Abstract
Neurodegeneration with brain iron accumulation (NBIA) is a genetically heterogeneous condition characterized by progressive dystonia with iron accumulation in the basal ganglia. How NBIA-associated mutations trigger iron overload remains poorly understood. After studying fibroblast cell lines from subjects carrying both known and unreported biallelic mutations in CRAT and REPS1, we ascribe iron overload to the abnormal recycling of transferrin receptor (TfR1) and the reduction of TfR1 palmitoylation in NBIA. Moreover, we describe palmitoylation as a hitherto unreported level of post-translational TfR1 regulation. A widely used antimalarial agent, artesunate, rescued abnormal TfR1 palmitoylation in cultured fibroblasts of NBIA subjects. These observations suggest therapeutic strategies aimed at targeting impaired TfR1 recycling and palmitoylation in NBIA.
Keywords: neurodegeneration with brain iron accumulation, NBIA, iron overload, transferrin receptor, palmitoylation, mitochondria, artesunate
Introduction
Neurodegeneration with brain iron accumulation (NBIA; NBIA1 [MIM: 234200], NBIA2A [MIM: 256600], NBIA3 [MIM: 606159], NBIA4 [MIM: 614298], NBIA5 [MIM: 300894], NBIA6 [MIM: 615643], SPG35 [MIM: 612319], KRS [MIM: 606693], Woodhouse-Sakati syndrome [MIM: 241080], and aceruloplasminemia [MIM: 604290]) is a genetically heterogeneous condition due to iron accumulation in the basal ganglia and manifests as a progressive extrapyramidal syndrome with dystonia, rigidity, and choreoathetosis. Mutations in 11 genes (PANK2 [MIM: 606157], PLA2G6 [MIM: 603604], COASY [MIM: 609855], FA2H [MIM: 611026], ATP13A2 [MIM: 610513], C2orf37 [MIM: 312515], WDR45 [MIM: 300526], C19ORF12 [MIM: 164297], CP [MIM: 117700], FTL [MIM: 134790], and GTPBP2 [MIM: 607434]) have been hitherto identified.1, 2 Mutations in PLA2G6, encoding a phospholipase A2, and PANK2, encoding the mitochondrial pantothenate kinase 2, are the most prevalent known cause of NBIA.
Only two genes, FTL (encoding the light subunit of ferritin) and CP (encoding ceruloplasmin, a metalloprotein binding plasma copper also involved in peroxidation of Fe(II) to Fe(III) transferrin (Tf)), are reportedly involved in iron metabolism. How the other NBIA proteins disturb iron metabolism remains poorly understood. On the other hand, iron has long been known to enter cells by either internalization of Tf receptor 1 (TfR1)-mediated endocytosis or non-Tf bound iron uptake (NTBI).3, 4 In living cells, newly imported iron is primarily used in the mitochondria and surpluses diverted to ferritin, a multimeric protein made up of two subunits, H-ferritin and L-ferritin. Homeostasis of cellular iron is post-transcriptionally regulated by iron-regulatory proteins (IRPs). When the level of cytosolic iron falls, cytosolic aconitase (ACO1) is converted into IRP (IRP1). IRP2, another IRP devoid of aconitase activity, is primarily regulated by iron-dependent degradation. IRPs bind the 3′ UTR of TfR1 mRNA, preventing it from degradation and allowing cells to import more iron. IRPs also bind the 5′ UTR of ferritin mRNAs, thereby inhibiting ferritin synthesis. In high-iron conditions, IRP1 is converted into aconitase and IRP2 is degraded, promoting iron storage and limiting iron entry. Up until now, this post-transcriptional regulation was the only mechanism reportedly known to maintain homeostatic levels of iron primarily via the control of TfR1 and ferritin amounts.5
After studying cultured skin fibroblasts of a series of subjects carrying mutations in both known and previously unreported genes, we report abnormal iron homeostasis and impaired palmitoylation and recycling of TfR1 as common features in NBIA, irrespective of the original defect. This observation paves the way for therapeutic strategies in this devastating group of diseases.
Material and Methods
Informed consent for diagnostic and research studies was obtained for all subjects in accordance with the Declaration of Helsinki protocols and approved by local institutional review boards in Paris.
Exome Sequencing
Exome sequencing was performed on genomic DNA (1 μg) isolated from blood leukocytes. Exons were captured by the in-solution enrichment methodology (SureSelect Human All Exon Kit v.3, Agilent) with the company’s biotinylated oligonucleotide probe library (Human All Exon v.3 50 Mb, Agilent). Each genomic DNA was then sequenced on a sequencer as 75 paired-end bases (Illumina HISEQ2000, Illumina). Image analyses and base calling were performed with Real Time Analysis Pipeline v.1.9 with default parameters (Illumina). Sequences were aligned to the human genome reference sequence (UCSC Genome Browser hg19 assembly). SNPs were called with the Consensus Assessment of Sequence and Variation (CASAVA) pipeline (v.1.8, Illumina) on the basis of the allele calls and read depth. Known SNPs reported in dbSNP, 1000 Genomes, the NHLBI Exome Sequencing Project (ESP) Exome Variant Server, and in-house SNP database—as well as intergenic, non-coding RNA, and UTR variants—were excluded.
Cell Culture
Skin fibroblasts were isolated from subjects 1 and 3 and from four NBIA subjects carrying biallelic PANK2, PLA2G6, C19ORF12, and FA2H mutations (Table S1). Fibroblasts were grown in Dulbecco’s modified Eagle’s medium (DMEM; Life Technologies) supplemented with 10% fetal bovine serum (FBS), 2 mM L-glutamine, 2.5 mM pyruvate, 100 μg/mL streptomycin, and 100 U/mL penicillin at 37°C. For treatment with ferric ammonium citrate (FAC), 90% confluent cells were incubated for 72 hr with or without 100 μM FAC in serum-free DMEM (i.e., Tf free). HeLa cells were grown in the same medium.
Modeling of the Human CRAT Variant
The 1.6 Å coordinate set for human CRAT (PDB: 1NM8) was used for mapping the Arg321 residue. Swiss-PdbViewer 3.7 was used for analyzing the structural insight into the CRAT variant and visualizing the structures.
β-Oxidation of Palmitoyl-CoA
Cells seeded in 6-well plates were incubated when confluent with 250 μM 13C16 -labeled palmitate, fatty-acid-free bovine serum albumin (palmitate/albumin molar ratio: 4/1) and 0.4 mmol/L carnitine for 6 hr. Cells were removed by trypsinization and, after being washed with PBS, stored at −80°C until analysis. For acylcarnitine analysis, pellets were resuspended in 60 μL deionized water and briefly sonicated. Mixed internal standards (20 μL 20 μM 2H9-L-carnitine, 4 μM 2H3 octanoylcarnitine, and 1 μM 2H3 octadecanoylcarnitine) were added to 45 μL cell homogenate. Proteins were precipitated by the addition of 400 μL ethanol, and the supernatant was extracted twice with 400 μL hexane. The aqueous layer was transferred in a vial, dried under nitrogen, and reconstituted with 100 μL acetonitrile/water (1:1 v/v, 0.025% formic acid). Acylcarnitines were analyzed on an API 3000 triple quadripole mass spectrometer (Applied Biosystems), which detected precursors of m/z 85 and quantified them by comparing deuterated internal standards. Proteins were measured by the Lowry method.
Immunoblotting
Cultured skin fibroblasts or HeLa cells were harvested on ice by being scraped in reducing (2 μM dithiothreitol [DTT] and denaturation at 95°C for 5 min) or non-reducing (no DTT and no heat denaturation) cell lysis buffer. SDS-PAGE was performed on whole-cell protein extracts in 10% acrylamide gel. Immunodetections were performed in PBS with 5% milk and 0.05% Tween20 (SIGMA) with the following antibodies: rabbit anti-CRAT (HPA022815, Sigma), mouse anti-REPS1 (ab69221), mouse anti-SOD1 (ab86454), mouse anti-TfR1 (136890, Zymed), rabbit anti-ferritin (ab75973), rabbit anti-IRP1 (ab126595), rabbit anti-IRP2 (ab80339), mouse anti-GRIM19 (ab110240), rabbit anti-catalase (ab16731), rabbit anti-FBXL5 (ab140175), rabbit anti-LC3B (ab51520), and mouse anti-actin antibodies (ab8226). HRP-conjugated goat anti-mouse IgG H&L (ab6789) and goat anti-mouse IgG H&L (ab6789) antibodies were used as secondary antibodies before electrochemiluminescence-based detection (SuperSignal West Dura, Thermo Fisher Scientific). Images were visualized and treated with a CCD camera (Bio-Rad) and ImageLab software (Bio-Rad). Quantification was performed with Bio-Rad imageLab 5.1 and Sigmaplot 13. Detection of carbonyl groups was performed with the OxyBlot Protein Oxidation Detection Kit (S7150, Chemicon) according to the manufacturer’s recommendations.
Knockdown of CRAT
HeLa cells were plated at 80% confluence in 6-well plates on the day before transfection. Cells were transfected with Dharmafect 1 (Dharmacon) and 50 nM siRNA per target in 10% FBS antibiotic-free OPTIMEM, as recommended by the manufacturer. Palmitoyl-CoA β-oxidation and immunoblotting were performed 2 days after transfection. Small interfering RNAs (siRNAs) purchased from Dharmacon were as follows: human carnitine O-acetyltransferase (CRAT [MIM: 600184]) siRNA (ON-TARGETplus SMARTpool L-009524-00-0005) and human non-targeting pool siRNA (ON-TARGETplus Non-targeting Pool).
Overexpression of CRAT and REPS1
The full-length CRAT and REPS1 (RALBP1-associated Eps-domain-containing 1 [MIM: 614825]) cDNAs were cloned in the pLenti7.3 expression vector. For virus production, HEK293FT cells were plated 3 days before transfection in DMEM supplemented with 10% of heat-inactivated FBS (10270106, Invitrogen) at 80% confluence. HEK293FT cells were co-transfected with 30 μL JetPRIME PolyPLUS reagents with the three packaging plasmids REV, VSVG, and PAX2 and the pLenti7.3 expression vector containing either CRAT or REPS1 cDNAs with a 1:3:6:10 DNA ratio. The supernatant containing the viral particles was collected every 12 hr during 3 days, centrifuged at 5,000 rpm for 5 min, filtered through a 0.8 μm filter, and centrifuged for 2 hr at 19,000 rpm at 4°C. Pellets were collected and resuspended in 200 μL DMEM. The fibroblasts were then transduced at 60%–70% confluence with a multiplicity of infection of 5.
Quantification of TFRC Transcripts
Total RNA was extracted with the RNeasy Mini Kit (QIAGEN) and DNase treated by the RNase-free DNase Set (QIAGEN) according to manufacturer’s protocol. The concentration and purity of total RNA were assessed with the NanoDrop 8000 spectrophotometer (Thermo Fisher Scientific) before being stored at −80°C. Then, mRNAs were reverse transcribed from 2 μg total RNA with the High-Capacity RNA-to-cDNA Kit (Thermo Fisher Scientific) and random priming according to the manufacturer’s instructions. Quantitative RT-PCR (qRT-PCR) was performed with digital droplet PCR (ddPCR) with the QX200 DropletDigital PCR System (Bio-Rad). Transferrin receptor (TFRC [MIM: 190010]) cDNAs were amplified with specific primers. β-glucuronidase (GUSB [GenBank: NM_000181.3]) was used for normalization. Primers are shown in Table S2. Data from triplicates were analyzed on a droplet flow cytometer (QX200 Droplet Reader) with QuantaSoft analysis software (Bio-Rad). TFRC expression levels were normalized to the mean copy numbers of the housekeeping gene GUSB.
Iron Content and Imaging Flow Cytometry (Imagestream)
Total iron contents were measured with a ferrozine-based iron assay modified from Barbeito et al.6 For imaging flow cytometry, fibroblasts (1 × 106 to 2 × 106) were starved for 1 hr in FBS-free DMEM, treated with 5 mM EDTA for harvesting them without disrupting TfR1 located at the cell surface (as trypsin does), washed three times with cold PBS, and then labeled with anti-TfR1 antibodies (A24)7 for 1 hr on ice to avoid TfR1 internalization. In this condition, only membrane-bound TfR1 was quantified. Secondary staining was performed with Alexa Fluor 488 goat anti-mouse antibody for 30 min on ice. Cells were washed and stained with Hoechst for 5 min in a total volume of 50 μL, and acquisitions were directly performed. Cell analysis was based on Hoechst-positive signal, allowing selection of living cells. Samples were run on an Imagestream ISX mkII (Amnis) that combines flow cytometry with detailed cell-imaging and functional studies, and a 40× magnification was used for all acquisitions. Data were acquired with INSPIRE software (Amnis) and analyzed with IDEAS software (v.6.2, Amnis). At least 20,000 events were collected in all experiments. Single-stain controls were run for each fluorochrome used, and spectral compensation was performed. Cells were gated for a single cell with the area and aspect ratio of the bright-field image and gated for focused cells with the gradient RMS feature. A specific mask was designed for analysis of the membrane localization of TfR1. Results were expressed as a mean pixel intensity value, which is the intensity normalized to surface area. Data analyses were performed with IDEAS software (Amnis). Data were compensated with a compensation matrix generated with singly stained samples. Gated data were used for generating histograms measuring fluorescence intensity (sum of all pixels in an image) at the plasma membrane with a specific mask. This mask was the result of a full bright-field mask minus a 10 pixel erode bright mask, resulting in a doughnut-like mask. Statistical analyses were performed with GraphPad Prism (v.5.0; GraphPad Software).
Confocal Microscopy
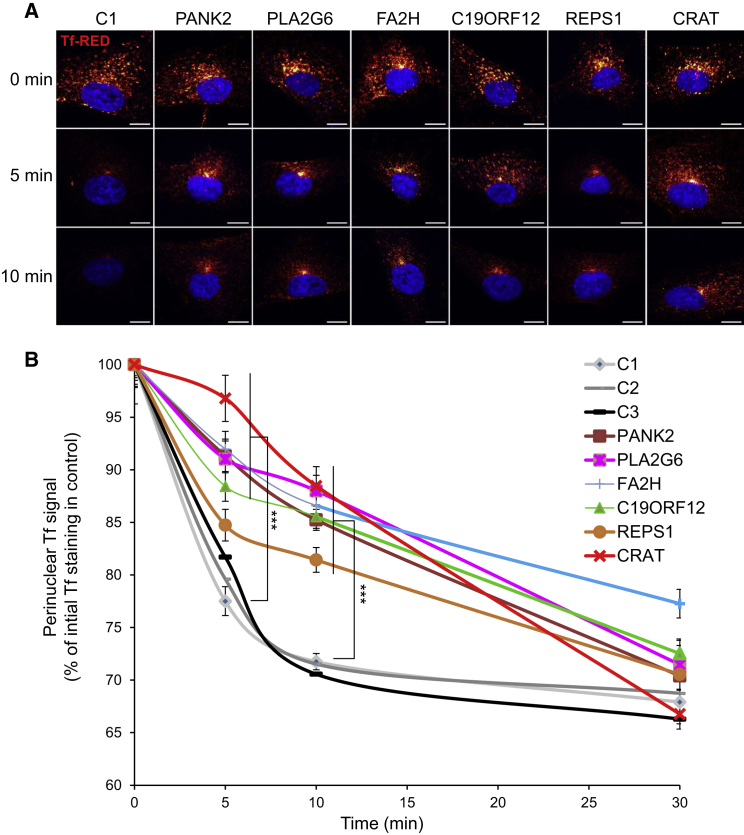
For Tf recycling, fibroblasts were spread at 40% confluence onto 12-well plates containing glass slides 48 hr before experiments. Cells were then starved in FBS-free DMEM for 1 hr at 37°C. Tf-RED (25 μg/mL) was then added for 30 min. Cells were washed three times with PBS and then incubated for 0, 5, 10, or 30 min in FBS-free DMEM. Finally, cells were washed in PBS and fixed in 4% cold paraformaldehyde (PFA). Slides were mounted in DAPI containing Diamond Prolong mounting media (Thermo Fisher Scientific). Slides were examined with a confocal laser microscope (Leica TCS SP8 SMD). Image analysis was performed with ImageJ software. For each independent experiment, at least 30 cells were collected per slide. Each field was selected from DAPI-positive staining, which was used for defining the nuclear region (NR). Each NR was enlarged by 10 pixels for the generation of the perinuclear region of interest (PNROI). The image calculator then applied PNROI masks on the RED channel to obtain quantitative mean fluorescence particle intensity. Only particles of at least 8 pixels were considered. Red fluorescence intensity thresholds were determined according to non-incubated Tf-RED cells.
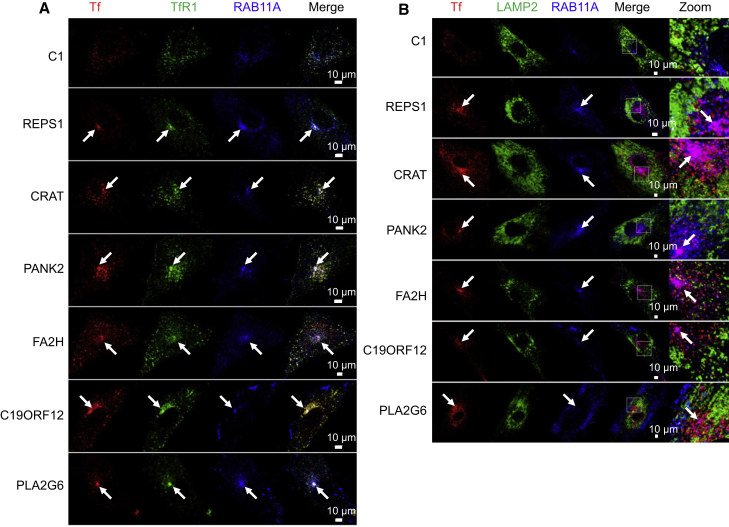
For Tf and TfR1 trafficking, fibroblasts were spread at 40% confluence onto 12-well plates containing glass-slides 48 hr before experiments. Cells were then starved in FBS-free DMEM for 1 hr at 37°C. Tf-RED (25 μg/mL) was then added for 30 min. Cells were washed three times with PBS and then incubated for 10 min in FBS-free DMEM. Finally, cells were washed in PBS and fixed in 4% cold PFA. Confocal microscopy was performed as previously described8 with rabbit anti-Rab11A (71-5300, Zymed), mouse anti-LAMP2 (ab25631), Alexa-Fluor-488-conjugated goat anti-mouse IgG (A-11001, Thermo Fisher Scientific), Alexa-Fluor-594-conjugated goat anti-rabbit IgG (A-11012, Thermo Fisher Scientific) and ProLong Diamond Antifade Mountant with DAPI (P36962, Thermo Fisher Scientific).
Palmitoylation Assay
TfR1 palmitoylation in cultured skin fibroblasts was modified from Ba et al.9 In brief, cells were lysed on ice in a DTT-free cell lysis buffer, and endogenous TfR1 was immunoprecipitated with mouse anti-TfR1 antibody (Zymed) with protein G magnetic beads (Bio-Rad). After being washed with PBS, beads were incubated overnight with 50 mM N-ethylmaleimide at 4°C, washed three times with PBS, and then incubated with 1 M hydroxylamine at room temperature for 2 hr. After three washes, the beads were incubated with 50 mM HPDP-Biotin (Thermo Fisher Scientific) in the dark for 2 hr. Samples were run in 7.5% acrylamide gel, and the biotin-labeled TfR1 level was determined by immunoblotting in non-reducing conditions with a goat anti-biotin antibody (Thermo Fisher Scientific), whereas immunoblotting with rabbit anti-TfR1 antibody (Abcam) was used as a loading control.
Statistics
All statistical analyses were performed with GraphPad Prism 5.0 (GraphPad Software) or SigmaPlot 11 (SYSTAT Software) with a two-tailed, unpaired t test or one-way ANOVA for multiple comparisons by the Holm-Sidak method (∗p < 0.05 and ∗∗∗p < 0.001).
Results
Abnormal Iron Content and Homeostasis in Cultured Fibroblasts of NBIA Subjects
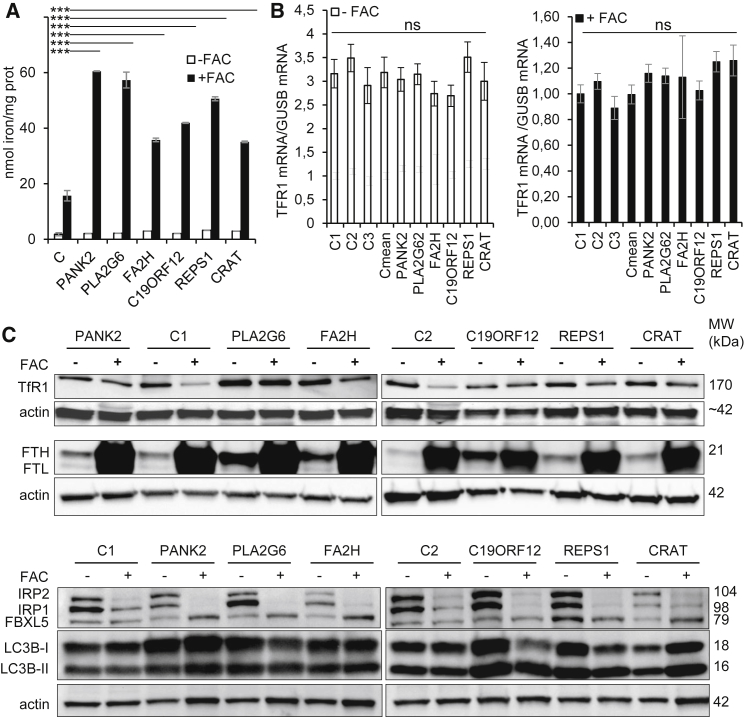
Iron content was quantified in cultured fibroblasts of NBIA subjects (Table S1) with a ferrozine-based colorimetric assay in either low-iron (−FAC; no FBS, i.e., devoid of Tf-bound iron) or high-iron (+FAC) conditions. FAC is a soluble NTBI form that permeates into cells opportunistically via resident transporters or endocytic pathways.4 In low-iron conditions (no FAC or FBS), relatively similar iron content was observed in fibroblasts of control and NBIA subjects (Figure 1A). After a 3 day incubation with FAC, all fibroblasts from NBIA subjects tested exhibited a major cellular iron increase (10- to 30-fold change), whereas control fibroblasts displayed an 8-fold increase (p < 0.001; Figure 1A). These data suggest abnormal iron homeostasis and a possible dysregulation of iron uptake in NBIA.
Figure 1.
Iron Content and Regulation of Iron Homeostasis in Fibroblasts of NBIA Subjects
(A) Iron quantification using the ferrozine-based colorimetric assay in control (C; mean of 3 controls) and NBIA subject fibroblasts grown in FBS-free DMEM and low-iron (−FAC) or high-iron (+FAC) conditions. Data represent the means ± SEM of three independent experiments. A Student-Newman-Keuls ANOVA multifactorial test was used (∗∗∗p < 0.001 [considered significant]; ns, non-significant).
(B) Post-transcriptional regulation of iron homeostasis in control and NBIA subject fibroblasts grown in FBS-free DMEM in −FAC or +FAC conditions. TFRC mRNAs were quantified by ddPCR and expressed as a ratio to GUSB mRNA. Data represent the means ± SEM of three independent experiments. The significance of variations among samples and controls was estimated by the one-way Holm-Sidak method.
(C) Steady-state levels of proteins involved in iron homeostasis. TfR1, IRP1, IRP2, FBXL5, and LC3B were studied in non-reducing conditions (7.5% acrylamide, no DTT, and no heat denaturation), and ferritin was studied in reducing conditions in control (C1 and C2) and NBIA subject fibroblasts grown in FBS-free DMEM in −FAC or +FAC conditions. Actin was used as a loading control. Immunoblotting quantification is presented in Figure S1.
Conversely, quantification by ddPCR of TFRC mRNAs failed to detect differences between control and subject cells grown in either −FAC or +FAC conditions, suggesting a normal downregulation of TFRC RNA transcripts in NBIA in +FAC conditions (Figure 1B). Immunoblot analyses of cultured fibroblasts of NBIA and control subjects grown in −FAC conditions detected similar levels of IRP2 and of a reduced form of IRP1, both of which represent reliable indicators of cell iron status. Moreover, a consistent increase in H- and L-ferritin reflected cellular iron overload in fibroblasts from NBIA subjects (Figure 1C and Figure S1). In control cells, exposure to high amounts of iron (+FAC) resulted in reduced expression of IRPs and consequently in reduced iron uptake concomitant with an increase in ferritins (H and L).
Most interestingly, in fibroblasts of NBIA subjects exposed to high amounts of iron (+FAC), TfR1 levels failed to decrease, whereas those of IRPs and ferritin responded similarly to those of control subjects. The same was observed with FBXL5, an iron-sensor protein reflecting the size of the labile iron pool (Figure 1C and Figure S1). Downregulation of TfR1 normally results from the combined decrease in TFRC transcripts5 and degradation of TfR1 in the lysosome.10 Considering that pre-translational regulation of TfR1 was unaffected in cells from NBIA subject, our results suggest non-canonical, post-translational regulation of TfR1 and its alteration in NBIA. Given that we consistently observed increased amounts of LC3B-II in fibroblasts from NBIA subjects exposed to high amounts of iron (Figure 1C and Figure S1), these data suggest deficient targeting of TfR1 to lysosomes and a reduced degradation of TfR1 in NBIA. Similar abnormal iron overload and TfR1 deregulation were also observed in fibroblasts of another subject (PLA2G6-2) with PLA2G6 mutations (c.[109C>T];[386T>C], p.[Arg37∗];[Leu129Pro]; Figures S3A–S3C).
Mutations in REPS1 and CRAT Are Associated with Abnormal Iron Homeostasis
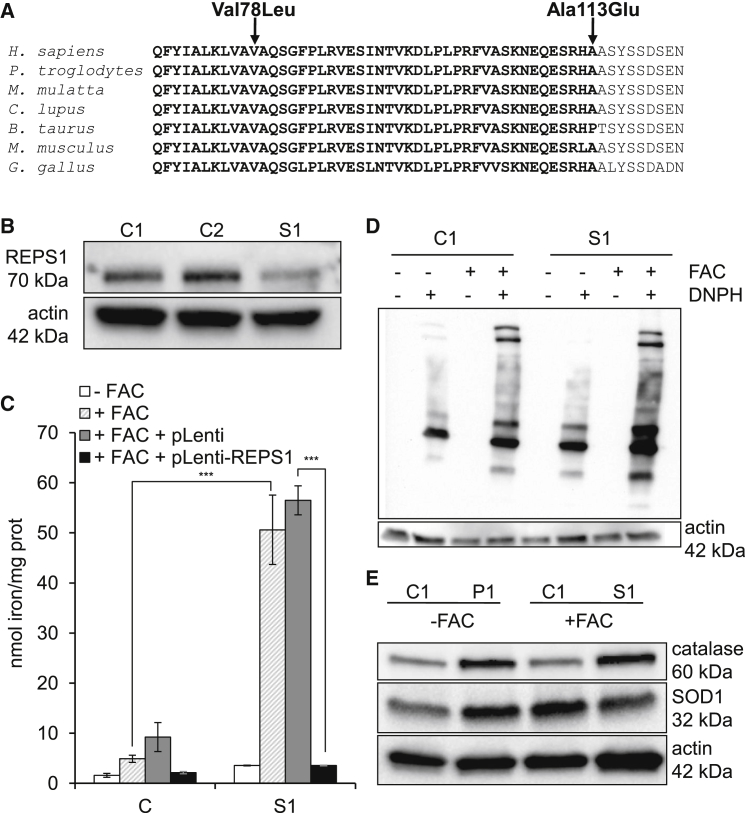
Exome sequencing in two sisters (subjects 1 and 2; see Supplemental Note) detected compound heterozygosity for REPS1 missense variations c.232G>C (p.Val78Leu) and c.338C>A (p.Ala113Glu) (dbSNP: rs201191394; ExAC Browser: A = 0.011%; Figure 2A), which are predicted to be deleterious by in silico analyses (Table S1). The variations co-segregated with the disease in the family and were absent from 110 NBIA subjects and 200 control chromosomes (data not shown). Immunoblot analysis detected low amounts of REPS1 in cultured fibroblasts of subject 1 (Figure 2B).
Figure 2.
REPS1 Variants
(A) Sequence alignment of REPS1 proteins from various species. The arrows indicate the amino acid changes. Bold letters indicate part of the first epsin 15 homology (EH1) domain.
(B) Immunoblot analysis of REPS1 in subject 1 (S1) and two control fibroblasts (C1 and C2). Actin was used as a loading control. Data represent three independent experiments.
(C) Iron quantification in control (C; mean of three control values) and S1 fibroblasts alone or transduced with pLenti7.3 with or without WT REPS1 cDNA by ferrozine-based colorimetric assay and after incubation with or without FAC (+FAC or −FAC, respectively) for 3 days. Data represent the means ± SEM of three independent experiments. Two-tailed Student’s paired or unpaired t tests were used as appropriate (∗p < 0.05, ∗∗∗p < 0.001).
(D) Protein oxidation status in −FAC or +FAC conditions in control and S1 fibroblasts. Proteins were treated with or without 2,4-dinitrophenylhydrazine (DNPH), a derivatization solution. Data represent three independent experiments.
(E) Immunoblot analysis of catalase and SOD1 in S1 and control (C1) fibroblasts in −FAC or +FAC conditions. Actin was used as a loading control. Data represent three independent experiments.
RalBP1 proteins, including REPS1, are involved in endocytosis and vesicle transport. REPS1, an endocytic adaptor localized in clathrin-coated pits, interacts with endocytic scaffold intersectin 1 (ITSN1), a key protein in clathrin-mediated endocytosis.11 REPS1 belongs to the Numb endocytic complex and helps recruit RalBP1 to this complex.12 It is ubiquitously present and conserved across species and contains two Epsin 15 homology domains (EH1 and EH2) involved in protein-protein interactions.13, 14 Both variants are located in the EH1 domain, a conserved region interacting with RalBP1 (Figure 2A). REPS1 also interacts with Rab11-FIP2, located at the membrane of early endosomes.15 Binding of REPS1 to Rab11-FIP2 induces the fusion of uncoated vesicles with the early endosome, forming the endosome recycling compartment.16
When incubated with FAC, fibroblasts with REPS1 mutations displayed a 10-fold increase in iron content similar to that of other NBIA strains (Figures 1A and 2C), and transduction of wild-type (WT) REPS1 cDNA returned iron content to control values in subject fibroblasts (Figure 2C). Labile iron is known to induce oxidative stress by the Fenton reaction. Oxyblot of subject 1 cultured fibroblasts detected slightly increased protein oxidation in −FAC conditions. By contrast, a major increase was observed in +FAC conditions (Figure 2D). Accordingly, steady-state levels of catalase and superoxide dismutase 1 (SOD1) were higher in subject 1 fibroblasts than in control cells in low-iron conditions (Figure 2E). Adding FAC to the culture medium increased protection against free-radical injury in control but not in subject 1 fibroblasts, suggesting that the maximum level of protection was already achieved in basal conditions. We conclude that biallelic REPS1 mutations cause NBIA by a mechanism that impairs endosome recycling.
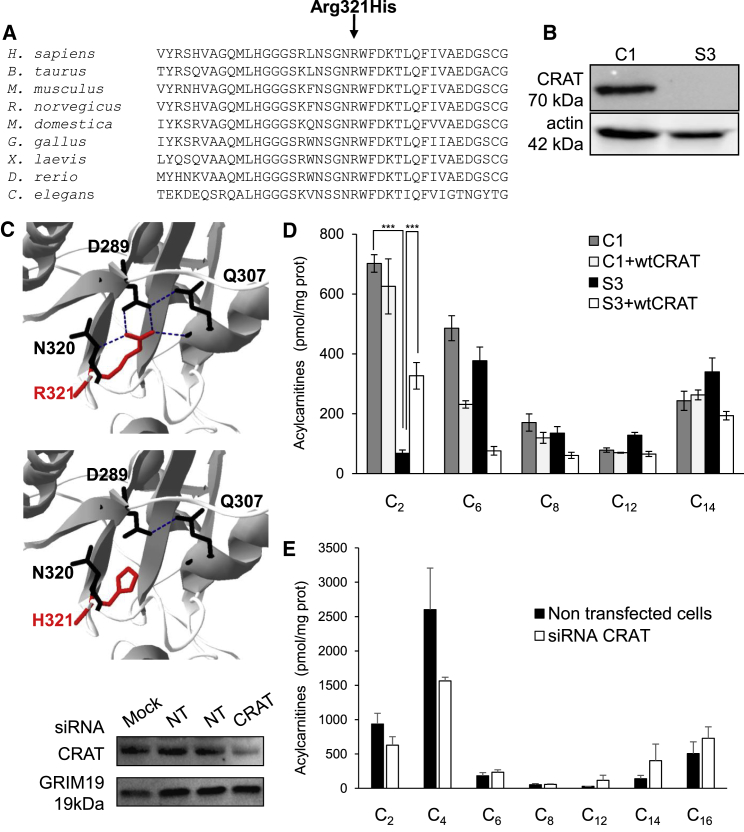
SNP genotyping and exome sequencing in subject 3 (see Supplemental Note) identified the homozygous CRAT variant c.962G>A (dbSNP: rs138665095; ExAC Browser: A = 0.0042%), which alters a highly conserved amino acid residue (p.Arg321His; Figure 3A). The variant was absent from 110 NBIA subject and 200 control chromosomes (data not shown). CRAT belongs to the carnitine acyltransferase family, which catalyzes the reversible transfer of acyl groups from carnitine to coenzyme A (CoA) and regulates the acyl-CoA/CoA ratio. CRAT also plays a crucial role in the transport of fatty acids for β-oxidation.17 Immunoblot analysis failed to detect CRAT in subject 3 fibroblasts (Figure 3B), and on the basis of the crystal structure, the p.Arg321His substitution was expected to destabilize the protein (Figure 3C), given that Arg321, the first residue of an α helix, is involved in the formation of hydrogen bonds with the surrounding amino acids.
Figure 3.
CRAT Variant
(A) Sequence alignment of CRAT proteins from various species. The arrow indicates the amino acid change.
(B) Immunoblot analysis of CRAT in control (C1) and subject 3 (S3) fibroblasts. Actin was used as a loading control.
(C) Localization of the p.Arg321>His (in red) substitutions in a crystallized model of human CRAT. The hydrogen (H) bonds (in blue) between Arg321 and Asp289, Gln307, and Asn320 are shown in the normal protein. The p.Arg321His change modified the H bonds with these three residues.
(D) Palmitoyl-CoA β-oxidation measured by U-13C16-labeled palmitate tracer experiments and mass spectrometry in control (C1) and S3 fibroblasts alone or transduced with pLenti7.3 WT CRAT cDNA.
(E) Knockdown of CRAT in HeLa cells by siRNAs. Left: immunoblot analysis showing the efficiency of knockdown (NT, non-targeted siRNA; GRIM19 was used as a loading control). Right: palmitoyl-CoA β-oxidation measured by U-13C16-labeled palmitate tracer experiments and mass spectrometry in HeLa cells knocked down or not for CRAT.
Data represent the means ± SEM of three independent experiments. Two-tailed Student’s paired or unpaired t tests were used as appropriate (∗p < 0.05, ∗∗∗p < 0.001). Immunoblots in (B) and (E) represent three independent experiments.
Studying β-oxidation of U-13C16 palmitate in cultured fibroblasts of subject 3 detected lower C2 levels than in control cells (Figure 3D), and knocking down CRAT expression by siRNA markedly reduced β-oxidation of labeled palmitoyl-CoA in HeLa cells and C2-C4 acylcarnitine derivatives in comparison with control levels (Figure 3E), a pattern similar to that observed in cultured fibroblasts of subject 3. Conclusive evidence for the disease-causing nature of the CRAT variation was provided by the lentiviral transduction of WT human CRAT cDNA into fibroblasts of subject 3, followed by a shift of palmitoyl-CoA β-oxidation toward control values (Figure 3D). Finally, when incubated with FAC, fibroblasts with CRAT mutations displayed a 10-fold increase in iron content similar to that of other NBIA strains (Figure 1A). We conclude that biallelic CRAT mutations cause NBIA by another mechanism in this family.
Impaired Tf Trafficking and TfR1 Recycling in Fibroblasts of NBIA Subjects
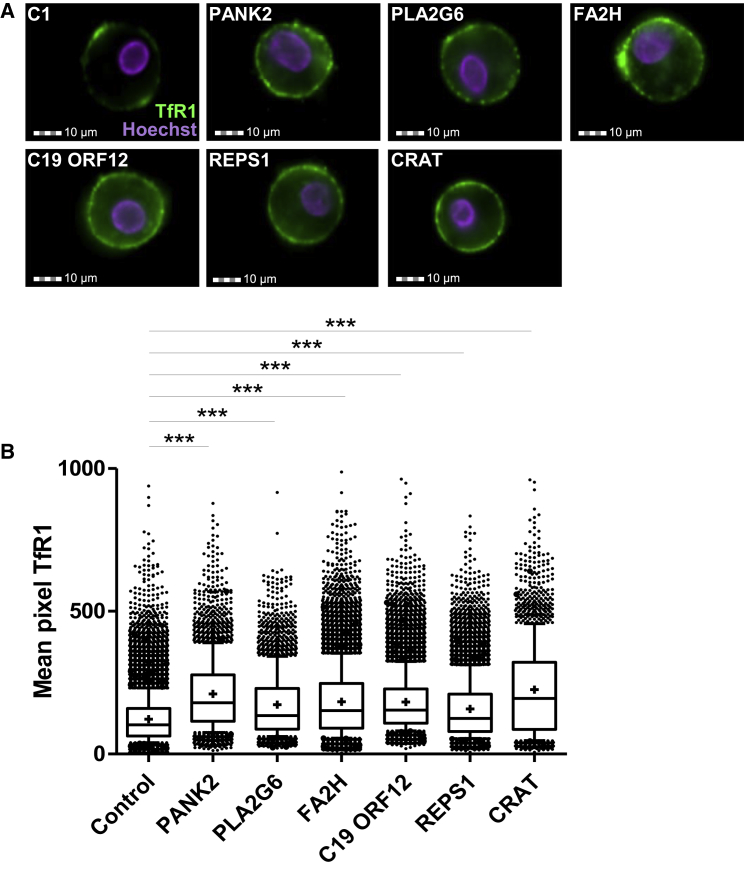
Next-generation imaging flow cytometry revealed increased amounts of TfR1 at the cell surface of NBIA subjects’ fibroblasts (Figure 4A), and quantification using IDEAS software confirmed increased TfR1 signal in affected individuals’ fibroblasts (Figure 4B). This result suggests that, despite iron overload, NBIA compromises the downregulation of iron uptake by TfR1 in cultured fibroblasts. Confocal microscopy using Tf-RED gave further support to this view. Indeed, perinuclear fluorescence after a 30 min Tf-RED pulse and chase showed a rapid decrease in Tf staining, ascribed to normal Tf recycling in control fibroblasts (Figures 5A and 5B). By contrast, Tf recycling was significantly delayed in NBIA subjects’ fibroblasts given that Tf signal failed to decline after the Tf-RED pulse. Moreover, Tf signal accumulated in the vicinity of the nucleus and persisted after 10 min and longer (Figure 5A). These results are consistent with an impairment of receptor-mediated endocytosis and endosome recycling in NBIA.
Figure 4.
Quantification of TfR1 Amounts at the Cell Surface of Control and NBIA Subject Fibroblasts
(A) Examples of TfR1 labeling in fibroblasts of control (C1) and NBIA subjects. Cell analysis was based on Hoechst-positive signal. Scale bar, 10 μm.
(B) Quantification of membrane-bound TfR1 signal on at least 20,000 fibroblasts was performed with IDEAS software (Amnis). The control value is the mean of three different control fibroblasts. Data represent the means ± SEM of three independent experiments (+ indicates the mean). The boxplots correspond to the first, median, and third quartiles. Two-tailed Student’s unpaired t tests or Mann-Whitney U tests were used as appropriate (∗∗∗p < 0.001 [considered significant]).
Figure 5.
Defective Tf Recycling in Fibroblasts of NBIA Subjects
(A) Tf-RED signal 0, 5, and 10 min after incubation of control (C1) and NBIA subject fibroblasts with Tf-RED in FBS-free medium for 30 min at 37°C. The figure was produced with the ImageJ plugin FigureJ.18 Scale bar, 10 μm.
(B) Relative mean fluorescence intensity of Tf-RED signal followed for 30 min in control (C1–C3) and NBIA subject fibroblasts as a percentage of Tf-RED initial staining in control cells. The apparently normal Tf signal at 30 min is due to the fact that the defined threshold for Tf detection in the perinuclear region at 0 min was higher than the Tf signal at the end of the experiment. The number of cells analyzed was higher than 30 in three independent experiments. Data represent the means ± SEM of three independent experiments. An ANOVA multifactorial test by the Holm-Sidak method was used (∗∗∗p < 0.001 [considered significant]).
Tf and TfR1staining colocalized and were evenly distributed in control cells (Figure 6A). Tf-TfR1 staining partly colocalized with RAB11A, a marker of the recycling compartment, or was located in lysosomes, given that Tf also colocalized with a lysosomal marker (LAMP2; Figure 6B). Moreover, Tf and TfR1 signals were more intense in NBIA subjects’ fibroblasts. They clustered in the perinuclear region and were associated with enhanced staining of RAB11A, suggesting an impairment of endosome recycling (Figure 6A). Of note, all NBIA subjects’ fibroblasts showed abnormally shaped, enlarged lysosomes with decreased Tf/LAMP2+ structures (Figure 6B), consistent with the increased LC3B-II signal detected by immunoblotting (Figure 1C) and suggestive of impaired TfR1 degradation. These results were also observed in another PLA2G6 subject’s fibroblast cell line (data not shown).
Figure 6.
Tf and TfR1 Trafficking in Fibroblasts of NBIA Subjects
(A) Tf, TfR1, and RAB11A localization in fibroblasts of control (C1) and NBIA subjects by confocal microscopy after 30 min of Tf-RED pulse followed by a 10 min chase. Scale bar, 10 μm. Arrows indicate the perinuclear colocalization of Tf, TfR1, and RAB11A.
(B) Tf, LAMP2, and RAB11A localization in fibroblasts of control (C1) and NBIA subjects after 30 min of Tf-RED pulse followed by a 10 min chase. Scale bar, 10 μm. Arrows indicate the perinuclear colocalization of Tf and RAB11A independently of LAMP2. These pictures represent seven experiments and three control fibroblasts.
TfR1 Palmitoylation in Fibroblasts of NBIA Subjects
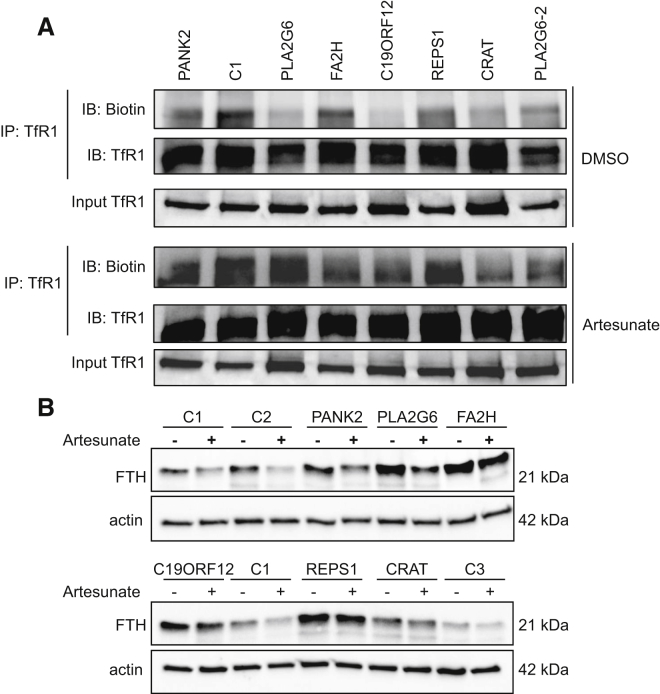
TfR1 is post-translationally modified by the covalent attachment of S-acyl via thioester bonds to Cys62 and Cys67,19 where palmitate is the predominant fatty acid. Considering that palmitoylation decreases cell-surface density and endocytosis of TfR19 and that acetyl-CoA is the sole donor of acetyl groups for acetyl transferases, we hypothesized that CoA deficiency related to PANK2 and CRAT mutations might reduce TfR1 palmitoylation. By studying TfR1 palmitoylation in PANK2- and CRAT-deficient fibroblasts by fatty acyl exchange chemistry, we showed a reduced palmitoylation of TfR1 in subject cells, suggesting that impaired CoA biosynthesis directly alters TfR1 palmitoylation (Figure 7A and Figure S2). Interestingly, this observation was not specific to PANK2 and CRAT strains, given that a major palmitoylation defect was also observed in other NBIA cell lines (particularly in C19ORF12 and PLA2G6 mutant fibroblasts; Figure 7A and Figure S2). Supplementation of fibroblasts with PANK2 and CRAT mutations with 25 μM CoA for 72 hr resulted in increased TfR1 palmitoylation (Figure S3D), suggesting that CoA depletion related to PANK2 and CRAT defects is directly related to TfR1 palmitoylation.
Figure 7.
A. TfR1 Palmitoylation in Fibroblasts of NBIA Subjects
Fibroblasts were treated with DMSO either alone or with 25 μM artesunate for 48 hr, and TfR1 was immunoprecipitated with mouse anti-TfR1 antibody for the palmitoylation assay. In each condition, the upper panel shows the palmitoylated amount of TfR1 (IB: biotin) and the immunoprecipitated amount of TfR1 (IB: TfR1). Input TfR1 was used as a loading control for each condition. IP: immunoprecipitation, IB: immunoblotting. Only one from three controls is presented here.
(B) Immunoblot analysis of ferritin (FTH) in control and NBIA subject fibroblasts treated or not with 25 μM artesunate. Immunoblotting quantification is presented in Figure S2.
Artesunate is known to inhibit liver and ovarian cancer growth by disrupting cellular iron homeostasis via the induction of TfR1 palmitoylation and the reduction of cell-surface TfR1.9 Artesunate (25 μM) enhanced TfR1 palmitoylation in fibroblasts from control and NBIA subjects (Figure 7A and Figure S2) and lowered steady-state levels of ferritin, reflecting a decrease in total iron content (Figure 7B and Figure S2).
Discussion
Transferrin-bound iron is known to be delivered to cells by TfR1 after internalization via clathrin-mediated endocytosis. Subsequent endosomal trafficking allows for its recycling to the plasma membrane. Iron stored in uncoated vesicles is normally released into the cytoplasm after vesicle acidification that occurs before this vesicle is fused with endosomes, the latter of which is mediated by the binding of REPS1 to RalBP1.
After studying cultured fibroblasts of NBIA subjects with known disease-causing biallelic mutations, we report increased cellular iron and abnormal TfR1 recycling as common features in NBIA. Increased cellular iron, with paradoxically enhanced steady-state TfR1 levels and delayed Tf or TfR1 recycling, was observed in all cell types tested, suggesting that this is a generic feature of NBIA. Hence, irrespective of the diversity of mutated genes and affected functions, it appears that iron overload in NBIA results from a single disease mechanism, i.e., abnormal TfR1 recycling. Of note, abnormal TfR1 recycling has been previously reported in phospholipase-A2-deficient models, where phospholipase A2 antagonists inhibited its recycling at the level of early sorting and late recycling endosomes.20, 21 Together, these results suggest that defective endosome recycling is a common disease mechanism in NBIA and that NBIA should be regarded as a trafficking disease, whatever the original defect.
An increased amount of TfR1 in the context of iron overload is paradoxical, because stored iron should normally downregulate TfR1. In keeping with this, increases in TfR1, albeit to a lesser extent, have been previously reported after PANK2 silencing in HeLa cells,22 whereas fibroblasts carrying PANK2 mutations failed to reduce TfR1.23 Different growth medium could explain these discrepancies. Here, post-transcriptional regulation of TfR1 by IRPs functioned normally, given that TFRC transcripts were efficiently downregulated in +FAC conditions. High amounts of TfR1 in NBIA are suggestive of post-translational dysfunction possibly related to impaired palmitoylation of the receptor. Several findings support defective palmitoylation in NBIA: (1) two genes, CRAT and PANK2, are involved in the CoA production required for palmitoylation, defective palmitate β-oxidation was found in CRAT-deficient fibroblasts, and CoA supplementation of CRAT- and PANK2-deficient fibroblasts increased TfR1 palmitoylation; (2) previous metabolic profiling experiments have shown decreased amounts of palmitic acid and reduced lipid biosynthesis in PANK2-deficient fibroblasts;24 (3) COASY-deficient fibroblasts displayed low amounts of acetyl and total CoA, which are expected to decrease palmitic acid;25 (4) phospholipase-A2-deficient mice displayed reduced palmitate β-oxidation;26 and finally (5), impaired CoA synthesis related to PANK2 mutations reportedly induced alterations in histone and tubulin acetylation.27 Another NBIA gene, FA2H, encodes a fatty acid hydroxylase, but no metabolic studies have been reported in FA2H subjects. Together, these data support the view that impaired palmitoylation of TfR1 is a common feature of NBIA. Therefore, we hypothesize that iron overload in NBIA results from (1) increased cell-surface amounts of TfR1 caused by impaired TfR1 palmitoylation and/or (2) the abnormal release of cytosolic iron stored in uncoated vesicles caused by defective endosome recycling.
TfR1 is known to be post-translationally modified by S-acylation, particularly palmitoylation, given that palmitate (C16:0) is the major lipid donor to S-acylated proteins. The amount of TfR1 palmitoylation controls cellular iron, given that substitutions at Cys62 and Cys67, the major sites of TfR1 palmitoylation, cause increased TfR1 internalization and iron overload.19 Conversely, dihydroartesunate, an artemisinin derivative widely used as an antimalarial drug and known to increase palmitoylation, has been shown to reduce membrane-associated TfR1 and decrease iron content.9 Although a number of endosome recycling proteins are palmitoylated, it is also possible that altered palmitoylation of other, as yet undetermined endosomal proteins could also alter TfR1 recycling, especially given that fibroblasts of NBIA subjects displayed enlarged lysosomes with decreased Tf/LAMP2+ structures. In keeping with this, phospholipase A2 is reportedly involved at early stages of membrane tubule formation in the trans-Golgi network28 and is known to regulate morphology and intraorganellar trafficking in the endoplasmic reticulum (ER)-Golgi intermediate compartment.29 Finally, abnormal recycling caused by REPS1 mutations could also slow down TfR1 trafficking and reduce palmitoylation, given that S-acyl transferases are exclusively located at the membrane surface and mainly localized to the ER and Golgi membranes.30
Palmitoylation is a rapid post-translational regulatory mechanism.31 TfR1 recycles over 100 times and undergoes several acylation-deacylation cycles.32 Because post-transcriptional downregulation of TfR1 in +FAC conditions requires several hours,33 given that the half-life of TfR1 is greater than 24 hr,34 post-translational TfR1 regulation by palmitoylation might represent an alternative means of rapidly modulating cellular iron content, a regulatory system that could be impaired in NBIA.
Abnormal palmitoylation has been occasionally reported to alter acylation enzymes in genetic diseases. Most of them result in neurological or psychiatric disorders (e.g., Huntington disease, Alzheimer disease, schizophrenia, bipolar disorder, X-linked intellectual disability, and ceroid lipofuscinosis),35 and models of Parkinson disease highlight the role of palmitoylation for normal neuronal function.36 Why mutations in NBIA genes, which are ubiquitously expressed, result in neurologically restricted phenotypes remains unanswered. At least 23 palmitoyl transferase genes are expressed in the brain, where they display various patterns of expression, and not all of their substrates have been hitherto identified. We hypothesize that the brain-restricted involvement of NBIA could result from hitherto unknown TfR1-specific palmitoyl transferases, the identification of which could help elucidate the disease mechanisms.
Finally, we have shown that artesunate improved TfR1 palmitoylation and decreased steady-state ferritin levels in fibroblasts of NBIA subjects. Artesunate has potent anticancer properties because it induces iron depletion, which is toxic for cancer cells.37 It is also used to treat malaria caused by Plasmodium falciparum. Although its safety profile and pharmacokinetics have not been assessed in neurodegenerative diseases, several millions of subjects have received artemisinin with very few side effects.38 Our data suggest that consideration should be given to this compound, as well as other drugs that increase TfR1 palmitoylation, as possible therapeutic approaches in the treatment of NBIA. This strategy might also have relevance to other, more common neurodegenerative disorders, including Parkinson disease, where brain iron accumulation is also reported.
Acknowledgments
This work was partially funded through the E-Rare project GENOMIT (01GM1207) and the Agence Nationale de la Recherche (ANR-10-IAHU-01). Whole-exome sequencing was funded by the GIS-Institut des Maladies Rares. Imaging and Amnis studies were performed at the Cell Imaging Platform and Cell Sorting Facility of the Imagine Institute and funded by the Imagine Foundation, ARC foundation, and GR-Ex LabEx. A.D. was supported by a fellowship from ApoPharma. We thank Olivier Pellé (Flow Cytometry Facility at INSERM UMR 1163) for assisting with cell sorting; Martial Saumier (BioRAD) for digital droplet PCR technical advice and support; Geneviève Courtois (Imagine Institute) for L3 maintenance, virus production, teaching, and advice; Sylvain Hanein (Imagine Institute) for assisting with confocal microscopy; Leona Enke for REPS1 Sanger sequencing; and Yanick Crow for helping with editing the manuscript. We acknowledge the use of bioresources of the Necker Imagine DNA biobank (BB-033-00065).
Published: February 1, 2018
Footnotes
Supplemental Data include a Supplemental Note, four figures, and two tables and can be found with this article online at https://doi.org/10.1016/j.ajhg.2018.01.003.
Accession Numbers
The REPS1 and CRAT mutations reported in this paper have been submitted to ClinVar under SUB3461990 and SUB3466483, respectively. Please contact the corresponding author for the final accession numbers.
Web Resources
Alamut Interpretation Software 2.0, http://alamut.interactivebiosoftware.com
ClinVar, http://www.ncbi.nlm.nih.gov/clinvar/
ExAC Browser, http://exac.broadinstitute.org/
Oligo Primer Analysis Software v.7, http://www.oligo.net
OMIM, http://www.omim.org
Primer-BLAST software, http://www.ncbi.nlm.nih.gov.gate2.inist.fr/tools/primer-blast
RSCB Protein Data Bank, https://www.rcsb.org/pdb/home/home.do
Swiss-PdbViewer 3.7, http://www.expasy.org/spdbv
Supplemental Data
References
- 1.Meyer E., Kurian M.A., Hayflick S.J. Neurodegeneration with brain iron accumulation: Genetic diversity and pathophysiological mechanisms. Annu. Rev. Genomics Hum. Genet. 2015;16:257–279. doi: 10.1146/annurev-genom-090314-025011. [DOI] [PubMed] [Google Scholar]
- 2.Jaberi E., Rohani M., Shahidi G.A., Nafissi S., Arefian E., Soleimani M., Rasooli P., Ahmadieh H., Daftarian N., KaramiNejadRanjbar M. Identification of mutation in GTPBP2 in patients of a family with neurodegeneration accompanied by iron deposition in the brain. Neurobiol. Aging. 2016;38:216.e11–216.e18. doi: 10.1016/j.neurobiolaging.2015.10.034. [DOI] [PubMed] [Google Scholar]
- 3.Lane D.J., Merlot A.M., Huang M.L., Bae D.H., Jansson P.J., Sahni S., Kalinowski D.S., Richardson D.R. Cellular iron uptake, trafficking and metabolism: Key molecules and mechanisms and their roles in disease. Biochim. Biophys. Acta. 2015;1853:1130–1144. doi: 10.1016/j.bbamcr.2015.01.021. [DOI] [PubMed] [Google Scholar]
- 4.Sohn Y.S., Ghoti H., Breuer W., Rachmilewitz E., Attar S., Weiss G., Cabantchik Z.I. The role of endocytic pathways in cellular uptake of plasma non-transferrin iron. Haematologica. 2012;97:670–678. doi: 10.3324/haematol.2011.054858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hentze M.W., Kühn L.C. Molecular control of vertebrate iron metabolism: mRNA-based regulatory circuits operated by iron, nitric oxide, and oxidative stress. Proc. Natl. Acad. Sci. USA. 1996;93:8175–8182. doi: 10.1073/pnas.93.16.8175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barbeito A.G., Levade T., Delisle M.B., Ghetti B., Vidal R. Abnormal iron metabolism in fibroblasts from a patient with the neurodegenerative disease hereditary ferritinopathy. Mol. Neurodegener. 2010;5:50. doi: 10.1186/1750-1326-5-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moura I.C., Lepelletier Y., Arnulf B., England P., Baude C., Beaumont C., Bazarbachi A., Benhamou M., Monteiro R.C., Hermine O. A neutralizing monoclonal antibody (mAb A24) directed against the transferrin receptor induces apoptosis of tumor T lymphocytes from ATL patients. Blood. 2004;103:1838–1845. doi: 10.1182/blood-2003-07-2440. [DOI] [PubMed] [Google Scholar]
- 8.Hanein S., Garcia M., Fares-Taie L., Serre V., De Keyzer Y., Delaveau T., Perrault I., Delphin N., Gerber S., Schmitt A. TMEM126A is a mitochondrial located mRNA (MLR) protein of the mitochondrial inner membrane. Biochim. Biophys. Acta. 2013;1830:3719–3733. doi: 10.1016/j.bbagen.2013.02.025. [DOI] [PubMed] [Google Scholar]
- 9.Ba Q., Zhou N., Duan J., Chen T., Hao M., Yang X., Li J., Yin J., Chu R., Wang H. Dihydroartemisinin exerts its anticancer activity through depleting cellular iron via transferrin receptor-1. PLoS ONE. 2012;7:e42703. doi: 10.1371/journal.pone.0042703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Byrne S.L., Buckett P.D., Kim J., Luo F., Sanford J., Chen J., Enns C., Wessling-Resnick M. Ferristatin II promotes degradation of transferrin receptor-1 in vitro and in vivo. PLoS ONE. 2013;8:e70199. doi: 10.1371/journal.pone.0070199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dergai O., Novokhatska O., Dergai M., Skrypkina I., Tsyba L., Moreau J., Rynditch A. Intersectin 1 forms complexes with SGIP1 and Reps1 in clathrin-coated pits. Biochem. Biophys. Res. Commun. 2010;402:408–413. doi: 10.1016/j.bbrc.2010.10.045. [DOI] [PubMed] [Google Scholar]
- 12.Krieger J.R., Taylor P., Gajadhar A.S., Guha A., Moran M.F., McGlade C.J. Identification and selected reaction monitoring (SRM) quantification of endocytosis factors associated with Numb. Mol. Cell. Proteomics. 2013;12:499–514. doi: 10.1074/mcp.M112.020768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu J., Zhou Z., Zeng L., Huang Y., Zhao W., Cheng C., Xu M., Xie Y., Mao Y. Cloning, expression and characterization of a novel human REPS1 gene. Biochim. Biophys. Acta. 2001;1522:118–121. doi: 10.1016/s0167-4781(01)00310-4. [DOI] [PubMed] [Google Scholar]
- 14.Yamaguchi A., Urano T., Goi T., Feig L.A. An Eps homology (EH) domain protein that binds to the Ral-GTPase target, RalBP1. J. Biol. Chem. 1997;272:31230–31234. doi: 10.1074/jbc.272.50.31230. [DOI] [PubMed] [Google Scholar]
- 15.Cullis D.N., Philip B., Baleja J.D., Feig L.A. Rab11-FIP2, an adaptor protein connecting cellular components involved in internalization and recycling of epidermal growth factor receptors. J. Biol. Chem. 2002;277:49158–49166. doi: 10.1074/jbc.M206316200. [DOI] [PubMed] [Google Scholar]
- 16.Boissel L., Fillatre J., Moreau J. Identification and characterization of the RLIP/RALBP1 interacting protein Xreps1 in Xenopus laevis early development. PLoS ONE. 2012;7:e33193. doi: 10.1371/journal.pone.0033193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jogl G., Hsiao Y.S., Tong L. Structure and function of carnitine acyltransferases. Ann. N Y Acad. Sci. 2004;1033:17–29. doi: 10.1196/annals.1320.002. [DOI] [PubMed] [Google Scholar]
- 18.Mutterer J., Zinck E. Quick-and-clean article figures with FigureJ. J. Microsc. 2013;252:89–91. doi: 10.1111/jmi.12069. [DOI] [PubMed] [Google Scholar]
- 19.Alvarez E., Gironès N., Davis R.J. Inhibition of the receptor-mediated endocytosis of diferric transferrin is associated with the covalent modification of the transferrin receptor with palmitic acid. J. Biol. Chem. 1990;265:16644–16655. [PubMed] [Google Scholar]
- 20.de Figueiredo P., Doody A., Polizotto R.S., Drecktrah D., Wood S., Banta M., Strang M.S., Brown W.J. Inhibition of transferrin recycling and endosome tubulation by phospholipase A2 antagonists. J. Biol. Chem. 2001;276:47361–47370. doi: 10.1074/jbc.M108508200. [DOI] [PubMed] [Google Scholar]
- 21.Doody A.M., Antosh A.L., Brown W.J. Cytoplasmic phospholipase A2 antagonists inhibit multiple endocytic membrane trafficking pathways. Biochem. Biophys. Res. Commun. 2009;388:695–699. doi: 10.1016/j.bbrc.2009.08.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Poli M., Derosas M., Luscieti S., Cavadini P., Campanella A., Verardi R., Finazzi D., Arosio P. Pantothenate kinase-2 (Pank2) silencing causes cell growth reduction, cell-specific ferroportin upregulation and iron deregulation. Neurobiol. Dis. 2010;39:204–210. doi: 10.1016/j.nbd.2010.04.009. [DOI] [PubMed] [Google Scholar]
- 23.Campanella A., Privitera D., Guaraldo M., Rovelli E., Barzaghi C., Garavaglia B., Santambrogio P., Cozzi A., Levi S. Skin fibroblasts from pantothenate kinase-associated neurodegeneration patients show altered cellular oxidative status and have defective iron-handling properties. Hum. Mol. Genet. 2012;21:4049–4059. doi: 10.1093/hmg/dds229. [DOI] [PubMed] [Google Scholar]
- 24.Leoni V., Strittmatter L., Zorzi G., Zibordi F., Dusi S., Garavaglia B., Venco P., Caccia C., Souza A.L., Deik A. Metabolic consequences of mitochondrial coenzyme A deficiency in patients with PANK2 mutations. Mol. Genet. Metab. 2012;105:463–471. doi: 10.1016/j.ymgme.2011.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dusi S., Valletta L., Haack T.B., Tsuchiya Y., Venco P., Pasqualato S., Goffrini P., Tigano M., Demchenko N., Wieland T. Exome sequence reveals mutations in CoA synthase as a cause of neurodegeneration with brain iron accumulation. Am. J. Hum. Genet. 2014;94:11–22. doi: 10.1016/j.ajhg.2013.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Song H., Wohltmann M., Bao S., Ladenson J.H., Semenkovich C.F., Turk J. Mice deficient in group VIB phospholipase A2 (iPLA2gamma) exhibit relative resistance to obesity and metabolic abnormalities induced by a Western diet. Am. J. Physiol. Endocrinol. Metab. 2010;298:E1097–E1114. doi: 10.1152/ajpendo.00780.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Siudeja K., Srinivasan B., Xu L., Rana A., de Jong J., Nollen E.A., Jackowski S., Sanford L., Hayflick S., Sibon O.C. Impaired Coenzyme A metabolism affects histone and tubulin acetylation in Drosophila and human cell models of pantothenate kinase associated neurodegeneration. EMBO Mol. Med. 2011;3:755–766. doi: 10.1002/emmm.201100180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schmidt J.A., Kalkofen D.N., Donovan K.W., Brown W.J. A role for phospholipase A2 activity in membrane tubule formation and TGN trafficking. Traffic. 2010;11:1530–1536. doi: 10.1111/j.1600-0854.2010.01115.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ben-Tekaya H., Kahn R.A., Hauri H.P. ADP ribosylation factors 1 and 4 and group VIA phospholipase A2 regulate morphology and intraorganellar traffic in the endoplasmic reticulum-Golgi intermediate compartment. Mol. Biol. Cell. 2010;21:4130–4140. doi: 10.1091/mbc.E10-01-0022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chamberlain L.H., Shipston M.J. The physiology of protein S-acylation. Physiol. Rev. 2015;95:341–376. doi: 10.1152/physrev.00032.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Magee A.I., Gutierrez L., McKay I.A., Marshall C.J., Hall A. Dynamic fatty acylation of p21N-ras. EMBO J. 1987;6:3353–3357. doi: 10.1002/j.1460-2075.1987.tb02656.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zaliauskiene L., Kang S., Brouillette C.G., Lebowitz J., Arani R.B., Collawn J.F. Down-regulation of cell surface receptors is modulated by polar residues within the transmembrane domain. Mol. Biol. Cell. 2000;11:2643–2655. doi: 10.1091/mbc.11.8.2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Müllner E.W., Kühn L.C. A stem-loop in the 3′ untranslated region mediates iron-dependent regulation of transferrin receptor mRNA stability in the cytoplasm. Cell. 1988;53:815–825. doi: 10.1016/0092-8674(88)90098-0. [DOI] [PubMed] [Google Scholar]
- 34.Zaliauskiene L., Fazio R.L., Kang S., Sparks K., Weaver C.T., Zinn K.R., Collawn J.F. Inhibition of T cell responses by transferrin-coupled competitor peptides. Immunol. Res. 2002;26:77–85. doi: 10.1385/IR:26:1-3:077. [DOI] [PubMed] [Google Scholar]
- 35.Chavda B., Arnott J.A., Planey S.L. Targeting protein palmitoylation: selective inhibitors and implications in disease. Expert Opin. Drug Discov. 2014;9:1005–1019. doi: 10.1517/17460441.2014.933802. [DOI] [PubMed] [Google Scholar]
- 36.Senyilmaz D., Virtue S., Xu X., Tan C.Y., Griffin J.L., Miller A.K., Vidal-Puig A., Teleman A.A. Regulation of mitochondrial morphology and function by stearoylation of TFR1. Nature. 2015;525:124–128. doi: 10.1038/nature14601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lai H.C., Singh N.P., Sasaki T. Development of artemisinin compounds for cancer treatment. Invest. New Drugs. 2013;31:230–246. doi: 10.1007/s10637-012-9873-z. [DOI] [PubMed] [Google Scholar]
- 38.Efferth T., Kaina B. Toxicity of the antimalarial artemisinin and its dervatives. Crit. Rev. Toxicol. 2010;40:405–421. doi: 10.3109/10408441003610571. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.