Abstract
Genome-wide association studies (GWASs) have identified more than 150 common genetic loci for breast cancer risk. However, the target genes and underlying mechanisms remain largely unknown. We conducted a cis-expression quantitative trait loci (cis-eQTL) analysis using normal or tumor breast transcriptome data from the Molecular Taxonomy of Breast Cancer International Consortium (METABRIC), The Cancer Genome Atlas (TCGA), and the Genotype-Tissue Expression (GTEx) project. We identified a total of 101 genes for 51 lead variants after combing the results of a meta-analysis of METABRIC and TCGA, and the results from GTEx at a Benjamini-Hochberg (BH)-adjusted p < 0.05. Using luciferase reporter assays in both estrogen-receptor positive (ER+) and negative (ER−) cell lines, we showed that alternative alleles of potential functional single-nucleotide polymorphisms (SNPs), rs11552449 (DCLRE1B), rs7257932 (SSBP4), rs3747479 (MRPS30), rs2236007 (PAX9), and rs73134739 (ATG10), could significantly change promoter activities of their target genes compared to reference alleles. Furthermore, we performed in vitro assays in breast cancer cell lines, and our results indicated that DCLRE1B, MRPS30, and ATG10 played a vital role in breast tumorigenesis via certain disruption of cell behaviors. Our findings revealed potential target genes for associations of genetic susceptibility risk loci and provided underlying mechanisms for a better understanding of the pathogenesis of breast cancer.
Keywords: breast cancer, GWAS, cis-eQTL, gene expression, risk variant, functional SNP, susceptibility gene, cell behavior, functional genomics, bioinformatics
Introduction
To date, genome-wide association studies (GWASs) have identified more than 150 genetic susceptibility loci associated with breast cancer risk.1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17 Approximately 90% of the single-nucleotide polymorphisms (SNPs) or variants initially identified by GWASs in these risk loci are located in intergenic, or non-coding, regions, and they are either not in linkage disequilibrium (LD) or have weak LD with coding variants. For the large majority of these risk variants, the mechanisms and biological relevance for their associations with breast cancer remain unclear. It is believed that most of these risk variants confer breast cancer pathogenesis by regulating the expression of genes, especially nearby genes.18, 19, 20, 21, 22 A recent study has shown that approximately 80% of the heritability of disease risk for 11 common diseases can be explained by variants in DNase I hypersensitivity sites, indicating that these variants, including the GWAS-identified risk variants, may play a regulatory role in gene expression.23
Large genomics data consortia, including the Molecular Taxonomy of Breast Cancer International Consortium (METABRIC), The Cancer Genome Atlas (TCGA), and the Genotype-Tissue Expression (GETx) project, have generated massive quantities of high-dimensional genomic data, including both matched genetic and transcriptome profiles from thousands of samples of breast cancer tumor tissue and normal tissue. These data provide an unprecedented opportunity for expression quantitative trait loci (eQTL) analysis, which evaluates the association of a variant genotype with gene expression levels measured in cells or tissues from individual subjects. Li et al. conducted a cis-eQTL analysis focused on 15 breast cancer index variants to identify potential nearby regulatory transcription factor (TF) targets.24 They subsequently expanded their cis-eQTL analysis to include risk loci for multiple cancer types using a subset of TCGA data.25 Recently, the GTEx project systematically identified thousands of eQTL target genes by evaluating the association between transcriptome variation and genome-wide variants across 43 types of normal tissues, including normal breast tissue from hundreds of individuals.26, 27 In another work, Castro and colleagues reported 36 TF regulons, described as a set of highly co-expressed genes regulated by potential TFs associated with breast cancer index variants, using variant and transcriptome data in breast tumor tissues from METABRIC.28 Most recently, Michailidou and colleagues reported 65 new breast cancer risk loci. They performed eQTL analysis using 458 tumor tissues from TCGA and 138 normal tissue samples from METABRIC.16 In addition to this large-scale analysis of index variants, over the past several years, we and other groups have conducted fine-mapping and cis-eQTL analyses to identify target genes in selected loci, including ESR1 (6q25),29, 30 IGFBP5 (2q35),31 FGFR2 (10q26),32, 33, 34 CCND1 (11q13),35 MAP3K1 (5q11),36 CASP8 (2q33),37 RCCD1 (15q26),2 TET2 (4q24),38 MYC (8q24),39 PTHLH (12p11),40 STXBP4 (17q22),41 HELQ (4q21),42 NRBF2 (10q21),43 and MRPS30 (5p12).44, 45.
While previous studies identified a large number of susceptibility gene candidates as described above, target genes for a large proportion of risk loci remain unknown. In addition, many candidate target genes were identified based on eQTL analysis at p < 0.05 in only one dataset; some false positive results can be ruled out only via independent replication using additional datasets. In particular, eQTL analysis has not been systematically performed to evaluate the associations of nearby genes and index variants using large-scale transcriptome data in tumor tissues from METABRIC. In the present study, we collected a total of 172 index variants for breast cancer risk at p < 5.0 × 10−8 from previous literature (Table S1). Using GWAS data from the Breast Cancer Association Consortium (BCAC), we identified a total of 159 breast cancer lead variants for these index variants, whereas they are not in LD (R2 < 0.1) (see Material and Methods, Table S1). We conducted a comprehensive cis-eQTL analysis of these variants to evaluate their associations with expression levels of nearby genes (1 Mb distance from the lead variant) in four transcriptome datasets from the METABRIC, TCGA, and GTEx project. Using luciferase reporter assays, we experimentally validated that alternative alleles of several functional SNPs could significantly change the promoter activities of target genes compared to their reference alleles. Using in vitro functional assays in breast cancer cell lines, our results further indicated that three candidate susceptibility genes play a vital role in breast tumorigenesis via certain disruption of cell behaviors. These findings provide additional insights into the understanding of regulatory mechanisms of genetic risk variants and genes for breast cancer development.
Material and Methods
Data Resources
We collected and characterized 172 index variants for breast cancer risk at p < 5.0 × 10−8 from previous literature (Table S1). We extracted 11,642 variants in strong LD with 172 index variants (R2 > 0.4). We retained any variants at p < 5.0 × 10−8 from the association results of the BCAC (122,977 breast cancer case subjects and 105,974 control subjects).16 If variants in the same locus were in LD (R2 > 0.1), only one variant with the best association was defined as the lead variant for the downstream analysis. In the end, we identified a total of 159 lead variants for the downstream analysis (Table S1).
We downloaded gene expression profiles generated by Illumina HT12 arrays in a total of 1,981 primary breast tumor tissues from Synapse (syn1757063) from the METABRIC project. The normalized gene expression and somatic copy alteration data were downloaded from the CbioPortal. The normalized gene expression has been described in a previous study.46 Genetic variant data, genotyped using array-based Affymetrix SNP 6.0 in a total of 1,992 samples, were downloaded from EBI (EGAD00010000164). A total of 1,895 tumor tissue samples with matched gene expression, somatic copy number alterations, and SNP data were included in our analysis.
For TCGA data, we downloaded RNA-seq V2 data (level 3), DNA methylation data, and somatic copy number alterations data from the CbioPortal. We also downloaded level 3 SNP data, genotyped using the Affymetrix SNP 6.0 array from TCGA’s data portal. A total of 536 tumor samples with matched gene expressions, DNA methylations, copy number alterations, and genetic variant data from European descendants were included. We also downloaded matched whole exome-seq and RNA-seq data in 494 tumor tissue samples from European descendants from the TCGA data portal.
We extracted cis-eQTL results for lead variants and nearby genes based on 251 normal breast tissues from the most recent GTEx database (v.7). We excluded results of long non-coding RNAs and ribosomal genes from our analysis. In total, we analyzed the association results for 147 variants (140 lead variants and 7 surrogate variants in strong LD, R2 > 0.8) and their nearby genes from the GTEx project. In addition, we also extracted significant cis-eQTL results for 72 lead variants and nearby genes at p < 0.05 based on 138 normal breast tissues from METABRIC from previous literature.16
Genotype Quality Control (QC)
We used the R package CRLMM to call the variant genotype for each probe from the original image array-based data in METABRIC.47 Only those probes of high quality, with intensity greater than 3,000 at a 95% calling rate, were included. From this METABRIC data and the level 3 TCGA data, genotype data of the nearby 1 Mb region for the 159 lead variants were extracted and then imputed with the 1000 Genomes Project data using Minimac.48 Only common variants (minor allele frequency > 0.05) with high imputation quality (R2 > 0.3) were included. We used a surrogate variant in strong LD (R2 > 0.8) instead of the lead variant if the lead variant failed to meet these criteria. In total, we included 147 variants (144 lead variants and three surrogate variants) from METABRIC and 155 variants (154 lead variants and one surrogate variant) from TCGA.
cis-eQTL Analysis
We used linear regression analysis to evaluate association between lead variants and expression levels of nearby genes (1 Mb distance to the lead variant). For the METABRIC and TCGA datasets, the normalized gene expression values were analyzed. To make the data conform better to the linear model for the eQTL analysis, we further transformed the gene expression levels across samples using an inverse normalizing transformation method. A full linear regression analysis was then performed to detect eQTLs, while adjusting for methylation and copy number alterations. For the METABRIC data, only copy number alterations were adjusted due to the lack of DNA methylation data in the tumor tissue samples. BH-adjusted p values were applied to determine final eQTL target genes.
Identification of Target Genes for Lead Variants
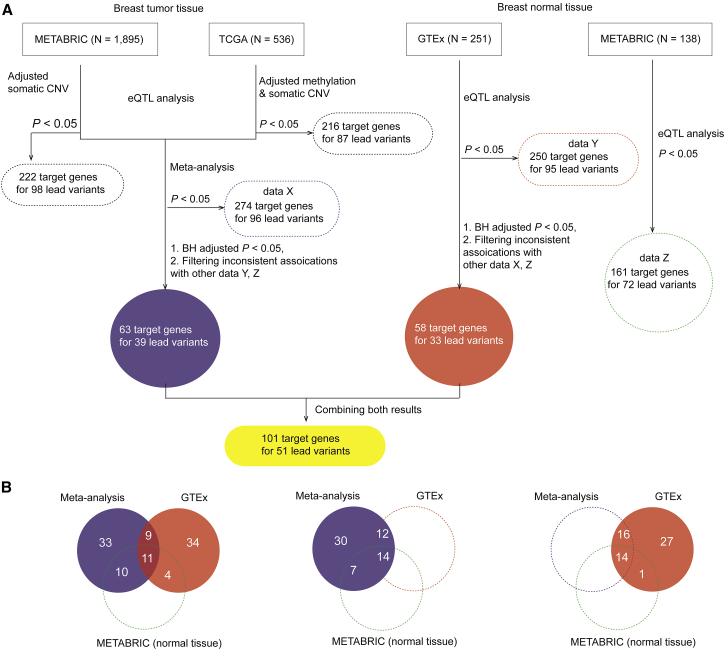
To increase the statistical power, we conducted a meta-analysis of eQTL results from tumor tissues from METABRIC and TCGA using the fixed-effects model.49 BH-adjusted p values were applied to determine eQTL target genes. In GTEx, we identified target genes using the same cutoff. In addition, we removed the target genes that had inconsistent associations in any other datasets at a less conservative unadjusted p < 0.05 (Figure 1). In the end, we identified final target genes for lead variants after combing the results from both meta-analysis and eQTL analysis of GTEx (Figure 1).
Figure 1.
A Workflow of Study Design
(A) A flow chart to illustrate the identification of target genes for GWAS lead variants based on cis-eQTL analysis using data from the METABRIC, TCGA, and GTEx datasets. The number in the dashed box indicates the total number of eQTL target genes that are identified by METABRIC, TCGA, GTEx, and meta-analysis at p < 0.05. The number in the highlighted blue and red box refers to the total of eQTL target genes that are identified using BH-adjusted p < 0.05 from a meta-analysis of tumor tissue results from METABRC and TCGA and normal tissues in GTEx, respectively. The number in the yellow box indicates the total number of genes after combing the results of the meta-analysis and the result in GTEx.
(B) The comparisons of the identified eQTL target genes with consistent associations across datasets (meta-analysis, GTEx, and normal tissues in METABRIC). From left to right, the number of the identified eQTL targets with consistent associations across the results from the meta-analysis and GTEx (BH-adjusted p < 0.05 for both) and normal tissue in METABRIC (unadjusted p < 0.05); from the meta-analysis (BH-adjusted p < 0.05), and GTEx and normal tissue in METABRIC (unadjusted p < 0.05 for both); from the meta-analysis (unadjusted p < 0.05), and GTEx (BH-adjusted p < 0.05) and normal tissue in METABRIC (unadjusted p < 0.05).
Pathway Enrichment Analysis
For the identified target genes, we examined their functional enrichment in the gene function category and biological pathways using the Ingenuity Pathway Analysis (IPA) tool. The most significant gene function categories and biological pathways were presented.
Functional Annotation
Functional annotation was performed using data from the Encyclopedia of DNA Elements (ENCODE) or the Roadmap Epigenomics Mapping Consortium (ROADMAP). We evaluated variants for potential functional significance using chromHMM annotation across nine ENCODE cell lines: HMEC, GM12878, H1-hESC, K562, HepG2, HSMM, HUVEC, NHEK, and NHLF.20 For each variant, we investigated whether or not it was mapped to functional regions (i.e., promoter or enhancer) using chromatin states annotation from the UCSC Genome Browser. The epigenetic signals of histone markers H3K4Me1, H3K4Me3, and H3K27Ac were also examined through layered histone tracks on all available ENCODE cell lines from the UCSC Genome Browser. DNase I hypersensitive sites were investigated in all available ENCODE cell lines and TF ChIP-seq datasets were also analyzed in a breast cancer cell line, MCF-7. Two publicly available tools, RegulomeDB50 and HaploReg v.3,51 were also used to evaluate the functional significance of each variant. The best candidate variant (searching from variants in LD with lead variants at R2 > 0.2) was prioritized and selected for functional assay, following the order of functional significance: TF binding, DNase Footprint, DNase peak, histone modification peak, and TF motif.
Chromatin-Chromatin Interaction Data Analysis
Experimentally derived chromatin interactions generated by 3C, 4C, 5C, Hi-C, and ChIA-PET were collected via 4DGenome.52 Hi-C data for MCF7 and MCF10A were obtained from published studies (GEO: GSE63525 and GSE66733).53, 54 ChIA-PET data for MCF7 (GEO: GSE39495) were downloaded from the ENCODE project. We directly extracted significant interactions processed by original studies. For raw interaction data available from the studies, we also processed and normalized the data by considering chromatin accessibility, nucleosome occupancy, alignability, and restriction site density. Significant interactions were identified using a ratio of observed to expected interactions more than the cutoff value, which was defined based on the background distribution.55 In addition, HDF5 interface rhdf5 (see Web Resources) was also used for processing the Hi-C data from the ENCODE project.
To analyze chromatin-chromatin interactions between the regions for functional variants in strong LD (R2 > 0.8) with lead variants and promoter regions of the identified candidate susceptibility genes, we examined ±250 bp nearby regions of functional variants and ±2 kb nearby regions of the gene transcription start site (TSS) (Figure S3).
Cell Culture and Transfection
Both estrogen-receptor-positive (ER+) MCF-7 and -negative (ER−) SK-BR3 cell lines were obtained from ATCC and cultured in Dulbecco’s Modified Eagle Medium (DMEM) (GIBCO 12430), supplemented with 10% fetal bovine serum (GIBCO 10099) and 1% penicillin/streptomycin (GIBCO 15140). Cells were maintained in a 37°C incubator with 5% CO2. Where appropriate, cells were plated into 6-well plates and transfected with 2 μg of pGL3-Basic/pGL3-Promoter vector constructions, along with 0.2 μg of pGL-TK plasmid using X-tremeGENE HP DNA Transfection Reagent (Roche 06365752001), according to the manufacturer’s instructions.
Plasmid Construction and Dual-Luciferase Reporter Assay
Luciferase reporter constructions for DCLRE1B, MRP30, and SSBP4 were generated by a polymerase chain reaction (PCR) using custom-designed primers, from which the genomic DNA were extracted from 293T cells (Table S2). The PCR products of these genes (at least 2 kb) were double-digested by the enzymes KpnI and HindIII (DCLRE1B), XhoI and HindIII (MRP30), and KpnI and HindIII (SSBP4) and then inserted into the pGL3-Basic vector. For the construction of the PAX9 and ATG10 expression vectors, the enhancer element containing the candidate functional SNP was introduced into the pGL3-Promoter vector by BamHI and Sall to construct PGL3-Promoter-PAX9 and PGL3-Promoter-ATG10. The promoter fragments of both genes were further subcloned into PGL3-Promoter-PAX9 and PGL3-Promoter-ATG10 by NheI and NcoI, and KpnI and NcoI, respectively (Table S2). All constructed target fragments were confirmed by sequencing. The minor allele of the individual SNP in the enhancer or promoter region for each gene construction was introduced into the plasmid using site-directed mutagenesis. For the DCLRE1B and MRPS30 expression vectors, the initiation codon ATG in the promoter regions of both genes were also mutated to TTG by the same procedure. We sequenced all constructed fragments to confirm variant incorporation. The dual-luciferase reporter assay was performed with the Dual-Luciferase Reporter Assay Kit (Promega E1910) following the manufacturer’s instructions. Briefly, both ER+ MCF-7 and ER− SK-BR3 cells transfected with luciferase reporter plasmids and pGL-TK transfection control plasmid were collected 24 hr post-transfection and lysed in a 1× Passive Lysis Buffer at room temperature for 25 min; then, 20 μL of the cell lysate were transferred into each well of a 96-well plate. 100 μL of LAR II were dispensed into each well, and firefly luciferase activity was measured; then 100 μL of Stop & Glo Reagent were dispensed into each well to measure the Renilla luciferase activity. We normalized firefly luciferase activity to Renilla luciferase activity to correct the potential effects of transfection efficiency or cell lysate preparation. All reporter assays were performed in triplicate and repeated in three independent experiments.
Gene Knockdown and Overexpression Experiments
SK-BR3 and MCF-7 cells were transfected with DCREL1B and MRPS30, respectively, using specifically designed small interfering RNA (siRNAs) (purchased from the Shanghai Genepharma company), which used a lipofectamine RNAiMAX transfection reagent (Invitrogen). We performed quantitative real-time PCR (RT-PCR) to verify siRNA knockdown efficiencies 1.5 days after transfection. The ATG10 overexpression and control vectors which carry puromycin selection marker were purchased from Youbio biotechology company. The MDA-MB-231 cells were transfected with these two plasmids using lipofectamine 3000 reagent (Invitrogen) and were further selected at a specific concentration of puromycin. After several weeks of growth selection, colonies were transferred to single-well plates. The expanded colonies confirmed by qRT-PCR assay were used for downstream experiments.
Quantitative RT-PCR Experiment
Quantitative RT-PCR experiments were performed as described previously.56 In brief, total mRNA were extracted using RNA plus reagent (TaKaRa Cat No. 9109), according to the manual, and cDNA was synthesized using a PrimeScript RT reagent kit with gDNA eraser (TaKaRa Cat No. RR047A). The quantitative RT-PCR primers used are listed in Table S3. All the PCR amplifications were performed in triplicate and repeated in three independent experiments. The relative expression levels of mRNAs were normalized to the expression levels of GAPDH.
Cell Proliferation, Cell Cycle, and Colony Formation Assays
To investigate whether knockdown or overexpression affects cell proliferation ability, siRNA transfected, overexpression stable cell lines, and control cell lines were seeded into 6-well plates in triplicate. The cell proliferations of these cells was measured by a Cell Counting kit-8 (CCK8) assay at different time points during the day, according to manufacturer’s instructions. For cell cycle analysis assays, the control and knockdown cells were first treated with different concentrations of MMC (0, 0.1, and 0.4 μM) for 4 hr, and detached with trypsin and fixed using 70% ethanol. Cell cycle analysis was used to determine the cell stage of each individual cell by a PI flow cytometry assay with a cell cycle analysis kit (from the Beyotime Institute of Biotechnology) according to the manufacturer’s instructions. The cells were trypsinized and plated into 6-well plates at a density of 200, 400, 800, or 1,000 cells per well according to cell types. 10–15 days later, these wells were washed with PBS three times and fixed with 4% paraformaldehyde for 30 min at room temperature, and stained with Coomassie Blue (Beyotime Biotechnology, Cat No. P0017B). Clones containing at least 50 cells were counted as one colony.
Allele-Specific Expression (ASE) Analysis using TCGA Data
We performed an ASE analysis for two targets, SSBP4 (rs7258465) and ZNF404 (rs1685191), which were selected based on the relative LD (R2 > 0.2) between exonic SNPs and lead SNPs, as well as top eQTL association signals. To measure ASE for each of both genes, we first determined an exonic SNP for each, located in a coding region of the target gene and in LD with the lead SNP, based on the European population in the 1000 Genomes project. We then extracted mapped reads for each exonic SNP using both whole exome-seq and RNA-seq data in 494 tumor tissue samples from European descendants in TCGA data portal. The total number of exome-seq and RNA-seq mapped reads were computed for reference and alternative alleles of the exonic SNPs using samtools and bcftools tools.57 We analyzed only samples containing heterozygous alleles of a exonic SNP: (1) at least 20 genomic DNA reads mapped to the surrogate SNP position, and (2) the ratio of mapped reads for reference and alternative alleles ranging from 0.2 to 0.8. The measurement of ASE difference for each exonic SNP in each sample was calculated using the differential expression ratio between reference and alternative allele. The significance of the ASE difference for overall samples was generated by a binominal test for the distribution of the ASE difference at p = 0.5.
Allele-Specific Expression Analysis using the Sequenom Technique
We performed experimental validation of ASE difference on two exonic SNPs, rs10405636 (SSBP4) and rs12977303 (ZNF404), using Sequenom MassARRAY in breast tumor adjacent and normal tissue samples in a cohort of 235 breast cancer patients, which were recruited as part of the Shanghai Breast Cancer Study.58 Total RNA was extracted from tissue specimens by homogenization in TRIzol solution (Invitrogen), phase separation, precipitation, and washing, following the manufacturer’s instructions. The quality and quantity of RNA was measured by spectrophotometric analysis. RNA was reverse-transcribed using a High Capacity cDNA Archive Kit (Applied Biosystems).29 To measure the ASE of each exonic SNP, we computed the ratio between reference and alternative allele abundance in cDNA in each of the adjacent normal breast tissues using the Sequenom allelotype genotyping approach. To filter samples possibly containing homozygous alleles of the exonic SNP, we included only those samples with ratios between 0.2 and 0.8. The significance of the ASE difference for overall samples was generated by a binominal test for evaluating the distribution of the ASE difference at p = 0.5.
Results
Identification of eQTL Target Genes in GWAS Risk Loci
To identify potential target genes for the 159 lead variants, we evaluated associations between lead variants and expression levels of nearby genes (±1 Mb distance) using four large-scale genetic and transcriptome datasets, including breast cancer tumor (n = 1,895) and normal tissue samples (n = 138) from METABRIC, tumor tissue samples from TCGA (n = 536), and normal tissue samples from the GTEx project (n = 251). The cis-eQTL analysis revealed hundreds of target genes detected at an unadjusted p < 0.05 significance threshold: 222 target genes for 98 lead variants and 161 target genes for 72 lead variants in tumor and normal tissue samples, respectively, from METABRIC, 216 target genes for 87 lead variants from TCGA, and 250 target genes for 95 lead variants from the GTEx project (Table S4). We further performed a meta-analysis of eQTL results of tumor tissue samples from both TCGA and METABRIC (see Material and Methods). We identified a total of 63 genes for 39 lead variants using a Benjamini-Hochberg (BH)-adjusted p < 0.05, after removing three genes with inconsistent associations in other datasets at p < 0.05 (Figure 1A). In GTEx, we identified a total of 58 target genes for 33 lead variants using the same criteria, after removing four genes with inconsistent associations (Figure 1A). In the end, we identified 101 target genes for 51 lead variants after we merged the results from both meta-analysis and eQTL analysis of GTEx (Figure 1A; Table S4). Of these 101 genes, consistent associations for a total of 44 genes (43.6%) were detected in at least one other dataset at p < 0.05 (Figure 1B). In particular, consistent associations for 20 genes (APOBEC3A, APOBEC3B, ARL17A, ATG10, ATP6AP1L, BBS2, BTN3A2, CTSW, FAM114A1, HAPLN4, L3MBTL3, LRRC37A, LRRC37A2, LRRC37A4P, MRPS30, OR2A7, PPM1K, SSBP4, SURF1, and ZNF404) were detected in both meta-analysis and GTEx (Figure 1B). We also confirmed a total of 41 previously reported genes and SNPs with consistent associations, including ZNF155, OR2A7,59 MRPS30,44, 45 FGF10,44 DCLRE1B,11 and others16 (Table S4).
An enrichment analysis in diseases and disorders using Ingenuity Pathway Analysis (IPA) revealed that these genes were the most significantly enriched in the cancer function category (p = 4.4 × 10−3); a total of eight genes, including WNT3, CASP8, ESR1, AKT1, POLR2L, FGF10, IGFBP5, and MUTYH, are well known to be involved in carcinogenesis and have been characterized accordingly by IPA. A functional enrichment analysis using IPA revealed that the top five significantly enriched networks were cell signaling, post-translational modification, protein synthesis, cell death and survival, and carbohydrate metabolism (p = 4.3 × 10−3).
Chromatin-Chromatin Interaction Analysis of Target Genes and Functional Variants
For the 51 lead variants identified as being associated with the 101 target genes, we performed extensive functional annotation in order to identify candidate functional variants (see Material and Methods). We evaluated and annotated the functional potential for a total of 1,184 variants in strong LD with the lead variants in the European population from the 1000 Genomes project (R2 > 0.8). Of 1,184 variants, 336 and 538 showed evidence of promoter and enhancer activities, respectively, with the epigenomic signals either in the ENCODE or the Roadmap project, based on the annotation of the HaploReg database.51 In particular, 146 and 49 showed evidence of promoter and enhancer activities in breast cancer cells, respectively (Table S5).
To directly search for evidence of regulatory variants associated with target genes identified from our eQTL analysis, we examined whether the above functional variants are located in the promoter or enhancer regions of these targets. We found that a total of 26 target genes are the nearest genes to the functional variants, which are located in the promoter or enhancer regions (Table S6). To further examine whether other target genes could interact with the functional variants via long distance cis-regulations, we analyzed the chromatin interaction data that were generated from multiple breast cancer and normal cells, including HMEC, MCF-7, and MCF-10 (see Material and Methods). We found that the additional 27 genes showed evidence of chromatin interactions between their promoter regions and functional variants (Table S6). Together, 53 (52.5%) of 101 target genes showed evidence of cis-regulation via promoter or enhancer-promoter interactions (Table S6).
Luciferase Reporter Assays for Functional SNPs and Target Genes
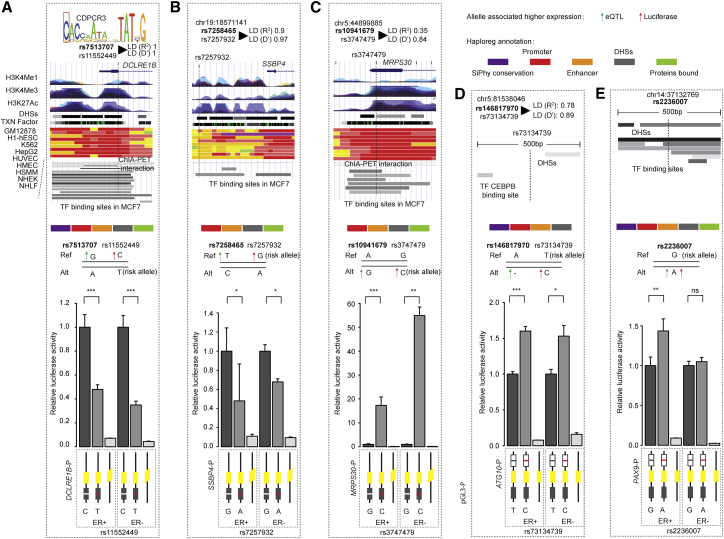
To further explore the regulatory mechanism of lead variants associated with their target genes, we focused on candidate functional variants that are located in the promoter or enhancer regions of their closest target genes. We selected the top five candidate functional SNPs for target genes, including rs11552449 (surrogate for lead SNP rs7513707; DCLRE1B), rs7257932 (surrogate for lead SNP rs7258465; SSBP4), rs3747479 (surrogate for lead SNP rs10941679; MRPS30), rs73134739 (surrogate for lead SNP rs146817970; ATG10), and rs35712350 (surrogate for lead SNP rs332529; ARRDC3) (see Material and Methods; Figure 2). Additionally, we selected one additional candidate functional SNP rs2236007 (PAX9), because this gene showed marginal association in meta-analysis with BH-adjusted p < 0.06 and p < 0.01 in GTEx. We next conducted luciferase reporter assays for these SNPs in both ER+ MCF-7 and ER− SK-BR3 breast cancer cell lines by introducing a fragment of the region containing functional SNPs into the luciferase reporter plasmids (see Material and Methods). Our results suggest that the fragment containing the alternative alleles significantly decreased the promoter activity of DCLRE1B and SSBP4 compared to the reference alleles in both cell lines (Figures 2A and 2B). For MRPS30 and ATG10, the fragment containing the alternative alleles significantly increased the promoter activity compared to the reference alleles in both cell lines (Figures 2C and 2D). For PAX9, the fragment containing the alternative allele significantly increased the promoter activity compared to the reference allele in ER+ MCF-7 cell lines. Although no statistical significance was observed in ER−, the same trend was observed (Figure 2E). These observations were all in line with the eQTL results described in the preceding section (Figures 1 and 2; Table S4). As an example, we observed that the reference allele C of rs11552449 was consistently associated with a higher expression level of its target DCLRE1B from the results of luciferase reporter assays and eQTL analysis when compared to alternative allele T (Figure 2A; Table S4). However, when investigating the promoter activities of ARRDC3, we failed to detect a significant difference between alternative and reference alleles (Figure S1).
Figure 2.
Alternative Alleles Affecting Target Genes’ Promoter Activity
Alternative allele of functional SNPs rs11552449 (surrogate for rs7513707) (DCLRE1B), rs7257932 (surrogate for rs7258465) (SSBP4), rs3747479 (surrogate for rs10941679) (MRPS30), rs73134739 (surrogate for rs146817970) (ATG10), and rs2236007 (PAX9) affecting promoter activities of target genes. From left to right: rs11552449 (DCLRE1B) (A), rs7257932 (SSBP4) (B), rs3747479 (MRPS30) (C), - rs73134739 (ATG10) (D) and rs2236007 (PAX9) (E). At the top of each panel: the epigenetic landscape of functional SNPs. From top to bottom, functional SNP in LD mapped to TF motif (if exists); LD R2 value between lead SNP (bold) and functional SNP in European population; RefSeq genes; layered H3K4Me1, H3K4Me3, and H3K27Ac histone modifications; DNase clusters; clustered ChIP-seq binding sites; annotation using chromatin states on the ENCODE cell lines; ChIA-PET interactions in MCF-7 cell; and TF binding sites. The signals of different layered histone modifications from the same ENCODE cell line are shown in the same color (the detailed color scheme for each ENCODE cell line is described in the UCSC Genome Browser). The red in chromatin states refers to active promoter. For the ChIA-PET track, black lines represent interactions with the promoter region, and gray lines represent chromatin interactions that do not involve the promoter region. The corresponding location of the variant is indicated by a dashed line. The HaploReg annotation51 for each functional variant is indicated in the top right panel. At the center of each panel (gray shadow box): the allele associated with higher gene expression is indicated by the upward pointing green (eQTL analysis) or red (luciferase reporter assay) arrow. At the bottom of each panel: the alternative allele of functional SNPs changing promoter activities using luciferase reporter assays in the ER+ MCF-7 and ER− SK-BR3 breast cancer cell lines. The fragment containing the reference allele of each SNP was cloned downstream for luciferase construct and an alternative allele was engineered into it. The alternative and reference alleles are indicated by the red and white lines, respectively. The error bars represent the standard deviation of promoter activities of target genes. A paired t test was performed to derive p value for each candidate functional SNP.
In Vitro Functional Assays for DCLRE1B, MRPS30, and ATG10
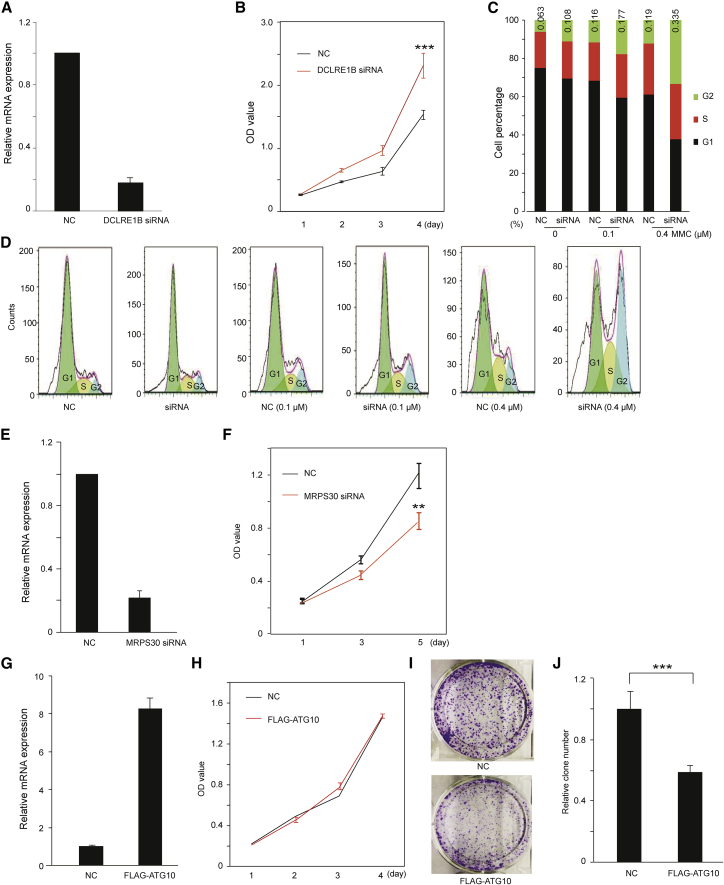
We performed in vitro functional assays in different breast cancer cell lines to investigate the biological function of the identified candidate susceptibility genes, including candidate tumor suppressors DCLRE1B and ATG10 and oncogene MRPS30, inferred from GWAS and eQTL results. Quantitative RT-PCR experiments were conducted to compare relative expression levels in three breast cancer cell lines: MCF-7, SK-BR3, and MDA-MB-231. All three genes exhibited the highest expression levels in MCF-7. DCLRE1B showed high expression in SK-BR3 cells, and ATG10 showed the lowest expression levels in MDA-MB-231 (Figure S2). Based on their relative expression levels in each cell line, we performed functional assays by knocking down genes in the cell line where the target gene was highly expressed or by overexpressing the gene in the cell line where the target gene was low expressed. Specifically, we designed the knockdown experiments for DCLRE1B in SK-BR3 and MRPS30 in MCF-7 cells via short interfering RNA (siRNA) and the overexpressed experiments for ATG10 in MDA-MB-23 by constructing a stable cell line (see Material and Methods).
Using quantitative RT-PCR in the SK-BR3 cells, we verified the high knockdown efficiency of DCLRE1B with approximately 80% of silencing endogenous transcripts (Figure 3A). The knockdown of DCLRE1B significantly increased breast cancer cell proliferation in the knockdown cells compared to the control cells (Figure 3B). In particular, we observed more significant knockdown cells stalled in G2 phase when they were treated with a higher concentration of mitomycin c (MMC), a reagent which induces cell cycle G2 arrest, by propidium iodide flow cytometry. For example, there is an approximate 2-fold increase in G2 cells for the DCLRE1B knockdown cells when treated with 0.4 μM mitomycin c (Figures 3C and 3D). Using quantitative RT-PCR in the MCF-7 cells, we also verified the high knockdown efficiency of MRPS30 with approximately 80% of silencing endogenous transcripts (Figure 3E). The knockdown of MRPS30 can significantly decrease cell viability in the knockdown cells compared to the control cells (Figure 3F).
Figure 3.
In Vitro Functional Assays for DCLRE1B, MRPS30, and ATG10
(A) Quantification of DCLRE1B knockdown efficiency in the SK-BR3 breast cancer cells using quantitative RT-PCR. The mRNA levels in both knockdown and control cells were measured in technical triplicates. “NC” refers to a normal control cell line with transfected control siRNA (A–F).
(B) After SK-BR3 cells were transfected with siRNA, cell viability assays were conducted in the control and knockdown cells using CCK8 assay at these time points: Days 1–4. “OD” refers to optical density, as measured by the assay.
(C) Cell cycle assays were performed in the control and knockdown cells treated with different concentrations of MMC by PI flow cytometry. Colors green, red, and black represent the cell stages of G2, S, and G1 cells, respectively.
(D) Distribution of relative cell percentage of G1, S, and G2 cells. Colors light green, light yellow, and light blue represent the relative percentage of G1, S, and G2 cells, respectively.
(E) Quantification of MRPS30 knockdown efficiency in the MCF-7 breast cancer cells using quantitative RT-PCR. The mRNA levels in both knockdown and control cells were measured in technical triplicates.
(F) After MCF-7 cells were transfected with siRNA, cell viability assays were conducted in control and knockdown cells using CCK8 assay at these time points: Days 1, 3, and 5.
(G) The relative expression levels of ATG10 in the MDA-MB-23 stable cells and non-target control cells measured by quantification RT-PCR. The mRNA levels in both overexpression and control cells were measured in technical triplicates. “NC” refers to a normal control cell line with transfected an empty vector (G–J).
(H) After constructing stable cells for ATG10 using the MDA-MB-23 cell lines, cell viabilities for these cells and control cells were measured using CCK8 at time points: 1, 2, 3, 4 days.
(I and J) After constructing stable cells for ATG10 using the MDA-MB-23 cell lines, these cells and control cells were reseeded after 12 days for colony formation assay. The colonies for these cells and control cells were imaged (I) and quantification (J) of the numbers of colonies in MDA-MB-23 stable cells and control cells.
p values were determined by t test from the comparison of knockdown and control cells for each time point. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; the error bars represent the standard deviation of the measurements from multiple replicates.
For ATG10, we generated FLAG-ATG10 overexpression stable MDA-MB-23 cell lines (see Material and Methods). Quantitative RT-PCR results verified that the expression levels of ATG10 were significantly increased by approximately 7-fold in the stable MDA-MB-23 cells compared to the control cells (Figure 3G). The overexpression of ATG10 can significantly decrease cell colony formation efficiency in the stable MDA-MB-23 cells compared to the control cells, while no significant difference was observed in cell viability between them (Figures 3H–3J).
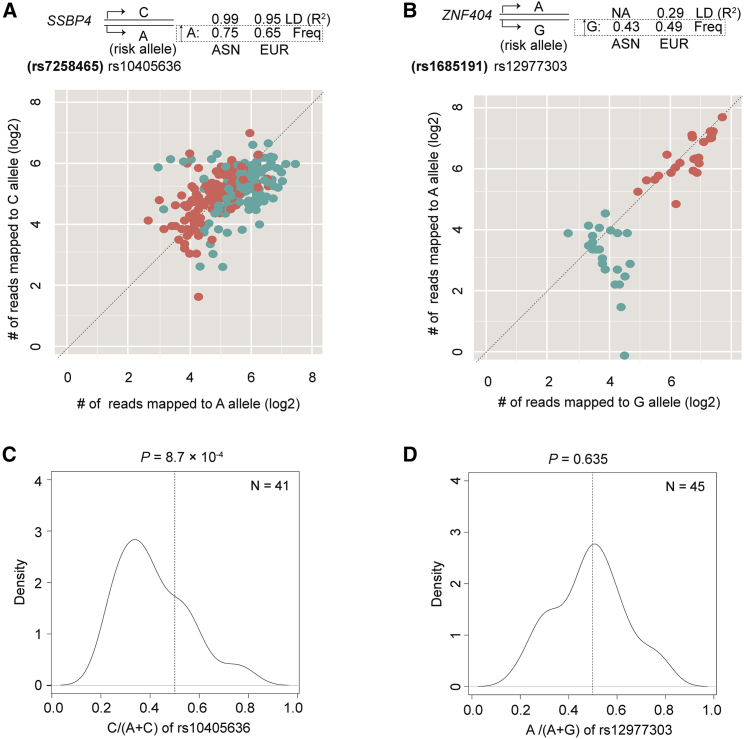
Allelic-Specific Expression Analysis for SSBP4 and ZNF404
We performed an ASE analysis on two selected genes, SSBP4 and ZNF404, via both computational and experimental validation (see Material and Methods). In the initial cis-eQTL analysis, the lead SNPs rs7258465 and rs1685191 were observed to be consistently associated with the expression levels of SSBP4 and ZNF404, respectively, in three datasets (Table S4). The alternative allele of rs4808801 was associated with decreased expressions of SSBP4, while the alternative allele rs1685191 was associated with an increased expression of ZNF404. We investigated the ASE for both genes using RNA-seq data in tumor tissue samples from 494 European descendants from TCGA (see Material and Methods). To measure the ASE for each gene, we searched for an exonic SNP in LD for each of the lead SNPs rs7258465 and rs1685191. The exonic SNPs rs10405636 (SSBP4; R2 = 0.95) and rs12977303 (ZNF404; LD R2 = 0.29) were identified as a surrogate for each gene (see Material and Methods). In accordance with our reported eQTL observations, we found that the SSBP4-expressed transcript was significantly biased to contain reference allele A relative to the alternative allele for rs10405636 (Figure 4A; binomial test p < 0.05 for both). For rs12977303, we observed that the ZNF404-expressed transcripts were significantly biased to contain the alternative allele A relative to reference allele G (Figure 4B; binomial test p < 0.05). As a background control, we did not observe the ASE of the exonic SNPs in these samples when analyzing the whole exome-seq data (Figures 4A and 4B). We further investigated the ASE for these exonic SNPs using the Sequenom allelotype technique in adjacent normal breast tissue samples from a cohort of 235 Chinese breast cancer patients (see Material and Methods). We identified a total of 41 samples containing heterozygous alleles for rs10405636, and 45 samples containing heterozygous alleles for rs12977303 (see Material and Methods). Consistent with the observation described previously, the SSBP4-expressed transcripts were significantly biased to contain the reference allele A for SNP rs10405636 (Figure 4C) (Wilcoxon test, p < 0.05 for both). No significant ASE was observed for SNP rs12977303 (ZNF404) (Figure 4D). Notably, this observation may be due to less LD between the lead SNP and surrogate SNP in the Asian population.
Figure 4.
Allelic-Specific Expression (ASE) Analysis for Two Targets, SSBP4 and ZNF404
(A and B) At the top of each panel, the exonic SNP rs10405636 (in LD with lead SNP rs7258465) and rs12977303 (in LD with rs1685191) were selected as surrogates for SSBP4 and ZNF404, respectively. The allele associated with higher expression is indicated by a black arrow based on the ASE analysis. Number of RNA-seq (blue) and exome-seq (red) mapped reads (log2 scale) containing alternative and reference alleles of (A) rs10405636 in SSBP4 and (B) rs12977303 in ZNF404 are plotted across tumor samples.
(C and D) Density plots indicate the fraction of expressed C allele relative to expression of both alleles of rs10405636 and A allele relative to expression f both alleles of rs12977303.
Discussion
Although a large number of genetic susceptibility loci have been identified for breast cancer risk, the mechanisms by which risk variants in these loci exert their functions remain largely unknown. In the present study, we comprehensively conducted a cis-eQTL analysis to identify target genes in these risk loci using four large-scale datasets, breast cancer tumor and normal tissue samples from METABRIC, tumor tissue samples from TCGA, and normal tissue samples from GTEx. Hundreds of associated genes were identified as potential targets at p < 0.05, including 101 genes identified with strong statistical evidence. We further selected potential functional variants for DCLRE1B, SSBP4, MRPS30, PAX9, and ATG10 for in vitro functional characterization using luciferase reporter assays in the ER+ MCF-7 and ER− SK-BR3 breast cancer cell lines and confirmed that alternative alleles of these SNPs could change the promoter activities of target genes. The above analyses provide strong evidence to identify candidate genes for further functional investigation in breast cancer cell behavior. In particular, our results indicate that DCLRE1B, MRPS30, and ATG10 play a vital role in breast tumorigenesis by their influence on basic cellular functions. The results from the eQTL analysis and in vitro experiments provide strong evidence for assessing the causality of the genes identified in our study. These findings provided additional insight into the genetic and biological basis for breast cancer development.
The reported target genes identified from eQTL analysis were further supported by additional evidence. For example, we observed that almost half of the identified target genes showed consistent association directions in at least two datasets. Meanwhile, more than half of candidate targets were found to be the nearest genes for functional variants in strong LD with lead variants, and some additional targets showed evidence of chromatin-chromatin interactions between their promoters and the regions where potential functional variants are located. Using luciferase reporter assays, we further experimentally confirmed that five functional variants could affect the promoter activities, providing direct evidence for potential regulatory mechanisms to link the variants to their target genes. We also evaluated two target genes by performing ASE analyses for both computational and experimental validation. All were replicated by ASE analyses using RNA-seq mapped reads in the TCGA data and one was further confirmed by ASE analyses of Sequenom allelotype genotyping of cDNA data in adjacent normal breast tissues. The advantage of using the ASE approach compared to eQTL analysis is that the effect of environmental or trans-acting factors on gene expression could be essentially eliminated by measuring expressed transcripts for each allele within the same sample.
Although previous studies have identified many candidate eQTL genes for lead variants, some of these candidate genes have been identified in limited sample size. In this study, we performed a metaanalysis of large-scale sample sizes with 1,895 samples from METABRIC and 536 samples from TCGA, which greatly improved the statistical power. Furthermore, we included eQTL results using data from 138 and 251 breast normal tissues from METABRIC and GTEx, respectively. Here, we reported candidate target genes with strong statistical evidence. Additionally, we analyzed 208 target genes for 86 lead variants with a significance level between unadjusted p < 0.05 and BH-adjusted p > 0.05 identified from a meta-analysis of eQTL results of tumor tissue from METABRIC and TCGA (Figure 1). Using functional data to examine variants in strong LD with the lead variants, we found that a total of 51 candidate genes (24.8%) showed evidence of cis-regulation via promoter or enhancer-promoter interactions. The results indicated that some target genes with less conservative p values may be dismissed due to our statistical cutoff.
Gene expression in tumor tissues could be influenced by both genetic and epigenetic variations or other somatic alterations. To account for a potential influence of these factors, we conducted a cis-eQTL analysis in TCGA data by adjusting methylation and somatic copy number alterations, following a previously reported approach.24 For the METABRIC data, no methylation data were available in the METABRIC samples, which prevented us from excluding the effect of this potential confounding factor. Notably, using data from TCGA, we performed eQTL analysis by adjusted methylation and somatic copy number alterations, and adjusted somatic copy number alterations only, respectively. We found a similar number of target genes were identified using the same statistical cutoff in both analyses, indicating that association detection is slightly affected without adjusted methylation (data not shown). The eQTL analysis could also be affected by other factors such as the effect size, data quality, experimental design, and tissue heterogeneity. For example, the eQTL results related to tumor subtypes (i.e., ESR1) may yield different results, depending on whether overall cancer cases or those of a particular tumor subtype are analyzed.16, 30 Notably, a particular lead variant may be a surrogate for multiple variants for breast cancer risk in the locus. The eQTL analysis for lead variants may not identify target genes for those surrogated variants when they are in weak LD, as was shown by a previous fine-mapping study.36 On the other hand, lead variants could be statistically excluded as candidate causative variants in some GWAS-identified loci. Nevertheless, the target genes identified based on the lead variants are still reliable, as most statistically causative variants are still expected to be in strong LD with them.
The identification of causal variants remains a challenge because many variants in strong LD are located in the same functional region. Identifying candidate target genes could be helpful for pinpointing functional variants for further in vitro functional experiments, as shown by the findings of the luciferase reporter assays for five functional variants. Many functional variants may not be located in the nearest eQTL genes, and it would be difficult to identify them. The findings from our study could provide data for designing in vitro functional assays (i.e., ChIA-PET and 5C chromatin interaction) to further explore the underlying mechanisms in breast cancer cell lines in the future. It should also be noted that we systematically annotated hundreds of potential functional variants for target genes. However, it is still a challenge to select variants for downstream functional assays to pinpoint the causative ones. In future studies, the dense genotype data from a fine-mapping study would be essential to statistically identify most likely causative variants. On the other hand, a massively parallel reporter assay (MPRA) approach may also be considered to simultaneously screen hundreds of variants for functional investigation.60
Our study suggests that a functional variant may affect multiple target genes, which is consistent with a previous observation.30 Some of the target genes of the same variant seem to share the same regulatory mechanism. As an example, a SNP rs720475 was found to be associated with one tandem duplication gene family, the OR2A (OR2A7 and OR2A20P) family.
In conclusion, we conducted a comprehensive cis-eQTL analysis for lead variants and nearby genes using data from four large-scale transcriptome datasets. We provided additional evidence that the associations of risk variants in many loci with breast cancer risk may be mediated through the regulation of eQTL target genes. Our study has discovered additional biological mechanisms for understanding genetic susceptibility risk loci and breast cancer risk and has provided additional insights into the genetic and biological basis for pathogenesis of this common cancer.
Acknowledgements
This work was supported in part by research development funds from Vanderbilt University Medical Center, the US National Institutes of Health grant R01 CA148667, and the grant from the National Natural Science Foundation of China (31470776). The sample preparation and ASE assays were performed at the Survey and Biospecimen Shared Resource, which was supported in part by the Vanderbilt-Ingram Cancer Center (P30 CA068485). We thank the METABRIC, TCGA, GTEx, ENCODE, and Roadmap for providing valuable data resources for the research. We also thank Regina Courtney and Jie Wu for laboratory assistance, and Kim Kreth and Marshal Younger for assistance with editing and manuscript preparation. The data analyses were conducted using the Advanced Computing Center for Research and Education (ACCRE) at Vanderbilt University.
Published: May 3, 2018
Footnotes
Supplemental Data include three figures and six tables and can be found with this article online at https://doi.org/10.1016/j.ajhg.2018.03.016.
Web Resources
Bioconductor, https://www.bioconductor.org/packages/release/bioc/html/rhdf5.html
cBioPortal for Cancer Genomics, http://www.cbioportal.org/
ENCODE, https://www.encodeproject.org/
GTEx Portal, https://www.gtexportal.org/home/
Synapse, https://www.synapse.org/
TCGA Portal, https://cancergenome.nih.gov/
UCSC Genome Browser, https://genome.ucsc.edu
Supplemental Data
References
- 1.Antoniou A.C., Wang X., Fredericksen Z.S., McGuffog L., Tarrell R., Sinilnikova O.M., Healey S., Morrison J., Kartsonaki C., Lesnick T., EMBRACE. GEMO Study Collaborators. HEBON. kConFab. SWE-BRCA. MOD SQUAD. GENICA A locus on 19p13 modifies risk of breast cancer in BRCA1 mutation carriers and is associated with hormone receptor-negative breast cancer in the general population. Nat. Genet. 2010;42:885–892. doi: 10.1038/ng.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cai Q., Long J., Lu W., Qu S., Wen W., Kang D., Lee J.Y., Chen K., Shen H., Shen C.Y. Genome-wide association study identifies breast cancer risk variant at 10q21.2: results from the Asia Breast Cancer Consortium. Hum. Mol. Genet. 2011;20:4991–4999. doi: 10.1093/hmg/ddr405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cox A., Dunning A.M., Garcia-Closas M., Balasubramanian S., Reed M.W., Pooley K.A., Scollen S., Baynes C., Ponder B.A., Chanock S., Kathleen Cunningham Foundation Consortium for Research into Familial Breast Cancer. Breast Cancer Association Consortium A common coding variant in CASP8 is associated with breast cancer risk. Nat. Genet. 2007;39:352–358. doi: 10.1038/ng1981. [DOI] [PubMed] [Google Scholar]
- 4.Easton D.F., Pooley K.A., Dunning A.M., Pharoah P.D., Thompson D., Ballinger D.G., Struewing J.P., Morrison J., Field H., Luben R., SEARCH collaborators. kConFab. AOCS Management Group Genome-wide association study identifies novel breast cancer susceptibility loci. Nature. 2007;447:1087–1093. doi: 10.1038/nature05887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gold B., Kirchhoff T., Stefanov S., Lautenberger J., Viale A., Garber J., Friedman E., Narod S., Olshen A.B., Gregersen P. Genome-wide association study provides evidence for a breast cancer risk locus at 6q22.33. Proc. Natl. Acad. Sci. USA. 2008;105:4340–4345. doi: 10.1073/pnas.0800441105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hunter D.J., Kraft P., Jacobs K.B., Cox D.G., Yeager M., Hankinson S.E., Wacholder S., Wang Z., Welch R., Hutchinson A. A genome-wide association study identifies alleles in FGFR2 associated with risk of sporadic postmenopausal breast cancer. Nat. Genet. 2007;39:870–874. doi: 10.1038/ng2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Long J., Cai Q., Shu X.O., Qu S., Li C., Zheng Y., Gu K., Wang W., Xiang Y.B., Cheng J. Identification of a functional genetic variant at 16q12.1 for breast cancer risk: results from the Asia Breast Cancer Consortium. PLoS Genet. 2010;6:e1001002. doi: 10.1371/journal.pgen.1001002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Long J., Cai Q., Sung H., Shi J., Zhang B., Choi J.Y., Wen W., Delahanty R.J., Lu W., Gao Y.T. Genome-wide association study in east Asians identifies novel susceptibility loci for breast cancer. PLoS Genet. 2012;8:e1002532. doi: 10.1371/journal.pgen.1002532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Long J., Delahanty R.J., Li G., Gao Y.T., Lu W., Cai Q., Xiang Y.B., Li C., Ji B.T., Zheng Y. A common deletion in the APOBEC3 genes and breast cancer risk. J. Natl. Cancer Inst. 2013;105:573–579. doi: 10.1093/jnci/djt018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Michailidou K., Beesley J., Lindstrom S., Canisius S., Dennis J., Lush M.J., Maranian M.J., Bolla M.K., Wang Q., Shah M., BOCS. kConFab Investigators. AOCS Group. NBCS. GENICA Network Genome-wide association analysis of more than 120,000 individuals identifies 15 new susceptibility loci for breast cancer. Nat. Genet. 2015;47:373–380. doi: 10.1038/ng.3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Michailidou K., Hall P., Gonzalez-Neira A., Ghoussaini M., Dennis J., Milne R.L., Schmidt M.K., Chang-Claude J., Bojesen S.E., Bolla M.K., Breast and Ovarian Cancer Susceptibility Collaboration. Hereditary Breast and Ovarian Cancer Research Group Netherlands (HEBON) kConFab Investigators. Australian Ovarian Cancer Study Group. GENICA (Gene Environment Interaction and Breast Cancer in Germany) Network Large-scale genotyping identifies 41 new loci associated with breast cancer risk. Nat. Genet. 2013;45:353–361. doi: 10.1038/ng.2563. e1–e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Purrington K.S., Slager S., Eccles D., Yannoukakos D., Fasching P.A., Miron P., Carpenter J., Chang-Claude J., Martin N.G., Montgomery G.W., GENICA Network Genome-wide association study identifies 25 known breast cancer susceptibility loci as risk factors for triple-negative breast cancer. Carcinogenesis. 2014;35:1012–1019. doi: 10.1093/carcin/bgt404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shi J., Sung H., Zhang B., Lu W., Choi J.Y., Xiang Y.B., Kim M.K., Iwasaki M., Long J., Ji B.T. New breast cancer risk variant discovered at 10q25 in East Asian women. Cancer Epidemiol. Biomarkers Prev. 2013;22:1297–1303. doi: 10.1158/1055-9965.EPI-12-1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Siddiq A., Couch F.J., Chen G.K., Lindström S., Eccles D., Millikan R.C., Michailidou K., Stram D.O., Beckmann L., Rhie S.K., Australian Breast Cancer Tissue Bank Investigators. Familial Breast Cancer Study. GENICA Consortium A meta-analysis of genome-wide association studies of breast cancer identifies two novel susceptibility loci at 6q14 and 20q11. Hum. Mol. Genet. 2012;21:5373–5384. doi: 10.1093/hmg/dds381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Turnbull C., Ahmed S., Morrison J., Pernet D., Renwick A., Maranian M., Seal S., Ghoussaini M., Hines S., Healey C.S., Breast Cancer Susceptibility Collaboration (UK) Genome-wide association study identifies five new breast cancer susceptibility loci. Nat. Genet. 2010;42:504–507. doi: 10.1038/ng.586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Michailidou K., Lindström S., Dennis J., Beesley J., Hui S., Kar S., Lemaçon A., Soucy P., Glubb D., Rostamianfar A., NBCS Collaborators. ABCTB Investigators. ConFab/AOCS Investigators Association analysis identifies 65 new breast cancer risk loci. Nature. 2017;551:92–94. doi: 10.1038/nature24284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Milne R.L., Kuchenbaecker K.B., Michailidou K., Beesley J., Kar S., Lindström S., Hui S., Lemaçon A., Soucy P., Dennis J., ABCTB Investigators. EMBRACE. GEMO Study Collaborators. HEBON. kConFab/AOCS Investigators. NBSC Collaborators Identification of ten variants associated with risk of estrogen-receptor-negative breast cancer. Nat. Genet. 2017;49:1767–1778. doi: 10.1038/ng.3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pierce B.L., Tong L., Chen L.S., Rahaman R., Argos M., Jasmine F., Roy S., Paul-Brutus R., Westra H.J., Franke L. Mediation analysis demonstrates that trans-eQTLs are often explained by cis-mediation: a genome-wide analysis among 1,800 South Asians. PLoS Genet. 2014;10:e1004818. doi: 10.1371/journal.pgen.1004818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Innocenti F., Cooper G.M., Stanaway I.B., Gamazon E.R., Smith J.D., Mirkov S., Ramirez J., Liu W., Lin Y.S., Moloney C. Identification, replication, and functional fine-mapping of expression quantitative trait loci in primary human liver tissue. PLoS Genet. 2011;7:e1002078. doi: 10.1371/journal.pgen.1002078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nicolae D.L., Gamazon E., Zhang W., Duan S., Dolan M.E., Cox N.J. Trait-associated SNPs are more likely to be eQTLs: annotation to enhance discovery from GWAS. PLoS Genet. 2010;6:e1000888. doi: 10.1371/journal.pgen.1000888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ward L.D., Kellis M. Interpreting noncoding genetic variation in complex traits and human disease. Nat. Biotechnol. 2012;30:1095–1106. doi: 10.1038/nbt.2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maurano M.T., Humbert R., Rynes E., Thurman R.E., Haugen E., Wang H., Reynolds A.P., Sandstrom R., Qu H., Brody J. Systematic localization of common disease-associated variation in regulatory DNA. Science. 2012;337:1190–1195. doi: 10.1126/science.1222794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gusev A., Lee S.H., Trynka G., Finucane H., Vilhjálmsson B.J., Xu H., Zang C., Ripke S., Bulik-Sullivan B., Stahl E., Schizophrenia Working Group of the Psychiatric Genomics Consortium. SWE-SCZ Consortium. Schizophrenia Working Group of the Psychiatric Genomics Consortium. SWE-SCZ Consortium Partitioning heritability of regulatory and cell-type-specific variants across 11 common diseases. Am. J. Hum. Genet. 2014;95:535–552. doi: 10.1016/j.ajhg.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li Q., Seo J.H., Stranger B., McKenna A., Pe’er I., Laframboise T., Brown M., Tyekucheva S., Freedman M.L. Integrative eQTL-based analyses reveal the biology of breast cancer risk loci. Cell. 2013;152:633–641. doi: 10.1016/j.cell.2012.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li Q., Stram A., Chen C., Kar S., Gayther S., Pharoah P., Haiman C., Stranger B., Kraft P., Freedman M.L. Expression QTL-based analyses reveal candidate causal genes and loci across five tumor types. Hum. Mol. Genet. 2014;23:5294–5302. doi: 10.1093/hmg/ddu228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Melé M., Ferreira P.G., Reverter F., DeLuca D.S., Monlong J., Sammeth M., Young T.R., Goldmann J.M., Pervouchine D.D., Sullivan T.J., GTEx Consortium Human genomics. The human transcriptome across tissues and individuals. Science. 2015;348:660–665. doi: 10.1126/science.aaa0355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Consortium G.T., GTEx Consortium Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348:648–660. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Castro M.A., de Santiago I., Campbell T.M., Vaughn C., Hickey T.E., Ross E., Tilley W.D., Markowetz F., Ponder B.A., Meyer K.B. Regulators of genetic risk of breast cancer identified by integrative network analysis. Nat. Genet. 2016;48:12–21. doi: 10.1038/ng.3458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sun Y., Ye C., Guo X., Wen W., Long J., Gao Y.T., Shu X.O., Zheng W., Cai Q. Evaluation of potential regulatory function of breast cancer risk locus at 6q25.1. Carcinogenesis. 2016;37:163–168. doi: 10.1093/carcin/bgv170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dunning A.M., Michailidou K., Kuchenbaecker K.B., Thompson D., French J.D., Beesley J., Healey C.S., Kar S., Pooley K.A., Lopez-Knowles E., EMBRACE. GEMO Study Collaborators. HEBON. kConFab Investigators Breast cancer risk variants at 6q25 display different phenotype associations and regulate ESR1, RMND1 and CCDC170. Nat. Genet. 2016;48:374–386. doi: 10.1038/ng.3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ghoussaini M., Edwards S.L., Michailidou K., Nord S., Cowper-Sal Lari R., Desai K., Kar S., Hillman K.M., Kaufmann S., Glubb D.M., Australian Ovarian Cancer Management Group. Australian Ovarian Cancer Management Group Evidence that breast cancer risk at the 2q35 locus is mediated through IGFBP5 regulation. Nat. Commun. 2014;4:4999. doi: 10.1038/ncomms5999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meyer K.B., Maia A.T., O’Reilly M., Teschendorff A.E., Chin S.F., Caldas C., Ponder B.A. Allele-specific up-regulation of FGFR2 increases susceptibility to breast cancer. PLoS Biol. 2008;6:e108. doi: 10.1371/journal.pbio.0060108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meyer K.B., O’Reilly M., Michailidou K., Carlebur S., Edwards S.L., French J.D., Prathalingham R., Dennis J., Bolla M.K., Wang Q., GENICA Network. kConFab Investigators. Australian Ovarian Cancer Study Group Fine-scale mapping of the FGFR2 breast cancer risk locus: putative functional variants differentially bind FOXA1 and E2F1. Am. J. Hum. Genet. 2013;93:1046–1060. doi: 10.1016/j.ajhg.2013.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhu X., Asa S.L., Ezzat S. Histone-acetylated control of fibroblast growth factor receptor 2 intron 2 polymorphisms and isoform splicing in breast cancer. Mol. Endocrinol. 2009;23:1397–1405. doi: 10.1210/me.2009-0071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.French J.D., Ghoussaini M., Edwards S.L., Meyer K.B., Michailidou K., Ahmed S., Khan S., Maranian M.J., O’Reilly M., Hillman K.M., GENICA Network. kConFab Investigators Functional variants at the 11q13 risk locus for breast cancer regulate cyclin D1 expression through long-range enhancers. Am. J. Hum. Genet. 2013;92:489–503. doi: 10.1016/j.ajhg.2013.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Glubb D.M., Maranian M.J., Michailidou K., Pooley K.A., Meyer K.B., Kar S., Carlebur S., O’Reilly M., Betts J.A., Hillman K.M., GENICA Network. kConFab Investigators. Norwegian Breast Cancer Study Fine-scale mapping of the 5q11.2 breast cancer locus reveals at least three independent risk variants regulating MAP3K1. Am. J. Hum. Genet. 2015;96:5–20. doi: 10.1016/j.ajhg.2014.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lin W.Y., Camp N.J., Ghoussaini M., Beesley J., Michailidou K., Hopper J.L., Apicella C., Southey M.C., Stone J., Schmidt M.K., GENICA Network. kConFab Investigators. Australian Ovarian Cancer Study Group. Breast and Ovarian Cancer Susceptibility (BOCS) Study Identification and characterization of novel associations in the CASP8/ALS2CR12 region on chromosome 2 with breast cancer risk. Hum. Mol. Genet. 2015;24:285–298. doi: 10.1093/hmg/ddu431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guo X., Long J., Zeng C., Michailidou K., Ghoussaini M., Bolla M.K., Wang Q., Milne R.L., Shu X.O., Cai Q., kConFab Investigators Fine-scale mapping of the 4q24 locus identifies two independent loci associated with breast cancer risk. Cancer Epidemiol. Biomarkers Prev. 2015;24:1680–1691. doi: 10.1158/1055-9965.EPI-15-0363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shi J., Zhang Y., Zheng W., Michailidou K., Ghoussaini M., Bolla M.K., Wang Q., Dennis J., Lush M., Milne R.L., Mervi Grip. kConFab Investigators Fine-scale mapping of 8q24 locus identifies multiple independent risk variants for breast cancer. Int. J. Cancer. 2016;139:1303–1317. doi: 10.1002/ijc.30150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zeng C., Guo X., Long J., Kuchenbaecker K.B., Droit A., Michailidou K., Ghoussaini M., Kar S., Freeman A., Hopper J.L., EMBRACE. behalf of GEMO Study Collaborators. HEBON. KConFab. AOCS Investigators Identification of independent association signals and putative functional variants for breast cancer risk through fine-scale mapping of the 12p11 locus. Breast Cancer Res. 2016;18:64. doi: 10.1186/s13058-016-0718-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wright F.A., Sullivan P.F., Brooks A.I., Zou F., Sun W., Xia K., Madar V., Jansen R., Chung W., Zhou Y.H. Heritability and genomics of gene expression in peripheral blood. Nat. Genet. 2014;46:430–437. doi: 10.1038/ng.2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hamdi Y., Soucy P., Adoue V., Michailidou K., Canisius S., Lemaçon A., Droit A., Andrulis I.L., Anton-Culver H., Arndt V., NBCS Collaborators. kConFab/AOCS Investigators Association of breast cancer risk with genetic variants showing differential allelic expression: Identification of a novel breast cancer susceptibility locus at 4q21. Oncotarget. 2016;7:80140–80163. doi: 10.18632/oncotarget.12818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Darabi H., McCue K., Beesley J., Michailidou K., Nord S., Kar S., Humphreys K., Thompson D., Ghoussaini M., Bolla M.K., German Consortium of Hereditary Breast and Ovarian Cancer. kConFab/AOCS Investigators Polymorphisms in a putative enhancer at the 10q21.2 breast cancer risk locus regulate NRBF2 expression. Am. J. Hum. Genet. 2015;97:22–34. doi: 10.1016/j.ajhg.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ghoussaini M., French J.D., Michailidou K., Nord S., Beesley J., Canisus S., Hillman K.M., Kaufmann S., Sivakumaran H., Moradi Marjaneh M., kConFab/AOCS Investigators. NBCS Collaborators Evidence that the 5p12 variant rs10941679 confers susceptibility to estrogen-receptor-positive breast cancer through FGF10 and MRPS30 regulation. Am. J. Hum. Genet. 2016;99:903–911. doi: 10.1016/j.ajhg.2016.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Quigley D.A., Fiorito E., Nord S., Van Loo P., Alnæs G.G., Fleischer T., Tost J., Moen Vollan H.K., Tramm T., Overgaard J. The 5p12 breast cancer susceptibility locus affects MRPS30 expression in estrogen-receptor positive tumors. Mol. Oncol. 2014;8:273–284. doi: 10.1016/j.molonc.2013.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Curtis C., Shah S.P., Chin S.F., Turashvili G., Rueda O.M., Dunning M.J., Speed D., Lynch A.G., Samarajiwa S., Yuan Y., METABRIC Group The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486:346–352. doi: 10.1038/nature10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Carvalho B.S., Louis T.A., Irizarry R.A. Quantifying uncertainty in genotype calls. Bioinformatics. 2010;26:242–249. doi: 10.1093/bioinformatics/btp624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Howie B., Fuchsberger C., Stephens M., Marchini J., Abecasis G.R. Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat. Genet. 2012;44:955–959. doi: 10.1038/ng.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Normand S.L. Meta-analysis: formulating, evaluating, combining, and reporting. Stat. Med. 1999;18:321–359. doi: 10.1002/(sici)1097-0258(19990215)18:3<321::aid-sim28>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 50.Boyle A.P., Hong E.L., Hariharan M., Cheng Y., Schaub M.A., Kasowski M., Karczewski K.J., Park J., Hitz B.C., Weng S. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. 2012;22:1790–1797. doi: 10.1101/gr.137323.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ward L.D., Kellis M. HaploReg v4: systematic mining of putative causal variants, cell types, regulators and target genes for human complex traits and disease. Nucleic Acids Res. 2016;44(D1):D877–D881. doi: 10.1093/nar/gkv1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Teng L., He B., Wang J., Tan K. 4DGenome: a comprehensive database of chromatin interactions. Bioinformatics. 2015;31:2560–2564. doi: 10.1093/bioinformatics/btv158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rao S.S., Huntley M.H., Durand N.C., Stamenova E.K., Bochkov I.D., Robinson J.T., Sanborn A.L., Machol I., Omer A.D., Lander E.S., Aiden E.L. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 2014;159:1665–1680. doi: 10.1016/j.cell.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Barutcu A.R., Lajoie B.R., McCord R.P., Tye C.E., Hong D., Messier T.L., Browne G., van Wijnen A.J., Lian J.B., Stein J.L. Chromatin interaction analysis reveals changes in small chromosome and telomere clustering between epithelial and breast cancer cells. Genome Biol. 2015;16:214. doi: 10.1186/s13059-015-0768-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Knight P.A., Ruiz D. A fast algorithm for matrix balancing. IMA J. Numer. Anal. 2013;33:1029–1047. [Google Scholar]
- 56.Ronchi D., Di Fonzo A., Lin W., Bordoni A., Liu C., Fassone E., Pagliarani S., Rizzuti M., Zheng L., Filosto M. Mutations in DNA2 link progressive myopathy to mitochondrial DNA instability. Am. J. Hum. Genet. 2013;92:293–300. doi: 10.1016/j.ajhg.2012.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gao Y.T., Shu X.O., Dai Q., Potter J.D., Brinton L.A., Wen W., Sellers T.A., Kushi L.H., Ruan Z., Bostick R.M. Association of menstrual and reproductive factors with breast cancer risk: results from the Shanghai Breast Cancer Study. Int. J. Cancer. 2000;87:295–300. doi: 10.1002/1097-0215(20000715)87:2<295::aid-ijc23>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 59.Cai Q., Zhang B., Sung H., Low S.K., Kweon S.S., Lu W., Shi J., Long J., Wen W., Choi J.Y., DRIVE GAME-ON Consortium Genome-wide association analysis in East Asians identifies breast cancer susceptibility loci at 1q32.1, 5q14.3 and 15q26.1. Nat. Genet. 2014;46:886–890. doi: 10.1038/ng.3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ulirsch J.C., Nandakumar S.K., Wang L., Giani F.C., Zhang X., Rogov P., Melnikov A., McDonel P., Do R., Mikkelsen T.S., Sankaran V.G. Systematic functional dissection of common genetic variation affecting red blood cell traits. Cell. 2016;165:1530–1545. doi: 10.1016/j.cell.2016.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.