ABSTRACT
Freeze-thaw stress causes various types of cellular damage, survival and/or proliferation defects, and metabolic alterations. However, the mechanisms underlying how cells cope with freeze-thaw stress are poorly understood. Here, model dough fermentations using two baker's yeast strains, 45 and YF, of Saccharomyces cerevisiae were compared after 2 weeks of cell preservation in a refrigerator or freezer. YF exhibited slow fermentation after exposure to freeze-thaw stress due to low cell viability. A DNA microarray analysis of the YF cells during fermentation revealed that the genes involved in oxidative phosphorylation were relatively strongly expressed, suggesting a decrease in the glycolytic capacity. Furthermore, we found that mRNA levels of the genes that encode the components of the proteasome complex were commonly low, and ubiquitinated proteins were accumulated by freeze-thaw stress in the YF strain. In the cells with a laboratory strain background, treatment with the proteasome inhibitor MG132 or the deletion of each transcriptional activator gene for the proteasome genes (RPN4, PDR1, or PDR3) led to marked impairment of model dough fermentation using the frozen cells. Based on these data, proteasomal degradation of freeze-thaw-damaged proteins may guarantee high cell viability and fermentation performance. We also found that the freeze-thaw stress-sensitive YF strain was heterozygous at the PDR3 locus, and one of the alleles (A148T/A229V/H336R/L541P) was shown to possess a dominant negative phenotype of slow fermentation. Removal of such responsible mutations could improve the freeze-thaw stress tolerance and the fermentation performance of baker's yeast strains, as well as other industrial S. cerevisiae strains.
IMPORTANCE The development of freezing technology has enabled the long-term preservation and long-distance transport of foods and other agricultural products. Fresh yeast, however, is usually not frozen because the fermentation performance and/or the viability of individual cells is severely affected after thawing. Here, we demonstrate that proteasomal degradation of ubiquitinated proteins is an essential process in the freeze-thaw stress responses of S. cerevisiae. Upstream transcriptional activator genes for the proteasome components are responsible for the fermentation performance after freezing preservation. Thus, this study provides a potential linkage between freeze-thaw stress inputs and the transcriptional regulatory network that might be functionally conserved in higher eukaryotes. Elucidation of the molecular targets of freeze-thaw stress will contribute to advances in cryobiology, such as freezing preservation of human cells, tissues, and embryos for medical purposes and breeding of industrial microorganisms and agricultural crops that adapt well to low temperatures.
KEYWORDS: baker's yeast, dough fermentation, freeze-thaw stress, proteasome, Saccharomyces cerevisiae
INTRODUCTION
Freezing preservation is one of the most widely used methods to maintain the quality and freshness of various raw materials, ingredients, in-process foods, and end products over long periods. During the freezing process, delay in chemical and metabolic reactions prevents the loss of the original flavor, texture, and nutritional values. Microbial food spoilage is also effectively blocked under frozen conditions. Nevertheless, intracellular and extracellular ice crystal formation and subsequent dehydration damage cellular structures and affect the frozen product's quality. Thawing is another critical phase, since frozen foods are subjected to damage by chemical and physical changes (1, 2). Fresh baker's yeast (e.g., liquid yeast and compressed yeast), which is the gas-forming ingredient in bakery products, is generally stored under refrigeration above the freezing point in order to retain the high gassing power (3–5). The breeding of baker's yeast strains that are suitable for freezing preservation may greatly improve the microbiological stability of fresh yeast.
Cell injury and adaptation under freeze-thaw stress in Saccharomyces cerevisiae, to which most baker's yeast strains belong, have been intensively studied over recent decades (6–8). Freeze-thawing directly or indirectly damages macromolecules, including cell membranes and proteins, and leads to the generation of reactive oxygen species, as many other stresses do. Thus, the functions of heat shock proteins and antioxidant enzymes are essential for high freezing tolerance. To decrease intracellular ice crystal formation, water efflux through the aquaporins and the intracellular accumulation of trehalose and glycerol are important as well. Proline shows cryoprotective activity that is similar to those of trehalose and glycerol (9–11), although proline is not physiologically accumulated in S. cerevisiae cells by freeze-thaw stress. While various protective factors and mechanisms have been analyzed as noted above, it is not yet known how S. cerevisiae cells effectively repair the damage from freeze-thawing, and such knowledge would make a significant contribution to the baking industry.
The ubiquitin-proteasome system is one of the major proteolytic machineries that have pivotal functions in the stress responses of eukaryotic cells (12). Proteins irreversibly damaged by stresses cannot be repaired, and they must therefore be removed by the proteasome complex after being labeled with polyubiquitin chains. The proteasome activity is at least partly controlled at the transcriptional level by a transcriptional activator, Rpn4p (12, 13). Rpn4p upregulates the proteasome genes with the consensus proteasome-associated control element (PACE) (5′-GGTGGCAAA-3′) sequence in the promoter region, and the proteasome degrades Rpn4p itself. This negative feedback loop (14) maintains the homeostatic expression level of the proteasome complex. However, under stress conditions in which polyubiquitinated proteins may accumulate, RPN4 gene expression is induced by the stress-responsive transcriptional activators Pdr1p and Pdr3p (Pdr1/3p), Yap1p, and Hsf1p (15–17). According to the evidence obtained to date, Rpn4p is required for the cellular resistance to heat shock, oxidative stress, DNA damage, proteotoxic stress, heavy metals, and other drugs, as well as for the extension of replicative life span (16–18). The significance of the Rpn4p-mediated proteasome gene expression under freeze-thaw stress has not yet been elucidated.
In this study, we investigated the effects of the freezing preservation of fresh yeast on model dough fermentation and the gene expression profile. Our results shed further light on the relationship between freeze-thaw stress and the proteasomal functions in S. cerevisiae, which may facilitate the design of novel high-performance yeast products.
RESULTS
Effects of freezing preservation on model dough fermentation using baker's yeast strains.
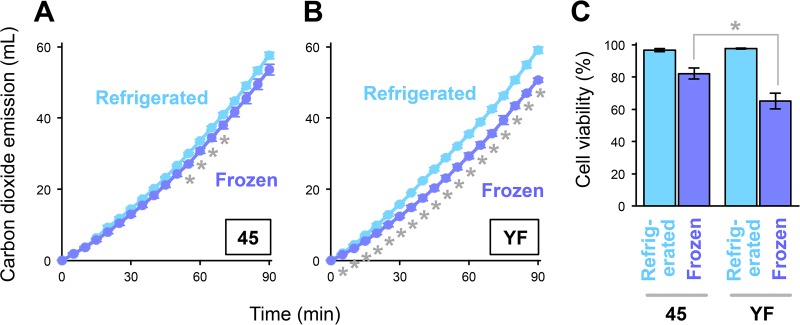
First, we compared the fermentation performances of two baker's yeast strains, 45 and YF, which may share an ancestor and have been used at TableMark Co., Ltd. Yeast pellets of each strain were used for the fermentation tests in a liquid medium after being refrigerated or frozen for 2 weeks. As shown in Fig. 1A, freezing did not have clear effects on the fermentation progression in strain 45, and the total carbon dioxide emission during 90 min of fermentation using frozen pellets was 93.8% of that using refrigerated pellets. In contrast to the case for strain 45, we observed slow fermentation using frozen pellets of strain YF (Fig. 1B). The freezing preservation of YF decreased the total carbon dioxide emission during the 90-min fermentation to 85.8% of the level in the refrigerated sample. It should be noted that the negative effect of freezing with YF was observed from the initial stage (within 5 min after inoculation) of model dough fermentation. The total carbon dioxide emissions of both strains were almost the same after refrigeration (only 3.5% higher in YF on average). Consistently, the cell viability after 2 weeks of freezing was significantly lower in YF than in 45, while refrigeration did not affect the cell viability (Fig. 1C). Based on these results, we concluded that YF is more susceptible to freezing preservation than 45.
FIG 1.
Phenotypes of strains 45 and YF after refrigeration or freezing preservation. (A and B) Progression of model dough fermentation. The graphs indicate the total carbon dioxide emission during the model dough fermentation tests using strains 45 (A) and YF (B). Light blue lines and blue lines show the means and standard deviations from three independent experiments using refrigerated cells and frozen cells, respectively. Asterisks represent significant differences (t test, P < 0.01). (C) Cell viability after 2 weeks of refrigeration or freezing preservation as determined by methylene blue staining. Data indicate the means and standard deviations from three independent experiments. Asterisks represent significant differences (t test, P < 0.05).
Effects of freezing preservation on gene expression profiles during model dough fermentation using baker's yeast strains.
To elucidate why YF decreased cell viability and exhibited slow fermentation after freezing preservation, we analyzed the transcriptomic profiles of both strains after refrigeration and after freezing. Total RNA samples were taken from the yeast cells 30 min after the onset of fermentation and used for a DNA microarray experiment. The genes differentially expressed between the frozen and refrigerated samples (see Table S1 in the supplemental material) were subjected to a gene ontology (GO) analysis using the GO Term Finder in the Saccharomyces Genome Database (SGD) (http://www.yeastgenome.org/cgi-bin/GO/goTermFinder.pl).
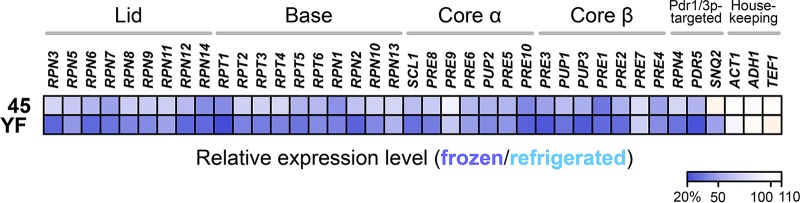
In strain 45, all of the enriched GO term categories showed P values higher than 1 × 10−10. In strain YF (Table 1), the genes that belong to five GO term categories associated with oxidative phosphorylation and aerobic respiration were strongly expressed after freeze-thaw stress. This may reflect the impaired glycolysis and alcoholic fermentation in this strain (19). In contrast, three GO term categories associated with proteasomal proteolysis were identified in YF as weakly expressed after freezing. As shown in Fig. 2, the expression levels of all 33 known proteasome genes, which encode 9 lid and 10 base subunits in the 19S regulatory particle and 7 α-ring and 7 β-ring subunits of the 20S catalytic core particle (12), were lower after freezing than after refrigerating in both strains. YF exhibited more prominent differences than 45 with no exception. The averaged relative expression level of all proteasome genes in YF was significantly lower than that in 45 (strain 45, 68.3% ± 9.8%; strain YF, 49.5% ± 9.6%; t test, P = 2.8 × 10−11).
TABLE 1.
GO analysis of genes differentially expressed (P < 1 × 10−10) between frozen and refrigerated samples of strain YF
| Expression during fermentation after freezing | GO term | Cluster frequency (%) | Background frequency (%) | P value |
|---|---|---|---|---|
| Strong | Oxidation-reduction process (GO:55114) | 46/355 (13.0) | 162/7,165 (2.3) | 1.72 × 10−20 |
| Energy derivation by oxidation of organic compounds (GO:15980) | 36/355 (10.1) | 135/7,165 (1.9) | 1.36 × 10−14 | |
| Generation of precursor metabolites and energy (GO:6091) | 38/355 (10.7) | 158/7,165 (2.2) | 7.42 × 10−14 | |
| Small-molecule metabolic process (GO:44281) | 85/355 (23.9) | 691/7,165 (9.6) | 2.87 × 10−13 | |
| Cellular respiration (GO:45333) | 27/355 (7.6) | 88/7,165 (1.2) | 3.77 × 10−12 | |
| Weak | Proteolysis involved in cellular protein catabolic process (GO:51603) | 43/351 (12.3) | 234/7,165 (3.3) | 1.57 × 10−11 |
| Modification-dependent protein catabolic process (GO:19941) | 41/351 (11.7) | 216/7,165 (3.0) | 2.13 × 10−11 | |
| Ubiquitin-dependent protein catabolic process (GO:6511) | 41/351 (11.7) | 216/7,165 (3.0) | 2.13 × 10−11 |
FIG 2.
Effects of freezing preservation on proteasome gene expression. The heat map indicates the relative expression levels of individual genes, which are represented by the percentages of frozen-sample data with respect to refrigerated-sample data of the DNA microarray analysis using the cells 30 min after the onset of model dough fermentation. The DNA microarray experiment was performed using a single sample for each strain/condition.
These results suggested that the proteasome gene expression is more severely disturbed in YF cells during fermentation after freezing preservation. We also observed that the RPN4 gene itself, which encodes a transcription activator for the proteasome genes, was apparently weakly expressed in YF (strain 45, 78.6%; strain YF, 45.3%) (Fig. 2). We thus speculated that the upstream transcription factors for RPN4 (Pdr1/3p, Hsf1p, and Yap1p) (15–17) are less activated in YF during fermentation after freeze-thaw stress. Pdr1/3p may be likely to be affected, since the different target genes of Pdr1/3p, such as the plasma membrane ATP-binding cassette (ABC) multidrug transporter genes PDR5 and SNQ2 (20), exhibited lower mRNA levels after freezing preservation in YF, as did the proteasome genes and RPN4 (Fig. 2). Known target genes of Hsf1p or Yap1p did not necessarily show similar defects (data not shown). Alternatively, degradation of RPN4 mRNA might also be enhanced in YF, which should be investigated in the future.
Evaluation of proteasomal function using baker's yeast strains.
To address the activities of proteasomal proteolysis, we analyzed the stress-induced accumulation of ubiquitinated proteins in the baker's yeast strains. YF exhibited larger amounts of ubiquitinated proteins than 45 upon heat shock, a representative stress that induces protein ubiquitination (Fig. 3A), and also immediately after 2-week freezing preservation (Fig. 3B). In the liquid medium for model dough fermentation, ubiquitinated proteins rapidly disappeared in both strains. These results suggested that the proteasomal degradation of ubiquitinated proteins under heat shock or freeze-thaw stress is impaired in YF. The slow growth of YF in the presence of 75 μM MG132 (a proteasome inhibitor) supported this idea (Fig. 3C).
FIG 3.
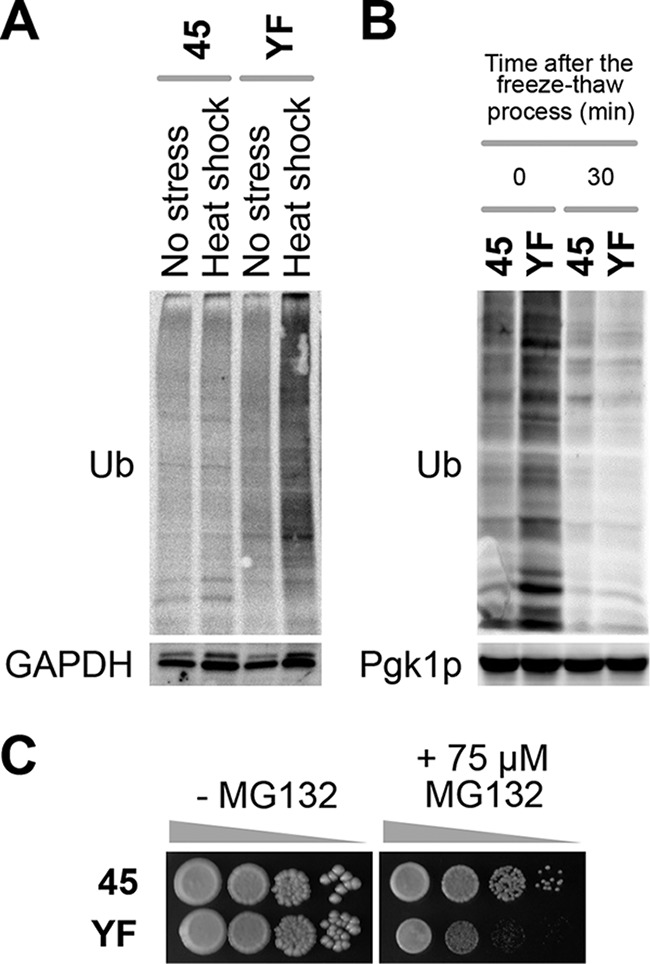
Assay of the proteasomal function of baker's yeast strains. (A) Ubiquitinated proteins upon heat shock. Cells were incubated at 42°C for 15 min for heat shock. The GAPDH level is shown as a protein loading control in Western blot analyses. Representative data from two repeated experiments are shown. (B) Ubiquitinated proteins after freeze-thaw process. Cells were frozen for 2 weeks and used for model dough fermentation. The Pgk1p level is shown as a protein loading control in Western blot analyses. Representative data from two repeated experiments are shown. (C) MG132 tolerance of baker's yeast strains. Approximately 107 cells (optical density at 600 nm [OD600] of 1) of strains 45 and YF were serially diluted to 101 to 103-fold (from left to right), spotted onto YPD plus DMSO (left panel) and YPD plus DMSO plus 75 μM MG132 (right panel) agar media, and incubated at 30°C for 2 days. Representative data from two repeated experiments are shown.
It should be noted that the slow-growth phenotype was observed when the MG132 concentration of the stock solution in dimethyl sulfoxide (DMSO) was 1 mM but not when it was 10 mM (data not shown). This raised the possibility that the high concentration of DMSO in the agar plate might solely cause the slow-growth phenotype. DMSO alone did not affect the cell growth of either strain, however. Thus, the growth inhibition of YF was presumably caused by a synergistic effect of MG132 and DMSO.
Effects of proteasomal function on model dough fermentation.
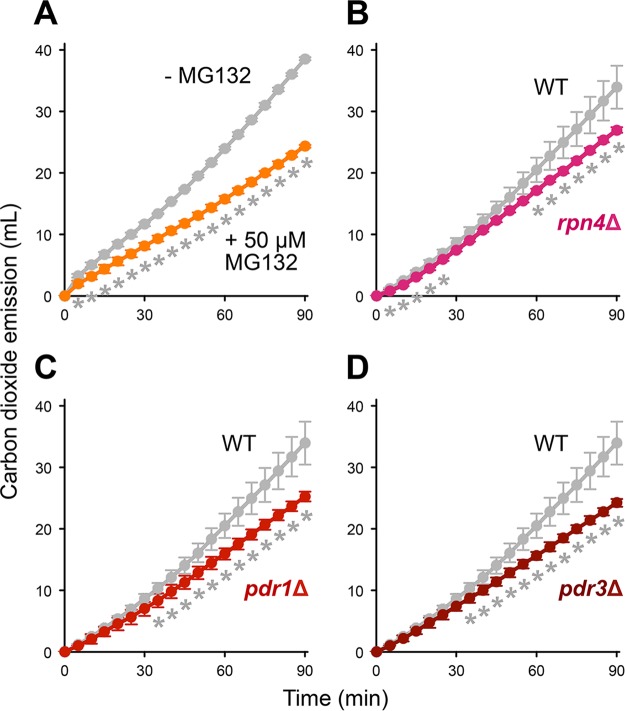
We next performed model dough fermentation tests using a laboratory strain, BY4741, and its gene disruptants (rpn4Δ, pdr1Δ, and pdr3Δ strains) to clarify the role of proteasomal function and proteasome gene expression in acquisition of freeze-thaw stress tolerance. When 50 μM MG132 was added to the liquid medium, the fermentation performance after freezing preservation was markedly decreased (Fig. 4A). The total carbon dioxide emission during 90 min of fermentation dropped to 63.1% upon MG132 treatment.
FIG 4.
Role of the proteasome in model dough fermentation after freezing preservation. (A) Effects of MG132 treatment. The graph indicates the total carbon dioxide emission during the model dough fermentation tests using the BY4741 wild-type strain in the absence (gray) and presence (orange) of 50 μM MG132. (B to D) Effects of proteasome gene expression. The graphs indicate the total carbon dioxide emission during the model dough fermentation tests using the BY4741 wild-type strain (gray) and the RPN4 (B)-, PDR1 (C)-, or PDR3 (D)-disrupted strain (red). The data shown are the means and standard deviations from two or three independent experiments using frozen cells. Asterisks represent significant differences (t test, P < 0.05). WT, wild type.
Loss of Rpn4p or Pdr1/3p, each of which upregulates the proteasome gene expression, also impaired fermentation after freezing preservation (Fig. 4B to D). The total carbon dioxide emission during 90 min of fermentation in these gene disruptants was 70 to 80% of the level in the wild-type strain. Accordingly, the proteasomal function and/or the proteasome gene expression through Rpn4p and Pdr1/3p is likely to be important to maintain high fermentation performance after freezing preservation.
Mutations involved in the freeze-thaw stress sensitivity of a baker's yeast strain.
To identify the cause of the low freeze-thaw stress tolerance observed in YF, we performed a whole-genome resequencing analysis. Nine nonsynonymous single-nucleotide polymorphisms (SNPs) were identified in the RPN4, PDR1, and PDR3 genes of strain YF (Fig. 5A) in comparison to the S288C reference genome. We identified five of these SNPs, i.e., homozygous C221T and A1279G in RPN4, heterozygous C53G and A280G in PDR1, and heterozygous A167G in PDR3 (corresponding to amino acid substitutions S74L and T427A in Rpn4p, T18R and T94A in Pdr1p, and Q56R in Pdr3p, respectively), in several other S. cerevisiae strains, according to SGD Align Strain Sequences (http://www.yeastgenome.org/cgi-bin/FUNGI/alignment.pl).
FIG 5.
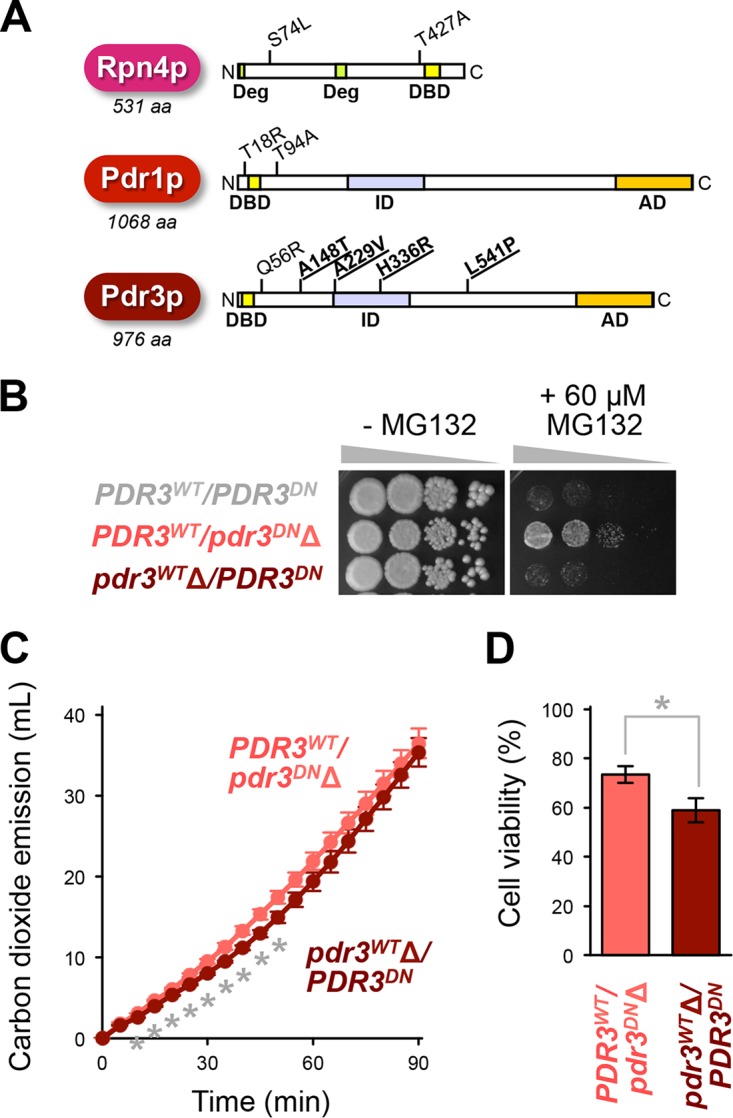
The dominant negative allele of PDR3 found in strain YF. (A) Nonsynonymous polymorphisms in the RPN4, PDR1, and PDR3 genes of YF. Amino acid substitutions unique to YF are indicated in bold and underlined. The colored boxes indicate the previously reported sequences and domains important for their function as transcriptional factors: Deg, degrons; DBD, DNA-binding motif; ID, inhibitory domain; AD, activation domain (29, 42). (B) MG132 tolerance of hemizygous pdr3Δ disruptants of YF. Approximately 107 cells (OD600 of 1) of parental YF (PDR3WT/PDR3DN) and hemizygous disruptants (PDR3WT/pdr3DNΔ and pdr3WTΔ/PDR3DN) were serially diluted to 101 to 103-fold (from left to right), spotted onto YPD plus DMSO (left panel) and YPD plus DMSO plus 60 μM MG132 (right panel) agar media, and incubated at 30°C for 1 day. Representative data from two repeated experiments are shown. (C) Effects of the PDR3DN allele on model dough fermentation after freezing preservation. The graph indicates the total carbon dioxide emission during the model dough fermentation tests using the PDR3WT/pdr3DNΔ (gray) and pdr3WTΔ/PDR3DN (black) strains. The data shown are the means and standard deviations from three independent experiments using frozen cells. Asterisks represent significant differences (t test, P < 0.01). WT, wild type; DN, dominant negative. (D) Cell viability after 2 weeks of freezing preservation as determined by methylene blue staining. Data indicate the means and standard deviations from three independent experiments. The asterisk represents a significant difference (t test, P < 0.05).
However, the other four heterozygous mutations, i.e., G442A, C686T, A1007G, and A1623T in PDR3 (corresponding to amino acid substitutions A148T, A229V, H336R, and L541P of Pdr3p, respectively), were unique to YF in our search of this database. Direct DNA sequencing of the PCR product amplified using a PDR3 gene-specific primer pair revealed that these four nucleotide polymorphisms were located on the identical allele, suggesting a high nonsynonymous mutation speed of this unique allele. In the reciprocal hemizygosity analysis of YF shown in Fig. 5B, deletion of the PDR3Q56R allele did not affect the growth (see pdr3WTΔ/PDR3DN), whereas deletion of the PDR3A148T/A229V/H336R/L541P allele increased the tolerance to MG132 (see PDR3WT/pdr3DNΔ).
As our findings indicated that this uniquely mutated allele has a dominant negative (DN) effect on the proteasomal function, it is referred to as PDR3DN in this study. In addition, the PDR3WT/pdr3DNΔ cells exhibited slightly faster fermentation after freezing preservation than the pdr3WTΔ/PDR3DN cells (Fig. 5C). The total carbon dioxide emission during 90 min fermentation using the pdr3WTΔ/PDR3DN strain was 6.4% lower than the level with the PDR3WT/pdr3DNΔ strain. The cell viability after 2 weeks of freezing was significantly lower in the pdr3WTΔ/PDR3DN strain than in the PDR3WT/pdr3DNΔ strain (Fig. 5D). Based on these results, it was confirmed that the PDR3DN allele is at least partly associated with the slow fermentation progression and the low cell viability of YF cells after exposure to freeze-thaw stress.
DISCUSSION
The control of proteasome gene expression in eukaryotic cells is complex. Although it has been speculated that cells effectively degrade the proteins damaged by freezing and/or thawing for adaptation, the significance of proteasome gene expression in freeze-thaw stress responses is not fully understood. In the yeast S. cerevisiae, the C2H2-type zinc finger transcriptional activator Rpn4p is required for the basal expression of the proteasome genes with a consensus PACE sequence (12, 13). Rpn4p is highly susceptible to proteasomal proteolysis as a substrate, and it thus regulates the proteasome gene expression in a negative-feedback manner. The basis of proteasome homeostasis is probably conserved in mammalian cells; the cap'n'collar (CNC) family bZIP transcription factor Nrf1 upregulates all proteasome subunits and mediates the recovery from inhibition of the proteasome (21–23), as yeast Rpn4p does. Thus, Rpn4p research may contribute to the elucidation of the mechanisms of human diseases that are caused by a defective Nrf1-mediated expression of the proteasome genes, such as neurodegenerative disorders.
In our study, freezing preservation led to low expression levels of the proteasome genes in two different strains of baker's yeast, and the extent of the decrease was correlated with the defects in fermentation performance after exposure to freeze-thaw stress. This finding suggests that the proteasome gene expression is one of the major targets damaged by freeze-thaw stress, highlighting an important role of the inductive pathway for the proteasome genes for cellular adaptation. Although we first focused on the RPN4 gene, the amino acid substitutions S74L and T427A identified in the freezing-sensitive strain did not significantly affect the tolerance to MG132 (data not shown).
The RPN4 gene is controlled not only by the proteasome activity but also at the transcriptional level by different transcription factors, Pdr1/3p, Yap1p, and Hsf1p (15–17). Homologous Pdr1/3p govern the gene expression of the multidrug efflux pumps for pleiotropic drug resistance, as well as that of RPN4 (24). Although Pdr1p plays a predominant role as a master regulator in most situations, the backup and extra functions of Pdr3p are also important. The expression of PDR3 (not PDR1) can be strongly induced through positive autoregulation by Pdr1/3p (25) when the activity of Pdr1p alone is deficient or insufficient. Moreover, Pdr3p and two other transcription factors (Yap1p and Hsf1p) for oxidative stress tolerance and heat shock response, respectively, comprise a transcriptional positive regulatory network to upregulate the genes that encode Rpn4p and each other (16, 26). Pdr3p participates in other transcriptional processes that do not involve Pdr1p, such as retrograde response signaling and DNA damage responses (27, 28). In our present investigation, deletion of PDR1 or PDR3 impaired model dough fermentation after freezing preservation to the same extent as disruption of the RPN4 gene, suggesting the importance of individual transcription factors in freeze-thaw stress responses (Fig. 6).
FIG 6.
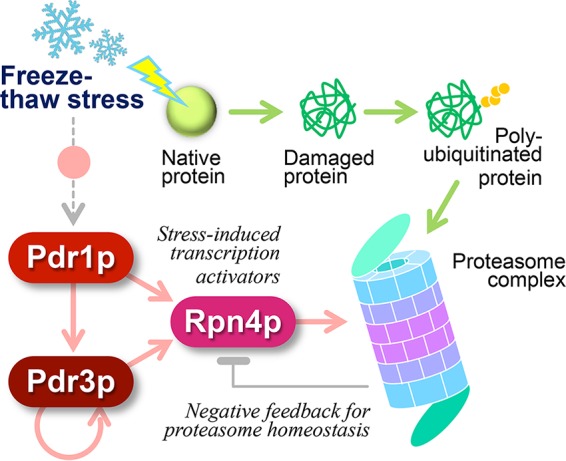
Proteasome gene expression as a freeze-thaw stress response in S. cerevisiae.
Although the PDR3 mutation responsible for the dominant negative phenotype of the freezing-sensitive baker's yeast strain has not yet been determined, two amino acid substitutions (i.e., A229V and H336R) in the conserved inhibitory domain (29) might be candidates for inhibition of the transcriptional induction activity. Alternatively, the responsible mutations might cause dominant negative inhibition by affecting the formation of Pdr3p homodimer and Pdr1/3p heterodimers (30), which may control the DNA-binding activity.
In the proteasome gene induction pathway mediated by Rpn4p and Pdr1/3p (Fig. 6), a question remains regarding freeze-thaw stress sensing. It was recently reported that the master regulator Pdr1p is activated by the molecular chaperones Ssz1p and Zuo1p (31), both of which are members of the conserved ribosome-associated complex (RAC) (32, 33). RAC assists the biogenesis of newly synthesized polypeptide chains at ribosomes and contributes not only to nascent protein folding but also to pleiotropic drug resistance and cold tolerance (33, 34). Thus, RAC might directly sense the nascent proteins damaged by freeze-thawing stress to upregulate the proteasome gene expression through Pdr1/3p and Rpn4p. This hypothesis should be addressed in future research.
Our present findings can be applied to the breeding of industrial strains that are optimal for the freezing preservation of fresh yeast products. Intriguingly, strain YF in this study was actually developed for superior fermentation performance in frozen doughs (data not shown), although it showed higher sensitivity to freezing preservation. Based on this finding, we should keep in mind that the stresses to be overcome during the freezing preservation of fresh yeast and during frozen dough fermentation are unlikely to be completely identical. It should be also noted that deletion of the RPN4 gene impaired model dough fermentation after freezing preservation (Fig. 4B) but led to slightly faster fermentation after refrigerating preservation compared to the wild-type strain (see Fig. S1 in the supplemental material).
Although the fermentation performance of the rpn4Δ strain in frozen doughs was not tested in this study, we speculate that the Pdr1/3p-Rpn4p pathway might be crucial for adaptation to freezing preservation but not necessarily for dough fermentation. This is consistent with our previous finding that unnecessary stress response mechanisms should be eliminated for high-efficiency fermentation (35, 36). Therefore, YF might have been selected as a strain that exhibits superior fermentation even after doughs are frozen, but the freeze-thaw tolerance of this strain as fresh yeast products is not high. If only the freezing preservation is considered, the resistance to the proteasome inhibitor MG132 in the presence of DMSO correlates with the freeze-thaw stress tolerance (Fig. 3C and 5B). The known cellular effects of DMSO on oxidative stress responses and on carbohydrate/nitrogen/lipid metabolism (37, 38) may be related to the proteasome gene expression via unknown mechanisms. Our findings thus provide clues for the development of a novel method for the breeding of freezing-tolerant industrial strains without the use of recombinant DNA technology.
MATERIALS AND METHODS
Yeast strains.
Two industrial baker's yeast strains (45 and YF) used at TableMark Co., Ltd., were used in this study. 45 is a traditional yeast strain previously used at TableMark Co., Ltd., but has never been published elsewhere. YF was also developed and patented at TableMark Co., Ltd. YF has been constructed for decades through chemical mutagenesis and crossbreeding and may partly share an ancestor with 45, although the detailed breeding information has been lost. The whole-genome resequencing of YF in this study may clarify the precise origin of this strain in the future. Since their fermentation abilities in frozen dough have been compared for a long time at TableMark Co., Ltd., we chose these two strains to be used in this study.
The S. cerevisiae laboratory strain BY4741 (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0) and its single-deletion mutants were provided by Euroscarf (Germany). Disruption of one PDR3 allele in diploid YF was performed using a PCR-based method (39) with oligonucleotide primers PDR3-DF (5′-CGA CAA CTG CAT CAG CAG TTT TAT TAA TTT TTT CTT ATT GCG TGA CCG CAC GGA TCC CCG GGT TAA TTA A-3′) and PDR3-DR (5′-GTG TCC CAT TTA CTA TGG TTA TGC TCT GCT TCC CTA TTT TCT TTG CGT TTG AAT TCG AGC TCG TTT AAA C-3′) and pFA6a-kanMX4 (39) as the template. Correct disruption of the PDR3WT or PDR3DN allele was confirmed by genomic PCR and direct DNA sequencing of the PCR product. Yeast cells were routinely grown in liquid YPD medium (1% yeast extract, 2% peptone, and 2% glucose) at 30°C, unless otherwise stated.
Yeast preservation and model dough fermentation tests.
For measurements of fermentation progression in model dough, yeast cells were precultured in YPD medium at 30°C and washed with distilled water. Wet yeast pellets were stored in the refrigerator (4°C) or in the freezer (−20°C) for 2 weeks. Approximately 0.4 g of yeast cells was inoculated into 30 ml of modified ASF medium (40), which consists of 23 g of sucrose, 2 g of urea, 1 g of ammonium sulfate, 0.8 g of magnesium sulfate, 1.6 mg of thiamine hydrochloride, 1.6 mg of pyridoxine hydrochloride, and 16 mg of nicotinic acid in a total of 350 ml of 67 mM potassium phosphate buffer (pH 5.6), and incubated at 30°C. When the laboratory strains were used, 70 mg of histidine, 70 mg of methionine, 70 mg of uracil, and 350 mg of leucine were also included in 350 ml of modified ASF medium to complement the auxotrophy. The course of the fermentation was continuously monitored by measuring the volume of evolved carbon dioxide gas using a Fermograph II apparatus (Atto). In preliminary experiments, we observed that 2-week refrigeration preservation did not affect the fermentation rates in either strain.
DNA microarray analysis of yeast cells during model dough fermentation.
Cells were sampled 30 min after inoculation into modified ASF medium. Total RNA extraction was performed using the RNeasy minikit (Qiagen). Cy3-labeled cRNA was prepared from 50 ng of total RNA using the low-input Quick Amp labeling kit (Agilent) according to the manufacturer's instructions, followed by purification with the RNeasy minikit (Qiagen). By using the gene expression hybridization kit (Agilent), Cy3-labeled cRNA were fragmented at 60°C for 30 min in a reaction volume of 24 μl containing 1× Agilent fragmentation buffer and 2× Agilent blocking agent. After the fragmentation reaction, 25 μl of 2× Agilent hybridization buffer was added to the sample, and cRNA was hybridized to the Agilent Technologies yeast (v2) gene expression 8 × 15K microarray for 17 h at 65°C in a rotating Agilent hybridization oven. After hybridization, the microarray was washed for 1 min at room temperature with wash buffer 1 and for 1 min at 37°C with wash buffer 2 of the gene expression wash buffer (Agilent), air dried immediately, and then scanned on the Agilent DNA microarray scanner (scan, resolution 5 μm; TIFF file dynamic change, 20 bit). The scanned images were analyzed with Feature Extraction Software 10.7.3.1 (Agilent), and the data were imported to the GeneSpring software (Agilent) and normalized by the 75-percentile shift. The microarray data were deposited to National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO). We performed GO analysis of the genes that were differentially expressed more than 2-fold between the refrigerated and frozen samples (listed in Table S1 in the supplemental material), using SGD GO Term Finder (http://www.yeastgenome.org/cgi-bin/GO/goTermFinder.pl).
Western blot analysis.
For the detection of ubiquitinated proteins upon heat shock, log-phase yeast cells grown in YPD medium at 30°C were shifted to 42°C for 15 min. For the detection of ubiquitinated proteins after freezing preservation, wet yeast pellets were stored in the freezer (−20°C) for 2 weeks and used for model dough fermentation as described above. Approximately 108 cells were collected, suspended in 10% trichloroacetic acid, and disrupted with glass beads in a Multibead Shocker (Yasui Kikai). Proteins in the whole-cell extracts were separated by SDS-PAGE (7.5% polyacrylamide), transferred to a polyvinylidene difluoride membrane, blocked with 3% powdered milk in Tris-buffered saline with Tween 20 (TBS-T) at 4°C overnight, and reacted with an antiubiquitin mouse antibody (P4D1; Santa Cruz Biotechnology) in Can Get signal immunoreaction enhancer solution 1 (Toyobo) at 1:2,000 dilutions for 60 min. As a protein loading control, an anti-glyceraldehyde-3-phosphate dehydrogenase (anti-GAPDH) rabbit antibody (Nordic Immunological Laboratories) or an anti-Pgk1p mouse antibody (Invitrogen) in Can Get signal immunoreaction enhancer solution 1 (Toyobo) at 1:10,000 dilutions was used as a primary antibody for 60 min. After several washing steps with TBS-T, the membrane was incubated for 40 min with horseradish peroxidase-conjugated anti-rabbit or anti-mouse IgG in Can Get signal immunoreaction enhancer solution 2 (Toyobo) at 1:2,000 dilutions as a secondary antibody. After several washing steps with TBS-T, the target proteins were visualized with the Pierce ECL Plus Western blotting substrate (Thermo Scientific) and detected using the Fuji LAS4000 imager (GE Healthcare).
Whole-genome resequencing analysis.
The whole genome of strain YF was analyzed by pair-end sequencing using the Illumina HiSeq 2500 sequencing system (Illumina) at Hokkaido System Science Co., Ltd. (Japan). This sequencing run yielded 45,740,770 high-quality filtered reads with 100-bp pair-end sequencing, providing approximately 308-fold genome coverage on average. The reads were mapped to the reference S. cerevisiae S288C genome using BWA, indexed with SAMtools, and realigned with GATK (see reference 41 for SAMtools and related tools). Duplicated read pairs were removed, and the remaining reads were realigned with GATK again. The final BAM file was deposited to NCBI BioProject. Based on this BAM file, variant calling was performed using SAMtools and BCFtools before variant annotation and filtering were performed using SnpEff.
Accession number(s).
The DNA microarray data for 45 and YF cells during model dough fermentation after 2 weeks of refrigeration or freezing can be accessed under GEO accession number GSE101071. Sequencing reads for the whole genome of YF can be accessed under BioProject accession number PRJNA392967.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Hisanori Endo (Fuji Foods Corporation) for providing the opportunity for collaboration. The DNA microarray analysis and the whole-genome resequencing experiment were carried out by Hokkaido System Science Co., Ltd.
We declare no conflicts of interest.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.00406-18.
REFERENCES
- 1.George RM. 1993. Freezing processes used in the food industry. Trends Food Sci Technol 4:134–138. doi: 10.1016/0924-2244(93)90032-6. [DOI] [Google Scholar]
- 2.Li B, Sun DW. 2002. Novel methods for rapid freezing and thawing of foods—a review. J Food Eng 54:175–182. doi: 10.1016/S0260-8774(01)00209-6. [DOI] [Google Scholar]
- 3.Hatano S, Udou M, Koga N, Honjoh K, Miyamoto T. 1996. Impairment of the glycolytic system and actin in baker's yeast during frozen storage. Biosci Biotechnol Biochem 60:61–64. doi: 10.1271/bbb.60.61. [DOI] [PubMed] [Google Scholar]
- 4.Ribotta PD, León AE, Añón MC. 2003. Effects of yeast freezing in frozen dough. Cereal Chem 80:454–458. doi: 10.1094/CCHEM.2003.80.4.454. [DOI] [Google Scholar]
- 5.O'Brien SS, Lindsay D, Von Holy A. 2008. Microbiological shelf-life studies on commercially manufactured yeast. J Food Qual 31:627–644. doi: 10.1111/j.1745-4557.2008.00225.x. [DOI] [Google Scholar]
- 6.Attfield PV. 1997. Stress tolerance: the key to effective strains of industrial baker's yeast. Nat Biotechnol 15:1351–1357. doi: 10.1038/nbt1297-1351. [DOI] [PubMed] [Google Scholar]
- 7.Shima J, Takagi H. 2009. Stress-tolerance of baker's-yeast (Saccharomyces cerevisiae) cells: stress-protective molecules and genes involved in stress tolerance. Biotechnol Appl Biochem 53:155–164. doi: 10.1042/BA20090029. [DOI] [PubMed] [Google Scholar]
- 8.Randez-Gil F, Córcoles-Sáez I, Prieto JA. 2013. Genetic and phenotypic characteristics of baker's yeast: relevance to baking. Annu Rev Food Sci Technol 4:191–214. doi: 10.1146/annurev-food-030212-182609. [DOI] [PubMed] [Google Scholar]
- 9.Takagi H, Iwamoto F, Nakamori S. 1997. Isolation of freeze-tolerant laboratory strains of Saccharomyces cerevisiae from proline-analogue-resistant mutants. Appl Microbiol Biotechnol 47:405–411. doi: 10.1007/s002530050948. [DOI] [PubMed] [Google Scholar]
- 10.Takagi H. 2008. Proline as a stress protectant in yeast: physiological functions, metabolic regulations, and biotechnological applications. Appl Microbiol Biotechnol 81:211–223. doi: 10.1007/s00253-008-1698-5. [DOI] [PubMed] [Google Scholar]
- 11.Tsolmonbaatar A, Hashida K, Sugimoto Y, Watanabe D, Furukawa S, Takagi H. 2016. Isolation of baker's yeast mutants with proline accumulation that showed enhanced tolerance to baking-associated stresses. Int J Food Microbiol 238:233–240. doi: 10.1016/j.ijfoodmicro.2016.09.015. [DOI] [PubMed] [Google Scholar]
- 12.Finley D, Ulrich HD, Sommer T, Kaiser P. 2012. The ubiquitin-proteasome system of Saccharomyces cerevisiae. Genetics 192:319–360. doi: 10.1534/genetics.112.140467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dohmen RJ, Willers I, Marques AJ. 2007. Biting the hand that feeds: Rpn4-dependent feedback regulation of proteasome function. Biochim Biophys Acta 1773:1599–1604. doi: 10.1016/j.bbamcr.2007.05.015. [DOI] [PubMed] [Google Scholar]
- 14.Xie Y, Varshavsky A. 2001. RPN4 is a ligand, substrate, and transcriptional regulator of the 26S proteasome: a negative feedback circuit. Proc Natl Acad Sci U S A 98:3056–3061. doi: 10.1073/pnas.071022298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Owsianik G, Balzi E, Ghislain M. 2002. Control of 26S proteasome expression by transcription factors regulating multidrug resistance in Saccharomyces cerevisiae. Mol Microbiol 43:1295–1308. doi: 10.1046/j.1365-2958.2002.02823.x. [DOI] [PubMed] [Google Scholar]
- 16.Hahn JS, Neef DW, Thiele DJ. 2006. A stress regulatory network for co-ordinated activation of proteasome expression mediated by yeast heat shock transcription factor. Mol Microbiol 60:240–251. doi: 10.1111/j.1365-2958.2006.05097.x. [DOI] [PubMed] [Google Scholar]
- 17.Wang X, Xu H, Ju D, Xie Y. 2008. Disruption of Rpn4-induced proteasome expression in Saccharomyces cerevisiae reduces cell viability under stressed conditions. Genetics 180:1945–1953. doi: 10.1534/genetics.108.094524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kruegel U, Robison B, Dange T, Kahlert G, Delaney JR, Kotireddy S, Tsuchiya M, Tsuchiyama S, Murakami CJ, Schleit J, Sutphin G, Carr D, Tar K, Dittmar G, Kaeberlein M, Kennedy BK, Schmidt M. 2011. Elevated proteasome capacity extends replicative lifespan in Saccharomyces cerevisiae. PLoS Genet 7:e1002253. doi: 10.1371/journal.pgen.1002253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rolland F, Winderickx J, Thevelein JM. 2002. Glucose-sensing and -signalling mechanisms in yeast. FEMS Yeast Res 2:183–201. [DOI] [PubMed] [Google Scholar]
- 20.Decottignies A, Lambert L, Catty P, Degand H, Epping EA, Moye-Rowley WS, Balzi E, Goffeau A. 1995. Identification and characterization of SNQ2, a new multidrug ATP binding cassette transporter of the yeast plasma membrane. J Biol Chem 270:18150–18157. doi: 10.1074/jbc.270.30.18150. [DOI] [PubMed] [Google Scholar]
- 21.Radhakrishnan SK, Lee CS, Young P, Beskow A, Chan JY, Deshaies RJ. 2010. Transcription factor Nrf1 mediates the proteasome recovery pathway after proteasome inhibition in mammalian cells. Mol Cell 38:17–28. doi: 10.1016/j.molcel.2010.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sha Z, Goldberg AL. 2014. Proteasome-mediated processing of Nrf1 is essential for coordinate induction of all proteasome subunits and p97. Curr Biol 24:1573–1583. doi: 10.1016/j.cub.2014.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bugno M, Daniel M, Chepelev NL, Willmore WG. 2015. Changing gears in Nrf1 research, from mechanisms of regulation to its role in disease and prevention. Biochim Biophys Acta 1849:1260–1276. doi: 10.1016/j.bbagrm.2015.08.001. [DOI] [PubMed] [Google Scholar]
- 24.Moye-Rowley WS. 2003. Transcriptional control of multidrug resistance in the yeast Saccharomyces. Prog Nucleic Acid Res Mol Biol 73:251–279. doi: 10.1016/S0079-6603(03)01008-0. [DOI] [PubMed] [Google Scholar]
- 25.Delahodde A, Delaveau T, Jacq C. 1995. Positive autoregulation of the yeast transcription factor Pdr3p, which is involved in control of drug resistance. Mol Cell Biol 15:4043–4051. doi: 10.1128/MCB.15.8.4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ma M, Liu ZL. 2010. Comparative transcriptome profiling analyses during the lag phase uncover YAP1, PDR1, PDR3, RPN4, and HSF1 as key regulatory genes in genomic adaptation to the lignocellulose derived inhibitor HMF for Saccharomyces cerevisiae. BMC Genomics 11:660. doi: 10.1186/1471-2164-11-660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Devaux F, Carvajal E, Moye-Rowley S, Jacq C. 2002. Genome-wide studies on the nuclear PDR3-controlled response to mitochondrial dysfunction in yeast. FEBS Lett 515:25–28. doi: 10.1016/S0014-5793(02)02387-6. [DOI] [PubMed] [Google Scholar]
- 28.Zhu Y, Xiao W. 2004. Pdr3 is required for DNA damage induction of MAG1 and DDI1 via a bi-directional promoter element. Nucleic Acids Res 32:5066–5075. doi: 10.1093/nar/gkh838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vermitsky JP, Edlind TD. 2004. Azole resistance in Candida glabrata: coordinate upregulation of multidrug transporters and evidence for a Pdr1-like transcription factor. Antimicrob Agents Chemother 48:3773–3781. doi: 10.1128/AAC.48.10.3773-3781.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mamnun YM, Pandjaitan R, Mahé Y, Delahodde A, Kuchler K. 2002. The yeast zinc finger regulators Pdr1p and Pdr3p control pleiotropic drug resistance (PDR) as homo- and heterodimers in vivo. Mol Microbiol 46:1429–1440. doi: 10.1046/j.1365-2958.2002.03262.x. [DOI] [PubMed] [Google Scholar]
- 31.Prunuske AJ, Waltner JK, Kuhn P, Gu B, Craig EA. 2012. Role for the molecular chaperones Zuo1 and Ssz1 in quorum sensing via activation of the transcription factor Pdr1. Proc Natl Acad Sci U S A 109:472–477. doi: 10.1073/pnas.1119184109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gautschi M, Lilie H, Fünfschilling U, Mun A, Ross S, Lithgow T, Rücknagel P, Rospert S. 2001. RAC, a stable ribosome-associated complex in yeast formed by the DnaK-DnaJ homologs Ssz1p and zuotin. Proc Natl Acad Sci U S A 98:3762–3767. doi: 10.1073/pnas.071057198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hundley H, Eisenman H, Walter W, Evans T, Hotokezaka Y, Wiedmann M, Craig E. 2002. The in vivo function of the ribosome-associated Hsp70, Ssz1, does not require its putative peptide-binding domain. Proc Natl Acad Sci U S A 99:4203–4208. doi: 10.1073/pnas.062048399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Conz C, Otto H, Peisker K, Gautschi M, Wölfle T, Mayer MP, Rospert S. 2007. Functional characterization of the atypical Hsp70 subunit of yeast ribosome-associated complex. J Biol Chem 282:33977–33984. doi: 10.1074/jbc.M706737200. [DOI] [PubMed] [Google Scholar]
- 35.Watanabe D, Araki Y, Zhou Y, Maeya N, Akao T, Shimoi H. 2012. A loss-of-function mutation in the PAS kinase Rim15p is related to defective quiescence entry and high fermentation rates of Saccharomyces cerevisiae sake yeast strains. Appl Environ Microbiol 78:4008–4016. doi: 10.1128/AEM.00165-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Watanabe D, Zhou Y, Hirata A, Sugimoto Y, Takagi K, Akao T, Ohya Y, Takagi H, Shimoi H. 2016. Inhibitory role of Greatwall-like protein kinase Rim15p in alcoholic fermentation via upregulating the UDP-glucose synthesis pathway in Saccharomyces cerevisiae. Appl Environ Microbiol 82:340–351. doi: 10.1128/AEM.02977-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang W, Needham DL, Coffin M, Rooker A, Hurban P, Tanzer MM, Shuster JR. 2003. Microarray analyses of the metabolic responses of Saccharomyces cerevisiae to organic solvent dimethyl sulfoxide. J Ind Microbiol Biotechnol 30:57–69. doi: 10.1007/s10295-002-0012-2. [DOI] [PubMed] [Google Scholar]
- 38.Sadowska-Bartosz I, Pączka A, Mołoń M, Bartosz G. 2013. Dimethyl sulfoxide induces oxidative stress in the yeast Saccharomyces cerevisiae. FEMS Yeast Res 13:820–830. doi: 10.1111/1567-1364.12091. [DOI] [PubMed] [Google Scholar]
- 39.Goldstein AL, McCusker JH. 1999. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast 15:1541–1553. doi:. [DOI] [PubMed] [Google Scholar]
- 40.Atkin L, Schultz AZ, Frey CN. 1945. Influence of dough constituents on fermentation. Cereal Chem 22:321–333. [Google Scholar]
- 41.Li H. 2011. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 27:2987–2993. doi: 10.1093/bioinformatics/btr509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang X, Xu H, Ha SW, Ju D, Xie Y. 2010. Proteasomal degradation of Rpn4 in Saccharomyces cerevisiae is critical for cell viability under stressed conditions. Genetics 184:335–342. doi: 10.1534/genetics.109.112227. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.