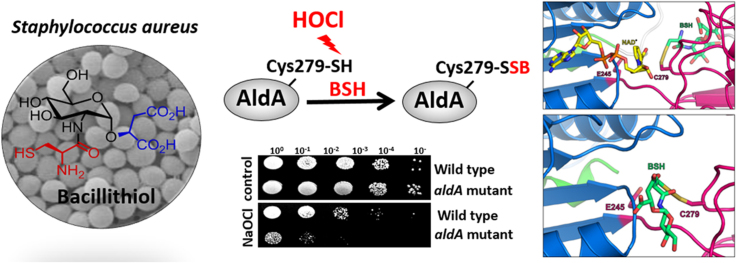
Abstract
Staphylococcus aureus produces bacillithiol (BSH) as major low molecular weight (LMW) thiol which functions in thiol-protection and redox-regulation by protein S-bacillithiolation under hypochlorite stress. The aldehyde dehydrogenase AldA was identified as S-bacillithiolated at its active site Cys279 under NaOCl stress in S. aureus. Here, we have studied the expression, function, redox regulation and structural changes of AldA of S. aureus. Transcription of aldA was previously shown to be regulated by the alternative sigma factor SigmaB. Northern blot analysis revealed SigmaB-independent induction of aldA transcription under formaldehyde, methylglyoxal, diamide and NaOCl stress. Deletion of aldA resulted in a NaOCl-sensitive phenotype in survival assays, suggesting an important role of AldA in the NaOCl stress defense. Purified AldA showed broad substrate specificity for oxidation of several aldehydes, including formaldehyde, methylglyoxal, acetaldehyde and glycol aldehyde. Thus, AldA could be involved in detoxification of aldehyde substrates that are elevated under NaOCl stress. Kinetic activity assays revealed that AldA is irreversibly inhibited under H2O2 treatment in vitro due to overoxidation of Cys279 in the absence of BSH. Pre-treatment of AldA with BSH prior to H2O2 exposure resulted in reversible AldA inactivation due to S-bacillithiolation as revealed by activity assays and BSH-specific Western blot analysis. Using molecular docking and molecular dynamic simulation, we further show that BSH occupies two different positions in the AldA active site depending on the AldA activation state. In conclusion, we show here that AldA is an important target for S-bacillithiolation in S. aureus that is up-regulated under NaOCl stress and functions in protection under hypochlorite stress.
Abbreviations: ADH, aldehyde dehydrogenase; BSH, bacillithiol; BSSB, oxidized bacillithiol disulfide; CFU, colony-forming unit; CD, catalytic domain; Co-BD, coenzyme-binding domain; DTT, dithiothreitol; EDTA, ethylenediaminetetraacetic acid; FA, formaldehyde; H2O2, hydrogen peroxide; HOCl, hypochloric acid; IPTG, isopropyl-β-D-thiogalactopyranoside; LB, Luria Bertani; LMW thiol, low molecular weight thiol; MD, molecular dynamics; MG, methylglyoxal; MHQ, 2-methylhydroquinone; MPO, myeloperoxidase; MRSA, methicillin-resistant Staphylococcus aureus; NADH, nicotinamide adenine dinucleotide; NADPH, nicotinamide adenine dinucleotide phosphate; NaOCl, sodium hypochlorite; NEM, N-ethylmaleimide; OD500, optical density at 500 nm; RCS, reactive chlorine species; RES, reactive electrophilic species; ROS, reactive oxygen species; SCV, small colony variant; SID, subunit interaction domain; X-gal, 5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside
Keywords: Staphylococcus aureus, AldA, Bacillithiol, Hypochlorite stress, MD simulations
Graphical abstract
Highlights
-
•
AldA is S-bacillithiolated at Cys279 under NaOCl stress in Staphylococcus aureus.
-
•
AldA is strongly induced under NaOCl and aldehyde stress and protects S. aureus under hypochlorite stress.
-
•
S-bacillithiolation functions in redox-regulation of AldA activity.
-
•
The position of BSH in the AldA active site depends on the Cys279 activation state.
1. Introduction
Staphylococcus aureus is a major human pathogen that causes local wound infections, but also life-threatening systemic and chronic infections, such as septicemia, endocarditis, necrotizing pneumonia and osteomyelitis [1], [2], [3]. Moreover, there is an increasing prevalence of hospital- and community-acquired methicillin-resistant S. aureus (MRSA) isolates that are often resistant to multiple antibiotics [4]. S. aureus quickly escapes to bactericidal action of new antibiotics and is therefore classified as ESKAPE pathogen by the “European Center of Disease Prevention and Control” [5]. The successful infection of S. aureus is mediated by a high diversity of virulence factors, such as toxins, proteases, lipases, superantigens, as well as efficient protection mechanisms against the host immune defense during invasion [6], [7]. During infections, S. aureus has to cope with the oxidative burst of activated macrophages and neutrophils, including reactive oxygen and chlorine species (ROS, RCS), such as hydrogen peroxide (H2O2) and the strong oxidant hypochloric acid (HOCl) [8], [9], [10], [11]. HOCl is generated in neutrophils from H2O2 and chloride by the myeloperoxidase (MPO) which is the main cause of bacterial killing [12], [13].
Apart from ROS and RCS, S. aureus is frequently exposed to reactive electrophile species (RES), such as quinones and aldehydes that originate from cellular metabolism, as secondary oxidation products from ROS and RCS as well as from external sources, such as antibiotics and host-defense components [11], [14], [15], [16], [17]. RES are α,β-unsaturated dicarbonyl compounds that have electron-deficient centers and can react with protein thiols via oxidation or thiol-S-alkylation chemistries [16], [17]. Methylglyoxal is an example for a highly toxic and reactive aldehyde produced as by-product from triose-phosphate intermediates during glycolysis [14], [15]. Methylglyoxal detoxification pathways and their regulatory mechanisms have been widely studied in E. coli and B. subtilis. E. coli utilises a glutathione (GSH)-dependent glyoxalase pathway and a GSH-independent pathway for methylglyoxal detoxification. In the glyoxalase pathway, methylglyoxal reacts spontaneously with GSH to form hemithioacetal which is converted by glyoxalase-I to S-lactoylglutathione. S-lactoylglutathione is the substrate for glyoxalase-II leading to lactate production [14], [18]. The glyoxalase gloA and the nemRA operon are induced by quinones, aldehydes and HOCl and regulated by the TetR-family NemR repressor in E. coli [19], [20], [21], [22]. GloA functions as glyoxalase in methylglyoxal detoxification and NemA is an FMN-dependent oxidoreductase involved in detoxification of quinones and aldehydes. Moreover, it was shown that methylglyoxal is produced as consequence of hypochlorite stress and that NemR confers protection to methylglyoxal and HOCl via control of the gloA-nemRA operon [20].
Gram-positive Firmicutes, such as Bacillus subtilis and S. aureus produce bacillithiol (BSH) as GSH surrogate which functions as protection mechanism against redox-active compounds and co-factor for thiol-dependent enzymes [23], [24]. Methylglyoxal detoxification in B. subtilis involves BSH-dependent and BSH-independent pathways [23], [25]. In the BSH-dependent glyoxalase pathway, BSH reacts with methylglyoxal to form BS-hemithioacetal which is converted to S-lactoyl-BSH by Glx-I and further by Glx-II to lactate [23], [25]. In addition, the thiol-dependent formaldehyde dehydrogenase AdhA confers protection under formaldehyde and methylglyoxal stress in B. subtilis which is controlled by the MerR/NmlR-like regulator AdhR [35]. However, the enzymatic pathways involved in detoxification of reactive aldehydes are unknown in S. aureus.
Recently, we identified the glycolytic glyceraldehyde-3-phosphate dehydrogenase GapDH as major S-bacillithiolated protein in S. aureus under NaOCl stress [26]. Apart from GapDH, the aldehyde dehydrogenase AldA was S-bacillithiolated at its active site Cys279 under NaOCl stress, which could function in detoxification of methylglyoxal or other aldehyde substrates. Here, we have studied the expression and function of AldA of S. aureus under formaldehyde, methylglyoxal and NaOCl stress. Transcriptional studies revealed an increased aldA transcription under aldehyde, NaOCl and diamide stress in S. aureus. In survival phenotype assays, the aldA mutant was more sensitive to NaOCl stress. Using biochemical activity assays, we provide evidence that S-bacillithiolation functions in redox-regulation of AldA activity. All-atom molecular dynamics (MD) simulations suggest that the location of BSH in the AldA active site depends on the Cys activation state in the apo- and holoenzyme structures. In conclusion, our results indicate that AldA plays an important role in the NaOCl stress defense and is redox-regulated by S-bacillithiolation in S. aureus.
2. Materials and methods
2.1. Bacterial strains, growth and survival assays
Bacterial strains, plasmids and primers are listed in Tables S1 and S2. For cloning and genetic manipulation, E. coli was cultivated in Luria Bertani (LB) medium. S. aureus COL was cultivated either in LB or RPMI medium as described previously [26]. For survival phenotype assays, S. aureus COL was grown in RPMI medium until an OD500 of 0.5, exposed to 2 mM formaldehyde, 4 mM methylglyoxal and 3.5 mM NaOCl stress and 10 µl of serial dilutions were spotted onto LB agar plates for 24 h to observe colonies. All complemented aldA deletion mutants with plasmid pRB473 were grown in the presence of 1% xylose and 10 µg/ml chloramphenicol. Sodium hypochlorite, diamide, dithiothreitol (DTT), hydrogen peroxide (H2O2, 35% w/v), formaldehyde, methylglyoxal and 2-methylhydroquinone (MHQ) were purchased from Sigma Aldrich.
2.2. RNA isolation and Northern blot analysis
For RNA isolation, S. aureus COL was cultivated in RPMI medium and treated with sub-lethal doses of 1 mM NaOCl, 0.75 mM formaldehyde (FA), 0.5 mM methylglyoxal (MG), 10 mM H2O2 and 50 µM MHQ for different times as described previously [26]. S. aureus COL cells were harvested before and after stress exposure and disrupted in lysis buffer [10 mM Tris-HCl, pH 8.0; 200 mM sodium chloride (NaCl); 3 mM ethylene diamine tetra acetic acid (EDTA)] with a Precellys24 ribolyzer. RNA was isolated using acid phenol extraction as described [26] and RNA quality was assessed using the Nanodrop. Northern blot hybridizations were performed with the digoxigenin-labelled aldA-specific antisense RNA probe synthesized in vitro using T7 RNA polymerase and the primer pairs aldA-for and aldA-rev (Table S2) as described [26], [27].
2.3. Cloning, expression and purification of His-tagged AldA and AldC279S mutant proteins in E. coli
The aldA gene was amplified from chromosomal DNA of S. aureus COL by PCR using primers aldA-for-NheI and aldA-rev-BamHI (Table S2), digested with NheI and BamHI and inserted into plasmid pET11b (Novagen) that was digested using the same enzymes to generate plasmid pET11b-aldA. For construction of pET11b expressing AldAC279S mutant protein, Cys279 was replaced by serine using PCR mutagenesis. Two first-round PCR reactions were performed using primer pairs aldA-for-NheI and aldA-C279S-Rev as well as primer pairs aldA-C279S-for and aldA-rev-BamHI (Table S2). The two first round PCR products were hybridized and subsequently amplified by a second round of PCR using primers aldA-for-NheI and aldA-rev-BamHI. The second-round PCR products were digested with NheI and BamHI and inserted into plasmid pET11b digested with the same enzymes to generate plasmid pET11b-aldAC279S. The correct aldA and aldAC279S sequences of the plasmids were confirmed by DNA sequencing. Plasmid pET11b-aldAC279S was also used for construction of the aldAC279S mutant in vivo and subcloned into the E. coli/S. aureus shuttle vector pRB473 as described below.
For expression and purification of His-tagged AldA and AldAC279S mutant protein, E. coli BL21(DE3) plysS was used expressing plasmids pET11b-aldA and pET11b-aldAC279S, respectively. Cultivation was performed in 1 l LB medium until the exponential growth phase at OD600 of 0.8 followed by addition of 1 mM isopropyl-β-D-thiogalactopyranoside (IPTG) for 3.5 h at 37 °C. Recombinant His-AldA and His-AldAC279S mutant proteins were purified after sonication of the E. coli cells in binding buffer (20 mM NaH2PO4, 500 mM NaCl, 20 mM imidazole, pH 7.4). Lysates were cleared from cell debris by repeated centrifugation and purification of the His-AldA and His-AldAC279S mutant proteins was performed by application of an imidazole gradient (0–500 mM) using His Trap™ HP Ni-NTA columns (5 ml; GE Healthcare, Chalfont St. Giles, UK) and the ÄKTA purifier liquid chromatography system (GE Healthcare) according to the instructions of the manufacturer. Purified proteins were extensively dialyzed against 10 mM Tris-HCl (pH 8.0), 100 mM NaCl, and stored on ice until usage.
2.4. Construction of the S. aureus COL aldA deletion mutant and the complemented aldA and aldAC279S mutant strains
The S. aureus COL ΔaldA deletion mutant was constructed by allelic replacement via the temperature-sensitive shuttle vector pMAD as described [28]. Briefly, for construction of the plasmids pMAD-ΔaldA, the 500 bp up- and downstream flanking gene regions of aldA were amplified using the primers aldA-pMAD-up-for/rev and aldA-pMAD-do-for/rev from S. aureus COL genomic DNA (Table S2). The aldA up- and downstream flanking regions were fused by overlap extension PCR and ligated into the BglII and SalI sites of plasmid pMAD. The pMAD constructs were electroporated into the restriction-negative and methylation-positive intermediate S. aureus RN4220 strain and further transferred to S. aureus COL by phage transduction using phage 80 [29]. Transductants were streaked out on LB agar with 10 µg/ml erythromycin and 40 µg/ml 5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside (X-gal) at 30 °C. Blue transductants with pMAD integrations were selected for plasmid excision by a heat shock as described [30]. Erythromycin-sensitive white colonies were selected on X-gal plates and screened for aldA deletions by PCR and DNA sequencing.
The complemented aldA and aldAC279S mutant strains were constructed using the pRB473 plasmid as described [31]. Briefly, aldA and aldAC279S sequences were amplified from plasmids pET11b-aldA and pET11b-aldAC279S using the primers aldA-pRB-for-BamHI and aldA-pRB-rev-KpnI. The PCR products were digested with BamHI and KpnI and inserted into the pRB473 plasmid that was digested using the same enzymes resulting in plasmids pRB473-aldA and pRB473-aldAC279S. The plasmids were transferred to the ΔaldA mutant via phage transduction as described [31].
2.5. AldA activity assays
AldA activity was monitored spectrophotometrically at 340 nm and 30 °C with the substrate and NAD+ as cofactor by the production of NADH using a CLARIOstar (BMG Labtech) spectrophotometer. The oxidation of different aldehyde substrates (formaldehyde, methylglyoxal, acetaldehyde and glycol aldehyde) was measured in an assay mixture containing 1.25 mM NAD+ and 2.5 µM AldA in reaction buffer (100 mM Tris-HCl, 1.25 mM EDTA, pH 7.5). After pre-incubation, the reaction was started by addition of the aldehyde substrates and NADH production was measured at 340 nm. The kinetic curves are presented as mean ± SEM from at least three independent experiments.
2.6. Western blot analysis
The purified His-AldA protein was separated using 12% SDS-PAGE and subjected to BSH-specific Western blot analysis using the polyclonal rabbit anti-BSH antiserum as described previously [32].
2.7. Molecular docking of the S-bacillithiolated AldA Cys279 active site
To model a covalent complex between BSH and the AldA Cys279 active site by molecular docking, the crystal structure of AldA from S. aureus was used as a receptor (PDB code 3TY7). The missing loop (residues 438-459) was modelled and fitted using MODELLER [33]. To identify the potential BSH binding site, FTMap solvent mapping calculations were performed [34] and two highest-occupancy binding sites were considered in the further calculations (Fig. 7EF). In the Q1 site, the NAD+ molecule has been fitted using crystal structures of the Pseudomonas fluorescens pfAMSDH co-crystallised with NAD+ (PDB code 4I1W). Then, the hydrogen atoms were added, and the charges for NAD+ molecule were assigned using AM1-BCC method [35]. The Cys279 thiol group was considered deprotonated. The BSH molecule was built, energy minimised (5000 cycles of steepest-descent minimisation), and the partial atomic charges were generated using AM1-BCC [35].
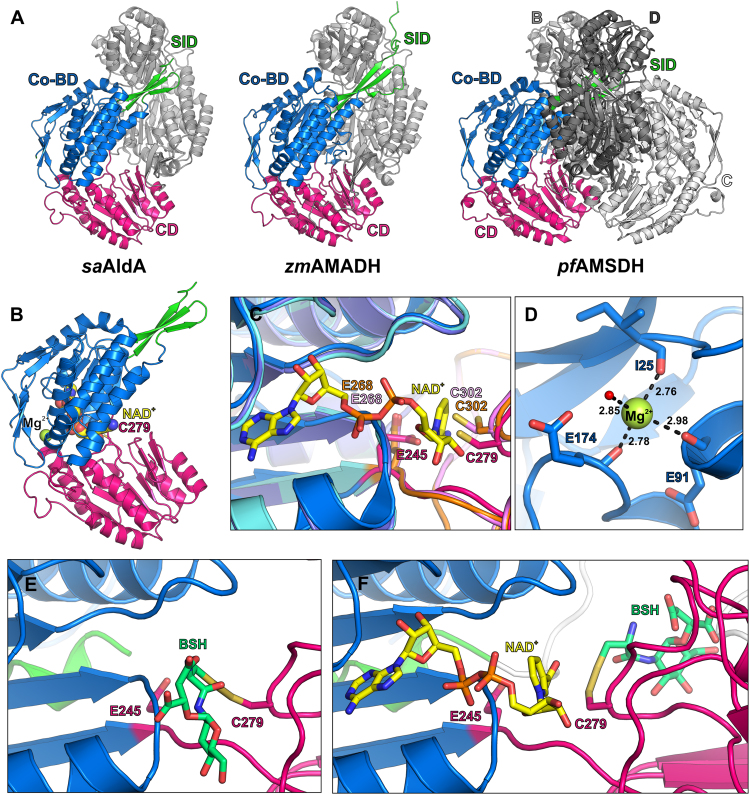
Fig. 7.
Structural insights into theS-bacillithiolatedsaAldA active site. (A) Structural overviews of dimeric saAldA (PDB ID: 3TY7), dimeric zmAMADH (PDB ID: 4I8P) and tetrameric pfAMSDH (PDB ID: 4I26). Dimers formed by chains A (colored by domain; coenzyme-binding domain [Co-BD] – blue; subunit interaction domain [SID] – green; catalytic domain [CD] – magenta) and B (grey) are oriented in the same way. The other dimer of the pfAMSDH tetramer (chains C and D) is shown in different shades of grey. (B) Model for NAD+ binding by saAldA obtained by superimposing a subunit of NAD+-bound pfAMSDH (PDB ID: 4I1W) on apo-saAldA. The modelled NAD+ (colored by atom type; carbon – yellow; oxygen – red; nitrogen – blue; phosphorus – orange) and a bound Mg2+ ion (lime green) are shown as spheres, the active site cysteine (C279) is shown as sticks (carbon – magenta; sulfur – yellow). (C) Active sites and NAD+-binding cavities of ADHs. A subunit of saAldA (colored as in A) was structurally aligned with subunits of apo-pfAMSDH (Co-BD – violet; CD – light pink) and of NAD+-bound pfAMSDH (Co-BD – cyan; CD – orange). NAD+ is shown as sticks (colored by atom type as in B). The catalytic cysteine residue is in the resting state in the apo-structures and in the attacking state in NAD+-bound pfAMSDH. (D) Interactions at the cation-binding site of saAldA. Red sphere – water oxygen. (E, F) The S-bacillithiolated active site pocket of the apo-saAldA (E) and holo-saAldA (F). A subunit of saAldA colored as in A, view as in C, NAD+ and BSH are shown as sticks, NAD+ is colored as in B, BSH colored by atom type (carbon – aquamarine; oxygen – red; nitrogen – blue; sulfur – yellow). The loop composed of residues 438-459 that is not present in saAldA structure (PDB ID: 3TY7) was modelled and is shown in white.
Molecular docking was performed using the University of California, San Francisco DOCK 6.8 suite [36] with grid scoring in an implicit solvent. The grid spacing was 0.25 Å, and the grid included 12 Å beyond the NAD+ modelled, which was subsequently removed for the pose Q2. The energy score was the sum of electrostatic and van der Waals contributions. To check the suitability of the methodology, the NAD+ was removed from the binding site, its translational and rotational degrees of freedom were altered and the molecule has been re-docked to the protein, in order to check whether the docking procedure was able to reproduce the native binding mode, as observed in related crystal structures. After the positive verification, the BSH molecule was docked to both Q1 (holo-enzyme with NAD+) and Q2 (apo-enzyme without NAD+) sites detected by FTMap [34].
During the docking calculations, the BSH molecule was subjected to 5000 cycles of molecular-mechanical energy minimisation at the protein-binding site. The number of maximum ligand orientations was 50,000. The constraint was the distance between sulfur atoms from the Cys279 thiol and the sulfur of BSH. The 25 best-scoring poses (BSH-protein complexes) were further analyzed by means of secondary rescoring using SeeSAR https://www.biosolveit.de/SeeSAR/ package with more accurate HYDE scoring function [37]. The best-scoring poses in Q1 and Q2 putative binding sites were subjected to all-atom MD simulations.
2.8. Molecular dynamics (MD) simulation of S-bacillithiolation
All simulations for the 5 studied systems: apo-enzyme, holo-enzyme (protein-NAD+), BSH-holo-enzyme (Q1), BSH-apo-enzyme (Q1), and BSH-apo-enzyme (Q2) were carried out using GROMACS2016.2 code [38], with Amber99SB-ILDN [39] force field for the duplexes and the TIP3P water model. Parameters for NAD+ and BSH were assigned by ACPYPE [40]. Obtained partial atomic charges were derived using the RESP methodology [41] and validated with the Gaussian09 programme [42] using HF/6-31G∗ basis set.
The temperature was kept constant at T = 300 K by using velocity rescaling with a coupling time of 0.1 ps. The pressure was kept constant at 1 bar using an isotropic coupling to Parrinello-Rahman barostat with a coupling time of 0.1 ps [43]. A cut-off of 1 nm was used for all non-bonded interactions. Long-range electrostatic interactions were treated with the particle-mesh Ewald [44] method using a grid spacing of 0.1 nm with cubic interpolation. All bonds between hydrogens and heavy atoms were constrained using the LINCS algorithm [45]. Each of the systems were immersed in a cubic TIP3P water box containing ∼ 115,000 atoms. Simulation units were maintained neutral by adding sodium and chloride counter ions (0.1 M concentration).
Prior to MD simulations, the systems undergone 50,000 steps of molecular mechanical energy minimisation. This was followed by 100 ps MD simulations, during which position constraints were used on all backbone atoms, heavy atoms of BSH and NAD+. After the following unrestrained equilibration phase (10 ns) the production runs were carried out for 50 ns, with an integration time step of 2 fs. The cut-off for non-bonded interactions was 0.1 nm. The atomic coordinates were saved every 100 ps. For the visual inspection of the results we used xmgrace [46] and UCSF Chimera [47] packages. Free binding energy calculations have been performed using the MMPBSA.py program from AmberTools package [48]. Binding energies have been calculated between BSH and the protein at the two different binding sites, as in Q1 and Q2, for the last 25 ns of the simulation.
3. Results
3.1. The aldehyde dehydrogenase AldA is strongly oxidized at its active site Cys279 due to S-bacillithiolation under NaOCl stress in S. aureus
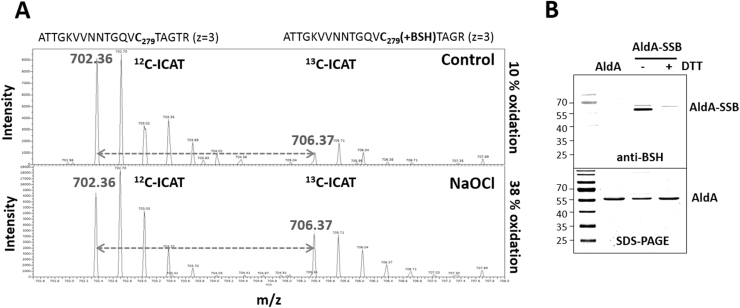
The aldehyde dehydrogenase AldA was previously identified as S-bacillithiolated at its catalytic active site Cys279 in S. aureus and Staphylococcus carnosus [26], [32]. In addition, both aldehyde dehydrogenases, GapDH and AldA displayed the highest oxidation increase of 29% under NaOCl stress in S. aureus using the thiol-redox proteomics approach OxICAT [26]. The OxICAT method is based on thiol-labelling of the reduced AldA Cys279 peptide with light 12C-ICAT reagent, followed by reduction of the Cys279-SSB peptide and its labelling with heavy 13C-ICAT reagent [49]. The percentage oxidation of the Cys279 peptide of AldA under control and NaOCl stress is reflected by the mass spectra of the ICAT-labelled peptide pair as quantified in the previous study [26] (Fig. 1A). The strong 29% oxidation increase of the active site Cys279 is shown here again which is caused by S-bacillithiolation [26]. To confirm that AldA can be S-bacillithiolated also in vitro, we expressed and purified His-tagged AldA from E. coli extracts. Purified AldA was treated with H2O2 after pre-exposure to 10-fold excess of BSH and the reversible S-bacillithiolation of AldA was verified using BSH-specific Western blot analyses in the absence and presence of DTT (Fig. 1B). The S-bacillithiolated AldA band is denoted with AldA-SSB. Next, we were interested to study the expression, function, redox-regulation and structural changes of AldA under NaOCl and aldehyde stress.
Fig. 1.
OxICAT analysis revealed a 29% increased oxidation of the AldA Cys279-peptide (A) and S-bacillithiolation of the AldA protein in vitro is shown by BSH-specific Western blot analysis (B). (A) The OxICAT mass spectrometry results from the previous study [26] are shown for the AldA-Cys279-peptide in S. aureus under control and 30 min after NaOCl stress. The reduced Cys279-peptides is labelled with light 12C-ICAT, followed by reduction of the S-bacillithiolated Cys279-peptide and labelling with heavy 13C-ICAT reagent. The Cys279-peptide was 10% oxidized in the control and 38% oxidized in the NaOCl stress sample indicating a 29% oxidation increase. (B) AldA is S-bacillithiolated in vitro by H2O2 in the presence of BSH as revealed by BSH-specific Western blots. Reduced purified AldA (40 µM) is pretreated with 10-fold molar excess of BSH (400 µM) and incubated with 10 mM H2O2 for 5 min. The S-bacillithiolated AldA was detected using non-reducing BSH-specific Western blot analysis. The loading control of AldA and S-bacillithiolated AldA (AldA-SSB) is shown as SDS-PAGE stained with Coomassie below the anti-BSH blot.
3.2. Transcription of aldA is induced SigmaB-independently under thiol-specific stress conditions by formaldehyde, NaOCl and diamide in S. aureus COL
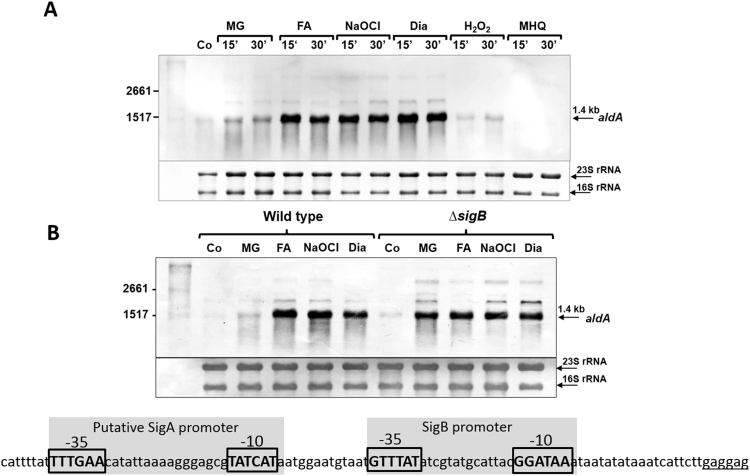
We used Northern blot analysis to study aldA transcription in S. aureus COL under different thiol-specific stress conditions, including sub-lethal doses of 1 mM NaOCl, 2 mM diamide, 0.75 mM formaldehyde, 0.5 mM methylglyoxal, 50 µM methylhydroquinone (MHQ) and 10 mM H2O2 (Fig. 2A). The Northern blot results revealed that aldA transcription is strongly induced in S. aureus COL wild type after exposure to formaldehyde, diamide and NaOCl stress, but less strongly under methylglyoxal stress (Fig. 2A). No significant induction of aldA was detected under MHQ and H2O2 treatment. These transcriptional results indicate that AldA could be involved in the hypochlorite stress defense or in detoxification of aldehydes. In previous microarray experiments, aldA was identified as member of the SigmaB general stress regulon, which responds to heat and salt stress (NaCl), MnCl2 and alkaline stress conditions in S. aureus [50], [51]. The sigB-dependent promoter sequence was mapped in the aldA regulatory upstream region (GTTTAT-N14-GGATAA) as promoter U1137.SigB.M2 previously (Fig. 2B) [52]. In the condition-dependent transcriptome of S. aureus NCTC8325-4 [53], the strongest aldA transcription was monitored during the stationary phase in rich LB and TSB medium as well as during plasma stress as visualized by the Aureowiki Expression data browser (http://genome.jouy.inra.fr/cgi-bin/aeb/viewdetail.py?id=NA_2184537_2185964_-1) [52].
Fig. 2.
Transcriptional induction of aldA under formaldehyde, methylglyoxal, NaOCl and diamide stress in S. aureus COL wild type (A) and in the sigB mutant (B). (A) RNA was isolated from S. aureus COL wild type under control conditions as well as after treatment with sub-lethal doses of 0.75 mM formaldehyde, 0.5 mM methylglyoxal, 1 mM NaOCl, 2 mM diamide, 10 mM H2O2 and 50 µM methylhydroquinone (MHQ) for 15 and 30 min and subjected to Northern blot analysis for aldA (SACOL2114) transcription. (B) For comparison of Northern blot analysis of aldA transcription between the wild type and the sigB mutant, RNA was isolated from S. aureus COL wild type and the sigB mutant after exposure to 0.75 mM formaldehyde, 0.5 mM methylglyoxal, 1 mM NaOCl and 2 mM diamide for 15 min. Transcription of aldA is similarly up-regulated under formaldehyde, NaOCl and diamide stress in the wild type (A) and in the sigB mutant (B) indicating a SigmaB-independent thiol-stress regulatory mechanism of aldA transcription. The known SigmaB-dependent promoter sequence and a putative SigA-dependent promoter in the aldA upstream regulatory region are shown below the Northern blot in (B). The methylene blue stain is the RNA loading control showing the abundant 16S and 23S rRNAs. The experiments were performed in 3 biological replicates.
To investigate whether the thiol-specific induction of aldA transcription by formaldehyde, diamide and NaOCl requires SigmaB, we performed Northern blot analysis with RNA isolated from a sigB deletion mutant in comparison to the wild type (Fig. 2B). The Northern blot results showed similar aldA transcriptional induction in the sigB mutant under NaOCl, diamide and formaldehyde stress compared to the wild type. Even a higher aldA transcription occurred under methylglyoxal stress in the sigB mutant. These results indicate that aldA transcription is subject to SigmaB-independent control mechanisms under thiol-specific stress conditions by an unknown thiol-specific transcription factor that remains to be elucidated. No additional SigA promoter was identified upstream of aldA previously [52], presumably because the conditions were different compared to our thiol-stress conditions. In previous studies, a refined consensus for SigA- and SigB-dependent promoter sequences was revealed based on 93% of S. aureus transcriptional units [52]. In the aldA regulatory region, a putative SigA-dependent promoter was identified upstream of the SigB promoter, which could drive the thiol-specific expression of aldA (Fig. 2B).
3.3. AldA plays important roles in the defense against NaOCl stress in S. aureus COL
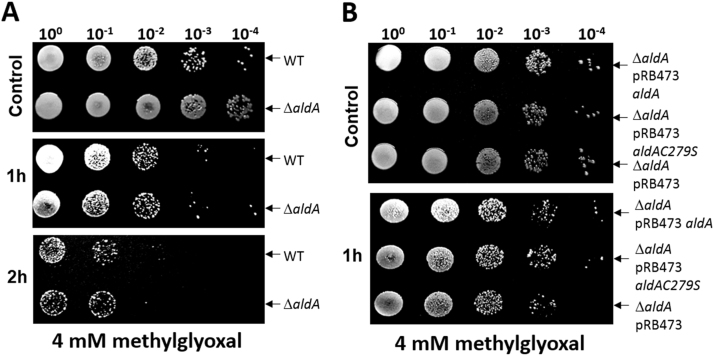
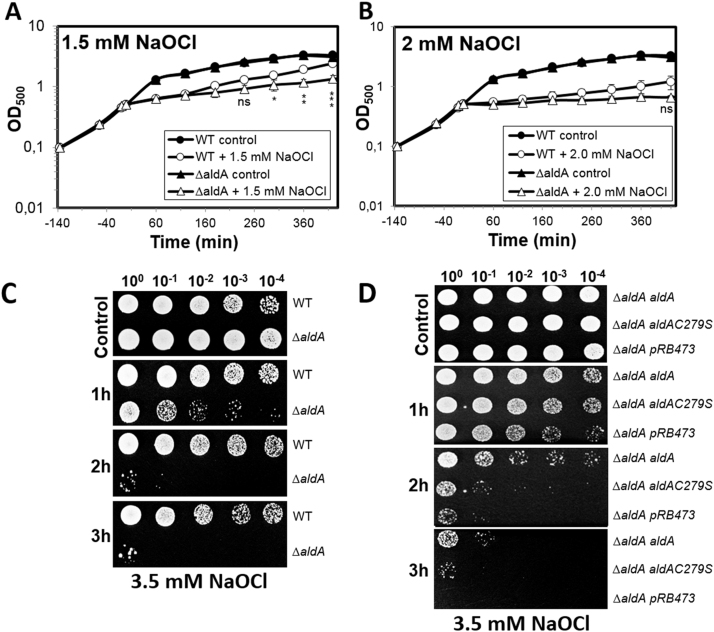
Next, we analyzed the role of AldA in protection under NaOCl and aldehyde stress in S. aureus. It was previously shown that methylglyoxal is produced in E. coli cells treated with HOCl [20]. Thus, AldA could function in methylglyoxal detoxification under HOCl stress also in S. aureus. AldA harbors a conserved active site Cys279 which is essential for its catalytic activity [54], [55], [56]. The function of AldA and the conserved Cys279 under methylglyoxal, formaldehyde and HOCl stress was analyzed in growth and survival assays of an aldA deletion mutant and its aldA and aldAC279S complemented strains (Fig. 3, Fig. 4, S1 and S2). The growth of the aldA mutant was not affected under sub-lethal formaldehyde and methylglyoxal stress in comparison to the wild type (Fig. S1). In addition, no significant phenotypes of the aldA mutant and the aldA complemented strains were detected in survival assays after exposure to 4 mM methylglyoxal (Fig. 3AB) and 2 mM formaldehyde stress (Fig. S2). However, the aldA mutant was significantly impaired in growth after exposure to sub-lethal concentrations of 1.5 mM NaOCl stress (Fig. 4A). In survival assays, the aldA mutant showed also a strongly decreased survival after treatment with 3.5 mM NaOCl (Fig. 4C). This survival defect of the ΔaldA mutant could be restored back to wild type level in the aldA complemented strain, but not in the aldAC279S mutant (Fig. 4D). This indicates that AldA is involved in protection of S. aureus against NaOCl stress and that Cys279 is essential for AldA activity in vivo.
Fig. 3.
AldA is not essential for the survival ofS. aureusunder methylglyoxal stress. For the survival phenotype assays, S. aureus COL wild-type (WT), the ΔaldA deletion mutant (A) and the aldA and aldAC279S complemented ΔaldA mutants (ΔaldA pRB473-aldA and ΔaldA pRB473-aldAC279S) (B) were grown in RPMI until an OD500 of 0.5 and treated with 4 mM methylglyoxal. Survival assays were performed by spotting 10 µl of serial dilutions after 1–3 h of NaOCl exposure onto LB agar plates. The experiments were performed in 3 biological replicates.
Fig. 4.
AldA is required for growth and survival under NaOCl stress inS. aureus. (A, B) Growth curves of S. aureus COL wild type and the aldA deletion mutant in RPMI medium after exposure to sublethal concentrations of 1.5 mM and 2 mM NaOCl stress at an OD540 of 0.5. The growth differences of the aldA mutant are significantly different compared to the wild type at 1.5 mM NaOCl. (C, D) For the survival phenotype assays, S. aureus COL wild-type (WT), the ΔaldA deletion mutant (C) and the aldA and aldAC279S complemented ΔaldA mutants (ΔaldApRB473-aldA and ΔaldA pRB473-aldAC279S) (D) were grown in RPMI until an OD500 of 0.5 and treated with 3.5 mM NaOCl. Survival assays were performed by spotting 10 µl of serial dilutions after 1–3 h of NaOCl exposure onto LB agar plates. Colonies were observed after overnight incubation of the LB plates at 37 °C. The active site Cys279 of AldA is required for NaOCl stress survival. The results for the growth curves and survival assays are from 5 biological replicate experiments. For the growth curves in Fig. 4AB, error bars represent the SEM and the statistics was calculated using a Student's unpaired two-tailed t-test by the graph prism software. Symbols are defined as follows: nsp > 0.05; *p ≤ 0.05; **p ≤ 0.01 and ***p ≤ 0.001.
3.4. AldA shows broad substrate specificity for oxidation of various aldehyde substrates, including formaldehyde and methylglyoxal in vitro
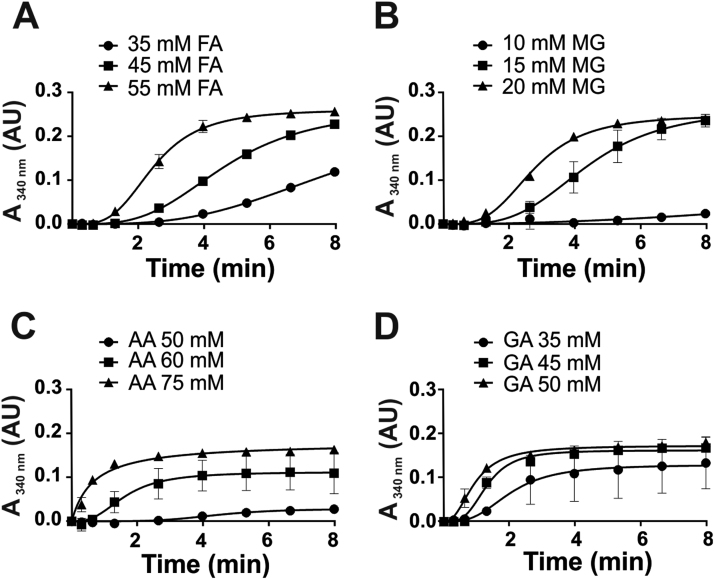
To study the function and substrate specificity of AldA in vitro, the catalytic activity was measured using different aldehyde substrates, including formaldehyde, methylglyoxal, glycol aldehyde and acetaldehyde in concentrations ranging from 0.5 to 100 mM. AldA activity was measured in a spectrophotometric assay in the presence of NAD+ as a cofactor with the different aldehyde substrates by monitoring the NADH production as absorbance increase at 340 nm. The AldA activity assays revealed increasing NADH production with increasing concentrations of all aldehyde substrates indicating that AldA has broad substrate specificities (Fig. 5). AldA showed the highest activities with 55 mM formaldehyde and 20 mM methylglyoxal, which could be possible substrates of AldA. Formaldehyde and methylglyoxal are oxidized to formate and lactate by AldA, resulting in NADH generation.
Fig. 5.
Purified AldA shows broad substrate specificity towards various aldehydesin vitro. The catalytic activity of the aldehyde dehydrogenase AldA was analyzed with increasing concentrations of different aldehyde substrates, including (A) formaldehyde (FA), (B) methylglyoxal (MG), (C) acetaldehyde (AA) and (D) glycol aldehyde (GA). Reduced AldA (2.5 µM) was incubated with different concentrations of aldehyde substrates ranging from 10 to 100 µM in reaction buffer (100 mM Tris HCl, 1.25 mM EDTA, pH 7.5). The oxidation of the aldehydes was measured in the presence of NAD+ as coenzyme and NADH generation was monitored at 340 nm using a spectrophotometer. The results are from 3 replicate experiments. Error bars represent the SEM.
To further confirm that Cys279 is the active site residue and essential for AldA activity, we used the purified AldAC279S mutant protein which was analyzed for formaldehyde and methylglyoxal oxidation in the AldA activity assays. However, the AldAC279S mutant protein did not show significant activity for formaldehyde and methylglyoxal oxidation in our activity assays (Fig. S4). This indicates that the conserved Cys279 is the active site residue and required for AldA activity as shown also for other homologs previously [54], [55], [57].
3.5. AldA is redox-regulated and protected by protein S-bacillithiolation under H2O2 stress in vitro
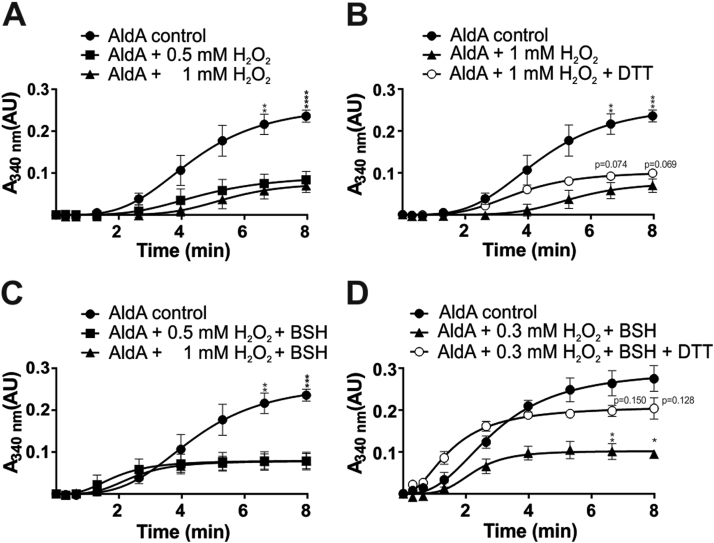
We were interested whether S-bacillithiolation inhibits AldA activity and protects the active site Cys279 against overoxidation in vitro. Using the spectrophotometric assay, AldA activity was measured after oxidative stress with 15 mM methylglyoxal as substrate and NAD+ as coenzyme by monitoring NADH generation at 340 nm. Treatment of AldA with 0.5–1 mM H2O2 resulted in a strong inactivation of its enzymatic activity (Fig. 6A). Inactivation of AldA with H2O2 alone was irreversible since AldA activity could not be restored after treatment with 10 mM DTT (Fig. 6B). These results indicate that the active site Cys279 of AldA is very sensitive to overoxidation by H2O2 in the absence of BSH. To assess the effect of S-bacillithiolation on AldA activity, the enzyme was pre-exposed to 0.3–0.5 mM BSH prior to oxidation with 0.3–1 mM H2O2 and the remaining AldA activity was measured in the spectrophotometric assay with 15 mM methylgyoxal as substrate. AldA activity was inhibited with 0.3–1 mM H2O2 after pre-treatment with 0.3–0.5 mM BSH (Fig. 6C). In this case, however, the activity of the oxidized AldA protein could be restored to 66% by DTT reduction indicating that AldA is subject to reversible S-bacillithiolation in the presence of BSH and H2O2 (Fig. 6D). S-bacillithiolation of AldA and its reversibility with DTT was further confirmed in BSH-specific Western blots (Fig. 1B). These results suggest that S-bacillithiolation protects the AldA active site Cys279 against overoxidation and functions in redox-regulation of AldA activity in vitro.
Fig. 6.
Inactivation of AldA ofS. aureusin response to H2O2in the absence and presence of BSHin vitro. Reduced AldA (30 µM) was oxidized with 0.3–1 mM H2O2 for 5 min in the absence (A, B) or presence of BSH (C, D) in reaction buffer (100 mM Tris HCl, 1.25 mM EDTA, pH 7.5). The AldA activities were measured with 15 mM methylglyoxal as substrate and NAD+ as coenzyme by monitoring NADH production at 340 nm using a spectrophotometer. To analyze the irreversible inactivation of AldA by H2O2 alone, AldA was treated with 1 mM H2O2 without BSH followed by reduction with 10 mM DTT (C). The reversibility of AldA S-bacillithiolation with 0.3 mM H2O2 and 0.3 mM BSH is shown after DTT-reduction resulting in 66% of regeneration of AldA activity (D). The S-bacilllithiolation of AldA and its reduction using DTT was further confirmed in BSH-specific Western blot analysis as shown in Fig. 1B. P-values were calculated as follows: p = 0,0012, p = 0,0001 for AldA control/0.5 mM H2O2 at 6.63 and 8 min, respectively (Fig. 6A); p = 0,0012, p = 0,0002 for AldA control/1 mM H2O2 at 6.63 and 8 min and p = 0,074, p = 0,069 for 1 mM H2O2/1 mM H2O2 + DTT at 6.63 and 8 min, respectively (Fig. 6B); p = 0,0021, p = 0,0008 for AldA control/0.5 mM H2O2 + BSH at 6.63 and 8 min, respectively (Fig. 6C); p = 0.003, p = 0.011 for 0.3 mM H2O2 + BSH/0.3 mM H2O2 + BSH + DTT at 6.63 and 8 min; p = 0.150, p = 0.128 for AldA control/0.3 mM H2O2 + BSH + DTT at 6.63 and 8 min, respectively (Fig. 6D). Symbols are defined as follows: nsp > 0.05; *p ≤ 0.05; **p ≤ 0.01; ***p ≤ 0.001; and ****p ≤ 0.0001. The results are from 3 replicate experiments. In all graphs, mean values are shown, error bars represent the SEM and p-values are calculated using a Student's unpaired two-tailed t-test by the graph prism software.
3.6. Structural comparison of AldA with other aldehyde dehydrogenases
We were further interested in the structure and the structural changes of AldA upon S-bacillithiolation. A crystal structure of S. aureus AldA (denoted as saAldA) has been determined by the Midwest Center for Structural Genomics (PDB 3TY7). For understanding the enzyme's catalytic mechanism, we performed structural homology searches for saAldA with the DALI server [58] (http://ekhidna.biocenter.helsinki.fi/dali_server/) and the PDBeFold (SSM) server (http://www.ebi.ac.uk/msd-srv/ssm/). SaAldA shows high homology to many other aldehyde dehydrogenases (ADHs) from bacteria, plants and humans. The root-mean-square deviations (r.m.s.d.’s) and sequence similarities of AldA's closest homologs are listed in Table S3.
In contrast to the tetrameric bacterial ADHs (pfAMSDH, saBADH, ecADH, paBADH), saAldA is a dimeric enzyme and thus more similar to plant ADHs that are also active as dimer (Fig. 7A). Regardless of the oligomerization state, the overall fold of a subunit is highly conserved among all ADH enzymes. Similarly as in other ADHs, a saAldA subunit is composed of a coenzyme (NAD+)-binding domain (Co-BD; residues 1-122, 137-244 and 439-464), a catalytic domain (CD; residues 245-438) and a subunit interaction domain (SID; residues 123-136 and 465-475; Fig. 7A).
In all ADHs, the active site harbors conserved Cys (C279 in saAldA) and glutamate (E245 in saAldA) residues (Fig. S5). The Cys residue can adopt two alternative conformations, a “resting” and “attacking” (Fig. 7C), depending on the enzyme activation state. In the apo-enzyme structure, the Cys residue is in the resting conformation, whereas upon NAD+ binding the Cys thiol moiety rotates away from the nicotinamide part of NAD+ and is closer to the substrate-binding pocket [54], [55], [56]. The Cys residue serves as a nucleophile during catalysis, leading to a covalent thioester-enzyme adduct with the substrate via a nucleophilic addition [54], [55], [57]. The conserved glutamate residue then serves as a base to activate a water molecule for hydrolysis of the thioester-enzyme intermediate [55], [59]. In addition to the Cys and glutamate residues, there are two other conserved residues, a lysine (K156 in saAldA) and a glutamate (E455 in saAldA), that are involved in a proton relay that allows the deprotonation of E245, and, as a consequence, proton abstraction from the hydrolytic water [56].
Another common feature of the ADHs is the presence of a cation-binding site located in the Co-BD (Fig. 7B, D). Co-BD is formed by the three main chain carbonyl groups of an isoleucine/valine (I25 in saAldA), a glutamate/aspartate (E91 in saAldA) and a glutamate residue (E173 in saAldA) [60], [61], [62]. The cation bound at this site is usually sodium or potassium, and it was reported that the enzyme activity is slightly higher in the presence of sodium [60]. In the saAldA structure, a magnesium ion is present at this site, most likely because magnesium was the only cation present in the crystallization solution. The role of the cation-binding site is to maintain the structural integrity of the protein and to stabilize a loop involved in binding of NAD+ [60], [61], [62].
The available saAldA structure represents the apo-enzyme. In contrast, the structures of plant ADHs and of paBADH contain the coenzyme NAD+. In the case of pfAMSDH, the structures of pfAMSDH/NAD+/intermediate complexes are also available [55]. Comparison of the apo, NAD+, NAD+/intermediate states shows that binding of the coenzyme or the formation of the intermediate does not influence the secondary structure elements within the enzyme, while rearrangements are observed in the side chains of residues involved in catalysis [54], [55]. In the ADHs, the NAD+ is bound in the hydrophobic pocket of the Co-BD. Only the nicotinamide nucleotide moiety is turned towards a negatively charged pocket, in which the catalytic cysteine residue is located (Fig. 7B). NAD+ is engaged in only few polar contacts with the enzyme [54], [62].
Although the overall structure, the active site and the cation-binding site are highly conserved among the ADHs, these enzymes show broad substrate specificities and the amino acid residues involved in substrate binding are different among the ADHs. Nevertheless, even a single ADH is able to use many different aldehydes as substrates. For example, slAMADH can oxidize many different aminoaldehydes [62]. Thus, differences in the substrate-binding residues determine differences in the still comparatively broad substrate spectra of the enzymes.
3.7. S-bacillithiolation of the AldA active site depends on the Cys activation state as revealed by molecular dynamics simulation
Next, we analyzed the structural changes of AldA upon S-bacillithiolation and used molecular docking and molecular dynamics simulations to model BSH into the active site of the apo- and holoenzyme structures (Fig. 7EF). The structure of saAldA apo-enzyme (PDB 3TY7) was superimposed with the NAD+ binding structure from Pseudomonas fluorescens pfAMSDH (PDB 4I1W) to model the NAD+ cofactor into the AldA active site pocket (Fig. 7C). We further noticed that in the saAldA dimeric structure, the loop composed of residues 438-459 is not present which was modelled into the saAldA holo-enzyme structure based on the structure of pfAMSDH (Fig. 7F). This loop in the saAldA holo-enzyme structure may interfere with the location of BSH at the active site. To model the S-bacillithiolated active site Cys279 in the saAldA apo- and holoenzyme structures, we applied an adapted molecular docking algorithm based on Steric Clashes-Alleviated Receptor (SCAR) approaches [63], which takes into account the possibility of bond formation between ligand and receptor. Molecular docking and atomistic molecular dynamics simulation of the covalent BSH enzyme complex resulted in two best-scoring poses for BSH in the apo-enzyme (Q2) or holo-enzyme complex (Q1) (Fig. 7EF). However, no overlap between BSH and the loop (aa438-459) in the holo-enzyme structure was found and there was still room for an aldehyde substrate. Interestingly, these two different BSH positions in the AldA active site depend on the Cys279 activation state in the presence or absence of the NAD+ cofactor (Fig. 7EF). In the apo-enzyme structure, Cys279 bound to BSH is still in "resting" position (Q2), while Cys279 is in the "attacking" position in the holo-enzyme (Q1). Thus, the location of BSH in the active site pocket depends on the Cys279 activation state in the presence or absence of NAD+. The Q2 pose of BSH at the apo-enzyme without NAD+ seems to be energetically more favorable since Q2 had much better energy score (-50.2 kJ/mol), while the Q1 position of the holo-enzyme had a lower energy score (-38.1 kJ/mol). This results were quantitatively supported by our all-atom MD simulation of the complexes and the follow-up MM-PBSA calculations: the interaction energy in the apo-enzyme complex with BSH in Q2 position was -24.8 +/- 15.4 kJ/mol, while the holo-enzyme complex with BSH in Q1 position had interaction energy of -19.7 +/- 10.0 kJ/mol.
We have further plotted the dihedral distribution of N-CA-CB-SG dihedral (rotation around the CA-CB bond) of Cys279 and the dihedral angle at the function of simulation time (Fig. S6). The results showed that Cys279 in the apo-enzyme has very different dihedral propensity than in the holo-enzyme in complex with NAD+. These data support that the apo-enzyme prefers the resting state position of Cys279 with BSH while the holo-enzyme favors the BSH complex with the thiol in the attacking state position.
In agreement with our previous GapDH results [26], S-bacillithiolation of the AldA apo- and holoenzyme active site does not require major structural changes. After 50 ns of MD simulations there was very little change in the backbone flexibility of the protein between different binding positions of BSH in the apo-enzyme (Q2) or the holo-enzyme (Q1) compared to the apo-enzyme without BSH (Fig. S7). This further confirms that BSH can undergo disulfide formation with the active site Cys279 at different positions without major conformational changes.
4. Discussion
S. aureus is a major human pathogen of hospital and community-acquired infections, ranging from local skin infections to life-threatening systemic and chronic infections. During infections, S. aureus is exposed to ROS, RCS and RES that are produced as first line of defense by activated macrophages and neutrophils or can be also encountered as consequence of antibiotics treatment [10], [11], [64]. Thus, the understanding of the adaptation mechanisms of S. aureus to infection conditions to avoid killing by ROS, RCS and RES is important for the discovery of new drug targets to combat multi-resistant S. aureus infections.
In our previous work, we have identified the aldehyde dehydrogenase AldA as one of the most strongly oxidized proteins in the thiol-redox proteome in S. aureus, which showed a 29% oxidation increase under NaOCl stress using the OxICAT analysis [26]. AldA uses a conserved active site Cys279 that was modified by S-bacillithiolation under NaOCl stress. Apart from AldA, the glyceraldehyde-3-phosphate dehydrogenase Gap was identified as S-bacillithiolated at its active site Cys151 under NaOCl stress. Thus, it is interesting to note that two functionally related aldehyde dehydrogenases are targets for oxidation at their active site Cys residues that both function in aldehyde oxidation. In this study, we demonstrated that AldA is specifically induced under thiol-specific stress conditions, such as NaOCl, diamide and formaldehyde stress. Expression of aldA was previously shown to be regulated by the alternative sigma factor SigmaB in response to heat shock, salt stress caused by NaCl and Mn2Cl as well as alkaline shock [50], [51]. Here, we have shown that the thiol-specific expression of aldA occurs SigmaB-independently. Thus, aldA seems to be double-controlled by SigmaB and another thiol-stress sensing regulator to allow adaptation to general stress and starvation as well as thiol-stress conditions.
SigmaB has been previously shown to play an important role under infection conditions and controls biofilm formation and several virulence factors, such as adhesins [65], [66]. The SigmaB regulon was induced after internalization of S. aureus by bronchial epithelial cells and required for intracellular growth as demonstrated by transcriptomics and proteomics [53], [67], [68]. Moreover, SigmaB has been implicated as central regulator in long-term persistence in human osteoblasts and controls the small colony variant (SCV) phenotype of persistent S. aureus infections [69], [70]. Thus, it might be possible that adaptation of S. aureus from acute to chronic and persistent infections requires SigmaB and AldA to cope and adapt to the stationary phase and thiol-specific stress conditions inside macrophages and neutrophils. This adaptation to thiol-stress conditions is particularly important for S. aureus to survive under conditions of long-term persistent and chronic infections.
In this work, we have shown that AldA is an important member of the SigmaB regulon that provides protection under NaOCl stress conditions as shown in survival assays. However, the thiol-specific induction of aldA transcription seems to be SigA-dependently since the same induction level was observed in the sigB mutant under thiol-stress. A putative SigA-promoter was observed upstream of the SigB-promoter indicating that aldA transcription might be controlled by SigB and SigA containing RNA polymerase (RNAP) from adjacent promoters. The stronger aldA induction in the sigB mutant under methylglyoxal stress could be explained by a higher affinity of SigA for the RNAP core enzyme compared to SigB and the lack of sigma factor competition in the sigB mutant [71]. Moreover, the thiol-stress-specific induction of aldA transcription might require additional transcriptional regulators that remain to be elucidated. In future studies, we also aim to investigate if AldA plays a role for the intracellular growth as well as persistence or chronic infections in S. aureus, which could require detoxification of toxic aldehydes to allow long-term survival.
To study the function of AldA and its redox-regulation under NaOCl stress in vitro, we purified the enzyme and determined its catalytic activities towards oxidation of various aldehydes. We could show that AldA has broad substrate specificities to oxidize formaldehyde, methylglyoxal, glycol aldehyde and acetaldehyde to their respective acids. The question arises about the physiological aldehyde substrate for AldA under in vivo conditions that are produced under infection conditions, such as under hypochlorite stress. Methylglyoxal was previously shown to be produced at higher levels under HOCl stress in E. coli [20]. Moreover, the gloA-nemRA operon was induced under methylglyoxal and HOCl stress, which functions as important HOCl and methylglyoxal defense mechanism [19], [20], [21], [22]. The FMN-dependent oxidoreductase NemA functions in detoxification of various electrophiles, such as aldehydes, N-ethylmaleimide and quinones and its up-regulation under HOCl stress indicates the link between HOCl and aldehyde stress. In our work, we could also show that AldA responds to aldehydes, diamide and NaOCl and hence could be involved in methylglyoxal detoxification in S. aureus as well. However, in growth and survival assays, no phenotypes of the aldA mutant were detected under formaldehyde and methylglyoxal stress. Since AldA showed broad substrate specificity towards various aldehydes in vitro, its natural substrates could be different aldehydes that remain to be elucidated.
Of note, AldA shares strong 57% sequence similarity to betaine aldehyde dehydrogenases from S. aureus, Pseudomonas aeruginosa and Spinacia oleracea. These enzymes function in oxidation of the toxic betaine aldehyde to glycine betaine which is a well-known compatible solute and accumulates in bacteria under osmotic stress conditions as osmoprotectant [72], [73]. Glycine betaine can be either taken up upon osmostress or synthesized from exogenously provided choline in a two oxidation steps via choline dehydrogenase (BetA) and betaine dehydrogenase (BetB) which are conserved in B. subtilis [72], [73] and S. aureus [54]. The human tissues are rich sources of choline and betaine and thus, S. aureus encounters toxic aldehydes produced from choline during colonization and internalization. For some bacteria, the importance of the choline oxidation pathway for survival and virulence has been already demonstrated [73], [74]. Of note, AldA is also induced under high osmolarity conditions provoked by NaCl stress in a SigmaB-dependent manner [50]. This could point to a possible function in the osmostress and thiol-stress response in S. aureus which remains to be elucidated. However, we could not detect AldA activity for oxidation of betaine aldehyde as substrate in vitro, indicating a different function of AldA in S. aureus (data not shown).
The catalytic activity of AldA depends on a highly conserved Cys279 active site which we identified as S-bacillithiolated under NaOCl stress in S. aureus [26]. Interestingly, this nucleophilic active site Cys residue was previously found oxidized to a mixed disulfide with beta-mercaptoethanol during protein crystallization of related betaine aldehyde dehydrogenases [54], [74]. These results confirm the redox-sensitivity of the active site Cys of AldA as shown in this work. Our results have further demonstrated that S-bacillithiolation functions in redox-regulation and inactivation of AldA activity under H2O2 stress. In the absence of BSH, the active site Cys279 was very sensitive to overoxidation as shown by its irreversible inactivation. In the presence of BSH, Cys279 was protected against overoxidation by the S-bacillithiolation as shown for the glyceraldehyde-3-phosphate dehydrogenase GapDH in S. aureus [26]. Both enzymes use a similar catalytic mechanism for the NAD+-dependent oxidation of the aldehyde substrate to generate the acid product [54], [55], [57]. In the catalytic mechanism of aldehyde dehydrogenase, the active site Cys was shown to adopt two conformations: the “attacking” or “resting” conformation depending on the presence or absence of the NAD+ cofactor. We used molecular docking and molecular dynamic simulations to model the S-bacillithiolated active site in the presence and absence of NAD+. In the apo-enzyme structure, BSH was bound to Cys279 in the resting state (Q2) position and occupied the cofactor-binding pocket. In the presence of NAD+, Cys279 was modified in the attacking state position (Q1) and BSH was repositioned close to the substrate-binding site.
In our previous docking approach with BSH at the Cys151 active site of GapDH, we found similar locations of BSH in the apo-enzyme and holo-enzyme structures related to the resting and attacking state. Thus, the highly flexible active site and the redox-sensitivity of the nucleophilic Cys residues facilitate their fast oxidation to the mixed disulfides with BSH. In both structural models, S-bacillithiolation of GapDH and AldA did not require major structural changes, which further explains their preferred formation of the BSH mixed disulfides. This flexible BSH position may ensure that catalytic active and resting AldA and GapDH enzymes can both be protected against overoxidation under NaOCl stress to ensure fast regeneration and reactivation of the enzymes.
Acknowledgements
This work was supported by an ERC Consolidator Grant (GA 615585) MYCOTHIOLOME and grants from the Deutsche Forschungsgemeinschaft (AN746/4-1 and AN746/4-2) within the SPP1710 on “Thiol-based Redox switches”, by the Research Training Group GRK1947 (Project C01) and by the SFB973 Project C08 to H.A. Protein crystal structure analysis was supported by an Alexander von Humboldt Post-doc Fellowship to A.P.-B.
Acknowledgments
Author disclosure statement
No competing financial interests exist.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.redox.2018.02.001.
Appendix A. Supplementary material
Supplementary material
References
- 1.Lowy F.D. Staphylococcus aureus infections. New Engl. J. Med. 1998;339:520–532. doi: 10.1056/NEJM199808203390806. [DOI] [PubMed] [Google Scholar]
- 2.Boucher H.W., Corey G.R. Epidemiology of methicillin-resistant Staphylococcus aureus. Clin. Infect. Dis.: Off. Publ. Infect. Dis. Soc. Am. 2008;46(Suppl. 5):S344–S349. doi: 10.1086/533590. [DOI] [PubMed] [Google Scholar]
- 3.Archer G.L. Staphylococcus aureus: a well-armed pathogen. Clin. Infect. Dis.: Off. Publ. Infect. Dis. Soc. Am. 1998;26:1179–1181. doi: 10.1086/520289. [DOI] [PubMed] [Google Scholar]
- 4.Livermore D.M. Antibiotic resistance in staphylococci. Intern. J. Antimicrob. Agents. 2000;16(Suppl. 1):S3–S10. doi: 10.1016/s0924-8579(00)00299-5. [DOI] [PubMed] [Google Scholar]
- 5.Pendleton J.N., Gorman S.P., Gilmore B.F. Clinical relevance of the ESKAPE pathogens. Expert Rev. Anti-Infect. Ther. 2013;11:297–308. doi: 10.1586/eri.13.12. [DOI] [PubMed] [Google Scholar]
- 6.Spaan A.N., van Strijp J.A.G., Torres V.J. Leukocidins: staphylococcal bi-component pore-forming toxins find their receptors. Nat. Rev. Microbiol. 2017 doi: 10.1038/nrmicro.2017.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dal Peraro M., van der Goot F.G. Pore-forming toxins: ancient, but never really out of fashion. Nat. Rev. Microbiol. 2016;14:77–92. doi: 10.1038/nrmicro.2015.3. [DOI] [PubMed] [Google Scholar]
- 8.Winterbourn C.C., Kettle A.J. Redox reactions and microbial killing in the neutrophil phagosome. Antioxid. Redox Signal. 2013;18:642–660. doi: 10.1089/ars.2012.4827. [DOI] [PubMed] [Google Scholar]
- 9.Winterbourn C.C., Kettle A.J., Hampton M.B. Reactive oxygen species and neutrophil function. Annu. Rev. Biochem. 2016;85:765–792. doi: 10.1146/annurev-biochem-060815-014442. [DOI] [PubMed] [Google Scholar]
- 10.Beavers W.N., Skaar E.P. Neutrophil-generated oxidative stress and protein damage in Staphylococcus aureus. Pathog. Dis. 2016;74 doi: 10.1093/femspd/ftw060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hillion M., Antelmann H. Thiol-based redox switches in prokaryotes. Biol. Chem. 2015;396:415–444. doi: 10.1515/hsz-2015-0102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hampton M.B., Kettle A.J., Winterbourn C.C. Involvement of superoxide and myeloperoxidase in oxygen-dependent killing of Staphylococcus aureus by neutrophils. Infect. Immun. 1996;64:3512–3517. doi: 10.1128/iai.64.9.3512-3517.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Klebanoff S.J., Kettle A.J., Rosen H., Winterbourn C.C., Nauseef W.M. Myeloperoxidase: a front-line defender against phagocytosed microorganisms. J. Leukoc. Biol. 2013;93:185–198. doi: 10.1189/jlb.0712349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Booth I.R., Ferguson G.P., Miller S., Li C., Gunasekera B., Kinghorn S. Bacterial production of methylglyoxal: a survival strategy or death by misadventure? Biochem. Soc. Trans. 2003;31:1406–1408. doi: 10.1042/bst0311406. [DOI] [PubMed] [Google Scholar]
- 15.Ferguson G.P., Totemeyer S., MacLean M.J., Booth I.R. Methylglyoxal production in bacteria: suicide or survival? Arch. Microbiol. 1998;170:209–218. doi: 10.1007/s002030050635. [DOI] [PubMed] [Google Scholar]
- 16.Farmer E.E., Davoine C. Reactive electrophile species. Curr. Opin. Plant Biol. 2007;10:380–386. doi: 10.1016/j.pbi.2007.04.019. [DOI] [PubMed] [Google Scholar]
- 17.Marnett L.J., Riggins J.N., West J.D. Endogenous generation of reactive oxidants and electrophiles and their reactions with DNA and protein. J. Clin. Invest. 2003;111:583–593. doi: 10.1172/JCI18022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ferguson G.P., Booth I.R. Importance of glutathione for growth and survival of Escherichia coli cells: detoxification of methylglyoxal and maintenance of intracellular K+ J. Bact. 1998;180:4314–4318. doi: 10.1128/jb.180.16.4314-4318.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gray M.J., Li Y., Leichert L.I., Xu Z., Jakob U. Does the transcription factor NemR use a regulatory sulfenamide bond to sense bleach? Antioxid. Redox Signal. 2015;23:747–754. doi: 10.1089/ars.2015.6346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gray M.J., Wholey W.Y., Parker B.W., Kim M., Jakob U. NemR is a bleach-sensing transcription factor. J. Biol. Chem. 2013;288:13789–13798. doi: 10.1074/jbc.M113.454421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee C., Shin J., Park C. Novel regulatory system nemRA-gloA for electrophile reduction in Escherichia coli K-12. Mol. Microbiol. 2013;88:395–412. doi: 10.1111/mmi.12192. [DOI] [PubMed] [Google Scholar]
- 22.Ozyamak E., de Almeida C., de Moura A.P., Miller S., Booth I.R. Integrated stress response of Escherichia coli to methylglyoxal: transcriptional readthrough from the nemRA operon enhances protection through increased expression of glyoxalase I. Mol. Microbiol. 2013;88:936–950. doi: 10.1111/mmi.12234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chandrangsu P., Loi V.V., Antelmann H., Helmann J.D. The role of bacillithiol in gram-positive Firmicutes. Antioxid. Redox Signal. 2017 doi: 10.1089/ars.2017.7057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Newton G.L., Rawat M., La Clair J.J., Jothivasan V.K., Budiarto T., Hamilton C.J., Claiborne A., Helmann J.D., Fahey R.C. Bacillithiol is an antioxidant thiol produced in Bacilli. Nat. Chem. Biol. 2009;5:625–627. doi: 10.1038/nchembio.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chandrangsu P., Dusi R., Hamilton C.J., Helmann J.D. Methylglyoxal resistance in Bacillus subtilis: contributions of bacillithiol-dependent and independent pathways. Mol. Microbiol. 2014;91:706–715. doi: 10.1111/mmi.12489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Imber M., Huyen N.T.T, Pietrzyk-Brzezinska A.J., Loi V.V., Hillion M., Bernhardt J., Thärichen T., Kolsek K., Saleh M., Hamilton C.J., Adrian L., Gräter F., Wahl M.C., Antelmann H. Protein S-Bacillithiolation functions in thiol protection and redox regulation of the glyceraldehyde-3-phosphate dehydrogenase gap in Staphylococcus aureus under hypochlorite stress. Antioxid. Redox Signal. 2017 doi: 10.1089/ars.2016.6897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wetzstein M., Völker U., Dedio J., Lobau S., Zuber U., Schiesswohl M., Herget C., Hecker M., Schumann W. Cloning, sequencing, and molecular analysis of the dnaK locus from Bacillus subtilis. J. Bact. 1992;174:3300–3310. doi: 10.1128/jb.174.10.3300-3310.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Arnaud M., Chastanet A., Debarbouille M. New vector for efficient allelic replacement in naturally nontransformable, low-GC-content, gram-positive bacteria. Appl. Environ. Microbiol. 2004;70:6887–6891. doi: 10.1128/AEM.70.11.6887-6891.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rosenblum E.D., Tyrone S. Serology, density, and morphology of Staphylococcal phages. J. Bact. 1964;88:1737–1742. doi: 10.1128/jb.88.6.1737-1742.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Müller M., Reiss S., Schlüter R., Mäder U., Beyer A., Reiss W., Marles-Wright J., Lewis R.J., Pförtner H., Völker U., Riedel K., Hecker M., Engelmann S., Pane-Farre J. Deletion of membrane-associated Asp23 leads to upregulation of cell wall stress genes in Staphylococcus aureus. Mol. Microbiol. 2014;93:1259–1268. doi: 10.1111/mmi.12733. [DOI] [PubMed] [Google Scholar]
- 31.Loi V.V., Harms M., Müller M., Huyen N.T.T., Hamilton C.J., Hochgräfe F., Pane-Farre J., Antelmann H. Real-time imaging of the bacillithiol redox potential in the human pathogen Staphylococcus aureus using a genetically encoded bacilliredoxin-fused redox biosensor. Antioxid. Redox Signal. 2017;26:835–848. doi: 10.1089/ars.2016.6733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chi B.K., Roberts A.A., Huyen T.T., Bäsell K., Becher D., Albrecht D., Hamilton C.J., Antelmann H. S-bacillithiolation protects conserved and essential proteins against hypochlorite stress in firmicutes bacteria. Antioxid. Redox Signal. 2013;18:1273–1295. doi: 10.1089/ars.2012.4686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Webb B., Sali A. Protein structure modeling with MODELLER. Methods Mol. Biol. 2017;1654:39–54. doi: 10.1007/978-1-4939-7231-9_4. [DOI] [PubMed] [Google Scholar]
- 34.Kozakov D., Grove L.E., Hall D.R., Bohnuud T., Mottarella S.E., Luo L., Xia B., Beglov D., Vajda S. The FTMap family of web servers for determining and characterizing ligand-binding hot spots of proteins. Nat. Prot. 2015;10:733–755. doi: 10.1038/nprot.2015.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Couch G.S., Hendrix D.K., Ferrin T.E. Nucleic acid visualization with UCSF Chimera. Nucleic Acids Res. 2006;34:e29. doi: 10.1093/nar/gnj031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Allen W.J., Balius T.E., Mukherjee S., Brozell S.R., Moustakas D.T., Lang P.T., Case D.A., Kuntz I.D., Rizzo R.C. DOCK 6: impact of new features and current docking performance. J. Comput. Chem. 2015;36:1132–1156. doi: 10.1002/jcc.23905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schneider N., Lange G., Hindle S., Klein R., Rarey M. A consistent description of HYdrogen bond and DEhydration energies in protein-ligand complexes: methods behind the HYDE scoring function. J. Comput. Mol. Des. 2013;27:15–29. doi: 10.1007/s10822-012-9626-2. [DOI] [PubMed] [Google Scholar]
- 38.Van Der Spoel D., Lindahl E., Hess B., Groenhof G., Mark A.E., Berendsen H.J. GROMACS: fast, flexible, and free. J. Comput. Chem. 2005;26:1701–1718. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- 39.Lindorff-Larsen K., Piana S., Palmo K., Maragakis P., Klepeis J.L., Dror R.O., Shaw D.E. Improved side-chain torsion potentials for the Amber ff99SB protein force field. Proteins. 2010;78:1950–1958. doi: 10.1002/prot.22711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sousa da Silva A.W., Vranken W.F. ACPYPE – AnteChamber PYthon Parser interfacE. BMC Res. Notes. 2012;5:367. doi: 10.1186/1756-0500-5-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dupradeau F.Y., Pigache A., Zaffran T., Savineau C., Lelong R., Grivel N., Lelong D., Rosanski W., Cieplak P. The R.E.D. tools: advances in RESP and ESP charge derivation and force field library building. Phys. Chem. Chem. Phys. 2010;12:7821–7839. doi: 10.1039/c0cp00111b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Frisch M.J., T G., Schlegel H.B., Scuseria G.E., Robb M.A., Cheeseman J.R. Vol. 34. Gaussian Inc.; Wallingford, CT: 2009. Gaussian 09, Revision A.02. [Google Scholar]
- 43.Parrinello M., Rahman A. Polymorphic transitions in single crystals: a new molecular dynamics method. J. Appl. Phys. 1981;52:7182–7190. [Google Scholar]
- 44.Darden T., York D., Pedersen L., J. Particle mesh Ewald: an N⋅log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993;98:10089–10092. [Google Scholar]
- 45.Hess B., Bekker H., Berendsen H.J.C., Fraaije J.G.E.M. LINCS: a linear constraint solver for molecular simulations. J. Comput. Chem. 1997;18:1463–1472. [Google Scholar]
- 46.Huang C.C., Meng E.C., Morris J.H., Pettersen E.F., Ferrin T.E. Enhancing UCSF Chimera through web services. Nucleic Acids Res. 2014;42:W478–484. doi: 10.1093/nar/gku377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 48.Miller B.R., 3rd, McGee T.D., Jr., Swails J.M., Homeyer N., Gohlke H., Roitberg A.E. MMPBSA.py: an efficient program for end-state free energy calculations. J. Chem. Theory Comput. 2012;8:3314–3321. doi: 10.1021/ct300418h. [DOI] [PubMed] [Google Scholar]
- 49.Leichert L.I., Gehrke F., Gudiseva H.V., Blackwell T., Ilbert M., Walker A.K., Strahler J.R., Andrews P.C., Jakob U. Quantifying changes in the thiol redox proteome upon oxidative stress in vivo. Proc. Natl. Acad. Sci. USA. 2008;105:8197–8202. doi: 10.1073/pnas.0707723105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pane-Farre J., Jonas B., Förstner K., Engelmann S., Hecker M. The sigmaB regulon in Staphylococcus aureus and its regulation. Int. J. Med. Microbiol. 2006;296:237–258. doi: 10.1016/j.ijmm.2005.11.011. [DOI] [PubMed] [Google Scholar]
- 51.Bischoff M., Dunman P., Kormanec J., Macapagal D., Murphy E., Mounts W., Berger-Bachi B., Projan S. Microarray-based analysis of the Staphylococcus aureus sigmaB regulon. J. Bact. 2004;186:4085–4099. doi: 10.1128/JB.186.13.4085-4099.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fuchs S., Mehlan H., Bernhardt J., Hennig A., Michalik S., Surmann K., Pane-Farre J., Giese A., Weiss S., Backert L., Herbig A., Nieselt K., Hecker M., Völker U., Mäder U. AureoWiki the repository of the Staphylococcus aureus research and annotation community. Int. J. Med. Microbiol. 2017 doi: 10.1016/j.ijmm.2017.11.011. [DOI] [PubMed] [Google Scholar]
- 53.Mäder U., Nicolas P., Depke M., Pane-Farre J., Debarbouille M., van der Kooi-Pol M.M., Guerin C., Derozier S., Hiron A., Jarmer H., Leduc A., Michalik S., Reilman E., Schaffer M., Schmidt F., Bessieres P., Noirot P., Hecker M., Msadek T., Völker U., van Dijl J.M. Staphylococcus aureus transcriptome architecture: from laboratory to infection-mimicking conditions. PLoS Genet. 2016;12:e1005962. doi: 10.1371/journal.pgen.1005962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Halavaty A.S., Rich R.L., Chen C., Joo J.C., Minasov G., Dubrovska I., Winsor J.R., Myszka D.G., Duban M., Shuvalova L., Yakunin A.F., Anderson W.F. Structural and functional analysis of betaine aldehyde dehydrogenase from Staphylococcus aureus. Act. Cryst. Sect. D. Biol. Cryst. 2015;71:1159–1175. doi: 10.1107/S1399004715004228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Huo L., Davis I., Liu F., Andi B., Esaki S., Iwaki H., Hasegawa Y., Orville A.M., Liu A. Crystallographic and spectroscopic snapshots reveal a dehydrogenase in action. Nat. Commun. 2015;6:5935. doi: 10.1038/ncomms6935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gonzalez-Segura L., Rudino-Pinera E., Munoz-Clares R.A., Horjales E. The crystal structure of a ternary complex of betaine aldehyde dehydrogenase from Pseudomonas aeruginosa provides new insight into the reaction mechanism and shows a novel binding mode of the 2'-phosphate of NADP+ and a novel cation binding site. J. Mol. Biol. 2009;385:542–557. doi: 10.1016/j.jmb.2008.10.082. [DOI] [PubMed] [Google Scholar]
- 57.Abriola D.P., Fields R., Stein S., MacKerell A.D., Jr, Pietruszko R. Active site of human liver aldehyde dehydrogenase. Biochemistry. 1987;26:5679–5684. doi: 10.1021/bi00392a015. [DOI] [PubMed] [Google Scholar]
- 58.Holm L., Rosenstrom P. Dali server: conservation mapping in 3D. Nucleic Acids Res. 2010;38:W545–549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.D'Ambrosio K., Pailot A., Talfournier F., Didierjean C., Benedetti E., Aubry A., Branlant G., Corbier C. The first crystal structure of a thioacylenzyme intermediate in the ALDH family: new coenzyme conformation and relevance to catalysis. Biochemistry. 2006;45:2978–2986. doi: 10.1021/bi0515117. [DOI] [PubMed] [Google Scholar]
- 60.Gruez A., Roig-Zamboni V., Grisel S., Salomoni A., Valencia C., Campanacci V., Tegoni M., Cambillau C. Crystal structure and kinetics identify Escherichia coli ydcW gene product as a medium-chain aldehyde dehydrogenase. J. Mol. Biol. 2004;343:29–41. doi: 10.1016/j.jmb.2004.08.030. [DOI] [PubMed] [Google Scholar]
- 61.Hurley T.D., Perez-Miller S., Breen H. Order and disorder in mitochondrial aldehyde dehydrogenase. Chem.-Biol. Interact. 2001;130–132:3–14. doi: 10.1016/s0009-2797(00)00217-9. [DOI] [PubMed] [Google Scholar]
- 62.Kopecny D., Koncitikova R., Tylichova M., Vigouroux A., Moskalikova H., Soural M., Sebela M., Morera S. Plant ALDH10 family: identifying critical residues for substrate specificity and trapping a thiohemiacetal intermediate. J. Biol. Chem. 2013;288:9491–9507. doi: 10.1074/jbc.M112.443952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ai Y., Yu L., Tan X., Chai X., Liu S. Discovery of covalent ligands via noncovalent docking by dissecting covalent docking based on a "Steric-Clashes Alleviating Receptor (SCAR)" strategy. J. Chem. Inform. Model. 2016;56:1563–1575. doi: 10.1021/acs.jcim.6b00334. [DOI] [PubMed] [Google Scholar]
- 64.Cheung A.L., Nishina K.A., Trotonda M.P., Tamber S. The SarA protein family of Staphylococcus aureus. Int. J. Biochem. Cell Biol. 2008;40:355–361. doi: 10.1016/j.biocel.2007.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mitchell G., Brouillette E., Seguin D.L., Asselin A.E., Jacob C.L., Malouin F. A role for sigma factor B in the emergence of Staphylococcus aureus small-colony variants and elevated biofilm production resulting from an exposure to aminoglycosides. Microb. Pathog. 2010;48:18–27. doi: 10.1016/j.micpath.2009.10.003. [DOI] [PubMed] [Google Scholar]
- 66.Mitchell G., Seguin D.L., Asselin A.E., Deziel E., Cantin A.M., Frost E.H., Michaud S., Malouin F. Staphylococcus aureus sigma B-dependent emergence of small-colony variants and biofilm production following exposure to Pseudomonas aeruginosa 4-hydroxy-2-heptylquinoline-N-oxide. BMC Microbiol. 2010;10:33. doi: 10.1186/1471-2180-10-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pförtner H., Wagner J., Surmann K., Hildebrandt P., Ernst S., Bernhardt J., Schurmann C., Gutjahr M., Depke M., Jehmlich U., Dhople V., Hammer E., Steil L., Völker U., Schmidt F. A proteomics workflow for quantitative and time-resolved analysis of adaptation reactions of internalized bacteria. Methods. 2013;61:244–250. doi: 10.1016/j.ymeth.2013.04.009. [DOI] [PubMed] [Google Scholar]
- 68.Michalik S., Depke M., Murr A., Gesell Salazar M., Kusebauch U., Sun Z., Meyer T.C., Surmann K., Pförtner H., Hildebrandt P., Weiss S., Palma Medina L.M., Gutjahr M., Hammer E., Becher D., Pribyl T., Hammerschmidt S., Deutsch E.W., Bader S.L., Hecker M., Moritz R.L., Mäder U., Völker U., Schmidt F. A global Staphylococcus aureus proteome resource applied to the in vivo characterization of host-pathogen interactions. Sci. Rep. 2017;7:9718. doi: 10.1038/s41598-017-10059-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tuchscherr L., Bischoff M., Lattar S.M., Noto Llana M., Pfortner H., Niemann S., Geraci J., Van de Vyver H., Fraunholz M.J., Cheung A.L., Herrmann M., Völker U., Sordelli D.O., Peters G., Löffler B. Sigma factor SigB is crucial to mediate Staphylococcus aureus adaptation during chronic infections. PLoS Pathog. 2015;11:e1004870. doi: 10.1371/journal.ppat.1004870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tuchscherr L., Löffler B. Staphylococcus aureus dynamically adapts global regulators and virulence factor expression in the course from acute to chronic infection. Curr. Genet. 2016;62:15–17. doi: 10.1007/s00294-015-0503-0. [DOI] [PubMed] [Google Scholar]
- 71.Rollenhagen C., Antelmann H., Kirstein J., Delumeau O., Hecker M., Yudkin M.D. Binding of sigma(A) and sigma(B) to core RNA polymerase after environmental stress in Bacillus subtilis. J. Bact. 2003;185:35–40. doi: 10.1128/JB.185.1.35-40.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Boch J., Kempf B., Bremer E. Osmoregulation in Bacillus subtilis: synthesis of the osmoprotectant glycine betaine from exogenously provided choline. J. Bact. 1994;176:5364–5371. doi: 10.1128/jb.176.17.5364-5371.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Boch J., Nau-Wagner G., Kneip S., Bremer E. Glycine betaine aldehyde dehydrogenase from Bacillus subtilis: characterization of an enzyme required for the synthesis of the osmoprotectant glycine betaine. Arch. Microbiol. 1997;168:282–289. doi: 10.1007/s002030050500. [DOI] [PubMed] [Google Scholar]
- 74.Lee J.J., Kim J.H., Kim D.G., Kim D.H., Simborio H.L., Min W.G., Rhee M.H., Lim J.H., Chang H.H., Kim S. Characterization of betaine aldehyde dehydrogenase (BetB) as an essential virulence factor of Brucella abortus. Vet. Microbiol. 2014;168:131–140. doi: 10.1016/j.vetmic.2013.10.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material