Abstract
The fungus Candida albicans colonizes oral cavity surfaces and is carried by up to 60% of human populations. Biofilm development by C. albicans may be modulated by oral streptococci, such as Streptococcus gordonii, S. oralis or S. mutans, so as to augment pathogenicity. In this study we sought to determine if the cell wall-associated secreted aspartyl proteinase Sap9 was necessary for hyphal adhesin functions associated with biofilm community development. A sap9Δ mutant of C. albicans SC5314 formed biofilms that were flatter, and contained fewer blastospores and more hyphal filaments than the parent strain. This phenotypic difference was accentuated under flow (shear) conditions and in the presence of S. gordonii. Dual-species biofilms of C. albicans sap9Δ with S. oralis, S. sanguinis, S. parasanguinis, S. mutans and Enterococcus faecalis all contained more matted hyphae and more bacteria bound to substratum compared to C. albicans wild type. sap9Δ mutant hyphae showed significantly increased cell surface hydrophobicity, ∼25% increased levels of binding C. albicans cell wall protein Als3, and reduced interaction with Eap1, implicating Sap9 in fungal cell–cell recognition. These observations suggest that Sap9 is associated with protein–receptor interactions between fungal cells, and with interkingdom communication in the formation of polymicrobial biofilm communities.
Keywords: Microbial interactions, mixed species communities, Streptococcus gordonii, Streptococcus mutans, oral cavity biofilms
This study shows that the fungus Candida albicans cell wall-associated Sap9 proteinase is required for control of hyphal filament formation and separation, and for regulation of mixed-species biofilm formation by C. albicans with oral Streptococcus bacteria.
INTRODUCTION
Candida albicans is an opportuntistic fungal parthogen that is found in the gut, oral cavity and genital tract microbiota in healthy individuals. Lowering of host immunity, immune dysfunction, corticosteroid use or administration of broad spectrum antibiotics can be sufficient for C. albicans transition to pathogen. This may lead to superficial infections, such as oropharyngeal candidiasis (thrush) and vulvovaginal candidiasis, or to life-threatening systemic infections. Candida albicans biofilm infections are common in subjects with prosthetic devices e.g. urinary or intravascular catheters, artificial joints or voice boxes (Ramage, Martinez and López-Ribot 2006).
Candida albicans biofilm formation on a range of surfaces in vitro and in vivo (Paulitsch et al.2009; Busscher et al.2010; Harriott et al.2010) occurs in several phases, beginning with the deposition of yeast-form cells onto a substratum and subsequent formation of hyphae (Nobile et al.2012). Hyphal filament production appears to be essential for biofilm formation (Banerjee et al.2013), with the transcriptional regulator protein Bcr1 (Dwivedi et al.2011; Fanning et al.2012) controlling expression of major cell wall-associated adhesins (Nobile et al.2006) involved in biofilm accretion (Nobile et al.2008). The C. albicans outer cell wall layer is comprised principally of mannoproteins that are embedded in a strong, flexible polysaccharide skeleton provided by β-(1,3)- and β-(1,6)-linked glucan chains and covalently-linked chitin (Gow et al.2011). The protein and carbohydrate components of the outer wall layers have both been implicated in adhesion to host surfaces (Herrero et al.2002) and in biofilm formation (Peltroche-Llacsahuanga et al.2006).
Glycosylphosphatidylinositol (GPI)-modified proteins, including adhesins such as Hwp1, Rbt1, Eap1 and the Als family of proteins (Hoyer et al.2008), have established roles in biofilm formation (Nobile et al.2006; Nobbs, Vickerman and Jenkinson 2010; Garcia et al.2011) and in interactions with host tissues and other biological cells (Finkel et al.2012). Glycosylation of these and other cell wall proteins (CWPs) is thought to be important for their functions (Richard et al.2002). N-linked glycans, attached to asparagine residues, carry a conserved core structure and highly branched outer mannose chains (Cutler 2001), while O-linked glycans are linear oligosaccharides of one to five α-1,2-linked mannose residues attached to serine or threonine residues (Buurman et al.1998). N- and O-linked glycosylations are mediated by a set of multifunctional mannosyltransferases (Mora-Montes et al.2010). Mnt1 and Mnt2 proteins are partially redundant α-1,2-mannosyltransferases that catalyse the addition of the second and third mannose residues in an O-linked mannose pentamer (Munro et al.2005). Candida albicans mutants deficient in production of Mnt1 and Mnt2 proteins are modified in cell wall structure (Munro et al.2005; Castillo et al.2011), reduced in adherence levels to human buccal epithelial cells (Buurman et al.1998), attenuated in virulence (Castillo et al.2011), deficient in binding oral Streptococcus gordonii (Dutton et al.2014) and abrogated in biofilm formation under a range of conditions (Dutton et al.2014).
Candida albicans expresses a family of 10 secreted aspartyl proteinases encoded by the SAP genes (Aoki et al.2011). Saps 1–8 are extracellular enzymes while Sap9 and Sap10 are GPI-modified and associated with the fungal cell wall (Monod et al.1998; Schild et al.2011). Candida albicans biofilms secrete more Saps than do planktonic cells, and SAP5 and SAP9 genes are upregulated in biofilms (Joo et al.2013) and in subjects with oral and vaginal candidosis (Naglik et al.2008). SAP9 and SAP10 are also upregulated in biofilms (Nailis et al.2010) and encode proteins that maintain cell surface integrity by processing CWPs such as Ywp1, Hwp1 and Rbt1, which are involved in biofilm formation (Schild et al.2011). Sap9 contributes to the process of adhesion to denture resin (Copping et al.2005; Albrecht et al.2006) and can inactivate human antimicrobial peptide cathelicidin LL-37 (Rapala-Kozik et al.2015). Sap9 may also regulate the activities of other Saps (Kitanovic et al.2005), and YPS1 in Saccharomyces cerevisiae (homologue of SAP9) is upregulated in response to cell wall stress (Krysan et al.2005) and to caspofungin (Copping et al.2005). SAP4-6 are predominantly expressed on hyphae (Naglik, Challacombe and Hube 2003) and bind integrins (Kumar et al.2015), while fungal cell surface Saps e.g. Sap6 (Kumar et al.2015) may themselves serve as ligands that are able to bind other cell surfaces independently of their proteolytic activities (Naglik, Challacombe and Hube 2003).
Many infections, especially those at mucosal membranes, involve two or more microbial species (Harriott and Noverr 2011). Candida albicans has been found in close association with oral streptococci in denture- and mucosal-related diseases (Campos et al.2008). The oral streptococci are primary colonizers of oral cavity surfaces and provide foundation biofilms to which oral microorganisms such as C. albicans attach, thus facilitating polymicrobial community development and persistence (Wright et al.2013). Streptococcus gordonii and S. oralis form dual species biofilms with C. albicans on salivary glycoprotein-coated surfaces (Bamford et al.2009) or on epithelium (Diaz et al.2012). Several species of oral streptococci promote growth, hyphae formation (Xu et al.2014a; Dutton et al.2015) and virulence of C. albicans (Xu et al.2014b; Bertolini et al.2015), while the fungus may augment streptococcal pathogenicity (Falsetta et al.2014; Xu et al.2014b).
In a recent study, we showed that O-mannosylation of CWPs was necessary for biofilm formation under a range of conditions, and for the development of biofilms with S. gordonii (Dutton et al.2014). One feature of the cell wall proteome from hyphae-forming cells of the mannosylation-deficient mnt1Δmnt2Δ mutant was that the aspartyl proteinase Sap9 was absent. We therefore investigated the possibility that aberrant biofilm formation by the C. albicans mnt1Δmnt2Δ mutant was the result of, at least in part, absence of Sap9. Utilizing a Δsap9/sap9 mutant of C. albicans, we show in this article that Sap9 proteinase is neither essential for biofilm formation nor for interactions with streptococci, but is involved in architectural design of monospecies and dual-species biofilms.
MATERIALS AND METHODS
Microbial strains and growth conditions
Candida albicans strains SC5314, CAI4[pClp10] (Murad et al.2000; Brand et al.2004) or M1018 Δsap9/sap9 (designated here sap9Δ) (Albrecht et al.2006) were cultivated on Sabouraud dextrose agar (Lab M) aerobically at 37°C. Bacterial strains S. gordonii Challis DL1, S. mutans Ingbritt, S. oralis 34, S. parasanguinis SK236, S. sanguinis SK36 or Enterococcus faecalis JH2-2 were cultivated anaerobically at 37°C on BHYN agar (L−1: 37 g brain heart infusion broth, 5 g yeast extract, 5 g neopeptone and 15 g agar). Saccharomyces cerevisiae strains expressing C. albicans CWPs (Nobbs, Vickerman and Jenkinson 2010) were cultivated aerobically at 30°C on CSM medium [L−1: 6.7 g Difco yeast nitrogen base, 0.77 g CSM Drop-out minus Ura (Formedium, Hunstanton, UK), 20 g glucose, 30 g agar]. Suspension cultures of C. albicans were grown in YPD medium (1% yeast extract, 2% peptone and 2% glucose) in conical flasks at 37°C with shaking (200-rpm). Bacterial strains were grown in BHY medium (L−1: 37 g brain heart infusion, 5 g yeast extract) in sealed glass bottles without shaking at 37°C. YPT medium (1 x Difco Yeast Nitrogen Base, 20 mM NaH2PO4-H3PO4 buffer pH 7.0, 0.1% Bacto-tryptone) supplemented with 0.4% glucose (YPT-Glc) was utilized to support growth of all microorganisms in planktonic cultures or biofilms.
Preparation of saliva
Collection of saliva from at least six healthy adult human subjects, who provided written informed consent, was approved by the National Research Ethics Committee South Central Oxford C. (# 08/H0606/87+5). Exclusion criteria were: pregnancy, lactation, gross caries, unstable periodontal disease, continuous medication, or antimicrobial medication within 7 days previously. Samples were pooled, mixed with 0.25 M dithiothreitol on ice for 10 min and clarified by centrifugation (8000 × g for 10 min). The supernatant was diluted to 10% with sterile water, filter sterilized (0.22 μm pore membrane) and aliquots were stored at −20°C.
Preparation of microbial cells
Candida albicans cells were grown for 16 h in YPD medium, harvested by centrifugation (5000 × g for 5 min), washed twice in YPT (no glucose) and suspended at optical density 600 nm (OD600) 1.0 (∼1 × 107 cells/ml). Streptococcus or E. faecalis cells were grown for 16 h in 10 ml YPT-Glc, harvested by centrifugation (5000 × g for 7 min) and washed twice with YPT. Bacteria were labelled with fluorescein isothiocyanate (FITC) as described previously (Dutton et al.2014) and suspended in YPT medium at OD600 0.5 (2 × 108 cells ml−1).
Candida albicans interactions with bacteria in planktonic phase
Portions (0.2 ml) of C. albicans cell suspension in YPT were added to glass tubes containing warm YPT-Glc medium (1.8 ml). Cultures were incubated at 37°C for 3 h with shaking at 220-rpm to induce hypha-formation. FITC-labelled bacterial cell suspension (1 ml) was then added and incubation continued at 37°C for 1 h. This period of incubation time was the most suitable for these experiments, allowing hyphae formation by ∼60% of the C. albicans cells and without extensive clumping of the hyphae, which occurred at later times. Samples (50 μl) were applied to microscope slides and visualized by light or fluorescence microscopy (Leica DMLB). Olympus Cell D software was utilized to determine hyphal cell lengths from at least 50 individual hyphal filaments over a minimum of 10 randomly-selected fields of view.
Cell surface hydrophobicity
Candida albicans cells were induced to form hyphae at 37°C in YPT-Glc medium as described above. Cells were harvested by centrifugation (5000 × g, 5 min) immediately after suspension (time zero) or after 3 h incubation, washed with distilled water and suspended at OD600 0.40 in YPT containing 2 M urea (Hazen, Plotkin and Klimas 1986). Portions of the suspensions (1 ml) in acid-washed glass tubes were overlaid with a mixture of toluene and hexadecane (0.2 ml of each), or with no hydrocarbon (control), and vortex-mixed for 2 min (6 × 20 s pulses). Once the phases had separated the OD600 of the lower aqueous layer was determined. Relative Cell surface hydrophobicity (CSH) was calculated as % change in OD600 = [(OD600 control – OD600 sample)/OD600 control)] × 100.
Planktonic phase interactions of S. cerevisiae with C. albicans
Saccharomyces cerevisiae BY4742 strains expressing C. albicans CWPs were grown in CSM for 16 h at 30°C with shaking at 220-rpm. Cells were collected by centrifugation (5000 × g, 5 min), washed and fluorescently labeled with FITC solution as described previously (Dutton et al.2014). One millilitre suspension of each strain in YPT-Glc (OD600 1.0, equivalent to 1 × 107 cells ml−1) was transferred to a glass tube containing C. albicans hyphae-induced cells (see above) and incubated at 30°C for 2 h with shaking at 220-rpm. The C. albicans cells were stained with Fluorescent brightener 28 (Calcofluor white; 0.5 μg ml−1), and portions of the suspensions were then visualized by transmitted light or fluorescence microscopy. Numbers of S. cerevisiae cells attached to hyphal filaments were counted over at least 10 fields of view (totalling 500–700 filaments) and expressed as a ratio of the number of S. cerevisiae cells bound/number of hyphal filaments.
Monospecies or dual-species biofilms of C. albicans and S. gordonii
Sterile 19 mm glass cover slips were incubated with 10% filter-sterilized saliva at 4°C for 16 h, and transferred into 12-well tissue culture plates containing 1.9 ml YPT-Glc and 0.1 ml C. albicans cell suspension (1 × 106 cells), prepared as described above. To visualize S. gordonii interactions with C. albicans hyphae in early biofilms, cover slips were incubated with C. albicans for 2 h at 37°C with gentle motion (50-rpm), and removed into wells containing fresh YPT-Glc medium (0.5 ml). Suspensions of S. gordonii (0.5 ml, 1 × 108 cells) were added and the cultures were incubated for a further 1 h. For comparative monospecies biofilms of C. albicans, culture medium alone (0.5 ml) was added. Cover slips were then washed, stained with crystal violet (Dutton et al.2014), and biofilms were visualized by light microscopy.
Preparation of biofilms for confocal scanning laser microscopy (CSLM)
Plastic culture dishes (Mat Tek, 35 mm diameter) with 14 mm No 1.0 cover slip base glass windows were incubated with 2 ml 10% saliva at 4°C for 16 h. The saliva was aspirated and 1.8 ml growth medium (YPT-Glc) was added to each dish followed by 0.2 ml C. albicans cell suspension (prepared as described above). For early phase biofilms, dishes were incubated in a humid environment at 37°C for 2 h with gentle motion (50-rpm). The culture suspensions were then aspirated and replaced with 1.8 ml warm YPT-Glc medium. FITC-labelled cell suspension (0.2 ml) of Streptococcus or Enterococcus was added, and the dishes were incubated at 37°C for a further 1 h. The bacterial cell suspensions were then aspirated, and 2 ml YPT medium containing Fluorescent brightener 28 (0.2 μg ml−1) was added to fluorescently stain C. albicans. The biofilms were visualized with a Leica SP5-AOBS confocal microscope attached to a Leica DM I6000 inverted epifluorescence microscope.
For 6 h biofilms, dishes containing C. albicans suspension, as described above, were incubated in a humid environment at 37°C for 1 h with gentle motion at 50-rpm. The culture suspensions were aspirated, replaced with YPT-Glc medium, and the dishes were incubated for a further 2 h. For dual-species biofilms, the C. albicans suspension was gently aspirated, replaced with 1.8 ml YPT-Glc medium, and 0.2 ml suspension of S. gordonii DL1 expressing green fluorescent protein (strain UB2549 GFPmut3b*) (Dutton et al.2014) was added. The dishes were incubated for a further 30 min, the S. gordonii culture suspension was aspirated, and 2 ml appropriate growth medium was added for further incubation at 37°C for 4 h. For monospecies biofilms exactly the same protocol was applied, with growth medium substituted for bacterial cell suspension. To process the samples for CSLM, the microbial cell suspensions were aspirated; the dishes were washed gently with sterile deionized water, and C. albicans cells were stained with Fluorescent brightener 28 (0.2 μg ml−1) just prior to CSLM, as above.
Flow cell biofilms
Flow cell units, consisting of two parallel chambers sealed with a glass cover slip, were prepared as described previously (Dutton et al.2014). The flow cells were injected with 0.5 ml 10% human saliva, and incubated at 4°C for 16 h to coat the inside surfaces of the chambers with salivary glycoproteins. The effluent line to a peristaltic pump was then connected and YPT-Glc medium was drawn through the flow cell for 15 min at a flow rate of 6 ml h−1. Candida albicans cell suspension in growth medium (0.2 ml) was injected into the flow cell chamber and incubated statically at 37°C for 1 h. The growth medium (containing 0.5 μg ml−1 Fluorescent brightener 28) was then drawn through the flow cell chamber at a rate of 6 ml h−1 for up to 16 h. For dual species biofilms with S. gordonii the medium flow was stopped after 2 h and S. gordonii UB2549 (expressing GFPmut3b*) cell suspension (0.2 ml) was injected into the chamber and incubated without medium flow at 37°C for 30 min. Flow of growth medium was then recommenced and continued for 4 h at a rate of 6 ml h−1. Biofilms were visualized by CSLM as noted above.
Image analysis
Volocity® software was utilized to prepare 3D images and to calculate biofilm heights (in μm). Imaris software version 7.5 for 3D interactive data visualization and management (Bitplane AG, Zurich, Switzerland) was used for calculations of biovolume (μm3).
Statistical analysis
All data are presented as the mean ± standard deviation (SD) of at least two independent experiments performed in triplicate. For normally distributed data, comparisons were tested with Student's t-test. The two-tailed Mann–Whitney U-test was used for comparisons between groups. P values <0.05 were considered statistically significant.
RESULTS
Sap9 aspartyl protease in C. albicans–S. gordonii interactions.
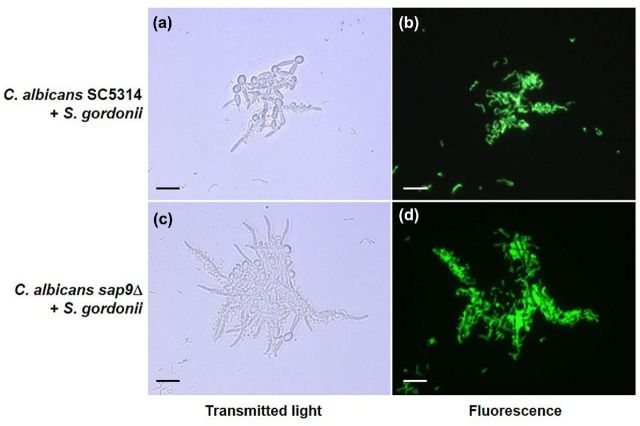
Deficient O-mannosylation in a C. albicans mnt1Δmnt2Δ mutant resulted in aberrant biofilm formation and reduced ability to bind S. gordonii cells (Dutton et al.2014). Proteome data showed that CWP Sap9 was consistently reduced in abundance from CWP extracts of the mnt1Δmnt2Δ mutant, so the possibility was considered that SAP9 expression might be necessary for C. albicans biofilm formation and interkingdom cell–cell contact. To test this, we examined a sap9Δ mutant (Albrecht et al.2006) for ability to form biofilms and to support adhesion of S. gordonii. Incidence of hyphal filament formation by the sap9Δ mutant under planktonic conditions was similar to C. albicans wild type, with ∼60% cells forming hyphae 3 h post-induction in YPT-Glc medium. The sap9Δ mutant hyphae tended to pack together, forming larger clumps overall than the wild type (Fig. 1). However, S. gordonii cells bound planktonically at similar levels to filaments formed by C. albicans wild type or sap9Δ mutant following 3 h hyphal induction (Fig. 1).
Figure 1.
Interactions of S. gordonii DL1 cells with C. albicans SC5314 wild type or sap9Δ mutant. Candida albicans cells were induced to form hyphae in YPT-Glc medium in suspension culture for 2 h at 37°C (planktonic conditions). Bacteria were fluorescently labelled with FITC (green) and were incubated for 1 h at 37°C with hyphae-forming cells in YPT-Glc. Transmitted light microscopic images (a, c) and corresponding fluorescence images (b, d) are shown. Scale bars = 20 μm.
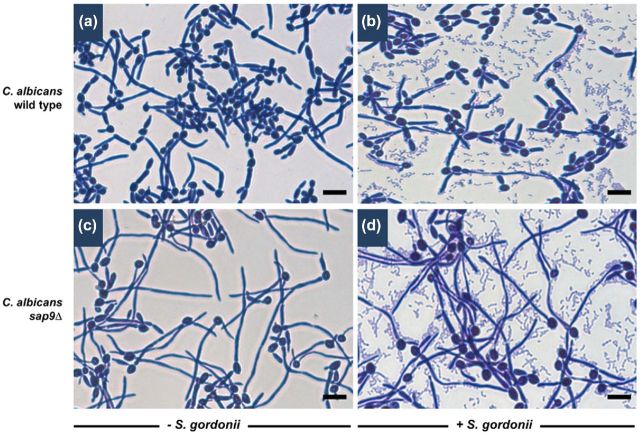
In 3 h monospecies biofilms, hyphae formed by the C. albicans sap9Δ mutant appeared longer on average than wild-type hyphae (Fig. 2), but this was not deemed to be statistically significant over multiple experiments. Binding patterns of S. gordonii cells to sap9Δ hyphae in dual species biofilms was not visibly different from wild type (Fig. 2). Biomass values measured by crystal violet stain assay (Dutton et al.2014) for C. albicans wild type or sap9Δ mutant biofilms were similar (data not shown). Therefore, absence of Sap9 from the mnt1Δmnt2Δ mutant CWPs would not alone account for the almost complete deficiencies in biofilm formation and in binding streptococci previously observed for the C. albicans mnt1Δmnt2Δ mutant (Dutton et al.2014).
Figure 2.
Biofilms formed by C. albicans SC5314 wild type or sap9Δ mutant in the presence or absence of S. gordonii. Biofilms were grown on saliva coated cover slips (as described in Materials and Methods) for 3 h in YPT-Glc, and the cells were then stained with crystal violet and visualized by transmitted light microscopy. Panels: a and b, C. albicans wild type; c and d, C. albicans sap9Δ. Bars = 10 μm.
Architecture of C. albicans wild type or sap9Δ mutant biofilms
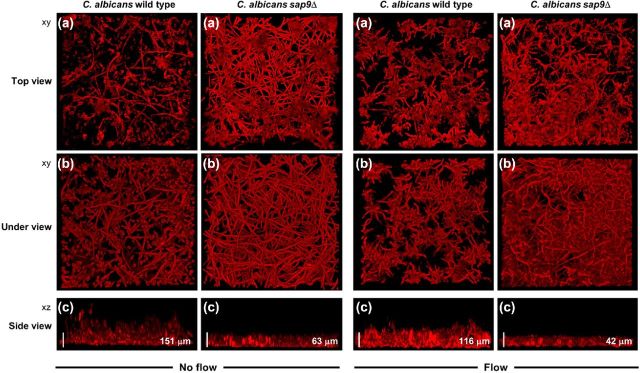
We then investigated by CSLM the architecture of C. albicans sap9Δ mutant biofilms under non-flow conditions and under conditions of flow to generate shear forces. Under both conditions, biofilms formed by the sap9Δ mutant were more compact than those formed by C. albicans wild type, with an average depth of 63 μm for the sap9Δ mutant compared with 151 μm for wild type (no flow), and 42 μm compared to 116 μm, respectively, under medium flow (Fig. 3). The hyphal filaments formed by the sap9Δ mutant strain presented a closely woven mat on the bottom and top surfaces of the biofilm (Fig. 3, panels a and b), while wild-type hyphal filaments at the top surface of the biofilm were extended into the environment (Fig. 3, panels a and c). Under flow conditions, the C. albicans biofilms consisted almost entirely of hyphal filaments (Fig. 3). Under no flow, there were approximately equal proportions of hyphal filaments and yeast morphology cells in wild-type biofilms, but substantially more hyphae in those of the sap9Δ mutant.
Figure 3.
CSLM images of monospecies biofilms formed by C. albicans SC5314 wild type or sap9Δ mutant strains under flow or non-flow conditions. Biofilms were prepared as described in Materials and Methods and grown for 6 h at 37°C in YPT-Glc under no flow or at a medium flow rate of 6 ml h−1. Left side group of six panels: monospecies biofilms of C. albicans wild type or sap9Δ, no flow. Right side group of six panels: monospecies biofilms of C. albicans wild type or sap9Δ mutant under medium flow. The image at the top of each group of six (a) is a representative xy stack of the biofilm assembled from top down. The second image down in each column (b) is a corresponding xy stack of the same biofilm assembled from bottom up, providing an underview aspect of the biofilm. The bottom image (c) in each case is the corresponding xz image showing organization and thickness (depth) of the biofilm. Note that the C. albicans sap9Δ mutant formed a more compact biofilm compared to wild type. The values shown in μm are average thickness (depth) measurements. Bars = 60 μm.
Architecture of C. albicans wild type- or sap9Δ mutant-S. gordonii biofilms
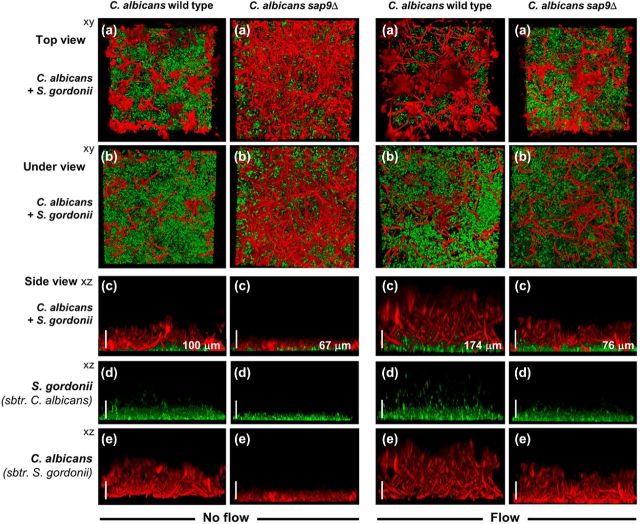
In dual species biofims, the compacted nature of the C. albicans sap9Δ mutant filament formation compared to wild type was more clearly visible (Fig. 4, panel c). The sap9Δ mutant biofilms were characterized by extensive hyphae formation across the surface substratum, as opposed to the wild-type structure in which the hyphal filaments extended into the environment (Fig. 4). In the presence of S. gordonii, bacterial cells could be visualized adhered to the extending hyphal filaments formed by C. albicans wild-type cells (Fig. 4, panels c and d), but remaining adhered to the substratum and within the hyphal filament mats of the sap9Δ mutant (Fig. 4). Under flow conditions, hyphal filaments formed by C. albicans wild-type cells extended at least twice as far into the environment than those produced by the sap9Δ mutant (Fig. 4).
Figure 4.
CSLM images of dual species biofilms formed by C. albicans SC5314 wild type or sap9Δ mutant strains with S. gordonii under flow or non-flow conditions. Biofilms were prepared as described in Materials and Methods and grown in YPT-Glc medium for 6 h at 37°C under no flow, or at a medium flow rate of 6 ml h−1. Left side group of 10 panels: dual species biofilms of C. albicans wild type or sap9Δ, no flow. Right side group of 10 panels: dual species biofilms of C. albicans wild type or sap9Δ with medium flow. The image at the top of each group of 6 (a) is a representative xy stack of the biofilm assembled from top down. The second image down (b) in each column is the corresponding xy stack of the biofilm assembled from bottom up, providing an under view aspect of the biofilm. The third image down (c) in each case is the corresponding xz image showing organization and thickness (depth) of the biofilm. The fourth (d) and fifth (e) images down show green S. gordonii only component of the biofilm (red channel subtracted) (d), and red C. albicans only component of the biofilm (green channel subtracted) (e). S. gordonii DL1 wild-type strain expressed Green Fluorescent Protein (GFP). The values shown in μm are average thickness (depth) measurements. Bars = 100 μm.
Candida albicans wild type or sap9Δ mutant biofilm development with streptococci
To determine if the effects of C. albicans SAP9 deletion were manifested in the presence of other oral streptocococci, early biofilms (3 h) were generated under non-flow conditions of C. albicans wild type or sap9Δ mutant with five different species of oral streptococci, and with E. faecalis. As previously observed, the C. albicans sap9Δ mutant formed hyphal filaments that lay more closely to the substratum, irrespective of the species of Gram-positive coccus present (Fig. 5). In all instances, it appeared that more bacterial cells were deposited upon the substratum within sap9Δ mutant biofilms compared to C. albicans wild type (Fig. 5). Streptococcus mutans and E. faecalis were least well incorporated into C. albicans biofilms. Strains of these two bacterial species have been shown to be weaker coaggregation partners of C. albicans compared to other species (Jenkinson, Lala and Shepherd 1990). These results suggested that the Sap9 proteinase might be beneficial in facilitating C. albicans competition with streptococci for colonization of salivary pellicle.
Figure 5.
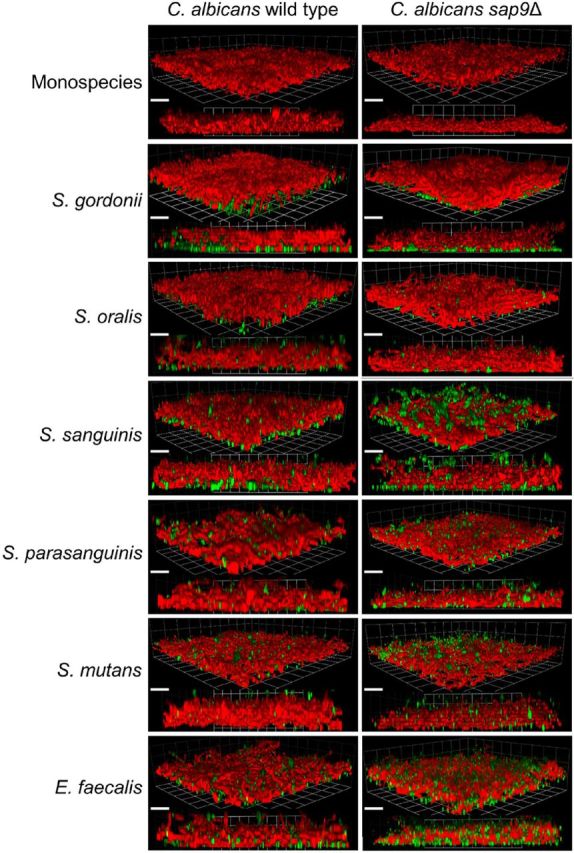
CSLM 3D-images and corresponding z-stack images of dual species biofilms formed by C. albicans SC5314 wild type or sap9Δ mutant strains with five species of viridans streptococci and with E. faecalis. Candida albicans biofilms were grown on saliva coated cover slips (as described in Materials and Methods) for 2 h in YPT-Glc, fluorescently labelled (FITC) bacteria were then added and biofilms were incubated for a further 1 h at 37°C. Candida albicans cells were stained with Fluorescent brightener 28 just prior to CSLM. The bacterial species employed are shown in the figure. Bacteria appear green, C. albicans cells appear red. Bars = 20 μm.
Cell surface hydrophobicity
Since a repeated observation was that the sap9Δ mutant formed hyphae that associated closely with each other and tended to form a biofilm mat, we investigated the possibility that this was related to increased CSH of the hyphal filaments. Accordingly, cells of wild type or sap9Δ mutant were suspended in hypha-inducing medium (YPT-Glc) and CSH measured by partition assay (Materials and Methods). Immediately after suspension the wild type and sap9Δ mutant cells exhibited low hydrophobicity (Fig. 6), but after 3 h incubation the sap9Δ mutant hyphae were substantially more hydrophobic compared to wild type (Fig. 6).
Figure 6.
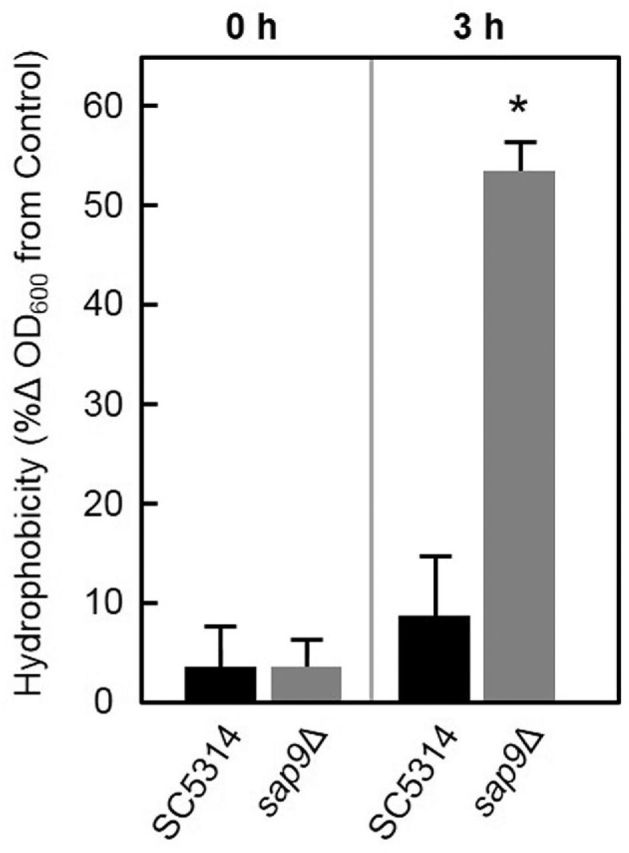
Cell surface hydrophobicity (CSH) of C. albicans SC5314 wild type and sap9Δ mutant immediately after suspension in hyphae-inducing YPT-Glc medium (0 h) and after 3 h incubation at 37°C when ∼60% cells were producing hyphal filaments. CSH was determined as described in Materials and Methods. Error bars are ± SD, n = 2. *Statistical significance relative to C. albicans wild type (P < 0.05).
Candida albicans wild type or sap9Δ mutant interactions with C. albicans CWPs
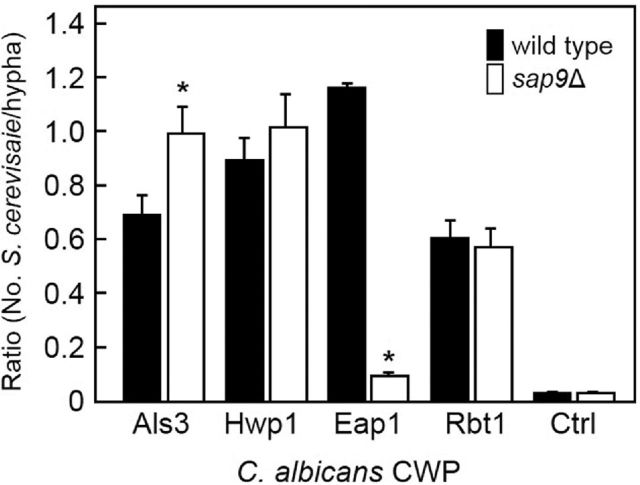
A proposed function of Sap9 proteinase is to modulate activities of other CWPs such as cell wall modifying enzymes (e.g. chitinase Cht2) (Schild et al.2011) or adhesins (Albrecht et al.2006). Proteinases may also mediate adhesion independently of proteolytic activity (Jarocki, Tacchi and Djordjevic 2015; Kumar et al.2015). To determine if the absence of Sap9 affected hyphal interactions with CWPs on other cells, S. cerevisiae strains expressing C. albicans adhesins Als3, Hwp1, Eap1 or Rbt1 were FITC-labelled and incubated for 2 h with hyphae-forming cells of C. albicans wild type or sap9Δ mutant. Analysis of images obtained from multiple fields of view (Fig. 7 shows some typical images) indicated that adherence levels of S. cerevisiae cells expressing Eap1 to C. albicans sap9Δ mutant hyphae were ∼90% reduced compared to wild type (Fig. 8). Conversely, binding levels of Als3-expressing S. cerevisiae cells were higher (by about 25%) to sap9Δ mutant hyphae than to wild-type hyphae (Fig. 8). There were no statistically significant differences between wild type and sap9Δ mutant in binding S. cerevisiae cells expressing Hwp1 or Rbt1, and <5% hyphae bound S. cerevisiae vector only control cells (Fig. 8). The differences in CWP activities shown by the sap9Δ mutant hyphae and their significantly increased hydrophobicity might explain, at least in part, the enhanced aggregation planktonically and intensified hyphal filament matting in biofilms.
Figure 7.
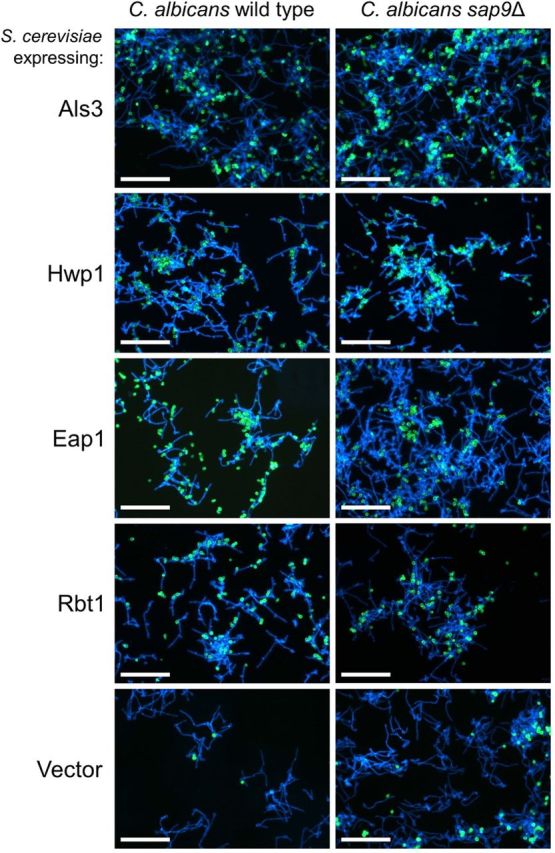
Interactions of C. albicans SC5314 wild type or sap9Δ mutant with S. cerevisiae strains expressing C. albicans CWPs. Saccharomyces cerevisiae cultures were grown to mid-exponential phase in CSM medium, the cells were FITC-labelled, and then mixed with C. albicans cells that had been induced to form hyphal filaments for 3 h at 37°C in YPT-Glc medium (see Materials and Methods). After 2 h incubation of cultures at 30°C with gentle agitation, Fluorescent brightener 28 (0.5 μg ml−1) was added to fluorescently label the C. albicans cells, and samples (10 μl) were removed onto glass microscope slides, cover slips applied, and visualized with a Leica microscope under fluorescence. Panels vertically indicate S. cerevisiae BY4742 cells expressing C. albicans CWPs Als3, Hwp1, Eap1 or Rbt1, and empty vector pBC542 control. S. cerevisiae green, C. albicans blue. Bars = 20 μm.
Figure 8.
Binding levels of S. cerevisiae expressing C. albicans CWPs to C. albicans SC5314 wild type or sap9Δ mutant hyphae. Candida albicans cells were induced to form hyphae for 3 h at 37°C and then incubated with FITC-labelled S. cerevisiae for 2 h (see legend to Fig. 7). Images are representative of typical field of view. Saccharomyces cerevisiae adherence is expressed as the ratio of numbers of bound S. cerevisiae cells: C. albicans hyphal filaments. Ctrl is S. cerevisiae empty vector pBC542 control. Error bars are ± SD, n = 2. *Statistical significance relative to C. albicans wild type (P < 0.05).
DISCUSSION
These studies were undertaken following the observations that in a biofilm-deficient mutant strain of C. albicans mnt1Δmnt2Δ, deleted in O-mannosylation enzymes Mnt1 and Mnt2 (Munro et al.2005); the cell wall proteome profile was lacking Sap9 (Dutton et al.2014). This CWP is one of a family of 10 aspartyl proteinases (Sap1-10) that are differentially expressed in C. albicans (Naglik, Challacombe and Hube 2003). SAP9 has been studied in detail but the precise role of this gene in growth and survival remains unclear. The main effects of SAP9 deletion are to reduce chitinase activity (Schild et al.2011), but deletion of SAP9 is also reported to result in increased adhesion to epithelial cells (Albrecht et al.2006). It has been suggested that Sap9 is able to trim cell surface proteins, on C. albicans or the host, to modulate receptor–ligand interactions or connections (Schild et al.2011). Our data show that SAP9 expression affects C. albicans biofilm structure, under non-flowing conditions and under medium flow. These are early biofilms (6 h) in a defined medium with glucose, and so are not conducive to matrix production under the conditions (Chandra et al. 2001). Sap9 deficiency led to formation of hyphal filaments with increased CSH that associated more tightly with the substratum and with each other. This could be consistent with the activity of secreted proteases normally shedding or modifying cell surface proteins such that hyphae are less self-adhesive, more able to extend into the environment, and more readily dispersed from the biofilm. However, based upon the phenotypic analyses of the sap9Δ mutant, loss of Sap9 alone does not explain the biofilm-forming deficiency of the C. albicans mnt1Δmnt2Δ mutant, or the inability of S. gordonii cells to interact with hyphae produced by the mnt1Δmnt2Δ mutant (Dutton et al.2014).
Sap9 has broad substrate specificity, and preferentially cleaves a peptide backbone at adjacent basic residues e.g. Lys-Arg or Arg-Arg when these are followed by an acidic amino acid residue (Aoki et al.2011; Laskay et al. 2014). However, cleavage events can occur independently of basic residues (Schild et al.2011). The enzyme has a near-neutral optimum pH and exhibits activity over a broad pH range. There is evidence that cleavage by Sap9 activates cell wall-associated Cht2 (chitinase) and alters the degree of cross-linking of glucan chains by Pir1 (Schild et al.2011). Overall effects of Sap9 proteinase are propsed to be (i) activation of cell wall proteins, (ii) rearrangement of cell wall components, (iii) shedding of substrate and (iv) direct adhesion independent of proteolytic activity.
We present evidence that loss of Sap9 affects interaction of hyphae with Eap1, so Sap9 could function as an adhesin in binding to Eap1, or be involved in regulating proteolytic trimming of Eap1 (or of a receptor for Eap1). Eap1 protein is upregulated in biofilms (Sherry et al.2014), is found on the surface of yeast and hyphal cells (Li et al.2007), binds plastic (Nobbs, Vickerman and Jenkinson 2010) and epithelial cells (Li and Palecek 2003), and promotes invasive growth (Li and Palecek 2008). However, binding to plastic and CSH do not necessarily correlate for C. albicans (Kennedy, Rogers and Yancey 1989). Eap1, like Hwp1 and Als family proteins, also contains functional amyloid-forming sequences and rapidly develops insoluble amyloids, thus contributing to cell–cell aggregation (Ramsook et al.2010). The mat-like biofilms of the C. albicans sap9Δ mutant could therefore be explained if a normal function of Sap9 is to assist with de-aggregation of hyphal filaments. This property would be consistent with the evidence that expression of SAP9 is constitutive in biofilms and shows similar relative expression levels across a range of biofilm models (Nailis et al. 2010). Although Eap1 is bound by streptococci (Nobbs, Vickerman and Jenkinson 2010), these bacteria also bind Als3 (Silverman et al.2010), which is thought to be a major target for interkindom interactions (Nobbs and Jenkinson, 2015).
Initial adherence in C. albicans biofilm formation is mediated at least in part by Als1 (Harriott and Noverr 2011) and ALS1 expression is under control of the transcriptional regulator Bcr1, as are the genes encoding Als3 and Hwp1, which mediate cell–cell interactions in biofilms (Nobile et al.2008). Cell wall protein Eap1 is required for biofilm formation in vitro and in vivo (Li et al.2007) and may be needed for initial layer formation on specific substrata (Nobbs, Vickerman and Jenkinson 2010). It has been suggested that Eap1 and Als proteins may play a role in environmental sensing as well as directly as adhesins (Fox, Shelton and Kruppa 2013).
Our observations would be consistent with the following interpretations. Sap9 is known to trim or cleave CWPs and may also regulate processing of other Saps (Monod et al.1998). Accordingly, Sap9 may bind directly to Eap1, or process C. albicans cell surface proteins that are recognized by Eap1. The reduced ability of sap9Δ mutant hyphae to interact with Eap1 on other cells would detrimentally impact early biofilm formation and reduce incorporation of blastospores (mother cells) into the early matrix (as seen in Fig. 3). This might then ameliorate salivary pellicle binding sites for streptococcal colonization (Fig. 4). Sap9 might normally also function to proteolytically cleave and thus destroy pellicle binding sites for streptococci. Thus, Sap9 potentially contributes to the competitive ability of C. albicans to grow and survive in oral microbial communities.
SAP9 gene inactivation also appears to augment expression of Als3 activity (Fig. 8). This promotes hyphal cell–cell aggregation and would assist in generating flatter and more compact hyphal-filament biofilm development under flow (shear) or non-flow conditions (Fig. 3). This is wholly in keeping with the observations that enhanced exposure of Als3 on hyphal filaments dramatically increases hydrophobicity (Beaussart et al.2012). The role of Sap9 here would be to modulate hyphal filament accretion and extrication, such that hyphae are free to spread into and monitor the external environment. Clearly though the increased Als3 activity does not significantly affect the levels of interaction between candidal hyphal filaments and streptococci, even though Als3 is a major adhesin for S. gordonii (Silverman et al.2010). Therefore, an Als3 modulation mechanism, which would function to disaggregate hyphae, might occur post-translationally and does not affect interactions of hyphae with streptococci. This in keeping with the evidence that hyphal cell–cell interactions involve, at least in part, functioning of amyloid-like sequences within CWPs (Ramsook et al.2010), while streptococcal interactions with Als3 appear to occur independently of amyloid formation (Nobbs, Vickerman and Jenkinson 2010; Bamford et al.2015). In summary, the results overall imply a subtle but physiologically, ecologically and pathogenically significant role for Sap9 in biofilm formation, cell–cell communication and interkingdom microbial biofilm formation.
Acknowledgments
We are most grateful to Bernhard Hube, Steven Bates and Carol Munro for the provision of strains; to Paul Kolenbrander and Rob Palmer for flow-cell design and training; and to Dominic Alibhai for skilled imaging assistance. We thank the Medical Research Council and Wolfson Foundation for establishing the Bioimaging Facility at the University of Bristol.
FUNDING
This work was supported by NIH/NIDCR grant no. R01 DE016690.
Conflicts of interest. None declared.
REFERENCES
- Albrecht A, Felk A, Pichova I, et al. Glycosylphosphatidyinositol-anchored proteases of Candida albicans target proteins necessary for both cellular processes and host-pathogen interactions. J Biol Chem. 2006;281:688–94. doi: 10.1074/jbc.M509297200. [DOI] [PubMed] [Google Scholar]
- Aoki W, Kitahara N, Miura N, et al. Comprehensive characterization of secreted aspartyl proteases encoded by a virulence gene family in Candida albicans. J Biochem. 2011;150:431–8. doi: 10.1093/jb/mvr073. [DOI] [PubMed] [Google Scholar]
- Bamford CV, d'Mello A, Nobbs AH, et al. Streptococcus gordonii modulates Candida albicans biofilm formation through intergeneric communication. Infect Immun. 2009;77:3696–704. doi: 10.1128/IAI.00438-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bamford CV, Nobbs AH, Barbour ME, et al. Funtional regions of Candida albicans hyphal cell wall protein Als3 that determine interaction with the oral bacterium Streptococcus gordonii. Microbiology. 2015;161:18–29. doi: 10.1099/mic.0.083378-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee M, Uppuluri P, Zhao XR, et al. Expression of UME6, a key regulator of Candida albicans hyphal development, enhances biofilm formation via Hgc1- and Sun41-dependent mechanisms. Eukaryot Cell. 2013;12:224–32. doi: 10.1128/EC.00163-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaussart A, Alsteens D, El-Kirat-Chatel S, et al. Single-molecule imaging and functional analysis of Als adhesins and mannans during Candida albicans morphogenesis. ACS Nano. 2012;6:10950–64. doi: 10.1021/nn304505s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertolini MM, Xu H, Sobue T, et al. Candida-streptococcal mucosal biofilms display distinct structural and virulence characteristics depending on growth conditions and hyphal morphotypes. Mol Oral Microbiol. 2015;30:307–22. doi: 10.1111/omi.12095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand A, MacCallum DM, Brown AJ, et al. Ectopic expression of URA3 can influence the virulence phenotypes and proteome of Candida albicans but can be overcome by targeted reintegration of URA3 at the RPS10 locus. Eukaryot Cell. 2004;3:900–9. doi: 10.1128/EC.3.4.900-909.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busscher HJ, Rinastiti M, Siswomihardjo W, et al. Biofilm formation on dental restorative and implant materials. J Dent Res. 2010;89:657–65. doi: 10.1177/0022034510368644. [DOI] [PubMed] [Google Scholar]
- Buurman ET, Westwater C, Hube B, et al. Molecular analysis of CaMnt1p, a mannosyl transferase important for adhesion and virulence of Candida albicans. Proc Natl Acad Sci USA. 1998;23:7670–5. doi: 10.1073/pnas.95.13.7670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos MS, Marchini L, Bernardes LA, et al. Biofilm microbial communities of denture stomatitis. Oral Microbiol Immunol. 2008;23:419–24. doi: 10.1111/j.1399-302X.2008.00445.x. [DOI] [PubMed] [Google Scholar]
- Castillo L, MacCallum DM, Brown AJ, et al. Differential regulation of kidney and spleen cytokine responses in mice challenged with pathology-standardized doses of Candida albicans mannosylation mutants. Infect Immun. 2011;79:146–52. doi: 10.1128/IAI.01004-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandra J, Kuhn DM, Mukherjee PK, et al. Biofilm formation by the fungalpathogen Candida albicans: development, architecture, and drug resistance. J Bacteriol. 2001;183:5385–94. doi: 10.1128/JB.183.18.5385-5394.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copping VM, Barelle CJ, Hube B, et al. Exposure of Candida albicans to antifungal agents affects expression of SAP2 and SAP9 secreted proteinase genes. J Antimicrob Chemother. 2005;55:645–54. doi: 10.1093/jac/dki088. [DOI] [PubMed] [Google Scholar]
- Cutler JE. N-glycosylation of yeast, with emphasis on Candida albicans. Med Mycol. 2001;39:75–86. [PubMed] [Google Scholar]
- Diaz PI, Xie Z, Sobue T, et al. Synergistic interaction between Candida albicans and commensal oral streptococci in a novel in vitro mucosal model. Infect Immun. 2012;80:620–32. doi: 10.1128/IAI.05896-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutton LC, Nobbs AH, Jepson K, et al. O-mannosylation in Candida albicans enables development of interkingdom biofilm communities. mBio. 2014;5:e00911. doi: 10.1128/mBio.00911-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutton LC, Paszkiewicz KH, Silverman RJ, et al. Transcriptional landscape of trans-kingdom communication between Candida albicans and Streptococcus gordonii. Mol Oral Microbiol. 2015;30 doi: 10.1111/omi.12111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwivedi P, Thompson A, Xie Z, et al. Role of Bcr1-activated genes hwp1 and hyr1 in Candida albicans oral mucosal biofilms and neutrophil evasion. PLoS One. 2011;6:e16218. doi: 10.1371/journal.pone.0016218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falsetta ML, Klein MI, Colonne PM, et al. Symbiotic relationship between Streptococcus mutans and Candida albicans synergizes virulence of plaque biofilms in vivo. Infect Immun. 2014;82:1968–81. doi: 10.1128/IAI.00087-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanning S, Xu W, Solis N, et al. Divergent targets of Candida albicans biofilm regulator Bcr1 in vitro and in vivo. Eukaryot Cell. 2012;11:896–904. doi: 10.1128/EC.00103-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkel JS, Xu W, Huang D, et al. Portrait of Candida albicans adherence regulators. PLoS Pathog. 2012;8:e1002525. doi: 10.1371/journal.ppat.1002525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox SJ, Shelton BT, Kruppa MD. Characterization of genetic determinants that modulate Candida albicans filamentation in the presence of bacteria. PLoS One. 2013;8:e71939. doi: 10.1371/journal.pone.0071939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia MC, Lee JT, Ramsook CB, et al. A role for amyloid in cell aggregation and biofilm formation. PLoS One. 2011;6:e17632. doi: 10.1371/journal.pone.0017632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gow NA, van de Veerdonk FL, Brown AJ, et al. Candida albicans morphogenesis and host defence: discriminating invasion from colonization. Nat Rev Microbiol. 2011;10:112–22. doi: 10.1038/nrmicro2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harriott MM, Lilly EA, Rodriguez TE, et al. Candida albicans forms biofilms on the vaginal mucosa. Microbiology. 2010;156:3635–44. doi: 10.1099/mic.0.039354-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harriott MM, Noverr MC. Importance of Candida-bacterial polymicrobial biofilms in disease. Trends Microbiol. 2011;19:557–63. doi: 10.1016/j.tim.2011.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazen KC, Plotkin BJ, Klimas DM. Influence of growth conditions on cell surface hydrophobicity of Candida albicans and Candida glabrata. Infect Immun. 1986;54:269–71. doi: 10.1128/iai.54.1.269-271.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrero AB, Uccelletti D, Hirschberg CB, et al. The Golgi GDPase of the fungal pathogen Candida albicans affects morphogenesis, glycosylation, and cell wall properties. Eukaryot Cell. 2002;1:420–31. doi: 10.1128/EC.1.3.420-431.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyer LL, Green CB, Oh SH, et al. Discovering the secrets of the Candida albicans agglutinin-like sequence (ALS) gene family-a sticky pursuit. Med Mycol. 2008;46:1–15. doi: 10.1080/13693780701435317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarocki VM, Tacchi JL, Djordjevic SP. Non-proteolytic functions of microbial proteases increase pathological complexity. Proteomics. 2015;15:1075–88. doi: 10.1002/pmic.201400386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkinson HF, Lala HC, Shepherd MG. Coaggregation of Streptococcus sanguis and other streptococci with Candida albicans. Infect Immun. 1990;58:1429–36. doi: 10.1128/iai.58.5.1429-1436.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joo MY, Shin JH, Jang H-C, et al. Expression of SAP5 and SAP9 in Candida albicans biofilms: comparison of bloodstream isolates with isolates from other sources. Med Mycol. 2013;51:892–6. doi: 10.3109/13693786.2013.824623. [DOI] [PubMed] [Google Scholar]
- Kennedy MJ, Rogers AL, Yancey RJ., Jr Environmental alteration and phenotypic regulation of Candida albicans adhesion to plastic. Infect Immun. 1989;57:3876–81. doi: 10.1128/iai.57.12.3876-3881.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitanovic A, Nguyen M, Vogl G, et al. Phosphatidylinositol 3-kinase VPS34 of Candida albicans is involved in filamentous growth, secretion of aspartic proteases, and intracellular detoxification. FEMS Yeast Res. 2005;5:431–9. doi: 10.1016/j.femsyr.2004.11.005. [DOI] [PubMed] [Google Scholar]
- Krysan DJ, Ting EL, Abeijon C, et al. Yapsins are a family of aspartyl proteases required for cell wall integrity in Saccharomyces cerevisiae. Eukaryot Cell. 2005;4:1364–74. doi: 10.1128/EC.4.8.1364-1374.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar R, Saraswat D, Tati S, et al. Novel aggregation properties of Candida albicans secreted aspartyl proteinase Sap6 mediate virulence in oral candidosis. Infect Immun. 2015;83:2614–26. doi: 10.1128/IAI.00282-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskay ÜA, Srzentic K, Monod M, et al. Extended bottom-up proteomics with secreted aspartic protease Sap9. J Proteomics. 2014;110:20–31. doi: 10.1016/j.jprot.2014.07.035. [DOI] [PubMed] [Google Scholar]
- Li F, Palecek SP. EAP1, a Candida albicans gene involved in binding human epithelial cells. Eukaryot Cell. 2003;2:1266–73. doi: 10.1128/EC.2.6.1266-1273.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Palecek SP. Distinct domains of the Candida albicans adhesin Eap1p mediate cell-cell and cell-substrate interactions. Microbiology. 2008;154:1193–203. doi: 10.1099/mic.0.2007/013789-0. [DOI] [PubMed] [Google Scholar]
- Li F, Svarovsky MJ, Karlsson AJ, et al. Eap1p, an adhesin that mediates Candida albicans biofilm formation in vitro and in vivo. Eukaryot Cell. 2007;6:931–9. doi: 10.1128/EC.00049-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monod M, Hube B, Hess D, et al. Differential regulation of SAP8 and SAP9, which encode two new members of the secreted aspartic proteinase family in Candida albicans. Microbiology. 1998;144:2731–7. doi: 10.1099/00221287-144-10-2731. [DOI] [PubMed] [Google Scholar]
- Mora-Montes HM, Bates S, Netea MG, et al. A multifunctional mannosyltransferase family in Candida albicans determines cell wall mannan structure and host-fungus interactions. J Biol Chem. 2010;285:12087–95. doi: 10.1074/jbc.M109.081513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro CA, Bates S, Buurman ET, et al. Mnt1p and Mnt2p of Candida albicans are partially redundant α-1,2-mannosyltransferases that participate in O-linked mannosylation and are required for adhesion and virulence. J Biol Chem. 2005;280:1051–60. doi: 10.1074/jbc.M411413200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murad AM, Lee RR, Broadbent D, et al. CIp10, an efficient and convenient integrating vector. Yeast. 2000;16:325–7. doi: 10.1002/1097-0061(20000315)16:4<325::AID-YEA538>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- Naglik JR, Challacombe SJ, Hube B. Candida albicans secreted aspartyl proteinases in virulence and pathogenesis. Microbiol Mol Biol Rev. 2003;67:400–28. doi: 10.1128/MMBR.67.3.400-428.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naglik JR, Moyes D, Makwana J, et al. Quantitative expression of the Candida albicans secreted aspartyl proteinase gene family in human and oral candidosis. Microbiology. 2008;154:3266–80. doi: 10.1099/mic.0.2008/022293-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nailis H, Kucharikovà S, Ricicovà M, et al. Real-time PCR expression profiling of genes encoding potential virulence factors in Candia albicans biofilms: identification of model-dependent and –independent gene expression. BMC Microbiol. 2010;10:114. doi: 10.1186/1471-2180-10-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobbs AH, Jenkinson HF. Interkingdom networking within the oral microbiome. Microbes Infect. 2015;17:484–92. doi: 10.1016/j.micinf.2015.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobbs AH, Vickerman MM, Jenkinson HF. Heterologous expression of Candida albicans cell wall-associated adhesins in Saccharomyces cerevisiae reveals differential specificities in adherence and biofilm formation and in binding oral Streptococcus gordonii. Eukaryot Cell. 2010;9:1622–34. doi: 10.1128/EC.00103-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobile CJ, Andes DR, Nett JE, et al. Critical role of Bcr1-dependent adhesins in C. albicans biofilm formation in vitro and in vivo. PLoS Pathog. 2006;2:e63. doi: 10.1371/journal.ppat.0020063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobile CJ, Fox EP, Nett JE, et al. A recently evolved transcriptional network controls biofilm development in Candida albicans. Cell. 2012;148:126–38. doi: 10.1016/j.cell.2011.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobile CJ, Schneider HA, Nett JE, et al. Complementary adhesin function in C. albicans biofilm formation. Curr Biol. 2008;18:1017–24. doi: 10.1016/j.cub.2008.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulitsch AH, Willinger B, Zsalatz B, et al. In-vivo Candida biofilms in scanning electron microscopy. Med Mycol. 2009;47:690–6. doi: 10.3109/13693780802635237. [DOI] [PubMed] [Google Scholar]
- Peltroche-Llacsahuanga H, Goyard S, d'Enfert C, et al. Protein O-mannosyltransferase isoforms regulate biofilm formation in Candida albicans. Antimicrob Agents Chemother. 2006;50:3488–91. doi: 10.1128/AAC.00606-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramage G, Martinez JP, López-Ribot JL. Candida biofilms on implanted materials: a clinically significant problem. FEMS Yeast Res. 2006;6:979–86. doi: 10.1111/j.1567-1364.2006.00117.x. [DOI] [PubMed] [Google Scholar]
- Ramsook CB, Tan C, Garcia MC, et al. Yeast cell adhesion molecules have functional amyloid-forming sequences. Eukaryot Cell. 2010;9:393–404. doi: 10.1128/EC.00068-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapala-Kozik M, Bochenska O, Zawrotniak M, et al. Inactivation of the antifungal and immunomodulatory properties of human cathelicidin LL-37 by aspartic proteases produced by the pathogenic yeast Candida albicans. Infect Immun. 2015;83:2518–30. doi: 10.1128/IAI.00023-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard M, Ibata-Ombetta S, Dromer F, et al. Complete glycosylphosphatidylinositol anchors are required in Candida albicans for full morphogenesis, virulence and resistance to macrophages. Mol Microbiol. 2002;44:841–53. doi: 10.1046/j.1365-2958.2002.02926.x. [DOI] [PubMed] [Google Scholar]
- Schild L, Heyken A, de Groot PWJ, et al. Proteolytic cleavage of covalently linked cell wall proteins by Candida albicans Sap9 and Sap10. Eukaryot Cell. 2011;10:98–109. doi: 10.1128/EC.00210-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherry L, Rajendran R, Lappin DF, et al. Biofilms formed by Candida albicans bloodstream isolates display phenotypic and transcriptional heterogeneity that are associated with resistance and pathogenicity. BMC Microbiol. 2014;14:182. doi: 10.1186/1471-2180-14-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman RJ, Nobbs AH, Vickerman MM, et al. Interaction of Candida albicans cell wall Als3 protein with Streptococcus gordonii SspB adhesin promotes development of mixed-species biofilm communities. Infect Immun. 2010;78:4644–52. doi: 10.1128/IAI.00685-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright CJ, Burns LH, Jack AA, et al. Microbial interactions in building of communities. Mol Oral Microbiol. 2013;28:83–101. doi: 10.1111/omi.12012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Jenkinson HF, Dongari-Bagtzoglou. Innocent until proven quilty: mechanisms and roles of Streptococcus-Candida interactions in oral health and disease. Mol Oral Microbiol. 2014a;29:99–116. doi: 10.1111/omi.12049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Sobue T, Thompson A, et al. Streptococcal co-infection augments Candida pathogenicity by amplifying the mucosal inflammatory response. Cell Microbiol. 2014b;16:214–31. doi: 10.1111/cmi.12216. [DOI] [PMC free article] [PubMed] [Google Scholar]