Abstract
Ras is a membrane-anchored signaling protein that serves as a hub for many signaling pathways and also plays a prominent role in cancer. The intrinsic behavior of Ras on the membrane has captivated the biophysics community in recent years, especially the possibility that it may form dimers. In this article, we describe results from a comprehensive series of experiments using fluorescence correlation spectroscopy and single-molecule tracking to probe the possible dimerization of natively expressed and fully processed K-Ras4B in supported lipid bilayer membranes. Key to these studies is the fact that K-Ras4B has its native membrane anchor, including both the farnesylation and methylation of the terminal cysteine, enabling detailed exploration of possible effects of cholesterol and lipid composition on K-Ras4B membrane organization. The results from all conditions studied indicate that full-length K-Ras4B lacks intrinsic dimerization capability. This suggests that any lateral organization of Ras in living cell membranes likely stems from interactions with other factors.
Introduction
Ras is a protein of paramount clinical importance with its oncogenic mutations correlated with 30% of all human cancers (1). Its principal function as a membrane-anchored GTPase is to serve as a molecular switch; when bound to GDP, Ras remains inert, but when GDP is exchanged to GTP, a conformational change allows interaction with downstream effectors and signal propagation. On the cell membrane surface, some reports have suggested Ras is ordered into clusters that may be central to signal propagation (2, 3). Additionally, direct molecular dimerization of Ras on the membrane has emerged in recent years as a possible underlying mechanism for the lateral organization (4, 5). The putative Ras dimer has also attracted attention as an alternative inhibition target for cancers involving hyperactivation of Ras, which to date has proven exceptionally difficult to control with conventional therapeutic drugs (6, 7). Although crystal structures of Ras indicate a possible dimer interface, dimers are not generally observed in solution, even at high Ras concentration (8, 9). The structure of Ras, however, may be different on the membrane (10, 11) and the possibility of a membrane-assisted dimerization or clustering mechanism has attracted much attention (9, 12, 13, 14, 15). Such mechanisms include membrane-mediated exposure of a dimerization interface and lipid-mediated interactions through the Ras membrane anchor.
Although studies of Ras in cell membranes have revealed nonuniform distributions (2, 13, 16, 17, 18), it is extremely difficult to conclusively determine if this organization is an intrinsic property of Ras or is the result of other interactions within the cellular environment (19, 20). A number of studies of purified Ras in lipid vesicles and supported bilayers have yielded mixed conclusions. Studies using fluorescent spectroscopic methods with N- and H-Ras with semisynthetic anchors in supported lipid bilayers reported dimerization (10, 11), but a subsequent study found Ras to be exceptionally susceptible to photosensitized oxidative cross linking, which could have affected the fluorescence experiments (21). K-Ras dimers were detected in solution by dynamic light scattering when bound to GTPγS, a sulfuric GTP analog (22). On the other hand, the globular domain of H-Ras was found to be incapable of dimerization when investigated with rotational anisotropy and NMR (23). Lacking in these studies, however, were the posttranslational modifications of Ras, which are known to be critical for proper membrane localization in cells and could introduce additional interactions within the membrane. Furthermore, possible effects of various lipid headgroups, chain chemistry, and the presence of cholesterol—a major component in the plasma membrane—were largely unexplored in these earlier in vitro studies.
To gain a clearer understanding of Ras dimerization on membranes, and to address potential roles of membrane lipids and cholesterol interacting with the Ras membrane anchor, we reconstituted natively purified K-Ras4B, wild-type, and tagged with eGFP, on supported lipid bilayers (Fig. 1). K-Ras4B is a Ras isoform associated with the highest incidence of oncogenic mutations (24). The purification and characterization of these constructs with complete posttranslational modifications has recently been accomplished, thus providing an opportunity for a more thorough investigation of K-Ras4B organization on membranes (25). K-Ras4B is composed of a globular G-domain, a positively charged hypervariable region, and posttranslational modifications including farnesylation and methylation of the terminal cysteine, which provide anchorage to the membrane (26). When introduced to supported lipid bilayers, K-Ras4B spontaneously inserts into the membrane and establishes equilibrium between membrane and solution (Fig. S1). Using this experimental platform, we systematically investigated how key parameters such as protein density on the membrane surface, nucleotide state, and lipid membrane composition (including cholesterol) modulate the behavior of K-Ras4B with quantitative spectroscopic measurements.
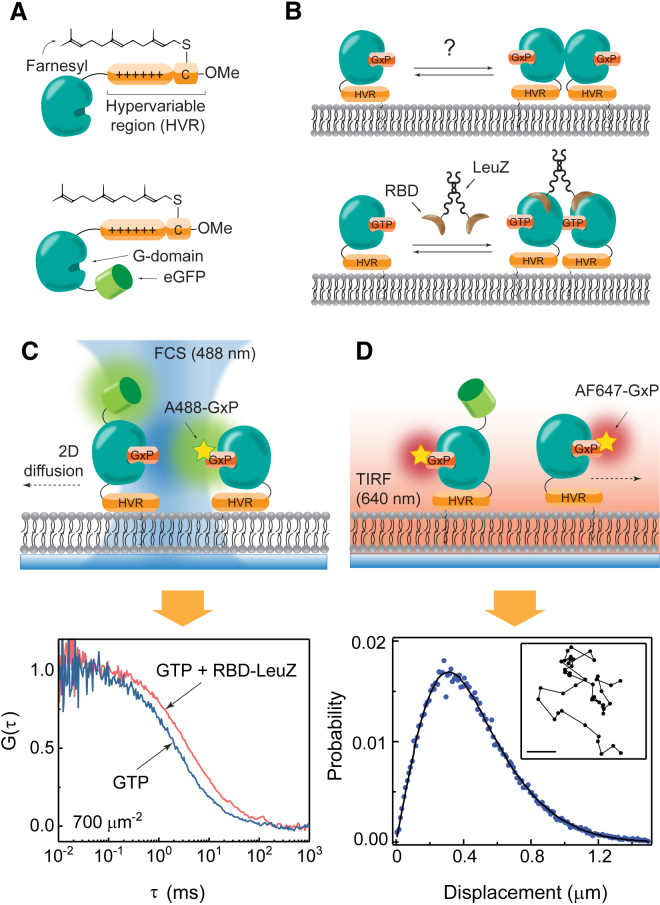
Figure 1.
Experimental design for K-Ras on SLBs. (A) Given here are processed K-Ras4B constructs used in these experiments: wild-type (top) and eGFP-labeled (bottom). (B) Cross-linked dimers formed by RBD fused to LeuZ were used as benchmarks for dimerization for the diffusion measurements. (C) Given here is the supported lipid bilayer-based experimental setup for FCS (top), and resulting autocorrelation traces for monomeric and cross-linked Ras (bottom). (D) Shown here is the TIRF microscopy setup for SMT (top), and step-size distribution (bottom). (Inset) This shows an example of SMT trajectories from which the step-size distributions are obtained; scale bar: 10 μm. To see this figure in color, go online.
Overall, the studies of fully processed K-Ras4B described here reveal universally monomeric behavior across a wide range of surface densities and membrane compositions. Even in cholesterol-containing membranes near a miscibility phase transition, strictly monomeric mobility is observed. Due to both the importance of Ras and its complexity on membranes, we have sought to acquire as comprehensive and quantitatively rigorous data as presently possible to address the question of its dimerization on membranes. These observations indicate that any dimerization or clustering of Ras that may exist in the cell membrane involves additional factors not present in these reconstituted experiments.
Materials and Methods
Protein preparation
K-Ras4B and eGFP-K-Ras4B. The complete protocol for purification of fully processed K-Ras4B has been published previously (25). For eGFP-K-Ras4B, the linker was GSG. The full amino acid sequence can be found in the Supporting Material. FNTA and FNTB, the human enzymes responsible for the first posttranslational modification (farnesylation), are introduced into the baculovirus/insect expression system by cloning the respective genes into the same baculovirus genome as the K-Ras4B expression construct. Insect proteins presumably carry out the subsequent posttranslational modification steps of C-terminal proteolysis (removing the C-terminal amino acid tripeptide of Val-Ile-Met and the hydroxymethylation of the C-terminal carboxyl group). K-Ras4B is expressed as the fusion protein His6-MBP-tev-K-Ras4B (His6, six-histidine affinity tag; MBP, maltose binding protein to enhance solubility; tev, TEV protease recognition site). Briefly, the expressed protein is captured from Tni-FNL cells (a cell line of Trichoplusia ni isolated at the Frederick National Laboratory) after lysis of the pellet (harvested 72 h postinfection from cultures incubated at 21°C) by immobilized metal-ion chromatography (IMAC). The fusion protein is buffer-exchanged to a lower salt and lower pH buffer (MES pH 6.0), bound to an SP sepharose column, and eluted with a moderate salt buffer. The fusion protein is then cleaved by the addition of a His6-TEV protease and simultaneously buffer-exchanged back to a neutral pH buffer. The final chromatographic step is to remove the His6-tagged contaminants from the TEV protease reaction (His6-TEV protease, noncleaved fusion protein, and His6-MBP) using another IMAC step (the non-His-tagged target protein has a low affinity for the IMAC resin but is well resolved from the contaminants in the column elution). The protein is dialyzed to remove the trace amounts of imidazole from the column elution, concentrated as necessary. The molecular weight of the processed K-Ras4B and eGFP-K-Ras4B was confirmed by SDS-PAGE and intact electrospray ionization mass spectrometry, shown in Fig. S1. The mass spectra for both K-Ras4B and eGFP-K-Ras4B show negligible abundance for unlipidated (−380 Da) species.
The Ras binding domain fused to leucine zipper (RBD-LeuZ) and the catalytic domain of Son of Sevenless (SOScat). Ras binding domain (RBD) (residues 56–131) of cRaf (accession number: BC018119) with N-terminal SNAP tag and Leucine zipper (LeuZ, amino acid sequence: YKQLEDKVEELASKNYHLENEVARLKKLVEF) was cloned into a modified pETM-33 vector (European Molecular Biology Laboratory, Heidelberg, Germany) using the KpnI/NotI sites. The expression vector fuses an N-terminal His6-GST-PreScission cleavage sequence (pr)-SNAP tag (SNAP) to RBD, creating His6-GST-pr-SNAP-LeuZ-RBD. SOScat Cys-lite (residues 566–1049 with the following mutations: C838A, C635A, C980S, and E718C) of human SOS1 (accession number: AK290228.1) was cloned in the expression vector pProEx HTb (Invitrogen, Carlsbad, CA) using NcoI/HindIII sites. The vector fuses an N-terminal His6 Tag-Tev sequence to SOScat. The detailed protein purification protocol for SOScat has been published previously (27). A similar protocol was also used for purification of RBD-LeuZ. RBD-LeuZ and SOScat were expressed in Escherichia coli (BL21 DE3) cells and purified using an N-terminal His6 tag. After elution from a HiTrap Chelating HP column (GE Healthcare, Little Chalfont, UK), the His6-GST tag for RBD-LeuZ and His6 tag for SOScat was cleaved by incubation of the protein with the TEV and PreScission protease, respectively. The cleavage reaction was carried out overnight at 4°C, while dialyzing in 50 mM PBS (pH 8.0), 300 mM NaCl and 0.4 mM 2-mercaptoethanol. The proteins were further purified by size-exclusion chromatography on a Superdex 75 column (GE Healthcare) that was equilibrated in gel-filtration buffer (20 mM Tris-HCl pH 8.0, 200 mM NaCl, 10% glycerol (v/v), and 1 mM tris(2-carboxyethyl)phosphine (Cat. No. C4706; Sigma-Aldrich, St. Louis, MO).
Supported lipid bilayer formation
Planar-supported lipid bilayers (SLBs) of varying compositions consisting primarily of 1,2-dioleoyl-sn-glycero-3-phosphocoline (DOPC, Cat. No. 850375C; Avanti Polar Lipids, Alabaster, AL) were prepared for fluorescence microscopy. The detailed protocol has been published previously (28). Briefly, SLBs were formed by fusion of small unilamellar vesicles, prepared by probe sonication, on glass substrates cleaned by Piranha etch (5 min in 3:1 H2SO4:H2O2). Other bilayer components such as phosphatidylinositol (4, 5)-bisphosphate (PIP2), 18:1-18:1 PS (DOPS), and 18:1-18:0 PS (SOPS) were also purchased from Avanti Polar Lipids (Cat. No. 850156P, 840046, and 840035C, respectively). For the visualization of the bilayers, a trace amount (0.005 mol %) of fluorescently labeled lipid, Texas Red DHPE (Cat. No. T1395MP; Thermo Fisher Scientific, Waltham, MA) was included. The bilayers were prepared in sticky-slide VI 0.4 microfluidic chambers (Cat. No. 80608; Ibidi, Munich, Germany) assembled with the glass slide substrate for imaging. The final buffer (pH 7.4) was composed of the following: 40 mM HEPES, 100 mM NaCl, 10 mM BME, and 0.1 mg/mL casein (all purchased from Sigma-Aldrich). BME is critical in preventing photosensitized cross linking that was found to affect Ras during fluorescence experiments (21). The integrity and fluidity of the bilayers and membrane-bound proteins was confirmed by fluorescence recovery after photobleaching or fluorescence correlation spectroscopy (FCS). All experiments were performed at room temperature, 23°C.
Supported lipid bilayer from ruptured giant unilamellar vesicles
Giant unilamellar vesicles (GUVs) were produced by electroformation, closely following a published protocol (29). The lipid compositions (either 10% DOPS vesicles: 0.39/0.1/0.2/0.3/0.01 DOPC/DOPS/DPPC/cholesterol/TR-DHPE, or 20% SOPS vesicles: 0.29/0.2/0.2/0.3/0.01 DOPC/SOPS/DPPC/cholesterol/TR-DHPE) were chosen such that the demixing transition temperature is below the room temperature (30). A quantity of 20 mmol of lipid mixtures in chloroform was deposited in the ITO-coated glass slides (30–40 Ω; Delta Technologies, Dallas, TX), and assembled into a capacitor using a rubber spacer and conductive copper tapes. GUV budding from lipid film was then induced by passing ±2.8 V sine waves at 10 Hz, provided by a waveform generator (Tektronics, Beaverton, OR), through the capacitor for 3 h in 250 mM sucrose solution (osmolarity 270 mOs) at 50°C to maximize homogeneous distribution of lipids. The resulting vesicles were then allowed to adhere and rupture onto Piranha-etched glass coverslip surfaces in HEPES buffer, and were imaged by Texas Red fluorescence.
Ras nucleotide exchange
Native nucleotides bound to K-Ras were exchanged by guanine nucleotide exchange factor SOS (31, 32). A quantity of 1 μM of K-Ras was incubated with 0.5 μM of catalytic domain of SOS, SOScat, in an excess amount of desired nucleotides (1 mM for nonfluorescent nucleotides, and 10 μM for fluorescent nucleotides). Nucleotide exchange reactions were carried out at least 3 h at room temperature.
FCS
The experimental setup for FCS has been described previously (21). Dual-color FCS measurements were performed on a home-built confocal system integrated into a TE2000 inverted microscope with a 100× oil immersion objective (PlanFluor, NA 1.30; Nikon, Melville, NY). The light source was a pulsed (100 ps pulse duration at 19.5 MHz repetition rate) supercontinuum laser (SuperK Extreme; NKT Photonics, Portland, OR), with the wavelengths selected by a combination of dichroic mirrors and bandpass filters, which were directed to the microscope via an optical fiber (Thorlabs, Newton, NJ) in the epifluorescence geometry. The average excitation power for a typical FCS measurement was 0.5 μW for blue light (488 ± 5 nm) and 1.0 μW for orange light (561 ± 5 nm) immediately before the objective. The fluorescent signals were collected by the objective and passed a 50-μm pinhole (Thorlabs) before they were split by a dichroic mirror (Thorlabs). Each signal was focused into 0.15 × 0.15 μm avalanche photodiode elements (Hamamatsu, Hamamatsu City, Japan), and subsequently processed by a hardware correlator (Correlator.com). The resulting autocorrelation G(τ) traces were fit to 2D Gaussian diffusion model (33),
| (1) |
where τ is the time delay, N is the number of particles in the focus, and τd is the correlation time. To calibrate the spot size of the confocal focus, N of a bilayer with a known surface density of fluorescent lipids of each color, BODIPY-FL-DHPE (Thermo Fisher Scientific) for the blue light and Texas Red-DHPE for the orange light were measured, which consistently yielded the radii of 0.20 ± 0.01 and 0.22 ± 0.01 μm for the blue and orange light, respectively. The diffusion coefficient D was calculated by using the relation
| (2) |
where w is the radius of the focus spot size.
Single-molecule total internal reflectance fluorescence microscopy
The experimental methods for single-molecule total internal reflectance fluorescence (TIRF) has been described previously (21, 32). TIRF images were acquired using an Eclipse Ti inverted microscope (Nikon) equipped with a 100× 1.49 NA oil immersion TIRF objective and an iXon EMCCD camera (Andor Technology, South Windsor, CT). Illumination sources for TIRF imaging in the form of 488- and 637-nm lasers (Coherent, Santa Clara, CA) were used, with ET500LP and ET525/50M filters (Chroma Technology, Bellows Falls, VT) for the 488-nm channel imaging, and ET660LP and ET700/75M filters for 637-nm channel imaging. To track K-Ras at the single-molecule level in a wide range of surface density, 100 pM of Alexa 647-GppNHp-loaded K-Ras was mixed with various concentrations of eGFP-K-Ras loaded with nonfluorescent GppNHp (typically up to 50 nM). TIRF intensity of the eGFP channel was used to estimate the overall density of K-Ras, and Alexa 647-GppNHp-loaded K-Ras was imaged for single-molecule tracking (SMT). Diffusion trajectories were analyzed with the softwares TrackMate (ImageJ plugins; National Institutes of Health, Bethesda, MD) and Igor Pro (WaveMetrics, Lake Oswego, OR). The step size distribution calculated from several thousands of trajectories was fit with the Brownian diffusion model as described by the following equation:
| (3) |
where D1 and D2 are different diffusion coefficients for fast and slow species, respectively; and α is a relative population for the fast species. For the single-species diffusion model, α is 1, thus eliminating the second term. Fitting residues for the single- and two-component model were monitored to determine the number of diffusion species. The diffusion coefficients and relative population of each component were calculated from the corresponding fitting (see the Supporting Material for more details).
Results and Discussion
We monitored the K-Ras4B (henceforth referred to simply as “Ras”) mobility on the membrane surface to detect dimerization. For all the lipid compositions studied here, membrane-anchored Ras exhibits unencumbered Brownian motion, as described by the Einstein relation,
| (4) |
where r is displacement, t is time, D is the diffusion coefficient, kB is the Boltzmann constant, T is the temperature, and λ is the drag coefficient. The diffusion coefficient of molecules or particles through a fluid is generally a function of size. In three dimensions, the familiar Stokes-Einstein relation (D = kBT/6πηr, where η is the fluid viscosity) is widely used as a basis for molecular and particle size determination. The situation in two dimensions, however, is more complex because no such simple scaling between mobility and size exists—a phenomenon known as the Stokes paradox (34, 35). Nonetheless, dimerization on membranes is robustly detectable by measuring changes in D resulting from the differing values for λ experienced by monomers and dimers. An additive effect in the frictional drags of dimers has been observed experimentally when well-defined and separated lipid anchors dictate protein diffusion (36, 37, 38). However, the actual values of λ generally depend on a complex convolution of many factors such as protein structure, degree of its interaction with the membrane, and the surrounding environment. Furthermore, although quantities such as protein radius have well-defined consequences on diffusion as described by Saffman and Delbrück (34), they are difficult to determine for Ras dimerization. For instance, it cannot be determined, based on currently available information, how much dimerization would increase the effective radius of the protein, given its irregular shape and ambiguous nature of the interaction with the membrane. Therefore, rather than attempting to calculate the theoretical diffusion coefficient of Ras dimers, we experimentally calibrate the differential mobility between a Ras monomer and dimer using a Ras binding domain of c-Raf fused with a leucine zipper (RBD-LeuZ) to induce dimerization in a controlled manner. RBD selectively binds to GTP-bound Ras and LeuZ is a constitutive dimer, therefore its presence creates a population of cross-linked Ras, providing a benchmark for dimerization (Fig. 1 B).
For quantitative measurement of Ras diffusion on SLBs, we employed FCS (Fig. 1 C) and SMT (Fig. 1 D) by TIRF microscopy. Although either method alone is, in principle, sufficient to obtain all of the conclusions derived here, they provide complementary information and we use both to obtain a more complete characterization of Ras mobility on the membrane. Additionally, two different labeling strategies were used to track Ras: eGFP fused at the N-terminus, or nucleotides labeled with extrinsic organic fluorophores, such as ATTO 488 (A488) or AlexaFluor 647 (AF647). The identity of the fluorescent label did not result in any detectable differences in the diffusion behavior of Ras, indicating that the eGFP label does not introduce a significant perturbation.
In FCS, the time-dependent fluorescence fluctuation due to molecules entering and exiting the confocal focus is recorded (Fig. 1 C). The autocorrelation function, G(τ), of these fluctuations of a mixed population in a 2D Gaussian focal spot is given by
| (5) |
where Ni, Bi, and τi are the number of particles, the molecular brightness, and the focus residence time for the ith species, respectively (33). The apparent surface density and diffusion can then be calculated for a calibrated spot size (Eq. 2). Equation 5 indicates that multiple diffusing species may in principle be resolved. In practice, however, the autocorrelation function of a mixed population of two species with diffusion coefficient within an order of magnitude, as is the case for Ras monomers and dimers, cannot be reliably resolved due to the relatively featureless and gently sloping lineshape of the autocorrelation function (see the Supporting Material). Thus, only the weighted average using single-species model, Eq. 1, is obtained in these experiments. Calibration experiments using the LeuZ dimer reveal that the mobility of monomer and dimer in this experimental system differ by a factor of 2.0 (see the Supporting Material). FCS is particularly advantageous for detection of dimers, even for those with extremely weak affinities, because the presence of a small fraction of brighter dimer contributes nonlinearly to the composite FCS data (see the Supporting Material). This is evident in Eq. 5, where G(τ) scales with the square of Bi. Therefore, even though the individual contributions from monomers and dimers to the FCS measurements cannot be separated, the measured average is extremely sensitive to a minute fraction of dimers. An additional advantage afforded by FCS is that the surface protein density is directly measured simultaneously with diffusion. As a large range of surface density needed to be examined due to the unknown dimerization affinity, this was particularly expedient.
In SMT, on the other hand, the trajectories of membrane-bound fluorescent molecules at single-molecule level are directly imaged (Fig. 1 D, top). The diffusion coefficients can be extracted by fitting the measured step-size distribution to the 2D Brownian diffusion model (Fig. 1 D, bottom). As a single-molecule technique, SMT can resolve two diffusing species with closer diffusion rates compared to FCS, provided that the camera has sufficient time resolution (20 ms in these studies). Additionally, SMT offers the possibility of detecting the timescale of transitions (e.g., between monomer and dimer) as well as the spectrum of different species present on the membrane. In combination, FCS and SMT provide a means of internal self-consistency checking (11).
Dimerization of Ras on the membrane surface is an essentially 2D reaction in which the fraction of dimers at equilibrium is a function of the overall Ras surface density. The 2D dissociation constant, Kd = [Ras]22D/[Ras⋅Ras]2D, is defined in terms of the equilibrium surface densities of Ras monomer, [Ras]2D, and Ras dimer, [Ras⋅Ras]2D. It is important to note that this 2D dissociation constant generally cannot be calculated from the corresponding 3D affinity. In the case of Ras as studied here, there are four factors that contribute to the actual 2D Kd: 1) translational entropy, referring to the reduction in spatial dimension from three to two, which is also sometimes called a concentration effect (this one can be easily calculated and generally favors dimerization); 2) rotational entropy, referring to the change in rotational degrees of freedom resulting from membrane anchorage (this may be estimated using protein atomic structure data (11, 39), but is complex and can be either positive or negative); 3) structural change, such as those that may result from sitting on the membrane surface (40, 41, 42); and 4) lipid anchor-mediated interactions (42, 43), in which attractive interactions are mediated through the solvation of the Ras membrane anchor by the surrounding membrane lipids.
As a general strategy in these experiments, we sought to map the 2D Ras-Ras binding curve as a function of Ras density to determine the 2D Kd. Example binding curves for a 2D binding reaction for a variety of 2D Kd values are plotted in Fig. 2 A. To provide a rough calibration relating to 3D affinities, if we consider only the translational entropy (concentration) effect, Kd values of 200 and 1.25 × 105 μm−2 are equivalent to 3D dissociation constants of 0.1 and 70 mM, respectively (see the Supporting Material for details). Note that a Ras surface density of 1.25 × 105 μm−2 corresponds to essentially dense packed on the membrane surface (44), so 2D Kd values in this range correspond to effectively no binding affinity. The density range accessible by FCS is determined by the autocorrelation signal-to-noise ratio, and spans from ∼10 to 1500 molecules/μm2 in this system (corresponding to 2–190 particles per near diffraction-limited focal radius of 200 nm).
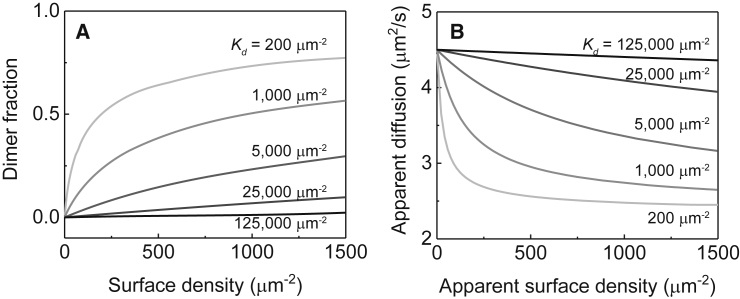
Figure 2.
Theoretical dynamic range for FCS in dimerization detection. (A) Fraction of dimers as a function of surface density for a series of 2D dissociation constants Kd are shown. (B) Calculated surface density-titration experiment from conditions given in (A), using experimentally determined diffusion coefficients for monomers and dimers, showing that even weak dimerization up to Kd = 25,000 μm−2 should produce a detectable change in diffusion.
FCS measurements reveal a weighted average of the mobility in a system containing a mixture of monomers and dimers. Calculations of the weighted averaged values of D, as would be reported by FCS (following Eq. 5 and based on the LeuZ dimer calibration experiment, detailed in the Supporting Material) corresponding to the 2D dimer binding curves in Fig. 2 A are plotted in Fig. 2 B. For this calculation, experimentally estimated values for the monomer and dimer diffusion coefficients (4.5 and 2.3 μm2/s) and presumed brightness ratio between dimer and monomer of 2 were used. In this system, FCS provides an excellent dynamic range to detect from strong to exceptionally weak dimerization.
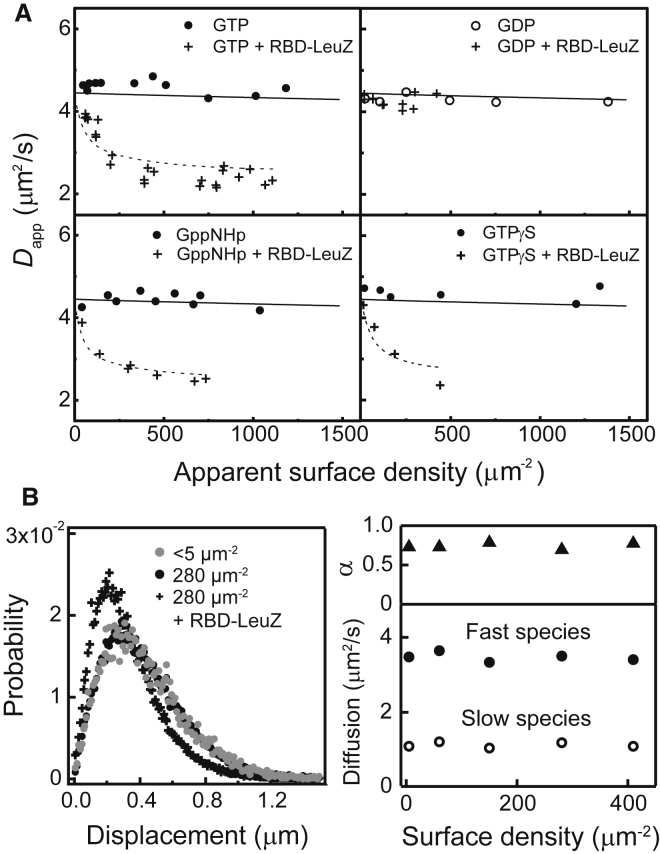
A series of density-dependent diffusion measurements of Ras by FCS (eGFP-labeled) on supported lipid bilayers are plotted alongside the LeuZ dimer calibration in Fig. 3 A. In these measurements, the supported membrane consisted of 20% PS, 80% DOPC, and a trace amount of Texas Red lipid probe, used as a benchmark standard to characterize the membrane integrity. PS is the major anionic lipid species found in the plasma membrane with the reported densities ranging between 10 and 20%. PS interacts directly with the hypervariable region of K-Ras4B and participates in stabilizing membrane association via electrostatic interactions (26). GDP, GTP, and nonhydrolyzable GTP analogs GppNHp and GTPγS were tested to see if dimerization is dependent on the activation state Ras, as has been previously reported (22). To ensure that Ras was in the desired nucleotide state, the catalytic domain of guanosine exchange factor Son of Sevenless (SOScat) was included with excess nucleotide in solution. The FCS measurements indicate that for Ras alone, its diffusion remained independent of the surface density of 1500 μm−2 at ∼4.5 μm2/s. The solid lines through these data represent calculated FCS for Kd = 1.25 × 105 μm−2, corresponding to no effective dimerization affinity. In contrast, GTP-, GppNHp-, and GTPγS-bound Ras shows a substantial decrease in diffusion in the presence of 0.5 μM RBD-LeuZ cross linker; the dotted lines represent calculated FCS data for fitted apparent Kd values, which ranged 300–400 μm−2. GDP-bound Ras is density-independent even with RBD-LeuZ, as it is incapable of interacting with RBD.
Figure 3.
Density titration diffusion measurement on PS bilayers. (A) FCS measurements of eGFP-K-Ras4B in excess of various guanosine nucleotides were titrated on SLBs containing 20% PS bilayers, with and without 0.5 μM RBD-LeuZ cross linker. (Solid lines) Shown here is the diffusion change if the dimerization occurs with Kd = 125,000 μm−2, which is essentially a dense-packed concentration. (Dotted lines) Shown here are fits for cross-linked Ras, with Kd values ranging between 300 and 400 μm−2. (B) (Left) Given here is the SMT step-size distribution for low- and high-density Ras, as well as high-density Ras with RBD-LeuZ. (Right) Given here are density-dependent diffusion coefficients D and fraction in faster species α for Ras without RBD-LeuZ. Both α and D of the two diffusing species are density-invariant, indicating that Ras dimerization does not occur under these circumstances.
The single-particle tracking results likewise illustrate that Ras by itself is monomeric on membrane surfaces. The step size distribution for Ras with and without RBD-LeuZ is plotted in Fig. 3 B (left). Ras shows essentially identical diffusion at low (<1 μm−2) and high (∼280 μm−2) surface densities. However, detailed analysis of the step size distribution, upon fitting to 2D Brownian diffusion, reveals the existence of two species of Ras on the membrane with differing mobilities: a major fast species with a diffusion coefficient of ∼3.5 μm2/s and minor slow species with ∼1 μm2/s (Fig. 3 B, right; also Fig. S8). Also, existence of the slow diffusing species is not due to dimerization. Even at extremely low total Ras surface density (<1 μm−2), where even high affinity dimers would not form, we observe both species in the mobility. In this extreme case, individual Ras molecules are several microns apart from each other, so they cannot interact to form dimers. Furthermore, the relative population for each diffusing state is density-invariant (as shown in Fig. 3 B, right). Based on these observations, we conclude that the two observed species likely reflect two different modes of protein-lipid interactions on the membrane surface, possibly with different numbers of anionic lipids making direct electrostatic contacts with the hypervariable region (9, 45, 46, 47). If these additional interactions reorient the protein and enhance the degree of immersion of the protein to the membrane, they may substantially slow down the diffusion, as observed. In the presence of RBD-LeuZ, Ras forms dimers and the mobility is significantly reduced, as is evident in the step size distribution (Fig. 3 B, left). The differences in the measured diffusion constants between the SMT and FCS are systematic and determined by accuracy of instrumental calibration, but both are within the range of values reported for comparable systems (11, 36, 48).
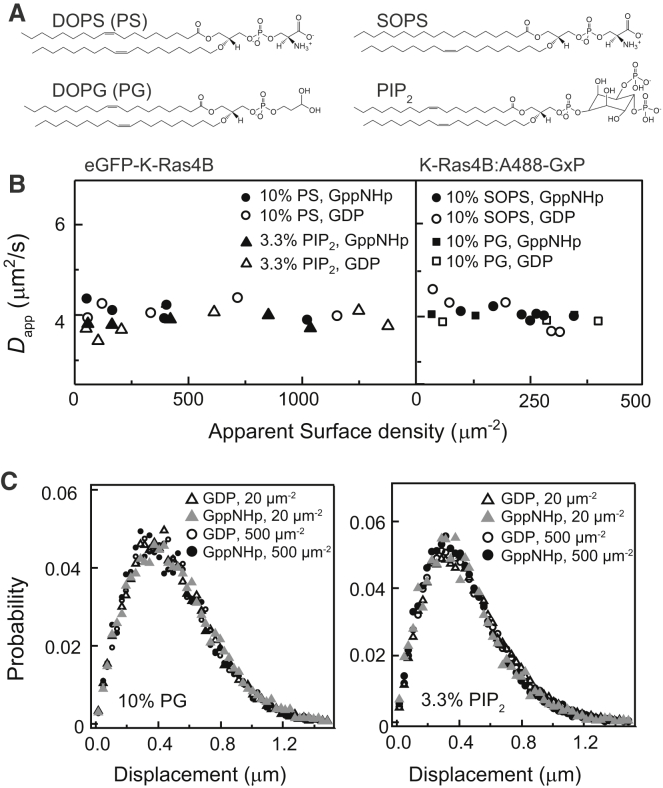
The diffusion behavior of Ras was also observed to be strictly monomeric in the presence of various types of lipids. The plasma membrane is a complex mixture of hundreds of phospholipid species as well as other components, and there have been reports suggesting that phospholipids with particular headgroups or chain chemistry may induce specific lateral organization of Ras. For example, it has been suggested that electrostatic lipid-protein interactions modulate orientation preference of K-Ras through charged membrane interaction surfaces in the G-domain, shifting the equilibrium toward specific dimer contacts (9, 14, 45); and asymmetric hydrocarbon chains corral Ras into clusters (49). We used DOPS (18:1-18:1), SOPS (18:0-18:1), DOPG, and PIP2, whose chemical structures are shown in Fig. 4 A, to see if any of these produced Ras dimers. For the SLBs containing these lipids, the compositions were adjusted such that the overall nominal charge is −10%. The lack of detectable change in diffusion as a function of Ras density by FCS measurements (Fig. 4 B) demonstrates that Ras is monomeric within the accessible density range on these membranes, regardless of the various headgroups, chain chemistry, or the nucleotide state. For PIP2, it has been reported that divalent cations such as Ca2+ can induce clustering of the lipids (50, 51). However, in our system, the presence of 1 mM Ca2+ had no effect on the diffusion of Ras. The SMT step size distributions were also density-invariant for DOPG and PIP2 bilayers when Ras is bound to GDP and GppNHp (Fig. 4 C).
Figure 4.
Effect of various lipids. (A) Chemical structures of anionic lipids used are shown. DOPS, SOPS, and DOPG have −1 nominal charge, and PIP2 has −3 nominal charge. SOPS has asymmetric chains (18:0-18:1), whereas the others have symmetric chains (18:1-18:1). (B) FCS measurements of eGFP-K-Ras (left) and wild-type K-Ras (right) on SLBs containing these lipids (charge normalized to −10%) display density-independent diffusion, indicating a monomeric behavior. (C) SMT step size distributions are identical for high- and low-density K-Ras for DOPG or PIP2 bilayers, regardless of the nucleotide states.
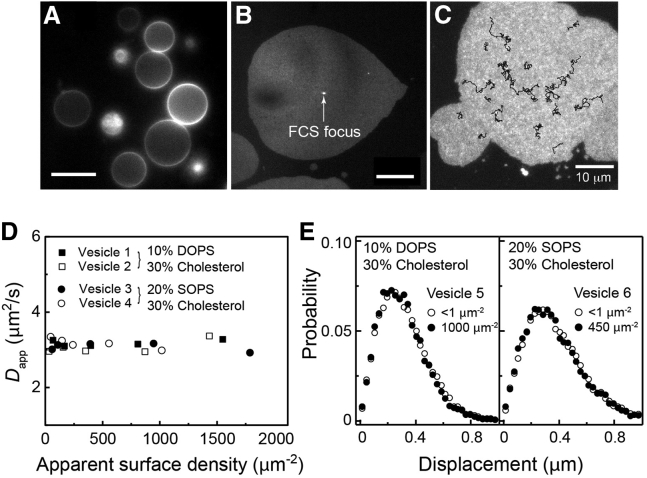
Finally, cholesterol was introduced to the lipid membranes, as it has been reported that cholesterol may facilitate segregation Ras in the plasma membrane (8, 49). Because it is difficult to make supported bilayers with cholesterol using the SUV rupture-fuse method, we instead used ruptured GUVs to create a planar patch of fluid bilayers (35, 52, 53, 54). Fig. 5 A shows a typical batch of intact GUVs composed of unsaturated lipids (DOPC), saturated lipids (DPPC), charged lipids (DOPS or SOPS), and cholesterol, and a trace amount of TR-DHPE for imaging. As the nominal demixing temperature for this composition is 19°C, they showed single macroscopic phase at room temperature (30). Fig. 5, B and C, displays ruptured GUVs displaying the FCS focus and SMT trajectories in the identical experimental setup as the SLB cases. Diffusion measurements once again indicate that even with cholesterol, there is no density-dependent diffusion change in either FCS (Fig. 5 D) or SMT (Fig. 5 E). Thus Ras remains monomeric at all densities in the cholesterol-containing membrane.
Figure 5.
Effect of cholesterol. (A) Intact GUVs were ruptured onto a glass surface to create patches of bilayers containing cholesterol for FCS (B, focus spot shown) and SMT (C, particle trajectories shown) diffusion measurements. Scale bar: 10 μm. (D) Shown here are FCS measurements for (B). (E) Shown here are SMT measurements for (C). K-Ras diffusion is density-independent for all cases.
Conclusion
In summary, we have shown that in supported lipid bilayers, K-Ras4B by itself remains monomeric over a wide range of surface densities in multiple types of membrane compositions. Careful diffusion measurements by FCS and SMT show that Ras has a density-independent diffusion with the value comparable to the lipids up to surface densities of nearly dense-packed. This means that the clusters and assemblies of Ras observed in live cell contexts are probably driven by interactions with other proteins, such as scaffolding proteins or downstream effectors. Another factor that may influence the lateral organization of Ras, but has not been experimentally examined here, is the topological membrane fluctuations. The supported lipid bilayers are held in a planar configuration by the glass support, and large membrane undulations are damped (55). However, it is not clear whether such large undulations would occur in cells, where they are expected to be damped by the cortical cytoskeleton.
Author Contributions
J.K.C., Y.K.L., and J.T.G. designed research and wrote the paper; J.K.C. and Y.K.L performed research and analyzed data; J.-P.D., W.K.G., S.A., and A.G.S. contributed new reagents.
Acknowledgments
We thank Jennifer Mehalko, Cammi Bittner, Zhaojing Meng, and Cheryl Habrukowich for technical support in the preparation and characterization of the K-Ras proteins, and Dr. Scott D. Hansen, Dr. Luke M. Oltrogge, and Dr. William Y.C. Huang for critical reading of the manuscript.
Primary support for this project was provided by the National Cancer Institute (NCI), National Institutes of Health (NIH), under contract U01CA202241 to J.T.G. Additional support to A.G.S. was provided by the National Cancer Institute (NCI)), National Institutes of Health (NIH) under contract HHSN261200800001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Editor: Arne Gericke.
Footnotes
Jean K. Chung and Young Kwang Lee contributed equally to this work.
Supporting Materials and Methods and nine figures are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(17)31205-5.
Supporting Material
References
- 1.Bollag G., McCormick F. Regulators and effectors of Ras proteins. Annu. Rev. Cell Biol. 1991;7:601–632. doi: 10.1146/annurev.cb.07.110191.003125. [DOI] [PubMed] [Google Scholar]
- 2.Plowman S.J., Muncke C., Hancock J.F. H-Ras, K-Ras, and inner plasma membrane raft proteins operate in nanoclusters with differential dependence on the actin cytoskeleton. Proc. Natl. Acad. Sci. USA. 2005;102:15500–15505. doi: 10.1073/pnas.0504114102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Prior I.A., Hancock J.F. Ras trafficking, localization and compartmentalized signalling. Semin. Cell Dev. Biol. 2012;23:145–153. doi: 10.1016/j.semcdb.2011.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Santos E. Dimerization opens new avenues into Ras signaling research. Sci. Signal. 2014;7:pe12. doi: 10.1126/scisignal.2005318. [DOI] [PubMed] [Google Scholar]
- 5.Chen M., Peters A., Nan X. Ras dimer formation as a new signaling mechanism and potential cancer therapeutic target. Mini Rev. Med. Chem. 2016;16:391–403. doi: 10.2174/1389557515666151001152212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stephen A.G., Esposito D., McCormick F. Dragging Ras back in the ring. Cancer Cell. 2014;25:272–281. doi: 10.1016/j.ccr.2014.02.017. [DOI] [PubMed] [Google Scholar]
- 7.Spencer-Smith R., Koide A., O’Bryan J.P. Inhibition of RAS function through targeting an allosteric regulatory site. Nat. Chem. Biol. 2017;13:62–68. doi: 10.1038/nchembio.2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Janosi L., Li Z., Gorfe A.A. Organization, dynamics, and segregation of Ras nanoclusters in membrane domains. Proc. Natl. Acad. Sci. USA. 2012;109:8097–8102. doi: 10.1073/pnas.1200773109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sayyed-Ahmad A., Cho K.J., Gorfe A.A. Computational equilibrium thermodynamic and kinetic analysis of K-Ras dimerization through an effector binding surface suggests limited functional role. J. Phys. Chem. B. 2016;120:8547–8556. doi: 10.1021/acs.jpcb.6b02403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Güldenhaupt J., Rudack T., Gerwert K. N-Ras forms dimers at POPC membranes. Biophys. J. 2012;103:1585–1593. doi: 10.1016/j.bpj.2012.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lin W.C., Iversen L., Groves J.T. H-Ras forms dimers on membrane surfaces via a protein-protein interface. Proc. Natl. Acad. Sci. USA. 2014;111:2996–3001. doi: 10.1073/pnas.1321155111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Abankwa D., Gorfe A.A., Hancock J.F. Ras membrane orientation and nanodomain localization generate isoform diversity. Proc. Natl. Acad. Sci. USA. 2010;107:1130–1135. doi: 10.1073/pnas.0903907107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cho K.J., Hancock J.F. Ras nanoclusters: a new drug target? Small GTPases. 2013;4:57–60. doi: 10.4161/sgtp.23145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Prakash P., Zhou Y., Gorfe A.A. Oncogenic K-Ras binds to an anionic membrane in two distinct orientations: a molecular dynamics analysis. Biophys. J. 2016;110:1125–1138. doi: 10.1016/j.bpj.2016.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou Y., Wong C.O., Hancock J.F. SIGNAL TRANSDUCTION. Membrane potential modulates plasma membrane phospholipid dynamics and K-Ras signaling. Science. 2015;349:873–876. doi: 10.1126/science.aaa5619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hancock J.F. Ras proteins: different signals from different locations. Nat. Rev. Mol. Cell Biol. 2003;4:373–384. doi: 10.1038/nrm1105. [DOI] [PubMed] [Google Scholar]
- 17.Nan X., Tamgüney T.M., Chu S. Ras-GTP dimers activate the mitogen-activated protein kinase (MAPK) pathway. Proc. Natl. Acad. Sci. USA. 2015;112:7996–8001. doi: 10.1073/pnas.1509123112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Šolman M., Ligabue A., Abankwa D. Specific cancer-associated mutations in the switch III region of Ras increase tumorigenicity by nanocluster augmentation. ELife. 2015;4:e08905. doi: 10.7554/eLife.08905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blazevits O., Mideksa Y.G., Abankwa D. Galectin-1 dimers can scaffold Raf-effectors to increase H-Ras nanoclustering. Sci. Rep. 2016;6:24165. doi: 10.1038/srep24165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guzmán C., Šolman M., Abankwa D. The efficacy of Raf kinase recruitment to the GTPase H-Ras depends on H-Ras membrane conformer-specific nanoclustering. J. Biol. Chem. 2014;289:9519–9533. doi: 10.1074/jbc.M113.537001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chung J.K., Lee Y.K., Groves J.T. Covalent Ras dimerization on membrane surfaces through photosensitized oxidation. J. Am. Chem. Soc. 2016;138:1800–1803. doi: 10.1021/jacs.5b12648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Muratcioglu S., Chavan T.S., Nussinov R. GTP-dependent K-Ras dimerization. Structure. 2015;23:1325–1335. doi: 10.1016/j.str.2015.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kovrigina E.A., Galiakhmetov A.R., Kovrigin E.L. The Ras G domain lacks the intrinsic propensity to form dimers. Biophys. J. 2015;109:1000–1008. doi: 10.1016/j.bpj.2015.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pylayeva-Gupta Y., Grabocka E., Bar-Sagi D. RAS oncogenes: weaving a tumorigenic web. Nat. Rev. Cancer. 2011;11:761–774. doi: 10.1038/nrc3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gillette W.K., Esposito D., Stephen A.G. Farnesylated and methylated KRAS4b: high yield production of protein suitable for biophysical studies of prenylated protein-lipid interactions. Sci. Rep. 2015;5:15916. doi: 10.1038/srep15916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ahearn I.M., Haigis K., Philips M.R. Regulating the regulator: post-translational modification of Ras. Nat. Rev. Mol. Cell Biol. 2011;13:39–51. doi: 10.1038/nrm3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sondermann H., Soisson S.M., Kuriyan J. Structural analysis of autoinhibition in the Ras activator Son of Sevenless. Cell. 2004;119:393–405. doi: 10.1016/j.cell.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 28.Lin W.C., Yu C.H., Groves J.T. Supported membrane formation, characterization, functionalization, and patterning for application in biological science and technology. Curr. Protoc. Chem. Biol. 2010;2:235–269. doi: 10.1002/9780470559277.ch100131. [DOI] [PubMed] [Google Scholar]
- 29.Schmid E.M., Richmond D.L., Fletcher D.A. Reconstitution of proteins on electroformed giant unilamellar vesicles. Methods Cell Biol. 2015;128:319–338. doi: 10.1016/bs.mcb.2015.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Veatch S.L., Keller S.L. Separation of liquid phases in giant vesicles of ternary mixtures of phospholipids and cholesterol. Biophys. J. 2003;85:3074–3083. doi: 10.1016/S0006-3495(03)74726-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Iversen L., Tu H.L., Groves J.T. Molecular kinetics. Ras activation by SOS: allosteric regulation by altered fluctuation dynamics. Science. 2014;345:50–54. doi: 10.1126/science.1250373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee Y.K., Low-Nam S.T., Groves J.T. Mechanism of SOS PR-domain autoinhibition revealed by single-molecule assays on native protein from lysate. Nat. Commun. 2017;8:15061. doi: 10.1038/ncomms15061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lakowicz J.R. Springer; New York, NY: 2006. Principles of Fluorescence Spectroscopy. [Google Scholar]
- 34.Saffman P.G., Delbrück M. Brownian motion in biological membranes. Proc. Natl. Acad. Sci. USA. 1975;72:3111–3113. doi: 10.1073/pnas.72.8.3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kaizuka Y., Groves J.T. Structure and dynamics of supported intermembrane junctions. Biophys. J. 2004;86:905–912. doi: 10.1016/S0006-3495(04)74166-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Knight J.D., Lerner M.G., Falke J.J. Single molecule diffusion of membrane-bound proteins: window into lipid contacts and bilayer dynamics. Biophys. J. 2010;99:2879–2887. doi: 10.1016/j.bpj.2010.08.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ziemba B.P., Falke J.J. Lateral diffusion of peripheral membrane proteins on supported lipid bilayers is controlled by the additive frictional drags of (1) bound lipids and (2) protein domains penetrating into the bilayer hydrocarbon core. Chem. Phys. Lipids. 2013;172-173:67–77. doi: 10.1016/j.chemphyslip.2013.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ziemba B.P., Knight J.D., Falke J.J. Assembly of membrane-bound protein complexes: detection and analysis by single molecule diffusion. Biochemistry. 2012;51:1638–1647. doi: 10.1021/bi201743a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu Y., Vendome J., Honig B. Transforming binding affinities from three dimensions to two with application to cadherin clustering. Nature. 2011;475:510–513. doi: 10.1038/nature10183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kapoor S., Triola G., Winter R. Revealing conformational substates of lipidated N-Ras protein by pressure modulation. Proc. Natl. Acad. Sci. USA. 2012;109:460–465. doi: 10.1073/pnas.1110553109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gorfe A.A., Babakhani A., McCammon J.A. Free energy profile of H-Ras membrane anchor upon membrane insertion. Angew. Chem. Int. Ed. Engl. 2007;46:8234–8237. doi: 10.1002/anie.200702379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gorfe A.A., Babakhani A., McCammon J.A. H-Ras protein in a bilayer: interaction and structure perturbation. J. Am. Chem. Soc. 2007;129:12280–12286. doi: 10.1021/ja073949v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gorfe A.A., Hanzal-Bayer M., McCammon J.A. Structure and dynamics of the full-length lipid-modified H-Ras protein in a 1,2-dimyristoylglycero-3-phosphocholine bilayer. J. Med. Chem. 2007;50:674–684. doi: 10.1021/jm061053f. [DOI] [PubMed] [Google Scholar]
- 44.Pai E.F., Kabsch W., Wittinghofer A. Structure of the guanine-nucleotide-binding domain of the Ha-Ras oncogene product p21 in the triphosphate conformation. Nature. 1989;341:209–214. doi: 10.1038/341209a0. [DOI] [PubMed] [Google Scholar]
- 45.Mazhab-Jafari M.T., Marshall C.B., Ikura M. Oncogenic and RASopathy-associated K-RAS mutations relieve membrane-dependent occlusion of the effector-binding site. Proc. Natl. Acad. Sci. USA. 2015;112:6625–6630. doi: 10.1073/pnas.1419895112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li Z.L., Buck M. Computational modeling reveals that signaling lipids modulate the orientation of K-Ras4A at the membrane reflecting protein topology. Structure. 2017;25:679–689.e2. doi: 10.1016/j.str.2017.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Janosi L., Gorfe A.A. Segregation of negatively charged phospholipids by the polycationic and farnesylated membrane anchor of K-Ras. Biophys. J. 2010;99:3666–3674. doi: 10.1016/j.bpj.2010.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ries J., Chiantia S., Schwille P. Accurate determination of membrane dynamics with line-scan FCS. Biophys. J. 2009;96:1999–2008. doi: 10.1016/j.bpj.2008.12.3888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhou Y., Prakash P., Hancock J.F. Lipid-sorting specificity encoded in K-Ras membrane anchor regulates signal output. Cell. 2017;168:239–251.e16. doi: 10.1016/j.cell.2016.11.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bilkova E., Pleskot R., Coskun Ü. Calcium directly regulates phosphatidylinositol 4,5-bisphosphate headgroup conformation and recognition. J. Am. Chem. Soc. 2017;139:4019–4024. doi: 10.1021/jacs.6b11760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Levental I., Christian D.A., Janmey P.A. Calcium-dependent lateral organization in phosphatidylinositol 4,5-bisphosphate (PIP2)- and cholesterol-containing monolayers. Biochemistry. 2009;48:8241–8248. doi: 10.1021/bi9007879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wong A.P., Groves J.T. Topographical imaging of an intermembrane junction by combined fluorescence interference and energy transfer microscopies. J. Am. Chem. Soc. 2001;123:12414–12415. doi: 10.1021/ja016677j. [DOI] [PubMed] [Google Scholar]
- 53.Parthasarathy R., Groves J.T. Protein patterns at lipid bilayer junctions. Proc. Natl. Acad. Sci. USA. 2004;101:12798–12803. doi: 10.1073/pnas.0402168101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Parthasarathy R., Yu C.H., Groves J.T. Curvature-modulated phase separation in lipid bilayer membranes. Langmuir. 2006;22:5095–5099. doi: 10.1021/la060390o. [DOI] [PubMed] [Google Scholar]
- 55.Kaizuka Y., Groves J.T. Hydrodynamic damping of membrane thermal fluctuations near surfaces imaged by fluorescence interference microscopy. Phys. Rev. Lett. 2006;96:118101. doi: 10.1103/PhysRevLett.96.118101. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.