Abstract
Chemotherapeutics cause the detachment and death of adherent cancer cells. When studying the proteome changes to determine the protein target and mechanism of action of anticancer drugs, the still-attached cells are normally used, whereas the detached cells are usually ignored. To test the hypothesis that proteomes of detached cells contain valuable information, we separately analyzed the proteomes of detached and attached HCT-116, A375, and RKO cells treated for 48 h with 5-fluorouracil, methotrexate and paclitaxel. Individually, the proteomic data on attached and detached cells had comparable performance in target and drug mechanism deconvolution, whereas the combined data significantly improved the target ranking for paclitaxel. Comparative analysis of attached versus detached proteomes provided further insight into cell life and death decision making. Six proteins consistently up- or downregulated in the detached versus attached cells regardless of the drug and cell type were discovered; their role in cell death/survival was tested by silencing them with siRNA. Knocking down USP11, CTTN, ACAA2, and EIF4H had anti-proliferative effects, affecting UHRF1 additionally sensitized the cells to the anticancer drugs, while knocking down RNF-40 increased cell survival against the treatments. Therefore, adding detached cells to the expression proteomics analysis of drug-treated cells can significantly increase the analytical value of the approach. The data have been deposited to the ProteomeXchange with identifier PXD007686.
Keywords: Cell death*, Mechanism of action, Cancer therapeutics, Mass Spectrometry, Label-free quantification, Drug targets*, Drug resistance, Cell survival, chemotherapeutic
Unlike the targeted screening in which a library of compounds is tested for binding to a protein, in phenotypic screening, mechanistically blind cell-based assays are used to identify compounds inducing a desired biological effect. Because in the latter approach the readout is a disease-relevant process and multiple targets can be covered in a single screening, the success rate for lead compound discovery is believed be higher than in the targeted approach. Indeed, 37% of the first-in-class compounds approved by FDA during years 1999–2008 were from phenotypic screening (compared with 23% from targeted screening), whereas targeted screening was more successful in follower drugs (51% versus 18% from phenotypic screening) (1, 2). Even though a more recent analysis has found a significant increase in 2008–2013 in the approval of first-in-class drugs discovered by target-based methods (3), phenotypic screening remains an important source of novel drug candidates. Given current interest in a thorough characterization of drug targets and mechanisms of action (MOAs)1, the targets and MOAs of these candidates need to be characterized by a set of sophisticated analytical methods. Also, although most FDA-approved drugs have a known target and MOA, there are also a multitude of promising experimental compounds for which no reliable mechanistic information is available (4, 5). Furthermore, the MOA of some compounds may have been wrongly assigned in the past, as targeted methods tend to ignore alternative targets and pathways (6). Thus, devising new methods for identification of the target and MOA deconvolution can help a great deal in anticancer drug discovery, where the desired effect is the cancer cell death.
Mass-spectrometry based proteomics has proved to be a useful tool in different stages of anticancer drug discovery. To assist deconvolution of protein targets and MOA of small molecule drugs, we have recently devised a mass-spectrometry-based method called Functional Identification of Target by Expression Proteomics (FITExP) (7). This method is based on the observation that in late stages of cancer cell death, the abundance change of the protein target of a small molecule is exceptionally strong. FITExP could pinpoint the known targets of several common anticancer agents, including paclitaxel (PCTL), doxorubicin, 5-fluorouracil (5-FU), methotrexate (MTX), raltitrexed and camptothecin, among more than 4000 proteins. This method is based on deduction of drug target solely from proteomic data, by sorting all proteins with respect to their regulation upon treatment and specificity of that regulation for a treatment. To introduce an internal positive control and increase the specificity by filtering out the generic proteins involved in cell death, several known drugs are added to the compound panel. It has been shown that specificity can be further enhanced by combining the data from several cell lines and considering only proteins that behave consistently in the panel (7). Besides the drug target identification, the benefit of FITExP is that the proteins with largest abundance changes and highest specificity could be mapped on known protein networks to reveal the drug MOA.
There is, however, a need to further improve the method by increasing the accuracy and specificity of target deconvolution. The potential for improvement lies in the fact that anticancer drugs applied to a culture of matrix-attached cancer cells cause cell detachment, and only then - cell death. When studying the proteome changes in FITExP and other proteomics approaches, the still-attached cells are usually used, whereas the detached cells are considered lacking structural integrity and are thus discarded. Many detached cells are indeed permeable to trypan blue and are therefore usually considered dead. However, when recultured in fresh media, the detached cell population can recover and grow, and thus the detached cell proteome may contain additional valuable information relevant to drug target and action. Because the detached cells are those that have been influenced most by the anticancer agent, their proteome changes might reflect the drug-induced processes even better than still-attached cells.
In this study, we attempted to test the above hypothesis and harvest additional information from detached cells as well as assess the relative value of this approach for FITExP. As a model system, we used HCT-116, A375 and RKO human cancer cells treated with 5-FU, MTX and PCTL at IC50 concentrations. The proteomics data from matrix-attached and detached cells were acquired separately and used as input for FITExP analysis. The accuracy of obtained information was assessed based on the known targets of the above drugs.
The data from detached cells proved nearly as valuable for FITExP as the attached cells about identification of drug targets and MOA. We could also identify several proteins regulated similarly in either attached or detached cells (i.e. in the dying or surviving states) regardless of the treatment and cell type. Testing the hypothesis that these proteins might be potentially involved in cell death and survival decisions, we investigated the effect of knocking-down such targets by siRNA on cell viability and sensitivity/resistance to the compounds.
EXPERIMENTAL PROCEDURES
Experimental Design and Statistical Rationale
The proteomics data is derived from three sets of samples prepared and analyzed by LC-MS/MS. All treatments were performed in three biological replicates, so that appropriate statistical analyses could be performed. In each part of the experiment, separate controls treated with the vehicle were included. Samples were analyzed in random order to reduce the “order of injection” effect. A total of 90 label-free analyses were performed with a 90 min gradient on a Q Exactive Plus mass spectrometer. In part 1, A375 and HCT116 cells were treated with 5-FU (24 samples), whereas in part 2, RKO cells were treated with 5-FU and A375 cells were treated with PCTL and MTX (30 samples). Finally, in part 3, RKO and HCT116 cells were treated with MTX and PCTL (36 samples). Quality check was performed by calculating the variation (CV) between the replicates as well as by building PCA models to verify the small overall spread between the replicates.
Cell Culture
Human colorectal carcinoma HCT116, colon carcinoma RKO and malignant melanoma A375 cells obtained from ATCC (Manassas, VA) were grown in Dulbecco's Modified Eagle's Medium (DMEM - Lonza, Wakersville, MD) supplemented with 10% FBS superior (Biochrom, Berlin, Germany), 2 mm l-glutamine (Lonza) and 100 units/ml penicillin/streptomycin (Thermo, Waltham, MA) at 37 °C in 5% CO2.
Determination of IC50 Values
The IC50 values (concentration at which 50% cytotoxicity occurs) were determined by MTT assay (8). Briefly, cells were seeded into 96 well plates at a density of 3000 cells per well and after 24 h of culturing, were treated with serial concentrations of the respective drug (0–100 μm) dissolved in DMSO. After the total treatment period of 48 h, the media were discarded and replaced with 100 μl of the fresh culture media. 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) was added to the cells, and the plates were incubated at 37 °C for 4 h. One hundred μl of a sodium dodecyl sulfate (SDS)-HCl solution (10 ml of 0.01 m HCl containing 1 g of SDS) were added to each well and the plate was incubated for 18 h. After mixing, the absorbance was read at 570 nm using Epoch Microplate Spectrophotometer (BioTek, Winooski, VT). The IC50 values were determined from the dose-response curves by calculating in Excel the concentration causing 50% absorbance reduction compared with untreated control. The final IC50 results are shown in supplemental Table S1.
Treatments and Cell Harvest
Cells were treated with 5-FU, MTX or PCTL at IC50 concentration in 25 cm2 flasks. Control cells were treated with the vehicle (highest DMSO concentration used in the treatments). After 48 h the supernatant was collected, and the attached cells were trypsinized for 5 min, after which they were harvested by centrifugation at 400 × g for 5 min. The above supernatant was centrifuged at 540 × g for 5 min, and the cells detached from the matrix were collected. Trypan blue dye exclusion assay was used to determine the percentage of collected viable cells in both attached and detached cell populations. The cell pellets were lysed and digested immediately. All samples were prepared in three biological replicates.
Lysis and Digestion
Digestion was performed according to our previously published workflow (9) with slight modifications. Lysis buffer consisting of 3% sodium deoxycholate (SDC) in Ambic buffer was added at the volume 4 times exceeding that of the cell pellet. After incubation at 80 °C for 10 min, samples were sonicated for 1.5 min (30% amplitude, 3s/3s on/off pulses) with a Branson sonicator (Thermo). The protein concentration was measured using Pierce BCA protein assay (Thermo). The extracted proteins were reduced with dithiothreitol (DTT) to the final concentration of 15 mm (60 °C for 20 min) and alkylated with 20 mm iodoacetamide (IAA) (room temperature for 30 min in the dark). SDC solution was diluted to concentration of 1.5% and mass spectrometry grade Lysyl endopeptidase (Wako Pure Chemical Industries, Osaka, Japan) was added at a ratio of 1:100 LysC to protein. Samples were incubated for 6 h, and thereafter diluted to 0.5% SDC. Modified sequencing grade trypsin (Promega, Madison, WI) was then added at a ratio of 1:100 trypsin to protein followed by overnight incubation. Subsequently, acetic acid was added to the final concentration of 5% and samples were left for 30 min at room temperature. The precipitates were removed by centrifugation at 20,000 × g over 15 min at room temperature. The supernatants were taken and cleaned using StageTips (Thermo) according to the manufacturer's protocol. Samples were dried using SpeedVac centrifugal evaporator and prior to LC-MS/MS analysis, 0.1% formic acid solution (Fluka) was added to the samples to achieve the concentration of 0.2 μg/μl.
Proteomics
Proteins digests (1 μg) were analyzed in a randomized order by LC-MS/MS. Samples were loaded onto a 50 cm column (EASY-Spray, 75 μm internal diameter (ID), PepMap C18, 2 μm beads, 100 Å pore size) connected to an Easy-nLC 1000 pump (Thermo Fisher Scientific) with buffer A (0.1% formic acid in water) and eluted with a 120-min gradient reaching from 2% to 30% of buffer B (98% ACN, 0.1% FA, 2% H2O) at a flow rate of 250 nL/min. Mass spectra were acquired with an Orbitrap Q Exactive Plus mass spectrometer (Thermo Fisher Scientific) in the data-dependent mode at a nominal resolution of 17,500, in the m/z range from 375 to 1700. Peptide fragmentation was performed via higher-energy collision dissociation (HCD) with energy set at 28 NCE. The ion selection abundance threshold was set at 0.1% with exclusion of singly charged ions.
Data Processing
The raw data from mass spectrometry were analyzed by MaxQuant, version 1.5.3.8 (10). The Andromeda search engine (11) searched MS/MS data against the International Protein Index database (human version 2014_02, 89054 entries). Mass tolerance for precursor ions was 20 ppm (initial search) and 4.5 ppm (main search) and the MS/MS mass tolerance was set at 20 ppm. Cysteine carbamidomethylation was used as a fixed modification, whereas N,Q-deamidation and methionine oxidation were selected as variable modifications. Trypsin/P and LysC/P were selected. No more than two missed cleavages were allowed. A 1% false discovery rate was used as a filter at both protein and peptide levels. For all other parameters, the default settings were used. Label-free quantification of peptides and proteins was performed. Protein abundances were normalized by the total protein abundance in each replicate. Only protein with at least two peptides were included in the final data set and all the contaminants were removed. Multivariate data analysis was performed using SIMCA (Version 14, UMetrics, Sweden) (12).
Network Mapping
The top consistently up- and downregulated proteins were mapped onto STRING v.10 (http://string-db.org) protein network analysis tool. Medium confidence threshold (0.4) was used to define protein-protein interactions. In-built gene set enrichment analysis with the whole genome as a background was used to identify enriched gene ontology terms and pathways.
siRNA Experiments
A pair of distinct functionally verified (when available) or pre-designed siRNAs were used to treat the three cell lines with and without test compounds. The siRNAs were purchased from Qiagen and are listed in supplemental Table S2.
In brief, HCT116, A375 or RKO cells were seeded at a density of 4000 cells/well in 96-well plates and allowed to reach ∼40% confluence. Afterward, 0.5 μl of lipofectamine 2000 (Thermo) mixed with serum-free medium was added to siRNA in serum-free medium (10 pmol siRNA per well), incubated for 15 min at room temperature and subsequently used to treat the cells. The siRNA treatments were done both in the presence and absence of the test compounds, with media being replaced with fresh full media after 4 h. Thereafter the cells were incubated for 44 h at 37 °C in a CO2 incubator. Cell viability was measured using the MTT assay, as explained above. The siRNAs were applied individually in four replicates.
For validation of the knockdowns, RKO cells were treated with 200 pmol siRNA in 6-well plates at a density of 200,000 cells per well, and samples were digested and prepared for proteomics analyses, as explained above. USP11 knockdown validation was performed in HCT116 cells. In all cases, the controls were treated with scrambled siRNA (Qiagen).
RESULTS
The trypan blue assay showed that 65% to 85% of the detached cells were permeable to the dye and thus most cells can be conventionally classified as dead. The population of the detached cells will be called “dying,” as opposed to the “surviving” or “living” attached population.
Life and Death Have the Greatest Impact on the Cellular Proteome
Overall, 57,255 peptides belonging to 5277 proteins were identified in all samples and 4885 proteins were quantified with at least two peptides. Considering only proteins with nonzero abundance values in all three replicates, the final data set contained 1950 proteins (supplemental Table S3). This number was found sufficient for our purposes, as we have previously shown that the abundances of even several hundred most abundant proteins may truthfully reflect the state of the dying cells (13).
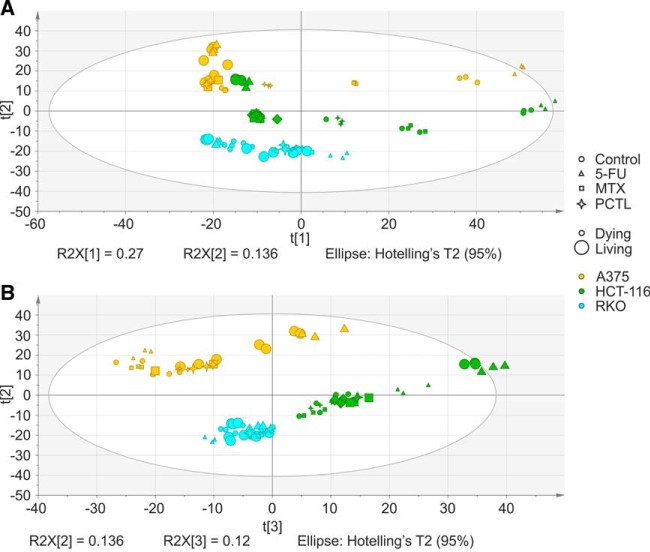
Using multivariate analysis, PCA models were built to quantify and visualize the differences between the samples. For 5-FU treatment, the first (main) component separated dying (detached) cells from surviving (attached) cells, whereas the second component separated the cell lines and the third one, treated cells from untreated controls (supplemental Fig. S1). For the whole data set, the first component separated dying cells from surviving cells and the second component separated the cell lines (Fig. 1). To analyze the effect responsible for separation of the samples in the third component, 50 proteins contributing most to the component were extracted and subjected to String analysis. These proteins mostly mapped to metabolic pathways (16 proteins, pathway ID in KEGG: 01100, p value = 4.47e-06), especially to cellular carbohydrate metabolic process (p value = 0.0111) such as galactose (p value = 0.00698), fructose and mannose (p value = 0.00698), but also to glutathione metabolism (p value = 0.0133).
Fig. 1.
Principal component analysis of the whole proteome data set (Q2 = 0.705). A, The first principal component separated dying (detached) versus living (attached) cells, whereas the second component separated the cell lines. B, The third component seems to separate the different states based on carbohydrate metabolic states such as galactose, fructose, mannose and glutathione metabolism.
Detached Cells Versus Attached Cells in Drug Target Deconvolution
Following our FITExP procedure, proteins were sorted and ranked based on their Regulation (fold changes upon treatment) in the detached and attached HCT116, A375, and RKO cells, separately. Specificity was calculated by dividing the median regulation of a protein in treatment A to the median regulation of the same protein in all other treatments. The overall ranking was done by summing the individual ranks in different cell lines within all detached and attached states. The data on matrix detached cells combined for all three cell lines was compared with that of attached cells to investigate which one can better identify the drug targets. Table I shows that combining the rankings of surviving cells with dying cells improved the deconvolution of drug targets, especially for 5-FU and PCTL.
Table I. The ranking of drug targets with Regulation and Specificity calculations in attached and detached cells, as well as combined ranking. The combined rankings were generated by summing up the individual rankings and ranking the sum.
| Drug | Known targets | Regulation |
Specificity |
Regulation + Specificity |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| La combined | Da combined | L & D combined | L combined | D combined | L & D combined | L combined | D combined | L & D combined | ||
| 5-FU | TYMS | 7 | 26 | 2 | 9 | 63 | 5 | 2 | 21 | 1 |
| MTX | DHFR | 1 | 16 | 2 | 1 | 1 | 1 | 1 | 2 | 1 |
| PCTL | TUBB2A | 19 | 77 | 11 | 113 | 17 | 19 | 15 | 16 | 3 |
| TUBB8 | 270 | 37 | 35 | 94 | 16 | 11 | 72 | 9 | 5 | |
| TUBB4B | 90 | 9 | 7 | 205 | 30 | 41 | 47 | 5 | 6 | |
| TUBB | 203 | 31 | 26 | 41 | 149 | 31 | 32 | 38 | 8 | |
| TUBA1B | 116 | 54 | 16 | 42 | 195 | 42 | 17 | 53 | 9 | |
| TUBA1A | 13 | 24 | 6 | 29 | 317 | 64 | 3 | 78 | 11 | |
| TUBA1C | 232 | 307 | 130 | 51 | 123 | 27 | 44 | 110 | 22 | |
| TUBB3 | 58 | 52 | 12 | 109 | 508 | 150 | 18 | 146 | 25 | |
a L = living cells; D = detached or dying cells.
Following this approach, thymidylate synthase (TYMS) was lifted from the 5th position for 5-FU to the top position, whereas dihydrofolate reductase (DHFR) remained at the 1st position for MTX, consistent with these proteins being known targets for these drugs. In the case of PCTL that targets beta-tubulins, eight tubulin subunits were identified among 25 top upregulated proteins, with six out of 11 top ranking proteins being tubulins, among which TUBB2A was the highest-ranking (3rd). In contrast, for living cells only, the eight tubulins were spread over 72 top positions. Therefore, combining the proteomics data from attached and detached cells improved the drug target identification procedure for two of the three compounds.
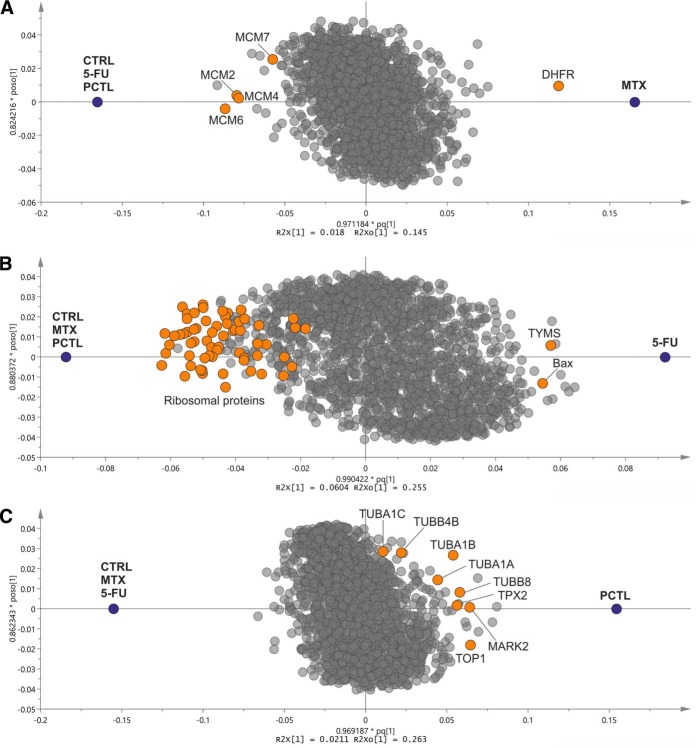
In order to visualize the specific response of drug targets and other involved proteins, we built orthogonal partial least squares (OPLS) models distinguishing the proteins contributing to a treatment versus all other states (treatments and controls). In such plots, proteins upregulated or downregulated specifically and consistently in all cell lines in response to a treatment are on the right and left side of the plot, respectively. As shown in Fig. 2, DHFR is contributing the most to the proteome response to MTX treatment, while several members of minichromosome maintenance protein (MCM) complex, a DNA helicase complex involved in the DNA replication process, are downregulated. Similarly, TYMS and tubulin subunits are among the top proteins substantially contributing to 5-FU and PCTL responses, respectively. A significant downregulation of ribosomal proteins could be seen in response to 5-FU, which is in line with both the FITExP analyses and our previously published results (14). For PCTL, a group of upregulated tubulins was preceded by Microtubule Nucleation Factor TPX2. Interestingly, TPX2 knockdown has been shown to sensitize pancreatic cancer cells to paclitaxel (15) and to be involved in the cellular effects of this drug (16). The upregulation of serine/threonine-protein kinase MARK2, which is involved in the regulation of cell polarity and microtubule dynamics, was also noted (17).
Fig. 2.
OPLS projections for A, MTX, B, 5-FU and C, PCTL versus all other compounds and controls in different cell lines. The drug targets and other mechanistically involved proteins are shown in color. (The variations in the x axis correspond to regulation and specificity and variations in the y axis arise from the orthogonal components in OPLS).
Detached Cells Versus Attached Cells: Dying Versus Surviving Processes
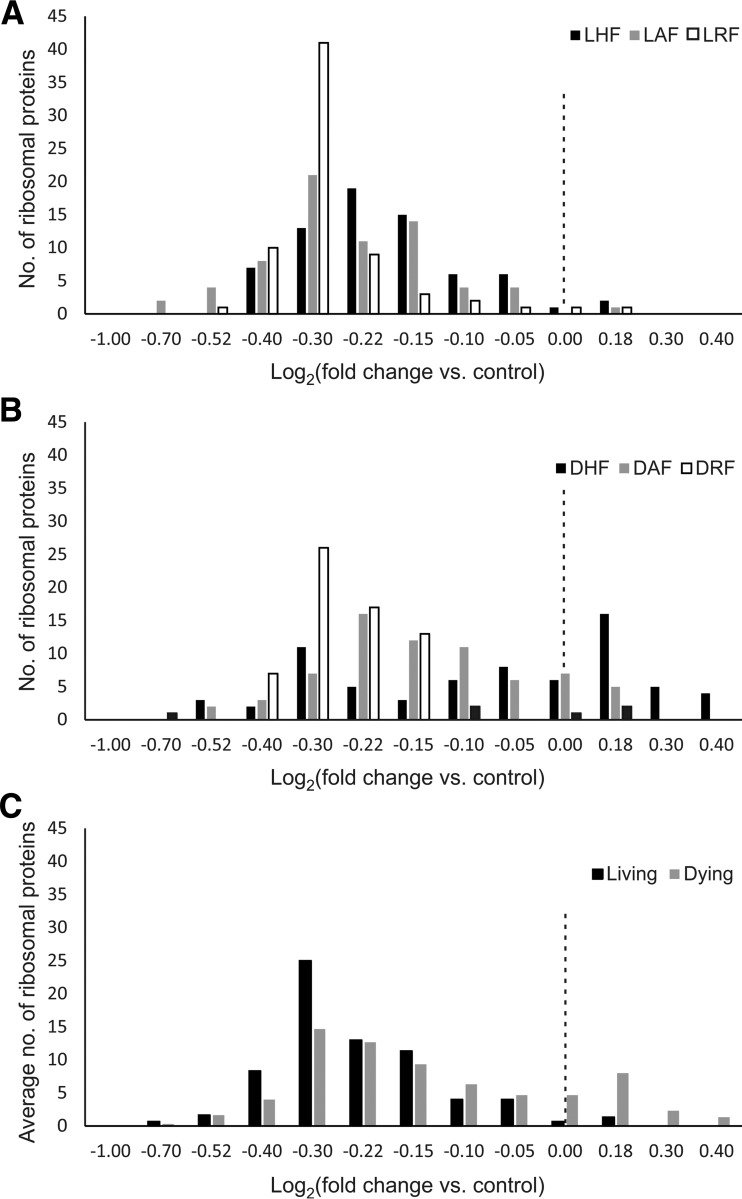
Differentially regulated proteins in cancer cells treated with anticancer agents are usually interpreted as being involved in cell death, whereas some of them may in fact be implicated in the survival process. Differentiation of the death processes from the survival pathways can be made by contrasting the regulated proteins in the detached versus attached cells. Such a comparison reveals, for instance, that the downregulation of ribosomal proteins upon 5-FU treatment is more universal in living cells compared with dying cells (Fig. 3). Of the 69 ribosomal proteins quantified in this study, on average 68 were downregulated in surviving cells compared with control, whereas a lower number of ribosomal proteins were downregulated on average in dying cells, and some were significantly upregulated (supplemental Table S4 and supplemental Fig. S2). The ribosome downregulation in attached cells -but to a lower extent in detached cells, might be part of a survival mechanism, halting cell division to survive 5-FU treatment.
Fig. 3.
Significant downregulation of ribosomal proteins upon 5-FU treatment in living cells compared with dying cells. A, The regulation frequency of ribosomal subunits in response to 5-FU in living cells and B, in dying cells. C, The average frequency of ribosomal subunits regulation in all three living cell lines versus dying ones. The dashed line represents a regulation of 1 fold (L = living, D = dying, H = HCT116 cells, A = A375 cells, R = RKO cells, F = 5-FU, M = methotrexate and P = paclitaxel).
In general, only eight ribosomal proteins (RPL4, RPL5, RPS15, RPL23, RPL26, RPS27, RPL27A, and RPS28) showed a >5% downregulation in dying versus surviving cells (the strongly coregulated RPS27 and RPS28—by a factor of two, the rest of the proteins—by ≤20%). Most of ribosomal proteins (55) showed a >5% upregulation in dying versus surviving cells, with the most upregulated (by 2.5-fold) being RPS17L and RPL21. Most ribosomal proteins coregulated with RPL21 across treatments and attachment status (59 proteins - strongly, with r≥0.7), and only 6 proteins (RPLP1, RPS15, RPS21, RPL23, RPS27, and RPS28) showed a r<0.4 correlation, of which RPS15, RPS27, and RPS28 manifested anti-correlation. The presence of the latter “renegade” ribosomal proteins is not unexpected: differences in ribosome stoichiometry between various cell types and cell states is a rather documented phenomenon and can be associated with cell fitness and different phases of growth (18). However, our finding of strong coregulation of RPS27 and RPS28 along with their anticorrelation with other ribosomal proteins in dying/surviving processes seems to be novel for literature as well as for protein-protein interaction databases. For instance, STRING database lists 10 interacting partners of RPS27, but RPS28 is not one of them. According to UniProt database, RPS27 has two binary interacting partners, one of which is MDM2. Previously, it has been shown that upon ribosomal stress, ribosomal proteins may bind to MDM2 and suppress MDM2-mediated p53 ubiquitination and degradation, leading to p53-dependent inhibition of cell division (19, 20) Therefore, 2-fold upregulation of RPS27 (and its coregulated partner RPS28) in attached cells versus detached cells may be a general mechanism blocking cell division.
Consistently Regulated Proteins in Attached Versus Detached Cells: Involvement in Life and Death Decisions
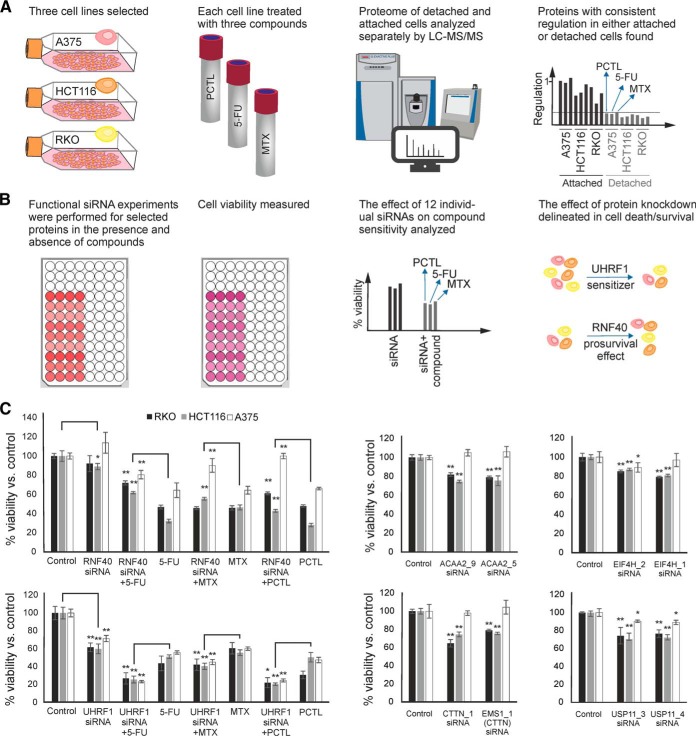
In treated detached cells versus treated attached cells, we found at least six proteins that were consistently regulated in the same direction (up- or down) regardless of the cell line and the anticancer agent used (Table II and supplemental Table S5). As a representative example, the regulation of EIF4H is depicted in Fig. 4. Three of these proteins are involved in the ubiquitination pathways and are all upregulated in dying cells compared with their surviving counterparts.
Table II. Selected proteins that are differentially expressed in treated detached vs. attached cells, regardless of the cell line and treatment.
| Gene name | Protein name | Number of peptides | Average ratio vs. control in attached cells | Average ratio vs. control in detached cells | Average ratio detached/attached | p-value (regulation in treated detached vs. attached cells) |
|---|---|---|---|---|---|---|
| EIF4H | Eukaryotic translation initiation factor 4H | 11 | 1.04 | 0.42 | 0.40 | 2.647E-06 |
| CTTN | Src substrate cortactin | 22 | 1.10 | 0.35 | 0.32 | 2.782E-06 |
| ACAA2 | 3-ketoacyl-CoA thiolase, mitochondrial | 19 | 0.97 | 2.48 | 2.56 | 0.0013 |
| RNF40 | E3 ubiquitin-protein ligase BRE1B | 20 | 0.86 | 2.12 | 2.47 | 0.0006 |
| UHRF1 | E3 ubiquitin-protein ligase UHRF1 | 26 | 0.41 | 0.92 | 2.24 | 0.0047 |
| USP11 | Ubiquitin carboxyl-terminal hydrolase 11 | 23 | 1.17 | 2.90 | 2.48 | 0.0100 |
Fig. 4.
EIF4H as an example of a protein downregulated in all types of detached treated cells treated with different anticancer agents. Error bars (standard deviations) are not available for DAF and DHF, because EIF4H was not quantified in 1–2 replicates (H = HCT116 cells, A = A375 cells, R = RKO cells, F = 5-FU, M = methotrexate and P = paclitaxel).
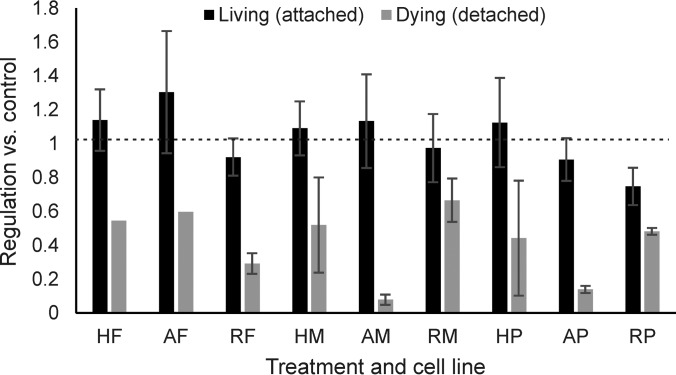
We hypothesized that such proteins might be involved in cell life and death decisions, and therefore analyzed the effect of their knock-down by siRNA on cell viability in the presence or absence of drugs. The rationale for selection of the candidates and the workflow for siRNA experiments have been shown in Fig. 5. Two different siRNAs were separately used for each protein. For five proteins out of six, knockdown reduced the cell viability, and in the case of UHRF1, increased the sensitivity to the tested drugs, whereas RNF-40 knockdown had a pro-survival effect (Fig. 5). Proteomics confirmed the successful knockdown of the selected protein in RKO or HCT116 cells in all 12 cases, with a downregulation factor of 3 to 6 (supplemental Fig. S3 and supplemental Table S6).
Fig. 5.
A, The rationale for selection of 6 proteins with potential effect on cell death or survival. B, Workflow for the functional follow-up siRNA experiments. C, The effect of siRNA knockdown on cell viability in the presence or absence of 5-FU, MTX, and PCTL. Knockdown of CTTN, USP11, ACAA2, and EIF4H had no effect on cell viability in the presence of drugs (error bars represent the standard deviations of three independent experiments in 4 replicates).
Regulated Proteins in Detached Versus Attached Cells for Untreated Controls
Even untreated control yielded a small number (≈2–10%) of detached cells because of natural death process. A multitude of proteins consistently regulated in untreated attached versus untreated detached cells were identified. Many upregulated proteins in the detached cells have been earlier associated with metastasis and cancer progression (supplemental Table S7). In STRING analysis, 30 most upregulated proteins in the detached cells mapped on energy-related pathways (supplemental Table S8, supplemental Fig. S4). The upregulated pathways were cellular respiration, electron transport chain, ATP metabolism and oxidative phosphorylation.
DISCUSSION
The unspecific response of cancer cells to an anticancer compound hampers the discerning of compound's target and MOA by expression proteomics, masking the true drug targets. In FITExP, merging proteomics data from multiple cell lines helped to increase the analysis specificity. The use of a panel of anticancer compounds enabled the Specificity calculations that further increased the analysis efficiency (7). However, for further improvements, additional dimensions of analysis were desirable. Such a dimension was hypothesized to be the detached state of the originally matrix-attached cells. Although dying in the present environment, many of these cells retain their structural integrity. Their proteomes differ most significantly from the surviving, still-attached cells (Fig. 1), which is not too surprising as death is arguably the most important event in cell's life cycle. Yet the expression levels of the drug targets turned out to be qualitatively and quantitatively similar to those in the attached cells. The specificity of FITExP analysis was improved when the rankings from the detached and attached cells were combined. Thus, the inclusion of the detached cells into the FITExP workflow represents further development of the method. The common practice in studying anticancer mechanisms, when still-attached cells are analyzed while the detached ones are often neglected and discarded, may need to be revised.
It is important to note that many, if not the majority, of the detached cells are not dead yet. Several groups, including ourselves, have observed that when the detached cells are recultured, they can start growing again and may form colonies on the plate. This observation supports the conclusion of the Nomenclature Committee on Cell Death (NCCD) that “defining life and death is more problematic than one would guess” and “the best biochemical marker of cell death is death itself” (21). Thus the detached cells should be considered to belong to the gray area between life and death, but not yet firmly committed to demise (in fact, the above NCCD report questions whether a strict border beyond which the cell is firmly committed to death, exists at all).
Although in dying cells, some downregulated proteins may have leaked out because of the loss of structural integrity of cellular membrane in a fraction of cells, the proteins found upregulated must have been really overexpressed. Furthermore, trypan blue is a relatively small molecule with a molecular weight of 873 Da, which is much smaller than the size of cellular proteins; therefore, permeability to trypan blue does not necessarily mean a total disruption of cell membrane (22). In untreated detached cells, the upregulated proteins mapped on energy-related pathways, which provides further insight into the assumed role of mitochondria in regulation/coordination of programmed cell death (23, 24). Two alternative explanations can be proposed: either mitochondria strive to produce more energy to drive the cell death process, or in a survival attempt, the cell tries to boost the ATP production to evade cell death (25).
Of a particular interest is the fact that on the PCA plot (Fig. 1) the proteomes of the surviving cells occupy much more compact areas than the corresponding proteomes of the dying cells, the latter showing large spread over the whole PCA plot. This finding can have intriguing and far-reaching consequences. The questions asking to be addressed are the following (among others). First, could this resistant phenotype be so different from the original cell as to qualify to be a different cell type (in other words, could drug-assisted near-death experience convert one cell type to another cell type)? In any case, will the surviving cells, if replated without a drug, possess epigenetic memory that would make them more resistant to the drug (26)? If yes, do these processes happen often in clinical practice and are they responsible in part for the disease relapse in cancer patients treated with chemotherapy?
In parallel, we identified several proteins that were either consistently up- or downregulated in treated detached versus attached cells, regardless of the cell type or anticancer agent used. We hypothesized that such proteins can be involved in cell life and death decisions, and, to test this hypothesis, knocked down six most promising candidates in the presence and absence of 5-FU, MTX and PCTL. As expected, all selected proteins had a certain effect on cell life and death outcomes. Although the knockdown of CTTN, USP11, ACAA2 and EIF4H had a general toxic effect with no impact on cell viability in the presence of drugs, UHRF1 sensitized the cells to the tested treatments and RNF40 gave the cells a pro-survival advantage against the treatments (Fig. 5). That three of the six proteins are involved in the ubiquitination pathways and are all upregulated in dying cells versus surviving cells is particularly interesting because of the role of the ubiquitin proteasome system in programmed cell death (27). Of the above proteins, E3 ubiquitin-protein ligase UHRF1 is perhaps the most interesting, as it seems to gradually become widely known as a biomarker for cancer (28). UHRF1 can contribute to tumor growth (29, 30) and is a primary regulator of cell senescence (31). UHRF1 has a key role in maintaining DNA methylation in mammalian cells (32) and its depletion is associated with cell cycle arrest, activation of DNA damage response and apoptosis (33). Downregulation of UHRF1 by several natural anticancer compounds has been documented (34). It is thus pretty interesting that comparative proteomics of dying and surviving cancer cells can pinpoint such an important potential drug target.
The sensitizing effect of UHRF1 knockdown might not necessarily be because of the protein itself but emerge from the cellular context. Because UHRF1 is a multifunctional protein involved in DNA methylation, chromatin modification and DNA repair, its knockdown may engage different mechanisms that ultimately result in higher sensitivity to anticancer compounds. For example, it has been shown that UHRF1 depletion can sensitize retinoblastoma and breast cancer cells to anticancer drugs via downregulation of XRCC4 and inhibition of MDR1 gene transcription, respectively (35, 36). Moreover, UHRF1 is involved in inactivation or degradation of several tumor suppressor genes, the expression of which could be restored upon UHRF1 knockdown (37, 38).
RNF40 forms an E3 ubiquitin-protein ligase complex with RNF20 that performs monoubiquitination of Lys-120 of histone H2B (39). This complex therefore plays a central role in gene regulation through the histone code. RNF40 knockdown has been shown to enhance estrogen-independent cell proliferation and activation of cell survival pathways in breast cancer (40). On the other hand, it has been suggested that RNF20 is a putative tumor suppressor, as its depletion increased cell migration and enhanced transformation and tumorigenesis. RNF20 is known to suppress the expression of several proto-oncogenes (41). These hypotheses offer tentative explanation of the observed effects of UHRF1 and RNF40 knockdown, but they need further testing.
Cell matrix interactions are required for providing essential signals for cell growth or survival of surface-attached cells. Cells detached from the surface usually die through anoikis or cell-detachment-induced apoptosis (42). Metastatic cells that colonize other organs in vivo, are however believed to somehow escape from anikosis (43). Thus, structural proteins may also relate to cell death decision-making process. Three structural proteins, keratin 2, 17, and 19, were found highly upregulated in untreated detached compared with attached cells. In detached HCT116, RKO, and A375 cells, keratin 2 was upregulated by 68-, 7-, and 54-fold, respectively; keratins 17 and 19 were upregulated less but still significantly. Keratin 2 is a late epithelial cell differentiation marker and has been associated with terminal cornification which involves keratinocyte activation, proliferation and keratinization (44). Keratin 2 binds to the tail domain of DSP (desmoplakin), which was also upregulated in our data (ranking 10th) and has been reported to anchor the intermediate filaments to the desmosomes (45). The intermediate filament vimentin protein, which is also involved in the keratinization process (46), was 20th most upregulated protein.
Many proteins found upregulated in the detached cells have been earlier associated with metastasis (supplemental Table S7). For example, cytokeratins immunostaining is often used to detect metastatic tumor cells (47). In a study by Uleberg et al., cytokeratin 2 had the strongest independent discriminatory power among other proteins identified by LC-MS/MS in cervical biopsies and could correctly classify grade 2 and 3 cervical intraepithelial neoplasia in 90% of cases (48). The coexpression of vimentin and keratin intermediate filaments in human breast cancer cells has been shown to lead to phenotypic interconversion and increased invasiveness of these cells (49). A similar study has shown that coexpression of vimentin and keratin intermediate filaments in human melanoma cells leads to increased motility (50). Among other upregulated proteins, ATP5I, TM9SF4, MAPRE2, CSPG4, MARCKSL1 and two integrins, namely ITGAV and ITGB1, have also been reported as markers of malignancy, invasion and/or metastatic phenotype (51–58). We hypothesize that downregulation of the above proteins might sensitize tumor cells to anikosis and hamper metastatic process. Testing this hypothesis experimentally goes however beyond the scope of the current study.
CONCLUSIONS
Our results revealed that transition to cell death has the greatest impact on cellular proteome that can be larger in relative magnitude than the difference between the cell lines of different cancer types or the changes induced by different treatments. The data also indicates that matrix detached cells are useful companions to attached cells in proteomics-based studies of drug target, MOA and cell death mechanisms. Characterization of these factors for a given compound can greatly reduce the time and labor in drug discovery, especially when phenotypic screening of compound libraries is used.
The approach has however several inherent limitations. First, it can be only used with cancer cells which are adherent in culture. Furthermore, the approach almost doubles the number of proteomics experiments needed to be performed in FITExP. Finally, the anticancer agents disrupting the integrity of cells create problems for this technique.
One of the most important findings of this study is that there are proteins that seem to be characteristic of cell death or survival regardless of the cell line and type of treatment. In this study, six such protein candidates were found, and their roles were validated by siRNA. Potentially, some of these proteins could represent targets for anticancer drug discovery.
DATA AVAILABILITY
Excel files containing the analyzed data are provided in Supplementary Materials. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository (59) with the data set identifier PXD007686.
Supplementary Material
Acknowledgments
We thank Marie Ståhlberg and Carina Palmberg for their assistance in different steps of the proteomic experiments.
Footnotes
* This work was supported by grants from Cancerfonden (CAN 2014/381 and 2016/546). Ülkü Güler Tokat gratefully acknowledges financial support from Scientific and Technological Research Council of Turkey (TÜBİTAK).
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- MOA
- mechanism of action
- 5-FU
- 5-fluorouracil
- DTT
- dithiothreitol
- FITExP
- Functional Identification of Target by Expression Proteomics
- IAA
- iodoacetamide
- LC-MS/MS
- liquid chromatography combined with tandem mass spectrometry
- MCM
- minichromosome maintenance protein
- MTX
- methotrexate
- OPLS
- orthogonal partial least square analysis
- PCA
- principal component analysis
- PCTL
- paclitaxel
- SDC
- sodium deoxycholate
- SDS
- sodium dodecyl sulfate.
REFERENCES
- 1. Lee J. A., Uhlik M. T., Moxham C. M., Tomandl D., and Sall D. J. (2012) Modern phenotypic drug discovery is a viable, neoclassic pharma strategy. J. Med. Chem. 55, 4527–4538 [DOI] [PubMed] [Google Scholar]
- 2. Swinney D. C., and Anthony J. (2011) How were new medicines discovered? Nature reviews Drug Discovery 10, 507–519 [DOI] [PubMed] [Google Scholar]
- 3. Eder J., Sedrani R., and Wiesmann C. (2014) The discovery of first-in-class drugs: origins and evolution. Nat. Rev. Drug discovery 13, 577–587 [DOI] [PubMed] [Google Scholar]
- 4. Holbeck S. L., Collins J. M., and Doroshow J. H. (2010) Analysis of food and drug administration–approved anticancer agents in the NCI60 panel of human tumor cell lines. Mol. Cancer Ther. 9, 1451–1460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Reinhold W. C., Sunshine M., Liu H., Varma S., Kohn K. W., Morris J., Doroshow J., and Pommier Y. (2012) CellMiner: a web-based suite of genomic and pharmacologic tools to explore transcript and drug patterns in the NCI-60 cell line set. Cancer Res. 72, 3499–3511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Somlyai G., Collins T. Q., Meuillet E. J., Hitendra P., D'Agostino D. P., and Boros L. G. (2017) Structural homologies between phenformin, lipitor and gleevec aim the same metabolic oncotarget in leukemia and melanoma. Oncotarget 8, 50187–50192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chernobrovkin A., Marin-Vicente C., Visa N., and Zubarev R. A. (2015) Functional Identification of Target by Expression Proteomics (FITExP) reveals protein targets and highlights mechanisms of action of small molecule drugs. Sci. Reports 5, 11176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ahmadian S., Barar J., Saei A. A., Fakhree M. A. A., and Omidi Y. (2009) Cellular toxicity of nanogenomedicine in MCF-7 cell line: MTT assay. J. Visualized Exp. 26, 1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Pirmoradian M., Budamgunta H., Chingin K., Zhang B., Astorga-Wells J., and Zubarev R. A. (2013) Rapid and deep human proteome analysis by single-dimension shotgun proteomics. Mol. Cell. Proteomics 12, 3330–3338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cox J., and Mann M. (2008) MaxQuant enables high peptide identification rates, individualized ppb-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 [DOI] [PubMed] [Google Scholar]
- 11. Cox J., Neuhauser N., Michalski A., Scheltema R. A., Olsen J. V., and Mann M. (2011) Andromeda: a peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 10, 1794–1805 [DOI] [PubMed] [Google Scholar]
- 12. Bylesjö M., Rantalainen M., Cloarec O., Nicholson J. K., Holmes E., and Trygg J. (2006) OPLS discriminant analysis: combining the strengths of PLS-DA and SIMCA classification. J. Chemometrics 20, 341–351 [Google Scholar]
- 13. Chernobrovkin A. L., and Zubarev R. A. (2016) How well can morphology assess cell death modality? A proteomics study. Cell Death Discovery 2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Marin-Vicente C., Lyutvinskiy Y., Romans Fuertes P., Zubarev R. A., and Visa N. (2013) The effects of 5-fluorouracil on the proteome of colon cancer cells. J. Proteome Res. 12, 1969–1979 [DOI] [PubMed] [Google Scholar]
- 15. Warner S. L., Stephens B. J., Nwokenkwo S., Hostetter G., Sugeng A., Hidalgo M., Trent J. M., Han H., and Von Hoff D. D. (2009) Validation of TPX2 as a potential therapeutic target in pancreatic cancer cells. Clin. Cancer Res. 15, 6519–6528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bian M., Fu J., Yan Y., Chen Q., Yang C., Shi Q., Jiang Q., and Zhang C. (2010) Short exposure to paclitaxel induces multipolar spindle formation and aneuploidy through promotion of acentrosomal pole assembly. Sci. China Life Sci. 53, 1322–1329 [DOI] [PubMed] [Google Scholar]
- 17. Drewes G., Ebneth A., Preuss U., Mandelkow E.-M., and Mandelkow E. (1997) MARK, a novel family of protein kinases that phosphorylate microtubule-associated proteins and trigger microtubule disruption. Cell 89, 297–308 [DOI] [PubMed] [Google Scholar]
- 18. Slavov N., Semrau S., Airoldi E., Budnik B., and van Oudenaarden A. (2015) Differential stoichiometry among core ribosomal proteins. Cell Reports 13, 865–873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhang Y., and Lu H. (2009) Signaling to p53: ribosomal proteins find their way. Cancer Cell 16, 369–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gilkes D. M., Chen L., and Chen J. (2006) MDMX regulation of p53 response to ribosomal stress. EMBO J. 25, 5614–5625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Galluzzi L., Bravo-San Pedro J., Vitale I., Aaronson S., Abrams J., Adam D., Alnemri E., Altucci L., Andrews D., and Annicchiarico-Petruzzelli M. (2015) Essential versus accessory aspects of cell death: recommendations of the NCCD 2015. Cell Death Differentiation 22, 58–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Strober W. (2001) Trypan blue exclusion test of cell viability. Curr. Protoc. Immunol. Appendix 3, Appendix 3B [DOI] [PubMed] [Google Scholar]
- 23. Kroemer G., and Reed J. C. (2000) Mitochondrial control of cell death. Nat. Med. 6, 513. [DOI] [PubMed] [Google Scholar]
- 24. Kroemer G. (1998) The mitochondrion as an integrator/coordinator of cell death pathways. Cell Death Differ. 5, 547–547 [DOI] [PubMed] [Google Scholar]
- 25. Ricci J. E., Waterhouse N., and Green D. R. (2003) Mitochondrial functions during cell death, a complex (I–V) dilemma. Cell Death Differ. 10, 488–492 [DOI] [PubMed] [Google Scholar]
- 26. Shaffer S. M., Dunagin M. C., Torborg S. R., Torre E. A., Emert B., Krepler C., Beqiri M., Sproesser K., Brafford P. A., and Xiao M. (2017) Rare cell variability and drug-induced reprogramming as a mode of cancer drug resistance. Nature 546, 431–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bernassola F., Ciechanover A., and Melino G. (2010) The ubiquitin proteasome system and its involvement in cell death pathways. Cell Death Differ. 17, 1. [DOI] [PubMed] [Google Scholar]
- 28. Ashraf W., Ibrahim A., Alhosin M., Zaayter L., Ouararhni K., Papin C., Hamiche T. A. A., Mély Y., Bronner C., and Mousli M. (2017) The epigenetic integrator UHRF1: on the road to become a universal biomarker for cancer. Oncotarget 8, 51946–51962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Jenkins Y., Markovtsov V., Lang W., Sharma P., Pearsall D., Warner J., Franci C., Huang B., Huang J., and Yam G. C. (2005) Critical role of the ubiquitin ligase activity of UHRF1, a nuclear RING finger protein, in tumor cell growth. Mol. Biol. Cell 16, 5621–5629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wang F., Yang Y.-Z., Shi C.-Z., Zhang P., Moyer M. P., Zhang H.-Z., Zou Y., and Qin H.-L. (2012) UHRF1 promotes cell growth and metastasis through repression of p16ink4a in colorectal cancer. Ann. Surg. Oncol. 19, 2753–2762 [DOI] [PubMed] [Google Scholar]
- 31. Jung H.-J., Byun H.-O., Jee B. A., Min S., Jeoun U-w Lee Y-K, Seo Y., Woo H. G., and Yoon G. (2017) The Ubiquitin-like with PHD and Ring Finger Domains 1 (UHRF1)/DNA Methyltransferase 1 (DNMT1) Axis Is a Primary Regulator of Cell Senescence. J. Biol. Chem. 292, 3729–3739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bostick M., Kim J. K., Estève P.-O., Clark A., Pradhan S., and Jacobsen S. E. (2007) UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science 317, 1760–1764 [DOI] [PubMed] [Google Scholar]
- 33. Tien A. L., Senbanerjee S., Kulkarni A., Mudbhary R., Goudreau B., Ganesan S., Sadler K. C., and Ukomadu C. (2011) UHRF1 depletion causes a G2/M arrest, activation of DNA damage response and apoptosis. Biochem. J. 435, 175–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Alhosin M., Sharif T., Mousli M., Etienne-Selloum N., Fuhrmann G., Schini-Kerth V. B., and Bronner C. (2011) Down-regulation of UHRF1, associated with re-expression of tumor suppressor genes, is a common feature of natural compounds exhibiting anti-cancer properties. J. Exp. Clin. Cancer Res. 30, 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Jin W., Liu Y., Xu S.-G. Yin W.-J. Li J.-J. Yang J.-M. and Shao Z.-M. (2010) UHRF1 inhibits MDR1 gene transcription and sensitizes breast cancer cells to anticancer drugs. Breast Cancer Res. Treatment 124, 39–48 [DOI] [PubMed] [Google Scholar]
- 36. He H., Lee C., and Kim J. K. (2018) UHRF1 depletion sensitizes retinoblastoma cells to chemotherapeutic drugs via downregulation of XRCC4. Cell Death Dis. 9, 164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Guan D., Factor D., Liu Y., Wang Z., and Kao H. (2013) The epigenetic regulator UHRF1 promotes ubiquitination-mediated degradation of the tumor-suppressor protein promyelocytic leukemia protein. Oncogene 32, 3819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Daskalos A., Oleksiewicz U., Filia A., Nikolaidis G., Xinarianos G., Gosney J. R., Malliri A., Field J. K., and Liloglou T. (2011) UHRF1-mediated tumor suppressor gene inactivation in nonsmall cell lung cancer. Cancer 117, 1027–1037 [DOI] [PubMed] [Google Scholar]
- 39. Zhu B., Zheng Y., Pham A.-D., Mandal S. S., Erdjument-Bromage H., Tempst P., and Reinberg D. (2005) Monoubiquitination of human histone H2B: the factors involved and their roles in HOX gene regulation. Mol. Cell 20, 601–611 [DOI] [PubMed] [Google Scholar]
- 40. Prenzel T., Begus-Nahrmann Y., Kramer F., Hennion M., Hsu C., Gorsler T., Hintermair C., Eick D., Kremmer E., and Simons M. (2011) Estrogen-dependent gene transcription in human breast cancer cells relies upon proteasome-dependent monoubiquitination of histone H2B. Cancer Res. 71, 5739–5753 [DOI] [PubMed] [Google Scholar]
- 41. Shema E., Tirosh I., Aylon Y., Huang J., Ye C., Moskovits N., Raver-Shapira N., Minsky N., Pirngruber J., and Tarcic G. (2008) The histone H2B-specific ubiquitin ligase RNF20/hBRE1 acts as a putative tumor suppressor through selective regulation of gene expression. Genes Development 22, 2664–2676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Frisch S. M., and Ruoslahti E. (1997) Integrins and anoikis. Curr. Opin. Cell Biol. 9, 701–706 [DOI] [PubMed] [Google Scholar]
- 43. Guadamillas M. C., Cerezo A., and del Pozo M. A. (2011) Overcoming anoikis–pathways to anchorage-independent growth in cancer. J. Cell Sci. 124, 3189–3197 [DOI] [PubMed] [Google Scholar]
- 44. Bloor B. K., Tidman N., Leigh I. M., Odell E., Dogan B., Wollina U., Ghali L., and Waseem A. (2003) Expression of keratin K2e in cutaneous and oral lesions: association with keratinocyte activation, proliferation, and keratinization. Am. J. Pathol. 162, 963–975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Broussard J. A., Yang R., Huang C., Nathamgari S. S. P., Beese A. M., Godsel L. M., Lee S., Zhou F., Sniadecki N. J., and Green K. J. (2017) The desmoplakin/intermediate filament linkage regulates cell mechanics. Mol. Biol. Cell 28, 3156–3164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Cheng F., Shen Y., Mohanasundaram P., Lindström M., Ivaska J., Ny T., and Eriksson J. E. (2016) Vimentin coordinates fibroblast proliferation and keratinocyte differentiation in wound healing via TGF-β–Slug signaling. Proc. Natl. Acad. Sci. USA 113, E4320–E4327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Bukholm I., Bondi J., Wiik P., Nesland J., Andersen S., Bakka A., and Bukholm G. (2003) Presence of isolated tumour cells in mesenteric lymph nodes predicts poor prognosis in patients with stage II colon cancer. Eur. J. Surgical Oncol. 29, 862–866 [DOI] [PubMed] [Google Scholar]
- 48. Uleberg K.-E., Munk A. C., Brede C., Gudlaugsson E., van Diermen B., Skaland I., Malpica A., Janssen E. A., Hjelle A., and Baak J. P. (2011) Discrimination of grade 2 and 3 cervical intraepithelial neoplasia by means of analysis of water soluble proteins recovered from cervical biopsies. Proteome Sci. 9, 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hendrix M., Seftor E. A., Seftor R., and Trevor K. T. (1997) Experimental coexpression of vimentin and keratin intermediate filaments in human breast cancer cells results in phenotypic interconversion and increased invasive behavior. Am. J Pathol. 150, 483. [PMC free article] [PubMed] [Google Scholar]
- 50. Chu Y.-W., Seftor E. A., Romer L. H., and Hendrix M. (1996) Experimental coexpression of vimentin and keratin intermediate filaments in human melanoma cells augments motility. Am. J. Pathol. 148, 63. [PMC free article] [PubMed] [Google Scholar]
- 51. Lozupone F., Perdicchio M., Brambilla D., Borghi M., Meschini S., Barca S., Marino M. L., Logozzi M., Federici C., and Iessi E. (2009) The human homologue of Dictyostelium discoideum phg1A is expressed by human metastatic melanoma cells. EMBO Reports 10, 1348–1354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Abiatari I., Gillen S., DeOliveira T., Klose T., Bo K., Giese N. A., Friess H., and Kleeff J. (2009) The microtubule-associated protein MAPRE2 is involved in perineural invasion of pancreatic cancer cells. Int. J. Oncol. 35, 1111–1116 [DOI] [PubMed] [Google Scholar]
- 53. Price M. A., Colvin Wanshura L. E., Yang J., Carlson J., Xiang B., Li G., Ferrone S., Dudek A. Z., Turley E. A., and McCarthy J. B. (2011) CSPG4, a potential therapeutic target, facilitates malignant progression of melanoma. Pigment Cell Melanoma Res. 24, 1148–1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Cattaruzza S., Nicolosi P. A., Braghetta P., Pazzaglia L., Benassi M. S., Picci P., Lacrima K., Zanocco D., Rizzo E., and Stallcup W. B. (2013) NG2/CSPG4–collagen type VI interplays putatively involved in the microenvironmental control of tumour engraftment and local expansion. J. Mol. Cell Biol. 5, 176–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Björkblom B., Padzik A., Mohammad H., Westerlund N., Komulainen E., Hollos P., Parviainen L., Papageorgiou A. C., Iljin K., and Kallioniemi O. (2012) c-Jun N-terminal kinase phosphorylation of MARCKSL1 determines actin stability and migration in neurons and in cancer cells. Mol. Cell. Biol. 32, 3513–3526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Waisberg J., De Souza Viana L., Affonso Junior R. J., Silva S. R., Denadai M. V., Margeotto F. B., De Souza C. S., Matos D. (2014) Overexpression of the ITGAV Gene Is Associated with Progression and Spread of Colorectal Cancer. Anticancer Res. 34, 5599–5607 [PubMed] [Google Scholar]
- 57. Wang X.-M., Li J., Yan M.-X., Liu L., Jia D.-S., Geng Q., Lin H.-C., He X.-H., Li J.-J., and Yao M. (2013) Integrative analyses identify osteopontin, LAMB3 and ITGB1 as critical pro-metastatic genes for lung cancer. PloS one 8, e55714. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 58. Patsialou A., Wang Y., Lin J., Whitney K., Goswami S., Kenny P. A., and Condeelis J. S. (2012) Selective gene-expression profiling of migratory tumor cells in vivo predicts clinical outcome in breast cancer patients. Breast Cancer Res. 14, R139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Vizcaíno J. A., Deutsch E. W., Wang R., Csordas A., Reisinger F., Rios D., Dianes J. A., Sun Z., Farrah T., and Bandeira N. (2014) ProteomeXchange provides globally coordinated proteomics data submission and dissemination. Nat. Biotechnol. 32, 223–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Excel files containing the analyzed data are provided in Supplementary Materials. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository (59) with the data set identifier PXD007686.