The longitudinal impact of apolipoprotein E ɛ4 on individual disease trajectories remains unclear. Mishra et al. show that group level effects on atrophy and β-amyloid are driven by ɛ4 carriers being more likely to develop Alzheimer’s disease, doing so earlier, and accumulating more β-amyloid even when matched for baseline pathology.
Keywords: apolipoprotein, PET, atrophy, amyloid, APOE
Abstract
While prior work reliably demonstrates that the APOE ɛ4 allele has deleterious group level effects on Alzheimer disease pathology, the homogeneity of its influence across the lifespan and spatially in the brain remains unknown. Further it is unclear what combinations of factors at an individual level lead to observed group level effects of APOE genotype. To evaluate the impact of the APOE genotype on disease trajectories, we examined longitudinal MRI and PET imaging in a cohort of 497 cognitively normal middle and older aged participants. A whole-brain regional approach was used to evaluate the spatial effects of genotype on longitudinal change of amyloid-β pathology and cortical atrophy. Carriers of the ɛ4 allele had increased longitudinal accumulation of amyloid-β pathology diffusely through the cortex, but the emergence of this effect across the lifespan differed greatly by region (e.g. age 49 in precuneus, but 65 in the visual cortex) with the detrimental influence already being evident in some regions in middle age. This increased group level effect on accumulation was due to a greater proportion of ɛ4 carriers developing amyloid-β pathology, on average doing so at an earlier age, and having faster amyloid-β accumulation even after accounting for baseline amyloid-β levels. APOE ɛ4 carriers displayed faster rates of structural loss in primarily constrained to the medial temporal lobe structures at around 50 years, although this increase was modest and proportional to the elevated disease severity in APOE ɛ4 carriers. This work indicates that influence of the APOE gene on pathology can be detected starting in middle age.
Introduction
Alzheimer’s disease is the most common cause of dementia in individuals above the age of 60. The most significant genetic modifier of late-onset Alzheimer disease is the apolipoprotein E (APOE) gene (Corder et al., 1993; Saunders et al., 1993; Strittmatter et al., 1993; Reiman et al., 1996). APOE is a protein that functions as a ligand in receptor-mediated endocytosis of lipoprotein particles in the CNS and the periphery (Holtzman and Herz, 2012). The ɛ4 allele of APOE increases the risk of developing Alzheimer’s disease and lowers the age of onset, while the ɛ2 allele is associated with a lower risk of Alzheimer’s disease (Corder et al., 1993; Reiman et al., 2009). The association between the presence of an APOE ɛ4 allele and the clinical risk of Alzheimer’s disease has been well established in the literature (Farrer et al., 1997). While the exact mechanism of the relationship is still an active area of research (Mahoney-Sanchez et al., 2016), one of the major effects of the APOE protein on Alzheimer’s disease risk is that it influences amyloid-β aggregation and clearance (Huynh et al., 2017).
The pathological hallmarks of Alzheimer’s disease are neurofibrillary tangles and amyloid-β plaques (Braak and Braak, 1991), and significant work has been done evaluating the effect of APOE ɛ4 allele on these biomarkers. Previous studies of pathology, CSF and PET imaging of the brain consistently find a relationship between APOE ɛ4 genotype and elevated amyloid-β burden (Schmechel et al., 1993; Morris et al., 2010; Rowe et al., 2010; Johnson et al., 2013; Mathis et al., 2013; Murphy et al., 2013; Villemagne and Rowe, 2013; Risacher et al., 2015; Gottesman et al., 2016). Whereas studies overwhelmingly find that the APOE ɛ4 genotype is associated with worse amyloid-β pathology, its effect on markers of neurodegeneration such as CSF tau, fludeoxyglucose PET, and structural MRI are less consistent. Studies have reported both an influence of the APOE ɛ4 allele (Reiman et al., 1996; Galasko et al., 1998; Liu et al., 2010; Jagust et al., 2012; Hostage et al., 2013; Lehmann et al., 2013; Roussotte et al., 2014) and no effect (Soininen et al., 1995; Jack et al., 1998; Reiman et al., 1998; Sunderland et al., 2004; Drzezga et al., 2009; Fan et al., 2010).
Perhaps most importantly, the fundamental interpretation of APOE ɛ4 effects on Alzheimer’s disease pathology has focused on general group effects while ignoring parallel ways such effects could arise. For example, it is unclear from the literature whether population level APOE ɛ4 effects on amyloid-β pathology are a result of a higher proportion of APOE ɛ4 carriers developing Alzheimer’s disease pathology, ɛ4 carriers developing Alzheimer’s disease pathology at an earlier age, whether ɛ4 carriers have an increased rate of amyloid-β accrual, or a combination of the three. These questions are critical to understanding the influence of APOE on Alzheimer’s disease as well as for clinical trials that are using ɛ4 genotype or PET amyloid-β levels to select participants (Reiman et al., 2011; Sperling et al., 2014). Disentangling these questions may be best addressed through the investigation of APOE ɛ4 effects using longitudinal data in a population that spans both middle and older age.
Reports on the relationship of APOE ɛ4 carrier status on longitudinal accumulation of amyloid-β pathology have been mixed, with studies showing that the APOE ɛ4 genotype increases the rate of change of amyloid-β pathology (Grimmer et al., 2010; Villemagne et al., 2011; Jack et al., 2013b), while others find no genotype effect (Vlassenko et al., 2011; Resnick et al., 2015). When examining APOE genotype effects across the lifespan, prior works have reported a significant ɛ4 and age interaction, (Fleisher et al., 2013; Scheinin et al., 2014; Jansen et al., 2015) although this is not always the case (Rodrigue et al., 2012).
The most common neurodegenerative biomarker studies with APOE have been structural MRI. Both cross-sectional (Hashimoto et al., 2001; Liu et al., 2010; Hostage et al., 2013; Manning et al., 2014) and longitudinal (Geroldi et al., 1999; Mori et al., 2002; Van De Pol et al., 2007; Morra et al., 2009; Risacher et al., 2010; Hostage et al., 2014; Manning et al., 2014) studies have reported significant effects of APOE genotype on hippocampal volume or cortical thickness in diseased populations, but other studies have shown no effect (Soininen et al., 1995; Jack et al., 1998; Reiman et al., 1998; Cohen et al., 2001; Lemaitre et al., 2005; Drzezga et al., 2009; Schuff et al., 2009; Fan et al., 2010; Leung et al., 2013). A similar pattern is seen within populations of only cognitively normal older adults, with studies finding differences between carriers and non-carriers on rates of change in medial temporal lobe structures (Moffat et al., 2000; Chen et al., 2007; Risacher et al., 2010; Lu et al., 2011; Cohen et al., 2013), while other work does not (Du et al., 2006; Taylor et al., 2014).
This heterogeneity in the literature may be attributed to inconsistencies in the populations studied, insufficient longitudinal follow-up, and relatively modest participant cohorts. The majority of the studies evaluating the effect of APOE ɛ4 on longitudinal changes in amyloid-β have either exclusively looked at impaired (Alzheimer’s disease or mild cognitive impairment) patients or combined healthy controls with cognitively impaired individuals (Jack et al., 2009, 2013b; Grimmer et al., 2010; Villemagne et al., 2011; Bilgel et al., 2016). Changes in Alzheimer’s disease pathology can begin decades before the onset of dementia (Bateman et al., 2012; Jack et al., 2013a). The focus of APOE studies on older and already impaired individuals has left the question of how the APOE ɛ4 genotype modulates early changes in Alzheimer’s disease pathology largely unanswered.
Pathology is not uniformly distributed, but has a spatial evolution in the brain as the disease progresses (Benzinger et al., 2013; Gordon et al., 2014). The majority of studies in the literature have looked at whole-brain summary measures of amyloid-β (Morris et al., 2010; Rowe et al., 2010; Johnson et al., 2013; Mathis et al., 2013; Risacher et al., 2015; Gottesman et al., 2016). Of those that have used a regional approach, some have not seen regional variability in ɛ4 effects (Murphy et al., 2013), while others have found effects in primarily frontal cortex regions (Reiman et al., 2009; Scheinin et al., 2014) or more posterior temporal-parietal regions (Fleisher et al., 2013). The APOE ɛ4 relationship on structural MRI has primarily focused on hippocampus or surrounding medial temporal lobe structures, with a minority of studies utilizing a whole-brain voxel-wise or regions of interest approach (Geroldi et al., 1999; Lu et al., 2011; Tosun et al., 2011; Hostage et al., 2014). While the literature demonstrates that the ɛ4 allele has an overall deleterious effect on amyloid-β pathology and neurodegenerative biomarkers, there are unanswered questions about where in the brain these effects spatially manifest.
The objective of this study was to utilize longitudinal data to characterize the effect of APOE ɛ4 on amyloid-β pathology as measured by PET and neurodegeneration as measured by structural MRI. Here, we attempt to understand the nature of the APOE ɛ4 effects by using a whole-brain regional approach to evaluate the spatial effects of APOE ɛ4 on longitudinal rates of change of pathology. Additionally, we explore whether ɛ4 carriers develop Alzheimer’s disease pathology at a higher proportion, at an earlier time point, or a faster rate than non-carriers. These analyses address shortcomings in the literature to better understand the role that the APOE ɛ4 genotype plays on regional pathology across the lifespan.
Materials and methods
Participants
Participants between the ages of 45 and 90 were included from ongoing studies on ageing and dementia from the Knight Alzheimer Disease Research Center at Washington University. Participants who had one or more ɛ4 allele were assigned a positive APOE ɛ4 status, while those with no ɛ4 allele were assigned a negative APOE ɛ4 status.
The longitudinal analyses of amyloid-β PET and structural MRI were analysed only in individuals who were cognitively normal at baseline [clinical dementia rating (CDR) of 0 (Morris, 1997)]. In this initial cohort 249 participants were identified that had two or more amyloid-β PET scans and 497 participants had two or more MRI sessions. Demographics for the entire cohort are shown in Table 1. Individuals with both an ɛ4 and an ɛ2 allele were excluded, yielding a final sample for statistical analyses of 242 with longitudinal PET and 482 with longitudinal MRI. Demographic comparisons between ɛ4 carrier and ɛ4 non-carrier groups are presented in the online Supplementary material. There were no significant demographic differences between carriers and non-carriers.
Table 1.
Longitudinal cohort demographics table for estimating rates of biomarker change
| PIB | MRI | |
|---|---|---|
| Participants, n | 249 | 497 |
| Age (SD) | 64.8 (9.4) | 66.8 (10.0) |
| Gender, n male (%) | 83 (33.3) | 189 (38.0) |
| Education (SD)a | 15.9 (2.5) | 15.8 (2.5) |
| MMSE (SD) | 29.1 (1.1) | 29.1 (1.1) |
| CDR Sum of Boxes (SD) | 0.01 (0.08) | 0.02 (0.10) |
| Scans, n, mean (SD) | 2.5 (0.6) | 3.1 (1.3) |
| Years of follow-up (SD) | 4.9 (2.1) | 5.6 (3.1) |
| n APOE ɛ4a (ɛ34 or ɛ44) (%) | 74 (29.7) | 150 (30.2) |
| ɛ22/ɛ23/ɛ33/ɛ24b/ɛ34/ɛ44, n | 3/30/135/7/63/11 | 4/59/269/15/127/23 |
| [%] | [1.2/12.0/54.2/2.8/25.3/4.4] | [0.8/11.9/54.1/3.0/25.6/4.6] |
| n that converted to CDR >0 | 13 (5.2%) | 89 (17.9%) |
aEducation values were not available for 23 individuals.
bIndividuals with both an ɛ2 and ɛ4 allele were excluded from analysis.
MMSE = Mini-Mental State Examination.
It was of additional interest to estimate population level frequencies of an abnormal amyloid-β PET scan across middle and older age. To do this we examined the baseline visits from the 242 cognitively normal individuals included in the longitudinal PET data, as well as another 333 (total n = 575) individuals with only one PET session. Individuals carrying both an ɛ4 and an ɛ2 were excluded from this sample. From this total population 443 were CDR = 0, 70 CDR = 0.5, 40 CDR = 1, 19 CDR = 2, and 3 CDR = 3. Cognitively impaired individuals were included in this one analyses as excluding them would erroneously underestimate the true percentages of the population with abnormal amyloid-β PET scans. Demographics for this cross-sectional cohort are shown in Table 2.
Table 2.
Cross-sectional demographics table for estimating population rates of amyloid-β positivity
| Participants, n | 575 |
| Age (SD) | 67.7 (9.9) |
| Gender, n male (%) | 239 (41.6) |
| CDR | 443 CDR 0, 70 CDR 0.5, 40 CDR 1, 19 CDR 2, 3 CDR 3 |
| ɛ22/ɛ23/ɛ33/ɛ34/ɛ44, n | 4/69/297/176/29 |
| [%] | [0.7/12.0/51.7/30.6/5.0] |
| n APOE ɛ4+ (ɛ34 or ɛ44) (%) | 205 (35.6) |
MRI acquisition and processing
Structural magnetization-prepared rapid gradient-echo (MPRAGE) images were acquired on either a 1.5 T (n = 336 sessions) or 3 T (n = 1199 sessions) Siemens. Scans had a resolution of either 1 × 1 × 1.25 mm or 1 × 1 × 1 mm. Structural scans were processed with FreeSurfer (Fischl, 2012) using the Desikan atlas. For each hemisphere, cortical thickness values were obtained for all FreeSurfer cortical regions of interest, and volumes were obtained for all FreeSurfer subcortical regions of interest. Cortical thickness was calculated as the shortest distance between the cortical grey/white boundary to the grey/CSF boundary (Fischl and Dale, 2000). All subcortical region volumes were adjusted for intracranial volume using a regression approach (Buckner et al., 2004). Left and right values for each region of interest were averaged together.
PIB PET acquisition and processing
Amyloid-β PET imaging was completed using 11C-Pittsburgh compound B (PIB). PET data from the 30–60-min post-injection window were analysed using regions of interests derived from Freesurfer (Fischl, 2012; Su et al., 2013) (PET Unified Pipeline, https://github.com/ysu001/PUP). Regional estimates were transformed into standardized uptake value ratios (SUVRs) with cerebellar cortex as the reference region. Partial volume correction was performed using a regional spread function technique (Rousset et al., 1998; Su et al., 2015). As with the MRI data, regions were averaged across hemispheres before being entered into statistical analyses. For an examination of population level frequencies of abnormal amyloid-β deposition, participants were additionally classified as having abnormal (amyloid-β positive) or normal (amyloid-β negative) levels using a previously established PIB mean cortical SUVR cut-off of 1.42 (Sutphen et al., 2015; Brier et al., 2016; Vlassenko et al., 2016; Mishra et al., 2017), which is the linear projection of a mean cortical binding potential (MCBP) cut-off previously established in this cohort of 0.18 (Mintun et al., 2006; Vlassenko et al., 2011; Gordon et al., 2015).
Genotyping
Genomic DNA was isolated from peripheral blood samples using standard procedures. APOE genotyping was performed as previously described (Talbot et al., 1994). The distribution of alleles was in Hardy-Weinberg equilibrium.
Statistical analysis
We used multivariate linear mixed effects models (LME) to evaluate longitudinal changes in amyloid-β PET and structural MRI. LME models provide a statistical approach that is flexible and can accommodate unequal numbers of measurement points or sampling intervals. Considering multiple regions of interest simultaneously in a multivariate approach provides the ability to account for correlations between regional measurements. Within each modality, we modelled the cortical and subcortical regions separately. One model fit the 34 FreeSurfer cortical regions simultaneously and one model fit the seven FreeSurfer subcortical regions simultaneously. For MRI, cortical thickness was used for the cortical regions’ model and intracranial volume-corrected volumes were used for the subcortical regions’ model. Models were fit on mean-centred and standard deviation (SD) scaled data. Within each model (e.g. cortical PIB) for each individual region of interest, the model included fixed effects of baseline age, time, and APOE ɛ4 status. To allow for non-linear associations with baseline age, we modelled baseline age as a restricted cubic spline with three knots. Restricted cubic spline functions allow for the flexibility to vary non-linearly without forcing the relationship into a particular polynomial fit and have been used previously for modelling imaging biomarkers with respect to age or time in longitudinal analyses (Vemuri et al., 2010; Jack et al., 2015; Jansen et al., 2015). In our final model we included the two terms for baseline age from the spline, APOE ɛ4 status, time, and all two-way and three-way interactions as fixed effects and also subject-varying slope and intercept random effect terms. Both the subject-level intercepts and the subject-level slopes were allowed to correlate across all brain regions included in a model.
To fit each model, we used the software package Stan (mc-stan.org) (Gelman et al., 2015; Carpenter et al., 2017) implemented in R using rstan version 2.14.1, to perform Markov Chain Monte Carlo (MCMC) analyses. A Bayesian approach was selected as the resultant credible intervals provide a richer, more informative estimate than classical confidence intervals obtained from null-hypothesis significance testing approaches. We used a normal distribution (mean = 0, SD = 5) as a prior for the fixed effects beta-estimates and used a normal distribution (mean = 0, SD = 1) as a prior for the random effects gamma-estimates of the scaled data. We used uniform priors for the variances and the lkj correlation density function (mu = 1.0) as a prior for the correlation matrix (Lewandowski et al., 2009). We implemented MCMC with eight chains of 10 000 iterations each (5000 warm-up, thinning 1 in 10 iterations) (Kruschke, 2014; Sorensen and Vasishth, 2016). Credible intervals were defined as the range between the 0.5% and 99.5% estimates to represent 99% credible intervals. To test for convergence, the Gelman-Rubin convergence statistic (Ř), the ratio of between-chain variance to within-chain variance was used (Gelman and Rubin, 1992). A value close to 1 indicates convergence. Ř for each parameter estimate was within 0.01 of 1.0. The code for the models is included in the Supplementary material.
We report the rate of change of PIB SUVR or structural MRI data as a function of baseline age for APOE ɛ4-positive and APOE ɛ4-negative individuals. The ages at which the slopes between groups diverge were determined as the first age at which the 99% credible interval of the distribution of differences between groups did not overlap zero. To improve reliability and avoid spurious points, a time point was only considered significant if the distribution continued to not overlap zero for 2 years. To understand whether ɛ4 carriers accumulate pathology at a faster rate relative to non-carriers at a similar point in the disease, both the PIB longitudinal rate of change in the precuneus and the volume longitudinal rate of change in the hippocampus were additionally evaluated as a function of baseline disease burden, measured by the mean cortical SUVR (mcSUVR) from the baseline PIB scan. A linear mixed effects model was fit including fixed effects of baseline mcSUVR, time, and APOE ɛ4 status, along with all two-way and three-way interactions and subject-varying slope and intercept random effect terms. To again allow for non-linear associations, mcSUVR was modelled as a restricted cubic spline with three knots. MCMC analyses were completed as previously described to derive PIB slope and volume slope estimates as a function of baseline PIB mcSUVR, with 99% credible intervals. As exploratory analyses, we additionally ran models for precuneus PIB and hippocampal volume stratifying ɛ4 carriers into homozygotes (ɛ44) and heterozygotes (ɛ43 or ɛ34) and including all appropriate additional terms in the linear mixed effects models. These results were considered exploratory rather than as main analyses due to the small number of homozygotes with PIB (n = 11) and MRI (n = 23) data.
Finally, to characterize abnormal levels of amyloid-β PET across the lifespan, the cross-sectional cohort of 575 individuals with at least one PET scan was used. Each participant was characterized as amyloid-β positive or negative based upon previously defined cut-offs. The proportion of all ɛ4 carriers and all non-carriers who were amyloid-β positive was determined as a function of age across the span of 42 to 89. The resultant curves were smoothed using a local regression method, using the loess method implemented in R, with a span = 0.7.
Results
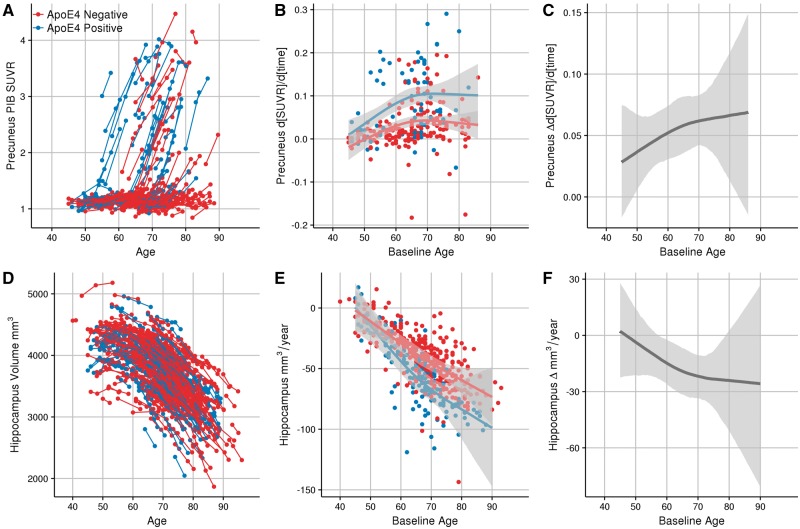
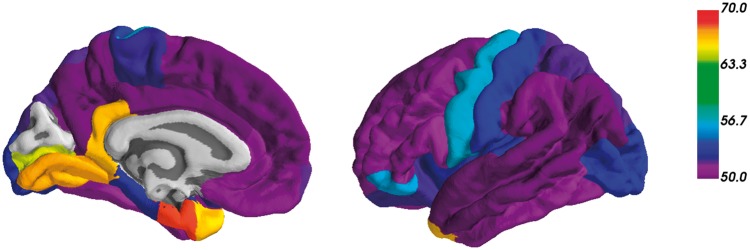
We found a statistically significant difference in the rate of amyloid-β accumulation in APOE ɛ4 carriers in comparison to non-carriers in 33/34 FreeSurfer cortical regions. Figure 1 shows an example of the results in the precuneus. Figure 1A shows a spaghetti plot of the model fits for the individual participant SUVR trajectories. Figure 1B shows the model fits for the SUVR rate of change as a function of baseline age and APOE genotype. Figure 1C shows the difference in the longitudinal rates of change of amyloid-β accumulation between the APOE ɛ4 carriers and non-carriers, along with the 99% credible intervals of this difference. The first point in this difference where credible intervals do not overlap 0 is the first baseline age where rates of accumulation significantly diverge between groups. In our data, APOE ɛ4 carriers demonstrate significantly accelerated accumulation of amyloid-β deposition in the precuneus, in comparison to non-carriers, at a baseline age of 49 years old. The difference in slopes between carriers and non-carriers is significantly greater than zero throughout the majority of the lifespan represented by our cohort. Figure 2 shows the age at which APOE ɛ4 carriers first have significantly greater longitudinal change in PIB binding than non-carriers for each of the cortical regions. These data are presented numerically in Table 3. In all cortical regions where there was a statistically significant difference between ɛ4 carriers and non-carriers, the earliest age of divergence in rates of amyloid-β accumulation between carriers and non-carriers varied between 49 and 68 years. Two out of seven of the FreeSurfer subcortical regions showed significant differences between groups in rates of amyloid-β plaque accumulation across the adult lifespan, the caudate (age 56) and the putamen (age 63). Graphs for all regions are depicted in the Supplementary material.
Figure 1.
Modelling longitudinal change in precuneus PIB (A–C) and hippocampal volume (D–F). A and D represent spaghetti plots of the model fits for the individual participant SUVR and volume trajectories, respectively. B and E show the longitudinal rates of change between the APOE ɛ4 carriers and non-carriers, along with the 99% credible intervals. For reference, individual random effect slope estimates are also plotted. C and F depict the difference in rate of biomarker change between APOE ɛ4 carriers and non-carriers across the course of the sampled lifespan. Shaded regions represent 99% credible intervals..
Figure 2.
Regional differences in emergence of PIB. The colour scale represents the first age where the rate of amyloid-β accrual in the cortical regions is significantly different between APOE ɛ4 carriers and non-carriers.
Table 3.
First age of accelerated PIB binding in ɛ4 carriers versus non-carriers
| Region of interest | Age of first detectable APOE effect |
|---|---|
| Cortical | |
| Inferior parietal cortex | 49.28 |
| Precuneus cortex | 49.32 |
| Middle temporal gyrus | 50.21 |
| Superior frontal gyrus | 50.29 |
| Superior temporal gyrus | 50.6 |
| Supramarginal gyrus | 50.69 |
| Caudal anterior cingulate cortex | 50.71 |
| Posterior cingulate cortex | 50.93 |
| Rostral anterior cingulate cortex | 50.94 |
| Banks of the superior temporal sulcus | 50.96 |
| Rostral middle frontal cortex | 50.99 |
| Medial orbitofrontal cortex | 51.1 |
| Pars triangularis | 51.29 |
| Caudal middle frontal cortex | 51.44 |
| Fusiform gyrus | 51.79 |
| Inferior temporal cortex | 51.89 |
| Pars opericularis | 52.06 |
| Superior parietal cortex | 52.32 |
| Frontal pole | 52.44 |
| Paracentral cortex | 53 |
| Lateral orbitofrontal cortex | 53.02 |
| Transverse temporal gyrus | 53.51 |
| Parahippocampal cortex | 53.67 |
| Insula cortex | 54.28 |
| Lateral occipital cortex | 54.5 |
| Postcentral gyrus | 54.71 |
| Pars orbitalis | 56.69 |
| Precentral gyrus | 57.19 |
| Pericalcarine cortex | 65.28 |
| Isthmus cingulate | 66.74 |
| Temporal pole | 67.41 |
| Lingual gyrus | 67.62 |
| Entorhinal cortex | 68.45 |
| Cuneus cortex | - |
| Subcortical | |
| Nucleus accumbens | - |
| Amygdala | - |
| Caudate | 56.76 |
| Hippocampus | - |
| Pallidum | - |
| Putamen | 64.04 |
| Thalamus | - |
In contrast to the effects on amyloid-β deposition, only 3/34 cortical regions showed a statistically significant difference in the rate of cortical atrophy, with carriers having faster rates of cortical thinning than non-carriers in the insula (age 47), parahippocampal cortex (age 51), and entorhinal cortex (age 71). All of these effects were very modest and bordered on significance (see regional graphs available in the Supplementary material). For the subcortical analyses 4/7 of the regions showed a significantly greater rate of subcortical atrophy in APOE ɛ4 carriers relative to non-carriers: the hippocampus, amygdala, putamen, and accumbens at ages of 57, 66, 70, and 71, respectively. Plots of the model fits of hippocampal volume are shown in Fig. 1D–F. The non-carriers’ hippocampal volumetric rate of change shows a linear acceleration throughout the lifespan, while APOE ɛ4 carriers demonstrate even greater accelerated cortical atrophy in midlife followed by a similar rate of acceleration to that of the non-carriers later in the lifespan (Fig. 1E). The slope difference between carriers and non-carriers significantly differs from zero between ∼55 to 80 years, after which there is not a significant difference in the rates (Fig. 1F). Exploratory analyses examining the effect of ɛ4 dosage are presented in Supplementary Fig. 1. These results indicate that ɛ4 homozygotes had earlier and greater rates of amyloid-β accumulation and hippocampal atrophy than APOE ɛ4 heterozygotes and non-carriers.
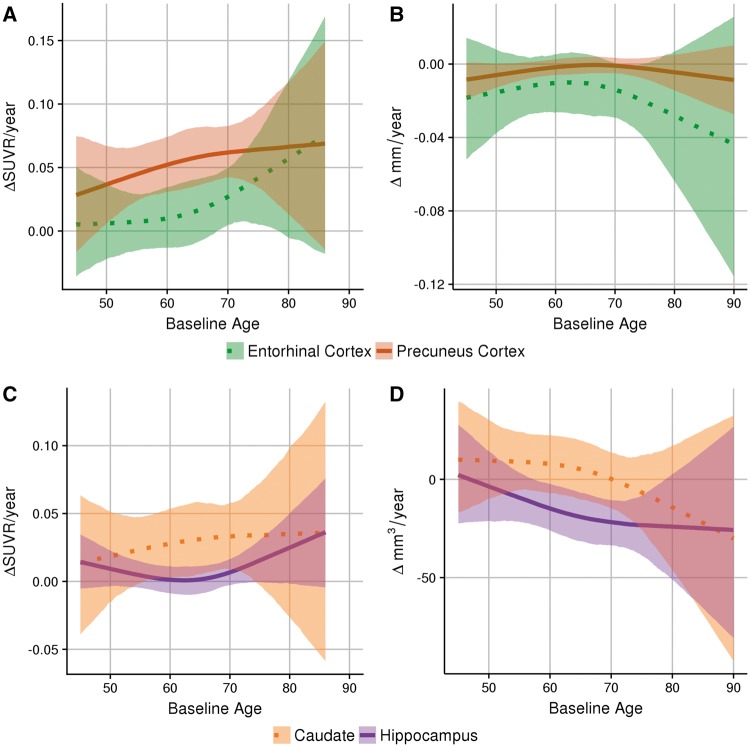
The temporal evolution of pathology as related to the APOE ɛ4 genotype differs regionally. Figure 3A and B shows the differences in the longitudinal rate of change of amyloid-β deposition and cortical atrophy, respectively, between carriers and non-carriers in two cortical regions prominently studied in Alzheimer’s disease. In comparison to the precuneus, which demonstrates early acceleration followed by a plateau in the rate of change of amyloid-β accumulation, the entorhinal cortex shows no difference in amyloid-β accumulation rates between carriers and non-carriers at younger ages but APOE ɛ4 carriers demonstrate increased rates of amyloid-β deposition in the entorhinal cortex at older ages. In contrast, the differences in cortical atrophy between carriers and non-carriers in the entorhinal cortex and precuneus are not significantly different across the lifespan sampled in this population.
Figure 3.
Regional differences in biomarker trajectories. Top: The difference in the longitudinal rates of change of PIB measurements (A) and cortical thickness (B) between APOE ɛ4 carriers and non-carriers in two cortical regions. Bottom: The difference in the longitudinal rates of change of PIB (C) and volume (D) between APOE ɛ4 carriers and non-carriers in two subcortical regions.
Figure 3C and D shows the differences in the longitudinal rate of change of amyloid-β accumulation and structural atrophy, respectively, between carriers and non-carriers in two subcortical regions. The differences in amyloid-β accumulation between carriers and non-carriers in the hippocampus and caudate did not differ significantly across the lifespan. When examining structural MRI, in the hippocampus ɛ4 carriers have accelerated volumetric loss early in the lifespan, but there is no significant difference in rate of change of volume loss between carriers and non-carriers in the caudate across the sampled lifespan.
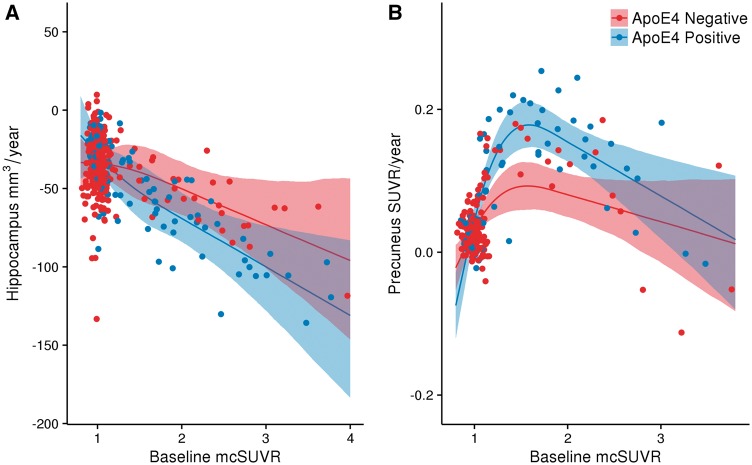
The difference in amyloid-β accumulation in the precuneus and atrophy in the hippocampus between APOE ɛ4 carriers and non-carriers was evaluated as a function of disease burden, measured by baseline PIB mean cortical SUVR. Figure 4A shows no difference between hippocampal atrophy as a function of disease burden in ɛ4 carriers, in comparison to non-carriers. Figure 4B demonstrates greater longitudinal amyloid-β accumulation as a function of disease burden in ɛ4 carriers, in comparison to non-carriers. These findings show that APOE ɛ4 carriers experience accelerated amyloid-β accumulation, but not atrophy, relative to non-carriers after matching individuals for baseline levels of pathology.
Figure 4.
Longitudinal change in PIB as a function of disease status. The longitudinal rates of change of PIB in the hippocampus (A) and the precuneus (B) for both APOE ɛ4 carriers and non-carriers are shown as a function of mean cortical SUVR from the baseline PIB assessment. Shaded areas represent 99% credible intervals.
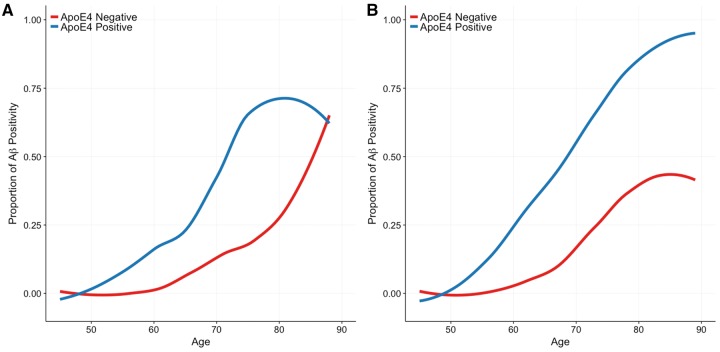
The population level frequencies of abnormal PIB levels stratified by genotype are shown across the ages of participants in the cross-sectional cohort (Fig. 5). At a population level, APOE ɛ4 carriers begin having elevated amyloid-β pathology at an earlier age, and at higher frequencies (steeper slope) after age 60. The proportion of APOE ɛ4 carriers that eventually develop pathological amyloid-β PET levels approaches 1 by age 90, while the proportion of non-carriers that develop pathological amyloid-β levels plateaus at less than 0.5, or half of the population.
Figure 5.
Characterization of abnormal levels of amyloid-β PET across the lifespan. The proportion of APOE ɛ4 carriers and non-carriers who are amyloid-β PET positive is depicted as function of age in (A) non-demented individuals only and (B) the combined sample of non-demented and demented individuals.
Discussion
The purpose of this study was to better characterize the role of the APOE ɛ4 allele on amyloid-β pathology and neurodegeneration across the lifespan of middle-aged to elderly cognitively normal adults. This study illustrates that the presence of the APOE ɛ4 allele results in increased longitudinal rates of change of amyloid-β plaque accumulation diffusely through the cortex. While APOE ɛ4 carriers show near ubiquitous accelerated amyloid-β accumulation throughout the brain relative to non-carriers, the temporal emergence of this pathological accumulation differs across brain regions (between ages 49 and 70). In addition, the present work is the first to show that APOE ɛ4 carriers have not only increased rates of amyloid-β accumulation earlier but also have faster rates of amyloid-β accumulation in comparison to non-carriers after adjusting for baseline levels of pathology. APOE ɛ4 effects on neurodegeneration were modest, with carriers displaying greater rates of structural loss in medial temporal lobe structures as early as 50 years.
The literature has shown that APOE ɛ4 carriers develop Alzheimer’s disease more often and earlier than non-carriers, and the APOE ɛ4 allele has deleterious effects on amyloid-β pathology. The majority of work on amyloid-β pathology has been cross-sectional (for a review see Fouquet et al., 2014), with a limited subset of work examining longitudinal change (Grimmer et al., 2010; Villemagne et al., 2011, 2013; Vlassenko et al., 2011; Jack et al., 2013b; Resnick et al., 2015; Bilgel et al., 2016). In general, longitudinal studies have found that APOE ɛ4 status did not significantly affect the rates of amyloid-β accumulation after adjusting for age (Vlassenko et al., 2011; Jack et al., 2013b; Villemagne et al., 2013; Resnick et al., 2015; Bilgel et al., 2016). Relatively modest longitudinal follow-ups, and study populations that contain older adults and demented individuals, but do not include middle-aged participants, may drive such null results. While prior work indicates there is an undeniable effect of the ɛ4 allele on group level measures of amyloid-β pathology, it is unclear how the ɛ4 allele produces this effect. It has remained to be shown whether these group differences can solely be attributed to a higher proportion of APOE ɛ4 carriers developing the disease, whether they develop it at an earlier age, or whether APOE ɛ4 carriers are developing pathology at a faster rate even relative to individuals with preclinical Alzheimer’s disease without the ɛ4 allele.
The current study found that at the group level ɛ4 carriers had greater longitudinal accumulation of amyloid-β than non-carriers and that this deposition accelerated with increasing baseline age. When considering population frequencies of PIB positivity (Fig. 5) we found that ɛ4 carriers had abnormal PIB scans earlier in time and at an elevated population frequency relative to non-carriers. Finally, in addition to looking at slope differences as a function of age, the present study shows that the accumulation of amyloid-β pathology as a function of baseline amyloid-β burden is also higher in APOE ɛ4 carriers than non-carriers (Fig. 4). This study examining cognitively normal middle-aged and older adults provides evidence that APOE ɛ4 positivity not only shifts the onset of amyloid-β deposition earlier, but also accelerates accrual of amyloid-β deposits once this process has begun. In contrast, previous work suggesting that APOE ɛ4 status primarily affects the age at which people begin to develop amyloid-β pathology, but not the rate of amyloid-β deposition (Jack et al., 2013b; Bilgel et al., 2016) has primarily been conducted using older adults and impaired individuals. Our findings suggest that genetic factors in Alzheimer’s disease may change the trajectory of disease progression, in contrast to the hypothesis that all individuals at a given disease burden will progress similarly. This may have important implications for clinical trial selection and therapeutic targets for APOE ɛ4 carriers.
The current study examined regional differences in the temporal evolution of amyloid-β accumulation in APOE ɛ4 carriers versus non-carriers. The vast majority of prior studies have used summary metrics in cross-sectional and longitudinal studies looking at APOE ɛ4 effects. In the present study, we consider the longitudinal rates of change of amyloid-β in all cortical and subcortical FreeSurfer anatomical regions of interest. The present study shows that the age at which APOE ɛ4 carriers display increased rates of accumulation in comparison to non-carriers is as early as 49 years old in regions that are affected early by amyloid-β pathology (i.e. precuneus). The presence of temporal differences in different regions introduces a novel aspect of genetic influences on late-onset Alzheimer’s disease.
The relationship between APOE ɛ4 and volumetric changes longitudinally has conflicting evidence in the current literature. Structural changes from neurodegeneration is a late biomarker change in the progression of Alzheimer’s disease (Bateman et al., 2012; Benzinger et al., 2013; Jack and Holtzman, 2013; Jack et al., 2013a). The present study shows that cognitively normal APOE ɛ4 carriers demonstrate more rapid volume loss in medial temporal lobe structures, in comparison to non-carriers, first detected around 60 years of age. This is consistent with prior work indicating significant effects of APOE genotype on structural MRI (Geroldi et al., 1999; Mori et al., 2002; Van De Pol et al., 2007; Morra et al., 2009; Risacher et al., 2010; Hostage et al., 2014; Manning et al., 2014) and in contrast to the studies that have shown no APOE ɛ4 effect on volumetric changes (Jack et al., 1998; Moffat et al., 2000; Du et al., 2006; Fan et al., 2010; Taylor et al., 2014).
Despite the large sample and high number of longitudinal visits the observed APOE ɛ4 effect on neurodegenerative biomarkers was quite subtle and the modest size of this effect may explain the heterogeneity in the field. The ɛ4 effect on neurodegenerative biomarkers may also be most prominent when including both middle-aged and older adults. Further, when looking at hippocampal atrophy as a function of baseline disease status, there was no difference between APOE ɛ4 carriers than non-carriers. These findings suggest that, in contrast with the APOE ɛ4 effect on amyloid-β, the neurodegeneration seen in both the APOE ɛ4 carriers and non-carriers was proportional to baseline levels of amyloid-β. This suggests that when considering populations matched for clinical severity or amyloid-β pathology, the ɛ4 genotype may have minimal effects on atrophy.
There are some limitations of our study. Our results reflect the detectable genotype differences given our sample. Larger samples with greater power would likely detect differences even earlier or detect effects in additional regions. Our results also focus on cognitively normal individuals. The effects of APOE genotype likely vary in demented cohorts. The earliest ages at which we can detect APOE ɛ4 carriers and non-carriers diverging are limited by the ages represented in our cohort, which begins at age 45. Including even younger ages could reveal that the influence of the ɛ4 allele begins even earlier in the lifespan. Prior work also indicates that the ɛ4 allele has a dose effect (Corder et al., 1993), with homozygotes having greater risk of developing Alzheimer’s disease. Even in our larger population with longitudinal MRI only 23 individuals were homozygous for the ɛ4 allele. This limits our ability to accurately model the interaction of ɛ4 dosage across the studied age ranges. Results from our exploratory analyses in the precuneus and hippocampus do suggest that ɛ4 homozygotes have greater and earlier amyloid-β accumulation and atrophy. However, our samples size is very modest. Future work integrating multiple cohorts would be able to better estimate how ɛ4 homozygosity or the ɛ2 alleles modify biomarker trajectories starting in middle age. Our cohort consisted of adults with longitudinal imaging biomarker data who were cognitively normal at baseline. The influence of the ɛ4 allele may be different as cognition declines. This study focused only on PIB PET and MRI. Evaluation of longitudinal changes of other imaging biomarkers of neurodegeneration, such as FDG and tau PET would be of interest as these may better predict cognition. Finally, our work represents the influence of the APOE ɛ4 allele in only one cohort of middle-aged and older adults. Replication of our results by other groups in an independent cohort would strengthen the interpretation of our findings.
In summary, the present study provides evidence that APOE ɛ4 is linked to increased rates of change of amyloid-β accumulation detectable in late middle-aged cognitively normal adults. In addition to APOE ɛ4 carriers having earlier amyloid-β accumulation, in comparison to non-carriers, they also have faster rates of accumulation as a function of baseline disease status. Through using a regional approach, the present study provides the first evidence that the temporal evolution of APOE ɛ4-related amyloid-β accumulation varies regionally. Finally, this study shows that cognitively normal APOE ɛ4 carriers show increased rates of volumetric change in medial temporal lobe structures as early as 60 years of age, in comparison to non-carriers.
Supplementary Material
Acknowledgements
Above all we thank the dedicated research participants and their families without which this work would not be possible.
Funding
Financial support was provided by NIH grants P01 AG003991, P50 AG005681, P01 AG026276, P30 NS098577, 1S10RR022984-01A1, 1S10OD018091, R01 EB009352, as well as gifts from the Charles and Joanne Knight Alzheimer Disease Research Center Support Fund, the David and Betty Farrell Medical Research Fund, the Daniel J Brennan Alzheimer Research Fund, the Thomas E. Brew Foundation Fund, the Barnes-Jewish Hospital Foundation, and the Fred Simmons and Olga Mohan Alzheimer Research Support Fund. Computations were performed using the Washington University Center for High Performance Computing.
Conflicts of interest
D.M.H. co-founded and is on the scientific advisory board of C2N Diagnostics. He consults for Genentech, AbbVie, Eli Lilly, Proclara, Glaxosmithkline, and Denali. Washington University receives research grants to the lab of D.M.H. from C2N Diagnostics, Eli Lilly, AbbVie, and Denali and to the laboratories of L.S.B. and J.C.M. from Eli Lilly and Avid Radiopharmaceuticals (a wholly owned subsidiary of Eli Lilly). All other authors report no conflicts.
Supplementary material
Supplementary material is available at Brain online.
Glossary
Abbreviations
- CDR
Clinical Dementia Rating
- PIB
11C-Pittsburgh compound B
- SUVR
standardized uptake value ratio
References
- Bateman RJ, Xiong C, Benzinger TLS, Fagan AM, Goate A, Fox NC, et al. Clinical and biomarker changes in dominantly inherited Alzheimer’s disease. N Engl J Med 2012; 367: 795–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benzinger TLS, Blazey T, Jack CR, Koeppe RA, Su Y, Xiong C, et al. Regional variability of imaging biomarkers in autosomal dominant Alzheimer’s disease. Proc Natl Acad Sci USA 2013; 110: E4502–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilgel M, An Y, Zhou Y, Wong DF, Prince JL, Ferrucci L, et al. Individual estimates of age at detectable amyloid onset for risk factor assessment. Alzheimers Dement 2016; 12: 373–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braak H, Braak E. Neuropathological stageing of Alzheimer-related changes. Acta Neuropathol 1991; 82: 239–59. [DOI] [PubMed] [Google Scholar]
- Brier M, Gordon B, Friedrichsen K, McCarthy J, Stern A, Christensen J, et al. Tau and AB imaging, CSF measures, and cognition in Alzheimers disease. Sci Transl Med 2016; 40: 135–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckner RL, Head D, Parker J, Fotenos AF, Marcus D, Morris JC, et al. A unified approach for morphometric and functional data analysis in young, old, and demented adults using automated atlas-based head size normalization: reliability and validation against manual measurement of total intracranial volume. Neuroimage 2004; 23: 724–38. [DOI] [PubMed] [Google Scholar]
- Carpenter B, Lee D, Brubaker MA, Riddell A, Gelman A, Goodrich B, et al. Stan: a probabilistic programming language. J Stat Softw 2017; 76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K, Reiman EM, Alexander GE, Caselli RJ, Gerkin R, Bandy D, et al. Correlations between apolipoprotein E ɛ4 gene dose and whole brain atrophy rates. Am J Psychiatry 2007; 164: 916–21. [DOI] [PubMed] [Google Scholar]
- Cohen AD, Mowrey W, Weissfeld LA, Aizenstein HJ, McDade E, Mountz JM, et al. Classification of amyloid-positivity in controls: comparison of visual read and quantitative approaches. Neuroimage 2013; 71: 207–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen RM, Small C, Lalonde F, Friz J, Sunderland T. Effect of apolipoprotein E genotype on hippocampal volume loss in aging healthy women. Neurology 2001; 57: 2223–8. [DOI] [PubMed] [Google Scholar]
- Corder E, Saunders A, Strittmatter W, Schmechel D, Gaskell P, Small G, et al. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer’s disease in late onset families. Science 1993; 261: 921–3. [DOI] [PubMed] [Google Scholar]
- Drzezga A, Grimmer T, Henriksen G, Mühlau M, Perneczky R, Miederer I, et al. Effect of APOE genotype on amyloid plaque load and gray matter volume in Alzheimer disease. Neurology 2009; 72: 1487–94. [DOI] [PubMed] [Google Scholar]
- Du AT, Schuff N, Chao LL, Kornak J, Jagust WJ, Kramer JH, et al. Age effects on atrophy rates of entorhinal cortex and hippocampus. Neurobiol Aging 2006; 27: 733–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan M, Liu B, Zhou Y, Zhen X, Xu C, Jiang T. Cortical thickness is associated with different apolipoprotein E genotypes in healthy elderly adults. Neurosci Lett 2010; 479: 332–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrer LA, Cupples LA, Haines JL, Hyman B, Kukull WA, Mayeux R, et al. Effects of age, sex, and ethnicity on the association between apolipoprotein E genotype and alzheimer disease. JAMA 1997; 278: 1349–56. [PubMed] [Google Scholar]
- Fischl B. FreeSurfer. Neuroimage 2012; 62: 774–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischl B, Dale AMM. Measuring the thickness of the human cerebral cortex from magnetic resonance images. Proc Natl Acad Sci USA 2000; 97: 11050–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleisher AS, Chen K, Liu X, Ayutyanont N, Roontiva A, Thiyyagura P, et al. Apolipoprotein E ɛ4 and age effects on florbetapir positron emission tomography in healthy aging and Alzheimer disease. Neurobiol Aging 2013; 34: 1–12. [DOI] [PubMed] [Google Scholar]
- Fouquet M, Besson FL, Gonneaud J, La Joie R, Chételat G. Imaging brain effects of APOE4 in cognitively normal individuals across the lifespan. Neuropsychol Rev 2014; 24: 290–9. [DOI] [PubMed] [Google Scholar]
- Galasko D, Chang L, Motter R, Clark CM, Kaye J, Knopman D, et al. High cerebrospinal fluid Tau and low amyloid β42 levels in the clinical diagnosis of Alzheimer disease and relation to apolipoprotein E genotype. Arch Neurol 1998; 55: 937–45. [DOI] [PubMed] [Google Scholar]
- Gelman A, Lee D, Guo J. Stan: a probabilistic programming language for Bayesian inference and optimization. J Educ Behav Stat 2015; 40: 530–43. [Google Scholar]
- Gelman A, Rubin DB. Inference from iterative simulation using multiple sequences linked references are available on JSTOR for this article: inference from iterative simulation using multiple sequences. Stat Sci 1992; 7: 457–72. [Google Scholar]
- Geroldi C, Pihlajamäki M, Laakso MP, Decarli C, Beltramello A, Bianchetti A, et al. APOE-epsilon4 is associated with less frontal and more medial temporal lobe atrophy in AD. Neurology 1999; 53: 1825–32. [DOI] [PubMed] [Google Scholar]
- Gordon BA, Blazey T, Benzinger TL. Regional variability in Alzheimer’s disease biomarkers. Future Neurol 2014; 9: 131–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon BA, Najmi S, Hsu P, Roe CM, Morris JC, Benzinger TLS. The effects of white matter hyperintensities and amyloid deposition on Alzheimer dementia. Neuroimage Clin 2015; 8: 246–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesman RF, Schneider ALC, Zhou Y, Chen X, Green E, Gupta N, et al. The ARIC-PET amyloid imaging study. Neurology 2016; 87: 473–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimmer T, Tholen S, Yousefi BH, Alexopoulos P, Frschler A, Frstl H, et al. Progression of cerebral amyloid load is associated with the apolipoprotein e ε4 genotype in Alzheimer’s disease. Biol Psychiatry 2010; 68: 879–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto M, Yasuda M, Tanimukai S, Matsui M, Hirono N, Kazui H, et al. Apolipoprotein E4 and the pattern of regional brain atrophy in Alzheimer’s disease. Neurology 2001: 1461–6. [DOI] [PubMed] [Google Scholar]
- Holtzman D, Herz J. Apolipoprotein E and apolipoprotein receptors: normal biology and roles in Alzheimer’s disease. Cold Spring Harb Perspect Med 2012; 2: a006312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hostage CA, Choudhury KR, Doraiswamy PM, Petrella JR. Mapping the effect of the apolipoprotein E genotype on 4-year atrophy rates in an Alzheimer disease—related brain. Radiology 2014; 271: 211–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hostage CA, Roy Choudhury K, Doraiswamy PM, Petrella JR, Simmons A. Dissecting the gene dose-effects of the APOE ɛ4 and ɛ2 alleles on hippocampal volumes in aging and Alzheimer’s disease. PLoS One 2013; 8: e54483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynh TV, Davis AA, Ulrich JD, Holtzman DM. Apolipoprotein E and Alzheimer’s disease: the influence of apolipoprotein E on amyloid-β and other amyloidogenic proteins. J Lipid Res 2017; 58: 824–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack CR, Holtzman DM. Biomarker modeling of alzheimer’s disease. Neuron 2013; 80: 1347–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack CR, Knopman DS, Jagust WJ, Petersen RC, Weiner MW, Aisen PS, et al. Tracking pathophysiological processes in Alzheimer’s disease: an updated hypothetical model of dynamic biomarkers. Lancet Neurol 2013a; 12: 207–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack CR, Lowe VJ, Weigand SD, Wiste HJ, Senjem ML, Knopman DS, et al. Serial PIB and MRI in normal, mild cognitive impairment and Alzheimer’s disease: implications for sequence of pathological events in Alzheimer’s disease. Brain 2009; 132: 1355–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack CR, Petersen RC, Xu YC, Obrien PC, Waring SC, Tangalos EG, et al. Hippocampal atrophy and apolipoprotein e genotype are independently associated with alzheimers-disease. Ann Neurol 1998; 43: 303–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack CR, Wiste HJ, Lesnick TG, Weigand SD, Knopman DS, Vemuri P, et al. Brain β-amyloid load approaches a plateau. Neurology 2013b; 80: 890–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack CR, Wiste HJ, Weigand SD, Knopman DS, Vemuri P, Mielke MM, et al. Age, sex, and APOE ɛ4 effects on memory, brain structure, and β-amyloid across the adult life span. JAMA Neurol 2015; 72: 511–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagust WJ, Landau SM; Alzheimer’s Disease Neuroimaging Initiative. Apolipoprotein E, not fibrillar β-amyloid, reduces cerebral glucose metabolism in normal aging. J Neurosci 2012; 32: 18227–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen WJ, Ossenkoppele R, Knol DL, Tijms BM, Scheltens P, Verhey FRJ, et al. Prevalence of cerebral amyloid pathology in persons without dementia: a meta-analysis. JAMA 2015; 313: 1924–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson KAA, Sperling RAA, Gidicsin CMM, Carmasin JSS, Maye JEE, Coleman REE, et al. Florbetapir (F18-AV-45) PET to assess amyloid burden in Alzheimer’s disease dementia, mild cognitive impairment, and normal aging. Alzheimers Dement 2013; 9: S72–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruschke JK. Doing Bayesian data analysis: a tutorial with R, JAGS, and Stan. 2nd edn. Amsterdam: Academic Press; 2014. [Google Scholar]
- Lehmann M, Ghosh PM, Madison C, Karydas A, Coppola G, O’Neil JP, et al. Greater medial temporal hypometabolism and lower cortical amyloid burden in ApoE4-positive AD patients. J Neurol Neurosurg Psychiatry 2013; 85: 266–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaitre H, Crivello F, Dufouil C, Grassiot B, Tzourio C, Alperovitch A, et al. No e4 gene dose effect on hippocampal atrophy in a large MRI database of healthy elderly subjects. Neuroimage 2005; 24: 1205–13. [DOI] [PubMed] [Google Scholar]
- Leung KK, Bartlett JW, Barnes J, Manning EN, Ourselin S, Fox NC. Cerebral atrophy in mild cognitive impairment and Alzheimer disease: rates and acceleration. Neurology 2013; 80: 648–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewandowski D, Kurowicka D, Joe H. Generating random correlation matrices based on vines and extended onion method. J Multivar Anal 2009; 100: 1989–2001. [Google Scholar]
- Liu Y, Paajanen T, Westman E, Wahlund LO, Simmons A, Tunnard C, et al. Effect of APOE ɛ4 allele on cortical thicknesses and volumes: the AddNeuroMed Study. J Alzheimers Dis 2010; 21: 947–66. [DOI] [PubMed] [Google Scholar]
- Lu PH, Thompson PM, Leow A, Lee GJ, Lee A, Yanovsky I, et al. NIH public access. J Alzheimers Dis 2011; 23: 433–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahoney-Sanchez L, Belaidi AA, Bush AI, Ayton S. The complex role of apolipoprotein E in Alzheimer’s disease: an overview and update. J Mol Neurosci 2016; 60: 325–35. [DOI] [PubMed] [Google Scholar]
- Manning EN, Barnes J, Cash DM, Bartlett JW, Leung KK, Ourselin S, et al. APOE ɛ4 is associated with disproportionate progressive hippocampal atrophy in AD. PLoS One 2014; 9: e97608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathis CA, Kuller LH, Klunk WE, Snitz BE, Price JC, Weissfeld LA, et al. In vivo assessment of amyloid-β deposition in nondemented very elderly subjects. Ann Neurol 2013; 73: 751–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mintun MA, Larossa GN, Sheline YI, Dence CS, Lee SY, Mach RH, et al. [11C]PIB in a nondemented population: potential antecedent marker of Alzheimer disease. Neurology 2006; 67: 446–52. [DOI] [PubMed] [Google Scholar]
- Mishra S, Gordon BA, Su Y, Christensen J, Friedrichsen K, Jackson K, et al. AV-1451 PET imaging of tau pathology in preclinical Alzheimer disease: defining a summary measure. Neuroimage 2017; 161: 171–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffat SD, Szekely CA, Zonderman AB, Kabani NJ, Resnick SM. Longitudinal change in hippocampal volume as a function of apolipoprotein E genotype. Neurology 2000; 55: 134–6. [DOI] [PubMed] [Google Scholar]
- Mori E, Lee K, Yasuda M, Hashimoto M, Kazui H, Hirono N, et al. Accelerated hippocampal atrophy in Alzheimer’s disease with apolipoprotein E epsilon4 allele. Ann Neurol 2002; 51: 209–14. [DOI] [PubMed] [Google Scholar]
- Morra JH, Tu Z, Apostolova LG, Green AE, Avedissian C, Madsen SK, et al. Automated mapping of hippocampal atrophy in 1-year repeat MRI data from 490 subjects with Alzheimer’s disease, mild cognitive impairment, and elderly controls. Neuroimage 2009; 45: S3–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris JC. Clinical dementia rating: a reliable and valid diagnostic and staging measure for dementia of the Alzheimer type. Int Psychogeriatr 1997; 9 (Suppl 1): 173–6; discussion 177–8. [DOI] [PubMed] [Google Scholar]
- Morris JC, Roe CM, Xiong C, Fagan AM, Goate AM, Holtzman DM, et al. APOE predicts amyloid-beta but not tau Alzheimer pathology in cognitively normal aging. Ann Neurol 2010; 67: 122–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy KR, Landau SM, Choudhury KR, Hostage CA, Shpanskaya KS, Sair HI, et al. Mapping the effects of ApoE4, age and cognitive status on 18F-florbetapir PET measured regional cortical patterns of beta-amyloid density and growth. Neuroimage 2013; 78: 474–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiman EM, Caselli RJ, Yun LS, Chen K, Bandy D, Minoshima S, et al. Preclinical evidence of alzheimer’s disease in persons homozygous for the. N Engl J Med 1996; 334: 752–8. [DOI] [PubMed] [Google Scholar]
- Reiman EM, Chen K, Liu X, Bandy D, Yu M, Lee W, et al. Fibrillar amyloid-beta burden in cognitively normal people at 3 levels of genetic risk for Alzheimer’s disease. Proc Natl Acad Sci USA 2009; 106: 6820–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiman EM, Langbaum JBS, Fleisher AS, Caselli RJ, Chen K, Ayutyanont N, et al. Alzheimer’s prevention initiative: a plan to accelerate the evaluation of presymptomatic treatments. J Alzheimers Dis 2011; 26 (Suppl 3): 321–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiman EM, Uecker A, Caselli RJ, Lewis S, Bandy D, De Leon MJ, et al. Hippocampal volumes in cognitively normal persons at genetic risk for Alzheimer’s disease. Ann Neurol 1998; 44: 288–91. [DOI] [PubMed] [Google Scholar]
- Resnick SM, Bilgel M, Moghekar A, An Y, Cai Q, Wnag MC, et al. Changes in Ab biomarkers and assocications with APOE genotype in two longitudinal cohorts. Neurobiol Aging 2015; 36: 2333–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risacher SL, Kim S, Nho K, Foroud T, Shen L, Petersen RC, et al. APOE effect on Alzheimer’s disease biomarkers in older adults with significant memory concern. Alzheimers Dement 2015; 11: 1417–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risacher SL, Shen L, West JD, Kim S, McDonald BC, Beckett LA, et al. Longitudinal MRI atrophy biomarkers: relationship to conversion in the ADNI cohort. Neurobiol Aging 2010; 31: 1401–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigue KM, Kennedy KM, Devous MD, Rieck JR, Hebrank AC, Diaz-Arrastia R, et al. β-Amyloid burden in healthy aging: regional distribution and cognitive consequences. Neurology 2012; 78: 387–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousset OG, Ma Y, Evans AC. Correction for partial volume effects in PET: principle and validation. J Nucl Med 1998; 39: 904–11. [PubMed] [Google Scholar]
- Roussotte FF, Gutman BA, Madsen SK, Colby JB, Narr KL, Thompson PM. The apolipoprotein E epsilon 4 allele is associated with ventricular expansion rate and surface morphology in dementia and normal aging. Neurobiol Aging 2014; 35: 1309–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe CC, Ellis KA, Rimajova M, Bourgeat P, Pike KE, Jones G, et al. Amyloid imaging results from the Australian Imaging, Biomarkers and Lifestyle (AIBL) study of aging. Neurobiol Aging 2010; 31: 1275–83. [DOI] [PubMed] [Google Scholar]
- Saunders AM, Strittmatter WJ, Schmechel D, George-Hyslop PH, Pericak-Vance MA, Joo SH, et al. Association of apolipoprotein E allele 4 with late-onset familial and sporadic Alzheimer’s disease. Neurology 1993; 43: 1467–72. [DOI] [PubMed] [Google Scholar]
- Scheinin NM, Wikman K, Jula A, Perola M, Vahlberg T, Rokka J, et al. Cortical 11C-PIB uptake is associated with age, APOE genotype, and gender in ‘healthy aging’. J Alzheimers Dis 2014; 41: 193–202. [DOI] [PubMed] [Google Scholar]
- Schmechel DE, Saunders AM, Strittmatter WJ, Crain BJ, Hulette CM, Joo SH, et al. Increased amyloid beta-peptide deposition in cerebral cortex as a consequence of apolipoprotein E genotype in late-onset Alzheimer disease. Proc Natl Acad Sci USA 1993; 90: 9649–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuff N, Woerner N, Boreta L, Kornfield T, Shaw LM, Trojanowski JQ, et al. MRI of hippocampal volume loss in early Alzheimer’s disease in relation to ApoE genotype and biomarkers. Brain 2009; 132: 1067–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soininen H, Partanen K, Pitkanen A, Hallikainen M, Tanninen T, Helisalmi S, et al. Decreased hippocampal volume asymmetry on MRIs in nondemented elderly subjects carrying the apolipoprotein E e4 allele. Neurology 1995; 45: 391–2. [DOI] [PubMed] [Google Scholar]
- Sorensen T, Vasishth S. Bayesian linear mixed models using Stan: a tutorial for psychologists, linguists, and cognitive scientists. Tutor Quant Methods Psychol 2016; 12: 175–200. [Google Scholar]
- Sperling RA, Rentz DM, Johnson KA, Karlawish J, Donohue M, Salmon DP, et al. The A4 study: stopping AD before symptoms begin? Sci Transl Med Med 2014; 6: 228fs13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strittmatter WJ, Saunders AM, Schmechel D, Pericak-Vance M, Enghild J, Salvesen GS, et al. Apolipoprotein E: high-avidity binding to beta-amyloid and increased frequency of type 4 allele in late-onset familial Alzheimer disease. Proc Natl Acad Sci USA 1993; 90: 1977–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Y, Blazey TM, Snyder AZ, Raichle ME, Marcus DS, Ances BM, et al. Partial volume correction in quantitative amyloid imaging. Neuroimage 2015; 107: 55–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Y, D’Angelo GM, Vlassenko AG, Zhou G, Snyder AZ, Marcus DS, et al. Quantitative analysis of PiB-PET with FreeSurfer ROIs. PLoS One 2013; 8: e73377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunderland T, Mirza N, Putnam KT, Linker G, Bhupali D, Durham R, et al. Cerebrospinal fluid β-amyloid1–42 and tau in control subjects at risk for Alzheimer’s disease: the effect of APOE ɛ4 allele. Biol Psychiatry 2004; 56: 670–6. [DOI] [PubMed] [Google Scholar]
- Sutphen CL, Jasielec MS, Shah AR, Macy EM, Xiong C, Vlassenko AG, et al. Longitudinal cerebrospinal fluid biomarker changes in preclinical Alzheimer disease during middle age. JAMA Neurol 2015; 72: 1029–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbot C, Lendon C, Craddock N, Shears S, Morris JC, Goate A. Protection against Alzheimer’s disease with apoe e2. Lancet 1994; 343: 1432–3. [DOI] [PubMed] [Google Scholar]
- Taylor JL, Scanlon BK, Farrell M, Hernandez B, Adamson MM, Ashford JW, et al. Neurobiology of aging APOE-epsilon4 and aging of medial temporal lobe gray matter in healthy adults older than 50 years. Neurobiol Aging 2014; 35: 2479–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tosun D, Schuff N, Shaw LM, Trojanowski JQ, Weiner MW. Relationship between CSF biomarkers of Alzheimer’s disease and rates of regional cortical thinning in ADNI data. Adv Alzheimers Dis 2011; 2: 127–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van De Pol LA, Van Der Flier WM, Korf ESC, Fox NC, Barkhof F, Scheltens P. Baseline predictors of rates of hippocampal atrophy in mild cognitive impairment. Neurology 2007; 69: 1491–7. [DOI] [PubMed] [Google Scholar]
- Vemuri P, Wiste HJ, Weigand SD, Knopman DS, Shaw LM, Trojanowski JQ, et al. Effect of apolipoprotein E on biomarkers of amyloid load and neuronal pathology in Alzheimer disease. Ann Neurol 2010; 67: 308–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villemagne VL, Burnham S, Bourgeat P, Brown B, Ellis KA, Salvado O, et al. Amyloid β deposition, neurodegeneration, and cognitive decline in sporadic Alzheimer’s disease: a prospective cohort study. Lancet Neurol 2013; 12: 357–67. [DOI] [PubMed] [Google Scholar]
- Villemagne VL, Pike KE, Chételat G, Ellis KA, Mulligan RS, Bourgeat P, et al. Longitudinal assessment of Aβ and cognition in aging and Alzheimer disease. Ann Neurol 2011; 69: 181–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villemagne VL, Rowe CC. Long night’s journey into the day: amyloid-beta imaging in Alzheimer’s disease. J Alzheimers Dis 2013; 33: S349–59. [DOI] [PubMed] [Google Scholar]
- Vlassenko AG, McCue L, Jasielec MS, Su Y, Gordon BA, Xiong C, et al. Imaging and cerebrospinal fluid biomarkers in early preclinical alzheimer disease. Ann Neurol 2016; 80: 379–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlassenko AG, Mintun MA, Xiong C, Sheline YI, Goate AM, Benzinger TLS, et al. Amyloid-beta plaque growth in cognitively normal adults: longitudinal [11C]Pittsburgh compound B data. Ann Neurol 2011; 70: 857–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.