Abstract
Nkx 3.1 is an evolutionarily conserved vertebrate homolog of the Drosophila Nk-3 homeodomain gene bagpipe that is expressed by a variety of cells during early mammalian development and has been shown to be a critical factor for prostate development and function. Previous studies utilizing a heterologous cell transfection strategy from our laboratory identified the smooth muscle γ-actin (SMGA) gene as a novel molecular target of Nkx 3.1 regulatory activity. In the studies presented here, SMGA gene activity and regulation were evaluated in normal and cancerous prostate epithelial cells. SMGA transcripts were demonstrated in prostate epithelia and SMGA mRNA levels were increased in androgen-responsive LNCaP cancer and normal prostate epithelial cells. SMGA gene transcriptional activity was androgen responsive in these cells and required a segment of the human SMGA promoter containing NKE and SRF (serum response factor) binding elements. This region of the human SMGA proximal promoter is well conserved across species and is synergistically activated by coexpression of Nkx 3.1 and SRF in heterologous CV-1 cells. SMGA transcription was not responsive to steroid in PC-3 prostate epithelial cancer cells, which do not express Nkx 3.1. However, SMGA transcription was influenced by expression of androgen receptor in these cells, a situation that allows the androgen-dependent expression of Nkx 3.1. Furthermore, SMGA gene activity was influenced by direct Nkx 3.1 expression in the PC-3 cells. Thus, SMGA gene activity in prostate epithelia is due, in part, to the androgen-dependent expression of Nkx 3.1. As such, our studies provide the initial description of Nkx 3.1 target gene regulatory activity in the prostate.
Keywords: Developmental gene regulation, Androgen receptor, Prostate cancer, Tissue-specific expression, Transcriptional regulation, Nk-homeodomain, Serum response factor
THE prostate is a ductal gland situated at the base of the bladder comprised of tall columnar epithelium surrounded by stroma that contributes secretory proteins to the seminal fluid. Normal prostate growth and differentiation requires inductive interactions between epithelial and stromal components. At all stages of prostate development as well as maturity, tissue interactions require functional androgen receptors, initially in the mesenchyme and subsequently in the epithelium (12,14). The reciprocal signaling interactions that are responsible for proper prostate development and maturation are likely to exert their effects through the alteration of gene expression patterns in the responding epithelium, but the molecular factors that are affected in this manner remain undefined.
Among the few regulatory genes known to be expressed in the prostate epithelium, the Nkx 3.1 homeobox gene is of particular interest as it maps to the minimal region of human chromosome 8p21 (17,46) that undergoes loss of heterozygosity in 60–80% of prostate tumors (11,42,43,45). Although Nkx 3.1 is expressed early in the sclerotome and a subset of vascular smooth muscle cells and later in the prostate, palatine gland, and restricted regions of the central nervous systems and lobular arteries of the kidney (5,39,41), many studies have ascribed a role for Nkx 3.1 in normal prostate development and function, with a potential role in prostate carcinogenesis. For example, Nkx 3.1 expression during embryogenesis appears to demarcate prospective prostate epithelium prior to prostate formation and continues to mark prostate epithelium during neonatal development (1,4). Furthermore, Nkx 3.1 null mutants generated by gene targeting display defects in ductal morphogenesis and secretory protein production. Finally, Nkx 3.1 loss results in epithelial hyperplasia and dysplasia that increases in severity with age, modeling a preneoplastic condition (1,4). Because of its described functions within the prostate, knowledge of the genes it regulates is of paramount importance for understanding prostate development, differentiation, and possibly the progression of carcinogenesis.
Smooth muscle gamma actin (SMGA) is the first identified molecular target for Nkx 3.1 regulatory activity (7). The combination of serum response factor (SRF) and Nkx 3.1 was shown to potentiate the synergistic activation of SMGA activity via a novel cis-element found within the first 100 bp of the promoter. The present studies examine the regulation of SMGA gene activity by SRF and Nkx 3.1 in prostate epithelia. We found that SMGA transcripts were present in the normal and cancerous prostate epithelia. Moreover, SMGA mRNA levels were upregulated in response to androgen treatment, and we demonstrate that the androgen-responsive appearance of SMGA gene activity in prostate epithelia is due to the androgen-dependent expression Nkx 3.1. The results of these studies are important because they provide the first description of an Nkx 3.1 target gene regulation in the prostate, which will aid in the understanding of prostate development, differentiation, and carcinogenesis.
MATERIALS AND METHODS
Cell Cultures
CV-1 cells (ATCC, catalog No. 70-CCL) were maintained at 37°C, 5% CO2 in Dulbecco’s minimal essential medium (DMEM) with 0.2% sodium bicarbonate, 1× antibiotic/antimycotic (GIBCO BRL), Kanamycin (0.1 mg/ml), and 10% fetal bovine serum. The human adenocarcinoma LNCaP cell line (ATCC catalog No. CRL 1740) was maintained in RPMI medium supplemented with 1 mM L-glutamine, 10 mM HEPES, pH 7.2, 0.25% glucose, 2 mM sodium pyruvate, 10% FBS, and 2% penicillin/streptomycin. The human adenocarcinoma PC-3 cell line (ATCC catalog No. CRL 1435) was maintained in RPMI medium supplemented with 10% FBS and 2% penicillin/streptomycin. Normal prostate epithelial cells (PrEC) (BioWhittaker, Walkersville, MD) were derived from a 17-year-old donor. The PrEC were maintained in the suggested medium in the Prostate Cell Systems protocol obtained from the supplier.
Northern Blot Analyses
Total RNA was obtained from prostate cell lines or normal prostate cells using the RNA STAT-60 (Tel Test β, Inc., Friendswood, TX) reagent. Equivalent quantities of RNA (5–10 μg) to be analyzed were denatured by glyoxal treatment, size fractionated on 1.2% agarose gels, and the RNAs within the gels then transferred to nylon membranes (Biotrans, ICN, Irvine, CA) as described previously (22). Duplicate RNA samples and RNA molecular weight markers run on the gel were cut away from transferred samples and ethidium bromide stained for photographing. The membranes containing RNAs were in cubated with the appropriate 32P-labeled probes [∼1–2 × 106 cpm/ml hybridization buffer for the 3′ UTR of the human SMGA mRNA (22) or coding sequences of the human Nkx 3.1 and SRF mRNA (7)] essentially as previously described by Kovacs and Zimmer (22). Nonspecific probe binding was washed from the membranes by incubation in 0.5× SSC, 0.5% SDS at 42°C with constant agitation and the membranes then exposed to BMR1 X-ray film (Kodak Inc., Rochester, NY) using Dupont Lightning Plus intensifying screens for a variety of times ranging from 1 to 48 h. Quantitative analysis of hybridizing signal was accomplished by scanning the blots with a Umax Astra 1200S scanner and analyzing the data with the Molecular Analyst software (Bio-Rad, Melville, NY). 18S and 28S rRNA within the same lane of the individual Northern blots were also quantitated and used to control for RNA loading.
DNA Transfections
CV-1 transfection experiments were performed using the Lipofectamine reagent (GIBCO BRL) as described previously (7) with the exception that cells were plated in 24-well dishes. DNA/liposome mixtures were added to each well of cells (in quadruplicate) and the cells incubated at 37°C, 5% CO2 for 5–6 h. Following the incubation, 1 ml of complete growth medium was added, and the cells were then allowed to incubate at 37°C, 5% CO2 for 48 h before harvesting.
LNCaP and PC-3 cells were also transfected using Lipofectamine reagent essentially as above. However, the medium used throughout the process was RPMI lacking phenol red but containing charcoal-stripped FBS. Following transfection, 1 ml of medium was added to each dish [steroid-stripped medium with either ethanol (EtOH) or R1881 (NEN, Boston, MA; made in 100% EtOH) to give a final concentration of 1% or 10 nM, respectively]. The cells were then incubated at 37°C, 5% CO2 for 48 h until harvesting.
Transfection of normal prostate epithelial cells was performed using methods similar to those used for LNCaP cells, except that the medium used after transfection was the normal growth medium with R1881 or EtOH to give a final concentration of 10 nM R1881 or 1% EtOH.
DNA Constructs
The plasmids encoding pCGN-SRF (8–10) and pCGN-Nkx 3.1 (7), as well as pCGN empty expression vector, have been described. The human SMGA gene was isolated in a P1 clone. Progressive 5′ deletions of the human SMGA promoter were obtained by PCR and cloned into pGL-3 Basic plasmid (Promega) driving the expression of the firefly luciferase reporter gene (6,23). DNAs were prepared by the Plasmid Midi Kit from Qiagen (Chatsworth, CA). The DNA samples were quantitated by absorbance at 260/280 nm using a Beckman Instruments DU Series 600 Spectrophotometer (Beckman Instruments).
Luciferase Assays
The level of promoter activity was evaluated by measurement of the firefly luciferase activity present in the measured sample volume using the Luciferase Assay System as outlined by the supplier (Promega) using a Turner Model 20 luminometer (Turner Designs, Sunnyvale, CA). The amount of firefly luciferase activity was normalized to the total microgram of protein in the sample (determined using the Bio-Rad Protein Assay System), and normalized activities from multiple experiments were averaged and plotted ± SEM.
Statistical Analyses
Statistical analyses for transfection and densitometric data were carried out using the Statistix for Windows software (Analytical Software, Tallahassee, FL). The analyses performed utilized the AOV method of comparison of the means for statistical significance at p < 0.05.
RESULTS
SMGA mRNA Content Is Androgen Responsive in Prostate Epithelial Cell Lines
We have previously shown that Nkx 3.1 and SRF selectively and synergistically activate SMGA gene transcriptional activity in a system, monkey kidney CV-1 cells, that does not normally express all three of these factors (7). Nkx 3.1 expression is found in the somites of early embryos (21), but in later stage embryos and adults is found in brain, kidney (particularly the blood vessels), duodenum, and cardiac outflow tract (17,21,36,39,41,48). However, the predominant place of expression seen for Nkx 3.1 is in the male urogenital system including the testis, seminal vesicle, and particularly within prostate ductal epithelial cells. To address whether SMGA and SRF are found in prostate epithelia, Northern blot analyses were carried out using an androgen-responsive prostate adenocarcinoma cell line, LNCaP (15,18,19,26). As shown in Figure 1A, all three transcripts are present in this prostate epithelial cell line. Because of the novelty of finding the presence of smooth muscle-specific SMGA transcripts in this epithelial cell type, control hybridization experiments were employed to demonstrate the isoform specificity of the SMGA probe used (data not shown). Nkx 3.1 expression in prostate epithelial cells has been demonstrated to be androgen dependent (5,17,21,36,39,41,48), and we observed that treatment of the cells with synthetic androgen, R1881, not only resulted in a dose-dependent increase in Nkx 3.1 transcripts, but also a 2.6-fold, statistically significant increase in the SMGA transcripts (Fig. 1). Further, as R1881 dosage increased there was a trend of increasing SMGA mRNA, parallel to Nkx 3.1 mRNA. Although present in LNCaP cells, SRF mRNA remained constant throughout androgen treatment (Fig. 1). These data show that all three transcripts (Nkx 3.1, SRF, SMGA) are present in prostate epithelial cell lines, and reveal the androgen-sensitive regulation of SMGA gene activity in these cells. The present findings, in addition to our previous in vitro studies (7), suggest an in vivo androgen-sensitive activation of SMGA gene activity via Nkx 3.1 and SRF within prostate epithelial cells.
Figure 1.

SMGA mRNA presence and androgen responsiveness within LNCaP cells. (A) Representative autoradiograms from Northern analyses using RNA isolated from LNCaP cells treated with either vehicle (lane 1) or increasing amounts of androgen (R1881; 0.1–10 nM) with 32P-labeled DNA probes representing human SMGA, SRF, or murine Nkx 3.1 are shown. (B) Striped bars represent each specific band appearing in (A) derived from cells cultured in the presence of vehicle, while the black bars represent those bands appearing in (A) in the presence of 10 nM R1881. The results are shown as the mean ± SE. *Statistical significance in the presence of R1881 versus its absence at p < 0.05.
SMGA Promoter Is Transcriptionally Activated in Androgen-Treated Prostate Epithelial Cells
We asked whether the SMGA gene was capable of being transcriptionally activated within the LNCaP cells to examine the possibility that Nkx 3.1 and SRF are activating SMGA transcription within these cells. We utilized transient transfection analyses of the prostate cancer cells in order to compare the activation obtained by different human SMGA promoter deletion fragments. There was rather strict maintenance of sequence identity over the initial ∼400 bp of SMGA promoter segments as well as maintenance of spacing of major cis-elements across species (Fig. 2A). Several human promoter fragments (Fig. 2B) were tested for activity within the LNCaP cells either in the absence or presence of androgen, and the results are shown as the fold induction by each construct over that obtained from a promoterless vector (Fig. 2C). HSMGA1, which contains a basic TATA sequence, caused an activation of SMGA promoter activity approximately two- to threefold over the pro-moterless construct in the absence or presence of R1881. Adding sequences containing the Nke/CArG1 element (7) resulted in only a slight increase in stimulated activity (approx. three- to fourfold), regardless of R1881 treatment. However, if sequences containing CArG/SRE2, which has been shown to strongly bind SRF (6), as well as a potential half-site ARE (androgen response element, −169 to −162, Fig. 2A) were added to the construct, there was an androgen-dependent, statistically significant increase in activity seven- to eightfold over the promoterless vector. Thus, SMGA is capable of being transcriptionally activated within LNCaP cells in an androgen-responsive manner.
Figure 2.
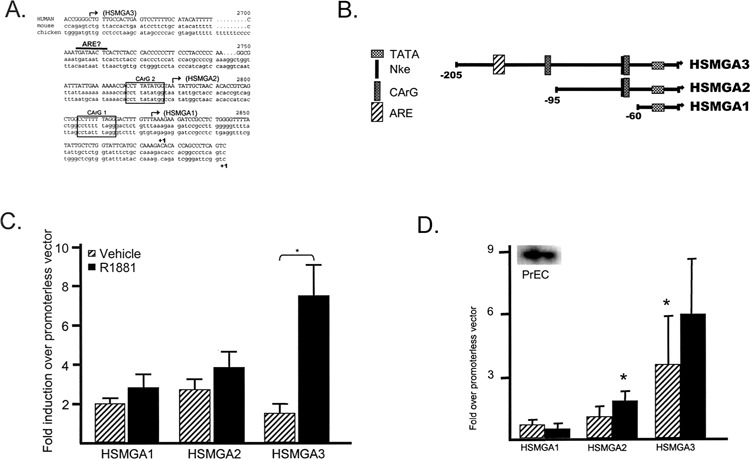
SMGA transcriptional activity within normal and cancerous prostate epithelial cells. (A) Sequences representing the first ∼200 bp of the avian, murine, and human SMGA promoter are compared. The two CArG elements are indicated by boxes, and the bent arrows represent the 5′ limit of human promoter sequences fused to the firefly luciferase reporter gene used in the present study. Also indicated is a putative half-site ARE found in the mammalian SMGA promoters. (B) The human SMGA promoter fragments linked to the firefly luciferase gene indicating the location of the TATA box, CArG, NKE element, and putative half-site ARE. (C) Luciferase results from the transfection of LNCaP cells with 1.2 μg of the indicated SMGA promoter construct. After transfection, the cells were either maintained in vehicle (striped bars) or 10 nM R1881 (black bars) before harvesting at 48 h for analysis. The results are shown as the fold induction by each promoter fragment over that obtained by the promoterless vector (pGL3-Basic). Results are derived from a minimum of six experiments performed in quadruplicate for each condition and are shown as the mean ± SEM. *Statistical significance at p < 0.05 for the activation obtained in the presence of R1881 over the activation obtained in its absence. (D) The luciferase activity results from the transfection of normal prostate epithelial cells with 1.2 μg of the indicated SMGA promoter construct. After transfection, the cells were maintained in either vehicle (striped bars) or R1881 (10 nM; black bars) before harvesting at 48 h for analysis. The results are shown as the fold induction by each promoter fragment over that obtained by the pGL3-Basic promoterless vector. The results represent four experiments performed in quadruplicate for each condition and are shown as the mean ± SEM. The inset is representative autoradiogram of slot blots consisting of 10 μg RNA isolated from normal prostate epithelial cells treated with 10 nM R1881 and hybridized with the 32P-labeled SMGA probe.
To distinguish whether the androgen-responsive SMGA gene regulation was a property of prostate epithelia or was due to anomalous effects of gene activation in cancer cell lines, we performed studies using normal prostate epithelial cells. Slot blot analyses of RNA isolated from normal epithelial cells (PrEC) demonstrate the presence of SMGA transcripts (Fig. 2D). We next analyzed SMGA transcriptional capacity within the normal prostate cells. Little activity was obtained until sequences containing CArG/SRE2 and a putative half-site ARE were added (HSMGA3), resulting in approximately four- to sixfold activation in PrEC depending on the absence or presence of androgen (Fig. 2D). SMGA promoter activity produced in the presence of androgen was consistently higher (approx. sixfold) than in nontreated cells (∼3.5-fold). Therefore, the presence of SMGA mRNA and the transcriptional capacity of the SMGA gene within prostate cells is not a property strictly attributed to the transformed phenotype.
Nkx 3.1 and Serum Response Factor Synergize to Activate Transcription From the Human SMGA Promoter
Transient transfection analyses using smooth muscle cells demonstrated similar activities of the equivalent human and chicken SMGA promoter fragments extending over the first four conserved CArG/SRE motifs (data not shown). Although SMGA promoter sequences are conserved across species (Fig. 2A), differences among these sequences are observed within the NK/CArG element previously demonstrated to be required for Nkx 3.1/SRF activation of the avian SMGA promoter. As shown in Figure 3, SRF induced the human SMGA promoter fragment equivalent to previously published analyses of the avian promoter (7) (HSMGA3) ∼5- to 10-fold (Fig. 3A), while Nkx 3.1 potentiated SMGA activity at ∼15- to 20-fold. However, the addition of SRF with Nkx 3.1 resulted in a synergistic activation of the human SMGA promoter similar to that seen for the chicken gene (Fig. 3A). Thus, these data demonstrate that the human SMGA promoter is synergistically regulated by SRF and Nkx 3.1.
Figure 3.
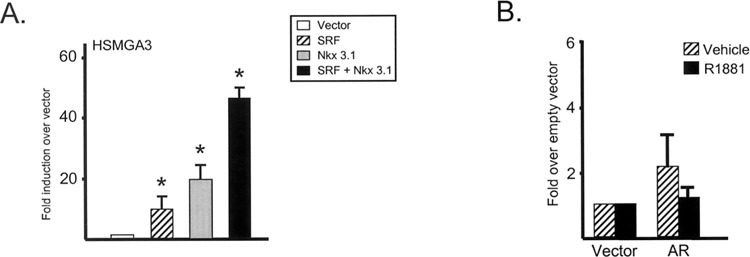
Human SMGA promoter is stimulated by SRF and Nkx 3.1 while androgen receptor is not sufficient for transcriptional activation. (A) The human −205 bp SMGA promoter fragment fused to the firefly luciferase reporter gene (0.3 μg) was transfected into CV-1 cells along with either empty CMV (open bar), SRF (striped bar), Nkx 3.1 (gray bar), or SRF in combination with Nkx 3.1 (black bar; both at 0.3 μg each). The transactivation reactions were balanced to 1.2 μg DNA content by the addition of empty CMV expression vector (pCG). The results are shown as the fold induction by each vector or combination of vectors over that obtained by the empty CMV expression vector (white bar). Results were derived from three experiments performed in quadruplicate for each condition and are shown as the mean ± SEM. *Statistical significance at p < 0.05 for the activation obtained by the condition indicated over that derived from the empty vector alone. (B) The HSMGA3 promoter fragment fused to luciferase (0.3 μg) was transfected into CV-1 cells along with either AR (0.3 μg) or the empty CMV expression vector are shown. After transfection, the cells were maintained in either vehicle (striped bars) or R1881 (black bars) for 48 h before harvesting for luciferase analyses. The results are shown as the fold induction of luciferase activity in the presence of AR over that obtained by the empty expression vector. The reactions were balanced to 1.2 μg DNA content by the addition of empty CMV promoter vector (pCG). The results were determined from a minimum of three experiments performed in quadruplicate for each condition and are shown as the mean ± SEM.
The human SMGA promoter fragment that was activated in an androgen-responsive manner within the PrECs contained sequences encompassing a putative half-site ARE (HSMGA3), which prompted us to study the requirement for this site for the androgen-responsive action on SMGA gene activity. It is known that AREs function through the binding of transcriptionally capacitated AR. As such, HSMGA3 was transiently transfected into CV-1 cells (which do not express AR, Nkx 3.1, or SMGA) along with AR. AR alone had no effect on the transcriptional capacity of HSMGA3 whether in the absence or presence of androgen (Fig. 3B). These results suggest that AR alone, and thus the half-site ARE, is not totally sufficient for directly activating androgen-responsive SMGA transcriptional activity.
SMGA Gene Activation in Prostate Epithelia Is Dependent on Nkx 3.1 Expression
PC-3 is a prostate cancer cell line that displays androgen-independent growth responses due to the lack of androgen receptor (26). Further, PC-3 cells demonstrate misregulation of genes characteristic of mature prostate epithelia, such as prostate-specific antigen (26,47). PC-3 cells were grown either in the absence or presence of increasing amounts of androgen before RNA was isolated for use in Northern blotting analyses to examine the expression of Nkx 3.1, SRF, and SMGA in prostate epithelia that no longer respond to androgen. As found with the LNCaP cells, both SMGA and SRF transcripts were present within this cell line (Fig. 4A). However, unlike the results for LNCaP cells, Nkx 3.1 transcripts were absent, and significantly, there was a lack of androgen-responsive induction of SMGA gene activity in these cells. These results imply that the androgen-responsive expression of SMGA gene activity within prostate cancer cells requires the action of AR and/or Nkx 3.1.
Figure 4.

AR and Nkx 3.1-mediated stimulation of HSMGA3 gene activity. (A) Representative autoradiograms from Northern analyses using RNA isolated from PC-3 cells treated with either vehicle (lane 1) or increasing amounts of androgen (R1881; 0.1–10 nM) with 32P-labeled DNA probes representing human SMGA, SRF, or murine Nkx 3.1 are shown. (B) The HSMGA3 promoter fragment fused to the firefly luciferase gene (0.3 μg) was transfected into PC-3 cells along with either AR (0.5 μg), Nkx 3.1 (0.5 μg), or the corresponding empty CMV expression vector. The results are shown as the fold induction seen by the presence of the various factors over that seen by the empty vector. The results are representative of a minimum of two experiments performed in triplicate for each condition and are shown as the mean ± SEM. *Statistical significance at p < 0.05 for the activation obtained by AR in the presence of androgen versus its absence. PC-3 cells were transfected with expression vector encoding human AR or the empty vector and the cultures then incubated with R1881 to examine the AR-dependent expression of Nkx 3.1 by RT-PCR (C) and Western blotting (D) analyses. (C) The relevant portion of a gel in which RNA from PC-3 cells transfected with empty vector (lanes 2 and 3) or an AR expression vector (lanes 4 and 5) were probed by RT-PCR using oligos specific for Nkx 3.1 mRNA (680-bp band) and GAPD (420-bp band). Quantitative analyses of three separate experiments revealed an increase of approximately fivefold (4.97 ± 0.05) of Nkx 3.1 mRNA in cells expressing human AR and incubated in 10 nM R1881. (D) Western blot of protein extracted from PC-3 cells transfected with a human AR expression vector or empty vector. Total cellular proteins were separated on 12.5% SDS-PAGE and transferred to nitrocellulose sheets. The blots were probed with an anti-Nkx 3.1 antibody (gift of Dr. Ed Gelmann) at a dilution of 1:10,000. The blots were developed with a chemilumenesence kit as previously described (7). Lanes 1 and 2 are proteins from control, vector-only cells while lanes 3 and 4 represent proteins derived from cells treated with a human AR expression vector. Lane 5 shows protein derived from Nkx 3.1-positive LNCaP cells as a positive control.
As shown in Figure 3B, androgen receptor alone was incapable of stimulating SMGA transcription when cotransfected into AR-negative cells (CV-1). Thus, we next pursued the possibility that the upregulation of androgen-responsive SMGA gene activity was due to the androgen-dependent increase in Nkx 3.1 transcripts. To address this, PC-3 cells were transiently transfected with HSMGA3 and AR and left in either the absence or presence of androgen. It has been demonstrated that PC-3 cells transfected with DNAs encoding functional androgen receptor can restore, to some degree, androgen-dependent transcriptional responses to target genes such as Nkx 3.1 (4). We verified the induction of Nkx 3.1 mRNA and protein expression in AR transfected PC-3 cells using RT-PCR and Western blotting (Fig. 4). The PC-3 cells were also transiently transfected with HSMGA3 along with Nkx 3.1 and left either in the absence or presence of androgen. Addition of Nkx 3.1 to the PC-3 cells resulted in an ∼1.5-fold increase in SMGA gene activity, regardless of the presence of androgen (Fig. 4B). Addition of AR to these cells resulted in an androgen-sensitive ∼1.5-fold increase in SMGA transcriptional activity (Fig. 4B). Taken together, our studies indicate that the androgen-sensitive activation of SMGA gene activity seen in prostate epithelia is due to the androgen-dependent increase in Nkx 3.1.
DISCUSSION
We have previously demonstrated in a heterologous transfection assay system that Nkx 3.1 and SRF can collaborate to activate transcription from the avian SMGA promoter (7). Although Nkx 3.1 is expressed within a variety of mesodermally derived cell types (21,39,41), it exhibits predominant expression within the male reproductive system, specifically within the urogenital system and prostate epithelia (41,48). Thus, a major goal of the present study was to determine if Nkx 3.1 and SRF regulated SMGA gene expression in prostate epithelia. Northern analyses of RNA isolated from a well-characterized human prostate adenocarcinoma cell line, LNCaP cells (15,18,19,26), demonstrated the presence of SRF and SMGA transcripts in cells treated with the synthetic androgen, R1881 (Fig. 1). An androgen-dependent regulation of Nkx 3.1 has been previously demonstrated in LNCaP and other androgen-responsive prostate epithelial cells (5,17,36,39,48). Our results demonstrated that although LNCaP cell SRF transcript levels were unaffected by androgen treatment, there was a dose-dependent increase in Nkx 3.1 transcripts and a concomitant, statistically significant, increase in SMGA transcripts in R1881-treated cells. These data indicated that an Nkx 3.1-dependent transcriptional activation of the SMGA gene is possible in androgen-stimulated prostate epithelial cells. Consistent with this hypothesis, a proximal segment of the human SMGA gene promoter was capable of androgen-responsive transcriptional activation in LNCaP and normal prostate epithelial cells (Fig. 2). The segment of the human SMGA promoter necessary for steroid-dependent transcriptional activation in prostate cells contains multiple cis-acting elements that include CArG/SRE and NKE motifs (Fig. 2), and is a well-conserved segment of DNA demonstrated to be synergistically acted upon by SRF and Nkx 3.1 (Fig. 3). Importantly, we show that increasing Nkx 3.1 expression in the androgen-independent prostate cell line PC-3, cells that do not express functional androgen receptors or Nkx 3.1 (5,39), increases SMGA gene transcription. Therefore, our results demonstrate that SMGA is a regulated gene in prostate epithelial cells, and appropriate expression of this gene occurs in prostate epithelia in part through an Nkx 3.1-dependent mechanism. Although a potentially unexpected result, this would explain recent observations of significant SMGA mRNA levels determined by expressed sequence tag (EST) analyses of cDNA libraries derived from normal and cancerous prostate epithelial (16,30–32). Because by our criteria SMGA is a product of the differentiated prostate epithelial cell, an understanding of mechanisms governing its expression will enhance comprehension of the genetic basis of prostate cell differentiation.
It is clear that cell-specific transcription of the SMGA gene requires complex interactions of factors upon multiple cis-acting DNA elements. A major element that influences SMGA transcription is the CArG/SRE, of which there are multiple representations in two separate, positive-acting domains of the SMGA promoter called the specifier and modulator (23). CArG/SRF-dependent stimulation of SMGA transcription occurs through the binding of SRF-containing nuclear complexes (6,7). Thus, our demonstration of prostate epithelial cell SRF expression indicates that such an SRF-dependent mechanism is operable within those cells.
The SMGA locus is highly conserved among all species examined to date (chick, mouse, human, rat), including sequences surrounding the gene as well as the structure of the gene (23). However, differences among these sequences have been noted. We observed that within the promoter segments of the mammalian SMGA genes there is a sequence (TGATAACT, −169 to −162, Fig. 2A) that is similar in structure to the cis-element responsible for androgen receptor-dependent transcriptional activation, the ARE (13). An ARE (or half-site ARE) is not apparent in the avian gene, which may indicate a difference in the regulatory potential of the mammalian and avian SMGA genes with regard to androgen responses. Our results show that the addition of the potential ARE sequence allowed the androgen-dependent activation of the human SMGA promoter in LNCaP cells (Fig. 2). However, cotransfection of CV-1 cells with the human promoter-luciferase reporter and a vector expressing human androgen receptor was unable to demonstrate an androgen-stimulated transcriptional response. Thus, androgen receptor alone is not able to stimulate androgen-responsive HSMGA promoter activity. This might be due to the inability of the ARE half-site within the human promoter to attract receptor binding. Steroid hormone response elements often contain a nearly cononical half-site of TGTTCT and another half-site of considerable deviation (30,38). Although the mammalian SMGA gene half-site ARE maintains a critical thymidine at position −4 (38), the existence of the nonconserved ARE segment has not been demonstrated. Thus, the androgen-dependent stimulation of SMGA transcription obtained through the nucleotides −205 to −90 of the human promoter may occur through interactions with the additional CArG sequence (CArG/SRE2), or the interactions of proteins that recognize or stabilize complexes formed with either or both of these DNA elements. In this regard, many coactivator proteins such as TIF2/SRC-1 (3,27), CBP/p300 families (48), and FHL2 (29,33) that have been shown to interact with the activating function (AF-2) domain of nuclear receptors in a ligand-dependent fashion, as well as SRF (20,24,25,34), may act in such a manner. Thus, it is possible that the ligand-dependent SMGA transcriptional activation in prostate epithelial cells includes a receptor-coactivator-SRF mechanism, which may not require the direct interaction of the androgen receptor with SMGA promoter elements. Although our work supports the hypothesis that the greatest part of the androgen-dependent SMGA transcriptional activation in prostate epithelial cells occurs from enhanced Nkx 3.1 expression, the ability of SRF to form higher order complexes with steroid receptor coactivation protein(s) may explain the increased SMGA transcription we observe when androgen receptor negative PC-3 cells were transfected with AR compared with transfection of these cells with Nkx 3.1 alone (Fig. 4). Furthermore, because SRF activity is dependent upon phosphorylation of specific residues (8–10,40,44), it is possible that the SRF in PC-3 cells does not allow for optimal interaction with Nkx 3.1. We are currently pursuing experiments to address if steroid receptor/coactivation protein complexes are capable of ligand-dependent transcriptional activation of genes (SMGA) through direct association with SRF.
An interesting finding from our work presented here is the misregulation of SMGA gene activation in the androgen-independent prostate cancer cell PC-3. Although SMGA transcripts were present in the LNCaP and PC-3 cell lines, we found higher basal levels of SMGA transcripts in PC-3 cells. Moreover, androgen treatment did not affect SMGA transcript levels within the PC-3 cells. This result underscores the requirement of androgen-dependent induction of Nkx 3.1 for the androgen-responsive SMGA gene regulation, as AR, and consequently Nkx 3.1, are not expressed in the PC-3 cells (26,33). The lack of androgen-responsive SMGA gene activity in these less differentiated, more aggressive cancer cells (26) suggests that the progression from an androgen-dependent into an androgen-independent prostate epithelial cell adenocarcinoma involves a homeodomain-dependent loss of SMGA gene regulation. One explanation for the misregulation of the SMGA gene within androgen-independent prostate cancer cells may indeed reside with the SRF/Nkx 3.1 complex. It has been demonstrated that SRF plays a primary role in the regulation of both cell cycle stimulatory and also differentiated stage-specific genes. The X-ray crystal structure of SRF bound to DNA (35) provides a possible explanation for how SRF may participate in these opposing events. The SRF MADS box has been shown to consist of several structures, including α-helices and β-sheets, which mediate various functions of the molecule (35). Importantly, one of the α-helices (αII helix) has been shown to contact growth factor-responsive ternary complex factors Elk-1 and SAP-1 (44) in order to interact with the SRE of the c-fos gene for transcriptional activation. These factors (Elk-1 and SAP-1) are unable to activate differentiated product genes that are dependent upon SRF, such as the α-cardiac actin (9) or SMGA genes (data not shown). It has been suggested that factors involved in controlling differentiation-specific gene activation in conjunction with SRF, such and Nkx 2.5 and Phox-1, require the amino-terminal region of the MADS box of SRF (9). While the exact interactions between differentiation factors and SRF have not been mapped, it is possible that they interact with the amino-terminal helix (αI), which may then alter the structure of the MADS box, effectively inhibiting the interaction with the ternary complex factors. A change in MADS box structure was noted in analyses of yeast SRF-related protein, MCMI, and MAT, a homeodomain protein that specifies mating type (40). In prostate epithelial cells, Nkx 3.1 might make contacts with the αI helix, which could inhibit the binding of the growth factor-regulated ternary complex proteins, thus maintaining the differentiated cell functions; when Nkx 3.1/SRF interactions are disrupted or lost (such as in androgen-independent prostate cancer), the ternary complex factor regulation of gene expression programs could predominate, resulting in altered cell cycle properties such as those observed in advanced stage prostate cancer. Therefore, the regulated expression of the SMGA gene within prostate epithelial cells serves as an excellent model/marker for the study of prostate cancer progression.
ACKNOWLEDGMENTS
This work represents partial fulfillment of the Ph.D. dissertation requirements for R. Fillmore and was supported by NIH grant RO1-Hl59956. The Nkx 3.1 antibody was the kind gift of Dr. Ed Gelmann, Georgetown University, Washington, DC.
REFERENCES
- 1. Abate-Shen C.; Shen M. M. Molecular genetics of prostate cancer. Genes Dev. 14:2410–2434; 2000. [DOI] [PubMed] [Google Scholar]
- 2. Bergerheim U. S.; Kunimi K.; Collins V. P.; Ekman P. Deletium mapping of chromosomes 8, 10, and 16 in human prostatic carcinoma. Genes Chromosomes Cancer 3:215–220; 1991. [DOI] [PubMed] [Google Scholar]
- 3. Bevan C. L.; Hoare S.; Claessens F.; Heery D. M.; Parker M. G. The AF1 and AF2 domains of the androgen receptor interaction with distinct regions of SRC1. Mol. Cell. Biol. 19:8383–8392; 1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bhatia-Gaur R.; Donjacour A. A.; Sciavolino P. J.; Kim M.; Desai N.; Young P.; Norton C. R.; Gridley T.; Cardiff R. D.; Cunha G. R.; Abate-Shen C.; Shen M. M. Roles for Nkx 3.1 in prostate development and cancer. Genes Dev. 13:966–977; 1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bieberich C. J.; Fujita K.; He W. W.; Jay G. Prostate-specific and androgen-dependent expression of a novel homeobox gene. J. Biol. Chem. 271:31779–31782; 1996. [DOI] [PubMed] [Google Scholar]
- 6. Browning C. L.; Culberson D. E.; Aragon I. V.; Fillmore R. A.; Croissant J. D.; Schwartz R. J.; Zimmer W. E. The developmentally regulated expression of serum response factor plays a key role in the control of smooth muscle-specific genes. Dev. Biol. 184:18–37; 1998. [DOI] [PubMed] [Google Scholar]
- 7. Carson J. A.; Fillmore R. A.; Schwartz R. J.; Zimmer W. E. The smooth muscle gamma actin gene promoter is a molecular target for mNKx 3-1, a vertebrate homologue of Drosophila bagpipe, and serum response factor. J. Biol. Chem. 275:39061–39072; 2001. [DOI] [PubMed] [Google Scholar]
- 8. Chen C.; Schwartz R. J. Identification of novel DNA binding targets and regulatory domains of a murine tinman homeodomain factor, Nkx-2.5. J. Biol. Chem. 270:15628–15633; 1995. [DOI] [PubMed] [Google Scholar]
- 9. Chen C.; Schwartz R. J. Recruitment of the tinman homologue Nkx-2.5 by serum response factor activates cardiac α-actin gene transcription. Mol. Cell. Biol. 16:6372–6384; 1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chen C. Y.; Croissant J.; Majesky M.; Topouzis S.; McQuinn T.; Frankovsky M.; Schwartz R. J. Activation of the cardiac α-actin promoter depends upon serum response factor, tinman homologue, Nkx-2.5, and intact serum response elements. Dev. Genet. 19:119–130; 1996. [DOI] [PubMed] [Google Scholar]
- 11. Cher M. L.; Bova G. S.; Moore D. H.; Small E. J.; Carroll P. R.; Pin S. S.; Epstein J. I.; Isaacs W. B.; Jensen R. H. Genetic alterations in untreated metastases and androgen independent prostate cancer detected by comparative genomic hybridization and allelotyping. Cancer Res. 56:3091–3102; 1996. [PubMed] [Google Scholar]
- 12. Chunha G. R. Growth factors as mediators of androgen action during male urogenital development. Prostate Suppl. 6:22–25; 1996. [PubMed] [Google Scholar]
- 13. Classens F.; Verrijdt G.; Shoemakers E.; Haelens A.; Peeters B.; Verhoeven G.; Rombauts W. Selective DNA binding by the androgen receptor as a mechanism for hormone-specific gene regulation. J. Steroid Biochem. Mol. Biol. 76:23–30; 2001. [DOI] [PubMed] [Google Scholar]
- 14. Cunha G. R.; Donjacour A. A.; Cooke P. S.; Mee S.; Bigsby R. M.; Higgins S. J.; Sugimura Y. The endocrinology and developmental biology of the prostate. Endocr. Rev. 8:338–362; 1987. [DOI] [PubMed] [Google Scholar]
- 15. Grant E. S.; Batchelor K. W.; Habib F. K. Androgen independence of primary epithelial cultures of the prostate is associated with a down-regulation of androgen receptor gene expression. The Prostate 29:339–349; 1996. [DOI] [PubMed] [Google Scholar]
- 16. Grouse L. H.; Munson P. J.; Nelson P. S. Sequence databases and microarrays as tools for identifying prostate cancer biomarkers. Urology 57:154–159; 2001. [DOI] [PubMed] [Google Scholar]
- 17. He W. W.; Sciavolino P. J.; Wing J.; Augustus M.; Hudson P.; Meissuer P. S.; Curtis R. T.; Shell B. K.; Bostwick D. G.; Tindall D. J.; Gelmann E. P.; Abate-Shen C.; Carter K. C. A novel human prostate-specific, androgen-regulated homeobox gene (NKX 3.1) that maps to 8p21, a region frequently deleted in prostate cancer. Genomics 43:69–77; 1997. [DOI] [PubMed] [Google Scholar]
- 18. Hedlund T. E.; Miller G. J. A serum-free defined medium capable of supporting growth of four established human prostatic carcinoma cell lines. The Prostate 24:221–228; 1994. [DOI] [PubMed] [Google Scholar]
- 19. Horoszewicz J. S.; Leong S. S.; Chu T. M.; Wajsman Z. L.; Friedman M.; Papsidero L.; Kim U.; Chai L. S.; Kakati S.; Arya S. K.; Sandberg A. A. In: Liss A. R., ed. Models for prostate cancer. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1980:115–132. [Google Scholar]
- 20. Kim H.; Kim J. H.; Lee J. W. Steroid receptor coactivator-1 interacts with serum response factor and coactivates serum response element-mediated transactivation. J. Biol. Chem. 273:28564–28567; 1998. [DOI] [PubMed] [Google Scholar]
- 21. Kos L.; Chiang C.; Mahon K. A. Mediolateral patterning of somites: Multiple axial signals, including Sonic hedgehog, regulate Nkx-3.1 expression. Mech. Dev. 70:25–34; 1998. [DOI] [PubMed] [Google Scholar]
- 22. Kovacs A. M.; Zimmer W. E. Molecular cloning and expression of the chicken smooth muscle γ-actin mRNA. Cell. Motil. Cytoskel. 24:67–81; 1993. [DOI] [PubMed] [Google Scholar]
- 23. Kovacs A. M.; Zimmer W. E. Cell specific transcription of the smooth muscle γ-actin gene requires both positive and negative acting cis-elements. Gene Expr. 7:115–129; 1998. [PMC free article] [PubMed] [Google Scholar]
- 24. Lee J. W.; Lee Y. L.; Na S. Y.; Jung D. J.; Lee S. K. Transcriptional coregulation of the nuclear receptor superfamily: Coactivators and corepressors. Cell. Mol. Life Sci. 58:289–297; 2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lee S. K.; Jung S. Y.; Kim Y. S.; Na S. Y.; Lee Y. C.; Lee J. W. Two distinct nuclear receptor interaction domains and CREβ-binding protein dependent transactivation function of activating signal cointegrator-2. Mol. Endocrinol. 5:241–254; 2000. [DOI] [PubMed] [Google Scholar]
- 26. Lim D. J.; Liu X.; Sutkowski D. M.; Braun E. J.; Lee C.; Kozlowski J. M. Growth of an androgen-sensitive human prostate cancer cell line, LNCaP, in nude mice. The Prostate 22:109–118; 1993. [DOI] [PubMed] [Google Scholar]
- 27. Liu Z.; Wong J.; Tsui S-Y.; Tsai M-J.; O’Malley B. W. Sequential recruitment of steroid receptor coactivator-1 (SRC-1) and p300 enhances progesterone receptor-dependent initiation and reinitiation of transcription from chromatin. Proc. Natl. Acad. Sci. USA 98:12426–12431; 2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. McNeal. In: Liss A. R., ed. Models for prostate cancer. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1980:149–160. [Google Scholar]
- 29. Muller J. M.; Isele U.; Metzger E.; Rempel A.; Moser M.; Pscherer A.; Breyer T.; Holubarsch C.; Buettner R.; Schule R. FHL2, a novel tissue-specific coactivator of the androgen receptor. EMBO J. 19:359–369; 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Nelson C. C.; Hendy S. C.; Shukin R. J.; Cheng H.; Bruchovsky N.; Koop B. F.; Rennie P. S. Determinants of DNA sequence specificity of the androgen, progesterone, and glucocorticoid receptors: Evidence for differential steroid receptor response elements. Mol. Endocrinol. 13:2090–2107; 1999. [DOI] [PubMed] [Google Scholar]
- 31. Nelson P. S.; Pritchard C.; Abbott D.; Clegg N. The human (PEDB) and mouse (mPEDB) prostate expression databases. Nucleic Acids Res. 30:218–220; 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Nelson P. S.; Stanford J. L.; Ostrander E. A. Prostate cancer research in the post-genome era. Epideomiol. Rev. 23:187–190; 2001. [DOI] [PubMed] [Google Scholar]
- 33. Nessler-Menardi C.; Jotova I.; Culig Z.; Eder I. E.; Pulz T.; Bartsch G.; Klocker H. Expression of androgen coregulatory proteins in prostate cancer and stromal-cell culture models. The Prostate 45:124–131; 2000. [DOI] [PubMed] [Google Scholar]
- 34. Nissen L. J.; Gelly J. C.; Hipskind R. A. Induction-independent recruitment of CREB-binding protein to the C-fos serum response element through interactions between the bromodomain and ELK-1. J. Biol. Chem. 276:5213–5221; 2001. [DOI] [PubMed] [Google Scholar]
- 35. Pelligrini L.; Song T.; Richmond T. J. Structure of serum response factor core bound to DNA. Nature 376:490–497; 1995. [DOI] [PubMed] [Google Scholar]
- 36. Prescott J. L.; Blok L.; Tindall D. J. Isolation and androgen regulation of the human homeobox cDNA, NKX 3.1. The Prostate 35:71–80; 1998. [DOI] [PubMed] [Google Scholar]
- 37. Ramirez S.; Ait-Si-Ali S.; Robin P.; Trouche D.; Harel-Bellan A. The CREB-binding protein (CBP) cooperates with the serum response factor for transactivation of the c-fos serum response element. J. Biol. Chem. 272:31016–31021; 1997. [DOI] [PubMed] [Google Scholar]
- 38. Schoenmakers E.; Verrijdt G.; Peeters B.; Verhoeven G.; Rombauts W.; Claessens F. Differences in DNA binding characteristics of the androgen and glucocorticoid receptors can determine hormone-specific responses. J. Biol. Chem. 275:12290–12297; 2000. [DOI] [PubMed] [Google Scholar]
- 39. Sciavolino P. J.; Abrams E. W.; Yang L.; Austenberg L. P.; Shen M. M.; Abate-Shen C. Tissue-specific expression of Murine Nkx 3.1 in the male urogenital system. Dev. Dyn. 209:127–138; 1997. [DOI] [PubMed] [Google Scholar]
- 40. Tan S.; Hunziken Y.; Pelligrini L.; Richmond T. Crystalization of the yeast MAT α-2/MCM1/DNA ternary complex: General methods and principals for protein-DNA cocrystalization. J. Mol. Biol. 297:947–959; 2000. [DOI] [PubMed] [Google Scholar]
- 41. Tanaka M.; Kasahara H.; Burtunkova S.; Schinke M.; Komuro I.; Inagaki H.; Lee Y.; Lyons G. E.; Izumo S. Vertebrate homologs of tinman and bagpipe: Roles of the homeobox genes in cardiovascular development. Dev. Gen. 22:239–249; 1998. [DOI] [PubMed] [Google Scholar]
- 42. Trapman J.; Cleutjens K. B. J. M. Androgen-regulated gene expression in prostate cancer. Cancer Biol. 8:29–36; 1997. [DOI] [PubMed] [Google Scholar]
- 43. Trapman J.; Sleddens H. F.; van der Weiden M. M.; Dingens W. N.; Konig J. J.; Schroder F. H.; Faber P. W.; Bosman F. T. Loss of heterozygosity of chromosome 8 microsatellite loci implicates a candidate tumor suppressor gene between the loci D8S87 and D8S133 in human prostate cancer. Cancer Res. 54:6061–6064; 1994. [PubMed] [Google Scholar]
- 44. Treisman R. Ternary complex factors: Growth factor regulated transcriptional activators. Curr. Opin. Genet. Dev. 4:96–101; 1994. [DOI] [PubMed] [Google Scholar]
- 45. Vocke C. D.; Pozza,tti R. O.; Bostwick D. G.; Florence C. D.; Jennings S. B.; Strup S. E.; Duray P. H.; Liotta L. A.; Emmert-Buck M. R.; Linehan W. M. Analysis of 99 micro-dissected prostate carcinomas reveals a high frequency of allelic loss on chromosome 8p12-21. Cancer Res. 56:2411–2416; 1996. [PubMed] [Google Scholar]
- 46. Voeller H. J.; Augustus M.; Madike V.; Bova G. S.; Carter K. C.; Gelmann E. P. Coding region of Nkx 3.1, a prostate-specific homeobox gene on 8p21 is not mutated in human prostate cancers. Cancer Res. 57:4455–4459; 1997. [PubMed] [Google Scholar]
- 47. Vollmer R. T.; Humphrey P. A. The relative importance of anatomic and PSA factors to outcomes after radical prostatectomy for prostate cancer. Am. J. Clin. Pathol. 116:864–870; 2001. [DOI] [PubMed] [Google Scholar]
- 48. Xu L. L.; Srikantan V.; Sesterhenn I. A.; Augustus M.; Dean R.; Moul J. W.; Carter K. C.; Srivastava S. Expression profile of an androgen regulated prostate specific homeobox gene NKX 3.1 in primary prostate cancer. J. Urol. 163:972–979; 2000. [PubMed] [Google Scholar]


