Abstract
Large-scale human genetics studies are ascertaining increasing proportions of populations as they continue growing in both number and scale. As a result, the amount of cryptic relatedness within these study cohorts is growing rapidly and has significant implications on downstream analyses. We demonstrate this growth empirically among the first 92,455 exomes from the DiscovEHR cohort and, via a custom simulation framework we developed called SimProgeny, show that these measures are in line with expectations given the underlying population and ascertainment approach. For example, within DiscovEHR we identified ∼66,000 close (first- and second-degree) relationships, involving 55.6% of study participants. Our simulation results project that >70% of the cohort will be involved in these close relationships, given that DiscovEHR scales to 250,000 recruited individuals. We reconstructed 12,574 pedigrees by using these relationships (including 2,192 nuclear families) and leveraged them for multiple applications. The pedigrees substantially improved the phasing accuracy of 20,947 rare, deleterious compound heterozygous mutations. Reconstructed nuclear families were critical for identifying 3,415 de novo mutations in ∼1,783 genes. Finally, we demonstrate the segregation of known and suspected disease-causing mutations, including a tandem duplication that occurs in LDLR and causes familial hypercholesterolemia, through reconstructed pedigrees. In summary, this work highlights the prevalence of cryptic relatedness expected among large healthcare population-genomic studies and demonstrates several analyses that are uniquely enabled by large amounts of cryptic relatedness.
Keywords: cryptic relatedness, pedigree reconstruction, relationship inference, identity by decent, compound heterozygous mutation phasing, de novo mutations, precision medicine, healthcare population-based genetic study, exome sequencing, family structure, familial hypercholesterolemia
Introduction
The number and scale of large human sequencing projects, including DiscovEHR,1 UK Biobank,2 the US government’s All of Us (part of the Precision Medicine Initiative),3 TOPMed (Web Resources), ExAC/gnomAD,4 and many others, is rapidly growing. Many of these studies are collecting samples from integrated healthcare populations that have accompanying phenotype-rich electronic health records (EHRs) with a goal of combining the EHRs and genomic sequence data to catalyze translational discoveries and precision medicine.1 These large-scale healthcare population-based genomic (HPG) studies are recruiting participants through healthcare systems where volunteers donate DNA and provide medically relevant metrics recorded in their EHRs. A major difference between the HPG study design and the design of traditional population-based studies is ascertainment, both in how participants are recruited and in the proportion of the population in a geographical area that participates (Figure 1A). Traditionally, the high expense of large-scale genetic studies and the limited resources of individual investigators have generated study populations exhibiting shallow ascertainment of individuals from a variety of geographical areas. To improve statistical power, researchers combine samples from many different collection centers into larger cohorts, and these cohorts are often merged into much larger consortiums consisting of tens to hundreds of thousands of individuals. Although the total number of individuals sampled is often high, these studies typically only sample a relatively small portion of individuals in any given geographic area. In contrast, planned and on-going HPG studies are sampling tens to hundreds of thousands of participants from individual healthcare systems.1
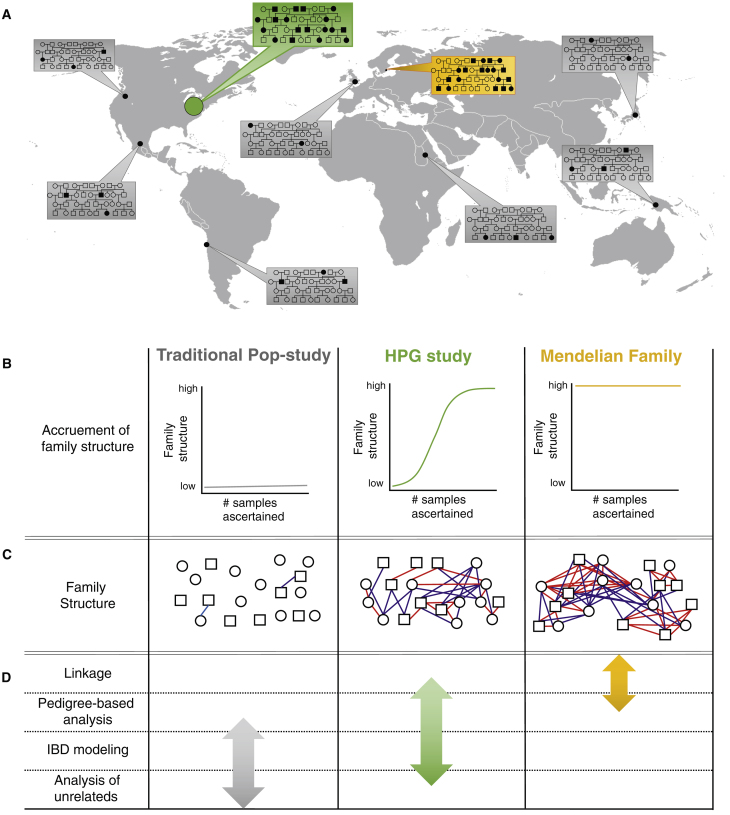
Figure 1.
Ascertaining a High Proportion of the Population in a Geographical Area Increases Family Structure and Impacts What Statistical-Analysis Approaches Should Be Used
(A) Traditional population-based studies (gray boxes) typically sample a small portion of individuals from several populations. HPG studies (green box) more densely sample individuals from one or more populations. Family-based studies (yellow box) heavily sample within extended families but do not sample nearly as many individuals as the other two study designs.
(B) The three study designs result in very different proportions of individuals in the cohort with one or more close relatives in the dataset.
(C) The three ascertainment approaches also result in very different amounts of family structure. Red and blue lines indicate first- and second-degree pairwise relationships, respectively. HPG studies are expected to contain a level of family structure between the other two designs.
(D) For this study, statistical-analysis approaches were binned into four categories on the basis of the level of family structure required to effectively use the approach. First column: “linkage” refers to traditional linkage analyses using one or more informative pedigrees; “pedigree-based analysis” refers to statistical methods beyond linkage that use pedigree structures within a larger cohort that includes unrelated individuals; “IBD modeling” refers to analyses that model the pairwise relationships between individuals without using the entire pedigree structure; “analysis of unrelateds” refers to analyses that assume all individuals in the cohort are unrelated. The amount of family structure impacts the approaches that can be used, and the arrows indicate the analysis ranges for which the three study designs are best suited.
The difference in these two ascertainment approaches results in different patterns of genetic relatedness among individuals in these cohorts. Relatedness is a continuum that manifests itself within a cohort in a variety of ways, depending on the population and how individuals are sampled from it. Because traditional population-based studies have generally collected samples from multiple geographical areas, they most commonly exhibit the broadest “class” of relatedness: population structure. Population structure (often referred to as “substructure” or “stratification”) within a genetic study results when the allele frequencies of different ancestral groups, or “genetic demes,” are more similar within than between demes. Genetic demes arise as a result of more-recent genetic isolation, drift, and migration patterns. Ascertainment of individuals within genetic demes can generate distant cryptic relatedness,5, 6 the second “class” of relatedness,7 defined here as third- to ninth-degree relatives. These distant relatives are unlikely to be identifiable from the EHR but are important because usually one or more large segments of their genomes are identical by descent, depending on their degree of relatedness and the recombination and segregation of alleles.8 Distant cryptic relatedness is usually limited in study cohorts built from small samplings of large populations, but the level of cryptic relatedness increases substantially as the effective population size decreases and the sample size increases. Finally, unless designed to collect families, traditional population-based studies typically have very little family structure: the third “class” of relatedness, consisting of first- and second-degree relationships2, 4, 9, 10, 11 (Figures 1B and 1C).
In contrast, the HPG study design enriches for family structure in several ways. First, HPG studies heavily sample from specific healthcare system regions, and the number of pairs of related individuals ascertained increases combinatorially as more individuals are sampled from a single region (Figure 1A). Second, families who live in the same geographic area most likely receive medical care from the same doctors at the same healthcare system because of referrals, shared insurance coverage, and convenience. Third, families who have visited a healthcare system for many years with multiple encounters will have extensive medical records, making them more likely to be included in a study than transient residents with brief medical records and fewer encounters. Both family structure and distant cryptic relatedness are more pronounced in populations with low migration rates.5 Conversely, confounding population substructure can be less of a factor in HPG studies if the sampled healthcare system’s population is a single homogeneous genetic deme.1 As a result, we expect to see more family structure in HPG studies compared to random ascertainment of a population. In this article, we focus on family structure and its prevalence in an HPG study by using both simulated and real data.
The increase in family structure within HPG studies has significant implications for the choice and execution of downstream analyses and must be considered thoughtfully.12, 13, 14, 15, 16, 17, 18 Some tools (e.g., principal component [PC] analysis) assume all individuals are unrelated, some (e.g., linear mixed models) effectively handle estimates of pairwise relationships, and others (e.g., linkage and TDT analyses) can directly leverage pedigree structures. The following description of some common analysis tools and their use cases is based on the varying levels of family structure within large population-based datasets (Figure 1D).
Removal of family structure (i.e., selectively excluding samples to eliminate relationships) is a viable option if a dataset has few closely related samples4, 9, 10, 11 and if the size of the unrelated subset is acceptable for the statistical analysis being performed. A number of methods exist to compute the maximally sized unrelated set of individuals.19, 20 However, this strategy reduces the sample size and power while discarding potentially valuable relationship information. In practice, the degree of information loss is unacceptable for many analyses if the dataset has even a moderate level of family structure.
Several methods that directly leverage the pairwise relationships have been developed. For example, researchers can explicitly model the relationships by using estimates of pairwise relationships (e.g., mixed models15, 21, 22, 23 and pedigree-free QTL linkage analysis24). Additionally, if a pedigree structure is needed for an analysis or visualization, close pairwise relationships can be used for reconstructing pedigree structures directly from the genetic data with tools such as PRIMUS25 and CLAPPER.26 Although estimated relationships and pedigrees are extremely useful, we echo Ko and Nielson’s26 caution regarding the use of estimated relationships and pedigrees with significant statistical uncertainty in analyses that are sensitive to inaccuracies in estimated relationships and pedigree structures. For example, first-degree and second-degree relationships are much more accurately estimated than more distant relationships. Furthermore, pedigrees where all individuals are connected by highly accurate first-degree relationships are much more likely to be correct than pedigrees connected only by more distant relationships.25
In this manuscript, we demonstrate the value of identifying family structure in a large clinical cohort as part of the DiscovEHR study. This cohort of 92,455 exomes originated from a collaborative, ongoing study by the Regeneron Genetics Center (RGC) and the Geisinger Health System (GHS) initiated in 2014.1 DiscovEHR is a dense sample of participants from a single healthcare system that serves a largely rural population with low migration rates in central Pennsylvania. We identify a tremendous amount of family structure within the DiscovEHR cohort, and our simulations project that 70%–80% of the individuals in our sequenced cohort will have a first- or second-degree relative as we continue sequencing up to 250K individuals. This has significant implications on downstream analyses but also affords us the opportunity to leverage the rich family structure through reconstructing pedigrees, phasing compound heterozygous mutation (CHM), and detecting de novo mutations (DNM).
Subjects and Methods
Individuals and Samples
We sequenced the exomes of 93,368 de-identified GHS participants who had given consent to be part of the MyCode Community Health Initiative.27 As part of this initiative, individuals agreed to provide blood and DNA samples for broad, future research, including genomic analyses as part of the Regeneron-GHS DiscovEHR collaboration1 and linking to data in the GHS EHR under a protocol approved by the Geisinger Institutional Review Board. The intended use of the data is for research into discovering gene-phenotype associations, and the data have not been used for re-identifying individuals or their family members. All analyses performed within this manuscript were done in concordance with the participants’ consent and IRB approval. Each participant has their exome linked to a corresponding de-identified EHR. A more detailed description of the first 50,726 sequenced individuals has been previously published.1, 27
The DiscovEHR study did not specifically target families as study participants but was implicitly enriched for adults who interact frequently with the healthcare system because of chronic health problems (and who might be related to each other) as well as participants from the Coronary Catheterization Laboratory and the Bariatric Service from GHS.
Sample Preparation, Sequencing, Variant Calling, and Sample QC
Sample preparation and sequencing for the first ∼61K samples have been previously described,1 and this set of samples is referred to in this manuscript as the “VCRome set.” The remaining set of ∼31K samples was prepared in the same process, except that in place of the NimbleGen probed capture, we used a slightly modified version of IDT’s xGen probes; we added supplemental probes to capture regions of the genome well covered by the NimbleGen VCRome capture reagent but poorly covered by the standard xGen probes. Captured fragments were bound to streptavidin-conjugated beads, and non-specific DNA fragments were removed by a series of stringent washes according to the manufacturer’s (IDT’s) recommended protocol. We refer to this second set of samples as the “xGen set.” Variant calls were produced with the Genome Analysis Toolkit (GATK; Web Resources). GATK was used for local realignment of the aligned, duplicate-marked reads of each sample around putative indels. We then used GATK’s HaplotypeCaller to process the INDEL-realigned, duplicate-marked reads to identify all exonic positions at which a sample varied from the genome reference in the genomic variant call format (gVCF). Genotyping was accomplished with GATK’s GenotypeGVCFs on each sample and a training set of 50 randomly selected samples outputting a single-sample variant call format (VCF) file identifying both single-nucleotide variants (SNVs) and indels as compared to the reference. We used the single-sample VCF files to create a pseudo-sample that contained all variable sites from the single-sample VCF files in both sets. We created independent pVCF files for the VCRome set by joint calling 200 single-sample gVCF files with the pseudo-sample to force a call or no-call for each sample at all variable sites across the two capture sets. We combined all 200-sample pVCF files to create the VCRome pVCF file and then repeated this process to create the xGen pVCF file. We then combined the VCRome and xGen pVCF files to create the union pVCF. We aligned sequence reads to GRCh38 and annotated variants by using Ensembl 85 gene definitions. We restricted the gene definitions to 54,214 transcripts, corresponding to 19,467 genes, that are protein-coding with an annotated start and stop. After the previously described sample QC process, 92,455 exomes remained for analysis.
Principal Components and Ancestry Estimation
We used PLINKv1.920 to merge the union datasets with HapMap328 and, on the basis of reference SNP cluster ID, kept only SNPs that were in both datasets. We restricted the analysis to high-quality common SNPs with minor-allele frequency > 10%, genotype missingness < 5%, and a Hardy-Weinberg Equilibrium p value > 0.00001 by applying the following PLINK filters: “--maf 0.1 --geno 0.05 --snps-only --hwe 0.00001.” We calculated PCs for the HapMap3 samples and then projected each sample in our dataset onto those PCs by using PLINK. We used the PCs for the HapMap3 samples to train a kernel density estimator (KDE) for each of the five ancestral super classes: African (AFR), admixed American (AMR), east Asian (EAS), European (EUR), and south Asian (SAS). We used the KDEs to calculate the likelihood that each sample belongs to each of the super classes. For each sample, we assigned the ancestral superclass on the basis of likelihoods. If a sample has two ancestral groups with a likelihood > 0.3, then we assigned AFR over EUR, AMR over EUR, AMR over EAS, SAS over EUR, and AMR over AFR; otherwise “UNKNOWN” (we did this to provide stringent estimates of the EUR and EAS populations and inclusive estimates for the more admixed populations in our dataset). If zero or more than two ancestral groups had a high enough likelihood, then the sample was assigned “UNKNOWN” for ancestry. Samples with unknown ancestry were excluded from the ancestry-based identity-by-descent (IBD) calculations.
IBD Estimation
Genome-wide IBD estimates are a metric used for quantifying the level of relatedness between pairs of individuals.24 We applied the same Hardy-Weinberg equilibrium, minor-allele frequency, and variant-level missingness that we applied during the PC analysis. Next, we used a two-pronged approach to obtain accurate IBD estimates from the DiscovEHR cohort exomes. First, we calculated IBD estimates among individuals within the same ancestral superclass (e.g., AMR, AFR, EAS, EUR, and SAS) as determined from our ancestry analysis. We calculated IBD estimates among all individuals by restricting pairs included as second-degree relatives to those with PI_HAT relatedness coefficients > 0.1875 and using the “--genome --min 0.1875” PLINK options. This approach allows for more accurate relationship estimates because all samples share similar ancestral alleles, but it is unable to predict relationships between individuals with different ancestral backgrounds, e.g., a child of a European father and Asian mother.
Second, in order to detect the first-degree relationships between individuals with different ancestries, we calculated IBD estimates among all individuals by restricting pairs included as first-degree relatives to those with PI_HAT relatedness coefficients >0.3. We then grouped individuals into first-degree family networks where network nodes are individuals and edges are first-degree relationships. We ran each first-degree family network through the prePRIMUS pipeline,25 which matches the ancestries of the samples to appropriate ancestral minor-allele frequencies to improve IBD estimation. This process accurately estimates first- and second-degree relationships among individuals within each family network (minimum PI_HAT of 0.15).
Finally, we combined the IBD estimates from the two previously described approaches by adding in any missing relationships from family-network-derived IBD estimates to the ancestry-based IBD estimates. This approach resulted in accurate IBD estimates out to second-degree relationships among all samples of similar ancestry and first-degree relationships among all samples.
IBD proportions for third-degree relatives are challenging to accurately estimate from a large exome sequencing dataset with diverse ancestral backgrounds because the analysis often results in an excess number of predicted third-degree relationships as a result of artificially inflated IBD estimates. We calculated IBD estimates out to third-degree relatives (defined by having a PI_HAT relatedness coefficients > 0.09875 during the ancestry-specific IBD analysis) to get a sense of how many third-degree relationships we might have in the DiscovEHR cohort, but these were not used in any of the phasing or pedigree-based analyses. For the relationship-based analyses reported in this paper, we only used high-confidence third-degree relationships we identified within first- and second-degree family networks.
During QC, before creating the final set of 92,455 individuals, we removed all identical pairs of samples (PI_HAT > 0.9) unless GHS was able to find evidence through a chart review that the two corresponding individuals appeared to be different people, share the same birthdate, and have one or more additional pieces of information indicating that they were related (e.g., same last name, shared parent, same address, or listed the other as a relative).
Pedigree Reconstruction
We reconstructed all first-degree family networks identified within the DiscovEHR cohort with PRIMUSv1.9.0.25 The combined IBD estimates were provided to PRIMUS along with the genetically derived sex and EHR reported age. We specified a relatedness cutoff of PI_HAT > 0.375 to limit the reconstruction to first-degree family networks and a minimum cutoff of 0.1875 to define second-degree networks.
Allele-Frequency-Based Phasing
We phased all bi-allelic variants from the VCRome and xGen exome datasets separately by using EAGLEv2.3.29 In order to parallelize our analysis, we divided the genome into overlapping segments of ∼40K variants with a minimum overlap of 500 variants and 250K base pairs. Because our goal was to phase putative compound heterozygous mutations within genes, we took care to have the segment breakpoints occur in intergenic regions.
We used the UCSC LiftOver program to lift-over EAGLE’s provided genetic_map_hg19.txt.gz file from hg19 to GRCh38 and removed all variants involving switched chromosomes or a change in relative order within a chromosome; such chromosomal changes resulted in failure of the centimorgan position to be continuously increasing when we sorted the variants on increasing chromosome position. In most cases, this QC step removed inversions around centromeres. In total, only 2,783 of the 3.3 million SNPs were removed from the genetic map file. We provided the data for each segment to EAGLE as PLINK-formatted files and ran them on DNAnexus with the described genetic map file running with 16 threads. We did not allow EAGLE to do any additional variant filtering on the basis of variant missingness, and we specified a genotype error rate of 0.01. These options were specified with the following EAGLE command line parameters:
“--geneticMapFile = genetic_map_hg19_withX.txt.GRCh38_liftover.txt.gz”
“--maxMissingPerIndiv 1”
“--genoErrProb 0.01”
“--numThreads = 16”
Compound Heterozygous Calling
Our goal was to obtain high-confidence CHM calls of putative loss-of-function variants (pLoFs) to identify humans with both copies of genes potentially knocked out or disrupted. We classify variants as pLoFs if they result in a frameshift, stop-codon gain, stop-codon loss, start-codon gain, start-codon loss, or splicing-acceptor or donor-altering variant. We created a second, expanded set of potentially harmful variants that included the pLOFs as well as likely disruptive missense variants, which are variants predicted to be deleterious by all five of the following methods: SIFT30 (damaging), PolyPhen2 HDIV31 (damaging and possibly damaging), PolyPhen2 HVAR (damaging and possibly damaging), LRT32 (deleterious), and MutationTaster33 (disease-causing automatic and disease-causing).
We identified rare (alternate allele frequency < 1%) potential compound heterozygous mutations (pCHMs) by testing all possible combinations of heterozygous pLoFs and/or deleterious missense variants within a gene of the same person. We excluded all variants that were out of Hardy-Weinberg equilibrium (p value < 10−15 calculated with PLINKv1.9), that exceeded 10% missingness within the individuals’ capture-specific dataset (i.e., VCRome or xGen sets), or that had another variant within 10 bp in the same individual. We also excluded SNPs with quality by depth (QD) < 3, alternate allele balance (AB) < 15%, or read depth < 7, and we excluded indels with QD < 5, AB < 20%, or read depth < 10. After filtering, we had 57,355 high-quality pCHMs, distributed among 36,739 individuals, that could knockout or disrupt normal function of both copies of a person’s gene if the pCHMs were phased in trans.
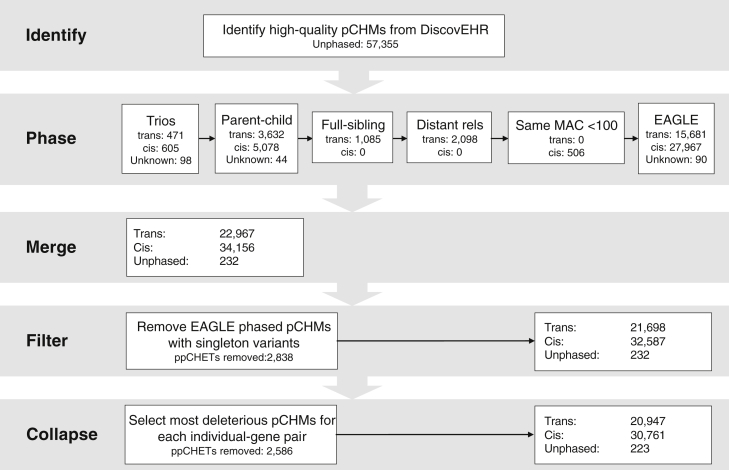
The next step was to phase the pCHMs. We used a combination of population allele-frequency-based phasing with EAGLE and pedigree- and relationship-based phasing to determine whether the pCHMs were in cis or trans. Figure 2 diagrams the pCHM phasing workflow we employed to obtain the most accurate phasing for each pCHM. Trios and relationships with individuals in both the VCRome and xGen datasets were used only if both variants in the pCHM were on both the VCRome and our modified xGen capture designs. Trio and relationship phasing proved to be more accurate than EAGLE phasing (Table S1), so we preferentially used the pedigree and relationship data for phasing. Table S2 describes the logic we used to determine the phase of the pCHMs for the different types of familial relationships. For all remaining pCHMs, we used the EAGLE-phased data described above. We excluded any EAGLE-phased pCHM where one or both of the variants were singletons because EAGLE’ s phasing accuracy with singletons was not significantly different from random guessing (Table S3). We found that if the two variants in the pCHM have the same minor-allele count (MAC) less than 100, then they are in cis (22 out of 22 occurrences in children of trios) in our dataset.
Figure 2.
Decision Cascade for Determining the Phase of Potential Compound Heterozygous Mutations (pCHMs) among the 92K DiscovEHR Participants
25.1% of pCHMs and 33.8% of the CHMs (trans) were phased with trio or relationship data.
We used the trio-phased pCHMs as the truth set to evaluate the overall phasing accuracy of EAGLE. EAGLE achieved a 91.1% accuracy when phasing rare pCHMs in the children in our reconstructed trios. However, having the parental haplotypes in the phasing dataset improved EAGLE’s accuracy of phasing the children’s pCHMs in comparison to not including parents in the cohort. Given that children of trios are a very small portion of the overall dataset, the 91.1% accuracy is an overestimate of the phasing accuracy for the overall dataset because most samples do not have first-degree relatives in the dataset. To obtain a good measure of accuracy for the EAGLE pCHM phasing across the entire cohort, we reran EAGLE on the entire dataset as before but excluded all first-degree relatives of one child in each nuclear family before phasing. We then compared the EAGLE-phased pCHMs to the trio-phased pCHMs to estimate EAGLE’s overall phasing accuracy, which we found to be 89.1% (Table S1).
Finally, if there were more than one pCHM within the same gene of an individual, then only the pCHM with the most deleterious profile was retained (Table S4). Using the approach outlined above, we were able to phase > 99% of all pCHMs and identify 20,947 rare CHMs that are predicted to alter function.
Compound Heterozygous Mutation Validation
We evaluated phasing accuracy by comparing phasing predictions to phasing done with trios and with Illumina reads. We performed Sanger validation on a subset of the incorrectly phased pCHMs to see whether the variants were false positive calls.
First, we evaluated the phasing accuracy of the pCHMs by using the trio phased pCHMs as truth. Given that the phasing approach for each familial relationship is performed independently from the trio phasing, we can get a good measure of phasing accuracy of each of the relationship classes as long as the pCHM carrier is a child in a trio. Table S1 shows that the accuracy of family-based phasing was 99.6% (1060/1064 pCHMs) for rare pCHMs. EAGLE phasing was less accurate, at 89.1% (766/860 pCHMs; Table S1). We evaluated EAGLE’s pCHM-phasing accuracy in different ranges of minor-allele frequency, and we found that EAGLE consistently attains an accuracy greater than 90% with a MAC greater than 9 and an accuracy around 77% for a MAC between 2 and 9 (Table S3). EAGLE phasing performed poorly with singletons.
Second, we attempted to validate 200 pCHMs with short (∼75 bp) Illumina reads by looking at the read stacks in the Integrative Genomics Viewer34 to see whether the two variants occur on the same read or independently. We were able to decisively phase 190 (115 cis and 79 trans; 126 EAGLE-phased and 74 pedigree- or relationship-phased) selected pCHMs by using short reads. The remaining ten showed read evidence of both cis and trans phasing, most likely because one or both of the variants were false positive calls. Visual validation showed an overall accuracy of 95.8% and 89.9% for pedigree and relationship phasing and EAGLE phasing, respectively (Table S5). Although the Illumina read-based validation results are in line with the trio validation results, we do note that the Illumina read-based validation accuracy results are lower than the accuracy of phasing with trios. The difference is most likely due to the enrichment for false-positive pCHMs in small problematic exon regions prone to sequencing and variant calling errors.
DNM Detection
We merged the results from two different approaches for detecting DNMs. The first method is TrioDeNovo,35 which reads likelihoods at each of the child’s variable sites in the child’s and parents’ genotype. One inputs these likelihoods into a Bayesian framework to calculate a posterior likelihood that a child’s variant is a DNM. The second program is DeNovoCheck (Web Resources), which is described in the supplemental methods of de Ligt et al.36 DeNovoCheck takes in a set of candidate DNMs. A variant was considered a candidate DNM if GATK called it a variant in the child but not in either parent. It then verifies the variant’s presence in the child and absence in both of the parents by examining the BAM files. We filtered these potential DNMs and evaluated a confidence level for each DNM in the union set by using a variety of QC metrics. Figure S1 illustrates this DNM-calling process, shows the variant filters we applied, and provides the criteria we used to classify each DNM as either low confidence, moderate confidence, or high confidence. We excluded all low-confidence and non-exonic DNMs from the summary results of this paper, but we considered them when doing visual validation to estimate the false-negative rate of excluding them. We also excluded the DNM calls for one extreme outlying participant who had an order of magnitude more DNMs called than any other participant.
Pedigree Estimation Based on Distant Relationships
Although we cannot know the true family history of the de-identified individuals in our cohort, we have used PRIMUS31 reconstructed pedigrees, ERSA distant-relationship estimates, and PADRE37 to connect the pedigrees to identify the best pedigree representation of the mutation carriers of a tandem duplication in LDLR.38 We used HumanOmniExpress array data (available for 25 out of the 37 carriers) to estimate the more distant relationships with the process described by Staples et. al.37 and used PADRE to connect the PRIMUS reconstructed pedigrees.
SimProgeny
We developed a forward simulation framework (SimProgeny) to simulate a wide variety of populations, including a population served by a healthcare system like GHS. SimProgeny also simulates sample ascertainment used by HPG studies (Figure S2). SimProgeny can simulate populations of millions of people dispersed across one or more sub-populations on the basis of user-specified population parameters (Table S6). Progressing year to year, the simulation creates couplings, births, separations, migrations, deaths, and movement between sub-populations on the basis of specified parameters. This process generates realistic pedigree structures and populations that represent a wide variety of HPG studies. The default values have been tuned so that the simulated population models the DiscovEHR cohort, but one can easily customize these parameters to model different populations by modifying the configuration file included with the SimProgeny code (available in the Web Resources).
In addition to modeling populations, SimProgeny simulates two ascertainment approaches to model selecting individuals from a population for a genetic study: random ascertainment and clustered sampling. Random ascertainment gives each individual in the population an equal chance of being ascertained without replacement. Clustered sampling is a way to enrich for close relatives by selecting an individual at random along with a number of their first- and second-degree relatives. One determines the number of first-degree relatives by sampling a value from a Poisson distribution with a user-specified first-degree ascertainment lambda (default is 0.2). The number of second-degree relatives is determined in the same way, and the default second-degree ascertainment lambda is 0.03.
Simulation of the Underlying DiscovEHR Population and Its Ascertainment
Our DiscovEHR simulations contained individual populations with starting sizes of 200K, 300K, 350K, 400K, 475K, 500K, and 550K. We tuned the SimProgeny parameters (Table S6) with publically available country-, state-, and county-level data as well as our own understanding of how individuals were ascertained through GHS consenting and sample collection. Sources for the selected parameters are available in Document S3. We reduced the immigration and emigration rates from the state-wide Pennsylvania average given that GHS primarily serves rural areas that tend to have lower migration rates than more urban areas. Simulations were run with a burn-in period of 120 years and then progressed for 101 years. Simulated populations grew by ∼15%, which is similar to the growth of the Pennsylvania population since the mid-20th century.
We performed both random and clustered ascertainment. For both ascertainment approaches, we shuffled the ascertainment order of the first 5% of the population (specified with the ordered_sampling_proportion parameter) to model the random sequencing order of the individuals in GHS biobank at the beginning of our collaboration. Although the selection of this parameter has no effect on random ascertainment and a negligible effect on the accumulation of pairwise relationships in clustered ascertainment, it does affect the proportion of individuals with one or more relatives in the clustered sampling dataset by creating an inflection point at 5% population ascertainment in the simulation results plots (Figures S3B and S3D). This inflection point would be less pronounced if we were to model the freeze process of the real data or model a smoother transition between sequencing samples from the biobank and newly ascertained individuals. Notably, the inflection point is more pronounced when values of lambda from the Poisson distribution are higher.
Results
Relationship Estimation and Relatedness in DiscovEHR
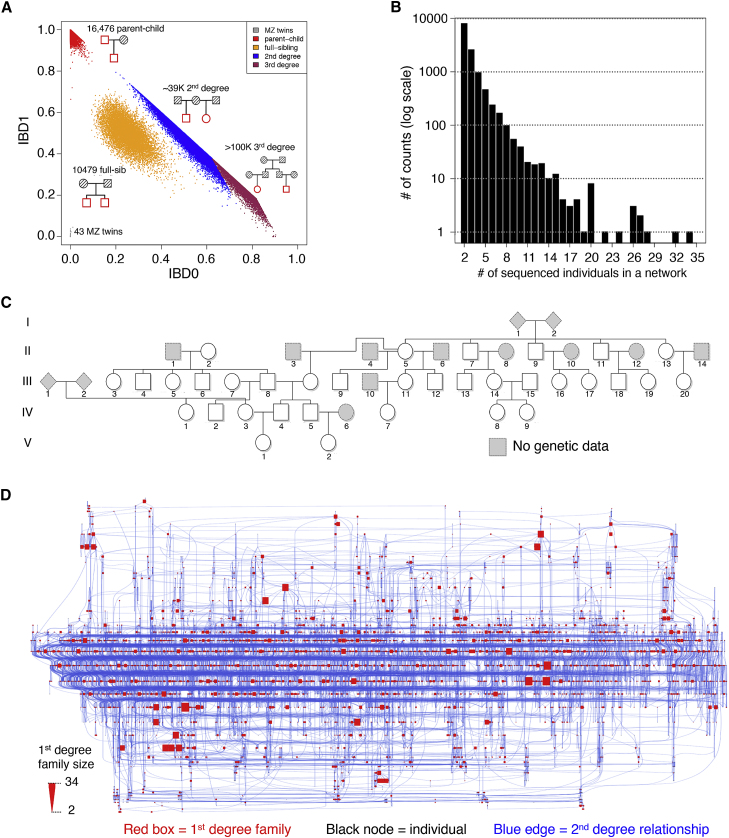
In the current dataset of 92,455 individuals, we identified 43 monozygotic twins, 16,476 parent-child relationships, 10,479 full-sibling relationships, and ∼39,000 second-degree relationships (Figure 3A). Next, we treated individuals as nodes and relationships as edges to generate undirected graphs. Using only first-degree relationships, we identified 12,594 connected components, which we refer to as first-degree family networks. Figure 3B shows the distribution in size of the first-degree family networks, which range from 2 to 25 sequenced individuals. Similarly, we found 10,173 second-degree family networks, the largest containing 19,968 individuals (∼22% of the overall dataset; Figure 3C). We were able to identify ∼5,300 third-degree relationships within the second-degree family networks. Using a lower IBD cutoff (PI_HAT > 0.09875) for the IBD estimations within ancestral groups without consideration of second-degree family networks, we found well over 100,000 third-degree relationships within the DiscovEHR cohort. Given that 95.9% of DiscovEHR individuals are of European ancestry (Table S7), it is not surprising that the vast majority (98.6%) of the pairwise relationships found were between two individuals of European ancestry (Table S8). Nonetheless, we identified many relationships between people of the same, non-European ancestry and between individuals with different ancestries; for example, there were several trios having one European parent, one East Asian parent, and a child whose ancestry was unassigned to a super-population because of the ad-mixed nature of his or her genome.
Figure 3.
First 92K Sequenced Individuals from the DiscovEHR Cohort Contain an Extensive Amount of Relatedness
(A) A plot of IBD0 versus IBD1 shows pairwise relationships segregating into different familial relationship classes. The IBD sharing distributions of second- and third-degree relationships overlap with each other, so a hard cutoff halfway between the two expected means was selected. Third-degree relationships are challenging to accurately estimate because of the technical limitations of exome data as well as the widening and overlapping variation around the expected mean IBD proportions of more distant relationship classes (e.g., fourth degree and fifth degree). We provided a lower-bound estimate of the number of third-degree relationships.
(B) The distribution of size of first-degree family networks ranges between 2 and 34 sequenced individuals, and the vast majority are smaller family networks.
(C) The largest reconsturcted first-degree family network consisting of 34 sequenced individuals; more than 99.98% of the first-degree family networks’ pedigree structures were reconstructed from the genetic data.
(D) The largest second-degree family network, consisting of 19,968 individuals (∼22% of the dataset), shows 4,062 first-degree family networks (represented as red boxes that are proportionally sized to the number of individuals in the network, including the network corresponding to the pedigree shown in [C]), and 5,584 additional individuals (black nodes) connected by 11,430 second-degree relationships (blue edges).
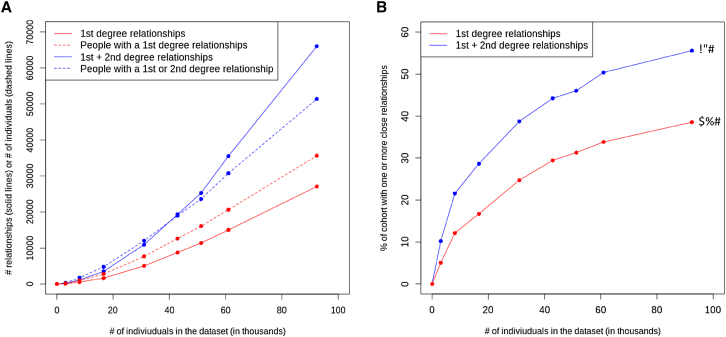
Importantly, we show both empirically (Figure 4A) and through simulation (Figure 5) that the rate of accumulating relatives far exceeds the rate of ascertaining samples. This is expected, given that there are combinatorially increasing numbers of possible pairwise relationships within the dataset as the size increases and that the likelihood that a previously unrelated individual in the dataset becomes involved in a newly identified relationship also increases. Currently, 39% of individuals in the DiscovEHR cohort have at least one first-degree relative in the dataset, and 56% of the participants have one or more first- or second-degree relatives in the dataset (Figure 4B).
Figure 4.
Accumulation of Relatedness within the DiscovEHR Cohort at Consecutive Data Freezes
(A) The number of pairwise relationships has grown rapidly.
(B) The proportion of individuals with a first- or second-degree relative identified in the cohort.
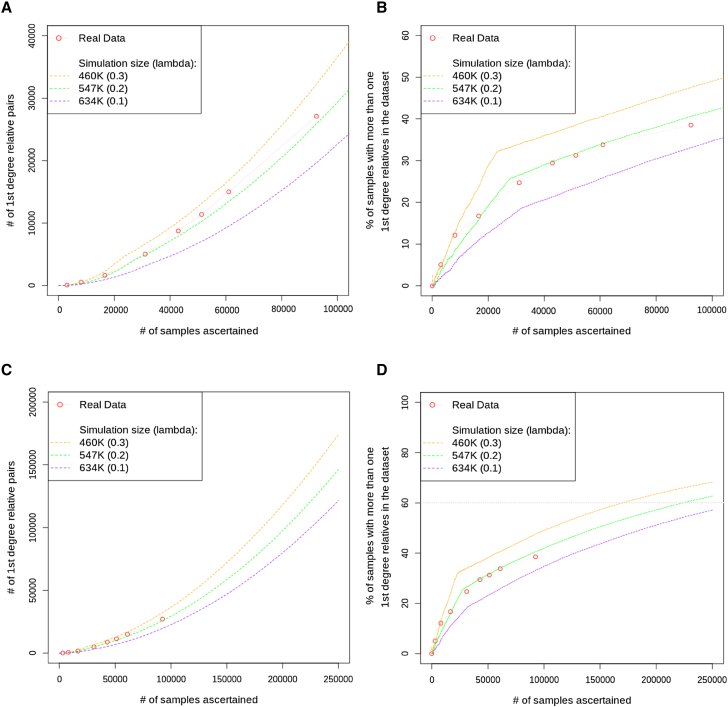
Figure 5.
Simulated Population and Ascertainment Fit to the Accumulation of First-Degree Relatedness in the DiscovEHR Cohort
The real data were calculated at periodic “freezes” indicated by punctuation points connected by the faint red line. Most simulation parameters were set on the basis of information about the real population demographics and the DiscovEHR ascertainment approach. However, two parameters were unknown and selected on the basis of their fit to the real data: (1) the effective population size from which samples were ascertained and (2) the increased chance that someone is ascertained given that a first-degree relative was previously ascertained, which we call “clustered ascertainment.” All panels show the same three simulated population sizes spanning the estimated effective population size. We simulated clustered ascertainment by randomly ascertaining an individual along with a Poisson-distributed random number of first-degree relatives (distributions’ lambdas are indicated in the legends).
(A) The accumulation of pairs of first-degree relatives as additional samples are ascertained.
(B) The proportion of the ascertained participants that have one or more first-degree relatives that have also been ascertained.
(C) Simulated ascertainment projections with upper and lower bounds of the number of first-degree relationships we expect with our current DiscovEHR ascertainment approach as we scale to our goal of 250K participants.
(D) Simulated projections with upper and lower bounds of the proportion of the ascertained participants that have one or more first-degree relatives that have also been ascertained.
Simulations with SimProgeny and Relatedness Projections
Prior to the launch of the DiscovEHR collaboration, it was unclear how much relatedness we should expect to see and how the amount of relatedness would compare to those seen in previous population-based genomic studies. However, it became clear early on that the cohort contained far more family structure than typically seen in population-based studies, and projections estimated that the proportion of the cohort involved in close relationships would eventually involve the majority of our dataset. Given the impact of this relatedness on downstream analyses, we set out to determine whether this amount of relatedness is expected, whether it is unique to our dataset, and how much it would grow as the sequenced cohort expands.
To answer these questions, we developed a flexible simulation framework (SimProgeny) to model a wide variety of study populations and sampling approaches in order to estimate the amount of relatedness researchers should expect to find for a given set of populations and sampling parameters. Although we apply this framework to the DiscovEHR cohort, it is flexible enough that it also can be applied to modeling shallower ascertainment of more transient populations.
We used SimProgeny to simulate the DiscovEHR population and the ascertainment of the first 92,455 participants. As expected, the simulations show that DiscovEHR participants were not randomly sampled from the population, but rather that the dataset is enriched for close relatives (Figure S4). Therefore, we used a clustered ascertainment approach (see Material and Methods) that more accurately models ascertainment from a healthcare system study population and the subsequent enrichment of close relatives observed in the real data (Figure 5). These simulation results suggest that the effective population size for the first 60K participants was ∼475K individuals, and a Poisson distribution with lambda of 0.2 most closely matches the enrichment of first-degree relatives. However, the departure of the real data line (Figure 4, faint red line) from the ∼475K simulation line (solid green line) at 90K ascertained samples suggests that the DiscovEHR cohort’s effective population size might have increased after ascertainment of the first 60K samples. These estimates are consistent with our knowledge that the majority of the first 30K–60K DiscovEHR participants reside in the counties surrounding the GHS headquarters in Danville and that the participant base subsequently expanded to more heavily include pockets of individuals from north-central and northeast rural Pennsylvania (Figure S5). Most notably, ascertainment was not evenly distributed across the entire GHS catchment area (containing >2.5 million individuals).
After identifying simulation parameters that reasonably fit the real data, we used SimProgeny to obtain a projection of the amount of first-degree relationships we should expect as DiscovEHR expands to our goal of 250K participants. If we continued to ascertain participants in the same way, we would expect to obtain ∼150K first-degree relationships (Figure 5C) involving ∼60% of DiscovEHR participants (Figure 5D). We then expanded our simulation analysis to include second-degree relationships, and the simulation results suggested that with 250K participants we should expect well over 200K combined first- and second-degree relationships involving over 70% of the individuals in DiscovEHR (Figure S3).
These projections of relatedness in DiscovEHR assume that we continue ascertaining participants in the same way we did for the first 60K–90K participants. However, if DiscovEHR expands ascertainment of participants to additional GHS clinics and hospitals in other regions, then these relatedness estimates are likely to drop because expanding the participant base increases the size of the effective sampling population and taps into new genetic demes or distant branches of the same demes. The relatedness will depend on the proportion of the total population we ascertain and the underlying regional population demographics, both of which can be simulated with SimProgeny.
Although SimProgeny is designed to reasonably model a real population, it has its limitations. For example, the Poisson distribution accurately models the clustered sampling of first-degree relationships we observe in our real data, but compared to DiscovEHR, it underestimates the clustering of second-degree relationships. Thus, there are bound to be nuances that cannot be easily modeled with a fixed distribution, and there are likely to be other confounding aspects to how participants were ascertained in the real dataset.
Regardless, our simulation results demonstrate a clear enrichment of relatedness in the DiscovEHR HPG study and provide key insights into the tremendous amount of relatedness we expect to see as we continue to ascertain additional participants, if we assume future ascertainment is reasonably well modeled by SimProgeny. These observations can also be extrapolated to other large HPG studies, and the flexibility built into the model provides the ability to tune the model to a wide variety of different populations and ascertainment approaches.
Leveraging Relatedness Instead of Treating It like a Nuisance
We reconstructed pedigree structures for 12,574 first-degree family networks in the DiscovEHR dataset by using the pedigree reconstruction tool PRIMUS, and we found that 98.9% of these pedigrees reconstructed unambiguously to a single pedigree structure when we considered IBD estimates and reported participant ages. These pedigrees include 2,192 nuclear families (1,841 trios, 297 quartets, 50 quintets, 3 sextets, and 1 septet). Table S9 shows a breakdown of the trios by ancestry. Figure 3C shows the largest first-degree pedigree, which contains 34 sequenced individuals. We have used these relationships and pedigrees in several ways, and we highlight three main applications in this section.
Compound Heterozygous Mutations
A major goal of human genetics is to better understand the function of every gene in the human genome. LoFs are a powerful tool that we can use to gain insight into gene function by analyzing the phenotypic effects when both copies of an individual’s gene are knocked out or disrupted (KOs). Rare (MAF < 1%) homozygous LoFs have been highlighted in recent large-scale sequencing studies and have been critical in identifying many gene-phenotype interactions.1, 4, 39, 40 Although rare CHMs of two heterozygous LoFs are functionally equivalent to rare homozygous KOs, they are more difficult to identify (particularly with short-read sequencing) and are rarely interrogated in large sequencing studies.1, 4, 39
We performed a survey of rare CHMs in the DiscovEHR cohort. First, we identified 57,355 high-quality pCHMs consisting of pairs of rare heterozygous variants that are either putative LoFs (pLoF; i.e., nonsense, frameshift, or splice-site mutations) or missense variants with strong evidence of being deleterious (see Material and Methods). Second, we phased the pCHMs by using a combination of allele-frequency-based phasing with EAGLE and pedigree-based phasing with the reconstructed pedigrees and relationship data (Figure 2). Trio validation indicated that EAGLE phased the pCHMs with an average of 89.1% accuracy (Table S1). However, because we had extensive pedigree and relationship data within this cohort, we were able to use them to phase 25.2% of the pCHMs and 33.8% of the trans CHMs with highly accurate trio and relationship phasing data (≥98.0%; Table S1), reducing inaccurate phasing of trans CHMs by approximately a third. The phased pCHMs spanned the entire frequency range from singletons to 1% MAF (Table S10).
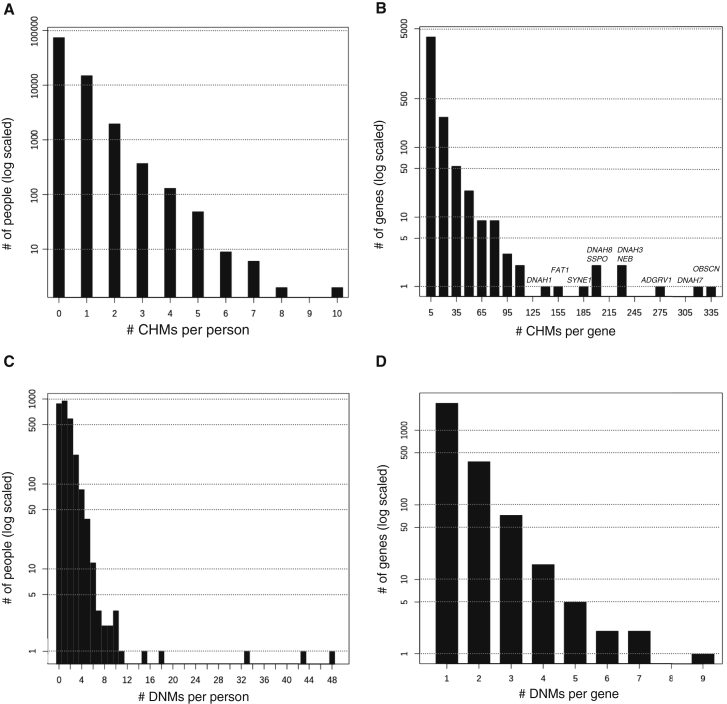
After processing, 40.3% of the pCHMs were phased in trans, yielding a high-confidence set of 20,947 rare, deleterious CHMs distributed among 17,533 of the 92K individuals (mean = 0.23 per person; max = 10 per person; Figure 6A). The median genomic distance between pCHM variants in cis (5,955 bp) was a little more than half the median distance between the pCHMs in trans (11,600 bp; Figure S6). Nearly a third of the CHMs involved at least one pLoF, and 8.9% of the CHMs consisted of two pLoF variants (Table S11). More than 4,216 of the 19,467 targeted genes contain one or more CHM carriers (Table S12), and 2,468 have more than one carrier (Figure 6B). ExAC pLI scores indicate that the ten genes with more than 125 CHM carriers are likely to be among the most LoF tolerant in the genome4 (Table S13), so it is no surprise that these genes would contain a higher number of CHMs.
Figure 6.
DiscovEHR Results for Compound Heterozygous Mutations and De Novo Mutations
(A) Distribution of the number of CHMs per individual in the DiscovEHR cohort.
(B) Distribution of the number of CHMs per gene. Names of genes with more than 125 CHMs are listed.
(C) Distribution of 3,415 exonic high- and moderate-confidence DNMs among the children of trios in the DiscovEHR cohort.
(D) The distribution of non-synonymous DNMs across the 2,802 genes with one or more DNM carriers.
In order to get a more robust set of genes where both copies of the gene are knocked out or disrupted in the same individual and to demonstrate the added value of CHMs, we combined the CHMs with the 6,560 rare (MAF < 1%) homozygous pLoFs found among the 92K DiscovEHR participants. pLoF-pLoF CHMs increased the number of genes that were knocked out in ≥1 and ≥20 individuals by 15% and 61%, respectively (Table S12). The benefit of including CHMs in a KO analysis is even more significant when we consider missense variants that are predicted to disrupt protein function. We found a combined 20,364 rare homozygous pLOFs and deleterious missense variants among the 92K participants. Carriers of homozygous pLoF or predicted deleterious missense variants provided a large number of genes that are predicted to be completely knocked out or disrupted. However, the inclusion of carriers of CHMs provided 26% more genes that are knocked out or disrupted in ≥1 individuals and 397% more genes knocked out or disrupted in ≥20 individuals (Table S12).
DNMs
DNMs are more likely to produce extreme phenotypes in humans than are other types of rare variations because DNMs occur sporadically and lack purifying selection. Many recent sequencing studies have shown that DNMs are a major driver in human genetic disease,36, 41, 42 demonstrating that DNMs are a valuable tool for better understanding gene function.
We used the nuclear families reconstructed from the 92K DiscovEHR participants to confidently call 3,415 moderate- and high-confidence exonic DNMs distributed among 1,783 of the 2,602 available children in trios (mean = 1.31; max = 48; Figure 6C). PolyPhen2 predicts 29.1% (n = 995) of the DNMs as “probably damaging” and an additional 9.2% (n = 316) as possibly damaging. The DNMs are distributed across 2,802 genes (Figure 6D), and TTN receives the most (nine). The most common types of DNM are nonsynonymous SNVs (58.5%), followed by synonymous SNVs (24.3%). Table 1 provides a complete breakdown of DNM types and shows that our proportions of DNMs falling into the different functional classes generally match those found in a recent study of DNMs in children with development disorders.41 We also observed an increase in the number of exonic DNMs with respect to both maternal (0.011 DNMs/year, p = 7.3x10−4; Poisson regression; Figure S7) and paternal (0.010 DNMs/year; p = 5.6x10−4) age at birth, consistent with other reports.41, 43, 44, 45 Notably, maternal and paternal age at birth are highly correlated in our dataset (ρ = 0.79; Figure S8); thus, the rates are not additive, and no significant difference identified either as a driving factor.
Table 1.
Breakdown by Functional Class of Moderate- and High-Confidence Exonic DNMs Found in the DiscovEHR Cohort alongside a Similar Breakdown for a Recent Developmental Delay Exome Study of 4,293 Trios
| Type of DNM | Number of DNMs | Percent of DNMs | Number in DDD Studya | Percent in DDD Studya |
|---|---|---|---|---|
| Nonsynonymous SNV | 1,996 | 58.5% | 4,797 | 57.8% |
| Synonymous SNV | 831 | 24.3% | 1,629 | 19.6% |
| Splicing | 153 | 4.5% | 671 | 8.1% |
| Non-frameshift deletion | 78 | 2.3% | 167 | 2.0% |
| Non-frameshift insertion | 55 | 1.6% | 28 | 0.3% |
| Frameshift | 187 | 5.5% | 603 | 7.3% |
| Stop-gain SNV | 112 | 3.3% | 402 | 4.8% |
| Stop-loss SNV | 3 | 0.1% | 7 | 0.1% |
The Deciphering Developmental Disorders Study (DDD).41 The DDD paper also reported 57 DNMs of other classes that were not included in our analysis or in this table; percentages were adjusted accordingly.
We attempted to perform visual validation of 23 high-confidence, 30 moderate-confidence, and 47 low-confidence DNMs spanning all functional classes. Eight moderate- and two low-confidence variants could not be confidently called as true- or false-positive DNMs. Of those remaining, 23/23 (100%) high-confidence, 19/22 (86%) moderate-confidence, and 12/43 (28%) low-confidence DNMs were validated as true positives. Visual validation also confirmed that the majority (40/49) of potential DNMs in individuals with >10 DNMs are most likely false-positive calls.
Variant and Phenotype Segregation in Pedigrees
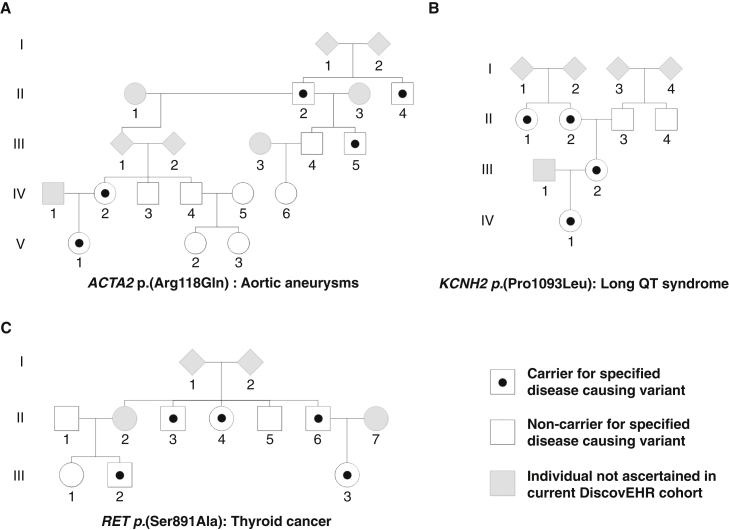
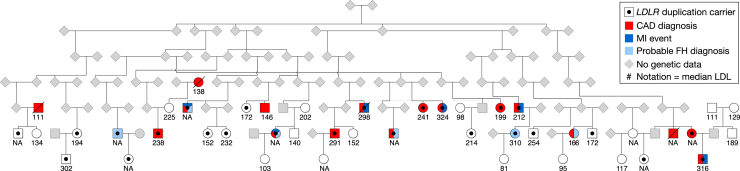
We have used the reconstructed pedigree data from among the 92K DiscovEHR participants to distinguish between rare population variation and familial variants and have leveraged it to identify highly penetrant disease variants segregating in families. Although this is not intended to be a survey of all known Mendelian-disease-causing variation transmitted through these pedigrees, we have identified a few illustrative examples, including familial aortic aneurysms (AAT6 [MIM: 611788]; Figure 7A), long QT syndrome (LQT2 [MIM: 613688]; Figure 7B), thyroid cancer (MTC [MIM: 155240]; Figure 7C), and familial hypercholesterolemia (FH [MIM: 143890]; Figure 8).38 The FH example is particularly interesting, given that we previously reported an FH-causing tandem duplication in LDLR [MIM: 606945].38 We have updated the CNV calls and found 37 carriers of the FH-causing tandem duplication among the 92K exomes, and we have reconstructed 30 out of the 37 carriers into a single extended pedigree. The carriers’ shared ancestral history provides evidence that they all inherited this duplication event from a common ancestor approximately six generations back. Although two of the seven remaining carriers are second-degree relatives to each other, genotyping array data was not available to confirm that the remaining seven carriers are also distantly related to the other carriers in Figure 8.
Figure 7.
Reconstructed Pedigree from DiscovEHR Demonstrates the Segregation of Known Disease-Causing Variants
Segregating variants include variants for (A) aortic aneurysms (ACTA2 [MIM: 102620], c.353G>A [p.Arg118Gln]; GenBank: NM_001613.2; Ensembl: ENST00000224784), (B) long QT syndrome (KCNH2 [MIM: 152427], c.3278C>T [p.Pro1093Leu]; GenBank: NM_000238.3; Ensembl: ENST00000262186), and (C) thyroid cancer (RET [MIM: 164761], c.2671T>G [p.Ser891Ala]; GenBank: NM_020630.4; Ensembl: ENST00000340058).
Figure 8.
Reconstructed Pedigree Prediction Containing 25/37 Carriers of the FH-Causing Tandem Duplication in LDLR
The pedigree also contains 20 non-carrier, related (first- or second-degree) individuals from the sequenced cohort. Carrier and non-carrier status was determined from the exome data from each individual. Elevated maximum LDL measurements (value under symbols) as well as increased prevalence of coronary artery disease (CAD, red fill) and pure hypercholesterolemia (ICD 272.0; blue) segregate with duplication carriers. Five additional carriers (not drawn) were found to be distant relatives (seventh- to ninth-degree relatives) of individuals in this pedigree.
Discussion
Sequencing studies continue to collect and sequence increasing proportions of human populations and are uncovering the extremely complex, intertwined nature of human relatedness. In the first 92K sequenced participants of the DiscovEHR cohort, we have identified ∼66K first- and second-degree relationships, reconstructed 12,574 pedigrees, and uncovered a second-degree family network of nearly 20,000 participants. The high level of cryptic relatedness within this dataset is certainly influenced by the underlying population structure of several close communities with relatively low migration rates. Studies in founder populations have already highlighted the complexity of relationships (Old Order Amish,46 Hutterites,47 and Ashkenazi Jews48), and recent studies of non-founder populations are reporting extensive levels of family structure (UK Biobank,49 NHANES,50 and AncestryDNA6). We observed family structure (first- and second-degree relationships) involving 55.6% of the DiscovEHR participants, and we expect family structure will involve a large proportion, if not a majority, of individuals in other large HPG studies. We have demonstrated through simulations and observations within our own data that we can obtain a large number of close familial relationships, nuclear families, and informative pedigrees within HPG studies. Although the underlying population structure and depth of ascertainment will vary between studies, we do believe that our observations in DiscovEHR will be applicable to other HPG studies, given that families tend to visit the same healthcare system and have similar genetic and environmental disease risks. The days of only having a handful of closely related individuals or samples in large sequencing cohorts are over, and we can no longer simply remove closely related pairs of individuals for our association studies on the assumption that they are only a small fraction of the overall cohort. Instead, we need to continue developing methods that are capable of leveraging the extensive relatedness of these rich cohorts and that can scale to accommodate growing population sizes and phenotype diversity of HPG studies.
In this study, we have demonstrated several ways to leverage family structure. First, we improved the phasing accuracy of rare CHMs. Although we did obtain accurate phasing of CHMs with EAGLE, our pedigree- and relationship-based phasing was far more accurate: this approach reduced the pCHM phasing error by approximately a third. We expect that the accuracy of the relationship-based phasing of pCHMs will be lower for variants with >1% MAF because phasing using the pairwise relationships assumes that if two variants appear together in two relatives, then they are in cis and have segregated together from a common ancestor. There is a higher chance that two independently segregating common variants will appear together in multiple people, which would result in their being incorrectly phased as cis by the algorithm. Therefore, common variants might be better phased using population allele frequencies with programs like EAGLE rather than phased using pairwise relationships.
Second, pedigree reconstruction using first-degree relationships within HPG studies provides highly accurate trios and other informative pedigree structures that can be leveraged for many use cases. We used the 2,602 reconstructed trios to find 3,415 DNMs and tracked known disease-causing mutations through extended pedigrees. Pedigrees and relationships are also particularly useful for tracking transmission of rare variants, providing increased confidence in variant calls, and allowing for the use of more traditional Mendelian genetic analyses. Pedigrees can be particularly useful when combined with follow-up chart reviews and the ability to re-contact participants and their family members.
As mentioned in the introduction, relationships and pedigrees with significant uncertainty should be used carefully in analyses that are sensitive to this uncertainty. The accuracy of an estimated relationship decreases as the relationship becomes more distant, and reconstructed pedigrees connected by these uncertain relationships should be used accordingly. First-degree relationships are extremely accurately estimated. Reconstructed pedigrees fully connected by first-degree relationships (e.g., nuclear families) have the highest level of certainty.25 Estimated pedigrees such as the one depicted in Figure 8 work well for depicting the transmission of disease-causing mutations in a family, but they are probably not appropriate for a linkage analysis because of the high level of uncertainty of the third- to ninth-degree relationships. We recommend validating significantly uncertain relationships and pedigree structures with independent data before using them in an analysis. If relationship and pedigree validation is not possible, then they can be used for prioritization of results that are directly validated with another approach.
We show that cryptic family structure in a large sequencing dataset can be an opportunity to harness a valuable, untapped source of genetic insights rather than a nuisance that must be managed during downstream analyses. As we enter the era of genome-based precision medicine, we see a critical need for additional innovative methods and tools that are capable of effectively mining the familial structure and distant relatedness contained within the ever-growing sequencing cohorts.
Declaration of Interests
The study was funded by Regeneron Pharmaceuticals. In addition to the Regeneron-affiliated authors being employed by and stockholders in Regeneron Pharmaceuticals, G.D.Y. is a cofounder and a member of the board of directors of Regeneron Pharmaceuticals. J.G.R., J.C.S., C.S., and L.H. are listed inventors on a related provisional patent application, filed by Regeneron Pharmaceuticals (62/555,597), which discloses the simulation framework and methods for identifying the familial relationships, phasing the pCHMs, and calling the DNMs. Additional information for reproducing the results described in the article is available upon reasonable request and subject to a data use agreement.
Acknowledgments
We thank the MyCode Community Health Initiative participants for their permission to use their health and genomics information in the DiscovEHR collaboration.
Published: May 3, 2018
Footnotes
Supplemental Data include 13 tables, eight figures, and one Excel file and can be found with this article online at https://doi.org/10.1016/j.ajhg.2018.03.012.
Web Resources
DeNovoCheck, https://sourceforge.net/projects/denovocheck
Github (SimProgeny), https://github.com/rgcgithub/SimProgeny
OMIM, https://www.omim.org
TopMed, https://www.nhlbiwgs.org
Supplemental Data
References
- 1.Dewey F.E., Murray M.F., Overton J.D., Habegger L., Leader J.B., Fetterolf S.N., O’Dushlaine C., Van Hout C.V., Staples J., Gonzaga-Jauregui C. Distribution and clinical impact of functional variants in 50,726 whole-exome sequences from the DiscovEHR study. Science. 2016;354:aaf6814. doi: 10.1126/science.aaf6814. [DOI] [PubMed] [Google Scholar]
- 2.Sudlow C., Gallacher J., Allen N., Beral V., Burton P., Danesh J., Downey P., Elliott P., Green J., Landray M. UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 2015;12:e1001779. doi: 10.1371/journal.pmed.1001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Collins F.S., Varmus H. A new initiative on precision medicine. N. Engl. J. Med. 2015;372:793–795. doi: 10.1056/NEJMp1500523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lek M., Karczewski K.J., Minikel E.V., Samocha K.E., Banks E., Fennell T., O’Donnell-Luria A.H., Ware J.S., Hill A.J., Cummings B.B., Exome Aggregation Consortium Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Henn B.M., Hon L., Macpherson J.M., Eriksson N., Saxonov S., Pe’er I., Mountain J.L. Cryptic distant relatives are common in both isolated and cosmopolitan genetic samples. PLoS ONE. 2012;7:e34267. doi: 10.1371/journal.pone.0034267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Han E., Carbonetto P., Curtis R.E., Wang Y., Granka J.M., Byrnes J., Noto K., Kermany A.R., Myres N.M., Barber M.J. Clustering of 770,000 genomes reveals post-colonial population structure of North America. Nat. Commun. 2017;8:14238. doi: 10.1038/ncomms14238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stevens E.L., Baugher J.D., Shirley M.D., Frelin L.P., Pevsner J. Unexpected relationships and inbreeding in HapMap phase III populations. PLoS ONE. 2012;7:e49575. doi: 10.1371/journal.pone.0049575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huff C.D., Witherspoon D.J., Simonson T.S., Xing J., Watkins W.S., Zhang Y., Tuohy T.M., Neklason D.W., Burt R.W., Guthery S.L. Maximum-likelihood estimation of recent shared ancestry (ERSA) Genome Res. 2011;21:768–774. doi: 10.1101/gr.115972.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fuchsberger C., Flannick J., Teslovich T.M., Mahajan A., Agarwala V., Gaulton K.J., Ma C., Fontanillas P., Moutsianas L., McCarthy D.J. The genetic architecture of type 2 diabetes. Nature. 2016;536:41–47. doi: 10.1038/nature18642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Locke A.E., Kahali B., Berndt S.I., Justice A.E., Pers T.H., Day F.R., Powell C., Vedantam S., Buchkovich M.L., Yang J., LifeLines Cohort Study. ADIPOGen Consortium. AGEN-BMI Working Group. CARDIOGRAMplusC4D Consortium. CKDGen Consortium. GLGC. ICBP. MAGIC Investigators. MuTHER Consortium. MIGen Consortium. PAGE Consortium. ReproGen Consortium. GENIE Consortium. International Endogene Consortium Genetic studies of body mass index yield new insights for obesity biology. Nature. 2015;518:197–206. doi: 10.1038/nature14177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Surendran P., Drenos F., Young R., Warren H., Cook J.P., Manning A.K., Grarup N., Sim X., Barnes D.R., Witkowska K., CHARGE-Heart Failure Consortium. EchoGen Consortium. METASTROKE Consortium. GIANT Consortium. EPIC-InterAct Consortium. Lifelines Cohort Study. Wellcome Trust Case Control Consortium. Understanding Society Scientific Group. EPIC-CVD Consortium. CHARGE+ Exome Chip Blood Pressure Consortium. T2D-GENES Consortium. GoT2DGenes Consortium. ExomeBP Consortium. CHD Exome+ Consortium Trans-ancestry meta-analyses identify rare and common variants associated with blood pressure and hypertension. Nat. Genet. 2016;48:1151–1161. doi: 10.1038/ng.3654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Santorico S.A., Edwards K.L. Challenges of linkage analysis in the era of whole-genome sequencing. Genet. Epidemiol. 2014;38(Suppl 1):S92–S96. doi: 10.1002/gepi.21832. [DOI] [PubMed] [Google Scholar]
- 13.Hu H., Roach J.C., Coon H., Guthery S.L., Voelkerding K.V., Margraf R.L., Durtschi J.D., Tavtigian S.V., Shankaracharya, Wu W. A unified test of linkage analysis and rare-variant association for analysis of pedigree sequence data. Nat. Biotechnol. 2014;32:663–669. doi: 10.1038/nbt.2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Price A.L., Zaitlen N.A., Reich D., Patterson N. New approaches to population stratification in genome-wide association studies. Nat. Rev. Genet. 2010;11:459–463. doi: 10.1038/nrg2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kang H.M., Sul J.H., Service S.K., Zaitlen N.A., Kong S.-Y., Freimer N.B., Sabatti C., Eskin E. Variance component model to account for sample structure in genome-wide association studies. Nat. Genet. 2010;42:348–354. doi: 10.1038/ng.548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sun L., Dimitromanolakis A. Identifying cryptic relationships. Methods Mol. Biol. 2012;850:47–57. doi: 10.1007/978-1-61779-555-8_4. [DOI] [PubMed] [Google Scholar]
- 17.Devlin B., Roeder K. Genomic control for association studies. Biometrics. 1999;55:997–1004. doi: 10.1111/j.0006-341x.1999.00997.x. [DOI] [PubMed] [Google Scholar]
- 18.Voight B.F., Pritchard J.K. Confounding from cryptic relatedness in case-control association studies. PLoS Genet. 2005;1:e32. doi: 10.1371/journal.pgen.0010032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Staples J., Nickerson D.A., Below J.E. Utilizing graph theory to select the largest set of unrelated individuals for genetic analysis. Genet. Epidemiol. 2013;37:136–141. doi: 10.1002/gepi.21684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chang C.C., Chow C.C., Tellier L.C., Vattikuti S., Purcell S.M., Lee J.J. Second-generation PLINK: Rising to the challenge of larger and richer datasets. Gigascience. 2015;4:7. doi: 10.1186/s13742-015-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang Z., Ersoz E., Lai C.-Q., Todhunter R.J., Tiwari H.K., Gore M.A., Bradbury P.J., Yu J., Arnett D.K., Ordovas J.M., Buckler E.S. Mixed linear model approach adapted for genome-wide association studies. Nat. Genet. 2010;42:355–360. doi: 10.1038/ng.546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang J., Zaitlen N.A., Goddard M.E., Visscher P.M., Price A.L. Advantages and pitfalls in the application of mixed-model association methods. Nat. Genet. 2014;46:100–106. doi: 10.1038/ng.2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kirkpatrick B., Bouchard-Côté A. Correcting for cryptic relatedness in genome-wide association studies. arXiv. 2016 1602.07956 q-bio.QM. [Google Scholar]
- 24.Day-Williams A.G., Blangero J., Dyer T.D., Lange K., Sobel E.M. Linkage analysis without defined pedigrees. Genet. Epidemiol. 2011;35:360–370. doi: 10.1002/gepi.20584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Staples J., Qiao D., Cho M.H., Silverman E.K., Nickerson D.A., Below J.E., University of Washington Center for Mendelian Genomics PRIMUS: rapid reconstruction of pedigrees from genome-wide estimates of identity by descent. Am. J. Hum. Genet. 2014;95:553–564. doi: 10.1016/j.ajhg.2014.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ko A., Nielsen R. Composite likelihood method for inferring local pedigrees. PLoS Genet. 2017;13:e1006963. doi: 10.1371/journal.pgen.1006963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Abul-Husn N.S., Manickam K., Jones L.K., Wright E.A., Hartzel D.N., Gonzaga-Jauregui C., O’Dushlaine C., Leader J.B., Lester Kirchner H., Lindbuchler D.M. Genetic identification of familial hypercholesterolemia within a single U.S. health care system. Science. 2016;354:aaf7000. doi: 10.1126/science.aaf7000. [DOI] [PubMed] [Google Scholar]
- 28.International HapMap 3 Consortium. Altshuler D.M., Gibbs R.A., Peltonen L., Dermitzakis E., Schaffner S.F., Yu F., Bonnen P.E., de Bakker P.I.W., Deloukas P. Integrating common and rare genetic variation in diverse human populations. Nature. 2010;467:52–58. doi: 10.1038/nature09298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Loh P.-R., Danecek P., Palamara P.F., Fuchsberger C., A Reshef Y., K Finucane H., Schoenherr S., Forer L., McCarthy S., Abecasis G.R. Reference-based phasing using the Haplotype Reference Consortium panel. Nat. Genet. 2016;48:1443–1448. doi: 10.1038/ng.3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kumar P., Henikoff S., Ng P.C. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat. Protoc. 2009;4:1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 31.Adzhubei I., Jordan D.M., Sunyaev S.R. Predicting functional effect of human missense mutations using PolyPhen-2. Curr. Protoc. Hum. Genet. 2013;7 doi: 10.1002/0471142905.hg0720s76. 10.1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chun S., Fay J.C. Identification of deleterious mutations within three human genomes. Genome Res. 2009;19:1553–1561. doi: 10.1101/gr.092619.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schwarz J.M., Cooper D.N., Schuelke M., Seelow D. MutationTaster2: Mutation prediction for the deep-sequencing age. Nat. Methods. 2014;11:361–362. doi: 10.1038/nmeth.2890. [DOI] [PubMed] [Google Scholar]
- 34.Robinson J.T., Thorvaldsdóttir H., Winckler W., Guttman M., Lander E.S., Getz G., Mesirov J.P. Integrative genomics viewer. Nat. Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wei Q., Zhan X., Zhong X., Liu Y., Han Y., Chen W., Li B. A Bayesian framework for de novo mutation calling in parents-offspring trios. Bioinformatics. 2015;31:1375–1381. doi: 10.1093/bioinformatics/btu839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.de Ligt J., Willemsen M.H., van Bon B.W.M., Kleefstra T., Yntema H.G., Kroes T., Vulto-van Silfhout A.T., Koolen D.A., de Vries P., Gilissen C. Diagnostic exome sequencing in persons with severe intellectual disability. N. Engl. J. Med. 2012;367:1921–1929. doi: 10.1056/NEJMoa1206524. [DOI] [PubMed] [Google Scholar]
- 37.Staples J., Witherspoon D.J., Jorde L.B., Nickerson D.A., Below J.E., Huff C.D., University of Washington Center for Mendelian Genomics PADRE: Pedigree-Aware Distant-Relationship Estimation. Am. J. Hum. Genet. 2016;99:154–162. doi: 10.1016/j.ajhg.2016.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maxwell E.K., Packer J.S., O’Dushlaine C., McCarthy S.E., Hare-Harris A., Staples J., Gonzaga-Jauregui C., Fetterolf S.N., Faucett W.A., Leader J.B. Profiling copy number variation and disease associations from 50,726 DiscovEHR Study exomes. bioRxiv. 2017 119461; 10.1101. [Google Scholar]
- 39.Saleheen D., Natarajan P., Armean I.M., Zhao W., Rasheed A., Khetarpal S.A., Won H.-H., Karczewski K.J., O’Donnell-Luria A.H., Samocha K.E. Human knockouts and phenotypic analysis in a cohort with a high rate of consanguinity. Nature. 2017;544:235–239. doi: 10.1038/nature22034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Narasimhan V.M., Hunt K.A., Mason D., Baker C.L., Karczewski K.J., Barnes M.R., Barnett A.H., Bates C., Bellary S., Bockett N.A. Health and population effects of rare gene knockouts in adult humans with related parents. Science. 2016;352:474–477. doi: 10.1126/science.aac8624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Deciphering Developmental Disorders Study Prevalence and architecture of de novo mutations in developmental disorders. Nature. 2017;542:433–438. doi: 10.1038/nature21062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fromer M., Pocklington A.J., Kavanagh D.H., Williams H.J., Dwyer S., Gormley P., Georgieva L., Rees E., Palta P., Ruderfer D.M. De novo mutations in schizophrenia implicate synaptic networks. Nature. 2014;506:179–184. doi: 10.1038/nature12929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kong A., Frigge M.L., Masson G., Besenbacher S., Sulem P., Magnusson G., Gudjonsson S.A., Sigurdsson A., Jonasdottir A., Jonasdottir A. Rate of de novo mutations and the importance of father’s age to disease risk. Nature. 2012;488:471–475. doi: 10.1038/nature11396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rahbari R., Wuster A., Lindsay S.J., Hardwick R.J., Alexandrov L.B., Turki S.A., Dominiczak A., Morris A., Porteous D., Smith B., UK10K Consortium Timing, rates and spectra of human germline mutation. Nat. Genet. 2016;48:126–133. doi: 10.1038/ng.3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wong W.S.W., Solomon B.D., Bodian D.L., Kothiyal P., Eley G., Huddleston K.C., Baker R., Thach D.C., Iyer R.K., Vockley J.G., Niederhuber J.E. New observations on maternal age effect on germline de novo mutations. Nat. Commun. 2016;7:10486. doi: 10.1038/ncomms10486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McKusick V.A., Hostetler J.A., Egeland J.A. Genetic studies of the Amish, background and potentialities. Bull Johns Hopkins Hosp. 1964;115:203–222. [PubMed] [Google Scholar]
- 47.Ober C., Abney M., McPeek M.S. The genetic dissection of complex traits in a founder population. Am. J. Hum. Genet. 2001;69:1068–1079. doi: 10.1086/324025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gusev A., Palamara P.F., Aponte G., Zhuang Z., Darvasi A., Gregersen P., Pe’er I. The architecture of long-range haplotypes shared within and across populations. Mol. Biol. Evol. 2012;29:473–486. doi: 10.1093/molbev/msr133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bycroft C., Freeman C., Petkova D., Band G., Elliott L.T., Sharp K., Motyer A., Vukcevic D., Delaneau O., O’Connell J. Genome-wide genetic data on ∼500,000 UK Biobank participants. bioRxiv. 2017 166298; 10.1101. [Google Scholar]
- 50.Malinowski J., Goodloe R., Brown-Gentry K., Crawford D.C. Cryptic relatedness in epidemiologic collections accessed for genetic association studies: experiences from the Epidemiologic Architecture for Genes Linked to Environment (EAGLE) study and the National Health and Nutrition Examination Surveys (NHANES) Front. Genet. 2015;6:317. doi: 10.3389/fgene.2015.00317. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.