Abstract
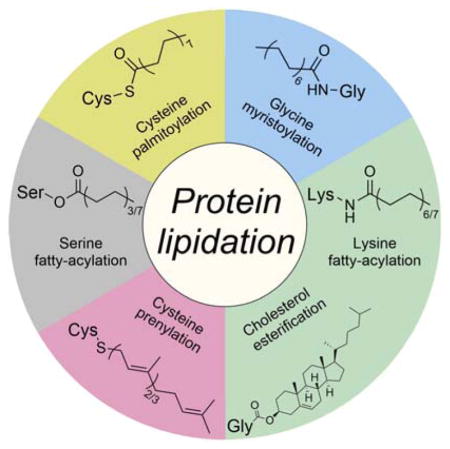
Protein lipidation, including cysteine prenylation, N-terminal glycine myristoylation, cysteine palmitoylation, and serine and lysine fatty acylation, occurs in many proteins in eukaryotic cells and regulates numerous biological pathways, such as membrane trafficking, protein secretion, signal transduction, and apoptosis. We provide a comprehensive review of protein lipidation, including descriptions of proteins known to be modified and the functions of the modifications, the enzymes that control them, and the tools and technologies developed to study them. We also highlight key questions about protein lipidation that remain to be answered, the challenges associated with answering such questions, and possible solutions to overcome these challenges.
Graphical Abstract
1. Introduction
Lipids are essential molecules that compose cellular membranes, which provide the barriers and boundaries needed for cells to survive and proliferate. This confinement of cellular materials by cellular membrane structures necessitates cellular communication (i.e., cell signaling and membrane trafficking) with the extracellular environment and among cellular membrane organelles. Cell signaling and membrane trafficking rely on proteins that are secreted into the environment, embedded in cellular membranes, and reversibly associated with membranes. Not surprisingly, nature also uses lipids to control and regulate membrane–protein interactions. These functions are achieved through two strategies. Certain proteins have evolved to bind specifically to certain lipid molecules. For example, some pleckstrin homology domains recognize specific phosphoinositides,1 and blood clotting factors recognize phosphatidylserine, which is found only in the inner leaflet of the plasma membrane.2 Another widely observed interaction strategy is the covalent modification of proteins by lipid molecules. These modifications are the focus of this review.
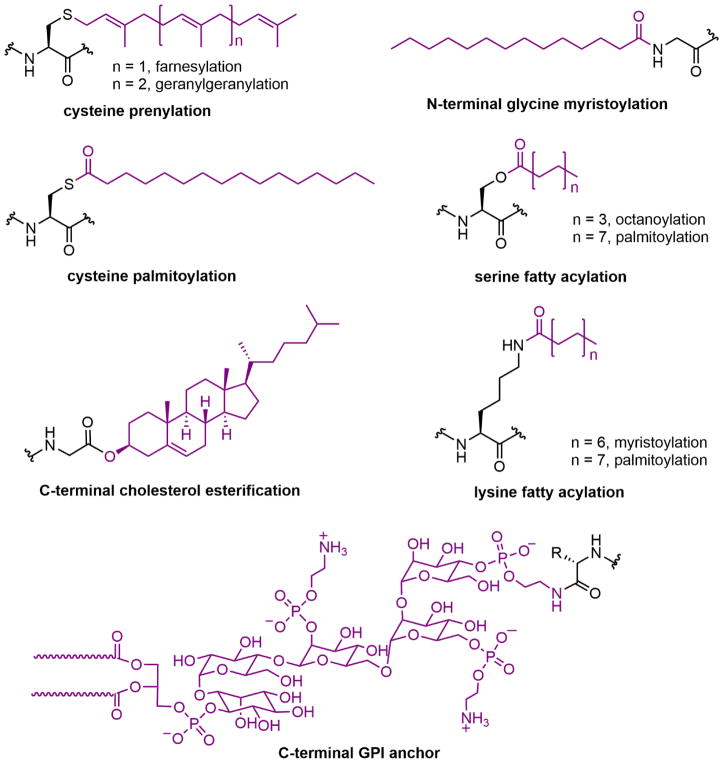
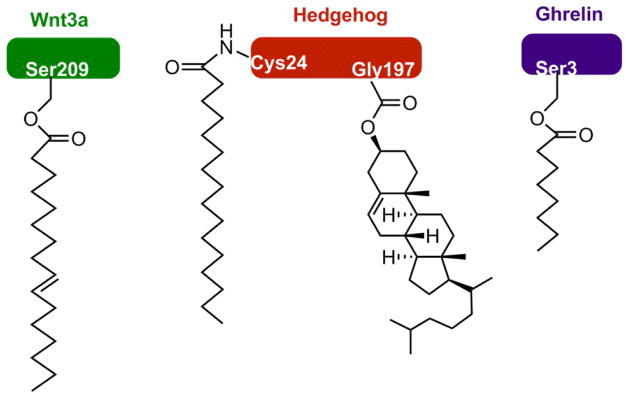
Lipidation occurs on numerous proteins and regulates many aspects of physiology. The effects of protein lipidation on cellular function are achieved by regulating protein–membrane interactions, and perhaps somewhat surprising, protein–protein interactions, protein stability, and enzymatic activities. The lipid moieties added to proteins can be either fatty acyl or polyisoprenyl groups, and the modifications typically occur on the nucleophilic side chains of proteins (e.g., cysteine, serine, and lysine) and the NH2 group at the N-termini of proteins (Figure 1). Two lipid modifications occur at the C-termini of certain extracellular-membrane-associated proteins: cholesterol esterification and glycosylphosphatidylinositol anchoring (see Figure 1). This review focuses on the direct modification of protein nucleophilic residues by lipid molecules. Glycosylphosphatidylinositol anchors, which are attached to proteins with a carbohydrate moiety via multiple enzymatic steps, are not discussed herein, but excellent books and reviews are available.3–5
Figure 1.
Lipid modifications of proteins. GPI, glycosylphosphatidylinositol.
The review is organized by the type of lipid modification that occurs on various nucleophilic groups. For each modification, we discuss the enzymes that control the modification, the modified proteins, the functions of the modification, and the tools or technologies that have been developed to study the modifications. Each section is independent; however, certain modifications, such as cysteine palmitoylation, depend on other modifications (cysteine prenylation or N-terminal glycine myristoylation). Therefore, the sections are ordered so that that the occurrence and functions of various modifications are easy to understand.
2. Protein Prenylation
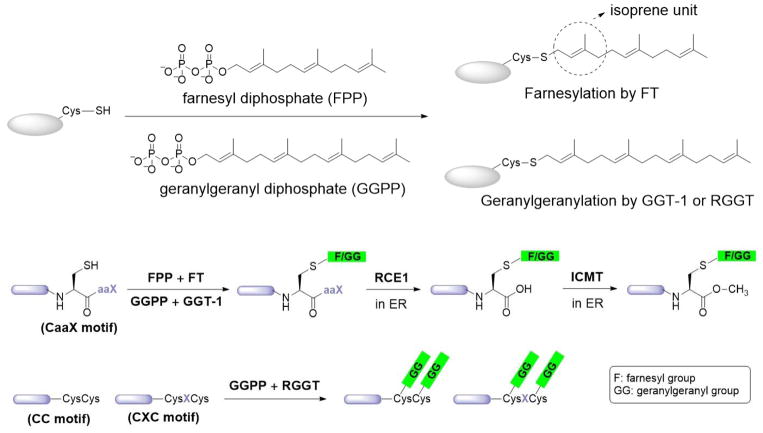
Prenylation is the addition of multiple isoprene units to cysteine residues near the C-termini of proteins. Up to 2% of the total cellular proteins in mammalian cells are prenylated.6 There are two types of prenylation—farnesylation and geranylgeranylation—which involve three and four isoprene units, respectively (Figure 2). The processes through which these modifications take place are also referred to in the literature as isoprenylation or polyisoprenylation. Technically, the most appropriate description is polyisoprenylation, but the simpler term prenylation is more popular and is therefore adopted here. The majority of prenylated proteins are geranylgeranylated proteins.6 The linkage between farnesyl or geranylgeranyl groups and cysteine residues is a thioether bond, which is more stable than ester and thioester bonds. The general belief is that this modification is irreversible, and no enzyme that reverses this modification in intact proteins has been identified. However, a prenylcysteine lyase is thought to be present in lysosomes7,8 and cleave the thioether bond of prenylcysteines in the degradation of prenylated proteins.
Figure 2.
Protein prenylation.
In 1989, several studies reported that Ras proteins and lamin B are farnesylated at cysteine residues.9,10 These studies showed that farnesylation occurs on a C-terminal CaaX sequence motif (C: cysteine, a: an aliphatic amino acid, X: any amino acid), which provided the initial paradigm with which to predict whether a protein will be prenylated. Soon thereafter, protein geranylgeranylation was discovered in HeLa cells and Chinese hamster ovary cells.11,12 Later, the C-terminal aaX was reported to be further cleaved by an endoplasmic reticulum (ER) protease, Ras-converting enzyme 1, or a-factor converting enzyme 1 after prenylation in the cytoplasm.13 The prenylated cysteine residue is then carboxylmethylated by another ER enzyme, isoprenylcysteine carboxylmethyltransferase (ICMT; see Figure 2).14
2.1. Protein Prenyltransferases
Three members of the protein prenyltransferase family are present in eukaryotes. Farnesyl transferase (FT) transfers the 15-carbon farnesyl group from farnesyl diphosphate (FPP) to substrate proteins. Geranylgeranyl transferase (GGT-1) catalyzes a similar reaction comprising the transfer of a 20-carbon geranylgeranyl group from geranylgeranyl diphosphate (GGPP). The substrate proteins of both FT and GGT-1 have typical C-terminal CaaX motifs for prenylation. Another protein prenyltransferase, Rab geranylgeranyl transferase (RGGT or GGT-2; see Figure 2), usually transfers two geranylgeranyl groups from GGPP to the C-terminal double-cysteine motif (CC or CXC) of Rab proteins.
2.1.1. FT and GGT-1
The first protein FT was isolated from rat brain in 1990.15 FPP, generated from mevalonate as an intermediate in the cholesterol biosynthetic pathway, was later shown to be the co-substrate of FT for p21Ras modification in vitro. Protein GGT-1 was also first identified from rat brain tissue as a modifier of Ras proteins.16 This study showed that GGT-1 has distinct selectivity for substrate proteins with C-terminal CaaL motifs rather than those with CaaM or CaaS motifs, which are preferred by FT. The authors also revealed that both FT and GGT-1 are heterodimers sharing a common α subunit with different β subunits. Further studies with recombinant rat FT and GGT-1 confirmed that the enzymes have the same α subunit of 48 kD and homologous β subunits of 46 kD and 43 kD, respectively.17–19
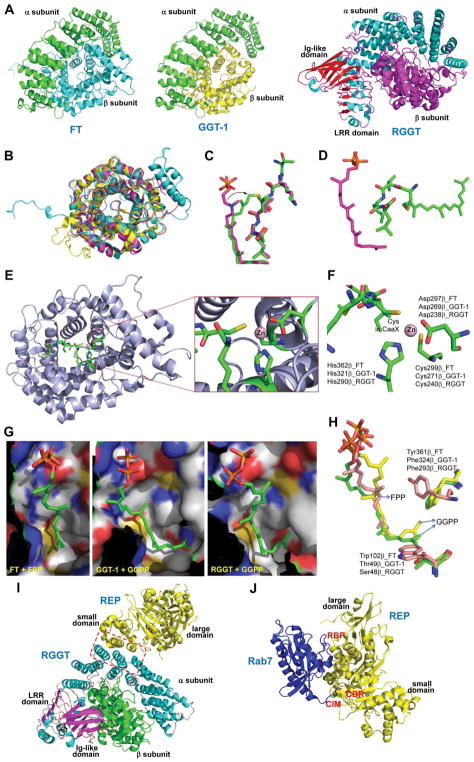
Crystal structures of rat FT and GGT-1 were solved in 1997 and 2003, respectively (Figure 3A)20,21 and showed that the major secondary structures of the α and β subunits are α-helices. In the α subunit, 14 of 15 α-helices are folded into seven successive helical hairpins and arranged in a double-layer super helix as a crescent-shaped domain that wraps around a portion of the β subunits. The β subunits of FT and GGT-1 share 25% sequence identity and have similar overall structures (Figure 3B) consisting of 14 and 13 α-helices, respectively. Twelve α-helices of the β subunits are folded into an unusual α-α barrel. Six parallel helices form the core of the barrel, and the other six form the outside of barrel, which is antiparallel to the inner core helices. One end of the barrel is blocked by the C-terminal loop of the β subunits, and the other end is open to the solvent and forms a deep hydrophobic pocket in the center of the barrel. This pocket has conserved aromatic residues that bind hydrophobic isoprene units of FPP and GGPP (Figure 3G).
Figure 3.
(A) Protein structures of FT (PDB ID 1FT1), GTT-1 (PDB ID 1N4P), and RGGT (PDB ID 1DCE). The α subunits (green) of FT and GTT-1 are identical. There are extra leucine-rich repeats (LRRs) and immunoglobulin (Ig)-like domains in the α subunit of RGGT (α-helices in cyan and β-sheets in red). (B) Superimposition of the β subunits of FT (cyan), GGT-1 (yellow), and RGGT (magenta) to show the structural homology. (C) The binding of substrates versus product in GGT-1. Geranylgeranyl diphosphate (GGPP; indicated by a GGPP analogue in magenta) rotates toward the cysteine in the CaaX peptide (PDB ID 1N4Q) to form the prenylated product (green; PDB ID 1N4R). (D) Simultaneous binding of GGPP (magenta) and the translocated prenylated product (green) at the active site of GGT-1 (PDB ID 1N4S). (E) The zinc binding site in the β subunit of FT (PDB ID 1D8D). (F) In FT, GGT-1, and RGGT, conserved residues in the β subunits of prenyltransferases bind to zinc, including an aspartate residue (Asp297β, Asp269β, and Asp238β, respectively), a cysteine residue (Cys299β, Cys271β, and Cys240β, respectively), and a histidine residue (His362β, His321β, and His290β, respectively). The zinc also coordinates with the cysteine residue of CaaX peptides. (G) Binding position of isoprenoid diphosphate in prenyltransferases, including FPP in FT (PDB ID 1FT2) and GGPP in GGT-1 (PDB ID 1N4P) and RGGT (PDB ID 3DST). (H) Comparison of isoprenoid diphosphate binding in FT (PDB ID 1FT2), GGT-I (PDB ID 1N4P), and RGGT (PDB ID 3DST). FPP (pink) with Trp102β and Tyr361β (pink) in FT, GGPP (green) with Thr49β and Phe324β (green) in GGT-1, and GGPP (yellow) with Ser48β and Phe293β (yellow) in RGGT. In FT, the bulky Trp102β residue occupies the space in which the fourth isoprene unit of GGPP binds in GGT-1 and RGGT. This residue determines the isoprenoid specificity. (I) Protein structure of the RGGT-REP-1 complex (PDB ID 1LTX). REP-1 is yellow. (J) Protein structure of the prenylated Rab7-REP-1 complex (PDB ID 1VG0). REP-1 is yellow and Rab7 is blue. All protein structures were made using PyMol with the PDB files.
The structures also reveal the location of the Zn2+ required for the enzymatic activities of FT and GGT-1.22,23 One zinc ion binds to the β subunit near the subunit interface (Figure 3E) and is coordinated by three conserved residues of the β subunit, Asp297β/Cys299β/His362β in FT and Asp269β/Cys271β/His321β in GGT-1 (Figure 3F).20,21 Ternary complex structures of FT or GGT-1 with peptide substrates and FPP or GGPP analogues show that the zinc ion is also coordinated with the cysteine thiol group in the C-terminal CaaX motif of the peptide substrates (Figure 3F),21,24 which is essential for the binding of CaaX peptides.
How do FT and GGT-1 achieve selectivity for FPP or GGPP? Binary complexes of FT with FPP and GGT-1 with GGPP provide clues about the mechanism for lipid length differentiation (Figure 3G).21,25 The diphosphate portion binds to a positively charged region at the top of the hydrophobic pocket near the subunit interface. The farnesyl portion of FPP binds in an extended conformation along one side of the hydrophobic pocket of the α-α barrel in the FT β subunit. The first three isoprene units of GGPP bind in a similar conformation within the GGT-1 β subunit, but the fourth isoprene unit is turned ~90° relative to the rest of the molecule. This positioning of the fourth isoprene unit indicates that Thr49β in GGT-1 is critical for lipid length discrimination because the corresponding position in FT is a bulky residue, Trp102β (Figure 3H). Phe324β in GGT-1 is also positioned near the fourth isoprene unit, whereas the corresponding residue in FT is Tyr361β. The hydroxyl group from Tyr361β might also help discriminate against GGPP in FT. Thus, steric hindrance in FT determines its preferential binding to FPP. A single mutation in FT, Trp102Thr, switches the co-substrate preference.21
The structures of GGT-1 in complex with the prenylated product reveal that GGPP rotates around the second isoprene unit to approach the thiol group of the cysteine in the CaaX peptide to generate the geranylgeranylated product while the other portion of isoprenoid retains its substrate binding position (Figure 3C). Product release from the GGT-1 active site requires the binding of fresh GGPP to displace the geranylgeranyl-peptide product (Figure 3D).21 The binding affinity of FPP for GGT-1 is much weaker and thus, FPP cannot efficiently displace the complex of GGT-1 and the geranylgeranylated product. This feature contributes to the isoprenoid substrate selectivity of GGT-1 for GGPP over FPP. However, RhoB is reportedly farnesylated and geranylgeranylated efficiently by GGT-1,26 which indicates that GGT-1 has the capability to transfer both farnesyl and geranylgeranyl groups, and the choice of prenylation may depend on the nature of the substrate proteins and relative concentrations of FPP and GGPP. The FPP and GGPP concentrations measured are similar in several human cancer cell lines (about 0.1 pmol/106 cells in K562 cells and 2.0 pmol/106 cells in MCF-7 cells).27 Notably, treating the cancer cells with a small molecule, zoledronic acid, dramatically increases the FPP concentration with minimal effects on GGPP concentration.27 The levels of FPP (0.9–3.7 ng/mg protein) and GGPP (3.7–27.8 ng/mg protein) in human brain tissue have also been determined and showed a significantly higher concentration of GGPP.28,29 Thus, certain conditions or biological environments may affect the ratio of farnesylation to geranylgeranylation.
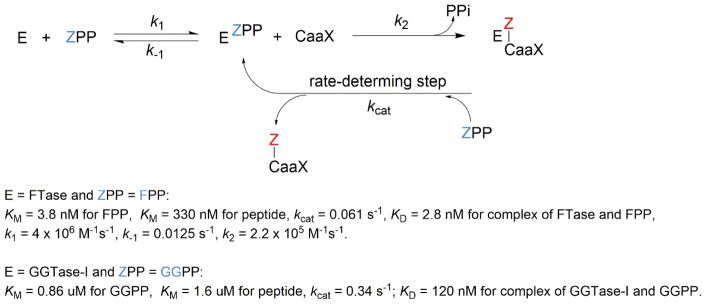
Based on kinetic studies15,30–34 and structures of FT in complex with substrates (FPP or its analogue and K-Ras4B C-terminal peptide) or products,20,24,25,35 an ordered sequential kinetic mechanism of farnesylation has been proposed (Figure 4). At the start of the reaction, a binary enzyme–substrate complex forms when FPP binds to the FT β subunit. Then, a ternary complex forms with the binding of the CaaX substrate. At the completion of the reaction, the farnesylated product remains in the active site until a new FPP displaces it; this step is the rate-limiting step.32,36,37 The resulting binary FT-FPP complex then enters the next round of the reaction. Geranylgeranylation catalyzed by GGT-1 is thought to follow the same reaction pathway, but detailed rate constants have not been reported.38 The results of a number of mechanistic studies that include stereochemical data and kinetic isotope effects data suggest that the transition states of FT- and GGT-catalyzed reactions have associative characteristics involving both the thiolate nucleophile and the diphosphate leaving group.39–42
Figure 4.
General reaction scheme with an ordered sequential kinetic mechanism for prenylation catalyzed by FT and GGT-1. The kinetics data for farnesylation and geranylgeranylation are from reference 22a and 23, respectively.
2.1.2. RGGT
RGGT (also called GGT-2) transfers two geranylgeranyl groups from GGPP to the C-terminal CC or CXC motifs in Rab proteins. RGGT has two subunits, a 60 kD α subunit and a 38 kD β subunit.43 Studies have shown that RGGT requires Rab escort proteins (REPs) to recruit substrate proteins for the geranylgeranylation reaction.43–45 Unlike FT and GGT-1, RGGT cannot catalyze reactions with short peptides containing a Rab C-terminal prenylation motif or recognize Rab proteins alone. Mammals have two REP proteins, REP-1 and REP-2. REP-1 is encoded on the X chromosome, and REP-1 mutations cause X-linked retinal degeneration (choroideremia). The substrate specificities of the REP proteins are essentially unknown, but Rab27a, a protein that accumulates in an unmodified form in choroideremia, cannot be efficiently modified with REP-2.46,47 Except in the retina, the presence of functional REP-2 largely compensates for the loss of REP-1 in choroideremia patients, which suggests that REP-1 and REP-2 have significantly overlapping functions. The first crystal structure of RGGT demonstrated that there are three domains in its α subunit (Figure 3A): a helical domain, an immunoglobulin (Ig)-like domain, and a leucine-rich repeat domain.48 RGGTα and FTα or GGT-1α have only 22% sequence identity according to structure-based alignment. The helical domain of RGGTα is structurally similar to the α subunit of FT and GGT-1 and forms a crescent-shaped super helix with 15 α-helices. The other domains, leucine-rich repeat domain and Ig-like domain, are unique in RGGTα, and their functions remain unknown.
Twelve α-helices in the β subunit of RGGT create an α-α barrel, which resembles the α-α barrels in FTβ and GTT-1β (Figure 3B). In the central pocket of the RGGT α-α barrel, Ser48β has the same functional role as Thr49β has in GGT-1 to accommodate GGPP, whereas Trp102β at the same position in FT prevents GGPP binding (Figure 3G and 3H).49
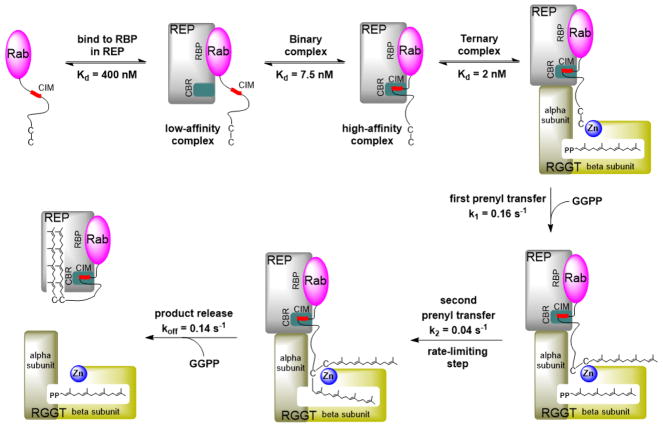
As shown by the structure of the RGGT–REP-1 complex (Figure 3I),50 REP-1 has two domains: a large domain consisting of four β–sheets and six α-helices, and a small domain with five α-helices. The interface between RGGT and REP-1 comprises two α-helices from the REP small domain and three α-helices from RGGTα. The interaction between RGGT and REP-1 is regulated by GGPP. Kinetics studies have demonstrated that REP-1 binds to RGGT with a Kd of 10 nM in the presence of GGPP,51 which is 100 times tighter than without GGPP.
The structure of monogeranylgeranylated Rab7 in complex with REP reveals that Rab7 binds to the Rab-binding platform (RBP) on the side of REP large domain, and the REP C-terminal binding region (CBR) associates with the Rab7 CBR-interacting motif (CIM) to form the binary complex (Figure 3J).47 Additional modeling experiments have shown that the prenylated C-terminus of Rab7 is harbored in the hydrophobic tunnel in the REP small domain to solubilize prenylated Rab7.47
Figure 5 shows the reaction pathway of Rab digeranylgeranylation by RGGT based on structural, computational, and biochemical studies.47,49–55 Rab and REP first form the binary complex, after which a high-affinity ternary complex of Rab-REP-RGGT is assembled via the interaction between the REP small domain and the RGGT α subunit. In this way, REP brings the Rab C-terminus to the active site of RGGT. Because RGGT does not bind its substrate peptide directly at the active site, the reaction is driven by concentration, and any cysteine presented by REP at the active site can be prenylated. This mechanism allows RGGT to modify more than 60 Rab proteins with unrelated C-terminal sequences. After the transfer of the first prenyl group from GGPP, a new GGPP molecule binds to the active site and displaces the substrate-conjugated isoprenoid. The mono-prenylated substrate is then conjugated with the second isoprenoid, and the resulting double-prenylated product is displaced by another new GGPP binding at the active site. The double-prenylated Rab C-terminus associates with the REP lipid-binding pocket and induces the conformational change in the REP small domain. Then REP dissociates from RGGT and translocates into the cell membrane.
Figure 5.
Reaction pathway of Rab digeranylgeranylation catalyzed by RGGT.
2.2. Protein Substrates of Prenyltransferases
Prenylation has been found only in eukaryotic cells, and most of the identified prenylated proteins are eukaryotic proteins. However, certain proteins from pathogenic bacteria can be prenylated by their hosts. Farnesylated proteins (substrates of FT) include Ras, Hdj2, nuclear lamins, and Rheb proteins.56 GGT-1 catalyzes the geranylgeranylation of Rac, RhoA, Cdc42, and the γ subunit of heterotrimeric G proteins.57 Most Rab proteins, with the exception of Rab8 and Rab13, are doubly geranylgeranylated by RGGT.58,59 Some proteins, such as K-Ras, N-Ras, and RhoB, are substrates of both FT and GGT-1.60,61 Prenylation Prediction Suite (http://mendel.imp.ac.at/PrePS/) is a Web-based tool that predicts whether a protein will be prenylated.
The originally discovered farnesylated and geranylgeranylated proteins provided the paradigm with which to identify protein substrates of prenylation. This paradigm is the C-terminal CaaX motif. Later studies with short peptides and FT or GGT-1 showed that a protein substrate is farnesylated by FT if the terminal “X” is serine, methionine, or glutamine, whereas the substrate is geranylgeranylated by GGT-1 if X is leucine.62,63 Later studies showed that this motif cannot fully describe the prenylated proteins or predict the prenylated substrates of FT or GGT-1.56,64
The screening of a CaaL peptide library for FT substrates revealed that FT can farnesylate a number of CaaL peptides,64 which is contrary to the CaaX paradigm describing CaaL as the canonical GGT-1 substrate sequence. Further screening with a large peptide library based on the human proteome identified two classes of FT substrates,56 one of which is farnesylated under multiple-turnover conditions and the other under single-turnover conditions. After the single-turnover substrate is modified by FT, the resulting product dissociates extremely slowly from the enzyme. Multiple-turnover substrates typically have CaaX sequences with phenylalanine, methionine, and glutamine at the X position, whereas the sequences of single-turnover substrates are more diverse. Computational techniques have also been applied to predict potential FT substrates65,66 and identified a novel substrate class with members that contain an acidic C-terminal residue (CaaD and CaaE).66 CVXX and CCXX peptide libraries were used to further probe the substrate specificity of rat FT and found several new sequences (e.g., CVIA, CVCS, CCIM, and CCVS) to be prenylation substrates.67 These studies demonstrate that FT can farnesylate a wide range of peptide substrates. Elucidating the physiological relevance of these findings will require additional research efforts to validate the protein substrates corresponding to these peptide substrates in vivo. Using a yeast-based screening system for FT, randomization of aaX residues in the CaaX sequence motif showed that the second “a” strongly prefers small hydrophobic residues, whereas the first a and X have relatively more relaxed specificities.68 This study further expanded the list of prenylated substrates.
Bacterial effector proteins with C-terminal CaaX motifs were also found to be prenylated by their host prenyltransferases. Salmonella-induced filament A from Salmonella typhimurium is geranylgeranylated at the C-terminal CCFL by mammalian host GGT-1.69 The farnesylation of Legionella pneumophila ankyrin B (ANKB) at the C-terminal of CVLC by the host FT anchors ANKB to the Legionella-containing vacuole for the intravacuolar proliferation of the bacterium.70 Additional effector proteins with CaaX motifs in L. pneumophila were later shown to be prenylated by the host to facilitate their targeting to host organelle membranes in the process of intracellular infection.71,72
Viral proteins containing the C-terminal CaaX motif can also be prenylated by host prenyltransferases. One example of clinical relevance is the large antigen of the hepatitis delta virus. The prenylation of the large antigen is key for virus assembly.73,74 Most important, prenylation inhibitors have been shown to depress viral particle formation,75 and a phase 2A clinical trial showed that the prenylation inhibitor lonafarnib significantly reduces hepatitis delta virus levels in humans.76
2.3. Chemical Probes for Protein Prenylation
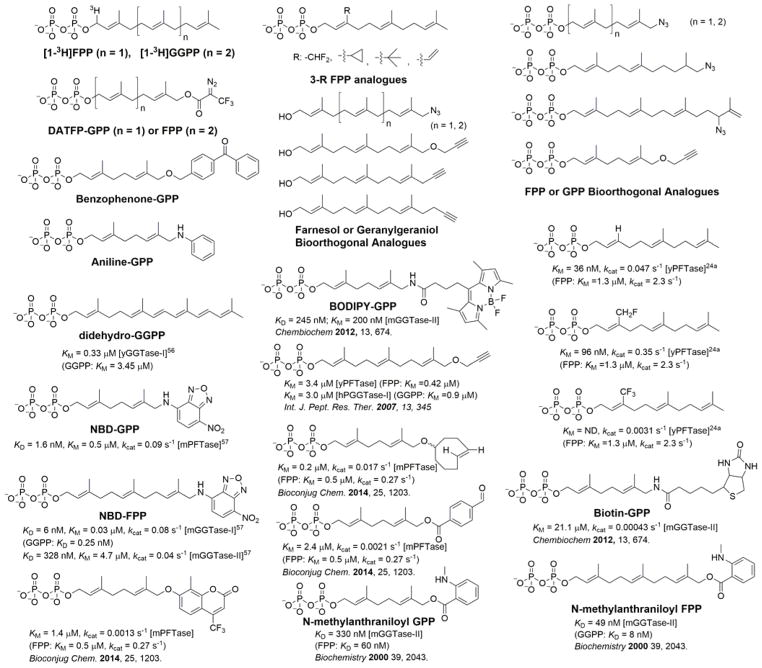
Since the discovery of prenylated proteins, various analogues of isoprenoid diphosphates have been synthesized and used to study the structures and reaction mechanisms of prenyltransferases and to visualize and identify prenylated proteins and prenyltransferases (Figure 6). Isotopic probes of FPP and GGPP including [1-3H]FPP and [1-3H]GGPP were originally used to validate the enzymatic activities of FT, GGT-1, and RGGT and elucidate their selectivity for peptide substrates. The photo-affinity probes [3H]-DATFP-FPP, [3H]-DATFP-GPP, [32P]DATFP-GPP, and benzophenone-GPP have also been applied to label FT enzymes.77–79 Methods using isotopic isoprenoid probes are usually not very sensitive and require long exposure time (days) for detection. Furthermore, these probes lack affinity tags for the isolation and identification of target proteins, which limits their applications. However, the isotopic native molecules [1-3H]FPP and [1-3H]GGPP have proved useful for validating whether the prenylated proteins identified in proteomics studies using other affinity probes are true substrates of FT, GGT-1, or RGGT. This confirmation is particularly critical because some studies have suggested that various farnesyl diphosphate analogues may differ in terms of protein substrate specificity and reaction rates with FT.80
Figure 6.
Chemical probes used to study protein prenylation.
Fluorine,81 vinyl,82 cyclopropyl, and tert-butyl groups83 have been incorporated into isoprenoid diphosphate analogues to study the farnesylation mechanism. As an immunogenic probe, an aniline-tagged isoprenoid diphosphate was shown to label several FT protein substrates in mammalian cells, which could be detected by the specific antibody raised against the aniline moiety.84 The corresponding aniline-tagged isoprenol, which is converted into the diphosphate in cells,85 was used to label cellular proteins metabolically before antibody-based detection.86
Fluorescent derivatives of isoprenoid diphosphate, such as didehydrogeranylgeranyl (ΔΔGG) diphosphate,87 7-nitro-benzo[1,2,5]oxadiazol-4-ylamino (NBD) FPP,88 and N-methylanthraniloyl isoprenoid diphosphate,89 have been designed as efficient isoprenoid donors for prenyltransferases and used in high-throughput fluorometric assays to screen potential inhibitors of in vivo protein trafficking. To facilitate the labeling and enrichment of prenylated proteins from biological samples, biotin-functionalized geranyl pyrophosphate has been applied to identify and analyze prenylated mammalian proteins with engineered prenyltransferases.90 Such probes can help elucidate the mechanisms through which protein prenylation is regulated and the therapeutic effects of various agents. Although fluorescent and biotin probes are convenient for in-gel detection, high-throughput assays, or affinity purification, their relative large and bulky conjugated functional groups may interfere with recognition by prenyltransferases and perturb signaling pathways.
With click chemistry now being widely applied in biological systems, bioorthogonal reporters of protein prenylation have been developed via the incorporation of small alkyne or azide groups into isoprenoid diphosphates (Figure 6).91–95 These probes can be efficiently incorporated into prenylated proteins in vitro and are easily conjugated to various functional tags for fluorescence detection or affinity purification. Furthermore, alkyne- or azide-labeled isoprenols are cell-permeable and can be used to label prenylated proteins metabolically in live cells (Figure 6).91,95–100 Studies using these probes indicated that the substrate specificity of prenyltransferases may depend on the bioorthogonal probes used, and alkynyl-isoprenoid probes are generally more sensitive than azido-isoprenoid probes.97 Studies of protein prenylation have historically focused on the Ras superfamily of G proteins. Proteomics studies using clickable probes have led to the identification of other proteins modified by prenylation, such as lamin B1, chaperonin DNAJA2, and zinc finger antiviral protein (ZAP).91,98,99 Recently, both alkyne-tagged isoprenols and isoprenoid diphosphates have been used to identify prenylated proteins in the malaria parasite Plasmodium falciparum.101,102
Notably, cross-reactivity is observed when prenyl probes are used to identify prenylomes in cells. For example, known geranylgeranylated protein Cdc42 was identified by using an FPP probe,91 and alkynyl-farnesol is utilized by all three cellular prenyltransferases.97 However, such cross-reactivity may also be physiologically relevant, as RhoB is reportedly farnesylated and geranylgeranylated efficiently by GGT-1.26
2.4. Functions of Prenylation
2.4.1. Membrane Association
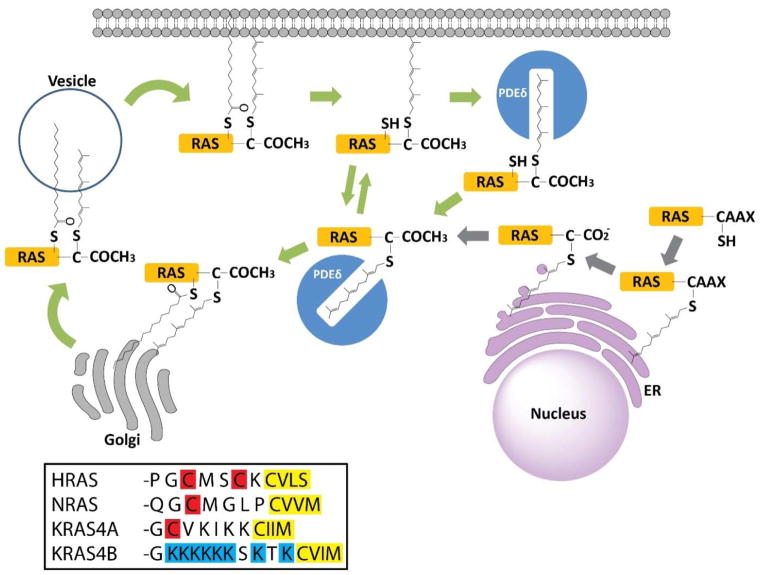
The prenyl group is hydrophobic and thus recruits soluble proteins to cellular membranes. In this mechanism, it is important to distinguish the plasma membrane from endomembranes (membranes of intracellular organelles such as the ER, Golgi, endosomes, lysosomes, and nucleus). Ras proteins were found to associate with the plasma membrane in a prenylation-dependent manner.9 Mutation of the prenylated cysteine residues or the blocking of isoprenoid biosynthesis abolished the prenylation of Ras proteins and their plasma membrane association. However, later studies suggested that prenylation is mainly responsible for targeting proteins to endomembranes.103 Specifically, the CaaX prenylation targets proteins to the ER and Golgi.103
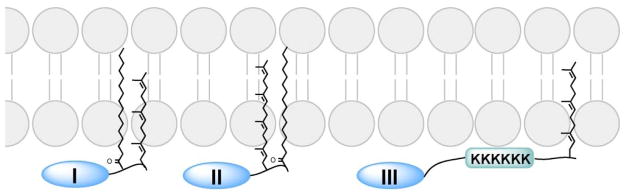
The endomembrane targeting of prenylation explains why many prenylated proteins with CaaX motifs require additional membrane targeting motifs for plasma membrane localization, including cysteine palmitoylation (which provides greater hydrophobic affinity to the membranes) and a polybasic domain (which interacts electrostatically with negatively charged phospholipid head groups on the inner leaflet of plasma membranes; Figure 7). These additional membrane-targeting motifs aid the translocation of these proteins from endomembranes to the plasma membrane. For example, H-Ras and N-Ras undergo both cysteine prenylation and cysteine palmitoylation at the C-terminus. Although 90% of wild-type (WT) H-Ras is associated with the plasma membrane, only 8% of a non-palmitoylated H-Ras mutant was found to do so,104 which indicates that both modifications are required for plasma membrane targeting. N-Ras has only one palmitoylated cysteine, but H-Ras contains two. Compared with the single cysteine palmitoylation on N-Ras, the double-cysteine palmitoylation on H-Ras reportedly promotes trans-Golgi localization.105
Figure 7.
Plasma membrane targeting involving prenylation and a second signal, including (I) upstream palmitoylation, (II) downstream palmitoylation, and (III) upstream polybasic domain (typically six lysine residues).
A similar model applies in the targeting of farnesylated proteins to other membrane organelles: farnesylation targets proteins to endomembranes, and other signals help target proteins to specific membrane organelles. For example, prelamin A requires both a C-terminal CSIM farnesylation motif and a nuclear localization signal to accumulate in the nuclear envelope for later endoproteolysis to generate mature lamin A.106 Another lamin protein, lamin B, also requires farnesylation to assemble into lamina and associate with the nuclear membrane during mitosis.107 Unlike lamin A, lamin B does not undergo endoproteolysis, and thus, mature lamin B retains the farnesylation. Lamin B1 farnesylation, but not lamin B2 farnesylation, is key for brain development and the formation of stable nuclear lamina in mice; a nonfarnesylated lamin B1 mutation led to death soon after birth.108 The farnesylation of the ZAP long isoform has been demonstrated to regulate the localization of the isoform to the lysosomes and late endosomes.98 Presumably, another signal is needed to target ZAP specifically to these organelles.
The process of protein prenylation with a CaaX motif typically requires three steps: prenylation, proteolysis, and carboxylmethylation. In vitro studies of K-Ras showed that only 20% of K-Ras is associated with membranes when K-Ras undergoes farnesylation without proteolysis and carboxylmethylation, whereas up to 80% of K-Ras is associated with membranes after the methylation step is completed.109 This result suggests that carboxylmethylation greatly enhances the membrane association of the farnesylated protein owing to the increase in hydrophobicity and the removal of the negative charge on the carboxylate group. Further studies demonstrated that carboxylmethylation has a much smaller effect on geranylgeranylated proteins.110,111 Notably, the membrane localization of Ras proteins is complicated and incompletely understood. For example, the small molecule fendiline reportedly promotes the intracellular membrane localization of K-Ras, but the mechanism remains unknown.112
Some Ras proteins have C-terminal CCaX motifs, including a brain-specific splice variant of Cdc42 (CCIF), RalA (CCIL), and RalB (CCLL). A recent study demonstrated that these proteins undergo prenylation on the first cysteine and palmitoylation on the second cysteine for stable anchoring in the plasma membrane (Figure 7). This reaction differs from and likely competes with the classical CaaX processing in which a sole prenylation is followed by proteolysis and carboxylmethylation.113
One of the potential advantages of having multiple membrane targeting motifs for membrane anchoring is the capacity for easy regulation of membrane associations. For example, K-Ras4B has a polybasic region containing six lysine residues upstream of the prenylation site (Figure 7). Alterations to this polybasic region significantly decrease the plasma membrane association of K-Ras4B.104,114 Phosphorylation on Ser181 within the region changes the electrostatic status of the protein by partially neutralizing the positive charge and thus destabilizes the electrostatic interaction between K-Ras4B and the plasma membrane. This change promotes the dissociation of K-Ras4B from the plasma membrane.115 In vitro studies using K-Ras4B and nanodiscs confirmed the effect of Ser181 phosphorylation and further demonstrated that farnesylated K-Ras4B prefers disordered lipid microdomains.116
Most Rab proteins have C-terminal CC or CXC motifs for digeranylgeranylation (Figure 7), which is more hydrophobic. In terms of membrane targeting, digeranylgeranylation seems only to target proteins to endomembranes, as most Rab proteins are targeted to specific intracellular membrane organelles, not the plasma membrane. However, the effects of digeranylgeranylation can differ from those of single geranylgeranylation. When the CC or CXC motif is replaced with a mono cysteine motif, Rab5a and Rab27a are mistargeted to the ER instead of to endosomes and melanosomes, respectively.117 Rab proteins with a CXC motif undergo terminal carboxylmethylation on prenylcysteine, whereas those with a CC motif do not.118 Although this methylation has no effect on the subcellular localization of Rab proteins,119 it might indicate that Rab proteins with CXC motifs need to pass through the ER for methylation by ICMT, whereas Rab proteins with CC motifs can be directly transferred to the target membrane without interacting with the ER. The localization of digeranylgeranylated Rab proteins to specific membrane organelles also requires an additional targeting signal in their protein sequences.106,120 Initially, the hypervariable C-terminal domains (HVDs) of Rab proteins120 were thought to help determine the appropriate subcellular localization of the proteins, but later experiments suggested that the situation is more complicated. Studies using semisynthetic Rab proteins (Rab1, Rab5, Rab7, Rab35) in which the HVDs were replaced with a polyethylene glycol linker have demonstrated that the HVDs of Rab1 and Rab5 are not required for Golgi and early endosome localization, respectively.121 By contrast, the HVD of Rab7 is key for late endosome and lysosome localization because this domain interacts with Rab-interacting lysosomal protein, which is a Rab7 effector. The HVD of Rab35 is also central to its plasma membrane localization owing to the presence of a polybasic sequence.121 Another study showed that interactions between Rab1A/Rab5A/Rab8A and their corresponding guanine nucleotide exchange factors (GEFs) play important roles in targeting the proteins to the correct intracellular membranes. Therefore, the correct targeting of Rab proteins is determined by prenylation; interactions with GEFs, effectors, and possibly other proteins; and negative charges on the plasma membrane.
2.4.2. Protein-Protein Interactions
Many studies of the Ras superfamily have demonstrated that prenylation is critical for protein-protein interactions. The farnesylation of yeast Ras2 increases the binding affinity to adenylyl cyclase 100-fold; however, the subsequent palmitoylation of Ras2 has little effect despite its importance for Ras2 membrane targeting.122 A recent study also showed that human Spindly, a mitotic checkpoint protein, requires farnesylation to target kinetochores via protein–protein interactions.123
Guanine nucleotides bound to Ras proteins are controlled by GEFs. One GEF, human SOS (hSOS1), forms a complex with farnesylated K-Ras4B, but not with unmodified K-Ras4B, to regulate the binding ofguanine nucleotides and response to growth factor stimulation.124 The polybasic domain of K-Ras4B is not required for the interaction with hSOS1. Other studies have emphasized that the prenylation of N-Ras is critical for the binding of N-Ras to both the active and allosteric sites of hSOS1.125 Interestingly, oncogenic K-Ras reportedly binds to the allosteric site of hSOS1, which promotes the activation of WT H-Ras and N-Ras.126 The farnesylation of Cdc42 is also central to the activation of Cdc42 by its GEF, Dock7.127
In vitro studies have shown that the geranylgeranylation of RhoA is important for interactions with the RhoA guanosine diphosphate (GDP) dissociation inhibitor (GDI) and GDP dissociation stimulator (GDS) but not GTPase activating proteins (GAPs).128 Geranylgeranylation is also required for the interaction between RhoA and IQ-motif-containing GTPase activating protein IQGAP1 to regulate RhoA functions in breast cancer cell proliferation and migration.129 IQGAP1 is likely an effector protein of RhoA because it functions downstream of RhoA.129
A short splice variant of small guanosine triphosphate (GTP)-binding protein guanine nucleotide dissociation stimulator, SmgGDS-558, selectively binds prenylated Rap1A to facilitate the trafficking of Rap1A to the plasma membrane,130 whereas the long splice variant SmgGDS-607 associates with non-prenylated Rap1A to regulate Rap1A entry into the prenylation pathway. This provides a regulatory mechanism for the prenylation of small GTPases.
In all the cases described above, it is unclear whether prenylation is involved in the protein-protein interaction directly or via indirect mechanisms, such as those affecting subcellular localization. By contrast, prenylation is directly involved in the protein-protein interactions described below for GDI proteins. RabGDIs specifically bind geranylgeranylated Rab proteins in their GDP-bound forms (but not their GTP-bound forms) to retrieve them from the target membranes after vesicular transport.131 This activity is central to the cellular recycling of Rab proteins for normal functioning. Similarly, RhoGDIs bind to and stabilize Rho proteins to regulate their cellular homeostasis.132
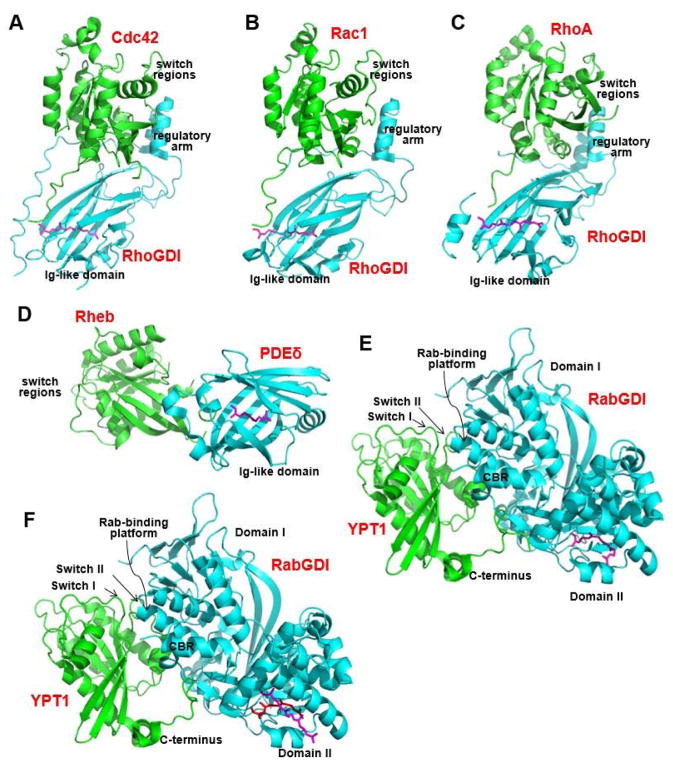
The structure of the Cdc42-RhoGDI complex demonstrates that a hydrophobic pocket exists between the two opposing β-sheets of the Ig-like domain of RhoGDI. This pocket binds the geranylgeranyl moiety of Cdc42 (Figure 8A),133 which changes the conformation of an α-helix (Rho insert) in Cdc42.134 The binding by RhoGDI also facilitates the extraction of Cdc42 from the cellular membrane. Additional structures of GDI complexed with Ras proteins further support the functional role of prenylation in the interaction between GDI and Ras proteins (Figure 8B and 8C).135–138 A GDI-like solubilizing factor, PDE6δ, can bind prenylated retinal PDE6 catalytic subunits,139 rhodopsin kinases,140 prostacyclin receptor,141 and Ras proteins.142 The C-terminal farnesyl moiety of Ras binds to a hydrophobic pocket in the Ig-like domain of PDE6δ, as demonstrated by crystal structures of the PDE6δ–Rheb complex (Figure 8D)143 and KRas4b–PDE6δ complex.144 Notably, PDE6δ lacks the regulatory arm required to interact with the switch regions of Rheb or Ras, which differs from the association of RhoGDI with Rho (compare Figure 8D to Figure 8A–8C). By binding to and solubilizing prenylated Ras proteins, PDE6δ may enhance the diffusion of these proteins into the cytoplasm and facilitate more effective trapping of both depalmitoylated Ras proteins at the Golgi and polycationic Ras proteins at the plasma membrane.144 Similarly, by binding to farnesylated or geranylgeranylated INPP5E, PDE6δ mediates the sorting of INPP5E into cilium.145
Figure 8.
Protein structures of guanosine diphosphate dissociation inhibitors (GDIs) in complex with prenylated proteins. (A) Prenylated Cdc42 (green)-RhoGDI (cyan) complex (PDB ID 1DOA), (B) prenylated Rac1 (green)–RhoGDI (cyan) complex (PDB ID 1HH4), (C) prenylated RhoA (green)-RhoGDI (cyan) complex (PDB ID 4F38), (D) prenylated Rheb (green)-PDEδ (cyan) complex (PDB ID 3T5G), (E) prenylated YPT1 (green)-RabGDI (cyan) complex (PDB ID 1UKV), and (F) doubly prenylated YPT1 (green)-RabGDI (cyan) complex (PDB ID 2BCG). CBR, C-terminal-binding region. The prenyl moiety is shown in purple or red. All protein structures were made using PyMOL with PDB files.
By contrast, the RabGDIs have a completely different fold from that of the RhoGDIs. RabGDIs have more than 440 amino acids and are larger than RhoGDIs, which have approximately 200 amino acids. No significant sequence homology exists between RabGDIs and RhoGDIs. In the structures of the prenylated YPT1-RabGDI complex and the doubly prenylated YPT1-RabGDI complex (Figure 8E and 8F), the Rab-binding platform and the C-terminal binding region in domain I of RabGDI interact with the Switch I/II regions and C-terminus of YPT1. Geranylgeranyl moieties are buried in the hydrophobic pocket formed by the α-helices of RabGDI domain II.
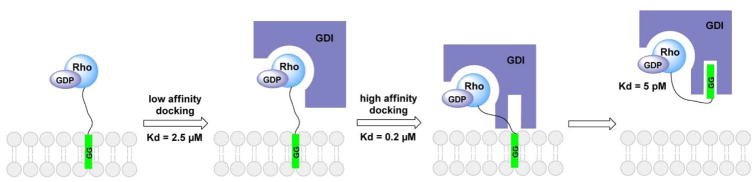
Quantitative analysis of the interaction between prenylated RhoA and RhoGDI has revealed that the extraction of Rho GTPase from membranes by RhoGDI is a thermodynamically favored passive process modulated by a series of progressively tighter complexes (Figure 9).135 RhoGDI initially binds RhoA to form a low-affinity complex. Then, the positively charged C-terminus of RhoA binds to the negatively charged residues at the C-terminus of RhoGDI, increasing the complex affinity. This complexation positions the C-terminus of RhoGDI near the membrane-buried geranylgeranyl moiety of RhoA and opens the lipid-binding pocket at the C-terminus of RhoGDI. Next, the geranylgeranyl moiety is transferred from the membrane to the lipid-binding pocket of RhoGDI, which forms a high-affinity complex that spontaneously dissociates from the membrane. RabGDI uses a similar mechanism to extract Rab proteins from membranes.146
Figure 9.
Mechanism of RhoA membrane extraction by RhoGDI. GG, geranylgeranyl group.
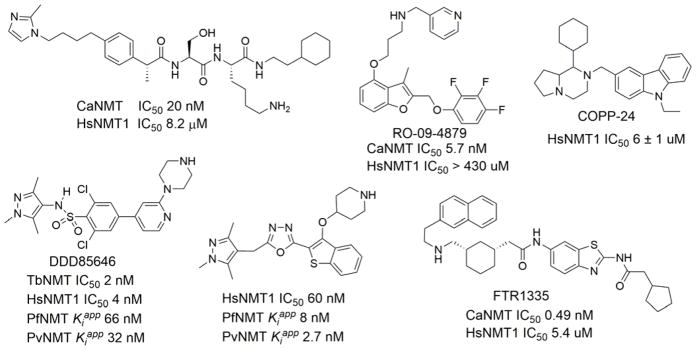
2.5. Prenyltransferase Inhibitors
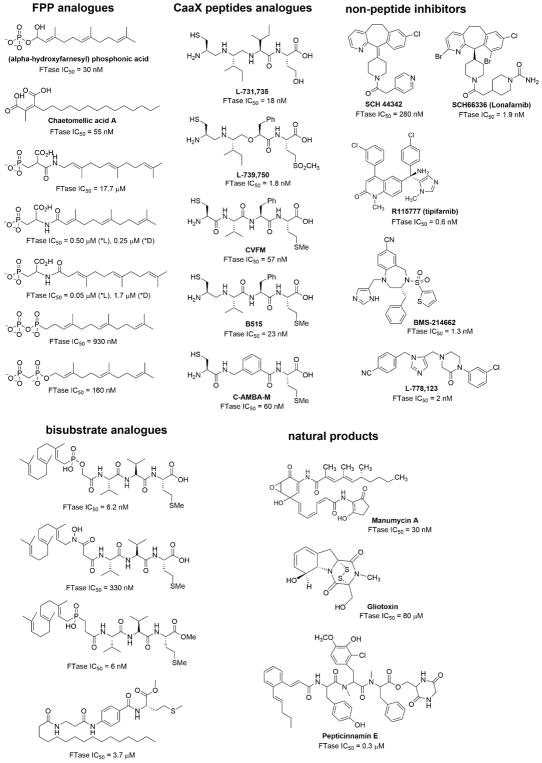
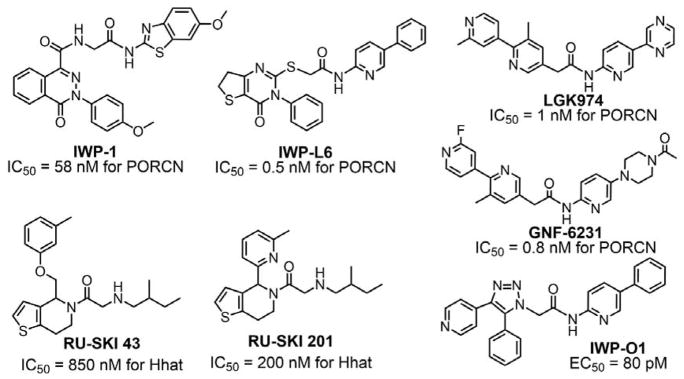
Because the oncogenic form of Ras requires farnesylation for activity, the inhibition of the farnesylation process may be a strategy to treat cancer. Thus, FT inhibitors have attracted attention,147 and many FT inhibitors have been reported (Figure 10). There are four types of FT inhibitors: FPP analogues, CaaX peptides analogues, bisubstrate analogues, and non-peptide inhibitors.148–152,104 104 104,103,147–151 Certain natural products have also been identified as FT inhibitors.
Figure 10.
Farnesyltransferase inhibitors.
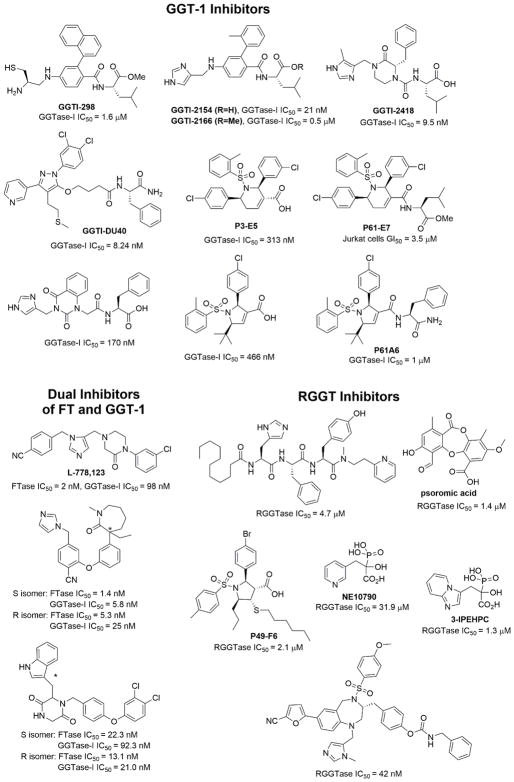
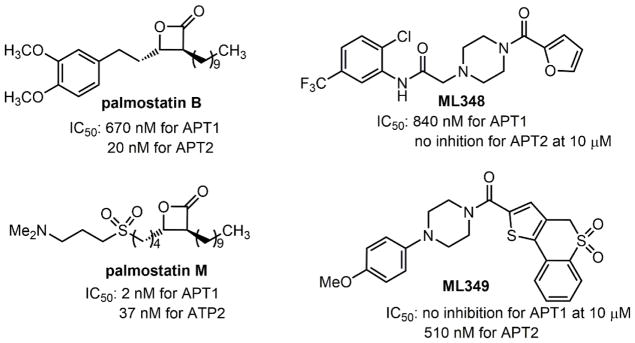
Although FT inhibitors generally have low toxicity, they lack efficacy in clinical trials,153 perhaps because GGT-1 compensates for the inhibited FT and carries out the geranylgeranylation of Ras proteins, thereby allowing the proliferation of cancer cells.154 Geranylgeranylated RalA transforms cells in several cancers,155 and geranylgeranylated RhoC is essential for cancer metastasis.156,157 These findings suggest that GGT-1 is a promising target for cancer treatment. Many specific GGT-1 inhibitors have been identified and show therapeutic effects (Figure 11).158–166 Dual inhibitors for FT and GGT-1167–169 and combination treatments using FT inhibitors with GGT-1 inhibitors or other agents153,170–174 have also been reported.
Figure 11.
Specific inhibitors of GGT-1 and RGGT and dual inhibitors of FT and GGT-1. IC50, half-maximal inhibitory concentration.
RGGT is overexpressed in several tumors and has an anti-apoptotic effect in some cancer cell lines.175 Studies have also demonstrated that RGGT is involved in tumor survival. Rab25, a substrate of RGGT, determines the aggressiveness of epithelial cancers.176 Other Rab proteins have elevated expression in various human cancers.177 However, only a few specific RGGT inhibitors (Figure 11) are available and they typically have low affinities.160,178–185
Another application of FT inhibitors is the treatment of parasitic diseases, including malaria (caused by Plasmodium falciparum),186–190 African sleeping sickness (caused by Trypanosoma brucei),191,192 Chagas disease (caused by Trypanosoma cruzi),193–196 and leishmaniasis (caused by Leishmania mexicana).197 The parasitic vectors of these diseases are hypothesized to lack GGT-1; therefore, FT inhibitors are sufficient to inhibit their growth. Antifungal198–202 and antiviral75,203–207 activities of FT inhibitors and GGT-1 inhibitors have also been explored.
Among the most promising clinical applications of FT inhibitors are the treatment of Hutchinson-Gilford progeria syndrome (HGPS) and hepatitis D. HGPS is a rare premature aging disease caused by mutations in the LMNA gene that encodes prelamin A and prelamin C.208 As described in section 2.4a, prelamin A is farnesylated and targeted to the nucleus, where it is proteolyzed to remove the C-terminal farnesylated peptide. The mutations that cause HGPS abolish the proteolysis step, which leads to premature aging. In one study, lonafarnib treatment increased body weight and lessened arterial stiffness in 25 children with HGPS.209 In another study, lonafarnib treatment increased mean survival by 1.6 years.210 Combining lonafarnib with pravastatin and zoledronic acid increased bone mineral density in patients with HGPS but offered no benefits beyond those of lonafarnib treatment alone.211
Hepatitis D is caused by the hepatitis delta virus, and no satisfactory treatment currently exists. As mentioned in section 2.2, the prenylation of the hepatitis delta virus large antigen is key for virus assembly,73,74 and prenylation inhibitors have been shown to inhibit virus particle formation.75 A proof of concept, randomized, double-blind, placebo-controlled phase 2A trial showed that lonafarnib significantly reduces hepatitis D viral load.76 Another trial to test lonafarnib in combination with ritonavir or PEGylated interferon α (PEG = polyethylene glycol) is ongoing (NCT02430194).
3. N-Terminal Glycine Myristoylation
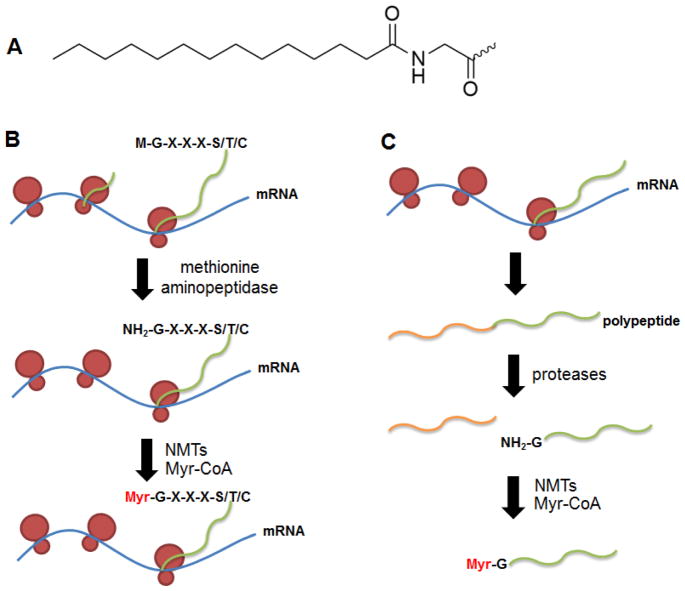
N-glycine myristoylation refers to the co- or post-translational attachment of a saturated 14-carbon fatty acyl group, myristoyl, to the N-terminal glycine of proteins via an amide bond (Figure 12). The consensus sequence required for the co-translational modification after removal of the first methionine residue by methionine aminopeptidase is Gly-XXX-Ser/Thr/Cys.212 N-Glycine myristoylation has also been reported as a post-translational modification for certain pro-apoptotic proteins.213 The cleavage of these proteins by caspases exposes an internal glycine for myristoylation (Figure 12). N-Glycine myristoylation plays essential roles in the targeting of proteins to desired subcellular localizations by mediating protein-protein and protein-membrane interactions. Owing to the diversity of substrate proteins modified, N-glycine myristoylation is critical for signal transduction, apoptosis, and virus-, protozoa-, and fungi-induced pathological processes.212,214 Therefore, this modification is a promising target for the development of anti-parasitic and antifungal drugs.215
Figure 12.
(A) Myristoyl modification at an N-terminal glycine residue. (B) Co-translational N-myristoyl modification. (C) Post-translational N-myristoyl modification.
3.1. N-Myristoyltransferase
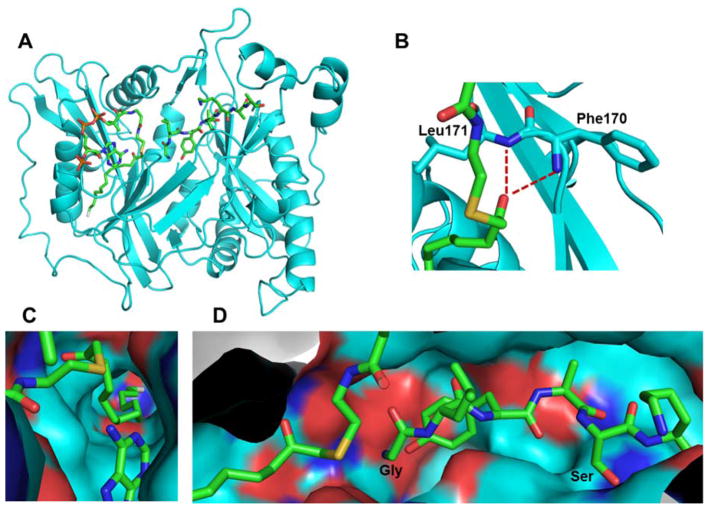
N-Glycine myristoylation is catalyzed by myristoyl-CoA: protein N-myristoyltransferase (NMT), which belongs to the GCN5-related N-acetyltransferase superfamily.216 NMT has been characterized extensively in many organisms, including mammals, insects, plants, parasites, yeast, and fungi. Saccharomyces cerevisiae and Candida albicans contain a single NMT, whereas Homo sapiens has two NMTs (NMT1 and NMT2).217 The X-ray crystal structures of S. cerevisiae NMT show that NMT is folded into a saddle-shaped β-sheet flanked by several α-helices (Figure 13A). Within this pseudo-two-fold symmetry, the N- and C-terminal halves of NMT contribute to the myristoyl-CoA and protein substrate binding sites, respectively.218,219
Figure 13.
(A) Crystal structure of S. cerevisiae NMT in complex with a non-hydrolyzable myristoyl-CoA analogue and a peptide substrate (PDB ID 1IID). (B) Phe170 and Leu171 form the oxyanion hole to stabilize the negative charge developed on the carbonyl oxygen of myristoyl-CoA during catalysis. (C) The hydrophobic myristoyl group binds in a deep pocket in NMT. (D) The peptide substrate recognition site of NMT, which explains the peptide sequence specificity of NMT. All protein structures were made using PyMOL with PDB files.
Kinetic and structural evidence suggests that NMT catalysis follows a sequential ordered Bi-Bi mechanism.220 The myristoyl-CoA initially binds the apo-NMT and induces a conformational change for peptide binding. After the formation of a ternary NMT - myristoyl-CoA - peptide complex, acyl transfer occurs via the attack of the N-terminal glycine at the thioester bond of myristoyl-CoA. Free CoA is then released, followed by the myristoylated peptide product.220 Several structures of the ternary complex have been reported221 and highlight several notable features. First, an oxyanion hole is formed by the main-chain amide bonds of Phe170 and Leu171 (Figure 13B). Second, the bent conformation around the C5 and C6 of myristoyl-CoA positions the end of the acyl chain in a deep pocket of the enzyme (Figure 13C). These features may provide the measurements of acyl chain length that result in the specificity toward myristoyl-CoA.218 The highly abundant palmitoyl-CoA is also capable of binding NMT; however, the catalytic efficiency is much lower than that of myristoyl-CoA.222 Finally, the structures provide an explanation for the peptide sequence selectivity of NMTs (Figure 13D). The amino group of the N-terminal glycine must rotate to the left to attack the carbonyl of myristoyl-CoA. A larger side chain group (if substituting glycine with other amino acids) may impede the rotation and thus the myristoylation.221 The serine side chain at position 5 interacts with a small hydrophilic pocket, which explains the preference for serine/threonine/cysteine at this position. By contrast, positions 2–4 are either solvent-exposed or accommodated by large pockets, which explains the lack of preference at these positions.
Several studies have shown that NMT is essential for the survival of mammals,223 fungi,224,225 flies,226 and parasites.227 In humans, NMT1 and NMT2 share approximately 76% sequence identity and have partially overlapping biological functions and substrate selectivity.217,223 S. cerevisiae and human NMTs are predominantly localized in the cytosol.228,229 The N-terminal region of human NMTs, which consists of polybasic amino acid sequences (K-box), is reported to be crucial for targeting to the ribosomes, where co-translational N-myristoylation modification occurs.230,231
NMT1, but not NMT2, is also critical for cell proliferation, whereas cell survival is likely regulated by both NMT1 and NMT2.223 NMT1 is essential for embryonic development and proper monocytic differentiation in mice,232,233 in which thymus-specific knockouts of NMT1 and NMT2 have been generated. NMT1 knockout significantly decreases T-cell numbers and T-cell receptor signaling, whereas NMT2 knockout has only minor effects.234 T-cell apoptosis increases most dramatically when both NMT1 and NMT2 are knocked out, but compared with NMT1 knockout, the knockout of NMT2 seems to have a stronger effect on apoptosis.234 An increase in the activity of both NMT1 and NMT2 has been observed in colonic and brain tumors.235
NMTs have been demonstrated to be substrates for caspases during apoptosis.236 The caspase cleavage of NMTs potentially regulates the localization of NMTs. The removal of a lysine cluster from NMT1 by caspase-3 or caspase-8 promotes the translocation of NMT1 from the ribosomal and membrane fractions to the cytosol. However, the caspase-3 cleavage of NMT2 leads to the relocalization NMT2 from the cytosol to the membrane fraction.236 The reasons for NMT-specific localization change during apoptosis require further investigation.
3.2. Proteins Modified by N-Glycine Myristoylation
Experimentally identified N-glycine myristoylated proteins can be classified into various functional classes such as signaling proteins (GTP-binding proteins, Ca2+-binding EF-hand proteins, and protein kinases), apoptotic proteins, and structural viral proteins. The modified mammalian proteins are summarized in Table 1.237
Table 1.
Myristoylated mammalian proteins
| Myristoylated proteins | Functions | Roles of N-myristoylation | First 20 amino acid sequences | Organisms | References |
|---|---|---|---|---|---|
|
| |||||
| Gelsolin (ctGelsolin, C-terminal after cleavage) | Actin-modulating protein | Anti-apoptotic activity | GLGLSYLSSH IANVERVPFD | Human | 238 |
|
| |||||
| Lunapark | Network formation of ER | ER morphology | MGGLFSRWRT KPSTVEVLES | Human | 239 |
|
| |||||
| Endothelial nitric oxide synthase (eNOS) | Nitric oxide production | Golgi membrane association | MGNLKSVAQE PGPPCGLGLG | Human | 240 |
|
| |||||
| Protein-associating with the carboxyl-terminal domain of ezrin (PACE-1) | Ezrin-binding partner | Golgi membrane association | MGSENSALKS YTLREPPFTL | Human | 241 |
|
| |||||
| Phosphoinositide 3-kinase regulatory subunit 4 (PI3K p150 subunit) | PI 3-Kinase adapter protein | Golgi membrane association | MGNQLAGIAP SQILSVESYF | Human | 242 |
|
| |||||
| 43 kDa acetylcholine receptor-associated protein of synapse (RAPSN) | Acetylcholine receptor clustering | Membrane association | MGQDQTKQQI EKGLQLYQSN | Mouse | 243 |
|
| |||||
| Adenylate kinase 1 | Energy homeostasis | Membrane association | MGCCVSSEPQ EEGGRKTGEK | Mouse | 244 |
|
| |||||
| ADP ribosylation factors (ARF) and ARF-like (ARL) | GTP binding protein | Membrane association | MGKVLSKIFG NKEMRILMLG MGILFTRIWR LFNHQEHKVI |
Human | 245,246 |
|
| |||||
| A-kinase anchoring protein 12 (Gravin, AKAP12) | Protein kinase A binding protein | Membrane association | MGAGSSTEQR SPEQPPEGSS | Human | 247 |
|
| |||||
| A-kinase anchoring protein 7 (AKAP 18) | Protein kinase A binding protein | Membrane association | MGQLCCFPFS RDEGKISEKN | Human | 248 |
|
| |||||
| 5′-AMP-activated protein kinase subunit beta-1 (AMPKβ) | scaffold for AMPK complex | Mitochondria association | MGNTSSERAA LERHGGHKTP | Human | 249 |
|
| |||||
| Annexin XIII | Ca2+/phospholipid binding proteins | Membrane association | MGNRHAKASS PQGFDVDRDA | Human | 250 |
|
| |||||
| Calcineurin B phosphatase subunit, (CHP1, p22) | Ca2+-binding proteins | Membrane association Microtubule targeting |
MGSRASTLLR DEELEEIKKE | Human | 251,252 |
|
| |||||
| Calmyrin | Ca2+-binding proteins | Membrane association | MGGSGSRLSK ELLAEYQDLT | Human | 253 |
|
| |||||
| Calpastatin (testis-specific isoform, tCAST) | Calpain inhibitor | Membrane association | MGQFLSSTFW EGSPAAVWQE | Mouse | 254 |
|
| |||||
| cAMP-dependent kinases (PKA catalytic subunit) | Serine/threonine kinases | Membrane association | MGNAAAAKKG SEQESVKEFL | Human | 255,256 |
|
| |||||
| cGMP-dependent kinase II | Serine/threonine kinases | Membrane association | MGNGSVKPKH SKHPDGHSGN | Human | 257 |
|
| |||||
| Charged multivesicular body protein 6 (CHMP6) | ESCRT-III protein | Membrane association | MGNLFGRKKQ SRVTEQDKAI | Human | 258 |
|
| |||||
| Erythrocyte membrane protein band 4.2 | Membrane stability maintenance | Membrane association | MGQALGIKSC DFQAARNNEE | Human | 259 |
|
| |||||
| Fibroblast growth factor receptor substrate 2 (FRS2) | Adapter protein | Membrane association | MGSCCSCPDK DTVPDNHRNK | Human | 260 |
|
| |||||
| Formin-like 1 (FMN1) | Cell morphology/cytoskeletal organization regulation | Membrane association | MGNAAGSAEQ PAGPAAPPPK | Human | 261 |
|
| |||||
| Fragile X mental retardation syndrome-related protein 2 (FXR2) | RNA binding protein | Axonal distribution | MGGLASGGDV EPGLPVEVRG | Human | 262 |
|
| |||||
| Golgi reassembly stacking protein 1 and 2 (GRASP65, GRASP55) | Golgi stacking | Membrane association/protein-protein interaction | MGLGVSAEQP AGGAEGFHLHMGSSQSVEIP GGGTEGYHVL | Human | 263,264 |
|
| |||||
| Golgi-associated plant pathogenesis-related protein 1 (GAPR-1) | Caveolin-interacting protein | Membrane association/protein-protein interaction | MGKSASKQFH NEVLKAHNEY | Human | 265 |
|
| |||||
| G-protein alpha subunits (Gα) | GTP binding protein | Membrane association | MGCTLSAEDK AAVERSKMID | Human | 247,266 |
|
| |||||
| hexokinase 1 variant in mammalian spermatozoa (HK1S) | Enzyme in glycolysis | Membrane association | MGQICQRESA TAAEKPKLHL | Human | 267 |
|
| |||||
| Myristoylated alanine-rich carboxy-kinase substrate (MARCKS) | Protein kinase C substrate | Membrane association | MGAQFSKTAA KGEAAAERPG | Human | 268,269 |
|
| |||||
| NADH ubiquinone dehydrogenase B18 | Enzyme in a respiratory electron transport chain | Membrane association | MGAHLARRYL GDASVEPDPL | Bovine | 270 |
|
| |||||
| Neuralized-like 1 (NEURL1) | E3 ubiquitin ligase | Membrane association | MGNNFSSVSS LQRGNPSRAS | Mouse | 271 |
|
| |||||
| Neuronal calcium sensor (NCS) proteins : | |||||
| Neuronal calcium sensor-1, | Ca2+-binding proteins | Membrane association | MGKSNSKLKP EVVEELTRKT | Human | 272–275 |
| Hippocalcin, | MGKQNSKLRP EMLQDLRENT | Human | |||
| Neurocalcin delta, | MGKQNSKLRP EVMQDLLEST | Human | |||
| Visinin-like protein-1 (VILIP-1) | MGKQNSKLRP EVLQDLREHT | Rat | |||
| Recoverin | MGNSKSGALS KEILEELQLN | Bovine | |||
| Guanylate cyclase-activating proteins (GCAPs) (GCAP-1, GCAP-2) | MGNVMEGKSV EELSSTECHQ | Human | |||
| MGQEFSWEEA EAAGEIDVAE | Human | ||||
|
| |||||
| P21-activated kinase 2 (ctPAK2) | Serine/threonine kinases | Membrane association | GAAKSLDK QKKKTKMTDE EI | Human | 276 |
|
| |||||
| Protein phosphatase 1A and 1B (PPM1A and PPM1B) | AMPKα dephosphorylation | Membrane association | MGAFLDKPKMEKHNAQGQGN MGAFLDKPKT EKHNAHGAGN |
Human | 277 |
|
| |||||
| Protein arginine N-methyltransferase 8 (PRMT8) | Arginine methylation | Membrane association | MGMKHSSRCL LLRRKMAENA | Human | 278 |
|
| |||||
| PSD-Zip70, FEZ1 | Synaptic plasticity | Membrane association | MGSVSSLISG HSLHSKHCRA | Rat | 279 |
|
| |||||
| Raftlin | Raft-linking protein | Membrane association | MGCGLNKLEK RDEKRPGNIY | Human | 280 |
|
| |||||
| Serine/threonine-protein kinase H1 (PSKH1) | Protein serine kinase | Membrane association | MGCGTSKVLP EPPKDVQLDL | Human | 281 |
|
| |||||
|
Src family of tyrosine kinases Blk, Fgr, Fyn, Hck, Lck, Lyn, Src, Yes |
Tyrosine-protein kinase | Membrane association | MGLLSSKRQV SEKGKGWSPV | Mouse | 282 |
| MGCVFCKKLE PVATAKEDAG | Mouse | ||||
| MGCVQCKDKE AAKLTEERDG | Mouse | ||||
| MGCMKSKFLQ VGGNTFSKTE | Human | ||||
| MGCVCSSNPE DDWMENIDVC | Mouse | ||||
| MGCIKSKRKD NLNDDEVDSK | Mouse | ||||
| MGSNKSKPKD ASQRRRSLEP | Mouse | ||||
| MGCIKSKENK SPAIKYTPEN | Mouse | ||||
|
| |||||
| Src-like-adapter 2 (SLAP-2) | Adapter protein | Membrane association | MGSLPSRRKS LPSPSLSSSV | Human | 283 |
|
| |||||
| SSeCKS (A-kinase anchor protein 12, AKAP12) | Protein kinase C substrate | Membrane association | MGAGSSTEQR SPEQPAESDT | Mouse | 284 |
|
| |||||
| T-lymphoma invasion and metastasis inducing protein 1 (TIAM1) | RHO-like protein activator | Membrane association | MGNAESQNVD HEFYGEKHAS | Mouse | 285 |
|
| |||||
| TRIF-related adaptor molecule (TRAM) | Protein adaptor in Toll-like receptor 4 signal transduction. | Membrane association | MGIGKSKINS CPLSLSWGKR | Human | 286 |
|
| |||||
| Actin (ct Actin, 15 kDa) | Cytoskeletal protein | Mitochondrial targeting | GQVITIGNER FRCPEALFQP | Human | 287 |
|
| |||||
| BH3-interacting domain death agonist (p15, ctBID) | Apoptotic protein | Mitochondrial targeting | GNRSSHSRLG RIEADSESQE | Human | 213 |
|
| |||||
| Dihydroceramide Delta 4-desaturase 1 (DES1) | Ceramide biosynthesis | Mitochondrial targeting | MGSRVSREDF EWVYTDQPHA | Human | 288 |
|
| |||||
| NADH cytochrome b-5 reductase (b5R) | Cholesterol biosynthesis | Mitochondrial targeting | MGAQLSTLSR VVLSPVWFVY | Rat | 289,290 |
|
| |||||
| 2′-5′-oligoadenylate synthetase 2 | Cellular innate antiviral response | n/a | MGNGESQLSS VPAQKLGWFI | Human | 291 |
|
| |||||
| Brain-specific protein kinase C substrate (BASP-1, CAP-23/NAP-22) | Protein kinase C substrate | Protein interaction Transcriptional repression |
MGSKLSKKKK GYNVNDEKAK | Rat | 292,293 |
|
| |||||
| Calcineurin B homologous proteins 3 (CHP3) | Ca2+-binding proteins | Protein stability | MGAAHSASEE VRELEGKTGF | Human | 294,295 |
|
| |||||
| Tumor suppressor candidate 2 (FUS1) | Tumor suppressor | Tumor suppressor activity | MGASGSKARG LWPFASAAGG | Human | 296 |
3.3. Functions of Glycine Myristoylation
3.3.1. Cellular Localization and Membrane Attachment
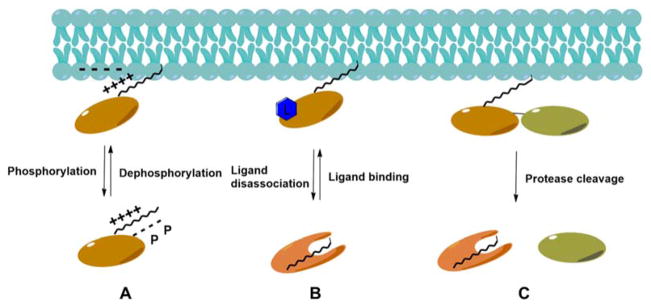
N-Glycine myristoylation mediates the targeting of modified proteins to various membranous locations (e.g., the plasma membrane, ER, Golgi complex, mitochondrial membranes, and nuclear envelope). However, glycine myristoylation alone is insufficient for membrane targeting, and another signal is typically required. This signal includes other proximate lipid modifications (e.g., cysteine palmitoylation or cysteine prenylation) and the presence of positively charged amino acid clusters.237 This requirement allows myristoylation to act as a “myristoyl switch” (Figure 14), in which the membrane association of myristoylated proteins is regulated by phosphorylation or ligands such as GTP and Ca2+.297 For example, the phosphorylation of myristoylated alanine-rich C kinase substrate (MARCKS) and Src stimulates membrane dissociation presumably by decreasing electrostatic interactions between the protein and the phospholipid membrane.298 On the contrary, GTP and Ca2+ have been shown to promote the membrane binding of myristoylated ADP ribosylation factors and recoverin, respectively.299–301 The binding of these ligands can induce conformational changes within proteins and results in the exposure of the N-myristoyl moiety for membrane association.297,300
Figure 14.
Myristoyl switch mechanisms. (A) The phosphorylation of N-glycine myristoylated protein stimulates membrane dissociation by interrupting the electrostatic interaction between proteins and the phospholipid. (B) Ligand binding enhances the membrane association of N-glycine myristoylated proteins. (C) Proteolysis triggers the release of N-glycine myristoylated protein from the membrane.
Proteolysis can also trigger a myristoyl switch.302 Human immunodeficiency virus (HIV)-1 Gag is initially synthesized in a 55 kDa precursor form (Pr55Gag), and the exposed myristoyl group promotes membrane binding. Upon cleavage by HIV-1 protease, the myristoyl moiety is sequestered and Gag is released from the membrane. The Gag myristoyl switch may not be induced by conformational changes as observed in other myristoyl switches, however.303 Instead, the synergistic interaction between Gag subdomains promotes the exposure of the myristoyl group and regulates membrane binding while protease cleavage of Gag decreases the cooperative effect and leads to the dissociation of Gag.
N-Glycine myristoylation also markedly increases the stability of hisactophilin, a membrane-binding protein in Dictyostelium discoideum.304 The modification also raises the protein dynamic (the rate of global protein folding and unfolding), which might facilitate conformational changes or myristoyl switching in hisactophilin.304
N-Glycine myristoylation functions not simply in membrane anchoring but also in the specific localization of certain transmembrane proteins. For example, NADH-cytochrome b5 reductase (b5R), an integral membrane protein, is dually targeted to the outer mitochondrial membrane and ER. The myristoylation of b5R is indispensable for targeting to the outer mitochondrial membrane, whereas a non-myristoylated mutant is localized to the ER.290 Notably, further study demonstrated that the myristoylation of b5R interferes with the recognition of the nascent peptide by the signal recognition particle, thereby preventing ER targeting.305
Another integral membrane protein that requires glycine myristoylation for localization is dihydroceramide Delta4-desaturase 1, an enzyme in the last step of de novo ceramide biosynthesis. In COS-7 cells, only the myristoyled form of this enzyme localizes to the mitochondria, which results in an increase in ceramide production. The non-myristoylatable mutant localizes primarily to the ER.288
The role of N-glycine myristoylation in controlling the cellular distribution of proteins has also been observed in yeast.295 Kimura and colleagues demonstrated that the N-glycine myristoylation of the Rpt2 subunit regulates the nuclear localization of the 26S proteasome, and the non-myristoylatable mutant of Rpt2 shifted the 26S proteasome into the cytoplasm without affecting its molecular assembly and peptidase activity.
3.3.2. Regulation of the Membrane Localization of Caspase Substrates in Apoptosis
The N-glycine myristoylation of some proteins occurs post-translationally. BID, a pro-apoptotic protein, was the first protein reported to undergo post-translational myristoylation.213 BID is cleaved by caspase-8 into a 7 kDa N-terminal fragment and a 15 kDa C-terminal fragment that remain associated as a complex. The exposed N-glycine of the BID C-terminal fragment is myristoylated to promote mitochondrial outer membrane targeting, thereby activating cytochrome C release and apoptosis, respectively.
Another caspase-cleaved protein, p21-activated kinase 2 (PAK2), is also post-translationally myristoylated.276 The myristoylation and the polybasic region are sufficient to relocalize the C-terminal of PAK2 (ctPAK2) from the cytosol to the plasma membrane and membrane ruffles. The overexpression of ctPAK2 has been shown to induce cell death.306 To investigate the role of myristoylation in apoptosis, the percentage of cell death was compared between myristoylatable and non-myristoylatable ctPAK2, the latter of which impaired the apoptotic effect. The non-myristoylatable mutant less efficiently activated Jun N-terminal kinase phosphorylation and signaling, a pathway known to be involved in apoptosis. To date, several caspase-cleaved proteins that undergo N-glycine myristoylation have been identified,238,287,307 and these findings emphasize the biological function of N-glycine myristoylation in the regulation of cell death.
3.3.3. Regulation of Protein-Protein Interaction
In addition to mediating protein localization and membrane targeting, N-glycine myristoylation plays a role in protein-protein interaction. Some of the examples described below are accompanied by structural evidence of this role. In examples that lack structural support, the effects on protein-protein interaction may be indirect.
CAP-23/NAP-22 is a brain-specific protein kinase C substrate involved in synaptic plasticity. The phosphorylation of CAP-23/NAP-22 by protein kinase C is regulated by calmodulin binding in a Ca2+-dependent manner. The myristoyl group and at least nine basic amino acids at the N-terminus are necessary for efficient interaction with calmodulin.292 A crystal structure of calmodulin in complex with the myristoylated CAP-23/NAP-22 N-terminal peptide shows that the myristoyl group is directly involved in calmodulin binding.308 The interaction between myristoylated alanine-rich C kinase substrate and calmodulin is also dependent on N-terminal myristoylation.309 However, the interaction between calmodulin and the HIV-1 Gag protein seems to occur independent of N-terminal myristoylation.310 Furthermore, the binding of calmodulin is thought to expose the N-terminal myristoyl group on Gag for membrane interaction.310 Notably, calmodulin also binds to farnesylated K-Ras4b in a nucleotide-independent manner. This interaction can occur even in the presence of negatively charged membranes, which suggests that calmodulin is able to extract K-Ras4b from membranes.311 By contrast, the PDE6δ–K-Ras4b interaction is less stable in the presence of negatively charged membranes, and thus it is unlikely that PDE6δ extracts K-Ras4b from membranes.312
Compared with the myristoylated form of the Goα protein, the non-myristoylated form shows decreased affinity for βγ subunits.313 The γ subunit of this protein is prenylated, and thus the increased binding affinity between α and βγ may be due to the targeting of both α and βγ to the membrane.
A role for N-glycine myristoylation in transcription has also been reported.293 The interaction of myristoylated CAP-23/NAP-22 (also called brain acid soluble protein 1 or BASP1) with PIP-2 is essential for the transcriptional corepression activity of Wilms’ tumor 1 (WT1), a transcriptional regulator involved in cell development. BASP1 binds to WT1 and mediates its transcriptional repression function. Notably, compared with WT BASP1, non-myristoylatable BASP1 shows significantly decreased transcriptional repression. The exact function of BASP1 myristoylation is unknown. However, non-myristoylatable BASP1 fails to recruit histone deacetylase (HDAC) 1 to the promoters of WT1 target genes and exhibits increased histone H3K9 acetylation,293 which suggests that myristoylation may regulate protein-protein interaction.
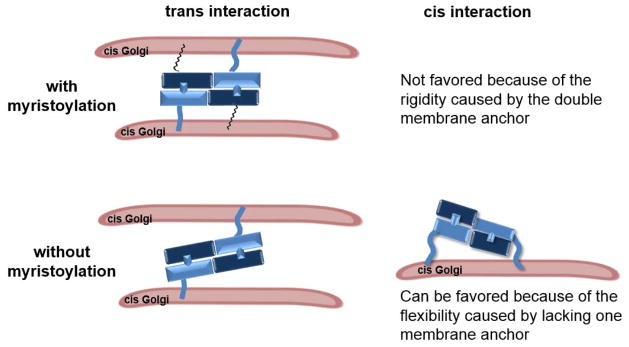
N-glycine myristoylation also regulates the Golgi membrane tethering process mediated by Golgi reassembly stacking protein (GRASP), which is required for the ribbon-like network of Golgi. GRASP undergoes myristoylation, and this modification is key to maintaining the structure of the Golgi network. The myristoylation of GRASP is thought to affect GRASP orientation and thus promote the trans interaction between GRASP proteins (a GRASP protein in one Golgi membrane interacting with a GRASP protein in a neighboring Golgi membrane) and prevent the cis interaction in the same membrane (Figure 15).314,315 A similar situation may explain the function of the myristoylation of Lunapark, a double-spanning integral membrane protein involved in ER network formation. The myristoylation of Lunapark is not required for specific membrane localization. Instead, the modification changes ER morphology by inducing polygonal tubular ER formation when the protein is overexpressed. This change is not observed for a non-myristoylated Lunapark mutant.239
Figure 15.
N-Glycine myristoylation may facilitate the trans interaction between Golgi reassembly stacking proteins by limiting conformational flexibility.
N-Glycine myristoylation has also been shown to mediate protein sorting into cilium. This process is mediated by two proteins, Uncoordinated 119a (Unc119a) and Unc119b.316 These proteins are homologous to PDE6δ, which binds to prenylated proteins (see section 2.4b). Notably, Unc119a and Unc119b recognize only myristoylated proteins, whereas PDE6δ recognizes only prenylated proteins.317 The structures of Unc119a and Unc119b in complex with the acylated peptides revealed that the recognition of myristoylated peptides by these proteins resembles that of prenylated peptides by PDE6δ.318,319 Notably, ADP ribosylation factor-like 2 and 3 release the bound prenylated and myristoylated proteins from PDE6δ and Unc119a and Unc119b, respectively, in a GTP-dependent manner.316,319
3.3.4. Regulation of Protein Stability
N-Glycine myristoylated calcineurin B homologous protein isoform 3 (CHP3) is a Ca2+ binding protein that plays a role in intracellular pH homeostasis by interacting with Na+/H+ exchanger (NHE1). CHP3 enhances the expression and stability of NHE1 at the cell surface through an unknown mechanism. N-myristoylation and the Ca2+ binding domain of CHP3 are not essential for interaction with NHE1.295 However, Gly2Ala and Ca2+ binding site CHP3 mutants decreased NHE1 half-life and exchange activity, which suggests that they are required for the stabilization of NHE1 at the plasma membrane and enhancement of Na+/H+ exchanger activity. Nevertheless, the underlying mechanism of this stabilizing effect by N-glycine myristoylation remains unknown and requires further investigation.295
3.3.5. Regulation of Enzymatic Activity
The best understood example of the regulation of enzymatic activity by myristoylation is the myristoyl switch that negatively regulates c-Abl tyrosine kinase activity. c-Abl is a member of the Src family of protein tyrosine kinases, which typically exist in an inactive state under resting conditions until activated through signaling.320 In addition to having a kinase domain, c-Src also has an SH2 and an SH3 domain. The SH2 domain binds to a phosphorylated tyrosine residue (pTyr527) and maintains c-Src in an inactive conformation. The SH3 domain binds a proline-rich sequence of c-Src and further locks c-Src in the inactive conformation. The activation of c-Src requires the binding of the SH2 domain to other phosphotyrosine residues, which unlocks the inactive conformation.320
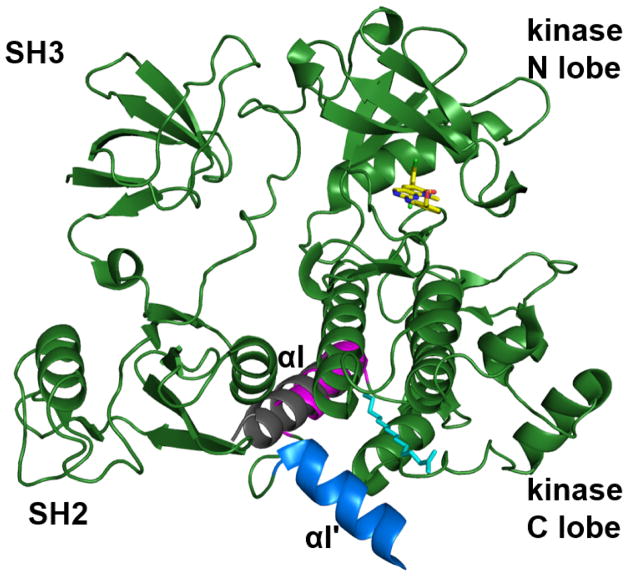
The c-Abl protein also has an SH2 and an SH3 domain N-terminal to the kinase domain. However, there is no pTyr corresponding to pTyr527 in c-Src. Thus, the mechanism through which c-Abl is maintained in an inactivate state is interesting: myristoylation of the N-terminal glycine plays a central role in maintaining this inactive form. Compared with the myristoylated form, unmyristoylated c-Abl is much more active.321 An X-ray crystal structure of a truncated c-Abl (containing the SH2, SH3, and kinase domains) with and without bound myristoyl peptide provides key insights on the regulation of c-Abl activity by myristoylation (Figure 16).322 The myristoyl group binds to a hydrophobic pocket in the C-lobe of the kinase domain, which triggers a conformational change in the C-terminal of the kinase domain. In the structure of c-Abl without bound myristoyl, an extended α-helix (αI, colored grey in Figure 16) prevents the binding of the SH2 domain to the kinase domain. In the myristoyl-bound state, the αI is separated into two short α-helices, αI (magenta in Figure 16) and αI′ (blue in Figure 16). The αI′ helix makes an abrupt turn to bind to the myristoyl group. These conformational changes lead to the docking of the SH2 domain onto the kinase domain and subsequent autoinhibition.322
Figure 16.
The myristoyl switch that regulates c-Abl activity. The c-Abl structure (PDB ID 1OPL) in complex with myristoyl and a kinase inhibitor is superimposed on the c-Abl structure without bound myristoyl (PDB ID 1M52). In the absence of myristoyl, an extended α-helix (αI, grey) prevents the binding of the SH2 domain to the kinase domain. In the myristoyl-bound state, the αI helix is separated into two shorter helices, αI (magenta) and αI′ (blue). The αI′ helix makes an abrupt turn to bind to the myristoyl group. This conformational change leads to the docking of the SH2 domain at the kinase domain and subsequent autoinhibition.
The Tyr kinase c-Src itself is also myristoylated. However, different from the regulation of c-Abl, myristoylation positively regulates c-Src kinase activity.214 The enhanced kinase activity of N-glycine-myristoylated c-Src is presumably due to a membrane attachment that orients c-Src favorably for kinase activity. The myristoylation of c-Src can also affect protein stability by regulating membrane association and facilitating ubiquitination and degradation mediated by the E3 ligase Cbl.214
3.4. Tools for the Study of Glycine Myristoylation
3.4.1. N-Myristoylation Predictive Tools
N-Glycine myristoylation predictive tools are bioinformatics methods that can predict potentially N-glycine myristoylated proteins. Three such tools are now available. The MYR Predictor (http://mendel.imp.univie.ac.at/myristate/) was first developed by Maurer-Stroh and co-workers.323 Based on known substrate sequences, crystal structures, and biochemical data of NMT, the motif for N-terminal myristoylation is 17 amino acids identified in three regions that (1) fit into the binding pocket, (2) interact with the NMT surface, and (3) form a hydrophilic linker. The second predictive tool, the Myristoylator (http://web.expasy.org/myristoylator/), predicts the N-terminal myristoylation of targets with neural network models trained to distinguish myristoylated and non-myristoylated proteins.324 The Myristoylator and MYR Predictor have similar error rates. Another software program, Terminator3 (http://www.isv.cnrs-gif.fr/terminator3/index.html), makes predictions based on pattern scanning.325 These predictive software tools require improvement in terms of sensitivity and accuracy.326
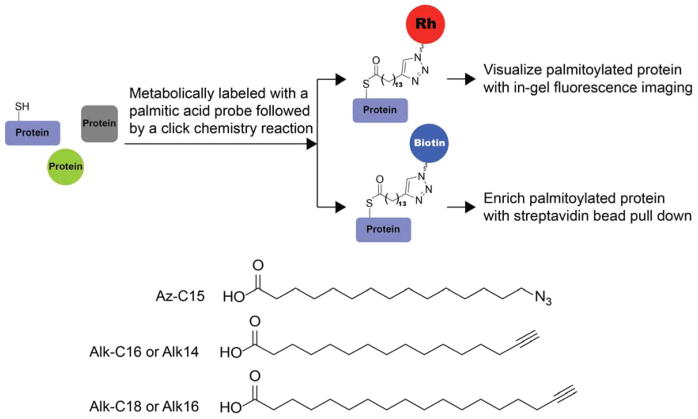
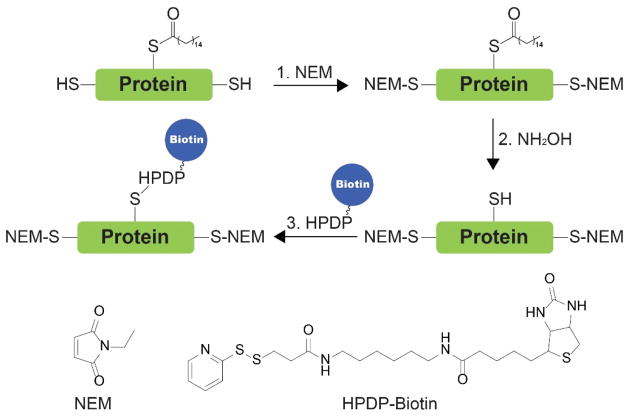
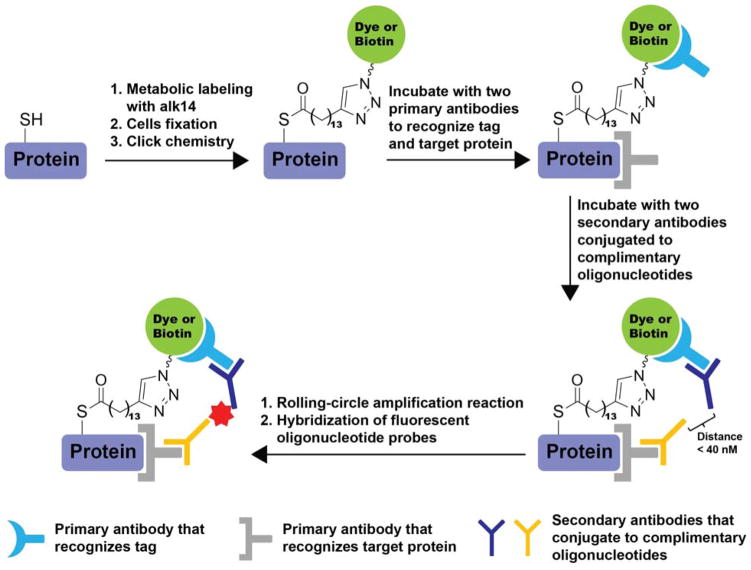
3.4.2. Chemical Tools for Detecting N-Myristoylation
Several approaches have been developed to detect N-glycine myristoylation in vivo and in vitro. The classic method uses radioactive-labeled fatty acids such as [3H]-myristic acid and [125I]-myristic acid, which are incorporated into cellular proteins, followed by the immunoprecipitation of target proteins and film exposure. This technique is typically time-consuming and insensitive. An alternative non-radioactive method has gained considerable attention since its development. This method uses ω-azido or ω-alkynyl myristate analogues as bioorthogonal probes to identify myristoylated proteins.327–329 These probes can be incorporated into proteins after addition into cultured cells, and the probe-modified proteins are then conjugated to fluorophores or biotin via the Staudinger ligation (for ω-azido probes) and the Huisgen cycloaddition reaction (for ω-alkynyl probes). The fluorophore- or biotin-conjugated myristoylated proteins can be detected via in-gel fluorescence after separation with sodium dodecyl sulfate polyacrylamide gel electrophoresis or western blot analysis.330
Several proteomics studies using bioorthogonal probes have been carried out to identify N-myristoylated proteins in various species, including T. brucei,331 Leishmania donovani,332 immortalized retinal pigment epithelial cells with and without herpes simplex virus (HSV) infection,333 CEMx174 cells with and without HIV infection,334 and HeLa cells with and without apoptosis.335 The study in HeLa cells is particularly notable because it uses NMT inhibitors in proteomics experiments to ensure that the identified proteins are indeed substrates of NMT. Furthermore, it compares the proteomics results with results predicted with the bioinformatics tools. This comparison shows that although the predication tools give largely correct predictions, some of the results are inconsistent with the proteomics results.335 The largest data set of experimentally validated human proteins myristoylated by NMT in living cells was obtained using a multifunctional enrichment reagent and NMT inhibitors.336
3.5. N-Glycine Myristoylation and Disease
3.5.1. NMT as a Target for Treating Fungal Infections and Parasitic Diseases
Several studies have shown that NMT is a potential target for antifungal225 and anti-parasite227,337,338 drugs because it is indispensable for the growth and viability of fungal and parasitic organisms. Moreover, compared with the myristoyl-CoA binding site, the peptide binding pocket of NMT is less well-conserved across species.339 The pocket can therefore be targeted for the development of selective NMT inhibitors. Several series of inhibitors (Figure 17) from high-throughput screening have been reported for NMTs in humans,340 parasites (P. falciparum, Leishmania sp., T brucei),338,341–344 and fungi.345–347
Figure 17.
Structures and half-maximal inhibitory concentration (IC50) values of representative inhibitors developed for NMTs in various species (CaNMT: Candida albicans NMT; HsNMT1/2, Homo sapiens NMT1/2; PfNMT, Plasmodium falciparum NMT; PvNMT, Plasmodium vivax NMT; and TbNMT: Trypanosoma brucei NMT).
Several peptidomimetic inhibitors were designed and synthesized to target Candida albicans NMT.348–350 These efforts lead to the development of an imidazole-substituted dipeptide that inhibits C. albicans NMT potently and selectively.348 RO-09-4879 and FTR1335, which are benzofuran346 and benzothiazole351,352 NMT inhibitors, respectively, were subsequently developed with high selectivity and promising properties as antifungal agents (Figure 17).
T. brucei NMT inhibitors have also been screened and developed.343 These pyrazole sulfonamide derived compounds strongly inhibit T. brucei NMT with selectivity over human NMT. Binding to the peptide substrate pocket of the enzyme, the inhibitor (DDD85646, Figure 17) kills T. brucei and cures trypanosomiasis in a mouse model of acute illness. These highly potent inhibitors thus pave the way for the development of therapeutic drugs for African sleeping sickness. These NMT inhibitors have also been used in proteomics studies to identify NMT substrate proteins in T. brucei331 and L. donovani.332
P. falciparum, a malaria parasite, contains a single NMT, and the inhibition of N-glycine myristoylation leads to the disruption of subcellular structure and cell death.338 Using bioorthogonal chemical probes and proteomics profiling of N-glycine myristoylated proteins, several P. falciparum NMT candidate substrates were identified with diverse biological functions, many of which are essential for parasite survival. Notably, enzyme inhibition using DDD85646, a compound originally developed for the T. brucei NMT, and a benzothiophene-containing compound (see Figure 17) results in the loss of inner membrane complex proteins required for parasite development and red blood cell invasion.338 NMT is therefore a promising target for the development of anti-malaria drugs.
3.5.2. NMT Inhibitors as Potential Cancer Treatments
NMT inhibitors have also been developed for cancer treatment. Myristoylated proteins are involved in cell signaling pathways and the apoptotic process (see the section 3.3 on the function of glycine myristoylation). Abnormalities in these proteins can lead to tumorigenesis. For example, N-glycine-myristoylated c-Src tyrosine kinase is activated in colon carcinoma.353 As mentioned in section 3.3e, N-glycine myristoylation can positively regulate c-Src kinase activity.214 Moreover, NMT expression and activity are increased in early stage rat and human colonic carcinogenesis.354 These results suggest that NMT might be a potential biomarker or target for colon cancer.355 Similarly, several studies have demonstrated that NMT expression is elevated in oral squamous cell carcinoma,356 gallbladder carcinoma,357 and brain tumors.235 Moreover, a cyclohexyl-octahydropyrrolo[1,2-a]pyrazine based NMT1 inhibitor, COPP-24 (Figure 17), has been shown to inhibit the proliferation of some tumor cancer cell lines.340 Another study showed that NMT inhibitors induce stress and an unfolded protein response in the ER, which led to apoptosis in several cancer cell lines.358
3.5.3. Viral and Microbial Utilization of Host Protein N-Glycine Myristoylation
Many viruses and bacteria exploit host N-glycine myristoylation systems for successful colonization. Several studies have shown that the N-myristoylation of certain viral proteins by host cell NMTs is critical for viral particle formation.359–361 The myristoylation of Gag, an HIV-1 structural protein, is crucial for viral replication and assembly.361 Also, the myristoylation of Nef (a virulence factor of lentiviruses) by NMT-1 facilitates viral replication.362 NMT1 and NMT2 have different specificities for the N-myristoylation of Gag and Nef.363 Therefore, NMTs have also been considered targets for antiviral drug development.
A study identified the novel demyristoylation activity of invasion plasmid antigen J (IpaJ) from the bacterial pathogen Shigella flexneri, which causes Golgi disruption in host cells.364 IpaJ is a cysteine protease that specifically recognizes and cleaves the amide bond after the N-myristoylated glycine residue. Several N-myristoylated proteins central in cell signaling and growth may be substrates for this enzyme. This discovery also suggests a new bacterial pathogenic mechanism that targets the N-glycine myristoylation of host cells.364
4. Cysteine Palmitoylation
Cysteine palmitoylation is the addition of a 16-carbon palmitoyl group via thioester bonds on protein cysteine residues (also known as S-palmitoylation; Figure 18). This reaction is highly reversible depending on the presence of enzymatic or non-enzymatic hydrolysis. Unlike other protein lipidations such as glycine N-myristoylation and cysteine prenylation, S-palmitoylation lacks a specific sequence motif. Thus, it is difficult to predict with precision which proteins will undergo the reaction. However, S-palmitoylation typically occurs on cysteines near or within a transmembrane domain or near a membrane-targeting PTM, such as prenylated cysteine or N-terminal myristoylated glycine.
Figure 18.
Reversible cysteine palmitoylation.
4.1. Palmitoyltransferases
4.1.1. Identification of the Cysteine Protein Acyltransferases
The covalent attachment of fatty acids to proteins was first observed in the early 1970s on a major structural protein found in bovine brain myelin.365,366 A later discovery that viral glycoproteins from the Sindbis virus contained a covalently linked palmitic acid on the side chain of an amino acid suggested that protein fatty acylation is prevalent.367 Additional protein substrates modified with palmitoyl groups were identified just a few years later, including G-protein-coupled receptors (GPCRs)368 and Ras proteins.369 The mechanism through which palmitoyl is attached to these protein substrates was not elucidated until 30 years after the first observation of the PTM. It is now known that the majority of protein palmitoylations are enzymatic events catalyzed by an evolutionarily conserved family of protein acyltransferases (PATs). These enzymes, which catalyze the attachment of a palmitoyl group to cysteine residues, were discovered in the early 2000s. Erf2–Erf4 were identified as an essential enzyme complex for the palmitoylation of Ras2 in S. cerevisiae.370 Erf2 or Erf4 alone cannot palmitoylate Ras2. The catalytic activity resides solely on Erf2, whereas Erf4 is required for the stable expression of Erf2. At the same time, Akr1 was identified as a PAT with activity against the yeast casein kinase Yck2.371 Erf2 and Akr1 share homology in a single domain, an aspartic acid-histidine-histidine-cysteine (DHHC) cysteine-rich domain (CRD), which is characteristic of palmitoyltransferases.
In 2004, the first mammalian protein with cysteine palmitoyltransferase activity was reported.372 The Golgi-apparatus-specific protein with the DHHC zinc finger domain (GODZ, also known as DHHC3) has PAT activity toward the γ-aminobutyric acid A receptor γ2 subunit and increases palmitoylation upon co-expression. DHHC3 palmitoylates the cytoplasmic loop domain of the γ2 subunit, which suggests that PAT activity functions in a cytosolic environment. Through a database search of the mouse and human genomes, 23 proteins were identified that have homology with the DHHC domain of DHHC3 (Table 2). Various members of this family have PAT activity.373 When transfected into COS7 cells, several DHHC enzymes increase the incorporation of 3H-palmitate into PSD-95, which suggests that the DHHC proteins are, in general, palmitoyltransferases.
Table 2.
Mammalian DHHC enzymes
| DHHC enzyme [Gene symbol] | Cellular localization | Size (# of a.a.) | Uniprot | Disease association | Known substrates | Tissue specificity, function, and other notes: |
|---|---|---|---|---|---|---|
| DHHC1 [zDHHC1] | ER, extracellular vesicular exosome | 485 | Q8WTX9 | NCDN374a | Expressed at high levels in fetal lung, kidney and heart. Expressed at lower levels in adult pancreas and lung. | |
| DHHC2 [zDHHC2] | plasma membrane, recycling endosome membranee | 367 | Q9UIJ5 | Lymph node metastasis and independently predicts an unfavorable prognosis in gastric adenocarcinoma patients; colorectal cancer.375 | Lck,376a R7BP (RGS7),377a CKAP4/p63,378a CD9 and CD151,379ad PSD-95373,380ab and GAP43,373b SNAP23/25,381b eNOS,382bd gp78,383b AKAP79/150384a | Ubiquitously expressed. Reduced expression in colorectal cancers with liver metastasis. |
| DHHC3 [zDHHC3] | Golgi | 327 | Q9NYG2 | (TARP γ-2, TARP γ-8, CNIH2, CaMKIIα, Syd-1, zyxin, TRPM8, TRPC1, HCRTR2),374b integrin α6/β4,385a TRAIL receptor-1,386a γ2 subunit of GABA(A),372bc NCDN,374a SNAP25/23,381b Gα,387a CSP,388b PI4KIIα,389a RGS4,390b STREX,391ac eNOS,382bd PICK1,392acd GluR1/2,393b PSD-95,373b CALHM1394, PPT1395b | DHHC3 and DHHC7 regulate GPCR-mediated signal transduction by controlling Gα localization to the plasma membrane. | |
| DHHC4 [zDHHC4] | Golgi, ER | 344 | Q9NPG8 | No known substrates | ||
| DHHC5 [zDHHC5] | Plasma membrane, dendrite | 715 | Q9C0B5 | Post-synaptic function affecting learning and memory.396 | δ-catenin,397a FLOT2,398a SSTR5,399adc GRIP1b,400acd STREX,391ac PLM401 | Notably, context-dependent fear conditioning in mice resulted in increased δ-catenin palmitoylation. Palmitoylated at non catalytic cysteine.402 First palmitoyl-transferase for a GPCR. Interaction with PSD-95 its PDZ domain Contains PDZ ligand. |
| DHHC6 [zDHHC6] | ER | 413 | Q9H6R6 | Calnexin,403a gp78,383b IP3R404 | ||
| DHHC7 [zDHHC7] | Golgi | 308 | Q9NXF8 | (TARP γ-8, CaMKIIα, Syd-1, NCDN)374b SNAP25/23,381b PSD-95,373b PI4KIIα,389a RGS4,390b Gα,387a CSP,388b estrogend, progesterone, and androgen receptors,405a STREX,391ac eNOS,382bd Glut4406a, Fas407b JAMC408a, Scribble,409 PPT1,395b NMNAT2410a | ||
| DHHC8 [zDHHC8] | Golgi, Cytoplasmic vesicle, mitochondrion | 765 | Q9ULC8 | Defects in the encoding gene may be linked to susceptibility to schizophrenia. Synaptic regulation. ZDHHC8 knockdown enhances radio-sensitivity and suppresses tumor growth in a mesothelioma mouse model. |
PICK1,392acd GRIP1b,400acd ABCA1,411b paralemmin-1,393 bd eNOS382bd | Phosphorylation regulated by PKMζ (brain-specific PKC variant protein kinase M). Contains PDZ ligand. |
| DHHC9 [zDHHC9] | Golgi, ER, cytoplasm | 364 | Q9Y397 | Mutations in the gene are associated with X-linked mental retardation. | H-Ras,412 N-Ras,412 STREX391ac | Forms a complex with Golgin subfamily A member 7 (GOLGA7, also known as GCP16). Highly expressed in kidney, skeletal muscle, brain, lung and liver. Absent in thymus, spleen and leukocytes. Highly expressed in microsatellite stable colon tumor. |
| DHHC10 [zDHHC11] | ER | 412 | Q9H8X9 | Gain of ZDHHC11 gene may be a potential biomarker for bladder cancer and non-small cell lung cancer. | NCDN374a | |
| DHHC11 [zDHHC23] | Plasma membrane | 409 | Q8IYP9 | KCNMA1,413a gp78383b | Interacts with nNOS.414 Expressed in the brain (at protein level), with highest levels in olfactory bulb, piriforc cortex and hippocampus. Highly expressed during the first week after birth.414 |
|
| DHHC12 [zDHHC12] | ER, Golgi | 267 | Q96GR4 | Alzheimer’s disease415 | No known substrates | |
| DHHC13 [zDHHC24] | ER416 | 284 | Q6UX98 | gp78383b | Contains a leucine rich partition. | |
| DHHC14 [zDHHC14] | ER9 | 488 | Q8IZN3 | Deletion may be linked to development delay417. Activation through chromosomal translocation in patients with acute biphenotypic leukemia.418 | No known substrates | Potential tumor suppressor. Decreased in testicular gerc cell tumors, prostate cancer.419 Overexpressed in Gastric Cancer.420 |
| DHHC15 [zDHHC15] | Golgi | 337 | Q96MV8 | Mutations in the gene cause X-linked mental retardation type 91.421 | CSP,388b SNAP25b,381b PSD-95,373b JNK3422 | Expressed in placenta, liver, lung, kidney, heart and brain.14 |
| DHHC16 [zDHHC16] | ER, cytoplasm423 | 377 | Q969W1 | PLN,424 DHHC6 | May be involved in ER stress-induced apoptosis regulation. Interact with c-ABl.425 |
|
| DHHC17 [zDHHC17] | Golgi, Golgi-associated vesicle membrane, cytoplasc vesicle membranes426 | 632 | Q8IUH5 | Memory and synaptic deficits in KO mice.427 | SNAP25/23,381b CSP,388b STREX,391ac ClipR-59,428 PSD-95,429b GAD65,429b SYT1,429b huntingtin,430a MPP1/p55,431a LCK,373b JNK3,432bd GLUR1,393b GLUR2,393b Caspase-6433 | Found in striatum, spring projection neurons and neurons affected by Huntington’s disease. Enriched in brain. High expression in the cortex, cerebellum, occipital lobe and caudate. Low expression in the spinal cord. Also detected in the testis, pancreas, heart and kidney, but not in the liver or lungs.426 ZDHHC17 is the only palmitoyl-transferase in erythrocytes.431 Contains 5 ankyrin repeat domains. Has transforming activity. Mediates Mg2+ transport. Low extracellular Mg2+ induces its level inside the Golgi and post Golgi membrane vesicles. Interacts with JNK434. |
| DHHC18 [zDHHC18] | Golgi | 388 | Q9NUE0 | H-Ras,373b Lck373b | Expressed ubiquitously | |
| DHHC19 [zDHHC19] | ER | 309 | Q8WVZ1 | R-Ras435b | Is prenylated.435 | |
| DHHC20 [zDHHC20] | Plasma membrane | 365 | Q5W0Z9 | EGFR,436 Peptide mimic of N terminal myristoylated proteins,437 δ-catenin,397b CALHM1394 | Overexpression of DHHC20 causes cellular transformation. Expressed mainly in lung388. Low expression in ovary, breast, and kidney tissue. High expression in thyroid, liver, colon and prostate tissue. | |
| DHHC21 [zDHHC21] | Golgi, plasma membrane | 265 | Q8IVQ6 | Loss of protein function result in delayed hair shaft differentiation and hyperplasia of interfollicular epidermis and sebaceous glands. | Estrogend, progesterone, and androgen receptors,405a PECAM-1,438a Fyn,439bc eNOS382ad | Expressed in epithelial tissue439 and hair follicles. |
| DHHC22 [zDHHC13] | Golgi, Golgi-associated vesicle membrane440, ER, intracellular membrane bound organelle | 622 | Q8IUH4 | ZDHHC13 deficient mice develop alopecia, amyloidosis and osteoporosis441 and have reduced bone mineral density.442 Huntington’s disease |
MT1-MMP,442bc huntingtin,443b CNFN444a | Contains 6 Ankyrin repeats. Interacts with huntingtin. Regulates fate specification of ectoderc and mesoderc cell lineages by modulating Smad6 activity.445 |
| DHHC24 [zDHHC22] | Golgi, ER | 263 | Q8N966 | KCNMA1,413a gp78383b | Gene expression microarray analysis shows zDHHC22 upregulated in cattle with MIMT1 deletion.446 |
- knockdown evidence
- overexpression evidence
= murine orthologue
= protein/substrate interaction
- localization may depend on cell type
4.1.2. Topology of Palmitoyltransferases
DHHC proteins are predicted to have a common topology comprising several trans-membrane domains (TMDs) and a conserved DHHC CRD active site on the cytosolic face (Figure 19). The number of TMDs ranges between four (DHHC1, DHHC2) and six (DHHC13, DHHC17). This conserved DHHC CRD is generally located in the middle of the enzyme on the cytoplasmic loop between TMD2 and TMD3. At the C- and N-terminal cytosolic domains, there is less homology among the family members. The variable domains include a predicted SH3 domain in DHHC6 and ankyrin repeats in DHHC13 and DHHC17. These variable domains and sequences at the N- and C-termini mediate protein–protein interactions, a key mechanism for the interaction of substrates and PATs. For example, DHHC17 and huntingtin interact through the ankyrin repeats on DHHC17.393 DHHC5 and DHHC8 interact with glutamate receptor-interacting protein 1b (GRIP1b) through the PDZ domains at the C-terminal end of DHHC5 and DHHC8.381,393,400 DHHC5 also interacts with cardiac phosphoprotein phospholemman via the C-terminal domain.401
Figure 19.
Predicted topology and domain structure of DHHCs. TMD, transmembrane domain.
Additionally, the DHHC family has long been annotated as zinc finger proteins and newer experimental evidence has demonstrated that DHHCs bind zinc ions. Zinc binds to the CRD of DHHCs and is crucial for enzyme stability. Generally, the DHHC CRD can be considered a stable core that is conserved among the family members, whereas the N- and C-termini are more disordered to allow for variable protein-protein interactions.401 These features are discussed in section 4.1c below. The lack of crystal structures of the catalytic domain currently limits our understanding of these enzymes.
4.1.3. Substrate Specificity of DHHCs
Many factors, such as potential protein interacting domains, the amino acid composition of the modification site, and cellular localization, determine the substrate specificity of PATs. These factors are discussed here.
In general, DHHCs have substrate specificity with some redundancies. When certain PATs are inactivated, a loss of modification occurs on specific proteins in yeast.447 Yeast Erf2p can palmitoylate substrates other than yeast Ras. However, the level of palmitoylation is weak (~5% of Ras palmitoylation). These results suggest that PATs can show strong preferences for specific substrates. In mammalian cells, co-expression studies confirm that specific DHHCs modify specific substrates: the palmitoylation of Lck, a tyrosine kinase, is increased upon overexpression of DHHC17 and DHHC18, that of SNAP-25b and Gαs by the overexpression of DHHC3 and DHHC7, Ras by DHHC18 and DHHC9, PSD-95 and GAP-43 by DHHC2 and DHHC15, and paralemmin by DHHC8.373,393,412 DHHC17 can also palmitoylate huntingtin, SNAP-25, PSD-95, GAD-46, and synaptotagmin I,429 and DHHC3 can palmitoylate endothelial nitric oxide synthase (NOS), GluR receptors, and GAP-43.382,448 GRIP1b palmitoylation is incompletely abolished when DHHC5 or DHHC8 is individually knocked down with short hairpin RNA, but double knockdown completely abolishes palmitoylation.400 In general, a palmitoylated substrate may be modified by more than one DHHC. Notably, although some DHHCs may appear to have highly specific substrate targets, such as DHHC19 and its only substrate R-Ras,449 the vast majority of palmitoylation events have yet to be assigned to the enzymatic activity of a specific DHHC. The closely related DHHC3 and DHHC7 have broad substrate specificities that allow for redundancies to be built into the regulation of protein palmitoylation. These redundancies may serve to ensure proper palmitoylation in the event that one DHHC is compromised.
The co-localization of a DHHC with its substrate ensures that the correct palmitoylation event occurs. DHHCs have distinct cellular localizations including the plasma membrane, ER, Golgi, and endosomal membranes. The exact mechanism through which DHHCs are properly sorted is unknown. However, several studies have advanced understanding of DHHC sorting and localization. The C-terminal portion of DHHC2 and DHHC15 regulate the localization of these two distinctly localized PATs.450 Swapping the C-terminal region of DHHC2 to DHHC15 altered the localization of the chimeric DHHC15 to regions similar to those of WT DHHC2. DHHC4 and DHHC6 were later found to sort to the ER through a canonical dilysine motif that interacts with coat protein complex 1. The five C-terminal amino acids containing the dilysine motif of DHHC4 or DHHC6 are also sufficient to relocalize the Golgi-specific DHHC3 to the ER.451
External stimuli may alter the localization of DHHC enzymes.380 In dendritic cells, palmitoylated PSD-95 localizes to the dendritic spine and, upon depalmitoylation, translocates to the shaft where it can be repalmitoylated by DHHC2-containing vesicles for shuttling back to the spine. When synaptic activity is blocked, DHHC2 relocalizes to the spine to increase PSD-95 palmitoylation levels to upregulate 2-amino-3-(hydroxy-5-methyl-4-isoxazole) propionic acid type glutamate receptor activity to maintain homeostasis. However, localization alone is insufficient to confer substrate specificity. For example, in human embryonic kidney 293T cells, up to 11 DHHCs are associated with the Golgi complex upon expression.416
The method through which DHHC substrate pairs have been identified has usually relied on what is known as the Fukata screen, in which individual DHHCs are ectopically overexpressed with a potential substrate. This process generates a panel of DHHCs capable of increasing the palmitoylation levels of the substrate. Next, the DHHCs are knocked down, and decreased palmitoylation after knockdown verifies the substrate–enzyme pair. However, the knockdown of a DHHC that can increase palmitoylation levels does not always result in complete or decreased palmitoylation. This phenomenon is likely attributable to the redundancies of the DHHCs. On the contrary, the overexpression of a DHHC could disrupt the fine localization of the enzyme.452 It was reported that decreased PSD-95 palmitoylation levels were not observed in DHHC3 knockout mice, whereas the ectopic expression of DHHC3 with PSD-95 increased palmitoylation levels in cells. Endogenous DHHC3 predominately localizes to the cis Golgi membranes, and the overexpression of DHHC3 disrupts the localization of endogenous DHHC3.452 Mislocalized enzymes that retain their activity could easily acylate substrates other than their natural substrates (false positives). This study highlighted that downside to the use of the Fukata screen for the identification of enzyme–substrate pairs, which is further complicated by the fact that single knockdown/knockout experiments do not always completely abolish substrate palmitoylation. Although there are robust examples of DHHC-substrate pairs, other substrates may be palmitoylated by several DHHCs. One example is N-Ras, in which palmitoylation decreases but persists at low levels in vivo when DHHC9 is knocked out.453 This divergence from a single enzyme - single substrate system highlights the complexity of protein palmitoylation and the challenges in elucidating the mechanism of palmitoylation regulation.
The variable N- and C- terminal domains of DHHCs play key roles in substrate specificity, whereas the conserved catalytic core contributes little.381 A chimeric DHHC15 construct containing the DHHC CRD of DHHC3 (DHHC15/3) failed to palmitoylate SNAP23, a substrate modified by WT DHHC3 but not WT DHHC15. This outcome suggests that the DHHC CRD of DHHC3 is insufficient to confer substrate specificity to SNAP23. DHHC17, also known as huntingtin-interacting protein 14 (HIP14), contains an ankyrin repeat domain that interacts with an N-terminal fragment of huntingtin.426 Although DHHC3 cannot interact with huntingtin, when the ankyrin repeat domain of DHHC17 is fused to DHHC3, the chimeric protein interacts with huntingtin and redistributes it to the perinuclear region through palmitoylation-dependent vesicular trafficking.393 This result and the DHHC15/3 chimera data suggest that substrate specificity is determined by the N- and C-termini of the enzyme. Additionally, DHHC23, also called neuronal NOS-interacting DHHC domain-containing protein, interacts with the PDZ domain on neuronal NOS through its PDZ-interacting EDIV motif.414 Several DHHCs contain PDZ-interacting domains that allow for enzyme–substrate interactions, which indicates that these DHHCs use such interactions to mediate substrate specificity.396,400 The interactions between a PAT and its substrate can be weak and transient, but increasing evidence suggests that stronger interactions exist, such as those between the ankyrin repeat of DHHC17 and huntingtin, DHHC3 and the γ-aminobutyric acid A receptor γ2 subunit,454 and DHHC8 and paralemmin.393
Crystal structures of DHHC–substrate complexes would shed invaluable insight on these interactions. Challenges inherent to the crystallization of membrane-bound proteins impede progress; however, several non-catalytic domains of DHHCs have been crystalized. The interaction of DHHC5 with its substrate, phosphoprotein phospholemman, has been studied and the binding site has been mapped to the disordered C-terminal tail of DHHC5.401 Another study 455 identified a unique ΨβXXQP motif in the substrates of DHHC17. This motif centers on glutamine and proline (QP) residues, whereas the other four residues are more variable. The motif is found in multiple DHHC17 and DHHC13 substrates and interacts with the ankyrin repeat domains found in these DHHCs. Crystal structures 456 of the ankyrin repeat domain of DHHC17 and a truncated form of Snap25b have elucidated the nature of the interaction, attributing it primarily to hydrogen bondings and hydrophobic interactions involving the QP motif of Snap25b. This QP dipeptide motif is present in all of the DHHC17 substrates, including Htt, and the loss of the QP motif in Htt disrupts DHHC17 binding.
The amino acid sequence in the vicinity of the palmitoylation site on the substrate is also important for PAT substrate recognition. The palmitoylation of PSD-95 has been shown to depend on the first 13 amino acids, MDCLCIVTTKKYR. The two modified cysteines are surrounded by hydrophobic residues (Leu4, Ile6, and Val7), and mutations of these amino acids to a hydrophilic serine residue result in mislocalization and much weaker palmitoylation, whereas mutations of the hydrophilic residues Asp2, Thr8, or Thr9 to alanine do not alter localization.457 SNAP23, which is not a substrate for DHHC15, can be acylated by DHHC15 when Cys79 (a residue close to the cysteine residue to be palmitoylated) is mutated to phenylalanine, because the resulting Cys79Phe mutant is highly similar to SNAP25b (a substrate for DHHC15) in terms of the number and configuration of cysteines in its CRD.381 Additional work further highlighted the importance of the secondary structure near the palmitoylation site.458 A 21 amino acid sequence enriched in aromatic amino acids, predicted to be an amphiphatic α-helix, near the Cys739 palmitoylation site of the sodium–calcium exchanger (NCX) is essential for NCX acylation. The most surprising discovery was the capability of this sequence to convert non-palmitoylated cysteines to bona fide modification sites when introduced adjacently, which demonstrates that fine structural elements exist to ensure that the correct cysteine is modified by the relatively promiscuous enzymatic activity of DHHCs. Thus, not only the amino acid sequences surrounding the palmitoylation site but also high-order structural elements on the substrate are critical.
There is limited evidence supporting the hypothesis that various DHHCs prefer particular types of substrates. For example the S. cerevisiae PAT Swf1 targets transmembrane proteins with juxtamembrane cysteine residues, whereas the substrates for Akr1 are mainly soluble proteins.447 The differential bias may be due simply to the small number of substrates identified for Akr1 and Swf1, however, and there is insufficient evidence for a definitive conclusion. Mammalian DHHCs may also be biased toward certain substrate types. Not surprisingly, substrates of the promiscuous DHHC3 and DHHC2 include both cytoplasmic and integral membrane proteins with various numbers of transmembrane domains (Table 2). In contrast to the involvement of DHHC2 and DHHC3 in many pathways, both DHHC15 and DHHC21 are less promiscuous and prefer cytosolic proteins in developmental singaling pathways as substrates. Tables 3–9 summarize known S-palmitoylated proteins according to whether they are cytoplasmic or transmembrane. Many substrate proteins are either integral membrane proteins or undergo prenylation or myristoylation that targets them to membranes in which DHHCs are localized. Notably, the reported palmitoylation sites of the majority (>95%) of palmitoylated single-pass integral membrane proteins are located either directly adjacent to or inside the annotated transmembrane domain. Furthermore, S-palmitoylation normally occurs close to the N-glycine myristoylation or C-terminal prenylation site for cytoplasmic proteins. The fact that S-palmitoylation occurs next to a transmembrane domain or another lipid modification is likely determined by the proximity of these sites to the DHHC active site. On the contrary, many S-palmitoylated proteins lack transmembrane domains or other lipid modifications that could recruit them to membrane-localized DHHCs (Table 8). These proteins may be recruited to membranes via interaction with membrane-localized proteins. For example, PSD-95 is recruited to synapses by the transmembrane protein ephrin-B3.459
Table 3.
Type I and III single-pass transmembrane proteins that undergo S-palmitoylationa
| Protein | Uniprot# | Palmitoylation sites | Modification site location | Sequence near the Cys | Function of palmitoylation | DHHC enzyme | Ref. |
|---|---|---|---|---|---|---|---|
| AATYK1 | Q6ZMQ8-1 | C52, C54, C57 | Cytoplasmic domain |
|
Required for endosomal localization | 525 | |
| BACE1 | P56817 | C474, C478, C482, C485 | C474, C478: inside TM domain; C482, C485: cytoplasmic domain |
|
Required for lipid rafts localization | 526 | |
| CD4 | P01730 | C419, 422 | Cytoplasmic end of TM domain |
|
Required for lipid rafts localization | 527 | |
| CHL1 | P70232 | C1102 | Inside TM domain |
|
Required for lipid rafts localization | 528 | |
| CPD | O75976 | C1317, C1321, C1323 | Inside TM domain |

|
Required for Golgi exit | 529 | |
| c-Met | P08581 | C882, C894 | Internal extracellular domain |

|
Regulates trafficking and stability | 530 | |
| DSG2 | Q14126 | C635, C637 | Cytoplasmic end of TM domain |
|
Required for regulating desmosome dynamics | 531 | |
| DR4 | O00220 | C261, C262, C263 | Extracellular end of TM domain |
|
Required for cell death signaling | DHHC3 | 532 |
| EGFR | P00533 | C1025, C1122 | Cytoplasmic C terminus |

|
Required for EGFR signaling | DHHC20 | 436 |
| FAS | P25445 | C199 | Cytoplasmic domain |
|
Required for cell death signaling | 533 | |
| FcγRIIa | P12318 | C208 | Cytoplasmic end of TM domain |

|
Required for lipid rafts localization | 534 | |
| IFNAR1 | P17181 | C463 | Cytoplasmic domain |
|
Required for Stat1 and Stat2 activation | 535 | |
| LAT | O43561 | C26, C29 | Cytoplasmic end of TM domain |
|
Required for lipid rafts localization | 536 | |
| LRP6 | O75581 | C1394, C1399 | Cytoplasmic end of TM domain |

|
Required for ER exit | 537 | |
| MICA | Q29983 | C306, C307 | Extracellular end of TM domain |
|
Required for plasma membrane localization | 538 | |
| MT1-MMP | P50281 | C574 | Cytoplasmic domain |

|
Required for promoting cell migration | DHHC22 | 539 |
| MUC1 | P15941 | C1184, C1186 | Cytoplasmic end of TM domain |
|
Required for recycling from endosomes to plasma membrane | 540 | |
| N-CAM | P13591 | C723, C729, C734, C740 | Cytoplasmic domain |
|
Required for plasma membrane localization | 541 | |
| Neurofascin | O94856 | C1236 | Cytoplasmic end of TM domain |
|
Required for targeting a specialized membrane microdomain | 542 | |
| Nicastrin | Q92542 | C689 | Cytoplasmic domain |

|
Required for lipid rafts localization, important for protein stability | 543 | |
| P0 glycoprotein | P06907 | C181 | Cytoplasmic domain |
|
Unknown | 544 | |
| p75(NTR) | P08138 | C278 | Cytoplasmic domain |

|
Required for cleavage by gamma-secretase | 545 | |
| PECAM-1 | P16284 | C622 | Cytoplasmic domain |
|
Required for efficient PECAM-1-mediated cytoprotection | DHHC21 | 546 |
| PLM | O00168 | C60, C62 | Cytoplasmic domain |
|
Required for inhibitory effect of PLM on the sodium pump | 547 | |
| PRCD | Q00LT1 | C2 | Cytoplasmic domain |
|
Enhance protein stability | DHHC3 | 548 |
| Syt VII | O43581 | C35, C38, C41 | C35, C38: Inside TM domain; C41: Cytoplasmic domain |
|
Required for lysosome localization | 549 | |
| TF | P13726 | C277 | Cytoplasmic end of TM domain |
|
Negative regulatory mechanism of Ser258 phosphorylation | 550 | |
| TGF-α | P01135 | C153, C154 | Cytoplasmic domain |
|
Required for TGF-α complex formation | 551 | |
| TMX | Q9H3N1 | C205, C207 | Cytoplasmic domain |
|
Required for targeting mitochondria-associated membrane | 552 |
Both type I and type III membrane proteins contain one transmembrane domain with a cytoplasmic C-terminus. Type I membrane proteins have a signal peptide that is cleaved in the mature form, whereas type III proteins do not.
Table 9.
Secreted extracellular proteins that undergo S-palmitoylation
| Protein | Uniprot# | Palmitoylation sites | Modification site location | Sequence near the Cys | Function of palmitoylation | Ref |
|---|---|---|---|---|---|---|
| ApoB | P04114 | C1112 | Internal domain |

|
Required for proper assembly of the hydrophobic core of lipoprotein particle | 668 |
| Mucin 2 | Q02817 | C12, C163, C166 | Internal domain |

|
Required for protein secretion | 669 |
Table 8.
S-palmitoylated proteins without other membrane-targeting signals (transmembrane domains, N-terminal glycine myristoylation, or C-terminal cysteine prenylation)
| Protein | Uniprot# | Palmitoylation sites | Modification site location | Sequence near the Cys | Function of palmitoylation | DHHC enzyme | Ref. |
|---|---|---|---|---|---|---|---|
| APT1, APT2 | O75608, O95372 | C2 | N terminus |
|
Required for plasma membrane localization | 504 | |
| ERBIN | Q96RT1 | C14, C16 | N terminus |
|
Required for plasma membrane localization | 627 | |
| Flotillin-1 | O75955 | C34 | N terminus |
|
Regulates IGF-1 signaling | 628 | |
| G14α | O95837 | C5, C6 | N terminus |
|
Required for plasma membrane localization | 629 | |
| G16α | Q9N2V6 | C9, C10 | N terminus |
|
Required for plasma membrane localization | 629 | |
| GAP43 | P17677 | C3, C4 | N terminus |
|
Required for plasma membrane localization | DHHC2 | 630 |
| Gqα | P50148 | C9, C10 | N terminus |
|
Required for plasma membrane localization | 631 | |
| Gα11 | P29992 | C9, C10 | N terminus |
|
Required for plasma membrane localization | ||
| Gα13 | Q14344 | C14, C18 | N terminus |
|
Required for plasma membrane localization | 632 | |
| GRIP1b | Q925T6-2 | C11 | N terminus |
|
Required for targeting dendritic endosomes | DHHC5 DHHC8 |
400 |
| LIMK1 | P53667 | C7, C8 | N terminus |
|
Regulates actin dynamics | 633 | |
| L-type calcium channel | Q08289 | C3, C4 | N terminus |
|
Required for targeting dendritic endosomes | 634 | |
| Melano-regulin | Not mapped | N terminus | LRTVCCCCGCECLE ERALPEKE | Required for targeting melanosome membrane | 635 | ||
| NOS2 | P35228 | C3 | N terminus |
|
Required for NOS2 activity | 636 | |
| p63RhoGEF | Q86VW2 | C23, C25, C26 | N terminus |
|
Required for plasma membrane localization | 637 | |
| PDE10A | Q9Y233 | C11 | N terminus |
|
Required for transport throughout dendritic processes | DHHC7 DHHC19 |
638 |
| PSD-95 | P78352 | C3, C5 | N terminus |
|
Modulate synaptic plasticity | DHHC2/3/7/15/17 | 639 |
| RGS16 | O15492 | C2, C12 | N terminus |
|
Required for lipid rafts localization | 640 | |
| AR8 | G4VV16 | C558, C560 | C terminus |
|
Required for plasma membrane localization | DHHC7 DHHC21 |
641 |
| GRK6 | P43250 | C561, C562, C565 | C terminus |

|
Increase kinase activity | 642 | |
| JNK3 | P53779 | C462, C463 | C terminus |
|
Required for Golgi complex localization | DHHC15 DHHC17 |
432 |
| R7BP | Q6MZT1 | C252, C253 | C terminus |
|
Required for nucleus to plasma membrane trafficking | DHHC2 | 643 |
| AKAP79/150 | P24588, D3YVF0 | C36, C129 | Internal domain |
|
Required for dendritic endosomal targeting | 644 | |
| AnkG | Q12955 | C70 | Internal domain |
|
Required for plasma membrane localization | 645 | |
| Bet3 | O43617 | C68 | Internal domain |

|
Required for vesicular trafficking | 646 | |
| Caveolin-1 | Q03135 | C133, C143, C156 | Internal domain |
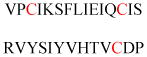
|
Unknown | 647 | |
| CSP | Q03751 | CRD domain (14 cysteines) | Internal domain |
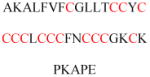
|
Required for plasma membrane localization | DHHC3/7/15/17 | 388 |
| DJ-1 | Q99497 | C46, C53, C106 | Internal domain |

|
Required for lipid rafts localization | 648 | |
| ERα | P03372 | C447 | Internal domain |
|
Regulates 17β-estradiol-induced ERα degradation and transcriptional activity | ZDHHC7 ZDHHC21 |
649 |
| Flotillin-1 | O75955 | C34 | Internal domain |
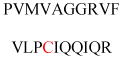
|
Required for plasma membrane localization | 650 | |
| GAD65 | Q05329 | C30, C45 | Internal domain |

|
Required for vesicle-associated trafficking | DHHC17 | 651 |
| HMGCS2 | P54868 | C166, C305 | Internal domain |
|
Enhance interaction with PPARalpha | 652 | |
| Huntingtin | P42858 | C212 | Internal domain |
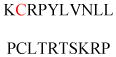
|
Required for Huntingtin activity | DHHC17 DHHC22 |
430 |
| KChIP2 | Q9NS61 | C45, C46 | Internal domain |
|
Required for cell surface protein expression | 653 | |
| MAP3K12 | Q12852 | C127 | Internal domain |
|
Regulates vesicle targeting | 654 | |
| Ndel1 | Q9GZM8 | C273 | Internal domain |
|
Required for cytoplasmic dynein activity | 655 | |
| nSMase2 | Q9NY59 | C53, C54, C59, C397, C398 | Internal domain |
|
Required for plasma membrane localization | 656 | |
| SCRIB | Q14160 | C4, C10 | Internal domain |
|
Required for tumor-suppressive activities | DHHC7 | 409 |
| SNAP25 | P60880 | C85, C88, C90, C92 | Internal domain |
|
Required for plasma membrane and endosomes localization | DHHC2 DHHC3 |
657 |
| Stomatin | P27105 | C30, C87 | Internal domain |

|
Required for plasma membrane localization | 658 | |
| TEAD1 | P28347 | C359 | Internal domain |
|
Regulates transcriptional output | 659 | |
| Tubulin | P68370 | C20, C213, C305, C376 | Internal domain |

|
Required for targeting microtubules and intracellular membranes | 660 | |
| PHB | P35232 | C69 | Internal domain |
|
Required for membrane translocation and interaction with EHD2 | 661 | |
| PLD | Q13393 | C240, C241 | Internal domain |

|
Required for targeting caveolin-enriched membrane | 662 | |
| PI4KIIα | Q9BTU6 | C174, C175, C177, C178 | Internal domain |
|
Required for catalytic activity | DHHC3 DHHC7 |
663 |
| RGS2 | P41220 | C106, C116, C199 | Internal domain |

|
Inhibit the GTPase-activating activity | 664 | |
| RGS7 | P49802 | C133 | Internal domain |

|
Required for plasma membrane localization | 665 | |
| RGS10 (isoform2) | O43665 | C60 | Internal domain |
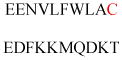
|
Required for RGS10 activity | 666 | |
| RPE65 | Q16518 | C231, C329, C330 | Internal domain |
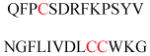
|
Required for isomerohydrolase activity of RPE65 | 667 |
PATs not only have broad specificity for protein substrates but also display broad specificity for the acyl-CoA co-substrate. Although palmitoyl-CoA (C16) is the preferred substrate, other long-chain acyl-CoAs such as myristoyl-CoA (C14) and stearoyl-CoA (C18) are also efficiently transferred by PATs. DHHC2 transfers acyl-CoAs with various chain lengths and degrees of saturation. DHHC15 also has a promiscuous fatty acyl-CoA substrate profile.373 The broad specificity indicates that cells can utilize various fatty acyl-CoA to modify the activity of PAT substrates depending on the metabolic state of the cell. DHHC3, however, exhibits a more stringent acyl-CoA substrate profile and efficiently transfers only C14 and C16 acyl groups.460 This specificity is independent of the protein substrate, which indicates a level of control to prevent the incorrect modification of DHHC substrates by other lipids. A more in-depth study of acyl-CoA substrate specificity 461 expanded previous studies by analyzing a larger number of acyl groups and DHHCs. The results supported the finding that each DHHC has individual acyl-CoA preferences. Surprisingly, DHHC3 and DHHC7, which have highly similar protein sequence, have different acyl-CoA substrate preferences: DHHC7 prefers the longer C18 groups whereas DHHC3 prefers shorter C14 and C16 groups. The determining factor was isolated through mutagenesis studies to be a single isoleucine in the third transmembrane domain of DHHC3. When the isoleucine on DHHC3 is mutated to serine, as found on DHHC7, the mutant utilizes C18 groups.
Notably, this review and the studies cited generally assume that palmitate is the acyl group being attached by the DHHC PATs. Although this attachment is the most likely event, a general lack of mass spectrometry (MS) data confirming the identity of the modification catalyzed by individual DHHCs leaves open the possibility that other acyl groups are being attached by this family of PATs. This possibility is supported by the observation that other fatty acids, such as arachidonate, eicosapentaenoate, palmitoleic acid, and stearic acid, reportedly attach to protein substrates through thioester bonds.462–465 Cysteines modified with 14:0, 18:0, 18:1, and 18:2 fatty acids were detected in bovine heart and liver tissue.466 S-acylation with stearate and arachidonate also occurs on the Gα subunit, myelin, the G2 protein of the Rift Valley fever virus, and the asialoglycoprotein receptor.464,465,467–469
Although many proteins are known to be palmitoylated, associating the modifications to the actions of specific DHHCs is difficult for several reasons, including PAT redundancy, the lack of clearly defined recognition sequences, difficulty associated with obtaining purified DHHCs, and the deconvolution of enzymatic versus non-enzymatic protein palmitoylation. However, it would not be surprising to find that most, if not all, cysteine S-palmitoylation events are mediated by DHHCs.
4.1.4. Mechanism of Palmitoylation
Cysteine palmitoylation forms a thioester bond that is similar in energy to the thioester bond in the palmitoyl donor, palmitoyl-CoA. Thus, the overall the reaction is energy-neutral, and no energy source (i.e., ATP) is needed. Indeed, purified PATs can directly modify their substrate using palmitoyl-CoA in the absence of an energy source.373
DHHCs themselves are autoacylated in vivo and in vitro.370,371,373 Incubating 3H-palmitoyl-CoA with partially purified yeast Erf2/Erf4 in the absence of Ras2 substrate results in the formation of 3H-labeled Erf2.370 Heat inactivation before the addition of 3H-palmitoyl-CoA abolishes the acyl-Erf2 intermediate. These results suggest that native Erf2 autoacylates. The formation of the intermediate depends on an intact DHHC domain. When the cysteine is mutated to serine, the resulting Erf2 C203S mutant cannot be acylated, which suggests that the cysteine is the site of palmitoylation.370 The acyl intermediate may be the active enzyme intermediate that transfers the acyl group to substrate proteins. Thus, the enzymatic mechanism likely has a two-step ping-pong mechanism (Figure 20). The first step is fast autoacylation, and in the slower second step, the palmitoyl is transferred to the substrate protein. Evidence to support the chemical and kinetic competence of this intermediate has been reported.460 In this study, purified DHHC2 and DHHC3 were labeled with 3H-palmitoyl-CoA in vitro and then re-purified to remove excess radioactive palmitoyl-CoA. The PAT was then incubated with a protein substrate. Over time, the signal was transferred from the PAT to the substrate protein, thereby directly demonstrating the transfer of the palmitoyl group from enzyme to substrate.
Figure 20.
Mechanism of DHHC-catalyzed cysteine palmitoylation.
Notably, the identity of the autoacylated cysteine remains unknown. Mutagenesis only shows that the cysteine in the DHHC domain is necessary for autoacylation because it is required for catalytic activity. A radioactive signal on the PAT was observed despite long incubation times with substrate proteins, which suggests that either 3H-palmitoyl is also located on a cysteine residue not involved in the catalytic transfer or the PAT is inactive.460,470 Additional results, such as X-ray crystal structures of the catalytic domain in complex with substrates, will greatly help to elucidate the catalytic mechanism of DHHCs.
When the His201 in Erf2, the first conserved histidine residue in the DHHC motif, is mutated to alanine, the resulting Erf2 H201A–Erf4 complex loses its PAT activity despite the formation of the acyl intermediate.370 This outcome suggests that His201 is involved in the transfer of the acyl group to the substrate but is not important for the formation of the acyl enzyme intermediate, which is not the case for all DHHCs. For example, Swf1 with a DQHC motif instead of the DHHC motif has partial activity.471 This motif also exists in the human DHHC13 protein that acylates the huntingtin protein. Surprisingly, in the yeast system, the overexpression of Swf1 mutants in which the catalytic cysteine of the DHHC motif is altered to arginine (DHHR) still results in increased palmitoylation of the Swf1 substrates Tlg1, Syn8, and Snc1. Most likely, the acyl-DHHC intermediate would not form with the Swf1 DHHR mutant.
The conserved CRDs of DHHCs contain many palmitoylated cysteine residues that are distinct from the catalytic cysteine. These cysteines are located downstream of the DHHC domain and form a unique motif, CCX(7–13)C(S/T).402 This motif is found in DHHC5, DHHC6, and DHHC8. The function of this modification on the DHHCs and its formation mechanism require further study. It could be a consequence of the auto-catalytic activity or the activity of another DHHC on DHHC5, DHHC6, and DHHC8. Indeed, DHHC6 is a downstream substrate for DHHC16 and the depalmitoylase APT2.472 When palmitoylated, but not when de-palmitoylated, DHHC6 has detectable activity. Notably, DHHC6 exists in multiple differentially palmitoylated states with variable activity and stability. This complex regulatory mechanism is reminiscent of that of protein phosphorylation and further highlights the importance of protein acylation.
One study demonstrated that DHHC2 and DHHC3 form homodimers that inhibit enzyme activity,473 which suggests that oligomerization may be a means to regulate DHHC PAT activity. Another potential regulatory mechanism is phosphorylation to turn DHHCs on or off. DHHC3 is regulated by the Src and fibroblast growth factor receptor tyrosine kinases. Compared with the WT, DHHC3 with the phosphorylated tyrosine sites mutated had a stronger interaction with neural cell adhesion molecule and further increased its palmitoylation levels.474
The interaction of DHHC with other non-substrate proteins also regulates the function and activity of DHHCs. DHHC9 requires Golgi complex-associated protein of 16 kDa (GCP16) for proper functioning. DHHC6, through its SH3 domain, reportedly associates with Selenoprotein K,404 which serves as a cofactor in a manner similar to that of GCP16. The DHHC–cofactor complex increases the palmitoylation of its substrates. The mechanism through which SelK interacts with DHHC6 to promote palmitoylation requires further study. The cofactor could stabilize the DHHC enzyme, as in the case of DHHC9 and GPCP16,412 or recruit the substrate to the complex.
The DHHC proteins bind zinc with specific cysteine residues in the CRD.475 Interestingly, these conserved cysteine residues can also be palmitoylated, which destabilizes the enzyme.476 The relationship between the zinc binding and palmitoylation of these cysteines is unknown but could be a potential regulatory mechanism.
4.1.5. Biological Function and Disease Relevance of DHHCs
Significant progress has been made in elucidating the functional role of palmitoylation, but the role of DHHC enzymes remains incompletely understood. Through knockdown and deletion studies, various biological functions have been attributed to specific DHHCs (Table 2). In general, most mutations are correlated with neurodegenerative diseases such as Huntington’s, Alzheimer’s, and schizophrenia. Other diseases such as cancer and developmental defects have also been attributed to various DHHCs. The biological functions of DHHCs are ultimately determined by the substrate proteins they modify and regulate. Because these substrates have not been completely identified in most cases, understanding of the biological functions of the DHHC enzymes remains limited. Redundancies among DHHCs, poor antibodies against endogenous DHHCs, and weak in vitro DHHC activity are a few of the obstacles that must be overcome to further elucidate DHHC function.
The most thoroughly studied case is the role of DHHC17 and HIP14L (DHHC13) in Huntington’s disease. These PATs were initially shown to interact and palmitoylate huntingtin through their ankyrin repeat domains. Disease mutations of huntingtin diminish interaction with PATs, which reduces palmitoylation and and ultimately causes cell death.426 Notably, when WT huntingtin levels are low, the degree to which DHHC17 itself is palmitoylated is significantly reduced. This decrease leads to defective enzymatic activity against known substrates SNAP25 and GluR1 in mice lacking one of the alleles coding for huntingtin, and the effect is even greater in cells treated with antisense oligos to degrade the huntingtin gene.443 Huntingtin likely acts as a protein scaffold to bring together DHHC17 and its substrates. Because most substrates of DHHC17 are involved in neurological processes, it is easy to see how the loss of normal huntingtin or DHHC17 function could result in neurological defects. Furthermore, mice deficient in DHHC17 exhibit a neurological and behavioral phenotype similar to that of patients with Huntington’s disease.477
These studies have generally shown that the palmitoylation of huntingtin is protective and that the inhibition of PAT–huntingtin interaction is necessary for the progression of Huntington’s disease. Although the exact mechanism through which DHHC17 contributes to Huntington’s disease remains to be established, a recent study showed that caspase-6, a cysteine protease involved in neurological disorders, is a substrate of DHHC17. Caspase-6 activity is inhibited by palmitoylation, and in DHHC17 −/− mice, decreased caspase-6 palmitoylation results in increased caspase-6 activity, which is reportedly required for the progression of Huntington’s disease.433 Additional mouse studies have demonstrated embryonic lethality in DHHC17 and DHHC13 knockout mice.478 These embryos have characteristics similar to those of huntingtin (−/−) embryos, such as a disorganized chorion. Although its mechanism remains to be elucidated, the lethality further emphasizes the importance of palmitoylation at various stages of development.
DHHC mutations are also associated with X-linked mental retardation,479 including X chromosome mutations in zDHHC9 and zDHHC15 (X-linked mental retardation type 91).421 It is not clear how deficiency in DHHC9 and DHHC15 leads to mental retardation, but it is not unexpected, because DHHC15 and DHHC9 substrates are involved in neural development (see Table 2).
Two studies have suggested that DHHC2 functions as a tumor suppressor. Reduced expression of the corresponding gene (zDHHC2) predicts a poor prognosis in gastric adenocarcinoma patients and is associated with lymph node metastasis.480 When zDHHC2 is knocked down, cytoskeleton-associated protein 4 (CKAP4) palmitoylation is significantly reduced, which decreases the capacity of antiproliferative factor to suppress proliferation and tumorigenesis. The interaction between CKAP4 and antiproliferative factor is mediated by the palmitoylation of CKAP4 by DHHC2, which explains its function as a tumor suppressor.481
The overexpression of DHHC14 is linked to gastric cancer. Gastric cancer tissue samples with higher levels of DHHC14 messenger RNA (mRNA) are associated with more aggressive tumor invasion in vivo. In vitro, DHHC14 activates gastric cancer cell migration and invasion, whereas cells with DHHC14 knockdown are relatively less invasive.482
Mice deficient in the zDHHC5 gene show a remarkable defective phenotype. Litter sizes are reduced by half, and the survivors are deficient in contextual fear conditioning. DHHC5 is also highly expressed in neural tissue and interacts with PSD-95 through the PDZ3 domain on PSD-95.396 These observations suggest that DHHC5 may be linked to post-synaptic function, learning, and memory. The effect of DHHC5 on learning and memory might be explained by the ability of DHHC5 to interact with and palmitoylate SSTR5, a GPCR expressed mainly in neural tissue but not in tissues such as the kidneys or liver.399 The exact function of palmitoylation on GPCRs is not well understood. Studies on rhodopsin have suggested that palmitoylation near the carboxyl-terminal tail at Cys322 and Cys323, which extends into the cytoplasm, induces the formation of a pseudo loop.483 The C-terminus of GPCRs is important for interaction with downstream signaling molecules such as receptor kinases, and the palmitoylation-dependent formation of the pseudo-loop could be a mechanism that regulates GPCR signaling484,485 through DHHCs. A study has also linked DHHC5 to non-small cell lung cancer.486 When zDHHC5 was knocked down, the cancer cells exhibited reduced cell proliferation, colony formation, and cell invasion, and could be rescued by overexpression of the WT DHHC5 but not the catalytically dead DHHS5. The phenotype was replicated in a mice tumor xenograft model in which DHHC5 knockdown inhibited tumor formation.486
A study in mice showed that a deletion of three base pairs resulting in the loss of a highly conserved phenylalanine in DHHC21 was sufficient for hair loss in mice.439 This single mutation resulted in the mislocalization and loss of catalytic activity of DHHC21. Re-introducing WT DHHC21 into the mice rescued the shiny and smooth coat phenotypes. The authors then showed that Fyn, a Src-family kinase involved in keratinocyte differentiation, is a substrate for DHHC21. The observed effects of Fyn mislocalization and reduced levels of Lef1, nuclear β-catenin, and Foxn1 in the DHHC21 mutant keratinocytes may explain the hair loss and differentiation phenotypes.439 DHHC21 is also linked to endothelial inflammation.487 This enzyme is required for the barrier response, and DHHC21-deficient mice are more resistant to injury. These effects are likely mediated by the palmitoylation of PLCβ1.487 Another study linked DHHC21 to vascular function in mice through the palmitoylation of the α1D adrenoceptor, the palmitoylation of which is required for receptor function.395
In mice, a nonsense mutation in the zDHHC13 gene results in the degradation of mRNA and phenotypes of amyloidosis, alopecia, and osteoporosis.441 The protein responsible for the osteoporosis phenotype is membrane type-1 matrix metalloproteinase (MT1-MMP), a factor that controls skeletal development. The palmitoylation of MT1-MMP by DHHC22 (encoded by zDHHC13) is required for its proper distribution and function in facilitating vascular endothelial growth factor expression. Osteocalcin expression is also associated with DHHC22-dependent MT1-MMP palmitoylation, which links DHHC22 to skeletal development through its palmitoylation activity on MT1-MMP.442 Other studies have linked DHHC22 to mitochondrial function and metabolism in mouse liver cells. A proteomics study identified 254 potential DHHC22 substrates. Among them, malonyl-CoA-acyl carrier protein transacylase and catenin delta are verified substrates.488 These findings were further confirmed in the hepatocytes of zDHHC13 knockout mice, which showed diminished mitochondrial function.488 DHHC22 also reportedly plays roles in hair anchoring and skin barrier integrity through its substrate cornifelin.444 The loss of zDHHC13 makes mice more susceptible to bacteria, which results in skin inflammation.487 Similarly, a spontaneous mouse mutation in zDHHC13 reportedly led to increased susceptibility to skin carcinogenesis.455
Mice deficient in zDHHC16 (Aph2) exhibit cardiomyopathy and cardiac defects such as bradycardia.424 The phenotype functions primarily through the DHHC16 substrate phospholamban (PLN). When PLN is palmitoylated, its interactions with protein kinase A and protein phosphatase 1 control the pentamer formation of PLN. In zDHHC16-deficient mice, PLN phosphorylation decreases, which inhibits PLN function. Surprisingly, the deleterious phenotype is alleviated to a degree in PLN−/− zDHHC16−/− mice. DHHC16 is also reportedly involved in the DNA damage response pathway; however, the mechanism has not been elucidated, and the effects are observed only when zDHHC16 is knocked out in mouse embryonic fibroblast cells.489
Genomic mapping studies in schizophrenia patients have identified multiple gene deletions that may be involved. zDHHC8 is a commonly observed deletion located in the chromosome 22q11 region490 that has been linked with schizophrenia. One potential substrate that may mediate the effects of zDHHC8 deletion is the ankyrin-G protein (ANK3). DHHC5 and DHHC8 are reportedly required for the palmitoylation and localization of ANK3,491 and other studies have linked ANK3 to schizophrenia.492 Another potential substrate is bCDC42, the overexpression of which restores dendritic spine cell density in adult 22q11 deletion mice.493 However, the association of zDHHC8 mutation with schizophrenia is controversial.490,494,495
Surprisingly, unlike the deletion of zDHHC17 or zDHHC5, the deletion of the broad-specificity DHHC3 or DHHC7 in mice does not result in obvious deleterious phenotypes.452 However, simultaneous knockout of DHHC3 and DHHC7 results in a drastic phenotype of reduced body and brain mass and perinatal lethality. This observation confirms to some degree the existence of functional redundancies for DHHC3 and DHHC7 and likely other DHHCs. DHHC7 knockout mice show increased glucose tolerance and hyperglycemia linked to the palmitoylation of Glut4.406 Additional evidence has linked DHHC7 to cell polarity and tumorigenesis through the palmitoylation of Scribble409 and to cell migration via junction adhesion molecule C.408
The palmitoylation of viral proteins is required for proper protein function as previously noted,496 but the transferases for these proteins have yet to be identified. The likelihood that viral proteins hijack the DHHCs of their target cells is high because viral proteins are known to hijack cellular machinery to ensure the survival of the virus. A recent example is the HSV-1 envelope protein UL20, which interacts with and serves as a substrate for DHHC3.497 Cells overexpressing catalytically dead DHHS3 have lower viral titers and altered UL20 localization. This hijacking is not limited to viruses. Bacterial pathogens have also been demonstrated to hijack host cells. The GobX protein from L. pneumophila and SspH2 from Salmonella are two examples of bacterial proteins that are palmitoylated inside host cells and require palmitoylation for proper localization.498
DHHC-mediated palmitoylation is also critical for calcium flux. IP3R, the receptor for inositol 1,4,5-triphosphate, is palmitoylated by the SelK–DHHC6 complex. Knockdown of DHHC6 disrupts IP3R-mediated Ca2+ flux, and mutagenesis of the IP3R palmitoylation sites decreases the function of the receptor. The electrogenic NCX1 is also regulated by palmitoylation.499
Generally, the deletion of a zDHHC gene and subsequent loss of the fine control of associated substrate palmitoylation is likely to be deleterious to cell homeostasis in healthy normal cells. The disruption of DHHC levels in malignant cells has not been well studied. One study reported that epidermal growth factor receptor (EGFR) signaling is increased in DHHC20-deficient cancer cells.436 Palmitoylation-deficient EGFR exhibited increased activation and downstream signaling, and the increased EGFR signaling sensitized the cells to EGFR inhibition and increased inhibitor-induced cell death.
In conclusion, DHHCs play vital roles in normal cellular functions and are involved in the development of neurological disease and cancer. Similar to protein kinases, most DHHCs are involved in signaling pathways, but the modifications DHHCs catalyze do not turn on their substrates but instead direct them to the correct cellular compartment. This mechanism is supported by the deleterious phenotypes observed when DHHCs are deleted or their catalytic activity is lost. How DHHCs themselved are regulated to control their catalytic activity is poorly understood and remains an exciting area of study.
4.2. Proteins That Catalyze Cysteine Depalmitoylation
Two known cytosolic acyl protein thioesterases, APT1 and APT2 (also called LYPLA1 and LYPLA2), are thought to be responsible for depalmitoylating many S-acylated proteins. APT1 was first reported to deacylate the α subunit of trimeric G proteins and the small GTPase H-Ras in vitro and when overexpressed in cells.500,501 Knockdown of APT1 also increases the acylation of overexpressed N-Ras.502 However, as described later in this section, APT1 and APT2 knockdown do not affect the acylation of endogenous Ras, which suggests that endogenous Ras is not a physiological substrate of APT1 and APT2.503 Notably, APT1 and APT2 themselves are also palmitoylated.504 APT1 can depalmitoylate both itself and APT2. Palmitoylation is proposed to target APT1 and APT2 to the plasma membrane, where they can deacylate other substrate proteins.504 However, another report suggested that the soluble unpalmitoylated APT deacylates substrate proteins on all membranes.505
The development and use of APT1 and APT2 inhibitors have provided further support for the roles of these acyl protein thioesterases. The first reported APT1/APT2 inhibitor was palmostatin B,502 and a more potent analogue, palmostatin M, has also been developed.506 However, later studies showed that palmostatin B and M are not specific for APT1 and APT2. They also inhibit other serine hydrolases according to the results of activity-based protein profiling.507 Thus, previous conclusions about the effects of palmostatins on APT1 and APT2 must be viewed with caution.
ML348 and ML349, which are more specific inhibitors for APT1 and APT2, respectively, have been developed (Figure 21) through high-throughput screening facilitated by activity-based protein profiling.508–510 Notably, ML348 is highly specific for APT1, and ML349 is highly specific for APT2.511,512 Thus, these inhibitors will be highly useful for dissecting the roles of APT1 and APT2. One study with ML348 and ML349 showed that APT1 and APT2 do not affect signaling downstream of N-Ras, thereby correcting a previous report obtained with nonspecific inhibitors.503 These APT1- and APT2-selective inhibitors have been used to demonstrate that APT2 depalmitoylates Scribble and affects its membrane localization.513
Figure 21.
Inhibitors for cysteine depalmitoylases APT1, APT2, and ABHD17.
The α/β-hydrolase domain 17 (ABHD17) family of proteins has been identified as a group of depalmitoylases. The knockdown of APT1 and APT2 affects the S-palmitoylation of huntingtin but not that of PSD-95 and N-Ras. They used the nonspecific inhibitor palmostatin B to profile novel serine hydrolase targets and discovered a family of uncharacterized ABHD17 proteins that catalyze the depalmitoylation of PSD-95 and N-Ras.498 Another group screened 38 mouse serine hydrolases and also found that ABHD17 members (ABHD17A, ABHD17B, and ABHD17C) are depalmitoylases of PSD-95.514 These studies broadened the family of depalmitoylase enzymes and suggest that even more proteins than previously thought can catalyze cysteine depalmitoylation.
4.3. Palmitoylation Inhibitors
Protein palmitoylation plays key roles in protein trafficking and is related to several diseases. Palmitoylation inhibitors can therefore be useful tools with which to study the function of palmitoylation or treat related diseases, and interest in their development is increasing. Currently available palmitoylation inhibitors can be categorized into two general types: lipid-based and non-lipid-based (Figure 22). The most commonly used lipid-based inhibitor is the non-selective 2-bromopalmitate (2BP). 2BP inhibits the palmitoylation of Src family kinases Fyn and Lck, Rho family kinases, and H-Ras.515–517 Cerulenin, initially discovered as a fatty acid synthase inhibitor, is also reportedly an S-palmitoylation inhibitor for CD36.518 Tunicamycin, an N-linked glycosylation inhibitor, also inhibits protein palmitoylation on substrates such as estrogen receptor α variant and Ca2+ channels.519,520
Figure 22.
Structures of reported palmitoylation inhibitors.
A high-throughput screening was used to identify several non-lipid-based palmitoylation inhibitors, which were reported to inhibit the Raf/Mek signaling pathway and suppress cancer cell proliferation.521 However, later studies using purified DHHCs showed that only one of the five compounds has inhibition activity and is less potent than 2BP.522
More efficient and selective inhibitors for cysteine palmitoylation are urgently needed. All current inhibitors are limited either by low inhibition potency or lack of selectivity. Although 2BP has historically been the most commonly used “palmitoylation inhibitor”, its noted off-target activity and toxicity are such that it could equally be considered the worst available inhibitor.523,524 In cells, 2BP is converted to its CoA form, which is a substrate for DHHCs and can lead to the labeling of substrate proteins.523 Thus, inhibitors that specifically target DHHCs are in great demand. Inhibitors that can distinguish different DHHCs would be even more useful. More efficient and selective inhibitors will greatly aid elucidation of the function of cysteine palmitoylation and its therapeutic potential.
4.4. Functions of Protein S-Palmitoylation
4.4.1. Proteins Known to Be S-Palmitoylated
We summarize proteins that are experimentally validated to be S-palmitoylated in Tables 3–9. The proteins are classified into these tables according to whether they contain other membrane-targeting signals, such as transmembrane domains, N-terminal glycine myristoylation or C-terminal prenylation. The tables clearly show that palmitoylation occurs on an extraordinarily diverse set of proteins, and unlike glycine myristoylation or cysteine prenylation, no consensus sequence exist for predicting which proteins undergo cysteine palmitoylation. The reported functions of S-palmitoylation are listed along with their references in the tables. A brief summary of the reported functions of cysteine palmitoylation is provided here.
4.4.2. Regulation of Protein Trafficking
Owing to the hydrophobicity of the acyl group, palmitoylated proteins normally associate with the membranes of various organelles and facilitate trafficking between these organelles. This section reviews two well-studied proteins, Ras and Cdc42, to illustrate the regulation of protein trafficking through palmitoylation.
Ras membrane trafficking is discussed in section 2.4a, but this section provides a more detailed picture. H-Ras, N-Ras, and K-Ras are the most well-known Ras genes in humans.670 Ras is a small GPTase that exists in a GTP-bound active state and a GDP-bound inactive state. GEFs activate Ras by catalyzing the exchange of GTP for GDP in Ras,671 whereas GAPs inactivate Ras by promoting the hydrolysis of GTP to GDP.672 Among the different PTMs that regulate Ras activity,673 lipidation acts mainly by controlling Ras trafficking. Ras proteins are prenylated at the C-terminal CaaX motif and subsequently cleaved and carboxylmethylated at the cysteine.13,14,674 Biochemical studies suggest that farnesylation cannot provide adequate binding affinity for the plasma membrane.110,111 Therefore, a second event is needed. This second event differs for the various members in Ras family. For H-Ras, N-Ras, and K-Ras4A, palmitoylation is this second event, and it occurs on cysteines near the CaaX motif after farnesylation (Figure 23). N-Ras and K-Ras4A each have only one cysteine (Cys181 and Cys180, respectively) near the CaaX motif (Figure 23), which is the palmitoylation site. H-Ras has two cysteines (Cys181 and Cys184) near the CaaX motif (Figure 23) and is dually palmitoylated.142
Figure 23.
C-terminal sequences of Ras family members and Ras trafficking.
After farnesylation and palmitoylation (known as dual lipidation), Ras is sorted into the vesicle and travel to the plasma membrane.675 In the case of H-Ras, the mono-palmitoylation of Cys181 is sufficient for plasma membrane localization, whereas the mono-palmitoylation of Cys184 leads to Golgi localization,676 which indicates that the Cys181 of H-Ras is more involved in targeting to the plasma membrane. Another member of the Ras family, K-Ras4B, has no cysteines for palmitoylation near the CaaX motif. However, K-Ras4B has eight lysines near the C-terminus that may interact electrostatically with the negatively charged plasma membrane for localization.104 After palmitoylation and localization, H-Ras, N-Ras, and K-Ras4A are depalmitoylated by acyl-protein thioesterases and return to their mono-lipidation states.502 Weak plasma membrane binding affinity results in the localization of Ras to the Golgi (facilitated by PDEδ142), in which it is re-acylated and sorted to the plasma membrane. This dynamic acylation–deacylation cycle therefore helps maintain the plasma membrane localization of Ras (Figure 23).677
Cdc42 belongs to the Rho GTPase family and regulates cell polarity, migration, and progression.678,679 Cdc42 has two isoforms. The ubiquitously expressed isoform 1 (aCdc42) contains a CaaX motif and is either farnesylated or geranylgeranylated. There is no additional cysteine near the CaaX motif, and thus, aCdc42 is not palmitoylated. Isoform 2 (bCdc42) is expressed specifically in the brain.680 It contains a unique CCaX motif in which the first cysteine is farnesylated. After farnesylation, two processing pathways become available. One pathway is the classical CaaX processing pathway. The protein is first farnesylated in the ER, followed by RCE1 and ICMT-mediated cleavage of aaX and carboxylmethylation of the terminal prenylcysteine. Then bCdc42 binds to RhoGDIα and travels to the plasma membrane.132 The other processing pathway bypasses the proteolysis step, and palmitoylation occurs on the second cysteine of the CCaX motif.113,681 Then, bCdc42 is localized to the Golgi and travels to the plasma membrane via vesicular transport. After plasma membrane localization, bCdc42 can be depalmitoylated and travel back to the Golgi via binding to RhoGDIα. In this second pathway, bCdc42 is dually lipidated followed by vesicle localization, a process similar to the one that Ras undergoes. However, it is unknown what mechanism determines the processing of Cdc42. Only some proteins with the CCaX motif undergo dual lipidation,113 which indicates that the CCaX motif is not a general feature of dual lipidation.
4.4.3. Regulation of Protein Stability
S-palmitoylation also regulates protein stability. The best-studied example is Tlg1 in yeast, which plays key roles in the regulation of protein recycling between endosomes and the Golgi.682,683 Tlg1 is palmitoylated by the yeast PAT Swf1.684 Palmitoylation retains Tlg1 on the trans Golgi network and endosome membranes and inhibits Tlg1 degradation. By contrast, the mutation of palmitoylation sites or inactivation of Swf1 results in Tlg1 ubiquitination and degradation, which are mediated by the Tlg1 E3 ligase Tul1.685 In this case, the function of palmitoylation is to prevent protein ubiquitylation and thus increase Tlg1 half-life and stability. Furthermore, the cellular localization of cysteine-mutated Tlg1 is similar to that of WT Tlg1,684 which indicates that palmitoylation does not influence the membrane association of Tlg1 but simply blocks Tlg1 ubiquitylation. However, the mechanism through which palmitoylation inhibits ubiquitylation is unknown. One hypothesis is that two contiguous aspartates are located in the transmembrane domain, which triggers quality control to ubiquitylate and degrade the protein because the negatively charged aspartate is incompatible with the membrane. When palmitoylation occurs on two adjacent cysteines, the long-chain fatty acyl group covers two aspartates and thus rescues the incompatibility. In addition to the regulation of Tlg1, the stability of several other proteins is regulated by S-palmitoylation. The palmitoylation of HIV receptor C-C chemokine receptor type 5 stabilizes the membrane expression of the receptor.572 The lack of palmitoylation of estrogen receptor-α results in more E2-dependent degradation.649 Palmitoylation prolongs the half-life of regulator of G protein signaling 4 (RGS4) more than 8-fold.390
4.4.4. Prevention of Unfolded Protein Response in the ER and Promotion of ER Exit
Low-density lipoprotein receptor-related protein 6 (LRP6) is a single-pass type I membrane protein. It is a co-receptor of Wnt and is required for the initiation of the Wnt/β-catenin signaling pathway.686,687 The palmitoylation of LRP6 occurs on Cys1394 and Cys1399 and is required for LRP6 exit from the ER.537 It has been proposed that palmitoylation allows LRP6 to avoid triggering ER quality control. Because LRP6 contains a 23 amino acid transmembrane domain, which is longer than the usual membrane thickness, the hydrophobicity of the extra residues is mismatched with the hydrophilic environment and thus can trigger the unfolded protein response. The palmitoylation of two juxtamembranous cysteines tilts the extra residues towards the membrane and thus avoids the mismatch.537 Yeast chitin synthase Chs3 must also be palmitoylated for ER export,688 and a study of amyloid precursor protein (APP) showed that the blocking of its palmitoylation causes nearly complete ER retention, which suggests that this reaction is required for the ER export of APP.689
4.4.5. Prevention of Protein Aggregation
Knowledge of the function of palmitoylation in protein aggregation comes from studies of Huntington’s disease, which is caused by the mutation of the huntingtin protein. In healthy individuals, the N-terminal region of the huntingtin protein contains 6–35 repeated glutamine residues (known as the polyQ region), whereas in patients with Huntington’s disease, the polyQ region expands to more than 40 repeated glutamines.690 These excess glutamines cause huntingtin aggregation, which is the primary marker of the disease.430 Huntingtin is palmitoylated on Cys214 by DHHC17,691 and compared with WT huntingtin, mutated huntingtin is reportedly palmitoylated at a much lower level.430 The overexpression of DHHC17 reduces huntingtin aggregation efficiently, whereas the knockdown of DHHC17 increases huntingtin aggregation and induces neuronal cell death. The involvement of palmitoylation by DHHC17 may provide new targets for the treatment of Huntington’s disease.
4.5. Techniques for Detecting Protein Palmitoylation
The study of protein cysteine palmitoylation has benefited significantly from technologies that can detect this process. To date, several methods have been developed to detect protein S-palmitoylation, thereby enabling the identification of palmitoylated proteins and the functional study of palmitoylation.
4.5.1. Radioactive-Isotope-Labeled Palmitic Acid
Using radioactive-isotope-labeled palmitic acid to label proteins metabolically was the earliest reported method for the detection of protein S-palmitoylation.692 After treatment with radiolabeled palmitic acid, radiolabeled palmitoyl-CoA forms in cells and used by PATs to modify target proteins. Immunoprecipitation followed by radioactivity monitoring allows the detection of palmitoylated proteins. Three radiolabeled palmitic acids are commonly used: 3H-, 14C- and 125I-palmitic acids.412,693,694 3H- and 14C-palmitic acids are structurally the same as endogenous palmitic acids and mimic palmitoylation accurately. However, the use of these radiolabeled palmitic acids requires long exposure times owing to the weak radioactive signals of 3H and 14C. 125I-palmitic acid has higher sensitivity, but the introduction of the iodo label significantly changes the structure of palmitic acid, and thus this probe may not be ideal.
4.5.2. Bioorthogonal Palmitic Acid Probes
To solve the problem of low sensitivity of 3H- and 14C-palmitic acids, bioorthogonal palmitic acid probes which contain a terminal azido or alkynyl group have been developed (Figure 24).695 Compared with radiolabeled palmitic acid probes, these bioorthogonal probes have high sensitivity and are more convenient to handle. Furthermore, combined with click chemistry, affinity probes such as biotin can be installed on proteins to allow the affinity purification and identification of modified proteins with MS. Currently, this method is broadly used with two types of probes: azido palmitic acid probes and alkynyl palmitic acid probes (Figure 24). Azido fatty acids with 15 carbons (Az-C15, Figure 24) primarily label S-palmitoylated proteins,327,696 whereas azido fatty acids with shorter carbon chains (e.g., Az-C12 and Az-C11) label only N-myristoylated proteins.696,697
Figure 24.
Bioorthogonal palmitic acid probes for the detection of protein palmitoylation.
Compared with radiolabeled probes, the azido fatty acid probes have significantly increased detection sensitivity. However, compared with alkyne probes, azido probes reportedly give higher background.698,352 Therefore, alkynyl fatty acid probes, which are structurally more similar to endogenous palmitic acid, have been developed to minimize background labeling. Alkynyl fatty acids with various carbon chain lengths can mimic a range of protein fatty acylations. For example, alkynyl fatty acids with 16 or 18 carbons (Alk-C16 or Alk14, Alk-C18 or Alk16) can label S-palmitoylated proteins,327–329 whereas alkynyl fatty acids with 12 carbons (Alk-C14 or Alk12) can label N-myristoylated proteins.329,697 However, there is overlap among proteins labeled by different probes: Alk12 can also label palmitoylated proteins, and Alk14 can also label myristoylated proteins.333,699 Therefore, analytical methods that can identify the modification site are helpful in determining which type of modification the probe is labeling and which enzymes may be responsible for that modification. A cleavable azido molecule was introduced to alkynyl fatty acid labeled proteins to facilitate the identification of modification sites.336 This molecule contains fluorescence and biotin tags for the visualization and enrichment of fatty-acylated proteins. It also bears a protease cleavage site and therefore can leave a hydrophilic and charged tag on fatty-acylated peptides after in vitro protease digestion. This method increases the hydrophilicity and ionization of fatty-acylated peptides and enables the direct identification of sites modified by fatty acid probes.
4.5.3. Acyl-Biotin Exchange
Acyl-biotin exchange (ABE) is an indirect method for detecting protein S-palmitoylation.700,701 The three-step ABE procedure is shown in Figure 25. The first step is to use N-ethylmaleimide to block all the free cysteines in proteins. Then, hydroxylamine is used to cleave the palmitoyl group from the modified cysteines. The third step is the use of biotin-N-[6-(biotinamido)hexyl]-3′-(2′-pyridyldithio)propionamide (biotin-HPDP) to label the relieved cysteines, followed by streptavidin pull-down and MS identification. Compared with palmitic acid probes, ABE has several advantages. Both the radiolabeled and bioorthogonal palmitic acid probes operate via metabolic labeling, which interferes with global metabolism status and may disrupt normal cell processes. ABE is not metabolic labeling, so it can detect protein S-palmitoylation under any conditions, including various stress conditions. In 2006, this method was used to profile global S-palmitoylated proteins in yeast, which was the first proteomics study of palmitoylation.447 Thirty-five new palmitoylated proteins were identified in this study. Furthermore, ABE is the most ideal method developed to date for the study of protein S-palmitoylation in animal tissues because it lacks a pre-treatment step and can monitor the dynamics of S-palmitoylation.681 By comparison, metabolic labeling with alkyne-tagged fatty acids and pulse-chase method can also be used to examine the dynamic of S-palmitoylation in cell culture,702 but it cannot be easily applied to study palmitoylation in animals.
Figure 25.
Procedure of ABE method for the detection of protein S-palmitoylation.
ABE also has limitations, however. Its most obvious drawback is the hydroxylamine treatment step, which removes the lipidation from cysteines and therefore obscures which form of lipidation (myristoylation, palmitoylation, or other acyl groups) is occurring on the cysteine residues. Certain S-palmitoylations may also be relatively resistant to hydroxylamine treatment; junction adhesion molecule C is one reported example.408 A variation of the ABE method called acyl-PEG exchange has been reported. In this method, a 5 kD or 10 kD PEG is added to S-acylated proteins instead of biotin. This mass tag allows the visualization of S-acylation level with western blots because the modified protein migrates more slowly than the unmodified protein.703 Ethylenediaminetetraacetic acid (EDTA) is necessary for effective hydroxylamine treatment, likely because it chelates metals that could oxidize the liberated cysteine residues.703
4.5.4. Imaging Palmitoylated Proteins in Cells
The three methods described above use biochemical approaches to detect protein palmitoylation after the lysing of cells or tissues. A fluorescence imaging method for tracking specific palmitoylated proteins in mammalian cells has also been developed.704 As shown in Figure 26, the method uses Alk14 (Alk-C16) metabolic labeling and click chemistry to install a tag on a target protein. Two primary antibodies are then used to recognize the target protein and tag, and two distinct secondary antibodies conjugated to oligonucleotides are used to bind specifically to the two primary antibodies. After this step, the two secondary antibodies form a closed circle because they bind to the same protein (distance between two secondary antibodies is <40 nm). A rolling-circle amplification reaction is performed, and then fluorescent oligonucleotide probes are added for hybridization and the signals that depend on the distance between the two secondary antibodies can be observed. Non-target proteins or non-palmitoylated proteins cannot be recognized by the primary antibodies, so the final hybridization cannot occur. Using this approach, the authors visualized the O-palmitoylation of Wnt3a in cells and successfully tracked the secretion pathway of the protein. However, because this method uses antibodies and click chemistry, which requires Cu(I), the cells must be fixed.
Figure 26.
Method for imaging palmitoylated proteins in cells.
4.5.5. Other Methods for the Detection of Protein Palmitoylation
Additional methods have been developed for the detection of protein palmitoylation. Difference gel electrophoresis based proteomics 705 detects slight differences in pI values and the relative mobility of palmitoylated and depalmitoylated proteinsMicellar electrokinetic chromatography was used identify GAP-43 palmitoylation in vitro.706 The separation of palmitoylated and unmodified GAP-43 peptides can be performed in less than 7 min. Gas chromatography-MS and liquid chromatography-MS methods have also been developed to directly identify and quantify palmitoylation and other lipidations by comparing the retention time and mass spectrum with standard samples.707,708 However, these methods require a large amount of protein, which limits their capacity to detect protein palmitoylation in cell lysates or tissues.
4.5.6. Software for the Prediction of Protein Palmitoylation
Several software programs have been developed for the prediction of protein palmitoylation. CSS-Palm 1.0 (CSS: clustering and scoring strategy) was the first model built for palmitoylation site prediction.709 NBA-Palm (NBA: naïve Bayes algorithm) is another program available for palmitoylation site prediction.710 CSS-Palm 1.0 has been updated to CSS-Palm 2.0 and used to predict the palmitoylation sites of 16 known palmitoylated proteins in budding yeast; these sites were subsequently validated experimentally.711 CSS-Palm 2.0 was used in global in silico screening and identifed neurochondrin/norbin as a novel palmitoylated protein.374 Yet another program for palmitoylation site prediction, CKSAAP-Palm (CKSAAP: composition of k-spaced amino acid pairs),712 has a sensitivity higher than that of CSS-Palm 2.0 for predicting palmitoylated proteins in budding yeast.
5. Lipidation on Other Residues
In addition to cysteine prenylation, N-terminal glycine myristoylation, and cysteine palmitoylation, several other lipid modifications of proteins have been reported, including serine O-acylation, N-terminal cysteine N-palmitoylation, lysine N-acylation, and C-terminal cholesterol esterification (Figure 27). Only a few proteins have been determined to undergo these modifications. Thus, the preferred sequence motifs and likelihood of these modifications occurring in other proteins are unknown. However, these lipid modifications clearly play important roles in the biological functions of the known proteins.
Figure 27.
Protein O- and N-acylation and protein C-terminal cholesterol esterification.
5.1. Serine Fatty Acylation of Wnt Proteins
Wnt proteins require acylation for secretion and activity. The Wnt family of secreted signaling proteins impacts virtually all aspects of developmental biology and is also essential during adulthood.713 In the canonical Wnt signaling pathway, Wnt binds to the Frizzled (Fz)–LRP complex, thus transducing a signal to Dishevelled and Axin. This signal leads to the inhibition of β-catenin degradation, and accumulated β-catenin then enters the nucleus and interacts with T-cell factor to regulate the transcription of certain genes.714
The first pure and active secretory Wnt protein (murine Wnt3a) was successfully isolated from cell culture medium in 2003.715 Triton-X-114 phase separation assays showed that most of the purified Wnt3a partitioned into the Triton-X-114 phase, which suggested that similar to integral membrane proteins, Wnt3a is highly hydrophobic. 3H-palmitate metabolic labeling further confirmed that Wnt3a is palmitoylated. MS analysis showed that Cys77 of Wnt3a is modified with a palmitate group. This cysteine residue is conserved among the Wnt family members. In 2006, it was reported that 3H-palmitate metabolic labeling of both the WT and a cysteine mutant (C77A) of Wnt3a were resistant to neutral hydroxylamine (pH 7.0), which was used to specifically cleave thioester linkages but leave oxyester and amide bonds intact.716 These observations suggest that Wnt3a undergoes another type of acylation. Truncation together with site-directed mutagenesis demonstrated that the conserved Ser209 residue of Wnt3a is required for acylation. Unexpectedly, a monounsaturated palmitoleoyl (C16:1) moiety was found to be attached to Ser209 via an oxyester linkage. Mutation of Ser209 yielded nonfunctional and poorly secreted Wnt3a protein. However, one of the Wnt proteins, Wnt8/WntD, lacks the corresponding serine but is secreted normally.716
Subsequently, an imaging method using click chemistry with bioorthogonal fatty acids and in situ proximity ligation was developed, which allowed the first visualization of acylated Wnt proteins in the cellular context.704 Their results demonstrated that Wnt3a is acylated only on Ser209 and not on the originally reported Cys77, consistent with the crystal structure of Wnt protein reported in 2012.717
5.1.1. Wnt Serine Acyltransferase and Its Inhibitors
Porcupine (PORCN), a member of the membrane-bound O-acyl-transferase (MBOAT) family,718 is thought to catalyze the transfer of acyl groups to the serine residue of Wnt proteins. Mutations in PORCN abrogate both the activity and the secretion of Wnt and result in early embryonic lethality in mice.719 Mutations in human PORCN lead to focal dermal hypoplasia, an X-linked developmental disorder.720 The catalytic mechanism of PORCN has not been established conclusively partly owing to its hydrophobic nature. However, the highly conserved histidine and asparagine residues among all 11 human MBOAT family members are considered putative catalytic sites.718 Mutations of the conserved His341 residue ablate the activity of PORCN, whereas the conserved Asn306 is not required for PORCN acyltransferase activity.721 Truncation of either the N- or C-terminal domain of PORCN causes destabilization and inactivity.722 PORCN is palmitoylated mainly at Cys187, which is likely catalyzed by DHHCs. A PORCN C187A mutant showed modestly increased fatty acylation and signaling activity of Wnt3a.704
Considering the broad biological roles of Wnt signaling, substantial effort has been made to develop potent agonists and antagonists of the Wnt signaling pathway. The most widely used small-molecule agonist inhibits glycogen synthase kinase 3,723 a component of the β-catenin destruction complex, thus leading to the stabilization of β-catenin and activation of its downstream gene transcription. To inhibit the Wnt signaling pathway, a synthetic chemical library was screened, which led to the identification of two classes of highly selective and powerful inhibitors: inhibitor of Wnt production 1 (IWP-1; Figure 28) and inhibitor of Wnt response.724 The former interacts with and inhibits PORCN specifically, and the latter abolishes the destruction of Axin proteins, which suppress Wnt signaling.724,725 Because PORCN is hypothesized to fatty-acylate Wnt proteins exclusively, the development of inhibitors similar to IWP-1 will allow the specific targeting of Wnt-involved biological processes without affecting others.
Figure 28.
Inhibitors targeting Porcupine (PORCN) and Hedgehog acyltransferase (Hhat).
Additional PORCN inhibitors have been developed, including IWP-L6,726 LGK974,727 and IWP-O1728 (Figure 28). IWP-L6 potently inhibited Wnt-mediated branching morphogenesis in cultured embryonic kidneys.726 LGK974 potently inhibited tumor growth in a murine mouse mammary tumor virus–Wnt1 breast cancer model and a human HN30 head and neck squamous cell carcinoma model, but it had no effect on cells from several other human cancer cell lines, such as brain cancer and colon cancer.727 GNF-6231 (Figure 28), a compound similar to LGK974, has also been reported.729
5.1.2. Extracellular Wnt Serine Deacylase
Unlike cysteine palmitoylation which usually undergoes multiple cycles of acylation and deacylation, the O-palmitoleate modification of Wnt was long thought to be irreversible given the presence of the more stable ester bond compared with a more labile thioester bond. In 2015, Notum, a secreted Wnt antagonist, was identified as the enzyme that deacylates the O-palmitoleic group of secreted Wnt protein.730 The crystal structure of catalytically inactive human Notum S232A in complex with a palmitoleoylated peptide derived from human Wnt7a shows that a large hydrophobic pocket accommodates the palmitoleoyl group. A “kink” in the monounsaturated hydrocarbon chain is positioned at the base of the cavity surrounded by Notum Ile291, Phe319, and Phe320. Notably, the lipid-binding cavity of Notum seems unable to accommodate saturated fatty acids (C14:0/C16:0). Kinetic and MS analyses further proved that Notum is an esterase using both Wnt peptide and protein substrates. Notum has a canonical α/β-hydrolase fold bearing the hallmark serine–histidine–aspartic acid catalytic triad, and it inactivates Wnt signaling by deacylating Wnt protein extracellularly and causing Wnt3a and Wnt5a to form oxidized oligomers.731 During development, Notum is required for neural and head induction via the inactivation of Wnt signaling pathway.
5.1.3. Functions of Wnt Serine Acylation
Wnt serine acylation is crucial for the binding of Wnt to its receptor. The structure of Xenopus Wnt8 in complex with its co-receptor Fz8 CRD in mice suggested that serine acylation is required for high-affinity interaction between Wnt and Fz8 (Figure 29). 717 Their study revealed two extending domains, an N-terminal domain and a C-terminal domain (see Figure 29) of Wnt. Ser187 is located at the tip of the N-terminal domain and is modified by a palmitoleoyl group, consistent with the results of a previous MS study. The palmitoleoyl group inserts deeply into a hydrophobic tunnel of the Fz8 CRD (see Figure 29). The conserved C-terminal domain of Wnt also interacts with a hydrophobic core of the Fz8 CRD. Notably, this structure revealed that instead of being palmitoylated, the conserved Cys77 residue forms a disulfide linkage, which supports the hypothesis that instead of being dually lipidated, Wnt proteins are lipidated only on the conserved serine residue.
Figure 29.
Crystal structure of Xenopus Wnt8 in complex with the Frizzled-8 (Fz8) cysteine-rich domain (CRD; PDB 4F0A). CTD, C-terminal domain; NTD, N-terminal domain.
Wnt serine acylation is critical for intracellular trafficking. Wnt is translated in the rough ER and then translocates into the ER lumen, in which glycosylation and fatty acylation are catalyzed by an oligosaccharyl transferase complex and PORCN, respectively. The acquisition of membrane-association allows modified Wnt to exit the ER for anterograde transport. In the Golgi complex, two cargo receptors, Wntless732–734 and p24,735,736 bind Wnt and escort it to the cell surface. Serine acylation is required for the interaction between Wntless and Wnt proteins.737,738 Wntless is recycled from the plasma membrane to the Golgi complex via endosome trafficking mediated by a retromer complex for the next round of Wnt secretion.739
Wnt serine acylation may also be important for extracellular transport. Lipoprotein particles are hypothesized to be long-range transporters of Wnt morphogen.740 In mammalian cells, Wnt3a co-fractionates with ApoB100 and associates with high- and low-density lipoproteins.741 The lipid modification on Wnt may contribute to the interaction with lipoproteins and further assembly into secretory particles.
5.2. N-terminal Cysteine N-Palmitoylation of Hedgehog
Hedgehog (Hh) signaling plays major roles in embryonic development and malignant tumorigenesis in pancreatic, gastric, and lung cancers. Mammals have three Hh family members, Sonic Hedgehog (Shh), Indian Hedgehog, and desert Hedgehog, among which Shh is the best studied. The Hh ligand binds to its transmembrane receptor, Patched, which then activates Smoothened, leading to the nuclear translocation of Gli transcription factors and activation of downstream gene expression. Hh proteins are initially synthesized as 45 kDa precursor proteins. Upon cleavage of an N-terminal signal peptide, Hh protein undergoes both C-terminal autoprocessing to incorporate a cholesterol modification and N-terminal cysteine palmitoylation via an amide linkage, thus generating a 19 kDa mature form of the Hh signaling molecule.742
Unlike the extensively studied cysteine palmitoylation via a labile thioester bond, Hh proteins are modified with a palmitoyl group at the N-terminal cysteine through a stable amide linkage. Two possible mechanisms have been proposed for this unique reaction (Figure 30). The first posits that palmitoylation initially occurs on the sulfhydryl group of the cysteine side chain. The thioester intermediate then rearranges to an amide linkage via an intramolecular S-to-N shift.743 The second mechanism proposes that N-terminal palmitoylation occurs directly via an enzymatic reaction similar to that of N-terminal myristoylation. The second model is supported by evidence that N-terminal-blocked Shh proteins cannot be palmitoylated and, more importantly, no thioester-linked palmitoylated intermediate has been detected.744 The first six amino acids of Hh are reportedly sufficient for palmitoylation by Hedgehog acyltransferase (Hhat).745 When the N-terminal cysteine is mutated to alanine, no acylation occurs, but the cysteine-to-serine mutant is acylated at reduced levels.745
Figure 30.
Two proposed mechanism for the N-palmitoylation of Hedgehog (Hh) proteins.
5.2.1. Hh Acyltransferase and Its Inhibitors
In 2001, three research groups discovered that in Drosophila melanogaster, the palmitoylation of Hh protein is catalyzed by a member of the MBOAT family called Skinny Hedgehog, Sightless Hedgehog, Central missing, or Rasp.746–748 The mammalian homolog of the corresponding Hh palmitoyltransferase is called Hhat. In Hhat-deficient mice, Hh proteins are not palmitoylated, and the mice exhibit impaired signaling activity evidenced by defects in neural tube formation and limb development.749 Moreover, the depletion of Hhat has been shown to reduce tumor growth in a mouse xenograft model of pancreatic cancer.750 Hhat is a ~50 kDa multiple-span transmembrane protein of the MBOAT family, and its enzymatic activity has been demonstrated with in vitro biochemical assays.744 A biochemical study also showed that the N- and C-terminal variable regions are central to Hhat stability and activity.751 Later studies showed that Hhat has 10 transmembrane domains and two re-entry loops. The catalytic histidine residue is in the loop on the luminal side, whereas the conserved aspartate residue is on a cytosolic loop.752,753
Several inhibitors have been developed to suppress the Hh signaling pathway, mostly by targeting downstream components including the Smoothened or Gli proteins.754 A high-throughput screen identified small-molecule inhibitors for Hhat, such as RU-SKI 43 (see Figure 28),755 which inhibits Hhat palmitoyl transferase activity specifically on Shh proteins both in vitro and in cells. However, later studies showed that RU-SKI 43 has off-target effects and that its cellular toxicity is unrelated to its effect on Hhat. By contrast, a new analogue, RU-SKI 201, is a specific inhibitor of Hhat with no off-target effects reported.756
5.2.2. Functions of Hh Palmitoylation
Hh palmitoylation is key for the proper secretion and signaling activity of Hh. Although mutation of the Shh N-terminal cysteine to serine (C25S) does not affect Shh localization in the lipid raft, the C25S mutant cannot form a soluble multimeric protein complex thought to be the major active component for Hh signaling.749 Notably, apart from lipidation, a conserved lysine/arginine residue in a predicted interaction interface has also been demonstrated to be crucial for Hh multimeric complex formation by contributing to electrostatic interactions.757 Furthermore, Hh oligomers co-localize with heparan sulfate proteoglycans on the surface of Hh-producing cells and assemble with lipoprotein particles, which mediate long-range Hh signaling activity and contribute to the formation of a morphogen concentration gradient during embryonic development.740,757
5.3. Cholesterol Modification of Hh
Apart from unusual N-terminal cysteine palmitoylation, Hedgehog proteins also undergo a unique auto-cleavage process that incorporates a cholesterol modification and releases the C-terminal domain.758,759 The two-step mechanism for Hh autoprocessing is similar to that of intein self-splicing proteins (Figure 31). First, the sulfhydryl group of a cysteine residue attacks the carbonyl of the preceding glycine residue to form a thioester linkage. Then, the labile thioester intermediate is attacked by the 3β-hydroxyl group of a cholesterol molecule to generate an oxyester bond and liberate the C-terminal autoprocessing domain.
Figure 31.
Mechanism of C-terminal autoprocessing of Hh proteins.
Both azido- and alkyne-modified cholesterol analogues have been synthesized and used to label modified Hh proteins.760,761 Compared with the azido analogue, the alkyne analogue is more efficient for labeling Hh.761 These analogues allow the installation of fluorescence or affinity probes for in-gel visualization and affinity purification.
C-terminal cholesterol modification is mainly responsible for the release of dually lipidated Hh proteins from the cell surface with the aid of Dispatched, a 12-pass transmembrane protein, and a secreted protein, Scube.762,763 Notably, Dispatched and Scube recognize different parts of the cholesterol molecule, which suggests a hand-off mechanism reminiscent of the transfer of free cholesterol between Niemann-Pick disease proteins NPC1 and NPC2 during the exit of cholesterol from late endosomes.762 However, cholesterol is not absolutely required for Hh signaling activity even though the absence of the modification reduces signaling potency.763 Moreover, several lines of evidence have shown that the cholesterol moiety is required for the short- and long-range distribution of Hh morphogen. Cholesterol incorporation restricts Hh diffusion by enhancing hydrophobic interactions with the plasma membranes of adjacent cells and thus increases short-range distribution.758 On the contrary, for long-range transport mediated by lipoprotein particle carriers,740 cholesterol modification contributes to the partitioning of Hh into particles and the formation of the soluble multimeric complex.764
5.4. Serine Octanoylation on Ghrelin
In 1999, the search for the ligand of growth hormone secretagogues receptor (GHSR) led to the discovery of a polypeptide ghrelin known as the “hunger hormone.”765 By binding to GHSR, ghrelin stimulates growth hormone release from the anterior pituitary and helps regulate energy homeostasis. The ghrelin gene is first transcribed into the 117-residue preproghrelin, which is then cleaved into the 94-residue proghrelin via the loss of the N-terminal signal peptide. Further processing of proghrelin yields a 28-residue ghrelin peptide that is released into the circulation.766
Ghrelin is the only mammalian peptide hormone known to be modified with an octanoyl group on the third serine residue,765 which is conserved from rats to humans. The initial report of ghrelin peptide indicated that only acylated ghrelin is functional and able to activate GHSR. Des-acyl ghrelin has long been considered a degradation product of acylated ghrelin. However, studies have shown that des-acyl ghrelin can antagonize acylated ghrelin and act as an independent hormone, likely via binding to its own receptor.766
In 2008, two research groups independently discovered the acyl transferase, ghrelin octanoyltransferase (GOAT), that adds the octanoyl group onto ghrelin. Similar to PORCN and Hhat, this enzyme belongs to the MBOAT family that resides in the ER.718 One group overexpressed all 16 mouse MBOAT family members and found that only the overexpression of GOAT dramatically increased the hydrophobicity of ghrelin.767 Another group discovered GOAT by knocking down potential MBOAT family proteins and monitoring the reduction in ghrelin octanoylation with matrix-assisted laser desorption ionization time-of-flight MS.768
Mutation of the conserved histidine or asparagine residue of GOAT completely abolishes its enzymatic activity. GOAT exhibits promiscuity toward various fatty acyl groups varying from C2 to C14.768 GOAT contains 11 transmembrane domains and one reentrant loop. Similar to Hhat, GOAT has a catalytic histidine in the ER lumen, whereas the asparagine residue is on a cytoplasmic loop.769 Purified GOAT can accept a minimal five-residue ghrelin peptide as a substrate, and the N-terminal glycine is required for recognition by GOAT.770
The identification of GOAT allowed the mechanism-based development of antagonists that could potentially prevent obesity. A pentapeptide inhibitor derived from the first five N-terminal amino acids of proghrelin was developed and further improved by replacing the oxyester linkage with a more stable amide linkage on the third serine residue.771 Later, developed a bi-substrate mimic peptide-based inhibitor, Go-CoA-Tat, was developed, which integrated the binding affinity of both substrates, octanoyl-CoA and ghrelin peptide, by linking them with a non-cleavable bridge.772 Go-CoA-Tat inhibited GOAT efficiently and selectively in mice, and the intraperitoneal administration of GO-CoA-Tat improved glucose intolerance and reduced weight gain in WT mice but not in ghrelin-deficient mice. Several non-peptide small-molecule inhibitors for GOAT have also been reported, including some triterpenoid compounds that act as covalent reversible inhibitors.773,774 However, the in vivo effects of these compounds have not been reported.
5.5. MBOATs
Members of the MBOAT enzyme family transfer fatty acyl groups to the hydroxyl moiety of either protein side chains or small hydrophobic lipid molecules. In 2000, Hofmann discovered the MBOAT family through bioinformatics analysis,718 thus leading to the subsequent identification of several other family members at the biochemical level. MBOAT family members contain multiple transmembrane domains and share two common putative catalytic residues: histidine and asparagine (Table 10). The active histidine residue is surrounded by a stretch of hydrophobic amino acids, whereas the asparagine site is embedded within a hydrophilic region. Both residues are highly conserved among MBOAT family members and required for enzymatic activities, with the exception of PORCN, in which the conserved asparagine is not required for activity.721 Human MBOAT family members can be characterized into three subclasses. Class I enzymes, including acyl-CoA cholesterol acyltransferase 1 and 2 (ACAT1/2)775,776 and diacylglycerol acyltransferase 1 (DGAT1), are mainly involved in neutral lipid biosynthesis.777 ACAT1/2 catalyze cholesterol esterification using oleoyl-coA and are potential drug targets for the treatment of Alzheimer’s disease. DGAT1 catalyzes the biosynthesis of retinyl esters, wax esters, and triacylglycerol.778 Class II MBOAT enzymes acylate protein substrates and consist of PORCN, Hhat, Hhat-like,779 and GOAT (see sections 5.1 and 5.2). The third subgroup belongs to the lysophospholipid acyltransferases (LPAT) family, which is involved in the phospholipid remodeling process. The fatty acid motif at the C2 position of a glycerolphospholipid can be cleaved by phospholipase A2 to produce lysophospholipid, which is reacylated by LPATs to diversify fatty acids at the C2 position. Lysophosphatidylethanolamine acyltransferase 1 (LPEAT1), lysophosphatidylcholine acyltransferase (LPCAT) 3, LPCAT4, and lysophosphatidylinositol acyltransferase 1 (LPIAT1) constitute the third class of MBOATs.780
Table 10.
Summary of membrane-bound O-acyl-transferase family members
| Protein | Gene | Alternative name | Subgroup | Functions | References |
|---|---|---|---|---|---|
| Acyl-CoA cholesterol acyltransferase 1 | ACAT1 | Neutral lipid biosynthesis | Cholesterol esterification | 775 | |
| Acyl-CoA cholesterol acyltransferase 2 | ACAT2 | Neutral lipid biosynthesis | Cholesterol esterification | 776 | |
| Diacylglycerol acyltransferase 1 | DGAT1 | Neutral lipid biosynthesis | Triglyceride synthesis | 778 | |
| Porcupine | PORCN | Protein fatty acylation | Fatty acylation of Wnt serine | 721 | |
| Hedgehog acyltransferase | HHAT | Protein fatty acylation | Fatty acylation of Hedgehog N-terminal cysteine | 746–748 | |
| Ghrelin octanoyltransferase | GOAT | MBOAT 4 | Protein fatty acylation | Fatty acylation of Ghrelin serine | 767,768 |
| Hedgehog acyltransferase-like | HHATL | MBOAT 3 | Protein fatty acylation | No enzymatic activity detected | 779 |
| Lysophosphatidylethanolamine acyltransferase 1 | LPEAT1 | MBOAT 1 | Membrane phospholipid remodeling | Fatty acylation of lysophospholipids | 780 |
| Lysophosphatidylcholine acyltransferase 3 | LPCAT3 | MBOAT 5 | Membrane phospholipid remodeling | Fatty acylation of lysophospholipids | 780 |
| Lysophosphatidylcholine acyltransferase 4 | LPCAT4 | MBOAT 2 | Membrane phospholipid remodeling | Fatty acylation of lysophospholipids | 780 |
| Lysophosphatidylinositol acyltransferase 1 | LPIAT1 | MBOAT 7 | Membrane phospholipid remodeling | Fatty acylation of lysophospholipids | 780 |
5.6. Histone Serine Palmitoylation
Histone H4 is reportedly palmitoylated on Ser47. Histone H4 serine palmitoylation occurs in a Ca2+-dependent manner, and LPCAT1 is the acyltransferase of histone serine palmitoylation.694 LPCAT1 acylates lysophosphatidylcholine to generate the pulmonary surfactant dipalmitoylphosphatidylcholine. Even though its name is similar to some of the MBOAT proteins mentioned above, LPCAT1 belongs to a different group of enzymes that contain only one transmembrane domain. Under normal conditions, LPCAT1 is found mainly in the cytosol. When the intracellular Ca2+ concentration increases, LPCAT1 translocates to the nucleus and promotes H4 palmitoylation, which is proposed to increase mRNA synthesis through an unknown mechanism.694
5.7. Lysine Acylation
Protein lysine residues are modified by many acyl groups from various acyl-CoA molecules produced during cellular metabolism, such as acetyl-CoA, propionyl-CoA, butyryl-CoA, succinyl-CoA, crotonyl-CoA, and long-chain fatty acyl-CoA.781 These modifications regulate various aspects of cell biology, most prominently epigenetics and metabolism. This section discusses the long-chain fatty acylation of protein lysine residues.
Escherichia coli hemolysin, a pore-forming toxin, undergoes lysine myristoylation,782 and this reaction is necessary for toxin activity. The myristoylation of hemolysin requires a specific acyl transferase that uses a myristoyl group tethered to the acyl carrier protein as the myristoyl donor.
In mammalian cells, the first protein reported to contain myristoyl lysine was tumor necrosis factor-alpha (TNFα).783 This discovery was made during the study of proteins that are myristoylated but lack an N-terminal glycine, which is the site for the well-known N-terminal glycine myristoylation. TNFα is a type II membrane protein with a single transmembrane domain. Lysine myristoylation occurs on Lys19 and Lys20 in the intracellular N-terminal domain. Similarly, interleukin-1 alpha is myristoylated on Lys82 and Lys83 in the propiece, catalyzed by an unidentified enzyme in monocyte lysate.784 Lens integral membrane protein aquaporin-0 reportedly undergoes lysine palmitoylation and oleylation. At present, the lysine acyltransferases remain unidentified.
The function of lysine fatty acylation in mammalian cells has been increasingly recognized owing to studies of a class of enzymes called sirtuins, which have begun to shed light on the function of this acylation. Sirtuins regulate many important biological processes, including transcription, metabolism, genome stability, and aging.785–787 They were thought to be NAD+-dependent protein lysine deacetylases.788 However, several of the seven mammalian sirtuins lack efficient deacetylase activity. Among them, SIRT5 is found to be an efficient desuccinylase and demalonylase,789 whereas SIRT6 can remove long-chain fatty acyl groups efficiently.790 One of the physiological substrates for the defatty-acylase activity of SIRT6 is TNFα.790 Defatty-acylation of TNFα on Lys19 and Lys20 by SIRT6 promotes the secretion of TNFα, which provides the first clue about the physiological function of lysine fatty acylation. One study has shown that the lysine fatty acylation of TNFα targets TNFα primarily to the lysosomes for degradation.791 However, the exact mechanism through which fatty acylation promotes this targeting remains unknown. Notably, although the original report suggested that TNFα is myristoylated, later studies of SIRT6-TNFα have suggested that palmitoylation might be more abundant because Alk14 produces stronger labeling than Alk12.
A more notable development is the finding that Ras-related protein R-Ras2 is fatty-acylated on lysine residues near the C-terminal, where the prenylated and palmitoylated cysteines reside.792 Similar to cysteine palmitoylation, lysine acylation promotes the plasma membrane targeting of R-Ras2. At the plasma membrane, R-Ras2 is more active and turns on the phosphatidylinositol 3-kinase (PI3K)/AKT signaling pathway, which leads to increased cell proliferation. Furthermore, R-Ras2 lysine fatty acylation can be reversed by SIRT6, a known tumor suppressor.792,793 The regulation of R-Ras2 and thus PI3K/AKT signaling may underlie this tumor suppressor role.792 The R-Ras2 lysine acylation study650 suggests that similar to cysteine acylation, lysine fatty acylation may have key biological functions.
Studies of sirutins have also suggested that lysine fatty acylation may be more abundant than previously thought. Data from our laboratory and others have shown that several mammalian sirtuins, such as SIRT1, SIRT2, and SIRT3, can remove long-chain fatty acyl groups with reasonable catalytic efficiency.794 A zinc-dependent histone deacetylase, HDAC8, also shows defatty-acylation activity in vitro.795 These sirtuins and HDAC may remove fatty acyl groups from various protein lysine residues in vivo, although the exact substrate proteins remain to be identified. A sirtuin from the malaria parasite was also demonstrated to prefer fatty acyl lysine over acetyl lysine, which suggests that protein lysine fatty acylation also occurs in this parasite.796
6. Concluding Remarks and Perspectives
This section highlights unaddressed fundamental questions in protein lipidation, the difficulties associated with addressing these questions, and potential solutions to overcoming these difficulties. Protein lipidation is clearly abundant and plays critical roles in biology. However, the detailed mechanistic understanding of the functions of lipidation is incomplete. Remarkable phenotypic observations have been made in many cases, but the fundamental mechanisms underlying these observations is lacking. When a lipid modification affects the activity of the protein being modified, the underlying mechanism is unknown in most cases. Does membrane association increase the chances of the protein interacting with its binding partners or substrates that are also membrane-associated? Or does the modification change the conformation of the protein, thereby affecting the binding of its partners or catalysis? Or is the modification directly involved in the binding interaction? Answering these questions requires structural information that may be difficult to obtain because many of the targets are membrane proteins or membrane-associated proteins. Although acquiring the structures of membrane proteins is becoming more tractable, it still requires tremendous effort. Therefore techniques such as hydrogen exchange MS797 may be more applicable. Technology that facilitates the preparation of membrane proteins, such as nanodiscs,798 will also be helpful for these studies.
Even for membrane targeting, specific questions must still be answered. For example, how can the same modification target different proteins to different organelles (e.g., N-terminal glycine myristoylation targets certain proteins to the mitochondria and others to the plasma membrane)? Do lipid modification and its local environment have intrinsic affinity for different membranes, or are different trafficking machineries engaged by the modified proteins? We do not think significant technical challenges are associated with addressing these questions. By contrast, the difficulty might be the complexity of the situation (e.g., the existence of different trafficking machineries).
The dynamic regulation of lipid modification is another area that has not been thoroughly investigated. For example, can the metabolic or nutritional status of a cell affect protein lipidation? Can certain signaling pathways affect lipid modifications? This area is a key research direction because it may provide another level of understanding of the physiological processes involving protein lipidation. As a reference point, the importance of protein phosphorylation would not have been appreciated without knowing the dynamics of protein phosphorylation. Our investigation of TNFα lysine fatty acylation indicates that the level of fatty acids in the cell medium can affect the secretion of TNFα, which suggests that the metabolic or nutritional status of cells can affect protein lipidation and therefore protein function.790 Currently, the most widely adapted technology to detect protein lipidation is metabolic labeling with chemical probes. However, this approach is difficult to apply to studies of the dynamic regulation of protein lipidation by metabolic or nutritional status. The use of chemical probes unavoidably changes the metabolic status of the cells, and these probes are challenging to use in whole animals. From this perspective, the most urgently needed tools are antibodies or their equivalents that can be used to detect protein lipidation. Currently, no antibodies are available for any of the lipid modifications discussed in this review, perhaps because the antigens used to immunize animals might nonspecifically associate with cell membranes and therefore cannot be effectively seen by the immune system. If this is true, then certain in vitro systems for antibody development (aptamers SELEX or phage display) may be useful alternatives to the development of such antibodies.
In addition to questions that are general to all lipid modifications, questions specific to certain lipidations also remain. Cysteine palmitoylation, by far the most complex of all protein lipid modifications, still requires elucidation. For example, the substrates and functions of each DHHC are largely unknown. No efficient inhibitors have been developed to determine whether any DHHCs can be pharmacologically targeted to treat human diseases (the most commonly used inhibitor to date is 2-BP, which has multiple drawbacks515,522,799,800). The reversibility of cysteine palmitoylation is also of great interest. Even though depalmitoylases have been reported, the extent to which cysteine palmitoylation is regulated by enzymatic depalmitoylation is unknown. For the MBOAT family of protein O-acyltransferases, a particularly compelling question is whether, similar to other lipidation enzymes, they have multiple substrate proteins. For lysine fatty acylation, pressing questions include how it occurs, how abundant it is, what biological functions it has, and how it achieves these functions.
Since the late 1990s, tremendous progress has been made in elucidating protein lipidation. Given the enormous body of knowledge accumulated and the availability of numerous tools and technologies in this field, progress in the coming decades will be even more impressive, and many of the questions raised in this review will be satisfactorily addressed.
Table 4.
Type II and IV single-pass transmembrane proteins that undergo S-palmitoylationa
| Protein | Uniprot# | Palmitoylation sites | Modification site location | Sequence near the Cys | Function of palmitoylation | DHHC enzyme | Ref. |
|---|---|---|---|---|---|---|---|
| Calnexin | P27824 | C503, C504 | Cytoplasmic end of TM domain |

|
Required for targeting mitochondria-associated membrane | DHHC6 | 553 |
| CKAP4 | Q07065 | C100 | Cytoplasmic domain |
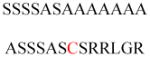
|
Required for antiproliferative factor-mediated signaling | DHHC2 | 481 |
| ECE-1a | Q28010 | C46 | Cytoplasmic domain |

|
Unknown | 554 | |
| FasL | P48023 | C82 | Cytoplasmic end of TM domain |
|
Required for cell death-inducing activity | 555 | |
| IFITM3 | Q01628 | C71, C72, C105 | Cytoplasmic domain |

|
Required for resistance to influenza virus | 556 | |
| PLSCR1 | O15162 | C184–186, C188, C189 | Cytoplasmic domain |
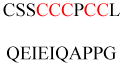
|
Required for nucleus to plasma membrane trafficking | 557 | |
| proSP-C | P11686 | C5, C6 | Cytoplasmic domain |
|
Required for ER exit | 558 | |
| Syntaxin 7 | O15400 | C239 | Cytoplasmic end of TM domain |
|
Required for cycles between endosomes and plasma membrane | 559 | |
| Syntaxin 8 | Q9UNK0 | C214 | Cytoplasmic end of TM domain |
|
Unknown | 559 | |
| TNF-α | P01375 | C30 | Cytoplasmic domain |
|
Required for lipid rafts localization | 560 |
Both type II and type IV membrane proteins contain one transmembrane domain with a cytoplasmic N-terminus. In type II membrane proteins, the transmembrane domain is close to the N-terminus, whereas in Type IV proteins, this domain is close to the C-terminus.
Table 5.
Multipass transmembrane proteins that undergo S-palmitoylation
| Protein | Uniprot# | Palmitoylation sites | Modification site location | Sequence near the Cys | Function of palmitoylation | DHHC enzyme | Ref. |
|---|---|---|---|---|---|---|---|
| α4 nAChR | P43681 | C273 | Intracellular loop between TM1 and TM2 |
|
Regulate cell surface protein expression | 561 | |
| β(1)AR | P08588 | C392, C393, C414 | Cytoplasmic C terminus |

|
Required for efficient internalization | 562 | |
| β(2)AR | P07550 | C341 | Cytoplasmic C terminus |
|
Required for binding to arrestin/PDE4D | 563 | |
| 5-HT4a receptor | P97288-2 | C328, C329, C386 | Cytoplasmic C terminus |
|
Regulate protein phosphorylation | 564 | |
| 5-HT(7) receptor | P34969 | C404, C438, C441 | Cytoplasmic C terminus |
|
Required for Gs-mediated activity of 5-HT(7) | 565 | |
| ABCA1 | O95477 | C3, C23, C1110, C1111 | C3: N-terminal cytoplasmic domain; C23, C1110, C1111: internal domain |

|
Required for plasma membrane localization | DHHC8 | 411 |
| ABCG1 | P45844 | C26, C150, C311, C390, C402 | Cytoplasmic N terminus |

|
Required for ABCG1-mediated cholesterol efflux | 566 | |
| APH-1 | Q96BI3 | C182, C245 | C182: internal domain; C245: C-terminal cytoplasmic domain |

|
Required for lipid rafts localization | 543 | |
| Barttin | Q8WZ55 | C54, C56 | Cytoplasmic domain |
|
Regulate channel activity | 567 | |
| Bradykinin B2 receptor | P30411 | C351, C356 | Cytoplasmic C terminus |
|
Required for the stabilization of the receptor-ligand complex | 568 | |
| BK channels | Q12791 | C53, C54, C56 | Intracellular loop between TM1 and TM2 |
|
Regulate cell surface protein expression | DHHC1 DHHC24 |
569 |
| Bovine rhodopsin | P02699 | C322, C323 | Cytoplasmic C terminus |
|
Required for phosphorylation by rhodopsin kinase | 570 | |
| CB(1) receptor | P21554 | C415 | Cytoplasmic C terminus |
|
Required for plasma membrane localization | 571 | |
| CCR5 | P51681 | C321, C323, C324 | Cytoplasmic C terminus |
|
Control CCR5 expression | 572 | |
| CD36 | P16671 | C3, C7, C464, C466 | C3, C7: Cytoplasmic N terminus; C464, C466: Cytoplasmic C terminus |
|
Required for ER to plasma membrane trafficking | 573 | |
| CD39 | P49961 | C13 | Cytoplasmic N terminus |
|
Required for targeting CD39 to caveolae | 574 | |
| CD81 | P60033 | C6, 9, 80, 89, 97, 104, 277, 228 | C6, C9: Cytoplasmic N terminus; C80: inside TM2; C89: Intracellular loop between TM2 and TM3; C97, C104: inside TM3; C227, C228: Cytoplasmic C terminus |
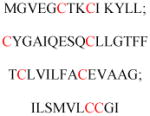
|
Required for association with tetraspanin-enriched microdomains | 575 | |
| CD82 | P27701 | C5, C74, C83, C251, C253 | C5: Cytoplasmic N terminus; C74, C83: Intracellular loop between TM2 and TM3; C251, C253: Cytoplasmic C terminus |
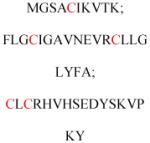
|
Required for motility- and invasiveness-inhibitory activity | 576 | |
| CD151 | P48509 | C11, C15, C242, C243 | C11, C15: Cytoplasmic N terminus; C242, C243: Cytoplasmic C terminus |
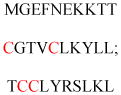
|
Required for tetraspanin-protein interactions | DHHC2 | 577 |
| Claudin-14 | O95500 | C104, C107 | Cytoplasmic C terminus |

|
Required for efficient tight-junction localization | 578 | |
| DATs | P23977 | C580 | Cytoplasmic C terminus |
|
Control DATs degradation | 579 | |
| Dopamine D1 receptor | P21728 | C347, C351 | Cytoplasmic C terminus |

|
Required for dopamine D1 receptor internalization | 580 | |
| Dopamine D4 receptor | P21917 | C467 | Cytoplasmic C terminus |
|
Enhance surface expression and endocytosis | 581 | |
| Dopamine transporter | Q01959 | C580 | Cytoplasmic C terminus |
|
Regulate transport capacity | 582 | |
| ENaC | Q9WU38 | C43, C557 | C43: Cytoplasmic N terminus; C557: Cytoplasmic C terminus |
|
Required for plasma membrane localization | 583 | |
| ETA | P25101 | C383, C385, C386, C387, C388 | Cytoplasmic C terminus |
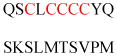
|
Required for ETA receptor activity | 584 | |
| ETB | P21451 | C402, C403, C405 | Cytoplasmic C terminus |
|
Required for coupling with Gi | 585 | |
| GLT-1 | P43006 | C38 | Cytoplasmic N terminus |
|
Required for glutamate uptake | 586 | |
| GluK2a | P42260 | C858, C871 | Cytoplasmic C terminus |

|
Required for interaction with 4.1N | 587 | |
| GluR1 | P42261 | C603, C829 | C603: intramembrane domain; C829: Cytoplasmic C terminus |
|
Required for intracellular trafficking | DHHC3 DHHC17 |
588 |
| GluR2 | P42262 | C610, C836 | C610: intramembrane domain; C836: Cytoplasmic C terminus |
|
Required for intracellular trafficking | DHHC3 DHHC17 |
588 |
| GluR6 | Q13002 | C858, C871 | Cytoplasmic C terminus |

|
Regulate phosphorylation state | 589 | |
| Gp78 | Q9UKV5 | RING finger (6 cysteines) | Intracellular loop between TM6 and TM7 |

|
Required for ER exit | DHHC2/6/11/13/24 | 383 |
| H2R | P17124 | C305 | Cytoplasmic C terminus |
|
Required for plasma membrane localization | 590 | |
| hIP | P43119 | C308, C311, C383 | Cytoplasmic C terminus |

|
Required for the interaction with Rab11 | 591 | |
| hPAR(2) | P55085 | C361 | Cytoplasmic C terminus |
|
Required for ERK1/2 activation and calcium signaling | 592 | |
| IFITM1 | Q9D103 | C49, C50, C83, C103 | C49, C50 is on extracellular domain, C83 is inside TM2 and C103 is cytoplasmic between TM1 and TM2 |

|
Regulate IFITM1 stability | DHHC5 | 593 |
| ITPR1 | Q14643 | C56, C849, C2214 | Cytoplasmic domain |

|
Regulate calcium flux | DHHC6 | 404 |
| LH/hCGR | P22888 | C643, C644 | Cytoplasmic C terminus |
|
Required for arrestin-mediated internalization | 594 | |
| LMP1 | P03230 | C78 | Inside TM domain |

|
Required for lipid rafts localization | 595 | |
| M2 receptor | P08172 | C457 | Cytoplasmic C terminus |

|
Enhances the ability to interact with G protein | 596 | |
| NCX1 | P32418 | C739 | Internal domain |
|
Regulate calcium homeostasis | 499 | |
| NMDARs | Q12879 | C848, C853, C870, C1214, C1217, C1236, C1239 | Cytoplasmic C terminus |

|
Required for cell surface protein expression | 597 | |
| NTSR-1 | P30989 | C381, C383 | Cytoplasmic C terminus |

|
Required for receptor-mediated mitogenic-signaling | 598 | |
| Kv1.1 ion channel | Q09470 | C243 | Internal domain |
|
Modulate voltage sensing | 599 | |
| P2X7R | Q99572 | C371, 373, 374, 477, 479, 482, 498, 499, 506, 572, 573 | Cytoplasmic C terminus |

|
Required for lipid rafts localization | 600 | |
| PAR2 | P55085 | C361 | Cytoplasmic C terminus |
|
Required for plasma membrane localization | 601 | |
| PMP22 | Q01453 | C85 | Inside TM2 domain |
|
Regulate epithelial cell morphology and migration | 602 | |
| Potassium channel Kv1.5 | P22460 | C593 | Cytoplasmic C terminus |
|
Required for plasma membrane localization | 603 | |
| Prostacyclin receptor | P43119 | C308, C311 | Cytoplasmic C terminus |
|
Required for coupling to G(s) | 604 | |
| Rhodopsin | P08100 | C322, C323 | Cytoplasmic C terminus |
|
Regulate Rhodopsin stability | 605 | |
| S1P | P21453 | C328 | Cytoplasmic C terminus |
|
Required for S1P internalization | 606 | |
| SCN5A | Q14524 | C981 | Internal cytoplasmic domain |
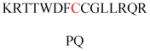
|
Increase channel availability and late sodium current activity | 607 | |
| SMS2 | Q8NHU3 | C331, C332, C343, C348 | Cytoplasmic C terminus |
|
Required for plasma membrane localization | 608 | |
| STING | Q86WV6 | C88, C91 | Inside TM domain |
|
Required for activation of STING | 609 | |
| STREX | Q08460-4 | C645, C646 | Cytoplasmic C terminus |

|
Required for plasma membrane localization | DHHC3/17/9/7/5 | 610 |
| SynDIG1 | Q9H7V2 | C191, C192 | Inside TM domain |
|
Regulate dendritic targeting in neurons | 611 | |
| TPβ (isoform2) | P21731 | C347 | Cytoplasmic C terminus |

|
Required for internalization | 612 | |
| TRH-R1 | P21761 | C335, C337 | Cytoplasmic C terminus |

|
Constrain TRH-R1 in an inactive conformation | 613 | |
| TRPP3 | Q9P0L9 | C38 | Cytoplasmic N terminus |
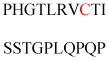
|
Required for the cationic channel function | ||
| V1aR | P37288 | C365, C366 | Cytoplasmic C terminus |

|
Regulated both phosphorylation and sequestration of the receptor | 614 | |
| V2 vasopressin receptor | P30518 | C341, C342 | Cytoplasmic C terminus |
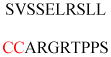
|
Enhances beta-arrestin recruitment | 615 | |
| Y(1) receptor | P21555 | C337 | Cytoplasmic C terminus |
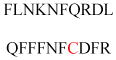
|
Required for coupling to G proteins | 616 |
Table 6.
S-Palmitoylated proteins that undergo N-terminal glycine myristoylation
| Protein | Uniprot# | Palmitoylation sites | Modification site location | Sequence near the Cys | Function of palmitoylation | DHHC enzyme | Ref. |
|---|---|---|---|---|---|---|---|
| eNOS | P29474 | C15, C26 | N terminus |
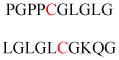
|
Enhance punctate Golgi membrane localization | 240 | |
| Fyn | P06241 | C3, C6 | N terminus |
|
Required for plasma membrane localization | DHHC21 | 617 |
| Globin X | Q5QSA9 | C3 | N terminus |
|
Required for plasma membrane localization | 618 | |
| Lck | P06239 | C3, C5 | N terminus |

|
Required for plasma membrane localization | DHHC2 | 619 |
| Lyn | P25911 | C3 | N terminus |
|
Required for plasma membrane localization | 617 | |
| Yes | P07947 | C3 | N terminus |
|
Required for plasma membrane localization | 617 | |
| PSKH1 | C3 | N terminus | MGCGTSKVLPEPPKDVQLDL | Required for Golgi membrane targeting | 281 | ||
| Gα i1, i2, i3 |
P63096 P04899 P08754 |
C3 | N terminus |

|
Required for signal transduction | 620 | |
| Gα(o) | P09471 | C3 | N terminus |
|
Depress susceptibility to GAP-43 activation | DHHC3 DHHC7 |
621 |
Table 7.
S-Palmitoylated proteins that undergo C-terminal prenylation
| Protein | Uniprot# | Palmitoylation sites | Modification site location | Sequence near the Cys | Function of palmitoylation | DHHC enzyme | Ref. |
|---|---|---|---|---|---|---|---|
| bCdc42 | P60953 | C189 | C terminus |
|
Required for plasma membrane localization | 113 | |
| cPLA(2)γ | Q9UP65 | C539 | C terminus |
|
Required for plasma membrane localization | 622 | |
| H-Ras | P01112 | C161, C164 | C terminus |
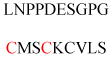
|
Required for plasma membrane localization | DHHC18 | 142 |
| K-Ras4A | P01116 | C160 | C terminus |
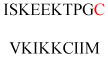
|
Required for plasma membrane localization | 142 | |
| N-Ras | P01111 | C161 | C terminus |
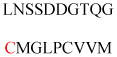
|
Required for plasma membrane localization | ZDHHC9-GOLGA7 complex | 142 |
| R-Ras | P10301 | C213 | C terminus |
|
Required for plasma membrane localization | DHHC19 | 449 |
| Rac1 | P63000 | C178 | C terminus |
|
Required for plasma membrane localization | 623 | |
| Rap2A/B/C | P10114, P61225, Q9Y3L5 | C176, C177 | C terminus |
|
Required for endosome localization | 624 | |
| RhoB | P62745 | C192 | C terminus |
|
Required for apoptotic activity | 625 | |
| Wrch-1 | Q7L0Q8 | C256 | C terminus |
|
Required for plasma membrane localization | 626 |
Acknowledgments
Work in our laboratory on protein lipidation is supported by NIH grants GM086703, GM098596, and DK107868.
Biographies
Hong Jiang is a currently Principle Investigator at Interdisciplinary Research Center on Biology and Chemistry (IRCBC), Chinese Academy of Sciences in Shanghai, China. He obtained his B.S. degree in chemistry from University of Science and Technology of China in 1998, and completed his Ph.D. in organic chemistry at University of Science and Technology of China with Professor You-Cheng Liu in 2003. After postdoctoral training with Professor T. J. Kappock at Washington University in St. Louis, he joined the group of Professor Hening Lin at Cornell University in 2006 focusing on chemical biology studies. In 2015, he received “Thousand Youth Talents Plan” from China and joined the faculty at IRCBC in Shanghai.
Xiaoyu Zhang obtained his B.S. degree in Pharmaceutical Sciences in 2008 and M.S. degree in Pharmaceutical Sciences in 2011 with Dr. Zhongjun Ma from Zhejiang University. He obtained his Ph.D. degree in 2017 from Cornell University under the guidance of Dr. Hening Lin. He is currently a postdoctoral research associate in the group of Dr. Benjamin Cravatt at the Scripps Research Institute.
Xiao Chen attended Iowa State University of Science and Technology and obtained his baccalaureate in Chemistry and a second baccalaureate in Genetics. He is attending Cornell University for his graduate studies.
Pornpun Aramsangtienchai obtained her B.Sc. and M.Sc. degrees in Biochemistry from Chulalongkorn University in Thailand. Then she received her Ph.D. in Biochemistry from Cornell University in 2016 under the supervision of Professor Hening Lin. Presently, she is a faculty member in Department of Biochemistry at Burapha University, Thailand. Her research interests center on glycobiology and enzymatic synthesis of oligo- and polysaccharides.
Zhen Tong was born in China and earned her B.S. degree in Chemical Biology in 2010 from Tsinghua University, Beijing, China. She obtained her Ph.D. degree in 2016 from Cornell University under the supervision of Dr. Hening Lin. Her doctoral research interests include the exploration of novel enzymatic activities and activation mechanism of SIRT7. She joined Bristol-Myers Squibb in 2016 working on biologics process development and commercial manufacturing support.
Hening Lin was born in China and obtained his B.S. in Chemistry in 1998 from Tsinghua University, Beijing, China. He obtained his PhD degree in 2003 from Columbia University under the guidance of Dr. Virginia Cornish. From 2003–2006, he was a Jane Coffin Childs postdoctoral fellow in Dr. Christopher Walsh’s lab at Harvard Medical School. He joined the faculty of Department of Chemistry and Chemical Biology at Cornell University as an assistant professor in 2006. He was promoted to associate professor in 2012 and professor in 2013. His lab works at the interface of chemistry, biology, and medicine. The research in his group focuses on NAD+-consuming enzymes that have important biological functions and human disease relevance, including poly(ADP-ribose) polymerases and sirtuins. His lab also works on the biosynthesis of diphthamide, the target of diphtheria toxin. His work is recognized by a Dreyfus New Faculty Award in 2006, the CAPA Distinguished Junior Faculty Award in 2011, the 2014 ACS Pfizer Award in Enzyme Chemistry, and the 2016 OKeanos-CAPA Senior Investigator Award. He has been a Howard Hughes Medical Institute investigator since 2015.
References
- 1.Lemmon MA, Ferguson KM. Signal-Dependent Membrane Targeting by Pleckstrin Homology (Ph) Domains. Biochem J. 2000;350(Pt 1):1–18. [PMC free article] [PubMed] [Google Scholar]
- 2.Lentz BR. Exposure of Platelet Membrane Phosphatidylserine Regulates Blood Coagulation. Prog Lipid Res. 2003;42:423–438. doi: 10.1016/s0163-7827(03)00025-0. [DOI] [PubMed] [Google Scholar]
- 3.Menon AK, Kinoshita T, Orlean PA, Tamanoi F. Glycosylphosphatidylinositol (Gpi) Anchoring of Proteins. Academic Press; 2009. [Google Scholar]
- 4.Zurzolo C, Simons K. Glycosylphosphatidylinositol-Anchored Proteins: Membrane Organization and Transport. Biochim Biophys Acta. 2016;1858:632–639. doi: 10.1016/j.bbamem.2015.12.018. [DOI] [PubMed] [Google Scholar]
- 5.Paladino S, Lebreton S, Zurzolo C. Trafficking and Membrane Organization of Gpi-Anchored Proteins in Health and Diseases. Curr Top Membr. 2015;75:269–303. doi: 10.1016/bs.ctm.2015.03.006. [DOI] [PubMed] [Google Scholar]
- 6.Epstein WW, Lever D, Leining LM, Bruenger E, Rilling HC. Quantitation of Prenylcysteines by a Selective Cleavage Reaction. Proc Natl Acad Sci US A. 1991;88:9668–9670. doi: 10.1073/pnas.88.21.9668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tschantz WR, Zhang L, Casey PJ. Cloning, Expression, and Cellular Localization of a Human Prenylcysteine Lyase. J Biol Chem. 1999;274:35802–35808. doi: 10.1074/jbc.274.50.35802. [DOI] [PubMed] [Google Scholar]
- 8.Tschantz WR, Digits JA, Pyun HJ, Coates RM, Casey PJ. Lysosomal Prenylcysteine Lyase Is a Fad-Dependent Thioether Oxidase. J Biol Chem. 2001;276:2321–2324. doi: 10.1074/jbc.C000616200. [DOI] [PubMed] [Google Scholar]
- 9.Hancock JF, Magee AI, Childs JE, Marshall CJ. All Ras Proteins Are Polyisoprenylated but Only Some Are Palmitoylated. Cell. 1989;57:1167–1177. doi: 10.1016/0092-8674(89)90054-8. [DOI] [PubMed] [Google Scholar]
- 10.Schafer WR, Kim R, Sterne R, Thorner J, Kim SH, Rine J. Genetic and Pharmacological Suppression of Oncogenic Mutations in Ras Genes of Yeast and Humans. Science. 1989;245:379–385. doi: 10.1126/science.2569235. [DOI] [PubMed] [Google Scholar]
- 11.Rilling HC, Breunger E, Epstein WW, Crain PF. Prenylated Proteins: The Structure of the Isoprenoid Group. Science. 1990;247:318–320. doi: 10.1126/science.2296720. [DOI] [PubMed] [Google Scholar]
- 12.Farnsworth CC, Gelb MH, Glomset JA. Identification of Geranylgeranyl-Modified Proteins in Hela Cells. Science. 1990;247:320–322. doi: 10.1126/science.2296721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boyartchuk VL, Ashby MN, Rine J. Modulation of Ras and a-Factor Function by Carboxyl-Terminal Proteolysis. Science. 1997;275:1796–1800. doi: 10.1126/science.275.5307.1796. [DOI] [PubMed] [Google Scholar]
- 14.Dai Q, Choy E, Chiu V, Romano J, Slivka SR, Steitz SA, Michaelis S, Philips MR. Mammalian Prenylcysteine Carboxyl Methyltransferase Is in the Endoplasmic Reticulum. J Biol Chem. 1998;273:15030–15034. doi: 10.1074/jbc.273.24.15030. [DOI] [PubMed] [Google Scholar]
- 15.Reiss Y, Goldstein JL, Seabra MC, Casey PJ, Brown MS. Inhibition of Purified P21ras Farnesyl:Protein Transferase by Cys-Aax Tetrapeptides. Cell. 1990;62:81–88. doi: 10.1016/0092-8674(90)90242-7. [DOI] [PubMed] [Google Scholar]
- 16.Seabra MC, Reiss Y, Casey PJ, Brown MS, Goldstein JL. Protein Farnesyltransferase and Geranylgeranyltransferase Share a Common Alpha Subunit. Cell. 1991;65:429–434. doi: 10.1016/0092-8674(91)90460-g. [DOI] [PubMed] [Google Scholar]
- 17.Chen WJ, Andres DA, Goldstein JL, Brown MS. Cloning and Expression of a Cdna Encoding the Alpha Subunit of Rat P21ras Protein Farnesyltransferase. Proc Natl Acad Sci US A. 1991;88:11368–11372. doi: 10.1073/pnas.88.24.11368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen WJ, Andres DA, Goldstein JL, Russell DW, Brown MS. Cdna Cloning and Expression of the Peptide-Binding Beta Subunit of Rat P21ras Farnesyltransferase, the Counterpart of Yeast Dpr1/Ram1. Cell. 1991;66:327–334. doi: 10.1016/0092-8674(91)90622-6. [DOI] [PubMed] [Google Scholar]
- 19.Zhang FL, Moomaw JF, Casey PJ. Properties and Kinetic Mechanism of Recombinant Mammalian Protein Geranylgeranyltransferase Type I. J Biol Chem. 1994;269:23465–23470. [PubMed] [Google Scholar]
- 20.Park HW, Boduluri SR, Moomaw JF, Casey PJ, Beese LS. Crystal Structure of Protein Farnesyltransferase at 2. 25 Angstrom Resolution. Science. 1997;275:1800–1804. doi: 10.1126/science.275.5307.1800. [DOI] [PubMed] [Google Scholar]
- 21.Taylor JS, Reid TS, Terry KL, Casey PJ, Beese LS. Structure of Mammalian Protein Geranylgeranyltransferase Type-I. EMBO J. 2003;22:5963–5974. doi: 10.1093/emboj/cdg571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reiss Y, Brown MS, Goldstein JL. Divalent Cation and Prenyl Pyrophosphate Specificities of the Protein Farnesyltransferase from Rat Brain, a Zinc Metalloenzyme. J Biol Chem. 1992;267:6403–6408. [PubMed] [Google Scholar]
- 23.Moomaw JF, Casey PJ. Mammalian Protein Geranylgeranyltransferase. Subunit Composition and Metal Requirements. J Biol Chem. 1992;267:17438–17443. [PubMed] [Google Scholar]
- 24.Long SB, Casey PJ, Beese LS. The Basis for K-Ras4b Binding Specificity to Protein Farnesyltransferase Revealed by 2 a Resolution Ternary Complex Structures. Structure. 2000;8:209–222. doi: 10.1016/s0969-2126(00)00096-4. [DOI] [PubMed] [Google Scholar]
- 25.Long SB, Casey PJ, Beese LS. Cocrystal Structure of Protein Farnesyltransferase Complexed with a Farnesyl Diphosphate Substrate. Biochemistry. 1998;37:9612–9618. doi: 10.1021/bi980708e. [DOI] [PubMed] [Google Scholar]
- 26.Armstrong SA, Hannah VC, Goldstein JL, Brown MS. Caax Geranylgeranyl Transferase Transfers Farnesyl as Efficiently as Geranylgeranyl to Rhob. J Biol Chem. 1995;270:7864–7868. doi: 10.1074/jbc.270.14.7864. [DOI] [PubMed] [Google Scholar]
- 27.Tong H, Kuder CH, Wasko BM, Hohl RJ. Quantitative Determination of Isopentenyl Diphosphate in Cultured Mammalian Cells. Anal Biochem. 2013;433:36–42. doi: 10.1016/j.ab.2012.09.001. [DOI] [PubMed] [Google Scholar]
- 28.Hooff GP, Volmer DA, Wood WG, Muller WE, Eckert GP. Isoprenoid Quantitation in Human Brain Tissue: A Validated Hplc-Fluorescence Detection Method for Endogenous Farnesyl- (Fpp) and Geranylgeranylpyrophosphate (Ggpp) Anal Bioanal Chem. 2008;392:673–680. doi: 10.1007/s00216-008-2306-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hooff GP, Patel N, Wood WG, Muller WE, Eckert GP, Volmer DA. A Rapid and Sensitive Assay for Determining Human Brain Levels of Farnesyl-(Fpp) and Geranylgeranylpyrophosphate (Ggpp) and Transferase Activities Using Uhplc-Ms/Ms. Anal Bioanal Chem. 2010;398:1801–1808. doi: 10.1007/s00216-010-4088-7. [DOI] [PubMed] [Google Scholar]
- 30.Pompliano DL, Rands E, Schaber MD, Mosser SD, Anthony NJ, Gibbs JB. Steady-State Kinetic Mechanism of Ras Farnesyl:Protein Transferase. Biochemistry. 1992;31:3800–3807. doi: 10.1021/bi00130a010. [DOI] [PubMed] [Google Scholar]
- 31.Pompliano DL, Schaber MD, Mosser SD, Omer CA, Shafer JA, Gibbs JB. Isoprenoid Diphosphate Utilization by Recombinant Human Farnesyl:Protein Transferase: Interactive Binding between Substrates and a Preferred Kinetic Pathway. Biochemistry. 1993;32:8341–8347. doi: 10.1021/bi00083a038. [DOI] [PubMed] [Google Scholar]
- 32.Furfine ES, Leban JJ, Landavazo A, Moomaw JF, Casey PJ. Protein Farnesyltransferase: Kinetics of Farnesyl Pyrophosphate Binding and Product Release. Biochemistry. 1995;34:6857–6862. doi: 10.1021/bi00020a032. [DOI] [PubMed] [Google Scholar]
- 33.Huang CC, Casey PJ, Fierke CA. Evidence for a Catalytic Role of Zinc in Protein Farnesyltransferase. Spectroscopy of Co2+-Farnesyltransferase Indicates Metal Coordination of the Substrate Thiolate. J Biol Chem. 1997;272:20–23. doi: 10.1074/jbc.272.1.20. [DOI] [PubMed] [Google Scholar]
- 34.Dolence JM, Cassidy PB, Mathis JR, Poulter CD. Yeast Protein Farnesyltransferase: Steady-State Kinetic Studies of Substrate Binding. Biochemistry. 1995;34:16687–16694. doi: 10.1021/bi00051a017. [DOI] [PubMed] [Google Scholar]
- 35.Long SB, Casey PJ, Beese LS. Reaction Path of Protein Farnesyltransferase at Atomic Resolution. Nature. 2002;419:645–650. doi: 10.1038/nature00986. [DOI] [PubMed] [Google Scholar]
- 36.Mathis JR, Poulter CD. Yeast Protein Farnesyltransferase: A Pre-Steady-State Kinetic Analysis. Biochemistry. 1997;36:6367–6376. doi: 10.1021/bi9629182. [DOI] [PubMed] [Google Scholar]
- 37.Tschantz WR, Furfine ES, Casey PJ. Substrate Binding Is Required for Release of Product from Mammalian Protein Farnesyltransferase. J Biol Chem. 1997;272:9989–9993. doi: 10.1074/jbc.272.15.9989. [DOI] [PubMed] [Google Scholar]
- 38.Stirtan WG, Poulter CD. Yeast Protein Geranylgeranyltransferase Type-I: Steady-State Kinetics and Substrate Binding. Biochemistry. 1997;36:4552–4557. doi: 10.1021/bi962579c. [DOI] [PubMed] [Google Scholar]
- 39.Dolence JM, Poulter CD. A Mechanism for Posttranslational Modifications of Proteins by Yeast Protein Farnesyltransferase. Proc Natl Acad Sci US A. 1995;92:5008–5011. doi: 10.1073/pnas.92.11.5008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mu Omer CA, Gibbs RA. On the Stereochemical Course of Human Protein-Farnesyl Transferase. J Am Chem Soc. 1996;118:1817–1823. [Google Scholar]
- 41.Clausen VA, Edelstein RL, Distefano MD. Stereochemical Analysis of the Reaction Catalyzed by Human Protein Geranylgeranyl Transferase. Biochemistry. 2001;40:3920–3930. doi: 10.1021/bi002011a. [DOI] [PubMed] [Google Scholar]
- 42.Lenevich S, Xu J, Hosokawa A, Cramer CJ, Distefano MD. Transition State Analysis of Model and Enzymatic Prenylation Reactions. J Am Chem Soc. 2007;129:5796–5797. doi: 10.1021/ja069119j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Armstrong SA, Seabra MC, Sudhof TC, Goldstein JL, Brown MS. Cdna Cloning and Expression of the Alpha and Beta Subunits of Rat Rab Geranylgeranyl Transferase. J Biol Chem. 1993;268:12221–12229. [PubMed] [Google Scholar]
- 44.Seabra MC, Brown MS, Slaughter CA, Sudhof TC, Goldstein JL. Purification of Component a of Rab Geranylgeranyl Transferase: Possible Identity with the Choroideremia Gene Product. Cell. 1992;70:1049–1057. doi: 10.1016/0092-8674(92)90253-9. [DOI] [PubMed] [Google Scholar]
- 45.Andres DA, Seabra MC, Brown MS, Armstrong SA, Smeland TE, Cremers FP, Goldstein JL. Cdna Cloning of Component a of Rab Geranylgeranyl Transferase and Demonstration of Its Role as a Rab Escort Protein. Cell. 1993;73:1091–1099. doi: 10.1016/0092-8674(93)90639-8. [DOI] [PubMed] [Google Scholar]
- 46.Larijani B, Hume AN, Tarafder AK, Seabra MC. Multiple Factors Contribute to Inefficient Prenylation of Rab27a in Rab Prenylation Diseases. J Biol Chem. 2003;278:46798–46804. doi: 10.1074/jbc.M307799200. [DOI] [PubMed] [Google Scholar]
- 47.Rak A, Pylypenko O, Niculae A, Pyatkov K, Goody RS, Alexandrov K. Structure of the Rab7:Rep-1 Complex: Insights into the Mechanism of Rab Prenylation and Choroideremia Disease. Cell. 2004;117:749–760. doi: 10.1016/j.cell.2004.05.017. [DOI] [PubMed] [Google Scholar]
- 48.Zhang H, Seabra MC, Deisenhofer J. Crystal Structure of Rab Geranylgeranyltransferase at 2. 0 a Resolution. Structure. 2000;8:241–251. doi: 10.1016/s0969-2126(00)00102-7. [DOI] [PubMed] [Google Scholar]
- 49.Guo Z, Wu YW, Das D, Delon C, Cramer J, Yu S, Thuns S, Lupilova N, Waldmann H, Brunsveld L, et al. Structures of Rabggtase-Substrate/Product Complexes Provide Insights into the Evolution of Protein Prenylation. EMBO J. 2008;27:2444–2456. doi: 10.1038/emboj.2008.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pylypenko O, Rak A, Reents R, Niculae A, Sidorovitch V, Cioaca MD, Bessolitsyna E, Thoma NH, Waldmann H, Schlichting I, et al. Structure of Rab Escort Protein-1 in Complex with Rab Geranylgeranyltransferase. Mol Cell. 2003;11:483–494. doi: 10.1016/s1097-2765(03)00044-3. [DOI] [PubMed] [Google Scholar]
- 51.Thoma NH, Iakovenko A, Goody RS, Alexandrov K. Phosphoisoprenoids Modulate Association of Rab Geranylgeranyltransferase with Rep-1. J Biol Chem. 2001;276:48637–48643. doi: 10.1074/jbc.M108241200. [DOI] [PubMed] [Google Scholar]
- 52.Wu YW, Goody RS, Abagyan R, Alexandrov K. Structure of the Disordered C Terminus of Rab7 Gtpase Induced by Binding to the Rab Geranylgeranyl Transferase Catalytic Complex Reveals the Mechanism of Rab Prenylation. J Biol Chem. 2009;284:13185–13192. doi: 10.1074/jbc.M900579200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Alexandrov K, Simon I, Yurchenko V, Iakovenko A, Rostkova E, Scheidig AJ, Goody RS. Characterization of the Ternary Complex between Rab7: Rep-1 and Rab Geranylgeranyl Transferase. Eur J Biochem. 1999;265:160–170. doi: 10.1046/j.1432-1327.1999.00699.x. [DOI] [PubMed] [Google Scholar]
- 54.Thoma NH, Iakovenko A, Kalinin A, Waldmann H, Goody RS, Alexandrov K. Allosteric Regulation of Substrate Binding and Product Release in Geranylgeranyltransferase Type Ii. Biochemistry. 2001;40:268–274. doi: 10.1021/bi002034p. [DOI] [PubMed] [Google Scholar]
- 55.Thoma NH, Niculae A, Goody RS, Alexandrov K. Double Prenylation by Rabggtase Can Proceed without Dissociation of the Mono-Prenylated Intermediate. J Biol Chem. 2001;276:48631–48636. doi: 10.1074/jbc.M106470200. [DOI] [PubMed] [Google Scholar]
- 56.Hougland JL, Hicks KA, Hartman HL, Kelly RA, Watt TJ, Fierke CA. Identification of Novel Peptide Substrates for Protein Farnesyltransferase Reveals Two Substrate Classes with Distinct Sequence Selectivities. J Mol Biol. 2010;395:176–190. doi: 10.1016/j.jmb.2009.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang FL, Casey PJ. Influence of Metal Ions on Substrate Binding and Catalytic Activity of Mammalian Protein Geranylgeranyltransferase Type-I. Biochem J. 1996;320(Pt 3):925–932. doi: 10.1042/bj3200925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Leung KF, Baron R, Seabra MC. Thematic Review Series: Lipid Posttranslational Modifications. Geranylgeranylation of Rab Gtpases. J Lipid Res. 2006;47:467–475. doi: 10.1194/jlr.R500017-JLR200. [DOI] [PubMed] [Google Scholar]
- 59.Joberty G, Tavitian A, Zahraoui A. Isoprenylation of Rab Proteins Possessing a C-Terminal Caax Motif. FEBS Lett. 1993;330:323–328. doi: 10.1016/0014-5793(93)80897-4. [DOI] [PubMed] [Google Scholar]
- 60.Zhang FL, Kirschmeier P, Carr D, James L, Bond RW, Wang L, Patton R, Windsor WT, Syto R, Zhang R, et al. Characterization of Ha-Ras, N-Ras, Ki-Ras4a, and Ki-Ras4b as in Vitro Substrates for Farnesyl Protein Transferase and Geranylgeranyl Protein Transferase Type I. J Biol Chem. 1997;272:10232–10239. doi: 10.1074/jbc.272.15.10232. [DOI] [PubMed] [Google Scholar]
- 61.Lebowitz PF, Casey PJ, Prendergast GC, Thissen JA. Farnesyltransferase Inhibitors Alter the Prenylation and Growth-Stimulating Function of Rhob. J Biol Chem. 1997;272:15591–15594. doi: 10.1074/jbc.272.25.15591. [DOI] [PubMed] [Google Scholar]
- 62.Moores SL, Schaber MD, Mosser SD, Rands E, O’Hara MB, Garsky VM, Marshall MS, Pompliano DL, Gibbs JB. Sequence Dependence of Protein Isoprenylation. J Biol Chem. 1991;266:14603–14610. [PubMed] [Google Scholar]
- 63.Yokoyama K, Goodwin GW, Ghomashchi F, Glomset JA, Gelb MH. A Protein Geranylgeranyltransferase from Bovine Brain: Implications for Protein Prenylation Specificity. Proc Natl Acad Sci US A. 1991;88:5302–5306. doi: 10.1073/pnas.88.12.5302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Krzysiak AJ, Aditya AV, Hougland JL, Fierke CA, Gibbs RA. Synthesis and Screening of a Caal Peptide Library Versus Ftase Reveals a Surprising Number of Substrates. Bioorg Med Chem Lett. 2010;20:767–770. doi: 10.1016/j.bmcl.2009.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Maurer-Stroh S, Koranda M, Benetka W, Schneider G, Sirota FL, Eisenhaber F. Towards Complete Sets of Farnesylated and Geranylgeranylated Proteins. PLoS Comput Biol. 2007;3:e66. doi: 10.1371/journal.pcbi.0030066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.London N, Lamphear CL, Hougland JL, Fierke CA, Schueler-Furman O. Identification of a Novel Class of Farnesylation Targets by Structure-Based Modeling of Binding Specificity. PLoS Comput Biol. 2011;7:e1002170. doi: 10.1371/journal.pcbi.1002170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang YC, Dozier JK, Beese LS, Distefano MD. Rapid Analysis of Protein Farnesyltransferase Substrate Specificity Using Peptide Libraries and Isoprenoid Diphosphate Analogues. ACS Chem Biol. 2014;9:1726–1735. doi: 10.1021/cb5002312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Stein V, Kubala MH, Steen J, Grimmond SM, Alexandrov K. Towards the Systematic Mapping and Engineering of the Protein Prenylation Machinery in Saccharomyces Cerevisiae. PLOS ONE. 2015;10:e0120716. doi: 10.1371/journal.pone.0120716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Reinicke AT, Hutchinson JL, Magee AI, Mastroeni P, Trowsdale J, Kelly AP. A Salmonella Typhimurium Effector Protein Sifa Is Modified by Host Cell Prenylation and S-Acylation Machinery. J Biol Chem. 2005;280:14620–14627. doi: 10.1074/jbc.M500076200. [DOI] [PubMed] [Google Scholar]
- 70.Price CT, Al-Quadan T, Santic M, Jones SC, Abu Kwaik Y. Exploitation of Conserved Eukaryotic Host Cell Farnesylation Machinery by an F-Box Effector of Legionella Pneumophila. J Exp Med. 2010;207:1713–1726. doi: 10.1084/jem.20100771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ivanov SS, Charron G, Hang HC, Roy CR. Lipidation by the Host Prenyltransferase Machinery Facilitates Membrane Localization of Legionella Pneumophila Effector Proteins. J Biol Chem. 2010;285:34686–34698. doi: 10.1074/jbc.M110.170746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Price CT, Jones SC, Amundson KE, Kwaik YA. Host-Mediated Post-Translational Prenylation of Novel Dot/Icm-Translocated Effectors of Legionella Pneumophila. Front Microbiol. 2010;1:131. doi: 10.3389/fmicb.2010.00131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lee CZ, Chen PJ, Lai MM, Chen DS. Isoprenylation of Large Hepatitis Delta Antigen Is Necessary but Not Sufficient for Hepatitis Delta Virus Assembly. Virology. 1994;199:169–175. doi: 10.1006/viro.1994.1109. [DOI] [PubMed] [Google Scholar]
- 74.Chang MF, Chen CJ, Chang SC. Mutational Analysis of Delta Antigen: Effect on Assembly and Replication of Hepatitis Delta Virus. J Virol. 1994;68:646–653. doi: 10.1128/jvi.68.2.646-653.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bordier BB, Marion PL, Ohashi K, Kay MA, Greenberg HB, Casey JL, Glenn JS. A Prenylation Inhibitor Prevents Production of Infectious Hepatitis Delta Virus Particles. J Virol. 2002;76:10465–10472. doi: 10.1128/JVI.76.20.10465-10472.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Koh C, Canini L, Dahari H, Zhao X, Uprichard SL, Haynes-Williams V, Winters MA, Subramanya G, Cooper SL, Pinto P, et al. Oral Prenylation Inhibition with Lonafarnib in Chronic Hepatitis D Infection: A Proof-of-Concept Randomised, Double-Blind, Placebo-Controlled Phase 2a Trial. Lancet Infect Dis. 2015;15:1167–1174. doi: 10.1016/S1473-3099(15)00074-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yokoyama K, McGeady P, Gelb MH. Mammalian Protein Geranylgeranyltransferase-I: Substrate Specificity, Kinetic Mechanism, Metal Requirements, and Affinity Labeling. Biochemistry. 1995;34:1344–1354. doi: 10.1021/bi00004a029. [DOI] [PubMed] [Google Scholar]
- 78.Yokoyama K, Trobridge P, Buckner FS, Van Voorhis WC, Stuart KD, Gelb MH. Protein Farnesyltransferase from Trypanosoma Brucei. A Heterodimer of 61- and 65-Kda Subunits as a New Target for Antiparasite Therapeutics. J Biol Chem. 1998;273:26497–26505. doi: 10.1074/jbc.273.41.26497. [DOI] [PubMed] [Google Scholar]
- 79.Turek-Etienne TC, Strickland CL, Distefano MD. Biochemical and Structural Studies with Prenyl Diphosphate Analogues Provide Insights into Isoprenoid Recognition by Protein Farnesyl Transferase. Biochemistry. 2003;42:3716–3724. doi: 10.1021/bi0266838. [DOI] [PubMed] [Google Scholar]
- 80.Jennings BC, Danowitz AM, Wang YC, Gibbs RA, Distefano MD, Fierke CA. Analogs of Farnesyl Diphosphate Alter Caax Substrate Specificity and Reactions Rates of Protein Farnesyltransferase. Bioorg Med Chem Lett. 2016;26:1333–1336. doi: 10.1016/j.bmcl.2015.12.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Huang C, Hightower KE, Fierke CA. Mechanistic Studies of Rat Protein Farnesyltransferase Indicate an Associative Transition State. Biochemistry. 2000;39:2593–2602. doi: 10.1021/bi992356x. [DOI] [PubMed] [Google Scholar]
- 82.Gibbs RA, Krishnan U, Dolence JM, Poulter CD. A Stereoselective Palladium Copper-Catalyzed Route to Isoprenoids - Synthesis and Biological Evaluation of 13-Methylidenefarnesyl Diphosphate. J Org Chem. 1995;60:7821–7829. [Google Scholar]
- 83.Mu Y, Gibbs RA, Eubanks LM, Poulter CD. Cuprate-Mediated Synthesis and Biological Evaluation of Cyclopropyl- and Tert-Butylfarnesyl Diphosphate Analogs. J Org Chem. 1996;61:8010–8015. doi: 10.1021/jo9614203. [DOI] [PubMed] [Google Scholar]
- 84.Troutman JM, Roberts MJ, Andres DA, Spielmann HP. Tools to Analyze Protein Farnesylation in Cells. Bioconjug Chem. 2005;16:1209–1217. doi: 10.1021/bc050068+. [DOI] [PubMed] [Google Scholar]
- 85.Onono F, Subramanian T, Sunkara M, Subramanian KL, Spielmann HP, Morris AJ. Efficient Use of Exogenous Isoprenols for Protein Isoprenylation by Mda-Mb-231 Cells Is Regulated Independently of the Mevalonate Pathway. J Biol Chem. 2013;288:27444–27455. doi: 10.1074/jbc.M113.482307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Onono FO, Morgan MA, Spielmann HP, Andres DA, Subramanian T, Ganser A, Reuter CW. A Tagging-Via-Substrate Approach to Detect the Farnesylated Proteome Using Two-Dimensional Electrophoresis Coupled with Western Blotting. Mol Cell Proteomics. 2010;9:742–751. doi: 10.1074/mcp.M900597-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Liu XH, Prestwich GD. Didehydrogeranylgeranyl (Delta Delta Gg): A Fluorescent Probe for Protein Prenylation. J Am Chem Soc. 2002;124:20–21. doi: 10.1021/ja0119144. [DOI] [PubMed] [Google Scholar]
- 88.Dursina B, Reents R, Delon C, Wu Y, Kulharia M, Thutewohl M, Veligodsky A, Kalinin A, Evstifeev V, Ciobanu D, et al. Identification and Specificity Profiling of Protein Prenyltransferase Inhibitors Using New Fluorescent Phosphoisoprenoids. J Am Chem Soc. 2006;128:2822–2835. doi: 10.1021/ja052196e. [DOI] [PubMed] [Google Scholar]
- 89.Owen DJ, Alexandrov K, Rostkova E, Scheidig AJ, Goody RS, Waldmann H. Chemo-Enzymatic Synthesis of Fluorescent Rab 7 Proteins: Tools to Study Vesicular Trafficking in Cells. Angew Chem Int Ed. 1999;38:509–512. doi: 10.1002/(SICI)1521-3773(19990215)38:4<509::AID-ANIE509>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 90.Nguyen UT, Guo Z, Delon C, Wu Y, Deraeve C, Franzel B, Bon RS, Blankenfeldt W, Goody RS, Waldmann H, et al. Analysis of the Eukaryotic Prenylome by Isoprenoid Affinity Tagging. Nat Chem Biol. 2009;5:227–235. doi: 10.1038/nchembio.149. [DOI] [PubMed] [Google Scholar]
- 91.Kho Y, Kim SC, Jiang C, Barma D, Kwon SW, Cheng J, Jaunbergs J, Weinbaum C, Tamanoi F, Falck J, et al. A Tagging-Via-Substrate Technology for Detection and Proteomics of Farnesylated Proteins. Proc Natl Acad Sci US A. 2004;101:12479–12484. doi: 10.1073/pnas.0403413101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Rose MW, Rose ND, Boggs J, Lenevich S, Xu J, Barany G, Distefano MD. Evaluation of Geranylazide and Farnesylazide Diphosphate for Incorporation of Prenylazides into a Caax Box-Containing Peptide Using Protein Farnesyltransferase. J Pept Res. 2005;65:529–537. doi: 10.1111/j.1399-3011.2005.00261.x. [DOI] [PubMed] [Google Scholar]
- 93.Duckworth BP, Zhang Z, Hosokawa A, Distefano MD. Selective Labeling of Proteins by Using Protein Farnesyltransferase. ChemBioChem. 2007;8:98–105. doi: 10.1002/cbic.200600340. [DOI] [PubMed] [Google Scholar]
- 94.Berry AF, Heal WP, Tarafder AK, Tolmachova T, Baron RA, Seabra MC, Tate EW. Rapid Multilabel Detection of Geranylgeranylated Proteins by Using Bioorthogonal Ligation Chemistry. ChemBioChem. 2010;11:771–773. doi: 10.1002/cbic.201000087. [DOI] [PubMed] [Google Scholar]
- 95.Palsuledesai CC, Ochocki JD, Markowski TW, Distefano MD. A Combination of Metabolic Labeling and 2d-Dige Analysis in Response to a Farnesyltransferase Inhibitor Facilitates the Discovery of New Prenylated Proteins. Mol Biosyst. 2014;10:1094–1103. doi: 10.1039/c3mb70593e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Chan LN, Hart C, Guo L, Nyberg T, Davies BS, Fong LG, Young SG, Agnew BJ, Tamanoi F. A Novel Approach to Tag and Identify Geranylgeranylated Proteins. Electrophoresis. 2009;30:3598–3606. doi: 10.1002/elps.200900259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Charron G, Tsou LK, Maguire W, Yount JS, Hang HC. Alkynyl-Farnesol Reporters for Detection of Protein S-Prenylation in Cells. Mol Biosyst. 2011;7:67–73. doi: 10.1039/c0mb00183j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Charron G, Li MM, MacDonald MR, Hang HC. Prenylome Profiling Reveals S-Farnesylation Is Crucial for Membrane Targeting and Antiviral Activity of Zap Long-Isoform. Proc Natl Acad Sci US A. 2013;110:11085–11090. doi: 10.1073/pnas.1302564110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.DeGraw AJ, Palsuledesai C, Ochocki JD, Dozier JK, Lenevich S, Rashidian M, Distefano MD. Evaluation of Alkyne-Modified Isoprenoids as Chemical Reporters of Protein Prenylation. Chem Biol Drug Des. 2010;76:460–471. doi: 10.1111/j.1747-0285.2010.01037.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Palsuledesai CC, Ochocki JD, Kuhns MM, Wang YC, Warmka JK, Chernick DS, Wattenberg EV, Li L, Arriaga EA, Distefano MD. Metabolic Labeling with an Alkyne-Modified Isoprenoid Analog Facilitates Imaging and Quantification of the Prenylome in Cells. ACS Chem Biol. 2016;11:2820–2828. doi: 10.1021/acschembio.6b00421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Gisselberg JE, Zhang L, Elias JE, Yeh E. The Prenylated Proteome of Plasmodium Falciparum Reveals Pathogen-Specific Prenylation Activity and Drug Mechanism-of-Action. Mol Cell Proteomics. 2016 doi: 10.1074/mcp.M116.064550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Suazo KF, Schaber C, Palsuledesai CC, Odom John AR, Distefano MD. Global Proteomic Analysis of Prenylated Proteins in Plasmodium Falciparum Using an Alkyne-Modified Isoprenoid Analogue. Sci Rep. 2016;6:38615. doi: 10.1038/srep38615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Choy E, Chiu VK, Silletti J, Feoktistov M, Morimoto T, Michaelson D, Ivanov IE, Philips MR. Endomembrane Trafficking of Ras: The Caax Motif Targets Proteins to the Er and Golgi. Cell. 1999;98:69–80. doi: 10.1016/S0092-8674(00)80607-8. [DOI] [PubMed] [Google Scholar]
- 104.Hancock JF, Paterson H, Marshall CJ. A Polybasic Domain or Palmitoylation Is Required in Addition to the Caax Motif to Localize P21ras to the Plasma Membrane. Cell. 1990;63:133–139. doi: 10.1016/0092-8674(90)90294-o. [DOI] [PubMed] [Google Scholar]
- 105.Lynch SJ, Snitkin H, Gumper I, Philips MR, Sabatini D, Pellicer A. The Differential Palmitoylation States of N-Ras and H-Ras Determine Their Distinct Golgi Subcompartment Localizations. J Cell Physiol. 2015;230:610–619. doi: 10.1002/jcp.24779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Holtz D, Tanaka RA, Hartwig J, McKeon F. The Caax Motif of Lamin a Functions in Conjunction with the Nuclear Localization Signal to Target Assembly to the Nuclear Envelope. Cell. 1989;59:969–977. doi: 10.1016/0092-8674(89)90753-8. [DOI] [PubMed] [Google Scholar]
- 107.Mical TI, Monteiro MJ. The Role of Sequences Unique to Nuclear Intermediate Filaments in the Targeting and Assembly of Human Lamin B: Evidence for Lack of Interaction of Lamin B with Its Putative Receptor. J Cell Sci. 1998;111(Pt 23):3471–3485. doi: 10.1242/jcs.111.23.3471. [DOI] [PubMed] [Google Scholar]
- 108.Jung HJ, Nobumori C, Goulbourne CN, Tu Y, Lee JM, Tatar A, Wu D, Yoshinaga Y, de Jong PJ, Coffinier C, et al. Farnesylation of Lamin B1 Is Important for Retention of Nuclear Chromatin During Neuronal Migration. Proc Natl Acad Sci US A. 2013;110:E1923–E1932. doi: 10.1073/pnas.1303916110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Hancock JF, Cadwallader K, Marshall CJ. Methylation and Proteolysis Are Essential for Efficient Membrane Binding of Prenylated P21k-Ras(B) EMBO J. 1991;10:641–646. doi: 10.1002/j.1460-2075.1991.tb07992.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Silvius JR, l’Heureux F. Fluorimetric Evaluation of the Affinities of Isoprenylated Peptides for Lipid Bilayers. Biochemistry. 1994;33:3014–3022. doi: 10.1021/bi00176a034. [DOI] [PubMed] [Google Scholar]
- 111.Shahinian S, Silvius JR. Doubly-Lipid-Modified Protein Sequence Motifs Exhibit Long-Lived Anchorage to Lipid Bilayer Membranes. Biochemistry. 1995;34:3813–3822. doi: 10.1021/bi00011a039. [DOI] [PubMed] [Google Scholar]
- 112.van der Hoeven D, Cho KJ, Ma X, Chigurupati S, Parton RG, Hancock JF. Fendiline Inhibits K-Ras Plasma Membrane Localization and Blocks K-Ras Signal Transmission. Mol Cell Biol. 2013;33:237–251. doi: 10.1128/MCB.00884-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Nishimura A, Linder ME. Identification of a Novel Prenyl and Palmitoyl Modification at the Caax Motif of Cdc42 That Regulates Rhogdi Binding. Mol Cell Biol. 2013;33:1417–1429. doi: 10.1128/MCB.01398-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Jackson JH, Li JW, Buss JE, Der CJ, Cochrane CG. Polylysine Domain of K-Ras 4b Protein Is Crucial for Malignant Transformation. Proc Natl Acad Sci US A. 1994;91:12730–12734. doi: 10.1073/pnas.91.26.12730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Bivona TG, Quatela SE, Bodemann BO, Ahearn IM, Soskis MJ, Mor A, Miura J, Wiener HH, Wright L, Saba SG, et al. Pkc Regulates a Farnesyl-Electrostatic Switch on K-Ras That Promotes Its Association with Bcl-Xl on Mitochondria and Induces Apoptosis. Mol Cell. 2006;21:481–493. doi: 10.1016/j.molcel.2006.01.012. [DOI] [PubMed] [Google Scholar]
- 116.Jang H, Abraham SJ, Chavan TS, Hitchinson B, Khavrutskii L, Tarasova NI, Nussinov R, Gaponenko V. Mechanisms of Membrane Binding of Small Gtpase K-Ras4b Farnesylated Hypervariable Region. J Biol Chem. 2015;290:9465–9477. doi: 10.1074/jbc.M114.620724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Gomes AQ, Ali BR, Ramalho JS, Godfrey RF, Barral DC, Hume AN, Seabra MC. Membrane Targeting of Rab Gtpases Is Influenced by the Prenylation Motif. Mol Biol Cell. 2003;14:1882–1899. doi: 10.1091/mbc.E02-10-0639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Smeland TE, Seabra MC, Goldstein JL, Brown MS. Geranylgeranylated Rab Proteins Terminating in Cys-Ala-Cys, but Not Cys-Cys, Are Carboxyl-Methylated by Bovine Brain Membranes in Vitro. Proc Natl Acad Sci US A. 1994;91:10712–10716. doi: 10.1073/pnas.91.22.10712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Bergo MO, Leung GK, Ambroziak P, Otto JC, Casey PJ, Gomes AQ, Seabra MC, Young SG. Isoprenylcysteine Carboxyl Methyltransferase Deficiency in Mice. J Biol Chem. 2001;276:5841–5845. doi: 10.1074/jbc.C000831200. [DOI] [PubMed] [Google Scholar]
- 120.Chavrier P, Gorvel JP, Stelzer E, Simons K, Gruenberg J, Zerial M. Hypervariable C-Terminal Domain of Rab Proteins Acts as a Targeting Signal. Nature. 1991;353:769–772. doi: 10.1038/353769a0. [DOI] [PubMed] [Google Scholar]
- 121.Li F, Yi L, Zhao L, Itzen A, Goody RS, Wu YW. The Role of the Hypervariable C-Terminal Domain in Rab Gtpases Membrane Targeting. Proc Natl Acad Sci US A. 2014;111:2572–2577. doi: 10.1073/pnas.1313655111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Kuroda Y, Suzuki N, Kataoka T. The Effect of Posttranslational Modifications on the Interaction of Ras2 with Adenylyl Cyclase. Science. 1993;259:683–686. doi: 10.1126/science.8430318. [DOI] [PubMed] [Google Scholar]
- 123.Moudgil DK, Westcott N, Famulski JK, Patel K, Macdonald D, Hang H, Chan GK. A Novel Role of Farnesylation in Targeting a Mitotic Checkpoint Protein, Human Spindly, to Kinetochores. J Cell Biol. 2015;208:881–896. doi: 10.1083/jcb.201412085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Porfiri E, Evans T, Chardin P, Hancock JF. Prenylation of Ras Proteins Is Required for Efficient Hsos1-Promoted Guanine Nucleotide Exchange. J Biol Chem. 1994;269:22672–22677. [PubMed] [Google Scholar]
- 125.Pechlivanis M, Ringel R, Popkirova B, Kuhlmann J. Prenylation of Ras Facilitates Hsos1-Promoted Nucleotide Exchange, Upon Ras Binding to the Regulatory Site. Biochemistry. 2007;46:5341–5348. doi: 10.1021/bi602353k. [DOI] [PubMed] [Google Scholar]
- 126.Jeng HH, Taylor LJ, Bar-Sagi D. Sos-Mediated Cross-Activation of Wild-Type Ras by Oncogenic Ras Is Essential for Tumorigenesis. Nat Commun. 2012;3:1168. doi: 10.1038/ncomms2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Zhou Y, Johnson JL, Cerione RA, Erickson JW. Prenylation and Membrane Localization of Cdc42 Are Essential for Activation by Dock7. Biochemistry. 2013;52:4354–4363. doi: 10.1021/bi301688g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Hori Y, Kikuchi A, Isomura M, Katayama M, Miura Y, Fujioka H, Kaibuchi K, Takai Y. Post-Translational Modifications of the C-Terminal Region of the Rho Protein Are Important for Its Interaction with Membranes and the Stimulatory and Inhibitory Gdp/Gtp Exchange Proteins. Oncogene. 1991;6:515–522. [PubMed] [Google Scholar]
- 129.Casteel DE, Turner S, Schwappacher R, Rangaswami H, Su-Yuo J, Zhuang S, Boss GR, Pilz RB. Rho Isoform-Specific Interaction with Iqgap1 Promotes Breast Cancer Cell Proliferation and Migration. J Biol Chem. 2012;287:38367–38378. doi: 10.1074/jbc.M112.377499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Berg TJ, Gastonguay AJ, Lorimer EL, Kuhnmuench JR, Li R, Fields AP, Williams CL. Splice Variants of Smggds Control Small Gtpase Prenylation and Membrane Localization. J Biol Chem. 2010;285:35255–35266. doi: 10.1074/jbc.M110.129916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Pfeffer SR, Dirac-Svejstrup AB, Soldati T. Rab Gdp Dissociation Inhibitor: Putting Rab Gtpases in the Right Place. J Biol Chem. 1995;270:17057–17059. doi: 10.1074/jbc.270.29.17057. [DOI] [PubMed] [Google Scholar]
- 132.Boulter E, Garcia-Mata R, Guilluy C, Dubash A, Rossi G, Brennwald PJ, Burridge K. Regulation of Rho Gtpase Crosstalk, Degradation and Activity by Rhogdi1. Nat Cell Biol. 2010;12:477–483. doi: 10.1038/ncb2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Hoffman GR, Nassar N, Cerione RA. Structure of the Rho Family Gtp-Binding Protein Cdc42 in Complex with the Multifunctional Regulator Rhogdi. Cell. 2000;100:345–356. doi: 10.1016/s0092-8674(00)80670-4. [DOI] [PubMed] [Google Scholar]
- 134.Abramovitz A, Gutman M, Nachliel E. Structural Coupling between the Rho-Insert Domain of Cdc42 and the Geranylgeranyl Binding Site of Rhogdi. Biochemistry. 2012;51:715–723. doi: 10.1021/bi201211v. [DOI] [PubMed] [Google Scholar]
- 135.Tnimov Z, Guo Z, Gambin Y, Nguyen UT, Wu YW, Abankwa D, Stigter A, Collins BM, Waldmann H, Goody RS, et al. Quantitative Analysis of Prenylated Rhoa Interaction with Its Chaperone, Rhogdi. J Biol Chem. 2012;287:26549–26562. doi: 10.1074/jbc.M112.371294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Pylypenko O, Rak A, Durek T, Kushnir S, Dursina BE, Thomae NH, Constantinescu AT, Brunsveld L, Watzke A, Waldmann H, et al. Structure of Doubly Prenylated Ypt1:Gdi Complex and the Mechanism of Gdi-Mediated Rab Recycling. EMBO J. 2006;25:13–23. doi: 10.1038/sj.emboj.7600921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Rak A, Pylypenko O, Durek T, Watzke A, Kushnir S, Brunsveld L, Waldmann H, Goody RS, Alexandrov K. Structure of Rab Gdp-Dissociation Inhibitor in Complex with Prenylated Ypt1 Gtpase. Science. 2003;302:646–650. doi: 10.1126/science.1087761. [DOI] [PubMed] [Google Scholar]
- 138.Grizot S, Faure J, Fieschi F, Vignais PV, Dagher MC, Pebay-Peyroula E. Crystal Structure of the Rac1-Rhogdi Complex Involved in Nadph Oxidase Activation. Biochemistry. 2001;40:10007–10013. doi: 10.1021/bi010288k. [DOI] [PubMed] [Google Scholar]
- 139.Cook TA, Ghomashchi F, Gelb MH, Florio SK, Beavo JA. Binding of the Delta Subunit to Rod Phosphodiesterase Catalytic Subunits Requires Methylated, Prenylated C-Termini of the Catalytic Subunits. Biochemistry. 2000;39:13516–13523. doi: 10.1021/bi001070l. [DOI] [PubMed] [Google Scholar]
- 140.Zhang H, Liu XH, Zhang K, Chen CK, Frederick JM, Prestwich GD, Baehr W. Photoreceptor Cgmp Phosphodiesterase Delta Subunit (Pdedelta) Functions as a Prenyl-Binding Protein. J Biol Chem. 2004;279:407–413. doi: 10.1074/jbc.M306559200. [DOI] [PubMed] [Google Scholar]
- 141.Wilson SJ, Smyth EM. Internalization and Recycling of the Human Prostacyclin Receptor Is Modulated through Its Isoprenylation-Dependent Interaction with the Delta Subunit of Cgmp Phosphodiesterase 6. J Biol Chem. 2006;281:11780–11786. doi: 10.1074/jbc.M513110200. [DOI] [PubMed] [Google Scholar]
- 142.Chandra A, Grecco HE, Pisupati V, Perera D, Cassidy L, Skoulidis F, Ismail SA, Hedberg C, Hanzal-Bayer M, Venkitaraman AR, et al. The Gdi-Like Solubilizing Factor Pdedelta Sustains the Spatial Organization and Signalling of Ras Family Proteins. Nat Cell Biol. 2012;14:148–158. doi: 10.1038/ncb2394. [DOI] [PubMed] [Google Scholar]
- 143.Ismail SA, Chen YX, Rusinova A, Chandra A, Bierbaum M, Gremer L, Triola G, Waldmann H, Bastiaens PI, Wittinghofer A. Arl2-Gtp and Arl3-Gtp Regulate a Gdi-Like Transport System for Farnesylated Cargo. Nat Chem Biol. 2011;7:942–949. doi: 10.1038/nchembio.686. [DOI] [PubMed] [Google Scholar]
- 144.Dharmaiah S, Bindu L, Tran TH, Gillette WK, Frank PH, Ghirlando R, Nissley DV, Esposito D, McCormick F, Stephen AG, et al. Structural Basis of Recognition of Farnesylated and Methylated Kras4b by Pdeδ. Proc Natl Acad Sci US A. 2016;113:E6766–E6775. doi: 10.1073/pnas.1615316113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Fansa EK, Kosling SK, Zent E, Wittinghofer A, Ismail S. Pde6delta-Mediated Sorting of Inpp5e into the Cilium Is Determined by Cargo-Carrier Affinity. Nat Commun. 2016;7:11366. doi: 10.1038/ncomms11366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Ignatev A, Kravchenko S, Rak A, Goody RS, Pylypenko O. A Structural Model of the Gdp Dissociation Inhibitor Rab Membrane Extraction Mechanism. J Biol Chem. 2008;283:18377–18384. doi: 10.1074/jbc.M709718200. [DOI] [PubMed] [Google Scholar]
- 147.Basso AD, Kirschmeier P, Bishop WR. Lipid Posttranslational Modifications. Farnesyl Transferase Inhibitors. J Lipid Res. 2006;47:15–31. doi: 10.1194/jlr.R500012-JLR200. [DOI] [PubMed] [Google Scholar]
- 148.Buser CA, Dinsmore CJ, Fernandes C, Greenberg I, Hamilton K, Mosser SD, Walsh ES, Williams TM, Koblan KS. High-Performance Liquid Chromatography/Mass Spectrometry Characterization of Ki4b-Ras in Psn-1 Cells Treated with the Prenyltransferase Inhibitor L-778:123. Anal Biochem. 2001;290:126–137. doi: 10.1006/abio.2000.4972. [DOI] [PubMed] [Google Scholar]
- 149.Hunt JT, Ding CZ, Batorsky R, Bednarz M, Bhide R, Cho Y, Chong S, Chao S, Gullo-Brown J, Guo P, et al. Discovery of (R)-7-Cyano-2,3,4, 5-Tetrahydro-1-(1h-Imidazol-4-Ylmethyl)-3- (Phenylmethyl)-4-(2-Thienylsulfonyl)-1h-1,4-Benzodiazepine (Bms-214662), a Farnesyltransferase Inhibitor with Potent Preclinical Antitumor Activity. J Med Chem. 2000;43:3587–3595. doi: 10.1021/jm000248z. [DOI] [PubMed] [Google Scholar]
- 150.Zujewski J, Horak ID, Bol CJ, Woestenborghs R, Bowden C, End DW, Piotrovsky VK, Chiao J, Belly RT, Todd A, et al. Phase I and Pharmacokinetic Study of Farnesyl Protein Transferase Inhibitor R115777 in Advanced Cancer. J Clin Oncol. 2000;18:927–941. doi: 10.1200/JCO.2000.18.4.927. [DOI] [PubMed] [Google Scholar]
- 151.Bishop WR, Bond R, Petrin J, Wang L, Patton R, Doll R, Njoroge G, Catino J, Schwartz J, Windsor W, et al. Novel Tricyclic Inhibitors of Farnesyl Protein Transferase. Biochemical Characterization and Inhibition of Ras Modification in Transfected Cos Cells. J Biol Chem. 1995;270:30611–30618. doi: 10.1074/jbc.270.51.30611. [DOI] [PubMed] [Google Scholar]
- 152.Fletcher S, Keaney EP, Cummings CG, Blaskovich MA, Hast MA, Glenn MP, Chang SY, Bucher CJ, Floyd RJ, Katt WP, et al. Structure-Based Design and Synthesis of Potent, Ethylenediamine-Based, Mammalian Farnesyltransferase Inhibitors as Anticancer Agents. J Med Chem. 2010;53:6867–6888. doi: 10.1021/jm1001748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Berndt N, Hamilton AD, Sebti SM. Targeting Protein Prenylation for Cancer Therapy. Nat Rev Cancer. 2011;11:775–791. doi: 10.1038/nrc3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Whyte DB, Kirschmeier P, Hockenberry TN, Nunez-Oliva I, James L, Catino JJ, Bishop WR, Pai JK. K- and N-Ras Are Geranylgeranylated in Cells Treated with Farnesyl Protein Transferase Inhibitors. J Biol Chem. 1997;272:14459–14464. doi: 10.1074/jbc.272.22.14459. [DOI] [PubMed] [Google Scholar]
- 155.Lim KH, Baines AT, Fiordalisi JJ, Shipitsin M, Feig LA, Cox AD, Der CJ, Counter CM. Activation of Rala Is Critical for Ras-Induced Tumorigenesis of Human Cells. Cancer Cell. 2005;7:533–545. doi: 10.1016/j.ccr.2005.04.030. [DOI] [PubMed] [Google Scholar]
- 156.Clark EA, Golub TR, Lander ES, Hynes RO. Genomic Analysis of Metastasis Reveals an Essential Role for Rhoc. Nature. 2000;406:532–535. doi: 10.1038/35020106. [DOI] [PubMed] [Google Scholar]
- 157.Hakem A, Sanchez-Sweatman O, You-Ten A, Duncan G, Wakeham A, Khokha R, Mak TW. Rhoc Is Dispensable for Embryogenesis and Tumor Initiation but Essential for Metastasis. Genes Dev. 2005;19:1974–1979. doi: 10.1101/gad.1310805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Carrico D, Blaskovich MA, Bucher CJ, Sebti SM, Hamilton AD. Design, Synthesis, and Evaluation of Potent and Selective Benzoyleneurea-Based Inhibitors of Protein Geranylgeranyltransferase-I. Bioorg Med Chem. 2005;13:677–688. doi: 10.1016/j.bmc.2004.10.053. [DOI] [PubMed] [Google Scholar]
- 159.Peterson YK, Kelly P, Weinbaum CA, Casey PJ. A Novel Protein Geranylgeranyltransferase-I Inhibitor with High Potency, Selectivity, and Cellular Activity. J Biol Chem. 2006;281:12445–12450. doi: 10.1074/jbc.M600168200. [DOI] [PubMed] [Google Scholar]
- 160.Watanabe M, Fiji HD, Guo L, Chan L, Kinderman SS, Slamon DJ, Kwon O, Tamanoi F. Inhibitors of Protein Geranylgeranyltransferase I and Rab Geranylgeranyltransferase Identified from a Library of Allenoate-Derived Compounds. J Biol Chem. 2008;283:9571–9579. doi: 10.1074/jbc.M706229200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Castellano S, Fiji HD, Kinderman SS, Watanabe M, Leon P, Tamanoi F, Kwon O. Small-Molecule Inhibitors of Protein Geranylgeranyltransferase Type I. J Am Chem Soc. 2007;129:5843–5845. doi: 10.1021/ja070274n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Chan LN, Fiji HD, Watanabe M, Kwon O, Tamanoi F. Identification and Characterization of Mechanism of Action of P61-E7, a Novel Phosphine Catalysis-Based Inhibitor of Geranylgeranyltransferase-I. PLOS ONE. 2011;6:e26135. doi: 10.1371/journal.pone.0026135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Zimonjic DB, Chan LN, Tripathi V, Lu J, Kwon O, Popescu NC, Lowy DR, Tamanoi F. In Vitro and in Vivo Effects of Geranylgeranyltransferase I Inhibitor P61a6 on Non-Small Cell Lung Cancer Cells. BMC Cancer. 2013;13:198. doi: 10.1186/1471-2407-13-198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Kazi A, Carie A, Blaskovich MA, Bucher C, Thai V, Moulder S, Peng H, Carrico D, Pusateri E, Pledger WJ, et al. Blockade of Protein Geranylgeranylation Inhibits Cdk2-Dependent P27kip1 Phosphorylation on Thr187 and Accumulates P27kip1 in the Nucleus: Implications for Breast Cancer Therapy. Mol Cell Biol. 2009;29:2254–2263. doi: 10.1128/MCB.01029-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Sun J, Qian Y, Chen Z, Marfurt J, Hamilton AD, Sebti SM. The Geranylgeranyltransferase I Inhibitor Ggti-298 Induces Hypophosphorylation of Retinoblastoma and Partner Switching of Cyclin-Dependent Kinase Inhibitors. A Potential Mechanism for Ggti-298 Antitumor Activity. J Biol Chem. 1999;274:6930–6934. doi: 10.1074/jbc.274.11.6930. [DOI] [PubMed] [Google Scholar]
- 166.Hamada M, Miki T, Iwai S, Shimizu H, Yura Y. Involvement of Rhoa and Ralb in Geranylgeranyltransferase I Inhibitor-Mediated Inhibition of Proliferation and Migration of Human Oral Squamous Cell Carcinoma Cells. Cancer Chemother Pharmacol. 2011;68:559–569. doi: 10.1007/s00280-010-1520-9. [DOI] [PubMed] [Google Scholar]
- 167.Martin NE, Brunner TB, Kiel KD, DeLaney TF, Regine WF, Mohiuddin M, Rosato EF, Haller DG, Stevenson JP, Smith D, et al. A Phase I Trial of the Dual Farnesyltransferase and Geranylgeranyltransferase Inhibitor L-778,123 and Radiotherapy for Locally Advanced Pancreatic Cancer. Clin Cancer Res. 2004;10:5447–5454. doi: 10.1158/1078-0432.CCR-04-0248. [DOI] [PubMed] [Google Scholar]
- 168.deSolms SJ, Ciccarone TM, MacTough SC, Shaw AW, Buser CA, Ellis-Hutchings M, Fernandes C, Hamilton KA, Huber HE, Kohl NE, et al. Dual Protein Farnesyltransferase-Geranylgeranyltransferase-I Inhibitors as Potential Cancer Chemotherapeutic Agents. J Med Chem. 2003;46:2973–2984. doi: 10.1021/jm020587n. [DOI] [PubMed] [Google Scholar]
- 169.Qiao Y, Gao J, Qiu Y, Wu L, Guo F, Lo KK, Li D. Design, Synthesis, and Characterization of Piperazinedione-Based Dual Protein Inhibitors for Both Farnesyltransferase and Geranylgeranyltransferase-I. Eur J Med Chem. 2011;46:2264–2273. doi: 10.1016/j.ejmech.2011.03.007. [DOI] [PubMed] [Google Scholar]
- 170.Mazet JL, Padieu M, Osman H, Maume G, Mailliet P, Dereu N, Hamilton AD, Lavelle F, Sebti SM, Maume BF. Combination of the Novel Farnesyltransferase Inhibitor Rpr130401 and the Geranylgeranyltransferase-1 Inhibitor Ggti-298 Disrupts Map Kinase Activation and G(1)-S Transition in Ki-Ras-Overexpressing Transformed Adrenocortical Cells. FEBS Lett. 1999;460:235–240. doi: 10.1016/s0014-5793(99)01355-1. [DOI] [PubMed] [Google Scholar]
- 171.Krzykowska-Petitjean K, Malecki J, Bentke A, Ostrowska B, Laidler P. Tipifarnib and Tanespimycin Show Synergic Proapoptotic Activity in U937 Cells. J Cancer Res Clin Oncol. 2012;138:537–544. doi: 10.1007/s00432-011-1131-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Li T, Guo M, Gradishar WJ, Sparano JA, Perez EA, Wang M, Sledge GW. A Phase Ii Trial of Capecitabine in Combination with the Farnesyltransferase Inhibitor Tipifarnib in Patients with Anthracycline-Treated and Taxane-Resistant Metastatic Breast Cancer: An Eastern Cooperative Oncology Group Study (E1103) Breast Cancer Res Treat. 2012;134:345–352. doi: 10.1007/s10549-012-2071-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Jawad M, Yu N, Seedhouse C, Tandon K, Russell NH, Pallis M. Targeting of Cd34+Cd38− Cells Using Gemtuzumab Ozogamicin (Mylotarg) in Combination with Tipifarnib (Zarnestra) in Acute Myeloid Leukaemia. BMC Cancer. 2012;12:431. doi: 10.1186/1471-2407-12-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Ghasemi S, Davaran S, Sharifi S, Asgari D, Abdollahi A, Shahbazi Mojarrad J. Comparison of Cytotoxic Activity of L778123 as a Farnesyltranferase Inhibitor and Doxorubicin against A549 and Ht-29 Cell Lines. Adv Pharm Bull. 2013;3:73–77. doi: 10.5681/apb.2013.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Lackner MR, Kindt RM, Carroll PM, Brown K, Cancilla MR, Chen C, de Silva H, Franke Y, Guan B, Heuer T, et al. Chemical Genetics Identifies Rab Geranylgeranyl Transferase as an Apoptotic Target of Farnesyl Transferase Inhibitors. Cancer Cell. 2005;7:325–336. doi: 10.1016/j.ccr.2005.03.024. [DOI] [PubMed] [Google Scholar]
- 176.Cheng KW, Lahad JP, Kuo WL, Lapuk A, Yamada K, Auersperg N, Liu J, Smith-McCune K, Lu KH, Fishman D, et al. The Rab25 Small Gtpase Determines Aggressiveness of Ovarian and Breast Cancers. Nat Med. 2004;10:1251–1256. doi: 10.1038/nm1125. [DOI] [PubMed] [Google Scholar]
- 177.Chia WJ, Tang BL. Emerging Roles for Rab Family Gtpases in Human Cancer. Biochim Biophys Acta. 2009;1795:110–116. doi: 10.1016/j.bbcan.2008.10.001. [DOI] [PubMed] [Google Scholar]
- 178.Tan KT, Guiu-Rozas E, Bon RS, Guo Z, Delon C, Wetzel S, Arndt S, Alexandrov K, Waldmann H, Goody RS, et al. Design, Synthesis, and Characterization of Peptide-Based Rab Geranylgeranyl Transferase Inhibitors. J Med Chem. 2009;52:8025–8037. doi: 10.1021/jm901117d. [DOI] [PubMed] [Google Scholar]
- 179.Guo Z, Wu YW, Tan KT, Bon RS, Guiu-Rozas E, Delon C, Nguyen TU, Wetzel S, Arndt S, Goody RS, et al. Development of Selective Rabggtase Inhibitors and Crystal Structure of a Rabggtase-Inhibitor Complex. Angew Chem Int Ed. 2008;47:3747–3750. doi: 10.1002/anie.200705795. [DOI] [PubMed] [Google Scholar]
- 180.Baron RA, Tavare R, Figueiredo AC, Blazewska KM, Kashemirov BA, McKenna CE, Ebetino FH, Taylor A, Rogers MJ, Coxon FP, et al. Phosphonocarboxylates Inhibit the Second Geranylgeranyl Addition by Rab Geranylgeranyl Transferase. J Biol Chem. 2009;284:6861–6868. doi: 10.1074/jbc.M806952200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Coxon FP, Helfrich MH, Larijani B, Muzylak M, Dunford JE, Marshall D, McKinnon AD, Nesbitt SA, Horton MA, Seabra MC, et al. Identification of a Novel Phosphonocarboxylate Inhibitor of Rab Geranylgeranyl Transferase That Specifically Prevents Rab Prenylation in Osteoclasts and Macrophages. J Biol Chem. 2001;276:48213–48222. doi: 10.1074/jbc.M106473200. [DOI] [PubMed] [Google Scholar]
- 182.Deraeve C, Guo Z, Bon RS, Blankenfeldt W, DiLucrezia R, Wolf A, Menninger S, Stigter EA, Wetzel S, Choidas A, et al. Psoromic Acid Is a Selective and Covalent Rab-Prenylation Inhibitor Targeting Autoinhibited Rabggtase. J Am Chem Soc. 2012;134:7384–7391. doi: 10.1021/ja211305j. [DOI] [PubMed] [Google Scholar]
- 183.McKenna CE, Kashemirov BA, Blazewska KM, Mallard-Favier I, Stewart CA, Rojas J, Lundy MW, Ebetino FH, Baron RA, Dunford JE, et al. Synthesis, Chiral High Performance Liquid Chromatographic Resolution and Enantiospecific Activity of a Potent New Geranylgeranyl Transferase Inhibitor, 2-Hydroxy-3-Imidazo[1,2-a]Pyridin-3-Yl-2-Phosphonopropionic Acid. J Med Chem. 2010;53:3454–3464. doi: 10.1021/jm900232u. [DOI] [PubMed] [Google Scholar]
- 184.Bon RS, Guo Z, Stigter EA, Wetzel S, Menninger S, Wolf A, Choidas A, Alexandrov K, Blankenfeldt W, Goody RS, et al. Structure-Guided Development of Selective Rabggtase Inhibitors. Angew Chem Int Ed. 2011;50:4957–4961. doi: 10.1002/anie.201101210. [DOI] [PubMed] [Google Scholar]
- 185.Stigter EA, Guo Z, Bon RS, Wu YW, Choidas A, Wolf A, Menninger S, Waldmann H, Blankenfeldt W, Goody RS. Development of Selective, Potent Rabggtase Inhibitors. J Med Chem. 2012;55:8330–8340. doi: 10.1021/jm300624s. [DOI] [PubMed] [Google Scholar]
- 186.Fletcher S, Cummings CG, Rivas K, Katt WP, Horney C, Buckner FS, Chakrabarti D, Sebti SM, Gelb MH, Van Voorhis WC, et al. Potent, Plasmodium-Selective Farnesyltransferase Inhibitors That Arrest the Growth of Malaria Parasites: Structure-Activity Relationships of Ethylenediamine-Analogue Scaffolds and Homology Model Validation. J Med Chem. 2008;51:5176–5197. doi: 10.1021/jm800113p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Kohring K, Wiesner J, Altenkamper M, Sakowski J, Silber K, Hillebrecht A, Haebel P, Dahse HM, Ortmann R, Jomaa H, et al. Development of Benzophenone-Based Farnesyltransferase Inhibitors as Novel Antimalarials. ChemMedChem. 2008;3:1217–1231. doi: 10.1002/cmdc.200800043. [DOI] [PubMed] [Google Scholar]
- 188.Nallan L, Bauer KD, Bendale P, Rivas K, Yokoyama K, Horney CP, Pendyala PR, Floyd D, Lombardo LJ, Williams DK, et al. Protein Farnesyltransferase Inhibitors Exhibit Potent Antimalarial Activity. J Med Chem. 2005;48:3704–3713. doi: 10.1021/jm0491039. [DOI] [PubMed] [Google Scholar]
- 189.Ohkanda J, Lockman JW, Yokoyama K, Gelb MH, Croft SL, Kendrick H, Harrell MI, Feagin JE, Blaskovich MA, Sebti SM, et al. Peptidomimetic Inhibitors of Protein Farnesyltransferase Show Potent Antimalarial Activity. Bioorg Med Chem Lett. 2001;11:761–764. doi: 10.1016/s0960-894x(01)00055-5. [DOI] [PubMed] [Google Scholar]
- 190.Chakrabarti D, Azam T, DelVecchio C, Qiu L, Park YI, Allen CM. Protein Prenyl Transferase Activities of Plasmodium Falciparum. Mol Biochem Parasitol. 1998;94:175–184. doi: 10.1016/s0166-6851(98)00065-6. [DOI] [PubMed] [Google Scholar]
- 191.Ohkanda J, Buckner FS, Lockman JW, Yokoyama K, Carrico D, Eastman R, de Luca-Fradley K, Davies W, Croft SL, Van Voorhis WC, et al. Design and Synthesis of Peptidomimetic Protein Farnesyltransferase Inhibitors as Anti-Trypanosoma Brucei Agents. J Med Chem. 2004;47:432–445. doi: 10.1021/jm030236o. [DOI] [PubMed] [Google Scholar]
- 192.Yokoyama K, Lin Y, Stuart KD, Gelb MH. Prenylation of Proteins in Trypanosoma Brucei. Mol Biochem Parasitol. 1997;87:61–69. doi: 10.1016/s0166-6851(97)00043-1. [DOI] [PubMed] [Google Scholar]
- 193.Buckner FS, Bahia MT, Suryadevara PK, White KL, Shackleford DM, Chennamaneni NK, Hulverson MA, Laydbak JU, Chatelain E, Scandale I, et al. Pharmacological Characterization, Structural Studies, and in Vivo Activities of Anti-Chagas Disease Lead Compounds Derived from Tipifarnib. Antimicrob Agents Chemother. 2012;56:4914–4921. doi: 10.1128/AAC.06244-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Kraus JM, Tatipaka HB, McGuffin SA, Chennamaneni NK, Karimi M, Arif J, Verlinde CL, Buckner FS, Gelb MH. Second Generation Analogues of the Cancer Drug Clinical Candidate Tipifarnib for Anti-Chagas Disease Drug Discovery. J Med Chem. 2010;53:3887–3898. doi: 10.1021/jm9013136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Kraus JM, Verlinde CL, Karimi M, Lepesheva GI, Gelb MH, Buckner FS. Rational Modification of a Candidate Cancer Drug for Use against Chagas Disease. J Med Chem. 2009;52:1639–1647. doi: 10.1021/jm801313t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Hucke O, Gelb MH, Verlinde CL, Buckner FS. The Protein Farnesyltransferase Inhibitor Tipifarnib as a New Lead for the Development of Drugs against Chagas Disease. J Med Chem. 2005;48:5415–5418. doi: 10.1021/jm050441z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Yokoyama K, Trobridge P, Buckner FS, Scholten J, Stuart KD, Van Voorhis WC, Gelb MH. The Effects of Protein Farnesyltransferase Inhibitors on Trypanosomatids: Inhibition of Protein Farnesylation and Cell Growth. Mol Biochem Parasitol. 1998;94:87–97. doi: 10.1016/s0166-6851(98)00053-x. [DOI] [PubMed] [Google Scholar]
- 198.Mabanglo MF, Hast MA, Lubock NB, Hellinga HW, Beese LS. Crystal Structures of the Fungal Pathogen Aspergillus Fumigatus Protein Farnesyltransferase Complexed with Substrates and Inhibitors Reveal Features for Antifungal Drug Design. Protein Sci. 2014;23:289–301. doi: 10.1002/pro.2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Hast MA, Nichols CB, Armstrong SM, Kelly SM, Hellinga HW, Alspaugh JA, Beese LS. Structures of Cryptococcus Neoformans Protein Farnesyltransferase Reveal Strategies for Developing Inhibitors That Target Fungal Pathogens. J Biol Chem. 2011;286:35149–35162. doi: 10.1074/jbc.M111.250506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Murthi KK, Smith SE, Kluge AF, Bergnes G, Bureau P, Berlin V. Antifungal Activity of a Candida Albicans Ggtase I Inhibitor-Alanine Conjugate. Inhibition of Rho1p Prenylation in C. Albicans. Bioorg Med Chem Lett. 2003;13:1935–1937. doi: 10.1016/s0960-894x(03)00320-2. [DOI] [PubMed] [Google Scholar]
- 201.Vallim MA, Fernandes L, Alspaugh JA. The Ram1 Gene Encoding a Protein-Farnesyltransferase Beta-Subunit Homologue Is Essential in Cryptococcus Neoformans. Microbiology. 2004;150:1925–1935. doi: 10.1099/mic.0.27030-0. [DOI] [PubMed] [Google Scholar]
- 202.Kelly R, Card D, Register E, Mazur P, Kelly T, Tanaka KI, Onishi J, Williamson JM, Fan H, Satoh T, et al. Geranylgeranyltransferase I of Candida Albicans: Null Mutants or Enzyme Inhibitors Produce Unexpected Phenotypes. J Bacteriol. 2000;182:704–713. doi: 10.1128/jb.182.3.704-713.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Amet T, Nonaka M, Dewan MZ, Saitoh Y, Qi X, Ichinose S, Yamamoto N, Yamaoka S. Statin-Induced Inhibition of Hiv-1 Release from Latently Infected U1 Cells Reveals a Critical Role for Protein Prenylation in Hiv-1 Replication. Microbes Infect. 2008;10:471–480. doi: 10.1016/j.micinf.2008.01.009. [DOI] [PubMed] [Google Scholar]
- 204.del Real G, Jimenez-Baranda S, Mira E, Lacalle RA, Lucas P, Gomez-Mouton C, Alegret M, Pena JM, Rodriguez-Zapata M, Alvarez-Mon M, et al. Statins Inhibit Hiv-1 Infection by Down-Regulating Rho Activity. J Exp Med. 2004;200:541–547. doi: 10.1084/jem.20040061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Bordier BB, Ohkanda J, Liu P, Lee SY, Salazar FH, Marion PL, Ohashi K, Meuse L, Kay MA, Casey JL, et al. In Vivo Antiviral Efficacy of Prenylation Inhibitors against Hepatitis Delta Virus. J Clin Invest. 2003;112:407–414. doi: 10.1172/JCI17704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Glenn JS, Marsters JC, Jr, Greenberg HB. Use of a Prenylation Inhibitor as a Novel Antiviral Agent. J Virol. 1998;72:9303–9306. doi: 10.1128/jvi.72.11.9303-9306.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Otto JC, Casey PJ. The Hepatitis Delta Virus Large Antigen Is Farnesylated Both in Vitro and in Animal Cells. J Biol Chem. 1996;271:4569–4572. doi: 10.1074/jbc.271.9.4569. [DOI] [PubMed] [Google Scholar]
- 208.Young SG, Yang SH, Davies BS, Jung HJ, Fong LG. Targeting Protein Prenylation in Progeria. Sci Transl Med. 2013;5:171ps173. doi: 10.1126/scitranslmed.3005229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Gordon LB, Kleinman ME, Miller DT, Neuberg DS, Giobbie-Hurder A, Gerhard-Herman M, Smoot LB, Gordon CM, Cleveland R, Snyder BD, et al. Clinical Trial of a Farnesyltransferase Inhibitor in Children with Hutchinson-Gilford Progeria Syndrome. Proc Natl Acad Sci US A. 2012;109:16666–16671. doi: 10.1073/pnas.1202529109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Gordon LB, Massaro J, D’Agostino RB, Sr, Campbell SE, Brazier J, Brown WT, Kleinman ME, Kieran MW. Impact of Farnesylation Inhibitors on Survival in Hutchinson-Gilford Progeria Syndrome. Circulation. 2014;130:27–34. doi: 10.1161/CIRCULATIONAHA.113.008285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Gordon LB, Kleinman ME, Massaro J, D’Agostino RB, Sr, Shappell H, Gerhard-Herman M, Smoot LB, Gordon CM, Cleveland RH, Nazarian A, et al. Clinical Trial of the Protein Farnesylation Inhibitors Lonafarnib, Pravastatin, and Zoledronic Acid in Children with Hutchinson-Gilford Progeria Syndrome. Circulation. 2016;134:114–125. doi: 10.1161/CIRCULATIONAHA.116.022188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Johnson DR, Bhatnagar RS, Knoll LJ, Gordon JI. Genetic and Biochemical Studies of Protein N-Myristoylation. Annu Rev Biochem. 1994;63:869–914. doi: 10.1146/annurev.bi.63.070194.004253. [DOI] [PubMed] [Google Scholar]
- 213.Zha J, Weiler S, Oh KJ, Wei MC, Korsmeyer SJ. Posttranslational N-Myristoylation of Bid as a Molecular Switch for Targeting Mitochondria and Apoptosis. Science. 2000;290:1761–1765. doi: 10.1126/science.290.5497.1761. [DOI] [PubMed] [Google Scholar]
- 214.Patwardhan P, Resh MD. Myristoylation and Membrane Binding Regulate C-Src Stability and Kinase Activity. Mol Cell Biol. 2010;30:4094–4107. doi: 10.1128/MCB.00246-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Tate EW, Bell AS, Rackham MD, Wright MH. N-Myristoyltransferase as a Potential Drug Target in Malaria and Leishmaniasis. Parasitology. 2014;141:37–49. doi: 10.1017/S0031182013000450. [DOI] [PubMed] [Google Scholar]
- 216.Dyda F, Klein DC, Hickman AB. Gcn5-Related N-Acetyltransferases: A Structural Overview. Annu Rev Biophys Biomol Struct. 2000;29:81–103. doi: 10.1146/annurev.biophys.29.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Giang DK, Cravatt BF. A Second Mammalian N-Myristoyltransferase. J Biol Chem. 1998;273:6595–6598. doi: 10.1074/jbc.273.12.6595. [DOI] [PubMed] [Google Scholar]
- 218.Bhatnagar RS, Futterer K, Farazi TA, Korolev S, Murray CL, Jackson-Machelski E, Gokel GW, Gordon JI, Waksman G. Structure of N-Myristoyltransferase with Bound Myristoylcoa and Peptide Substrate Analogs. Nat Struct Biol. 1998;5:1091–1097. doi: 10.1038/4202. [DOI] [PubMed] [Google Scholar]
- 219.Weston SA, Camble R, Colls J, Rosenbrock G, Taylor I, Egerton M, Tucker AD, Tunnicliffe A, Mistry A, Mancia F, et al. Crystal Structure of the Anti-Fungal Target N-Myristoyl Transferase. Nat Struct Biol. 1998;5:213–221. doi: 10.1038/nsb0398-213. [DOI] [PubMed] [Google Scholar]
- 220.Rudnick DA, McWherter CA, Rocque WJ, Lennon PJ, Getman DP, Gordon JI. Kinetic and Structural Evidence for a Sequential Ordered Bi Bi Mechanism of Catalysis by Saccharomyces Cerevisiae Myristoyl-Coa:Protein N-Myristoyltransferase. J Biol Chem. 1991;266:9732–9739. [PubMed] [Google Scholar]
- 221.Farazi TA, Waksman G, Gordon JI. Structures of Saccharomyces Cerevisiae N-Myristoyltransferase with Bound Myristoylcoa and Peptide Provide Insights About Substrate Recognition and Catalysis. Biochemistry. 2001;40:6335–6343. doi: 10.1021/bi0101401. [DOI] [PubMed] [Google Scholar]
- 222.Bhatnagar RS, Schall OF, Jackson-Machelski E, Sikorski JA, Devadas B, Gokel GW, Gordon JI. Titration Calorimetric Analysis of Acylcoa Recognition by Myristoylcoa:Protein N-Myristoyltransferase. Biochemistry. 1997;36:6700–6708. doi: 10.1021/bi970311v. [DOI] [PubMed] [Google Scholar]
- 223.Ducker CE, Upson JJ, French KJ, Smith CD. Two N-Myristoyltransferase Isozymes Play Unique Roles in Protein Myristoylation, Proliferation, and Apoptosis. Mol Cancer Res. 2005;3:463–476. doi: 10.1158/1541-7786.MCR-05-0037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Duronio RJ, Towler DA, Heuckeroth RO, Gordon JI. Disruption of the Yeast N-Myristoyl Transferase Gene Causes Recessive Lethality. Science. 1989;243:796–800. doi: 10.1126/science.2644694. [DOI] [PubMed] [Google Scholar]
- 225.Weinberg RA, McWherter CA, Freeman SK, Wood DC, Gordon JI, Lee SC. Genetic Studies Reveal That Myristoylcoa:Protein N-Myristoyltransferase Is an Essential Enzyme in Candida Albicans. Mol Microbiol. 1995;16:241–250. doi: 10.1111/j.1365-2958.1995.tb02296.x. [DOI] [PubMed] [Google Scholar]
- 226.Ntwasa M, Aapies S, Schiffmann DA, Gay NJ. Drosophila Embryos Lacking N-Myristoyltransferase Have Multiple Developmental Defects. Exp Cell Res. 2001;262:134–144. doi: 10.1006/excr.2000.5086. [DOI] [PubMed] [Google Scholar]
- 227.Price HP, Menon MR, Panethymitaki C, Goulding D, McKean PG, Smith DF. Myristoyl-Coa:Protein N-Myristoyltransferase, an Essential Enzyme and Potential Drug Target in Kinetoplastid Parasites. J Biol Chem. 2003;278:7206–7214. doi: 10.1074/jbc.M211391200. [DOI] [PubMed] [Google Scholar]
- 228.McIlhinney RA, McGlone K. Immunocytochemical Characterization and Subcellular Localization of Human Myristoyl-Coa: Protein N-Myristoyltransferase in Hela Cells. Exp Cell Res. 1996;223:348–356. doi: 10.1006/excr.1996.0090. [DOI] [PubMed] [Google Scholar]
- 229.Knoll LJ, Levy MA, Stahl PD, Gordon JI. Analysis of the Compartmentalization of Myristoyl-Coa:Protein N-Myristoyltransferase in Saccharomyces Cerevisiae. J Biol Chem. 1992;267:5366–5373. [PubMed] [Google Scholar]
- 230.Glover CJ, Hartman KD, Felsted RL. Human N-Myristoyltransferase Amino-Terminal Domain Involved in Targeting the Enzyme to the Ribosomal Subcellular Fraction. J Biol Chem. 1997;272:28680–28689. doi: 10.1074/jbc.272.45.28680. [DOI] [PubMed] [Google Scholar]
- 231.Takamune N, Kuroe T, Tanada N, Shoji S, Misumi S. Suppression of Human Immunodeficiency Virus Type-1 Production by Coexpression of Catalytic-Region-Deleted N-Myristoyltransferase Mutants. Biol Pharm Bull. 2010;33:2018–2023. doi: 10.1248/bpb.33.2018. [DOI] [PubMed] [Google Scholar]
- 232.Yang SH, Shrivastav A, Kosinski C, Sharma RK, Chen MH, Berthiaume LG, Peters LL, Chuang PT, Young SG, Bergo MO. N-Myristoyltransferase 1 Is Essential in Early Mouse Development. J Biol Chem. 2005;280:18990–18995. doi: 10.1074/jbc.M412917200. [DOI] [PubMed] [Google Scholar]
- 233.Shrivastav A, Varma S, Lawman Z, Yang SH, Ritchie SA, Bonham K, Singh SM, Saxena A, Sharma RK. Requirement of N-Myristoyltransferase 1 in the Development of Monocytic Lineage. J Immunol. 2008;180:1019–1028. doi: 10.4049/jimmunol.180.2.1019. [DOI] [PubMed] [Google Scholar]
- 234.Rampoldi F, Bonrouhi M, Boehm ME, Lehmann WD, Popovic ZV, Kaden S, Federico G, Brunk F, Grone HJ, Porubsky S. Immunosuppression and Aberrant T Cell Development in the Absence of N-Myristoylation. J Immunol. 2015;195:4228–4243. doi: 10.4049/jimmunol.1500622. [DOI] [PubMed] [Google Scholar]
- 235.Lu Y, Selvakumar P, Ali K, Shrivastav A, Bajaj G, Resch L, Griebel R, Fourney D, Meguro K, Sharma RK. Expression of N-Myristoyltransferase in Human Brain Tumors. Neurochem Res. 2005;30:9–13. doi: 10.1007/s11064-004-9680-9. [DOI] [PubMed] [Google Scholar]
- 236.Perinpanayagam MA, Beauchamp E, Martin DD, Sim JY, Yap MC, Berthiaume LG. Regulation of Co- and Post-Translational Myristoylation of Proteins During Apoptosis: Interplay of N-Myristoyltransferases and Caspases. FASEB J. 2013;27:811–821. doi: 10.1096/fj.12-214924. [DOI] [PubMed] [Google Scholar]
- 237.Maurer-Stroh S, Gouda M, Novatchkova M, Schleiffer A, Schneider G, Sirota FL, Wildpaner M, Hayashi N, Eisenhaber F. Myrbase: Analysis of Genome-Wide Glycine Myristoylation Enlarges the Functional Spectrum of Eukaryotic Myristoylated Proteins. Genome Biol. 2004;5:R21. doi: 10.1186/gb-2004-5-3-r21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Sakurai N, Utsumi T. Posttranslational N-Myristoylation Is Required for the Anti-Apoptotic Activity of Human Tgelsolin, the C-Terminal Caspase Cleavage Product of Human Gelsolin. J Biol Chem. 2006;281:14288–14295. doi: 10.1074/jbc.M510338200. [DOI] [PubMed] [Google Scholar]
- 239.Moriya K, Nagatoshi K, Noriyasu Y, Okamura T, Takamitsu E, Suzuki T, Utsumi T. Protein N-Myristoylation Plays a Critical Role in the Endoplasmic Reticulum Morphological Change Induced by Overexpression of Protein Lunapark, an Integral Membrane Protein of the Endoplasmic Reticulum. PLOS ONE. 2013;8:e78235. doi: 10.1371/journal.pone.0078235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Liu J, Hughes TE, Sessa WC. The First 35 Amino Acids and Fatty Acylation Sites Determine the Molecular Targeting of Endothelial Nitric Oxide Synthase into the Golgi Region of Cells: A Green Fluorescent Protein Study. J Cell Biol. 1997;137:1525–1535. doi: 10.1083/jcb.137.7.1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Sullivan A, Uff CR, Isacke CM, Thorne RF. Pace-1, a Novel Protein That Interacts with the C-Terminal Domain of Ezrin. Exp Cell Res. 2003;284:224–238. doi: 10.1016/s0014-4827(02)00054-x. [DOI] [PubMed] [Google Scholar]
- 242.Panaretou C, Domin J, Cockcroft S, Waterfield MD. Characterization of P150, an Adaptor Protein for the Human Phosphatidylinositol (Ptdins) 3-Kinase. Substrate Presentation by Phosphatidylinositol Transfer Protein to the P150.Ptdins 3-Kinase Complex. J Biol Chem. 1997;272:2477–2485. doi: 10.1074/jbc.272.4.2477. [DOI] [PubMed] [Google Scholar]
- 243.Musil LS, Carr C, Cohen JB, Merlie JP. Acetylcholine Receptor-Associated 43k Protein Contains Covalently Bound Myristate. J Cell Biol. 1988;107:1113–1121. doi: 10.1083/jcb.107.3.1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Collavin L, Lazarevic D, Utrera R, Marzinotto S, Monte M, Schneider C. Wt P53 Dependent Expression of a Membrane-Associated Isoform of Adenylate Kinase. Oncogene. 1999;18:5879–5888. doi: 10.1038/sj.onc.1202970. [DOI] [PubMed] [Google Scholar]
- 245.D’Souza-Schorey C, Stahl PD. Myristoylation Is Required for the Intracellular Localization and Endocytic Function of Arf6. Exp Cell Res. 1995;221:153–159. doi: 10.1006/excr.1995.1362. [DOI] [PubMed] [Google Scholar]
- 246.Lin CY, Li CC, Huang PH, Lee FJ. A Developmentally Regulated Arf-Like 5 Protein (Arl5), Localized to Nuclei and Nucleoli, Interacts with Heterochromatin Protein 1. J Cell Sci. 2002;115:4433–4445. doi: 10.1242/jcs.00123. [DOI] [PubMed] [Google Scholar]
- 247.Suzuki T, Moriya K, Nagatoshi K, Ota Y, Ezure T, Ando E, Tsunasawa S, Utsumi T. Strategy for Comprehensive Identification of Human N-Myristoylated Proteins Using an Insect Cell-Free Protein Synthesis System. Proteomics. 2010;10:1780–1793. doi: 10.1002/pmic.200900783. [DOI] [PubMed] [Google Scholar]
- 248.Fraser ID, Tavalin SJ, Lester LB, Langeberg LK, Westphal AM, Dean RA, Marrion NV, Scott JD. A Novel Lipid-Anchored a-Kinase Anchoring Protein Facilitates Camp-Responsive Membrane Events. EMBO J. 1998;17:2261–2272. doi: 10.1093/emboj/17.8.2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Liang J, Xu ZX, Ding Z, Lu Y, Yu Q, Werle KD, Zhou G, Park YY, Peng G, Gambello MJ, et al. Myristoylation Confers Noncanonical Ampk Functions in Autophagy Selectivity and Mitochondrial Surveillance. Nat Commun. 2015;6:7926. doi: 10.1038/ncomms8926. [DOI] [PubMed] [Google Scholar]
- 250.Wice BM, Gordon JI. A Strategy for Isolation of Cdnas Encoding Proteins Affecting Human Intestinal Epithelial Cell Growth and Differentiation: Characterization of a Novel Gut-Specific N-Myristoylated Annexin. J Cell Biol. 1992;116:405–422. doi: 10.1083/jcb.116.2.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Timm S, Titus B, Bernd K, Barroso M. The Ef-Hand Ca(2+)-Binding Protein P22 Associates with Microtubules in an N-Myristoylation-Dependent Manner. Mol Biol Cell. 1999;10:3473–3488. doi: 10.1091/mbc.10.10.3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Perrino BA, Martin BA. Ca(2+)- and Myristoylation-Dependent Association of Calcineurin with Phosphatidylserine. J Biochem. 2001;129:835–841. doi: 10.1093/oxfordjournals.jbchem.a002927. [DOI] [PubMed] [Google Scholar]
- 253.Stabler SM, Ostrowski LL, Janicki SM, Monteiro MJ. A Myristoylated Calcium-Binding Protein That Preferentially Interacts with the Alzheimer’s Disease Presenilin 2 Protein. J Cell Biol. 1999;145:1277–1292. doi: 10.1083/jcb.145.6.1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Li S, Goldberg E. A Novel N-Terminal Domain Directs Membrane Localization of Mouse Testis-Specific Calpastatin. Biol Reprod. 2000;63:1594–1600. doi: 10.1095/biolreprod63.6.1594. [DOI] [PubMed] [Google Scholar]
- 255.Pepperkok R, Hotz-Wagenblatt A, Konig N, Girod A, Bossemeyer D, Kinzel V. Intracellular Distribution of Mammalian Protein Kinase a Catalytic Subunit Altered by Conserved Asn2 Deamidation. J Cell Biol. 2000;148:715–726. doi: 10.1083/jcb.148.4.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Gaffarogullari EC, Masterson LR, Metcalfe EE, Traaseth NJ, Balatri E, Musa MM, Mullen D, Distefano MD, Veglia G. A Myristoyl/Phosphoserine Switch Controls Camp-Dependent Protein Kinase Association to Membranes. J Mol Biol. 2011;411:823–836. doi: 10.1016/j.jmb.2011.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Vaandrager AB, Ehlert EM, Jarchau T, Lohmann SM, de Jonge HR. N-Terminal Myristoylation Is Required for Membrane Localization of Cgmp-Dependent Protein Kinase Type Ii. J Biol Chem. 1996;271:7025–7029. doi: 10.1074/jbc.271.12.7025. [DOI] [PubMed] [Google Scholar]
- 258.Yorikawa C, Shibata H, Waguri S, Hatta K, Horii M, Katoh K, Kobayashi T, Uchiyama Y, Maki M. Human Chmp6, a Myristoylated Escrt-Iii Protein, Interacts Directly with an Escrt-Ii Component Eap20 and Regulates Endosomal Cargo Sorting. Biochem J. 2005;387:17–26. doi: 10.1042/BJ20041227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Risinger MA, Dotimas EM, Cohen CM. Human Erythrocyte Protein 4.2, a High Copy Number Membrane Protein, Is N-Myristylated. J Biol Chem. 1992;267:5680–5685. [PubMed] [Google Scholar]
- 260.Kouhara H, Hadari YR, Spivak-Kroizman T, Schilling J, Bar-Sagi D, Lax I, Schlessinger J. A Lipid-Anchored Grb2-Binding Protein That Links Fgf-Receptor Activation to the Ras/Mapk Signaling Pathway. Cell. 1997;89:693–702. doi: 10.1016/s0092-8674(00)80252-4. [DOI] [PubMed] [Google Scholar]
- 261.Han Y, Eppinger E, Schuster IG, Weigand LU, Liang X, Kremmer E, Peschel C, Krackhardt AM. Formin-Like 1 (Fmnl1) Is Regulated by N-Terminal Myristoylation and Induces Polarized Membrane Blebbing. J Biol Chem. 2009;284:33409–33417. doi: 10.1074/jbc.M109.060699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Stackpole EE, Akins MR, Fallon JR. N-Myristoylation Regulates the Axonal Distribution of the Fragile X-Related Protein Fxr2p. Mol Cell Neurosci. 2014;62:42–50. doi: 10.1016/j.mcn.2014.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Barr FA, Puype M, Vandekerckhove J, Warren G. Grasp65, a Protein Involved in the Stacking of Golgi Cisternae. Cell. 1997;91:253–262. doi: 10.1016/s0092-8674(00)80407-9. [DOI] [PubMed] [Google Scholar]
- 264.Kuo A, Zhong C, Lane WS, Derynck R. Transmembrane Transforming Growth Factor-Alpha Tethers to the Pdz Domain-Containing, Golgi Membrane-Associated Protein P59/Grasp55. EMBO J. 2000;19:6427–6439. doi: 10.1093/emboj/19.23.6427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Eberle HB, Serrano RL, Fullekrug J, Schlosser A, Lehmann WD, Lottspeich F, Kaloyanova D, Wieland FT, Helms JB. Identification and Characterization of a Novel Human Plant Pathogenesis-Related Protein That Localizes to Lipid-Enriched Microdomains in the Golgi Complex. J Cell Sci. 2002;115:827–838. doi: 10.1242/jcs.115.4.827. [DOI] [PubMed] [Google Scholar]
- 266.Chen CA, Manning DR. Regulation of G Proteins by Covalent Modification. Oncogene. 2001;20:1643–1652. doi: 10.1038/sj.onc.1204185. [DOI] [PubMed] [Google Scholar]
- 267.Kumar S, Parameswaran S, Sharma RK. Novel Myristoylation of the Sperm-Specific Hexokinase 1 Isoform Regulates Its Atypical Localization. Biol Open. 2015;4:1679–1687. doi: 10.1242/bio.012831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Arbuzova A, Schmitz AA, Vergeres G. Cross-Talk Unfolded: Marcks Proteins. Biochem J. 2002;362:1–12. doi: 10.1042/0264-6021:3620001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Taniguchi H, Manenti S. Interaction of Myristoylated Alanine-Rich Protein Kinase C Substrate (Marcks) with Membrane Phospholipids. J Biol Chem. 1993;268:9960–9963. [PubMed] [Google Scholar]
- 270.Walker JE, Arizmendi JM, Dupuis A, Fearnley IM, Finel M, Medd SM, Pilkington SJ, Runswick MJ, Skehel JM. Sequences of 20 Subunits of Nadh:Ubiquinone Oxidoreductase from Bovine Heart Mitochondria. Application of a Novel Strategy for Sequencing Proteins Using the Polymerase Chain Reaction. J Mol Biol. 1992;226:1051–1072. doi: 10.1016/0022-2836(92)91052-q. [DOI] [PubMed] [Google Scholar]
- 271.Koutelou E, Sato S, Tomomori-Sato C, Florens L, Swanson SK, Washburn MP, Kokkinaki M, Conaway RC, Conaway JW, Moschonas NK. Neuralized-Like 1 (Neurl1) Targeted to the Plasma Membrane by N-Myristoylation Regulates the Notch Ligand Jagged1. J Biol Chem. 2008;283:3846–3853. doi: 10.1074/jbc.M706974200. [DOI] [PubMed] [Google Scholar]
- 272.O’Callaghan DW, Ivings L, Weiss JL, Ashby MC, Tepikin AV, Burgoyne RD. Differential Use of Myristoyl Groups on Neuronal Calcium Sensor Proteins as a Determinant of Spatio-Temporal Aspects of Ca2+ Signal Transduction. J Biol Chem. 2002;277:14227–14237. doi: 10.1074/jbc.M111750200. [DOI] [PubMed] [Google Scholar]
- 273.Spilker C, Dresbach T, Braunewell KH. Reversible Translocation and Activity-Dependent Localization of the Calcium-Myristoyl Switch Protein Vilip-1 to Different Membrane Compartments in Living Hippocampal Neurons. J Neurosci. 2002;22:7331–7339. doi: 10.1523/JNEUROSCI.22-17-07331.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Dizhoor AM, Ericsson LH, Johnson RS, Kumar S, Olshevskaya E, Zozulya S, Neubert TA, Stryer L, Hurley JB, Walsh KA. The Nh2 Terminus of Retinal Recoverin Is Acylated by a Small Family of Fatty Acids. J Biol Chem. 1992;267:16033–16036. [PubMed] [Google Scholar]
- 275.Hwang JY, Koch KW. Calcium- and Myristoyl-Dependent Properties of Guanylate Cyclase-Activating Protein-1 and Protein-2. Biochemistry. 2002;41:13021–13028. doi: 10.1021/bi026618y. [DOI] [PubMed] [Google Scholar]
- 276.Vilas GL, Corvi MM, Plummer GJ, Seime AM, Lambkin GR, Berthiaume LG. Posttranslational Myristoylation of Caspase-Activated P21-Activated Protein Kinase 2 (Pak2) Potentiates Late Apoptotic Events. Proc Natl Acad Sci US A. 2006;103:6542–6547. doi: 10.1073/pnas.0600824103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Chida T, Ando M, Matsuki T, Masu Y, Nagaura Y, Takano-Yamamoto T, Tamura S, Kobayashi T. N-Myristoylation Is Essential for Protein Phosphatases Ppm1a and Ppm1b to Dephosphorylate Their Physiological Substrates in Cells. Biochem J. 2013;449:741–749. doi: 10.1042/BJ20121201. [DOI] [PubMed] [Google Scholar]
- 278.Lee J, Sayegh J, Daniel J, Clarke S, Bedford MT. Prmt8, a New Membrane-Bound Tissue-Specific Member of the Protein Arginine Methyltransferase Family. J Biol Chem. 2005;280:32890–32896. doi: 10.1074/jbc.M506944200. [DOI] [PubMed] [Google Scholar]
- 279.Konno D, Ko JA, Usui S, Hori K, Maruoka H, Inui M, Fujikado T, Tano Y, Suzuki T, Tohyama K, et al. The Postsynaptic Density and Dendritic Raft Localization of Psd-Zip70, Which Contains an N-Myristoylation Sequence and Leucine-Zipper Motifs. J Cell Sci. 2002;115:4695–4706. doi: 10.1242/jcs.00127. [DOI] [PubMed] [Google Scholar]
- 280.Saeki K, Miura Y, Aki D, Kurosaki T, Yoshimura A. The B Cell-Specific Major Raft Protein, Raftlin, Is Necessary for the Integrity of Lipid Raft and Bcr Signal Transduction. EMBO J. 2003;22:3015–3026. doi: 10.1093/emboj/cdg293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Brede G, Solheim J, Stang E, Prydz H. Mutants of the Protein Serine Kinase Pskh1 Disassemble the Golgi Apparatus. Exp Cell Res. 2003;291:299–312. doi: 10.1016/j.yexcr.2003.07.009. [DOI] [PubMed] [Google Scholar]
- 282.Resh MD. Myristylation and Palmitylation of Src Family Members: The Fats of the Matter. Cell. 1994;76:411–413. doi: 10.1016/0092-8674(94)90104-x. [DOI] [PubMed] [Google Scholar]
- 283.Holland SJ, Liao XC, Mendenhall MK, Zhou X, Pardo J, Chu P, Spencer C, Fu A, Sheng N, Yu P, et al. Functional Cloning of Src-Like Adapter Protein-2 (Slap-2), a Novel Inhibitor of Antigen Receptor Signaling. J Exp Med. 2001;194:1263–1276. doi: 10.1084/jem.194.9.1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Gelman IH, Lee K, Tombler E, Gordon R, Lin X. Control of Cytoskeletal Architecture by the Src-Suppressed C Kinase Substrate, Ssecks. Cell Motil Cytoskeleton. 1998;41:1–17. doi: 10.1002/(SICI)1097-0169(1998)41:1<1::AID-CM1>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 285.Michiels F, Stam JC, Hordijk PL, van der Kammen RA, Ruuls-Van Stalle L, Feltkamp CA, Collard JG. Regulated Membrane Localization of Tiam1, Mediated by the Nh2-Terminal Pleckstrin Homology Domain, Is Required for Rac-Dependent Membrane Ruffling and C-Jun Nh2-Terminal Kinase Activation. J Cell Biol. 1997;137:387–398. doi: 10.1083/jcb.137.2.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Rowe DC, McGettrick AF, Latz E, Monks BG, Gay NJ, Yamamoto M, Akira S, O’Neill LA, Fitzgerald KA, Golenbock DT. The Myristoylation of Trif-Related Adaptor Molecule Is Essential for Toll-Like Receptor 4 Signal Transduction. Proc Natl Acad Sci US A. 2006;103:6299–6304. doi: 10.1073/pnas.0510041103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Utsumi T, Sakurai N, Nakano K, Ishisaka R. C-Terminal 15 Kda Fragment of Cytoskeletal Actin Is Posttranslationally N-Myristoylated Upon Caspase-Mediated Cleavage and Targeted to Mitochondria. FEBS Lett. 2003;539:37–44. doi: 10.1016/s0014-5793(03)00180-7. [DOI] [PubMed] [Google Scholar]
- 288.Beauchamp E, Tekpli X, Marteil G, Lagadic-Gossmann D, Legrand P, Rioux V. N-Myristoylation Targets Dihydroceramide Delta4-Desaturase 1 to Mitochondria: Partial Involvement in the Apoptotic Effect of Myristic Acid. Biochimie. 2009;91:1411–1419. doi: 10.1016/j.biochi.2009.07.014. [DOI] [PubMed] [Google Scholar]
- 289.Murakami K, Yubisui T, Takeshita M, Miyata T. The Nh2-Terminal Structures of Human and Rat Liver Microsomal Nadh-Cytochrome B5 Reductases. J Biochem. 1989;105:312–317. doi: 10.1093/oxfordjournals.jbchem.a122659. [DOI] [PubMed] [Google Scholar]
- 290.Borgese N, Aggujaro D, Carrera P, Pietrini G, Bassetti M. A Role for N-Myristoylation in Protein Targeting: Nadh-Cytochrome B5 Reductase Requires Myristic Acid for Association with Outer Mitochondrial but Not Er Membranes. J Cell Biol. 1996;135:1501–1513. doi: 10.1083/jcb.135.6.1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Sarkar SN, Bandyopadhyay S, Ghosh A, Sen GC. Enzymatic Characteristics of Recombinant Medium Isozyme of 2′-5′ Oligoadenylate Synthetase. J Biol Chem. 1999;274:1848–1855. doi: 10.1074/jbc.274.3.1848. [DOI] [PubMed] [Google Scholar]
- 292.Takasaki A, Hayashi N, Matsubara M, Yamauchi E, Taniguchi H. Identification of the Calmodulin-Binding Domain of Neuron-Specific Protein Kinase C Substrate Protein Cap-22/Nap-22. Direct Involvement of Protein Myristoylation in Calmodulin-Target Protein Interaction. J Biol Chem. 1999;274:11848–11853. doi: 10.1074/jbc.274.17.11848. [DOI] [PubMed] [Google Scholar]
- 293.Toska E, Campbell HA, Shandilya J, Goodfellow SJ, Shore P, Medler KF, Roberts SG. Repression of Transcription by Wt1-Basp1 Requires the Myristoylation of Basp1 and the Pip2-Dependent Recruitment of Histone Deacetylase. Cell Rep. 2012;2:462–469. doi: 10.1016/j.celrep.2012.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Gutierrez-Ford C, Levay K, Gomes AV, Perera EM, Som T, Kim YM, Benovic JL, Berkovitz GD, Slepak VZ. Characterization of Tescalcin, a Novel Ef-Hand Protein with a Single Ca2+-Binding Site: Metal-Binding Properties, Localization in Tissues and Cells, and Effect on Calcineurin. Biochemistry. 2003;42:14553–14565. doi: 10.1021/bi034870f. [DOI] [PubMed] [Google Scholar]
- 295.Zaun HC, Shrier A, Orlowski J. N-Myristoylation and Ca2+ Binding of Calcineurin B Homologous Protein Chp3 Are Required to Enhance Na+/H+ Exchanger Nhe1 Half-Life and Activity at the Plasma Membrane. J Biol Chem. 2012;287:36883–36895. doi: 10.1074/jbc.M112.394700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Uno F, Sasaki J, Nishizaki M, Carboni G, Xu K, Atkinson EN, Kondo M, Minna JD, Roth JA, Ji L. Myristoylation of the Fus1 Protein Is Required for Tumor Suppression in Human Lung Cancer Cells. Cancer Res. 2004;64:2969–2976. doi: 10.1158/0008-5472.can-03-3702. [DOI] [PubMed] [Google Scholar]
- 297.McLaughlin S, Aderem A. The Myristoyl-Electrostatic Switch: A Modulator of Reversible Protein-Membrane Interactions. Trends Biochem Sci. 1995;20:272–276. doi: 10.1016/s0968-0004(00)89042-8. [DOI] [PubMed] [Google Scholar]
- 298.Swierczynski SL, Blackshear PJ. Myristoylation-Dependent and Electrostatic Interactions Exert Independent Effects on the Membrane Association of the Myristoylated Alanine-Rich Protein Kinase C Substrate Protein in Intact Cells. J Biol Chem. 1996;271:23424–23430. doi: 10.1074/jbc.271.38.23424. [DOI] [PubMed] [Google Scholar]
- 299.Liu Y, Kahn RA, Prestegard JH. Structure and Membrane Interaction of Myristoylated Arf1. Structure. 2009;17:79–87. doi: 10.1016/j.str.2008.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Tanaka T, Ames JB, Harvey TS, Stryer L, Ikura M. Sequestration of the Membrane-Targeting Myristoyl Group of Recoverin in the Calcium-Free State. Nature. 1995;376:444–447. doi: 10.1038/376444a0. [DOI] [PubMed] [Google Scholar]
- 301.Desmeules P, Grandbois M, Bondarenko VA, Yamazaki A, Salesse C. Measurement of Membrane Binding between Recoverin, a Calcium-Myristoyl Switch Protein, and Lipid Bilayers Byafm-Based Force Spectroscopy. Biophys J. 82:3343–3350. doi: 10.1016/S0006-3495(02)75674-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Hermida-Matsumoto L, Resh MD. Human Immunodeficiency Virus Type 1 Protease Triggers a Myristoyl Switch That Modulates Membrane Binding of Pr55(Gag) and P17ma. J Virol. 1999;73:1902–1908. doi: 10.1128/jvi.73.3.1902-1908.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Tang C, Loeliger E, Luncsford P, Kinde I, Beckett D, Summers MF. Entropic Switch Regulates Myristate Exposure in the Hiv-1 Matrix Protein. Proc Natl Acad Sci US A. 2004;101:517–522. doi: 10.1073/pnas.0305665101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Smith MT, Meissner J, Esmonde S, Wong HJ, Meiering EM. Energetics and Mechanisms of Folding and Flipping the Myristoyl Switch in the {Beta}-Trefoil Protein, Hisactophilin. Proc Natl Acad Sci US A. 2010;107:20952–20957. doi: 10.1073/pnas.1008026107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Colombo S, Longhi R, Alcaro S, Ortuso F, Sprocati T, Flora A, Borgese N. N-Myristoylation Determines Dual Targeting of Mammalian Nadh-Cytochrome B5 Reductase to Er and Mitochondrial Outer Membranes by a Mechanism of Kinetic Partitioning. J Cell Biol. 2005;168:735–745. doi: 10.1083/jcb.200407082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Koeppel MA, McCarthy CC, Moertl E, Jakobi R. Identification and Characterization of Ps-Gap as a Novel Regulator of Caspase-Activated Pak-2. J Biol Chem. 2004;279:53653–53664. doi: 10.1074/jbc.M410530200. [DOI] [PubMed] [Google Scholar]
- 307.Martin DD, Ahpin CY, Heit RJ, Perinpanayagam MA, Yap MC, Veldhoen RA, Goping IS, Berthiaume LG. Tandem Reporter Assay for Myristoylated Proteins Post-Translationally (Trampp) Identifies Novel Substrates for Post-Translational Myristoylation: Pkcepsilon, a Case Study. FASEB J. 2012;26:13–28. doi: 10.1096/fj.11-182360. [DOI] [PubMed] [Google Scholar]
- 308.Matsubara M, Nakatsu T, Kato H, Taniguchi H. Crystal Structure of a Myristoylated Cap-23/Nap-22 N-Terminal Domain Complexed with Ca2+/Calmodulin. EMBO J. 2004;23:712–718. doi: 10.1038/sj.emboj.7600093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Matsubara M, Titani K, Taniguchi H, Hayashi N. Direct Involvement of Protein Myristoylation in Myristoylated Alanine-Rich C Kinase Substrate (Marcks)-Calmodulin Interaction. J Biol Chem. 2003;278:48898–48902. doi: 10.1074/jbc.M305488200. [DOI] [PubMed] [Google Scholar]
- 310.Ghanam RH, Fernandez TF, Fledderman EL, Saad JS. Binding of Calmodulin to the Hiv-1 Matrix Protein Triggers Myristate Exposure. J Biol Chem. 2010;285:41911–41920. doi: 10.1074/jbc.M110.179093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Sperlich B, Kapoor S, Waldmann H, Winter R, Weise K. Regulation of K-Ras4b Membrane Binding by Calmodulin. Biophys J. 2016;111:113–122. doi: 10.1016/j.bpj.2016.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Weise K, Kapoor S, Werkmuller A, Mobitz S, Zimmermann G, Triola G, Waldmann H, Winter R. Dissociation of the K-Ras4b/Pdedelta Complex Upon Contact with Lipid Membranes: Membrane Delivery Instead of Extraction. J Am Chem Soc. 2012;134:11503–11510. doi: 10.1021/ja305518h. [DOI] [PubMed] [Google Scholar]
- 313.Linder ME, Pang IH, Duronio RJ, Gordon JI, Sternweis PC, Gilman AG. Lipid Modifications of G Protein Subunits. Myristoylation of Go Alpha Increases Its Affinity for Beta Gamma. J Biol Chem. 1991;266:4654–4659. [PubMed] [Google Scholar]
- 314.Bachert C, Linstedt AD. Dual Anchoring of the Grasp Membrane Tether Promotes Trans Pairing. J Biol Chem. 2010;285:16294–16301. doi: 10.1074/jbc.M110.116129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315.Heinrich F, Nanda H, Goh HZ, Bachert C, Losche M, Linstedt AD. Myristoylation Restricts Orientation of the Grasp Domain on Membranes and Promotes Membrane Tethering. J Biol Chem. 2014;289:9683–9691. doi: 10.1074/jbc.M113.543561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316.Wright KJ, Baye LM, Olivier-Mason A, Mukhopadhyay S, Sang L, Kwong M, Wang W, Pretorius PR, Sheffield VC, Sengupta P, et al. An Arl3-Unc119-Rp2 Gtpase Cycle Targets Myristoylated Nphp3 to the Primary Cilium. Genes Dev. 2011;25:2347–2360. doi: 10.1101/gad.173443.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317.Mejuch T, van Hattum H, Triola G, Jaiswal M, Waldmann H. Specificity of Lipoprotein Chaperones for the Characteristic Lipidated Structural Motifs of Their Cognate Lipoproteins. ChemBioChem. 2015;16:2460–2465. doi: 10.1002/cbic.201500355. [DOI] [PubMed] [Google Scholar]
- 318.Zhang H, Constantine R, Vorobiev S, Chen Y, Seetharaman J, Huang YJ, Xiao R, Montelione GT, Gerstner CD, Davis MW, et al. Unc119 Is Required for G Protein Trafficking in Sensory Neurons. Nat Neurosci. 2011;14:874–880. doi: 10.1038/nn.2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Jaiswal M, Fansa EK, Kösling SK, Mejuch T, Waldmann H, Wittinghofer A. Novel Biochemical and Structural Insights into the Interaction of Myristoylated Cargo with Unc119 Protein and Their Release by Arl2/3. J Biol Chem. 2016;291:20766–20778. doi: 10.1074/jbc.M116.741827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320.Boggon TJ, Eck MJ. Structure and Regulation of Src Family Kinases. Oncogene. 2004;23:7918–7927. doi: 10.1038/sj.onc.1208081. [DOI] [PubMed] [Google Scholar]
- 321.Hantschel O, Nagar B, Guettler S, Kretzschmar J, Dorey K, Kuriyan J, Superti-Furga G. A Myristoyl/Phosphotyrosine Switch Regulates C-Abl. Cell. 2003;112:845–857. doi: 10.1016/s0092-8674(03)00191-0. [DOI] [PubMed] [Google Scholar]
- 322.Nagar B, Hantschel O, Young MA, Scheffzek K, Veach D, Bornmann W, Clarkson B, Superti-Furga G, Kuriyan J. Structural Basis for the Autoinhibition of C-Abl Tyrosine Kinase. Cell. 2003;112:859–871. doi: 10.1016/s0092-8674(03)00194-6. [DOI] [PubMed] [Google Scholar]
- 323.Maurer-Stroh S, Eisenhaber B, Eisenhaber F. N-Terminal N-Myristoylation of Proteins: Prediction of Substrate Proteins from Amino Acid Sequence. J Mol Biol. 2002;317:541–557. doi: 10.1006/jmbi.2002.5426. [DOI] [PubMed] [Google Scholar]
- 324.Bologna G, Yvon C, Duvaud S, Veuthey AL. N-Terminal Myristoylation Predictions by Ensembles of Neural Networks. Proteomics. 2004;4:1626–1632. doi: 10.1002/pmic.200300783. [DOI] [PubMed] [Google Scholar]
- 325.Martinez A, Traverso JA, Valot B, Ferro M, Espagne C, Ephritikhine G, Zivy M, Giglione C, Meinnel T. Extent of N-Terminal Modifications in Cytosolic Proteins from Eukaryotes. Proteomics. 2008;8:2809–2831. doi: 10.1002/pmic.200701191. [DOI] [PubMed] [Google Scholar]
- 326.Traverso JA, Giglione C, Meinnel T. High-Throughput Profiling of N-Myristoylation Substrate Specificity across Species Including Pathogens. Proteomics. 2013;13:25–36. doi: 10.1002/pmic.201200375. [DOI] [PubMed] [Google Scholar]
- 327.Charron G, Zhang MM, Yount JS, Wilson J, Raghavan AS, Shamir E, Hang HC. Robust Fluorescent Detection of Protein Fatty-Acylation with Chemical Reporters. J Am Chem Soc. 2009;131:4967–4975. doi: 10.1021/ja810122f. [DOI] [PubMed] [Google Scholar]
- 328.Martin BR, Cravatt BF. Large-Scale Profiling of Protein Palmitoylation in Mammalian Cells. Nat Methods. 2009;6:135–138. doi: 10.1038/nmeth.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 329.Hannoush RN, Arenas-Ramirez N. Imaging the Lipidome: Omega-Alkynyl Fatty Acids for Detection and Cellular Visualization of Lipid-Modified Proteins. ACS Chem Biol. 2009;4:581–587. doi: 10.1021/cb900085z. [DOI] [PubMed] [Google Scholar]
- 330.Hannoush RN, Sun J. The Chemical Toolbox for Monitoring Protein Fatty Acylation and Prenylation. Nat Chem Biol. 2010;6:498–506. doi: 10.1038/nchembio.388. [DOI] [PubMed] [Google Scholar]
- 331.Wright MH, Paape D, Price HP, Smith DF, Tate EW. Global Profiling and Inhibition of Protein Lipidation in Vector and Host Stages of the Sleeping Sickness Parasite Trypanosoma Brucei. ACS Infect Dis. 2016;2:427–441. doi: 10.1021/acsinfecdis.6b00034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332.Wright MH, Paape D, Storck EM, Serwa RA, Smith DF, Tate EW. Global Analysis of Protein N-Myristoylation and Exploration of N-Myristoyltransferase as a Drug Target in the Neglected Human Pathogen Leishmania Donovani. Chem Biol. 2015;22:342–354. doi: 10.1016/j.chembiol.2015.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333.Serwa RA, Abaitua F, Krause E, Tate EW, O’Hare P. Systems Analysis of Protein Fatty Acylation in Herpes Simplex Virus-Infected Cells Using Chemical Proteomics. Chem Biol. 2015;22:1008–1017. doi: 10.1016/j.chembiol.2015.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 334.Colquhoun DR, Lyashkov AE, Ubaida Mohien C, Aquino VN, Bullock BT, Dinglasan RR, Agnew BJ, Graham DR. Bioorthogonal Mimetics of Palmitoyl-Coa and Myristoyl-Coa and Their Subsequent Isolation by Click Chemistry and Characterization by Mass Spectrometry Reveal Novel Acylated Host-Proteins Modified by Hiv-1 Infection. Proteomics. 2015;15:2066–2077. doi: 10.1002/pmic.201500063. [DOI] [PubMed] [Google Scholar]
- 335.Thinon E, Serwa RA, Broncel M, Brannigan JA, Brassat U, Wright MH, Heal WP, Wilkinson AJ, Mann DJ, Tate EW. Global Profiling of Co- and Post-Translationally N-Myristoylated Proteomes in Human Cells. Nat Commun. 2014;5:4919. doi: 10.1038/ncomms5919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 336.Broncel M, Serwa RA, Ciepla P, Krause E, Dallman MJ, Magee AI, Tate EW. Multifunctional Reagents for Quantitative Proteome-Wide Analysis of Protein Modification in Human Cells and Dynamic Profiling of Protein Lipidation During Vertebrate Development. Angew Chem Int Ed. 2015;54:5948–5951. doi: 10.1002/anie.201500342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 337.Herrera LJ, Brand S, Santos A, Nohara LL, Harrison J, Norcross NR, Thompson S, Smith V, Lema C, Varela-Ramirez A, et al. Validation of N-Myristoyltransferase as Potential Chemotherapeutic Target in Mammal-Dwelling Stages of Trypanosoma Cruzi. PLOS Negl Trop Dis. 2016;10:e0004540. doi: 10.1371/journal.pntd.0004540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 338.Wright MH, Clough B, Rackham MD, Rangachari K, Brannigan JA, Grainger M, Moss DK, Bottrill AR, Heal WP, Broncel M, et al. Validation of N-Myristoyltransferase as an Antimalarial Drug Target Using an Integrated Chemical Biology Approach. Nat Chem. 2014;6:112–121. doi: 10.1038/nchem.1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 339.Kishore NS, Wood DC, Mehta PP, Wade AC, Lu T, Gokel GW, Gordon JI. Comparison of the Acyl Chain Specificities of Human Myristoyl-Coa Synthetase and Human Myristoyl-Coa:Protein N-Myristoyltransferase. J Biol Chem. 1993;268:4889–4902. [PubMed] [Google Scholar]
- 340.French KJ, Zhuang Y, Schrecengost RS, Copper JE, Xia Z, Smith CD. Cyclohexyl-Octahydro-Pyrrolo[1,2-a]Pyrazine-Based Inhibitors of Human N-Myristoyltransferase-1. J Pharmacol Exp Ther. 2004;309:340–347. doi: 10.1124/jpet.103.061572. [DOI] [PubMed] [Google Scholar]
- 341.Bowyer PW, Gunaratne RS, Grainger M, Withers-Martinez C, Wickramsinghe SR, Tate EW, Leatherbarrow RJ, Brown KA, Holder AA, Smith DF. Molecules Incorporating a Benzothiazole Core Scaffold Inhibit the N-Myristoyltransferase of Plasmodium Falciparum. Biochem J. 2007;408:173–180. doi: 10.1042/BJ20070692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 342.Goncalves V, Brannigan JA, Whalley D, Ansell KH, Saxty B, Holder AA, Wilkinson AJ, Tate EW, Leatherbarrow RJ. Discovery of Plasmodium Vivax N-Myristoyltransferase Inhibitors: Screening, Synthesis, and Structural Characterization of Their Binding Mode. J Med Chem. 2012;55:3578–3582. doi: 10.1021/jm300040p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 343.Frearson JA, Brand S, McElroy SP, Cleghorn LA, Smid O, Stojanovski L, Price HP, Guther ML, Torrie LS, Robinson DA, et al. N-Myristoyltransferase Inhibitors as New Leads to Treat Sleeping Sickness. Nature. 2010;464:728–732. doi: 10.1038/nature08893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 344.Brand S, Cleghorn LA, McElroy SP, Robinson DA, Smith VC, Hallyburton I, Harrison JR, Norcross NR, Spinks D, Bayliss T, et al. Discovery of a Novel Class of Orally Active Trypanocidal N-Myristoyltransferase Inhibitors. J Med Chem. 2012;55:140–152. doi: 10.1021/jm201091t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 345.Masubuchi M, Ebiike H, Kawasaki K, Sogabe S, Morikami K, Shiratori Y, Tsujii S, Fujii T, Sakata K, Hayase M, et al. Synthesis and Biological Activities of Benzofuran Antifungal Agents Targeting Fungal N-Myristoyltransferase. Bioorg Med Chem. 2003;11:4463–4478. doi: 10.1016/s0968-0896(03)00429-2. [DOI] [PubMed] [Google Scholar]
- 346.Masubuchi M, Kawasaki K, Ebiike H, Ikeda Y, Tsujii S, Sogabe S, Fujii T, Sakata K, Shiratori Y, Aoki Y, et al. Design and Synthesis of Novel Benzofurans as a New Class of Antifungal Agents Targeting Fungal N-Myristoyltransferase. Part 1. Bioorg Med Chem Lett. 2001;11:1833–1837. doi: 10.1016/s0960-894x(01)00319-5. [DOI] [PubMed] [Google Scholar]
- 347.Sheng C, Xu H, Wang W, Cao Y, Dong G, Wang S, Che X, Ji H, Miao Z, Yao J, et al. Design, Synthesis and Antifungal Activity of Isosteric Analogues of Benzoheterocyclic N-Myristoyltransferase Inhibitors. Eur J Med Chem. 2010;45:3531–3540. doi: 10.1016/j.ejmech.2010.03.007. [DOI] [PubMed] [Google Scholar]
- 348.Devadas B, Freeman SK, Zupec ME, Lu HF, Nagarajan SR, Kishore NS, Lodge JK, Kuneman DW, McWherter CA, Vinjamoori DV, et al. Design and Synthesis of Novel Imidazole-Substituted Dipeptide Amides as Potent and Selective Inhibitors of Candida Albicans Myristoylcoa:Protein N-Myristoyltransferase and Identification of Related Tripeptide Inhibitors with Mechanism-Based Antifungal Activity. J Med Chem. 1997;40:2609–2625. doi: 10.1021/jm970094w. [DOI] [PubMed] [Google Scholar]
- 349.Devadas B, Zupec ME, Freeman SK, Brown DL, Nagarajan S, Sikorski JA, McWherter CA, Getman DP, Gordon JI. Design and Syntheses of Potent and Selective Dipeptide Inhibitors of Candida Albicans Myristoyl-Coa:Protein N-Myristoyltransferase. J Med Chem. 1995;38:1837–1840. doi: 10.1021/jm00011a001. [DOI] [PubMed] [Google Scholar]
- 350.Nagarajan SR, Devadas B, Zupec ME, Freeman SK, Brown DL, Lu HF, Mehta PP, Kishore NS, McWherter CA, Getman DP, et al. Conformationally Constrained [P-(Omega-Aminoalkyl)Phenacetyl]-L-Seryl-L-Lysyl Dipeptide Amides as Potent Peptidomimetic Inhibitors of Candida Albicans and Human Myristoyl-Coa:Protein N-Myristoyl Transferase. J Med Chem. 1997;40:1422–1438. doi: 10.1021/jm9608671. [DOI] [PubMed] [Google Scholar]
- 351.Ebara S, Naito H, Nakazawa K, Ishii F, Nakamura M. Ftr1335 Is a Novel Synthetic Inhibitor of Candida Albicans N-Myristoyltransferase with Fungicidal Activity. Biol Pharm Bull. 2005;28:591–595. doi: 10.1248/bpb.28.591. [DOI] [PubMed] [Google Scholar]
- 352.Yamazaki K, Kaneko Y, Suwa K, Ebara S, Nakazawa K, Yasuno K. Synthesis of Potent and Selective Inhibitors of Candida Albicans N-Myristoyltransferase Based on the Benzothiazole Structure. Bioorg Med Chem. 2005;13:2509–2522. doi: 10.1016/j.bmc.2005.01.033. [DOI] [PubMed] [Google Scholar]
- 353.Talamonti MS, Roh MS, Curley SA, Gallick GE. Increase in Activity and Level of Pp60c-Src in Progressive Stages of Human Colorectal Cancer. J Clin Invest. 1993;91:53–60. doi: 10.1172/JCI116200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 354.Magnuson BA, Raju RV, Moyana TN, Sharma RK. Increased N-Myristoyltransferase Activity Observed in Rat and Human Colonic Tumors. J Natl Cancer Inst. 1995;87:1630–1635. doi: 10.1093/jnci/87.21.1630. [DOI] [PubMed] [Google Scholar]
- 355.Selvakumar P, Lakshmikuttyamma A, Shrivastav A, Das SB, Dimmock JR, Sharma RK. Potential Role of N-Myristoyltransferase in Cancer. Prog Lipid Res. 2007;46:1–36. doi: 10.1016/j.plipres.2006.05.002. [DOI] [PubMed] [Google Scholar]
- 356.Shrivastav A, Sharma AR, Bajaj G, Charavaryamath C, Ezzat W, Spafford P, Gore-Hickman R, Singh B, Copete MA, Sharma RK. Elevated N-Myristoyltransferase Activity and Expression in Oral Squamous Cell Carcinoma. Oncol Rep. 2007;18:93–97. [PubMed] [Google Scholar]
- 357.Rajala RV, Radhi JM, Kakkar R, Datla RS, Sharma RK. Increased Expression of N-Myristoyltransferase in Gallbladder Carcinomas. Cancer. 2000;88:1992–1999. [PubMed] [Google Scholar]
- 358.Thinon E, Morales-Sanfrutos J, Mann DJ, Tate EW. N-Myristoyltransferase Inhibition Induces Er-Stress, Cell Cycle Arrest, and Apoptosis in Cancer Cells. ACS Chem Biol. 2016;11:2165–2176. doi: 10.1021/acschembio.6b00371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 359.Hill BT, Skowronski J. Human N-Myristoyltransferases Form Stable Complexes with Lentiviral Nef and Other Viral and Cellular Substrate Proteins. J Virol. 2005;79:1133–1141. doi: 10.1128/JVI.79.2.1133-1141.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 360.Rein A, McClure MR, Rice NR, Luftig RB, Schultz AM. Myristylation Site in Pr65gag Is Essential for Virus Particle Formation by Moloney Murine Leukemia Virus. Proc Natl Acad Sci US A. 1986;83:7246–7250. doi: 10.1073/pnas.83.19.7246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 361.Bryant M, Ratner L. Myristoylation-Dependent Replication and Assembly of Human Immunodeficiency Virus 1. Proc Natl Acad Sci US A. 1990;87:523–527. doi: 10.1073/pnas.87.2.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 362.Takamune N, Gota K, Misumi S, Tanaka K, Okinaka S, Shoji S. Hiv-1 Production Is Specifically Associated with Human Nmt1 Long Form in Human Nmt Isozymes. Microbes Infect. 2008;10:143–150. doi: 10.1016/j.micinf.2007.10.015. [DOI] [PubMed] [Google Scholar]
- 363.Seaton KE, Smith CD. N-Myristoyltransferase Isozymes Exhibit Differential Specificity for Human Immunodeficiency Virus Type 1 Gag and Nef. J Gen Virol. 2008;89:288–296. doi: 10.1099/vir.0.83412-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 364.Burnaevskiy N, Fox TG, Plymire DA, Ertelt JM, Weigele BA, Selyunin AS, Way SS, Patrie SM, Alto NM. Proteolytic Elimination of N-Myristoyl Modifications by the Shigella Virulence Factor Ipaj. Nature. 2013;496:106–109. doi: 10.1038/nature12004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 365.Braun PE, Radin NS. Interactions of Lipids with a Membrane Structural Protein from Myelin. Biochemistry. 1969;8:4310–4318. doi: 10.1021/bi00839a014. [DOI] [PubMed] [Google Scholar]
- 366.Stoffyn P, Folch-Pi J. On the Type of Linkage Binding Fatty Acids Present in Brain White Matter Proteolipid Apoprotein. Biochem Biophys Res Commun. 1971;44:157–161. doi: 10.1016/s0006-291x(71)80172-9. [DOI] [PubMed] [Google Scholar]
- 367.Schmidt MF, Bracha M, Schlesinger MJ. Evidence for Covalent Attachment of Fatty Acids to Sindbis Virus Glycoproteins. Proc Natl Acad Sci US A. 1979;76:1687–1691. doi: 10.1073/pnas.76.4.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 368.O’Brien PJ, Zatz M. Acylation of Bovine Rhodopsin by [3h]Palmitic Acid. J Biol Chem. 1984;259:5054–5057. [PubMed] [Google Scholar]
- 369.Chen ZQ, Ulsh LS, DuBois G, Shih TY. Posttranslational Processing of P21 Ras Proteins Involves Palmitylation of the C-Terminal Tetrapeptide Containing Cysteine-186. J Virol. 1985;56:607–612. doi: 10.1128/jvi.56.2.607-612.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 370.Lobo S, Greentree WK, Linder ME, Deschenes RJ. Identification of a Ras Palmitoyltransferase in Saccharomyces Cerevisiae. J Biol Chem. 2002;277:41268–41273. doi: 10.1074/jbc.M206573200. [DOI] [PubMed] [Google Scholar]
- 371.Roth AF, Feng Y, Chen L, Davis NG. The Yeast Dhhc Cysteine-Rich Domain Protein Akr1p Is a Palmitoyl Transferase. J Cell Biol. 2002;159:23–28. doi: 10.1083/jcb.200206120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 372.Keller CA, Yuan X, Panzanelli P, Martin ML, Alldred M, Sassoè-Pognetto M, Lüscher B. The Γ2 Subunit of Gabaa Receptors Is a Substrate for Palmitoylation by Godz. J Neurosci. 2004;24:5881–5891. doi: 10.1523/JNEUROSCI.1037-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 373.Fukata M, Fukata Y, Adesnik H, Nicoll RA, Bredt DS. Identification of Psd-95 Palmitoylating Enzymes. Neuron. 2004;44:987–996. doi: 10.1016/j.neuron.2004.12.005. [DOI] [PubMed] [Google Scholar]
- 374.Oku S, Takahashi N, Fukata Y, Fukata M. In Silico Screening for Palmitoyl Substrates Reveals a Role for Dhhc1/3/10 (Zdhhc1/3/11)-Mediated Neurochondrin Palmitoylation in Its Targeting to Rab5-Positive Endosomes. J Biol Chem. 2013;288:19816–19829. doi: 10.1074/jbc.M112.431676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 375.Oyama T, Miyoshi Y, Koyama K, Nakagawa H, Yamori T, Ito T, Matsuda H, Arakawa H, Nakamura Y. Isolation of a Novel Gene on 8p21. 3–22 Whose Expression Is Reduced Significantly in Human Colorectal Cancers with Liver Metastasis. Genes Chromosomes Cancer. 2000;29:9–15. doi: 10.1002/1098-2264(2000)9999:9999<::aid-gcc1001>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- 376.Zeidman R, Buckland G, Cebecauer M, Eissmann P, Davis DM, Magee AI. Dhhc2 Is a Protein S -Acyltransferase for Lck. Mol Membr Biol. 2011;28:473–486. doi: 10.3109/09687688.2011.630682. [DOI] [PubMed] [Google Scholar]
- 377.Jia L, Linder ME, Blumer KJ. Gi/O Signaling and the Palmitoyltransferase Dhhc2 Regulate Palmitate Cycling and Shuttling of Rgs7 Family-Binding Protein. J Biol Chem. 2011;286:13695–13703. doi: 10.1074/jbc.M110.193763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 378.Zhang J, Planey SL, Ceballos C, Stevens SM, Keay SK, Zacharias DA. Identification of Ckap4/P63 as a Major Substrate of the Palmitoyl Acyltransferase Dhhc2: a Putative Tumor Suppressor, Using a Novel Proteomics Method. Mol Cell Proteomics. 2008;7:1378–1388. doi: 10.1074/mcp.M800069-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 379.Sharma C, Yang XH, Hemler ME. Dhhc2 Affects Palmitoylation, Stability, and Functions of Tetraspanins Cd9 and Cd151. Mol Biol Cell. 2008;19:3415–3425. doi: 10.1091/mbc.E07-11-1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 380.Noritake J, Fukata Y, Iwanaga T, Hosomi N, Tsutsumi R, Matsuda N, Tani H, Iwanari H, Mochizuki Y, Kodama T, et al. Mobile Dhhc Palmitoylating Enzyme Mediates Activity-Sensitive Synaptic Targeting of Psd-95. J Cell Biol. 2009;186:147–160. doi: 10.1083/jcb.200903101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 381.Greaves J, Gorleku OA, Salaun C, Chamberlain LH. Palmitoylation of the Snap25 Protein Family Specificity and Regulation by Dhhc Palmitoyl Transferases. J Biol Chem. 2010;285:24629–24638. doi: 10.1074/jbc.M110.119289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 382.Fernández-Hernando C, Fukata M, Bernatchez PN, Fukata Y, Lin MI, Bredt DS, Sessa WC. Identification of Golgi-Localized Acyl Transferases That Palmitoylate and Regulate Endothelial Nitric Oxide Synthase. J Cell Biol. 2006;174:369–377. doi: 10.1083/jcb.200601051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 383.Fairbank M, Huang K, El-Husseini A, Nabi IR. Ring Finger Palmitoylation of the Endoplasmic Reticulum Gp78 E3 Ubiquitin Ligase. FEBS Lett. 2012;586:2488–2493. doi: 10.1016/j.febslet.2012.06.011. [DOI] [PubMed] [Google Scholar]
- 384.Woolfrey KM, Sanderson JL, Dell’Acqua ML. The Palmitoyl Acyltransferase Dhhc2 Regulates Recycling Endosome Exocytosis and Synaptic Potentiation through Palmitoylation of Akap79/150. J Neurosci. 2015;35:442–456. doi: 10.1523/JNEUROSCI.2243-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 385.Sharma C, Rabinovitz I, Hemler ME. Palmitoylation by Dhhc3 Is Critical for the Function, Expression, and Stability of Integrin Alpha6beta4. Cell Mol Life Sci. 2012;69:2233–2244. doi: 10.1007/s00018-012-0924-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 386.Oh Y, Jeon YJ, Hong GS, Kim I, Woo HN, Jung YK. Regulation in the Targeting of Trail Receptor 1 to Cell Surface Via Godz for Trail Sensitivity in Tumor Cells. Cell Death Differ. 2012;19:1196–1207. doi: 10.1038/cdd.2011.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 387.Tsutsumi R, Fukata Y, Noritake J, Iwanaga T, Perez F, Fukata M. Identification of G Protein Alpha Subunit-Palmitoylating Enzyme. Mol Cell Biol. 2009;29:435–447. doi: 10.1128/MCB.01144-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 388.Greaves J, Salaun C, Fukata Y, Fukata M, Chamberlain LH. Palmitoylation and Membrane Interactions of the Neuroprotective Chaperone Cysteine-String Protein. J Biol Chem. 2008;283:25014–25026. doi: 10.1074/jbc.M802140200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 389.Lu D, Sun HQ, Wang H, Barylko B, Fukata Y, Fukata M, Albanesi JP, Yin HL. Phosphatidylinositol 4-Kinase Iialpha Is Palmitoylated by Golgi-Localized Palmitoyltransferases in Cholesterol-Dependent Manner. J Biol Chem. 2012;287:21856–21865. doi: 10.1074/jbc.M112.348094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 390.Wang J, Xie Y, Wolff DW, Abel PW, Tu Y. Dhhc Protein-Dependent Palmitoylation Protects Regulator of G-Protein Signaling 4 from Proteasome Degradation. FEBS Lett. 2010;584:4570–4574. doi: 10.1016/j.febslet.2010.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 391.Tian L, McClafferty H, Jeffries O, Shipston MJ. Multiple Palmitoyltransferases Are Required for Palmitoylation-Dependent Regulation of Large Conductance Calcium- and Voltage-Activated Potassium Channels. J Biol Chem. 2010;285:23954–23962. doi: 10.1074/jbc.M110.137802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 392.Thomas GM, Hayashi T, Huganir RL, Linden DJ. Dhhc8-Dependent Pick1 Palmitoylation Is Required for Induction of Cerebellar Long-Term Synaptic Depression. J Neurosci. 2013;33:15401–15407. doi: 10.1523/JNEUROSCI.1283-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 393.Huang K, Sanders S, Singaraja R, Orban P, Cijsouw T, Arstikaitis P, Yanai A, Hayden MR, El-Husseini A. Neuronal Palmitoyl Acyl Transferases Exhibit Distinct Substrate Specificity. FASEB J. 2009;23:2605–2615. doi: 10.1096/fj.08-127399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 394.Taruno A, Sun H, Nakajo K, Murakami T, Ohsaki Y, Kido MA, Ono F, Marunaka Y. Post-Translational Palmitoylation Controls the Voltage Gating and Lipid Raft Association of Calhm1 Channel. J Physiol. 2017 doi: 10.1113/JP274164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 395.Segal-Salto M, Sapir T, Reiner O. Reversible Cysteine Acylation Regulates the Activity of Human Palmitoyl-Protein Thioesterase 1 (Ppt1) PLOS ONE. 2016;11:e0146466. doi: 10.1371/journal.pone.0146466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 396.Li Y, Hu J, Hofer K, Wong AM, Cooper JD, Birnbaum SG, Hammer RE, Hofmann SL. Dhhc5 Interacts with Pdz Domain 3 of Post-Synaptic Density-95 (Psd-95) Protein and Plays a Role in Learning and Memory. J Biol Chem. 2010;285:13022–13031. doi: 10.1074/jbc.M109.079426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 397.Brigidi GS, Sun Y, Beccano-Kelly D, Pitman K, Mobasser M, Borgland SL, Milnerwood AJ, Bamji SX. Palmitoylation of Delta-Catenin by Dhhc5 Mediates Activity-Induced Synapse Plasticity. Nat Neurosci. 2014;17:522–532. doi: 10.1038/nn.3657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 398.Li Y, Martin BR, Cravatt BF, Hofmann SL. Dhhc5 Protein Palmitoylates Flotillin-2 and Is Rapidly Degraded on Induction of Neuronal Differentiation in Cultured Cells. J Biol Chem. 2012;287:523–530. doi: 10.1074/jbc.M111.306183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 399.Kokkola T, Kruse C, Roy-Pogodzik EM, Pekkinen J, Bauch C, Honck HH, Hennemann H, Kreienkamp HJ. Somatostatin Receptor 5 Is Palmitoylated by the Interacting Zdhhc5 Palmitoyltransferase. FEBS Lett. 2011;585:2665–2670. doi: 10.1016/j.febslet.2011.07.028. [DOI] [PubMed] [Google Scholar]
- 400.Thomas GM, Hayashi T, Chiu SL, Chen CM, Huganir RL. Palmitoylation by Dhhc5/8 Targets Grip1 to Dendritic Endosomes to Regulate Ampa-R Trafficking. Neuron. 2012;73:482–496. doi: 10.1016/j.neuron.2011.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 401.Howie J, Reilly L, Fraser NJ, Vlachaki Walker JM, Wypijewski KJ, Ashford ML, Calaghan SC, McClafferty H, Tian L, Shipston MJ, et al. Substrate Recognition by the Cell Surface Palmitoyl Transferase Dhhc5. Proc Natl Acad Sci US A. 2014;111:17534–17539. doi: 10.1073/pnas.1413627111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 402.Yang W, Di Vizio D, Kirchner M, Steen H, Freeman MR. Proteome Scale Characterization of Human S-Acylated Proteins in Lipid Raft-Enriched and Non-Raft Membranes. Mol Cell Proteomics. 2010;9:54–70. doi: 10.1074/mcp.M800448-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 403.Lakkaraju AK, Abrami L, Lemmin T, Blaskovic S, Kunz B, Kihara A, Dal Peraro M, van der Goot FG. Palmitoylated Calnexin Is a Key Component of the Ribosome–Translocon Complex. EMBO J. 2012;31:1823–1835. doi: 10.1038/emboj.2012.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 404.Fredericks GJ, Hoffmann FW, Rose AH, Osterheld HJ, Hess FM, Mercier F, Hoffmann PR. Stable Expression and Function of the Inositol 1,4,5-Triphosphate Receptor Requires Palmitoylation by a Dhhc6/Selenoprotein K Complex. Proc Natl Acad Sci US A. 2014;111:16478–16483. doi: 10.1073/pnas.1417176111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 405.Pedram A, Razandi M, Deschenes RJ, Levin ER. Dhhc-7 and -21 Are Palmitoylacyltransferases for Sex Steroid Receptors. Mol Biol Cell. 2012;23:188–199. doi: 10.1091/mbc.E11-07-0638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 406.Du K, Murakami S, Sun Y, Kilpatrick CL, Luscher B. Dhhc7 Palmitoylates Glucose Transporter 4 (Glut4) and Regulates Glut4 Membrane Translocation. J Biol Chem. 2017;292:2979–2991. doi: 10.1074/jbc.M116.747139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 407.Rossin A, Durivault J, Chakhtoura-Feghali T, Lounnas N, Gagnoux-Palacios L, Hueber AO. Fas Palmitoylation by the Palmitoyl Acyltransferase Dhhc7 Regulates Fas Stability. Cell Death Differ. 2015;22:643–653. doi: 10.1038/cdd.2014.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 408.Aramsangtienchai P, Spiegelman NA, Cao J, Lin H. S-Palmitoylation of Junctional Adhesion Molecule C Regulates Its Tight Junction Localization and Cell Migration. J Biol Chem. 2017;292:5325–5334. doi: 10.1074/jbc.M116.730523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 409.Chen B, Zheng B, DeRan M, Jarugumilli GK, Fu J, Brooks YS, Wu X. Zdhhc7-Mediated S-Palmitoylation of Scribble Regulates Cell Polarity. Nat Chem Biol. 2016;12:686–693. doi: 10.1038/nchembio.2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 410.Milde S, Coleman MP. Identification of Palmitoyltransferase and Thioesterase Enzymes That Control the Subcellular Localization of Axon Survival Factor Nicotinamide Mononucleotide Adenylyltransferase 2 (Nmnat2) J Biol Chem. 2014;289:32858–32870. doi: 10.1074/jbc.M114.582338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 411.Singaraja RR, Kang MH, Vaid K, Sanders SS, Vilas GL, Arstikaitis P, Coutinho J, Drisdel RC, El-Husseini Ael D, Green WN, et al. Palmitoylation of Atp-Binding Cassette Transporter A1 Is Essential for Its Trafficking and Function. Circ Res. 2009;105:138–147. doi: 10.1161/CIRCRESAHA.108.193011. [DOI] [PubMed] [Google Scholar]
- 412.Swarthout JT, Lobo S, Farh L, Croke MR, Greentree WK, Deschenes RJ, Linder ME. Dhhc9 and Gcp16 Constitute a Human Protein Fatty Acyltransferase with Specificity for H- and N-Ras. J Biol Chem. 2005;280:31141–31148. doi: 10.1074/jbc.M504113200. [DOI] [PubMed] [Google Scholar]
- 413.Tian L, McClafferty H, Knaus HG, Ruth P, Shipston MJ. Distinct Acyl Protein Transferases and Thioesterases Control Surface Expression of Calcium-Activated Potassium Channels. J Biol Chem. 2012;287:14718–14725. doi: 10.1074/jbc.M111.335547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 414.Saitoh F, Tian QB, Okano A, Sakagami H, Kondo H, Suzuki T. Nidd, a Novel Dhhc-Containing Protein, Targets Neuronal Nitric-Oxide Synthase (Nnos) to the Synaptic Membrane through a Pdz-Dependent Interaction and Regulates Nnos Activity. J Biol Chem. 2004;279:29461–29468. doi: 10.1074/jbc.M401471200. [DOI] [PubMed] [Google Scholar]
- 415.Mizumaru C, Saito Y, Ishikawa T, Yoshida T, Yamamoto T, Nakaya T, Suzuki T. Suppression of App-Containing Vesicle Trafficking and Production of Beta-Amyloid by Aid/Dhhc-12 Protein. J Neurochem. 2009;111:1213–1224. doi: 10.1111/j.1471-4159.2009.06399.x. [DOI] [PubMed] [Google Scholar]
- 416.Ohno Y, Kihara A, Sano T, Igarashi Y. Intracellular Localization and Tissue-Specific Distribution of Human and Yeast Dhhc Cysteine-Rich Domain-Containing Proteins. Biochim Biophys Acta. 2006;1761:474–483. doi: 10.1016/j.bbalip.2006.03.010. [DOI] [PubMed] [Google Scholar]
- 417.Michelson M, Ben-Sasson A, Vinkler C, Leshinsky-Silver E, Netzer I, Frumkin A, Kivity S, Lerman-Sagie T, Lev D. Delineation of the Interstitial 6q25 Microdeletion Syndrome: Refinement of the Critical Causative Region. Am J Hum Genet. 2012;158A:1395–1399. doi: 10.1002/ajmg.a.35361. [DOI] [PubMed] [Google Scholar]
- 418.Yu L, Reader JC, Chen C, Zhao XF, Ha JS, Lee C, York T, Gojo I, Baer MR, Ning Y. Activation of a Novel Palmitoyltransferase Zdhhc14 in Acute Biphenotypic Leukemia and Subsets of Acute Myeloid. Leukemia. 2011;25:367–371. doi: 10.1038/leu.2010.271. [DOI] [PubMed] [Google Scholar]
- 419.Yeste-Velasco M, Mao X, Grose R, Kudahetti SC, Lin D, Marzec J, Vasiljević N, Chaplin T, Xue L, Xu M, et al. Identification of Zdhhc14 as a Novel Human Tumour Suppressor Gene. J Pathol. 2014;232:566–577. doi: 10.1002/path.4327. [DOI] [PubMed] [Google Scholar]
- 420.Anami K, Oue N, Noguchi T, Sakamoto N, Sentani K, Hayashi T, Hinoi T, Okajima M, Graff JM, Yasui W. Search for Transmembrane Protein in Gastric Cancer by the Escherichia Coli Ampicillin Secretion Trap: Expression of Dsc2 in Gastric Cancer with Intestinal Phenotype. J Pathol. 2010;221:275–284. doi: 10.1002/path.2717. [DOI] [PubMed] [Google Scholar]
- 421.Mansouri MR, Marklund L, Gustavsson P, Davey E, Carlsson B, Larsson C, White I, Gustavson KH, Dahl N. Loss of Zdhhc15 Expression in a Woman with a Balanced Translocation T(X;15)(Q13.3;Cen) and Severe Mental Retardation. Eur J Hum Gen. 2005;13:970–977. doi: 10.1038/sj.ejhg.5201445. [DOI] [PubMed] [Google Scholar]
- 422.Yang G, Cynader MS. Regulation of Protein Trafficking: Jnk3 at the Golgi Complex. Cell Cycle. 2014;13:5–6. doi: 10.4161/cc.27019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 423.Zhang F, Di Y, Li J, Shi Y, Zhang L, Wang C, He X, Liu Y, Wan D, Huo K, et al. Molecular Cloning and Characterization of Human Aph2 Gene, Involved in Ap-1 Regulation by Interaction with Jab1. Biochim Biophys Acta. 2006;1759:514–525. doi: 10.1016/j.bbaexp.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 424.Zhou T, Li J, Zhao P, Liu H, Jia D, Jia H, He L, Cang Y, Boast S, Chen YH, et al. Palmitoyl Acyltransferase Aph2 in Cardiac Function and the Development of Cardiomyopathy. Proc Natl Acad Sci US A. 2015;112:15666–15671. doi: 10.1073/pnas.1518368112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 425.Li B, Cong F, Tan CP, Wang SX, Goff SP. Aph2, a Protein with a Zf-Dhhc Motif, Interacts with C-Abl and Has Pro-Apoptotic Activity. J Biol Chem. 2002;277:28870–28876. doi: 10.1074/jbc.M202388200. [DOI] [PubMed] [Google Scholar]
- 426.Singaraja RR, Hadano S, Metzler M, Givan S, Wellington CL, Warby S, Yanai A, Gutekunst CA, Leavitt BR, Yi H, et al. Hip14, a Novel Ankyrin Domain-Containing Protein, Links Huntingtin to Intracellular Trafficking and Endocytosis. Hum Mol Genet. 2002;11:2815–2828. doi: 10.1093/hmg/11.23.2815. [DOI] [PubMed] [Google Scholar]
- 427.Milnerwood AJ, Parsons MP, Young FB, Singaraja RR, Franciosi S, Volta M, Bergeron S, Hayden MR, Raymond LA. Memory and Synaptic Deficits in Hip14/Dhhc17 Knockout Mice. Proc Natl Acad Sci US A. 2013;110:20296–20301. doi: 10.1073/pnas.1222384110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 428.Ren W, Sun Y, Du K. Dhhc17 Palmitoylates Clipr-59 and Modulates Clipr-59 Association with the Plasma Membrane. Mol Cell Biol. 2013;33:4255–4265. doi: 10.1128/MCB.00527-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 429.Huang K, Yanai A, Kang R, Arstikaitis P, Singaraja RR, Metzler M, Mullard A, Haigh B, Gauthier-Campbell C, Gutekunst CA, et al. Huntingtin-Interacting Protein Hip14 Is a Palmitoyl Transferase Involved in Palmitoylation and Trafficking of Multiple Neuronal Proteins. Neuron. 2004;44:977–986. doi: 10.1016/j.neuron.2004.11.027. [DOI] [PubMed] [Google Scholar]
- 430.Yanai A, Huang K, Kang R, Singaraja RR, Arstikaitis P, Gan L, Orban PC, Mullard A, Cowan CM, Raymond LA, et al. Palmitoylation of Huntingtin by Hip14 Is Essential for Its Trafficking and Function. Nat Neurosci. 2006;9:824–831. doi: 10.1038/nn1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 431.Łach A, Grzybek M, Heger E, Korycka J, Wolny M, Kubiak J, Kolondra A, Bogusławska DM, Augoff K, Majkowski M, et al. Palmitoylation of Mpp1 (Membrane-Palmitoylated Protein 1)/P55 Is Crucial for Lateral Membrane Organization in Erythroid Cells. J Biol Chem. 2012;287:18974–18984. doi: 10.1074/jbc.M111.332981. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 432.Yang G, Zhou X, Zhu J, Liu R, Zhang S, Coquinco A, Chen Y, Wen Y, Kojic L, Jia W, et al. Jnk3 Couples the Neuronal Stress Response to Inhibition of Secretory Trafficking. Sci Signal. 2013;6:ra57. doi: 10.1126/scisignal.2003727. [DOI] [PubMed] [Google Scholar]
- 433.Skotte NH, Sanders SS, Singaraja RR, Ehrnhoefer DE, Vaid K, Qiu X, Kannan S, Verma C, Hayden MR. Palmitoylation of Caspase-6 by Hip14 Regulates Its Activation. Cell Death Differ. 2017;24:433–444. doi: 10.1038/cdd.2016.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 434.Yang G, Cynader MS. Palmitoyl Acyltransferase Zd17 Mediates Neuronal Responses in Acute Ischemic Brain Injury by Regulating Jnk Activation in a Signaling Module. J Neurosci. 2011;31:11980–11991. doi: 10.1523/JNEUROSCI.2510-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 435.Baumgart F, Corral-Escariz M, Pérez-Gil J, Rodríguez-Crespo I. Palmitoylation of R-Ras by Human Dhhc19: a Palmitoyl Transferase with a Caax Box. Biochim Biophys Acta. 2010;1798:592–604. doi: 10.1016/j.bbamem.2010.01.002. [DOI] [PubMed] [Google Scholar]
- 436.Runkle KB, Kharbanda A, Stypulkowski E, Cao XJ, Wang W, Garcia BA, Witze ES. Inhibition of Dhhc20-Mediated Egfr Palmitoylation Creates a Dependence on Egfr Signaling. Mol Cell. 2016;62:385–396. doi: 10.1016/j.molcel.2016.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 437.Draper JM, Smith CD. Dhhc20: A Human Palmitoyl Acyltransferase That Causes Cellular Transformation. Molec Membrane Biol. 2010;27:123–136. doi: 10.3109/09687681003616854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 438.Marin EP, Derakhshan B, Lam TT, Davalos A, Sessa WC. Endothelial Cell Palmitoylproteomic Identifies Novel Lipid-Modified Targets and Potential Substrates for Protein Acyl Transferases. Circ Res. 2012;110:1336–1344. doi: 10.1161/CIRCRESAHA.112.269514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 439.Mill P, Lee AWS, Fukata Y, Tsutsumi R, Fukata M, Keighren M, Porter RM, McKie L, Smyth I, Jackson IJ. Palmitoylation Regulates Epidermal Homeostasis and Hair Follicle Differentiation. PLoS Genet. 2009;5:e1000748. doi: 10.1371/journal.pgen.1000748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 440.Goytain A, Hines RM, Quamme GA. Huntingtin-Interacting Proteins, Hip14 and Hip14l, Mediate Dual Functions, Palmitoyl Acyltransferase and Mg2+ Transport. J Biol Chem. 2008;283:33365–33374. doi: 10.1074/jbc.M801469200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 441.Saleem AN, Chen YH, Baek HJ, Hsiao YW, Huang HW, Kao HJ, Liu KM, Shen LF, Song IW, Tu CPD, et al. Mice with Alopecia, Osteoporosis, and Systemic Amyloidosis Due to Mutation in Zdhhc13, a Gene Coding for Palmitoyl Acyltransferase. PLoS Genet. 2010;6:e1000985. doi: 10.1371/journal.pgen.1000985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 442.Song IW, Li WR, Chen LY, Shen LF, Liu KM, Yen JJY, Chen YJ, Chen YJ, Kraus VB, Wu JY, et al. Palmitoyl Acyltransferase, Zdhhc13, Facilitates Bone Mass Acquisition by Regulating Postnatal Epiphyseal Development and Endochondral Ossification: A Mouse Model. PLOS ONE. 2014;9:e92194. doi: 10.1371/journal.pone.0092194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 443.Huang K, Sanders SS, Kang R, Carroll JB, Sutton L, Wan J, Singaraja R, Young FB, Liu L, El-Husseini A, et al. Wild-Type Htt Modulates the Enzymatic Activity of the Neuronal Palmitoyl Transferase Hip14. Hum Mol Genet. 2011;20:3356–3365. doi: 10.1093/hmg/ddr242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 444.Liu KM, Chen YJ, Shen LF, Haddad AN, Song IW, Chen LY, Chen YJ, Wu JY, Yen JJ, Chen YT. Cyclic Alopecia and Abnormal Epidermal Cornification in Zdhhc13-Deficient Mice Reveal the Importance of Palmitoylation in Hair and Skin Differentiation. J Invest Dermatol. 2015;135:2603–2610. doi: 10.1038/jid.2015.240. [DOI] [PubMed] [Google Scholar]
- 445.Chen X, Shi W, Wang F, Du Z, Yang Y, Gao M, Yao Y, He K, Wang C, Hao A. Zinc Finger Dhhc-Type Containing 13 Regulates Fate Specification of Ectoderm and Mesoderm Cell Lineages by Modulating Smad6 Activity. Stem Cells Dev. 2014;23:1899–1909. doi: 10.1089/scd.2014.0068. [DOI] [PubMed] [Google Scholar]
- 446.Flisikowski K, Venhoranta H, Bauersachs S, Hanninen R, Furst RW, Saalfrank A, Ulbrich SE, Taponen J, Lohi H, Wolf E, et al. Truncation of Mimt1 Gene in the Peg3 Domain Leads to Major Changes in Placental Gene Expression and Stillbirth in Cattle. Biol Reprod. 2012;87:140. doi: 10.1095/biolreprod.112.104240. [DOI] [PubMed] [Google Scholar]
- 447.Roth AF, Wan J, Bailey AO, Sun B, Kuchar JA, Green WN, Phinney BS, Yates JR, Davis NG. Global Analysis of Protein Palmitoylation in Yeast. Cell. 2006;125:1003–1013. doi: 10.1016/j.cell.2006.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 448.Stellwagen D, Beattie EC, Seo JY, Malenka RC. Differential Regulation of Ampa Receptor and Gaba Receptor Trafficking by Tumor Necrosis Factor-A. J Neurosci. 2005;25:3219–3228. doi: 10.1523/JNEUROSCI.4486-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 449.Baumgart F, Corral-Escariz M, Perez-Gil J, Rodriguez-Crespo I. Palmitoylation of R-Ras by Human Dhhc19, a Palmitoyl Transferase with a Caax Box. Biochim Biophys Acta. 2010;1798:592–604. doi: 10.1016/j.bbamem.2010.01.002. [DOI] [PubMed] [Google Scholar]
- 450.Greaves J, Carmichael JA, Chamberlain LH. The Palmitoyl Transferase Dhhc2 Targets a Dynamic Membrane Cycling Pathway: Regulation by a C-Terminal Domain. Mol Biol Cell. 2011;22:1887–1895. doi: 10.1091/mbc.E10-11-0924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 451.Gorleku OA, Barns AM, Prescott GR, Greaves J, Chamberlain LH. Endoplasmic Reticulum Localization of Dhhc Palmitoyltransferases Mediated by Lysine-Based Sorting Signals. J Biol Chem. 2011;286:39573–39584. doi: 10.1074/jbc.M111.272369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 452.Kilpatrick CL, Murakami S, Feng M, Wu X, Lal R, Chen G, Du K, Luscher B. Dissociation of Golgi-Associated Dhhc-Type Zinc Finger Protein (Godz)- and Sertoli Cell Gene with a Zinc Finger Domain-Beta (Serz-Beta)-Mediated Palmitoylation by Loss of Function Analyses in Knock-out Mice. J Biol Chem. 2016;291:27371–27386. doi: 10.1074/jbc.M116.732768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 453.Liu P, Jiao B, Zhang R, Zhao H, Zhang C, Wu M, Li D, Zhao X, Qiu Q, Li J, et al. Palmitoylacyltransferase Zdhhc9 Inactivation Mitigates Leukemogenic Potential of Oncogenic Nras. Leukemia. 2016;30:1225–1228. doi: 10.1038/leu.2015.293. [DOI] [PubMed] [Google Scholar]
- 454.Keller CA, Yuan X, Panzanelli P, Martin ML, Alldred M, Sassoe-Pognetto M, Luscher B. The Gamma2 Subunit of Gaba(a) Receptors Is a Substrate for Palmitoylation by Godz. J Neurosci. 2004;24:5881–5891. doi: 10.1523/JNEUROSCI.1037-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 455.Lemonidis K, Sanchez-Perez MC, Chamberlain LH. Identification of a Novel Sequence Motif Recognized by the Ankyrin Repeat Domain of Zdhhc17/13 S-Acyltransferases. J Biol Chem. 2015;290:21939–21950. doi: 10.1074/jbc.M115.657668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 456.Verardi R, Kim JS, Ghirlando R, Banerjee A. Structural Basis for Substrate Recognition by the Ankyrin Repeat Domain of Human Dhhc17 Palmitoyltransferase. Structure. 2017 doi: 10.1016/j.str.2017.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 457.El-Husseini AE, Craven SE, Chetkovich DM, Firestein BL, Schnell E, Aoki C, Bredt DS. Dual Palmitoylation of Psd-95 Mediates Its Vesiculotubular Sorting, Postsynaptic Targeting, and Ion Channel Clustering. J Cell Biol. 2000;148:159–172. doi: 10.1083/jcb.148.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 458.Plain F, Congreve SD, Yee RSZ, Kennedy J, Howie J, Kuo CW, Fraser NJ, Fuller W. An Amphipathic Alpha-Helix Directs Palmitoylation of the Large Intracellular Loop of the Sodium/Calcium Exchanger. J Biol Chem. 2017;292:10745–10752. doi: 10.1074/jbc.M116.773945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 459.Hruska M, Henderson NT, Xia NL, Le Marchand SJ, Dalva MB. Anchoring and Synaptic Stability of Psd-95 Is Driven by Ephrin-B3. Nat Neurosci. 2015;18:1594–1605. doi: 10.1038/nn.4140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 460.Jennings BC, Linder ME. Dhhc Protein S-Acyltransferases Use Similar Ping-Pong Kinetic Mechanisms but Display Different Acyl-Coa Specificities. J Biol Chem. 2012;287:7236–7245. doi: 10.1074/jbc.M111.337246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 461.Greaves J, Munro KR, Davidson SC, Riviere M, Wojno J, Smith TK, Tomkinson NC, Chamberlain LH. Molecular Basis of Fatty Acid Selectivity in the Zdhhc Family of S-Acyltransferases Revealed by Click Chemistry. Proc Natl Acad Sci US A. 2017;114:E1365–E1374. doi: 10.1073/pnas.1612254114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 462.Casey WM, Gibson KJ, Parks LW. Covalent Attachment of Palmitoleic Acid (C16:1 Delta 9) to Proteins in Saccharomyces Cerevisiae. Evidence for a Third Class of Acylated Proteins. J Biol Chem. 1994;269:2082–2085. [PubMed] [Google Scholar]
- 463.Fujimoto T, Stroud E, Whatley RE, Prescott SM, Muszbek L, Laposata M, McEver RP. P-Selectin Is Acylated with Palmitic Acid and Stearic Acid at Cysteine 766 through a Thioester Linkage. J Biol Chem. 1993;268:11394–11400. [PubMed] [Google Scholar]
- 464.Hallak H, Muszbek L, Laposata M, Belmonte E, Brass LF, Manning DR. Covalent Binding of Arachidonate to G Protein Alpha Subunits of Human Platelets. J Biol Chem. 1994;269:4713–4716. [PubMed] [Google Scholar]
- 465.Muszbek L, Laposata M. Covalent Modification of Proteins by Arachidonate and Eicosapentaenoate in Platelets. J Biol Chem. 1993;268:18243–18248. [PubMed] [Google Scholar]
- 466.DeMar JC, Jr, Anderson RE. Identification and Quantitation of the Fatty Acids Composing the Coa Ester Pool of Bovine Retina, Heart and Liver. J Biol Chem. 1997;272:31362–31368. doi: 10.1074/jbc.272.50.31362. [DOI] [PubMed] [Google Scholar]
- 467.Bizzozero OA, McGarry JF, Lees MB. Acylation of Endogenous Myelin Proteolipid Protein with Different Acyl-Coas. J Biol Chem. 1987;262:2138–2145. [PubMed] [Google Scholar]
- 468.Zeng FY, Kaphalia BS, Ansari GA, Weigel PH. Fatty Acylation of the Rat Asialoglycoprotein Receptor. The Three Subunits from Active Receptors Contain Covalently Bound Palmitate and Stearate. J Biol Chem. 1995;270:21382–21387. doi: 10.1074/jbc.270.36.21382. [DOI] [PubMed] [Google Scholar]
- 469.Okubo K, Hamasaki N, Hara K, Kageura M. Palmitoylation of Cysteine 69 from the Cooh-Terminal of Band 3 Protein in the Human Erythrocyte Membrane. Acylation Occurs in the Middle of the Consensus Sequence of F--I-Iiclavl Found in Band 3 Protein and G2 Protein of Rift Valley Fever Virus. J Biol Chem. 1991;266:16420–16424. [PubMed] [Google Scholar]
- 470.Mitchell DA, Mitchell G, Ling Y, Budde C, Deschenes RJ. Mutational Analysis of Saccharomyces Cerevisiae Erf2 Reveals a Two-Step Reaction Mechanism for Protein Palmitoylation by Dhhc Enzymes. J Biol Chem. 2010;285:38104–38114. doi: 10.1074/jbc.M110.169102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 471.Gonzalez Montoro A, Chumpen Ramirez S, Valdez Taubas J. The Canonical Dhhc Motif Is Not Absolutely Required for the Activity of the Yeast S-Acyltransferases Swf1 and Pfa4. J Biol Chem. 2015;290:22448–22459. doi: 10.1074/jbc.M115.651356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 472.Abrami L, Dallavilla T, Sandoz PA, Demir M, Kunz B, Savoglidis G, Hatzimanikatis V, van der Goot FG. Identification and Dynamics of the Human Zdhhc16-Zdhhc6 Palmitoylation Cascade. Elife. 2017;6 doi: 10.7554/eLife.27826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 473.Lai J, Linder ME. Oligomerization of Dhhc Protein S-Acyltransferases. J Biol Chem. 2013;288:22862–22870. doi: 10.1074/jbc.M113.458794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 474.Lievens PM, Kuznetsova T, Kochlamazashvili G, Cesca F, Gorinski N, Galil DA, Cherkas V, Ronkina N, Lafera J, Gaestel M, et al. Zdhhc3 Tyrosine Phosphorylation Regulates Neural Cell Adhesion Molecule Palmitoylation. Mol Cell Biol. 2016;36:2208–2225. doi: 10.1128/MCB.00144-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 475.Gonzalez Montoro A, Quiroga R, Valdez Taubas J. Zinc Co-Ordination by the Dhhc Cysteine-Rich Domain of the Palmitoyltransferase Swf1. Biochem J. 2013;454:427–435. doi: 10.1042/BJ20121693. [DOI] [PubMed] [Google Scholar]
- 476.Gottlieb CD, Zhang S, Linder ME. The Cysteine-Rich Domain of the Dhhc3 Palmitoyltransferase Is Palmitoylated and Contains Tightly Bound Zinc. J Biol Chem. 2015;290:29259–29269. doi: 10.1074/jbc.M115.691147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 477.Sutton LM, Sanders SS, Butland SL, Singaraja RR, Franciosi S, Southwell AL, Doty CN, Schmidt ME, Mui KKN, Kovalik V, et al. Hip14l-Deficient Mice Develop Neuropathological and Behavioural Features of Huntington Disease. Hum Mol Genet. 2012;22:452–465. doi: 10.1093/hmg/dds441. [DOI] [PubMed] [Google Scholar]
- 478.Sanders SS, Hou J, Sutton LM, Garside VC, Mui KK, Singaraja RR, Hayden MR, Hoodless PA. Huntingtin Interacting Proteins 14 and 14-Like Are Required for Chorioallantoic Fusion During Early Placental Development. Dev Biol. 2015;397:257–266. doi: 10.1016/j.ydbio.2014.11.018. [DOI] [PubMed] [Google Scholar]
- 479.Raymond FL, Tarpey PS, Edkins S, Tofts C, O’Meara S, Teague J, Butler A, Stevens C, Barthorpe S, Buck G, et al. Mutations in Zdhhc9, Which Encodes a Palmitoyltransferase of Nras and Hras, Cause X-Linked Mental Retardation Associated with a Marfanoid Habitus. Am J Hum Genet. 2007;80:982–987. doi: 10.1086/513609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 480.Yan SM, Tang JJ, Huang CY, Xi SY, Huang MY, Liang JZ, Jiang YX, Li YH, Zhou ZW, Ernberg I, et al. Reduced Expression of Zdhhc2 Is Associated with Lymph Node Metastasis and Poor Prognosis in Gastric Adenocarcinoma. PLOS ONE. 2013;8:e56366. doi: 10.1371/journal.pone.0056366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 481.Planey SL, Keay SK, Zhang CO, Zacharias DA. Palmitoylation of Cytoskeleton Associated Protein 4 by Dhhc2 Regulates Antiproliferative Factor-Mediated Signaling. Mol Biol Cell. 2009;20:1454–1463. doi: 10.1091/mbc.E08-08-0849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 482.Oo HZ, Sentani K, Sakamoto N, Anami K, Naito Y, Uraoka N, Oshima T, Yanagihara K, Oue N, Yasui W. Overexpression of Zdhhc14 Promotes Migration and Invasion of Scirrhous Type Gastric Cancer. Oncol Rep. 2014;32:403–410. doi: 10.3892/or.2014.3166. [DOI] [PubMed] [Google Scholar]
- 483.Qanbar R, Bouvier M. Role of Palmitoylation/Depalmitoylation Reactions in G-Protein-Coupled Receptor Function. Pharmacol Ther. 2003;97:1–33. doi: 10.1016/s0163-7258(02)00300-5. [DOI] [PubMed] [Google Scholar]
- 484.Rasmussen SG, Choi HJ, Rosenbaum DM, Kobilka TS, Thian FS, Edwards PC, Burghammer M, Ratnala VR, Sanishvili R, Fischetti RF, et al. Crystal Structure of the Human Beta2 Adrenergic G-Protein-Coupled Receptor. Nature. 2007;450:383–387. doi: 10.1038/nature06325. [DOI] [PubMed] [Google Scholar]
- 485.Palczewski K, Kumasaka T, Hori T, Behnke CA, Motoshima H, Fox BA, Trong IL, Teller DC, Okada T, Stenkamp RE, et al. Crystal Structure of Rhodopsin: A G Protein-Coupled Receptor. Science. 2000;289:739–745. doi: 10.1126/science.289.5480.739. [DOI] [PubMed] [Google Scholar]
- 486.Tian H, Lu JY, Shao C, Huffman KE, Carstens RM, Larsen JE, Girard L, Liu H, Rodriguez-Canales J, Frenkel EP, et al. Systematic Sirna Screen Unmasks Nsclc Growth Dependence by Palmitoyltransferase Dhhc5. Mol Cancer Res. 2015;13:784–794. doi: 10.1158/1541-7786.MCR-14-0608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 487.Chen LY, Yang-Yen HF, Tsai CC, Thio CL, Chuang HL, Yang LT, Shen LF, Song IW, Liu KM, Huang YT, et al. Protein Palmitoylation by Zdhhc13 Protects Skin against Microbial-Driven Dermatitis. J Invest Dermatol. 2017;137:894–904. doi: 10.1016/j.jid.2016.12.011. [DOI] [PubMed] [Google Scholar]
- 488.Shen LF, Chen YJ, Liu KM, Haddad ANS, Song IW, Roan HY, Chen LY, Yen JJY, Chen YJ, Wu JY, et al. Role of S-Palmitoylation by Zdhhc13 in Mitochondrial Function and Metabolism in Liver. Sci Rep. 2017;7:2182. doi: 10.1038/s41598-017-02159-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 489.Cao N, Li JK, Rao YQ, Liu H, Wu J, Li B, Zhao P, Zeng L, Li J. A Potential Role for Protein Palmitoylation and Zdhhc16 in DNA Damage Response. BMC Mol Biol. 2016;17:12. doi: 10.1186/s12867-016-0065-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 490.Mukai J, Liu H, Burt RA, Swor DE, Lai WS, Karayiorgou M, Gogos JA. Evidence That the Gene Encoding Zdhhc8 Contributes to the Risk of Schizophrenia. Nat Genet. 2004;36:725–731. doi: 10.1038/ng1375. [DOI] [PubMed] [Google Scholar]
- 491.He M, Abdi KM, Bennett V. Ankyrin-G Palmitoylation and Betaii-Spectrin Binding to Phosphoinositide Lipids Drive Lateral Membrane Assembly. J Cell Biol. 2014;206:273–288. doi: 10.1083/jcb.201401016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 492.Wirgenes KV, Tesli M, Inderhaug E, Athanasiu L, Agartz I, Melle I, Hughes T, Andreassen OA, Djurovic S. Ank3 Gene Expression in Bipolar Disorder and Schizophrenia. Br J Psychiatry. 2014;205:244–245. doi: 10.1192/bjp.bp.114.145433. [DOI] [PubMed] [Google Scholar]
- 493.Moutin E, Nikonenko I, Stefanelli T, Wirth A, Ponimaskin E, De Roo M, Muller D. Palmitoylation of Cdc42 Promotes Spine Stabilization and Rescues Spine Density Deficit in a Mouse Model of 22q11.2 Deletion Syndrome. Cereb Cortex. 2016 doi: 10.1093/cercor/bhw183. [DOI] [PubMed] [Google Scholar]
- 494.Ota VK, Gadelha A, Assunção IB, Santoro ML, Christofolini DM, Bellucco FT, Santos-Filho AF, Ottoni GL, Lara DR, Mari JJ, et al. Zdhhc8 Gene May Play a Role in Cortical Volumes of Patients with Schizophrenia. Schizophr Res. 2013;145:33–35. doi: 10.1016/j.schres.2013.01.011. [DOI] [PubMed] [Google Scholar]
- 495.Xu M, St Clair D, He L. Testing for Genetic Association between the Zdhhc8 Gene Locus and Susceptibility to Schizophrenia: An Integrated Analysis of Multiple Datasets. Am J Med Genet. 2010;153B:1266–1275. doi: 10.1002/ajmg.b.31096. [DOI] [PubMed] [Google Scholar]
- 496.Veit M, Siche S. S-Acylation of Influenza Virus Proteins: Are Enzymes for Fatty Acid Attachment Promising Drug Targets? Vaccine. 2015;33:7002–7007. doi: 10.1016/j.vaccine.2015.08.095. [DOI] [PubMed] [Google Scholar]
- 497.Wang S, Mott KR, Wawrowsky K, Kousoulas KG, Luscher B, Ghiasi H. Binding of Hsv-1 Ul20 to Godz Affects Its Palmitoylation and Is Essential for Infectivity and Proper Targeting and Localization of Ul20 and Gk. J Virol. 2017 doi: 10.1128/JVI.00945-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 498.Lin DT, Conibear E. Abhd17 Proteins Are Novel Protein Depalmitoylases That Regulate N-Ras Palmitate Turnover and Subcellular Localization. Elife. 2015;4:e11306. doi: 10.7554/eLife.11306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 499.Reilly L, Howie J, Wypijewski K, Ashford ML, Hilgemann DW, Fuller W. Palmitoylation of the Na/Ca Exchanger Cytoplasmic Loop Controls Its Inactivation and Internalization During Stress Signaling. FASEB J. 2015;29:4532–4543. doi: 10.1096/fj.15-276493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 500.Yeh DC, Duncan JA, Yamashita S, Michel T. Depalmitoylation of Endothelial Nitric-Oxide Synthase by Acyl-Protein Thioesterase 1 Is Potentiated by Ca2+-Calmodulin. J Biol Chem. 1999;274:33148–33154. doi: 10.1074/jbc.274.46.33148. [DOI] [PubMed] [Google Scholar]
- 501.Duncan JA, Gilman AG. A Cytoplasmic Acyl-Protein Thioesterase That Removes Palmitate from G Protein A Subunits and P21ras. J Biol Chem. 1998;273:15830–15837. doi: 10.1074/jbc.273.25.15830. [DOI] [PubMed] [Google Scholar]
- 502.Dekker FJ, Rocks O, Vartak N, Menninger S, Hedberg C, Balamurugan R, Wetzel S, Renner S, Gerauer M, Scholermann B, et al. Small-Molecule Inhibition of Apt1 Affects Ras Localization and Signaling. Nat Chem Biol. 2010;6:449–456. doi: 10.1038/nchembio.362. [DOI] [PubMed] [Google Scholar]
- 503.Vujic I, Sanlorenzo M, Esteve-Puig R, Vujic M, Kwong A, Tsumura A, Murphy R, Moy A, Posch C, Monshi B, et al. Acyl Protein Thioesterase 1 and 2 (Apt-1, Apt-2) Inhibitors Palmostatin B, Ml348 and Ml349 Have Different Effects on Nras Mutant Melanoma Cells. Oncotarget. 2016;7:7297–7306. doi: 10.18632/oncotarget.6907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 504.Kong E, Peng S, Chandra G, Sarkar C, Zhang Z, Bagh MB, Mukherjee AB. Dynamic Palmitoylation Links Cytosol-Membrane Shuttling of Acyl-Protein Thioesterase-1 and Acyl-Protein Thioesterase-2 with That of Proto-Oncogene H-Ras Product and Growth-Associated Protein-43. J Biol Chem. 2013;288:9112–9125. doi: 10.1074/jbc.M112.421073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 505.Vartak N, Papke B, Grecco Hernan E, Rossmannek L, Waldmann H, Hedberg C, Bastiaens Philippe IH. The Autodepalmitoylating Activity of Apt Maintains the Spatial Organization of Palmitoylated Membrane Proteins. Biophys J. 2014;106:93–105. doi: 10.1016/j.bpj.2013.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 506.Hedberg C, Dekker FJ, Rusch M, Renner S, Wetzel S, Vartak N, Gerding-Reimers C, Bon RS, Bastiaens PIH, Waldmann H. Development of Highly Potent Inhibitors of the Ras-Targeting Human Acyl Protein Thioesterases Based on Substrate Similarity Design. Angew Chem Int Ed. 2011;50:9832–9837. doi: 10.1002/anie.201102965. [DOI] [PubMed] [Google Scholar]
- 507.Rusch M, Zimmermann TJ, Burger M, Dekker FJ, Gçrmer K, Triola G, Brockmeyer A, Janning P, Bçttcher T, Sieber SA, et al. Identification of Acyl Protein Thioesterases 1 and 2 as the Cellulartargets of the Ras-Signaling Modulators Palmostatin B and M. Angew Chem Int Ed. 2011;50:9838–9842. doi: 10.1002/anie.201102967. [DOI] [PubMed] [Google Scholar]
- 508.Adibekian A, Martin BR, Chang JW, Hsu KL, Tsuboi K, Bachovchin DA, Speers AE, Brown SJ, Spicer T, Fernandez-Vega V, et al. Probe Reports from the Nih Molecular Libraries Program. National Center for Biotechnology Information (US); Bethesda (MD): 2010. [PubMed] [Google Scholar]
- 509.Adibekian A, Martin BR, Chang JW, Hsu KL, Tsuboi K, Bachovchin DA, Speers AE, Brown SJ, Spicer T, Fernandez-Vega V, et al. Probe Reports from the Nih Molecular Libraries Program. National Center for Biotechnology Information (US); Bethesda (MD): 2010. [PubMed] [Google Scholar]
- 510.Hulce JJ, Joslyn C, Speers AE, Brown SJ, Spicer T, Fernandez-Vega V, Ferguson J, Cravatt BF, Hodder P, Rosen H. Probe Reports from the Nih Molecular Libraries Program. National Center for Biotechnology Information (US); Bethesda (MD): 2010. [Google Scholar]
- 511.Adibekian A, Martin BR, Chang JW, Hsu KL, Tsuboi K, Bachovchin DA, Speers AE, Brown SJ, Spicer T, Fernandez-Vega V, et al. Confirming Target Engagement for Reversible Inhibitors in Vivo by Kinetically Tuned Activity-Based Probes. J Am Chem Soc. 2012;134:10345–10348. doi: 10.1021/ja303400u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 512.Won SJ, Davda D, Labby KJ, Hwang SY, Pricer R, Majmudar JD, Armacost KA, Rodriguez LA, Rodriguez CL, Chong FS, et al. Molecular Mechanism for Isoform-Selective Inhibition of Acyl Protein Thioesterases 1 and 2 (Apt1 and Apt2) ACS Chem Biol. 2016;11:3374–3382. doi: 10.1021/acschembio.6b00720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 513.Hernandez JL, Davda D, Cheung See Kit M, Majmudar JD, Won SJ, Gang M, Pasupuleti SC, Choi AI, Bartkowiak CM, Martin BR. Apt2 Inhibition Restores Scribble Localization and S-Palmitoylation in Snail-Transformed Cells. Cell Chem Bio. 24:87–97. doi: 10.1016/j.chembiol.2016.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 514.Yokoi N, Fukata Y, Sekiya A, Murakami T, Kobayashi K, Fukata M. Identification of Psd-95 Depalmitoylating Enzymes. J Neurosci. 2016;36:6431–6444. doi: 10.1523/JNEUROSCI.0419-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 515.Webb Y, Hermida-Matsumoto L, Resh MD. Inhibition of Protein Palmitoylation, Raft Localization, and T Cell Signaling by 2-Bromopalmitate and Polyunsaturated Fatty Acids. J Biol Chem. 2000;275:261–270. doi: 10.1074/jbc.275.1.261. [DOI] [PubMed] [Google Scholar]
- 516.Coleman RA, Rao P, Fogelsong RJ, Bardes ES. 2-Bromopalmitoyl-Coa and 2-Bromopalmitate: Promiscuous Inhibitors of Membrane-Bound Enzymes. Biochim Biophys Acta. 1992;1125:203–209. doi: 10.1016/0005-2760(92)90046-x. [DOI] [PubMed] [Google Scholar]
- 517.Chenette EJ, Abo A, Der CJ. Critical and Distinct Roles of Amino- and Carboxyl-Terminal Sequences in Regulation of the Biological Activity of the Chp Atypical Rho Gtpase. J Biol Chem. 2005;280:13784–13792. doi: 10.1074/jbc.M411300200. [DOI] [PubMed] [Google Scholar]
- 518.Jochen AL, Hays J, Mick G. Inhibitory Effects of Cerulenin on Protein Palmitoylation and Insulin Internalization in Rat Adipocytes. Biochim Biophys Acta. 1995;1259:65–72. doi: 10.1016/0005-2760(95)00147-5. [DOI] [PubMed] [Google Scholar]
- 519.Hurley JH, Cahill AL, Currie KP, Fox AP. The Role of Dynamic Palmitoylation in Ca2+ Channel Inactivation. Proc Natl Acad Sci US A. 2000;97:9293–9298. doi: 10.1073/pnas.160589697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 520.Li L, Haynes MP, Bender JR. Plasma Membrane Localization and Function of the Estrogen Receptor Alpha Variant (Er46) in Human Endothelial Cells. Proc Natl Acad Sci US A. 2003;100:4807–4812. doi: 10.1073/pnas.0831079100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 521.Ducker CE, Griffel LK, Smith RA, Keller SN, Zhuang Y, Xia Z, Diller JD, Smith CD. Discovery and Characterization of Inhibitors of Human Palmitoyl Acyltransferases. Mol Cancer Ther. 2006;5:1647–1659. doi: 10.1158/1535-7163.MCT-06-0114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 522.Jennings BC, Nadolski MJ, Ling Y, Baker MB, Harrison ML, Deschenes RJ, Linder ME. 2-Bromopalmitate and 2-(2-Hydroxy-5-Nitro-Benzylidene)-Benzo[B]Thiophen-3-One Inhibit Dhhc-Mediated Palmitoylation in Vitro. J Lipid Res. 2009;50:233–242. doi: 10.1194/jlr.M800270-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 523.Davda D, El Azzouny MA, Tom CT, Hernandez JL, Majmudar JD, Kennedy RT, Martin BR. Profiling Targets of the Irreversible Palmitoylation Inhibitor 2-Bromopalmitate. ACS Chem Biol. 2013;8:1912–1917. doi: 10.1021/cb400380s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 524.Zheng B, DeRan M, Li X, Liao X, Fukata M, Wu X. 2-Bromopalmitate Analogues as Activity-Based Probes to Explore Palmitoyl Acyltransferases. J Am Chem Soc. 2013;135:7082–7085. doi: 10.1021/ja311416v. [DOI] [PubMed] [Google Scholar]
- 525.Tsutsumi K, Tomomura M, Furuichi T, Hisanaga S. Palmitoylation-Dependent Endosomal Localization of Aatyk1a and Its Interaction with Src. Genes Cells. 2008;13:949–964. doi: 10.1111/j.1365-2443.2008.01219.x. [DOI] [PubMed] [Google Scholar]
- 526.Vetrivel KS, Meckler X, Chen Y, Nguyen PD, Seidah NG, Vassar R, Wong PC, Fukata M, Kounnas MZ, Thinakaran G. Alzheimer Disease Abeta Production in the Absence of S-Palmitoylation-Dependent Targeting of Bace1 to Lipid Rafts. J Biol Chem. 2009;284:3793–3803. doi: 10.1074/jbc.M808920200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 527.Fragoso R, Ren D, Zhang X, Su MW, Burakoff SJ, Jin YJ. Lipid Raft Distribution of Cd4 Depends on Its Palmitoylation and Association with Lck, and Evidence for Cd4-Induced Lipid Raft Aggregation as an Additional Mechanism to Enhance Cd3 Signaling. J Immunol. 2003;170:913–921. doi: 10.4049/jimmunol.170.2.913. [DOI] [PubMed] [Google Scholar]
- 528.Tian N, Leshchyns’ka I, Welch JH, Diakowski W, Yang H, Schachner M, Sytnyk V. Lipid Raft-Dependent Endocytosis of Close Homolog of Adhesion Molecule L1 (Chl1) Promotes Neuritogenesis. J Biol Chem. 2012;287:44447–44463. doi: 10.1074/jbc.M112.394973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 529.Kalinina EV, Fricker LD. Palmitoylation of Carboxypeptidase D. Implications for Intracellular Trafficking. J Biol Chem. 2003;278:9244–9249. doi: 10.1074/jbc.m209379200. [DOI] [PubMed] [Google Scholar]
- 530.Coleman DT, Gray AL, Kridel SJ, Cardelli JA. Palmitoylation Regulates the Intracellular Trafficking and Stability of C-Met. Oncotarget. 2016;7:32664–32677. doi: 10.18632/oncotarget.8706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 531.Roberts BJ, Svoboda RA, Overmiller AM, Lewis JD, Kowalczyk AP, Mahoney MG, Johnson KR, Wahl JK., 3rd Palmitoylation of Desmoglein 2 Is a Regulator of Assembly Dynamics and Protein Turnover. J Biol Chem. 2016 doi: 10.1074/jbc.M116.739458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 532.Rossin A, Derouet M, Abdel-Sater F, Hueber AO. Palmitoylation of the Trail Receptor Dr4 Confers an Efficient Trail-Induced Cell Death Signalling. Biochem J. 2009;419:185–192. 182–192. doi: 10.1042/BJ20081212. [DOI] [PubMed] [Google Scholar]
- 533.Chakrabandhu K, Herincs Z, Huault S, Dost B, Peng L, Conchonaud F, Marguet D, He HT, Hueber AO. Palmitoylation Is Required for Efficient Fas Cell Death Signaling. EMBO J. 2007;26:209–220. doi: 10.1038/sj.emboj.7601456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 534.Barnes NC, Powell MS, Trist HM, Gavin AL, Wines BD, Hogarth PM. Raft Localisation of Fcgammariia and Efficient Signaling Are Dependent on Palmitoylation of Cysteine 208. Immunol Lett. 2006;104:118–123. doi: 10.1016/j.imlet.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 535.Claudinon J, Gonnord P, Beslard E, Marchetti M, Mitchell K, Boularan C, Johannes L, Eid P, Lamaze C. Palmitoylation of Interferon-Alpha (Ifn-Alpha) Receptor Subunit Ifnar1 Is Required for the Activation of Stat1 and Stat2 by Ifn-Alpha. J Biol Chem. 2009;284:24328–24340. doi: 10.1074/jbc.M109.021915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 536.Levental I, Lingwood D, Grzybek M, Coskun U, Simons K. Palmitoylation Regulates Raft Affinity for the Majority of Integral Raft Proteins. Proc Natl Acad Sci US A. 2010;107:22050–22054. doi: 10.1073/pnas.1016184107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 537.Abrami L, Kunz B, Iacovache I, van der Goot FG. Palmitoylation and Ubiquitination Regulate Exit of the Wnt Signaling Protein Lrp6 from the Endoplasmic Reticulum. Proc Natl Acad Sci US A. 2008;105:5384–5389. doi: 10.1073/pnas.0710389105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 538.Aguera-Gonzalez S, Gross CC, Fernandez-Messina L, Ashiru O, Esteso G, Hang HC, Reyburn HT, Long EO, Vales-Gomez M. Palmitoylation of Mica, a Ligand for Nkg2d, Mediates Its Recruitment to Membrane Microdomains and Promotes Its Shedding. Eur J Immunol. 2011;41:3667–3676. doi: 10.1002/eji.201141645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 539.Anilkumar N, Uekita T, Couchman JR, Nagase H, Seiki M, Itoh Y. Palmitoylation at Cys574 Is Essential for Mt1-Mmp to Promote Cell Migration. FASEB J. 2005;19:1326–1328. doi: 10.1096/fj.04-3651fje. [DOI] [PubMed] [Google Scholar]
- 540.Kinlough CL, McMahan RJ, Poland PA, Bruns JB, Harkleroad KL, Stremple RJ, Kashlan OB, Weixel KM, Weisz OA, Hughey RP. Recycling of Muc1 Is Dependent on Its Palmitoylation. J Biol Chem. 2006;281:12112–12122. doi: 10.1074/jbc.M512996200. [DOI] [PubMed] [Google Scholar]
- 541.Little EB, Edelman GM, Cunningham BA. Palmitoylation of the Cytoplasmic Domain of the Neural Cell Adhesion Molecule N-Cam Serves as an Anchor to Cellular Membranes. Cell Adhes Commun. 1998;6:415–430. doi: 10.3109/15419069809109150. [DOI] [PubMed] [Google Scholar]
- 542.Ren Q, Bennett V. Palmitoylation of Neurofascin at a Site in the Membrane-Spanning Domain Highly Conserved among the L1 Family of Cell Adhesion Molecules. J Neurochem. 1998;70:1839–1849. doi: 10.1046/j.1471-4159.1998.70051839.x. [DOI] [PubMed] [Google Scholar]
- 543.Cheng H, Vetrivel KS, Drisdel RC, Meckler X, Gong P, Leem JY, Li T, Carter M, Chen Y, Nguyen P, et al. S-Palmitoylation of Gamma-Secretase Subunits Nicastrin and Aph-1. J Biol Chem. 2009;284:1373–1384. doi: 10.1074/jbc.M806380200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 544.Bizzozero OA, Fridal K, Pastuszyn A. Identification of the Palmitoylation Site in Rat Myelin P0 Glycoprotein. J Neurochem. 1994;62:1163–1171. doi: 10.1046/j.1471-4159.1994.62031163.x. [DOI] [PubMed] [Google Scholar]
- 545.Underwood CK, Reid K, May LM, Bartlett PF, Coulson EJ. Palmitoylation of the C-Terminal Fragment of P75(Ntr) Regulates Death Signaling and Is Required for Subsequent Cleavage by Gamma-Secretase. Mol Cell Neurosci. 2008;37:346–358. doi: 10.1016/j.mcn.2007.10.005. [DOI] [PubMed] [Google Scholar]
- 546.Sardjono CT, Harbour SN, Yip JC, Paddock C, Tridandapani S, Newman PJ, Jackson DE. Palmitoylation at Cys595 Is Essential for Pecam-1 Localisation into Membrane Microdomains and for Efficient Pecam-1-Mediated Cytoprotection. Thromb Haemost. 2006;96:756–766. [PubMed] [Google Scholar]
- 547.Tulloch LB, Howie J, Wypijewski KJ, Wilson CR, Bernard WG, Shattock MJ, Fuller W. The Inhibitory Effect of Phospholemman on the Sodium Pump Requires Its Palmitoylation. J Biol Chem. 2011;286:36020–36031. doi: 10.1074/jbc.M111.282145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 548.Murphy J, Kolandaivelu S. Palmitoylation of Progressive Rod-Cone Degeneration (Prcd) Regulates Protein Stability and Localization. J Biol Chem. 2016;291:23036–23046. doi: 10.1074/jbc.M116.742767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 549.Flannery AR, Czibener C, Andrews NW. Palmitoylation-Dependent Association with Cd63 Targets the Ca2+ Sensor Synaptotagmin Vii to Lysosomes. J Cell Biol. 2010;191:599–613. doi: 10.1083/jcb.201003021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 550.Dorfleutner A, Ruf W. Regulation of Tissue Factor Cytoplasmic Domain Phosphorylation by Palmitoylation. Blood. 2003;102:3998–4005. doi: 10.1182/blood-2003-04-1149. [DOI] [PubMed] [Google Scholar]
- 551.Shum L, Turck CW, Derynck R. Cysteines 153 and 154 of Transmembrane Transforming Growth Factor-Alpha Are Palmitoylated and Mediate Cytoplasmic Protein Association. J Biol Chem. 1996;271:28502–28508. doi: 10.1074/jbc.271.45.28502. [DOI] [PubMed] [Google Scholar]
- 552.Lynes EM, Bui M, Yap MC, Benson MD, Schneider B, Ellgaard L, Berthiaume LG, Simmen T. Palmitoylated Tmx and Calnexin Target to the Mitochondria-Associated Membrane. EMBO J. 2012;31:457–470. doi: 10.1038/emboj.2011.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 553.Lynes EM, Raturi A, Shenkman M, Ortiz Sandoval C, Yap MC, Wu J, Janowicz A, Myhill N, Benson MD, Campbell RE, et al. Palmitoylation Is the Switch That Assigns Calnexin to Quality Control or Er Ca2+ Signaling. J Cell Sci. 2013;126:3893–3903. doi: 10.1242/jcs.125856. [DOI] [PubMed] [Google Scholar]
- 554.Schweizer A, Loffler BM, Rohrer J. Palmitoylation of the Three Isoforms of Human Endothelin-Converting Enzyme-1. Biochem J. 1999;340(Pt 3):649–656. [PMC free article] [PubMed] [Google Scholar]
- 555.Guardiola-Serrano F, Rossin A, Cahuzac N, Luckerath K, Melzer I, Mailfert S, Marguet D, Zornig M, Hueber AO. Palmitoylation of Human Fasl Modulates Its Cell Death-Inducing Function. Cell Death Dis. 2010;1:e88. doi: 10.1038/cddis.2010.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 556.Yount JS, Karssemeijer RA, Hang HC. S-Palmitoylation and Ubiquitination Differentially Regulate Interferon-Induced Transmembrane Protein 3 (Ifitm3)-Mediated Resistance to Influenza Virus. J Biol Chem. 2012;287:19631–19641. doi: 10.1074/jbc.M112.362095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 557.Wiedmer T, Zhao J, Nanjundan M, Sims PJ. Palmitoylation of Phospholipid Scramblase 1 Controls Its Distribution between Nucleus and Plasma Membrane. Biochemistry. 2003;42:1227–1233. doi: 10.1021/bi026679w. [DOI] [PubMed] [Google Scholar]
- 558.ten Brinke A, Batenburg JJ, Gadella BM, Haagsman HP, Vaandrager AB, van Golde LM. The Juxtamembrane Lysine and Arginine Residues of Surfactant Protein C Precursor Influence Palmitoylation Via Effects on Trafficking. Am J Respir Cell Mol Biol. 2001;25:156–163. doi: 10.1165/ajrcmb.25.2.4423. [DOI] [PubMed] [Google Scholar]
- 559.He Y, Linder ME. Differential Palmitoylation of the Endosomal Snares Syntaxin 7 and Syntaxin 8. J Lipid Res. 2009;50:398–404. doi: 10.1194/jlr.M800360-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 560.Poggi M, Kara I, Brunel JM, Landrier JF, Govers R, Bonardo B, Fluhrer R, Haass C, Alessi MC, Peiretti F. Palmitoylation of Tnf Alpha Is Involved in the Regulation of Tnf Receptor 1 Signalling. Biochim Biophys Acta. 2013;1833:602–612. doi: 10.1016/j.bbamcr.2012.11.009. [DOI] [PubMed] [Google Scholar]
- 561.Amici SA, McKay SB, Wells GB, Robson JI, Nasir M, Ponath G, Anand R. A Highly Conserved Cytoplasmic Cysteine Residue in the Alpha4 Nicotinic Acetylcholine Receptor Is Palmitoylated and Regulates Protein Expression. J Biol Chem. 2012;287:23119–23127. doi: 10.1074/jbc.M111.328294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 562.Zuckerman DM, Hicks SW, Charron G, Hang HC, Machamer CE. Differential Regulation of Two Palmitoylation Sites in the Cytoplasmic Tail of the Beta1-Adrenergic Receptor. J Biol Chem. 2011;286:19014–19023. doi: 10.1074/jbc.M110.189977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 563.Liu R, Wang D, Shi Q, Fu Q, Hizon S, Xiang YK. Palmitoylation Regulates Intracellular Trafficking of Beta2 Adrenergic Receptor/Arrestin/Phosphodiesterase 4d Complexes in Cardiomyocytes. PLOS ONE. 2012;7:e42658. doi: 10.1371/journal.pone.0042658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 564.Ponimaskin E, Dumuis A, Gaven F, Barthet G, Heine M, Glebov K, Richter DW, Oppermann M. Palmitoylation of the 5-Hydroxytryptamine4a Receptor Regulates Receptor Phosphorylation, Desensitization, and Beta-Arrestin-Mediated Endocytosis. Mol Pharmacol. 2005;67:1434–1443. doi: 10.1124/mol.104.008748. [DOI] [PubMed] [Google Scholar]
- 565.Kvachnina E, Dumuis A, Wlodarczyk J, Renner U, Cochet M, Richter DW, Ponimaskin E. Constitutive Gs-Mediated, but Not G12-Mediated, Activity of the 5-Hydroxytryptamine 5-Ht7(a) Receptor Is Modulated by the Palmitoylation of Its C-Terminal Domain. Biochim Biophys Acta. 2009;1793:1646–1655. doi: 10.1016/j.bbamcr.2009.08.008. [DOI] [PubMed] [Google Scholar]
- 566.Gu HM, Li G, Gao X, Berthiaume LG, Zhang DW. Characterization of Palmitoylation of Atp Binding Cassette Transporter G1: Effect on Protein Trafficking and Function. Biochim Biophys Acta. 2013;1831:1067–1078. doi: 10.1016/j.bbalip.2013.01.019. [DOI] [PubMed] [Google Scholar]
- 567.Steinke KV, Gorinski N, Wojciechowski D, Todorov V, Guseva D, Ponimaskin E, Fahlke C, Fischer M. Human Clc-K Channels Require Palmitoylation of Their Accessory Subunit Barttin to Be Functional. J Biol Chem. 2015;290:17390–17400. doi: 10.1074/jbc.M114.631705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 568.Pizard A, Blaukat A, Michineau S, Dikic I, Muller-Esterl W, Alhenc-Gelas F, Rajerison RM. Palmitoylation of the Human Bradykinin B2 Receptor Influences Ligand Efficacy. Biochemistry. 2001;40:15743–15751. doi: 10.1021/bi011600t. [DOI] [PubMed] [Google Scholar]
- 569.Jeffries O, Geiger N, Rowe IC, Tian L, McClafferty H, Chen L, Bi D, Knaus HG, Ruth P, Shipston MJ. Palmitoylation of the S0–S1 Linker Regulates Cell Surface Expression of Voltage- and Calcium-Activated Potassium (Bk) Channels. J Biol Chem. 2010;285:33307–33314. doi: 10.1074/jbc.M110.153940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 570.Karnik SS, Ridge KD, Bhattacharya S, Khorana HG. Palmitoylation of Bovine Opsin and Its Cysteine Mutants in Cos Cells. Proc Natl Acad Sci US A. 1993;90:40–44. doi: 10.1073/pnas.90.1.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 571.Oddi S, Dainese E, Sandiford S, Fezza F, Lanuti M, Chiurchiu V, Totaro A, Catanzaro G, Barcaroli D, De Laurenzi V, et al. Effects of Palmitoylation of Cys(415) in Helix 8 of the Cb(1) Cannabinoid Receptor on Membrane Localization and Signalling. Br J Pharmacol. 2012;165:2635–2651. doi: 10.1111/j.1476-5381.2011.01658.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 572.Percherancier Y, Planchenault T, Valenzuela-Fernandez A, Virelizier JL, Arenzana-Seisdedos F, Bachelerie F. Palmitoylation-Dependent Control of Degradation, Life Span, and Membrane Expression of the Ccr5 Receptor. J Biol Chem. 2001;276:31936–31944. doi: 10.1074/jbc.M104013200. [DOI] [PubMed] [Google Scholar]
- 573.Thorne RF, Ralston KJ, de Bock CE, Mhaidat NM, Zhang XD, Boyd AW, Burns GF. Palmitoylation of Cd36/Fat Regulates the Rate of Its Post-Transcriptional Processing in the Endoplasmic Reticulum. Biochim Biophys Acta. 2010;1803:1298–1307. doi: 10.1016/j.bbamcr.2010.07.002. [DOI] [PubMed] [Google Scholar]
- 574.Koziak K, Kaczmarek E, Kittel A, Sevigny J, Blusztajn JK, Schulte Am Esch J, 2nd, Imai M, Guckelberger O, Goepfert C, Qawi I, et al. Palmitoylation Targets Cd39/Endothelial Atp Diphosphohydrolase to Caveolae. J Biol Chem. 2000;275:2057–2062. doi: 10.1074/jbc.275.3.2057. [DOI] [PubMed] [Google Scholar]
- 575.Zhu YZ, Luo Y, Cao MM, Liu Y, Liu XQ, Wang W, Wu DG, Guan M, Xu QQ, Ren H, et al. Significance of Palmitoylation of Cd81 on Its Association with Tetraspanin-Enriched Microdomains and Mediating Hepatitis C Virus Cell Entry. Virology. 2012;429:112–123. doi: 10.1016/j.virol.2012.03.002. [DOI] [PubMed] [Google Scholar]
- 576.Zhou B, Liu L, Reddivari M, Zhang XA. The Palmitoylation of Metastasis Suppressor Kai1/Cd82 Is Important for Its Motility- and Invasiveness-Inhibitory Activity. Cancer Res. 2004;64:7455–7463. doi: 10.1158/0008-5472.CAN-04-1574. [DOI] [PubMed] [Google Scholar]
- 577.Yang X, Claas C, Kraeft SK, Chen LB, Wang Z, Kreidberg JA, Hemler ME. Palmitoylation of Tetraspanin Proteins: Modulation of Cd151 Lateral Interactions, Subcellular Distribution, and Integrin-Dependent Cell Morphology. Mol Biol Cell. 2002;13:767–781. doi: 10.1091/mbc.01-05-0275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 578.Van Itallie CM, Gambling TM, Carson JL, Anderson JM. Palmitoylation of Claudins Is Required for Efficient Tight-Junction Localization. J Cell Sci. 2005;118:1427–1436. doi: 10.1242/jcs.01735. [DOI] [PubMed] [Google Scholar]
- 579.Foster JD, Vaughan RA. Palmitoylation Controls Dopamine Transporter Kinetics, Degradation, and Protein Kinase C-Dependent Regulation. J Biol Chem. 2011;286:5175–5186. doi: 10.1074/jbc.M110.187872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 580.Kong MM, Verma V, O’Dowd BF, George SR. The Role of Palmitoylation in Directing Dopamine D1 Receptor Internalization through Selective Endocytic Routes. Biochem Biophys Res Commun. 2011;405:445–449. doi: 10.1016/j.bbrc.2011.01.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 581.Zhang X, Kim KM. Palmitoylation of the Carboxyl-Terminal Tail of Dopamine D4 Receptor Is Required for Surface Expression, Endocytosis, and Signaling. Biochem Biophys Res Commun. 2016;479:398–403. doi: 10.1016/j.bbrc.2016.09.094. [DOI] [PubMed] [Google Scholar]
- 582.Moritz AE, Rastedt DE, Stanislowski DJ, Shetty M, Smith MA, Vaughan RA, Foster JD. Reciprocal Phosphorylation and Palmitoylation Control Dopamine Transporter Kinetics. J Biol Chem. 2015;290:29095–29105. doi: 10.1074/jbc.M115.667055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 583.Mueller GM, Maarouf AB, Kinlough CL, Sheng N, Kashlan OB, Okumura S, Luthy S, Kleyman TR, Hughey RP. Cys Palmitoylation of the Beta Subunit Modulates Gating of the Epithelial Sodium Channel. J Biol Chem. 2010;285:30453–30462. doi: 10.1074/jbc.M110.151845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 584.Horstmeyer A, Cramer H, Sauer T, Muller-Esterl W, Schroeder C. Palmitoylation of Endothelin Receptor A. Differential Modulation of Signal Transduction Activity by Post-Translational Modification. J Biol Chem. 1996;271:20811–20819. doi: 10.1074/jbc.271.34.20811. [DOI] [PubMed] [Google Scholar]
- 585.Okamoto Y, Ninomiya H, Tanioka M, Sakamoto A, Miwa S, Masaki T. Palmitoylation of Human Endothelinb. Its Critical Role in G Protein Coupling and a Differential Requirement for the Cytoplasmic Tail by G Protein Subtypes. J Biol Chem. 1997;272:21589–21596. doi: 10.1074/jbc.272.34.21589. [DOI] [PubMed] [Google Scholar]
- 586.Huang K, Kang MH, Askew C, Kang R, Sanders SS, Wan J, Davis NG, Hayden MR. Palmitoylation and Function of Glial Glutamate Transporter-1 Is Reduced in the Yac128 Mouse Model of Huntington Disease. Neurobiol Dis. 2010;40:207–215. doi: 10.1016/j.nbd.2010.05.027. [DOI] [PubMed] [Google Scholar]
- 587.Copits BA, Swanson GT. Kainate Receptor Post-Translational Modifications Differentially Regulate Association with 4.1n to Control Activity-Dependent Receptor Endocytosis. J Biol Chem. 2013;288:8952–8965. doi: 10.1074/jbc.M112.440719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 588.Yang G, Xiong W, Kojic L, Cynader MS. Subunit-Selective Palmitoylation Regulates the Intracellular Trafficking of Ampa Receptor. Eur J Neurosci. 2009;30:35–46. doi: 10.1111/j.1460-9568.2009.06788.x. [DOI] [PubMed] [Google Scholar]
- 589.Pickering DS, Taverna FA, Salter MW, Hampson DR. Palmitoylation of the Glur6 Kainate Receptor. Proc Natl Acad Sci US A. 1995;92:12090–12094. doi: 10.1073/pnas.92.26.12090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 590.Fukushima Y, Saitoh T, Anai M, Ogihara T, Inukai K, Funaki M, Sakoda H, Onishi Y, Ono H, Fujishiro M, et al. Palmitoylation of the Canine Histamine H2 Receptor Occurs at Cys(305) and Is Important for Cell Surface Targeting. Biochim Biophys Acta. 2001;1539:181–191. doi: 10.1016/s0167-4889(01)00104-5. [DOI] [PubMed] [Google Scholar]
- 591.Reid HM, Mulvaney EP, Turner EC, Kinsella BT. Interaction of the Human Prostacyclin Receptor with Rab11: Characterization of a Novel Rab11 Binding Domain within Alpha-Helix 8 That Is Regulated by Palmitoylation. J Biol Chem. 2010;285:18709–18726. doi: 10.1074/jbc.M110.106476. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 592.Botham A, Guo X, Xiao YP, Morice AH, Compton SJ, Sadofsky LR. Palmitoylation of Human Proteinase-Activated Receptor-2 Differentially Regulates Receptor-Triggered Erk1/2 Activation, Calcium Signalling and Endocytosis. Biochem J. 2011;438:359–367. doi: 10.1042/BJ20101958. [DOI] [PubMed] [Google Scholar]
- 593.Hach JC, McMichael T, Chesarino NM, Yount JS. Palmitoylation on Conserved and Nonconserved Cysteines of Murine Ifitm1 Regulates Its Stability and Anti-Influenza a Virus Activity. J Virol. 2013;87:9923–9927. doi: 10.1128/JVI.00621-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 594.Munshi UM, Peegel H, Menon KM. Palmitoylation of the Luteinizing Hormone/Human Chorionic Gonadotropin Receptor Regulates Receptor Interaction with the Arrestin-Mediated Internalization Pathway. Eur J Biochem. 2001;268:1631–1639. doi: 10.1046/j.1432-1327.2001.02032.x. [DOI] [PubMed] [Google Scholar]
- 595.Higuchi M, Izumi KM, Kieff E. Epstein-Barr Virus Latent-Infection Membrane Proteins Are Palmitoylated and Raft-Associated: Protein 1 Binds to the Cytoskeleton through Tnf Receptor Cytoplasmic Factors. Proc Natl Acad Sci US A. 2001;98:4675–4680. doi: 10.1073/pnas.081075298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 596.Hayashi MK, Haga T. Palmitoylation of Muscarinic Acetylcholine Receptor M2 Subtypes: Reduction in Their Ability to Activate G Proteins by Mutation of a Putative Palmitoylation Site, Cysteine 457, in the Carboxyl-Terminal Tail. Arch Biochem Biophys. 1997;340:376–382. doi: 10.1006/abbi.1997.9906. [DOI] [PubMed] [Google Scholar]
- 597.Mattison HA, Hayashi T, Barria A. Palmitoylation at Two Cysteine Clusters on the C-Terminus of Glun2a and Glun2b Differentially Control Synaptic Targeting of Nmda Receptors. PLOS ONE. 2012;7:e49089. doi: 10.1371/journal.pone.0049089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 598.Heakal Y, Woll MP, Fox T, Seaton K, Levenson R, Kester M. Neurotensin Receptor-1 Inducible Palmitoylation Is Required for Efficient Receptor-Mediated Mitogenic-Signaling within Structured Membrane Microdomains. Cancer Biol Ther. 2011;12:427–435. doi: 10.4161/cbt.12.5.15984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 599.Gubitosi-Klug RA, Mancuso DJ, Gross RW. The Human Kv1.1 Channel Is Palmitoylated, Modulating Voltage Sensing: Identification of a Palmitoylation Consensus Sequence. Proc Natl Acad Sci US A. 2005;102:5964–5968. doi: 10.1073/pnas.0501999102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 600.Gonnord P, Delarasse C, Auger R, Benihoud K, Prigent M, Cuif MH, Lamaze C, Kanellopoulos JM. Palmitoylation of the P2×7 Receptor, an Atp-Gated Channel, Controls Its Expression and Association with Lipid Rafts. FASEB J. 2009;23:795–805. doi: 10.1096/fj.08-114637. [DOI] [PubMed] [Google Scholar]
- 601.Adams MN, Christensen ME, He Y, Waterhouse NJ, Hooper JD. The Role of Palmitoylation in Signalling, Cellular Trafficking and Plasma Membrane Localization of Protease-Activated Receptor-2. PLOS ONE. 2011;6:e28018. doi: 10.1371/journal.pone.0028018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 602.Zoltewicz SJ, Lee S, Chittoor VG, Freeland SM, Rangaraju S, Zacharias DA, Notterpek L. The Palmitoylation State of Pmp22 Modulates Epithelial Cell Morphology and Migration. ASN Neuro. 2012;4:409–421. doi: 10.1042/AN20120045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 603.Jindal HK, Folco EJ, Liu GX, Koren G. Posttranslational Modification of Voltage-Dependent Potassium Channel Kv1.5: Cooh-Terminal Palmitoylation Modulates Its Biological Properties. Am J Physiol Heart Circ Physiol. 2008;294:H2012–2021. doi: 10.1152/ajpheart.01374.2007. [DOI] [PubMed] [Google Scholar]
- 604.Miggin SM, Lawler OA, Kinsella BT. Palmitoylation of the Human Prostacyclin Receptor. Functional Implications of Palmitoylation and Isoprenylation. J Biol Chem. 2003;278:6947–6958. doi: 10.1074/jbc.M210637200. [DOI] [PubMed] [Google Scholar]
- 605.Maeda A, Okano K, Park PS, Lem J, Crouch RK, Maeda T, Palczewski K. Palmitoylation Stabilizes Unliganded Rod Opsin. Proc Natl Acad Sci US A. 2010;107:8428–8433. doi: 10.1073/pnas.1000640107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 606.Ohno Y, Ito A, Ogata R, Hiraga Y, Igarashi Y, Kihara A. Palmitoylation of the Sphingosine 1-Phosphate Receptor S1p Is Involved in Its Signaling Functions and Internalization. Genes Cells. 2009;14:911–923. doi: 10.1111/j.1365-2443.2009.01319.x. [DOI] [PubMed] [Google Scholar]
- 607.Pei Z, Xiao Y, Meng J, Hudmon A, Cummins TR. Cardiac Sodium Channel Palmitoylation Regulates Channel Availability and Myocyte Excitability with Implications for Arrhythmia Generation. Nat Commun. 2016;7:12035. doi: 10.1038/ncomms12035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 608.Tani M, Kuge O. Sphingomyelin Synthase 2 Is Palmitoylated at the Cooh-Terminal Tail, Which Is Involved in Its Localization in Plasma Membranes. Biochem Biophys Res Commun. 2009;381:328–332. doi: 10.1016/j.bbrc.2009.02.063. [DOI] [PubMed] [Google Scholar]
- 609.Mukai K, Konno H, Akiba T, Uemura T, Waguri S, Kobayashi T, Barber GN, Arai H, Taguchi T. Activation of Sting Requires Palmitoylation at the Golgi. Nat Commun. 2016;7:11932. doi: 10.1038/ncomms11932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 610.Zhou X, Wulfsen I, Korth M, McClafferty H, Lukowski R, Shipston MJ, Ruth P, Dobrev D, Wieland T. Palmitoylation and Membrane Association of the Stress Axis Regulated Insert (Strex) Controls Bk Channel Regulation by Protein Kinase C. J Biol Chem. 2012;287:32161–32171. doi: 10.1074/jbc.M112.386359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 611.Kaur I, Yarov-Yarovoy V, Kirk LM, Plambeck KE, Barragan EV, Ontiveros ES, Diaz E. Activity-Dependent Palmitoylation Controls Syndig1 Stability, Localization, and Function. J Neurosci. 2016;36:7562–7568. doi: 10.1523/JNEUROSCI.4859-14.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 612.Reid HM, Kinsella BT. Palmitoylation of the Tpbeta Isoform of the Human Thromboxane A2 Receptor. Modulation of G Protein: Effector Coupling and Modes of Receptor Internalization. Cell Signal. 2007;19:1056–1070. doi: 10.1016/j.cellsig.2006.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 613.Du D, Raaka BM, Grimberg H, Lupu-Meiri M, Oron Y, Gershengorn MC. Carboxyl Tail Cysteine Mutants of the Thyrotropin-Releasing Hormone Receptor Type 1 Exhibit Constitutive Signaling: Role of Palmitoylation. Mol Pharmacol. 2005;68:204–209. doi: 10.1124/mol.105.012641. [DOI] [PubMed] [Google Scholar]
- 614.Hawtin SR, Tobin AB, Patel S, Wheatley M. Palmitoylation of the Vasopressin V1a Receptor Reveals Different Conformational Requirements for Signaling, Agonist-Induced Receptor Phosphorylation, and Sequestration. J Biol Chem. 2001;276:38139–38146. doi: 10.1074/jbc.M106142200. [DOI] [PubMed] [Google Scholar]
- 615.Charest PG, Bouvier M. Palmitoylation of the V2 Vasopressin Receptor Carboxyl Tail Enhances Beta-Arrestin Recruitment Leading to Efficient Receptor Endocytosis and Erk1/2 Activation. J Biol Chem. 2003;278:41541–41551. doi: 10.1074/jbc.M306589200. [DOI] [PubMed] [Google Scholar]
- 616.Holliday ND, Cox HM. Control of Signalling Efficacy by Palmitoylation of the Rat Y1 Receptor. Br J Pharmacol. 2003;139:501–512. doi: 10.1038/sj.bjp.0705276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 617.Sato I, Obata Y, Kasahara K, Nakayama Y, Fukumoto Y, Yamasaki T, Yokoyama KK, Saito T, Yamaguchi N. Differential Trafficking of Src, Lyn, Yes and Fyn Is Specified by the State of Palmitoylation in the Sh4 Domain. J Cell Sci. 2009;122:965–975. doi: 10.1242/jcs.034843. [DOI] [PubMed] [Google Scholar]
- 618.Blank M, Wollberg J, Gerlach F, Reimann K, Roesner A, Hankeln T, Fago A, Weber RE, Burmester T. A Membrane-Bound Vertebrate Globin. PLOS ONE. 2011;6:e25292. doi: 10.1371/journal.pone.0025292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 619.Yurchak LK, Sefton BM. Palmitoylation of Either Cys-3 or Cys-5 Is Required for the Biological Activity of the Lck Tyrosine Protein Kinase. Mol Cell Biol. 1995;15:6914–6922. doi: 10.1128/mcb.15.12.6914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 620.Linder ME, Middleton P, Hepler JR, Taussig R, Gilman AG, Mumby SM. Lipid Modifications of G Proteins: Alpha Subunits Are Palmitoylated. Proc Natl Acad Sci US A. 1993;90:3675–3679. doi: 10.1073/pnas.90.8.3675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 621.Yang H, Wan L, Song F, Wang M, Huang Y. Palmitoylation Modification of Galpha(O) Depresses Its Susceptibility to Gap-43 Activation. Int J Biochem Cell Biol. 2009;41:1495–1501. doi: 10.1016/j.biocel.2008.12.011. [DOI] [PubMed] [Google Scholar]
- 622.Tucker DE, Stewart A, Nallan L, Bendale P, Ghomashchi F, Gelb MH, Leslie CC. Group Ivc Cytosolic Phospholipase A2gamma Is Farnesylated and Palmitoylated in Mammalian Cells. J Lipid Res. 2005;46:2122–2133. doi: 10.1194/jlr.M500230-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 623.Navarro-Lerida I, Sanchez-Perales S, Calvo M, Rentero C, Zheng Y, Enrich C, Del Pozo MA. A Palmitoylation Switch Mechanism Regulates Rac1 Function and Membrane Organization. EMBO J. 2012;31:534–551. doi: 10.1038/emboj.2011.446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 624.Uechi Y, Bayarjargal M, Umikawa M, Oshiro M, Takei K, Yamashiro Y, Asato T, Endo S, Misaki R, Taguchi T, et al. Rap2 Function Requires Palmitoylation and Recycling Endosome Localization. Biochem Biophys Res Commun. 2009;378:732–737. doi: 10.1016/j.bbrc.2008.11.107. [DOI] [PubMed] [Google Scholar]
- 625.Wang DA, Sebti SM. Palmitoylated Cysteine 192 Is Required for Rhob Tumor-Suppressive and Apoptotic Activities. J Biol Chem. 2005;280:19243–19249. doi: 10.1074/jbc.M411472200. [DOI] [PubMed] [Google Scholar]
- 626.Berzat AC, Buss JE, Chenette EJ, Weinbaum CA, Shutes A, Der CJ, Minden A, Cox AD. Transforming Activity of the Rho Family Gtpase, Wrch-1, a Wnt-Regulated Cdc42 Homolog, Is Dependent on a Novel Carboxyl-Terminal Palmitoylation Motif. J Biol Chem. 2005;280:33055–33065. doi: 10.1074/jbc.M507362200. [DOI] [PubMed] [Google Scholar]
- 627.Izawa I, Nishizawa M, Hayashi Y, Inagaki M. Palmitoylation of Erbin Is Required for Its Plasma Membrane Localization. Genes Cells. 2008;13:691–701. doi: 10.1111/j.1365-2443.2008.01198.x. [DOI] [PubMed] [Google Scholar]
- 628.Jang D, Kwon H, Jeong K, Lee J, Pak Y. Essential Role of Flotillin-1 Palmitoylation in the Intracellular Localization and Signaling Function of Igf-1 Receptor. J Cell Sci. 2015;128:2179–2190. doi: 10.1242/jcs.169409. [DOI] [PubMed] [Google Scholar]
- 629.Pedone KH, Hepler JR. The Importance of N-Terminal Polycysteine and Polybasic Sequences for G14alpha and G16alpha Palmitoylation, Plasma Membrane Localization, and Signaling Function. J Biol Chem. 2007;282:25199–25212. doi: 10.1074/jbc.M610297200. [DOI] [PubMed] [Google Scholar]
- 630.Skene JH, Virag I. Posttranslational Membrane Attachment and Dynamic Fatty Acylation of a Neuronal Growth Cone Protein, Gap-43. J Cell Biol. 1989;108:613–624. doi: 10.1083/jcb.108.2.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 631.Wedegaertner PB, Chu DH, Wilson PT, Levis MJ, Bourne HR. Palmitoylation Is Required for Signaling Functions and Membrane Attachment of Gq Alpha and Gs Alpha. J Biol Chem. 1993;268:25001–25008. [PubMed] [Google Scholar]
- 632.Bhattacharyya R, Wedegaertner PB. Galpha 13 Requires Palmitoylation for Plasma Membrane Localization, Rho-Dependent Signaling, and Promotion of P115-Rhogef Membrane Binding. J Biol Chem. 2000;275:14992–14999. doi: 10.1074/jbc.M000415200. [DOI] [PubMed] [Google Scholar]
- 633.George J, Soares C, Montersino A, Beique JC, Thomas GM. Palmitoylation of Lim Kinase-1 Ensures Spine-Specific Actin Polymerization and Morphological Plasticity. Elife. 2015;4:e06327. doi: 10.7554/eLife.06327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 634.Chien AJ, Gao T, Perez-Reyes E, Hosey MM. Membrane Targeting of L-Type Calcium Channels. Role of Palmitoylation in the Subcellular Localization of the Beta2a Subunit. J Biol Chem. 1998;273:23590–23597. doi: 10.1074/jbc.273.36.23590. [DOI] [PubMed] [Google Scholar]
- 635.Wu XS, Martina JA, Hammer JA., 3rd Melanoregulin Is Stably Targeted to the Melanosome Membrane by Palmitoylation. Biochem Biophys Res Commun. 2012;426:209–214. doi: 10.1016/j.bbrc.2012.08.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 636.Navarro-Lerida I, Corvi MM, Barrientos AA, Gavilanes F, Berthiaume LG, Rodriguez-Crespo I. Palmitoylation of Inducible Nitric-Oxide Synthase at Cys-3 Is Required for Proper Intracellular Traffic and Nitric Oxide Synthesis. J Biol Chem. 2004;279:55682–55689. doi: 10.1074/jbc.M406621200. [DOI] [PubMed] [Google Scholar]
- 637.Aittaleb M, Nishimura A, Linder ME, Tesmer JJ. Plasma Membrane Association of P63 Rho Guanine Nucleotide Exchange Factor (P63rhogef) Is Mediated by Palmitoylation and Is Required for Basal Activity in Cells. J Biol Chem. 2011;286:34448–34456. doi: 10.1074/jbc.M111.273342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 638.Charych EI, Jiang LX, Lo F, Sullivan K, Brandon NJ. Interplay of Palmitoylation and Phosphorylation in the Trafficking and Localization of Phosphodiesterase 10a: Implications for the Treatment of Schizophrenia. J Neurosci. 2010;30:9027–9037. doi: 10.1523/JNEUROSCI.1635-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 639.Zhang X, Liu X, Wang D, Liu H, Hao W. Conditioned Place Preference Associated with Level of Palmitoylation of Psd-95 in Rat Hippocampus and Nucleus Accumbens. Neuropsychobiology. 2011;64:211–218. doi: 10.1159/000327603. [DOI] [PubMed] [Google Scholar]
- 640.Hiol A, Davey PC, Osterhout JL, Waheed AA, Fischer ER, Chen CK, Milligan G, Druey KM, Jones TL. Palmitoylation Regulates Regulators of G-Protein Signaling (Rgs) 16 Function. I. Mutation of Amino-Terminal Cysteine Residues on Rgs16 Prevents Its Targeting to Lipid Rafts and Palmitoylation of an Internal Cysteine Residue. J Biol Chem. 2003;278:19301–19308. doi: 10.1074/jbc.M210123200. [DOI] [PubMed] [Google Scholar]
- 641.Yang X, Guo Z, Sun F, Li W, Alfano A, Shimelis H, Chen M, Brodie AM, Chen H, Xiao Z, et al. Novel Membrane-Associated Androgen Receptor Splice Variant Potentiates Proliferative and Survival Responses in Prostate Cancer Cells. J Biol Chem. 2011;286:36152–36160. doi: 10.1074/jbc.M111.265124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 642.Stoffel RH, Inglese J, Macrae AD, Lefkowitz RJ, Premont RT. Palmitoylation Increases the Kinase Activity of the G Protein-Coupled Receptor Kinase, Grk6. Biochemistry. 1998;37:16053–16059. doi: 10.1021/bi981432d. [DOI] [PubMed] [Google Scholar]
- 643.Drenan RM, Doupnik CA, Boyle MP, Muglia LJ, Huettner JE, Linder ME, Blumer KJ. Palmitoylation Regulates Plasma Membrane-Nuclear Shuttling of R7bp, a Novel Membrane Anchor for the Rgs7 Family. J Cell Biol. 2005;169:623–633. doi: 10.1083/jcb.200502007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 644.Keith DJ, Sanderson JL, Gibson ES, Woolfrey KM, Robertson HR, Olszewski K, Kang R, El-Husseini A, Dell’acqua ML. Palmitoylation of a-Kinase Anchoring Protein 79/150 Regulates Dendritic Endosomal Targeting and Synaptic Plasticity Mechanisms. J Neurosci. 2012;32:7119–7136. doi: 10.1523/JNEUROSCI.0784-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 645.He M, Jenkins P, Bennett V. Cysteine 70 of Ankyrin-G Is S-Palmitoylated and Is Required for Function of Ankyrin-G in Membrane Domain Assembly. J Biol Chem. 2012;287:43995–44005. doi: 10.1074/jbc.M112.417501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 646.Kummel D, Walter J, Heck M, Heinemann U, Veit M. Characterization of the Self-Palmitoylation Activity of the Transport Protein Particle Component Bet3. Cell Mol Life Sci. 2010;67:2653–2664. doi: 10.1007/s00018-010-0358-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 647.Cenedella RJ, Neely AR, Sexton P. Multiple Forms of 22 Kda Caveolin-1 Alpha Present in Bovine Lens Cells Could Reflect Variable Palmitoylation. Exp Eye Res. 2006;82:229–235. doi: 10.1016/j.exer.2005.06.016. [DOI] [PubMed] [Google Scholar]
- 648.Kim KS, Kim JS, Park JY, Suh YH, Jou I, Joe EH, Park SM. Dj-1 Associates with Lipid Rafts by Palmitoylation and Regulates Lipid Rafts-Dependent Endocytosis in Astrocytes. Hum Mol Genet. 2013;22:4805–4817. doi: 10.1093/hmg/ddt332. [DOI] [PubMed] [Google Scholar]
- 649.La Rosa P, Pesiri V, Leclercq G, Marino M, Acconcia F. Palmitoylation Regulates 17beta-Estradiol-Induced Estrogen Receptor-Alpha Degradation and Transcriptional Activity. Mol Endocrinol. 2012;26:762–774. doi: 10.1210/me.2011-1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 650.Morrow IC, Rea S, Martin S, Prior IA, Prohaska R, Hancock JF, James DE, Parton RG. Flotillin-1/Reggie-2 Traffics to Surface Raft Domains Via a Novel Golgi-Independent Pathway. Identification of a Novel Membrane Targeting Domain and a Role for Palmitoylation. J Biol Chem. 2002;277:48834–48841. doi: 10.1074/jbc.M209082200. [DOI] [PubMed] [Google Scholar]
- 651.Rush DB, Leon RT, McCollum MH, Treu RW, Wei J. Palmitoylation and Trafficking of Gad65 Are Impaired in a Cellular Model of Huntington’s Disease. Biochem J. 2012;442:39–48. doi: 10.1042/BJ20110679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 652.Kostiuk MA, Keller BO, Berthiaume LG. Palmitoylation of Ketogenic Enzyme Hmgcs2 Enhances Its Interaction with Pparalpha and Transcription at the Hmgcs2 Ppre. FASEB J. 2010;24:1914–1924. doi: 10.1096/fj.09-149765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 653.Takimoto K, Yang EK, Conforti L. Palmitoylation of Kchip Splicing Variants Is Required for Efficient Cell Surface Expression of Kv4.3 Channels. J Biol Chem. 2002;277:26904–26911. doi: 10.1074/jbc.M203651200. [DOI] [PubMed] [Google Scholar]
- 654.Holland SM, Collura KM, Ketschek A, Noma K, Ferguson TA, Jin Y, Gallo G, Thomas GM. Palmitoylation Controls Dlk Localization, Interactions and Activity to Ensure Effective Axonal Injury Signaling. Proc Natl Acad Sci US A. 2016;113:763–768. doi: 10.1073/pnas.1514123113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 655.Shmueli A, Segal M, Sapir T, Tsutsumi R, Noritake J, Bar A, Sapoznik S, Fukata Y, Orr I, Fukata M, et al. Ndel1 Palmitoylation: A New Mean to Regulate Cytoplasmic Dynein Activity. EMBO J. 2010;29:107–119. doi: 10.1038/emboj.2009.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 656.Tani M, Hannun YA. Neutral Sphingomyelinase 2 Is Palmitoylated on Multiple Cysteine Residues. Role of Palmitoylation in Subcellular Localization. J Biol Chem. 2007;282:10047–10056. doi: 10.1074/jbc.M611249200. [DOI] [PubMed] [Google Scholar]
- 657.Greaves J, Chamberlain LH. Differential Palmitoylation Regulates Intracellular Patterning of Snap25. J Cell Sci. 2011;124:1351–1360. doi: 10.1242/jcs.079095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 658.Snyers L, Umlauf E, Prohaska R. Cysteine 29 Is the Major Palmitoylation Site on Stomatin. FEBS Lett. 1999;449:101–104. doi: 10.1016/s0014-5793(99)00417-2. [DOI] [PubMed] [Google Scholar]
- 659.Chan P, Han X, Zheng B, DeRan M, Yu J, Jarugumilli GK, Deng H, Pan D, Luo X, Wu X. Autopalmitoylation of Tead Proteins Regulates Transcriptional Output of the Hippo Pathway. Nat Chem Biol. 2016;12:282–289. doi: 10.1038/nchembio.2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 660.Zhao Z, Hou J, Xie Z, Deng J, Wang X, Chen D, Yang F, Gong W. Acyl-Biotinyl Exchange Chemistry and Mass Spectrometry-Based Analysis of Palmitoylation Sites of in Vitro Palmitoylated Rat Brain Tubulin. Protein J. 2010;29:531–537. doi: 10.1007/s10930-010-9285-x. [DOI] [PubMed] [Google Scholar]
- 661.Ande SR, Mishra S. Palmitoylation of Prohibitin at Cysteine 69 Facilitates Its Membrane Translocation and Interaction with Eps 15 Homology Domain Protein 2 (Ehd2) Biochem Cell Biol. 2010;88:553–558. doi: 10.1139/o09-177. [DOI] [PubMed] [Google Scholar]
- 662.Han JM, Kim Y, Lee JS, Lee CS, Lee BD, Ohba M, Kuroki T, Suh PG, Ryu SH. Localization of Phospholipase D1 to Caveolin-Enriched Membrane Via Palmitoylation: Implications for Epidermal Growth Factor Signaling. Mol Biol Cell. 2002;13:3976–3988. doi: 10.1091/mbc.E02-02-0100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 663.Barylko B, Mao YS, Wlodarski P, Jung G, Binns DD, Sun HQ, Yin HL, Albanesi JP. Palmitoylation Controls the Catalytic Activity and Subcellular Distribution of Phosphatidylinositol 4-Kinase Ii{Alpha} J Biol Chem. 2009;284:9994–10003. doi: 10.1074/jbc.M900724200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 664.Ni J, Qu L, Yang H, Wang M, Huang Y. Palmitoylation and Its Effect on the Gtpase-Activating Activity and Conformation of Rgs2. Int J Biochem Cell Biol. 2006;38:2209–2218. doi: 10.1016/j.biocel.2006.06.015. [DOI] [PubMed] [Google Scholar]
- 665.Takida S, Fischer CC, Wedegaertner PB. Palmitoylation and Plasma Membrane Targeting of Rgs7 Are Promoted by Alpha O. Mol Pharmacol. 2005;67:132–139. doi: 10.1124/mol.104.003418. [DOI] [PubMed] [Google Scholar]
- 666.Castro-Fernandez C, Janovick JA, Brothers SP, Fisher RA, Ji TH, Conn PM. Regulation of Rgs3 and Rgs10 Palmitoylation by Gnrh. Endocrinology. 2002;143:1310–1317. doi: 10.1210/endo.143.4.8713. [DOI] [PubMed] [Google Scholar]
- 667.Takahashi Y, Moiseyev G, Chen Y, Ma JX. The Roles of Three Palmitoylation Sites of Rpe65 in Its Membrane Association and Isomerohydrolase Activity. Invest Ophthalmol Vis Sci. 2006;47:5191–5196. doi: 10.1167/iovs.06-0614. [DOI] [PubMed] [Google Scholar]
- 668.Zhao Y, McCabe JB, Vance J, Berthiaume LG. Palmitoylation of Apolipoprotein B Is Required for Proper Intracellular Sorting and Transport of Cholesteroyl Esters and Triglycerides. Mol Biol Cell. 2000;11:721–734. doi: 10.1091/mbc.11.2.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 669.Wei X, Yang Z, Rey FE, Ridaura VK, Davidson NO, Gordon JI, Semenkovich CF. Fatty Acid Synthase Modulates Intestinal Barrier Function through Palmitoylation of Mucin 2. Cell Host Microbe. 2012;11:140–152. doi: 10.1016/j.chom.2011.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 670.Barbacid M. Ras Genes. Annu Rev Biochem. 1987;56:779–827. doi: 10.1146/annurev.bi.56.070187.004023. [DOI] [PubMed] [Google Scholar]
- 671.Chardin P, Camonis JH, Gale NW, van Aelst L, Schlessinger J, Wigler MH, Bar-Sagi D. Human Sos1: A Guanine Nucleotide Exchange Factor for Ras That Binds to Grb2. Science. 1993;260:1338–1343. doi: 10.1126/science.8493579. [DOI] [PubMed] [Google Scholar]
- 672.Mittal R, Ahmadian MR, Goody RS, Wittinghofer A. Formation of a Transition-State Analog of the Ras Gtpase Reaction by Ras-Gdp, Tetrafluoroaluminate, and Gtpase-Activating Proteins. Science. 1996;273:115–117. doi: 10.1126/science.273.5271.115. [DOI] [PubMed] [Google Scholar]
- 673.Ahearn IM, Haigis K, Bar-Sagi D, Philips MR. Regulating the Regulator: Post-Translational Modification of Ras. Nat Rev Mol Cell Biol. 2012;13:39–51. doi: 10.1038/nrm3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 674.Casey PJ, Solski PA, Der CJ, Buss JE. P21ras Is Modified by a Farnesyl Isoprenoid. Proc Natl Acad Sci US A. 1989;86:8323–8327. doi: 10.1073/pnas.86.21.8323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 675.Rocks O, Peyker A, Kahms M, Verveer PJ, Koerner C, Lumbierres M, Kuhlmann J, Waldmann H, Wittinghofer A, Bastiaens PI. An Acylation Cycle Regulates Localization and Activity of Palmitoylated Ras Isoforms. Science. 2005;307:1746–1752. doi: 10.1126/science.1105654. [DOI] [PubMed] [Google Scholar]
- 676.Roy S, Plowman S, Rotblat B, Prior IA, Muncke C, Grainger S, Parton RG, Henis YI, Kloog Y, Hancock JF. Individual Palmitoyl Residues Serve Distinct Roles in H-Ras Trafficking, Microlocalization, and Signaling. Mol Cell Biol. 2005;25:6722–6733. doi: 10.1128/MCB.25.15.6722-6733.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 677.Rocks O, Gerauer M, Vartak N, Koch S, Huang ZP, Pechlivanis M, Kuhlmann J, Brunsveld L, Chandra A, Ellinger B, et al. The Palmitoylation Machinery Is a Spatially Organizing System for Peripheral Membrane Proteins. Cell. 2010;141:458–471. doi: 10.1016/j.cell.2010.04.007. [DOI] [PubMed] [Google Scholar]
- 678.Heasman SJ, Ridley AJ. Mammalian Rho Gtpases: New Insights into Their Functions from in Vivo Studies. Nat Rev Mol Cell Biol. 2008;9:690–701. doi: 10.1038/nrm2476. [DOI] [PubMed] [Google Scholar]
- 679.Jaffe AB, Hall A. Rho Gtpases: Biochemistry and Biology. Annu Rev Cell Dev Biol. 2005;21:247–269. doi: 10.1146/annurev.cellbio.21.020604.150721. [DOI] [PubMed] [Google Scholar]
- 680.Marks PW, Kwiatkowski DJ. Genomic Organization and Chromosomal Location of Murine Cdc42. Genomics. 1996;38:13–18. doi: 10.1006/geno.1996.0586. [DOI] [PubMed] [Google Scholar]
- 681.Kang R, Wan J, Arstikaitis P, Takahashi H, Huang K, Bailey AO, Thompson JX, Roth AF, Drisdel RC, Mastro R, et al. Neural Palmitoyl-Proteomics Reveals Dynamic Synaptic Palmitoylation. Nature. 2008;456:904–909. doi: 10.1038/nature07605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 682.Coe JG, Lim AC, Xu J, Hong W. A Role for Tlg1p in the Transport of Proteins within the Golgi Apparatus of Saccharomyces Cerevisiae. Mol Biol Cell. 1999;10:2407–2423. doi: 10.1091/mbc.10.7.2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 683.Siniossoglou S, Pelham HR. An Effector of Ypt6p Binds the Snare Tlg1p and Mediates Selective Fusion of Vesicles with Late Golgi Membranes. EMBO J. 2001;20:5991–5998. doi: 10.1093/emboj/20.21.5991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 684.Valdez-Taubas J, Pelham H. Swf1-Dependent Palmitoylation of the Snare Tlg1 Prevents Its Ubiquitination and Degradation. EMBO J. 2005;24:2524–2532. doi: 10.1038/sj.emboj.7600724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 685.Reggiori F, Pelham HR. A Transmembrane Ubiquitin Ligase Required to Sort Membrane Proteins into Multivesicular Bodies. Nat Cell Biol. 2002;4:117–123. doi: 10.1038/ncb743. [DOI] [PubMed] [Google Scholar]
- 686.Koduri V, Blacklow SC. Requirement for Natively Unstructured Regions of Mesoderm Development Candidate 2 in Promoting Low-Density Lipoprotein Receptor-Related Protein 6 Maturation. Biochemistry. 2007;46:6570–6577. doi: 10.1021/bi700049g. [DOI] [PubMed] [Google Scholar]
- 687.Kikuchi A, Yamamoto H, Kishida S. Multiplicity of the Interactions of Wnt Proteins and Their Receptors. Cell Signal. 2007;19:659–671. doi: 10.1016/j.cellsig.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 688.Lam KK, Davey M, Sun B, Roth AF, Davis NG, Conibear E. Palmitoylation by the Dhhc Protein Pfa4 Regulates the Er Exit of Chs3. J Cell Biol. 2006;174:19–25. doi: 10.1083/jcb.200602049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 689.Bhattacharyya R, Barren C, Kovacs DM. Palmitoylation of Amyloid Precursor Protein Regulates Amyloidogenic Processing in Lipid Rafts. J Neurosci. 2013;33:11169–11183. doi: 10.1523/JNEUROSCI.4704-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 690.Landles C, Bates GP. Huntingtin and the Molecular Pathogenesis of Huntington’s Disease. Fourth in Molecular Medicine Review Series. EMBO Rep. 2004;5:958–963. doi: 10.1038/sj.embor.7400250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 691.Huang K, Yanai A, Kang R, Arstikaitis P, Singaraja RR, Metzler M, Mullard A, Haigh B, Gauthier-Campbell C, Gutekunst CA, et al. Huntingtin-Interacting Protein Hip14 Is a Palmitoyl Transferase Involved in Palmitoylation and Trafficking of Multiple Neuronal Proteins. Neuron. 2004;44:977–986. doi: 10.1016/j.neuron.2004.11.027. [DOI] [PubMed] [Google Scholar]
- 692.Schlesinger MJ, Magee AI, Schmidt MF. Fatty Acid Acylation of Proteins in Cultured Cells. J Biol Chem. 1980;255:10021–10024. [PubMed] [Google Scholar]
- 693.Peseckis SM, Deichaite I, Resh MD. Iodinated Fatty Acids as Probes for Myristate Processing and Function. Incorporation into Pp60v-Src. J Biol Chem. 1993;268:5107–5114. [PubMed] [Google Scholar]
- 694.Zou C, Ellis BM, Smith RM, Chen BB, Zhao Y, Mallampalli RK. Acyl-Coa:Lysophosphatidylcholine Acyltransferase I (Lpcat1) Catalyzes Histone Protein O-Palmitoylation to Regulate Mrna Synthesis. J Biol Chem. 2011;286:28019–28025. doi: 10.1074/jbc.M111.253385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 695.Wang Q, Chan TR, Hilgraf R, Fokin VV, Sharpless KB, Finn MG. Bioconjugation by Copper(I)-Catalyzed Azide-Alkyne [3 + 2] Cycloaddition. J Am Chem Soc. 2003;125:3192–3193. doi: 10.1021/ja021381e. [DOI] [PubMed] [Google Scholar]
- 696.Hang HC, Geutjes EJ, Grotenbreg G, Pollington AM, Bijlmakers MJ, Ploegh HL. Chemical Probes for the Rapid Detection of Fatty-Acylated Proteins in Mammalian Cells. J Am Chem Soc. 2007;129:2744–2745. doi: 10.1021/ja0685001. [DOI] [PubMed] [Google Scholar]
- 697.Heal WP, Wickramasinghe SR, Leatherbarrow RJ, Tate EW. N-Myristoyl Transferase-Mediated Protein Labelling in Vivo. Org Biomol Chem. 2008;6:2308–2315. doi: 10.1039/b803258k. [DOI] [PubMed] [Google Scholar]
- 698.Speers AE, Cravatt BF. Profiling Enzyme Activities in Vivo Using Click Chemistry Methods. Chem Biol. 2004;11:535–546. doi: 10.1016/j.chembiol.2004.03.012. [DOI] [PubMed] [Google Scholar]
- 699.Wilson JP, Raghavan AS, Yang Y-Y, Charron G, Hang HC. Proteomic Analysis of Fatty-Acylated Proteins in Mammalian Cells with Chemical Reporters Reveals S-Acylation of Histone H3 Variants. Mol Cell Proteomics. 2011;10:M110.001198. doi: 10.1074/mcp.M110.001198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 700.Drisdel RC, Green WN. Labeling and Quantifying Sites of Protein Palmitoylation. Biotechniques. 2004;36:276–285. doi: 10.2144/04362RR02. [DOI] [PubMed] [Google Scholar]
- 701.Wan J, Roth AF, Bailey AO, Davis NG. Palmitoylated Proteins: Purification and Identification. Nat Protoc. 2007;2:1573–1584. doi: 10.1038/nprot.2007.225. [DOI] [PubMed] [Google Scholar]
- 702.Martin BR, Wang C, Adibekian A, Tully SE, Cravatt BF. Global Profiling of Dynamic Protein Palmitoylation. Nat Methods. 2011;9:84–89. doi: 10.1038/nmeth.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 703.Percher A, Ramakrishnan S, Thinon E, Yuan X, Yount JS, Hang HC. Mass-Tag Labeling Reveals Site-Specific and Endogenous Levels of Protein S-Fatty Acylation. Proc Natl Acad Sci U S A. 2016;113:4302–4307. doi: 10.1073/pnas.1602244113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 704.Gao X, Hannoush RN. Single-Cell Imaging of Wnt Palmitoylation by the Acyltransferase Porcupine. Nat Chem Biol. 2014;10:61–68. doi: 10.1038/nchembio.1392. [DOI] [PubMed] [Google Scholar]
- 705.DeKroon RM, Robinette JB, Osorio C, Jeong JS, Hamlett E, Mocanu M, Alzate O. Analysis of Protein Posttranslational Modifications Using Dige-Based Proteomics. Methods Mol Biol. 2012;854:129–143. doi: 10.1007/978-1-61779-573-2_9. [DOI] [PubMed] [Google Scholar]
- 706.Borland LM, Allbritton NL. Use of Micellar Electrokinetic Chromatography to Measure Palmitoylation of a Peptide. J Chromatogr B Analyt Technol Biomed Life Sci. 2008;875:451–458. doi: 10.1016/j.jchromb.2008.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 707.McClure M, DeLucas LJ, Wilson L, Ray M, Rowe SM, Wu X, Dai Q, Hong JS, Sorscher EJ, Kappes JC, et al. Purification of Cftr for Mass Spectrometry Analysis: Identification of Palmitoylation and Other Post-Translational Modifications. Protein Eng Des Sel. 2012;25:7–14. doi: 10.1093/protein/gzr054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 708.Sorek N, Yalovsky S. Analysis of Protein S-Acylation by Gas Chromatography-Coupled Mass Spectrometry Using Purified Proteins. Nat Protoc. 2010;5:834–840. doi: 10.1038/nprot.2010.33. [DOI] [PubMed] [Google Scholar]
- 709.Zhou F, Xue Y, Yao X, Xu Y. Css-Palm: Palmitoylation Site Prediction with a Clustering and Scoring Strategy (Css) Bioinformatics. 2006;22:894–896. doi: 10.1093/bioinformatics/btl013. [DOI] [PubMed] [Google Scholar]
- 710.Xue Y, Chen H, Jin C, Sun Z, Yao X. Nba-Palm: Prediction of Palmitoylation Site Implemented in Naive Bayes Algorithm. BMC Bioinform. 2006;7:458. doi: 10.1186/1471-2105-7-458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 711.Ren J, Wen L, Gao X, Jin C, Xue Y, Yao X. Css-Palm 2.0: An Updated Software for Palmitoylation Sites Prediction. Protein Eng Des Sel. 2008;21:639–644. doi: 10.1093/protein/gzn039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 712.Wang XB, Wu LY, Wang YC, Deng NY. Prediction of Palmitoylation Sites Using the Composition of K-Spaced Amino Acid Pairs. Protein Eng Des Sel. 2009;22:707–712. doi: 10.1093/protein/gzp055. [DOI] [PubMed] [Google Scholar]
- 713.Willert K, Nusse R. Wnt Proteins. Cold Spring Harb Perspect Biol. 2012;4:a007864. doi: 10.1101/cshperspect.a007864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 714.Clevers H, Nusse R. Wnt/Beta-Catenin Signaling and Disease. Cell. 2012;149:1192–1205. doi: 10.1016/j.cell.2012.05.012. [DOI] [PubMed] [Google Scholar]
- 715.Willert K, Brown JD, Danenberg E, Duncan AW, Weissman IL, Reya T, Yates JR, 3rd, Nusse R. Wnt Proteins Are Lipid-Modified and Can Act as Stem Cell Growth Factors. Nature. 2003;423:448–452. doi: 10.1038/nature01611. [DOI] [PubMed] [Google Scholar]
- 716.Takada R, Satomi Y, Kurata T, Ueno N, Norioka S, Kondoh H, Takao T, Takada S. Monounsaturated Fatty Acid Modification of Wnt Protein: Its Role in Wnt Secretion. Dev Cell. 2006;11:791–801. doi: 10.1016/j.devcel.2006.10.003. [DOI] [PubMed] [Google Scholar]
- 717.Janda CY, Waghray D, Levin AM, Thomas C, Garcia KC. Structural Basis of Wnt Recognition by Frizzled. Science. 2012;337:59–64. doi: 10.1126/science.1222879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 718.Hofmann K. A Superfamily of Membrane-Bound O-Acyltransferases with Implications for Wnt Signaling. Trends Biochem Sci. 2000;25:111–112. doi: 10.1016/s0968-0004(99)01539-x. [DOI] [PubMed] [Google Scholar]
- 719.Barrott JJ, Cash GM, Smith AP, Barrow JR, Murtaugh LC. Deletion of Mouse Porcn Blocks Wnt Ligand Secretion and Reveals an Ectodermal Etiology of Human Focal Dermal Hypoplasia/Goltz Syndrome. Proc Natl Acad Sci US A. 2011;108:12752–12757. doi: 10.1073/pnas.1006437108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 720.Wang X, Reid Sutton V, Omar Peraza-Llanes J, Yu Z, Rosetta R, Kou YC, Eble TN, Patel A, Thaller C, Fang P, et al. Mutations in X-Linked Porcn, a Putative Regulator of Wnt Signaling, Cause Focal Dermal Hypoplasia. Nat Genet. 2007;39:836–838. doi: 10.1038/ng2057. [DOI] [PubMed] [Google Scholar]
- 721.Proffitt KD, Virshup DM. Precise Regulation of Porcupine Activity Is Required for Physiological Wnt Signaling. J Biol Chem. 2012;287:34167–34178. doi: 10.1074/jbc.M112.381970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 722.Rios-Esteves J, Haugen B, Resh MD. Identification of Key Residues and Regions Important for Porcupine-Mediated Wnt Acylation. J Biol Chem. 2014 doi: 10.1074/jbc.M114.561209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 723.Bregman H, Williams DS, Atilla GE, Carroll PJ, Meggers E. An Organometallic Inhibitor for Glycogen Synthase Kinase 3. J Am Chem Soc. 2004;126:13594–13595. doi: 10.1021/ja046049c. [DOI] [PubMed] [Google Scholar]
- 724.Chen B, Dodge ME, Tang W, Lu J, Ma Z, Fan CW, Wei S, Hao W, Kilgore J, Williams NS, et al. Small Molecule-Mediated Disruption of Wnt-Dependent Signaling in Tissue Regeneration and Cancer. Nat Chem Biol. 2009;5:100–107. doi: 10.1038/nchembio.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 725.Dodge ME, Moon J, Tuladhar R, Lu J, Jacob LS, Zhang L-s, Shi H, Wang X, Moro E, Mongera A, et al. Diverse Chemical Scaffolds Support Direct Inhibition of the Membrane-Bound O-Acyltransferase Porcupine. J Biol Chem. 2012;287:23246–23254. doi: 10.1074/jbc.M112.372029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 726.Wang X, Moon J, Dodge ME, Pan X, Zhang L, Hanson JM, Tuladhar R, Ma Z, Shi H, Williams NS, et al. The Development of Highly Potent Inhibitors for Porcupine. J Med Chem. 2013;56:2700–2704. doi: 10.1021/jm400159c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 727.Liu J, Pan S, Hsieh MH, Ng N, Sun F, Wang T, Kasibhatla S, Schuller AG, Li AG, Cheng D, et al. Targeting Wnt-Driven Cancer through the Inhibition of Porcupine by Lgk974. Proc Natl Acad Sci US A. 2013;110:20224–20229. doi: 10.1073/pnas.1314239110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 728.You L, Zhang C, Yarravarapu N, Morlock L, Wang X, Zhang L, Williams NS, Lum L, Chen C. Development of a Triazole Class of Highly Potent Porcn Inhibitors. Bioorg Med Chem Lett. 2016;26:5891–5895. doi: 10.1016/j.bmcl.2016.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 729.Cheng D, Liu J, Han D, Zhang G, Gao W, Hsieh MH, Ng N, Kasibhatla S, Tompkins C, Li J, et al. Discovery of Pyridinyl Acetamide Derivatives as Potent, Selective, and Orally Bioavailable Porcupine Inhibitors. ACS Med Chem Lett. 2016;7:676–680. doi: 10.1021/acsmedchemlett.6b00038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 730.Kakugawa S, Langton PF, Zebisch M, Howell SA, Chang TH, Liu Y, Feizi T, Bineva G, O’Reilly N, Snijders AP, et al. Notum Deacylates Wnt Proteins to Suppress Signalling Activity. Nature. 2015;519:187–192. doi: 10.1038/nature14259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 731.Zhang X, Cheong SM, Amado NG, Reis AH, MacDonald BT, Zebisch M, Jones EY, Abreu JG, He X. Notum Is Required for Neural and Head Induction Via Wnt Deacylation, Oxidation, and Inactivation. Dev Cell. 2015;32:719–730. doi: 10.1016/j.devcel.2015.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 732.Banziger C, Soldini D, Schutt C, Zipperlen P, Hausmann G, Basler K. Wntless, a Conserved Membrane Protein Dedicated to the Secretion of Wnt Proteins from Signaling Cells. Cell. 2006;125:509–522. doi: 10.1016/j.cell.2006.02.049. [DOI] [PubMed] [Google Scholar]
- 733.Bartscherer K, Pelte N, Ingelfinger D, Boutros M. Secretion of Wnt Ligands Requires Evi, a Conserved Transmembrane Protein. Cell. 2006;125:523–533. doi: 10.1016/j.cell.2006.04.009. [DOI] [PubMed] [Google Scholar]
- 734.Goodman RM, Thombre S, Firtina Z, Gray D, Betts D, Roebuck J, Spana EP, Selva EM. Sprinter: A Novel Transmembrane Protein Required for Wg Secretion and Signaling. Development. 2006;133:4901–4911. doi: 10.1242/dev.02674. [DOI] [PubMed] [Google Scholar]
- 735.Buechling T, Chaudhary V, Spirohn K, Weiss M, Boutros M. P24 Proteins Are Required for Secretion of Wnt Ligands. EMBO Rep. 2011;12:1265–1272. doi: 10.1038/embor.2011.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 736.Port F, Hausmann G, Basler K. A Genome-Wide Rna Interference Screen Uncovers Two P24 Proteins as Regulators of Wingless Secretion. EMBO Rep. 2011;12:1144–1152. doi: 10.1038/embor.2011.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 737.Herr P, Basler K. Porcupine-Mediated Lipidation Is Required for Wnt Recognition by Wls. Dev Biol. 2012;361:392–402. doi: 10.1016/j.ydbio.2011.11.003. [DOI] [PubMed] [Google Scholar]
- 738.Coombs GS, Yu J, Canning CA, Veltri CA, Covey TM, Cheong JK, Utomo V, Banerjee N, Zhang ZH, Jadulco RC, et al. Wls-Dependent Secretion of Wnt3a Requires Ser209 Acylation and Vacuolar Acidification. J Cell Sci. 2010;123:3357–3367. doi: 10.1242/jcs.072132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 739.Belenkaya TY, Wu Y, Tang X, Zhou B, Cheng L, Sharma YV, Yan D, Selva EM, Lin X. The Retromer Complex Influences Wnt Secretion by Recycling Wntless from Endosomes to the Trans-Golgi Network. Dev Cell. 2008;14:120–131. doi: 10.1016/j.devcel.2007.12.003. [DOI] [PubMed] [Google Scholar]
- 740.Panakova D, Sprong H, Marois E, Thiele C, Eaton S. Lipoprotein Particles Are Required for Hedgehog and Wingless Signalling. Nature. 2005;435:58–65. doi: 10.1038/nature03504. [DOI] [PubMed] [Google Scholar]
- 741.Neumann S, Coudreuse DY, van der Westhuyzen DR, Eckhardt ER, Korswagen HC, Schmitz G, Sprong H. Mammalian Wnt3a Is Released on Lipoprotein Particles. Traffic. 2009;10:334–343. doi: 10.1111/j.1600-0854.2008.00872.x. [DOI] [PubMed] [Google Scholar]
- 742.Buglino JA, Resh MD. Palmitoylation of Hedgehog Proteins. Vitam Horm. 2012;88:229–252. doi: 10.1016/B978-0-12-394622-5.00010-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 743.Mann RK, Beachy PA. Novel Lipid Modifications of Secreted Protein Signals. Annu Rev Biochem. 2004;73:891–923. doi: 10.1146/annurev.biochem.73.011303.073933. [DOI] [PubMed] [Google Scholar]
- 744.Buglino JA, Resh MD. Hhat Is a Palmitoylacyltransferase with Specificity for N-Palmitoylation of Sonic Hedgehog. J Biol Chem. 2008;283:22076–22088. doi: 10.1074/jbc.M803901200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 745.Hardy RY, Resh MD. Identification of N-Terminal Residues of Sonic Hedgehog Important for Palmitoylation by Hedgehog Acyltransferase. J Biol Chem. 2012;287:42881–42889. doi: 10.1074/jbc.M112.426833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 746.Amanai K, Jiang J. Distinct Roles of Central Missing and Dispatched in Sending the Hedgehog Signal. Development. 2001;128:5119–5127. doi: 10.1242/dev.128.24.5119. [DOI] [PubMed] [Google Scholar]
- 747.Chamoun Z, Mann RK, Nellen D, von Kessler DP, Bellotto M, Beachy PA, Basler K. Skinny Hedgehog, an Acyltransferase Required for Palmitoylation and Activity of the Hedgehog Signal. Science. 2001;293:2080–2084. doi: 10.1126/science.1064437. [DOI] [PubMed] [Google Scholar]
- 748.Lee JD, Treisman JE. Sightless Has Homology to Transmembrane Acyltransferases and Is Required to Generate Active Hedgehog Protein. Curr Biol. 2001;11:1147–1152. doi: 10.1016/s0960-9822(01)00323-2. [DOI] [PubMed] [Google Scholar]
- 749.Chen MH, Li YJ, Kawakami T, Xu SM, Chuang PT. Palmitoylation Is Required for the Production of a Soluble Multimeric Hedgehog Protein Complex and Long-Range Signaling in Vertebrates. Genes Dev. 2004;18:641–659. doi: 10.1101/gad.1185804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 750.Petrova E, Matevossian A, Resh MD. Hedgehog Acyltransferase as a Target in Pancreatic Ductal Adenocarcinoma. Oncogene. 2014 doi: 10.1038/onc.2013.575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 751.Buglino JA, Resh MD. Identification of Conserved Regions and Residues within Hedgehog Acyltransferase Critical for Palmitoylation of Sonic Hedgehog. PLOS ONE. 2010;5:e11195. doi: 10.1371/journal.pone.0011195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 752.Matevossian A, Resh MD. Membrane Topology of Hedgehog Acyltransferase. J Biol Chem. 2015;290:2235–2243. doi: 10.1074/jbc.M114.625764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 753.Konitsiotis AD, Jovanovic B, Ciepla P, Spitaler M, Lanyon-Hogg T, Tate EW, Magee AI. Topological Analysis of Hedgehog Acyltransferase, a Multipalmitoylated Transmembrane Protein. J Biol Chem. 2015;290:3293–3307. doi: 10.1074/jbc.M114.614578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 754.Stanton BZ, Peng LF, Maloof N, Nakai K, Wang X, Duffner JL, Taveras KM, Hyman JM, Lee SW, Koehler AN, et al. A Small Molecule That Binds Hedgehog and Blocks Its Signaling in Human Cells. Nat Chem Biol. 2009;5:154–156. doi: 10.1038/nchembio.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 755.Petrova E, Rios-Esteves J, Ouerfelli O, Glickman JF, Resh MD. Inhibitors of Hedgehog Acyltransferase Block Sonic Hedgehog Signaling. Nat Chem Biol. 2013;9:247–249. doi: 10.1038/nchembio.1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 756.Rodgers UR, Lanyon-Hogg T, Masumoto N, Ritzefeld M, Burke R, Blagg J, Magee AI, Tate EW. Characterization of Hedgehog Acyltransferase Inhibitors Identifies a Small Molecule Probe for Hedgehog Signaling by Cancer Cells. ACS Chem Biol. 2016;11:3256–3262. doi: 10.1021/acschembio.6b00896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 757.Vyas N, Goswami D, Manonmani A, Sharma P, Ranganath HA, VijayRaghavan K, Shashidhara LS, Sowdhamini R, Mayor S. Nanoscale Organization of Hedgehog Is Essential for Long-Range Signaling. Cell. 2008;133:1214–1227. doi: 10.1016/j.cell.2008.05.026. [DOI] [PubMed] [Google Scholar]
- 758.Porter JA, Ekker SC, Park WJ, von Kessler DP, Young KE, Chen CH, Ma Y, Woods AS, Cotter RJ, Koonin EV, et al. Hedgehog Patterning Activity: Role of a Lipophilic Modification Mediated by the Carboxy-Terminal Autoprocessing Domain. Cell. 1996;86:21–34. doi: 10.1016/s0092-8674(00)80074-4. [DOI] [PubMed] [Google Scholar]
- 759.Porter JA, Young KE, Beachy PA. Cholesterol Modification of Hedgehog Signaling Proteins in Animal Development. Science. 1996;274:255–259. doi: 10.1126/science.274.5285.255. [DOI] [PubMed] [Google Scholar]
- 760.Heal WP, Jovanovic B, Bessin S, Wright MH, Magee AI, Tate EW. Bioorthogonal Chemical Tagging of Protein Cholesterylation in Living Cells. Chem Commun (Camb) 2011;47:4081–4083. doi: 10.1039/c0cc04710d. [DOI] [PubMed] [Google Scholar]
- 761.Ciepla P, Konitsiotis AD, Serwa RA, Masumoto N, Leong WP, Dallman MJ, Magee AI, Tate EW. New Chemical Probes Targeting Cholesterylation of Sonic Hedgehog in Human Cells and Zebrafish. Chem Sci. 2014;5:4249–4259. doi: 10.1039/c4sc01600a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 762.Tukachinsky H, Kuzmickas RP, Jao CY, Liu J, Salic A. Dispatched and Scube Mediate the Efficient Secretion of the Cholesterol-Modified Hedgehog Ligand. Cell Rep. 2012;2:308–320. doi: 10.1016/j.celrep.2012.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 763.Burke R, Nellen D, Bellotto M, Hafen E, Senti KA, Dickson BJ, Basler K. Dispatched, a Novel Sterol-Sensing Domain Protein Dedicated to the Release of Cholesterol-Modified Hedgehog from Signaling Cells. Cell. 1999;99:803–815. doi: 10.1016/s0092-8674(00)81677-3. [DOI] [PubMed] [Google Scholar]
- 764.Briscoe J, Chen Y, Jessell TM, Struhl G. A Hedgehog-Insensitive Form of Patched Provides Evidence for Direct Long-Range Morphogen Activity of Sonic Hedgehog in the Neural Tube. Mol Cell. 2001;7:1279–1291. doi: 10.1016/s1097-2765(01)00271-4. [DOI] [PubMed] [Google Scholar]
- 765.Kojima M, Hosoda H, Date Y, Nakazato M, Matsuo H, Kangawa K. Ghrelin Is a Growth-Hormone-Releasing Acylated Peptide from Stomach. Nature. 1999;402:656–660. doi: 10.1038/45230. [DOI] [PubMed] [Google Scholar]
- 766.Lim CT, Kola B, Korbonits M. The Ghrelin/Goat/Ghs-R System and Energy Metabolism. Rev Endocr Metab Disord. 2011;12:173–186. doi: 10.1007/s11154-011-9169-1. [DOI] [PubMed] [Google Scholar]
- 767.Yang J, Brown MS, Liang G, Grishin NV, Goldstein JL. Identification of the Acyltransferase That Octanoylates Ghrelin, an Appetite-Stimulating Peptide Hormone. Cell. 2008;132:387–396. doi: 10.1016/j.cell.2008.01.017. [DOI] [PubMed] [Google Scholar]
- 768.Gutierrez JA, Solenberg PJ, Perkins DR, Willency JA, Knierman MD, Jin Z, Witcher DR, Luo S, Onyia JE, Hale JE. Ghrelin Octanoylation Mediated by an Orphan Lipid Transferase. Proc Natl Acad Sci US A. 2008;105:6320–6325. doi: 10.1073/pnas.0800708105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 769.Taylor MS, Ruch TR, Hsiao PY, Hwang Y, Zhang P, Dai L, Huang CR, Berndsen CE, Kim MS, Pandey A, et al. Architectural Organization of the Metabolic Regulatory Enzyme Ghrelin O-Acyltransferase. J Biol Chem. 2013;288:32211–32228. doi: 10.1074/jbc.M113.510313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 770.Darling JE, Zhao F, Loftus RJ, Patton LM, Gibbs RA, Hougland JL. Structure-Activity Analysis of Human Ghrelin O-Acyltransferase Reveals Chemical Determinants of Ghrelin Selectivity and Acyl Group Recognition. Biochemistry. 2015;54:1100–1110. doi: 10.1021/bi5010359. [DOI] [PubMed] [Google Scholar]
- 771.Yang J, Zhao TJ, Goldstein JL, Brown MS. Inhibition of Ghrelin O-Acyltransferase (Goat) by Octanoylated Pentapeptides. Proc Natl Acad Sci US A. 2008;105:10750–10755. doi: 10.1073/pnas.0805353105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 772.Barnett BP, Hwang Y, Taylor MS, Kirchner H, Pfluger PT, Bernard V, Lin YY, Bowers EM, Mukherjee C, Song WJ, et al. Glucose and Weight Control in Mice with a Designed Ghrelin O-Acyltransferase Inhibitor. Science. 2010;330:1689–1692. doi: 10.1126/science.1196154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 773.Garner AL, Janda KD. A Small Molecule Antagonist of Ghrelin O-Acyltransferase (Goat) Chem Commun (Camb) 2011;47:7512–7514. doi: 10.1039/c1cc11817j. [DOI] [PubMed] [Google Scholar]
- 774.McGovern-Gooch KR, Mahajani NS, Garagozzo A, Schramm AJ, Hannah LG, Sieburg MA, Chisholm JD, Hougland JL. Synthetic Triterpenoid Inhibition of Human Ghrelin O-Acyltransferase: The Involvement of a Functionally Required Cysteine Provides Mechanistic Insight into Ghrelin Acylation. Biochemistry. 2017;56:919–931. doi: 10.1021/acs.biochem.6b01008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 775.Chang CC, Miyazaki A, Dong R, Kheirollah A, Yu C, Geng Y, Higgs HN, Chang TY. Purification of Recombinant Acyl-Coenzyme A:Cholesterol Acyltransferase 1 (Acat1) from H293 Cells and Binding Studies between the Enzyme and Substrates Using Difference Intrinsic Fluorescence Spectroscopy. Biochemistry. 2010;49:9957–9963. doi: 10.1021/bi1013936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 776.Cases S, Novak S, Zheng YW, Myers HM, Lear SR, Sande E, Welch CB, Lusis AJ, Spencer TA, Krause BR, et al. Acat-2, a Second Mammalian Acyl-Coa:Cholesterol Acyltransferase. Its Cloning, Expression, and Characterization. J Biol Chem. 1998;273:26755–26764. doi: 10.1074/jbc.273.41.26755. [DOI] [PubMed] [Google Scholar]
- 777.Cases S, Smith SJ, Zheng YW, Myers HM, Lear SR, Sande E, Novak S, Collins C, Welch CB, Lusis AJ, et al. Identification of a Gene Encoding an Acyl Coa:Diacylglycerol Acyltransferase, a Key Enzyme in Triacylglycerol Synthesis. Proc Natl Acad Sci US A. 1998;95:13018–13023. doi: 10.1073/pnas.95.22.13018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 778.Yen CL, Monetti M, Burri BJ, Farese RV., Jr The Triacylglycerol Synthesis Enzyme Dgat1 Also Catalyzes the Synthesis of Diacylglycerols Waxes and Retinyl Esters. J Lipid Res. 2005;46:1502–1511. doi: 10.1194/jlr.M500036-JLR200. [DOI] [PubMed] [Google Scholar]
- 779.Abe Y, Kita Y, Niikura T. Mammalian Gup1, a Homolog of Saccharomyces Cerevisiae Glycerol Uptake/Transporter 1, Acts as a Negative Regulator for N-Terminal Palmitoylation of Sonic Hedgehog. FEBS J. 2008;275:318–331. doi: 10.1111/j.1742-4658.2007.06202.x. [DOI] [PubMed] [Google Scholar]
- 780.Shindou H, Hishikawa D, Harayama T, Yuki K, Shimizu T. Recent Progress on Acyl Coa: Lysophospholipid Acyltransferase Research. J Lipid Res. 2009;50(Suppl):S46–51. doi: 10.1194/jlr.R800035-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 781.Lin H, Su X, He B. Protein Lysine Acylation and Cysteine Succination by Intermediates of Energy Metabolism. ACS Chem Biol. 2012;7:947–960. doi: 10.1021/cb3001793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 782.Stanley P, Koronakis V, Hughes C. Acylation of Escherichia Coli Hemolysin: A Unique Protein Lipidation Mechanism Underlying Toxin Function. Microbiol Mol Biol Rev. 1998;62:309–333. doi: 10.1128/mmbr.62.2.309-333.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 783.Stevenson FT, Bursten SL, Locksley RM, Lovett DH. Myristyl Acylation of the Tumor Necrosis Factor Alpha Precursor on Specific Lysine Residues. J Exp Med. 1992;176:1053–1062. doi: 10.1084/jem.176.4.1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 784.Stevenson FT, Bursten SL, Fanton C, Locksley RM, Lovett DH. The 31-Kda Precursor of Interleukin 1 Alpha Is Myristoylated on Specific Lysines within the 16-Kda N-Terminal Propiece. Proc Natl Acad Sci US A. 1993;90:7245–7249. doi: 10.1073/pnas.90.15.7245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 785.Chalkiadaki A, Guarente L. Sirtuins Mediate Mammalian Metabolic Responses to Nutrient Availability. Nat Rev Endocrinol. 2012 doi: 10.1038/nrendo.2011.225. advance online publication. [DOI] [PubMed] [Google Scholar]
- 786.Houtkooper RH, Pirinen E, Auwerx J. Sirtuins as Regulators of Metabolism and Healthspan. Nat Rev Mol Cell Biol. 2012;13:225–238. doi: 10.1038/nrm3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 787.Haigis MC, Sinclair DA. Mammalian Sirtuins: Biological Insights and Disease Relevance. Annu Rev Pathol. 2010;5:253–295. doi: 10.1146/annurev.pathol.4.110807.092250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 788.Imai S-i, Armstrong CM, Kaeberlein M, Guarente L. Transcriptional Silencing and Longevity Protein Sir2 Is an Nad-Dependent Histone Deacetylase. Nature. 2000;403:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- 789.Du J, Zhou Y, Su X, Yu J, Khan S, Jiang H, Kim J, Woo J, Kim JH, Choi BH, et al. Sirt5 Is an Nad-Dependent Protein Lysine Demalonylase and Desuccinylase. Science. 2011;334:806–809. doi: 10.1126/science.1207861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 790.Jiang H, Khan S, Wang Y, Charron G, He B, Sebastian C, Du J, Kim R, Ge E, Mostoslavsky R, et al. Sirt6 Regulates Tnf-α Secretion through Hydrolysis of Long-Chain Fatty Acyl Lysine. Nature. 2013;496:110–113. doi: 10.1038/nature12038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 791.Jiang H, Zhang X, Lin H. Lysine Fatty Acylation Promotes Lysosomal Targeting of Tnf-Alpha. Sci Rep. 2016;6:24371. doi: 10.1038/srep24371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 792.Zhang X, Spiegelman NA, Nelson OD, Jing H, Lin H. Sirt6 Regulates Ras-Related Protein R-Ras2 by Lysine Defatty-Acylation. Elife. 2017;6 doi: 10.7554/eLife.25158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 793.Sebastián C, Zwaans Bernadette MM, Silberman Dafne M, Gymrek M, Goren A, Zhong L, Ram O, Truelove J, Guimaraes Alexander R, Toiber D, et al. The Histone Deacetylase Sirt6 Is a Tumor Suppressor That Controls Cancer Metabolism. Cell. 2012;151:1185–1199. doi: 10.1016/j.cell.2012.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 794.Feldman JL, Baeza J, Denu JM. Activation of the Protein Deacetylase Sirt6 by Long-Chain Fatty Acids and Widespread Deacylation by Mammalian Sirtuins. J Biol Chem. 2013;288:31350–31356. doi: 10.1074/jbc.C113.511261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 795.Aramsangtienchai P, Spiegelman NA, He B, Miller SP, Dai L, Zhao Y, Lin H. Hdac8 Catalyzes the Hydrolysis of Long Chain Fatty Acyl Lysine. ACS Chem Biol. 2016;11:2685–2692. doi: 10.1021/acschembio.6b00396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 796.Zhu AY, Zhou Y, Khan S, Deitsch KW, Hao Q, Lin H. Plasmodium Falciparum Sir2a Preferentially Hydrolyzes Medium and Long Chain Fatty Acyl Lysine. ACS Chem Biol. 2012;7:155–159. doi: 10.1021/cb200230x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 797.Konermann L, Pan J, Liu YH. Hydrogen Exchange Mass Spectrometry for Studying Protein Structure and Dynamics. Chem Soc Rev. 2011;40:1224–1234. doi: 10.1039/c0cs00113a. [DOI] [PubMed] [Google Scholar]
- 798.Schuler MA, Denisov IG, Sligar SG. Nanodiscs as a New Tool to Examine Lipid-Protein Interactions. Methods Mol Biol. 2013;974:415–433. doi: 10.1007/978-1-62703-275-9_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 799.Pedro MP, Vilcaes AA, Tomatis VM, Oliveira RG, Gomez GA, Daniotti JL. 2-Bromopalmitate Reduces Protein Deacylation by Inhibition of Acyl-Protein Thioesterase Enzymatic Activities. PLOS ONE. 2013;8:e75232. doi: 10.1371/journal.pone.0075232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 800.Coleman RA, Rao P, Fogelsong RJ, Bardes ESG. 2-Bromopalmitoyl-Coa and 2-Bromopalmitate: Promiscuous Inhibitors of Membrane-Bound Enzymes. Biochim Biophys Acta. 1992;1125:203–209. doi: 10.1016/0005-2760(92)90046-x. [DOI] [PubMed] [Google Scholar]