by boxes. Details of the genes are given in Supplementary File S2. (C) Correlations of gene expression ratios obtained by RNA-seq and qPCR. The qPCR log2-value of the expression ratio (transgenic lines-inoculated/WT-inoculated) (y-axis) was plotted against the value from the RNA-seq (x-axis). All qPCR data were collected from three biological replicates and three technical replicates for each sample, and RNA-seq data were obtained from one pooled sample.
GO analysis reveals the defense mechanisms of OsPGIP2 transgenic lines to S. sclerotiorum
Gene ontology (GO) term annotations were performed to identify any significant enrichment in each term relative to the entire proteome (see Supplementary Fig. S4B, C). Although T45B#2 and 7-5D showed a similar pathway to enhanced SSR resistance, comprising 17 up- and 92 down-regulated shared genes, genes in the same category were largely different (Fig. 3B, Supplementary Fig. S5). In 7-5D compared with 7-5WT, we identified three up- and six down-regulated receptor-like protein kinases (RLKs), which participate in intracellular protein interactions and signal transduction, and function in PAMP recognition (Zhang et al., 2013) (Supplementary Fig. S5, Fig. 3B). However, comparing transgenic and wild-type in the T45 background, we identified three up- and three down-regulated RLKs, and these DEGs are different from the RLKs in the 7-5 background (Supplementary Fig. S5, Fig. 3B). Eight hormone-related genes were up-regulated in T45B#2 compared to the T45WT line, including JAZ1 (jasmonate-zim-domain protein 1) and seven auxin-associated genes (Fig. 3B). For signal transduction, three mitogen-activated protein (MAP) kinases were down-regulated and MAP kinases (MKK5) were up-regulated in the 7-5D transgenic line compared to 7-5WT (Supplementary Fig. S5, Fig. 3B). After S. sclerotiorum infection, defense-related genes and transcription factors (TFs) also were induced in the OsPGIP2-expressing lines; for example, plant defensin 1.3 (PDF1.3), pathogenesis-related 5 (PR-5), and WRKYs were induced in the 7-5D and T45B#2 transgenic lines (Fig. 3B, Supplementary S5). Within the ‘cell wall reconstruction’ pathway, the most prominent changes in expression included the up-regulation of xyloglucan endotransglucosylase/hydrolase protein 24 (XTH24) and pectinesterase family proteins PME61, and the down-regulation of arabinogalactan (AGP20) in 7-5D (Fig. 3B, Supplementary S5). These cell wall-modifying enzymes modulate cell wall extensibility and plasticity during both developmental and stress-response programs (Le Gall et al., 2015). Based on the distribution of all the DEGs, up- and down-regulated genes in the transgenic lines responding to S. sclerotiorum were classified into plant defense-response pathways, including serine/threonine kinase and RLKs, calcium and ion transporting, peroxidases/catalases, signal transduction/hormone-related, pathogen-related defense response, transcription-related, cell wall reconstruction, cytochrome/glutathione/histone (CYP/GSH/histone), and carbon source metabolism and transport (Fig. 3B, Supplementary S5).
S. sclerotiorum infection induces H2O2 accumulation in OsPGIP2 transgenic lines
As a second messenger, ROS directly and indirectly regulates the innate immune system of plants to enhance defense against disease (Foyer and Noctor, 2011). In our experiments, more peroxidase and catalase genes were differently expressing in transgenic and control plants after S. sclerotiorum inoculation (Fig. 3B, Supplementary S5). The difference in ROS (H2O2-derived) production between transgenic and wild-type plants was detected by DAB staining. Inoculated sites on transgenic 7-5D and T45B#2 leaves infiltrated with S. sclerotiorum were more strongly stained by DAB than controls (Fig. 4A), indicating that H2O2 accumulation in OsPGIP2-expressing leaves was strongly induced by infection. After S. sclerotiorum inoculation, the expression of ROS-related enzymes such as PER21 (peroxidase 21, BnaC03g20530D), PER54 (BnaA10g24230D), and CAT1 (catalase 1, BnaA07g11370D) increased significantly in both the transgenic and wild-type plants, but this increase was more significant in the OsPGIP2 transgenic plants than in the wild-type controls (Fig. 4B, C). These data indicated that OsPGIP2 transgenic plants accumulated more H2O2 to increase innate immune signaling and to enhance plant resistance to S. sclerotiorum.
Fig. 4.
H2O2 accumulation and the expression levels of H2O2-associated genes in response to S. sclerotiorum inoculation in OsPGIP2-overexpressing and wild-type lines of rapeseed. (A) Detection of H2O2 accumulation by DAB staining in B. napus leaves after S. sclerotiorum inoculation. The images show leaves at 72 h post-infection with either mock solution or S. sclerotiorum (S). Seedlings with 9–10 leaves were used and the inoculation assays were conducted on detached leaves. Three biological replicates were used in this experiment. Scale bars are 500 μm. (B, C) Expression levels of PER21 (BnaC03g20530D), PER54 (BnaA10g24230D), and CAT1 (BnaA07g11370D) in wild-type (WT) and OsPGIP2-expressing transgenic lines inoculated with S. sclerotiorum as determined by qPCR and normalized to BnACTIN7 and UBC21. Data are means (±SD) and are representative of three biological and three technical replicates. dpi, days post-inoculation.
OsPGIP2 directly interacts with SsPG3 and SsPG6
To further understand the mechanism of S. sclerotiorum resistance by OsPGIP2 gene regulation, we measured gene expression in the stems of plants infected with S. sclerotiorum by qPCR. In 7-5 transgenic lines, the expression of OsPGIP2 peaked at 5 dpi and then decreased; however, in T45 transgenic lines, the expression of OsPGIP2 gradually increased over time (Fig. 5A). Furthermore, we found that the relative expression of the S. sclerotiorum SsPG genes changed at the necrotic site of the infected B. napus. The expressions of SsPG1, SsPG3, SsPG5, and SsPG6 were significantly lower in the transgenic than in the wild-type lines (Fig. 5B). These results indicated that the enhanced S. sclerotiorum resistance in transgenic OsPGIP2 lines might be due to a mechanism that reduces the expression of S. sclerotiorum pathogenic genes.
Fig. 5.
OsPGIP2 interacts with SsPG3 and SsPG6 when transiently expressed in N. benthamiana leaves. (A) The expression levels of OsPGIP2 in transgenic and wild-type (WT) lines inoculated with S. sclerotiorum. qPCR results were normalized to BnACTIN7 and UBC21. Data are means (±SD) and are representative of three biological and three technical replicates. (B) Expression levels of S. sclerotiorum SsPGs in transgenic and WT plants after inoculation. qPCR results were normalized to S. sclerotiorum 28S ribosomal DNA and histone H3. Three biological and three technical replicates were used in this experiment. (C–G) Transient expression assays to examine the interaction between OsPGIP2 and SsPGs. OsPGIP2 did not interact with SsPG1 (C); it interacted with SsPG3 (D); it did not interact with SsPG5 (E); it interacted with SsPG6 (F); positive control (G). Two biological replicates and two technical replicates were used in this experiment. The results of the interactions are presented on the right of each panel. The left of each panel shows the interaction between C-terminal luciferase (LUC) and the N-terminal target gene (top), and between the N-terminal LUC and the C-terminal target gene (bottom).
The expression of S. sclerotiorum SsPGs has been associated with the establishment and development of infection (Li et al., 2004). Based on the hypothesis of a ‘PG–PGIP’ interaction (Benedetti et al., 2011; Gutierrez, 2012), we hypothesized that OsPGIP2 could improve SSR resistance in B. napus by interacting with the PGs secreted by S. sclerotiorum. To examine this hypothesis, the affinities between OsPGIP2 and SsPGs were predicted using PPA-Pred2. The results showed that the affinities were higher in OsPGIP2–SsPG3 and OsPGIP2–SsPG6 than in OsPGIP2–SsPGl and OsPGIP2–SsPG5, suggesting that OsPGIP2 interacts more easily with SsPG3 and SsPG6 (see Supplementary Table S3). We cloned OsPGIP2 into the C-terminus and SsPGs into the N-terminus of the luciferase reporter gene, and then co-transfected into N. benthamiana leaves using an A. tumefaciens-mediated transient expression system. Fluorescence observations indicated that SsPG3 and SsPG6 interacted with OsPGIP2, but SsPG1 and SsPG5 did not (Fig. 5C–G), indicating that OsPGIP2 might interact with SsPG3 and SsPG6 in vivo to enhance S. sclerotiorum resistance.
Changes in cell wall monosaccharide content in the stems of OsPGIP2-expressing rapeseed
Plant cell walls largely consist of the polysaccharides cellulose, hemicellulose, pectin, and lignin. The cell wall structural profile was found to be altered during the development of transgenic VvPGIP1 tobacco leaves (Nguema-Ona et al., 2013). In our study, RNA-seq data showed that some cell wall synthetic and degrading enzymes were up-regulated in the transgenic OsPGIP2 lines. Thus, we hypothesized that OsPGIP2 overexpression might affect the composition of plant cell walls, and hence we determined the monosaccharide composition of rapeseed stem cell walls by HPLC (Table 3, and see Supplementary Fig. S8). The hemicellulose side-chain substitution rate (xylose/arabinose, Xyl/Ara) is also included in Table 3. Interestingly, although arabinose is not the major side-chain substitution for xylan in dicot plants, the xylose content of OsPGIP2-expressing lines in the flowering period was significantly lower than that of wild-type controls (Table 3). However, with flower development, the content of xylose dramatically increased in OsPGIP2-expressing lines compared to the wild-type. In 7-5WT and T45WT, the arabinose content from the flowering to final flowering stages gradually decreased. In contrast, arabinose contents gradually increased in the 7-5 and T45 transgenic lines during these developmental stages. The content of cellulose was evaluated based on the glucose content, and we found that transgenic lines had higher glucose and hemicellulose contents than the wild-type lines. Because lignin has an important role in cell wall remodeling, its content was further examined. As flowering progressed, the content of lignin in the stem increased in all the tested lines. In addition, except for T45B#1 and T45B#2, there were no significant differences in lignin content between the transgenic and control lines in the two flowering stages, indicating that overexpressing OsPGIP2 had a low effect on lignin content (Table 3).
Table 3.
Cell wall composition profile (%, mass ratio) of OsPGIP2-overexpressing rapeseed stem samples
| Glu | Xyl | Gal | Ara | Man | Xyl/Ara | Hemicellulose | Total lignin | |
|---|---|---|---|---|---|---|---|---|
| Flowering period | ||||||||
| 7-5WT | 27.47 ± 0.17 | 10.58 ± 0.09 | 0.0531 ± 0.0004 | 1.84 ± 0.04 | 2.06 ± 0.04 | 5.78 ± 0.18 | 14.51 ± 0.03 | 11.46 ± 0.04 |
| 7-5C | 30.46 ± 0.29** | 6.96 ± 0.09** | 0.0535 ± 0.0009 | 1.81 ± 0.10 | 2.79 ± 0.07* | 3.86 ± 0.17** | 11.61 ± 0.26** | 11.14 ± 0.63 |
| 7-5D | 30.35 ± 0.23** | 7.66 ± 0.06** | 0.0519 ± 0.0011 | 1.65 ± 0.27 | 2.86 ± 0.11* | 4.65 ± 0.22* | 12.21 ± 0.12* | 12.59 ± 0.75 |
| 7-5G | 30.48 ± 0.38** | 7.08 ± 0.05** | 0.0584 ± 0.0087 | 1.55 ± 0.25 | 2.83 ± 0.16* | 4.60 ± 0.10* | 11.51 ± 0.08** | 13.13 ± 0.11 |
| T45WT | 26.91 ± 0.27 | 7.76 ± 0.06 | 0.0540 ± 0.0047 | 1.83 ± 0.01 | 1.88 ± 0.10 | 4.25 ± 0.02 | 11.52 ± 0.05 | 14.51 ± 0.13 |
| T45B#1 | 27.33 ± 0.28 | 6.37 ± 0.11* | 0.0480 ± 0.0047 | 1.65 ± 0.04 | 2.05 ± 0.53 | 3.87 ± 0.15* | 10.11 ± 0.06* | 15.91 ± 0.47 |
| T45B#2 | 27.62 ± 0.33 | 4.91 ± 0.24** | 0.0528 ± 0.0016 | 1.72 ± 0.03 | 2.20 ± 0.03** | 2.04 ± 0.09** | 8.88 ± 0.24** | 13.05 ± 0.86 |
| T45C | 27.69 ± 0.15 | 5.47 ± 0.14** | 0.0550 ± 0.0028 | 1.79 ± 0.13 | 2.07 ± 0.44 | 3.04 ± 0.12** | 9.39 ± 0.11* | 13.24 ± 0.50 |
| Final flowering period | ||||||||
| 7-5WT | 30.39 ± 0.95 | 10.97 ± 0.77 | 0.0517 ± 0.0015 | 1.63 ± 0.04 | 2.56 ± 0.13 | 6.78 ± 0.61 | 15.21 ± 0.62 | 18.37 ± 0.04 |
| 7-5C | 32.51 ± 0.28** | 13.25 ± 0.06** | 0.0510 ± 0.0009 | 1.85 ± 0.05 | 3.05 ± 0.02* | 7.17 ± 0.19** | 18.20 ± 0.05** | 20.03 ± 0.10 |
| 7-5D | 31.65 ± 0.64* | 11.39 ± 0.03 | 0.0530 ± 0.0021 | 1.72 ± 0.03 | 3.29 ± 0.12** | 6.64 ± 0.11* | 16.43 ± 0.13* | 20.57 ± 0.53 |
| 7-5G | 31.32 ± 0.22* | 14.12 ± 0.14** | 0.0521 ± 0.0013 | 2.53 ± 0.05** | 3.19 ± 0.09** | 5.60 ± 0.05* | 19.89 ± 0.25*** | 20.91 ± 0.05 |
| T45WT | 31.74 ± 0.11 | 8.47 ± 0.17 | 0.0524 ± 0.0011 | 0.66 ± 0.13 | 1.91 ± 0.10 | 12.91 ± 0.27 | 11.09 ± 0.12 | 17.28 ± 0.22 |
| T45B#1 | 32.92 ± 0.78* | 11.67 ± 0.13** | 0.0532 ± 0.0025 | 2.48 ± 0.27** | 2.80 ± 0.06** | 4.71 ± 0.18** | 17.00 ± 0.04*** | 20.10 ± 0.29* |
| T45B#2 | 32.67 ± 0.24* | 10.65 ± 0.18** | 0.0547 ± 0.0015 | 2.19 ± 0.11** | 2.59 ± 0.05* | 4.88 ± 0.10** | 15.69 ± 0.46** | 22.30 ± 0.14** |
| T45C | 32.96 ± 0.71* | 10.43 ± 0.20** | 0.0554 ± 0.0033 | 2.08 ± 0.10** | 3.00 ± 0.12*** | 5.04 ± 0.04** | 15.56 ± 0.18** | 19.84 ± 1.24 |
Glu, glucose; Xyl, xylose; Gal, galactose; Ara, arabinose; Man, mannose. Total lignin includes acid-insoluble and acid-soluble lignin. Analyses were performed using three biological replicates for wild-type and transgenic plants. Data are means ±SD. Significance was tested by one-way ANOVA: ***P<0.001, **P<0.01, *P<0.05. 7-5WT and T45WT are the wild-type controls.
Discussion
The resistance of rapeseed to S. sclerotiorum obtained by using transgenic technology has mainly been based on observations of resistance in detached leaves in in vitro experiments (Wang et al., 2014; Liu et al., 2015). In the present study, transgenic OsPGIP2-overexpressing lines were created in two B. napus backgrounds and resulted in rapeseed plants with increased resistance to S. sclerotiorum, as indicated by survival tests after spraying with fungal hyphae and by the assessment of three consecutive years of stem inoculation with S. sclerotiorum in the field. Because transgenic OsPGIP2 lines showed significantly smaller lesions than wild-type plants in detached-leaf inoculation experiments at 72 but not at 48 hpi, the function of OsPGIP2 in increasing SSR resistance might have its major effect during the expansion of the infection. Thus, RNA-seq of transgenic B. napus was performed at 3 d after S. sclerotiorum infection.
When plants are infected with S. sclerotiorum, they show lower 1000-seed weight and lower seed quality than non-infected plants in soybean (Danielson et al., 2004). Although we did not focus on the yield difference between the transgenic and wild-type lines, we also detected a reduction in the 1000-seed weight and significant effects on seed quality traits after S. sclerotiorum inoculation. Lower expression of GSL biosynthesis genes was related to lower GSL contents in the 7-5 and T45 transgenic lines after inoculation (see Supplementary Fig. S9A and Supplementary File S3). However, Wu et al. (2016) compared gene transcription between resistant and susceptible B. napus lines inoculated with S. sclerotiorum and found that biosynthesis of indolic GSL was more intensely induced in the resistant line than in the susceptible line, and these results are consistent with the expression of indolic GSL biosynthesis genes. Zhao and Meng (2003) found that the contents of aliphatic GSL and 3-indolyl-methyl GSL in seeds were in a positive relationship with S. sclerotiorum resistance on leaves at the seedling stage and on stems of maturing plants, respectively. In our present study, when comparing between low disease pressure and field stem inoculation, the wild-type seeds had significant changes in the content of GSL but this was not the case in the transgenic lines (Supplementary Fig. S9B). This means that the transgenic OsPGIP2 rapeseed lines did not show additional induction of GSL production against SSR, indicating that OsPGIP2 led to S. sclerotiorum resistance in transgenic lines through a mechanism that did not involve accumulation of GSL. Thus, there seems to be no direct correlation between disease resistance due to the OsPGIP2 gene and synthesis of GSLs.
The ‘PG–PGIP’ interaction plays a key role in determining protein–protein recognition in plant–pathogen interactions. The pathogenic PGs from S. sclerotiorum are critical to both the establishment and expansion of infection (Li et al., 2004). Bashi et al. (2013) demonstrated that BnPGIP1 could interact with SsPG6 in vitro, and, in the present study, we showed that OsPGIP2 could interact with SsPG3 and SsPG6 but not with SsPG1 and SsPG5 in vivo, indicating that the inhibitory activity of every PGIP is specific to a range of PGs from different fungal pathogens. For instance, the purified PGIP1 of Malus domestica inhibits the PGs from Colletotrichum lupine, Botryosphaeria obtusa, and Diaporthe ambigua, but not the A. niger PG (Oelofse et al., 2006); PvPGIP1, PvPGIP2, and PvPGIP4 inhibit a PG from A. niger (Leckie et al., 1999; D’Ovidio et al., 2004) but PvPGIP3 does not (D’Ovidio et al., 2004). However, PvPGIP2-expressing wheat lines infected with Claviceps purpurea do not show a clear reduction of disease symptoms due to the lack of function of inhibiting PGs from C. purpurea (Volpi et al., 2013). Although the overexpression of OsPGIP2 significantly increased S. sclerotiorum resistance in transgenic B. napus lines compared with wild-type lines, their resistance did not increase to the level shown by ZS11, indicating that increasing the expression of PGIP alone is not sufficient to greatly increase SSR resistance in rapeseed. In particular, S. sclerotiorum has evolved a variety of pathogenic factors, such as oxalic acid (OA), cerato-platanin protein 1 (CP1), SsCUTA (cutinase), and fungal integrin-like protein (SsITL) (Heard et al., 2015; Yang et al., 2018). Thus, although overexpressing OsPGIP2 in rapeseed must involve the ‘PG–PGIP’ interaction, it is also necessary to find additional mechanisms of interaction between rapeseed and S. sclerotiorum, and to combine PGIP with other defense-response genes, such as PR-1 that interacts with SsCP1 in the apoplast (Yang et al., 2018), in order to improve the degree of rapeseed resistance (Bashi et al., 2013).
Biochemical and RNA-seq analyses of OsPGIP2-expressing rapeseed lines revealed that the content of cell wall monosaccharides and polysaccharide synthases or hydrolases were specifically up-regulated in the transgenic lines. Endotransglucosylases/hydrolases (XTHs) are believed to be important for regulating cell wall strength, extensibility, and tissue integrity (Saladié et al., 2006). Cellulose synthases (CESAs) are involved in the synthesis of primary and secondary cell wall cellulose (Chundawat et al., 2011). In tobacco, VvPGIP1 overexpression results in the down-regulation of a group of XTHs and in a reduction of XTH enzyme activity in uninfected transgenic tobacco (Alexandersson et al., 2011). However, in our present study, the cytoskeletal genes CESA6 and XTH24 were specifically up-regulated in the transgenic OsPGIP2 lines, which had higher cellulose contents than the wild-type lines, indicating that OsPGIP2 has a direct or indirect effect on cell wall modification, possibly leading to changes in cell wall monosaccharide metabolism. In addition, during the process of infection in plants, intracellular soluble sugar provides a source of nutrition for colonization by the pathogens (Doidy et al., 2012). Thus, the enhancement of the metabolism and transport of carbon compounds in the transgenic OsPGIP2 lines also demonstrated competition for nutrients between the plant and pathogen. Indeed, increased invertase activity and plant monosaccharide-importer expression may result in pathogens having reduced access to sugars during infection (Sutton et al., 2007).
Reactive oxygen species are important mediators in PAMP-triggered immunity (PTI), contributing to defense responses during plant–pathogen interactions (Baxter et al., 2014). The defense response is mediated by oxidative waves of ROS that activate signal transduction through phosphorylation cascades, accompanied by hormonal signaling and by the expression of defense-related genes (Baxter et al., 2014). In A. thaliana, peroxidases have been identified as major contributors to ROS production during responses to fungal elicitors (Daudi et al., 2012). In our present study, elevated H2O2 accumulation and expression of peroxidases were detected in the leaves of OsPGIP2-expressing lines after inoculation. Although ROS plays a role in resistance during early infection, more H2O2 may promote disease during later infection stages of S. sclerotiorum (Williams et al., 2011). Therefore, to reduce H2O2 toxicity to plants, CAT1s and glutathione metabolic processes have been induced in transgenic lines, and this induction at the core of the plant antioxidant system protects ROS signaling and contributes to the removal of O2– to protect the plant from developing lesions due to ROS toxicity (Foyer and Noctor, 2011).
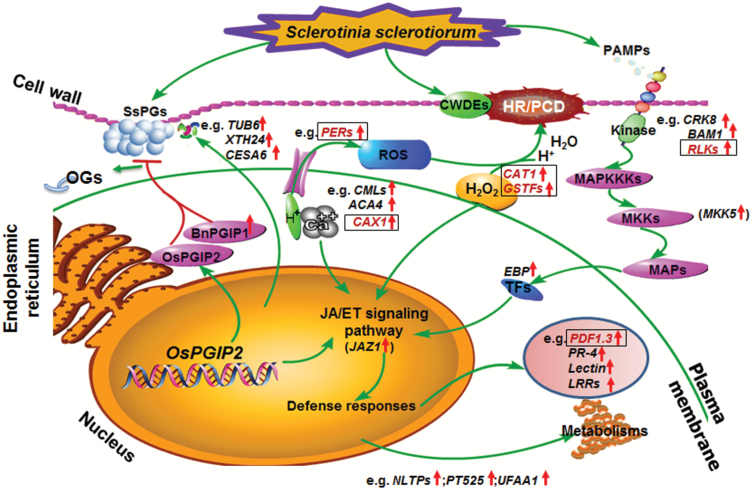
Although the mechanism by which OsPGIP2 reduces SSR development is still largely unknown, the RNA-seq results presented here hint at a possible molecular mechanism. Based on changes in gene expression in response to infection in the transgenic rapeseed lines, we propose a model for OsPGIP2-conferred resistance to S. sclerotiorum in these plants (Fig. 6). After S. sclerotiorum infection, PAMP perception and a series of signal transductions are initiated, and then followed by the activation of several B. napus defenses aimed at delaying S. sclerotiorum development. These include: (i) increasing ROS (H2O2-derived) to active the innate immune pathway (Fig. 4); (ii) reinforcing cell walls to inhibit the growth of S. sclerotiorum (Table 3); (iii) increasing the accumulation of OsPGIP2 to inhibit SsPG3 and SsPG6 (Fig. 5); and increasing the accumulation of BnPGIP1 to inhibit SsPG6 (Bashi et al., 2013). As a result of the synergistic actions of these genes, the transgenic rapeseed plants have a stronger resistance to S. sclerotiorum than wild-type plants at both the seedling and adult stages. In summary, the increased resistance observed in transgenic OsPGIP2-expressing lines suggests that overexpression can activate the basal defense pathway, which provides a molecular basis for the application of OsPGIP2 to develop plants resistant to fungal pathogens in other important crops.
Fig. 6.
A model for the molecular mechanism by which OsPGIP2 confers S. sclerotiorum resistance through increased activation of defense mechanisms. Upward arrows next to gene names highlight the up-regulation of differentially expressed genes, as revealed in RNA-seq analysis. Genes highlighted in boxes are the common up-regulated DEGs in the 7-5 and T45 backgrounds. Gene details are given in Supplementary File S2. CWDEs, cell wall-degrading enzymes; HR/PCD, hypersensitive response/programmed cell death; MAPs, mitogen-activated proteins; MAPKKKs, MAP-kinase-kinase kinases; MKKs, MAP-kinase kinases; OGs, oligogalacturonides; PAMPs, pathogen-associated molecular patterns; PGIP, polygalacturonase inhibitor protein; ROS, reactive oxygen species; SsPGs, S. sclerotiorum polygalacturonases; TFs, transcription factors.
Supplementary data
Supplementary data are available at JXB online.
Fig. S1. Analysis of OsPGIP2 gene expression in transgenic lines by molecular identification methods.
Fig. S2. The phenotypes of seedlings and adult plants of T4 transgenic and wild-type lines.
Fig. S3. Seed quality traits of T4 transgenic lines following inoculation with S. sclerotiorum.
Fig. S4. Clustered heatmap and GO enrichment analysis of differentially expressed genes between transgenic and wild-type lines of B. napus responsive to S. sclerotiorum.
Fig. S5. Clustered heatmap analysis of down-regulated differentially expressed genes between transgenic and wild-type lines of B. napus responsive to S. sclerotiorum.
Fig. S6. qPCR confirmation of the differentially expressed genes between 7-5 transgenic and wild-type lines after inoculation.
Fig. S7. qPCR confirmation of the differentially expressed genes between T45 transgenic and wild-type lines after inoculation.
Fig. S8. Determination of cell wall monosaccharides in the transgenic lines at different stages by HPLC.
Fig. S9. Expression of GSL biosynthesis genes and the seed content of GSL.
Table S1. Field testing data for disease severity for individual rapeseed lines in three consecutive years.
Table S2. Number of clean sequence reads that mapped to the S. sclerotiorum 1980 genome.
Table S3. The affinity between OsPGIP2 and S. sclerotiorum PGs.
Table S4. List of primers used in this study.
File S1. List of all differentially expressed gene annotations and FPKM values.
File S2. Up- and down-regulated differentially expressed genes in defense related pathways.
File S3. Transcript abundance of GSL genes for transgenic and wild-type lines.
Acknowledgements
We thank for Dr Jian Wu for maintaining the S. sclerotiorum fungus and Dr Liaoxun Lu for the pCAMIBA1300-35S:OsPGIP2 vector. This work was supported by the National Natural Science Foundation of China (no. 31401413), the National Key Research and Development Plan of China (2016YFD0101300), and the Technological innovation project of HuBei (2016ABA084).
References
- Agüero CB, Uratsu SL, Greve C, Powell AL, Labavitch JM, Meredith CP, Dandekar AM. 2005. Evaluation of tolerance to Pierce’s disease and Botrytis in transgenic plants of Vitis vinifera L. expressing the pear PGIP gene. Molecular Plant Pathology 6, 43–51. [DOI] [PubMed] [Google Scholar]
- Alizadeh N, Babaiahary A, Assadi A, Valizadeh M. 2006. The effects of Sclerotinia stem rot of oilseed rape on the production and quality of extracted oil. Journal of Science & Technology of Agriculture & Natural Resources 3, 485–495. [Google Scholar]
- Akhgari AB, Motallebi M, Zamani MR. 2012. Bean polygalacturonase-inhibiting protein expressed in transgenic Brassica napus inhibits polygalacturonase from its fungal pathogen Rhizoctonia solani. Plant Protection Science 48, 1–9. [Google Scholar]
- Alexandersson E, Becker JV, Jacobson D, Nguema-Ona E, Steyn C, Denby KJ, Vivier MA. 2011. Constitutive expression of a grapevine polygalacturonase-inhibiting protein affects gene expression and cell wall properties in uninfected tobacco. BMC Research Notes 4, 493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amselem J, Cuomo CA, van Kan JA, et al. 2011. Genomic analysis of the necrotrophic fungal pathogens Sclerotinia sclerotiorum and Botrytis cinerea. PLoS Genetics 7, e1002230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aziz A, Gauthier A, Bézier A, Poinssot B, Joubert JM, Pugin A, Heyraud A, Baillieul F. 2007. Elicitor and resistance-inducing activities of beta-1,4 cellodextrins in grapevine, comparison with beta-1,3 glucans and alpha-1,4 oligogalacturonides. Journal of Experimental Botany 58, 1463–1472. [DOI] [PubMed] [Google Scholar]
- Bashi ZD, Rimmer SR, Khachatourians GG, Hegedus DD. 2013. Brassica napus polygalacturonase inhibitor proteins inhibit Sclerotinia sclerotiorum polygalacturonase enzymatic and necrotizing activities and delay symptoms in transgenic plants. Canadian Journal of Microbiology 59, 79–86. [DOI] [PubMed] [Google Scholar]
- Baxter A, Mittler R, Suzuki N. 2014. ROS as key players in plant stress signalling. Journal of Experimental Botany 65, 1229–1240. [DOI] [PubMed] [Google Scholar]
- Benedetti M, Bastianelli E, Salvi G, Lorenzo GD, Caprari C. 2011. Artificial evolution corrects a repulsive amino acid in polygalacturonase inhibiting proteins (PGIPs). Journal of Plant Pathology 93, 89–95. [Google Scholar]
- Benedetti M, Pontiggia D, Raggi S, Cheng Z, Scaloni F, Ferrari S, Ausubel FM, Cervone F, De Lorenzo G. 2015. Plant immunity triggered by engineered in vivo release of oligogalacturonides, damage-associated molecular patterns. Proceedings of the National Academy of Sciences, USA 112, 5533–5538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boland GJ, Hall R. 1994. Index of plant hosts of Sclerotinia sclerotiorum. Canadian Journal of Plant Pathology 16, 93–108. [Google Scholar]
- Boller T, Felix G. 2009. A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annual Review of Plant Biology 60, 379–406. [DOI] [PubMed] [Google Scholar]
- Bolton MD, Thomma BP, Nelson BD. 2006. Sclerotinia sclerotiorum (Lib.) de Bary: biology and molecular traits of a cosmopolitan pathogen. Molecular Plant Pathology 7, 1–16. [DOI] [PubMed] [Google Scholar]
- Borras-Hidalgo O, Caprari C, Hernandez-Estevez I, Lorenzo GD, Cervone F. 2012. A gene for plant protection: expression of a bean polygalacturonase inhibitor in tobacco confers a strong resistance against Rhizoctonia solani and two oomycetes. Frontiers in Plant Science 3, 268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalhoub B, Denoeud F, Liu S, et al. 2014. Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science 345, 950–953. [DOI] [PubMed] [Google Scholar]
- Chotechung S, Somta P, Chen J, Yimram T, Chen X, Srinives P. 2016. A gene encoding a polygalacturonase-inhibiting protein (PGIP) is a candidate gene for bruchid (Coleoptera: bruchidae) resistance in mungbean (Vigna radiata). Theoretical and Applied Genetics 129, 1673–1683. [DOI] [PubMed] [Google Scholar]
- Chowdhury J, Henderson M, Schweizer P, Burton RA, Fincher GB, Little A. 2014. Differential accumulation of callose, arabinoxylan and cellulose in nonpenetrated versus penetrated papillae on leaves of barley infected with Blumeria graminis f. sp. hordei. New Phytologist 204, 650–660. [DOI] [PubMed] [Google Scholar]
- Chundawat SP, Beckham GT, Himmel ME, Dale BE. 2011. Deconstruction of lignocellulosic biomass to fuels and chemicals. Annual Review of Chemical and Biomolecular Engineering 2, 121–145. [DOI] [PubMed] [Google Scholar]
- Danielson GA, Nelson BD, Helms TC. 2004. Effect of Sclerotinia stem rot on yield of soybean inoculated at different growth stages. Plant Disease 88, 297–300. [DOI] [PubMed] [Google Scholar]
- Daudi A, Cheng Z, O’Brien JA, Mammarella N, Khan S, Ausubel FM, Bolwell GP. 2012. The apoplastic oxidative burst peroxidase in Arabidopsis is a major component of pattern-triggered immunity. The Plant Cell 24, 275–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doidy J, Grace E, Kühn C, Simon-Plas F, Casieri L, Wipf D. 2012. Sugar transporters in plants and in their interactions with fungi. Trends in Plant Science 17, 413–422. [DOI] [PubMed] [Google Scholar]
- Dong X, Ji R, Guo X, Foster SJ, Chen H, Dong C, Liu Y, Hu Q, Liu S. 2008. Expressing a gene encoding wheat oxalate oxidase enhances resistance to Sclerotinia sclerotiorum in oilseed rape (Brassica napus). Planta 228, 331–340. [DOI] [PubMed] [Google Scholar]
- D’Ovidio R, Mattei B, Roberti S, Bellincampi D. 2004. Polygalacturonases, polygalacturonase-inhibiting proteins and pectic oligomers in plant–pathogen interactions. Biochimica et Biophysica Acta 1696, 237–244. [DOI] [PubMed] [Google Scholar]
- Doyle J. 1990. Isolation of plant DNA from fresh tissue. Focus 12, 13–15. [Google Scholar]
- Dupont J, Reverchon M, Cloix L, Froment P, Ramé C. 2006. Transcriptional analysis of calcium-dependent and calcium-independent signalling pathways induced by oligogalacturonides. Journal of Experimental Botany 57, 2847–2865. [DOI] [PubMed] [Google Scholar]
- Falak I, Tulsieram L, Patel J, Charne D. 2011. Sclerotinia-resistant Brassica and methods for development of resistance to Sclerotinia. United States Patent no. US 7939722 B2. [Google Scholar]
- Fan C, Cai G, Qin J, Li Q, Yang M, Wu J, Fu T, Liu K, Zhou Y. 2010. Mapping of quantitative trait loci and development of allele-specific markers for seed weight in Brassica napus. Theoretical and Applied Genetics 121, 1289–1301. [DOI] [PubMed] [Google Scholar]
- Federici L, Di Matteo A, Fernandez-Recio J, Tsernoglou D, Cervone F. 2006. Polygalacturonase inhibiting proteins: players in plant innate immunity?Trends in Plant Science 11, 65–70. [DOI] [PubMed] [Google Scholar]
- Ferrari S, Savatin DV, Sicilia F, Gramegna G, Cervone F, Lorenzo GD. 2013. Oligogalacturonides: plant damage-associated molecular patterns and regulators of growth and development. Frontiers in Plant Science 4, 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foyer CH, Noctor G. 2011. Ascorbate and glutathione: the heart of the redox hub. Plant Physiology 155, 2–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fróes A, Macrae A, Rosa J, Franco M, Souza R, Soares R, Coelho R. 2012. Selection of a Streptomyces strain able to produce cell wall degrading enzymes and active against Sclerotinia sclerotiorum. Journal of Microbiology 50, 798–806. [DOI] [PubMed] [Google Scholar]
- Glawischnig E. 2007. Camalexin. Phytochemistry 68, 401–406. [DOI] [PubMed] [Google Scholar]
- Gutierrez G. 2012. SPR and differential proteolysis/MS provide further insight into the interaction between PGIP2 and EPGs. Fungal Biology 116, 737–746. [DOI] [PubMed] [Google Scholar]
- Heard S, Brown NA, Hammond-Kosack K. 2015. An interspecies comparative analysis of the predicted secretomes of the necrotrophic plant pathogens Sclerotinia sclerotiorum and Botrytis cinerea. PLoS ONE 10, e0130534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegedus DD, Li R, Buchwaldt L, Parkin I, Whitwill S, Coutu C, Bekkaoui D, Rimmer SR. 2008. Brassica napus possesses an expanded set of polygalacturonase inhibitor protein genes that are differentially regulated in response to Sclerotinia sclerotiorum infection, wounding and defense hormone treatment. Planta 228, 241–253. [DOI] [PubMed] [Google Scholar]
- Huangfu H, Guan C, Jin F, Yin C. 2014. Prokaryotic expression and protein function of Brassica napus PGIP2 and its genetic transformation. Plant Biotechnology Reports 8, 171–181. [Google Scholar]
- Hwang BH, Bae H, Lim HS, Kim KB, Kim SJ, Im MH, Park BS, Kim DS, Kim J. 2010. Overexpression of polygalacturonase-inhibiting protein 2 (PGIP2) of Chinese cabbage (Brassica rapa ssp pekinensis) increased resistance to the bacterial pathogen Pectobacterium carotovorum ssp carotovorum. Plant Cell, Tissue and Organ Culture 103, 293–305. [Google Scholar]
- Jang S, Lee B, Kim C, Kim SJ, Yim J, Han JJ, Lee S, Kim SR, An G. 2003. The OsFOR1 gene encodes a polygalacturonase-inhibiting protein (PGIP) that regulates floral organ number in rice. Plant Molecular Biology 53, 357–369. [DOI] [PubMed] [Google Scholar]
- Jiang Y, Fu X, Wen M, Wang F, Tang Q, Tian Q, Luo K. 2013. Overexpression of an nsLTPs-like antimicrobial protein gene (LJAMP2) from motherwort (Leonurus japonicus) enhances resistance to Sclerotinia sclerotiorum in oilseed rape (Brassica napus). Physiological and Molecular Plant Pathology 82, 81–87. [Google Scholar]
- Jonoubi P, Mousavi A, Majd A, Salmanian AH, Javaran MJ, Daneshian J. 2005. Efficient regeneration of Brassica napus L. hypocotyls and genetic transformation by Agrobacterium tumefaciens. Biologia Plantarum 49, 175–180. [Google Scholar]
- Juge N. 2006. Plant protein inhibitors of cell wall degrading enzymes. Trends in Plant Science 11, 359–367. [DOI] [PubMed] [Google Scholar]
- Kalunke RM, Tundo S, Benedetti M, Cervone F, De Lorenzo G, D’Ovidio R. 2015. An update on polygalacturonase-inhibiting protein (PGIP), a leucine-rich repeat protein that protects crop plants against pathogens. Frontiers in Plant Science 6, 146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Chauhan JS, Kumar A. 2010. Screening for erucic acid and glucosinolate content in rapeseed-mustard seeds using near infrared reflectance spectroscopy. Journal of Food Science and Technology 47, 690–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Gall H, Philippe F, Domon JM, Gillet F, Pelloux J, Rayon C. 2015. Cell wall metabolism in response to abiotic stress. Plants 4, 112–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leckie F, Mattei B, Capodicasa C, Hemmings A, Nuss L, Aracri B, De Lorenzo G, Cervone F. 1999. The specificity of polygalacturonase-inhibiting protein (PGIP): a single amino acid substitution in the solvent-exposed beta-strand/beta-turn region of the leucine-rich repeats (LRRs) confers a new recognition capability. The Embo Journal 18, 2352–2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecourieux D, Ranjeva R, Pugin A. 2006. Calcium in plant defence-signalling pathways. New Phytologist 171, 249–269. [DOI] [PubMed] [Google Scholar]
- Li R, Rimmer R, Buchwaldt L, Sharpe AG, Séguin-Swartz G, Hegedus DD. 2004. Interaction of Sclerotinia sclerotiorum with Brassica napus: cloning and characterization of endo- and exo-polygalacturonases expressed during saprophytic and parasitic modes. Fungal Genetics and Biology 41, 754–765. [DOI] [PubMed] [Google Scholar]
- Li S, Zhao B, Yuan D, Duan M, Qian Q, Tang L, Wang B, Liu X, Zhang J, Wang J. 2013. Rice zinc finger protein DST enhances grain production through controlling Gn1a/OsCKX2 expression. Proceedings of the National Academy of Sciences, USA 110, 3167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Wang M, Wen J, Yi B, Shen J, Ma C, Tu J, Fu T. 2015. Overexpression of barley oxalate oxidase gene induces partial leaf resistance to Sclerotinia sclerotiorum in transgenic oilseed rape. Plant Pathology 64, 1407–1416. [Google Scholar]
- Liu S, Wang H, Zhang J, Fitt BD, Xu Z, Evans N, Liu Y, Yang W, Guo X. 2005. In vitro mutation and selection of doubled-haploid Brassica napus lines with improved resistance to Sclerotinia sclerotiorum. Plant Cell Reports 24, 133–144. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2–△△Ct method. Methods 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Lu L, Zhou F, Zhou Y, Fan X, Ye S, Wang L, Chen H, Lin Y. 2012. Expression profile analysis of the polygalacturonase-inhibiting protein genes in rice and their responses to phytohormones and fungal infection. Plant Cell Reports 31, 1173–1187. [DOI] [PubMed] [Google Scholar]
- Matern U, Grimmig B, Kneusel RE. 2011. Plant cell wall reinforcement in the disease-resistance response: molecular composition and regulation. Canadian Journal of Botany 73, 511–517. [Google Scholar]
- Maulik A, Sarkar AI, Devi S, Basu S. 2012. Polygalacturonase-inhibiting proteins–leucine-rich repeat proteins in plant defence. Plant Biology 14, 22–30. [DOI] [PubMed] [Google Scholar]
- Meng X, Zhang S. 2013. MAPK cascades in plant disease resistance signaling. Annual Review of Phytopathology 51, 245–266. [DOI] [PubMed] [Google Scholar]
- Mengiste T. 2012. Plant immunity to necrotrophs. Annual Review of Phytopathology 50, 267–294. [DOI] [PubMed] [Google Scholar]
- Nguema-Ona E, Moore JP, Fagerström AD, Fangel JU, Willats WG, Hugo A, Vivier MA. 2013. Overexpression of the grapevine PGIP1 in tobacco results in compositional changes in the leaf arabinoxyloglucan network in the absence of fungal infection. BMC Plant Biology 13, 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson P. 2010. Plant pathogen resistance. WIPO Patent no. WO2010023491A2. [Google Scholar]
- Oelofse D, Dubery IA, Meyer R, Arendse MS, Gazendam I, Berger DK. 2006. Apple polygalacturonase inhibiting protein1 expressed in transgenic tobacco inhibits polygalacturonases from fungal pathogens of apple and the anthracnose pathogen of lupins. Phytochemistry 67, 255–263. [DOI] [PubMed] [Google Scholar]
- Osorio S, Castillejo C, Quesada MA, Medina-Escobar N, Brownsey GJ, Suau R, Heredia A, Botella MA, Valpuesta V. 2008. Partial demethylation of oligogalacturonides by pectin methyl esterase 1 is required for eliciting defence responses in wild strawberry (Fragaria vesca). The Plant Journal 54, 43–55. [DOI] [PubMed] [Google Scholar]
- Peliter AJ, Hatfield RD, Grau CR. 2009. Soybean stem lignin concentration relates to resistance to Sclerotinia sclerotiorum. Plant Disease 93, 149–154. [DOI] [PubMed] [Google Scholar]
- Ridley BL, O’Neill MA, Mohnen D. 2001. Pectins: structure, biosynthesis, and oligogalacturonide-related signaling. Phytochemistry 57, 929–967. [DOI] [PubMed] [Google Scholar]
- Robert-Seilaniantz A, Grant M, Jones JD. 2011. Hormone crosstalk in plant disease and defense: more than just jasmonate–salicylate antagonism. Annual Review of Phytopathology 49, 317–343. [DOI] [PubMed] [Google Scholar]
- Saladié M, Rose JK, Cosgrove DJ, Catalá C. 2006. Characterization of a new xyloglucan endotransglucosylase/hydrolase (XTH) from ripening tomato fruit and implications for the diverse modes of enzymic action. The Plant Journal 47, 282–295. [DOI] [PubMed] [Google Scholar]
- Schacht T, Unger C, Pich A, Wydra K. 2011. Endo- and exopolygalacturonases of Ralstonia solanacearum are inhibited by polygalacturonase-inhibiting protein (PGIP) activity in tomato stem extracts. Plant Physiology and Biochemistry 49, 377–387. [DOI] [PubMed] [Google Scholar]
- Seybold H, Trempel F, Ranf S, Scheel D, Romeis T, Lee J. 2014. Ca2+ signalling in plant immune response: from pattern recognition receptors to Ca2+ decoding mechanisms. New Phytologist 204, 782–790. [DOI] [PubMed] [Google Scholar]
- Sharma P, Meena PD, Verma PR, Saharan GS, Mehta N, Singh D, Kumar A. 2015. Sclerotinia sclerotiorum (Lib) de Bary causing Sclerotinia rot in oilseed Brassicas: a review. Journal of Oilseed Brassica 6, 1–44. [Google Scholar]
- Sluiter A, Hames B, Ruiz RO, Scarlata C, Sluiter J, Templeton D. 2004. Determination of structural carbohydrates and lignin in biomass. Technical Report. Denver, CO: US Department of Energy, National Renewable Energy Laboratory. [Google Scholar]
- Sparkes IA, Runions J, Kearns A, Hawes C. 2006. Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nature Protocols 1, 2019–2025. [DOI] [PubMed] [Google Scholar]
- Sutton PN, Gilbert MJ, Williams LE, Hall JL. 2007. Powdery mildew infection of wheat leaves changes host solute transport and invertase activity. Physiologia Plantarum 129, 787–795. [Google Scholar]
- Szankowski I, Briviba K, Fleschhut J, Schönherr J, Jacobsen HJ, Kiesecker H. 2003. Transformation of apple (Malus domestica Borkh.) with the stilbene synthase gene from grapevine (Vitis vinifera L.) and a PGIP gene from kiwi (Actinidia deliciosa). Plant Cell Reports 22, 141–149. [DOI] [PubMed] [Google Scholar]
- Torres MA. 2010. ROS in biotic interactions. Physiologia Plantarum 138, 414–429. [DOI] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. 2012. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nature Protocols 7, 562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volpi C, Raiola A, Janni M, Gordon A, O’Sullivan DM, Favaron F, D’Ovidio R. 2013. Claviceps purpurea expressing polygalacturonases escaping PGIP inhibition fully infects PvPGIP2 wheat transgenic plants but its infection is delayed in wheat transgenic plants with increased level of pectin methyl esterification. Plant Physiology and Biochemistry 73, 294–301. [DOI] [PubMed] [Google Scholar]
- Wang A, Wei X, Rong W, Dang L, Du LP, Qi L, Xu HJ, Shao Y, Zhang Z. 2015. GmPGIP3 enhanced resistance to both take-all and common root rot diseases in transgenic wheat. Functional & Integrative Genomics 15, 375–381. [DOI] [PubMed] [Google Scholar]
- Wang L, Feng Z, Wang X, Wang X, Zhang X. 2010. DEGseq: an R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics 26, 136–138. [DOI] [PubMed] [Google Scholar]
- Wang Z, Fang H, Chen Y, Chen K, Li G, Gu S, Tan X. 2014. Overexpression of BnWRKY33 in oilseed rape enhances resistance to Sclerotinia sclerotiorum. Molecular Plant Pathology 15, 677–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei L, Jian H, Lu K, Filardo F, Yin N, Liu L, Qu C, Li W, Du H, Li J. 2016. Genome-wide association analysis and differential expression analysis of resistance to Sclerotinia stem rot in Brassica napus. Plant Biotechnology Journal 14, 1368–1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams B, Kabbage M, Kim HJ, Britt R, Dickman MB. 2011. Tipping the balance: Sclerotinia sclerotiorum secreted oxalic acid suppresses host defenses by manipulating the host redox environment. PLoS Pathogens 7, e1002107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Cai G, Tu J, Li L, Liu S, Luo X, Zhou L, Fan C, Zhou Y. 2013. Identification of QTLs for resistance to Sclerotinia stem rot and BnaC.IGMT5.a as a candidate gene of the major resistant QTL SRC6 in Brassica napus. PloS ONE 8, e67740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Zhao Q, Yang Q, Liu H, Li Q, Yi X, Cheng Y, Guo L, Fan C, Zhou Y. 2016. Comparative transcriptomic analysis uncovers the complex genetic network for resistance to Sclerotinia sclerotiorum in Brassica napus. Scientific Reports 6, 19007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang G, Tang L, Gong Y, Xie J, Fu Y, Jiang D, Li G, Collinge DB, Chen W, Cheng J. 2018. A cerato-platanin protein SsCP1 targets plant PR1 and contributes to virulence of Sclerotinia sclerotiorum. New Phytologist 217, 739–755. [DOI] [PubMed] [Google Scholar]
- Young MD, Wakefield MJ, Smyth GK, Oshlack A. 2010. Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biology 11, R14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Fraiture M, Kolb D, Löffelhardt B, Desaki Y, Boutrot FF, Tör M, Zipfel C, Gust AA, Brunner F. 2013. Arabidopsis receptor-like protein30 and receptor-like kinase suppressor of BIR1-1/EVERSHED mediate innate immunity to necrotrophic fungi. The Plant Cell 25, 4227–4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Meng J. 2003. Detection of loci controlling seed glucosinolate content and their association with Sclerotinia resistance in Brassica napus. Plant Breeding 122, 19–23. [Google Scholar]
- Zhao J, Wang J, An L, Doerge RW, Chen ZJ, Grau CR, Meng J, Osborn TC. 2007. Analysis of gene expression profiles in response to Sclerotinia sclerotiorum in Brassica napus. Planta 227, 13–24. [DOI] [PubMed] [Google Scholar]
- Zurbriggen MD, Carrillo N, Tognetti VB, Melzer M, Peisker M, Hause B, Hajirezaei MR. 2009. Chloroplast-generated reactive oxygen species play a major role in localized cell death during the non-host interaction between tobacco and Xanthomonas campestris pv. vesicatoria. The Plant Journal 60, 962–973. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.