Abstract Abstract
The monotypic genus Neoaquastroma (Parabambusicolaceae, Pleosporales) was introduced for a microfungus isolated from a collection of dried stems of a dicotyledonous plant in Thailand. In this paper, we introduce two novel species, N. bauhiniae and N. krabiense, in this genus. Their asexual morphs comprise conidiomata with aseptate and hyaline conidia. Neoaquastroma bauhiniae has ascomata, asci and ascospores that are smaller than those of N. krabiense. Descriptions and illustrations of N. bauhiniae and N. krabiense are provided and the two species compared with the type species of the genus, N. guttulatum. Evidence for the introduction of the new taxa is also provided from phylogenetic analysis of a combined dataset of partial LSU, SSU, ITS and tef1 sequence data. The phylogenetic analysis revealed a distinct lineage for N. bauhiniae and N. krabiense within the family Parabambusicolaceae.
Keywords: Dothideomycetes, holomorph, Massarineae, saprotrophs, Southeast Asia
Introduction
Thailand is a highly biodiverse country in the tropics with hot and humid climate (MacKinnon et al. 1986, Marod and Kutintara 2012). Although the fungal diversity in Thailand has been relatively well-studied (Rostrup 1902, Schumacher 1982, Hyde 1989, Jones 2000, Jones et al. 2006, Suetrong et al. 2009), the number of species being discovered is steadily growing due to increasing activities in studying microfungi in a large variety of terrestrial and aquatic ecosystems (Mapook et al. 2016, Phukhamsakda et al. 2016, Dai et al. 2017, Doilom et al. 2017, Phukhamsakda et al. 2017).
The family Parabambusicolaceae was introduced for a distinct phylogenetic lineage in the suborder Massarineae (Pleosporales) (Tanaka et al. 2015). Species of Parabambusicolaceae are characterised by pseudothecioid ascomata with or without stromatic tissues, papillate to apapillate ostioles, clavate to fusiform asci and hyaline or brown phragmospores (Liu et al. 2015, Tanaka et al. 2015, Li et al. 2016, Wanasinghe et al. 2017). The asexual morphs are sporodochial or Monodictys-like (Tanaka et al. 2015, Ariyawansa et al. 2015). Currently, there are seven known genera in this family; Aquastroma, Monodictys-like spp., Multilocularia, Multiseptospora, Neoaquastroma, Parabambusicola (with P. bambusina as generic type) and Pseudomonodictys (Tanaka and Harada 2003, Wijayawardene et al. 2017, 2018). The genus Neoaquastroma Wanas., E.B.G. Jones & K.D. Hyde, has been introduced from a dead twig of a herbaceous plant collected in Northern Thailand and been typifed with N. guttulatum Wanas., E.B.G. Jones & K.D. Hyde (Wanasinghe et al. 2017). The genus is characterised by immersed, glabrous pseudothecia, short, papillate, fissitunicate, clavate asci and ellipsoidal to subfusiform, multi-septate hyaline phragmospores, surrounded by a mucilaginous sheath (Wanasinghe et al. 2017). Molecular phylogenetic analysis using ribosomal DNA (LSU, SSU and ITS) and translation elongation factor 1-alpha (tef1) sequence data support it as a distinct genus in Parabambusicolaceae.
The purpose of this study is to describe two new species of Neoaquastroma from collections of dicotyledonous plants in Thailand. Phylogenetic analysis of combined of LSU, SSU, ITS and tef1 sequence data are provided.
Materials and methods
Sample collection, morphological study and isolation
Fresh specimens were collected from northern and southern part of Thailand during 2015–2017. The specimens were packed into brown paper bags for transport to the laboratory. Pure cultures were obtained from single ascospores on malt extract agar (MEA; 62 g/l) in distilled water following the method of Chomnunti et al. (2014). Cultures were incubated at 25 °C for up to 8 weeks. Induction of asexual reproduction has been adapted from Tanaka and Harada (2003) by placing agar squares with mycelia on water agar placed with sterile rice straw pieces. The plates were incubated at room temperature (25 °C) with the standard light cycles, 12 hrs in the light followed by 12 hrs in the dark for about eight weeks until the fructifications were produced. Type specimens are deposited in Mae Fah Luang University (MFLU) herbarium and isotypes are deposited at the Kunming Institute of Botany, Academia Sinica Herbarium (HKAS), China. Ex-type living cultures are deposited at the Mae Fah Luang Culture Collection (MFLUCC) and duplicates at the International Collection of Microorganisms and Plants (ICMP), New Zealand. Faces of fungi numbers (Jayasiri et al. 2015) and MycoBank number (http://www.MycoBank.org) are provided. Samples were examined under a Nikon ECLIPSE 80i compound microscope and photographed with a Canon 600D digital camera fitted to the microscope. Measurements were made using Tarosoft (R) Image Frame Work programme and photo-plates were made by using Adobe Photoshop CS6 Extended version 10.0 software (Adobe Systems, United States).
DNA extraction, amplification and sequencing
DNA was extracted from mycelium by using Biospin Fungus Genomic DNA Extraction Kit (BioFlux) (Hangzhou, P. R. China) and gene extraction kit (Bio Basic Inc., Canada). PCR amplification was carried out using primers LROR/LR5 for the nuclear ribosomal large subunit 28S rDNA gene (LSU), NS1/NS4 for the nuclear ribosomal small subunit 18S rDNA gene (SSU) and ITS5/ITS4 for internal transcribed spacer rDNA region (ITS1, 5.8S rDNA and ITS2); partial fragments of the translation elongation factor 1-alpha (tef1) gene region was amplified using primers EF1-983F and EF1-2218R (Vilgalys and Hester 1990, White et al. 1990, Carbone and Kohn 1999). Primer sequences are available at the WASABI database at the AFTOL website (aftol.org). Amplification reactions for LSU, SSU and ITS followed Phukhamsakda et al. (2016). The PCR thermal cycle programme for EF1-983F and EF1-2218R (Carbone and Kohn 1999) for translation elongation factor 1-alpha (tef1) was set for denaturation at 96 °C for 2 min, followed by 40 cycles of denaturation at 96 °C for 45 sec, annealing at 54 °C for 30 sec and extension at 72 °C for 1.30 min, with a final extension step at 72 °C for 5 min. Genomic DNA and PCR amplification products were checked on 1% agarose gel. PCR products were purified as described in Wendt et al. (2017), sequences were generated by Shanghai Sangon Biological Engineering Technology & Services Co. (Shanghai, P.R. China) and sequencing services at Helmholtz Centre For Infection Research (HZI, Braunschweig, Germany).
Sequence alignment and phylogenetic analysis
SeqMan v. 7.0.0 (DNASTAR, Madison, WI) was used to assemble consensus sequences. Sequences of closely related strains were retrieved using BLAST searches against GenBank (http://www.ncbi.nlm.nih.gov). We also included the strains from Wanasinghe et al. (2017) and these are listed in Table 1. Sequences were aligned with MAFTT version 7.220 (Katoh et al. 2013) online sequence alignment tools (mafft.cbrc.jp/alignment/server), with minimal adjustment of the ambiguous nucleotides by visual examination and manually corrected in AliView programme (Larsson 2014). Leading or trailing gaps exceeding from primer binding site were trimmed from the alignments prior to tree building and alignment gaps were treated as missing data. The concatenation of the multigene alignment was created in MEGA 6 (Tamura et al. 2013).
Table 1.
Taxa used in the phylogenetic analysis and their corresponding culture collections, and accession numbers used in this study.
| Taxon | Culture accession number(s)1,2 | GenBank accession numbers | References | |||
|---|---|---|---|---|---|---|
| LSU | SSU | ITS | tef1 | |||
| Aquastroma magniostiolata | CBS 139680T = MAFF 243824 | AB807510 | AB797220 | LC014540 | AB808486 | Tanaka et al. 2015 |
| Aquilomyces patris | CBS 135661T | KP184041 | KP184077 | KP184002 | – | Knapp et al. 2015 |
| Aquilomyces rebunensis | CBS 139684T | AB807542 | AB797252 | AB809630 | AB808518 | Tanaka et al. 2015 |
| Bambusicola massarinia | MFLUCC 11-0389T | JX442037 | JX442041 | NR_121548 | – | Dai et al. 2012 |
| Clypeoloculus akitaensis | CBS 139681T | AB807543 | AB797253 | AB809631 | AB808519 | Tanaka et al. 2015 |
| Corynespora cassiicola | CBS 100822T | GU301808 | GU296144 | – | GU349052 | Schoch et al. 2009 |
| Corynespora smithii | CABI 5649b | GU323201 | – | FJ852597 | GU349018 | Schoch et al. 2009 |
| Falciformispora lignatilis | BCC 21117 | GU371826 | GU371834 | KF432942 | GU371819 | Schoch et al. 2009 |
| Falciformispora senegalensis | CBS 196.79T | KF015631 | KF015636 | KF015673 | KF015687 | Ahmed et al. 2014 |
| Falciformispora tompkinsii | CBS 200.79T | KF015625 | KF015639 | NR_132041 | KF015685 | Ahmed et al. 2014 |
| Helicascus elaterascus | A22-5A = HKUCC 7769 | AY787934 | AF053727 | – | – | Tanaka et al. 2015 |
| Massarina eburnea | CBS 473.64 | GU301840 | GU296170 | – | GU349040 | Zhang et al. 2009 |
| Monodictys sp. | KH 331 = MAFF 243826 | AB807553 | AB797263 | – | AB808529 | Tanaka et al. 2015 |
| Monodictys sp. | JO 10 = MAFF 243825 | AB807552 | AB797262 | – | AB808528 | Tanaka et al. 2015 |
| Morosphaeria ramunculicola | BCC 18404 | GQ925853 | GQ925838 | – | – | Suetrong et al. 2009 |
| Morosphaeria velatispora | BCC 17059T | GQ925852 | GQ925841 | – | – | Suetrong et al. 2010 |
| Multilocularia bambusae | MFLUCC 11-0180T | KU693438 | KU693442 | KU693446 | – | Li et al. 2016 |
| Multiseptospora thailandica | MFLUCC 11-0183T | KP744490 | KP753955 | KP744447 | – | Liu et al. 2015 |
| Multiseptospora thailandica | MFLUCC 11-0204 | KU693440 | KU693444 | KU693447 | KU705659 | Liu et al. 2015 |
| Multiseptospora thailandica | MFLUCC 12-0006 | KU693441 | KU693445 | KU693448 | KU705660 | Liu et al. 2015 |
| Multiseptospora thysanolaenae | MFLUCC 11-0238T | KU693439 | KU693443 | – | KU705658 | Li et al. 2016 |
| Neoaquastroma bauhiniae | MFLUCC 16-0398T | MH023319 | MH023315 | MH025952 | MH028247 | This study |
| Neoaquastroma bauhiniae | MFLUCC 17-2205 | MH023320 | MH023316 | MH025953 | MH028248 | This study |
| Neoaquastroma krabiense | MFLUCC 16-0419T | MH023321 | MH023317 | MH025954 | MH028249 | This study |
| Neoaquastroma guttulatum | MFLUCC 14-0917T | KX949740 | KX949741 | KX949739 | KX949742 | Wanasinghe et al. 2017 |
| Palmiascoma gregariascomum | MFLUCC 11-0175T | KP744495 | KP753958 | KP744452 | – | Liu et al. 2015 |
| Parabambusicola bambusina | KH 139 = MAFF 243823 | AB807537 | AB797247 | LC014579 | AB808512 | Tanaka et al. 2015 |
| Parabambusicola bambusina | H 4321 = MAFF 239462 | AB807536 | AB797246 | LC014578 | AB808511 | Tanaka et al. 2015 |
| Parabambusicola bambusina | KT 2637 = MAFF 243822 | AB807538 | AB797248 | LC014580 | AB808513 | Tanaka et al. 2015 |
| Pseudomonodictys tectonae | MFLUCC 12-0552 | KT285573 | KT285574 | – | KT285571 | Ariyawansa et al. 2015 |
| Stagonospora pseudocaricis | CBS 135132 | KF251762 | – | KF251259 | – | Quaedvlieg et al. 2013 |
| Trematosphaeria pertusa | CBS 122368ET | FJ201990 | FJ201991 | NR_132040 | KF015701 | Zhang et al. 2008 |
1 Abbreviations: BCC: BIOTEC Culture Collection, Bangkok, Thailand; CABI: Centre for Agriculture and Biosciences International, Egham, UK; CBS: CBS-KNAW Collections, Westerdijk Fungal Biodiversity Institute, Utrecht, The Netherlands; CPC: Culture collection of Pedro Crous, housed at CBS; HKUCC: The University of Hong Kong Culture Collection; HHUF: Herbarium of Hirosaki University, Fungi; JCM: The Japan Collection of Microorganisms, Japan; JK: J. Kohlmeyer; JO: J. Onodera; KH: K. Hirayama; KT: K. Tanaka; MAFF: Ministry of Agriculture, Forestry and Fisheries, Japan; MFLU: Mae Fah Luang University herbarium, MFLUCC: Mae Fah Luang University Culture Collection, Chiang Rai, Thailand.
2 Status of the strains: (T) ex-type, (ET) ex-epitype. The strains generated in this study are given in bold.
Maximum likelihood analyses (ML), including 1,000 bootstrap replicates, was performed using RAxML (Stamatakis 2014) as implemented in raxmlGUI version v.1.3.1 (Silvestro and Michalak 2011). The search strategy was set to rapid bootstrapping. The analysis was carried out with the general time reversible (GTR) model for nucleotide substitution and a discrete gamma-distributed with four rate categories (O’Meara et al. 2006, Stamatakis et al. 2008). The bootstrap replicates were summarised on to the best scoring tree.
The best fitting substitution model for each single gene partition and the concatenated data set was determined in MrModeltest 2.3 (Nylander 2004) for Bayesian inference posterior probabilities (PP). In our analysis, GTR+I+G model was used for each partition. The Bayesian inference posterior probabilities (PP) distribution (Zhaxybayeva and Gogarten 2002) was estimated by Markov Chain Monte Carlo sampling (MCMC) in MrBayes v. 3.2.2 (Huelsenbeck and Ronquist 2001). Six simultaneous Markov chains were run for 1,000,000 generations and trees were sampled every 100th generation, thus 10,000 trees were obtained. The suitable burn-in phases were determined by traces inspected in Tracer version 1.6 (Rambaut et al. 2014). Based on the tracer analysis, the first 1,000 trees representing 10% of burn-in phase of the analyses were discarded. While the remaining trees were used for calculating posterior probabilities in the majority rule consensus tree (critical value for the topological convergence diagnostic set to 0.01).
Phylogenetic trees and data files were visualised in FigTree v. 1.4 (Rambaut and Drummond 2008). The phylogram with bootstrap values and/or posterior probabilities on the branches are presented in Fig. 1 by using graphical options available in Adobe Illustrator CS v. 6. All sequences generated in this study were submitted to GenBank. The finalised alignment and tree were deposited in TreeBASE, submission ID: 22419 (http://www.treebase.org/). Maximum likelihood bootstrap values equal to or greater than 70% with Bayesian Posterior Probabilities (PP) equal or greater than 0.90 are presented below or above each node (Fig. 1).
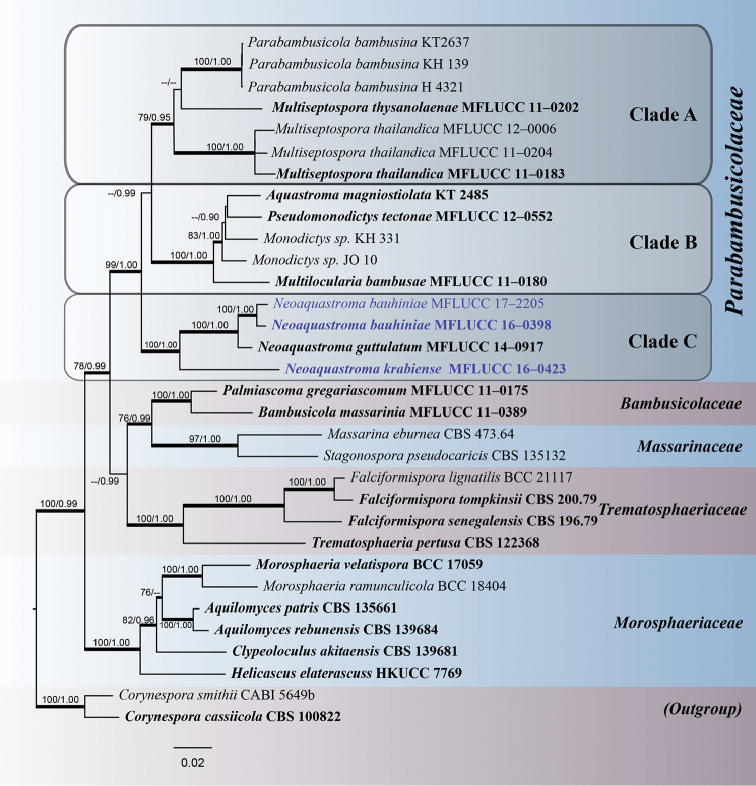
Figure 1.
The best scoring RAxML tree based on a combined partial LSU, SSU, ITS and tef1 gene datasets. Bootstrap values (BS) from maximum likelihood (ML, left) of more than 70% BS and Bayesian posterior probabilities (PP, right) greater than 0.90 are given above or below the nodes. The tree is rooted with Corynespora smithii (CABI 5649b) and C. cassiicola (CBS 100822) in Corynesporaceae. The species, determined in this study, are indicated in blue. The ex-type and references strains are indicated in bold. Hyphens (-) represent support values less than 70% BS/0.90 PP. Thick branches represent significant support values from all analyses (BS ≥ 70%/PP ≥ 0.95).
Results
Phylogenetic analyses
The phylogenetic tree included 32 taxa representing six families from the suborder Massarineae. The phylogenetic trees from each individual data sets were initially generated, these were not significantly different (data not shown) and therefore combined data sets were performed. The combined dataset consisting 3,554 nucleotide characters, of which 1,001 characters corresponded to LSU, 1,038 characters to SSU, 508 characters to ITS and 929 characters to tef1. Corynespora smithii (CABI 5649b) and C. cassiicola (CBS 100822) are used as outgroup taxa. An insertion in the SSU rDNA region of isolates Aquilomyces rebunensis Tanaka & K. Hiray. (CBS 139684), Clypeoloculus akitaensis Tanaka & K. Hiray. (CBS 139681) and Trematosphaeria pertusa Fuckel (CBS 122368) were excluded from the analysis prior to tree building. The best scoring tree from maximum likelihood analysis was selected with a final likelihood value of – In: 23014.934293 and the result is presented in Fig. 1. Phylogenetic trees obtained from maximum likelihood and Bayesian analyses yielded trees with similar overall topology as that of previous work (Tanaka et al. 2015, Liu et al. 2015, Wanasinghe et al. 2017).
In this study, the family Parabambusicolaceae received high support in the phylogenetic analysis. While within the family, the taxa are separated into three subclades (Fig. 1). Parabambusicola bambusina, the generic type, clustered with Multiseptospora with high support. However, M. thysanolaenae (MFLUCC 11-0202) formed a sister taxon with Parabambusicola bambusina (Clade A). Aquastroma magniostiolata (CBS 139680) and Multilocularia bambusae (MFLUCC 11-0180) formed a clade with the hyphomycetes strains of Monodictys spp. and Pseudomonodictys tectonae (MFLUCC 12-0552) with high support in all computational methods (Clade B). Neoaquastroma formed a basal clade (Clade C), with N. bauhiniae (MFLUCC 16-0398, 17-2205) and N. krabiense (MFLUCC 16-0423) clustered with the type species N. guttulatum, with strong support (100% ML /1.00 PP). We describe the new taxa based on agreement in support for all computational methods (Jeewon and Hyde 2016). The new sequence data is deposited in GenBank (Table 1).
Taxonomy
Neoaquastroma bauhiniae
C. Phukhams. & K.D. Hyde sp. nov.
824673
Figure 2.
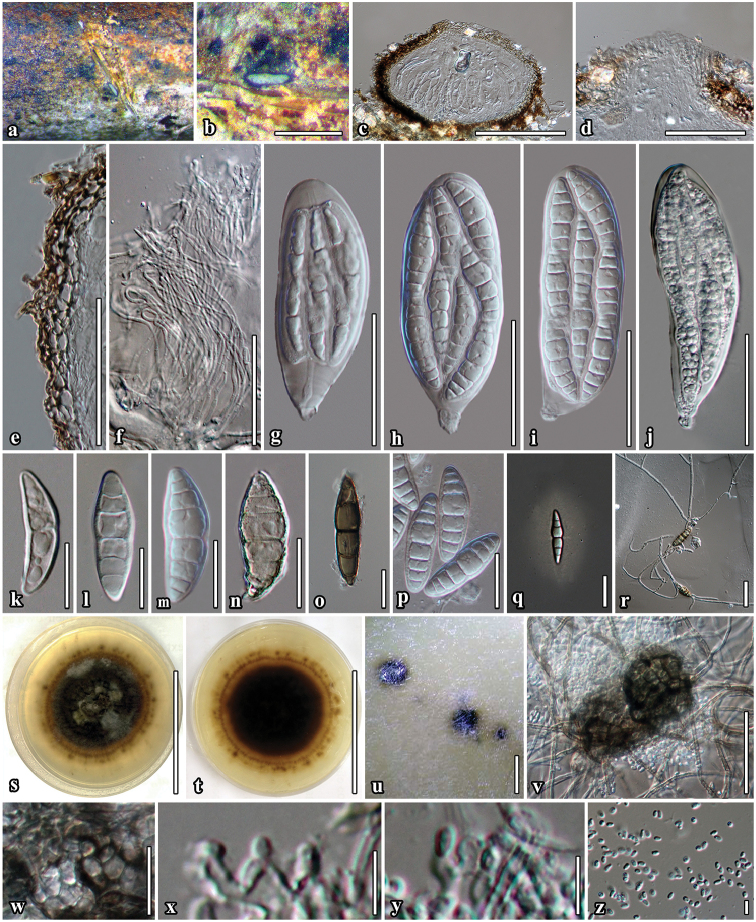
Neoaquastroma bauhiniae (MFLU 17-0002, holotype) a Appearance of ascomata on host surface b Close up of ascoma c Section of ascoma d Ostiolar canal e Section of partial peridium layer f Pseudoparaphyses g–j Development state of asci j Asci produced in culture k–p Development state of ascospores; (n, o Senescent spores m, p ascospores in 5% of KOH reagent); q Ascospores stained with India ink, sheath surrounding the entire ascospore r Germinated ascospore s, t Culture character on MEA u Conidiomata forming on agar on rice straw media after 8 weeks v Immature conidiomata w Conidiomatal wall x, y Conidiogenous cells and developing conidia z Conidia j, m Asci and ascospore in culture (on rice straw). Scale bars: 500 µm (b); 100 µm (c, v); 50 µm (d–j); 20 µm (k–r, w); 5 µm (x–z).
Etymology.
Name refers the host from which this fungus was isolated.
Type material.
THAILAND. Phrae Province: Song District, on dead twigs of Bauhinia variegata L. (Fabaceae), 25 July 2015, C. Phukhamsakda, S1-11, MFLU 17-0002 (holotype), MFLUCC 16-0398 = ICMP 21572 (ex-type living culture).
Description.
Saprobic on dead twigs of Bauhinia variegata L. Sexual morph. Ascomata 113–190 μm high × 170–307 μm diam. (x̄ = 160 × 260 μm, n = 10), semi-immersed to immersed, solitary, scattered, subglobose to compressed, coriaceous, brown to dark brown, rough-walled, with short hyphae projecting from peridium, ostiolate. Ostiole 33 × 85 μm diam., centrally located, papillate, periphysoid. Peridium 8–25 μm wide (x̄ = 17, n = 30), with cells 3–8 μm wide, composed of 3 layers of reddish-brown to dark brown, cells of textura angularis, inner layer composed of hyaline gelatinous cells. Hamathecium composed of numerous, dense, long, 1–2.4 μm (x̄ = 1.7 μm, n = 50), narrow, filiform, transversely septate, branched, anastomosing, cellular psedoparaphyses. Asci 53–116 × 26–43 μm (x̄ = 98 × 37 μm, n = 30), 8-spored, bitunicate, fissitunicate, oboviod to oblong, with furcate pedicel, with ocular chamber visible when immature. Ascospores 37–46 × 9–16 μm (x̄ = 43 × 13 μm, n = 50), bi-seriate or overlapping, broad fusiform, narrow towards the apex, initially hyaline, becoming brown to dark brown at maturity, 4–7-transversely euseptate, constricted at the septa, with cell above central septum wider, rough-walled, indentations present, surrounded by 7–12 μm wide, mucilaginous sheath. Asexual morph coelomycetous. Pycnidia produced on mycelium in water agar. Conidiomata 33−49 μm high × 92–108 μm wide diam., pycnidial, dark brown to black, covered by dense vegetative hyphae, globose, in agar immersed to superficial, uniloculate, solitary to scattered, ostiolate. Conidiomatal wall thin, brown to black-walled with cells of textura angularis. Conidiophores reduced to conidiogenous cells. Conidiogenous cells 3−4 × 2−3.5 μm, enteroblastic, phialidic, integrated, oblong, hyaline, formed from the inner layer of pycnidium wall. Conidia 2–4 × 1.5–2 μm (x̄ = 3 × 1.7 μm, n = 100), broad-oblong to oval, hyaline, aseptate, smooth-walled.
Culture characteristics.
Colonies on MEA, reaching 50 mm diam. after 4 weeks at 25 °C. Culture dark olive-green with black centre, with dense mycelia, circular, flat, umbonate, rough surface, dull, fimbriate, radially furrowed, covered with white aerial mycelium; mycelium strongly radiating into agar, yellow pigment diffusing in the agar; reverse black with radiating brown mycelium. Sexual and asexual morphs formed in culture. Morphology of sexual phase similar to those on substrate.
Additional material examined.
THAILAND, Phrae Province, Song District, on dead twigs of Bauhinia variegata L. (Fabaceae), 25 July 2015, C Phukhamsakda, S1-11 (isotype in HKAS, under the code of HKAS 99513); ibid., on dead twigs of Bauhinia purpurea L. (Fabaceae), 5 May 2016, C Phukhamsakda, S1_03_16, ex-paratype living culture, MFLUCC 17-2205.
Distribution.
Phrae Province, Thailand.
Notes.
Neoaquastroma bauhiniae is similar to N. krabiense, but the ascomata, asci and ascospores are smaller and the species also has a thinner peridium with 4−7 septate hyaline ascospores. Thus, Neoaquastroma bauhiniae is introduced as a second species in Neoaquastroma based on its unique morphology coupled with high support values from the phylogenetic analysis (100% ML/1.00 PP, Fig. 1). Tanaka et al. (2015) only described the asexual morph in Parabambusicola to produce spermatia. We now obtained a single spore isolate which produces both sexual and asexual morphs in culture. The asexual morph of Neoaquastroma bauhiniae produced pycnidial conidiomata with hyaline conidia (Fig. 2, u–z).
Neoaquastroma krabiense
C. Phukhams. & K.D. Hyde sp. nov.
824674
Figure 3.
Neoaquastroma krabiense (MFLU 17-0003, holotype) a Barringtonia acutangula (L.) Gaertn specimens b Appearance of ascomata on host surface c Close up of ascomata d Ascomata forming on rice straw on WA after 8 weeks e, f Section of ascoma g Ostiolar canal h Section of partial peridium layer i Hyaline pseudoparaphyses j–m Asci n–s Hyaline ascospores with visible mucilaginous sheath q Ascospores stained in Indian ink to show sheath u Germinated ascospore v, w Culture characteristics on MEA x, y Conidiomata forming in culture after 8 weeks z Conidiomatal wall aa–ad Conidiogenous cells and developing conidia ae Conidia n–p Ascospores in 5% of KOH reagent m, r Asci and ascospore in culture (on rice straw). Scale bars: 500 µm (c–e); 200 µm (f, x); 50 µm (g–m, y), 20 µm (n–u, z); 5 µm (aa-af); 20 mm (v–w).
Etymology.
Name refers the location where this fungus was collected.
Type material.
THAILAND, Krabi Province: Meuang district, on dead twigs of Barringtonia acutangula (Lecythidaceae), 16 December 2015, C. Phukhamsakda, Kr015, MFLU 17-0003 (holotype), MFLUCC 16-0419 = ICMP 21572 (ex-type living culture).
Description.
Saprobic on dead twigs of Barringtonia acutangula (L.) Gaertn. Sexual morph. Ascomata 404–498 μm high × 290–319 μm diam. (x̄ = 426 × 300 μm, n = 10), immersed in bark, solitary, scattered or sometimes gregarious, compressed globose, with a flattened base, coriaceous, black to dark brown, smooth, papillate, ostiolate. Ostiole 137–146 μm high × 117–154 μm diam. (x̄ = 143 × 137 μm, n = 10), centrally located, oblong, filled with hyaline periphysoid. Peridium 45–73 μm wide (x̄ = 56, n = 30), cell width 3–12 (x̄ = 8 μm, n = 40) composed of 6–10(–13 at base) layers of blackish-brown to dark brown, with cells of textura angularis, outer layer heavily pigmented, inner layer composed of hyaline gelatinous cells. Hamathecium composed of numerous, dense, long, 1.6–2.4 μm (x̄ = 2 μm, n = 50), broad, filiform, transversely septate, branched, anastomosing, cellular pseudoparaphyses. Asci 95–169 × 29–45 μm (x̄ = 135 × 35 μm, n = 25), 8-spored, bitunicate, fisitunicate, oboviod to clavate, with furcate pedicel, ocular chamber clearly visible when immature. Ascospores 50–64 × 9–18 μm (x̄ = 57 × 13 μm, n = 50), bi-seriate or overlapping, fusiform, narrow towards the apex, hyaline, 5–8-transversely septate, constricted at the septa, cell above central septum slightly wider, rough-walled, indentations present when mature, granulate when stained with India ink, surrounded by 3–9 μm wide, mucilaginous sheath. Asexual morph coelomycetous, formed on rice straw agar. Conidiomata 84−90 μm high × 73–89 μm wide., pycnidial, uniloculate, confluent or scattered, superficial, covered with dense vegetative hyphae, globose, dark brown to black. Conidiomatal wall thin, brown to black-walled cells of textura angularis. Conidiophores reduced to conidiogenous cells. Conidiogenous cells 3−5 × 1.5−4 μm, enteroblastic, phialidic, integrated, broad-cylindrical to oblong, hyaline, formed from the inner layer of pycnidium wall. Conidia 2–4 × 1.5–2.5 μm (x̄ = 3 × 2 μm, n = 60), ellipsoidal to oblong, hyaline, aseptate, smooth-walled.
Culture characteristics.
Colonies on MEA, reaching 50 mm diam. after four weeks at 25 °C. Culture grey, becoming dark-olive brown after four weeks, of dense mycelia, colonies circular, flat, umbonate, raised from the agar in the centre, surface rough, dull, covered with aerial mycelium, white mycelium radiating into the agar, pale orange pigment diffusing in the agar; reverse black, dense, circular, with irregular, fimbriate margin. Sexual and asexual morphs formed in culture. Morphology of sexual phase similar to those on the substrates.
Additional material examined.
THAILAND, Krabi Province, Meuang district, on dead twigs of Barringtonia acutangula (Lecythidaceae), 16 December 2015, C. Phukhamsakda, Kr015, (isotype in HKAS, under the code of HKAS 99512).
Distribution.
Krabi Province, Thailand
Notes.
Neoaquastroma krabiense was collected in the southern part of Thailand on dead twigs of Barringtonia acutangula. It is placed in Neoaquastroma based on its morphology of both sexual and asexual morph and close phylogenetic affinity to other species of Neoaquastroma. Neoaquastroma krabiense is distinct in that it has a flattened ascomata base and larger and more slender asci and ascospores than N. guttulatum and N. bauhiniae. The species formed an asexual morph in culture (Fig. 3, m) as pycnidial conidiomata with hyaline conidia (Fig. 3, x-ae).
Discussion
In the present study, we introduce two new species of Neoaquastroma, as N. bauhiniae and N. krabiense. The descriptions were made from fungi isolated from dicotyledonous plants in Thailand. The new species are introduced based on multi-locus phylogeny coupled with morphology that support their placement within Parabambusicolaceae.
Parabambusicolaceae is typified with Parabambusicola Tanaka & K. Hiray. The type of the genus was described originally as Massarina bambusina Teng (Teng, 1936) from bamboo. The family is characterised by ascomata surrounded by stromatic tissues and multiseptate, clavate to fusiform and hyaline ascospores (Tanaka and Harada 2003, Tanaka et al. 2015). The asexual morph in Parabambusicolaceae can be coelomycetous or hyphomycetous. Sporodochia or pycnidia with hyaline conidia are formed in Parabambusicola and Neoaquastroma (Tanaka et al. 2015, this study), while hyphomyceteous structures are known from Pseudomonodictys and Monodictys spp. (Ariyawansa et al. 2015, Tanaka et al. 2015).
Neoaquastroma was introduced as a distinct genus in Parabambusicolaceae, with N. guttulatum as the type species (Wanasinghe et al. 2017). The genus resembles Parabambusicola and Multiseptospora, but form distinct lineages in phylogenetic studies (Liu et al. 2015, Tanaka et al. 2015, Wanasinghe et al. 2017). Parabambusicola and Neoaquastroma are similar in their morphology. The differentiation between Multiseptospora, Neoaquastroma and Parabambusicola is predominantly based on the morphology of ascospores, particularly with the size and number of septa.
In the phylogenetic analyses of Wanasinghe et al. (2017), Parabambusicolaceae clustered into three clades, where Neoaquastroma guttulatum (MFLUCC 14-0917) clustered with Aquastroma magniostiolata (KT 2485), Multilocularia bambusae (MFLUCC 11-0180), Monodictys sp. (JO 10, KH 331) and Pseudomonodictys tectonae (MFLUCC 12-0552) with high statistical support. In this study, Neoaquastroma forms a separate clade, sister to Multiseptospora and Parabambusicola. This is probably due to limited taxon sampling.
Supplementary Material
Acknowledgements
The authors would like to thank the Royal Golden Jubilee PhD Program under Thailand Research Fund (RGJ) and the German Academic Exchange Service (DAAD) for a joint TRF-DAAD (PPP 2017–2018) academic exchange grant to K.D. Hyde and M. Stadler and the RGJ for a personal grant to C. Phukhamsakda (The scholarship no. PHD/0020/2557 to study towards a PhD). Dr. Shaun Pennycook for checking and suggesting Latin names of the new taxa. Dr. Rajesh Jeewon for his suggestions on the phylogenetic analysis.
Citation
Phukhamsakda C, Bhat DJ, Hongsanan S, Xu J-C, Stadler M, Hyde KD (2018) Two novel species of Neoaquastroma (Parabambusicolaceae, Pleosporales) with their phoma-like asexual morphs. MycoKeys 34: 47–62. https://doi.org/10.3897/mycokeys.34.25124
References
- Ahmed SA, Van De Sande WW, Stevens DA, Fahal A, Van Diepeningen AD, Menken SB, de Hoog GS. (2014) Revision of agents of black-grain eumycetoma in the order Pleosporales. Persoonia 33: 141–154. http://doi.org/10.3767/003158514X684744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariyawansa HA, Hyde KD, Jayasiri SC, Buyck B, Chethana T, Dai DQ, Dai YC, Daranagama DA, Jayawardena RS, Luecking R, Ghobad-Nejhad M. (2015) Fungal diversity notes 111–252 – taxonomic and phylogenetic contributions to fungal taxa. Fungal Diversity 75(1): 27–274. http://dx.doi.org/10.1007/s13225-015-0346-5 [Google Scholar]
- Carbone I, Kohn LM. (1999) A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia 91(3): 553–556. http://doi.org/10.2307/3761358 [Google Scholar]
- Dai DQ, Bhat DJ, Liu JK, Chukeatirote E, Zhao RL, Hyde KD. (2012) Bambusicola, a new genus from bamboo with asexual and sexual morphs. Cryptogamie Mycologie 33(3): 363–379. https://doi.org/10.7872/crym.v33.iss3.2012.363 [Google Scholar]
- Dai DQ, Phookamsak R, Wijayawardene NN, Li WJ, Bhat DJ, Xu JC, Taylor JE, Hyde KD, Chukeatirote E. (2017) Bambusicolous fungi. Fungal Diversity 82(1): 1–105. https://doi.org/10.1007/s13225-016-0367-8 [Google Scholar]
- Huelsenbeck JP, Ronquist F. (2001) MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17: 754–755. https://doi.org/10.1093/bioinformatics/17.8.754 [DOI] [PubMed] [Google Scholar]
- Hyde KD. (1989) Ecology of tropical marine fungi. Hydrobiologia 178: 199–208. https://doi.org/10.1007/BF00006027 [Google Scholar]
- Jaklitsch WM, Olariaga I, Voglmayr H. (2016) Teichospora and the Teichosporaceae. Mycological Progress 15(31): 1–20. https://doi.org/10.1007/s11557-016-1171-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayasiri SC, Hyde KD, Ariyawansa HA, Bhat J, Buyck B, Cai L, Dai YC, Abd-Elsalam KA, Ertz D, Hidayat I, Jeewon R. (2015) The Faces of fungi database: fungal names linked with morphology, molecular and human attributes. Fungal Diversity 74(1): 3–18. https://doi.org/10.1007/s13225-015-0351-8 [Google Scholar]
- Jeewon R, Hyde KD. (2016) Establishing species boundaries and new taxa among fungi: recommendations to resolve taxonomic ambiguities. Mycosphere 7(11): 1669–1677. https://doi.org/10.5943/mycosphere/7/11/4 [Google Scholar]
- Jones EBG. (2000) Marine fungi: some factors influencing biodiversity. Fungal Diversity 4: 53–73. [Google Scholar]
- Jones EBG, Pilantanapak A, Chatmala I, Sakayaroj J, Phongpaichit S, Choeyklin R. (2006) Thai marine fungal diversity. Songklanakarin Journal of Science and Technology 28: 687–708. [Google Scholar]
- Katoh K, Standley K. (2013) MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Molecular Biology and Evolution 30(4): 772–780. http://dx.doi.org/10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knapp DG, Kovács GM, Zajta E, Groenewald JZ, Crous PW. (2015) Dark septate endophytic pleosporalean genera from semiarid areas. Persoonia 35: 87–100. https://doi.org/10.3767/003158515X687669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson A. (2014) AliView: a fast and lightweight alignment viewer and editor for large data sets. Bioinformatics 30(22): 3276–3278. http://doi.org/10.1093/bioinformatics/btu531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li GJ, Hyde KD, Zhao RL, Hongsanan S, Abdel-Aziz FA, Abdel-Wahab MA, Alvarado P, Alves-Silva G, Ammirati JF, Ariyawansa HA, Baghela A. (2016) Fungal diversity notes 253–366: taxonomic and phylogenetic contributions to fungal taxa. Fungal Diversity 78(1): 1–237. http://doi.org/10.1007/s13225-016-0366-9 [Google Scholar]
- Liu JK, Hyde KD, Jones EG, Ariyawansa HA, Bhat DJ, Boonmee S, Maharachchikumbura SS, McKenzie EH, Phookamsak R, Phukhamsakda C, Shenoy BD. (2015) Fungal Diversity Notes 1–110: Taxonomic and phylogenetic contributions to fungal species. Fungal Diversity 72(1): 1–197. http://doi.org/10.1007/s13225-015-0324-y [Google Scholar]
- MacKinnon J, MacKinnon K, Child G, Thorsell J. (1986) Managing Protected Areas in the Tropics IUCN, Gland, Switzerland and Cambridge, UK, 295.
- Mapook A, Hyde KD, Dai DQ, Li J, JONES EG, Bahkali AH, Boonmee S. (2016) Muyocopronales, ord nov, (Dothideomycetes, Ascomycota) and a reappraisal of Muyocopron species from northern Thailand. Phytotaxa 265: 225–237. http://doi.org/10.11646/phytotaxa.265.3.3 [Google Scholar]
- Marod D, Kutintara U. (2012) Biodiversity observation and monitoring in Thailand. The Biodiversity Observation Network in the Asia-Pacific Region. Springer, Japan, 53–63. https://doi.org/10.1007/978-4-431-54032-8_5
- Doilom M, Dissanayake AJ, Wanasinghe DN, Boonmee S, Liu JK, Bhat DJ, Taylor JE, Bahkali AH, McKenzie EH, Hyde KD. (2017) Microfungi on Tectona grandis (teak) in Northern Thailand. Fungal Diversity 82(1): 107–182. http://doi.org/10.1007/s13225-016-0368-7 [Google Scholar]
- MycoBank (2018) MycoBank. http://www.MycoBank.org. [Accessed in March 2018]
- Nylander JAA. (2004) MrModeltest v2 Program distributed by the author. Evolutionary Biology Centre, Uppsala University.
- O’Meara BC, Ané C, Sanderson MJ, Wainwright PC. (2006) Testing for different rates of continuous trait evolution using likelihood. Evolution 60: 922–933. https://doi.org/10.1111/j.0014-3820.2006.tb01171.x [PubMed] [Google Scholar]
- Phukhamsakda C, Ariyawansa HA, Phillips AJ, Wanasinghe DN, Bhat DJ, McKenzie EH, Singtripop C, Camporesi E, Hyde KD. (2016) Additions to Sporormiaceae: Introducing two novel genera, Sparticola and Forliomyces, from Spartium. Cryptogamie, Mycologie 37(1): 75–97. https://doi.org/10.7872/crym/v37.iss1.2016.75 [Google Scholar]
- Phukhamsakda C, Bhat DJ, Hongsanan S, Tibpromma S, Yang JB, Promputtha I. (2017) Magnicamarosporium diospyricola sp. nov. (Sulcatisporaceae) from Thailand. Mycosphere 8(4): 512–520. https://doi.org/10.5943/mycosphere/8/4/3 [Google Scholar]
- Phukhamsakda C, Hongsanan S, Ryberg M, Ariyawansa HA, Chomnunti P, Bahkali AH, Hyde KD. (2016) The evolution of Massarineae with Longipedicellataceae fam. nov. Mycosphere 7(11): 1713–1731. https://doi.org/10.5943/mycosphere/7/11/7 [Google Scholar]
- Quaedvlieg W, Verkley GJ, Shin HD, Barreto RW, Alfenas AC, Swart WJ, Groenewald JZ, Crous PW. (2013) Sizing up Septoria. Studies in Mycology 75: 307–339. https://doi.org/10.3114/sim0017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut A, Drummond A. (2008) FigTree: Tree figure drawing tool, version 12 2 Institute of Evolutionary Biology, University of Edinburgh
- Rambaut A, Suchard MA, Xie D, Drummond AJ. (2014) Tracer v16, Available from http://beast.bio.ed.ac.uk/Tracer
- Rostrup E. (1902) Flora of Koh Chang Contributions to the knowledge of the vegetation of the gulf of Siam. Fungi Botanisk Tidsskrift 24: 355–363. https://doi.org/10.5962/bhl.title.55188 [Google Scholar]
- Schoch CL, Crous PW, Groenewald JZ, Boehm EW, Burgess TI, De Gruyter J, De Hoog GS, Dixon LJ, Grube M, Gueidan C, Harada Y. (2009) A class-wide phylogenetic assessment of Dothideomycetes. Studies in Mycology 64: 1–15. https://doi.org/10.3114/sim.2009.64.01 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher T. (1982) Ascomycetes from Northern Thailand. Nordic Journal of Botany 2: 257–263. https://doi.org/10.1111/j.1756-1051.1982.tb01187.x [Google Scholar]
- Silvestro D, Michalak I. (2011) raxmlGUI: a graphical front-end for RAxML. Organisms Diversity & Evolution 12(4): 335–337. https://doi.org/10.1007/s13127-011-0056-0 [Google Scholar]
- Stamatakis A. (2006) RAXML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22: 2688–2690. https://doi.org/10.1093/bioinformatics/btl446 [DOI] [PubMed] [Google Scholar]
- Stamatakis A. (2014) RAxML Version 8: A tool for Phylogenetic Analysis and Post-Analysis of Large Phylogenies. Bioinformatics 30: 1312–1313. https://doi.org/10.1093/bioinformatics/btu033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A, Hoover P, Rougemont J. (2008) A rapid bootstrap algorithm for the RAxML web servers. Systematic biology 57: 758–771. https://doi.org/10.1080/10635150802429642 [DOI] [PubMed] [Google Scholar]
- Suetrong S, Schoch CL, Spatafora JW, Kohlmeyer J, Volkmann-Kohlmeyer B, Sakayaroj J, Phongpaichit S, Tanaka K, Hirayama K, Jones EB. (2009) Molecular systematics of the marine Dothideomycetes. Studies in Mycology 64: 155–173. https://doi.org/10.3114/sim.2009.64.09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swofford DL. (2002) PAUP*: phylogenetic analysis using parsimony (* and other methods) Sunderland, MA. https://doi.org/10.1002/0471650129.dob0522
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. (2013) MEGA6: Molecular Evolutionary Genetics Analysis Version 60. Molecular Biology and Evolution 30: 2725–2729. https://doi.org/10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K, Harada Y. (2003) Pleosporales in Japan (1): the genus Lophiostoma. Mycoscience 44(2): 85–96. https://doi.org/10.1007/s10267-002-0085-9 [Google Scholar]
- Tanaka K, Harada Y. (2003) Pleosporales in Japan (3) The genus Massarina. Mycoscience 44(3): 173–185. https://doi.org/10.1007/s10267-003-0102-7 [Google Scholar]
- Tanaka K, Hirayama K, Yonezawa H, Sato G, Toriyabe A, Kudo H, Hashimoto A, Matsumura M, Harada Y, Kurihara Y, Shirouzu T. (2015) Revision of the Massarineae (Pleosporales, Dothideomycetes). Studies in Mycology 82: 75–136. https://doi.org/10.1016/j.simyco.2015.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng SC. (1936) Additional fungi from China II. Sinensia 7: 490–527. [Google Scholar]
- Vilgalys R, Hester M. (1990) Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. Journal of bacteriology 172(8): 4238–4246. https://doi.org/10.1128/jb.172.8.4238-4246.1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanasinghe DN, Hyde KD, Konta S, To-Anun C, Jones EG. (2017) Saprobic Dothideomycetes in Thailand: Neoaquastroma gen. nov. (Parabambusicolaceae) introduced based on morphological and molecular data. Phytotaxa 302: 133–144. http://doi.org/10.11646/phytotaxa.302.2.3 [Google Scholar]
- Wendt L, Sir EB, Kuhnert E, Heitkämper S, Lambert C, Hladki AI, Romero AI, Luangsa-ard JJ, Srikitikulchai P, Peršoh D, Stadler M. (2018) Resurrection and emendation of the Hypoxylaceae, recognised from a multigene phylogeny of the Xylariales. Mycological Progress 17(1–2): 115–154. http://doi.org/10.1007/s11557-017-1311-3 [Google Scholar]
- White TJ, Bruns T, Lee SJ, Taylor JW. (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR protocols: a guide to methods and applications 18(1): 315–322. https://doi.org/10.1016/B978-0-12-372180-8.50042-1 [Google Scholar]
- Wijayawardene NN, Hyde KD, Lumbsch HT, Liu JK, Maharachchikumbura SS, Ekanayaka AH, Tian Q, Phookamsak R. (2018) Outline of Ascomycota: 2017. Fungal Diversity 88(1): 167–263. http://doi.org/10.1007/s13225-018-0394-8 [Google Scholar]
- Wijayawardene NN, Hyde KD, Rajeshkumar KC, Hawksworth DL, Madrid H, Kirk PM, Braun U, Singh RV, Crous PW, Kukwa M, Lücking R. (2017) Notes for genera-Ascomycota. Fungal Diversity 86(1): 1–594. http://doi.org/10.1007/s13225-017-0386-0 [Google Scholar]
- Zhang Y, Fournier J, Pointing SB, Hyde KD. (2008) Are Melanomma pulvis-pyrius and Trematosphaeria pertusa congeneric?. Fungal Diversity 33: 47–60. http://doi.org/10.13140/2.1.3875.1364 [Google Scholar]
- Zhang Y, Wang HK, Fournier J, Crous PW, Jeewon R, Pointing SB, Hyde KD. (2009) Towards a phylogenetic clarification of Lophiostoma/Massarina and morphologically similar genera in the Pleosporales. Fungal Diversity 38: 225–251. [Google Scholar]
- Zhaxybayeva O, Gogarten JP. (2002) Bootstrap, Bayesian probability and maximum likelihood mapping: exploring new tools for comparative genome analyses. BMC genomics 3(1): 1−4. https://doi.org/10.1186/1471-2164-3-4 [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.