Abstract
The nuclear envelope (NE) is one of the least characterized structures of eukaryotic cells. The study of its functional roles is hampered by the small number of proteins known to be specifically located to it. Here, we present a comprehensive characterization of the NE proteome. We applied different fractionation procedures and isolated protein subsets derived from distinct NE compartments. We identified 148 different proteins by 16-benzyl dimethyl hexadecyl ammonium chloride (16-BAC) gel electrophoresis and matrix-assisted laser desorption ionization (MALDI) mass spectrometry; among them were 19 previously unknown or noncharacterized. The identification of known proteins in particular NE fractions enabled us to assign novel proteins to NE substructures. Thus, our subcellular proteomics approach retains the screening character of classical proteomic studies, but also allows a number of predictions about subcellular localization and interactions of previously noncharacterized proteins. We demonstrate this result by showing that two novel transmembrane proteins, a 100-kDa protein with similarity to Caenorhabditis elegans Unc-84A and an unrelated 45-kDa protein we named LUMA, reside in the inner nuclear membrane and likely interact with the nuclear lamina. The utility of our approach is not restricted to the investigation of the NE. Our approach should be applicable to the analysis of other complex membrane structures of the cell as well.
The identification of predicted gene products at the protein level bridges the gap between genome sequencing data and protein function, and is referred to as “functional genomics” (1, 2). In this respect, the combination of subcellular fractionation and mass spectrometrical techniques (“subcellular proteomics”) is a powerful strategy for the initial identification of previously unknown protein components and for their assignment to particular subcellular structures (3, 4).
The nuclei of eukaryotic cells contain several compartments defined by their morphological appearance in electron microscopy and by the distribution of a limited number of marker proteins (5). Because the structural and functional organization of nuclei seems to be intimately linked to the epigenetic control of gene expression, the characterization of such nuclear compartments at the molecular level is of great importance (6). The nuclear envelope (NE) is one of the least characterized compartments of the nucleus. It comprises an outer and inner nuclear membrane (ONM and INM, respectively), the pore membrane, the nuclear pore complexes, and the nuclear lamina (7). These subcompartments differ with respect to their protein components, but a thorough molecular characterization has not yet been achieved. Furthermore, a two-dimensional separation of NE membrane proteins by isoelectric focusing and SDS/PAGE fails because the separation system discriminates against integral membrane proteins (8). Therefore, the characterization of the NE at the protein level has to overcome general analytical challenges; the NE contains several compartments enriched in different subsets of integral membrane proteins and multiprotein complexes. Many of the integral membrane proteins interact with a stable network of structural proteins and cannot be extracted by detergents (7), a problem that is relevant for the analysis of any other complex membrane protein structure. Thus, finding a strategy capable of overcoming these problems would be of general interest and find broad application.
By using a subcellular proteomics approach designed for the analysis of integral membrane proteins, we screened for new nuclear membrane proteins in NE fractions prepared from nuclei isolated from neuro 2a (N2a) cells. We used different extraction schemes and achieved isolation of subsets of proteins derived from different NE compartments. One hundred twenty-nine different known proteins were identified by matrix-assisted laser desorption ionization (MALDI) mass spectrometry. In addition, we identified 19 gene products that had not been previously detected at the protein level. The known proteins provided a framework that enabled us to assign the novel ones to different NE substructures. To independently confirm the subcellular localization of some of these proteins, we used indirect immunofluorescence of transiently transfected cells in combination with confocal laser scanning microscopy. Two previously unknown proteins are apparently integral membrane proteins of the INM. One of these proteins with a mass of 100 kDa is related to Caenorhabditis elegans Unc-84A. The other protein, which we named LUMA, has a mass of 45 kDa and displays no similarity to any known proteins.
Our work demonstrates the power of the subcellular proteomics approach. The large percentage of new or up to now noncharacterized proteins detected in our study underlines the need for further efforts in the investigation of the NE.
Methods
Cell Culture and Transfection.
Mouse neuroblastoma N2a cells were cultured in DMEM containing 10% FBS, 100 μg⋅ml−1 streptomycin, and 100 μg⋅ml−1 penicillin at 37°C in a humidified atmosphere with 5% CO2. COS-7 cells were cultured at the same conditions on 13-mm glass coverslips. Monolayer cultures at 50% confluency were transfected by using SuperFect (Qiagen, Chatsworth, CA) according to the manufacturer's manual.
Preparation of Nuclei and NE, and Fractionation of NE.
Nuclei and NE from N2a cells were prepared as described (9). Briefly, the nuclei (5–8 mg of protein) were suspended in 40 ml of ice cold TP buffer (10 mM Tris⋅HCl, pH 7.4/10 mM NaH2PO4 /Na2HPO4, pH 7.4/1 mM PMSF/10 μg⋅ml−1 aprotinin/10 μg⋅ml−1 leupeptin) containing 250 μg ml−1 Heparin, 1 mM Na3VO4, 10 mM NaF, and 400 units of Benzon Nuclease (Merck). After stirring for 90 min at 4°C, nuclear envelopes were sedimented by centrifugation (10,000 × g) for 30 min at 4°C and resuspended in STM 0.25 buffer (20 mM Tris⋅HCl, pH 7.4/0.25 M Sucrose/5 mM MgSO4/1 mM Na3VO4/1 mM PMSF/10 μg⋅ml−1 aprotinin/10 μg⋅ml−1 leupeptin). Protein concentrations were determined according to Bradford (10). For further fractionation, the NE preparation in STM 0.25 (200 μg protein per sample) was supplemented either with Triton X-100 [TX-100; final concentration 0.5% (wt/vol)], with urea and Na2CO3 (final concentration 4 M and 0.1 M, respectively), or NaCl (final concentration 1 M). The samples were incubated for 15 min in a shaker at 4°C. TX-100-resistant material, as well as the NaCl-washed sample, was pelleted at 13,000 rpm in a Biofuge (Heraeus). Chaotrope-resistant material was pelleted at 50,000 rpm in a Beckman tabletop ultracentrifuge. Protein concentrations were determined according to Bradford (10). For further fractionation, the NE preparation in STM 0.25 (200 μg protein per sample) was supplemented either with TX-100 [final concentration 0.5% (wt/vol)], with urea and Na2CO3 (final concentration 4 M and 0.1 M, respectively), or with NaCl (final concentration 1 M). The samples were incubated for 15 min in a shaker at 4°C. TX-100-resistant material, as well as the NaCl-washed NE, was pelleted at 13,000 rpm. Chaotrope-resistant material was pelleted at 50,000 rpm in a Beckman tabletop ultracentrifuge.
Two-Dimensional Gel Electrophoresis, In-Gel Digestion, and MALDI-MS Sample Preparation.
NE proteins were separated by two-dimensional 16-benzyldimethyl-n-hexadecyl ammonium chloride (16-BAC)-SDS/PAGE as described (11). Minigels (1.5 mm thickness) were used in both dimensions. After two-dimensional separation of the NE proteins, all protein spots that could be visualized by Coomassie G-250 staining were manually cut from the gel by using pipet tips. The in-gel digestion and MALDI-MS measurements on digest supernatants were carried out as previously described (12). The eluates of the gel pieces were dried, desalted by C18 ZipTips (Millipore) by using a stepwise elution of 1–2 μl of each 30%, 50%, and 70% (vol/vol) methanol/5% formic acid. These samples were prepared for MALDI-MS by using a sandwich matrix preparation as described (13). In some cases, N-terminal peptide derivatization by chlorosulfonyl acetyl chloride was performed (14).
MALDI-MS and Database Search.
The mass spectrometric measurements were performed on a Bruker Reflex MALDI-time of flight (TOF) mass spectrometer (Bruker Daltonik, Bremen, Germany) equipped with an ion gate and pulsed ion extraction. Post source decay fragment ion spectra were obtained by using the FAST method (Bruker Daltonik). The peptide mass fingerprint spectra were matched to the NCBI nonredundant database entries by using the program profound at http://www.proteometrics.com. The mass tolerance was set to 50 ppm; one missed cleavage site was tolerated and the search was restricted to mammalian proteins. A size cut-off was set to about 200% of the apparent molecular weight of the protein as estimated from the gels. The proteins were regarded as identified according to the significance criteria of the search program. A new search was performed with the unmatched peptide masses to identify comigrating proteins. Post source decay (PSD) spectra were matched to NCBI nonredundant database entries or to dbEST-entries by using the program pepfrag at http://www.proteometrics.com. Expressed sequence tag (EST) sequence data were translated into a hypothetical protein sequence by using the program protein machine (http://www2.ebi.ac.uk/translate/). Subsequently, peptide mass fingerprint spectra were reinvestigated to assign more peptide masses to predicted peptides corresponding to the same EST. blast searches were performed at http://www.ncbi.nlm.nih.gov/blast/. To identify potential protein domain signature sequences and putative transmembrane regions in the novel or noncharacterized proteins, the programs smart (http://smart.embl-heidelberg.de), pfam hmm (http://pfam.wustl.edu/hmmsearch.shtml), and protscale (http://www.expasy.ch/cgi-bin/protscale.pl?l) were used.
RNA Isolation and cDNA Synthesis.
Total RNA from 5 × 106 N2a cells was isolated by using the SV Total RNA Isolation System (Promega) according to the manufacturer's manual. Total RNA (5 μg) was incubated with 200 ng oligo(dT)17 primer for 10 min at 65°C; then the RT buffer (GIBCO), 15 nmol dNTPs, 0.3 μmol DTT, RnaseOut, and 300 units of Reverse Transcriptase (GIBCO) were added to a total volume of 30 μl. After 1 h incubation at 42°C, the reaction was supplemented with 6 μl of Second Strand Buffer (GIBCO) and 5 units of RnaseH (GIBCO) and incubated for another 30 min at 37°C.
DNA Constructs.
The obtained cDNA was used as template in a PCR reaction with complementary sense (5′-GCCACAATGGCCGCGAATTATTCCAGTACC) and antisense (5′-CTCCAACTTTTTGGCTGGCACCCGTGTCCG) primers, which resulted in amplification of a 1200-bp DNA fragment [corresponding to the cDNA encoding the putative protein PID (protein identifier) 12836214]. KIAA0810 was amplified from human cDNA clone HK05647 (kindly provided by T. Nagase, Kazusa DNA Research Institute, Japan) with the sense primer 5′-GCCACAATGCGGCGGCGCGAGGCAGTATGG and the antisense primer 5′-CTTGACAGGTTCGCCATGAACTCTGAACCG.
PCR products were cloned into a eukaryotic expression plasmid by using the pcDNA3.1/V5-His TOPO TA Cloning Kit (Invitrogen) according to the manufacturer's manual. The correctness of all PCR-derived sequences was confirmed by sequencing.
Immunofluorescence.
Immunofluorescence studies were carried out to examine the subcellular distribution of the recombinant proteins. COS-7 cells were transferred into 24-well plates containing sterilized glass cover slips and transfected. Approximately 19 h later, cells were fixed and probed with a mouse monoclonal anti-V5 antibody (Invitrogen) as primary antibody at 1:500 dilution and a Cy2-conjugated goat anti-mouse (Jackson ImmunoResearch), diluted 1:500, as secondary antibody. Fluorescence images were obtained with a confocal laser scanning microscope (LSM 510, Carl Zeiss Jena GmbH, Jena, Germany).
Results
Enrichment of Protein Subsets Derived from Distinct NE Compartments.
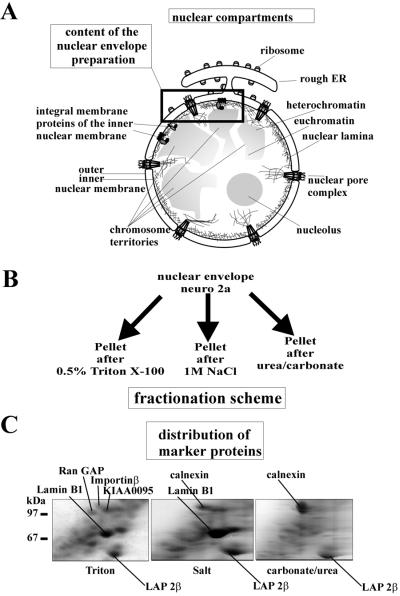
The initial NE preparation contains the nuclear membranes, nuclear pore complexes, the nuclear lamina with the lamina-interacting nuclear matrix, and associated proteins. It represents 6% of the total nuclear protein (refs. 15 and 16; Fig. 1A). We used the 16-BAC-SDS/PAGE system for the separation of NE proteins because it is capable of representing integral membrane proteins. The fractionation of the initial NE preparation served two goals. First, we aimed at reducing the sample complexity to account for the limited resolution of the 16-BAC-SDS/PAGE gels. Second, by applying different fractionation procedures (Fig. 1B), we prepared subsets of NE proteins derived from distinct NE compartments. We furthermore focused on the INM because of its unique protein composition. Therefore, our strategy was to isolate and analyze fractions enriched in lamina-associated polypeptide 2β (LAP 2β). LAP 2β is an integral membrane protein of the INM; it interacts with the nuclear lamina and chromatin and is resistant to limited extraction by TX-100 (17, 18). The fractionation was characterized according to distribution of compartment-specific marker proteins: lamin B1 (nuclear lamina), calnexin [ONM/endoplasmic reticulum (ER)], and LAP 2β (INM, Fig. 1C).
Figure 1.
Preparation of NE subfractions. (A) Illustrates schematically the architecture of the nucleus of a eukaryotic cell with emphasis on the NE compartments. The frame indicates the substructures present in the NE preparation. (B) Fractionation scheme applied to the fractionation of the NE. (C) Distribution of the marker proteins calnexin (ONM/ER membrane), LAP 2β (INM), and lamin B1 (nuclear lamina) throughout the different NE subfractions. Calnexin is absent from the TX-100-resistant fraction; lamin B1 is almost absent from the chaotrope-resistant fraction.
LAP 2β was observed as a major protein spot in all fractions. Calnexin was absent from the TX-100-resistant fraction, but was a major protein in the chaotrope-resistant fraction where, in contrast, the amount of the lamina marker lamin B1 was significantly reduced. These data indicate that TX-100 treatment removes the ONM/ER membrane, and that the chaotrope treatment removes the nuclear lamina. All three marker proteins were abundant in the salt-washed fraction.
Identification of the Protein Ensembles Present in the NE Subfractions.
In total, 148 different proteins were identified. Nineteen proteins (which have never been identified at the protein level) were gene products corresponding to ESTs or full-length ORFs without any annotations regarding their function and subcellular localization. We identified 79 different proteins in the TX-100-resistant fraction, 73 in the salt-resistant fraction, and 72 in the chaotrope-resistant fraction. Seventy-nine of these proteins appeared only in one fraction.
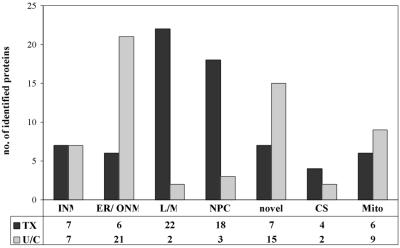
Consistent with the distribution of the marker proteins, we observed characteristic differences for the entire sets of proteins in the TX-100-resistant fraction as compared with the chaotrope-resistant fraction (Fig. 2; see also corresponding Tables 5 and 6, which are published as supporting information on the PNAS web site, www.pnas.org). We found most striking differences for the distribution of nucleoporins, for the proteins of the nuclear lamina and the attached scaffold, and the ER/ONM proteins. The distribution of INM proteins differed only in the abundance of emerin (see below). The presence of mitochondrial proteins in both fractions is most probably due to a minor contamination of the NE preparation.
Figure 2.
Distribution of NE proteins on the different fractions. Selected proteins detected in the TX-100-resistant NE fraction (Tx) and in the chaotrope-resistant fraction (U/C) grouped according to their subcellular localization. See also Tables 5 and 6 for the identity of the proteins. INM, inner nuclear membrane; ER/ONM, endoplasmic reticulum/outer nuclear membrane; L/M, nuclear lamina and attached protein scaffold; NPC, nuclear pore complex; CS, cytoskeleton; Mito, mitochondria. Note the differences in the distribution of ER/ONM proteins, L/M proteins, and NPC proteins.
Annotated gel pictures and complete lists of the identified proteins are provided in Figs. 6–8 and Tables 2–6, which are published as supporting information on the PNAS web site, www.pnas.org.
Proteins of the TX-100-Resistant Fraction.
The TX-100-resistant fraction contains almost all known nuclear pore complex proteins that, according to literature data (19), face the nucleoplasma or reside in the pore membrane (see Table 5 for details). Thus, the subcellular assignment of these proteins based on our proteomic approach is in agreement with results obtained by classical cell biological methods. Also, a number of known RNA binding proteins and nuclear matrix structural proteins were detected. This indicates that some nuclear matrix components are tightly attached to the nuclear lamina.
Surprisingly, an oligosaccharyl transferase supposedly located in the ONM/ER membrane and two proteins, Sec22b- and Sec13-related protein, both involved in vesicle trafficking, were also found in this fraction.
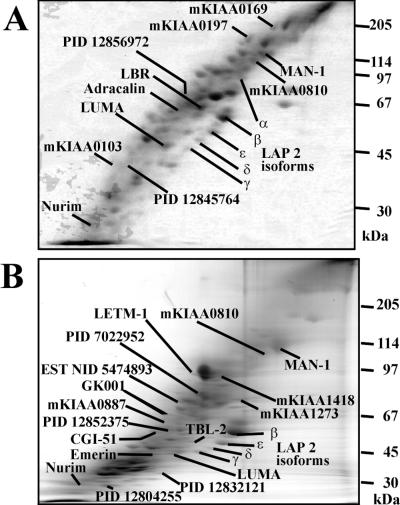
Most of the known integral membrane proteins of the INM were detected in this fraction, namely nurim, lamin B receptor, transmembrane LAP 2 splice variants β, γ, δ, and ɛ, and MAN-1 (Fig. 3A). LAP 2γ and δ were identified at the protein level. Emerin was absent from this fraction, although it has been described to interact with lamin A in vitro (20). LAP 1 was not found in any fraction although it was suggested to be a major integral membrane protein of the INM (21). The murine orthologue of MAN-1 was detected as a protein migrating at an apparent molecular weight of 110 kDa, which indicates that the nonredundant database contains only a fragment of the murine sequence (22). The presence of MAN-1 and nurim in the TX-100-extracted fraction suggests that both proteins interact with the nuclear lamina or other stable components.
Figure 3.
INM and novel NE proteins as separated by 16-BAC-SDS/PAGE. (A) Proteins of the TX-100-resistant fraction. (B) Proteins of the chaotrope-resistant fraction.
Detection of Murine KIAA0810 and LUMA, Two Novel Integral Membrane Proteins in the TX-100-Resistant Fraction.
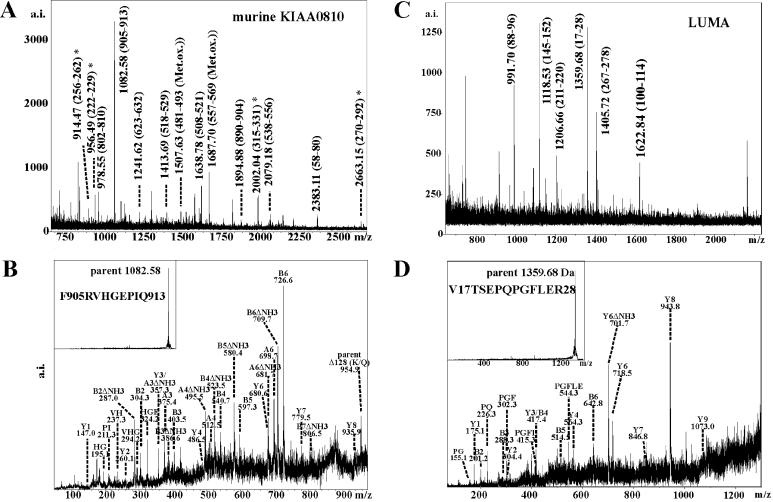
We identified two previously unknown putative integral membrane proteins. The murine homologue of KIAA0810 encoded by the Institute of Physical and Chemical Research (Japan) (RIKEN) cDNA clone ID 15962 [Functional Annotation of Mouse (FANTOM) database at http://www.gsc.riken.go.jp/e/FANTOM] was identified at an apparent molecular mass of ≈100 kDa. Different databases contain conflicting sequence data with respect to hypothetical proteins corresponding to the KIAA0810 cDNA. Based on the peptide mass fingerprint data, we were able to discriminate between the protein corresponding to the cDNA sequence contained in the FANTOM database and the hypothetical protein PID 12852531, which lacks amino acid residues 221–344 of the FANTOM sequence. The protein expressed in N2a cells corresponds to sequence ID 15962 in the FANTOM database (Fig. 4A). The protein is partially homologous to the C. elegans Unc-84A protein (23). Based on the primary structure analysis, murine KIAA0810 is predicted to contain a C2H2 zinc finger domain and three to four putative transmembrane segments.
Figure 4.
Identification of murine KIAA0810 and LUMA by MALDI-MS. (A) Peptide mass fingerprint of murine KIAA0810. Some of the peptides matching the predicted protein sequence are indicated. Peptides labeled by asterisks correspond to the amino acid sequence stretch present in the translated cDNA clone ID15962 of the FANTOM database, but missing in the putative protein PID 12852531. (B) PSD fragment ion spectrum of the peptide of MH+ = 1082.58 Da corresponding to the sequence F905RVHGEPIQ913 of murine KIAA0810. B-ions are N-terminal; Y′′-ions are C-terminal. These predominant ion types arise from backbone fragmentation within the amide bonds but still contain one terminus of the parent ion. Because of the presence of an internal arginine residue, strong signals of −17-Da satellites of B-ions (loss of ammonia) are observed and indicated as “ΔNH3.” (C) Peptide mass fingerprint of LUMA; some peptides matching the predicted amino acid sequence are indicated. (D) PSD fragment ion spectrum of the peptide of MH+ = 1359.68 Da corresponding to the sequence V17TSEPQPGFLER28 of LUMA. Proline-directed decay can be observed giving rise to a characteristic internal B-ion series starting with one of the prolines. Because of this decay, large B- or Y′′-ions exceeding the positions of the prolines are hardly detected.
We also identified a putative integral membrane protein of 45 kDa (PID 12836214) which, based on primary structure analysis, contains three to four putative transmembrane segments. We name this protein LUMA.
Novel Non-Membrane-Spanning Proteins of the TX-100-Resistant Fraction.
Apart from murine KIAA0810 and LUMA, we detected several other previously unknown proteins. For example, the murine homologue of KIAA0103 (≈40 kDa) was found in this fraction. KIAA0103 may represent, based on its similarity to Nicotiana tabacum protein PID 10799830 and C. elegans PID 3319457, a novel putative O-linked glycosyl transferase. Another interesting protein found was adracalin (PID 7021151). Adracalin was described at the genome level as being mutated in triple A syndrome (24).
Four more as yet undetected proteins of this group were found. Their features, as deduced from their primary structures, are listed in Table 1.
Table 1.
Features of the previously noncharacterized proteins
| Protein/PID | kDa | TX-r | U/C-r | TM | Domains/similarities |
|---|---|---|---|---|---|
| KIAA0810 | 100 | Yes | Yes | 3–4 | C2H2 zinc finger/UNC-84A |
| LUMA | 45 | Yes | Yes | 3–4 | — |
| LAP 2γ | 40 | Yes | Yes | 1 | LEM domain |
| LAP 2δ | 45 | Yes | Yes | 1 | LEM domain |
| KIAA0169 | 200 | Yes | — | — | — |
| gene trap locus 13 | 160 | Yes | — | — | — |
| 12856972 | 75 | Yes | — | — | — |
| adracalin | 60 | Yes | — | — | WD40 |
| 12845764 | 36 | Yes | — | — | WD40 |
| KIAA0103 | 40 | Yes | — | — | TPR repeats/putative OGT |
| KIAA1418 | 94 | — | Yes | 1 | — |
| LETM1 | 83 | — | Yes | 1 | EF-hand, leucine zipper |
| KIAA1273 | 67 | — | Yes | — | Triple A domain |
| GK001 | 56 | — | Yes | ? | — |
| KIAA0887 | 52 | — | Yes | — | UBX domain |
| 7022952 | 63 | — | Yes | ? | DnaJ domain |
| CGI-51 | 52 | — | Yes | ? | — |
| 12852375 | 60 | — | Yes | 1–3 | — |
| 5474893 | ∼65 | — | Yes | ? | — |
| TBL 2 | 50 | — | Yes | — | WD40 |
| 12832121 | 26 | — | Yes | — | — |
| 12804255 | 23 | — | Yes | — | G-patch |
Summary of novel proteins detected in the TX-100-resistant and in the chaotrope-resistant fraction. The programs smart, pfam hmm, and protscale were used for primary structure analysis (see Methods). TX-r, TX-100-resistant fraction; U/C-r, chaotrope-resistant fraction; TM, putative transmembrane regions; OGT, O-linked glycosyl transferase; PID, protein identifier.
Proteins of the Chaotrope-Resistant Fraction.
The chaotrope-resistant NE fraction was highly enriched in ER/ONM proteins (Fig. 2). We also detected the known INM proteins (Fig. 3B), including emerin, which had been absent from the TX-100-resistant fraction. Murine KIAA0810 was identified, and its identity was further confirmed by PSD sequencing of the peptide MH+ = 1082.58 Da. The peptide corresponds to amino acid residues F905RVHGEPIQ913 (Fig. 4B). LUMA was detected in spot 81 (Fig. 4C). Its identity was confirmed by PSD sequencing of several peptides, e.g., of the peptide MH+ = 1359.60 Da corresponding to V17TSEPQPGFLER28 (Fig. 4D).
Several other as yet undetected proteins were found (Fig. 3B). Their predicted molecular features are listed in Table 1.
Subcellular Localization of KIAA 0810 and LUMA as Assessed by Indirect Immunofluorescence on Transiently Transfected Cells.
KIAA0810 and LUMA are two putative integral membrane proteins present in the TX-100-resistant NE fraction and thus are likely to represent novel integral membrane proteins of the INM. To confirm their subcellular localization independently, we performed indirect immunofluorescence on transiently transfected cells. Human KIAA0810 was amplified by PCR from its cDNA clone, yielding a 2500-bp fragment. This fragment was cloned into pcDNA3.1/V5-His, a eukaryotic expression vector providing a C-terminal His6- and V5-tag. LUMA was amplified from the cDNA of N2a cells, yielding a 1200-bp fragment that was also ligated into the pcDNA3.1/V5-His vector. The plasmids were transfected into COS-7 cells, and the distribution of the proteins was examined by anti-V5 immunofluorescence studies.
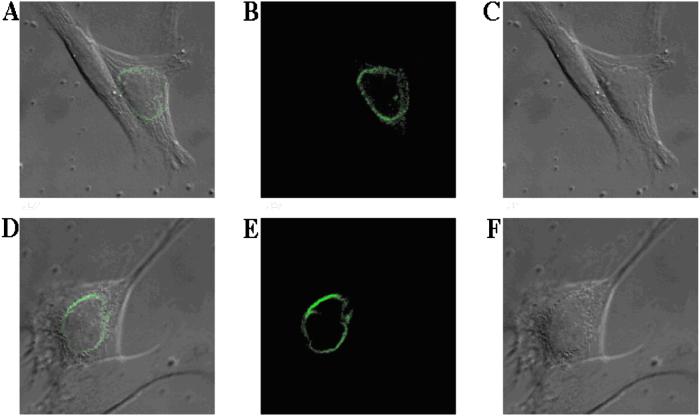
Indirect immunofluorescence detected by confocal laser scanning microscopy both with KIAA0810-transfected COS 7 cells (Fig. 5 A–C) and LUMA cDNA-transfected COS 7 cells (Fig. 5 D–F) revealed rim-like fluorescence patterns around the nucleus typical for integral membrane proteins of the INM (25).
Figure 5.
Overexpressed KIAA0810 and LUMA give rise to a rim-like staining around the nucleus as visualized by indirect immunofluorescence: COS-7 cells were transfected with a V5-tagged KIAA0810 (A, B, and C) and LUMA (D, E, and F) cDNA plasmids. Immunofluorescence studies were carried out with permeabilized cells grown on glass cover slips, as described under Methods. Cells were treated with an anti-V5 monoclonal antibody and then incubated with a Cy2-linked goat anti-mouse IgG secondary antibody. Fluorescence images were obtained with a confocal laser scanning microscope. The overlay pictures (A and D) are composed of immunofluorescence (B and E) and differential interference contrast (C and F) images, which were recorded in parallel. Each picture is representative of at least three independent experiments.
Discussion
There has been no previous systematic investigation of the NE proteome. By applying different one-step fractionation procedures to the NE preparation, we achieved two important goals: first, we reduced the sample complexity and thus compensated for the limited resolution of the 16-BAC-SDS/PAGE gels. Second, the different NE fractions contained characteristic subsets of known proteins derived from defined NE compartments. These proteins provide a structural framework of known proteins that allow us to assign unknown or noncharacterized proteins to one of the NE substructures and to predict some of their properties.
For example, according to the framework we obtained, novel putative integral membrane proteins detected in the TX-100-resistant fraction are likely to reside in the INM. We prove the validity of this prediction by the identification of two novel integral membrane proteins of the INM-murine KIAA0810 and LUMA. Both contain several putative transmembrane sequences and are resistant to limited extraction by TX-100. When overexpressed in COS-7 cells, they display a rim-like staining around the nucleus, as expected for integral INM proteins.
Our comprehensive characterization of the NE proteome also reveals previously unreported aspects of known INM proteins. MAN-1 and nurim were detected in the TX-100-resistant fraction, which is consistent with possible interactions with the nuclear lamina. Emerin was only abundant in the chaotrope-resistant fraction and therefore might be only weekly associated with the nuclear membranes. None of the LAP 1 isoforms could be detected. This observation argues against LAP 1 being a ubiquitous major integral membrane protein of the INM, at least in N2a cells.
The detection of previously unknown or noncharacterized molecules at the protein level confirms that they are indeed expressed in cells and are not merely predicted putative proteins deduced from sequences obtained by the diverse genome projects. In addition, their apparent molecular mass, combined with peptide mass fingerprint data, makes it possible to pick the truly expressed gene product among the many predicted from partly redundant and more or less complete cDNA sequences, as demonstrated in the case of murine KIAA0810. Furthermore, the source from which the protein was identified can serve as an experimental system by which the endogenous protein can be investigated.
Taken together, our study demonstrates the power of the subcellular proteomics approach and provides not only the basis for the detailed functional characterization of the nuclear envelope, but also opens new possibilities for the investigation of other complex membrane structures.
Supplementary Material
Acknowledgments
We thank Dr. T. Nagase, Kazusa DNA Research Institute, Chiba, Japan, for providing the KIAA0810 cDNA. We thank Doris Krück for culturing N2a cells and for the preparation of nuclei, and Inka Fedke for the preparation of DNA. This work was supported by the Deutsche Forschungsgemeinschaft (grants Hu 146-4 and Hu 146-22) and the Fonds der Chemischen Industrie.
Abbreviations
- N2a
neuro 2a
- NE
nuclear envelope
- INM
inner nuclear membrane
- ONM
outer nuclear membrane
- ER
endoplasmic reticulum
- LAP
lamina-associated polypeptide
- 16-BAC
16-benzyl dimethyl hexadecyl ammonium chloride
- PSD
post source decay
- PID
protein identifier
- TX-100
Triton X-100
- MALDI
matrix-assisted laser desorption ionization
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Pandey A, Mann M. Nature (London) 2000;405:837–846. doi: 10.1038/35015709. [DOI] [PubMed] [Google Scholar]
- 2.Andersen J S, Mann M. FEBS Lett. 2000;480:25–31. doi: 10.1016/s0014-5793(00)01773-7. [DOI] [PubMed] [Google Scholar]
- 3.Bell A W, Ward M A, Freeman H N, Choudhary J S, Blackstock W P, Lewis A P, Fazel A, Gushue J N, Paiement J, Palcy S, et al. J Biol Chem. 2000;276:5152–5165. doi: 10.1074/jbc.M006143200. [DOI] [PubMed] [Google Scholar]
- 4.Neubauer G, King A, Rappsilber J, Calvio C, Watson M, Ajuh P, Sleeman J, Lamond A, Mann M. Nat Genet. 1998;20:46–50. doi: 10.1038/1700. [DOI] [PubMed] [Google Scholar]
- 5.Lamond A I, Earnshaw W C. Science. 1998;280:547–553. doi: 10.1126/science.280.5363.547. [DOI] [PubMed] [Google Scholar]
- 6.Misteli T. Science. 2001;291:843–847. doi: 10.1126/science.291.5505.843. [DOI] [PubMed] [Google Scholar]
- 7.Gant T M, Wilson K L. Annu Rev Cell Dev Biol. 1997;13:669–695. doi: 10.1146/annurev.cellbio.13.1.669. [DOI] [PubMed] [Google Scholar]
- 8.Santoni V, Molloy M, Rabilloud T. Electrophoresis. 2000;21:1054–1070. doi: 10.1002/(SICI)1522-2683(20000401)21:6<1054::AID-ELPS1054>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 9.Dreger M, Otto H, Neubauer G, Mann M, Hucho F. Biochemistry. 1999;38:9426–9434. doi: 10.1021/bi990645f. [DOI] [PubMed] [Google Scholar]
- 10.Bradford M M. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 11.Hartinger J, Stenius K, Hogemann D, Jahn R. Anal Biochem. 1996;240:126–133. doi: 10.1006/abio.1996.0339. [DOI] [PubMed] [Google Scholar]
- 12.Shevchenko A, Wilm M, Vorm O, Mann M. Anal Chem. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- 13.Watty A, Neubauer G, Dreger M, Zimmer M, Wilm M, Burden S J. Proc Natl Acad Sci USA. 2000;97:4585–4590. doi: 10.1073/pnas.080061997. . (First Published April 18, 2000; 10.1073/pnas.080061997) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Keough T, Youngquist R S, Lacey M P. Proc Natl Acad Sci USA. 1999;96:7131–7136. doi: 10.1073/pnas.96.13.7131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Emig S, Schmalz D, Shakibaei M, Buchner K. J Biol Chem. 1995;270:13787–13793. doi: 10.1074/jbc.270.23.13787. [DOI] [PubMed] [Google Scholar]
- 16.Otto H, Buchner K, Beckmann R, Hilbert R, Hucho F. Neurochem Int. 1992;21:409–414. doi: 10.1016/0197-0186(92)90192-t. [DOI] [PubMed] [Google Scholar]
- 17.Foisner R, Gerace L. Cell. 1993;73:1267–1279. doi: 10.1016/0092-8674(93)90355-t. [DOI] [PubMed] [Google Scholar]
- 18.Furukawa K, Pante N, Aebi U, Gerace L. EMBO J. 1995;14:1626–1636. doi: 10.1002/j.1460-2075.1995.tb07151.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Allen T D, Cronshaw J M, Bagley S, Kiseleva E, Goldberg M W. J Cell Sci. 2000;113:1651–1659. doi: 10.1242/jcs.113.10.1651. [DOI] [PubMed] [Google Scholar]
- 20.Clements L, Manilal S, Love D R, Morris G E. Biochem Biophys Res Commun. 2000;267:709–714. doi: 10.1006/bbrc.1999.2023. [DOI] [PubMed] [Google Scholar]
- 21.Maison C, Pyrpasopoulou A, Theodoropoulos P A, Georgatos S D. EMBO J. 1997;16:4839–4850. doi: 10.1093/emboj/16.16.4839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin F, Blake D L, Callebaut I, Skerjanc I S, Holmer L, McBurney M W, Paulin-Levasseur M, Worman H J. J Biol Chem. 2000;275:4840–4847. doi: 10.1074/jbc.275.7.4840. [DOI] [PubMed] [Google Scholar]
- 23.Malone C J, Fixsen W D, Horvitz H R, Han M. Development. 1999;126:3171–3181. doi: 10.1242/dev.126.14.3171. [DOI] [PubMed] [Google Scholar]
- 24.Tullio-Pelet A, Salomon R, Hadj-Rabia S, Mugnier C, de Laet M H, Chaouachi B, Bakiri F, Brottier P, Cattolico L, Penet C, et al. Nat Genet. 2000;26:332–335. doi: 10.1038/81642. [DOI] [PubMed] [Google Scholar]
- 25.Wilson K L. Trends Cell Biol. 2000;10:125–129. doi: 10.1016/s0962-8924(99)01708-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.