Abstract
Clinical and epidemiological data suggest that asthma and allergic diseases are associated and may share a common genetic etiology. We analyzed genome-wide single-nucleotide polymorphism (SNP) data for asthma and allergic diseases in 33,593 cases and 76,768 controls of European ancestry from the UK Biobank. Two publicly available independent genome wide association studies (GWAS) were used for replication. We have found a strong genome-wide genetic correlation between asthma and allergic diseases (rg = 0.75, P = 6.84×10−62). Cross trait analysis identified 38 genome-wide significant loci, including 7 novel shared loci. Computational analysis showed that shared genetic loci are enriched in immune/inflammatory systems and tissues with epithelium cells. Our work identifies common genetic architectures shared between asthma and allergy and will help to advance our understanding of the molecular mechanisms underlying co-morbid asthma and allergic diseases.
Asthma is a chronic respiratory syndrome that is characterized by abnormal and inflamed mucosa of the airways, wheezing, and shortness of breath. Allergic diseases are immune responses for allergies, such as allergic rhinitis and atopic dermatitis (eczema). Asthma is a heterogeneous disease that can be either allergic or non-allergic1. Allergic asthma (including IgE-mediated sub-type), allergic rhinitis and eczema can be viewed as an immune response to foreign antigen and often associates with immunoglobulin E (IgE)-mediated inflammation2,3. Genetic studies offer a structured means of understanding the causes of asthma and allergic diseases, as well as identifying targets that can be used to treat the syndrome4–8.
Clinical and epidemiological studies suggest that asthma and allergy are associated9,10. Several studies have identified allergic diseases, such as allergic rhinitis and eczema, with the prevalence of them in asthmatic patients being up to 90%9,11. These studies demonstrate that the coexistence of asthma and allergy is frequent, and that allergy usually precedes asthma. Also our previous epigenetic study has identified methylation loci linked to asthma and allergy via IgE pathway12.
One hypothesis to account for the similarity in symptoms for these conditions is that they share a common genetic etiology. Cotsapas et al discovered nearly half of loci in genome-wide association studies (GWAS) of an individual immune related disease influence risk to at least two diseases, indicating the shared genetic architecture of immune-mediated inflammatory and autoimmune diseases13. As each of the shared or similar risk factors has strong genetic influences on disease risk, the observed clustering of multiple risk factors could be due to an overlap in the causal genes and pathways14–21. In addition, grouping variants by the traits they influence should provide insight into the specific biological processes underlying comorbidity and disease risk. Clinical and epidemiological studies have found asthma and allergic diseases can occur either in the same individual or in closely related family members22–25, suggesting potential pleiotropic effect. Heritability has been estimated at varying between 35% and 95% for asthma22,26 and 33% and 91% for allergic rhinitis, 71% and 84% for atopic dermatitis (eczema), and 34% and 84% for serum IgE levels, which suggests that the genetic contribution for each may be significant22. While Hinds and colleagues investigated the shared genetic etiology for 38 allergic diseases including asthma27, our understanding has been limited to only 16 genome-wide shared susceptibility loci which were based on a self-reported phenotype. Additionally, a recent larger study has identified a shared genetic origin among three allergic disease using a mixed population of self-reported and doctor diagnosed phenotypes; however a standard fixed effect meta-analysis model was applied without considering artificial inflation due to heterogeneous effect among different sub-studies and phenotype definitions28.
In order to increase our knowledge of shared genetic determinants influencing doctor diagnosed allergic diseases and asthma, and to potentially discover novel loci, we investigated the genetic commonality among asthma and allergic diseases (hay fever/allergic rhinitis or eczema). We conducted a large scale GWAS analysis based on these traits to explore genetic correlations and shared genetic components among these diseases using data from the UK Biobank, which is the largest and most complete European Biobank available at present. Replication was performed in two independent publically available GWAS studies, the GABRIEL asthma GWAS study5 and the EArly Genetics & Lifecourse Epidemiology (EAGLE) eczema consortium study29.
RESULTS
Genetic correlation among immune related traits
There was a strong positive genetic correlation between asthma and allergic diseases (rg = 0.75, P = 6.84×10−62). When extending our genetic correlation analysis to rheumatoid arthritis30, Crohn’s disease and ulcerative colitis31; we did not find significant genetic correlation between asthma and these other immune traits and confirmed the high genetic correlation between Crohn’s disease and ulcerative colitis (rg = 0.57, P = 7.98×10−23) (Table 1). In a functional partitioned genetic correlation analysis, we observed significant genetic correlation in 11 of 12 functional categories with only exception being repressed regions (Figure 1 and Supplementary Table 1). This empirical evidence of a shared genetic etiology for asthma and allergic diseases encourages the investigation of a common pathophysiology, particularly in specific functional categories. Estimates of SNP-based heritability on the observed scale using GWAS summary statistics were 7.2% (s.e. 0.7%) for asthma and 7.5% (s.e. 0.7%) for allergic diseases (Supplementary Table 2).
Table 1.
Genetic correlation between immune diseases
| Phenotype | Asthma | Allergic diseases | Rheumatoid arthritis | Crohn’s disease | Ulcerative colitis | Asthma (GABRIEL) | Eczema (EAGLE) |
|---|---|---|---|---|---|---|---|
| Asthma (N=90,853) | 1 | 0.7455 | 0.0942 | 0.0428 | −0.0514 | 0.6649 | 0.4138 |
| Allergic diseases (N=102,453) | 6.84×10−62 | 1 | 0.0042 | 0.1415 | 0.0611 | 0.5008 | 0.5308 |
| Rheumatoid arthritis (N=58,284) | 0.0793 | 0.9259 | 1 | 0.067 | 0.1003 | 0.1001 | −0.0463 |
| Crohn’s disease (N=69,268) | 0.5925 | 0.0371 | 0.318 | 1 | 0.5711 | 0.0604 | 0.1190 |
| Ulcerative colitis (N=72,647) | 0.4942 | 0.3721 | 0.1032 | 7.98×10−23 | 1 | −0.0289 | −0.0121 |
| Asthma (GABRIEL) (N=26,475) | 3.52×10−25 | 3.24×10−15 | 0.1312 | 0.5005 | 0.7338 | 1 | 0.5310 |
| Eczema (EAGLE) (N=54,632) | 1.29×10−9 | 4.04×10−15 | 0.5846 | 0.1982 | 0.9134 | 6.05×10−6 | 1 |
Upper off-diagonal shows LD score regression estimates of genetic correlation score (Rg, ranges from −1 to 1). Lower off-diagonal shows corresponding genetic correlation P value.
Figure 1.
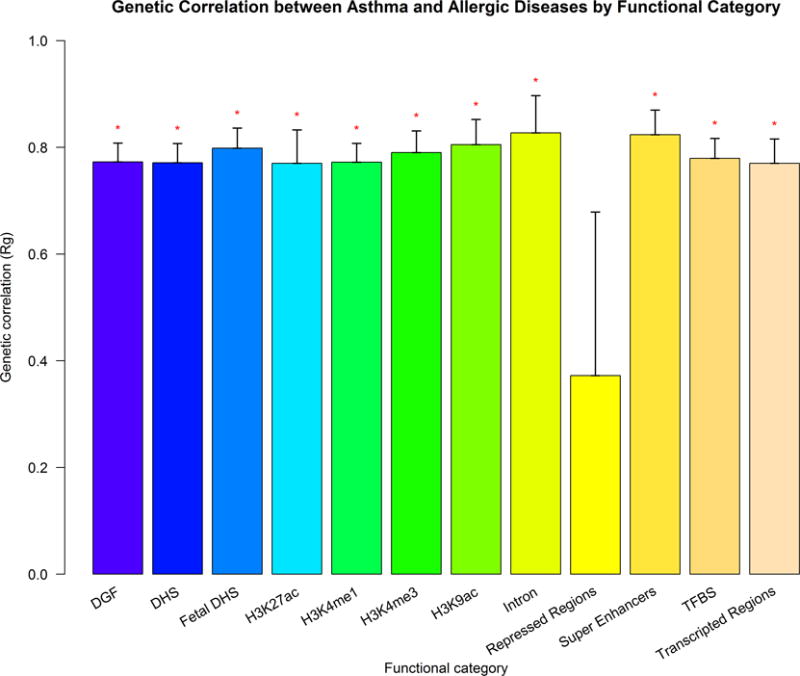
Partitioned genetic correlation between asthma and allergic diseases in the UK Biobank. Vertical axis represents the genetic correlation estimate Rg (standard error), horizontal axis represents 12 functional categories. Asterisk represents significance (P<0.05). DGF: DNaseI Digital Genomic Footprinting; DHS: DNase I hypersensitivity Site; TFBS: Transcription Factor Binding Sites.
Genome-wide association in UK Biobank
The phenotype-genotype association test was carried out on 110,361 samples and 7,488,535 SNPs from UK Biobank after quality control. The genomic control parameter λ was 1.13 for asthma and 1.14 for allergic diseases (Supplementary Fig. 1 and 2)32–34. The intercept (s.e.) from LD score regression (LDSC) for asthma and allergy were 1.0279 (0.0074) and 1.0242 (0.0054) respectively. We identified 32 genome-wide significant (P < 5×10−8) independent loci for asthma and 33 for allergic diseases (See Online Method, Supplementary Table 3 and 4, Supplementary Fig. 3 and 4).
Figure 2.
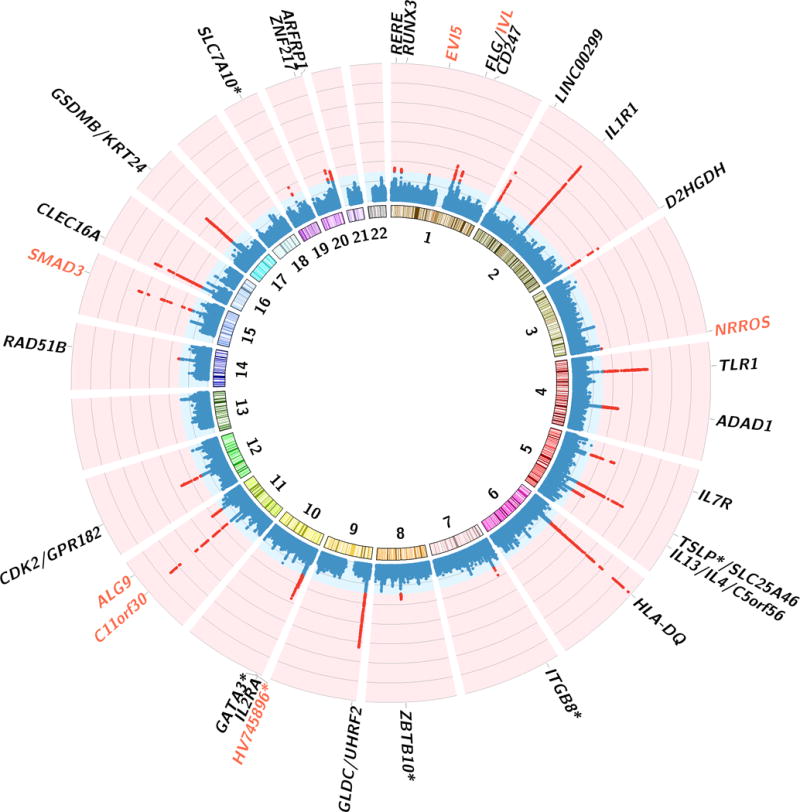
Circus Manhattan plot of cross trait meta-analysis. The first layer of the plot illustrates chromosome position and the second layer illustrates the representative genes of significant loci. Red genes are novel shared genes between asthma and allergic diseases The inside layer illustrates the significance level -log10(P-value) of asthma and allergic diseases’ shared markers from cross trait meta-analysis. Red dots means genome-wide significant (P < 5×10−8). A total of 38 independent loci were identified to be associated with both diseases. Genes at loci in close proximity were assigned one gene label separated by a slash. Sample size for asthma is 90,853, for allergic diseases is 102,453.
For asthma, we confirmed most of the previously reported association loci, including IL1R1, IL1RL1, IL18R1, IL13, SLC25A46, HLA regions, SMAD3, GSDMB and many others (Supplementary Table 3). Out of the 32 independent loci reported here, 6 are novel associations with asthma and 2 of these fall within protein-coding gene bodies. For allergic diseases, genes such as HLA-DQB, C11orf30, FLG, SLC25A46, TMEM232, CAMK4, TSLP, WDR36, CLEC16A, DEXI, HLA regions and many others were confirmed in our analysis (Supplementary Table 4). Out of the 33 independent loci identified for allergic diseases, 8 are novel, of which 7 were mapped to protein-coding genes. Six index SNPs were found to show genome wide significance in both asthma and allergic diseases (rs34290285 on D2HGDH and GAL3ST2, rs7705653 on SLC25A46 and TMEM232, rs12413578 near HV745896, rs7936070 on C11orf30, rs56062135 on SMAD3 and rs10414065 near SLC7A10), and 22 loci contains the same protein-coding genes between asthma and allergic diseases, although they do not share the same peak SNP.
Among the six loci with overlapping index SNPs, 4 were mapped to protein-coding genes. TMEM232 and SLC25A46 (index SNP: rs7705653, Supplementary Fig. 5 and 6) encode transmembrane protein 232 that belongs to the tetraspanin family and a solute carrier protein involved in transport of metabolites respectively. C11orf30 (index SNP: rs7936070, Supplementary Fig. 7 and 8) is associated with total serum IgE levels in asthma35; and the C11orf30 locus increases susceptibility to poly-sensitization36. SMAD3 (index SNP: rs56062135, Supplementary Fig. 9 and 10) is involved in the development of asthmatic inflammation due to the response of immune cells, such as cytokines and T-helper 2 (Th2) cells, which are known to be important in generating an inflammation that characterizes asthma and allergic disease37. A fourth variant, rs34290285, is the index SNP within locus that contained two relatively new genes implicated in asthma and allergic diseases, D2HGDH and GAL3ST2 (Supplementary Fig. 11 and 12), which are responsible for encoding metabolism related D-2hydroxyglutarate dehydrogenase enzyme38 and tumor metastasis related Galactose-3-O-sulfotransferase 2 enzyme39.
Cross trait meta-analysis
Given the strong genetic correlation and similarity of SNP effect sizes for asthma and allergic diseases (Supplementary Fig. 13), we improved our power to detect shared loci between asthma and allergy by performing a cross trait analysis. When doing so, we identified 38 loci that contained SNPs with P < 5×10−8 (Fig. 2 and Table 2). The genomic control parameter λ was 1.31 in the cross trait meta-analysis (Supplementary Fig. 14) and Figure 2 displays the Manhattan plot of these results.
Table 2.
Summary of the 38 genomic loci associated with the asthma and allergic disease in cross trait meta-analysis
| SNP | N | CHR | Peak SNP Position |
Ref Allele |
Alt Allele |
Asthma | Allergy | UK
Biobank Meta-analysis |
Replication | Genes Within
Clumping Range |
|||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P | P | OR | ASSET P | GABRIEL P | EAGLE P | CPMA P | |||||||
| rs9273374 | 65 | 6 | 32626614 | A | G | 1.31×10−35 | 1.17×10−4 | 0.84 | 7.87×10−35 | 9.27×10−3** | 0.0331 | NA*** | HLA-DOB,HLA-DQA1,HLA-DQA2,HLA-DQB1,HLA-DQB2,HLA-DRB1,HLA-DRB6,TAP2 |
| rs7936070 | 77 | 11 | 76293527 | T | G | 2.17×10−21 | 4.40×10−22 | 1.08 | 2.81×10−28 | 1.06×10−5 | 1.27×10−12 | 3.77×10−15 | C11orf30,LOC100506127,PRKRIR |
| rs72823641 | 151 | 2 | 102936159 | A | T | 7.43×10−18 | 4.27×10−24 | 0.89 | 1.58×10−27 | 1.86×10−9 | 1.50×10−3 | 4.67×10−10 | IL1R1,IL1RL1,IL1RL2,IL18R1,IL18RAP,MIR4772, SLC9A2,SLC9A4 |
| rs56062135 | 25 | 15 | 67455630 | T | C | 2.62×10−23 | 6.23×10−13 | 1.16 | 1.56×10−22 | 7.46×10−7 | 0.2354 | 1.37×10−5 | SMAD3 |
| rs9775039 | 109 | 9 | 6177453 | A | G | 7.41×10−23 | 4.84×10−8 | 1.18 | 4.42×10−22 | 2.38×10−12 | 0.1900 | 8.11×10−11 | GLDC,IL33,TPD52L3, UHRF2 |
| rs36045143 | 259 | 16 | 11224966 | G | A | 9.05×10−17 | 8.94×10−17 | 0.93 | 1.83×10−21 | 1.81×10−3 | 0.1057 | 6.90×10−3 | CLEC16A,DEXI |
| rs1837253 | 1 | 5 | 110401872 | T | C | 1.81×10−20 | 1.07×10−13 | 0.93 | 4.38×10−21 | 3.03×10−10 | 0.5307 | 2.09×10−8 | TSLP* |
| rs7705653 | 349 | 5 | 110142816 | G | A | 4.58×10−10 | 1.88×10−20 | 1.14 | 1.12×10−19 | 1.39×10−3 | 3.00×10−3 | 2.00×10−3 | SLC25A46,TMEM232 |
| rs28393318 | 200 | 4 | 38784267 | G | A | 2.61×10−10 | 6.67×10−20 | 0.92 | 2.14×10−19 | 3.92×10−3 | 0.0596 | 8.10×10−3 | FAM114A1,MIR574,TLR1,TLR6,TLR10 |
| rs869402 | 225 | 17 | 38068043 | C | T | 7.02×10−18 | 6.70×10−4 | 1.12 | 4.15×10−17 | 1.14×10−16 | 0.0247 | 8.30×10−16 | ERBB2,GRB7,GSDMA,GSDMB,IKZF3,LRRC3C,MIEN1,MIR4728,ORMDL3,PGAP3,PNMT,STARD3,TCAP, ZPBP2 |
| rs34290285 | 73 | 2 | 242698640 | A | G | 3.89×10−11 | 1.54×10−15 | 0.93 | 5.17×10−17 | 0.0240# | 0.1619 | 0.082 | D2HGDH,GAL3ST2 |
| rs10174949 | 59 | 2 | 8442248 | A | G | 6.08×10−10 | 5.72×10−16 | 0.94 | 1.70×10−16 | 5.77×10−3 | 6.91×10−6 | 3.53×10−6 | LINC00299 |
| rs9911533 | 104 | 17 | 38775476 | C | T | 2.08×10−07 | 1.65×10−16 | 0.92 | 9.70×10−16 | 0.8210 | 0.0302 | 0.3335 | KRT24,KRT222,SMARCE1 |
| rs12413578 | 24 | 10 | 9049253 | T | C | 3.27×10−14 | 4.63×10−10 | 0.91 | 1.09×10−14 | 0.3080 | 0.0855 | 0.3470 | HV745896* |
| rs6881270 | 71 | 5 | 35879095 | T | C | 7.87×10−6 | 2.61×10−15 | 0.91 | 1.53×10−14 | 0.2700 | 2.40×10−3 | 0.0195 | CAPSL,IL7R,LOC100506406,SPEF2,UGT3A1 |
| rs10876864 | 61 | 12 | 56401085 | G | A | 1.43×10−12 | 8.49×10−10 | 1.05 | 1.41×10−13 | 0.1880 | 0.0407 | 0.1396 | CDK2,ERBB3,IKZF4,PA2G4,RAB5B,RPL41,RPS26,SUOX,ZC3H10 |
| rs1059513 | 15 | 12 | 57489709 | C | T | 1.18×10−11 | 1.62×10−09 | 0.92 | 7.65×10−13 | 9.85×10−3 | 1.74×10−5 | 1.34×10−5 | GPR182,MYO1A,NAB2,RDH16,SDR9C7,STAT6,TAC3, TMEM194A,ZBTB39 |
| rs56267605 | 196 | 4 | 123363109 | C | A | 3.29×10−10 | 6.59×10−10 | 1.05 | 2.56×10−12 | 9.02×10−3 | 1.64×10−5 | 1.17×10−5 | ADAD1,IL2,IL21, IL21-AS1,KIAA1109 |
| rs61816766 | 10 | 1 | 152319572 | C | T | 3.10×10−8 | 3.07×10−11 | 1.14 | 4.63×10−12 | 0.970** | 8.17×10−21 | NA*** | C1orf68,CRCT1,CRNN,FLG,FLG-AS1,FLG2,HRNR,
LCE2A,LCE2B,LCE2C,LCE2D,LCE3A,LCE3B,LCE3C,LCE3D,LCE3E, LCE4A,LCE5A,NBPF18P,RPTN,S100A11,TCHH, TCHHL1 |
| rs61839660 | 47 | 10 | 6094697 | T | C | 6.86×10−4 | 3.98×10−12 | 1.12 | 2.30×10−11 | 0.2650 | 0.0226 | 0.1157 | IL2RA,RBM17 |
| rs2706362 | 96 | 5 | 131925187 | C | T | 1.27×10−11 | 1.73×10−7 | 1.06 | 3.75×10−11 | 8.14×10−8 | 9.76×10−14 | 2.84×10−18 | IL13,RAD50 |
| rs1214598 | 33 | 1 | 167426424 | A | G | 6.66×10−8 | 4.54×10−10 | 0.95 | 5.14×10−11 | 4.04×10−4 | 0.194 | 3.19×10−3 | CD247 |
| rs659529 | 192 | 11 | 111436896 | T | A | 3.45×10−7 | 1.20×10−10 | 0.95 | 6.03×10−11 | 0.9940 | 0.9530 | 1.70×10−3 | ALG9,BTG4,C11orf1,C11orf88,FDXACB1,LAYN,MIR34B,MIR34C,PPP2R1B,SIK2 |
| rs2766664 | 14 | 20 | 52171241 | A | G | 1.72×10−3 | 1.41×10−11 | 1.08 | 8.07×10−11 | 0.3540 | 2.25×10−5 | 4.00×10−3 | LOC101927770,ZNF217 |
| rs2169282 | 42 | 9 | 6350235 | A | G | 3.13×10−11 | 1.30×10−5 | 1.09 | 1.80×10−10 | 0.0172 | 0.7321 | 0.2030 | GLDC,UHRF2 |
| rs10074523 | 132 | 5 | 132060583 | C | A | 4.15×10−11 | 1.13×10−5 | 1.1 | 2.37×10−10 | 1.22×10−3 | 2.01×10−10 | 4.52×10−11 | CCNI2,IL4,KIF3A,SEPT8 |
| rs10414065 | 5 | 19 | 33721455 | T | C | 6.78×10−9 | 2.74×10−8 | 0.91 | 2.63×10−10 | 0.0740 | 0.249 | 0.2690 | SLC7A10* |
| rs6461503 | 89 | 7 | 20560996 | T | C | 2.13×10−9 | 8.16×10−8 | 1.05 | 3.19×10−10 | 6.65×10−3 | 0.0133 | 3.60×10−3 | ITGB8* |
| rs3208007 | 133 | 20 | 62322288 | T | C | 9.27×10−9 | 2.81×10−8 | 0.95 | 3.22×10−10 | 0.9310 | 7.40×10−6 | 4.00×10−3 | ARFRP1,LIME1,RTEL1, RTEL1-TNFRSF6B, SLC2A4RG,STMN3,TNFRSF6B,ZBTB46,ZGPAT |
| rs10795656 | 36 | 10 | 8595839 | A | G | 2.10×10−6 | 3.19×10−10 | 1.05 | 4.07×10−10 | 2.93×10−3 | 0.6890 | 0.0488 | GATA3* |
| rs2548992 | 88 | 5 | 131808668 | A | G | 7.96×10−11 | 5.40×10−4 | 1.1 | 4.54×10−10 | 3.68×10−6 | 0.5090 | 1.00×10−3 | C5orf56,IRF1,LOC553103,MIR3936,P4HA2,PDLIM4, SLC22A4,SLC22A5 |
| rs2136016 | 95 | 8 | 81300681 | G | A | 3.72×10−10 | 3.57×10−4 | 1.09 | 2.11×10−9 | 5.03×10−3 | 9.40×10−6 | 4.13×10−6 | ZBTB10* |
| rs61815704 | 24 | 1 | 152893891 | G | C | 4.08×10−7 | 5.51×10−8 | 1.14 | 5.16×10−9 | 0.2520** | 3.91×10−20 | NA*** | IVL,LCE1A,LCE1B,LCE1C, LCE1D,LCE6A,SMCP, SPRR1A,SPRR1B,SPRR2A, SPRR2B,SPRR2D,SPRR2E, SPRR2F,SPRR3,SPRR4 |
| rs301805 | 31 | 1 | 8481016 | T | G | 6.01×10−8 | 3.36×10−7 | 0.96 | 6.43×10−9 | 0.0591 | 8.02×10−5 | 3.00×10−3 | RERE |
| rs742230 | 19 | 1 | 25251424 | G | A | 5.95×10−6 | 1.27×10−8 | 0.96 | 1.02×10−8 | 0.3030 | 0.0287 | 0.1538 | RUNX3 |
| rs8008961 | 66 | 14 | 68752643 | T | C | 2.14×10−7 | 3.01×10−7 | 1.05 | 1.24×10−8 | 4.05×10−3 | 0.2360 | 0.0266 | RAD51B |
| rs4916533 | 9 | 3 | 196373582 | T | C | 2.79×10−7 | 3.62×10−7 | 0.93 | 1.66×10−8 | 0.6070 | 0.0667 | 0.4690 | NRROS |
| rs12743520 | 77 | 1 | 93037112 | A | C | 3.02×10−3 | 6.87×10−9 | 0.93 | 3.83×10−8 | 0.0460 | 0.7340 | 0.7340 | EVI5 |
The nearest gene to this locus.
The index SNP is missing in replication dataset, thus the best available proxy (highest LD) from the replication data within 500 KB distance of the index SNP was used. The best proxy SNPs, rs9273374: rs3135344 (R2=0.36); rs61816766: rs9050 (R2=0.44); rs61815704: rs61811907 (R2=0.77);
No common SNP between GABRIEL and EAGLE for CPMA analysis.
The imputation accuracy (r2pred) for rs34290285 from ImpG equals to 0.54, lower than ImpG standard criteria r2pred>0.6.
N, number of SNPs clumped with peak SNP; CHR, chromosome; OR: odds ratio
The strongest association signal was observed on chromosome 6 at HLA-DQ region (rs9273374, Pmeta = 7.87×10−35), the first asthma-susceptibility locus identified40, and extended haplotypes encompassing HLA-DQ and HLA-DR have been studied for their effects on specific allergen sensitization27,41. The second strongest signal was mapped to C11orf30 on chromosome 11q13 (rs7936070, Pmeta = 2.81×10−28), a gene associated with total serum IgE levels and increased susceptibility to poly-sensitization. This SNP shows a significant association with both asthma and allergic diseases in each respective single trait GWAS model. The third strongest signal was observed on IL1R1 genes (rs72823641, Pmeta = 1.58×10−27)42. Cytokine receptors have an impact on many cytokine induced immune and inflammatory responses, such as asthma and allergy5,29,43. The fourth strongest signal observed on SMAD3 (rs56062135, Pmeta = 1.56×10−22) is another loci found in both asthma and allergic diseases, which is known to have a role in immune cell response37. We have also identified the FLG (Filaggrin) gene (rs61816766, Pmeta = 4.63×10−12) to be associated with both asthma and allergic diseases, which is crucial for maintaining normal skin layer function44. In general, our cross-trait results show that most identified loci are significant for both asthma and allergic disease, suggesting that it is these shared genetic loci driving the overall significant positive genetic correlation.
GTEx tissue enrichment analysis of asthma and allergic diseases shared genes
To understand whether the 38 identified loci are enriched for expression in certain tissue types, we performed a tissue specific expression analysis (TSEA) using the GTEx pilot data45. We identified five independent tissues that demonstrated significantly enriched expression (after Benjamin-Hochberg correction) of cross-trait associated genes (Fig. 3). The most strongly enriched tissue was part of the integumentary system (skin). The other significantly enriched tissues included esophagus, vagina, lung and whole blood. We also found that the identified shared genes between asthma and allergic diseases have significant cross tissue heritability between each pair of tissues, with skin and lung showing notably high cross tissue heritability (Supplementary Fig. 15).
Figure 3.
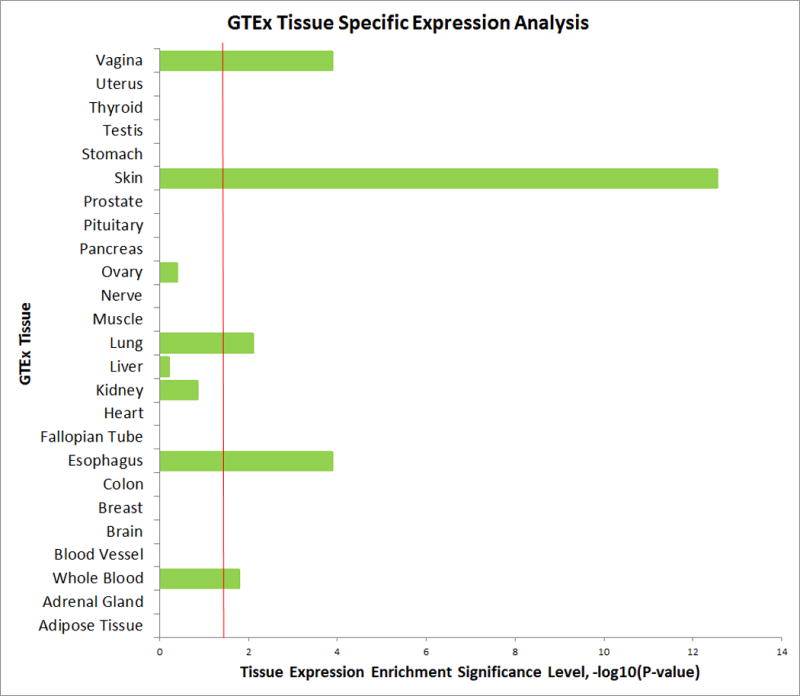
GTEx Tissue Specific Expression Analysis (TSEA). The horizontal axis illustrates tissue expression enrichment P-values after Benjamin-Hochberg correction. The vertical axis illustrates 25 independent tissue types from GTEx.
Replication analysis in two independent asthma and eczema studies
In our replication analysis, we have confirmed a significant level of genetic correlation between asthma and eczema by using two independent GWAS data: the GABRIEL study5 and EAGLE study29. LDSC results showed there was a strong positive genetic correlation between asthma (GABRIEL) and eczema (EAGLE) (rg = 0.53, P = 6.05×10−6) (Table 1). We have then used Cross-Phenotype Meta-Analysis (CPMA) to identify genes with shared genetic signals between asthma and one allergic disease, eczema. Twenty-two loci demonstrated genome-wide significance. Of these, genes HLA-DQB, LCE5A, FLG, IL6R, IL1RL1, IL13, IL4, ACSL6, TSLP, IL33, C11orf30, GSDMB and some others were replicated when comparing with the UK Biobank results (Supplementary Table 5 and Supplementary Fig. 16). Some genes were found as suggestive even though the association signal did not reach genome-wide significance. This includes RERE (rs11581328, Pmeta = 4.47×10−6), SMAD3 (rs744910, Pmeta = 1.46×10−7) and CLEC16A (rs2041733, Pmeta = 2.32×10−6) (Supplementary Table 6). While the phenotypes in our replication phase are not exactly the same as the UK Biobank’s, these results underscore the emerging overlap in genetic bases of asthma and allergy.
Overrepresentation Enrichment Analysis
GO analysis highlighted several biological processes significantly enriched in the shared genes between asthma and allergic diseases, including epithelium development, keratinization, skin development and T helper 1 type immune response (Supplementary Table 7). Additional analyses of KEGG pathways found that sharing association signals for asthma and allergic disease were significantly enriched in immune related pathway (Supplementary Table 8), which is consistent with a recent study28.
Fine-mapping and colocalization analysis identify shared causal variants
A list of credible set of SNPs for each locus was shown in Supplementary Table 9. A colocalization analysis showed that 33 out of 38 loci share causal variants between asthma and allergic diseases, consistent with being driven by the same signal from meta-analysis (Supplementary Table 10).
DISCUSSION
In this study, we have shown a strong genetic correlation between asthma and allergic diseases (hay fever/allergic rhinitis or eczema), as well as identified specific loci that are shared between diseases. Furthermore, we have replicated our finding in two independent GWAS studies5,29. Finally, we have found genetic loci shared between asthma and allergic diseases were significantly enriched in several tissues, including skin, esophageal, vagina, lung and whole blood tissue expression.
Most of our identified shared loci are consistent with two recently published studies17,28. In total, 7 novel loci were identified that are shared between asthma and allergic diseases (Figure 2). Of these, 6 loci were mapped within a gene body. All of these loci have significant single tissue expression quantitative trait loci (eQTL) mainly in the skin, lung, esophagus system, thyroid and whole blood (Supplementary Table 11). Given that GTEx eQTLs are pervasive at many loci, we have tried to determine whether our association signals colocalized with eQTL signals from 8 relevant GTEx tissues. Several important genes showed strong evidence of colocalization (H4>0.5), such as RERE and GSDMB in epithelium and immune related tissues. On the other hand, even though the HLA regions were shown colocalize in all 8 tissues, we note that the extended linkage disequilibrium in the HLA region may not provide sufficient resolution to colocalize relevant eQTL results with the cross-disease associations. Further experiments are required to elucidate the association between asthma/allergy and expression of HLA region. A relatively new region we identified as having an association signal with both asthma and allergic disease was 2q14.3 (index SNP: rs34290285), which contains the genes D2HGDH and GAL3ST2. Both of these genes have significant single tissue eQTL from multiple tissues including lung, esophagus mucosa, and skin (Supplementary Table 12 and 13). Notably, the lead variant in this locus, rs34290285, was only found to colocalize with multiple tissues through GAL3ST2, not through D2HGDH (Supplementary Table 14). GAL3ST2 encodes tumor metastasis related Galactose-3-O-sulfotransferase 2 enzyme39 and is expressed in the human small airway epithelium46.
We found genes that are associated with IgE function can consist of another crucial pathway for atopic diseases, such as asthma and eczema. We have found that C11orf30 on chromosome 11 (Supplementary Table 3-5) falls in an association locus shared between asthma and allergic diseases. C11orf30 was reported to be associated with total serum IgE levels and increases susceptibility to poly-sensitization35. When individuals are exposed to allergen, IgE is released from the immune system and travel to local organs or tissues to release type 2 cytokines such as IL-4,IL-13 and other inflammatory mediators47. These cytokines can further cause allergic diseases and asthma, such as reddish/dry skin and coughing, wheezing or shortness of breath48,49.
Clinical and epidemiologic observations have shown that immune-mediated inflammatory and autoimmune diseases can be associated due to overlap of genes13. Indeed, our LDSC analysis showed high genome-wide genetic correlation between asthma and allergic diseases and identified shared genes between them. However, we did not find significant genetic correlation between asthma and three other immune related diseases, rheumatoid arthritis, Crohn’s disease and ulcerative colitis, suggesting that the patterns of genetic architecture among immune related traits can still be distinct. IgE-mediated hypersensitivity may underlie allergic asthma and other allergic diseases. Rheumatoid arthritis belongs to immune complex mediated hypersensitivity, whereas Crohn’s disease and ulcerative colitis belong to delayed cell-mediated hypersensitivity. This may explain why we identified strong genetic correlation for the traits within each hypersensitivity category. Different types of hypersensitivity are characterized by different types of gene sets.
Though the association between respiratory diseases like asthma and skin tissue diseases may, at first, seem counterintuitive, exploring the link between the two apparently distinct phenotypes from a gene function perspective reveals a noticeable overlap in underlying genetics and molecular pathways in their etiologies. We have confirmed that the FLG gene on chromosome 1 was associated with both allergic diseases and asthma. FLG is important for the formation of the stratum corneum and also for the hydration of this barrier layer44. People who have mutations in the FLG gene can be sensitive to external allergens and produce dry and flaky skin44. Meanwhile it can activate a strong allergic immune response through peripheral blood to many organs, such as lung, causing the inflammation in the lungs that results in shortness of breath44.
Our finding of in whole blood tissue enrichment indicated that these diseases were caused by some malfunction of the shared immune system. Genes such as CD247 and RUNX3 play important roles in T-helper cells, CD8+ T cells, NK and DCs50 and were previously reported as risk genes for many immune-related inflammatory diseases, such as asthma, Crohn’s disease, ulcerative colitis and eczema50–53. Additionally whole blood is responsible for production of anti-inflammatory cytokines and circulating them in the whole body54.
Additional tissue enrichment in skin, esophagus, vagina, and lung suggests that the shared pathway between asthma and allergic diseases might have significant functions extending beyond whole blood. Skin, esophagus, vagina and lung tissues are all characterized by various type of epithelium or mucosa55,56, which share similarities in terms of functions and mechanisms. For example, the epithelial cells lining the airway, lungs and skin serve as the primary defense to inflammatory stimuli and antigens infiltrating these tissues through inducing cell signaling pathways in response to contact with the irritants. Indeed, a hallmark of asthma and allergic rhinitis is allergic sensitization by means of epithelial activation in the respiratory tract57,58. Also epithelial cells can transdifferentiate into motile mesenchymal cells, which can behave like stem cells and differentiate into multiple cell lineages and migrate to damaged tissues to repair59. Thus, the enrichment of asthma and allergy genes expressed in epithelial components is not specific to a certain organ or tissue; rather, it seems to be generalizable across epithelium types.
It is worth noting that a recently published study28 identified 136 independent risk variants spanning 73 novel loci that were shared between 3 allergic diseases. Some of their identified loci are consistent with ours, such as shared genes in the HLA region, D2HGDH, IL1RL1, RERE, RUNX3 and many others. Additionally, they have noted enrichment of results in whole blood and lung tissue, as well as immune related pathways. However, this study was based on a mixed population of self-reported and doctor diagnosed allergic diseases, whereas our study was based only on doctor diagnosed phenotype. Such complex phenotype background could introduce heterogeneity between studies. Additionally, a standard inverse-variance-weighted fixed-effects meta-analysis model was used to combine the association results from each sub-study, which has limited ability to account for heterogeneous effect. In comparison with Ferreira et al, our study has several strengths and presents additional novel results. First, we included an independent cohort to replicate our findings of shared loci between asthma and allergy. Second, we demonstrated both genome-wide LDSC and partitioned LDSC to assess the overall genetic correlation and functionally categorized genetic correlation between asthma and allergy. Third, we conducted cross trait meta-analysis using ASSET, which is robust to heterogeneous effect and overlap samples between two phenotypes. Finally, we found skin as one of the major tissue that was significantly enriched between asthma and allergy. We further showed significant cross tissue heritability for identified genes among enriched tissues and found that such connection might be due to similar epithelial cell components.
We also acknowledge the limitations of our study. First, our allergic symptom phenotype contained both allergic rhinitis/hay fever and eczema. The allergy genes we found may not be associated with a specific type of allergy. However, since allergic rhinitis and eczema are both IgE-mediated hypersensitivity and characterized by epithelial cells, they might share highly similar physiology60,61. Also, one cannot exclude the potential for misclassification of allergy phenotypes since participants in the UK Biobank may not report a condition they have. Second, our UK Biobank cohort traits are not independent. There are shared cases between asthma and allergic diseases and completely shared controls. However, our sensitivity analysis showed that LDSC analysis is robust to overlap samples under different scenarios (Supplementary Fig. 17 and Supplementary Table 15). Additionally, in our meta-analysis, we have applied methods that can account for overlapping samples and replicated our results in two independent studies to demonstrate the robustness of shared loci. We demonstrated the effect estimates of 38 loci were not influenced after excluding asthma and allergic diseases overlapped subjects in meta-analysis, though significance level were lowered due to smaller sample size (Supplementary Fig. 18-19 and Supplementary Table 16). Given the high genetic correlation between asthma and allergy GWAS within Caucasian population in the UK Biobank, population heterogeneity among GABRIEL and UK Biobank Caucasian could give a slightly lower value for the cross-population genetic correlation for asthma. For example, cross-population Rg for the same disease (asthma) could be smaller than cross-disease Rg within the same population, especially when the two diseases share substantial genetic correlation such as asthma and allergy. Since the GABRIEL study was based on childhood asthma, and the UK Biobank is based on adult asthma, the genetic effect between these two types of asthma may also lead to Rg differences. Finally, even after imputing the GABRIEL study summary statistics, there were still ~1 million SNPs less than the UK Biobank dataset. A smaller number of SNPs from the GABRIEL asthma GWAS could also affect the Rg estimation and provide fewer overlapping SNPs for both traits in the replication data compared to UK Biobank. This is also the reason that only a fraction of the shared genes was found in replication analysis.
These shared genetic components can provide opportunity to investigate asthma and allergy co-morbidity on molecular and functional level (Supplementary Table 17). Drugs targeting shared genetic loci might benefit treatment of both diseases. Our findings can also inspire patients and doctors to be more cautious about co-morbidity of asthma and allergic diseases in the clinical practice.
Understanding the genetic overlap between asthma and allergic diseases can be beneficial to treat disease outcomes more efficiently. Our GWAS study has highlighted shared genetic pathway in immune and inflammatory systems and epithelium tissues by asthma and allergic diseases. This work reinforces the idea that asthma and allergy implicate shared common biological processes and open new avenues for treatment.
ONLINE METHODS
Study population and design
Study participants were from the UK Biobank study, described in detail elsewhere62–64. In brief, the UK Biobank is a prospective study of > 500,000 people living in the UK. All people in the National Health Service registry who were aged 40–69 years and living <25 miles from a study center were invited to participate from 2006–2010. In total, 503,325 participants were recruited from over 9.2 million mailed invitations. For the current analysis, individuals of non-white ethnicity were excluded to avoid confounding effects. All participants provided informed consent to the UK Biobank.
To identify genetic variants that contribute to doctor-diagnosed asthma and allergic diseases (detailed phenotype information described in Supplementary Note) and link them with other conditions, we performed GWAS using phenotype measures in UK Biobank participants (N = 150,059). We removed 39,698 samples that are non-white, related, withdrew from UK Biobank or are missing genotype information. Thus, a total of 110,361 European descents with high-quality genotyping and complete phenotype/covariate data were used for these analyses, including 25,685 allergic diseases subjects (hay fever/allergic rhinitis or eczema, some with doctor diagnosed asthma), 14,085 asthma subjects and 76,768 controls for the analysis. Key demographic characteristics for each population was shown in Supplementary Table 18.
Furthermore, we have included summary statistics for a replication analysis from an independent asthma GWAS (GABRIEL study, 10,365 cases 16,110 controls)5 and an independent eczema GWAS (EAGLE eczema consortium, 13,287 cases and 41,345 controls)29.
Data summary, quality control (QC) and imputation
Detailed genotyping and QC procedures of UK Biobank were described previously (http://biobank.ctsu.ox.ac.uk/) and in Supplementary Note. In summary, after QC procedures were applied, the current released UK Biobank data contains 806,466 SNPs that passed SNP QC in at least one batch. Post-imputation QC was performed as previously outlined (http://biobank.ctsu.ox.ac.uk/); and genotype imputation procedure provided a total of 73,355,677 SNPs. In this study, we only focused on common variants for the analysis with minor allele frequency (MAF) > 1% due to higher imputation accuracy of common variants in UK Biobank65. In addition, we performed stringent QC standards by PLINK 1.90.66. We selected variants that did not deviate from Hardy-Weinberg Equilibrium (HWE) (P > 1×10−12), per variant missing call rates<5%, per-sample missing rate<40% and an imputation quality score (INFO) > 0.8. Quantile-Quantile (QQ) plots were produced and checked for each phenotype. R package GenABEL was used to compute genomic inflation values. A total of 7,488,535 SNPs passed QC on whole genome and with complete information in both phenotypes, which were eligible for statistical association analyses.
Genome-wide association analysis
We performed additive logistic regression genetic association analysis adjusting for age, sex, genotyping array and ten ancestry principal components using PLINK 1.9066 to assess association between phenotype and genotype on each individual disease. After association analysis, we applied PLINK clumping function (parameters: –clump-p1 5e-8 –clump-p2 1e-5 –clump-r2 0.2 –clump-kb 500) to determine top loci that are independent to each other, i.e. variants with p-value less than 1×10−5, has R2 more than 0.2 and less than 500 kb away from the peak will be assigned to that peak’s clump. The top loci that were annotated to the same gene were then further combined as one independent loci for downstream analysis. We have used the GWAS catalog (https://www.ebi.ac.uk/gwas, search date: 10/18/2017) to identify previously reported genes by other GWAS studies.
LD score regression analysis
Post-GWAS genome-wide genetic correlation analysis by LD score regression (LDSC)67 was conducted using UK Biobank summary statistics overlap with HapMap3 variants as recommended68. LDSC estimates genetic correlation between the true causal effects of two traits (ranging from −1 to 1) from summary statistics using the facts that the GWAS effect size estimate for each SNP incorporates the effects of all SNPs in linkage disequilibrium with that SNP. SNPs in a high linkage disequilibrium region would have higher χ2 statistics than SNPs in a low linkage disequilibrium region, and a similar relationship is observed when single-study test statistics are replaced with the product of the z scores from two studies of traits with some correlation67. LDSC applied a self-estimated intercept during the analysis to account for shared subjects between studies67.
Partitioned genetic correlation
We estimated the genetic correlation between asthma and allergic diseases within functional categories using partitioned LDSC69. Twelve functional categories were used including transcribed region, repressed region, intron, transcription factor binding sites (TFBS), super-enhancer, DNaseI digital genomic footprinting (DGF) region, DNase I hypersensitivity sites (DHSs), fetalDHS, and histone marks H3K4me1, H3K27ac, H3K4me3, H3K9ac from Roadmap Epigenomics Project70. Recalculated LD scores for SNPs classified in each particular annotation were used for estimating the asthma-allergy genetic correlation within that functional group.
Cross trait meta-analysis
After assessing genetic correlations among all traits, we applied cross trait GWAS meta-analysis by using the R package Association analysis based on SubSETs (ASSET) to combine the association evidence for asthma and allergic diseases at individual variants71. This method combines effect estimate and standard error of the GWAS summary statistics to test hypothesis of association between the SNP with any subset of studies. It can also account the correlation among studies/subjects that might arise due to shared subjects across distinct studies or due to correlation among related traits in the same study by using case-control overlap matrices. We used 1-sided ASSET assuming a same direction of association for asthma and allergic diseases.
Fine mapping credible set analysis
We identified a credible set of causal variants at each of the 38 shared loci that met asthma-allergic disease cross trait meta-analysis criteria using the Bayesian fine-mapping algorithm FM-summary (see URLs). For each locus, we extracted variants within 500kb of the index SNP. The FM-summary program maps the primary signal and uses a flat prior with steepest descent approximation. The details of this method are described elsewhere.72–74.
Colocalization analysis
We used the R package coloc75 to determine whether asthma and allergic disease association signals co-localized at shared loci and whether shared loci co-localized with GTEx (version 7) eQTL in 8 relevant tissues, including skin (not sun exposed), skin (sun exposed), esophagus mucosa, esophagus muscularis, esophagus gastroesophageal junction, lung, vagina, whole blood. We extracted summary association data for variants within 500kb of the index SNP at each of the 38 shared loci and calculated the probability that the two traits share one common causal variant (H4). Loci with probability greater than 0.5 were considered to colocalize.
GTEx Tissue Specific Expression Analysis
In order to determine if shared asthma and allergy genes are overly expressed in disease-relevant tissues, we conducted tissue specific expression analysis (TSEA)76,77. Gene lists were generated from lists obtained from analyses for asthma, allergic diseases and cross-disease, and by including those which have a matching HUGO Gene Nomenclature Committee (HGNC) name. Genes identified in this way were analyzed for tissue enrichment using publicly available expression data from pilot phase of the Genotype-Tissue Expression project (GTEx)45 (www.gtexportal.org, 1/31/2013). In the GTEx project, postmortem samples from a wide variety of tissues and donors have been used for bulk RNA sequencing according to a unified protocol. All samples were sequenced using Illumina 76 base-pair paired-end reads. The pilot phase of the GTEx project consists of 1,839 samples derived from 189 post-mortem subjects, including samples from 45 different tissues, with some tissues offering multiple ‘sub-tissue’ types. TSEA averaged collapsed reads per kilobase per million mapped reads (RPKM) values for the sub-tissue types (25 ‘whole-tissue’ types). We filtered for unique HGNC IDs (n = 213 shared genes mapped by identified loci from meta-analysis result existing in tissue expression dataset). For each tissue, transcripts from the processed GTEx transcripts that are specifically expressed or enriched have been identified by using the TSEA pSI R package function to calculate the specificity index probability (pSI). Significance level of shared genes between asthma and allergic diseases enriched in each tissue were identified by by Fisher’s exact test with Benjamini-Hochberg correction.
Replication analysis
In order to confirm our findings, we have further included two public available independent GWAS data: asthma GWAS, the GABRIEL consortium study5 and eczema GWAS, the EArly Genetics & Lifecourse Epidemiology (EAGLE) eczema consortium study29, for LD score regression analysis, cross trait analysis and GTEx tissue expression enrichment analysis. We have used LiftOver tool (see URLs) to convert asthma GWAS reference genome from hg18 to hg19. ImpG-Summary was used to impute asthma GWAS summary statistics to the 1000 Genomes Project variants (phase 3 release v5)78. SNPs with imputation quality r2pred < 0.6 were removed. Cross trait analysis between two replication GWAS datasets was conducted using R package Cross-Phenotype Meta-Analysis (CPMA), which tests for the hypothesis that each independent SNP has multiple phenotype association and combines the empirical evidence based on their P-values13.
Overrepresentation Enrichment Analysis
In order to understand the biological insights of the shared genes, we have used the WebGestalt tool79 to assess overrepresented enrichment of the identified shared gene set between asthma and allergic diseases in KEGG80 pathways and Gene Ontology (GO) biological process81,82.
Cross tissue heritability of gene expression
The GTEx project45 (phs000424.v6.p1) was also used to quantify the cis SNP cross-tissue heritability ( ) of gene expression across a range of tissues. All estimates were obtained using restricted maximum likelihood (REML) as implemented in Genome-wide Complex Trait Analysis (GCTA)83. Only common variants that fall within 1 MB of the transcription start site of a gene were considered in deriving these estimates. Enrichment of was determined by comparing average for a given gene list to 1,000,000 random gene lists of equal size in the same tissue pair. Detailed information of our method are described in Supplementary Note.
Sensitivity analysis
To estimate the effect of using non-allergic asthma individuals, we performed additive logistic regression analysis adjusting for the same covariates as the primary analysis to assess the association between phenotype and genotype on each individual disease excluding allergic asthma (N = 6,177). We carried out cross-disease heritability analysis and meta-analysis to identify the loci that are associated with both diseases. Due to the large reduction in sample size of asthma cases and allergic diseases cases, the power of detecting an association between phenotypes and SNP is expected to decrease. Furthermore, in order to know if overlap samples can bias genetic correlation estimates, we performed additional sensitivity analysis. We randomly split controls into two equal sample size datasets (N=38,384 for each), and used real disease proportions of asthma and allergic diseases with two distinct control datasets. Details of overlap sample sensitivity analysis design can be found in Supplementary Fig. 17. Using the same set of cases, we also compared the genetic correlation estimates via using disease free controls versus using non-asthma or non-allergy controls (Supplementary Table 15).
Data availability
Summary GWAS statistics will be made available at the website (http://lianglab.rc.fas.harvard.edu/AsthmaAllergyHeritability/).
Supplementary Material
Acknowledgments
This research has been conducted using the UK Biobank Resource under application number 16549. We would like to thank the participants and researchers from the UK Biobank who significantly contributed or collected data. We thank to GABRIEL consortium and EAGLE consortium for providing GWAS summary statistic data. We also thank to W. Cookson and M. Moffatt their clinical advice; D. Chasman, V. Anttila, S. Gazal, H. Shi, Y. Feng and M. Chen for their statistical advice. This study was supported by grants R01HL060710 (DCC), R56HL134356 (DCC), R01HL114769 (QL), AAF15-0097 (QL), R00MH101367 (PHL) from National Heart, Lung, and Blood Institute (NHLBI), National Institutes of Health, American Asthma Foundation and National Institute of Mental Health.
Footnotes
URLs. UK Biobank, http://biobank.ctsu.ox.ac.uk/; 1000 Genomes Project, http://www.1000genomes.org/; PLINK, https://www.cog-genomics.org/plink2; ASSET, https://bioconductor.org/packages/release/bioc/html/ASSET.html; CPMA, http://coruscant.itmat.upenn.edu/software.html; GenABEL, http://www.genabel.org/packages/GenABEL; FM-summary, https://github.com/hailianghuang/FM-summary; Coloc tool, https://github.com/chr1swallace/coloc; Roadmap Epigenomics, http://www.roadmapepigenomics.org/; GABRIEL consortium, https://www.cng.fr/gabriel/index.html; EAGLE consortium, http://data.bris.ac.uk/data; LiftOver, http://genome.sph.umich.edu/wiki/LiftOver; ImpG, https://github.com/huwenboshi/ImpG; LDSC, https://github.com/bulik/ldsc; GCTA, http://cnsgenomics.com/software/gcta; GTEx, http://www.gtexportal.org; TSEA, http://genetics.wustl.edu/jdlab/tsea/; WebGestalt, http://www.webgestalt.org/option.php; GWAS catalog, https://www.ebi.ac.uk/gwas; LocusZoom, http://locuszoom.org
Methods, data availability and associated references are available in the online version of the paper
Note: Any Supplementary Information is available in the online version of the paper.
AUTHOR CONTRIBUTIONS
Z.Z., L.L., P.H.L., Q.L., D.C.C. designed the study.
Z.Z., M.C., W.C., P.R.L. performed the statistical analysis.
Z.Z., M.C., L.L., W.C., Q.L. wrote the manuscript.
All authors helped interpret the data, reviewed and edited the final paper, and approved the submission.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
ORCID:
Zhaozhong Zhu: 0000-0001-5662-1541
Liming Liang: 0000-0001-8261-3174
References
- 1.Johansson SG, et al. Revised nomenclature for allergy for global use: Report of the Nomenclature Review Committee of the World Allergy Organization, October 2003. J Allergy Clin Immunol. 2004;113:832–6. doi: 10.1016/j.jaci.2003.12.591. [DOI] [PubMed] [Google Scholar]
- 2.Lenz HJ. Management and preparedness for infusion and hypersensitivity reactions. Oncologist. 2007;12:601–9. doi: 10.1634/theoncologist.12-5-601. [DOI] [PubMed] [Google Scholar]
- 3.Wallace DV, et al. The diagnosis and management of rhinitis: an updated practice parameter. J Allergy Clin Immunol. 2008;122:S1–84. doi: 10.1016/j.jaci.2008.06.003. [DOI] [PubMed] [Google Scholar]
- 4.Torgerson DG, et al. Meta-analysis of genome-wide association studies of asthma in ethnically diverse North American populations. Nat Genet. 2011;43:887–92. doi: 10.1038/ng.888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moffatt MF, et al. A large-scale, consortium-based genomewide association study of asthma. N Engl J Med. 2010;363:1211–21. doi: 10.1056/NEJMoa0906312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bunyavanich S, et al. Integrated genome-wide association, coexpression network, and expression single nucleotide polymorphism analysis identifies novel pathway in allergic rhinitis. BMC Med Genomics. 2014;7:48. doi: 10.1186/1755-8794-7-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ramasamy A, et al. A genome-wide meta-analysis of genetic variants associated with allergic rhinitis and grass sensitization and their interaction with birth order. J Allergy Clin Immunol. 2011;128:996–1005. doi: 10.1016/j.jaci.2011.08.030. [DOI] [PubMed] [Google Scholar]
- 8.Pillai SG, et al. A genome-wide association study in chronic obstructive pulmonary disease (COPD): identification of two major susceptibility loci. PLoS Genet. 2009;5:e1000421. doi: 10.1371/journal.pgen.1000421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leynaert B, Neukirch F, Demoly P, Bousquet J. Epidemiologic evidence for asthma and rhinitis comorbidity. J Allergy Clin Immunol. 2000;106:S201–5. doi: 10.1067/mai.2000.110151. [DOI] [PubMed] [Google Scholar]
- 10.Brauer M, et al. Air pollution from traffic and the development of respiratory infections and asthmatic and allergic symptoms in children. Am J Respir Crit Care Med. 2002;166:1092–8. doi: 10.1164/rccm.200108-007OC. [DOI] [PubMed] [Google Scholar]
- 11.Pariente PD, LePen C, Los F, Bousquet J. Quality-of-life outcomes and the use of antihistamines in a French national population-based sample of patients with perennial rhinitis. Pharmacoeconomics. 1997;12:585–95. doi: 10.2165/00019053-199712050-00009. [DOI] [PubMed] [Google Scholar]
- 12.Liang L, et al. An epigenome-wide association study of total serum immunoglobulin E concentration. Nature. 2015;520:670–4. doi: 10.1038/nature14125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cotsapas C, et al. Pervasive sharing of genetic effects in autoimmune disease. PLoS Genet. 2011;7:e1002254. doi: 10.1371/journal.pgen.1002254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Criswell LA, et al. Analysis of families in the multiple autoimmune disease genetics consortium (MADGC) collection: the PTPN22 620W allele associates with multiple autoimmune phenotypes. Am J Hum Genet. 2005;76:561–71. doi: 10.1086/429096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cross-Disorder Group of the Psychiatric Genomics, C. Identification of risk loci with shared effects on five major psychiatric disorders: a genome-wide analysis. Lancet. 2013;381:1371–9. doi: 10.1016/S0140-6736(12)62129-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Global Lipids Genetics, C. Discovery and refinement of loci associated with lipid levels. Nat Genet. 2013;45:1274–83. doi: 10.1038/ng.2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pickrell JK, et al. Detection and interpretation of shared genetic influences on 42 human traits. Nat Genet. 2016;48:709–17. doi: 10.1038/ng.3570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lane JM, et al. Genome-wide association analyses of sleep disturbance traits identify new loci and highlight shared genetics with neuropsychiatric and metabolic traits. Nat Genet. 2016 doi: 10.1038/ng.3749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hobbs BD, et al. Genetic loci associated with chronic obstructive pulmonary disease overlap with loci for lung function and pulmonary fibrosis. Nat Genet. 2017;49:426–432. doi: 10.1038/ng.3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Emdin CA, et al. Genetic Association of Waist-to-Hip Ratio With Cardiometabolic Traits, Type 2 Diabetes, and Coronary Heart Disease. JAMA. 2017;317:626–634. doi: 10.1001/jama.2016.21042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hammerschlag AR, et al. Genome-wide association analysis of insomnia complaints identifies risk genes and genetic overlap with psychiatric and metabolic traits. Nat Genet. 2017;49:1584–1592. doi: 10.1038/ng.3888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ober C, Yao TC. The genetics of asthma and allergic disease: a 21st century perspective. Immunol Rev. 2011;242:10–30. doi: 10.1111/j.1600-065X.2011.01029.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cookson W. The immunogenetics of asthma and eczema: a new focus on the epithelium. Nat Rev Immunol. 2004;4:978–88. doi: 10.1038/nri1500. [DOI] [PubMed] [Google Scholar]
- 24.Belsky DW, et al. Polygenic risk and the development and course of asthma: an analysis of data from a four-decade longitudinal study. Lancet Respir Med. 2013;1:453–61. doi: 10.1016/S2213-2600(13)70101-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Holgate ST. The epidemic of allergy and asthma. Nature. 1999;402:B2–4. doi: 10.1038/35037000. [DOI] [PubMed] [Google Scholar]
- 26.Duffy DL, Martin NG, Battistutta D, Hopper JL, Mathews JD. Genetics of asthma and hay fever in Australian twins. Am Rev Respir Dis. 1990;142:1351–8. doi: 10.1164/ajrccm/142.6_Pt_1.1351. [DOI] [PubMed] [Google Scholar]
- 27.Hinds DA, et al. A genome-wide association meta-analysis of self-reported allergy identifies shared and allergy-specific susceptibility loci. Nat Genet. 2013;45:907–11. doi: 10.1038/ng.2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ferreira MA, et al. Shared genetic origin of asthma, hay fever and eczema elucidates allergic disease biology. Nat Genet. 2017 doi: 10.1038/ng.3985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Paternoster L, et al. Multi-ancestry genome-wide association study of 21,000 cases and 95,000 controls identifies new risk loci for atopic dermatitis. Nat Genet. 2015;47:1449–56. doi: 10.1038/ng.3424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Okada Y, et al. Genetics of rheumatoid arthritis contributes to biology and drug discovery. Nature. 2014;506:376–81. doi: 10.1038/nature12873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jostins L, et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491:119–24. doi: 10.1038/nature11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Devlin B, Roeder K. Genomic control for association studies. Biometrics. 1999;55:997–1004. doi: 10.1111/j.0006-341x.1999.00997.x. [DOI] [PubMed] [Google Scholar]
- 33.de Bakker PI, et al. Practical aspects of imputation-driven meta-analysis of genome-wide association studies. Hum Mol Genet. 2008;17:R122–8. doi: 10.1093/hmg/ddn288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang J, et al. Genomic inflation factors under polygenic inheritance. Eur J Hum Genet. 2011;19:807–12. doi: 10.1038/ejhg.2011.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li X, et al. The C11orf30-LRRC32 region is associated with total serum IgE levels in asthmatic patients. J Allergy Clin Immunol. 2012;129:575–8. 578 e1–9. doi: 10.1016/j.jaci.2011.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Amaral AF, et al. The locus C11orf30 increases susceptibility to poly-sensitization. Allergy. 2015;70:328–33. doi: 10.1111/all.12557. [DOI] [PubMed] [Google Scholar]
- 37.Anthoni M, et al. Smad3 -signalling and Th2 cytokines in normal mouse airways and in a mouse model of asthma. Int J Biol Sci. 2007;3:477–85. doi: 10.7150/ijbs.3.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Struys EA, et al. Mutations in the D-2-hydroxyglutarate dehydrogenase gene cause D-2-hydroxyglutaric aciduria. Am J Hum Genet. 2005;76:358–60. doi: 10.1086/427890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Seko A, Nagata K, Yonezawa S, Yamashita K. Down-regulation of Gal 3-O-sulfotransferase-2 (Gal3ST-2) expression in human colonic non-mucinous adenocarcinoma. Jpn J Cancer Res. 2002;93:507–15. doi: 10.1111/j.1349-7006.2002.tb01285.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marsh DG, Meyers DA, Bias WB. The epidemiology and genetics of atopic allergy. N Engl J Med. 1981;305:1551–9. doi: 10.1056/NEJM198112243052603. [DOI] [PubMed] [Google Scholar]
- 41.Moffatt MF, et al. Association between quantitative traits underlying asthma and the HLA-DRB1 locus in a family-based population sample. Eur J Hum Genet. 2001;9:341–6. doi: 10.1038/sj.ejhg.5200636. [DOI] [PubMed] [Google Scholar]
- 42.Dale M, Nicklin MJ. Interleukin-1 receptor cluster: gene organization of IL1R2, IL1R1, IL1RL2 (IL-1Rrp2), IL1RL1 (T1/ST2), and IL18R1 (IL-1Rrp) on human chromosome 2q. Genomics. 1999;57:177–9. doi: 10.1006/geno.1999.5767. [DOI] [PubMed] [Google Scholar]
- 43.Gudbjartsson DF, et al. Sequence variants affecting eosinophil numbers associate with asthma and myocardial infarction. Nat Genet. 2009;41:342–7. doi: 10.1038/ng.323. [DOI] [PubMed] [Google Scholar]
- 44.McLean WH. The allergy gene: how a mutation in a skin protein revealed a link between eczema and asthma. F1000 Med Rep. 2011;3:2. doi: 10.3410/M3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Consortium, G.T. The Genotype-Tissue Expression (GTEx) project. Nat Genet. 2013;45:580–5. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hackett NR, et al. RNA-Seq quantification of the human small airway epithelium transcriptome. BMC Genomics. 2012;13:82. doi: 10.1186/1471-2164-13-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Novak N, Kraft S, Bieber T. IgE receptors. Curr Opin Immunol. 2001;13:721–6. doi: 10.1016/s0952-7915(01)00285-0. [DOI] [PubMed] [Google Scholar]
- 48.Wynn TA. Type 2 cytokines: mechanisms and therapeutic strategies. Nat Rev Immunol. 2015;15:271–82. doi: 10.1038/nri3831. [DOI] [PubMed] [Google Scholar]
- 49.Barnes PJ. Th2 cytokines and asthma: an introduction. Respir Res. 2001;2:64–5. doi: 10.1186/rr39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lotem J, et al. Runx3 at the interface of immunity, inflammation and cancer. Biochim Biophys Acta. 2015;1855:131–43. doi: 10.1016/j.bbcan.2015.01.004. [DOI] [PubMed] [Google Scholar]
- 51.Laprise C. The Saguenay-Lac-Saint-Jean asthma familial collection: the genetics of asthma in a young founder population. Genes Immun. 2014;15:247–55. doi: 10.1038/gene.2014.12. [DOI] [PubMed] [Google Scholar]
- 52.Guo C, et al. Chromatin immunoprecipitation and association study revealed a possible role of Runt-related transcription factor 3 in the ulcerative colitis of Chinese population. Clin Immunol. 2010;135:483–9. doi: 10.1016/j.clim.2010.01.004. [DOI] [PubMed] [Google Scholar]
- 53.Esparza-Gordillo J, et al. A functional IL-6 receptor (IL6R) variant is a risk factor for persistent atopic dermatitis. J Allergy Clin Immunol. 2013;132:371–7. doi: 10.1016/j.jaci.2013.01.057. [DOI] [PubMed] [Google Scholar]
- 54.Meijer H, Reinecke J, Becker C, Tholen G, Wehling P. The production of anti-inflammatory cytokines in whole blood by physico-chemical induction. Inflamm Res. 2003;52:404–7. doi: 10.1007/s00011-003-1197-1. [DOI] [PubMed] [Google Scholar]
- 55.Presland RB, Dale BA. Epithelial structural proteins of the skin and oral cavity: function in health and disease. Crit Rev Oral Biol Med. 2000;11:383–408. doi: 10.1177/10454411000110040101. [DOI] [PubMed] [Google Scholar]
- 56.Ganesan S, Comstock AT, Sajjan US. Barrier function of airway tract epithelium. Tissue Barriers. 2013;1:e24997. doi: 10.4161/tisb.24997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang Y, Bai C, Li K, Adler KB, Wang X. Role of airway epithelial cells in development of asthma and allergic rhinitis. Respir Med. 2008;102:949–55. doi: 10.1016/j.rmed.2008.01.017. [DOI] [PubMed] [Google Scholar]
- 58.Rackley CR, Stripp BR. Building and maintaining the epithelium of the lung. J Clin Invest. 2012;122:2724–30. doi: 10.1172/JCI60519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol. 2014;15:178–96. doi: 10.1038/nrm3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Alonso L, Fuchs E. Stem cells of the skin epithelium. Proc Natl Acad Sci U S A. 2003;100(Suppl 1):11830–5. doi: 10.1073/pnas.1734203100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pawankar R, Mori S, Ozu C, Kimura S. Overview on the pathomechanisms of allergic rhinitis. Asia Pac Allergy. 2011;1:157–67. doi: 10.5415/apallergy.2011.1.3.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Collins R. What makes UK Biobank special? Lancet. 2012;379:1173–4. doi: 10.1016/S0140-6736(12)60404-8. [DOI] [PubMed] [Google Scholar]
- 63.Sudlow C, et al. UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 2015;12:e1001779. doi: 10.1371/journal.pmed.1001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Allen NE, Sudlow C, Peakman T, Collins R, Biobank UK. UK biobank data: come and get it. Sci Transl Med. 2014;6(224ed4) doi: 10.1126/scitranslmed.3008601. [DOI] [PubMed] [Google Scholar]
- 65.McCarthy S, et al. A reference panel of 64,976 haplotypes for genotype imputation. Nat Genet. 2016;48:1279–83. doi: 10.1038/ng.3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Purcell S, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bulik-Sullivan B, et al. An atlas of genetic correlations across human diseases and traits. Nat Genet. 2015;47:1236–41. doi: 10.1038/ng.3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bulik-Sullivan BK, et al. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet. 2015;47:291–5. doi: 10.1038/ng.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Finucane HK, et al. Partitioning heritability by functional annotation using genome-wide association summary statistics. Nat Genet. 2015;47:1228–35. doi: 10.1038/ng.3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bernstein BE, et al. The NIH Roadmap Epigenomics Mapping Consortium. Nat Biotechnol. 2010;28:1045–8. doi: 10.1038/nbt1010-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bhattacharjee S, et al. A subset-based approach improves power and interpretation for the combined analysis of genetic association studies of heterogeneous traits. Am J Hum Genet. 2012;90:821–35. doi: 10.1016/j.ajhg.2012.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gormley P, et al. Meta-analysis of 375,000 individuals identifies 38 susceptibility loci for migraine. Nat Genet. 2016;48:856–66. doi: 10.1038/ng.3598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Schizophrenia Working Group of the Psychiatric Genomics, C. Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–7. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wellcome Trust Case Control, C et al. Bayesian refinement of association signals for 14 loci in 3 common diseases. Nat Genet. 2012;44:1294–301. doi: 10.1038/ng.2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wallace C. Statistical testing of shared genetic control for potentially related traits. Genet Epidemiol. 2013;37:802–13. doi: 10.1002/gepi.21765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Dougherty JD, Schmidt EF, Nakajima M, Heintz N. Analytical approaches to RNA profiling data for the identification of genes enriched in specific cells. Nucleic Acids Res. 2010;38:4218–30. doi: 10.1093/nar/gkq130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Xu X, Wells AB, O’Brien DR, Nehorai A, Dougherty JD. Cell type-specific expression analysis to identify putative cellular mechanisms for neurogenetic disorders. J Neurosci. 2014;34:1420–31. doi: 10.1523/JNEUROSCI.4488-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pasaniuc B, et al. Fast and accurate imputation of summary statistics enhances evidence of functional enrichment. Bioinformatics. 2014;30:2906–14. doi: 10.1093/bioinformatics/btu416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wang J, Duncan D, Shi Z, Zhang B. WEB-based GEne SeT AnaLysis Toolkit (WebGestalt): update 2013. Nucleic Acids Res. 2013;41:W77–83. doi: 10.1093/nar/gkt439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ashburner M, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–9. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gene Ontology, C. Gene Ontology Consortium: going forward. Nucleic Acids Res. 2015;43:D1049–56. doi: 10.1093/nar/gku1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lee SH, Yang J, Goddard ME, Visscher PM, Wray NR. Estimation of pleiotropy between complex diseases using single-nucleotide polymorphism-derived genomic relationships and restricted maximum likelihood. Bioinformatics. 2012;28:2540–2. doi: 10.1093/bioinformatics/bts474. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Summary GWAS statistics will be made available at the website (http://lianglab.rc.fas.harvard.edu/AsthmaAllergyHeritability/).


