Abstract
Genetic variation modulating risk of sporadic Parkinson disease (PD) has been primarily explored through genome-wide association studies (GWASs). However, like many other common genetic diseases, the impacted genes remain largely unknown. Here, we used single-cell RNA-seq to characterize dopaminergic (DA) neuron populations in the mouse brain at embryonic and early postnatal time points. These data facilitated unbiased identification of DA neuron subpopulations through their unique transcriptional profiles, including a postnatal neuroblast population and substantia nigra (SN) DA neurons. We use these population-specific data to develop a scoring system to prioritize candidate genes in all 49 GWAS intervals implicated in PD risk, including genes with known PD associations and many with extensive supporting literature. As proof of principle, we confirm that the nigrostriatal pathway is compromised in Cplx1-null mice. Ultimately, this systematic approach establishes biologically pertinent candidates and testable hypotheses for sporadic PD, informing a new era of PD genetic research.
Keywords: mouse, Parkinson disease, substantia nigra, single-cell RNA sequencing, dopaminergic neurons, genome-wide association studies, Complexin 1, gene regulatory networks
Introduction
The most commonly used genetic tool today for studying complex disease is the genome-wide association study (GWAS). As a strategy, GWASs were initially hailed for the insight they might provide into the genetic architecture of common human disease risk. Indeed, the collective data from GWASs since 2005 have revealed a trove of variants and genomic intervals associated with an array of phenotypes.1 The majority of variants identified in GWASs are located in non-coding DNA2 and are enriched for characteristics denoting regulatory DNA.2, 3 This regulatory variation is expected to impact expression of a nearby gene, leading to disease susceptibility.
Traditionally, the gene closest to the lead SNP has been prioritized as the gene most likely to be affected by the disease variation. However, recent studies show that disease-associated variants can act on more distally located genes, invalidating genes that were previously extensively studied.4, 5 The inability to systematically connect common variation with the genes impacted limits our capacity to elucidate potential therapeutic targets and can waste valuable research efforts.
Although GWASs are inherently agnostic to the context in which disease-risk variation acts, the biological impact of common functional variation has been shown to be cell context dependent.2, 6 Extending these observations, Pritchard and colleagues recently demonstrated that although genes need only to be expressed in disease-relevant cell types to contribute to risk, those expressed preferentially or exclusively therein contribute more per SNP.7 Thus, accounting for the cellular and gene regulatory network (GRN) contexts within which variation acts may better inform the identification of impacted genes. These principles have not yet been applied systematically to many of the traits for which GWAS data exist. We have chosen Parkinson disease (PD) as a model complex disorder for which a significant body of GWAS data remains to be explored biologically in a context-dependent manner.
PD is the most common progressive neurodegenerative movement disorder. Incidence of PD increases with age, affecting an estimated 1% worldwide beyond 70 years of age.8, 9 The genetic underpinnings of non-familial or sporadic PD have been studied through the use of GWASs with recent meta-analyses highlighting 49 loci associated with sporadic PD susceptibility.10, 11 While a small fraction of PD GWAS loci contain genes known to be mutated in familial PD (SNCA and LRRK2),12, 13 most indicted intervals do not contain a known mutated gene or genes. Although PD ultimately affects multiple neuronal centers, preferential degeneration of DA neurons in the SN leads to functional collapse of the nigrostriatal pathway and loss of fine motor control. The preferential degeneration of SN DA neurons in relation to other mesencephalic DA neurons has driven research interest in the genetic basis of selective SN vulnerability in PD. Consequently, one can reasonably assert that a significant fraction of PD-associated variation likely mediates its influence specifically within the SN.
In an effort to illuminate a biological context in which PD GWAS results could be better interpreted, we undertook single-cell RNA-seq (scRNA-seq) analyses of multiple DA neuronal populations in the brain, including ventral midbrain DA neurons. This analysis defined the heterogeneity of DA populations over developmental time in the brain, revealing gene expression profiles specific to discrete DA neuron subtypes. These data further facilitated the definition of GRNs active in DA neuron populations including the SN. With these data, we establish a framework to systematically prioritize candidate genes in all 49 PD GWAS loci and begin exploring their pathological significance.
Material and Methods
Animals
The Th:EGFP BAC transgenic mice (Tg(Th-EGFP)DJ76Gsat/Mmnc) used in this study were generated by the GENSAT Project and were purchased through the Mutant Mouse Resource & Research Centers (MMRRC) Repository. Mice were maintained on a Swiss Webster (SW) background with female SW mice obtained from Charles River Laboratories. The Tg(Th-EGFP)DJ76Gsat/Mmnc line was primarily maintained through matings between Th:EGFP-positive, hemizygous male mice and wild-type SW females (dams). Timed matings for cell isolation were similarly established between hemizygous male mice and wild-type SW females. The observation of a vaginal plug was defined as embryonic day 0.5 (E0.5). All work involving mice (husbandry, colony maintenance, and euthanasia) were reviewed and pre-approved by the institutional care and use committee.
Cplx1 knockout mice and wild-type littermates used for immunocytochemistry were taken from a colony established in Cambridge using founders from mutant mouse lines that were obtained from the Max-Planck-Institute for Experimental Medicine (Gottingen, Germany). Cplx1 mice in this colony have been backcrossed onto a C57BL/6J inbred background for at least ten generations. All experimental procedures were licensed and undertaken in accordance with the regulations of the UK Animals (Scientific Procedures) Act 1986. Housing, rearing, and genotyping of mice has been described in detail previously.14, 15 Mice were housed in hard-bottomed polypropylene experimental cages in groups of 5–10 mice in a housing facility maintained at 21°C–23°C with relative humidity of 55% ± 10%. Mice had ad libitum access to water and standard dry chow. Because homozygous knockout Cplx1 mice have ataxia, they have difficulty in reaching the hard pellets in the food hopper and drinking from the water bottles. Lowered waterspouts were provided and access to normal laboratory chow was improved by providing mash (made by soaking 100 g of chow pellets in 230 mL water for 60 min until the pellets were soft and fully expanded) on the floor of the cage twice daily. Cplx1 genotyping to identify mice with a homozygous (Cplx1−/−) or heterozygous (Cplx1+/−) deletion of Cplx1 was conducted as previously described,14 using DNA prepared from tail biopsies.
Dissection of Embryonic 15.5 (E15.5) Brains
At 15.5 days after the timed mating, pregnant dams were euthanized and the entire litter of E15.5 embryos were dissected out of the mother and immediately placed in chilled Eagle’s Minimum Essential Media (EMEM). Individual embryos were then decapitated and heads were placed in fresh EMEM on ice. Embryonic brains were removed and placed in Hank’s Balanced Salt Solution (HBSS) without Mg2+ and Ca2+ and manipulated while on ice. The brains were immediately observed under a fluorescent stereomicroscope and EGFP+ brains were selected. EGFP+ regions of interest in the forebrain (hypothalamus) and the midbrain were then dissected and placed in HBSS on ice. This process was repeated for each EGFP+ brain. Brain regions from four EGFP+ mouse pups were pooled together for dissociation.
Dissection of Postnatal Day 7 (P7) Brains
After timed matings, pregnant females were sorted into their own cages and checked daily for newly born pups. The morning the pups were born was considered postnatal day 0 (P0). Once the mice were aged to P7, all the mice from the litter were euthanized and the brains were then quickly dissected and placed in HBSS without Mg2+ and Ca2+ on ice. As before, the brains were observed under a fluorescent microscope, EGFP+ status for P7 mice was determined, and EGFP+ brains were retained. For each EGFP+ brain, the entire olfactory bulb was first resected and placed in HBSS on ice. Immediately thereafter, the EGFP+ forebrain and midbrain regions for each brain were resected and also placed in distinct containers of HBSS on ice. Brain regions from five EGFP+ P7 mice were pooled together for dissociation.
Generation of Single-Cell Suspensions from Brain Tissue
Resected brain tissues were dissociated using papain (Papain Dissociation System, Worthington Biochemical Corporation; Cat#: LK003150) following the trehalose-enhanced protocol reported by Saxena et al.16 with the following modifications. The dissociation was carried out at 37°C in a sterile tissue culture cabinet and RNase inhibitor was added to all solutions. During dissociation, all tissues at all time points were triturated every 10 min using a sterile Pasteur pipette. For E15.5 tissues, this was continued for no more than 40 min. For P7, this was continued for up to 1.5 hr or until the tissue appeared to be completely dissociated.
Additionally, for P7 tissues, after dissociation but before cell sorting, the cell pellets were passed through a discontinuous density gradient in order to remove cell debris that could impede cell sorting. This gradient was adapted from the Worthington Papain Dissociation System kit. Briefly, after completion of dissociation according to the Saxena protocol,16 the final cell pellet was resuspended in DNase dilute albumin-inhibitor solution, layered on top of 5 mL of albumin-inhibitor solution, and centrifuged at 70 × g for 6 min. The supernatant was then removed.
Fluorescence-Activated Cell Sorting (FACS) and Single-Cell Collection
For each time point-region condition, pellets were resuspended in 200 μL of media without serum comprised of DMEM/F12 without phenol red, 5% trehalose (w/v), 25 μM AP-V, 100 μM kynurenic acid, and 10 μL of 40 U/μL RNase inhibitor (RNasin Plus RNase Inhibitor, Promega) at room temperature. The resuspended cells were then passed through a 40 μM filter and introduced into a FACS machine (Beckman Coulter MoFlo Cell Sorter or Becton Dickinson FACSJazz). Viable cells were identified via propidium iodide staining, and individual neurons were sorted based on their fluorescence directly into lysis buffer in individual wells of 96-well plates for single-cell sequencing (2 μL Smart-Seq2 lysis buffer + RNase inhibitor, 1 μL oligo-dT primer, and 1 μL dNTPs) according to Picelli et al.17 Blank wells were used as negative controls for each plate collected. Upon completion of a sort, the plates were briefly spun in a tabletop microcentrifuge and snap-frozen on dry ice. Single-cell lysates were subsequently kept at −80°C until cDNA conversion.
Single-Cell Reverse Transcription, Library Prep, and Sequencing
Library preparation and amplification of single-cell samples were performed using a modified version of the Smart-Seq2 protocol.17 Briefly, 96-well plates of single cell lysates were thawed to 4°C, heated to 72°C for 3 min, then immediately placed on ice. Template switching first-strand cDNA synthesis was performed as described above using a 5′-biotinylated TSO oligo. cDNAs were amplified using 20 cycles of KAPA HiFi PCR and 5′-biotinylated ISPCR primer. Amplified cDNA was cleaned with a 1:1 ratio of Ampure XP beads and approximately 200 pg was used for a one-quarter standard sized Nextera XT tagmentation reaction. Tagmented fragments were amplified for 14 cycles and dual indexes were added to each well to uniquely label each library. Concentrations were assessed with Quant-iT PicoGreen dsDNA Reagent (Invitrogen) and samples were diluted to ∼2 nM and pooled. Pooled libraries were sequenced on the Illumina HiSeq 2500 platform to a target mean depth of ∼8.0 × 105 50-bp paired-end fragments per cell at the Hopkins Genetics Research Core Facility.
RNA Sequencing and Alignment
For all libraries, paired-end reads were aligned to the mouse reference genome (mm10) supplemented with the Th-EGFP+ transgene contig, using HISAT218 with default parameters except: -p 8. Aligned reads from individual samples were quantified against a reference transcriptome (GENCODE vM8)19 supplemented with the addition of the EGFP transcript. Quantification was performed using cuffquant20 with default parameters and the following additional arguments:–no-update-check –p 8. Normalized expression estimates across all samples were obtained using cuffnorm20 with default parameters.
Single-Cell RNA Data Analysis
Expression Estimates
Gene-level and isoform-level FPKM (fragments per kilobase of transcript per million) values produced by cuffquant20 and the normalized FPKM matrix from cuffnorm were used as input for the Monocle 2 single-cell RNA-seq framework21 in R/Bioconductor.22 Genes were annotated using the Gencode vM8 release.19 A CellDataSet (cds) was then created using Monocle 2 (v2.2.0)21 containing the gene FPKM table, gene annotations, and all available metadata for the sorted cells. All cells labeled as negative controls and empty wells were removed from the data. Relative FPKM values for each cell were converted to estimates of absolute mRNA counts per cell (RPC) using the Monocle 2 Census algorithm23 using the Monocle function “relative2abs().” After RPCs were inferred, a new cds was created using the estimated RNA copy numbers with the expression Family set to “negbinomial.size()” and a lower detection limit of 0.1 RPC.
QC Filtering
After expression estimates were inferred, the cds containing a total of 473 cells was run through Monocle 2's “detectGenes()” function with the minimum expression level set at 0.1 transcripts. The following filtering criteria were then imposed on the entire dataset:
-
(1)
Number of expressed genes: The number of expressed genes detected in each cell in the dataset was plotted and the high and low expressed gene thresholds were set based on observations of each distribution. Only those cells that expressed between 2,000 and 10,000 genes were retained.
-
(2)
Cell mass: Cells were then filtered based on the total mass of RNA in the cells calculated by Monocle 2. Again, the total mass of the cell was plotted and mass thresholds were set based on observations from each distribution. Only those cells with a total cell mass between 100,000 and 1,300,000 fragments mapped were retained.
-
(3)
Total RNA copies per cell: Cells were then filtered based on the total number of RNA transcripts estimated for each cell. Again, the total RNA copies per cell was plotted and RNA transcript thresholds were set based on observations from each distribution. Only those cells with a total mRNA count between 1,000 and 40,000 RPCs were retained.
A total of 410 individual cells passed these initial filters. Outliers found in subsequent, reiterative analyses described below were analyzed and removed, resulting in a final cell number of 396.
Log Distribution QC
Analysis using Monocle 2 relies on the assumption that the expression data being analyzed follows a log-normal distribution. Comparison to this distribution was performed after initial filtering prior to continuing with analysis and was observed to be well fit.
Reiterative Single-Cell RNA Data Analysis
After initial filtering described above, the entire cds as well as subsets of the cds based on “age” and “region” of cells were created for recursive analysis. Regardless of how the data were subdivided, all data followed a similar downstream analysis workflow.
Determining Number of Cells Expressing Each Gene
The genes to be analyzed for each iteration were filtered based on the number of cells that expressed each gene. Genes were retained if they were expressed in >5% of the cells in the dataset being analyzed. These were designated “expressed_genes.” For example, when analyzing all cells collected together (n = 410), a gene had to be expressed in 20.5 cells (410 × 0.05 = 20.5) to be included in the analysis. In contrast, when analyzing P7 MB cells (n = 80), a gene had to be expressed in just four cells (80 × 0.05 = 4). This was done to include genes that may define rare populations of cells that could be present in any given population.
Monocle Model Preparation
The data were prepared for Monocle analysis by retaining only the expressed genes that passed the filtering described above. Size factors were estimated using the Monocle 2 “estimateSizeFactors()” function. Dispersions were estimated using the “estimateDispersions()” function.
High Variance Gene Selection
Genes that have a high biological coefficient of variation (BCV) were identified by first calculating the BCV by dividing the standard deviation of expression for each expressed gene by the mean expression of each expressed gene. A dispersion table was then extracted using the “dispersionTable()” function from Monocle 2. Genes with a mean expression > 0.5 transcripts and a “dispersion_empirical” ≥ 1.5∗dispersion_fit or 2.0∗dispersion_fit were identified as “high variance genes.”
Principal Component Analysis (PCA)
PCA was run using the R “prcomp()” function on the centered and scaled log2 expression values of the “high variance genes.” PC1 and PC2 were visualized to scan the data for outliers as well as bias in the PCs for age, region, or plates on which the cells were sequenced. If any visual outliers in the data were observed, those cells were removed from the original subsetted cds and all filtering steps above were repeated. Once there were no visual outliers in PC1 or PC2, a screeplot was used to determine the number of PCs that contributed most significantly to the variation in the data. This was manually determined by inspecting the screeplot and including only those PCs that occur before the leveling-off of the plot.
t-Distributed Stochastic Neighbor Embedding (t-SNE) and Clustering
Once the number of significant PCs was determined, t-SNE24 was used to embed chosen PC dimensions in a 2D space for visualization. This was done using the “tsne()” function available through the tsne package (v.0.1-3) in R with “whiten = FALSE.” The parameters “perplexity” and “max_iter” were tested with various values and set according to what was deemed to give the cleanest clustering of the data.
After dimensionality reduction via t-SNE, the number of clusters was determined in an unbiased manner by fitting multiple Gaussian distributions over the 2D t-SNE projection coordinates using the R package ADPclust.25 t-SNE plots were visualized using a custom R script. The number of genes expressed and the total mRNAs for each cluster were then compared.
Differential Expression Analyses
In order to find differentially expressed genes between brain DA populations at each age, the E15.5 and P7 datasets were annotated with regional cluster identity (“subset cluster”). Differential expression analysis was performed using the “differentialGeneTest()” function from Monocle 2 that uses a likelihood ratio test to compare a vector generalized additive model (VGAM) using a negative binomial family function to a reduced model in which one parameter of interest has been removed. In practice, the following model was fit: “∼subset.cluster” for E15.5 or P7 dataset. Genes were called as significantly differentially expressed if they had a q value (Benjamini-Hochberg corrected p value) < 0.05.
Cluster-Specific Marker Genes
In order to identify differentially expressed genes that were “specifically” expressed in a particular subset cluster, R code calculating the Jensen-Shannon-based specificity score from the R package cummeRbund26 was used similarly to what was described in Burns et al.27
Briefly, the mean RPC within each cluster for each expressed gene as well as the percentage of cells within each cluster that express each gene at a level >1 transcript were calculated. The “.specificity()” function from the cummeRbund package was then used to calculate and identify the cluster with maximum specificity of each gene’s expression. Details of this specificity metric can be found in Molyneaux et al.28
To identify subset cluster-specific genes, the distribution of specificity scores for each subset cluster was plotted and a specificity cutoff was chosen so that only the “long right tail” of each distribution was included (i.e., genes with a specificity score above the cutoff chosen). Within each iterative analysis, the same cutoff was used for each cluster or region (specificity ≥ 0.3 or 0.4 depending on time point analyzed). Once the specificity cutoff was chosen, genes were further filtered by retaining only genes that were expressed in ≥40% of cells within the subset cluster that the gene was determined to be specific for.
Gene Set Enrichment Analyses
Gene set enrichment analyses were performed in two separate ways depending upon the situation. A Gene Set Enrichment Analysis (GSEA) PreRanked analysis was performed when a ranked list (e.g., genes ranked by PC1 loadings) using GSEA software available from the Broad Institute (v2.2.4).29, 30 Ranked gene lists were uploaded to the GSEA software and a “GSEAPreRanked” analysis was performed with the following settings: Number of Permutations = 1,000, Collapse dataset to gene symbols = true, Chip platform(s) = GENE_SYMBOL.chip, and Enrichment statistic = weighted. Analysis was performed against Gene Ontology (GO) collections from MSigDB, including c2.all.v5.2.symbols and c5.all.v5.2.symbols. Top ten gene sets were reported for each analysis (Table S1 for outliers and Figure 1C for time points). Figures and tables displaying the results were produced using custom R scripts.
Figure 1.
scRNA-Seq Analysis of Isolated Cells Allows Their Separation by Developmental Time
(A) Diagram of scRNA-seq experimental procedures for isolating and sequencing EGFP+ cells. Timeline adapted from Barallobre et al.41
(B) Principal component analysis (PCA) on all cells collected using genes with highly variant transcriptional profiles. The cells that were included are those that passed quality control measures. The greatest source of variation (PC1) is explained by the time point at which the cells were collected, not the region from which the cells were collected.
(C) The top ten Gene Ontology (GO) gene sets enriched in genes with positive (red) and negative (green) PC1 loadings from the PCA plot in (B). Gene sets are arranged by normalized enrichment scores (NES) and all gene sets displayed had a false discovery rate (FDR) q value ≤ 0.05.
(D) A t-distributed stochastic neighbor embedding (t-SNE) plot of all collected cells that passed quality control measures colored by regional identity. E15.5 cells cluster together while P7 cells cluster primarily by regional identity.
(E) A t-SNE plot of all collected cells colored by subset cluster identity. Through iterative analysis, time point-regions collected can be separated into multiple subpopulations (13 in total).
Abbreviations: midbrain, MB; forebrain, FB; olfactory bulb, OB; fluorescence activated cell sorting, FACS.
Unranked GSEA analyses for lists of genes were performed using hypergeometric tests from the R package clusterProfiler implemented through the functions “enrichGO()”, “enrichKEGG()”, and “enrichPathway()” with pvalueCutoff set at 0.01, 0.1, 0.1, respectively, with default settings.31 These functions were implemented through the “compareCluster()” function.
Weighted Gene Co-Expression Network Analysis (WGCNA)
WGCNA was performed in R using the WGCNA package (v1.51)32, 33 following established pipelines laid out by the package authors. Briefly, log2(Transcript +1) expression counts for all genes expressed in ≥20 cells (n = 12,628) in all P7 neurons were used and outliers were removed. The soft threshold (power) for WGCNA was determined by calculating the scale free topology model fit for a range of powers (1:10, 12, 14, 16, 18, 20) using the WGCNA function “pickSoftThreshold()” setting the networkType = “signed.” A power of 10 was chosen. Network adjacency was then calculated using the WGCNA function “adjacency()” with the following settings: power = 10 and type = “signed.” Adjacency calculations were used to then calculate topological overlap using the WGCNA function “TOMsimilarity()” with the following settings: TOMtype = “signed.” Distance was then calculated by subtracting the topological overlap from 1. Hierarchical clustering was then performed on the distance matrix and modules were identified using the “cuttreeDynamic()” function from the dynamicTreeCut package34 with the following settings: deepSplit = T; pamRespectsDendro = FALSE, and minClusterSize = 20. This analysis initially identified 18 modules. Eigengenes for each module were then calculated using the “moduleEigengenes()” function and each module was assigned a color. Two modules (“grey” and “turquoise”) were removed at this point. Turquoise was removed because it contained 11,567 genes or all the genes that could not be grouped with another module. Grey was removed because it contained only four genes, falling below the minimum set module size of 20. Significance of correlations between module eigengenes and subset cluster identity was calculated using the Student asymptotic p value for correlations employed by the WGCNA “corPvalueStudent()” function. Gene set enrichments for modules were determined by using the clusterProfiler R package.31 The correlations between the t-SNE position of a cell and the module eigengenes were calculated using custom R scripts.
Prioritizing Genes in PD GWAS Loci
Topologically Associated Domain (TAD) and Megabase (Mb) Gene Data
The data for human TAD boundaries were obtained from human embryonic stem cell (hESC) Hi-C data35 and converted from human genome hg18 to hg38 using the liftOver tool from UCSC Genome Browser. PD GWAS SNP locations in hg38 were intersected with the TAD information to identify TADs containing a PD GWAS SNP. The data for ±1 Mb regions surrounding PD GWAS SNPs was obtained by taking PD GWAS SNP locations in hg38 and adding or subtracting 1 × 106 from each location. All hg38 Ensembl (v.87) genes that fell within the TADs or megabase regions were then identified by using the biomaRt R package.36, 37 All genes were then annotated with PD locus and SNP information. Mouse homologs for all genes were identified using human to mouse homology data from Mouse Genome Informatics (MGI). Gene homologs were manually annotated using the MGI database if a homolog was found to exist. The TAD and megabase tables were then combined to create a final PD GWAS locus-gene table.
PD GWAS Loci Gene Scoring
Genes within PD GWAS loci were initially scored using two gene lists: genes with an average expression ≥ 0.5 transcripts in the SN cluster in our data (points = 1; number of genes = 6,126) and genes with an average expression ≥ 0.5 transcripts in the SN population in La Manno et al.38 (points = 1; number of genes = 5,406). La Manno et al. data (GSE76381_MouseAdultDAMoleculeCounts.cef.txt.gz) was accessed via the Gene Expression Omnibus (GEO: GSE76381). Further prioritization was accomplished by using three gene lists: genes that were differentially expressed between P7 subset clusters (points = 1); genes found to be “specifically” expressed in the P7 MB SN cluster (points = 1); and genes found in the WGCNA modules that are enriched for PD gene sets (points = 1). Expression in the SN cluster was considered the most important feature and was weighted as such through the use of two complementary datasets with genes found to be expressed in both receiving priority. Furthermore, a piece of external data, the probability of being loss-of-function (LoF) intolerant (pLI) scores for each gene from the ExAC database,39 was added to the scores in order to rank loci that were left with ≥2 genes in the loci after the initial scoring. pLI scores were downloaded March 30, 2017 (fordist_cleaned_exac_r03_march16_z_pli_rec_null_data.txt).
In Situ Hybridization Data
In situ hybridization data were downloaded from the Allen Institute through the Allen Brain Atlas (Web Resources). The image used in Figure 4A was obtained from the Reference Atlas at the Allen Brain Atlas. URLs for all Allen Brain Atlas in situ data analyzed and downloaded for SN marker genes (Figure 4B) are available in Table S5. Data for SN expression in situ data for PD GWAS genes (Figure 5B) were obtained from the following experiments: 1056 (Th), 79908848 (Snca), 297 (Crhr1), 74047915 (Atp6v1d), 72129244 (Mmp16), and 414 (Cntn1). Data accessed on 03/02/17.
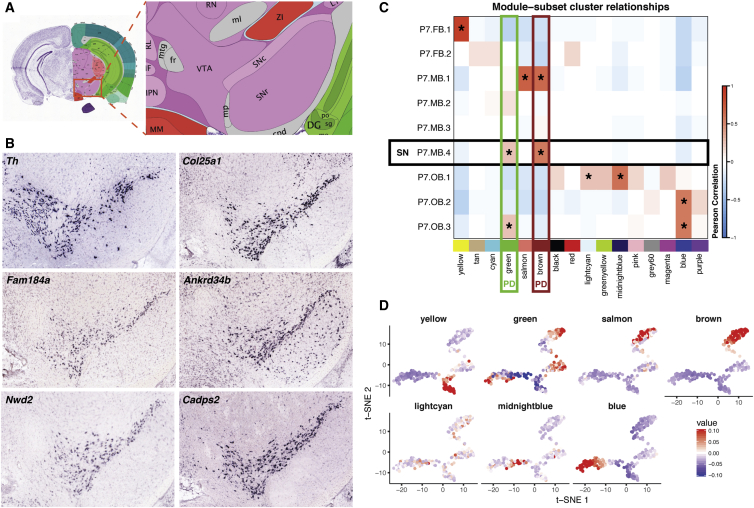
Figure 4.
Genetic Markers and Gene Modules Reveal Context-Specific SN DA Biology
(A) Reference Atlas diagram from the Allen Brain Atlas (ABA) of the P56 mouse ventral midbrain. Important abbreviations include: VTA, ventral tegmental area; SNc, substantia nigra pars compacta; SNr, substantia nigra pars reticulata.
(B) Confirmation of SN DA neuron marker genes through the use of ABA in situ hybridization data. Coronal, P56 mouse in situ data were explored in order to confirm the expression of 25 previously uncharacterized SN markers. Th expression in P56 mice was used as an anatomical reference during analysis.
(C) Correlation heatmap of the Pearson correlation between module eigengenes and P7 Th+ subset cluster identity. Modules are represented by their assigned colors at the bottom of the matrix. Modules that had a positive correlation with a subset cluster and had a correlation p value less than the Bonferroni corrected significance level (p < 3.5 × 10−4) contain an asterisk. SN cluster (P7.MB.4) identity is denoted by a black rectangle. Modules (“green” and “brown”) that were enriched for the “Parkinson’s Disease” KEGG gene set are labeled with “PD.”
(D) The eigengene value for each P7 neuron in the seven WGCNA modules shown to be significantly positively associated with a subset cluster overlaid on the t-SNE plot of all P7 neurons (Figure 2A). Plotting of eigengenes confirms strict spatial restriction of module association. Only the “lightcyan” module does not seem to show robust spatial restriction.
Figure 5.
Context-Specific SN DA Data Allow for the Prioritization of Genes in PD GWAS Loci
(A) A locus plot displaying 4-megabase regions in the human genome (hg38) centered on PD GWAS SNPs in six loci. Genes are displayed as boxes on their appropriate strand. Genes are shaded by their prioritization score and gene names are displayed for genes with a score of 3 or higher in each locus.
(B) In situ hybridization from the Allen Brain Atlas (ABA) of five prioritized genes along with Th for an anatomical reference. Coronal, P56 mouse in situ data were used.
(C) Boxplots displaying expression of prioritized genes from the MAPT locus (Figure 5A; Table 2). ±1.5× interquartile range is represented by the whiskers on the boxplots. Data points beyond 1.5× interquartile range are considered as outliers and plotted as black points.
(D) Representative light microscopy images of Th+ innervation density in the striatum of WT and Cplx1 knockout (KO) mice. Scale bar, 1 mm.
(E) Boxplots comparing the level of Th+ striatum innervation between WT and Cplx1 KO mice. DAB staining density was measured in 35 μm, horizontal sections in WT mice (mice = 3, sections = 16) and Cplx1 KO mice (mice = 8, sections = 40). Each point in the boxplot represents the average signal from a stained, 35 μm section. Statistical analyses were performed between conditions with section averages in order to preserve observed variability (WT n = 16, Cplx1 KO n = 40). A two-sample t test revealed that Th+ innervation density was significantly lower in Cplx1 KO mice (t = 6.4395, df = 54, p = 3.386 × 10−8). Data points outside of 1.5× interquartile range, represented by the whiskers on the boxplots, are considered as outliers and plotted as black points.
Single-Molecule In Situ Hybridization (smFISH)
For in situ hybridization experiments, untimed pregnant Swiss Webster mice were ordered from Charles River Laboratories (Crl:CFW(SW)). Mice were maintained as previously described. Pups were considered P0 on the day of birth. At P7, the pups were decapitated, the brain was quickly removed, and the brain was then washed in 1× PBS. The intact brain was then transferred to a vial containing freshly prepared 4% PFA in 1× PBS and incubated at 4°C for 24 hr. After 24 hr, brains were removed from PFA and washed three times in 1× PBS. The brains were then placed in a vial with 10% sucrose at 4°C until the brains sunk to the bottom of the vial (usually ∼1 hr). After sinking, brains were immediately placed in a vial containing 30% sucrose at 4°C until once again sinking to the bottom of the vial (usually overnight). After cryoprotection, the brains were quickly frozen in optimal cutting temperature (O.C.T.) compound (Tissue-Tek) on dry ice and stored at −80°C until use. Brains were sectioned at a thickness of 14 μm and mounted on Superfrost Plus microscope slides (Fisherbrand, Cat. # 12-550-15) with two sections per slide. Sections were then dried at room temperature for at least 30 min and then stored at −80°C until use.
RNAscope in situ hybridization (Advanced Cell Diagnostics) was used to detect single RNA transcripts. RNAscope probes were used to detect Th (C1; Cat No. 317621, Lot: 17073A), Slc6a3 (C2; Cat No. 315441-C2, Lot: 17044A), Lhx9 (C3; Cat No. 495431-C3, Lot: 17044A), and Ldb2 (C3; Cat No. 466061-C3, Lot: 17044A). The RNAscope Fluorescent Multiplex Detection kit (Cat No. 320851) and the associated protocol provided by the manufacturer were used, with slight modifications. Briefly, frozen tissues were removed from −80°C and equilibrated at room temperature for 5 min. Slides were then washed at room temperature in 1× PBS for 3 min with agitation, then immediately washed in 100% ethanol by moving the slides up and down 5–10 times. The slides were then allowed to dry at room temperature and hydrophobic barriers were drawn using a hydrophobic pen (ImmEdge Hydrophobic Barrier PAP Pen, Vector Laboratories, Cat. # H-4000) around the tissue sections. The hydrophobic barrier was allowed to dry overnight. After drying, the tissue sections were treated with RNAscope Protease IV at room temperature for 30 min and then slides were washed in 1× PBS. Approximately 100 μL of multiplex probe mixtures (C1, Th; C2, Slc6a3; and C3, one of Lhx9 or Ldb2) containing either approximately 96 μL C1: 2 μL C2: 2 μL C3 (Th:Slc6a3:Lhx9) or 96 μL C1: 0.6 μL C2: 2 μL C3 (Th:Slc6a3:Ldb2) were applied to appropriate sections. Both mixtures provided adequate in situ signals. Sections were then incubated at 40°C for 2 hr in the ACD HybEZ oven. Sections were then sequentially treated with the RNAscope Multiplex Fluorescent Detection Reagents kit solutions AMP 1-FL, AMP 2-FL, AMP 3-FL, and AMP 4 Alt B-FL, with washing in between each incubation, according to manufacturer’s recommendations. Sections were then treated with DAPI provided with the RNAscope Multiplex Fluorescent Detection Reagents kit. One drop of Prolong Gold Antifade Mountant (Invitrogen, Cat # P36930) was then applied to each section and a coverslip was then placed on the slide. The slides were then stored in the dark at 4°C overnight before imaging. Slides were further stored at 4°C throughout imaging. Manufacturer-provided positive and negative controls were performed alongside experimental probe mixtures according to manufacturer’s protocols. Four sections that encompassed relevant populations in the P7 ventral MB (SN, ventral tegmental area [VTA], etc.) were chosen for each combination of RNAscope smFISH probes and subsequent analyses.
smFISH Confocal Microscopy
RNAscope fluorescent in situ experiments were analyzed using the Nikon A1 confocal system equipped with a Nikon Eclipse Ti inverted microscope running Nikon NIS-Elements AR 4.10.01 64-bit software. Images were captured using a Nikon Plan Apo λ 60×/1.40 oil immersion lens with a common pinhole size of 19.2 μM, a pixel dwell of 28.8 μs, and a pixel resolution of 1,024 × 1,024. DAPI, FITC, Cy3, and Cy5 channels were used to acquire RNAscope fluorescence. Positive and negative control slides using probe sets provided by the manufacturer were used in order to calibrate laser power, offset, and detector sensitivity, for all channels in all experiments performed.
smFISH Image Analysis and Processing
Confocal images were saved as .nd2 files. Images were then processed in ImageJ as follows. First, the .nd2 files were imported into ImageJ and images were rotated in order to reflect a ventral midbrain orientation with the ventral side of the tissue at the bottom edge. Next, the LUT ranges were adjusted for the FITC (range: 0–2,500), Cy3 (range: 0–2,500), and Cy5 (range: 0–1,500) channels. All analyzed images were set to the same LUT ranges. Next, the channels were split and merged back together to produce a “composite” image. Scale bars were then added. Cells of interest were then demarcated and duplicated and the channels were split.
Immunohistochemistry and Quantification of Th Striatum Staining in Cplx1−/− Mice
Mice (n = 8 Cplx1−/−; n = 3 WT littermates; ages between 4 and 7.5 weeks) were euthanized and their brains fresh-frozen on powdered dry ice. Brains were sectioned at 35 μm and sections were mounted onto Superfrost-plus glass slides (VWR International). Sections were peroxidase inactivated, and one in every ten sections was processed immunohistochemically for tyrosine hydroxylase. Sections were incubated in primary anti-tyrosine hydroxylase antibody (AB152, Millipore) used at 1/2,000 dilution in 1% normal goat serum in phosphate-buffered saline and 0.2% Triton X-100 overnight at 4°C. Antigens were visualized using a horseradish peroxidase-conjugated anti-rabbit second antibody (Vector, PI-1000, 1/2,000 dilution) and by using diaminobenzidine (DAB; Sigma). The slides were stored in the dark (in black slide boxes) at room temperature (21°C).
Images of stained striatum were taken using a Nikon AZ100 microscope equipped with a 2× lens (Nikon AZ Plan Fluor, NA 0.2, WD45), a Nikon DS-Fi2 camera, and NIS-Elements AR 4.5 software. Appropriate zoom and light exposure were determined before imaging and kept constant for all slides and sections. Density of TH+ DAB staining was measured using ImageJ software. Briefly, images were imported into ImageJ and the background was subtracted (default 50 pixels with “light background” selected). Next, images were converted to 8-bit and the image was inverted. Five measurements of density were taken for each side of a striatum in a section along with a density measurement from adjacent, unstained cortex. Striosomes were avoided during measuring when possible. Striatal measurements had background (defined as staining in the adjacent cortex in a section) subtracted. The mean section measurements (intensity/pixels squared) for each brain were calculated and represented independent measurements of the same brain. Variances were compared between the WT and KO populations. A two-sample t test was then used to compare WT versus Cplx1−/− section densities with the following parameters in R using the “t.test()” function: alternative = “two-sided,” var.equal = “T.”
Results
scRNA-Seq Characterization Defines DA Neuronal Subpopulation Heterogeneity
In order to characterize DA neuron molecular phenotypes, we undertook scRNA-seq on cells isolated from distinct anatomical locations of the mouse brain over developmental time. We used FACS to retrieve single DA neurons from the Tg(Th-EGFP)DJ76Gsat BAC transgenic mouse line, which expresses EGFP under the control of the tyrosine hydroxylase (Th) locus.40 We microdissected both midbrain (MB) and forebrain (FB) from E15.5 mice, extending our analyses to MB, FB, and olfactory bulb (OB) in P7 mice (Figure 1A). Brains from four and five mice were pooled for E15.5 and P7, respectively. E15.5 and P7 time points were chosen based on their representation of stable MB DA populations, either after neuron birth (E15.5) or between periods of programmed cell death (P7) (Figure 1A).41
Quality control and outlier analysis identified 396 high-quality cell transcriptomes to be used in our analyses. We initially sequenced RNA from 473 single cells to an average depth of ∼8 × 105 50-bp paired-end fragments per cell. Using Monocle 2, we converted normalized expression estimates into estimates of RNA copies per cell.23 Cells were filtered based on the distributions of total mass, total number of mRNAs, and total number of expressed genes per cell (Figures S1A–S1C; detailed in Material and Methods). After QC, 410 out of 473 cells were retained. Using principal component analysis (PCA) as part of the iterative analysis described below, we identified and removed 14 outliers determined to be astrocytes, microglia, or oligodendrocytes (Figure S1E; Table S1), leaving 396 cells (∼79 cells/time point-region; Figure S1D).
To confirm that our methods can discriminate between different populations of neurons, we first explored differences between time points. In order to do this, we identified genes with highly variable transcriptional profiles and performed PCA. As anticipated, we observed that the greatest source of variation was between developmental ages (Figure 1B). Genes associated with negative PC1 loadings (E15.5 cells) were enriched for gene sets consistent with mitotically active neuronal, undifferentiated precursors (Figure 1C). In contrast, genes associated with positive PC1 loadings (P7 cells) were enriched for ontology terms associated with mature, post-mitotic neurons (Figure 1C). This initial analysis establishes our capacity to discriminate among biological classes present in our data using PCA as a foundation.
Recursive Analysis of scRNA-Seq Data Reveals 13 DA Neuron Subtypes
We set out to identify clusters of single cells within time points and anatomical regions. Following a workflow similar to the recently described “dpFeature” procedure,42 we identified highly variable genes and performed PCA using those gene transcriptional profiles. We selected the PCs that described the most variance in the data and used t-SNE24 to further elucidate the relationships between our cells. We then identified clusters of cells in an unsupervised manner using local Gaussian densities.25 The steps taken in this analysis were performed in a recursive manner for both time points across all regions to further explore heterogeneity (see Material and Methods).
Analysis of all cells revealed E15.5 cells from both MB and FB cluster together (Figure 1D), supporting the notion that they are less differentiated. By contrast, cells isolated at P7 mostly cluster by anatomical region, suggesting progressive functional divergence with time (Figure 1D). The recursive analysis performed across all time points and regions revealed a total of 13 clusters (E15.5 FB.1-2, MB.1-2; P7 OB.1-3, FB.1-2, MB.1-4; Figure 1E), demonstrating the diversity of DA neuron subtypes and providing a framework upon which to evaluate the biological context of genetic association signals across closely related cell types. Using known markers, we confirmed that all clusters expressed high levels of pan-neuronal markers (Snap25, Eno2, and Syt1) (Figure S2A). By contrast, we observed scant evidence of astrocyte (Aldh1l1, Slc1a3, Aqp4, and Gfap; Figure S2A) or oligodendrocyte markers (Mag, Mog, and Mbp; Figure S2A), thus confirming we successfully isolated our intended substrate, Th+ neurons.
scRNA-Seq Reveals Biologically and Temporally Discriminating Transcriptional Signatures
With subpopulations of DA neurons defined by our data, we set out to assign a biological identity to each cluster. To do this, we identified differentially expressed genes between clusters within each time point, then identified marker genes for each cluster within each time point (see Material and Methods; Table S2). Since the age of the mice constituted the greatest source of variation in the data (Figure 1B), we undertook differential expression analyses and downstream analyses separately for each time point.
Among the four clusters identified at E15.5, two were represented in t-SNE space as a single large group that included cells from both MB and FB (E15.MB.1, E15.FB.1), leaving two smaller clusters that were comprised solely of MB or FB cells (Figure S3A). Both E15.MB.1 and E15.FB.1 show markers consistent with neuroblast populations (Tables 1 and S3). The isolated MB cluster (E15.MB.2; Figures S3A and S3C) specifically expressed Foxa1, Lmx1a, Pitx3, and Nr4a2 and thus likely represents a post-mitotic DA neuron population43 (Tables 1, S2, and S3). Similarly, the discrete E15.FB.2 cluster expressed markers of post-mitotic FB/hypothalamic neurons (Figures S3A and S3B), including Six3, Six3os1, Sst, and Npy (Tables 1, S2, and S3). These embryonic data did not discriminate between cells populating known domains of DA neurons, such as the SN or ventral tegmental area (VTA).
Table 1.
Summary of Cell Population Identities
| Age | Cluster | Identity |
|---|---|---|
| E15.5 | FB.1 | forebrain neuroblast |
| FB.2 | post-mitotic forebrain Th+ neurons | |
| MB.1 | midbrain neuroblast | |
| MB.2 | post-mitotic midbrain DA neuron | |
| P7 | FB.1 | arcuate nucleus neuroendocrine Th+ neurons |
| FB.2 | mixture of arcuate nucleus Th+ subtypes | |
| MB.1 | ventral tegmental area (VTA) | |
| MB.2 | postnatal neuroblast | |
| MB.3 | periaqueductal gray area (PAG) | |
| MB.4 | substantia nigra (SN) | |
| OB.1 | least mature Th+ neurons | |
| OB.2 | progressively maturing Th+ neurons | |
| OB.3 | most mature Th+ neurons |
Summary of the identities of cell populations identified through recursive scRNA-seq analysis of E15.5 and P7 DA neurons. 13 cell populations are described, each with their age, cell cluster name, and biological identity. Additional information can be found in Table S3.
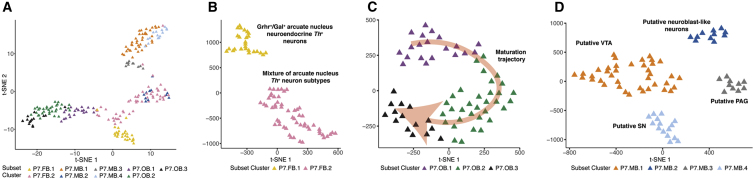
By contrast, P7 cells mostly cluster by anatomical region and each region has defined subsets (Figures 1D, 1E and 2A). Analysis of P7 FB revealed two distinct cell clusters (Figure 2B). Expression of the neuropeptides Gal and Ghrh and the Gsx1 transcription factor place P7.FB.1 cells in the arcuate nucleus (Tables 1, S2, and S3).44, 45, 46 The identity of P7.FB.2, however, was less clear, although subsets of cells therein did express other arcuate nucleus markers for Th+/Ghrh− neuronal populations, e.g., Onecut2, Arx, Prlr, Slc6a3, and Sst (Figure S3D; Table S3).46 All three identified OB clusters (Figure 2C) express marker genes of OB DA neuronal development or survival (Tables S2 and S3; Figure S3E).47 It has previously been reported that Dcx expression diminishes with neuronal maturation48 and Snap25 marks mature neurons.49 We observe that these OB clusters seem to reflect this continuum of maturation wherein expression of Dcx diminishes and Snap25 increases with progression from P7.OB1 to OB3 (Figure S3E). This pattern is mirrored by a concomitant increase in OB DA neuron fate specification genes (Figure S3E).47, 50 In addition, we identified four P7 MB DA subset clusters (Figure 2D). Marker gene analysis confirmed that three of the clusters correspond to DA neurons from the VTA (Otx2 and Neurod6; P7.MB.1),51, 52 the periaqueductal gray area (PAG; Vip and Pnoc; P7.MB.3),53, 54 and the SN (Sox6, Aldh1a7, Ndnf, Serpine2, Rbp4, and Fgf20; P7.MB.4)38, 51, 55, 56 (Tables 1, S2, and S3). These data are consistent with recent scRNA-seq studies of similar populations.38, 57 Through this marker gene analysis, we successfully assigned a biological identity to 12/13 clusters (Table 1).
Figure 2.
Subclusters of P7 Th+ Neurons Are Identified Based on Marker Gene Analyses
(A) A t-SNE plot of all P7 neurons collected colored by subset cluster identity. The neurons mostly cluster by regional identity.
(B) t-SNE plot of P7 FB neurons. P7 FB neurons cluster into two distinct populations.
(C) t-SNE plot of P7 OB neurons. P7 OB neurons cluster into three populations. These populations represent a trajectory of Th+ OB maturation (Table S3) as indicated by the red arrow.
(D) A t-SNE plot of P7 MB neurons. P7 MB neurons cluster into four clusters: the substantia nigra (SN), the ventral tegmental area (VTA), the periaqueductal gray area (PAG), and a neuroblast-like population.
Multiplex, smFISH Confirms the Existence of a Putative Postnatal Neuroblast Population
The only cluster without a readily assigned identity was P7.MB.2. This population of P7 MB DA neurons, P7.MB.2 (Figure 2D), is likely a neuroblast-like population based on marker gene analysis (Tables 1 and S3). Like the overlapping E15.MB.1 and E15.FB.1 clusters (Figure S3A), this cluster preferentially expresses markers of neuronal precursors/differentiation/maturation (Table S3). In addition to sharing markers with the neuroblast-like E15.MB.1 cluster, P7.MB.2 exhibits gene expression consistent with embryonic mouse neuroblast populations38 as well as cell division and neuron development58, 59, 60, 61, 62 (Tables S2 and S3). Consistent with the hypothesis, this population displayed lower levels of both Th and Slc6a3, markers of mature DA neurons, than the terminally differentiated and phenotypically discrete P7 MB DA neuron populations of the VTA, SN, and PAG (Figure 3A).
Figure 3.
Multiplex, smFISH Confirms the Existence of a Putative Postnatal Neuroblast Population
(A) Boxplots displaying the expression of four genes (Th, Slc6a3, Lhx9, and Ldb2) across all subclusters identified. E15.MB.1 and P7.MB.2 labels are bold due to similar expression profile of displayed genes (Tables S2 and S3). ±1.5× interquartile range is represented by the whiskers on the boxplots. Data points beyond 1.5× interquartile range are considered as outliers and plotted as black points.
(B) Representative image of multiplex single molecule fluorescent in situ hybridization (smFISH) for Th, Slc6a3, and Lhx9 in the mouse ventral midbrain. Zoomed-in panels represent cell populations observed. Scale bar, 50 μm.
(C) Representative image of multiplex smFISH for Th, Slc6a3, and Ldb2, in the mouse ventral midbrain. Zoomed-in panels represent cell populations observed. Scale bar, 50 μm.
(D) Diagram of ventral midbrain summarizing the results of smFISH. Th+/Slc6a3−/Lhx9+ and Th+/Slc6a3−/Ldb2+ cells are both found in the dorsal SN.
Abbreviations: NB, neuroblast; SN, substantia nigra; VTA, ventral tegmental area; IPN, interpeduncular nucleus.
With this hypothesis in mind, we sought to ascertain the spatial distribution of P7.MB.2 DA neurons through multiplex, smFISH for Th (pan-P7 MB DA neurons), Slc6a3 (P7.MB.1, P7.MB.3, P7.MB.4), and one of the neuroblast marker genes identified through our analysis, either Lhx9 or Ldb2 (P7.MB.2) (Figure 3A). In each experiment, we scanned the ventral midbrain for cells that were Th+/Slc6a3− and positive for the third gene. Th+/Slc6a3−/Lhx9+ cells were found scattered in the dorsal SN pars compacta (SNpc) along with cells expressing Lhx9 alone (Figures 3B and 3D). Expression of Ldb2 was found to have a similar pattern to Lhx9, with Th+/Slc6a3−/Ldb2+ cells found in the dorsal SNpc (Figures 3C and 3D). Expression of Lhx9 and Ldb2 was low or non-existent in Th+/Slc6a3+ cells in the SNpc (Figures 3B and 3C). Importantly, cells expressing these markers express Th at lower levels than Th+/Slc6a3+ neurons (Figures 3B and 3C), consistent with our scRNA-seq data (Figure 3A). Thus, with the resolution of the spatial distribution of this neuroblast-like P7 MB DA population, we assign biological identity to each defined brain DA subpopulation (Table 1).
SN-Specific Transcriptional Profiles and GRNs Highlight Its Association with PD
Overall, our analyses allowed us to successfully separate and identify 13 brain DA neuronal populations present at E15.5 and P7, including SN DA neurons (Table 1). Motivated by the clinical relevance of SN DA neurons to PD, we set out to understand what makes them transcriptionally distinct from the other MB DA neuron populations.
In order to look broadly at neuronal subtypes, we evaluated expression of canonical markers of other neuronal subtypes in our Th+ neuron subpopulations. We noted that Th and EGFP were inconsistently detected in some E15.5 clusters (Figure S4A). This likely reflects lower Th transcript abundance at this developmental state, but sufficient expression of the EGFP reporter to permit FACS collection (Figure S4B). The expression of other DA markers, Ddc and Slc18a2, mirror Th expression, while Slc6a3 expression is more spatially and temporally restricted (Figure S4A). The SN cluster displays robust expression of all explored canonical DA markers (Figure S4A). Multiple studies have demonstrated that Th+ neurons may also express markers characteristic of other major neuronal subtypes.63, 64, 65 We found that all but the SN and PAG showed expression of either GABAergic (Gad1/Gad2/Slc32a1) or glutamatergic (Slc17a6) markers (Figure S4A). This neurotransmitter specificity may represent a valuable avenue for exploring the preferential vulnerability of the SN in PD.
Next, we postulated that genes whose expression defined the P7 SN DA neuron cluster might illuminate their preferential vulnerability in PD. We identified 110 SN-specific genes, by first finding all differentially expressed genes between P7 subset clusters and then using the Jensen-Shannon distance to identify cluster specific genes (see Material and Methods; Table S2). Prior reports confirm the expression of 49 of the 110 SN-specific genes (∼45%) in postnatal SN (Table S4). We then sought evidence to confirm or exclude SN expression for the remaining 61 genes (55%). Of these, 25/61 (∼41%) were detected in the adult SN by in situ hybridization (ISH) of coronal sections in adult (P56) mice (Allen Brain Atlas, ABA), including Col25a1, Fam184a, Ankrd34b, Nwd2, and Cadps2 (Figures 4A and 4B; Table S5). Only 4/61 genes, for which ISH data existed in the ABA, lacked clear evidence of expression in the adult SN (Table S5). The ABA lacked coronal ISH data on 32/61 genes, so we were unable to confirm their presence in the SN. Collectively, we identified 110 postnatal SN DA marker genes and confirmed the expression of those genes in the adult rodent SN for 74 (67%) of them, including 25 previously uncharacterized markers of this clinically relevant cell population.
We next asked whether we could identify significant relationships between cells defined as being P7 SN DA neurons and distinctive transcriptional signatures in our data. In order to do this, we performed weighted gene co-expression network analysis (WGCNA).32, 33 WGCNA learns modules of genes with similar expression patterns across individual cells. By using expression data for all expressed genes in our P7 DA neuron dataset, we identify 16 co-expressed gene modules (Figure S5; Table S6). By calculating pairwise correlations between modules and P7 subset cluster identity, we reveal that 7/16 modules are significantly and positively correlated (Bonferroni corrected p < 3.5 × 10−4) with at least one subset cluster (Figure 4C). We graphically represented the eigenvalues for each module in each cell in P7 t-SNE space, confirming that a majority of these significant modules (6/7), except for “lightcyan,” displayed robust spatial, isotype enrichment (Figure 4D).
In order to identify the biological relevance of these modules, each module was tested for enrichment for Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways, Gene Ontology (GO) gene sets, and Reactome gene sets. Two modules, the “brown” and “green” modules, were significantly associated with the Parkinson disease KEGG pathway gene set (Figure 4C; Table S7). Interestingly, the “brown” module was also significantly correlated with the P7 VTA population (P7.MB.1) and enriched for addiction gene sets (Table S7), highlighting the link between VTA DA neurons and addiction.66 Strikingly, only the P7 SN cluster was significantly correlated with both PD-enriched modules (Figure 4C). This specific correlation suggests that these gene modules may play a role in the preferential susceptibility of the SN in PD.
Integrating SN DA Neuron-Specific Data Enables Prioritization of Genes within PD-Associated Intervals
With these context-specific data in hand, we posited that SN DA neuron-specific genes and the broader gene co-expression networks that correlate with SN DA neurons might be used to prioritize genes that may be affected by disease-associated variation within PD GWAS loci. Such a strategy would be agnostic to prior biological evidence and independent of genic position relative to the lead SNP, the traditional method used to prioritize causative genes.
To investigate pertinent genes within PD GWAS loci, we identified all human genes within topologically associated domains (TADs) and 2-megabase (Mb) intervals encompassing each PD-associated lead SNP. TADs were chosen because regulatory DNA impacted by GWAS variation is more likely to act on genes within their own TAD.67 While topological data do not exist for SN DA neurons, we used TAD boundaries from hESCs as a proxy, as TADs are generally conserved across cell types.35 To improve our analyses, we also selected ±1 Mb intervals around each lead SNP, thus including the upper bounds of reported enhancer-promoter interactions.68, 69 All PD GWAS SNPs interrogated were identified by the most recent meta-analyses (49 SNPs in total),10, 11 implicating a total of 1,751 unique genes (both protein coding and non-coding; Table S8). We then identified corresponding one-to-one mouse to human homologs (1,009/1,751; ∼58%), primarily through the Mouse Genome Informatics (MGI) homology database.
To prioritize these genes in GWAS loci, we developed a gene-centric score that integrates our data as well as data in the public domain. We began by intersecting the PD loci genes with our scRNA-seq data as well as previously published SN DA expression data,38 identifying 430 genes (430/1,009; ∼43%) with direct evidence of expression in SN DA neurons in at least one dataset. These 49 PD loci are significantly enriched for genes expressed in SN DA neurons when compared to randomly selected GWAS loci (Figure S6A). Each PD-associated interval contained ≥1 of those SN-expressed genes (Table S8); this is more than what is expected from 49 random GWAS loci (Figure S6B). Emphasizing the need for a novel, systematic strategy, in 20/49 GWA intervals (∼41%), the most proximal gene to the lead SNP was not detectably expressed in mouse SN DA neuron populations (Tables S8 and S9). Three loci contained only one SN DA-expressed gene: Mmp16 (rs60298754 locus, Figure 5A), Tsnax (rs10797576 locus), and Satb1 (rs4073221 locus). The number of PD loci with only one gene expressed is slightly less than expected from 49 random GWAS loci (Figure S6C). The relevance of these candidate genes to neuronal function/dysfunction is well supported.70, 71, 72, 73 This establishes gene expression in a relevant tissue as a powerful tool in the identification of genes impacted by disease variation.
In order to prioritize likely disease-associated genes in the remaining 46 loci, we scored genes on three criteria: whether genes were identified as specific markers for the P7.MB.4 (SN) cluster (Table S2), whether the genes were differentially expressed between all P7 DA neuron populations, and whether the genes were included in PD gene set enriched and SN-correlated gene modules uncovered in WGCNA (Table S6). This strategy facilitated further prioritization of a single gene in 21 additional loci including the rs356182 (SNCA), rs76904798 (PDZRN4), and rs11158026 (GCH1) loci (Figure 5A; Tables 2 and S9). Importantly, using this approach we indict the gene implicated in familial PD alpha-synuclein (SNCA) as responsible for the observed PD association with rs356182 (Figure 5A; Tables 2 and S9). Thus, by using context-specific data alone, we were able to prioritize a single candidate gene in roughly half (24/49, ∼49%) of PD-GWAS associated loci.
Table 2.
Summary of the Systematic Scoring of Genes in 49 GWAS Loci Associated with PD
| Lead SNP | Top Candidate Genes | Prioritized by |
|---|---|---|
| rs6430538 | UBXN4;CCNT2;R3HDM1;RAB3GAP1 | SN expression; pLI |
| rs14235 | MAPK3;VKORC1; BOLA2B | SN expression; differential expression; pLI |
| rs11724635 | CPEB2 | SN expression; differential expression |
| rs11060180 | ARL6IP4 | SN expression; differential expression |
| rs8118008 | ATRN; NOP56; MRPS26; C20orf27;IDH3B | SN expression; differential expression; pLI |
| rs3793947 | DLG2;CCDC90B | SN expression; differential expression; pLI |
| rs6812193 | G3BP2;CCNI;CDKL2 | SN expression; differential expression; pLI |
| rs591323 | FGF20; ZDHHC2; TUSC3; MICU3; MTMR7 | SN expression; differential expression; SN specific; pLI |
| rs35749011 | KCNN3 | SN expression; differential expression; SN specific; WGCNA module |
| rs11158026 | GCH1 | SN expression; differential expression; SN specific; WGCNA module |
| rs199347 | RAPGEF5 | SN expression; differential expression |
| rs9275326 | ATP6V1G2 | SN expression; differential expression; WGCNA module |
| rs117896735 | PRDX3;NANOS1;INPP5F;SFXN4 | SN expression; differential expression; pLI |
| rs7077361 | FAM171A1 | SN expression; differential expression |
| rs115185635 | CHMP2B | SN expression; differential expression |
| rs76904798 | PDZRN4 | SN expression; differential expression; WGCNA module |
| rs17649553 | CRHR1; NSF; MAPT | SN expression; differential expression; pLI |
| rs12637471 | DCUN1D1; ABCC5; PARL | SN expression; pLI |
| rs329648 | OPCML | SN expression; differential expression |
| rs60298754 | MMP16 | SN expression |
| rs34016896 | B3GALNT1 | SN expression; differential expression |
| rs823118 | LRRN2; KLHDC8A; SRGAP2 | SN expression; differential expression; pLI |
| rs12456492 | RIT2;SYT4 | SN expression; differential expression; pLI |
| rs10797576 | TSNAX | SN expression |
| rs356182 | SNCA | SN expression; differential expression; WGCNA module |
| rs62120679 | UQCR11 | SN expression; differential expression; WGCNA module |
| rs11868035 | COPS3; NT5M | SN expression; differential expression; pLI |
| rs1474055 | STK39;B3GALT1 | SN expression; differential expression; pLI |
| rs34311866 | MAEA; CPLX1; ATP5I; TMEM175 | SN expression; differential expression; WGCNA module; pLI |
| rs1555399 | VTI1B; ATP6V1D | SN expression; differential expression; pLI |
| rs2823357 | HSPA13 | SN expression |
| rs2414739 | TLN2; RORA | SN expression; pLI |
| rs143918452 | NISCH; PCBP4; SPCS1; SMIM4 | SN expression; differential expression; pLI |
| rs78738012 | ANK2; CAMK2D | SN expression; differential expression; pLI |
| rs601999 | DNAJC7; ATP6V0A1; ACLY; PSME3; CNP; RPL27; VAT1; COA3; HAP1 | SN expression; differential expression; pLI |
| rs11343 | SYT17 | SN expression; differential expression; WGCNA module |
| rs2740594 | FAM167A | SN expression; differential expression; SN specific; WGCNA module |
| rs2694528 | NDUFAF2 | SN expression |
| rs10906923 | FAM171A1 | SN expression; differential expression |
| rs8005172 | ZC3H14 | SN expression |
| rs34043159 | RPL31; CREG2 | SN expression; differential expression; pLI |
| rs4653767 | SRP9; PSEN2; PARP1 | SN expression; pLI |
| rs12497850 | SMARCC1; PRKAR2A; RHOA; NICN1; UQCRC1; APEH; TCTA; TMA7; GPX1; IMPDH2; QARS; SHISA5; WDR6 | SN expression; differential expression; pLI |
| rs4073221 | SATB1 | SN expression |
| rs353116 | SCN3A; CSRNP3 | SN expression; differential expression; pLI |
| rs13294100 | BNC2 | SN expression; differential expression; SN specific; WGCNA module |
| rs2280104 | CHMP7; DMTN | SN expression; differential expression; pLI |
| rs4784227 | TOX3; AKTIP | SN expression; differential expression; WGCNA module; pLI |
| rs9468199 | ZSCAN26 | SN expression |
Scoring was carried out at described in the Results and Material and Methods. Candidate genes are presented for each of 49 PD GWAS loci analyzed. Information for each PD GWAS locus is presented including the lead SNP for each locus, the prioritized genes in each locus, and which data prioritized the top genes. Detailed scoring for each gene can be found in Table S9.
Furthermore, at loci in which a single gene did not emerge, we identified dosage-sensitive genes by considering the probability of being loss-of-function (LoF) intolerant (pLI) metric from the ExAC database.39, 74 Since most GWAS variation is predicted to impact regulatory DNA and in turn impact gene expression, it follows that genes in GWAS loci that are more sensitive to dosage levels may be more likely to be candidate genes. With that in mind, the pLI for each gene was used to further “rank” the genes within loci where a single gene was not prioritized. For those loci, including rs17649553 and rs8118008 loci (Figure 5A), we report a group of top-scoring candidate genes (Tables 2 and S9). Expression of prioritized genes in the adult SN adds to the validity of the genes identified as possible candidates (Figure 5B).
Two particularly interesting examples that emerge from this scoring are found at the rs17649553 and rs34311866 loci. The rs17649553 locus contains MAPT, which has previously been implicated in multiple neurodegenerative phenotypes, including PD (MIM: 168600). We instead prioritize CRHR1 and NSF before it (Table 2). We detect Mapt and Nsf expression consistently across all assayed DA neurons (Figure 5C). By contrast, expression of Crhr1, encoding the corticotropin releasing hormone receptor 1, is restricted to P7 DA neurons in the SN and the more mature OB neuronal populations (Figure 5C). Similarly, at the rs34311866 locus, our data show that although all three proximal genes are expressed in the SN (TMEM175, GAK, DGKQ), the adjacent CPLX1 was one of the prioritized genes (Tables 2 and S9).
There are multiple lines of evidence that strengthen CPLX1 as a candidate. Expression of CPLX1 is elevated in the brains of individuals with PD and Cplx1 is elevated in the brains of mice overexpressing SNCA with a familial PD mutation, c.157G>A (p.Ala53Thr) (GenBank: NM_000345.3).75, 76 Additionally, mice deficient in Cplx1 display an early-onset, cerebellar ataxia along with prolonged motor and behavioral phenotypes.14, 15 However, the impact of Cplx1 deficiency on the integrity of the nigrostriatal pathway, to date, has not been explored. In order to confirm CPLX1 as a candidate, we performed immunohistochemistry (IHC) for Th in the Cplx1 knockout mouse model (Tables S10 and S11).14, 15, 77 We measured the density of Th+ innervation in the striatum of Cplx1−/− mice and controls (Figure 5D, Table S10) and found that Cplx1−/− mice had significantly lower Th+ staining in the striatum (p value = 3.385 × 10−8; Figure 5E). This indicates that Cplx1 KO mice have less Th+ fiber innervation and a compromised nigrostriatal pathway, supporting its biological significance in MB DA populations and to PD.
Discussion
Midbrain DA neurons in the SN have been the subject of intense research since being definitively linked to PD nearly 100 years ago.78 While degeneration of SN DA neurons in PD is well established, they represent only a subset of brain DA populations. It remains unknown why nigral DA neurons are particularly vulnerable. We set out to explore this question using scRNA-seq. Recently, others have used scRNA-seq to characterize the mouse MB, including DA neurons.38 Here, we extend these data significantly, extensively characterizing the transcriptomes of multiple brain DA populations longitudinally and discovering GRNs associated with specific populations.
A Postnatal MB Th+ Cell Type Is a Putative Progenitor-like MB DA Neuron
Our analysis of embryonic and postnatal MB Th+ neurons revealed a population of neurons, present at both embryonic and postnatal time points (E15.MB.1 and P7.MB.2), that share expressed genes indicative of MB DA neuron progenitors. While progenitor cell populations in the ventral MB have been previously characterized at embryonic time points,38 the existence of a postnatal MB progenitor neuron population has not been noted in previous single-cell studies.38, 57 Notably, previous studies characterized postnatal neurons marked by transgenes under Slc6a3 regulatory control. Given that we demonstrate this marker to be absent from P7.MB.2 cluster, it follows that this population would likely have been overlooked. By contrast, our use of Th left this population available for discovery. We show through smFISH that specific markers for this population place it in the dorsal portion of the SN at P7.
One may speculate regarding the function of a postnatal MB progenitor population. While beyond the scope of this paper, some clues may be found in the literature about Th+ neuron development. Studies of SN DA neuron development in mice have shown that there are two periods of programmed cell death, with peak apoptosis occurring at P2 and P14 (Figure 1A).79 Paradoxically, even though there are high levels of cell death at these points, the actual number of Th+ neurons in the mouse SN does not decrease.79, 80 It has been shown that this can be explained by increasing levels of Th in cells over time, leading to “new” neurons appearing that are able to be immunostained.79 These results have led to the suggestion that there is a “phenotypic maturation” of MB DA neurons during the early postnatal time period.79 This phenomenon may explain the presence of our “progenitor-like” MB DA neurons at P7, which display much lower levels of Th than other populations.
Prioritization of Genes within PD GWAS Loci Identifies Genes that May Contribute to Common PD Susceptibility
Our data facilitate the iterative and biologically informed prioritization of gene candidates for all PD-associated genomic intervals. In practice, the gene closest to the lead SNP identified within a GWAS locus is frequently treated as the prime candidate gene, often without considering tissue-dependent context. Our study overcomes this by integrating genomic data derived from specific cell contexts with analyses that are agnostic to one another. We posit that genes pertinent to PD are likely expressed within SN DA neurons. This hypothesis is consistent with the recent description of the “omnigenic” nature of common disease, wherein variation impacting genes expressed in a disease tissue explain the vast majority of risk.7
First, we identify intervals that reveal one primary candidate, i.e., those that harbor only one SN-expressed gene. Next, we examine those intervals with many candidates and prioritize based on a cumulative body of biological evidence. In total, we prioritize 5 or fewer candidates in 47/49 (∼96%) PD GWAS loci studied, identifying a single gene in 24 loci (24/49; ∼49%) and 3 or fewer genes in ∼84% of loci (41/49). Ultimately this prioritization reduces the candidate gene list for PD GWAS loci dramatically from 1,751 genes to 112 genes.
The top genes we identify in three PD loci (rs356182, SNCA; rs591323, FGF20; rs11158026, GCH1) have been directly associated with PD, MB DA development, and MB DA function56 (MIM: 163890, 128230). Furthermore, our prioritization of CPLX1 in the rs34311866 locus is supported by multiple lines of evidence. Additionally, we demonstrate that the integrity of the nigrostriatal pathway is disrupted in Cplx1 knockout mice. Dysregulation of CPLX1 RNA is also a biomarker in individuals with pre-PD prodromal phenotypes harboring the PARK4 mutation (SNCA gene duplication).81 These results validate our approach and strengthen the argument for the use of context-specific data in pinpointing candidate genes in GWAS loci.
In light of the recently described “omnigenic” hypothesis of complex traits, we anticipate that risk variants may impact common cellular pathways within this primary impacted cell population. Consistent with this, many of the genes prioritized (Table 2) have been shown to impact mitochondrial biology,82, 83, 84, 85, 86 the dysfunction of which has been extensively implicated in PD.87 The prioritized genes may represent “core” genes that in turn can affect the larger mitochondrial-associated regulatory networks active in the disease-relevant cell type (SN DA neurons). One such gene we identify is PARL (presenilin-associated rhomboid like). PARL encodes a protease that cleaves PINK1, which has been implicated in PD pathology.88, 89, 90 Further, recently a variant in PARL has been associated, but not definitively linked, with early-onset PD (MIM: 607858).
While our method successfully prioritized one gene with a known role in familial PD (SNCA), we do not prioritize LRRK2, another familial PD-associated gene harbored within a PD GWAS locus (rs76904798 locus). LRRK2 is not prioritized simply because it is not detectably expressed in our SN DA neuronal population. This is expected as numerous studies have reported little to no LRRK2 expression in Th+ MB DA neurons both in mice and humans.91, 92 Instead, our method prioritizes PDZRN4. This result does not necessarily argue against the potential relevance of LRRK2 but instead provides an additional candidate that may contribute to PD susceptibility. Further, we acknowledge that our focus on SN neurons risks overlooking variants whose immediate functional context lies in other cells, yielding non-cell-autonomous influence on the SN (see discussion below). This same logic should be noted for two other PD-associated loci (rs35749011 and rs17649553), wherein our scoring prioritizes different genes (KCNN3 and CRHR1/NSF, respectively) than one previously implicated in PD (GBA and MAPT) (MIM: 168600). Notably, KCNN3, CRHR1, and NSF all have previous biological evidence making them plausible candidates.93, 94, 95
Comparison of PD Gene Prioritization Schemes
Studying disease-relevant tissue has proven to be essential for elucidating the genetic architecture underlying GWA signals;2 our scoring method relies upon data from the most overtly relevant cell type to PD, SN DA neurons. While this study was under consideration for publication, Chang and colleagues11 endeavored to prioritize PD GWAS loci using publically available data. Although their pipeline strives to be “neuro-centric,” it is not predicated on the biological relevance of candidates to SN DA neurons.
Through comparison of the two scoring paradigms, the methods agree on at least one gene in 17/44 (∼39%) jointly scored loci, including SNCA (Table S12), bolstering the evidence for those candidate genes. However, we see ∼44% (31/71) of the genes prioritized by Chang et al.11 are not expressed in either of the SN DA expression datasets used in our scoring scheme (Table S12), including LRRK2 (addressed above). One prime example of this discrepancy is the rs12637471 locus. Chang et al.11 identify MCCC1 to be the prime candidate in the locus. However, we find that MCCC1 is not expressed in SN DA neurons (Table S8). Instead, we prioritize PARL, which has an established role in a PD pathogenesis pathway.88, 89
Our focus on disease-relevant cell type data also leads us to identify genes previously implicated in neurodegeneration, which make obvious candidates. As described, in the rs34311866 locus, we identify CPLX1 and functionally confirm its relevance. We also identify ATRN (attractin) in the rs8118008 locus. Loss of Atrn has been shown to cause age-related neurodegeneration of SN DA neurons in rats,96, 97 making it an ideal candidate. Neither gene is identified using other metrics11 (Table S12).
Despite this success, we acknowledge several notable caveats. First, not all genes in PD-associated human loci have identified mouse homologs via the MGI homology database used. The majority of genes without identified mouse homologs are classified as non-coding genes which include microRNAs (miRNAs), long non-coding RNAs (lncRNAs), and pseudogenes (Figure S7). Thus, it remains possible that we may have overlooked the contribution of some human non-coding genes whose biology cannot be comprehensively queried in this study.
Second, we assume that identified genetic variation acts in a manner that is at least preferential, if not exclusive, to SN DA neurons. It is possible that genetic variation contributing to risk of PD may be acting in other cell types. While SN DA neurons are primarily affected in PD, other cell types, especially microglia and astrocytes, have been shown to play a role in PD.98, 99 In addition, both of these cell types have been implicated in PD through the process of neuroinflammation.98, 99 Intriguingly, PD-causing mutations in LRRK2 have been shown to affect microglia and play a role in neuroinflammation.100 The expression and function of LRRK2 in microglia instead of SN DA neurons could be another explanation as to why LRRK2 is not prioritized in our scoring system. While out of the scope of this paper, future work will be needed to assess whether glial transcriptional landscapes or genes modulated by neuroinflammation could explain some genetic signals underlying sporadic PD.
These caveats notwithstanding, our strategy sets the stage for a new generation of independent and combinatorial functional evaluation of gene candidates for PD-associated genomic intervals. Emerging studies, including ours, highlight the need for strategies that can systematically identify biologically pertinent gene candidates. Such strategies are necessary for the community to take full advantage of the immense body of GWAS data now in the public domain. We demonstrate the potential power of integrating scRNA-seq data from disease-relevant populations to illuminate corresponding GWASs and facilitate systematic prioritization and testing of gene candidates within risk loci.
Acknowledgments
The authors wish to thank Stephen M. Brown for implementation and optimization of smFISH, Dr. Zhiguang Zheng and Mrs. Wendy Leavens for excellent technical support with the Cplx1 knockout mice and immunohistochemistry, and Drs. Kerstin Reim and Niels Brose for the gift of the founder mice for the Cambridge Cplx1 knockout mice colony. This research was supported in part by US NIH grants R01 NS62972 and MH106522 to A.S.M. and a grant from CHDI Inc. to A.J.M.
Published: March 1, 2018
Footnotes
Supplemental Data include 7 figures and 12 tables and can be found with this article online at https://doi.org/10.1016/j.ajhg.2018.02.001.
Contributor Information
Loyal A. Goff, Email: loyalgoff@jhmi.edu.
Andrew S. McCallion, Email: andy@jhmi.edu.
Accession Numbers
Single-cell RNA-sequencing data is available at the Gene Expression Omnibus (GEO) under the accession number GSE108020.
Web Resources
ACDBio, https://acdbio.com/
ADPclust, https://cran.r-project.org/web/packages/ADPclust/vignettes/ADPclust.html
Allen Brain Atlas, http://www.brain-map.org/
Allen Mouse Brain Atlas, http://mouse.brain-map.org/static/atlas
biomaRt R package, https://bioconductor.org/packages/release/bioc/html/biomaRt.html
Charles River Laboratories, https://www.criver.com/
clusterProfiler, https://guangchuangyu.github.io/clusterProfiler/
cuffnorm and cuffquant, http://cole-trapnell-lab.github.io/cufflinks/
CummeRbund, http://compbio.mit.edu/cummeRbund/
ExAC Browser, http://exac.broadinstitute.org/
GENCODE, http://www.gencodegenes.org
GitHub repository containing code and documentation, https://github.com/pwh124/sc-da-parkinsons
GWAS Catalog, http://www.ebi.ac.uk/gwas/
Hi-C project, http://chromosome.sdsc.edu/mouse/hi-c/download.html
ImageJ, https://imagej.nih.gov/ij/
MGI Human-to-Mouse homolog data (accessed 07/07/2017), http://www.informatics.jax.org/downloads/reports/HOM_MouseHumanSequence.rpt
MMRRC, http://mmrrc.org/
Monocle 2, http://cole-trapnell-lab.github.io/monocle-release/
Mouse Genome Informatics, http://www.informatics.jax.org/
OMIM, http://www.omim.org/
R statistical software, http://www.r-project.org/
Tsne R package, https://github.com/jdonaldson/rtsne
UCSC Genome Browser, http://genome.ucsc.edu
WGCNA, http://labs.genetics.ucla.edu/horvath/CoexpressionNetwork/Rpackages/WGCNA/
Supplemental Data
Provides additional information for Table 1.
This includes the “Lead SNP” associated with each locus, the “Closest Genes” to the lead SNP, and whether or not the closest genes are expressed (“Closest Gene Expressed”). This also has meta-data for genes in each locus including: the number of human genes (“num_genes”), the number of genes expressed in either of the SN DA scRNA-seq datasets used in scoring (“num_expressed_either”), the number of genes expressed in both SN DA scRNA-seq datasets using in scoring (“num_expressed_both”), the number of genes that had a one-toone mouse homolog (“num_homolog”), and the number of genes that did not have a oneto- one mouse homolog (“num_no_homolog”).
References
- 1.Visscher P.M., Brown M.A., McCarthy M.I., Yang J. Five years of GWAS discovery. Am. J. Hum. Genet. 2012;90:7–24. doi: 10.1016/j.ajhg.2011.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Maurano M.T., Humbert R., Rynes E., Thurman R.E., Haugen E., Wang H., Reynolds A.P., Sandstrom R., Qu H., Brody J. Systematic localization of common disease-associated variation in regulatory DNA. Science. 2012;337:1190–1195. doi: 10.1126/science.1222794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Farh K.K., Marson A., Zhu J., Kleinewietfeld M., Housley W.J., Beik S., Shoresh N., Whitton H., Ryan R.J., Shishkin A.A. Genetic and epigenetic fine mapping of causal autoimmune disease variants. Nature. 2015;518:337–343. doi: 10.1038/nature13835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smemo S., Tena J.J., Kim K.H., Gamazon E.R., Sakabe N.J., Gómez-Marín C., Aneas I., Credidio F.L., Sobreira D.R., Wasserman N.F. Obesity-associated variants within FTO form long-range functional connections with IRX3. Nature. 2014;507:371–375. doi: 10.1038/nature13138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gupta R.M., Hadaya J., Trehan A., Zekavat S.M., Roselli C., Klarin D., Emdin C.A., Hilvering C.R.E., Bianchi V., Mueller C. A genetic variant associated with five vascular diseases is a distal regulator of endothelin-1 gene expression. Cell. 2017;170:522–533.e15. doi: 10.1016/j.cell.2017.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee D., Gorkin D.U., Baker M., Strober B.J., Asoni A.L., McCallion A.S., Beer M.A. A method to predict the impact of regulatory variants from DNA sequence. Nat. Genet. 2015;47:955–961. doi: 10.1038/ng.3331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boyle E.A., Li Y.I., Pritchard J.K. An expanded view of complex traits: from polygenic to omnigenic. Cell. 2017;169:1177–1186. doi: 10.1016/j.cell.2017.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Rijk M.C., Tzourio C., Breteler M.M., Dartigues J.F., Amaducci L., Lopez-Pousa S., Manubens-Bertran J.M., Alpérovitch A., Rocca W.A. Prevalence of parkinsonism and Parkinson’s disease in Europe: the EUROPARKINSON Collaborative Study. European Community Concerted Action on the Epidemiology of Parkinson’s disease. J. Neurol. Neurosurg. Psychiatry. 1997;62:10–15. doi: 10.1136/jnnp.62.1.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pringsheim T., Jette N., Frolkis A., Steeves T.D. The prevalence of Parkinson’s disease: a systematic review and meta-analysis. Mov. Disord. 2014;29:1583–1590. doi: 10.1002/mds.25945. [DOI] [PubMed] [Google Scholar]
- 10.Nalls M.A., Pankratz N., Lill C.M., Do C.B., Hernandez D.G., Saad M., DeStefano A.L., Kara E., Bras J., Sharma M., International Parkinson’s Disease Genomics Consortium (IPDGC) Parkinson’s Study Group (PSG) Parkinson’s Research: The Organized GENetics Initiative (PROGENI) 23andMe. GenePD. NeuroGenetics Research Consortium (NGRC) Hussman Institute of Human Genomics (HIHG) Ashkenazi Jewish Dataset Investigator. Cohorts for Health and Aging Research in Genetic Epidemiology (CHARGE) North American Brain Expression Consortium (NABEC) United Kingdom Brain Expression Consortium (UKBEC) Greek Parkinson’s Disease Consortium. Alzheimer Genetic Analysis Group Large-scale meta-analysis of genome-wide association data identifies six new risk loci for Parkinson’s disease. Nat. Genet. 2014;46:989–993. doi: 10.1038/ng.3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chang D., Nalls M.A., Hallgrímsdóttir I.B., Hunkapiller J., van der Brug M., Cai F., Kerchner G.A., Ayalon G., Bingol B., Sheng M., International Parkinson’s Disease Genomics Consortium. 23andMe Research Team A meta-analysis of genome-wide association studies identifies 17 new Parkinson’s disease risk loci. Nat. Genet. 2017;49:1511–1516. doi: 10.1038/ng.3955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Puschmann A. Monogenic Parkinson’s disease and parkinsonism: clinical phenotypes and frequencies of known mutations. Parkinsonism Relat. Disord. 2013;19:407–415. doi: 10.1016/j.parkreldis.2013.01.020. [DOI] [PubMed] [Google Scholar]
- 13.Klein C., Westenberger A. Genetics of Parkinson’s disease. Cold Spring Harb. Perspect. Med. 2012;2:a008888. doi: 10.1101/cshperspect.a008888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Glynn D., Drew C.J., Reim K., Brose N., Morton A.J. Profound ataxia in complexin I knockout mice masks a complex phenotype that includes exploratory and habituation deficits. Hum. Mol. Genet. 2005;14:2369–2385. doi: 10.1093/hmg/ddi239. [DOI] [PubMed] [Google Scholar]
- 15.Glynn D., Sizemore R.J., Morton A.J. Early motor development is abnormal in complexin 1 knockout mice. Neurobiol. Dis. 2007;25:483–495. doi: 10.1016/j.nbd.2006.10.011. [DOI] [PubMed] [Google Scholar]
- 16.Saxena A., Wagatsuma A., Noro Y., Kuji T., Asaka-Oba A., Watahiki A., Gurnot C., Fagiolini M., Hensch T.K., Carninci P. Trehalose-enhanced isolation of neuronal sub-types from adult mouse brain. Biotechniques. 2012;52:381–385. doi: 10.2144/0000113878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Picelli S., Faridani O.R., Björklund Å.K., Winberg G., Sagasser S., Sandberg R. Full-length RNA-seq from single cells using Smart-seq2. Nat. Protoc. 2014;9:171–181. doi: 10.1038/nprot.2014.006. [DOI] [PubMed] [Google Scholar]
- 18.Kim D., Langmead B., Salzberg S.L. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods. 2015;12:357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mudge J.M., Harrow J. Creating reference gene annotation for the mouse C57BL6/J genome assembly. Mamm. Genome. 2015;26:366–378. doi: 10.1007/s00335-015-9583-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Trapnell C., Roberts A., Goff L., Pertea G., Kim D., Kelley D.R., Pimentel H., Salzberg S.L., Rinn J.L., Pachter L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Trapnell C., Cacchiarelli D., Grimsby J., Pokharel P., Li S., Morse M., Lennon N.J., Livak K.J., Mikkelsen T.S., Rinn J.L. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol. 2014;32:381–386. doi: 10.1038/nbt.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huber W., Carey V.J., Gentleman R., Anders S., Carlson M., Carvalho B.S., Bravo H.C., Davis S., Gatto L., Girke T. Orchestrating high-throughput genomic analysis with Bioconductor. Nat. Methods. 2015;12:115–121. doi: 10.1038/nmeth.3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qiu X., Hill A., Packer J., Lin D., Ma Y.A., Trapnell C. Single-cell mRNA quantification and differential analysis with Census. Nat. Methods. 2017;14:309–315. doi: 10.1038/nmeth.4150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Van Der Maaten L., Hinton G. Visualizing data using t-SNE. J. Mach. Learn. Res. 2008;9:2579–2605. [Google Scholar]
- 25.Wang X.-F., Xu Y. Fast clustering using adaptive density peak detection. Stat. Methods Med. Res. 2017;26:2800–2811. doi: 10.1177/0962280215609948. [DOI] [PubMed] [Google Scholar]
- 26.Trapnell C., Hendrickson D.G., Sauvageau M., Goff L., Rinn J.L., Pachter L. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat. Biotechnol. 2013;31:46–53. doi: 10.1038/nbt.2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Burns J.C., Kelly M.C., Hoa M., Morell R.J., Kelley M.W. Single-cell RNA-Seq resolves cellular complexity in sensory organs from the neonatal inner ear. Nat. Commun. 2015;6:8557. doi: 10.1038/ncomms9557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Molyneaux B.J., Goff L.A., Brettler A.C., Chen H.H., Brown J.R., Hrvatin S., Rinn J.L., Arlotta P. DeCoN: genome-wide analysis of in vivo transcriptional dynamics during pyramidal neuron fate selection in neocortex. Neuron. 2015;85:275–288. doi: 10.1016/j.neuron.2014.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., Mesirov J.P. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mootha V.K., Lindgren C.M., Eriksson K.F., Subramanian A., Sihag S., Lehar J., Puigserver P., Carlsson E., Ridderstråle M., Laurila E. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 31.Yu G., Wang L.-G., Han Y., He Q.-Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Langfelder P., Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Langfelder P., Horvath S. Fast R functions for robust correlations and hierarchical clustering. J. Stat. Softw. 2012;46:1–17. [PMC free article] [PubMed] [Google Scholar]
- 34.Langfelder P., Zhang B., Horvath S. Defining clusters from a hierarchical cluster tree: the Dynamic Tree Cut package for R. Bioinformatics. 2008;24:719–720. doi: 10.1093/bioinformatics/btm563. [DOI] [PubMed] [Google Scholar]
- 35.Dixon J.R., Selvaraj S., Yue F., Kim A., Li Y., Shen Y., Hu M., Liu J.S., Ren B. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Durinck S., Moreau Y., Kasprzyk A., Davis S., De Moor B., Brazma A., Huber W. BioMart and Bioconductor: a powerful link between biological databases and microarray data analysis. Bioinformatics. 2005;21:3439–3440. doi: 10.1093/bioinformatics/bti525. [DOI] [PubMed] [Google Scholar]
- 37.Durinck S., Spellman P.T., Birney E., Huber W. Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nat. Protoc. 2009;4:1184–1191. doi: 10.1038/nprot.2009.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.La Manno G., Gyllborg D., Codeluppi S., Nishimura K., Salto C., Zeisel A., Borm L.E., Stott S.R.W., Toledo E.M., Villaescusa J.C. Molecular diversity of midbrain development in mouse, human, and stem cells. Cell. 2016;167:566–580. doi: 10.1016/j.cell.2016.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lek M., Karczewski K.J., Minikel E.V., Samocha K.E., Banks E., Fennell T., O’Donnell-Luria A.H., Ware J.S., Hill A.J., Cummings B.B., Exome Aggregation Consortium Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Heintz N. Gene expression nervous system atlas (GENSAT) Nat. Neurosci. 2004;7 doi: 10.1038/nn0504-483. 483–483. [DOI] [PubMed] [Google Scholar]
- 41.Barallobre M.J., Perier C., Bové J., Laguna A., Delabar J.M., Vila M., Arbonés M.L. DYRK1A promotes dopaminergic neuron survival in the developing brain and in a mouse model of Parkinson’s disease. Cell Death Dis. 2014;5:e1289. doi: 10.1038/cddis.2014.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Qiu X., Mao Q., Tang Y., Wang L., Chawla R., Pliner H.A., Trapnell C. Reversed graph embedding resolves complex single-cell trajectories. Nat. Methods. 2017;14:979–982. doi: 10.1038/nmeth.4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Arenas E., Denham M., Villaescusa J.C. How to make a midbrain dopaminergic neuron. Development. 2015;142:1918–1936. doi: 10.1242/dev.097394. [DOI] [PubMed] [Google Scholar]
- 44.Björklund A., Dunnett S.B. Dopamine neuron systems in the brain: an update. Trends Neurosci. 2007;30:194–202. doi: 10.1016/j.tins.2007.03.006. [DOI] [PubMed] [Google Scholar]
- 45.Li H., Zeitler P.S., Valerius M.T., Small K., Potter S.S. Gsh-1, an orphan Hox gene, is required for normal pituitary development. EMBO J. 1996;15:714–724. [PMC free article] [PubMed] [Google Scholar]
- 46.Campbell J.N., Macosko E.Z., Fenselau H., Pers T.H., Lyubetskaya A., Tenen D., Goldman M., Verstegen A.M., Resch J.M., McCarroll S.A. A molecular census of arcuate hypothalamus and median eminence cell types. Nat. Neurosci. 2017;20:484–496. doi: 10.1038/nn.4495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Agoston Z., Heine P., Brill M.S., Grebbin B.M., Hau A.C., Kallenborn-Gerhardt W., Schramm J., Götz M., Schulte D. Meis2 is a Pax6 co-factor in neurogenesis and dopaminergic periglomerular fate specification in the adult olfactory bulb. Development. 2014;141:28–38. doi: 10.1242/dev.097295. [DOI] [PubMed] [Google Scholar]
- 48.Francis F., Koulakoff A., Boucher D., Chafey P., Schaar B., Vinet M.C., Friocourt G., McDonnell N., Reiner O., Kahn A. Doublecortin is a developmentally regulated, microtubule-associated protein expressed in migrating and differentiating neurons. Neuron. 1999;23:247–256. doi: 10.1016/s0896-6273(00)80777-1. [DOI] [PubMed] [Google Scholar]
- 49.Gokce O., Stanley G.M., Treutlein B., Neff N.F., Camp J.G., Malenka R.C., Rothwell P.E., Fuccillo M.V., Südhof T.C., Quake S.R. Cellular taxonomy of the mouse striatum as revealed by single-cell RNA-seq. Cell Rep. 2016;16:1126–1137. doi: 10.1016/j.celrep.2016.06.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vergaño-Vera E., Díaz-Guerra E., Rodríguez-Traver E., Méndez-Gómez H.R., Solís Ó., Pignatelli J., Pickel J., Lee S.H., Moratalla R., Vicario-Abejón C. Nurr1 blocks the mitogenic effect of FGF-2 and EGF, inducing olfactory bulb neural stem cells to adopt dopaminergic and dopaminergic-GABAergic neuronal phenotypes. Dev. Neurobiol. 2015;75:823–841. doi: 10.1002/dneu.22251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Panman L., Papathanou M., Laguna A., Oosterveen T., Volakakis N., Acampora D., Kurtsdotter I., Yoshitake T., Kehr J., Joodmardi E. Sox6 and Otx2 control the specification of substantia nigra and ventral tegmental area dopamine neurons. Cell Rep. 2014;8:1018–1025. doi: 10.1016/j.celrep.2014.07.016. [DOI] [PubMed] [Google Scholar]
- 52.Viereckel T., Dumas S., Smith-Anttila C.J., Vlcek B., Bimpisidis Z., Lagerström M.C., Konradsson-Geuken Å., Wallén-Mackenzie Å. Midbrain gene screening identifies a new mesoaccumbal glutamatergic pathway and a marker for dopamine cells neuroprotected in Parkinson’s disease. Sci. Rep. 2016;6:35203. doi: 10.1038/srep35203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kozicz T., Vigh S., Arimura A. The source of origin of PACAP- and VIP-immunoreactive fibers in the laterodorsal division of the bed nucleus of the stria terminalis in the rat. Brain Res. 1998;810:211–219. doi: 10.1016/s0006-8993(98)00692-1. [DOI] [PubMed] [Google Scholar]
- 54.Darland T., Heinricher M.M., Grandy D.K. Orphanin FQ/nociceptin: a role in pain and analgesia, but so much more. Trends Neurosci. 1998;21:215–221. doi: 10.1016/s0166-2236(97)01204-6. [DOI] [PubMed] [Google Scholar]
- 55.Cai H., Liu G., Sun L., Ding J. Aldehyde Dehydrogenase 1 making molecular inroads into the differential vulnerability of nigrostriatal dopaminergic neuron subtypes in Parkinson’s disease. Transl. Neurodegener. 2014;3:27. doi: 10.1186/2047-9158-3-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Itoh N., Ohta H. Roles of FGF20 in dopaminergic neurons and Parkinson’s disease. Front. Mol. Neurosci. 2013;6:15. doi: 10.3389/fnmol.2013.00015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Poulin J.F., Zou J., Drouin-Ouellet J., Kim K.Y.A., Cicchetti F., Awatramani R.B. Defining midbrain dopaminergic neuron diversity by single-cell gene expression profiling. Cell Rep. 2014;9:930–943. doi: 10.1016/j.celrep.2014.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Uhde C.W., Vives J., Jaeger I., Li M. Rmst is a novel marker for the mouse ventral mesencephalic floor plate and the anterior dorsal midline cells. PLoS ONE. 2010;5:e8641. doi: 10.1371/journal.pone.0008641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ng S.Y., Bogu G.K., Soh B.S., Stanton L.W. The long noncoding RNA RMST interacts with SOX2 to regulate neurogenesis. Mol. Cell. 2013;51:349–359. doi: 10.1016/j.molcel.2013.07.017. [DOI] [PubMed] [Google Scholar]
- 60.Ellis B.C., Molloy P.L., Graham L.D. CRNDE: A long non-coding RNA involved in CanceR Neurobiology, and DEvelopment. Front. Genet. 2012;3:270. doi: 10.3389/fgene.2012.00270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lin M., Pedrosa E., Shah A., Hrabovsky A., Maqbool S., Zheng D., Lachman H.M. RNA-Seq of human neurons derived from iPS cells reveals candidate long non-coding RNAs involved in neurogenesis and neuropsychiatric disorders. PLoS ONE. 2011;6:e23356. doi: 10.1371/journal.pone.0023356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Guttman M., Donaghey J., Carey B.W., Garber M., Grenier J.K., Munson G., Young G., Lucas A.B., Ach R., Bruhn L. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477:295–300. doi: 10.1038/nature10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Morales M., Margolis E.B. Ventral tegmental area: cellular heterogeneity, connectivity and behaviour. Nat. Rev. Neurosci. 2017;18:73–85. doi: 10.1038/nrn.2016.165. [DOI] [PubMed] [Google Scholar]
- 64.Everitt B.J., Hökfelt T., Wu J.Y., Goldstein M. Coexistence of tyrosine hydroxylase-like and gamma-aminobutyric acid-like immunoreactivities in neurons of the arcuate nucleus. Neuroendocrinology. 1984;39:189–191. doi: 10.1159/000123977. [DOI] [PubMed] [Google Scholar]
- 65.Asmus S.E., Cocanougher B.T., Allen D.L., Boone J.B., Brooks E.A., Hawkins S.M., Hench L.A., Ijaz T., Mayfield M.N. Increasing proportions of tyrosine hydroxylase-immunoreactive interneurons colocalize with choline acetyltransferase or vasoactive intestinal peptide in the developing rat cerebral cortex. Brain Res. 2011;1383:108–119. doi: 10.1016/j.brainres.2011.01.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pascoli V., Terrier J., Hiver A., Lüscher C. Sufficiency of mesolimbic dopamine neuron stimulation for the progression to addiction. Neuron. 2015;88:1054–1066. doi: 10.1016/j.neuron.2015.10.017. [DOI] [PubMed] [Google Scholar]
- 67.Dekker J., Marti-Renom M.A., Mirny L.A. Exploring the three-dimensional organization of genomes: interpreting chromatin interaction data. Nat. Rev. Genet. 2013;14:390–403. doi: 10.1038/nrg3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lettice L.A., Heaney S.J.H., Purdie L.A., Li L., de Beer P., Oostra B.A., Goode D., Elgar G., Hill R.E., de Graaff E. A long-range Shh enhancer regulates expression in the developing limb and fin and is associated with preaxial polydactyly. Hum. Mol. Genet. 2003;12:1725–1735. doi: 10.1093/hmg/ddg180. [DOI] [PubMed] [Google Scholar]
- 69.Benko S., Fantes J.A., Amiel J., Kleinjan D.J., Thomas S., Ramsay J., Jamshidi N., Essafi A., Heaney S., Gordon C.T. Highly conserved non-coding elements on either side of SOX9 associated with Pierre Robin sequence. Nat. Genet. 2009;41:359–364. doi: 10.1038/ng.329. [DOI] [PubMed] [Google Scholar]
- 70.Yong V.W., Power C., Forsyth P., Edwards D.R. Metalloproteinases in biology and pathology of the nervous system. Nat. Rev. Neurosci. 2001;2:502–511. doi: 10.1038/35081571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li Z., Wu Y., Baraban J.M. The Translin/Trax RNA binding complex: clues to function in the nervous system. Biochim. Biophys. Acta. 2008;1779:479–485. doi: 10.1016/j.bbagrm.2008.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Close J., Xu H., De Marco García N., Batista-Brito R., Rossignol E., Rudy B., Fishell G. Satb1 is an activity-modulated transcription factor required for the terminal differentiation and connectivity of medial ganglionic eminence-derived cortical interneurons. J. Neurosci. 2012;32:17690–17705. doi: 10.1523/JNEUROSCI.3583-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Brichta L., Shin W., Jackson-Lewis V., Blesa J., Yap E.L., Walker Z., Zhang J., Roussarie J.P., Alvarez M.J., Califano A. Identification of neurodegenerative factors using translatome-regulatory network analysis. Nat. Neurosci. 2015;18:1325–1333. doi: 10.1038/nn.4070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Doan R.N., Bae B.I., Cubelos B., Chang C., Hossain A.A., Al-Saad S., Mukaddes N.M., Oner O., Al-Saffar M., Balkhy S., Homozygosity Mapping Consortium for Autism Mutations in human accelerated regions disrupt cognition and social behavior. Cell. 2016;167:341–354. doi: 10.1016/j.cell.2016.08.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Basso M., Giraudo S., Corpillo D., Bergamasco B., Lopiano L., Fasano M. Proteome analysis of human substantia nigra in Parkinson’s disease. Proteomics. 2004;4:3943–3952. doi: 10.1002/pmic.200400848. [DOI] [PubMed] [Google Scholar]
- 76.Gispert S., Kurz A., Brehm N., Rau K., Walter M., Riess O., Auburger G. Complexin-1 and Foxp1 expression changes are novel brain effects of alpha-synuclein pathology. Mol. Neurobiol. 2015;52:57–63. doi: 10.1007/s12035-014-8844-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kielar C., Sawiak S.J., Navarro Negredo P., Tse D.H.Y., Morton A.J. Tensor-based morphometry and stereology reveal brain pathology in the complexin1 knockout mouse. PLoS ONE. 2012;7:e32636. doi: 10.1371/journal.pone.0032636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Parent M., Parent A. Substantia nigra and Parkinson’s disease: a brief history of their long and intimate relationship. Can. J. Neurol. Sci. 2010;37:313–319. doi: 10.1017/s0317167100010209. [DOI] [PubMed] [Google Scholar]
- 79.Jackson-Lewis V., Vila M., Djaldetti R., Guegan C., Liberatore G., Liu J., O’Malley K.L., Burke R.E., Przedborski S. Developmental cell death in dopaminergic neurons of the substantia nigra of mice. J. Comp. Neurol. 2000;424:476–488. doi: 10.1002/1096-9861(20000828)424:3<476::aid-cne6>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- 80.Lieb K., Andersen C., Lazarov N., Zienecker R., Urban I., Reisert I., Pilgrim C. Pre- and postnatal development of dopaminergic neuron numbers in the male and female mouse midbrain. Brain Res. Dev. Brain Res. 1996;94:37–43. doi: 10.1016/0165-3806(96)00063-6. [DOI] [PubMed] [Google Scholar]
- 81.Lahut S., Gispert S., Ömür Ö., Depboylu C., Seidel K., Domínguez-Bautista J.A., Brehm N., Tireli H., Hackmann K., Pirkevi C. Blood RNA biomarkers in prodromal PARK4 and rapid eye movement sleep behavior disorder show role of complexin 1 loss for risk of Parkinson’s disease. Dis. Model. Mech. 2017;10:619–631. doi: 10.1242/dmm.028035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hildick-Smith G.J., Cooney J.D., Garone C., Kremer L.S., Haack T.B., Thon J.N., Miyata N., Lieber D.S., Calvo S.E., Akman H.O. Macrocytic anemia and mitochondriopathy resulting from a defect in sideroflexin 4. Am. J. Hum. Genet. 2013;93:906–914. doi: 10.1016/j.ajhg.2013.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Islam M.M., Suzuki H., Yoneda M., Tanaka M. Primary structure of the smallest (6.4-kDa) subunit of human and bovine ubiquinol-cytochrome c reductase deduced from cDNA sequences. Biochem. Mol. Biol. Int. 1997;41:1109–1116. doi: 10.1080/15216549700202191. [DOI] [PubMed] [Google Scholar]
- 84.Plovanich M., Bogorad R.L., Sancak Y., Kamer K.J., Strittmatter L., Li A.A., Girgis H.S., Kuchimanchi S., De Groot J., Speciner L. MICU2, a paralog of MICU1, resides within the mitochondrial uniporter complex to regulate calcium handling. PLoS ONE. 2013;8:e55785. doi: 10.1371/journal.pone.0055785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wonsey D.R., Zeller K.I., Dang C.V. The c-Myc target gene PRDX3 is required for mitochondrial homeostasis and neoplastic transformation. Proc. Natl. Acad. Sci. USA. 2002;99:6649–6654. doi: 10.1073/pnas.102523299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Curran J.E., Jowett J.B.M., Abraham L.J., Diepeveen L.A., Elliott K.S., Dyer T.D., Kerr-Bayles L.J., Johnson M.P., Comuzzie A.G., Moses E.K. Genetic variation in PARL influences mitochondrial content. Hum. Genet. 2010;127:183–190. doi: 10.1007/s00439-009-0756-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Winklhofer K.F., Haass C. Mitochondrial dysfunction in Parkinson’s disease. Biochim. Biophys. Acta. 2010;1802:29–44. doi: 10.1016/j.bbadis.2009.08.013. [DOI] [PubMed] [Google Scholar]
- 88.Shi G., Lee J.R., Grimes D.A., Racacho L., Ye D., Yang H., Ross O.A., Farrer M., McQuibban G.A., Bulman D.E. Functional alteration of PARL contributes to mitochondrial dysregulation in Parkinson’s disease. Hum. Mol. Genet. 2011;20:1966–1974. doi: 10.1093/hmg/ddr077. [DOI] [PubMed] [Google Scholar]
- 89.Jin S.M., Lazarou M., Wang C., Kane L.A., Narendra D.P., Youle R.J. Mitochondrial membrane potential regulates PINK1 import and proteolytic destabilization by PARL. J. Cell Biol. 2010;191:933–942. doi: 10.1083/jcb.201008084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Deas E., Plun-Favreau H., Gandhi S., Desmond H., Kjaer S., Loh S.H.Y., Renton A.E.M., Harvey R.J., Whitworth A.J., Martins L.M. PINK1 cleavage at position A103 by the mitochondrial protease PARL. Hum. Mol. Genet. 2011;20:867–879. doi: 10.1093/hmg/ddq526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Galter D., Westerlund M., Carmine A., Lindqvist E., Sydow O., Olson L. LRRK2 expression linked to dopamine-innervated areas. Ann. Neurol. 2006;59:714–719. doi: 10.1002/ana.20808. [DOI] [PubMed] [Google Scholar]
- 92.Higashi S., Moore D.J., Colebrooke R.E., Biskup S., Dawson V.L., Arai H., Dawson T.M., Emson P.C. Expression and localization of Parkinson’s disease-associated leucine-rich repeat kinase 2 in the mouse brain. J. Neurochem. 2007;100:368–381. doi: 10.1111/j.1471-4159.2006.04246.x. [DOI] [PubMed] [Google Scholar]
- 93.Soden M.E., Jones G.L., Sanford C.A., Chung A.S., Güler A.D., Chavkin C., Luján R., Zweifel L.S. Disruption of dopamine neuron activity pattern regulation through selective expression of a human KCNN3 mutation. Neuron. 2013;80:997–1009. doi: 10.1016/j.neuron.2013.07.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Abuirmeileh A., Harkavyi A., Kingsbury A., Lever R., Whitton P.S. The CRF-like peptide urocortin greatly attenuates loss of extracellular striatal dopamine in rat models of Parkinson’s disease by activating CRF(1) receptors. Eur. J. Pharmacol. 2009;604:45–50. doi: 10.1016/j.ejphar.2008.11.009. [DOI] [PubMed] [Google Scholar]
- 95.Simunovic F., Yi M., Wang Y., Macey L., Brown L.T., Krichevsky A.M., Andersen S.L., Stephens R.M., Benes F.M., Sonntag K.C. Gene expression profiling of substantia nigra dopamine neurons: further insights into Parkinson’s disease pathology. Brain. 2009;132:1795–1809. doi: 10.1093/brain/awn323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ueda S., Aikawa M., Ishizuya-Oka A., Yamaoka S., Koibuchi N., Yoshimoto K. Age-related dopamine deficiency in the mesostriatal dopamine system of zitter mutant rats: regional fiber vulnerability in the striatum and the olfactory tubercle. Neuroscience. 2000;95:389–398. doi: 10.1016/s0306-4522(99)00451-0. [DOI] [PubMed] [Google Scholar]
- 97.Nakadate K., Noda T., Sakakibara S., Kumamoto K., Matsuura T., Joyce J.N., Ueda S. Progressive dopaminergic neurodegeneration of substantia nigra in the zitter mutant rat. Acta Neuropathol. 2006;112:64–73. doi: 10.1007/s00401-006-0058-8. [DOI] [PubMed] [Google Scholar]
- 98.Perry V.H. Innate inflammation in Parkinson’s disease. Cold Spring Harb. Perspect. Med. 2012;2:a009373. doi: 10.1101/cshperspect.a009373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Booth H.D.E., Hirst W.D., Wade-Martins R. The role of astrocyte dysfunction in Parkinson’s disease pathogenesis. Trends Neurosci. 2017;40:358–370. doi: 10.1016/j.tins.2017.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Russo I., Bubacco L., Greggio E. LRRK2 and neuroinflammation: partners in crime in Parkinson’s disease? J. Neuroinflammation. 2014;11:52. doi: 10.1186/1742-2094-11-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Provides additional information for Table 1.
This includes the “Lead SNP” associated with each locus, the “Closest Genes” to the lead SNP, and whether or not the closest genes are expressed (“Closest Gene Expressed”). This also has meta-data for genes in each locus including: the number of human genes (“num_genes”), the number of genes expressed in either of the SN DA scRNA-seq datasets used in scoring (“num_expressed_either”), the number of genes expressed in both SN DA scRNA-seq datasets using in scoring (“num_expressed_both”), the number of genes that had a one-toone mouse homolog (“num_homolog”), and the number of genes that did not have a oneto- one mouse homolog (“num_no_homolog”).