Abstract
Oxidative stress and the resulting damage to genomic DNA are inevitable consequences of endogenous physiological processes, and they are amplified by cellular responses to environmental exposures. One of the most frequent reactions of reactive oxygen species with DNA is the oxidation of guanine to pre-mutagenic 8-oxo-7,8-dihydroguanine (8-oxoG). Despite the vulnerability of guanine to oxidation, vertebrate genes are primarily embedded in GC-rich genomic regions, and over 72% of the promoters of human genes belong to a class with a high GC content. In the promoter, 8-oxoG may serve as an epigenetic mark, and when complexed with the oxidatively inactivated repair enzyme 8-oxoguanine DNA glycosylase 1, provide a platform for the coordination of the initial steps of DNA repair and the assembly of the transcriptional machinery to launch the prompt and preferential expression of redox-regulated genes. Deviations/variations from this artful coordination may be the etiological links between guanine oxidation and various cellular pathologies and diseases during ageing processes.
Abbreviations: ROS, reactive oxygen species; 8-oxoG, 7, 8-dihydro-8-oxoguanine; OGG1, 8-oxoguanine DNA glycosylase 1; BER, base excision repair; OGG1-BER, OGG1-initiated DNA base excision repair; IIR, innate immune response; TNFα, Tumor necrosis factor alpha; NF-κB, Nuclear Factor kappa B; Sp1, specificity protein1; AP-1, activator protein 1; STAT1, signal transducer and activator of transcription 1; (Me)C, methylation of cytosine; MBPs, methyl-CpG-binding proteins
Keywords: OGG1, 8-oxoguanine, Gene expression regulation, Epigenetic, DNA methylation
Graphical abstract
Binding of OGG1 to genomic 8-oxoguanine facilitates gene expression. Proposed actions of OGG1 on open chromatin region (left), OGG1 binding to its substrate facilitates DNA occupancy of transcription factors (NF-κB is shown as example) and promote transcription. If guanine oxidation occurs at closed chromatin region (right), binding of OGG1 could block the interaction of methyl binding proteins (BMPs) with their substrates, subsequently recruit transcription machinery components to activate transcription.
1. Introduction
The generation of reactive oxygen species (ROS) by various endogenous physiological processes and/or environmental agents is inevitable, and they cause damage to cellular macromolecules. While proteins, lipids and RNA having oxidative damage are usually subjected to degradation and recycling, DNA lesions should be repaired to maintain genomic integrity [1], [2]. Oxidative DNA damage includes oxidized bases, oxidized sugar fragments, abasic/apurinic/apyrimidinic (AP) sites and single-strand breaks [3], [4]. Closely spaced single-strand breaks generated during the repair of oxidized bases can result in DNA double-strand breaks [3], [4]. ROS primarily damage guanine because it has the lowest oxidation potential (midpoint potential is −1.17 mV on a nickel hydrogen electrode) among the DNA bases [5], [6]. Guanine's oxidized product 7,8-dihydro-8-oxoguanine (8-oxoG) is the most predominant DNA oxidative lesion in the genome [5], [6], [7]. It is estimated that up to 100,000 8-oxoG lesions can be formed daily in DNA per cell [8]; thus, it is considered as a biomarker of oxidative stress [9], [10]. 8-OxoG is mutagenic because it may pair with adenine instead of cytosine, resulting in a G:C to T:A transversion during DNA replication. [1], [9], [10], [11], [12]. In eukaryotic cells, guanine lesions (including 8-oxoG and its open-ring product 2,6-diamino-4-hydroxy-5-formamidopyrimidine, FapyG) are primarily repaired by 8-oxoG DNA glycosylase 1 (OGG1), a functional homologue of Escherichia coli protein MutM, through stepwise base excision repair (BER) pathway [1], [7], [13], [14], [15], [16]. The process of recognition and repair of 8-oxoG by OGG1 is well understood and reviewed [7], [17], and it represents one of the oldest DNA BER pathways, which constitutes a major area in studies of DNA repair mechanisms and led to the 2015 Nobel Prize in Chemistry.
Even though the direct experimental evidence is lacking, oxidation product(s) of guanine have often been linked to various ageing-associated conditions, including tissue and organ dysfunction, carcinogenesis, and neurodegenerative and cardiovascular diseases [1], [7], [18], [19], [20]. OGG1 is a multifunctional protein, when complexed with its repair product free base 8-oxoG, activates Ras family GTPases and downstream signaling, and this has been extensively documented and reviewed in the context of innate and adaptive inflammatory processes [7], [20], [21], [22]. This review discusses present views on the utilization of a transient OGG1-DNA complex on genomic DNA by transcriptional machinery during gene expression to enable prompt cellular responses in oxidatively stressed cells.
2. The modulation of OGG1's enzymatic activity
OGG1 initiated BER (OGG1-BER) entails multiple steps including lesion recognition, flipping the substrate from DNA double helix into the base-binding pocket (active site) of OGG1 and site-specific changes in DNA structure [23], [24]. As a bifunctional DNA glycosylase, OGG1 excises base(s) and then cleaves the phosphodiester bond 3′ of the damaged base through its AP lyase activity. This leaves a blocking residue attached to the deoxyribose upstream of the nick, which is removed by apurinic/apyrimidic endonuclease 1 (APE1). The resulting nucleotide gap is filled in by DNA polymerase β, and the nick is sealed by DNA ligase III bound to the presumptive scaffold protein X-ray repair cross complementing 1 (XRCC1) [4], [7], [20].
Protein–protein interactions may be the major mode of regulating OGG1 activity (Fig. 1 left). For example, the AP lyase activity of OGG1, which creates a nick in the DNA backbone, was increased with the addition of APE-1 in in vitro assays [25], [26]. However, recent studies suggested that the in cellulo activity of OGG1 has a primarily monofunctional mode of removing 8-oxoG bases only, while APE-1 enhances this activity by stimulating the release of OGG1 from AP sites [27], [28]. XRCC1, a scaffold protein in the BER pathway, also interacts with and stimulates many DNA glycosylases including OGG1 [29], [30]. Another study documented that OGG1 binds directly to poly(ADP-ribose) polymerase 1, a DNA-damage sensing protein that is involved in DNA repair. This interaction is enhanced by oxidative stress; however, binding with and modification by PARP1 decreases the BER function of OGG1 [31]. Cut homeobox 1 (CUX1) and CUX2, as transcriptional activators of many genes involved in the DNA damage response, have been implicated in cancer as having both tumor suppression and oncogenic potentials. CUX1 and CUX2 interact with and stimulate the DNA glycosylase and AP lyase activities of OGG1 without involving their transcriptional function [32], [33]. SATB1, a genome organizer and transcriptional regulator containing two CUT domains, was recently shown to interact with OGG1, stimulating two of its enzymatic activities [34], which needs to be further studied in the physiological context.
Fig. 1.
The repair activity of OGG1 is tightly controlled by protein-protein interactions and post-translational modifications. APE1, apurinic/apyrimidic (AP) endonuclease 1; XRCC1, X-ray repair cross complementing 1; CUX1, Cut homeobox 1; HDAC, histone deacetylase; PARylation, Poly(ADP-ribosyl)ation; PARP1, Poly(ADP-ribose) polymerase 1; OGT, O-GlcNAc transferase. Red, up-regulation; Green, down-regulation; Black, no change.
Moreover, OGG1 activity is also regulated by posttranslational modifications (Fig. 1 right). OGG1 is acetylated in vivo by histone acetyltransferases, such as cyclic AMP response element-binding protein/p300 complex (CBP/p300), and the acetylation at Lys338/Lys341 significantly increases OGG1 activity. OGG1 interacts with class I histone deacetylases, which might be responsible for its deacetylation [35], [36], [37]. OGG1 also physically interacts with the protein kinases CDK4, c-ABL and PKC [38], [39]. The phosphorylation of OGG1 by CDK4 increases its 8-oxoG incision activity and affects AP lyase activity, whereas the phosphorylation induced by PKC and c-ABL does not affect OGG1 activity. The distinct functional outcomes of serine/threonine versus tyrosine phosphorylation may indicate that the activation of different signal transduction pathways modulates OGG1 activity based on the actual needs of the cells [38], [39]. Interestingly, OGG1 is highly O-GlcNAcylated in diabetic mice compared with controls. In vitro experiments demonstrated that O-GlcNAcylation inhibits OGG1 activity [40]. Hyperglycaemia is conventionally associated with the over-production of ROS; thus, the significance of the inhibition of OGG1 activity through O-GlcNAcylation needs to be elucidated. Furthermore, studies showed that oxidative stress decreases the activity of OGG1, but it is reestablished once the cellular redox status is normalized [41], [42], [43]. Of note, the reduced state of the redox-sensitive residues of OGG1, cysteine (Cys) is important for its glycosylase activity [41]. Recently, in tumor necrosis factor alpha (TNF-α)-exposed cells, the oxidation of cysteine residue(s) to sulfenic acid, along with the impaired base excision activity of OGG1, was observed. Human OGG1 contains seven cysteine residues, of which, the one(s) subject to oxidation need to be further investigated.
It is seemingly paradoxical that OGG1-BER activity is impaired in cells under oxidative stress, during which levels of oxidized guanines are enhanced and effective excision/repair is needed. OGG1's enzymatic inactivation was temporally correlated with an increase in intracellular ROS levels, as well as 8-oxoG accumulation in the genome, primarily at the promoter regions of a number of genes [44], [45]. Significant increases in mRNAs levels from pro-inflammatory genes were concomitant, suggesting that the transient inactivation of OGG1 and accumulation of 8-oxoG in the promoter region may have important roles in transcriptional regulation [45], [46].
3. Aberrant gene expression in the absence of OGG1
To study the roles of OGG1 in carcinogenesis and ageing processes, Ogg1-null mice were developed [47], [48]. Unexpectedly, under normal conditions, the lack of OGG1 activity and consequent supraphysiological levels of genomic 8-oxoG do not affect the embryonic development or life span of Ogg1-/- mice, and the animals showed no marked changes in pathology or in tumor frequency [49]. Mitochondrial DNA isolated from the liver of Ogg1-null mutant animals contains a >20-fold increase in the 8-oxoG level compared with that in wild-type animals, with no detectable changes in maximal respiration rates or mitochondrial ROS generation [50]. However, Ogg1-/- mice showed a decreased serum lgG2a level (Th1 response) in response to bacterial infection, and increased resistance to lipopolysaccharide (LPS)-induced inflammation and organ dysfunction, coupled with lower levels of the chemokine Mip-1 alpha and Th1 cytokines interleukin (Il)-12 and Tnf-α [51], [52]. Likewise, after an ovalbumin challenge, Ogg1 knockout mice, compared with wild-type mice, exhibit less inflammatory cell infiltration and decreased oxidative stress in the lungs. The phenotype includes decreased levels of Il-4, Il-6, Il-10 and Il-17 and the accumulation of inflammatory cells in lung tissue [53]. In addition, the siRNA-mediated deficient expression of Ogg1 in the airway epithelium results in a lower inflammatory response after allergen challenge in sensitized mice, as shown by the decreased expression of the Th2 cytokines (Il-4, Il-5 and Il-13), eosinophilia, and airway hyper-responsiveness [54]. Moreover, Ogg1 silencing in airway epithelial cells or mice lungs significantly decreases the expression of innate immune-response (IIR) cytokines/chemokines (e.g., Cxcl2, Tnf and IL-1β) induced by TNF-α exposure, and thus, the reduced infiltration of neutrophils in bronchoalveolar lavage fluid [46]. Recently, LPS-induced activation of the primary splenocytes obtained from two different Ogg1−/− mouse strains was analyzed. The induction of Tnf-α expression was reduced in splenocytes (in particular macrophages) of both Ogg1−/− strains [55]. These observations suggest potential roles for OGG1 in regulating gene expression in immune responses. Additionally, key genes of fatty acid oxidation, including carnitine palmitoyl transferase-1 and the integral transcriptional co-activator Pgc-1α, as well as multiple genes involved in the tricarboxylic acid cycle's metabolism were significantly down-regulated in livers, resulting in the Ogg1-null mice being susceptible to obesity and metabolic dysfunction [56]. Taken together, role of OGG1 in gene expression modulation has been revealed in the context of various physiopathologic processes.
The transcriptional regulation of pro-inflammatory genes is regulated, at least partly, by ROS-mediated signaling [57], [58], [59], [60]. Many, if not all, pro-inflammatory genes have high GC-content promoters, which usually contain multiple nuclear factor kappa B (NF-κB) and specificity protein1 (Sp1) binding sites. In addition, the binding motifs for these transcription factors (TFs) also contain runs of guanines. Thus, OGG1, as a DNA repair enzyme, having a beneficial effect on pro-inflammatory gene expression under oxidative stress, might be conventionally explained by the repair of 8-oxoG ensures the recognition and binding of TFs to their cis elements. However, given that abasic sites and strand cleavage are intermediates in the BER pathway, inevitably, the prompt and efficient repair of 8-oxoGs (or 2,6-diamino-4-hydroxy-5-formamidopyrimidines) in a high GC-content region may lead to a decrease in promoter integrity and, therefore, the impaired activation of the relevant transcriptome. The timely ROS-mediated confrontation with OGG1-BER activity [41], [42] is obviously an adaption to avoid such damage to promoters, which in turn ensures the prompt expression of a “ROS-response-ome”.
4. OGG1 alters the binding of transcription factors to oxidized guanine-containing DNA
Previous studies have addressed the role of ROS in the redox status of reactive Cys residues located within the DNA-binding domain of TFs, which may control the transcriptional activity of these TFs [(e.g., NF-κB, activator protein 1 (AP-1), transcriptional activator Myb (Myb), cyclic adenosine 3,5-monophosphate response element-binding protein (CREB), early growth response protein 1 (Egr-1), hypoxia inducible factor 1 alpha (HIF-1α) or tumor protein p53 (TP53)] [61], [62], [63], [64], [65]; however, the oxidation of DNA may serve as an essential manner by which ROS signals are sensed and the transcription from the redox-responsive genes is regulated. The role of guanine lesions located within G-rich binding motifs of TFs (such as NF-κB and Sp1) has been investigated by utilizing synthetic DNA and electrophoretic mobility shift assays (EMSAs) [66], [67], [68], yet the conclusions remain controversial. In one study, substitution of 8-oxoG had no effect on the binding of recombinant p50, regardless of the lesion position (5′-G1G2G3G4ACTTTCCC-3′) [66]; while another study utilizing the same motif claimed a significant decrease in p50 binding when the lesion was located at G2, a significant increase in binding when the lesion was at G1, and minimal effects when lesions were at G3 and G4 [68]. The crystal structures of the p50/p50 homodimer and p50/p65 heterodimer of NF-κB bound to the canonical motifs revealed that all of the guanines are conserved and are the conduits by which protein subunits make contact with DNA [69], [70], [71]. Thus, it appears that the lesions within the motif are not favorable for NF-κB sequence recognition and DNA occupancy. Moreover, when a study reported that a single 8-oxoG may be sufficient to inhibit the binding of recombinant Sp1 [66], others used nuclear extracts from HeLa cells to perform an EMSA, and showed the importance of the core (G2–G6) of the GC box (5′-G1G2G3G4C5G6G7G8G9-3′) but not the other guanine residues (G1, G7–9) [67]. Other TFs also recognize GC-rich consensus sequences (e.g., early growth response protein 1 (Egr-1: 5′-GCGGGGGCG-3′) and nuclear respiratory factor 1 (NRF-1: 5′-YGCGCAYGCGCR-3′)) [72], [73], [74], [75]. However, the effect of guanine oxidation within these motifs on the binding of the cognate TFs has not been addressed. The role of guanine damage in the binding of AP-1, despite its lack of a G-rich (5′-TGACTCA-3′) binding consensus was also studied, and the placements of 8-oxoG had an effect on recombinant AP-1 DNA occupancy [66].
Although the influence of 8-oxoG on TF recognition of the G-rich consensus has drawn attention, the potential roles of repair enzymes, including OGG1, have rarely been considered. OGG1 continuously scans for damaged bases in the DNA duplex and efficiently recognizes and binds to its substrates [76], [77], [78], [79]. This raises the question of whether the binding affinity of TFs with 8-oxoG-containing motifs will be altered by the involvement of OGG1 in the assays. The competition between TFs and DNA repair enzymes for oxidatively damaged promoter sites has been examined [68], [80]. CREB1 controls approximately 25% of the mammalian transcriptome through binding to its consensus CRE (5’-TGACGTCA-3’) sequence. 8-OxoG within the CpG islet of the CRE site by itself did not affect total CREB1 binding or dimerization. Binding and particularly dimerization of CREB1 were lowered along with an increase in OGG1 concentration, regardless of the order in which CREB1 and OGG1 were supplied to lesion-containing substrates. The decreased binding was due to the physical presence of OGG1 blocking the approach of the TF to the CRE site rather than the existence of any AP site product [80] because OGG1 has a very low turnover rate on substrate-containing DNA [25], [81]. Another study documented that NF-κB binding might shield guanine lesions from recognition and repair by BER enzymes [68]. In that study, the p50 transcription factor protein was added to an 8-oxoG-modified κB site-containing probe before the addition of Fpg, the OGG1 functional analog in the Escherichia coli, and the addition of the TF shielded these lesions from the cleavage by the DNA glycosylase. The authors proposed that this occurs for the period required for transcription [68].
In vitro biochemical analyses may have suggested that the existence of the DNA repair enzyme interferes with the access of TFs to a damage-containing promoter [80], or that entry into the repair pathway of the glycosylases could be diminished by the binding of TFs [68]. However, how does a cell prioritize the engagement of a particular site with a TF versus a DNA repair enzyme? Two scenarios should be kept in mind: first, the binding of TFs to consensus motifs is normally proscribed by the energetic cost of DNA bending and twisting, whereas a universal feature of most DNA repair glycosylases, without forming sequence-specific contacts with the DNA substrate, is free energy for DNA bending during base flipping, which is fundamentally different from TF binding [82], [83]. Second, while many TFs undergo nuclear translocation upon oxidative stress, DNA glycosylases, as housekeeping gene products, constitutively dwell in the nucleus and are committed to their duties on chromatin.
Additionally, the situation might become more complicated if glycosylase is taken into consideration. DNA glycosylases, including OGG1, interact with their substrate through a set of amino acids that make extensive contact with the DNA backbone at both the 5′- and the 3′-ends of the lesions along with the opposite strand's base cytosine [24], [77]. For example, the crystal structure of the OGG1–DNA complex shows that the OGG1 footprint covers several nucleotides. Thus the binding of TFs to their cognate sequences could be impaired by the occupancy of OGG1 even if the lesion is located outside of (but within the OGG1 “foot-print” to) the motif. The oxidation of the guanines surrounding a motif inevitably adds a layer of complexity, which has been considered in recent studies (described below) [45], [84]. Collectively, sophisticated strategies must be evolved in a cell to guarantee prompt expression from stress-responding genes without interference from repair enzymes during the homing of the relevant TFs to their consensus sequences in 8-oxoG-containing promoters.
5. Interaction of OGG1 with promoter-contained 8-oxoGmodulates the transcription of NF-κB target genes
The effects of 8-oxoG in coding sequences on transcription have been independently investigated by Drs. Wallace and Khobta. They found that guanine lesions do not constitute a significant barrier to transcription; however, OGG1-BER-generated repair intermediate(s) do [28], [85], [86]. Recently, the outcomes of oxidative base lesions in non-coding regions have gained interest. A series of studies shed light on the role of promoter-located 8-oxoG along with its cognate repair protein OGG1 in the regulation of IIR gene expression [45], [46], [84]. The rapid (within 30 min) and robust expression of pro-inflammatory mediators (e.g., IL-1β, TNF, CCL20, CXCL1 and CXCL2) was induced by pro-inflammatory agents or the ROS inducer glucose oxidase. In parallel, the global 8-oxoG content, as well as the level of 8-oxoG in promoters (surprisingly not evenly along the entire gene bodies), also reached the peak. Meanwhile, the inactivation of OGG1 through cysteine oxidation to sulfenic acid was observed [45]. The coincident high levels of 8-oxoG in promoter regions and the up-regulated pro-inflammatory gene expression implied that formation of guanine lesions might be advantageous for the initiation of gene transcription in cellulo.
5.1. Cooperation between OGG1 and NF-κB represents a sub-group of NF-κB-driven transcriptomes
To further explore whether OGG1 binding to 8-oxoG in chromatinized DNA can modulate gene transcription, chromatin immuno-precipitation (ChIP) and ChIP sequencing were performed. Immediately after exposure (15 and 30 min) to TNF-α, OGG1 was preferentially enriched on gene regulatory regions [45]. A genome-wide analysis revealed that at 30 min post-exposure, OGG1 enrichment sites, possibly linked to the location of its substrate 8-oxoG, are inclined to be in non-coding regions, such as promoters, introns and intergenic regions (Fig. 2). Unexpectedly, the distribution on genomic regions of OGG1 peaks was similar to that of NF-κB, for which sequence-specific binding in the genome is well documented [87]. The correlation of the location of OGG1 with that of NF-κB may directly link OGG1 to transcriptional regulation. A system-level analysis revealed that gene products from OGG1-enriched promoters are linked to the regulation of cellular processes, including response to oxidative stress, signal transduction, regulation of transcription from RNA pol II promoters, cellular homeostasis, and immune responses. Intriguingly, a subgroup of OGG1-ChIP-ed genes was positively correlated with the regulation of NF-κB nuclear import, NF-κB signaling and its sequence binding (data submitted). Importantly, OGG1 enrichment on gene regulatory regions was functional for corresponding to changes in mRNA levels.
Fig. 2.
Genome-wide distribution of OGG1 in response to stimuli. Flag-OGG1-expressing cells were exposed to TNF-α for 30 min. ChIP-ed DNA was sequenced (GSE #: GSE75652) and the enrichment of OGG1 was analyzed. The diagram illustrates the genome-wide enrichment of OGG1 within 1–10 kb from transcription start sites (TSS).
The binding of a DNA repair enzyme to a substrate-containing promoter region under oxidative stress may not be surprising; however, an OGG1 knock-down markedly impaired the recruitment of NF-κB to promoter regions and significantly modulated expression from NF-κB-dependent genes [45], [46]; in parallel, OGG1 interacts with TF IID, site-specific TFs Sp1 and NF-κB, and these interactions were blocked by the administration of a ROS scavenger [45], [46], which is intriguing. Additionally, a system-level analysis showed that immediately after TNF-α exposure, within the innate immune pathway, 673 out of 840 OGG1-enriched genes were common with those of NF-κB-enriched (data submitted). Because NF-κB is a central player in prompt stress responses [88], [89], the emerging role of OGG1 in transcriptional regulation in response to a ROS burst appears to be linked to a sub-group of NF-κB-driven genes.
5.2. Positional value of 8-oxoG for the DNA occupancy of NF-κB
To gain insight into the positional value of 8-oxoG for the interaction of NF-κB with its consensus in the presence of OGG1, we extensively examined DNA segments containing NF-κB motifs and corresponding to the native sequences in the proximal region of the human TNF and mouse Cxcl2 promoters. Guanines within and outside NF-κB motifs were individually substituted with 8-oxoG, and EMSAs were performed. In the presence of recombinant OGG1 or the nuclear extract from OGG1-expressing cells, the binding of NF-κB to DNA containing 8-oxoG, located several nucleotides upstream from the motif, was consistently enhanced (Fig. 3a). However, 8-oxoG being within or closely proximal to the NF-κB binding site decreased NF-κB's DNA occupancy, which may involve either 8-oxoG itself or the interference of OGG1 [45], [84]. Importantly, through the binding of 8-oxoG 8–11 bp upstream from the motif, OGG1 accelerated NF-κB's docking, primarily in its hetero-dimeric form [45]. A recent study analyzed the surrounding sequences (10 bp up- and downstream from the motif) of 70 functional human κB sites. The frequencies of guanine at −8 and −10 bases are more than 40%, apparently higher than other positions (Fig. 3b) [90], supporting the idea that a properly positioned guanine and its oxidation may be utilized for the homing of TFs (such as NF-κB) to their motif beaconed by substrate-bound OGG1.
Fig. 3.
Positional value of 8-oxoG for the DNA occupancy of NF-κB. (a) Binding to 8-oxoG 8–11 bp upstream from the motif, OGG1 accelerates NF-κB occupancy [45]. Sequence of the DNA fragment containing a NF-κB motif used in the EMSA assay is derived from the human TNF promoter. 8-oxoG substitution was carried out individually (shown in red). (b) Frequency of G at each base in the surrounding sequences 10-bp up- and downstream from the motifs of 70 functional human κB sites (data provided by Wang et al. [89]). The average frequency of one of four nucleobases at each position is considered as 25%.
Thus, the proposed mechanism by which OGG1 through binding with 8-oxoG in promoter region modulates the transcription of NF-κB target genes is as follows: upon the engagement of OGG1 with its substrate, the successive events, including the intrusion of its amino acid residues into the DNA helix, extraction of 8-oxoG from the DNA, insertion of the damaged base into OGG1 active-site pocket and un-stacking the opposite cytosine from the DNA helix [24], [75], [76] result in a sharp (~70°) bending of the DNA duplex, inducing DNA architectural change and create a specific interface in DNA that allows the prompt recognition of motifs by NF-κB and the assembly of the transcriptional initiation complex [91] (Fig. 4).
Fig. 4.
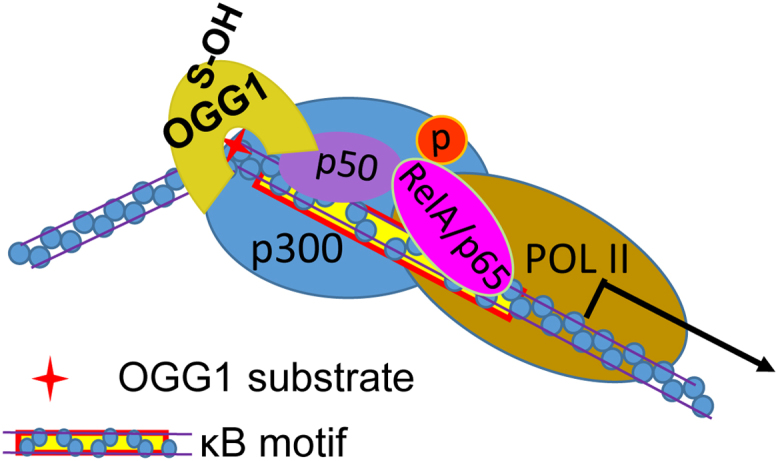
Enzymatically inactive OGG1 binding to promoter-contained 8-oxoG facilitates transcription factor binding and the assembly of the transcriptional machinery. OGG1 binding to its substrate bends DNA and induces allosteric alteration of DNA, that facilitates NF-κB occupancy and the assembly of the transcription machinery.
Nevertheless, the coincident prompt expression of the ROS-responding and NF-κB-driven genes with the accumulation of 8-oxoG in the promoter regions raises another question of how cells can control the positional oxidation of guanine. Promoters driving the expression of immune- or stress- responsive genes, such as TNF, ILs, CCLs, CXCLs and p21 (WAF1/Cip1), contain multiple copies of NF-κB and/or Sp1 motifs in tandem [46], [92], [93], [94], [95]. One could hypothesize that the copy number of such TF motifs correlates with the magnitude of promoter activation in response to stimuli. However, it can be speculated that some of the TF consensus sequences could be ensured to be protected by the chromatin structure, thus staying undamaged. These consensus sequences have appropriately positioned 8-oxoGs nearby, and through interactions with OGG1, TFs (e.g., NF-κB) may rapidly home to their binding motifs in chromatin owing to the cognate repair enzyme's efficient recognition of the substrate.
6. Epigenetic role of guanine oxidation in gene expression
The compositional pattern of the human genome shows a high level of heterogeneity. GC-poor content represents approximately 63% of the genome, but the density of genes is much greater in the GC-rich fractions. Additionally, the transcriptional activity of genes is correlated with the GC content of the genome [96], [97]. Genes densely distributed in high GC-content regions are inclined to be actively transcribed, whereas those sparsely distributed in GC-poor regions are usually occasionally transcribed (such as in a tissue-dependent or developmentally regulated manner) [96] (Fig. 5). Moreover, despite the vulnerability of guanine and the mutagenicity of 8-oxoG, the vertebrate genome has an evolutionarily conserved high GC content in promoter regions. A genomic-wide survey revealed that 72% of human gene promoters belong to a class with a high GC content [98]. One may readily link this evolutionarily conserved property of the genome to the benefits of the GC content in controlling of gene expression. Indeed, considerable evidence suggests that base oxidation in DNA is not only a carcinogenic risk factor but also plays regulatory functions as an epigenetic mark [22], [99].
Fig. 5.

Transcriptionally active genes are embedded in a GC-rich fraction of chromatin[95]. Upper panel, actively or constitutively transcribed genes are densely distributed in high GC-content regions. Lower panel, genes sparsely distributed in GC-poor regions are transcribed in a tissue-dependent or developmentally regulated manner.
6.1. Biased guanine oxidation in the genome
Increasing evidence suggests that guanine oxidation does not occur randomly but exhibits a strong distributional bias in the genome. The DNA double helix, containing a π-stacked array of heterocyclic base pairs, is favorable for the migration of charge over long distances through a multistep hopping reaction with all guanines as carriers of positive charges [100], [101], [102]. Based on the G–G stacking rule, the most readily oxidized sites in one-electron oxidation of duplex DNA are the guanine residues located 5′ of guanine runs [103]. Genes containing G:C-rich sequences outside of the coding area (poly-G:C domains) may act as sinks for positive charges. Charge transport can occur over distances of up to 200 Angstroms away from the initial site of oxidant injury [104], [105]. The ability of DNA molecules to transport charges over long distances could provide a strategy for funneling damage to particular sites in the genome [106]. Electron transfer through DNA can occur under conditions of oxidative stress and therefore, biological consequences are highly likely [101], [107].
In addition to in vitro studies that proposed the preferential oxidation of guanine in a 5'-GG-3' sequence context, efforts have also been made to reveal the in vivo existence of 8-oxoG at the genome-wide scale [108], [109], [110], [111]. In situ immunofluorescence of 8-oxoG on human metaphase chromosomes showed that regions with a high frequency of recombination and single nucleotide polymorphisms within chromosomal regions are preferentially located with a high density of 8-oxoG [111]. In addition, an 8-oxoG antibody (Ab) was utilized to enrich lesion-containing fragments from isolated and sonicated genomic DNA, and microarray and next-generation sequencing (NGS) were carried out [109], [110]. The microarray results revealed that low levels of 8-oxoG are generated in genetic regions compared with “gene deserts” (such as lamina-associated domains), which supports the hypothesis that GC-rich sequences outside of coding areas may act as sinks for positive charges to protect the fidelity of genetic information under oxidative stress [1], [106]. NGS provided a higher resolution showing that most of the oxidized guanine bases were localized to promoters, which is in line with data from OGG1 ChIP Sequencing (Section 4). This indicated a high correlation of OGG1-enriched peaks with promoter regions [45], [109]. The drawbacks of the application of 8-oxoG Ab, including that the resolution is not high enough to determine precise genomic elements; and the binding affinity is significantly impacted by DNA secondary structures, introducing considerable bias in the data obtained, were overcome by a deliberate approach recently developed in Dr. Burrows’ laboratory [108]. An unusual property of 8-oxoG is that its redox potential (~ 600 mV) is much lower than that of guanine, allowing for the selective oxidation of 8-oxoG by a mild oxidant [112]. The oxidation of 8-oxoG yields an electrophilic intermediate that can be trapped with a primary amine nucleophile to form a stable amine-conjugated product amine-terminated biotin (BTN) for streptavidin enrichment [113]. Affinity purification rather than immunoprecipitation provided ~ 0.15 kb resolution to map 8-oxoG in the mouse genome [108]. Gene promoters and untranslated regions harbor more 8-oxoG-enriched sites, which were correlated with reactive 5′-GG-3′ sequences as shown in in vitro studies from past decades [108].
6.2. The roles of OGG1 in transcriptional regulation implicit in the biased oxidation of guanine
Considering the promptitude of pro-inflammatory gene activation in IIR and the coincident high 8-oxoG levels, transiently hampered OGG1–BER activity, and the timely preservation of promoter integrity [44], [45], [46], we propose that, in response to an oxidative burst, guanine oxidation in a GC-rich promoter has a cis effect, whereas OGG1 acts as a trans factor, whose oxidation is deemed to be a reversible posttranslational modification. The oxidation of cysteine thiol (RSH/RS−) by ROS leads to the formation of highly reactive sulfenic acid (RSOH), which can react either with another thiol to form a disulfide bond (RSSR) or with GSH to become S-glutathionylated (RSSG). These oxidative modifications are reversible, and the reduction is catalyzed by the Trx and/or Grx system [114]. The timely binding of oxidatively inactivated OGG1 and the consequent allosteric alteration of DNA during the pre-excision step of BER [115], [116] facilitate the homing of TFs (e.g., NF-κB and signal transducer and activator of transcription 1; STAT1) [45], [46], [117] or co-activator (e.g., CBP/p300) [35] to their binding sites, and thereby the assembly of the transcriptional machinery is promoted. When redox balance is re-established, OGG1 regains its enzymatic activity through the reduction of its oxidized cysteine, and thus the damaged base is excised to prevent mutation(s) in promoter regions (Fig. 6). Although additional studies are required, we deduced that after a rapid boost, the fall in the mRNA levels of pro-inflammatory genes, explained by others as a consequence of programmed NF-kB oscillation [118], may be partly due to the introduction of the repair intermediates (AP-sites, strand breaks) into the promoter by OGG1 after the redox-permitted restoration of its enzymatic activity [28], [85].
Fig. 6.
Oxidative stress-mediated inactivation of OGG1 enzymatic activity converts the OGG1–DNA complex as part of transcriptional machinery. Under the physiological redox state, OGG1 fulfills its repair function in the DNA BER pathway (left panel). Upon oxidative stress OGG1's enzymatic function is compromised by cysteine modifications, and the OGG1-8-oxoG-DNA complex is utilized by NF-κB-centered transcriptional machinery, leading to the transcription of target genes (middle panel). When redox balance is re-established, OGG1 regains its repair function and, by introducing repair intermediates into the promoter, may disrupt the actions of transcriptional machinery.
Studies have also addressed the post-8-oxoG-excision mechanisms by which OGG1 plays a key role in transcriptional activation [109], [119], [120], [121]. The localized demethylation of histone H3 by demethylase LSD1 produces ROS, which generates 8-oxoG and recruites OGG1. BER-introduced strand breaks attract topoisomerase II, which seems to be important for long-range changes in DNA topology and is essential for estrogen-induced gene expression [119], [120]. In addition, G-rich DNA structures such as G-quadruplexes are highly susceptible to oxidation [122], [123]. An increased 8-oxoG enrichment in G-quadruplexes has been affirmed by NGS technologies [108], [109]. The enhanced expression of vascular endothelial growth factor (VEGF) or endonuclease III-like protein 1 (NTHL1) was observed when 8-oxoG was formed in G-quadruplex sequences in the promoters [109], [121]. The induction of transcription requires OGG1 and an enzymatically inactive APE1. Following OGG1's removal of a base lesion, the yielded AP site enables the melting of the duplex to adopt a G-quadruplex fold that is crucial for gene expression [124]. APE1 binds but inefficiently cleaves AP sites, which induces transcription most likely with the aid of other activating factors [121].
The conservation of G-rich promoters and the biased occurrence of guanine modifications led to the hypothesis that guanines in promoter regions not only have a sacrificial role in maintaining the fidelity of coding sequences but also play an active role in gene regulation. The oxidation of specific nucleobases, as well as the redox-restricted base repair may be utilized by trans factors, allowing the preferential selection of the transcription from ROS-responding genes. Given this possibility, the compromise in OGG1's enzymatic activity under oxidative stress conditions through interactions with or modification by other proteins (section 1, Fig. 1) may imply the functional regulation of OGG1 in other cellular processes other than DNA repair, such as transcription. Accordingly, the co-localization of OGG1 with RNA Pol II and active chromatin marker H3K4me along with the increase in the 8-oxoG level, which led to the elucidation of the preferential repair of active chromosome regions [125], may also suggest a transcription-directing role for OGG1. Different interpretations of the interactions of OGG1 with RNA Pol II and active chromatin marker [125], or with RNA Pol II and TFs [46] indicate that in the open chromatin, an elaborate coordination and reciprocity between two biological processes, transcription initiation and DNA repair, exists both depending on the engagement of OGG1 with its substrate. In support, we observed an intriguing bidirectional promotion of the DNA binding of OGG1 or NF-κB in the presence of one another [84].
ROS introduce both guanine lesion(s) in DNA and activate cell signaling to cause posttranslational modification to modulate activity of the cognate repair enzyme. Because the loss or regain of catalytic activity of OGG1 is redox-restricted, the timely restriction in OGG1's excision function after grip of its substrate appears to regulate transcriptional response to oxidative stress. A delay in 8-oxoG repair prevents the guanine runs-containing promoters being “chopped”, ensuring the prompt actions of transcriptional machineries for IIR gene expression [45], [46], [55]. On the other hand, expression of large numbers genes are partly controlled by G-quadruplexes in their promoters, and activation of such genes such as VEGF or NTHL1 requires quadruplex structure, which is linked to 8-oxoG excision and AP-site formation by OGG1-BER [109], [121]. Taken these data together, one may conclude that the deviations from accurate cellular responses to oxidative stress may be due to unscheduled actions of OGG1-BER at oxidized guanines, which might be the etiological link of 8-oxoG to various cellular pathologies and diseases.
6.3. Can guanine oxidation unlock the transcriptional repression imparted by cytosine methylation?
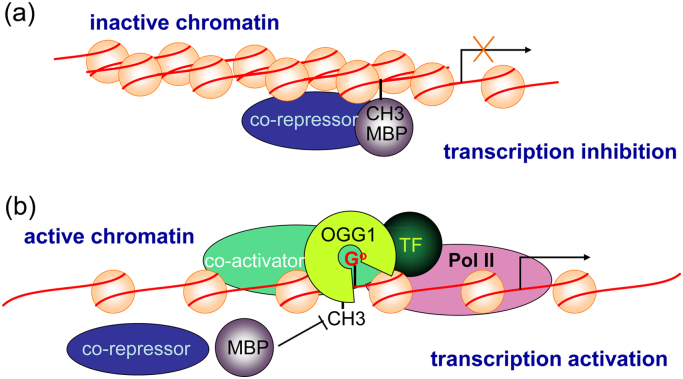
The nonrandom occurrence of guanine oxidation may be affected by epigenetic DNA modifications, for instance, the methylation of cytosine ((Me)C). Guanine reactivity towards oxidants may be targeted to methylated (Me)CpG sequences due to the lowered ionization potential of the guanine base when paired with (Me)C [126], [127]. Linking cytosine methylation to guanine oxidation would really validate the epigenetic role of the latter. DNA methylation, a classical epigenetic marker, appears to be predominantly confined to cytosine in the CpG dinucleotide context [128]. Cytosine methylation is usually associated with a repressed chromatin state and the inhibition of gene expression [128]. The repression may either result from the direct effects on TF binding, or may be indirectly caused by repressor protein(s), methyl-CpG-binding proteins (MBPs) [128], [129], recognizing methyl-CpG and eliciting the repressive potential of methylated DNA [130], [131]. MBPs may also recruit transcriptional co-repressor molecules [132], [133], [134], [135]. A series of studies by Dr. Sowers’ group documented that the recognition elements of MBPs include the guanine's O6 and N7 atoms present in the major groove [136], and the oxidation of guanine converts the N7 position from a hydrogen bond acceptor to a hydrogen bond donor and replaces the 8-proton with an oxygen atom, potentially interfering with the recognition of the methyl-CpG dinucleotide by MBPs [137]. EMSAs revealed that the oxidation of guanine to 8-oxoG significantly inhibits binding of the methyl-CpG-binding domain of methyl-CpG-binding protein 2 (MeCP2) to the oligonucleotide duplex [137]. Although a role of OGG1 has not been indicated, the inhibition of MBPs binding to substrates may be amplified by the involvement of OGG1. Moreover, because of OGG1's DNA bending/twisting and its physical interaction with the components of the transcription machinery [46], the promoter may be released from the repression in an intracellular scenario, although further experimental evidence is needed (Fig. 7).
Fig. 7.
OGG1-DNA complex at 8-oxoG may unlock the transcriptional repression caused by cytosine methylation. (a) MBPs recognize methyl-CpG and recruite co-repressor molecules to silence transcription. (b) Binding to oxidized guanine in the opposite strand, OGG1 may interfere the interaction of MBPs with their substrates, and recruite transcriptional machinery components to activate transcription.
It has been assumed that to activate the transcription of genes locked by cytosine methylation, cells need to eliminate the methyl group from (Me)C. The removal of (Me)C has been observed in the genome of mouse primordial germ cells during development and in cells or tissues of patients with inflammatory disorders [138], [139], [140]. Tremendous effort has been devoted to unveiling the mechanisms of DNA demethylation; however, while passive DNA demethylation, which refers to the loss of the methyl group from 5-methyl cytosine (5meC) when DNMT1 is inhibited or absent during successive rounds of DNA replication, is generally understood and accepted, the subject of active DNA demethylation remains controversial [141], [142]. Various mechanisms include the enzymatic removal of the methyl group of 5meC, direct excision of 5meC through BER, deamination of 5meC to thymidine (T) followed by BER of the T-G mismatch through nucleotide excision repair, oxidative demethylation and radical S-adenosylmethionine (SAM)-based demethylation [141], [142]. Recently, enzymatic oxidation of 5meC followed by BER has emerged as a plausive mechanism of active removal of (Me)C. The family of dioxygenases commonly known as Ten-eleven translocation (Tet) proteins are responsible for the enzymatic oxidation of 5mC into 5-hydroxymethylcytosine (5hmC), 5-formylcytosine (5fC) and 5-carboxylcytosine (5caC), which are recognized and excised by the DNA glycosylase TDG through the BER pathway [143]. The interference of 8-oxoG in the binding of MBPs indicates that, at least, a group of genes might utilize the engagement of OGG1 with oxidized guanine to promptly evade the repression posed by methylated cytosine, passing by methyl group elimination, to implement the rapid cellular response to ROS. Guanine oxidation is instant and non-enzymatic; thus, OGG1 complexed with its substrate might be the most rapid and economical strategy for cells to unlock the repression caused by methylated cytosine. While oxidative stress persists, OGG1 may further recruit TET1 for enzymatic catalysis of 5meC into 5hmC; thus the BER pathway could be utilized to transfer the DNA oxidation signal to downstream DNA demethylation enzymes [144]. In this scenario, whether OGG1-TET interaction could be used for gene transcription is enigmatic.
7. Conclusions
Among bio-macromolecules, guanine in the DNA is the primary target of ROS; however, its oxidation has not yet been appreciated as a way to transmit ROS signals, other than as oxidative damage to be repaired. As discussed here, guanine may serve as a ROS sensor, and the resulting product is an epigenetic mark. 8-OxoG itself does not induce the structural alterations in DNA; however it may serve as a ligand for OGG1 and together they play a role in the regulation of gene expression. In this context, OGG1 may function as an adaptor to facilitate the DNA occupancy of TFs at target gene promoters. While links between deficient OGG1-BER and susceptibility to cancer and accelerated ageing have been documented, we propose that it may not be the accumulation and mutagenicity of 8-oxoG itself but deviations/variations from the coordination between the OGG1-initiated repair process and transcriptional regulation could be the etiological link. Although further studies are needed, pharmacological modulation of OGG1 activity by small molecules could have clinical utilities in the prevention and amelioration of the unscheduled decline of cell/tissue functions and degenerative diseases during ageing processes.
Acknowledgements
This work was supported by grants from the National Natural Science Foundation of China (Grant nos. 31571339 and 31371293 to XB), the Program for Introducing Talent to Universities (Grant no. B07017 to X.B), and United States National Institute of Environmental Health and Sciences (Grant no. RO1 ES018948 to IB); United States National Institute of Allergic and Infectious Diseases (Grant no. AI062885, to IB). We thank Mardelle Susman (Department of Microbiology and Immunology, Institute of Human Infections and Immunity) for her scientific input and for critically editing the manuscript.
Acknowledgments
Conflicts of interest
None.
Contributor Information
Xueqing Ba, Email: baxq755@nenu.edu.cn.
Istvan Boldogh, Email: sboldogh@utmb.edu.
References
- 1.Radak Z., Boldogh I. 8-Oxo-7,8-dihydroguanine: links to gene expression, aging, and defense against oxidative stress. Free Radic. Biol. Med. 2010;49:587–596. doi: 10.1016/j.freeradbiomed.2010.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Friedberg E.C. DNA damage and repair. Nature. 2003;421:436–440. doi: 10.1038/nature01408. [DOI] [PubMed] [Google Scholar]
- 3.Hegde M.L., Hazra T.K., Mitra S. Early steps in the DNA base excision/single-strand interruption repair pathway in mammalian cells. Cell Res. 2008;18:27–47. doi: 10.1038/cr.2008.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hegde M.L., Izumi T., Mitra S. Oxidized base damage and single-strand break repair in mammalian genomes: role of disordered regions and posttranslational modifications in early enzymes. Prog. Mol. Biol. Transl. Sci. 2012;110:123–153. doi: 10.1016/B978-0-12-387665-2.00006-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burrows C.J., Muller J.G. Oxidative nucleobase modifications leading to strand scission. Chem. Rev. 1998;98:1109–1152. doi: 10.1021/cr960421s. [DOI] [PubMed] [Google Scholar]
- 6.Candeias L.P., Steenken S. Reaction of HO* with guanine derivatives in aqueous solution: formation of two different redox-active OH-adduct radicals and their unimolecular transformation reactions. Properties of G(-H)*. Chemistry. 2000;6:475–484. doi: 10.1002/(sici)1521-3765(20000204)6:3<475::aid-chem475>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 7.Ba X., Aguilera-Aguirre L., Rashid Q.T., Bacsi A., Radak Z., Sur S., Hosoki K., Hegde M.L., Boldogh I. The role of 8-oxoguanine DNA glycosylase-1 in inflammation. Int. J. Mol. Sci. 2014;15:16975–16997. doi: 10.3390/ijms150916975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lindahl T., Barnes D.E. Repair of endogenous DNA damage. Cold Spring Harb. Symp. Quant. Biol. 2000;65:127–133. doi: 10.1101/sqb.2000.65.127. [DOI] [PubMed] [Google Scholar]
- 9.Dizdaroglu M. Formation of an 8-hydroxyguanine moiety in deoxyribonucleic acid on gamma-irradiation in aqueous solution. Biochemistry. 1985;24:4476–4481. doi: 10.1021/bi00337a032. [DOI] [PubMed] [Google Scholar]
- 10.Grollman A.P., Moriya M. Mutagenesis by 8-oxoguanine: an enemy within. Trends Genet. 1993;9:246–249. doi: 10.1016/0168-9525(93)90089-z. [DOI] [PubMed] [Google Scholar]
- 11.Akiyama M., Maki H., Sekiguchi M., Horiuchi T. A specific role of MutT protein: to prevent dG.dA mispairing in DNA replication. Proc. Natl. Acad. Sci. USA. 1989;86:3949–3952. doi: 10.1073/pnas.86.11.3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shibutani S., Takeshita M., Grollman A.P. Insertion of specific bases during DNA synthesis past the oxidation-damaged base 8-oxodG. Nature. 1991;349:431–434. doi: 10.1038/349431a0. [DOI] [PubMed] [Google Scholar]
- 13.Krokan H.E., Bjoras M. Base excision repair. Cold Spring Harb. Perspect. Biol. 2013;5:a012583. doi: 10.1101/cshperspect.a012583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.David S.S., O'Shea V.L., Kundu S. Base-excision repair of oxidative DNA damage. Nature. 2007;447:941–950. doi: 10.1038/nature05978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dizdaroglu M., Kirkali G., Jaruga P. Formamidopyrimidines in DNA: mechanisms of formation, repair, and biological effects. Free Radic. Biol. Med. 2008;45:1610–1621. doi: 10.1016/j.freeradbiomed.2008.07.004. [DOI] [PubMed] [Google Scholar]
- 16.Michaels M.L., Pham L., Cruz C., Miller J.H. MutM, a protein that prevents G.C----T.A transversions, is formamidopyrimidine-DNA glycosylase. Nucleic Acids Res. 1991;19:3629–3632. doi: 10.1093/nar/19.13.3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hazra T.K., Das A., Das S., Choudhury S., Kow Y.W., Roy R. Oxidative DNA damage repair in mammalian cells: a new perspective. DNA Repair. 2007;6:470–480. doi: 10.1016/j.dnarep.2006.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hart R.W., Setlow R.B. Correlation between deoxyribonucleic acid excision-repair and life-span in a number of mammalian species. Proc. Natl. Acad. Sci. USA. 1974;71:2169–2173. doi: 10.1073/pnas.71.6.2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ames B.N., Shigenaga M.K., Hagen T.M. Oxidants, antioxidants, and the degenerative diseases of aging. Proc. Natl. Acad. Sci. USA. 1993;90:7915–7922. doi: 10.1073/pnas.90.17.7915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ba X., Aguilera-Aguirre L., Sur S., Boldogh I. 8-Oxoguanine DNA glycosylase-1-driven DNA base excision repair: role in asthma pathogenesis. Curr. Opin. Allergy Clin. Immunol. 2015;15:89–97. doi: 10.1097/ACI.0000000000000135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aguilera-Aguirre L., Bacsi A., Radak Z., Hazra T.K., Mitra S., Sur S., Brasier A.R., Ba X., Boldogh I. Innate inflammation induced by the 8-oxoguanine DNA glycosylase-1-KRAS-NF-kappaB pathway. J. Immunol. 2014;193:4643–4653. doi: 10.4049/jimmunol.1401625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Seifermann M., Epe B. Oxidatively generated base modifications in DNA: not only carcinogenic risk factor but also regulatory mark? Free Radic. Biol. Med. 2016 doi: 10.1016/j.freeradbiomed.2016.11.018. [DOI] [PubMed] [Google Scholar]
- 23.Izumi T., Wiederhold L.R., Roy G., Roy R., Jaiswal A., Bhakat K.K., Mitra S., Hazra T.K. Mammalian DNA base excision repair proteins: their interactions and role in repair of oxidative DNA damage. Toxicology. 2003;193:43–65. doi: 10.1016/s0300-483x(03)00289-0. [DOI] [PubMed] [Google Scholar]
- 24.Bruner S.D., Norman D.P., Verdine G.L. Structural basis for recognition and repair of the endogenous mutagen 8-oxoguanine in DNA. Nature. 2000;403:859–866. doi: 10.1038/35002510. [DOI] [PubMed] [Google Scholar]
- 25.Hill J.W., Hazra T.K., Izumi T., Mitra S. Stimulation of human 8-oxoguanine-DNA glycosylase by AP-endonuclease: potential coordination of the initial steps in base excision repair. Nucleic Acids Res. 2001;29:430–438. doi: 10.1093/nar/29.2.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saitoh T., Shinmura K., Yamaguchi S., Tani M., Seki S., Murakami H., Nojima Y., Yokota J. Enhancement of OGG1 protein AP lyase activity by increase of APEX protein. Mutat. Res. 2001;486:31–40. doi: 10.1016/s0921-8777(01)00078-7. [DOI] [PubMed] [Google Scholar]
- 27.Dalhus B., Forsbring M., Helle I.H., Vik E.S., Forstrom R.J., Backe P.H., Alseth I., Bjoras M. Separation-of-function mutants unravel the dual-reaction mode of human 8-oxoguanine DNA glycosylase. Structure. 2011;19:117–127. doi: 10.1016/j.str.2010.09.023. [DOI] [PubMed] [Google Scholar]
- 28.Allgayer J., Kitsera N., Bartelt S., Epe B., Khobta A. Widespread transcriptional gene inactivation initiated by a repair intermediate of 8-oxoguanine. Nucleic Acids Res. 2016;44:7267–7280. doi: 10.1093/nar/gkw473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marsin S., Vidal A.E., Sossou M., Menissier-de Murcia J., Page F. Le, Boiteux S., de Murcia G., Radicella J.P. Role of XRCC1 in the coordination and stimulation of oxidative DNA damage repair initiated by the DNA glycosylase hOGG1. J. Biol. Chem. 2003;278:44068–44074. doi: 10.1074/jbc.M306160200. [DOI] [PubMed] [Google Scholar]
- 30.Campalans A., Moritz E., Kortulewski T., Biard D., Epe B., Radicella J.P. Interaction with OGG1 is required for efficient recruitment of XRCC1 to base excision repair and maintenance of genetic stability after exposure to oxidative stress. Mol. Cell Biol. 2015;35:1648–1658. doi: 10.1128/MCB.00134-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Noren Hooten N., Kompaniez K., Barnes J., Lohani A., Evans M.K. Poly(ADP-ribose) polymerase 1 (PARP-1) binds to 8-oxoguanine-DNA glycosylase (OGG1) J. Biol. Chem. 2011;286:44679–44690. doi: 10.1074/jbc.M111.255869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ramdzan Z.M., Vadnais C., Pal R., Vandal G., Cadieux C., Leduy L., Davoudi S., Hulea L., Yao L., Karnezis A.N., Paquet M., Dankort D., Nepveu A. RAS transformation requires CUX1-dependent repair of oxidative DNA damage. PLoS Biol. 2014;12:e1001807. doi: 10.1371/journal.pbio.1001807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pal R., Ramdzan Z.M., Kaur S., Duquette P.M., Marcotte R., Leduy L., Davoudi S., Lamarche-Vane N., Iulianella A., Nepveu A. CUX2 protein functions as an accessory factor in the repair of oxidative DNA damage. J. Biol. Chem. 2015;290:22520–22531. doi: 10.1074/jbc.M115.651042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kaur S., Coulombe Y., Ramdzan Z.M., Leduy L., Masson J.Y., Nepveu A. Special AT-rich sequence-binding Protein 1 (SATB1) functions as an accessory factor in base excision repair. J. Biol. Chem. 2016;291:22769–22780. doi: 10.1074/jbc.M116.735696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bhakat K.K., Mokkapati S.K., Boldogh I., Hazra T.K., Mitra S. Acetylation of human 8-oxoguanine-DNA glycosylase by p300 and its role in 8-oxoguanine repair in vivo. Mol. Cell Biol. 2006;26:1654–1665. doi: 10.1128/MCB.26.5.1654-1665.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Radak Z., Bori Z., Koltai E., Fatouros I.G., Jamurtas A.Z., Douroudos I.I., Terzis G., Nikolaidis M.G., Chatzinikolaou A., Sovatzidis A., Kumagai S., Naito H., Boldogh I. Age-dependent changes in 8-oxoguanine-DNA glycosylase activity are modulated by adaptive responses to physical exercise in human skeletal muscle. Free Radic. Biol. Med. 2011;51:417–423. doi: 10.1016/j.freeradbiomed.2011.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kang L., Zhao W., Zhang G., Wu J., Guan H. Acetylated 8-oxoguanine DNA glycosylase 1 and its relationship with p300 and SIRT1 in lens epithelium cells from age-related cataract. Exp. Eye Res. 2015;135:102–108. doi: 10.1016/j.exer.2015.02.005. [DOI] [PubMed] [Google Scholar]
- 38.Dantzer F., Luna L., Bjoras M., Seeberg E. Human OGG1 undergoes serine phosphorylation and associates with the nuclear matrix and mitotic chromatin in vivo. Nucleic Acids Res. 2002;30:2349–2357. doi: 10.1093/nar/30.11.2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hu J., Imam S.Z., Hashiguchi K., de Souza-Pinto N.C., Bohr V.A. Phosphorylation of human oxoguanine DNA glycosylase (alpha-OGG1) modulates its function. Nucleic Acids Res. 2005;33:3271–3282. doi: 10.1093/nar/gki636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cividini F., Scott B.T., Dai A., Han W., Suarez J., Diaz-Juarez J., Diemer T., Casteel D.E., Dillmann W.H. O-GlcNAcylation of 8-Oxoguanine DNA Glycosylase (Ogg1) Impairs Oxidative Mitochondrial DNA Lesion Repair in Diabetic Hearts. J. Biol. Chem. 2016;291:26515–26528. doi: 10.1074/jbc.M116.754481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bravard A., Vacher M., Gouget B., Coutant A., de Boisferon F.H., Marsin S., Chevillard S., Radicella J.P. Redox regulation of human OGG1 activity in response to cellular oxidative stress. Mol. Cell Biol. 2006;26:7430–7436. doi: 10.1128/MCB.00624-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bravard A., Campalans A., Vacher M., Gouget B., Levalois C., Chevillard S., Radicella J.P. Inactivation by oxidation and recruitment into stress granules of hOGG1 but not APE1 in human cells exposed to sub-lethal concentrations of cadmium. Mutat. Res. 2010;685:61–69. doi: 10.1016/j.mrfmmm.2009.09.013. [DOI] [PubMed] [Google Scholar]
- 43.Morreall J., Limpose K., Sheppard C., Kow Y.W., Werner E., Doetsch P.W. Inactivation of a common OGG1 variant by TNF-alpha in mammalian cells. DNA Repair. 2015;26:15–22. doi: 10.1016/j.dnarep.2014.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jamaluddin M., Wang S., Boldogh I., Tian B., Brasier A.R. TNF-alpha-induced NF-kappaB/RelA Ser(276) phosphorylation and enhanceosome formation is mediated by an ROS-dependent PKAc pathway. Cell Signal. 2007;19:1419–1433. doi: 10.1016/j.cellsig.2007.01.020. [DOI] [PubMed] [Google Scholar]
- 45.Pan L., Zhu B., Hao W., Zeng X., Vlahopoulos S.A., Hazra T.K., Hegde M.L., Radak Z., Bacsi A., Brasier A.R., Ba X., Boldogh I. Oxidized guanine base lesions function in 8-oxoguanine DNA glycosylase-1-mediated epigenetic regulation of nuclear factor kappaB-driven gene expression. J. Biol. Chem. 2016;291:25553–25566. doi: 10.1074/jbc.M116.751453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ba X., Bacsi A., Luo J., Aguilera-Aguirre L., Zeng X., Radak Z., Brasier A.R., Boldogh I. 8-oxoguanine DNA glycosylase-1 augments proinflammatory gene expression by facilitating the recruitment of site-specific transcription factors. J. Immunol. 2014;192:2384–2394. doi: 10.4049/jimmunol.1302472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Klungland A., Rosewell I., Hollenbach S., Larsen E., Daly G., Epe B., Seeberg E., Lindahl T., Barnes D.E. Accumulation of premutagenic DNA lesions in mice defective in removal of oxidative base damage. Proc. Natl. Acad. Sci. USA. 1999;96:13300–13305. doi: 10.1073/pnas.96.23.13300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Minowa O., Arai T., Hirano M., Monden Y., Nakai S., Fukuda M., Itoh M., Takano H., Hippou Y., Aburatani H., Masumura K., Nohmi T., Nishimura S., Noda T. Mmh/Ogg1 gene inactivation results in accumulation of 8-hydroxyguanine in mice. Proc. Natl. Acad. Sci. USA. 2000;97:4156–4161. doi: 10.1073/pnas.050404497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Arai T., Kelly V.P., Minowa O., Noda T., Nishimura S. High accumulation of oxidative DNA damage, 8-hydroxyguanine, in Mmh/Ogg1 deficient mice by chronic oxidative stress. Carcinogenesis. 2002;23:2005–2010. doi: 10.1093/carcin/23.12.2005. [DOI] [PubMed] [Google Scholar]
- 50.de Souza-Pinto N.C., Eide L., Hogue B.A., Thybo T., Stevnsner T., Seeberg E., Klungland A., Bohr V.A. Repair of 8-oxodeoxyguanosine lesions in mitochondrial dna depends on the oxoguanine dna glycosylase (OGG1) gene and 8-oxoguanine accumulates in the mitochondrial dna of OGG1-defective mice. Cancer Res. 2001;61:5378–5381. [PubMed] [Google Scholar]
- 51.Touati E., Michel V., Thiberge J.M., Ave P., Huerre M., Bourgade F., Klungland A., Labigne A. Deficiency in OGG1 protects against inflammation and mutagenic effects associated with H. pylori infection in mouse. Helicobacter. 2006;11:494–505. doi: 10.1111/j.1523-5378.2006.00442.x. [DOI] [PubMed] [Google Scholar]
- 52.Mabley J.G., Pacher P., Deb A., Wallace R., Elder R.H., Szabo C. Potential role for 8-oxoguanine DNA glycosylase in regulating inflammation. FASEB J. 2005;19:290–292. doi: 10.1096/fj.04-2278fje. [DOI] [PubMed] [Google Scholar]
- 53.Li G., Yuan K., Yan C., Fox J., 3rd, Gaid M., Breitwieser W., Bansal A.K., Zeng H., Gao H., Wu M. 8-Oxoguanine-DNA glycosylase 1 deficiency modifies allergic airway inflammation by regulating STAT6 and IL-4 in cells and in mice. Free Radic. Biol. Med. 2012;52:392–401. doi: 10.1016/j.freeradbiomed.2011.10.490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bacsi A., Aguilera-Aguirre L., Szczesny B., Radak Z., Hazra T.K., Sur S., Ba X., Boldogh I. Down-regulation of 8-oxoguanine DNA glycosylase 1 expression in the airway epithelium ameliorates allergic lung inflammation. DNA Repair. 2013;12:18–26. doi: 10.1016/j.dnarep.2012.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Seifermann M., Ulges A., Bopp T., Melcea S., Schafer A., Oka S., Nakabeppu Y., Klungland A., Niehrs C., Epe B. Role of the DNA repair glycosylase OGG1 in the activation of murine splenocytes. DNA Repair. 2017;58:13–20. doi: 10.1016/j.dnarep.2017.08.005. [DOI] [PubMed] [Google Scholar]
- 56.Sampath H., Vartanian V., Rollins M.R., Sakumi K., Nakabeppu Y., Lloyd R.S. 8-Oxoguanine DNA glycosylase (OGG1) deficiency increases susceptibility to obesity and metabolic dysfunction. PLoS One. 2012;7:e51697. doi: 10.1371/journal.pone.0051697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Choudhary S., Boldogh I., Brasier A.R. Inside-Out Signaling Pathways from Nuclear Reactive Oxygen Species Control Pulmonary Innate Immunity. J. Innate Immun. 2016;8:143–155. doi: 10.1159/000442254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cachat J., Deffert C., Hugues S., Krause K.H. Phagocyte NADPH oxidase and specific immunity. Clin. Sci. 2015;128:635–648. doi: 10.1042/CS20140635. [DOI] [PubMed] [Google Scholar]
- 59.Kalyan S., Kabelitz D. When neutrophils meet T cells: beginnings of a tumultuous relationship with underappreciated potential. Eur. J. Immunol. 2014;44:627–633. doi: 10.1002/eji.201344195. [DOI] [PubMed] [Google Scholar]
- 60.Sen C.K., Packer L. Antioxidant and redox regulation of gene transcription. FASEB J. 1996;10:709–720. doi: 10.1096/fasebj.10.7.8635688. [DOI] [PubMed] [Google Scholar]
- 61.Xanthoudakis S., Miao G., Wang F., Pan Y.C., Curran T. Redox activation of Fos-Jun DNA binding activity is mediated by a DNA repair enzyme. EMBO J. 1992;11:3323–3335. doi: 10.1002/j.1460-2075.1992.tb05411.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nishi T., Shimizu N., Hiramoto M., Sato I., Yamaguchi Y., Hasegawa M., Aizawa S., Tanaka H., Kataoka K., Watanabe H., Handa H. Spatial redox regulation of a critical cysteine residue of NF-kappa B in vivo. J. Biol. Chem. 2002;277:44548–44556. doi: 10.1074/jbc.M202970200. [DOI] [PubMed] [Google Scholar]
- 63.Huang R.P., Adamson E.D. Characterization of the DNA-binding properties of the early growth response-1 (Egr-1) transcription factor: evidence for modulation by a redox mechanism. DNA Cell Biol. 1993;12:265–273. doi: 10.1089/dna.1993.12.265. [DOI] [PubMed] [Google Scholar]
- 64.Huang L.E., Arany Z., Livingston D.M., Bunn H.F. Activation of hypoxia-inducible transcription factor depends primarily upon redox-sensitive stabilization of its alpha subunit. J. Biol. Chem. 1996;271:32253–32259. doi: 10.1074/jbc.271.50.32253. [DOI] [PubMed] [Google Scholar]
- 65.Seo Y.R., Kelley M.R., Smith M.L. Selenomethionine regulation of p53 by a ref1-dependent redox mechanism. Proc. Natl. Acad. Sci. USA. 2002;99:14548–14553. doi: 10.1073/pnas.212319799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ghosh R., Mitchell D.L. Effect of oxidative DNA damage in promoter elements on transcription factor binding. Nucleic Acids Res. 1999;27:3213–3218. doi: 10.1093/nar/27.15.3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ramon O., Sauvaigo S., Gasparutto D., Faure P., Favier A., Cadet J. Effects of 8-oxo-7,8-dihydro-2′-deoxyguanosine on the binding of the transcription factor Sp1 to its cognate target DNA sequence (GC box) Free Radic. Res. 1999;31:217–229. doi: 10.1080/10715769900300781. [DOI] [PubMed] [Google Scholar]
- 68.Hailer-Morrison M.K., Kotler J.M., Martin B.D., Sugden K.D. Oxidized guanine lesions as modulators of gene transcription. Altered p50 binding affinity and repair shielding by 7,8-dihydro-8-oxo-2′-deoxyguanosine lesions in the NF-kappaB promoter element. Biochemistry. 2003;42:9761–9770. doi: 10.1021/bi034546k. [DOI] [PubMed] [Google Scholar]
- 69.Berkowitz B., Huang D.B., Chen-Park F.E., Sigler P.B., Ghosh G. The x-ray crystal structure of the NF-kappa Bp50. p65 heterodimer bound to the interferon beta -kappa B site. J. Biol. Chem. 2002;277:24694–24700. doi: 10.1074/jbc.M200006200. [DOI] [PubMed] [Google Scholar]
- 70.Chen F.E., Huang D.B., Chen Y.Q., Ghosh G. Crystal structure of p50/p65 heterodimer of transcription factor NF-kappaB bound to DNA. Nature. 1998;391:410–413. doi: 10.1038/34956. [DOI] [PubMed] [Google Scholar]
- 71.Ghosh G., van Duyne G., Ghosh S., Sigler P.B. Structure of NF-kappa B p50 homodimer bound to a kappa B site. Nature. 1995;373:303–310. doi: 10.1038/373303a0. [DOI] [PubMed] [Google Scholar]
- 72.Evans M.J., Scarpulla R.C. NRF-1: a trans-activator of nuclear-encoded respiratory genes in animal cells. Genes Dev. 1990;4:1023–1034. doi: 10.1101/gad.4.6.1023. [DOI] [PubMed] [Google Scholar]
- 73.Scarpulla R.C. Transcriptional paradigms in mammalian mitochondrial biogenesis and function. Physiol. Rev. 2008;88:611–638. doi: 10.1152/physrev.00025.2007. [DOI] [PubMed] [Google Scholar]
- 74.Sukhatme V.P. The Egr transcription factor family: from signal transduction to kidney differentiation. Kidney Int. 1992;41:550–553. doi: 10.1038/ki.1992.79. [DOI] [PubMed] [Google Scholar]
- 75.Christy B., Nathans D. DNA binding site of the growth factor-inducible protein Zif268. Proc. Natl. Acad. Sci. USA. 1989;86:8737–8741. doi: 10.1073/pnas.86.22.8737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Dunn A.R., Kad N.M., Nelson S.R., Warshaw D.M., Wallace S.S. Single Qdot-labeled glycosylase molecules use a wedge amino acid to probe for lesions while scanning along DNA. Nucleic Acids Res. 2011;39:7487–7498. doi: 10.1093/nar/gkr459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Banerjee A., Santos W.L., Verdine G.L. Structure of a DNA glycosylase searching for lesions. Science. 2006;311:1153–1157. doi: 10.1126/science.1120288. [DOI] [PubMed] [Google Scholar]
- 78.Nelson S.R., Dunn A.R., Kathe S.D., Warshaw D.M., Wallace S.S. Two glycosylase families diffusively scan DNA using a wedge residue to probe for and identify oxidatively damaged bases. Proc. Natl. Acad. Sci. USA. 2014;111:E2091–E2099. doi: 10.1073/pnas.1400386111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Banerjee A., Yang W., Karplus M., Verdine G.L. Structure of a repair enzyme interrogating undamaged DNA elucidates recognition of damaged DNA. Nature. 2005;434:612–618. doi: 10.1038/nature03458. [DOI] [PubMed] [Google Scholar]
- 80.Moore S.P., Kruchten J., Toomire K.J., Strauss P.R. Transcription factors and DNA repair enzymes compete for damaged promoter sites. J. Biol. Chem. 2016;291:5452–5460. doi: 10.1074/jbc.M115.672733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zharkov D.O., Rosenquist T.A., Gerchman S.E., Grollman A.P. Substrate specificity and reaction mechanism of murine 8-oxoguanine-DNA glycosylase. J. Biol. Chem. 2000;275:28607–28617. doi: 10.1074/jbc.M002441200. [DOI] [PubMed] [Google Scholar]
- 82.Stivers J.T., Site-specific DNA damage recognition by enzyme-induced base flipping. Prog. Nucleic Acid Res. Mol. Biol. 2004;77:37–65. doi: 10.1016/S0079-6603(04)77002-6. [DOI] [PubMed] [Google Scholar]
- 83.Roberts R.J., Cheng X. Base flipping. Annu. Rev. Biochem. 1998;67:181–198. doi: 10.1146/annurev.biochem.67.1.181. [DOI] [PubMed] [Google Scholar]
- 84.Pan L., Hao W., Zheng X., Zeng X., Ahmed Abbasi A., Boldogh I., Ba X. OGG1-DNA interactions facilitate NF-kappaB binding to DNA targets. Sci. Rep. 2017;7:43297. doi: 10.1038/srep43297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kitsera N., Stathis D., Luhnsdorf B., Muller H., Carell T., Epe B., Khobta A. 8-Oxo-7,8-dihydroguanine in DNA does not constitute a barrier to transcription, but is converted into transcription-blocking damage by OGG1. Nucleic Acids Res. 2011;39:5926–5934. doi: 10.1093/nar/gkr163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kathe S.D., Shen G.P., Wallace S.S. Single-stranded breaks in DNA but not oxidative DNA base damages block transcriptional elongation by RNA polymerase II in HeLa cell nuclear extracts. J. Biol. Chem. 2004;279:18511–18520. doi: 10.1074/jbc.M313598200. [DOI] [PubMed] [Google Scholar]
- 87.Yang J., Mitra A., Dojer N., Fu S., Rowicka M., Brasier A.R. A probabilistic approach to learn chromatin architecture and accurate inference of the NF-kappaB/RelA regulatory network using ChIP-Seq. Nucleic Acids Res. 2013;41:7240–7259. doi: 10.1093/nar/gkt493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Karin M., Greten F.R. NF-kappaB: linking inflammation and immunity to cancer development and progression. Nat. Rev. Immunol. 2005;5:749–759. doi: 10.1038/nri1703. [DOI] [PubMed] [Google Scholar]
- 89.Siebenlist U., Franzoso G., Brown K. Structure, regulation and function of NF-kappa B. Annu. Rev. Cell Biol. 1994;10:405–455. doi: 10.1146/annurev.cb.10.110194.002201. [DOI] [PubMed] [Google Scholar]
- 90.Wang T., Li J., Ding K., Zhang L., Che Q., Sun X., Dai Y., Sun W., Bao M., Wang X., Yang L., Li Z. The CpG dinucleotide adjacent to a kappaB site affects NF-kappaB function through its methylation. Int. J. Mol. Sci. 2017;18 doi: 10.3390/ijms18030528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Travers A.A. Why bend DNA? Cell. 1990;60:177–180. doi: 10.1016/0092-8674(90)90729-x. [DOI] [PubMed] [Google Scholar]
- 92.Crispin J.C., Tsokos G.C. Transcriptional regulation of IL-2 in health and autoimmunity. Autoimmun. Rev. 2009;8:190–195. doi: 10.1016/j.autrev.2008.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Trede N.S., Tsytsykova A.V., Chatila T., Goldfeld A.E., Geha R.S. Transcriptional activation of the human TNF-alpha promoter by superantigen in human monocytic cells: role of NF-kappa B. J. Immunol. 1995;155:902–908. [PubMed] [Google Scholar]
- 94.Sowa Y., Orita T., Hiranabe-Minamikawa S., Nakano K., Mizuno T., Nomura H., Sakai T. Histone deacetylase inhibitor activates the p21/WAF1/Cip1 gene promoter through the Sp1 sites. Ann. N.Y. Acad. Sci. 1999;886:195–199. doi: 10.1111/j.1749-6632.1999.tb09415.x. [DOI] [PubMed] [Google Scholar]
- 95.Kuprash D.V., Udalova I.A., Turetskaya R.L., Kwiatkowski D., Rice N.R., Nedospasov S.A. Similarities and differences between human and murine TNF promoters in their response to lipopolysaccharide. J. Immunol. 1999;162:4045–4052. [PubMed] [Google Scholar]
- 96.D'Onofrio G., Jabbari K., Musto H., Alvarez-Valin F., Cruveiller S., Bernardi G. Evolutionary genomics of vertebrates and its implications. Ann. N.Y. Acad. Sci. 1999;870:81–94. doi: 10.1111/j.1749-6632.1999.tb08867.x. [DOI] [PubMed] [Google Scholar]
- 97.Gautier C. Compositional bias in DNA. Curr. Opin. Genet. Dev. 2000;10:656–661. doi: 10.1016/s0959-437x(00)00144-1. [DOI] [PubMed] [Google Scholar]
- 98.Saxonov S., Berg P., Brutlag D.L. A genome-wide analysis of CpG dinucleotides in the human genome distinguishes two distinct classes of promoters. Proc. Natl. Acad. Sci. USA. 2006;103:1412–1417. doi: 10.1073/pnas.0510310103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Fleming A.M., Burrows C.J. 8-Oxo-7,8-dihydroguanine, friend and foe: Epigenetic-like regulator versus initiator of mutagenesis. DNA Repair. 2017 doi: 10.1016/j.dnarep.2017.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Hall D.B., Holmlin R.E., Barton J.K. Oxidative DNA damage through long-range electron transfer. Nature. 1996;382:731–735. doi: 10.1038/382731a0. [DOI] [PubMed] [Google Scholar]
- 101.Giese B., Amaudrut J., Kohler A.K., Spormann M., Wessely S. Direct observation of hole transfer through DNA by hopping between adenine bases and by tunnelling. Nature. 2001;412:318–320. doi: 10.1038/35085542. [DOI] [PubMed] [Google Scholar]
- 102.Nunez M.E., Hall D.B., Barton J.K. Long-range oxidative damage to DNA: effects of distance and sequence. Chem. Biol. 1999;6:85–97. doi: 10.1016/S1074-5521(99)80005-2. [DOI] [PubMed] [Google Scholar]
- 103.Saito I., Takayama M., Nakamura T., Sugiyama H., Komeda Y., Iwasaki M. The most electron-donating sites in duplex DNA: guanine-guanine stacking rule. Nucleic Acids Symp. Ser. 1995:191–192. [PubMed] [Google Scholar]
- 104.Genereux J.C., Boal A.K., Barton J.K. DNA-mediated charge transport in redox sensing and signaling. J. Am. Chem. Soc. 2010;132:891–905. doi: 10.1021/ja907669c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Merino E.J., Boal A.K., Barton J.K. Biological contexts for DNA charge transport chemistry. Curr. Opin. Chem. Biol. 2008;12:229–237. doi: 10.1016/j.cbpa.2008.01.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Giese B. Long-distance electron transfer through DNA. Annu Rev. Biochem. 2002;71:51–70. doi: 10.1146/annurev.biochem.71.083101.134037. [DOI] [PubMed] [Google Scholar]
- 107.Giese B. Electron transfer in DNA. Curr. Opin. Chem. Biol. 2002;6:612–618. doi: 10.1016/s1367-5931(02)00364-2. [DOI] [PubMed] [Google Scholar]
- 108.Ding Y., Fleming A.M., Burrows C.J. Sequencing the mouse genome for the oxidatively modified base 8-Oxo-7,8-dihydroguanine by OG-seq. J. Am. Chem. Soc. 2017;139:2569–2572. doi: 10.1021/jacs.6b12604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Pastukh V., Roberts J.T., Clark D.W., Bardwell G.C., Patel M., Al-Mehdi A.B., Borchert G.M., Gillespie M.N. An oxidative DNA "damage" and repair mechanism localized in the VEGF promoter is important for hypoxia-induced VEGF mRNA expression. Am. J. Physiol. Lung Cell Mol. Physiol. 2015;309:L1367–L1375. doi: 10.1152/ajplung.00236.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Yoshihara M., Jiang L., Akatsuka S., Suyama M., Toyokuni S. Genome-wide profiling of 8-oxoguanine reveals its association with spatial positioning in nucleus. DNA Res. 2014;21:603–612. doi: 10.1093/dnares/dsu023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ohno M., Miura T., Furuichi M., Tominaga Y., Tsuchimoto D., Sakumi K., Nakabeppu Y. A genome-wide distribution of 8-oxoguanine correlates with the preferred regions for recombination and single nucleotide polymorphism in the human genome. Genome Res. 2006;16:567–575. doi: 10.1101/gr.4769606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Fleming A.M., Burrows C.J. Formation and processing of DNA damage substrates for the hNEIL enzymes. Free Radic. Biol. Med. 2016 doi: 10.1016/j.freeradbiomed.2016.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Hosford M.E., Muller J.G., Burrows C.J. Spermine participates in oxidative damage of guanosine and 8-oxoguanosine leading to deoxyribosylurea formation. J. Am. Chem. Soc. 2004;126:9540–9541. doi: 10.1021/ja047981q. [DOI] [PubMed] [Google Scholar]
- 114.Cremers C.M., Jakob U. Oxidant sensing by reversible disulfide bond formation. J. Biol. Chem. 2013;288:26489–26496. doi: 10.1074/jbc.R113.462929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Koval V.V., Kuznetsov N.A., Zharkov D.O., Ishchenko A.A., Douglas K.T., Nevinsky G.A., Fedorova O.S. Pre-steady-state kinetics shows differences in processing of various DNA lesions by Escherichia coli formamidopyrimidine-DNA glycosylase. Nucleic Acids Res. 2004;32:926–935. doi: 10.1093/nar/gkh237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Fedorova O.S., Nevinsky G.A., Koval V.V., Ishchenko A.A., Vasilenko N.L., Douglas K.T. Stopped-flow kinetic studies of the interaction between Escherichia coli Fpg protein and DNA substrates. Biochemistry. 2002;41:1520–1528. doi: 10.1021/bi011524u. [DOI] [PubMed] [Google Scholar]
- 117.Kim H.S., Kim B.H., Jung J.E., Lee C.S., Lee H.G., Lee J.W., Lee K.H., You H.J., Chung M.H., Ye S.K. Potential role of 8-oxoguanine DNA glycosylase 1 as a STAT1 coactivator in endotoxin-induced inflammatory response. Free Radic. Biol. Med. 2016;93:12–22. doi: 10.1016/j.freeradbiomed.2015.10.415. [DOI] [PubMed] [Google Scholar]
- 118.Nelson D.E., Ihekwaba A.E., Elliott M., Johnson J.R., Gibney C.A., Foreman B.E., Nelson G., See V., Horton C.A., Spiller D.G., Edwards S.W., McDowell H.P., Unitt J.F., Sullivan E., Grimley R., Benson N., Broomhead D., Kell D.B., White M.R. Oscillations in NF-kappaB signaling control the dynamics of gene expression. Science. 2004;306:704–708. doi: 10.1126/science.1099962. [DOI] [PubMed] [Google Scholar]
- 119.Perillo B., Ombra M.N., Bertoni A., Cuozzo C., Sacchetti S., Sasso A., Chiariotti L., Malorni A., Abbondanza C., Avvedimento E.V. DNA oxidation as triggered by H3K9me2 demethylation drives estrogen-induced gene expression. Science. 2008;319:202–206. doi: 10.1126/science.1147674. [DOI] [PubMed] [Google Scholar]
- 120.Amente S., Bertoni A., Morano A., Lania L., Avvedimento E.V., Majello B. LSD1-mediated demethylation of histone H3 lysine 4 triggers Myc-induced transcription. Oncogene. 2010;29:3691–3702. doi: 10.1038/onc.2010.120. [DOI] [PubMed] [Google Scholar]
- 121.Fleming A.M., Ding Y., Burrows C.J. Oxidative DNA damage is epigenetic by regulating gene transcription via base excision repair. Proc. Natl. Acad. Sci. USA. 2017;114:2604–2609. doi: 10.1073/pnas.1619809114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Zhou J., Liu M., Fleming A.M., Burrows C.J., Wallace S.S. Neil3 and NEIL1 DNA glycosylases remove oxidative damages from quadruplex DNA and exhibit preferences for lesions in the telomeric sequence context. J. Biol. Chem. 2013;288:27263–27272. doi: 10.1074/jbc.M113.479055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Zhou J., Fleming A.M., Averill A.M., Burrows C.J., Wallace S.S. The NEIL glycosylases remove oxidized guanine lesions from telomeric and promoter quadruplex DNA structures. Nucleic Acids Res. 2015;43:4039–4054. doi: 10.1093/nar/gkv252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Rhodes D., Lipps H.J. G-quadruplexes and their regulatory roles in biology. Nucleic Acids Res. 2015;43:8627–8637. doi: 10.1093/nar/gkv862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Amouroux R., Campalans A., Epe B., Radicella J.P. Oxidative stress triggers the preferential assembly of base excision repair complexes on open chromatin regions. Nucleic Acids Res. 2010;38:2878–2890. doi: 10.1093/nar/gkp1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Kawai K., Wata Y., Hara M., Tojo S., Majima T. Regulation of one-electron oxidation rate of guanine by base pairing with cytosine derivatives. J. Am. Chem. Soc. 2002;124:3586–3590. doi: 10.1021/ja016530s. [DOI] [PubMed] [Google Scholar]
- 127.Ming X., Matter B., Song M., Veliath E., Shanley R., Jones R., Tretyakova N. Mapping structurally defined guanine oxidation products along DNA duplexes: influence of local sequence context and endogenous cytosine methylation. J. Am. Chem. Soc. 2014;136:4223–4235. doi: 10.1021/ja411636j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Klose R.J., Bird A.P. Genomic DNA methylation: the mark and its mediators. Trends Biochem. Sci. 2006;31:89–97. doi: 10.1016/j.tibs.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 129.Tate P.H., Bird A.P. Effects of DNA methylation on DNA-binding proteins and gene expression. Curr. Opin. Genet. Dev. 1993;3:226–231. doi: 10.1016/0959-437x(93)90027-m. [DOI] [PubMed] [Google Scholar]
- 130.Hendrich B., Bird A. Identification and characterization of a family of mammalian methyl-CpG binding proteins. Mol. Cell Biol. 1998;18:6538–6547. doi: 10.1128/mcb.18.11.6538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Boyes J., Bird A. DNA methylation inhibits transcription indirectly via a methyl-CpG binding protein. Cell. 1991;64:1123–1134. doi: 10.1016/0092-8674(91)90267-3. [DOI] [PubMed] [Google Scholar]
- 132.Jones P.L., Veenstra G.J., Wade P.A., Vermaak D., Kass S.U., Landsberger N., Strouboulis J., Wolffe A.P. Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nat. Genet. 1998;19:187–191. doi: 10.1038/561. [DOI] [PubMed] [Google Scholar]
- 133.Nan X., Ng H.H., Johnson C.A., Laherty C.D., Turner B.M., Eisenman R.N., Bird A. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature. 1998;393:386–389. doi: 10.1038/30764. [DOI] [PubMed] [Google Scholar]
- 134.Zhang Y., Ng H.H., Erdjument-Bromage H., Tempst P., Bird A., Reinberg D. Analysis of the NuRD subunits reveals a histone deacetylase core complex and a connection with DNA methylation. Genes Dev. 1999;13:1924–1935. doi: 10.1101/gad.13.15.1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Bird A.P. Functions for DNA methylation in vertebrates. Cold Spring Harb. Symp. Quant. Biol. 1993;58:281–285. doi: 10.1101/sqb.1993.058.01.033. [DOI] [PubMed] [Google Scholar]
- 136.Lao V.V., Darwanto A., Sowers L.C. Impact of base analogues within a CpG dinucleotide on the binding of DNA by the methyl-binding domain of MeCP2 and methylation by DNMT1. Biochemistry. 2010;49:10228–10236. doi: 10.1021/bi1011942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Valinluck V., Tsai H.H., Rogstad D.K., Burdzy A., Bird A., Sowers L.C. Oxidative damage to methyl-CpG sequences inhibits the binding of the methyl-CpG binding domain (MBD) of methyl-CpG binding protein 2 (MeCP2) Nucleic Acids Res. 2004;32:4100–4108. doi: 10.1093/nar/gkh739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Hackett J.A., Sengupta R., Zylicz J.J., Murakami K., Lee C., Down T.A., Surani M.A., Germline DNA demethylation dynamics and imprint erasure through 5-hydroxymethylcytosine. Science. 2013;339:448–452. doi: 10.1126/science.1229277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Lim E., Rothenberg M.E. Demethylation of the human eotaxin-3 gene promoter leads to the elevated expression of eotaxin-3. J. Immunol. 2014;192:466–474. doi: 10.4049/jimmunol.1302454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Zhang F.F., Santella R.M., Wolff M., Kappil M.A., Markowitz S.B., Morabia A. White blood cell global methylation and IL-6 promoter methylation in association with diet and lifestyle risk factors in a cancer-free population. Epigenetics. 2012;7:606–614. doi: 10.4161/epi.20236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Ooi S.K., Bestor T.H. The colorful history of active DNA demethylation. Cell. 2008;133:1145–1148. doi: 10.1016/j.cell.2008.06.009. [DOI] [PubMed] [Google Scholar]
- 142.Wu S.C., Zhang Y. Active DNA demethylation: many roads lead to Rome. Nat. Rev. Mol. Cell Biol. 2010;11:607–620. doi: 10.1038/nrm2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Xu G.L., Walsh C.P. Enzymatic DNA oxidation: mechanisms and biological significance. BMB Rep. 2014;47:609–618. doi: 10.5483/BMBRep.2014.47.11.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Zhou X., Zhuang Z., Wang W., He L., Wu H., Cao Y., Pan F., Zhao J., Hu Z., Sekhar C., Guo Z. OGG1 is essential in oxidative stress induced DNA demethylation. Cell Signal. 2016;28:1163–1171. doi: 10.1016/j.cellsig.2016.05.021. [DOI] [PubMed] [Google Scholar]