Abstract
Worldwide, breast cancer is the most frequent neoplasm and the second leading cause of cancer death among females. It dominates in both developed and developing countries and represents a major public health problem. The etiology is multifactorial and involves exogenous agents as well as endogenous factors. Although they account for only a small fraction of the breast cancer burden, mutations in the BRCA1 and BRCA2 genes are known to confer a high risk predisposition. Mutations in moderate/low-penetrance genes may also contribute to breast cancer risk. Previous studies have shown that mutations in the CHEK2 gene are involved in breast cancer susceptibility due to its impact on DNA repair processes and replication checkpoints. This study was conducted to evaluate the frequencies of three germline mutations in CHEK2 gene (c.1100delC, R145W and I157T) in breast cancers in Rwanda. Using direct DNA sequencing, we analyzed 41 breast cancer patients and 42 normal breast controls but could not detect any positives. CHEK2 mutations may be a rare event in Rwandan population and may only play a minor if an role in breast cancer predisposition among familial and sporadic cases.
Keywords: Breast cancer, CHEK2, mutations, Rwanda
Introduction
Breast cancer is the most prevalent and the second leading cause of cancer death among females worldwide. It prevails in both developed and developing countries and represents a real public health problem (Farmer et al., 2010; Brinton et al., 2014; Ferlay et al., 2015). In Rwanda, although the breast cancer is most diagnosed and treated with an estimated rate of 12.3/100,000 women, it’s still under characterized (Mody et al., 2013).
Breast cancer is a complex and heterogeneous disease associated with clinical, pathological and biological factors including age, gender, ethnicity, reproductive and hormonal factors, past history of breast cancer, exposure to ionizing radiation, environmental and lifestyle factors and, family history and genetic factors. BRCA1 and BRCA2 are the most implicated genes in hereditary breast cancer development. These 2 genes with high penetrance susceptibility play an important role in genetic predisposition to breast/ovarian cancer. Germline mutations in BRCA1/2 genes are known to be responsible of hereditary breast cancer susceptibility with the cumulative average risks of developing breast cancer by the age of 70 years of 60% (Henouda et al., 2016). During the last decades, BRCA1/2 mutations were the main molecular markers used to characterize the individual genetic risk factors and to establish the susceptibility to develop breast cancer (Zaridze, 2008). Currently, advances in genomic technology have allowed the development of breast cancer susceptibility gene panels for germline genetic testing of patients, including p53, PTEN, ATM, HRAS1, BRIP1, PALB2 and CHEK2 with low and moderate penetrance (Rinella et al., 2013).
Cell cycle checkpoint kinase 2 gene (CHEK2 or CHK2) has been identified to be involved in breast cancer susceptibility among familial breast cancer cases due to its implication in DNA repair processes and replication checkpoints (Meijers-Heijboer et al., 2002; Kriege et al., 2014). Three germline mutations in CHEK2 gene have been widely studied: 1100delC, R145W and I157T and are widely accepted to be associated with breast cancer (Desrichard et al., 2011; Elamrani et al., 2014). The 1100delC mutation is the most studied and is associated with defective reduced protein CHEK2 which lacks kinase activity (Chen et al., 2008), the missense mutations R145W and I157T, with a less penetrance than 1100delC mutation (Cybulski et al., 2011), lead to unstable mutant proteins and to the deleterious binding of CHECK2 protein to p53, BRCA1 and Cdc25A (Lee et al., 2001; Li et al., 2002; Kilpivaara et al., 2006).
This preliminary study was planned to evaluate the frequencies of the three CHEK2 mutations (c.1100delC, R145W and I157T) in a case-control study of 41 breast cancer patients and 42 normal breast controls to evaluate the implication of these mutations in predisposing Rwandan women to breast cancer.
Materials and Methods
Study population
CHEK2 mutations were screened on a cohort of 41 Rwandan breast cancer patients recruited in 2016 at Rwanda Military Hospital and King Faisal Hospital, both located at Kigali - Rwanda and 42 normal cases under 45 years old. Breast cancer cases have been chosen according to the following criteria: age at diagnosis < 45 years for sporadic cases; one or more first degree relatives with breast cancer and/or other cancer for familial cases. From each breast cancer case, clinical and pathological data were collected: in patients group, 40 were females and 1 male while in controls 39 were females and 3 males; the mean age of participants was 40 (26-60) and 25 (18-33) for patients and controls, respectively. Family history of breast cancer was reported in 13 patients. Among breast cancer cases, 35 have been diagnosed with an invasive ductal carcinoma and 6 were not specified. Among cases, 25 had the right breast affected, 15 had the left affected breast and for 1 patient the tumor side was not specified. Fresh 5 ml of peripheral blood were collected into EDTA tube and were stored at -20°C before DNA isolation. The protocol of this study was approved by the Rwanda National Ethics Committee (197/RNEC/2015) and informed consent was obtained from each participant.
CHEK2 DNA amplification
Genomic DNA was isolated from peripheral blood samples by using a commercial kit (Isolate II Genomic DNA Kit, BIOLINE) according to manufacturer’s instructions. DNA obtained was immediately used for PCR amplification or stored at −20°C until use. DNA from cases and controls was screened for 1100delC, R145W and I157T mutations. Mutations detection was done by PCR amplification and direct DNA sequencing as previously described (Kuusisto et al., 2011). Two regions of CHEK2 exon 10 were amplified for mutations detection using specific PCR primers: H4/A5 for 1100delC (Cybulski et al., 2004) and B5/D11 for R145W and I157T mutations (Friedrichsen et al., 2004).
DNA was amplified in a final volume of 25 μl containing 1.5 mM MgCl2, 200 μM of each dNTP, 50 pmol of each primer, 0.5 units Taq DNA polymerase and 50 ng genomic DNA in 1x reaction buffer. PCR primers and the amplimers’ sizes are reported in Table 1.
Table 1.
Sequences of Primers Used for CHEK2 DNA Amplification
| Mutation (s) | Primer | Sequence 5’-3’ | PCR product size |
|---|---|---|---|
| c.1100delC | H4 | TTAATTTAAGCAAAATTAAATGTC | 556 bp |
| A5 | GGCATGGTGGTGTGCATC | ||
| I157T and R145W | B5 | AAAGGTTCCATTGCCACTGT | 409 bp |
| D11 | TTGCCTTCTTAGGCTATTTTCC |
The mixture was first denatured at 95°C for 7 min. Then, thirty-five cycles of PCR were performed with denaturation at 95°C for 30 s, primer annealing for 30 s at corresponding Tm and primer extension for 1 min at 72°C. At the end of the last cycle, the mixture was incubated at 72°C for 7 min. For every set of reactions, a negative control in which DNA template was omitted from the amplification mixture is included. Amplicons were visualized after electrophoretic fractionation in 1 % agarose gel in 0.5 X TBE buffer and staining with ethidium bromide.
CHEK2 DNA sequencing
Amplicons were purified using the Exo SaP-IT clean up system (USB, USA) and were sequenced in both forward and reverse strands on an ABI 3130XL DNA analyzer (Applied Biosystems, Foster city, CA, USA), using Big Dye® Terminator v3.1 Cycle Sequencing Kit that includes dideoxynucleotides labelled with four fluorochromes of different colours (Applied Biosystems, Foster city, CA, USA). The obtained chromatograms were manually edited to ensure sequence accuracy and were compared with the wild type reference sequence of CHEK2 gene available in Genatlas database.
Results
Among the breast cancer cases, 31.7% had a cancer family history (13/41). In this cohort, family history with breast cancer prevails and was reported in 11 cases, with first and second degree family history. The other 2 cases showed a family history with cervical cancer and liver cancer. These results are in agreement with previously reported data worldwide. Indeed, it is estimated that 10%–30% of breast cancer cases are associated with familial factors, but only 5%–10% of breast cancer cases are identified to be inheritable (Friedrichsen et al., 2004; Carroll et al., 2008; Apostolou and Fostira, 2013; Economopoulou et al., 2015). Moreover, all cases with cancer family history were female and mainly diagnosed at young age (Table 2). Our results are in concordance with the observation of other investigators that the effect of family history on breast cancer risk was strongest for women under 50. Indeed, large studies conducted worldwide converge to the fact that cases with family history, particularly BRCA1/2 or check2 mutations carriers, were significantly younger, giving arise that genetic factors may play a role in affecting rates of early onset breast cancer (Pharoah, 1997; Okobia, 2006, Assi, 2013; Azim, 2014).
Table 2.
Characteristics of the Population with Cancer Family History
| BC cases | Sex | Age | Age at 1st diagnosis | Affected family members | Type of cancer |
|---|---|---|---|---|---|
| 1 | F | 48 | 48 | Mother | Breast cancer |
| 2 | F | 42 | 41 | Sister | Breast cancer |
| 3 | F | 51 | 49 | Maternal cousin | Breast cancer |
| 4 | F | 58 | 58 | Sister | Breast cancer |
| 5 | F | 26 | 25 | Maternal aunt | Breast cancer |
| 6 | F | 34 | 34 | Sister | Breast cancer |
| 7 | F | 28 | 28 | One paternal aunt and one maternal aunt | Breast cancer |
| 8 | F | 37 | 37 | Sister and maternal aunt | Breast cancer |
| 9 | F | 45 | 45 | Mother | Breast cancer |
| 10 | F | 38 | 37 | Two maternal aunts | Breast cancer |
| 11 | F | 53 | 51 | Paternal aunt | Breast cancer |
| 12 | F | 30 | 29 | Maternal aunt | Cervical cancer |
| 13 | F | 60 | 59 | Mother | Liver cancer |
The prevalence of cases with breast cancer family, with respect to limited number of cases, is higher as compared to other Sub-Saharan African countries (Awadelkarim et al., 2007; Okobia et al., 2006). This could be explained by the few number of genetic studies conducted in Sub-Saharan countries and the limited data available on the familial and hereditary history of breast cancer cases in this region (Brinton et al., 2014).
Genetic predisposition to develop breast cancer is widely studied and discussed, and mainly focused on the analysis of BRCA1 and BRCA2 genes only, considered as high-penetrance genes. Currently, it’s widely accepted that a high number of genes are eligible of testing and a well-known association with breast cancer development is well documented. These breast cancer predisposition genes, including ATM, BARD1, BRIP1, CDH1, TP53, NBN, PALB2, PTEN, STK11 and CHEK2, are considered as moderate/low-penetrance according to the breast cancer risks they present (Tung et al., 2016). However, there’s evidence that this classification is evolutive and dynamic, and can changes from moderate to high-penetrance character especially when studied in a specific population (Antoniou et al., 2014).
Discussion
The last decades have given specifically a great interest was given specifically to the association between CHEK2 gene and breast cancer development and mutational status of CHEK2 was assessed in many populations around the world (Cybulski et al., 2011). In African countries, and to the best of our knowledge, mutational status of CHEK2 gene in breast cancer was evaluated only in Morocco (Elamrani et al., 2014; Marouf et al., 2015), Tunisia (Riahi et al., 2017) and South Africa (Francies et al., 2015). There are no related studies in Rwanda or any other country in Sub-Saharan Africa. Therefore, we have planned to conduct this case–control study to assess the mutational status of CHEK2 in Rwandese population and the interest was focused on three CHEK2 variants that are known to affect protein function (c.1100delC, R145W and I157T).
Both cancer cases and controls were successfully amplified and sequenced. Figure I illustrates examples of obtained electropherograms. Results clearly showed that 1100delC, R145W and I157T germilne mutations are absent in both breast cancer cases, with and without family history, and in controls. Our results are in agreement with previously reported data in many countries, including Morocco (Elamrani et al., 2014; Marouf et al., 2015) and Tunisia (Riahi et al., 2017).
Figure 1.
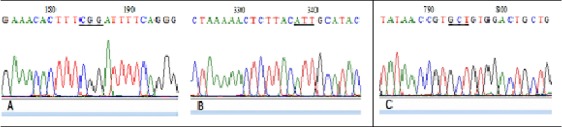
The Sequence Results of CHEK2. Sequence electropherogram showing the position (underlined) of the screened mutations in CHEK2 gene. DNA sequences blasted in Genatlas database has shown the absence of R145W mutation (CGG to TGG) (A), the absence of I157T (ATT to ACT) (B) and the absence of 1100delC (deletion of C in GCT) (C).
It’s widely accepted that the spectrum of mutations in the CHEK2 gene varies between populations, and some of them exhibit high frequencies and may contribute to differences in cancer risk between populations and allow to genetically stratifying the population (Leedom et al., 2016).
Several previously published studies reported an elevated frequency of the CHEK2 1100delC variant in specific populations. In USA, 1100delC mutation was reported in 1.2% for cases and 0.4% of matched (Friedrichsen et al., 2004). Similar results were reported in UK and The Netherlands; 1.3% and 2.5% for cases and 0.3% and 1.2% for controls, respectively (Meijers-Heijboer et al., 2002). 1100delC truncating mutation was also found in Europe and the highest frequency has been found in patients from the North and the West of Europe, as compared to the southern countries exhibiting the lowest frequencies (Italy and Spain) (Caligo et al., 2004; Osorio et al., 2004). However, to our knowledge, this mutation was not reported in Asia. Of particular interest, 1100delC was reported twice in South Africa and was detected in white women coming certainly from Europe or North America (Francies et al., 2015). Our findings confirm and consolidate the hypothesis of the 1100delC frequency gradient from Northern and Western populations to the Mediterranean and southern populations (Martínez-Bouzas et al., 2007; Elamrani et al., 2014). This genetic specification has been already described in other known diseases, such as the delta F508 cystic fibrosis mutation, from European populations to the Mediterranean populations (Estivill et al., 1997). This potential gradient may be caused by a common founder mutation in North-West European and North American populations (Caligo et al., 2004).
In our study, CHEK2 I157T and R145W, affecting respectively the kinase activity of the CHEK2 protein and its binding to BRCA and p53, were also absent in Rwandan samples, both cases and controls. These two missense mutations were not detected in many populations around the world. In USA, Friedrichsen et al., (2004) have reported no I157T and R145W CHEK2 mutations, suggesting the absence of correlation between the R145W and I157T CHEK2 variants and breast cancer risk. The same results were reported in African countries including Morocco (Elamrani et al., 2014; Marouf et al., 2015), Tunisia (Riahi et al., 2017) and South Africa (Francies et al., 2015).
In a multi-population study, Schutte et al., (2003) have found that I157T was absent in patients and controls from the United Kingdom and The Nederland but present in 2 cases and 1 control from the United States of America, whereas R145W mutations was absent in all specimens. However, I157T was identified in 22/996 cases (2.2%) vs. 3/486 controls (0.6%) in the German population and in 24/424 cases (5.7%) vs. 4/307 controls (1.3%) in the Byelorussian cohorts, suggesting an ethnically specification of these variant and the moderate associated risk for developing breast cancer.
The 11 breast cancer cases with familial history from Rwanda don’t harbor any point mutation in CHEK2 associated with breast cancer development and could be a good candidate for exploring genetic predisposition by analyzing BRCA1 and BRAC2 mutations.
In conclusion, the absence of CHEK2 variants in our cases study highlights that 1100delC, R145W and I175T CHEK2 mutations are rare events suggesting a no correlation between these germline mutations and breast cancer risk in Rwanda. Thus, for breast cancer practical clinics and early diagnosis, the use of CHEK2 germline mutations as breast cancer susceptibility biomarker should not be recommended for routine use in Rwanda.
Conflict of interest
The authors declare no conflict of interest.
Acknowledgements
We thank the Rwanda Military Hospital and King Faical Hospital staff for collaboration and the staff of Unité de Biologie et Recherche Médicale, Centre National de l’Energie, des Sciences et des Techniques Nucléaires (CNESTEN) for technical assistance. Special thanks to study participants.
References
- Antoniou AC, Casadei S, Heikkinen T, et al. Breast-cancer risk in families with mutations in PALB2. N Engl J Med. 2014;371:497–506. doi: 10.1056/NEJMoa1400382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apostolou P, Fostira F. Hereditary breast cancer:the era of new susceptibility genes. Bio Med Res Int. 2013;2013:747318. doi: 10.1155/2013/747318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assi HA, Khoury KE, Dbouk H, et al. Epidemiology and prognosis of breast cancer in young women. J Thorac Dis. 2013;5:2–8. doi: 10.3978/j.issn.2072-1439.2013.05.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azim HA, Partridge AH. Biology of breast cancer in young women. Breast Cancer Res. 2014;16:427–35. doi: 10.1186/s13058-014-0427-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awadelkarim KD, Aceto G, Veschi S, et al. BRCA1 and BRCA2 status in a central Sudanese series of breast cancer patients:Interactions with genetic, ethnic and reproductive factors. Breast Cancer Res Treat. 2007;102:189–99. doi: 10.1007/s10549-006-9303-z. [DOI] [PubMed] [Google Scholar]
- Brinton LA, Figueroa JD, Awuah B, et al. Breast cancer in sub-saharan Africa:opportunities for prevention. Breast Cancer Res Treat. 2014;144:467–78. doi: 10.1007/s10549-014-2868-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caligo MA, Agata S, Aceto G, et al. The CHEK2 c.1100delC mutation plays an irrelevant role in breast cancer predisposition in Italy. Hum Mutat. 2004;24:100–1. doi: 10.1002/humu.20051. [DOI] [PubMed] [Google Scholar]
- Carroll JC, Cremin C, Allanson J, et al. Hereditary breast and ovarian cancers. Can Fam Physician. 2008;54:1691–2. [PMC free article] [PubMed] [Google Scholar]
- Chen W, Yurong S, Liansheng N. Breast cancer low-penetrance allele 1100delC in the CHEK2 gene:not present in the Chinese familial. Adv Therapy. 2008;25:496–501. doi: 10.1007/s12325-008-0057-3. [DOI] [PubMed] [Google Scholar]
- Cybulski C, Górski B, Huzarski T, et al. CHEK2 is a multiorgan cancer susceptibility gene. Am J Hum Genet. 2004;75:1131–5. doi: 10.1086/426403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cybulski C, Wokołorczyk D, Jakubowska A, et al. Risk of breast cancer in women with a CHEK2 mutation with and without a family history of breast cancer. J Clin Oncol. 2011;29:3747–52. doi: 10.1200/JCO.2010.34.0778. [DOI] [PubMed] [Google Scholar]
- Desrichard A, Bidet Y, Uhrhammer N, Bignon YJ. CHEK2 contribution to hereditary breast cancer in non-BRCA families. Breast Cancer Res. 2011;13:R119. doi: 10.1186/bcr3062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Economopoulou P, Dimitriadis G, Psyrri A. Beyond BRCA:New hereditary breast cancer susceptibility genes. Cancer Treat Rev. 2015;41:1–8. doi: 10.1016/j.ctrv.2014.10.008. [DOI] [PubMed] [Google Scholar]
- Elamrani A, Moumad K, Attaleb M, et al. Absence of CHEK2 1100delC, R145W and I157T mutations in breast cancer in a Moroccan population. Breast Cancer Res Treat. 2014;2:6–9. [Google Scholar]
- Estivill X, Bancclls C, Ramos C, the Biomed CF Mutation Analysis Consortium. Geographic distribution and regional origin of 272 cystic fibrosis mutations in European populations. Hum Mutat. 1997;10:135–54. doi: 10.1002/(SICI)1098-1004(1997)10:2<135::AID-HUMU6>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- Farmer P, Frenk J, Knaul FM, et al. Expansion of cancer care and control in countries of low and middle income:a call to action. Lancet. 2010;376:1186–93. doi: 10.1016/S0140-6736(10)61152-X. [DOI] [PubMed] [Google Scholar]
- Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide:sources, methods and major patterns in Globocan 2012. Int J Cancer. 2015;136:359–86. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- Francies FZ, Wainstein T, De Leeneer K, et al. BRCA1, BRCA2 and PALB2 mutations and CHEK2 c.1100delC in different South African ethnic groups diagnosed with premenopausal and/or triple negative breast cancer. BMC Cancer. 2015;15:912. doi: 10.1186/s12885-015-1913-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrichsen DM, Malone KE, Doody DR, Daling JR, Ostrander EA. Frequency of CHEK2 mutations in a population based, case-control study of breast cancer in young women. Breast Cancer Res. 2004;6:629–35. doi: 10.1186/bcr933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henouda S, Bensalem A, Reggad R, et al. Contribution of BRCA1 and BRCA2 germline mutations to early algerian breast cancer. Disease Markers. 2016;2016:7869095. doi: 10.1155/2016/7869095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilpivaara O, Alhopuro P, Vahteristo P, Aaltonen LA, Nevanlinna H. CHEK2 I157T associates with familial and sporadic colorectal cancer. J Med Gene. 2006;43:34. doi: 10.1136/jmg.2005.038331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriege M, Hollestelle A, Jager A, et al. Survival and contralateral breast cancer in CHEK2 1100delC breast cancer patients:impact of adjuvant chemotherapy. Br J Cancer. 2014;111:1004–13. doi: 10.1038/bjc.2014.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuusisto KM, Bebel A, Vihinen M, Schleutker J, Sallinen SL. Screening for BRCA1, BRCA2, CHEK2, PALB2, BRIP1, RAD50, and CDH1 mutations in high-risk Finnish BRCA1/2-founder mutation-negative breast and/or ovarian cancer individuals. Breast Cancer Res. 2011;13:20. doi: 10.1186/bcr2832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SB, Kim SH, Bell DW, et al. Advances in brief destabilization of CHK2 by a missense mutation associated with. In Vitro. 2001;60:8062–7. [PubMed] [Google Scholar]
- Leedom TP, LaDuca H, McFarland R, et al. Breast cancer risk is similar for CHEK2 founder and non-founder mutation carriers. Cancer Genet. 2016;209:403–7. doi: 10.1016/j.cancergen.2016.08.005. [DOI] [PubMed] [Google Scholar]
- Li J, Williams BL, Haire LF, et al. Structural and functional versatility of the FHA domain in DNA-damage signaling by the tumor suppressor kinase Chk2. Mol Cell. 2002;9:1045–54. doi: 10.1016/s1097-2765(02)00527-0. [DOI] [PubMed] [Google Scholar]
- Marouf C, Hajji O, Diakité B, et al. The CHEK2 1100delC allelic variant is not present in familial and sporadic breast cancer cases from Moroccan population. Springerplus. 2015;4:38. doi: 10.1186/s40064-014-0778-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Bouzas C, Beristain E, Guerra I, et al. CHEK2 1100delC is present in familial breast cancer cases of the Basque Country [2] Breast Cancer Res and Treat. 2007;103:111–3. doi: 10.1007/s10549-006-9351-4. [DOI] [PubMed] [Google Scholar]
- Meijers-Heijboer H, van den Ouweland A, Klijn J, et al. Low-penetrance susceptibility to breast cancer due to CHEK2 * 1100delC in noncarriers of BRCA1 or BRCA2 mutations. Nat Genet. 2002;31:55–9. doi: 10.1038/ng879. [DOI] [PubMed] [Google Scholar]
- Mody GN, Nduaguba A, Ntirenganya F, Riviello R. Characteristics and presentation of patients with breast cancer in Rwanda. Am J Surg. 2013;205:409–13. doi: 10.1016/j.amjsurg.2013.01.002. [DOI] [PubMed] [Google Scholar]
- Okobia M, Bunker C, Zmuda J, et al. Case-control study of risk factors for breast cancer in Nigerian women. Int J Cancer. 2006;119:2179–85. doi: 10.1002/ijc.22102. [DOI] [PubMed] [Google Scholar]
- Osorio A, Rodríguez-López R, Díez O, et al. The breast cancer low-penetrance allele 1100delC in the CHEK2 gene is not present in Spanish familial breast cancer population. Int J Cancer. 2004;108:54–6. doi: 10.1002/ijc.11414. [DOI] [PubMed] [Google Scholar]
- Pharoah PD, Day NE, Duffy S, et al. Family history and the risk of breast cancer:a systematic review and meta-analysis. Int J Cancer. 1997;71:800–9. doi: 10.1002/(sici)1097-0215(19970529)71:5<800::aid-ijc18>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- Riahi A, Chabouni-Bouhamed H, Kharrat M. Prevalence of BRCA1 and BRCA2 large genomic rearrangements in Tunisian high risk breast/ovarian cancer families:Implications for genetic testing. Cancer Genet. 2017;210:22–7. doi: 10.1016/j.cancergen.2016.11.002. [DOI] [PubMed] [Google Scholar]
- Rinella ES, Shao Y, Yackowski L, et al. Genetic variants associated with breast cancer risk for Ashkenazi Jewish women with strong family histories but no identifiable BRCA1/2 mutation. Hum Genet. 2013;132:523–36. doi: 10.1007/s00439-013-1269-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schutte M, Seal S, Barfoot R, et al. Variants in CHEK2 other than 1100delC do not make a major contribution to breast cancer susceptibility. Am J Hum Genet. 2003;72:1023–8. doi: 10.1086/373965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tung N, Lin NU, Kidd J, et al. Frequency of germline mutations in 25 cancer susceptibility genes in a sequential series of patients with breast cancer. J Clin Oncol. 2016;34:1460–8. doi: 10.1200/JCO.2015.65.0747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaridze DG. Molecular epidemiology of cancer. Biochemistry (Moscow) 2008;73:532–42. doi: 10.1134/s0006297908050064. [DOI] [PubMed] [Google Scholar]


