Abstract
The spatial distribution of genetic variation within proteins is shaped by evolutionary constraint and provides insight into the functional importance of protein regions and the potential pathogenicity of protein alterations. Here, we comprehensively evaluate the 3D spatial patterns of human germline and somatic variation in 6,604 experimentally derived protein structures and 33,144 computationally derived homology models covering 77% of all human proteins. Using a systematic approach, we quantify differences in the spatial distributions of neutral germline variants, disease-causing germline variants, and recurrent somatic variants. Neutral missense variants exhibit a general trend toward spatial dispersion, which is driven by constraint on core residues. In contrast, germline disease-causing variants are generally clustered in protein structures and form clusters more frequently than recurrent somatic variants identified from tumor sequencing. In total, we identify 215 proteins with significant spatial constraints on the distribution of disease-causing missense variants in experimentally derived protein structures, only 65 (30%) of which have been previously reported. This analysis identifies many clusters not detectable from sequence information alone; only 12% of proteins with significant clustering in 3D were identified from similar analyses of linear protein sequence. Furthermore, spatial analyses of mutations in homology-based structural models are highly correlated with those from experimentally derived structures, supporting the use of computationally derived models. Our approach highlights significant differences in the spatial constraints on different classes of mutations in protein structure and identifies regions of potential function within individual proteins.
Keywords: protein structure, genetic variation, spatial distribution, clustering, constraint, 3D model, evolution, ClinVar, COSMIC, gNOMAD
Introduction
Patterns of genetic variation along the human genome provide insight into functional and evolutionary constraints on different loci. A lack of common genetic variation in a locus is often indicative of functional constraint, suggesting that sequence changes negatively influence reproductive fitness.1 The first systematic examinations of fully sequenced human genomes established consistently stronger constraint (i.e., less genetic variation) in protein-coding regions compared to non-coding sequences.2, 3, 4, 5 Furthermore, early candidate gene-sequencing studies identified lower rates of non-synonymous variation than synonymous variation within protein-coding regions,6 highlighting the increased constraint on protein-altering mutations. Quantifying these patterns of constraint improved the ability to identify functional regions and interpret the phenotypic effects of genetic mutations.7, 8 Building on exome-sequencing data from tens of thousands of individuals, we are now able to quantify constraint on a large scale.
Recently developed methods have analyzed the frequency of variation in coding regions to provide estimates of gene-level constraint based on intolerance to variation.8, 9 However, the proteins encoded by these genes are often composed of multiple structural domains that perform distinct functions. Constraint on missense variation differs between structural domains; some are highly constrained, while others are more tolerant of variation.10, 11 Also, mutations to spatially distinct regions within the same protein often influence risk for different diseases.12 While gene-level approaches identify strongly constrained genes in which variation is likely pathogenic, these assessments do not identify specific protein regions and functions that are constrained and may overlook genes with different levels of constraint across their folded structures.
Analysis of the spatial distribution of missense variants in proteins can identify specific regions relevant to protein function and disease.13, 14 For example, structural analyses of tumor-derived somatic mutations have identified spatial clusters of mutations in many proteins.15, 16, 17, 18, 19 These clusters often overlap known functional regions of oncogenes and tumor suppressors and can assist in identifying functional driver mutations. Germline mutations also display non-random spatial patterns of constraint. Post-translational modification (PTM) sites cluster in 3D protein structures and constraint on germline variation at PTM sites is strongest in clustered PTMs.20 Protein-protein interaction (PPI) interfaces are also depleted for common missense variation,21 but enriched for disease-causing germline missense variation, in particular missense variants causing recessive disease.22 Several algorithms have recently been developed to identify somatic mutation hotspots, with some targeting heterogeneous clusters of multiple mutated sites15, 16 and others seeking small clusters of a few highly recurrent mutations.17, 18, 19 Most somatic mutation clustering approaches incorporate cancer-specific assumptions into their methodologies13 that limit application to other variants. Furthermore, these existing approaches have largely focused on finding clusters, rather than quantifying spatial constraint.
The recent abundance of human population-based sequencing studies2, 7, 23 paired with growth in the number of solved structures deposited in the Protein Data Bank (PDB) facilitates the systematic spatial analysis of functional constraint on naturally occurring germline and somatic variation in protein structure. In this article, we describe the comprehensive mapping of millions of human genetic variants into 6,604 experimentally derived structures and 33,144 computationally derived homology models of human proteins. We then introduce an analytical method for quantifying and comparing spatial distributions of genetic variation within protein space. The algorithm can be applied to any type of variation, identifies both significant clustering and dispersion of variants, and can incorporate relevant residue-level annotations as weights. Using this method, we identify significant differences between synonymous, missense, and pathogenic variation that reflect patterns of constraint on protein structure and function.
Material and Methods
Genetic Variant and Structural Datasets
We analyzed single-nucleotide variants (SNVs) from Genome Aggregation Database7 (gnomAD), ClinVar (01-07-2016), and COSMIC v.74. Variant consequences and annotations were determined using v82 of the Ensembl Variant Effect Predictor for genomic build GRCh37.24 Synonymous SNVs in gnomAD were included for comparison with gnomAD missense SNVs. All other datasets were filtered to include only missense SNVs. For all analyses involving gnomAD data, amino acids with median sequencing coverage less than 30× were identified.25 All variants mapped to those amino acids were excluded from all gnomAD analyses, and no variants were assigned to those amino acids during permutation.
Genetic variants were mapped into representative protein structures using Ensembl26 transcript models, which were matched with UniProt27 accession and Protein Data Bank28 (PDB, 01-07-2017) IDs using cross-reference tables provided by UniProt. PDB structures were included if they were determined through X-ray crystallography or solution NMR and contained at least 20 amino acids. Reference protein sequences were aligned with observed sequences in the PDB using SIFTS.29 Discrepancies were corrected by Needleman-Wunsch pairwise alignment with Biopython.30, 31 Computational homology models from ModBase32 (Human 2013 and 2016) were used to extend coverage of the proteome.
To reduce redundancy, each structural dataset was independently reduced to a minimally overlapping set of protein structures or homology models following an approach similar to Kamburov et al.16 For each dataset, we iteratively selected the structure/model that provided the greatest coverage of the target protein, omitting structures with >10% sequence overlap with the existing set. For structures/models with similar sequence coverage, we selected the highest-quality structure (by resolution for the PDB and the ModBase Quality Score for ModBase).
In comparisons between the PDB and ModBase, only structure-model pairs with >95% sequence overlap were included to limit the effects of sequence coverage on observed spatial differences. We also excluded models for which the solved structure was used as a template from the comparison. All other models in the minimally overlapping subset were used to extend coverage for spatial analyses.
The evolutionary conservation of each protein was quantified as the average residue level conservation of the protein across species as quantified by the Jensen-Shannon divergence applied to HSSP alignments.33 The tolerance of each protein to functional genetic variation was quantified by the residual variation intolerance score (RVIS).9 The evolutionary age of each protein was taken from the ProteinHistorian PPODv4_PTHR7-OrthoMCL_wagner1.0 database.34 The proportion of disorder per protein was calculated from disordered region annotations in MOBIdb.35 The relationship between spatial statistics and each feature was measured with linear regression analysis using the python package scipy.stats.linregress.
Quantifying and Comparing the Spatial Distributions of Protein-Coding Mutations
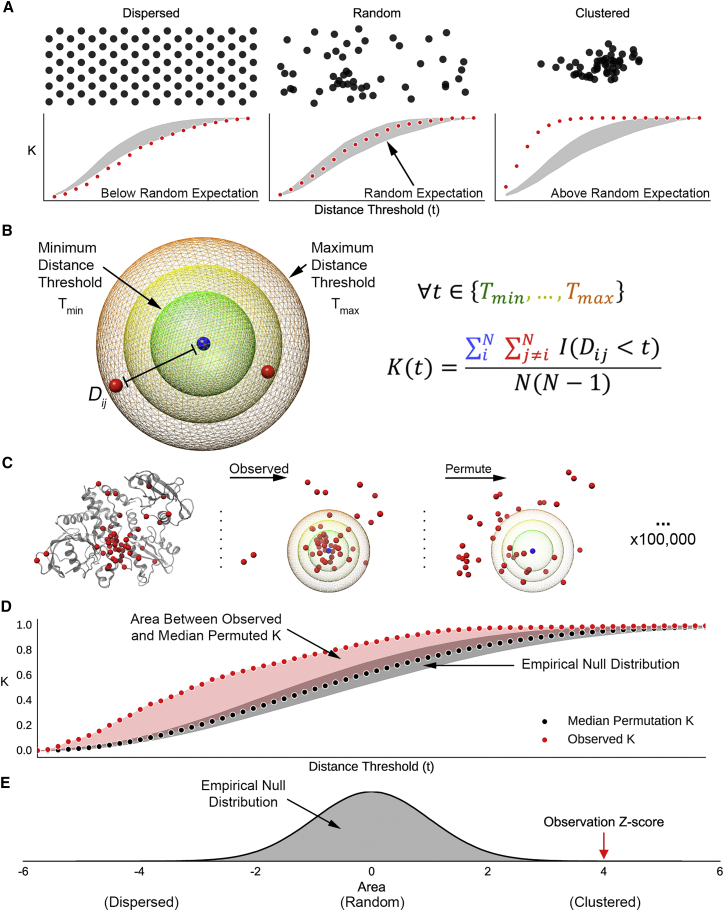
We developed a framework for evaluating hypotheses about the spatial distributions of genetic variants in protein structures based on Ripley’s K, a spatial descriptive statistic commonly used in ecology and epidemiology.36, 37, 38 Ripley’s K quantifies the spatial heterogeneity of a set of variants by comparing the proportion of variants within a given distance from one another to the expected proportion under a random spatial distribution. Variants are considered clustered if the proportion of neighbors exceeds the expectation and dispersed if the number of neighbors is lower than the expectation. K can be calculated across a range of distance thresholds (t), enabling the identification of clustering or dispersion at different scales (Figure 1A). We define K as
where N is the number of variants in the protein structure, Dij is the Euclidean distance between variants i and j, and I is an indicator function that evaluates to 1 when Dij is less than the distance threshold t and 0 otherwise. The denominator normalizes for the number of variant pairs considered. As a result, K can be interpreted as the proportion of variant pairs within distance t of one another. This normalization also allows for comparison between proteins with different variant counts. Distance thresholds larger than the approximate size of a functional domain (45Å for structures, 100 amino acids for sequence) were not considered. Variant positions were defined as the centroid of the reference amino acid (Figure 1B).
Figure 1.
Schematic of Our Framework for Evaluating the Spatial Distribution of Genetic Variants
(A) Spatial distributions can diverge from random in two ways; they may have fewer neighbors than expected by chance (dispersed) or more neighbors than expected by chance (clustered). Example distributions are illustrated in reference to a random spatial distribution in 2D. Below each set of points, the resulting K statistic at multiple distance thresholds (red) is plotted in reference to the expected K distribution under a random distribution (gray). K values below the range expected at random indicate dispersion, and K values above indicate clustering.
(B) Definition of the K statistic. For a range of distance thresholds (t), the number of variants neighboring each variant is computed and normalized by the total number of variant pairs. The indicator function I evaluates to 1 when two variants are neighbors (the distance between them [Dij] is less than t) and 0 otherwise.
(C) The observed K values are evaluated in reference to an empirical null distribution generated from 100,000 random permutations of variant locations within the protein structure.
(D) The spatial distribution trend for each protein is summarized by calculating the area between the observed K values (red points) and the median permuted K values (black points).
(E) This process is repeated for the K values resulting from each permuted set to generate an empirical null distribution. From this distribution, we calculate a Z-score and p value for the observed area. Positive Z-scores indicate clustering, negative Z-scores indicate dispersion, and Z-scores near zero indicate a lack of spatial constraint.
Missense variants can be observed only at the positions of amino acids in a protein structure, so complete spatial randomness is not a valid null model for randomly distributed variants (Figure 1C). To account for these constraints, we calculate an empirical null distribution of K through 100,000 random permutations of variant positions within the structure. Two-tailed p values are derived from the proportion of permuted K values more extreme than the observed K value. Lastly, Z-scores are calculated to quantify the direction (clustering or dispersion) and magnitude of the effect.
To evaluate the spatial distribution of real-valued attributes (e.g., evolutionary conservation and solvent accessibility), we compute a weighted form of the statistic, which we define as
where wj is the weight associated with protein position j. We evaluate the significance of the weighted K by permuting the weights over fixed amino acid positions and empirically computing p values as previously described. This statistic assesses whether the weights are spatially non-random (clustered or dispersed) beyond what is explained by their positions alone.
To summarize spatial patterns across distance scales into a protein-level summary statistic, we compute the area between the observed K curve and an empirical null K curve using Simpson’s rule (Figure 1D). This process is repeated for each round of permutations to generate an empirical null distribution. From this distribution, we calculate a permutation p value and Z-score for the area between observed and randomized K curves (Figure 1E). Positive Z-scores indicate clustering, negative Z-scores indicate dispersion, and Z-scores near zero indicate spatial randomness (e.g., a lack of spatial constraint). We control the false discovery rate (FDR) at 10% by computing q values from the protein-summary p value distribution in each analysis39 (see Web Resources). This summarization method captures the general spatial tendencies for each protein.
Automated Identification and Manual Review of Mutation Clustering in Previous Literature
To estimate the proportion of novel germline and somatic clustering patterns identified by our methodology, we performed an automated search and manual review of abstracts from PubMed. For each experimentally derived protein structure with significant clustering of ClinVar pathogenic or COSMIC recurrent somatic variants, we identified the primary citation from the Protein Data Bank for any solved structure of that protein, then queried all PubMed Central abstracts citing those publications. We filtered this set of abstracts to those containing cluster-related keywords. We then manually reviewed the remaining abstracts (N = 218) to assess whether they described a cluster of naturally occurring pathogenic variants within protein structure (Table S3). Clusters were not considered novel if either of the two expert reviewers flagged any abstract citing that protein structure.
Results
Quantifying Constraint on Spatial Patterns of Genetic Variation
We mapped genetic variants from three large variant datasets into a representative subset of 6,604 experimentally derived human protein structures from the Protein Data Bank28 (representing 5,209 distinct proteins) and 33,144 computationally derived homology models from ModBase32 (representing 17,984 distinct proteins). We considered the spatial distribution of 1,380,872 synonymous and 2,260,141 missense variants from exome sequencing of 138,632 diverse unrelated adults from the Genome Aggregation Database7 (gnomAD), 19,274 pathogenic and likely pathogenic missense variants from ClinVar,40 and 725,267 recurrent somatic missense variants (observed in at least two human tumor samples) from the Catalogue of Somatic Mutations in Cancer41 (COSMIC).
To quantify and contrast patterns of spatial constraint on different variant sets, we developed a statistic for evaluating deviations from a random spatial distribution based on Ripley’s K (see Material and Methods). Spatial distributions can diverge from random in two ways; variants may have fewer neighbors than expected by chance (dispersed) or more neighbors than expected by chance (clustered) (Figure 1A). This method identifies clustering and dispersion at any distance scale by quantifying the density of variation in increasingly larger neighborhoods (Figure 1B). To determine the significance of an observed variant distribution, we use a permutation procedure that accounts for the background distribution of amino acids in the protein structure (Figures 1C–1E; Material and Methods). From these permutations, we also derive a Z-score-based statistic that quantifies the magnitude of clustering (positive value) or dispersion (negative value) relative to random expectation (Figures 1D and 1E). This approach allows for direct comparisons across structurally distinct proteins. We required at least three variants from a dataset be present in a protein structure or model to be analyzed; we report the total number of structures and models meeting this criteria for each analysis.
To evaluate the use of homology models to extend structural coverage of the proteome, we compared the results from PDB and ModBase on shared proteins. We found that when both experimentally derived and computationally predicted structural models were available for a protein (>95% sequence overlap and excluding models for which the solved structure was used as a template; N = 3,316), the spatial analysis results were highly correlated (Figure S1). Relative to the PDB, the ModBase results displayed low recall but very high precision (Table S1). Thus, analysis of computational models often has less power but produces few false positives. For all analyses, we report the results on solved structures and predicted models separately. To reduce redundancy, the PDB-overlapping ModBase models were excluded from all other analyses.
Synonymous and Missense Variants Have Different Spatial Distributions
Synonymous genetic variants can have non-neutral effects, e.g., by influencing alternative splicing, mRNA stability, or translational efficiency; however, they ultimately result in an identical translated sequence for a given template mRNA and rarely influence the folded protein.42, 43 Thus, we hypothesized that synonymous variants are not subject to significant spatial constraint in protein structure. Consistent with this hypothesis, synonymous variants from gnomAD are nearly randomly distributed in protein structure (Figure 2A, PDB: median Z = 0.1, ModBase: median Z = 0.06) and deviated from a random distribution in only 2 of the 34,178 structures tested (PDB: 2P64 and 4RWT). These results were stable across distinct CATH structural architectures (Figure S2), indicating that synonymous variation is generally unconstrained in the context of protein structure.
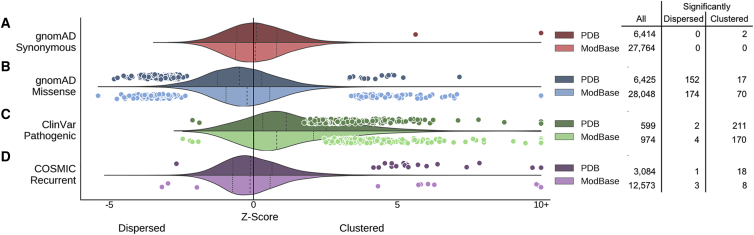
Figure 2.
Synonymous, Missense, and Disease-Associated Protein-Coding Variants Have Significantly Different Spatial Constraints
Each panel summarizes the spatial constraints on a different variant set. For each set, the distribution of summary Z-scores is plotted as a violin plot, with experimentally derived protein structures plotted above the center axis and computationally predicted homology models plotted below the center axis. The Z-scores of proteins with spatial distributions significantly different from random (by permutation, FDR < 0.1) are overlaid as points. Positive and negative Z-scores indicate clustering and dispersion, respectively. Summary statistics and all p values are provided in Table S2.
(A) Synonymous variants from gnomAD are approximately randomly distributed, as indicated by Z-score distributions with median near 0.
(B) In contrast, missense variants from gnomAD trend toward spatial dispersion, but many structures exhibit significant variant clustering.
(C) Pathogenic missense variants from ClinVar are the most strongly clustered variant set, with significant clustering in 381 structures/models.
(D) COSMIC recurrent somatic missense variants are also nearly randomly distributed, but 26 structures/models exhibit significant clustering.
In contrast, the spatial distribution of missense variants is constrained by the functional consequences of amino acid substitutions.13, 44, 45 Thus, we hypothesized that missense variants are non-randomly distributed within protein structure. In particular, we expected missense variants from gnomAD to be enriched in regions tolerant of amino acid substitution and depleted in regions of functional or structural importance. As expected, missense variants displayed significant constraint on their spatial distribution (Figure 2B). We identified 326 structural models with significant evidence of dispersed missense variants and 87 structural models with significant evidence of spatial clustering (Figure 2B, Table S1). There was a strong overall trend toward spatial dispersion (PDB: median Z = –0.49, ModBase: median Z = –0.22). Missense variation is therefore subject to significant spatial constraint within protein structure.
Previous analyses of missense variants reported enrichment for missense variants at the protein surface.44 Therefore, we hypothesized that the strong trend toward spatial dispersion of gnomAD missense variants is due to selective constraint against variation in the core residues of many proteins, which can destabilize the protein structure and disrupt function. We investigated the relationship between spatial dispersion and relative solvent accessibility (RSA) and found that residues with high RSA are significantly spatially dispersed (Figure S3). Furthermore, residues with neutral missense variants were more solvent accessible than residues overall and significantly dispersed missense variants were more solvent accessible than missense variants overall (Figure S4). In contrast, significantly clustered missense variants were no more or less solvent accessible than all residues, suggesting that clustered missense variants are found in many structural contexts and reflect intolerance to amino acid substitution in diverse structural domains. The significant spatial dispersion of missense variants demonstrates the prevalence of well-known patterns such as widespread constraint on the protein core and greater tolerance of missense variation at the protein surface.44
Germline Pathogenic Missense Variants Are Significantly Clustered in Protein Structure
Amino acids that are evolutionarily conserved across diverse species (and thus likely functional) are spatially constrained and significantly clustered within protein structure (Figure S5).46, 47 Because deleterious mutations often impact evolutionarily conserved amino acids44 and many studies have identified clustering of disease-causing mutations in specific proteins, we hypothesized that missense variants causing heritable diseases would commonly be spatially clustered. Indeed, germline pathogenic missense variants from ClinVar were the most clustered of all variant datasets analyzed (Figure 2C, PDB: median Z = 1.14, ModBase: median Z = 0.81); 35% of PDB structures (211 of 599) and 17% of ModBase models (170 of 974) with at least three ClinVar pathogenic variants exhibited significant clustering at FDR < 10%. Through automated search and manual review of the literature, we estimate that approximately 70% of the identified pathogenic clusters are previously unreported and may provide novel insight into disease mechanisms (Table S3).
Missense variants causing dominant and recessive diseases can usually be attributed to gain and loss of function, respectively.48 Protein sequence analyses have revealed that loss-of-function variants can disrupt numerous critical elements of a protein structure, while gain-of-function variants are limited to a smaller subset of regions with functional potential.48 We evaluated whether this relationship holds for protein structure using the dataset of dominant and recessive variants from the Human Gene Mutation Database (HGMD)49 curated by Turner et al.48 Both dominant and recessive variants are significantly clustered in structure (Figure S6); however, dominant variants are clustered at shorter distances (median peak significance: 8Å) than recessive variants (median peak significance: 14Å) indicating more focal clustering. The smaller clusters formed by dominant variants support the hypothesis that gain-of-function mutations are limited to specific sites with functional potential, while loss-of-function mutations more generally disrupt regions of functional importance. In summary, the frequent clustering of germline pathogenic missense variants underscores the spatial constraint on protein-coding variation and likely highlights regions of protein structures that are functionally and clinically relevant.
Recurrent Somatic Mutations Are Clustered in a Small Subset of Protein Structures
Several studies of tumor-derived somatic mutations have identified clustering in both sequence and structure that may highlight protein regions important for tumorigenesis.14, 15, 16, 17, 18, 19 We hypothesized that recurrent somatic mutations identified from tumor samples would exhibit patterns of spatial constraint similar to germline pathogenic missense variants. Surprisingly, we found that recurrent somatic mutations from COSMIC exhibited a weak overall trend toward spatial dispersion (Figure 2D; PDB: median Z = –0.11, ModBase: median Z = –0.12). Consistent with previous studies, we also identified significant clustering in a small fraction of protein structures (18 of 3,084, 0.6%) and models (12 of 9,346, 0.1%). This set consists of 25 unique proteins and includes many known cancer proteins,50 12 of which have been identified by at least one previous study of somatic mutation clustering,15, 16, 17, 18, 19 and one of which was identified from our manual review of the literature. To our knowledge, somatic mutation clustering in the remaining 12 proteins has not been previously reported: AR, CCDC160, COMP, CREBBP, DDX3X, ITLN2, MROH2B, PCDHAC1, SEZ6, SIRPA, SMO, and TET2 (Figure S7).
Neutral and Pathogenic Missense Variants Have Distinct Spatial Patterns
Given broad evidence of spatial constraint on both putatively neutral and pathogenic variants, we hypothesized that neutral and pathogenic distributions are spatially complementary—with functionally important regions depleted of neutral variants and enriched for pathogenic variants. To test this, we evaluated whether proteins with clustering (or dispersion) of neutral variants from gnomAD were also likely to exhibit clustering (or dispersion) of germline pathogenic variants from ClinVar (Figure 3A) or recurrent somatic mutations from COSMIC (Figure 3B).
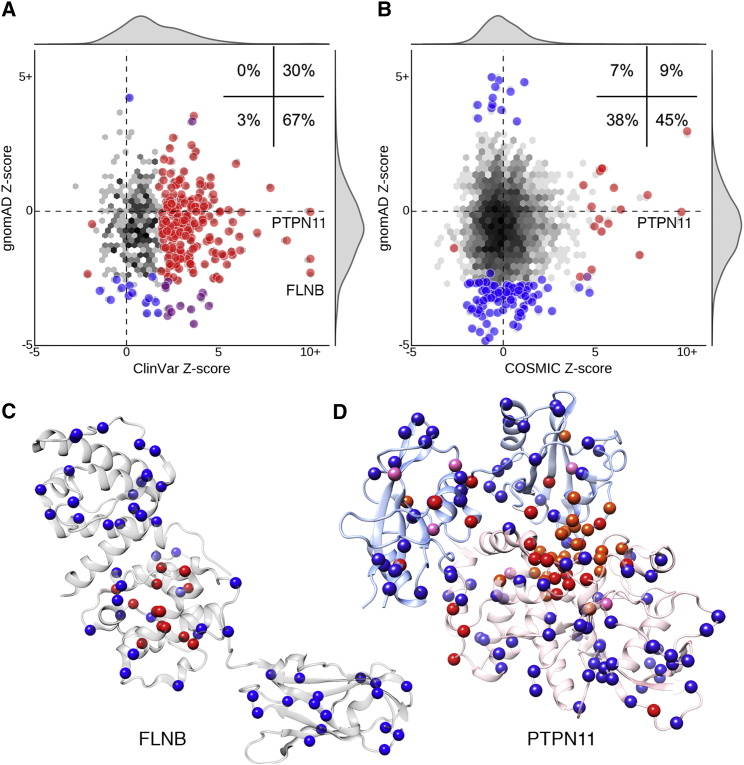
Figure 3.
Pathogenic and Neutral Missense Variants Have Distinct Spatial Distributions
(A and B) Comparison of the gnomAD missense Z-scores against ClinVar pathogenic (A) and COSMIC recurrent somatic (B) univariate Z-scores for experimentally derived protein structures. The inset reports the percentage of significant structures in each quadrant. The distribution over all structures is shown as a density plot, with black indicating higher density (log-scale). Large circles indicate structures with significant spatial distributions of either set of variants (two-sided permutation p value, FDR < 10%). Circles are colored red if the structure exhibits significant constraint on the variant set plotted on the x-axis, blue for significant contraint on the y-axis variant set, and purple if there is significant on both.
(C) Pathogenic variants (red) in FLNB (PDB: 4B7L) are clustered in the second calponin-homology domain, responsible for actin binding; neutral variants (blue) are distributed throughout the structure.
(D) Germline disease-causing (red) and recurrent somatic (pink) missense variants in PTPN11 (PDB: 5I6V) are clustered and frequently overlapping (orange) at the structural interface of the PTP (pink ribbon) and SH2 (blue ribbon) domains.
Over all proteins, there was no significant linear relationship between gnomAD-derived and ClinVar-derived Z-scores (Spearman’s rho = –0.02, p = 0.61; Figure 3A). The majority (67%) of proteins with significant evidence of spatial constraint exhibit clustering of germline pathogenic variation on a background of dispersed neutral variation (Figure 3A, lower right). Meanwhile, some (30%) exhibit significant germline pathogenic clustering on a background of modest neutral clustering, and a small fraction (3%) of proteins show trends toward significant dispersion of both. No protein exhibits significant clustering of neutral variants in the context of dispersed pathogenic variants.
Filamin-B (FLNB), a protein that links the cellular membrane to the actin cytoskeleton, illustrates the most common spatial pattern: dispersion of neutral missense variation and clustering of pathogenic missense variation. Pathogenic variation is clustered in the second calponin-homology (CH2) domain; CH2 is responsible for actin binding (Figure 3C). While complete loss of FLNB causes the recessive syndrome spondylocarpotarsal synostosis (SCT [MIM: 272460]), missense variants in the CH2 domain cause autosomal-dominant atelosteogenesis, types I and III (AO1 [MIM: 108720], AO3 [MIM: 108721]), and Larsen syndrome (LRS [MIM: 150250]). Missense variants in CH2 have been shown to increase actin binding affinity, suggesting a gain-of-function disease mechanism.51 The spatial dispersion of neutral missense variants indicates that substitutions to the core of the protein are likely destabilizing and thus may cause FLNB loss of function.
There was also no significant linear relationship between gnomAD-derived and COSMIC-derived Z-scores (Spearman’s rho = 0.02, p = 0.20; Figure 3B). As for germline variants, the most common scenario was significantly clustered recurrent somatic mutations on a background of dispersed neutral variation (45%), but significantly clustered recurrent somatic mutations rarely coincided with significant neutral missense variant distributions (Figure 3B, right). For example, recurrent somatic mutations in PTPN11 (MIM: 176876), which encodes the protein tyrosine-protein phosphatase non-receptor type 11 (SHP-2), are clustered at the structural interface between the protein tyrosine phosphatase (PTP) and Src-homology 2 (SH2) domains (Figure 3D). Germline pathogenic missense variants at this interface are associated with LEOPARD syndrome (LPRD1 [MIM: 151100]), Noonan syndrome (NS1 [MIM: 163950]), and increased risk for juvenile myelomonocytic leukemia (JMML [MIM: 607785]). Somatic mutations to PTPN11 are often found in leukemias and several solid tumors.52 The relative orientation of the PTP and SH2 domains determines whether SHP-2 is in its active or inactive state. Disease-causing mutations have been shown to disrupt the interaction interface, with mutations causing NS1 leading to a more energetically favorable active state relative to wild-type53 (gain-of-function) and mutations causing LPRD1 resulting in an inactive state54 (dominant negative). It has been proposed that the association with Noonan syndrome may be mediated by disruption of a cluster of phosphorylation sites.20 Despite significant clustering of germline and somatic pathogenic variants, neutral missense variants in SHP-2 are randomly spatially distributed throughout the structure. Overall, these results demonstrate consistent, uncorrelated differences in the spatial constraint on neutral missense and pathogenic variants, indicating that when considered broadly across all proteins, patterns of neutral variation are not strongly predictive of the spatial constraint on known pathogenic variants.
Analysis of Protein Structure Reveals Significant Patterns of Spatial Constraint Not Identified from Protein Sequence
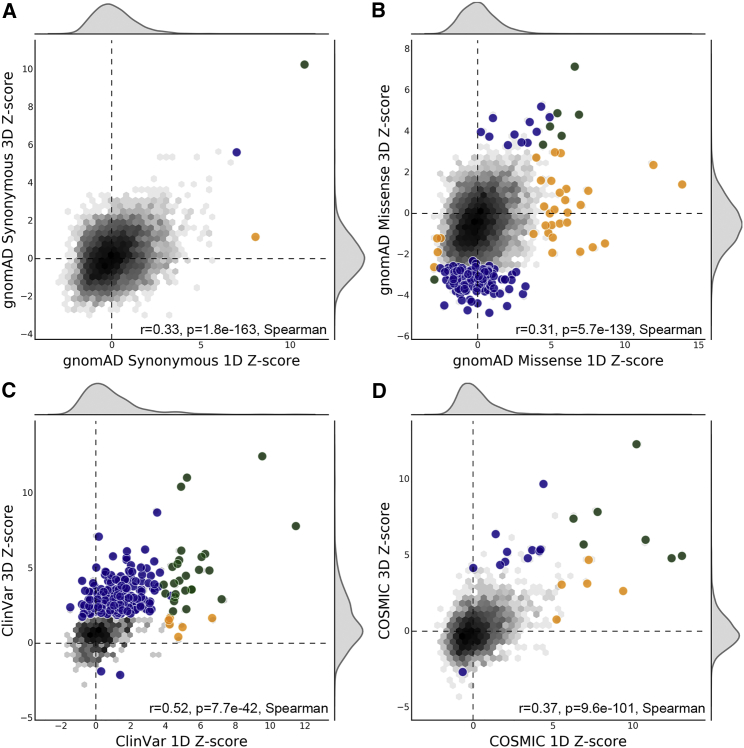
Experimentally derived protein structures are available for approximately 22% of human proteins. Computationally derived homology models expand coverage (of at least part of the protein) to 77%, but there are thousands of human proteins for which we do not have reliable structural information. The linear protein sequence is available for all proteins but does not represent the functional context of the protein. Thus, we hypothesized that significant spatial patterns within the three-dimensional protein structure may not be identifiable from protein sequence alone. We repeated our analysis using the protein sequence of each experimentally derived protein structure to compute the linear K statistic and measured the overall correlation and predictive performance compared to structure-based K analyses. There is little overlap in the proteins identified as significantly constrained by each analysis (Table 1). Sequence-based analyses of missense variation recalled at most 37% of the significant spatial patterns identified in protein structure, suggesting that many significant spatial patterns in protein structure are introduced by protein folding. Conversely, the observed precision in each analysis (between 0.18 and 0.81) indicates that significant spatial patterns of variants in protein sequence are often disrupted in the folded protein structure. Overall, the statistics for sequence and structure are correlated (Spearman’s rho between 0.31 and 0.52), but proteins without significant constraint in either sequence or structure drive this pattern (Figure 4). These results demonstrate that sequence-based analyses do not accurately predict significant spatial constraint on missense variation in protein structure.
Table 1.
Protein Sequence Is a Poor Predictor of Spatial Patterns in Protein Structure
| N |
Significant Proteins |
Performance |
||||
|---|---|---|---|---|---|---|
| Structure | Sequence | Both | Precision | Recall | ||
| gnomAD synonymous | 6,413 | 2 | 2 | 1 | 0.50 | 0.50 |
| gnomAD missense | 6,425 | 169 | 38 | 7 | 0.18 | 0.04 |
| ClinVar pathogenic | 589 | 213 | 32 | 26 | 0.81 | 0.12 |
| COSMIC recurrent | 3,052 | 19 | 12 | 7 | 0.58 | 0.37 |
Structural analysis identified more significant constraint than sequence analysis for all missense variant datasets. Precision and recall were calculated by treating structure-derived results as truth and sequence-derived results as predictions.
Figure 4.
Protein Sequence Is a Poor Predictor of Spatial Patterns in Protein Structure
The Ripley’s K Z-score for significant spatial constraint on each protein in the PDB set computed over its 3D structure is contrasted with the K Z-score computed using its 1D sequence for each variant dataset: (A) gnomAD synonymous, (B) gnomAD missense, (C) ClinVar, and (D) COSMIC. Axes are scaled independently for each comparison. The distribution over all structures is shown as a density plot, with black indicating higher density. Large circles indicate structures with spatial distributions significantly different from random; circles are colored blue if significant in the structural analysis, yellow if significant in the sequence analysis, and green if significant in both analyses. The sequence- and structure-derived Z-scores are correlated for each variant dataset (Spearman’s rho between 0.31 and 0.52), but sequence analysis identified very few proteins with significant spatial distributions in protein structure (Table 1).
Variant Spatial Patterns Are Similar across Proteins with Different Evolutionary Origins, Tolerance to Variation, and Amounts of Disorder
Many functional, evolutionary, and structural factors could influence the distribution of genetic variants across protein structures. To evaluate the impact of such factors, we used linear regression analysis to quantify the relationship between the spatial distribution of variants in a protein and its (1) evolutionary origin, (2) residue-level conservation across species, (3) intolerance to variation in humans, and (4) amount of structural disorder. The spatial distributions observed show very little association with the evolutionary history of the proteins considered (Figures S9A–S11A); the greatest proportion of variance explained (R2) in the spatial statistics by any evolutionary metric is only 0.009 by intolerance to variation (as quantified by RVIS) with germline missense variants. Though the magnitudes of all the associations are very small, a few achieve statistical significant due to the large sample size. Neutral missense variants are slightly more constrained in proteins with markers of functional importance: evolutionary conservation (R2 = 0.004; p = 3.17 × 10−42) and protein intolerance to variation (R2 = 0.009; p = 2.57 × 10−14). Pathogenic missense variants are not significantly associated with any of these evolutionary metrics. Furthermore, the significant spatial patterns observed in our variant analyses held when analyzing proteins at the extremes of these evolutionary metrics (Figures S9B–S11B). Thus, the trends in the spatial patterns of genetic variation identified here are present across proteins with diverse evolutionary origins and levels of genetic variation.
Many proteins contain dynamic mobile regions that may not adopt a single stable structural conformation.55, 56 These disordered regions are often critical to protein function but may not be present in or accurately represented by available PDB and ModBase structures. To evaluate the influence of protein disorder on our spatial analyses, we calculated the proportion of each protein annotated as disordered by MOBIdb35 and tested its correlation with spatial patterns. The amount of disordered sequence is not substantially correlated with our spatial metrics; the greatest proportion of variance explained was only 0.002 for recurrent somatic variants (Figure S12A). Due to the large sample size, these modest effects achieved statistical significance for synonymous (R2 = 0.0008; p = 0.0025) and recurrent (R2 = 0.002; p = 0.001) somatic variants. Furthermore, the overall spatial patterns are similar across proteins with high and low disorder (Figure S12B). This suggests that our observations are robust to differences in levels of disorder. With the increasing understanding of mobile and disordered protein regions, adapting our spatial statistics to account for disorder is a promising area for future work.
Discussion
By projecting millions of variants observed in human populations into three-dimensional protein structures, we comprehensively quantified patterns of spatial constraint on human genetic variation within its functional and evolutionary context. As expected, synonymous variants are nearly randomly distributed within protein structures. In contrast, missense variants exhibit significant dispersion in some proteins and significant clustering in others, reflecting the diversity of constraints on protein structure and function. The spatial dispersion of missense variants is often driven by intolerance to substitutions in the protein core. Germline pathogenic missense variants display evidence of spatial clustering in more than three quarters of protein structures and models, and hundreds of proteins exhibit significantly more variant clustering than expected in the absence of constraint. In contrast, significant clustering of recurrent somatic mutations was identified in relatively few proteins. Finally, we demonstrate that protein sequence is a poor substitute for protein structure in the analysis of variant spatial distributions in 3D and that our findings are robust to differences in protein evolutionary origins, overall levels of genetic variation, and the amount of protein disorder.
Several studies have examined the spatial clustering of somatic mutations within protein structures.15, 16, 17, 18, 19 The number of proteins exhibiting somatic mutation clustering varies between studies: Kamburov et al. identified only 17 proteins with significant somatic clustering, while Meyer et al. identified 75 proteins with high-scoring somatic clusters (Figure S7). Our analysis of the Protein Data Bank and ModBase identified 25 proteins with significantly clustered recurrent somatic mutations from COSMIC, of which 12 had been previously identified. The variation between methods is attributable to differences in many aspects of the studies, including the algorithms, mutation cluster definitions, limits on cluster size, and the genetic and structural datasets considered. Prior approaches focused on the identification of clusters of somatic variants, and thus they may not have identified other patterns of spatial constraint, such as dispersion. Key advances of our approach to characterizing spatial distributions include identification of both significant clustering and dispersion (at any scale) compared to an appropriate null distribution and avoiding domain-specific assumptions. As a result, our method captures additional patterns of spatial constraint on genetic variation over all proteins. This may consequently reduce its power to identify some somatic mutation clusters detected by cancer-focused approaches, in particular those that detect clusters of two highly recurrent mutations. However, we note that our method identifies a similar number of proteins as other studies aimed at identifying proteins with significant overall clustering of somatic mutations15, 16 (Figure S7).
The mutation datasets considered also influence the power of different methods to detect spatial patterns. For our analysis of somatic mutations in cancer, we selected the COSMIC dataset for consistency with our use of ClinVar, a submission-based database of pathogenic germline variants, and to maximize the number of available variants for analysis. However, the use of a submission-based system introduces the potential for reporting bias into the representation of proteins and mutations. In contrast, the Cancer Genome Atlas (TCGA) provides consistent, whole-exome sequencing data from many cancer studies and tumor types but has smaller sample size. We attempted to analyze recurrent somatic mutations from 18 TCGA studies in solved protein structures, but most structures did not satisfy our inclusion criteria (three or more recurrent somatic mutations), so we instead analyzed all somatic TCGA mutations. We identified three proteins with significant clustering (including two known cancer proteins, TP53 and STK11). There was no significant difference in the overall distribution of COSMIC and TCGA results (Figure S8), suggesting that bias in the COSMIC dataset did not critically affect our overall findings.
The stronger clustering of germline disease-causing variation compared to recurrent somatic variants may reflect differences in spatial constraint and phenotypic effects of variation outside of the germline.12 There are likely differences in variant tolerance between germline and somatic contexts; germline variants are present in all tissues and are subject to many powerful constraints throughout development. In contrast, somatic variants influence only a subset of tissues and developmental time points and thus may be tolerated in contexts that would be lethal in the germline.12 Alternatively, germline and somatic differences may be attributable to relaxed constraint within the tumor context, which is already highly dysregulated. While we limited our analyses to recurrent somatic mutations (observed in multiple tumors), this dataset likely still contains some neutral passenger mutations, which may further explain the overall similarity between the somatic and neutral missense variant results.
By characterizing both clustering and dispersion, we identified spatial patterns of genetic variation that have not been previously described. For example, our comparative analysis identified 3% of proteins with significant spatial dispersion of both neutral and pathogenic germline missense variants. This interesting group of proteins includes enzymes, activators, chaperones, and inhibitors with many intermolecular interactions. Furthermore, these proteins harbor variants associated with reduced rather than abolished activity, which may be related to their frequent annotation as likely loss-of-function intolerant genes.7
These and other spatial patterns we detected provide a useful perspective from which to study protein function and the phenotypic effects of coding variation; however, there are limitations to our approach. First, high-quality protein structural information is available for only ∼25% of human proteins, and available protein structures often do not cover the entire protein sequence, leaving much of the proteome inaccessible to spatial analyses. Computationally derived homology models extend partial coverage to 77% of human proteins, and these models are often sufficiently accurate to enable evaluation of spatial patterns of variation. However, there is still bias in the proteins available for structural analysis. For example, it is more difficult to experimentally determine the structure of membrane proteins than soluble proteins, reducing both the number of solved protein structures and the availability of structural templates for homology modeling.57 Intrinsically disordered proteins are also less represented within structural databases, due to their lack of a stable tertiary structure. Structural models are also often lacking for the multiple isoforms known to exist for many proteins. When this information is available, our methodology can contrast patterns in alternative isoforms, different 3D conformations, and protein complexes, but our current analyses focus on a minimally overlapping subset of protein structures and homology models representing canonical isoforms of human proteins. These structures are only a subset of the dynamic and biologically relevant conformations adopted by proteins. Nonetheless, they are informative representations of the functional context of missense variation, and by analyzing them, we identified significant spatial patterns that were not found in analyses of linear sequence.
Another challenge is the incomplete knowledge of all pathogenic variants within a protein. We used germline disease-causing missense variants from the curated ClinVar database, a submission-based resource that may also include some incorrect disease assignments. Most variants in ClinVar are linked to rare Mendelian diseases, and thus may represent an extreme that does not generalize to variants influencing complex diseases. We anticipate that mapping pathogenic variants across homologous protein families, and potentially even from model organisms, will significantly increase the number of human proteins with sufficient numbers of variants for spatial analysis. It will also be valuable to examine the spatial distribution of protein-coding mutations associated with complex disease.
Finally, we consider missense variants from the gnomAD dataset to be putatively neutral. Although gnomAD excludes individuals with severe pediatric disease and is not enriched for pathogenic variants,7 the dataset likely does include variants that contribute to late-onset and complex diseases. Nonetheless, this variant set reflects the largest population-level assessment of coding sequence variation, and the resulting comparisons are a representative, comprehensive, and informative quantification of spatial patterns of genetic variation in protein structure.
In summary, we provide a consistent statistical framework in which to identify significant constraint on genetic variation in protein structures and identify significant differences in the spatial distribution of synonymous, non-synonymous, and pathogenic protein-coding variation. We identify hundreds of proteins with significant clustering of germline disease-causing missense variants, the majority of which have not been previously reported in the literature. Structural analysis of these spatial clusters has the potential to uncover previously unknown disease etiologies and suggest potential drug targets. More broadly, our results indicate that selective constraint influences the spatial distribution of missense variation in protein structures and support the use of large reference datasets to highlight regions of functional importance and disease relevance.
To facilitate further analyses, we provide ASTRID, a web-interface for viewing the structural locations of all gnomAD, ClinVar, and COSMIC variants, along with the results of all spatial analyses, in the representative set of 6,604 experimentally derived human protein structures and 33,144 computationally derived homology models (see Web Resources).
Acknowledgments
R.M.S. was supported by the NIH (T32 EY021453) and a SPORE grant from the Vanderbilt-Ingram Cancer Center. J.M. was supported by the NIH (R01 GM080403, R01 GM099842, R01 HL122010). W.S.B. was supported by the NIH (U54 AG052427, UF01 AG07133). J.A.C. was supported by institutional funds and a Vanderbilt Ambassadors Discovery Grant in Cancer Research. This work was conducted in part using the resources of the Advanced Computing Center for Research and Education at Vanderbilt University, Nashville, TN. These funding bodies had no part in the design of the study or collection, analysis, and interpretation of data or in writing the manuscript. We thank Jonathan Sheehan and Greg Sliwoski for helpful discussions. The authors would like to thank the Genome Aggregation Database (gnomAD) and the groups that provided exome and genome variant data to this resource. A full list of contributing groups can be found at http://gnomad.broadinstitute.org/about.
Published: February 15, 2018
Footnotes
Supplemental Data include 12 figures and 3 tables and can be found with this article online at https://doi.org/10.1016/j.ajhg.2018.01.017.
Contributor Information
William S. Bush, Email: wsb36@case.edu.
John A. Capra, Email: tony.capra@vanderbilt.edu.
Web Resources
ASTRID, http://astrid.icompbio.net
OMIM, http://www.omim.org/
qvalue, http://github.com/nfusi/qvalue
RCSB Protein Data Bank, http://www.rcsb.org/pdb/home/home.do
Supplemental Data
References
- 1.Bustamante C.D., Fledel-Alon A., Williamson S., Nielsen R., Hubisz M.T., Glanowski S., Tanenbaum D.M., White T.J., Sninsky J.J., Hernandez R.D. Natural selection on protein-coding genes in the human genome. Nature. 2005;437:1153–1157. doi: 10.1038/nature04240. [DOI] [PubMed] [Google Scholar]
- 2.Abecasis G.R., Altshuler D., Auton A., Brooks L.D., Durbin R.M., Gibbs R.A., Hurles M.E., McVean G.A., 1000 Genomes Project Consortium A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boyko A.R., Williamson S.H., Indap A.R., Degenhardt J.D., Hernandez R.D., Lohmueller K.E., Adams M.D., Schmidt S., Sninsky J.J., Sunyaev S.R. Assessing the evolutionary impact of amino acid mutations in the human genome. PLoS Genet. 2008;4:e1000083. doi: 10.1371/journal.pgen.1000083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tennessen J.A., Bigham A.W., O’Connor T.D., Fu W., Kenny E.E., Gravel S., McGee S., Do R., Liu X., Jun G., Broad GO. Seattle GO. NHLBI Exome Sequencing Project Evolution and functional impact of rare coding variation from deep sequencing of human exomes. Science. 2012;337:64–69. doi: 10.1126/science.1219240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fu W., O’Connor T.D., Jun G., Kang H.M., Abecasis G., Leal S.M., Gabriel S., Rieder M.J., Altshuler D., Shendure J., NHLBI Exome Sequencing Project Analysis of 6,515 exomes reveals the recent origin of most human protein-coding variants. Nature. 2013;493:216–220. doi: 10.1038/nature11690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cargill M., Altshuler D., Ireland J., Sklar P., Ardlie K., Patil N., Shaw N., Lane C.R., Lim E.P., Kalyanaraman N. Characterization of single-nucleotide polymorphisms in coding regions of human genes. Nat. Genet. 1999;22:231–238. doi: 10.1038/10290. [DOI] [PubMed] [Google Scholar]
- 7.Lek M., Karczewski K.J., Minikel E.V., Samocha K.E., Banks E., Fennell T., O’Donnell-Luria A.H., Ware J.S., Hill A.J., Cummings B.B., Exome Aggregation Consortium Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Samocha K.E., Robinson E.B., Sanders S.J., Stevens C., Sabo A., McGrath L.M., Kosmicki J.A., Rehnström K., Mallick S., Kirby A. A framework for the interpretation of de novo mutation in human disease. Nat. Genet. 2014;46:944–950. doi: 10.1038/ng.3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Petrovski S., Wang Q., Heinzen E.L., Allen A.S., Goldstein D.B. Genic intolerance to functional variation and the interpretation of personal genomes. PLoS Genet. 2013;9:e1003709. doi: 10.1371/journal.pgen.1003709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peterson T.A., Nehrt N.L., Park D., Kann M.G. Incorporating molecular and functional context into the analysis and prioritization of human variants associated with cancer. J. Am. Med. Inform. Assoc. 2012;19:275–283. doi: 10.1136/amiajnl-2011-000655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nehrt N.L., Peterson T.A., Park D., Kann M.G. Domain landscapes of somatic mutations in cancer. BMC Genomics. 2012;13(Suppl 4):S9. doi: 10.1186/1471-2164-13-S4-S9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lahiry P., Torkamani A., Schork N.J., Hegele R.A. Kinase mutations in human disease: interpreting genotype-phenotype relationships. Nat. Rev. Genet. 2010;11:60–74. doi: 10.1038/nrg2707. [DOI] [PubMed] [Google Scholar]
- 13.Porta-Pardo E., Kamburov A., Tamborero D., Pons T., Grases D., Valencia A., Lopez-Bigas N., Getz G., Godzik A. Comparison of algorithms for the detection of cancer drivers at subgene resolution. Nat. Methods. 2017;14:782–788. doi: 10.1038/nmeth.4364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Araya C.L., Cenik C., Reuter J.A., Kiss G., Pande V.S., Snyder M.P., Greenleaf W.J. Identification of significantly mutated regions across cancer types highlights a rich landscape of functional molecular alterations. Nat. Genet. 2016;48:117–125. doi: 10.1038/ng.3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stehr H., Jang S.-H.J., Duarte J.M., Wierling C., Lehrach H., Lappe M., Lange B.M.H. The structural impact of cancer-associated missense mutations in oncogenes and tumor suppressors. Mol. Cancer. 2011;10:54. doi: 10.1186/1476-4598-10-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kamburov A., Lawrence M.S., Polak P., Leshchiner I., Lage K., Golub T.R., Lander E.S., Getz G. Comprehensive assessment of cancer missense mutation clustering in protein structures. Proc. Natl. Acad. Sci. USA. 2015;112:E5486–E5495. doi: 10.1073/pnas.1516373112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meyer M.J., Lapcevic R., Romero A.E., Yoon M., Das J., Beltrán J.F., Mort M., Stenson P.D., Cooper D.N., Paccanaro A., Yu H. mutation3D: cancer gene prediction through atomic clustering of coding variants in the structural proteome. Hum. Mutat. 2016;37:447–456. doi: 10.1002/humu.22963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tokheim C., Bhattacharya R., Niknafs N., Gygax D.M., Kim R., Ryan M., Masica D.L., Karchin R. Exome-scale discovery of hotspot mutation regions in human cancer using 3D protein structure. Cancer Res. 2016;76:3719–3731. doi: 10.1158/0008-5472.CAN-15-3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Niu B., Scott A.D., Sengupta S., Bailey M.H., Batra P., Ning J., Wyczalkowski M.A., Liang W.-W., Zhang Q., McLellan M.D. Protein-structure-guided discovery of functional mutations across 19 cancer types. Nat. Genet. 2016;48:827–837. doi: 10.1038/ng.3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reimand J., Wagih O., Bader G.D. Evolutionary constraint and disease associations of post-translational modification sites in human genomes. PLoS Genet. 2015;11:e1004919. doi: 10.1371/journal.pgen.1004919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nishi H., Nakata J., Kinoshita K. Distribution of single-nucleotide variants on protein-protein interaction sites and its relationship with minor allele frequency. Protein Sci. 2016;25:316–321. doi: 10.1002/pro.2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guo Y., Wei X., Das J., Grimson A., Lipkin S.M., Clark A.G., Yu H. Dissecting disease inheritance modes in a three-dimensional protein network challenges the “guilt-by-association” principle. Am. J. Hum. Genet. 2013;93:78–89. doi: 10.1016/j.ajhg.2013.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Abecasis G.R., Auton A., Brooks L.D., DePristo M.A., Durbin R.M., Handsaker R.E., Kang H.M., Marth G.T., McVean G.A., 1000 Genomes Project Consortium An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McLaren W., Pritchard B., Rios D., Chen Y., Flicek P., Cunningham F. Deriving the consequences of genomic variants with the Ensembl API and SNP Effect Predictor. Bioinformatics. 2010;26:2069–2070. doi: 10.1093/bioinformatics/btq330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bentley D.R., Balasubramanian S., Swerdlow H.P., Smith G.P., Milton J., Brown C.G., Hall K.P., Evers D.J., Barnes C.L., Bignell H.R. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;456:53–59. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cunningham F., Amode M.R., Barrell D., Beal K., Billis K., Brent S., Carvalho-Silva D., Clapham P., Coates G., Fitzgerald S. Ensembl 2015. Nucleic Acids Res. 2015;43:D662–D669. doi: 10.1093/nar/gku1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.UniProt Consortium UniProt: a hub for protein information. Nucleic Acids Res. 2015;43:D204–D212. doi: 10.1093/nar/gku989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N., Weissig H., Shindyalov I.N., Bourne P.E. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Velankar S., Dana J.M., Jacobsen J., van Ginkel G., Gane P.J., Luo J., Oldfield T.J., O’Donovan C., Martin M.-J., Kleywegt G.J. SIFTS: structure integration with function, taxonomy and sequences resource. Nucleic Acids Res. 2013;41:D483–D489. doi: 10.1093/nar/gks1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cock P.J.A., Antao T., Chang J.T., Chapman B.A., Cox C.J., Dalke A., Friedberg I., Hamelryck T., Kauff F., Wilczynski B., de Hoon M.J. Biopython: freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics. 2009;25:1422–1423. doi: 10.1093/bioinformatics/btp163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Needleman S.B., Wunsch C.D. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J. Mol. Biol. 1970;48:443–453. doi: 10.1016/0022-2836(70)90057-4. [DOI] [PubMed] [Google Scholar]
- 32.Pieper U., Webb B.M., Barkan D.T., Schneidman-Duhovny D., Schlessinger A., Braberg H., Yang Z., Meng E.C., Pettersen E.F., Huang C.C. ModBase, a database of annotated comparative protein structure models, and associated resources. Nucleic Acids Res. 2011;39:D465–D474. doi: 10.1093/nar/gkq1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Capra J.A., Singh M. Predicting functionally important residues from sequence conservation. Bioinformatics. 2007;23:1875–1882. doi: 10.1093/bioinformatics/btm270. [DOI] [PubMed] [Google Scholar]
- 34.Capra J.A., Williams A.G., Pollard K.S. ProteinHistorian: tools for the comparative analysis of eukaryote protein origin. PLoS Comput. Biol. 2012;8:e1002567. doi: 10.1371/journal.pcbi.1002567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Piovesan D., Tabaro F., Paladin L., Necci M., Mičetić I., Camilloni C., Davey N., Dosztányi Z., Mészáros B., Monzon A.M. MobiDB 3.0: more annotations for intrinsic disorder, conformational diversity and interactions in proteins. Nucleic Acids Res. 2017;46:D471–D476. doi: 10.1093/nar/gkx1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dixon P.M. Ripley’s K function. Encycl. Environmetrics. 2002;3:1796–1803. [Google Scholar]
- 37.Gaines K.F., Bryan A.L., Dixon P.M. The effects of drought on foraging habitat selection of breeding wood storks in coastal Georgia. Waterbirds. 2000;23:64–73. [Google Scholar]
- 38.Diggle P.J., Chetwynd A.G. Second-order analysis of spatial clustering for inhomogeneous populations. Biometrics. 1991;47:1155–1163. [PubMed] [Google Scholar]
- 39.Storey J.D., Tibshirani R. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. USA. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Landrum M.J., Lee J.M., Benson M., Brown G., Chao C., Chitipiralla S., Gu B., Hart J., Hoffman D., Hoover J. ClinVar: public archive of interpretations of clinically relevant variants. Nucleic Acids Res. 2016;44(D1):D862–D868. doi: 10.1093/nar/gkv1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Forbes S.A., Beare D., Gunasekaran P., Leung K., Bindal N., Boutselakis H., Ding M., Bamford S., Cole C., Ward S. COSMIC: exploring the world’s knowledge of somatic mutations in human cancer. Nucleic Acids Res. 2015;43:D805–D811. doi: 10.1093/nar/gku1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hunt R.C., Simhadri V.L., Iandoli M., Sauna Z.E., Kimchi-Sarfaty C. Exposing synonymous mutations. Trends Genet. 2014;30:308–321. doi: 10.1016/j.tig.2014.04.006. [DOI] [PubMed] [Google Scholar]
- 43.Sauna Z.E., Kimchi-Sarfaty C. Understanding the contribution of synonymous mutations to human disease. Nat. Rev. Genet. 2011;12:683–691. doi: 10.1038/nrg3051. [DOI] [PubMed] [Google Scholar]
- 44.de Beer T.A., Laskowski R.A., Parks S.L., Sipos B., Goldman N., Thornton J.M. Amino acid changes in disease-associated variants differ radically from variants observed in the 1000 genomes project dataset. PLoS Comput. Biol. 2013;9:e1003382. doi: 10.1371/journal.pcbi.1003382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gong S., Blundell T.L. Structural and functional restraints on the occurrence of single amino acid variations in human proteins. PLoS ONE. 2010;5:e9186. doi: 10.1371/journal.pone.0009186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schueler-furman O., Baker D. Conserved residue clustering and protein structure prediction. Proteins. 2003;52:225–235. doi: 10.1002/prot.10365. [DOI] [PubMed] [Google Scholar]
- 47.Madabushi S., Yao H., Marsh M., Kristensen D.M., Philippi A., Sowa M.E., Lichtarge O. Structural clusters of evolutionary trace residues are statistically significant and common in proteins. J. Mol. Biol. 2002;316:139–154. doi: 10.1006/jmbi.2001.5327. [DOI] [PubMed] [Google Scholar]
- 48.Turner T.N., Douville C., Kim D., Stenson P.D., Cooper D.N., Chakravarti A., Karchin R. Proteins linked to autosomal dominant and autosomal recessive disorders harbor characteristic rare missense mutation distribution patterns. Hum. Mol. Genet. 2015;24:5995–6002. doi: 10.1093/hmg/ddv309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stenson P.D., Ball E.V., Mort M., Phillips A.D., Shiel J.A., Thomas N.S.T., Abeysinghe S., Krawczak M., Cooper D.N. Human Gene Mutation Database (HGMD): 2003 update. Hum. Mutat. 2003;21:577–581. doi: 10.1002/humu.10212. [DOI] [PubMed] [Google Scholar]
- 50.Futreal P.A., Coin L., Marshall M., Down T., Hubbard T., Wooster R., Rahman N., Stratton M.R. A census of human cancer genes. Nat. Rev. Cancer. 2004;4:177–183. doi: 10.1038/nrc1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sawyer G.M., Clark A.R., Robertson S.P., Sutherland-Smith A.J. Disease-associated substitutions in the filamin B actin binding domain confer enhanced actin binding affinity in the absence of major structural disturbance: Insights from the crystal structures of filamin B actin binding domains. J. Mol. Biol. 2009;390:1030–1047. doi: 10.1016/j.jmb.2009.06.009. [DOI] [PubMed] [Google Scholar]
- 52.Chakravarty D., Gao J., Phillips S.M., Kundra R., Zhang H., Wang J., Rudolph J.E., Yaeger R., Soumerai T., Nissan M.H. OncoKB: a precision oncology knowledge base. JCO Precis. Oncol. 2017;1:1–16. doi: 10.1200/PO.17.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tartaglia M., Mehler E.L., Goldberg R., Zampino G., Brunner H.G., Kremer H., van der Burgt I., Crosby A.H., Ion A., Jeffery S. Mutations in PTPN11, encoding the protein tyrosine phosphatase SHP-2, cause Noonan syndrome. Nat. Genet. 2001;29:465–468. doi: 10.1038/ng772. [DOI] [PubMed] [Google Scholar]
- 54.Kontaridis M.I., Swanson K.D., David F.S., Barford D., Neel B.G. PTPN11 (Shp2) mutations in LEOPARD syndrome have dominant negative, not activating, effects. J. Biol. Chem. 2006;281:6785–6792. doi: 10.1074/jbc.M513068200. [DOI] [PubMed] [Google Scholar]
- 55.Dyson H.J., Wright P.E. Intrinsically unstructured proteins and their functions. Nat. Rev. Mol. Cell Biol. 2005;6:197–208. doi: 10.1038/nrm1589. [DOI] [PubMed] [Google Scholar]
- 56.Oldfield C.J., Dunker A.K. Intrinsically disordered proteins and intrinsically disordered protein regions. Annu. Rev. Biochem. 2014;83:553–584. doi: 10.1146/annurev-biochem-072711-164947. [DOI] [PubMed] [Google Scholar]
- 57.Carpenter E.P., Beis K., Cameron A.D., Iwata S. Overcoming the challenges of membrane protein crystallography. Curr. Opin. Struct. Biol. 2008;18:581–586. doi: 10.1016/j.sbi.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.