Abstract
Many factors have been characterized as essential for vesicle trafficking, including a number of proteins commonly referred to as soluble N-ethylmaleimide-sensitive factor adaptor protein receptor (SNARE) components. The Arabidopsis genome contains a remarkable number of SNAREs. In general, the vesicle fusion machinery appears highly conserved. However, whereas some classes of yeast and mammalian genes appear to be lacking in Arabidopsis, this small plant genome has gene families not found in other eukaryotes. Very little is known about the precise function of plant SNAREs. By contrast, the intracellular localization of and interactions between a large number of plant SNAREs have been determined, and these data are discussed in light of the phylogenetic analysis.
An essential function for all eukaryotic cells is to control the traffic of proteins and lipids through the endomembrane system (for review, see Sanderfoot and Raikhel, 1999). Most protein cargo first enters the endomembrane system at the endoplasmic reticulum (ER) before moving on to the Golgi apparatus. After sequential passage through the stacks of the Golgi, proteins arrive at the trans-Golgi network (TGN). At the TGN, cargo is sorted into vesicles destined for various endosomal organelles including the prevacuolar compartment (PVC) or vacuole, or is targeted to the plasma membrane (PM) for secretion. Each of these compartments must maintain some independence and a unique protein content, while accommodating a vast amount of cargo in transit to another destination.
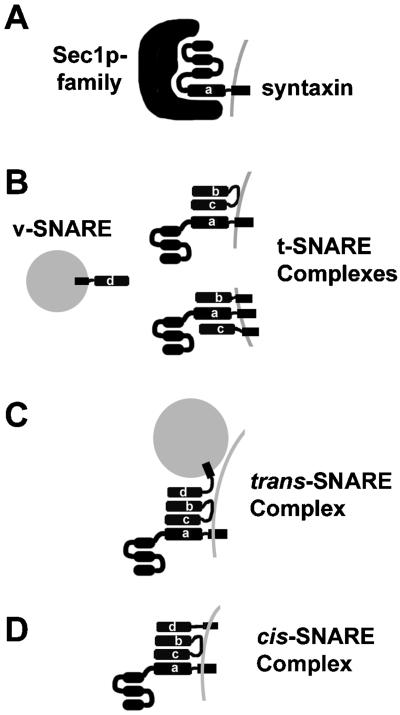
Many factors including proteins and lipids are thought to be involved in packaging cargo into vesicles and delivering them to different organelles. A large diverse group of proteins called soluble N-ethylmaleimide-sensitive factor (NSF) adaptor proteins (SNAPs) receptors (SNAREs) and their associated proteins are vital for this process. The term SNARE describes the activity of these proteins as binding partners of two proteins, NSF and SNAPs. It is now clear that the role of SNAREs is much deeper than simply as receptors for SNAPs. As diagrammed in Figure 1, the role of the SNAREs appears to be in the assembly of a four-helix SNARE bundle (often called a trans-SNARE complex or a SNAREpin), which probably drives the fusion of transport vesicles with target membranes. Three of the helices form a “t-SNARE complex” on the target membrane. One of the helices is always derived from a member of the syntaxin family of SNAREs, whereas the other two helices come from other types of SNAREs. A fourth helix (the v-SNARE) is derived from the vesicle membrane, and assembly of this helix into the t-SNARE complex is very energetically favorable and is sufficient to drive fusion the membranes in vitro (Weber et al., 1998; McNew et al., 2000). Following vesicle fusion the four-helix bundle exists as a cis-SNARE complex where all four proteins are found on the same membrane. It is at this point that NSF and SNAPs bind onto the cis-SNARE complex and the ATPase activity of NSF is used to disassemble the complex, freeing the individual members for additional rounds of vesicle fusion.
Figure 1.
SNAREs in vesicle fusion. A, Syntaxins are usually found in an inactive state associated with a member of the Sec1p family. In some manner the Sec1 protein and other effectors activate the syntaxin, allowing it to associate with other SNAREs to form a t-SNARE complex. B, Four SNARE helicies are required for vesicle fusion to occur (labeled the a-, b-, c-, and d- helices as described in Scales et al., 2000). The a-helix is always contributed by a syntaxin residing on a target membrane. At certain targeting steps (e.g. at the PM) the syntaxin associates with a member of the SNAP25 class of SNAREs, which contributes a b- and c- helix to the t-SNARE complex (B, top right). Other steps, including at most intracellular membranes, the syntaxin associates with two other SNAREs who each contribute a b- or a c-helix to the t-SNARE complex (B, bottom right). C, Vesicle fusion likely occurs when the v-SNARE (or d-helix) from the vesicle assembles into a trans-SNARE complex (or SNAREpin) with the t-SNARE complex (here just showing the SNAP25-containing complex). For simplicity a single trans-SNARE complex is drawn; it is possible that the concerted action of many trans-SNARE complexes forming simultaneously may be required for a vesicle to fuse. D, The four-helix cis-SNARE complex results, where all four proteins are on the same membrane. The cis-SNARE complex is a binding site for SNAPs and NSF, proteins that disassociate the SNARE complex, freeing the individual SNAREs for future fusion events.
The mixing of synthetic liposomes containing just SNAREs can result in fusion of membranes in vitro (Weber et al., 1998; McNew et al., 2000); however, this process must be highly regulated in vivo. This regulation is the role of proteins of the Rab and Sec1 families of proteins. Sec1 proteins directly bind to syntaxins and regulate their activity and conformation (for review, see Hanson, 2000). The Rab proteins do not interact directly with the SNAREs, and instead function through various effectors, as well as interacting with the Sec1 proteins (Gonzalez and Scheller, 1999). As they do not interact directly with the SNAREs, the Rab family of proteins will be covered in a separate analysis.
The availability of the complete sequence of several model eukaryotes has provided useful information on the overall conservation of the vesicle fusion machinery. Many important distinctions are also found, notably the absence of certain proteins in some eukaryotes, an increase in the number of SNAREs in multicellular eukaryotes, and the presence of novel proteins found only in particular eukaryotes. The genome sequence of the model plant Arabidopsis has provided the first look at the complete complement of SNARE components in plants.
Because it is now clear that a single SNARE may function at multiple targeting events and may be required on the vesicle (as a v-SNARE) in one case and on the target membrane (as a t-SNARE) in another (McNew et al., 2000), definition of a particular SNARE as a v- or t-SNARE is now inaccurate. It is fortunate that the particular types of SNAREs appear to be conserved across the eukaryotes and we can therefore discuss these SNAREs as orthologous groups rather than describing a particular protein as a t- or v-SNARE. Here we will first describe the syntaxin family, a group of SNAREs that are always found as part of the t-SNARE complex. We will then describe the other SNAREs groups, proteins that are in some cases part of the t-SNARE complex or that in other cases function as v-SNAREs. In addition, we will also discuss the Sec1p family of proteins and those proteins involved in breaking up the cis-SNARE complex (SNAPs, NSF, and its homologs).
SYNTAXINS
Syntaxins can be recognized by sequence homology and by common structural features. For example, all are membrane proteins most commonly anchored to membranes via their C-terminal transmembrane domains, although some are anchored by post-translational addition of lipids to a C-terminal Cys residue. Adjacent to the membrane anchor is a coiled-coil domain centered on a Glu residue. At the N terminus of the protein are three other α-helical domains, which form a bundle in the crystal structure of mammalian syntaxin 1 (see Fig. 1A; Fernandez et al., 1998; Misura et al., 2000). Other syntaxins are predicted to contain these N-terminal helices and this has been confirmed by structural studies in some cases (Fiebig et al., 1999).
Syntaxins have been well studied in a few eukaryotes, and several isoforms are known to localize to distinct compartments where they function in vesicle fusion (for review, see Sanderfoot and Raikhel, 1999). All syntaxins can be recognized by the high degree of sequence homology in their coiled-coil domain, however, certain syntaxins form groups where the sequence homology is extended throughout the entire protein (i.e. groups of orthologs). It is interesting that when the functions of the members of a particular orthologous group are examined they each seem to have a common function or at least intracellular localization. Although sophisticated algorithms have been used in the past to identify SNAREs in sequence databases (Weimbs et al., 1998), BLAST searches (Altschul et al., 1997) can be used to distinguish syntaxins from other proteins, including other SNAREs (Adavani et al., 1998; Holthuis et al., 1998; Simonsen et al., 1998; Steegmaier et al., 1998; Sanderfoot et al., 1999).
Yeast (Saccharomyces cerevisiae) is known to contain eight syntaxins (for review, see Pelham, 1999), two of which share high sequence identity and are known to be functionally redundant (Aalto et al., 1993). These syntaxins minimally define the known compartments of the yeast secretory pathway and can serve as a basis for classification of syntaxins from other eukaryotes. Although some syntaxins have a broad localization, and in fact, often cycle between target organelles, a steady-state localization for the yeast syntaxins can be defined. Ufe1p resides on the ER, Sed5p on the Golgi, Tlg2p on the late Golgi, Pep12p on the PVC, Vam3p on the vacuole, Tlg1p on the endosome, and the redundant pair of Sso1p and Sso2p (hereafter Sso1/2p) on the PM (see Pelham, 1999 and refs. therein).
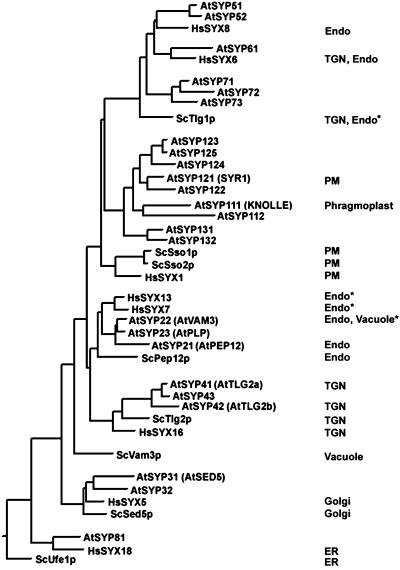
Based upon our analysis of the sequence databases for several eukaryotes with complete or near-complete genome sequences, Caenorhabiditis elegans (The C. elegans Sequencing Consortium, 1998) contains nine syntaxins, the fruit fly (Adams et al., 2000) contains 10, and humans contain at least 15. Similar analyses carried out by others (Pelham, 1999; Koushika and Nonet, 2000; Littleton, 2000) have yielded comparable results. The completed genome of Arabidopsis has revealed 24 syntaxins (The Arabidopsis Genome Initiative, 2000). In a general sense, animals and plants have syntaxins that are orthologous to one of the yeast syntaxins; and where it has been examined, they have a similar localization and/or function (see below). Often, the multicellular eukaryotes have multiple paralogs of the single yeast syntaxins (i.e. genes having higher sequence identity to a gene within the same genome than to the ortholog from a different organism). In a converse manner, none of the multicellular eukaryotes contain an ortholog of the yeast vacuolar syntaxin Vam3p (Pelham, 1999; Littleton, 2000; Fig. 3). On the other hand, multicellular eukaryotes have evolved novel groups that yeast appears to lack. For example, plants, fruit fly, and mammals have elaborated the Tlg1p group to create the Syntaxin 6/10 subgroup and the Syntaxin 8 subgroup; plants appear to have a third novel group of Tlg1p-like syntaxins.
Figure 3.
Phylogenetic analysis of eukaryotic syntaxins. Full-length sequences of the Arabidopsis (At), yeast (Sc), and selected human (Hs) syntaxins were acquired from GenBank and aligned as described in Figure 2. Where it is known, the intracellular localization of the indicated syntaxins is given (see text for references). Because the nature of the endosomal compartments (i.e. early, late, prevacuolar, etc.) can vary considerably depending on the cell type we use the general term endosome (Endo) for those syntaxins localized to these types of endomembranes. Often, different researchers have found conflicting results with the same syntaxin and these are noted by an asterisk. More information on these sequences, as well as further alignments with other eukaryotic syntaxins, can be acquired from http://www.msu.edu/∼sanderfo/atsnare.htm.
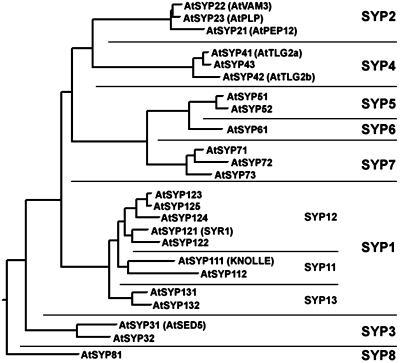
Some of the 24 syntaxins of Arabidopsis have been previously examined and have been named according to various (often conflicting and potentially confusing) criteria. To synchronize the nomenclature we have developed a system for naming the plant syntaxins using the name syntaxin of plants (SYP), and here we apply this system to the syntaxins of Arabidopsis. The syntaxins of Arabidopsis (and of other plants, see below) cluster into eight groups based upon sequence homology—groups that we named SYP1 to SYP8 (Fig. 2). As can be seen in Figure 3, each of these groups is paralogous, having a higher degree of homology with members of their own group than with orthologous syntaxins from fungi or animals.
Figure 2.
Arabidopsis syntaxin groups. The protein sequences of Arabidopsis syntaxins were aligned using default parameters of the CLUSTAL algorithm of the MEGALIGN program in the DNASTAR package. The resulting phylogenetic tree revealed the presence of eight groups (SYP1–8), which were numbered according to various criteria (see text). For those Arabidopsis syntaxins that have been previously published, the prior name is given in parenthesis. The accession numbers for all the Arabidopsis SNAREs can be acquired from http://www.msu.edu/∼sanderfo/atsnare.htm
Within most of the groups the individual members share between 50% and 80% protein sequence identity. In the SYP3 family only approximately 44% identity is found, consistent with the members of this group being more highly diverged. The members of the SYP1 group were found to have a large range of sequence identities (between 34% and 63% protein sequence identity among each other), and the members seemed to cluster into three subgroups. To further examine this finding we examined the structure of each syntaxin gene locus to determine the location of introns within each gene (Fig. 4). We found that the SYP1 group consisted of three gene families, each of which corresponded to the subgroups suggested by sequence identity. A gene family is defined here as a paralogous group of genes that share a common splicing pattern (and presumably, arose by way of gene duplication). We, therefore, subdivided the SYP 1 group into three subgroups (1, 2, and 3; or SYP11, SYP12, and SYP13), and named the individual members sequentially within the subgroups (i.e. SYP111, SYP112… ) in the order of prior publication or of deposition into sequence databases. With the exception of the SYP6 and SYP8 groups, which contained only a single member, we found that each of the other groups of Arabidopsis syntaxins was represented by a gene family (Fig. 4). The two members of the SYP3 group share a somewhat similar splicing pattern, especially near the 3′ end of the gene (Fig. 4) and are likely a gene family whose members have diverged significantly. As with the SYP1 subgroups, we named each member of the various groups sequentially (i.e. SYP21, SYP22… ) in the order of prior publication or of deposition into sequence databases. It should be noted that 20 of the 24 Arabidopsis syntaxins are known to be expressed, as judged by the existence of a cloned cDNA or a cognate expressed sequence tag (EST; see Table I). This suggests that this large number is not due to multiple pseudogenes. The complete complement of Arabidopsis syntaxins is listed in Table I.
Figure 4.
Gene structure of the Arabidopsis syntaxins. The cDNA sequence of each Arabidopsis syntaxin is represented schematically as a line with the position of the introns indicated by the triangles. White triangles within each group indicate an identical intron position within a particular gene family and black triangles indicate introns in a position not found in other members of the gene family.
Table I.
Syntaxins of Arabidopsis
| Name | Previous Namea | Genomic Locusb | AGI IDc | Intracellular Localizationd | Expressed Gene?e | Notef |
|---|---|---|---|---|---|---|
| AtSYP111 | KNOLLE | F22O13.4 | At1g08350 | Phragmoplast | Yes | 1 |
| AtSYP112 | T30D6.23 | At2g18260 | ? | |||
| AtSYP121 | AtSYR1 | F26K24.11 | At3g11820 | PM | Yes | 2 |
| AtSYP122 | F22O6.220 | At3g52400 | Yes | 3 | ||
| AtSYP123 | F4C21.26 | At4g03330 | Yes | |||
| AtSYP124 | T1F9.22 | At1g55410 | ? | |||
| AtSYP125 | T28P6.10 | At1g10980 | ? | |||
| AtSYP131 | F20H23.28 | At3g03800 | ? | |||
| AtSYP132 | T22D6.20 | At5g08080 | ? | |||
| AtSYP21 | AtPEP12 | F5E19.180 | At5g16830 | PVC | Yes | 4 |
| AtSYP22 | AtVAM3 | MSD23.4 | At5g46860 | PVC or vac | Yes | 5 |
| AtSYP23 | AtPLP | dl14901c | At4g17730 | Yes | 6 | |
| AtSYP31 | AtSED5 | MJJ3.17 | At5g05760 | Yes | ||
| AtSYP32 | K7M2.10 | At3g24350 | Yes | |||
| AtSYP41 | AtTLG2a | F2P16.240 | At5g26980 | TGN | Yes | 7 |
| AtSYP42 | AtTLG2b | T10M13.19 | At4g** | TGN | Yes | 7 |
| AtSYP43 | F18C1.4 | At3g05710 | Yes | 8/9 | ||
| AtSYP51 | F3O9.4 | At1g15930 | Yes | 9 | ||
| AtSYP52 | F20B17.2 | At1g73260 | Yes | 9 | ||
| AtSYP61 | F3M18.7 | At1g27550 | Yes | 9 | ||
| AtSYP71 | F11F8.33 | At3g09740 | Yes | 9 | ||
| AtSYP72 | F18N11.40 | At3g45280 | Yes | 9 | ||
| AtSYP73 | F2A19.50 | At3g61450 | Yes | 9 | ||
| AtSYP81 | F19C24.5 | At1g47920 | Yes |
Name given to previously published Arabidopsis syntaxins, see notes for references. See Tables I and II for accession nos. and associated protein sequences.
Designation of gene locus on annotated bacterial artificial chromosome.
Systematic designation given to gene by Arabidopsis Genome Initiative, At4g03330 indicates the 333rd annotated gene (counting from top) on chromosome 4. Genes marked by a double asterisk were not assigned nos. during annotation.
Intracellular localization, if known; see note for reference.
Indicates whether gene is expressed, based on cloned cDNA (see note for reference) or presence of cognate EST.
1, Lukowitz et al., 1996; 2, Leyman et al., 1999; 3, X. Gansel and L. Sticher, GenBank accession no. AJ245407; 4, Bassham et al., 1995; 5, Sato et al., 1997; Sanderfoot et al., 1999; 6, Zheng et al., 1999a; 7, Bassham et al., 2000; 8, X. Gansel and L. Sticher, GenBank accession no. AJ245408; 9, A.A. Sanderfoot and N.V. Raikhel, unpublished data.
When compared with other eukaryotic syntaxins the members of the SYP1 group show highest homology with yeast Sso1/2p or mammalian Syntaxin 1, which reside on the PM (Aalto et al., 1993; Bennett et al., 1993). However, compared with the other syntaxin groups (see below), this association of SYP1 with the PM syntaxins of other organisms is relatively weak. On the other hand, one member of the SYP1 group is known to localize to the PM (Leyman et al., 1999) and the others seem to have roles somewhat analogous to those of the PM syntaxins, suggesting this association may be real. AtSYP111 or KNOLLE was first characterized as a seedling lethal gene (Lukowitz et al., 1996; and see Lethality below). KNOLLE is expressed only during cell division and is found at the phragmoplast (Lauber et al., 1997). AtSYP121 (AtSYR1) is found on the PM of guard cells and may play a role in abscisic acid signaling (Leyman et al., 1999). Of the other four members of this gene family (AtSYP122–125), two are known to be expressed, but their function or localization remains to be determined. The AtSYP131/132 group has not been examined.
The AtSYP2 family has three members orthologous to Pep12p and to mammalian syntaxins 7 and 13. Yeast Pep12p is found on the late endosome/PVC (Becherer et al., 1996), whereas syntaxins 7 and 13 are found on distinct endosomal structures and likely have distinct functions (Prekeris et al., 1999). AtSYP21 (AtPEP12) is found on the PVC in Arabidopsis (Conceição et al., 1997; Sanderfoot et al., 1998). AtSYP22 (AtVAM3) has been reported to localize to the vacuole in the shoot apical meristem (Sato et al., 1997). However, we have shown that it is found only on the PVC, colocalized with AtSYP21 in vegetative cells (Sanderfoot et al., 1999). Whether this syntaxin is found on different membranes in different cell types is currently being investigated. AtSYP23 is known to be expressed (Zheng et al., 1999a), though its localization has not been examined.
The AtSYP3 family contains two members orthologous to Sed5p and mammalian syntaxin 5, which localize to the Golgi (Hardwick and Pelham, 1993; Dascher et al., 1994). Both members are known to be expressed, although the localization of either has yet to be reported.
The AtSYP4 family has three members that are orthologs of Tlg2p and mammalian syntaxin 16, syntaxins of the late Golgi/TGN (Abeliovich et al., 1998; Holthuis et al., 1998; Séron et al., 1998; Simonsen et al., 1998). Two members of this group, AtSYP41 (AtTLG2a) and AtSYP42 (AtTLG2b), have been examined, both localizing to the TGN; however, they do not colocalize, but instead are found on distinct domains of the TGN (Bassham et al., 2000).
Like fruit flies and mammals, plants have elaborated the Tlg1p-group. The two members of the SYP5 family are orthologous to syntaxin 8, a protein found preferentially on the late endosomes in mammalian cells (Prekeris et al., 1999). The SYP6 family (one member) is related to syntaxins 6 and 10. Syntaxin 6 is a protein involved in TGN to late endosome trafficking in mammalian cells that is found preferentially on the TGN (Bock et al., 1997). Syntaxin 10 is paralogous to Syntaxin 6 and is also found in the TGN, though its functions have not been examined (Tang et al., 1998). We have recently begun to characterize the members of these groups (SYP5 and SYP6), and preliminary results suggest that these syntaxins have locations similar to their eukaryotic orthologs (A.A. Sanderfoot and N.V. Raikhel, unpublished data), though their functions remain unclear. The SYP7 family (three members) does not appear to have an ortholog among the yeast or animal syntaxins, and this group may be unique to plants.
The SYP8 family has a single member and is related to ER-localized syntaxins such as Ufe1p and syntaxin 18 (Lewis et al., 1997; Hatsuzawa et al., 2000). An EST is found for AtSYP81, but its localization has not been examined.
That Arabidopsis syntaxins are encoded in gene families appears not to be indicative of redundancy. In fact, research has indicated that the individual members of the families may have distinct localizations (Bassham et al., 2000) and expression patterns (Zheng et al., 1999a) and are likely to posses distinct functions as well. Gene disruption experiments of individual Arabidopsis syntaxins (i.e. AtSYP21, AtSYP22, AtSYP41, and AtSYP42) indicate that loss of a single member of a gene family is lethal (A.A. Sanderfoot and N.V. Raikhel, unpublished data). These results suggest that members of these gene families have diverged such that each has a unique essential function. Further work is needed to define the exact functions of each Arabidopsis syntaxin, yet these studies should provide much useful information about the functioning of the plant endomembrane system.
OTHER SNAREs
Other SNAREs besides syntaxins are required for vesicle fusion. The members of these groups contribute helices to the four-helix bundle at various trafficking steps throughout the endomembrane system (for review, see Scales et al., 2000). Often, the members of these groups may have roles in more than a single trafficking event and may interact with many different syntaxins (i.e. be a part of several different t-SNARE complexes). Even more confusing, the same SNARE that is part of a t-SNARE complex at one trafficking step can act as a v-SNARE in another. Thus classification as to function in these groups is difficult. It is fortunate that many of these groups appear to be conserved across eukaryotes and they can be classified by sequence homology. Just as was seen for the syntaxins above, these SNAREs are often encoded by small gene families made of two to seven members in Arabidopsis. As with the syntaxins, these gene families generally share identical splicing patterns (data not shown). A complete list of these SNAREs can be found in Table II.
Table II.
Other SNAREs of Arabidopsisa
| Name | Previous Name | Genomic Locus | AGI ID | Intracellular Localization | Expressed Gene? | Noteb |
|---|---|---|---|---|---|---|
| SNAP25 Group | ||||||
| AtSNAP33 | AtSNAP33 | MAF19.6 | At5g61210 | PM | Yes | 1 |
| AtSNAP29 | AtSNAP29 | MXM12.4 | At5g07880 | Yes | 2 | |
| AtSNAP30 | – | F16A14.10 | At1g13530 | ? | ||
| Vti1p Group | ||||||
| AtVTI11 | AtVTI1a | MUL8.190 | At5g38510 | TGN, PVC | Yes | 3 |
| AtVTI12 | AtVTI1b | T24P13.5 | At1g25740 | TGN, PVC | Yes | 3 |
| AtVTI13 | – | MXE2.6 | At3g29100 | Yes | ||
| Gos1p Group | ||||||
| AtGOS11 | – | F7H2.21 | At1g15590 | Yes | ||
| AtGOS12 | – | F4L23.29 | At2g45200 | Yes | ||
| Bet1p Group | ||||||
| AtBET11 | – | F9D24.80 | At3g58170 | Yes | ||
| AtBET12 | – | dl3265w | At4g14450 | ? | ||
| Membrin Group | ||||||
| AtMEMB11 | – | T1J8.8 | At2g36900 | Yes | ||
| AtMEMB12 | – | MXI22.9 | At5g** | ? | ||
| Novel plant Group | ||||||
| AtNPSN11 | – | T4C15.14 | At2g35190 | Yes | ||
| AtNPSN12 | – | F11A17.20 | At1g44640 | Yes | ||
| AtNPSN13 | – | MTO12.3 | At3g17440 | ? | ||
| VAMPS | ||||||
| AtSEC22 | – | F12F1.27 | At1g11610 | Yes | ||
| AtYKT61 | – | K21L19.5 | At5g58060 | Yes | ||
| AtYKT62 | – | MCK7.5 | At5g58180 | ? | ||
| AtVAMP711 | VAMP7C | F10N7.4 | At4g32150 | Yes | 4 | |
| AtVAMP712 | – | T22F11.7 | At2g25340 | Yes | ||
| AtVAMP713 | – | F2I11.40 | At5g11150 | ? | ||
| AtVAMP714 | – | MWD9.16 | At5g22360 | ? | ||
| AtVAMP721 | SAR1 | T1G11.1 | At1g04630 | Yes | 5 | |
| AtVAMP722 | VAMP7B | F25I18.14 | At2g33120 | Yes | 6 | |
| AtVAMP723 | – | F25I18.15 | At2g33110 | Yes | ||
| AtVAMP724 | – | dl3930c | At4g15780 | Yes | ||
| AtVAMP725 | – | F24L7.19 | At2g32670 | ? | ||
| AtVAMP726 | – | F13M7.25 | At1g04650 | ? | ||
| AtVAMP727 | – | F24B22.260 | At3g54300 | ? |
Headings and definitions as given in Table I.
1, X. Gansel and L. Sticher, GenBank accession no. X92419; 2, C. Dickey and S. Bednarek, personal communication; 3, Zheng et al., 1999b; 4, D.M. Nikoloff and C. Somerville, GenBank accession no. AF025333; 5, M. Schena and R.W. Davis, GenBank accession no. M90418; 6, D.M. Nikoloff and C. Somerville, GenBank accession no. AF025332.
SNAP25-Like SNAREs
The SNAP25 class of SNAREs was first described in the mammalian neuron. SNAP25 contributes two helices (one N-terminal and one C-terminal) to the t-SNARE complex of the mammalian neuronal synaptic membrane (McNew et al., 2000). SNAP25 is bound to the membrane by post-translational lipid addition; however, other members of the SNAP25 class are found to associate with membranes without addition of lipid anchors (Steegmaier et al., 1998). Mammalian cells have at least three members of this group: at the PM, there is one neuron-specific and one ubiquitous SNAP25; a third member functions at other endomembranes (Oyler et al., 1989; Ravichandran et al., 1996; Steegmaier et al., 1998). Yeast have two members of the SNAP25 class. Sec9p interacts with Sso1/2p at the PM as part of general secretion (Brennwald et al., 1994), whereas Spo20p is a sporulation-specific protein (Neiman et al., 2000). Arabidopsis has three genes that encode proteins of the SNAP25 group, one of which (AtSNAP33) has been found to localize to the PM and likely functions in vesicle secretion (X. Gansel and L. Sticher, personal communication).
Vti1p-, Gos1p-, and Membrin-Like SNAREs
In yeast, Vti1p was first described as a v-SNARE involved in TGN-to-PVC trafficking (Fischer von Mollard et al., 1997). Further work has indicated that this SNARE functions in many additional pathways such as intra-Golgi trafficking (Fischer von Mollard et al., 1997; Lupashin et al., 1997), as well as all three vesicle trafficking steps to the yeast vacuole where it is part of a vacuolar t-SNARE complex (Fischer von Mollard and Stevens, 1999; Fukuda et al., 2000). Mammals contain two Vti1p orthologs (Fischer von Mollard and Stevens, 1998; Xu et al., 1998), whereas C. elegans and fruit fly contain one (Littleton, 2000). Arabidopsis contains three orthologs, two of which probably have distinct functions in vesicle trafficking (Zheng et al., 1999b; Bassham et al., 2000).
Yeast Gos1p is believed to have a role in intra-Golgi trafficking, probably as part of a t-SNARE complex with the Golgi syntaxin Sed5p (McNew et al., 1998), though the precise role of this SNARE remains unclear. Mammals have a Gos1p-ortholog called GS28 (Subramaniam et al., 1997), whereas Arabidopsis has two. Trafficking between the ER and Golgi in yeast requires the action of the two SNAREs: Bet1p is the v-SNARE for this step, whereas Bos1p is part of a t-SNARE complex that includes Sec22p (see VAMPs) and the Golgi syntaxin Sed5p (Newman et al., 1990; Parlati et al., 2000). Arabidopsis has two orthologs of Bet1p, however, no homologs of Bos1p are found. In mammals it is believed that the SNARE membrin has taken on the role of Bos1p (Hay et al., 1998). This is likely to be true in plants as well, since Arabidopsis has two membrin orthologs. In addition, Arabidopsis has a novel SNARE gene family of three members that has no counterparts in other eukaryotes.
VAMPs
VAMPs (also called synaptobrevins) are anchored to membranes by a C-terminal transmembrane domain or by post-translational addition of lipids. Adjacent to the membrane anchor is a coiled-coil domain centered on an Arg residue. Mammals contain many VAMP isoforms that play roles in many different trafficking steps, usually as v-SNAREs. Yeast encodes for only five, including a redundant pair. Sec22p is required for ER-to-Golgi (where it is part of the t-SNARE complex; Parlati et al., 2000) and Golgi-to-ER trafficking (probably as a v-SNARE; Newman et al., 1990; Sacher et al., 1997), Ykt6p is involved in intra-Golgi and perhaps vacuolar trafficking (McNew et al., 1997; Ungermann et al., 1999; Fukada et al., 2000), Nyv1p is required for homotypic vacuole fusion as a v-SNARE (Nichols et al., 1997; Fukada et al., 2000), and the redundant pair of Snc1/2p are the v-SNAREs involved in secretion (Protopopov et al., 1993; McNew et al., 2000). Arabidopsis encodes 14 VAMP isoforms (see Table II).
Arabidopsis seems to lack a VAMP of the Snc1/2p-class (orthologous with VAMP 1/synaptobrevin 1 in mammals). VAMPs of this class are known to be required for secretion (Protopopov et al., 1993; McMahon and Sudhof, 1995). Whether another Arabidopsis VAMP-homolog has replaced the Snc1/2p-type protein in this capacity should be examined.
Equally unique is the observation that the only ortholog of Sec22p in Arabidopsis actually encodes a protein with a C-terminal coiled-coil centered on a Val residue instead of an Arg. This gene is expressed in Arabidopsis; however, whether it can function in the role of a Sec22p-type protein remains to be determined. If not, it seems that Arabidopsis lacks two vital classes of VAMP isoforms.
Arabidopsis does have a gene family of two members encoding Ykt6p-orthologs. In a similar manner, a gene family of four members encodes VAMP7 orthologs, a mammalian VAMP isoform probably involved in endosomal/lysosomal trafficking (Advani et al., 1999). It is interesting that Arabidopsis encodes a distinct gene family of seven members that also show homology to VAMP7, including some genes that encode proteins lacking membrane anchors. The roles of this large number of potential VAMP7 homologs remains unstudied.
NSF AND SNAPs
The SNARE complex is remarkably stable once formed and requires a great deal of energy to disassemble (Fasshauer et al., 1998). It is believed that this is the role of two soluble proteins, NSF and SNAP. The SNARE complex serves as a binding site for SNAP, which then recruits NSF. The ATPase activity of NSF then serves to disassemble the SNARE complex, freeing the components for subsequent pairing and fusion events. As in other eukaryotes NSF is a single-copy gene in Arabidopsis (see Table III). The yeast genome encodes a single SNAP, called Sec17p, whereas the mammalian genome codes for three types of SNAPs: α-, β-, and γ-SNAP. Arabidopsis encodes two isoforms of the α-SNAP type and one of the γ-SNAP type (see Table III).
Table III.
SNARE interacting proteinsa
| Groups | Gene Names | Genomic Locus | AGI ID | Intracellular Localization | Expressed Gene? | Noteb |
|---|---|---|---|---|---|---|
| α-SNAP | α-SNAP1 | T5P19.100 | At3g56450 | Yes | ||
| α-SNAP2 | F18O21.150 | At3g56190 | Yes | |||
| γ-SNAP | γ-SNAP | F9F13.60 | At4g20410 | Yes | ||
| NSF | NSF | T1J1.4 | At4g04910 | Yes | ||
| CDC48 | AtCDC48a | F8A24.11 | At3g09840 | Yes | 1 | |
| AtCDC48b | F19B11.22 | At2g03670 | Yes | |||
| AtCDC48c | F4P13.15 | At3g01610 | ? | |||
| Sec1p-Family | ||||||
| Sec1p-Group | KEULE | F5O11.8 | At1g12080 | Phragmoplast | Yes | 2 |
| AtSEC1a | F22M8.14 | At1g01980 | Yes | 2 | ||
| AtSEC1b | F16J13.190 | At4g12120 | Yes | 2 | ||
| Vps33p-Group | AtVPS33 | F28P10.160 | At3g54860 | Yes | ||
| Vps45p-Group | AtVPS45 | T14N5.4 | At1g70890 | TGN | Yes | 3 |
| Sly1p-Group | AtSLY1 | T27K22.15 | At2d17980 | Yes |
Headings and definitions as given in Table I.
A second large ATPase, CDC48 (p97 in mammals), is known to interact with certain SNAREs as part of specialized membrane fusion events where vesicles derived from the same organelle fuse (called homotypic fusion; Patel et al., 1998; Rabouille et al., 1998). Three orthologs are encoded in Arabidopsis and at least two are expressed (see Table III). Although the precise functions of these factors have not yet been determined, AtCDC48 has been localized to the phragmoplast in dividing cells, suggesting some role in cytokinesis (C. Dickey and S. Bednarek, personal communication).
Sec1p Family
The Sec1p-family is another well-conserved group of large peripheral membrane proteins. In neuronal cells Sec1 proteins positively regulate vesicle fusion by inducing conformational changes in syntaxins (for review, see Hanson, 2000); however, the precise role of Sec1 proteins remains unclear. Yeast contains four members of this family. Sec1p functions at the PM, forming complexes with Sso1/2p and somehow controlling the activity of these syntaxins in secretion (Aalto et al., 1991, 1993). A second member, Sly1p, functions in traffic in both directions between the ER and Golgi (Ossig et al., 1991). Vps45p plays roles in anterograde transport to the PVC through interactions with the prevacuolar syntaxin Pep12p (Peterson et al., 1999) and in an alternate pathway to the vacuole in concert with the TGN syntaxin Tlg2p (Abeliovich et al., 1999). Vps33p is found on the vacuolar membrane and functions in several trafficking steps to that organelle through interactions with the vacuolar syntaxin Vam3p (Darsow et al., 1997).
Arabidopsis contains six members of the Sec1p family (see Table III and Fig. 4). Three are orthologous to Sec1p. Among these is KEULE, which is required for cytokinesis rather than secretion (Assaad et al., 2001) Perhaps the other Sec1 orthologues function in secretion. A single ortholog of Sly1p is found in Arabidopsis, perhaps functioning in the early secretory pathway. Arabidopsis also has a single gene encoding a Vps33p-ortholog. This is interesting considering that no Vam3p-type syntaxin is found in Arabidopsis. It is likely that this protein has adopted a new function in plants, though this has yet to be investigated. What has been investigated is the function of AtVPS45, a Vps45p-homolog from Arabidopsis. Expression of this gene in yeast is able to complement all the defects associated with a vps45Δ mutation (Bassham and Raikhel, 1998), suggesting that this Arabidopsis protein can interact with the yeast TGN-to-PVC trafficking machinery (including the PVC syntaxin Pep12p). However, in Arabidopsis we have found that this protein is found only at the TGN and does not interact with any of the Arabidopsis PVC syntaxins (AtSYP21 or AtSYP22); instead, AtVPS45 was found to only interact with the TGN syntaxins of the SYP4 gene family (Bassham et al., 2000). It is clear that the function of AtVPS45 is different in plants and is the subject of ongoing research. These results clearly indicate that simple homology and even complementation of yeast mutants is not proof of conserved function, a finding that has been noted for other plant genes as well (Bassham and Raikhel, 2000).
There is a total of six Sec1 proteins compared with 24 syntaxins. It is clear that there are not enough Sec1 proteins to individually bind to so many syntaxins. This is similar to the situation found in mammalian cells, and to a lesser extent in yeast. Though this has not been studied in great detail, it is believed that a single Sec1 protein can interact specifically with a number of syntaxins (Abeliovich et al., 1999; Peterson et al., 1999; Bassham et al., 2000).
THE LETHALITY OF VESICLE TRAFFICKING MUTANTS
Although analyses have been carried out on the intracellular localization of Arabidopsis SNAREs and related proteins, little is known about the functions of the corresponding genes. In fact, until recently the only vesicle trafficking mutants of Arabidopsis were, surprisingly, ones defective in cytokinesis. Building a structure as complex as a cell wall in the brief period of time between anaphase and telophase requires a rapid and efficient mobilization of resources. This is achieved by the targeting and fusion of Golgi-derived vesicles at the equator of a dividing cell: the vesicle contents build the cell wall and the vesicle membranes form the flanking PMs. A number of Arabidopsis mutants affect the execution of cytokinesis (F.F. Assaad, U. Mayer, and G. Jürgens, unpublished data). These mutants all have similar phenotypes, characterized by multinucleate cells with gapped or incomplete cross walls. Two cytokinesis-defective genes of Arabidopsis have been cloned: KNOLLE (AtSYP111) encodes a cytokinesis-specific syntaxin (Lukowitz et al., 1996) and KEULE encodes a cytokinesis-related Sec1p family member (Assaad et al., 2001). KNOLLE and KEULE interact genetically (Waizzenegger et al., 2000) and have been shown to bind to each other in vitro (Assaad et al., 2001). KEULE and KNOLLE show some similarity to groups of Sec1 or syntaxin genes involved in exocytosis. Although this might appear surprising for proteins involved in cytokinesis, it is consistent with the idea that cytokinesis in plants is a specialized form of exocytosis. Further analysis of these and related genes will shed light on the as yet elusive biological function of the Sec1-syntaxin complex in the novel context of cell cycle progression.
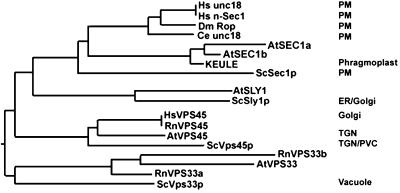
It is to be noted in this context, however, that lethality in plants comes in three different forms: seedling lethality, embryo lethality, and gametophytic lethality. Thus, although keule and knolle mutants complete embryogenesis, germinate, and die as seedlings, keule-knolle double mutants are embryo lethal and die during embryogenesis as bags of nuclei with no cross walls (Waizzenegger et al., 2000). The striking phenotype of the double mutant compared with the attenuated phenotypes of the single mutants suggests that other Arabidopsis genes are partially redundant to KEULE and KNOLLE, allowing the corresponding cytokinesis-defective mutants to muddle through life until the seedling stage. In a consistent manner, phylogenetic analysis places KEULE and KNOLLE in subfamilies with several members: KEULE has two close paralogues, and KNOLLE has eight (see Figs. 1 and 5).
Figure 5.
Phylogenetic analysis of Sec1 proteins from several eukaryotes. Protein sequences from representative Sec1p family members from yeast (Sc), Arabidopsis (At), Human (Hs), Rat (Rattus novernicus; Rn), fruit fly (Dm), and C. elegans (Ce) were acquired from GenBank and aligned as described in Figure 2. Where it is known the intracellular localization of Sec1 proteins is indicated. Note that although the closest homologs of AtSec1a, AtSec1b, and KEULE are the animal exocytic Sec1 proteins, the three Arabidopsis Sec1 proteins cluster as a somewhat separate group and may have evolved to play novel roles (such as cytokinesis in the case of KEULE).
Gene disruptions in syntaxins other than KNOLLE are also lethal (A.A. Sanderfoot and N.V. Raikhel, unpublished data). In general, the severe lethality of vesicle trafficking mutants renders their genetic characterization difficult. The isolation and characterization of conditional mutants would greatly further our understanding of the plant SNAREs.
HOW GOOD A MODEL IS ARABIDOPSIS FOR OTHER PLANTS?
Limited genomic sequencing as well as several extensive EST projects have been carried out in other plants. Examination of these databases reveals that the patterns outlined above for Arabidopsis also hold in other plants such as soybean, tomato, rice, and maize; namely, the presence of small gene families encoding SNARE components and the presence of syntaxins and other SNARE groups not found in animals or yeast. Investigation of the functions of these proteins in other plants, especially in the monocots such as rice and maize, may provide useful information that will help clarify the functions of these essential genes.
WHY SO MANY SNAREs IN SUCH A COMPACT GENOME?
The large number of SNAREs in the Arabidopsis genome is unprecedented: Arabidopsis has at least two times more syntaxins than worms or flies, although the genomes are of similar complexity. In a similar manner, Arabidopsis will likely have more syntaxins than humans, which have greater than 10-fold more DNA. Although the majority of these “extra” SNAREs come in gene families, several lines of evidence suggest that they are not functionally redundant. In cases where it has been studied, gene disruptions of syntaxins are lethal (Lukowitz et al., 1996; A.A. Sanderfoot and N.V. Raikhel, unpublished data), indicating that each SNARE fulfills a unique essential function. Furthermore, although the two highly conserved syntaxins, AtSYP41 and AtSYP42, localize to the TGN, they are in fact on distinct domains of this network. Of the 24 syntaxin genes, at least 20 are expressed and therefore unlikely to be pseudogenes.
But why so many? It appears that some of these proteins have specialized in the plant-specific method of cytokinesis (Lauber et al., 1997; Assaad et al., 2001), whereas others appear to have taken on roles in hormone signaling (SYP121/SYR1; Leyman et al., 1999). Since it is known that protein targeting in plant cells can be polarized to particular domains of the PM (Steinmann et al., 1999), it is possible that other “extra” SNAREs have specialized into these roles. These speculations require further analysis of the Arabidopsis SNAREs before one can explain why this modest plant has evolved such a sophisticated and differentiated vesicle trafficking machinery.
ACKNOWLEDGMENTS
We thank those many researchers who provided results prior to publication. We also thank Diane Bassham, John Froehlich, and Jason Bock for discussions and critical reading of the manuscript. We acknowledge many helpful comments during the development of a systematic nomenclature system for the syntaxins from members of the BioSci Arabidopsis newsgroup, the Secretory Group mailing list, the curators of The Arabidopsis Information Resource (www.Arabidopsis.org), and especially Mike Blatt.
Footnotes
A.A.S. was supported by a National Institute of Health postdoctoral fellowship (no. GM 18861). F.F.A. was supported by the Deutsche Forschungsgemeinschaft (grant no. AS110/2–1). N.V.R. was supported by funds from the U.S. Department of Energy (grant no. DE–FG02–91ER–20021) and by the National Science Foundation (grant no. MCB–9507030).
LITERATURE CITED
- Aalto MK, Ronne H, Keranen S. Yeast syntaxins Sso1p and Sso2p belong to a family of related membrane proteins that function in vesicular transport. EMBO J. 1993;12:4095–4104. doi: 10.1002/j.1460-2075.1993.tb06093.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aalto MK, Ruohonen L, Hosono K, Keranen S. Cloning and sequencing of the yeast Saccharomyces cerevisiae SEC1 gene localized on chromosome IV. Yeast. 1991;7:643–650. doi: 10.1002/yea.320070613. [DOI] [PubMed] [Google Scholar]
- Abeliovich H, Darsow T, Emr SD. Cytoplasm to vacuole trafficking of aminopeptidase I requires a t-SNARE-Sec1p complex composed of Tlg2p and Vps45p. EMBO J. 1999;18:6005–6016. doi: 10.1093/emboj/18.21.6005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abeliovich H, Grote E, Novick P, Ferro-Novick S. Tlg2p, a yeast syntaxin homolog that resides on the Golgi and endocytic structures. J Biol Chem. 1998;273:11719–11727. doi: 10.1074/jbc.273.19.11719. [DOI] [PubMed] [Google Scholar]
- Adams MD. The genome sequence of Drosophila melanogaster. Science. 2000;287:2185–2195. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- Advani RJ, Hae-Rahn B, Bock JB, Chao DS, Doung Y-C, Prekeris R, Yoo J-S, Scheller RH. Seven novel mammalian SNARE proteins localize to distinct membrane compartments. J Biol Chem. 1998;273:10317–10324. doi: 10.1074/jbc.273.17.10317. [DOI] [PubMed] [Google Scholar]
- Advani RJ, Yang B, Prekeris R, Lee KC, Klumperman J, Scheller RH. VAMP-7 mediates vesicular transport from endosomes to lysosomes. J Cell Biol. 1999;146:765–776. doi: 10.1083/jcb.146.4.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assaad FF, Huet Y, Mayer U, Jürgens G (2001) The cytokinesis gene KEULE encodes a sec1 protein which binds the syntaxin KNOLLE. J Cell Biol (in press) [DOI] [PMC free article] [PubMed]
- Bassham DC, Gal S, Conceição AS, Raikhel NV. An Arabidopsis syntaxin homologue isolated by functional complementation of a yeast pep12 mutant. Proc Natl Acad Sci USA. 1995;92:7262–7266. doi: 10.1073/pnas.92.16.7262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassham DC, Raikhel NV. An Arabidopsis VPS45p homolog implicated in protein transport to the vacuole. Plant Physiol. 1998;117:407–415. doi: 10.1104/pp.117.2.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassham DC, Raikhel NV. Plants are not just green yeast. Plant Physiol. 2000;122:999–1002. doi: 10.1104/pp.122.4.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassham DC, Sanderfoot AA, Kovaleva V, Zheng H, Raikhel NV. AtVPS45 complex formation at the trans-Golgi network. Mol Biol Cell. 2000;11:2251–2265. doi: 10.1091/mbc.11.7.2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becherer KA, Reider SE, Emr SD, Jones EW. Novel syntaxin homologue, Pep12p, required for the sorting of lumenal hydrolases to the lysosome-like vacuole of yeast. Mol Biol Cell. 1996;7:579–594. doi: 10.1091/mbc.7.4.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett MK, Garcia-Arras JE, Elferink K, Peterson K, Fleming AM, Hazuka CD, Scheller RH. The syntaxin family of vesicular transport receptors. Cell. 1993;74:863–873. doi: 10.1016/0092-8674(93)90466-4. [DOI] [PubMed] [Google Scholar]
- Bock JB, Klumperman J, Davanger S, Scheller RH. Syntaxin 6 functions in trans-Golgi network vesicle trafficking. Mol Biol Cell. 1997;8:1261–1271. doi: 10.1091/mbc.8.7.1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennwald P, Kearns B, Champion K, Keranen S, Bankaitis V, Novick P. Sec9 is a SNAP-25-like component of a yeast SNARE complex that may be the effector of Sec4 function in exocytosis. Cell. 1994;79:245–258. doi: 10.1016/0092-8674(94)90194-5. [DOI] [PubMed] [Google Scholar]
- Conceição AdS, Marty-Mazars D, Bassham DC, Sanderfoot AA, Marty F, Raikhel NV. The syntaxin homolog AtPEP12p resides on a late post-Golgi compartment in plants. Plant Cell. 1997;9:571–582. [PMC free article] [PubMed] [Google Scholar]
- Darsow T, Rieder SE, Emr SD. A multispecificity syntaxin homologue, Vam3p, essential for autophagic and biosynthetic protein transport to the vacuole. J Cell Biol. 1997;138:517–529. doi: 10.1083/jcb.138.3.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dascher C, Matteson J, Balch WE. Syntaxin 5 regulates endoplasmic reticulum to Golgi transport. J Biol Chem. 1994;269:29363–29366. [PubMed] [Google Scholar]
- Fasshauer D, Eliason WK, Brunger AT, Jahn R. Identification of a minimal core of the synaptic SNARE complex sufficient for reversible assembly and disassembly. Biochemistry. 1998;37:10354–10362. doi: 10.1021/bi980542h. [DOI] [PubMed] [Google Scholar]
- Feiler HS, Desprez T, Santoni V, Kronenberger J, Caboche M, Traas J. The higher plant Arabidopsis thaliana encodes a functional CDC48 homologue which is highly expressed in dividing and expanding cells. EMBO J. 1995;14:5626–5637. doi: 10.1002/j.1460-2075.1995.tb00250.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez I, Ubach J, Dulubova I, Zhang X, Sudhof TC, Rizo J. Three-dimensional structure of an evolutionarily conserved N-terminal domain of syntaxin 1A. Cell. 1998;94:841–849. doi: 10.1016/s0092-8674(00)81742-0. [DOI] [PubMed] [Google Scholar]
- Fiebig KM, Rice LM, Pollack E, Brunger AT. Folding intermediates of SNARE complex assembly. Nat Struct Biol. 1999;6:117–123. doi: 10.1038/5803. [DOI] [PubMed] [Google Scholar]
- Fischer von Mollard G, Nothwehr SF, Stevens TH. The yeast v-SNARE Vti1p mediates two vesicle transport pathways through interactions with the t-SNAREs Sed5p and Pep12p. J Cell Biol. 1997;137:1511–1524. doi: 10.1083/jcb.137.7.1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer von Mollard G, Stevens TH. A human homolog can functionally replace the yeast vesicle-associated SNARE Vti1p in two vesicle transport pathways. J Biol Chem. 1998;273:2624–2630. doi: 10.1074/jbc.273.5.2624. [DOI] [PubMed] [Google Scholar]
- Fischer von Mollard G, Stevens TH. The Saccharomyces cerevisiae v-SNARE Vti1p is required for multiple membrane transport pathways to the vacuole. Mol Biol Cell. 1999;10:1719–1732. doi: 10.1091/mbc.10.6.1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda R, McNew JA, Weber T, Parlati F, Engel T, Nickel W, Rothman JE, Söllner TH Functional architecture of an intracellular membrane t-SNARE. Nature 407: 198–202 [DOI] [PubMed]
- Gonzalez L, Scheller RH. Regulation of membrane trafficking: structural insights from a Rab/effector complex. Cell. 1999;96:755–758. doi: 10.1016/s0092-8674(00)80585-1. [DOI] [PubMed] [Google Scholar]
- Hanson PI. Sec1 gets a grip on syntaxin. Nat Struct Biol. 2000;7:347–349. doi: 10.1038/75103. [DOI] [PubMed] [Google Scholar]
- Hardwick KG, Pelham HR. SED5 encodes a 39-kD integral membrane protein required for vesicular transport between the ER and the Golgi complex. J Cell Biol. 1993;119:513–521. doi: 10.1083/jcb.119.3.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatsuzawa K, Hirose H, Tani K, Yamamoto A, Scheller RH, Tagaya M. Syntaxin 18, a SNAP receptor that functions in the endoplasmic reticulum, intermediate compartment, and cis-Golgi vesicle trafficking. J Biol Chem. 2000;275:13713–13720. doi: 10.1074/jbc.275.18.13713. [DOI] [PubMed] [Google Scholar]
- Hay JC, Klumperman J, Oorschot V, Steegmaier M, Kuo CS, Scheller RH. Localization, dynamics, and protein interactions reveal distinct roles for ER and Golgi SNAREs. J Cell Biol. 1998;141:1489–502. doi: 10.1083/jcb.141.7.1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holthuis JCM, Nichols BJ, Dhruvakumar S, Pelham HR. Two syntaxin homologues in the TGN/endosomal system of yeast. EMBO J. 1998;17:113–126. doi: 10.1093/emboj/17.1.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koushika SP, Nonet ML. Sorting and transport in C. elegans: a model system with a sequenced genome. Curr Opin Cell Biol. 2000;12:517–523. doi: 10.1016/s0955-0674(00)00125-3. [DOI] [PubMed] [Google Scholar]
- Lauber MH, Waizenegger I, Steinmann T, Schwarz H, Mayer U, Hwang I, Lukowitz W, Jurgens G. The Arabidopsis KNOLLE protein is a cytokinesis-specific syntaxin. J Cell Biol. 1997;139:1485–1493. doi: 10.1083/jcb.139.6.1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis MJ, Rayner JC, Pelham HR. A novel SNARE complex implicated in vesicle fusion at the endoplasmic reticulum. EMBO J. 1997;16:3017–3024. doi: 10.1093/emboj/16.11.3017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leyman B, Geelen D, Quintero FJ, Blatt MR. A tobacco syntaxin with a role in hormonal control of guard cell ion channels. Science. 1999;283:537–540. doi: 10.1126/science.283.5401.537. [DOI] [PubMed] [Google Scholar]
- Littleton JT. A genomic analysis of membrane trafficking and neurotransmitter release in Drosophila. J Cell Biol. 2000;150:F77–F82. doi: 10.1083/jcb.150.2.f77. [DOI] [PubMed] [Google Scholar]
- Lukowitz W, Mayer U, Jurgens G. Cytokinesis in the Arabidopsis embryo involves the syntaxin-related KNOLLE gene product. Cell. 1996;84:61–71. doi: 10.1016/s0092-8674(00)80993-9. [DOI] [PubMed] [Google Scholar]
- Lupashin VV, Pokrovskaya ID, McNew JA, Waters MG. Characterization of a novel yeast SNARE protein implicated in Golgi retrograde traffic. Mol Biol Cell. 1997;8:2659–2676. doi: 10.1091/mbc.8.12.2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon HT, Sudhof TC. Synaptic core complex of synaptobrevin, syntaxin, and SNAP25 forms high affinity α-SNAP binding site. J Biol Chem. 1995;270:2213–2217. doi: 10.1074/jbc.270.5.2213. [DOI] [PubMed] [Google Scholar]
- McNew JA, Coe JG, Sogaard M, Zemelman BV, Wimmer C, Hong W, Sollner TH. Gos1p, a Saccharomyces cerevisiae SNARE protein involved in Golgi transport. FEBS Lett. 1998;435:89–95. doi: 10.1016/s0014-5793(98)01044-8. [DOI] [PubMed] [Google Scholar]
- McNew JA, Parlati F, Fukuda R, Johnston J, Paz K, Paumet F, Söllner TH, Rothman JH. Compartmental specificity of cellular membrane fusion encoded in SNARE proteins. Nature. 2000;407:153–159. doi: 10.1038/35025000. [DOI] [PubMed] [Google Scholar]
- McNew JA, Sogaard M, Lampen NM, Machida S, Ye RR, Lacomis L, Tempst P, Rothman JE, Sollner TH. Ykt6p, a prenylated SNARE essential for endoplasmic reticulum-Golgi transport. J Biol Chem. 1997;272:17776–17783. doi: 10.1074/jbc.272.28.17776. [DOI] [PubMed] [Google Scholar]
- Misura KM, Scheller RH, Weis WI. Three-dimensional structure of the neuronal-Sec1-syntaxin 1a complex. Nature. 2000;404:355–362. doi: 10.1038/35006120. [DOI] [PubMed] [Google Scholar]
- Neiman AM, Katz L, Brennwald PJ. Identification of domains required for developmentally regulated SNARE function in Saccharomyces cerevisiae. Genetics. 2000;155:1643–1655. doi: 10.1093/genetics/155.4.1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman AP, Shim J, Ferro-Novick S. BET1, BOS1, and SEC22 are members of a group of interacting yeast genes required for transport from the endoplasmic reticulum to the Golgi complex. Mol Cell Biol. 1990;10:3405–3414. doi: 10.1128/mcb.10.7.3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols BJ, Ungermann C, Pelham HR, Wickner WT, Haas A. Homotypic vacuolar fusion mediated by t- and v-SNAREs. Nature. 1997;387:199–202. doi: 10.1038/387199a0. [DOI] [PubMed] [Google Scholar]
- Ossig R, Dascher C, Trepte HH, Schmitt HD, Gallwitz D. The yeast SLY gene products, suppressors of defects in the essential GTP-binding Ypt1 protein, may act in endoplasmic reticulum-to-Golgi transport. Mol Cell Biol. 1991;11:2980–2993. doi: 10.1128/mcb.11.6.2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oyler GA, Higgins GA, Hart RA, Battenberg E, Billingsley M, Bloom FE, Wilson MC. The identification of a novel synaptosomal-associated protein, SNAP-25, differentially expressed by neuronal subpopulations. J Cell Biol. 1989;109:3039–3052. doi: 10.1083/jcb.109.6.3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parlati F, McNew JA, Fukuda R, Miller R, Söllner TH, Rothman JH. Topological restriction of SNARE-dependent membrane fusion. Nature. 2000;407:194–198. doi: 10.1038/35025076. [DOI] [PubMed] [Google Scholar]
- Patel SK, Indig FE, Olivieri N, Levine ND, Latterich M. Organelle membrane fusion: a novel function for the syntaxin homolog Ufe1p in ER membrane fusion. Cell. 1998;92:611–620. doi: 10.1016/s0092-8674(00)81129-0. [DOI] [PubMed] [Google Scholar]
- Pelham HR. SNAREs and the secretory pathway: lessons from yeast. Exp Cell Res. 1999;247:1–8. doi: 10.1006/excr.1998.4356. [DOI] [PubMed] [Google Scholar]
- Peterson MR, Burd CG, Emr SD. Vac1p coordinates Rab and phosphatidylinositol 3-kinase signaling in Vps45p-dependent vesicle docking/fusion at the endosome. Curr Biol. 1999;9:159–162. doi: 10.1016/s0960-9822(99)80071-2. [DOI] [PubMed] [Google Scholar]
- Prekeris R, Yang B, Oorschot V, Klumperman J, Scheller RH. Differential roles of syntaxin 7 and syntaxin 8 in endosomal trafficking. Mol Biol Cell. 1999;10:3891–3908. doi: 10.1091/mbc.10.11.3891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Protopopov V, Govindan B, Novick P, Gerst JE. Homologs of the synaptobrevin/VAMP family of synaptic vesicle proteins function on the late secretory pathway in S. cerevisiae. Cell. 1993;74:855–861. doi: 10.1016/0092-8674(93)90465-3. [DOI] [PubMed] [Google Scholar]
- Rabouille C, Kondo H, Newman R, Hui N, Freemont P, Warren G. Syntaxin 5 is a common component of the NSF- and p97-mediated reassembly pathways of Golgi cisternae from mitotic Golgi fragments in vitro. Cell. 1998;92:603–610. doi: 10.1016/s0092-8674(00)81128-9. [DOI] [PubMed] [Google Scholar]
- Ravichandran V, Chawla A, Roche PA. Identification of a novel syntaxin- and synaptobrevin/VAMP-binding protein, SNAP-23, expressed in non-neuronal tissues. J Biol Chem. 1996;271:13300–13303. doi: 10.1074/jbc.271.23.13300. [DOI] [PubMed] [Google Scholar]
- Sacher M, Stone S, Ferro-Novick S. The synaptobrevin-related domains of Bos1p and Sec22p bind to the syntaxin-like region of Sed5p. J Biol Chem. 1997;272:17134–17138. doi: 10.1074/jbc.272.27.17134. [DOI] [PubMed] [Google Scholar]
- Sanderfoot AA, Ahmed SU, Marty-Mazars D, Rapoport I, Kirchhausen T, Marty F, Raikhel NV. A putative vacuolar cargo receptor partially colocalizes with AtPEP12p on a prevacuolar compartment in Arabidopsis roots. Proc Natl Acad Sci USA. 1998;95:9920–9925. doi: 10.1073/pnas.95.17.9920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderfoot AA, Kovaleva V, Zheng H, Raikhel NV. The t-SNARE AtVAM3p resides on the prevacuolar compartment in Arabidopsis root cells. Plant Physiol. 1999;121:929–938. doi: 10.1104/pp.121.3.929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderfoot AA, Raikhel NV. The specificity of vesicle trafficking: coat proteins and SNAREs. Plant Cell. 1999;11:629–642. doi: 10.1105/tpc.11.4.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato MH, Nakamura N, Ohsumi Y, Kouchi H, Kondo M, Hara-Nishimura I, Nishimura M, Wada Y. The AtVAM3 encodes a syntaxin-related molecule implicated in the vacuolar assembly in Arabidopsis thaliana. J Biol Chem. 1997;272:24530–24535. doi: 10.1074/jbc.272.39.24530. [DOI] [PubMed] [Google Scholar]
- Scales SJ, Bock J, Scheller R. The specifics of membrane fusion. Nature. 2000;407:144–146. doi: 10.1038/35025176. [DOI] [PubMed] [Google Scholar]
- Séron K, Tieaho V, Prescianotto-Baschong C, Aust T, Blondel M-O, Guillaud P, Devilliers G, Rossanese OW, Glick BS, Riezman H, Keränen S, Hauguenauer-Tsapis R. A yeast t-SNARE involved in endocytosis. Mol Biol Cell. 1998;9:2873–2889. doi: 10.1091/mbc.9.10.2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonsen A, Bremnes B, Rønning E, Aasland R, Stenmark H. Syntaxin 16, a putative Golgi t-SNARE. Eur J Cell Biol. 1998;75:223–231. doi: 10.1016/S0171-9335(98)80116-7. [DOI] [PubMed] [Google Scholar]
- Steegmaier M, Yang B, Yoo JS, Huang B, Shen M, Yu S, Luo Y, Scheller RH. Three novel proteins of the syntaxin/SNAP-25 family. J Biol Chem. 1998;273:34171–34179. doi: 10.1074/jbc.273.51.34171. [DOI] [PubMed] [Google Scholar]
- Steinmann T, Geldner N, Grebe M, Mangold S, Jackson CL, Paris S, Galweiler L, Palme K, Jurgens G. Coordinated polar localization of auxin efflux carrier PIN1 by GNOM ARF GEF. Science. 1999;286:316–318. doi: 10.1126/science.286.5438.316. [DOI] [PubMed] [Google Scholar]
- Subramaniam VN, Loh E, Hong W. N-Ethylmaleimide-sensitive factor (NSF) and alpha-soluble NSF attachment proteins (SNAP) mediate dissociation of GS28-syntaxin 5 Golgi SNAP receptors (SNARE) complex. J Biol Chem. 1997;272:25441–25444. doi: 10.1074/jbc.272.41.25441. [DOI] [PubMed] [Google Scholar]
- Tang BL, Low DY, Tan AE, Hong W. Syntaxin 10: a member of the syntaxin family localized to the trans-Golgi network. Biochem Biophys Res Commun. 1998;242:345–350. doi: 10.1006/bbrc.1997.7966. [DOI] [PubMed] [Google Scholar]
- The Arabidopsis Genome Initiaitive (2000) Analysis of the genome of the flowering plant Arabidopsis thaliana. Nature (in press) [DOI] [PubMed]
- The C. elegans Sequencing Consortium. Genome sequence of the nematode C. elegans: a platform for investigating biology. Science. 1998;282:2012–2018. doi: 10.1126/science.282.5396.2012. [DOI] [PubMed] [Google Scholar]
- Ungermann C, Fischer von Mollard G, Jensen ON, Margolis N, Stevens TH, Wickner W. Three v-SNAREs and two t-SNAREs, present in a pentameric cis-SNARE complex on isolated vacuoles, are essential for homotypic fusion. J Cell Biol. 1999;145:1435–1442. doi: 10.1083/jcb.145.7.1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waizzenegger I, Lukowitz W, Assaad F, Mayer U, Jurgens G (2000) The Arabidopsis KNOLLE and KEULE genes interact to promote fusion of cytokinetic vesicles during cell plate formation. Curr Biol (in press) [DOI] [PubMed]
- Weber T, Zemelman BV, McNew JA, Westermann B, Gmachl M, Parlati F, Sollner TH, Rothman JE. SNAREpins: minimal machinery for membrane fusion. Cell. 1998;92:759–772. doi: 10.1016/s0092-8674(00)81404-x. [DOI] [PubMed] [Google Scholar]
- Weimbs T, Low SH, Chapin SJ, Mostov KE, Bucher P, Hofmann K. A conserved domain is present in different families of vesicular fusion proteins: a new superfamily. Proc Natl Acad Sci USA. 1998;94:3046–3051. doi: 10.1073/pnas.94.7.3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Wong SH, Tang BL, Subramaniam VN, Zhang T, Hong WJ. A 29-kilodalton Golgi soluble N-ethylmaleimide-sensitive factor attachment protein receptor (Vti1-rp2) implicated in protein trafficking in the secretory pathway. J Biol Chem. 1998;273:21783–21789. doi: 10.1074/jbc.273.34.21783. [DOI] [PubMed] [Google Scholar]
- Zheng H, Bassham DC, Conceição AS, Raikhel NV. The syntaxin family of proteins in Arabidopsis: a new syntaxin homologue shows polymorphism between two ecotypes. J Exp Bot. 1999a;50:915–924. [Google Scholar]
- Zheng H, Fischer von Mollard G, Kovaleva V, Stevens TH, Raikhel NV. The plant vesicle-associated SNARE AtVTI1a likely mediates vesicle transport from the trans-Golgi network to the prevacuolar compartment. Mol Biol Cell. 1999b;10:2251–2264. doi: 10.1091/mbc.10.7.2251. [DOI] [PMC free article] [PubMed] [Google Scholar]