Significance
Thanks to improved screening technologies, many small “premalignant” tumors are now detected and treated, which reduces late-stage disease and cancer mortality, but also leads to overdiagnosis. Indeed, it remains uncertain if the starts of benign and malignant tumors differ. Here, we combine mathematical multiscale models with multiregional sequencing to infer that the starts of benign and some malignant tumors differ. The abnormal cell mobility eventually needed for invasion and metastasis is already expressed at the start of growth for some malignant tumors but not benign tumors. Therefore, some malignant tumors start with more invasive potential (“born to be bad”), and this early abnormal cell mobility can potentially be used to identify patients more likely to benefit from aggressive treatment.
Keywords: tumor evolution, cellular mobility, intratumor heterogeneity, cancer modeling
Abstract
A growing body of evidence suggests that a subset of human cancers grows as single clonal expansions. In such a nearly neutral evolution scenario, it is possible to infer the early ancestral tree of a full-grown tumor. We hypothesized that early tree reconstruction can provide insights into the mobility phenotypes of tumor cells during their first few cell divisions. We explored this hypothesis by means of a computational multiscale model of tumor expansion incorporating the glandular structure of colorectal tumors. After calibrating the model to multiregional and single gland data from 19 human colorectal tumors using approximate Bayesian computation, we examined the role of early tumor cell mobility in shaping the private mutation patterns of the final tumor. The simulations showed that early cell mixing in the first tumor gland can result in side-variegated patterns where the same private mutations could be detected on opposite tumor sides. In contrast, absence of early mixing led to nonvariegated, sectional mutation patterns. These results suggest that the patterns of detectable private mutations in colorectal tumors may be a marker of early cell movement and hence the invasive and metastatic potential of the tumor at the start of the growth. In alignment with our hypothesis, we found evidence of early abnormal cell movement in 9 of 15 invasive colorectal carcinomas (“born to be bad”), but in none of 4 benign adenomas. If validated with a larger dataset, the private mutation patterns may be used for outcome prediction among screen-detected lesions with unknown invasive potential.
Evolution to cancer progresses through the stepwise accumulation of selective driver mutations that confer new capabilities (1, 2). Tumors are clonal expansions that start from single progenitors, and their growth can be described with ancestral trees with billions of tips that represent present-day tumor cells (Fig. 1). Mutations that arise before tumor expansion are public and present in all tumor cells, whereas mutations that arise during growth are subclonal or private and present in a subset of tumor cells only. Multiregional sequencing studies have shown that intratumoral heterogeneity (ITH) is very common in human tumors (3–7), which implies that many private mutations arise during growth.
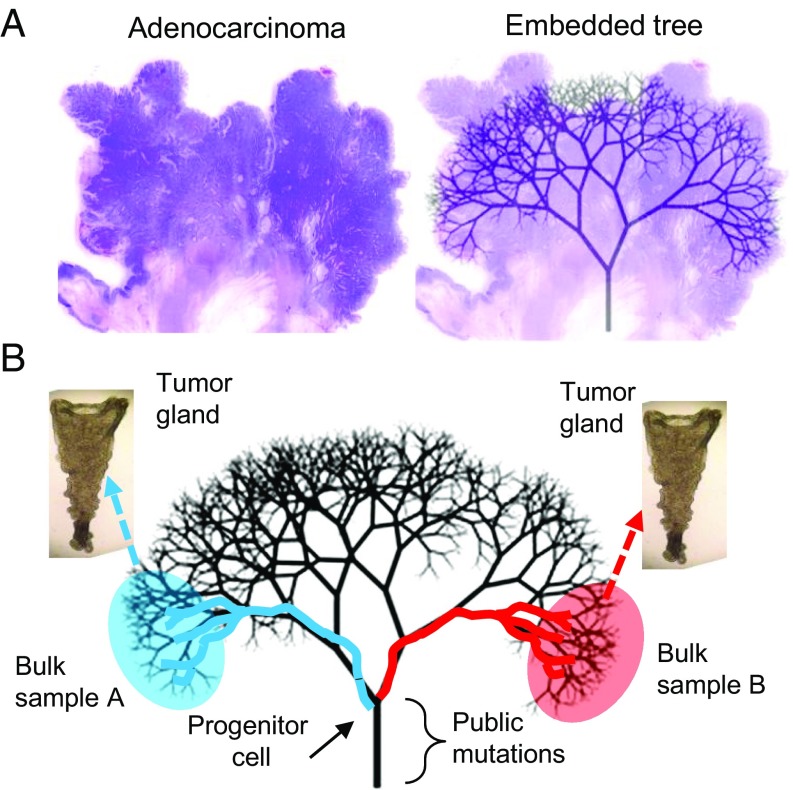
Fig. 1.
Tumor ancestral trees are physically embedded within their tumors. (A) The typical colorectal adenocarcinoma is spherical, and its cells are compartmentalized into glands. Present-day tumor cells coalesce back in time to progressively fewer ancestors. The final common ancestor is the progenitor cell. Cell mobility is relatively limited in solid glandular tumors, so a cell on one side of a large tumor will tend to remain on that side. Hence, the ancestral tree is physically embedded within the tumor. (B) Only a portion of the embedded tree can be sampled because the tree has billions of tips. With neutral evolution and a single expansion, sampling glands from opposite tumor sides misses most later branches but reliably samples earlier branches that coalesce to the progenitor cell. Because of growth, only mutations acquired early during growth attain allelic frequencies detectable by current sequencing technologies. Hence, most of the detectable information recorded by private mutations reflect the cell divisions and movements during the first few divisions after a tumor starts to grow.
Tumor ancestries can be reconstructed by sampling different parts of a tumor and comparing their mutations. Because it is impractical to sample every part of a tumor, such trees only reconstruct a small fraction of its ancestry. The sampling strategy is critical because the topography of an ancestral tree is physically embedded within a solid tumor where cell mobility is restricted (Fig. 1). For primary colorectal tumors, which are usually spherical, their embedded ancestral trees can be visualized as the branches of a tree. Notably, although peripheral tree branches and tips change during growth, branches near the trunk remain stable.
Although all branches of a tumor cannot be reconstructed due to practical limitations, it is possible to gain insight into the main branches of the tree close to the root by sampling from opposite tumor sides (Fig. 1). This early tree has only a few branches and represents the genealogy of the tumor during its early growth phase. Importantly, private mutations that occur early during growth are more likely to be detectable than later mutations because they are present in more final tumor cells (8, 9). This relationship between when a mutation occurs during growth and its detectability in the final tumor is critical because current exome-sequencing technologies have sensitivities of ∼10% (10). Indeed, the low sensitivity requires a mutation to be present in at least 20% of the cells in a sample if its locus is diploid. For an exponential expansion, private mutations that occur after the first few divisions will have frequencies undetectable by exome sequencing, and their branches are less likely to be sampled. Hence, only the earliest cell division branches can be reliably sampled and reconstructed.
Through reconstruction of the early ancestral tree of a clinical tumor specimen, we can gain insights into the phenotype of the founding cell and its immediate progeny. Specifically, we hypothesize that early tree reconstruction based on multiregional sampling of the final tumor may record cell movement during the early stages of the tumorigenesis. More precisely, in absence of early cell movement, we expect private mutations in the final tumor to cluster spatially, in alignment with the respective branches of the early tree. On the other hand, if early cell movement is present, we expect private mutations to be side-variegated in the final tumor and present on branches that cross sides of the early tree.
To explore this hypothesis, we developed a stochastic multiscale model of colorectal tumor expansions. We specifically incorporated the glandular structure of colorectal tumors because glands physically partition a tumor into neighborhoods of related cells. Based on the model simulations, we made several predictions, which we then tested against genomic data from multiregional and single gland data from 19 human colorectal tumors. Our findings indicate that the topography of mutations in the final tumor can provide insights into the early tumor phenotype.
Results
Model Validation.
The model presented here is an extension of the computational model in ref. 6 and explicitly accounts for the spatial configuration of cells inside the glands (Fig. 2A). First, we fit the model to multiregional sequencing data from 19 human colorectal tumor specimens using approximate Bayesian computation (ABC). Posterior parameter distributions for the mutation burst rate, the number of stem cells per gland, and the cellular mobility, as well as posterior checks that illustrate goodness-of-fit, are found in SI Text.
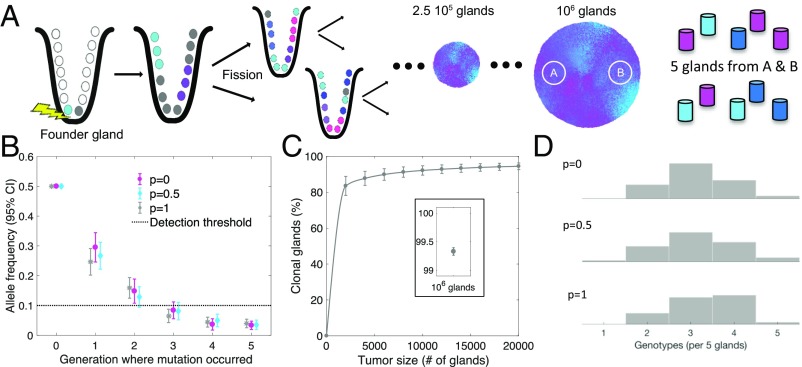
Fig. 2.
Simulations reveal the critical role of early events during tumorigenesis in defining final tumor ITH. (A) The first cancer cell in the founder gland initiates the exponential growth phase of the tumor through cell divisions followed by consecutive gland fission (34)—that is, the division of one gland into two daughter glands. Tumors are simulated until they contain 1 million cancer glands (∼10 billion cells). Two bulk samples from opposite sides are extracted from the final tumor, and five glands are sampled at random from each bulk sample. (B) Only private mutations that arise during the first few generations are detectable in a bulk sample of the final tumor. The mean allelic frequency of a private mutation generated after the second generation drops below the next-generation sequencing detection threshold of 10% (dotted line); the strength of cell mobility (p) does not substantially influence the outcome. (C) During tumor growth, glands quickly become clonal with respect to detectable mutations. Once the tumor reaches a size of 10,000 glands, ∼90% are already clonal; at a size of 106 glands, virtually all glands are clonal (C, Inset). (D) Due to the absence of selective sweeps, neutral evolution predicts high local ITH. Histograms show the distribution of unique gland genotypes per five glands sampled from the left bulk region for p = 0, p = 0.5, and p = 1, respectively (each histogram based on 100 simulated tumors). Simulation parameters (unless otherwise specified): n = 16, λ = 1.13, p = 1.
Next, we used the model to predict several features of tumor growth. As expected in a neutral evolution scenario, only mutations acquired by the second growth division could be reliably detected in the final tumor (Fig. 2B), because private mutations acquired during later divisions had allelic frequencies of <10%, the detectability threshold for current exome sequencing (10). In particular, this indicates that most detectable ITH in the final tumor is due to private mutations acquired during the first few divisions of growth. At the single gland level, ITH disappeared rapidly because, through exponential expansion, most glands eventually originated from only one of the early genotypes, and their cells had the same public and private mutations (Fig. 2C). Because of the star-shaped genealogies of the neutral evolution dynamics, substantial ITH was maintained within bulk regions of the tumor as adjacent glands often had distinctly different genotypes (Fig. 2D).
The above model predictions were corroborated by the data from human colorectal tumors. In all 19 specimens, private mutations differed between opposite sides (Fig. 3A and Dataset S1), indicating at least two different subclones per tumor. However, many private mutations had subclonal mutation frequencies (Fig. 3B), suggesting more than one subclone per bulk region. Since the neutral model indicates that these mutations arose early during tumorigenesis (Fig. 2B), mutation rates may have been elevated early during growth (11). Consistent with the simulations (Fig. 2B), genotypes often differed between glands isolated from the same tumor side (Fig. 3A), with an average of 2.4 different gland subclones (range 1–4) per bulk tumor region (Table 1). Equally consistent with the simulations (Fig. 2C), individual glands were clonal populations as demonstrated by comparable mutation frequencies of public and private mutations (Fig. 3B and Dataset S1). In summary, the proposed multiscale model of neutral evolution was in agreement with key features observed in the human tumors.
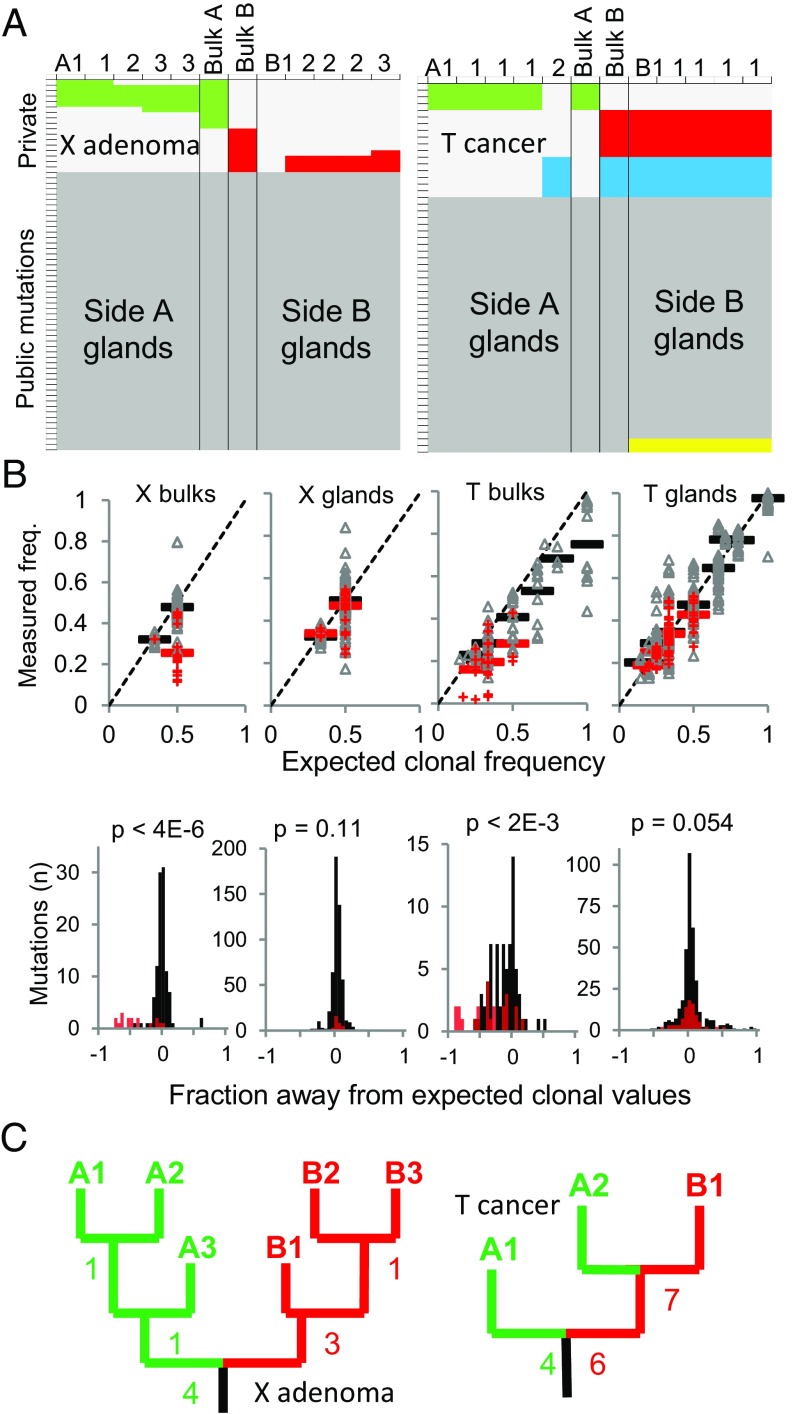
Fig. 3.
ITH between opposite sides of tumors X and T. Data for other tumors are provided in Dataset S1. (A) Targeted DNA resequencing of bulk and individual gland samples defines public (dark gray) and private mutations. Private mutations are either side-specific (green is side A or left; red is side B or right) or found on both tumor sides (blue). Yellow panels indicate inferred mutation losses based on homoplasy (Methods). (B) Based on ploidy, public mutations (black) are near their expected clonal frequencies (freq.) in both bulk and gland specimens. Private mutations (red) are at lower than expected clonal frequencies in bulk specimens but indistinguishable from the public mutations in the gland specimens, indicating that ITH disappears within glands because they are composed of cells with identical genotypes. (C) Gland trees based on the gland data from A. Numbers indicate how many additional private mutations separate nodes. Each branch tip represents one of the unique gland genotypes.
Table 1.
Colorectal tumors
| Tumor | Type | Size, cm | Glands (unique R, L) | Drivers: public, private | Side mixing |
| K | Adx | 6.0 | 12 (4,2) | 1, 1 | No |
| S | Adx | 6.0 | 8 (1,1) | 3, 0 | No |
| P | Adx | 3.5 | 7 (1,1) | 3, 0 | No |
| X | Adx | 2.5 | 10 (3,3) | 3, 0 | No |
| A | Cx S1 | 5.6 | 9 (4,4) | 2, 1 | No |
| O | Cx S3 | 9.5 | 11 (3,2) | 1, 0 | No |
| C | Cx S3 | 6.4 | 10 (3,3) | 1, 0 | No |
| H | Cx S4 | 4.0 | 10 (2,3) | 1, 0 | No |
| G | Cx S3 | 3.5 | 10 (2,3) | 5, 0 | No |
| J | Cx S3 | 5.0 | 10 (2,2) | 1, 1 | No |
| M | Cx S2 | 3.0 | 14 (3,3) | 4, 0 | Yes |
| N | Cx S1 | 2.3 | 10 (1,3) | 4, 0 | Yes |
| T | Cx S3 | 5.7 | 10 (2,1) | 1, 0 | Yes |
| W* | Cx S1 | 3.4 | 10 (2,2) | 5, 0 | Yes |
| U | Cx S2 | 3.9 | 10 (1,1) | 2, 0 | Yes† |
| D | Cx S1 | 2.0 | 10 (2,4) | 2, 2 | Yes |
| F | Cx S1 | 1.8 | 10 (2,4) | 1, 0 | Yes |
| E‡ | Cx S1 | 6.1 | 10 (3,3) | 4, 5 | Yes |
| R | Cx S1 | 3.5 | 10 (3,3) | 1, 0 | Yes |
| Avg | — | — | 2.4§ | 2.4, 0.12 | — |
Adx, adenoma; Cx, carcinoma; L, left; R, right.
MSI+.
Bulk only.
POLE mutant.
Per bulk region.
Reconstructing Early Ancestral Trees.
Both the simulations and the experimental data indicated stereotypic private mutation topographies, where ITH was consistently present between tumor sides, sometimes present within small bulk regions, and inevitably disappeared within individual tumor glands. In consequence, the clonal gland genotypes could be used to reconstruct ancestral trees that coalesced to single common ancestors (Fig. 3C and Fig. S1). Indeed, the early ancestral trees reflected past cell divisions because the genotype of each clonal gland represents the genotype of a single cell. Based on the detectability thresholds, these trees reflected the earliest growth divisions (i.e., the base of the full ancestral tree) (Fig. 1).
One way to distinguish selection from neutral evolution is to compare the relative frequencies of private and public driver mutations. With neutral evolution, selection for growth is conferred by driver mutations in the progenitor cell, and therefore most driver mutations are public mutations. With selection during growth, each branch of the early ancestral tree should have a unique driver mutation, and therefore most driver mutations should be private mutations. While at least one canonical driver mutation was identified per tumor, and an average of 2.9 public drivers per tree trunk, most individual branches lacked private driver mutations with an average of only 0.25 drivers per branch (Table 1). For the 17 nonmutator tumors (excluding tumors E and W; Table 1), there was an average of 2.4 public drivers per trunk and only 0.04 private drivers per branch. Therefore, we found little evidence that tree branching was due to the selection of new private driver mutations because most driver mutations were public mutations and most branches lacked driver mutations.
The Starting Points of Benign and Malignant Tumors May Be Different.
If driver mutations are infrequently acquired during tumor growth, the early growth of the final tumor largely depends on the drivers present in the founding cell. This reasoning further implies that phenotypes of benign and malignant progenitor cells differ. A fundamental phenotypic difference between benign and malignant tumors is cellular mobility. Similar to normal tissue growth, as exemplified by the patch-like distributions of G6PDH-stained crypts in normal human colon (12), the cells of benign tumors remain localized. In contrast, the cells of malignant tumors invade the surrounding tissue and seed metastases to distant organs. To explore potential fingerprints of progenitor cell mobility in the final tumor specimen, we simulated different degrees of cell mobility during growth (Fig. 4).
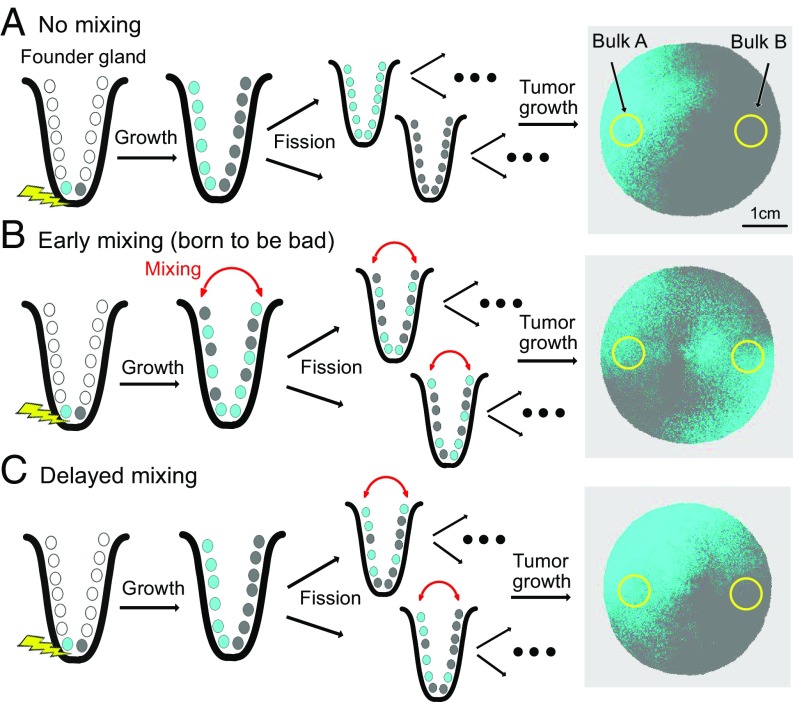
Fig. 4.
Born to be bad. (A) The phenotype of the founding cell dictates the subsequent distribution of private mutations: a lack of cell mobility (p = 0) leads to side-segregated hemispheric distributions of private mutations. (B) Strong cell mobility (p = 1) leads to cell mixing in the first gland (born to be bad) and variegated distributions in the final tumor. (C) If the onset of a mobile cell phenotype is delayed to the second gland, private mutations remain segregated, similar to A. Simulation parameters (unless otherwise specified): n = 16; λ = 1; p = 1; final tumor size: 106 glands.
In the benign phenotype scenario without cellular mobility (Fig. 4A), daughter cells remained adjacent after division of the mother cell and did not intermix (i.e., move between sides of the first tumor gland). Consequently, the mutation topography of the final tumor was hemispheric, and the same private mutation was rarely detected when sampling opposite tumor sides (Fig. 5A; p = 0). In the malignant scenario (Fig. 4B), abnormal cell movement was simulated by allowing daughter cells to randomly move and intermix after cell division. Daughter cells with the same private mutation could thus move to opposite tumor sides during the early growth phase. Further growth and expansion resulted in the same private mutation on opposite tumor sides (Fig. 5A; p > 0) because the final tumor was essentially a larger, expanded version of its smaller state. The timing of cell intermixing was critical because it had to occur in the first tumor gland to be reliably detectable in the final tumor. Cell intermixing after the first gland division rarely led to the same private mutation on opposite final tumor sides because increasing spatial separation hindered hemispheric mutation cross-over.
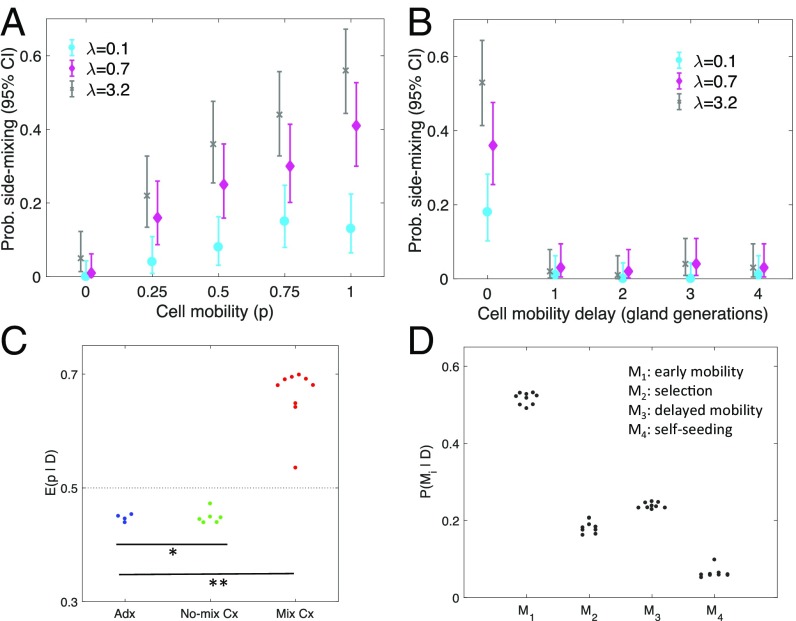
Fig. 5.
The footprints of early cell mobility. (A) The degree of cell mobility (p) strongly influences the probability (Prob.) of side-mixing (i.e., of finding one or more private mutations on both sides of the tumor). The probability to observing side-mixing is sensitive to the mutation burst rate (λ). (B) The importance of early cell mobility is emphasized by simulations where the onset of a mobile cell phenotype is delayed to the first, second, third, and fourth gland generation, respectively. Even with high cellular mobility (p = 1) and a high mutation burst rate (λ = 3.2), the probability of mixing is negligible if onset of a mobile cell phenotype is delayed beyond the founding gland. (C) The model is individually fit to each of the 19 human colorectal tumor samples, and the posterior mean of the cell mixing strength p is shown (dotted line is the prior mean of 0.5). In contrast to adenomas (Adx) and nonmixing carcinomas (No-mix Cx), there is evidence for cellular mobility in the mixing carcinomas (Mix Cx). *P < 0.03 [significant differences (Wilcoxon rank-sum test) between Mix Cx and Adx]; **P < 0.001 [significant differences (Wilcoxon rank-sum test) between Mix Cx and No-Mix Cx]. (D) For the mixing carcinomas, Bayesian model selection is used to compute the marginal posterior probabilities of the early cell mobility model (M1), a model with selection (M2), a model with delayed onset of cell mobility (M3), and a model with self-seeding of cells across the tumor mass (M4).
Experimentally, none of the four adenomas had private mutations on opposite tumor sides (Table 1), indicating a lack of early abnormal cell intermixing. In contrast, private mutations were detected on both sides of 9 of 15 cancers. The abnormal movements of daughter cells to opposite sides of their first tumor glands are reflected by trees where private mutations acquired on one tumor side branch over to the other tumor side (Fig. 3C and Fig. S1). Results from statistical inference corroborated the role of early cell mobility in explaining these findings. The posterior mean of the mixing strength p was elevated in 9 of 15 carcinomas and reduced in all four adenomas (Fig. 5C). Similarly, the posterior probability of a complete lack of cellular mobility (p = 0) was ∼5% in 8 of 15 carcinomas and ∼25% in adenomas (Fig. 5D). Because cell mixing was assumed to be stochastic, mixing did not inevitably result in variegated mutation distributions, and simulations showed that even with extensive mixing (p = 1), the probability to detect a private mutation in both bulk regions did not exceed 60% (Fig. 5A). Finally, Bayesian model selection suggested that early cell mobility provided a more likely mechanism for the side-mixing of carcinomas compared with models where mixing could arise through alternative mechanisms, e.g., selection, delayed onset of cell mobility, and tumor self-seeding (Fig. 5D).
Discussion
Due to the impracticability of longitudinal sampling, the evolutionary dynamics of human tumors can rarely be observed directly. Instead, an attractive systematic method to study human tumorigenesis is to reconstruct their ancestries from mutations, which is facilitated by the widespread ITH in most human tumors (3–7). Tumor phylogenies can be reconstructed with many different types of data, sampling schemes, and methods that have different strengths and weaknesses (13). Real tumor trees are complex and bushy because there are billions of cells or tips (Fig. 1). Because most tips cannot be experimentally sampled, inferred trees are limited representations of the past history, and it is often unclear or unstated what exact events are reconstructed. Several studies have previously modeled tumor growth and the role of cell migration (14–17), including simulations that describe how early cell mixing determines the topography of mutations in the final tumor (18). Here, we combined simulations with experimental spatial sampling of natural tumor subclones (glands) to infer that much of the ancestral information recorded by easily detectable somatic mutations comes from the early divisions when a tumor starts to grow. We then showed that the spatial distribution of private mutations in the final tumor could provide insights into the founding phenotype, particularly with respect to cellular mobility.
Abnormal cellular mobility constitutes one of several phenotypic traits that distinguish benign from malignant tumors. Indeed, important hallmarks of tumor malignancy are invasion and metastasis, which both require abnormal cell mobility. The model of stepwise sequential selection implies that increasingly more malignant phenotypes arise during tumor growth due to the acquisition of additional private driver mutations. In contrast, under the model of neutral evolution, the founding cancer cells already have a malignant phenotype, and additional private driver mutations are rarely acquired during growth. Thus, the first founding cell of a cancer should already have the mobile phenotype needed for eventual invasion and metastasis. Our results suggest that some cells present in the founding tumor gland already express a phenotype of abnormal cell mobility. The latter can be detected because mobile and immobile phenotypes leave different footprints in the final tumor. Mobile cells born next to each other may end up on opposite sides of the final tumor, whereas immobile cell phenotypes are more likely to cluster together in the final tumor. Based on our early tree reconstruction and statistical inference of the cellular mobility parameter, there was evidence of early cell mobility in more than half the invasive colorectal carcinomas, but none in the benign adenomas. The small sample size precludes definite conclusions, and based on the simulations, even if there was abnormal cell mobility in all of the cancers, such mobility could be detected by our sampling in only about half the cancers. Nevertheless, these observations suggest that benign and malignant tumors may have different mobility patterns early on and that their footprints remain visible in the clinically detected tumor specimen.
The observed patterns of side-mixing in many carcinomas and lack thereof in adenomas could potentially be attributable to factors other than early cell mobility. First, selection during growth could reduce heterogeneity and hence increase the likelihood to observe side-mixing in the final tumor through the formation of clonal patches. Second, cellular mobility may not be present in all cells, but be delayed to a later mutation. Third, side-mixing may be caused by tumor self-seeding whereby cells are seeded to opposite sides of primary tumor via the circulation (19). However, formal model selection (Fig. 5D) favored the early cell mobility model to explain the observed patterns.
Prior studies using some of the same tumors as described in this study indicated that many human colorectal tumors grow through neutral evolution as single Big Bang expansions (6). Such single neutral expansions (Fig. 1) emphasize the relationship between spatial tumor growth and phylogenetic branching. Neutral evolution is relatively easy to simulate because all tumor cells have similar fitness, which is conferred by the public driver mutations in the progenitor cell. The subdivision of tumor cells into small glands may inherently limit selection during growth because competition between cells is limited to neighboring cells. Consistent with neutral evolution and single expansions, canonical driver mutations were predominately public mutations and most tree branches lacked private drivers. Potentially, other unmeasured private alterations could confer selection. However, consistent with single star-shaped expansions, small bulk tumor areas often contained individual glands with different private mutations, indicating that selection was at best very weak. Any selective mechanism compatible with our data would require private drivers to be as common as branches on the underlying tree and yet be sufficiently weak to preserve the local ITH between individual glands.
The single expansion hypothesis of neutral evolution provides a very simple model of tumorigenesis because no further driver mutations are needed once the tumors start to grow. The early expression of abnormal cell mobility is consistent with neutral evolution and the hypothesis that a cancer progenitor already has the driver mutations necessary for malignancy—they are “born to be bad”—and can help explain the occurrence of metastases very early during tumorigenesis (20, 21). Another clinically relevant observation is that neutrality maximizes ITH (5) and thus the potential for variants that may be responsible for subsequent therapeutic resistance. Other tumor types such as breast, lung, and kidney cancers show evidence of selection during growth (3, 4, 7, 22), but at least subsets of these tumors have mutation frequencies consistent with neutral evolution (23). To some degree, selection likely occurs in most tumors, and further evolution in response to new environmental stresses such as chemotherapy is likely to collapse and remodel a tumor tree (24).
In summary, our findings further corroborate that most clinically detectable colorectal tumors are single Big Bang expansions (6), where highly capable progenitors start with the drivers sufficient for tumorigenesis. We demonstrated the possibility to infer certain phenotypic traits of the progenitor cells based on their spatial footprints in the clinical tumor specimen. If validated in a larger dataset, these footprints could potentially be used for outcome prediction in small screen-detected lesions with unknown invasive potential, where early cell mixing could herald a much greater future propensity for invasion and metastasis.
Methods
Multiscale Model of Colorectal Tumor Growth.
The cells of the human colon are partitioned into a large number of individual glands (crypts), arranged in a regular, locally 2D lattice (25). Each gland contains ∼2,000 cells, and growth occurs through crypt fission in the normal colon (26). Tumor glands are larger (10,000–50,000 cells), and growth also occurs through gland fission, resulting in tumors filled with glands (i.e., adenomas and adenocarcinomas). Although clinical tumors appear spherical, if unfolded, the locally flat arrangement of glands can be recovered. Consequently, we modeled tumor growth in two spatial dimensions by arranging the glands on a regular lattice. Each gland was assumed to contain n spatially arranged stem cells responsible for renewal of the gland. Transit-amplifying and fully differentiated cells were not modeled because they only remain in the gland for a few weeks and are unlikely to contribute substantially to the process of carcinogenesis. Tumorigenesis was initiated by a single founding tumor cell. As the founding cell and its progeny kept dividing, they accumulated mutations and replaced the resident population of noncancerous stem cells. To allow for high mutational activity during tumor initiation (11), cells acquired mutations in bursts of size M ∼ Poisson (λ). Because mutations arising after the second generation are rarely detectable (Fig. 2B), mutations were only tracked until the fifth generation. Once the tumor cells made up the entire stem cell population of the first tumor gland, cell mobility was modeled by having each stem cell exchange its position with a neighboring cell with probability p. After cell mixing, the gland underwent fission, and each of the new daughter glands inherited half of the mother gland’s stem cells. The two daughter glands then grew back to carrying capacity through repeated stem cell division before undergoing fission themselves. Space for new glands was created by pushing neighboring glands toward the periphery of the tumor. Through repeated cell division and gland fission, the tumor grew in a spatially structured and spherical manner (Fig. 4). After birth of the first tumor cell, growth can broadly be divided into two main phases: an exponential growth phase from first gland to carrying capacity and a maintenance phase where birth and death are in equilibrium and the tumor no longer grows in size. Because mutations arising during the latter phase are unlikely to reach detectable allelic frequencies (they are trapped within glands), we only modeled the exponential expansion phase. Tumor growth was simulated until a final tumor size of 750,000 glands (∼1010 tumor cells, ∼3 × 3 × 1.5 cm). All simulations were performed in MATLAB (Version 2017a; The MathWorks, Inc.). Details are found in SI Text.
Statistical Inference.
Statistical inference on the model parameters n (number of stem cells per gland), λ (mutation burst rate), and p (cellular mobility) was performed by using a rejection sampling-based version of ABC (27, 28). Parameter space was discretized over plausible ranges, and 100 tumors were simulated on a total of 15,000 grid points that covered the plausible range in parameter space. The distance between simulated and real tumors was ascertained based on the L2 distance of multivariate summary statistics. Rejection sampling was then applied to obtain approximate posterior distributions for the model parameters. Posterior checks were used to evaluate goodness of fit. Bayesian model selection through rejection sampling on the joint model–parameter space (29) was performed to compare the early cell mobility model against alternative modes with selection, delayed cell mobility, and self-seeding, respectively. Details are found in SI Text.
Data from Human Tumor Specimens.
We sampled small (∼0.5 cm3) bulk regions from opposite sides of 4 colorectal adenomas and 15 colorectal adenocarcinomas. From each bulk sample, DNA was isolated, and whole-exome sequencing was performed. Mutations detected on both tumor sides were labeled as public, and mutations detected only on a single side were labeled as private. Up to five individual glands per bulk were used for targeted deep resequencing of candidate mutations as identified by the whole-exome sequencing (Table 1).
Tumors.
Tumors and matched normal colon (Table 1) were obtained fresh at the Norris Comprehensive Cancer Center, and bulk samples (∼0.5 cm3) from opposite sides (arbitrarily called A and B) were procured, as described (6). Some data from 10 of the tumors have been published (6, 8) and have the same names, except tumors R and G in this study are new. Individual tumor glands were isolated by an EDTA washout (>95% epithelial cell purity). Bulk samples consisted of hundreds of individual tumor glands. All samples were obtained during normal clinical care and deidentified, with written approval by the Institutional Review Board of the University of Southern California Keck School of Medicine.
DNA sequencing.
DNA was isolated from the bulk specimens and exome-sequenced by using TruSeq or Nextera Rapid Capture kits (Illumina) on HiSeq2000 or NextSeq500 platforms (Illumina). Files were processed by using the Galaxy public website (30) using BWA for alignment, followed by RmDup, Realigner Target Creator, Count Covariates, and Table Recalibration. Mutations were called by using MuTect (10) at standard high-confidence settings. Average sequencing depth at the called mutations was 54 reads in the tumors and 45 reads in the normal colon. Further filtering removed mutations with frequencies <10%. For bulk samples, a mutation was considered public if it was detected on both tumor sides and private if detected on only one side. For the gland samples, a mutation was considered public if found in all glands and private if found in only some glands. The information obtained from the gland sequencing superseded the bulk data because with cell mixing, a private mutation can be found on both tumor sides, and sometimes a private mutation was detected on both sides after targeted DNA resequencing. Targeted DNA resequencing (AmpliSeq followed by IonTorrent) was performed on subsets of the mutations for bulk samples and individual glands. Average resequencing depth was ∼1,500 reads per mutation, with a minimum of 20 reads per mutation. A mutation was considered present if its frequency was >5%.
Determination of public and private mutation clonality.
Gland chromosome copy number (CN) was determined for three to five glands per tumor side by using high-density SNP arrays (Illumina) and Paired Parent-Specific Circular Binary Segmentation (31) as described (6). For most mutations, ploidy at the locus could be inferred because CN was constant in all glands on a side. Mutations where CN at the locus was variable were not used. A two-sided t test was used to determine if private mutation frequencies were significantly different from public mutation frequencies. Fraction away from expected clonal values was calculated as measured frequency minus expected clonal frequency divided by expected clonal frequency.
Driver mutations.
Driver mutations were defined as nonsynonymous mutations in 29 significantly mutated genes in colorectal carcinomas (excluding TTN from figure 1B in ref. 32) or in any of the 572 consensus COSMIC mutations (Census_allMon Mar 21 17-49-11 2016) that truncated a designated tumor suppressor gene or had a high functional impact as judged by MutationAssessor (33).
Tree inference.
The gland trees were manually reconstructed by using the targeted DNA resequencing data from individual glands. Reconstruction was facilitated by the clonal nature of all sampled glands. The primary consideration was homoplasy avoidance (i.e., avoiding the scenario whereby a unique mutation arises multiple times in a tree). For 10 of the 19 trees, homoplasy could be avoided by evoking gland-specific loss of a mutation (yellow panels in Fig. 3A and Dataset S1) at a small number of loci (n = 1–6). In many cases, loss of a single stretch of DNA could account for the loss of multiple loci because they were neighbors on a chromosome.
Supplementary Material
Acknowledgments
We thank Prof. Rick Durrett (Duke University) for fruitful discussions on model development. This work was supported by National Institutes of Health Grants CA185016, CA196569, P30CA014089, K99CA207872; National Science Foundation Grant DMS 1614838; and Swiss National Science Foundation Grant P300P2-154583.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1716552115/-/DCSupplemental.
References
- 1.Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61:759–767. doi: 10.1016/0092-8674(90)90186-i. [DOI] [PubMed] [Google Scholar]
- 2.Nowell PC. The clonal evolution of tumor cell populations. Science. 1976;194:23–28. doi: 10.1126/science.959840. [DOI] [PubMed] [Google Scholar]
- 3.de Bruin EC, et al. Spatial and temporal diversity in genomic instability processes defines lung cancer evolution. Science. 2014;346:251–256. doi: 10.1126/science.1253462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gerlinger M, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366:883–892. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ling S, et al. Extremely high genetic diversity in a single tumor points to prevalence of non-Darwinian cell evolution. Proc Natl Acad Sci USA. 2015;112:E6496–E6505. doi: 10.1073/pnas.1519556112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sottoriva A, et al. A Big Bang model of human colorectal tumor growth. Nat Genet. 2015;47:209–216. doi: 10.1038/ng.3214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yates LR, et al. Subclonal diversification of primary breast cancer revealed by multiregion sequencing. Nat Med. 2015;21:751–759. doi: 10.1038/nm.3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kang H, et al. Many private mutations originate from the first few divisions of a human colorectal adenoma. J Pathol. 2015;237:355–362. doi: 10.1002/path.4581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Durrett R. Population genetics of neutral mutations in exponentially growing cancer cell populations. Ann Appl Probab. 2013;23:230–250. doi: 10.1214/11-aap824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cibulskis K, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013;31:213–219. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhao J, et al. Early mutation bursts in colorectal tumors. PLoS One. 2017;12:e0172516. doi: 10.1371/journal.pone.0172516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Novelli M, et al. X-inactivation patch size in human female tissue confounds the assessment of tumor clonality. Proc Natl Acad Sci USA. 2003;100:3311–3314. doi: 10.1073/pnas.0437825100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schwartz R, Schäffer AA. The evolution of tumour phylogenetics: Principles and practice. Nat Rev Genet. 2017;18:213–229. doi: 10.1038/nrg.2016.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Poleszczuk J, Hahnfeldt P, Enderling H. Evolution and phenotypic selection of cancer stem cells. PLOS Comput Biol. 2015;11:e1004025. doi: 10.1371/journal.pcbi.1004025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martens EA, Kostadinov R, Maley CC, Hallatschek O. Spatial structure increases the waiting time for cancer. New J Phys. 2011;13:115014. doi: 10.1088/1367-2630/13/11/115014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hallatschek O, Fisher DS. Acceleration of evolutionary spread by long-range dispersal. Proc Natl Acad Sci USA. 2014;111:E4911–E4919. doi: 10.1073/pnas.1404663111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Manem VS, Kohandel M, Komarova NL, Sivaloganathan S. Spatial invasion dynamics on random and unstructured meshes: Implications for heterogeneous tumor populations. J Theor Biol. 2014;349:66–73. doi: 10.1016/j.jtbi.2014.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Waclaw B, et al. A spatial model predicts that dispersal and cell turnover limit intratumour heterogeneity. Nature. 2015;525:261–264. doi: 10.1038/nature14971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim MY, et al. Tumor self-seeding by circulating cancer cells. Cell. 2009;139:1315–1326. doi: 10.1016/j.cell.2009.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klein CA. Parallel progression of primary tumours and metastases. Nat Rev Cancer. 2009;9:302–312. doi: 10.1038/nrc2627. [DOI] [PubMed] [Google Scholar]
- 21.Naxerova K, et al. Origins of lymphatic and distant metastases in human colorectal cancer. Science. 2017;357:55–60. doi: 10.1126/science.aai8515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sun R, et al. Between-region genetic divergence reflects the mode and tempo of tumor evolution. Nat Genet. 2017;49:1015–1024. doi: 10.1038/ng.3891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Williams MJ, Werner B, Barnes CP, Graham TA, Sottoriva A. Identification of neutral tumor evolution across cancer types. Nat Genet. 2016;48:238–244. doi: 10.1038/ng.3489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Robertson-Tessi M, Anderson AR. Big Bang and context-driven collapse. Nat Genet. 2015;47:196–197. doi: 10.1038/ng.3231. [DOI] [PubMed] [Google Scholar]
- 25.Baker AM, et al. Quantification of crypt and stem cell evolution in the normal and neoplastic human colon. Cell Rep. 2014;8:940–947. doi: 10.1016/j.celrep.2014.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Humphries A, Wright NA. Colonic crypt organization and tumorigenesis. Nat Rev Cancer. 2008;8:415–424. doi: 10.1038/nrc2392. [DOI] [PubMed] [Google Scholar]
- 27.Marjoram P, Molitor J, Plagnol V, Tavare S. Markov chain Monte Carlo without likelihoods. Proc Natl Acad Sci USA. 2003;100:15324–15328. doi: 10.1073/pnas.0306899100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sottoriva A, Spiteri I, Shibata D, Curtis C, Tavaré S. Single-molecule genomic data delineate patient-specific tumor profiles and cancer stem cell organization. Cancer Res. 2013;73:41–49. doi: 10.1158/0008-5472.CAN-12-2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Toni T, Stumpf MP. Simulation-based model selection for dynamical systems in systems and population biology. Bioinformatics. 2010;26:104–110. doi: 10.1093/bioinformatics/btp619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Afgan E, et al. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2016 update. Nucleic Acids Res. 2016;44:W3–W10. doi: 10.1093/nar/gkw343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Olshen AB, et al. Parent-specific copy number in paired tumor-normal studies using circular binary segmentation. Bioinformatics. 2011;27:2038–2046. doi: 10.1093/bioinformatics/btr329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cancer Genome Atlas Network Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reva B, Antipin Y, Sander C. Predicting the functional impact of protein mutations: Application to cancer genomics. Nucleic Acids Res. 2011;39:e118. doi: 10.1093/nar/gkr407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Garcia SB, Park HS, Novelli M, Wright NA. Field cancerization, clonality, and epithelial stem cells: The spread of mutated clones in epithelial sheets. J Pathol. 1999;187:61–81. doi: 10.1002/(SICI)1096-9896(199901)187:1<61::AID-PATH247>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.