Abstract
c-Myc regulates cellular proliferation, differentiation, and apoptosis. Temporal expression of c-Myc during all-trans-retinoic acid (RA)-mediated neural differentiation in murine embryonic stem cell (ES) was investigated. Correlation to the modulation of dimerizing partners Max and Mad that may influence c-Myc signaling and transcription regulation was elucidated for the first time in these cells. In RA-treated cells, increase in c-myc mRNA was detected by reverse transcriptase polymerase chain reaction on days 11 and 14 of differentiation compared with the vehicle-treated controls. The results were further corroborated by ribonuclease protection assay (RPA). Western blots revealed an increase in c-Myc protein only on day 14 of differentiation in RA-treated cells. Increases in max and mad gene transcription detected by RPA at times of elevated c-Myc in RA-treated ES cells suggest that a transient increase in c-Myc protein expression may influence differential dimerization of Myc partners needed for signaling in the neural differentiation of ES cells.
Keywords: Murine embryonic stem cells, c-Myc, Differentiation, Retinoic acid, Max, Mad
ONCOGENIC transcription factor c-Myc regulates cellular proliferation, differentiation, cellular adhesion, metabolism, and apoptosis (17). Activation of c-Myc by different pathways has been noted in oncogenic transformation of cells. Normally expressed c-Myc is extensively regulated. However, deregulation is the cardinal feature in malignant transformations of cells. The confirmation that c-Myc regulates the cell cycle comes from data relating to the increase in the doubling time of nullizygous c-myc cells (29).
Several studies have suggested that c-Myc plays an important role during embryonic development and growth (38). Homozygous deletion of the c-myc gene leads to embryonic lethality at 10.5 days postcoitum (20). It has been noted that c-myc expression in mice is highest in the proliferating tissues of mesodermal origin. Conversely, ectodermal and endodermal tissues express little or no c-myc (24,49,57). Several studies have reported a decrease in c-myc mRNA with the initiation of differentiation and growth arrest (35,47,55). However, in some cell types a decrease in c-myc expression is noted on terminal differentiation (13,27,48). Overexpression of c-myc leads to nonregulation and consequently a reduced requirement for growth factors, accelerated cell division, and increased cell size (32,50,52). Consequently, elevated expression of c-myc can impede differentiation in many cell types (18). In addition, exposure of antisense oligonucleotide to c-myc leads to growth arrest and terminal differentiation in F9 and erythroleukemic HL-60 cells (28,30). Contrary to the above findings, c-myc RNA has been reported to undergo a transient increase in differentiating lens cells (43). Lens maturation studies in transgenic mice have also shown that differentiation can proceed in the presence of elevated levels of c-myc (41).
The mechanism by which c-Myc influences differentiation has not been fully elucidated. It has been speculated that c-Myc, in conjunction with the Max family of proteins, promotes the activation of genes that control proliferation (4,10) and that the process is inhibited by Mad (6). Max/Max and Max/Mad heterodimers have been thought to repress c-Myc targets and promote differentiation (2,33).
All-trans-retinoic acid (RA) is an important metabolite of retinol (vitamin A) that mediates epidermal as well as bone growth, differentiation, reproductive, and immune functions (14) by its interaction with various retinoid receptors (RAR) and retinoid X receptors (RXR) (37). The retinoid–receptor complexes influence gene regulation by inducing or repressing gene transcription. Murine embryonic stem cells (ES) were chosen to examine the influence of RA on c-Myc expression during differentiation and apoptosis. Embryonic cells are derived from the inner cell mass of 4-day blastocysts and can be maintained in an undifferentiated state by growing them on fibroblast feeder layers (22). The ES cells respond to various internal and external signals of proliferation and differentiation, thus mimicking the in vivo differentiation process. All-trans-RA has been used extensively as a differentiating agent in ES neuronal models (8,53) because of its ability to mediate proliferation, differentiation, and cell death by regulating gene expression (39).
Because retinoids play an important role in proliferation, differentiation, and morphogenesis, the aim of this study was to investigate RA-mediated modulation of the Myc family of genes, which appear to be the key players in the cellular decision-making process between differentiation and proliferation during cellular growth. Carcinogenesis entails dedifferentiation and reverting back to embryonic stages. A basic understanding of embryonic development requires an appreciation of the complexity involved in Myc signaling and facilitates understanding the loss of orderly control during oncogenic transformation. Our study demonstrates the temporal pattern of c-myc, max, and mad expression during RA-mediated differentiation in murine embryonic stem cells.
MATERIALS AND METHODS
ES Cell Culture and RA Treatment
Murine embryonic stem cells (ES-D3, ATCC # 1934-CRL) were maintained on mouse fibroblast feeder layers, (STO, ATCC # 1503-CRL) treated with 10 μg/ml mitomycin c, in Dulbecco’s modified Eagle’s medium. The medium was supplemented with 15% Knock-Out™ Serum Replacement (Gibco, Life Technologies, Grand Island, NY), 10 μM β-mercaptoethanol, and 1000 IU/ml leukemia inhibitory factor (LIF; Sigma, St. Louis, MO). The medium was changed every day. The ES cells were passaged every 2 days to maintain an undifferentiated state. To induce differentiation, 2 × 105 ES cells were plated in a monolayer in the absence of feeder layers and LIF on six-well plates (Falcon, Becton Dickinson Labware, Franklin Lakes, NJ), which was counted as day 1 of differentiation. It has been published previously that no difference had been noted in differentiation potential between ES cells grown in monolayer or suspension form (22). All-trans-retinoic acid (Sigma) dissolved in 85% ethanol at a concentration of 10−6 M was added to the medium on days 8, 9, and 10 of differentiation. Our preliminary experiments have indicated the optimal dose and duration of treatment with RA for neural differentiation of these cells. The control medium was treated with equal volume of 85% ethanol that was added to the experimental medium. The total volume of ethanol in the culture medium did not exceed 0.15% and did not influence the differentiation process.
c-myc mRNA Expression by Reverse Transcriptase-Polymerase Chain Reaction (RT-PCR)
ES cells were grown in the absence of fibroblasts and LIF in six-well plates at a density of 2 × 105. The cells were harvested at 0 (undifferentiated), 12, 24, 36, and 48 h after differentiation in the absence of RA. To look at cellular response in the presence of RA, ES cells seeded at similar density were treated on days 8, 9, and 10 of differentiation with 10−6 M RA and harvested on days 11, 14, 17, and 21 of differentiation to investigate modulation of c-myc genes in response to RA. Control cells were treated with equal volume of 85% ethanol. One milliliter of TRI Reagent® LS (Molecular Research, Cincinnati, OH) was added to the cell suspension, and total RNA was extracted according to the manufacturer’s protocol. Three micrograms of total RNA was used to synthesize first-strand cDNA using Superscript II reverse transcriptase (Life Technologies). Samples equivalent to 1 μl of the first-strand reaction cDNA were then used as a template for amplification in 50 μl PCR reaction. Two microliters of dimethyl sulphoxide (DMSO) (Fisher, Fair Lawn, NJ) was added to c-myc reaction to enhance PCR specificity. The sense and antisense primers for β-actin and c-myc were chosen by the Primer3 program (Whitehead Institute, Cambridge, MA). These were 5′-ATGGATGACGATAT CGCT-3′ and 5′-ATGAGGTAGTCTGTCAGGT-3′, 5′-ATCTGCGACGAGGAAGAGAA-3′ and 5′-ATCGCAGATGAAGCTCTGGT-3′ for β-actin and c-myc, respectively. The DNA was initially denatured for 5 min at 95°C. The following parameters were used for amplification: 95°C for 30 s, annealing temperature of 48°C (β-actin) and 55°C (c-myc) for 30 s, 72°C for 1 min and run for 25 cycles and 33 cycles for β-actin and c-myc, respectively. Initial trials were run to ensure that the genes are amplified in the exponential phase (Fig. 1, inset). β-actin was used as a positive control, and negative controls with respective primers and no cDNA in the PCR reactions were used. After amplification, 10 μl of PCR products was mixed with 10× DNA dye [5 mg/ml bromophenol blue, 50% glycerol, 100 mM Tris, 20 mM NaCl, 1 mM ethylenediaminetetraacetic acid (EDTA)] in 10:1 ratio, and run on 2.0% agarose gel stained with ethidium bromide for electrophoretic separation. The gels were photographed in a backlighted UV transilluminator. Photographs were scanned and quantified on UN-SCAN-IT™ automated digitizing system (Silk Scientific, Inc., Orem, UT). Total pixel count for c-myc was normalized to β-actin. All experiments were repeated several times with the number of independent replicates (n = 3) for each investigated time point.
Figure 1.
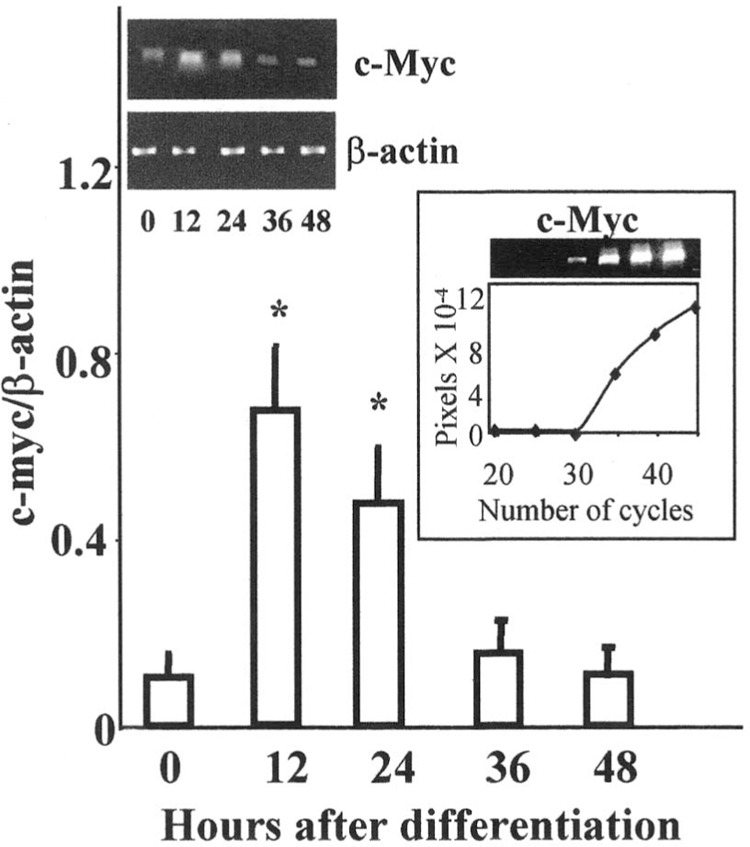
Increased c-myc mRNA expression in spontaneously dividing ES cells at 12 and 24 h as detected by RT-PCR. The mRNA expression was normalized to house keeping gene β-actin at 0 = undifferentiated, 12, 24, 36, and 48 h after initiation of differentiation. The products were amplified at 33 cycles in the exponential phase of PCR reaction and the gel representative of exponential amplification of c-myc is shown (inset). The results are expressed as mean ± SEM (n = 3). The photograph of representative gel shows β-actin and c-myc mRNA at respective times. *Significantly different from undifferentiated group at p ≤ 0.05.
Determination of c-Myc Protein by Western Blot
The control and RA-treated ES cells were lysed on ice for 30 min in lysis buffer (54). Cell lysates were centrifuged at 14,000 rpm at 4°C for 15 min to remove cell debris. The aliquots of supernatants were stored at −80°C and assayed using Bradford reagent (Sigma) for detection of protein. Fifteen micrograms of the supernatant protein from each sample was heated with 4× lauryl sulphate (SDS) sample buffer at 95°C for 5 min and separated on 10% SDS-polyacyramide gel electrophoretically. Subsequently, the proteins were transferred onto 0.45-μm pore size nitrocellulose membranes for 90 min and blocked overnight with 5% milk in Tween-Tris buffer saline (TBS). Nitrocellulose membranes were then exposed to c-Myc primary monoclonal antibody (Santa Cruz Biotechnologies, Santa Cruz, CA). The membranes were incubated for 1 h with horseradish peroxide-conjugated polyclonal goat anti-mouse IgG at 1:5000 dilutions. Between each exposure, nitrocellulose membranes were washed for 30 min with Tween-TBS. c-Myc was visualized autoradiographically by enhanced chemiluminescent substrate (ECL® Amersham Pharmacia, Piscataway, NJ) after 60 min of exposure. The radiographs were scanned and digitized using UN-SCAN-IT™ automated digitizing system as mentioned for PCR gels above. All experiments were repeated several times with the number of replicates (n = 3) for each investigated time point.
Interaction of Max and Mad With c-Myc During RA-Mediated Cellular Differentiation by Ribonuclease Protection Assay (RPA)
Ribonuclease protection assay for c-myc-max/mad family was undertaken to simultaneously analyze their gene expression during ES differentiation. RNase protection assay is a sensitive and quantitative method of measuring the expression level of several genes simultaneously. The assay was performed with Pharmingen’s Riboquant, multiprobe RNase assay custom kit (Pharmingen, San Diego, CA). ES cells were grown at a density of 2 × 105 cells in six-well plates. The cells were harvested at undifferentiated = 0, 24, and 48 h after differentiation in the absence of RA. To investigate RA-mediated differentiation, cells plated in similarly densities were treated with RA as mention before and harvested at days 11, 14, 17, and 21 of differentiation. One milliliter of TRI Reagent® LS (Molecular Research, Cincinnati, OH) was added to the cell suspension and total RNA was extracted according to the manufacturer’s protocol. Total RNA from three wells was pooled together and quantified in the spectrophotometer at A260 and 40 μg aliquoted and stored at −80°C. α-32P-labeled antisense RNA probe was synthesized according to manufacturer’s protocol from custom DNA templates and quantified in a liquid scintillating counter (1214 Rackbeta, Pharmacia, Finland). The probe was diluted with hybridizing buffer at strengths of 4 × 105/μl counts per minute and hybridized overnight to the target RNA previously extracted from RA- and vehicle-treated ES cells. Appropriate positive and yeast tRNA negative controls, provided by the manufacturer, were simultaneously hybridized. Subsequently the free probe and single-stranded RNA were treated with RNAse A to destroy single-stranded RNA. The cRNA–mRNA complexes were purified and electrophoresed on denaturing polyacrylamide gels. The gels were transferred onto filter papers and dried at 80°C for 1 h and quantified by autoradiography after 16 h of exposure. During the duration of exposure, the gels were stored at −80°C.
Statistical Analysis and Replications
All cell culture experiments were repeated several times using at least three independent replicates. Pooled RNA from three wells was used for RPA. The RPA was repeated twice for consistency. Mean ± SEM of a representative experiment is presented in the results. The difference between control and treated samples was analyzed using Student’s t-test assuming equal variances. The error bars represent the SEM. A probability of p ≤ 0.05 was considered significant.
RESULTS
Expression of c-myc Gene by RT-PCR and RPA
Mouse ES cells can be induced to differentiate into cells exhibiting neural morphology with RA, whereas absence of RA leads to differentiation into mesodermal tissue. To answer the question of temporal pattern of c-Myc expression during ES neural differentiation, c-myc gene and protein expression was investigated by RT-PCR, RPA, and Western blot. However, because Max and Mad essentially regulate c-Myc signaling, it was important to elucidate the pattern of gene expression of the dimerizing partners simultaneously; the subsequent experiments were therefore performed by RPA. Undifferentiated ES cells grown in the presence of fibroblast feeder layers and leukemia inhibitory factor (LIF) express basal quantities of c-myc gene. Changes in the level of c-myc were noted upon induction of spontaneous differentiation in the absence of feeder layers, LIF, or RA. Figure 1 shows expression of c-myc gene normalized to β-actin (house keeping gene) at undifferentiated = 0, 12, 24, 36, and 48 h of differentiation. A transient increase in the level of c-myc mRNA was noted at 12 and 24 h after initiation of differentiation and subsequently a decline was observed by 48 h as detected by RT-PCR (Fig. 1). The results obtained by RT-PCR were corroborated by RPA. Total RNA was extracted from the ES cells, hybridized to template RNA, and treated with RNase A, as explained in Materials and Methods. The assay revealed an increased expression of c-myc RNA after 24 h of initiation of differentiation, which was also found to be statistically significant (p ≤ 0.05) compared with the undifferentiated controls harvested at 0 h. Similar to the observations of RT-PCR reactions, after 48 h of initiation of differentiation c-myc mRNA levels returned to basal values of their undifferentiated phenotype as noted by RPA (Fig. 2A, B).
Figure 2.
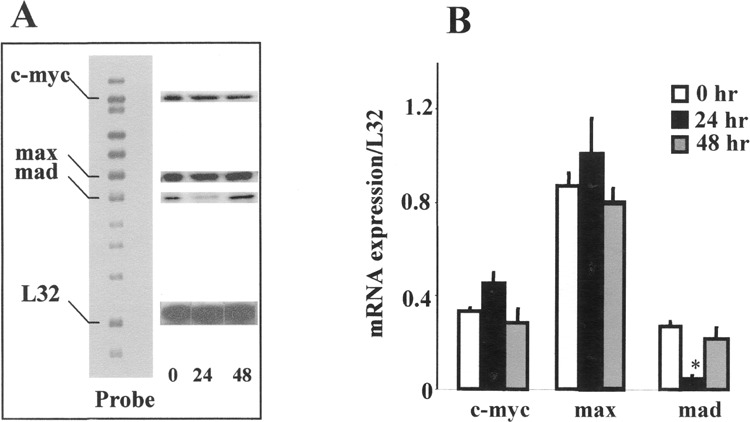
Differential expression of c-myc, max, and mad mRNA expression by RNase protection assay (RPA). (A) Representative gel of RPA showing only c-myc, max, and mad (indicated on the left) mRNA expression at 0 = undifferentiated, 24 and 48 h. L32 represents the housekeeping gene. A nonhybridized probe set was run as a size marker. (B) The relative mRNA expression normalized to housekeeping gene L32 at 0 = undifferentiated, 24 and 48 h after differentiation. The results are expressed as mean ± SEM (n = 2). *Significantly different from undifferentiated group at p ≤ 0.05.
When ES cells were allowed to differentiate in the presence of vehicle only, they differentiated into fibroblasts, endodermal, and mesodermal cell types. No neural differentiation was seen in the absence of RA. Modulation of c-myc gene expression during RA-mediated differentiation was also examined by RT-PCR and RPA. The ES cells were exposed to 1 μM RA on days 8, 9, and 10 of differentiation. Differentiation in the presence of RA leads to neural cell types. Interestingly, a distinct response was observed upon RA exposure by RT-PCR. The ratio of c-myc RNA, normalized to β-actin, was elevated in RA-treated cells relative to the ethanol-treated controls on all days. However, the increased gene expression on day 11 was significant (p ≤ 0.05) (Fig. 3A, B). Following RA exposure, an increase in c-myc mRNA expression was also detected by RPA in RA-treated cells compared with the concurrent control for the respective days (11, 14, 17, and 21) (Fig. 4A, B). The difference in gene expression between RA- and vehicle-treated cells was found to be statistically significant on days 11 and 14 of differentiation. Maximal amount of RNA expression was seen on day 14 in RA-treated cells, and a subsequent decline was seen from days 17 to 21. Additionally, even during the declining phase of c-myc expression, about two-to threefold higher level of c-myc RNA was detected in RA-treated cells from concurrent controls on days 17 and 21.
Figure 3.
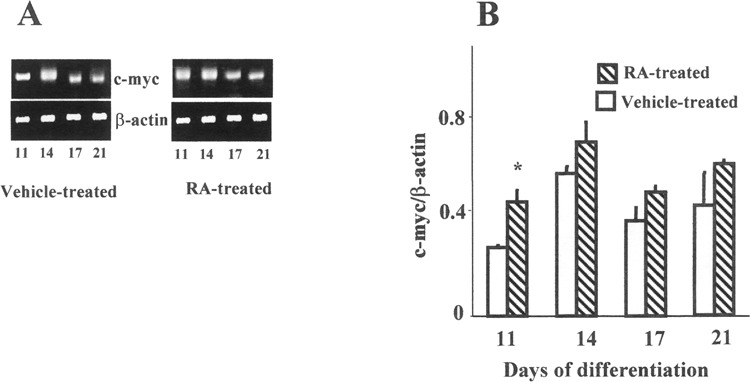
Modulation of c-myc mRNA expression by RT-PCR during RA-mediated ES neural differentiation. The cells were treated with RA on days 8, 9, and 10 of differentiation. (A) The photographs of representative gels show β-actin and c-myc mRNA on days 11, 14, 17, and 21 of differentiation. (B) The relative c-myc gene expression was normalized to β-actin. The results are expressed as mean ± SEM (n = 3). *Significantly different from concurrent control p ≤ 0.05.
Figure 4.
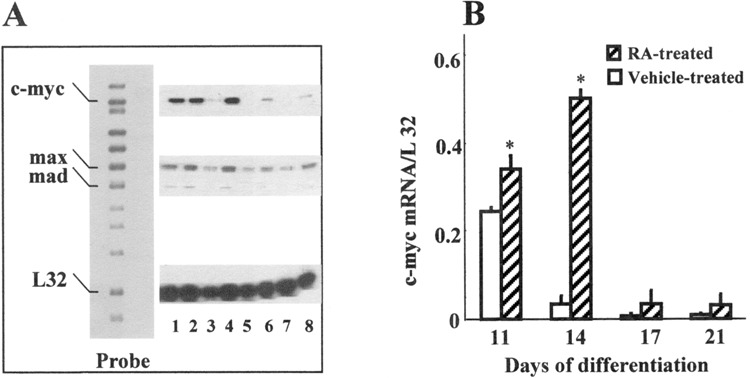
Alteration in c-myc expression during RA-mediated ES differentiation. (A) Representative gel of RNase protection assay. A nonhybridized probe was run as a size marker. The names of the genes are listed on the left. L32 represents the housekeeping gene. Treatment groups are indicated as control: (lane 1) 11-day vehicle-treated, (lane 3) 14-day vehicle-treated, (lane 5) 17-day vehicle-treated, and (lane 7) 21-day vehicle-treated. RA-exposed: (lane 2) 11-day RA-treated, (lane 4) 14-day RA-treated, (lane 6) 17-day RA-treated, and (lane 8) 21-day RA-treated. (B) The relative mRNA expression is indicated against L32. Pooled RNA from three independent wells was used for the assay, and the results are expressed as mean ± SEM (n = 2). *Significantly different from concurrent control at p ≤ 0.05.
Expression of c-Myc Protein
Upon initial spontaneous differentiation, c-Myc protein expression paralleled the gene expression and an increased amount of translated product was detected 36 h after differentiation, relative to the undifferentiated controls (Fig. 5A). Additionally, differentiation in the presence of RA on days 8, 9, and 10, which predominantly promotes the formation of cells resembling neural morphology, revealed a significant increase in the level of c-Myc protein on day 14 of differentiation, compared with the vehicle-treated 4:32 controls. Thereafter, a gradual decline was noted in the expressed protein. No difference in the level of protein expression was noted in the presence or absence of RA on subsequent days (Fig. 5B).
Figure 5.
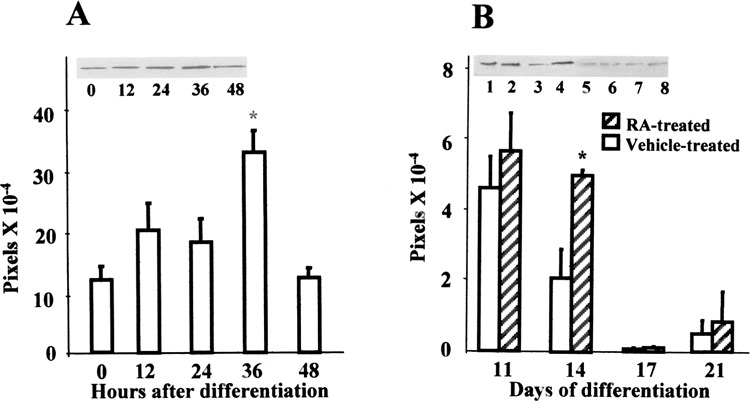
Alteration in c-Myc protein expression during differentiation by Western blot. (A) Fifteen micrograms of cytosolic cell lysate was electrophoretically separated and transferred onto nitrocellulose membranes. After incubation with primary and secondary antibody, the proteins were visualized autoradiographically with chemiluminiscent reagents and quantified as pixels. Results expressed are mean ± SEM (n = 3). Representative gel shows c-Myc expression at 0 = undifferentiated, 12, 24, 36, and 48 h after initiation of differentiation. *Significantly different from undifferentiated group at p ≤ 0.05. (B) Alteration in c-Myc protein expression during RA-mediated ES differentiation on days 11, 14, 17, and 21 of differentiation. The results are expressed as mean ± SEM (n = 3). *Significantly different from concurrent control at p ≤ 0.05. Representative photograph of gel is shown. Treatment groups are indicated as control: (lane 1) 11-day vehicle-treated, (lane 3) 14-day vehicle-treated, (lane 5) 17-day vehicle-treated, and (lane 7) 21-day vehicle-treated. RA-exposed: (lane 2) 11-day RA-treated, (lane 4) 14-day RA-treated, (lane 6) 17-day RA-treated, and (lane 8) 21-day RA-treated.
Expression of Related Myc Family of Genes Encoding for Dimerizing Partners Max and Mad by RPA
After having established that RA treatment during ES differentiation could modulate c-myc gene and protein levels, concurrent expression of the dimerizing partners of c-Myc was investigated to understand the implications of upregulation of c-Myc. No difference in max mRNA was detected by RPA upon initial differentiation at 24 and 48 h (Fig. 2). In vehicle-treated ES cells, not in RA-treated cells, max RNA expression declined twofold on day 14 from day 11 mRNA levels and subsequently remained steady throughout the period of differentiation. Increased expression of max mRNA was noted on all days in RA-treated ES cells compared with the concurrent vehicle-treated cells, and the increase was statistically significant (p ≤ 0.05) on days 11, 14, and 21 of differentiation (Fig. 4A, Fig. 6). About a threefold induction of max was noted on day 14 compared with concurrent vehicle-treated controls.
Figure 6.
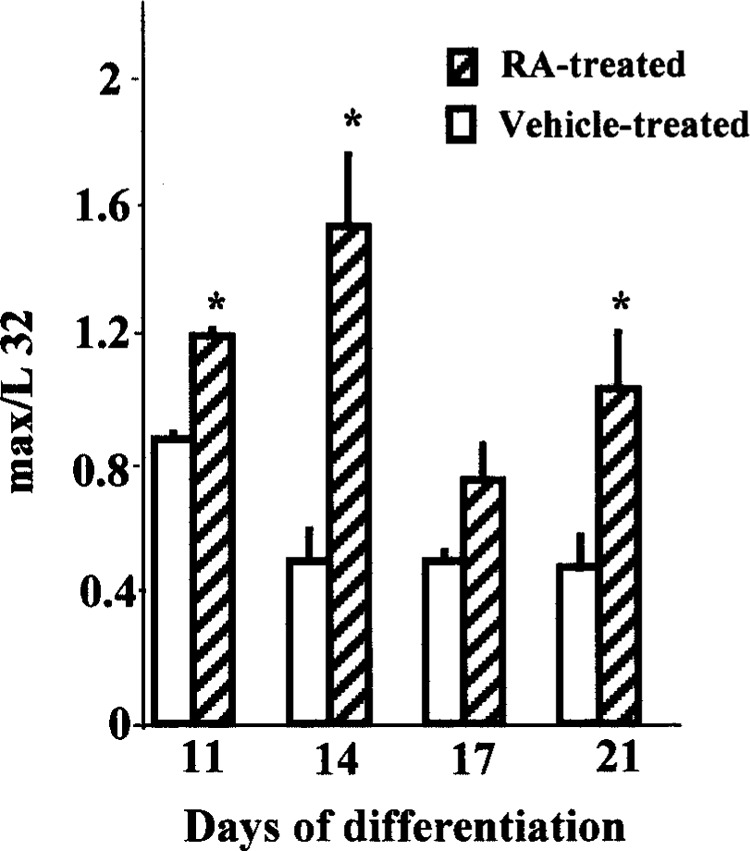
Alterations in max expression during RA-mediated ES differentiation. The relative mRNA expression is indicated against L32. Pooled RNA from three independent wells was used for the assay, and the results of a representative experiment are expressed as mean ± SE (n = 2). *Significantly different from concurrent control at p ≤ 0.05.
Contrary to max expression, a significant decline in the level of mad mRNA was noted at 24 h compared with the undifferentiated cells (Fig. 2A, B). However, increased mad mRNA was detected at 11 and 14 days upon RA treatment compared with the concurrent vehicle-treated cells. In addition, the level of mad mRNA expression was found to be significantly elevated from concurrent vehicle-treated cells on day 14 of differentiation (Fig. 4A, Fig. 7). Furthermore, nonquantifiable to very low levels of mad transcripts were detected on subsequent days of differentiation in both RA- and vehicle-treated cells.
Figure 7.
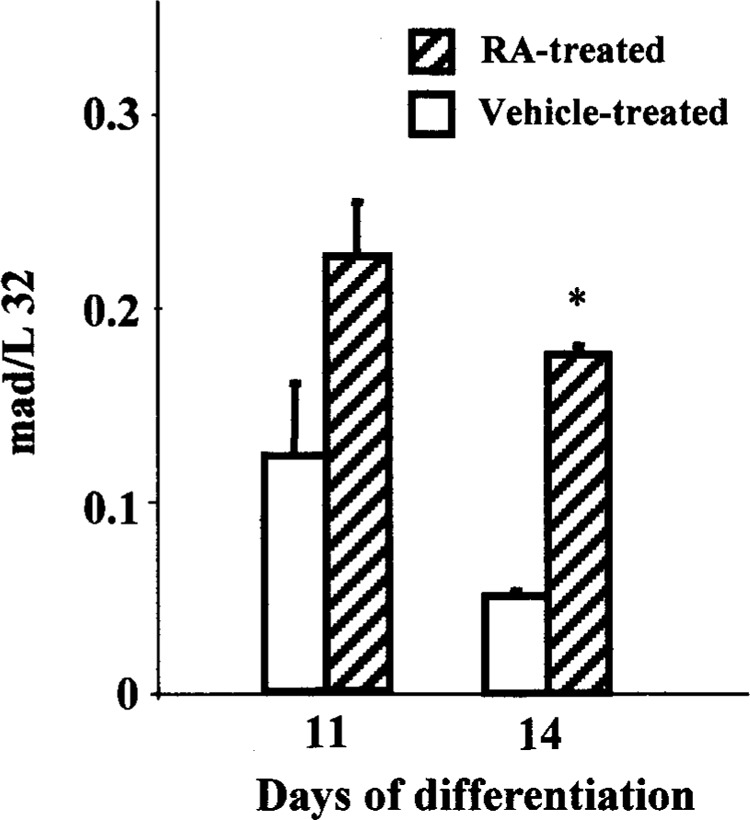
Alterations in mad expression during RA-mediated ES differentiation. The relative mRNA expression is indicated against L32. Pooled RNA from three independent wells was used for the assay, and the results are expressed as mean ± SE (n = 2). *Significantly different from concurrent control at p ≤ 0.05.
DISCUSSION
The results of this study show that RA can effectively modulate Myc family gene expression during ES neural differentiation. Undifferentiated ES cells express Myc and its dimerizing partners, Max and Mad. Upon spontaneous differentiation in the absence of feeder layer and LIF, an increase in c-myc mRNA and a decline in mad mRNA was observed at 24 h. No change in the level of max was noted. Exposure to RA at days 8, 9, and 10 during ES neural differentiation causes distinct changes in the Myc family of genes. The most prominent effect in response to RA exposure was an upregulation of c-myc gene and protein expression on day 14 of differentiation and a gradual decrease in their levels. That RA exposure can effectively modulate Myc family at specific stages of ES neural differentiation is suggested by increased presence of c-myc, max, and mad mRNA at specific times during ES neural differentiation.
A biphasic response was observed during differentiation of ES cells. Upon initiation of differentiation, even in the absence of differentiating-inducing agents, c-myc mRNA showed a transient increase in 12–24 h as detected by RT-PCR. This finding was also corroborated by RPA where an increase in c-myc mRNA levels was observed at 24 h and returned to basal levels of undifferentiated state at 48 h. This initial peak of increased expression of c-myc has been correlated to commitment to differentiate rather than RA exposure in P19 cells (12,42). Modulation of c-myc expression was also noted during RA-mediated differentiation, where the ES cells were exposed to 1 μM of RA on days 8, 9, and 10 of differentiation. A distinct temporal response to RA exposure was seen on days 11 and 14 compared with the ethanol-treated controls in the form of increased c-myc mRNA expression as detected by RT-PCR and RPA. A similar biphasic response to RA treatment has been previously elicited in mouse embryonal carcinoma cells that were induced to differentiate into neurons and astrocytes by RA (51). Increased c-myc mRNA has also been noted during mouse brain development (57). Increased biphasic expression of c-myc mRNA also has been reported in erythroleukemic cells with DMSO and hexamethylene bisacetamide (40,44). Although the overwhelming body of evidence suggests that overexpression of c-Myc causes cellular proliferation and repression of differentiation, contrary to the common findings of decreased c-Myc expression during differentiation, several studies have reported high levels of c-Myc during differentiation in a variety of cell lines (19,23,56).
There was a gradual decline in the overall message from days 17 to 21 when the ES cells were morphologically differentiated in control and RA-treated cells. However, the level of mRNA was elevated in RA-treated cells only, suggesting increased c-myc mRNA in neural cells. There is mounting evidence that c-Myc may not be required for terminal differentiation and that its expression is insufficient by itself to suppress the differentiated phenotype (26). Increased c-myc noted in RA-treated cells may be required for maintenance and metabolism of newly formed neural cells.
An increased level of c-myc transcription was paralleled by increased protein observed at 36 h after initiation of differentiation, in the absence of feeder layers, LIF, or RA. However, upon RA exposure, increased levels of c-Myc protein was observed only on day 14 of differentiation and not on all the times of elevated c-myc mRNA. Increased c-myc gene expression is not followed by increased detection of protein by Western blot at similar times. This observation may be caused by increased c-Myc degradation, as c-Myc is a short-lived nuclear protein. However, increased availability of c-Myc could be indicative of increased stability or translation control (45). On day 14 in RA-treated cells, increased availability of c-Myc protein may provide the necessary switching signal during ES cell neural differentiation in conjunction with other associated proteins.
Investigation of c-myc gene and protein expression alone can only partially be correlated to ES proliferation and differentiation because Myc family is a group of transcription factors that act together to influence apoptosis, differentiation, and growth arrest. Contrasting its expression with other interacting partners can better elucidate the implications of c-Myc upregulation during RA-mediated neural differentiation. The c-Myc signaling effectively requires dimerizing partners Max and Mad, as c-Myc by itself is unable to bind to DNA. In contrast to c-Myc, Max is more stable and exhibits a longer half-life (9). The role of Max in differentiation is controversial. It is generally agreed that Max expression does not change during cell cycle or differentiation (36). However, downregulation of max mRNA and protein in erythroid differentiation (21,25) has been noted. Overexpression of Max may lead to reduced growth and delayed differentiation (16). Interestingly, Max overexpression in neuroblastoma cells lines can enhance RA-mediated growth arrest and differentiation (46). No change in max mRNA was noted between undifferentiated ES cells and initial differentiation at 24 and 48 h. However, increased expression of max mRNA levels was noted by RPA on RA exposure on days 11, 14, 17, and 21. About a threefold increase in Max transcripts was seen on day 14 of differentiation upon RA exposure, coinciding with increased c-myc mRNA expression detected by RPA. Increased Max may be required in conjunction with increased c-Myc for transcriptional activation in differentiating neural cells. C-Myc regulates the cell cycle primarily in G1 phase or G1/S transition and has been known to repress differentiation and cell adhesion while actively promoting cellular proliferation (1). C-Myc binds to the DNA in association with its dimerizing partner Max (11). Max-Myc heterodimers can effectively bind to specific DNA sequences to promote transcription, whereas c-Myc alone is unable to bind to DNA. Max lacks a transactivation domain and Max/Max homodimers can effectively block c-Myc function (3,34). Furthermore, because of a longer half-life, Max interaction with the Mad family of proteins may logically influence c-Myc function.
We also noted an initial decrease in mad transcription after 24 h of differentiation. At this time we do not know the significance of transient decrease in mad mRNA during the early part of spontaneous differentiation. However, increased c-myc gene expression in the presence of Max is indicative of a Myc-Max heterodimer-driven trans-activation process. Increased mad mRNA was observed during RA-mediated differentiation only on days 11 and 14. The relative amount of mad mRNA was very low compared with c-myc and max. Coinciding with increased c-myc mRNA and protein, about a 3.5-fold increase in mad mRNA expression was observed by RPA during ES differentiation in the presence of RA on day 14 compare with ethanol-treated controls. The findings suggest that RA exposure during ES differentiation upregulates Myc-associated partners for optimal signaling. Evidence has been presented that accumulation of Mad/Max complexes during development of mouse embryo (15), myeloid leukemic cells (5), and keratinocytes (31) may facilitate differentiation. Accordingly, transcriptional properties of c-Myc may be influenced by binding with the Mad family of proteins, which in turn recruit sin3 and other co-repressors that compete for c-Myc binding sites and preclude c-Myc/Max-mediated gene regulation (2,7). Consequently, increased expression of Mad proteins has been associated with cellular differentiation and growth arrest. It can be envisioned that Mad and c-Myc inversely regulate differentiation, and upregulation of Mad in RA-treated cells may provide the signal for ES differentiation. It is likely that Mad acts as a switch between proliferation and differentiation, and retinoids may act as a master controller of the cellular fate in the decision-making process. The virtual absence of Mad at later stages of differentiation and is indicative of increased probability of Myc-Max or Max-Max dominated response.
The correlation of c-Myc expression to cellular proliferation and differentiation is fraught with variations, depending upon cell type and differentiating agent used, and is incomplete without investigating the modulation of selected Myc-associated partners. Our results show that RA can effectively modulate the expression of c-myc gene and protein during ES neural differentiation. We have also shown the ability of RA to effectively influence the c-Myc signaling process by differentially regulating the expression of genes encoding for c-Myc dimerizing partners, namely Max and Mad, suggesting that Myc family plays an important role during RA-mediated ES neural differentiation.
REFERENCES
- 1. Amati B.; Alevizopoulos K.; Vlach J. Myc and the cell cycle. Front. Biosci. 3:D250–D268; 1998. [DOI] [PubMed] [Google Scholar]
- 2. Amati B.; Land H. Myc-Max-Mad: A transcription factor network controlling cell cycle progression, differentiation and death. Curr. Opin. Genet. Dev. 4:102–108; 1994. [DOI] [PubMed] [Google Scholar]
- 3. Amati B.; Dalton S.; Brooks M. W.; Littlewood T. D.; Evan G. I.; Land H. Transcriptional activation by the human c-Myc oncoprotein in yeast requires interaction with Max. Nature 359:423–426; 1992. [DOI] [PubMed] [Google Scholar]
- 4. Amati B.; Littlewood T. D.; Evan G. I.; Land H. The c-Myc protein induces cell cycle progression and apoptosis through dimerization with Max. EMBO J. 12:5083–5087; 1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ayer D. E.; Eisenman R. N. A switch from Myc:Max to Mad:Max heterocomplexes accompanies monocyte/macrophage differentiation. Genes Dev. 7:2110–2119; 1993. [DOI] [PubMed] [Google Scholar]
- 6. Ayer D. E.; Kretzner L.; Eisenman R. N. Mad: A heterodimeric partner for Max that antagonizes Myc transcriptional activity. Cell 72:211–222; 1993. [DOI] [PubMed] [Google Scholar]
- 7. Ayer D. E.; Lawrence Q. A.; Eisenman R. N. Mad-Max transcriptional repression is mediated by ternary complex formation with mammalian homologs of yeast repressor Sin3. Cell 80:767–776; 1995. [DOI] [PubMed] [Google Scholar]
- 8. Bain G.; Kitchens D.; Yao M.; Huettner J. E.; Gottlieb D. I. Embryonic stem cells express neuronal properties in vitro. Dev. Biol. 168:342–357; 1995. [DOI] [PubMed] [Google Scholar]
- 9. Berberich S. J.; Cole M. D. Casein kinase II inhibits the DNA-binding activity of Max homodimers but not Myc/Max heterodimers. Genes Dev. 6:166–176; 1992. [DOI] [PubMed] [Google Scholar]
- 10. Blackwood E. M.; Eisenman R. N. Regulation of Myc:Max complex formation and its potential role in cell proliferation. Tohoku J. Exp. Med. 168:195–202; 1992. [DOI] [PubMed] [Google Scholar]
- 11. Blackwood E. M.; Kretzner L.; Eisenman R. N. Myc and Max function as a nucleoprotein complex. Curr. Opin. Genet. Dev. 2:227–235; 1992. [DOI] [PubMed] [Google Scholar]
- 12. Campione-Piccardo J.; Craig J.; Sun J. J.; McBurney M. W. Commitment in a murine embryonal carcinoma cell line during differentiation induced by retinoic acid. Exp. Cell Res. 156:544–552; 1985. [DOI] [PubMed] [Google Scholar]
- 13. Campisi J.; Gray H. E.; Pardee A. B.; Dean M.; Sonenshein G. E. Cell-cycle control of c-myc but not c-ras expression is lost following chemical transformation. Cell 2:241–247; 1984. [DOI] [PubMed] [Google Scholar]
- 14. Chambon P. A decade of molecular biology of retinoic acid receptors. FASEB J. 10:940–954; 1996. [PubMed] [Google Scholar]
- 15. Chin L.; Schreiber-Agus N.; Pellicer I.; Chen K.; Lee H. W.; Dudast M.; Cordon-Cardo C.; DePinho R. A. Contrasting roles for Myc and Mad proteins in cellular growth and differentiation. Proc. Natl. Acad. Sci. USA 92:8488–8492; 1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cogliati T.; Dunn B. K.; Bar-Ner M.; Cultraro C. M.; Segal S. Transfected wild-type and mutant max regulate cell growth and differentiation of murine erythroleukemia cells. Oncogene 8:1263–1268; 1993. [PubMed] [Google Scholar]
- 17. Cole M. D. The myc oncogene: Its role in transformation and differentiation. Annu. Rev. Genet. 20:361–384; 1986. [DOI] [PubMed] [Google Scholar]
- 18. Coppola J. A.; Cole M. D. Constitutive c-myc onco-gene expression blocks mouse erythroleukaemia cell differentiation but not commitment. Nature 320:760–763; 1986. [DOI] [PubMed] [Google Scholar]
- 19. Craig R. W.; Buchan H. L.; Civin C. I.; Kastan M. B. Altered cytoplasmic/nuclear distribution of the c-myc protein in differentiating ML-1 human myeloid leukemia cells. Cell Growth Differ. 4:349–357; 1993. [PubMed] [Google Scholar]
- 20. Davis A. C.; Wims M.; Spotts G. D.; Hann S. R.; Bradley A. A null c-myc mutation causes lethality before 10.5 days of gestation in homozygotes and reduced fertility in heterozygous female mice. Genes Dev. 7:671–682; 1993. [DOI] [PubMed] [Google Scholar]
- 21. Delgado M. D.; Lerga A.; Canelles M.; Gomez-Casares M. T.; Leon J. Differential regulation of Max and role of c-Myc during erythroid and myelomonocytic differentiation of K562 cells. Oncogene 10: 1659–1665; 1995. [PubMed] [Google Scholar]
- 22. Deotchman T. C.; Eistetter H.; Katz M.; Schmidt W.; Kemler R. The in-vitro development of blastcyst-derived embryonic stem cell lines: Formation of visceral yolk sac, blood island and myocardium. J. Embryol. Exp. Morphol. 87:27–45; 1985. [PubMed] [Google Scholar]
- 23. Dotto G. P.; Gilman M. Z.; Maruyama M.; Wein-berg R. A. c-myc and c-fos expression in differentiating mouse primary keratinocytes. EMBO J. 11:2853– 2857; 1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Downs K. M.; Martin G. R.; Bishop J. M. Contrasting patterns of myc and N-myc expression during gastrulation of the mouse embryo. Genes Dev. 3:860–869; 1989. [DOI] [PubMed] [Google Scholar]
- 25. Dunn B. K.; Cogliati T.; Cultraro C. M.; Bar-Ner M.; Segal S. Regulation of murine Max (Myn) parallels the regulation of c-Myc in differentiating murine erythroleukemia cells. Cell Growth Differ. 8:847–854; 1994. [PubMed] [Google Scholar]
- 26. Endo T.; Nadal-Ginard B. Transcriptional and post-transcriptional control of c-myc during myogenesis: Its mRNA remains inducible in differentiated cells and does not suppress the differentiated phenotype. Mol. Cell. Biol. 6:1412–1421; 1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gonda T. J.; Metcalf D. Expression of myb, myc and fos proto-oncogenes during the differentiation of a murine myeloid leukaemia. Nature 310:249–251; 1984. [DOI] [PubMed] [Google Scholar]
- 28. Griep A. E.; Westphal H. Antisense Myc sequences induce differentiation of F9 cells. Proc. Natl. Acad. Sci. USA 85:6806–6810; 1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hanson K. D.; Shichiri M.; Follansbee M. R.; Sed-ivy J. M. Effects of c-myc expression on cell cycle progression. Mol. Cell. Biol. 14:5748–5755; 1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Holt J. T.; Redner R. L.; Nienhuis A. W. An oligomer complementary to c-myc mRNA inhibits proliferation of HL-60 promyelocytic cells and induces differentiation. Mol. Cell. Biol. 8:963–973; 1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hurlin P. J.; Foley K. P.; Ayer D. E.; Eisenman R. N.; Hanahan D.; Arbeit J. M. Regulation of Myc and Mad during epidermal differentiation and HPV-associated tumorigenesis. Oncogene 11:2487–2501; 1995. [PubMed] [Google Scholar]
- 32. Karn J.; Watson J. V.; Lowe A. D.; Green S. M.; Vedeckis W. Regulation of cell cycle duration by c-myc levels. Oncogene 6:773–787; 1989. [PubMed] [Google Scholar]
- 33. Kato G. J.; Dang C. V. Function of the c-Myc oncoprotein. FASEB J. 6:3065–3072; 1992. [DOI] [PubMed] [Google Scholar]
- 34. Kretzner L.; Blackwood E. M.; Eisenman R. N. Myc and Max proteins possess distinct transcriptional activities. Nature 359:426–429; 1992. [DOI] [PubMed] [Google Scholar]
- 35. Lachman H. M.; Skoultchi A. I. Expression of c-myc changes during differentiation of mouse erythroleukaemia cells. Nature 310:592–594; 1984. [DOI] [PubMed] [Google Scholar]
- 36. Larsson L. G.; Pettersson M.; Oberg F.; Nilsson K.; Luscher B. Expression of mad, mxi1, max and c-myc during induced differentiation of hematopoietic cells: Opposite regulation of mad and c-myc. Oncogene 9: 1247–1252; 1994. [PubMed] [Google Scholar]
- 37. Leid M.; Kastner P.; Chambon P. Multiplicity generates diversity in the retinoic acid signaling pathways. Trends Biochem. Sci. 17:427–433; 1992. [DOI] [PubMed] [Google Scholar]
- 38. Lemaitre J. M.; Buckle R. S.; Mechali M. c-Myc in the control of cell proliferation and embryonic development. Adv. Cancer Res. 70:95–144; 1996. [DOI] [PubMed] [Google Scholar]
- 39. Lotan R. Effects of Vitamin A and its analogs (retinoids) on normal and neoplastic cells. Biochim. Biophys. Acta 605:33–91; 1980. [DOI] [PubMed] [Google Scholar]
- 40. Mechti N.; Piechaczyk M.; Blanchard J. M.; Marty L.; Bonnieu A.; Jeanteur P. Transcriptional and post-transcriptional regulation of c-myc expression during the differentiation of murine erythroleukemia Friend cells. Nucleic Acids Res. 14:9653–9666; 1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Morgenbesser S. D.; Schreiber-Agus N.; Bidder M.; Mahon K. A.; Overbeek P. A.; Horner J.; DePinho R. A. Contrasting roles for c-Myc and L-Myc in the regulation of cellular growth and differentiation in vivo. EMBO J. 14:743–756; 1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Mummery C. L.; van den Brink C. E.; de Laat S. W. Commitment to differentiation induced by retinoic acid in P19 embryonal carcinoma cells is cell cycle dependent. Dev. Biol. 121:10–19; 1987. [DOI] [PubMed] [Google Scholar]
- 43. Nath P.; Getzenberg R.; Beebe D.; Pallansch L.; Zelenka P. c-myc mRNA is elevated as differentiating lens cells withdraw from the cell cycle. Exp. Cell. Res. 169:215–222; 1987. [DOI] [PubMed] [Google Scholar]
- 44. Nepveu A.; Marcu K. B.; Skoultchi A. I.; Lachman H. M. Contributions of transcriptional and post-transcriptional mechanisms to the regulation of c-myc expression in mouse erythroleukemia cells. Genes Dev. 9:938–945; 1987. [DOI] [PubMed] [Google Scholar]
- 45. Parkin N.; Darveau A.; Nicholson R.; Sonenberg N. cis-acting translational effects of the 5′ noncoding region of c-myc mRNA. Mol. Cell. Biol. 8:2875–2883; 1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Peverali F. A.; Orioli D.; Tonon L.; Ciana P.; Bunone G.; Negri M.; Della-Valle G. Retinoic acid-induced growth arrest and differentiation of neuroblastoma cells are counteracted by N-myc and enhanced by max overexpressions. Oncogene 12:457–462; 1996. [PubMed] [Google Scholar]
- 47. Reitsma P. H.; Rothberg P. G.; Astrin S. M.; Trial J.; Bar-Shavit Z.; Hall A.; Teitelbaum S. L.; Kahn A. J. Regulation of myc gene expression in HL-60 leukaemia cells by a vitamin D metabolite. Nature 306: 492–494; 1983. [DOI] [PubMed] [Google Scholar]
- 48. Resnitzky D.; Yarden A.; Zipori D.; Kimchi A. Autocrine beta-related interferon controls c-myc suppression and growth arrest during hematopoietic cell differentiation. Cell 6:31–40; 1986. [DOI] [PubMed] [Google Scholar]
- 49. Schmid P.; Schulz W. A.; Hameister H. Dynamic expression pattern of the myc protooncogene in midgestation mouse embryos. Science 243:226–229; 1989. [DOI] [PubMed] [Google Scholar]
- 50. Sorrentino V.; Drozdoff V.; McKinney M. D.; Zeitz L.; Fleissner E. Potentiation of growth factor activity by exogenous c-myc expression. Proc. Natl. Acad. Sci. USA 83:8167–8171; 1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. St-Arnaud R.; Nepveu A.; Marcu K. B.; McBurney M. W. Two transient increases in c-myc gene expression during neuroectodermal differentiation of mouse embryonal carcinoma cells. Oncogene 3:553–559; 1988. [PubMed] [Google Scholar]
- 52. Stern D. F.; Roberts A. B.; Roche N. S.; Sporn M. B.; Weinberg R. A. Differential responsiveness of myc and rastransfected cells to growth factors: Selective stimulation of myc-transfected cells by epidermal growth factor. Mol. Cell. Biol. 6:870–877; 1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. van Inzen W. G.; Peppelenbosch M. P.; van den Brand M. W.; Tertoolen L. G.; de Laat S. W. Neuronal differentiation of embryonic stem cells. Biochim. Biophys. Acta 1312:21–26; 1996. [DOI] [PubMed] [Google Scholar]
- 54. Wali A.; Strayer D. S. Regulation of p53 gene expression by a poxviral transcription factor. Virology 224: 63–72; 1996. [DOI] [PubMed] [Google Scholar]
- 55. Westin E. H.; Wong-Steal F.; Gilman E. P.; Dalla-Favera R.; Papas T. S.; Lautenberger J. A.; Eva A.; Reddy E. P.; Tronick S. R.; Aaronson S. A.; Gallo R. C. Expression of cellular homologues of retroviral oncogenes in human hematopoietic cells. Proc. Natl. Acad. Sci. USA 79:2490–2494; 1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Younus J.; Gilchrest B. A. Modulation of mRNA levels during human keratinocyte differentiation. J. Cell Physiol. 152:232–239; 1992. [DOI] [PubMed] [Google Scholar]
- 57. Zimmerman K. A.; Yancopoulos G. D.; Collum R. G.; Smith R. K.; Kohl N. E.; Denis K. A.; Nau M. M.; Witte O. N.; Toran-Allerand D.; Gee C. E. Differential expression of myc family genes during murine development. Nature 319:780–783; 1986. [DOI] [PubMed] [Google Scholar]


