ABSTRACT
Recent studies on the role of T cells in Zika virus (ZIKV) infection have shown that T cell responses to Asian ZIKV infection are important for protection, and that previous dengue virus (DENV) exposure amplifies the protective T cell response to Asian ZIKV. Human T cell responses to African ZIKV infection, however, remain unexplored. Here, we utilized the modified anthrax toxin delivery system to develop a flavivirus enzyme-linked immunosorbent spot (ELISPOT) assay. Using human ZIKV and DENV samples from Senegal, West Africa, our results demonstrate specific and cross-reactive T cell responses to nonstructural protein 3 (NS3). Specifically, we found that T cell responses to NS3 protease are ZIKV and DENV specific, but responses to NS3 helicase are cross-reactive. Sequential sample analyses revealed immune responses sustained many years after infection. These results have important implications for African ZIKV/DENV vaccine development, as well as for potential flavivirus diagnostics based on T cell responses.
IMPORTANCE The recent Zika virus (ZIKV) epidemic in Latin America and the associated congenital microcephaly and Guillain-Barré syndrome have raised questions as to why we have not recognized these distinct clinical diseases in Africa. The human immunologic response to ZIKV and related flaviviruses in Africa represents a research gap that may shed light on the mechanisms contributing to protection. The goal of our study was to develop an inexpensive assay to detect and characterize the T cell response to African ZIKV and DENV. Our data show long-term specific and cross-reactive human immune responses against African ZIKV and DENV, suggesting the usefulness of a diagnostic based on the T cell response. Additionally, we show that prior flavivirus exposure influences the magnitude of the T cell response. The identification of immune responses to African ZIKV and DENV is of relevance to vaccine development.
KEYWORDS: ZIKV, DENV, flavivirus, T cells, Senegal, West Africa, T cell immunity
INTRODUCTION
Dengue virus (DENV), serotypes 1 to 4, and Zika virus (ZIKV) belong to the family Flaviviridae (1). These viruses contain a single-stranded, positive-sense RNA genome encoding a polyprotein that is cleaved into 10 polypeptides: three structural proteins, capsid (C), premembrane (prM), and envelope (E), and seven nonstructural proteins (NS) proteins, NS1, NS2a, NS2b, NS3, NS4a, NS4b, and NS5 (2, 3).
More recently, regions where DENV is endemic have been experiencing ZIKV outbreaks. Discovered in Uganda in 1947, ZIKV had caused sporadic disease throughout Africa and Asia with limited clinical consequences (4–6). ZIKV remained obscure until the 2007 Micronesia outbreak, with subsequent larger epidemics in 2013 to 2014 and 2015 to 2016 in French Polynesia and Latin America, respectively (7–9). In April 2015, ZIKV was detected for the first time in the Western Hemisphere in Brazil, and by January 2016 reports of the virus had spread throughout the Americas and the Caribbean (10–12).
Phylogenetic analyses have distinguished African and Asian ZIKV lineages, with Asian ZIKV being responsible for the recent outbreaks (5, 13, 14). As with DENV, both Asian and African ZIKV are transmitted to humans by Aedes mosquitoes (15). Additional modes of transmission for Asian ZIKV, including sexual and maternal-fetal routes, have been shown (16, 17). Recent evidence also points to the capacity of Asian ZIKV to cause severe neuropathology, including disorders of fetal brain development and Guillain-Barré syndrome (11, 18, 19). Given the recent epidemic in Latin America with the previously unrecognized neuropathology in Africa, the distinctions between the pathogenesis of African and Asian ZIKV have taken on new significance.
The role of the immune system in DENV infection has been extensively investigated (20, 21). A number of recent studies have sought to understand the consequences of DENV and ZIKV interactions, with conflicting results. Some studies have suggested that previous DENV infection protects against ZIKV through DENV-induced cross-reactive neutralizing antibodies (22–25); in contrast, other studies have shown that cross-reactive antibodies enhance infection of ZIKV, supporting the antibody-dependent enhancement phenomenon (23, 26–28). There is a need to better understand the evolution and maintenance of immune responses to these related viruses in people.
While the role of T cells continues to be debated in DENV infection, recent work has found evidence of a protective role in ZIKV infection. In silico analysis has identified T cell reactivity conserved across all flaviviruses, including the ZIKV nonstructural proteins, namely, NS3 and NS5 (29). Studies in mice have identified immunodominant and protective roles for T cells that were either ZIKV specific or cross-reactive for DENV and ZIKV (30). One study involving ZIKV-infected humans found poorly cross-reactive memory T cells, even in individuals that were previously exposed to DENV (24). Another study demonstrated more rapid and stronger T cell responses to the structural proteins of Asian ZIKV in ZIKV-positive, DENV-negative individuals (31). To date, no study has examined the role human T cells play in African ZIKV and DENV infections.
The aforementioned T cell studies examined the role of T cells in ZIKV infection by peptide stimulation or a T cell library method. Bacterial toxins represent an alternative strategy for the delivery of antigen into the cytosol of host cells for major histocompatibility complex (MHC) processing, an essential step in T cell activation (32). A modified, nontoxic form of the Bacillus anthracis (anthrax) toxin has been shown to translocate protein antigens to both the MHC class I and class II pathways (33). This antigen delivery system exploits the exotoxin properties of anthrax. The toxin produced by anthrax is tripartite, composed of protective antigen (PA), edema factor (EF), and lethal factor (LF). Terminally truncated LF (LFn; lacking 255 amino acids) has been shown to be nontoxic and contain the information necessary to translocate into the cytosol any heterologous protein that can be stably fused to it (34). Proteins delivered by this system will be processed by either the MHC class I or class II pathway for T cell presentation.
Various studies have successfully employed the modified anthrax toxin delivery system to stimulate specific T cell responses. One study demonstrated in vitro generation of T cell epitopes against HIV-1 V3 by fusion of LFn to the Gp-120 portion of the HIV-1 envelope (35). The utility of LFn and its large coding capacity was demonstrated with its fusion to HIV-1 p24 and nef and their use in the induction of strong T cell responses in mice and humans (36). LFn-HIV-1-p24C and -gag vaccine candidates demonstrated the capacity to elicit cell-mediated immune responses in Chinese rhesus macaques (37). We previously developed an LFn enzyme-linked immunosorbent spot (ELISPOT) assay to assess HIV-2-specific T cell responses using peripheral blood mononuclear cells (PBMCs) from HIV-2-infected subjects to demonstrate increased anti-HIV-2 gag cellular responses compared to those of other T cell assay methodologies (38).
In this study, we applied the LFn ELISPOT assay to analyze T cell responses in African ZIKV and DENV infections. To assess the modified anthrax toxin antigen delivery system, we fused the subdomains, protease and helicase, of African ZIKV and DENV NS3 to LFn. The LFn-ZIKV and -DENV protease and helicase recombinant proteins were expressed and used as antigens to PBMCs isolated from human West African ZIKV and DENV infections in an ELISPOT assay. We report, for the first time, both specific and cross-reactive, long-term responses to African ZIKV and DENV.
RESULTS
Generation of LFn-ZIKV-protease and -helicase and LFn-DENV-protease and -helicase.
We aimed to define the T cell response to African ZIKV and DENV in samples previously collected from female sex workers (FSW) reporting to a local clinic in Dakar, Senegal, West Africa. We focused the design of the LFn fusion antigens on NS3, the conserved and preferential T cell target demonstrated in multiple flavivirus studies (39–41). Prior to the 2007 and 2009 DENV3 outbreaks, human infections in Senegal were primarily caused by DENV2 (42, 43). The PBMC samples used in this study were collected between 1992 and 2004; therefore, we fused the coding sequences corresponding to the NS3 subdomains, protease and helicase, of African ZIKV and DENV2 to LFn (Fig. 1A).
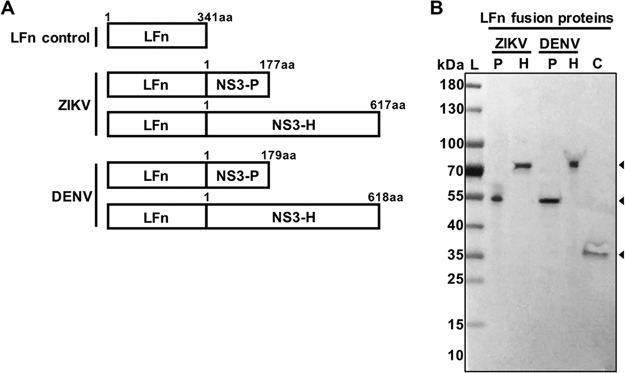
FIG 1.
(A) Schematic representation of the LFn control and LFn-ZIKV and -DENV fusion proteins. aa, amino acid. (B) Western blot analysis of the purified proteins LFn-ZIKV-NS3-protease (ZIKV P), LFn-ZIKV-NS3-helicase (ZIKV H), LFn-DENV-NS3-protease (DENV P), and LFn-DENV-NS3-helicase (DENV H) C, LFn control. Arrowheads indicate the size of the proteins. Molecular size marker units are kilodaltons.
One-step purification by affinity chromatography of LFn-ZIKV- and -DENV-NS3-protease (LFn-ZIKV- and -DENV-NS3-P; 56-kDa), LFn-ZIKV- and -DENV-NS3-helicase (LFn-ZIKV- and -DENV-NS3-H; 77-kDa), and the LFn control (37-kDa) produced soluble and stable proteins. The purified LFn fusion proteins were recognized by rabbit polyclonal antibodies to LFn (Fig. 1B).
Cohort characteristics.
We recently reported on the continued human transmission of African ZIKV in Senegal and Nigeria using samples collected from three different cohorts over a 25-year period; the presence of ZIKV in these locales had previously not been appreciated (44). Two hundred twenty-four plasma samples from the Senegalese HIV-1/2 female sex worker cohort were available for ZIKV serology, and of them, 118 samples were also available for DENV serology.
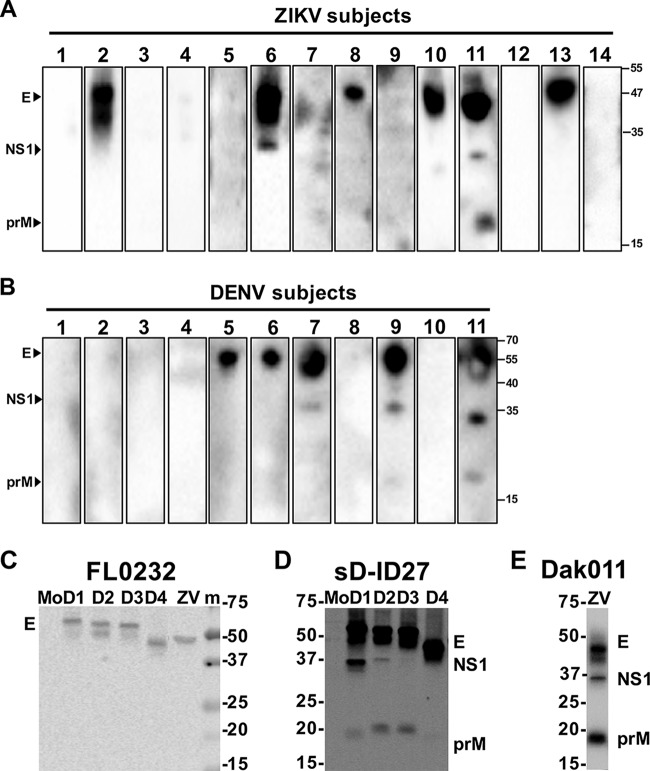
Characteristics associated with the samples included year of sample collection and patient age, temperature, and HIV serology (Table 1). Of 224 total plasma samples tested for ZIKV IgM antibodies, 14 tested positive (6.3%); all 14 ZIKV IgM-positive samples tested negative for DENV IgM. A total of 118 out of the 224 plasma samples were available for DENV serology. Of the plasma samples tested for DENV IgM antibodies, 11 tested positive (9.3); all 11 DENV IgM-positive samples tested negative for ZIKV IgM. In order to determine previous flavivirus exposure in the IgM-positive subjects, we assessed IgG antibody responses to prM, E, and/or NS1 by Western blotting in the most recent available plasma sample that preceded the ZIKV or DENV IgM-positive sample. Six out of 14 (42.8%) subjects who tested positive for ZIKV IgM and 5 out of 11 (45.5%) subjects who tested positive for DENV IgM demonstrated previous flavivirus exposure (Fig. 2A and B). Control Western blotting with a monoclonal antibody (MAb) or with confirmed ZIKV and DENV immune sera were performed on the ZIKV and DENV1-4 Vero cell lysates (Fig. 2C to E).
TABLE 1.
Cohort characteristics, ZIKV and DENV serology, and RT-PCR results
| Characteristic(s) and serology | Value(s) for Senegal FSW cohort (n = 224) |
|---|---|
| Sample date range | 1992–2004 |
| Age, yr, median (range) | 38 (21–66) |
| Avg temp, °C | 37.8 |
| HIV status [no. (%)] | |
| HIV negative | 148 (66.1) |
| HIV-1 positive | 40 (17.9) |
| HIV-2 positive | 25 (11.2) |
| HIV dual positive | 11 (4.8) |
| Serology and RT-PCR, no. positive/total no. tested (%) | |
| ZIKV | |
| ZIKV IgM | 14/224 (6.3) |
| ZIKV RT-PCR | 2/14 (14.3) |
| Prior flavivirus exposure | 6/14 (42.8) |
| DENV | |
| DENV IgM | 11/118 (9.3) |
| DENV RT-PCR | 0/11 (0) |
| Prior flavivirus exposure | 5/11 (45.5) |
FIG 2.
Antibody responses to different ZIKV or DENV proteins. Preinfection sera from ZIKV (A) or DENV (B) IgM-positive subjects were subjected to Western blot analysis using lysates derived from African ZIKV (unpublished strain)- or DENV2 (NGC strain)-infected Vero cells. (C) FL0232, a mouse anti-E monoclonal antibody recognizing similar flaviviral E protein levels, was used to standardize comparable amounts of E proteins from different virus-infected Vero cell lysates. (D) sD-ID27 is a confirmed case of secondary DENV1 infection at 3 months postinfection run on DENV1-4 Vero cell lysates. (E) Dak010 is a confirmed case of African ZIKV infection run on an African ZIKV Vero cell lysate. Arrowheads indicate PrM, E, and NS1 proteins recognized. Molecular size marker units are kilodaltons. Mo, mock-infected; m, molecular mass marker; D1, DENV1; D2, DENV2; D3, DENV3; D4, DENV4; ZV, ZIKV.
Logistic regression using sample year (1988 to 1999 and 2000 to 2004), rainy season (June to October), age, HIV serology, and body temperature as potential predictors found no association with ZIKV IgM positivity and increased odds of DENV IgM positivity with temperatures of ≥38°C only (odds ratio, 11.45 [95% confidence interval [CI], 2.98 to 44.10]; P < 0.001) (Table 2). ZIKV or DENV nucleic acid could not be successfully amplified by reverse transcription-PCR (RT-PCR) for any of the new samples that tested positive for ZIKV or DENV IgM.
TABLE 2.
Association between ZIKV or DENV IgM-positive results and patient characteristics
| Parameter | Frequencya | Odds ratio (95% CI) | P value |
|---|---|---|---|
| ZIKV IgM | |||
| Sample year | |||
| 1988–1999 | 4/51 (7.8) | Reference | |
| 2000–2004 | 10/173 (5.8) | 0.72 (0.22–2.40) | 0.594 |
| Season | |||
| Dry (Nov-May) | 8/137 (5.8) | Reference | |
| Rainy (Jun-Oct) | 6/87 (6.9) | 1.19 (0.40–3.57) | 0.750 |
| Temp, °C | |||
| 37.5–37.9 | 10/171 (5.9) | Reference | |
| 38.0–39.4 | 4/53 (7.6) | 1.31 (0.39–4.38) | 0.656 |
| Age, yr (range, 21–66) | 0.95 (0.88–1.02) | 0.143 | |
| HIV serology | |||
| Negative | 12/148 (8.1) | Reference | |
| Positive | 2/76 (2.6) | 0.31 (0.07–1.41) | 0.128 |
| DENV IgM | |||
| Sample yr | |||
| 1988–1999 | 5/51 (9.8) | Reference | |
| 2000–2004 | 7/173 (4.1) | 0.39 (0.12–1.28) | 0.120 |
| Season | |||
| Dry (Nov-May) | 9/137 (6.6) | Reference | |
| Rainy (Jun-Oct) | 3/87 (3.5) | 0.51 (0.13–1.93) | 0.320 |
| Temp, °C | |||
| 37.5–37.9 | 3/171 (1.8) | Reference | |
| 38.0–39.4 | 9/53 (17.0) | 11.45 (2.98–44.10) | <.001 |
| Age, yr (range, 21–66) | 0.99 (0.92–1.07) | 0.845 | |
| HIV serology | |||
| Negative | 6/148 (4.1) | Reference | |
| Positive | 69/76 (7.9) | 2.03 (0.63–6.52) | 0.235 |
Frequency data are number of cases positive for Zika virus by Zika virus-specific IgM or for dengue virus by dengue virus-specific IgM divided by total number of patients. Values in parentheses are percentages.
LFn-ZIKV-protease and -helicase and LFn-DENV-protease and -helicase elicit virus-specific T cells responses.
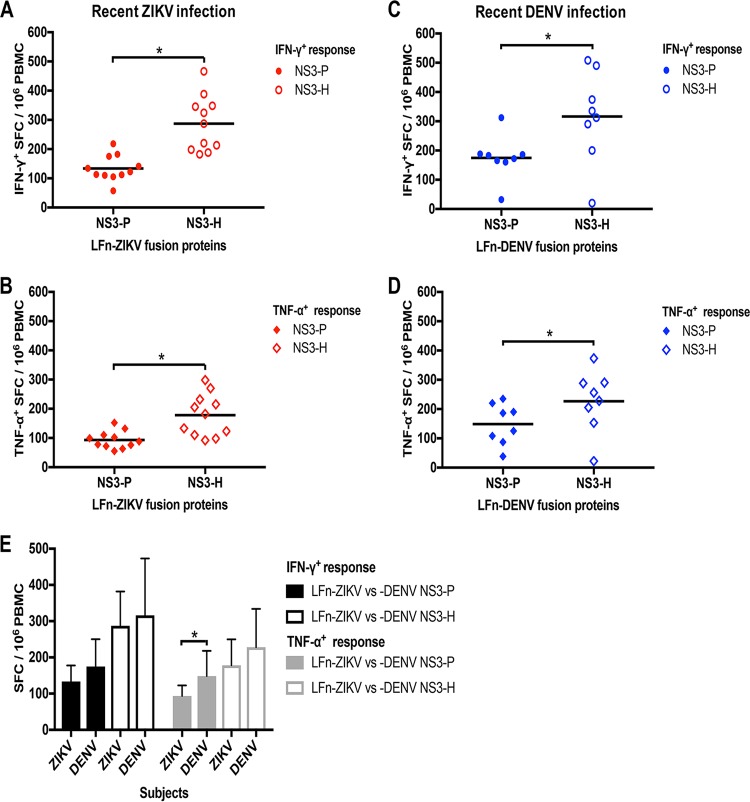
To assess the magnitude of T cell responses during ZIKV and DENV infection, we first measured the response by homologous LFn fusion antigen stimulation of PBMCs in a gamma interferon (IFN-γ) and tumor necrosis factor alpha (TNF-α) ELISPOT assay from IgM-positive subjects (presumed acute or recent phase of infection). PBMC samples collected during the recent phase of infection were available for analysis from 11 out of 14 ZIKV and 8 out of 11 DENV IgM-positive subjects. Responses to LFn-ZIKV-NS3-P and LFn-ZIKV-NS3-H compared to those of the LFn control were demonstrated in 11 out of 11 ZIKV IgM-positive samples (100%) (Table 3 and Fig. 3A and B); the frequency of IFN-γ+ and TNF-α+ T cells responsive to LFn-ZIKV-NS3-P ranged from 57 to 218 spot-forming cells (SFC)/106 PBMCs and 55 to 152 SFC/106 PBMCs, respectively, while the frequency responsive to LFn-ZIKV-NS3-H ranged from 182 to 466 SFC/106 PBMCs and 92 to 298 SFC/106 PBMCs, respectively. T cell responses to LFn-DENV-NS3-P and LFn-DENV-NS3-H compared to the LFn control were demonstrated in 7 out 8 DENV IgM-positive samples (87.5%) (Table 3 and Fig. 3C and D); the frequency of IFN-γ+ and TNF-α+ T cells responsive to LFn-DENV-NS3-P ranged from 32 to 312 SFC/106 PBMCs and 38 to 235 SFC/106 PBMCs, respectively, while the frequency responsive to LFn-DENV-NS3-H ranged from 20 to 508 SFC/106 PBMCs and 22 to 373 SFC/106 PBMCs, respectively. DENV subject 3 demonstrated IFN-γ+ and TNF-α+ T cell responses that were well below the positive threshold. Overall, IFN-γ+ and TNF-α+ responses to LFn-ZIKV/DENV-NS3-H were significantly stronger than responses to LFn-ZIKV/DENV-NS3-P. Both IFN-γ+ and TNF-α+ T cell responses were greater in subjects who tested positive for DENV IgM compared to those who tested positive for ZIKV IgM; this difference was statistically significant for TNF-α+ responses to LFn-DENV-NS3-P (Fig. 3E).
TABLE 3.
Characteristics of ZIKV and DENV LFn fusion proteins during recent phase of infection
| Subject | HIV status | Flavivirus immune status | Infection PBMC collection yra | No. of SFC/106 PBMC |
|||
|---|---|---|---|---|---|---|---|
| IFN-γ+ |
TNF-α+ |
||||||
| LFn ZIKV or DENV NS3-P | LFn ZIKV or DENV NS3-H | LFn ZIKV or DENV NS3-P | LFn ZIKV or DENV NS3-H | ||||
| ZIKV | |||||||
| 1 | Negative | No | 1998 | 57 | 188 | 55 | 92 |
| 2 | Negative | Yes | 1999 | 182 | 466 | 63 | 205 |
| 3 | HIV dual | No | 2000 | 105 | 220 | 78 | 182 |
| 4 | Negative | No | 2000 | 122 | 213 | 99 | 123 |
| 5 | HIV-1 | No | 2000 | 141 | 182 | 132 | 110 |
| 6 | Negative | Yes | 2001 | 112 | 345 | 102 | 298 |
| 7 | Negative | No | 2001 | 134 | 287 | 152 | 133 |
| 8 | Negative | Yes | 2002 | 175 | 324 | 110 | 270 |
| 9 | Negative | No | 2003 | 113 | 198 | 72 | 98 |
| 10 | Negative | Yes | 2003 | 218 | 348 | 76 | 215 |
| 11 | Negative | Yes | 2003 | 110 | 388 | 88 | 232 |
| 12 | Negative | No | NA | ||||
| 13 | HIV dual | Yes | NA | ||||
| 14 | Negative | No | NA | ||||
| DENV | |||||||
| 1 | Negative | No | 1999 | 172 | 335 | 186 | 205 |
| 2 | HIV-2 | No | 1999 | 165 | 290 | 87 | 153 |
| 3 | Negative | No | 2000 | 32 | 20 | 38 | 22 |
| 4 | Negative | No | 2000 | 186 | 312 | 235 | 228 |
| 5 | Negative | Yes | 2000 | 183 | 374 | 125 | 266 |
| 6 | Negative | Yes | 2001 | 188 | 490 | 190 | 288 |
| 7 | Negative | Yes | 2002 | 312 | 508 | 220 | 373 |
| 8 | HIV-1 | No | 2002 | 160 | 200 | 108 | 290 |
| 9 | HIV-1 | Yes | NA | ||||
| 10 | Negative | No | NA | ||||
| 11 | Negative | Yes | NA | ||||
NA, not applicable.
FIG 3.
Ex vivo T cell reactivity to ZIKV or DENV LFn fusion proteins in the recent phase of ZIKV or DENV infection. PBMC samples from ZIKV IgM-positive subjects collected during the recent phase of infection were subjected to homologous LFn NS3 protease and helicase protein stimulation in IFN-γ (A) and TNF-α (B) LFn ELISPOT ex vivo experiments. LFn-ZIKV-NS3-P IFN-γ, red-shaded circle; LFn-ZIKV-NS3-H IFN-γ, red-outlined circle; LFn-ZIKV-NS3-P TNF-α, red-shaded diamond; LFn-ZIKV-NS3-H TNF-α, red-outlined diamond. PBMC samples from DENV IgM-positive subjects collected during the recent phase of infection were subjected to homologous LFn NS3 protease and helicase protein stimulation in IFN-γ (C) and TNF-α (D) LFn ELISPOT ex vivo experiments. LFn-DENV-NS3-P IFN-γ, blue-shaded circle; LFn-DENV-NS3-H IFN-γ, blue-outlined circle; LFn-DENV-NS3-P TNF-α, blue-shaded diamond; LFn-DENV-NS3-H TNF-α, blue-outlined diamond. Responses are expressed as the number of secreting cells per 106 PBMCs and are considered positive if the net numbers of spot-forming cells (SFC) per 106 are ≥55, are greater than four times the mean background, and are three standard deviations above the background. (E) Comparison of IFN-γ ZIKV versus DENV NS3 protease (black-shaded bars) and helicase (black-outlined bars) and TNF-α ZIKV versus DENV NS3 protease (gray-shaded bars) and helicase (gray-outlined bars) T cell responses in ZIKV and DENV subjects. Magnitude of responses is expressed as geometric positive mean, and statistical analysis was performed with Mann-Whitney U test. *, P < 0.05.
IgG antibody responses are sustained after ZIKV or DENV infection.
We then assessed sequential antibody responses in available plasma samples by IgG Western blotting. Sequential date-matched plasma and PBMC samples were available for analysis in 6 ZIKV and 4 DENV IgM-positive subjects.
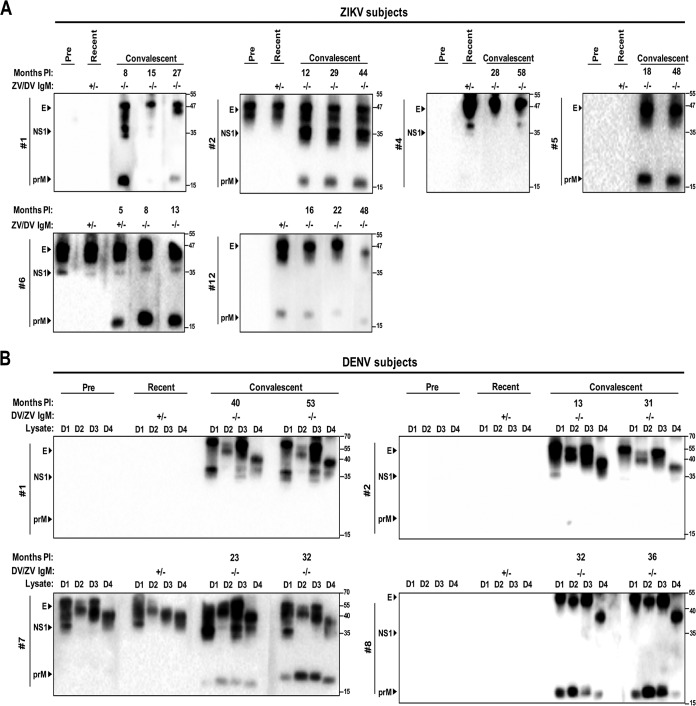
Western blot analysis using ZIKV- or DENV-infected Vero cell lysates was carried out to test for IgG antibody responses to several ZIKV or DENV proteins, including prM, E, and NS1. Two out of 6 ZIKV subjects demonstrated previous flavivirus exposure prior to testing positive for ZIKV IgM (ZIKV patients 2 and 6; 33.3%) (Fig. 4A). During the recent phase of infection, ZIKV patients 2, 4, 6, and 12 had detectable IgG antibodies to ZIKV prM, E, and/or NS1, but ZIKV patients 1 and 5 did not. Probing plasma collected months up to almost 5 years after infection (convalescent phase to later time points postinfection), antibody responses to prM, E, and/or NS1 were detectable in all cases, with some responses waning over time (ZIKV subject 1, prM and NS1; ZIKV subject 4, NS1; ZIKV subject 12, prM and E). With the exception of ZIKV subject 6, who tested ZIKV IgM positive at the first convalescent time point (5 months after infection), all convalescent-phase sera tested negative for ZIKV and DENV IgM.
FIG 4.
Sequential antibody responses to different ZIKV or DENV proteins in ZIKV or DENV subjects. Preinfection, recent-, and convalescent-phase sera from ZIKV (A) or DENV (B) IgM-positive subjects were subjected to Western blot analysis using lysates derived from ZIKV- or DENV-infected Vero cells. Arrowheads indicate PrM, E, and NS1 proteins recognized. Molecular size marker units are kilodaltons. Dates are given in month/year format. Lysates include ZIKV African strain (unpublished), DENV1 Hawaii strain (D1), DENV2 NGC strain (D2), DENV3 H87 strain (D3), and DENV4 H241 strain (D4). Months PI, months postinfection. ZV/DV IgM, results for ZIKV (ZV) or DENV (DV) IgM serology expressed as positive (+) or negative (−).
Among the DENV cases, 1 out of 4 subjects demonstrated previous flavivirus exposure prior to testing positive for DENV IgM (DENV subject 7; 25%); this subject was also the only one to demonstrate detectable IgG in the recent phase of infection (Fig. 4B). In all cases, IgG antibody responses to DENV prM, E, and/or NS1 were sustained months up to over 4.5 years after infection. As described above, all convalescent-phase sera from the DENV subjects tested negative for DENV and ZIKV IgM.
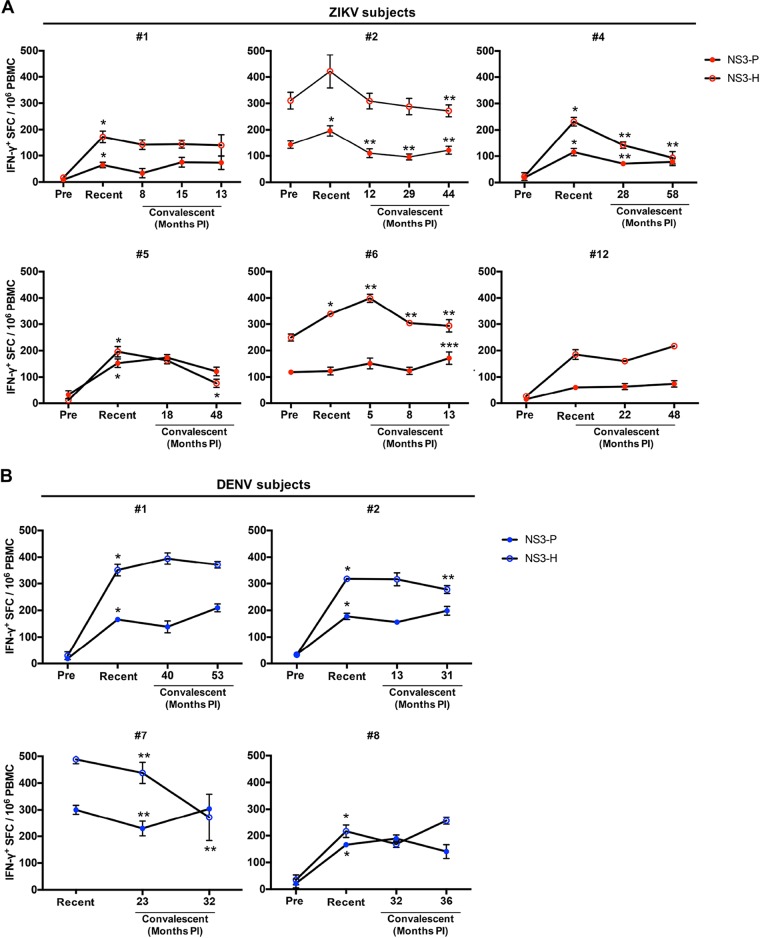
T cell responses are sustained after ZIKV or DENV infection.
We further examined the T cell responses by IFN-γ ELISPOT assay in sequential date-matched PBMC samples. In all ZIKV cases, robust T cell responses to LFn-ZIKV-NS3-P or LFn-ZIKV-NS3-H were detected at dynamic levels throughout the time points tested (Fig. 5A). Peak responses were observed in the recent phase of infection for ZIKV subjects 1 (NS3-P and -H), 2 (NS3-P), 4 (NS3-P and -H), 5 (NS3-P and -H), and 6 (NS3-H). T cell responses waned over time but were sustained well above the negative cutoff for subjects 2 (NS3-P and -H), 4 (NS3-P and -H), 5 (NS3-H), and 6 (NS3-P). For subject 6, a peak response was observed 13 months postinfection. PBMCs were not available at the time of IgM positivity for ZIKV subject 12; however, this subject demonstrated trends of responses during the recent phase of infection that were sustained throughout the convalescent time points tested. Only the two subjects with antibody responses demonstrating previous flavivirus exposure (ZIKV subjects 2 and 6) demonstrated preinfection T cell responses, and their T cell responses were generally stronger than responses in the other subjects.
FIG 5.
Sequential IFN-γ T cell reactivity to ZIKV or DENV LFn fusion proteins in ZIKV or DENV subjects. Preinfection, recent-, and convalescent-phase PBMC samples from ZIKV (A) or DENV (B) IgM-positive subjects were subjected to homologous LFn NS3 protease (ZIKV, red-shaded circle; DENV, blue-shaded circle) and helicase (ZIKV, red-outlined circle; DENV, blue-outlined circle) protein stimulation in IFN-γ LFn ELISPOT ex vivo experiments. Responses for each time point are expressed as the number of secreting cells per 106 PBMCs. Months PI, months postinfection. *, preinfection versus recent, P < 0.05; **, recent versus convalescent time point 1, P < 0.05; ***, convalescent time point 2 versus time point 3, P < 0.05.
Similarly, in all DENV cases, robust and dynamic T cell responses to LFn-DENV-NS3-P or LFn-DENV-NS3-H were detected (Fig. 5B). For DENV subjects 1, 2, and 8, peak responses to LFn-DENV-NS3-P and -H were observed during the recent phase of infection. T cell responses significantly waned over time for subjects 2 (NS3-H) and 7 (NS3-P and -H), while T cell responses in subjects 1 and 8 were sustained throughout the time points tested. DENV subject 7, the only one of the 4 tested who demonstrated previous flavivirus exposure, generally had the strongest T cell responses.
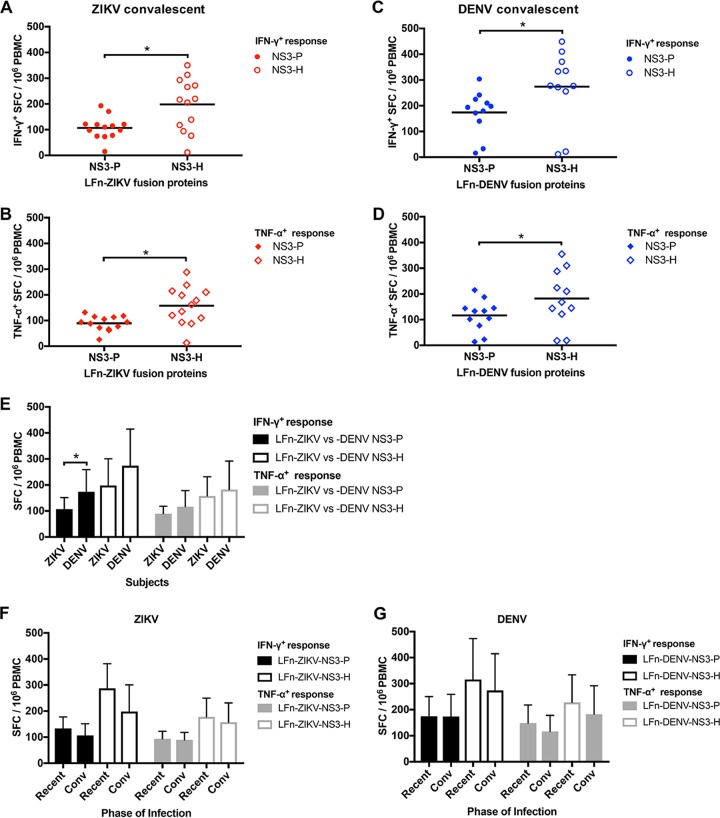
While sequential analysis was not possible for a majority of the subjects, we examined the IFN-γ and TNF-α T cell responses using available PBMC samples collected during the convalescent phase of infection for 13 ZIKV and 11 DENV subjects. T cell responses to LFn-ZIKV-NS3-P and LFn-ZIKV-NS3-H compared to those of the LFn control were demonstrated in 12 out of 13 samples (92.3%) (Table 4 and Fig. 6A and B). ZIKV subject 14 demonstrated IFN-γ+ and TNF-α+ T cell responses that were well below the positive threshold. T cell responses to LFn-DENV-NS3-P and LFn-DENV-NS3-H compared to the LFn control were demonstrated in 9 out of 11 samples from the DENV subjects (81.8%) (Fig. 6C and D). DENV subjects 3 and 10 demonstrated IFN-γ+ and TNF-α+ T cell responses that were well below the positive threshold. Overall, IFN-γ and TNF-α convalescent-phase T cell responses to LFn-ZIKV/DENV-NS3-H were significantly stronger than responses to LFn-ZIKV/DENV-NS3-P. Both IFN-γ and TNF-α T cell responses were greater in subjects who tested positive for DENV IgM than in those who tested positive for ZIKV IgM; this difference was statistically significant for IFN-γ responses to LFn-DENV-NS3-P (Fig. 6E). Additionally, in both the ZIKV and DENV cases, IFN-γ and TNF-α T cell responses to LFn-ZIKV/DENV-NS3-P and LFn-ZIKV/DENV-NS3-H showed a trend toward stronger responses in the recent phase of infection compared to those during convalescence, although this was not statistically significant (Fig. 6F and G). Importantly, all ZIKV and DENV date-matched convalescent-phase serum samples tested negative for ZIKV and DENV IgM (Table 4).
TABLE 4.
Characteristics of ZIKV and DENV LFn fusion proteins during convalescence
| Subject | HIV status | Flavivirus immune status | Infection PBMC collection yr | No. of SFC/106 PBMC |
|||
|---|---|---|---|---|---|---|---|
| IFN-γ+ |
TNF-α+ |
||||||
| LFn ZIKV or DENV NS3-P | LFn ZIKV or DENV NS3-H | LFn ZIKV or DENV NS3-P | LFn ZIKV or DENV NS3-H | ||||
| ZIKV | |||||||
| 1 | Negative | No | 1998 | 57 | 188 | 55 | 92 |
| 2 | Negative | Yes | 1999 | 182 | 466 | 63 | 205 |
| 3 | HIV dual | No | 2000 | 105 | 220 | 78 | 182 |
| 4 | Negative | No | 2000 | 122 | 213 | 99 | 123 |
| 5 | HIV-1 | No | 2000 | 141 | 182 | 132 | 110 |
| 6 | Negative | Yes | 2001 | 112 | 345 | 102 | 298 |
| 7 | Negative | No | 2001 | 134 | 287 | 152 | 133 |
| 8 | Negative | Yes | 2002 | 175 | 324 | 110 | 270 |
| 9 | Negative | No | 2003 | 113 | 198 | 72 | 98 |
| 10 | Negative | Yes | 2003 | 218 | 348 | 76 | 215 |
| 11 | Negative | Yes | 2003 | 110 | 388 | 88 | 232 |
| 12 | Negative | No | NA | ||||
| 13 | HIV dual | Yes | NA | ||||
| 14 | Negative | No | NA | ||||
| DENV | |||||||
| 1 | Negative | No | 1999 | 172 | 335 | 186 | 205 |
| 2 | HIV-2 | No | 1999 | 165 | 290 | 87 | 153 |
| 3 | Negative | No | 2000 | 32 | 20 | 38 | 22 |
| 4 | Negative | No | 2000 | 186 | 312 | 235 | 228 |
| 5 | Negative | Yes | 2000 | 183 | 374 | 125 | 266 |
| 6 | Negative | Yes | 2001 | 188 | 490 | 190 | 288 |
| 7 | Negative | Yes | 2002 | 312 | 508 | 220 | 373 |
| 8 | HIV-1 | No | 2002 | 160 | 200 | 108 | 290 |
| 9 | HIV-1 | Yes | NA | ||||
| 10 | Negative | No | NA | ||||
| 11 | Negative | Yes | NA | ||||
FIG 6.
Ex vivo T cell reactivity to ZIKV or DENV LFn fusion proteins in ZIKV or DENV convalescence. PBMC samples from ZIKV IgM-positive subjects collected during the convalescent phase of infection were subjected to homologous LFn NS3 protease and helicase protein stimulation in IFN-γ (A) and TNF-α (B) LFn ELISPOT ex vivo experiments. LFn-ZIKV-NS3-P IFN-γ, red-shaded circle; LFn-ZIKV-NS3-H IFN-γ, red-outlined circle; LFn-ZIKV-NS3-P TNF-α, red-shaded diamond; LFn-ZIKV-NS3-H TNF-α, red-outlined diamond. PBMC samples from DENV IgM-positive subjects collected during the convalescent phase of infection were subjected to homologous LFn NS3 protease and helicase protein stimulation in IFN-γ (C) and TNF-α (D) LFn ELISPOT ex vivo experiments. LFn-DENV-NS3-P IFN-γ, blue-shaded circle; LFn-DENV-NS3-H IFN-γ, blue-outlined circle; LFn-DENV-NS3-P TNF-α, blue-shaded diamond; LFn-DENV-NS3-H TNF-α, blue-outlined diamond. Responses are expressed as the number of secreting cells per 106 PBMCs and are considered positive if the net numbers of spot-forming cells (SFC) per 106 are ≥55, are greater than four times the mean background, and are three standard deviations above the background. Comparison of IFN-γ+ ZIKV/DENV NS3 protease (black-shaded bars) and helicase (black-outlined bars) and TNF-α+ ZIKV/DENV NS3 protease (gray-shaded bars) and helicase (gray-outlined bars) T cell responses in ZIKV and DENV subjects (E) and between recent and convalescent (Conv) phases of ZIKV (F) or DENV (G) infection. The magnitudes of responses are expressed as geometric positive means ± standard deviations, and statistical analyses were performed with Mann-Whitney U test. *, P < 0.05.
ZIKV- or DENV-specific and cross-reactive T cell responses.
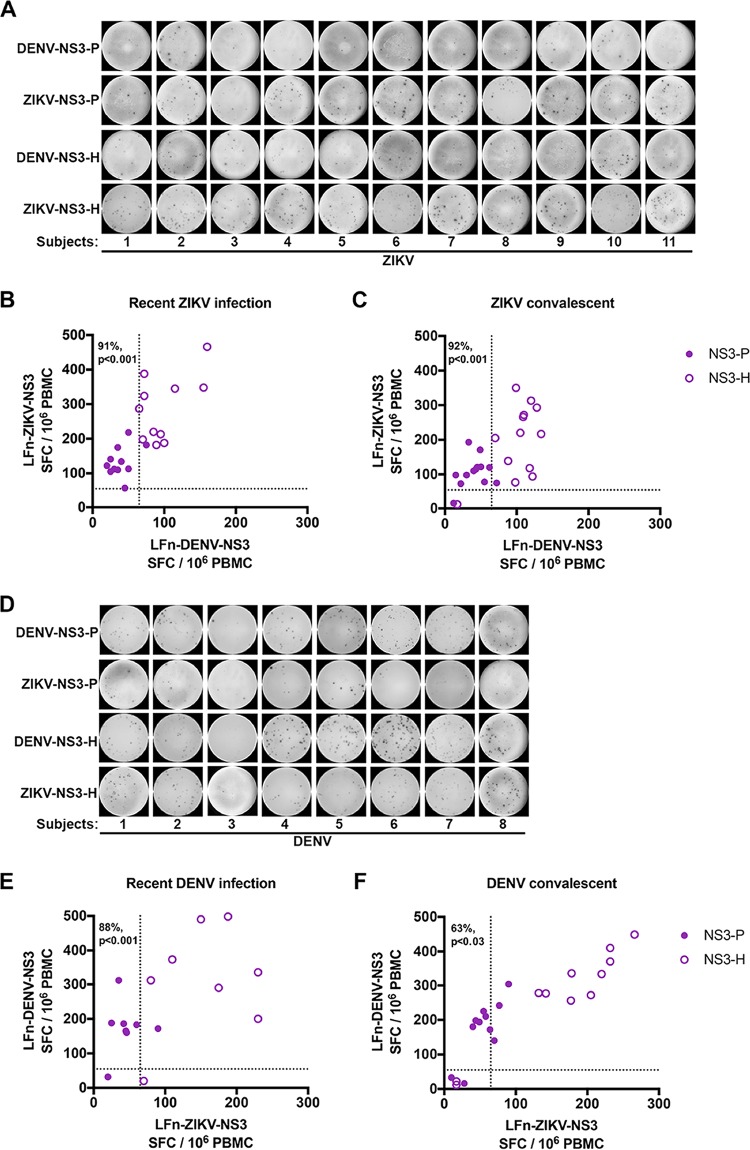
To assess specific and/or cross-reactive T cell responses between ZIKV and DENV among the subjects, PBMCs collected during the recent and convalescent phases of infection were stimulated with homologous and heterologous LFn fusion antigens in an IFN-γ ELISPOT assay.
PBMC samples were individually stimulated with LFn-ZIKV-NS3-P, LFn-DENV-NS3-P, LFn-ZIKV-NS3-H, and LFn-DENV-NS3-H. At the time of ZIKV IgM positivity, T cell responses to NS3 protease were largely ZIKV specific, even in cases where individuals had previous flavivirus exposure (Fig. 7A and B), while responses to NS3 helicase were largely cross-reactive between ZIKV and DENV (91% specific for protease versus 0% specific for helicase; P < 0.001); both LFn-ZIKV-NS3-H and LFn-DENV-NS3-H elicited significant responses. Similarly, ZIKV convalescent T cell responses to NS3 protease were specific for ZIKV but cross-reactive to NS3 helicase (92% specific for protease versus 0% specific for helicase; P < 0.001) (Fig. 7C). Although PBMCs were not available for ZIKV subject 14 and could not be evaluated, convalescent responses for this subject were not observed by ELISPOT assay; this same subject also demonstrated no TNF-α response to LFn-ZIKV-NS3-P or LFn-DENV-NS3-H (Table 3), suggesting false ZIKV IgM positivity.
FIG 7.
T cell-specific and cross-reactive NS3 responses in the recent and convalescent phases of ZIKV or DENV infections. Recent- and convalescent-phase PBMC samples from ZIKV or DENV subjects were treated with homologous and heterologous LFn-ZIKV and -DENV NS3 protease and helicase proteins, and the specific IFN-γ+ and TNF-α+ T cells were detected by LFn ELISPOT ex vivo experiments. (A to C) Representative image and magnitude of recent ZIKV/DENV infection-phase responses (A and B) and magnitude of convalescent-phase responses (C) from ZIKV subjects. (D to F) Representative image and magnitude of recent DENV/ZIKV infection-phase responses (D and E) and magnitude of convalescent responses from DENV subjects (F). Shown is the average magnitude of response of individual subjects to either LFn-ZIKV or -DENV NS3 protease (purple-shaded circle) and helicase (purple-outlined circle) proteins. Dotted lines represent the cutoff value. *, P < 0.05.
Consistent with these results, at the time of DENV IgM positivity and at the convalescent time point, T cell responses to NS3 protease were DENV specific but were largely cross-reactive between DENV and ZIKV to NS3 helicase (recent, 88% specific for protease versus 0% specific for helicase, P value of <0.001; convalescent, 63% specific for protease versus 0% specific for helicase, P value of <0.03) (Fig. 7D to F). There were a few exceptions in which NS3 protease responses were cross-reactive between DENV and ZIKV, particularly during the convalescent phase. Responses for DENV subjects 3 and 10 were not observed by ELISPOT assay at the recent and convalescent phases of infection (PBMCs were not available for DENV subject 10 at the acute recent phase of infection and could not be evaluated); these results corroborate the unobserved TNF-α response to LFn-ZIKV/DENV-NS3P or LFn-ZIKV/DENV-NS3-H for these subjects (Tables 3 and 4), suggesting false DENV IgM positivity.
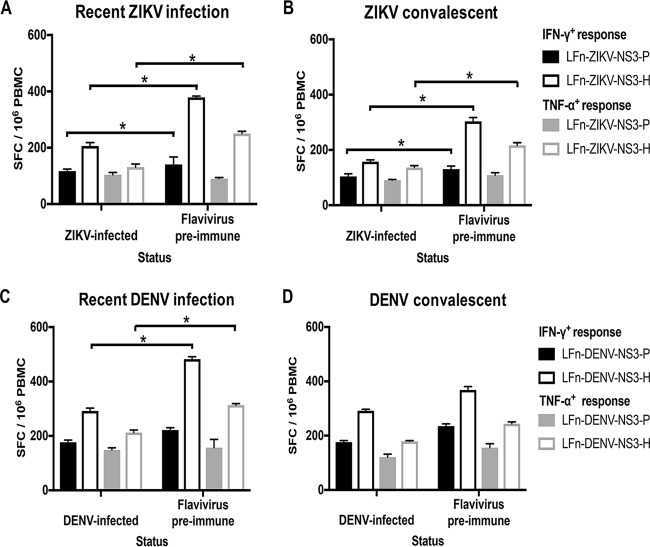
Previous flavivirus exposure is associated with enhanced ZIKV or DENV T cell responses.
We further evaluated the impact of previous flavivirus exposure on the frequency and magnitude of the T cell response in PBMC samples that were collected during both the recent and convalescent phases of infection. At the time the subjects tested positive for ZIKV IgM, responses for 6 subjects who demonstrated flavivirus preimmunity (flavivirus preimmune) and 5 who did not (primary ZIKV infected) were included in the analysis. Additionally, responses corresponding to the convalescent phase of infection were available for analysis for 5 subjects with flavivirus preimmunity and 8 primary ZIKV-infected subjects. ZIKV subject 14 was excluded from the analysis due to the suspected ZIKV IgM false positivity. During both recent and convalescent phases of infection, LFn-ZIKV-NS3-P and LFn-ZIKV-NS3-H elicited significantly stronger IFN-γ T cell responses in subjects with flavivirus preimmunity (Fig. 8A). LFn-ZIKV-NS3-P elicited slightly increased TNF-α T cell responses in subjects who experienced a primary ZIKV infection at the IgM-positive date, although the difference was not statistically significant. LFn-ZIKV-NS3-H also elicited significantly stronger TNF-α T cell responses in subjects with flavivirus preimmunity during both the recent and convalescent phases of infection (Fig. 8B).
FIG 8.
Impact of prior flavivirus infection on the ZIKV or DENV T cell response. Comparison of recent (A)- and convalescent (B)-phase IFN-γ+ ZIKV NS3 protease (black shaded bars) and helicase (black unshaded bars) and TNF-α+ ZIKV NS3 protease (gray shaded bars) and helicase (gray unshaded bars) T cell responses from presumed primary ZIKV-infected and flavivirus preimmune groups of the ZIKV subjects. Comparison of recent (C)- and convalescent (D)-phase IFN-γ+ DENV NS3 protease (black shaded bars) and helicase (black unshaded bars) and TNF-α+ DENV NS3 protease (gray shaded bars) and helicase (gray unshaded bars) T cell responses from presumed primary DENV-infected and flavivirus preimmune groups of the DENV subjects. Magnitude of response for each group is expressed as geometric positive means ± standard deviations, and statistical analyses were performed with Mann-Whitney U test. *, P < 0.05.
At the time the subjects tested positive for DENV IgM, responses for 3 flavivirus preimmune subjects and 4 primary DENV-infected subjects were included in the analysis; DENV subject 3 was excluded due to the suspected DENV IgM false positivity. Additionally, responses corresponding to the convalescent phase of infection were available for analysis for 4 subjects with flavivirus preimmunity and 5 primary DENV-infected subjects; DENV subjects 3 and 10 were excluded from the analysis due to suspected DENV IgM false positivity. During both the recent and convalescent phases of infection, LFn-ZIKV-NS3-P and LFn-ZIKV-NS3-H elicited stronger IFN-γ and TNF-α T cell responses in subjects with flavivirus preimmunity; the difference was statistically significant for the responses to LFn-DENV-NS3-H at the time the DENV-infected subjects tested DENV IgM positive (Fig. 8C and D).
DISCUSSION
For decades, human ZIKV infection has remained geographically limited to Africa and Asia and has gone largely unnoticed due to its mild clinical outcomes with symptoms similar to those of other acute febrile diseases endemic to the same regions (5). However, the recent rapid emergence of ZIKV in the Pacific and the Americas and Caribbean have greatly heightened awareness of the virus. A retrospective analysis of the 2014 French Polynesian ZIKV outbreak identified associations between infection and adult Guillain-Barré syndrome, an autoimmune disease causing acute or subacute flaccid paralysis (18, 45). The 2015 ZIKV outbreak, which started in Brazil, highlighted the association of infection with other severe neurologic manifestations, including microcephaly and congenital malformations in infants born to infected mothers that had previously not been recognized (19).
The mechanisms that cause these severe ZIKV disease outcomes remain unknown; however, associations between infection and neurologic symptoms are being elucidated. Asian ZIKV strains demonstrated neurotropism, causing cellular death in human neural progenitor cells (NPC) and immature cortical neurons, thereby affecting neurogenesis (16, 46). Despite the absence of reported associations between ZIKV infection and neuropathology in Africa, several African strains were also shown to infect and impair growth of human NPC (47). Efforts are needed to examine similarities and differences in infection and outcomes between the African and Asian ZIKV lineages.
The human immunologic response to ZIKV and DENV infections represents a critical research gap that may shed light on the observance of severe ZIKV-associated disease outcomes in some individuals but not in others. Identifying immune responses with a protective profile is also useful for vaccine development. Therefore, the goal of the present study was to develop a novel modified anthrax-based ELISPOT assay utilizing the LFn-ZIKV-NS3-P/H and LFn-DENV-NS3-P/H priming systems to measure the T cell response to African ZIKV and DENV infections. There are 4 important findings from our study: (i) robust IFN-γ and TNF-α T cell responses against African ZIKV and DENV NS3, (ii) ZIKV- or DENV-specific T cell responses to NS3 protease but cross-reactive responses to NS3 helicase, (iii) long-term dynamic antibody and T cell responses, and (iv) stronger T cell responses in patients with flavivirus preimmunity.
We initially screened plasma collected from febrile patients between 1992 and 2004 from a natural-history HIV Senegalese FSW cohort for the presence of ZIKV and DENV IgM antibodies. Consistent with our previous findings, the overall seroprevalence of ZIKV IgM in this study remained approximately 6.3% (44). Our serology testing also revealed a DENV IgM seroprevalence of 9.3%. These results provide evidence that both African ZIKV and DENV are endemic to Senegal and cause infection in humans.
Activation of CD4+ and CD8+ T cells has been extensively examined in several flavivirus infections, with responses preferentially targeting epitopes within the nonstructural proteins, namely, NS3, NS4b, and NS5 (39–41). These studies have also shown that CD8+ and CD4+ T cell reactivity in Sri Lankan and Nicaraguan populations were HLA linked and associated with increased or decreased disease susceptibility and were also associated with responses of lower quality, breadth, and magnitude (40, 48, 49). A recent study, however, demonstrated that a significant number of CD4+ and CD8+ T cell responses in ZIKV-positive individuals were directed against the structural proteins, and that previous DENV exposure amplifies these responses (31). A study using HLA transgenic mice infected with ZIKV revealed immunodominant and protective roles for DENV cross-reactive CD8+ T cells (30). Whether adaptive HLA-linked T cell responses are clinically beneficial or detrimental during human ZIKV infections is currently not known.
We report, for the first time, significant IFN-γ and TNF-α T cell responses against African ZIKV and DENV NS3 during both the recent and convalescent phases of infection. We found that for both ZIKV and DENV, responses against NS3 helicase were significantly stronger than those against NS3 protease during both the recent and convalescent phases. In general, T cell responses against DENV NS3 were stronger than responses against ZIKV NS3. Both for subjects with ZIKV and those with DENV infection, T cell responses generally were stronger during the recent phase of infection than responses measured during convalescence (Fig. 6F and G).
A study on secondary DENV infection in Thai children demonstrated NS3-specific T cell responses that peaked during early convalescence (50). In another study on Vietnamese children, NS3-specific T cell responses were undetectable until after the development of plasma leakage that occurs in dengue hemorrhagic fever in the first few days of illness (51). In most cases, our sequential T cell analysis of ZIKV and DENV infections demonstrated peak frequencies of NS3-specific T cells during the recent phase and several responses to either NS3 protease or helicase that were sustained throughout the time points tested. A possible explanation for these differences is that NS3-specific T cell responses differ between African and Thai or Vietnamese patients. Interestingly, in 2 ZIKV (subjects 1 and 5) and 3 DENV (subjects 1, 2, and 8) cases, IgG was not detectable during the recent phase of infection; however, strong T cell responses were mounted.
In regions of West Africa where flaviviruses are endemic and different viruses, including ZIKV, DENV, West Nile virus, and Yellow Fever virus, cocirculate, multiple sequential infections are common. Therefore, it is possible that low levels of infection by these viruses lead to preexisting flavivirus immunity that impacts the subsequent immune response. Our IgG serology demonstrates significant flavivirus exposure. Among the subjects who tested ZIKV IgM positive, 42.8% (6/14) had experienced a previous flavivirus infection. Similarly, among the subjects who tested DENV IgM positive, 41.6% (5/12) experienced a previous flavivirus infection. Due to the high number of cross-reactive flaviviruses circulating in Senegal, determining the previous flavivirus infections in these subjects is outside the scope of the present study. However, these results suggest that humans residing in Senegal experience multiple infections by flaviviruses throughout their lives.
Studies examining the impact of T cell responses to prior DENV infections on the outcome of the current infection have been inconclusive with respect to the pathogenic or protective consequences (39, 40). Our results demonstrate that preexisting flavivirus immunity is associated with enhanced T cell responses. For both ZIKV and DENV infections, subjects with flavivirus preimmunity demonstrated stronger responses than those experiencing a primary infection. Whether the enhanced responses result in protection or more severe disease remains to be determined.
Our analysis of T cell cross-reactivity between ZIKV and DENV demonstrates ZIKV- or DENV-specific responses to NS3 protease in particular during the recent stage but cross-reactive responses to NS3 helicase during both the recent and convalescent phases of infection. While we did not define the epitopes targeted by T cells during ZIKV or DENV infections, the higher percent amino acid identity between the ZIKV and DENV helicases (71%) versus the ZIKV and DENV proteases (53%) might help explain the ZIKV/DENV cross-reactive T cell responses to the helicase. These results have several important implications. NS3 helicase cross-reactivity between ZIKV and DENV suggests that the T cell response is protective in individuals experiencing a secondary ZIKV infection with preexisting DENV immunity or vice versa. Because cross-reactive responses were observed during the convalescent phase of infection, the T cell response may be protective even if the secondary infection occurs months to years after the primary infection. Finally, the combination of specific and cross-reactive responses to NS3 elicited by the LFn priming system has diagnostic potential. Diagnosis of flaviviruses is complicated by two major factors: (i) the endemicity of related flaviviruses which cross-react in typical serologic tests, and (ii) the fact that the diagnostic usefulness of nucleic acid tests is limited to the recent phase of infection. Our novel modified anthrax-based flavivirus ELISPOT assay has the potential to be developed into a specific T cell-based diagnostic with detection capabilities well beyond the recent phase of infection.
This study has a number of limitations. First, our study population is comprised entirely of women and is not necessarily generalizable to men, and the ZIKV and DENV sample sizes are small. Second, it was not within the scope of our study to identify the virus that caused the primary infection or if there were multiple prior infections. Given the low level of endemic infection by ZIKV and DENV in West Africa, prospective studies using specific diagnostic tools to actively screen for these infections are needed. Third, the plasma and PBMC samples used in this study were cryopreserved for an extended period of time, from 12 to almost 20 years. Future studies are needed to validate these observations using fresh samples. Fourth, the focus of the T cell response was on ZIKV and DENV NS3. While we observed strong responses to NS3, there are likely other proteins that are being targeted by T cells, which may correlate with disease severity. Finally, due to the limited amount of PBMC available for each patient, we were unable to define the CD8+- and CD4+-specific T cell responses.
In summary, while this is the first characterization of human T cell responses to African ZIKV and DENV, it raises new questions and highlights the need for further investigation to better understand the immune mechanisms and consequences related to the high cross-reactivity among these related viruses and strains. This study has identified responses to NS3 with specific and cross-reactive phenotypes that could be detected during both the recent and convalescent phases of infection. These results suggest the potential for development of a simple and low-cost African ZIKV and DENV diagnostic assay based on T cell responses. Our analysis of prior flavivirus exposure on the T cell response to ZIKV and DENV demonstrates enhanced responses in those with flavivirus preimmunity. This finding has critical implications for development of vaccines against ZIKV and DENV, which should include the induction of ZIKV- and DENV-specific and ZIKV/DENV cross-reactive T cell responses and should also consider specific responses between the African and Asian viral lineages.
MATERIALS AND METHODS
Clinical samples and ethics statement.
Plasma and peripheral blood mononuclear cell (PBMC) samples were obtained from a female sex worker (FSW) cohort in Dakar, Senegal. Self-identified FSW visiting a health care clinic in Dakar underwent annual blood tests for multiple sexually transmitted infections, including HIV-1/2. This natural-history HIV-1/2 cohort helped define some of the first in vivo characteristics of HIV-2 infection in West Africa; epidemiological and clinical details of this cohort have been reported elsewhere (52).
Clinical visit records from the cohort were queried for recorded fever of ≥37.5°C. Of these, excess stored plasma samples that were collected within 7 days of the fever visit were used in serological testing. PBMCs were separated from whole blood collected in EDTA tubes by Ficoll-Hypaque gradient density (Organon Technika, Durham, NC) and cryopreserved in freezing medium (10% dimethyl sulfoxide [DMSO; Sigma-Aldrich, St. Louis, MO], 90% fetal bovine serum [FBS; Medicorp, Montreal, Quebec, Canada]) at −80°C overnight prior to transfer to liquid nitrogen. The plasma and PBMC samples included in the study were prospectively collected between 1992 and 2004.
The Harvard T. H. Chan School of Public Health Institutional Review Board (IRB) and the local research ethics committees at Cheikh Anta Diop University, Dakar, Senegal, approved the primary studies under which the samples and data were collected. All patients provided informed consent for the original collection of samples. Excess samples and corresponding data were banked and anonymized. This study received an exemption determination from the Harvard Longwood Medical Area IRB.
Construction and expression of LFn fusion antigens.
Commercially synthesized amino acid fragments encoding the NS3 protease and helicase of African ZIKV(consensus sequence based on the GenBank accession numbers KU955594.1, KF268948.1, KF268949.1, KF268950.1, KU955591.1, KU955592.1, and KU955595.1) and DENV2 (consensus based on the GenBank accession numbers EF105384.1, EF105383.1, EF105389.1, EF105390.1, and EF457904.1) were cloned into the LFn expression plasmid (pET15bLFn). The pET15bLFn plasmid contains a T7 promoter, histidine tag (His6), and the terminal domain of the LF (LFn; 255 amino acids). pET15bLFn containing the coding sequence of African ZIKV and DENV2 was transformed into E. coli BLR (DE3) (Millipore, Medford, MA) for expression. Selected clones were sequenced to verify the reading frame, and clones containing the correct sequence were used for protein expression.
The LFn-DENV and -ZIKV fusion antigens were expressed upon isopropyl-β-d-thiogalactopyranoside (IPTG; Thermo Fisher Scientific, Rockford, IL) induction in Luria broth containing carbenicillin and chloramphenicol. Cells were pelleted by centrifugation and resuspended in imidazole (1 mM) binding buffer (Novagen, Madison, WI) in the presence of protease inhibitor cocktail (Boehringer Mannheim, Framingham, MA). Cell pellets were sonicated and centrifuged at 4°C, and the supernatants were loaded in an equilibrated nickel-charged column for affinity purification based on the His6 tag present in the amino terminus of the fusion protein. The bound proteins were eluted in 125 to 200 mM imidazole, desalted with a Sephadex G-25 M column (Sigma-Aldrich, St. Louis, MO), and eluted in phosphate-buffered saline (PBS). The PBS-eluted proteins were passed through an endotoxin-removing gel, Detoxi-Gel (Thermo Fisher Scientific, Rockford, IL). Protein concentrations were determined and samples were stored at −80°C. pET15bLFn was expressed and purified as described above for use as a control.
Serology and RT-PCR.
Plasma samples were screened for the presence of IgM antibodies by ZV-IgM enzyme-linked immunosorbent assay (ELISA) (MyBioSource, San Diego, CA) or DENV detect IgM capture ELISA (InBios, Seattle, WA) according to the manufacturer's instructions. Using internal controls, assay performances were monitored, and cutoffs were determined as specified by the manufacturer for individual kits.
RNA was extracted from serum using the QIAamp viral RNA kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. Samples were tested by RT-PCR for ZIKV as previously described (5). Samples were tested by RT-PCR for DENV as previously described (53).
Western blotting.
Briefly, for Western blotting C6/36 or Vero cells infected with DENV1 (Hawaii strain), DENV2 (NGC strain), DENV3 (H87 strain), DENV4 (H241 strain), or African ZIKV (strain unpublished) were lysed when cytopathic effects were observed in 20% of cells with 1% NP-40 lysis buffer (100 mM Tris [pH 7.5], 150 mM NaCl, 20 mM EDTA, 1% NP-40, 0.5% sodium deoxycholate) containing protease inhibitors (Roche Diagnostics), followed by centrifugation at 20,000 × g at 4°C for 30 min to obtain cell lysates, as previously described (54). Aliquots of cell lysates were added to nonreducing buffer (final concentrations of 2% SDS, 0.5 M Tris [pH 6.8], 20% glycerol, 0.001% bromophenol blue) and subjected to 12% PAGE and Western blot analysis using patient serum (1:100) as primary antibody and anti-human IgG horseradish peroxidase (HRP) (1:2,500; Thermo Fisher Scientific, Rockford, IL) as secondary antibody. Visualization was performed using SuperSignal Femto chemiluminescent substrate (Thermo Fisher Scientific, Rockford, IL) per the manufacturer's instructions and with a molecular imager (Chemi Doc XRS+ imaging system; Bio-Rad Technologies, Hercules, CA).
Ex vivo ELISPOT assay.
Ninety-six-well polyvinylidene difluoride (PVDF)-backed MultiScreenHTS (MSIP) microtiter plates (Millipore, Medford, MA) were treated with 100 μl of 90% methanol for 30 s and washed 5 times with sterile PBS (Sigma-Aldrich, St. Louis, MO). Plates were coated with 100 μl of capture antibodies in PBS at the following concentrations: 15 μg/ml anti-human IFN-γ MAb 1-D1K and 3 μg/ml anti-human TNF-α MAb TNF3/4 (Mabtech, Cincinnati, OH). Plates containing capture antibodies were incubated overnight at 4°C. Plates were blocked with 100 μl/well of 1% bovine serum albumin (BSA; Sigma-Aldrich, St. Louis, MO) in PBS for 1 h and then washed 6 times with PBS. Cryopreserved PBMCs were thawed in R10 medium (RPMI, 20% FBS) and incubated overnight at 37°C. PBMCs were washed two times with PBS and seeded in triplicate at 2 × 105 cells/well in a final volume of 100 μl/well. Fusion LFn-ZIKV and -DENV proteins were added to each well at a 2.5-μg/ml final concentration. As a positive control, PBMCs were stimulated with 5 μg/ml of phytohemagglutinin (PHA; Sigma-Aldrich, St. Louis, MO). As a negative control, wells received LFn with no fused protein at a 2.5-μg/ml final concentration.
After incubation for 28 to 32 h at 37°C in 5% CO2, the cells were discarded and plates were washed three times with PBS and again three times with PBS with 0.05% Tween 20 (PBST; Bio-Rad Technologies, Hercules, CA) to remove cells. The detection Ab were added at 100 μl/well in PBST, 1% BSA at the following concentrations: 1 μg/ml anti-human IFN-γ MAb 7-B6-1 and 0.5 μg/ml anti-human TNF-α MAb TNF5 (Mabtech, Cincinnati, OH). Plates were incubated overnight at 4°C. Plates were washed six times with PBST and then incubated for 2 h at room temperature with 100 μl/well of a mixture with the following enzymatic conjugate: 0.5 μg/ml streptavidin alkaline phosphatase (Jackson ImmunoResearch Laboratories, West Grove, PA).
To develop the spots, plates were washed four times with PBST, three times with PBS, and one time with water. Vector Blue substrate solution (Vector Laboratories, Burlingame, CA) was added at 100 μl/well for 5 to 15 min before rinsing with water and air drying. Digitized images were analyzed for spots using a CTL ImmunoSpot reader (Cellular Technology Limited, Cleveland, OH) or by counting spots using a stereozoom microscope (×20 magnification). ZIKV- or DENV-specific spots were calculated by subtracting the mean of the negative-control values of the replicates from the mean values of the specific stimulation. Positive responses had to be greater than four times the mean background, three standard deviations above the background, and ≥55 SFC/106 PBMCs.
Statistical analysis.
Statistical analyses were performed using the programs Prism 7 (GraphPad Software, San Diego, CA) and Stata v. 13.1 (College Station, TX). Data were expressed as geometric positive means ± standard deviations. Data comparisons were conducted using the Wilcoxon rank-sum and Fisher's exact tests, as relevant. A threshold of a P value of ≤0.05 was considered statistically significant.
ACKNOWLEDGMENTS
We thank Seema Thakore Meloni for help with statistical analysis and Nicholas Kushner and Neal Touzjian for technical advice with the modified anthrax delivery system. We also thank Ying Kai Chan for critical readings of the manuscript.
The Senegalese FSW cohort was supported in part by the National Institutes of Health (NIH) (grant numbers AI301795 and CA39805 to P.J.K.) and the U.S. Department of Defense (grant number DAMD-17-87-7072 to P.J.K.). This work was supported in part by the National Institutes of Allergy and Infectious Diseases, NIH (grant number R01AI110769-01 to W.-K.W.).
We have no conflicts of interest to declare.
REFERENCES
- 1.Lazear HM, Diamond MS. 2016. Zika virus: new clinical syndromes and its emergence in the Western Hemisphere. J Virol 90:4864–4875. doi: 10.1128/JVI.00252-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kuno G, Chang GJ. 2007. Full-length sequencing and genomic characterization of Bagaza, Kedougou, and Zika viruses. Arch Virol 152:687–696. doi: 10.1007/s00705-006-0903-z. [DOI] [PubMed] [Google Scholar]
- 3.Knipe DM, Howley PM. 2013. Fields virology, 6th ed Wolters Kluwer/Lippincott Williams & Wilkins Health, Philadelphia, PA. [Google Scholar]
- 4.Dick GW, Kitchen SF, Haddow AJ. 1952. Zika virus I isolations and serological specificity. Trans R Soc Trop Med Hyg 46:509–520. doi: 10.1016/0035-9203(52)90042-4. [DOI] [PubMed] [Google Scholar]
- 5.Faye O, Freire CC, Iamarino A, Faye O, de Oliveira JV, Diallo M, Zanotto PM, Sall AA. 2014. Molecular evolution of Zika virus during its emergence in the 20(th) century. PLoS Negl Trop Dis 8:e2636. doi: 10.1371/journal.pntd.0002636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Simpson DI. 1964. Zika virus infection in man. Trans R Soc Trop Med Hyg 58:335–338. doi: 10.1016/0035-9203(64)90200-7. [DOI] [PubMed] [Google Scholar]
- 7.Duffy MR, Chen TH, Hancock WT, Powers AM, Kool JL, Lanciotti RS, Pretrick M, Marfel M, Holzbauer S, Dubray C, Guillaumot L, Griggs A, Bel M, Lambert AJ, Laven J, Kosoy O, Panella A, Biggerstaff BJ, Fischer M, Hayes EB. 2009. Zika virus outbreak on Yap Island, Federated States of Micronesia. N Engl J Med 360:2536–2543. doi: 10.1056/NEJMoa0805715. [DOI] [PubMed] [Google Scholar]
- 8.Musso D, Nilles EJ, Cao-Lormeau VM. 2014. Rapid spread of emerging Zika virus in the Pacific area. Clin Microbiol Infect 20:O595–O596. doi: 10.1111/1469-0691.12707. [DOI] [PubMed] [Google Scholar]
- 9.Musso D. 2015. Zika virus transmission from French Polynesia to Brazil. Emerg Infect Dis 21:1887. doi: 10.3201/eid2110.151125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Campos GS, Bandeira AC, Sardi SI. 2015. Zika virus outbreak, Bahia, Brazil. Emerg Infect Dis 21:1885–1886. doi: 10.3201/eid2110.150847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zanluca C, Melo VC, Mosimann AL, Santos GI, Santos CN, Luz K. 2015. First report of autochthonous transmission of Zika virus in Brazil. Mem Inst Oswaldo Cruz 110:569–572. doi: 10.1590/0074-02760150192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hennessey M, Fischer M, Staples JE. 2016. Zika virus spreads to new areas–region of the Americas, May 2015-January 2016. MMWR Morb Mortal Wkly Rep 65:55–58. doi: 10.15585/mmwr.mm6503e1. [DOI] [PubMed] [Google Scholar]
- 13.Haddow AD, Schuh AJ, Yasuda CY, Kasper MR, Heang V, Huy R, Guzman H, Tesh RB, Weaver SC. 2012. Genetic characterization of Zika virus strains: geographic expansion of the Asian lineage. PLoS Negl Trop Dis 6:e1477. doi: 10.1371/journal.pntd.0001477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lanciotti RS, Kosoy OL, Laven JJ, Velez JO, Lambert AJ, Johnson AJ, Stanfield SM, Duffy MR. 2008. Genetic and serologic properties of Zika virus associated with an epidemic, Yap State, Micronesia, 2007. Emerg Infect Dis 14:1232–1239. doi: 10.3201/eid1408.080287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Musso D, Gubler DJ. 2016. Zika virus. Clin Microbiol Rev 29:487–524. doi: 10.1128/CMR.00072-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brasil P, Pereira JP Jr, Moreira ME, Ribeiro Nogueira RM, Damasceno L, Wakimoto M, Rabello RS, Valderramos SG, Halai UA, Salles TS, Zin AA, Horovitz D, Daltro P, Boechat M, Raja Gabaglia C, Carvalho de Sequeira P, Pilotto JH, Medialdea-Carrera R, Cotrim da Cunha D, Abreu de Carvalho LM, Pone M, Machado Siqueira A, Calvet GA, Rodrigues Baiao AE, Neves ES, Nassar de Carvalho PR, Hasue RH, Marschik PB, Einspieler C, Janzen C, Cherry JD, Bispo de Filippis AM, Nielsen-Saines K. 2016. Zika virus infection in pregnant women in Rio de Janeiro. N Engl J Med 375:2321–2334. doi: 10.1056/NEJMoa1602412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hills SL, Russell K, Hennessey M, Williams C, Oster AM, Fischer M, Mead P. 2016. Transmission of Zika virus through sexual contact with travelers to areas of ongoing transmission–continental United States, 2016. MMWR Morb Mortal Wkly Rep 65:215–216. doi: 10.15585/mmwr.mm6508e2. [DOI] [PubMed] [Google Scholar]
- 18.Oehler E, Watrin L, Larre P, Leparc-Goffart I, Lastere S, Valour F, Baudouin L, Mallet H, Musso D, Ghawche F. 2014. Zika virus infection complicated by Guillain-Barre syndrome–case report, French Polynesia, December 2013. Euro Surveill 19:20720. doi: 10.2807/1560-7917.ES2014.19.9.20720. [DOI] [PubMed] [Google Scholar]
- 19.Calvet G, Aguiar RS, Melo ASO, Sampaio SA, de Filippis I, Fabri A, Araujo ESM, de Sequeira PC, de Mendonca MCL, de Oliveira L, Tschoeke DA, Schrago CG, Thompson FL, Brasil P, Dos Santos FB, Nogueira RMR, Tanuri A, de Filippis AMB. 2016. Detection and sequencing of Zika virus from amniotic fluid of fetuses with microcephaly in Brazil: a case study. Lancet Infect Dis 16:653–660. doi: 10.1016/S1473-3099(16)00095-5. [DOI] [PubMed] [Google Scholar]
- 20.Rothman AL. 2011. Immunity to dengue virus: a tale of original antigenic sin and tropical cytokine storms. Nat Rev Immunol 11:532–543. doi: 10.1038/nri3014. [DOI] [PubMed] [Google Scholar]
- 21.Screaton G, Mongkolsapaya J, Yacoub S, Roberts C. 2015. New insights into the immunopathology and control of dengue virus infection. Nat Rev Immunol 15:745–759. doi: 10.1038/nri3916. [DOI] [PubMed] [Google Scholar]
- 22.Barba-Spaeth G, Dejnirattisai W, Rouvinski A, Vaney MC, Medits I, Sharma A, Simon-Loriere E, Sakuntabhai A, Cao-Lormeau VM, Haouz A, England P, Stiasny K, Mongkolsapaya J, Heinz FX, Screaton GR, Rey FA. 2016. Structural basis of potent Zika-dengue virus antibody cross-neutralization. Nature 536:48–53. doi: 10.1038/nature18938. [DOI] [PubMed] [Google Scholar]
- 23.Dejnirattisai W, Supasa P, Wongwiwat W, Rouvinski A, Barba-Spaeth G, Duangchinda T, Sakuntabhai A, Cao-Lormeau VM, Malasit P, Rey FA, Mongkolsapaya J, Screaton GR. 2016. Dengue virus sero-cross-reactivity drives antibody-dependent enhancement of infection with Zika virus. Nat Immunol 17:1102–1108. doi: 10.1038/ni.3515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stettler K, Beltramello M, Espinosa DA, Graham V, Cassotta A, Bianchi S, Vanzetta F, Minola A, Jaconi S, Mele F, Foglierini M, Pedotti M, Simonelli L, Dowall S, Atkinson B, Percivalle E, Simmons CP, Varani L, Blum J, Baldanti F, Cameroni E, Hewson R, Harris E, Lanzavecchia A, Sallusto F, Corti D. 2016. Specificity, cross-reactivity, and function of antibodies elicited by Zika virus infection. Science 353:823–826. doi: 10.1126/science.aaf8505. [DOI] [PubMed] [Google Scholar]
- 25.Swanstrom JA, Plante JA, Plante KS, Young EF, McGowan E, Gallichotte EN, Widman DG, Heise MT, de Silva AM, Baric RS. 2016. Dengue virus envelope dimer epitope monoclonal antibodies isolated from dengue patients are protective against Zika virus. mBio 7:e01123-. doi: 10.1128/mBio.01123-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Paul LM, Carlin ER, Jenkins MM, Tan AL, Barcellona CM, Nicholson CO, Michael SF, Isern S. 2016. Dengue virus antibodies enhance Zika virus infection. Clin Transl Immunol 5:e117. doi: 10.1038/cti.2016.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Priyamvada L, Quicke KM, Hudson WH, Onlamoon N, Sewatanon J, Edupuganti S, Pattanapanyasat K, Chokephaibulkit K, Mulligan MJ, Wilson PC, Ahmed R, Suthar MS, Wrammert J. 2016. Human antibody responses after dengue virus infection are highly cross-reactive to Zika virus. Proc Natl Acad Sci U S A 113:7852–7857. doi: 10.1073/pnas.1607931113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bardina SV, Bunduc P, Tripathi S, Duehr J, Frere JJ, Brown JA, Nachbagauer R, Foster GA, Krysztof D, Tortorella D, Stramer SL, Garcia-Sastre A, Krammer F, Lim JK. 2017. Enhancement of Zika virus pathogenesis by preexisting antiflavivirus immunity. Science 356:175–180. doi: 10.1126/science.aal4365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu X, Vaughan K, Weiskopf D, Grifoni A, Diamond MS, Sette A, Peters B. 2016. Identifying candidate targets of immune responses in Zika virus based on homology to epitopes in other flavivirus species. PLoS Curr 8:ecurrents.outbreaks.9aa2e1fb61b0f632f58a098773008c4b. doi: 10.1371/currents.outbreaks.9aa2e1fb61b0f632f58a098773008c4b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wen J, Tang WW, Sheets N, Ellison J, Sette A, Kim K, Shresta S. 2017. Identification of Zika virus epitopes reveals immunodominant and protective roles for dengue virus cross-reactive CD8+ T cells. Nat Microbiol 2:17036. doi: 10.1038/nmicrobiol.2017.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grifoni A, Pham J, Sidney J, O'Rourke PH, Paul S, Peters B, Martini SR, de Silva AD, Ricciardi MJ, Magnani DM, Silveira CGT, Maestri A, Costa PR, de-Oliveira-Pinto LM, de Azeredo EL, Damasco PV, Phillips E, Mallal S, de Silva AM, Collins M, Durbin A, Diehl SA, Cerpas C, Balmaseda A, Kuan G, Coloma J, Harris E, Crowe JE Jr, Stone M, Norris PJ, Busch M, Vivanco-Cid H, Cox J, Graham BS, Ledgerwood JE, Turtle L, Solomon T, Kallas EG, Watkins DI, Weiskopf D, Sette A. 4 October 2017. Prior dengue virus exposure shapes T cell immunity to Zika virus in humans. J Virol doi: 10.1128/JVI.01469-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Goletz TJ, Klimpel KR, Leppla SH, Keith JM, Berzofsky JA. 1997. Delivery of antigens to the MHC class I pathway using bacterial toxins. Hum Immunol 54:129–136. doi: 10.1016/S0198-8859(97)00081-5. [DOI] [PubMed] [Google Scholar]
- 33.Milne JC, Blanke SR, Hanna PC, Collier RJ. 1995. Protective antigen-binding domain of anthrax lethal factor mediates translocation of a heterologous protein fused to its amino- or carboxy-terminus. Mol Microbiol 15:661–666. doi: 10.1111/j.1365-2958.1995.tb02375.x. [DOI] [PubMed] [Google Scholar]
- 34.Ballard JD, Collier RJ, Starnbach MN. 1996. Anthrax toxin-mediated delivery of a cytotoxic T-cell epitope in vivo. Proc Natl Acad Sci U S A 93:12531–12534. doi: 10.1073/pnas.93.22.12531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Goletz TJ, Klimpel KR, Arora N, Leppla SH, Keith JM, Berzofsky JA. 1997. Targeting HIV proteins to the major histocompatibility complex class I processing pathway with a novel gp120-anthrax toxin fusion protein. Proc Natl Acad Sci U S A 94:12059–12064. doi: 10.1073/pnas.94.22.12059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lu Y, Friedman R, Kushner N, Doling A, Thomas L, Touzjian N, Starnbach M, Lieberman J. 2000. Genetically modified anthrax lethal toxin safely delivers whole HIV protein antigens into the cytosol to induce T cell immunity. Proc Natl Acad Sci U S A 97:8027–8032. doi: 10.1073/pnas.97.14.8027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liao Q, Strong AJ, Liu Y, Liu Y, Meng P, Fu Y, Touzjian N, Shao Y, Zhao Z, Lu Y. 2012. HIV vaccine candidates generate in vitro T cell response to putative epitopes in Chinese-origin rhesus macaques. Vaccine 30:1601–1608. doi: 10.1016/j.vaccine.2011.12.117. [DOI] [PubMed] [Google Scholar]
- 38.Sarr AD, Lu Y, Sankale JL, Eisen G, Popper S, Mboup S, Kanki PJ, Cao H. 2001. Robust HIV type 2 cellular immune response measured by a modified anthrax toxin-based enzyme-linked immunospot assay. AIDS Res Hum Retrovir 17:1257–1264. doi: 10.1089/088922201750461311. [DOI] [PubMed] [Google Scholar]
- 39.Duangchinda T, Dejnirattisai W, Vasanawathana S, Limpitikul W, Tangthawornchaikul N, Malasit P, Mongkolsapaya J, Screaton G. 2010. Immunodominant T-cell responses to dengue virus NS3 are associated with DHF. Proc Natl Acad Sci U S A 107:16922–16927. doi: 10.1073/pnas.1010867107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weiskopf D, Angelo MA, de Azeredo EL, Sidney J, Greenbaum JA, Fernando AN, Broadwater A, Kolla RV, De Silva AD, de Silva AM, Mattia KA, Doranz BJ, Grey HM, Shresta S, Peters B, Sette A. 2013. Comprehensive analysis of dengue virus-specific responses supports an HLA-linked protective role for CD8+ T cells. Proc Natl Acad Sci U S A 110:E2046–E2053. doi: 10.1073/pnas.1305227110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Turtle L, Bali T, Buxton G, Chib S, Chan S, Soni M, Hussain M, Isenman H, Fadnis P, Venkataswamy MM, Satishkumar V, Lewthwaite P, Kurioka A, Krishna S, Shankar MV, Ahmed R, Begum A, Ravi V, Desai A, Yoksan S, Fernandez S, Willberg CB, Kloverpris HN, Conlon C, Klenerman P, Satchidanandam V, Solomon T. 2016. Human T cell responses to Japanese encephalitis virus in health and disease. J Exp Med 213:1331–1352. doi: 10.1084/jem.20151517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Diallo M, Ba Y, Sall AA, Diop OM, Ndione JA, Mondo M, Girault L, Mathiot C. 2003. Amplification of the sylvatic cycle of dengue virus type 2, Senegal, 1999-2000: entomologic findings and epidemiologic considerations. Emerg Infect Dis 9:362–367. doi: 10.3201/eid0903.020219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Traore-Lamizana M, Zeller H, Monlun E, Mondo M, Hervy JP, Adam F, Digoutte JP. 1994. Dengue 2 outbreak in southeastern Senegal during 1990: virus isolations from mosquitoes (Diptera: Culicidae). J Med Entomol 31:623–627. [DOI] [PubMed] [Google Scholar]
- 44.Herrera BB, Chang CA, Hamel DJ, Mboup S, Ndiaye D, Imade G, Okpokwu J, Agbaji O, Bei AK, Kanki PJ. 2017. Continued transmission of Zika virus in humans in West Africa, 1992-2016. J Infect Dis 215:1546–1550. doi: 10.1093/infdis/jix182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cao-Lormeau VM, Blake A, Mons S, Lastere S, Roche C, Vanhomwegen J, Dub T, Baudouin L, Teissier A, Larre P, Vial AL, Decam C, Choumet V, Halstead SK, Willison HJ, Musset L, Manuguerra JC, Despres P, Fournier E, Mallet HP, Musso D, Fontanet A, Neil J, Ghawche F. 2016. Guillain-Barre syndrome outbreak associated with Zika virus infection in French Polynesia: a case-control study. Lancet 387:1531–1539. doi: 10.1016/S0140-6736(16)00562-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cugola FR, Fernandes IR, Russo FB, Freitas BC, Dias JL, Guimaraes KP, Benazzato C, Almeida N, Pignatari GC, Romero S, Polonio CM, Cunha I, Freitas CL, Brandao WN, Rossato C, Andrade DG, Faria DP, Garcez AT, Buchpigel CA, Braconi CT, Mendes E, Sall AA, Zanotto PM, Peron JP, Muotri AR, Beltrao-Braga PC. 2016. The Brazilian Zika virus strain causes birth defects in experimental models. Nature 534:267–271. doi: 10.1038/nature18296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liang Q, Luo Z, Zeng J, Chen W, Foo SS, Lee SA, Ge J, Wang S, Goldman SA, Zlokovic BV, Zhao Z, Jung JU. 2016. Zika virus NS4A and NS4B proteins deregulate Akt-mTOR signaling in human fetal neural stem cells to inhibit neurogenesis and induce autophagy. Cell Stem Cell 19:663–671. doi: 10.1016/j.stem.2016.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.de Alwis R, Bangs DJ, Angelo MA, Cerpas C, Fernando A, Sidney J, Peters B, Gresh L, Balmaseda A, de Silva AD, Harris E, Sette A, Weiskopf D. 2016. Immunodominant dengue virus-specific CD8+ T cell responses are associated with a memory PD-1+ phenotype. J Virol 90:4771–4779. doi: 10.1128/JVI.02892-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Grifoni A, Angelo MA, Lopez B, O'Rourke PH, Sidney J, Cerpas C, Balmaseda A, Silveira CGT, Maestri A, Costa PR, Durbin AP, Diehl SA, Phillips E, Mallal S, De Silva AD, Nchinda G, Nkenfou C, Collins MH, de Silva AM, Lim MQ, Macary PA, Tatullo F, Solomon T, Satchidanandam V, Desai A, Ravi V, Coloma J, Turtle L, Rivino L, Kallas EG, Peters B, Harris E, Sette A, Weiskopf D. 2017. Global assessment of dengue virus-specific CD4(+) T cell responses in dengue-endemic areas. Front Immunol 8:1309. doi: 10.3389/fimmu.2017.01309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mongkolsapaya J, Dejnirattisai W, Xu XN, Vasanawathana S, Tangthawornchaikul N, Chairunsri A, Sawasdivorn S, Duangchinda T, Dong T, Rowland-Jones S, Yenchitsomanus PT, McMichael A, Malasit P, Screaton G. 2003. Original antigenic sin and apoptosis in the pathogenesis of dengue hemorrhagic fever. Nat Med 9:921–927. doi: 10.1038/nm887. [DOI] [PubMed] [Google Scholar]
- 51.Dung NT, Duyen HT, Thuy NT, Ngoc TV, Chau NV, Hien TT, Rowland-Jones SL, Dong T, Farrar J, Wills B, Simmons CP. 2010. Timing of CD8+ T cell responses in relation to commencement of capillary leakage in children with dengue. J Immunol 184:7281–7287. doi: 10.4049/jimmunol.0903262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kanki PJ, Travers KU, S MB, Hsieh CC, Marlink RG, Gueye NA, Siby T, Thior I, Hernandez-Avila M, Sankale JL, Hsieh C-C, Hernandez-Avila M, Ndoye I. 1994. Slower heterosexual spread of HIV-2 than HIV-1. Lancet 343:943–946. [DOI] [PubMed] [Google Scholar]
- 53.Alm E, Lesko B, Lindegren G, Ahlm C, Soderholm S, Falk KI, Lagerqvist N. 2014. Universal single-probe RT-PCR assay for diagnosis of dengue virus infections. PLoS Negl Trop Dis 8:e3416. doi: 10.1371/journal.pntd.0003416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lai CY, Tsai WY, Lin SR, Kao CL, Hu HP, King CC, Wu HC, Chang GJ, Wang WK. 2008. Antibodies to envelope glycoprotein of dengue virus during the natural course of infection are predominantly cross-reactive and recognize epitopes containing highly conserved residues at the fusion loop of domain II. J Virol 82:6631–6643. doi: 10.1128/JVI.00316-08. [DOI] [PMC free article] [PubMed] [Google Scholar]