Significance
Hormone-like peptides derived from small coding genes (<100 amino acids) have not been extensively characterized in relation to abiotic stress tolerance. Focusing on 17 salinity stress-inducible small coding genes in Arabidopsis, we showed that four genes conferred increased salinity stress tolerance when overexpressed in transgenic plants. One of the four genes (AtPROPEP3) was found to induce salinity stress tolerance by treatment with a 13-peptide (KPTPSSGKGGKHN) fragment, providing unique functional evidence for enhanced salinity stress tolerance in plants in response to a peptide treatment. Although the 13-peptide fragment shares homology with known peptides associated with immune response, the other peptides may encode unique hormone-like peptides associated with salinity stress tolerance.
Keywords: peptide hormone, small coding gene, abiotic stress, salt tolerance, Arabidopsis
Abstract
Peptides encoded by small coding genes play an important role in plant development, acting in a similar manner as phytohormones. Few hormone-like peptides, however, have been shown to play a role in abiotic stress tolerance. In the current study, 17 Arabidopsis genes coding for small peptides were found to be up-regulated in response to salinity stress. To identify peptides leading salinity stress tolerance, we generated transgenic Arabidopsis plants overexpressing these small coding genes and assessed survivability and root growth under salinity stress conditions. Results indicated that 4 of the 17 overexpressed genes increased salinity stress tolerance. Further studies focused on AtPROPEP3, which was the most highly up-regulated gene under salinity stress. Treatment of plants with synthetic peptides encoded by AtPROPEP3 revealed that a C-terminal peptide fragment (AtPep3) inhibited the salt-induced bleaching of chlorophyll in seedlings. Conversely, knockdown AtPROPEP3 transgenic plants exhibited a hypersensitive phenotype under salinity stress, which was complemented by the AtPep3 peptide. This functional AtPep3 peptide region overlaps with an AtPep3 elicitor peptide that is related to the immune response of plants. Functional analyses with a receptor mutant of AtPep3 revealed that AtPep3 was recognized by the PEPR1 receptor and that it functions to increase salinity stress tolerance in plants. Collectively, these data indicate that AtPep3 plays a significant role in both salinity stress tolerance and immune response in Arabidopsis.
Some of the peptides encoded by small coding genes are secreted from cells and translocated to other cells. These secreted peptides, referred to as peptide hormones (1–4), play an essential role in plant development; similar to the role played by phytohormones. Examples include phytosulfokine (PSK), a growth factor related to cell proliferation; rapid alkalinization factor (RALF), which regulates root growth; CLAVATA3 (CLV3), which is associated with the differentiation of stem cells; LUREs, which guides pollen tube growth; STOMAGEN, which is related to stomatal development; and casparian strip integrity factor (CIF), which is related to the formation of the casparian strip diffusion barrier, among others (5–12). Several hormone-like peptides have been reported to function as long-distance signal molecules in organ-to-organ communication (13, 14). C-terminally encoded peptide (CEP1) has been shown to regulate root development in plants subjected to nitrate starvation (15) and also functions in root-to-shoot signaling. Recently, CLAVATA3/ESR-related 25 (CLE25) peptide has been shown to transmit water-deficiency signals from root to shoot and functions for dehydration stress tolerance as well (16). Although CEP1 or CLE25 responses are dependent on environmental conditions, most peptide hormones have been reported to play a role in plant growth and development. Peptide hormones are secreted from cells and bind to a receptor protein in a targeted cell, resulting in the induction of a physiological response in plants (2, 17). Therefore, treatment of plants with synthetic peptides that were manufactured based on the coding sequence of the small coding genes produces the similar phenotypic effect observed in plants in which these small coding genes have been overexpressed (ox) (8, 18, 19).
Peptide hormones related to wound response and biotic stresses have been reported in plants (20–22). Systemin, CEP, and the C terminus of a cysteine-rich secretory protein antigen 5, and genes within the pathogenesis-related 1 proteins (CAP) superfamily are associated with salinity stress response (23–25). When overexpressed in plants, systemin functions as a positive regulator of salinity stress tolerance, whereas both CEP and CAP-derived peptide (CAPE) function as negative regulators. Despite the knowledge gained from the aforementioned studies, very little is known pertaining to the role of peptide hormones in abiotic stress responses such as drought, heat, salinity, and cold stress. Therefore, in the present study which characterized hormone-like peptides in Arabidopsis, we focused on salinity stress as a representative abiotic stress. Salinity stress, which has a significant negative impact on crop yields, is a global problem (26–28). Salinity stress has two unique phases consisting of (i) osmotic and (ii) ionic stress (27). When salts accumulate to high concentrations in the soil, plants lose water due to osmotic stress and they suffer growth inhibition. Consequently, both shoot growth and leaf size are significantly reduced (29). Plants coordinate the intracellular levels of compatible solutes to maintain osmotic balance, cell structure, and water flux (27). In plants, the accumulation of Na+ or Cl− to high levels results in the reduction of enzyme activity due to ionic toxicity (30, 31). Several genes function in the salinity stress response such as salt overly sensitive (SOS1–6), histidine kinase transporter (HKT), and Na+/H+ exchanger (NHX), which are sodium (Na+) and potassium (K+) transporters. SOS1–3, HKT, and NHX are associated with controlling the ion flux of Na+/H+ (32–38). SOS4 encodes pyridoxal (PL) kinase, which is a cofactor that regulates Na+ and K+ ion transport (39). SOS5 functions in relation to cell wall expansion and SOS6 is involved with osmotic stress tolerance. Mutations within these genes result in hypersensitive phenotypes against salinity stress (40, 41). Thus, these phenotypes support the conclusion that plants induce salinity stress tolerance by regulating ion homeostasis and cell expansion.
To date, however, there have been few reports of hormone-like peptides associated with salinity stress tolerance. It is plausible that the functional roles of many small coding genes (<100 amino acids) have not been well annotated (42, 43). Since large-scale overexpression studies of small coding genes have been previously used to successfully identify many hormone-like peptides associated with plant development (42, 44–46), we generated transgenic plants overexpressing individual salinity-inducible small coding genes in the present study. Four of the small coding genes conferred high levels of salt tolerance when overexpressed in the aforementioned transgenic plants. Additional functional characterizations were performed for one of the four small coding genes (AtPROPEP3) that conferred increased salt tolerance by generating AtPROPEP3 knockdown plants, confirming the essential role of AtPROPEP3 in salt tolerance. Lastly, the functional segment of the AtPep3 peptide was identified, which was responsible for inducing salt tolerance in Arabidopsis plants.
Results
Stress Tolerance of Transgenic Plants Subjected to Salt Stress.
Analysis of microarray data (46) for annotated and nonannotated small coding genes (<150 aa) revealed more than a fourfold increase in expression in response to salinity conditions for 81 of the small coding genes. Among the salinity-inducible genes, 19 genes were assigned to categories such as enzyme, transcription factor, and heat shock lipoprotein, etc. (SI Appendix, Table S1). Among the remaining 53 small coding genes and 9 unannotated small coding genes, representing gene candidates encoding hormone-like peptides, we manually selected 15 small coding genes and 2 unannotated small coding genes based upon the presence of secreted signal amino acid sequences and high homology across other plant species (SI Appendix, Fig. S1). However, for these selections, we did not impose any bias with respect to the amount of fold changes in response to salt treatment (SI Appendix, Fig. S1).
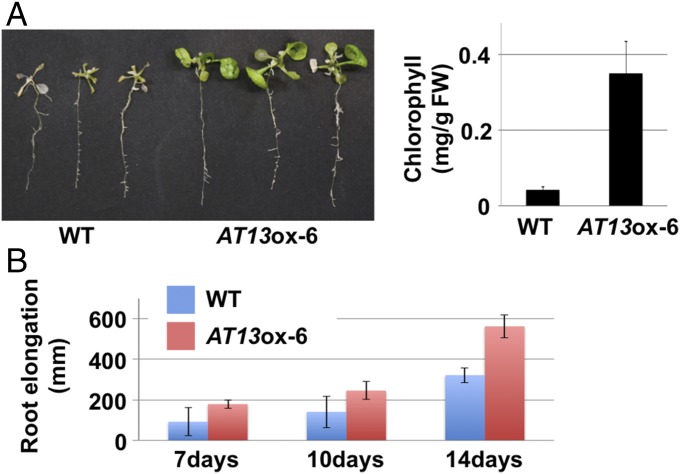
To determine whether salt stress-induced small coding genes play an integral role in salinity tolerance, we performed salinity stress screening using overexpression (ox) plant lines corresponding to the small coding gene candidates. As indicated in SI Appendix, Fig. S2 and Table S2, more than two lines overexpressing 13 of the 17 small coding genes exhibited higher levels of survivorship than the wild-type (WT, Col-0) plants (survival ratio ox/WT > 2). To further evaluate the effect of these small coding genes in salinity stress tolerance, root growth was evaluated in homozygous material (T3 generation) of the ox lines under salinity stress conditions. Results indicated that primary root length in AT5-, AT12-, AT13-, and AT23-ox plants was more than 1.5 times longer than in WT plants (P value <0.05 using a two-tailed t test, SI Appendix, Fig. S3). Our initial microarray data indicated that these four genes had a higher level of induction in response to salinity stress than the other 13 small coding genes (P value = 0.0003, SI Appendix, Fig. S4). Among the four small coding genes, AT13 mRNA exhibited the highest induction (>100-fold) in response to salt stress. A liquid culture-based salinity stress (150 mM NaCl) assay was also conducted in which AT13-ox and WT plants were compared. Similar to the previous screening assay, AT13-ox plants exhibited higher salinity stress tolerance than WT plants (Fig. 1A). In addition, chlorophyll content in AT13-ox and WT plants was also measured, indicating a significantly higher chlorophyll content in AT13-ox plants than WT plants (Fig. 1A). A time-course evaluation of root growth in WT and AT13-ox plants over a 14-d period revealed significant differences between the plants by day 14 (Fig. 1B). Collectively, these observations support the supposition that AT13 plays an important role in salinity stress tolerance in Arabidopsis. To provide further corroboration, additional functional analyses of AT13 were performed.
Fig. 1.
AtPROPEP3/AT13-ox plants exhibit high levels of salinity stress tolerance. (A) Photograph of AtPROPEP3/AT13-ox and WT plants after the liquid culture-based salinity stress test and measurement of chlorophyll content per fresh weight (FW). Ten-day-old plants were treated with MS medium amended with 150 mM NaCl and then grown for 10 d. (B) Measurement of primary root length in AtPROPEP3/AT13-ox (red bar) and WT (blue bar) plants after the plate-based salinity stress test. One-week-old plants were transferred to an agar plate containing MS medium amended with 150 mM NaCl and then grown in a vertical orientation.
AtPROPEP3/AT13, a Member of the AtPROPEP Family, Plays an Important Role in Salinity Stress Tolerance.
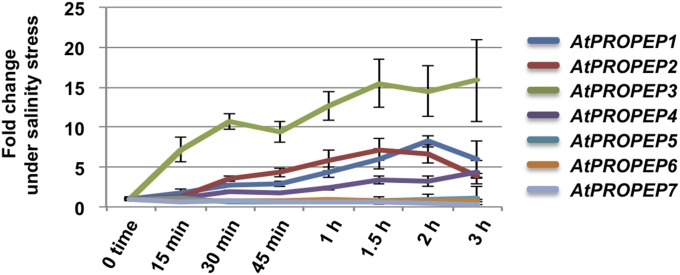
Although AT13 was previously identified as AtPROPEP3, its functional role in relation to salt stress tolerance was not known. AtPROPEP3/AT13 is a member of the AtPROPEP1–8 gene family, whose functions are related to the plant immune system (21, 47). AtPROPEP1–8 genes encode elicitor peptides (AtPep1–8) that have been functionally confirmed to induce defense-related responses in plants via spray applications of synthetic AtPep1–8 peptides (47, 48). Data from the initial microarray analysis indicated that AtPROPEP3/AT13 showed higher expression under salinity stress relative to control conditions (SI Appendix, Fig. S5). Furthermore, in our previously published microarray data, AtPROPEP3/AT13 exhibited the highest level of expression among all of the AtPROPEP gene family members in plants subjected to salt stress (SI Appendix, Fig. S6). When the expression levels were confirmed with qRT-PCR, AtPROPEP3/AT13 mRNA showed the greatest induction, which was more than 15 times at 1.5 h after NaCl treatment. On the other hand, AtPROPEP1 and -2 were ∼7–8 times greater after the same salinity treatment compared with controls. When expression levels were compared 15 min after NaCl treatment, AtPROPEP3 showed more than 7 times greater expression, while AtPROPEP1 and -2 were 1.8 and 1.3 times, respectively. Thus, AtPROPEP3 showed the most rapid response among the AtPROPEP gene family after salinity stress treatment (Fig. 2). Collectively, these data support the hypothesis that AtPROPEP3/AT13 plays an important role in inducing salinity stress tolerance.
Fig. 2.
Expression level of AtPROPEPs under salinity stress treatment. Fold changes of AtPROPEP genes were analyzed by qRT-PCR analysis. Two-week-old wild-type plants were treated with 200 mM NaCl and collected over a 3-h time-coursed sampling period.
AT13 Peptide Enhances Salinity Stress Tolerance.
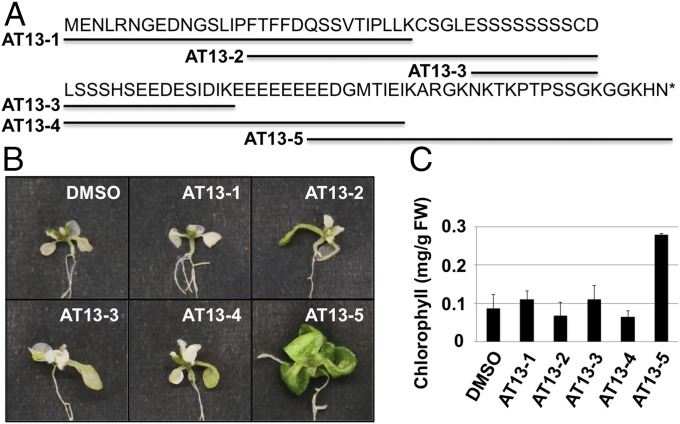
To determine whether or not the AtPep3/AT13 peptide encoded by AtPROPEP3/AT13 plays a functional role in salinity stress tolerance, a variety of synthetic peptide fragments were applied and salinity stress tolerance was evaluated using a liquid culture-based salinity stress assay. The structures of the various synthetic peptides are presented in Fig. 3A. To identify the functional region(s) within AtPROPEP3/AT13, peptide fragments (∼30 amino acids), with overlapping regions of 15 amino acids, were synthesized and used to treat plants. Summarized data for treatment with the synthetic peptides are presented in SI Appendix, Table S3. Seven-day-old WT Arabidopsis plants were pretreated with each peptide for a period of 3 d. Subsequently, both 150 mM NaCl and 10 µM of peptide fragments were administered at the same time in a liquid culture-based salinity stress assay. Arabidopsis plants treated with the AT13-5 peptide fragment exhibited the highest level of salinity stress tolerance in relative comparison with plants treated with the other peptide fragments or the untreated controls (Fig. 3B and SI Appendix, Fig. S7). Additionally, the chlorophyll content in seedlings treated with AT13-5 was significantly higher than the untreated control or seedlings treated with the other peptide fragments (Fig. 3C).
Fig. 3.
AtPep3/AT13 peptide induced salinity stress tolerance. (A) Full-length amino acid sequence of AT13 and synthetic peptide fragments used in the salt treatment assay. Overlapping peptide fragments covering the entire AT13 protein (∼30 amino acids in length) were synthesized. Sequence details are provided in SI Appendix, Table S3. (B and C) Images of representative plants after each peptide/salt treatment and chlorophyll content per fresh weight (FW) in plants after each peptide/salt treatment, respectively. Seven-day-old plants were pretreated with 10 µM of a peptide fragment for 3 d (peptide pretreatment) followed by a combined treatment with the peptide plus 150 mM NaCl. All treatments were carried out in liquid culture. Plants were grown for 10 d and chlorophyll content was subsequently assessed.
Functional Roles of AT13/AtPep3 Peptides Relative to Salinity Stress.
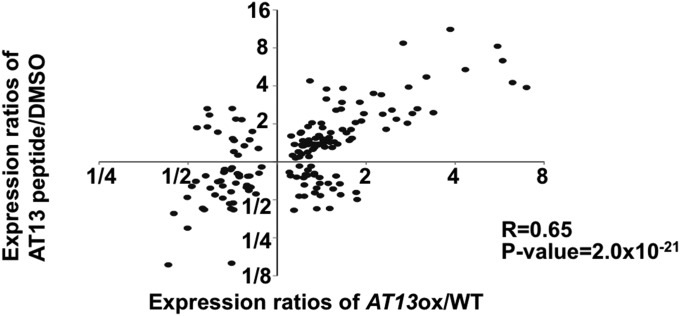
Treatment of plants with the AtPep3/AT13 peptide resulted in a phenotype similar to the one observed in AtPROPEP3/AT13-ox plants (Figs. 1 and 3). Therefore, a microarray analysis of AtPROPEP3/AT13-ox, AT13-5 peptide-treated, and WT plants was conducted to identify genes that were induced by the AT13-5 peptide. The analysis focused on 165 genes that were up- or down-regulated in both AtPROPEP3/AT13-ox and AT13-5 peptide-treated plants compared with WT plants (SI Appendix, Table S4) [false discovery rate (FDR) <0.05]. A salinity-inducible gene, AT1g17710, was up-regulated in both the AtPROPEP3/AT13-ox and AtPep3/AT13 peptide-treated plants and this response was subsequently confirmed by qRT-PCR (SI Appendix, Fig. S8 and Table S4). Although overrepresented gene ontology (GO) categories in AtPROPEP3/AT13-ox and AT13-5 peptide-treated plants were examined, none of the typical salinity or abiotic stress responsive genes were identified in either AtPROPEP3/AT13-ox or AT13-5 peptide-treated plants (SI Appendix, Table S5). Nevertheless, both up- and down-regulated genes exhibited a significant linear correlation in overexpressing and peptide-treated plants (r = 0.65, P value = 2.0 × 10−21), indicating that similar sets of genes were altered in both sets of plants (Fig. 4). These data indicate that the AT13-5 peptide treatment has a similar effect as AtPROPEP3/AT13-ox at the genomic level.
Fig. 4.
Comparison of gene expression in AtPROPEP3/AT13-ox and WT plants treated with the AT13-5 peptide. Log values of gene expression ratios of AtPROPEP3/AT13-ox and AT13-5 peptide-treated plants were plotted. Vertical and horizontal plots indicate alterations in gene expression patterns in AtPROPEP3/AT13-ox and AT13-5 peptide-treated plants, respectively.
To address the functional roles of AtPROPEP3/AT13 in salinity stress responses, we examined the common genes regulated by salinity stress conditions (2 h and 6 h) in our previous microarray and the 165 genes regulated by treatment with the AT13-5 peptide and AtPROPEP3/AT13-ox. Out of 165 genes, most of the genes (157/165 = 95%) were regulated by salinity stress treatment as well, indicating that unknown salinity-induced genes are substantially regulated by AtPROPEP3/AT13. Additionally, genes related to other phytohormones, such as ABA, JA, and SA, were not particularly altered (SI Appendix, Table S6).
As shown in Fig. 5 and SI Appendix, Fig. S9, AtPROPEP3/AT13 RNAi plants exhibited a salt-sensitive phenotype when exposed to 125 mM NaCl. This response was quantitatively verified by measuring chlorophyll content (Fig. 5 and SI Appendix, Fig. S9). The ability to complement the AtPROPEP3/AT13 RNAi plants by applying synthetic AT13-5 peptide was also examined. When RNAi plants were treated with 100 nM of AT13-5 peptide, the salt-sensitive phenotype of RNAi plants was complemented and these plants exhibited salinity stress tolerance relative to nontreated plants (Fig. 5 and SI Appendix, Fig. S9). As confirmed by qRT-PCR, one of the genes regulated by AtPROPEP3/AT13, AT1g17710, was down-regulated in RNAi plants (SI Appendix, Fig. S8). These data indicated that the AtPep3/AT13 peptide was essential for the induction of salinity stress tolerance.
Fig. 5.
Phenotype of AtPROPEP3/AT13 RNAi plants subjected to salinity stress. (Left) Representative images of WT, AtPROPEP3/AT13 RNAi, and AtPROPEP3/AT13 RNAi + AT13-5-treated (100 nM) plants subjected to salt stress in a liquid culture-based salinity stress test. (Right) Chlorophyll content per fresh weight (FW) in seedlings after the salt stress test. The salinity stress test utilized 125 mM NaCl and plants were grown for 10 d before measuring the chlorophyll content.
High levels of salinity stress tolerance were conferred both when the AtPROPEP3/AT13 coding sequence was overexpressed and also when plants received treatment with the AtPep3/AT13 peptide (Figs. 1 and 3). In plants, salinity stress occurs in two phases: (i) osmotic and (ii) Na+ ion stress. When osmotic stress tolerance was evaluated after application of the AtPep3/AT13 peptide, no significant differences were observed between the peptide treatment and the control (SI Appendix, Fig. S10). These data suggested that the AtPep3/AT13 peptide might function to confer salinity stress tolerance in relation to the ionic stress induced by Na+.
Identification of the Minimal Functional Fragment for Inducing Salinity Stress Tolerance.
A dose–response test was conducted to determine the lowest concentration of AT13-5 peptide that was sufficient enough to induce salinity stress tolerance. The results revealed that 10 nM of the AT13-5 peptide was still capable of inducing a high level of salt tolerance relative to untreated plants (Fig. 6), suggesting that the AtPep3/AT13 peptide functions at low concentrations in a similar effective range as other known peptide hormones. In addition, the data suggest that the AT13-5 peptide fragment contains the functional region of the AtPep3/AT13 peptide. Furthermore, to identify the causal functional region of the AtPep3/AT13 peptide for salinity stress tolerance, we carried out a peptide application test using several different lengths of the AT13 peptide, including the AtPep3 fragment as previously reported (21) (SI Appendix, Fig. S11 and Table S3). According to the test, 13-peptide (KPTSSGKGGKHN), AT13 (11–23) was the minimum size for activity under our experimental conditions (SI Appendix, Fig. S12).
Fig. 6.
Dose–response test of the AtPep3/AT13 peptide affected in salinity stress tolerance. (Left) Representative images of plants from the AT13-5 dose–response test. (Right) Chlorophyll content per fresh weight (FW) in plants after each peptide/salt treatment. Plants were treated with different concentrations of AT13-5 peptides for 3 d (peptide pretreatment) before being treated with medium containing the different concentrations of the peptide plus 150 mM NaCl. The presented data represent the average of three biological replicates. DMSO treatment was used as a control.
When using synthetic peptides, contamination risks associated with the preparation of peptide batches have been previously reported (49). To minimize the potential for additional effects contributed from other peptide impurities, a bioactivity examination was performed with high-purity (>90%) synthetic peptides of Pep3 and AT13 (11–23) (SI Appendix, Table S3). As shown in SI Appendix, Fig. S13, WT plants exhibited high salinity tolerance when treated with both of the purified peptides.
To detect the AtPep3/AT13 peptide in planta, we performed mass spectrometric analysis in either NaCl-treated or nontreated plants (SI Appendix, Fig. S14). According to the LC-MS/MS analysis, we identified a “TKPTPSSGKG” fragment, which overlapped with the minimum functional size of the AtPep3/AT13 peptide, from both NaCl-treated and nontreated plants (SI Appendix, Fig. S14 A and B). When comparing the areas of detected peaks of Pep3 from NaCl-treated and control samples by LC-MS analysis, the NaCl-treated sample was more than two times greater than the nontreated control (SI Appendix, Fig. S14 C and D). Collectively, these data suggest that this functional region of the AtPep3/AT13 peptide increased under salinity stress conditions and is capable of inducing salinity stress tolerance in plants.
Common Pathway for Salinity Stress Tolerance and Immune Response.
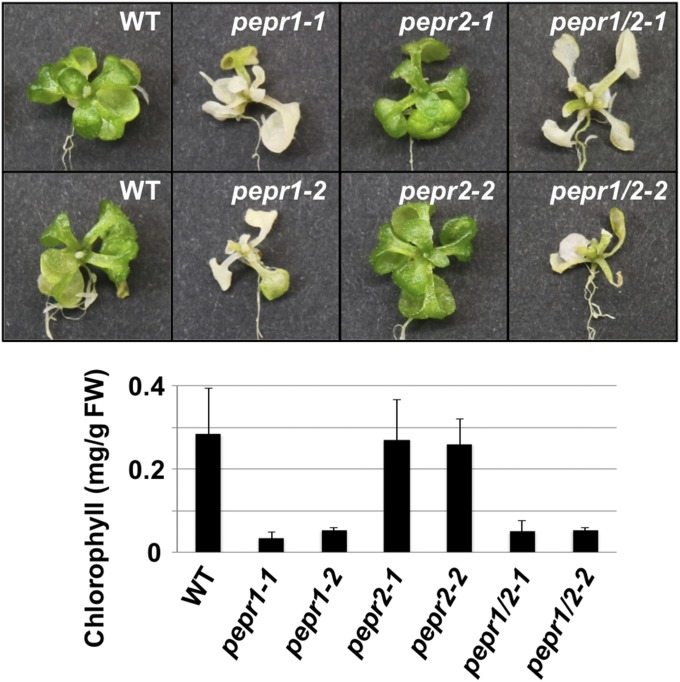
AtPep3 is a member of the Pep family and is an elicitor peptide for immune response in plants (21). Infections from both pathogens or bacteria have been shown to up-regulate AtPROPEP1–7 genes belonging to the Pep family. The binding of AtPeps encoded by AtPROPEP1–7 with two membrane-receptor kinase genes (PEPR1 and -2) as ligands (47, 48, 50) is functionally important for the immune response in Arabidopsis. The AtPep3/AT13 peptide has already been reported to preferentially bind to the PEPR1 receptor (48). In the present study, we demonstrated that the AtPep3/AT13 peptide plays an important role in plant salinity stress tolerance (Fig. 3). To elucidate whether the salinity stress tolerance conferred by treatment with the AtPep3/AT13 peptide is dependent upon a similar interaction of with PEPR1 or -2, as observed in the immune response of plants, we examined tolerance to salinity stress using single or double mutant plants of pepr1 and -2. When pepr1, -2, and double mutants were treated with only 125 mM NaCl, and did not receive an AtPep3/AT13 peptide treatment, all mutants exhibited a salinity sensitive phenotype compared with WT (SI Appendix, Fig. S15). When the AtPep3/AT13 peptide was applied to these mutants under the 125 mM NaCl stress condition, only pepr2 single mutants exhibited salinity stress tolerance, while both pepr1 and double mutants exhibited sensitivity to the salinity stress (Fig. 7 and SI Appendix, Fig. S16). These data indicated that the pathway between AtPep3/AT13 peptide and PEPR1 plays an important role for salinity stress tolerance. Thus, it is likely that salinity stress tolerance and immune response are induced by a shared pathway among the AtPep3/AT13 peptide and PEPR1.
Fig. 7.
Phenotype of pepr mutant plants subjected to salinity stress. (Top) Images of representative plants after each peptide/salt treatment. (Bottom) Chlorophyll content per fresh weight (FW) in plants after each peptide/salt treatment. Seven-day-old single or double receptor mutant plants were pretreated with 100 nM of a peptide fragment for 3 d (peptide pretreatment) followed by a combined treatment with the peptide plus 150 mM NaCl. All treatments were carried out in liquid culture. Plants were grown for 10 d and chlorophyll content was subsequently assessed. The presented data represent the average of three biological replicates.
Discussion
Hormone-like peptides have been recently identified in plants (1–3) and only a few have been shown to play a role in abiotic stress tolerance. In the present study, we demonstrated that AT13-ox plants exhibited a high level of survivorship compared with WT plants. In addition, we isolated function to the peptide level and confirmed that exogenous application of synthetic AtPep3 peptide was capable of enhancing salinity tolerance of plants.
Although AT13 was previously identified, its functional role in relation to salt stress tolerance was not known. AtPROPEP3/AT13 is a member of the AtPROPEP1–8 gene family, whose functions are related to the plant immune system (21). We confirmed that the AtPROPEP3/AT13 gene was induced by treatment with salinity stress and that AtPROPEP3/AT13 showed the highest induction among the gene family (Fig. 2). These data suggested that AtPROPEP3/AT13 functions as a trigger for salinity stress response.
Exogenous treatment of plants with a predicted mature peptide of AtPep3 (EIKARGKNKTKPTPSSGKGGKHN) induced biotic stress tolerance. These results were in accordance to what was observed for treatment with AtPep1, -2, and -4–7 (47, 48). In the present study, a larger fragment of AtPep3 was applied and found to induce salinity stress tolerance. To determine whether the functional region of the peptide was similar for inducing tolerance to both salinity and biotic stresses, we examined salinity stress tolerance after treatment with a size range of synthetic AtPep3/AT13 peptides (Fig. 3 and SI Appendix, Fig. S11 and Table S3). We identified the minimal fragment (KPTPSSGKGGKHN) which effectively induced salinity stress tolerance, and importantly, this fragment overlapped with the efficacious region which also conferred tolerance to biotic stress (SI Appendix, Figs. S11 and S12). We partially identified the endogenous AtPep3/AT13 peptide (TKPTPSSGKG) from plants by LC-MS/MS analysis (SI Appendix, Fig. S14). LC-MS/MS analysis revealed that the AtPep3/AT13 peptide was increased by NaCl treatment (SI Appendix, Fig. S14). Taken together with the increased expression of AtPROPEP3/AT13 mRNA under NaCl-treated conditions (as shown in Fig. 2), these data suggest that the AtPep3/AT13 peptide accumulated under salinity stress conditions and functioned for salinity stress tolerance in plants. In addition, our present data determined that the C-terminal region of the AtPep3/AT13 peptide plays an essential role for inducing tolerance to both salinity and biotic stress.
Treatment with the AtPep3/AT13 peptide induces both salinity stress and biotic stress tolerance within a similar concentration range to other peptide hormones. Specifically, a dose–response test revealed that application with 10 nM of the AtPep3/AT13 peptide still induced a high level of salinity stress tolerance in WT Arabidopsis plants (Fig. 6 and SI Appendix, Fig. S13). With respect to immunity response, treatment with either AtPep1 or AtPep3 peptides at comparably low concentrations (1–10 nM) induced the expression of defense-responsive genes (21, 48, 51). Taken together, it is likely that, similar to other peptide hormones, that the AtPep3/AT13 peptide functions in plants at a very low concentration within intercellular spaces.
Interestingly, AtPROPEP1–7 genes, including AtPROPEP3/AT13, do not contain signals for secretion in their N-terminal regions. It is not clear how AtPep1–7, which is encoded by AtPROPEP1–7, functions within plant intercellular spaces. AtPeps are thought to increase within cytoplasm after pathogen or bacterial infection. Subsequently, cell membranes are damaged and AtPeps are released and then recognized by PEPR1 and -2 receptors which are localized on the cell surface (48, 50, 52, 53). Indeed, similar to the plant immune response, we found that the AtPep3/AT13 peptide was recognized by PEPR1 but not by PEPR2; resulting in the induction of salinity stress tolerance in WT plants (Fig. 7). AtPeps, especially AtPep3, were highly induced by salinity stress and were likely release after the occurrence of cellular damage resultant from the stress. After release, the AtPeps were recognized by specific receptors shared between the common pathway that could be activated to elicit immune response and function to induce salinity stress tolerance in plants. These observations support the conclusion that both salinity and biotic stress result in comparable types of cellular damage, resulting in similar cellular responses that are mediated by AtPeps.
At the present time, specifics pertaining to the molecular signaling of salinity tolerance by the AtPep3/AT13 peptide are still not known. The AT1g17710 gene (PEPC1), was up-regulated in both the AtPROPEP3/AT13-ox and AtPep3/AT13 peptide-treated plants, and conversely, down-regulated in RNAi plants (SI Appendix, Fig. S8 and Table S4). The PEPC1 gene possesses a pyridoxal phosphate phosphatase-related domain which may likely catalyze pyridoxal 5-phosphate (PLP) to pyridoxal, which is an active form of vitamin B6. PLP is also recognized as a cofactor of several enzymes and can bind to ion channels and may therefore regulate Na+ influx (54, 55). It is possible that the regulation of Na+ influx by activated AtPep3/AT13 peptide might be the basis for the observed salinity stress tolerance observed in the present study, which is opposite of the response reported in the salt overly sensitive 4 mutant (54, 56, 57).
In this study, out of 81 up-regulated small coding genes, four candidates of sORF overexpressing plants exhibited high levels of salinity stress tolerance. Although we only examined the functional role of AT13, it is likely that the other three genes may also be associated to a new signaling network in relation to salinity stress tolerance. In a similar investigation, Chien et al. (25) also examined the salinity stress response within the CAP gene family consisting of small coding genes. Among the CAP genes, Chien et al. identified a secreted small peptide (AtCAPE1) which exhibited a salinity stress-tolerant phenotype within the knockout mutant. It is important to note that no similar genes were affected by the overexpression of AT13 or the knockout of AtCAPE1. Taken together, it appears that multiple small coding genes play various functional roles within salinity stress-related signal cascades.
Materials and Methods
Plant material and growth conditions, generation of AtPROPEP3/AT13-overexpressing and RNAi plant lines, salinity stress assay, liquid culture-based salinity stress test, measurements of chlorophyll content, peptide design and treatment assay, microarray analysis, immunoprecipitation, sample preparation for MS/MS analysis and LC-MS/MS analysis, and database searching are described in SI Appendix, Supporting Text.
Supplementary Material
Acknowledgments
We thank Dr. Fumiyoshi Myouga for advice on the experimental design of the chlorophyll measurement studies; Toshie Kita, Emiri Kuwabara, and Miyuki Ebashi for their technical assistance; and Rie Kurata for her excellent technical assistance. This work was supported by the Program for Promotion of Basic and Applied Research for Innovations in Bio-Oriented Industries (K.H. and M.H.-T.); RIKEN, FY2015 Incentive Research Projects (to K.N.); Grants-in-Aid for Scientific Research 15K14421, 15H02433, 17H03727, 15H02433, and 18H02420 (to K.H.), 17K07690 (to K.N.), and 18H04791 and 18H04705 (to M. Seki); and research grants from the Takeda Science Foundation (to K.H.), the Sumitomo Foundation (to K.H.), Kurume Research Park (to K.H.), the Asahi Glass Foundation (to K.H.), and Japan Science and Technology, Core Research for Evolutional Science and Technology [JPMJCR11B3 (to K.H.) and JPMJCR13B4 (to M. Seki)].
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE78515).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1719491115/-/DCSupplemental.
References
- 1.Matsubayashi Y, Sakagami Y. Peptide hormones in plants. Annu Rev Plant Biol. 2006;57:649–674. doi: 10.1146/annurev.arplant.56.032604.144204. [DOI] [PubMed] [Google Scholar]
- 2.Matsubayashi Y. Posttranslationally modified small-peptide signals in plants. Annu Rev Plant Biol. 2014;65:385–413. doi: 10.1146/annurev-arplant-050312-120122. [DOI] [PubMed] [Google Scholar]
- 3.Tavormina P, De Coninck B, Nikonorova N, De Smet I, Cammue BP. The plant peptidome: An expanding repertoire of structural features and biological functions. Plant Cell. 2015;27:2095–2118. doi: 10.1105/tpc.15.00440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andrews SJ, Rothnagel JA. Emerging evidence for functional peptides encoded by short open reading frames. Nat Rev Genet. 2014;15:193–204. doi: 10.1038/nrg3520. [DOI] [PubMed] [Google Scholar]
- 5.Matsubayashi Y, Sakagami Y. Phytosulfokine, sulfated peptides that induce the proliferation of single mesophyll cells of Asparagus officinalis L. Proc Natl Acad Sci USA. 1996;93:7623–7627. doi: 10.1073/pnas.93.15.7623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pearce G, Moura DS, Stratmann J, Ryan CA., Jr RALF, a 5-kDa ubiquitous polypeptide in plants, arrests root growth and development. Proc Natl Acad Sci USA. 2001;98:12843–12847. doi: 10.1073/pnas.201416998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ito Y, et al. Dodeca-CLE peptides as suppressors of plant stem cell differentiation. Science. 2006;313:842–845. doi: 10.1126/science.1128436. [DOI] [PubMed] [Google Scholar]
- 8.Whitford R, Fernandez A, De Groodt R, Ortega E, Hilson P. Plant CLE peptides from two distinct functional classes synergistically induce division of vascular cells. Proc Natl Acad Sci USA. 2008;105:18625–18630. doi: 10.1073/pnas.0809395105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Okuda S, et al. Defensin-like polypeptide LUREs are pollen tube attractants secreted from synergid cells. Nature. 2009;458:357–361. doi: 10.1038/nature07882. [DOI] [PubMed] [Google Scholar]
- 10.Hara K, et al. Epidermal cell density is autoregulated via a secretory peptide, EPIDERMAL PATTERNING FACTOR 2 in Arabidopsis leaves. Plant Cell Physiol. 2009;50:1019–1031. doi: 10.1093/pcp/pcp068. [DOI] [PubMed] [Google Scholar]
- 11.Sugano SS, et al. Stomagen positively regulates stomatal density in Arabidopsis. Nature. 2010;463:241–244. doi: 10.1038/nature08682. [DOI] [PubMed] [Google Scholar]
- 12.Nakayama T, et al. A peptide hormone required for Casparian strip diffusion barrier formation in Arabidopsis roots. Science. 2017;355:284–286. doi: 10.1126/science.aai9057. [DOI] [PubMed] [Google Scholar]
- 13.Okamoto S, Shinohara H, Mori T, Matsubayashi Y, Kawaguchi M. Root-derived CLE glycopeptides control nodulation by direct binding to HAR1 receptor kinase. Nat Commun. 2013;4:2191. doi: 10.1038/ncomms3191. [DOI] [PubMed] [Google Scholar]
- 14.Okamoto S, Suzuki T, Kawaguchi M, Higashiyama T, Matsubayashi Y. A comprehensive strategy for identifying long-distance mobile peptides in xylem sap. Plant J. 2015;84:611–620. doi: 10.1111/tpj.13015. [DOI] [PubMed] [Google Scholar]
- 15.Tabata R, et al. Perception of root-derived peptides by shoot LRR-RKs mediates systemic N-demand signaling. Science. 2014;346:343–346. doi: 10.1126/science.1257800. [DOI] [PubMed] [Google Scholar]
- 16.Takahashi F, et al. A small peptide modulates stomatal control via abscisic acid in long-distance signalling. Nature. 2018;556:235–238. doi: 10.1038/s41586-018-0009-2. [DOI] [PubMed] [Google Scholar]
- 17.Tör M, Lotze MT, Holton N. Receptor-mediated signalling in plants: Molecular patterns and programmes. J Exp Bot. 2009;60:3645–3654. doi: 10.1093/jxb/erp233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ohyama K, Ogawa M, Matsubayashi Y. Identification of a biologically active, small, secreted peptide in Arabidopsis by in silico gene screening, followed by LC-MS-based structure analysis. Plant J. 2008;55:152–160. doi: 10.1111/j.1365-313X.2008.03464.x. [DOI] [PubMed] [Google Scholar]
- 19.Stenvik GE, et al. The EPIP peptide of INFLORESCENCE DEFICIENT IN ABSCISSION is sufficient to induce abscission in arabidopsis through the receptor-like kinases HAESA and HAESA-LIKE2. Plant Cell. 2008;20:1805–1817. doi: 10.1105/tpc.108.059139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ryan CA, Pearce G. Systemins: A functionally defined family of peptide signals that regulate defensive genes in Solanaceae species. Proc Natl Acad Sci USA. 2003;100:14577–14580. doi: 10.1073/pnas.1934788100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huffaker A, Pearce G, Ryan CA. An endogenous peptide signal in Arabidopsis activates components of the innate immune response. Proc Natl Acad Sci USA. 2006;103:10098–10103. doi: 10.1073/pnas.0603727103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Narvaez-Vasquez J, Orozco-Cardenas ML. Systemins and AtPeps: Defense-related peptide signals. In: Schaller A, editor. Induced Plant Resistance to Herbivory. Springer Science; New York: 2008. pp. 313–328. [Google Scholar]
- 23.Orsini F, et al. Systemin-dependent salinity tolerance in tomato: Evidence of specific convergence of abiotic and biotic stress responses. Physiol Plant. 2010;138:10–21. doi: 10.1111/j.1399-3054.2009.01292.x. [DOI] [PubMed] [Google Scholar]
- 24.Delay C, Imin N, Djordjevic MA. CEP genes regulate root and shoot development in response to environmental cues and are specific to seed plants. J Exp Bot. 2013;64:5383–5394. doi: 10.1093/jxb/ert332. [DOI] [PubMed] [Google Scholar]
- 25.Chien PS, Nam HG, Chen YR. A salt-regulated peptide derived from the CAP superfamily protein negatively regulates salt-stress tolerance in Arabidopsis. J Exp Bot. 2015;66:5301–5313. doi: 10.1093/jxb/erv263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Flowers TJ. Improving crop salt tolerance. J Exp Bot. 2004;55:307–319. doi: 10.1093/jxb/erh003. [DOI] [PubMed] [Google Scholar]
- 27.Munns R, Tester M. Mechanisms of salinity tolerance. Annu Rev Plant Biol. 2008;59:651–681. doi: 10.1146/annurev.arplant.59.032607.092911. [DOI] [PubMed] [Google Scholar]
- 28.Roy SJ, Negrão S, Tester M. Salt resistant crop plants. Curr Opin Biotechnol. 2014;26:115–124. doi: 10.1016/j.copbio.2013.12.004. [DOI] [PubMed] [Google Scholar]
- 29.Munns R, Passioura JB, Guo J, Chazen O, Cramer GR. Water relations and leaf expansion: Importance of time scale. J Exp Bot. 2000;51:1495–1504. doi: 10.1093/jexbot/51.350.1495. [DOI] [PubMed] [Google Scholar]
- 30.Munns R. Comparative physiology of salt and water stress. Plant Cell Environ. 2002;25:239–250. doi: 10.1046/j.0016-8025.2001.00808.x. [DOI] [PubMed] [Google Scholar]
- 31.Davenport R, James RA, Zakrisson-Plogander A, Tester M, Munns R. Control of sodium transport in durum wheat. Plant Physiol. 2005;137:807–818. doi: 10.1104/pp.104.057307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu SJ, Ding L, Zhu JK. SOS1, a genetic locus essential for salt tolerance and potassium acquisition. Plant Cell. 1996;8:617–627. doi: 10.1105/tpc.8.4.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhu JK, Liu J, Xiong L. Genetic analysis of salt tolerance in arabidopsis. Evidence for a critical role of potassium nutrition. Plant Cell. 1998;10:1181–1191. doi: 10.1105/tpc.10.7.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang Q, et al. Overexpression of SOS (Salt Overly Sensitive) genes increases salt tolerance in transgenic Arabidopsis. Mol Plant. 2009;2:22–31. doi: 10.1093/mp/ssn058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Halfter U, Ishitani M, Zhu JK. The Arabidopsis SOS2 protein kinase physically interacts with and is activated by the calcium-binding protein SOS3. Proc Natl Acad Sci USA. 2000;97:3735–3740. doi: 10.1073/pnas.040577697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mäser P, et al. Altered shoot/root Na+ distribution and bifurcating salt sensitivity in Arabidopsis by genetic disruption of the Na+ transporter AtHKT1. FEBS Lett. 2002;531:157–161. doi: 10.1016/s0014-5793(02)03488-9. [DOI] [PubMed] [Google Scholar]
- 37.Barragán V, et al. Ion exchangers NHX1 and NHX2 mediate active potassium uptake into vacuoles to regulate cell turgor and stomatal function in Arabidopsis. Plant Cell. 2012;24:1127–1142. doi: 10.1105/tpc.111.095273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schroeder JI, et al. Using membrane transporters to improve crops for sustainable food production. Nature. 2013;497:60–66. doi: 10.1038/nature11909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shi H, Xiong L, Stevenson B, Lu T, Zhu JK. The Arabidopsis salt overly sensitive 4 mutants uncover a critical role for vitamin B6 in plant salt tolerance. Plant Cell. 2002;14:575–588. doi: 10.1105/tpc.010417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shi H, Kim Y, Guo Y, Stevenson B, Zhu JK. The Arabidopsis SOS5 locus encodes a putative cell surface adhesion protein and is required for normal cell expansion. Plant Cell. 2003;15:19–32. doi: 10.1105/tpc.007872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhu J, et al. A cellulose synthase-like protein is required for osmotic stress tolerance in Arabidopsis. Plant J. 2010;63:128–140. doi: 10.1111/j.1365-313X.2010.04227.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hanada K, Zhang X, Borevitz JO, Li WH, Shiu SH. A large number of novel coding small open reading frames in the intergenic regions of the Arabidopsis thaliana genome are transcribed and/or under purifying selection. Genome Res. 2007;17:632–640. doi: 10.1101/gr.5836207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hanada K, et al. sORF finder: A program package to identify small open reading frames with high coding potential. Bioinformatics. 2010;26:399–400. doi: 10.1093/bioinformatics/btp688. [DOI] [PubMed] [Google Scholar]
- 44.Wen J, Lease KA, Walker JC. DVL, a novel class of small polypeptides: Overexpression alters Arabidopsis development. Plant J. 2004;37:668–677. doi: 10.1111/j.1365-313x.2003.01994.x. [DOI] [PubMed] [Google Scholar]
- 45.Hara K, Kajita R, Torii KU, Bergmann DC, Kakimoto T. The secretory peptide gene EPF1 enforces the stomatal one-cell-spacing rule. Genes Dev. 2007;21:1720–1725. doi: 10.1101/gad.1550707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hanada K, et al. Small open reading frames associated with morphogenesis are hidden in plant genomes. Proc Natl Acad Sci USA. 2013;110:2395–2400. doi: 10.1073/pnas.1213958110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bartels S, et al. The family of Peps and their precursors in Arabidopsis: Differential expression and localization but similar induction of pattern-triggered immune responses. J Exp Bot. 2013;64:5309–5321. doi: 10.1093/jxb/ert330. [DOI] [PubMed] [Google Scholar]
- 48.Yamaguchi Y, Huffaker A, Bryan AC, Tax FE, Ryan CA. PEPR2 is a second receptor for the Pep1 and Pep2 peptides and contributes to defense responses in Arabidopsis. Plant Cell. 2010;22:508–522. doi: 10.1105/tpc.109.068874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mueller K, et al. Contamination risks in work with synthetic peptides: flg22 as an example of a pirate in commercial peptide preparations. Plant Cell. 2012;24:3193–3197. doi: 10.1105/tpc.111.093815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yamaguchi Y, Pearce G, Ryan CA. The cell surface leucine-rich repeat receptor for AtPep1, an endogenous peptide elicitor in Arabidopsis, is functional in transgenic tobacco cells. Proc Natl Acad Sci USA. 2006;103:10104–10109. doi: 10.1073/pnas.0603729103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Krol E, et al. Perception of the Arabidopsis danger signal peptide 1 involves the pattern recognition receptor AtPEPR1 and its close homologue AtPEPR2. J Biol Chem. 2010;285:13471–13479. doi: 10.1074/jbc.M109.097394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Huffaker A, Ryan CA. Endogenous peptide defense signals in Arabidopsis differentially amplify signaling for the innate immune response. Proc Natl Acad Sci USA. 2007;104:10732–10736. doi: 10.1073/pnas.0703343104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yamaguchi Y, Huffaker A. Endogenous peptide elicitors in higher plants. Curr Opin Plant Biol. 2011;14:351–357. doi: 10.1016/j.pbi.2011.05.001. [DOI] [PubMed] [Google Scholar]
- 54.González E, Danehower D, Daub ME. Vitamer levels, stress response, enzyme activity, and gene regulation of Arabidopsis lines mutant in the pyridoxine/pyridoxamine 5′-phosphate oxidase (PDX3) and the pyridoxal kinase (SOS4) genes involved in the vitamin B6 salvage pathway. Plant Physiol. 2007;145:985–996. doi: 10.1104/pp.107.105189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fitzpatrick TB, et al. Vitamin deficiencies in humans: Can plant science help? Plant Cell. 2012;24:395–414. doi: 10.1105/tpc.111.093120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tambasco-Studart M, et al. Vitamin B6 biosynthesis in higher plants. Proc Natl Acad Sci USA. 2005;102:13687–13692. doi: 10.1073/pnas.0506228102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xia J, et al. Nitric oxide negatively regulates AKT1-mediated potassium uptake through modulating vitamin B6 homeostasis in Arabidopsis. Proc Natl Acad Sci USA. 2014;111:16196–16201. doi: 10.1073/pnas.1417473111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.