ABSTRACT
Activation of the inflammatory transcription factor NF-κB in tumor-associated macrophages (TAMs) is assumed to contribute to tumor promotion. However, whether and how NF-κB drives the antitumor macrophages to become pro-tumorigenic have not been determined in any cancer type yet. Similarly, how TAMs repress CD8+ cytotoxic T lymphocytes (CTLs) remains largely unknown, although their importance in regulatory T (Treg) cell regulation and tumor promotion has been well appreciated. Here, using an endogenous lung cancer model we uncover a direct crosstalk between TAMs and CTLs. TAMs suppress CTLs through the T-cell inhibitory molecule B7x (B7-H4/B7S1) in a cell-cell contact manner, whereas CTLs kill TAMs in a tumor antigen-specific manner. Remarkably, TAMs secrete the known T-cell suppressive cytokine interleukin-10 (IL-10) to activate, but not to repress, CTLs. Notably, one major role of cell-intrinsic NF-κB RelA is to drive TAMs to suppress CTLs for tumor promotion. It induces B7x expression in TAMs directly, and restricts IL-10 expression indirectly by repressing expression of the NF-κB cofactor Bcl3 and subsequent Bcl3/NF-κB1-mediated transcription of IL-10. It also renders TAMs resistant to CTLs by up-regulating anti-apoptotic genes. These studies help understand how immunity is shaped in lung tumorigenesis, and suggest a RelA-targeted immunotherapy for this deadliest cancer.
KEYWORDS: B7x/B7-H4/B7S1, immune checkpoint, IL-10, lung cancer, NF-κB, RelA/p65, tumor-associated macrophage
Introduction
A causal link between inflammation and cancer has been recently established in several types of cancer including lung cancer, the leading cause of cancer-related deaths in both men and women.1–4 However, the normal role of immunity is to clear pathogens and damaged or transformed cells. Although it remains largely unknown how the antitumor immunity becomes tumor-promoting inflammation during tumorigenesis, macrophages, which are mature myeloid cells and can be simply defined as F4/80+ cells in mice and CD68+ cells in humans, have recently been suggested to be main culprits.5 Macrophages are the most abundant immune cells around and within tumors. Increase of macrophages or total myeloid cells in lung tissues or even in uninvolved lymph nodes or blood has been found to correlate with the risk, progression, chemoresistance and poor prognosis of lung cancer in humans.6–8 On the other hand, depletion of macrophages decreases both the tumor number and size in a mouse model of lung cancer.9 It is generally believed that tumor-associated macrophages (TAMs) contribute to tumor development mainly through secretion of various cytokines/chemokines and many other molecules, promoting tumor cell growth and protecting tumor cells from the immune system and in particular CD8+ cytotoxic T lymphocytes (CTLs).
Our current knowledge on TAM suppression of CTLs is largely derived from studies on the role of TAMs in secreting cytokines and chemokines to recruit and activate regulatory T (Treg) cells. Whether and how TAMs directly suppress CTLs are yet to be determined in lung or any other cancer type. Moreover, the intrinsic signaling pathways that switch TAMs from their original pro-inflammatory and anti-tumorigenic M1 phenotype to an anti-inflammatory and pro-tumorigenic M2 phenotype also remain largely unknown.
To address these fundamental questions in the fields of cancer and immunology, we investigated whether RelA functions as an intrinsic driver of macrophages for switching immunity from tumor immunosurveillance to tumor-promoting inflammation. RelA, also known as p65, is the prototypical member of the nuclear factor-κB (NF-κB) family of transcription factors, a central mediator of immune and inflammatory responses.10 Currently, the role of NF-κB and in particular its prototypical member RelA, in tumor immunology has rarely been studied. So far, there is only one report directly studying the role of myeloid RelA in tumor. It showed that by inducing expression of the cytokine tumor necrosis factor alpha (TNFα), myeloid RelA is required to promote proliferation of a xenografted mouse lung cancer cell line.11 However, the roles of RelA in TAM-mediated tumor development, progression, or immune evasion are yet to be investigated. Particularly, whether and how RelA transforms TAMs from anti-tumorigenic into pro-tumorigenic to repress CTLs is yet to be investigated. In addition, xenograft models using established cancer cell lines cannot recapitulate the natural coevolution of cancer and stromal cells as well as their intricate interplay in the complex microenvironment. Especially, they cannot address the roles of immune cells and how different immune cells interact in the early stages of tumorigenesis.
In this study, using endogenously arising lung tumor model, we examined the impact of selective deletion of RelA from myeloid cells on lung inflammation, tumor development and progression. We also investigated the molecular and cellular mechanisms by which RelA drives TAMs to transform immunity from tumor immunosurveillance to tumor-promoting inflammation, with a focus on the role of RelA in the TAM/CTL interaction. Moreover, we examined the activation status of macrophage RelA in human and mouse primary lung cancer tissues, as well as the association between macrophage RelA activation and lung cancer patient survival.
Results
RelA activation in lung myeloid cells is associated with poor survival of lung cancer patients
To address the role of RelA in myeloid cell regulation and lung cancer, we performed immunocytochemistry (IHC) to examine whether RelA is activated in myeloid cells associated with mouse lung tumors induced by the tobacco carcinogen urethane, a well-accepted and widely used model for human lung cancer. As expected, myeloid cells were readily detected surrounding mouse lung tumors induced by urethane, whereas very few myeloid cells were found in the lung tissues from untreated mice (Fig. 1A). Notably, myeloid cells associated with lung tumors, but not those in normal lung tissues, exhibited strong nuclear expressions of RelA, indicating high RelA activation in those cells. This was confirmed by immunofluorescent (IF) staining of mouse bronchioalveolar lavage (BAL) cells using RelA and F4/80 antibodies (Fig. 1B). The finding is highly relevant to human lung cancer, because the same phenotypes were observed in human lung tissues (Fig. 1C). Remarkably, high RelA activation in tumor-associated myeloid cells correlated with poor survival of lung cancer patients (Fig. 1D). These data suggest that RelA activation may function as an intrinsic force of myeloid cells and in particular lung macrophages to promote lung cancer.
Figure 1.
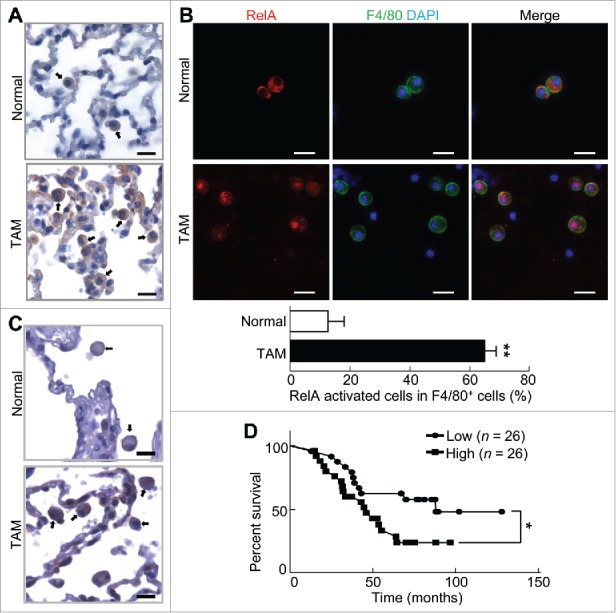
Myeloid RelA activation is associated with poor lung cancer patient survival. (A) IHC staining of RelA in lung tissues from untreated mice or mice with lung tumors induced by urethane. Arrows indicate myeloid cells. Scale bar: 20 μm. (B) IF staining of RelA and F4/80 in BAL cells from untreated mice or mice with lung tumors induced by urethane. Scale bar: 20 μm. Data are means ± SD (n = 3 mice, 3–5 images per mouse). (C) IHC staining of RelA in human lung cancer tissues and matched normal human lung tissues. Arrows indicate myeloid cells. Scale bar: 20 μm. (D) Kaplan-Meier survival curve showing the association between pulmonary myeloid RelA activation and lung cancer patient survival. p = 0.033 (Log-rank test); p = 0.048 (Gehan-Breslow-Wilcoxon test); p = 0.046 (Cox proportional hazards model).
Deletion of myeloid RelA leads to decreased TAMs and increased CTL activation
To examine the role of myeloid RelA activation in lung inflammation and tumorigenesis, we generated RelAflx/flx/lysozyme M-Cre (RelAΔMye) mice, in which RelA is selectively deleted from myeloid cells and in particular macrophages (Fig. 2A–C). RelAΔMye mice were healthy and showed no apparent abnormalities in myeloid cell development, lung size or morphology (data not shown). RelAΔMye mice and wildtype (WT) control mice were treated with urethane for lung carcinogenesis as described in Materials and Methods and then subjected to careful examinations of lung inflammation and cancer.
Figure 2.
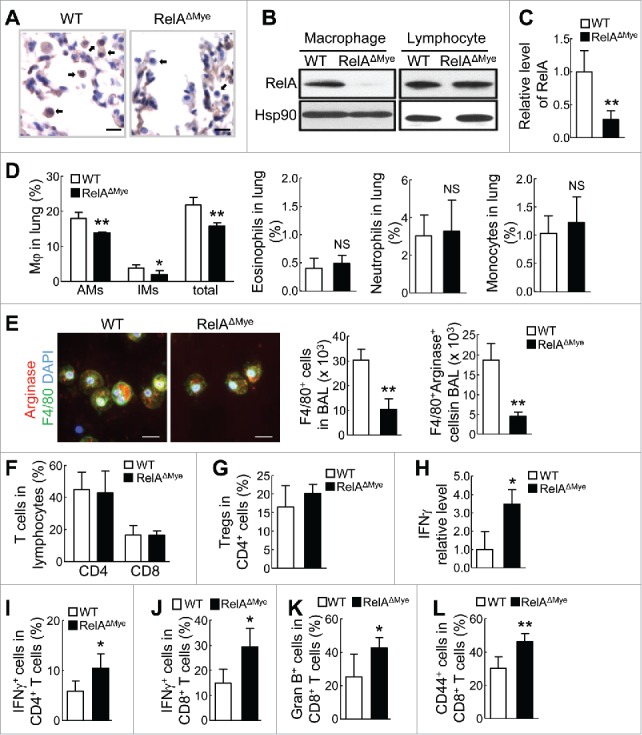
Macrophages are decreased and CTL activation is increased in lungs of RelAΔMye mice treated with urethane. (A) IHC staining of RelA in lung tissues from urethane-treated WT or RelAΔMye mice. Arrows indicate myeloid cells. Scale bar: 20 μm. (B) Immunoblotting of RelA and Hsp90 in peritoneal macrophages and splenic lymphocytes from urethane-treated WT or RelAΔMye mice. (C) qPCR showing RelA mRNA levels in BAL cells from urethane-treated WT or RelAΔMye mice. Data are mean ± SD (n ≥ 4). (D) Flow cytometry (FACS) of alveolar MACs (CD11b+CD11c+F4/80+Siglec-F+), interstitial MACs (CD11b+CD11c−F4/80+Siglec-F−), eosinophils (CD11b+CD11c−F4/80+Siglec-F+), neutrophils (CD11b+Ly6G+) and monocytes (CD11b+Ly6G−Ly6C+) from urethane-treated WT or RelAΔMye mice. Data are means ± SD (n ≥ 3). (E) IF co-staining of F4/80 and arginase in BAL cells from urethane-treated WT or RelAΔMye mice. Scale bar: 20 μm. Data are means ± SD (n = 3 mice, 4–5 images per mouse). (F-G and I-L) FACS of CD4+ and CD8+ T cells (F), Treg cells (G), IFNγ+ CD4+ T cells (I), IFNγ+ CD8+ T cells (J), granzyme B+ CD8+ T cells (K), and CD44+ CD8+ T cells (L) in lungs of urethane-treated WT or RelAΔMye mice. Data are means ± SD (n ≥ 3 for (G) and (K), n ≥ 4 for others). (H) qPCR showing IFNγ mRNA levels in BAL cells of urethane-treated WT or RelAΔMye mice. Data are means ± SD (n ≥ 4). For (C-L), *p < 0.05, **p < 0.01, Student's t test.
Compared to WT mice, RelAΔMye mice had significantly fewer alveolar MACs (AMs) and interstitial MACs (IMs) in their lungs (Fig. 2D; and Supplementary Fig. S1A). On the other hand, other major myeloid cells, such as eosinophils, neutrophils and monocytes, were comparable in WT and RelAΔMye mice (Fig. 2D; and Supplementary Fig. S1A and B). Since over 95% of F4/80+ cells in the lungs were macrophages and all lung macrophages were F4/80+, we simply used F4/80 as a lung macrophage marker for further IF assays. In support of the FACS data, IF staining of F4/80 showed significant decrease of macrophages in the BAL fluids (BALF) from urethane-treated RelAΔMye mice (Fig. 2E). In line with the fact that TAMs mainly exhibit M2 phenotype, most lung macrophages in either the RelAΔMye or WT mice were positively stained for arginase, a hallmark of mouse M2 macrophages. However, arginase+ macrophages were significantly decreased in the RelAΔMye mice. These data together suggest that RelA is mainly involved in the regulation of lung macrophages in lung tumorigenesis.
Although the numbers of lung CD4+ T cells or CD8+ T cells or Treg cells were comparable in those mice, interestingly, BAL cells from the RelAΔMye mice expressed much higher level of the antitumor cytokine interferon-γ (IFNγ) (Fig. 2F-H; and Supplementary Fig. S1C and D). Consistent with the fact that activated type 1 CD4+ T-helper (Th1) cells and CD8+ T cells are major sources of IFNγ, significantly more lung CD4+ and CD8+ T cells in the RelAΔMye mice were IFNγ+ (Fig. 2I and J; and Supplementary Fig. S1E and F). Moreover, more lung CD8+ T cells in the RelAΔMye mice expressed the activation markers granzyme B (Gran B) and CD44 (Fig. 2K and L; and Supplementary Fig. S1G and H). Altogether, these data suggest that RelA is required for lung macrophages to repress CTL activation in lung tumorigenesis.
Repression of CTL activation is a major mechanism underlying RelA-driven pro-tumorigenic action of macrophages
In association with decreased lung macrophages and increased CTL activation, interestingly, urethane-treated RelAΔMye mice developed fewer and smaller lung tumors (Fig. 3A and B). Histopathological analysis indicated that compared to WT mice, RelAΔMye mice had significantly fewer atypical adenomatous hyperplasia (AAH), adenomas (AD) and adenocarcinomas (AC) in their lungs (Fig. 3C). Not surprisingly, the decreased lung tumor induction in RelAΔMye mice was associated with increased apoptosis of tumor cells (Fig. 3D). These data suggest that through repressing tumor cell apoptosis by CTLs, macrophage RelA contributes to both the initiation and progression of lung cancer.
Figure 3.
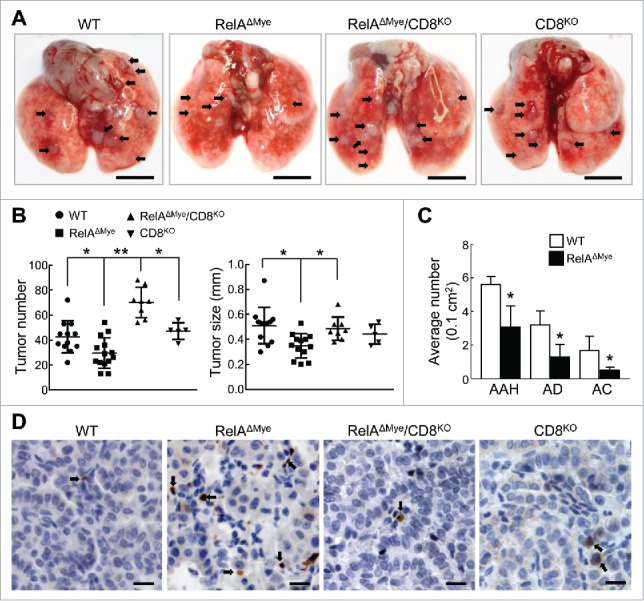
CTL activation plays a causative role in decreased lung tumorigenesis in RelAΔMye mice treated with urethane. (A) Representatives of lungs from the indicated mice treated with urethane. Arrows indicate tumors. Scale bar: 5 mm. (B) Lung tumor number and size in the indicated mice treated with urethane. Data are means ± SD. *p < 0.05, **p < 0.01, one way ANOVA/Tukey's multiple comparisons test. (C) H&E staining showing numbers of AAH, AD and AC in lung sections (per 0.1 cm2) of the indicated mice treated with urethane. Data shown are means ± SD (n ≥ 3; *, p < 0.05, Student's t test). (D) IHC staining of cleaved caspase-3 in lung tumors from the indicated mice treated with urethane. Scale bar: 20 μm.
To directly examine the role of the increased CTL activity in the decreased lung tumorigenesis in RelAΔMye mice, we generated RelAΔMye/CD8KO mice. Consistent with previous studies showing that CD8 is needed only for the development of CTLs but not CD4+ T cells,12 CD8 deletion had no obvious effect on lung CD4+ T cells (data not shown). Notably, deletion of CD8+ T cells led to a complete recovery of lung tumorigenesis by urethane in RelAΔMye mice, as evidenced by the increased lung tumor number and size in urethane-treated RelAΔMye/CD8KO mice (Fig. 3A and B). The apoptosis rate of tumor cells in the urethane-treated RelAΔMye/CD8KO mice was also reversed to the level in the WT control mice (Fig. 3D). It should be pointed out that CD8+ T-cell deletion had no obvious effect on lung tumorigenesis or tumor cell survival in the WT mice (Fig. 3A and B), which further supports our previous finding that CTLs are repressed by RelA-driven macrophages in lung tumorigenesis (Fig. 2 above and Fig. 4–7 below). It should also be pointed out that although the tumor sizes were comparable, RelAΔMye/CD8KO mice developed more lung tumors than CD8KO mice, suggesting a CTL-independent, tumor suppressive role for RelA in macrophages. Nevertheless, our data clearly indicate that one major role of macrophage-intrinsic RelA in tumor promotion is to drive these important immune cells to suppress CTL activation.
Figure 4.
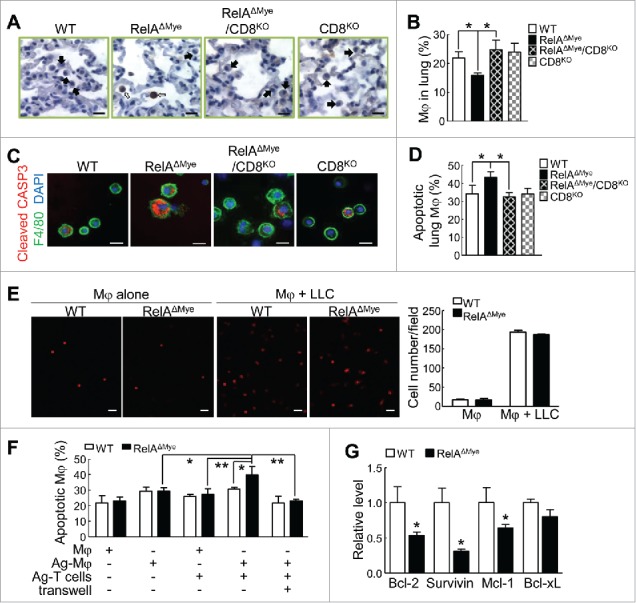
RelA is required for protection of pulmonary macrophages from CTLs in lung tumorigenesis. (A) IHC staining of cleaved caspase-3 in lung tissues from the indicated mice treated with urethane. Filled arrows and open arrows indicate normal and apoptotic myeloid cells, respectively. Scale bar: 20 μm. (B) FACS of macrophages in the lungs of the indicated mice treated with urethane. (C) IF co-staining of cleaved caspase-3 and F4/80 in BAL cells from the indicated mice treated with urethane. Scale bar: 20 μm. (D) FACS assays showing apoptosis rates of lung macrophages in the lungs from the indicated mice treated with urethane. Apoptotic macrophages (Annexin V+F4/80+ cells) were represented as the percentage of total macrophages (F4/80+ cells). (E) In vitro cell transwell migration assays showing similar migration to lung cancer cells for RelAKO and WT macrophages. PI-stained migrated macrophages were counted and represented as cell number per field. Scale bar: 100 µm. (F) FACS showing the apoptosis of tumor antigen pulsed or un-pulsed WT or RelAKO macrophages co-cultured with tumor antigen activated CD8+ T cells in normal plates or transwell plates. (G) qPCR showing the mRNA levels of the indicated genes in WT or RelAKO macrophages. For (B) and (D-G), data are means ± SD (n ≥ 3), *p < 0.05, **p < 0.01, one way ANOVA/Tukey's multiple comparisons test (B, D, and F) or Student's t test (E and G).
Figure 5.
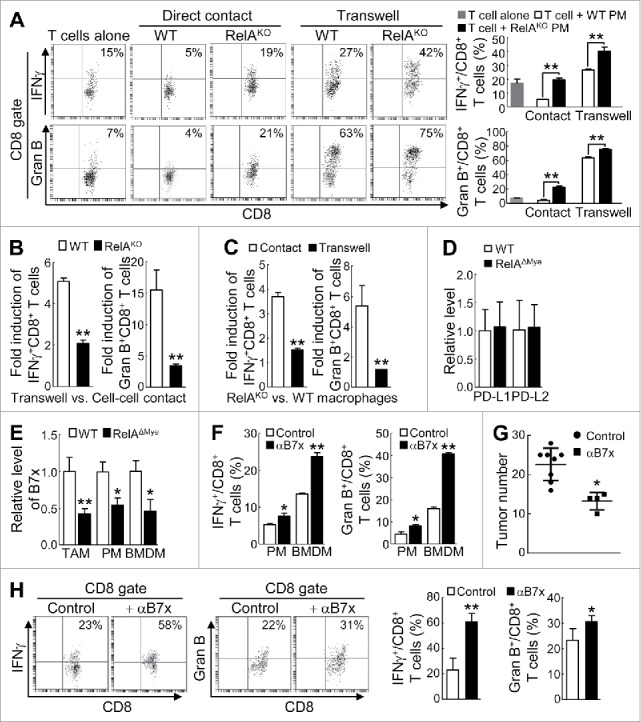
RelA induces B7x to arm macrophages for cell-cell contact inhibition of CTLs in lung tumorigenesis. (A) FACS of IFNγ+ or granzyme B+ cells in CD8+ T cells co-cultured with WT or RelAKO PMs in normal plates or transwell plates with LCCM containing anti-CD3 and anti-CD28 antibodies. (B) Fold induction of IFNγ+ or granzyme B+ CD8+ T cells by co-cultured WT or RelAKO macrophages under transwell versus normal cell-cell contact culture conditions. (C) RelAKO versus WT macrophage induction (fold) of IFNγ+ or granzyme B+ CD8+ T cells under normal cell-cell contact or transwell culture conditions. (D) qPCR showing PD-L1 and PD-L2 mRNA levels in TAMs from urethane-treated WT or RelAΔMye mice. (E) qPCR showing B7x mRNA levels in WT or RelAKO TAMs, LCCM cultured PMs and BMDMs. (F) FACS of IFNγ+ or granzyme B+ CD8+ T cells in T cells co-cultured with WT PMs or BMDMs in LCCM in normal plates with or without B7x neutralizing antibodies. (G and H) Lung tumor numbers (G) and FACS of IFNγ+ CD8+ T cells and granzyme B+ CD8+ T cells in lungs (H) of WT mice administered with B7x neutralizing antibodies or control antibodies after urethane treatment. In (A-H), data are means ± SD (n ≥ 4 for (G), n ≥ 3 for others). *, p < 0.05, **, p < 0.01, Student's t test.
Figure 6.
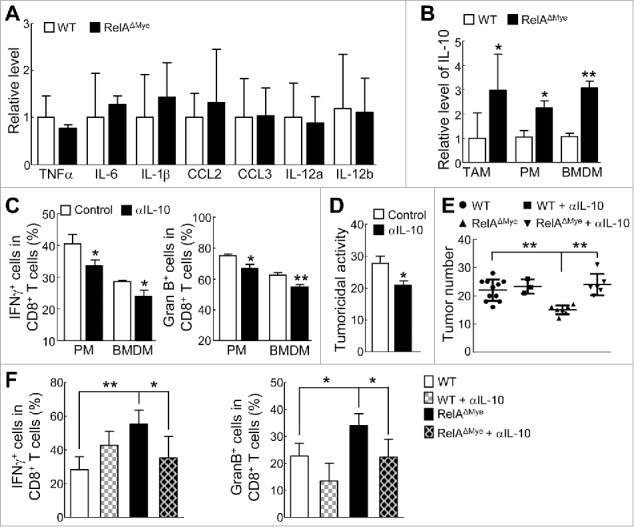
IL-10 activates CTL and RelA represses IL-10 to restrict macrophage-mediated paracrine activation of CTLs in lung tumorigenesis. (A) qPCR analysis showing RNA levels of the indicated cytokine/chemokine in TAMs from urethane-treated WT or RelAΔMye mice. (B) qPCR showing IL-10 RNA levels in WT or RelAKO TAMs, as well as PMs and BMDMs cultured with LCCM. (C) FACS of IFNγ+ or granzyme B+ cells in CD8+ T cells co-cultured in LCCM in transwell plates with RelAKO macrophages in the presence or absence of IL-10 neutralizing antibodies. (D) In vitro tumoricidal activity of T cells co-cultured in LCCM in transwell plates with RelAKO macrophages in the presence or absence of IL-10 neutralizing antibodies. (E) Lung tumor numbers in WT and RelAΔMye mice administered with IL-10 neutralizing antibodies or control antibodies after urethane treatment. (F) FACS of IFNγ+ CD8+ T cells and granzyme B+ CD8+ T cells in the lungs from WT and RelAΔMye mice administered with IL-10 neutralizing antibodies or control antibodies after urethane treatment. In (A-F), data are means ± SD (n ≥3). *, p < 0.05, **, p < 0.01, Student's t test (A-D), or one way ANOVA/Tukey's multiple comparisons test (E and F).
Figure 7.
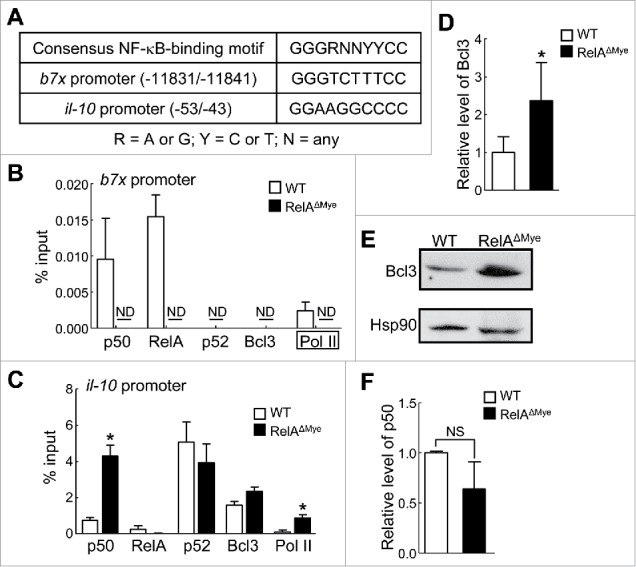
In TAMs RelA induces B7x directly by binding to B7x promoter while represses IL-10 indirectly via repressing Bcl3 transcription and Bcl3/p50-mediated IL-10 transcription. (A) Putative NF-κB-binding motifs within the b7x and il-10 promoters. (B and C) ChIP of the indicated proteins bound to the b7x (B) and il-10 (C) promoters in WT or RelAKO macrophages cultured in LCCM. ND, not detected. Primers used for Pol II pull-down in (B) were different, as the NF-κB-binding site within the b7x promoter was far upstream from the transcription site, see (A). (D and F) qPCR showing Bcl3 (D) and p50 (F) mRNA levels in pulmonary macrophages from WT or RelAΔMye mice treated with urethane. (E) Immunoblotting of Bcl3 and Hsp90 in WT and RelAKO macrophages cultured in LCCM. In (B-D) and (F), data are means ± SD (n ≥3). *, p < 0.05, **, p < 0.01, Student's t test.
RelA protects TAMs from CTL killing in lung tumorigenesis by repressing CTL activation and inducing cell survival genes
In line with the decrease in lung macrophages in urethane-treated RelAΔMye mice, our IHC and IF staining of cleaved caspase 3 (marker of apoptosis) as well as FACS assays showed dramatically increased apoptosis of lung macrophages in those mice (Fig. 4A-D; and Supplementary Fig. S2). On the other hand, RelA knockout (RelAKO) or WT macrophages exhibited high but comparable migration toward lung tumor cells (Fig. 4E). These data suggest that RelA is required for the survival but not migration of lung macrophages in lung tumorigenesis.
We assumed that the high lung CTL activation accounts for the high apoptosis of TAMs in urethane-treated RelAΔMye mice, given the potent cytocidal activity of CTLs. Currently, it remains unknown whether and how CTLs kill TAMs in any cancer type. Indeed, CD8 deletion completely blocked the increased apoptosis of lung macrophages in urethane-treated RelAΔMye mice, leading to a full recovery of lung macrophages (Fig. 4A-D; and Supplementary Fig. S2). These data suggest that the decrease of lung macrophages in urethane-treated RelAΔMye mice was fully mediated by CTL killing.
To validate the in vivo studies and to investigate the molecular mechanisms by which CTLs specifically recognize and kill TAMs, we tested the potential role of tumor antigens, because one important function of macrophages is to act as professional antigen-presenting cells to induce the antigen-specific cytotoxicity of CTLs. Indeed, macrophages pulsed with lung tumor lysate (Ag-Mφ) could induce strong activation of CD8+ T cells in vitro (Supplementary Fig. S3A). We used Ag-Mφ-activated CD8+ T cells (Ag-T cells) to directly co-culture with RelAKO and WT macrophages with or without pulse treatments. Ag-T cells did not significantly increase the apoptosis of macrophages without tumor antigen pulse, no matter of RelA expression status (Fig. 4F; and Supplementary Fig. S3B). They also failed to significantly increase the apoptosis of WT macrophages pulsed with tumor antigens. However, they did significantly increase the apoptosis of tumor antigen-pulsed RelAKO macrophages. The apoptosis of tumor antigen-pulsed RelAKO macrophages induced by co-cultured Ag-T cells was completely blocked when their direct contact was blocked using transwell chambers for their co-culture. These data suggest that in addition to killing tumor cells, CTLs exert tumor antigen-specific cytotoxicity toward TAMs, which is inhibited by RelA activation in TAMs.
Given its transcription activity, we examined whether RelA induces anti-apoptotic genes to make TAMs resistant to CTLs. Indeed, several important cell survival genes that are known transcription targets of NF-κB, including Bcl-2, survivin and Mcl-1, were significantly decreased in RelAKO macrophages, in comparison to WT cells (Fig. 4G). Another well-known NF-κB target survival gene, Bcl-xL, however, was not significantly decreased. These data suggest that in addition to arming TAMs to suppress CTL activation, RelA also renders those important pro-tumorigenic immune cells resistant to CTL cytotoxicity by inducing expression of multiple cell survival genes.
RelA promotes TAMs to directly repress CTL activation in lung tumorigenesis by inducing B7x and restricting IL-10 expression
To investigate the molecular mechanisms by which RelA drives TAMs to repress CTL activation for lung cancer promotion, we compared the effect of RelAKO and WT macrophage co-culture on CD8+ T-cell activation. We used both peritoneal macrophages (PMs) and bone marrow-derived macrophages (BMDMs) for the studies. To mimic lung tumor environment, lung cancer conditioned medium (LCCM) was used for the in vitro co-culture. We first tested whether the in vitro system recapitulates the in vivo situation. Indeed, direct co-culture with WT PMs or BMDMs decreased while with RelAKO PMs or BMDMs increased CD8+ T-cell activation stimulated by CD3 and CD28 antibodies (Fig. 5A and Supplementary Fig. S4). Similarly, RelAKO macrophages pulsed with tumor antigens induced higher activation of naïve CD8+ T cells, in comparison to tumor antigen-pulsed WT macrophages (Supplementary Fig. S5).
We then used the authentic in vitro system to examine the contributions of direct cell-cell contact and paracrine signaling in TAM repression of CTL activation. Remarkably, blockade of direct contact between LCCM-polarized WT PMs or BMDMs and CD3/CD28-stimulated CD8+ T cells by culturing them in different transwell chambers led to dramatically increased activation of CD8+ T cells (Fig. 5A and Supplementary Fig. S4). These data suggest that the net outcome of paracrine signals from macrophages is actually to induce CD8+ T-cell activation, and that cell-cell contact signals from macrophages are able to overturn the paracrine activation resulting in an overall repression of CD8+ T cells. It seems that RelA is involved in both confining the paracrine activation and inducing the cell-cell contact inhibition of CD8+ T cells by TAMs. Although blockade of RelAKO macrophage/CD8+ T-cell contact further increased CD8+ T-cell activation, also in a dramatic way, the increased CD8+ T-cell activation was significantly lower than that by WT macrophages (Fig. 5B). The difference in CD8+ T-cell activation by RelAKO macrophages and WT macrophages was also much smaller under this cell-cell contact free condition (Fig. 5C).
To investigate the molecular mechanisms of how RelA induces the cell-cell contact inhibition of CTLs by TAMs, we examined the expression levels of PD-L1 (also known as B7-H1), PD-L2, B7x (also called B7-H4 or B7S1) and VSIG4, known cell surface inhibitors of T-cell activation, in the TAMs isolated from urethane-treated WT or RelAΔMye mice. While PD-L1 and PD-L2 expression was comparable and VSIG4 was undetectable (Fig. 5D, and data not shown), B7x was significantly decreased in RelAKO TAMs (Fig. 5E). The same phenomena were observed in PMs and BMDMs cultured with LCCM in vitro (Fig. 5E). Importantly, B7x blockade by a specific neutralizing antibody significantly released CD8+ T cells from WT macrophage-mediated suppression (Fig. 5F). To validate the in vitro data and also to further determine the significance of B7x induction by RelA in lung tumorigenesis, we used specific neutralizing antibodies to block B7x in WT mice in lung tumorigenesis by urethane (Supplementary Fig. S6). Blockade of B7x led to significantly fewer lung tumors in WT mice (Fig. 5G). In inverse association with the lung tumorigenesis, blockade of B7x significantly increased the activation of pulmonary CD8+ T cells in the WT mice (Fig. 5H). These data indicate that RelA induces B7x expression on the cell surface of TAMs to repress CTL activation in lung tumorigenesis.
To identify RelA-targeted secretory molecule(s) in TAMs that are involved in restricting TAM paracrine activation of CTLs, we compared the expression levels of a large panel of cytokines in WT and RelAKO TAMs from urethane-treated mice, including the T-cell activating cytokine interleukin-12 (IL-12) and the T-cell suppressing cytokine IL-10. Among those cytokines we examined, only IL-10 was statistically differentially expressed in RelAKO and WT TAMs (Fig. 6A and B). However, IL-10 was increased, instead of being decreased, in RelAKO macrophages, suggesting that IL-10 may actually contribute to CD8+ T-cell activation. Indeed, biological inhibition of IL-10 using a specific neutralizing antibody prevented the activation and tumoricidal activity of CD8+ T cells by RelAKO macrophages (Fig. 6C and D). On the other hand, IL-10 activated CD8+ T cells and increased their tumoricidal activity (Supplementary Fig. S7). Consistent with the in vitro data, in vivo blockade of IL-10 led to significantly decreased pulmonary CTL activation and increased lung tumors in RelAΔMye mice but not in WT mice (Fig. 6E and F). Taken together, these data clearly suggest that RelA induces B7x and represses IL-10 in TAMs to prevent CTL activation respectively in cell-cell contact dependent and independent manners, thereby promoting lung tumorigenesis.
RelA directly binds to the b7x promotor for B7x induction in TAMs and restricts IL-10 expression indirectly via repressing Bcl-3/p50
To examine the molecular mechanisms by which RelA induces B7x and represses IL-10 expression in TAMs for CTL suppression and lung tumor promotion, we performed chromatin immunoprecipitation (ChIP) assays to examine whether RelA directly binds to the b7x and il-10 promoters. Our computer-based promoter analysis revealed that both b7x and il-10 promoters contained a putative NF-κB-binding site (Fig. 7A). We also included NF-κB1 p50, NF-κB2 p52, and Bcl3 in our assays. p50 and p52 usually form heterodimers with and are functional partners of RelA, while Bcl3 is the functional partner of p50 or p52 homodimers in inducing gene transcription.13 We found that RelA and p50, but not p52 or Bcl3, bound to the NF-κB-binding site within the b7x promoter in WT macrophages cultured with LCCM (Fig. 7B). However, like p52 and Bcl3, p50 also failed to do so in RelAKO macrophages cultured with LCCM. Consistently, RNA polymerase II (Pol II) bound to the b7x promoter only in WT but not in RelAKO macrophages. These data suggest that RelA forms heterodimers with and recruits p50 onto the b7x promoter to induce B7x expression in TAMs for CTL suppression and lung cancer promotion.
Whereas abundant p52 and relatively low levels of Bcl3 and p50 bound to the il-10 promoter, RelA was hardly detected at the promoter in the LCCM-cultured WT macrophages (Fig. 7C). These data were consistent with previous studies showing that the p52/Bcl3 or p50/Bcl3 complex, but not RelA, bound to the il-10 promoter in a murine macrophage cell line in response to bacterial lipopolysaccharide.14 In the RelAKO macrophages cultured with LCCM, however, p50, but not p52, was significantly increased at the il-10 promoter. Bcl3 was also increased at the il-10 promoter. Further, significantly more Pol II occupied the il-10 promoter in the RelAKO macrophages. These data suggest that RelA represses p50/Bcl3 promoter recruitment and thereby indirectly restricts IL-10 expression in TAMs in lung tumorigenesis.
To investigate the mechanisms by which RelA represses the promoter recruitment of the p50/Bcl3 complex for IL-10 repression in TAMs, we compared the expression levels of p50 and Bcl3 in WT and RelAKO macrophages cultured in LCCM. Different from previous reports showing that over-expressed NF-κB, particularly RelA, induces p50 and Bcl3 expression,15, 16 we found that while the expression of p50 was not statistically different, Bcl3 was significantly increased in RelAKO macrophages in comparison to WT macrophages (Fig. 7D-F). These data suggest that RelA restriction of Bcl3/p50-dependent IL-10 transcription in TAMs for CTL suppression in lung tumorigenesis is indirectly through repressing Bcl3 expression.
Discussion
One of the most fundamental and challenging questions in the cancer and immunology fields is how the immunity in cancer patients is transformed from tumor immunosurveillance to tumor-promoting inflammation. In particular, it remains largely unknown how the antitumor macrophages become pro- tumorigenic to repress the antitumor CTLs, and whether and how this pathogenic process can be blocked or even reversed in cancer patients for cancer treatment. The knowledge is direly needed for successful development of effective cancer immunotherapy. Currently, the overall clinical benefit of cancer immunotherapy is still very limited, particularly for solid tumors, although it is being touted as a breakthrough in cancer treatment.17 Our studies uncover a direct counterbalance between TAMs and CTLs through previously unidentified mechanisms, and identify RelA as a main culprit tipping the balance toward tumor development. Notably, deletion of RelA from macrophages boosts CTL antitumor immunity and suppresses lung cancer in a mouse model. These findings are highly relevant to human lung cancer, because RelA is constitutively activated in lung macrophages in lung cancer patients and the RelA activation correlates with poor patient survival.
Most studies on tumor immunology have used tumor implants. Although informative, these studies cannot resemble the natural coevolution of cancer and immune cells. They also cannot address how different immune cells interact to control tumor initiation and progression. Using an endogenously arising mouse lung cancer model, our studies reveal some important but previously unidentified functions for CTLs and TAMs. We find that in addition to their well-known tumoricidal activity, CTLs also kill pro-tumorigenic TAMs in a tumor antigen-specific manner to suppress lung cancer indirectly. On the other hand, TAMs exert a complex role in CTL activation under lung cancer conditions, although the final effect is to repress CTLs. Our current knowledge on macrophage repression of CTLs is very limited and largely relies on the paracrine induction of Treg cells through secretion of IL-10 and transforming growth factor β (TGFβ). Notably, we find that IL-10 actually activates CTLs to suppress lung cancer. Currently, this important function of IL-10 has been commonly ignored and drowned out by its well-known anti-inflammatory function, although previous studies from us and others also showed that IL-10 has the potential to stimulate CD8+ T cells.3, 18–20 Interestingly, TAMs repress CTL activation in a cell-cell contact manner and this cell-cell contact inhibition dominates TAM paracrine activation of CTLs resulting in the net repression of CTLs during lung tumorigenesis.
We also uncover, for the first time, the detailed mechanisms by which RelA drives TAMs to prevent CTL activation for lung cancer promotion. We find that while directly inducing B7x in TAMs to mediate the cell-cell contact inhibition of CTLs, RelA limits IL-10 expression in TAMs to prevent the paracrine activation of CTLs indirectly via repressing Bcl3 expression and subsequent Bcl3/p50- mediated IL-10 transcription. Of note, RelA is dispensable for the expressions of several other well- known T-cell activation inhibitors, such as PD-L1, PD-L2 and VSIG4, in the TAMs. This identifies B7x as a unique RelA target gene that is important for TAMs to directly suppress CTLs for lung tumorigenesis. RelA also induces the expressions of the cell survival genes Bcl-2, Mcl-1 and survivin, rendering TAMs resistant to CTLs and cancer stress. Although the main role of cell intrinsic RelA is to drive macrophages to suppress CTLs for tumor promotion, RelA also has CTL-independent and even antitumor functions in macrophages. In this regard, after urethane treatment RelAΔMye/CD8KO mice developed lung tumors not only more than RelAΔMye mice but also more than WT and CD8KO mice. Mechanistically, we found that RelA is almost absolutely required for TAMs to express inducible nitric oxide synthase (iNOS), an enzyme responsible for the production of the tumoricidal molecule nitric oxide (NO) (data not shown). Decreased NO production by RelAKO TAMs could contribute to the increased lung tumorigenesis in RelAΔMye/CD8KO mice. It is noteworthy that RelAΔMye/CD8KO mice have comparable numbers of TAMs relative to WT and CD8KO mice. Moreover, increased IL-10 due to RelA deletion in macrophages may also contribute to the increased lung tumorigenesis in RelAΔMye/CD8KO mice. While one key function of IL-10 is to activate CTLs for tumor suppression, particularly when the immune checkpoint inhibitory signal is lost (such as in RelAΔMye mice in which the expression of B7x in TAMs is repressed), it also stimulates the survival and proliferation of pre-cancerous and cancer cells in RelAΔMye/CD8KO mice and other conditions that CTLs are inhibited.
It should be pointed out that it currently remains largely unknown whether and how RelA and other NF-κB members regulate immune cells and in particular macrophages in tumor pathogenesis. Except the xenograft study suggesting that macrophage RelA is required to induce TNFα expression for the proliferation of an established mouse lung cancer cell line,11 only two studies have indirectly linked macrophage NF-κB to tumorigenesis by using mice in which IKKβ, a known NF-κB- activating kinase,13 is selectively deleted from myeloid cells. Similar to the xenograft study, however, these two studies only suggested that macrophage IKKβ is indispensable for the carcinogenesis of colon and ovarian cancers by inducing IL-6 or repressing IL-12, respectively.21, 22 They did not examine the role of macrophage IKKβ in tumor immunology. Moreover, IKKβ activates many other signaling pathways important for immunity and cancer, besides RelA and other NF-κB members.23 Indeed, different from those studies, we find that RelA is not required for IL-6 or IL-12 regulation but controls a different set of genes in TAMs for lung cancer promotion. This could be due to the redundant functions with RelA of other NF-κB members or the roles of NF-κB-unrelated targets of IKKβ in IL-6 and IL-12 regulation. It could also be that IKKβ and RelA work as an integrated signaling unit but regulate different genes in different cancers.
Our findings provide a molecular and cellular basis to target RelA for the prevention and treatment of lung cancer. RelA has long been a target of great interest for cancer therapy, given its causative role in lung and many other cancers. However, no clinically practical way is currently available to specifically target NF-κB in cancer cells. Systemic inhibition of NF-κB will be too toxic to cancer patients, given the physiological importance of the NF-κB, particularly RelA. In fact, the function of NF-κB is very complex, and individual NF-κB members have different or even opposite functions, although functional redundancy does exist among them. In this regard, we have recently shown that in contrast to RelA, the NF-κB1 precursor p105 functions as a lung tumor suppressor.24 Since deletion of RelA from myeloid cells suppresses both the initiation and progression of lung cancer, targeting myeloid RelA may provide a novel, feasible and effective approach for lung cancer therapy. It should be pointed out that myeloid RelA could be efficiently knocked down in cancer patients by RelA siRNAs conjugated with the Toll-like Receptor 9 (TLR9) agonists CpG oligonucleotides.25 RelA-based immunotherapy can be used independently or combined with conventional chemoradiotherapies and immunotherapies to treat lung cancer. This is particularly important and interesting, given the critical role of tumor-associated myeloid cells and in particular macrophages in the resistance to chemo- and radio- and immuno-therapies of cancer cells as well as our findings showing the central role of RelA in the functions of TAMs. Moreover, lung tumors are known to be highly resistant to these classical therapies.
In summary, the data presented here demonstrate several novel mechanisms underlying the counterattack between pulmonary macrophages and CD8+ T cells, and identify RelA as a major culprit to drive lung macrophages to tilt the balance toward lung cancer development and progression (Fig. 8). Whereas activated CD8+ T cells have the potential to kill TAMs and tumor cells in a tumor antigen- specific manner, RelA arms TAMs to suppress CD8+ T-cell activation by directly inducing B7x and indirectly restricting IL-10 expression through repressing Bcl3 expression and subsequent Bcl3/p50 transcription of IL-10. RelA also induces several cell survival genes to render TAMs resistant to CD8+ T cells. Remarkably, myeloid RelA deletion reverts these pathogenic processes, leading to lung cancer suppression. These preclinical studies greatly increase our understanding of lung cancer immunology and more importantly, suggest a novel, feasible and effective RelA-based immunotherapy for lung cancer prevention and treatment. Given the important roles of tumor-associated myeloid cells in many other solid tumors, these studies are highly relevant to the cancer field at large.
Figure 8.
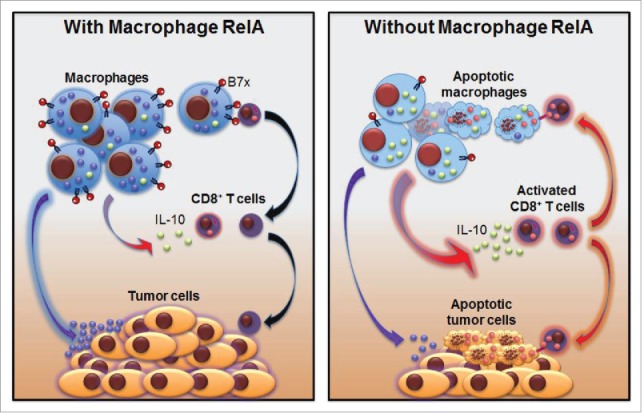
Modeling the roles of macrophage RelA in lung cancer biology and therapy.
Materials and Methods
Animals and lung carcinogenesis
Lysozyme M-Cre mice, CD8KO mice (Jackson laboratory), and RelAflx/flx mice26 were backcrossed to FVB/N mice for more than ten generations, and then used to generate the experimental mice for lung carcinogenesis as described previously.27 Briefly, mice were intraperitoneally (i.p.) injected with urethane (1 mg/g body weight, Sigma-Aldrich, St. Louis, MO, USA) once a week for six weeks. Mice were sacrificed for lung inflammation and tumor examinations at eight weeks post urethane treatment, except for those also treated with B7x and IL-10 neutralizing antibodies or their control antibodies, which were sacrificed at three weeks post urethane treatment. Surface tumors in mouse lungs were counted by three blinded readers under a dissecting microscope, and tumor diameters were measured by microcalipers. For B7x antibody treatment, WT mice were i.p. injected with B7x or control antibodies (200 μg) three times in the first, second and sixth week and two times in the third, fourth and fifth week after the third urethane injection. For IL-10 antibody treatment, RelAΔMye mice were i.p. injected with IL-10 or control antibodies (500 μg) twice weekly after the third urethane injection. All animals were maintained and used according to protocols approved by Institutional Animal Care and Use Committee of the University of Pittsburgh. B7x antibodies were generated and purified from the hybridoma cell line 1H3 as described previously.28 IL-10 antibodies were purchased from BioXCell, West Lebanon, NH, USA.
BAL
Upon sacrifice, mice lungs were lavaged with phosphate buffered saline as described.29 The recovered BAL fluids were centrifuged.
IF analysis
Cells were fixed, permeabilized, and subsequently incubated with the indicated primary antibodies, followed by FITC- or TRITC-conjugated secondary antibodies.30 Cells were also counterstained with DAPI for nuclear staining. Stained proteins and their subcellular localizations were detected using a fluorescence microscope.31 Antibodies used are listed in Supplementary Table S1.
Histology and IHC and human lung tumor tissue array assays
Lung tissues were excised, fixed in formalin, embedded in paraffin, and cut into 4-μm-thick sections. Sections were stained with H&E, or subjected to sequential incubations with the indicated primary antibodies, biotinylated secondary antibodies and streptavidin-HRP.24 The human lung tumor tissue arrays were described previously.4 Briefly, at least 50 myeloid cells were counted in randomly selected microscopic fields in each sample. Cells with obvious nuclear staining of RelA were scored as 2, otherwise, scored as 1. Scores were averaged and used for the cutoff of high (≥1.5) and low (<1.5) RelA activation. Antibodies used are listed in Supplementary Table S1.
Quantitative polymerase chain reaction (qPCR) analysis
The indicated cells were subjected to RNA extraction, RNA reverse transcription and qPCR as described.32, 33 Primers for qPCR are listed in Supplementary Table S2.
FACS analysis
The indicated cells were fixed with paraformaldehyde (2%) and permeablized with saponin (0.5%), or directly treated with the indicated antibodies. Data were acquired using FACSCalibur (BD Biosciences) and analyzed using CellQuest software (Becton Dickinson) as described.34 Antibodies used are listed in Supplementary Table S1.
Peritoneal macrophage (PM) and bone marrow-derived macrophage (BMDM) generation
Five days after intraperitoneal (i.p.) injection of 2 mL of 3% thioglycollate (Sigma-Aldrich, St. Louis, MO, USA), ice-cold phosphate-buffered saline (PBS) was injected into peritoneal cavity. Peritoneum was gently and completely massaged, and PBS was then recovered from peritoneal cavity. Peritoneal cells were centrifuged and cultured for 24 hours. Non-adherent cells were washed away and the remaining cells were PMs. For BMDM preparation, bone marrow cells were flushed from femurs of mice and cultured for 8 days with 10 ng/mL macrophage colony-stimulating factor (M-CSF). Adherent cells were BMDMs and non-attached cells were washed away. The purities of PMs and BMDMs were > 90% as confirmed by FACS analysis.
In vitro transwell migration assays
Essentially as described before,4 the indicated macrophages were plated in the upper chamber of transwell (with 8-μm pore size membrane) coated with Matrigel (BD Biosciences, Bedford, MA, USA). The lower chambers were seeded with murine Lewis lung carcinoma (LLC) cells. Cells were incubated for 24 hours. Nonmigrated cells were scraped from the upper surface of the membrane with a cotton swab, and migrated cells remaining on the lower surface were stained with propidium iodide (PI).
In vitro tumor antigen-dependent and -independent activation of T cells by macrophages
For tumor antigen-dependent T-cell activation assay, BMDMs from WT or RelAΔMye mice were pulsed with LLC lysate (1:3 ratio) for 18 hours, and then co-cultured with splenic CD3+ or CD8+ T cells from WT mice (1:5 ratio) in the presence of CD40L (0.5 μg/mL) and IL-2 (50 U/mL) for 4 days, followed by FACS analysis to detect granzyme B+ and IFNγ+ CD8+ T cells. For tumor antigen-independent T-cell activation assay, splenic CD3+ or CD8+ T cells from WT mice and macrophages from WT or RelAΔMye mice (2:1 ratio) were co-cultured in LLC cell-conditioned medium with or without B7x or IL-10 neutralizing antibodies in normal plates or in different chambers of transwell plates (0.4 μm pore size) for 3 days, followed by FACS analysis to detect granzyme B+ and IFNγ+ CD8+ T cells. CD3e (10 μg/mL) and CD28 (2 μg/mL) antibodies were also added in the co-culture.
In vitro CTL killing of macrophages and tumor cells
RelAKO or WT BMDMs with or without tumor antigen pulse were co-cultured with T cells activated by tumor antigen-pulsed BMDMs (1:2 ratio) for 2 days, followed by FACS analysis to detect apoptotic macrophages. For T-cell tumoricidal activity assays, T cells activated by CD3 and CD28 antibodies were treated in vitro with IL-10 or co-cultured with RelAKO macrophages in transwell plates in the presence or absence of IL-10 neutralizing antibodies for three days and then co-cultured with LLC cells stably expressing luciferase (LLC-Luc) in 10:1 ratio in normal plates for 8 hours, followed by detection of luciferase in the culture medium.
ChIP assays
ChIP assays were performed essentially as described.35 Primers for ChIP assays are listed in Supplementary Table S3.
Statistical analysis
Data were reported as mean ± standard deviation (SD). Student's t test (two tailed) and one way ANOVA/Tukey's multiple comparisons test were used to assess significance of differences between two groups and multiple comparisons, respectively. Gehan-Breslow-Wilcoxon test and log-rank test were used to compare overall patient survival between high and low myeloid RelA activation groups. Multivariate survival analysis was also performed using Cox's proportional hazards model to statistically consider and adjust the potential effect of other clinical factors, such as age and tumor stage. The p values < 0.05 and 0.01 were considered statistically significant and highly statistically significant, respectively.36
Supplementary Material
Funding Statement
This work was supported by the HHS | National Institutes of Health (NIH).
Disclosure of potential conflicts of interest
The authors declare no potential conflicts of interest.
Acknowledgments
This study was financially supported in part by the National Institute of Health (NIH)/National Cancer Institute (NCI) grants R01 CA172090, R21 CA175252, R21 CA189703, P30 CA047904, and P50 CA090440, as well as the American Lung Association (ALA) Lung Cancer Discovery Award.
Authors' contributions
Conception and design: Z. Qu, G. Xiao
Development of methodology: L. Li, L. Han, F. Sun, J. Zhou, Z. Qu, G. Xiao
Acquisition of data (provided animals, acquired and managed patients, provided facilities, etc.): L. Li, L. Han, F. Sun, J. Zhou
Analysis and interpretation of data (e.g., statistical analysis, biostatistics, computational analysis): L. Li, L. Han, F. Sun, J. Zhou, Z. Qu, G. Xiao
Writing, review, and/or revision of the manuscript: Z. Qu, G. Xiao
Administrative, technical, or material support (i.e., reporting or organizing data, constructing databases): L. Li, L. Han, F. Sun, J. Zhou, K. C. Ohaegbulam, X. Tang, X. Zang, Z. Qu, G. Xiao
Study supervision: Z. Qu, G. Xiao
References
- 1.Shalapour S, Karin M. Immunity, inflammation, and cancer: an eternal fight between good and evil. J Clin Invest. 2015;125:3347–55. doi: 10.1172/JCI80007. PMID:26325032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017 . CA Cancer J Clin. 2017;67:7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 3.Qu Z, Sun F, Zhou J, Li L, Shapiro SD, Xiao G. Interleukin-6 Prevents the Initiation but Enhances the Progression of Lung Cancer. Cancer Res. 2015;75:3209–15. doi: 10.1158/0008-5472.CAN-14-3042. PMID:26122841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhou JJ, Qu ZX, Sun F, Han L, Li LW, Yan SP, Stabile LP, Chen LF, Siegfried JM, Xiao G, et al.. Myeloid STAT3 Promotes Lung Tumorigenesis by Transforming Tumor Immunosurveillance into Tumor-Promoting Inflammation. Cancer Immunol Res. 2017;5:257–68. doi: 10.1158/2326-6066.CIR-16-0073. PMID:28108629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Noy R, Pollard JW. Tumor-associated macrophages: from mechanisms to therapy. Immunity. 2014;41:49–61. doi: 10.1016/j.immuni.2014.06.010. PMID:25035953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pei BX, Sun BS, Zhang ZF, Wang AL, Ren P. Interstitial tumor-associated macrophages combined with tumor-derived colony-stimulating factor-1 and interleukin-6, a novel prognostic biomarker in non-small cell lung cancer. J Thorac Cardiovasc Surg. 2014;148:1208–16 e2. doi: 10.1016/j.jtcvs.2014.05.003. [DOI] [PubMed] [Google Scholar]
- 7.Sun J, Mao Y, Zhang YQ, Guo YD, Mu CY, Fu FQ, Zhang XG. Clinical significance of the induction of macrophage differentiation by the costimulatory molecule B7-H3 in human non-small cell lung cancer. Oncol lett. 2013;6:1253–60. doi: 10.3892/ol.2013.1586. PMID:24179504. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 8.Zhang W, Pal SK, Liu X, Yang C, Allahabadi S, Bhanji S, Figlin RA, Yu H, Reckamp KL. Myeloid clusters are associated with a pro-metastatic environment and poor prognosis in smoking-related early stage non-small cell lung cancer. PloS one. 2013;8:e65121. doi: 10.1371/journal.pone.0065121. PMID:23717691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zaynagetdinov R, Sherrill TP, Polosukhin VV, Han W, Ausborn JA, McLoed AG, McMahon FB, Gleaves LA, Degryse AL, et al.. A critical role for macrophages in promotion of urethane-induced lung carcinogenesis. J immunol. 2011;187:5703–11. doi: 10.4049/jimmunol.1100558. PMID:22048774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xiao G, Rabson AB, Young W, Qing G, Qu Z. Alternative pathways of NF-kappaB activation: a double-edged sword in health and disease. Cytokine Growth Factor Rev. 2006;17:281–93. doi: 10.1016/j.cytogfr.2006.04.005. PMID:16793322. [DOI] [PubMed] [Google Scholar]
- 11.Li D, Beisswenger C, Herr C, Hellberg J, Han G, Zakharkina T, Voss M, Wiewrodt R, Bohle RM, Menger MD, et al.. Myeloid cell RelA/p65 promotes lung cancer proliferation through Wnt/beta-catenin signaling in murine and human tumor cells. Oncogene. 2014;33:1239–48. doi: 10.1038/onc.2013.75. PMID:23563178. [DOI] [PubMed] [Google Scholar]
- 12.Fung-Leung WP, Schilham MW, Rahemtulla A, Kundig TM, Vollenweider M, Potter J, et al.. CD8 is needed for development of cytotoxic T cells but not helper T cells. Cell. 1991;65:443–9. doi: 10.1016/0092-8674(91)90462-8. PMID:1673361. [DOI] [PubMed] [Google Scholar]
- 13.Xiao G, Fu J. NF-kappaB and cancer: a paradigm of Yin-Yang. Am J Cancer Res. 2011;1:192–221. PMID:21969033. [PMC free article] [PubMed] [Google Scholar]
- 14.Wang VY, Huang W, Asagiri M, Spann N, Hoffmann A, Glass C, Ghosh G. The transcriptional specificity of NF-kappaB dimers is coded within the kappaB DNA response elements. Cell Rep. 2012;2:824–39. doi: 10.1016/j.celrep.2012.08.042. PMID:23063365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ge B, Li O, Wilder P, Rizzino A, McKeithan TW. NF-kappa B regulates BCL3 transcription in T lymphocytes through an intronic enhancer. J Immunol. 2003;171:4210–8. doi: 10.4049/jimmunol.171.8.4210. [DOI] [PubMed] [Google Scholar]
- 16.Ten RM, Paya CV, Israel N, Le Bail O Mattei MG, Virelizier JL, Kourilsky P, Israël A. The characterization of the promoter of the gene encoding the p50 subunit of NF-kappa B indicates that it participates in its own regulation. EMBO J. 1992;11:195–203. PMID:1740105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Couzin-Frankel J. Breakthrough of the year 2013. Cancer immunotherapy. Science. 2013;342:1432–3. doi: 10.1126/science.342.6165.1432. PMID:24357284. [DOI] [PubMed] [Google Scholar]
- 18.Chen WF, Zlotnik A. IL-10: a novel cytotoxic T cell differentiation factor. J Immunol. 1991;147:528–34. [PubMed] [Google Scholar]
- 19.MacNeil IA, Suda T, Moore KW, Mosmann TR, Zlotnik A. IL-10, a novel growth cofactor for mature and immature T cells. J Immunol. 1990;145:4167–73. [PubMed] [Google Scholar]
- 20.Mumm JB, Emmerich J, Zhang X, Chan I, Wu L, Mauze S, Blaisdell S, Basham B, Dai J, Grein J, et al.. IL-10 elicits IFNgamma-dependent tumor immune surveillance. Cancer cell. 2011;20:781–96. doi: 10.1016/j.ccr.2011.11.003. PMID:22172723. [DOI] [PubMed] [Google Scholar]
- 21.Greten FR, Eckmann L, Greten TF, Park JM, Li ZW, Egan LJ, Kagnoff MF, Karin M. IKKbeta links inflammation and tumorigenesis in a mouse model of colitis-associated cancer. Cell. 2004;118:285–96. doi: 10.1016/j.cell.2004.07.013. PMID:15294155. [DOI] [PubMed] [Google Scholar]
- 22.Hagemann T, Lawrence T, McNeish I, Charles KA, Kulbe H, Thompson RG, Robinson SC, Balkwill FR "Re-educating" tumor-associated macrophages by targeting NF-kappaB. J Exp Med. 2008;205:1261–8. doi: 10.1084/jem.20080108. PMID:18490490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chariot A. The NF-kappaB-independent functions of IKK subunits in immunity and cancer. Trends Cell Biol. 2009;19:404–13. doi: 10.1016/j.tcb.2009.05.006. PMID:19648011. [DOI] [PubMed] [Google Scholar]
- 24.Sun F, Qu Z, Xiao Y, Zhou J, Burns TF, Stabile LP, Siegfried JM, Xiao G. NF-kappaB1 p105 suppresses lung tumorigenesis through the Tpl2 kinase but independently of its NF-kappaB function. Oncogene. 2016;35:2299–310. doi: 10.1038/onc.2015.299. PMID:26300007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kortylewski M, Swiderski P, Herrmann A, Wang L, Kowolik C, Kujawski M, Lee H, Scuto A, Liu Y, Yang C, et al.. In vivo delivery of siRNA to immune cells by conjugation to a TLR9 agonist enhances antitumor immune responses. Nat Biotechnol. 2009;27:925–32. doi: 10.1038/nbt.1564. PMID:19749770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Steinbrecher KA, Harmel-Laws E, Sitcheran R, Baldwin AS. Loss of epithelial RelA results in deregulated intestinal proliferative/apoptotic homeostasis and susceptibility to inflammation. J Immunol. 2008;180:2588–99. doi: 10.4049/jimmunol.180.4.2588. [DOI] [PubMed] [Google Scholar]
- 27.Zhou J, Qu Z, Yan S, Sun F, Whitsett JA, Shapiro SD, Xiao G. Differential roles of STAT3 in the initiation and growth of lung cancer. Oncogene. 2015;34:3804–14. doi: 10.1038/onc.2014.318. PMID:25284582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jeon H, Vigdorovich V, Garrett-Thomson SC, Janakiram M, Ramagopal UA, Abadi YM, Lee JS, Scandiuzzi L, Ohaegbulam KC, Chinai JM, et al.. Structure and cancer immunotherapy of the B7 family member B7x. Cell Rep. 2014;9:1089–98. doi: 10.1016/j.celrep.2014.09.053. PMID:25437562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sun F, Xiao G, Qu Z. Murine Bronchoalveolar Lavage. Bio-protocol. 2017;7: e2287. doi: 10.21769/BioProtoc.2287. PMID:29082285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yan P, Fu J, Qu Z, Li S, Tanaka T, Grusby MJ, Xiao G. PDLIM2 suppresses human T-cell leukemia virus type I Tax-mediated tumorigenesis by targeting Tax into the nuclear matrix for proteasomal degradation. Blood. 2009;113:4370–80. doi: 10.1182/blood-2008-10-185660. PMID:19131544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen M, Sun F, Han L, Qu Z. Kaposi's sarcoma herpesvirus (KSHV) microRNA K12-1 functions as an oncogene by activating NF-kappaB/IL-6/STAT3 signaling. Oncotarget. 2016;7:33363–73. PMID:27166260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fu J, Qu Z, Yan P, Ishikawa C, Aqeilan RI, Rabson AB, Xiao G. The tumor suppressor gene WWOX links the canonical and noncanonical NF-kappaB pathways in HTLV-I Tax-mediated tumorigenesis. Blood. 2011;117:1652–61. doi: 10.1182/blood-2010-08-303073. PMID:21115974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Qu Z, Yan P, Fu J, Jiang J, Grusby MJ, Smithgall TE, Xiao G. DNA methylation-dependent repression of PDZ-LIM domain-containing protein 2 in colon cancer and its role as a potential therapeutic target. Cancer Res. 2010;70:1766–72. doi: 10.1158/0008-5472.CAN-09-3263. PMID:20145149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Qu Z, Fu J, Ma H, Zhou J, Jin M, Mapara MY, Grusby MJ, Xiao G. PDLIM2 restricts Th1 and Th17 differentiation and prevents autoimmune disease. Cell Biosci. 2012;2:23. doi: 10.1186/2045-3701-2-23. PMID:22731402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Qing G, Qu Z, Xiao G. Endoproteolytic processing of C-terminally truncated NF-kappaB2 precursors at kappaB-containing promoters. Proc Natl Acad Sci U S A. 2007;104:5324–9. doi: 10.1073/pnas.0609914104. PMID:17363471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Qu Z, Fu J, Yan P, Hu J, Cheng SY, Xiao G. Epigenetic repression of PDZ-LIM domain-containing protein 2: implications for the biology and treatment of breast cancer. J Biol Chem. 2010;285:11786–92. doi: 10.1074/jbc.M109.086561. PMID:20185823. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


