Abstract
Background & Aims
Individuals with monogenic disorders of phagocyte function develop chronic colitis that resembles Crohn’s Disease (CD). We tested for associations between mutations in genes encoding NADPH oxidases, neutrophil function, and phenotypes of CD in pediatric patients.
Methods
We performed whole-exome sequence analysis to identify mutations in genes encoding NADPH oxidases (such as CYBA, CYBB, NCF1, NCF2, NCF4, RAC1, and RAC2) using DNA from 543 pediatric patients with inflammatory bowel diseases. Blood samples were collected from an additional 129 pediatric patients with CD and 26 children without IBD (controls); we performed assays for neutrophil activation, reactive oxygen species (ROS) production, and bacteria uptake and killing. Whole-exome sequence analysis was performed using DNA from 46 of the children with CD to examine associations with NADPH gene mutations; RNA sequence analyses were performed using blood cells from 46 children with CD to test for variations in neutrophil gene expression associated with ROS production.
Results
We identified 26 missense mutations in CYBA, CYBB, NCF1, NCF2, and NCF4. Patients with CD who carried mutations in these genes were 3-fold more likely to have perianal disease (P=.0008) and stricturing complications (P=.002) than children with CD without these mutations. Among patients with CD with none of these mutations, 9% had undergone abdominal surgery; among patients with mutations in these NADPH oxidase genes, 31% had undergone abdominal surgery (P=.0004). A higher proportion of neutrophils from children with CD had low ROS production (47%) than from controls (15%) among the 129 patients tested for ROS (P=.002). Minor alleles of the NADPH genes were detected in 7% of children with CD whose neutrophils produced normal levels of ROS vs 38% of children whose neutrophils produced low levels of ROS (P=.009). Neutrophils that produced low levels of ROS had specific alterations in genes that regulate glucose metabolism and anti-microbial responses.
Conclusions
We identified missense mutations in genes that encode NADPH oxidases in children with CD; these associated with a more aggressive disease course and reduced ROS production by neutrophils from the patients.
Keywords: WES, IBD, neutrophil oxidative burst, genetic variant
Short summary
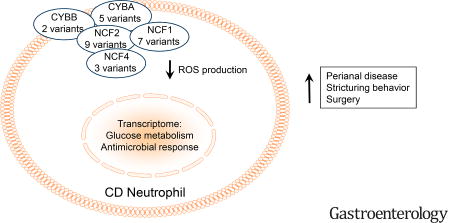
A subset of pediatric Crohn’s Disease patients exhibit damaging mutations in core NADPH oxidase genes which are associated with reduced neutrophil reactive oxygen species production and a more aggressive clinical course.
Introduction
The development of chronic intestinal inflammation similar to Crohn Disease (CD) during the first decade of life in children with Chronic Granulomatous Disease (CGD) has suggested that loss-of-function in phagocyte reactive oxygen species (ROS) production is likely to be a fundamental mechanism of pediatric CD pathogenesis.1–3 CGD patients are notable for predominantly distal colonic and perianal disease which is often refractory to standard medical approaches.4, 5 Genome-wide Association Studies (GWAS) of CD have linked pathways including autophagy and ROS production to disease pathogenesis.6–8 Rare missense mutations in the CYBB, NCF1, NCF2, and NCF4 genes which regulate neutrophil NADPH oxidase function and ROS production have been reported in children with very early onset (VEO, <age 6) IBD.9, 10 However, an assessment of the association between NADPH oxidase gene mutations, neutrophil ROS production and gene expression, and clinical phenotype in an older cohort of pediatric-onset IBD patients had not been performed.
A variety of defects in neutrophil function have been described in both pediatric and adult onset CD.9–12 These include reduced adhesion, chemotaxis, phagocytosis, and ROS production. Recent studies have defined the fundamental role which cellular metabolism plays in immune cell function.13 Neutrophils have been shown to rely critically upon glycolysis to mount effective anti-microbial responses.14–16 Conversely, defects in ROS production in response to immune stimuli have in turn been shown to result in impaired glucose metabolism and overall cell function. While the global pattern of gene expression has recently been defined in circulating neutrophils isolated from healthy adults, this had not been defined relative to neutrophil function including ROS production in children with IBD.17
We hypothesized that rare genetic variants cause neutrophil dysfunction and thereby contribute to disease pathogenesis in pediatric CD. In the current study we have tested for associations between neutrophil ROS production, clinical phenotypes, and rare missense mutations in the core genes which comprise the NADPH oxidase complex. Whole exome sequencing was performed in pediatric IBD patients and potentially damaging missense mutations in CYBA, CYBB, NCF1, NCF2, NCF4, RAC1, and RAC2 were validated by Sanger Sequencing. Peripheral blood neutrophil ROS production and the associated global pattern of gene expression were measured in pediatric IBD patients and controls. We identified a subset of pediatric CD patients characterized by low normal neutrophil ROS production, which was in turn associated with rare NADPH oxidase gene missense mutations, alterations in neutrophil gene pathways for glucose metabolism and antimicrobial responses, and a more aggressive disease course.
Materials and Methods
Whole exome sequencing
Subjects for whole exome sequencing (WES) were selected from the multi-center Crohn’s and Colitis Foundation (CCF) sponsored RISK inception cohort study and the National Institute of Health (NIH) sponsored Emory African-American gene discovery study. The clinical and demographic features of the WES group are summarized in Supplemental Table 1. Patients were diagnosed with CD, UC or inflammatory bowel disease-undetermined (IBD-U).18, 19 None of the participants carried a diagnosis of CGD or other primary immune deficiency disorder.
Exome sequencing data analysis
The raw sequence reads were mapped relative to the human genome reference sequence (hg38) and variants were called using PEmapper/PECaller.20 Unique variants were functionally annotated using SeqAnt version 2.0 (Beta 3, https://seqant.genetics.emory.edu/) which reports the variant’s type, functional classification (nonsense, replacement, silent, 5’ or 3’ UTR, intronic, intergenic), presence in databases like dbSNP and Combined Annotation-Dependent Depletion (CADD) scores.21 To solidify our confidence in prioritizing deleterious variants based on functional consequence and allele frequency, we used the Exome Aggregation Consortium (ExAC) reference dataset, which provides a resource for the estimation of allele frequency of coding variants (http://exac.broadinstitute.org/).22
Filtering and validation of variants
In addition to the standard variant quality controls, we filtered all variants by minor allele frequency (MAF) < 5% and nonsynonymous variants by CADD score greater than 10 based on our initial SeqAnt annotation to identify causal variants.21 A variant was predicted to be pathogenic if the scaled C-score calculated by the software was above 10, a score indicative of the variant being within the top 10% of deleterious substitutions.21 Updates to the CADD database in SeqAnt resulted in some variants being re-annotated with lower CADD score. These variants were validated by Sanger sequencing to confirm bioinformatics analysis.
Neutrophil Function Studies
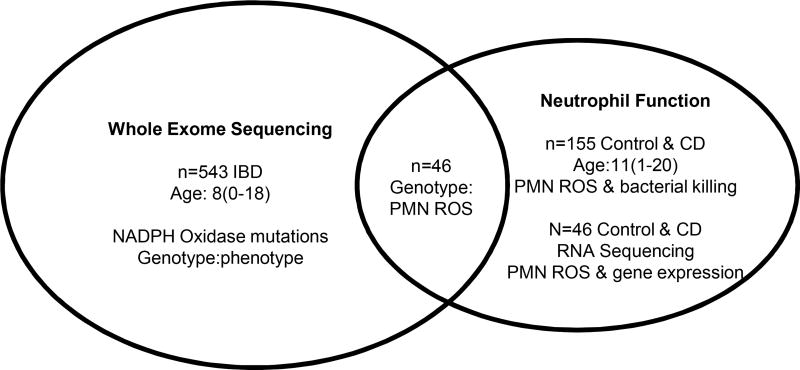
Subjects for neutrophil function studies were enrolled at the Cincinnati site and included 129 CD patients and 26 non-IBD controls. 46 of the CD patients also had WES completed (Fig.1). The clinical and demographic features of the neutrophil function group are summarized in Supplemental Table 3. Peripheral blood neutrophil CD64 index (Trillium Diagnostics, LLC, #Leuko64), and CD11B (BectonDickinson, #340937) and formyl peptide receptor 1 (FPR1) (R&D Systems, #FAB3744F) cell surface abundance, were determined by flow cytometry using our published methods.12 ROS production was measured in neutrophils in whole blood using the dihydrorhodamine 123 dye (Molecular Probes, #D632) following FMLP (Sigma, #F3506) stimulation.12 Phagocytosis and killing of Staph. aureus was measured in adherent neutrophils using the acridine orange (Polysciences, Inc, #04539) method as per the CCHMC Clinical Immunology laboratory standard operating procedure for the Phagocytic Microbiocidal Assay P285-701.12 The expected normal range for the assay is 6–15/cell for phagocytic capacity and >80% for percent killing for S. aureus 502A.
Figure 1. Flow Chart for Patient Populations with Whole Exome Sequencing and Neutrophil Function Studies.
Whole exome sequencing (WES) was completed for 543 pediatric IBD patients, with a median (range) age at diagnosis of 8(0–18) years. These data defined NADPH oxidase gene mutations in this population, and associations with clinical phenotype. Neutrophil function studies including measures of ROS production and bacterial killing were completed for 129 pediatric CD patients with a median (range) age at diagnosis of 11(1–20) years, and 26 non-IBD controls. 46 CD patients had both WES and neutrophil function testing completed to test for associations between NADPH oxidase gene mutations and neutrophil ROS production and bacterial killing. A sub-group of CD patients (n=40) and non-IBD controls (n=6) with neutrophil function testing completed also had RNASeq performed to define the neutrophil global pattern of gene expression in patients stratified by ROS production.
Statistical Analysis for Neutrophil Function Studies
CD cases for functional assays were defined as CD patients who carried the candidate variant of interest which could affect the functional read-out including CYBA, CYBB, NCF1, NCF2, and NCF4. CD controls for the functional assays were defined as not carrying any potentially damaging variants in the pathway genes for the variant of interest. Statistical analyses were performed using GraphPad PRISM© Version 7. Continuous variables were analyzed using unpaired t test, two sample t test, Mann-Whitney test, one-way ANOVA with Bonferroni multiple comparison test, Kruskal-Wallis with Dunn’s test for multiple comparisons, or linear test-for-trend. Discrete variables were analyzed using Fisher’s exact test or chi-square test. A p-value <0.05 was considered significant.
Sample Size for Neutrophil Function Studies
A significant effect upon neutrophil ROS production was defined as a reduction in the mean (SD) MFI from 600(200) in CD disease controls lacking a specific genetic variant to 400(150) in CD cases carrying a specific genetic variant. 15 subjects carrying the variant of interest, compared to 15 subjects not carrying the variant, would be required to test for this difference with >80% power and ≤ 0.05.
RNA Sequencing and Analysis
Total purified neutrophil RNA was amplified using the Ovation RNA-Seq System v2 (NuGEN) according to the manufacturer’s protocol. The libraries were prepared with the Nextera XT DNA Sample Preparation kit (Illumina Technologies). Libraries were sequenced on the Illumina HiSeq2500 following the manufacturer's protocol with paired end, 75 bp reads (Illumina, San Diego, CA). Expression values were compared between low ROS neutrophils from CD patients, normal ROS neutrophils from CD patients, and normal ROS neutrophils from controls. The analysis was carried out using two pipelines (Suppl. Fig. 3). The first pipeline determined genes differentially regulated between low ROS CD neutrophils vs controls and between normal ROS CD neutrophils and controls using unpaired Welch’s t-tests (assuming unequal variance) with significance at p<0.05 and fold change >1.5. Gene lists were overlapped using a Venn diagram (bioinfogp.cnb.csic.es/tools/venny/index.html) to identify gene sets specific to and shared between low and normal ROS neutrophils compared to controls (Suppl. Fig. 4). The second pipeline directly compared low and normal ROS neutrophils within CD, applying the same tests and cutoffs used in the first pipeline (411 differentially regulated transcripts).
Ontological Analysis
In order to determine processes and pathways contributing to increased disease severity associated with low ROS CD neutrophils, overlapping and specific transcripts were assessed at the mechanism level. Genes up- and down-regulated from each analysis were submitted to ToppGene and ToppCluster for assessment of ontological enrichments (toppgene.cchmc.org, toppcluster.cchmc.org), focusing on molecular functions, biological processes, and pathways.23, 24 Ontological maps were generated through Toppcluster and Cytoscape.25 The full analytic pipeline is described in Suppl. Fig. 3.
Study approval
The patient-based studies were approved by the Institutional Review Boards at each of the RISK and Emory African-American gene discovery study sites including Emory and CCHMC, and consent was obtained from parents and adult subjects and assent from pediatric subjects age 11 and above.
Results
Missense Mutations in Core NADPH Oxidase Genes Detected in Early Onset IBD Patients
Potentially damaging genetic variants in the protein coding regions of CYBA, CYBB, NCF1, NCF2, NCF4, RAC1, and RAC2 were determined using whole exome sequencing of 543 early onset pediatric IBD patients (Fig. 1 and Suppl. Table 1). As summarized in Table 1, we identified 26 potentially damaging heterozygous or homozygous missense mutations in CYBA, CYBB, NCF1, NCF2, and NCF4. Three variants (CYBA R164H, NCF1 R90C, and NCF1 A149E) are novel. The IBD case minor allele frequency (MAF) for each of the remaining variants did not differ from control MAF obtained from publically available databases. The MAF from dbSNP are shown in Table 1 for individuals of European ancestry; the MAF reported for each of these variants in the ExAC database for individuals of European, African, or East/South Asian ancestry are included in Suppl. Table 4.
Table 1.
Missense Mutations in Core NADPH Oxidase Genes
| Gene | Chrom | Position | Ref | Alt | AA change |
CADD | dbSNP MAF |
|---|---|---|---|---|---|---|---|
| CYBA | chr16 | 88643450 | C | T | R164H | 35 | - |
| CYBA | chr16 | 88646184 | C | T | A101T# | 24.9 | 0.000009 |
| CYBA | chr16 | 88646816 | C | T | V76M# | 24 | 0.000795 |
| CYBA | chr16 | 88646828 | A | G | Y72H | 11.7 | 0.689776 |
| CYBA | chr16 | 88647125 | T | G | K60T# | 3.32 | 0.003791 |
| CYBB | chrX | 37798966 | G | A | R229H | 26.7 | 0.000788 |
| CYBB | chrX | 37804069 | G | C | G364R | 23 | 0.003901 |
| NCF1 | chr7 | 74777318 | C | T | R42W | 31 | 0.000062 |
| NCF1 | chr7 | 74779274 | G | A | G83R# | 20.2 | 0.008833 |
| NCF1 | chr7 | 74779295 | C | T | R90C | 31 | - |
| NCF1 | chr7 | 74779319 | T | G | C98G | 22.9 | 0.000712 |
| NCF1 | chr7 | 74779322 | A | G | S99G# | - | - |
| NCF1 | chr7 | 74780830 | C | A | A149E | - | - |
| NCF1 | chr7 | 74783529 | G | A | W193* | 36 | 0.000646 |
| NCF2 | chr1 | 183560204 | G | A | P454S# | 0.75 | 0.004066 |
| NCF2 | chr1 | 183563229 | T | A | N419I# | 28.4 | 0.005324 |
| NCF2 | chr1 | 183563301 | C | T | R395Q | 23.4 | 0.001036 |
| NCF2 | chr1 | 183563302 | G | A | R395W | 34 | 0.015222 |
| NCF2 | chr1 | 183563455 | C | T | R386Q# | 13.4 | 0.002271 |
| NCF2 | chr1 | 183563531 | T | A | T361S# | 26.6 | 0.001892 |
| NCF2 | chr1 | 183566954 | A | G | V297A# | 22.9 | 0.004115 |
| NCF2 | chr1 | 183574503 | G | A | A162V | 35 | 0.000025 |
| NCF2 | chr1 | 183590217 | C | T | R38Q# | 34 | 0.001408 |
| NCF4 | chr22 | 36865055 | C | A | T85N# | 7.59 | 0.002867 |
| NCF4 | chr22 | 36870523 | C | T | R151C | 35 | 0.000056 |
| NCF4 | chr22 | 36875672 | C | T | T216M | 33 | 0.000647 |
Chrom: chromosome; AA: amino acid; CADD: Combined Annotation-Dependent Depletion score; MAF: minor allele frequency;
: variant included in Table 2 analysis
Rare NADPH Oxidase Gene Mutation Carriage is Associated with Higher Rates of Stricturing Complications and Surgery
We tested for associations between NADPH oxidase gene mutation carriage and clinical phenotype in 228 CD patients with both WES and prospective clinical data for a minimum of 36 months from diagnosis. Thirteen percent carried from 1–3 variants as indicated in Table 1. For each of these we considered heterozygous minor allele carriage as being variant positive, with the exception of the more common NCF1 S99G variant for which homozygous carriage was considered positive. These included one patient each who carried CYBA V76M, CYBA A101T, NCF2 V297A, NCF2 P454S, NCF2 R386Q, or NCF4 T85N, two patients who carried NCF2 N419I, three patients who carried NCF2 R38Q or NCF2 T361S, five patients who carried NCF1 G83R, six patients who carried CYBA K60T, and seven patients who were homozygous for NCF1 S99G. As shown in Table 2, age-of-onset, sex, race, CYBA Y72H homozygous variant carriage, disease location, and disease duration did not vary between the groups. Treatment exposures for corticosteroids (65% vs 62%), mesalamine (44% vs 38%), immune modulators (78% vs 69%) and anti-TNF (56% vs 45%) also did not differ between the CD patients without or with NADPH oxidase gene mutation carriage, respectively. However, CD patients with NADPH oxidase gene mutation carriage were three times more likely to exhibit perianal disease (p=0.0008) and experience stricturing complications (p=0.002). Consistent with this, the frequency of abdominal surgeries increased from 9% in CD patients with no variants to 31% in those carrying NADPH oxidase gene mutations (p=0.0004). Forty-six of the CD patients with WES also had neutrophil function testing (Fig. 1). We therefore next asked whether any of the missense mutations summarized in Table 1 would be associated with a reduction in neutrophil ROS production.
Table 2.
Clinical and Demographic Characteristics of CD Patients Stratified by Core NADPH Oxidase Gene Mutation Carriage
| No mutation n=199 |
Mutation n=29 |
|
|---|---|---|
| Age-of-onset (years) | 9(1–15) | 9(5–12) |
| Male | 120(60) | 18(62) |
| White | 158(88) | 24(86) |
| CYBA Y72H homozygote | 79(40) | 13(45) |
| Ileal location | 29(15) | 6(21) |
| Colon-only location | 45(24) | 7(25) |
| Ileo-colonic location | 114(61) | 15(54) |
| Disease duration (months) | 36(36–156) | 36(36–137) |
| Perianal disease | 15(8) | 8(28)*** |
| Stricturing Behavior | 17(9) | 8(28)** |
| Penetrating Behavior | 7(4) | 2(7) |
| Abdominal Surgery | 17(9) | 9(31)*** |
N-Formyl Peptide Induced Neutrophil Reactive Oxygen Species Production is Reduced in a Subset of Pediatric CD Patients
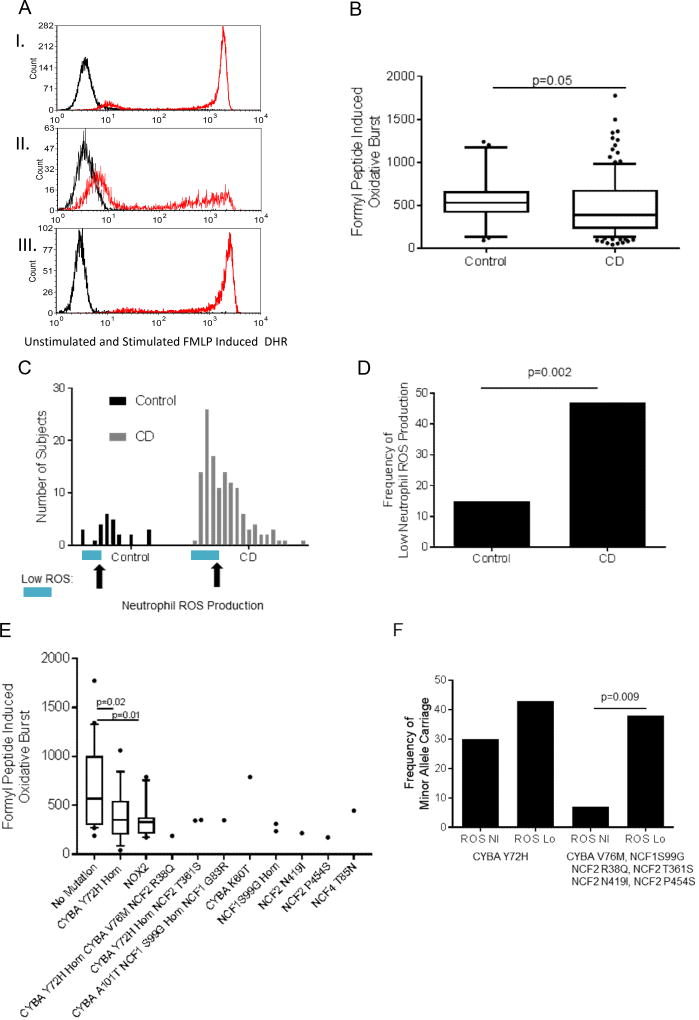
We first asked whether neutrophil ROS production would vary between pediatric CD patients and healthy age-matched controls. The clinical and demographic features of the participants used for neutrophil function testing are summarized in Suppl. Table 3. N-formyl peptides including N-formyl-methionyl-leucyl-phenylalanine (FMLP) released by both bacteria and damaged host tissues are a highly relevant physiologic stimulus for neutrophil ROS production in IBD.26 Peripheral blood samples were stimulated with FMLP and neutrophil ROS production was measured by flow cytometry. A representative FACS plot for unstimulated (black) and stimulated (red) control and CD samples is shown in Figure 2A. Median (IQR) FMLP induced neutrophil ROS production was higher in healthy controls (513(409,671)) compared to CD patients (391(221,684)) (Figure 2B, p=0.05). Histograms demonstrated that a subset of CD patients exhibited reduced ROS production compared to the control distribution, defined as a DHR MFI of less than 360, or the 15th percentile for controls (Figure 2C). This resulted in 47% of CD patients being classified as having low ROS production compared to 15% of controls (Figure 2D, p= 0.002). However, this low level of ROS production in a subset of CD patients was still significantly higher than that measured for X-linked or autosomal recessive CGD patients in the CCHMC Clinical Immunology lab (Suppl. Fig. 1).
Figure 2. N-Formyl Peptide Induced Neutrophil ROS Production and Core NADPH Oxidase Gene Mutations in Crohn Disease Patients.
Peripheral blood samples were stimulated with N-formyl peptide (FMLP) and neutrophil ROS production was measured by flow cytometry using the dihydrorhodamine 123 (DHR) dye. A) Representative FACS plot for unstimulated (black) and stimulated (red) I) control, II) low ROS CD and III) high ROS CD samples are shown. B) FMLP induced neutrophil ROS production measured by the DHR MFI in healthy controls (n=26), and Crohn Disease patients (CD, n=129) is shown as the median (IQR). Differences between the groups was tested by unpaired t-test of log transformed values. C) Histograms demonstrating the number of subjects with each level of FMLP induced neutrophil ROS production in groups of 100 for the DHR MFI is shown, with the point for a DHR MFI of 360, the 15th percentile within controls, on the x-axis for each group indicated by the arrows, and those with low ROS production indicated by the blue bar. D) The frequency of subjects with low FMLP induced ROS production defined by a DHR MFI less than 360 is shown. Differences between groups was tested using the chi2 test. E) FMLP induced neutrophil ROS production is shown as the median (IQR) for CD patients lacking any potentially damaging variants in core NADPH oxidase genes (no mutation), or patients carrying the indicated missense mutations. Differences between the no mutation group (n=20) and the groups homozygous for the CYBA Y72H variant (n=16) or carrying one of the other NADPH oxidase gene mutations (NOX2, n=10) were tested by the unpaired t-test. For each of the other missense mutations, only 1–3 patient samples were tested, with the results shown for each. F) The frequency of minor allele carriage is shown for CD patients stratified by a FMLP induced neutrophil ROS MFI of 360, ROS Nl versus ROS Lo. Differences between the ROS Nl versus ROS Lo groups were tested by chi2 test for CYBA Y72H homozygous carriage alone, or for carriage of CYBA V76M, NCF1 S99G, NCF2 R38Q, NCF2 T361S, NCF2 N419I, or NCF2 P454S.
NADPH Oxidase Gene Mutations are Associated with Low FMLP Induced Neutrophil ROS Production
Neither cell surface abundance of the formyl peptide receptor (FPR1), nor the activation marker CD64, varied with neutrophil ROS production (Suppl. Table 5 and Suppl. Fig. 2). We did observe a modest reduction in cell surface abundance of the neutrophil adhesion molecule CD11B in CD samples with low ROS production (Suppl. Table 5). Similarly, neither clinical disease activity nor treatment exposures differed between the CD sub-groups with low versus normal ROS production (Supplemental Table 6). These data confirmed that the observed reductions in FMLP induced ROS production were not simply due to differences in cellular FPR1 expression or activation state, or concurrent disease activity or treatment exposures. We therefore asked whether CD patients with low neutrophil ROS production would be more likely to carry genetic variants in the core NADPH oxidase complex genes shown in Table 1.
FMLP induced neutrophil ROS production is shown in Figure 2E as the median (IQR) for CD patients lacking any potentially damaging variants in these genes (no mutation), or patients carrying the indicated missense mutations. Median(IQR) FMLP induced ROS production was reduced from 571(301,1005) in the group carrying no NADPH oxidase gene mutations (n=20), to 350(198,545) in the group homozygous for the CYBA Y72H variant (n=16, p=0.02), and to 329(211,377) in the group carrying at least one of the other NADPH oxidase gene mutations shown (NOX2 group, n=10, p=0.01). For each of the rare NADPH oxidase gene missense mutations, only 1–3 patient samples were tested, with results shown for each of the ten subjects in Fig 2E and Suppl. Table 7. Consistent with results in the larger group with WES (Table 2), neither age-of-onset, sex, race, nor disease location varied with NADPH oxidase gene mutation carriage in the sub-group with neutrophil function testing completed (Suppl. Table 7). However, CD patients carrying rare NADPH oxidase gene missense mutations were three-fold more likely to experience a stricturing complication (p=0.03) and two-fold more likely to require abdominal surgery (p=0.04, Suppl. Table 7).
Note that the subjects with heterozygous missense mutations in CYBA V76M, NCF2 R38Q, and NCF2 T361S were also homozygous for the CYBA Y72H missense mutation. We then asked if there was evidence for enrichment of these mutations in patients with low ROS production. The frequency of minor allele carriage is shown in Figure 2F for CD patients stratified by FMLP induced ROS production of 360, classified as ROS Nl versus ROS Lo. Differences between the ROS Nl versus ROS Lo groups are shown for CYBA Y72H homozygous carriage alone, or for carriage of the rare CYBA V76M, NCF1 S99G, NCF2 R38Q, NCF2 T361S, NCF2 N419I, or NCF2 P454S missense mutations. As shown in Figure 2F, the frequency of minor allele carriage for these rare potentially damaging missense mutations increased from 7% in subjects with normal FMLP induced ROS production to 38% in subjects with low FMLP induced ROS production (p=0.009).
NADPH Oxidase Gene Mutations are Associated with Reduced Bacterial Killing
Intact neutrophil ROS production is required for killing of catalase positive bacteria including Staph. Aureus. We therefore next asked whether Staph. Aureus phagocytosis or killing would vary between CD patients with high versus low FMLP induced neutrophil ROS production. Median (IQR) Staph. Aureus phagocytosis defined as the number of bacteria engulfed per neutrophil did not vary between healthy controls (7(6,9)) and CD patients (7(6,8)). Median (IQR) Staph. Aureus killing defined as the percentage of intra-cellular bacteria killed also did not vary between healthy controls (91(86,92)) and CD patients (86(78,91)). Moreover, we did not observe a difference in either Staph. Aureus phagocytosis or killing in the low ROS production CD sub-group defined by FMLP stimulation responses (Suppl. Table 5).
Finally, we asked whether bacterial uptake or killing would vary with NADPH oxidase gene mutation carriage within CD patients. Median(IQR) bacterial uptake per neutrophil did not vary across the CD sub-groups, and was equal to 7(6,8) in the group carrying no NADPH oxidase gene mutations, 7(6,10) in the group homozygous for the CYBA Y72H variant, and 6(5,6) in the group carrying at least one of the other NADPH oxidase gene mutations shown. However, the frequency of bacteria killed per neutrophil was reduced in CD patients with NADPH oxidase gene mutation carriage, decreasing from a median(IQR) of 88(82,89) in the group carrying no NADPH oxidase gene mutations, to 78(77,89) in the group homozygous for the CYBA Y72H variant (p=0.08), and to 79(71,87) in the group carrying at least one of the other NADPH oxidase gene mutations shown (p=0.04). Collectively, these data demonstrated a specific association between potentially damaging missense mutations in CYBA, NCF1, and NCF2, and both low neutrophil ROS production and reduced bacterial killing.
Alterations in Gene Expression Pathways Regulating Glucose Metabolism and Anti-microbial Responses in Low ROS CD Neutrophils
With the exception of a modest increase in RAC1 expression, gene expression for core NADPH oxidase genes did not vary between low and normal ROS CD neutrophils (Suppl. Table 8). We therefore next employed RNA sequencing to test for global differences in gene expression and associated biologic pathways between neutrophils isolated from CD patients with low ROS production, compared to both healthy controls and CD patients with normal ROS production. We employed two analytic pipelines to test this as illustrated in Suppl. Figure 3.
Pipeline 1: Normal ROS CD neutrophils and low ROS CD neutrophils vs controls
We identified 834 and 618 transcripts differentially regulated when comparing low and normal ROS CD neutrophils to control neutrophils, respectively (Suppl. Table 9). Of these, 272 downregulated and 105 upregulated transcripts were common between low and normal ROS CD neutrophils, leaving 241 transcripts specifically dysregulated in normal ROS CD neutrophils (195 down and 46 up compared to control) and 457 transcripts specific to low ROS CD neutrophils (186 down and 271 up compared to controls; Suppl. Fig. 4). Ontological enrichments were identified for the 151 transcripts up in normal ROS CD vs control neutrophils and the 376 transcripts up in low ROS CD vs control neutrophils, and again for the 467 transcripts down in normal ROS CD vs control neutrophils and the 458 transcripts down in low ROS CD vs control neutrophils. Shared transcripts were represented in shared ontological enrichments between normal and low ROS CD neutrophils, whereas transcripts specific to normal or low ROS CD neutrophils were represented through unshared ontological enrichments.
In normal and low ROS CD neutrophils we observed the expected and predominant upregulation of inflammatory and immune signaling, specifically through I-kappaB/NF-kappaB, toll-like receptor, and cytokine signaling/response, potentially through the MAPK/ERK axes (Suppl. Fig. 5A). Specific to low ROS neutrophils in CD, an additional set of inflammatory/immune processes were upregulated, in addition to antimicrobial pathways and reactive oxygen species metabolism. In the shared downregulated signals we identify further signals supporting dysregulated immune and inflammatory signaling (Suppl. Fig. 5B). Further, neutrophil energy generation and sources appeared altered, with overrepresentation of ontologies related to mitochondrial energy dynamics (OXPHOS and ATP synthesis) and glucose production and tolerance. Specific to low ROS CD neutrophils, antimicrobial pathways were downregulated with further indication of mitochondrial dysfunction (response to oxidative stresses and fatty acid oxidation) and glucose/carbohydrate transport, metabolism, and homeostasis.
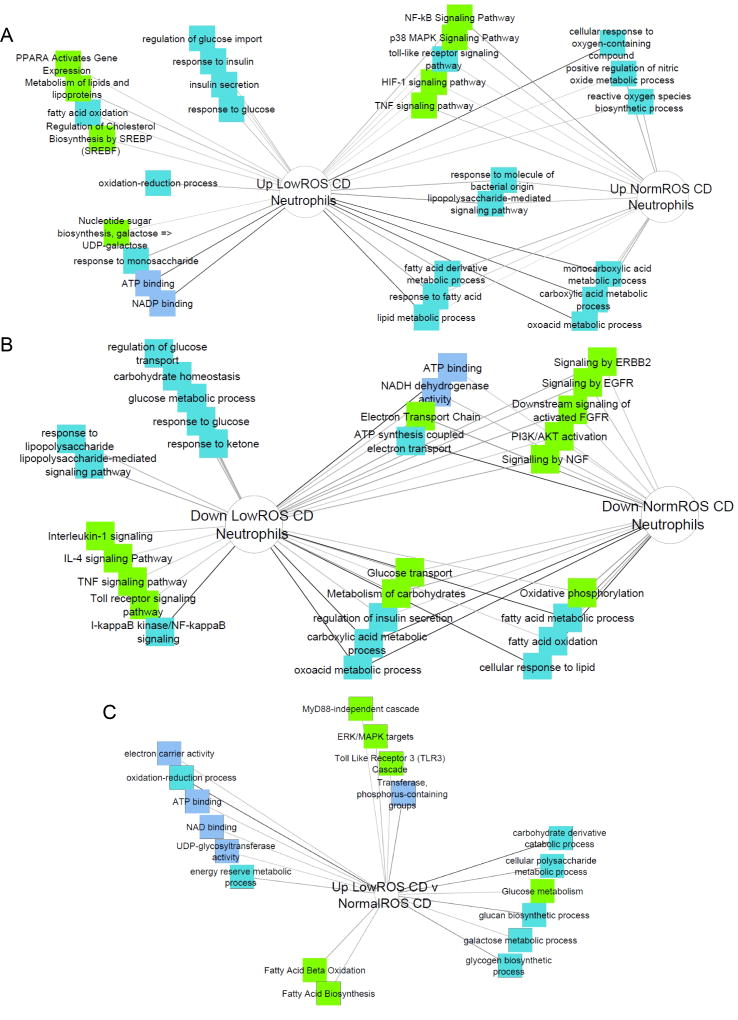
Because of the critical role of glycolysis in neutrophil antimicrobial function, genes up- and down-regulated in normal or low ROS CD vs controls were then ranked based on functional enrichment with glycolysis-related genes (Suppl. Table 10, Suppl. Figure 6). In normal ROS CD neutrophils, 19 upregulated transcripts and 78 downregulated transcripts showed significant functional enrichment, while in low ROS CD neutrophils 67 upregulated transcripts and 77 downregulated transcripts exhibited functional enrichment (Suppl. Table 10). Ontological enrichments of upregulated genes indicated a shared enhancement of HIF-1, TNF, NFKb, and TNF signaling with evidence of increasing reliance on fatty acids as a fuel source (lipid metabolism, fatty acid metabolic process, and response to fatty acid; Fig. 3A)). Additionally, metabolism and biosynthesis or reactive oxygen species and nitric oxide showed upregulation. Upregulated ontologies specific to low ROS CD neutrophils indicate mishandling of glucose (regulation of glucose import, response to insulin, insulin secretion, response to glucose) with further reliance on alternate fuel sources (NADP binding, ATP binding, response to monosaccharide, nucleotide sugar biosynthesis, fatty acid oxidation, lipid metabolism and biosynthesis). Shared down ontologies included indicators of mitochondrial dysfunction (OXPHOS, ATP synthesis coupled electron transport, electron transport chain, fatty acid oxidation) and enrichment of energy-related processes/pathways (glucose transport, carbohydrate metabolism, insulin secretion, and fatty acid/lipid processes (Fig. 3B). Downregulated processes specific to low ROS CD neutrophils included further indications of enhanced glucose dysregulation (carbohydrate homeostasis, glucose metabolism and response, and glucose transport), and antimicrobial pathways. Key pathways, enrichment values, and gene lists from glycolysis-trained functional enrichment analysis are presented in Suppl. Table 11.
Figure 3. Metabolic and Immune Functions Differentially Expressed in CD Neutrophils with Normal and Low ROS Production.
Candidate gene prioritization through functional enrichment analysis, using genes differentially regulated between normal ROS CD neutrophils and controls as the test set and a glycolysis gene list as the training set, ranked test genes in the order of their importance in context of glycolytic functions. Genes with significant glycolytic functional enrichment were submitted to ToppCluster to identify mechanistic enrichments. A) Pathways and mechanisms enriched in genes that were upregulated in low ROS CD neutrophils vs control neutrophils or normal ROS CD neutrophils vs control neutrophils with significant ranking in glycolysis candidate gene analysis (part of the first pipeline). B) Pathways and mechanisms enriched in genes that were downregulated in low ROS CD neutrophils vs control neutrophils or normal ROS CD neutrophils vs control neutrophils with significant ranking in glycolysis candidate gene analysis (part of the first pipeline). C) Pathways and mechanisms enriched in genes that were upregulated in low ROS CD neutrophils vs normal ROS CD neutrophils with significant ranking in glycolysis candidate gene analysis (second pipeline).
Pipeline 2: Low ROS CD neutrophils vs Normal ROS CD neutrophils
We identified 367 upregulated and 44 downregulated transcripts in low ROS CD neutrophils compared to normal ROS CD neutrophils (Suppl. Table 9). Ontologies enriched in the up- and down-regulated gene sets confirmed dysregulation of glucose and carbohydrate processes (polysaccharide biosynthesis, carbohydrate biosynthesis/metabolism, fatty acid activation, and ATP binding; Suppl. Fig. 7) and reliance on alternate energy sources (glucan and glycogen metabolism, and fatty acid beta-oxidation). As before, up- and down-regulated genes were assessed for functional overlap with 90 glycolysis genes, where we observed significance for 67 upregulated in low ROS genes (Suppl. Table 10). Functional analysis of these genes further implicate dysregulated glucose metabolism (carbohydrate catabolism/metabolism, glucose metabolism) paired with indicators of dependence on mitochondrial, non-glucose energy (OXPHOS, ATP binding, and fatty acid oxidation/biosynthesis; Fig. 3C). Key pathways, enrichment values, and gene lists from glycolysis-trained functional enrichment analysis are presented in Suppl. Table 11.
Core Genes Distinguishing Low ROS CD Neutrophils from Normal ROS CD Neutrophils
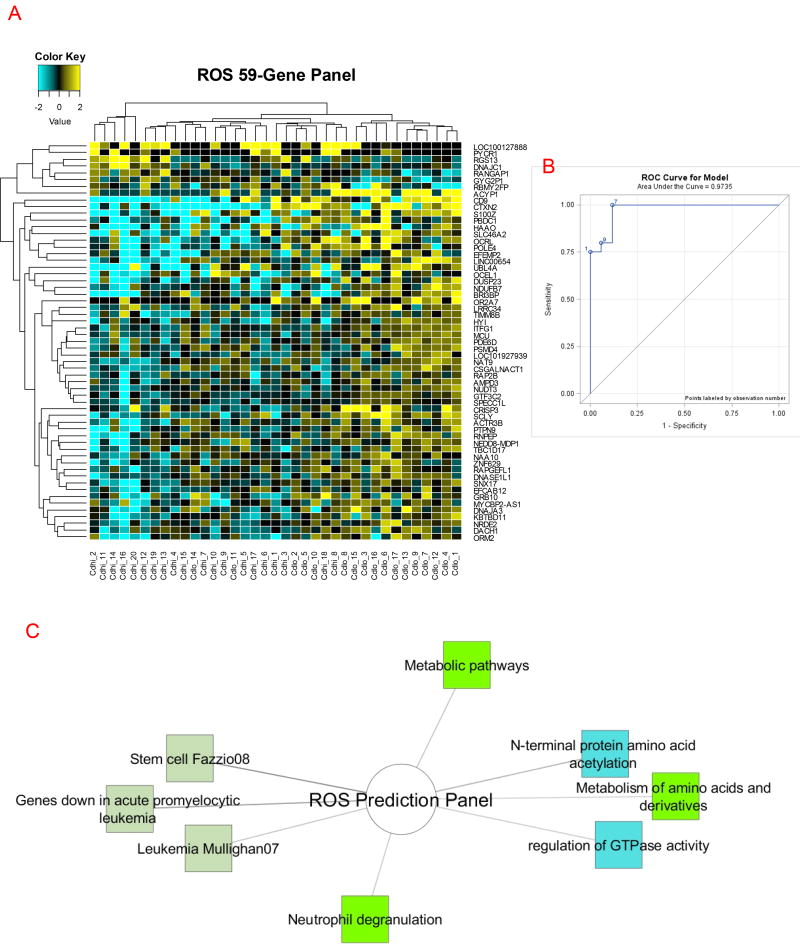
We directly compared low ROS CD neutrophils to normal ROS CD neutrophils using an unpaired T-test assuming unequal variance and identified a core gene panel of 59 genes (7 down- and 52 up-regulated genes with p<0.01 and FC>1.5; Supplemental Table 12). We generated a heatmap using hierarchical clustering of genes and samples (Figure 4A), and observed consistent separation of low and normal ROS CD neutrophils into three clusters. Two clusters with homogenous across-sample expression of the 59 genes segregated perfectly by ROS status, with a larger cluster exhibiting a distribution of low and high ROS CD neutrophils that correlated with the expression pattern’s transition between the two homogeneous clusters. Further, we tested the predictive power of the gene panel using a Support Vector Machine algorithm and found 94.1% accuracy, with AUC for ROC analysis of 0.97 (Fig. 4B). The 52 genes upregulated in Low ROS CD neutrophils were submitted for ontological enrichment analysis using ToppCluster.cchmc.org and Cytoscape, where we focused on pathways (bright green nodes), biological processes (blue nodes), and overlap with previously published gene sets from microarray studies (light green nodes; Figure 4C). We observe enrichments in amino acid metabolism, neutrophil degranulation, and overlap with gene sets from leukemia and stem cell microarray studies. When considering potentially damaging genetic variation in these genes, 50 of 59 genes have variants with CADD score of 10 or higher (Suppl. Table 13). In total, 368 variants were extracted, of which 275 are known dbSNP variants, while 93 are not in dbSNP and are therefore novel variants. These genetic variants may therefore also contribute to differences in neutrophil ROS production and antimicrobial function in CD.
Figure 4. Core Genes and Functions which Distinguish CD Neutrophils with Normal and Low ROS Production.
A 59-gene ROS panel was generated using an unpaired T-test to compared low ROS CD neutrophils to normal ROS CD neutrophils, assuming unequal variance with significance cutoffs p<0.05 and FC>1.5. A) Hierarchical clustering of the 59-gene panel predictive of Low ROS status in CD neutrophils generated using a Pearson’s Centered distance metric and Average linkage rule to cluster both samples and genes. B) ROC analysis of the discriminant power of the 59-gene panel. C) The 52 genes upregulated in low ROS CD neutrophils were submitted for ontological enrichment analysis through ToppCluster.cchmc.org and Cytoscape, with pathways (bright green), biological processes (bright blue), and previously published microarray gene sets (light green) presented.
Discussion
The development of intestinal inflammation in children with CGD has suggested that loss-of-function in neutrophil antimicrobial pathways is likely to be a fundamental mechanism of pediatric CD pathogenesis.2–5, 27 Rare variants in the CYBB, NCF1, NCF2 and NCF4 genes have previously been reported in VEO IBD.9, 10 We hypothesized that pediatric CD would be influenced by mutations affecting neutrophil ROS production. We identified a subset of patients characterized by low normal neutrophil ROS production, which was in turn associated with rare NADPH oxidase gene missense mutations, alterations in neutrophil gene expression pathways for glucose metabolism and anti-microbial responses, and a more aggressive disease course.
Genetic factors account for 20% to 35% of the variation in plasma hydrogen peroxide production.28 Resequencing analysis of CYBA, CYBB, NCF2, and NCF4 demonstrated patterns of genetic diversity compatible with pathogen-driven natural selection.29 This mechanism has also been demonstrated for CD risk genes including NOD2 and IL23R.7 We identified 26 potentially damaging mutations in CYBA, CYBB, NCF1, NCF2, and NCF4 in 543 pediatric IBD patients. The median (range) age at diagnosis for CD patients in our phenotypic analysis was 9(5–12) years, demonstrating that NADPH oxidase gene mutations have important consequences beyond the VEO (< 6 years) age group. We replicated a prior report of CYBB G364R, NCF1 R90H, NCF2 R38Q, NCF2 N419I, and NCF2 P454S variants in a study which included 122 VEOIBD patients.9, 10 Similarly, a study which utilized WES in 43 VEOIBD patients also reported the CYBB G364R and NCF1 G83R variants, defining an enrichment of deleterious rare variants in genes regulating innate immune function in VEOIBD.30 We replicate and extend these prior findings in defining genetic variation in the NADPH oxidase complex in older pediatric CD patients, suggesting a predominant role in regulating disease behavior.
We confirmed that homozygous carriage of the common CYBA Y72H missense mutation was associated with reduced ROS production.31–33 For each of the other missense mutations, only 1–3 patient samples were tested, with low ROS production detected for six of these. The frequency of minor allele carriage for CYBA V76M, NCF1 S99G, NCF2 R38Q, NCF2 T361S, NCF2 N419I, or NCF2 P454S was increased five-fold in CD subjects with low ROS production, supporting an important functional effect.34 Results agreed with those reported for reduced neutrophil ROS production in adult CD patients with NCF4 risk allele carriage, and VEOIBD patients with several of the same NCF2 mutations.9, 35 Further validation of functional effects of these NADPH oxidase gene variants will now be required, prioritized by the clinical and functional associations identified in the current study.
The majority of CD patients with low ROS production did not exhibit a potentially damaging variant in the core NADPH oxidase genes tested. This could be due to upstream variants in the FPR1 signaling pathway.26, 36 However, it is likely that additional mechanisms beyond protein coding mutations control the ROS response in CD. While several NADPH oxidase genes are regulated by expression quantitative trait loci (eQTL) in primary human neutrophils, our data did not support a difference in NADPH oxidase gene expression underlying differences in ROS production.17 Alternately, factors present in the injured gut such as lipid peroxidation products may also suppress neutrophil ROS production via post-translational modification of key enzymes.14
Many CGD patients develop severe distal colonic and rectal inflammation with perianal involvement.4, 5 A prior study in adult-onset CD linked reduced neutrophil ROS production to colon-only location.11 We found that CD patients with NADPH oxidase gene mutation carriage were three times more likely to exhibit perianal disease and experience stricturing complications. The frequency of abdominal surgery increased from 9% in CD patients with no variants to 31% in those carrying NADPH oxidase gene mutations. These differences in rates of surgery were not associated with overall differences in disease location or medication treatment exposures. These results are in agreement with the prior report in VEOIBD patients, which linked three of the same missense mutations in NCF2 to low ROS production.9, 10, 35 Moreover, the phenotype of more aggressive disease was also consistent with exacerbation of murine colitis previously reported with p40(phox) deficiency. This was attributed to defects in neutrophil dependent epithelial restitution.37 We have extended these prior findings to a larger group of patients, prioritizing several additional missense mutations for functional characterization, and showing an association with more aggressive small bowel, perianal, and colonic CD.
A prior study in adult onset CD also did not detect a reduction in overall neutrophil E. Coli killing, despite a similar reduction in ROS production.11 It should be noted that even the low ROS CD neutrophils in the current study did not display the profound reduction typically associated with CGD, and so it is not surprising that low ROS production alone was not associated with a reduction in Staph. Aureus killing. However, we did detect a modest reduction in bacterial killing associated with NADPH oxidase gene mutation carriage. ROS also modulate metabolic and cell signaling pathways which are intimately linked.15, 38, 39 Neutrophils are dependent upon glycolysis for mounting effective anti-microbial responses, and conversely ROS production is required to up-regulate glycolysis.14–16 We identified for the first time a suppression of gene expression pathways regulating glucose metabolism and anti-microbial responses in low ROS CD neutrophils. Deletion of Ncf2 in mice leads to hyper-activation of cell signaling pathways, via inactivation of ROS dependent tyrosine phosphatases. This may explain the somewhat paradoxical hyper-inflammatory state of CGD phagocytes, which may drive gut inflammation via inflammasome activation.38, 39 IL-1 receptor blockade has been shown to reduce inflammation in CGD.40 Whether this is also the case in low ROS CD neutrophils is not known, and is the subject of ongoing studies. Alternately, strategies to specifically boost glycolysis and associated antimicrobial responses may represent a novel approach for this subset of patients.
Our studies add to a growing body of literature regarding genetic variation and reduced epithelial and phagocyte ROS production in both pediatric and adult-onset IBD.34 A recent report characterized missense mutations in the intestinal epithelial NADPH oxidases DUOX2 and NOX1 in children with VEOIBD.41 We have replicated several missense mutations in core phagocyte NADPH oxidase genes previously reported in VEOIBD patients, and increased the number of validated mutations in this pathway linked to reduced ROS production in primary neutrophils.9 Importantly, NADPH oxidase gene mutations were associated with higher rates of refractory CD. These data advance our understanding of pathogenesis, and may inform novel therapeutic approaches including targeting of endogenous ROS regulators to boost cellular metabolic responses.42 However, routine genetic testing and screening for low ROS production at this stage should still likely be reserved for VEOIBD patients, or those with atypical features such as recurrent infections, in which CGD is suspected.
Supplementary Material
Acknowledgments
Grant Support: This work was supported by the Gene Analysis and Flow Cytometry cores of the National Institutes of Health (NIH)-supported Cincinnati Children’s Hospital Research Foundation Digestive Health Center (1P30DK078392-01), the Crohn’s and Colitis Foundation (LAD & SK), and NIH grant R01 DK098231 (LAD & SK) and P30 DK043351 (RJK).
Dr. Denson has received grant support from Janssen, and holds a patent for the use of GM-CSF auto-antibodies as a diagnostic test in IBD. Dr. Dubinsky has served as a consultant for Abbvie, Takeda, Janssen, and Pfizer and has received research support from Abbvie, Janssen, and Prometheus. Dr. Guthery has received research support in the last year from Regeneron Pharmaceuticals. Dr. Leleiko or his immediate family have equity interests in Celgene, Vericel, Ionis, Vertex, and Alnylam, and he has served as a consultant for CRICO.
The study sponsor did not play any role in the study design or in the collection, analysis, and interpretation of data.
Abbreviations
- B1
inflammatory behavior
- B2 or B3
stricturing or internal penetrating behavior
- CADD
Combined Annotation-Dependent Depletion
- CD
Crohn Disease
- CGD
Chronic Granulomatous Disease
- E1E2
left-sided Ulcerative Colitis
- E3E4
extensive Ulcerative Colitis
- eQTL
expression quantitative trait loci
- FMLP
N-formyl-methionyl-leucyl-phenylalanine
- FPR1
formyl peptide receptor 1
- GM-CSF
Granulocyte-Macrophage Colony Stimulating Factor
- IBD
Inflammatory Bowel Disease
- IQR
inter-quartile range
- L1
ileum-only location
- L2
colon-only location
- L3
ileo-colonic location
- MAF
minor allele frequency
- NADPH
nicotinamide-adenine dinucleotide phosphate
- PGA
physician’s global assessment
- PMA
phorbol myristate acetate
- ROC
receiver operator characteristic
- ROS
reactive oxygen species
- VEOIBD
very early onset Inflammatory Bowel Disease
- WES
whole exome sequencing
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest statement: The authors have no financial arrangement(s) with a company whose product figures prominently in the submitted manuscript or with a company making a competing product. The remaining authors have nothing to declare.
Competing Interest: None to declare
Author Contributions:
Study concept and design: LD, IJ, RK, DC, BA, YH, MZ, SK
Acquisition of data: LD, IJ, KS, DO, AD, KQ, AL, AP, RB, KL, KJ, TW, AG, RB, JN, JH, WC, BK, MH, SS, SG, MD, NL, AO, CS, MZ, SK
Analysis and interpretation of data: LD, IJ, YH, RK, KS, AD, DC, CS, MD, MZ, DO, BA, SK
Drafting of the manuscript: LD, IJ, RK, KS, SK
Critical revision of the manuscript for important intellectual content: LD, IJ, RK, KS, DC, RJX, MZ, JH, SK
Obtained funding: LD, RJX, SK
Administrative, technical, or material support: IJ, AL, AP, RB, KL, KJ, KS, AD, KM
Study supervision: LD, DC, MD, MZ, SK
Writing assistance: not applicable
References
- 1.Damen GM, van Krieken JH, Hoppenreijs E, et al. Overlap, common features, and essential differences in pediatric granulomatous inflammatory bowel disease. J Pediatr Gastroenterol Nutr. 2010;51:690–7. doi: 10.1097/MPG.0b013e3181dc0d73. [DOI] [PubMed] [Google Scholar]
- 2.Marciano BE, Rosenzweig SD, Kleiner DE, et al. Gastrointestinal involvement in chronic granulomatous disease. Pediatrics. 2004;114:462–8. doi: 10.1542/peds.114.2.462. [DOI] [PubMed] [Google Scholar]
- 3.Marks DJ, Miyagi K, Rahman FZ, et al. Inflammatory bowel disease in CGD reproduces the clinicopathological features of Crohn's disease. Am J Gastroenterol. 2009;104:117–24. doi: 10.1038/ajg.2008.72. [DOI] [PubMed] [Google Scholar]
- 4.Alvarez-Downing MM, Kamal N, Inchauste SM, et al. The role of surgery in the management of patients with refractory chronic granulomatous disease colitis. Dis Colon Rectum. 2013;56:609–14. doi: 10.1097/DCR.0b013e3182781504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Khangura SK, Kamal N, Ho N, et al. Gastrointestinal Features of Chronic Granulomatous Disease Found During Endoscopy. Clin Gastroenterol Hepatol. 2016;14:395–402. e5. doi: 10.1016/j.cgh.2015.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cutler DJ, Zwick ME, Okou DT, et al. Dissecting Allele Architecture of Early Onset IBD Using High-Density Genotyping. PLoS One. 2015;10:e0128074. doi: 10.1371/journal.pone.0128074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jostins L, Ripke S, Weersma RK, et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491:119–24. doi: 10.1038/nature11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rioux JD, Xavier RJ, Taylor KD, et al. Genome-wide association study identifies new susceptibility loci for Crohn disease and implicates autophagy in disease pathogenesis. Nat Genet. 2007;39:596–604. doi: 10.1038/ng2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dhillon SS, Fattouh R, Elkadri A, et al. Variants in nicotinamide adenine dinucleotide phosphate oxidase complex components determine susceptibility to very early onset inflammatory bowel disease. Gastroenterology. 2014;147:680–689. e2. doi: 10.1053/j.gastro.2014.06.005. [DOI] [PubMed] [Google Scholar]
- 10.Muise AM, Xu W, Guo CH, et al. NADPH oxidase complex and IBD candidate gene studies: identification of a rare variant in NCF2 that results in reduced binding to RAC2. Gut. 2012;61:1028–35. doi: 10.1136/gutjnl-2011-300078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hayee B, Rahman FZ, Tempero J, et al. The neutrophil respiratory burst and bacterial digestion in Crohn's disease. Dig Dis Sci. 2011;56:1482–8. doi: 10.1007/s10620-010-1426-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jurickova I, Collins MH, Chalk C, et al. Paediatric Crohn disease patients with stricturing behaviour exhibit ileal granulocyte-macrophage colony-stimulating factor (GM-CSF) autoantibody production and reduced neutrophil bacterial killing and GM-CSF bioactivity. Clin Exp Immunol. 2013;172:455–65. doi: 10.1111/cei.12076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Loftus RM, Finlay DK. Immunometabolism: Cellular Metabolism Turns Immune Regulator. J Biol Chem. 2016;291:1–10. doi: 10.1074/jbc.R115.693903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chacko BK, Wall SB, Kramer PA, et al. Pleiotropic effects of 4-hydroxynonenal on oxidative burst and phagocytosis in neutrophils. Redox Biol. 2016;9:57–66. doi: 10.1016/j.redox.2016.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Griffiths HR, Gao D, Pararasa C. Redox regulation in metabolic programming and inflammation. Redox Biol. 2017;12:50–57. doi: 10.1016/j.redox.2017.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thompson AA, Dickinson RS, Murphy F, et al. Hypoxia determines survival outcomes of bacterial infection through HIF-1alpha dependent re-programming of leukocyte metabolism. Sci Immunol. 2017;2 doi: 10.1126/sciimmunol.aal2861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Naranbhai V, Fairfax BP, Makino S, et al. Genomic modulators of gene expression in human neutrophils. Nat Commun. 2015;6:7545. doi: 10.1038/ncomms8545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brant SR, Okou DT, Simpson CL, et al. Genome-Wide Association Study Identifies African-Specific Susceptibility Loci in African Americans With Inflammatory Bowel Disease. Gastroenterology. 2017;152:206–217. e2. doi: 10.1053/j.gastro.2016.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kugathasan S, Denson LA, Walters TD, et al. Prediction of complicated disease course for children newly diagnosed with Crohn's disease: a multicentre inception cohort study. Lancet. 2017;389:1710–1718. doi: 10.1016/S0140-6736(17)30317-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johnston HR, Chopra P, Wingo TS, et al. PEMapper and PECaller provide a simplified approach to whole-genome sequencing. Proc Natl Acad Sci U S A. 2017;114:E1923–E1932. doi: 10.1073/pnas.1618065114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shetty AC, Athri P, Mondal K, et al. SeqAnt: a web service to rapidly identify and annotate DNA sequence variations. BMC Bioinformatics. 2010;11:471. doi: 10.1186/1471-2105-11-471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lek M, Karczewski KJ, Minikel EV, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–91. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen J, Bardes EE, Aronow BJ, et al. ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res. 2009;37:W305–11. doi: 10.1093/nar/gkp427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kaimal V, Bardes EE, Tabar SC, et al. ToppCluster: a multiple gene list feature analyzer for comparative enrichment clustering and network-based dissection of biological systems. Nucleic Acids Res. 2010;38:W96–102. doi: 10.1093/nar/gkq418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dorward DA, Lucas CD, Chapman GB, et al. The role of formylated peptides and formyl peptide receptor 1 in governing neutrophil function during acute inflammation. Am J Pathol. 2015;185:1172–84. doi: 10.1016/j.ajpath.2015.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dabritz J, Weinhage T, Varga G, et al. Reprogramming of monocytes by GM-CSF contributes to regulatory immune functions during intestinal inflammation. J Immunol. 2015;194:2424–38. doi: 10.4049/jimmunol.1401482. [DOI] [PubMed] [Google Scholar]
- 28.Lacy F, Kailasam MT, O'Connor DT, et al. Plasma hydrogen peroxide production in human essential hypertension: role of heredity, gender, and ethnicity. Hypertension. 2000;36:878–84. doi: 10.1161/01.hyp.36.5.878. [DOI] [PubMed] [Google Scholar]
- 29.Tarazona-Santos E, Machado M, Magalhaes WC, et al. Evolutionary dynamics of the human NADPH oxidase genes CYBB, CYBA, NCF2, and NCF4: functional implications. Mol Biol Evol. 2013;30:2157–67. doi: 10.1093/molbev/mst119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ostrowski J, Paziewska A, Lazowska I, et al. Genetic architecture differences between pediatric and adult-onset inflammatory bowel diseases in the Polish population. Sci Rep. 2016;6:39831. doi: 10.1038/srep39831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bedard K, Attar H, Bonnefont J, et al. Three common polymorphisms in the CYBA gene form a haplotype associated with decreased ROS generation. Hum Mutat. 2009;30:1123–33. doi: 10.1002/humu.21029. [DOI] [PubMed] [Google Scholar]
- 32.Taylor RM, Dratz EA, Jesaitis AJ. Invariant local conformation in p22phox p.Y72H polymorphisms suggested by mass spectral analysis of crosslinked human neutrophil flavocytochrome b. Biochimie. 2011;93:1502–9. doi: 10.1016/j.biochi.2011.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wyche KE, Wang SS, Griendling KK, et al. C242T CYBA polymorphism of the NADPH oxidase is associated with reduced respiratory burst in human neutrophils. Hypertension. 2004;43:1246–51. doi: 10.1161/01.HYP.0000126579.50711.62. [DOI] [PubMed] [Google Scholar]
- 34.O'Neill S, Brault J, Stasia MJ, et al. Genetic disorders coupled to ROS deficiency. Redox Biol. 2015;6:135–56. doi: 10.1016/j.redox.2015.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Somasundaram R, Deuring JJ, van der Woude CJ, et al. Linking risk conferring mutations in NCF4 to functional consequences in Crohn's disease. Gut. 2012;61:1097. doi: 10.1136/gutjnl-2011-301344. author reply 1097-8. [DOI] [PubMed] [Google Scholar]
- 36.Zhang Y, Syed R, Uygar C, et al. Evaluation of human leukocyte N-formylpeptide receptor (FPR1) SNPs in aggressive periodontitis patients. Genes Immun. 2003;4:22–9. doi: 10.1038/sj.gene.6363900. [DOI] [PubMed] [Google Scholar]
- 37.Conway KL, Goel G, Sokol H, et al. p40phox expression regulates neutrophil recruitment and function during the resolution phase of intestinal inflammation. J Immunol. 2012;189:3631–40. doi: 10.4049/jimmunol.1103746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kobayashi SD, Voyich JM, Braughton KR, et al. Gene expression profiling provides insight into the pathophysiology of chronic granulomatous disease. J Immunol. 2004;172:636–43. doi: 10.4049/jimmunol.172.1.636. [DOI] [PubMed] [Google Scholar]
- 39.Rieber N, Hector A, Kuijpers T, et al. Current concepts of hyperinflammation in chronic granulomatous disease. Clin Dev Immunol. 2012;2012:252460. doi: 10.1155/2012/252460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.de Luca A, Smeekens SP, Casagrande A, et al. IL-1 receptor blockade restores autophagy and reduces inflammation in chronic granulomatous disease in mice and in humans. Proc Natl Acad Sci U S A. 2014;111:3526–31. doi: 10.1073/pnas.1322831111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hayes P, Dhillon S, O'Neill K, et al. Defects in NADPH Oxidase Genes NOX1 and DUOX2 in Very Early Onset Inflammatory Bowel Disease. Cell Mol Gastroenterol Hepatol. 2015;1:489–502. doi: 10.1016/j.jcmgh.2015.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Graham DB, Becker CE, Doan A, et al. Functional genomics identifies negative regulatory nodes controlling phagocyte oxidative burst. Nat Commun. 2015;6:7838. doi: 10.1038/ncomms8838. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.