Abstract
Helicase-like transcription factor (HLTF) is a central mediator of the DNA damage response and maintains genome stability by regressing stalled replication forks. The N-terminal HIRAN domain binds specifically to the 3′-end of single-stranded DNA (ssDNA), and disrupting this function interferes with fork regression in vitro as well as replication fork progression in cells under replication stress. Here, we investigated the mechanism by which the HIRAN-ssDNA interaction facilitates fork remodeling. Our results indicated that HIRAN capture of a denatured nascent leading 3′-end directs specific binding of HLTF to forks. DNase footprinting revealed that HLTF binds to the parental duplex ahead of the fork and at the leading edge behind the fork. Moreover, we found that the HIRAN domain is important for initiating regression of forks when both nascent strands are at the junction, but is dispensable when forks contain ssDNA regions on either template strand. We also found that HLTF catalyzes fork restoration from a partially regressed structure in a HIRAN-dependent manner. Thus, HIRAN serves as a substrate-recognition domain to properly orient the ATPase motor domain at stalled and regressed forks and initiates fork remodeling by guiding formation of a four-way junction. We discuss how these activities compare with those of two related fork remodelers, SWI/SNF-related, matrix-associated, actin-dependent regulator of chromatin, subfamily A–like 1 (SMARCAL1) and zinc finger RANBP2 type–containing 3 (ZRANB3) to provide insight into their nonredundant roles in DNA damage tolerance.
Keywords: DNA replication, DNA repair, DNA damage response, DNA helicase, nucleic acid enzymology, DNA damage response, DNA repair, fork reversal, replication restart, stalled replication fork
Introduction
The accurate and complete replication of DNA is crucial for maintaining genomic stability and for cell survival. Multiple forms of replication stress, including DNA damage, difficult to replicate sequences, DNA secondary structures, and protein-DNA and RNA-DNA complexes, inhibit progression of the replication fork (1). Polymerase stalling can cause uncoupling of DNA synthesis and unwinding activities (2), leading to an accumulation of single-stranded DNA (ssDNA)2 that is vulnerable to nuclease cleavage or formation of aberrant DNA structures (3). Stalled replication and a failure of the cell to respond to replication stress leads to genomic instability, chromosomal rearrangements, mutations, cell death, and a number of human diseases (1). As a way to avoid the genomic instability associated with fork stalling and arrest, cells possess DNA damage tolerance (DDT) pathways that maintain fork progression, facilitate replication restart, and promote the completion of DNA replication (4–6). Translesion synthesis (TLS) involves specialized DNA polymerases that bypass DNA lesions in an error-prone manner, whereas the template switching pathway relies on an undamaged template for error-free synthesis past a replication blockage.
One template switching mechanism by which stalled forks may be rescued or avoided is fork regression (also known as fork reversal), in which the two parental strands are re-annealed and the two nascent daughter strands are unpaired from their templates and repaired together to form a four-way junction (Fig. 1A). Fork reversal has been observed as a response to Top1 poisoning in yeast, mouse, and human cells, to replication stress caused by a prolonged S-phase in mouse embryonic stem cells, and as a response to other forms of replication stress (7–12). Reversed forks are important replication intermediates that maintain genomic integrity in several ways (12). In addition to initiating template switching to allow for error-free DNA synthesis using the undamaged sister chromatid, fork regression may limit the accumulation of vulnerable DNA structures generated by polymerase stalling. Additionally, fork reversal may facilitate DNA repair through recombination pathways or by sequestering the lesion back into the context of duplex DNA.
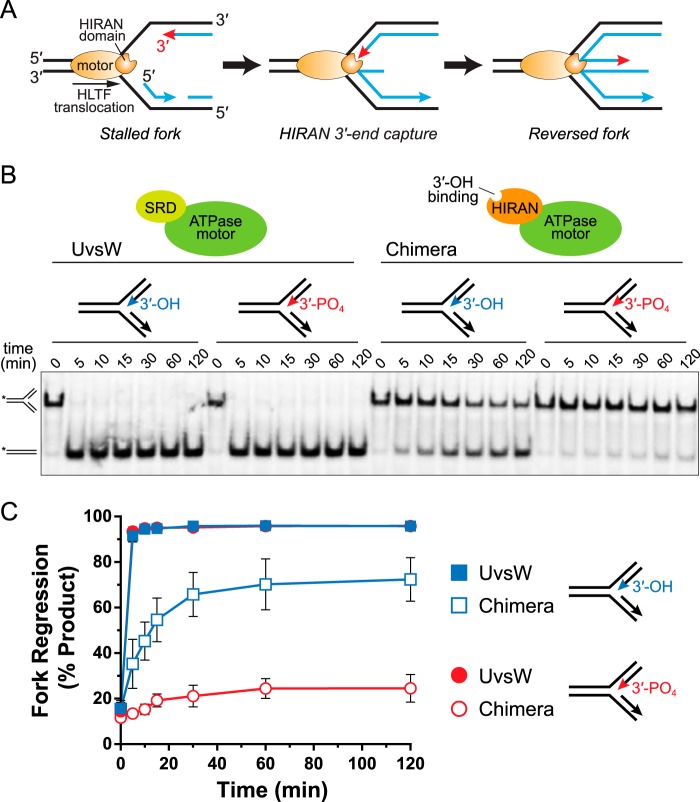
Figure 1.
HIRAN interacts with the nascent 3′-end to drive fork regression. A, model of fork regression by HLTF. B, representative native gel showing time-dependent fork regression by WT UvsW and a HIRAN-UvsW chimera against model fork substrates containing either a native 3′-OH or capped 3′-PO4 on the nascent leading strand. 32P labels are indicated by asterisks. SRD, substrate recognition domain. C, quantitation of data from three independent experiments (mean ± S.D.).
Helicase-like transcription factor (HLTF) serves to promote DDT in mammalian cells (13–15). Inactivation of HLTF leads to increased UV and MMS sensitivity in cells and alters the progression of replicating forks under replication stress (15, 16). Like its yeast homologue Rad5, HLTF functions as a RING E3 ubiquitin ligase that works together with Rad18 and Mms2-Ubc13 complexes to polyubiquitinate proliferating cell nuclear antigen (PCNA), activating the template switching pathway (14, 17). HLTF contains a DNA-dependent ATPase motor domain related to the SWI/SNF2-family chromatin remodelers that enables translocation on dsDNA and regression of model replication forks in vitro (16). HLTF's dsDNA translocation activity can displace proteins that might be found at a stalled replication fork from DNA (16, 18). HLTF has also been reported to form D-loops without the assistance of Rad51 in vitro (18). Both the ubiquitin ligase and ATPase activities of HLTF and Rad5 are required for DDT (16, 19–21).
Two other SNF2-related DNA translocases—SMARCAL1 and ZRANB3—have been identified to catalyze fork regression in vitro and to be important for DDT and maintenance of genomic stability (12, 22–32). Inactivation of these proteins leads to a sensitivity to genotoxic agents, as well as defective fork restart, increased double-strand breaks, and sister chromatid exchange events in response to replication stress (23–25, 28, 30, 31). Outside their ATPase motor domains, HLTF, SMARCAL1, and ZRANB3 are distinguished by substrate recognition domains (SRDs) that impart specificity for a particular DNA structure found at a stalled fork (32–35). Despite the importance of these enzymes to DDT, the mechanisms by which their motors and SRDs catalyze fork remodeling and how this activity is tied to their cellular roles are not well understood.
HLTF contains a HIRAN (HIP116, Rad5p, N-terminal) domain that serves as the SRD through its ability to bind specifically to the 3′-end of ssDNA (33, 36, 37). Crystal structures of HIRAN bound to DNA showed that HIRAN adopts a modified oligonucleotide/oligosaccharide-fold that clamps the two 3′-terminal nucleobases and forms a binding pocket for the 3′-hydroxyl group. Mutation of the interacting residues impaired DNA binding by HIRAN in solution (33, 36, 37). Importantly, site-directed incorporation of these mutations into full-length HLTF impaired in vitro fork regression but not ATPase, ubiquitin ligase, or translocation activities, and also prevented complementation of replication fork progression and UV sensitivity defects in HLTF-depleted cells (33, 37). Moreover, deletion of HIRAN from HLTF recapitulates these defects. Depletion of the HLTF ortholog from human cells, Arabidopsis (Rad5A), and Schizosaccharomyces pombe (Rad8) caused sensitivity to DNA-damaging agents and defects in DNA replication and cell growth, none of which were complemented by ΔHIRAN mutants (37–39). Thus, HIRAN's DNA 3′-end–binding activity is essential to HLTF function.
Despite its importance to DDT, the mechanism by which HLTF remodels stalled forks, and the roles of HIRAN and motor domains in this process, is unknown. From our previous mutational analysis of HIRAN function in fork reversal and replication fork progression, we proposed a model in which recognition and remodeling of stalled forks by HLTF is accomplished through the combined activities of the dsDNA translocase motor and HIRAN 3′-end–binding domains (33). In this model, HLTF facilitates reannealing of parental strands by binding and translocating along the unreplicated template toward the stalled fork, eventually destabilizing the nascent duplexes, after which capture of the destabilized 3′-end of the nascent leading strand by the HIRAN domain would facilitate efficient unwinding of the leading arm and/or annealing of the nascent strands (Fig. 1A). This model has not been tested and it is not yet clear how HLTF engages a fork to catalyze fork remodeling. Specifically, we do not know how HLTF is positioned at a stalled replication fork, how the protein deals with ssDNA gaps that would be present on leading and lagging strands of stalled and normal forks, or the ability of the HIRAN domain to denature the 3′-end of the nascent leading strand. We tested these aspects of our model biochemically using HIRAN deletion mutants previously shown to impair HLTF function in cells (37–39) to better understand how the HIRAN and ATPase domains engage fork structures to facilitate efficient fork regression.
Results
HIRAN specifically interacts with the 3′-end of the nascent leading strand
We previously showed that efficient fork regression by HLTF depends on a functional HIRAN domain and on a free 3′-hydroxyl group on the nascent leading strand of a model replication fork, which suggested a direct interaction between HIRAN and the 3′-end of the nascent leading strand (Fig. 1A) (33). Specifically, HLTF fork regression activity is reduced either by amino acid substitutions within the DNA-binding cleft of HIRAN or by phosphorylation of the 3′-end of the nascent leading strand, both of which inhibit ssDNA binding by the HIRAN domain. Moreover, this reduction in fork regression when blocking the leading 3′-end is HLTF specific, as a 3′-phosphorylated substrate did not impair fork reversal by SMARCAL1 or T4 phage UvsW, which share a similar SRD unrelated to HIRAN (33). To verify that a direct interaction between HIRAN and the 3′-end of the nascent leading strand is responsible for the reduced activity of HLTF against a 3′-blocked substrate, we constructed a HIRAN-UvsWmotor chimera protein by replacing UvsW's N-terminal SRD with HLTF's HIRAN domain (Fig. S1) and tested fork regression activity against substrates containing 3′-OH (unblocked) and 3′-PO4 (blocked) nascent leading strands (Fig. 1B). In this assay, we monitor regression as a time-dependent formation of annealed template strands from a homologous fork containing a mismatch at the junction to prevent spontaneous regression. We previously showed that removal of UvsW's SRD from the motor domain abrogates fork regression activity, and that this activity can be partially restored by fusion of the structurally homologous SRD from SMARCAL1 onto the UvsW motor (34). Similarly, the HIRAN-UvsWmotor chimera showed only a modest reduction in activity against an unblocked substrate (Fig. 1, B and C), indicating that HIRAN acts independently of the motor domain to facilitate fork regression. However, whereas 3′-phosphorylation of the nascent leading strand had no effect on WT UvsW, blocking the 3′-end abrogated fork regression activity of the HIRAN-UvsWmotor chimera (Fig. 1, B and C). We therefore conclude that HIRAN serves as the SRD of HLTF through a specific interaction with the 3′-end of the nascent leading strand.
HIRAN captures unpaired 3′-ends from duplex DNA
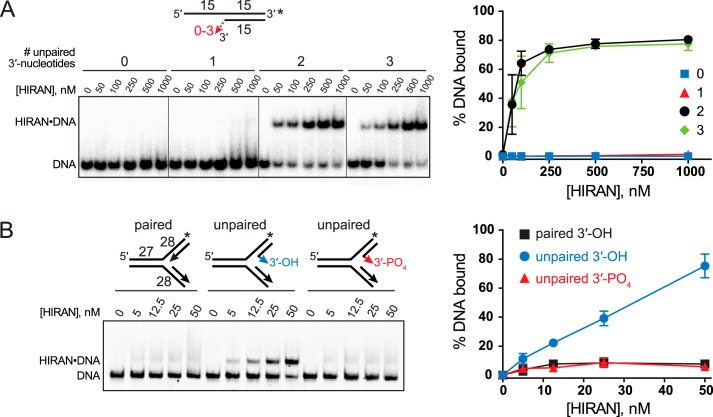
Crystal structures of HIRAN showed that the protein clamps two stacked nucleobases from the 3′-terminal nucleotides in a conformation that precludes base pairing to a complementary strand (33, 36, 37). This specificity for ssDNA and HIRAN's association with the leading end of the nascent strand prompted us to explore whether HIRAN has the ability to denature the 3′-end on its own, or whether it facilitates fork regression by capturing the 3′-end of an already denatured duplex. We tested the ability of HIRAN to bind to the 3′-end in the context of dsDNA using an electrophoretic mobility shift assay (EMSA). Purified HIRAN domain was added to DNA substrates containing 0, 1, 2, or 3 unpaired 3′-nucleotides across from a 5′-overhang (Fig. 2A). The 3′-end of the overhang strand was capped with a fluorescein molecule to prevent any spurious binding there (33). Consistent with the crystal structures, binding was only observed to DNA that contained at least two unpaired 3′-nucleotides. No binding was observed when the 3′-end was fully base paired or frayed by only one nucleotide (Fig. 2A), indicating that the HIRAN-DNA–binding energy is not sufficient to denature the 3′-end away from the template strand.
Figure 2.
HIRAN binds unpaired DNA ends in the context of duplex DNA. A and B, EMSAs of purified HIRAN domain binding to DNA containing zero, one, two, or three unpaired 3′-nucleotides within dsDNA (A) or to DNA forks that contain either fully paired or two unpaired nucleotides at the 3′-end of the nascent leading strand (B). Numbers with the DNA schematics refer to the number of nucleotides, and asterisks represent the location of fluorescein tags. Quantitation of three experiments (mean ± S.D.) is shown to the right of a representative gel.
We next examined the ability of HIRAN to bind to DNA ends in the context of a DNA fork structure. Because HIRAN cannot access a paired DNA end, we hypothesized that any apparent binding of HIRAN to a fork would occur only if the nascent leading 3′-end were frayed. To test this possibility, we performed an EMSA with purified HIRAN against fork structures containing a nascent leading strand that was either fully base paired or that contained two unpaired nucleotides at the 3′-end. Consistent with our previous results, binding was observed only to the fork with a frayed leading strand, and this binding was abolished by phosphorylation of the frayed end (Fig. 2B). These data confirm that HIRAN only recognizes an unpaired 3′-end, even in the context of a fork. Moreover, we conclude that HIRAN has no affinity for the structure of the fork itself, inconsistent with a previous report (37). The binding to fork structures used in that study may have resulted from partially denatured or frayed fork substrates.
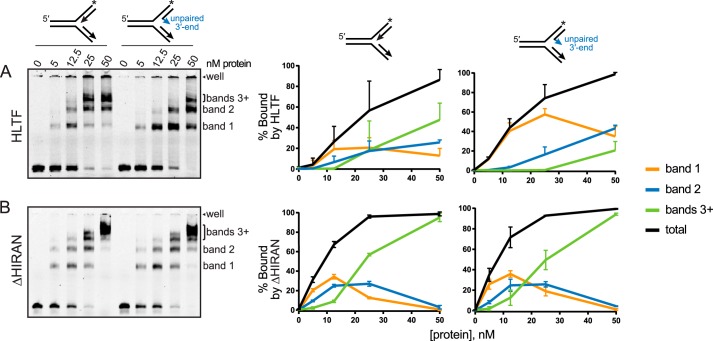
HIRAN enforces a specificity to HLTF at forks containing a frayed nascent leading strand
HLTF was previously shown to bind to various DNA junction structures with similar affinity, suggesting that the full-length protein lacks specificity for a particular fork (16). However, we hypothesized that the interaction between HIRAN and the 3′-end of a frayed nascent leading strand would impose a specificity to HLTF. We tested this by comparing binding of HLTF and a deletion mutant lacking the HIRAN domain (ΔHIRAN) to fully paired and partially frayed fork structures (Fig. 3). Consistent with the previous study (16), our EMSAs showed a protein-dependent appearance of three to four discrete bands corresponding to multiple protein-DNA complexes, likely containing one or more proteins bound per DNA because the dsDNA translocase motor domain would be capable of binding any of the three arms. However, the presence of the frayed nascent leading 3′-end resulted in a significant accumulation in the highest mobility complex (band 1) and a concomitant decrease in the lowest mobility complex (bands 3+) for the full-length protein (Fig. 3A), suggesting that this structural feature favors a stoichiometric HLTF-DNA complex. This increase in band 1 was not observed from the ΔHIRAN construct (Fig. 3B), indicating that the HIRAN domain imparts a specificity to HLTF for the 3′-end of the nascent leading strand in these model replication forks. Consistent with this conclusion, removing the HIRAN domain resulted in an accumulation of the lower mobility complexes (bands 3+) relative to the others (Fig. 3B).
Figure 3.
Effect of HIRAN on HLTF-fork binding. A and B, EMSAs of HLTF (A) or ΔHIRAN (B) binding to 25 nm DNA forks containing fully paired or two unpaired nucleotides at the 3′-end of the nascent leading strand. Bands representing discrete protein-DNA complexes are labeled to the right of each gel. Quantitation of the percentage of each shifted band and the total from three experiments (mean ± S.D.) is shown to the right of a representative gel. The DNA remaining in the well (topmost band of each gel) was not included in the quantification. Asterisks represent the location of fluorescein tags.
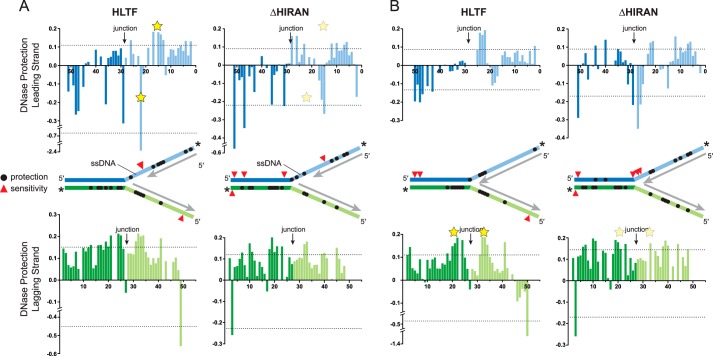
HLTF binds to the parental duplex ahead of the fork and tracks with the 3′ leading end
Our model of fork regression by HLTF postulates that the motor domain translocates on the unreplicated parental duplex toward the fork whereas the HIRAN domain engages a displaced 3′-end of the nascent leading strand (Fig. 1A). To probe how HLTF is positioned at a DNA fork, we performed DNase footprinting on immobile forks containing two unpaired nucleotides at the 3′-end of the nascent leading strand to promote the stoichiometric 1:1 complex (Fig. 4 and Fig. S2). We tested both full-length HLTF and the HLTF-ΔHIRAN mutant against two forks that differed by the presence (Fig. 4A) or absence (Fig. 4B) of a 10-nucleotide ssDNA gap on the leading template to alter the position of the nascent 3′-end with respect to the junction. Full-length HLTF showed a virtually identical pattern of nuclease protection on the lagging templates across the junctions of both forks that spanned 8–17 nucleotides along the parental duplexes and 4–5 nucleotides on the unwound arms behind the junction (Fig. 4, A and B). There was no protection of the leading strand of the parental duplex ahead of either fork, but instead was moderately sensitized to nuclease cleavage upon addition of HLTF. Deletion of the HIRAN domain delocalized the protection on the lagging template away from the junction on both gapped and nongapped forks (Fig. 4, A and B). Moreover, the ΔHIRAN mutant modestly protected the leading strand upstream of the nongapped fork (Fig. 4B), providing additional evidence that the motor has reduced specificity for this fork without the HIRAN domain.
Figure 4.
DNase footprint of HLTF bound to model forks. A and B, quantitation of HLTF (WT and ΔHIRAN mutant) footprinting on gapped (A) and nongapped (B) fork substrates. Bar graphs plot the amount of nuclease protection (positive value) or sensitivity (negative value) as a function of nucleotide position relative to the fluorescein label along the leading (blue) or lagging (green) strand. Dashed lines signify 0.5 S.D. from the mean protection/sensitivity. Yellow stars indicate significant differences in protection between HLTF and ΔHIRAN in each fork. The location of significant protection and sensitivity sites are shown as black circles and red triangles, respectively, on the schematic diagrams. Asterisks indicate the position of the fluorescein labels on each strand, and unlabeled nascent strands are shown in gray. Representative footprinting gels are shown in Fig. S2.
Protection of the leading template behind the junction tracked with the position of the 3′-end of the nascent strand in a HIRAN-dependent manner. On the ssDNA gapped fork, HLTF protection of a four- to five-nucleotide region and strong hypersensitivity of a single nucleotide was observed in the base paired and ssDNA regions, respectively, of the leading template across from the nascent 3′-end (Fig. 4A). Strikingly, this pattern of protection and sensitivity opposite the 3′-end was not evident in the ΔHIRAN mutant, consistent with HIRAN engagement with the frayed 3′-end of the nascent leading strand. When the nascent 3′-end abutted the junction in the nongapped fork, HLTF protection of the leading template coincided with the position of the nascent 3′-end (Fig. 4B). This pattern did not significantly change by deleting the HIRAN domain (Fig. 4B). Interestingly, ΔHIRAN caused nuclease sensitivity directly at the nongapped junction, although the significance of this is unclear considering that a small amount of sensitivity is also evident at the junction of the gapped fork when treated with HLTF and ΔHIRAN proteins (Fig. 4A). Nonetheless, these results are consistent with HLTF positioned on the lagging strand ahead of the fork, and show that engagement of the unwound templates behind the fork corresponds to the position of the stalled nascent strand on the leading arm and to the position of the junction on the lagging arm.
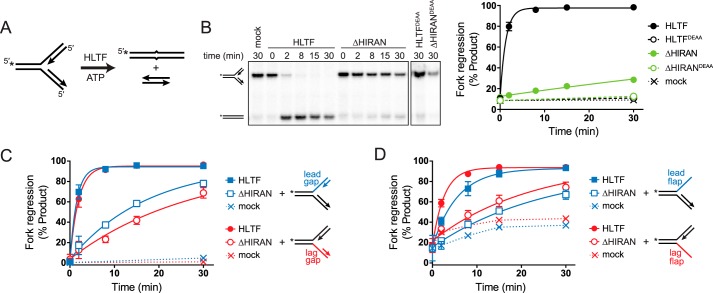
HIRAN facilitates initial formation of the four-way junction
To investigate the manner in which remodeling of the leading strand by the HIRAN domain facilitates fork regression, we compared fork regression activities of HLTF and ΔHIRAN against forks containing ssDNA gaps on either the leading or lagging templates immediately behind the junction (Fig. 5). These substrates model replication forks that have stalled from impediments on the leading or lagging strands, and also provide a way to ascertain the importance of the HIRAN and motor domains in annealing and denaturing parental and nascent strands. The presence of nascent strands annealed right up to the junction present a barrier to initial formation of the four-way junction from the three-way fork (40), and thus moving the ends of the nascent strands from the junction to create an ssDNA region in the template would lower the barrier to this transition. We hypothesized that if the HIRAN domain played a role in denaturing the nascent arm, then deletion of the HIRAN domain would affect regression activity of a fully paired fork more than an ssDNA gapped fork. Indeed, it was previously shown that deletion of the HIRAN domain from HLTF abolished regression of a fully base-paired fork without affecting DNA-dependent ATPase or DNA translocation activities (37). Consistent with those results, under our experimental conditions in a standard regression assay using a model fork with both nascent strands abutted against a junction (Fig. 5A), the ΔHIRAN mutant severely diminished HLTF's robust regression activity (Fig. 5B) without impairing ATPase activity (Fig. S3A). We confirmed that the small amount of residual regression activity present in our purified ΔHIRAN preparation was attributed to the HLTF motor domain, as incorporation of an ATPase-dead D557A/E558A (DEAA) mutation abolished fork regression activity by both HLTF and ΔHIRAN (Fig. 5B and Fig. S3A).
Figure 5.
HIRAN is not required for regression of forks containing ssDNA at the junction. A, schematic of fork regression assay. A model fork substrate containing a two-nucleotide mismatch at the junction to prevent spontaneous branch migration is incubated with HLTF and ATP to yield annealed template and nascent duplexes. 32P labels are indicated by asterisks. B, representative native gel of time-dependent fork regression by HLTF and ΔHIRAN against a fork substrate with fully paired nascent arms. Endpoints of reactions containing ATPase-dead HLTFDEAA or ΔHIRANDEAA proteins, or of a mock reaction with no protein are shown as negative controls. The plot shows quantitation of three experiments (mean ± S.D.). Data were fit by a single exponential function. C and D, quantitation of three time-dependent fork regression reactions (mean ± S.D.) with HLTF and ΔHIRAN using forks in which the leading or lagging nascent strands have been shortened (C) or removed (D). Mock reactions contain no enzyme. Representative gels for panels C and D are shown in Fig. S3.
Incorporation of a 30-nucleotide ssDNA gap on either leading or lagging template did not significantly affect fork regression activity by HLTF as compared with the fully paired fork (Fig. 5C and Fig. S3C). The ΔHIRAN mutant reduced HLTF activity against the gapped fork substrates, but this reduction of activity was strikingly weaker than that of the fully paired fork (Fig. 5C). These data suggest that HIRAN assists in the transition from three-way fork to four-way junction by displacing the nascent strands away from the template. Consistent with this interpretation, we found that the ΔHIRAN mutant had greater activity against a fork containing an unpaired nascent 3′-end as compared with the fully paired fork (Fig. S3B). Interestingly, complete removal of either nascent strand caused a modest (2- to 5-fold) reduction in the rate of regression by full-length HLTF as compared with the four-stranded forks (Fig. 5D and Fig. S3, D and E), indicating that HLTF prefers a four-stranded fork and that the nascent strands are not required for fork regression. Consistent with the result obtained with the gapped substrates, the ΔHIRAN mutant showed only a very modest reduction in activity against these partial forks (Fig. 5D), further supporting a model in which HIRAN's main function is to promote the transition from a three-way to a four-way junction. Removal of the nascent leading strand appeared to have a slightly greater effect than removal of the lagging strand, but this difference can be attributed to the inherent instability of these flap substrates and not to an HLTF-mediated effect because the same effect was observed in the rates of spontaneous fork regression (Fig. 5D).
In the cell, ssDNA present at a stalled fork would be bound by replication protein A (RPA), which interacts with other replication factors to promote restart. SMARCAL1 is recruited to forks through a direct interaction with RPA (23–27). RPA imposes a bias on SMARCAL1 toward forks containing a leading strand gap (41, 42). On the same gapped substrates used in Fig. 5C, SMARCAL1 activity was stimulated when RPA was bound to the leading strand gap and inhibited by RPA on the lagging strand. In contrast, we found that RPA binding to these ssDNA regions modestly inhibited regression by HLTF, consistent with a previous study (42), without imposing a specificity for a leading or lagging gapped fork (Fig. S4A). These data suggest that HLTF and SMARCAL1 use different mechanisms for fork recognition and remodeling.
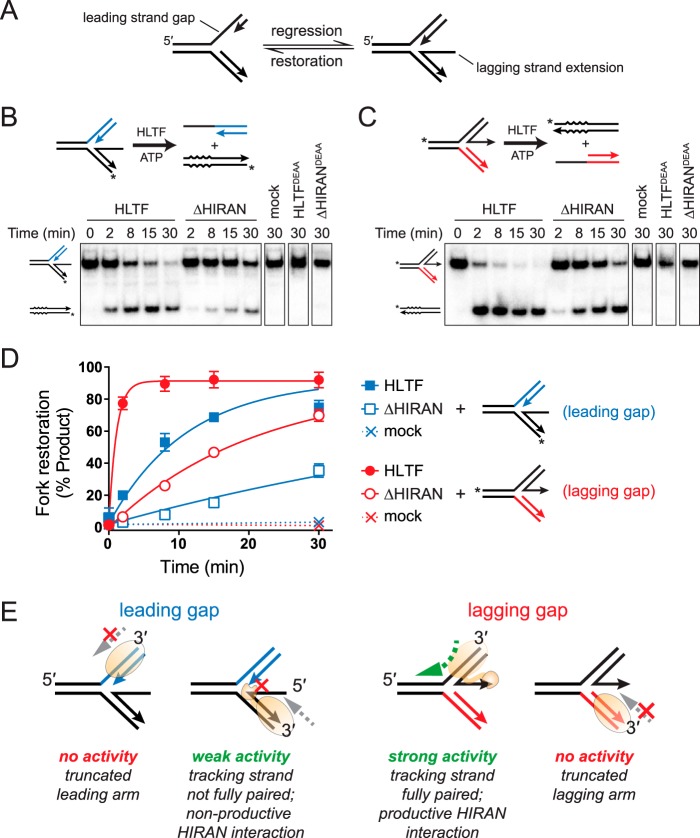
HLTF catalyzes fork restoration
In addition to their ability to regress stalled forks, SMARCAL1 and ZRANB3 have been shown to catalyze fork restoration in vitro, whereby a partially regressed fork is pushed back toward the three-way replication fork structure (Fig. 6A) (41). We tested whether HLTF also possesses this activity using the previously established assay with the same substrates used for SMARCAL1 and ZRANB3 (41). In this assay, partially regressed fork substrates containing either nascent leading or lagging strand ssDNA extensions are used to resemble regressed forks with leading or lagging strand gaps, respectively (Fig. 6A). We found that HLTF catalyzed ATP-dependent restoration of both forks, with significantly greater activity on the fork containing an extended leading strand (Fig. 6, B–D). Under the assumption that HLTF drives restoration by translocating 3′-5′ (16) toward the fork on either leading or lagging arm, we would predict restoration of the lagging gap substrate to be most productive because translocation along the fully base-paired leading arm would keep the protein engaged to dsDNA throughout the annealing reaction (Fig. 6E). In contrast, the weaker activity of the leading gap substrate can be explained by the fact that a protein tracking 3′-5′ along the nascent lagging stand would eventually encounter the ssDNA tail, which would impede dsDNA translocation until it becomes annealed. We also found that the ΔHIRAN mutant reduced restoration of both substrates, indicating that HIRAN plays a role in this process (Fig. 6, B–D). The residual activity of the ΔHIRAN mutant further indicates that HIRAN is not essential to branch migration. Although the molecular basis for the effect of HIRAN on the restoration reaction is not entirely clear, we speculate that HIRAN interaction with the nascent leading 3′-end stimulates annealing of the leading arm and inhibits annealing of the lagging arm (Fig. 6E). Consistent with this, occluding access to the nascent leading 3′-end by binding of RPA inhibited restoration of the lagging gap fork, whereas RPA binding to the 5′ ssDNA lagging strand extension had no effect on restoration of the leading gap fork (Fig. S4B).
Figure 6.
HLTF catalyzes fork restoration. A, schematic of fork restoration. B and C, restoration of partially regressed fork substrates containing ssDNA tails on the lagging (B) or leading (C) strands. Assay reaction schematics are shown above each representative gel. Restoration activity is defined by annealing of the ssDNA tail to the complementary parental strand (which contains four base mismatches to prevent spontaneous annealing), subsequently denaturing the parental duplex. Leading and lagging arms are noncomplementary to prevent regression. Asterisks denote 32P labels. Representative gels show time-dependent accumulation of products for HLTF and ΔHIRAN proteins, as well as 30-min endpoints of reactions containing no enzyme (mock) or ATPase-dead HLTFDEAA or ΔHIRANDEAA proteins. D, quantitation of data shown in panels B and C (n = 3, mean ± S.D.). E, schematic model for fork restoration by HLTF. Fork restoration is driven by 3′-5′ translocation HLTF on leading or lagging duplex arms toward the fork, and can only be supported when the protein has a full duplex on which to translocate.
Discussion
This work provides experimental evidence for and clarifies some aspects of our previous model for fork binding and remodeling by HLTF (33). DNase footprinting reveals that HLTF engages all three arms of a stalled fork, both ahead of and behind the junction. The preferential DNase protection of the lagging template ahead of the fork is consistent with the dsDNA motor domain translocating in a 3′-5′ direction (16) toward the fork. Translocation into the junction would conceivably promote annealing of the template strands, which would be facilitated by prior or concomitant unwinding of daughter duplexes. Indeed, our data show that the motor domain alone is capable of catalyzing regression of forks, but only when they contain ssDNA regions at the junction that lowers the barrier to reannealing of template strands. We provide three pieces of evidence to support the conclusion that HIRAN facilitates four-way junction formation by remodeling the nascent leading strand from its template. First, the motor domain is unable to regress a fully paired fork in the absence of the HIRAN–3′-end interaction. Second, deletion of the HIRAN domain reduced fork regression activity against a lagging gap fork more than against a leading gap fork, indicating that HIRAN is more important to regression when the end of the nascent leading strand is directly at the fork. Third, HLTF binding to a 3′-frayed fork causes a specific DNase protection and sensitivity pattern along the leading template that tracks with the position of the nascent 3′-end and that depends on the presence of the HIRAN domain, indicating that the leading arm at the junction is remodeled by the HIRAN–3′ interaction.
We now know that the HIRAN domain requires an already frayed 3′-end to bind. How the nascent 3′-end is initially denatured is a remaining question. In our in vitro assays, we speculate that the activity of the HLTF ATPase motor pushing into the junction is sufficient to destabilize and eventually denature the 3′-end. In the cell, a stalled fork containing a partially unpaired or frayed leading 3′-end is likely to be formed by nucleotide misincorporation by DNA polymerase encountering a damaged template, and/or by dissociation of the DNA polymerase from the template (43). Given that the HIRAN–3′-end interaction facilitates but is not required for fork regression, we cannot rule out the possibility that the HIRAN domain also protects a frayed leading end in addition to its role in initiating four-way junction formation. In vitro binding data and co-crystal structures of HIRAN with different DNA sequences are consistent with accommodation of any nucleobase in the 3′-binding pocket (33, 36, 37) and thus we do not expect HIRAN to have a sequence preference for the frayed 3′-end.
The precise roles of HLTF, SMARCAL1, and ZRANB3 and the need for three related remodelers in the cell remain to be determined. However, the use of the same assays and substrates here and in previous work (41) enables a direct comparison of how the unique SRD of each protein imparts a specificity for a particular stalled fork or DNA intermediate generated during fork remodeling. Differences in footprinting on the parental duplex ahead of the fork suggest that HLTF and SMARCAL1 track with opposite polarity on the lagging and leading strands, respectively (Fig. S2C). This difference in strand tracking is consistent with activation of SMARCAL1, but not HLTF, by RPA on leading strand gaps (41). It also may suggest that SMARCAL1 processes forks stalled on the leading strand, whereas HLTF processes lagging strand stalls. Because lesions on the discontinuously replicated lagging strand are less of a threat than impediments on the leading strand, this strand bias helps to explain the stronger cellular phenotypes observed from SMARCAL1 deficiency (32). It is also intriguing to speculate that regression of forks stalled on the leading strand by SMARCAL1 creates a substrate recognized by HLTF by pushing the junction closer to the nascent leading strand. Such a sequential operation of each protein would be consistent with a specific recognition of nonduplex DNA structures by the SMARCAL1 HARP domain (34) and the specific interaction with the nascent 3′-end by HIRAN.
In addition to the template strand bias, there is a marked difference in the intrinsic and RPA-dependent regression and restoration activities of HLTF, SMARCAL1, and ZRANB3 (41) (Fig. S4C). SMARCAL1 regression of forks is enhanced by RPA bound to the leading template and inhibited by RPA on the lagging template, despite a slight intrinsic preference to regress forks containing an ssDNA gap on the lagging arm. In contrast, neither HLTF nor ZRANB3 shows a preference for regression of forks containing leading and lagging gaps. As noted above, this difference is consistent with putative operation of SMARCAL1 on the leading strand template. Interestingly, whereas HLTF regression is modestly inhibited by RPA bound to the lagging template, ZRANB3 is inhibited by RPA on the leading strand, suggesting a preference for a particular type of stalled fork by these two proteins. Similar differences are evident in fork restoration activities, in which HLTF and ZRANB3 are similar to one another and markedly different from SMARCAL1. Most strikingly, SMARCAL1 restores leading and lagging gapped forks equally unless bound by RPA, which dramatically stimulates restoration of a lagging gapped fork. In contrast, HLTF and ZRANB3 show an intrinsic preference to restore forks containing a gap on the lagging strand, and this preference is inhibited by RPA. Because RPA coating of ssDNA at a stalled fork is an early response to replication stress, the RPA stimulation of fork remodeling activities of SMARCAL1, but not HLTF and ZRANB3, further suggests that SMARCAL1 operates earlier in the replication stress response than HLTF and ZRANB3. Thus, a more comprehensive picture of the differences and similarities in remodeling activities of HLTF, SMARCAL1, and ZRANB3 is starting to emerge that will facilitate further studies into their nonredundant roles in the cell.
Experimental procedures
Protein purification
The HIRAN-UvsW chimera (Fig. S1) was constructed by the overlap extension PCR method, in which the gene sequences corresponding to the HIRAN domain (HLTF residues 55–180) and the UvsW motor (residues 83–503) were inserted into a pBG101 expression vector (Vanderbilt Center for Structural Biology). The chimera was expressed as a His6-GST–tagged protein in Escherichia coli BL-21 RIL cells at 16 °C for 16 h. Cells were lysed using an Avestin Emulsifier C3 Homogenizer in 50 mm Tris-HCl, pH 7.5, 500 mm NaCl, 20% glycerol, 0.01% Nonidet P-40, 0.1% Triton X-100, 0.1 mm PMSF, and an EDTA-free protease inhibitor tablet (Roche). The clarified lysate was passed over a Ni-NTA column, eluted with lysis buffer containing 500 mm imidazole, and the His6-GST tag removed by treatment with rhinovirus 3C (PreScission) protease at 4 °C. The ionic strength of the sample was reduced to 300 mm NaCl by dilution with 50 mm Tris-HCl, pH 7.5, 20% glycerol, 28 mm 2-mercaptoethanol buffer, and then purified over a heparin-Sepharose column, eluted by a 300–2000 mm NaCl gradient. The protein was then purified over a Superdex S200 size-exclusion column (GE Healthcare) in 50 mm Tris-HCl, pH 7.5, 20% glycerol, 500 mm NaCl, and 1 mm DTT, concentrated using an Amicon Ultra 3 kDa concentrator and stored at −80 °C.
The HIRAN domain from human HLTF was purified as previously described (33). Full-length HLTF was cloned into a pFastBac 438-C vector (Addgene) and expressed as a His6-MBP–tagged protein from baculovirus-infected Hi5 insect cells. Cells were harvested 48 h after infection and lysed using a Dounce homogenizer in Buffer A (50 mm Tris-HCl, pH 7.5, 500 mm NaCl, 10% glycerol, 0.01% Nonidet P-40) supplemented with 1 μg/ml aproptin, 5 μg/ml leupeptin, 2 μg/ml pepstatin A, and 20 mm imidazole. The clarified lysate was incubated with Ni-NTA resin and eluted with 300 mm imidazole in Buffer A. The protein sample was incubated with amylose resin in 50 mm Tris-HCl, pH 7.5, 300 mm NaCl, 10% glycerol, and 0.01% Nonidet P-40 and eluted by on-column cleavage of the His6-MBP tag by tobacco etch virus protease at 4 °C. Imidazole was added to a final concentration of 30 mm and repassed through a nickel affinity column. The sample was concentrated using an Amicon Ultra 30 kDa concentrator, buffer exchanged into 50 mm Tris-HCl, pH 7.5, 250 mm NaCl, 20% glycerol, 0.5 mm EDTA, and 1 mm tris(2-carboxyethyl)phosphine (TCEP), and stored at −80 °C.
The HLTFDEAA construct was prepared by cloning human HLTF into pBG101, followed by mutation of the codons corresponding to Asp-557 (GAT→GCT) and Glu-558 (GAA→GCG) using the QuikChange mutagenesis kit (Agilent). Protein was overexpressed with an N-terminal His6-GST tag in E. coli Rosetta cells at 16 °C for 16 h. Cells were lysed using an Avestin Emulsifier C3 Homogenizer in Buffer A containing 5 mm imidazole, 1 μg/ml aproptin, 5 μg/ml leupeptin, and 2 μg/ml pepstatin A and purified by Ni-NTA affinity chromatography using 500 mm imidazole in Buffer A. The protein sample was incubated with GSH resin and eluted by an on-column cleavage by rhinovirus 3C (PreScission) protease at 4 °C. The sample was diluted to 250 mm NaCl by addition of a Tris-HCl, pH 7.5, 20% glycerol, 1 mm TCEP buffer and purified from heparin-Sepharose column using a 250–2000 mm NaCl gradient. Protein was concentrated and stored in the same manner as WT HLTF.
The ΔHIRAN mutant (residues 181–1009) gene was cloned into pFastBac-HTb vector (Invitrogen) and expressed as a His6-tagged protein in baculovirus-infected Hi5 insect cells. Cells were lysed 48 h after infection using a Dounce homogenizer in Buffer A and protein purified by Ni-NTA affinity chromatography, eluting with 300 mm imidazole in Buffer A. The sample was diluted to 250 mm NaCl by addition of a Tris-HCl, pH 7.5, 20% glycerol, 1 mm TCEP buffer and purified by heparin-Sepharose with a 200–1000 mm NaCl gradient elution. Protein was concentrated, buffer exchanged into 50 mm Tris-HCl, pH 7.5, 250 mm NaCl, 20% glycerol, 0.5 mm EDTA, and 1 mm TCEP, and stored at −80 °C. ΔHIRANDEAA mutant was constructed by introducing D557A and D558A mutations into the ΔHIRAN/pFastBac vector, and purified the same as ΔHIRAN protein.
DNA substrate preparation
Oligodeoxynucleotides used for DNA binding, footprinting, and enzymatic assays are shown in Table S1 in the supporting data. Annealing reactions were carried out in 1× SSC buffer (15 mm sodium citrate pH 7.0 and 150 mm NaCl) and decreasing the temperature from 95° to 25 °C at 1°/min using a thermal cycler. Duplexes containing 5′-overhangs used in HIRAN-binding experiments were generated by annealing oligonucleotide ov_F, which contained a 5′-32P and a 3′-6-carboxyfluorescein (FAM), to either ov_0, ov_1, ov_2, or ov_3 oligonucleotide in a 1:1 ratio. Heterologous forks used in EMSA and DNase footprinting studies were generated by annealing all four oligonucleotides using a 1.2-fold molar excess of each unlabeled strand. Fork regression substrates were generated by annealing leading (A/B or 48/50) and lagging (C/D or 52/53) arms of the fork separately. The two duplexed arms were then combined using a 2-fold molar excess of the unlabeled arm and incubated at 37 °C for 30 min. Fork restoration substrates were generated by annealing three arms of the fork (R1/R2/R3 or R1/R2/R5), followed by addition of 5-fold excess of the R5 or R6 oligo and incubation at 37 °C for 30 min. With the exception of 48/50/52/53, fork substrates were PAGE purified using a 6% 0.5× Tris borate-EDTA (TBE) DNA Retardation Gel (Invitrogen). Substrates were excised from the gel and electroeluted in 0.5× TBE, concentrated, and stored at −20 °C.
DNA binding
All DNA-binding reactions were carried out for 20 min at 25 °C using 0–50 nm protein and either 5 nm (5′-overhang duplex) or 25 nm (fork) DNA. HIRAN binding was performed in 20 mm Tris-HCl, pH 8.3, 100 mm NaCl, 2 mm DTT, and 0.1 mm EDTA and electrophoresed on a 5% 29:1 (acrylamide:bis-acrylamide) 0.5× TBE gel. HLTF (WT and ΔHIRAN) binding was performed in 20 mm Tris-HCl, pH 7.76, 3% glycerol, 0.5 mm CaCl2, 5 mm MgCl2, 0.2% Nonidet P-40 and 0.05 mm TCEP and electrophoresed on a 5% 79:1 (acrylamide:bis-acrylamide) 0.5× TBE 3% glycerol gel. Gels were run with 0.5× TBE at 200 V for 0.5–1 h. Gels were either phosphorimaged ([32P]DNA) or directly fluorimaged at 532 nm excitation and 526 nm emission wavelengths on a Typhoon Trio variable mode imager. Band intensities were quantified with GelAnalyzer and data were plotted using GraphPad Prism 6.
Fork regression and restoration assays
Fork remodeling reactions were carried out at 37 °C and contained 10 nm HLTF, 1 nm 32P-labeled fork substrate, 2 mm ATP, 40 mm Tris-HCl, pH 7.8, 50 mm NaCl, 5 mm MgCl2, and 1 mm TCEP. At various time points, 10 μl of the reaction were stopped by adding 320 milliunits Proteinase K (Sigma) and electrophoresed on a 6% 19:1 (acrylamide:bis-acrylamide) 1× TBE gel at 6 watts for 1–1.5 h. Gels were phosphorimaged and quantified as above.
DNase footprinting
100 nm fork substrate, in which either the leading or lagging template strand contained a 3′-6-carboxyfluorescein label (Table S1 and Fig. S2) was incubated for 20 min at 25 °C with 0, 25, 50, 100, or 200 nm HLTF in 20 mm Tris-HCl, pH 7.8, 3% glycerol, 0.5 mm CaCl2, 5 mm MgCl2, and 0.05 mm TCEP. DNA was digested with 80 milliunits Benzonase (Sigma) for 2 min at 25 °C. The digestion reaction was stopped by addition of Proteinase K, denatured with loading buffer (80% formamide, 10 mm EDTA) at 70° for 5 min and electrophoresed on a 15% 19:1 (acrylamide:bis-acrylamide) 1× TBE 7 m urea denaturing gel for 3 h. Gels were fluorimaged and quantified as above. The changes in band intensities between only the 0 and 200 nm HLTF samples were determined for each band and normalized to the band intensities from the 0 HLTF sample. The mean changes were calculated separately for positive and negative slopes. Bands with a normalized slope of 0.5 S.D. from the mean were labeled as significant.
ATPase activity assays
5 nm HLTF was incubated with 0, 5, or 30 nm fork DNA substrate in 10-μl reactions containing 40 mm Tris-HCl, pH 7.8, 5 mm MgCl2, 50 mm NaCl, 1 mm TCEP, and 1.67 μm [γ–32P]ATP at 37 °C for 30 min. Reactions were stopped by adding EDTA to a final concentration of 25 mm. ATP and Pi were separated by TLC, in which a 1-μl reaction aliquot was spotted on PEI cellulose F TLC plates (Millipore) and resolved in 0.25 m LiCl/1 M formic acid for 30 min. Plates were phosphorimaged (20-min exposure) and quantified as above.
Author contributions
D. A. C. and B. F. E. conceptualization; D. A. C. data curation; D. A. C., B. H. G., and B. F. E. formal analysis; D. A. C. and B. F. E. validation; D. A. C. and B. H. G. investigation; D. A. C. and B. H. G. methodology; D. A. C. writing-original draft; D. A. C., B. H. G., and B. F. E. writing-review and editing; B. F. E. supervision; B. F. E. funding acquisition; B. F. E. project administration.
Supplementary Material
Acknowledgments
We thank Miaw-Sheue Tsai for baculovirus production and insect cell expression of HLTF proteins (supported by Structural Cell Biology of DNA Repair Machines Grant P01CA092584 from the National Institutes of Health). We also thank Karlene Cimprich, David Cortez, and members of the Cortez laboratory for comments on the manuscript and for helpful discussions. Additional core facilities were supported by the Vanderbilt-Ingram Cancer Center, which was supported by National Institutes of Health Grant P30CA068485.
This work was supported by the National Institutes of Health Grants R01GM117299 and P01CA092584. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Figs. S1–S4 and Table S1.
- ssDNA
- single-stranded DNA
- DDT
- DNA damage tolerance
- HLTF
- helicase-like transcription factor
- SRD
- substrate recognition domain
- EMSA
- electrophoretic mobility shift assay
- RPA
- replication protein A
- Ni-NTA
- nickel-nitrilotriacetic acid
- TCEP
- tris(2-carboxyethyl)phosphine
- Buffer A
- 50 mm Tris-HCl, pH 7.5, 500 mm NaCl, 10% glycerol, 0.01% Nonidet P-40
- TBE
- Tris borate-EDTA.
References
- 1. Zeman M. K., and Cimprich K. A. (2014) Causes and consequences of replication stress. Nat. Cell Biol. 16, 2–9 10.1038/ncb2897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Byun T. S., Pacek M., Yee M. C., Walter J. C., and Cimprich K. A. (2005) Functional uncoupling of MCM helicase and DNA polymerase activities activates the ATR-dependent checkpoint. Genes Dev. 19, 1040–1052 10.1101/gad.1301205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Berti M., and Vindigni A. (2016) Replication stress: Getting back on track. Nat. Struct. Mol. Biol. 23, 103–109 10.1038/nsmb.3163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Friedberg E. C. (2005) Suffering in silence: The tolerance of DNA damage. Nat. Rev. Mol. Cell Biol. 6, 943–953 10.1038/nrm1781 [DOI] [PubMed] [Google Scholar]
- 5. Chang D. J., and Cimprich K. A. (2009) DNA damage tolerance: When it's OK to make mistakes. Nat. Chem. Biol. 5, 82–90 10.1038/nchembio.139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Branzei D., and Psakhye I. (2016) DNA damage tolerance. Curr. Opin. Cell Biol. 40, 137–144 10.1016/j.ceb.2016.03.015 [DOI] [PubMed] [Google Scholar]
- 7. Ray Chaudhuri A., Hashimoto Y., Herrador R., Neelsen K. J., Fachinetti D., Bermejo R., Cocito A., Costanzo V., and Lopes M. (2012) Topoisomerase I poisoning results in PARP-mediated replication fork reversal. Nat. Struct. Mol. Biol. 19, 417–423 10.1038/nsmb.2258 [DOI] [PubMed] [Google Scholar]
- 8. Ahuja A. K., Jodkowska K., Teloni F., Bizard A. H., Zellweger R., Herrador R., Ortega S., Hickson I. D., Altmeyer M., Mendez J., and Lopes M. (2016) A short G1 phase imposes constitutive replication stress and fork remodelling in mouse embryonic stem cells. Nat. Commun. 7, 10660 10.1038/ncomms10660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zellweger R., Dalcher D., Mutreja K., Berti M., Schmid J. A., Herrador R., Vindigni A., and Lopes M. (2015) Rad51-mediated replication fork reversal is a global response to genotoxic treatments in human cells. J. Cell Biol. 208, 563–579 10.1083/jcb.201406099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Follonier C., Oehler J., Herrador R., and Lopes M. (2013) Friedreich's ataxia–associated GAA repeats induce replication-fork reversal and unusual molecular junctions. Nat. Struct. Mol. Biol. 20, 486–494 10.1038/nsmb.2520 [DOI] [PubMed] [Google Scholar]
- 11. Neelsen K. J., Zanini I. M., Herrador R., and Lopes M. (2013) Oncogenes induce genotoxic stress by mitotic processing of unusual replication intermediates. J. Cell Biol. 200, 699–708 10.1083/jcb.201212058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Neelsen K. J., and Lopes M. (2015) Replication fork reversal in eukaryotes: From dead end to dynamic response. Nat. Rev. Mol. Cell Biol. 16, 207–220 10.1038/nrm3935 [DOI] [PubMed] [Google Scholar]
- 13. Motegi A., Liaw H.-J., Lee K.-Y., Roest H. P., Maas A., Wu X., Moinova H., Markowitz S. D., Ding H., Hoeijmakers J. H., and Myung K. (2008) Polyubiquitination of proliferating cell nuclear antigen by HLTF and SHPRH prevents genomic instability from stalled replication forks. Proc. Natl. Acad. Sci. U.S.A. 105, 12411–12416 10.1073/pnas.0805685105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Unk I., Hajdú I., Fátyol K., Hurwitz J., Yoon J.-H., Prakash L., Prakash S., and Haracska L. (2008) Human HLTF functions as a ubiquitin ligase for proliferating cell nuclear antigen polyubiquitination. Proc. Natl. Acad. Sci. U.S.A. 105, 3768–3773 10.1073/pnas.0800563105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lin J.-R., Zeman M. K., Chen J.-Y., Yee M.-C., and Cimprich K. A. (2011) SHPRH and HLTF act in a damage-specific manner to coordinate different forms of postreplication repair and prevent mutagenesis. Mol. Cell 42, 237–249 10.1016/j.molcel.2011.02.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Blastyák A., Hajdú I., Unk I., and Haracska L. (2010) Role of double-stranded DNA translocase activity of human HLTF in replication of damaged DNA. Mol. Cell. Biol. 30, 684–693 10.1128/MCB.00863-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Burkovics P., Sebesta M., Balogh D., Haracska L., and Krejci L. (2014) Strand invasion by HLTF as a mechanism for template switch in fork rescue. Nucleic Acids Res. 42, 1711–1720 10.1093/nar/gkt1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Achar Y., Balogh D., and Haracska L. (2011) Coordinated protein and DNA remodeling by human HLTF on stalled replication fork. Proc. Natl. Acad. Sci. U.S.A., 108, 14073–14078 10.1073/pnas.1101951108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Choi K., Batke S., Szakal B., Lowther J., Hao F., Sarangi P., Branzei D., Ulrich H. D., and Zhao X. (2015) Concerted and differential actions of two enzymatic domains underlie Rad5 contributions to DNA damage tolerance. Nucleic Acids Res., 43, 2666–2677 10.1093/nar/gkv004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Minca E., and Kowalski D. (2010) Multiple Rad5 activities mediate sister chromatid recombination to bypass DNA damage at stalled replication forks. Mol. Cell 38, 649–661 10.1016/j.molcel.2010.03.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ortiz-Bazán M. Á., Gallo-Fernández M., Saugar I., Jiménez-Martín A., Vázquez M. V., and Tercero J. A. (2014) Rad5 plays a major role in the cellular response to DNA damage during chromosome replication. Cell Rep., 9, 460–468 10.1016/j.celrep.2014.09.005 [DOI] [PubMed] [Google Scholar]
- 22. Yusufzai T., and Kadonaga J. T. (2008) HARP is an ATP-driven annealing helicase. Science, 322, 748–750 10.1126/science.1161233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bansbach C. E., Bétous R., Lovejoy C. A., Glick G. G., and Cortez D. (2009) The annealing helicase SMARCAL1 maintains genome integrity at stalled replication forks. Genes Dev. 23, 2405–2414 10.1101/gad.1839909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ciccia A., Bredemeyer A. L., Sowa M. E., Terret M.-E., Jallepalli P. V., Harper J. W., and Elledge S. J. (2009) The SIOD disorder protein SMARCAL1 is an RPA-interacting protein involved in replication fork restart. Genes Dev. 23, 2415–2425 10.1101/gad.1832309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yuan J., Ghosal G., and Chen J. (2009) The annealing helicase HARP protects stalled replication forks. Genes Dev. 23, 2394–2399 10.1101/gad.1836409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yusufzai T., Kong X., Yokomori K., and Kadonaga J. T. (2009) The annealing helicase HARP is recruited to DNA repair sites via an interaction with RPA. Genes Dev. 23, 2400–2404 10.1101/gad.1831509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Postow L., Woo E. M., Chait B. T., and Funabiki H. (2009) Identification of SMARCAL1 as a component of the DNA damage response. J. Biol. Chem., 284, 35951–35961 10.1074/jbc.M109.048330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yusufzai T., and Kadonaga J. T. (2010) Annealing helicase 2 (AH2), a DNA-rewinding motor with an HNH motif. Proc. Natl. Acad. Sci. U.S.A. 107, 20970–20973 10.1073/pnas.1011196107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bétous R., Mason A. C., Rambo R. P., Bansbach C. E., Badu-Nkansah A., Sirbu B. M., Eichman B. F., and Cortez D. (2012) SMARCAL1 catalyzes fork regression and Holliday junction migration to maintain genome stability during DNA replication. Genes Dev. 26, 151–162 10.1101/gad.178459.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ciccia A., Nimonkar A. V., Hu Y., Hajdu I., Achar Y. J., Izhar L., Petit S. A., Adamson B., Yoon J. C., Kowalczykowski S. C., Livingston D. M., Haracska L., and Elledge S. J. (2012) Polyubiquitinated PCNA recruits the ZRANB3 translocase to maintain genomic integrity after replication stress. Mol. Cell, 47, 396–409 10.1016/j.molcel.2012.05.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yuan J., Ghosal G., and Chen J. (2012) The HARP-like domain-containing protein AH2/ZRANB3 binds to PCNA and participates in cellular response to replication stress. Mol. Cell 47, 410–421 10.1016/j.molcel.2012.05.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Poole L. A., and Cortez D. (2017) Functions of SMARCAL1, ZRANB3, and HLTF in maintaining genome stability. Crit. Rev. Biochem. Mol. Biol. 52, 696–714 10.1080/10409238.2017.1380597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kile A. C., Chavez D. A., Bacal J., Eldirany S., Korzhnev D. M., Bezsonova I., Eichman B. F., and Cimprich K. A. (2015) HLTF's ancient HIRAN domain binds 3′ DNA ends to drive replication fork reversal. Mol. Cell 58, 1090–1100 10.1016/j.molcel.2015.05.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mason A. C., Rambo R. P., Greer B., Pritchett M., Tainer J. A., Cortez D., and Eichman B. F. (2014) A structure-specific nucleic acid-binding domain conserved among DNA repair proteins. Proc. Natl. Acad. Sci. U.S.A. 111, 7618–7623 10.1073/pnas.1324143111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Badu-Nkansah A., Mason A. C., Eichman B. F., and Cortez D. (2016) Identification of a substrate recognition domain in the replication stress response protein zinc finger ran-binding domain-containing protein 3 (ZRANB3). J. Biol. Chem. 291, 8251–8257 10.1074/jbc.M115.709733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hishiki A., Hara K., Ikegaya Y., Yokoyama H., Shimizu T., Sato M., and Hashimoto H. (2015) Structure of a novel DNA-binding domain of helicase-like transcription factor (HLTF) and its functional implication in DNA damage tolerance. J. Biol. Chem. 290, 13215–13223 10.1074/jbc.M115.643643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Achar Y. J., Balogh D., Neculai D., Juhasz S., Morocz M., Gali H., Dhe-Paganon S., Venclovas Č., Haracska L. (2015) Human HLTF mediates postreplication repair by its HIRAN domain-dependent replication fork remodelling. Nucleic Acids Res. 43, 10277–10291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ding L., and Forsburg S. L. (2014) Essential domains of Schizosaccharomyces pombe Rad8 required for DNA damage response. G3 (Bethesda) 4, 1373–1384 10.1534/g3.114.011346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kobbe D., Kahles A., Walter M., Klemm T., Mannuss A., Knoll A., Focke M., and Puchta H. (2016) AtRAD5A is a DNA translocase harboring a HIRAN domain which confers binding to branched DNA structures and is required for DNA repair in vivo. Plant J. 88, 521–530 10.1111/tpj.13283 [DOI] [PubMed] [Google Scholar]
- 40. Manosas M., Perumal S. K., Bianco P. R., Bianco P., Ritort F., Benkovic S. J., and Croquette V. (2013) RecG and UvsW catalyse robust DNA rewinding critical for stalled DNA replication fork rescue. Nat. Commun. 4, 2368 10.1038/ncomms3368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Bétous R., Couch F. B., Mason A. C., Eichman B. F., Manosas M., and Cortez D. (2013) Substrate-selective repair and restart of replication forks by DNA translocases. Cell Rep. 3, 1958–1969 10.1016/j.celrep.2013.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bhat K. P., Bétous R., and Cortez D. (2015) High-affinity DNA-binding domains of replication protein A (RPA) direct SMARCAL1-dependent replication fork remodeling. J. Biol. Chem. 290, 4110–4117 10.1074/jbc.M114.627083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Fernandez-Leiro R., Conrad J., Yang J.-C., Freund S. M., Scheres S. H. , and Lamers M. H. (2017) Self-correcting mismatches during high-fidelity DNA replication. Nat. Struct. Mol. Biol. 24, 140–143 10.1038/nsmb.3348 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.