Abstract
Bacillus thuringiensis Cry toxins are insecticidal proteins used widely for pest control. They are lethal to a restricted range of insects via specific interactions with insect receptors such as the ABC transporter subfamily members C2 (ABCC2) and C3 (ABCC3). However, it is still unclear how these different receptors contribute to insect susceptibility to Cry1A toxins. Here, we investigated the differences between the silkworm (Bombyx mori) ABCC2 (BmABCC2_S) and ABCC3 (BmABCC3) receptors in mediating Cry toxicity. Compared with BmABCC2_S, BmABCC3 exhibited 80- and 267-fold lower binding affinities to Cry1Aa and Cry1Ab, respectively, and these decreased affinities correlated well with the lower receptor activities of BmABCC3 for these Cry1A toxins. To identify the amino acid residues responsible for these differences, we constructed BmABCC3 variants containing a partial amino acid replacement with extracellular loops (ECLs) from BmABCC2_S. Replacing three amino acids from ECL 1 or 3 increased BmABCC3 activity toward Cry1Aa and enabled its activity toward Cry1Ab. Meanwhile, BmABCC2_S and BmABCC3 exhibited no receptor activities for Cry1Ca, Cry1Da, and Cry3Bb, correlating with markedly lower binding affinities for these Cry toxins. ABCC2 from a Cry1Ab-resistant B. mori strain (BmABCC2_R), which has a tyrosine insertion in ECL 2, displayed 93-fold lower binding affinity to Cry1Ab compared with BmABCC2_S but maintained high binding affinity to Cry1Aa. These results indicate that the Cry toxin–binding affinities of ABCC transporters are largely linked to the level of Cry susceptibility of ABCC-expressing cells and that the ABCC ECL structures determine the specificities to Cry toxins.
Keywords: Bacillus, bacterial toxin, ABC transporter, insect, Bombyx mori, Bacillus thuringiensis, Cry toxin, ABC transporter C2 (ABCC2), ABC transporter C3 (ABCC3), binding affinity
Introduction
Cry toxin, a pore-forming toxin produced by the insect bacterial pathogen Bacillus thuringiensis, exhibits specific toxicity restricted to certain types of insects. The specificity against target insects is largely determined by the specific interaction between the Cry toxin and the membrane protein receptors expressed in the apical membrane of insect midgut cells (1). The receptor–toxin interaction promotes pore formation of the toxin on the cell membrane, leading to loss of membrane integrity, cell swelling, necrotic cell death, and, finally, the death of the insect (2).
A gene for the ATP-binding cassette (ABC)3 transporter subfamily member C2 (ABCC2) in lepidopteran insects has been identified as a gene responsible for high resistance against Cry1A toxins (3). Using heterologous expression systems of Sf9 cells and Xenopus oocytes, we demonstrated previously that silkworm (Bombyx mori) ABCC2 from a Cry1A-susceptible strain Ringetsu (BmABCC2_S) mediates high susceptibilities to Cry1A toxins by allowing the induction of powerful pore formation activity compared with other Cry1A toxin receptors, including cadherin-like protein and aminopeptidase N1 (4, 5). Additionally, we showed a high-affinity interaction (KD ≈ 10−10 m) between the Cry1Aa toxin and BmABCC2_S (6). However, little is known regarding the binding of other Cry toxins to BmABCC2_S; therefore, it remains unclear whether the specificities of BmABCC2_S for Cry1A toxins are determined by the binding affinity.
ABCC3, a paralog of ABCC2, mediates the toxicities of Cry1 toxins (7–10). ABCC2 and ABCC3 are unique to lepidopteran insects and have not been found in other insects (10), suggesting that these lepidopteran-specific ABCC transporters largely determine the specificity of Cry1A for lepidopteran insects. However, the binding of Cry1A toxins to ABCC3 and the extent to which ABCC3, compared with ABCC2, contributes to Cry toxin intoxication have yet to be elucidated.
ABCC2 and ABCC3 are predicted to have six extracellular loops (ECLs). We showed that, among these ECLs, 770DYWL773 of ECL 4 in BmABCC2_S is required for receptor activity for Cry1Aa, suggesting that the residues in ECL 4 are a part of the interaction sites of BmABCC2_S with Cry1Aa (11). Accordingly, differences in the ECL amino acid residues, especially residues in ECL 4, might generate different receptor activities between ABCC2 and ABCC3. Unlike most Cry1A-resistant lepidopteran strains, Cry1Ab-resistant silkworm strains do not lack the ABCC2 protein; instead, they express the molecule with a tyrosine insertion in ECL 2 at position 234 (12). Cry1Ab did not induce cell swelling of Sf9 cells expressing the tyrosine-inserted BmABCC2 (BmABCC2_R) from the resistant strain C2, whereas Cry1Aa did (4). Additionally, a three-alanine substitution in the center of ECL 2 containing five amino acid residues did not affect the receptor function of BmABCC2 for Cry1A toxins, including Cry1Ab (13). Therefore, it is unlikely that ECL 2 functions as a direct interaction site of BmABCC2 to Cry1A toxins, and the extra tyrosine insertion probably causes inhibition of some processes leading to pore formation. However, binding analysis of Cry1Ab to BmABCC2_R is required to clarify whether BmABCC2_R lacks binding affinity to Cry1Ab.
In the present study, we investigated the correlation between the receptor activities and binding affinities of ABCC transporters to Cry toxins. We compared silkworm ABCC2 (BmABCC2_S) and ABCC3 (BmABCC3) with regard to Cry toxin specificities, expression levels in midgut cells, and binding affinities with Cry toxins. Additionally, we investigated the binding affinities of Cry1A toxins to BmABCC2_R. Last, we identified amino acid residues in the ECLs that generate differences in receptor activities between BmABCC2_S and BmABCC3 in Cry toxin intoxication. Our results suggest that the receptor activities of these ABC transporters for Cry toxins are largely determined by the binding affinity with Cry toxins derived from structural differences in the ECLs.
Results
Cry toxin specificities of BmABCC2 and BmABCC3
To investigate the Cry toxin specificities of BmABCC2 and BmABCC3, bioassays against Cry toxins using HEK293T cells transiently expressing BmABCC2_S or BmABCC3 were performed. First, expression levels of BmABCC2 or BmABCC3 in HEK293T cells were evaluated using enhanced GFP (EGFP), which was fused to the C termini of the ABC transporters. The relative fluorescence ratio (EGFP/4′,6-diamidino-2-phenylindole (DAPI)) was equivalent between BmABCC2_S- and BmABCC3-expressing cells (Fig. 1), indicating that the per-cell expression levels of BmABCC2_S and BmABCC3 were almost the same.
Figure 1.
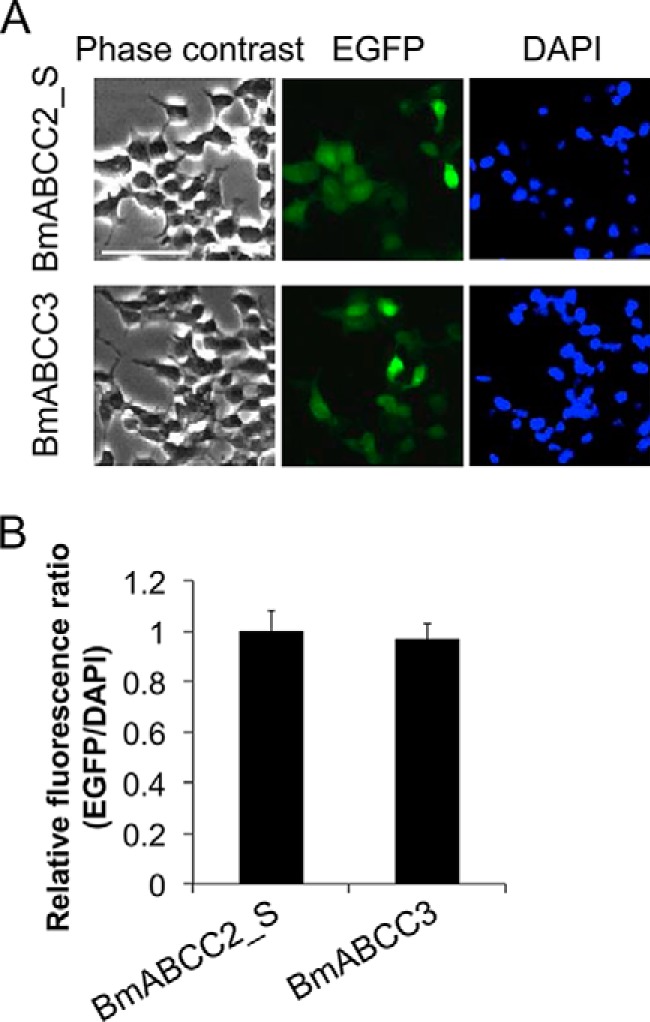
Expression levels of BmABCC2 and BmABCC3 in transfected HEK293T cells. A, HEK293T cells seeded on a 6-well plate were transfected with 2 μg of vector for 2 h. EGFP, which was fused to the C termini of the BmABCC3 mutants, and DAPI, which stains nuclear DNA, were visualized under a fluorescence microscope after 48 h, as described under “Experimental procedures.” Scale bar, 40 μm. B, the relative fluorescence ratio (GFP/DAPI) indicating the per-cell expression levels of BmABCC2_S or BmABCC3 was calculated using the fluorescence intensities from three fields of view, including images in Fig. S3. Error bars, S.E.
We tested the susceptibilities of BmABCC2_S- or BmABCC3-expressing cells against four lepidopteran-specific and B. mori larvicidal Cry toxins (Cry1Aa, Cry1Ab, Cry1Ca, and Cry1Da) and a coleopteran-specific Cry3Bb toxin. Cells expressing BmABCC2_S or BmABCC3 swell in response to Cry toxins when the ABC transporter serves as a Cry toxin receptor, as in the case of BmABCC2_S-expressing cells in the presence of Cry1Aa (4). Such cell swelling directly reflects the cytotoxicity of the Cry1 toxins (14). BmABCC2_S-expressing cells began to swell when incubated for 1 h in the presence of 1 nm Cry1Aa, whereas cells transiently expressing BmABCC3 started swelling in the presence of 100 nm Cry1Aa toxin (Fig. 2A). BmABCC2_S-expressing cells responded to 100 nm Cry1Ab, but BmABCC3-expressing cells showed no swelling when incubated with up to 4.5 μm Cry1Ab (Fig. 2B). For quantitative evaluation of difference in receptor activities, we conducted a lactose dehydrogenase (LDH) release assay. Results confirmed that receptor activities of BmABCC3 for Cry1Aa and Cry1Ab are lower than those of BmABCC2_S (Fig. 3). No cell swelling was observed in either cell type in the presence of a 1 μm concentration of the other Cry toxins, including Cry1Ca, Cry1Da, and Cry3Bb (Fig. 2C). These results suggest that BmABCC2_S and BmABCC3 have different receptor activities for Cry1A toxins; however, neither receptor functions as a receptor for Cry1Ca and Cry1Da.
Figure 2.
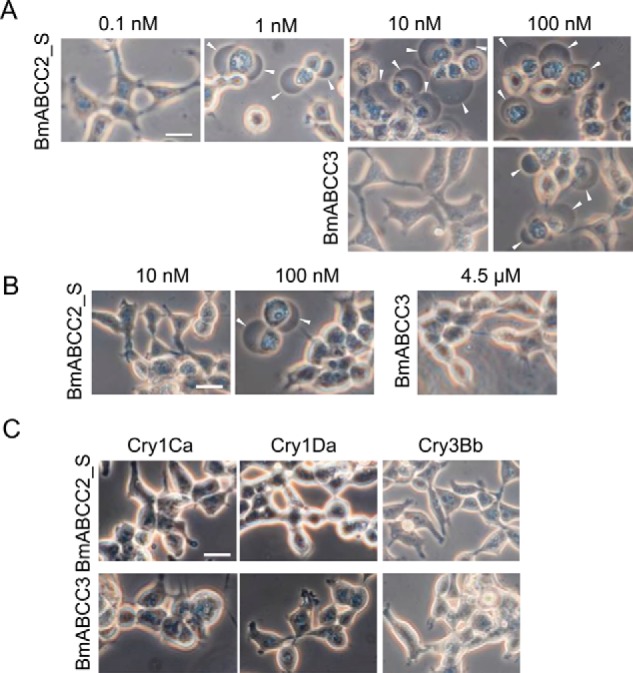
Cry toxin specificities of HEK293T cells expressing BmABCC2_S or BmABCC3. Transfected HEK293T cells seeded on a glass cover were incubated with Cry1Aa (A), Cry1Ab (B), and 1 μm Cry1Ca, Cry1Da, and Cry3Bb (C) at 37 °C for 1 h. Arrowheads, cells swollen in response to the toxins. Scale bar, 20 μm.
Figure 3.
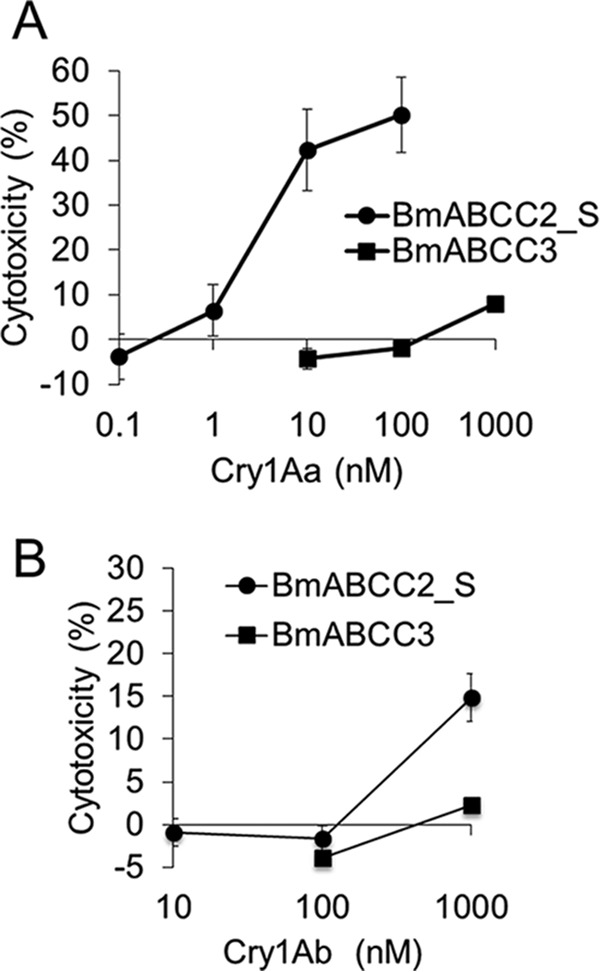
LDH release of HEK293T cells expressing BmABCC2_S or BmABCC3 in response to Cry1A toxins. Cells were seeded onto 96-well plates and incubated with Cry toxin solution for 1 h. The cytotoxicity (%) was calculated using the absorbance values with the equation, cytotoxicity (%) = (experimental value − low control)/(high control − low control) × 100, where low and high controls indicate the values when incubated with HBSS buffer and 2% Triton X-100/HBSS, respectively. Error bars, S.E. (n = 4).
Expression levels of BmABCC2 and BmABCC3 in the midgut of silkworm larvae
Using a heterologous expression system, BmABCC3 showed markedly lower receptor activity for Cry1Aa compared with BmABCC2 (Fig. 2A); however, the activities of the ABC transporters involved in larval susceptibility to the Cry1Aa toxin should be influenced by the expression levels in midgut cells. Hence, we conducted quantitative RT-PCR using cDNA prepared from midgut total RNA from each instar silkworm larva. The mRNA expression levels of the two ABC transporters increased from the first instar to the fourth instar and then decreased in the fifth instar, with similar expression patterns in the larval stages (Fig. 4). The mRNA expression levels of BmABCC3 in the second and third instar larvae were 3- and 2.2-fold higher than those of BmABCC2, respectively; however, the mRNA expression levels of these molecules in the other instar larvae were almost the same (Fig. 4). Therefore, even in the second and third instar B. mori larvae, the contribution of BmABCC2 to Cry1Aa susceptibility is likely to be higher than BmABCC3, with ∼100-fold higher receptor activity observed in BmABCC2-expressing HEK293T cells than in BmABCC3-expressing cells (Figs. 2 and 3).
Figure 4.
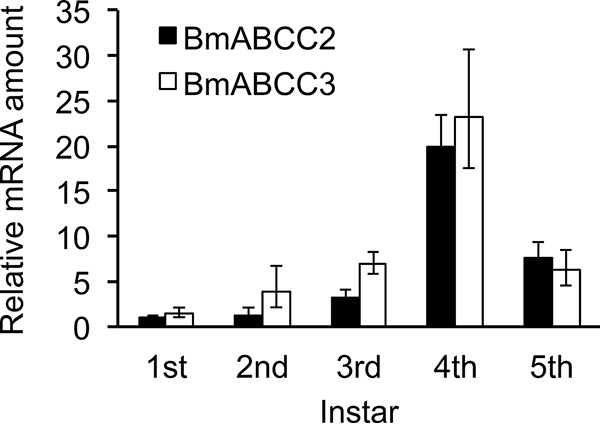
Expression levels of BmABCC2_S and BmABCC3 in midgut tissues from silkworm larvae by quantitative RT-PCR. The y axis shows the relative amount of mRNA, with the mRNA amount of BmABCC2_S in the first instar defined as 1. Midgut tissues were sampled from day 2 larvae from the first to fifth instars. Error bars, S.E. (n = 3).
Binding affinities of Cry toxins to BmABCC2_S, BmABCC3, and BmABCC2_R
To elucidate whether the differences in toxin specificities and receptor activities between the two ABC transporters depended on binding kinetics, we employed surface plasmon resonance (SPR) analysis of BmABCC2_S or BmABCC3 association with and dissociation from Cry toxins. Additionally, we carried out SPR analysis of BmABCC2_R with Cry1A toxins. BmABCC2_S, BmABCC3, and BmABCC2_R proteins were prepared using a baculovirus expression system and purified with anti-FLAG tag antibody–conjugated gels, as reported previously (6). The purities of the proteins were confirmed by silver staining and Western blotting using anti-FLAG tag antibody after separation by SDS-PAGE (Fig. 5). Because bands of contaminating proteins were detected even after affinity purification, we investigated whether these proteins affect results of binding analysis. Proteins that nonspecifically bound to affinity gels conjugated with anti-FLAG tag antibody were collected from a membrane fraction from a 2-liter culture of EGFP-alone–expressing Sf9 cells and immobilized on the Biacore sensor chip. We observed no Cry toxins binding (400 nm Cry1Aa and Cry1Ab) to the nonspecific contaminating proteins derived from Sf9 cells (Fig. S1), suggesting that the nonspecific binding included in the sensorgrams was negligible, and partially purified proteins were worth using for binding analysis.
Figure 5.
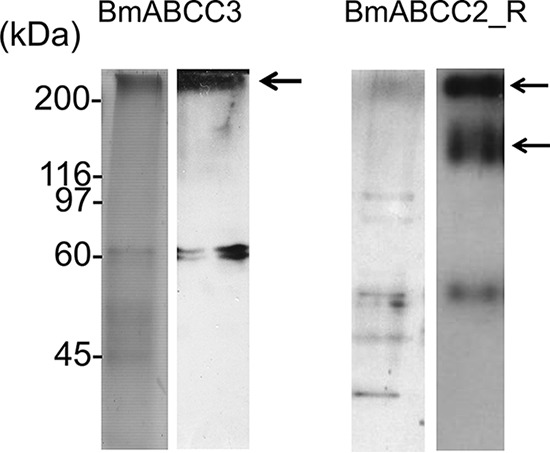
Purity of BmABCC3 and BmABCC2_R used for SPR analysis. Purified BmABCC3 and BmABCC2_R proteins were visualized using silver staining (left) and Western blotting with anti-FLAG antibody (right) following SDS-PAGE. The arrows indicate bands of BmABCC3-FLAG (300 kDa) and BmABCC2_R (150 and 300 kDa). Although the theoretical molecular mass of BmABCC3 and BmABCC2_R is ∼150 kDa, the band at 300 kDa was consistently observed, probably representing a putative dimer or different glycosylated form of this protein.
Cry1Aa toxin showed rapid association but slow dissociation with the BmABCC3-immobilized sensor chip (Fig. 6), as we showed previously using a BmABCC2_S-immobilized sensor chip (6). With repetitive trials of fitting and careful observations using BIAevaluation version 4.1 software, we found that the Langmuir 1:1 binding model did not fit the sensorgrams, particularly during the dissociation phase (Fig. S2). Instead, the two-state and bivalent binding models fit better than did the Langmuir 1:1 binding model with regard to the sensorgram of Cry1Aa (Fig. S2). The X2 value was too high using the Langmuir 1:1 binding model but was reasonable when the two-state and bivalent binding models were applied (Fig. S2). Because no obvious oligomers were observed in the Cry1Aa toxin solution (15), it is likely that Cry1Aa toxin binds to BmABCC2_S or BmABCC3 with a 1:1 stoichiometry, and the bivalent binding model seems unlikely. Other sensorgrams against other toxins also showed the same fitting pattern to the three models. For these reasons, we used the two-state binding model for curve fitting.
Figure 6.
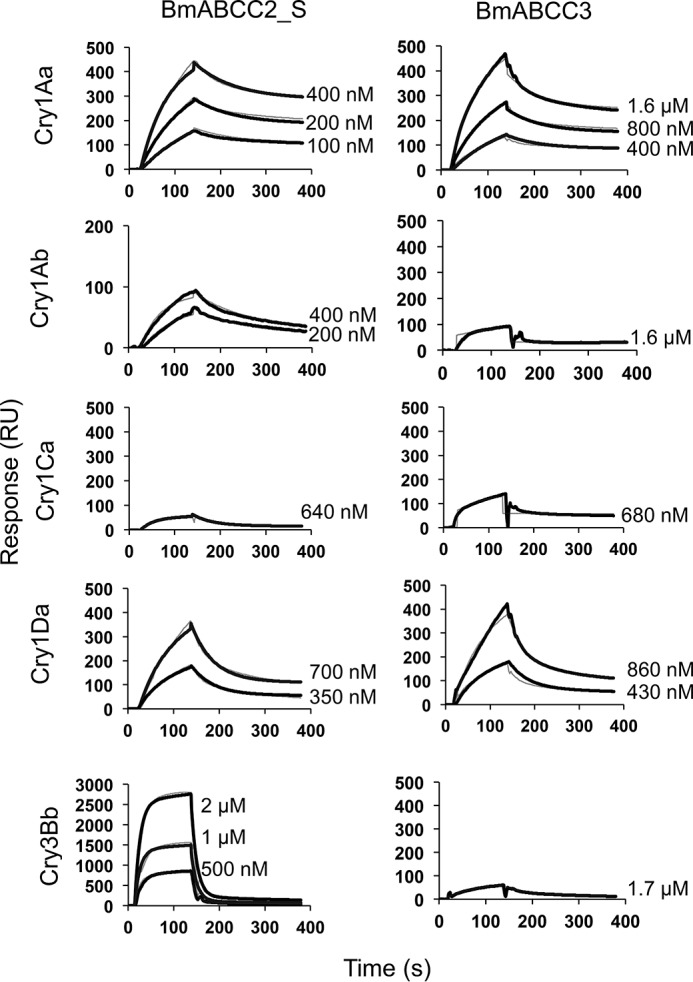
Cry toxin association with and dissociation from BmABCC2_S and BmABCC3 evaluated by SPR. Recombinant BmABCC2_S and BmABCC3 with FLAG tag were purified using an anti-FLAG tag antibody–conjugated gel and immobilized on a CM5 sensor chip. Cry toxins were injected over the sensor chip of Biacore J as analytes. The thick black lines indicate the actual response curves, whereas the thin gray lines show the two-state reaction models indicated by BIAevaluation software. Some of the model curves are difficult to discern, as the actual response curves overlap the models, indicating a good fit.
The dissociation constant (KD) was calculated using kinetic parameters obtained from the curve fitting of the sensorgrams in Fig. 6. The KD values of Cry1Aa to BmABCC2_S and BmABCC3 were 4.30 × 10−10 and 3.42 × 10−8 m, respectively (Table 1), indicating that Cry1Aa bound to BmABCC2_S with an 80-fold higher affinity than that bound to BmABCC3. The KD values of Cry1Ab to BmABCC2_S and BmABCC3 were 2.37 × 10−10 and 6.85 × 10−8 m, respectively (Table 1), indicating that Cry1Ab bound to BmABCC2_S with a 267-fold higher affinity than that bound to BmABCC3. These results suggest that the difference in the binding affinity of Cry1A toxins is mainly responsible for the differences in the Cry1Aa and Cry1Ab susceptibilities of BmABCC2_S- and BmABCC3-expressing cells observed in Fig. 2. The KD values for the binding of Cry1Aa and Cry1Ab on ABCC2_S are similar (Table 1), as well as the KD values for the binding of the same toxins on ABCC3, although Cry1Ab is less toxic than Cry1Aa (Fig. 2). This result indicates that the different toxicities of Cry1Aa and Cry1Ab against the same ABC transporter were not based on differences in binding affinity.
Table 1.
KD values of Cry toxins to silkworm ABC transporters
| Cry1Aa | Cry1Ab | Cry1Ca | Cry1Da | Cry3Bb | |
|---|---|---|---|---|---|
| BmABCC2_S | 4.30 × 10−10 | 2.57 × 10−10 | 1.71 × 10−7 | 2.30 × 10−6 | 1.96 × 10−5 |
| BmABCC3 | 3.42 × 10−8 | 6.85 × 10−8 | 3.92 × 10−7 | 4.18 × 10−4 | 3.98 × 10−8 |
| BmABCC2_R | 2.82 × 10−10 | 2.39 × 10−8 | NDa | ND | ND |
a ND, not done.
Both Cry1Ca and Cry1Da were active against silkworm larvae but not against BmABCC2- or BmABCC3-expressing cells (Fig. 2C). Cry1Ca showed relatively low binding affinity (∼10−7 m) to both BmABCC2 and BmABCC3 (Table 1). Cry1Da showed even lower binding affinity (>10−6 m) to both BmABCC2 and BmABCC3 (Table 1). Cry3Bb toxin, which is not toxic to B. mori larvae or cells expressing BmABCC2 and BmABCC3 (Fig. 2C), showed markedly lower binding affinity to BmABCC2 (∼10−5 m) but high affinity to BmABCC3 (∼10−8 m) (Table 1). However, the association rate of Cry3Bb to BmABCC3 was 10–1000-fold lower compared with those of active toxins against BmABCC2_S or BmABCC3 (e.g. Cry1Aa binding to BmABCC2_S and BmABCC3 or Cry1Ab binding to BmABCC2) (Table S2), suggesting that the minimal amount of Cry3Bb binding attained during incubation resulted in no toxicity to BmABCC3-expressing cells.
With regard to BmABCC2_R, Cry1Aa showed almost the same high binding affinity (∼10−10 m) as that bound to WT BmABCC2 (Fig. 7 and Table 1). In contrast, Cry1Ab showed a 93-fold lower binding affinity (∼10−8 m) to BmABCC2_R than to BmABCC2 (Fig. 7 and Table 1), suggesting that the tyrosine insertion in ECL 2 reduced the binding affinity of Cry1Ab to BmABCC2.
Figure 7.
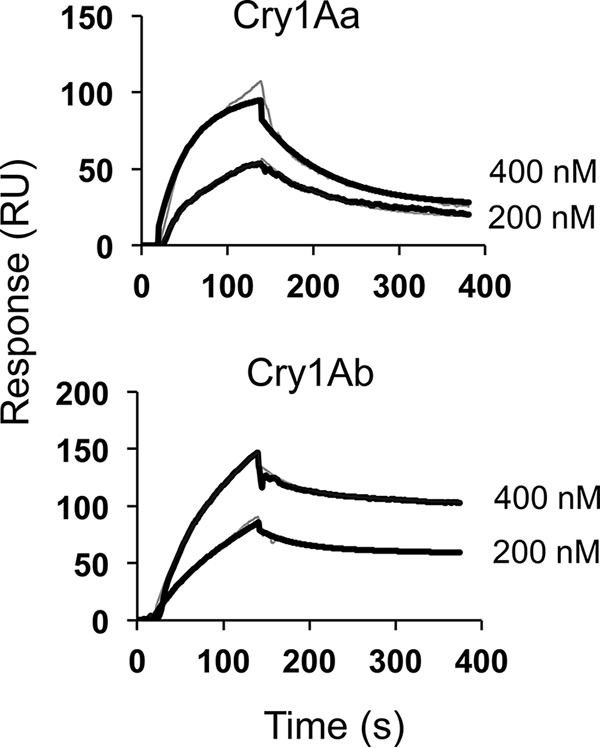
Cry1A toxin association with and dissociation from BmABCC2_R evaluated by SPR. Recombinant BmABCC2_R purified with anti-FLAG tag antibody–conjugated gel was immobilized on a CM5 sensor chip. Cry toxins were injected over the sensor chip of Biacore J as analytes. The thick black lines indicate the actual response curves, whereas the thin gray lines show the two-state reaction models indicated by the BIAevaluation software. Some of the model curves are difficult to discern, as the actual response curves overlap the models, indicating a good fit.
Identification of amino acid residues generating different Cry1A receptor activities in BmABCC2 and BmABCC3
As the tyrosine insertion in ECL 2 of BmABCC2_R caused decreased binding affinity to Cry1Ab (Table 1), structural differences between BmABCC2 and BmABCC3 in six ECLs were expected to generate differences in binding affinities and receptor activities for Cry1A toxins. Thus, when the ECLs of BmABCC3 were partially substituted for the ECLs of BmABCC2, the receptor activity of the mutant BmABCC3 for Cry1A toxins was expected to be higher than that of the WT BmABCC3. According to the predicted transmembrane topology using Phobius, ECLs 1 and 4 have long loops that consist of 18 and 35 amino acid residues, respectively, in BmABCC2 (Fig. 8). Based on results from our recent study (11) (Fig. 8), we focused on the sites 120AELLSYWSVEAT131 in ECL 1 and 770DYWLSFWTN778 in ECL 4 as candidate regions responsible for the receptor activity of BmABCC2 for Cry1Aa. We generated three ECL 1 mutants (Ala122, 126SYWS129, 131EAT133) and one ECL 4 mutant (792LSF794) of BmABCC3. Additionally, we generated BmABCC3 mutants with ECL 2, 3, 5, or 6 that were identical to those from BmABCC2. The expression levels of these mutants were evaluated using the EGFP/DAPI fluorescence ratio (Fig. S3), and no apparent differences in expression levels (maximum 1.3-fold difference) were observed.
Figure 8.

Alignment of amino acid sequences in the ECLs from BmABCC2_S and BmABCC3. The extracellular loops of BmABCC2_S were predicted by Phobius (http://phobius.sbc.su.se/) (see Footnote 4). Red, identical amino acid residues; black, nonidentical residues. The black squares indicate amino acid residues responsible for the receptor function of BmABCC2_S for the Cry1Aa toxin, as suggested by Tanaka et al. (11).
We evaluated the susceptibility of HEK293T cells expressing the BmABCC3 mutants against 10 nm Cry1Aa and 500 nm Cry1Ab. As controls, BmABCC2-expressing cells were swollen in these toxin concentrations, whereas the BmABCC3-expressing cells showed no swelling (Fig. 9). Among the cells expressing the BmABCC3 ECL mutants, cells expressing two BmABCC3 mutants, 131EAT133 (ECL 1) and 363YIS365 (ECL 3), were swollen in the presence of 10 nm Cry1Aa and 500 nm Cry1Ab (Fig. 9). The LDH assay also revealed that the two mutations confer BmABCC3 higher receptor activities for Cry1Aa and Cry1Ab (Fig. 10). Note that receptor activities of the mutants for Cry1Aa were still lower than that of BmABCC2_S; those for Cry1Ab were comparable with or even higher than that of BmABCC2_S (Fig. 10). These results suggest that these amino acid residues in ECL 1 and 3 are included in the regions responsible for the receptor activity. To clarify whether the higher receptor activities of BmABCC3 mutants were based on higher binding affinity to Cry1A toxins, we further conducted binding analysis of BmABCC3 mutants. The recombinant BmABCC3 mutant proteins were partially purified (Fig. 11A). The KD values of Cry1Aa and Cry1Ab to BmABCC3 131EAT133 calculated from parameters shown in Table S3 were 6.6- and 2.3-fold lower than those to WT BmABCC3, respectively (Fig. 11B and Table 2). Conversely, the KD values of Cry1Aa and Cry1Ab to BmABCC3 363YIS365 were rather 4.2- and 12.6-fold higher than those to WT BmABCC3, respectively (Fig. 11B and Table 2). These results indicate that higher binding affinities of Cry1A toxins largely contribute to higher receptor activity of BmABCC3 131EAT133. Meanwhile, other factor(s) might underlie the mechanism of higher receptor activities of 363YIS365.
Figure 9.
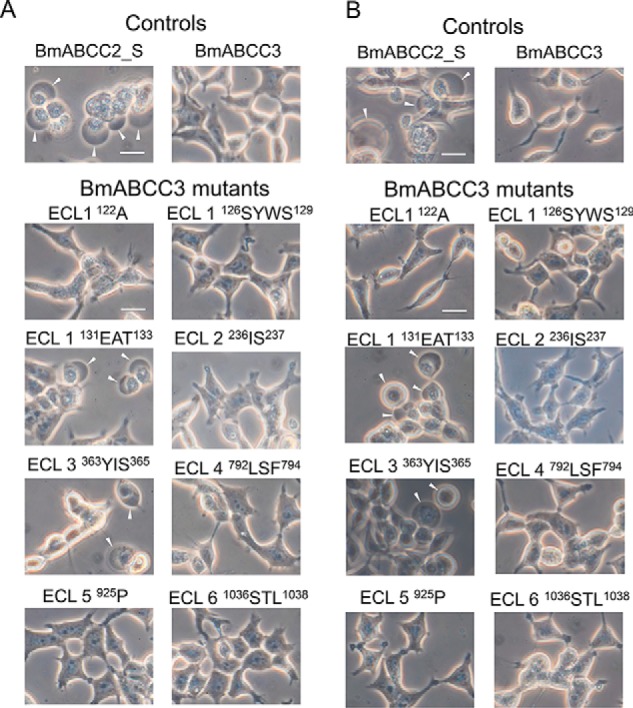
Cry1A susceptibilities of HEK293T cells expressing BmABCC3 mutants with partial replacement of the amino acid residues in the ECLs with the comparative amino acid residues from BmABCC2_S. Cells expressing WT BmABCC2_S, BmABCC3, or BmABCC3 mutants were incubated with 10 nm Cry1Aa (A) or 500 nm Cry1Ab (B) at 37 °C for 1 h. Arrowheads, cells swollen in response to toxins. Scale bar, 20 μm.
Figure 10.
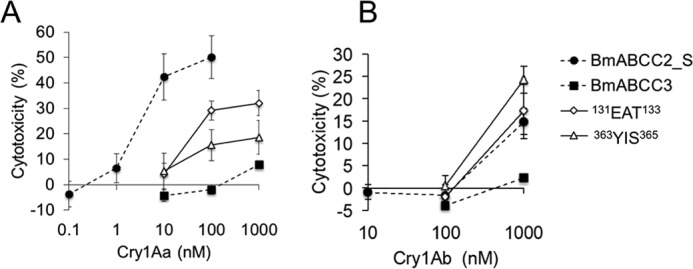
LDH release of HEK293T cells expressing BmABCC3 mutants (131EAT133 and 363YIS365) in response to Cry1Aa (A) and Cry1Ab (B) toxins. Cells were seeded onto 96-well plates and incubated with Cry toxin solution for 1 h. The cytotoxicity (%) was calculated using the absorbance values with the equation, cytotoxicity (%) = (experimental value − low control)/(high control − low control) × 100, where low and high controls indicate the values when incubated with HBSS buffer and 2% Triton X-100/HBSS, respectively. Error bars, S.E. (n = 4). The data of WT ABCC transporters are derived from Fig. 3.
Figure 11.
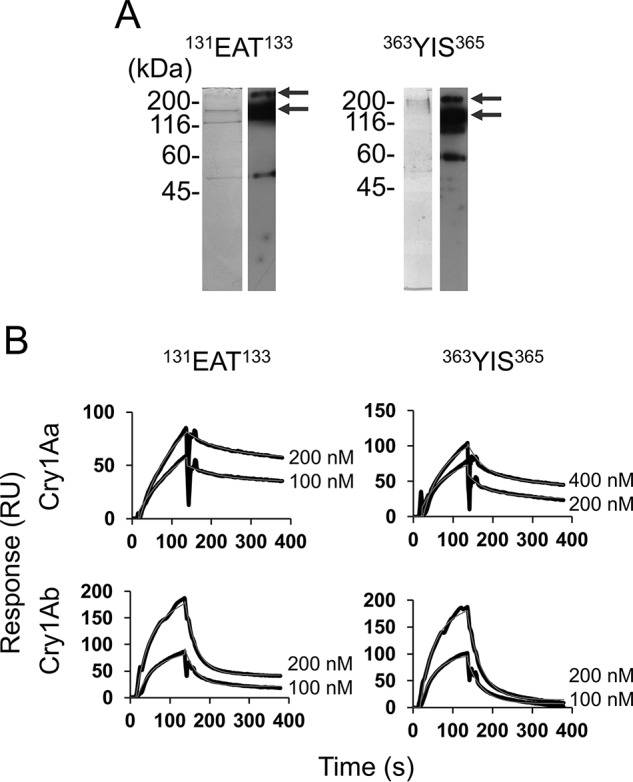
Cry1A toxin association with and dissociation from BmABCC3 mutants (131EAT133 and 363YIS365) evaluated by SPR. A, purity of BmABCC3 mutants used for SPR analysis. Partially purified BmABCC3 mutant proteins were visualized using silver staining (left) and Western blotting with anti-FLAG antibody (right) following SDS-PAGE. The arrows indicate bands of BmABCC3 131EAT133-FLAG and BmABCC3 363YIS365-FLAG (150 and 300 kDa). B, recombinant BmABCC3 mutants with FLAG tag were partially purified using an anti-FLAG tag antibody–conjugated gel and immobilized on a CM5 sensor chip. Cry toxins were injected over the sensor chip of Biacore J as analytes. The thick black lines indicate the actual response curves, whereas the thin gray lines show the two-state reaction models indicated by BIAevaluation software. Some of the model curves are difficult to discern, as the actual response curves overlap the models, indicating a good fit.
Table 2.
KD values of Cry toxins to BmABCC3 mutants
| Cry toxin |
KD(mut) |
KD(WT)/KD(mut) |
||
|---|---|---|---|---|
| Cry1Aa | Cry1Ab | Cry1Aa | Cry1Ab | |
| ECL1 129EAT131 | 5.13 × 10−9 | 2.98 × 10−8 | 6.6 | 2.3 |
| ECL3 363YIS365 | 1.43 × 10−7 | 8.70 × 10−7 | 0.24 | 0.08 |
Discussion
Although BmABCC3 is a functional receptor for Cry1Aa, its toxicity-mediating activity for Cry1Aa was markedly lower than that of BmABCC2_S (Figs. 2A and 3), and this difference was considered to be due to lower binding affinity (Table 1). There were only small differences in the expression levels between the two ABC transporters in the midgut (Fig. 4), suggesting that the expression level of BmABCC3 cannot compensate for the lower receptor function of BmABCC3 compared with BmABCC2. Therefore, the contribution of BmABCC3 to B. mori larval susceptibility to Cry1Aa is much lower than that of BmABCC2_S. However, a synergistic effect with a cadherin-like protein may increase the level of contribution of BmABCC3 to Cry1Aa intoxication; such synergy was previously shown between BmABCC2_S and B. mori cadherin-like protein, BtR175 (4), and between HevABCC2 and HevCaLP (16). Chen et al. (8) showed that Spodoptera litura ABCC3 conferred higher Cry1Ac susceptibility with co-expression of Helicoverpa armigera cadherin-like protein. However, the levels of synergistic effect are not predictable using in vitro assays, as the susceptibility to Cry1Ab conferred by BmABCC2 itself was 100-fold lower than that conferred to Cry1Aa in Sf9 cells, whereas the susceptibility conferred by co-expression of BmABCC2 and BtR175 was almost the same for Cry1Aa and Cry1Ab (4). Therefore, to evaluate the precise level of contribution, including the synergistic effect of BmABCC2 or BmABCC3 with a cadherin-like receptor in larval susceptibility, bioassays using BmABCC2- or BmABCC3-knockout silkworm larvae are indispensable.
In contrast to Cry1Aa, Cry1Ab showed lower and no activity to cells expressing BmABCC2_S and BmABCC3, respectively (Figs. 2 (A and B) and 3), although the binding affinities of Cry1Ab to the ABC transporters were comparable with those of Cry1Aa (Table 1). These results suggest that, whereas the binding amounts might appear similar between Cry1Aa and Cry1Ab, the efficiencies of pore formation might differ between the toxins. Differences in the structure of the binding sites between Cry1Aa and Cry1Ab toxins might generate differences in the efficiency of pore formation. Our recent study indicated a phenomenon analogous to this scenario. Several Cry1Aa mutants with mutations in domain II loop 2 or 3 preserved high binding affinity to BmABCC2_S but showed markedly lower toxicity to BmABCC2_S-expressing Sf9 cells than did the WT Cry1Aa toxin (6). We hypothesized that the differences in amino acid residues in the domain II loops of Cry1A toxins affect not only the binding affinity to BmABCC2_S but also the process of pore formation after receptor binding (6). In fact, the amino acid sequences of domain II loops are not conserved between Cry1Aa and Cry1Ab, and the differences might generate the differences in toxicity to ABC transporter-expressing cells. Further analyses are needed to elucidate the role of domain II loops after receptor binding and how the event contributes to pore formation.
Neither BmABCC2 nor BmABCC3 functioned as a receptor for Cry1Ca and Cry1Da (Fig. 2C), although both toxins kill B. mori larvae. This observation agrees with a previous report indicating that Cry1A-resistant strains did not show cross-resistance to these two toxins (17, 18). Our current results indicated that SeABCC2 and SeABCC3 did not serve as receptors for the two toxins (10). We showed that Cry1Ca and Cry1Da bound to BmABCC2 and BmABCC3 with markedly lower affinity than did Cry1Aa (Table 1), indicating that these toxins cannot form pores via BmABCC2 and BmABCC3 binding due to low binding affinity.
A Cry1Ab-resistant silkworm strain C2 exhibited a more than 2800-fold resistance to Cry1Ab compared with a susceptible strain, Ringetsu (12). Cry1Ab bound to BmABCC2_R with a 93-fold lower affinity than to BmABCC2_S (Table 1), indicating that an extra tyrosine insertion in ECL 2 decreased the binding affinity of BmABCC2 to Cry1Ab, and this insertion could cause high resistance in the BmABCC2_R strain. The possibility that the tyrosine insertion in ECL 2 affected other processes of pore formation after binding to BmABCC2_R cannot be excluded, as it is unclear whether the >1000-fold resistance of the C2 strain can be explained simply by a 93-fold reduction in binding affinity.
Replacement of the amino acid residues in ECL 1 and 3 of BmABCC3 with those of BmABCC2 increased the receptor activity of BmABCC3 for Cry1Aa (Fig. 9). Additionally, these replacements conferred BmABCC3 receptor activity for Cry1Ab on BmABCC3 (Fig. 9). The substitution in ECL 1 (131EAT133) increased the binding affinity of Cry1A toxins (Table 2), suggesting that these amino acid residues are involved in binding to Cry1A toxins. Our previous study (11) screened regions of BmABCC2_S that are responsible for its receptor activity for Cry1Aa using a cell-swelling assay with BmABCC2_S mutants. Deletion mutant analyses suggested that BmABCC2_S ECL 1 (120AELLSYWSVEAT131), including 129EAT131, and ECL 4 (770DYWLSFWTN778) are important regions to exhibit the receptor activity for Cry1Aa, whereas alanine-scanning analyses suggested that only 770DYWL773 in ECL 4 affects the receptor activity (11). Overall, to know the precise toxin-binding sites in ABCC2 and ABCC3, binding analysis using alanine-substituted ABCC transporter mutants is further required at least. On the other hand, ECL 3 is unlikely to interact directly with Cry1Aa, as three alanine substitutions in BmABCC2_S ECL 3 did not affect the receptor function of BmABCC2_S with respect to Cry1Aa (11). However, the substitution in ECL 3 (363YIS365) unexpectedly resulted in a decrease of binding affinities to Cry1A toxins (Table 2), suggesting that the substitution indirectly affected binding affinities in the same scenario as the tyrosine insertion in ECL 2 of BmABCC2_R (Table 1). Our data suggest that binding affinities do not always correlate with the higher receptor activities of the mutants, and there should be other factor(s) that confer high receptor activities. For example, the substitution might have removed a factor of steric hindrance that restricted the mode of action of Cry1A toxins after binding to ABCC3, such as toxin insertion into the cell membrane.
This study used BmABCC3 ECL mutants to investigate whether the ECLs of ABCC transporters are responsible for determining Cry toxin specificities. However, data using site-directed mutants cannot adequately clarify which ECLs constitute the proper interaction sites of BmABCC2 and BmABCC3 to Cry1A toxins. Structural analyses of the insect ABC transporters and Cry toxin complexes using methods such as X-ray diffraction might be informative with regard to the receptor–toxin interaction and the mechanism of Cry toxin–induced pore formation in ABC transporters. Our results suggested candidate structures for co-crystallization analysis with Cry1A toxins.
Experimental procedures
Cry toxin preparation
Cry1Aa, Cry1Ab, Cry1Da, and Cry3Bb toxins were produced as recombinant proteins from Escherichia coli as described previously (4, 10). Cry1Ca toxin was produced by a B. thuringiensis recombinant stain (19). The inclusion bodies were solubilized and activated as described elsewhere (6). The toxin concentration was determined by densitometry using Alpha-DigiDocTM (Alpha Innotech, San Leandro, CA).
cDNA cloning of BmABCC2 and BmABCC3
Bombyx mori hybrid strain Kinshu × Showa was purchased from Ueda Sanshu Ltd. (Ueda, Japan). Total RNA was isolated from larval midgut tissue using ISOGEN II (NIPPON GENE, Tokyo, Japan) and used for cDNA synthesis as template. The cDNAs were synthesized using ReverTra Ace® (TOYOBO, Osaka, Japan). BmABCC2_S cDNA already cloned in our previous study (4) was amplified by PCR. BmABCC3 cDNA sequence (AK378482.1) was obtained from the full-length cDNA clone database in KAIKObase (20). The amplified cDNAs using primer sets shown in Table S1 were cloned into the EcoRV site of pcDNA3.1 vector (Thermo Fisher Scientific, Tokyo, Japan) using GeneArt® Seamless Cloning and Assembly Enzyme Mix (Thermo Fisher Scientific). Enhanced GFP was introduced downstream of BmABCC2 and BmABCC3. Constructs of BmABCC3 ECL mutants were prepared by site-directed mutagenesis using inverse PCR. Primers are shown in Table S1.
Expression of BmABCC2 and BmABCC3 in HEK293T cells and cell swelling assay with Cry toxins
HEK293T cells were cultured and transfected as described previously (10). Briefly, HEK293T cells were transfected with the vectors constructed in the previous section using Opti-MEM® (Thermo Fisher Scientific) containing polyethyleneimine (PEI Max, Polysciences, Inc., Warrington, PA) for 2 h. The media were changed to a fresh Dulbecco's modified Eagle's medium, and the cells were incubated for 48 h at 37 °C. After incubation, the coverglass was placed on a slide glass (Matsunami Glass, Osaka, Japan) filled with activated Cry toxin solution diluted in Hanks' buffered saline solution (HBSS; 137 mm NaCl, 5.4 mm KCl, 0.3 mm Na2HPO4, 0.4 mm KH2PO4, 4.2 mm NaHCO3, 1.3 mm CaCl2, 0.5 mm MgCl2, and 0.4 mm MgSO4, pH 7.4). After 60 min of incubation in a CO2 incubator at 37 °C, the cells were observed under phase-contrast microscopy. To compare expression levels of ABC transporters in transfected HEK293T cells, cells were fixed with 4% formaldehyde/HBSS for 10 min and permeabilized with 0.1% Triton X-100/HBSS for 10 min and subsequently incubated with 1 μg/ml DAPI for 5 min. Intensity of fluorescence emitted from EGFP and DAPI was measured using the MetaView® imaging system (Universal Imaging Co., Westchester, PA) under microscopy BX53 (Olympus, Tokyo, Japan). The release of LDH was detected as described elsewhere (10). In short, Cells expressing BmABCC2_S, BmABCC3, or BmABCC3 mutants seeded on a 96-well plate were treated with Cry toxins for 1 h. The LDH release was measured using an LDH Cytotoxicity Detection Kit (Takara Bio, Shiga, Japan).
Quantitative RT-PCR
Total RNA of midgut from each instar B. mori larvae was isolated with ISOGEN II (NIPPON GENE, Tokyo, Japan). The single strand cDNA was prepared using the PrimeScriptTM RT reagent Kit (TaKaRa bio), and the concentrations were determined using Nanodrop ND-1000 (Thermo Fisher Scientific). Quantitative RT-PCR was performed using SYBR® Premix Ex TaqTM II (TaKaRa bio) on a StepOnePlusTM (Applied Biosystems, CA). Quantitative PCRs were performed in technical triplicate. The relative expression levels were calculated using ΔΔCt and normalized using B. mori actin A3. The primers were shown in Table S1.
Preparation of BmABCC proteins using a baculovirus expression system
BmABCC proteins for SPR analysis were produced in Sf9 cells using a baculovirus expression system. To generate transfer vectors for recombinant Autographa californica nuclear polyhedrosis viruses (AcNPV), including AcNPV-BmABCC2_S-FLAG, AcNPV-BmABCC2_R-FLAG, and AcNPV-BmABCC3-FLAG, cDNAs of silkworm ABC transporters were amplified by PCR and cloned into pBac4x-1 vector (Merck Millipore) in which EGFP was previously introduced as an expression marker downstream of a polyhedrin promoter (4) different from that of BmABCC cDNAs. 3× FLAG tag was introduced downstream of BmABCC cDNAs using overlap extension PCR or the same restriction site of pBAC4x-1 vector. Primers were shown in Table S1. Recombinant AcNPVs were generated using the BacMagicTM kit (Merck Millipore) according to the manufacturer's instructions. Sf9 cells were infected with the recombinant virus and cultured for 3 days. An approximately 2-liter culture of Sf9 cells expressing BmABCC-FLAG was used for preparing a membrane fraction that was subsequently solubilized with n-dodecyl-β-d-maltoside (Dojindo, Tokyo, Japan) and purified using anti-FLAG® M2 Affinity Gel (Sigma-Aldrich) as described elsewhere (6). With regard to WT BmABCC2 protein, the same lot as purified BmABCC2-FLAG reported by Adegawa et al. (6) was used for SPR analysis. Purity was analyzed by silver staining (silver staining kit, APRO Science, Tokushima, Japan) and Western blotting using monoclonal ANTI-FLAG M2 antibody (Sigma-Aldrich).
SPR analysis
Partially purified ABCC transporters and mutants were immobilized on a CM5 sensor chip (GE Healthcare, Chalfont, UK) using the amine-coupling method. Measurement and analysis of binding kinetics were performed using Biacore J equipped with BIAevaluation version 4.0 (GE Healthcare) as described previously (6).
Alignment of amino acid sequences and transmembrane topology prediction
The amino sequence alignment was carried out using CLC sequence viewer version 7.5 (CLC bio, Aarhus, Denmark). Transmembrane topology of BmABCC2_S was predicted by Phobius (http://phobius.sbc.su.se/)4 (21).
Author contributions
H. E. and R. S. conceived of and designed the experiments, analyzed the data, and wrote the manuscript. H. E., S. T., S. A., and F. I. performed the experiments. H. T. and S. K. contributed reagents, materials, and analysis tools.
Supplementary Material
Acknowledgments
We thank anonymous reviewers for their insightful suggestions and comments.
This research was financially supported by Japan Society for the Promotion of Science (JSPS) KAKENHI Grants 15H02837 (to R. S.) and 15J08610 (to H. E.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Tables S1–S3 and Figs. S1–S3.
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.
- ABC
- ATP-binding cassette
- ABCC2
- ABC transporter subfamily C2
- ABCC3
- ABC transporter subfamily C3
- ECL
- extracellular loop
- EGFP
- enhanced green fluorescent protein
- DAPI
- 4′,6-diamidino-2-phenylindole
- SPR
- surface plasmon resonance
- AcNPV
- A. californica nuclear polyhedrosis viruses
- HEK293T
- human embryonic kidney 293 T
- LDH
- lactate dehydrogenase
- HBSS
- Hanks' buffered saline solution.
References
- 1. Pigott C. R., and Ellar D. J. (2007) Role of receptors in Bacillus thuringiensis crystal toxin activity. Microbiol. Mol. Biol. Rev. 71, 255–281 10.1128/MMBR.00034-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bravo A., Likitvivatanavong S., Gill S. S., and Soberón M. (2011) Bacillus thuringiensis: a story of a successful bioinsecticide. Insect Biochem. Mol. Biol. 41, 423–431 10.1016/j.ibmb.2011.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Heckel D. G. (2012) Learning the ABCs of Bt: ABC transporters and insect resistance to Bacillus thuringiensis provide clues to a crucial step in toxin mode of action. Pesticide Biochem. Physiol. 104, 103–110 10.1016/j.pestbp.2012.05.007 [DOI] [Google Scholar]
- 4. Tanaka S., Miyamoto K., Noda H., Jurat-Fuentes J. L., Yoshizawa Y., Endo H., and Sato R. (2013) The ATP-binding cassette transporter subfamily C member 2 in Bombyx mori larvae is a functional receptor for Cry toxins from Bacillus thuringiensis. FEBS J. 280, 1782–1794 10.1111/febs.12200 [DOI] [PubMed] [Google Scholar]
- 5. Tanaka S., Endo H., Adegawa S., Kikuta S., and Sato R. (2016) Functional characterization of Bacillus thuringiensis Cry toxin receptors explain resistance in insects. FEBS J. 283, 4474–4490 10.1111/febs.13952 [DOI] [PubMed] [Google Scholar]
- 6. Adegawa S., Nakama Y., Endo H., Shinkawa N., Kikuta S., and Sato R. (2017) The domain II loops of Bacillus thuringiensis Cry1Aa form an overlapping interaction site for two Bombyx mori larvae functional receptors, ABC transporter C2 and cadherin-like receptor. Biochim. Biophys. Acta 1865, 220–231 10.1016/j.bbapap.2016.11.011 [DOI] [PubMed] [Google Scholar]
- 7. Park Y., González-Martínez R. M., Navarro-Cerrillo G., Chakroun M., Kim Y., Ziarsolo P., Blanca J., Cañizares J., Ferré J., and Herrero S. (2014) ABCC transporters mediate insect resistance to multiple Bt toxins revealed by bulk segregant analysis. BMC Biol. 12, 46 10.1186/1741-7007-12-46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chen Z., He F., Xiao Y., Liu C., Li J., Yang Y., Ai H., Peng J., Hong H., and Liu K. (2015) Endogenous expression of a Bt toxin receptor in the Cry1Ac-susceptible insect cell line and its synergistic effect with cadherin on cytotoxicity of activated Cry1Ac. Insect Biochem. Mol. Biol. 59, 1–17 10.1016/j.ibmb.2015.01.014 [DOI] [PubMed] [Google Scholar]
- 9. Guo Z., Kang S., Chen D., Wu Q., Wang S., Xie W., Zhu X., Baxter S. W., Zhou X., Jurat-Fuentes J. L., and Zhang Y. (2015) MAPK signaling pathway alters expression of midgut ALP and ABCC genes and causes resistance to Bacillus thuringiensis Cry1Ac toxin in diamondback moth. PLoS Genet. 11, e1005124 10.1371/journal.pgen.1005124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Endo H., Tanaka S., Imamura K., Adegawa S., Kikuta S., and Sato R. (2017) Cry toxin specificities of insect ABCC transporters closely related to lepidopteran ABCC2 transporters. Peptides 98, 86–92 10.1016/j.peptides.2017.04.003 [DOI] [PubMed] [Google Scholar]
- 11. Tanaka S., Endo H., Adegawa S., Iizuka A., Imamura K., Kikuta S., and Sato R. (2017) Bombyx mori ABC transporter C2 structures responsible for the receptor function of Bacillus thuringiensis Cry1Aa toxin. Insect Biochem. Mol. Biol. 91, 44–54 10.1016/j.ibmb.2017.11.002 [DOI] [PubMed] [Google Scholar]
- 12. Atsumi S., Miyamoto K., Yamamoto K., Narukawa J., Kawai S., Sezutsu H., Kobayashi I., Uchino K., Tamura T., Mita K., Kadono-Okuda K., Wada S., Kanda K., Goldsmith M. R., and Noda H. (2012) Single amino acid mutation in an ATP-binding cassette transporter gene causes resistance to Bt toxin Cry1Ab in the silkworm, Bombyx mori. Proc. Natl. Acad. Sci. U.S.A. 109, E1591–E1598 10.1073/pnas.1120698109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tanaka S., Miyamoto K., Noda H., Endo H., Kikuta S., and Sato R. (2016) Single amino acid insertions in extracellular loop 2 of Bombyx mori ABCC2 disrupt its receptor function for Bacillus thuringiensis Cry1Ab and Cry1Ac but not Cry1Aa toxins. Peptides 78, 99–108 10.1016/j.peptides.2016.01.006 [DOI] [PubMed] [Google Scholar]
- 14. Endo H., Azuma M., Adegawa S., Kikuta S., and Sato R. (2017) Water influx via aquaporin directly determines necrotic cell death induced by the Bacillus thuringiensis Cry toxin. FEBS Lett. 591, 56–64 10.1002/1873-3468.12506 [DOI] [PubMed] [Google Scholar]
- 15. Obata F., Kitami M., Inoue Y., Atsumi S., Yoshizawa Y., and Sato R. (2009) Analysis of the region for receptor binding and triggering of oligomerization on Bacillus thuringiensis Cry1Aa toxin. FEBS J. 276, 5949–5959 10.1111/j.1742-4658.2009.07275.x [DOI] [PubMed] [Google Scholar]
- 16. Bretschneider A., Heckel D. G., and Pauchet Y. (2016) Three toxins, two receptors, one mechanism: mode of action of Cry1A toxins from Bacillus thuringiensis in Heliothis virescens. Insect Biochem. Mol. Biol. 76, 109–117 10.1016/j.ibmb.2016.07.008 [DOI] [PubMed] [Google Scholar]
- 17. Tabashnik B. E., Malvar T., Liu Y. B., Finson N., Borthakur D., Shin B. S., Park S. H., Masson L., de Maagd R. A., and Bosch D. (1996) Cross-resistance of the diamondback moth indicates altered interactions with domain II of Bacillus thuringiensis toxins. Appl. Environ. Microbiol. 62, 2839–2844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhao J. Z., Li Y. X., Collins H. L., Cao J., Earle E. D., and Shelton A. M. (2001) Different cross-resistance patterns in the diamondback moth (Lepidoptera: Plutellidae) resistant to Bacillus thuringiensis toxin Cry1C. J. Econ. Entomol. 94, 1547–1552 10.1603/0022-0493-94.6.1547 [DOI] [PubMed] [Google Scholar]
- 19. Masson L., Mazza A., Sangadala S., Adang M. J., and Brousseau R. (2002) Polydispersity of Bacillus thuringiensis Cry1 toxins in solution and its effect on receptor binding kinetics. Biochim. Biophys. Acta 1594, 266–275 10.1016/S0167-4838(01)00312-0 [DOI] [PubMed] [Google Scholar]
- 20. Suetsugu Y., Futahashi R., Kanamori H., Kadono-Okuda K., Sasanuma S., Narukawa J., Ajimura M., Jouraku A., Namiki N., Shimomura M., Sezutsu H., Osanai-Futahashi M., Suzuki M. G., Daimon T., Shinoda T., et al. (2013) Large scale full-length cDNA sequencing reveals a unique genomic landscape in a lepidopteran model insect, Bombyx mori. G3 3, 1481–1492 10.1534/g3.113.006239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Käll L., Krogh A., and Sonnhammer E. L. L. (2007) Advantages of combined transmembrane topology and signal peptide prediction–the Phobius web server. Nucleic Acids Res. 35, W429–W432 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


