Abstract
The origin recognition complex (ORC) is the DNA replication initiator protein in eukaryotes. We have reconstituted a functional recombinant Drosophila ORC and compared activities of the wild-type and several mutant ORC variants. Drosophila ORC is an ATPase, and our studies show that the ORC1 subunit is essential for ATP hydrolysis and for ATP-dependent DNA binding. Moreover, DNA binding by ORC reduces its ATP hydrolysis activity. In vitro, ORC binds to chromatin in an ATP-dependent manner, and this process depends on the functional AAA+ nucleotide-binding domain of ORC1. Mutations in the ATP-binding domain of ORC1 are unable to support cell-free DNA replication. However, mutations in the putative ATP-binding domain of either the ORC4 or ORC5 subunits do not affect either of these functions. We also provide evidence that the Drosophila ORC6 subunit is directly required for all of these activities and that a large pool of ORC6 is present in the cytoplasm, cytologically proximal to the cell membrane. Studies reported here provide the first functional dissection of a metazoan initiator and highlight the basic conserved and divergent features among Drosophila and budding yeast ORC complexes.
Keywords: DrosophilaORC, DNA replication, ATPase activity, DNA binding
The initiation of DNA replication in higher eukaryotes occurs at thousands of sites along chromosomes, and significant progress has been made in understanding how proteins work to regulate this event such that each region replicates exactly once per cell cycle (1). The origin recognition complex (ORC), a heteromeric six-subunit protein, is a central component for DNA replication. ORC binds to DNA at replication origin sites and serves in some way as a scaffold for assembly of other key initiation factors such as cdc6, cdt1, MCM complex, and cdc45 (2). How these factors eventually lead to a melting of the duplex strands and an engagement of the DNA polymerase alpha is still unknown. A deeper understanding of how the cell cycle machinery triggers initiation and couples this event to signal transduction pathways requires a finer dissection of the initiation process. The initiator role of ORC in the DNA replication process was first discovered in Saccharomyces cerevisiae (3), where special sequences of about 100 base pairs serve as origin (ori) sites (4). Within these elements, a short 11-base pair consensus sequence serves as a core binding site for ORC.
Although there is a gratifying conservation of such protein factors in different organisms, a surprising divergence for the cis-acting origin sequences almost certainly underscores evolutionary changes in the way DNA replication is regulated and the addition of roles for replication in sculpting functions of the chromosome. For example, in Saccharomyces pombe, ori sequences appear to be largely asymmetric stretches of AT that do not show consensus sequence elements. DNA binding by ORC may depend on a special N-terminal domain of the ORC4 subunit that contains multiple A/T hook motifs that can each recognize such DNA elements (5). In metazoans, the search for DNA sequence definitions of ori sites has been hampered by the lack of convenient genetic or biochemical assays for critical cis-acting motifs (6). To further increase the complexity, a developmental program regulates origin selection functions in temporal and tissue-specific ways (7, 8), and DNA elements spaced over large distances participate in replicator activity (9, 10). Biochemical studies on ORC activity in metazoans will help to clarify some of the questions, and divergences in subunit structure or function may provide clues as to how origin selection and replication regulation evolved in eukaryotes.
All characterized eukaryotic ORCs contain three polypeptides that are members of the large group of ATPases known as the AAA family (11). The proteins of the AAA+ subclass to which ORC1, ORC4, and ORC5 belong are thought to bind ATP, which causes conformational change that will in turn result in a conformational change in an interacting partner. How ORC or the highly homologous Cdc6/18 proteins fit into this paradigm is not understood. The ORC1 subunit in the S. cerevisiae complex must bind ATP to bind ori DNA specifically, but ATP binding of the other ORC-AAA+ subunits in the replication process is not essential (12). It is interesting to note that the ORC4 protein in all eukaryotes, except for the well characterized budding yeast homologue, preserves both the Walker A and B motifs of the general nucleotide-binding Rossman fold present in the AAA family.
The Drosophila (13) and Homo sapiens (14) ORC6 subunits are homologues and are similar in size to the S. pombe counterpart (15). However, S. cerevisiae ORC6 is considerably larger (≈48,000 kDa versus ≈27,000 kDa) and does not share amino acid homology. Moreover, the human homologue does not seem to be tightly associated with the other subunits (14, 16), and no data directly establishing its role in DNA replication have been reported for any system. In S. cerevisiae, the gene encoding ORC6 is essential for viability and cell cycle progression (17), but the protein is not important for ori recognition in DNA-binding assays. A partial complex missing this subunit behaves as does wild type (18).
The six-subunit ORC has been described in Drosophila (13, 19), and biochemical and genetic data support its role as an initiator protein. Mutants, homozygous for ORC2, ORC3, or ORC5, all die in larval stages as large maternal ORC stores are depleted. In the terminal stages, there is a dramatic decrease in DNA replication and cellular proliferation (20–23). Furthermore, females harboring hypomorphic mutations in ORC2 are sterile because they do not amplify the chorion genes in follicle cells, where ORC is localized at four discrete amplification foci (24–26). Purified ORC can bind to the DNA fragments containing these genetically defined elements (24). In previous studies (13), we have shown that the replication of chromatin in a Drosophila cell-free system depends on ORC and that a six-subunit recombinant ORC complex restores activity to depleted fractions. The recombinant ORC also shows a specific activity for replication equivalent to the endogenous or partially purified embryonic complex. We now use recombinant complexes to address which of the potential ATP binding and hydrolysis sites in the subunits are required for various functions and provide direct evidence for the role of ORC6 in DNA replication. However, a large free pool of ORC6 exists in cultured cells and in early embryos, and its peripheral cytoplasmic membrane localization raises the possibility of distinct cellular roles for this protein.
Methods
Purification of Recombinant DmORC.
Baculovirus-expressed wild-type and mutant DmORC complexes were purified from High5 cells (Invitrogen). We used PCR-based mutagenesis methods to create the following mutants in ORC subunits: ORC-1A (K604A), ORC-1B (DE684/685AA), ORC-4A (K62A), ORC-4B (EE147/148AA), and ORC-5A (K47A). All mutant genes were verified by sequencing to confirm that only desired changes were made (for primer sequences see http://www.ocf.berkeley.edu∼ pembwl/supplement). Recombinant baculoviruses were generated by using the BAC-TO-BAC expression system (GIBCO/BRL). ORC1 wild-type and mutant proteins contained a 6 × His N-terminal tag. High5 cells were infected for 72 h, and extracts of the nuclear pellet were prepared with 0.4 M (NH4)2SO4. Nuclear extracts were precipitated with 0.3 mg/ml (NH4)2SO4, and the resulting pellet was redissolved in 50 mM Na2HPO4/NaH2PO4, pH 7.8/300 mM NaCl/5 mM NaF/2 mM imidazole/10% glycerol/2 mM β-ME/0.2 mM PMSF and subjected to nickel-chelate chromatography by using Ni-NTA agarose (Qiagen). Peak fractions were pooled and precipitated with 0.3 mg/ml (NH4)2SO4. The pellet was redissolved in buffer A (25 mM Hepes, pH 7.6/1 mM EDTA/1 mM EGTA/0.05% Nonidet P-40/5 mM NaF/10% glycerol/1 mM DTT/0.2 mM PMSF)/300 mM KCl and fractionated by gel-filtration (Superdex 200), anion-exchange (MonoQ), and cation-exchange (MonoS) chromatography by using FPLC (Amersham Pharmacia). For subsequent experiments (except ATP hydrolysis assays), the MonoS material was further purified on a 4-ml 15–35% glycerol gradient containing buffer A/300 mM KCl. Centrifugation was carried out in an SW-60 rotor at 42,000 rpm for 16 h, by using an L8-80M ultracentrifuge (Beckman). The fractionation of Drosophila extracts was described before (19). In brief, the crude nuclear extract from Drosophila embryos was subjected first to heparin–Sepharose purification and subsequently fractionated on Sephacryl S300 (Amersham Pharmacia) by using HEMG buffer (25 mM Hepes, pH 7.5/0.5 mM EDTA/5 mM MgCl2/10% glycerol) with 0.2 M KCl. Fraction sizes were 8 ml.
ATPase Assays.
Reactions were carried out in 25 μl of ATPase buffer (45 mM Hepes, pH 7.6/150 mM KCl/5 mM MgOAc/1 mM EDTA/1 mM EGTA/0.02% Nonidet P-40/1 mM DTT/0.12 mg/ml BSA) containing 10 μM ATP (including 1 μCi of α-[32P]ATP) and 600 ng (60 nM) purified ORC, in the absence or presence of 160 nM of a 324-bp ori-β DNA fragment. This fragment corresponds to region +929 to +1,253 relative to the s18 transcription start. The DNA was amplified by PCR from pT2 (gift from Allan Spradling, Carnegie Institute, Baltimore) by using the following primer pair: 5′-CCAAGCGATACTTTGAGCC-3′, 5′-GTGATTACTAGTCACATAC-3′. Reactions were incubated at room temperature and stopped at indicated time points by spotting 1 μl of the reaction on a PEI-cellulose TLC plate (Sigma). The plate was developed in 0.6 M Na2HPO4/NaH2PO4, pH 3.5, and analyzed on a PhosphoImager (Fuji).
DNA Binding Assays.
Electrophoretic mobility shift assays.
Binding reactions were carried out in 15 μl of 25 mM Hepes, pH 7.6/60 mM KCl/5 mM MgCl2/0.4 mM EDTA/0.4 mM EGTA/0.1% Nonidet P-40/10% glycerol/1 mM DTT/0.12 mg/ml BSA. Each reaction contained 3 μl of glycerol gradient purified DmORC (≈60–80 ng, ≈10 nM final), 20 μg/ml poly(dG-dC)⋅poly(dG-dC) (Amersham Pharmacia), ≈5 fmol (≈0.33 nM) end-labeled ori-β DNA, and ATP as indicated in the figure legends. Reactions were set up on ice and incubated at room temperature for 15 min. Half of each reaction was loaded on a 4% native polyacrylamide gel. Electrophoresis was performed at 75 V for 5 h at room temperature, by using 12.5 mM Tris/95 mM glycine/0.5 mM EDTA, pH 8.3 as running buffer. The gel was dried on Whatman paper and exposed to x-ray film.
Chromatin binding.
Drosophila egg extracts and demembranated Xenopus sperm DNA were prepared as described (ref. 13 and references therein). Xenopus sperm DNA was incubated with Drosophila egg extract without addition of Xenopus membrane fraction for 30 min at room temperature. The samples were then diluted with 10 V of buffer containing 100 mM KCl/2.5 mM MgCl2/50 mM Hepes-KOH (pH 7.5)/0.2% Triton X-100. The diluted samples were layered on top of 30% sucrose in the same buffer and sedimented at 10,000 × g for 15 min essentially as described (27). The pellets were washed once with buffer. Chromatin-associated proteins were extracted by dissolving pellets in SDS/PAGE sample buffer and passing the solution through a 25-gauge needle. Proteins were separated on a polyacrylamide SDS gel and subjected to a Western blotting reaction using antibody against ORC2.
In Vitro DNA Replication.
Drosophila egg extract preparation was based on procedure previously described (13, 28). Briefly, embryos (0–2 h) were washed with extraction buffer, cold treated, and homogenized. The homogenate was centrifuged for 20 min at 20,000 rpm in TLA100 Beckmann rotor. Middle layer was collected and recentrifuged. The supernatant was collected and used immediately. The extracts were supplemented with ATP regenerating system and 1 μl of Xenopus membrane fraction prepared as in ref. 29. DNA template was added to a final concentration of 1–10 ng/μl. For the DNA synthesis experiment, 5 μCi of labeled dCTP was added. For density substitution experiments (30), in addition to labeled dCTP, BrdUTP was added to a concentration of 1 mM. The reactions were stopped, and DNA was extracted and loaded onto a CsCl density gradient. The gradient was spun in a Beckman 50Ti rotor at 36,000 rpm at 2°C for 40 h. Fractions were collected and counted by Cerenkov radiation.
Green Fluorescent Protein (GFP)–ORC Constructs and Immunofluorescence.
ORC genes were fused with wild-type GFP at the N terminus and subcloned into inducible pMT/V5-B vector (Invitrogen). Transfection of the Drosophila L2 Schneider cells was done according to manufacturer (Invitrogen) recommendations. Cells were fixed by using 1–3% paraformaldehyde and subsequently subjected to immunofluorescent microscopy (Carl Zeiss Axioplan, 100× magnification). Confocal microscopy was performed by using a Carl Zeiss LSM 510 microscope. Immunostaining of the Drosophila embryos was performed as described (31) by using affinity-purified antibody raised against Drosophila ORC2 and ORC6 proteins.
Results
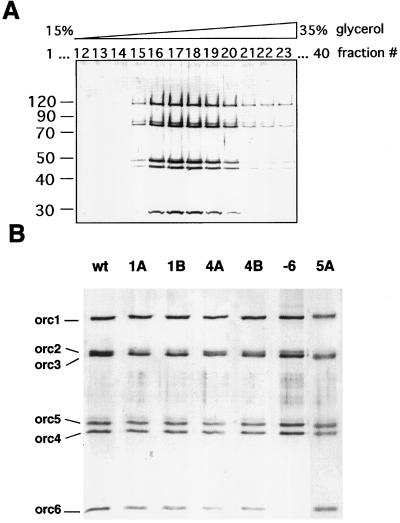
Six different mutant complexes and wild-type recombinant ORC were prepared as described above. For each case, simultaneous expression of the wild-type or mutant genes in the baculovirus expression system resulted in complexes that could be purified to homogeneity through four chromatographic steps, and the mutant complexes assembled and exhibited no chromatographic differences during the purification. In a final step, the pooled peak fractions from the MonoS profile were subjected to glycerol-gradient sedimentation, and the complexes displayed sedimentation behavior as reported (ref. 19; Fig. 1A). Fig. 1B shows a silver-stained gel for each of the mutant complexes and wild-type ORC used in the subsequent studies.
Figure 1.
(A) Silver-stained gel of glycerol-gradient fractions. Fifteen micrograms of recombinant DmORC were fractionated on a 4-ml glycerol gradient; 100-μl fractions were collected from the top of the gradient, and 10 μl of the peak containing fractions were separated on a 9% SDS/polyacrylamide gel and stained with silver. Fractions 16 to 19 were pooled and used for subsequent studies. (B) Silver-stained gel of recombinant purified wild-type and mutant DmORC proteins. Left to right: wild-type complex, ORC1A (K604A) mutant complex, ORC1B (DE684/685AA) mutant complex, ORC4A (K62A) mutant complex, ORC4B (EE147/148AA) mutant complex, ORC5A (K47A) mutant complex, and ORCΔ6 complex deficient for ORC6 subunit. In each lane, 100 ng of proteins were loaded.
DNA Binding and ATP Hydrolysis of Wild-Type and Mutant Complexes.
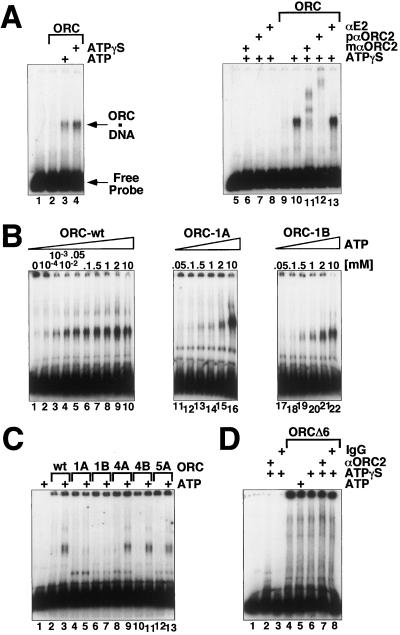
The best understood functions of the yeast ORC are its DNA-binding and ATP hydrolysis functions, so we began our comparative studies there. The bulk of recombinant (or purified embryonic) ORC DNA binding activity is nonspecific and ATP-independent (data not shown). However, we found, as have others (24), that this ATP-independent DNA binding activity could be titrated away with sufficient amount of carrier DNA when the carrier DNA was in a range 50–100 molar excess to the probe DNA. The amount of ATP-dependent DNA–protein complex (Fig. 2A) was slightly increased in the presence of a nonhydrolyzable ATP analogue. The electrophoretic mobility of the complex was slowed when ORC2 antibodies but not a control monoclonal anti-BPV E2 antibody were used in the reaction (Fig. 2A). The specificity of this ATP-dependent DNA binding activity of DmORC will be described in greater depth elsewhere. For the purposes of these studies, it is important to note that with increasing ORC concentration (16–100 nM), other discrete complexes assemble, indicative of multiple sites for ORC binding on the DNA fragment. Furthermore, the affinity of ORC for the chorion gene ori-β DNA fragment, used in Fig. 2, was no better than for any other A/T-rich DNA fragment, such as the ends of the Drosophila P element. ORC was at best two to three times more tightly bound to A/T-rich DNA fragments than to other “control” DNAs with lower A-T content (D.R., E. Beall, and M.B., unpublished data). At physiologically relevant ATP concentrations (10 μM to 1 mM), the wild-type ORC binds to DNA ≈10–50-fold better than either the ORC1A or ORC1B mutant complex (Fig. 2B). Data shown in Fig. 2C reveal that mutations in either the Walker A or B motif of ORC4 or the Walker A motif of ORC5 have no effect on the formation of ATP-dependent DNA–protein complex. These experiments lead us to infer that the recombinant Drosophila ORC, like the recombinant S. cerevisiae homologue, requires only the ORC1 component of the complex to bind ATP for tight DNA interactions. However, the complex missing the ORC6 subunit does not form an ATP-dependent DNA–protein complex (Fig. 2D).
Figure 2.
ATP-dependent DNA binding of wild-type and mutant ORCs. Binding to a radiolabeled ori-β fragment (see Materials and Methods) was monitored in electrophoretic mobility shift assays. (A) ORC-wt was tested for DNA binding without ATP (lanes 2 and 9), with 0.5 mM ATP (lane 3), or with 0.5 mM ATPγS (lanes 4 and 10–13). Addition of monoclonal (lane 11) or of affinity-purified polyclonal (lane 12) antibodies against ORC2, but not a control antibody (lane 13), supershift the observed ATP-dependent protein–DNA complex. Controls: lanes 1 and 5, no protein; lanes 6–8, antibodies without addition of ORC. Arrows indicate the positions of unbound DNA and ORC–DNA complexes in the gel. (B) ORC-wt (lanes 1–10), ORC-1A (lanes 11–16), and ORC1B (lanes 17–22) were tested for DNA binding in the presence of increasing concentrations of ATP as indicated. (C) Wild-type ORC (lanes 2 and 3), ORC1A (lanes 4 and 5), ORC1B (lanes 6 and 7), ORC4A (lanes 8 and 9), ORC4B (lanes 10 and 11), or ORC5A (lanes 12 and 13) was incubated in the absence (lanes 2, 4, 6, 8, 10, and 12) or presence (lanes 3, 5, 7, 9, 11, and 13) of 50 μM ATP. (D) ORC composed only of subunits ORC1–5 (ORCΔ6) was tested for DNA binding in the absence (lane 4) or presence of 0.5 mM ATP (lane 5) or ATPγS (lane 6) and in the presence of ATPγS and either affinity-purified polyclonal antibodies against ORC2 (lane 7) or control rabbit IgG (lane 8).
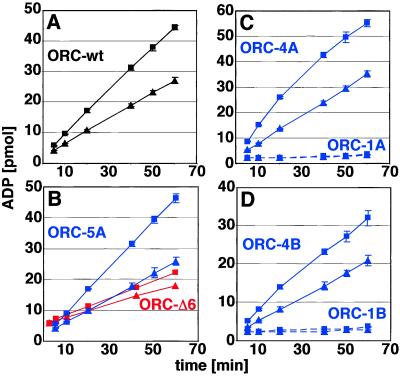
Kinetic analysis of ATP hydrolysis with multiple independent wild-type (wt) ORC preparations showed a Km of 1.92 μM (±0.27) and a Vmax of 0.4 mol ATP hydrolyzed per min per mol of complex. Fig. 3A shows that binding to DNA has a small (2-fold) but measurable effect on slowing the rate of ATP hydrolysis by ORC. In these experiments, ATP was not limiting, and we titrated the Ori-β DNA to its maximal effect. In the absence of any carrier DNA, the saturation is reached at an approximate 2.5-fold molar excess of DNA to ORC. Klemm et al. (12) reported an 8-fold reduction in the hydrolysis rate of DNA-bound yeast ORC; it is likely that these differences reflect divergences in the structure of the DNA–protein–ATP complex. Complexes harboring similar mutations in either ORC4 or ORC5 hydrolyzed ATP with equivalent kinetics to wild type, all displaying Km values and Vmax within the experimental error range of wild type. Consistent with our DNA-binding experiments, we found that the ATP-hydrolysis rate for these mutant complexes was slowed by DNA similar to the effect observed for the wild-type ORC (Fig. 3). In contrast, ORC1A or ORC1B mutants had severely crippled enzymatic activity, too close to background to measure any kinetic parameters (Fig. 3 C and D). The ORC–Δ6 complex was able to hydrolyze ATP at reduced levels, but this activity was unaffected by DNA, consistent with the finding that ORC6 is critical for formation of an ATP-dependent ternary complex (Fig. 3B).
Figure 3.
ATPase activity of recombinant wild-type and mutant DmORC proteins. DNA inhibits the ATPase activity of ORC. ORC-wt (A), ORC5A (B, upper two lanes), ORCΔ6 (B, lower two lanes), ORC4A (C, upper two lines), ORC1A (C, lower two lines), ORC4B (D, upper two lines), and ORC1B (D, lower two lines) were incubated with α-[32P]ATP in the absence (■) or presence (▴) of ori-β-DNA. At indicated time points, reactions were stopped, and the amount of ADP produced was determined by TLC and quantified by PhosphoImager analysis. The average of two independent experiments and ranges are shown.
Function of Mutant Complexes in DNA Replication.
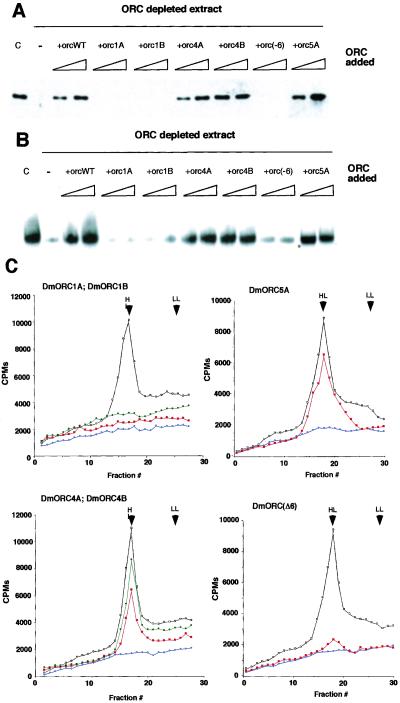
Chromatin binding assays were performed by using both mutant and wt ORC in extracts depleted of membranes. As replication doesn't initiate in such extracts, we were measuring only chromatin binding and not something that might be affected by the process of DNA replication itself. For these experiments Drosophila preblastula embryo extracts were immunodepleted of ORC by using antibody raised against ORC2 and ORC6. The effectiveness of immunodepletion was verified by immunoblotting (data not shown). Demembranated sperm chromatin was added to the depleted extracts, and the binding activities of mutant and wild-type recombinant DmORC were compared with the endogenous Drosophila ORC. Treatment of the extracts with Apyrase abolished ORC-chromatin binding (data not shown), thus we infer that the binding process requires ATP. We relied on the endogenous ATP levels (which are estimated to be at 30–50 μM; ref. 28) to mediate tight chromatin binding. Proteins associated with the chromosomes were separated from the unbound proteins by sedimentation. The results obtained via this assay parallel those obtained by the gel-shift experiments. Recombinant wt ORC, ORC4A, ORC4B, and ORC5A complexes associated with the chromatin with apparently the same efficiency as did endogenous protein, whereas the ORC1A, ORC1B, and ORCΔ6 complexes were severely crippled (Fig. 4A).
Figure 4.
(A) Chromatin binding of Drosophila wild-type and mutant ORC proteins. Demembranated Xenopus sperm DNA was incubated for 30 min in Drosophila extract depleted of the membranes. Where indicated, extracts were immunodepleted for ORC by using antibodies raised against ORC2 and ORC6. Add back experiments were performed by addition of 50 or 150 ng of recombinant wild-type or mutant ORC. Lane C is endogenous ORC binding to Xenopus chromatin. Chromatin-associated proteins were extracted with SDS/PAGE sample buffer, separated on a polyacrylamide gel, and subjected to a Western blotting reaction using antibody against ORC2. (B) DNA replication in Drosophila extracts. Xenopus sperm DNA was incubated for 1 h in Drosophila extract (with membranes) at a concentration of 2–5 ng/μl in a presence of [32P]dCTP. Where indicated, extracts were depleted for ORC by using antibodies raised against ORC2 and ORC6. Add back experiment was performed by addition to depleted extracts of 50 or 150 ng of recombinant ORC proteins. (C) Density substitution analysis of replicated DNA. Demembranated Xenopus sperm DNA was incubated for 1 h in Drosophila egg extract at a concentration of 10 ng/μl in a presence of BrdUTP and [32P]dCTP. DNA was extracted and subjected to centrifugation through gradient of CsCl. Xenopus sperm DNA in ORC-depleted extracts (blue) and after addition of 150 ng of recombinant wild-type (black) or recombinant mutant ORC (red, Walker A mutants or ORCΔ6 complex; green, Walker B mutations) are presented on density profiles. LL indicates the position of light–light DNA; HL shows the position of heavy–light DNA.
We used two independent measures of DNA replication competence for accessing the abilities of the mutant complexes to restore activity to depleted extracts. In the first assay, we detected labeled precursor incorporation into high molecular DNA by autoradiography of gels after electrophoresis (Fig. 4B) or in a second assay after CsCl density gradient separation of DNA that was replicated in extracts with the density label precursor BrdUrd (Fig. 4C). As anticipated from the DNA and chromatin binding results, the ORC1A, ORC1B, and ORCΔ6 complexes were essentially inactive by at least 10–20-fold below the activity of wt recombinant ORC in restoring replication to the extracts. The ORC4A, ORC4B, and ORC5A mutants were effective in reconstitution but were in multiple experiments between 50 and 90% of wild-type complex.
ORC6 Is a Peripheral Membrane and Nuclear Protein.
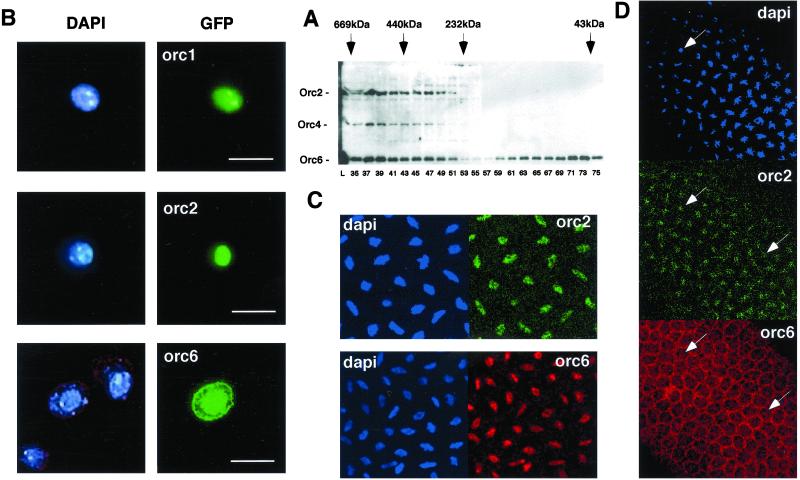
In previous work, we had concluded that the bulk of the subunits of the Drosophila ORC biochemically behaved as a complex (13, 19). We used ORC2 antibodies to track ORC in fractions from 0–12-h embryo extracts after gel-filtration chromatography. We found two broad zones containing ORC. The highest apparent molecular weight fractions containing all ORC subunits were pooled and purified (19). A smaller complex was also detected that was apparently without ORC-1 (32). However, when following ORC6 using ORC6-specific antibodies, we detected a pool of ORC6 devoid of other ORC subunits (Fig. 5A). No other ORC subunits were found in a form unassociated with other ORC proteins (data not shown). We estimate that this free pool is at least one-half of the total ORC6 protein present in these extracts. Given the important role that Drosophila ORC6 plays in cell-free replication and the other activities of ORC that we have analyzed, it was of interest to ask whether this separate pool of ORC6 is localized with the other ORC subunits in the cell.
Figure 5.
(A) Crude Drosophila egg extract separated on S300 Sephacryl column. Proteins from indicated fractions were separated on SDS gel and subjected to Western blot analysis using antibody raised against ORC2, ORC4, and ORC6. Markers used during fractionations were thyroglobulin (669 kDa), ferritin (440 kDa), catalase (232 kDa), and ovalbumin (43 kDa). (B) Localization of Drosophila ORC subunits in L2 cells. GFP-tagged Drosophila ORC1, ORC2, and ORC6 gene constructs were transiently transfected into Drosophila L2 cells. After 48 h, cells were fixed by using 1–3% paraformaldehyde and subsequently subjected to immunofluorescent microscopy (Carl Zeiss Axioplan, 100× magnification). (Scale Bar, 10 μm.) In vivo localization of Drosophila ORC subunits in early embryos before (C) and after (D) cellularization. Confocal microscopy was performed by using a Carl Zeiss LSM 510 microscope (40× and 100× magnification). Immunostaining of the Drosophila embryos was performed by using affinity-purified antibody raised against Drosophila ORC2 and ORC6 proteins. Arrows indicate the same cells within the Drosophila embryo shown here.
Transient ectopic expression of ORC1 or ORC2 GFP-fusion proteins in cultured cells showed a distinct nuclear localization; in unexpected contrast, the GFP-ORC6 fusion protein was found both in the nucleus and cytoplasm (Fig. 5B). The ORC6 cytoplasmic signal seems to be closely associated, in various focal planes, with the cytoplasmic membranes. These experiments rely on overexpression, so we probed this issue further by direct immunofluorescence of endogenous levels of the ORC proteins in Drosophila embryos. Before the onset of cellularization, ORC6 protein localizes only with ORC2 in the nuclear space of both interphase and mitotic cells (Fig. 5C). However, after cellularization, ORC6 seems to localize in the cytoplasm and nucleus (Fig. 5D). The signals for ORC6 could be blocked by preincubating the affinity-purified antibodies with recombinant ORC6 proteins and are clearly distinct from the ORC2 pattern. Further work will be required to judge whether the cytoplasmic pool of ORC6 is truly membrane associated, but it is worth noting that the carboxyl terminus of Drosophila ORC6 contains a predicted leucine-zipper region that could be involved in mediating multiple heterologous protein–protein interactions.
Discussion
The biochemical studies presented here rely on our ability to reconstitute wild-type and mutant DmORC proteins from baculovirus-infected cells. This approach was necessary to obtain sufficient amounts of homogeneous material to detect contributions of mutant subunits for specific functions. An important finding is that the Drosophila ORC complex likely uses mechanisms for binding DNA that are similar to those reported for the budding yeast homologue. Of the three potential ATP binding proteins in ORC, only ORC1 seems to be critical for establishing a tight ternary complex with DNA and for binding to chromatin. Similarly only mutations in the ATP binding domains of ORC1 critically affect a single round of DNA replication in cell-free extracts. Additional experimentation needs to be done to test the roles of the conserved domains in ORC4 and ORC5. Particularly intriguing is the wide conservation of the GKT (Walker A motif) and D (D/EE) (Walker B motif) in the ORC4 subunit. Such domains may be critical for recycling ORC for subsequent rounds of replication or for other activities of the complex in heterochromatin formation or putative check-point control. Drosophila ORC is an ATPase, and again ORC1 seems to play the critical role for ATP hydrolysis, as mutants in the putative ATPase domains of ORC4 and ORC5 do not affect the kinetic parameters of the mutant complex. Nevertheless, it is possible that in the presence of other bound factors, ATP binding or hydrolysis by the other subunits plays some critical role. ATP hydrolysis by any subunit does not seem important for DNA-binding activity. ADP could not mediate such a DNA–protein complex (data not shown), and ATPγS was better at forming a ternary complex than ATP. X-ray crystallographic structure models for several AAA+ proteins have been solved, and a common fold has been observed. We may then use the crystal structure model (33) of an archael Cdc6 ortholog as a guide for the ATP-binding structures of ORC1. In the nucleotide-binding domain of this protein family, both the GKT and the DE motifs contribute to nucleotide affinity. In fact, Klemm et al. (34) have recently shown that similar mutants in the amino-part of the Walker B motif of the S. cerevisiae ORC1 are defective for ATP binding, in contrast to mutations at the carboxyl end of the B motif that are competent for such activity. Moreover, the solvent-exposed surfaces present in these parts of the ORC1 protein may influence interactions with other partners, yielding a mutant complex with altered functions. Our studies of the ATPase activity of DmORC indicate that turnover is slower when ORC is bound to DNA, but the effect is significantly less than that observed for the budding yeast complex. Divergence in the way in which these proteins interact with DNA is also highlighted by the critical role that the Drosophila ORC6 protein plays in ATP-dependent DNA binding. Perhaps, given the lack of amino acid homologies found between the ScORC6 and DmORC6 proteins, it is dangerous to consider each to be homologues.
Asano and Wharton (35) have shown that overexpression of ORC1 trans-genes in Drosophila can alter DNA replication patterns. This overexpression leads to detectable levels of BrdUrd incorporation in normally quiescent cells or increased levels of replication in follicle cells normally amplifying the chorion genes. They found that similar ectopic expression of an ORC1A mutant (ORC1K604E) had no phenotype. Our biochemical results with the ORC1A mutant K604A predict that their mutation might have a dominant negative effect on DNA replication in vivo. It is possible that the mutant gene would not be antimorphic by virtue of its not being able to compete with a wild-type ORC1 protein for assembly into complex. Leaving this point aside, one idea favored by these authors is that ORC1 is limiting for replication in some cellular environments and, for example, complexes containing solely ORC2–6 wait for ORC1 for activation. These pools may or may not be bound to chromosomal DNA. Recent work in mammalian systems indicates that ORC1 may be more loosely associated with chromatin than is ORC2. ORC2, presumably with some of the subunits, can be pelletted with the chromosomes (reviewed in ref. 10; see also ref. 16). Our results reveal that intact ORC needs ATP and functional ORC1 to bind tightly to chromatin. Are all of these data compatible, assuming a conservation in basic binding properties for ORC between mammals and Drosophila? Perhaps, in the absence of ORC1 other subunits mediate another sort of chromatin association. More complex notions are possible, including the interaction of unknown chromatin binding proteins that serve to tether a complex lacking ORC1 to the ori sites.
Results presented here and by others suggest that ORC6 is another subunit that may play important and perhaps dynamic roles in regulating replication activity (36). Our data show that ORC6 is an essential component of the complex per se and may be directly involved in DNA binding and other replication functions or needed for proper ORC assembly. In H. sapiens extracts, ORC6 is not found associated with other ORC subunits (14, 16), but when expressed in the baculovirus system with the other ORC genes, the protein does join a six-subunit complex (16). We are intrigued by the high levels of free ORC6 in embryonic and cultured cell extracts. A considerable fraction of this pool as judged by cytological methods is cytoplasmic, and the protein is perhaps associated with or proximal to the cytoplasmic membranes. It is possible that this localization enables ORC6 to participate in functions unrelated to DNA replication per se, as has been suggested for the “latheo” gene product, which is ORC3 (22). Latheo seems to be involved in ion transport at neuromuscular junctions. Data now exist for both the budding yeast and for the Drosophila ORC, which directly indicate that all of the subunits are critical for DNA replication function, and complex models involving traffic of subsets of ORC subunits can be the subject of future work.
Acknowledgments
We thank Eileen Beall for help in DNA binding assays and valuable comments on the manuscript. This work was supported by National Institutes of Health Grant CA-30490.
Abbreviations
- ORC
origin recognition complex
- ori
origin
- wt
wild type
- GFP
green fluorescent protein
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Diffley J F X. Curr Biol. 2001;11:R367–R370. doi: 10.1016/s0960-9822(01)00196-8. [DOI] [PubMed] [Google Scholar]
- 2.Kelly T J, Brown G W. Annu Rev Biochem. 2000;69:829–880. doi: 10.1146/annurev.biochem.69.1.829. [DOI] [PubMed] [Google Scholar]
- 3.Bell S P, Stillman B. Nature (London) 1992;357:128–134. doi: 10.1038/357128a0. [DOI] [PubMed] [Google Scholar]
- 4.Campbell J L, Newlon C S. Chromosomal DNA Replication. Plainview, NY: Cold Spring Harbor Lab. Press; 1991. [Google Scholar]
- 5.Chuang R Y, Kelly T J. Proc Natl Acad Sci USA. 1999;96:2656–2661. doi: 10.1073/pnas.96.6.2656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gilbert D M. Curr Opin Genet Dev. 1998;8:194–199. doi: 10.1016/s0959-437x(98)80141-x. [DOI] [PubMed] [Google Scholar]
- 7.Spradling A C. Genes Dev. 1999;13:2619–2623. doi: 10.1101/gad.13.20.2619. [DOI] [PubMed] [Google Scholar]
- 8.Edgar B A, Orr-Weaver T L. Cell. 2001;105:297–306. doi: 10.1016/s0092-8674(01)00334-8. [DOI] [PubMed] [Google Scholar]
- 9.Lu L, Zhang H, Tower J. Genes Dev. 2001;15:134–146. doi: 10.1101/gad.822101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cimbora D M, Schübeler D, Reik A, Hamilton J, Francastel C, Epner E M, Groudine M. Mol Cell Biol. 2000;20:5581–5591. doi: 10.1128/mcb.20.15.5581-5591.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Neuwald A F, Aravind L, Spouge J L, Koonin E V. Genome Res. 1999;9:27–43. [PubMed] [Google Scholar]
- 12.Klemm R D, Austin R J, Bell S P. Cell. 1997;88:493–502. doi: 10.1016/s0092-8674(00)81889-9. [DOI] [PubMed] [Google Scholar]
- 13.Chesnokov I, Gossen M, Remus D, Botchan M. Genes Dev. 1999;13:1289–1296. doi: 10.1101/gad.13.10.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dhar S K, Dutta A. J Biol Chem. 2000;201:279. doi: 10.1074/jbc.M006069200. [DOI] [PubMed] [Google Scholar]
- 15.Moon K Y, Kong D, Lee J K, Raychaudhuri S, Hurwitz J. Proc Natl Acad Sci USA. 1999;96:12367–12372. doi: 10.1073/pnas.96.22.12367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vashee S, Simancek P, Challberg M D, Kelly T J. J Biol Chem. 2001;276:26666–26673. doi: 10.1074/jbc.M102493200. [DOI] [PubMed] [Google Scholar]
- 17.Li J J, Herskowitz I. Science. 1993;262:1870–1874. doi: 10.1126/science.8266075. [DOI] [PubMed] [Google Scholar]
- 18.Lee D G, Bell S P. Mol Cell Biol. 1997;17:7159–7168. doi: 10.1128/mcb.17.12.7159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gossen M, Pak D T, Hansen S K, Acharya J K, Botchan M R. Science. 1995;270:1674–1677. doi: 10.1126/science.270.5242.1674. [DOI] [PubMed] [Google Scholar]
- 20.Landis G, Kelley R, Spradling A C, Tower J. Proc Natl Acad Sci USA. 1997;94:3888–3892. doi: 10.1073/pnas.94.8.3888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Loupart M L, Krause S A, Heck M S. Curr Biol. 2000;10:1547–1556. doi: 10.1016/s0960-9822(00)00844-7. [DOI] [PubMed] [Google Scholar]
- 22.Pinto S, Quintana D G, Smith P, Mihalek R M, Hou Z H, Boynton S, Jones C J, Hendricks M, Velinzon K, Wohlschlegel J A, et al. Neuron. 1999;23:45–54. doi: 10.1016/s0896-6273(00)80752-7. [DOI] [PubMed] [Google Scholar]
- 23.Pflumm M F, Botchan M R. Development (Cambridge, UK) 2001;128:1697–1707. doi: 10.1242/dev.128.9.1697. [DOI] [PubMed] [Google Scholar]
- 24.Austin R J, Orr-Weaver T L, Bell S P. Genes Dev. 1999;13:2639–2649. doi: 10.1101/gad.13.20.2639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Calvi B R, Lilly M A, Spradling A C. Genes Dev. 1998;12:734–744. doi: 10.1101/gad.12.5.734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Royzman I, Austin R J, Bosco G, Bell S P, Orr-Weaver T L. Genes Dev. 1999;13:827–840. doi: 10.1101/gad.13.7.827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kubota Y, Mimura S, Nishimoto S, Takisawa H, Nojima H. Cell. 1995;81:601–609. doi: 10.1016/0092-8674(95)90081-0. [DOI] [PubMed] [Google Scholar]
- 28.Crevel G, Cotterill S. EMBO J. 1991;10:4361–4369. doi: 10.1002/j.1460-2075.1991.tb05014.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sheehan M A, Mills A D, Sleeman A M, Laskey R A, Blow J J. J Cell Biol. 1988;106:1–12. doi: 10.1083/jcb.106.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Blow J J, Laskey R A. Cell. 1986;47:577–587. doi: 10.1016/0092-8674(86)90622-7. [DOI] [PubMed] [Google Scholar]
- 31.Pak D T, Pflumm M, Chesnokov I, Huang D W, Kellum R, Marr J, Romanowski P, Botchan M R. Cell. 1997;91:311–323. doi: 10.1016/s0092-8674(00)80415-8. [DOI] [PubMed] [Google Scholar]
- 32.Pak D T S. Ph.D. thesis. Berkeley: Univ. of California; 1996. [Google Scholar]
- 33.Liu J, Smith C L, DeRyckere D, DeAngelis K, Martin G S, Berger J. Mol Cell. 2000;6:637–648. doi: 10.1016/s1097-2765(00)00062-9. [DOI] [PubMed] [Google Scholar]
- 34.Klemm R D, Bell S P. Proc Natl Acad Sci USA. 2001;98:8361–8367. doi: 10.1073/pnas.131006898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Asano M, Wharton R P. EMBO J. 1999;18:2435–2448. doi: 10.1093/emboj/18.9.2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nguyen V Q, Co C, Li J J. Nature (London) 2001;411:1068–1073. doi: 10.1038/35082600. [DOI] [PubMed] [Google Scholar]