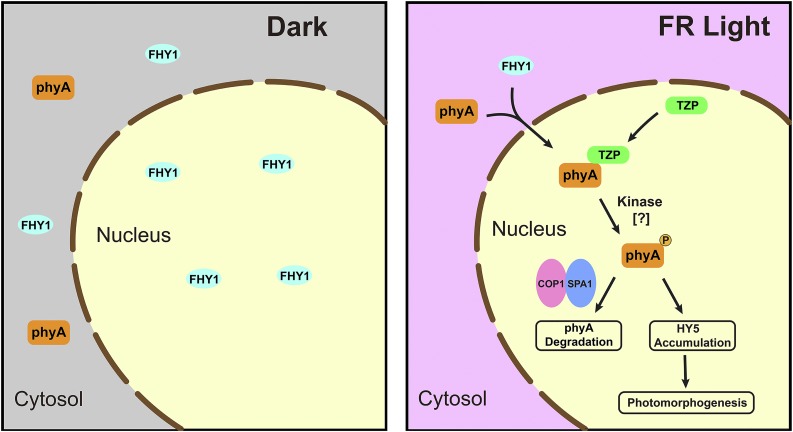
TZP is a positive regulator of phyA signaling and is required for the formation of a phosphorylated nuclear form of phyA in far-red light.
Abstract
Phytochrome A (phyA) is the primary plant photoreceptor responsible for perceiving and mediating various responses to far-red (FR) light and is essential for survival in canopy shade. In this study, we identified two Arabidopsis thaliana mutants that grew longer hypocotyls in FR light. Genetic analyses showed that they were allelic and their FR phenotypes were caused by mutations in the gene named TANDEM ZINC-FINGER/PLUS3 (TZP), previously shown to encode a nuclear protein involved in blue light signaling and phyB-dependent regulation of photoperiodic flowering. We show that the expression of TZP is dramatically induced by light and that TZP proteins are differentially modified in different light conditions. Furthermore, we show that TZP interacts with both phyA and FAR-RED ELONGATED HYPOCOTYL1 (FHY1) and regulates the abundance of phyA, FHY1, and ELONGATED HYPOCOTYL5 proteins in FR light. Moreover, our data indicate that TZP is required for the formation of a phosphorylated form of phyA in the nucleus in FR light. Together, our results identify TZP as a positive regulator of phyA signaling required for phosphorylation of the phyA photoreceptor, thus suggesting an important role of phosphorylated phyA in inducing the FR light response.
INTRODUCTION
Phytochromes are photoreceptors that most strongly absorb red (R) and far-red (FR) light (600–750 nm), but also absorb the blue (B)/UV-A wavelengths (320–500 nm) (Li et al., 2011). The Arabidopsis thaliana genome encodes five phytochrome proteins, designated phytochrome A (phyA) to phyE, which are generally categorized into two groups: light-labile type I (phyA) and light-stable type II (phyB to phyE) (Sharrock and Quail, 1989; Li et al., 2011). PhyA is the most abundant phytochrome in etiolated seedlings, whereas phyB is most abundant in light-grown plants. PhyA is the primary photoreceptor responsible for mediating photomorphogenic responses in FR light, whereas phyB is the predominant phytochrome regulating deetiolation responses in R light (Franklin and Quail, 2010; Li et al., 2011). Phytochromes exist in two distinct but interconvertible forms: the R light-absorbing Pr form and the FR light-absorbing Pfr form, and the Pfr form is generally considered to be the biologically active form (Li et al., 2011). Phytochromes are synthesized in their inactive Pr form in the cytosol; upon light irradiation, they are converted to their biologically active Pfr form and translocate into the nucleus, where they trigger a signaling cascade that alters the expression of many target genes and ultimately leads to the modulation of many biological responses (Jiao et al., 2007; Franklin and Quail, 2010; Li et al., 2011).
Type I and type II phytochromes have virtually identical photophysical properties, based on which they are expected to have an action peak in R light (Eichenberg et al., 2000; Possart and Hiltbrunner, 2013). However, the phenotype of phyA mutants indicates that the action peak of phyA is in FR, even though only ∼2% of phyA is in the Pfr form in FR light (Mancinelli, 1994; Rausenberger et al., 2011). Genetic screens initiated in the early 1990s have identified many mutants that are defective in FR light responses. The far-red elongated hypocotyl (fhy) mutants, including fhy1 to fhy3, were first reported in 1993 (Whitelam et al., 1993). This pioneering report showed that the FHY2 locus was allelic to PHYA (Whitelam et al., 1993); FHY1 and FHY3 were cloned later as separate loci (Desnos et al., 2001; Wang and Deng, 2002). When the FHY3 gene was cloned, sequence alignments revealed that FHY3 shares high sequence similarity with FAR-RED IMPAIRED RESPONSE1 (FAR1), a previously identified phyA signaling component (Hudson et al., 1999; Wang and Deng, 2002). Subsequent studies identified a homolog of FHY1, named FHY1-LIKE (FHL), based on its sequence homology to FHY1 (Zhou et al., 2005). Later studies indicated that FHY1 and FHL are essential for nuclear accumulation of light-activated phyA and subsequent light responses (Hiltbrunner et al., 2005, 2006; Rösler et al., 2007; Genoud et al., 2008), whereas FHY3 and FAR1 are transposase-derived transcription factors acting together to directly activate the transcription of FHY1 and FHL, thus indirectly regulating phyA nuclear accumulation and responses (Lin et al., 2007, 2008). Recently, it was proposed that phyA-specific properties, such as the rapid degradation of the Pfr form and the interaction with the nuclear transport proteins FHY1 and FHL, shift the action peak of phyA from R to FR light (Rausenberger et al., 2011).
In addition to the above-mentioned factors regulating phyA nuclear accumulation, numerous signaling intermediates have been identified that are either specific for phyA or shared by type II phytochromes or multiple types of photoreceptors. ELONGATED HYPOCOTYL5 (HY5), a bZIP family transcription factor, acts as a key positive regulator of photomorphogenic development under a wide spectrum of wavelengths, including FR, R, B, and UV-B, by binding directly to the promoters of a large number of light-responsive genes (Koornneef et al., 1980; Oyama et al., 1997; Osterlund et al., 2000; Ulm et al., 2004; Lee et al., 2007; Li et al., 2010; Zhang et al., 2011). A group of basic helix-loop-helix (bHLH) class transcription factors, also known as PHYTOCHROME INTERACTING FACTORs (PIFs), have been shown to accumulate in dark-grown seedlings and together act as constitutive repressors of photomorphogenesis, while light-activated phytochromes induce rapid phosphorylation and degradation of these repressors (Ni et al., 1998, 2013, 2014, 2017; Duek and Fankhauser, 2005; Al-Sady et al., 2006; Castillon et al., 2007; Leivar et al., 2008). LONG HYPOCOTYL IN FAR-RED1 (HFR1) and LONG AFTER FAR-RED LIGHT1 (LAF1) are atypical bHLH and R2R3-MYB transcription factors, respectively, that both act as positive regulators of phyA signaling (Fairchild et al., 2000; Fankhauser and Chory, 2000; Soh et al., 2000; Ballesteros et al., 2001; Jang et al., 2007). CONSTITUTIVE PHOTOMORPHOGENIC1 (COP1) interacts with SUPPRESSOR OF phyA-105 (SPA) proteins to form E3 ligase complexes, targeting several key positive regulators of light signaling for degradation, such as HY5 (Osterlund et al., 2000), HY5 HOMOLOG (HYH; Holm et al., 2002), LAF1 (Seo et al., 2003), HFR1 (Duek et al., 2004; Jang et al., 2005; Yang et al., 2005), and the phytochromes (Seo et al., 2004; Jang et al., 2010). It was recently shown that light-activated phyA and phyB interact with SPA proteins, leading to the disruption and inactivation of the COP1/SPA complexes and promotion of photomorphogenesis (Sheerin et al., 2015).
Here, we report the identification of two mutants that develop longer hypocotyls in FR light, both of whose phenotypes are caused by mutations in TANDEM ZINC-FINGER/PLUS3 (TZP). TZP was previously identified as the locus regulating morning-specific growth by quantitative trait locus mapping between Arabidopsis ecotypes Bay-0 and Shahdara (Loudet et al., 2008). A later study showed that TZP is localized to transcriptionally active nuclear photobodies to directly activate FLOWERING LOCUS T expression and promote flowering by interacting with phyB (Kaiserli et al., 2015). Here, by systematically analyzing the phenotypes of two tzp mutants, we show that TZP not only acts as a positive regulator of FR light responses, but also as a negative regulator of blue light signaling. Moreover, our data indicate that TZP interacts with both phyA and FHY1, and regulates phyA protein abundance and phosphorylation in FR light. Therefore, our study demonstrates that TZP is a signaling component closely tied to the action of phytochrome photoreceptors.
RESULTS
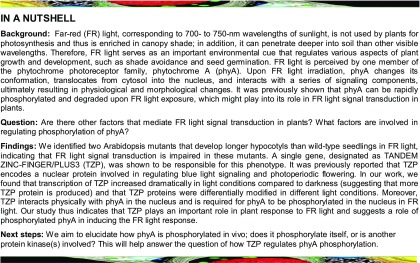
Identification of Two tzp Alleles Displaying Long Hypocotyls in FR Light
To discover novel components involved in phyA signaling, we screened a mutant library of ∼25,000 activation-tagged Arabidopsis lines (Qin et al., 2003) for mutants displaying altered response to FR light. The screen initially produced ∼50 mutants whose hypocotyls were either longer or shorter than the wild type. Two mutants, both growing longer hypocotyls in FR light, were selected for further investigation in this study. When grown under different fluence rates of continuous FR light, these two mutants always grew significantly longer hypocotyls than the wild type (Supplemental Figure 1). Genetic analyses showed that both were recessive mutants and, interestingly, their F1 hybrids showed long hypocotyls, indicating that they were allelic (Figure 1A). We therefore conducted whole-genome DNA sequencing of these two mutants and found that only one gene, At5g43630, encoding an 831-amino acid nuclear protein designated as TZP in previous publications (Loudet et al., 2008; Kaiserli et al., 2015), harbored mutations in both mutants. In one mutant, an A-to-T substitution at position 1063 from the start codon changed Lys-355 (AAG) to a premature stop codon (TAG), while in another mutant, DNA duplication and structural rearrangement occurred in the third exon, resulting in the disruption of TZP transcription (Figure 1B; Supplemental Figure 2). We therefore named these two mutants tzp-1 (rearrangement) and tzp-2 (point mutation), respectively.
Figure 1.
Genetic and Molecular Characterization of the Two tzp Mutants.
(A) Genetic analysis showing that the two FR-deficient mutants are allelic.
(B) Schematic illustration of the exon-intron structure of TZP. Exons are shown as black boxes, 5′- and 3′-untranslated regions (UTRs) are shown as gray boxes, and introns are shown as lines. The mutations in tzp-1 and tzp-2 are also shown.
(C) The long hypocotyl phenotype of tzp-1 and tzp-2 mutants in FR light was fully rescued by transformation with TZPp:TZP-3flag. The phenotypes of two representative complementation lines of tzp-1 and tzp-2 are shown.
(D) Phenotypes of Col, tzp-1, tzp-2, and two representative tzp complementation lines grown in blue light for 4 d.
(E) Hypocotyl lengths of Col, tzp-1, tzp-2, and two representative tzp complementation lines grown in B light for 4 d. Error bars represent sd from 15 seedlings. ***P < 0.001 (Student’s t test) for the indicated pairs of seedlings.
Bar = 1 mm in (A), (C), and (D).
To further confirm that the phenotypes of both mutants were caused by mutations in TZP, we generated the TZPp:TZP-3flag construct and transformed it into tzp-1 and tzp-2 mutants, respectively. Multiple homozygous lines were obtained, and the long hypocotyl phenotypes of tzp-1 and tzp-2 were both fully rescued by transgenic TZP-3flag (Figure 1C; Supplemental Figure 3), demonstrating that the mutations of TZP were responsible for the long hypocotyl phenotypes of tzp-1 and tzp-2 mutants in FR light.
It was previously reported that the 35S:TZP-GFP seedlings displayed longer hypocotyls in blue light but did not show any significant changes in FR and R light (Kaiserli et al., 2015). To comprehensively understand the role of TZP in light signaling, we also analyzed the phenotypes of tzp mutants and tzp complementation lines grown in darkness (D), R, B, and white (W) light conditions. Notably, the tzp mutants did not show any differences compared with the wild type in D, R, and W light conditions (Supplemental Figure 4). However, consistent with the function of TZP to promote hypocotyl growth in blue light (Kaiserli et al., 2015), both tzp mutants displayed significantly shorter hypocotyls under blue light, and introduction of TZP-3flag under the control of the native TZP promoter successfully rescued the short hypocotyl phenotype of tzp mutants in blue light (Figures 1D and 1E), indicating that TZP negatively regulates blue light signaling although it positively regulates FR light responses.
TZP Regulates Anthocyanin Accumulation and Light-Responsive Gene Expression in FR Light
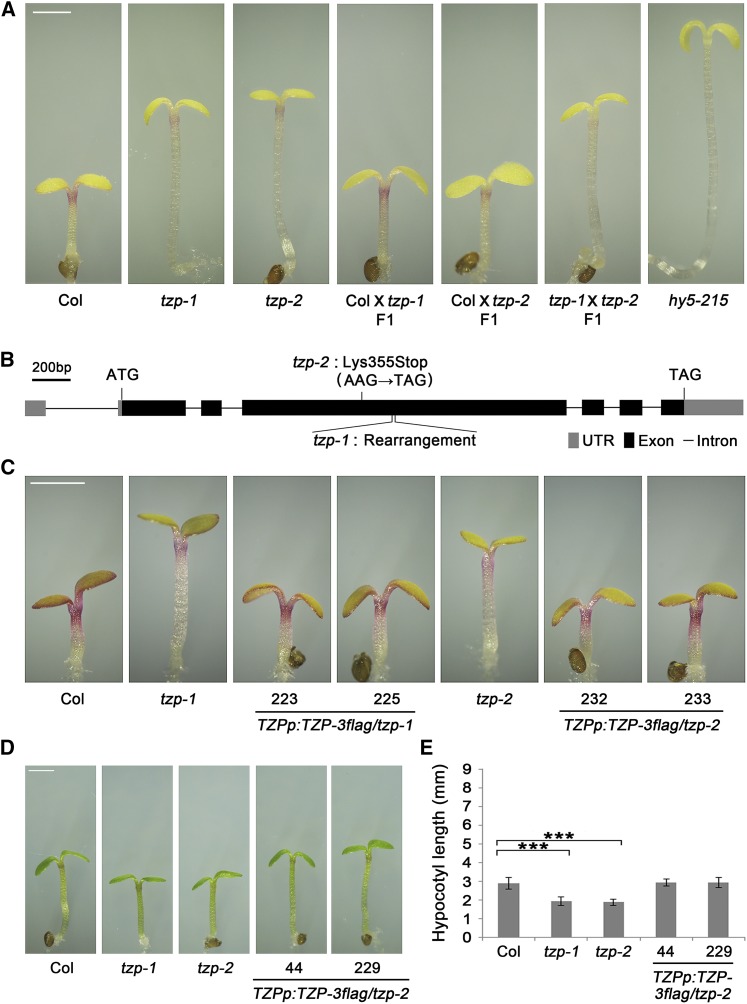
Next, we asked whether TZP is involved in other FR light-regulated responses, e.g., anthocyanin accumulation and FR block of greening (Barnes et al., 1996), in addition to regulating hypocotyl elongation. Our data indicated that the tzp mutants accumulated less anthocyanin (Figure 2A), but did not show any defect in FR block of greening (Supplemental Figure 5), indicating that TZP regulates some, but not all, branches of FR light response.
Figure 2.
TZP Regulates Anthocyanin Accumulation and Light-Responsive Gene Expression in FR Light.
(A) Anthocyanin contents of 4-d-old FR-grown Col, tzp-1, tzp-2, and one tzp complementation line seedlings. Values are means ± sd of three pools of seedlings.
(B) RT-qPCR analysis showing the expression of six anthocyanin biosynthetic genes (CHS, CHI, F3H, F3′H, DFR, and LDOX) in 4-d-old FR-grown Col, tzp-1, tzp-2, and one tzp complementation line seedlings.
(C) RT-qPCR analysis showing the expression of several phyA signaling components (PHYA, FHY1, FHL, HY5, and HYH) in 4-d-old FR-grown Col, tzp-1, tzp-2, and one tzp complementation line seedlings.
(D) RT-qPCR analysis showing that IAA19 is upregulated but ELIP2 is downregulated in tzp mutants in FR light.
Error bars in (B) to (D) represent sd of three technical replicates.
As anthocyanin is synthesized in multiple steps catalyzed by several enzymes including chalcone synthase (CHS), chalcone isomerase (CHI), flavonone 3-hydroxylase (F3H), flavonoid 3′-hydroxylase (F3′H), dihydroflavonol 4-reductase (DFR), and leucoanthocyanidin dioxygenase (LDOX) (Shin et al., 2007), we next examined whether the expression of genes encoding these enzymes was changed in tzp mutants. Our RT-qPCR data showed that all six genes were downregulated in two tzp mutants, but their expression was largely restored in the tzp complementation line (Figure 2B), suggesting that TZP regulates anthocyanin accumulation through modulating the expression of these anthocyanin biosynthetic genes.
We also examined whether TZP regulates the expression of other light-responsive genes in FR light. Interestingly, our RT-qPCR data indicated that the expression of PHYA was not changed in tzp mutants; however, FHY1 and FHL were evidently upregulated, whereas HY5 and HYH were slightly downregulated in tzp mutants in FR light (Figure 2C). The expression of PHYB, PIF1, PIF3, and PIF5 was not obviously changed in tzp mutants in FR light, whereas PIF4 was slightly upregulated in tzp mutants (Supplemental Figure 6A). We also examined the expression of several other types of light-responsive genes and found that INDOLE-3-ACETIC ACID INDUCIBLE19 (IAA19), involved in auxin signaling (Tatematsu et al., 2004), was upregulated, whereas EARLY LIGHT-INDUCED PROTEIN2 (ELIP2), regulating chlorophyll synthesis (Tzvetkova-Chevolleau et al., 2007), was downregulated in tzp mutants in FR light (Figure 2D). In addition, DWARF4 (DWF4), encoding a key enzyme in brassinosteroid biosynthesis (Choe et al., 1998), EXTENSIN3 (EXT3), encoding a structural glycoprotein required for cell wall assembly (Cannon et al., 2008), EXPANSIN2 (EXP2), and XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE17 (XTH17), encoding the key enzymes implicated in cell wall loosening and cell elongation (Cosgrove, 2005), were previously shown to be regulated by light (Jing et al., 2013; Shi et al., 2013), but their expression was not evidently changed in tzp mutants in FR light (Supplemental Figure 6B). Collectively, our data indicate that TZP regulates a portion of light-responsive genes in FR light, consistent with its proposed role as a transcriptional regulator (Kaiserli et al., 2015).
TZP Expression Is Induced by Light
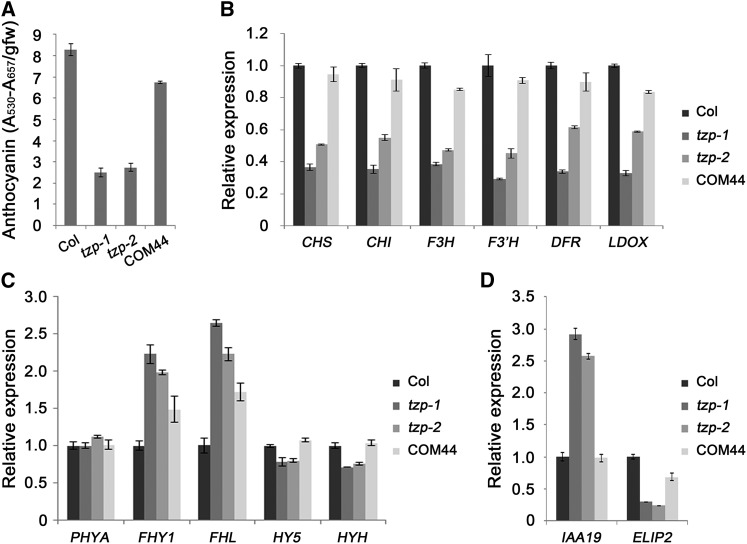
Next, we investigated whether TZP expression is regulated by light. We compared TZP transcript levels in wild-type and two tzp mutant seedlings grown under different light conditions by RT-qPCR. Our data showed that in the wild type, TZP transcript levels were significantly elevated in light compared with D, with at least a 10-fold induction in FR, B, and W light (Figure 3A). Moreover, our RT-qPCR results showed that the TZP transcript levels in tzp-2 mutant seedlings were about half of those in wild-type seedlings grown in D, FR, B, and W (except for R) light, respectively (Figure 3A), indicating that TZP positively regulated its own expression. The tzp-1 mutant is a null allele of TZP since no TZP transcripts were detected in all tested light conditions (Figure 3A).
Figure 3.
The Expression of TZP Is Dramatically Induced by Light.
(A) RT-qPCR analysis showing the relative expression of TZP in 4-d-old Col, tzp-1, and tzp-2 mutants grown in D or continuous FR, R, B, and W light conditions.
(B) RT-qPCR analysis showing that FR, R, and B-induced expression of TZP is mediated through phyA, phyB, and cry1/cry2, respectively.
(C) GUS staining of 4-d-old homozygous TZPp:GUS transgenic seedlings grown in D or continuous FR, R, B, and W light. More than 10 independent homozygous TZPp:GUS transgenic seedlings were stained for GUS activity, and the results of one representative line are shown. Bar = 1 mm.
Error bars in (A) and (B) represent sd of three technical replicates.
We then examined the TZP expression levels in the wild type and phytochrome (i.e., phyA and phyB) and cryptochrome mutants (i.e., cry1, cry2, and cry1 cry2) grown in FR, R, and B light conditions, respectively. Our RT-qPCR data showed that FR and R-induced TZP expression was abolished in phyA and phyB mutants, respectively (Figure 3B), indicating that phyA and phyB mediate FR and R-induced TZP expression, respectively. In blue light, however, TZP expression was decreased in cry1 or cry2 single mutants compared with that in the wild type, but further decreased in cry1 cry2 double mutants (Figure 3B), indicating that both cry1 and cry2 mediate blue light-induced TZP expression.
To further characterize how light changes the spatial expression of TZP, we generated TZPp:GUS lines in which expression of the GUS gene was controlled by the native TZP promoter. GUS activity assays indicated that GUS was widely expressed in all seedling tissues in the light including cotyledons, hypocotyls, and roots (Figure 3C). Low levels of GUS activity were also observed in dark-grown seedlings, consistent with the low expression level of TZP in darkness revealed by RT-qPCR assays (Figure 3A).
Light Regulates the Modifications of TZP Proteins
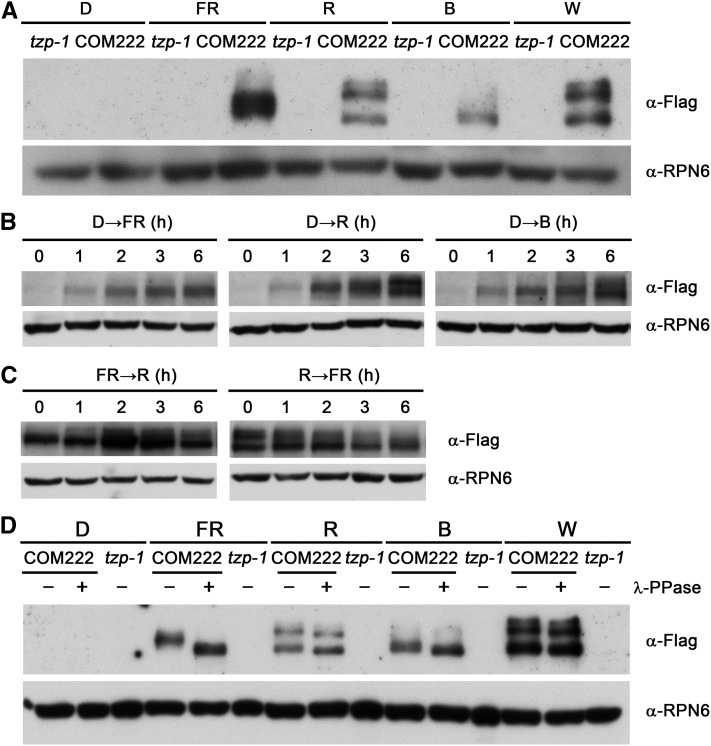
We then examined whether TZP protein accumulation is also regulated by light. The tzp-1 and tzp complementation line seedlings were grown in D, FR, R, B, and W light for 4 d and then harvested and subjected to immunoblotting. TZP proteins only accumulated in light, and, surprisingly, different patterns of TZP proteins were observed in different light conditions (Figure 4A). Notably, the pattern of TZP in W light is most close to that in R light (Figure 4A). The fact that no bands were observed in tzp-1 seedlings grown in the same conditions indicates that the various forms observed in tzp complementation seedlings in FR, R, B, and W light are not cross-reacting bands.
Figure 4.
TZP Proteins Are Differentially Modified in Different Light Conditions.
(A) Immunoblots showing TZP protein levels in 4-d-old tzp-1 and homozygous tzp complementation seedlings grown in D or continuous FR, R, B, and W light.
(B) Immunoblots showing that TZP proteins accumulated in different patterns after the 4-d-old etiolated tzp complementation seedlings were transferred to FR, R, or B light conditions for the indicated time periods.
(C) Immunoblots showing that the distinct patterns of TZP proteins under FR and R light could be completely interconverted after 6 h of exposure to R and FR light, respectively.
(D) TZP is a phosphoprotein in light. Total proteins extracted from 4-d-old tzp-1 and homozygous tzp complementation seedlings grown in D or continuous FR, R, B, and W light conditions were treated without (−) or with (+) λ-PPase for 1 h and then analyzed by immunoblots probed with the anti-flag antibodies.
Anti-RPN6 was used as a sample loading control.
To further confirm that different patterns of TZP proteins were indeed specifically regulated by different light, the tzp complementation seedlings were first grown in D for 4 d, then transferred to FR, R, and B light for the indicated time periods ranging from 1 to 6 h, and then harvested and subjected to immunoblotting. We observed that TZP proteins accumulated within 1 h of light exposure, and there were no obvious differences in band patterns in 2 h after the etiolated tzp complementation seedlings were transferred to FR, R, or B light (Figure 4B). However, the band patterns of TZP proteins started to differ at 3 h, and different patterns were finally formed at 6 h of exposure to FR, R, and B light (Figure 4B). This observation was further supported by the transfer between FR and R light, under which light responses are solely regulated by phytochromes. It was shown that the distinct patterns of TZP proteins under FR and R light could be completely interconverted after 6 h of exposure to R and FR light, respectively (Figure 4C). Taken together, our data demonstrate that TZP protein patterns are differentially regulated by different light conditions.
To investigate whether protein phosphorylation contributed to the formation of various forms of TZP in light, we treated the total proteins extracted from tzp complementation seedlings grown in FR, R, B, and W light with or without protein phosphatase. Our immunoblot data indicated that although no specific bands of TZP proteins were obviously absent after phosphatase treatment, all TZP forms observed in light conditions became more or less smaller, and in particular, the molecular weight of TZP in FR light displayed a dramatic decrease after phosphatase treatment (Figure 4D). Collectively, our data indicate that TZP is a phosphoprotein in light (especially in FR) and that different bands observed in tzp complementation seedlings likely represent differentially modified forms of TZP in different light conditions.
Genetic Relationship between TZP and phyA/phyB
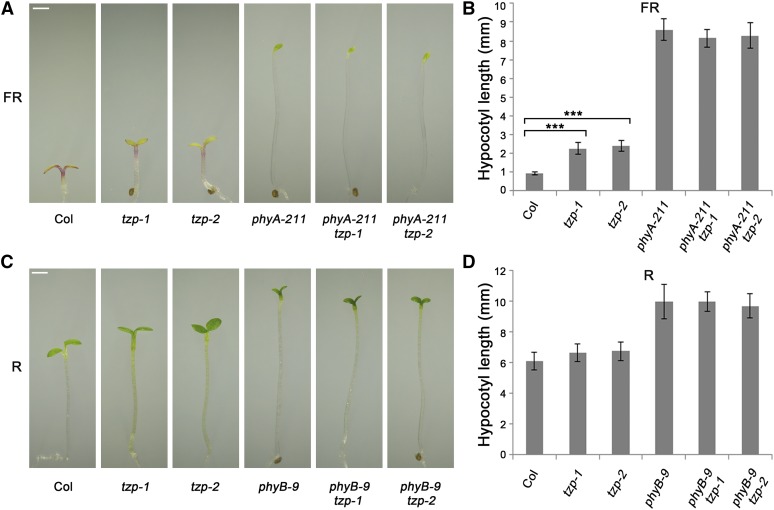
To determine the genetic relationships between TZP and phyA and phyB, we generated double mutants of tzp-1 and tzp-2 with phyA-211 and phyB-9. We grew the phyA-211 tzp-1 and phyA-211 tzp-2 mutants in continuous FR light for 4 d and observed that the hypocotyl lengths of phyA-211 tzp-1 and phyA-211 tzp-2 mutant seedlings were indistinguishable from those of phyA-211 single mutants (Figures 5A and 5B). Similar observations were also made when we compared phyB-9 tzp-1 and phyB-9 tzp-2 with phyB-9 mutants grown in continuous R light (Figures 5C and 5D). Together, our data suggest that phyA and phyB are epistatic to tzp under FR and R light, respectively.
Figure 5.
Genetic Analyses Showing That phyA and phyB Are Epistatic to tzp.
(A) and (B) Phenotype (A) and hypocotyl lengths (B) of Col, tzp-1, tzp-2, phyA-211, phyA-211 tzp-1, and phyA-211 tzp-2 seedlings grown for 4 d in FR light.
(C) and (D) Phenotype (C) and hypocotyl lengths (D) of Col, tzp-1, tzp-2, phyB-9, phyB-9 tzp-1, and phyB-9 tzp-2 seedlings grown for 4 d in R light.
Bar = 1 mm in (A) and (C). Error bars in (B) and (D) represent sd from 15 seedlings. ***P < 0.001 (Student’s t test) for the indicated pairs of seedlings.
TZP Interacts with phyA and FHY1
It was previously shown that TZP interacts with full-length apo-phyB but not apo-phyA in GAL4 yeast two-hybrid assays (Kaiserli et al., 2015). When we extracted the chromophore phycocyanobilin from Spirulina and added it to the GAL4 yeast system to allow the phytochromes to form Pr or Pfr forms after FR or R light pulse treatments, we also observed that TZP only interacted with holo-phyB but not with holo-phyA (Supplemental Figure 7).
To further investigate whether TZP could interact with phyA, we employed the LexA yeast two-hybrid system. We generated bait vectors expressing either the N-terminal, C-terminal, PAS-related domain (designated as C1), or histidine kinase-related domain (designated as C2) of phyA and phyB apoproteins fused to the LexA DNA binding domain and prey vectors expressing either the full-length, N-terminal, or the C-terminal domains of TZP fused to the activation domain (AD) (Figures 6A and 6B). Intriguingly, we found that the C-terminal domains of both phyA and phyB interacted with the full-length and N-terminal domains of TZP in this system (Figure 6C). In particular, we observed that the histidine kinase-related domain (C2) of phyA (but not C2 of phyB) interacted with the C-terminal domain of TZP (Figure 6C). This interaction was confirmed by firefly luciferase complementation imaging (LCI) assays (Figure 6D).
Figure 6.
TZP Interacts with phyA.
(A) Schematic diagram of bait proteins (PHYA/B-N, PHYA/B-C, PHYA/B-C1, and PHYA/B-C2 fused with LexA DNA binding domains). NTE, N-terminal extension; PRD, PAS-related domain; HKRD, histidine kinase-related domain; H, hinge.
(B) Schematic diagram of prey proteins (TZP, TZP-N, and TZP-C fused with AD domains). ZF, zinc finger.
(C) Yeast two-hybrid assays showing that the C-terminal domains of phyA and phyB both interacted with TZP.
(D) LCI assays showing that PHYA-C2 interacted with TZP-C in plant cells. Bar = 1 cm.
(E) In vitro pull-down of PHYA-C2 (the HKRD domain) with TZP-C2 (the PLUS3 domain). The 6×His-tagged PHYA-C2 proteins pulled down with GST-TZP-C1, GST-TZP-C2 or GST were detected by anti-His antibody. Input, 6% of the purified 6×His-tagged target proteins used in pull-down assays.
(F) Co-IP assays showing that TZP interacted with phyA and phyB in vivo. Left, the tzp-1 and homozygous tzp complementation seedlings were first grown in FR for 4 d and then the total proteins were extracted and treated with 5-min R light or with 5-min R light followed by 5-min FR light (R+FR) before immunoprecipitation. Right, the tzp-1 and homozygous tzp complementation seedlings were first grown in R for 4 d and then the total proteins were extracted and treated with 5-min FR light or with 5-min FR light followed by 5-min red light (FR+R) before immunoprecipitation. After light treatments, total proteins were incubated with anti-flag M2 Affinity Gel (Sigma-Aldrich). The total and precipitated proteins were subjected to immunoblot analyses with antibodies against flag, phyA, phyB, and RPN6, respectively. The asterisk indicates a cross-reacting band recognized by our anti-phyB antibodies.
Since the C-terminal domain of TZP includes the tandem zinc-finger and PLUS3 domains, we performed in vitro pull-down assays to further investigate which domain was responsible for interacting with phyA-C2. Our data showed that GST-TZP-C2 (i.e., PLUS3 domain), but not GST-TZP-C1 (i.e., tandem zinc-finger domain) or GST alone, was able to pull down the C2 domain of phyA (Figure 6E). Collectively, these data indicated that phyA interacted with TZP in vitro and in yeast cells and that the PLUS3 domain of TZP and the histidine kinase-related domain of phyA mediated their interaction.
To confirm the physical interaction between TZP and phyA in vivo, we conducted coimmunoprecipitation (co-IP) assays using tzp complementation seedlings grown 4 d in FR light. To determine which form (Pr or Pfr) of phyA associated with TZP more strongly, total proteins were exposed to 5 min of R light (for conversion to the Pfr form) or 5 min of R light exposure immediately followed by 5 min of FR light pulse (for conversion back to the Pr form). As shown in Figure 6F, phyA coprecipitated with TZP-3flag but not in tzp mutant seedlings, and the observation that similar amounts of phyA were coprecipitated by the anti-flag antibody in both light pulse treatments indicated that the Pfr and Pr forms of phyA associated with TZP with similar affinity. We also grew the tzp complementation seedlings in R for 4 d and then conducted light pulse treatments and the co-IP assays to examine the in vivo association of TZP with phyB. Our data showed that phyB also coprecipitated with TZP-3flag, consistent with a previous study (Kaiserli et al., 2015). Moreover, we also observed that both the Pfr and Pr forms of phyB associated with TZP similarly in yeast cells and in vivo (Figure 6F; Supplemental Figure 7B).
Because FHY1/FHL physically interact with phyA and play an important role in importing light-activated phyA into the nucleus, we next asked whether TZP could interact with FHY1. Our yeast two-hybrid and LCI assay data both showed that both the N- and C-terminal domains of TZP could interact with FHY1 (Supplemental Figures 8A and 8B); interestingly, the PLUS3 domain of TZP responsible for interacting with phyA could also interact with FHY1 (Supplemental Figure 8C). Collectively, our data demonstrate that TZP interacts with both phyA and FHY1.
TZP Regulates the Protein Abundance of phyA, FHY1, and HY5 in FR
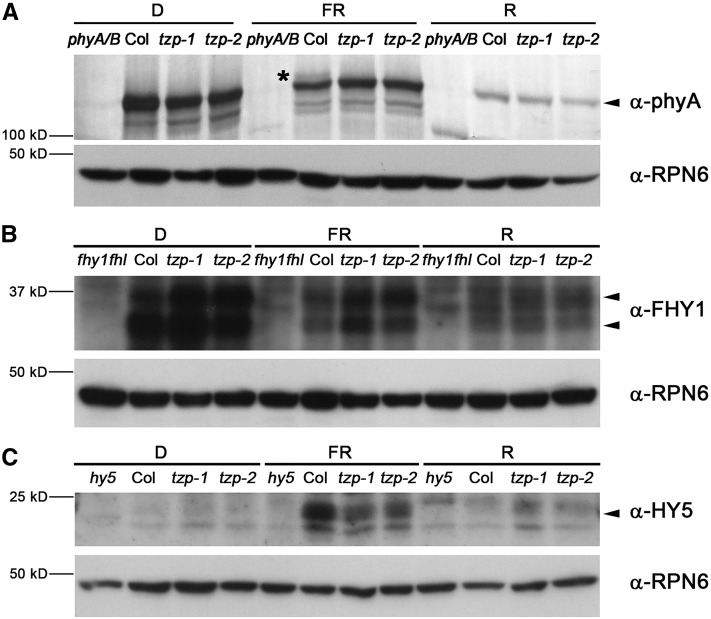
We next studied whether TZP could regulate the abundance of phyA and phyB proteins. To address this question, we performed immunoblot assays to examine phyA and phyB protein levels in wild-type and two tzp mutant seedlings grown in D, continuous FR, or R light for 4 d. Our data showed that although phyB levels did not change significantly in the tzp mutants in R light (Supplemental Figure 9), more phyA proteins accumulated in both tzp mutant seedlings in FR (Figure 7A), indicating that TZP negatively regulates phyA protein abundance in FR light. We also compared the FHY1 protein levels in the wild type and tzp mutants grown in D or continuous FR or R light. As reported in our previous studies (Shen et al., 2009; Li et al., 2010), our FHY1 antibodies always recognize two endogenous FHY1 bands in immunoblots, and our data showed that TZP negatively regulates FHY1 protein levels in FR light (Figure 7B), consistent with an increase of FHY1 transcript levels in tzp mutants (Figure 2C).
Figure 7.
TZP Regulates the Protein Abundance of phyA, FHY1, and HY5 in FR Light.
(A) Immunoblot data showing the phyA protein levels in 4-d-old phyA phyB, Col, tzp-1, and tzp-2 seedlings grown in D, continuous FR, or R light. The asterisk and arrowhead represent the phosphorylated and unphosphorylated phyA forms, respectively (Saijo et al., 2008).
(B) Immunoblot data showing the FHY1 protein levels in 4-d-old fhy1 fhl, Col, tzp-1, and tzp-2 seedlings grown in D, continuous FR, or R light. As reported in our previous studies (Shen et al., 2009; Li et al., 2010), our FHY1 antibodies recognized two endogenous FHY1 bands in immunoblots.
(C) Immunoblot data showing the HY5 protein levels in 4-d-old hy5, Col, tzp-1, and tzp-2 seedlings grown in D, continuous FR, or R light.
Anti-RPN6 was used as a sample loading control.
HY5 is a pivotal transcription factor promoting photomorphogenesis, and it was shown that the abundance of HY5 protein is directly correlated with the extent of photomorphogenic development (Osterlund et al., 2000). We therefore examined HY5 protein levels in wild-type and tzp mutant seedlings grown in D or continuous FR or R light. Our data showed that fewer HY5 proteins accumulated in the tzp mutants in FR light (Figure 7C), consistent with the tzp mutant phenotype of defective FR light signaling. Taken together, our data indicate that TZP regulates the protein abundance of phyA, FHY1, and HY5 in FR light.
TZP Regulates phyA Phosphorylation in the Nucleus in FR Light
Nuclear translocation of phyA upon light exposure is a prerequisite for phyA signaling, and this process has been shown to be controlled by FHY1 and FHL (Hiltbrunner et al., 2005, 2006; Lin et al., 2007; Rösler et al., 2007; Genoud et al., 2008; Rausenberger et al., 2011). Our observations made from confocal microscopy indicated that phyA-GFP entered the nucleus and formed nuclear bodies in both the wild type and tzp mutants after 10 min of R light or 30 min of FR light treatments (Supplemental Figure 10), implying that TZP may not be involved in regulating light-induced translocation of phyA into the nucleus. Instead, we noticed that a slower-migrating band of phyA in the wild type under FR light, previously shown to be a phosphorylated form of phyA in vivo (Saijo et al., 2008), was absent in both tzp mutant seedlings (Figure 7A). The reappearance of phosphorylated phyA form in FR-grown tzp complementation lines (Supplemental Figure 11) indicates that TZP is involved in regulating the formation of this phosphorylated phyA form.
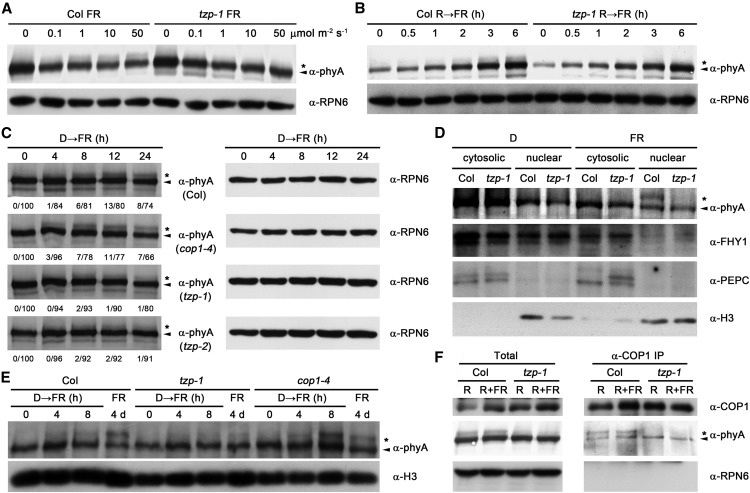
To understand the functional significance of phosphorylated phyA in FR light response, we first grew wild-type and tzp mutant seedlings under different fluence rates of FR light for 4 d. Our immunoblot data showed that the abundance of phosphorylated phyA form was increased with higher fluence rates of FR light; however, it was hardly visible in tzp mutants (Figure 8A; Supplemental Figure 12A). In addition, transfer of 4-d-old R-grown wild-type seedlings to FR light rapidly induced the formation of phosphorylated phyA form; again, this induction was not observed in tzp mutants (Figure 8B; Supplemental Figure 12B). We also transferred 4-d-old etiolated wild-type and tzp seedlings to FR light for different times and compared the occurrence of the phosphorylated phyA form. It was previously shown that COP1 preferentially targets phosphorylated phyA for 26S proteasome-mediated degradation (Saijo et al., 2008); therefore, cop1-4 seedlings were also included in this assay. Our immunoblot data showed that the phosphorylated phyA form appeared in both tzp mutants after 8 h of FR light exposure, but was evidently less abundant than in the wild type at 12 and 24 h (Figure 8C). By contrast, the phosphorylated phyA form emerged obviously earlier and was more abundant at early time points (4–8 h) in the cop1-4 mutant than in the wild type (Figure 8C), consistent with our previous report (Saijo et al., 2008). Collectively, our data demonstrate that the abundance of phosphorylated phyA increases with higher intensities of FR light and with longer exposure to FR light, but this FR-induced accumulation of phosphorylated phyA is greatly impaired without the presence of TZP.
Figure 8.
TZP Regulates phyA Phosphorylation in FR Light.
(A) Immunoblot data showing the accumulation of phosphorylated and unphosphorylated phyA forms in 4-d-old Col and tzp-1 mutant seedlings grown in different fluence rates of FR light.
(B) Immunoblot data showing the accumulation of phosphorylated and unphosphorylated phyA forms in Col and tzp-1 mutant seedlings after the 4-d-old R light-grown seedlings were transferred to FR light for the indicated time periods.
(C) Immunoblot data showing the accumulation of phosphorylated and unphosphorylated phyA forms in Col, cop1-4, tzp-1, and tzp-2 seedlings after the 4-d-old etiolated seedlings were transferred to FR light for the indicated time periods. Relative band intensities were quantified and normalized for each anti-phyA blot, and the ratio of the phosphorylated-to-unphosphorylated form of phyA is shown.
(D) Immunoblot analysis of purified nuclear and cytoplasmic fractions from 4-d-old Col and tzp-1 mutant seedlings grown in D or continuous FR light. PEPC and H3 were used as cytosolic and nuclear markers, respectively.
(E) Immunoblot data showing the accumulation of phosphorylated and unphosphorylated phyA forms in the nuclear fractions from 4-d-old Col, tzp-1, and cop1-4 seedlings grown in D or continuous FR light, or after the 4-d-old etiolated seedlings were transferred to FR light for 4 and 8 h, respectively. Anti-H3 was used as a sample loading control.
(F) Co-IP assays showing that both phosphorylated and unphosphorylated forms of phyA coprecipitated at similar levels with COP1 in 4-d-old FR-grown Col seedlings, but only unphosphorylated phyA coprecipitated with COP1 in 4-d-old FR-grown tzp-1 mutant seedlings.
The asterisk and arrowhead represent the phosphorylated and unphosphorylated phyA forms, respectively (Saijo et al., 2008). Anti-RPN6 was used as a sample loading control in (A) to (C) and (F).
TZP was shown to be exclusively localized in the nucleus in light conditions (Kaiserli et al., 2015). To investigate whether TZP regulates phyA phosphorylation in the nucleus, we performed nuclear-cytoplasmic fractionation assays using 4-d-old wild-type and tzp mutant seedlings grown in darkness or FR light. Our subsequent immunoblot data indicated that phosphorylated phyA accumulated to high levels only in the nuclei of FR-grown wild-type seedlings, but not in the cytosol of FR-grown or in the nuclei of dark-grown wild-type seedlings (Figure 8D). Evidently, the formation of phosphorylated phyA was greatly impaired in the nuclei of FR-grown tzp mutant seedlings (Figure 8D). We then extracted the nuclear proteins from wild-type, tzp-1, and cop1-4 mutant seedlings grown in D or FR light for 4 d and from the 4-d-old etiolated seedlings exposed to FR light for 4 or 8 h. Our immunoblot data showed that phosphorylated phyA increasingly accumulated in the nuclei of wild-type seedlings upon longer exposure of FR light (Figure 8E). Moreover, the abundance of phosphorylated phyA in the nuclei of tzp-1 mutant seedlings was evidently lower than in those of wild-type and cop1-4 mutant seedlings in response to the same FR light treatments (Figure 8E). Collectively, our data indicate that TZP is involved in regulating phyA phosphorylation in the nucleus.
It was previously shown that COP1 associates with both phosphorylated and unphosphorylated forms of phyA in FR light (Saijo et al., 2008). To investigate whether TZP regulates the association between COP1 and phyA in vivo, we performed co-IP assays using wild-type and tzp-1 mutant seedlings grown in FR light for 4 d. Consistent with the report by Saijo et al. (2008), both phosphorylated and unphosphorylated forms of phyA coprecipitated at similar levels with COP1 in wild-type seedlings (Figure 8F). However, only unphosphorylated phyA coprecipitated with COP1 in tzp-1 mutant seedlings (Figure 8F). Notably, our data showed that similar levels of unphosphorylated phyA coprecipitated with COP1 in wild-type and in tzp-1 mutant seedlings and that the interaction between phyA (both phosphorylated and unphosphorylated forms) and COP1 was not obviously regulated by R/FR pulse exposure (Figure 8F). Together, our data further substantiated that TZP indeed regulates phyA phosphorylation in FR light, indicating that the pool of phyA interacting with COP1 is altered without the presence of TZP.
DISCUSSION
Genetic screens conducted in the early 1990s led to the identification of multiple key components of the phyA signaling pathway, e.g., FHY1 and FHY3. Elucidation of the action mechanisms of these key components has answered several important questions related to phyA signaling, such as how nuclear accumulation of phyA is controlled (Hiltbrunner et al., 2005, 2006; Lin et al., 2007), why the action spectrum of phyA is shifted to FR light (Rausenberger et al., 2011), etc. However, many other important questions still remain to be answered. For example, how is phyA phosphorylated, what factors are involved in regulating phyA phosphorylation, and what are the exact functions of phosphorylated phyA? Undoubtedly, identification of new components and dissection of their action mechanisms will contribute to a better understanding of phyA signaling. However, possibly because previous mutant screens were nearly saturated, novel components of phyA signaling based on mutant screens have been rarely reported in the last decade. In this study, by screening a new mutant library, we identified two mutant alleles of TZP, both of which displayed longer hypocotyls in FR light. Our subsequent genetic and biochemical analyses indicated that TZP not only interacts with phyA and FHY1, but also regulates phyA phosphorylation and protein abundance in FR light. Therefore, our mutant screen efforts led to the identification of TZP as a regulator of phyA signaling.
TZP was initially identified by quantitative trait locus mapping as the causative locus regulating morning-specific growth (Loudet et al., 2008). That study also discovered that TZP played a role in mediating blue light-induced hypocotyl elongation (Loudet et al., 2008), and a later study uncovered a role for TZP in regulating photoperiodic flowering via a phyB-dependent signal transduction pathway (Kaiserli et al., 2015). A more recent proteomic study employing affinity purification and mass spectrometry methods revealed that TZP is associated with the evening complex composed of EARLY FLOWERING4 (ELF4), ELF3, and LUX ARRHYTHMO (LUX) (Huang et al., 2016). However, the functions of TZP were only deduced by the phenotypes of TZP overexpression lines, and the lack of tzp mutants has prevented a thorough and complete evaluation of TZP functions in plants. In this study, by systematically analyzing the phenotypes of two tzp mutant alleles, we discovered that TZP acts as a positive regulator of FR light signaling but as a negative regulator of blue light signaling. It should be noted that the role of TZP in blue light is consistent with the conclusion of the reports that studied TZP overexpression lines (Loudet et al., 2008; Kaiserli et al., 2015); however, the involvement of TZP in phyA signaling has only been shown by the phenotypes of the tzp mutants. How TZP plays opposite roles in FR and blue light signaling remain to be answered by further investigation; however, the distinct modifications of TZP proteins in FR, R, and B light may provide a biochemical basis for TZP to play different roles in different light conditions.
It was established long ago that phytochromes are phosphoproteins (Wong et al., 1986; Biermann et al., 1994), and the phosphorylation sites of oat (Avena sativa) phyA have been investigated since the 1980s. Three serine residues of oat phyA, i.e., Ser-8 and Ser-18 in the N-terminal extension region and Ser-599 in the hinge region, have been well characterized to be phosphorylation sites (Wong et al., 1986; McMichael and Lagarias, 1990; Lapko et al., 1997, 1999). The phosphorylation and dephosphorylation of phyA have been suggested to either regulate phyA stability (Stockhaus et al., 1992; Jordan et al., 1996, 1997; Casal et al., 2002; Han et al., 2010) or regulate the interactions of phyA with downstream signal transducers (Kim et al., 2004; Ryu et al., 2005). However, most of the previous studies on phyA phosphorylation were conducted using oat phyA as a model. A recent study from our group first detected an in vivo phosphorylated form of Arabidopsis phyA protein (Saijo et al., 2008). It was suggested that phosphorylated phyA may be a preferred substrate for the COP1/SPA complex-mediated degradation (Saijo et al., 2008). Our data support this notion because in FR light, the expression of PHYA was not changed in tzp mutants (Figure 2C), but the formation of phosphorylated phyA form was greatly impaired, which may consequently lead to a slower turnover of phyA proteins and thus higher levels of unphosphorylated phyA accumulated in tzp mutants than in the wild type in FR light (Figures 7A and 8A; Supplemental Figure 12A).
Moreover, several lines of evidence suggest that the phosphorylated phyA form plays an important role in inducing FR light responses. First, our data showed that the abundance of phosphorylated phyA increased with higher intensities of FR light (Figure 8A; Supplemental Figure 12A) and with longer exposure to FR light (Figures 8B and 8C; Supplemental Figure 12B). Under these conditions, the actions of phyA are increasingly photoactivated. Therefore, there is a good correlation between the abundance of phosphorylated phyA and the degree of phyA response. Second, although more unphosphorylated phyA proteins accumulated in tzp mutants in FR light, the tzp mutants showed phenotypes defective in phyA response. The HY5 protein level, a molecular marker of the extent of photomorphogenesis, also indicated that phyA signaling was impaired in tzp mutants (Figure 7C). These data suggest that the unphosphorylated phyA might not be as efficient or robust as phosphorylated phyA in initiating FR light responses. Third, it was recently reported that light-activated phyA interacts with SPA proteins to disrupt the direct interaction between COP1 and SPA proteins and thus inactivate the COP1/SPA complexes, leading to rapid accumulation of transcription factors (such as HY5) and initiation of photomorphogenesis (Sheerin et al., 2015). In addition, it was also suggested that COP1 and SPAs may still independently interact with phyA (Sheerin et al., 2015). Thus, the interaction between the pool of phyA (both phosphorylated and unphosphorylated forms) and COP1 may define a critical step in phyA signaling, not only for the inactivation of the COP1/SPA complexes and initiation of phyA response, but also for phyA degradation, an important phyA-specific property required for shifting the action peak of phyA from R to FR light (Rausenberger et al., 2011). Our co-IP data showed that COP1 associated with both phosphorylated and unphosphorylated phyA forms in wild-type seedlings, but only with unphosphorylated phyA in tzp-1 mutant seedlings in FR light (Figure 8F). These data indicate that the pool of phyA interacting with COP1 is altered without the presence of TZP, which may lead to altered phyA degradation (slower turnover of unphosphorylated phyA) and insufficient inactivation of the COP1/SPA complexes (thus, less HY5 accumulation). Collectively, our study suggests that in addition to being a preferred substrate of the COP1/SPA complexes, the phosphorylated phyA form is essential for HY5 accumulation and phyA signaling (Figure 9).
Figure 9.
A Working Model Depicting That TZP-Regulated phyA Phosphorylation in the Nucleus Is Essential for HY5 Accumulation and phyA Signaling.
In the absence of light, phyA is synthesized in the cytosol in its inactive Pr form. FHY1 is localized in both cytoplasm and nucleus (Shen et al., 2005) but does not interact with the Pr form of phyA (Hiltbrunner et al., 2005). Upon light exposure, the Pr form of phyA is converted into the active Pfr form, which can interact with FHY1 and is then imported into the nucleus by FHY1 (with FHL playing a minor role). In the nucleus, phyA interacts with TZP and is phosphorylated by itself or with a certain protein kinase(s). The phosphorylated phyA form may serve as a preferred substrate for the COP1/SPA complex-mediated degradation (Saijo et al., 2008) but may also play an essential role in promoting HY5 accumulation and FR light response.
TZP was shown to be a nuclear protein and proposed to be a transcriptional regulator (Kaiserli et al., 2015). However, how TZP regulates phyA phosphorylation remains obscure. Since PHYA expression was not changed (Figure 2C) but phyA protein pattern was altered (an increase of unphosphorylated phyA but a decrease of phosphorylated phyA) in tzp mutants in FR light, it seems likely that TZP regulates phyA phosphorylation and abundance posttranscriptionally in FR light. Evidently, the identification of protein kinases responsible for phosphorylating phyA will help to answer the question. Many reports indicate that phytochromes themselves may function as light-regulated serine/threonine kinases (Wong et al., 1986; Yeh and Lagarias, 1998; Shin et al., 2016) and can phosphorylate several substrates, including histone H1, PHYTOCHROME KINASE SUBSTRATE1 (PKS1), cryptochromes, AUX/IAA proteins, FHY1, PIFs, and themselves in vitro (Wong et al., 1989; Ahmad et al., 1998; Fankhauser et al., 1999; Colón-Carmona et al., 2000; Shen et al., 2009; Shin et al., 2016). However, whether phytochromes are responsible for the multisite phosphorylation pattern of PIFs in vivo still remains controversial (Ni et al., 2017). Moreover, although the two phosphorylation sites in the N-terminal extension region of oat phyA (Ser-8 and Ser-18) were suggested to be autophosphorylated by phyA itself in vitro, Ser-599 in the hinge region of oat phyA was not autophosphorylated by phyA (Kim et al., 2004; Han et al., 2010), indicating that other kinases are responsible for phosphorylating the sites in the hinge region. In terms of Arabidopsis phyA, no phosphorylation sites have been identified so far, and how many phosphorylation sites contribute to produce this phosphorylated phyA form in vivo remains currently unknown. The extremely low level of phosphorylated phyA in Arabidopsis may explain why limited data have been published so far regarding phosphorylation of Arabidopsis phyA.
To summarize, our work reveals that TZP is a positive regulator of phyA signaling. Together with the findings that TZP is involved in phyB-dependent regulation of FT expression (Kaiserli et al., 2015) and that TZP negatively regulates blue light signaling (Figures 1D and 1E; Loudet et al., 2008; Kaiserli et al., 2015), it is evident that TZP plays pleiotropic roles in light signaling. Further investigation and ultimate elucidation of action mechanisms of TZP in different light conditions will undoubtedly shed more light on the molecular mechanisms of light signal transduction in plants.
METHODS
Plant Materials and Growth Conditions
The wild-type Arabidopsis thaliana used in this study was the Columbia (Col) ecotype, unless otherwise indicated. The phyA-211 (Reed et al., 1994), phyB-9 (Reed et al., 1993), cop1-4 (McNellis et al., 1994), cry1-304 (Mockler et al., 1999), cry2-1 (Guo et al., 1998), cry1 cry2 (Mockler et al., 1999), fhy1-3 fhl-1 (Rösler et al., 2007), and hy5-215 (Oyama et al., 1997) mutants were of the Col ecotype and have been described previously. The tzp-1 and tzp-2 mutants were obtained by screening a mutant library made up of ∼25,000 activation-tagged Arabidopsis lines (Qin et al., 2003) for mutants showing altered FR light response. The phyA-211 tzp-1, phyA-211 tzp-2, phyB-9 tzp-1, and phyB-9 tzp-2 were generated by genetic crossing. The growth conditions and light sources were as described previously (Li et al., 2010). The fluence rates of the light growth chambers (Percival Scientific) were 50 μmol m–2 s–1 for W light, 50 μmol m–2 s–1 for FR light, 12 μmol m–2 s–1 for R light, and 3 μmol m–2 s–1 for B light.
Plasmid Construction and Generation of Transgenic Arabidopsis Plants
To generate the LexA-PHYA-N, LexA-PHYA-C, LexA-PHYA-C1, and LexA-PHYA-C2 constructs, the fragments were amplified by PCR with the respective pairs of primers shown in Supplemental Table 1 and then cloned into the BamHI-XhoI sites of the pLexA vector (Clontech), respectively. To generate the LexA-PHYB-N, LexA-PHYB-C, LexA-PHYB-C1, and LexA-PHYB-C2 constructs, the fragments were amplified by PCR with the respective pairs of primers shown in Supplemental Table 1 and then cloned into the EcoRI-NotI (for LexA-PHYB-N), BamHI-NotI (for LexA-PHYB-C), EcoRI-BamHI (for LexA-PHYB-C1), or BamHI-XhoI (for LexA-PHYB-C2) sites of the pLexA vector (Clontech), respectively. To generate the AD-TZP, AD-TZP-N, and AD-TZP-C constructs (for LexA yeast two-hybrid system), the coding sequences of TZP, TZP-N, and TZP-C were amplified by PCR with the respective pairs of primers shown in Supplemental Table 1 and then cloned into the EcoRI-XhoI sites of the pB42AD vector (Clontech), respectively.
To generate the PHYA-BD and PHYB-BD constructs, the full-length coding sequences of PHYA and PHYB were amplified by PCR with the respective pairs of primers shown in Supplemental Table 1 and then cloned into the BamHI-NotI sites and NotI site of the D153 vector (Shimizu-Sato et al., 2002), respectively. To generate the AD-TZP and AD-FHY1 constructs (for GAL4 yeast two-hybrid system), the full-length coding sequences of TZP and FHY1 were amplified by PCR with the respective pairs of primers shown in Supplemental Table 1 and then cloned into the EcoRI-XhoI sites of the pGADT7 vector (Clontech), respectively.
The 6His-FHY1 construct was described previously (Shen et al., 2005). To generate the constructs expressing 6His-PHYA-C2 and 6His-PHYB-C2, the PCR fragments were cloned into the BamHI-SalI and BamHI-XhoI sites of the pET-28a vector (Novagen), respectively. To generate the constructs expressing GST-TZP-C1 and GST-TZP-C2, the PCR fragments were cloned into the EcoRI-XhoI sites of the pGEX-4T-1 vector (Amersham Biosciences), respectively.
To generate PHYA-C2-nLuc and FHY1-nLuc, the PCR fragments of PHYA and FHY1 were cloned into the BamHI-SalI sites of the 35S:nLuc vector (Chen et al., 2008), respectively. To generate cLuc-TZP-N and cLuc-TZP-C, the corresponding PCR fragments of TZP were cloned into the KpnI-BamHI sites of the 35S:cLuc vector (Chen et al., 2008), respectively.
To generate the TZPp:TZP-3flag construct, we first modified the pCGN1547 vector by inserting more cloning sites into the multiple cloning site region, resulting in the pCGN1547-JL vector. The coding sequence for 3×flag was ligated into the pCGN1547-JL vector by annealing the synthesized complementary oligonucleotides with overhangs at both ends. The full-length coding sequence of TZP was then inserted into the pCGN1547-JL vector upstream of the 3×flag coding sequence, resulting in pCGN1547-TZP-3flag. Next, PCR reactions were performed using pCGN1547-TZP-3flag as the template and the pair of primers shown in Supplemental Table 1, and then cloned into the BamHI-KpnI sites of the pCAMBIA1300 vector. The TZP promoter (3.3 kb upstream of the ATG including the first intron of TZP) was amplified by PCR from Arabidopsis genomic DNA (Col) using the pair of primers shown in Supplemental Table 1 and then cloned into the PstI-XbaI sites of the pCAMBIA1300 vector, resulting in TZPp:TZP-3flag.
To generate the TZPp:GUS construct, the TZP promoter was amplified by PCR using TZPp:TZP-3flag as the template and the pair of primers shown in Supplemental Table 1 and then cloned into the XbaI-BamHI sites of the pBI101 vector.
To generate the PHYAp:PHYA-GFP construct, the pCF225 vector (PHYAp:PHYA-ST2; Trupkin et al., 2007) was first cut with XbaI and BamHI to release the PHYA-ST2 coding sequence. Then, the coding sequence of PHYA was amplified by PCR using the pair of primers listed in Supplemental Table 1, digested with SpeI and BamHI, and ligated into the pCF225 vector digested with XbaI and BamHI, producing the PHYAp:PHYA vector. The reverse primer used to amplify the PHYA coding sequence contains a NotI site upstream of the BamHI site; therefore, the NotI site was introduced into the PHYAp:PHYA construct. Finally, the GFP coding sequence was amplified by PCR using 35S:GFP-FHY1 (Shen et al., 2005) as template and the pair of primers listed in Supplemental Table 1 and then cloned into the NotI-BamHI sites of PHYAp:PHYA, producing the PHYAp:PHYA-GFP vector.
To generate the TZPp:TZP-3flag and TZPp:GUS transgenic plants, the corresponding constructs were transformed into Agrobacterium tumefaciens (strain GV3101) and then into Arabidopsis plants (Col for TZPp:GUS, and tzp-1 and tzp-2 mutants for TZPp:TZP-3flag) by the floral dip method (Clough and Bent, 1998). To generate the PHYAp:PHYA-GFP, PHYAp:PHYA-GFP tzp-1, and PHYAp:PHYA-GFP tzp-2 transgenic plants, the PHYAp:PHYA-GFP vector was transformed into Agrobacterium (strain LBA4404) and then into Col or tzp-1 and tzp-2 mutant plants.
All of the primers used to generate the above-mentioned constructs are listed in Supplemental Table 1 online, and all of the constructs were confirmed by sequencing prior to usage in various assays.
Yeast Two-Hybrid Assays
For LexA-based yeast two-hybrid assays, the respective combinations of LexA- and AD- fusion plasmids were cotransformed into the yeast strain EGY48, which already contains the reporter plasmid p8op:LacZ (Clontech); for GAL4-based yeast two-hybrid assays, the respective combinations of GAL4 BD- and AD-fusion plasmids were cotransformed into the yeast strain Y187, which harbors the URA3:GAL1UAS-GAL1TATA-lacZ reporter in its genome. For plate assays, transformants were grown on SD/-Ura-His-Trp dropout plates containing X-gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) for blue color development. Yeast transformation was conducted as described in the Yeast Protocols Handbook (Clontech), and liquid assays were performed as described previously (Shimizu-Sato et al., 2002).
RT-qPCR
Total RNA was extracted from Arabidopsis seedlings using the RNeasy plant mini kit (Tiangen). The cDNAs were synthesized from 1 μg total RNA using RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher Scientific) according to the manufacturer’s instructions. Real-time qPCR analysis was performed using PowerUp SYBR Green PCR Master Mix (Thermo Fisher Scientific) with a 7300 Real-Time PCR detection system (Thermo Fisher Scientific). qPCR was performed in triplicate for each sample, and the expression levels were normalized to that of a ubiquitin gene. The primers used for RT-qPCR are listed in Supplemental Table 1.
GUS Staining
More than 10 independent Arabidopsis transgenic lines homozygous for a single copy of the reporter gene were analyzed for their GUS activity, and the results of a representative transgenic line are shown. The GUS activity analysis was performed as described previously (Jefferson et al., 1987).
Chlorophyll and Anthocyanin Measurements
Measurement of chlorophyll content was performed as described previously (Porra, 2002). Briefly, Arabidopsis seedlings were weighed and ground in liquid nitrogen. Chlorophyll was extracted from powdered samples with 80% acetone in water, and chlorophyll concentration was calculated after measuring the absorption at 663 and 645 nm.
Measurement of anthocyanin content was performed as described previously (Zhang et al., 2014). Briefly, homogenized samples were incubated overnight in 0.3 mL of 1% HCl in methanol at 4°C and extracted using an equal volume of chloroform after the addition of 0.2 mL of water. The quantity of anthocyanins was determined by spectrophotometric measurement of the aqueous phase (A530–A657) and normalized to the fresh weight of each sample.
Preparation of Recombinant Proteins
All constructs were transformed into Escherichia coli BL21 (DE3) cells that were treated with isopropyl-β-d-thiogalactoside to induce fusion protein expression. The GST fusion proteins were purified with Glutathione Sepharose 4B beads (GE Healthcare), and the 6×His-fusion proteins were purified with nickel-nitrilotriacetic acid beads (Qiagen).
In Vitro Pull-Down Assays
For in vitro binding, 2 µg of purified recombinant bait proteins (GST-TZP-C1, GST-TZP-C2, and GST) and 2 µg of prey proteins (6×His-PHYA-C2 or 6×His-FHY1) were added to 1 mL of binding buffer containing 50 mM Tris-HCl, pH 7.5,100 mM NaCl, 0.2% glycerol, and 0.6% Triton X-100. After incubation at 4°C for 2 h, Glutathione Sepharose 4B beads (GE Healthcare) were then added and incubated for a further 2 h. After washing six times with the binding buffer, pulled-down proteins were eluted in 2× SDS loading buffer at 95°C for 15 min, separated on 10% SDS-PAGE gels, and detected by immunoblotting.
LCI Assays
Transient LCI assays in Nicotiana benthamiana were performed as described previously (Chen et al., 2008). Briefly, Agrobacterium (strain GV3101) bacteria containing indicated constructs were infiltrated into young but fully expanded leaves of the 7-week-old N. benthamiana plants using a needleless syringe. After infiltration, plants were grown under 16-h-light/8-h-dark for 3 d. Before imaging, the abaxial sides of leaves were sprayed with 1 mM luciferin, and a CCD camera (1300B; Roper) was used to capture the LUC signal at −110°C with 30-min exposures.
Nuclear-Cytoplasmic Fractionation
Nuclear fractionation was performed essentially as previously described (Wang et al., 2011) with the following modifications. Briefly, 500 mg of Arabidopsis seedlings (Col and tzp-1) grown in darkness or FR light for 4 d were ground to a fine powder in liquid nitrogen, thawed in 2 mL of precooled (4°C) lysis buffer (20 mM Tris-HCl, pH 7.4, 25% glycerol, 20 mM KCl, 2 mM EDTA, 2.5 mM MgCl2, 250 mM sucrose, and 5 mM DTT) supplemented with 1× protease inhibitor cocktail (Roche), and filtered twice through a double layer of Miracloth (Merck Millipore). The flow-through was spun at 1500g for 10 min at 4°C, and the supernatant, consisting of the cytoplasmic fraction, was centrifuged at 13,000g for 15 min at 4°C and collected. The pellet, containing the nuclear fraction, was washed four times with 2 mL of nuclear resuspension buffer NRBT (20 mM Tris-HCl, pH 7.4, 25% glycerol, 2.5 mM MgCl2, and 0.2% Triton X-100) and centrifuged for 2 min at 1500g, 4°C. The pellet was resuspended in 300 μL of precooled NRBT2 (250 mM sucrose, 10 mM Tris-HCl, pH 7.5, 10 mM MgCl2, 1% Triton X-100, and 0.035% β-mercaptoethanol) supplemented with 1× protease inhibitor cocktail (Roche), and carefully overlaid on top of 300 μL of NRBT3 (1.7 M sucrose, 10 mM Tris-HCl, pH 7.5, 2 mM MgCl2, 0.15% Triton X-100, and 0.035% β-mercaptoethanol) supplemented with 1× protease inhibitor cocktail (Roche). These were centrifuged for 45 min at 13,000g, 4°C, and the final nuclear pellet was resuspended in 2× SDS loading buffer. As quality controls for the fractionation, PEPC protein was detected and used as a cytoplasmic marker, and histone H3 was probed and used as a nuclear marker.
Co-IP and Immunoblotting
Arabidopsis seedlings were homogenized in an extraction buffer containing 50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 10 mM MgCl2, 1 mM EDTA, 0.1% Nonidet P-40, 1 mM PMSF, 1× MG132, and 1× complete protease inhibitor cocktail (Roche). For co-IP experiments, the homozygous TZPp:TZP-3flag/tzp-2 transgenic seedlings were grown first in FR or R light for 4 d and then collected and homogenized in 1 mL of protein extraction buffer and centrifuged twice for 15 min at 12,000g, 4°C. Then, the extracts were treated with the indicated combinations of R/FR light pulses. Of the 1000 μL of supernatant for each sample, 100 μL was reserved as total, and the remaining was incubated with an anti-flag M2 Affinity Gel (Sigma-Aldrich) for 2 h at 4°C. The beads were then washed four times with protein extraction buffer, and the immunoprecipitated proteins were analyzed by immunoblotting as previously described (Shen et al., 2005). Primary antibodies used in this study include anti-phyA (Shen et al., 2009), anti-FHY1 (Shen et al., 2005), anti-HY5 (Li et al., 2010), anti-RPN6 (Chen et al., 2010), anti-COP1 (Saijo et al., 2008), anti-flag (Sigma-Aldrich), anti-PEPC (Agrisera), anti-H3 (Abcam), anti-GST (Sigma-Aldrich), and anti-His (Sigma-Aldrich) antibodies.
The anti-phyB polyclonal antibodies were made by Beijing Protein Innovation. Briefly, 6His-PHYB-C2 proteins were first expressed in E. coli and then purified and used as antigens to immunize rabbits for the production of polyclonal antiserum. Antigen affinity purified anti-phyB antibodies were used in immunoblots.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: TZP (At5g43630), PHYA (At1g09570), FHY1 (At2g37678), FHL (At5g02200), HY5 (At5g11260), HYH (At3g17609), PHYB (At2g18790), PIF1 (At2g20180), PIF3 (At1g09530), PIF4 (At2g43010), PIF5 (At3g59060), COP1 (At2g32950), CHS (At5g13930), CHI (At3g55120), F3H (At3g51240), F3′H (At5g07990), DFR (At5g42800), LDOX (At4g22880), IAA19 (At3g15540), ELIP2 (At4g14690), DWF4 (At3g50660), XTH17 (At1g65310), EXP2 (At5g05290), and EXT3 (At1g21310).
Supplemental Data
Supplemental Figure 1. Phenotypes of two tzp mutant seedlings grown under different fluence rates of continuous FR light.
Supplemental Figure 2. Genotyping of the two tzp mutants.
Supplemental Figure 3. Characterization of the tzp complementation lines.
Supplemental Figure 4. Phenotypes of the tzp mutants and tzp complementation lines in darkness, continuous red, and white light conditions.
Supplemental Figure 5. The tzp mutants did not show any defect in FR block of greening.
Supplemental Figure 6. The expression of some light-responsive genes in tzp mutants in FR light.
Supplemental Figure 7. TZP interacts with both Pr and Pfr forms of holo-phyB in yeast cells.
Supplemental Figure 8. TZP interacts with FHY1.
Supplemental Figure 9. TZP does not regulate the protein abundance of phyB.
Supplemental Figure 10. Nuclear translocation and nuclear body formation of phyA-GFP in tzp mutant background after R and FR light treatments.
Supplemental Figure 11. The phosphorylated phyA form is present in FR-grown tzp complementation lines.
Supplemental Figure 12. The formation of phosphorylated phyA form is impaired in tzp-2 mutant seedlings in response to FR light.
Supplemental Table 1. Summary of primers used in this study.
Acknowledgments
We thank Peter Quail for D153 vector, Christian Fankhauser for pCF225 vector, and Shuhua Yang for suggestions and comments on the project. This work was supported by grants from the National Key Research and Development Program of China (2016YFD0100404 and 2017YFD0102001), the National Natural Science Foundation of China (31770321, 31371221, and 31171564), and the Recruitment Program of Global Youth Experts of China.
AUTHOR CONTRIBUTIONS
J.L. designed research. S.Z., C.L., Y.Z., X.W., H.L., Z.F., and D.J. performed research. H.C., G.Q., H.G., and L.-J.Q. contributed new methods/materials. J.L., D.K., and X.W.D.analyzed data. J.L., W.T., and X.W.D. wrote the article.
References
- Ahmad M., Jarillo J.A., Smirnova O., Cashmore A.R. (1998). The CRY1 blue light photoreceptor of Arabidopsis interacts with phytochrome A in vitro. Mol. Cell 1: 939–948. [DOI] [PubMed] [Google Scholar]
- Al-Sady B., Ni W., Kircher S., Schäfer E., Quail P.H. (2006). Photoactivated phytochrome induces rapid PIF3 phosphorylation prior to proteasome-mediated degradation. Mol. Cell 23: 439–446. [DOI] [PubMed] [Google Scholar]
- Ballesteros M.L., Bolle C., Lois L.M., Moore J.M., Vielle-Calzada J.P., Grossniklaus U., Chua N.H. (2001). LAF1, a MYB transcription activator for phytochrome A signaling. Genes Dev. 15: 2613–2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes S.A., Nishizawa N.K., Quaggio R.B., Whitelam G.C., Chua N.H. (1996). Far-red light blocks greening of Arabidopsis seedlings via a phytochrome A-mediated change in plastid development. Plant Cell 8: 601–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biermann B.J., Pao L.I., Feldman L.J. (1994). Pr-specific phytochrome phosphorylation in vitro by a protein kinase present in anti-phytochrome maize immunoprecipitates. Plant Physiol. 105: 243–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon M.C., Terneus K., Hall Q., Tan L., Wang Y., Wegenhart B.L., Chen L., Lamport D.T., Chen Y., Kieliszewski M.J. (2008). Self-assembly of the plant cell wall requires an extensin scaffold. Proc. Natl. Acad. Sci. USA 105: 2226–2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casal J.J., Davis S.J., Kirchenbauer D., Viczian A., Yanovsky M.J., Clough R.C., Kircher S., Jordan-Beebe E.T., Schäfer E., Nagy F., Vierstra R.D. (2002). The serine-rich N-terminal domain of oat phytochrome a helps regulate light responses and subnuclear localization of the photoreceptor. Plant Physiol. 129: 1127–1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillon A., Shen H., Huq E. (2007). Phytochrome Interacting Factors: central players in phytochrome-mediated light signaling networks. Trends Plant Sci. 12: 514–521. [DOI] [PubMed] [Google Scholar]
- Chen H., Zou Y., Shang Y., Lin H., Wang Y., Cai R., Tang X., Zhou J.M. (2008). Firefly luciferase complementation imaging assay for protein-protein interactions in plants. Plant Physiol. 146: 368–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Huang X., Gusmaroli G., Terzaghi W., Lau O.S., Yanagawa Y., Zhang Y., Li J., Lee J.H., Zhu D., Deng X.W. (2010). Arabidopsis CULLIN4-damaged DNA binding protein 1 interacts with CONSTITUTIVELY PHOTOMORPHOGENIC1-SUPPRESSOR OF PHYA complexes to regulate photomorphogenesis and flowering time. Plant Cell 22: 108–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe S., Dilkes B.P., Fujioka S., Takatsuto S., Sakurai A., Feldmann K.A. (1998). The DWF4 gene of Arabidopsis encodes a cytochrome P450 that mediates multiple 22alpha-hydroxylation steps in brassinosteroid biosynthesis. Plant Cell 10: 231–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Colón-Carmona A., Chen D.L., Yeh K.C., Abel S. (2000). Aux/IAA proteins are phosphorylated by phytochrome in vitro. Plant Physiol. 124: 1728–1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosgrove D.J. (2005). Growth of the plant cell wall. Nat. Rev. Mol. Cell Biol. 6: 850–861. [DOI] [PubMed] [Google Scholar]
- Desnos T., Puente P., Whitelam G.C., Harberd N.P. (2001). FHY1: a phytochrome A-specific signal transducer. Genes Dev. 15: 2980–2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duek P.D., Fankhauser C. (2005). bHLH class transcription factors take centre stage in phytochrome signalling. Trends Plant Sci. 10: 51–54. [DOI] [PubMed] [Google Scholar]
- Duek P.D., Elmer M.V., van Oosten V.R., Fankhauser C. (2004). The degradation of HFR1, a putative bHLH class transcription factor involved in light signaling, is regulated by phosphorylation and requires COP1. Curr. Biol. 14: 2296–2301. [DOI] [PubMed] [Google Scholar]
- Eichenberg K., Bäurle I., Paulo N., Sharrock R.A., Rüdiger W., Schäfer E. (2000). Arabidopsis phytochromes C and E have different spectral characteristics from those of phytochromes A and B. FEBS Lett. 470: 107–112. [DOI] [PubMed] [Google Scholar]
- Fairchild C.D., Schumaker M.A., Quail P.H. (2000). HFR1 encodes an atypical bHLH protein that acts in phytochrome A signal transduction. Genes Dev. 14: 2377–2391. [PMC free article] [PubMed] [Google Scholar]
- Fankhauser C., Chory J. (2000). RSF1, an Arabidopsis locus implicated in phytochrome A signaling. Plant Physiol. 124: 39–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fankhauser C., Yeh K.C., Lagarias J.C., Zhang H., Elich T.D., Chory J. (1999). PKS1, a substrate phosphorylated by phytochrome that modulates light signaling in Arabidopsis. Science 284: 1539–1541. [DOI] [PubMed] [Google Scholar]
- Franklin K.A., Quail P.H. (2010). Phytochrome functions in Arabidopsis development. J. Exp. Bot. 61: 11–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genoud T., Schweizer F., Tscheuschler A., Debrieux D., Casal J.J., Schäfer E., Hiltbrunner A., Fankhauser C. (2008). FHY1 mediates nuclear import of the light-activated phytochrome A photoreceptor. PLoS Genet. 4: e1000143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H., Yang H., Mockler T.C., Lin C. (1998). Regulation of flowering time by Arabidopsis photoreceptors. Science 279: 1360–1363. [DOI] [PubMed] [Google Scholar]
- Han Y.J., Kim H.S., Kim Y.M., Shin A.Y., Lee S.S., Bhoo S.H., Song P.S., Kim J.I. (2010). Functional characterization of phytochrome autophosphorylation in plant light signaling. Plant Cell Physiol. 51: 596–609. [DOI] [PubMed] [Google Scholar]
- Hiltbrunner A., Viczián A., Bury E., Tscheuschler A., Kircher S., Tóth R., Honsberger A., Nagy F., Fankhauser C., Schäfer E. (2005). Nuclear accumulation of the phytochrome A photoreceptor requires FHY1. Curr. Biol. 15: 2125–2130. [DOI] [PubMed] [Google Scholar]
- Hiltbrunner A., Tscheuschler A., Viczián A., Kunkel T., Kircher S., Schäfer E. (2006). FHY1 and FHL act together to mediate nuclear accumulation of the phytochrome A photoreceptor. Plant Cell Physiol. 47: 1023–1034. [DOI] [PubMed] [Google Scholar]
- Holm M., Ma L.G., Qu L.J., Deng X.W. (2002). Two interacting bZIP proteins are direct targets of COP1-mediated control of light-dependent gene expression in Arabidopsis. Genes Dev. 16: 1247–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H., Alvarez S., Bindbeutel R., Shen Z., Naldrett M.J., Evans B.S., Briggs S.P., Hicks L.M., Kay S.A., Nusinow D.A. (2016). Identification of evening complex associated proteins in Arabidopsis by affinity purification and mass spectrometry. Mol. Cell. Proteomics 15: 201–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson M., Ringli C., Boylan M.T., Quail P.H. (1999). The FAR1 locus encodes a novel nuclear protein specific to phytochrome A signaling. Genes Dev. 13: 2017–2027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang I.C., Yang J.Y., Seo H.S., Chua N.H. (2005). HFR1 is targeted by COP1 E3 ligase for post-translational proteolysis during phytochrome A signaling. Genes Dev. 19: 593–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang I.C., Yang S.W., Yang J.Y., Chua N.H. (2007). Independent and interdependent functions of LAF1 and HFR1 in phytochrome A signaling. Genes Dev. 21: 2100–2111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang I.C., Henriques R., Seo H.S., Nagatani A., Chua N.H. (2010). Arabidopsis PHYTOCHROME INTERACTING FACTOR proteins promote phytochrome B polyubiquitination by COP1 E3 ligase in the nucleus. Plant Cell 22: 2370–2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson R.A., Kavanagh T.A., Bevan M.W. (1987). GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6: 3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y., Lau O.S., Deng X.W. (2007). Light-regulated transcriptional networks in higher plants. Nat. Rev. Genet. 8: 217–230. [DOI] [PubMed] [Google Scholar]
- Jing Y., Zhang D., Wang X., Tang W., Wang W., Huai J., Xu G., Chen D., Li Y., Lin R. (2013). Arabidopsis chromatin remodeling factor PICKLE interacts with transcription factor HY5 to regulate hypocotyl cell elongation. Plant Cell 25: 242–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan E.T., Cherry J.R., Walker J.M., Vierstra R.D. (1996). The amino-terminus of phytochrome A contains two distinct functional domains. Plant J. 9: 243–257. [DOI] [PubMed] [Google Scholar]
- Jordan E.T., Marita J.M., Clough R.C., Vierstra R.D. (1997). Characterization of regions within the N-terminal 6-kilodalton domain of phytochrome A that modulate its biological activity. Plant Physiol. 115: 693–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiserli E., Páldi K., O’Donnell L., Batalov O., Pedmale U.V., Nusinow D.A., Kay S.A., Chory J. (2015). Integration of light and photoperiodic signaling in transcriptional nuclear foci. Dev. Cell 35: 311–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.I., Shen Y., Han Y.J., Park J.E., Kirchenbauer D., Soh M.S., Nagy F., Schäfer E., Song P.S. (2004). Phytochrome phosphorylation modulates light signaling by influencing the protein-protein interaction. Plant Cell 16: 2629–2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef M., Rolff E., Spruit C.J.P. (1980). Genetic control of light-inhibited hypocotyl elongation in Arabidopsis thaliana (L.). Heynh. Z. Pflanzenphysiol. 100: 147–160. [Google Scholar]
- Lapko V.N., Jiang X.Y., Smith D.L., Song P.S. (1997). Posttranslational modification of oat phytochrome A: phosphorylation of a specific serine in a multiple serine cluster. Biochemistry 36: 10595–10599. [DOI] [PubMed] [Google Scholar]
- Lapko V.N., Jiang X.Y., Smith D.L., Song P.S. (1999). Mass spectrometric characterization of oat phytochrome A: isoforms and posttranslational modifications. Protein Sci. 8: 1032–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J., He K., Stolc V., Lee H., Figueroa P., Gao Y., Tongprasit W., Zhao H., Lee I., Deng X.W. (2007). Analysis of transcription factor HY5 genomic binding sites revealed its hierarchical role in light regulation of development. Plant Cell 19: 731–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P., Monte E., Oka Y., Liu T., Carle C., Castillon A., Huq E., Quail P.H. (2008). Multiple phytochrome-interacting bHLH transcription factors repress premature seedling photomorphogenesis in darkness. Curr. Biol. 18: 1815–1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Li G., Gao S., Martinez C., He G., Zhou Z., Huang X., Lee J.H., Zhang H., Shen Y., Wang H., Deng X.W. (2010). Arabidopsis transcription factor ELONGATED HYPOCOTYL5 plays a role in the feedback regulation of phytochrome A signaling. Plant Cell 22: 3634–3649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Li G., Wang H., Deng X.W. (2011). Phytochrome signaling mechanisms. The Arabidopsis Book 9: e0148, doi/10.1199/tab.0148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin R., Ding L., Casola C., Ripoll D.R., Feschotte C., Wang H. (2007). Transposase-derived transcription factors regulate light signaling in Arabidopsis. Science 318: 1302–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin R., Teng Y., Park H.J., Ding L., Black C., Fang P., Wang H. (2008). Discrete and essential roles of the multiple domains of Arabidopsis FHY3 in mediating phytochrome A signal transduction. Plant Physiol. 148: 981–992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loudet O., Michael T.P., Burger B.T., Le Metté C., Mockler T.C., Weigel D., Chory J. (2008). A zinc knuckle protein that negatively controls morning-specific growth in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 105: 17193–17198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancinelli A.L. (1994). The physiology of phytochrome action. In Photomorphogenesis in Plants, Kendrick R.E., Kronenberg G.H.M., eds (Dordrecht, The Netherlands: Kluwer Academic Publishers; ), pp. 211–269. [Google Scholar]
- McMichael R.W. Jr., Lagarias J.C. (1990). Phosphopeptide mapping of Avena phytochrome phosphorylated by protein kinases in vitro. Biochemistry 29: 3872–3878. [DOI] [PubMed] [Google Scholar]
- McNellis T.W., von Arnim A.G., Araki T., Komeda Y., Miséra S., Deng X.W. (1994). Genetic and molecular analysis of an allelic series of cop1 mutants suggests functional roles for the multiple protein domains. Plant Cell 6: 487–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mockler T.C., Guo H., Yang H., Duong H., Lin C. (1999). Antagonistic actions of Arabidopsis cryptochromes and phytochrome B in the regulation of floral induction. Development 126: 2073–2082. [DOI] [PubMed] [Google Scholar]
- Ni M., Tepperman J.M., Quail P.H. (1998). PIF3, a phytochrome-interacting factor necessary for normal photoinduced signal transduction, is a novel basic helix-loop-helix protein. Cell 95: 657–667. [DOI] [PubMed] [Google Scholar]
- Ni W., Xu S.L., Chalkley R.J., Pham T.N., Guan S., Maltby D.A., Burlingame A.L., Wang Z.Y., Quail P.H. (2013). Multisite light-induced phosphorylation of the transcription factor PIF3 is necessary for both its rapid degradation and concomitant negative feedback modulation of photoreceptor phyB levels in Arabidopsis. Plant Cell 25: 2679–2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni W., Xu S.L., Tepperman J.M., Stanley D.J., Maltby D.A., Gross J.D., Burlingame A.L., Wang Z.Y., Quail P.H. (2014). A mutually assured destruction mechanism attenuates light signaling in Arabidopsis. Science 344: 1160–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni W., Xu S.L., González-Grandío E., Chalkley R.J., Huhmer A.F.R., Burlingame A.L., Wang Z.Y., Quail P.H. (2017). PPKs mediate direct signal transfer from phytochrome photoreceptors to transcription factor PIF3. Nat. Commun. 8: 15236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterlund M.T., Hardtke C.S., Wei N., Deng X.W. (2000). Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature 405: 462–466. [DOI] [PubMed] [Google Scholar]
- Oyama T., Shimura Y., Okada K. (1997). The Arabidopsis HY5 gene encodes a bZIP protein that regulates stimulus-induced development of root and hypocotyl. Genes Dev. 11: 2983–2995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porra R.J. (2002). The chequered history of the development and use of simultaneous equations for the accurate determination of chlorophylls a and b. Photosynth. Res. 73: 149–156. [DOI] [PubMed] [Google Scholar]
- Possart A., Hiltbrunner A. (2013). An evolutionarily conserved signaling mechanism mediates far-red light responses in land plants. Plant Cell 25: 102–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin G., et al. (2003). Obtaining and analysis of flanking sequences from T-DNA transformants of Arabidopsis. Plant Sci. 165: 941–949. [Google Scholar]
- Rausenberger J., Tscheuschler A., Nordmeier W., Wüst F., Timmer J., Schäfer E., Fleck C., Hiltbrunner A. (2011). Photoconversion and nuclear trafficking cycles determine phytochrome A’s response profile to far-red light. Cell 146: 813–825. [DOI] [PubMed] [Google Scholar]
- Reed J.W., Nagpal P., Poole D.S., Furuya M., Chory J. (1993). Mutations in the gene for the red/far-red light receptor phytochrome B alter cell elongation and physiological responses throughout Arabidopsis development. Plant Cell 5: 147–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed J.W., Nagatani A., Elich T.D., Fagan M., Chory J. (1994). Phytochrome A and phytochrome B have overlapping but distinct functions in Arabidopsis development. Plant Physiol. 104: 1139–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rösler J., Klein I., Zeidler M. (2007). Arabidopsis fhl/fhy1 double mutant reveals a distinct cytoplasmic action of phytochrome A. Proc. Natl. Acad. Sci. USA 104: 10737–10742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu J.S., et al. (2005). Phytochrome-specific type 5 phosphatase controls light signal flux by enhancing phytochrome stability and affinity for a signal transducer. Cell 120: 395–406. [DOI] [PubMed] [Google Scholar]
- Saijo Y., Zhu D., Li J., Rubio V., Zhou Z., Shen Y., Hoecker U., Wang H., Deng X.W. (2008). Arabidopsis COP1/SPA1 complex and FHY1/FHY3 associate with distinct phosphorylated forms of phytochrome A in balancing light signaling. Mol. Cell 31: 607–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo H.S., Yang J.Y., Ishikawa M., Bolle C., Ballesteros M.L., Chua N.H. (2003). LAF1 ubiquitination by COP1 controls photomorphogenesis and is stimulated by SPA1. Nature 423: 995–999. [DOI] [PubMed] [Google Scholar]
- Seo H.S., Watanabe E., Tokutomi S., Nagatani A., Chua N.H. (2004). Photoreceptor ubiquitination by COP1 E3 ligase desensitizes phytochrome A signaling. Genes Dev. 18: 617–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharrock R.A., Quail P.H. (1989). Novel phytochrome sequences in Arabidopsis thaliana: structure, evolution, and differential expression of a plant regulatory photoreceptor family. Genes Dev. 3: 1745–1757. [DOI] [PubMed] [Google Scholar]
- Sheerin D.J., Menon C., zur Oven-Krockhaus S., Enderle B., Zhu L., Johnen P., Schleifenbaum F., Stierhof Y.D., Huq E., Hiltbrunner A. (2015). Light-activated phytochrome A and B interact with members of the SPA family to promote photomorphogenesis in Arabidopsis by reorganizing the COP1/SPA complex. Plant Cell 27: 189–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y., Feng S., Ma L., Lin R., Qu L.J., Chen Z., Wang H., Deng X.W. (2005). Arabidopsis FHY1 protein stability is regulated by light via phytochrome A and 26S proteasome. Plant Physiol. 139: 1234–1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y., Zhou Z., Feng S., Li J., Tan-Wilson A., Qu L.J., Wang H., Deng X.W. (2009). Phytochrome A mediates rapid red light-induced phosphorylation of Arabidopsis FAR-RED ELONGATED HYPOCOTYL1 in a low fluence response. Plant Cell 21: 494–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H., Zhong S., Mo X., Liu N., Nezames C.D., Deng X.W. (2013). HFR1 sequesters PIF1 to govern the transcriptional network underlying light-initiated seed germination in Arabidopsis. Plant Cell 25: 3770–3784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu-Sato S., Huq E., Tepperman J.M., Quail P.H. (2002). A light-switchable gene promoter system. Nat. Biotechnol. 20: 1041–1044. [DOI] [PubMed] [Google Scholar]
- Shin A.Y., Han Y.J., Baek A., Ahn T., Kim S.Y., Nguyen T.S., Son M., Lee K.W., Shen Y., Song P.S., Kim J.I. (2016). Evidence that phytochrome functions as a protein kinase in plant light signalling. Nat. Commun. 7: 11545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J., Park E., Choi G. (2007). PIF3 regulates anthocyanin biosynthesis in an HY5-dependent manner with both factors directly binding anthocyanin biosynthetic gene promoters in Arabidopsis. Plant J. 49: 981–994. [DOI] [PubMed] [Google Scholar]
- Soh M.S., Kim Y.M., Han S.J., Song P.S. (2000). REP1, a basic helix-loop-helix protein, is required for a branch pathway of phytochrome A signaling in Arabidopsis. Plant Cell 12: 2061–2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockhaus J., Nagatani A., Halfter U., Kay S., Furuya M., Chua N.H. (1992). Serine-to-alanine substitutions at the amino-terminal region of phytochrome A result in an increase in biological activity. Genes Dev. 6: 2364–2372. [DOI] [PubMed] [Google Scholar]
- Tatematsu K., Kumagai S., Muto H., Sato A., Watahiki M.K., Harper R.M., Liscum E., Yamamoto K.T. (2004). MASSUGU2 encodes Aux/IAA19, an auxin-regulated protein that functions together with the transcriptional activator NPH4/ARF7 to regulate differential growth responses of hypocotyl and formation of lateral roots in Arabidopsis thaliana. Plant Cell 16: 379–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trupkin S.A., Debrieux D., Hiltbrunner A., Fankhauser C., Casal J.J. (2007). The serine-rich N-terminal region of Arabidopsis phytochrome A is required for protein stability. Plant Mol. Biol. 63: 669–678. [DOI] [PubMed] [Google Scholar]
- Tzvetkova-Chevolleau T., Franck F., Alawady A.E., Dall’Osto L., Carrière F., Bassi R., Grimm B., Nussaume L., Havaux M. (2007). The light stress-induced protein ELIP2 is a regulator of chlorophyll synthesis in Arabidopsis thaliana. Plant J. 50: 795–809. [DOI] [PubMed] [Google Scholar]
- Ulm R., Baumann A., Oravecz A., Máté Z., Adám E., Oakeley E.J., Schäfer E., Nagy F. (2004). Genome-wide analysis of gene expression reveals function of the bZIP transcription factor HY5 in the UV-B response of Arabidopsis. Proc. Natl. Acad. Sci. USA 101: 1397–1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Deng X.W. (2002). Arabidopsis FHY3 defines a key phytochrome A signaling component directly interacting with its homologous partner FAR1. EMBO J. 21: 1339–1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Ye R., Xin Y., Fang X., Li C., Shi H., Zhou X., Qi Y. (2011). An importin β protein negatively regulates MicroRNA activity in Arabidopsis. Plant Cell 23: 3565–3576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitelam G.C., Johnson E., Peng J., Carol P., Anderson M.L., Cowl J.S., Harberd N.P. (1993). Phytochrome A null mutants of Arabidopsis display a wild-type phenotype in white light. Plant Cell 5: 757–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong Y.S., Cheng H.C., Walsh D.A., Lagarias J.C. (1986). Phosphorylation of Avena phytochrome in vitro as a probe of light-induced conformational changes. J. Biol. Chem. 261: 12089–12097. [PubMed] [Google Scholar]
- Wong Y.S., McMichael R.W., Lagarias J.C. (1989). Properties of a polycation-stimulated protein kinase associated with purified Avena phytochrome. Plant Physiol. 91: 709–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Lin R., Sullivan J., Hoecker U., Liu B., Xu L., Deng X.W., Wang H. (2005). Light regulates COP1-mediated degradation of HFR1, a transcription factor essential for light signaling in Arabidopsis. Plant Cell 17: 804–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh K.C., Lagarias J.C. (1998). Eukaryotic phytochromes: light-regulated serine/threonine protein kinases with histidine kinase ancestry. Proc. Natl. Acad. Sci. USA 95: 13976–13981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., He H., Wang X., Wang X., Yang X., Li L., Deng X.W. (2011). Genome-wide mapping of the HY5-mediated gene networks in Arabidopsis that involve both transcriptional and post-transcriptional regulation. Plant J. 65: 346–358. [DOI] [PubMed] [Google Scholar]
- Zhang H., Zhao X., Li J., Cai H., Deng X.W., Li L. (2014). MicroRNA408 is critical for the HY5-SPL7 gene network that mediates the coordinated response to light and copper. Plant Cell 26: 4933–4953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q., Hare P.D., Yang S.W., Zeidler M., Huang L.F., Chua N.H. (2005). FHL is required for full phytochrome A signaling and shares overlapping functions with FHY1. Plant J. 43: 356–370. [DOI] [PMC free article] [PubMed] [Google Scholar]