Abstract
Although the anticancer properties of oligomeric proanthocyanidins (OPCs) from grape seeds have been well recognized, the molecular mechanisms by which they exert anticancer effects are poorly understood. In this study, through comprehensive RNA-sequencing-based gene expression profiling in multiple colorectal cancer cell lines, we for the first time illuminate the genome-wide effects of OPCs from grape seeds in colorectal cancer. Our data revealed that OPCs affect several key cancer-associated genes. In particular, genes involved in cell cycle and DNA replication were most significantly and consistently altered by OPCs across multiple cell lines. Intriguingly, our in vivo experiments showed that OPCs were significantly more potent at decreasing xenograft tumor growth compared with the unfractionated grape seed extract (GSE) that includes the larger polymers of proanthocyanidins. These findings were further confirmed in colorectal cancer patient-derived organoids, wherein OPCs more potently inhibited the formation of organoids compared with GSE. Furthermore, we validated alteration of cell cycle and DNA replication-associated genes in cancer cell lines, mice xenografts as well as patient-derived organoids. Overall, this study provides an unbiased and comprehensive look at the mechanisms by which OPCs exert anticancer properties in colorectal cancer.
A comprehensive analysis of gene expression and pathway alterations that are mediated by OPCs present in GSE was identified by genome-wide RNA sequencing in colorectal cancer cells and validated animal models and patient-derived organoid models.
Introduction
Flavonoids are a heterogeneous group of polyphenolic compounds with a C6–C3–C6 backbone, which is present abundantly in several fruits and vegetables (1,2). Proanthocyanidins are the most common subtype of flavonoids, encompassing flavanol-3-ol monomers catechin and epicatechin, their esters of gallic acid and oligomers (Figure 1A). Grape seed extract (GSE) contains an abundance of the monomeric, dimeric and trimeric proanthocyanidins termed ‘Oligomeric proanthocyanidins (OPCs),’ and small amounts of the larger and biologically unavailable polymers (Figure 1A) (3). Benefits of the dietary intake of proanthocyanidins from grape seeds are multitudinous including protection against cardiovascular diseases and type 2 diabetes (1,4,5). However, based upon emerging evidence, GSE is now recognized for its anticancer effects, which it wields by inhibiting cellular proliferation, inducing apoptosis, arresting cell cycle and inhibiting metastatic processes (6–11). Previous studies have elegantly identified several molecular targets of proanthocyanidins, such as Bax/Bcl-2, MMPs and TNF-α (7–10,12); however, a more comprehensive interrogation of various signaling pathways and key genes that mediate the anticancer activity of proanthocyanidins remains unclear. Herein, for the first time, we attempt to cognize the underlying molecular mechanisms of the anticancer properties of OPCs by identifying its downstream targets by performing genome-wide RNA sequencing in a panel of colorectal cancer cell lines. This was followed by KEGG pathway analysis, wherein we identified several key cancer-associated pathways that were regulated by OPCs, particularly cell cycle and DNA replication pathways. To further confirm the biological consequences of in vitro findings, we also validated the regulation of cell-cycle genes by OPCs in vivo in a xenograft animal model.
Figure 1.
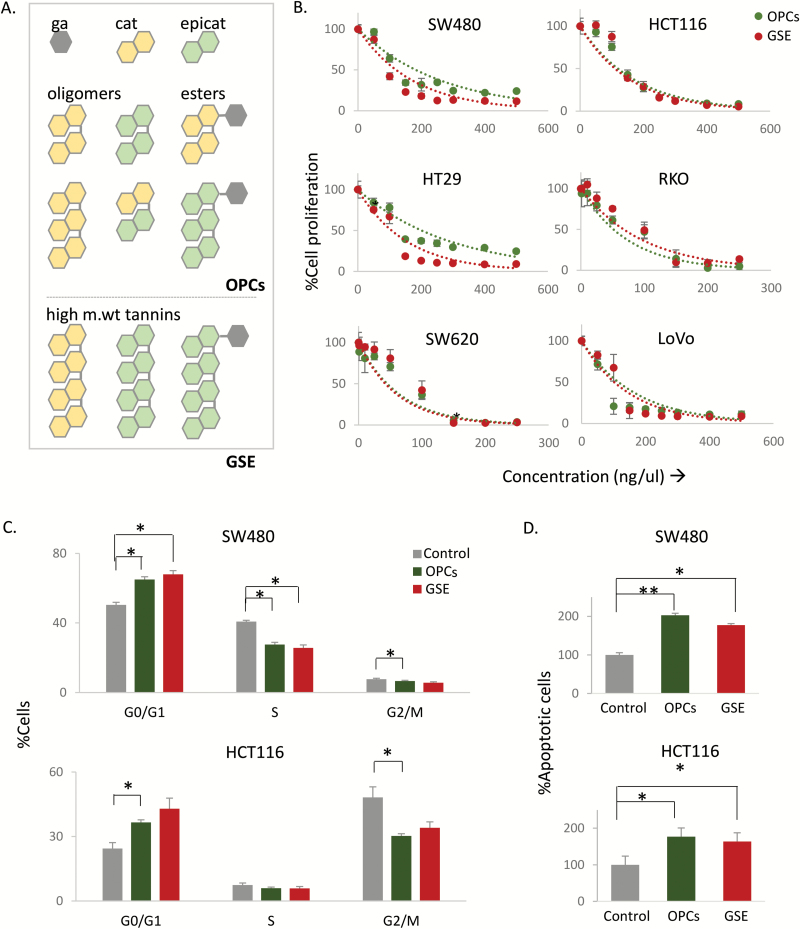
OPCs and GSE exert antitumorigenic properties in colorectal cancer cell lines. (A) Structural representation of basic monomeric units of proanthocyanidins. (B) Effect of OPCs and GSE on proliferation of six colorectal cancer cell lines treated for 72 h with 0–500 ng/ul of OPCs. (C) Cell-cycle analysis in HCT116 and SW480 cells treated with 100 ng/ul of OPCs or GSE. (D) Apoptosis in HCT116 and SW480 cells treated with 100 ng/ul of OPCs or GSE, shown as fold change with respect to control cells treated with vehicle (DMSO). *P < 0.05, **P < 0.01, ***P < 0.001 compared with control treatments.
In this study, we also endeavored to understand whether systemic absorption of smaller versus larger OPCs present in the GSEs has any influence on their efficacy as potential anticancer compounds. It is well recognized that bioavailability of molecules decreases with increased degree of polymerization of the precursor molecules. Hence, it is believed that the oligomeric fractions with a lesser degree of polymerization (e.g. ˂5) may have enhanced biological activity, while the larger proanthocyanidins often are not completely absorbed and are excreted intact out of the body (13,14). Contrarily, some studies claim that the larger proanthocyanidins may be metabolized to absorbable monomers by the gut microbiota (15); however, studies using proanthocyanidins from cocoa showed no such depolymerization (16). To clarify this, we compared the anticancer efficacy of OPCs from grape seeds and the unfractionated GSE containing the larger polymers side-by-side in both in vitro and in vivo experimental settings.
Another distinctive aspect of our study is the use of organoids derived from colorectal cancer patient tumors as a model to validate the anticancer properties and the underlying molecular mechanisms of OPCs. Organoids are extolled as a near-physiological experimental model in therapeutic drug development as they closely recapitulate the 3D tumor architecture containing both differentiated and cancer stem cells (17).
Overall, this study provides important, previously unrecognized insights into the molecular mechanisms underlying anticancer activities exerted by grape seed-derived OPCs, in cell lines, animal models and patient tumor organoids. Taken together, these results highlight the anticancer activity of short-chain OPCs present in GSEs and provide a rationale for their use as cancer-preventative compounds.
Materials and methods
Cell culture and materials
Colorectal cancer cell lines HCT116, SW480, SW620, HT29, Caco-2, RKO and LoVo were purchased from American Type Culture Collection (Manassas, VA). All cell lines were tested and authenticated using a panel of genetic and epigenetic markers in June 2017. The cells were grown in Iscove’s Modified Dulbecco’s Medium (IMDM; Gibco, Carlsbad, CA), supplemented with 10% fetal bovine serum, 1% penicillin and streptomycin and maintained at 37°C in a humidified incubator at 5% CO2.
Unfractionated GSE that includes both small oligomeric (monomers, dimers and trimers) proanthocyanidins and larger polymers and the OPCs fraction (VX1 extract, Berkem, France) that is devoid of larger proanthocyanidins were used in this study. For treatments, OPCs and GSE were dissolved in DMSO and diluted to appropriate experimental concentrations in culture medium.
Patient-derived tumor organoids
Fresh tumor tissues were obtained from colorectal cancer patients enrolled at the Baylor University Medical Center, Dallas, following written informed consent from the patients. The study was approved by the Institutional Review Board of Baylor Scott and White Research Institute, Dallas, TX. Colorectal cancer organoids were cultured using a modified protocol described previously (18). Briefly, following excision, tumors were maintained in a medium containing Dulbecco's modified Eagle's medium (DMEM)-F12 (Gibco) supplemented with 1% HEPES (Sigma-Aldrich), 1% l-glutamine (Gibco), 10% fetal bovine serum (FBS) (Gibco), 2% penicillin/streptomycin (Sigma-Aldrich) and 10 μM of Rho-associated protein kinase (ROCK) inhibitor Y-27632 (R&D Systems). Tissues were minced and digested with collagenase solution (5 ml of above medium with 75 μl collagenase, 124 ug/ml dispase type II and 0.2% Primocen) for 30 min and then filtered through a 70-μm filter (Corning). Cells were pelleted by centrifugation (200g for 10 min) and then suspended in Matrigel (BD Biosciences, Franklin Lake, NJ). Fifteen microliters of the cell-Matrigel suspension were placed in the center of 24-well plate and polymerized. A 1:1 mixture of L-WRN-conditioned medium and DMEM/F12 medium (Gibco) supplemented with 20% FBS (Gibco), 2 mM l-glutamine (Gibco), 0.2% Primocen, 10 μM of ROCK inhibitor Y-27632 (R&D Systems), 10 μM of TGF-β type I receptor inhibitor SB431542 (R&D Systems) and 5% penicillin/streptomycin (Sigma-Aldrich) were added to the well and replaced every 2 days. L-WRN is a modified Wnt3a-secreting mouse L (L-Wnt3a) cell line that also secretes Wnt, R-spondin and noggin, factors essential for the propagation and maintenance of intestinal epithelial cells (19). ROCK inhibitor Y27632 and TGF-β type I receptor inhibitor SB431542 help in the early growth of developing primary organoids. For treatments, appropriate concentration of OPCs or GSE was added to the culture medium.
Viability and cell proliferation
Cells were plated in 96-well dishes at a density of 2000 cells/well in DMEM supplemented with 5% FBS and antibiotics and allowed to attach overnight. Cell viability was measured using trypan blue exclusion test in cells treated with different concentrations (10, 100, 500, 1000 ng/ul) of OPCs or GSE for 24 h and read on Countess II cell counter (ThermoFisher Scientific). Cell proliferation was measured in cells incubated with various concentrations of OPCs or GSE for 72 h using WST-1 assay (Sigma-Aldrich) per manufacturer’s instructions.
Cell cycle and apoptosis analysis
Cells plated in 24-well dishes in DMEM media supplemented with 5% FBS were treated with OPCs or GSE at 100 ng/ul for 48 h in quadruplicates. Cell cycle and apoptosis assays were performed using Muse kits (MCH100105, MCH100106 Millipore) on Muse Cell Analyzer (Millipore) per manufacturer’s instructions.
Cell migration assays
To perform migration assays, 5000 cells/well were plated on the polycarbonate membrane inserts in transwell polycarbonate membrane dishes (3421, Corning) in 100 μl DMEM + 1% FBS media ± 100 ng/ul of OPCs/GSE). Five hundred microliters of DMEM + 10% FBS media were added to the bottom chamber of the plate to induce chemotaxis. Cells were allowed to migrate for 20 h and then stained using Three-step stain (3300, ThermoFisher Scientific) per manufacturer’s protocol. The stained cells were observed and counted under the microscope. The average of the counts from the biological replicates was plotted.
To perform the wound-scratch assay, 20 000 cells/well were plated in 12-well dishes in DMEM +5% FBS + 1% penicillin/streptomycin. The cells were allowed to attach overnight and then treated with vehicle/OPCs/GSE at 100 ng/ul. A straight line was scratched using a pipet tip in each well 4 h after treatment. Images were taken immediately (0 h) and 24 h after scratching. Distance migrated by the cells was calculated as a percentage of the width of the gap at 24 h with respect to that at the time of scratch (0 h). The average of three biological replicates is plotted.
mRNA expression analysis
RNA from HCT116, SW480, SW620, RKO and HT29 cells treated for 18 h or organoids treated for 7 days with DMSO (vehicle)/OPCs (100 ng/ul)/GSE (100 ng/ul) was isolated using mRNeasy kit (Qiagen). RNA from mice xenograft tumors collected in RNAlater solution (Qiagen) were extracted using mRNeasy Kit (Qiagen) following the manufacturer’s instructions. Extracted RNA was used as a template for cDNA synthesis using high-capacity cDNA Reverse Transcription Kit (ThermoFisher Scientific) according to manufacturer’s protocol. RT-qPCR was performed using SensiFAST SYBR mix (Bioline, London, UK) using the primer sequences listed in Supplementary Table 2, available at Carcinogenesis Online. All RT-qPCR target genes were calculated using ΔΔCt method normalized to β-actin.
Genome-wide RNA-sequencing analysis
RNA from cell lines treated with DMSO or 100 ng/ul of OPCs or GSE in duplicates were single-end sequenced as described. NGS library construction was performed using the TruSeq RNA Library Kit (Illumina) with up to 1 µg of total RNA input according to manufacturer’s protocol. The quality of individual libraries was assessed using the High Sensitivity DNA Kit (Agilent). Libraries were pooled together using a Pippin HT instrument (Sage Science). Efficiency of size selection was assessed using a High Sensitivity DNA Kit (Agilent). Pooled libraries were quantitated via qPCR using the KAPA Library Quantification Kit, Universal (KAPA Biosystems) prior to sequencing on an Illumina HighSeq 2500 with single-end 75 base read lengths. For the analysis of RNA-seq, Fastq files were trimmed using Flexbar to remove 3′ bases with quality score lower than 30 before alignment as described previously (20). The trimmed reads were mapped to human genome version GRCH38 downloaded from GENCODE (21) using HISAT2 (22) to generate alignment files in bam format. Samtools name-sorted bam files (23) were processed using htseq-count to summarize gene level counts as described previously (24). DESeq2 was used for differential gene expression analysis of RNA-seq read counts (25). All sequencing data has been deposited to the GEO database (GSE109607).
Meta-analysis was performed using Stouffer’s P-value combination method (26) on OPCs and GSE to identify genes that are homogenously up- or downregulated independently. KEGG pathway analysis was performed, and the cellular pathways commonly regulated by both OPCs and GSE with a P < 0.05 were identified, and the top 20 pathways were plotted. A similar meta-analysis approach was used to identify the cellular pathways that were uniquely regulated by OPCs or GSE. Genes that were uniquely regulated by OPCs of GSE were identified by Venn comparison. KEGG pathway analysis was done with these unique genes for OPCs and GSE, and pathways with P < 0.05 were plotted.
Xenograft animal experiments
Seven-week-old male athymic nude mice (Envigo, Houston, TX) were housed under controlled conditions of light and fed ad libitum. Approximately, 1 × 106 HCT116 cells were suspended in Matrigel matrix (BD Biosciences) and subcutaneously injected into mice using 27-gauge needle (n = 11 per group). Mice were then orally gavaged with vehicle (water) or OPCs (dissolved in water) or GSE (dissolved in water) at two different concentrations—50 and 100 mg/kg body weight—daily for the first week and on alternative days for the second week. Tumor size was measured every day by calipers for 13 days. Tumor volume was calculated using the following formula: 1/2 (length × width × width). The animal protocol was approved by the Institutional Animal Care and Use Committee, Baylor Scott & White Research Institute, Dallas, Texas.
Statistical analysis
All data were expressed as mean ± SEM with statistical significance indicated when P < 0.05. Statistical comparisons between control and treatment groups were determined using paired t-test.
Results
OPCs exert antitumorigenic effects in colorectal cancer cells
GSE is comprised of oligomers (OPCs) and larger polymers and esters of proanthocyanidins catechin and epicatechin (Figure 1A). Since it is believed that there may be differences between the bioactivity of smaller versus larger polymers from the GSE, we performed a head-to-head comparison of the antitumorigenic properties of GSE and OPCs, which are the smaller polymeric fraction of GSE, in colorectal cancer cells. Trypan blue exclusion test in colorectal cancer cell lines HCT116 and SW480 treated with OPCs or GSE for 24 h showed they were well tolerated with no observable cytotoxicity up to 1000 ng/ul (Supplementary Figure 1, available at Carcinogenesis Online). We thereafter compared the antiproliferative properties of GSE and OPCs in six colorectal cancer cell lines that broadly represent the common microsatellite and mutational statuses in colorectal cancer, namely SW480, SW620, HT-29, HCT116, RKO and LoVo (Supplementary Table 1, available at Carcinogenesis Online). Interestingly, both OPCs and GSE inhibited cell proliferation in all six cell lines regardless of their genetic features (Figure 1B). The inhibition of cell proliferation by both OPCs and GSE were dose dependent with an IC50 value of ~100 ng/ul for all cells.
We observed that both OPCs and GSE induced cell-cycle arrest in HCT116 and SW480 cells. When treated with 100 ng/ul of OPCs and GSE for 20 h, there was a significant increase in the percentage of cells in the G0/G1 phase and a corresponding decrease in S and G2/M phases of cell cycle in HCT116 and SW480 cells (Figure 1C). Additionally, in agreement with previous studies in other colorectal cancer cells (11,27,28), both OPCs and GSE induced apoptosis in HCT116 and SW480 cells at 100 ng/ul, as measured using annexin V- and 7-AAD-based assay (Figure 1D).
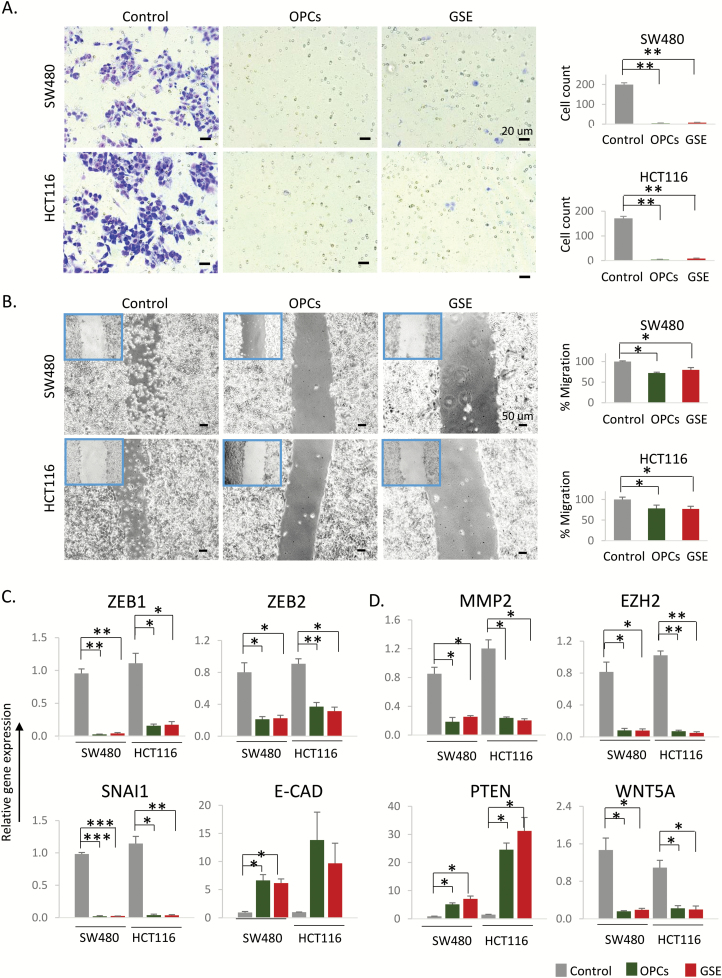
We next assessed the ability of OPCs and GSE to inhibit cell migration, a process critical for cancer progression and metastasis. To achieve this, we performed a transwell migration assay using polycarbonate membrane inserts, where the cells were plated on the top chamber in the presence or absence of 100 ng/ul of OPCs or GSE. The bottom chamber was filled with tissue culture media containing higher concentration of FBS to induce chemotaxis. We observed that the cells incubated with OPCs and GSE almost completely lost their ability to migrate through the polycarbonate membrane (Figure 2A). We subsequently confirmed that OPCs and GSE block cell migration by wound-healing assay (Figure 2B). We noticed that, while untreated cells migrated toward each other to decrease the gap 24 h post scratch, cells treated with OPCs and GSE did not show significant change in migration. In support of these results, EMT markers, Zeb1, Zeb2 and Snai1, were all significantly downregulated, and epithelial marker E-cadherin was upregulated by both OPCs and GSE in HCT116 and SW480 cells (Figure 2C). In addition, OPCs and GSE treatment downregulated the expression of several genes associated with cancer cell migration, namely MMP2, EZH2 and WNT5A, and upregulated tumor suppressor gene PTEN (Figure 2D).
Figure 2.
OPCs and GSE inhibit cancer cell migration. (A) Left: Representative images of cells that migrated through the permeable membrane in a transwell migration plate, 6 h after treatment with OPCs or GSE at 100 ng/ul. Right: Count of cells that migrated through the membrane, shown as an average of triplicates. (B) Left: Representative images of monolayer of SW480 and HCT116 cells treated with 100 ng/ul OPCs or GSE, 6 h post scratch. Right: Quantification of migrated cells from triplicate treatments. (C) mRNA expression of EMT markers Zeb1, Zeb2, Snai1 and E-Cad in OPCs/GSE-treated HCT116 and SW480 cells, shown as fold change with respect to control cells treated with vehicle (DMSO). (D) mRNA expression of genes associated with cancer cell migration. *P < 0.05, **P < 0.01, ***P < 0.001 compared with control treatments.
These data taken together corroborate that both OPCs and GSE affect the hallmark capabilities of cancer and its progression: they inhibit cell proliferation and cell migration and induce apoptosis and cell-cycle arrest.
OPCs inhibit multiple cancer-associated pathways
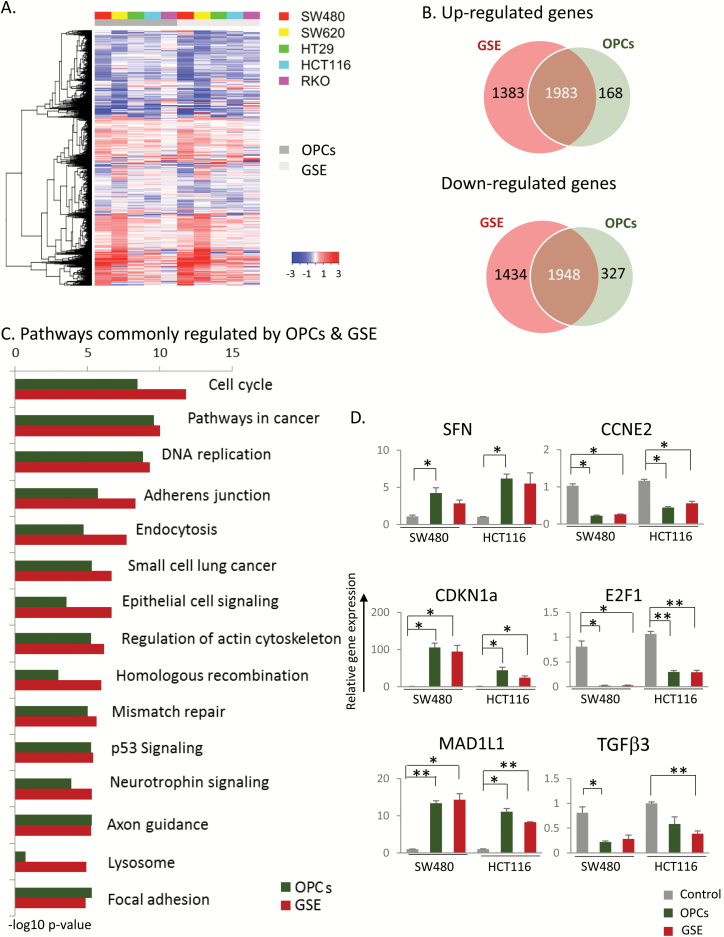
We next examined the genome-wide changes in gene expression induced by OPCs and GSE in five colorectal cancer cell lines SW480, HT29, SW620, HCT116 and RKO by RNA sequencing (Figure 3A). Analysis of differential expression of genes relative to untreated cells revealed a total of 6748 genes that were dysregulated (3366 upregulated, 3382 downregulated; P < 0.05) by GSE and 4426 genes by OPCs (2153 upregulated, 2275 downregulated; P < 0.05; Figure 3B). Of these, 3931 genes (35% of all affected genes by either OPCs or GSE) were commonly dysregulated by both OPCs and GSE. A total of 2817 genes (1383 upregulated, 1434 downregulated) were uniquely regulated by GSE but not by OPCs, and 495 genes (168 upregulated, 327 downregulated) were uniquely regulated by OPCs.
Figure 3.
OPCs and GSE inhibit multiple cancer-associated pathways. (A) Heatmap for five different cancer cell lines showing genes that were differentially expressed in cells treated with 100 ng/ul OPCs or GSE compared with control treated cells. (B) Venn diagram displaying the overlap between quantified mRNAs. (C) Ranking of pathways based on P-values that were identified by KEGG pathway analysis. (D) qPCR validation of top genes in cell cycle in SW480 and HCT116 cells, normalized to β-actin, shown as fold change with respect to control cells treated with vehicle (DMSO). *P < 0.05, **P < 0.01, ***P < 0.001 compared with control treatments.
KEGG pathway analysis revealed that the subset of genes involved in cell cycle (P = 3.63e−09 for OPCs, 1.66e−12 for GSE) and DNA replication (P = 1.49e−09 for OPCs, 5.18e−10 for GSE) were the most significantly altered pathways across all cell lines (Figure 3C). Among other key cancer-associated pathways that were identified as modulated by both OPCs and GSE were p53 signaling (P = 5.9e−06 for OPCs, 4.22e−06 for GSE) and the DNA mismatch repair (P = 1e−05 for OPCs, 2.45e−06 for GSE; Figure 3C). Metabolic pathways such as selanoamine acid metabolism (P = 0.003), O-glycan biosynthesis (P = 0.004) and alanine aspartate and glutamate metabolism (P = 0.005) were the top pathways identified by meta-analysis to be uniquely regulated by OPCs (Supplementary Figure 2A, left, available at Carcinogenesis Online). On the other hand, lysosome-associated pathway (P = 0.0003) and glycosphingolipid biosynthesis (P = 0.0005) were the top pathways identified to be uniquely regulated by GSE (Supplementary Figure 2A, right, available at Carcinogenesis Online).
OPCs inhibit cell cycle and DNA replication-associated pathways
A closer look at the two most commonly regulated pathways by OPCs and GSE, namely cell-cycle regulation and DNA replication, revealed the downregulation of several well-characterized oncogenes as well as the upregulation of tumor suppressors (Supplementary Figure 3, available at Carcinogenesis Online). To further validate the RNA-sequencing results, we first ordered the cell-cycle genes by the number of cell lines in which their expression was homogenously increased or decreased by at least 2-fold. We selected the top three upregulated genes, namely Stratifin (SFN), CDKN1A and MAD1L1, and the top three downregulated genes TGFβ3, E2F1 and CCNE with P-values <0.05 (meta-analysis; Supplementary Figure 3A, available at Carcinogenesis Online, genes highlighted in blue box) for validation by qPCR in SW480 and HCT116 cells treated for 18 h with 100 ng/ul of OPCs or GSE. Consistent with our RNA-sequencing results, the mRNA levels of TGFβ3, E2F1 and CCNE were downregulated, and SFN, CDKN1A and MAD1L1 were upregulated in both SW480 and HCT116 cells (Figure 3D), further confirming the effect of OPCs and GSE on the regulation of cell cycle.
The tumor growth inhibitory effects of OPCs validated in an animal model
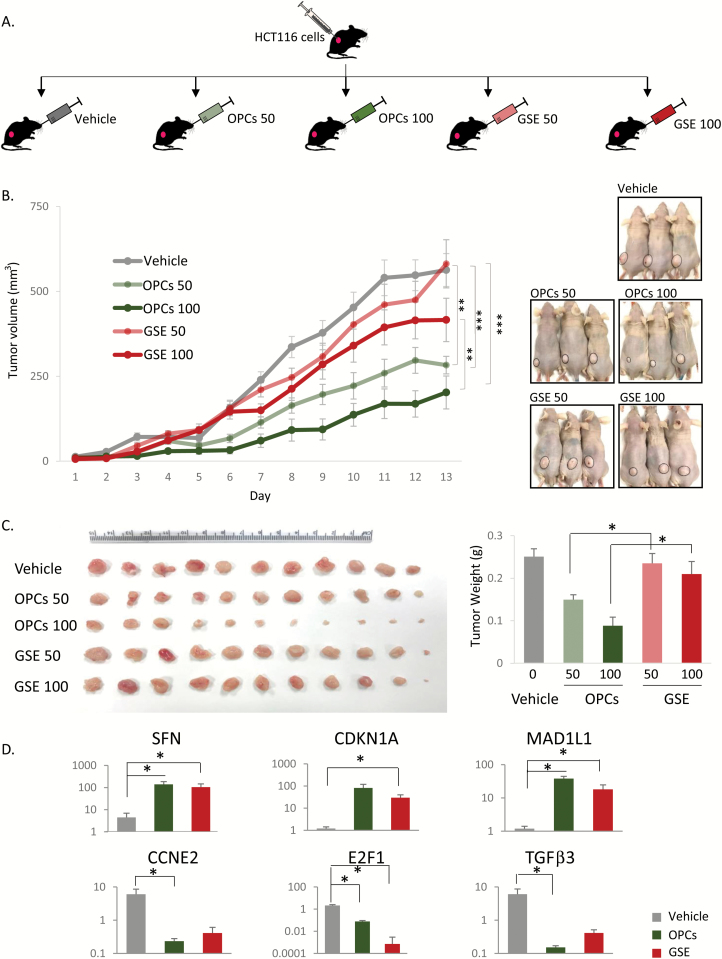
To evaluate the effect of OPCs and GSE in vivo, tumor growth was followed in athymic mice with subcutaneous xenografts of HCT116 cells that were administered OPCs or GSE (Figure 4A). There was no significant change in the weight of the mice during the course of the treatment (data not shown). While the tumors continued to grow in mice that were administered vehicle (saline), tumor growth in OPCs- or GSE-administered mice was significantly attenuated (Figure 4B). Intriguingly, OPCs at both concentrations were significantly more effective in decreasing tumor growth than GSE (Figure 4B). Tumor weight measurements were also consistent with the observation that OPCs were a more potent inhibitor of tumor growth than GSE (Figure 4C). Furthermore, in line with results obtained in cell lines, OPCs and GSE decreased expression of oncogenes TGFβ3, E2F1 and CCNE2 and increased expression of tumor suppressors SFN, CDKN1A and MAD1L1 in mouse xenograft tumors (Figure 4D).
Figure 4.
(A) Schematic of OPCs gavaging of mice xenografted with HCT116 cells. Mice were subcutaneously injected with HCT116 cells in Matrigel and after 2 days, orally gavaged with OPCs or GSE (indicated dose is in mg/kg) every day for the first week and then alternative day for the second week. (B) Left: Progressive tumor volume in mice orally gavaged with OPCs. Right: Representative mice images from different treatment groups on day 13 (the day of sacrifice) showing change in tumor volume. (C) Left: Xenograft tumor collected from sacrificed mice at the end of 13 day treatments. Right: Quantification of tumor weights from different treatment groups. (D) qPCR analysis of mRNA expression of genes normalized to control (‘vehicle’) group. *P < 0.05, **P < 0.01, ***P < 0.001.
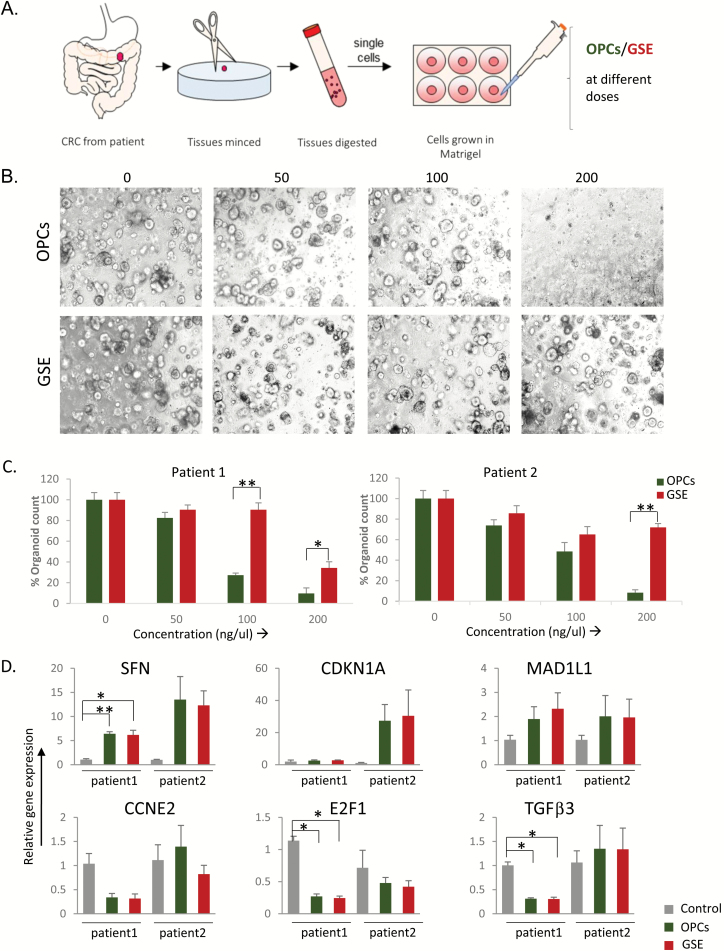
OPCs inhibit tumor growth in patient-derived organoids
To further confirm our in vitro and in vivo observations, we generated tumor organoids from colorectal cancer patients and examined the effects of both OPCs and GSE. Organoid cultures derived from human colorectal cancer specimens were treated with different doses of OPCs or GSE for a week (Figure 5A). Both OPCs and GSE decreased the growth of organoids considerably, as evident from the decrease in the number (Figure 5B and C) and the size of the organoids (Supplementary Figure 4, available at Carcinogenesis Online). Interestingly, OPCs abolished organoid growth at a lower concentration of 200 ng/ul than GSE (Figure 5B and C). Furthermore, consistent with our results so far, expression of oncogenes TGFβ3, E2F1 and CCNE2 were increased, and tumor suppressors SFN, CDKN1A and MAD1L1 were decreased by OPCs and GSE in these organoids (Figure 5D), corroborating that OPCs and GSE exert antitumorigenic functions by attenuating cell cycle.
Figure 5.
OPCs suppress growth of organoids derived from human colorectal tumors. (A) Schematic protocol of primary culture of tumors derived from patients. (B) Representative images showing tumor organoid cultures derived from a colorectal cancer patient, treated with 0–200 µg/ml of OPCs and GSE. (C) Bar graph showing a decrease in spheroid count with OPCs or GSE treatments in two separate patient-derived organoids. (D) mRNA expression levels of genes in patient-derived organoids treated with OPCs and GSE, normalized to control (DMSO treated) organoids. *P < 0.05, **P < 0.01, ***P < 0.001.
Discussion
There is a growing armory of therapeutics that target mechanisms of cancer pathogenesis. Most of these therapeutics are designed to deliberately target specific molecular targets, the benefit, in principle being fewer off-target effects and toxicity. However, due to the redundancy in cellular pathways, therapeutics that target only a specific molecule or pathway only transiently inhibit tumor growth. Eventually, tumor relapses by activating alternative cellular pathways (29) or shifting its dependency from one tumor capability to another. The emergence of EGFR mutations in colorectal cancer patients treated with cetuximab (30) and increased invasiveness and metastasis in tumors treated with antiangiogenic therapies are examples of adaptive shifts that help tumors overcome therapeutics targeting specific genes or pathways (31). Alternatively, simultaneously blocking multiple cancer-associated pathways is a viable approach to inhibit tumor growth and progression without relapse. Several nutraceuticals including curcumin, green tea and GSE are sought for their ability to impede tumor growth by simultaneously blocking several cancer-associated pathways without cellular toxicity. In our current study, we show the possibility of using OPCs derived from grape seeds to target multiple pathways that impart tumor pathogenicity.
One of the fundamental traits of cancer cells is to proliferate chronically by progressing unchecked through defective checkpoints in the cell cycle. Our RNA-sequencing data revealed that OPCs affected the expression of several key genes involved in cell cycle and DNA replication. Particularly, the top genes involved in cell cycle that OPCs downregulated across all cell lines are well-established oncogenes: Cyclin E2 (CCNE2) and E2F1 regulate entry of the cells into the DNA-synthesis phase of the cell cycle and are commonly overexpressed in tumor-derived cells (32–34). TGFβ3 is a key contributor to the malignant phenotype in cancers and studied as a therapeutic target (35). Correspondingly, the top genes that were upregulated, namely SFN, CDKN1A and MAD1L1, have recognized roles as tumor suppressors. Stratifin and CDKN1A, gene encoding p21 protein, coordinate to inhibit G2/M progression and exert antitumorigenic properties by inhibiting angiogenesis and inducing apoptosis (36,37). The third most upregulated gene by OPCs, MAD1L1, functions as a spindle-assembly checkpoint during mitosis. Subsequently, we were able to demonstrate the regulation of these genes by qPCR in all three models that were used in the study, namely cell lines, mice xenograft tumors and patient-derived organoids.
In addition, we have demonstrated that OPCs inhibit metastasis-associated processes, consistent with previous findings in breast, prostate and other cancers (38–42). In this study, we confirmed the inhibition of cell migration and modulation of EMT-associated genes by OPCs in colorectal cancer cells HCT116 and SW480. We also identified previously unreported pathways implicated in cancer cell motility, namely axon guidance, focal adhesion and regulation of actin (43–45) were affected by OPCs. All these data strongly suggest the inhibition of cancer cell migration and metastasis by OPCs and warrants further investigation.
A main advantage of OPCs as a tumor-inhibiting agent is its ability to simultaneously block multiple hallmark tumor capabilities (46), specifically cellular proliferation, progression through cell cycle and cellular migration, and induce apoptosis without causing cellular toxicity. To date, no single clinical therapeutic has the ability to effectively block multiple oncogenic pathways. Drug combinations to optimally target complex signaling networks and feedback loops in cancer have limited clinical utility due to toxicities. Also, the choice of drug combinations is contingent upon the biologic and genetic features of the tumor cell (47). Interestingly, OPCs inhibited the cellular proliferation of a gamut of colorectal cancer cells with a variety of genetic mutations and biological features without any observable cellular toxicity. Based on our mice studies, the human equivalent dose at which OPCs effectively decrease tumor growth is 240 mg per day, which is a well tolerated and safe oral dose as determined in clinical trials with breast cancer patients (48). These findings demonstrate the promise of OPCs as a therapeutic candidate against different types of colorectal cancer and necessitate its development into a clinical therapeutic.
One intriguing finding in our study was that unlike in cell lines (Figure 1B), OPCs more potently inhibited tumor growth in mice xenografts than GSE (Figure 4B and C). The observed lower efficacy of GSE in vivo may potentially be due to the poorer absorption of the larger procyanidins in GSE through the gut barrier (14), a concept that must be systematically addressed in future studies. Moreover, larger procyanidins can complex with proteins, carbohydrates and other macromolecules to become less digestible complexes, further decreasing their efficacy (15,49). Although there is evidence of the larger polycyanidins metabolized by the colonic microbiota to make them partially bioavailable, our mice experimental data clearly show better efficacy of OPCs compared with GSE (50,51). Similarly, the poorer efficacy of GSE compared with OPCs in patient-derived organoid models may be due to the inability of the larger polycyanidins to penetrate the Matrigel to reach the organoids (17). Collectively, we not only successfully demonstrated the anticancer effects of OPCs in colorectal cancer patient-derived organoids but also validated the modulation of several cell cycle-associated genes, namely SFN, CDKN1A, MAD1L, CCNE2, E2F1 and TGFβ3.
A limitation of using OPCs clinically is its broad targeting of multiple cellular pathways. While blocking multiple pathways may be effective in targeting cancer cells (52), it may also result in undesired off-target effects. In our experiments, no toxicity was observed in cell lines or upon oral administration in mice for 2 weeks. However, further evaluation of the bioavailability, metabolism and toxicity of OPCs and detailed pharmacodynamic studies in human are necessary to further understand its effects on human health.
In summary, we for the first time showed that OPCs from grape seeds inhibit colorectal cancer growth through multiple cellular pathways, especially by modulating key cell-cycle genes. In addition to validating our results in animal models, we also validated the anticancer effects of OPCs in a preclinical ex vivo patient tumor organoids—something which is quite attractive and not been explored previously. Secondly, we showed that OPCs has higher efficacy in tumor inhibition in mice xenografts than GSE, possibly due to better bioavailability. Consequently, as OPCs block various oncogenic pathways, it could be utilized for preventing the emergence of acquired resistance in cancer patients undergoing targeted therapies. Collectively, these findings will be important for ongoing preclinical development of OPCs and other botanical therapeutics that can target multiple cellular pathways.
Supplementary material
Supplementary material can be found at Carcinogenesis online.
Funding
The present work was supported by the CA72851, CA181572, CA184792, CA187956 and CA202797 grants from the National Cancer Institute, National Institute of Health; RP140784 from the Cancer Prevention Research Institute of Texas; grants from the Sammons Cancer Center and Baylor Foundation, as well as funds from the Baylor Scott and White Research Institute, Dallas, TX, USA.
Acknowledgements
We would like to thank Dr Roshni Roy for her assistance with data analysis.
Conflict of Interest Statement: None declared.
Abbreviations
- DMEM
Dulbecco's modified Eagle's medium
- FBS
fetal bovine serum
- GSE
grape seed extract
- OPCs
oligomeric proanthocyanidins
References
- 1. Smeriglio A., et al. (2017)Proanthocyanidins and hydrolysable tannins: occurrence, dietary intake and pharmacological effects. Br. J. Pharmacol., 174, 1244–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Rasmussen S.E., et al. (2005)Dietary proanthocyanidins: occurrence, dietary intake, bioavailability, and protection against cardiovascular disease. Mol. Nutr. Food Res., 49, 159–174. [DOI] [PubMed] [Google Scholar]
- 3. Downing L.E., et al. (2015)A grape seed procyanidin extract ameliorates fructose-induced hypertriglyceridemia in rats via enhanced fecal bile acid and cholesterol excretion and inhibition of hepatic lipogenesis. PLoS One, 10, e0140267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wang X., et al. (2014)Flavonoid intake and risk of CVD: a systematic review and meta-analysis of prospective cohort studies. Br. J. Nutr., 111, 1–11. [DOI] [PubMed] [Google Scholar]
- 5. Banihani S., et al. (2013)Pomegranate and type 2 diabetes. Nutr. Res., 33, 341–348. [DOI] [PubMed] [Google Scholar]
- 6. Gollucke A.P., et al. (2013)Use of grape polyphenols against carcinogenesis: putative molecular mechanisms of action using in vitro and in vivo test systems. J. Med. Food, 16, 199–205. [DOI] [PubMed] [Google Scholar]
- 7. Sieniawska E. (2015)Activities of tannins—from in vitro studies to clinical trials. Nat. Prod. Commun., 10, 1877–1884. [PubMed] [Google Scholar]
- 8. Skrovankova S., et al. (2015)Bioactive compounds and antioxidant activity in different types of berries. Int. J. Mol. Sci., 16, 24673–24706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Nile S.H., et al. (2014)Edible berries: bioactive components and their effect on human health. Nutrition, 30, 134–144. [DOI] [PubMed] [Google Scholar]
- 10. Ouédraogo M., et al. (2011)An overview of cancer chemopreventive potential and safety of proanthocyanidins. Nutr. Cancer, 63, 1163–1173. [DOI] [PubMed] [Google Scholar]
- 11. Kaur M., et al. (2006)Grape seed extract inhibits in vitro and in vivo growth of human colorectal carcinoma cells. Clin. Cancer Res., 12, 6194–6202. [DOI] [PubMed] [Google Scholar]
- 12. Gollucke A.P., et al. (2013)Polyphenols: a nutraceutical approach against diseases. Recent Pat. Food. Nutr. Agric., 5, 214–219. [DOI] [PubMed] [Google Scholar]
- 13. Ou K.Q., et al. (2014)Absorption and metabolism of proanthocyanidins. J. Funct. Foods, 7, 43–53. [Google Scholar]
- 14. Wiese S., et al. (2015)Comparative biokinetics and metabolism of pure monomeric, dimeric, and polymeric flavan-3-ols: a randomized cross-over study in humans. Mol. Nutr. Food Res., 59, 610–621. [DOI] [PubMed] [Google Scholar]
- 15. Zhang L., et al. (2016)The absorption, distribution, metabolism and excretion of procyanidins. Food Funct., 7, 1273–1281. [DOI] [PubMed] [Google Scholar]
- 16. Ellam S., et al. (2013)Cocoa and human health. Annu. Rev. Nutr., 33, 105–128. [DOI] [PubMed] [Google Scholar]
- 17. Fatehullah A., et al. (2016)Organoids as an in vitro model of human development and disease. Nat. Cell Biol., 18, 246–254. [DOI] [PubMed] [Google Scholar]
- 18. Miyoshi H., et al. (2013)In vitro expansion and genetic modification of gastrointestinal stem cells in spheroid culture. Nat. Protoc., 8, 2471–2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Miyoshi H., et al. (2012)Wnt5a potentiates TGF-β signaling to promote colonic crypt regeneration after tissue injury. Science, 338, 108–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Dodt M., et al. (2012)FLEXBAR—flexible barcode and adapter processing for next-generation sequencing platforms. Biology (Basel), 1, 895–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Harrow J., et al. (2012)GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res., 22, 1760–1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kim D., et al. (2015)HISAT: a fast spliced aligner with low memory requirements. Nat. Methods, 12, 357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Li H., et al. ; 1000 Genome Project Data Processing Subgroup. (2009)The sequence alignment/map format and SAMtools. Bioinformatics, 25, 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Anders S., et al. (2015)HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics, 31, 166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Love M.I., et al. (2014)Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol., 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Dewey M. (2017)metap: Meta-Analysis of Significance Values https://CRAN.R-project.org/package=metap (16 August 2017, date last accessed).
- 27. Hsu C.P., et al. (2009)Mechanisms of grape seed procyanidin-induced apoptosis in colorectal carcinoma cells. Anticancer Res., 29, 283–289. [PubMed] [Google Scholar]
- 28. Nomoto H., et al. (2004)Chemoprevention of colorectal cancer by grape seed proanthocyanidin is accompanied by a decrease in proliferation and increase in apoptosis. Nutr. Cancer, 49, 81–88. [DOI] [PubMed] [Google Scholar]
- 29. Widakowich C., et al. (2007)Review: side effects of approved molecular targeted therapies in solid cancers. Oncologist, 12, 1443–1455. [DOI] [PubMed] [Google Scholar]
- 30. Arena S., et al. (2015)Emergence of multiple EGFR extracellular mutations during cetuximab treatment in colorectal cancer. Clin. Cancer Res., 21, 2157–2166. [DOI] [PubMed] [Google Scholar]
- 31. Folkman J., et al. (2004)Cancer without disease. Nature, 427, 787. [DOI] [PubMed] [Google Scholar]
- 32. Odajima J., et al. (2016)Proteomic landscape of tissue-specific cyclin E functions in vivo. PLoS Genet., 12, e1006429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Xie L., et al. (2017)E2F2 induces MCM4, CCNE2 and WHSC1 upregulation in ovarian cancer and predicts poor overall survival. Eur. Rev. Med. Pharmacol. Sci., 21, 2150–2156. [PubMed] [Google Scholar]
- 34. Sheldon L.A. (2017)Inhibition of E2F1 activity and cell cycle progression by arsenic via retinoblastoma protein. Cell Cycle, 16, 2058–2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Seystahl K., et al. (2017)Biological role and therapeutic targeting of TGF-β3in glioblastoma. Mol. Cancer Ther., 16, 1177–1186. [DOI] [PubMed] [Google Scholar]
- 36. Liu P., et al. (2017)Sulforaphane exerts anti-angiogenesis effects against hepatocellular carcinoma through inhibition of STAT3/HIF-1α/VEGF signalling. Sci. Rep., 7, 12651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Arcidiacono P., et al. (2017)Antitumor activity and expression profiles of genes induced by sulforaphane in human melanoma cells. Eur. J. Nutr. doi:10.1007/s00394-017-1527-7. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Vaid M., et al. (2015)Therapeutic intervention of proanthocyanidins on the migration capacity of melanoma cells is mediated through PGE2 receptors and β-catenin signaling molecules. Am. J. Cancer Res., 5, 3325–3338. [PMC free article] [PubMed] [Google Scholar]
- 39. Zheng H.L., et al. (2015)Oligomer procyanidins (F2) isolated from grape seeds inhibits tumor angiogenesis and cell invasion by targeting HIF-1α in vitro. Int. J. Oncol., 46, 708–720. [DOI] [PubMed] [Google Scholar]
- 40. Sun Q., et al. (2011)Grape seed proanthocyanidins inhibit the invasive potential of head and neck cutaneous squamous cell carcinoma cells by targeting EGFR expression and epithelial-to-mesenchymal transition. BMC Complement. Altern. Med., 11, 134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sun Q., et al. (2012)Grape seed proanthocyanidins inhibit the invasiveness of human HNSCC cells by targeting EGFR and reversing the epithelial-to-mesenchymal transition. Plos One, 7, e31093. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 42. Vayalil P.K., et al. (2012)Proanthocyanidins from grape seeds inhibit expression of matrix metalloproteinases in human prostate carcinoma cells, which is associated with the inhibition of activation of MAPK and NF kappa B (Retraction vol 25, pg 987, 2004). Carcinogenesis, 33, 1121. [DOI] [PubMed] [Google Scholar]
- 43. Amodeo V., et al. (2017)A PML/Slit axis controls physiological cell migration and cancer invasion in the CNS. Cell Rep., 20, 411–426. [DOI] [PubMed] [Google Scholar]
- 44. Idoux-Gillet Y., et al. (2018)Slug/Pcad pathway controls epithelial cell dynamics in mammary gland and breast carcinoma. Oncogene, 37, 578–588. [DOI] [PubMed] [Google Scholar]
- 45. Qiao Y., et al. (2017)YAP regulates actin dynamics through ARHGAP29 and promotes metastasis. Cell Rep., 19, 1495–1502. [DOI] [PubMed] [Google Scholar]
- 46. Hanahan D., et al. (2000)The hallmarks of cancer. Cell, 100, 57–70. [DOI] [PubMed] [Google Scholar]
- 47. Kummar S., et al. (2010)Utilizing targeted cancer therapeutic agents in combination: novel approaches and urgent requirements. Nat. Rev. Drug Discov., 9, 843–856. [DOI] [PubMed] [Google Scholar]
- 48. Brooker S., et al. (2006)Double-blind, placebo-controlled, randomised phase II trial of IH636 grape seed proanthocyanidin extract (GSPE) in patients with radiation-induced breast induration. Radiother. Oncol., 79, 45–51. [DOI] [PubMed] [Google Scholar]
- 49. Ou K., et al. (2015)Depolymerisation optimisation of cranberry procyanidins and transport of resultant oligomers on monolayers of human intestinal epithelial Caco-2 cells. Food Chem., 167, 45–51. [DOI] [PubMed] [Google Scholar]
- 50. Espín J.C., et al. (2013)Biological significance of urolithins, the gut microbial ellagic acid-derived metabolites: the evidence so far. Evid. Based. Complement. Alternat. Med., 2013, 270418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kamiloglu S., et al. (2016)Bioaccessibility of polyphenols from plant-processing byproducts of black carrot (Daucus carota L.). J. Agric. Food Chem., 64, 2450–2458. [DOI] [PubMed] [Google Scholar]
- 52. Motzer R.J., et al. (2006)Activity of SU11248, a multitargeted inhibitor of vascular endothelial growth factor receptor and platelet-derived growth factor receptor, in patients with metastatic renal cell carcinoma. J. Clin. Oncol., 24, 16–24. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.