ABSTRACT
In Escherichia coli, many recombinant proteins are produced in the periplasm. To direct these proteins to this compartment, they are equipped with an N-terminal signal sequence so that they can traverse the cytoplasmic membrane via the protein-conducting Sec translocon. Recently, using the single-chain variable antibody fragment BL1, we have shown that harmonizing the target gene expression intensity with the Sec translocon capacity can be used to improve the production yields of a recombinant protein in the periplasm. Here, we have studied the consequences of improving the production of BL1 in the periplasm by using a proteomics approach. When the target gene expression intensity is not harmonized with the Sec translocon capacity, the impaired translocation of secretory proteins, protein misfolding/aggregation in the cytoplasm, and an inefficient energy metabolism result in poor growth and low protein production yields. The harmonization of the target gene expression intensity with the Sec translocon capacity results in normal growth, enhanced protein production yields, and, surprisingly, a composition of the proteome that is—besides the produced target—the same as that of cells with an empty expression vector. Thus, the single-chain variable antibody fragment BL1 can be efficiently produced in the periplasm without causing any notable detrimental effects to the production host. Finally, we show that under the optimized conditions, a small fraction of the target protein is released into the extracellular milieu via outer membrane vesicles. We envisage that our observations can be used to design strategies to further improve the production of secretory recombinant proteins in E. coli.
IMPORTANCE The bacterium Escherichia coli is widely used to produce recombinant proteins. Usually, trial-and-error-based screening approaches are used to identify conditions that lead to high recombinant protein production yields. Here, for the production of an antibody fragment in the periplasm of E. coli, we show that an optimization of its production is accompanied by the alleviation of stress. This indicates that the monitoring of stress responses could be used to facilitate enhanced recombinant protein production yields.
KEYWORDS: Escherichia coli, recombinant protein production, periplasm, Sec translocon, proteomics
INTRODUCTION
The bacterium Escherichia coli is widely used to produce recombinant proteins (1, 2). In the oxidizing environment of the periplasm of E. coli, the disulfide bond formation (Dsb) system catalyzes the formation of disulfide bonds (3). Therefore, disulfide bond-containing recombinant proteins, such as antibody fragments and many peptide hormones, are often produced in the periplasm to enable folding into their native conformation (4, 5). In addition, it is easier to isolate proteins from the periplasm than from whole-cell lysates (6).
To reach the periplasm, recombinant proteins are equipped with an N-terminal signal sequence that guides them to the Sec translocon, which is the major protein-conducting channel in the cytoplasmic membrane assisting the biogenesis of both membrane and secretory proteins (7). Secretory proteins are translocated in a mostly unfolded state through the Sec translocon (8). Two pathways can guide secretory proteins to the Sec translocon, the posttranslational SecB-targeting pathway and the cotranslational signal recognition particle (SRP)-targeting pathway (9). The nature of the signal sequence is decisive for the choice of the targeting pathway (10–12). The DsbA signal sequence, which directs secretory proteins to the Sec translocon in an SRP-dependent fashion, is widely used for the production of recombinant secretory proteins (13–15). The idea behind the use of a signal sequence promoting cotranslational targeting to produce a recombinant protein in the periplasm is that, given the Sec translocon capacity is sufficient, it avoids the retention of the target in the cytoplasm due to (mis)folding. Upon translocation of the target across the cytoplasmic membrane, the signal sequence is clipped off by a leader peptidase (7, 16). In the periplasm, in addition to the Dsb system, various catalysts can assist the folding process (17).
When attempting to obtain high yields of a recombinant protein, the goal is usually set that the gene encoding the recombinant protein is expressed at the highest level possible. The rationale behind this approach is that the more mRNA that is synthesized, the more recombinant protein that will be produced. With this rationale in mind, the BL21(DE3) strain was engineered, which is now one of the E. coli strains most widely used to produce recombinant proteins (2). In strain BL21(DE3), the expression of the gene encoding the target protein is driven by the bacteriophage T7 RNA polymerase (RNAP), which transcribes eight times faster than E. coli RNAP (18–20). T7 RNAP specifically recognizes the T7 promoter, which drives the expression of the target gene from a plasmid (18, 20). The gene encoding the T7 RNAP is under the control of the not-well-titratable isopropyl-β-d-1-thiogalactopyranoside (IPTG)-inducible lacUV5 promoter, which is a strong variant of the wild-type lac promoter (21, 22). Recently, we have shown for membrane and secretory proteins that an adjustment of the expression level of the target gene is in most instances critical to enhance protein production yields (23–25). Adjusting the expression level of the target gene was done using the Lemo setup (25). The Lemo setup is based on the pLemo plasmid, which contains the gene encoding the T7 lysozyme under the control of the titratable l-rhamnose promoter (25, 26). The T7 lysozyme is a natural inhibitor of the T7 RNAP (27, 28). Thus, the Lemo setup enables us to precisely modulate the activity of the T7 RNAP and consequently target the gene expression level by simply adding different amounts of l-rhamnose to the culture medium.
The consequences of the production of membrane proteins under nonoptimized conditions in E. coli BL21(DE3) have been studied in quite some detail (29). Membrane protein production under nonoptimized conditions leads to saturation of the Sec translocon capacity (24, 25, 29). The saturation of the Sec translocon capacity leads to, e.g., decreased accumulation levels of many secretory and membrane proteins, the accumulation of precursors of secretory proteins in the cytoplasm, the induction of the heat shock or σ32 response due to the aggregation/misfolding of proteins in the cytoplasm, the inefficient production of ATP due to decreased accumulation levels of enzymes of the tricarboxylic acid (TCA) cycle, and the production of acetate due to an increased capacity of the Pta pathway (25, 29). Membrane protein production under nonoptimized conditions eventually leads to the accumulation of mutations that lower or abolish target gene expression (30–32). The consequences of optimizing the production of membrane proteins in E. coli have been studied using the BL21(DE3)-derived C41(DE3) and C43(DE3) strains rather than the aforementioned Lemo setup (25, 33). The C41(DE3) and C43(DE3) strains have accumulated mutations that strongly reduce the expression intensity of t7rnap, which for most membrane proteins, leads to higher production yields and less, but still notable, stress (25, 31, 34). This is in line with the assumption that recombinant protein production affects the physiology of the cell per se (35). The consequences of secretory protein production under both nonoptimized and optimized conditions have hardly been studied.
Here, to study the effects of optimizing the production of a recombinant protein in the E. coli periplasm, the proteomes of E. coli cells producing a model single-chain variable antibody fragment under nonoptimized and Lemo setup-based optimized conditions were characterized.
RESULTS
Optimizing the production of the scFv BL1 in the E. coli periplasm using the Lemo setup.
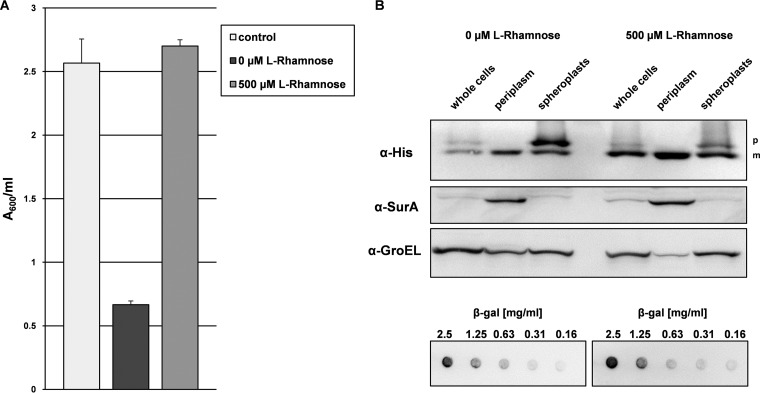
Recently, we have shown that the production of the model single-chain variable antibody fragment (scFv) BL1 in the periplasm of E. coli can be readily optimized by modulating its gene expression level using the Lemo setup (23). The DsbA signal sequence was used to target the scFv BL1 via the SRP-targeting pathway to the Sec translocon in the cytoplasmic membrane (14). We showed that target protein production is lowest, with a considerable part of the protein not secreted, at an l-rhamnose concentration of 0 μM, which is comparable to using cells without pLemo, and highest, with most of the target secreted, at an l-rhamnose concentration of 500 μM. Enhancing the production of the scFv BL1 in the periplasm using the Lemo setup results not only in the formation of more biomass but also in considerably more secreted target protein per unit of biomass (A600) (Fig. 1A and B, top, left lanes). As a control, cells with an empty expression vector were cultured at an l-rhamnose concentration of 500 μM (Fig. 1A). The A600 values of control cells and cells producing the scFv BL1 at 500 μM l-rhamnose show that biomass formation is actually not affected when the target protein is produced at the optimized condition (Fig. 1A).
FIG 1.
Periplasmic production of the scFv BL1 under nonoptimized and optimized conditions. Production of the scFv BL1 was induced with IPTG in Tuner(DE3) cells harboring pLemo in the absence and presence of 500 μM l-rhamnose. Tuner(DE3) cells harboring pLemo and an empty expression vector cultured in the presence of 500 μM l-rhamnose were used as a control. (A) Four hours after induction, biomass formation was monitored by measuring A600. (B, top) Levels of the precursor form (p) and the mature form (m) of the scFv BL1 were monitored using SDS-PAGE followed by immunoblotting in whole cells cultured in the absence and presence of 500 μM l-rhamnose using an α-His antibody recognizing the C-terminal His tag of the scFv BL1. Cells were converted into spheroplasts and the periplasmic fractions were isolated. Levels of the scFv BL1 were also monitored in the periplasmic fractions and spheroplasts. The periplasmic protein SurA and the cytoplasmic protein GroEL were used as markers to monitor the spheroplasting efficiency and the isolation of the periplasmic fractions. (B, bottom) PVDF membranes containing decreasing amounts of β-galactosidase were incubated with whole-cell lysate. Binding of the scFv BL1 present in whole-cell lysates to the β-galactosidase was detected using an α-His antibody recognizing the C-terminal His tag of the scFv BL1.
Converting cells producing the target protein at l-rhamnose concentrations of 0 and 500 μM l-rhamnose into spheroplasts and the subsequent isolation of the periplasmic fractions corroborated that cells cultured under optimized conditions produce considerably more secreted scFv BL1 (Fig. 1B, middle and right lanes).
Previously, using the Sec translocon-dependent secretory proteins MalE and OmpA and the cytoplasmic inclusion body-associated protein B (IbpB) as markers, it was shown by means of immunoblotting that nonoptimized scFv BL1 production hampers the secretion of secretory proteins via the Sec translocon and leads to protein aggregate formation in the cytoplasm (23). This indicates that the Sec translocon capacity is saturated under these conditions (23). However, under the optimized production condition, there was no indication that the Sec translocon capacity was saturated and any protein aggregate formation occurred (23). Since it is generally assumed that recombinant protein production affects the physiology of the cell per se and that monitoring the accumulation levels of only a few proteins gives a very limited insight into the physiology of the cell, and as a consequence, into the effects of the Lemo-based optimization process, we decided to characterize the consequences of optimizing the production of the scFv BL1 using a proteomics approach.
Consequences of scFv BL1 production under nonoptimized conditions.
To study the consequences of the production of the scFv BL1 under nonoptimized conditions, cells producing the scFv BL1 in the absence of any target gene regulation, i.e., without any l-rhamnose, were isolated by centrifugation and subsequently lysed. Proteins in the cell lysate were precipitated using acetone and subsequently solubilized and digested in-solution. The digested material was analyzed using label-free mass spectrometry (see Materials and Methods for a detailed description of the experimental setup used). As a reference, digested proteins from cells with an empty expression vector were used. We could identify and quantify 1,111 proteins in each sample, and the accumulation level of a protein was considered to be changed if the false-discovery-rate (q)-corrected P value was lower than 0.05. The accumulation levels of 778 of the 1,111 identified proteins were affected by the production of the scFv BL1 under nonoptimized conditions. In Table S1 in the supplemental material, all the quantified proteins are listed. In addition, immunoblotting was used to corroborate some of the mass spectrometry-based data as well as to cover some proteins that were not identified using mass spectrometry.
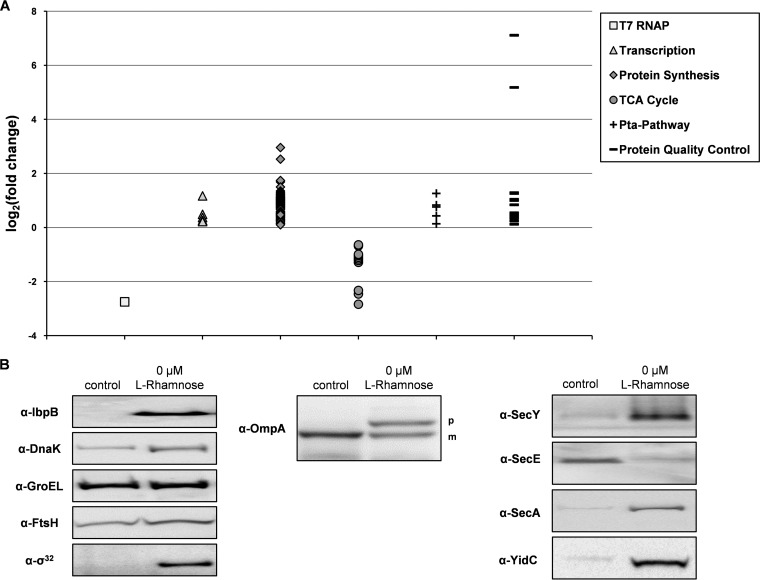
In the cells producing the scFv BL1 under nonoptimized conditions, the accumulation level of the T7 RNAP, which drives target gene expression, was considerably lower than in the control (Fig. 2A; Table S2). The accumulation levels of many proteins involved in the synthesis of proteins were increased (Fig. 2A; Table S2). The accumulation levels of all enzymes involved in the TCA cycle were decreased. In contrast, the levels of enzymes that are part of the Pta pathway were increased (Fig. 2A; Table S2).
FIG 2.
Consequences of the nonoptimized production of the scFv BL1. Production of the scFv BL1 was induced with IPTG in Tuner(DE3) cells harboring pLemo in the absence of l-rhamnose. Tuner(DE3) cells harboring pLemo and an empty expression vector were used as a reference. The proteomes of aforementioned cells were analyzed using a combination of label-free mass spectrometry and immunoblotting. (A) Mass spectrometry was used to determine fold changes in the abundance of T7 RNAP and proteins involved in transcription, protein synthesis, the TCA cycle, the Pta pathway, and protein quality control. Fold changes are plotted as log2 (fold change) on the y axis (see Table S2 in the supplemental material). (B, left) Accumulation levels of the σ32-regulated proteins IbpB, DnaK, GroEL, and FtsH along with σ32 itself were monitored by immunoblotting. (B, middle) The presence of the precursor form (p) and mature form (m) of OmpA was probed by immunoblotting. (B, right) The accumulation levels of the Sec translocon components SecY, SecE, SecA, and YidC were monitored by immunoblotting as well.
The accumulation levels of many σ32-regulated chaperones and proteases, e.g., DnaK, GroEL, IbpB, and FtsH, were increased (Fig. 2A and B, left; Table S2). Furthermore, the accumulation levels of many secretory proteins, e.g., OmpA, were decreased (Fig. 2A and B, middle; see Tables S3 and S4). For two secretory proteins, the scFv BL1 and OmpA, it could be shown that their precursors accumulate in the cytoplasm (Fig. 1B and 2B, middle). The effects on the accumulation levels of cytoplasmic membrane proteins were less dramatic; many were actually unaffected or even increased (Tables S3 and S4).
Previously, we proposed that the accumulation of precursors of secretory proteins in the cytoplasm under nonoptimized scFv BL1 production conditions is due to the saturation of the Sec translocon capacity (23). Therefore, we specifically monitored the accumulation levels of the core Sec translocon constituents, SecY, SecE, and SecA, and the auxiliary Sec translocon component YidC (7) (Fig. 2B, right). Notably, YidC assists the biogenesis of membrane proteins in conjunction with the Sec translocon but can also function as an independent entity assisting the biogenesis of membrane proteins (36–38). SecY and SecE are both integral membrane proteins and form a functional entity in a 1-to-1 stoichiometry. SecY levels were increased, whereas SecE levels were decreased. SecA is the peripheral ATP-dependent motor of the Sec translocon and its levels were increased (39, 40). Finally, the accumulation level of YidC was increased. Unfortunately, it was not possible to separate the cytoplasmic membrane fraction of cells producing the scFv BL1 under nonoptimized conditions from the outer membrane fraction and protein aggregates using sucrose density gradient centrifugation, so that the observations based on whole-cell lysates could not be verified at the membrane level (data not shown).
Altogether, the production of the scFv BL1 under nonoptimized conditions has a major impact on the composition of the proteome of the producing cells. Next, we analyzed the proteome composition of cells producing the scFv BL1 under Lemo setup-based optimized conditions.
Consequences of optimizing the production of the scFv BL1 using the Lemo setup.
Recently, we have shown that the E. coli-based production of the scFv BL1 can be optimized using the Lemo setup (23). As shown in Fig. 1, the addition of 500 μM l-rhamnose to cells producing the scFv BL1 leads to improved production yields in the periplasm per unit of biomass, and in stark contrast to the nonoptimized production of the scFv BL1, there is no notable effect on biomass formation.
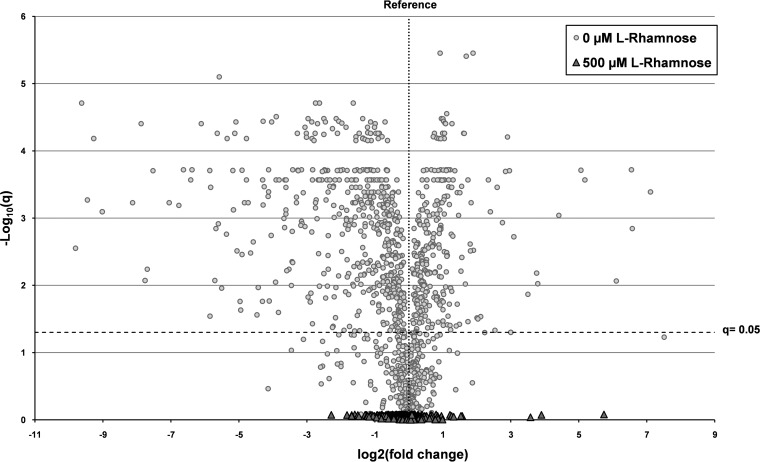
Here, we analyzed the proteome of cells producing the scFv BL1 under optimized conditions using label-free mass spectrometry (see Material and Methods). As a reference, the proteome of cells harboring an empty expression vector was used. We quantified 1,111 proteins and to our surprise, there were no significant differences in the proteome composition of the cells producing the scFv BL1 under Lemo-based optimized conditions and that of the cells with an empty expression vector that was used as a reference (Fig. 3; see Table S5). To put this observation in perspective, we included in the same figure a comparison of the proteome of cells producing the scFv BL1 under nonoptimized conditions with that of cells harboring an empty expression vector (Fig. 3).
FIG 3.
Consequences of optimizing the production of the scFv BL1 using the Lemo setup. The scFv BL1 was produced in Tuner(DE3) cells harboring pLemo cultured in the presence of 500 μM l-rhamnose and the absence of l-rhamnose (see Fig. 1). Tuner(DE3) cells harboring pLemo and an empty expression vector cultured in the presence of 500 μM l-rhamnose were used as a reference (see Fig. 1). The proteome composition of cells was analyzed using label-free mass spectrometry. The q values, plotted as −log10 on the y axis, are plotted against the relative fold changes in protein abundance, plotted as log2 on the x axis (see Table S5 in the supplemental material). The horizontal dashed line indicates the significance threshold; q values below the line represent changes that are not considered to be significant, whereas q values above the line represent changes that are considered to be significant.  , proteins in cells producing the scFv BL1 under optimized conditions;
, proteins in cells producing the scFv BL1 under optimized conditions;  , proteins in cells producing the scFv BL1 under nonoptimized conditions.
, proteins in cells producing the scFv BL1 under nonoptimized conditions.
Altogether, the Lemo-based optimization of the production of the scFv BL1 in E. coli results in unimpaired growth, strongly enhanced protein production levels, and—besides the produced scFv BL1—no notable effects on the proteome composition of the cells producing the protein.
Outer membrane vesicle-mediated release of the scFv BL1 into the extracellular medium.
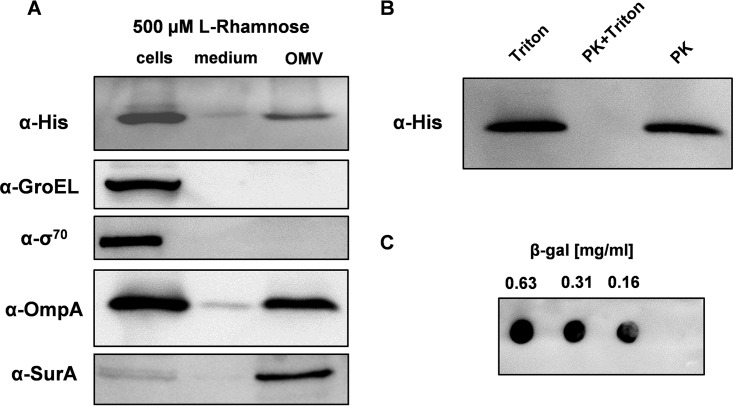
It has been reported that proteins produced in the periplasm of E. coli often end up in the extracellular medium (6, 41–43). Therefore, we monitored if the scFv BL1 can be detected in the culture medium of cells producing the protein. The scFv BL1 could only be detected in the culture medium of cells producing the scFv BL1 under optimized conditions (Fig. 4A). Two cytoplasmic markers, the chaperone GroEL and the sigma factor σ70, could not be detected in the culture medium, indicating that no notable cell lysis had occurred (Fig. 4A). In contrast, the outer membrane protein OmpA could be detected in the culture medium (Fig. 4A). These observations suggested that the processed form of the scFv BL1 in the culture medium was somehow associated with outer membrane vesicles (OMVs). Indeed, when OMVs were isolated, the scFv BL1 and OmpA signals were strongly enriched and also the periplasmic marker SurA could now be readily detected, whereas the cytoplasmic markers GroEL and σ70 still could not be detected.
FIG 4.
Release of the scFv BL1 in the extracellular medium. The scFv BL1 was produced in Tuner(DE3) cells harboring pLemo cultured in the presence of 500 μM l-rhamnose. The culture medium was cleared from whole cells by centrifugation followed by filtration. The OMVs in the spent medium were subsequently isolated by ultracentrifugation. (A) Levels of the scFv BL1, the cytoplasmic markers GroEL and σ70, the outer membrane marker OmpA, and the periplasmic marker SurA were monitored in whole cells, the spent medium, and isolated OMVs using SDS-PAGE followed by immunoblotting. The scFv BL1 was detected using an α-His antibody recognizing the C-terminal His tag of the scFv BL1. (B) The localization of the scFv BL1 associated with OMVs was examined using a proteinase K protection assay. OMVs were treated with either Triton X-100 (left lane), Triton X-100 and proteinase K (middle lane), or proteinase K only (right lane). Samples were analyzed using SDS-PAGE followed by immunoblotting and the scFv BL1 was detected using an α-His antibody recognizing the C-terminal His tag of the scFv BL1. (C) The proper folding of the scFv BL1 was assessed using a dot blot assay. Decreasing concentrations of β-galactosidase were spotted onto a PVDF membrane and incubated with Triton X-100-lysed OMVs. The scFv BL1 bound to the β-galactosidase was detected using an α-His antibody recognizing the C-terminal His tag of the scFv BL1.
The size of the OMVs isolated from the culture medium of cells producing the scFv BL1 under optimized conditions was monitored using dynamic light scattering (44). Almost all OMVs had a diameter of around 30 nm, which is in keeping with the diameter reported in previous reports (45, 46). Subsequently, to monitor how the scFv BL1 was associated with the OMVs, they were treated with the detergent Triton X-100 to solubilize the OMV membrane, the detergent Triton X-100 and proteinase K, and proteinase K alone (Fig. 4B). Subsequently, the scFv BL1 present in the three different samples was detected by means of immunoblotting. Only when OMVs were treated with both Triton X-100 and proteinase K was it not possible to detect the scFv BL1. This indicates that the scFv BL1 resides in the lumen of the OMVs. By using Triton X-100-solubilized OMVs, it was shown in a dot blot assay that the scFv BL1 could bind to its substrate, β-galactosidase (Fig. 4C).
Altogether, the use of Lemo-based optimized production conditions for the scFv BL1 resulted in the release of a small fraction of the target protein into the extracellular medium via OMVs.
DISCUSSION
In E. coli, many recombinant proteins, in particular, ones containing disulfide bonds, are produced in the periplasm (4–6). Recently, by using the scFv BL1 as a model secretory recombinant protein, we have shown that being able to precisely tune the expression intensity of the gene encoding the target protein can be instrumental for optimizing the production yields of a protein in the periplasm (23). So far, the consequences of the production of proteins in the periplasm and the optimization of this process have been hardly studied. Therefore, in this study, we characterized the proteomes of cells producing the scFv BL1 under nonoptimized and optimized conditions (23).
We found that the accumulation levels of T7 RNAP, which drives the expression of the gene encoding DsbA BL1, are considerably lower in cells producing the scFv BL1 under nonoptimized protein production conditions than in the control cells harboring an empty expression vector. Recently, it has been shown that E. coli BL21(DE3) rapidly accumulates mutations weakening or abolishing the expression of t7rnap or the activity of T7 RNAP in response to the stress caused by nonoptimized recombinant protein production (30–32, 34). Thus, the decreased T7 RNAP accumulation levels indicate that cells producing the scFv BL1 under nonoptimized conditions are also accumulating the aforementioned mutations.
The decreased accumulation levels of the enzymes involved in the TCA cycle and the increased levels of enzymes of the Pta pathway indicate that cells producing the scFv BL1 under nonoptimized conditions synthesize ATP inefficiently and produce acetate, which is toxic (47–49). These consequences of the production of recombinant proteins, including membrane proteins, have been described before and appear to be generic (29). Also, the induction of the σ32 response due to protein misfolding/aggregation in the cytoplasm has been described before for the production of recombinant proteins and is in keeping with the accumulation of precursors of secretory proteins upon scFv BL1 production under nonoptimized conditions (23).
Previously, we proposed that the production of the scFv BL1 under nonoptimized conditions leads to the saturation of Sec translocon capacity, similar as to what happens if a membrane protein is produced under nonoptimized conditions (23). Surprisingly, under nonoptimized conditions, the accumulation levels of the core Sec translocon components SecY and SecA are increased, whereas the accumulation level of the other core Sec translocon component, SecE, is decreased. The nonoptimized production of a membrane protein does not affect SecY and SecE levels, but only SecA levels are increased (29). Since SecY and SecE form a complex in a 1-to-1 stoichiometry that constitutes the core of the protein-conducting channel of the Sec translocon and SecY cannot function properly without SecE (50–52), our observations indicate that there are actually fewer functional Sec translocons available under the nonoptimized scFv BL1 production condition. It has been shown that cells experiencing Sec translocon capacity problems respond by synthesizing more SecA in an attempt to alleviate the protein translocation stress (53). This explains the increased SecA levels under nonoptimized conditions. We do not have a ready explanation for the increased SecY levels and the decreased SecE levels. As mentioned before, the nonoptimized production of membrane proteins does not affect SecY and SecE accumulation levels (29). This suggests that the nature of the saturation of the Sec translocon capacity caused by the nonoptimized production of a membrane protein is different from that of the saturation of the Sec translocon caused by the nonoptimized production of a secretory protein.
The observation that the accumulation levels of a considerable number of membrane proteins are not affected or are even increased when the scFv BL1 is produced under nonoptimized conditions may be explained by the increased accumulation levels of YidC (54). Although under normal conditions, most membrane proteins are integrated into the cytoplasmic membrane via the Sec translocon assisted by YidC, it has been shown that the integration of membrane proteins into the membrane can also be mediated by only YidC if no Sec translocon is available (55). Thus, the increased levels of YidC may very well compensate for the lack of sufficient Sec translocon capacity. Interestingly, when the biogenesis of membrane proteins in Bacillus subtilis is hampered, it responds by increasing the synthesis of one of its two YidC homologs in order to keep the biogenesis and membrane integration of membrane proteins going (56, 57).
Under the optimized production condition, the scFv BL1 was also present in the extracellular medium. Notably, in E. coli, the extracellular localization of recombinant secreted proteins has been observed repeatedly, but how these proteins end up in the extracellular medium is still enigmatic (6, 41–43). Here, we have shown that the release of the scFv BL1 in the extracellular medium is mediated by OMVs, which are naturally released from Gram-negative bacteria (58).
Altogether, the Lemo setup-based optimization of the production of the scFv BL1 in the periplasm of E. coli leads to a situation where the target is produced efficiently without causing any notable consequences at the level of the proteome. This surprising observation indicates that in protein production screens, monitoring stress holds great potential for optimizing production yields. Furthermore, we envisage that engineering approaches can be used to promote the OMV-mediated release of secreted recombinant proteins in the extracellular medium, thereby facilitating the isolation of recombinant proteins.
MATERIALS AND METHODS
Strain and plasmids.
The Lemo setup was used in this study for the production of the scFv BL1 (25, 59). However, rather than BL21(DE3) transformed with pLemo, Tuner(DE3) (Novagen) transformed with pLemo was used (23). Tuner(DE3) is a BL21(DE3)-derived strain lacking the gene encoding β-galactosidase, which is the protein that is recognized by the model recombinant protein, the scFv BL1, used in this study. The genetic information encoding the scFv BL1 was fused at the 5′ end to the genetic information encoding the DsbA signal sequence and at the 3′ end to the genetic information encoding a His6 tag. The DsbA signal sequence directs BL1 to the Sec translocon, and the His6 tag enables the detection of both the precursor form and the processed form of the scFv BL1 by means of immunoblotting (23). The gene encoding DsbA BL1 was expressed from a pET28a+-derived vector as described before (23). As a control, an empty vector was used as described before (23).
Culture conditions.
Cells were grown aerobically at 30°C and 200 rpm in lysogeny broth (LB; Difco) supplemented with 50 μg/ml kanamycin (to maintain the overexpression vector) and 30 μg/ml chloramphenicol (to maintain pLemo). Cells were grown either in the absence of l-rhamnose or the presence of 500 μM l-rhamnose. At an A600 of ∼0.4, the expression of the gene encoding DsbA BL1 was induced by adding 400 μM IPTG (final concentration) for 4 h. IPTG was also added to control cells harboring the “empty” vector. Growth was monitored by measuring the A600 with a UV-1601 spectrophotometer (Shimadzu). The standard deviations shown in Fig. 1 were calculated from at least three biologically independent experiments.
Subcellular fractionation.
Spheroplasts were generated and the periplasmic fraction isolated as described below. Cells were incubated in ice-cold osmotic shock buffer (75 A600 units/ml in 20 mM Tris-HCl [pH 8.0], 2.5 mM EDTA, 30% [wt/vol] sucrose) for 1 h. After 1 h, the cell suspension was diluted eight times using 20 mM Tris-HCl (pH 8.0) and incubated on ice for at least 30 min. Spheroplasts were pelleted at 10,000 × g for 10 min. The supernatant, which represents the periplasm, was concentrated by trichloroacetic acid precipitation. All steps were carried out on ice at 4°C. Protein concentrations were determined by using a bicinchoninic acid (BCA) assay (Pierce).
SDS-PAGE and immunoblotting.
Whole-cell lysates (0.1 A600 units), periplasmic fractions (5 μg of protein), spheroplasts (5 μg of protein), and outer membrane vesicles (1 μg of protein) were analyzed by SDS-PAGE using standard polyacrylamide gels followed by immunoblotting as described before (25). BL1 was detected using a horseradish peroxidase (HRP)-conjugated α-His antibody (Thermo Fisher) recognizing its C-terminal His tag (23). The anti-SurA, -IbpB, -FtsH, -OmpA, -SecY, -SecE, -SecA, and -YidC rabbit polyclonal antisera used were from our serum collection. The anti-GroEL rabbit polyclonal and anti-DnaK mouse monoclonal antibodies were from Sigma, and the anti-σ32 and -σ70 mouse monoclonal antibodies were from Neoclone. The incubation with a primary antibody was performed in a manner depending on its source, followed by an incubation with a secondary HRP-conjugated goat α-rabbit or α-mouse antibody (Bio-Rad). Proteins were visualized using the ECL system (GE Healthcare) according to the instructions of the manufacturer and a Fuji LAS-1000 charge-coupled-device (CCD) camera.
BL1 activity assay.
The proper folding of the scFv BL1 was assayed by the recognition of its substrate, E. coli β-galactosidase, using a dot blot assay and whole-cell lysate as described previously (23). In short, different amounts of β-galactosidase were spotted on a polyvinylidene difluoride (PVDF) membrane, and the membrane was subsequently incubated with cell lysate. Binding of the scFv BL1 was visualized using an HRP-conjugated α-His antibody (Pierce), the ECL system (GE Healthcare), and a Fuji LAS-1000 CCD camera.
Mass spectrometry, protein identification, and quantitative proteomics.
E. coli cells were pelleted and subsequently lysed using a solubilization buffer consisting of 2% SDS, 50 mM NaCl, 50 mM ammonium bicarbonate, and probe sonication. The proteins were isolated using acetone precipitation and resuspended in 2% SDS. The protein concentrations were determined by the Pierce BCA protein assay kit (Thermo Fisher). Ten micrograms of each sample was reduced, alkylated, and digested in-solution according to Ytterberg et al. (60). After zip-tip cleanup (Merck Millipore, Ltd.), 1 μg of each sample was separated using C18 RP columns coupled on-line to a tandem mass spectrometer using liquid chromatography (LC-MS/MS). The chromatographic separation was achieved using an acetonitrile (ACN)-water solvent system containing 0.2% formic acid. The gradient was set up as following: 5 to 40% ACN in 89 min, 40 to 95% ACN for 5 min, and 95% ACN for 8 min, all at a flow rate of 300 nl/min. The samples were analyzed by an LTQ Orbitrap Velos mass spectrometer equipped with collision-induced dissociation (CID), higher energy collisional dissociation (HCD), and electron-transfer dissociation (ETD) MS/MS (Thermo Fisher Scientific). The spectra were acquired with a resolution of 60,000 in MS mode, and the top 5 precursors were selected for fragmentation using CID. Mass lists were extracted from the raw data using Raw2MGF v2.1.3 and combined into one file using Cluster MGF v2.1.1, programs that are part of the Quanti workflow (61). The data were searched against a concatenated version of the E. coli complete proteome database (UP000000625; downloaded from www.uniprot.org in June 2016), using the Mascot search engine v2.5.1 (Matrix Science Ltd., UK). The database also included 13 non-E. coli proteins: the scFv BL1 without signal sequence, the scFv BL1 fused to the DsbA signal sequence, bovine serum albumin, bovine α-S1-casein, bovine β-lactoglobulin, the kanamycin and the chloramphenicol resistance proteins, T7 lysozyme, T7 RNA polymerase, three keratins, and trypsin. In total, the database consisted of 8,628 sequences containing 2,722,180 amino acid residues. The following parameters were used: tryptic digestion (maximum, 2 missed cleavages); carbamidomethylation (C) as a fixed modification; deamidation (N/Q), oxidation (M), and pyroglutamate (Q) as variable modifications; 10 ppm as the precursor tolerance; and 0.25 Da as the fragment mass tolerance. The threshold for 1% false-discovery rate (FDR) was calculated to a peptide score of 15.
The quantification was done using the Quanti workflow, which is quantification software based on extracted ion chromatograms (61). In short, after searching the combined mgf data against the E. coli complete proteome, the resulting data file and the raw files were uploaded into Quanti v2.5.4.3. The following parameters were used: score threshold, 15; mass tolerance, 10 ppm; minimum peptides/protein, 2; maximum allowed deviation in retention time, 5 min; rt order, 50; only “charge deconvolution” and “use best mascot peptide” were used. The quantitative values were further processed by multiplying the values with the reference abundance and normalizing each sample to the median of the summed intensities for all the samples. Pairwise comparisons between treatments were performed using the log10-transformed normalized protein intensities. The log2 ratios were calculated using the mean from the two treatments compared, and Student's t tests were used to estimate the P values for the comparisons. To account for false positives during multiple testing, q values (i.e., FDR-adjusted P values) were also calculated (62). The raw data, mgf files, and result files have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the data set identifier PXD008777 (username, reviewer05073@ebi.ac.uk; password, ekhO1Mp3 [www.ebi.ac.uk/pride/archive/]) (63).
OMV isolation and characterization.
Cells (250 ml of culture) were harvested and spun at 7,000 × g for 15 min. The supernatant, i.e., the spent medium, was subsequently filtered (0.45-μm pore size) to remove any residual bacteria. To verify that the filtrate did not contain any bacteria, 100 μl of filtered material was plated on an LB agar plate not containing any antibiotics. OMVs were harvested from the spent medium by ultracentrifugation (100,000 × g for 1 h at 4°C) and resuspended in ice-cold phosphate-buffered saline (PBS) for further analysis (see also “SDS-PAGE and immunoblotting”).
The size distribution of the OMVs was determined by dynamic light scattering using a Zetasizer Nano ZS (Malvern Instruments Ltd.) as described previously (44). To monitor how the scFv BL1 was associated with the OMVs, a proteinase K assay was used. In short, harvested OMVs were resuspended in 100 μl reaction buffer (50 mM Tris-HCl [pH 7.5], 1 mM CaCl2). Fractions of 30 μl were treated with Triton X-100 (final concentration, 0.5%), proteinase K (0.05 mg/ml), or both and incubated for 30 min at 30°C. Subsequently, the protease inhibitor phenylmethylsulfonyl fluoride (PMSF; final concentration, 5 mM) was added, and the samples were immediately frozen. Subsequently, the samples were analyzed by SDS-PAGE followed by immunoblotting as described above. The activity of the scFv BL1 colocalized with OMVs was monitored as described above with Triton X-100 solubilized OMVs.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants from the Swedish Research Council and the Swedish Foundation for Strategic Research to J.-W.D.G.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.00270-18.
REFERENCES
- 1.Makino T, Skretas G, Georgiou G. 2011. Strain engineering for improved expression of recombinant proteins in bacteria. Microb Cell Fact 10:32. doi: 10.1186/1475-2859-10-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rosano GL, Ceccarelli EA. 2014. Recombinant protein expression in Escherichia coli: advances and challenges. Front Microbiol 5:172. doi: 10.3389/fmicb.2014.00172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Denoncin K, Collet JF. 2013. Disulfide bond formation in the bacterial periplasm: major achievements and challenges ahead. Antioxid Redox Signal 19:63–71. doi: 10.1089/ars.2012.4864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Marco A. 2009. Strategies for successful recombinant expression of disulfide bond-dependent proteins in Escherichia coli. Microb Cell Fact 8:26. doi: 10.1186/1475-2859-8-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Marco A. 2012. Recent contributions in the field of the recombinant expression of disulfide bonded proteins in bacteria. Microb Cell Fact 11:129. doi: 10.1186/1475-2859-11-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mergulhão FJM, Summers DK, Monteiro GA. 2005. Recombinant protein secretion in Escherichia coli. Biotechnol Adv 23:177–202. doi: 10.1016/j.biotechadv.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 7.du Plessis DJF, Nouwen N, Driessen AJM. 2011. The Sec translocase. Biochim Biophys Acta 1808:851–865. doi: 10.1016/j.bbamem.2010.08.016. [DOI] [PubMed] [Google Scholar]
- 8.Arkowitz RA, Joly JC, Wickner W. 1993. Translocation can drive the unfolding of a preprotein domain. EMBO J 12:243–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Valent QA, Scotti PA, High S, de Gier JW, von Heijne G, Lentzen G, Wintermeyer W, Oudega B, Luirink J. 1998. The Escherichia coli SRP and SecB targeting pathways converge at the translocon. EMBO J 17:2504–2512. doi: 10.1093/emboj/17.9.2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim J, Rusch S, Luirink J, Kendall DA. 2001. Is Ffh required for export of secretory proteins? FEBS Lett 505:245–248. doi: 10.1016/S0014-5793(01)02784-3. [DOI] [PubMed] [Google Scholar]
- 11.Lee HC, Bernstein HD. 2001. The targeting pathway of Escherichia coli presecretory and integral membrane proteins is specified by the hydrophobicity of the targeting signal. Proc Natl Acad Sci U S A 98:3471–3476. doi: 10.1073/pnas.051484198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Valent QA, de Gier JW, von Heijne G, Kendall DA, ten Hagen-Jongman CM, Oudega B, Luirink J. 1997. Nascent membrane and presecretory proteins synthesized in Escherichia coli associate with signal recognition particle and trigger factor. Mol Microbiol 25:53–64. doi: 10.1046/j.1365-2958.1997.4431808.x. [DOI] [PubMed] [Google Scholar]
- 13.Hegde RS, Bernstein HD. 2006. The surprising complexity of signal sequences. Trends Biochem Sci 31:563–571. doi: 10.1016/j.tibs.2006.08.004. [DOI] [PubMed] [Google Scholar]
- 14.Schierle CF, Berkmen M, Huber D, Kumamoto C, Boyd D, Beckwith J. 2003. The DsbA signal sequence directs efficient, cotranslational export of passenger proteins to the Escherichia coli periplasm via the signal recognition particle pathway. J Bacteriol 185:5706–5713. doi: 10.1128/JB.185.19.5706-5713.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Steiner D, Forrer P, Stumpp MT, Plückthun A. 2006. Signal sequences directing cotranslational translocation expand the range of proteins amenable to phage display. Nat Biotechnol 24:823–831. doi: 10.1038/nbt1218. [DOI] [PubMed] [Google Scholar]
- 16.Zwizinski C, Wickner W. 1980. Purification and characterization of leader (signal) peptidase from Escherichia coli. J Biol Chem 255:7973–7977. [PubMed] [Google Scholar]
- 17.Merdanovic M, Clausen T, Kaiser M, Huber R, Ehrmann M. 2011. Protein quality control in the bacterial periplasm. Annu Rev Microbiol 65:149–168. doi: 10.1146/annurev-micro-090110-102925. [DOI] [PubMed] [Google Scholar]
- 18.Chamberlin M, McGrath J, Waskell L. 1970. New RNA polymerase from Escherichia coli infected with bacteriophage T7. Nature 228:227–231. doi: 10.1038/228227a0. [DOI] [PubMed] [Google Scholar]
- 19.Iost I, Guillerez J, Dreyfus M. 1992. Bacteriophage T7 RNA polymerase travels far ahead of ribosomes in vivo. J Bacteriol 174:619–622. doi: 10.1128/jb.174.2.619-622.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Studier FW, Moffatt BA. 1986. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol 189:113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- 21.Silverstone AE, Arditti RR, Magasanik B. 1970. Catabolite-insensitive revertants of lac promoter mutants. Proc Natl Acad Sci U S A 66:773–779. doi: 10.1073/pnas.66.3.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wanner BL, Kodaira R, Neidhardt FC. 1977. Physiological regulation of a decontrolled lac operon. J Bacteriol 130:212–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schlegel S, Rujas E, Ytterberg AJ, Zubarev RA, Luirink J, de Gier JW. 2013. Optimizing heterologous protein production in the periplasm of E. coli by regulating gene expression levels. Microb Cell Fact 12:24. doi: 10.1186/1475-2859-12-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schlegel S, Löfblom J, Lee C, Hjelm A, Klepsch MM, Strous M, Drew D, Slotboom DJ, de Gier JW. 2012. Optimizing membrane protein overexpression in the Escherichia coli strain Lemo21(DE3). J Mol Biol 423:648–659. doi: 10.1016/j.jmb.2012.07.019. [DOI] [PubMed] [Google Scholar]
- 25.Wagner S, Klepsch MM, Schlegel S, Appel A, Draheim R, Tarry M, Högbom M, van Wijk KJ, Slotboom DJ, Persson JO, de Gier JW. 2008. Tuning Escherichia coli for membrane protein overexpression. Proc Natl Acad Sci U S A 105:14371–14376. doi: 10.1073/pnas.0804090105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Giacalone M, Gentile A, Lovitt B, Berkley N, Gunderson C, Surber M. 2006. Toxic protein expression in Escherichia coli using a rhamnose-based tightly regulated and tunable promoter system. Biotechniques 40:355–364. doi: 10.2144/000112112. [DOI] [PubMed] [Google Scholar]
- 27.Cheng X, Zhang X, Pflugrath JW, Studier FW. 1994. The structure of bacteriophage T7 lysozyme, a zinc amidase and an inhibitor of T7 RNA polymerase. Proc Natl Acad Sci U S A 91:4034–4038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Studier FW. 1991. Use of bacteriophage T7 lysozyme to improve an inducible T7 expression system. J Mol Biol 219:37–44. doi: 10.1016/0022-2836(91)90855-Z. [DOI] [PubMed] [Google Scholar]
- 29.Wagner S, Baars L, Ytterberg AJ, Klußmeier A, Wagner CS, Nord O, Nygren PÅ, van Wijk KJ, de Gier JW. 2007. Consequences of membrane protein overexpression in Escherichia coli. Mol Cell Proteomics 6:1527–1550. doi: 10.1074/mcp.M600431-MCP200. [DOI] [PubMed] [Google Scholar]
- 30.Baumgarten T, Schlegel S, Wagner S, Löw M, Eriksson J, Bonde I, Herrgård MJ, Heipieper HJ, Nørholm MHH, Slotboom DJ, de Gier JW. 2017. Isolation and characterization of the E. coli membrane protein production strain Mutant56(DE3). Sci Rep 7:45089. doi: 10.1038/srep45089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schlegel S, Genevaux P, de Gier JW. 2015. De-convoluting the genetic adaptations of E. coli C41(DE3) in real time reveals how alleviating protein production stress improves yields. Cell Rep 10:1758–1766. doi: 10.1016/j.celrep.2015.02.029. [DOI] [PubMed] [Google Scholar]
- 32.Vethanayagam JGG, Flower AM. 2005. Decreased gene expression from T7 promoters may be due to impaired production of active T7 RNA polymerase. Microb Cell Fact 4:3. doi: 10.1186/1475-2859-4-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Klepsch MM, Persson JO, de Gier JW. 2011. Consequences of the overexpression of a eukaryotic membrane protein, the human KDEL receptor, in Escherichia coli. J Mol Biol 407:532–542. doi: 10.1016/j.jmb.2011.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kwon S-K, Kim SK, Lee D-H, Kim JF. 2015. Comparative genomics and experimental evolution of Escherichia coli BL21(DE3) strains reveal the landscape of toxicity escape from membrane protein overproduction. Sci Rep 5:16076. doi: 10.1038/srep16076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu M, Feng X, Ding Y, Zhao G, Liu H, Xian M. 2015. Metabolic engineering of Escherichia coli to improve recombinant protein production. Appl Microbiol Biotechnol 99:10367–10377. doi: 10.1007/s00253-015-6955-9. [DOI] [PubMed] [Google Scholar]
- 36.Scotti PA, Urbanus ML, Brunner J, de Gier JW, von Heijne G, van der Does C, Driessen AJM, Oudega B, Luirink J. 2000. YidC, the Escherichia coli homologue of mitochondrial Oxa1p, is a component of the Sec translocase. EMBO J 19:542–549. doi: 10.1093/emboj/19.4.542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Serek J, Bauer-Manz G, Struhalla G, van den Berg L, Kiefer D, Dalbey R, Kuhn A. 2004. Escherichia coli YidC is a membrane insertase for Sec-independent proteins. EMBO J 23:294–301. doi: 10.1038/sj.emboj.7600063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.van der Laan M, Bechtluft P, Kol S, Nouwen N, Driessen AJM. 2004. F1F0 ATP synthase subunit c is a substrate of the novel YidC pathway for membrane protein biogenesis. J Cell Biol 165:213–222. doi: 10.1083/jcb.200402100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Economou A, Wickner W. 1994. SecA promotes preprotein translocation by undergoing ATP-driven cycles of membrane insertion and deinsertion. Cell 78:835–843. doi: 10.1016/S0092-8674(94)90582-7. [DOI] [PubMed] [Google Scholar]
- 40.Matsumoto G, Yoshihisa T, Ito K. 1997. SecY and SecA interact to allow SecA insertion and protein translocation across the Escherichia coli plasma membrane. EMBO J 16:6384–6393. doi: 10.1093/emboj/16.21.6384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fu XY. 2010. Extracellular accumulation of recombinant protein by Escherichia coli in a defined medium. Appl Microbiol Biotechnol 88:75–86. doi: 10.1007/s00253-010-2718-9. [DOI] [PubMed] [Google Scholar]
- 42.Ong RM, Goh KM, Mahadi NM, Hassan O, Rahman RNZRA, Illias RM. 2008. Cloning, extracellular expression and characterization of a predominant beta-CGTase from Bacillus sp. G1 in E. coli. J Ind Microbiol Biotechnol 35:1705–1714. doi: 10.1007/s10295-008-0462-2. [DOI] [PubMed] [Google Scholar]
- 43.Ukkonen K, Veijola J, Vasala A, Neubauer P. 2013. Effect of culture medium, host strain and oxygen transfer on recombinant Fab antibody fragment yield and leakage to medium in shaken E. coli cultures. Microb Cell Fact 12:73. doi: 10.1186/1475-2859-12-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Baumgarten T, Vazquez J, Bastisch C, Veron W, Feuilloley MGJ, Nietzsche S, Wick LY, Heipieper HJ. 2012. Alkanols and chlorophenols cause different physiological adaptive responses on the level of cell surface properties and membrane vesicle formation in Pseudomonas putida DOT-T1E. Appl Microbiol Biotechnol 93:837–845. doi: 10.1007/s00253-011-3442-9. [DOI] [PubMed] [Google Scholar]
- 45.Daleke-Schermerhorn MH, Felix T, Soprova Z, ten Hagen-Jongman CM, Vikström D, Majlessi L, Beskers J, Follmann F, de Punder K, van der Wel NN, Baumgarten T, Pham TV, Piersma SR, Jiménez CR, van Ulsen P, de Gier JW, Leclerc C, Jong WSP, Luirink J. 2014. Decoration of outer membrane vesicles with multiple antigens by using an autotransporter approach. Appl Environ Microbiol 80:5854–5865. doi: 10.1128/AEM.01941-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Manning AJ, Kuehn MJ. 2011. Contribution of bacterial outer membrane vesicles to innate bacterial defense. BMC Microbiol 11:258. doi: 10.1186/1471-2180-11-258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jensen EB, Carken S. 1990. Production of recombinant human growth hormone in Escherichia coli: expression of different precursors and physiological effects of glucose, acetate, and salts. Biotechnol Bioeng 36:1–11. doi: 10.1002/bit.260360102. [DOI] [PubMed] [Google Scholar]
- 48.Nakano K, Rischke M, Sato S, Märkl H. 1997. Influence of acetic acid on the growth of Escherichia coli K-12 during high-cell-density cultivation in a dialysis reactor. Appl Microbiol Biotechnol 48:597–601. doi: 10.1007/s002530051101. [DOI] [PubMed] [Google Scholar]
- 49.Eiteman MA, Altman E. 2006. Overcoming acetate in Escherichia coli recombinant protein fermentations. Trends Biotechnol 24:530–536. doi: 10.1016/j.tibtech.2006.09.001. [DOI] [PubMed] [Google Scholar]
- 50.Brundage L, Hendrick JP, Schiebel E, Driessen AJM, Wickner W. 1990. The purified E. coli integral membrane protein SecY/E is sufficient for reconstitution of SecA-dependent precursor protein translocation. Cell 62:649–657. doi: 10.1016/0092-8674(90)90111-Q. [DOI] [PubMed] [Google Scholar]
- 51.Taura T, Baba T, Akiyama Y, Ito K. 1993. Determinants of the quantity of the stable SecY complex in the Escherichia coli cell. J Bacteriol 175:7771–7775. doi: 10.1128/jb.175.24.7771-7775.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Traxler B, Murphy C. 1996. Insertion of the polytopic membrane protein MalF is dependent on the bacterial secretion machinery. J Biol Chem 271:12394–12400. doi: 10.1074/jbc.271.21.12394. [DOI] [PubMed] [Google Scholar]
- 53.Nakatogawa H, Murakami A, Ito K. 2004. Control of SecA and SecM translation by protein secretion. Curr Opin Microbiol 7:145–150. doi: 10.1016/j.mib.2004.01.001. [DOI] [PubMed] [Google Scholar]
- 54.Kuhn A, Koch H-G, Dalbey RE. 7 March 2017. Targeting and insertion of membrane proteins. EcoSal Plus doi: 10.1128/ecosalplus.ESP-0012-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Welte T, Kudva R, Kuhn P, Sturm L, Braig D, Müller M, Warscheid B, Drepper F, Koch H-G. 2012. Promiscuous targeting of polytopic membrane proteins to SecYEG or YidC by the Escherichia coli signal recognition particle. Mol Biol Cell 23:464–479. doi: 10.1091/mbc.E11-07-0590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chiba S, Ito K. 2015. MifM monitors total YidC activities of Bacillus subtilis, including that of YidC2, the target of regulation. J Bacteriol 197:99–107. doi: 10.1128/JB.02074-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chiba S, Lamsa A, Pogliano K. 2009. A ribosome-nascent chain sensor of membrane protein biogenesis in Bacillus subtilis. EMBO J 28:3461–3475. doi: 10.1038/emboj.2009.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schwechheimer C, Kuehn MJ. 2015. Outer-membrane vesicles from Gram-negative bacteria: biogenesis and functions. Nat Rev Microbiol 13:605–619. doi: 10.1038/nrmicro3525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Martineau P, Jones P, Winter G. 1998. Expression of an antibody fragment at high levels in the bacterial cytoplasm. J Mol Biol 280:117–127. doi: 10.1006/jmbi.1998.1840. [DOI] [PubMed] [Google Scholar]
- 60.Ytterberg AJ, Peltier JB, van Wijk KJ. 2006. Protein profiling of plastoglobules in chloroplasts and chromoplasts. A surprising site for differential accumulation of metabolic enzymes. Plant Physiol 140:984–997. doi: 10.1104/pp.105.076083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lyutvinskiy Y, Yang H, Rutishauser D, Zubarev RA. 2013. In silico instrumental response correction improves precision of label-free proteomics and accuracy of proteomics-based predictive models. Mol Cell Proteomics 12:2324–2331. doi: 10.1074/mcp.O112.023804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol 57:289–300. [Google Scholar]
- 63.Vizcaíno JA, Csordas A, del-Toro N, Dianes JA, Griss J, Lavidas I, Mayer G, Perez-Riverol Y, Reisinger F, Ternent T, Xu Q-W, Wang R, Hermjakob H. 2016. 2016 update of the PRIDE database and its related tools. Nucleic Acids Res 44:D447–D456. doi: 10.1093/nar/gkv1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.