Abstract
Large-scale, population-based genomic studies have provided a context for modern medical genetics. Among such studies, however, African populations have remained relatively underrepresented. The breadth of genetic diversity across the African continent argues for an exploration of local genomic context to facilitate burgeoning disease mapping studies in Africa. We sought to characterize genetic variation and to assess population substructure within a cohort of HIV-positive children from Botswana—a Southern African country that is regionally underrepresented in genomic databases. Using whole-exome sequencing data from 164 Batswana and comparisons with 150 similarly sequenced HIV-positive Ugandan children, we found that 13%–25% of variation observed among Batswana was not captured by public databases. Uncaptured variants were significantly enriched (p = 2.2 × 10−16) for coding variants with minor allele frequencies between 1% and 5% and included predicted-damaging non-synonymous variants. Among variants found in public databases, corresponding allele frequencies varied widely, with Botswana having significantly higher allele frequencies among rare (<1%) pathogenic and damaging variants. Batswana clustered with other Southern African populations, but distinctly from 1000 Genomes African populations, and had limited evidence for admixture with extra-continental ancestries. We also observed a surprising lack of genetic substructure in Botswana, despite multiple tribal ethnicities and language groups, alongside a higher degree of relatedness than purported founder populations from the 1000 Genomes project. Our observations reveal a complex, but distinct, ancestral history and genomic architecture among Batswana and suggest that disease mapping within similar Southern African populations will require a deeper repository of genetic variation and allelic dependencies than presently exists.
Keywords: Africa, genomics, population genetics, HIV, AIDS, genetic mapping
Introduction
Genomic studies have played a crucial role in enhancing knowledge of genetic variation between and within populations1 and have presented a new lens through which to view the genetic basis of disease.1, 2, 3 Such surveys have consistently indicated a broad genetic diversity and complex ancestry among African populations, characterized by migration out of Africa and subsequent back migration, leading to gene flow between and within both African and non-African populations.3, 4, 5, 6, 7, 8, 9
Language and geographical distance have been shown to have the strongest correlation with differences in genetic variation between populations.4, 5, 6, 9, 10, 11, 12 For instance, Bantu-speaking populations—the largest language grouping in sub-Saharan Africa—appear separate from non-Bantu groups such as the Khoisan on principal component and FST analysis and have been shown to have differing levels of recent return-admixture events within their genomes, particularly among coastal populations such as those in Kenya.3, 6, 7, 9, 11, 13, 14 Southern African populations also separate from—and exhibit varying patterns of admixture when contrasted with—East and West African populations.3, 6, 9, 10, 11, 15 Despite this marked diversity in African populations, ascertainment bias in genotyping technologies and limited sampling have meant that African populations remain vastly underrepresented and poorly characterized at the genome level, a stark oversight in an era of large-scale genomics.3, 9, 16
Genomic underrepresentation is particularly true of Southern African populations,1, 3, 9, 17 especially when compared to populations from West and East Africa.1, 18 This lack of data adversely impacts the interpretation of medical genomic and disease association studies in these groups.2, 3, 19, 20, 21 The Southern African country of Botswana provides such an example; it is one of several Southern African countries occupying a large geographical region of Africa, sharing its borders with Namibia, Zimbabwe, Zambia, and South Africa (Figure 1). The people of Botswana—collectively referred to as Batswana—represent an amalgam of populations from multiple ethnicities, languages, and ancestry groups (Figure 1).22, 23, 24, 25, 26 Historically, the southeastern and central areas of the country, which include the capital Gaborone, were largely populated by ethnic groups speaking Setswana, a term used to describe a cluster of Sotho-Tswana-derived, and closely related, Bantu languages.23 Migrant Southeastern and Western Bantu groups interacted with Khoisan hunter-gatherers and pastoralists along migration routes northward and southward, respectively, into modern day Botswana.25, 26, 27 The Batswana thus share deep ancestral roots with established African ethnic groups; however, a subsequent history of cultural customs that included polygynous marriage, often-disputed patrilineal succession, and tribal schisms22, 24, 28, 29 and the potential admixture of Bantu ancestry individuals with Khoisan30, 31 and, to a limited extent, Eurasian ancestry individuals22 means that the genetic ancestry among Batswana is difficult to extrapolate from other African ethnic groups. Further complicating definitions of ethnic identity, anecdotal reports indicate that ethnic determination among Batswana is defined on a patrilineal basis, defaulting to the mother’s ethnicity for births that occur out of wedlock. Notwithstanding the aforementioned cultural complexities, previous genetic surveys of Batswana have been limited to a small number of geographically limited individuals, mainly of Khoisan ancestry, and included only a handful of loci.30, 31, 32 Therefore, the broader genetic variation and substructure among the Batswana remains largely undescribed.33
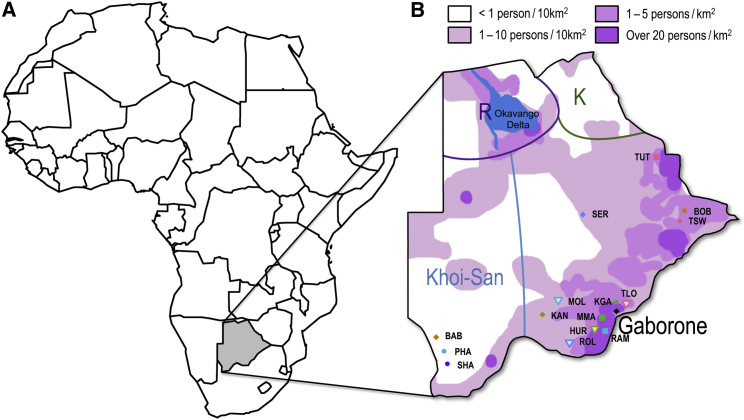
Figure 1.
Geographical Location of Botswana in Africa
(A) The location of Botswana (gray-shaded) on the African continent.
(B) The approximate regions within Botswana where self-reported ethnicities represented in the study are traditionally located. Abbreviations: TUT, Bakalanga; SHO, Shona; BOB, Babirwa; TSW, Batswapong; SER, Bangwato; KKRA, Bakgatla-Ba-Kgafela; TLO, Batlokwa; MOL, Bakwena; MMA, Bakgatla-Ba-Mmanaana; HUR, Bahurutshe; KAN, Bangwaketse; ROL, Barolong; RAM, Balete; BAB, Babolaongwe; SHA, Bashaga; PHA, Baphaleng. Color shades give the approximate population density across the country. Regions marked ‘K’ and ‘R’ correspond to the primary concentrations of individuals belonging to the K and R Guthrie groups.
The Collaborative African Genomics Network (CAfGEN), under the auspices of the Human Heredity and Health in Africa (H3Africa) Consortium (Web Resources),34, 35 has among its primary goals the use of genomics to study pediatric HIV and TB disease progression in the sub-Saharan countries of Uganda, Botswana, and, more recently, Swaziland. The ability to utilize the wealth of genetic diversity within these countries to better understand phenotypic variability in HIV and other prevalent diseases is highly desirable;1, 3, 16 however, the present dearth of available population-level genomic data30, 31, 32 makes attaining such a goal challenging. Therefore, to fulfil the primary aims of CAfGEN, we sought to provide a reference framework for medically relevant genomics studies in our Southern African population of Botswana by characterizing genetic variation and assessing genetic substructure within our cohort. We used whole-exome sequencing (WES),36 complemented in part by genome-wide SNP genotyping, from 164 pediatric HIV-affected case subjects from Botswana to assess genetic variation and population substructure based on self-reported ethnicity and Guthrie language groups. WES is attractive for variant characterization in underrepresented populations37 due to its comparatively low cost,38 its amenability to unbiased variant discovery, and the relative ease of interpretation of the molecular impact of discovered variants.39, 40 We compared the frequency and burden of rare variants within the Botswana population with data from public databases and 150 pediatric CAfGEN case subjects from the East African country of Uganda—recruited under the same protocol and sequenced on the same platform—and contextualized population-level variation and admixture with publicly available datasets.
Subjects and Methods
Study Samples
Samples were collected as part of a study of pediatric HIV disease progression within CAfGEN,35 and all participants were HIV positive. The 164 Batswana participants were recruited through the Botswana-Baylor Children’s Clinical Centre of Excellence—a tertiary pediatric HIV referral center in Gaborone, the capital of Botswana, which has the largest density of the country’s population of ∼2 million (Figure 1). The 150 Ugandan participants were recruited at the Baylor College of Medicine Children’s Foundation in Kampala, Uganda, under the same consent and protocol, and both cohort groups underwent the same genomic studies at the same time (Table S1). Approvals for the study were obtained from the institutional review boards of each of the participating institutions in CAfGEN. Genetic variation among the population of Uganda is well-represented among the ∼2,000 samples from the African Genome Variation Project (AGVP)3 and is more broadly proxied by the East African Luhya in Webuye, Kenya (LWK) from the 1000 Genomes project;1 both of these datasets are currently included among public databases. For our analyses, therefore, the use of the Uganda dataset was predominantly as a sequencing control to account for potential differences in disease ascertainment and capture platform between the Botswana dataset and available public data.
Self-reported ethnicity data were recorded for all participants and this was used to infer language group for each ethnic group from Botswana using the Ethnologue website database (Web Resources) as per the Guthrie classification of Bantu languages.23 The 164 Botswana participants included 18 self-reported ethnicities and 8 Guthrie Bantu language classes (Table S2). More than half of all Botswana participants self-reported belonging to ethnic groups associated with the Tswana language, with most groups falling into the Guthrie S-class of languages, which are associated with the Southern Bantu; one participant self-reported as Herero, which is associated with the Western Bantu.
DNA Processing and Sequencing
DNA was collected from whole blood after informed consent and was quantified for quality control (QC) using a Thermo Scientific NanoDrop 2000 and underwent Picogreen quantification on the Tecan Genios Basic plate reader. Sequencing was undertaken at the Baylor College of Medicine Human Genome Sequencing Center (HGSC). Paired-end 100 bp read libraries were prepared and sequenced on an Illumina HiSeq 2500 machine after whole-exome capture using the VCRome v.2.1 capture kit41 with an in-house spike-in called Panel-Killer v.2 (PKv2) for 209 of the 314 samples. The final batch of 105 Uganda samples was captured using VCRome v.2.1 with a newer spike-in (PKv3). Both spike-in versions are designed to capture low-coverage targets in VCRome v.2.1. At least 96% of the bases targeted were covered at a depth >20× and the sequences were aligned to the human genome reference build 37 using BWA-mem (v0.75).42 Filtered variants were then imported into Variant Tools v2.6.143 and were further annotated using data within publicly available databases: 1000 Genomes,1 dbSNP (see Web Resources), Exome Aggregation Consortium (ExAC),44 and ANNOVAR.45
Joint Calling of Sequence Data
Variants were jointly called across Botswana and Uganda samples using the Genome Analysis Toolkit (GATK v.3.5.0) Haplotype caller.46, 47 Sequence variant quality was assessed using SnpEff (v.4.22, build 2015-12-05)48 and VCFTools (v.01.1.12).49 Variants were initially filtered to have a minimum depth of 10×, Phred quality score > 30, and genotype quality score > 20; thereafter, we removed variants with >5% missingness. A total of 600,965 high-quality variants remained after QC and filtering, inclusive of 194,186 exonic variants. Variants utilized had a median of average depth of 92× and transition/transversion (TiTv) ratios within expected ranges:38 2.49 for non-exonic variants and 3.10 for exonic variants. In Uganda the median depth of sequencing coverage per-sample was 72×; similarly, TiTv ratios were within the expected range: 2.56 for non-exonic variants and 3.11 for exonic variants.
Exon Definition and Coding Variant Annotation
To define exons for the WES data, we used Variant Tools (v.2.6.1) and selected the UCSC KnownGene exon definition from the KnownGene database.50 Exonic data were then annotated on the basis of exon coding sequence start and end sites as defined within the database. Additional annotation was conducted on the coding sequence variants with the online version of the Variant Effect Predictor (VEP) tool (Ensembl GRCh37 release 88),51 using the following parameters: exon and exon position defined; biotype defined; APPRIS, which accounts for alternative splicing; Consensus Coding Sequence identifier; Condel,52 which is a consensus score of deleteriousness from SIFT53 and PolyPhen-2;54 and canonical transcripts. Annotations were then visualized with R package ggplot2 v.2.1055 within the R statistical software v.3.2.4 (see Web Resources). Uncaptured low-frequency variants were selected using Variant Tools and annotated using VEP (Ensembl GRCh37 release 88).
BeadChip Microarray Genotyping
Concurrently with exome sequencing, samples from both populations were also genotyped for quality control using the Illumina HumanCoreExome-24 BeadChip kit v.1.0, which has 547,644 markers of which 265,919 are exonic. There was at least 90% self-concordance in variant genotype calls between sequencing and array platforms in both country cohorts, which was the minimum self-concordance ratio QC threshold. SNP genotyping quality control was performed using PLINK v.1.90b3.36.56 No samples were excluded on the basis of excessive heterozygosity, which had been defined as samples exceeding 5 standard deviations of the mean heterozygosity. A set of independent SNPs for ancestry inference was obtained using linkage disequilibrium (LD)-based pruning (r2 > 0.2) and variants out of Hardy-Weinberg equilibrium (p < 0.0001) and samples with close familial relationship (PI_HAT > 0.1), limited to one of a pair of sisters (with PI_HAT > 0.5) from Botswana, were removed prior to analysis.
Principal Component Analysis
Principal component analyses (PCA) were used to distinguish cohort individuals among global populations represented in publicly available data. Data were merged with 1000 Genomes phase 3 data for 2,504 individuals, representing 5 continental ancestries, as well as genotyping and sequence data from the AGVP for 86 Sotho and 100 Zulu individuals, respectively. Shared markers between Batswana and 1000 Genomes populations were identified and extracted using Variant Tools (v.2.6.1)43 and the resulting variants were exported to PLINK57 v.1.9 to subset the datasets to the shared markers. QC was undertaken for each population before merging the datasets. Retained markers had a minor allele frequency (MAF) > 0.05, passed the Hardy-Weinberg equilibrium test at a p value of 0.0001, and were deemed independent on the basis of pairwise LD pruning using a window of 1,000 base pairs advanced by 100 SNPs at a time and an r-squared coefficient of 0.2 (see Table S4). The median PI_HAT, assessed in PLINK, for the Sotho and Zulu datasets were 0 and 0.0136, respectively, with no pair of individuals above a PI_HAT value of 0.1. Plots were then visualized with R package ggplot2 v.2.1055 within the R statistical software v.3.2.4. Comparisons of the Botswana cohort with close African populations was undertaken after excluding Afro-Caribbean in Barbados (ACB) and the African American in Southwest USA (ASW) individuals from the 1000 Genomes African populations and retaining the Sotho and Zulu AGVP populations.
Substructure Analysis within the Botswana Population (Batswana)
To assess substructure within the Botswana cohort, we followed the same QC pipeline as the PCA analysis. Independent autosomal markers pruned by LD (r-squared coefficient of 0.2) in windows of 1,000 base pairs advanced 100 SNPs at a time were used for the analysis with the SNPRelate v.1.2.0 package58 in R v.3.2.4. Botswana ethnic groups were defined per their self-reported ethnicities. The Guthrie classification of Bantu languages was used to infer language associated with the self-reported ancestry to have a secondary definition of ethnic groups23 (see Ethnologue website).
Weir and Cockerham’s Fst
Differentiation between the Botswana and the 1000 Genomes phase 3 populations using Weir and Cockerham’s fixation index estimator was assessed with the SNPRelate v.1.2.0 package58 in R v.3.2.4. Only biallelic autosomal SNPs that were shared between the Botswana population and the reference dataset (1000 Genomes) were used for this analysis. We used the default Weir and Cockerham 84 estimator option to run the analysis.
Admixture Analysis
ADMIXTURE v.1.3.0 (see Web Resources) was used to estimate ancestral clusters within the cohort given the potential of genetic admixture between the African, Khoisan, Asian, and European ancestries that are present within the population of Botswana. This software models K, the number of ancestral populations that best represent the data in the model. We used the default cross validation parameters for determining the best estimate of K using the 5-fold cross validation settings. We assessed structure between Batswana and African populations in the 1000 Genomes project and two Southern African populations from the African Genome Variation Project (AGVP) (Sotho and Zulu). The input was 26,763 WES autosomal markers at the default cross validation parameter. The estimation was from K = 1 to K = 8 and the CV error estimation minimized at K = 3. Markers were pruned for LD in PLINK57 using a window of 50 SNPs advanced by 5 SNPs at a time and an r-squared coefficient of 0.2. Plots were visualized with the web tool Pophelper v.1.1.10.59 We then assessed potential admixture within the Batswana using another unsupervised clustering model based on African, Asian, and European data from the 1000 Genomes using default parameters.
Comparison of Pathogenic Variants between Botswana and ExAC AFR Samples
The allele frequencies of pathogenic variants among Botswana samples were compared with that of samples in the Exome Aggregation Consortium (ExAC; Web Resources) denoted as “African” (AFR). To classify the pathogenic variants, we first filtered all variants found in Botswana samples with MAF > 5% in any super-population in 1000 Genomes, ExAC, or Exome Sequencing Project (ESP). After removing common variants, a variant was classified as pathogenic if it was annotated as “Pathogenic” in HGMD60 (version March 2014) or “Disease-causing mutation” (DM) in Clinvar61 (version September 2017). In total, 485 pathogenic variants were discovered in 164 Botswana samples, of which 64 were not captured among African samples in ExAC (n = 5,203). The number of captured pathogenic variants in the rare (with ExAC AFR allele frequency f < 0.1%), low frequency (0.1% < f < 1%), and intermediate frequency (1% < f < 5%) ranges were 67, 183, and 171, respectively.
Identical-by-Descent (IBD) and Inbreeding Analyses
To assess IBD across populations, variant call format (vcf.) files for each population were intersected with the target region bed files using BEDTools62 v.2.16.2. Only autosomes were used in this analysis. Phasing information was removed from vcf. files of the 1000 Genomes Phase 3 populations, and each population under study was then phased using Beagle63 4.1 with default burn-in iterations and 15 phasing iterations. In the subsequent stage, 15 independent runs of the Beagle64 4.1 IBD calling algorithm were executed on the phased target region data. For each of the runs, a random seed was generated between 1 and 10,000. We limited the IBD length to a minimum of 3 cM in each run to reduce the effects of incorrect phasing and genotyping errors in IBD length estimation. Beagle’s utilities tool ibdmerge was used to combine the output of the 15 Beagle IBD runs. To normalize the population-wide IBD length comparisons for differences in the sample sizes of each population, we used the normalization factor employed by Nakatsuka et al.,65 in which the sum of all the IBD segment lengths in a population are divided by (2nC2)-n, where n is the sample size. VCFTools (v.0.1.12) was used to compute the sample wise inbreeding coefficients and LROH (long runs of homozygosity) analysis.
Correlation between Variant Classification and African Ancestry
To determine the correlation between Southern African admixture and the number of variants classified as pathogenic and/or deleterious in each of the Botswana samples, we used the classification criteria of Kessler et al.66 Specifically, among 202,547 exonic variants called in 164 Botswana samples, we filtered 17,292 (8.5%) common variants (MAF > 5% in any super-population in 1000 Genomes, ExAC, and ESP) in genes related to diseases as reported in HGMD60 (version March 2014) and Clinvar61 (version September 2017). For variants not in these genes, we first filtered 10,982 (5.4%) variants that were not protein changing or RNA splicing from gene-based annotations (refGene, knownGene, ensGene) using Annovar (July 2017) and then further filtered 59,246 (29%) common variants with MAF > 2%. Variants passing the above filters were further classified as “pathogenic” if they were annotated “Pathogenic” in HGMD60 or “Disease-causing mutation” in Clinvar,61 and as “deleterious” if they were predicted as deleterious among at least 2 of 11 in silico predictors or if they were nonsense or splicing variants. We found 308 (0.15%) variants classified as pathogenic and deleterious (PAV), 223 (0.11%) pathogenic but non-deleterious (PAV Non-del.), 21,405 (11%) non-pathogenic but deleterious (NAV Del.), and 93,091 (46%) non-pathogenic and non-deleterious variants (NAV Non-Del.). We evaluated the correlation between the number of variants belonging to each classification in each Botswana samples with the proportion of Southern African ancestry estimated using ADMIXTURE with K = 2, which was the optimal cross-validation for number of sub-populations within the Botswana cohort.
Statistical Comparisons
The prop.test() function in R, which uses a chi-square test with a continuity correction, was used to test whether the proportion of ExAC uncaptured low-frequency variants were significantly different between the two CAfGEN groups. The cut-off for significance was set at 0.05.
Results
Uncaptured Low-Frequency Variation Is Characteristic of the Botswana Population
We observed that between 15% and 25% of sequence variants observed among Batswana were not represented in dbSNP141 (15.9%) or 1000 Genomes phase 3 (25.1%). By comparison, in the Uganda cohort, sequenced on the same platform, the proportion of uncaptured variants was smaller, particularly in the 1000 Genomes database (13.1% not in dbSNP; 12.4% not in 1000 Genomes); this presumably reflects the better overall representation of African genomes in dbSNP than 1000 Genomes and the comparatively better representation of East African versus Southern African variation in both databases. Among these uncaptured variants (n = 312,920), 2.6% were common (with MAF > 5%) and the majority (n = 257,562; 82.3%) were non-coding. Among the 194,186 coding variants in Batswana, 191,758 overlapped UCSC KnownGene50 and ENSMBL51 exon definitions, of which 19.0% (n = 36,432) were not represented within dbSNP 141 and approximately a quarter (26.6%; n = 50,955) were not observed in 1000 Genomes phase 3 (Table S3). When we compared our coding SNVs with those in the larger Exome Aggregation Consortium (ExAC) database, 16.7% of variants (n = 32,077) were not observed and the majority of these were rare, often singleton, SNVs; however, in Botswana 19.6% (n = 6,294) of ExAC uncaptured SNVs had minor allele frequencies between 1% and 5% (which we refer to as low-frequency variants); this was significantly more than that observed in Uganda (9.1%; n = 2,330; χ2 p < 2.2 × 10−16) and vastly different than the comparable proportions of captured variants (database variants found in cohort datasets) in the two countries (Figures 2A and 2B).
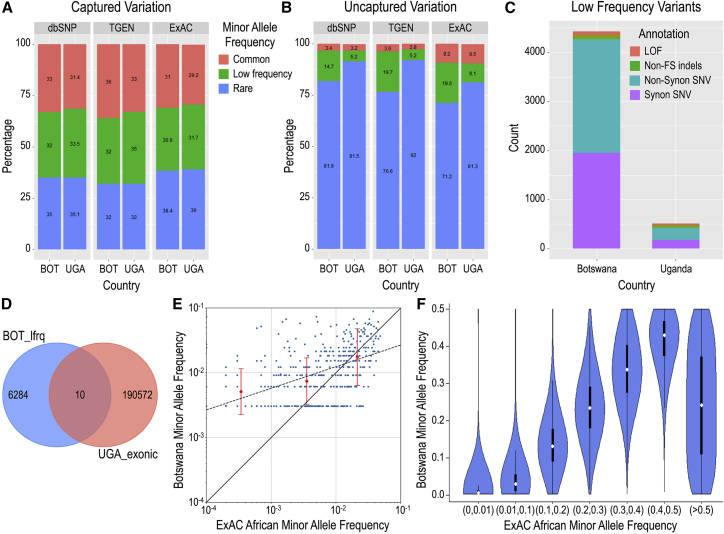
Figure 2.
Genetic Variation within the CAfGEN Cohorts
(A) CAfGEN coding variant representation captured within public databases.
(B) CAfGEN coding variation uncaptured in public databases. Abbreviations: dbSNP, Database of Single Nucleotide Polymorphisms; TGEN, 1000 Genomes phase3.
(C) Annotation of uncaptured low-frequency variants (minor allele frequency [MAF] 0.01–0.05) within Botswana and Uganda populations. Abbreviations: LOF, putative loss-of-function; non-FS indels, non-frameshifting insert-deletions; non-synon SNV, non-synonymous single-nucleotide variants.
(D) Overlap of uncaptured low-frequency variants in Botswana (BOT_lowfrq) and all exonic variants in Uganda (UGA_exonic).
(E) Comparison of allele frequencies of ClinVar and HGMD pathogenic and damaging variants among Batswana versus ExAC Africans (AFR). Red bars represent the mean (central point) ± 3 standard deviations (whiskers) for allele frequencies of all Botswana pathogenic variants in each ExAC AFR frequency (x-axis) bin range (<0.001, 0.001–0.009, 0.01–0.05).
(F) Violin plot showing minor allele frequencies of ExAC African (AFR) variants (x axis) among Botswana (y axis). The median allele frequency is shown as a white circle, with the 25th and 75th centiles as black bars, and the 5th and 95th centiles as whiskers.
More than half of these uncaptured, low-frequency, coding variants were either missense or putative loss-of-function (LOF; splice variants, stop-gains, stop-losses, frameshifts) variants (Figure 2C) and the majority were unique to Botswana (Figure 2D). Of the 2,458 uncaptured, missense, or putative LOF variants in Botswana, 193 (7.8%) were predicted to be both deleterious and probably/possibly damaging by SIFT53 and PolyPhen-2,54 respectively. Low-frequency putative LOF or predicted-damaging missense variants were found in 184 unique genes, of which 45 (24%) are associated with a known Mendelian phenotype, including PYGM and PHKG2 (glycogen storage disease types V and IX [MIM: 232600 and 613027]) and TGM6 (spinocerebellar ataxia, type 35 [MIM: 613908]) (Table S5). The majority of remaining genes were not associated with known human disease phenotypes.
When we focused on overlapping variation—SNVs observed in both public references and in our data—putative LOF variants among Batswana showed a similar pattern to uncaptured LOF variants: 44% were non-singleton, with one-quarter (25.2%) of these being observed in more than three individuals (Figure S1). In fact, when we compared the allele frequencies of uncommon (MAF < 5%) variants between two datasets, we found that while the allele frequencies were concordant between two datasets for MAF∼1%, for rare predicted-pathogenic variants with MAF < 0.1% (mean allele frequency f = 0.034%) in ExAC AFR populations, the allele frequencies in Botswana samples were significantly elevated (mean f = 0.51%, p = 1.24 × 10−29) (Figure 2E). Across the full range of MAFs observed among ExAC “African” groups, the Botswana cohort also exhibited a broad range of corresponding MAFs (Figure 2F); for instance, among ExAC variants reported as rare (MAF < 1%) in African groups, 1.2% (n = 1,924) had a MAF > 5% in Botswana, whereas only 0.4% (n = 692) had MAF > 5% in Uganda (p < 2.2 × 10−16).
The Population of Botswana Illustrates Regional Distinctions among African Populations
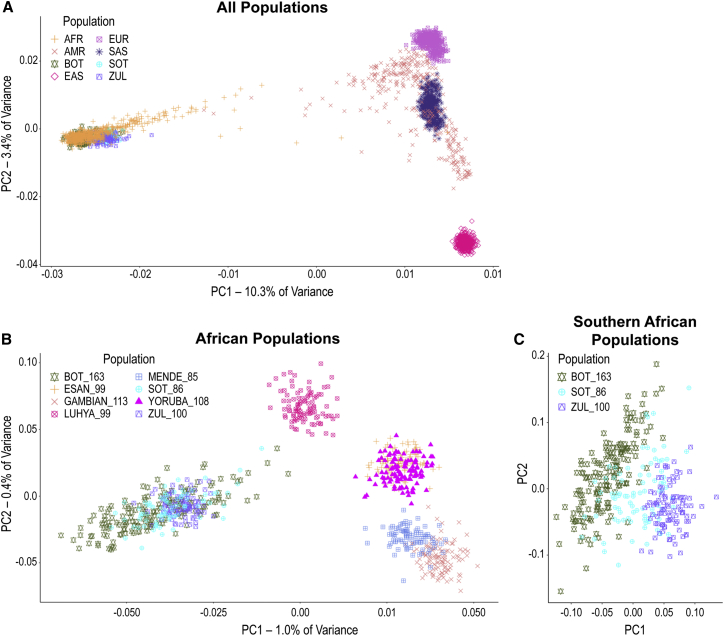
Next, we sought to place the genetic variation observed in the Botswana cohort within the context of other global populations. Using common markers (MAF > 0.05) shared with 1000 Genomes phase 3 data as well as Sotho genotyping and Zulu sequence data from the African Genome Variation Project (AGVP), the Botswana population was found to cluster closely with the other African populations on principal components 1 and 2 (Figure 3A). When we focused on solely African populations in 1000 Genomes phase 3, however, the Botswana cohort and the other Southern African populations clearly separated from West and East African populations on principal component 1 (Figure 3B). Additionally, when we focused on the Southern African populations alone, we found that Botswana clustered separately from Zulu, with the Sotho being intermediate between the two (Figure 3C). Quantitative assessments of inter-population genetic distance using Weir and Cockerham’s fixation index (FST) confirmed these observations, with higher FST values being observed between Botswana and 1000 Genomes non-African populations than between Botswana and other African populations (Figure S2).
Figure 3.
Principal Component Analysis of Batswana, 1000 Genomes, and Southern African Populations
(A) Botswana in the context of global 1000 Genomes and AGVP populations using common (MAF > 0.05) shared biallelic autosomal markers. Each symbol is an individual.
(B) Analysis of “African Populations” shows separation of Southern African populations from East and West African groups.
(C) “Southern African Populations” analysis was restricted to Batswana, Sotho, and Zulu (AGVP) and shows separation between the three groups with partial overlap of individuals at the margins of these clusters.
Abbreviations: BOT, Batswana; SOT, Sotho; ZUL, Zulu; AFR, African; AMR, Admixed American; EAS, East Asian; EUR, European; the number of individuals sampled follows the underscore (_).
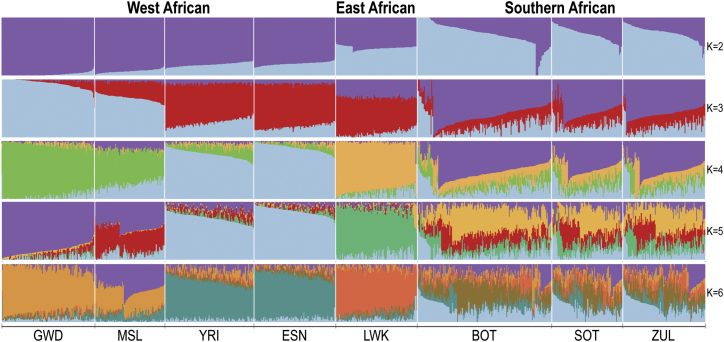
Given the complex historical ancestry of the Batswana and the apparent genetic isolation in comparison with East and West African populations, we then assessed evidence of structure between our cohort and populations represented in 1000 Genomes under a maximum likelihood-based model using ADMIXTURE v.1.3.0 (see Subjects and Methods). Using 501,963 WES autosomal markers, we compared West African, East African, and Southern African population groups (Figure 4) from 1000 Genomes and AGVP. This cross-validation error for the model was minimized at K = 3 (see Subjects and Methods), with clusters K2 and K3 separating the populations into West and East, with Batswana and the other Southern African populations appearing closer to the East African Luhya (LWK) population. Cluster K4 distinguished West African populations and at cluster K5 Batswana and the other Southern African populations had a component distinct from both the LWK and the West African populations. Cluster K6 demonstrated the higher level of sub-structure within the Southern African populations with respect to their West African and East African counterparts from 1000 Genomes; this highlights the level of structure that exists between our populations and the reference populations found within databases. When we included non-African ancestral groups (European, South Asian, and East Asian populations) in the model, we observed minimal Eurasian ancestral components within the Botswana population, suggesting minimal admixture between our cohort and these population groups (Figure S3).
Figure 4.
Admixture Analysis of Botswana and African Populations
The x axis shows the populations that were included in the ADMIXTURE unsupervised clustering maximum likelihood model. The width of each column represents the number of individuals within the listed population. Each row is independent of the other rows and the colors reflect the different genomic proportions that are derived from the African populations included in the model. The y axis represents a proportion ranging from 0 to 1. More uniform color within a given column suggests a genome composed of fewer contributing components, while increased color suggests an individual genome made up of multiple components from the surveyed populations. Abbreviations: GWD, Gambians in the Western Divisions of the Gambia; MSL, Mende in Sierra Leone; YRI, Yoruba in Nigeria; ESN, Esan in Nigeria; BOT, Batswana; SOT, Sotho from the African Genome Variation Project; ZUL, Zulu from the African Genome Variation Project.
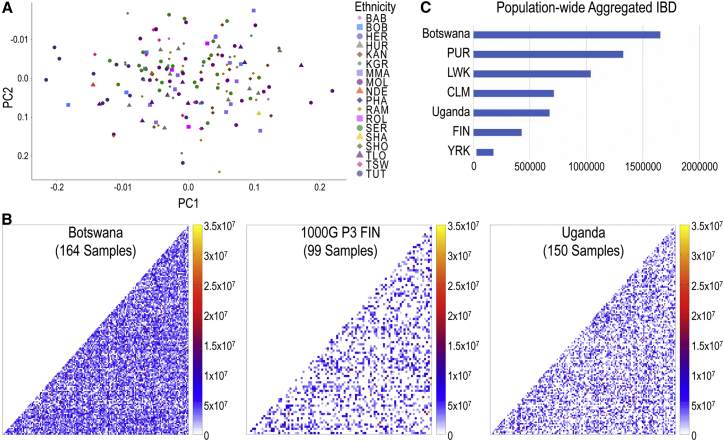
Batswana Show Minimal Population Stratification, but a High Degree of Relatedness
We then assessed substructure within the Botswana population based on self-reported ethnicity and Guthrie language. Somewhat surprisingly, given strong self-identification of ethnic affiliations within the country, the resulting PCA plot showed little evidence for genetic substructure within the Botswana population on the first two principal components (PCs) (1.56% of variance; Figure 5A). Self-reported ethnicities were distributed in a cline along PC1 with non-Sotho-Tswana groups at the edges. When we repeated this analysis using the language associated with individuals’ self-reported ancestry, there were again no easily distinguishable clusters (Figure S4A); the majority of individuals clustered on a cline along principal component 2, with individuals that speak Kalanga (a non-Sotho-Tswana language) tending toward the extremes of the cline, which was consistent with expectations based on self-reported ethnicity.
Figure 5.
Population Stratification and IBD in the Botswana Population
(A) PCA analysis of Botswana population; each point indicates an individual defined by their self-reported ancestry.
(B) Mean length of pairwise IBD segments shared within Botswana, 1000 Genomes phase 3 FIN, and the Uganda populations. Each dot represents a pairwise IBD comparison between two individuals; scale has been normalized to the same value (based on FIN) for all the plots.
(C) Aggregate IBD sharing normalized by population sample size for 1000 Genomes populations (PUR, Puerto Ricans from Puerto Rico; LWK, Luhya in Webuye, Kenya; CLM, Colombians from Medellin, Colombia; FIN, Finnish in Finland; YRI, Yoruba in Ibadan, Nigeria) and CAfGEN populations (Botswana and Uganda). YRI populations serve as a control for low IBD sharing, as they have the largest effective population size among African populations in 1000 Genomes.
Given the lack of substructure within Botswana, we proceeded to assess segments of the genome shared identical-by-descent (IBD) using the exome data from our two cohorts. Consistent with the PCA analyses, we did not find evidence of stratification within Botswana using IBD; however, when we considered the degree of pairwise IBD sharing among Botswana samples alongside our Uganda cohort and the Finnish population from phase 3 of the 1000 Genomes project (FIN), we found that Botswana had the longest shared IBD tracks (total and normalized IBD lengths of 8.84 × 1010 and 1.65 × 106, respectively) of the three populations, with Uganda also having longer normalized shared segment lengths (total 3.01 × 1010 and mean 6.74 × 105) than the purportedly founder Finnish population of 1000 Genomes (total 8.30 × 109 and mean 4.28 × 105) (Figure 5B). Even after normalizing for sample size (Subjects and Methods), IBD sharing in Botswana was still 4–5 times higher than that in FIN and was substantially greater than among the PUR, CLM, FIN, and LWK populations from 1000 Genomes (Figure 5C), which are known to have among the smallest effective population sizes among 1000 Genomes populations1 (see Web Resources for IBD). IBD sharing in Uganda was closest to the CLM population, but was still greater than FIN.
To assess the possible contribution of consanguinity and inbreeding to these results, we also calculated the sample-wise inbreeding coefficient for the populations used in Figure 5C; however, we did not observe significant differences in sample-wise inbreeding coefficients (Figure S4B), and analysis of runs of homozygosity did not demonstrate any extended segments in the Botswana population. These results suggest that both of our HIV-positive pediatric cohorts, but Botswana in particular, have a smaller effective population size than African populations currently represented in 1000 Genomes.
Discussion
We provide data derived from high-depth sequencing coverage of the coding regions of a Southern African population, sampling more than 160 individuals (>320 chromosomes). In contrast to previous surveys of genetic variation from the region, which have often utilized smaller sample sizes and relatively isolated population groups, we evaluated individuals from a highly populous urban center and utilized exome-wide sequencing data;10, 13, 67 this afforded us a view of uncaptured and rare variation that was not evident from previous characterizations.
Our Botswana population was found to harbor a significant proportion of variants that are not represented in public databases, and among variants that are represented, there are wide discrepancies with respect to minor allele frequencies. These observations present practical lessons for disease mapping efforts in Africans. Both rare-variant enrichment models (in which disease variants are expected to be relatively rare in the population but highly enriched among individuals at the extremes of disease68) and Mendelian disease mapping (in which very rare or novel variation is associated with causal variation) rely upon allele frequencies observed in large public databases1, 18, 20, 69, 70, 71 to define “rare” or “novel.” Our study suggests that identifying and interpreting such variants in exomes of individuals with African, and particularly Southern African and Botswana, ancestry using current iterations of available public databases will be challenging;1 however, viewed in the context of our population-level sequence data, many of the uncaptured variants occurred at frequencies that would make them less likely to be considered “damaging” or “deleterious.” Further, among captured variants, several uncommon, putatively damaging variants in ClinVar61 and HGMD60 were found to have an appreciably higher MAF in Botswana, making their pathogenicity more suspect, at least under a dominant mode of inheritance.
African ancestry has been shown to be positively correlated with the number of damaging variants identified in a given individual over most variant classes except predicted-deleterious variants annotated as pathogenic.66 We observed the same trend in our data (Subjects and Methods): significant correlation between Southern African admixture proportion and the number of neutral variants in the genome (r = 0.831, p = 6.21 × 10−43 for non-pathogenic, non-deleterious variants (NAV Non-del.) and r = 0.689, p = 2.72 × 10−24 for NAV Deleterious) and no significant correlation with the number of damaging variants (r = 0.038, p = 0.631 for pathogenic deleterious variants (PAV Del). and r = 0.055, p = 0.484 for PAV Non-deleterious) (Figure S5). Thus, at an individual exome level, current clinical filtering procedures will still result in multiple candidate variants to be reviewed and validated in persons of Southern African ancestry. These results bolster the assertion that the discovery of medically relevant genetic variants in African populations will likely require sequenced-based characterization of genetic variation in the respective, relevant populations.17, 20, 37, 70
The successful mapping of complex disease traits over the past decade has exploited linkage disequilibrium-derived haplotype proxies to provide genome-wide coverage using common variants. Given more disparate patterns of LD among African populations, the more common uncaptured variants identified here are unlikely to be well represented on genotyping assays designed using non-African populations, and the widely varying allele frequencies of common variants in our cohorts compared to public databases suggest that imputation of variants in Botswana may also be sub-optimal. Much has been recently made of the “genomic gap” between genome-wide studies among African (and other underrepresented) populations versus that in European groups.72, 73, 74 Our results suggest that redressing this gap will require additional investments in under-studied, high-yield populations in order to ascertain variant markers within representative populations.3, 9, 16, 17, 20, 70 Ongoing efforts to produce representative genotyping platforms aimed at African34 and African ancestry75, 76 populations are thus laudable and auger well for maximizing disease-variant genetic mapping within Africa and the wider genomics community.
The tremendous ethnic diversity within African countries is typically cited as a hindrance for conducting genomic studies within Africa,77 as the potential for population stratification creates a challenge for adequately powering association studies.78, 79 By contrast, we observed minimal evidence for substructure in our Botswana cohort, despite multiple self-reported language groups and ethnicities; this suggests that conducting traditional genome-wide association studies (GWASs) within the Botswana population will be feasible even with more modest sample sizes. This lack of substructure is likely to reflect, in part, the complex social ancestry of the country, which includes a shared history of succession breakaways and intermarriage, particularly between Sotho-Tswana ethnic groups,22, 24, 25, 28 along with historical, political, and cultural practices of assimilating large groups from different Bantu language clusters under a single umbrella ethnicity.26, 80 For example, the Balete, despite their large size and Nguni origins, were assimilated26 into Sotho-Tswana groups and now speak Tswana, which is a different language group from the Nguni Bantu language they spoke prior to their assimilation. The presence of non-Tswana-speaking ethnicities at the margins of the clines observed in our data may thus represent the remnants of past Sotho-Tswana endogamy.22, 24 These anthropological groups have since merged within modern-day Gaborone, a populous, urban city in which current marriage and mating practices17 mean that self-reported ethnic affiliations are more social constructs than reflective of the highly stratified genetic ancestry found in more rural Botswana.33 This separates our work from previous genetic surveys.
In fact, compared to other African populations, Batswana not only clustered distinctly from—and showed little evidence of admixture with—East and West African population genomes, but also had a much higher degree of relatedness than even known founder populations. This relatedness is likely to be a major factor contributing to the significantly higher number of low-frequency variants observed in our cohort. Despite this, the data provided little evidence for excessive inbreeding; therefore, we postulate that demographic events and genetic drift are the main contributors to the distinctiveness of the Botswana cohort.31 As noted above, our cohort included a large number of Southern Bantu, whose migration patterns,11 shared history,22, 26, 27, 81 and assimilation of other ethnic groups11, 17, 24 following their split from East and West African Bantu populations are likely to have contributed to the distinct Batswana ancestry. In addition, at its height in the mid 1990s, HIV prevalence in Botswana for the general population was close to 25% and closer to 40% in pregnant women (UNAIDS in Web Resources). Given the high mortality rate of HIV before the widespread use of antiretroviral therapy (ART), it is possible that the excessive lengths of the genome found to be IBD in this childhood cohort are the consequence of the HIV epidemic, with the effects of this population drift being more manifest in the relatively small population of Botswana (∼2 million) than in the large population of Uganda (∼40 million), even though HIV levels were higher in Uganda at the peak of the epidemic. Irrespective of the underlying factors driving this particular observation, the resulting extensive shared segments would be expected, paradoxically, to make disease mapping in Botswana significantly easier than in other African populations.
The clinical center in Botswana is the largest regional pediatric HIV referral center in the country; however, our local ascertainment meant that we did not have any participants from the genetically isolated Khoisan people6, 30, 31 (who were also not represented in our ADMIXTURE analysis) or Western Bantu related ethnicities—both more populous in the western and northern regions of the country (Figure 1).23 It is still plausible, however, that admixture with the Khoisan also contributes to the relative distinctness of the Botswana population10, 13, 17, 30 and that wider sampling of the population would reveal still greater isolation. Although our population is biased by HIV status, infection with HIV is known to cut across all strata of the population in Botswana; we thus do not expect our overall results to differ significantly if our study was conducted in a non-HIV-positive cohort. To the best of our knowledge, there is no other currently available similar sequence data from Botswana to provide further context for our findings, and we note that these observations were also unique with respect to our similarly recruited pediatric HIV cohort from Uganda.
The genetic architecture of the Botswana population described here underscores the complex ancestry of Southern African populations and reinforces recent suggestions that reliance on ethnic, tribal, or language group labels as indicators of study feasibility may be overstated, particularly among urban African populations.17 As one of the first large-scale deep-sequencing studies in sub-Saharan Africa, our results also emphasize the need to characterize fine-scale genome variation among underrepresented African populations; this is imperative to better facilitate Mendelian and complex-trait mapping among those who harbor a significant burden of global disease. Doing so promises to both uncover the allelic architecture needed to interpret genomic studies on the continent and provide a deeper understanding of population movements that are fundamental to human history.
Acknowledgments
This study makes use of data generated by the African Partnership for Chronic Disease Research. A full list of the investigators and funders who contributed to the generation of the data is available from APCDR website (see Web Resources). The authors would like to acknowledge the participating families, as well as the contributions of Kennedy Sichone in participant recruitment and sample acquisition, Nancy Hall and Roa Sadat in sample inventory and management, and Adam Gillum with manuscript figures. This work was supported by a Collaborative Center grant (Collaborative African Genomics Network [CAfGEN]) to G.A., G. Mardon, A.K., M.J., and S.W.M., from the National Institutes of Health (grant U54AI110398), and from the Center for Globalization at Baylor College of Medicine to G. Mardon. N.A.H. was also supported by a Clinical Scientist Development Award from the Doris Duke Charitable Foundation (grant 2013096).
Published: April 26, 2018
Footnotes
Supplemental Data include five figures and five tables and can be found with this article online at https://doi.org/10.1016/j.ajhg.2018.03.010.
Accession Numbers
AGVP Datasets: EGAS00001000960/TBA (AGV curated all WGS vcf. files), GAS00001000960/EGAD00001001663 (AGV allele frequencies vcf. files). CAfGEN exome sequence datasets (BAMs and vcfs) are being made publicly available via the European Genome Archive (EGA) in accordance with guidelines agreed upon by the Human Health and Heredity in Africa (H3Africa) Consortium.
Web Resources
1000 Genomes, http://www.internationalgenome.org/
1000 Genomes IBD Segment Methods, http://ftp.1000genomes.ebi.ac.uk/vol1/ftp/release/20130502/supporting/ibd_by_pair/20150129_IBD_segment_methods.pdf
Admixture Manual, https://www.genetics.ucla.edu/software/admixture/admixture-manual.pdf
APCDR, https://www.apcdr.org
dbNSFP, http://varianttools.sourceforge.net/Annotation/DbNSFP
Ethnologue, https://www.ethnologue.com
European Genome-phenome Archive (EGA), https://www.ebi.ac.uk/ega
ExAC Browser, http://exac.broadinstitute.org/
H3Africa, https://h3africa.org/
OMIM, http://www.omim.org/
Pophelper, http://pophelper.com/SUWRS
R statistical software, https://www.r-project.org/
UNAIDS, http://aidsinfo.unaids.org
Supplemental Data
References
- 1.Auton A., Brooks L.D., Durbin R.M., Garrison E.P., Kang H.M., Korbel J.O., Marchini J.L., McCarthy S., McVean G.A., Abecasis G.R., 1000 Genomes Project Consortium A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baker J.L., Shriner D., Bentley A.R., Rotimi C.N. Pharmacogenomic implications of the evolutionary history of infectious diseases in Africa. Pharmacogenomics J. 2017;17:112–120. doi: 10.1038/tpj.2016.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gurdasani D., Carstensen T., Tekola-Ayele F., Pagani L., Tachmazidou I., Hatzikotoulas K., Karthikeyan S., Iles L., Pollard M.O., Choudhury A. The African Genome Variation Project shapes medical genetics in Africa. Nature. 2015;517:327–332. doi: 10.1038/nature13997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reed F.A., Tishkoff S.A. African human diversity, origins and migrations. Curr. Opin. Genet. Dev. 2006;16:597–605. doi: 10.1016/j.gde.2006.10.008. [DOI] [PubMed] [Google Scholar]
- 5.Tishkoff S.A., Reed F.A., Friedlaender F.R., Ehret C., Ranciaro A., Froment A., Hirbo J.B., Awomoyi A.A., Bodo J.M., Doumbo O. The genetic structure and history of Africans and African Americans. Science. 2009;324:1035–1044. doi: 10.1126/science.1172257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schlebusch C.M., Skoglund P., Sjödin P., Gattepaille L.M., Hernandez D., Jay F., Li S., De Jongh M., Singleton A., Blum M.G. Genomic variation in seven Khoe-San groups reveals adaptation and complex African history. Science. 2012;338:374–379. doi: 10.1126/science.1227721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Currie T.E., Meade A., Guillon M., Mace R. Cultural phylogeography of the Bantu languages of sub-Saharan Africa. Proc. Biol. Sci. 2013;280:20130695. doi: 10.1098/rspb.2013.0695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hellenthal G., Busby G.B.J., Band G., Wilson J.F., Capelli C., Falush D., Myers S. A genetic atlas of human admixture history. Science. 2014;343:747–751. doi: 10.1126/science.1243518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Busby G.B., Band G., Si Le Q., Jallow M., Bougama E., Mangano V.D., Amenga-Etego L.N., Enimil A., Apinjoh T., Ndila C.M., Malaria Genomic Epidemiology Network Admixture into and within sub-Saharan Africa. eLife. 2016;5:e15266. doi: 10.7554/eLife.15266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schuster S.C., Miller W., Ratan A., Tomsho L.P., Giardine B., Kasson L.R., Harris R.S., Petersen D.C., Zhao F., Qi J. Complete Khoisan and Bantu genomes from southern Africa. Nature. 2010;463:943–947. doi: 10.1038/nature08795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li S., Schlebusch C., Jakobsson M. Genetic variation reveals large-scale population expansion and migration during the expansion of Bantu-speaking peoples. Proc. Biol. Sci. 2014;281:1448. doi: 10.1098/rspb.2014.1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Novembre J., Johnson T., Bryc K., Kutalik Z., Boyko A.R., Auton A., Indap A., King K.S., Bergmann S., Nelson M.R. Genes mirror geography within Europe. Nature. 2008;456:98–101. doi: 10.1038/nature07331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Petersen D.C., Libiger O., Tindall E.A., Hardie R.A., Hannick L.I., Glashoff R.H., Mukerji M., Fernandez P., Haacke W., Schork N.J., Hayes V.M., Indian Genome Variation Consortium Complex patterns of genomic admixture within southern Africa. PLoS Genet. 2013;9:e1003309. doi: 10.1371/journal.pgen.1003309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pickrell J.K., Patterson N., Loh P.-R., Lipson M., Berger B., Stoneking M., Pakendorf B., Reich D. Ancient west Eurasian ancestry in southern and eastern Africa. Proc. Natl. Acad. Sci. USA. 2014;111:2632–2637. doi: 10.1073/pnas.1313787111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim H.L., Ratan A., Perry G.H., Montenegro A., Miller W., Schuster S.C. Khoisan hunter-gatherers have been the largest population throughout most of modern-human demographic history. Nat. Commun. 2014;5:5692. doi: 10.1038/ncomms6692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rotimi C.N., Tekola-Ayele F., Baker J.L., Shriner D. The African diaspora: history, adaptation and health. Curr. Opin. Genet. Dev. 2016;41:77–84. doi: 10.1016/j.gde.2016.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.May A., Hazelhurst S., Li Y., Norris S.A., Govind N., Tikly M., Hon C., Johnson K.J., Hartmann N., Staedtler F., Ramsay M. Genetic diversity in black South Africans from Soweto. BMC Genomics. 2013;14:644. doi: 10.1186/1471-2164-14-644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Altshuler D.M., Gibbs R.A., Peltonen L., Altshuler D.M., Gibbs R.A., Peltonen L., Dermitzakis E., Schaffner S.F., Yu F., Peltonen L., International HapMap 3 Consortium Integrating common and rare genetic variation in diverse human populations. Nature. 2010;467:52–58. doi: 10.1038/nature09298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mersha T.B., Abebe T. Self-reported race/ethnicity in the age of genomic research: its potential impact on understanding health disparities. Hum. Genomics. 2015;9:1. doi: 10.1186/s40246-014-0023-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dopazo J., Amadoz A., Bleda M., Garcia-Alonso L., Alemán A., García-García F., Rodriguez J.A., Daub J.T., Muntané G., Rueda A. 267 Spanish exomes reveal population-specific differences in disease-related genetic variation. Mol. Biol. Evol. 2016;33:1205–1218. doi: 10.1093/molbev/msw005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chapman S.J., Hill A.V. Human genetic susceptibility to infectious disease. Nat. Rev. Genet. 2012;13:175–188. doi: 10.1038/nrg3114. [DOI] [PubMed] [Google Scholar]
- 22.Sillery A. Botswana: A Short Political History. In: Kirk-Greene A.H.M., editor. Bungay: Methuen & Co Ltd; 1974. [Google Scholar]
- 23.Batibo H.M. A lexicostatistical survey of the Bantu language of Botswana. S. Afr. J. Afr. Lang. 1998;18:22–28. [Google Scholar]
- 24.Tlou T. The nature of Batswana states: towards a theory of Botswana traditional government - the Batawana case. In: Edge W.A., Lekorwe M.H., editors. Botswana: Politics and Society. J.L van Schaik; Pretoria: 1998. p. 22. [Google Scholar]
- 25.Gulbrandsen Ø. The rise of the North-Western Tswana kingdoms: on the dynamics of interaction between internal relations and external forces. Africa. 1993;63:550–582. [Google Scholar]
- 26.Schapera I., Comaroff J., Kuper A. The Tswana. In: Forde D., editor. Plymoth: Clarke, Doble & Brendon; 1953. [Google Scholar]
- 27.van Waarden C. The Late Iron Age. In: Lane P., Reid A., Segobye A., editors. Ditswammung, the Archeology of Botswana. Pula Press; Gaborone: 1998. pp. 115–160. [Google Scholar]
- 28.Ngcongo L.D. Gaborone; 1978. Origins of the Tswana. [Google Scholar]
- 29.Matemba Y.H. The pre-colonial political history of Bakgatla ba ga Mmanaana of Botswana, c. 1600-1881. Botsw. Notes Rec. 2003;2003:53–67. [Google Scholar]
- 30.Pickrell J.K., Patterson N., Barbieri C., Berthold F., Gerlach L., Güldemann T., Kure B., Mpoloka S.W., Nakagawa H., Naumann C. The genetic prehistory of southern Africa. Nat. Commun. 2012;3:1143. doi: 10.1038/ncomms2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.González-Santos M., Montinaro F., Oosthuizen O., Oosthuizen E., Busby G.B.J., Anagnostou P., Destro-Bisol G., Pascali V., Capelli C. Genome-wide snp analysis of southern african populations provides new insights into the dispersal of bantu-Speaking groups. Genome Biol. Evol. 2015;7:2560–2568. doi: 10.1093/gbe/evv164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barbieri C., Butthof A., Bostoen K., Pakendorf B. Genetic perspectives on the origin of clicks in Bantu languages from southwestern Zambia. Eur. J. Hum. Genet. 2013;21:430–436. doi: 10.1038/ejhg.2012.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tau T., Wally A., Fanie T.P., Ngono G.L., Mpoloka S.W., Davison S., D’Amato M.E. Genetic variation and population structure of Botswana populations as identified with AmpFLSTR Identifiler short tandem repeat (STR) loci. Sci. Rep. 2017;7:6768. doi: 10.1038/s41598-017-06365-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rotimi C., Abayomi A., Abimiku A., Adabayeri V.M., Adebamowo C., Adebiyi E., Ademola A.D., Adeyemo A., Adu D., Affolabi D., H3Africa Consortium Research capacity. Enabling the genomic revolution in Africa. Science. 2014;344:1346–1348. doi: 10.1126/science.1251546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mlotshwa B.C., Mwesigwa S., Mboowa G., Williams L., Retshabile G., Kekitiinwa A., Wayengera M., Kyobe S., Brown C.W., Hanchard N.A. The collaborative African genomics network training program: a trainee perspective on training the next generation of African scientists. Genet. Med. 2017;19:826–833. doi: 10.1038/gim.2016.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Belkadi A., Pedergnana V., Cobat A., Itan Y., Vincent Q.B., Abhyankar A., Shang L., El Baghdadi J., Bousfiha A., Alcais A., Exome/Array Consortium Whole-exome sequencing to analyze population structure, parental inbreeding, and familial linkage. Proc. Natl. Acad. Sci. USA. 2016;113:6713–6718. doi: 10.1073/pnas.1606460113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tang D., Anderson D., Francis R.W., Syn G., Jamieson S.E., Lassmann T., Blackwell J.M. Reference genotype and exome data from an Australian Aboriginal population for health-based research. Sci. Data. 2016;3:160023. doi: 10.1038/sdata.2016.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Carson A.R., Smith E.N., Matsui H., Brækkan S.K., Jepsen K., Hansen J.-B., Frazer K.A. Effective filtering strategies to improve data quality from population-based whole exome sequencing studies. BMC Bioinformatics. 2014;15:125. doi: 10.1186/1471-2105-15-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Warr A., Robert C., Hume D., Archibald A., Deeb N., Watson M. Exome Sequencing: Current and Future Perspectives. G3 (Bethesda) 2015;5:1543–1550. doi: 10.1534/g3.115.018564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lelieveld S.H., Veltman J.A., Gilissen C. Novel bioinformatic developments for exome sequencing. Hum. Genet. 2016;135:603–614. doi: 10.1007/s00439-016-1658-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bainbridge M.N., Wang M., Wu Y., Newsham I., Muzny D.M., Jefferies J.L., Albert T.J., Burgess D.L., Gibbs R.A. Targeted enrichment beyond the consensus coding DNA sequence exome reveals exons with higher variant densities. Genome Biol. 2011;12:R68. doi: 10.1186/gb-2011-12-7-r68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li H. (2013). Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. (2013). arXiv. 1303.3997.
- 43.Wang G.T., Peng B., Leal S.M. Variant association tools for quality control and analysis of large-scale sequence and genotyping array data. Am. J. Hum. Genet. 2014;94:770–783. doi: 10.1016/j.ajhg.2014.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lek M., Karczewski K.J., Minikel E.V., Samocha K.E., Banks E., Fennell T., O’Donnell-Luria A.H., Ware J.S., Hill A.J., Cummings B.B., Exome Aggregation Consortium Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang K., Li M., Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M., DePristo M.A. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Van Der Auwera G.A., Carneiro M.O., Hartl C., Poplin R., Del Angel G., Levy-Moonshine A., Jordan T., Shakir K., Roazen D., Thibault From FastQ data to high confidence varant calls: the Genome Analysis Toolkit best practices pipeline. Curr. Protoc. Bioinformatics. 2014;43:1–33. doi: 10.1002/0471250953.bi1110s43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cingolani P., Platts A., Wang L., Coon M., Nguyen T., Wang L., Land S.J., Lu X., Ruden D.M. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 2012;6:80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Danecek P., Auton A., Abecasis G., Albers C.A., Banks E., DePristo M.A., Handsaker R.E., Lunter G., Marth G.T., Sherry S.T., 1000 Genomes Project Analysis Group The variant call format and VCFtools. Bioinformatics. 2011;27:2156–2158. doi: 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hsu F., Kent W.J., Clawson H., Kuhn R.M., Diekhans M., Haussler D. The UCSC known genes. Bioinformatics. 2006;22:1036–1046. doi: 10.1093/bioinformatics/btl048. [DOI] [PubMed] [Google Scholar]
- 51.McLaren W., Gil L., Hunt S.E., Riat H.S., Ritchie G.R.S., Thormann A., Flicek P., Cunningham F. The Ensembl Variant Effect Predictor. Genome Biol. 2016;17:122. doi: 10.1186/s13059-016-0974-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.González-Pérez A., López-Bigas N. Improving the assessment of the outcome of nonsynonymous SNVs with a consensus deleteriousness score, Condel. Am. J. Hum. Genet. 2011;88:440–449. doi: 10.1016/j.ajhg.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kumar P., Henikoff S., Ng P.C. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat. Protoc. 2009;4:1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 54.Adzhubei I.A., Schmidt S., Peshkin L., Ramensky V.E., Gerasimova A., Bork P., Kondrashov A.S., Sunyaev S.R. A method and server for predicting damaging missense mutations. Nat. Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wickham H. Springer-Verlag New York; 2009. Elegant Graphics for Data Analysis. [Google Scholar]
- 56.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A.R., Bender D., Maller J., Sklar P., de Bakker P.I., Daly M.J., Sham P.C. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chang C.C., Chow C.C., Tellier L.C., Vattikuti S., Purcell S.M., Lee J.J. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience. 2015;4:7. doi: 10.1186/s13742-015-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zheng X., Levine D., Shen J., Gogarten S.M., Laurie C., Weir B.S. A high-performance computing toolset for relatedness and principal component analysis of SNP data. Bioinformatics. 2012;28:3326–3328. doi: 10.1093/bioinformatics/bts606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Francis R.M. pophelper: an R package and web app to analyse and visualize population structure. Mol. Ecol. Resour. 2017;17:27–32. doi: 10.1111/1755-0998.12509. [DOI] [PubMed] [Google Scholar]
- 60.Stenson P.D., Mort M., Ball E.V., Shaw K., Phillips A., Cooper D.N. The Human Gene Mutation Database: building a comprehensive mutation repository for clinical and molecular genetics, diagnostic testing and personalized genomic medicine. Hum. Genet. 2014;133:1–9. doi: 10.1007/s00439-013-1358-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Landrum M.J., Lee J.M., Riley G.R., Jang W., Rubinstein W.S., Church D.M., Maglott D.R. ClinVar: public archive of relationships among sequence variation and human phenotype. Nucleic Acids Res. 2014;42:D980–D985. doi: 10.1093/nar/gkt1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Quinlan A.R., Hall I.M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Browning S.R., Browning B.L. Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. Am. J. Hum. Genet. 2007;81:1084–1097. doi: 10.1086/521987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Browning B.L., Browning S.R. Improving the accuracy and efficiency of identity-by-descent detection in population data. Genetics. 2013;194:459–471. doi: 10.1534/genetics.113.150029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nakatsuka N., Moorjani P., Rai N., Sarkar B., Tandon A., Patterson N., Bhavani G.S., Girisha K.M., Mustak M.S., Srinivasan S. The promise of discovering population-specific disease-associated genes in South Asia. Nat. Genet. 2017;49:1403–1407. doi: 10.1038/ng.3917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kessler M.D., Yerges-Armstrong L., Taub M.A., Shetty A.C., Maloney K., Jeng L.J.B., Ruczinski I., Levin A.M., Williams L.K., Beaty T.H., Consortium on Asthma among African-ancestry Populations in the Americas (CAAPA) Challenges and disparities in the application of personalized genomic medicine to populations with African ancestry. Nat. Commun. 2016;7:12521. doi: 10.1038/ncomms12521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fagny M., Patin E., MacIsaac J.L., Rotival M., Flutre T., Jones M.J., Siddle K.J., Quach H., Harmant C., McEwen L.M. The epigenomic landscape of African rainforest hunter-gatherers and farmers. Nat. Commun. 2015;6:10047. doi: 10.1038/ncomms10047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cirulli E.T., Goldstein D.B. Uncovering the roles of rare variants in common disease through whole-genome sequencing. Nat. Rev. Genet. 2010;11:415–425. doi: 10.1038/nrg2779. [DOI] [PubMed] [Google Scholar]
- 69.Wong L.P., Ong R.T.H., Poh W.T., Liu X., Chen P., Li R., Lam K.K., Pillai N.E., Sim K.S., Xu H. Deep whole-genome sequencing of 100 southeast Asian Malays. Am. J. Hum. Genet. 2013;92:52–66. doi: 10.1016/j.ajhg.2012.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Scott E.M., Halees A., Itan Y., Spencer E.G., He Y., Azab M.A., Gabriel S.B., Belkadi A., Boisson B., Clark A.G., Greater Middle East Variome Consortium. Alkuraya F.S., Casanova J.L., Gleeson J.G. Characterization of Greater Middle Eastern genetic variation for enhanced disease gene discovery. Nat. Genet. 2016;48:1071–1076. doi: 10.1038/ng.3592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Petrovski S., Goldstein D.B. Unequal representation of genetic variation across ancestry groups creates healthcare inequality in the application of precision medicine. Genome Biol. 2016;17:157. doi: 10.1186/s13059-016-1016-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Need A.C., Goldstein D.B. Next generation disparities in human genomics: concerns and remedies. Trends Genet. 2009;25:489–494. doi: 10.1016/j.tig.2009.09.012. [DOI] [PubMed] [Google Scholar]
- 73.Popejoy A.B., Fullerton S.M. Genomics is failing on diversity. Nature. 2016;538:161–164. doi: 10.1038/538161a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bustamante C.D., Burchard E.G., De la Vega F.M. Genomics for the world. Nature. 2011;475:163–165. doi: 10.1038/475163a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mathias R.A., Taub M.A., Gignoux C.R., Fu W., Musharoff S., O’Connor T.D., Vergara C., Torgerson D.G., Pino-Yanes M., Shringarpure S.S., CAAPA A continuum of admixture in the Western Hemisphere revealed by the African Diaspora genome. Nat. Commun. 2016;7:12522. doi: 10.1038/ncomms12522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Johnston H.R., Hu Y.-J., Gao J., O’Connor T.D., Abecasis G.R., Wojcik G.L., Gignoux C.R., Gourraud P.A., Lizee A., Hansen M., CAAPA Consortium Identifying tagging SNPs for African specific genetic variation from the African Diaspora Genome. Sci. Rep. 2017;7:46398. doi: 10.1038/srep46398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Novembre J., Ramachandran S. Perspectives on human population structure at the cusp of the sequencing era. Annu. Rev. Genomics Hum. Genet. 2011;12:245–274. doi: 10.1146/annurev-genom-090810-183123. [DOI] [PubMed] [Google Scholar]
- 78.Berger M., Stassen H.H., Köhler K., Krane V., Mönks D., Wanner C., Hoffmann K., Hoffmann M.M., Zimmer M., Bickeböller H., Lindner T.H. Hidden population substructures in an apparently homogeneous population bias association studies. Eur. J. Hum. Genet. 2006;14:236–244. doi: 10.1038/sj.ejhg.5201546. [DOI] [PubMed] [Google Scholar]
- 79.Tian C., Gregersen P.K., Seldin M.F. Accounting for ancestry: population substructure and genome-wide association studies. Hum. Mol. Genet. 2008;17(R2):R143–R150. doi: 10.1093/hmg/ddn268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wilmsen E.N. Mutable identities: moving beyond ethnicity in Botswana. J. South. Afr. Stud. 2002;28:825–841. [Google Scholar]
- 81.Morton F. Settlements, landscapes and identities among the Tswana of the western transvaal and eastern Kalahari before 1820. S. Afr. Archaeol. Bull. 2013;68:15–26. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.