Abstract
Pluripotent stem cells (PSCs) lie at the heart of modern regenerative medicine due to their properties of unlimited self-renewal in vitro and their ability to differentiate into cell types representative of the three embryonic germ layers—mesoderm, ectoderm and endoderm. The derivation of induced PSCs bypasses ethical concerns associated with the use of human embryonic stem cells and also enables personalized cell-based therapies. To exploit their regenerative potential, it is essential to have a firm understanding of the molecular processes associated with their induction from somatic cells. This understanding serves two purposes: first, to enable efficient, reliable and cost-effective production of excellent quality induced PSCs and, second, to enable the derivation of safe, good manufacturing practice-grade transplantable donor cells. Here, we review the reprogramming process of somatic cells into induced PSCs and associated mechanisms with emphasis on self-renewal, epigenetic control, mitochondrial bioenergetics, sub-states of pluripotency, naive ground state, naive and primed. A meta-analysis identified genes expressed exclusively in the inner cell mass and in the naive but not in the primed pluripotent state. We propose these as additional biomarkers defining naive PSCs.
This article is part of the theme issue ‘Designer human tissue: coming to a lab near you’.
Keywords: iPSC, epigenetics, pluripotency, naive, primed, urine
1. Background
The life span of an organism is characterized by continuous development and maturation of early embryonic stages into later stages leading to adulthood. It is a dream to revert or at least halt the ageing process, which is often accompanied by devastating degenerative diseases. Contrary to the normal developmental path, Yamanaka and his colleagues reported in 2006 for the first time the successful reprogramming of adult mouse tail fibroblasts into pluripotent stem cells (PSCs) that closely resembled pluripotent embryonic stem cells (ESCs) derived from the inner cell mass (ICM) of a blastocyst [1]. In other words, they induced the early properties of 3.5-day-old embryos in adult cells, and at face value it seems as if the ageing process or at least maturation had been reversed. The significance of this ground-breaking achievement is that these so-called induced pluripotent stem cells (iPSCs) hold great promise for regenerative therapies that exceeds that of the ESCs, because iPSCs allow personalized medicine and bypass ethical concerns of destroying human embryos.
2. A historical perspective of cellular reprogramming
The foundation underlying the concept of totipotency, pluripotency and multipotency is based on the experiments described by Hans Driesch. In 1891, he showed that separating two-cell stage sea urchin embryos through vigorous shaking in calcium-free water resulted in two new whole embryos [2, pp. 61–63]. Later, in 1962, Sir John Gurdon succeeded in transferring the nuclei of small intestinal epithelial cells into enucleated unfertilized eggs and obtained tadpoles [3]. This demonstrated that the adult nuclei do contain the information for the development of tadpoles. Since then, numerous animals have been cloned using somatic nuclear transfer, including Dolly the sheep [4], thus substantiating that somatic nuclei do have the potential and plasticity to adopt other functional nuclear states. Moreover, these experiments suggested that the oocyte contains factors that change the nuclear state back into a totipotent state. Additional clues on totipotency and pluripotency that paved the way for the current protocols for induction of pluripotency were obtained from embryonal carcinoma cells (ECCs) and ESCs. ECCs were isolated in 1964 from teratocarcinomas [5]. When fused with somatic cells, they could reprogram them into a pluripotent state [6]. ESCs were established by isolating cells from the ICM of blastocysts in 1981 [7,8]. Eleven years later, embryonic germ cells (EGCs) were isolated from primordial germ cells [9,10]. All these cell types proved to be capable of reprogramming somatic cells upon fusion [11,12]. These experiments indicated that not only oocytes but also ECCs, EGCs and pluripotent ESCs contained ‘reprogramming factors'. Investigation of these cell types enabled the identification and characterization of several key genes (e.g. OCT3/4, NANOG, SOX2, SALL4, ESRRB, DPPA4) associated with the pluripotent state [13–15].
Meanwhile, other groups dedicated their efforts to cell fate conversion studies, which did not employ somatic nuclear transfer or cell fusion, but overexpression of transcription factors. Myogenic transcription factor (MyoD) was the first transcription factor to be overexpressed to achieve a cell fate conversion, also called ‘direct reprogramming’ or ‘trans-differentiation’. Overexpressed in fibroblasts, MyoD trans-differentiated the cells into myoblasts in vitro [16].
Many reports followed using various cell types and trans-differentiating them into other cell types (e.g. fibroblasts into hepatocyte-like cells or neural stem cells) [17–19]. It was concluded that in spite of the power of a single transcription factor to alter a specific cell identity, it is still limited within the boundaries of the germ layer of origin. It cannot direct a cell state to cross these boundaries unless overexpressed simultaneously with other key transcription factors.
Based on these insights and those obtained from the field of somatic nuclear transfer, Yamanaka and colleagues speculated that overexpressing key ESC-associated factors could convert somatic cells into PSCs. To identify key factors of the pluripotent state that might revert cell fate, they screened publically available databases of expressed sequence tags in somatic tissues and ESCs [13]. They referred to those expressed uniquely in ESCs as ESC-associated transcripts (ECATs). Among the genes they identified and studied are NANOG, DPPA2, DPPA4 [13,20]. Finally, they chose 24 candidates that played important roles in ESCs or were highly expressed in pluripotent ESCs including ECATs. The strategy that they adopted was to use G418-resistant clones among Fbx15neo/neo mouse embryonic fibroblasts (MEFs) [1]. Fbx15 (ECAT3) is expressed in ESCs, and not in somatic cells [21], which means that ESCs and potential ESC-like cells would be resistant to G418, but not MEFs. They transduced the 24 candidate genes individually and as a cocktail. The individual transduction did not lead to any G418-resistant colonies. The cocktail transduction, however, generated 22 colonies. These colonies were similar to ESCs in terms of morphology, differentiation potential, expression profiles and epigenetic profiles. They named these cells iPSCs. To reduce the necessary factors, Yamanaka and his group eliminated some of them during further rounds of transduction and finally identified Oct3/4, Sox2, Klf4, c-Myc (OSKM) as essential and sufficient to generate iPSCs [1]. This combination is routinely referred to as the ‘Yamanaka Cocktail’. A year later, Yamanaka and colleagues generated human iPSCs using the same cocktail [22]. Within the same year, James Thomson's group independently also reported the generation of human iPSCs, but using a different cocktail, namely OCT3/4, SOX2, NANOG, LIN28—referred to as the ‘Thomson Cocktail’ [23].
3. Delivery of the reprogramming factors into somatic cells
Ever since the identification of the Yamanaka and the Thomson reprogramming ‘Cocktails’, researchers have struggled to optimize their delivery into cells (table 1). In the very early protocols, retroviruses or lentiviruses were used as they allow efficient and stable transduction [22,23]. However, in both cases the transgenes integrate into the host genome, potentially inactivating tumour suppressor genes or activating proto-oncogenes. Here, the most critical of the reprogramming factors is c-MYC, an oncogene that can lead to tumour development. Fortunately, it can be substituted by the less-tumorigenic L-MYC [36].
Table 1.
Methods for delivering the reprogramming factors.
| reprogramming factor delivery | considerations | references | |
|---|---|---|---|
| integrative | retrovirus (ssRNA) | high efficiency integrates into genome: activation of proto-oncogenes/ disruption of tumour suppressor genes possible only infects dividing cells reactivation of viral genes possible |
[22] |
| lentivirus (ssRNA) | high efficiency infects also non-dividing cells integrates into genome: activation of proto-oncogenes/disruption of tumour suppressor genes possible reactivation of viral genes possible |
[23] | |
| not integrative | adenovirus (dsDNA) | low efficiency several rounds of infection necessary |
[24] |
| sendai virus (ssRNA) | high efficiency replicating virus has to be removed from iPSCs by negative selection |
[25] | |
| pSin plasmid | low efficiency several transfections necessary integrates occasionally |
[26] | |
| minicircle DNA | low efficiency several transfections necessary integrates occasionally does not contain any bacterial genes |
[27] | |
| piggyBac transposon |
traceless excision possible integrates into genome at specific integration sites: some effect transcription units excision may affect endogenous piggyBac elements reintegration possible |
[28,29] | |
| sleepingbeauty transposon |
traceless excision possible transposase allows efficient removal of transposon integrates into genome at specific integration sites: some effect transcription units reintegration possible |
[30] | |
| oriP/EBNA1 based episomal plasmids | low efficiency self-replicative |
[31] | |
| synthetic modified mRNA | increasing efficiency with more elaborate mRNA synthesis methods repeated transfections necessary because of rapid mRNA degradation |
[32] | |
| VEE RF-RNA | self-replicating RNA that can be easily eliminated | [33] | |
| miRNA | very low efficiency repeated transfections necessary because of rapid miRNA degradation |
[34] | |
| cell penetrating peptide-coupled protein | very low efficiency repeated transfections necessary because of rapid mRNA degradation |
[35] |
iPSCs derived by integrative methods are not suitable for clinical applications due to the above-mentioned risks, and several non-integrative methods for reprogramming have been developed to circumvent these problems. Some involve quite tedious removal of the transfecting agent, for example sendai virus or transposon-based methods [25,28,30]. Others, such as transfection of minicircle DNA, pSin plasmid, RNA or protein, require repeated transfections of the reprogramming factors, inducing unwanted additional stress to the cells [26,27,32,35]. Episomal-based reprogramming is the most convenient and widely used protocol and iPSCs derived using this system have already entered clinical trials for macular degeneration [37].
Not introducing DNA at all increases the safety of reprogramming, but is usually less efficient. mRNA or proteins are quite unstable and require several rounds of transfection to successfully reprogramme somatic cells [32,35]. Apart from increased cell stress, another drawback of these methods is the fact that the stoichiometry of the reprogramming factors cannot be controlled, which is a crucial factor for successful reprogramming. To bypass this drawback, Yoshioka et al. [33] developed a synthetic polycistronic and self-replicating mRNA molecule containing all four factors. Here, one transfection is sufficient and the relative abundance of each factor can be modulated by changing its position within the RNA molecule. The synthetic RNA only remains in the cells as long as their innate immune reaction against it is suppressed. The latest development in reprogramming techniques focuses on a method where the reprogramming factors do not even have to enter the cells. In this regard, Blanchard et al. [38] recently described antibodies that are able to stimulate intracellular pathways that ultimately activate the same target genes as the reprogramming factors. However, so far it has not been possible to simultaneously replace all four factors by antibodies.
4. Improving the efficiency of inducing pluripotency
iPSC derivation is a very inefficient process with several bottlenecks. To identify these, we previously analysed pluripotency-associated gene regulatory networks and associated biological processes governed by OCT4, SOX2, KLF4 and c-MYC [39]. This analysis unveiled diverse processes such as innate immunity, response to free radical generation-reactive oxygen species (ROS), hypoxia, oxidative DNA damage, p53 activation, senescence, apoptosis, epithelial mesenchymal transition (EMT) and epigenetic modification impinging on the efficiency of iPSC derivation. Distinct cell permeable and non-immunogenic small molecules have been employed to improve the reprogramming process by modulating the above-mentioned biological and metabolic processes, signalling pathways and epigenetic modifications (table 2). These small molecules usually enhance the reprogramming efficiency and can sometimes even replace one of the pluripotency factors. However, for most of these molecules we do not know which off-target effects they might have, if they act directly or indirectly to repress or activate known targets and in which additional pathways they might be involved [84]. The advantages, on the other hand, are that small molecules are only needed temporarily and can be removed after successful reprogramming. In addition, they are standardized, cost-effective, and easy to handle and to synthesize [47,62].
Table 2.
Small molecule-based signalling pathway modulators and cytokines supporting efficient iPSC derivation.
| small molecule | mechanism | function | references |
|---|---|---|---|
| 5-azacytidine (AZA) | DNA methyltransferase inhibitor | promotion of reprogramming c-Myc replacement |
[40] |
| 8-Br-cAMP | human cAMP-dependent protein kinase activator | promotion of reprogramming | [41] |
| A83-01 | TGF-β receptor ALK5/4/7 inhibitor | promotion of reprogramming | [42] |
| AM580 | retinoic acid receptor a agonist | promotion of reprogramming | [43,44] |
| compound B6 | AKT-mediated inhibitor of GSK3-b | promotion of reprogramming | [45] |
| compound B4 | ALK4 inhibitor | promotion of reprogramming | [45] |
| compound B8 | IP3K inhibitor | promotion of reprogramming | [45] |
| compound B10 | P38 kinase inhibitor | promotion of reprogramming | [45] |
| DAPT | γ-secretase inhibitor | promotion of reprogramming | [46] |
| DZNep | histone methyltransferase EZH2 inhibitor | promotion of reprogramming | [47] |
| DNP | oxidative phosphorylation uncoupler | promotion of reprogramming | [42] |
| EPZ004777 | DOT1 L inhibitor | promotion of reprogramming | [48] |
| fructose 2,6-bisphosphate | phosphofructokinase 1 activator | promotion of reprogramming | [42] |
| LiCl | GSK-3b inhibitor, LSD1 inhibitor | promotion of reprogramming | [49] |
| N-oxaloylglycine | prolyl-4-hydroxylase inhibitor | promotion of reprogramming | [42] |
| NaB (sodium butyrate) | HDAC inhibitor | promotion of reprogramming | [50] |
| nicotinamide | inhibition of the H3K79 histone methyltransferase DOT1 L | promotion of reprogramming | [51,52] |
| PD0325901 | potent MEK1 and MEK2 inhibitor | promotion of reprogramming inhibits growth of non-iPS cell colonies |
[53] |
| PS48 | PDK1 activator | promotion of reprogramming | [42] |
| prostaglandin | cAMP agonists | promotion of reprogramming | [47] |
| quercetin | hypoxia-inducible factor pathway activator | promotion of reprogramming | [42] |
| rapamycin | mTOR inhibitor activating autophagy |
promotion of reprogramming | [54] |
| RG108 | DNA methyltransferase inhibitor | promotion of reprogramming Sox2 (with BIX) or Oct3/4 substitute |
[55] |
| RSC133 | DMNT inhibitor | promotion of reprogramming | [56] |
| rolipram | cAMP agonists | promotion of reprogramming | [47] |
| SAHA | HDAC inhibitor | promotion of reprogramming | [40] |
| SB431542 | TGF-β receptor ALK5/4/7 inhibitor | promotion of reprogramming | [57] |
| SGC0496 | DOT1 L inhibitor | promotion of reprogramming | [58] |
| SMER28 | autophagy modulator | promotion of reprogramming | [59] |
| Tranylcypromine (Parnate) | lysine-specific demethylase 1 inhibitor | promotion of reprogramming | [57] |
| TSA | HDAC inhibitor | promotion of reprogramming | [40] |
| TTNPB | retinoic acid receptor ligand | promotion of reprogramming | [47] |
| VPA | HDAC inhibitor | promotion of reprogramming | [60,61] |
| Y-27632 | ROCK inhibitor | promotion of reprogramming | [62] |
| 2-Me-5HT | 5-HT3 agonist | Oct3/4 substitute | [47] |
| BIX-01294 | G9a histone lysine methyltransferase inhibitor | Oct3/4 substitute | [53,63] |
| D4476 | mouse CK1 inhibitor | Oct3/4 substitute | [47] |
| Forskolin (FSK) | PKA activator, mouse cAMP agonist | Oct3/4 substitute | [47] |
| O4I1 (modifications 1-11) | unknown | Oct3/4 substitute | [64] |
| O4I2 (modifications 1-21) | unknown | Oct3/4 substitute | [65] |
| OAC1 | activates Oct3/4 and Nanog promoters | Oct3/4 substitute | [45,66] |
| OAC2 | activates Oct3/4 and Nanog reporters | Oct3/4 substitute | [45] |
| BayK | L-channel calcium agonist | Sox2 substitute | [55] |
| CHIR99021 | GSK-3 inhibitor Wnt pathway activator |
Sox2 substitute | [60] |
| Dasatinib | Src family tyrosine kinase inhibitor | Sox2 substitute | [67] |
| iPY | Src family tyrosine kinase inhibitor | Sox2 substitute | [67] |
| LY-364947 | TGF-b inhibitor | Sox2 substitute | [67] |
| PP1 | Src family tyrosine kinase inhibitor | Sox2 substitute | [67] |
| Repsox (E-616452) | TGFβ receptor I kinase inhibitor | Sox2 substitute | [68] |
| Kenpaullone | GSK-3 and CDK1/cyclin B inhibitor | Klf4 substitute | [69] |
| AMI5 | protein arginine methyltransferase (PRMT) inhibitor | Sox2, Klf4 substitute (with A-83-01) | [70] |
| Hh-Ag 1.5 | Smo agonist activating MAPK and SHH pathways | Sox2 and Nestin induction | [59] |
| Oxysterol | sonic hedgehog signalling activator | Sox2, Klf4, and C-Myc substitute | [71] |
| Purmorphamine | hedgehog activator | Sox2, Klf4, and C-Myc substitute differentiation inducer to neural stem cells |
[71,72] |
| Shh | sonic hedgehog signalling | Sox2, Klf4 and C-Myc substitute | [71] |
| (±) BayK 8644 | L-type calcium channel agonist | increase in spontaneous beating frequency promotion of reprogramming maintenance of pluripotency |
[73] |
| JNJ-10198409 | PDGFR-a and PDGFR-b inhibitor | increase in spontaneous beating frequency differentiation inducer to cardiomyocytes |
[74] |
| SU16F | PDGFR inhibitor | increase in spontaneous beating frequency promotion of reprogramming differentiation inducer to cardiomyocytes |
[74] |
| ATRA | retinoic acid receptor agonist | differentiation inducer to extra-embryonic lineage | [75,76] |
| AS8351 | epigenetic modifications modulation of a specific JmjC-KDM |
differentiation inducer to cardiomyocytes | [77] |
| JAK inhibitor 1 (JI1) | JAK–STAT pathway inhibitor | differentiation inducer to cardiomyocytes | [78] |
| SC1 (pluripotin) | dual inhibitor of extracellular signal-regulated kinase 1 and RasGAP/inhibitor of the ERK1 and Ras-GAP signalling pathways | differentiation inducer to cardiomyocytes | [73,74] |
| SU5402 | FGFR, VEGFR and PDGFR inhibitor | differentiation inducer to cardiovascular progenitor cells | [79] |
| Poly(I:C) | toll-like receptor 3 agonist | differentiation inducer to endothelial cells | [80] |
| Go 6983 | broad spectrum protein kinase C inhibitor | differentiation inducer to neurons | [62] |
| I-BET151 | BET bromodomain inhibitor | differentiation inducer to neurons | [81] |
| ISX9 | neurogenic agent | differentiation inducer to neurons | [81] |
| LDN193189 | BMP receptor ALK2 and ALK3 antagonists | differentiation inducer to neurons | [59,82] |
| Pifithrin-α (PFT-α) | p53 inhibitor | differentiation inducer to neurons | [83] |
| SAG | hedgehog activator smoothened agonist | differentiation inducer to neurons | [46] |
| SP600125 | JNK inhibitor | differentiation inducer to neurons | [62] |
| Thiazovivin (Tzv) | selective ROCK inhibitor | differentiation inducer to neurons | [46] |
5. Phases of pluripotency induction
Key events that were initially discovered in the reprogramming process included the gradual downregulation of Thy1, a fibroblast marker, followed by the activation of pluripotency markers like alkaline phosphatase, stage-specific antigen 1 (SSEA-1 mouse; SSEA3&4 human) and, later, the activation of endogenous Oct3/4 and Nanog [85,86]. It has been demonstrated that c-Myc is responsible for the loss of somatic expression patterns, while the function of Oct3/4, Sox2 and Nanog lies in the induction of the pluripotency gene regulatory network, whereby Sox2 expression occurs only very late in the reprogramming process [87]. Samavarchi-Tehrani et al. [88] dissected the reprogramming process in mice into three distinct subsequent phases: the initiation phase, the maturation phase and finally the stabilization phase. The initiation phase is dominated by an immediate mesenchymal-to-epithelial transition (MET) that is driven by bone morphogenetic protein (BMP) signalling and the induction of miRNA-200 family members [88,89]. However, Teshigawara et al. [90] showed that during human iPSC induction, the onset of MET occurs later and that it is preceded by OCT3/4 activation. The authors generated intermediately reprogrammed stem cells (iRSCs), which, in contrast with partially reprogrammed iPSCs, can resume the reprogramming process depending on the cell density. The progression from the intermediate state to the iPSC state is through MET, which appears to be a cell cycle-dependent checkpoint leading to the primed state, rather than the naive state like in mice.
Following the initiation phase, the maturation phase is characterized by the activation of endogenous key pluripotency genes such as Oct3/4, Nanog and Sall4. It is noteworthy that only a subset of pluripotency genes is induced in the maturation phase. Finally, in the stabilization phase pluripotency markers such as Lin28a, Dppa2, Dppa4, Utf1 and others are induced. It was recognized early on that silencing of the OSKM transgene is crucial for complete and successful reprogramming [91], to release the stabilization phase and express the full pluripotency network as well as its maintenance network. This is in agreement with the observation that key pluripotency-regulating factors such as Oct3/4 and Nanog are dose-dependent and that their overexpression or depletion leads to differentiation [92–94]. In the maturation phase, the cells' (chromatin) state changes to such a degree that transgene expression is essential for colony growth. In the stabilization phase, transgene silencing is essential for consolidation and maintenance of the pluripotent state [95].
It seems that the transition from the maturation phase to the stabilization phase is critical and constitutes the bottleneck of iPSC generation. This limitation will have a direct impact on the percentage of partially reprogrammed cells in vitro. The stabilization phase relies on the successful activation of downstream factors of OSKM and on the silencing of the transgenes, which otherwise destabilize the pluripotent state [95]. The activation of downstream targets of OSKM factors is in line with a report from Jaenisch and his colleagues describing that somatic cell nuclear transfer (SCNT) fails when genes such as Dppa1-5 and Pramel 4-7 fail to be expressed [96]. The factors identified in both reports are the well-known key pluripotency-regulating markers Oct3/4, Sox2, Nanog, Klf4, Sall4, etc. as well as genes associated with the cytoskeleton, or involved in chromosomal architecture and segregation, which allow the maintenance of the PSCs [88]. Although few in number, there are increasing reports demonstrating that in addition to the rewiring of the genetic circuitry and epigenetic remodelling, cytoskeletal remodelling is also significant, occurring upon PSC differentiation, upon acquisition of pluripotency of reprogrammed somatic cells or during PSC maintenance [95,97,98].
Another class of factors associated with pluripotency and, hence, requiring reverse regulation upon iPSC induction are miRNAs. Anokye-Danso et al. [99] in 2011 were the first to show that the microRNA cluster miR302/367 is capable of reprogramming mouse and human fibroblasts into iPSCs, with miR-367 being involved in activation of OCT4 expression. In addition, miR-302b and miR-372 are involved in the induction of MET, thus promoting one of the first steps during reprogramming [100].
6. Epigenetic basis of cellular reprogramming
Developmental processes are particularly dependent on the coordinated and sequential regulation of genetic programmes, which are reliant on epigenetic mechanisms. PSCs are, in general, euchromatic and become increasingly heterochromatic with progressing cell fate commitment. Hence, the induction of iPSCs requires the reverse process. Several effective reprogramming cocktails have been developed using epigenetic modulators such as SAHA-PiPs [101], NaButyrate [102], Parnate [57], valproic acid (VPA) [41,103,104] or 5-azacytidine [40], all of which induce chromatin de-condensation and/or de-methylation, and thereby activate Oct3/4, Nanog and Sox2 expression (table 2). Indeed, the de-methylation of the promoter regions of the endogenous key pluripotency markers such as Oct3/4 and Nanog is essential to enable complete reprogramming [1,105–107].
It has now been widely accepted that the overall euchromatic state of developmental genes in PSCs is bivalent, harbouring active (H3K4me3) and inactive (H3K27me3) histone marks. They resolve into one or the other direction during differentiation according to the needs of the mature cell type [108,109]. Interestingly, it seems that in the naive ground state bivalent domains tend to be occupied by H3K4me3 and only gain the repressive histone mark upon differentiating into naive PSCs [110].
7. Mitochondrial bioenergetics and the induction of pluripotency
The ability of PSCs to self-renew in part occurs via the fine-tuning of pathways associated with cellular senescence, such as the p53 and the mitochondrial/oxidative stress pathways [111–113]. Interestingly, upon the induction of pluripotency in somatic cells, the mitochondria revert to a pre-implantation embryo and ESC-like state. Mitochondria within PSCs, unlike somatic cells, lack well-defined cristae and generate lower levels of ATP but have an increased lactate production, thus implying a dependence on anaerobic respiration for energy supply and activation of the hypoxia-inducible pathway to overcome the decrease in mitochondria function [114].
8. Pluripotency—naive and primed
Mouse ESCs (mESCs) were derived from the ICM of a blastocyst in 1981. However, it took more than 17 years until James Thomson and his colleagues were able to derive human ESCs (hESCs) in 1998 [115,116]. Such a delay would imply that the two species might have different PSC characteristics based on major mechanistic discrepancies. Indeed, mESC colonies are dome-shaped, while hESCs grow as flat colonies. mESCs depend on Lif signalling to activate the Jak/Stat3 pathway; this pathway seems dispensable for hESCs [117]. Although treatment of hESCs with LIF leads to STAT3 phosphorylation and nuclear translocation, this fails to maintain the pluripotent state. Instead, hPSCs require the addition of FGF2 as well as TGFβ and ACTIVIN A [115,118]. In due course, additional features were discovered to be different, such as X-chromosome activation/inactivation (XaXa/XaXi), methylation status, strength of pluripotency as measured by high versus low chimaera contribution ability, and sensitivity of primed PSCs to single cell dissociation [119–121].
It turned out that such discrepancies between mESCs and hESCs are attributable to different developmental stages, namely naive and primed, rather than species-specific differences. The most striking support for this new concept was provided by Tesar et al. and Brons et al. who isolated murine PSCs from the epiblasts (EpiSCs) from post-implantation embryos that shared defined features with human ICM-derived ESCs [122,123]. Since then, mESCs have been described as naive, and the mEpiSCs as primed, referring to their distinct developmental stages. This immediately triggered the quest for the human naive pluripotent state, raising the questions: what happens to the human pre-implantation ICM during the process of isolation? In other words, assuming that the human naive state exists, what induces its differentiation from the naive to the primed state upon isolation?
9. Pluripotent sub-states: naive and naive ground state
Austin Smith and his group identified another sub-state of naive pluripotency, which seems to have captured the core regulatory circuit of pluripotency and is, therefore, termed the naive ground state [124]. When cultured with Lif and with the two inhibitors of Mek/Erk signalling and GSK3ß signalling (2i-Lif), murine ESCs are not reliant on external signalling anymore, including JAK/STAT3 signalling. In addition, they do not express, as in the traditional culture medium containing serum and Lif, developmental and differentiation-associated genes, but rather genes related to glycolysis, lipid, vesicle biology and metabolism, which is in agreement with the Warburg-effect of pluripotent cells, and they appear more pluripotent as judged by higher contribution to chimerism [110,124].
The significance of the Lif-2i culture condition is that it de-differentiates the naive PSCs even further, which are cultured in the Lif-serum conditions into naive ground state PSCs [110]. Naive ground state PSCs do not only exhibit higher chimera contribution, hypomethylation and distinct transcriptome and epigenetic signatures, but most importantly, the Lif-2i culture condition allows the derivation of mESCs from any mouse strain. The serum–Lif condition allowed the derivation only from the 129 strain. This strongly suggests that 2i-Lif allows the pluripotent state to be governed by its core regulatory circuits and is independent of any permissive strain-specific background mutations like in the mouse strain 129. Furthermore, the 2i-Lif culture conditions allowed for the first time the isolation of rat ESCs [125,126].
Indeed, Marks et al. [110] investigated the naive sub-states, naive and naive ground state of mESCs in both conditions, serum-Lif and 2i-Lif, respectively. The states are readily interconvertible upon culture medium change. They discovered that in comparison to the cultivation in 2i-Lif, many upregulated genes are involved in the differentiation of the different germ layers. The 2i-Lif, however, revealed the expression of many metabolism-associated and vesicle-associated pathways, in particular lysosome biology, indicating the existence of further unravelled mechanisms associated with the induction and maintenance of pluripotency.
While the murine naive pluripotent state per se and its sub-states are subjected to intense investigation, the human naive state has yet to be identified. It is currently being hunted and although several groups have reported to have captured the human naive pluripotent state [127–129], these different groups do not use the same cell culture conditions. This might or might not reflect the plasticity of the supposedly generated naive PSCs.
While at first glance it appeared that species differences account for the different characteristics of murine and human ESCs, which in fact was the difference between the naive and the primed state, it now appears puzzling why murine and supposedly human naive PSCs are very different in their culture condition requirements bearing in mind the astonishing similarity of murine and human primed PSCs. Therefore, it might be justified to expect that human naive stem cells resemble murine naive stem cells very closely, and hence the right culture conditions for robust human naive (ground) state derivation and maintenance have not been found yet. For instance, the murine naive ground state PSCs require 2i-Lif, which results in robust culture across laboratories. Also, new ESCs can be derived by the isolation of the ICM in this condition. So far, however, the culture conditions for human naive PSCs contain factors typical for the primed state such as Rock inhibitor, FGF2, ACTIVIN A or they require the transient and simultaneous overexpression of KLF2 or NANOG [127–132]. In the light of these very diverse culture conditions and the inclusion of factors that are required in the primed state, it seems as if the claimed human naive state is in fact still not fully dedifferentiated. Therefore, key questions remain unanswered: what are the authentic differences between the murine and the human blastocysts in terms of developmental timing? What happens to human ICM in contrast with the murine ICM upon isolation? Why does it appear to differentiate? One potential explanation could be the sensitivity to oxygen. Indeed, it has been reported that there is a reciprocal relationship between body size and oxygen tension in the fallopian tubes and uteri of monkeys, rabbits and hamsters [133]. As there is a huge difference in body size in mice and human, one possible explanation could be that the murine embryo is not affected upon ICM isolation in vitro, because it was exposed to higher oxygen tension in the murine reproductive tract. By contrast, the human body size makes oxygen diffusion more difficult, resulting in decreased oxygen tension, which could be a differentiation cue upon ICM isolation. Indeed, none of the present studies has applied oxygen tension below 5% [127–132].
10. Comparative meta-analysis of the transcriptomes of naive and primed pluripotent stem cells with isolated inner cell mass cells
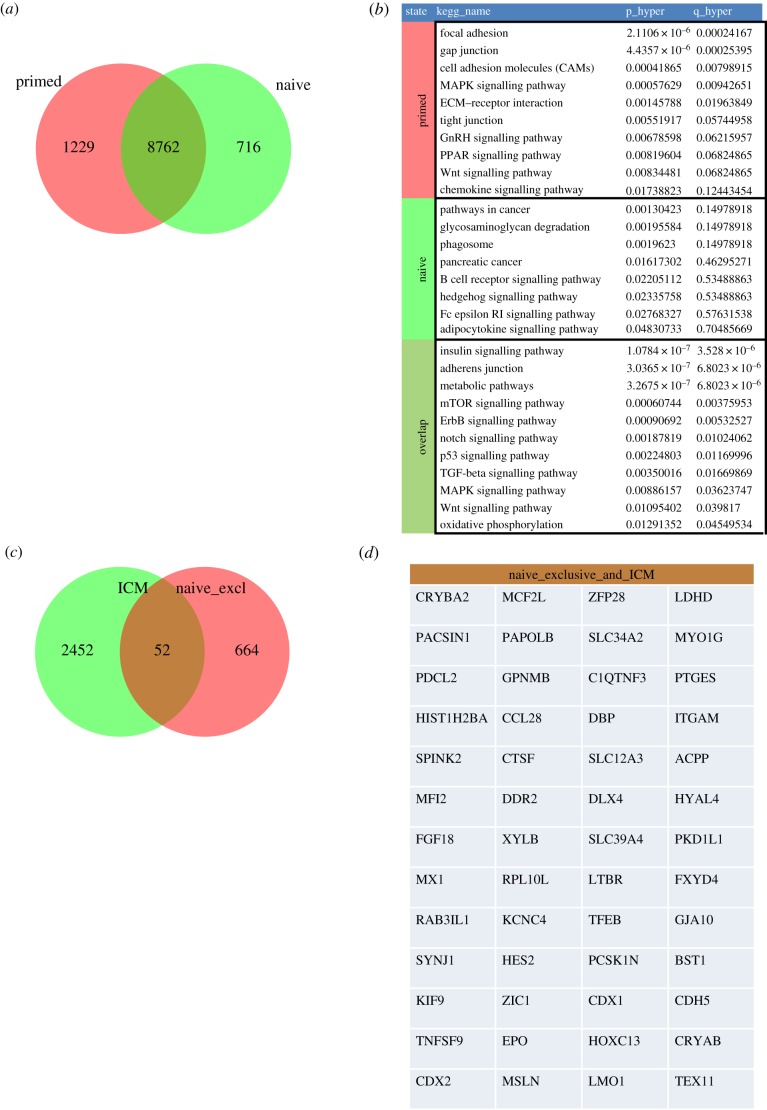
To better understand the molecular basis of the naive ground state, we compared it to the ICM in the human blastocyst. For this comparison we used datasets of deep sequencing measurements from human ESCs H9 and ‘reset’ H9 from Takashima et al. [129]. Takashima et al. converted human ESCs to a state they call ‘reset’ via short-term expression of the transcription factors NANOG and KLF2. Reset cells can be kept continuously in a self-renewal state by inhibition of ERK and protein kinase C. We annotated H9 as ‘primed’ and reset H9 as ‘naive’. Figure 1a shows a Venn diagram of genes expressed (FPKM: fragments per kilobase of transcript per million mapped reads > 1) in the naive and primed state in the data from Takashima et al. [129]. We identified 8762 genes as expressed in common in both states; 1229 genes were expressed exclusively in the primed state and 716 genes exclusively in the naive state. Figure 1b shows relevant KEGG pathways [135] over-represented—as calculated via the hypergeometric test—in these subsets of the Venn diagram. Within the overlap were—as expected—many pathways associated with pluripotency such as Wingless-Type MMTV Integration Site Family (WNT) and Notch signalling. In the exclusively primed subset, there is a tendency for tight junction and adhesion-related pathways, while the naive exclusive subset was enriched with genes associated with cancer. In figure 1c, the 716 genes exclusively expressed in the naive state were further compared with genes expressed (background tag: percentile of background spot intensities greater than 0.95) in isolated ICMs [134]. Here, 2452 genes are expressed exclusively in the ICM, 664 exclusively in the naive state and 52 genes are expressed in both (electronic supplementary material, table S1).
Figure 1.
A gene signature defining the naive state. Genes expressed in the primed and naive state in deep sequencing data from Takashima et al. [129] were compared (a) and the most relevant over-represented KEGG pathways exclusively in naive, primed and common in naive and primed are listed in (b). ECM, extracellular matrix. Genes exclusively expressed in the naive state were compared to genes expressed in the inner cell mass (ICM) from Adjaye et al. [134] (c) and are proposed as the marker signature of the naive state (d).
Figure 1d shows the common signature of genes that we propose as putative and additional markers defining the naive state. Furthermore, associations with pluripotency or differentiation have been ascribed to some of the genes (e.g. ZIC1, TEX11, FGF18, ZFP28, CDX1 and CDX2) [136].
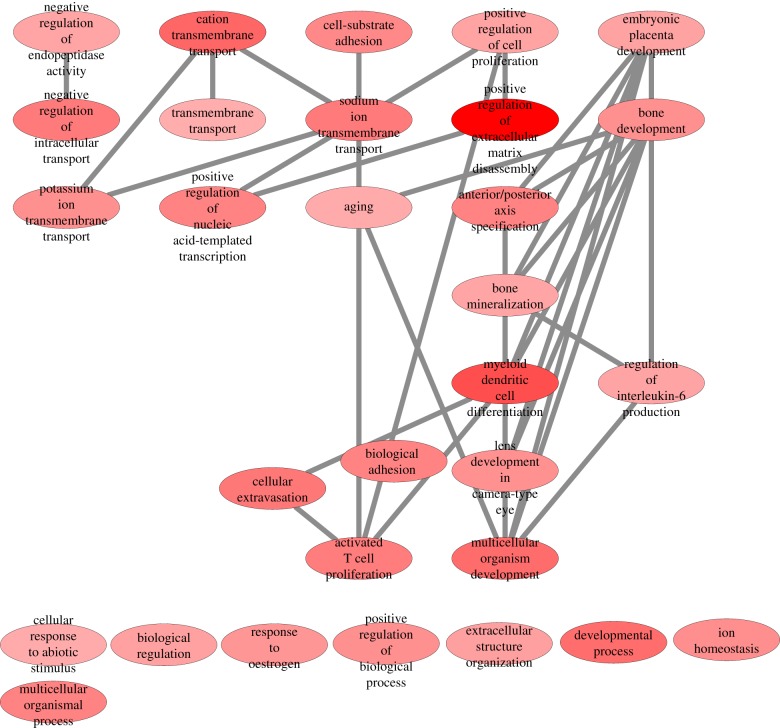
The naive state gene signature can be further characterized by significantly over-represented gene ontologies (GOs) as shown in figure 2. These GOs were identified by employing the R package GOstats [137] and summarized to a network using the REVIGO tool [138] and Cytoscape [139]. GO terms in dark red have a greater significance for over-representation; highly similar GO terms are connected. In this network, embryonic placenta development emerges as a major hub connecting many other developmental GO terms such as bone development, lens development in the camera-type eye and multicellular organism development. Though speculative, these GO terms would imply that naive PSCs have a broader developmental potential than their primed counterpart. The full list of GO terms and corresponding genes is presented in electronic supplementary material, table S2.
Figure 2.
Gene ontology (GO) network of the gene signature defining the naive state. Significantly over-represented GOs for the naive state gene signature were determined via the R package GOstats [137] and summarized to a network via the REVIGO tool [138] and Cytoscape [139]. The colour reflects the significance of over-representation of a GO term. The darker the red of a GO term node, the lower is its p-value. GO terms determined as highly similar by the REVIGO tool are connected. In this network, embryonic placenta development emerges as a major hub connecting many other developmental GO terms such as bone development, lens development in the camera-type eye and multicellular organism development. (Online version in colour.)
11. Conclusion and summary
Since the initial publication describing the derivation of iPSCs from fibroblasts, several inroads have been made leading to an increase in our understanding of the biological, molecular and biochemical processes associated with the successful derivation of iPSCs. Distinct states of pluripotency—primed and naive—have been actively debated. To date the naive state of pluripotency, in the human in particular, is still ‘work in progress’ as there remains a lack of a well-defined robust protocol that works reproducibly across laboratories and somatic cell types, in contrast with the primed state.
The shift from retroviral and lentiviral to non-integrative (episomal-based plasmids) means of delivery of the reprogramming factors has been monumental and even enabled the transfer of iPSC technology to the clinic for treating macular degeneration [37]. We believe the quest now is to identify non-invasive means of obtaining a well-defined and characterized cell population permissible for reprogramming. In this regard, urine has been shown to be an ideal source. For example, retroviruses, episomal-based plasmids and sendai viruses in combination with small molecule-mediated pathway modulations have been employed to successfully derive iPSCs from a not well-defined cell population within urine [140–142]. As a benchmark, SIX2-and SSEA4-positive renal progenitor cells expressing the CYP2D6 *4/*17 variant and of known human leukocyte antigen were isolated from urine and reprogrammed using episomal-based plasmids omitting pathway (TGFβ, MEK and GSK3β) inhibition [143].
Supplementary Material
Supplementary Material
Data accessibility
This article has no additional data.
Authors' contributions
J.A. and R.A.-D. conceived the concept of this review. All authors contributed to the writing of the manuscript. N.G. and S.F. provided the tables. W.W. and J.A. performed and interpreted the meta-analysis and provided the figures. J.A. performed final edits and gave the final approval of the manuscript prior to submission.
Competing interests
We declare we have no competing interests.
Funding
J.A. acknowledges funding from the Medical Faculty, Heinrich-Heine-University Düsseldorf. N.G. is funded by the Research Commission of the Medical Faculty, Heinrich-Heine-University Düsseldorf. R.A.-D. is funded by the King Faisal Specialist Hospital and Research Centre Riyadh.
References
- 1.Takahashi K, Yamanaka S. 2006. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663–676. ( 10.1016/j.cell.2006.07.024) [DOI] [PubMed] [Google Scholar]
- 2.Gilbert SF. 2010. Developmental biology. Sunderland, MA: Sinauer Associates, Inc. [Google Scholar]
- 3.Gurdon JB. 1962. The developmental capacity of nuclei taken from intestinal epithelium cells of feeding tadpoles. J. Embryol. Exp. Morphol. 10, 622–640. [PubMed] [Google Scholar]
- 4.Wilmut I, Schnieke AE, McWhir J, Kind AJ, Campbell KH. 1997. Viable offspring derived from fetal and adult mammalian cells. Nature 385, 810–813. ( 10.1038/385810a0) [DOI] [PubMed] [Google Scholar]
- 5.Kleinsmith LJ, Pierce GB Jr. 1964. Multipotentiality of single embryonal carcinoma cells. Cancer Res. 24, 1544–1551. [PubMed] [Google Scholar]
- 6.Miller RA, Ruddle FH. 1976. Pluripotent teratocarcinoma–thymus somatic cell hybrids. Cell 9, 45–55. ( 10.1016/0092-8674(76)90051-9) [DOI] [PubMed] [Google Scholar]
- 7.Martin GR. 1981. Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc. Natl Acad. Sci. USA 78, 7634–7638. ( 10.1073/pnas.78.12.7634) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Evans MJ, Kaufman MH. 1981. Establishment in culture of pluripotential cells from mouse embryos. Nature 292, 154–156. ( 10.1038/292154a0) [DOI] [PubMed] [Google Scholar]
- 9.Matsui Y, Zsebo K, Hogan BL. 1992. Derivation of pluripotential embryonic stem cells from murine primordial germ cells in culture. Cell 70, 841–847. ( 10.1016/0092-8674(92)90317-6) [DOI] [PubMed] [Google Scholar]
- 10.Resnick JL, Bixler LS, Cheng L, Donovan PJ. 1992. Long-term proliferation of mouse primordial germ cells in culture. Nature 359, 550–551. ( 10.1038/359550a0) [DOI] [PubMed] [Google Scholar]
- 11.Tada M, Tada T, Lefebvre L, Barton SC, Surani MA. 1997. Embryonic germ cells induce epigenetic reprogramming of somatic nucleus in hybrid cells. EMBO J. 16, 6510–6520. ( 10.1093/emboj/16.21.6510) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cowan CA, Atienza J, Melton DA, Eggan K. 2005. Nuclear reprogramming of somatic cells after fusion with human embryonic stem cells. Science 309, 1369–1373. ( 10.1126/science.1116447) [DOI] [PubMed] [Google Scholar]
- 13.Mitsui K, Tokuzawa Y, Itoh H, Segawa K, Murakami M, Takahashi K, Maruyama M, Maeda M, Yamanaka S. 2003. The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell 113, 631–642. ( 10.1016/S0092-8674(03)00393-3) [DOI] [PubMed] [Google Scholar]
- 14.Scholer HR, Balling R, Hatzopoulos AK, Suzuki N, Gruss P. 1989. Octamer binding proteins confer transcriptional activity in early mouse embryogenesis. EMBO J. 8, 2551–2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yuan H, Corbi N, Basilico C, Dailey L. 1995. Developmental-specific activity of the FGF-4 enhancer requires the synergistic action of Sox2 and Oct-3. Genes Dev. 9, 2635–2645. ( 10.1101/gad.9.21.2635) [DOI] [PubMed] [Google Scholar]
- 16.Davis RL, Weintraub H, Lassar AB. 1987. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell 51, 987–1000. ( 10.1016/0092-8674(87)90585-X) [DOI] [PubMed] [Google Scholar]
- 17.Sekiya S, Suzuki A. 2011. Direct conversion of mouse fibroblasts to hepatocyte-like cells by defined factors. Nature 475, 390–393. ( 10.1038/nature10263) [DOI] [PubMed] [Google Scholar]
- 18.Thier M, et al. 2012. Direct conversion of fibroblasts into stably expandable neural stem cells. Cell Stem Cell 10, 473–479. ( 10.1016/j.stem.2012.03.003) [DOI] [PubMed] [Google Scholar]
- 19.Huang P, et al. 2014. Direct reprogramming of human fibroblasts to functional and expandable hepatocytes. Cell Stem Cell 14, 370–384. ( 10.1016/j.stem.2014.01.003) [DOI] [PubMed] [Google Scholar]
- 20.Nakamura T, Nakagawa M, Ichisaka T, Shiota A, Yamanaka S. 2011. Essential roles of ECAT15-2/Dppa2 in functional lung development. Mol. Cell. Biol. 31, 4366–4378. ( 10.1128/MCB.05701-11) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tokuzawa Y, Kaiho E, Maruyama M, Takahashi K, Mitsui K, Maeda M, Niwa H, Yamanaka S. 2003. Fbx15 is a novel target of Oct3/4 but is dispensable for embryonic stem cell self-renewal and mouse development. Mol. Cell. Biol. 23, 2699–2708. ( 10.1128/MCB.23.8.2699-2708.2003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. 2007. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131, 861–872. ( 10.1016/j.cell.2007.11.019) [DOI] [PubMed] [Google Scholar]
- 23.Yu J, et al. 2007. Induced pluripotent stem cell lines derived from human somatic cells. Science 318, 1917–1920. ( 10.1126/science.1151526) [DOI] [PubMed] [Google Scholar]
- 24.Stadtfeld M, Nagaya M, Utikal J, Weir G, Hochedlinger K. 2008. Induced pluripotent stem cells generated without viral integration. Science 322, 945–949. ( 10.1126/science.1162494) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fusaki N, Ban H, Nishiyama A, Saeki K, Hasegawa M. 2009. Efficient induction of transgene-free human pluripotent stem cells using a vector based on Sendai virus, an RNA virus that does not integrate into the host genome. Proc. Jpn Acad. Ser. B 85, 348–362. ( 10.2183/pjab.85.348) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Si-Tayeb K, Noto FK, Sepac A, Sedlic F, Bosnjak ZJ, Lough JW, Duncan SA. 2010. Generation of human induced pluripotent stem cells by simple transient transfection of plasmid DNA encoding reprogramming factors. BMC Dev. Biol. 10, 81 ( 10.1186/1471-213X-10-81) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Narsinh KH, Jia F, Robbins RC, Kay MA, Longaker MT, Wu JC. 2011. Generation of adult human induced pluripotent stem cells using nonviral minicircle DNA vectors. Nat. Protoc. 6, 78–88. ( 10.1038/nprot.2010.173) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Woltjen K, et al. 2009. piggyBac transposition reprograms fibroblasts to induced pluripotent stem cells. Nature 458, 766–770. ( 10.1038/nature07863) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kaji K, Norrby K, Paca A, Mileikovsky M, Mohseni P, Woltjen K. 2009. Virus-free induction of pluripotency and subsequent excision of reprogramming factors. Nature 458, 771–775. ( 10.1038/nature07864) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grabundzija I, et al. 2013. Sleeping beauty transposon-based system for cellular reprogramming and targeted gene insertion in induced pluripotent stem cells. Nucleic Acids Res. 41, 1829–1847. ( 10.1093/nar/gks1305) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yu J, Hu K, Smuga-Otto K, Tian S, Stewart R, Slukvin II, Thomson JA. 2009. Human induced pluripotent stem cells free of vector and transgene sequences. Science 324, 797–801. ( 10.1126/science.1172482) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Warren L, et al. 2010. Highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mRNA. Cell Stem Cell 7, 618–630. ( 10.1016/j.stem.2010.08.012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yoshioka N, et al. 2013. Efficient generation of human iPSCs by a synthetic self-replicative RNA. Cell stem cell 13, 246–254. ( 10.1016/j.stem.2013.06.001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miyoshi N, et al. 2011. Reprogramming of mouse and human cells to pluripotency using mature microRNAs. Cell stem cell 8, 633–638. ( 10.1016/j.stem.2011.05.001) [DOI] [PubMed] [Google Scholar]
- 35.Kim D, et al. 2009. Generation of human induced pluripotent stem cells by direct delivery of reprogramming proteins. Cell Stem Cell 4, 472–476. ( 10.1016/j.stem.2009.05.005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nakagawa M, Takizawa N, Narita M, Ichisaka T, Yamanaka S. 2010. Promotion of direct reprogramming by transformation-deficient Myc. Proc. Natl Acad. Sci. USA 107, 14 152–14 157. ( 10.1073/pnas.1009374107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mandai M, et al. 2017. Autologous induced stem-cell-derived retinal cells for macular degeneration. N Engl. J. Med. 376, 1038–1046. ( 10.1056/NEJMoa1608368) [DOI] [PubMed] [Google Scholar]
- 38.Blanchard JW, Xie J, El-Mecharrafie N, Gross S, Lee S, Lerner RA, Baldwin KK. 2017. Replacing reprogramming factors with antibodies selected from combinatorial antibody libraries. Nat. Biotechnol. 35, 960–968. ( 10.1038/nbt.3963) [DOI] [PubMed] [Google Scholar]
- 39.Mah N, et al. 2011. Molecular insights into reprogramming-initiation events mediated by the OSKM gene regulatory network. PLoS ONE 6, e24351 ( 10.1371/journal.pone.0024351) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huangfu D, Maehr R, Guo W, Eijkelenboom A, Snitow M, Chen AE, Melton DA. 2008. Induction of pluripotent stem cells by defined factors is greatly improved by small-molecule compounds. Nat. Biotechnol. 26, 795–797. ( 10.1038/nbt1418) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang Y, Adjaye J. 2011. A cyclic AMP analog, 8-Br-cAMP, enhances the induction of pluripotency in human fibroblast cells. Stem Cell Rev. 7, 331–341. ( 10.1007/s12015-010-9209-3) [DOI] [PubMed] [Google Scholar]
- 42.Zhu S, Li W, Zhou H, Wei W, Ambasudhan R, Lin T, Kim J, Zhang K, Ding S. 2010. Reprogramming of human primary somatic cells by OCT4 and chemical compounds. Cell Stem Cell 7, 651–655. ( 10.1016/j.stem.2010.11.015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nashun B, Hill PW, Hajkova P. 2015. Reprogramming of cell fate: epigenetic memory and the erasure of memories past. EMBO J. 34, 1296–1308. ( 10.15252/embj.201490649) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang W, et al. 2011. Rapid and efficient reprogramming of somatic cells to induced pluripotent stem cells by retinoic acid receptor gamma and liver receptor homolog 1. Proc. Natl Acad. Sci. USA 108, 18 283–18 288. ( 10.1073/pnas.1100893108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li W, et al. 2012. Identification of Oct4-activating compounds that enhance reprogramming efficiency. Proc. Natl Acad. Sci. USA 109, 20 853–20 858. ( 10.1073/pnas.1219181110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang L, et al. 2015. Small molecules efficiently reprogram human astroglial cells into functional neurons. Cell Stem Cell 17, 735–747. ( 10.1016/j.stem.2015.09.012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hou P, et al. 2013. Pluripotent stem cells induced from mouse somatic cells by small-molecule compounds. Science 341, 651–654. ( 10.1126/science.1239278) [DOI] [PubMed] [Google Scholar]
- 48.Ye J, et al. 2016. Pluripotent stem cells induced from mouse neural stem cells and small intestinal epithelial cells by small molecule compounds. Cell Res. 26, 34–45. ( 10.1038/cr.2015.142) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang Q, et al. 2011. Lithium, an anti-psychotic drug, greatly enhances the generation of induced pluripotent stem cells. Cell Res. 21, 1424–1435. ( 10.1038/cr.2011.108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cheng L, Hu W, Qiu B, Zhao J, Yu Y, Guan W, Wang M, Yang W, Pei G. 2014. Generation of neural progenitor cells by chemical cocktails and hypoxia. Cell Res. 24, 665–679. ( 10.1038/cr.2014.32) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Son MJ, Son MY, Seol B, Kim MJ, Yoo CH, Han MK, Cho YS. 2013. Nicotinamide overcomes pluripotency deficits and reprogramming barriers. Stem Cells 31, 1121–1135. ( 10.1002/stem.1368) [DOI] [PubMed] [Google Scholar]
- 52.Onder TT, et al. 2012. Chromatin-modifying enzymes as modulators of reprogramming. Nature 483, 598–602. ( 10.1038/nature10953) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shi Y, Do JT, Desponts C, Hahm HS, Schöler HR, Ding S. 2008. A combined chemical and genetic approach for the generation of induced pluripotent stem cells. Cell Stem Cell 2, 525–528. ( 10.1016/j.stem.2008.05.011) [DOI] [PubMed] [Google Scholar]
- 54.Chen T, Shen L, Yu J, Wan H, Guo A, Chen J, Long Y, Zhao J, Pei G. 2011. Rapamycin and other longevity-promoting compounds enhance the generation of mouse induced pluripotent stem cells. Aging Cell 10, 908–911. ( 10.1111/j.1474-9726.2011.00722.x) [DOI] [PubMed] [Google Scholar]
- 55.Shi Y, Desponts C, Do JT, Hahm HS, Schöler HR, Ding S. 2008. Induction of pluripotent stem cells from mouse embryonic fibroblasts by Oct4 and Klf4 with small-molecule compounds. Cell Stem Cell 3, 568–574. ( 10.1016/j.stem.2008.10.004) [DOI] [PubMed] [Google Scholar]
- 56.Lee J, et al. 2012. A novel small molecule facilitates the reprogramming of human somatic cells into a pluripotent state and supports the maintenance of an undifferentiated state of human pluripotent stem cells. Angew. Chem. Int. Ed. Engl. 51, 12 509–12 513. ( 10.1002/anie.201206691) [DOI] [PubMed] [Google Scholar]
- 57.Li W, et al. 2009. Generation of human-induced pluripotent stem cells in the absence of exogenous Sox2. Stem Cells 27, 2992–3000. ( 10.1002/stem.240) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhao Y, et al. 2015. A XEN-like state bridges somatic cells to pluripotency during chemical reprogramming. Cell 163, 1678–1691. ( 10.1016/j.cell.2015.11.017) [DOI] [PubMed] [Google Scholar]
- 59.Zhang M, et al. 2016. Pharmacological reprogramming of fibroblasts into neural stem cells by signaling-directed transcriptional activation. Cell Stem Cell 18, 653–667. ( 10.1016/j.stem.2016.03.020) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li Y, et al. 2011. Generation of iPSCs from mouse fibroblasts with a single gene, Oct4, and small molecules. Cell Res. 21, 196–204. ( 10.1038/cr.2010.142) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hawkins KE, et al. 2017. Human amniocytes are receptive to chemically induced reprogramming to pluripotency. Mol. Ther. 25, 427–442. ( 10.1016/j.ymthe.2016.11.014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hu W, et al. 2015. Direct conversion of normal and Alzheimer's disease human fibroblasts into neuronal cells by small molecules. Cell Stem Cell 17, 204–212. ( 10.1016/j.stem.2015.07.006) [DOI] [PubMed] [Google Scholar]
- 63.Kubicek S, et al. 2007. Reversal of H3K9me2 by a small-molecule inhibitor for the G9a histone methyltransferase. Mol. Cell 25, 473–481. ( 10.1016/j.molcel.2007.01.017) [DOI] [PubMed] [Google Scholar]
- 64.Cheng X, et al. 2015. Identification of 2-[4-[(4-methoxyphenyl)methoxy]-phenyl]acetonitrile and derivatives as potent Oct3/4 inducers. J. Med. Chem. 58, 4976–4983. ( 10.1021/acs.jmedchem.5b00144) [DOI] [PubMed] [Google Scholar]
- 65.Cheng X, et al. 2015. Ethyl 2-((4-chlorophenyl)amino)thiazole-4-carboxylate and derivatives are potent inducers of Oct3/4. J. Med. Chem. 58, 5742–5750. ( 10.1021/acs.jmedchem.5b00226) [DOI] [PubMed] [Google Scholar]
- 66.Esteban MA, et al. 2010. Vitamin C enhances the generation of mouse and human induced pluripotent stem cells. Cell Stem Cell 6, 71–79. ( 10.1016/j.stem.2009.12.001) [DOI] [PubMed] [Google Scholar]
- 67.Staerk J, et al. 2011. Pan-Src family kinase inhibitors replace Sox2 during the direct reprogramming of somatic cells. Angew. Chem. Int. Ed. Engl. 50, 5734–5736. ( 10.1002/anie.201101042) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ichida JK, et al. 2009. A small-molecule inhibitor of tgf-β signaling replaces Sox2 in reprogramming by inducing Nanog. Cell Stem Cell 5, 491–503. ( 10.1016/j.stem.2009.09.012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lyssiotis CA, et al. 2009. Reprogramming of murine fibroblasts to induced pluripotent stem cells with chemical complementation of Klf4. Proc. Natl Acad. Sci. USA 106, 8912–8917. ( 10.1073/pnas.0903860106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yuan X, Wan H, Zhao X, Zhu S, Zhou Q, Ding S. 2011. Brief report: combined chemical treatment enables Oct4-induced reprogramming from mouse embryonic fibroblasts. Stem Cells 29, 549–553. ( 10.1002/stem.594) [DOI] [PubMed] [Google Scholar]
- 71.Moon JH, et al. 2011. Reprogramming fibroblasts into induced pluripotent stem cells with Bmi1. Cell Res. 21, 1305–1315. ( 10.1038/cr.2011.107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zheng J, et al. 2016. A combination of small molecules directly reprograms mouse fibroblasts into neural stem cells. Biochem. Biophys. Res. Commun. 476, 42–48. ( 10.1016/j.bbrc.2016.05.080) [DOI] [PubMed] [Google Scholar]
- 73.Park G, et al. 2015. Conversion of mouse fibroblasts into cardiomyocyte-like cells using small molecule treatments. Biomaterials 54, 201–212. ( 10.1016/j.biomaterials.2015.02.029) [DOI] [PubMed] [Google Scholar]
- 74.Cao N, et al. 2016. Conversion of human fibroblasts into functional cardiomyocytes by small molecules. Science 352, 1216–1220. ( 10.1126/science.aaf1502) [DOI] [PubMed] [Google Scholar]
- 75.Jagtap S, Meganathan K, Wagh V, Natarajan K, Hescheler J, Sachinidis A. 2013. All-trans retinoic acid and basic fibroblast growth factor synergistically direct pluripotent human embryonic stem cells to extraembryonic lineages. Stem Cell Res. 10, 228–240. ( 10.1016/j.scr.2012.12.002) [DOI] [PubMed] [Google Scholar]
- 76.Christman JK. 2002. 5-Azacytidine and 5-aza-2'-deoxycytidine as inhibitors of DNA methylation: mechanistic studies and their implications for cancer therapy. Oncogene 21, 5483–5495. ( 10.1038/sj.onc.1205699) [DOI] [PubMed] [Google Scholar]
- 77.Xu J, Du Y, Deng H. 2015. Direct lineage reprogramming: strategies, mechanisms, and applications. Cell Stem Cell 16, 119–134. ( 10.1016/j.stem.2015.01.013) [DOI] [PubMed] [Google Scholar]
- 78.Efe JA, Hilcove S, Kim J, Zhou H, Ouyang K, Wang G, Chen J, Ding S. 2011. Conversion of mouse fibroblasts into cardiomyocytes using a direct reprogramming strategy. Nat. Cell Biol. 13, 215–222. ( 10.1038/ncb2164) [DOI] [PubMed] [Google Scholar]
- 79.Kattman SJ, Witty AD, Gagliardi M, Dubois NC, Niapour M, Hotta A, Ellis J, Keller G. 2011. Stage-specific optimization of activin/nodal and BMP signaling promotes cardiac differentiation of mouse and human pluripotent stem cell lines. Cell Stem Cell 8, 228–240. ( 10.1016/j.stem.2010.12.008) [DOI] [PubMed] [Google Scholar]
- 80.Sayed N, Wong WT, Ospino F, Meng S, Lee J, Jha A, Dexheimer P, Aronow BJ, Cooke JP. 2015. Transdifferentiation of human fibroblasts to endothelial cells: role of innate immunity. Circulation 131, 300–309. ( 10.1161/CIRCULATIONAHA.113.007394) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Li X, et al. 2015. Small-molecule-driven direct reprogramming of mouse fibroblasts into functional neurons. Cell Stem Cell 17, 195–203. ( 10.1016/j.stem.2015.06.003) [DOI] [PubMed] [Google Scholar]
- 82.Mikkelsen TS, et al. 2008. Dissecting direct reprogramming through integrative genomic analysis. Nature 454, 49–55. ( 10.1038/nature07056) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Dai P, Harada Y, Takamatsu T. 2015. Highly efficient direct conversion of human fibroblasts to neuronal cells by chemical compounds. J. Clin. Biochem. Nutr. 56, 166–170. ( 10.3164/jcbn.15-39) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hoelder S, Clarke PA, Workman P. 2012. Discovery of small molecule cancer drugs: successes, challenges and opportunities. Mol. Oncol. 6, 155–176. ( 10.1016/j.molonc.2012.02.004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Brambrink T, Foreman R, Welstead GG, Lengner CJ, Wernig M, Suh H, Jaenisch R. 2008. Sequential expression of pluripotency markers during direct reprogramming of mouse somatic cells. Cell Stem Cell 2, 151–159. ( 10.1016/j.stem.2008.01.004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Stadtfeld M, Maherali N, Breault DT, Hochedlinger K. 2008. Defining molecular cornerstones during fibroblast to iPS cell reprogramming in mouse. Cell Stem Cell 2, 230–240. ( 10.1016/j.stem.2008.02.001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sridharan R, Tchieu J, Mason MJ, Yachechko R, Kuoy E, Horvath S, Zhou Q, Plath K. 2009. Role of the murine reprogramming factors in the induction of pluripotency. Cell 136, 364–377. ( 10.1016/j.cell.2009.01.001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Samavarchi-Tehrani P, Golipour A, David L, Sung HK, Beyer TA, Datti A, Woltjen K, Nagy A, Wrana JL. 2010. Functional genomics reveals a BMP-driven mesenchymal-to-epithelial transition in the initiation of somatic cell reprogramming. Cell Stem Cell 7, 64–77. ( 10.1016/j.stem.2010.04.015) [DOI] [PubMed] [Google Scholar]
- 89.Buganim Y, Faddah DA, Cheng AW, Itskovich E, Markoulaki S, Ganz K, Klemm SL, van Oudenaarden A, Jaenisch R. 2012. Single-cell expression analyses during cellular reprogramming reveal an early stochastic and a late hierarchic phase. Cell 150, 1209–1222. ( 10.1016/j.cell.2012.08.023) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Teshigawara R, Hirano K, Nagata S, Ainscough J, Tada T. 2016. OCT4 activity during conversion of human intermediately reprogrammed stem cells to iPSCs through mesenchymal–epithelial transition. Development 143, 15–23. ( 10.1242/dev.130344) [DOI] [PubMed] [Google Scholar]
- 91.Okita K, Ichisaka T, Yamanaka S. 2007. Generation of germline-competent induced pluripotent stem cells. Nature 448, 313–317. ( 10.1038/nature05934) [DOI] [PubMed] [Google Scholar]
- 92.Niwa H, Miyazaki J, Smith AG. 2000. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat. Genet. 24, 372–376. ( 10.1038/74199) [DOI] [PubMed] [Google Scholar]
- 93.Chambers I, Colby D, Robertson M, Nichols J, Lee S, Tweedie S, Smith A. 2003. Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell 113, 643–655. ( 10.1016/S0092-8674(03)00392-1) [DOI] [PubMed] [Google Scholar]
- 94.Loh KM, Lim B. 2011. A precarious balance: pluripotency factors as lineage specifiers. Cell Stem Cell 8, 363–369. ( 10.1016/j.stem.2011.03.013) [DOI] [PubMed] [Google Scholar]
- 95.Golipour A, David L, Liu Y, Jayakumaran G, Hirsch CL, Trcka D, Wrana JL. 2012. A late transition in somatic cell reprogramming requires regulators distinct from the pluripotency network. Cell Stem Cell 11, 769–782. ( 10.1016/j.stem.2012.11.008) [DOI] [PubMed] [Google Scholar]
- 96.Bortvin A, Eggan K, Skaletsky H, Akutsu H, Berry DL, Yanagimachi R, Page DC, Jaenisch R. 2003. Incomplete reactivation of Oct4-related genes in mouse embryos cloned from somatic nuclei. Development 130, 1673–1680. ( 10.1242/dev.00366) [DOI] [PubMed] [Google Scholar]
- 97.Kim M.-H, Kino-oka M. 2014. Maintenance of undifferentiated state of human induced pluripotent stem cells through cytoskeleton-driven force acting to secreted fibronectin on a dendrimer-immobilized surface. J. Biosci. Bioeng. 118, 716–722. ( 10.1016/j.jbiosc.2014.05.011) [DOI] [PubMed] [Google Scholar]
- 98.Boraas LC, Guidry JB, Pineda ET, Ahsan T. 2016. Cytoskeletal expression and remodeling in pluripotent stem cells. PLoS ONE 11, e0145084 ( 10.1371/journal.pone.0145084) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Anokye-Danso F, et al. 2011. Highly efficient miRNA-mediated reprogramming of mouse and human somatic cells to pluripotency. Cell stem cell 8, 376–388. ( 10.1016/j.stem.2011.03.001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Subramanyam D, Lamouille S, Judson RL, Liu JY, Bucay N, Derynck R, Blelloch R. 2011. Multiple targets of miR-302 and miR-372 promote reprogramming of human fibroblasts to induced pluripotent stem cells. Nat. Biotechnol. 29, 443–448. ( 10.1038/nbt.1862) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Pandian GN, Nakano Y, Sato S, Morinaga H, Bando T, Nagase H, Sugiyama H. 2012. A synthetic small molecule for rapid induction of multiple pluripotency genes in mouse embryonic fibroblasts. Sci. Rep. 2, Article number 544 ( 10.1038/srep00544) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zhang Z, Gao Y, Gordon A, Wang ZZ, Qian Z, Wu W-S. 2011. Efficient generation of fully reprogrammed human iPS cells via polycistronic retroviral vector and a new cocktail of chemical compounds. PLoS ONE 6, e26592 ( 10.1371/journal.pone.0026592) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Huangfu D, Osafune K, Maehr R, Guo W, Eijkelenboom A, Chen S, Muhlestein W, Melton DA. 2008. Induction of pluripotent stem cells from primary human fibroblasts with only Oct4 and Sox2. Nat. Biotechnol. 26, 1269–1275. ( 10.1038/nbt.1502) [DOI] [PubMed] [Google Scholar]
- 104.Moschidou D, et al. 2012. Valproic acid confers functional pluripotency to human amniotic fluid stem cells in a transgene-free approach. Mol. Ther. 20, 1953–1967. ( 10.1038/mt.2012.117) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Deng J, et al. 2009. Targeted bisulfite sequencing reveals changes in DNA methylation associated with nuclear reprogramming. Nat. Biotechnol. 27, 353–360. ( 10.1038/nbt.1530) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kim K, et al. 2010. Epigenetic memory in induced pluripotent stem cells. Nature 467, 285–290. ( 10.1038/nature09342) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Lister R, et al. 2009. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature 462, 315–322. ( 10.1038/nature08514) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Bernstein BE, et al. 2006. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125, 315–326. ( 10.1016/j.cell.2006.02.041) [DOI] [PubMed] [Google Scholar]
- 109.Azuara V, et al. 2006. Chromatin signatures of pluripotent cell lines. Nat. Cell Biol. 8, 532–538. ( 10.1038/ncb1403) [DOI] [PubMed] [Google Scholar]
- 110.Marks H, et al. 2012. The transcriptional and epigenomic foundations of ground state pluripotency. Cell 149, 590–604. ( 10.1016/j.cell.2012.03.026) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Prigione A, Fauler B, Lurz R, Lehrach H, Adjaye J. 2010. The senescence-related mitochondrial/oxidative stress pathway is repressed in human induced pluripotent stem cells. Stem Cells 28, 721–733. ( 10.1002/stem.404) [DOI] [PubMed] [Google Scholar]
- 112.Armstrong L, Tilgner K, Saretzki G, Atkinson SP, Stojkovic M, Moreno R, Przyborski S, Lako M. 2010. Human induced pluripotent stem cell lines show stress defense mechanisms and mitochondrial regulation similar to those of human embryonic stem cells. Stem Cells 28, 661–673. ( 10.1002/stem.307) [DOI] [PubMed] [Google Scholar]
- 113.Folmes CD, et al. 2011. Somatic oxidative bioenergetics transitions into pluripotency-dependent glycolysis to facilitate nuclear reprogramming. Cell Metab. 14, 264–271. ( 10.1016/j.cmet.2011.06.011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Prigione A, et al. 2014. HIF1α modulates cell fate reprogramming through early glycolytic shift and upregulation of PDK1-3 and PKM2. Stem Cells 32, 364–376. ( 10.1002/stem.1552) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, Jones JM. 1998. Embryonic stem cell lines derived from human blastocysts. Science 282, 1145–1147. ( 10.1126/science.282.5391.1145) [DOI] [PubMed] [Google Scholar]
- 116.Greber B, Lehrach H, Adjaye J. 2007. Fibroblast growth factor 2 modulates transforming growth factor β signaling in mouse embryonic fibroblasts and human ESCs (hESCs) to support hESC self-renewal. Stem Cells 25, 455–464. ( 10.1634/stemcells.2006-0476) [DOI] [PubMed] [Google Scholar]
- 117.Daheron L, Opitz SL, Zaehres H, Lensch WM, Andrews PW, Itskovitz-Eldor J, Daley GQ. 2004. LIF/STAT3 signaling fails to maintain self-renewal of human embryonic stem cells. Stem Cells 22, 770–778. ( 10.1634/stemcells.22-5-770) [DOI] [PubMed] [Google Scholar]
- 118.Peterson H, Abu Dawud R, Garg A, Wang Y, Vilo J, Xenarios I, Adjaye J. 2013. Qualitative modeling identifies IL-11 as a novel regulator in maintaining self-renewal in human pluripotent stem cells. Front. Physiol. 4, 303 ( 10.3389/fphys.2013.00303) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Gaspar-Maia A, Alajem A, Meshorer E, Ramalho-Santos M. 2011. Open chromatin in pluripotency and reprogramming. Nat. Rev. Mol. Cell Biol. 12, 36–47. ( 10.1038/nrm3036) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Watanabe K, et al. 2007. A ROCK inhibitor permits survival of dissociated human embryonic stem cells. Nat. Biotechnol. 25, 681–686. ( 10.1038/nbt1310) [DOI] [PubMed] [Google Scholar]
- 121.Chen G, Hou Z, Gulbranson DR, Thomson JA. 2010. Actin–myosin contractility is responsible for the reduced viability of dissociated human embryonic stem cells. Cell Stem Cell 7, 240–248. ( 10.1016/j.stem.2010.06.017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Tesar PJ, Chenoweth JG, Brook FA, Davies TJ, Evans EP, Mack DL, Gardner RL, McKay RD. 2007. New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature 448, 196–199. ( 10.1038/nature05972) [DOI] [PubMed] [Google Scholar]
- 123.Brons IG, et al. 2007. Derivation of pluripotent epiblast stem cells from mammalian embryos. Nature 448, 191–195. ( 10.1038/nature05950) [DOI] [PubMed] [Google Scholar]
- 124.Ying QL, Wray J, Nichols J, Batlle-Morera L, Doble B, Woodgett J, Cohen P, Smith A. 2008. The ground state of embryonic stem cell self-renewal. Nature 453, 519–523. ( 10.1038/nature06968) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Li P, et al. 2008. Germline competent embryonic stem cells derived from rat blastocysts. Cell 135, 1299–1310. ( 10.1016/j.cell.2008.12.006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Buehr M, et al. 2008. Capture of authentic embryonic stem cells from rat blastocysts. Cell 135, 1287–1298. ( 10.1016/j.cell.2008.12.007) [DOI] [PubMed] [Google Scholar]
- 127.Gafni O, et al. 2013. Derivation of novel human ground state naive pluripotent stem cells. Nature 504, 282–286. ( 10.1038/nature12745) [DOI] [PubMed] [Google Scholar]
- 128.Theunissen TW, et al. 2014. Systematic identification of culture conditions for induction and maintenance of naive human pluripotency. Cell Stem Cell. 15, 471–487. ( 10.1016/j.stem.2014.07.002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Takashima Y, et al. 2014. Resetting transcription factor control circuitry toward ground-state pluripotency in human. Cell 158, 1254–1269. ( 10.1016/j.cell.2014.08.029) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Chan YS, et al. 2013. Induction of a human pluripotent state with distinct regulatory circuitry that resembles preimplantation epiblast. Cell Stem Cell 13, 663–675. ( 10.1016/j.stem.2013.11.015) [DOI] [PubMed] [Google Scholar]
- 131.Valamehr B, et al. 2014. Platform for induction and maintenance of transgene-free hiPSCs resembling ground state pluripotent stem cells. Stem Cell Rep. 2, 366–381. ( 10.1016/j.stemcr.2014.01.014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Ware CB, et al. 2014. Derivation of naive human embryonic stem cells. Proc. Natl Acad. Sci. USA 111, 4484–4489. ( 10.1073/pnas.1319738111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Fischer B, Bavister BD. 1993. Oxygen tension in the oviduct and uterus of rhesus monkeys, hamsters and rabbits. J. Reproduct. Fertil. 99, 673–679. ( 10.1530/jrf.0.0990673) [DOI] [PubMed] [Google Scholar]
- 134.Adjaye J, et al. 2005. Primary differentiation in the human blastocyst: comparative molecular portraits of inner cell mass and trophectoderm cells. Stem Cells 23, 1514–1525. ( 10.1634/stemcells.2005-0113) [DOI] [PubMed] [Google Scholar]
- 135.Kanehisa M, Furumichi M, Tanabe M, Sato Y, Morishima K. 2017. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 45, D353–D361. ( 10.1093/nar/gkw1092) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Rao J, et al. 2016. Stepwise clearance of repressive roadblocks drives cardiac induction in human ESCs. Cell Stem Cell 18, 554–556. ( 10.1016/j.stem.2016.03.008) [DOI] [PubMed] [Google Scholar]
- 137.Falcon S, Gentleman R. 2007. Using GOstats to test gene lists for GO term association. Bioinformatics 23, 257–258. ( 10.1093/bioinformatics/btl567) [DOI] [PubMed] [Google Scholar]
- 138.Supek F, Bosnjak M, Skunca N, Smuc T. 2011. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS ONE 6, e21800 ( 10.1371/journal.pone.0021800) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. 2003. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504. ( 10.1101/gr.1239303) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Zhou T, et al. 2011. Generation of induced pluripotent stem cells from urine. J. Am. Soc. Nephrol. 22, 1221–1228. ( 10.1681/ASN.2011010106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Sauer V, et al. 2016. Human urinary epithelial cells as a source of engraftable hepatocyte-like cells using stem cell technology. Cell Transplant. 25, 2221–2243. ( 10.3727/096368916X692014) [DOI] [PubMed] [Google Scholar]
- 142.Si-Tayeb K, et al. 2016. Urine-sample-derived human induced pluripotent stem cells as a model to study PCSK9-mediated autosomal dominant hypercholesterolemia. Dis. Model. Mech. 9, 81–90. ( 10.1242/dmm.022277) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Bohndorf M, Ncube A, Spitzhorn LS, Enczmann J, Wruck W, Adjaye J. 2017. Derivation and characterization of integration-free iPSC line ISRM-UM51 derived from SIX2-positive renal cells isolated from urine of an African male expressing the CYP2D6 *4/*17 variant which confers intermediate drug metabolizing activity. Stem Cell Res. 25, 18–21. ( 10.1016/j.scr.2017.10.004) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This article has no additional data.