Abstract
We have analyzed the pattern of core histone acetylation across 250 kb of the telomeric region of the short arm of human chromosome 16. This gene-dense region, which includes the α-globin genes and their regulatory elements embedded within widely expressed genes, shows marked differences in histone acetylation between erythroid and non-erythroid cells. In non-erythroid cells, there was a uniform 2- to 3-fold enrichment of acetylated histones, compared with heterochromatin, across the entire region. In erythroid cells, an ≈100-kb segment of chromatin encompassing the α genes and their remote major regulatory element was highly enriched in histone H4 acetylated at Lys-5. Other lysines in the N-terminal tail of histone H4 showed intermediate and variable levels of enrichment. Similar broad segments of erythroid-specific histone acetylation were found in the corresponding syntenic regions containing the mouse and chicken α-globin gene clusters. The borders of these regions of acetylation are located in similar positions in all three species, and a sharply defined 3′ boundary coincides with the previously identified breakpoint in conserved synteny between these species. We have therefore demonstrated that an erythroid-specific domain of acetylation has been conserved across several species, encompassing not only the α-globin genes but also a neighboring widely expressed gene. These results contrast with those at other clusters and demonstrate that not all genes are organized into discrete regulatory domains.
An important question in the postgenome era concerns the relationship between chromosome structure and function, in particular whether genes and the cis-acting sequences regulating their expression are organized into discrete chromosomal domains. To date, such domains have been variously defined by nuclease sensitivity, histone acetylation, replication timing, and DNA methylation (1). Specialized elements flanking such domains are thought to demarcate the transition between active and inactive chromatin and may isolate cis-acting elements within the domain from the influence of those outside (2–4). It has been suggested that some, but not all, boundary elements co-map with nuclear matrix attachment sites (3). Such independently regulated domains are proposed to be a regular feature of genome organization in eukaryotic chromosomes (2, 3).
The human α-globin cluster provides a challenge to this model. The embryonic (ζ) and fetal/adult (α) α-like genes lie within a gene dense region close to the telomere of the short arm of chromosome 16 (5). Although the ζ and α genes are expressed in a tissue- and developmental stage-specific manner, they are closely flanked by widely expressed genes. Curiously, their major regulatory element (αMRE) lies 40 kb upstream of the ζ-α cluster within the intron of a widely expressed gene (C16orf35) (6, 7). Clearly, this arrangement is not consistent with a model in which independently regulated genes are arranged into structurally discrete domains. Nevertheless, it does not rule out a more complex arrangement in which there are overlapping, independently regulated chromosomal domains that do not interact with each other because of the specificity of the cis-acting elements contained within them (8).
Previous studies have attempted to define a regulatory domain containing the human α-globin cluster. A search for DNase1 hypersensitive sites (HSs) throughout the terminal region of chromosome 16 (5, 6) identified erythroid-specific HSs associated with the αMRE (HS −40) and the promoters of the α-like genes (Fig. 1). Additional sites map between HS −40 and the α cluster but no erythroid-specific HSs have been found upstream or downstream of these sites (9). However, transgenes including a 70-kb segment of DNA spanning all of these erythroid HSs (GG1/GG2 in Fig. 1) are expressed suboptimally in transgenic mice (10), suggesting that the smallest region of the chromosome that will allow full regulation of the α-globin cluster extends beyond this 70-kb segment. Studies defining patterns of methylation (9, 11), replication timing (12), matrix attachment regions (13), and sequences defining nuclear position (14) have not defined any putative domain.
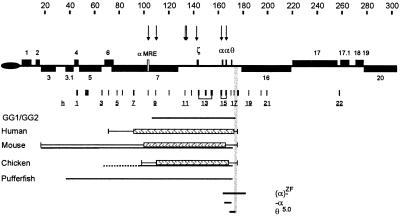
Figure 1.
The telomeric region of human chromosome 16. Telomeric repeats (TTAGGG)n are represented by an oval. Genes surrounding the α cluster are shown as gray boxes and numbered as in ref. 50. The direction of transcription toward the centromere (above the line) or telomere (below the line) is indicated. The αMRE is shown as an open box. Erythroid-specific DNase1 HSs are represented as arrows above the chromosome. The scale is in kilobases. Probes used to assess the pattern of core histone acetylation are shown as underlined numbered boxes below the chromosome (for clarity not all probes are numbered in the figure). The extent of the previously described 70-kb construct (GG1/GG2) used to make transgenic mice (10) is shown as a horizontal line. The domains of acetylation (hatched box) in human, mouse, and chicken are shown, and the regions in which a transition occurs from hyperacetylated to normally acetylated chromatin are indicated by thin horizontal lines. The segments corresponding to conserved syntenic regions in mouse and chicken and pufferfish are shown as continuous horizontal lines below the acetylation domains. The dashed line in the chicken cluster represents DNA that has been mapped but not sequenced. The region containing the 3′ acetylation boundary is indicated by gray shading, and the extents of deletions described in the text are shown. The extent of the θ5.0 deletion (168,996–169,014 to 173,925–173,943; there are 19 bases identical at both ends because the recombination occurred between two Alu repeats) was accurately determined as part of this study (see Materials and Methods).
Recently, DNA sequence comparisons of the α-globin clusters in human, mouse, chicken, and pufferfish (15) have defined an ≈135- to 155-kb region of conserved synteny containing all of the erythroid-specific DNase1 HSs (Fig. 1). At the telomeric end of the α-globin cluster, synteny extends for 67–87 kb upstream of the αMRE. At the centromeric end, synteny in all species ends abruptly beyond the last globin-like gene (θ1 in human). The simplest interpretation is that this segment of conserved synteny, corresponding to ≈155 kb in the human cluster, contains all sequences required for α-globin gene regulation and may correspond to or include the regulatory domain.
To examine this further we have studied the pattern of histone acetylation throughout the terminal region of chromosome 16, including the putative α-globin domain. Reversible acetylation of core histone tails plays an important role in modifying chromatin structure and regulating gene expression, affecting interactions at specific nucleosomes (e.g., at promoters and enhancers) and having long range effects throughout the chromosome (16). In general, inactive chromatin is associated with hypoacetylated histones and active chromatin is hyperacetylated. Previous studies have characterized tissue-specific domains of acetylated histones across the β-globin (17–21) and the GH (22, 23) gene clusters, which are consistent with the widely accepted chromosomal domain model.
Here, we show that, although the cis-acting sequences regulating α-globin expression overlap at least one widely expressed gene (C16orf35), a discrete, broad domain of histone acetylation spanning ≈100 kb forms specifically in human erythroid cells. Similar domains of histone acetylation are also seen spanning the α-globin clusters in the erythroid cells of mouse and chicken. In all three species, these domains are contained within the segment of conserved synteny between them. These data unexpectedly define a discrete erythroid-specific domain of acetylation across the α-globin cluster and extend the models relating chromosomal organization to gene expression.
Materials and Methods
Primary Cells and Cell Lines.
Cell lines examined included the human erythroleukemia cell line K562 (24), a mouse erythroleukemia (MEL) cell line 585 (6), Epstein–Barr virus (EBV)-transformed human lymphocyte cell lines, human primary skin fibroblasts, and the chicken erythroblast cell line HD3 (25). Primary human erythroid cells were obtained from the peripheral blood of normal donors (26).
Antibodies.
Polyclonal antisera to H3 and H4 acetylated at specific sites were raised in rabbits by immunization with synthetic peptides corresponding to sequences in the N-terminal domains of these histones containing acetyl-lysine residues at defined positions (27). Each antiserum was affinity purified (28).
Preparation and Immunoprecipitation of Chromatin (ChIP).
In each ChIP experiment, soluble chromatin fractions (S1 and S2) were isolated from ≈2 × 108 cells (28, 29). DNA from the input, unbound, and bound fractions was obtained by phenol/chloroform extraction and analyzed by 1.2% agarose gel electrophoresis. The specificity and efficiency of ChIP assays was determined by analyzing proteins from input, unbound, and bound fractions by electrophoresis in SDS-polyacrylamide gels. Resolved proteins were transferred to Hybond-C nitrocellulose (Amersham Pharmacia), and histones H3 and H4 were detected by immunostaining (29).
DNA samples, quantified by scintillation counting of [3H]thymidine, were analyzed by slot blots (28). Southern blots of human, mouse, and chicken genomic DNA were included in each experiment to ensure an appropriately low noise/signal ratio. Slot blot signals were quantified by PhosphorImager (Molecular Dynamics) by using imagequant software, and the ratios of bound and unbound DNA fractions were calculated. Quantitations were corrected by estimating the quantity of DNA in each slot after rehybridizing with random primer-labeled genomic DNA or by [3H]thymidine counting.
Preparation of DNA Probes.
The positions of probes corresponding to fragments from human, mouse, and chicken clusters are shown in the figures. Their coordinates with reference to the human (GenBank no. AE005175), mouse (GenBank nos. AY016021 and AY016022), and chicken (GenBank no. AY016022) sequences are available in Table 1, which is published as supporting information on the PNAS web site, www.pnas.org. Probes used as controls were the heterochromatin alphoid repeat het 266 (29), het 947 (28), rDNA probe R4, the Y repeat DYZ2 probe pHY2.1 (30), γ-globin (31), and α-tubulin (28).
Characterization of a Deletion (θ5.0) Lying Close to the 3′ Boundary.
Hematologic analysis and mapping of the α-globin cluster by Southern blot analysis of an unreported patient (AP) was consistent with the previously described θ5.0 deletion (32). Gap PCR with Expand Long Template PCR System (Roche) and the primers θ1-F (167,581–167,604) and θ2-R (174,516–174,495) (Table 2) produced an ≈2-kb band. This fragment was sequenced to determine precisely the position of the breakpoints by using the primers θ1-F, θ3-F, θ4-F, θ5-F, and θ6-R (Table 2).
Results
Histone Acetylation Throughout the Terminal Region of the Short Arm of Chromosome 16 in Non-Erythroid Cells.
We used a previously described chromatin immunoprecipitation (ChIP) protocol (28) to study histone acetylation throughout the terminal ≈250 kb of chromosome 16. Oligonucleosomes were prepared by micrococcal nuclease digestion of nuclei, and soluble chromatin was immunoprecipitated without further purification of the nucleosomes. Although the resolution by using this approach is less than achieved by analyzing mono- and di-nucleosomes, it provides very similar results (19) and is appropriate for characterizing the broad domains of acetylation being sought in this study. Chromatin was immunoprecipitated with polyclonal antibodies that distinguish histone H4 (H4) isoforms acetylated at Lys-16, Lys-12, Lys-8, or Lys-5 (33, 34). We also used an antibody directed against the N-terminal tails of histone H3 (H3) acetylated at Lys-14 (27). Previous analyses with these antibodies have shown that H4 associated with both centromeric and telomeric heterochromatin is underacetylated. By contrast, H4 acetylation of three nonexpressed, tissue-specific genes (β-globin, human GH, and pro-insulin) and two widely expressed genes (c-myc and α-tubulin), in EBV-transformed lymphoblasts, showed a 2- to 3-fold higher level of acetylation than heterochromatin (28).
ChIP experiments were performed on EBV-transformed lymphoblasts and primary fibroblasts, which express many of the genes (e.g., gene nos. 5, 6, 7, 16, and 17 in Fig. 1) in this region of chromosome 16, but not the globin genes (5, 35). Slot blots containing input, unbound, and bound (immunoprecipitated) fractions of DNA from the ChIP experiment were analyzed with unique sequence probes (Fig. 1) located throughout the terminal region of chromosome 16 and previously characterized control sequences (Fig. 2). Each cell line was examined in three entirely independent ChIP experiments to evaluate the reproducibility of the assay. As previously noted (28), the chromatin associated with such genes was consistently enriched (2- to 3-fold) for all forms of acetylated histones compared with heterochromatin (het 266 and DYZ2), but there were no differences between expressed and nonexpressed genes in the regions analyzed (Figs. 2 a and b, 3, and 4a).
Figure 2.
The patterns of core histone acetylation across the human α-globin cluster with the probes used set out below annotated as in Fig. 1 and described in detail in Table 1, which is published as supporting information on the PNAS web site, www.pnas.org. The scale is in kilobases. Acetylation of histone H4 at Lys-16 (white boxes and dashed line) and Lys-5 (black circles and black line) in (a) human EBV-transformed B lymphocytes, (b) fibroblasts, (c) K562, and (d) primary human erythroid progenitors. Levels of acetylation at control loci were established independently for each experiment and include the α-tubulin genes (T), a probe to heterochromatin repeat sequences (het 266) (H), human rDNA (R), a probe directed to a heterochromatic Y chromosome repeat (D), which is also present on the X chromosome, and a probe to the human γ-globin gene (G). The putative boundaries of acetylation are indicated by vertical black lines. Gray lines indicate the position of the erythroid-specific HSs.
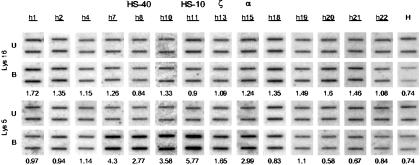
Figure 3.
Slot blots used to calculate the ratios (corrected for loading) of bound/unbound chromatin in the ChIP assays for primary erythroid cells. The probes used are annotated as in Fig. 1. The ratio of bound(B)/unbound(U) chromatin at each point (average of three or four doubling dilutions) by using antibodies against H4 Lys-5 and Lys-16 is shown below. Examples of the controls by using the het 266 probe (H) for the corresponding experiment are shown. The values plotted in Figs. 2 and 4 are the averages of these and other ChIP experiments.
Figure 4.
Analysis of acetylation of the core histone H4 at Lys-16 (black), Lys-12 (gray), Lys-8 (white), Lys-5 (red), and H3 at Lys-14 (blue) in EBV-transformed lymphoblasts (a) and primary erythroblasts (b). The probes correspond to the annotation in Fig. 1. Error bars correspond to +1 SD. For full analyses of similar data for each experiment described here, see Fig. 7.
Histone Acetylation Across the α-globin Cluster in Human Erythroid Cells.
By using the same experimental approach, we analyzed histone acetylation in an erythroid cell line (K562) and primary human erythroblasts (26). The clearest change in histone acetylation was seen by using antibodies against H4 Lys-16 and H4 Lys-5 (Fig. 2 c and d). Acetylation of H4 Lys-16 appeared unchanged, compared with non-erythroid cells, whereas acetylation of H4 Lys-5 was enriched between 2- and 6-fold (Fig. 2 c and d). The broad segment of chromatin enriched in acetylated H4 Lys-5 was quite discrete (coordinates ≈72,000–176,000) with well defined telomeric (≈72,000–91,000) and centromeric (≈173,000–176,000) borders. Analysis of other isoforms of acetylated histone H4 showed intermediate levels of enrichment across this region (Fig. 4b). Acetylated histone H3 (Lys-14) was also enriched in some, but not all, areas within this region.
This erythroid-specific pattern of acetylation was similar in K562 and primary erythroid cells, although an additional peak of acetylation (coordinate ≈92,000–93,000) was seen in primary erythroid cells. Although these experiments were not designed to look at the globin genes at high resolution (see above), we noted that, in K562 cells, which express embryonic and fetal globins, acetylation of regions around the ζ gene were higher than those around the α genes. In primary adult erythroid cells, the opposite was found (Fig. 2 c and d), thus correlating globin gene expression and histone acetylation.
The Erythroid Domain of Histone Acetylation Is Contained Within the Region of Conserved Synteny Between the Human and Mouse α-globin Clusters.
The region of conserved synteny between the human and mouse α-globin clusters has been defined (15). Whereas the human gene cluster lies close to the 16p telomere, the mouse cluster is located at an interstitial site on chromosome 11. In the region corresponding to the telomeric end of the human α cluster, conserved synteny ends beyond the IL-9 receptor pseudogene (gene no. 3 in Fig. 1). At the centromeric end, conserved synteny ends abruptly beyond the θ1 gene (no. 15 in Fig. 1), and the orthologue of Luc7L (gene no. 16 in Fig. 1) lies on mouse chromosome 17.
To investigate further the significance of the pattern of histone acetylation observed in the human cluster, we asked whether the region of conserved synteny also contained a domain of histone hyperacetylation in the mouse. By using the same panel of antibodies to perform ChIP assays in mouse erythroleukemia cells, we identified a similar, well-defined domain of H4 Lys-5 acetylation between coordinates 2,000–142,000 of the mouse α-globin cluster (Fig. 5). The pattern of H4 Lys-12 acetylation appeared similar to Lys-16; Lys-8 showed similar levels of enrichment to Lys-5 (Fig. 7). As before, the borders of the acetylation domain were well defined and located in very similar positions to those in the human cluster (Figs. 1 and 5). This phenomenon was particularly clear at the border corresponding to the centromeric end of the human cluster, which was analyzed in seven entirely independent ChIP experiments (Fig. 5b).
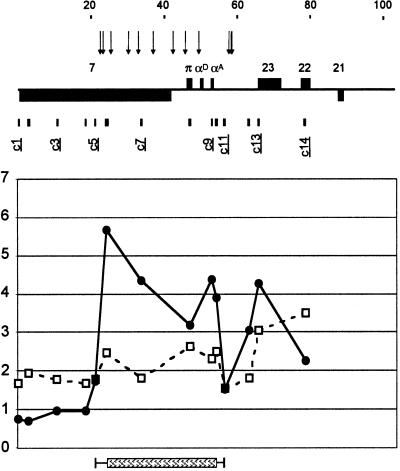
Figure 5.
The pattern of histone H4 acetylation across the murine α-globin locus in mouse erythroleukemia cells. (a) Above, the structure of the murine locus is set out as previously described (15). The globin genes are annotated, and other orthologous genes correspond to the human genes set out in Fig. 1 (e.g., murine no. 7 is the orthologue of human no. 7). The probes used to establish the patterns of core histone acetylation across the murine αglobin cluster are described in Table 1. The scale is in kilobases. The asterisk denotes a hypersensitive site that is assumed to be present but has not been experimentally demonstrated. Acetylation of the core histone H4 at Lys-16 (white boxes and dashed line) and Lys-5 (black circles and black line) is shown. The levels of acetylation for other N-terminal tail lysines are displayed in Fig. 7. Levels of acetylation at control loci were established independently for each experiment and include the α-tubulin genes (T) and a probe (het 947) to heterochromatin repeat sequences (H). (b) The levels of acetylation of Lys-5 and Lys-16 across the putative 3′ boundary were assessed in seven entirely independent experiments. The results obtained with a probe (m8: 123,063–124,463 and 135,985–137,367) to the α-globin gene are shown as a diagonally striped box and those by using a probe beyond the putative boundary (m9: 141886–142293) as a vertically striped box. Vertical lines indicate ±1 SD.
It therefore seems likely that this region of conserved synteny, which contains all known erythroid-specific HSs and the entire segment of histone acetylation in both human and mouse, contains the α-globin regulatory domain.
The Erythroid Domain of Histone Acetylation Also Corresponds to the Region of Conserved Synteny in Chicken.
It has been shown previously that the chicken α-globin gene (αD) is associated with acetylated core histones in erythrocytes (18). Here, we studied acetylation across the entire chicken α-globin cluster (Fig. 6). In this case, the extent of conserved synteny with the telomeric region of the human α-globin cluster has not been fully defined. However, as for the mouse and pufferfish (15), conservation of synteny at the centromeric end of the human α-globin cluster ends abruptly beyond the globin genes. By using HD3 chicken erythroid progenitor cells, we again defined a domain of H4 Lys-5 acetylation spanning a similar region of the α-globin cluster (21,479–56,000) with a boundary corresponding to the well-defined break in synteny downstream of the α genes. The patterns of acetylation observed at Lys-8 and Lys-12 were similar to Lys-5 although less consistently enriched (see Fig. 7). Acetylation of H3 Lys-14 showed a similar pattern to that of H4 Lys-5. Therefore, these patterns of histone acetylation are similar to those observed in other species, although the differences in the degree of acetylation of the various H4 isoforms was less prominent in the chicken cluster.
Figure 6.
The pattern of histone H4 acetylation across the chicken α-globin locus in HD3 cells. Above, the structure of the chicken locus is set out as described in ref. 15. The previously mapped positions of erythroid-specific DNase I hypersensitive sites are shown by vertical arrows. The globin genes are annotated, and other orthologous genes correspond to the human genes set out in Fig. 1 (e.g., chicken no. 7 is the orthologue of human no. 7). The probes used to establish the patterns of core histone acetylation across the chicken α-globin cluster are set out in Table 1. The scale is in kilobases. Acetylation of the core histone H4 at Lys-16 (white boxes and dashed line) and Lys-5 (black circles and black line) is shown. The levels of acetylation for other N-terminal tail lysines are displayed in Fig. 7.
Discussion
Histone acetylation may decrease the stability of the compacted 30-nm chromatin fiber (36) and thereby destabilize the higher order structures through which chromatin is folded into the chromosome (37). Unfolding of chromatin may facilitate many nuclear processes (transcription, replication, recombination, methylation, and DNA repair) and be reflected in a variety of indirect assays and observations (accessibility to endonucleases, timing of replication, nuclear position, and patterns of DNA methylation). By using a ChIP assay, we have shown that, in fibroblasts and lymphoblasts, chromatin in the most telomeric 250-kb region of chromosome 16 is uniformly enriched in acetylated histones when compared with centric (het 266), interstitial (DYZ2), and the telomeric (TTAGGG)n heterochromatin reported by Johnson et al. (28). This finding is consistent with observations on other euchromatic regions, suggesting that this segment of 16p is a region of “open,” early replicating chromatin, containing a high density of unmethylated CpG islands and widely expressed genes (5, 12, 14).
Consistent with previous observations (28, 29), we found that, in non-erythroid cells, there are no differences in the levels of any of the H3 or H4 acetylated isoforms when comparing expressed (e.g., nos. 5, 6, 7, 16, and 17) and nonexpressed genes (e.g., globin genes). Therefore, as for other areas of euchromatin, nucleosomes monoacetylated at Lys-16 may reflect the potential for transcription rather than the transcriptional process itself. Whereas widely expressed genes are transcribed from 16p in its uniformly acetylated state in non-erythroid cells, the globin genes remain silent. This may result from the lack of the tissue-specific transcriptional activators (e.g., GATA-1) known to be involved in globin gene expression but may also involve active repression of the α-globin genes.
In erythroid cells, in which the globin genes are expressed at very high levels, we observed considerable enrichment of acetylated histones across a broad segment of ≈100 kb of the human α-globin cluster, including all currently known cis-acting elements and almost all of the gene (C16orf35, gene no. 7) lying upstream. Although the entire domain becomes acetylated in erythroid cells, there is variation from one region to another. At least part of this pattern could be explained by the recruitment of HATs to erythroid-specific cis-acting elements (e.g., αMRE and the ζ and α promoters) either by transcriptional activators (e.g., GATA-1) and coactivators (e.g., p300/CBP) or as part of the basal transcription machinery (e.g., TFIID or pol II holoenzyme). It is interesting that the highest peak of acetylation is associated with a previously described erythroid-specific hypersensitive site (HS-10) for which no functional role has yet been identified. Other regions of high acetylation include the αMRE, the erythroid specific HS −33, the ζ genes in K562 cells, and α genes in primary erythroid cells, consistent with their relative activities in these cells. We also noted that regions located as far as 10 kb from any known erythroid-specific HSs (e.g., probe h10) showed an enrichment in acetylated histones. This suggests that either HATs are recruited to multiple sites throughout the domain or that they are recruited as part of one or more complexes, assembled where the peaks of acetylation occur, that move through the domain (38) in erythroid cells.
The acetylation of specific H4 isoforms in erythroid cells was of interest. Whereas acetylation of Lys-16 appeared unchanged, or slightly depleted compared with non-erythroid cells, Lys-5 appeared most enriched, with Lys-8 and –12 showing intermediate levels of enrichment. Does this represent a specific acetylation of Lys-5 or does it more simply reflect the overall level of histone acetylation? In general, the N-terminal lysines in H4 are acetylated in a defined order: Lys-16 followed by Lys-8 and –12, with Lys-5 being acetylated last (39). A simple increase in acetylation of all terminal lysines moving the equilibrium toward the tetra-acetylated form would certainly result in enrichment of Lys-5. However, unless Lys-16 in this region is fully acetylated in both erythroid and non-erythroid cells, we would have also expected to see further enrichment of this isoform in erythroid cells. Therefore the N-terminal lysines may not be acetylated in this strictly defined order in all regions of the genome, and there may be specific acetylation of H4 Lys-5 (and to a lesser extent Lys-8 and Lys-12) in erythroid cells, possibly by a specific HAT activity. This pattern could represent part of an erythroid-specific histone code (40, 41) for the α cluster.
Very similar patterns of acetylation were observed in the corresponding syntenic regions of the mouse and chicken globin loci, suggesting that the process by which this domain is created has been conserved throughout evolution. Perhaps the most striking conclusion from these comparative analyses was the similar locations of the borders of the acetylated domain. In the region corresponding to the telomeric end of the domain in the human cluster, there appeared to be a border lying between the C16orf35 (gene no. 7) and MPG (gene no. 6) genes. At the centromeric end, there was a striking correspondence between the end of the acetylation domain (coordinates ≈173,000–176,000 of the human α cluster) and the previously described breakpoint in synteny between the α-globin clusters of human, mouse, chicken, and pufferfish (coordinates 171,000–171,186), which, in man, separates the α cluster from the Luc7L gene (gene no. 16).
There are several interesting aspects to the region spanning and adjacent to this segment of the α-globin cluster. In the species studied, the break in synteny in this region has arisen by different, independent recombination events in evolution, suggesting that it marks one end of the α-globin domain (15). Furthermore, it could be a preferred site of recombination; many deletions that cause α thalassemia have breakpoints in or close to this area (42). In fluorescence in situ hybridization (FISH)-based replication studies, we noted that sequences spanning this region separate later than any other segments in the terminal region of chromosome 16, suggesting that this is a point where sister chromatids might be strongly attached to each other (12). This finding may be of interest because of two recent reports of a role for cohesin in boundary function in Saccharomyces cerevisiae (43, 44). All of these observations suggest that this region of the chromosome may be somewhat different from the flanking regions.
Because this region includes a transition in the levels of acetylation of nucleosomes, by analogy to previous observations in the chicken β-globin cluster (4, 45–47), it may represent a chromosomal domain boundary. It is therefore of interest that we recently described an ≈18-kb deletion (coordinates 164,044–182,396) spanning this area (α-ZF in Fig. 1), which silences expression of the remaining structurally normal α-globin gene. During development, the CpG island associated with this α-globin gene becomes methylated and insensitive to endonucleases, demonstrating that the normal chromatin structure around this α gene is perturbed by the α-ZF deletion (48). It appears that the structurally normal α gene is inactivated by a negative chromosomal position effect. This finding is consistent with the view that sequences in this region normally provide a barrier to as yet undetermined effect(s) of the gene (Luc7L, gene no. 16) and/or chromatin downstream. This region may be further defined by other deletions, including the θ5.0 (32) characterized further here, removing sequences flanking the putative boundary that appear not to exert any position effects on the α genes (Fig. 1).
This study has shown that one of the features associated with the model describing chromosomal domains can be applied to the α-globin cluster even though this domain overlaps at least one widely expressed gene (C16orf35, gene no. 7) lying upstream of the cluster. Expression of this gene appears not to affect α-globin expression in non-erythroid cells, and its own expression appears to be unaffected in human and mouse by activation of the αMRE and the globin genes (9), with extension of the domain of erythroid-specific acetylation across most of its length. Presumably the multiprotein complexes regulating expression of these two genes do not interact and therefore are “insulated” from each other by a quite different mechanism from that proposed in the classical domain model (8). For example, acetylation of H4 Lys-5 may provide a signal involved in erythroid expression that is irrelevant to other genes embedded in the same region.
To date, all transgenes containing the α genes and their regulatory element have been expressed suboptimally (49), but this study has shown that none of them contains the entire domain of acetylation and its boundaries. Clearly an important extension of these findings will be to evaluate transgenes containing the entire putative domain.
Supplementary Material
Acknowledgments
We thank Dr. C. Tufarelli for probes and unpublished information on the mouse α cluster. This work was supported by the Medical Research Council and Wellcome Trust (E.A. and B.T.).
Abbreviations
- αMRE
major regulatory element
- HS
hypersensitive site
- EBV
Epstein–Barr virus
- ChIP
chromatin immunoprecipitation
References
- 1.Wolffe A, editor. Chromatin: Structure and Function. London: Academic; 1995. [Google Scholar]
- 2.Bell A C, Felsenfeld G. Curr Opin Genet Dev. 1999;9:191–198. doi: 10.1016/S0959-437X(99)80029-X. [DOI] [PubMed] [Google Scholar]
- 3.Udvardy A. EMBO J. 1999;18:1–8. doi: 10.1093/emboj/18.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bell A C, West A G, Felsenfeld G. Science. 2001;291:447–450. doi: 10.1126/science.291.5503.447. [DOI] [PubMed] [Google Scholar]
- 5.Flint J, Thomas K, Micklem G, Raynham H, Clark K, Doggett N A, King A, Higgs D R. Nat Genet. 1997;15:252–257. doi: 10.1038/ng0397-252. [DOI] [PubMed] [Google Scholar]
- 6.Higgs D R, Wood W G, Jarman A P, Sharpe J, Lida J, Pretorius I-M, Ayyub H. Genes Dev. 1990;4:1588–1601. doi: 10.1101/gad.4.9.1588. [DOI] [PubMed] [Google Scholar]
- 7.Vyas P, Vickers M A, Picketts D J, Higgs D R. Genomics. 1995;29:679–689. doi: 10.1006/geno.1995.9951. [DOI] [PubMed] [Google Scholar]
- 8.Dillon N, Sabbattini P. BioEssays. 2000;22:657–665. doi: 10.1002/1521-1878(200007)22:7<657::AID-BIES8>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 9.Vyas P, Vickers M A, Simmons D L, Ayyub H, Craddock C F, Higgs D R. Cell. 1992;69:781–793. doi: 10.1016/0092-8674(92)90290-s. [DOI] [PubMed] [Google Scholar]
- 10.Gourdon G, Sharpe J A, Wells D, Wood W G, Higgs D R. Nucleic Acids Res. 1994;22:4139–4147. doi: 10.1093/nar/22.20.4139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Craddock C F, Vyas P, Sharpe J A, Ayyub H, Wood W G, Higgs D R. EMBO J. 1995;14:1718–1726. doi: 10.1002/j.1460-2075.1995.tb07161.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smith Z E, Higgs D R. Hum Mol Genet. 1999;8:1373–1386. doi: 10.1093/hmg/8.8.1373. [DOI] [PubMed] [Google Scholar]
- 13.Jarman A P, Higgs D R. EMBO J. 1988;7:3337–3344. doi: 10.1002/j.1460-2075.1988.tb03205.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brown K E, Amoils S, Horn J M, Buckle V J, Higgs D R, Merkenschlager M, Fisher A G. Nat Cell Biol. 2001;3:602–606. doi: 10.1038/35078577. [DOI] [PubMed] [Google Scholar]
- 15.Flint J, Tufarelli C, Peden J, Clark K, Daniels R J, Hardison R, Miller W, Philipsen S, Tan-Un K C, McMorrow T, Frampton J, Alter B, Frischauf A-M, Higgs D R. Hum Mol Genet. 2001;10:371–382. doi: 10.1093/hmg/10.4.371. [DOI] [PubMed] [Google Scholar]
- 16.Grunstein M. Nature (London) 1997;389:349–352. doi: 10.1038/38664. [DOI] [PubMed] [Google Scholar]
- 17.Hebbes T R, Thorne A W, Crane-Robinson C. EMBO J. 1988;7:1395–1402. doi: 10.1002/j.1460-2075.1988.tb02956.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hebbes T R, Clayton A L, Thorne A W, Crane-Robinson C. EMBO J. 1994;13:1823–1830. doi: 10.1002/j.1460-2075.1994.tb06451.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Litt M D, Simpson M, Recillas-Targa F, Prioleau M-N, Felsenfeld G. EMBO J. 2001;20:2224–2235. doi: 10.1093/emboj/20.9.2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schübeler D, Francastel C, Cimbora D M, Reik A, Martin D I K, Groudine M. Genes Dev. 2000;14:940–950. [PMC free article] [PubMed] [Google Scholar]
- 21.Forsberg E C, Downs K M, Christensen H M, Im H, Nuzzi P A, Bresnick E H. Proc Natl Acad Sci USA. 2000;97:14494–14499. doi: 10.1073/pnas.97.26.14494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Elefant F, Cooke N E, Liebhaber S A. J Biol Chem. 2000;275:13827–13834. doi: 10.1074/jbc.275.18.13827. [DOI] [PubMed] [Google Scholar]
- 23.Elefant F, Su Y, Liebhaber S A, Cooke N E. EMBO J. 2000;19:6814–6822. doi: 10.1093/emboj/19.24.6814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Andersson L C, Nilsson K, Gahmberg C G. Int J Cancer. 1979;23:143–147. doi: 10.1002/ijc.2910230202. [DOI] [PubMed] [Google Scholar]
- 25.Beug H, Doederlein G, Freudenstein C, Graf T. J Cell Physiol. 1982;1:195–207. doi: 10.1002/jcp.1041130427. [DOI] [PubMed] [Google Scholar]
- 26.Fibach E, Manor D, Oppenheim A, Rachmilewitz E A. Blood. 1989;73:100–103. [PubMed] [Google Scholar]
- 27.White D A, Belyaev N D, Turner B M. Methods. 1999;19:417–424. doi: 10.1006/meth.1999.0878. [DOI] [PubMed] [Google Scholar]
- 28.Johnson C A, O'Neill L P, Mitchell A, Turner B M. Nucleic Acids Res. 1998;26:994–1001. doi: 10.1093/nar/26.4.994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.O'Neill L P, Turner B M. EMBO J. 1995;14:3946–3957. doi: 10.1002/j.1460-2075.1995.tb00066.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gibbons R J, McDowell T L, Raman S, O'Rourke D M, Garrick D, Ayyub H, Higgs D R. Nat Genet. 2000;24:368–371. doi: 10.1038/74191. [DOI] [PubMed] [Google Scholar]
- 31.Morley B J, Abbot C A, Wood W G. Blood. 1991;78:1355–1363. [PubMed] [Google Scholar]
- 32.Fei Y J, Fujita S, Huisman T H J. Br J Haematol. 1988;68:249–254. doi: 10.1111/j.1365-2141.1988.tb06197.x. [DOI] [PubMed] [Google Scholar]
- 33.Turner B M, Fellows G. Eur J Biochem. 1989;179:131–139. doi: 10.1111/j.1432-1033.1989.tb14530.x. [DOI] [PubMed] [Google Scholar]
- 34.Turner B M. Exp Cell Res. 1989;182:206–214. doi: 10.1016/0014-4827(89)90292-9. [DOI] [PubMed] [Google Scholar]
- 35.Vickers M A, Vyas P, Harris P C, Simmons D L, Higgs D R. Proc Natl Acad Sci USA. 1993;90:3437–3441. doi: 10.1073/pnas.90.8.3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wolffe A P, Hayes J J. Nucleic Acids Res. 1999;27:711–720. doi: 10.1093/nar/27.3.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tse C, Sera T, Wolffe A P, Hansen J C. Mol Cell Biol. 1998;18:4629–4638. doi: 10.1128/mcb.18.8.4629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wittschieben B O, Otero G, de Bizemont T, Fellows J, Erdjument-Bromage H, Ohba R, Li Y, Allis C D, Tempst P, Svejstrup J Q. Mol Cell. 1999;4:123–128. doi: 10.1016/s1097-2765(00)80194-x. [DOI] [PubMed] [Google Scholar]
- 39.Thorne A W, Kmiciek D, Mitchelson K, Sautiere P, Crane-Robinson C. Eur J Biochem. 1990;93:701–713. doi: 10.1111/j.1432-1033.1990.tb19390.x. [DOI] [PubMed] [Google Scholar]
- 40.Strahl B D, Allis C D. Nature (London) 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 41.Turner B M. BioEssays. 2000;22:836–845. doi: 10.1002/1521-1878(200009)22:9<836::AID-BIES9>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 42.Nicholls R D, Fischel-Ghodsian N, Higgs D R. Cell. 1987;49:369–378. doi: 10.1016/0092-8674(87)90289-3. [DOI] [PubMed] [Google Scholar]
- 43.Donze D, Adams C R, Rine J, Kamakaka R T. Genes Dev. 1999;13:698–708. doi: 10.1101/gad.13.6.698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Laloraya S, Guacci V, Koshland D. J Cell Biol. 2000;151:1047–1056. doi: 10.1083/jcb.151.5.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Prioleau M N, Nony P, Simpson M, Felsenfeld G. EMBO J. 1999;18:4035–4048. doi: 10.1093/emboj/18.14.4035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Saitoh N, Bell A C, Recillas-Targa F, West A G, Simpson M, Pikaart M, Felsenfeld G. EMBO J. 2000;19:2315–2322. doi: 10.1093/emboj/19.10.2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bell A C, West A G, Felsenfeld G. Cell. 1999;98:387–396. doi: 10.1016/s0092-8674(00)81967-4. [DOI] [PubMed] [Google Scholar]
- 48.Barbour V M, Tufarelli C, Sharpe J A, Smith Z E, Ayyub H, Heinlein C A, Sloane-Stanley J, Indrak K, Wood W G, Higgs D R. Blood. 2000;96:800–807. [PubMed] [Google Scholar]
- 49.Higgs D R, Sharpe J A, Wood W G. Semin Hematol. 1998;35:93–104. [PubMed] [Google Scholar]
- 50.Daniels R J, Peden J F, Lloyd C, Horsley S W, Clark K, Tufarelli C, Kearney L, Buckle V J, Doggett N A, Flint J, Higgs D R. Hum Mol Genet. 2001;10:339–352. doi: 10.1093/hmg/10.4.339. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.