Abstract
Wnt/β-catenin signaling activity is maintained in homeostasis by an expanding list of molecular determinants. However, the molecular components and the regulatory mechanisms involved in its fine-tuning remain to be determined. Here, we identified C9orf140, a tumor-specific protein, as a novel Axin1-interacting protein by tandem-affinity purification and mass spectrometry. We further showed that C9orf140 is a negative regulator of Wnt/β-catenin signaling in cultured cells as well as in zebrafish embryos. It functions upstream of β-catenin, outcompetes PP2A for binding to Axin1, influences the balance between phosphorylation and de-phosphorylation of β-catenin, and ultimately compromises Wnt3A-induced β-catenin accumulation. Interestingly, Wnt-induced C9orf140 expression via β-catenin. We propose that C9orf140 mediates a negative feedback loop of Wnt/β-catenin signaling by interacting with Axin1. Our results advance the current understanding of the exquisite control of Wnt/β-catenin signaling cascade, and provide evidence of the new role of C9orf140.
Introduction
The canonical Wnt signaling pathway coordinates cell-fate specification and has a key role in normal metazoan development and survival [1]. Dysregulated Wnt signaling results in developmental deficits and contributes to multiple human malignancies, including cancer [2]. A key feature of the canonical Wnt signaling is the regulation of β-catenin stabilization by a cytoplasmic destruction complex, composed of Axin1, adenomatous polyposis coli (APC), casein kinase 1 (CK1), and glycogen synthase kinase 3 (GSK3) [3]. Without Wnt signals, a low level of cytoplasmic β-catenin was kept through captured by Axin1 in the destruction complex, phosphorylated by CK1 and GSK3, and then ubiquitinated by E3 ubiquitin ligase SCFβ-TrCP [2]. In the process, protein phosphatases (PP) such as PP2A may act as antagonists of CK1 and GSK3, dephosphorylating β-catenin [4, 5]. With Wnt ligand stimulation, the SCFβ-TrCP -mediated ubiquitination of β-catenin was suppressed, which leads to the stabilization of β-catenin and the transcriptional activation of Wnt target genes [2]. Among all other core components of the destruction complex, Axin1 is the central scaffold and the rate-limiting factor, because the other components regulate Wnt/β-catenin signaling by directly interacting with Axin1 [6]. Given the important role of Wnt/β-catenin signaling in maintenance of cell and tissue homeostasis, novel regulators that interact with Axin1 and the regulatory mechanisms involved in their fine-tuning should be determined [6].
Here, we identified a novel Axin1-interacting protein named C9orf140, also known as p42.3 and suppressor APC domain containing 2 (SAPCD2). Functional analyses in cultured cells and zebrafish demonstrate that C9orf140 negatively regulates Wnt/β-catenin signaling at the destruction complex level. Moreover, the expression of C9orf140 is induced by Wnt through β-catenin. Our results suggest that C9orf140 induced by Wnt functions in a negative feedback loop by outcompeting PP2A for binding to Axin1, thus leading to a significant decrease in β-catenin protein levels and the transcript levels of Wnt target genes.
Results
C9orf140 is a novel Axin1-interacting factor
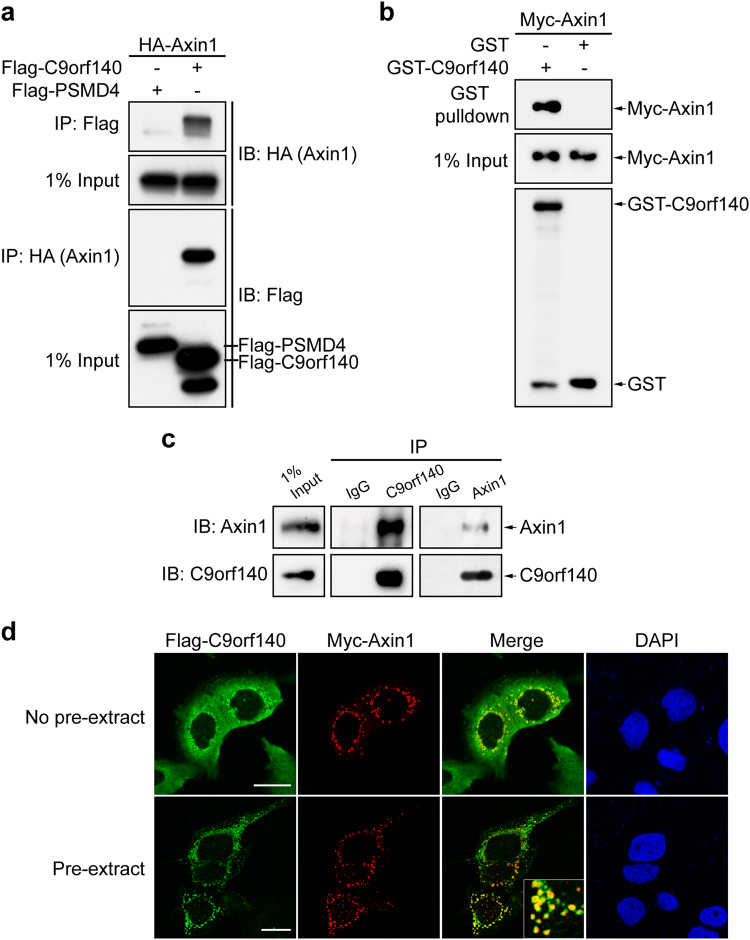
To understand how the Wnt/β-catenin signaling pathway is regulated, we set out to define its molecular makeup using proteomic approaches. By coupling tandem-affinity purification with mass spectrometry analysis, we have successfully identified C9orf140, an uncharacterized protein, as a novel factor that resides in the Axin1 protein complex (Supplementary Table 1). We further carried out a reciprocal co-immunoprecipitation experiment (Fig. 1a) and a GST pulldown experiment (Fig. 1b), respectively, which confirmed the interaction between ectopically expressed C9orf140 and Axin1. Furthermore, we found that C9orf140 also interacted with endogenous Axin1 (Fig. 1c). Together, these data suggested that C9orf140 is a novel Axin1-interacting protein.
Fig. 1.
C9orf140 is a novel Axin1-interacting protein. a Confirmation of the C9orf140–Axin1 interaction by using co-immunoprecipitation. The 293T cells were co-transfected with Flag-C9orf140/Flag-PSMD4 and HA-Axin1 for 24 h. Cell lysates were subjected to immunoprecipitation with an anti-Flag antibody and an anti-HA antibody. Co-precipitating proteins were visualized by western blotting. The unrelated protein PSMD4 was used as a negative control. b The purified GST or GST-C9orf140-fusion protein bound to agarose beads was added to the lysate of 293T cells ectopically expressing Myc-Axin1. After GST affinity purification, protein complexes were washed and detected by western blot analysis with anti-Myc or anti-GST as indicated. GST protein was used as a negative control. c HeLa cell lysates were immunoprecipitated with rabbit anti-Axin1 antibody, rabbit anti-C9orf140 antibody and control rabbit IgG, then immunoblotted with antibodies as indicated. d C9orf140 co-localizes with Axin1. HeLa cells were transfected with Flag-C9orf140 and Myc-Axin1. The cells were then fixed and stained with anti-Flag antibody (green), anti-Myc antibody (red), and DAPI (blue). No pre-extract: cells were first fixed and then permeabilized as described in the Materials and methods; Pre-extract: cells were first permeabilized and then fixed as described in the Materials and Methods. Bar, 20 μm
To support this hypothesis, we performed indirect immunofluorescence studies to examine the sub-cellular localization of C9orf140 and Axin1 through overexpressing Flag-C9orf140 and Myc-Axin1 in the cells. Without pre-extraction treatment, we found that C9orf140 was primarily diffused in the cytoplasm and partly formed distinct cytoplasmic puncta, which seemed to co-localize with those of Axin1, in HeLa cells (Fig. 1d, upper panel). To better show their co-localization at the puncta, we removed the cytosolic proteins through firstly permeabilizing and then fixing the cells. After the pre-extraction treatment, we found that C9orf140 formed striking puncta which overlapped with Axin1 puncta perfectly, as indicated by the yellow signals in the merged images (Fig. 1d, lower panel). The co-localization of C9orf140 and Axin1 was also detected in 293T cells (Supplementary Fig 1). It seems that C9orf140 is not sequestered by Axin1, because its localization did not change when Flag-C9orf140 was transfected into HeLa cells alone or together with Axin1 (data not shown).
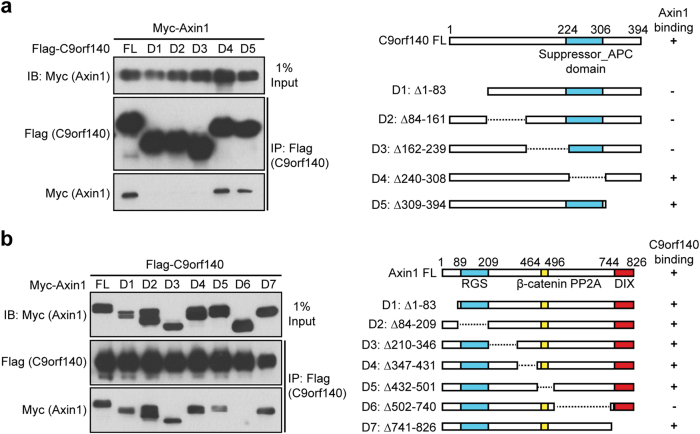
The N-terminal of C9orf140 ranging from amino acids 1–239 is crucial for its interaction with Axin1
C9orf140 has been reported to have an EF-hand domain generally found in the tumor-related calcium-binding protein S100 family at the N-terminal and a coil–coil (CC) domain involved in protein interactions at the C-terminal [7]. Additionally, the regulatory network of C9orf140 was established by bioinformatics methods based on these two domains [8]. Through a BLAST search in the NCBI database, we analyzed the protein sequence of C9orf140 and found no obvious domain or motif except for a suppressor_APC domain at its C-terminus, which has nuclear export activity as well as many different intracellular functions (Fig. 2a). To determine the domains responsible for C9orf140’s interaction with Axin1, we generated a series of deletion mutants of C9orf140 on the basis of the above motif analyses and co-transfected these constructs with Axin1 into 293T cells. We found that amino acids 1–239 of C9orf140 are necessary for the interaction with Axin1 (Fig. 2a).
Fig. 2.
Determination of the domains responsible for the interaction between C9orf140 and Axin1. a Identification of the C9orf140 domain responsible for interacting with Axin1. The 293T cells were transfected with Myc-Axin1 and a series of Flag-tagged C9orf140 deletion mutants as depicted in the diagrams. Cell lysates were immunoprecipitated with an anti-Flag antibody, and the precipitates and whole-cell lysates were then analyzed by western blotting. b Identification of the Axin1 domain responsible for interacting with C9orf140. Flag-C9orf140 was co-transfected into 293T cells with Myc-Axin1 or a series of Myc-tagged Axin1 mutants. Flag-C9orf140 was immunoprecipitated, and the precipitates and cell lysates were then subjected to western blot analysis
Using similar methods, we further studied the region of Axin1 responsible for its binding to C9orf140. Seven deletion mutants of Axin1 were generated on the basis of its identified functional domains [9, 10]. Only the D6 deletion mutant (lacking the PP2A binding region) lost the ability to bind to C9orf140 (Fig. 2b), thus indicating that the domain comprising amino acids 502–740 of Axin1 is crucial for its interaction with C9orf140.
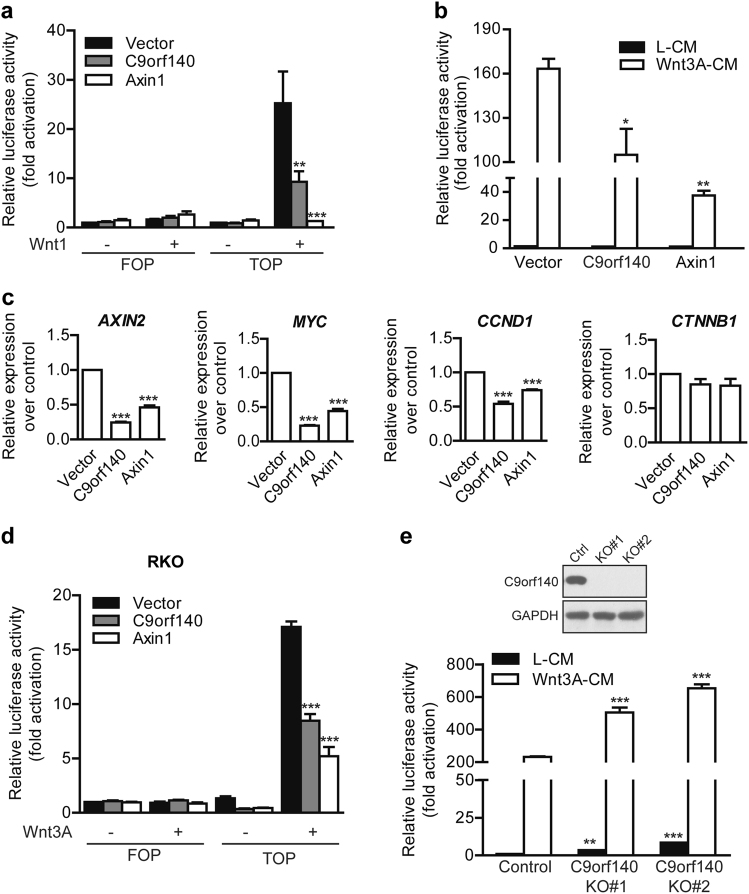
C9orf140 is a negative regulator of Wnt/β-catenin signaling in cultured cells
Axin1 plays pivotal roles in canonical Wnt signaling by directly interacting with other core components of the destruction complex and regulating the β-catenin protein level [1]. Because C9orf140 interacted with Axin1, we investigated whether C9orf140 might also play a role in regulating canonical Wnt signaling. TOP luciferase reporter (TOPFlash) which contains TCF-binding/LEF-binding sites upstream a luciferase reporter gene was widely used to evaluate β-catenin-mediated transcriptional activation, while FOP luciferase reporter which has mutant TCF/LEF-binding sites was used as a negative control. Using TOPFlash luciferase assays, we found that C9orf140 overexpression strongly suppressed TOP luciferase reporter activity induced by the transfection of Wnt1 from 25-fold to 11-fold but had no effect on FOP luciferase reporter activity (Fig. 3a). The group of Axin1 overexpression was used as a positive control (Fig. 3a). Similar results were obtained when Wnt3A-CM was used as the stimulus (Fig. 3b). To confirm the results obtained in the synthetic TOP luciferase reporter assays, we further examined the effects of C9orf140 overexpression on the transcript levels of well-defined Wnt target genes, including AXIN2 [11], MYC [12], and CCND1 [13]. Similarly to Axin1 overexpression, C9orf140 overexpression significantly decreased Wnt3A-induced transcriptional upregulation of AXIN2, MYC, and CCND1 but had no effect on the transcription of CTNNB1 (Fig. 3c). The suppression of Wnt-induced signaling by overexpression of C9orf140 was reproduced in RKO cells (Fig. 3d).
Fig. 3.
C9orf140 is a negative regulator of Wnt/β-catenin signaling. a The 293T cells were transfected with C9orf140, Axin1, or empty vector, together with TCF optimal (TOP) or mutant negative control (FOP) luciferase reporter, TK-Renilla reporter and Wnt1. The firefly luciferase activity measured by using the Dual-Luciferase Reporter Assay System was normalized to the Renilla luciferase activity. Data are presented as average fold activation relative to empty vector-treated cells +SD in the FOP-transfected or TOP-transfected group. b TOPFlash luciferase assays were performed in 293T cells transfected with C9orf140, Axin1, or empty vector. Luciferase activities were measured after treatment with control L cell-conditioned medium (L-CM) or Wnt3A-conditioned medium (Wnt3A-CM) for 18 h. Data are normalized to empty vector-transfected cells and presented as the mean + SD. c Real-time PCR for AXIN2, MYC, CCND1 and CTNNB1, normalized to GAPDH expression, in 293T cells transfected and treated as in (b). The ratio of normalized gene expression in Wnt3A-CM-treated cells relative to the L-CM-treated cells, in empty vector-transfected cells, was set to 1. The values of other groups were normalized to this value and are presented as the mean + SD. d RKO cells were transfected with C9orf140, Axin1, or empty vector, together with TOP or FOP luciferase reporter and TK-Renilla reporter. Luciferase activities were measured after treatment with L-CM or Wnt3A-CM for 18 h. Data are presented as average fold activation relative to empty vector-treated cells + SD in FOP-transfected or TOP-transfected group. e TOPFlash luciferase assays were performed in control 293T cells or two C9orf140 knockout cell lines. Luciferase activities were measured after treatment with L-CM or Wnt3A-CM for 18 h. Data are normalized to L-CM-treated control cells and presented as the mean + SD. The upper panel shows western blot verification of endogenous C9orf140 knockout by CRISPR/Cas9 in 293T cells. All results in the figure are representative of at least three experiments performed in duplicate. Statistical significance was calculated by one-way ANOVA, and significance was defined as ***p < 0.001, **p < 0.01, or *p < 0.05
To unambiguously confirm the inhibitory effect of C9orf140 on Wnt/β-catenin signaling, we generated C9orf140 knockout 293T cells by using the CRISPR/Cas9 system. Two C9orf140 knockout cell lines verified by genomic sequencing and protein immunoblotting were chosen and used in TOPFlash luciferase assays. The basal level of TOP luciferase reporter activity had a 3.5-fold and 8.5-fold increase in C9orf140 KO#1 and KO#2 293T cells, respectively, as compared to the activity in control 293T cells (Fig. 3e). Moreover, Wnt3A-induced TOP luciferase reporter activity was further enhanced in C9orf140 KO cell lines (Fig. 3e). Together, these results indicated that C9orf140 is a negative regulator of Wnt/β-catenin signaling.
The Wnt signaling plays important roles in the regulation of cell proliferation and epithelial–mensenchymal transition (EMT). We next addressed the role of C9orf140 as a negative regulator of Wnt/β-catenin signaling during Wnt-controlled cell proliferation and EMT. In 293T cells, we found that C9orf140 strongly inhibited cell proliferation and treatment with recombinant Wnt3A caused a further decrease of the cell proliferative capability (Supplementary Fig 2a). Additionally, C9orf140 overexpression reversed Wnt-induced EMT of MCF7 cells by enhancing the expression of E-cadherin and reducing the expression of N-cadherin and β-catenin (Supplementary Fig 2b and 2c). These results reinforce our notion that C9orf140 is a negative regulator of Wnt signaling, functionally relevant for Wnt-controlled cell proliferation and EMT.
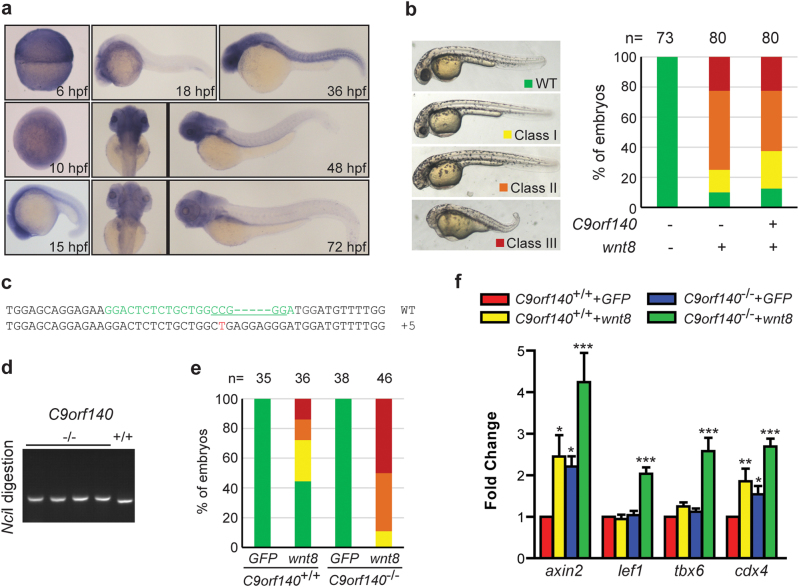
C9orf140 is a negative regulator of Wnt/β-catenin signaling in zebrafish embryogenesis
To further confirm the function of endogenous C9orf140 as a negative regulator of canonical Wnt signaling, we cloned a zebrafish C9orf140 homolog. An in situ hybridization analysis showed that C9orf140 was ubiquitously expressed in the early developmental stages (Fig. 4a). However, as development proceeded, C9orf140 was predominantly expressed in the central nervous system, blood, liver, and intestine (Fig. 4a). We further performed gain-of-function experiments in zebrafish embryos. Ectopic activation of Wnt/β-catenin signaling by injection of Wnt8 mRNA led to anterior truncations, such as a headless and a small eye phenotype [14]. However, co-injection of C9orf140 mRNA did not significantly (χ2 = 3.574, p > 0.05) restore the phenotypes though slightly increasing the proportion of the embryos with wild type (WT) and small eye phenotypes (Fig. 4b).
Fig. 4.
C9orf140 negatively regulates Wnt/β-catenin signaling in zebrafish. a Spatial and temporal expression of C9orf140 in zebrafish embryogenesis. The embryos at the indicated developmental stages were fixed for in situ hybridization with a C9orf140 probe. Both dorsal (left) and lateral (right) views are shown at 48 and 72 h post fertilization (hpf). b Inhibition of Wnt/β-catenin signaling by C9orf140 in zebrafish. Embryos were injected with wnt8 mRNA (5 pg) alone or together with C9orf140 mRNA (150 pg). The phenotypes of the injected embryos at 36 hpf were classified into four categories, as shown in the left panel. WT wild type; class I: small eyes; class II: headless; class III: epiboly defect or even more severe phenotype. Percentages of embryos displaying specific phenotypes are shown in the right panel. Statistical significance was calculated by Pearson χ2 test. c The sequencing results for the mutant C9orf140 allele. The CRISPR target sequence for zebrafish C9orf140 is in green and mutations in red. The NciI site used for genotype screening and identification is underlined. A 5-bp insertion in C9orf140 was identified in the C9orf140 mutant zebrafish line. d Genotypic identification of the C9orf140 mutant zebrafish lines. Genomic DNA extracted from the caudal fin was used as the template to amplify C9orf140 containing the CRISPR target sequence. The PCR products were digested by NciI and separated by electrophoresis. e Disruption of zebrafish C9orf140 facilitates Wnt signaling activation. Embryos from TU or C9orf140 -/- fish were injected with wnt8 (2 pg) or GFP (100 pg) and scored for the phenotypes shown in (b). Statistical significance was calculated by Pearson χ2 test. f The embryos injected as described in (e) were collected at the shield stage. The axin2, lef1, tbx6, and cdx4 expression levels were detected by real-time PCR and normalized to β-actin gene expression. The normalized value in the TU + GFP group was set to 1, and the values of other groups were normalized to this value. The experiment was independently performed three times, and similar results were obtained. The data shown are derived from a representative experiment expressed as the mean + SD. Statistical significance was calculated by one-way ANOVA, and significance was defined as ***p < 0.001, **p < 0.01, or *p < 0.05
We next generated C9orf140-deficient zebrafish by using the CRISPR/Cas9 system (Fig. 4c,d) and found that embryos in which endogenous C9orf140 was disrupted were hypersensitive to activation of Wnt/β-catenin signaling by Wnt8 mRNA injection with significant difference (χ2 = 35.9, p < 0.001) (Fig. 4e). To further support the phenotype data, we performed real-time PCR analysis to determine the effects of C9orf140 knockout on Wnt target gene expression. The results showed that the mRNA levels of axin2, lef1 [15], tbx6 and cdx4 (ventrolateral mesodermal marker and Wnt target genes) [16] were further induced by injection of wnt8 mRNA in C9orf140 −/− compared C9orf140 +/+ embryos (Fig. 4f). Together, these results indicated a conserved role of C9orf140 as a negative regulator of Wnt/β-catenin signaling in the early development of zebrafish.
C9orf140 negatively regulates Wnt/β-catenin signaling at the destruction complex level by compromising Wnt3A-induced β-catenin accumulation
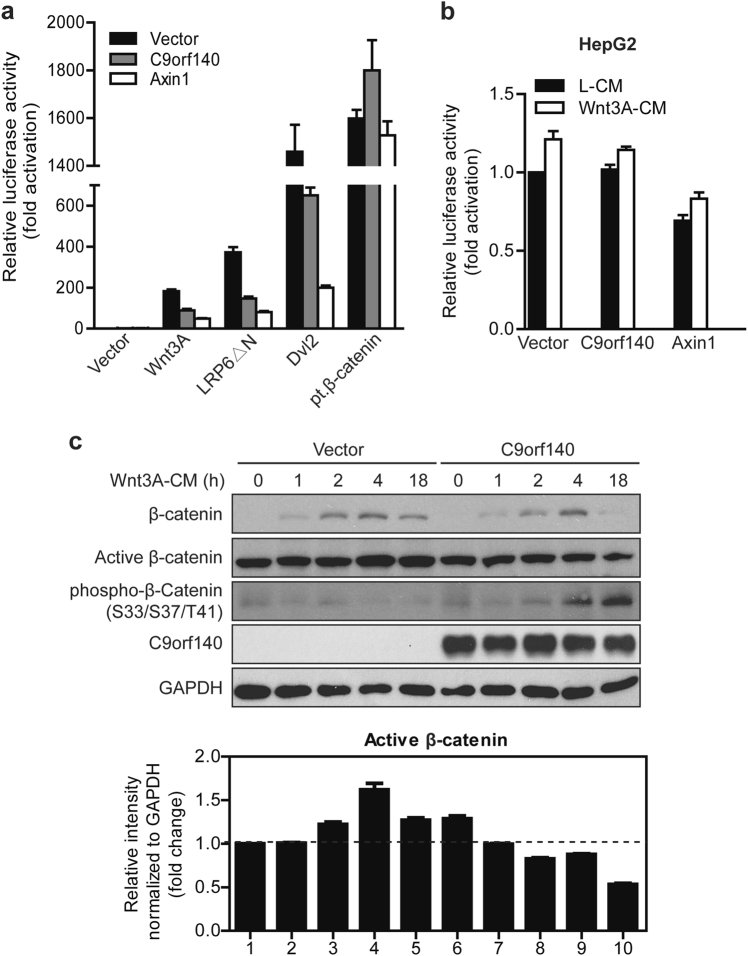
Epistasis experiments were performed to position C9orf140 within the Wnt cascade. Strikingly, similarly to Axin1 overexpression, C9orf140 overexpression in 293T and RKO cells inhibited TOP luciferase reporter activity driven by ectopic expression of Wnt3A (ligand), LRP6ΔN (active form of co-receptor), or Dvl2 (Wnt signaling effector) but not pt.β-catenin, a degradation-resistant form of β-catenin (Fig. 5a; Supplementary Fig 3). These results suggested that C9orf140 functions at the destruction complex level. To further support this hypothesis, we tested the effect of C9orf140 on the constitutive Wnt signaling observed in HepG2 cells. These cells contain a truncated β-catenin allele lacking the GSK3 phosphorylation sites and encode a constitutively active β-catenin [17, 18]. As expected, the C9orf140 overexpression-mediated inhibitory effect on TOP luciferase reporter activity was abolished in HepG2 cells (Fig. 5b), thus suggesting that C9orf140 functions upstream of β-catenin. However, Axin1 still had a weak inhibitory effect, which may be due to its positive regulatory roles downstream of β-catenin except for its well-known function as a negative regulator [19]. HepG2 cells were further activated by Wnt3A-CM (Fig. 5b), possibly because of the remaining WT β-catenin allele.
Fig. 5.
C9orf140 negatively regulates Wnt/β-catenin signaling at the destruction complex level by compromising Wnt3A-induced β-catenin accumulation. a Positioning C9orf140 in the Wnt pathway by epistatic analysis. Wnt/β-catenin signaling was induced by transfection of 293T cells with Wnt3A, LRP6ΔN, Dvl2, or pt.β-catenin. TOP luciferase reporter and TK-Renilla reporter, together with C9orf140, Axin1, or empty vector, were co-transfected as indicated. Data were normalized to empty vector-transfected cells and are presented as the mean + SD. b TOPFlash luciferase assays were performed in HepG2 cells transfected with C9orf140, Axin1, or empty vector. Luciferase activities were measured after treatment with L-CM or Wnt3A-CM for 18 h. Data are normalized to empty vector-transfected cells and presented as the mean + SD. c Overexpression of C9orf140 decreases β-catenin accumulation. RKO cells were first transfected with empty vector or C9orf140, treated with Wnt3A-CM, and finally harvested at the indicated times. Cell lysates were subjected to western blot analysis for the indicated proteins (upper panel). The lower panel shows quantitative analysis of active β-catenin levels normalized to GAPDH
Because the regulation of β-catenin protein level is the key feature of Wnt/β-catenin signaling, and C9orf140 acts as a negative regulator upstream of β-catenin, we further tested whether overexpression of C9orf140 might decrease β-catenin protein level in cells. We chose RKO cells because these cells have low cellular levels of β-catenin and no plasma membrane-associated β-catenin, thus allowing for analysis of cytoplasmic β-catenin stability. Under resting conditions, these cells had a minimal amount of cytosolic β-catenin, and β-catenin expression was strongly induced by Wnt3A-CM (Fig. 5c). We performed a time course experiment of Wnt3A-induced β-catenin stabilization in empty vector-transfected or C9orf140-transfected cells and observed that overexpression of C9orf140 significantly decreased cytosolic β-catenin accumulation (Fig. 5c). The decrease in β-catenin protein was unlikely to be at the transcriptional level, because the mRNA level of CTNNB1 did not change when C9orf140 was overexpressed (Fig. 3c). Therefore, we further detected the phosphorylation status of β-catenin and found that overexpression of C9orf140 significantly decreased non-phospho-β-catenin (active β-catenin) levels but increased phosphorylated β-catenin levels (Fig. 5c). These findings suggested that C9orf140 may regulate the phosphorylation and de-phosphorylation of β-catenin and influence β-catenin stability, thus leading to β-catenin degradation and inactivation of Wnt/β-catenin signaling.
C9orf140 outcompetes PP2A for binding to Axin1
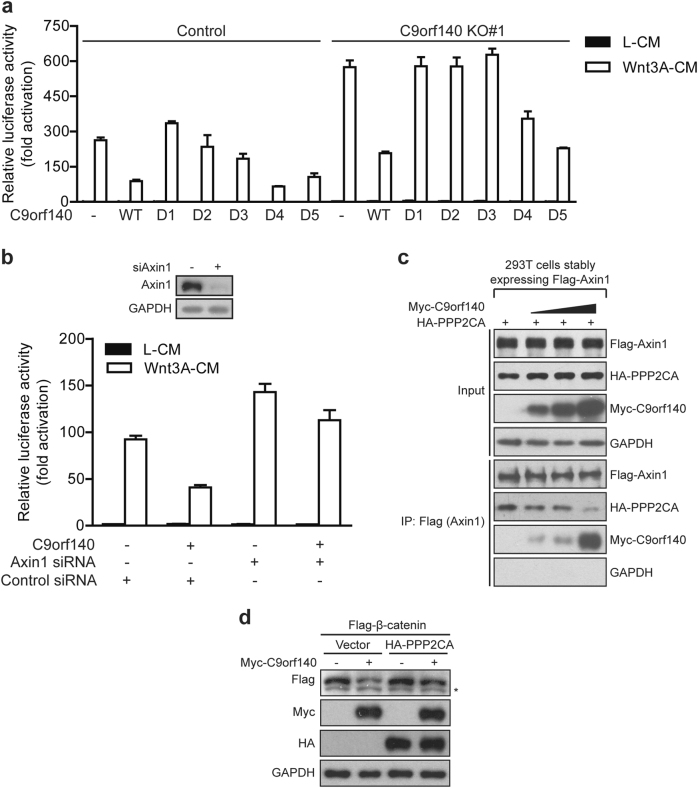
How does C9orf140 regulate the phosphorylation and de-phosphorylation of β-catenin, and inhibit Wnt/β-catenin signaling at the destruction complex level? To answer this question, we first sought to determine the functional domain of C9orf140. By using TOPFlash luciferase assays, we found that overexpression of D4 or D5 deletion mutants of C9orf140 in control 293T cells exhibited inhibitory effects on Wnt3A-induced TOP luciferase reporter activity similar to those of the WT, whereas overexpression of the D1, D2, or D3 deletion mutants of C9orf140 had almost no inhibitory effect (Fig. 6a). Similar results were obtained in C9orf140 KO cells (Fig. 6a). These results indicated that amino acids 1–239 of C9orf140 are necessary for its negative regulation of Wnt/β-catenin signaling. Because this region is also necessary for binding to Axin1, we hypothesized that the inhibitory effect of C9orf140 on Wnt/β-catenin signaling might depend on its interaction with Axin1. To test this hypothesis, we knocked down Axin1 and found that this knockdown eliminated the inhibitory effect of C9orf140 on Wnt3A-induced TOP luciferase reporter activity (Fig. 6b). Together, the data supported the hypothesis that C9orf140 functionally depends on Axin1 in regulating Wnt/β-catenin signaling.
Fig. 6.
C9orf140 outcompetes binding of PP2A to Axin1. a The region around amino acids 1–239 of C9orf140 is necessary for negative regulation of Wnt/β-catenin signaling. TOPFlash luciferase assays were performed in control and C9orf140 KO#1 293T cells transfected with empty vector, C9orf140 WT or a series of Flag-tagged C9orf140 deletion mutants. Luciferase activities were measured after treatment with L-CM or Wnt3A-CM for 18 h. Data were normalized to empty vector-transfected control 293T cells and are presented as the mean + SD. b The inhibitory effect of C9orf140 on Wnt/β-catenin signaling depends on Axin1. The 293T cells were first reverse transfected with control or Axin1 siRNA at the time of seeding cells, then subjected to cDNA transfection after adherence and treatment with L-CM or Wnt3A-CM. Data were normalized to L-CM-treated control cells. The data shown are derived from a representative experiment expressed as the mean + SD. The upper panel shows western blot verification of the validation of Axin1 siRNA. c Binding of PPP2CA to Axin1 is an alternative to the C9orf140–Axin1 interaction. The 293T cells stably expressing Flag-Axin1 were transfected with limiting amounts of HA-PPP2CA and increasing amounts of Myc-C9orf140 as indicated, and the lysates were subjected to anti-Flag immunoprecipitation. d The 293T cells were transfected with Flag-β-catenin, HA-PPP2CA, and Myc-C9orf140 as indicated for 24 h, and lysates were subjected to western blotting. *Non-specific band
Axin1 is the central scaffold of the destruction complex, and it directly interacts with many components in Wnt/β-catenin signaling, such as LRP, Dvl, PP2A, APC, CK1, GSK3, and β-catenin. Does C9orf140 function by competing with other Axin1-associated proteins for Axin1 binding and then affect Wnt/β-catenin signaling? We observed that C9orf140 bound to the domain spanning amino acids 502–740 of Axin1, which is the core binding region of PP2A. PP2A is a heterotrimeric protein comprising a catalytic subunit named PPP2CA, a structural subunit termed PR65/A and variable regulatory B subunits, and PPP2CA is generally responsible for its binding to Axin1 [20, 21]. Given these biochemical interactions, we hypothesized that the binding of PP2A to Axin1 might be outcompeted by C9orf140. This hypothesis prompted us to investigate the effect of increasing amounts of C9orf140 on the binding of PPP2CA to Axin1 by co-immunoprecipitation. Indeed, we found that the gradually increased amounts of C9orf140 dramatically weakened the interaction between PPP2CA and Axin1 (Fig. 6c).
PP2A has multiple and opposing roles in the Wnt/β-catenin signaling pathway, owing to the different associated regulatory subunits and substrates [22]. Recently, PP2A has been reported to counteract the action of GSK3 or CK1 in the destruction complex, dephosphorylate β-catenin and stabilize β-catenin, and therefore positively regulate Wnt/β-catenin signaling [4, 5]. Because C9orf140 outcompetes PP2A for binding to Axin1, and PP2A dephosphorylates β-catenin and stabilizes β-catenin, a potential explanation for the decreased β-catenin and non-phospho-β-catenin levels after C9orf140 overexpression may be that C9orf140 blocks PP2A-mediated de-phosphorylation and stability of β-catenin by binding with Axin1. To confirm these hypotheses, we ectopically expressed β-catenin together with C9orf140 and PPP2CA. As expected, overexpression of C9orf140 or PPP2CA significantly decreased or increased β-catenin levels, respectively (Fig. 6d). Furthermore, ectopically expressed C9orf140 overrode the effect of PPP2CA overexpression in 293T cells, causing a drastic decrease in β-catenin protein level.
β-catenin mediates the induction of C9orf140 by Wnt3A
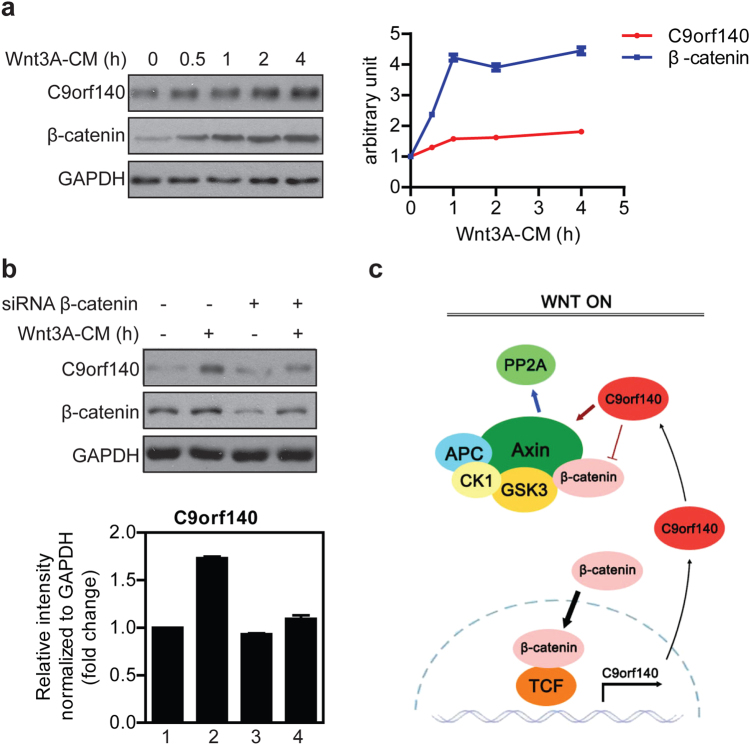
We stimulated 293T cells with Wnt3A-CM and monitored C9orf140 and β-catenin protein at different times by western blot analysis. As expected, the accumulation of β-catenin was detectable as early as 30 min after Wnt treatment (Fig. 7a). Notably, Wnt3A-CM also triggered a substantial increase in C9orf140 protein levels (Fig. 7a). These results were further confirmed by immunofluorescence staining in both in HeLa (Supplementary Fig 4a) and 293T cells (Supplementary Fig 4b).
Fig. 7.
Negative feedback loop of Wnt/β-catenin signaling through upregulation of C9orf140. a The 293T cells were treated with Wnt3A-CM and harvested at the indicated times. Cell lysates were subjected to western blot analysis for the indicated proteins. The right panel shows quantitative analysis of β-catenin and C9orf140 levels normalized to GAPDH using Image Lab software (Bio-Rad). Data are presented as average fold induction relative to the control ± SD. b The 293T cells were first transfected with 20 nM negative control or β-catenin siRNA and then treated with Wnt3A-CM for 18 h. Cells were harvested 48 h after transfection and lysed for western blot analysis (upper panel). The lower panel shows quantitative analysis of C9orf140 levels normalized to GAPDH. c A model depicting the proposed mechanism for negative feedback regulation of Wnt/β-catenin signaling by C9orf140
Interestingly, we found that the protein level of C9orf140, along with that of β-catenin, increased after Wnt treatment (Fig. 7a, right panel). These findings prompted us to further investigate whether β-catenin might mediate the induction of C9orf140 by Wnt3A. Indeed, knockdown of β-catenin strongly compromised the Wnt3A-mediated induction of C9orf140 protein (Fig. 7b).
Discussion
C9orf140 is a newly identified tumor-specific gene and a cell cycle-dependent gene that is highly expressed in many human tumor cell lines and primary tumor tissues [7, 23]. Previous findings have suggested that C9orf140 may be used as a tumor biomarker and a prospective target for antitumor drugs. In this paper, our findings provide a new perspective on the role of C9orf140 by identifying it as a novel Axin1-interacting protein and a negative regulator of Wnt/β-catenin signaling in cultured cells. Here, we showed that C9orf140 functions upstream of β-catenin, and its role is dependent on its interaction with Axin1. When C9orf140 was overexpressed, PPP2CA, the catalytic subunit of PP2A, detached from Axin1, and the balance was shifted toward the phosphorylation of β-catenin; at last, β-catenin was tended to degrade and Wnt/β-catenin signaling was inhibited (Fig. 7c). These findings also broadened our knowledge of the control of Wnt/β-catenin signaling cascade and may contribute to the development of a novel target for anticancer therapy.
C9orf140 has previously been reported to be an oncogene playing important roles in tumorigenesis by promoting cell proliferation, cell migration, and invasion [23]. We believe that C9orf140, which is upregulated in cancer, is a protein with multifunctions that promotes tumorigenesis through Wnt/β-catenin signaling-independent mechanisms. Furthermore, the inhibition of Wnt/β-catenin signaling identified in this study appears to be the major one of a series of roles of C9orf140. In line with our findings, DDK4, a Wnt inhibitor, has been shown to be induced by TCF/β-catenin, to be upregulated in human colon cancer and to promote the migratory, invasive, and proangiogenic potential of neoplastic cells [24]. Moreover, the Wnt inhibitors secreted frizzled-related proteins (SFRPs), which are upregulated in glioma-derived cell lines, accelerate the growth of malignant glioma cells but inhibit its migration [25]. Additionally, Axin2, which regulates Wnt/β-catenin signaling in a negative feedback loop, not only acts as a tumor suppressor mutated in cancers but also functions as an oncogene overexpressed in cancers [11, 26–28]. Similarly, C9orf140 may function as a tumor suppressor, oncogene, or both in various cancers. Further studies are needed to elucidate the relationship between C9orf140 and human cancer and its role in tumorigenesis.
Interestingly, we found that C9orf140 is not only a regulator of Wnt/β-catenin signaling but also a putative target of the Wnt pathway, in a process mediated through β-catenin. Because the knockdown of β-catenin dramatically decreased the transcriptional activity of the C9orf140 promoter [29] and several putative TCF/β-catenin-binding sites were found in the 5’ flanking region of C9orf140 (data not shown), we hypothesized that the TCF/β-catenin complex might bind to the TCF/LEF consensus binding sites on the C9orf140 promoter and consequently mediate its Wnt3A-induced activation (Fig. 7c). However, Wnt/β-catenin/TCF signaling is not the only regulatory mechanism for C9orf140 expression. Recently, STAT5 has been reported to directly interact with EZH2 and β-catenin to transactivate C9orf140 gene expression in CRC, thus indicating that other signaling pathway such as the JAK/STAT pathway may also be involved in the regulation of C9orf140 expression and its functions [29]. Cui et al. have shown that miR-29a inhibits the expression of C9orf140 both at the mRNA and protein levels by directly binding to its 3’UTR in gastric cancer [30]. Moreover, H. pylori and inflammatory cytokine tumor necrosis factor alpha stimulate C9orf140 gene expression in normal gastric epithelium cells (GES-1), although the regulatory mechanism is unknown [31].
Negative and positive feedback loops are important ways to dynamically regulate developmental signaling and have been identified in many well-described signaling pathways [32]. For example, the Wnt-induced target genes AXIN2, Dickkopf1 (DKK1), and Naked constitute negative feedback loops that serve to overcome excessive Wnt/β-catenin signaling [6]. Here, we demonstrated that upregulation of C9orf140 constitutes a novel mechanism of negative feedback regulation that silences Wnt/β-catenin signaling (Fig. 7c). The overexpression of C9orf140 in cancers may be either an effect of constitutive Wnt activation or a defense mechanism restricting the duration or spread of the Wnt/β-catenin signaling after the initial stimulus.
In conclusion, we demonstrate that β-catenin mediates the induction of C9orf140 by Wnt3A and that C9orf140 is a negative regulator of Wnt/β-catenin signaling, which disturbs the interaction between PP2A and Axin1, thus leading to β-catenin degradation and inactivation of Wnt/β-catenin signaling (Fig. 7c). Our findings represent an initiative to further dissecting the Wnt/beta-catenin signaling pathway and provide new insights into the role of C9orf140.
Materials and methods
Plasmids and antibodies
Full-length cDNAs encoding human C9orf140, Axin1, Wnt3A, and PPP2CA was cloned into pDONR201 using Gateway technology (Invitrogen) and were subsequently transferred to Gateway-compatible destination vectors for bacterial or mammalian expression studies. Deletion mutants of C9orf140 and Axin1 were prepared using site-directed mutagenesis and were verified by sequencing. The expression plasmids for LRP6ΔN, Dvl2, and pt.β-catenin were purchased from Addgene.
Rabbit anti-C9orf140 polyclonal antibodies were raised against maltose-binding protein (MBP)-C9orf140 and purified using a column conjugated with GST-C9orf140. Antibody specificity was confirmed by western blot experiments. Other antibodies used in this work included anti-Axin1 (Cell Signaling, #2074), anti-β-catenin (Santa Cruz, sc-7199), anti-active-β-catenin (Millipore, 05-665), anti-phospho-β-catenin (S33/S37/T41) (Cell Signaling, #9561), anti-E-cadherin (Cell Signaling, #3195), anti-N-cadherin (BD Biosciences, 610920), anti-GAPDH (Santa Cruz, sc-32233), anti-HA (Cell Signaling, #3724), anti-Myc (Cell signaling, #2278), anti-Flag (Sigma, F7425), and anti-GST (Santa Cruz, sc-138).
Cell culture, transfection, and luciferase assay
HeLa (ATCC, CCL-2), 293T (ATCC, CRL-3216), RKO (ATCC, CRL-2577), MCF7 (ATCC, HTB-22), and HepG2 (ATCC, HB-8065) cells were maintained at 37 °C in DMEM supplemented with 10% FBS. The cells were tested negative for mycoplasma contamination. Selection and passage of stable cell lines was performed with 2 μg/mL puromycin. Wnt3A-producing L cells (ATCC, CRL-2647) or control L cells (ATCC, CRL-2648) were cultured, and Wnt3A-conditioned or control-conditioned medium was prepared according to the Product Information Sheet (ATCC). Transient cDNA transfections were performed using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s protocol. SiRNAs targeting Axin1 or β-catenin or control siRNA with recommended amounts of DharmaFECT (Dharmacon) were transfected, and their respective sequences have been described previously [7, 33].
For TOPFlash luciferase assay, about 2 × 105 cells were plated into one well from 24-well plates and then transfected with 50 ng of the TCF optimal (TOP) or mutant negative control (FOP) luciferase reporter, 10 ng of the pRL-TK vector (Promega), and 50 ng of the expression vector (if needed) by using Lipofectamine 2000. After 24 h, the cells were lysed, and the luciferase activities were detected and analyzed as previously described [34]. For experiments involving Wnt3A stimulation, cells were transfected as described above, and the medium was then replaced with Wnt3A-conditioned or control-conditioned medium 6 h after transfection and subsequently cultured for 18 h. Then, the cells were lysed, and the luciferase activities were measured as described above.
Generation of knockout cells with CRISPR/Cas9 genome editing
The first coding exon of C9orf140 was chosen for guide RNA design using the CRISPR design tool at http://crispr.mit.edu/. The guide sequence used in this study was 5’-GCTGGCAGGGCACCGACGCG-3’. Annealed oligos were cloned into the PX330 expression vector (Addgene) containing the human codon-optimized Cas9, and the construct was then transfected into 293T cells using Lipofectamine 2000. Cells were digested 24 h after transfection, diluted into approximately one cell/well, and plated into 96-well plates. Single clones were expanded and screened by western blotting, genomic sequencing, and functional assays.
RNA extraction, cDNA synthesis, and real-time PCR
Cells and zebrafish embryos were harvested for total RNA extraction using TRIzol Reagent (Invitrogen). After DNase I (New England Biolabs) treatment, 2 μg of total RNA for each sample was reverse-transcribed to complementary DNA (cDNA) using M-MLV Reverse Transcriptase (Promega). The primers used were as follows:
human GAPDH-F: 5′-GGCTGCTTTTAACTCTGGTA-3′;
GAPDH-R: 5′-ACTTGATTTTGGAGGGATCT-3′;
AXIN2-F: 5′-CTGGCTCCAGAAGATCACAAAG-3′;
AXIN2-R: 5′-ATCTCCTCAAACACCGCTCCA-3′;
CCND1-F: 5′-TCGCCACCTGGATGCTGGAGGTCTG-3′;
CCND1-R: 5′-CACCAGGAGCAGCTCCATTTGCAGCAG-3′;
CTNNB1-F: 5′-GAGCCTGCCATCTGTGCTCT-3′;
CTNNB1-R: 5′-ACGCAAAGGTGCATGATTTG-3′;
MYC-F: 5′-CCTACCCTCTCAACGACAGCAGC-3′;
MYC-R: 5′-CTCTGACCTTTTGCCAGGAG-3′;
zebrafish axin2-F: 5′-TCCAATGGACGAAAGGAAAGA-3′;
axin2-R: 5′-TGAAGTGTCCGAGGGTAAGG-3′;
lef1-F: 5′-CCCAGAACGTCGAATAAGGT-3′;
lef1-R: 5′-TGTGATGTGAGAACCAACCC-3′;
cdx4-F: 5′-CTCCGGGACCAGTTTCCTAT-3′;
cdx4-R: 5′-CTCCTTCGTTCTCGTTTTGC-3′;
tbx6-F: 5′-CAAGCTGGATTTGACTGCAA-3′;
tbx6-R: 5′-GGGGTTTGTGAAGGCTGATA-3′;
β-actin-F: 5′-GGATTCGCTGGAGATGATG-3′;
β-actin-R: 5′-AGCTGTCTTTCCGTCCCATAC-3′.
Real-time PCR was performed using GoTaq qPCR Mater Mix (Promega) on a CFX ConnectTM Real-Time System (Bio-Rad) according to the manufacturers’ instructions. All samples were performed in triplicate, and the data were calculated and analyzed according to the 2−ΔCT (or 2−ΔΔCT) method [35].
Immunoprecipitation and western blotting
Cells were collected in NETN buffer (20 mM Tris-HCl pH 8.0, 100 mM NaCl, 1 mM EDTA, 0.5% NP-40). For detection of endogenous interactions, cells were treated with 10 μM MG132 for 4 h before lysis. Protein samples were centrifuged at 13,000 × g rpm for 10 min at 4 °C after lysis. Ten percent of the supernatant was kept as input, whereas the leftover was incubated with protein A/G-agarose beads conjugated with antibodies for 6 h at 4 °C. Immunocomplexes were washed four times with NETN buffer before boiling in SDS sample buffer for 10 min. Eluted proteins were then separated on SDS-PAGE and transferred to an Immobilon-P PVDF Transfer Membrane (Millipore) followed by blocking in 5% milk for 1 h, incubating with the indicated primary antibodies overnight and subsequently incubating with secondary antibodies (Cell Signaling) for 1 h. The Signals were detected by chemiluminescence (Cell Signaling).
GST pulldown assay
GST and GST-C9orf140 were expressed from Rosetta bacterial cells by using standard procedures and were subsequently incubated with glutathione Sepharose 4B (GE Healthcare) at 4 °C with agitation overnight. Beads were then washed and resuspended with NETN buffer. Each lysate from 293T cells was first mixed with agarose beads conjugated with ~30 μg of GST fusion protein and then incubated for 5 h with gentle rotation at 4 °C. The beads were washed four times with NETN buffer, and the eluted protein samples were further subjected to western blot analysis for the indicated proteins.
Immunofluorescence staining
Cells grown as monolayers on coverslips were first fixed in 4% paraformaldehyde for 20 min and then permeabilized in 0.5% Triton X-100 for 1 min or treated with the reverse protocol. After washes with PBS, the cells were blocked in 5% FBS for 1 h and incubated with the indicated antibodies followed by DAPI staining. Images were acquired using an Olympus BX51 fluorescence microscope and the Leica TCS-SP2 Spectral Confocal System.
Zebrafish microinjection, CRISPR/Cas9 mutagenesis, and in situ hybridization
The TU line zebrafish used in the study was maintained and handled according to the standard protocols (zfin.org). Animal experiments were approved by the Guangzhou Institutes of Biomedicine and Health Ethical Committee. Embryos were produced by random pair-mating of fish raised. Zebrafish C9orf140 (NM_212911.1) and Wnt8 ORF1 (NM_130946.3) coding regions were cloned into the pCS2 + vector for mRNA synthesis respectively, and capped mRNA was synthesized by using the mMessage mMachine kit (Ambion). For RNA injections, a volume of 2 nL mRNA was injected into the yolk of the embryo at the one-cell stage.
The plasmids of pCS2-Cas9 and pT7-gRNA for generating Cas9 mRNA and guide RNAs were kindly provided by Dr. Bo Zhang (Peking University). The target site of zebrafish C9orf140 gRNA (5’-GGACTCTCTGCTGGCCGGGA-3’), which was designed with the online tool ZIFIT Targeter (http://zifit.partners.org/zifit/CSquare9Nuclease.aspx) was inserted into BbsI-digested pT7-gRNA vector. A mixture of guide RNA (20 ng/µl) synthesized using a MAXIscript T7 Kit (Ambion) and Cas9 mRNA (200 ng/µl) prepared using a mMessage mMachine kit was injected into one-cell stage embryos. The putative mutation was screened by PCR using the C9orf140 genotype primers 5′-GCTGAAGCAGATGAAGGAGC-3′ and 5′-TCTGCCCGATGAGCCGCTGT-3′ followed by NciI digestion, and further confirmed by genomic DNA sequencing.
The complete ORF of C9orf140 was inserted into the pGEM-T vector for a template of the C9orf140 probe. Digoxigenin-UTP-labeled antisense RNA probes were generated by in vitro transcription using a DIG RNA Labeling Kit (Roche) according to the manufacturer’s instructions. Whole-mount in situ hybridization was performed as described [36].
Statistical analysis
The experiments were performed at least in triplicate in three biological replicates, and the data were expressed as the mean + SD. One-way analysis of variance (ANOVA) followed by Dunnett’s test and unpaired two-tailed Student’s t-test were used to analyze the statistical significances among different groups. Significance was defined as ***p < 0.001, **p < 0.01 or *p < 0.05. The data shown in Fig. 5b,e were analyzed by Pearson χ2 test, and statistical significance was defined as p < 0.05. SPSS software was used for statistical analysis. No statistical method was used to pre-determine sample size and no samples were excluded for any analysis. For animal studies, no randomization was used and no blinding was done.
Electronic supplementary material
a marked up version of revised supplementary information
Acknowledgements
We thank Dr. Shengcai Lin for providing the Axin1 and Wnt1 constructs used in this study and Dr. Hans Clevers for providing crucial reagents. This work was supported by the Natural Science Foundation of Guangdong Province [Research Team Project No. 2015A030312005 to YD]; Training program for outstanding young teachers in higher education institutions of Guangdong Province [No. YQ2014027 to JJ]; the National Natural Science Foundation of China [No. 31672611 to YD]; and the Pearl River S&T Nova Program of Guangzhou [No. 201610010063 to JJ].
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
The online version of this article (10.1038/s41388-018-0166-7) contains supplementary material, which is available to authorized users.
References
- 1.Clevers H. Wnt/beta-catenin signaling in development and disease. Cell. 2006;127:469–80. doi: 10.1016/j.cell.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 2.Clevers H, Nusse R. Wnt/beta-catenin signaling and disease. Cell. 2012;149:1192–205. doi: 10.1016/j.cell.2012.05.012. [DOI] [PubMed] [Google Scholar]
- 3.Gao C, Xiao G, Hu J. Regulation of Wnt/beta-catenin signaling by posttranslational modifications. Cell Biosci. 2014;4:13. doi: 10.1186/2045-3701-4-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Su Y, Fu C, Ishikawa S, Stella A, Kojima M, Shitoh K, et al. APC is essential for targeting phosphorylated beta-catenin to the SCFbeta-TrCP ubiquitin ligase. Mol Cell. 2008;32:652–61. doi: 10.1016/j.molcel.2008.10.023. [DOI] [PubMed] [Google Scholar]
- 5.Zhang W, Yang J, Liu Y, Chen X, Yu T, Jia J, et al. PR55 alpha, a regulatory subunit of PP2A, specifically regulates PP2A-mediated beta-catenin dephosphorylation. J Biol Chem. 2009;284:22649–56. doi: 10.1074/jbc.M109.013698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.MacDonald BT, Tamai K, He X. Wnt/beta-catenin signaling: components, mechanisms, and diseases. Dev Cell. 2009;17:9–26. doi: 10.1016/j.devcel.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xu X, Li W, Fan X, Liang Y, Zhao M, Zhang J, et al. Identification and characterization of a novelp42.3 gene as tumor-specific and mitosis phase-dependent expression in gastric cancer. Oncogene. 2007;26:7371–9. doi: 10.1038/sj.onc.1210538. [DOI] [PubMed] [Google Scholar]
- 8.Zhang J, Lu C, Shang Z, Xing R, Shi L, Lv Y. p42.3 gene expression in gastric cancer cell and its protein regulatory network analysis. Theor Biol Med Model. 2012;9:53. doi: 10.1186/1742-4682-9-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fagotto F, Jho E, Zeng L, Kurth T, Joos T, Kaufmann C, et al. Domains of axin involved in protein-protein interactions, Wnt pathway inhibition, and intracellular localization. J Cell Biol. 1999;145:741–56. doi: 10.1083/jcb.145.4.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Luo W, Lin SC. Axin: a master scaffold for multiple signaling pathways. Neurosignals. 2004;13:99–113. doi: 10.1159/000076563. [DOI] [PubMed] [Google Scholar]
- 11.Lustig B, Jerchow B, Sachs M, Weiler S, Pietsch T, Karsten U, et al. Negative feedback loop of Wnt signaling through upregulation of conductin/axin2 in colorectal and liver tumors. Mol Cell Biol. 2002;22:1184–93. doi: 10.1128/MCB.22.4.1184-1193.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.He TC, Sparks AB, Rago C, Hermeking H, Zawel L, da Costa LT, et al. Identification of c-MYC as a target of the APC pathway. Science. 1998;281:1509–12. doi: 10.1126/science.281.5382.1509. [DOI] [PubMed] [Google Scholar]
- 13.Tetsu O, McCormick F. Beta-catenin regulates expression of cyclin D1 in colon carcinoma cells. Nature. 1999;398:422–6. doi: 10.1038/18884. [DOI] [PubMed] [Google Scholar]
- 14.Kelly GM, Greenstein P, Erezyilmaz DF, Moon RT. Zebrafish wnt8 and wnt8b share a common activity but are involved in distinct developmental pathways. Development. 1995;121:1787–99. doi: 10.1242/dev.121.6.1787. [DOI] [PubMed] [Google Scholar]
- 15.Hagemann AI, Kurz J, Kauffeld S, Chen Q, Reeves PM, Weber S, et al. In vivo analysis of formation and endocytosis of the Wnt/beta-catenin signaling complex in zebrafish embryos. J Cell Sci. 2014;127:3970–82. doi: 10.1242/jcs.148767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li Z, Nie F, Wang S, Li L. Histone H4 Lys 20 monomethylation by histone methylase SET8 mediates Wnt target gene activation. Proc Natl Acad Sci USA. 2011;108:3116–23. doi: 10.1073/pnas.1009353108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carruba G, Cervello M, Miceli MD, Farruggio R, Notarbartolo M, Virruso L, et al. Truncated form of beta-catenin and reduced expression of wild-type catenins feature HepG2 human liver cancer cells. Ann N Y Acad Sci. 1999;886:212–6. doi: 10.1111/j.1749-6632.1999.tb09419.x. [DOI] [PubMed] [Google Scholar]
- 18.Azzolin L, Zanconato F, Bresolin S, Forcato M, Basso G, Bicciato S, et al. Role of TAZ as mediator of Wnt signaling. Cell. 2012;151:1443–56. doi: 10.1016/j.cell.2012.11.027. [DOI] [PubMed] [Google Scholar]
- 19.Major MB, Roberts BS, Berndt JD, Marine S, Anastas J, Chung N, et al. New regulators of Wnt/beta-catenin signaling revealed by integrative molecular screening. Sci Signal. 2008;1:ra12. doi: 10.1126/scisignal.2000037. [DOI] [PubMed] [Google Scholar]
- 20.Hsu W, Zeng L, Costantini F. Identification of a domain of Axin that binds to the serine/threonine protein phosphatase 2A and a self-binding domain. J Biol Chem. 1999;274:3439–45. doi: 10.1074/jbc.274.6.3439. [DOI] [PubMed] [Google Scholar]
- 21.Janssens V, Goris J. Protein phosphatase 2A: a highly regulated family of serine/threonine phosphatases implicated in cell growth and signalling. Biochem J. 2001;353:417–39. doi: 10.1042/bj3530417. [DOI] [Google Scholar]
- 22.Kimelman D, Xu W. beta-catenin destruction complex: insights and questions from a structural perspective. Oncogene. 2006;25:7482–91. doi: 10.1038/sj.onc.1210055. [DOI] [PubMed] [Google Scholar]
- 23.Cao WJ, Mao LL, Zheng JN, Pei DS. p42.3: an abductor of cell cycle. Anticancer Agents Med Chem. 2015;15:157–62. doi: 10.2174/1871520614666140818200202. [DOI] [PubMed] [Google Scholar]
- 24.Pendas-Franco N, Garcia JM, Pena C, Valle N, Palmer HG, Heinaniemi M, et al. DICKKOPF-4 is induced by TCF/beta-catenin and upregulated in human colon cancer, promotes tumour cell invasion and angiogenesis and is repressed by 1alpha,25-dihydroxyvitamin D3. Oncogene. 2008;27:4467–77. doi: 10.1038/onc.2008.88. [DOI] [PubMed] [Google Scholar]
- 25.Roth W, Wild-Bode C, Platten M, Grimmel C, Melkonyan HS, Dichgans J, et al. Secreted Frizzled-related proteins inhibit motility and promote growth of human malignant glioma cells. Oncogene. 2000;19:4210–20. doi: 10.1038/sj.onc.1203783. [DOI] [PubMed] [Google Scholar]
- 26.Li S, Wang C, Liu X, Hua S, Liu X. The roles of AXIN2 in tumorigenesis and epigenetic regulation. Fam Cancer. 2015;14:325–31. doi: 10.1007/s10689-014-9775-7. [DOI] [PubMed] [Google Scholar]
- 27.Wu ZQ, Brabletz T, Fearon E, Willis AL, Hu CY, Li XY, et al. Canonical Wnt suppressor, Axin2, promotes colon carcinoma oncogenic activity. Proc Natl Acad Sci USA. 2012;109:11312–7. doi: 10.1073/pnas.1203015109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jho EH, Zhang T, Domon C, Joo CK, Freund JN, Costantini F. Wnt/beta-catenin/Tcf signaling induces the transcription of Axin2, a negative regulator of the signaling pathway. Mol Cell Biol. 2002;22:1172–83. doi: 10.1128/MCB.22.4.1172-1183.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weng YR, Yu YN, Ren LL, Cui Y, Lu YY, Chen HY, et al. Role of C9orf140 in the promotion of colorectal cancer progression and mechanisms of its upregulation via activation of STAT5, beta-catenin and EZH2. Carcinogenesis. 2014;35:1389–98. doi: 10.1093/carcin/bgu057. [DOI] [PubMed] [Google Scholar]
- 30.Cui Y, Su WY, Xing J, Wang YC, Wang P, Chen XY, et al. MiR-29a inhibits cell proliferation and induces cell cycle arrest through the downregulation ofp42.3 in human gastric cancer. PloS ONE. 2011;6:e25872. doi: 10.1371/journal.pone.0025872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen P, Cui Y, Fu QY, Lu YY, Fang JY, Chen XY. Positive relationship betweenp42.3 gene and inflammation in chronic non-atrophic gastritis. J Dig Dis. 2015;16:568–74. doi: 10.1111/1751-2980.12282. [DOI] [PubMed] [Google Scholar]
- 32.Freeman M. Feedback control of intercellular signalling in development. Nature. 2000;408:313–9. doi: 10.1038/35042500. [DOI] [PubMed] [Google Scholar]
- 33.Major MB, Camp ND, Berndt JD, Yi X, Goldenberg SJ, Hubbert C, et al. Wilms tumor suppressor WTX negatively regulates WNT/beta-catenin signaling. Science. 2007;316:1043–6. doi: 10.1126/science/1141515. [DOI] [PubMed] [Google Scholar]
- 34.Dong L, Chen Q, Liu X, Wen J, Jiang J, Deng Y. Role of specificity protein 1, hepatocyte nuclear factor 1alpha, and pregnane X receptor in the basal and rifampicin-induced transcriptional regulation of porcine cytochrome P450 3A46. Drug Metab Dispos. 2015;43:1458–67. doi: 10.1124/dmd.115.065565. [DOI] [PubMed] [Google Scholar]
- 35.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 36.Chen Y, Wu B, Xu L, Li H, Xia J, Yin W, et al. A SNX10/V-ATPase pathway regulates ciliogenesis in vitro and in vivo. Cell Res. 2012;22:333–45. doi: 10.1038/cr.2011.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
a marked up version of revised supplementary information