Summary
The mammary gland (MG) is composed of basal cells (BCs) and luminal cells (LCs). While it is generally believed that MG arises from embryonic multipotent progenitors (EMPs), it remains unclear when lineage restriction occurs and what are the mechanisms responsible for the switch from multipotency to unipotency during MG morphogenesis. Here, we performed multicolor lineage tracing and assessed the fate of single progenitors and demonstrated the existence of a developmental switch from multipotency to unipotency during embryonic MG development. Molecular profiling and single cell RNA-seq revealed that EMPs express a unique hybrid basal and luminal signature and the factors associated with the different lineages. Sustained p63 expression in EMPs promotes unipotent BC fate and was sufficient to reprogram adult LCs into BCs by promoting an intermediate hybrid multipotent like state. Altogether, this study identifies the timing and the mechanisms mediating the early lineage segregation of multipotent progenitors during MG development.
Introduction
The mammary gland (MG) is a branched epithelium that produces the milk during lactation. The MG is composed of two main lineages: the basal cells (BCs), which are surrounding the inner luminal cells (LCs). The LCs can be subdivided into estrogen receptor (esr1 or ER) positive and ER negative ductal cells, and alveolar cells that produce the milk1.
The MG derives from the ectoderm around embryonic day 10.5 (E10.5). At E13, the MG placodes invaginate to form buds that continue to sprout until E16, when they start to branch. By E18.5, the epithelium forms a rudimentary ductal structure. From E18.5, the MG grows proportionally to the body size until puberty when the estrogen stimulates the rapid growth and further branching of the MG. During pregnancy and lactation, MG further develops and gives rise to alveolar LCs that differentiate into milk producing cells. At the end of the lactation, the MG involutes and goes back to its virgin appearance, ready to undergo a new cycle of growth for the next pregnancy1.
Lineage tracing experiments demonstrate that postnatal pubertal development and adult remodelling are mediated by unipotent basal and luminal progenitors/stem cells,2–12. Whereas multicolour clonal analysis combined with statistical modeling demonstrate the unipotency of adult BCs and LCs10–12, such experimental approaches have never been undertaken so far during MG development. Lineage tracing of keratin 14 (K14) expressing cells that compose the embryonic MG at E17 demonstrated that both basal and luminal lineages arise from K14-expressing cells during embryonic development8 and suggest the existence of embryonic multipotent progenitors (EMPs). However, these experiments cannot discriminate whether the apparent multipotency of embryonic MG arises from the labelling of distinct pools of already pre-committed BCs and LCs or whether EMPs are truly multipotent at the single cell level. In addition, it remains unclear when the basal and luminal lineage segregation occurs and what are the mechanisms responsible for the switch from multipotency to unipotency during MG morphogenesis.
Here, using multicolour clonal analysis in mice, we demonstrate the multipotency of EMPs and the existence of a switch from multipotency to unipotency that occurs during embryonic MG development. Using molecular profiling and single cell RNA sequencing, we demonstrate that multipotency is associated with a hybrid basal and luminal gene expression signature. Finally we show the key role of p63 in promoting BC fate in EMPs.
Results
Clonal analysis demonstrates the switch from multipotency to unipotency during MG development
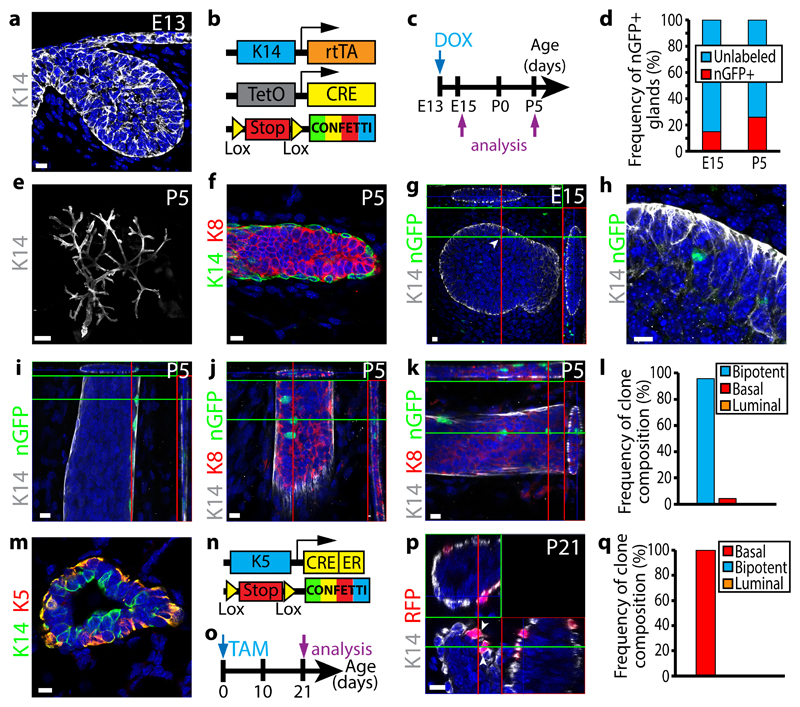
To assess whether MG arises from early multipotent progenitors or from a mixture of different lineage restricted progenitors, we performed clonal analysis using lineage tracing experiments at the early stages of MG development, when K14 is homogenously expressed in all MG cells (Fig. 1a). To this end, we generated K14rtTA/TetO-Cre/Rosa-Confetti mice (Fig. 1b) and titrated the dose of doxycycline that lead to a clonal labelling of the MG. Among the four colours of the confetti reporter system, the nGFP is much less frequently recombined than the other fluorescent proteins10, 13, 14. Consequently, nGFP can be used to further ensure clonal labelling in lineage tracing experiments. By administrating 1μg/g of mouse of Doxycycline to pregnant mice at E13 by intravenous injection (IV), we found that about 80% of the MGs were not labelled by nGFP (Fig. 1c, d). Similar proportions of MGs (20%) were labelled with nGFP two days after the injection (E15) and at postnatal day 5 (P5) when MG has branched and basal and luminal lineages are clearly separated (Fig. 1d-f). At E15, MG contained one to three nGFP+ cells spatially close to each other (Fig. 1g-h), consistent with the clonal expression of nGFP. Interestingly, at P5, almost all nGFP clones contained both BCs and LCs, even though BCs and LCs could be relatively distant to each other (Fig. 1i-l). These data clearly show that the MG initially develops through multipotent progenitors.
Figure 1. Clonal analysis demonstrates the switch from multipotency to unipotency during MG development.
a, Confocal imaging of immunostaining of K14 in embryonic MG (E13) (8 embryos). b, Genetic strategy used to target Confetti expression in K14-expressing cells. c, Protocol used to study the fate of cells targeted during embryogenesis. d, Graph representing the fraction of glands containing nGFP+ cells at E15 (n= 73 from 8 embryos) and P5 (n=85 from 13 mice). e-f, Confocal imaging of immunostaining of K14 in P5 postnatal MG at low magnification (e) and of K14 and K8 in P5 postnatal MG (f) (85 glands from 13 mice). g-h, Confocal imaging of immunostaining of K14 and nGFP in E15 K14rtTA/TetO-Cre/Rosa-Confetti embryo induced at E13 with 1μg/g of DOX shows clonal induction in mammary buds (arrow) (g) and zoom onto the labelled cell (h) (73 glands from 8 embryos). i-k, Confocal imaging of immunostaining of K14, nGFP and/or K8 in P5 K14rtTA/TetO-Cre/Rosa-Confetti glands induced at E13 with 1μg/g of DOX shows the presence of isolated BCs (i), isolated LCs (j) and adjacent BCs and LCs (k) (85 glands from 13 mice). l, Graph representing the frequency of nGFP clone composition observed in P5 K14rtTA/TetO-Cre/Rosa-Confetti mice induced at E13 with 1μg/g of DOX (n=85 gland from 13 mice). m, Confocal imaging of immunostaining of K14 and K5 in P5 MG (3 mice). n, Genetic strategy used to target Confetti expression in K5-expressing cells. o, Protocol used to study the fate of cells targeted at birth. p, Confocal imaging of immunostaining of K14 and nGFP in P21 K5CreER/Rosa-Confetti mice induced at birth with 50μg of TAM (12 glands from 3 mice). q, Graph representing the frequency of clone composition observed in P21 K5CreER/Rosa-Confetti mice induced at birth with 50μg of TAM (n=36 clones from 12 glands from 3 mice). g, i, j, k, p represent orthogonal projections of 3D stacks. Scale bars, 10 μm, except E : 500μm. Arrowheads represent labelled nGFP cells at E15 (g) and labelled BCs at P21 (p). See Supplementary Table1 for source data related to d, l and q.
It has previously been shown that pubertal development and adult MG homeostasis are mediated by distinct lineage restricted stem cells2–12. However, it is still unclear when the lineage restriction between BCs and LCs occurs. At P1, keratin 5 (K5) was specifically expressed in the outer layer of the MG (Fig. 1m). Clonal lineage tracing of BCs using K5CreERT2/Rosa-Confetti mice at P1 lead to the exclusive labelling of BCs (Fig. 1n-q), showing that at P1 all BCs are already unipotent.
Hybrid adult basal and luminal gene expression in EMPs
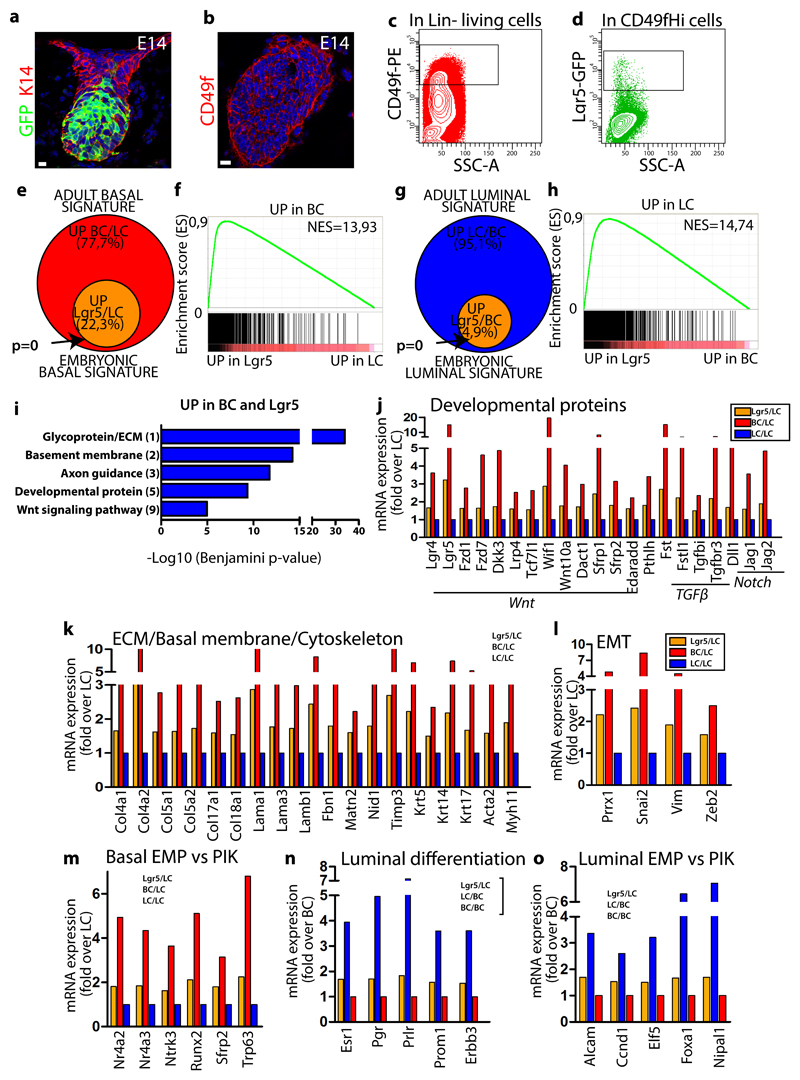
To understand the molecular mechanisms regulating embryonic multipotency, we developed a strategy to FACS isolate EMPs at E14, enabling to compare their transcriptome to adult BCs and LCs. To this end, we used Lgr5-GFP reporter mice15 to specifically isolate EMPs (CD49f Hi/Lgr5-GFP Hi) from the underlying mesenchyme and surrounding epidermis16 and compared to their transcriptome to adult BCs (CD24+CD29Hi) and LCs (CD24+CD29Lo)17(Fig. 2a-d and Supplementary Figure 1). To determine to which extent EMPs resemble to adult BCs, we compared the transcriptome of EMPs with LCs and defined the genes upregulated by 1.5 fold in EMPs (embryonic basal signature) and assessed which genes upregulated in this signature were also upregulated in adult BCs (when compared to LCs) by more than 1.5 fold (adult basal signature). A high proportion of the genes of the adult basal signature were also upregulated in EMPs as compared to LCs (22,3%) (Fig. 2e). Lgr5 isolated EMPs expressed a number of genes previously shown to be enriched in laser capture embryonic MG18. Gene set enrichment analysis (GSEA) confirmed the high enrichment of adult basal genes in EMPs (Fig. 2f). Interestingly, when EMPs were compared to adult BCs (embryonic luminal signature) and assessed for the expression of markers of the adult luminal signature (genes upregulated by 1.5 in adult LCs when compared to BCs), EMPs also expressed luminal specific genes (Fig. 2g). GSEA confirmed the enrichment of adult luminal genes in EMPs (Fig. 2h). These data demonstrate that, at the population level at E14, EMPs express genes of the basal and the luminal lineages.
Figure 2. Transcriptional profiling of EMPs reveals their compound basal and luminal gene signature.
a-b, Confocal imaging of immunostaining of K14 and GFP in Lgr5-IRES-GFP embryo at E14 (a) and of CD49f in wild type E14 embryo (b) (5 embryos analysed). c-d, FACS analysis of CD49f and GFP expression in Lin- epithelial cells (c) and Lin-CD49fHi mammary cells (d) in E14 Lgr5-GFP embryos (5 embryos analysed). e, Venn diagram showing the overlap between the genes upregulated by 1.5 fold in BCs compared to LCs (adult basal signature) and in Lgr5 cells compared to LCs (embryonic basal signature). f, GSEA of the upregulated genes in BCs (vs LCs) with the genes upregulated in Lgr5 cells (vs LCs), showing the enrichment of the basal signature in Lgr5 cells. g, Venn diagram showing the overlap between the genes upregulated in LCs compared to BCs (adult luminal signature) and in Lgr5 cells compared to BCs (embryonic luminal signature). h, GSEA of the upregulated genes in LCs (vs BCs) with the genes upregulated in Lgr5 cells (vs BCs), showing the enrichment of the LC signature in Lgr5 cells. i, Gene ontology (GO) analysis of genes upregulated >1.5-fold in both BCs and Lgr5 cells compared to LCs. Histograms represent –log10 of Benjamini score. j-m, Graph representing mRNA expression measured by microarray analysis of upregulated genes in FACS-isolated BCs and Lgr5 cells (fold over LC), showing the transcriptional priming of BC genes in Lgr5 cells. n-o, Graph representing mRNA expression measured by microarray analysis of upregulated genes in FACS-isolated LCs and Lgr5 cells (fold over BC), showing the transcriptional priming of LC genes in Lgr5 cells. Analysis presented in e-o are derived from the fold change ratio of the mean of Lgr5 microarray data (n=3) over the mean of BC (n=2) or LC (n=2) microarray data. Enrichment P-value in e, g derived from hypergeometric test performed with R software without adjustment (n= 2958 and 3999 genes respectively for BC/LC and LC/BC signatures). Scale bars, 10 μm.
Gene ontology (GO) analysis of the genes belonging to the basal signature and expressed in EMPs revealed a high enrichment for developmental proteins; Wnt, Edar Pth, TGFβ, and Notch pathways (Fig. 2i,j and Supplementary Fig. 2a), which are well known to regulate MG morphogenesis and adult maintenance3, 5, 19–27. Extracellular matrix (ECM) genes were also strongly enriched (Fig. 2i,k and Supplementary Fig. 2a), suggesting that these progenitors contribute to the formation of their own niche. In addition, EMPs were enriched in developmental proteins, including Trp63, a key transcription factor (TF) known to be essential for MG embryonic development and expressed in adult BCs28–30, and epithelial to mesenchymal transition (EMT) genes (Fig. 2l), a feature associated with embryonic MG31 and adult BCs32. EMPs also expressed cell adhesion and cytoskeleton molecules including basal keratins, members of the planar cell polarity pathway, growth factors binding and axon guidance molecules (Fig. 2i, k and Supplementary Fig. 2a,b), which may regulate the growth and the branching of MG. Oncogenic Pik3ca expression in adult BCs or LCs promotes the activation of a multipotent program17. Interestingly, EMP basal signature encompassed a high number of genes (31,2%, 124 out of 398 genes) previously associated with oncogenic Pik3ca-induced multipotency signature (Fig. 2m-o) 17.
GO analysis of the genes of the adult luminal signature expressed in the EMPs revealed that EMPs were highly enriched for genes regulating cell cycle and mitosis (Supplementary Fig. 2c-e), and the well-known key regulators promoting MG luminal lineage specification and maintenance 18, 33–38, together with known ER+ and ER- luminal markers (Fig. 2n) that characterize luminal lineages9, 39. A significant proportion of the EMP luminal signature was common with the oncogenic Pik3ca-induced multipotent gene signature (14.3%, 20 out of 140 genes) (Fig. 2o).
Single cell RNA-seq uncovers the hybrid EMP signature
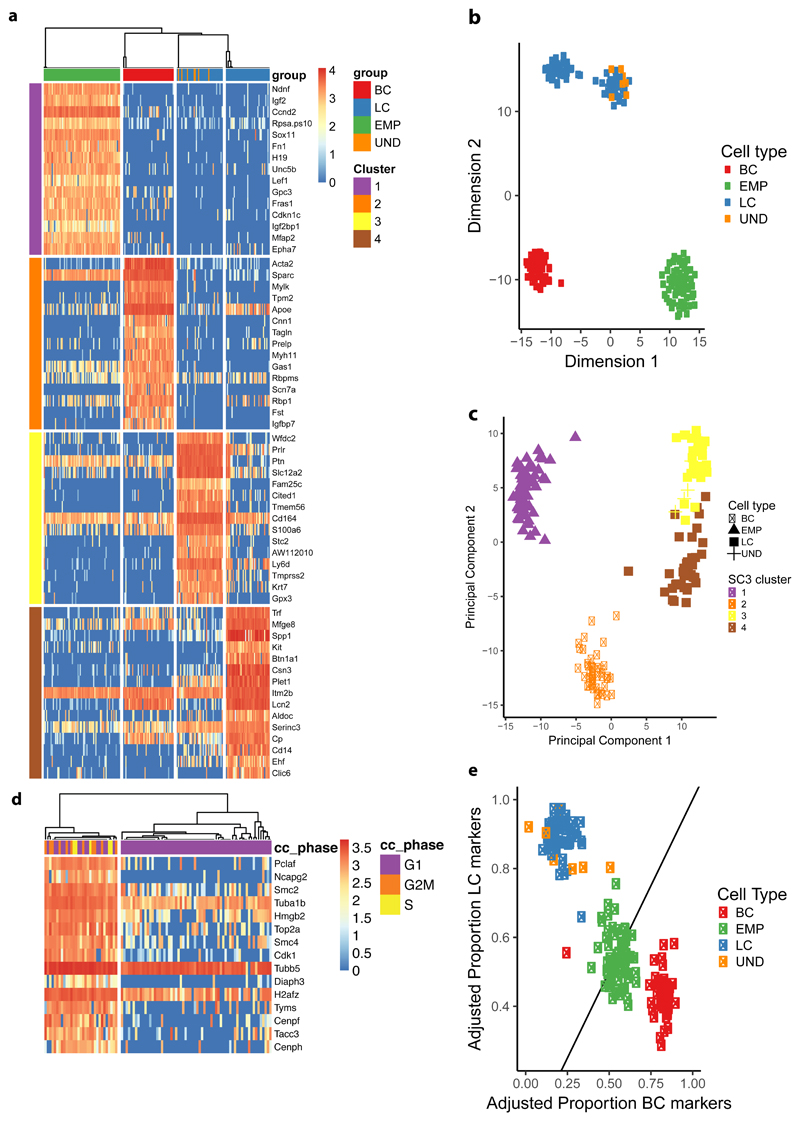
To further define the molecular features associated with the multipotency of EMPs at the early stages of MG morphogenesis, we performed single cell RNA-seq (sc RNA-seq) following a SMARTseq2-based approach on FACS-isolated cells. After a very stringent quality control, 69 single EMPs at E14, 51 adult BCs and 73 adult LCs were retained for downstream analyses. Unsupervised clustering analysis using the SC3 method40 showed that these cells could be individualized into four main clusters specific for the EMPs, adult BCs and both Esr1+ and Esr1- LCs (Fig. 3a and Supplementary Fig. 3). EMPs were specifically enriched for genes that regulate proliferation, signalling, and previously reported to be expressed in breast cancers41–47, whereas BCs and LCs expressed classical basal and luminal genes respectively. Gene ontology analysis of the single cell signature of EMPs, similarly to the microarray analysis, showed high enrichment for developmental proteins, differentiation, transcription, axon guidance, Wnt signalling and ECM genes (Supplementary Fig. 3). EMPs at E14 did not express Sox10, contrasting with its expression in late embryonic development (E18.5) and adult MG 31, 38 (Supplementary Fig. 3d). The four main clusters found by SC3 were also found by dimensionality reduction using T-SNE and PCA (Fig. 3b, c), demonstrating the robustness of these clusters. The first component in the PCA reflected 21% of total variance and could be attributed to the difference between the embryonic and adult cell types. Consistent to what we found in microarray analysis, BCs more closely resemble EMPs along PC1. The second component of the PCA constitutes 12% of total variance and represents transcriptional differences between adult LCs and BCs. The third (5% of total variance) and fourth (2% of total variance) components represent variance attributed to the Esr1+ and Esr1- status of the LCs.
Figure 3. Single cell RNA-seq shows the EMP hybrid gene signature.
a, Unsupervised clustering using SC3 on EMPs (n=68), adult BC (n=45) and LCs (n=73) using clustering parameter k=4. Heatmaps of the top 15 marker genes for each cluster and their corresponding normalized expression are displayed (AUC > 0.8 and Wilcoxon signed rank test FDR adjusted p-value < 0.01). Columns represent single cells, colour-coded by their respective lineage. UND (undetermined significance) represents few FACS isolated CD29HiCD24+ cells with LC gene signature, which probably represent errors during cell sorting. b-c, Dimensionality reduction using t-Distributed Stochastic Neighbor Embedding (b) and Principal Component Analysis (c), every dot (n=193) represents one cell with the colour representing either cell-type or the assigned SC3 cluster represented in (a) respectively. d, SC3 clustering of EMPs (n=68) using clustering parameter k=2. Heatmap of top 15 marker genes for each cluster and their corresponding normalized expression are displayed (AUC > 0.9 and Wilcoxon signed rank test FDR adjusted p-value < 0.01). Columns represent single cells, colour-coded by their assigned cell-cycle phase. e, Scatter plot with the X-axis representing the adjusted proportion of BC-specific marker genes detected by SC3 (n=53) and the Y-axis LC-specific marker genes (n=47). Marker genes were selected to be expressed in at least 75% of the respective cell type and in less than 50% of the opposite cell type. The proportion of expressed markers is computed as the fraction of markers with > 0 expression over the total number of markers. Every dot (n=193) represents one cell and are colour-coded according to cell type.
We then assessed whether EMPs constitute a transcriptionally homogeneous population expressing a hybrid LC/BC gene signature or are composed of distinct pre-committed subpopulations of cells with distinct gene signatures corresponding to BC or LC progenitors. Only scRNA-seq can discriminate between these two scenarios. We performed SC3 unsupervised clustering on the EMPs only and found two subclusters (Fig. 3d). One subcluster comprises the majority of EMPs, expressing no significant cluster-specific marker genes. The second subcluster represents a proliferative subpopulation of EMPs expressing marker genes associated with cell cycle. To avoid aberrant clustering, these cycling cells were omitted from further analysis. Very interestingly, only EMPs and not adult LCs or BCs, expressed a hybrid transcriptional signature comprising markers for both LC and BC lineages (Fig. 3e, Supplementary Fig. 4). Lowering the stringency of quality controls of scRNA-seq pipeline such as the number expressed genes per cell may result in the appearance of few adult LC and BCs with artefactual hybrid gene expression profile (Supplementary Fig. 5).
We also found that few Lgr5 expressing cells at E14 were clustering in a separate mesenchymal cluster distinct from all the other EMPs. Some of these cells did not express any epithelial markers including K14, K5 or K8 and correspond to stromal cells of the embryonic mammary gland, which were not eliminated by the α6-integrin/CD49f FACS gating strategy that separates embryonic mammary epithelial from mesenchymal Lgr5-expressing cells. However, few epithelial cells, as demonstrated by their keratin expression, were also expressing a higher level of the mesenchymal genes, and thus corresponding to an EMT hybrid state (Supplementary Fig. 6a).
By looking at the proportion of exclusively expressed lineage-specific markers in EMPs, we observed no clear subpopulations of EMPs expressing a higher proportion of either sets of genes (Fig. 3e, Supplementary Fig. 6), suggesting that EMPs were not committed to basal or luminal lineage at E14.
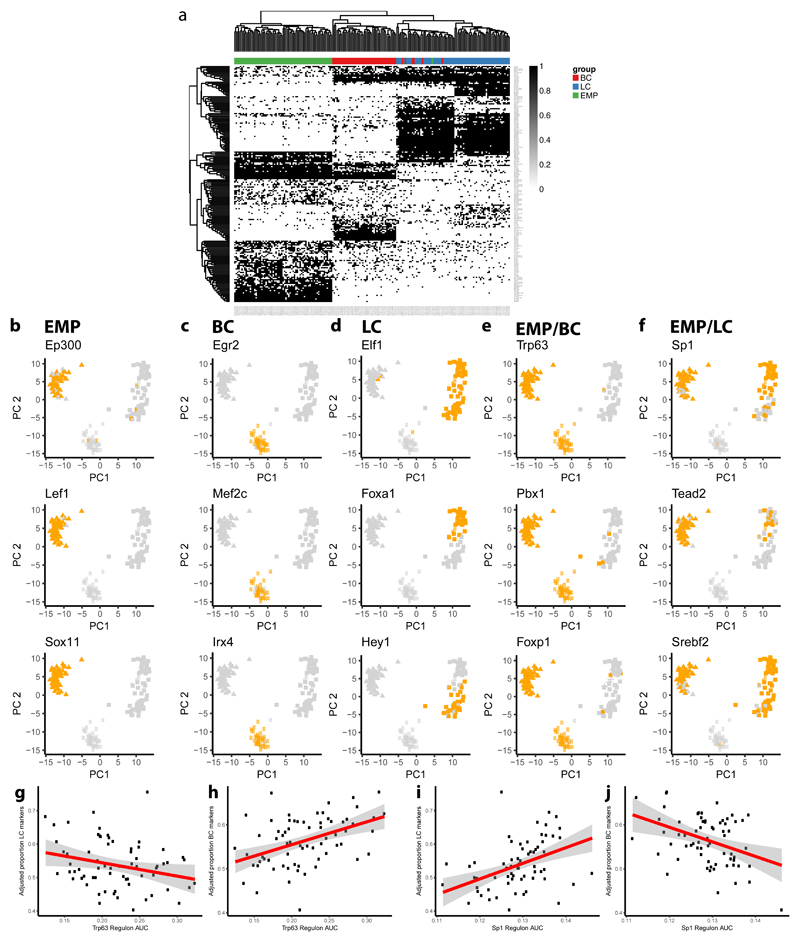
To further assess the regulatory mechanisms associated with the different cell states, we performed gene regulatory network analysis using SCENIC48, which allows identifying regulatory modules by inferring co-expression between TFs and genes containing the respective TF binding motif in their regulatory regions. These modules or regulons are then assessed in each cell to ascertain their activity and infer cell specific states. By hierarchical clustering cells according to the binary activity of regulons, we recapitulated the clusters previously found by SC3. We identified regulons active only in EMPs, only in LCs, only in BCs, in both EMPs and LCs and in EMPs and BCs (Fig. 4a-f). We then investigated the correlation between the degree of regulon activity, by the number of their expressed target genes, and basal or luminal marker expression. Interestingly, in EMP population we identified TFs such as p63 for BC or Sp1 for LC, which showed opposite correlations with regards to LC and BC marker expression, suggesting their role as cell lineage regulators (Fig. 4g-j).
Figure 4. SCENIC analysis of EMP, LC and BC scRNA-seq data.
a, Binary activity matrix for regulons inferred by SCENIC: Regulons were determined to be active (black) if they exceeded a manually adjusted AUC regulon-specific threshold or inactive under this threshold (white). Columns represent cells (n=193) colour-coded by cell type, rows represent regulons. Hierarchical clustering is performed and clusters of regulons can be observed specific for each cell population but also shared between the different cell populations. Only regulons with an absolute correlation with any other regulon > 0.3 and at least active in 1% of cells are shown. b-f, PCA plots showing the binary activity of regulons inferred by SCENIC: PCA was performed on scRNAseq data on the normalized expression values of top 500 most variable genes. Every dot (n=193) represents a single cell whereas the activity of the respective regulon is colour-coded as active (orange) or inactive (light grey) for each cell. Examples of regulons are grouped by their respective populations: only active in EMPs (b), only active in BCs (c), only active in LCs (d), active in both EMPs and BCs but not active in LCs (e), active in both EMPs and BCs but not in LCs (f). g-j, Scatter plots depicting the linear relationship in the EMP population (n=68) between regulon activity and the adjusted proportion of specific LC (g, i) and BC markers (h, j) for Trp63 (g, h) and Sp1 regulons (i, j). Regulon activity is measured as the regulon AUC which is a function of the number of inferred target genes of that regulon being expressed (0 meaning no genes, and 1 meaning all genes being expressed). The red line represents a linear model fitted using the lm function in R, the grey area represents the 95% confidence interval.
Asymmetrical expression of basal and luminal markers marks the early step of lineage segregation
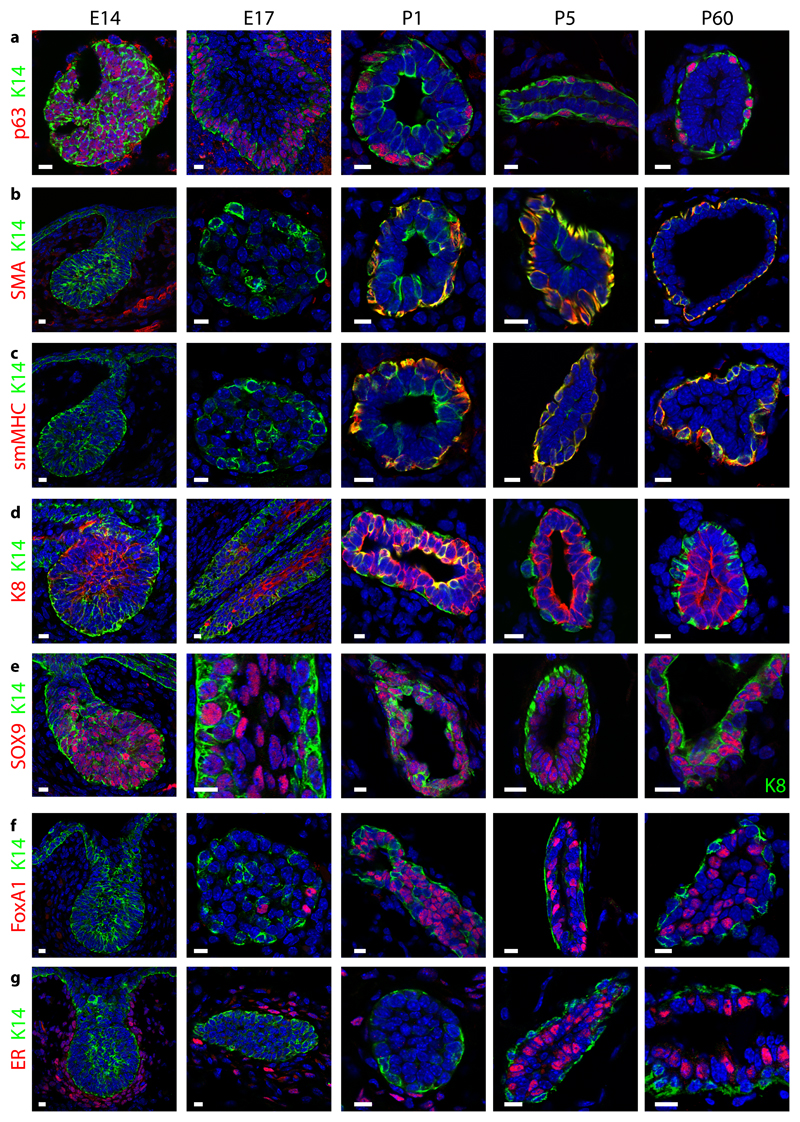
To get further insight into the mechanisms that regulate cellular heterogeneity and lineage segregation during MG morphogenesis, we assessed by immunofluorescence the temporal expression of basal and luminal markers differentially regulated at the bulk mRNA level in EMPs, adult BCs and LC. At E14, basal (K14 and p63) and luminal (K8 and Sox9) markers were expressed in all cells of the MG, although lower levels of K8 were already visible along the outer region of the embryonic MG (Fig. 5a-e). At E17, the cells of the outer layer of the embryonic MG expressed p63 and lower levels of K8, whereas the inner cells of the embryonic MG expressed higher levels of K8 and no longer p63 (Fig. 5a, d). Sox9, which is expressed by a fraction of adult LCs9, 10, 49 was still expressed by BCs and LCs at this stage of embryonic development (Fig. 5e). Foxa1 and ER were only observed at the protein level in a fraction of LCs during late embryogenesis for Foxa1 and in the postnatal MG for ER (Fig. 5f, g). Basal markers SMA and smMHC (Fig. 5b,c), which were detected at E14 at the mRNA level, were only detected at the protein level at P1 in outer BCs.
Figure 5. Asymmetrical expression of basal and luminal markers marks the early step of MG lineage segregation.
Confocal imaging of immunostaining of K14 and p63 (a), SMA (b), smMHC (c), K8 (d), Sox9 (e), FoxA1 (f) and ER (g) in E14, E17, P1, P5 and P60 MG shows the temporality of cellular heterogeneity during MG development (except in e where the MG at P60 is represented with K8 staining). Representative images from 3 independent mice analysed per time point. Scale bars, 10 μm.
p63 promotes unipotent BC fate in EMPs
To further dissect the molecular mechanisms associated with MG lineage segregation, we assessed whether p63 controls the switch from multipotency to BC unipotency during MG development. p63 deletion during embryonic development leads to the absence of MG formation28, 29, demonstrating the essential early function of p63 during MG specification but preventing to study the role of p63 in MG lineage segregation. Overexpression or ShRNA knock down of ΔNp63 in primary culture of mouse mammary epithelial cells (MMEC) in vitro showed that ΔNp63 decreases the expression of LC markers and increases BC marker expression, suggesting that ΔNp63 regulates directly or indirectly BC characteristics. Furthermore, transplantation of MMEC overexpressing ΔNp63 reduced the proportion of LCs in the MG outgrowth, suggesting that downregulation of ΔNp63 is required for proper luminal lineage differentiation50. However, it remains unclear whether the spatial restriction of ΔNp63 expression during MG embryonic development is important to promote BC fate.
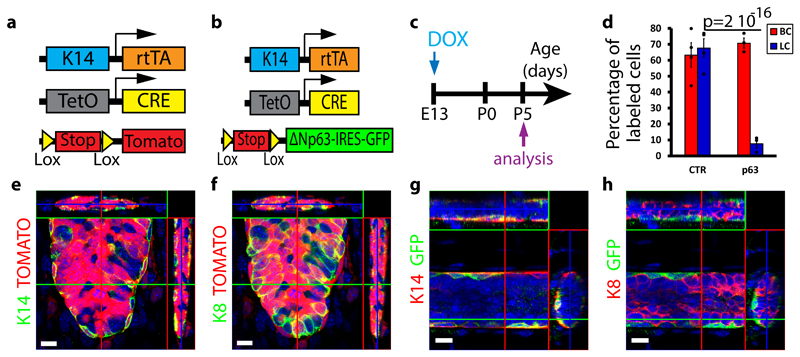
To address this question, we used a genetic approach allowing sustaining p63 expression in the outer and inner cells of the embryonic MGs51. Dox administration to K14rtTA/TetO-Cre/Rosa-tdTomato at E13 led to the labelling of the same proportion of BCs (Tomato+/K14+) and LCs (Tomato+/K8+) (70%) at P5. In contrast, Dox administration to K14rtTA/TetO-Cre/Rosa-ΔNp63-IRES-GFP mice led to the labelling of 70% of BCs (GFP+/K14+) and only 10% of LCs (Fig. 6a-h), demonstrating that most LCs arise from the rare EMPs that did not express the p63 transgene.
Figure 6. p63 promotes unipotent BC fate in EMPs.
a-b, Scheme summarizing the genetic strategy used to target tdTomato (a) or ΔNp63-IRES-GFP (b) expression in K14-expressing cells at E13. c, Scheme summarizing the protocol used to study the fate of cells targeted during embryogenesis using K14rtTA/TetO-Cre/Rosa-tdTomato or Rosa-ΔNp63-IRES-GFP mice. d, Graph representing the mean percentage of labelled BCs and LCs in control versus p63-overexpressing mice. Respectively n=4 and n=3 independent mice were analysed in K14rtTA/TetO-Cre/Rosa-tdTomato and in Rosa-ΔNp63-IRES-GFP mice. Individual data points are represented as dots. Error bars, s.e.m. P-value derived from Fisher exact test without adjustment. See Supplementary Table 1 for source data related to d. e-f, Confocal imaging of immunostaining of K14 (e) or K8 (f) and Tomato in K14rtTA/TetO-Cre/Rosa-tdTomato mice induced at E13 with 15μg/g of DOX. Representative images from 4 mice analysed. g-h, Confocal imaging of immunostaining of K14 (g) or K8 (h) and GFP in K14rtTA/TetO-Cre/Rosa-ΔNp63-IRES-GFP mice induced at E13 with 15μg/g of DOX. Representative images from 3 mice analysed. e-h represent orthogonal projections of 3D stacks. Scale bars, 10 μm.
Reprogramming of adult LC into BCs by ΔNp63
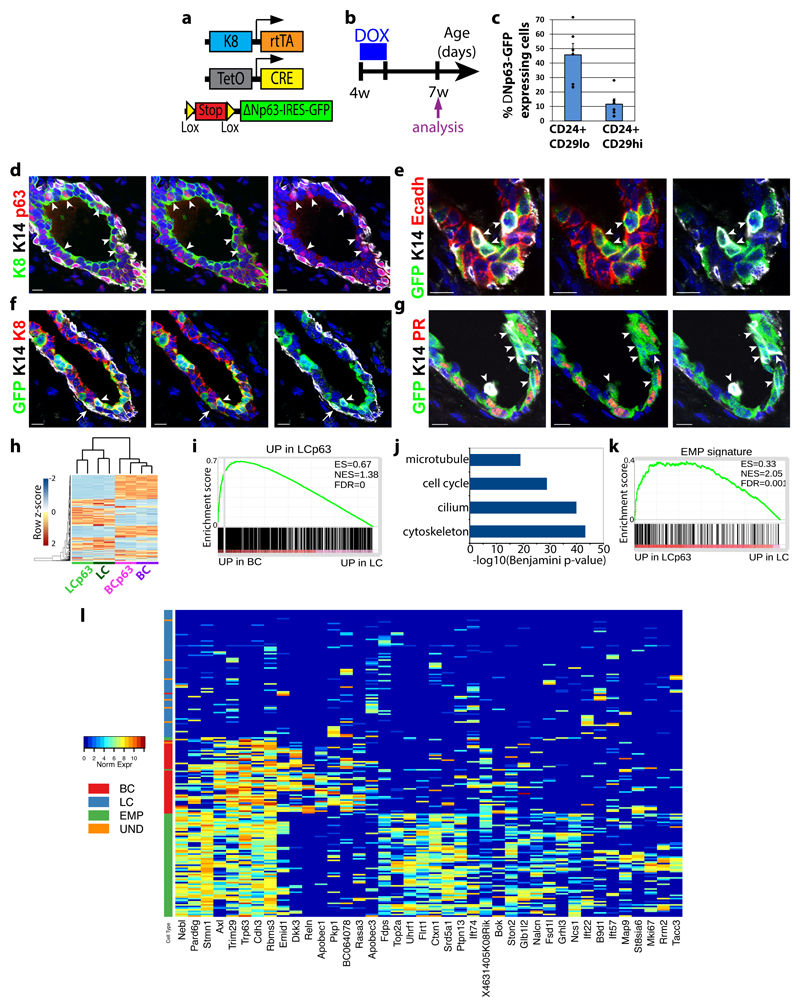
We next examined whether ΔNp63 could reprogram adult LC into BC lineage, and if so by which molecular mechanisms. To express ΔNp63 in adult LCs, we administered Dox for 7 consecutive days to 4 weeks old K8rtTA/TetO-Cre/Rosa-ΔNp63-IRES-GFP females (Fig. 7a,b), and analysed the presence of GFP in BC and LC by FACS 2 weeks after the last Dox administration. Interestingly, we found that ΔNp63 expression in adult LCs was sufficient to reprogram a fraction of ΔNp63 expressing LCs into BCs (Fig. 7c), demonstrating the important plasticity of LCs and the master regulatory function of ΔNp63 to promote BC fate in vivo.
Figure 7. In vivo reprogramming of adult LC into BC by p63.
a, Genetic strategy to target ΔNp63-IRES-GFP expression in LCs. b, Protocol used to study the fate of cells targeted using K8rtTA/TetO-Cre/Rosa-ΔNp63-IRES-GFP mice. c, Percentage of GFP-labelled LCs (CD24+ CD29lo) and newly formed BCs (CD24+ CD29Hi) following expression of ΔNp63-IRES-GFP in adult LCs (n=6 mice) (mean + sem). Dots, individual data points. See Supplementary Table 1 for source data.
d-g, Immunofluorescence of p63, K14 and K8 (d), GFP, K14 and Ecadh (e), GFP, K14 and K8 (f) and GFP, K14 and PR (g) 2 weeks following p63-IRES-GFP expression in LCs. Representative images from 6 mice. h, Heatmap representing the similarities between the different BC (CD24+ CD29Hi) and LC populations (CD24+ CD29lo) populations (n=2 RNAseq datafor each population). The top 500 most variable genes across the 8 samples are plotted in the heatmap. The dendrogram shows hierarchical gene expression clustering of BCs and LCs with or without p63 overexpression. Blue and red correspond to low and high expressed genes, respectively. The two major branches of the tree perfectly discriminate between LCs and BCs and between WT and ΔNp63 cells. i, GSEA of the genes upregulated by p63 in LCs (vs WT LCs) with upregulated genes in BCs (vs LCs), showing the enrichment of basal genes in p63 upregulated genes in LCs. j, Bar chart of Benjamini corrected enrichment p-value of the first four functional annotation clusters for the 902 genes overexpressed in ΔNp63 expressing LC compared to WT LCs. k, GSEA of the upregulated genes in EMPs (vs LCs) with the genes upregulated by p63 in LCs (vs WT LCs), showing the enrichment of the EMP signature in p63 upregulated genes in LCs. l, Heatmap representing genes overexpressed in ΔNp63 expressing LC bulk RNA-seq data. The top 200 overexpressed genes are chosen and the 40 genes with the highest variance amongst the single cells are plotted in the heatmap. Columns represent single cells colour-coded by cell type. Colours in the heatmap represent normalized expression values. RNA-seq analysis in i-k are derived from the means of two RNAseq datasets per condition.
To gain further insights into the cellular mechanisms that accompanied LC fate transition upon ectopic p63 expression, we performed immunofluorescence characterisation of the MG during the cellular reprogramming. In good accordance with the FACS data, p63 immunostaining showed that p63 was expressed in about half of the LCs (Fig 7d). Interestingly, we found that some LCs expressing p63-IRES-GFP co-expressed basal and luminal markers (Fig 7e-g and Supplementary Fig 7). Most of these hybrid cells were still located in the inner cells together with the other LCs, and still presented the round shape of LCs (Fig 7e-g). More rarely at this stage, some of these p63 targeted LCs were located in the outer part of the mammary epithelium, lost the expression of LC markers, expressed BC markers and presented BC elongated morphology (Fig. 7f). Although p63-IRES-GFP was initially evenly expressed in ER/PR+ and negative LCs, the LCs that co-expressed basal and luminal markers did not express Foxa1 or PR (Fig 7g and Supplementary Fig. 7d), suggesting that LCs need to shut down their hormone receptor differentiation program to undergo cell fate reprograming upon ΔNp63 expression.
To understand the molecular mechanisms by which ΔNp63 reprograms adult LCs into BCs, we performed bulk RNA-seq of FACS isolated LCs and BCs upon ΔNp63 expression in adult MGs. Gene clustering demonstrated that the newly generated basal like cells upon ΔNp63 expression in LCs resemble molecularly to WT BCs (Fig. 7h). Venn diagram and GSEA also showed that a very significant fraction of the LC upregulated genes upon ΔNp63 expression belongs to the BCs signature and among the 48 p63 inferred target genes by SCENIC, 18 were overexpressed in LCs expressing ΔNp63 (p=1.349e-06 given a random sampling of genes, Fig. 7i and Supplementary Fig. 7), supporting the notion that ΔNp63 expression in LC induced a transient hybrid multipotent state before the de novo generation of BC. Gene ontology analysis showed that the genes upregulated by ΔNp63 in LCs are highly enriched for cytoskeleton, cilium biogenesis and cell division genes (Fig. 67). Interestingly, scRNAseq EMP signature was also enriched in adult LCs expressing ΔNp63 (Fig. 7k). A subset of the most upregulated genes in adult LCs expressing ΔNp63 versus WT LCs were expressed in the single-cell EMP data but not in BCs, further suggesting that the reprograming of adult LC following ΔNp63 expression lead to a multipotent embryonic like hybrid state before giving rise to fully reprogrammed BCs (Fig. 7l).
Discussion
Here, using clonal analysis during MG development, we formally demonstrate that MG initially develops from EMPs that rapidly switch from multipotency to unipotency during the course of embryonic development. Similar conclusions were made from lineage tracing experiments using Notch1-CreER at different stages of embryogenesis in a related study by Fre and colleagues52. This rapid switch from multipotency to unipotency may explain the very rare and large bipotent clones dispersed from the nipple region to the distal part of the epithelial tree found in lineage tracing induced by random recombination12.
Microarray and scRNA-seq analyses indicate that EMPs are associated with a hybrid signature that overlaps with both basal and luminal lineages. The greater resemblance of EMPs with BCs may explain why BCs are multipotent in transplantation assays4, 7, 8. The hybrid gene expression of EMPs at E14 sharing similarities with both BCs and LCs is consistent with their multipotent fate at this stage of embryonic development.
Reactivation of multipotency is associated with the early stages of mammary tumour initiation and the development of tumour heterogeneity. Our data showing that EMPs express the same genes as expressed during the reactivation of multipotency and cell fate changes induced by oncogenic Pik3ca expression 17 support the notion that the mechanisms associated with multipotency during tumorigenesis recapitulate at least partially the genetic program that regulates multipotency during embryonic MG development. Moreover many genes found to be specifically expressed in EMPs by scRNA-seq such as Sox11, Stmn1 and Mdk are expressed in human breast cancers with poor prognosis41–46, further suggesting that the reactivation of an EMP gene expression program of during tumorigenesis is essential for tumour growth and invasion.
Temporal in situ characterization reveals that the early signs of lineage segregation visible at the protein level consist of a restricted expression of p63 together with a decreased expression of K8 in the outermost layer of the embryonic MG in contact with the stroma. By sustaining the expression of p63 in EMPs, we found that p63 promotes the differentiation of EMPs into BC during MG development. Similarly, p63 overexpression in adult MMEC decreases the formation of LC upon transplantation of these cells into the mammary mesenchyme50. Expression of p63 in adult LC is sufficient to reprogram these cells into BC, further demonstrating that p63 acts as a master regulator BC fate. It has been proposed that p63 and Notch signalling act in an opposite manner to promote respectively BC and LC fates50, 52. It is thus possible that the downregulation of K8 and the restricted expression of p63 at the outer layer are the consequences of an inhibition of Notch signalling in these cells.
Ectopic expression of ΔNp63 in adult LCs induces the reprogramming of these cells into basal like cells. This cell fate change is a progressive process rather than a direct trans-differentiation event that undergoes through a hybrid state, reminiscent of the EMP. Our molecular analysis identifies a specific gene program regulated by p63 sufficient to convert adult LC into a BC and that encompassed many genes expressed by EMPs, further suggesting that the cell fate reprogramming of LC into a BC by p63 reactivate some features of the embryonic multipotent genetic program.
Although our study uncovers the importance of hybrid gene expression in regulating the multipotent state during embryonic development and p63 mediated reprograming of adult LC into BC and defines the BC molecular program controlled by p63 during these conditions, many important questions remain unanswered. Further studies will be needed to better understand the molecular mechanisms that regulate the switch from multipotency to unipotency occurring during MG development. What are the intrinsic and extrinsic signals that act upstream and sustain the expression of p63 and inhibit the expression of luminal genes in the inner part of the MG at the early stage of lineage segregation? What are the key p63 target genes that regulate BC fate in EMP and the reactivation of multipotency in adult unipotent luminal SC? What the role of the other genes beside p63 that are specifically expressed by EMPs and regulate multipotency during development? As p63 is also expressed in the basal lineage of other epithelial tissues53, 54, the key role of p63 in regulating the switch from multipotency to unipotency is likely to be conserved across different organs. Defect in lineage segregation and regulation of multipotency by p63 may potentially explain some pathological features of the different developmental syndromes affecting these tissues including the MG that are associated with p63 mutations in humans55.
In conclusion, our study identifies the temporal switch from multipotency to unipotency that occurs during MG development and the molecular mechanisms that regulate cell fate transition and lineage segregation during this process. The paradigm and molecular mechanisms uncovered here have important implications for the understanding of multipotency, unipotency and lineage segregation in other tissues and during tumorigenesis.
Material and Methods
Mice
Lgr5-EGFP-IRES-CreER15 and Rosa-tdTomato56 mice were obtained from the Jackson Laboratory. Rosa-Confetti13 mice were provided by H. Clevers; K14rtTA transgenic mice57 were provided by Elaine Fuchs; TetO-Cre mice58 were provided by Andreas Nagy; Rosa26-ΔNp63-IRES-GFP mice51 were provided by Wim Declercq. The generation of K5CreER and of K8rtTA were previously described8, 10. Mice colonies were maintained in a certified animal facility in accordance with European guidelines. The experiments were approved by the local ethical committee (CEBEA) under protocols #477 and #527. The study is compliant with all relevant ethical regulations regarding animal research. Mice were analysed at embryonic stages E14, E15 and E17, during postnatal development at P1, P5, P21, P60, 7w and in adult mice (over 8w), as indicated in figure legends.
Targeting Confetti, tdTomato or ΔNp63-IRES-GFP expression in the MG
For clonal lineage tracing, K14rtTA/TetO-Cre/Rosa-Confetti embryos were induced at E13 by intravenous (IV) injection in the tail vein of the pregnant mother with 1μg/g of Doxycycline (diluted in sterile PBS, Sigma cat#D9891) and at E15 or P5. K5CreER/Rosa-Confetti pups at P1 were induced by intraperitoneal (IP) injection of 50μg of Tamoxifen (diluted in sunflower seed oil, Sigma cat#T5648) and killed 21 days later. For K14rtTA/TetO-Cre/Rosa-tdTomato or K14rtTA/TetO-Cre/Rosa-ΔNp63-IRES-GFP embryos were induced at E13 by IV injection of the pregnant mother with 15μg/g of Doxycycline (diluted in sterile PBS) and killed at P5. K8rtTA/TetO-Cre/Rosa-ΔNp63-IRES-GFP female mice were induced at 4w with Doxycyclin 10g/kg in diet (Bio-Serv), 2g/l in drinking water (AG Scientific cat#D2545) and 3 intraperitoneal injections of 2 mg in 200μl PBS during 7 days.
MG whole-mount processing and immunostaining
For MG processing at E15, whole embryos were collected and fixed in PFA 4% for 2h at room temperature (RT) or overnight (O/N) at 4°C. The following day, the whole skin (of female embryos) containing mammary buds was dissected and stained as detailed below. For MG processing at P5, thoracic and inguinal MGs were dissected and fixed in PFA 4% for 2h at RT or O/N at 4°C, then stained. For MG processing at P21, inguinal glands were dissected and enzymatically digested in HBSS (GIBCO) + 300U/ml collagenase (Sigma cat#C0130) + 300μg/ml hyaluronidase (Sigma cat#4272) for 20min at 37°C under shaking. Glands were fixed in PFA 4% for 2h at RT. Confetti samples were washed in ammonium chloride (NH4Cl 0,5M in PBS) for 20min, followed by washes in PBS. For WM immunostaining, all samples were incubated in blocking buffer for 3h (bovine serum albumin (BSA) 1%, horse serum (HS) 5%, TritonX 0,8% in PBS) at RT. The different primary antibodies were incubated O/N at RT and washed for 1h at RT with PBS 0,2% Tween20 before incubation with secondary antibodies diluted in blocking buffer at 1:400 for 5h at RT. The following primary antibodies were used: anti-K14 (rabbit or chicken, 1:1000, Thermo), anti-K8 (rat, 1:500, Developmental Studies Hybridoma Bank, University of Iowa), anti-GFP (chicken, 1:500, ab13970, Abcam). The following secondary antibodies were used: anti-rabbit, anti-rat, anti-chicken conjugated to AlexaFluor488 (Molecular Probes), Rhodamine Red-X or Cy5 (JacksonImmunoResearch). Nuclei were stained with a Hoechst solution (1:1000 in PBS 0,2% Tween20) for 30min and washed for another hour in PBS 0,2% Tween20 before mounting on slides in DAKO mounting medium supplemented with 2,5% Dabco (Sigma).
MG immunofluorescence on sections
Whole embryos and newborn pups collected at E13, E14, E17 and dissected MGs from P5 and adult mice were pre-fixed in PFA 4% for 2h at RT or directly embedded in OCT and kept at -80°C. Pre-fixed tissues were washed in PBS, incubated overnight in 30% sucrose in PBS at 4°C and embedded in OCT and kept at -80°C. Sections of 10μm were cut using a HM560 Microm cryostat (Mikron Instruments). Tissue sections were fixed in PFA 4% for 10 min at RT (for non pre-fixed sections only) and incubated in blocking buffer (BSA 1%, HS 5%, Triton-X 0.2% in PBS) for 1h at RT. The different primary antibodies were incubated overnight at 4°C. Sections were then rinsed in PBS and incubated with the corresponding secondary antibodies diluted at 1:400 in blocking buffer for 1h at RT. The following primary antibodies were used: anti-GFP (chicken, 1:1000, ab13970, Abcam), anti-K8 (rat, 1:1000, Troma-I, Developmental Studies Hybridoma Bank, University of Iowa), anti-K14 (rabbit or chicken, 1:1000, Thermo), anti-K5 (rabbit, 1:1000, PRB-160P-0100, Covance), anti-CD49f-PE (rat, 1:100, clone GoH3, eBiosciences), anti-p63 (rabbit, 1:500, clone EPR5701, Abcam), anti-SMA-Cy3 (mouse, 1:500, clone 1A4, Sigma), anti-smMHC (rabbit, 1:100, BT562, Biomedical Technologies), anti-Sox9 (rabbit, 1:5000, AB5535, Millipore), anti-FoxA1 (rabbit, 1:100, clone EPR10881, Abcam), anti-ER (rabbit, 1:300, sc542, Santa Cruz), anti-PR (Rabbit 1/250, MA5-14505, ThermoFisher Scientific), anti-Ecadherin (Rat, 1/500, 14-3249-82, Ebioscience). The following secondary antibodies, diluted 1:400, were used: anti-rabbit (A21206), anti-rat (A21208), anti-chicken (A11039) conjugated to AlexaFluor488 (Molecular Probes), anti-rabbit (711-295-152), anti-rat (712-295-153), anti-chicken (703-295-155) Rhodamine Red-X or anti-rabbit (711-605-152), anti-rat (712-605-153), anti-chicken (703-605-155) Cy5 (JacksonImmunoResearch). Nuclei were stained with Hoechst solution (1:2000) and slides were mounted in DAKO mounting medium supplemented with 2.5% Dabco (Sigma).
Microscope image acquisition
Confocal images were acquired at RT using a Zeiss LSM780 confocal microscope fitted on an Axiovert M200 inverted microscope equipped with a C-Apochromat (40X, NA=1.2) water immersion objective (Carl Zeiss Inc.). Optical sections 1024 x 1024 pixels, were collected sequentially for each fluorochrome. The data-sets generated were merged and displayed with the ZEN 2 software.
Quantification of % GFP labelled glands and clone composition
Whole mounts of E15 and P5 K14rtTA/TetO-Cre/Rosa-Confetti MG were analysed by confocal microscopy. For E15 embryos, 73 MGs (n=8 embryos, 1 litter) were analysed, among which 11 were positive for the presence of nGFP cells (16%). For P5 mice, 85 MGs (n=13 mice, 4 litters) were analysed, among which 22 were positive for the presence of nGFP cells (26%). At P5, clones were scored according to their keratin expression into basal (K14+), luminal (K8+), and bipotent clones (K14+/K8+). Whole mounts of P21 K5CreER/Rosa-Confetti MG were analysed by confocal microscopy. At P21, 36 unipotent basal clones were analysed (from 12 MGs, n=3 mice). See Supplementary table 1 for further details.
Quantification of % labelled cells
Whole-mounts stained for K14, K8, and GFP were analysed by confocal microscopy. 2417, 2234 and 2249 total cells from 3 different mice were quantified in K14rtTA/TetO-Cre/Rosa-ΔNp63-IRES-GFP; and 2428, 2031, 2315 and 2601 cells from 4 different mice were analysed in K14rtTA/TetO-Cre/Rosa-tdTomato. Proportion of labelled LCs in K14rtTA/TetO-Cre/Rosa-tdTomato or K14rtTA/TetO-Cre/Rosa-ΔNp63-IRES-GFP was quantified as the ratio of K8+ Tomato+ or GFP+ LCs over total K8+ LCs, whereas the proportion of labelled BCs was quantified as the ratio of K14+ Tomato+ or GFP+ BCs over K14+ total BCs.
Mammary cell preparation
E14 Lgr5-GFP embryos were collected and the whole skin containing the MGs was dissected. Tissues were digested in 300U/ml collagenase (Sigma cat#C0130) + 300μg/ml hyaluronidase (Sigma cat#4272) diluted in HBSS for 1h30 at 37°C under shaking. EDTA at a final concentration of 5mM was added for 3min, followed by two washes in 10% FBS/PBS and 2% FBS/PBS before filtration through a 40μm mesh. Adult MGs were dissected and the lymph nodes removed. Tissues were briefly washed in HBSS, and chopped in 1mm3 pieces. Chopped tissues were digested in HBSS + 300 U/ml collagenase (Sigma cat#C0130) + 300μg/ml hyaluronidase (Sigma cat#4272) for 2h at 37°C under agitation. Physical dissociation using a P1000 pipette was done every 15mins throughout the enzymatic digestion. EDTA 5mM was added for 5 minutes, followed by 0,25% Trypsin-EGTA for 1 min before filtration through a 70-μm mesh, and 2 successive washes in 2% FBS/PBS.
Cell labelling, flow cytometry and sorting
Samples were incubated in 250μl of 2% FBS/PBS with fluorochrome-conjugated primary antibodies for 30min, with shaking every 10min. Primary antibodies were washed with 2% FBS/PBS, and cells were resuspended in 2.5mg/ml DAPI (Invitrogen D1306) before analysis. The following primary antibodies were used: APC-conjugated anti-CD45 (1:100, clone 30-F11, eBiosciences), APC-conjugated anti-CD31 (1:100, clone 390, eBiosciences), APC-conjugated anti-CD140a (1:100, clone APA5, eBiosciences) and PE-conjugated anti-CD49f (1:200, clone GoH3, eBiosciences) for embryos; PECy7-conjugated anti-CD24 (1:100, clone M1/69, BD Biosciences), APC-conjugated anti-CD29 (1:100, clone eBioHMb1-1, eBiosciences), PE-conjugated anti-CD45 (1:100, clone 30-F11, eBiosciences), PE-conjugated anti-CD31 (1:100, clone MEC 13.33, BD Biosciences), PE-conjugated anti-CD140a (1:100, clone APA5, eBiosciences) for adult MGs. Data analysis and cell sorting was performed on a FACSAria sorter using the FACS DiVa software (BD Biosciences). Dead cells were excluded with DAPI; CD45+, CD31+ and CD140a+ cells were excluded (Lin+) before analysis of the GFP+ cells.
Microarray processing and analysis
Sorted CD49f Hi Lgr5-GFP Hi cells (2000 cells per sample, n=3 biological replicates) were collected directly in 45 μl of lysis buffer (20mM DTT, 10mM Tris–HCl pH 7.4, 0.5% SDS, 0.5 mg/ml proteinase K). Samples were then lysed at 65°C for 15 min and frozen. RNA isolation, amplification and microarray were performed in the Functional Genomics Core, Barcelona. cDNA synthesis, library preparation and amplification were performed as described59. Microarrays were then performed on Mouse Genome 430 PM strip Affymetrix array at IRB Functional Genomics Core (Barcelona, Spain). All the results were normalized using the RMA normalization algorithm using R-bioconductor affy package with standard parameters60, 61. Cross experiment normalization was further performed to eliminate the batch effect using non-parametric empirical Bayes frameworks for adjusting data implemented in ComBat function of the Surrogate Variable Analysis package (SVA) in R-bioconductor62. The transcriptional profiles of Lgr5 cells were normalized with the transcriptional profiles adult BCs arising from K5CreER/Rosa-YFP mice and LCs arising from adult K8CreER/Rosa-YFP (n=2 for each sample, previously described in 17). Only probes upregulated or downregulated by at least 1.5 fold were considered in the analysis. Genes up- or down-regulated were defined as having at least one probe displaying a 1.5 fold change. Venn diagrams were generated using Venny 2.0. The hypergeometric p-value associated with each comparison between two signatures (calculated using R statistical tool) corresponds to the probability to observe an intersection of a given size by chance only, knowing the number of genes tested on a microarray.
RNAseq and analysis of bulk samples
40000 LCs and 5000 BCs were isolated by FACS as described above and collected into kit lysis buffer. RNA was extracted using absolutely RNA nanoprep kit (Stratagene). RNA quality was checked using a Bioanalyzer 2100 (Agilent technologies). Indexed cDNA libraries were obtained using the Ovation Solo RNA-Seq System (NuGen) following manufacturer recommendation. The multiplexed libraries (18 pM) were loaded on flow cells and sequences were produced using a NovaSeq 6000 S2 Reagent Kit (200 cycles) from a NovaSeq 6000 System (Illumina). Approximately 19 million of paired-end reads per sample were mapped against the mouse reference genome (GRCm38/mm10) using STAR software to generate read alignments for each sample. Annotations Mus_musculus.GRCm38.87.gtf were obtained from ftp.Ensembl.org. After transcripts assembling, gene level counts were obtained using HTSeq. Fold change of mean gene expression for the duplicates were used to calculate the level of differential gene expression. Heatmap of the 500 most variable genes across the 8 samples and corresponding clustering dendrogram were drawn with heatmap.2 function of the R package gplots. Euclidian distance with complete linkage agglomeration method was used for clustering.
GSEA analysis
For Fig 2, GSEA analysis was performed using ranked fold change of probe expression values between Lgr5 and BCs or LCs and genes upregulated in LCs or BCs for the displayed dataset63. For Fig.6, GSEA analysis were performed using ranked fold change of gene expression values between BCs and LCs or between LCΔNp63 and LCs for genes expressed with at least 10 reads per 20 millions of aligned reads in RNAseq counts and genes upregulated in LCΔNp63 compared to LCs or EMPs signature from single cell.
Gene ontology analysis of the multipotent signatures of the Venn diagrams
Genes up-regulated in each subset of the Venn Diagrams were tested for enrichment in each Gene Ontology class using the DAVID web server64, 65. Statistically significant enrichments correspond to those presenting a corrected P-value (e.g. Bonferoni or Benjamini) smaller or equal to 0.05 although some genes involved in non-statistically significant GO classes were plotted for their biological relevance.
scRNA-seq
scRNA-seq were generated using a modified Smartseq-2 protocol66. 1μL lysis buffer in 384 well PCR plates for cell sorting was prepared with 0.4% v/v Triton-X lysis buffer, 2.5mM dNTPs, 2.5μM oligo-dT30-VN and ERCC controls at a final dilution of 1:100 million. Reverse transcription and PCR was performed according to the Smartseq-2 protocol with reduced volumes: 1μl of reverse transcription mix instead of 5.7μl and 3μl PCR Master mix instead of 12.5μl. cDNA was amplified for 24 cycles and cleaned using HighPrep PCR beads (MagBio Genomics) at a 0.8x ratio on a Hamilton STAR (Hamilton Germany GmbH) liquid handler, eluted in 30μl buffer EB (Qiagen) and transferred to 384_PP acoustic plates (LabCyte). DNA quantification was performed with Picogreen assay (Thermofisher) and a subset of samples were selected for quality control using a Agilent 2100 BioAnalyser (Agilent Technologies) using a High Sensitivity DNA kit. After initial quality control 24 samples were discarded with cDNA yields of less than 21ng. These samples were replaced with 24 cells from another 384 well plate from the same cell sort by plate reformatting with an acoustic dispenser (LabCyte Echo 525).
Library preparation continued from Smartseq-2
Next generation sequencing library preparation was performed using a Nextera XT DNA library preparation kit with volumes reduced by one-tenth (Illumina) using an acoustic dispenser. In brief, 100pg of cDNA in a volume of 500nL was tagmented by adding 1.5μl Tn5-buffer mix and incubating for 10 min at 55°C. Tagmented samples were barcoded with Nextera index sets A - D and amplified with 11 cycles of PCR. After PCR, all samples were pooled and cleaned using HighPrep PCR beads at a 0.6x ratio. Library pools were eluted in buffer EB and quality control performed using an Agilent 2100 BioAnalyser and High Sensitiviy DNA chip before adjusting to a concentration of 4nM. The diluted pools were quantified using a KAPA qPCR library quantification kit on a LightCycler 480 (Roche) before a final dilution to 2nM. The pool of 384 samples was sequenced on 2 lanes of a HiSeq2500 in high output mode v4 chemistry with 1x100bp read length.
Single-cell bioinformatics analysis
Sequencing reads were trimmed for adapter sequences using cutadapt (version 1.13) and reads were aligned to the GRCm38 reference genome including ERCC sequences using STAR with default parameters (version 2.5.2b)67. The expression count matrix was generated using HTSeq (version 0.6.0)68 on GENCODE M12 transcript annotations and counts for each protein coding gene were collapsed. Quality control was performed using the scater R package (version 1.2.0)69. Cells that complied with one of the following conditions were excluded: had fewer than 105 counts, showed expression of fewer than 2500 unique genes, had more than 20% counts belonging to ERCC sequences, had more than 8% counts belonging to mitochondrial sequences. BCs and EMPs that showed no expression of neither K5 nor K14, and LCs that did not express K8 were further excluded. Cells coming from a row F of the 384-well plate which showed systematic mixing of LC and BC markers were excluded from further analysis due to a likely pipetting error. Out of the 377 samples which passed sequencing, 221 passed quality control. Genes for which less than 20 counts were observed across the complete dataset were excluded from further analysis. Read counts were normalized using scran with default parameters (version 1.2.2). Clustering using the SC3 R package (version 1.3.18)40 and PCA was performed using the prcomp function in R, plots were generated using the ggplot2 R package (version 2.2.1). We chose k=4 (all cells), k=2 (for EMP-only and LC vs BC clustering) for SC3 as this best represented the heterogeneity in our dataset and recapitulated the studied cell lineages. For cluster marker gene discovery we set the thresholds to all genes with an AUC higher than 0.8 and p-value lower than 0.01. BC/LC specific markers were determined by filtering marker genes identified by SC3 on non-EMP cells and retaining only marker genes which were expressed in 75% of the respective population and less than 50% of the opposite population. The adjusted proportion of specific markers for each cell was computed by counting the number of specific LC/BC markers over the total number of LC/BC specific markers and correcting for differences in sensitivity by modelling the linear relationship between the adjusted proportion of markers detected and the total number of genes detected for each cell. Heatmaps were generated using a modified version of the gplots R package (3.0.1). Cell-cycle phase was automatically assigned using the scran package and cells not in G1 phase were excluded from further analysis. Gene regulatory network analysis was performed using SCENIC48 using default parameters. Regulon AUC thresholds for binary activity determination were manually assessed and adjusted for 425 regulons expressed. Pearson correlations between regulon AUCs and the adjusted proportion of specific LC/BC markers was computed and the corresponding p-value was FDR corrected using the Benjamini-Hochberg method. To determine the enrichment of p63 inferred target genes in p63 overexpressing LC cells we computed the probability under a binomial model with p=0.12 (fraction of genes with 1.5 fold enrichment in ΔNp63 expressing LC cells compared to WT LC cells) for which 18 (or more) out of 48 trials were a success.
Statistics and Reproducibility
All experiments were repeated at least three times with similar results unless a different number of repeats is stated in the legend. Method used, P values and N numbers are indicated in the figure legends. No statistical method was used to predetermine sample size. All experimental mice used in this study were females of mixed genetic backgrounds. No animals were excluded from the study. No method of randomization was used. The investigators were not blinded to allocation during experiments or outcome assessment.
Data availability
Microarray, RNAseq and single cell RNA sequencing data that support the findings of this study have been deposited in the Gene Expression Omnibus (GEO) under accession code GSE109711. Previously published microarray data that were re-analysed here are available under accession code GSE69290.
Source data for Figure 1d, 1l, 1q, 6d and 7c have been provided as Supplementary Table 1.
Supplementary Material
Acknowledgments
We thank the animal house facility from the ULB (Erasme campus). Sequencing was performed at the Brussels Interuniversity Genomics High Throughput core (www.brightcore.be) and the Genomics Core Leuven. We thank Nina Dedoncker for help in single-cell RNA-seq library construction. C.B. is an investigator of WELBIO, A.W. is supported by a FNRS fellowship. M.F. is supported by a Télévie fellowship. A.V.K. is Maître de Recherches of the FNRS. A.S., D.B. and T.V. are supported by KU Leuven (SymBioSys, PFV/10/016), Stichting Tegen Kanker (2015-143) and FWO (Postdoctoral Fellow number 12W7318N, [PEGASUS]2 Marie Skłodowska-Curie Fellow number 12O5617N). We thank our colleagues who provided us with reagents, which are cited in the text. We thank J-M. Vanderwinden for his help with confocal imaging. This work was supported by the FNRS, a research grant from the Fondation Contre le Cancer, the ULB fondation, the Fond Gaston Ithier, the Télévie, the foundation Bettencourt Schueller, the foundation Baillet Latour, and the European Research Council (EXPAND).
Footnotes
Author Contributions
A.W. and C.B. designed the experiments and performed data analysis. A.W. S.M, M.F., A.C. and A.V.K performed the biological experiments. A.B. performed GSEA analysis. A.S. and T.V. performed single cell RNA seq data analysis and provided the related figures and methods, D.B. and T.V. performed single cell RNA seq processing and sequencing, and provided the related methods, A.D. provided technical support. C.D. provided technical support for cell sorting. A.W., A.S., C.B., A.V.K. prepared the figures. C.B. wrote the manuscript.
Financial and Non-Financial Competing Interests
The authors declare to have no financial and non-financial competing interests.
References
- 1.Watson CJ, Khaled WT. Mammary development in the embryo and adult: a journey of morphogenesis and commitment. Development. 2008;135:995–1003. doi: 10.1242/dev.005439. [DOI] [PubMed] [Google Scholar]
- 2.de Visser KE, et al. Developmental stage-specific contribution of LGR5(+) cells to basal and luminal epithelial lineages in the postnatal mammary gland. J Pathol. 2012;228:300–309. doi: 10.1002/path.4096. [DOI] [PubMed] [Google Scholar]
- 3.Lafkas D, et al. Notch3 marks clonogenic mammary luminal progenitor cells in vivo. J Cell Biol. 2013;203:47–56. doi: 10.1083/jcb.201307046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Prater MD, et al. Mammary stem cells have myoepithelial cell properties. Nat Cell Biol. 2014;16:942–950. doi: 10.1038/ncb3025. 941–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rodilla V, et al. Luminal progenitors restrict their lineage potential during mammary gland development. PLoS Biol. 2015;13:e1002069. doi: 10.1371/journal.pbio.1002069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tao L, van Bragt MP, Laudadio E, Li Z. Lineage tracing of mammary epithelial cells using cell-type-specific cre-expressing adenoviruses. Stem cell reports. 2014;2:770–779. doi: 10.1016/j.stemcr.2014.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van Amerongen R, Bowman AN, Nusse R. Developmental Stage and Time Dictate the Fate of Wnt/beta-Catenin-Responsive Stem Cells in the Mammary Gland. Cell Stem Cell. 2012;11:387–400. doi: 10.1016/j.stem.2012.05.023. [DOI] [PubMed] [Google Scholar]
- 8.Van Keymeulen A, et al. Distinct stem cells contribute to mammary gland development and maintenance. Nature. 2011;479:189–193. doi: 10.1038/nature10573. [DOI] [PubMed] [Google Scholar]
- 9.Wang C, Christin JR, Oktay MH, Guo W. Lineage-Biased Stem Cells Maintain Estrogen-Receptor-Positive and -Negative Mouse Mammary Luminal Lineages. Cell Rep. 2017;18:2825–2835. doi: 10.1016/j.celrep.2017.02.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wuidart A, et al. Quantitative lineage tracing strategies to resolve multipotency in tissue-specific stem cells. Genes Dev. 2016;30:1261–1277. doi: 10.1101/gad.280057.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Scheele CL, et al. Identity and dynamics of mammary stem cells during branching morphogenesis. Nature. 2017;542:313–317. doi: 10.1038/nature21046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Davis FM, et al. Single-cell lineage tracing in the mammary gland reveals stochastic clonal dispersion of stem/progenitor cell progeny. Nature communications. 2016;7:13053. doi: 10.1038/ncomms13053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Snippert HJ, et al. Intestinal crypt homeostasis results from neutral competition between symmetrically dividing Lgr5 stem cells. Cell. 2010;143:134–144. doi: 10.1016/j.cell.2010.09.016. [DOI] [PubMed] [Google Scholar]
- 14.Lescroart F, et al. Early lineage restriction in temporally distinct populations of Mesp1 progenitors during mammalian heart development. Nat Cell Biol. 2014;16:829–840. doi: 10.1038/ncb3024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barker N, et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003–1007. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- 16.Trejo CL, Luna G, Dravis C, Spike BT, Wahl GM. Lgr5 is a marker for fetal mammary stem cells, but is not essential for stem cell activity or tumorigenesis. NPJ breast cancer. 2017;3:16. doi: 10.1038/s41523-017-0018-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Van Keymeulen A, et al. Reactivation of multipotency by oncogenic PIK3CA induces breast tumour heterogeneity. Nature. 2015;525:119–123. doi: 10.1038/nature14665. [DOI] [PubMed] [Google Scholar]
- 18.Wansbury O, et al. Transcriptome analysis of embryonic mammary cells reveals insights into mammary lineage establishment. Breast Cancer Res. 2011;13:R79. doi: 10.1186/bcr2928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Biggs LC, Mikkola ML. Early inductive events in ectodermal appendage morphogenesis. Semin Cell Dev Biol. 2014;25–26:11–21. doi: 10.1016/j.semcdb.2014.01.007. [DOI] [PubMed] [Google Scholar]
- 20.Boras-Granic K, Chang H, Grosschedl R, Hamel PA. Lef1 is required for the transition of Wnt signaling from mesenchymal to epithelial cells in the mouse embryonic mammary gland. Dev Biol. 2006;295:219–231. doi: 10.1016/j.ydbio.2006.03.030. [DOI] [PubMed] [Google Scholar]
- 21.Chu EY, et al. Canonical WNT signaling promotes mammary placode development and is essential for initiation of mammary gland morphogenesis. Development. 2004;131:4819–4829. doi: 10.1242/dev.01347. [DOI] [PubMed] [Google Scholar]
- 22.Hiremath M, Wysolmerski J. Parathyroid hormone-related protein specifies the mammary mesenchyme and regulates embryonic mammary development. J Mammary Gland Biol Neoplasia. 2013;18:171–177. doi: 10.1007/s10911-013-9283-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Howard BA, Lu P. Stromal regulation of embryonic and postnatal mammary epithelial development and differentiation. Semin Cell Dev Biol. 2014;25–26:43–51. doi: 10.1016/j.semcdb.2014.01.004. [DOI] [PubMed] [Google Scholar]
- 24.Wysolmerski JJ, McCaughern-Carucci JF, Daifotis AG, Broadus AE, Philbrick WM. Overexpression of parathyroid hormone-related protein or parathyroid hormone in transgenic mice impairs branching morphogenesis during mammary gland development. Development. 1995;121:3539–3547. doi: 10.1242/dev.121.11.3539. [DOI] [PubMed] [Google Scholar]
- 25.Raafat A, et al. Expression of Notch receptors, ligands, and target genes during development of the mouse mammary gland. J Cell Physiol. 2011;226:1940–1952. doi: 10.1002/jcp.22526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sale S, Lafkas D, Artavanis-Tsakonas S. Notch2 genetic fate mapping reveals two previously unrecognized mammary epithelial lineages. Nat Cell Biol. 2013;15:451–460. doi: 10.1038/ncb2725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Robinson GW. Cooperation of signalling pathways in embryonic mammary gland development. Nat Rev Genet. 2007;8:963–972. doi: 10.1038/nrg2227. [DOI] [PubMed] [Google Scholar]
- 28.Mills AA, et al. p63 is a p53 homologue required for limb and epidermal morphogenesis. Nature. 1999;398:708–713. doi: 10.1038/19531. [DOI] [PubMed] [Google Scholar]
- 29.Yang A, et al. p63 is essential for regenerative proliferation in limb, craniofacial and epithelial development. Nature. 1999;398:714–718. doi: 10.1038/19539. [DOI] [PubMed] [Google Scholar]
- 30.Forster N, et al. Basal cell signaling by p63 controls luminal progenitor function and lactation via NRG1. Dev Cell. 2014;28:147–160. doi: 10.1016/j.devcel.2013.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dravis C, et al. Sox10 Regulates Stem/Progenitor and Mesenchymal Cell States in Mammary Epithelial Cells. Cell Rep. 2015;12:2035–2048. doi: 10.1016/j.celrep.2015.08.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ye X, et al. Distinct EMT programs control normal mammary stem cells and tumour-initiating cells. Nature. 2015;525:256–260. doi: 10.1038/nature14897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Choi YS, Chakrabarti R, Escamilla-Hernandez R, Sinha S. Elf5 conditional knockout mice reveal its role as a master regulator in mammary alveolar development: failure of Stat5 activation and functional differentiation in the absence of Elf5. Dev Biol. 2009;329:227–241. doi: 10.1016/j.ydbio.2009.02.032. [DOI] [PubMed] [Google Scholar]
- 34.Oakes SR, Hilton HN, Ormandy CJ. The alveolar switch: coordinating the proliferative cues and cell fate decisions that drive the formation of lobuloalveoli from ductal epithelium. Breast Cancer Res. 2006;8:207. doi: 10.1186/bcr1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bernardo GM, et al. FOXA1 is an essential determinant of ERalpha expression and mammary ductal morphogenesis. Development. 2010;137:2045–2054. doi: 10.1242/dev.043299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Balko JM, et al. The receptor tyrosine kinase ErbB3 maintains the balance between luminal and basal breast epithelium. Proc Natl Acad Sci U S A. 2012;109:221–226. doi: 10.1073/pnas.1115802109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kendrick H, et al. Transcriptome analysis of mammary epithelial subpopulations identifies novel determinants of lineage commitment and cell fate. BMC Genomics. 2008;9:591. doi: 10.1186/1471-2164-9-591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Spike BT, et al. A mammary stem cell population identified and characterized in late embryogenesis reveals similarities to human breast cancer. Cell Stem Cell. 2012;10:183–197. doi: 10.1016/j.stem.2011.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Van Keymeulen A, et al. Lineage-Restricted Mammary Stem Cells Sustain the Development, Homeostasis, and Regeneration of the Estrogen Receptor Positive Lineage. Cell Rep. 2017;20:1525–1532. doi: 10.1016/j.celrep.2017.07.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kiselev VY, et al. SC3: consensus clustering of single-cell RNA-seq data. Nat Methods. 2017;14:483–486. doi: 10.1038/nmeth.4236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ibusuki M, et al. Midkine in plasma as a novel breast cancer marker. Cancer Sci. 2009;100:1735–1739. doi: 10.1111/j.1349-7006.2009.01233.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kuang XY, et al. Stathmin and phospho-stathmin protein signature is associated with survival outcomes of breast cancer patients. Oncotarget. 2015;6:22227–22238. doi: 10.18632/oncotarget.4276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Prochazkova I, et al. Targeted proteomics driven verification of biomarker candidates associated with breast cancer aggressiveness. Biochim Biophys Acta. 2017;1865:488–498. doi: 10.1016/j.bbapap.2017.02.012. [DOI] [PubMed] [Google Scholar]
- 44.Saal LH, et al. Poor prognosis in carcinoma is associated with a gene expression signature of aberrant PTEN tumor suppressor pathway activity. Proc Natl Acad Sci U S A. 2007;104:7564–7569. doi: 10.1073/pnas.0702507104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shepherd JH, et al. The SOX11 transcription factor is a critical regulator of basal-like breast cancer growth, invasion, and basal-like gene expression. Oncotarget. 2016;7:13106–13121. doi: 10.18632/oncotarget.7437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zvelebil M, et al. Embryonic mammary signature subsets are activated in Brca1-/- and basal-like breast cancers. Breast Cancer Res. 2013;15:R25. doi: 10.1186/bcr3403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fernandez-Garcia B, et al. Expression and prognostic significance of fibronectin and matrix metalloproteases in breast cancer metastasis. Histopathology. 2014;64:512–522. doi: 10.1111/his.12300. [DOI] [PubMed] [Google Scholar]
- 48.Aibar S, et al. SCENIC: single-cell regulatory network inference and clustering. Nat Methods. 2017;14:1083–1086. doi: 10.1038/nmeth.4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Malhotra GK, et al. The role of Sox9 in mouse mammary gland development and maintenance of mammary stem and luminal progenitor cells. BMC Dev Biol. 2014;14:47. doi: 10.1186/s12861-014-0047-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yalcin-Ozuysal O, et al. Antagonistic roles of Notch and p63 in controlling mammary epithelial cell fates. Cell Death Differ. 2010;17:1600–1612. doi: 10.1038/cdd.2010.37. [DOI] [PubMed] [Google Scholar]
- 51.Latil M, et al. Cell-Type-Specific Chromatin States Differentially Prime Squamous Cell Carcinoma Tumor-Initiating Cells for Epithelial to Mesenchymal Transition. Cell Stem Cell. 2017;20:191–204.e195. doi: 10.1016/j.stem.2016.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fre S. Notch1 clonal analysis reveals the existence of unipotent stem cells that retain long-term plasticity in the embryonic mammary gland. Nat Cell Biol. 2018 doi: 10.1038/s41556-018-0108-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Signoretti S, et al. p63 is a prostate basal cell marker and is required for prostate development. Am J Pathol. 2000;157:1769–1775. doi: 10.1016/S0002-9440(10)64814-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kurita T, Medina RT, Mills AA, Cunha GR. Role of p63 and basal cells in the prostate. Development. 2004;131:4955–4964. doi: 10.1242/dev.01384. [DOI] [PubMed] [Google Scholar]
- 55.van Bokhoven H, McKeon F. Mutations in the p53 homolog p63: allele-specific developmental syndromes in humans. Trends Mol Med. 2002;8:133–139. doi: 10.1016/s1471-4914(01)02260-2. [DOI] [PubMed] [Google Scholar]
- 56.Madisen L, et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci. 2010;13:133–140. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nguyen H, Rendl M, Fuchs E. Tcf3 governs stem cell features and represses cell fate determination in skin. Cell. 2006;127:171–183. doi: 10.1016/j.cell.2006.07.036. [DOI] [PubMed] [Google Scholar]
- 58.Perl AK, Wert SE, Nagy A, Lobe CG, Whitsett JA. Early restriction of peripheral and proximal cell lineages during formation of the lung. Proc Natl Acad Sci U S A. 2002;99:10482–10487. doi: 10.1073/pnas.152238499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gonzalez-Roca E, et al. Accurate expression profiling of very small cell populations. PLoS One. 2010;5:e14418. doi: 10.1371/journal.pone.0014418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gautier L, Cope L, Bolstad BM, Irizarry RA. affy--analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20:307–315. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- 61.Huber W, et al. Orchestrating high-throughput genomic analysis with Bioconductor. Nat Methods. 2015;12:115–121. doi: 10.1038/nmeth.3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Leek JT, Johnson WE, Parker HS, Jaffe AE, Storey JD. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics. 2012;28:882–883. doi: 10.1093/bioinformatics/bts034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 65.Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Picelli S, et al. Smart-seq2 for sensitive full-length transcriptome profiling in single cells. Nat Methods. 2013;10:1096–1098. doi: 10.1038/nmeth.2639. [DOI] [PubMed] [Google Scholar]
- 67.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Anders S, Pyl PT, Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.McCarthy DJ, Campbell KR, Lun AT, Wills QF. Scater: pre-processing, quality control, normalization and visualization of single-cell RNA-seq data in R. Bioinformatics. 2017;33:1179–1186. doi: 10.1093/bioinformatics/btw777. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Microarray, RNAseq and single cell RNA sequencing data that support the findings of this study have been deposited in the Gene Expression Omnibus (GEO) under accession code GSE109711. Previously published microarray data that were re-analysed here are available under accession code GSE69290.
Source data for Figure 1d, 1l, 1q, 6d and 7c have been provided as Supplementary Table 1.