Significance
Cytokines are produced in response to microbial threat and aid in the recruitment and activation of immune cells to protect the host. Using complementary in vitro and in vivo approaches, we have defined a cytokine network involving IL-36γ, IL-23, and IL-22 that is induced following intestinal damage and is critical for antimicrobial activity, tissue repair, and host survival. Our data identify IL-36γ/IL-36 receptor signaling as a central upstream driver of the IL-23/IL-22/antimicrobial peptide (AMP) pathway during intestinal injury and advance the concept that IL-36γ and IL-23 are fundamentally linked to repair of acute barrier damage. These findings provide new mechanistic insight into how the host commandeers proinflammatory cytokines for tissue repair and highlight the potential for manipulating the IL-36/IL-23/IL-22/AMP network in treating acute intestinal damage.
Keywords: innate immunity, interleukin, inflammatory bowel disease, repair
Abstract
The gut epithelium acts to separate host immune cells from unrestricted interactions with the microbiota and other environmental stimuli. In response to epithelial damage or dysfunction, immune cells are activated to produce interleukin (IL)-22, which is involved in repair and protection of barrier surfaces. However, the specific pathways leading to IL-22 and associated antimicrobial peptide (AMP) production in response to intestinal tissue damage remain incompletely understood. Here, we define a critical IL-36/IL-23/IL-22 cytokine network that is instrumental for AMP production and host defense. Using a murine model of intestinal damage and repair, we show that IL-36γ is a potent inducer of IL-23 both in vitro and in vivo. IL-36γ–induced IL-23 required Notch2-dependent (CD11b+CD103+) dendritic cells (DCs), but not Batf3-dependent (CD11b−CD103+) DCs or CSF1R-dependent macrophages. The intracellular signaling cascade linking IL-36 receptor (IL-36R) to IL-23 production by DCs involved MyD88 and the NF-κB subunits c-Rel and p50. Consistent with in vitro observations, IL-36R– and IL-36γ–deficient mice exhibited dramatically reduced IL-23, IL-22, and AMP levels, and consequently failed to recover from acute intestinal damage. Interestingly, impaired recovery of mice deficient in IL-36R or IL-36γ could be rescued by treatment with exogenous IL-23. This recovery was accompanied by a restoration of IL-22 and AMP expression in the colon. Collectively, these data define a cytokine network involving IL-36γ, IL-23, and IL-22 that is activated in response to intestinal barrier damage and involved in providing critical host defense.
At mucosal surfaces, particularly the intestine, epithelial cells form a physical and functional barrier that protects the host from the unrestricted barrage of microbial and environmental stimuli (1, 2). Compromises in the epithelial barrier due to damage or dysfunction can result in activation of underlying immune cells. Once activated, innate and adaptive immune cells display enhanced antimicrobial activity and promote epithelial proliferation, repair of the damaged barrier, and resolution of inflammation (3). However, if the insult persists, or if repair processes are ineffective, chronic intestinal inflammation as seen in human inflammatory bowel disease (IBD) may ensue (4). Therefore, delineating the specific mechanisms involved in efficient tissue repair processes in the damaged intestine may provide insight into therapeutic strategies for the treatment of these inflammatory conditions.
Interleukin (IL)-22 is a key cytokine that links intestinal immune activation to epithelial repair and barrier protection following damage (3, 5). IL-22 is expressed by numerous immune cells, including type 3 innate lymphoid cells (ILC3), natural killer (NK) cells, neutrophils, and Th17 and Th22 cells (6). Intestinal epithelial cells express the IL-22R complex, and binding of IL-22 results in the induction of mucins, antimicrobial peptides (AMPs), and antiapoptotic pathways that collectively aid in limiting bacterial encroachment while promoting epithelial proliferation, wound healing, and repair (7). Mice that lack the ability to produce IL-22 following administration of dextran sodium sulfate (DSS) or Citrobacter rodentium are grossly unable to repair barrier damage or control pathogenic bacterial expansion (8–10). These data suggest that IL-22 plays a nonredundant function in mucosal barrier defense (11, 12).
Investigations into how IL-22 is regulated have led to the identification of IL-23 as one of the most potent inducers of this cytokine. Systemic administration of bacterial flagellin was shown to rapidly induce IL-23 production by intestinal Toll-like receptor 5 (TLR5)–expressing CD103+CD11b+ dendritic cells (DCs) and subsequent IL-22 expression (13). Additionally, stimulation of intestinal ILC3s, NK cells, neutrophils, and Th17 cells with IL-23 potently induces IL-22 production (6). Similarly, loss of IL-23 signaling in vivo during DSS-induced colitis completely abrogates colonic IL-22 expression and results in exacerbated disease (10). Furthermore, IL-23p19–deficient mice fail to produce IL-22, which leads to overgrowth of segmented filamentous bacteria (14). Collectively, these studies demonstrate an important role for the IL-23/IL-22 axis in barrier protection and control of bacteria, yet the upstream regulators of this critical pathway are incompletely undefined.
Among the many immunological factors produced in response to intestinal damage, IL-1 superfamily cytokines appear to play a major role in the inflammatory program (15). IL-1β, IL-18, and IL-33 are all induced during experimental colitis and are believed to contribute to the pathogenesis of IBD, but they may also be involved in tissue protection (16–18). Similarly, IL-36 cytokines, the more recently described members of the IL-1 superfamily, appear to potently induce inflammatory responses and regulate mucosal immunity (19, 20). We and others have reported that IL-36 cytokines are expressed in the intestine during inflammation (21–26) in response to stimulation by the microbiota (22). Once expressed, IL-36 ligands are involved in the activation of innate and adaptive immune cells and stromal cells that can exacerbate intestinal inflammation, and also play an instrumental role in resolution of intestinal damage (22, 23, 27, 28). This bifunctional effect of the IL-36/IL-36 receptor (IL-36R) axis during intestinal inflammation likely depends on the inducing stimuli, extent of tissue damage, and timing and chronicity of expression. In response to robust intestinal barrier destruction, IL-36R signals augment the inflammatory cascade early on, which appears to be linked to subsequent tissue protection and repair (22, 23). However, the specific pathways via which IL-36R signaling controls host defense and barrier protection remain to be elucidated.
In this report, we define a critical IL-36/IL-23/IL-22 cytokine network that is instrumental for AMP production and host defense following intestinal damage. Using a murine model of colonic damage and inflammation, we show that IL-36γ is a potent inducer of IL-23 production both in vitro and in vivo. IL-36γ–induced IL-23 was highly dependent upon Notch2-dependent (CD11b+CD103+) DCs, but not CSFR1-dependent macrophages or Batf3-dependent (CD11b−CD103+) DCs. The intracellular signaling cascade linking IL-36R signaling to IL-23 production from DCs involved MyD88 and the NF-κB subunits c-Rel and p50. Consistent with in vitro observations, IL-36R–deficient mice exhibited dramatically reduced IL-23 and IL-22/AMP levels, and these mice consequently failed to recover from acute intestinal damage. Interestingly, impaired recovery of mice deficient in IL-36R or IL-36γ could be completely rescued by treatment with exogenous IL-23. This recovery was accompanied by a restoration of IL-22 and AMP expression in the colon. Collectively, these data define a cytokine network involving IL-36γ, IL-23, and IL-22 that is activated in response to intestinal barrier damage and involved in providing critical host defense.
Results
IL-36R Deficiency Results in Impaired IL-23 and IL-22 Expression in the Colons of DSS-Treated Mice.
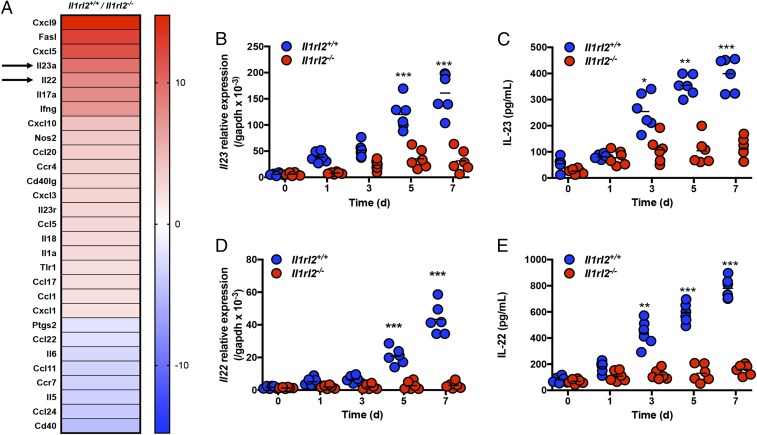
Recently, IL-36R signaling has been implicated in healing of mucosal damage (22, 23, 29), and our group demonstrated that IL-36R–deficient mice have impaired IL-22 production, and consequently fail to recover from acute intestinal damage. To begin exploring potential mechanisms of how IL-36R signaling induces IL-22 expression, we performed a PCR array on total colonic tissues isolated from Il1rl2+/+ and Il1rl2−/− mice at day 5 of DSS treatment. The array analysis revealed that the expression of IL-23 and IL-22 mRNA was approximately ninefold and approximately sevenfold higher, respectively, in Il1rl2+/+ mice compared with Il1rl2−/− mice (Fig. 1A). Given that IL-23 is a potent inducer of IL-22 (6), we postulated that impaired IL-22 expression in Il1rl2−/− mice may be associated with a lack of IL-23. To explore the link between IL-36R signaling, IL-23, and IL-22, we first examined the expression of IL-23 and IL-22 in Il1rl2+/+ and Il1rl2−/− mice during the course of DSS treatment (5 d of DSS followed by 2 d of regular water). Quantitative PCR and ELISA analysis of colonic tissue revealed that DSS-induced expression of IL-23 and IL-22 mRNA and protein was significantly higher in colonic tissue isolated from Il1rl2+/+ mice compared with Il1rl2−/− mice (Fig. 1 B–E). Additionally, following DSS treatment, the peak of IL-36γ expression preceded that of IL-23 and IL-22 at day 3, followed by robust IL-23 expression on day 5 and IL-22 expression on day 7 (SI Appendix, Fig. S1A). Collectively, these data suggest that signaling via IL-36R is involved in optimal IL-23 and IL-22 expression during DSS-induced damage.
Fig. 1.
IL-36R deficiency results in impaired IL-23 and IL-22 expression in the colons of DSS-treated mice. (A) PCR array gene expression analyses from colon tissues of Il1rl2+/+ and Il1rl2−/− mice treated with DSS for 5 d. The time course of IL-23 mRNA (B) and protein (C) expression in colons from Il1rl2+/+ and Il1rl2−/− mice treated with DSS is shown. The time course of IL-22 mRNA (D) and protein (E) expression in colons from Il1rl2+/+ and Il1rl2−/− mice treated with DSS is shown. Data are representative of three independent experiments with three to four mice per group. All data are presented as mean ± SEM. *P < 0.5; **P < 0.05; ***P < 0.001.
IL-36γ–Induced IL-22 Production in Colonic Explants from DSS-Treated Mice Is IL-23–Dependent.
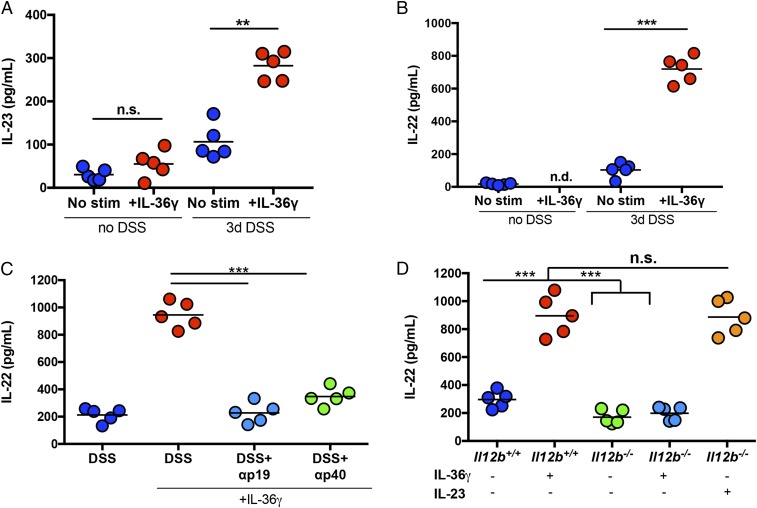
Next, we determined whether IL-23 is required for IL-36γ–induced IL-22 expression in colonic explants from DSS-treated mice. We focused our studies on IL-36γ as it is the predominant IL-36 ligand produced in the colon of mice during DSS-induced damage (22). Colonic explants isolated from healthy (non–DSS-treated), wild-type (WT) mice and stimulated with IL-36γ in vitro showed no detectable increases in either IL-23 or IL-22. However, colonic explants isolated from DSS-treated mice on day 3, a time when endogenous IL-36γ mRNA expression is highest (22) (SI Appendix, Fig. S1A), responded to exogenous IL-36γ stimulation by inducing IL-23 (approximately threefold) and IL-22 (approximately fivefold) compared with unstimulated controls (Fig. 2 A and B). Of note, IL-36α and IL-36β were also capable of inducing IL-23 and IL-22 (SI Appendix, Fig. S1 B and C). These data further correlated with a strong induction of IL-36R (Il1rl2) mRNA expression at day 3 following DSS treatment (SI Appendix, Fig. S1D). Having established that IL-36γ is an inducer of IL-23, we next assessed whether IL-36γ–induced IL-22 was IL-23–dependent. Indeed, antibody-mediated blockade of IL-23p19 (αp19) or IL-12/23p40 (αp40) was able to significantly reduce the ability of IL-36γ to induce IL-22 in colonic explants from DSS-treated mice (Fig. 2C). Similarly, while colonic explants from DSS-treated IL-12/23p40-sufficient mice (Il12b+/+) produced high levels of IL-22 in response to IL-36γ stimulation, explants obtained from DSS-treated IL-12/23p40–deficient mice (Il12b−/−) produced significantly less IL-22 under these conditions. This defect in IL-36γ–induced IL-22 production in Il12b−/− explant cultures was reversible by the addition of exogenous IL-23 (Fig. 2D). These results highlight a functional IL-36γ/IL-23/IL-22 cytokine network in colonic tissue from DSS-treated mice.
Fig. 2.
IL-36γ–induced IL-22 production in colonic explants from DSS-treated mice is IL-23–dependent. (A and B) Colonic explants from control (no DSS) or 3-d DSS-treated (3 d DSS) WT mice were cultured for 60 h in the presence or absence of IL-36γ. Supernatants were analyzed for IL-23 (A) and IL-22 (B) by ELISA. (C) Colonic explants from 3-d DSS-treated WT mice were stimulated with IL-36γ and αp19 or αp40 antibodies for 60 h, and IL-22 expression was assessed by ELISA. (D) Colonic explants from 3-d DSS-treated WT (Il12b+/+) or Il12b−/− mice stimulated with IL-36γ or IL-23 for 60 h and IL-22 expression were assessed by ELISA. Data are representative of two independent experiments with four to five mice per group. All data are presented as mean ± SEM (one-way ANOVA with Tukey’s multiple comparison test: **P < 0.01; ***P < 0.001). n.d., not detected; n.s., not significant; stim, stimulation.
Notch2-Dependent DCs Are Required for IL-36γ–Induced IL-23 and IL-22 Expression and Recovery from Acute Colonic Damage.
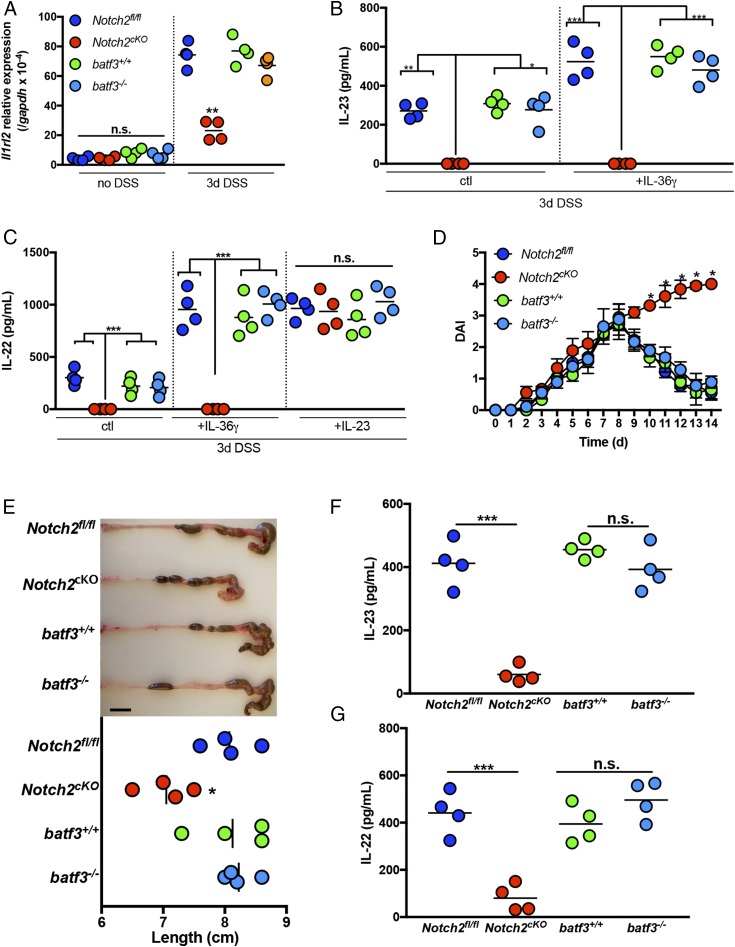
Having established IL-23 as a key intermediary between IL-36γ and IL-22, we next examined whether specific antigen-presenting cell subsets may be involved in IL-23 induction in response to IL-36γ. Intestinal DCs are categorized into two main populations: Notch2-dependent DCs and Batf3-dependent DCs (30). Within these subsets, CD103+CD11b+ DCs have been reported to accumulate in the intestines during DSS-induced colitis (31). To determine if either of these DC subsets is involved in IL-36γ–induced IL-23 production, we used Notch2-floxed mice that had been crossed with CD11c-Cre mice to generate mice with a deletion of Notch2 in the DC lineage (Notch2cKO), as well as Batf3-deficient mice (batf3−/−). Initially, we examined the expression of IL-36R (Il1rl2) mRNA in colonic tissue isolated from these mice following treatment with DSS for 3 d. Consistent with SI Appendix, Fig. S1B, we found that Il1rl2 was strongly induced in the colons of DSS-treated WT (batf3+/+) mice, as well as in batf3−/− mice. Conversely, the induction of Il1rl2 was significantly reduced in the colons of DSS-treated Notch2cKO mice, compared with control mice (Notchfl/fl) (Fig. 3A). Next, colonic explants from DSS-treated batf3−/− and Notch2cKO mice, and their respective controls, were stimulated in vitro with IL-36γ, and IL-23, as well as IL-22, expression was assessed by ELISA. While batf3−/− mice exhibited normal induction of IL-23 and IL-22 in response to IL-36γ, Notch2cKO mice completely failed to produce IL-23 and IL-22 in the presence of IL-36γ, compared with Notchfl/fl controls. Furthermore, the addition of exogenous IL-23 to Notch2cKO explant cultures was sufficient to restore IL-22 production in these cultures to normal levels (Fig. 3 B and C). Thus, IL-36γ–induced IL-23 appears to be dependent upon Notch2-dependent DCs in vitro. Of note, macrophages did not appear to play a significant role in IL-36γ–induced IL-23 expression during DSS, since treatment of mice with αCSF-1R antibody to deplete macrophages (32) had no detectable effect on the ability of IL-36γ to induce IL-23 or IL-22 in colonic explant cultures (SI Appendix, Fig. S2).
Fig. 3.
Notch2-dependent DCs are required for IL-36γ–induced IL-23 and IL-22 expression and recovery from colonic damage. (A) IL-36R (Il1rl2) mRNA expression was analyzed by qPCR in colon tissue isolated from DSS-treated batf3+/+, batf3−/−, Notch2fl/fl, and Notch2cKO mice directly ex vivo. (B and C) Colonic explants from DSS-treated mice were cultured for 60 h in the presence or absence of IL-36γ or IL-23. Supernatants were analyzed for IL-23 (B) and IL-22 (C) expression by ELISA. (D) DAI of batf3+/+, batf3−/−, Notch2fl/fl, and Notch2cKO mice treated with DSS for 5 d, followed by normal water. (E) Image and colon length from mice treated as in D, at day 14. (Scale bar, 1 cm.) The expression of IL-23 (F) and IL-22 (G) in colon tissues from DSS mice at day 5 is shown. Data are representative of two independent experiments with three to four mice per group. All data are presented as mean ± SEM (one-way ANOVA with Tukey’s multiple comparison test: *P < 0.5; **P < 0.05; ***P < 0.001). n.s., not significant.
To investigate whether Notch2-dependent DCs are also necessary for host recovery from DSS-induced intestinal damage in vivo, the disease activity index (DAI) of batf3+/+, batf3−/−, Notch2fl/fl, and Notch2cKO mice was compared following administration of DSS in the drinking water for 5 d, followed by normal water thereafter. While batf3+/+, batf3−/−, and Notch2fl/fl mice were all able to recover normally from DSS-induced intestinal damage, Notch2cKO mice were defective in colonic repair and had higher DAI scores (Fig. 3D), shorter colon length (Fig. 3E), and significantly reduced levels of IL-23 and IL-22 in colons directly ex vivo (Fig. 3 F and G). Together, these data highlight Notch2-dependent DCs as a critical cellular source of IL-23 in response to IL-36γ stimulation.
IL-36γ Induces IL-23 via Signaling Through MyD88, c-Rel, and NF-κBp50.
MyD88 is an adaptor protein known to induce signaling through TLRs as well as IL-1 family receptors. To begin to define the signaling cascade linking IL-36R signaling to IL-23 expression in DCs, we generated bone marrow-derived DCs (BMDCs) from WT (myd88+/+) and MyD88-deficient (myd88−/−) mice and cultured them in the absence or presence of IL-36γ. Upon stimulation of myd88+/+ BMDCs with IL-36γ, IL-23 was robustly expressed, while myd88−/− BMDCs completely failed to induce IL-23 protein secretion in response to IL-36γ stimulation (SI Appendix, Fig. S3).
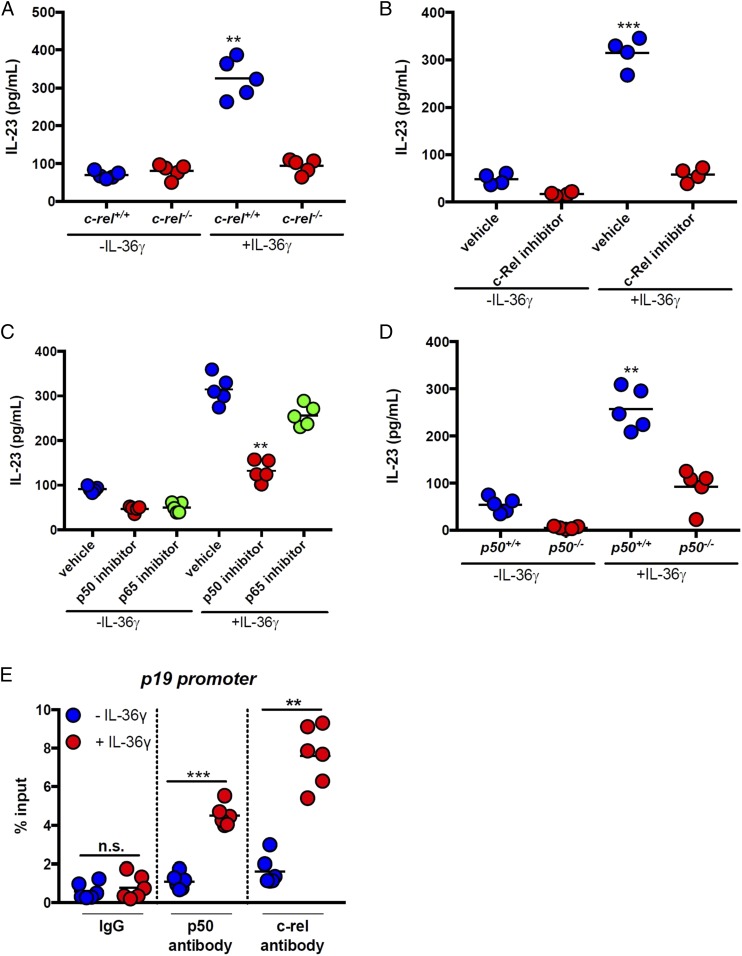
We next explored the involvement of the NF-κB pathway in IL-36γ–induced IL-23 expression. Since previous studies have implicated c-Rel in the expression of IL-23p19 to form functional IL-23 (33), we investigated the effects of c-Rel deficiency on IL-36γ–induced IL-23 expression by using BMDCs isolated from c-rel+/+ and c-rel−/− mice. Following stimulation with IL-36γ for 6 h, we observed a strong induction of IL-23 from c-rel+/+ BMDCs, but no increase over baseline in c-rel−/− cultures (Fig. 4A). Similarly, treatment with the c-Rel inhibitor IT-603 nearly completely abolished IL-36γ–induced IL-23 expression (Fig. 4B).
Fig. 4.
IL-36γ induces IL-23 via signaling through c-Rel and NF-κBp50. (A) BMDCs were generated from c-rel+/+ and c-rel−/− mice and cultured in the presence or absence of IL-36γ for 24 h, and IL-23 was assessed by ELISA. (B and C) WT BMDCs were cultured in the presence or absence of IL-36γ for 24 h, and IL-23 was assessed by ELISA. (B) Some cultures were pretreated with the c-Rel inhibitor (IT-603) or with vehicle control (DMSO) for 1 h. (C) Some cultures were pretreated with p50 or p65 inhibitor peptides or control peptides for 1 h. (D) BMDCs were generated from p50+/+ and p50−/− mice and cultured in the presence or absence of IL-36γ for 24 h, and IL-23 was assessed by ELISA. (E) ChIP assays for p50 and c-Rel binding to the p19 promoter in BMDCs treated with IL-36γ for 8 h. Data in A–D are representative of at least two independent experiments with n = 5 mice. Data in E are the combined data of two independent experiments with three replicates per experiment. All data are presented as mean ± SEM (one-way ANOVA with Tukey’s multiple comparison test: **P < 0.01; ***P < 0.001). n.s., not significant.
Similar to the other components of the NF-κB family of transcription factors, c-Rel complexes with proteins to facilitate downstream gene expression. Complexes of c-Rel can be either c-Rel/c-Rel, c-Rel/p50, or c-Rel/p65, so we sought to determine which NF-κB proteins besides c-Rel may be involved in IL-36γ–induced IL-23 expression. BMDCs from WT mice that were stimulated with IL-36γ induced robust secretion of IL-23, and this effect was significantly inhibited (∼60%) by p50 inhibitor peptide, but not by p65 inhibitor peptide (Fig. 4C). Furthermore, BMDCs generated from NF-κBp50–deficient mice (p50−/−) and stimulated with IL-36γ showed significantly lower IL-23 expression (approximately threefold) compared with those from WT (p50+/+) mice (Fig. 4D). We next performed ChIP assays to assess p50 and c-Rel binding to the IL-23p19 promoter in BMDCs treated with IL-36γ. As shown in Fig. 4E, there was a significant increase in c-Rel and p50 binding to the IL-23p19 promoter in response to treatment of BMDCs with IL-36γ for 8 h. Collectively, these results demonstrate that MyD88, c-Rel, and NF-κBp50 are part of a signaling cascade downstream of IL-36R that is involved in IL-23 expression by DCs.
Systemic IL-23 Administration Promotes Recovery from DSS-Induced Intestinal Damage in IL-36R– and IL-36γ–Deficient Mice in Association with Restoring IL-22 and AMP Production.
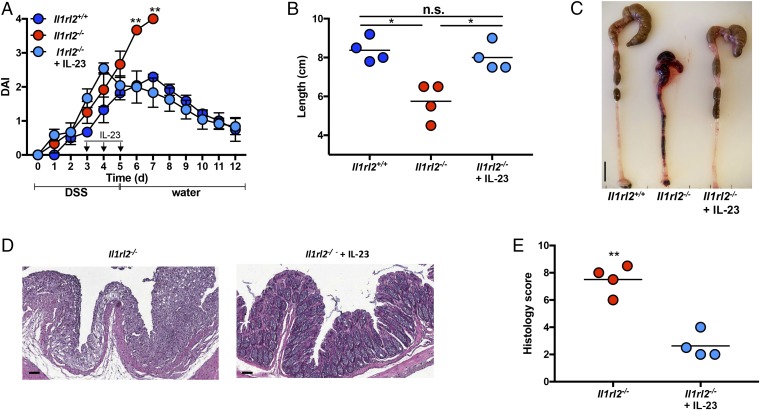
Since IL-36γ–induced IL-22 production in colonic explants from DSS-treated mice was IL-23–dependent (Fig. 2A), we next explored whether administration of IL-23 could rescue defective resolution of DSS-induced colonic damage in Il1rl2−/− mice in association with restoring IL-22 and AMP expression. DSS-treated Il1rl2−/− mice received either PBS or IL-23 (0.25 μg) at days 3, 4, and 5; DSS was discontinued at day 5, and mice were switched to regular drinking water to monitor recovery from DSS-induced intestinal damage. Strikingly, systemic administration of IL-23 to DSS-treated Il1rl2−/− mice was sufficient to promote full resolution of intestinal damage as DAI, colon length, and histology were similar to those observed in DSS-treated Il1rl2+/+ mice (Fig. 5).
Fig. 5.
Systemic IL-23 administration induces resolution of DSS-induced colonic damage in Il1rl2−/− mice. (A) DAI of Il1rl2+/+ and Il1rl2−/− mice treated with DSS for 5 d, followed by normal water for 7 d, in the presence or absence of IL-23. (B and C) Image and length of colons from mice treated as in A. (Scale bar, 1 cm.) The H&E staining (D) and histology scoring (E) of colon sections from mice treated as in A are shown. (Scale bar, 100 μm.) Data are representative of three independent experiments with four to five mice per group. All data are presented as mean ± SEM (one-way ANOVA with Tukey’s multiple comparison test: *P < 0.05; **P < 0.01). n.s., not significant.
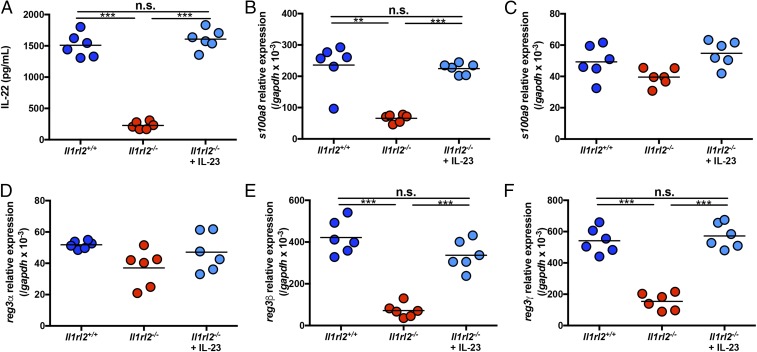
DSS induces massive damage to the intestinal epithelial barrier that allows microbes from the gut lumen to enter the underlying lamina propria. The physiological immune response to this damage is the induction of IL-22 and AMPs, including S100A8, S100A9, and members of the Reg3 family (Reg3α, Reg3β, and Reg3γ) (2). Since IL-22 and AMPs are critically important in resolution of DSS-induced intestinal damage, we next examined if Il1rl2−/− mice were defective in AMP expression following treatment with DSS and if this could be reversed by IL-23 administration. Following DSS treatment for 5 d, Il1rl2+/+ mice expressed high levels of IL-22 and AMPs, particularly S100A8, Reg3β, and Reg3γ. Consistent with their inability to induce IL-22 (Fig. 6A), Il1rl2−/− mice were significantly impaired in S100A8, Reg3β, and Reg3γ mRNA expression in response to DSS treatment, and this defect was nearly completely reversible with administration of IL-23. Indeed, delivery of IL-23 to Il1rl2−/− mice was sufficient to induce S100A8, Reg3β, and Reg3γ to the normal levels detected in Il1rl2+/+ mice (Fig. 6 B–F).
Fig. 6.
Systemic IL-23 administration induces IL-22 and AMPs and rescues Il1rl2−/− mice from DSS-induced colonic damage. (A) IL-22 protein expression in colons from Il1rl2+/+ and Il1rl2−/− mice treated with DSS for 5 d in the presence or absence of IL-23. S100A8 (B), S100A9 (C), Reg3α (D), Reg3β (E), and Reg3γ (F) mRNA expression is shown in colons isolated from mice as in A. Data are representative of two independent experiments with five to six mice per group. All data are presented as mean ± SEM (one-way ANOVA with Tukey’s multiple comparison test: **P < 0.01; ***P < 0.001). n.s., not significant.
Having observed that Notch2cKO mice, like Il1rl2−/− mice, failed to recover from DSS-induced intestinal damage and failed to express IL-23 and IL-22 in vitro, we next attempted to rescue these mice by injecting IL-23. The administration of IL-23 to Notch2cKO mice was able to significantly reduce DAI and restore colon length (SI Appendix, Fig. S4 A and B), while also normalizing tissue architecture and histology scores to levels detected in Notch2fl/fl mice (SI Appendix, Fig. S4 C and D). IL-23 treatment of Notch2cKO mice also induced IL-22 expression to similar levels as seen in control Notch2fl/fl mice (SI Appendix, Fig. S4E). Furthermore, delivery of IL-23 to Notch2cKO mice was sufficient to induce Reg3β and Reg3γ (Fig. 4 F and G).
We next sought to determine if our observations using Il1rl2−/− mice were predominantly due to loss of IL-36γ signaling or if other IL-36 agonist cytokines (IL-36α and IL-36β) could be playing a role. To do so, we first treated IL-36γ–deficient mice (Il1f9−/−) and control mice (Il1f9+/+) with DSS, and at day 3, colonic tissues were harvested and analyzed for IL-36α, IL-36β, and IL-36γ expression. In the absence of DSS, IL-36α and IL-36β were not detectable in Il1f9+/+ and Il1f9−/− mice, while DSS treatment of Il1f9+/+ mice led to the robust expression of IL-36γ, and only very low levels of IL-36α and undetectable levels of IL-36β expression. As expected, Il1f9−/− mice had undetectable levels of IL-36γ and did not appear to induce IL-36α or IL-36β to compensate for the loss of IL-36γ (SI Appendix, Fig. S5A). Interestingly, Il1f9−/− mice appeared to phenocopy Il1rl2−/− mice in response to DSS treatment in that they exhibited grossly impaired IL-23 and IL-22 production in colonic explants compared with control Il1f9+/+ mice, and this defect could be overcome by the addition of IL-36γ (SI Appendix, Fig. S5 B and C). To extend these in vitro observations to the in vivo setting, Il1f9−/− mice were treated with DSS in the presence or absence of IL-23 administration, as shown in Fig. 6. Similar to effects observed in Il1rl2−/− mice, treatment of Il1f9−/− mice with IL-23 was able to significantly reduce DAI (SI Appendix, Fig. S6A) and to normalize colon length (SI Appendix, Fig. S6 B and C) and histological damage to those seen in control Il1f9+/+ mice (SI Appendix, Fig. S6 D and E). IL-23 treatment of Il1f9−/− mice further restored IL-22 (SI Appendix, Fig. S7A) and AMP expression, particularly S100A8, Reg3β, and Reg3γ, back to levels observed in Il1f9+/+ mice (SI Appendix, Fig. S7 B–F). Collectively, these results demonstrate that delivery of IL-23 to Il1rl2−/− mice and Il1f9−/− mice is sufficient to restore IL-22 and AMP expression and recovery from acute intestinal damage.
Since intercellular tight junctions are essential for maintaining the integrity and function of the intestinal barrier in the steady state and following damage (34), we next assessed whether the mRNA expression of the tight junction components occludin and claudin 2 was affected by the loss of IL-36 signaling. Following treatment with DSS for 5 d, both Il1rl2−/− and Il1f9−/− mice exhibited significantly reduced occludin and claudin 2 mRNA expression compared with WT controls. Additionally, IL-23 administration was able to restore occludin and claudin 2 mRNA expression to WT levels (SI Appendix, Fig. S8). These data suggest that signaling via IL-36R not only induces IL-23–dependent mucosal protection via IL-22 and AMP expression but may also help to reseal the damaged intestinal epithelial barrier via effects on tight junctions.
Given the dynamic cross-talk between the mucosal immune system and the gut microbiota (35–37), combined with the established involvement of IL-23, IL-22, and AMPs in controlling the microbiota (38, 39), we next explored the contribution of the IL-36/IL-36R axis in regulating microbiota composition. In the steady state, we observed that Il1rl2−/− mice had a significant increase in several flagellated bacterial groups, including Clostridium clusters XIVa and XI and Oscillibacter, and significant decreases in the nonflagellated bacterial groups Bacteroides, Prevotella, and Lactobacillus (SI Appendix, Fig. S9A). Interestingly, most of these changes were further augmented upon DSS treatment (SI Appendix, Fig. S9B). These observations support a potential role for the IL-36/IL-23/IL-22/AMP axis in control of the microbiota during health and disease, and future studies using 16S rRNA sequencing should further clarify the full extent to which this cytokine axis influences the microbiota.
Discussion
In this study, we provide evidence demonstrating that the IL-36/IL-36R pathway acts as a key upstream inducer of IL-23/IL-22/AMP–dependent colonic tissue repair. While IL-36 ligands are well appreciated to promote chronic inflammation and contribute to pathological tissue damage (18, 19, 40), their role in mediating tissue protection in response to acute insult is newly emerging (22, 23, 29). We and others have recently reported that IL-36R–deficient mice treated with DSS have reduced signs of intestinal inflammation during the damage phase of disease yet are impaired in mucosal healing (21–23). These data suggest that the proinflammatory functions of the IL-36 pathway are intimately linked to epithelial regeneration, tissue repair, and healing of intestinal damage and act as part of a feedback loop that then limits further production of proinflammatory factors and pathological inflammation. IL-36 cytokines may likewise function to promote wound repair at other barrier surfaces, such as the skin. In this regard, a recent report observed that IL-36γ was induced in a model of skin injury and that signaling via IL-36R promoted wound healing via the induction of Reg3γ (29). Whether or not IL-23 and/or IL-22 was also involved in IL-36R–mediated wound repair in this skin model remains an open question.
The IL-36/IL-23/IL-22 inflammatory cytokine cascade in response to DSS-mediated intestinal injury is a highly orchestrated process that involves numerous innate immune cell subsets. Early following DSS-induced damage, inflammatory monocytes/macrophages are a main source of IL-36γ in response to components of the microbiota (22); however, keratinocytes (41), myofibroblasts (27, 28), and other cells types may also be important sources of IL-36 cytokines (25). While many cell types express IL-36R, we found that IL-36R expression was dramatically increased early following DSS treatment at a time that coincided with accumulation of CD11b+CD103+ DCs. Furthermore, in Notch2cKO mice that lack CD11b+CD103+ DCs, IL-36R expression was not increased following DSS treatment suggesting that Notch2-dependent CD103+CD11b+ DCs are recruited into the inflamed colon, where they express high levels of IL-36R and produce IL-23 in response to IL-36γ stimulation. These data are consistent with several reports demonstrating that CD103+CD11b+ DCs are a main source of IL-23 (13, 42), and now directly link IL-36R signaling to IL-23 production by these cells. Following secretion of IL-23, numerous cell types in the colon express IL-23R (43–45) and are capable of producing IL-22 (3, 6, 46). While ILC3s are the most well-documented IL-22 producers in the gut (5, 47, 48), activated neutrophils (10, 22, 49) and NK cells (8) can also produce IL-22 in response to IL-23 stimulation, and the relative contribution of IL-22 from these sources during intestinal damage and repair remains unclear. Additionally, IL-23R signaling directly in intestinal epithelial cells was recently shown to induce Reg3β and CXCL1 expression, as well as the recruitment and activation of IL-22–producing neutrophils (38). Regardless of the source, IL-22 is a potent inducer of epithelial proliferation, mucus production, and AMP expression, all of which support efficient intestinal tissue repair (11, 50, 51).
Several lines of evidence are consistent with the IL-36γ/IL-36R axis playing a central role in IL-23/IL-22/AMP–dependent resolution of acute intestinal damage. First, IL-36R– and IL-36γ–deficient mice are grossly impaired in their ability to recover from DSS-induced intestinal damage, and this phenotype can be rescued by treatment with IL-23, as well as an IL-22–inducing aryl-hydrocarbon receptor agonist (22). Second, mice deficient in IL-23 (52) and IL-22 (8) appear to phenocopy the defective tissue repair in response to DSS that we observed in IL-36R– and IL-36γ–deficient mice. Third, Notch2cKO mice, which lack CD103+CD11b+ DCs, are also defective in colonic repair, an effect that can also be rescued by treatment with IL-23. Of note, the defective repair in Notch2cKO mice was not as profound as that in IL-36R– and IL-36γ–deficient mice, suggesting that other cell types aside from CD103+CD11b+ DCs may also be involved in IL-36γ–induced IL-23 production.
Both IL-36 cytokines and IL-23 are potent inflammatory cytokines that function in a context-dependent manner. In models of acute barrier damage that predominantly involve innate immune activation, the proresolution functions of these cytokines likely dominate over their proinflammatory effects on T cells, and the net result may be beneficial to the host. Alternatively, in chronic conditions where T cells play a major role, proinflammatory effects of IL-36 cytokines and IL-23 may dominate over barrier protective effects and exacerbate disease pathology. This appears evident during skin and intestinal inflammation in which CD4+ T cells are involved (20, 25, 53–56). Importantly, monoclonal antibody-mediated blockade of the p40 subunit of IL-12 and IL-23 is approved for the treatment of moderately to severely active Crohn’s disease, and specific IL-23 blockers are showing efficacy in clinical trials (57). Thus, our data demonstrating that the IL-36/IL-36R axis augments IL-23 expression in the intestine may inform on potential therapeutic targeting of IL-36 cytokines and/or IL-36R as a novel strategy to limit proinflammatory effects of IL-23 during human IBD.
The context-dependent role of IL-36 cytokines in inducing proinflammatory responses that lead to intestinal barrier protection appears to be an emerging paradigm for members of the IL-1 family cytokines (16, 17, 58). While IL-1α augments colonic inflammation, IL-1β is involved in restitution of the epithelial barrier and resolution of acute colonic damage (59). Similarly, NLRP6, ASC, caspase-1, and IL-18 are all protective in the DSS model of colitis (60, 61). The alarmin IL-33 can also promote intestinal tissue protection via the amphiregulin/EGF receptor pathway and act on ST2-expressing regulatory T cells to promote their function in suppression of colitis (62). IL-37 is an atypical member of the IL-1 family in that it functions as an inhibitor of innate inflammation and immunity, yet it still functions to protect from colitis in mice (63, 64). Our data indicate that IL-36γ and IL-36R are rapidly induced following acute colonic damage and orchestrate a key inflammatory process involving CD103+CD11b+ DCs, IL-23, IL-22, and AMPs, which ultimately functions to resolve colonic damage and provide host protection (SI Appendix, Fig. S9). These findings have potential implications for the treatment of intestinal inflammatory conditions, including IBD, where the beneficial effects of IL-36 and/or IL-23 blockade may be limited by concomitant interference with tissue repair processes. Therefore, a combined therapeutic approach aimed at inhibiting proinflammatory cytokines while augmenting tissue repair mechanisms may afford optimal treatment for chronic intestinal inflammation.
Materials and Methods
Mice.
The following mice were obtained from The Jackson Laboratory: WT C57BL/6 (B6 WT), B6.129S(C)-Batf3tm1Kmm/J (batf3−/−), CD11c-cre [B6.Cg-Tg(Itgax-cre)1-1Reiz/J], Notch2f/f (B6.129S-Notch2tm3Grid/J), B6.129P-Nfkb1tm1Bal/J (p50−/−), and B6.129P2(SJL)-Myd88tm1.1Defr/J (myd88−/−). IL-36R−/− mice (Il1rl2−/−) on the C57BL/6 background (backcrossed more than nine generations) were originally provided by Amgen. To generate IL-36γ−/− (Il1f9−/−) mice, sperm from IL-36γ−/− mice was obtained from the Knockout Mouse Project repository (University of California, Davis), and heterozygous Il1f9+/− founder mice were generated by the Mouse Transgenic and Gene Targeting Core facility at Emory University. Il1f9+/− mice were subsequently bred to generate Il1f9−/− mice on the C57BL/6 background (backcrossed more than nine generations). Notch2cKO mice were generated as previously described (30). Animal protocols were approved by the Institutional Animal Care and Use Committee of Georgia State University.
DSS Model of Colitis.
Mice were treated with 2.5–3% (wt/vol) DSS (MP Biomedicals; molecular weight: 36,000–50,000) in their drinking water for 5 d and then switched to regular drinking water. Mice were monitored daily for signs of disease, and DAI and histology scoring was performed as previously described (22).
Colonic Explants.
Colon tissue was harvested from mice, opened longitudinally, and washed in PBS plus Tween-20. Biopsy punches (3 mm; Integra Miltex) were used to excise sections of the colon, which were placed in 96-well plates and cultured with RPMI-1640 supplemented with 10% FBS and penicillin/streptomycin. Recombinant IL-23 (R&D Systems) or recombinant IL-36γ (R&D Systems) was added to each well at 20 ng/mL and 100 ng/mL, respectively. For gene expression analysis, tissues were collected and processed for downstream applications 6 h following stimulation. For protein analysis, supernatant from the tissues were collected 60 h after stimulation.
ELISA.
IL-22 and IL-23 secretion was measured in cell-free culture supernatants using IL-22 and IL-23 ELISA kits (R&D Systems) according to the manufacturer’s protocols.
In Vivo Administration of IL-23.
Recombinant mouse IL-23 was purchased from R&D Systems. Il1rl2−/− and Il1f9−/− mice received either PBS or 0.25 μg of IL-23 via i.p. injection at days 3, 4, and 5 of DSS treatment.
Histology.
Colons were fixed in 10% formalin. Paraffin embedding, sectioning, hematoxylin/eosin staining, and slide scanning were performed at the University of Michigan Pathology Core.
Statistical Analysis.
All statistical analyses were performed with GraphPad Prism software, version 7.0. One-way ANOVA and Tukey’s multiple comparison test or Student’s t test were used to determine significance (*P < 0.05; **P < 0.01; ***P < 0.001).
Supplementary Material
Acknowledgments
We thank Dr. Ifor R. Williams for critical discussion and Dr. Bruce Horwitz for c-Rel–deficient bone marrow. This work was supported by NIH Grants 1R01DK097256 (to T.L.D.) and 1R01DK107739 (to D.M. and T.L.D.), Japanese Society for the Promotion of Science KAKENHI Grants 17H07013 and 18K15128 (to A.H.), and Crohn’s and Colitis Foundation Research Fellowship Award (to A.H.). A.D.M. is a recipient of a Veterans Affairs Research Career Science Award.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. F.C. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1718902115/-/DCSupplemental.
References
- 1.Martini E, Krug SM, Siegmund B, Neurath MF, Becker C. Mend your fences: The epithelial barrier and its relationship with mucosal immunity in inflammatory bowel disease. Cell Mol Gastroenterol Hepatol. 2017;4:33–46. doi: 10.1016/j.jcmgh.2017.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Peterson LW, Artis D. Intestinal epithelial cells: Regulators of barrier function and immune homeostasis. Nat Rev Immunol. 2014;14:141–153. doi: 10.1038/nri3608. [DOI] [PubMed] [Google Scholar]
- 3.Sonnenberg GF, Fouser LA, Artis D. Border patrol: Regulation of immunity, inflammation and tissue homeostasis at barrier surfaces by IL-22. Nat Immunol. 2011;12:383–390. doi: 10.1038/ni.2025. [DOI] [PubMed] [Google Scholar]
- 4.Turner JR. Intestinal mucosal barrier function in health and disease. Nat Rev Immunol. 2009;9:799–809. doi: 10.1038/nri2653. [DOI] [PubMed] [Google Scholar]
- 5.Sonnenberg GF, Fouser LA, Artis D. Functional biology of the IL-22-IL-22R pathway in regulating immunity and inflammation at barrier surfaces. Adv Immunol. 2010;107:1–29. doi: 10.1016/B978-0-12-381300-8.00001-0. [DOI] [PubMed] [Google Scholar]
- 6.Dudakov JA, Hanash AM, van den Brink MR. Interleukin-22: Immunobiology and pathology. Annu Rev Immunol. 2015;33:747–785. doi: 10.1146/annurev-immunol-032414-112123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sugimoto K, et al. IL-22 ameliorates intestinal inflammation in a mouse model of ulcerative colitis. J Clin Invest. 2008;118:534–544. doi: 10.1172/JCI33194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zenewicz LA, et al. Innate and adaptive interleukin-22 protects mice from inflammatory bowel disease. Immunity. 2008;29:947–957. doi: 10.1016/j.immuni.2008.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zheng Y, et al. Interleukin-22 mediates early host defense against attaching and effacing bacterial pathogens. Nat Med. 2008;14:282–289. doi: 10.1038/nm1720. [DOI] [PubMed] [Google Scholar]
- 10.Zindl CL, et al. IL-22-producing neutrophils contribute to antimicrobial defense and restitution of colonic epithelial integrity during colitis. Proc Natl Acad Sci USA. 2013;110:12768–12773. doi: 10.1073/pnas.1300318110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eidenschenk C, Rutz S, Liesenfeld O, Ouyang W. Role of IL-22 in microbial host defense. Curr Top Microbiol Immunol. 2014;380:213–236. doi: 10.1007/978-3-662-43492-5_10. [DOI] [PubMed] [Google Scholar]
- 12.Rutz S, Wang X, Ouyang W. The IL-20 subfamily of cytokines–From host defence to tissue homeostasis. Nat Rev Immunol. 2014;14:783–795. doi: 10.1038/nri3766. [DOI] [PubMed] [Google Scholar]
- 13.Kinnebrew MA, et al. Interleukin 23 production by intestinal CD103(+)CD11b(+) dendritic cells in response to bacterial flagellin enhances mucosal innate immune defense. Immunity. 2012;36:276–287. doi: 10.1016/j.immuni.2011.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shih VF, et al. Homeostatic IL-23 receptor signaling limits Th17 response through IL-22-mediated containment of commensal microbiota. Proc Natl Acad Sci USA. 2014;111:13942–13947. doi: 10.1073/pnas.1323852111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garlanda C, Dinarello CA, Mantovani A. The interleukin-1 family: Back to the future. Immunity. 2013;39:1003–1018. doi: 10.1016/j.immuni.2013.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lopetuso LR, Chowdhry S, Pizarro TT. Opposing functions of classic and novel IL-1 family members in gut health and disease. Front Immunol. 2013;4:181. doi: 10.3389/fimmu.2013.00181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bamias G, Corridoni D, Pizarro TT, Cominelli F. New insights into the dichotomous role of innate cytokines in gut homeostasis and inflammation. Cytokine. 2012;59:451–459. doi: 10.1016/j.cyto.2012.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bamias G, Cominelli F. Cytokines and intestinal inflammation. Curr Opin Gastroenterol. 2016;32:437–442. doi: 10.1097/MOG.0000000000000315. [DOI] [PubMed] [Google Scholar]
- 19.Gabay C, Towne JE. Regulation and function of interleukin-36 cytokines in homeostasis and pathological conditions. J Leukoc Biol. 2015;97:645–652. doi: 10.1189/jlb.3RI1014-495R. [DOI] [PubMed] [Google Scholar]
- 20.Walsh PT, Fallon PG. The emergence of the IL-36 cytokine family as novel targets for inflammatory diseases. Ann N Y Acad Sci. 2016;1417:23–34. doi: 10.1111/nyas.13280. [DOI] [PubMed] [Google Scholar]
- 21.Russell SE, et al. IL-36α expression is elevated in ulcerative colitis and promotes colonic inflammation. Mucosal Immunol. 2016;9:1193–1204. doi: 10.1038/mi.2015.134. [DOI] [PubMed] [Google Scholar]
- 22.Medina-Contreras O, et al. Cutting edge: IL-36 receptor promotes resolution of intestinal damage. J Immunol. 2016;196:34–38. doi: 10.4049/jimmunol.1501312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Scheibe K, et al. IL-36R signalling activates intestinal epithelial cells and fibroblasts and promotes mucosal healing in vivo. Gut. 2016;66:823–838. doi: 10.1136/gutjnl-2015-310374. [DOI] [PubMed] [Google Scholar]
- 24.Nishida A, et al. Increased expression of interleukin-36, a member of the interleukin-1 cytokine family, in inflammatory bowel disease. Inflamm Bowel Dis. 2016;22:303–314. doi: 10.1097/MIB.0000000000000654. [DOI] [PubMed] [Google Scholar]
- 25.Boutet MA, et al. Distinct expression of interleukin (IL)-36α, β and γ, their antagonist IL-36Ra and IL-38 in psoriasis, rheumatoid arthritis and Crohn’s disease. Clin Exp Immunol. 2016;184:159–173. doi: 10.1111/cei.12761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Friedrich M, Tillack C, Wollenberg A, Schauber J, Brand S. IL-36γ sustains a proinflammatory self-amplifying loop with IL-17C in anti-TNF-induced psoriasiform skin lesions of patients with Crohn’s disease. Inflamm Bowel Dis. 2014;20:1891–1901. doi: 10.1097/MIB.0000000000000198. [DOI] [PubMed] [Google Scholar]
- 27.Takahashi K, et al. Interleukin (IL)-1β is a strong inducer of IL-36γ expression in human colonic myofibroblasts. PLoS One. 2015;10:e0138423. doi: 10.1371/journal.pone.0138423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kanda T, et al. Interleukin(IL)-36α and IL-36γ induce proinflammatory mediators from human colonic subepithelial myofibroblasts. Front Med (Lausanne) 2015;2:69. doi: 10.3389/fmed.2015.00069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jiang Z, et al. IL-36gamma induced by the TLR3-SLUG-VDR axis promotes wound healing via REG3A. J Invest Dermatol. 2017;137:2620–2629. doi: 10.1016/j.jid.2017.07.820. [DOI] [PubMed] [Google Scholar]
- 30.Satpathy AT, et al. Notch2-dependent classical dendritic cells orchestrate intestinal immunity to attaching-and-effacing bacterial pathogens. Nat Immunol. 2013;14:937–948. doi: 10.1038/ni.2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Denning TL, et al. Functional specializations of intestinal dendritic cell and macrophage subsets that control Th17 and regulatory T cell responses are dependent on the T cell/APC ratio, source of mouse strain, and regional localization. J Immunol. 2011;187:733–747. doi: 10.4049/jimmunol.1002701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.MacDonald KP, et al. An antibody against the colony-stimulating factor 1 receptor depletes the resident subset of monocytes and tissue- and tumor-associated macrophages but does not inhibit inflammation. Blood. 2010;116:3955–3963. doi: 10.1182/blood-2010-02-266296. [DOI] [PubMed] [Google Scholar]
- 33.Carmody RJ, Ruan Q, Liou HC, Chen YH. Essential roles of c-Rel in TLR-induced IL-23 p19 gene expression in dendritic cells. J Immunol. 2007;178:186–191. doi: 10.4049/jimmunol.178.1.186. [DOI] [PubMed] [Google Scholar]
- 34.Luissint AC, Parkos CA, Nusrat A. Inflammation and the intestinal barrier: Leukocyte-epithelial cell interactions, cell junction remodeling, and mucosal repair. Gastroenterology. 2016;151:616–632. doi: 10.1053/j.gastro.2016.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Round JL, Mazmanian SK. The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol. 2009;9:313–323. doi: 10.1038/nri2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhou L, Sonnenberg GF. Essential immunologic orchestrators of intestinal homeostasis. Sci Immunol. 2018;3:eaao1605. doi: 10.1126/sciimmunol.aao1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hooper LV, Littman DR, Macpherson AJ. Interactions between the microbiota and the immune system. Science. 2012;336:1268–1273. doi: 10.1126/science.1223490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Aden K, et al. Epithelial IL-23R signaling licenses protective IL-22 responses in intestinal inflammation. Cell Rep. 2016;16:2208–2218. doi: 10.1016/j.celrep.2016.07.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zenewicz LA, et al. IL-22 deficiency alters colonic microbiota to be transmissible and colitogenic. J Immunol. 2013;190:5306–5312. doi: 10.4049/jimmunol.1300016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Palomo J, Dietrich D, Martin P, Palmer G, Gabay C. The interleukin (IL)-1 cytokine family–Balance between agonists and antagonists in inflammatory diseases. Cytokine. 2015;76:25–37. doi: 10.1016/j.cyto.2015.06.017. [DOI] [PubMed] [Google Scholar]
- 41.Li N, et al. Alarmin function of cathelicidin antimicrobial peptide LL37 through IL-36γ induction in human epidermal keratinocytes. J Immunol. 2014;193:5140–5148. doi: 10.4049/jimmunol.1302574. [DOI] [PubMed] [Google Scholar]
- 42.Schlitzer A, et al. IRF4 transcription factor-dependent CD11b+ dendritic cells in human and mouse control mucosal IL-17 cytokine responses. Immunity. 2013;38:970–983. doi: 10.1016/j.immuni.2013.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Buonocore S, et al. Innate lymphoid cells drive interleukin-23-dependent innate intestinal pathology. Nature. 2010;464:1371–1375. doi: 10.1038/nature08949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Morrison PJ, Ballantyne SJ, Kullberg MC. Interleukin-23 and T helper 17-type responses in intestinal inflammation: From cytokines to T-cell plasticity. Immunology. 2011;133:397–408. doi: 10.1111/j.1365-2567.2011.03454.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Awasthi A, et al. Cutting edge: IL-23 receptor gfp reporter mice reveal distinct populations of IL-17-producing cells. J Immunol. 2009;182:5904–5908. doi: 10.4049/jimmunol.0900732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zenewicz LA, Flavell RA. Recent advances in IL-22 biology. Int Immunol. 2011;23:159–163. doi: 10.1093/intimm/dxr001. [DOI] [PubMed] [Google Scholar]
- 47.Vivier E, Spits H, Cupedo T. Interleukin-22-producing innate immune cells: New players in mucosal immunity and tissue repair? Nat Rev Immunol. 2009;9:229–234. doi: 10.1038/nri2522. [DOI] [PubMed] [Google Scholar]
- 48.Colonna M. Interleukin-22-producing natural killer cells and lymphoid tissue inducer-like cells in mucosal immunity. Immunity. 2009;31:15–23. doi: 10.1016/j.immuni.2009.06.008. [DOI] [PubMed] [Google Scholar]
- 49.Chen F, et al. mTOR mediates IL-23 induction of neutrophil IL-17 and IL-22 production. J Immunol. 2016;196:4390–4399. doi: 10.4049/jimmunol.1501541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Parks OB, Pociask DA, Hodzic Z, Kolls JK, Good M. Interleukin-22 signaling in the regulation of intestinal health and disease. Front Cell Dev Biol. 2016;3:85. doi: 10.3389/fcell.2015.00085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mizoguchi A. Healing of intestinal inflammation by IL-22. Inflamm Bowel Dis. 2012;18:1777–1784. doi: 10.1002/ibd.22929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Becker C, et al. Cutting edge: IL-23 cross-regulates IL-12 production in T cell-dependent experimental colitis. J Immunol. 2006;177:2760–2764. doi: 10.4049/jimmunol.177.5.2760. [DOI] [PubMed] [Google Scholar]
- 53.Mahil SK, et al. An analysis of IL-36 signature genes and individuals with IL1RL2 knockout mutations validates IL-36 as a psoriasis therapeutic target. Sci Transl Med. 2017;9:eaan2514, and erratum (2017) 9:eaar6600. doi: 10.1126/scitranslmed.aan2514. [DOI] [PubMed] [Google Scholar]
- 54.Harusato A, et al. IL-36γ signaling controls the induced regulatory T cell-Th9 cell balance via NFκB activation and STAT transcription factors. Mucosal Immunol. 2017;10:1455–1467. doi: 10.1038/mi.2017.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dietrich D, Gabay C. Inflammation: IL-36 has proinflammatory effects in skin but not in joints. Nat Rev Rheumatol. 2014;10:639–640. doi: 10.1038/nrrheum.2014.156. [DOI] [PubMed] [Google Scholar]
- 56.Gresnigt MS, van de Veerdonk FL. Biology of IL-36 cytokines and their role in disease. Semin Immunol. 2013;25:458–465. doi: 10.1016/j.smim.2013.11.003. [DOI] [PubMed] [Google Scholar]
- 57.Deepak P, Sandborn WJ. Ustekinumab and anti-interleukin-23 agents in Crohn’s disease. Gastroenterol Clin North Am. 2017;46:603–626. doi: 10.1016/j.gtc.2017.05.013. [DOI] [PubMed] [Google Scholar]
- 58.Bamias G, Dinarello CA, Rivera-Nieves J. Innate cytokines dictate the fate of acute intestinal inflammation. Gastroenterology. 2015;148:248–250. doi: 10.1053/j.gastro.2014.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bersudsky M, et al. Non-redundant properties of IL-1α and IL-1β during acute colon inflammation in mice. Gut. 2014;63:598–609. doi: 10.1136/gutjnl-2012-303329. [DOI] [PubMed] [Google Scholar]
- 60.Elinav E, et al. NLRP6 inflammasome regulates colonic microbial ecology and risk for colitis. Cell. 2011;145:745–757. doi: 10.1016/j.cell.2011.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Seregin SS, et al. NLRP6 function in inflammatory monocytes reduces susceptibility to chemically induced intestinal injury. Mucosal Immunol. 2017;10:434–445. doi: 10.1038/mi.2016.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Monticelli LA, et al. IL-33 promotes an innate immune pathway of intestinal tissue protection dependent on amphiregulin-EGFR interactions. Proc Natl Acad Sci USA. 2015;112:10762–10767. doi: 10.1073/pnas.1509070112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dinarello CA, et al. Suppression of innate inflammation and immunity by interleukin-37. Eur J Immunol. 2016;46:1067–1081. doi: 10.1002/eji.201545828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.McNamee EN, et al. Interleukin 37 expression protects mice from colitis. Proc Natl Acad Sci USA. 2011;108:16711–16716. doi: 10.1073/pnas.1111982108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.