Abstract
Schizophrenia (SCZ), bipolar disorder (BD) and recurrent major depressive disorder (rMDD) are common psychiatric illnesses. All have been associated with lower cognitive ability, and show evidence of genetic overlap and substantial evidence of pleiotropy with cognitive function and neuroticism. Disrupted in schizophrenia 1 (DISC1) protein directly interacts with a large set of proteins (DISC1 Interactome) that are involved in brain development and signaling. Modulation of DISC1 expression alters the expression of a circumscribed set of genes (DISC1 Regulome) that are also implicated in brain biology and disorder. Here we report targeted sequencing of 59 DISC1 Interactome genes and 154 Regulome genes in 654 psychiatric patients and 889 cognitively-phenotyped control subjects, on whom we previously reported evidence for trait association from complete sequencing of the DISC1 locus. Burden analyses of rare and singleton variants predicted to be damaging were performed for psychiatric disorders, cognitive variables and personality traits. The DISC1 Interactome and Regulome showed differential association across the phenotypes tested. After family-wise error correction across all traits (FWERacross), an increased burden of singleton disruptive variants in the Regulome was associated with SCZ (FWERacross P=0.0339). The burden of singleton disruptive variants in the DISC1 Interactome was associated with low cognitive ability at age 11 (FWERacross P=0.0043). These results identify altered regulation of schizophrenia candidate genes by DISC1 and its core Interactome as an alternate pathway for schizophrenia risk, consistent with the emerging effects of rare copy number variants associated with intellectual disability.
Introduction
Schizophrenia (SCZ), bipolar disorder (BD) and recurrent major depressive disorder (rMDD) affect tens of millions of people worldwide. These disorders are moderately heritable and family history is a strong predictor of risk. Genome-wide association studies (GWAS), structural variant analyses and genome sequencing studies have identified that common single-nucleotide variants (SNVs), low penetrant rare SNVs, moderate to high penetrant copy number variants (CNVs) and potentially causal de novo mutations each play a role in the genetic etiology of SCZ and BD and, to a lesser extent, in rMDD.1, 2, 3, 4
There is now strong evidence for shared genetic risk across traditional diagnostic boundaries supporting the observation of ‘mixed’ diagnoses families.5, 6 GWAS studies capture common, ancient variation and point to an additive, polygenic architecture that transcends psychiatric diagnoses to predict cognitive ability variables.1, 7, 8 Lower cognitive function, both premorbid and post-onset, has been associated with these disorders, and recently polygenic risk score analysis has suggested a small, but significant, genetic correlation between risk for major mental illness and cognitive ability.9
In a complementary fashion to common variants identified in GWAS, rare variants have been identified that segregate with psychiatric disorder in a quasi-Mendelian manner and impact upon normal cognitive function.10, 11 One such example is a balanced t(1;11) (q42;q14) translocation in the Disrupted in Schizophrenia 1 (DISC1) gene, which was identified in a large Scottish pedigree highly burdened with SCZ, BD and rMDD.12 Independent reports of linkage and association have since reported evidence for region-wide association of DISC1 variants, or more commonly haplotypes, with these and other psychiatric disorders as well as for cognitive and neuropsychological traits.13, 14, 15 Although DISC1 itself is not a GWAS significant finding, its interactor PDE4B and regulated gene NRXN1 are reported as significant.16, 17 Convergent functional genomics approaches integrating the functional and genotypic data continue to support involvement of DISC1 disruption in schizophrenia and related biological pathways.18, 19
Recently, we reported deep sequencing of the DISC1 locus (528 kb) in 1542 samples that identified 2010 rare variants, of which ~60% were novel.17 We identified a common intronic variant with region-wide association for rMDD, and a rare missense mutation (R37W), previously reported in a SCZ case,20 in an individual with rMDD and in additional family members with mental disorders.17 Burden analysis also identified nominal associations with measures of depressed mood and cognitive ability at age 11, age 70 and cognitive ageing (change in cognitive ability between age 11 and 70).17 Motivated by these findings, we hypothesized that further insights might emerge from a directly comparable study of the DISC1 pathway genes.21, 22, 23, 24
Molecular studies have shown that DISC1 functions as a scaffold protein that is critical in cell signaling, neuronal development and ontogenesis through multiple protein-protein interactions.25, 26 DISC1-interacting partners (DISC1 Interactome) are enriched for proteins known to be involved in neural proliferation, migration, signaling and synaptic function.14, 21, 22, 24, 27 Positive case-control associations have been reported with psychiatric disorders for the following DISC1 Interactome genes: ATF4, CIT, NDE1, PCM1, PDE4B, PDE4D and YWHAE.14 In addition, structural rearrangements of PDE4B and NDE1 have been reported in patients with SCZ.28, 29 Sequencing studies in SCZ have examined the burden of variants in DISC1 and its interacting partners with mixed results. Moens et al.30 sequenced DISC1 and ten interacting genes in 486 SCZ cases and 514 controls and observed an excess of rare missense variants in affected individuals. Kenny et al.31 sequenced the exons of 215 synaptic genes including DISC1 and 22 interaction partners in 147 individuals with autism (ASD), 273 with SCZ and 287 controls. There was no enrichment of loss-of-function (LoF) mutations in the subset of DISC1-interacting genes, but singleton LoF variants were identified in DISC1, DMD and TRAF3IP1 in patients with ASD. Although recent exome based studies of SCZ did not analyze a set of DISC1-interacting genes directly, an over representation of de novo and rare variation was observed in genes sets such as ARC-associated scaffold protein complex at the postsynaptic density.32 This finding suggests that the DISC1 Interactome may be enriched in genetic risk factors that function through perturbing neuronal development and function.32
In addition to the DISC1 Interactome, disruption of the expression of DISC1, or its interactors, has been shown to alter the expression of a further set of genes (DISC1 Regulome).23, 33, 34 This gene set is enriched for neurodevelopmental, synaptogenic and sensory perception genes as well as for registered drug targets for psychiatric and neurological disorders.23 We selected DISC1 Regulome genes on the basis of prior evidence of genetic association of common variants with psychiatric illness, as well as either: haplotype or model dependent expression patterns correlated with DISC1 expression,35 DISC1 alleles23 or with DISC1 Interactome genes; or proteins that directly interact with other Interactome gene proteins (Supplementary Information: gene selection).
Here we report the targeted sequencing of 59 DISC1 Interactome genes (including TSNAX and DISC1) and 154 DISC1 Regulome genes in the same cohort of subjects in which the full DISC1 locus (528 kb) was sequenced and reported.17 As for our previous study, we report the gene-wide and gene-set burden analysis of rare variants and singletons in the DISC1 Interactome and Regulome with respect to psychiatric disorders, associated personality traits, and cognitive variables.
Materials and methods
The materials and methods are described in full in the Supplementary Information. Briefly, we analyzed 1543 DNA samples comprising 654 cases (241 SCZ, 221 BD and 192 rMDD) from Scottish hospital patients and 889 community-dwelling, generally healthy older people from the Lothian Birth Cohort of 1936 (LBC1936), as described previously.17
A total of 213 genes were selected for sequence analysis (Table 1, Supplementary Information: gene selection, Supplementary Table S1). The DISC1 locus (DISC1, TSNAX and TSNAX-DISC1) and 56 direct DISC1 protein-protein interactors defined the DISC1 Interactome gene set. A total of 154 additional genes related to DISC1 expression from previous microarray analyzes comprised the DISC1 Regulome gene set. Genomic regions comprising ~11.7 Mb (3.3 Mb exons) were captured using a custom solution capture probe set (Roche NimbleGen, Pleasanton, CA, USA). Each sample capture was sequenced using a HiSeq2000 sequencer (Illumina, San Diego, CA, USA). Sequence reads were aligned to the human NCBI Build 36 (hg18) reference using BWA.36 Variant calling was performed using GATK,37 and high-quality SNVs were filtered by standardized filtering parameters. Using PLINK,38 we applied data quality control filters as described previously17, 39 to exclude samples and SNVs that introduce bias (Supplementary Figures S2–S5). Sanger sequencing was used to optimize the quality control filters and exclude all identified false positive SNVs from further analysis. SNVs were matched to hg19 coordinates using liftOver from UCSC, and ANNOVAR40 was used for variant annotation based on the human reference genome hg19 (RefSeq). The coding variants were grouped into three mutation classes, similar to previous analyses,4, 32 based on predicted functional effects: Disruptive, nonsense and splice site variants; NSstrict, disruptive plus missense variants predicted as damaging by all five algorithms (SIFT,41 PolyPhen2 HumDiv and HumVar,42 LRT,43 and MutationTaster44); NSbroad, disruptive plus missense variants predicted as damaging by at least one of the algorithms above; NSstrict is therefore a subset of NSbroad. The burden and accumulation rate of rare variants (MAF<1%) and singletons in these mutation classes was assessed in each of the case cohorts and a combined cohort of all diagnoses; and for cognitive measures: cognitive ability at ages 11 and 70, change in cognitive ability, crystallized cognitive ability and general cognitive ability; and personality traits: neuroticism, anxiety and depression (Supplementary Information: phenotypes). The gene set and gene-wide burden analyses for all genes containing more than one rare variant were performed using the R package ‘SKAT’.45 Exact Poisson tests were performed in R to evaluate the accumulation rates of singletons and rare variants under a Poisson distribution in cases compared to controls in each functional mutation class. To control for multiple testing and reduce the risk of false positives, a bootstrap resampling approach (n=10 000) was used to estimate the significance of all tests (Family-Wise Error Rate, FWER), taking into account multiple tests within each diagnosis (FWERwithin) and across all diagnoses (FWERacross)(Supplementary Information: rare variant burden analysis).
Table 1. DISC1 (Number 1) Interactome and Regulome.
| DISC1 Interactome | DISC1 Regulome | |||||
|---|---|---|---|---|---|---|
| AKAP9 | KCNQ1 | ACTG2 | CYB5R2 | HNRNPK | NSUN6 | SORT1 |
| AP4B1 | KIF3A | ADCY1 | DLG2 | HOOK3 | NUDT1 | SOX10 |
| AP4M1 | KIFAP3 | ADCY3 | DLGAP1 | HSPA2 | NUMBa | SRSF6 |
| APP | MACF1 | AKT1 | DMRT2 | ID1 | OGFR | STAT5A |
| ATF4 | MAP1Ab | ANKRD16 | DTNBP1 | IL9R | OLIG2 | SV2B |
| ATF5 | NDE1 | APPL2 | DUSP6 | ITPR1 | OPTN | SYN1 |
| ATF7IP | NDEL1 | ARC | DYNLL1 | JUNB | PAK3 | SYN2 |
| CCDC88A | OLFM1 | ARHGEF11 | EEF1E1 | KIF5Ca | PCBP1 | SYP |
| CDC5L | PAFAH1B1 | ATP6V0A1 | EEF2K | LIMA1 | PCM1a | TACR1 |
| CDK5RAP3 | PCNT | BANK1 | EGR2 | LITAF | PDE4A | TGFBI |
| CEP290 | PDE4B | BBS4 | EGR3 | LPIN1 | PITPNM1 | THOP1 |
| CITb | PDE4D | BCRa | EGR4 | MARCH3 | POU6F1 | TIMM8A |
| CLIC1 | RAD21 | BDNF | ERBB2 | MCPH1 | PPARGC1A | TMEM163 |
| CLU | RANBP9 | BEX2 | ERBB3 | MDFIC | PPP4R2 | TNFAIP8L2 |
| CTNNB1 | SH3BP5 | BLZF1 | ERBB4a | MEG3 | PRKCB | TNFRSF11B |
| DCTN1 | SMC3 | BRD1a | FHIT | MGLL | PRR18 | TP53RK |
| DCTN2 | SNX6 | C5orf48 | GABBR1 | MMP7 | PRRC2A | TRIOBP |
| DISC1b | SYNE1 | CACNA1C | GABRA5 | MTPAP | RAB3A | TSNAXIP1a |
| DIXDC1 | TNIK | CALM3 | GJA5a | MYRIP | RGS4 | TUBB3 |
| DPYSL3 | TRAF3IP1 | CAMK1D | GNB1L | NAPB | RHOA | UBL3 |
| DSTb | TRIO | CAMK4 | GRIA1 | NCOR2 | RPS6KA2 | UQCRC1a |
| EIF3H | TSNAX | CCND2 | GRIA2a | NDN | SCHIP1 | USP7 |
| EXOC4 | TSNAX-DISC1 | CCNE1 | GRIA3 | NEK1 | SERPINI1 | VASH2 |
| FBXO41 | TUBB | CETP | GRIN1 | NFKB2a | SKAP1 | VPS45 |
| FEZ1 | TUBG1 | CHMP1A | GRIN2Aa | NFKBIA | SLC17A7 | VRK2 |
| GNB1 | WNT3A | CHRNA5 | GRIN2B | NOS1 | SLC1A2a | WDFY1 |
| GRB2 | YWHAE | CLINT1 | GRM3 | NOS1AP | SLC41A2 | XPOT |
| GRIPAP1 | YWHAZ | COMMD1 | HERPUD1 | NOTCH4 | SMC1A | ZHX2 |
| GSK3B | CORO1B | HIPK2 | NRG1 | SNAP91 | ZIC1a | |
| ITSN1 | CPLX2 | HIST1H2BN | NRXN1a | SNPH | ZNF140 | |
| KALRN | CTTN | HLA-DQA1a | NRXN3 | SORCS3 | ||
Genes with a nominally significant burden p-values for schizophrenia (16 of 154 genes in the DISC1 Regulome). These gene level results did not survive family-wise error rate correction across all tests.
Genes with a nominally significant burden p-values for cognitive ability at age 11 (4 of 59 genes in the DISC1 Interactome).
Results
Targeted sequencing and genetic discovery in 213 DISC1 Interactome and Regulome genes
A total of 1464 samples (95%) were sequenced to a minimum coverage depth of 20x across at least 80% of the targeted bases (Supplementary Table S2). Coverage was uniform across all sample groups (Supplementary Figure S1). Following sequence- and variant-based quality filters, 196 080 SNVs in 1446 samples (211 cases of SCZ, 169 cases of rMDD, 195 cases of BD, and 871 controls from the LBC1936) remained for further analyses (Supplementary Table S2 and Supplementary Figures S2–S5). Of the 196 080 SNVs, 78% have a MAF less than 1%. Only 40% are reported in the 1000 Genome Project European subset (Supplementary Table S3). On the basis of RefSeq functional annotations using ANNOVAR, 169 905 SNVs mapped to introns, 5410 to 3′ or 5′ UTRs, and 4523 to coding regions. Of the 4523 exonic variants, 1893 were functionally classified with respect to coding potential as silent variants, 2569 as missense, and 41 as nonsense. A further 24 SNVs were annotated as splice site variants. SNVs showing greater functional impacts on protein function are more likely to be rare: 100% of nonsense and 92% of splice site variants have MAF <1%, compared to 79% of silent and 78% of intronic variants.
Analysis of genetic variation in the DISC1 Interactome and Regulome with psychiatric illness
Rare functional variant analysis in the DISC1 Interactome
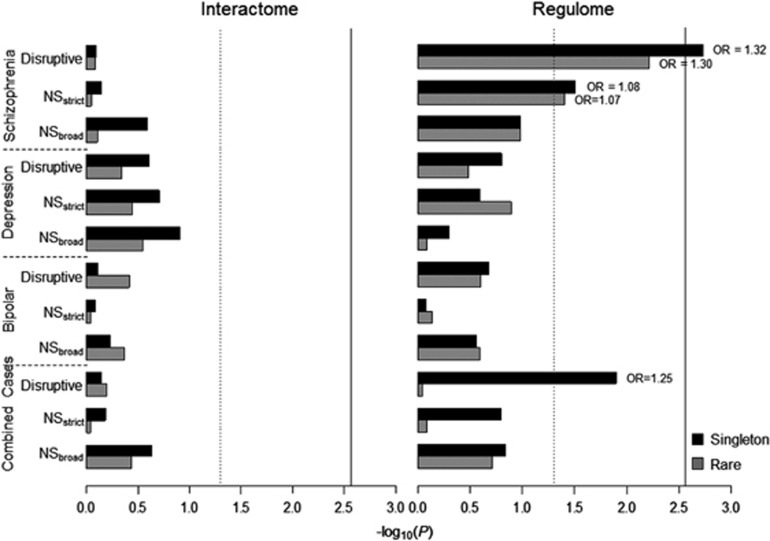
There was no significant burden of rare disruptive, NSstrict or NSbroad variants in SCZ, BD, or rMDD nor in a combined cohort of all diagnoses compared to controls in the DISC1 Interactome (Supplementary Table S6). There was a nominal association of fewer rare disruptive variants in SCZ (unadjusted P=0.0188), but no significant difference between the accumulation rate of rare variants for any diagnosis after Family-Wise Error Rate (FWER) correction (Supplementary Table S7). None of the proportions of NSstrict and NSbroad rare or singleton variants deviated from the null hypothesis after FWERacross correction.
The gene-wide burden of non-synonymous coding changes was nominally, but not significantly increased in psychiatric disorders (unadjusted P=0.0048–0.0488) for several DISC1 Interactome genes. None survived correction for multiple testing (Supplementary Table S8).
Rare functional variant analysis in the DISC1 Regulome
We analyzed the burden and accumulation rates of rare and singleton functional variants in the DISC1 Regulome. For SCZ compared to control samples, we observed a significantly increased burden of singleton disruptive variants (unadjusted P=0.0019, FWERwithin P=0.0069, FWERacross P=0.0339, OR=1.3162, SE=0.0941; Figure 1 and Supplementary Table S9), and a nominally higher accumulation rate (4.13-fold, unadjusted P=9.00 × 10−4, FWERwithin P=0.0185, FWERacross P=0.0965, Supplementary Table S10). In addition, the accumulation rate of rare disruptive variants, as opposed to singleton disruptive variants, was 3.47-fold higher in SCZ cases than in healthy controls and remained significant after multiple test correction (unadjusted P=1.68 × 10−6, FWERwithin P=1.00 × 10−4, FWERacross P=0.0022, Supplementary Table S10). Unlike singleton disruptive variants, although the burden of rare disruptive variants in SCZ was nominally significant, and survived FWER correction for all tests within the trait, it did not meet the threshold for tests across all traits (unadjusted P=0.0061, FWERwithin P=0.0228, FWERacross P=0.0863, Supplementary Table S9). We also observed a nominally higher proportion and burden of NSstrict singleton and rare variants in SCZ and disruptive singleton and rare variants in combined cases compared to controls, but none survived FWER for all tests across all traits (Supplementary Tables S9 and S10). There was no evidence for an increased overall burden in rMDD, BD or combined cases compared to controls after FWER correction across all traits.
Figure 1.
Gene set burden analysis of rare functional variants for case-control traits. Case-control gene set burden analysis of singletons and rare variants (MAF<1%) in the DISC1 Interactome (left) and Regulome (right). x-axis represents –log10(P), vertical dashed line: P=0.05, vertical solid line: FWERacross P=0.05; Odds ratio (OR) is labeled for the significant tests with P<0.05. Disruptive mutations, which included nonsense and splice site variants; NSstrict, Non-synonymous strict damaging mutations which included disruptive variants plus missense variants predicted as damaging by all five algorithms (PolyPhen2 HumDiv and HumVar, SIFT, LRT and MutationTaster); NSbroad, Non-synonymous broad damaging mutations which included disruptive plus missense variants predicted as damaging by at least one of the algorithms above.
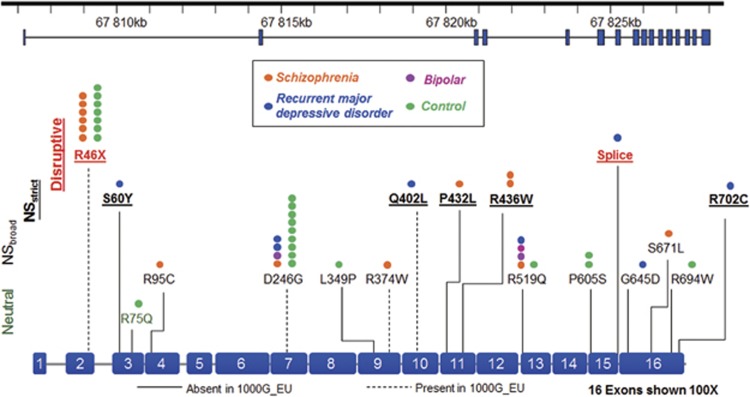
At the gene-wide level, Translin-associated factor X interacting protein 1 (TSNAXIP1) showed greater burden of NSstrict singletons in rMDD (unadjusted P=1.29 × 10−4, FWERwithin P=0.0253) and NSstrict rare variants in SCZ (unadjusted P=2.22 × 10−4, FWERwithin P=0.0410, Supplementary Table S11) compared to controls. However, these results did not survive correction for all tests (rMDD FWERacross P=0.0864, SCZ FWERacross P=0.1600). TSNAXIP1 has 16 exons encoding 712 amino acids. We validated 17 rare coding variants in TSNAXIP1 in all carriers, including 1 splice site, 1 nonsense and 15 missense variants (Figure 2 and Supplementary Table S12). Of these 17 rare substitutions, 4 were previously reported in the 1000 Genomes Project European subset. In total, 7 rare variants in TSNAXIP1 including 2 disruptive and 5 predicted damaging missense variants contributed to the gene burden analysis of NSstrict variants in rMDD and SCZ. In a ‘leave-one-out’ approach, we determined that the nonsense mutation at chr16:66405794 (rs146214814, p.R46X) located in exon 2, contributed most to the higher burden of NSstrict variants in SCZ. Relative to controls, this disruptive variant had a 3.58-fold higher allele frequency in SCZ (0.0146 vs 0.0041) and was not observed in any other mental illness cohort. Further information on the neurobiology of TSNAXIP1 is given in Supplementary Information: TSNAXIP1.
Figure 2.
Translin-associated factor X interacting protein 1 (TSNAXIP1) rare functional variants. TSNAXIP1 exon structure and mutations. Definitions of Disruptive (red), NSstrict (underlined) and NSbroad mutations are given in Figure 1. Neutral variants (green) were defined as missense variants predicted as not damaging by any of the five predictive algorithms. Dash lines represent the variants present in the European subset of the 1000 Genomes Project (1000G_EU), and the solid lines represent the variants present in the 1000G_EU. The number of circles represents the number of samples carry the rare variant.
Burden analysis of coding variants on quantitative cognitive ability and personality traits associated with psychiatric illness
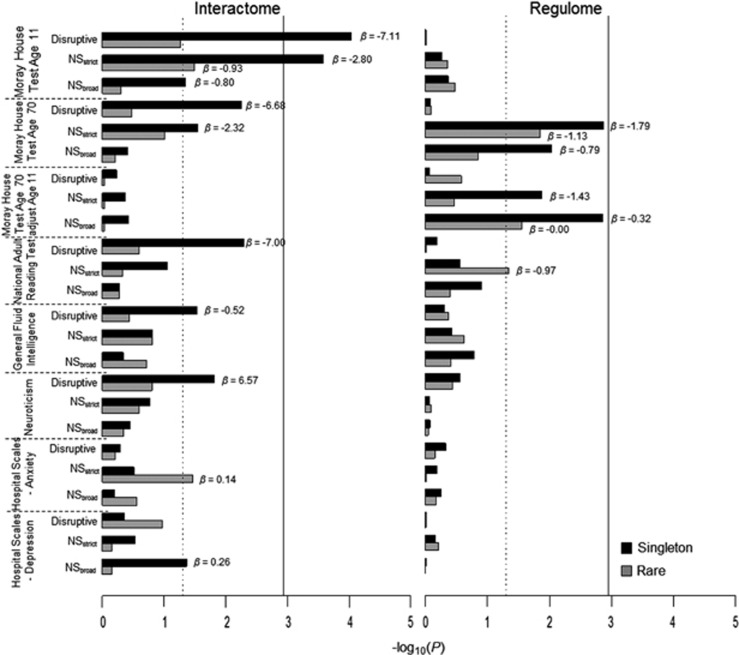
We found that a significantly higher burden of singleton disruptive variants in the DISC1 Interactome was associated with lower cognitive ability assessed by Moray House Test (MHT) scores at age 11 (unadjusted P=9.35 × 10−5, FWERwithin P=0.0005, FWERacross P=0.0043, β=−7.1141, SE=3.6863; Figure 3 and Supplementary Table S13). The burden of NSstrict singletons in the Interactome gene set was associated with lower MHT scores at age 11 (unadjusted P=0.0003, FWERwithin P=0.0017, FWERacross P=0.0122, β=−2.7865, SE=1.2877). In addition, although these did not survive FWERacross correction, nominally significant associations in the burden of disruptive singletons were observed with MHT scores at age 70 (unadjusted P=0.0056, β=−6.6785), National Adult Reading Test (unadjusted P=0.0051, β=−6.9970) and General Fluid Intelligence (unadjusted P=0.0293, β=−0.5152). Interestingly, there were nominally significant associations between the burden of rare functional variants and increased symptoms of neuroticism (Disruptive singletons: unadjusted P=0.0154, β=6.5671), anxiety (NSstrict rare variants: unadjusted P=0.0349, β=0.1394) and depression (NSbroad singletons: unadjusted P=0.0431, β=0.2587). At the gene-wide level, no association was found between the variability in cognitive ability or personality scores and the burden of damaging or disruptive variants in any specific gene of the DISC1 Interactome after FWERacross correction (Supplementary Table S14).
Figure 3.
Gene set burden analysis of rare functional variants for quantitative traits. Quantitative trait gene set burden analysis of singletons and rare variants (MAF<1%) in the DISC1 Interactome (left) and Regulome (right). x-axis represents –log10(P), vertical dashed line: P=0.05, vertical solid line: FWERacross P=0.05; Effect size (β, Beta) is labeled for the significant tests with P<0.05. See phenotype descriptions in Supplementary Information for all quantitative traits. Moray House Test is the mental ability test used in the LBC1936 cohort. A priori hypothesis is that an increased burden of rare predicted damaging variants would reduce scores for cognitive variables and increase scores for personality traits. The definition of the variant classes is given in Figure 1.
In the analysis of the DISC1 Regulome, we observed a burden of NSstrict singletons associated with lower MHT scores at age 70 (unadjusted P=0.0014, β=−1.7895; Figure 3 and Supplementary Table S15) that withstood FWER correction for all tests within the trait (FWERwithin P=0.0079), but not all tests across all traits (FWERacross P=0.0609). The burdens of NSstrict and NSbroad variants were nominally significantly associated with greater decrease in cognitive ability between the ages of 11 and 70 (NSstrict singletons: unadjusted P=0.0131, β=−1.4338; NSbroad singletons: unadjusted P=0.0014, β=−0.3175; NSbroad rare variants: unadjusted P=0.0280, β=−0.0010). At the gene-wide level, the strongest association with cognitive function was observed with rare and singleton NSstrict variants in CACNA1C, but this did not pass FWERacross correction (Supplementary Table S16).
Discussion
Encouraging progress towards delineating the genetic architecture of psychiatric disorders has been made and roles for both common, rare and de novo mutations established. Rare variants of high impact can provide valuable mechanistic insight. Recent case-control deep sequencing studies indicate that in individuals with SCZ rare loss-of-function variants are enriched in genes related to synaptic function,31 in target genes of the FMPRP32 and in genes known to be associated with SCZ.46 The biological impacts of several DISC1 missense variants identified through deep sequencing have been demonstrated.30, 47 We previously reported the discovery of rare disruptive DISC1 variants in individuals with psychiatric illness and demonstrated the biological impact of the p.R37W variant.17 Here we report the association of both clinical diagnoses and cognitive ability with rare variants in the DISC1 Interactome and the DISC1 Regulome.
Before discussing these positive findings, we first consider some limitations of the study. Although the sample size was large by current standards, these numbers are modest in size for comprehensive rare variant detection.17, 48 We were unable to perform sex-specific analyses in our study given our sample size. Such analyses may be important in our understanding of the relationships between genetic variants and gene expression particularly in psychiatric illness, given reports of sex-specific differences in gene expression in the brain49, 50 but also due to reports of sex-specific differences in association of variants and haplotypes in DISC1,51, 52, 53 the success of the CONVERGE strategy that relied on mapping loci for severe depression within a female-only cohort54 and the differences in disease presentation between sexes that have likewise been reported.55 Burden analysis increases the power of analyses in such small samples, but the rules for annotating rare variants as ‘damaging’ are far from foolproof: biological validation is required. Last, but not least, whole genome sequencing of all 1543 individuals, while ideal, was beyond the scope of our resources. Targeted capture sequencing was a practical option, but it is likely that relevant variants will have been missed by virtue of poor capture. It is also almost certainly the case that our list of bona fide DISC1 interactors is incomplete, and that contra wise, not all members of the Regulome that met our inclusion criteria will be regulated by DISC1 in practice.
Acknowledging these limitations, there were findings of note. No association was seen between rare variants in the DISC1 Interactome and any psychiatric diagnosis. There was, however, a significant excess of singleton disruptive variants in the DISC1 Regulome associated with SCZ, but not with BD or rMDD. We have shown that disruptive and NSstrict singleton variants in the DISC1 Interactome show significant association with cognitive ability at age 11. These classes of variants are also nominally associated in the DISC1 Regulome with cognitive ability at age 70 and change in cognitive ability between age 11 and 70. The DISC1 Regulome gene set was assembled from genes that show both i) altered expression in response to genetic variation in DISC1 or its interactors, or are themselves protein interactors of the core complex, and ii) evidence of association with psychiatric illness from candidate gene studies, or some of the earliest genome-wide association studies.1, 56, 57 We note that in this study, we found nominal association of rare Regulome variants with both increased schizophrenia risk and lower adult cognitive ability, particularly in older age. This mirrors the observation of association with common variants from the DISC1 Regulome in GWAS studies.8, 9, 58, 59 Overall, the patterns of associations seen across diagnostic and cognitive traits in the DISC1 Interactome and Regulome are consistent with the hypothesis that genetic disruption of DISC1 or its direct interactors has a proximal effect on cognitive ability and a distal effect, through regulation of gene expression, on schizophrenia risk in later life. Indeed, we have shown previously that disruption of the Disc1 gene in mice results in altered expression of Nrxn1,33 a gene in which copy number variation, common variants and rare variants are associated with schizophrenia.60, 61, 62 The hypothesis of a distal effect of variants in the Interactome on disease risk is also consistent with the recent association of copy number variants linked to intellectual disability with schizophrenia in a much larger sample.63
Four genes in the Interactome gene set, DISC1, CIT, DST and MAP1A, were nominally associated at the gene level with cognitive ability at age 11 (Table 1). CIT and MAP1A are known interactors with DLG4, PSD-95, which in turn interacts with the cytoplasmic tail of NMDA receptor subunits and with shaker-type potassium channels, regulating the ratio of excitatory to inhibitory synapses in the hippocampus.64, 65, 66 Because of the complex network multiplicity of protein interactions identified by the DISC1 Interactome gene set, we cannot conclude that our association with cognitive ability at age 11 is specific, or restricted to, the DISC1 Interactome or indeed that this phenotype is the only one likely to be associated with rare functional variants in this set of genes. This important caveat also applies not only to the DISC1 Regulome gene set, in which 16 genes were nominally associated at the gene level with schizophrenia (Table 1), but likely to the majority of brain-expressed gene sets. None of the gene level associations survived FWER correction across all tests.
To better understand the distinctive patterns of association between the DISC1 Interactome and Regulome, and their relationship to previously published mental health-related gene sets, we performed gene ontology (GO) enrichment analyses (Supplementary Tables S17, 18 and Supplementary Figures S9). The DISC1 Interactome is significantly enriched for proteins involved in regulation of nervous system development, microtubule cytoskeleton organization, and vesicle localization (Supplementary Tables S17, 18 and Supplementary Figures S9). Our findings suggest that disruptive singletons in these biological processes may make significant contributions to variability in cognitive function. These processes have also been associated with intellectual disability.67 Together, these datasets suggest that there is a spectrum of effect sizes or penetrance associated with genetic variants in this pathway. In contrast, the DISC1 Regulome is enriched for genes involved in synaptic transmission and glutamate-gated ion channel activity, reflecting the regulation by the DISC1 Interactome of these processes and their importance as inferred from GWAS. The specificity of the association between single disruptive Regulome variants and SCZ in our sample suggests a greater impact of glutamate dysregulation in this disorder than BD or rMDD. A role for DISC1 in glutamate-related processes has previously been suggested in both a mouse model and in the t(1;11) translocation family.68, 69 Comparison of the GO terms associated with both the DISC1 Interactome and Regulome reveals largely independent GO term associations with a very limited set of intersecting terms focused on negative regulation of cellular process, protein binding, and cell projections (Supplementary Figures S9).
In conclusion, and despite the limitations, these findings provide further genetic evidence to support the impact of both DISC1-interacting proteins and genes whose expression is modulated by genetic variants in the DISC1 pathway on schizophrenia.
Acknowledgments
This study was supported by a gift from T and V Stanley and a grant from NIH (R01MH102068). We thank the LBC1936 participants and team members who contributed to these studies. Phenotype collection was supported by Age UK (The Disconnected Mind project). The work was undertaken by The University of Edinburgh Centre for Cognitive Ageing and Cognitive Epidemiology, part of the cross council Lifelong Health and Wellbeing Initiative (MR/K026992/1). Funding from the BBSRC and Medical Research Council (MRC) is gratefully acknowledged. ST thanks supports from the Howard University startup funds (U100193) and Junior Faculty Writing & Creative Works Summer Academy. ST acknowledges Professor Fatimah Jackson for critical comments.
Author contributions
DJP and WRM designed the study, are PIs on the grant funding, supervised the study and supported the analysis. ST, SM, PT, VM, MLC and MK carried out the bioinformatics analysis. MK, SM and ST organized and conducted targeted resequencing and variant validation. ST, PT, SM, DJP and WRM wrote the manuscript, and all authors critically revised the manuscript.
Footnotes
Supplementary Information accompanies the paper on the Molecular Psychiatry website (http://www.nature.com/mp)
WRM has participated in Illumina sponsored meetings over the past 4 years, and received travel reimbursement and an honorarium for presenting at these events. Illumina had no role in decisions relating to the study/work to be published, the data collection and analysis of the data, and the decision to publish. WRM has participated in Pacific Biosciences sponsored meetings over the past 3 years and received travel reimbursement for presenting at these events. WRM is a founder and shareholder of Orion Genomics. WRM is a member of the scientific Advisory Board of RainDance, Inc. The remaining authors declare no conflicts of interest.
Supplementary Material
References
- Purcell SM, Wray NR, Stone JL, Visscher PM, O’Donovan MC, Sullivan PF et al. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature 2009; 460: 748–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SH, DeCandia TR, Ripke S, Yang J, Sullivan PF, Goddard ME et al. Estimating the proportion of variation in susceptibility to schizophrenia captured by common SNPs. Nat Genet 2012; 44: 247–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripke S, O’Dushlaine C, Chambert K, Moran JL, Kahler AK, Akterin S et al. Genome-wide association analysis identifies 13 new risk loci for schizophrenia. Nat Genet 2013; 45: 1150–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy SE, Gillis J, Kramer M, Lihm J, Yoon S, Berstein Y et al. De novo mutations in schizophrenia implicate chromatin remodeling and support a genetic overlap with autism and intellectual disability. Mol Psychiatry 2014; 19: 652–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesman II, Laursen TM, Bertelsen A, Mortensen PB. Severe mental disorders in offspring with 2 psychiatrically ill parents. Arch Gen Psychiatry 2010; 67: 252–257. [DOI] [PubMed] [Google Scholar]
- Lichtenstein P, Yip BH, Bjork C, Pawitan Y, Cannon TD, Sullivan PF et al. Common genetic determinants of schizophrenia and bipolar disorder in Swedish families: a population-based study. Lancet 2009; 373: 234–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray NR, Lee SH, Mehta D, Vinkhuyzen AAE, Dudbridge F, Middeldorp CM. Research Review: Polygenic methods and their application to psychiatric traits. J Child Psychol Psychiatry Allied Discip 2014; 55: 1068–1087. [DOI] [PubMed] [Google Scholar]
- McIntosh AM, Gow A, Luciano M, Davies G, Liewald DC, Harris SE et al. Polygenic risk for schizophrenia is associated with cognitive change between childhood and old age. Biol Psychiatry 2013; 73: 938–943. [DOI] [PubMed] [Google Scholar]
- Hagenaars SP, Harris SE, Davies G, Hill WD, Liewald DCM, Ritchie SJ et al. Shared genetic aetiology between cognitive functions and physical and mental health in UK Biobank (N=112 151) and 24 GWAS consortia. Mol Psychiatry 2016; 21: 1624–1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marioni RE, Penke L, Davies G, Huffman JE, Hayward C, Deary IJ. The total burden of rare, non-synonymous exome genetic variants is not associated with childhood or late-life cognitive ability. Proc Biol Sci 2014; 281: 20140117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefansson H, Meyer-Lindenberg A, Steinberg S, Magnusdottir B, Morgen K, Arnarsdottir S et al. CNVs conferring risk of autism or schizophrenia affect cognition in controls. Nature 2014; 505: 361–366. [DOI] [PubMed] [Google Scholar]
- Blackwood DH, Fordyce A, Walker MT St, Clair DM, Porteous DJ, Muir WJ. Schizophrenia and affective disorders—cosegregation with a translocation at chromosome 1q42 that directly disrupts brain-expressed genes: clinical and P300 findings in a family. Am J Hum Genet 2001; 69: 428–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carless MA, Glahn DC, Johnson MP, Curran JE, Bozaoglu K, Dyer TD et al. Impact of DISC1 variation on neuroanatomical and neurocognitive phenotypes. Mol Psychiatry 2011; 16: 1096–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradshaw NJ, Porteous DJ. DISC1-binding proteins in neural development, signalling and schizophrenia. Neuropharmacology 2012; 62: 1230–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnstone M, Thomson PA, Hall J, McIntosh AM, Lawrie SM, Porteous DJ. DISC1 in schizophrenia: genetic mouse models and human genomic imaging. Schizophr Bull 2011; 37: 14–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomppo L, Ekelund J, Lichtermann D, Veijola J, Ja M. DISC1 Conditioned GWAS for psychosis proneness in a Large Finnish Birth Cohort. PLoS One 2012; 7: 1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson PA, Parla JS, McRae AF, Kramer M, Ramakrishnan K, Yao J et al. 708 Common and 2010 rare DISC1 locus variants identified in 1542 subjects: analysis for association with psychiatric disorder and cognitive traits. Mol Psychiatry 2014; 19: 668–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayalew M, Le-Niculescu H, Levey DF, Jain N, Changala B, Patel SD et al. Convergent functional genomics of schizophrenia: from comprehensive understanding to genetic risk prediction. Mol Psychiatry 2012; 17: 887–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahoun T, Trossbach SV, Brandon NJ, Korth C, Howes OD. The impact of Disrupted-in-Schizophrenia 1 (DISC1) on the dopaminergic system: a systematicreview. Transl Psychiatry 2017; 7: e1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song W, Li W, Feng J, Heston LL, Scaringe WA, Sommer SS. Identification of high risk DISC1 structural variants with a 2% attributable risk for schizophrenia. Biochem Biophys Res Commun 2008; 367: 700–706. [DOI] [PubMed] [Google Scholar]
- Boxall R, Porteous DJ, Thomson PA. DISC1 and huntington’s disease - Overlapping pathways of vulnerability to neurological disorder? PLoS One 2011; 6: 1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camargo LM, Collura V, Rain JC, Mizuguchi K, Hermjakob H, Kerrien S et al. Disrupted in Schizophrenia 1 Interactome: evidence for the close connectivity of risk genes and a potential synaptic basis for schizophrenia. Mol Psychiatry 2007; 12: 74–86. [DOI] [PubMed] [Google Scholar]
- Hennah W, Porteous D. The DISC1 pathway modulates expression of neurodevelopmental, synaptogenic and sensory perception genes. PLoS ONE 2009; 4: e4906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson PA, Malavasi ELV, Grünewald E, Soares DC, Borkowska M, Millar JK. DISC1 genetics, biology and psychiatric illness. Front Biol 2013; 8: 1–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozeki Y, Tomoda T, Kleiderlein J, Kamiya A, Bord L, Fujii K et al. Disrupted-in-Schizophrenia-1 (DISC-1): mutant truncation prevents binding to NudE-like (NUDEL) and inhibits neurite outgrowth. Proc Natl Acad Sci USA 2003; 100: 289–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar JK, Christie S, Porteous DJ. Yeast two-hybrid screens implicate DISC1 in brain development and function. Biochem Biophys Res Commun 2003; 311: 1019–1025. [DOI] [PubMed] [Google Scholar]
- Brandon NJ, Sawa A. Linking neurodevelopmental and synaptic theories of mental illness through DISC1. Nat Rev Neurosci 2011; 12: 707–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar JK, Pickard BS, Mackie S, James R, Christie S, Buchanan SR et al. DISC1 and PDE4B Are Interacting Genetic Factors in Schizophrenia That Regulate cAMP Signaling. Science 2005; 310: 1187–1191. [DOI] [PubMed] [Google Scholar]
- Ingason A, Rujescu D, Cichon S, Sigurdsson E, Sigmundsson T, Pietilainen OP et al. Copy number variations of chromosome 16p13.1 region associated with schizophrenia. Mol Psychiatry 2011; 16: 17–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moens LN, De Rijk P, Reumers J, Van den Bossche MJ, Glassee W, De Zutter S et al. Sequencing of DISC1 pathway genes reveals increased burden of rare missense variants in schizophrenia patients from a northern Swedish population. PLoS ONE 2011; 6: e23450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenny EM, Cormican P, Furlong S, Heron E, Kenny G, Fahey C et al. Excess of rare novel loss-of-function variants in synaptic genes in schizophrenia and autism spectrum disorders. Mol Psychiatry 2014; 19: 872–879. [DOI] [PubMed] [Google Scholar]
- Purcell SM, Moran JL, Fromer M, Ruderfer D, Solovieff N, Roussos P et al. A polygenic burden of rare disruptive mutations in schizophrenia. Nature 2014; 506: 185–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown SM, Clapcote SJ, Millar JK, Torrance HS, Anderson SM, Walker R et al. Synaptic modulators Nrxn1 and Nrxn3 are disregulated in a Disc1 mouse model of schizophrenia. Mol Psychiatry 2011; 16: 585–587. [DOI] [PubMed] [Google Scholar]
- Katsel P, Tan W, Abazyan B, Davis KL, Ross C, Pletnikov MV et al. Expression of mutant human DISC1 in mice supports abnormalities in differentiation of oligodendrocytes. Schizophr Res 2011; 130: 238–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown SM, Clapcote SJ, Millar JK, Torrance HS, Anderson SM, Walker R et al. Synaptic modulators Nrxn1 and Nrxn3 are disregulated in a Disc1 mouse model of schizophrenia. Mol Psychiatry 16: 585–587. [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009; 25: 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 2010; 20: 1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 2007; 81: 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson CA, Pettersson FH, Clarke GM, Cardon LR, Morris AP, Zondervan KT. Data quality control in genetic case-control association studies. Nat Protoc 2010; 5: 1564–1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 2010; 38: e164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng PC, Henikoff S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res 2003; 31: 3812–3814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P et al. A method and server for predicting damaging missense mutations. Nat Methods 2010; 7: 248–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun S, Fay JC. Identification of deleterious mutations within three human genomes. Genome Res 2009; 19: 1553–1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz JM, Rodelsperger C, Schuelke M, Seelow D. MutationTaster evaluates disease-causing potential of sequence alterations. Nat Methods 2010; 7: 575–576. [DOI] [PubMed] [Google Scholar]
- Wu MC, Lee S, Cai T, Li Y, Boehnke M, Lin X. Rare-variant association testing for sequencing data with the sequence kernel association test. Am J Hum Genet 2011; 89: 82–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X, Zhang B, Liu W, Paciga S, He W, Lanz TA et al. A survey of rare coding variants in candidate genes in schizophrenia by deep sequencing. Mol Psychiatry 2014; 19: 857–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh KK, De Rienzo G, Drane L, Mao Y, Flood Z, Madison J et al. Common DISC1 polymorphisms disrupt Wnt/GSK3β signaling and brain development. Neuron 2011; 72: 545–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiezun A, Garimella K, Do R, Stitziel NO, Benjamin M, Mclaren PJ et al. Exome sequencing and the genetic basis of complex traits. Nat Genet 2012; 44: 623–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le-Niculescu H, Case NJ, Hulvershorn L, Patel SD, Bowker D, Gupta J et al. Convergent functional genomic studies of ω-3 fatty acids in stress reactivity, bipolar disorder and alcoholism. Transl Psychiatry 2011; 1: e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gozes I. Sexual divergence in activity-dependent neuroprotective protein impacting autism, schizophrenia, and Alzheimer’s disease. J Neurosci Res 2017; 95: 652–660. [DOI] [PubMed] [Google Scholar]
- Schumacher J, Laje G, Abou Jamra R, Becker T, Mühleisen TW, Vasilescu C et al. The DISC locus and schizophrenia: evidence from an association study in a central European sample and from a meta-analysis across different European populations. Hum Mol Genet 2009; 18: 2719–2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mühle C, Kreczi J, Rhein C, Richter-Schmidinger T, Alexopoulos P, Doerfler A et al. Additive sex-specific influence of common non-synonymous DISC1 variants on amygdala, basal ganglia, and white cortical surface area in healthy young adults. Brain Struct Funct 2016; 222: 881–894. [DOI] [PubMed] [Google Scholar]
- Thomson PA, Wray NR, Millar JK, Evans KL, Hellard S, Le, Condie A et al. Association between the TRAX/DISC locus and both bipolar disorder and schizophrenia in the Scottish population. Mol Psychiatry 2005; 10: 616. [DOI] [PubMed] [Google Scholar]
- CONVERGE consortium. Sparse whole-genome sequencing identifies two loci for major depressive disorder. Nature 2015; 523: 588–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kokras N, Dalla C. Preclinical sex differences in depression and antidepressant response: Implications for clinical research. J Neurosci Res 2017; 95: 731–736. [DOI] [PubMed] [Google Scholar]
- The International Schizophrenia Consortium, International T, Consortium S. Rare chromosomal deletions and duplications increase risk of schizophrenia. Nature 2008; 455: 237–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira MAR, O’Donovan MC, Meng YA, Jones IR, Ruderfer DM, Jones L et al. Collaborative genome-wide association analysis supports a role for ANK3 and CACNA1C in bipolar disorder. Nat Genet 2008; 40: 1056–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard L, Tansey KE, Rai D, Jones P, Ripke S, Chambert KD et al. Evidence of common genetic overlap between schizophrenia and cognition. Schizophr Bull 2016; 42: 832–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill WD, Davies G, Liewald DC, McIntosh AM, Deary IJ, Deary IJ. Age-dependent pleiotropy between general cognitive function and major psychiatric disorders. Biol Psychiatry 2016; 80: 266–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardiñas AF, Holmans P, Pocklington AJ, Escott-Price V, Ripke S, Carrera N et al. Common schizophrenia alleles are enriched in mutation-intolerant genes and maintained by background selection. bioRxiv 2016. doi: https://doi.org/10.1101/068593.
- Kirov G. CNVs in neuropsychiatric disorders. Hum Mol Genet 2015; 24: R45–R49. [DOI] [PubMed] [Google Scholar]
- Suárez-Rama JJ, Arrojo M, Sobrino B, Amigo J, Brenlla J, Agra S et al. Resequencing and association analysis of coding regions at twenty candidate genes suggest a role for rare risk variation at AKAP9 and protective variation at NRXN1 in schizophrenia susceptibility. J Psychiatr Res 2015; 66–67: 38–44. [DOI] [PubMed] [Google Scholar]
- Rees E, Kendall K, Pardiñas AF, Legge SE, Pocklington A, Escott-Price V et al. Analysis of intellectual disability copy number variants for association with schizophrenia. JAMA Psychiatry 2016; 73: 963–969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Xu T-X, Hallett PJ, Watanabe M, Grant SGN, Isacson O et al. PSD-95 uncouples dopamine-glutamate interaction in the D1/PSD-95/NMDA receptor complex. J Neurosci 2009; 29: 2948–2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E, Niethammer M, Rothschild A, Jan YN, Sheng M. Clustering of Shaker-type K+ channels by interaction with a family of membrane-associated guanylate kinases. Nature 1995; 378: 85–88. [DOI] [PubMed] [Google Scholar]
- Prange O, Wong TP, Gerrow K, Wang YT, El-Husseini A. A balance between excitatory and inhibitory synapses is controlled by PSD-95 and neuroligin. Proc Natl Acad Sci USA 2004; 101: 13915–13920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Q, Liu J, Fang A, Li R, Bai Y, Kriegstein AR et al. The dynamics of neuronal migration. Adv Exp Med Biol 2014; 800: 25–36. [DOI] [PubMed] [Google Scholar]
- Dawson N, Kurihara M, Thomson DM, Winchester CL, McVie A, Hedde JR et al. Altered functional brain network connectivity and glutamate system function in transgenic mice expressing truncated Disrupted-in-Schizophrenia 1. Transl Psychiatry 2015; 5: e569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson PA, Duff B, Blackwood DHR, Romaniuk L, Watson A, Whalley HC et al. Balanced translocation linked to psychiatric disorder, glutamate, and cortical structure/function. NPJ Schizophr 2016; 2: 16024. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.