Abstract
Liver disease is now a major cause of morbidity and mortality among persons infected with the human immunodeficiency virus (HIV). An increasing body of evidence suggests that HIV infection is associated with exacerbated liver fibrosis and that HIV has the ability to infect several hepatic cell types. Despite the recognized existence of genetically distinct subpopulations of HIV in the central nervous system and genital tract, viral diversity and compartmentalization in the liver have not been explored extensively. Therefore, phylogenetic analysis was performed on full-length env and nef sequences for four patients. Distinct clustering of viral variants was observed for all patients in both areas of the genome. Statistical evidence of HIV compartmentalization in the liver was demonstrated in 85.4% of comparisons. Signature sequence analysis identified several liver-specific amino acids in all patients. Thus, the current study demonstrates statistically significant evidence for HIV compartmentalization in the liver. Additionally, these data suggest that the hepatic microenvironment harbors unique selective pressures that drive viral adaptation.
Introduction
Chronic liver disease is a major cause of non-AIDS-related mortality in HIV-infected individuals.1–3 This can partially be attributed to coinfection with hepatitis C virus (HCV) and HIV, which share similar transmission routes. An estimated seven million people are HIV/HCV coinfected worldwide.4 Coinfection is associated with acceleration of liver fibrosis and cirrhosis, elevated HCV RNA levels, and decreased HCV treatment response rates compared to HCV monoinfected individuals.5 Additionally, HIV infection—with and without viral hepatitis coinfection—is associated with increased liver enzyme levels and markers for liver fibrosis.6,7 Increasing evidence indicates a direct correlation between HIV RNA levels and liver fibrosis in persons infected with HIV, even in the absence of viral hepatitis, antiretroviral therapy (ART), and alcohol use.8,9 A number of other clinical presentations of liver disease are associated with HIV monoinfection, including noncirrhotic portal hypertension and nonalcoholic fatty liver disease.10 These studies suggest that HIV itself may play a role in liver disease, despite other potential confounders.
Emerging data suggest that HIV has the ability to infect cells of the liver, including endothelial sinusoidal cells, stellate cells, Kupffer cells, and hepatocytes.11–16 Collectively, these data indicate that HIV is present in several intrahost microenvironments, each harboring its own unique selective pressures, thus contributing to the extensive sequence diversity of HIV. However, to our knowledge, HIV diversity in the liver has been explored in only one previous study.17 Therefore, the current analysis utilized complementary phylogenetic and statistical approaches to examine intrapatient HIV variability and compartmentalization in the liver.
Materials and Methods
All sequence data from patients HIVAms198, AM, AZ, and DY were previously published without phylogenetic or statistical evaluation of viral compartmentalization.18,19 All tissue samples were obtained postmortem from patients who died of AIDS-related causes. For the current analysis of viral compartmentalization, alignments were performed using the neighbor-joining (NJ) approach as implemented in Clustal X2.20 The statistical robustness and reliability of the branching order within each phylogenetic tree were confirmed by bootstrap analysis using 1,000 replicates.21 The Viral Epidemiology Signature Pattern Analysis (VESPA) program was used to determine the frequency of each amino acid in liver-derived viral variants versus viral variants from other tissues for each individual (www.hiv.lanl.gov/content/sequence/VESPA/vespa.html).22 Only amino acid signatures above an 80% threshold were considered significant. All nucleotide sequences were translated to amino acids using Molecular Evolutionary Genetics Analysis software version 5 (MEGA5).23 Compartmentalization of viral variants from each tissue type was assessed using Mantel's test as previously described for HIV.24,25
Results
Phylogenetic analyses were performed on full-length HIV gp120 and nef sequences for patients AM, AZ, and DY, and on V3 loop sequences for patient HIVAms198. A single intrapatient neighbor-joining tree was compiled including sequence data from all tissues for subject HIVAms198 (Fig. 1A). Interestingly, liver-derived sequences formed a single monophyletic cluster that contained no non-liver-derived sequences. Peripheral blood mononuclear cell (PBMC)-derived sequences were interspersed throughout the phylogenetic tree but were not closely related to the liver sequences, suggesting that liver-derived sequences were not the result of peripheral lymphocyte infiltration. Neighbor-joining phylogenetic trees including sequence data from all tissues were created for HIV gp120 and nef sequences for patients AZ and DY (Figs. 1B and C and 3A and B). Due to tree size constraints, phylogenetic trees for HIV gp120 and nef sequences from patient AM are shown as paired comparisons of the liver and one other tissue (Figs. 2 and 4). Distinct clustering of liver variants was observed in patients AM, AZ, and DY as well. Evidence of viral compartmentalization was supported further by Mantel's tests results, with a p-value<0.05 for 41 of 48 comparisons involving the liver (85.4%) (Table 1).
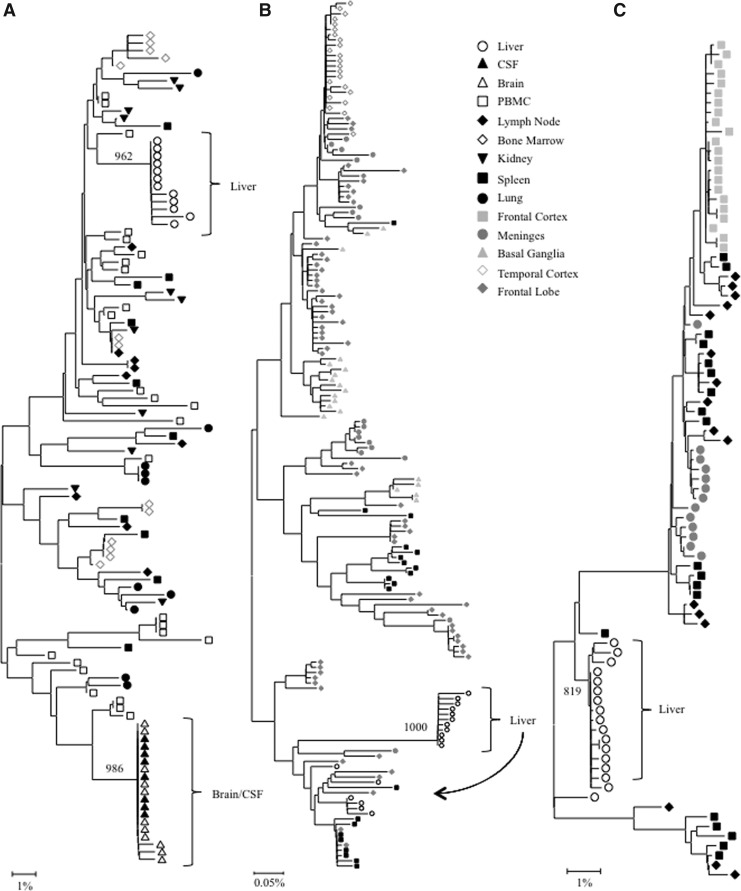
FIG. 1.
Intrapatient phylogenetic trees of (A) V3 loop variants from patient HIVams198, (B) gp120 variants from patient DY, and (C) gp120 variants from patient AZ. Shown in the lower left corner is a bar depicting the percent genetic distance for each tree. Compartmentalization is observed only in the liver and brain tissues. PBMC, peripheral blood mononuclear cells; CSF, cerebrospinal fluid. Only relevant bootstrap values greater than 700 out of 1,000 are shown. Sequence data for patient HIVams198 were originally reported in Van't Wout et al.19 Sequence data for patients AZ and DY were originally reported in Lamers et al.18
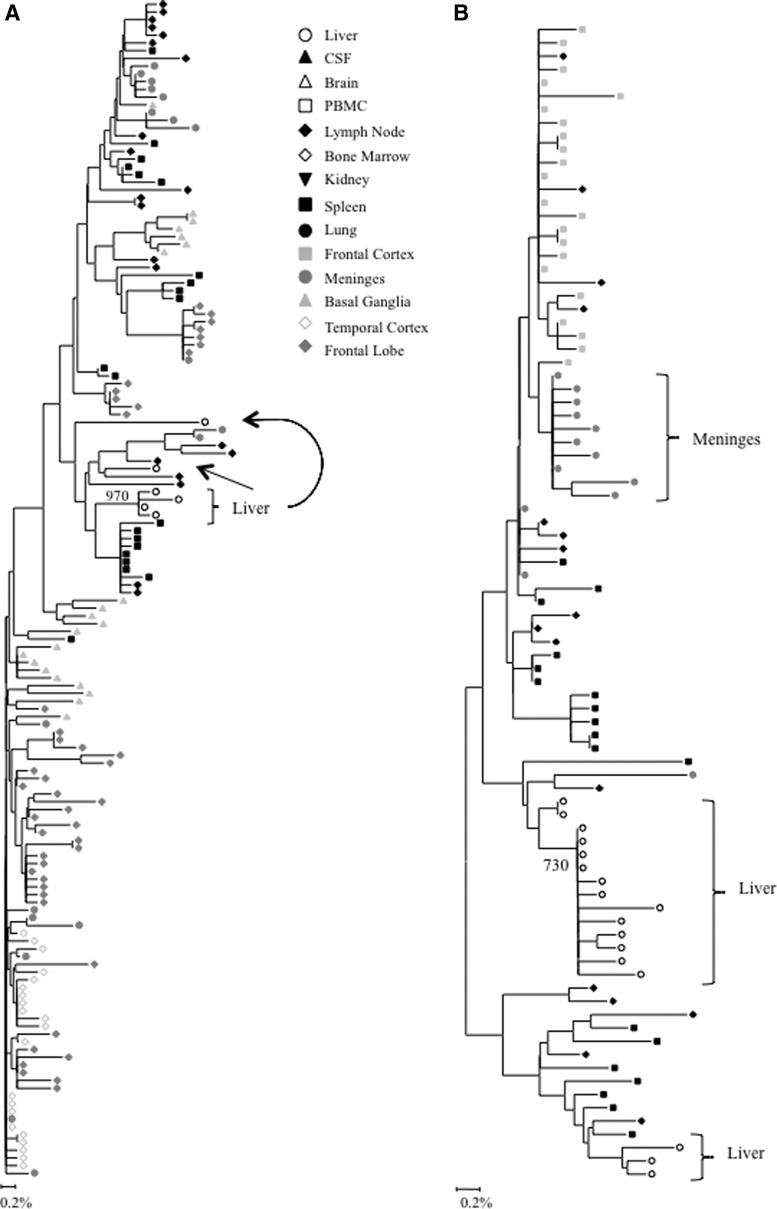
FIG. 3.
Intrapatient phylogenetic trees of (A) nef variants from patient DY and (B) nef variants from patient AZ. Shown in the lower left corner is a bar depicting the percent genetic distance for each tree. Only relevant bootstrap values greater than 700 out of 1,000 are shown. Sequence data were originally reported in Lamers et al.18
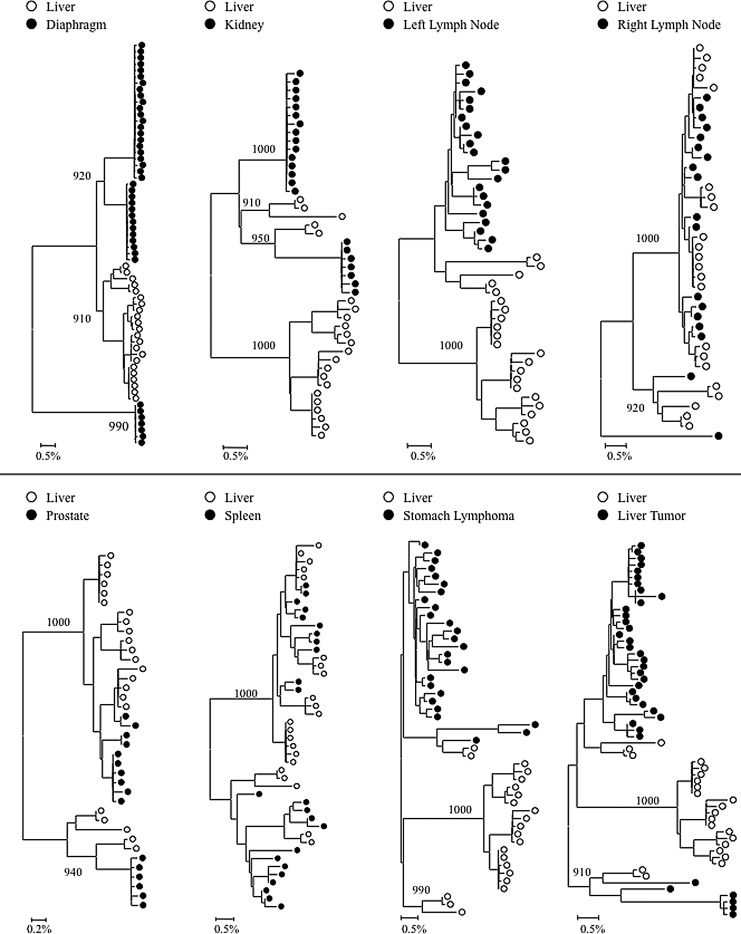
FIG. 2.
Intrapatient phylogenetic trees of gp120 variants from patient AM. Shown in the lower left corner is a bar depicting the percent genetic distance for each tree. Only relevant bootstrap values greater than 700 out of 1,000 are shown. Sequence data were originally reported in Lamers et al.18
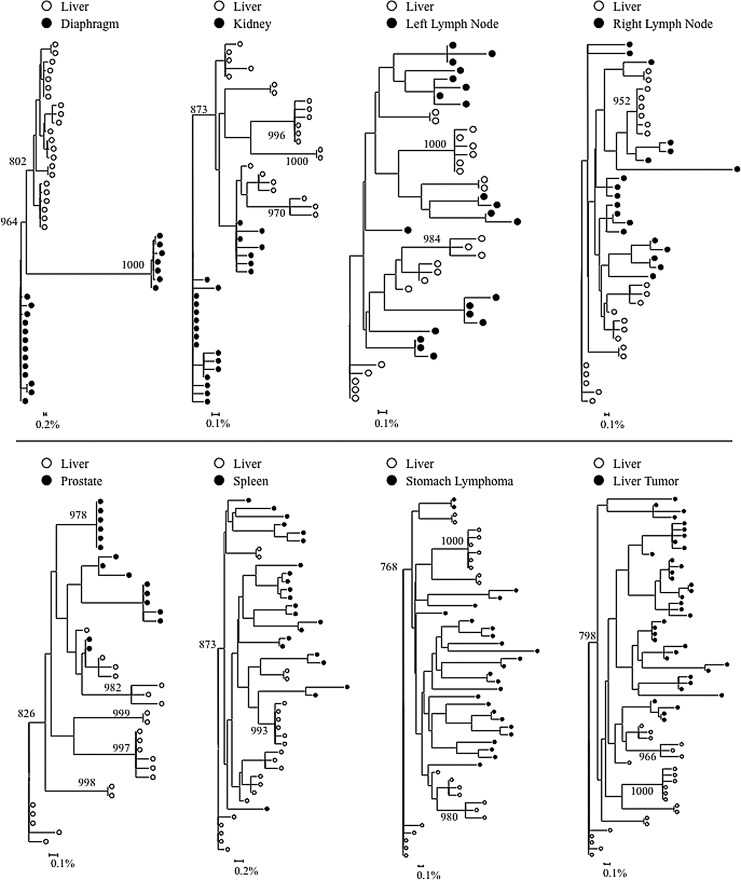
FIG. 4.
Intrapatient phylogenetic trees of nef variants from patient AM. Shown in the lower left corner is a bar depicting the percent genetic distance for each tree. Only relevant bootstrap values greater than 700 out of 1,000 are shown. Sequence data were originally reported in Lamers et al.18
Table 1.
Mantel's Test for Compartmentalization of V3 Loop, gp120, and nef Variants Compared to the Liver
| V3 loop | gp120 | nef | |||||
|---|---|---|---|---|---|---|---|
| Tissue being compared to liver | HIVams198 | DY | AZ | AM | DY | AZ | AM |
| Basal ganglia | — | 0.0001 | — | — | 0.0002 | — | — |
| Bone marrow | 0.0002 | — | — | — | — | — | — |
| Brain (unspecified) | 0.0001 | — | — | — | — | — | — |
| CSF | 0.0001 | — | — | — | — | — | — |
| Diaphragm | — | — | — | 0.0062 | — | — | 0.0001 |
| Frontal cortex | — | — | 0.0001 | — | — | 0.0001 | — |
| Frontal lobe GM | — | 0.0001 | — | — | 0.0002 | — | — |
| Frontal lobe WM | — | 0.0001 | — | — | 0.0001 | — | — |
| Kidney | 0.0014 | — | — | 0.0001 | — | — | 0.0091 |
| Lung | 0.0001 | — | — | — | — | — | — |
| Lymph node | 0.0002 | 0.0002 | 0.0001 | — | NS | 0.0001 | — |
| Left lymph node | — | — | — | 0.0001 | — | — | 0.0108 |
| Right lymph node | — | — | — | NS | — | — | NS |
| Liver tumor 1 | — | — | — | 0.0001 | — | — | 0.0027 |
| Liver tumor 2 | — | — | — | 0.0001 | — | — | 0.0009 |
| Meninges | — | 0.0001 | 0.0001 | — | 0.0010 | 0.0001 | — |
| PBMC | 0.0050 | — | — | — | — | — | — |
| Prostate | — | — | — | NS | — | — | 0.0011 |
| Spleen | 0.0017 | 0.0001 | 0.0012 | 0.0071 | 0.0046 | 0.0042 | NS |
| Stomach lymphoma | — | — | — | 0.0001 | — | — | NS |
| Temporal cortex | — | 0.0001 | — | — | 0.0001 | — | — |
Sequence data for patient HIVams198 were originally reported by Van't Wout et al.19 Sequence data for patients AM, AZ, and DY were originally reported in Lamers et al.18
CSF, cerebrospinal fluid; PBMC, primary blood mononuclear cells; GM, gray matter; WM, white matter; NS, not significant; dash, no data available; p-values<0.05 were considered statistically significant.
Analysis of V3 loop sequences
Phylogenetic analysis demonstrated that V3 loop sequences from the brain and cerebral spinal fluid (CSF) exhibited distinct phylogenetic clustering. Likewise, V3 loop variants from the liver formed a distinct monophyletic group supported by a high bootstrap value (962 out of 1,000). No further compartmentalization was observed for any V3 loop sequences from the kidney, lung, lymph node, bone marrow, spleen, and PBMCs (Fig. 1A). Thus, HIV quasispecies compartmentalization was observed in the liver and brain/CSF only for patient HIVAms198. Evidence for significant V3 loop compartmentalization was further supported by Mantel's test, in which all pairwise genetic distance comparisons of V3 loop sequences involving the liver had a p-value<0.05 (Table 1). Signature sequence analysis identified specific amino acid differences between V3 loop variants from the liver and all other tissues (Supplementary Table S1; Supplementary Data are available online at www.liebertpub.com/aid). The frequency of amino acid differences ranged from 2 in the liver–spleen comparison to 4 in liver–bone marrow, liver–brain, liver–lung, and liver–lymph node comparisons (mean=3.4) (Supplementary Table S1). The most notable residue change occurred in the conserved GPGR tip of the V3 loop, where a single mutation (R315K) was found only in the liver, brain, and CSF variants (Supplementary Table S1). Two other possible function-altering mutations that serve as coreceptor binding sites were found in the V3 loop—I309M in the brain and N325D in the brain, CSF, and lung.26
Analysis of gp120 sequences
Intrapatient phylogenetic trees including gp120 sequences from all available tissues were created for patients DY and AZ (Fig. 1B and C). Both patients exhibited distinct phylogenetic clustering of gp120 variants from the liver supported by high bootstrap values (1,000 for DY and 819 for AZ). Similar clustering was observed with temporal cortex variants from patient DY and frontal cortex variants from patient AZ. Phylogenetic trees containing paired comparisons involving the liver were created for patient AM. Viral variants from the liver and diaphragm were particularly distinct from one another, as complete separation between these tissues was observed (Fig. 2). All pairings, except liver–right lymph node and liver–spleen showed evidence of compartmentalization supported by high bootstrap values (Fig. 2). Despite the lack of visible phylogenetic separation between the liver and spleen variants from patient AM, Mantel's test comparing these two tissues was statistically significant (p=0.0071) (Table 1). In the analysis of gp120 sequences, Mantel's test confirmed significant evidence of compartmentalization for 17 of 20 comparisons involving the liver (85%) (Table 1). Signature sequence analysis highlighted multiple amino acid differences in HIV gp120 from the liver versus all other tissues for patient DY (range=21–41, mean=30.3), AZ (range=42–46, mean=43.3), and AM (range=2–50, mean=29.3) (Supplementary Tables S2–4). Liver sequence divergence was most apparent in patient AZ, as 42 of 48 total residue differences between liver-derived variants and variants derived from other tissues were found in all three paired comparisons. The liver–diaphragm comparison from patient AM had 50 amino acid substitutions, the highest of all paired comparisons (Supplementary Table S4). Several amino acid differences between gp120 variants from the liver and other tissues occurred in functional areas, including the LDI/LDV tripeptide in the V2 loop, several CD4 contact residues and coreceptor binding sites, the GPGR tip of the V3 loop, the CD4 binding loop, a lectin DC-SIGN binding site, monoclonal antibody glycosylation binding sites, and gp120 contact residues with gp41.27–30
Analysis of nef sequences
Complete or near complete separation of nef variants from the liver was observed in phylogenetic analyses of sequences from patients DY and AZ (Fig. 3). The statistical robustness of liver variant branching order was supported by high bootstrap values (970 for DY and 730 for AM). Distinct phylogenetic clustering was also demonstrated by nef variants from the meninges of patient AZ. No further phylogenetic evidence of compartmentalization was observed for any other tissue evaluated from patients DY and AZ (Fig. 3). Neighbor-joining trees were completed for paired comparisons involving the liver for patient AM (Fig. 4). Complete or near complete separation of viral variants was observed for liver–diaphragm, liver–kidney, liver–prostate, and liver–liver tumor comparisons. Although there was no visible phylogenetic separation of variants between the liver and left lymph node variants from patient AM, Mantel's test suggested that sequences from the liver shared more genetic identity with each other than with sequences from the left lymph node (p=0.0108) (Table 1). Furthermore, Mantel's test provided statistically significant evidence of viral compartmentalization for 16 of 20 HIV nef comparisons involving the liver (80 %) (Table 1). Signature sequence analysis of nef identified multiple amino acid substitutions between liver-derived variants and variants from all other tissues for patients DY (range=2–5, mean=3.1), AZ (range=1–2, mean=1.5), and AM (range=1–8, mean=2) (Supplementary Table S5A–C). The two comparisons with the highest frequency of amino acid differences were the liver–diaphragm comparison from patient AM (eight substitutions) and the liver–frontal lobe white matter comparison from patient DY (five substitutions). Several residue changes occurred in the nef core domain structure and the conserved SH3 binding pocket.31
Discussion
A very limited number of studies have examined HIV variability in the liver. Van't Wout et al. performed a phylogenetic analysis on HIV proviral DNA from multiple tissues, including the liver, of one patient who died of AIDS-related causes and observed that HIV variants from the liver clearly clustered separately from variants of all other tissues analyzed. As well, there was no phylogenetic clustering observed between PBMC-derived and liver-derived HIV sequences, suggesting that HIV compartmentalization in the liver was not due to infiltrating lymphocytes. Despite this, the authors concluded that the monophyletic grouping of liver variants was the result of low proviral copy numbers, as all liver sequences were generated from a single PCR.19 Only one other study to date has predicted liver-specific amino acid variations in env.17 However, that study included only partial env sequences and, therefore, was unable to identify liver-specific signature sequences outside of the region amplified. Thus, in the current study, we sought to confirm these findings in an independent dataset, analyze the entire env gene, and examine compartmentalization of HIV nef sequences in the liver. Our results demonstrate that phylogenetically distinct HIV variants from the liver exist in three other patients who have died of AIDS-related causes. These findings were further supported statistically by Mantel's test. Collectively, these data suggest that viral compartmentalization may occur more frequently in the liver than previously recognized.
Signature sequence analysis identified specific amino acids in both gp120 and nef that were unique to the liver. While a bioinformatics approach cannot specifically determine if the predicted amino acid variations are relevant in the context of a replicating virus, several of the predicted amino acid variations were identified only in liver-derived HIV sequences, thus strengthening the body of evidence that suggests viral compartmentalization in the liver. Importantly, many amino acid substitutions were found in conserved areas of gp120. This suggests that they did not simply occur as a result of random sequence variability; rather, there may be unique selective pressures acting on HIV in the liver that are not found elsewhere in the body. For example, CD4-independent HIV interaction with cells is known to occur through gp120 binding of C-type lectin receptors, such as the mannose receptor or the DC-SIGN receptor on macrophages and dendritic cells.32,33 Indeed, even minor mutations in gp120 can drastically alter the requirement for entry receptors and cell tropism. Such mutations may be the result of strong selective pressures on HIV resulting in liver-specific viral variants.
Compartmentalization can also occur without adaptive mutation, when the selective pressures in separate organs differ such that the fitness of a particular preexisting viral subpopulation is greater in one organ than another. The predicted amino acid variations discovered in this study may have arisen from adaptive mutations or selective pressures on an existent viral subpopulation, as both mechanisms can result in viral compartmentalization. The lack of in vitro analysis in the current study does not allow for the determination of specific cell types that may support HIV replication in the liver. However, compelling evidence indicates that HIV has the ability to infect endothelial sinusoidal cells, stellate cells, Kupffer cells, and hepatocytes.11–16 Studies also demonstrate that the gp120 antigen may engage apoptotic pathways without direct viral infection.34 Both direct infection of liver cells and antigen activation of liver cells may contribute to viral compartmentalization in the liver. Future in vitro studies are warranted to determine the exact mechanisms by which HIV interacts with specific liver cell types and to confirm the role of organ-specific signature sequences.
Recent data suggest a role for HIV in liver disease.35,36 This study provides evidence of HIV compartmentalization in the liver and identifies signature amino acids that may characterize liver-specific quasispecies. These data strongly suggest that HIV variants differ in their ability to infect various cell types. Furthermore, HIV mutation may be required for attachment, entry, and replication within a given cell type. Thus, these investigations may provide additional insight into the immune response and selective pressures unique to the hepatic microenvironment.
There are several limitations to the current analysis including the small number of patients, the lack of sequence data outside the env and nef genes, and limited information of whether other comorbid conditions may impact HIV diversity and/or compartmentalization. As noted in our previous study,17 cross-sectional analyses do not permit a detailed examination of liver-specific HIV variants over time. Similarly, the cloning strategy utilized—while robust–may not have amplified all minor variants present in a given tissue/cell type. Thus, additional studies are now warranted in larger patient populations at varying stages of liver disease progression. It is recommended that future studies concerning HIV compartmentalization in the liver compare viral variants from (1) HIV monoinfected individuals with no history of liver complication, (2) HIV monoinfected individuals with liver disease, and (3) HIV/HCV coinfected individuals. However, it should be noted that due to the rapid adaptation of HIV to its host, organ-specific (including liver-specific) signature sequences may differ between patients. The phenotypic implications of liver-specific HIV mutations should be further examined, as they may aid in the development of liver-specific treatment modalities.
Supplementary Material
Acknowledgments
The authors would like to thank Ms. Eleanor Powell for her review of this manuscript and the University Honors Program for funding this summer undergraduate research project and for facilitating interactions between College of Medicine faculty and undergraduate students.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Bica I, McGovern B, Dhar R, et al.: Increasing mortality due to end-stage liver disease in patients with human immunodeficiency virus infection. Clin Infect Dis 2001;32:492–497 [DOI] [PubMed] [Google Scholar]
- 2.Smith C, Sabin CA, Lundgren JD, et al.: Factors associated with specific causes of death amongst HIV-positive individuals in the D:A:D Study. AIDS 2010;24(10):1537–1548 [DOI] [PubMed] [Google Scholar]
- 3.Tedaldi E, Baker R, Moorman A, et al.: Influence of coinfection with hepatitis C virus on morbidity and mortality due to human immunodeficiency virus infection in the era of highly active antiretroviral therapy. Clin Infect Dis 2003;36:363–367 [DOI] [PubMed] [Google Scholar]
- 4.Soriano V, Barreiro P, and Sherman K: The changing epidemiology of liver disease in HIV patients. AIDS Rev 2013;15(1):25–31 [PubMed] [Google Scholar]
- 5.Kim AY. and Chung R: Coinfection with HIV-1 and HCV—a one-two punch. Gastroenterology 2009;137(3):795–814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Price JC, Seaberg EC, Badri S, Witt MD, D'Acunto K, and Thio C: HIV monoinfection is associated with increased aspartate aminotransferase-to-platelet ratio index, a surrogate marker for hepatic fibrosis. J Infect Dis 2012;205(6):1005–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sterling RK, Chiu S, Snider K, and Nixon D: The prevalence and risk factors for abnormal liver enzymes in HIV-positive patients without hepatitis B or C coinfections. Digest Dis Sci 2007;53(5):1375–1382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blackard JT, Welge JA, Taylor LE, et al.: HIV mono-infection is associated with FIB-4–a noninvasive index of liver fibrosis–in women. Clin Infect Dis 2011;52(5):674–680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Forrester JE, Rhee MS, McGovern BH, Sterling RK, Knox TA, and Terrin N: The association of HIV viral load with indirect markers of liver injury. J Viral Hepatitis 2012;19(2):202–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Crane M, Iser D, and Lewin S: Human immunodeficiency virus infection and the liver. World J Hepatol 2012;4(3):91–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cao YZ, Friedman-Kein AE, Huang YX, et al.: CD4-independent, productive human immunodeficiency virus type 1 infection of hepatoma cell lines in vitro. J Virol 1990;64(6):2553–2559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Housset C, Lamas E, Courgnaud V, et al.: Presence of HIV-1 in human parenchymal and non-parenchymal liver cells in vivo. J Hepatol 1993;19(2):252–256 [DOI] [PubMed] [Google Scholar]
- 13.Hufert FT, Schmitz J, Schreiber M, Schmitz H, Racz P, and Laer Dv: Human Kupffer cels infected with HIV-1 in vivo. J Acquir Immune Defic Syndr 1993;6(7):772–777 [PubMed] [Google Scholar]
- 14.Kong L, Cardona Maya W, Moreno-Fernandez ME, et al.: Low-level HIV infection of hepatocytes. Virol J 2012;9(1):157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tuyama AC, Hong F, Saiman Y, et al.: Human immunodeficiency virus (HIV)-1 infects human hepatic stellate cells and promotes collagen I and monocyte chemoattractant protein-1 expression: Implications for the pathogenesis of HIV/hepatitis C virus-induced liver fibrosis. Hepatology 2010;52(2):612–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xiao P, Usami O, Suzuki Y, et al.: Characterization of a CD4-independent clinical HIV-1 that can efficiently infect human hepatocytes through chemokine (C-X-C motif) receptor 4. AIDS 2008;22(14):1749–1757 [DOI] [PubMed] [Google Scholar]
- 17.Blackard JT, Ma G, Martin CM, Rouster SD, Shata MT, and Sherman K: HIV variability in the liver and evidence of possible compartmentalization. AIDS Res Hum Retroviruses 2011;27(10):1117–1126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lamers SL, Salemi M, Galligan DC, et al.: Extensive HIV-1 intra-host recombination is common in tissues with abnormal histopathology. PLoS One 2009;4(3):e5065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van't Wout AB, Ran LJ, Kuiken CL, Kootstra NA, Pals ST, and Schuitemaker H: Analysis of the temporal relationship between human immunodeficiency virus type 1 quasispecies in sequential blood samples and various organs obtained at autopsy. J Virol 1998;72(1):488–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Larkin MA, Blackshields G, Brown NP, et al.: Clustal W and Clustal X version 2.0. Bioinformatics 2007;2007(23):21. [DOI] [PubMed] [Google Scholar]
- 21.Felsenstein J: Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985;39:783–791 [DOI] [PubMed] [Google Scholar]
- 22.Korber B. and Myers G: Signature pattern analysis: A method for assessing viral sequence relatedness. AIDS Res Hum Retroviruses 1992;8(9):1549–1560 [DOI] [PubMed] [Google Scholar]
- 23.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, and Kumar S: MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 2011;28(10):2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Collins KR, Quiñones-Mateu ME, Wu M, et al.: Human immunodeficiency virus type 1 (HIV-1) quasispecies at the sites of Mycobacterium tuberculosis infection contribute to systemic HIV-1 heterogeneity. J Virol 2002;76(4):1697–1706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Poss M, Rodrigo AG, Gosink JJ, et al.: Evolution of envelope sequences from the genital tract and peripheral blood of women infected with clade A human immunodeficiency virus type 1. J Virol 1998;72(10):8240–8451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Korber B. and Gnanakaran S: The implications of patterns in HIV diversity for neutralizing antibody induction and susceptibility. Curr Opin HIV AIDS 2009;4(5):408–417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Arthos J, Cicala C, Martinelli E, et al.: HIV-1 envelope protein binds to and signals through integrin alpha4beta7, the gut mucosal homing receptor for peripheral T cells. Nat Immunol 2008;9(3):301–309 [DOI] [PubMed] [Google Scholar]
- 28.Zhou T, Georgiev I, Wu X, et al.: Structural basis for broad and potent neutralization of HIV-1 by antibody VRC01. Science 2010;329(5993):811–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sanders RW, Venturi M, Schiffner L, et al.: The mannose-dependent epitope for neutralizing antibody 2G12 on human immunodeficiency virus type 1 glycoprotein gp120. J Virol 2002;76(14):7293–7305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hong PW, Nguyen S, Young S, Su SV, and Lee B: Identification of the optimal DC-SIGN binding site on human immunodeficiency virus type 1 gp120. J Virol 2007;81(15):8325–8336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lamers SL, Poon AF, and McGrath M: HIV-1 nef protein structures associated with brain infection and dementia pathogenesis. PLoS One 2011;6(2):e16659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nguyen DG. and Hildreth J: Involvement of macrophage mannose receptor in the binding and transmission of HIV by macrophages. Eur J Immunol 2003;33(2):483–493 [DOI] [PubMed] [Google Scholar]
- 33.Geijtenbeek T, Kwon DS, van Vliet SJ, et al.: DC-SIGN, a dendritic cell-specific HIV-1 binding protein that enhances trans-infection of T cells. Cell 2000;100(3):587–597 [DOI] [PubMed] [Google Scholar]
- 34.Popik W. and Pitha P: Exploitation of cellular signaling by HIV-1: Unwelcome guests with master keys that signal their entry. Virology 2000;276(1):1–6 [DOI] [PubMed] [Google Scholar]
- 35.Blackard JT. and Sherman K: HCV/ HIV co-infection: Time to re-evaluate the role of HIV in the liver? J Viral Hepatitis 2008;15(5):323–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bansal M. and Blackard J: Effects of HIV on liver cell populations. Springer Science+Business Media, New York, 2012 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.