Abstract
Phosphatase and tensin homolog (PTEN) is a tumor suppressor frequently mutated in diverse cancers. Germline PTEN mutations are also associated with a range of clinical outcomes, including PTEN hamartoma tumor syndrome (PHTS) and autism spectrum disorder (ASD). To empower new insights into PTEN function and clinically relevant genotype-phenotype relationships, we systematically evaluated the effect of PTEN mutations on lipid phosphatase activity in vivo. Using a massively parallel approach that leverages an artificial humanized yeast model, we derived high-confidence estimates of functional impact for 7,244 single amino acid PTEN variants (86% of possible). We identified 2,273 mutations with reduced cellular lipid phosphatase activity, which includes 1,789 missense mutations. These data recapitulated known functional findings but also uncovered new insights into PTEN protein structure, biochemistry, and mutation tolerance. Several residues in the catalytic pocket showed surprising mutational tolerance. We identified that the solvent exposure of wild-type residues is a critical determinant of mutational tolerance. Further, we created a comprehensive functional map by leveraging correlations between amino acid substitutions to impute functional scores for all variants, including those not present in the assay. Variant functional scores can reliably discriminate likely pathogenic from benign alleles. Further, 32% of ClinVar unclassified missense variants are phosphatase deficient in our assay, supporting their reclassification. ASD-associated mutations generally had less severe fitness scores relative to PHTS-associated mutations (p = 7.16 × 10−5) and a higher fraction of hypomorphic mutations, arguing for continued genotype-phenotype studies in larger clinical datasets that can further leverage these rich functional data.
Keywords: PTEN, PTEN hamartoma tumor syndrome, autism spectrum disorder, deep mutation scanning, cancer, Cowden syndrome, genotype-phenotype, tumor suppressor, variants of uncertain significance, protein function
Introduction
Recent large-scale exome-sequencing studies have highlighted the abundance of protein-coding variation in the human population.1 It remains challenging to predict variant pathogenicity and clinical outcomes, especially for genes with pleiotropic effects. With most rare variants private to a single family or individual, using traditional approaches to establish pathogenicity such as variant segregation within a pedigree or identification in independent patients is infeasible. Even for well-studied genes, hundreds of variants are currently defined as variants of uncertain significance (VUS). Moreover, purely computational approaches still suffer from high false positive rates2 and subjective interpretations that limit the clinical utility of these predictions.
To address these challenges for genes of clinical importance, one proposed approach is to prospectively measure the functional effects of all possible mutations, allowing these empirical data to be integrated into the clinical assessment of novel rare variants.3, 4 Historically, these types of functional assays have been conducted in a serial nature, which limits scalability and often only within a portion of the protein of interest. While there are some notable examples of whole-gene brute force saturation mutagenesis, e.g., TP535 (MIM: 191170), new more scalable experimental paradigms are being developed that allow the functional dissection of the effects of thousands of genetic mutations in parallel.6 These approaches leverage recent advances in DNA synthesis and sequencing technologies and have proven particularly valuable in understanding the effects of mutations in cancer-associated genes.7, 8
With these issues in mind, we have developed a saturation mutagenesis approach to comprehensively assess the effect of nonsynonymous mutations on the lipid phosphatase activity of phosphatase and tensin homolog (PTEN [MIM: 601728]). PTEN antagonizes the phosphoinositide 3-kinase (PI3K) signaling pathway through its lipid phosphatase activity toward the signaling lipid phosphatidylinositol (3,4,5)-trisphosphate (PIP3).9, 10 In mice, loss of this activity increases tumor susceptibility in a dose-dependent manner.11 This observation led to a continuum model for PTEN’s role in cancer development, with the level of phenotypic severity tightly coupled to the level of lipid phosphatase activity.12
Germline PTEN mutations are associated with a range of clinical outcomes, including autism spectrum disorder (ASD [MIM: 605309])13, 14, 15 and tumor predisposition phenotypes collectively known as PTEN hamartoma tumor syndrome (PHTS).16, 17, 18 Germline mutation carriers often share the common feature of increased head size or macrocephaly.19 However, there is substantial variability in the neurological and tumor phenotypes present in these individuals. PHTS is an umbrella term that encompasses Cowden syndrome (MIM: 158350), Bannayan-Riley-Ruvalcaba syndrome (MIM: 153480), and PTEN-related Proteus sy3ndrome (MIM: 176920).20 PHTS-affected individuals typically present with macrocephaly and hamartomatous polyps and have an extremely high life-time risk of cancer.20 PTEN mutations have been identified in macrocephaly cohorts of individuals with formal ASD diagnoses or developmental delay (DD)/intellectual disability (ID)13, 21, 22 as well as idiopathic ASD.14, 23, 24
It is currently impossible to predict the phenotypic outcome of a given PTEN mutation. Even predicting whether a PTEN mutation will have a pathogenic effect is still challenging. This is exemplified by the fact that a majority of missense variants (131/241, 54%) in ClinVar are considered VUS and seven additional variants have inconsistent pathogenicity reported across laboratories. Recent evidence from functional assays on a limited number of mutations and using diverse models, including humanized yeast,25 cultured human cells,26 and in vivo mouse neurons,27 suggest that mutations identified in individuals with ASD or DD without obvious PHTS features tend to have hypomorphic lipid phosphatase activity, while PHTS-associated mutations more frequently show complete loss of lipid phosphatase activity. Further supporting this hypomorphic hypothesis, the distributions of mutation types are consistent with ASD-associated mutations being generally less severe, with reported missense mutations three to four times as common in ASD compared with PHTS.26, 28 These findings, as well as the established genotype-phenotype relationships for PTEN in cancer, led us to hypothesize that, at the population level, ASD-associated PTEN variants are hypomorphic compared to PHTS-associated PTEN variants.
To systematically test this hypothesis and improve our ability to interpret the functional effects of any PTEN mutation, we modified a previously validated humanized yeast model for massively parallel functional testing of the effects of PTEN mutations on lipid phosphatase activity in vivo.25, 29 Given that yeast do not signal through PIP3-dependent pathways,30 this model system challenges PTEN protein variants to act on their preferred substrate in a cellular environment, but removes the confounding signaling and regulatory milieu present in mammalian cells. Accordingly, the model is more sensitive than in vitro assays in which PTEN dephosphorylates a water-soluble substrate.31 The utility of the yeast model for measuring lipid phosphatase activity has been demonstrated through validation of mutation effects on downstream Akt1 activation in mammalian cells, exhibiting complete concordance for the variants tested.31
With this system, we analyzed the functional effect of 86% of all possible single amino acid alterations. Overlaying these data onto PTEN secondary and tertiary structures recapitulated many known or predicted structure-function and biochemical relationships but also revealed surprising patterns of mutational tolerance. We discovered that several residues within the catalytic pocket are surprisingly tolerant to mutation and identified residues that are critical for membrane interaction. Moreover, we demonstrate that these functional fitness scores have clinical utility by showing that they can outperform in silico-based approaches in characterizing likely pathogenic and benign variants. Finally, we provide compelling support for the existence of germline PTEN genotype-phenotype relationships that should be further explored in larger longitudinal clinical cohorts.
Material and Methods
PTEN Saturation Mutagenesis
We obtained wild-type PTEN sequence from GenBank (NM_000314.6). All protein variants are reported relative to the corresponding 403 amino acid protein (GenPept: NP_000305). Our mutagenesis approach was similar to the mutagenesis by integrated tiles (MITE) approach.32 We designed a series of DNA “tiles” that were complementary to wild-type PTEN except for one codon (Figure S1A). At this single codon, each molecule bore a substitution to the yeast-optimized codon for each non-wild-type amino acid, the yeast-preferred stop codon, or an in-frame, single codon deletion. Additionally, each set of “tiles” contained unique DNA adapters on either end to allow PCR retrieval of individual tiles from the pool (using primers with prefix: PTEN_sliceprimer, Table S1). These DNA tiles were synthesized as 130-mers (prefix: PTENTile) as part of a 12,000-feature oligo pool by CustomArray. For each tile, we designed inverse PCR primers that linearized the pYES2-PTEN wild-type sequence, excluding the portion encoded by the corresponding tile. Following amplification, the tile PCR products were incorporated into the appropriate linear pYES2-PTEN by SLiCE-mediated recombination.33 SLiCE reactions were 10 μL and consisted of 100 ng of linearized vector with 15 ng of tile DNA, along with 1× SLiCE buffer and 1× SLiCE extract. SLiCE extract and buffer were prepared as described previously.34 Reactions were incubated for 60 min at 37°C, then diluted 1:10 in water, and 2.5 μL used to electroporate 50 μL of NEB 10-beta electrocompetent E. coli. Transformation reactions were plated on LB agar plates with 100 mg/mL carbenecillin (GoldBio) and grown overnight at 37°C. Colonies were collected and plasmids isolated with the QIAprep Spin Miniprep Kit (QIAGEN).
Yeast Selection Experiments
Plasmid libraries were normalized and pooled into four mega-pools, each representing saturation mutagenesis for one quadrant (quadrants 1–3 = 100 codons, quadrant 4 = 103 codons). 1 μg of each mega-pool was transformed into the S. cerevisiae strain YPH-499, which already contained YCpLG-p110α-CAAX, using the Li-Ac/SS carrier DNA/PEG method.35 More than 50,000 colony forming units were generated per reaction. Colonies for each quadrant were pooled and grown overnight in SC-glucose –leu –ura (synthetic complete medium lacking leucine and uracil, using glucose as carbon source), pelleted and frozen down in 15% glycerol at −80°C.
Selection experiments began with overnight outgrowth of frozen stocks in SC-raffinose –leu –ura (raffinose neither induces nor represses GAL1/10 promoter). Following outgrowth, 25 or 30 million cells (replicate A or B) were pelleted for each quadrant as the “input” sample and frozen at −20°C. Then, 25 or 30 million cells were seeded into three cultures of 50 mL SC-galactose –leu –ura. Cultures were incubated at 30°C with 185 rpm shaking. After 24 and 36 hr of growth, cell concentrations were measured with a TC-20 Automated Cell Counter and 20 million cells (for each replicate) were passaged into fresh medium. At 48 hr, samples of 20 million cells were spun down with 13,000 × g for 30 s, medium withdrawn, and frozen at −20°C.
Library Prep and Sequencing
Plasmid DNA was isolated from pelleted cells (input and 48 hr) with Zymoprep Yeast Plasmid Miniprep II kit (Zymo Research). Stage-one PCR was performed in 25 μL reactions using: 5 ng of plasmid DNA, primers pYES2-PTEN_Q[1-4][F/R]_S1 (containing partial Illumina TruSeq adaptors) at 0.5 μM, 1× KAPA HiFi Hotstart Readymix (KHF), and 1× SYBR Green. Reactions were monitored by qPCR with cycling conditions: [95°C 3 min (98°C 20 s, 55°C 30 s, 72°C 15 s, plate read, 72°C 8 s) × 28–36 cycles]. Reactions were removed during or immediately following exponential phase of amplification. Stage-two PCR was then performed in 25 μL reactions using: 1 μL of uncleaned stage-one product, custom Illumina dual index TruSeq primers (prefixes: S2, i7) at 0.5 μM, 1× KHF, and 1× SYBR Green (Table S1). Reactions were monitored by qPCR with cycling conditions: [95°C 3 min (98°C 20 s, 55°C 15 s, 72°C 15 s, plate read, 72°C 8 s) × 6 cycles]. Reaction products were checked on a 1.5% agarose gel, purified using NucleoSpin PCR Clean-up (Machery-Nagel), and concentrations were measured using a Nanodrop 1000 Spectrophotometer. Samples were normalized and combined into a common pool that was sequenced across multiple runs using paired-end 300 base-pair reads on the Illumina MiSeq platform (v.3 reagent kit).
Sequencing Data Analysis
Paired-end reads were merged with PEAR36 and common priming sequences were trimmed from the 5′ and 3′ ends using cutadapt.37 For each quadrant, a purely wild-type sample was sequenced in order to identify sequencing error profiles. Counts of error reads were normalized to wild-type counts, and then this normalized amount of reads were removed from all experimental samples.7 Sequence variants were identified and counted with custom python scripts. These raw variant counts files were analyzed with Enrich2 v.1.2.038 to calculate scores and standard errors for each variant. If the 95% confidence interval (based on the standard error) of the fitness score was ≤1, the variant was considered high-confidence. If the 95% confidence interval was >1 but the measurements from each biological replicate were concordant (both lower or both higher than the 95% bound of the synonymous distribution), the variant was also considered high-confidence.
Mutation Collation
We considered any PTEN missense or nonsense (excluding frameshifting insertions or deletions [indels]) mutation in the gnomAD database1 (accessed 11/19/17) to be benign, with the exception of two variants that are considered pathogenic in ClinVar (p.Lys289Glu and p.Arg173His). We considered single-residue missense mutations from ClinVar (accessed 09/30/17) that were considered either pathogenic or likely pathogenic, were submitted with criteria, and had no conflicting reports to be pathogenic. We collected ASD-associated variants from SFARI Gene39 (accessed 10/09/17) and the literature. We collected PHTS-associated mutations from the literature. A mutation was considered ASD/DD associated if the report did not include symptoms of PHTS and the mutation had not been reported in another individual with PHTS. If an individual had ASD/DD and PHTS features, or was observed in multiple individuals representing both presentations, we considered it PHTS. We used Mann-Whitney U test to compare fitness scores (including high and low confidence) ASD/DD and PHTS variants. For any clinical mutation that was a frameshifting indel, the fitness score of the nonsense mutation from the corresponding position was used.
Protein Positional Features and Modeling
Conservation values were acquired from Consurf DB40 with default settings. Relative solvent exposure was calculated with GETAREA web tool.41 A position was considered exposed if its ratio of side-chain surface area to random-coil surface area exceeded 50, intermediate if the ratio was between 20 and 50, and buried if its ratio was less than 20. Secondary structure assignments were enumerated with STRIDE.42 Pymol was used to generate representations based on known partial crystal structure (PDB: 1D5R). Clustering was performed on the 326 positions with all 19 missense mutations measured (including high and low confidence). Clustering was performed with scipy.cluster.hierarchy.linkage, method = “ward.”
Mutation Effect Predictors
We obtained Provean and SIFT predictions from Provean Protein (v.1.1.3) with default settings. For Provean, we considered “deleterious” predictions as pathogenic and “neutral” predictions as benign. For SIFT, we considered “damaging” predictions as pathogenic and “tolerated” predictions as benign. We obtained PolyPhen-2 predictions from the PolyPhen-2 batch query web server. For PolyPhen-2, “probably damaging” or “possibly damaging” predictions were considered pathogenic, whereas “benign” predictions were considered benign.
Results
Establishing a Massively Parallel Functional Assay for PTEN Lipid Phosphatase Activity
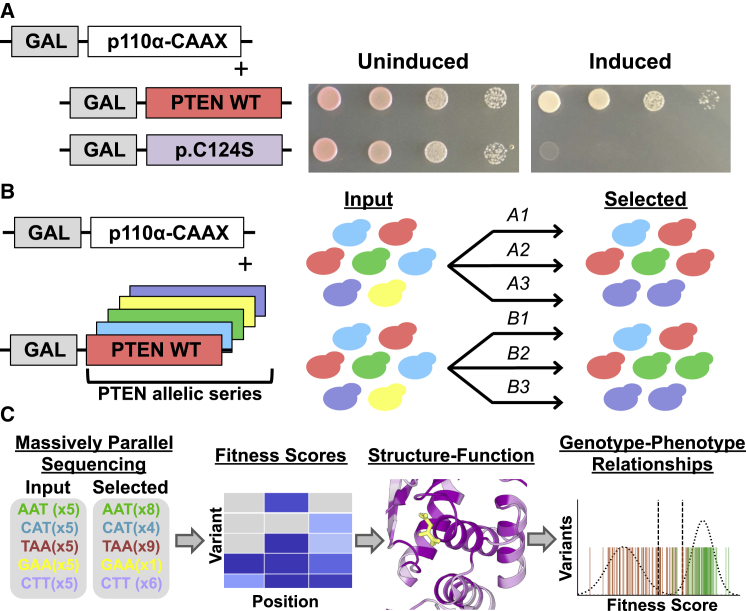
We leveraged an artificial humanized yeast model in order to assess the relative phosphatase activity of PTEN variants.25, 29 In this system, the human PI3K catalytic subunit p110α (encoded by PIK3CA [MIM: 171834]) is expressed in Saccharomyces cerevisiae and artificially directed to the membrane by a C-terminal prenylation box motif.29 At the membrane, p110α is able to catalyze the conversion of the essential pool of phosphatidylinositol (4,5)-bisphosphate (PIP2) to PIP3, which potently inhibits growth through cytoskeletal disruption.29 Upon induction of gene expression, cells proliferate at a rate that is proportional to the ability of the PTEN variant to convert PIP3 to PIP2.31 Co-expression of wild-type PTEN, but not catalytically dead mutants, e.g., p.Cys124Ser, catalyzes the reverse reaction, restoring the PIP2 pool and allowing the yeast to grow and survive (Figure 1A). Moreover, growth rate provides a quantitative surrogate of lipid phosphatase activity with partial loss-of-function mutations showing intermediate growth phenotypes.25
Figure 1.
A Framework for Massively Parallel Functional Testing of PTEN Mutations
(A) Humanized yeast model for evaluating the effect of PTEN mutations on lipid phosphatase activity. Exogenous expression of the catalytic subunit of human PI3K with a membrane-targeting prenylation box motif (p110α-CAAX) in yeast is toxic. However, co-expression of human PTEN wild-type, but not catalytically dead PTEN p.Cys124Ser, can rescue growth. Both genes are under the control of a galactose inducible promoter (GAL).
(B and C) Modifications to allow massively parallel variant assessment.
(B) We generated a comprehensive PTEN allelic series, introduced these variants into yeast en masse, and subjected them to p110α-CAAX-mediated selection in liquid culture. We performed two biological replicates, each consisting of three technical replicates.
(C) We collected input and selected time points and subjected these to deep sequencing. We used read counts to calculate fitness scores and used these scores to highlight structure-function insights as well as genotype-phenotype relationships.
We made several modifications to this system that allowed for massively parallel testing of preprogrammed mutations. First, to allow for parallel testing, rather than serial plating of single mutations, we modified the assay to support complex populations of PTEN-bearing yeast in liquid culture and sequencing as a readout of growth (Figures 1B, 1C, and S1). We then introduced the yeast-preferred codon for each non-wild-type amino acid, stop codon, and single residue deletion at all PTEN codons en masse, utilizing a homologous recombination-based mutagenesis approach (Material and Methods, Figures 1B and S2A, Table S1).32, 33 To allow direct sequencing of each mutagenized region, mutational space was separated into ∼300 base-pair quadrants (Figure S2A).
We transformed two independent yeast populations with our mutagenesis library. Sequencing of naive yeast libraries indicated that 95% of all intended mutations were present (Figures 1B and S2A). No position had less than 33% mutational coverage. Mutation dropout was largely confined to a single oligo pool in the C2 domain of the protein, which repeatedly performed poorly. We then performed selection experiments on these two independent yeast populations, each with three selection replicates (Figure 1B). We calculated natural log-scaled and wild-type normalized fitness scores for each variant, along with standard error-based confidence intervals (Material and Methods, Figure S2B).38 Score estimates were generated for 8,012 (95% of intended) PTEN nonsynonymous mutations and between mutational libraries fitness scores were highly correlated (Pearson’s r = 0.76, Tables 1 and S2, Figures S3A and S3B). The distribution of fitness effects illustrates two major populations corresponding to likely damaging and wild-type-like mutations (Figure S3A). Based on low standard error or replicate concordance, scores for 7,244 mutations (86% of intended) were classified as high confidence (Material and Methods, Tables 1 and S2, Figure S3C). Mutations were classified as wild-type-like if their cumulative fitness score was within the 95th percentile (two-tailed) of observed synonymous mutations (Figure S3D). We identified 2,273 likely damaging mutations (31%) and 4,872 wild-type like mutations (67%) (Table 1). We also observed 99 mutations that performed better than wild-type (1%), which was within what was expected due to chance based on the total number of wild-type-like variants. Among the likely damaging missense mutations, 1,249/1,789 (70%) fell within the observed distribution for programmed premature truncations (excluding C-terminal tail), with the remainder having intermediate phenotypes in this assay.
Table 1.
Summary of PTEN Mutagenesis and High-Confidence Effect Classifications
| Mut. Type |
Mutagenesis Summarya |
HC Classificationsb |
||||||
|---|---|---|---|---|---|---|---|---|
| Designed | Created | HC | Total < WTc | Trunc.-liked | Hypo.e | WT-likef | >WTg | |
| Missense | 7,657 | 7,260 (0.95) | 6,564 (0.86) | 1,789 (0.27) | 1,249 (0.19) | 540 (0.08) | 4,679 (0.71) | 96 (0.015) |
| A.A. del | 403 | 377 (0.94) | 340 (0.84) | 193 (0.57) | 168 (0.49) | 25 (0.07) | 144 (0.42) | 3 (0.007) |
| Trunc. | 403 | 375 (0.93) | 340 (0.84) | 291 (0.86) | 284 (0.84) | 7 (0.02) | 49 (0.14)h | 0 (–) |
| Total | 8,463 | 8,012 (0.95) | 7,244 (0.86) | 2,273 (0.31) | 1,701 (0.23) | 572 (0.08) | 4,872 (0.67) | 99 (0.014) |
Abbreviations: A.A. del, single amino acid deletion; HC, high-confidence; Hypo., hypomorphic; Mut., mutation; Trunc., truncation; WT, wild-type.
Numbers in parentheses represent the fraction of designed variants.
Numbers in parentheses represent the fraction of high-confidence variants.
Total < WT: less than wild-type; variants with scores less than or equal to −1.11, the lower 95th percentile (two-tailed) for synonymous variants.
Trunc.-like: truncation-like; subset of less than wild-type variants with scores less than or equal to −2.13, the upper 95th percentile (two-tailed) of nonsense mutations at positions 1-349.
Hypo: hypomorphic; subset of less than wild-type variants with scores between −2.13 and −1.11, the upper truncation and lower synonymous 95th percentiles (two-tailed).
WT-like: wild-type like; variants with scores between −1.11 and 0.89, the 95th percentile (two-tailed) of synonymous variants.
>WT: greater than wild-type; variants with scores exceeding 0.89, the upper 95th percentile (two-tailed) of synonymous variants.
48 of these truncating mutations fall within regulatory tail, positions 352–403.
High-Resolution Mutation Data Reveal Structure-Function Insights
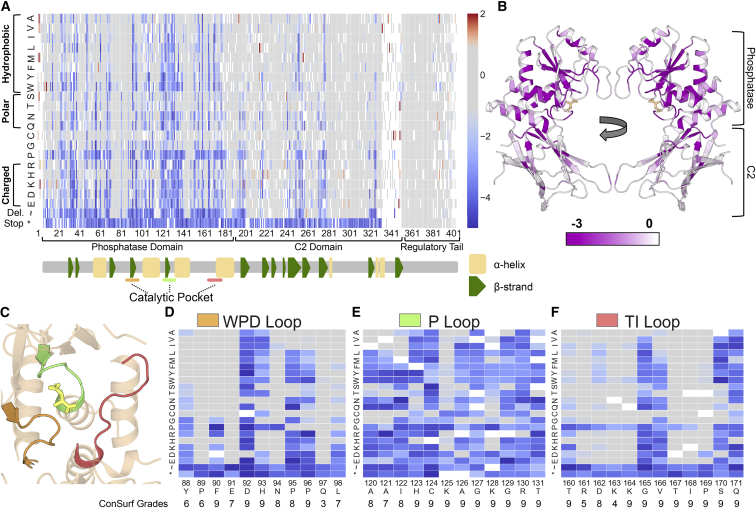
Using the high-confidence data, we first analyzed structure-function relationships, including known or predicted functional domains. Our complete sequence function map recapitulates many known features of PTEN biochemistry. For example, early truncating mutations are uniformly damaging through the phosphatase and C2 domain, but are tolerated in the regulatory tail (Figure 2A).31 Overlaying the median fitness score of each position onto the partial crystal structure of PTEN (including residues 7–285 and 310–353) reveals strong intolerance of positions in the phosphatase domain, especially those positions near the catalytic pocket (Figure 2B). The median fitness scores are also correlated with evolutionary conservation (Spearman, ρ = 0.58, Figure S3E). When compared to positions in alpha helices and beta strands, unstructured positions are very tolerant to mutation (Figure S3F).
Figure 2.
High-Resolution Map of the Functional Effects of PTEN Mutations
(A) Heatmap schematic showing high-confidence fitness scores for 7,244 PTEN missense, nonsense, or in-frame deletion mutations (86% of possible). Columns are each protein position and amino acids are listed in rows ordered according to biophysical characteristics. Variants with fitness scores within the 95th percentile (two-sided) of synonymous wild-type like mutations are colored gray. Variants with fitness scores lower than the synonymous distribution are colored blue while variants with higher fitness scores are colored red. The major protein domains, as well as the secondary structure features, are indicated in the track below the heatmap (α-helices as yellow rectangles and β-strands as green pentagons).
(B) Ribbon diagram of PTEN crystal structure with residues colored by average fitness score. Darker purple corresponds to more damaging scores.
(C) Ribbon diagram highlighting the crystal structure of the PTEN catalytic pocket, composed of the WPD (orange), P (green), and TI-loops (salmon).
(D–F) The fitness scores of mutations at the residues composing the three catalytic pocket loops. Beneath each position is the Consurf grade (Material and Methods), which represents the relative evolutionary conservation, with nine being the most conserved and one being the least conserved.
The catalytic pocket of PTEN is composed of the WPD, P, and TI loops (Figure 2C). This motif has sequence homology to dual specificity protein phosphatases, especially within the signature motif (123-HisCysXXGlyXXArg-130).43 Arg130 is a hot spot for somatic cancer-associated mutations with multiple different missense and truncations frequently reported.44 We observed that this critical position was intolerant to all mutations (Figure 2E). Compared to other phosphatases, PTEN also has unique sequence features in order to accommodate the highly acidic and bulky PIP3 substrate. Residues His93, Lys125, and Lys128 impart a basic character on the pocket,43 the importance of which is demonstrated by the mutational intolerance at these positions (Figures 2D and 2E). Asp92 is a critical residue for PTEN catalysis, but its exact role remains uncertain.25, 45 We find that the only substitution with wild-type-like activity is asparagine. Additionally, the PTEN catalytic pocket is larger compared to other dual specificity phosphatases.43 The Cowden-associated p.Gly129Glu variant has been shown to abolish lipid phosphatase while preserving protein phosphatase activity.46 Our data show that Gly129 is intolerant to all mutations except to alanine and serine, the two next smallest amino acids (Figure 2E). Unexpectedly, despite their presence in the catalytic pocket, several residues in the WPD and TI loops are highly tolerant to mutations (Figures 2D–2F), highlighting the power of functional data to delineate truly functional from non-functional alterations within highly conserved protein domains.
PTEN associates with the plasma membrane through multiple domains. A PIP2 binding motif in the phosphatase domain (residues 6–15) is rich in positively charged amino acids and allosterically promotes catalysis upon PIP2 binding.47, 48 An additional positively charged residue, Arg47, contributes to this interaction.49 Our data suggest that Arg15, Lys13, and Arg47 are the most critical of the positively charged residues in this motif (Figure S4A).50 Additionally, an intramolecular regulatory interaction between the C-terminal tail and the phosphatase domain is controlled by phosphorylation at four sites in the tail, in mammalian cells.51 We find that individual phosphomimetic substitutions at these sites are insufficient to decrease activity in our assay (Figure S4B).
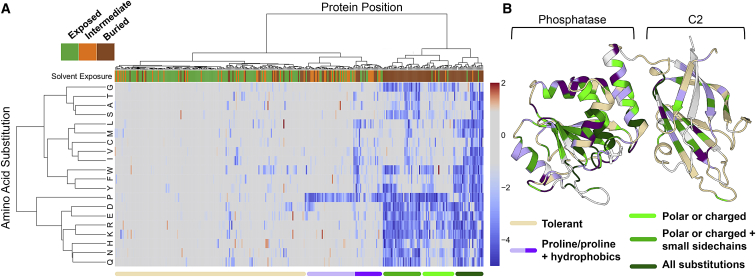
Protein Positions Cluster into Stereotyped Patterns of Mutational Sensitivity
In order to identify patterns of mutational sensitivity among PTEN positions and amino acid substitutions, we performed hierarchical clustering with all positions at which we measured effects of all missense substitutions (including high and low confidence, n = 326, Figure 3A). We found that positions clustered into two major clades, corresponding to positions broadly tolerant/intolerant to proline or highly sensitive positions. We identified solvent exposure as a highly discriminatory feature between sensitive and tolerant clades, with 80/88 (91%) positions in the sensitive clade being in buried positions, while only 44/170 (26%) are buried in the tolerant clade (Figure 3A). The tolerant clade splits into two major groups with a sub-clade broadly tolerant to all substitutions (beige) and a second sub-clade where positions are sensitive either to proline alone or to proline and hydrophobic residues (purple). The proline-sensitive positions generally are part of secondary structures that are not buried in the hydrophobic core (Figure 3A). The sensitive clade positions split into three groups (green shaded sub-clades), which differ in their tolerance to charged, polar, or hydrophobic residues. The dark green clade represents the most constrained positions and includes positions 92, 123, 124, and 130, all of which are in the catalytic pocket and critical for catalysis. Overlaying the sub-clade assignment of each position onto the crystal structure highlights the intolerance of mutations within the hydrophobic core of the phosphatase domain. Many of the solvent-exposed positions in the C2 domain are tolerant to mutation (Figure 3B).
Figure 3.
Hierarchical Clustering Reveals Patterns of Mutational Tolerance among Protein Positions and Amino Acid Substitutions
(A) Hierarchical clustering of the 326 sites with all missense mutations measured. Clustering was performed by positions and amino acid substitutions (positions are columns and amino acid substitutions are rows). Overlaid on this heatmap is a top track showing the solvent exposure of each position in the crystal structure, with solvent-exposed positions colored green, intermediate positions orange, and buried positions brown. We identified two major clades, which partitioned into five sub-clades with prevailing characteristics indicated and represented in the bottom track. We further divided the purple clade to reflect major differences in mutational tolerance.
(B) Ribbon diagram of PTEN crystal structure with residues colored according to clade assignment.
Clustering by amino acid substitutions recapitulated known functional relationships with proline correlated poorly with other substitutions (Figure 3A). We sought to leverage these patterns of correlation to predict the fitness scores of mutations that were not present in our mutagenesis library or that were low confidence.52 We developed a heuristic for using only the most closely correlated observed substitutions53 at the site of interest to compute an “informed position average” (Figure S5A). We combined this with several other prediction-based, evolutionary, and biophysical features to train and test a random forest regression algorithm on our high-confidence measurements (Material and Methods, Figures S5B and S5C, Table S6).52 We used 10-fold cross validation to confirm that this approach can predict unseen data with high confidence (Pearson’s r = 0.80, Figure S5E). We further performed a downsampling analysis to assess the expected accuracy of imputing scores at different levels of saturation, finding that reductions of 10%–20% (65.8%–74% of saturation) achieve similar performance (Figure S5F). Finally, we generated imputations for all variants that were absent from our library or measured with low confidence (Figure S6 and Table S2).
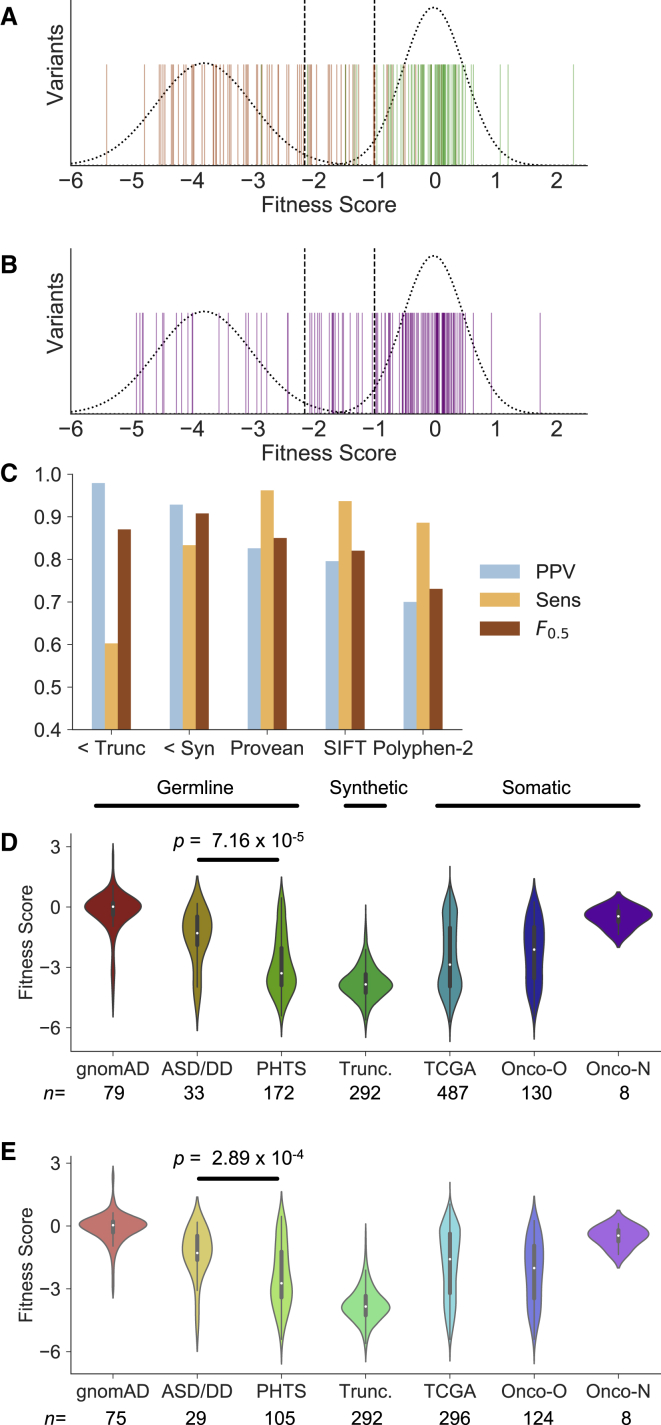
Fitness Scores Discriminate between Likely Pathogenic and Benign Alleles
To determine whether our empirically determined fitness scores were informative for discriminating between germline likely pathogenic and benign alleles, we collected germline missense mutations reported as pathogenic or likely pathogenic from ClinVar54 and rare variants from gnomAD,1 excluding p.Arg173His and p.Lys289Glu that are reported pathogenic in ClinVar (Material and Methods, Tables S3 and S4). Fitness scores alone discriminated pathogenic from benign germline alleles (Figure 4A). We found that the F0.5 score, which weights predictive value (PPV) over sensitivity, reaches its maximum at a cutoff based on the synonymous distribution (≤−1, ∼95th percentile, PPV = 0.93, sensitivity 0.83) and outperforms several in silico mutation effect prediction algorithms (Figures 4C and S7). PPV was maximized (0.98) at a more conservative cutoff based on the 95th percentile of the truncation distribution but with reduced sensitivity (0.60) (Figures 4A and 4C). Given the high PPV of our scores, we evaluated distribution of fitness scores among ClinVar missense VUS (Figure 4B). We found that 21/127 (17%) VUS with high-confidence data met the strict truncation-based cutoff and 41/127 (32%) met the synonymous cutoff, suggesting that fitness scores could be used to reclassify a major fraction of VUS.
Figure 4.
Fitness Scores Discriminate between Likely Pathogenic and Benign Variants and Support Genotype-Phenotype Relationships
(A) Fitness scores for missense variants considered pathogenic or likely pathogenic in ClinVar (orange) and putatively benign variants from gnomAD (green). Dashed curves represent truncation (left) and synonymous (right) distributions. Dashed lines at −2.15 and −1 represent the approximate 95% two-tailed distribution of truncations (before regulatory tail) and synonymous mutations, respectively.
(B) Fitness scores of ClinVar VUS (purple), with truncation and synonymous distributions and 95% limits.
(C) To test the ability of fitness scores to discriminate likely pathogenic from likely benign missense mutations, we calculated positive predictive value (PPV), sensitivity, and F0.5 scores for our fitness scores (“<Trunc” represents the threshold at −2.15, “<Syn” represents the threshold at −1). In these tests, a true positive represented a ClinVar pathogenic allele having a fitness score less than or equal to the threshold. We compared the performance of the fitness scores at these two thresholds with in silico pathogenicity predictors for missense mutations using their default thresholds (Material and Methods).
(D) Fitness scores of all curated mutations associated with the indicated phenotype (Material and Methods).
(E) Fitness scores of only curated missense mutations associated with the indicated phenotype (programmed truncations also shown for clarity). Abbreviations: ASD/DD, autism spectrum disorder/developmental delay; PHTS, PTEN hamartoma tumor syndrome; TCGA, The Cancer Genome Atlas; Onco-O, OncoKB mutations considered “oncogenic” or “likely oncogenic;” Onco-N, OncoKB mutations considered “likely neutral.”
PTEN mutations are extremely frequent in somatic cancer. We extracted nonsynonymous mutations from The Cancer Genome Atlas (TCGA) and observed a multimodal and wide distribution of fitness scores (Figures 4D and 4E, Table S5). This is likely due to the presence of both driver and passenger mutations in these data. Similar to the germline analysis, to test whether fitness scores could discriminate somatic mutations that are likely pathogenic, we evaluated mutations from Onco-KB, a precision oncology database with expert annotation of somatic mutations (Table S5).55 We found that fitness scores of PTEN mutations considered “oncogenic” or “likely oncogenic” were substantially more negative than those considered “likely neutral” (Figures 4D and 4E). Of the missense likely oncogenic, 86/124 (69%) and 56/124 (45%) were below the synonymous and truncation thresholds, respectively. In contrast, of the eight variants considered likely neutral (all missense), only one (p.Ala121Val) had a fitness score marginally below the synonymous cutoff (fitness score, −1.3). Taken together, these findings emphasize the ability of empirically determined fitness scores to discriminate between pathogenic and benign human alleles, in both the germline and somatic setting.
Finally, we evaluated potential genotype-phenotype relationships for germline PTEN mutations. We first compared the fitness scores of PTEN mutations associated with various clinical presentations acquired from multiple sources (Material and Methods, Figure 4D, Table S5). We found that, as a population, fitness scores of nonsynonymous mutations exclusively reported in ASD/DD-affected cohorts were less severe than PHTS-associated mutations (Mann-Whitney U-test, two-sided, p = 7.16 × 10−5). Comparing only the missense, we found that this significant difference persists (Mann-Whitney U-test, two-sided, p = 2.89 × 10−4), indicating that the mutation type alone does not drive these differences (Figure 4E). We found 12/29 (41%) and 21/105 (20%) of the ASD and PHTS missense mutation fell within the hypomorphic activity range, respectively. Overall, these data provide strong support for the hypothesis that ASD/DD-associated mutations often retain hypomorphic PTEN phosphatase activity.
Discussion
Massively multiplexed functional assays represent a promising approach to understanding the effect of mutations on protein function, which can provide immediate insights into structure-function relationships and clinical interpretation. Modifying a humanized yeast assay that uses growth to read out relative phosphatase activity, we were able to assess the functional effects of human PTEN mutations on a massive scale. Our approach yielded high-confidence measurements of 86% of the possible single-residue nonsynonymous mutations. A limited number of human proteins have been subjected to full-length massively multiplexed functional assessment and very few have been assayed at the depth we achieved.7, 8, 52, 56, 57, 58, 59, 60 Similar approaches could be used with this model to the study of various aspects of the PI3K/Akt pathway at scale, including mutations in PIK3CA/B31 (p110α/β [PIK3CB (MIM: 602925)]), PIK3R161 (p85α [MIM: 171833]), and AKT162 (MIM: 164730), as well as drug screening for PIK3CA inhibitors.63
Several features of the data support the validity of these function estimates and their relevance to human health. We observed high correlation between biological replicates and recapitulated known features of PTEN function. For example, there were no pathogenic mutations within our curated clinical dataset in the C-terminal tail. The set of early terminating mutations confirm that the minimal catalytic unit includes the phosphatase and C2 domains, but not the C-terminal tail.31 Likewise, we found that position Cys124, which takes part directly in phosphatase catalysis, and position Arg130, which is a hotspot for cancer mutations, are completely mutation intolerant. Additionally, we found that mutations are not well tolerated within the loops forming the catalytic pocket or residues mediating interactions with PIP2. Finally, we found that proline was the most damaging substitution, consistent with a recent meta-analysis of massively multiplexed experiments53 and decades of biochemistry.64
While the humanized yeast system faithfully reports on intrinsic lipid phosphatase activity, mutations that functionally disrupt protein-protein interactions, subcellular localization, post-translational modifications, or function through a dominant-negative mechanism65 in mammalian cells will not be captured. We observed 99 variants with greater than wild-type like activity, none of which were present in curated pathogenic datasets. While it is possible that some of these variants increased PTEN activity, the number of variants of this class does not exceed what we would expect under the null assumption of wild-type-like activity. PTEN has relatively low thermostability66 and protein destabilization is a known mechanism for PTEN loss-of-function.26, 67 A concurrent functional screen assaying protein stability found ∼1/4th of mutations alter steady-state stability.59 Six mutations that destabilized PTEN in breast cancer cell lines also decreased steady-state abundance in this yeast model,31 suggesting that mutations affecting thermostability will be detected in our screen. However, our sensitivity to detect destabilizing mutations is unknown, as is whether mutations specifically altering the rate of proteasome-mediated degradation68 will be reported on. We believe that independently assaying these important factors at similar scale would provide useful complementary insights into PTEN function.
We discovered that approximately half of all positions in PTEN were broadly tolerant to substitutions, suggesting that they are not required for lipid phosphatase activity. While there is a degree of correlation between the median fitness score and the evolutionary conservation of each position, we identified positions within the highly conserved catalytic pocket and elsewhere in the protein that are highly tolerant to specific mutations. This is in apparent contradiction with PTEN’s high evolutionary conservation (99.75% identity between human and mouse28) and constraint in humans.1 This suggests that many PTEN positions are potentially under selection due to phosphatase-independent functions.
Our high-resolution mutation data empowered unique insights into PTEN biochemistry and structure. The substitution p.Gly129Glu is a well-known Cowden-associated variant that disrupts lipid phosphatase activity while maintaining protein phosphatase activity.46 We found that substitutions to alanine and serine are tolerated at this position, while mutations to bulkier residues are damaging. This suggests that there is a size limit for the amino acid that occupies this position. Asp92 matches the position of aspartic acid in the WPD loop of PTP1B, which acts as a general acid in the catalytic mechanism.45 Asp92 is a critical residue in the PTEN catalytic pocket, but its role in the reaction mechanism remains uncertain.25, 45, 69 Our data support previous findings that all mutations except p.Asp92Asn are strongly damaging.25 However, the p.Asp92Asn variant has been reported in an individual with ASD, indicating that it still may have a clinical effect.70 Similar to our findings, Rodríguez-Escudero and colleagues found in the yeast assay p.Asp92Asn had growth rescue similar to wild-type, but partial activity relative to wild-type using an indirect fluorescence indicator of PIP3 levels or an in vitro phosphatase assay.25 Combined, these data are consistent with the p.Asp92Asn variant retaining partial activity. We propose that p.Asp92Asn could be showing wild-type-like activity in our assay through asparagine deamidation, which is a spontaneous, intramolecular reaction that can result in the conversion of asparagine to aspartic acid.71 In biochemical systems and mammalian cells, this spontaneous conversion may not be sufficient to fully rescue PTEN activity.
Similar to previous studies,5, 72 we used hierarchical clustering to look for patterns among the positions and amino acid substitutions. We found that PTEN positions fall into a few stereotyped patterns of mutational tolerance and that a critical determinant of mutational tolerance is the relative solvent exposure of the position. These findings are consistent with a recent meta-analysis of similar experiments.73 We leveraged the correlation among amino acid substitutions, along with several other features, to generate a random forest regression model that could accurately predict the fitness scores of unseen mutations and create a comprehensive functional map encompassing the effects of all possible single nonsynonymous mutations. To guide future studies of similar proteins, we performed a downsampling analysis of the training data and found that for similar accuracy, ∼70% mutation saturation would likely be sufficient. Moreover, proline substitutions predict poorly and should be directly assayed.
A critical hurdle for the application of massively multiplexed functional assays is bridging the gap between molecular phenotype and human phenotype.74 We found that fitness scores are able to discriminate between likely pathogenic and benign human alleles in both the germline and somatic condition. On this basis, we expect that these scores will be of tremendous clinical value for reclassifying VUS4 and also for predicting the effects of private alleles that remain to be identified. A major question related to PTEN genetics is whether genotype-phenotype relationships can explain the heterogeneity in clinical presentation for carriers of germline mutations. Our comprehensive dataset provides strong evidence that the mutations associated with ASD/DD are hypomorphic for lipid phosphatase activity and are significantly more active than the mutations that lead to PHTS. This suggests that distinct biological mechanisms underlie the differential presentations, and understanding these differences will be critical for the eventual treatment of these disorders. While it is possible that these different mechanisms are the direct result of lipid phosphatase activity at the plasma membrane, ASD-associated mutations may specifically disrupt another of PTEN’s cellular functions.75, 76 Supporting this idea, some ASD-associated mutations are excluded from the nucleus and lead to neuronal hypertrophy, but this phenotype can be rescued by artificial direction to the nucleus.77
While massively parallel functional data are a significant advance for understanding function-specific mutation effects, further untangling complex genotype-phenotype relationships will require similar advances in clinical genetics databases with standardized descriptors of clinical presentations and symptoms.28 Our study was limited by both the number of publicly available mutations and associated clinical information. Since there are no coding variants considered benign in ClinVar, we used PTEN variants in the gnomAD database as a proxy for likely benign mutations. While these mutations are on average wild-type-like, we recognize that this is an imperfect approach and it is possible that some of the variants in gnomAD are pathogenic. We excluded variants that were only in ClinVar from our genotype-phenotype analysis because of their ambiguous annotation and lack of clinical data. For example, 17% of the pathogenic/likely pathogenic mutation submissions had no indicating condition provided and 36% of all missense entries use the ambiguous term “hereditary cancer-predisposing syndrome.” Requiring submitters to provide more information in a consistent way will maximize the utility of massively multiplexed functional data. Finally, it is still unclear whether individuals ascertained for neurological phenotypes as children will have a higher risk to develop PHTS-like or cancer presentations later in life.78 Moving forward, large-scale sequencing efforts that permit longitudinal assessment as well as patient re-contact will be instrumental. A new initiative, SPARK, aims to partner with 50,000 individuals with ASD and their families to create the largest genetically characterized ASD cohort to date.79 It is likely that hundreds of new PTEN mutation carriers will be identified in SPARK and would be available for re-contact and detailed prospective study.
We demonstrate that comprehensively assaying the molecular phenotypes of thousands of mutations to a human protein can yield clinically relevant insights, even for proteins with pleiotropic effects. Future efforts that combine multiple functional modalities and rich clinical datasets may allow for the precision needed to fully realize personalized genomic medicine.
Acknowledgments
We thank R. Pulido for providing yeast expression constructs and YPH-499 yeast strain and Y. Zhang for providing E. coli strain used for generating the SLiCE reagent. We thank Y. Jia and the Oregon National Primate Research Center Molecular & Cell Biology Core for technical assistance. We thank A.C. Adey, E. Fombonne, D.M. Fowler, A.F. Rubin, J. Mester, J. Weile, J. Savage, U. Shinde, J. Zonana, G. Mandel, P.J. Stork, and K.M. Wright for helpful discussions. We thank Martha Atherton and the Atherton Foundation for their support of the NARSAD awards. This work was supported by a NARSAD Young Investigator Grant from the Brain and Behavior Research Foundation through the NARSAD-Atherton Foundation Young Investigator Award (22935 to B.J.O.), a Sloan Research Fellowship in Neurosciences (Alfred P. Sloan Foundation, FG-2015-65608 to B.J.O.), and internal funds (B.J.O.). T.L.M received support through a training grant supported by National Institute of Diabetes and Digestive and Kidney Diseases of the National Institutes of Health under award number T32DK007680. T.L.M. is an ARCS scholar (Achievement Rewards for College Scientists Foundation, Inc., Oregon Chapter) and B.J.O. is a Klingenstein-Simons Fellow (Esther A. & Joseph Klingenstein Fund, Simons Foundation).
Published: April 26, 2018
Footnotes
Supplemental Data include seven figures and six tables and can be found with this article online at https://doi.org/10.1016/j.ajhg.2018.03.018.
Accession Numbers
The accession number for the DNA sequencing data reported in this paper is SRA: SRP134135.
Web Resources
gnomAD Browser, http://gnomad.broadinstitute.org/
OMIM, http://www.omim.org/
OncoKB, http://oncokb.org/
PolyPhen-2, http://genetics.bwh.harvard.edu/pph2/
PROVEAN, http://provean.jcvi.org
PyMOL, https://pymol.org/2
RCSB Protein Data Bank, http://www.rcsb.org/pdb/home/home.do
Sequence Read Archive (SRA), http://www.ncbi.nlm.nih.gov/sra
SFARI Gene, https://gene.sfari.org/autdb/
TCGA Portal, https://cancergenome.nih.gov/
Supplemental Data
References
- 1.Lek M., Karczewski K.J., Minikel E.V., Samocha K.E., Banks E., Fennell T., O’Donnell-Luria A.H., Ware J.S., Hill A.J., Cummings B.B., Exome Aggregation Consortium Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sun S., Yang F., Tan G., Costanzo M., Oughtred R., Hirschman J., Theesfeld C.L., Bansal P., Sahni N., Yi S. An extended set of yeast-based functional assays accurately identifies human disease mutations. Genome Res. 2016;26:670–680. doi: 10.1101/gr.192526.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Starita L.M., Ahituv N., Dunham M.J., Kitzman J.O., Roth F.P., Seelig G., Shendure J., Fowler D.M. Variant interpretation: functional assays to the rescue. Am. J. Hum. Genet. 2017;101:315–325. doi: 10.1016/j.ajhg.2017.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Richards S., Aziz N., Bale S., Bick D., Das S., Gastier-Foster J., Grody W.W., Hegde M., Lyon E., Spector E., ACMG Laboratory Quality Assurance Committee Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kato S., Han S.-Y., Liu W., Otsuka K., Shibata H., Kanamaru R., Ishioka C. Understanding the function-structure and function-mutation relationships of p53 tumor suppressor protein by high-resolution missense mutation analysis. Proc. Natl. Acad. Sci. USA. 2003;100:8424–8429. doi: 10.1073/pnas.1431692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fowler D.M., Fields S. Deep mutational scanning: a new style of protein science. Nat. Methods. 2014;11:801–807. doi: 10.1038/nmeth.3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brenan L., Andreev A., Cohen O., Pantel S., Kamburov A., Cacchiarelli D., Persky N.S., Zhu C., Bagul M., Goetz E.M. Phenotypic characterization of a comprehensive set of MAPK1/ERK2 missense mutants. Cell Rep. 2016;17:1171–1183. doi: 10.1016/j.celrep.2016.09.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Starita L.M., Young D.L., Islam M., Kitzman J.O., Gullingsrud J., Hause R.J., Fowler D.M., Parvin J.D., Shendure J., Fields S. Massively parallel functional analysis of BRCA1 RING domain variants. Genetics. 2015;200:413–422. doi: 10.1534/genetics.115.175802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Worby C.A., Dixon J.E. PTEN. Annu. Rev. Biochem. 2014;83:641–669. doi: 10.1146/annurev-biochem-082411-113907. [DOI] [PubMed] [Google Scholar]
- 10.Song M.S., Salmena L., Pandolfi P.P. The functions and regulation of the PTEN tumour suppressor. Nat. Rev. Mol. Cell Biol. 2012;13:283–296. doi: 10.1038/nrm3330. [DOI] [PubMed] [Google Scholar]
- 11.Alimonti A., Carracedo A., Clohessy J.G., Trotman L.C., Nardella C., Egia A., Salmena L., Sampieri K., Haveman W.J., Brogi E. Subtle variations in Pten dose determine cancer susceptibility. Nat. Genet. 2010;42:454–458. doi: 10.1038/ng.556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Berger A.H., Knudson A.G., Pandolfi P.P. A continuum model for tumour suppression. Nature. 2011;476:163–169. doi: 10.1038/nature10275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Varga E.A., Pastore M., Prior T., Herman G.E., McBride K.L. The prevalence of PTEN mutations in a clinical pediatric cohort with autism spectrum disorders, developmental delay, and macrocephaly. Genet. Med. 2009;11:111–117. doi: 10.1097/GIM.0b013e31818fd762. [DOI] [PubMed] [Google Scholar]
- 14.O’Roak B.J., Vives L., Fu W., Egertson J.D., Stanaway I.B., Phelps I.G., Carvill G., Kumar A., Lee C., Ankenman K. Multiplex targeted sequencing identifies recurrently mutated genes in autism spectrum disorders. Science. 2012;338:1619–1622. doi: 10.1126/science.1227764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Butler M.G., Dasouki M.J., Zhou X.-P., Talebizadeh Z., Brown M., Takahashi T.N., Miles J.H., Wang C.H., Stratton R., Pilarski R., Eng C. Subset of individuals with autism spectrum disorders and extreme macrocephaly associated with germline PTEN tumour suppressor gene mutations. J. Med. Genet. 2005;42:318–321. doi: 10.1136/jmg.2004.024646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liaw D., Marsh D.J., Li J., Dahia P.L.M., Wang S.I., Zheng Z., Bose S., Call K.M., Tsou H.C., Peacocke M. Germline mutations of the PTEN gene in Cowden disease, an inherited breast and thyroid cancer syndrome. Nat. Genet. 1997;16:64–67. doi: 10.1038/ng0597-64. [DOI] [PubMed] [Google Scholar]
- 17.Marsh D.J., Dahia P.L.M., Zheng Z., Liaw D., Parsons R., Gorlin R.J., Eng C. Germline mutations in PTEN are present in Bannayan-Zonana syndrome. Nat. Genet. 1997;16:333–334. doi: 10.1038/ng0897-333. [DOI] [PubMed] [Google Scholar]
- 18.Padberg G.W., Schot J.D.L., Vielvoye G.J., Bots G.T.A.M., de Beer F.C. Lhermitte-Duclos disease and Cowden disease: a single phakomatosis. Ann. Neurol. 1991;29:517–523. doi: 10.1002/ana.410290511. [DOI] [PubMed] [Google Scholar]
- 19.Mester J.L., Tilot A.K., Rybicki L.A., Frazier T.W., 2nd, Eng C. Analysis of prevalence and degree of macrocephaly in patients with germline PTEN mutations and of brain weight in Pten knock-in murine model. Eur. J. Hum. Genet. 2011;19:763–768. doi: 10.1038/ejhg.2011.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eng C. University of Washington; Seattle: 1993. PTEN Hamartoma Tumor Syndrome. [Google Scholar]
- 21.Buxbaum J.D., Cai G., Chaste P., Nygren G., Goldsmith J., Reichert J., Anckarsäter H., Rastam M., Smith C.J., Silverman J.M. Mutation screening of thePTEN gene in patients with autism spectrum disorders and macrocephaly. Am. J. Med. Genet. Part B Neuropsychiatr. Genet. 2007;144B:484–491. doi: 10.1002/ajmg.b.30493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McBride K.L., Varga E.A., Pastore M.T., Prior T.W., Manickam K., Atkin J.F., Herman G.E. Confirmation study of PTEN mutations among individuals with autism or developmental delays/mental retardation and macrocephaly. Autism Res. 2010;3:137–141. doi: 10.1002/aur.132. [DOI] [PubMed] [Google Scholar]
- 23.C Yuen R.K., Merico D., Bookman M., L Howe J., Thiruvahindrapuram B., Patel R.V., Whitney J., Deflaux N., Bingham J., Wang Z. Whole genome sequencing resource identifies 18 new candidate genes for autism spectrum disorder. Nat. Neurosci. 2017;20:602–611. doi: 10.1038/nn.4524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.O’Roak B.J., Stessman H.A., Boyle E.A., Witherspoon K.T., Martin B., Lee C., Vives L., Baker C., Hiatt J.B., Nickerson D.A. Recurrent de novo mutations implicate novel genes underlying simplex autism risk. Nat. Commun. 2014;5:5595. doi: 10.1038/ncomms6595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rodríguez-Escudero I., Oliver M.D., Andrés-Pons A., Molina M., Cid V.J., Pulido R. A comprehensive functional analysis of PTEN mutations: implications in tumor- and autism-related syndromes. Hum. Mol. Genet. 2011;20:4132–4142. doi: 10.1093/hmg/ddr337. [DOI] [PubMed] [Google Scholar]
- 26.Spinelli L., Black F.M., Berg J.N., Eickholt B.J., Leslie N.R. Functionally distinct groups of inherited PTEN mutations in autism and tumour syndromes. J. Med. Genet. 2015;52:128–134. doi: 10.1136/jmedgenet-2014-102803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vogt D., Cho K.K.A., Lee A.T., Sohal V.S., Rubenstein J.L.R. The parvalbumin/somatostatin ratio is increased in Pten mutant mice and by human PTEN ASD alleles. Cell Rep. 2015;11:944–956. doi: 10.1016/j.celrep.2015.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leslie N.R., Longy M. Inherited PTEN mutations and the prediction of phenotype. Semin. Cell Dev. Biol. 2016;52:30–38. doi: 10.1016/j.semcdb.2016.01.030. [DOI] [PubMed] [Google Scholar]
- 29.Rodríguez-Escudero I., Roelants F.M., Thorner J., Nombela C., Molina M., Cid V.J. Reconstitution of the mammalian PI3K/PTEN/Akt pathway in yeast. Biochem. J. 2005;390:613–623. doi: 10.1042/BJ20050574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cid V.J., Rodríguez-Escudero I., Andrés-Pons A., Romá-Mateo C., Gil A., den Hertog J., Molina M., Pulido R. Assessment of PTEN tumor suppressor activity in nonmammalian models: the year of the yeast. Oncogene. 2008;27:5431–5442. doi: 10.1038/onc.2008.240. [DOI] [PubMed] [Google Scholar]
- 31.Rodrı I., Gil A., Blanco A., Vega A., Cid J., Valiente M., Torres J., Ripoll F., Cervera J., Pulido R. In vivo functional analysis of the counterbalance of hyperactive phosphatidylinositol 3-kinase p110 catalytic oncoproteins. Cancer Res. 2007;67:9731–9739. doi: 10.1158/0008-5472.CAN-07-1278. [DOI] [PubMed] [Google Scholar]
- 32.Melnikov A., Rogov P., Wang L., Gnirke A., Mikkelsen T.S. Comprehensive mutational scanning of a kinase in vivo reveals substrate-dependent fitness landscapes. Nucleic Acids Res. 2014;42:e112. doi: 10.1093/nar/gku511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang Y., Werling U., Edelmann W. SLiCE: a novel bacterial cell extract-based DNA cloning method. Nucleic Acids Res. 2012;40:e55. doi: 10.1093/nar/gkr1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang Y., Werling U., Edelmann W. Seamless Ligation Cloning Extract (SLiCE) cloning method. Methods Mol. Biol. 2014;1116:235–244. doi: 10.1007/978-1-62703-764-8_16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gietz R.D., Schiestl R.H. High-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat. Protoc. 2007;2:31–34. doi: 10.1038/nprot.2007.13. [DOI] [PubMed] [Google Scholar]
- 36.Zhang J., Kobert K., Flouri T., Stamatakis A. PEAR: a fast and accurate Illumina Paired-End reAd mergeR. Bioinformatics. 2014;30:614–620. doi: 10.1093/bioinformatics/btt593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. 2011;17:10–12. [Google Scholar]
- 38.Rubin A.F., Gelman H., Lucas N., Bajjalieh S.M., Papenfuss A.T., Speed T.P., Fowler D.M. A statistical framework for analyzing deep mutational scanning data. Genome Biol. 2017;18:150. doi: 10.1186/s13059-017-1272-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Abrahams B.S., Arking D.E., Campbell D.B., Mefford H.C., Morrow E.M., Weiss L.A., Menashe I., Wadkins T., Banerjee-Basu S., Packer A. SFARI Gene 2.0: a community-driven knowledgebase for the autism spectrum disorders (ASDs) Mol. Autism. 2013;4:36. doi: 10.1186/2040-2392-4-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ashkenazy H., Abadi S., Martz E., Chay O., Mayrose I., Pupko T., Ben-Tal N. ConSurf 2016: an improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Res. 2016;44(W1) doi: 10.1093/nar/gkw408. W344-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fraczkiewicz R., Braun W. Exact and efficient analytical calculation of the accessible surface areas and their gradients for macromolecules. J. Comput. Chem. 1998;19:319–333. [Google Scholar]
- 42.Heinig M., Frishman D. STRIDE: a web server for secondary structure assignment from known atomic coordinates of proteins. Nucleic Acids Res. 2004;32 doi: 10.1093/nar/gkh429. W500-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee J.O., Yang H., Georgescu M.M., Di Cristofano A., Maehama T., Shi Y., Dixon J.E., Pandolfi P., Pavletich N.P. Crystal structure of the PTEN tumor suppressor: implications for its phosphoinositide phosphatase activity and membrane association. Cell. 1999;99:323–334. doi: 10.1016/s0092-8674(00)81663-3. [DOI] [PubMed] [Google Scholar]
- 44.Forbes S.A., Beare D., Boutselakis H., Bamford S., Bindal N., Tate J., Cole C.G., Ward S., Dawson E., Ponting L. COSMIC: somatic cancer genetics at high-resolution. Nucleic Acids Res. 2017;45(D1):D777–D783. doi: 10.1093/nar/gkw1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xiao Y., Yeong Chit Chia J., Gajewski J.E., Sio Seng Lio D., Mulhern T.D., Zhu H.-J., Nandurkar H., Cheng H.-C. PTEN catalysis of phospholipid dephosphorylation reaction follows a two-step mechanism in which the conserved aspartate-92 does not function as the general acid--mechanistic analysis of a familial Cowden disease-associated PTEN mutation. Cell. Signal. 2007;19:1434–1445. doi: 10.1016/j.cellsig.2007.01.021. [DOI] [PubMed] [Google Scholar]
- 46.Myers M.P., Pass I., Batty I.H., Van der Kaay J., Stolarov J.P., Hemmings B.A., Wigler M.H., Downes C.P., Tonks N.K. The lipid phosphatase activity of PTEN is critical for its tumor supressor function. Proc. Natl. Acad. Sci. USA. 1998;95:13513–13518. doi: 10.1073/pnas.95.23.13513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Das S., Dixon J.E., Cho W. Membrane-binding and activation mechanism of PTEN. Proc. Natl. Acad. Sci. USA. 2003;100:7491–7496. doi: 10.1073/pnas.0932835100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Campbell R.B., Liu F., Ross A.H. Allosteric activation of PTEN phosphatase by phosphatidylinositol 4,5-bisphosphate. J. Biol. Chem. 2003;278:33617–33620. doi: 10.1074/jbc.C300296200. [DOI] [PubMed] [Google Scholar]
- 49.Wei Y., Stec B., Redfield A.G., Weerapana E., Roberts M.F. Phospholipid binding sites of PTEN: exploring the mechanism of PIP 2 activation. J. Biol. Chem. 2014;290:1592–1606. doi: 10.1074/jbc.M114.588590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gil A., Rodríguez-Escudero I., Stumpf M., Molina M., Cid V.J., Pulido R. A functional dissection of PTEN N-terminus: implications in PTEN subcellular targeting and tumor suppressor activity. PLoS ONE. 2015;10:e0119287. doi: 10.1371/journal.pone.0119287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vazquez F., Ramaswamy S., Nakamura N., Sellers W.R. Phosphorylation of the PTEN tail regulates protein stability and function. Mol. Cell. Biol. 2000;20:5010–5018. doi: 10.1128/mcb.20.14.5010-5018.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Weile J., Sun S., Cote A.G., Knapp J., Verby M., Mellor J.C., Wu Y., Pons C., Wong C., van Lieshout N. A framework for exhaustively mapping functional missense variants. Mol. Syst. Biol. 2017;13:957. doi: 10.15252/msb.20177908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gray V.E., Hause R.J., Fowler D.M. Analysis of large-scale mutagenesis data to assess the impact of single amino acid substitutions. Genetics. 2017;207:53–61. doi: 10.1534/genetics.117.300064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Landrum M.J., Lee J.M., Benson M., Brown G., Chao C., Chitipiralla S., Gu B., Hart J., Hoffman D., Hoover J. ClinVar: public archive of interpretations of clinically relevant variants. Nucleic Acids Res. 2016;44(D1):D862–D868. doi: 10.1093/nar/gkv1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chakravarty D., Gao J., Phillips S.M., Kundra R., Zhang H., Wang J., Rudolph J.E., Yaeger R., Soumerai T., Nissan M.H. OncoKB: A Precision Oncology Knowledge Base. JCO Precis Oncol. 2017;2017:1–16. doi: 10.1200/PO.17.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kitzman J.O., Starita L.M., Lo R.S., Fields S., Shendure J. Massively parallel single-amino-acid mutagenesis. Nat. Methods. 2015;12:203–206. doi: 10.1038/nmeth.3223. 4, 206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Findlay G.M., Boyle E.A., Hause R.J., Klein J.C., Shendure J. Saturation editing of genomic regions by multiplex homology-directed repair. Nature. 2014;513:120–123. doi: 10.1038/nature13695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fowler D.M., Araya C.L., Fleishman S.J., Kellogg E.H., Stephany J.J., Baker D., Fields S. High-resolution mapping of protein sequence-function relationships. Nat. Methods. 2010;7:741–746. doi: 10.1038/nmeth.1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Matreyek K.A., Starita L.M., Stephany J.J., Martin B., Chiasson M.A., Gray V.E., Kircher M., Khechaduri A., Dines J.N., Hause R.J. Multiplex assessment of protein variant abundance by massively parallel sequencing. bioRxiv. 2018 doi: 10.1038/s41588-018-0122-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Majithia A.R., Tsuda B., Agostini M., Gnanapradeepan K., Rice R., Peloso G., Patel K.A., Zhang X., Broekema M.F., Patterson N., UK Monogenic Diabetes Consortium. Myocardial Infarction Genetics Consortium. UK Congenital Lipodystrophy Consortium Prospective functional classification of all possible missense variants in PPARG. Nat. Genet. 2016;48:1570–1575. doi: 10.1038/ng.3700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Oliver M.D., Fernández-Acero T., Luna S., Rodríguez-Escudero I., Molina M., Pulido R., Cid V.J. Insights into the pathological mechanisms of p85α mutations using a yeast-based phosphatidylinositol 3-kinase model. Biosci. Rep. 2017;37:37. doi: 10.1042/BSR20160258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rodríguez-Escudero I., Andrés-Pons A., Pulido R., Molina M., Cid V.J. Phosphatidylinositol 3-kinase-dependent activation of mammalian protein kinase B/Akt in Saccharomyces cerevisiae, an in vivo model for the functional study of Akt mutations. J. Biol. Chem. 2009;284:13373–13383. doi: 10.1074/jbc.M807867200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fernández-Acero T., Rodríguez-Escudero I., Vicente F., Monteiro M.C., Tormo J.R., Cantizani J., Molina M., Cid V.J. A yeast-based in vivo bioassay to screen for class I phosphatidylinositol 3-kinase specific inhibitors. J. Biomol. Screen. 2012;17:1018–1029. doi: 10.1177/1087057112450051. [DOI] [PubMed] [Google Scholar]
- 64.Henikoff S., Henikoff J.G. Amino acid substitution matrices from protein blocks. Proc. Natl. Acad. Sci. USA. 1992;89:10915–10919. doi: 10.1073/pnas.89.22.10915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Papa A., Wan L., Bonora M., Salmena L., Song M.S., Hobbs R.M., Lunardi A., Webster K., Ng C., Newton R.H. Cancer-associated PTEN mutants act in a dominant-negative manner to suppress PTEN protein function. Cell. 2014;157:595–610. doi: 10.1016/j.cell.2014.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Johnston S.B., Raines R.T. Conformational stability and catalytic activity of PTEN variants linked to cancers and autism spectrum disorders. Biochemistry. 2015;54:1576–1582. doi: 10.1021/acs.biochem.5b00028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Redfern R.E., Daou M.-C., Li L., Munson M., Gericke A., Ross A.H. A mutant form of PTEN linked to autism. Protein Sci. 2010;19:1948–1956. doi: 10.1002/pro.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yang J.-M., Schiapparelli P., Nguyen H.-N., Igarashi A., Zhang Q., Abbadi S., Amzel L.M., Sesaki H., Quiñones-Hinojosa A., Iijima M. Characterization of PTEN mutations in brain cancer reveals that pten mono-ubiquitination promotes protein stability and nuclear localization. Oncogene. 2017;36:3673–3685. doi: 10.1038/onc.2016.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chia J.Y.-C., Gajewski J.E., Xiao Y., Zhu H.-J., Cheng H.-C. Unique biochemical properties of the protein tyrosine phosphatase activity of PTEN-demonstration of different active site structural requirements for phosphopeptide and phospholipid phosphatase activities of PTEN. Biochim. Biophys. Acta. 2010;1804:1785–1795. doi: 10.1016/j.bbapap.2010.05.009. [DOI] [PubMed] [Google Scholar]
- 70.Krumm N., Turner T.N., Baker C., Vives L., Mohajeri K., Witherspoon K., Raja A., Coe B.P., Stessman H.A., He Z.-X. Excess of rare, inherited truncating mutations in autism. Nat. Genet. 2015;47:582–588. doi: 10.1038/ng.3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Geiger T., Clarke S. Deamidation, isomerization, and racemization at asparaginyl and aspartyl residues in peptides. Succinimide-linked reactions that contribute to protein degradation. J. Biol. Chem. 1987;262:785–794. [PubMed] [Google Scholar]
- 72.Melamed D., Young D.L., Gamble C.E., Miller C.R., Fields S. Deep mutational scanning of an RRM domain of the Saccharomyces cerevisiae poly(A)-binding protein. RNA. 2013;19:1537–1551. doi: 10.1261/rna.040709.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gray V.E., Hause R.J., Luebeck J., Shendure J., Fowler D.M. Quantitative missense variant effect prediction using large-scale mutagenesis data. Cell Syst. 2018;6:116–124.e3. doi: 10.1016/j.cels.2017.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Shendure J., Fields S. Massively parallel genetics. Genetics. 2016;203:617–619. doi: 10.1534/genetics.115.180562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chen Z.H.H., Zhu M., Yang J., Liang H., He J., He S., Wang P., Kang X., McNutt M.A.A., Yin Y., Shen W.H. PTEN interacts with histone H1 and controls chromatin condensation. Cell Rep. 2014;8:2003–2014. doi: 10.1016/j.celrep.2014.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Liang H., Chen X., Yin Q., Ruan D., Zhao X., Zhang C., McNutt M.A., Yin Y. PTENβ is an alternatively translated isoform of PTEN that regulates rDNA transcription. Nat. Commun. 2017;8:14771. doi: 10.1038/ncomms14771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fricano-Kugler C.J., Getz S.A., Williams M.R., Zurawel A.A., DeSpenza T., Frazel P.W., Li M., O’Malley A.J., Moen E.L., Luikart B.W. Nuclear-excluded autism-associated PTEN mutations dysregulate neuronal growth. Biol. Psychiatry. 2017 doi: 10.1016/j.biopsych.2017.11.025. Published online December 2, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tan M.-H., Mester J.L., Ngeow J., Rybicki L.A., Orloff M.S., Eng C. Lifetime cancer risks in individuals with germline PTEN mutations. Clin. Cancer Res. 2012;18:400–407. doi: 10.1158/1078-0432.CCR-11-2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Feliciano P., Daniels A.M., Green Snyder L., Beaumont A., Camba A., Esler A., Gulsrud A.G., Mason A., Gutierrez A., Nicholson A. SPARK (Simons Foundation Powering Autism Research for Knowledge): a US cohort of 50,000 families to accelerate autism research. Neuron. 2018;97:488–493. doi: 10.1016/j.neuron.2018.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.