Abstract
Although DNA replication and repair in bacteria have been extensively studied for many decades, in recent years the development of single-molecule microscopy has provided a new perspective on these fundamental processes. Because single-molecule imaging super-resolves the nanometer-scale dynamics of molecules, and because single-molecule imaging is sensitive to heterogeneities within a sample, this nanoscopic microscopy technique measures the motions, localizations, and interactions of proteins in real time without averaging ensemble observations, both in vitro and in vivo. In this Review, we provide an overview of several recent single-molecule fluorescence microscopy studies on DNA replication and repair. These experiments have shown that, in both Escherichia coli and Bacillus subtilis the DNA replication proteins are highly dynamic. In particular, even the highly processive replicative DNA polymerases—E. coli Pol III and B. subtilis PolC—exchange to and from the replication fork on the scale of a few seconds. Furthermore, single-molecule investigations of the DNA mismatch repair (MMR) pathway have measured the complex interactions between MMR proteins, replication proteins, and DNA. Single-molecule imaging will continue to improve our understanding of fundamental processes in bacteria including DNA replication and repair.
Graphical abstract
Introduction
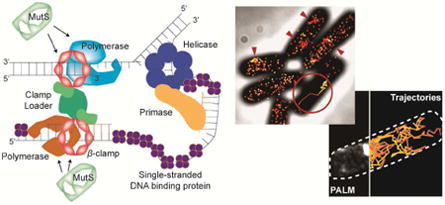
Understanding how genomes are accurately copied and maintained during the cell cycle is of fundamental importance to all of biology. When cells fail to replicate their DNA accurately, numerous disease states or even cell death can occur in multicellular organisms, while in bacteria, DNA replication errors have the potential to affect cell fitness and viability [1,2]. Therefore it is critical for cells to replicate their DNA accurately and efficiently. In all cells, this process is accomplished by replisomes, which are multi-protein machines containing all components necessary to duplicate genomic DNA [3,4]. The architecture and dynamics of the Escherichia coli bacterial replisome has been extensively characterized (Fig. 1a) [5-8]. The E. coli replisome contains the Pol III holoenzyme (HE), a molecular machine composed of ten different proteins grouped into several functional subassemblies. The Pol III core is the catalytic subassembly important for replication; it has three subunits: α, which has DNA polymerase activity, ε, which has proofreading activity, and θ, which is involved in aiding ε [9,10]. The Pol III* subassembly is the core plus the clamp loading complex, which includes τ/γ, δ, δ′, ψ and χ [11]. Finally, the Pol III HE is Pol III* plus the β sliding clamp encoded by dnaN. The β clamp enhances processivity and increases the DNA synthesis rate up to a thousand fold by holding the DNA polymerase on the template strand to increase the number of nucleotides synthesized per binding event [12]. The multiprotein clamp loader complex is required to open and close the β clamps around DNA for loading and unloading during replication. Apart from the Pol III HE, the DNA helicase (DnaB) is essential and required for DNA replication. The helicase is linked to Pol III HE through τ. With the help of the replication initiation proteins DnaA and DnaC, the E. coli replisome assembles at oriC to establish bi-directional replication forks [13] that then track along DNA until replication is complete [6].
Figure 1. Molecular architecture of the DNA replication fork.
(a) The architecture of the E. coli DNA replication fork. Reproduced from eLife 2017, 6:10.7554 [46]. Creative commons. (b) The architecture of the B. subtilis DNA replication fork.
A replisome that exhibits a different mechanism is found in Bacillus subtilis, a prototypical Gram-positive bacterium (Fig. 1b). Rather than tracking along the DNA strands, the B. subtilis replisome is relatively stationary during replication, and the template DNA is pulled into the replisome for duplication [14,15]. Unlike E. coli, the B. subtilis replisome has two distinct replicative DNA polymerases: PolC and DnaE. Reconstitution of the B. subtilis replisome in vitro has shown that PolC is responsible for all leading strand synthesis and most of the lagging DNA strand synthesis, while DnaE is used to extend the lagging strand RNA primer before handing off to PolC [16]. The B. subtilis DNA replication mechanism has some features that are more reminiscent of DNA replication in eukaryotic systems than what is found in E. coli. For example, in human lagging strand replication, the Pol α polymerase extends RNA primers before handing off to Pol δ [17,18].
Because high fidelity replication is so important, cells have also developed mechanisms to detect and repair DNA damage and replication errors shortly after they occur. For example, DNA mismatch repair (MMR) is triggered when a DNA replication error is detected [19]. This highly conserved process is important for replication fidelity despite the low frequency of replication error occurrence (∼1 error per 33,000,000 base pairs) [20]. In B. subtilis, MutS is responsible for detecting base-pairing errors, and once a mismatch is detected, the MutL endonuclease is recruited for incision of the error-containing strand followed by removal of the incised strand [21-23]. Biophysical studies have suggested that MutS homologs can target a mismatch by one-dimensional scanning along DNA [24].
Biochemical methods and bulk fluorescence imaging have been used to understand how cells maintain their genomic integrity [6,16,25-32]. However, such methods generally lose the heterogeneity of a system by averaging over all observations. Understanding all the complex and highly dynamic aspects of DNA replication and repair requires quantitative biophysical tools, and molecular resolution is necessary to characterize the dynamics of all subpopulations, to understand detailed kinetics, and to uncover intricate mechanisms that are relevant in vivo. Single-molecule microscopy is a great solution to this problem because it captures the different behaviors of every molecule [33] and provides nanometer-scale resolution [34], even in living cells [35]. By using photoactivatable or photoswitchable fluorescent labels, one can observe and image the fluorescence from individual single emitters, and each isolated molecule image can be analyzed to obtain the nanometer-scale locations and trajectory of that molecule [36-38]. Importantly, single-molecule microscopy can be easily applied to bacteria in vivo [39-41], making it now possible to visualize DNA replication and repair in real time in living bacterial cells. In this review, we provide an overview of some recent studies on bacterial DNA replication and repair using single-molecule microscopy. The ability to quantitatively characterize a heterogeneous dynamic system and to provide molecular resolution has greatly deepened our understanding of biology.
DNA replication at the single-molecule level
Several recent papers have explored DNA replication from a single-molecule perspective. Importantly, though replicative DNA polymerases have long been known to be highly processive based on in vitro results [42], single-molecule biophysics and live-cell experiments have recently revealed that the bacterial replicative polymerases are far more dynamic than previously anticipated. Single-molecule stoichiometry distributions have suggested replisome instability [43], and three separate investigations have measured polymerase dwell times of only a few seconds at the replisome [44-46].
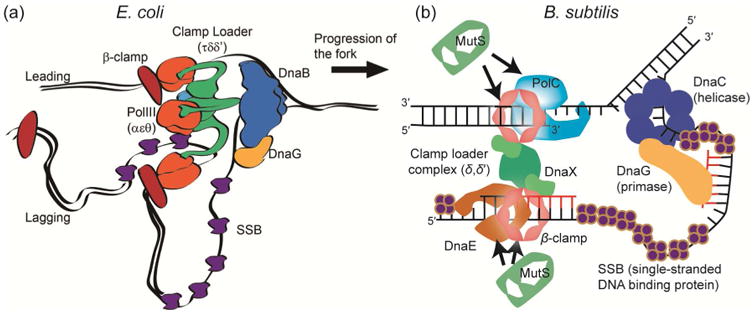
Liao et al. tracked single B. subtilis PolC molecules to determine dwell timescales in living cells expressing PolC fused to the photoactivatable fluorescent protein PAmCherry [44]. Based on photoactivation of single copies of PolC-PAmCherry [47,48], each molecule was studied individually with super-resolved localization precision to determine the dwell times of PolC molecules at foci (red arrows in Fig. 2a), as well as the apparent diffusion coefficients for three-dimensional PolC trajectories (Fig. 2a inset). The PolC foci are most often observed at the quarter-cell positions (Fig. 2b), and are still observed in B. subtilis cells treated with HPUra, which arrests DNA replication. We further analyzed the trajectories to measure the apparent diffusion coefficient of each molecule, and measured that, though many PolC molecules diffuse at a rate slower than 0.1 μm2/s (Fig. 2c inset), even the slowest PolC molecules are not completely stationary relative to the localization precision of the immobile PolC in fixed cells (red dashed line in Fig. 2c inset), indicating that all molecules have some mobility during the course of their measured > 300 ms trajectories. This PolC motion is unchanged upon HPUra treatment.
Figure 2. DNA polymerase localization, dynamics, and exchange in living bacterial cells.
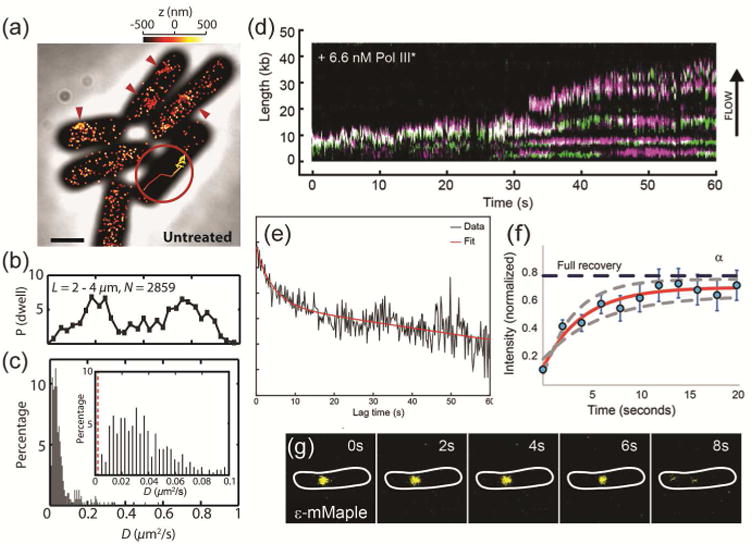
(a) 3D super-resolution reconstruction image and a 3D single-molecule trajectory (inset) of PolC-PAmCherry in B. subtilis. The molecules are dynamic and enriched at foci (red arrows). (b) Localization probabilities of B. subtilis PolC-PAmCherry dwelling events along the longitudinal cell axis in 2859 cells. (c) Distribution of PolC-PAmCherry diffusion coefficients, D, in B. subtilis. Even the slowest moving molecules (inset) are more mobile than stationary PolC-PAmCherry molecules measured in fixed cells (red dashed line). (a) – (c) Reprinted from Biophys J, 111:2562-2569, Copyright 2016, with permission from Elsevier [44]. (d) Kymographs of the distributions of red Pol III* (magenta) and green Pol III* (green) show rapid and frequent exchange of E. coli Pol III* on individual DNA molecules. (e) Exponential fit (red) to the cross-correlation function of 1210 pairs of τ-YPet and ε-mKate2 foci in living E. coli cells gives a 4 ± 2 s exchange time scale. (d) – (e) Reproduced from eLife 2017, 6:e23932 [45]. Creative commons. (f) FRAP recovery for the α subunit in E. coli cells. A fit to a reaction-diffusion model (red line) indicates a 4 ± 2 s bound-time for this Pol III subunit. (g) The ε subunit remains replisome-associated for 10 ± 0.7 s in E. coli. (f) – (g) Reproduced from eLife 2017, 6:10.7554 [46]. Creative commons.
In this study, on average 61 PolC copies were observed in each cell, whereas only 3 PolC copies were found at each replication fork. A similar disparity has also been reported in E. coli, where there are on average 40 copies of Pol III core enzymes per cell [10], but again only 3 Pol III copies at each replication fork [7]. The difference between these numbers is explained by the highly dynamic nature of PolC, which enables it to affect replication and to play multiple roles in the cell, including assisting in DNA resynthesis during repair elsewhere in the cell. We hypothesize from a biophysical perspective that the additional PolC molecules form a reservoir that can efficiently exchange with active PolC at the replication fork. We characterized the magnitude of this exchange by measuring the dwell time of PolC-PAmCherry in B. subtilis cells, i.e., the length of time over which a molecule does not move significantly between observations. Time-lapse 3D single-molecule tracking accounted for fluorescence photobleaching by introducing a variable dark time delay to extend trajectories and separate the PolC dissociation rate from the PAmCherry photobleaching rate. The PolC dwell time in untreated cells is 2.79 ± 0.47 s, consistent with the time needed to synthesize one Okazaki fragment [16], indicating that—at least for lagging strand synthesis—a PolC molecule will synthesize a single Okazaki fragment each time it is recruited to the replisome. HPUra treatment reduces this dwell time to 0.97 ± 0.04 s. Overall, these results demonstrate that B. subtilis PolC is highly mobile and undergoes dynamic exchange at the replisome. Furthermore, the results for HPUra treatment indicate that PolC mobility is mediated by protein-protein or protein-DNA interactions, and not by active DNA replication.
These dynamical behaviors are not limited to B. subtilis, and recently, single-molecule investigations have observed rapid exchange dynamics for the E. coli replicative polymerase [42]. In vitro experiments with T4 bacteriophage have shown stable replisome assembly capable of entire genome duplication [49], while other in vitro studies have shown rapid polymerase exchange [50]. To understand the dynamics of E. coli replication in vivo, Lewis et al. investigated Pol III* with a green or red SNAP tag label during in vitro rolling-circle DNA amplification and demonstrated frequent exchanges between the molecules (green to magenta transitions in Fig. 2d) during ongoing DNA replication [45]. Furthermore, the exchange time measured was found to be concentration-dependent, which can account for the discrepancies among current and previous studies. Lewis et al. extended this study by monitoring polymerase exchange through cross-correlation of the fluorescence intensities of τ-YPet and ε-mKate2 at the E. coli replisome in live cells (Fig. 2e). From the cross-correlation function, the authors measured the characteristic exchange time of Pol III* during coupled DNA replication in living E. coli cells to be 4 ± 2 s. Furthermore, correlation with in vitro measurements of the concentration-dependent exchange time determined an effective Pol III* concentration of ∼23 nM per cell.
Beattie et al. also measured rapid Pol III* exchange in living E. coli based on single-molecule FRAP of DNA polymerase in active replisomes [46]. In this study, the authors separately characterized the different subunits and found replisome-bound times of 3 – 6 s for the α (Fig. 2f), ε, τ, δ, and χ subunits. The β clamp remains associated for a longer 36 ± 21 s, while the DnaB helicase is highly stationary, remaining DNA-bound for tens of minutes. The authors then tracked single replisome proteins fused to the photoswitchable fluorescent protein mMaple, and used time lapses and long exposure times to confirm that most of the replisome components are in frequent exchange (with bound times of tens of seconds; Fig. 2g) except for DnaB, the helicase, whose bound time is thousands of seconds long.
Overall, these studies give new insight into DNA replication in live cells, where the highly processive DNA polymerase can also exchange on the scale of seconds. In the end, the newly revealed dynamics of DNA Pol III or PolC underscore a dynamic process that may help the replisome circumvent barriers to progression or more easily exchange with translesion DNA polymerases for the bypass of noncoding bases. Furthermore, recent work has shown that DNA replication and DNA repair are indeed intimately coupled [51].
Single-molecule investigations of DNA mismatch repair (MMR)
DNA mismatch repair (MMR) is a highly conserved process which corrects DNA replication errors that have evaded proofreading by replicative polymerases. MMR is initiated by MutS, which finds and identifies DNA base-pair mismatches. Recently, Liao et al. characterized the dynamics of B. subtilis MutS by localizing and tracking single MutS-PAmCherry fusions in living cells (Fig. 3a) [51]. Localization probability density maps show that MutS foci colocalize with the replisome, marked by fluorescent fusions to the β clamp or the clamp loader protein DnaX (Fig. 3b). The replication protein localization pattern is unchanged after the cells are treated with 2-aminopurine (2-AP), which induces mismatch formation (Fig. 3c), indicating that the positioning of MutS at the replisome is independent of the number or density of DNA mismatches that occur in cells. Single-molecule microscopy, which is sensitive to even transient dwelling events, was critical in this observation as previous bulk fluorescence microscopy studies could not visualize MutS focus formation in more than 5 – 8% of cells in the absence of an exogenous mutagen [27]. These new observations demonstrate that MutS constantly monitors the site of DNA replication, even at low spontaneous level of mismatches.
Figure 3. Single-molecule imaging yields the molecular mechanism for B. subtilis MutS.

(a) Photoactivated localization microscopy (PALM) reconstruction (lower left) and single-molecule trajectories of MutS-PAmCherry (right) in a live B. subtilis cell (inset). The red arrow indicates a region of MutS accumulation and the dashed line indicates the cell boundary. Scale bar: 1 μm. (b) – (c) Localization probability density maps of DnaX-mCitrine (upper; blue-green) and MutS-PAmCherry (lower; red-yellow) within a normalized cell that is (b) untreated, or (c) treated with the mismatch-forming drug 2-AP. (d) Diffusion coefficients of B. subtilis MutS-PAmCherry as a function of separation distance from the nearest replisome show the molecules slowing near the replication fork. (e) Schematic of the first four steps of MMR: replisome binding, mismatch recognition, ATPase activity, and MutL recruitment, each of which is blocked in one of four mutant strains to determine the relationship between DNA MMR and DNA replication. Reprinted with permission from Proc. Natl. Acad. Sci. USA 2015, 112:E6898–E6906 [51].
By analyzing each single-molecule trajectory, we further found that the average apparent diffusion coefficient for MutS-PAmCherry decreases near the replisome (blue curve in Fig. 3d) and that MutS-PAmCherry has an average dwell time at the replisome of 188 ms, consistent with constant exchange of MutS at the replisome, even in the absence of a mismatch. Moreover, 2-AP increases the rate of MutS-PAmCherry diffusion, and decreases the proportion of MutS that is slowed down at the replisome (red curve in Fig. 3d), consistent with the transition of MutS to a fast sliding clamp conformation following mismatch binding [52].
Finally, through single-molecule measurements in cells with a series of protein variants that sequentially blocked MMR steps by: (1) removing β-clamp binding affinity (MutS800), (2) disabling mismatch recognition (MutS[F30A]), (3) inhibiting ATPase and nucleotide binding activity (MutS[K608M]), and (4) preventing MutS dissociation from the vicinity of the mismatch (ΔmutL), the molecular mechanism of MMR in B. subtilis was revealed (Fig. 3e). In particular, these studies demonstrated that MutS is recruited to the replisome prior to mismatch binding to scan local DNA in anticipation of errors, and that mismatch repair occurs at the replisome. Overall, B. subtilis MMR and DNA replication are intimately coupled in vivo.
The search mechanism of MutS and MutL in mismatch repair was further investigated for the E. coli proteins [53]. Liu et al. directly visualized MutS/MutL interactions by following single molecules of MutS, MutL, and the MutH endonuclease as they move along tethered DNA strands with total-internal reflection (TIRF) microscopy. The benefit of this in vitro system is that it is based on well-defined, mismatched DNA sequences, and ATP levels can be controlled. Based on the diffusion kinetics of red MutS-AF647 and green MutL-Cy3 molecules along the mismatched DNA, the authors observed the formation of MutS/MutL complexes. These complexes last for an average of 43 ± 3 s, after which MutS and MutL dissociate by one of four different mechanisms (Fig. 4a). On the other hand, ATP binding by MutL was found to produce a stable MutS-MutL sliding clamp on mismatched DNA. Overall, these results point to a mechanism in which E. coli MMR employs a cascade of stable ATP-bound sliding clamps (the MutS/MutL/MutH search complex) to modulate 1D diffusion mechanics along the DNA.
Figure 4. Mismatch repair in E. coli.
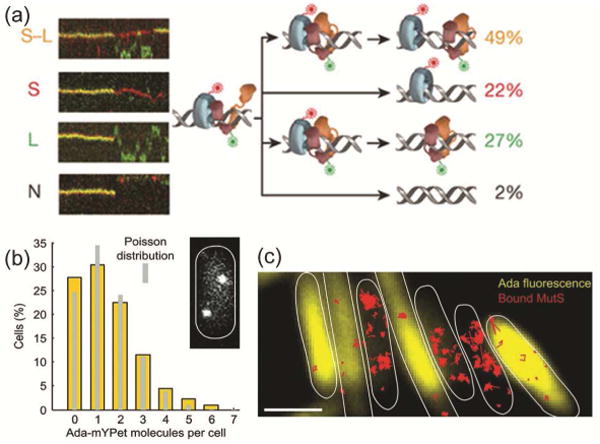
(a) Four different types of E. coli MutS/MutL complex dissociations (from top to bottom: oscillation dissociation-association, MutL dissociation, MutS dissociation, and simultaneous dissociation) observed in kymographs of E. coli MutS-AF647 (two-tone blue protein with red dye label) and MutL-Cy3 (two-tone orange protein with green dye label) diffusion in vitro on doubly-tethered mismatched DNAs. Reprinted from reference [53] by permission from Macmillan Publishers Ltd.: Nature 539: 583-587, Copyright 2016. (b) Single-molecule counting of Ada-mYPet in single E. coli cells measures the copy number and cell-to-cell heterogeneity of this DNA repair protein. Inset: a representative cell with two Ada-mYPet copies. (c) Cells with high Ada-mYPet expression (yellow) show fewer immobile MutS-PAmCherry molecules (red). Scale bars: 2 μm. Panels ‘b’ and ‘c’ are from Uphoff et al., Science 2016, 351:1094-1097 [54]. Reprinted by permission with permission from AAAS.
The action of other mutagens was further explored in E. coli by Uphoff et al. who measured the adaptive response to alkylation by methyl methanesulfonate (MMS) as a function of the expression level of the DNA repair protein Ada [54]. Based on single-molecule counting in single-cell experiments, these studies uncovered a previously unmeasurable heterogeneity in the abundance of Ada. The study found on average 1 fluorescent Ada-mYPet copy per cell; moreover the copy number per cell was Poisson distributed and 20 – 30 % of cells expressed no Ada at all (Fig. 4b). The authors further tested the effect of heterogeneity of Ada expression on MMR using the dynamics of MutS-PAmCherry trajectories as a readout of MMR activity. In these two-color microscopy experiments, cells with low Ada-mYPet expression showed more immobile MutS-PAmCherry molecules, which may indicate increased MutS activity due to the formation of lesions that are refractory to MMR repair in the absence of Ada (Fig. 4c).
Conclusions
Single-molecule methods continue to reveal new insight into the dynamical nature of DNA replication and repair in living cells. Interestingly, the dwelling of PolC and Pol III at the replisome is similar to the behavior that has been observed for the DNA mismatch repair protein MutS, which is coupled to DNA replication. The new advances in single-molecule imaging of DNA replication and repair described in this Review indicate that the activities of bacterial replisomal and repair proteins may be regulated in cells by coordinating and modulating the dynamics of protein recruitment, binding, and unbinding at the site of DNA synthesis.
Highlights.
Single-molecule microscopy resolves nm-scale positioning in vitro and in cells.
DNA replication proteins exchange rapidly at the bacterial replication fork.
DNA mismatch repair involves dynamic protein and DNA interactions.
Acknowledgments
This work was supported by NIH grant R01 GM107312 to L.A.S. J.W.S. was supported in part by the NIH National Research Service Award T32 GM007544.
Footnotes
Conflicts of interest: none
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Friedberg EC, Walker GC, Siede W, Wood RD, Schultz RA, Ellenberger T. DNA Repair and Mutagenesis. Second. ASM Press; 2006. [Google Scholar]
- 2.Lenhart JS, Schroeder JW, Walsh BW, Simmons LA. DNA Repair and Genome Maintenance in Bacillus subtilis. Microbiol Mol Biol Rev. 2012;76:530–564. doi: 10.1128/MMBR.05020-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Johnson A, O'Donnell M. Cellular DNA replicases: components and dynamics at the replication fork. Annu Rev Biochem. 2005;74:283–315. doi: 10.1146/annurev.biochem.73.011303.073859. [DOI] [PubMed] [Google Scholar]
- 4.Burgers PMJ, Kunkel TA. Eukaryotic DNA Replication Fork. Annu Rev Biochem. 2017;86:417–438. doi: 10.1146/annurev-biochem-061516-044709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.O'Donnell M. Replisome architecture and dynamics in Escherichia coli. J Biol Chem. 2006;281:10653–10656. doi: 10.1074/jbc.R500028200. [DOI] [PubMed] [Google Scholar]
- 6.Reyes-Lamothe R, Possoz C, Danilova O, Sherratt DJ. Independent Positioning and Action of Escherichia coli Replisomes in Live Cells. Cell. 2008;133:90–102. doi: 10.1016/j.cell.2008.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reyes-Lamothe R, Sherratt DJ, Leake MC. Stoichiometry and architecture of active DNA replication machinery in Escherichia coli. Science. 2010;328:498–501. doi: 10.1126/science.1185757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McHenry CS. DNA Replicases from a Bacterial Perspective. Annu Rev Biochem. 2011;80:403–436. doi: 10.1146/annurev-biochem-061208-091655. [DOI] [PubMed] [Google Scholar]
- 9.Welch MM, McHenry CS. Cloning and identification of the product of the dnaE gene of Escherichia coli. J Bacteriol. 1982;152:351–356. doi: 10.1128/jb.152.1.351-356.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maki H, Kornberg A. The polymerase subunit of DNA polymerase III of Escherichia coli. II Purification of the alpha subunit, devoid of nuclease activities. J Biol Chem. 1985;260:12987–12992. [PubMed] [Google Scholar]
- 11.Jeruzalmi D, O'Donnell M, Kuriyan J. Crystal structure of the processivity clamp loader gamma (gamma) complex of E. coli DNA polymerase III. Cell. 2001;106:429–441. doi: 10.1016/s0092-8674(01)00463-9. [DOI] [PubMed] [Google Scholar]
- 12.Mizrahi V, Henrie R, Marlier J, Johnson K, Benkovic S. Rate-limiting steps in the DNA polymerase I reaction pathway. Biochemistry. 1985;24:4010–4018. doi: 10.1021/bi00336a031. [DOI] [PubMed] [Google Scholar]
- 13.Chodavarapu S, Kaguni JM. Replication Initiation in Bacteria. The Enzymes. 2016;39:1–30. doi: 10.1016/bs.enz.2016.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lemon KP, Grossman AD. Localization of bacterial DNA polymerase: evidence for a factory model of replication. Science. 1998;282:1516–1519. doi: 10.1126/science.282.5393.1516. [DOI] [PubMed] [Google Scholar]
- 15.Sawitzke J, Austin S. An analysis of the factory model for chromosome replication and segregation in bacteria. Mol Microbiol. 2001;40:786–794. doi: 10.1046/j.1365-2958.2001.02350.x. [DOI] [PubMed] [Google Scholar]
- 16.Sanders GM, Dallmann HG, McHenry CS. Reconstitution of the B. subtilis Replisome with 13 Proteins Including Two Distinct Replicases. Mol Cell. 2010;37:273–281. doi: 10.1016/j.molcel.2009.12.025. [DOI] [PubMed] [Google Scholar]
- 17.Kunkel TA, Burgers PM. Dividing the workload at a eukaryotic replication fork. Trends Cell Biol. 2008;18:521–527. doi: 10.1016/j.tcb.2008.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kunkel TA. Balancing eukaryotic replication asymmetry with replication fidelity. Curr Opin Chem Biol. 2011;15:620–626. doi: 10.1016/j.cbpa.2011.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Modrich P. Mechanisms in eukaryotic mismatch repair. J Biol Chem. 2006;281:30305–30309. doi: 10.1074/jbc.R600022200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee H, Popodi E, Tang H, Foster PL. Rate and molecular spectrum of spontaneous mutations in the bacterium Escherichia coli as determined by whole-genome sequencing. Proc Natl Acad Sci U S A. 2012;109:E2774–E2783. doi: 10.1073/pnas.1210309109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kadyrov FA, Dzantiev L, Constantin N, Modrich P. Endonucleolytic function of MutL alpha in human mismatch repair. Cell. 2006;126:297–308. doi: 10.1016/j.cell.2006.05.039. [DOI] [PubMed] [Google Scholar]
- 22.Pillon MC, Lorenowicz JJ, Uckelmann M, Klocko AD, Mitchell RR, Chung YS, Modrich P, Walker GC, Simmons LA, Friedhoff P, Guarne A. Structure of the Endonuclease Domain of MutL: Unlicensed to Cut. Mol Cell. 2010;39:145–151. doi: 10.1016/j.molcel.2010.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iyer R, Pluciennik A, Burdett V, Modrich P. DNA mismatch repair: Functions and mechanisms. Chem Rev. 2006;106:302–323. doi: 10.1021/cr0404794. [DOI] [PubMed] [Google Scholar]
- 24.Gorman J, Wang F, Redding S, Plys AJ, Fazio T, Wind S, Alani EE, Greene EC. Single-molecule imaging reveals target-search mechanisms during DNA mismatch repair. Proc Natl Acad Sci U S A. 2012;109:E3074–E3083. doi: 10.1073/pnas.1211364109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smith B, Grossman A, Walker G. Visualization of mismatch repair in bacterial cells. Mol Cell. 2001;8:1197–1206. doi: 10.1016/s1097-2765(01)00402-6. [DOI] [PubMed] [Google Scholar]
- 26.Kleczkowska H, Marra G, Lettieri T, Jiricny J. hMSH3 and hMSH6 interact with PCNA and colocalize with it to replication foci. Genes Dev. 2001;15:724–736. doi: 10.1101/gad.191201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lenhart JS, Sharma A, Hingorani MM, Simmons LA. DnaN clamp zones provide a platform for spatiotemporal coupling of mismatch detection to DNA replication. Mol Microbiol. 2013;87:553–568. doi: 10.1111/mmi.12115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hombauer H, Campbell CS, Smith CE, Desai A, Kolodner RD. Visualization of Eukaryotic DNA Mismatch Repair Reveals Distinct Recognition and Repair Intermediates. Cell. 2011;147:1040–1053. doi: 10.1016/j.cell.2011.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Klocko AD, Schroeder JW, Walsh BW, Lenhart JS, Evans ML, Simmons LA. Mismatch repair causes the dynamic release of an essential DNA polymerase from the replication fork. Mol Microbiol. 2011;82:648–663. doi: 10.1111/j.1365-2958.2011.07841.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pluciennik A, Burdett V, Lukianova O, O'Donnell M, Modrich P. Involvement of the beta Clamp in Methyl-directed Mismatch Repair in Vitro. J Biol Chem. 2009;284:32782–32791. doi: 10.1074/jbc.M109.054528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lemon K, Grossman A. Movement of replicating DNA through a stationary replisome. Mol Cell. 2000;6:1321–1330. doi: 10.1016/s1097-2765(00)00130-1. [DOI] [PubMed] [Google Scholar]
- 32.Meile J, Wu L, Ehrlich S, Errington J, Noirot P. Systematic localisation of proteins fused to the green fluorescent protein in Bacillus subtilis: Identification of new proteins at the DNA replication factory. Proteomics. 2006;6:2135–2146. doi: 10.1002/pmic.200500512. [DOI] [PubMed] [Google Scholar]
- 33*.Monachino E, Spenkelink LM, van Oijen AM. Watching cellular machinery in action, one molecule at a time. J Cell Biol. 2017;216:41–51. doi: 10.1083/jcb.201610025. This Review outlines how single-molecule approaches have increased our understanding of the dynamic behavior of complex multiprotein systems both in vitro and in vivo. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yildiz A, Selvin PR. Fluorescence imaging with one nanometer accuracy: application to molecular motors. Acc Chem Res. 2005;38:574–582. doi: 10.1021/ar040136s. [DOI] [PubMed] [Google Scholar]
- 35*.Tuson HH, Biteen JS. Unveiling the Inner Workings of Live Bacteria Using Super-Resolution Microscopy. Anal Chem. 2015;87:42–63. doi: 10.1021/ac5041346. This Review provides an overview of live-cell single-molecule imaging methods and reviews how single-molecule and super-resolution microscopy has been applied to measure, image, and understand sub-cellular biology in bacteria over the last decade. [DOI] [PubMed] [Google Scholar]
- 36.Moerner WE. Single-molecule mountains yield nanoscale images. Nature Meth. 2006;3:781–782. doi: 10.1038/nmeth1006-781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Betzig E, Patterson GH, Sougrat R, Lindwasser OW, Olenych S, Bonifacino JS, Davidson MW, Lippincott-Schwartz J, Hess HF. Imaging intracellular fluorescent proteins at nanometer resolution. Science. 2006;313:1642–1645. doi: 10.1126/science.1127344. [DOI] [PubMed] [Google Scholar]
- 38.Hess ST, Girirajan TPK, Mason MD. Ultra-high resolution imaging by fluorescence photoactivation localization microscopy. Biophys J. 2006;91:4258–4272. doi: 10.1529/biophysj.106.091116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Elf J, Li GW, Xie XS. Probing transcription factor dynamics at the single-molecule level in a living cell. Science. 2007;316:1191–1194. doi: 10.1126/science.1141967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yu J, Xiao J, Ren X, Lao K, Xie XS. Probing gene expression in live cells, one protein molecule at a time. Science. 2006;311:1600–1603. doi: 10.1126/science.1119623. [DOI] [PubMed] [Google Scholar]
- 41.Biteen JS, Thompson MA, Tselentis NK, Bowman GR, Shapiro L, Moerner WE. Super-resolution imaging in live Caulobacter crescentus cells using photoswitchable EYFP. Nat Methods. 2008;5:947–949. doi: 10.1038/NMETH.1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kornberg A, Baker TA. DNA Replication. Second. University Science Books; 2005. [Google Scholar]
- 43.Mangiameli SM, Merrikh CN, Wiggins PA, Merrikh H. Transcription leads to pervasive replisome instability in bacteria. eLife. 2017;6:e19848. doi: 10.7554/eLife.19848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44**.Liao Y, Li Y, Schroeder JW, Simmons LA, Biteen JS. Single-Molecule DNA Polymerase Dynamics at a Bacterial Replisome in Live Cells. Biophys J. 2016;111:2562–2569. doi: 10.1016/j.bpj.2016.11.006. This paper uses photobleaching-assisted microscopy, three-dimensional superresolution imaging, and single-particle tracking to measure the organization and dynamics of the essential replicative polymerase, PolC, in living Bacillus subtilis cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45**.Lewis JS, Spenkelink LM, Jergic S, Wood EA, Monachino E, Horan NP, Duderstadt KE, Cox MM, Robinson A, Dixon NE, van Oijen AM. Single-molecule visualization of fast polymerase turnover in the bacterial replisome. eLife. 2017;6:e23932. doi: 10.7554/eLife.23932. This paper measures the single-molecule exchange dynamics of the replicative DNA Pol III holoenzyme during rolling-circle DNA amplification in vitro and via single-molecule fluorescence recovery after photobleaching in living Escherichia coli cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46**.Beattie TR, Kapadia N, Nicolas E, Uphoff S, Wollman AJ, Leake MC, Reyes-Lamothe R. Frequent exchange of the DNA polymerase during bacterial chromosome replication. eLife. 2017;6:10.7554. doi: 10.7554/eLife.21763. This paper uses single-molecule fluorescence recovery after photobleaching in living Escherichia coli cells to measure the exchange time of multiple fluorescently labeled replisome subunits. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Subach FV, Patterson GH, Manley S, Gillette JM, Lippincott-Schwartz J, Verkhusha VV. Photoactivatable mCherry for high-resolution two-color fluorescence microscopy. Nat Methods. 2009;6:153–159. doi: 10.1038/nmeth.1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Manley S, Gillette JM, Patterson GH, Shroff H, Hess HF, Betzig E, Lippincott-Schwartz J. High-density mapping of single-molecule trajectories with photoactivated localization microscopy. Nat Methods. 2008;5:155–157. doi: 10.1038/nmeth.1176. [DOI] [PubMed] [Google Scholar]
- 49.Kaboord BF, Benkovic SJ. Accessory proteins function as matchmakers in the assembly of the T4 DNA polymerase holoenzyme. Curr Biol. 1995;5:149–157. doi: 10.1016/s0960-9822(95)00036-4. [DOI] [PubMed] [Google Scholar]
- 50.Yang J, Zhuang Z, Roccasecca RM, Trakselis MA, Benkovic SJ. The dynamic processivity of the T4 DNA polymerase during replication. Proc Natl Acad Sci U S A. 2004;101:8289–8294. doi: 10.1073/pnas.0402625101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51**.Liao Y, Schroeder JW, Gao B, Simmons LA, Biteen JS. Single-molecule motions and interactions in live cells reveal target search dynamics in mismatch repair. Proc Natl Acad Sci U S A. 2015;112:E6898–E6906. doi: 10.1073/pnas.1507386112. This paper tracks single molecules of the mismatch repair protein MutS in living Bacillus subtilis cells and measures changes in these dynamics in several key MutS mutants and following DNA synthesis arrest to provide mechanistic insight into the searching behavior of MutS during repair. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jeong C, Cho WK, Song KM, Cook C, Yoon TY, Ban C, Fishel R, Lee JB. MutS switches between two fundamentally distinct clamps during mismatch repair. Nat Struct Mol Biol. 2011;18:379–385. doi: 10.1038/nsmb.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53**.Liu J, Hanne J, Britton BM, Bennett J, Kim D, Lee J, Fishel R. Cascading MutS and MutL sliding clamps control DNA diffusion to activate mismatch repair. Nature. 2016;539:583–587. doi: 10.1038/nature20562. This paper measures the assembly of the Escherichia coli MutS-MutL-MutH search complex in an in vitro tethered DNA assay to determine the assembly dynamics and mechanism in DNA mismatch repair. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54**.Uphoff S, Lord ND, Okumus B, Potvin-Trottier L, Sherratt DJ, Paulsson J. Stochastic activation of a DNA damage response causes cell-to-cell mutation rate variation. Science. 2016;351:1094–1097. doi: 10.1126/science.aac9786. This paper couples single-molecule detection and tracking with multi-generational single-cell imaging in a microfluidic device to measure the heterogeneous response of E. coli cells to DNA damage as a function of adaptive response activity. [DOI] [PMC free article] [PubMed] [Google Scholar]


