Abstract
We describe a method based on a latent Dirichlet allocation model for predicting functional effects of noncoding genetic variants in a cell-type- and/or tissue-specific way (FUN-LDA). Using this unsupervised approach, we predict tissue-specific functional effects for every position in the human genome in 127 different tissues and cell types. We demonstrate the usefulness of our predictions by using several validation experiments. Using eQTL data from several sources, including the GTEx project, Geuvadis project, and TwinsUK cohort, we show that eQTLs in specific tissues tend to be most enriched among the predicted functional variants in relevant tissues in Roadmap. We further show how these integrated functional scores can be used for (1) deriving the most likely cell or tissue type causally implicated for a complex trait by using summary statistics from genome-wide association studies and (2) estimating a tissue-based correlation matrix of various complex traits. We found large enrichment of heritability in functional components of relevant tissues for various complex traits, and FUN-LDA yielded higher enrichment estimates than existing methods. Finally, using experimentally validated functional variants from the literature and variants possibly implicated in disease by previous studies, we rigorously compare FUN-LDA with state-of-the-art functional annotation methods and show that FUN-LDA has better prediction accuracy and higher resolution than these methods. In particular, our results suggest that tissue- and cell-type-specific functional prediction methods tend to have substantially better prediction accuracy than organism-level prediction methods. Scores for each position in the human genome and for each ENCODE and Roadmap tissue are available online (see Web Resources).
Keywords: functional genomics, noncoding variation, prediction of functional effect
Introduction
Understanding the functional consequences of noncoding genetic variation is one of the most important problems in human genetics. Comparative genomics studies suggest that most of the mammalian conserved and recently adapted regions consist of noncoding elements.1, 2, 3 Furthermore, most of the loci identified in genome-wide association studies (GWASs) fall in noncoding regions and are likely to be involved in gene regulation in a cell-type- and tissue-specific manner.4 Noncoding variants are also known to play an important role in cancer. Somatic variants in noncoding regions can act as drivers of tumor progression, and germline noncoding variants can act as risk alleles.5 Thus, improved understanding of tissue-specific functional effects of noncoding variants will have implications for multiple diseases and traits.
Prediction of the functional effects of genetic variation is difficult for several reasons. To begin with, there is no single definition of function. As previously discussed,6 there are several possible definitions depending on whether one considers genetic, evolutionary conservation, or biochemical perspectives. These different approaches each have limitations and vary substantially with respect to the specific genomic regions that they predict to be functional. In particular, the genetic approach, which is based on experimental evaluation of the phenotypic consequence of a sequence alteration (e.g., through measurement of the impact of individual alleles on gene expression in a particular context, massively parallel reporter assays [MPRAs],7 and CRISPR/Cas-9 mediated in situ saturating mutagenesis8), is currently laborious, has modest throughput, and can miss elements that lead to phenotypic effects only in rare cells or specific contexts. The evolutionary approach relies on accurate multispecies alignment, which makes it challenging to identify certain functional elements, such as distal regulatory elements, known to evolve rapidly, although recently several approaches have been developed for primate- or even human-specific elements.9 An additional limitation of the evolutionary approach is that it is not sensitive to tissue or cell type. Finally, the biochemical approach adopted by projects such as ENCODE3 and Roadmap Epigenomics,10 although helpful in identifying potentially regulatory elements in specific contexts, does not provide definitive proof of function given that the observed biochemical signatures can occur stochastically and in general are not completely correlated with function. Besides the difficulty in precisely defining function, a challenge is that the use of functional genomics features from ENCODE and Roadmap Epigenomics (e.g., chromatin immunoprecipitation sequencing [ChIP-seq] and DNase I hypersensitive site [DHS] signals) are mostly useful for predicting the effects of variants in cis-regulatory elements, such as promoters, enhancers, silencers, and insulators. Other classes of functional variants, for example, those with effects on post-transcriptional regulation by alteration of RNA secondary structure or RNA-protein interactions, would be missed by these features.
Recently, several computational approaches have been proposed for the prediction of functional effects of genetic variation in noncoding regions of the genome on the basis of epigenetic and evolutionary conservation features.2, 11, 12, 13, 14, 15, 16 These predictions are at the organism level and are not specific to particular cell types or tissues. Here, we are interested in predicting functional effects of genetic variants in specific cell types and tissues. The ENCODE and Roadmap Epigenomics projects have profiled various epigenetic features, including histone modifications and chromatin accessibility, genome-wide in more than a hundred different cell types and tissues. Histone modifications are chemical modifications of the DNA-binding histone proteins and influence transcription as well as other DNA processes. Particular histone modifications have characteristic genomic distributions.17 For example, trimethylation of histone H3 lysine 4 (H3K4me3) is associated with promoter regions, monomethylation of histone H3 lysine 4 (H3K4me1) is associated with enhancer regions, and acetylation of histone H3 lysine 27 (H3K27ac) and of histone H3 lysine 9 (H3K9ac) are associated with increased activation of enhancer and promoter regions.10 Repressive marks include trimethylation of histone H3 lysine 27 (H3K27me3) and trimethylation of histone H3 lysine 9 (H3K9me3), both associated with inactive promoters of protein-coding genes; H3K27me3 is found in facultatively repressed genes by Polycomb-group factors, whereas H3K9me3 is found in heterochromatin regions corresponding to constitutively repressed genes.18 Dozens of chromatin marks have been assayed in large numbers of different cell types and tissues, and studying them individually is inefficient.
Several unsupervised approaches exist for the integration of these epigenetic features in specific cell types and tissues. Such integrative approaches reflect the belief that epigenetic features interact with one another to control gene expression. One class of methods attempts to segment the genome into non-overlapping segments, representing major patterns of chromatin marks, and labels these segments by using a small set of labels, such as active transcription start site, enhancer, strong transcription, weak transcription, quiescent, etc. This class includes methods such as ChromHMM10, 19, 20 and Segway,21 which are based on hidden Markov models (HMMs) and dynamic Bayesian networks (DBNs), respectively. ChromHMM is based on the complete pooling of data from multiple tissues and fitting a single model to this superdataset, whereas Segway is based on fitting separate models to data from each tissue (no pooling). Various extensions of these early segmentation approaches have been proposed. Several approaches have focused on better modeling the read count data by using Poisson-lognormal and negative multinomial distributions,22, 23 whereas others have focused on better modeling of the correlations among related cell types and tissues.24, 25, 26 Yet another approach attempts to improve the HMM parameter estimation procedure in ChromHMM by replacing the expectation-maximization algorithm with a spectral learning procedure.27 Another class of methods focuses exclusively on predicting functional effects of variants rather than segmenting the genome as discussed above. A recent method in this class, GenoSkyline,28 is based on fitting a two-component mixture model of multivariate Bernoulli distributions to epigenetic data for each tissue separately and then computing a posterior probability that each variant is in the functional class. Recently, several supervised approaches have been proposed as well, and these include deltaSVM29 and cepip.30 Although supervised approaches can be more efficient than the unsupervised ones when high-quality, unbiased labeled data are available for training, unsupervised approaches as proposed here can provide more robust, less biased functional predictions across a large number of tissues and cell types when such unbiased labeled data are scarce, as it is the case now.
We introduce here an integrated functional score that combines different epigenetic features in specific cell types and tissues. Our model is based on the latent Dirichlet allocation (LDA) model,31 a generative probabilistic model that is often used in the topic modeling literature and allows joint modeling of data from multiple cell types and tissues. In our context, the latent functional classes correspond to latent topics in the topic modeling setting, the various tissues correspond to different documents, and the tissue-specific variant scores correspond to words in a document. The proposed LDA model has several advantages. First, our method makes no distributional assumptions on the data, allowing us to avoid various data transformations employed by other approaches (such as binary peak calling or dichotomization) and facilitating the integration of annotation data on the original scale (e.g., quantitative, binary, etc.). Second, because the model is fit jointly to data from multiple cell types and tissues, cross-tissue comparisons are meaningful. Third, relative to existing methods, our method can improve the precision of locating functional variants. Fourth, even though we provide only functional scores in the tissues and cell types available in Roadmap, it is easy to perform functional prediction in additional cell types and tissues once the model has been fit to the original Roadmap data. Furthermore, although we regard FUN-LDA as primarily an approach for performing cell-type- and tissue-specific functional prediction, we additionally assign functional variants to “active promoter” or “active enhancer” elements.
In the Results section, we demonstrate the usefulness of our predictions through several validation experiments. In summary, we present the following results: (1) we provide cell-type- and tissue-specific functional predictions for every possible position in the hg19 human genome build (UCSC Genome Browser) for 127 cell types and tissues in Roadmap, (2) we provide a global view of the sharing of predicted functional variants across a large number of cell types and tissues and show that predicted functional variants that fall in enhancers are more likely to be tissue specific than those that fall in promoters, (3) we show that expression quantitative trait loci (eQTLs) identified in specific tissues from several sources (Genotype-Tissue Expression [GTEx] project, Geuvadis, and TwinsUK) tend to be most enriched among the predicted functional variants in a relevant Roadmap tissue, (4) we use these cell-type- and tissue-specific scores in conjunction with summary statistics from 21 GWASs to identify the most likely causal cell type or tissue implicated for a particular trait and estimate a tissue-based correlation matrix among these complex traits, and (5) we use experimentally validated functional variants in the literature and variants possibly implicated in disease by previous studies to rigorously compare FUN-LDA with state-of-the-art tissue- and cell-type-specific functional annotation methods, such as GenoSkyline,28 ChromHMM,19 Segway,21 IDEAS,26 deltaSVM29 (when available), and cepip,30 as well as organism-level functional prediction methods, such as CADD,11 Eigen,13 DANN,14 DeepSea,15 and LINSIGHT.16
Material and Methods
LDA Model for Functional Annotation
We propose an application of the LDA model,31 a generative probabilistic model, in the setting of functional genomics annotations with the goal of computing posterior probabilities that variants belong to different functional classes.
Let us assume that we have a set of m genetic variants in the training set together with a set of k functional annotations. For each variant i, we have k tissue-specific functional scores: Xi = (Xi1, …, Xik). Let X = (X1, …, Xm) be the set of (continuous) functional scores for all the variants. These scores are epigenetic features (histone modifications and DNase) from ENCODE and Roadmap Epigenomics across a varied set of tissues and cell types. Let l be the number of tissues and mj be the number of variants with tissue j annotations in the training set . For each variant i ≤ m in the training set, we denote by ti the corresponding tissue (i.e., the annotations corresponding to this variant are for tissue ti). For each tissue, the variants’ scores are represented as a mixture over latent functional classes, where each functional class is characterized by a distribution over variant scores. In what follows, for ease of presentation, we assume only two latent functional classes, but the number of classes can be chosen to be greater than two (see Choosing the Number of Functional Classes in the LDA Model according to the Perplexity Measure for a discussion on the choice of the number of functional classes). We let C = (C1, …, Cm) denote the set of indicator variables for all the variants, where Ci = 1 if variant i belongs to the first functional class and Ci = 0 otherwise. We are not able to observe C.
Let α = (α0, α1) be the hyperparameter vector with α0 and α1 > 0. We assume that the functional annotation data have been generated from the following generative model:
-
(1)
For each tissue j, choose (1 − πj, πj) ∼ Dir(α0, α1).
-
(2)
Given πj, for each variant i with ti = j, choose a class Ci ∼ Bern(πj).
-
(3)
Given C1, …, Cm, X1, …, Xm are independently generated such that each Xi is generated from the appropriate multivariate distribution: F1 if Ci = 1 and F0 otherwise.
Here, π = (π1, … πl) and C are latent variables. We want to calculate the posterior probability that each variant i is in the first functional class,
and the densities f0 and f1. Also, we want to estimate the hyperparameter α = (α0, α1) empirically by using X. For a given tissue, the conditional density of (π, C) given X and α is
For the numerator, we have
This is easy to compute. However, the denominator is not. For the denominator, we have
There are 2m terms in the summation, so this is difficult to compute for moderately large m. We propose instead to use a variational approach as previously described.31 In the variational inference approach, we first introduce a family of distributions {q(·,· | a, w)} over the latent variables (π, C) with its own variational parameters α = (α0, α1) and w (these are tissue-specific parameters).
Then
where q(π | a) is the density of Dir(a), and q(Ci | wi) is the probability mass function of Bern(wi) for i = 1 … m.
Using Jensen’s inequality, we have
Note that L(a, w | α) is a lower bound on the log likelihood. So instead of maximizing the log likelihood directly, we maximize this lower bound with respect to the variational parameters a and w, as well as the hyperparameter α. It can be shown that log p(X | α) − L(a, w | α) is the Kullback-Leibler (KL) divergence between the true posterior p(π, C | α, X) and the variational posterior q(π, C | a, w) with respect to q(π, C | a, w). Therefore, by maximizing L(a, w | α) with respect to a and w, we minimize the KL divergence between the variational posterior probability and the true posterior probability. Then we can estimate P(Ci = 1 | α, X) by wi for each variant i. Below, we describe the variational inference algorithm.
Variational Inference Algorithm
Assume the initial state (w1, …, wm, f0, f1, α). The algorithm proceeds as follows:
Step 1: Kernel Density Estimation
Fit a multivariate kernel density estimate for each annotation and component separately ( and for each annotation s = 1, …, k) by weighting variants by component membership probability. Specifically, for any and s = 1, …, k, we let
and
The scaled kernel , where K(·) is taken to be the probability density function of a standard normal, and the bandwidth parameter hs is chosen to be
according to a rule of thumb from Silverman,32 where SDs and IQRs are the standard deviation and interquartile range of annotation s, respectively. Then,
Step 2: Variational Step
For each tissue j, we obtain wi for all variants i with ti = j and by maximizing the lower bound on the marginal likelihood of X, i.e., L(a, w | α), with respect to a and w. Details are shown in the Supplemental Data.
This results in the following iterative algorithm:
where , and is the gamma function.
Step 3: Newton-Raphson Algorithm for Estimating the Hyperparameters α
Obtain the empirical Bayes estimate of α = (α0, α1) by maximizing the bound L(a, w | α) with the Newton-Raphson algorithm, where a and w are from step 2. That is, we find optimal α by iterating
where H(α) is the Hessian matrix evaluated at current α.
The gradient has this form:
The Hessian matrix takes the following form:
LDA Implementation
We have implemented the above algorithm in an R package, FUNLDA. In our implementation, we assume a symmetric Dirichlet prior, with α = 1, corresponding to a uniform distribution. For training purposes, we select 4,000 random positions in each of the 127 tissues. The positions are chosen among 9,254,335 single-nucleotide polymorphisms (SNPs) with a minor allele count greater than 5 in European samples from the 1000 Genomes Project. We have also looked at other ways to select variants in the training set (e.g., randomly from across the entire genome or with enrichment near genes), and the results were similar, suggesting that our predictions are robust to the choice of variants used in the training sets. The number of outer iterations in the variational inference algorithm is 250, and the number of inner iterations is 200.
We compute FUN-LDA by fitting the LDA model with nine classes to valley scores for the four activating histone modifications (H3K4me1, H3K4me3, H3K9ac, and H3K27ac) and quantitative DNase. For the histone modifications and DNase, we start with the negative log10 of the Poisson p value of ChIP-seq or DNase counts relative to expected background counts, as output by ChromImpute.20 The valley scores are computed as previously reported;33 for every window of 25 bp, we calculate the maximum score for the two regions from −100 to −500 bp and from 100 to 500 bp. If the score at the 25 bp window is less than 90% of the minimum of those two maxima, we set the value in that window to that minimum. Otherwise, we set the value in that 25 bp window to 0. For each variant, we get a set of nine posterior probabilities that the variant is in a specific functional class. To get a functional score, we sum the posterior probabilities for the active functional classes, namely active promoters and active enhancers (Figure S1 and Table S1).
Prediction in a New Tissue
Once the LDA model has been fit to the epigenetic data for cell types and tissues available in Roadmap, making predictions for a new cell type or tissue is easy. Basically, one only needs to run the iterative algorithm in step 2 of the variational inference algorithm on the epigenetic data for the new tissue.
Choosing the Number of Functional Classes in the LDA Model according to the Perplexity Measure
Choosing the number of functional classes in the LDA model is not straightforward. Too few classes can be insufficient and can lower the accuracy of the resulting classifier. Too many classes can lead to an overly complex model and is subject to overfitting.
Heuristic methods exist on the basis of computing the perplexity of a model with a given number of clusters on held-out datasets. Perplexity is used in information theory to describe how well a statistical model fits the data. The lower the perplexity, the better the model and its generalization performance. In our case, if we let be the log-likelihood for a held-out set of variants for each tissue ti, the perplexity is defined as
where l is the total number of tissues, and mi is the number of variants for tissue ti. Evaluating the perplexity measure directly is computationally intractable (the computation of the likelihood for each tissue involves a summation over terms, where K is the number of classes), and therefore we use the lower bound on the log-likelihood, i.e., L(a, w | α) (see Supplemental Data), to derive an upper bound on the perplexity. This upper bound on the perplexity is referred to as the variational Bayesian bound on the perplexity. In the large data limit, the bound on the log perplexity evaluated on the training data converges to the Bayesian information criterion (BIC) for the model.34 If the training and testing datasets are assumed to come from the same distributions, then the variational Bayesian bound on the log perplexity converges to the BIC.
Alternative Functional Annotation Methods Used in Our Comparisons
We compare our approach with the following state-of-the-art functional annotation methods.
Tissue- and Cell-Type-Specific Functional Prediction Methods
Individual Histone Modifications and DNase Scores
Instead of integrating the various epigenetic marks, one can use the individual scores to predict functional variants. For the histone modifications and DNase, we use the negative log10 of the Poisson p value of ChIP-seq or DNase counts relative to expected background counts, as output by ChromImpute.20 In addition, for DNase, we also use narrow peaks and gapped peaks (defined as broad peaks that contain at least one strong narrow peak).
GenoSkyline: Multivariate Bernoulli Mixture Models
A simpler mixture model than the LDA described here is the two-component mixture model , where f0 and f1 are the probability densities for each of the components, and π is a mixing parameter. We can fit such a model to data from each tissue separately and calculate posterior probabilities that each variant is in the “functional” class given the observed scores X, i.e., . For tractability, it is often assumed that the individual scores are conditionally independent given the functional class. Such a two-component multivariate Bernoulli mixture model using dichotomized data from peak-calling algorithms—an approach called GenoSkyline—has been proposed previously.28
ChromHMM
ChromHMM19 is a method for chromatin-state discovery and characterization through the integration of multiple chromatin datasets. The underlying algorithm is a multivariate HMM that produces a segmentation of the genome; each segment is assigned a putative function on the basis of enrichment analyses of different biological states in these segments. The ChromHMM 25-state model20 is based on 12 marks and, like ours, uses imputed data: H3K4me1, H3K4me2, H3K4me3, H3K9ac, H3K27ac, H4K20me1, H3K79me2, H3K36me3, H3K9me3, H3K27me3, H2A.Z, and DNase. ChromHMM is based on the complete pooling of data from multiple tissues and fitting a single model to this superdataset.
Segway
Like ChromHMM, Segway21 is a genome segmentation approach based on a DBN model. Segway is based on fitting separate models to data from each tissue. Segmentations for most of the cell types and tissues in Roadmap have been recently generated.35
IDEAS
IDEAS25 is an integrative and discriminative epigenome annotation algorithm that, like ChromHMM and Segway, segments the genome and assigns each segment a specific functional class. Unlike ChromHMM and Segway, IDEAS models the correlations both along the genome and across cell types. Segmentations for all 127 cell types and tissues in Roadmap have been produced with IDEAS.26
deltaSVM
deltaSVM29 is a new sequence-based computational method that predicts the effect of regulatory variation by using a classifier (gkm-SVM) that encodes cell-specific regulatory sequence vocabularies. The induced change in the gkm-SVM score, deltaSVM, quantifies the effect of variants.
cepip
By connecting large-scale epigenome profiles to eQTLs across human tissues and cell types, Li et al.30 identified combinations of chromatin features that are predictive of a variant’s regulatory potential. They developed a joint likelihood framework to measure the probability that genetic variants are functional in a context-dependent manner.
Organism-Level Functional Prediction Methods
CADD
CADD11 is based on a supervised approach (support vector machine) to training a discriminative model. That is, it begins with two sets of variants—one labeled deleterious and the other labeled benign—and fits a model that best separates the two sets. It selects benign variants by comparing the human genome to the inferred genome of the most recent shared human-chimpanzee ancestor. Alleles that are not found in the common ancestor and that are fixed in the human genome are assumed to be mostly benign. These are compared with de novo variants generated randomly according to models of mutation rates across the genome.
Eigen
Eigen13 is an unsupervised spectral approach for scoring variants and does not make use of labeled training data. Eigen uses a variety of functional annotations in both coding and noncoding regions and combines them into one single measure of functional importance. Eigen produces estimates of predictive accuracy for each functional annotation score and subsequently uses these estimates of accuracy to derive the aggregate functional score.
DANN
DANN14 uses the same feature set and training data as CADD to train a deep neural network (DNN). Unlike CADD, which trains a linear kernel support vector machine to differentiate between likely benign and likely deleterious variants and therefore cannot capture non-linear relationships among the features, DNNs can capture non-linear and interactive relationships among features.
LINSIGHT
LINSIGHT16 is a statistical method that combines a generalized linear model for functional genomic data with a probabilistic model of molecular evolution to predict the fitness consequences of mutations.
DeepSea
DeepSea15 is a deep-learning-based approach for predicting the chromatin effects of sequence alterations with single-nucleotide sensitivity.
PhyloP
PhyloP36 quantifies evolutionary conservation at individual sites.
Generalized Jaccard Index of Overlap
We are interested in computing a similarity measure of predicted functional variants in two different tissues. Because the distribution of posterior probabilities in any one tissue is highly bimodal (where most of the mass is at 0 and a small proportion of variants have posterior probabilities close to 1; in other words, we are dealing with sparse binary data), a natural measure of similarity is the Jaccard measure of overlap, defined as follows. If X = (x1, …, xk) and Y = (y1, …, yk) are two vectors with xi and yi ≥ 0, respectively (e.g., vectors of posterior probabilities that variants are in the functional components for two different tissues), then the generalized Jaccard index of overlap is defined as
When X and Y are binary vectors, then the Jaccard index of overlap is simply the size of the intersection divided by the size of the union of the two sets. The closer it is to 1, the more overlap there is between the two sets. A Jaccard index of 0 means no overlap.
Promoter and Tissue-Specific Enhancer Regions
The promoter region of a protein-coding gene is defined as the union of the regions 2,500 bases upstream of any protein-coding transcripts for the gene, as defined by GENCODE version 24. For enhancer regions, we use the Roadmap Stringent enhancer list available at the Reg2Map website.
eQTL Enrichment
Let G1, …, G44 be the 44 GTEx tissues with at least 70 samples (Table S2) and R1, …, R127 be the 127 Roadmap tissues. For a given tissue in GTEx Gi, we are interested in identifying the Roadmap tissue Rj with a higher enrichment in eQTLs from Gi than other tissues in Roadmap.
Let
Note that the number of eQTLs in GTEx tissue Gi is a weighted count (an eQTL is weighted by the inverse of the number of GTEx tissues in which the variant is eQTL) such that . This way, eQTLs that are unique to tissue Gi are given higher weight than eQTLs that are shared across many tissues. For GTEx tissue Gi, to test whether there is an enrichment in the functional component of Roadmap tissue Rj, we compare with
The null hypothesis is versus . We apply a two-sample proportion test for each Roadmap tissue Rj and report the Roadmap tissue with minimum p value in Table 1. Also reported in Table 1 is the enrichment ratio .
Table 1.
Enrichment of eQTLs from Different Sources (GTEx, Geuvadis, and TwinsUK Cohort) among Functional Variants Predicted by FUN-LDA in Tissues and Cell Types in Roadmap Epigenomics
| Tissue | Roadmap Epigenomics Name | −log10(p) | −log10(FDR.p) | Enrichment Ratio |
|---|---|---|---|---|
| GTEx | ||||
| Whole blood | primary neutrophils from peripheral blood | 189.72 | 185.92 | 1.38 |
| Cells – transformed fibroblasts | muscle satellite cultured cells | 62.69 | 59.59 | 1.22 |
| Cells – EBV-transformed lymphocytes | GM12878 lymphoblastoid cells | 37.74 | 35.23 | 1.48 |
| Liver | liver | 31.82 | 29.45 | 1.68 |
| Muscle – skeletal | skeletal muscle male | 19.42 | 17.26 | 1.13 |
| Heart – left ventricle | fetal heart | 15.83 | 13.74 | 1.22 |
| Esophagus – mucosa | esophagus | 12.78 | 10.74 | 1.15 |
| Pancreas | pancreas | 10.84 | 8.89 | 1.25 |
| Colon – transverse | rectal mucosa donor 31 | 10.46 | 8.52 | 1.23 |
| Artery – tibial | stomach smooth muscle | 7.74 | 5.87 | 1.10 |
| Esophagus muscularis | stomach smooth muscle | 6.74 | 4.91 | 1.11 |
| Thyroid | fetal intestine small | 5.96 | 4.19 | 1.06 |
| Skin – sun exposed (lower leg) | foreskin keratinocyte primary cells skin03 | 5.47 | 3.73 | 1.07 |
| Spleen | primary B cells from peripheral blood | 5.35 | 3.61 | 1.22 |
| Artery – aorta | aorta | 5.28 | 3.57 | 1.14 |
| Brain – hippocampus | brain cingulate gyrus | 5.10 | 3.42 | 1.38 |
| Small intestine – terminal ileum | fetal intestine large | 5.04 | 3.37 | 1.37 |
| Heart – atrial appendage | fetal heart | 4.90 | 3.25 | 1.15 |
| Adipose – subcutaneous | adipose nuclei | 4.74 | 3.11 | 1.07 |
| Colon – sigmoid | colon smooth muscle | 4.62 | 2.99 | 1.19 |
| Brain – caudate (basal ganglia) | brain substantia nigra | 4.17 | 2.60 | 1.18 |
| Nerve – tibial | brain hippocampus middle | 4.11 | 2.56 | 1.05 |
| Adrenal gland | fetal adrenal gland | 3.94 | 2.42 | 1.14 |
| Skin – not sun exposed (suprapubic) | foreskin keratinocyte primary cells skin03 | 3.56 | 2.07 | 1.09 |
| Brain – putamen (basal ganglia) | brain substantia nigra | 3.36 | 1.90 | 1.23 |
| Brain – cerebellum | primary T cells from cord blood | 3.20 | 1.78 | 1.11 |
| Brain – cerebellar hemisphere | brain angular gyrus | 3.08 | 1.67 | 1.12 |
| Stomach | stomach mucosa | 3.02 | 1.63 | 1.13 |
| Lung | primary hematopoietic stem cells G-CSF-mobilized male | 2.42 | 1.13 | 1.05 |
| Adipose – visceral (omentum) | primary T helper cells from peripheral blood | 2.00 | 0.82 | 1.08 |
| Brain – nucleus accumbens (basal ganglia) | H9 cells | 1.80 | 0.67 | 1.17 |
| Pituitary | ES-I3 cells | 1.75 | 0.64 | 1.13 |
| Brain – cortex | primary natural killer cells from peripheral blood | 1.61 | 0.54 | 1.11 |
| Esophagus – gastresophageal junction | primary B cells from peripheral blood | 1.49 | 0.46 | 1.09 |
| Artery – coronary | primary B cells from peripheral blood | 1.35 | 0.38 | 1.10 |
| Brain – hypothalamus | primary natural killer cells from peripheral blood | 1.26 | 0.33 | 1.16 |
| Brain – frontal cortex (BA9) | H1 cells | 1.08 | 0.22 | 1.11 |
| Brain – anterior cingulate cortex (BA24) | primary natural killer cells from peripheral blood | 1.00 | 0.19 | 1.12 |
| Geuvadis | ||||
| Lymphoblastoid cell line | GM12878 lymphoblastoid cells | 9.27 | 7.35 | 1.13 |
| TwinsUK | ||||
| Blood | primary neutrophils from peripheral blood | 7.30 | 5.46 | 1.42 |
| Fat | mesenchymal stem cell derived adipocyte cultured cells | 6.26 | 4.45 | 1.21 |
| Skin | foreskin keratinocyte primary cells skin02 | 3.99 | 2.45 | 1.18 |
| Lymphoblastoid cell line | GM12878 lymphoblastoid cells | 3.08 | 1.67 | 1.08 |
The top Roadmap tissue (with smallest enrichment p value) is given for each eQTL tissue, along with the p value from a two-sample proportion test, the FDR-adjusted p value (FDR.p), and the enrichment ratio (see Material and Methods). Tests that are significant at an FDR of 0.05 have −log10(FDR.p) > 1.30.
The eQTLs that we used in these analyses are all significantly associated SNP-gene pairs for eGenes in each of these 44 GTEx tissues and were obtained by the permutation-threshold-based approach described by the GTEx Consortium37 (see Web Resources for more details). For the follow-up study making use of eQTLs from Geuvadis and the TwinsUK cohort, we used the lead eQTLs, i.e., those most strongly associated with gene expression (publicly available for download from Brown et al.38).
Stratified LD Score Regression Approach to Identifying the Tissue of Interest
The stratified linkage disequilibrium (LD)-score regression approach1 uses two sets of SNPs, reference SNPs and regression SNPs. The regression SNPs are SNPs that are used in a regression of chi-square statistics from GWASs against the LD scores of those regression SNPs. The LD score of a regression SNP is a numeric score that captures the amount of genetic variation tagged by the SNP. Here, following Finucane et al.,39 we use HapMap3 SNPs as regression SNPs because of their high imputation quality and use SNPs with a minor allele count greater than 5 in the 379 European samples from the 1000 Genomes Project as reference SNPs.2 We first compute tissue-specific scores by using each of our methods for the 9,254,335 reference SNPs.
In the stratified LD-score regression approach, a linear model is used to model a quantitative phenotype yi for an individual i:
Here, G is some set of SNPs, Xij is the standardized genotype of individual i at SNP j, βj is the effect size of SNP j, and is mean-zero noise. In this framework, β, the vector of all βj, is modeled as a mean-zero random vector with independent entries, and the variance of βj depends on the functional categories included in the model. We have a set of functional categories, C1, …, CC, and the variance of a SNP’s effect size will depend on which functional categories it belongs to:
Here, is the per-SNP contribution to heritability of SNPs in category CC. Lindblad-Toh et al.1 show that under this model, can be estimated through the following equation:
Here, is the chi-square statistic for SNP j from a GWAS, N is the sample size from that study, and l(j, c) is the LD score of SNP j with respect to category Cc, . This equation therefore allows for the estimation of via the regression of the chi-square statistics from a GWAS on the LD scores of the regression SNPs.
Here, we extend the stratified LD score by allowing SNPs to be assigned to a category Cc probabilistically, i.e., we assume a probability pkc that SNP k belongs to category Cc and therefore that the variance of its effect size is affected by its membership in that category. This involves only minor changes to the above equations, namely, we have that
where pjc is the probability that SNP j belongs to category Cc, and as above,
although now , where pkc is the probability that SNP k belongs to category Cc. We can therefore still estimate via the regression of the chi-square statistics from a GWAS on the LD scores of the regression SNPs, but in calculating these LD scores, we weight the squared correlation of a SNP k with a regression SNP j by the probability that SNP k belongs to a particular category.
For each tissue and phenotype and each of our functional scores, we fit a separate LD-score regression model, including the LD score derived from the posterior probability that each regression SNP is in the functional component in that tissue, to estimate the per-SNP contribution of SNPs belonging to that component to heritability. To control for overlap of the tissue-specific functional score with other functional categories, we use the same 54 baseline categories used in Lindblad-Toh et al.,1 which represent various non-tissue-specific annotations, including histone-modification measurements combined across tissues, measurements of open chromatin, and super enhancers.
Assessing Pairwise Correlations among 21 Complex Traits
Our aim here is to calculate a correlation matrix of 21 phenotypes on the basis of the Z scores from the LD-score regression procedure and a p value corresponding to each pair of phenotypes. From the LD-score regression approach, we obtain a matrix of Z scores corresponding to 127 (p = 127) tissues and 21 (q = 21) phenotypes. The main issue we need to account for when computing the correlations and the p values is that the tissues are correlated.
Let Zij be the Z score corresponding to the ith tissue and jth phenotype, and let Zi = (Zi1, …, Ziq) and Zj = (Z1j, …, Zpj) be the row and column vectors, respectively, of matrix Z. Given that the elements of Z are Z scores, we assume Zi ∼ N(0, Σq) and Zj ∼ N(0, Σp).
Estimation of the Correlation Matrix
We aim to estimate Σq, but the problem is that Zi values are not independent. To solve the problem, we propose the following perturbation method.
Let B be the number of perturbation replicates. For the bth replicate, we generate p independent random variables from N(0, 1), αb1, …, αbp. Let
It can be shown that cov(Xb) = Σq and cov(Xb, Xb′) = 0 for any 1 ≤ b and b′ ≤ B, so we are able to use the uncorrelated perturbation samples X1, …, XB to approximate Σq and the corresponding correlation matrix Pq. We take B = 100,000.
p Values Corresponding to All Pairs of Phenotypes
For pairs from an uncorrelated bivariate normal distribution, the sampling distribution of a certain function of Pearson’s correlation coefficient follows Student’s t distribution with degrees of freedom M − 2, where M is the number of uncorrelated random variables. Specifically, if the underlying variables have a bivariate normal distribution, the variable
follows a Student’s t distribution with degrees of freedom M − 2.
In our case, the number of uncorrelated random variables M depends on the correlation structure of the 127 tissues. M can be understood as the effective number of tissues. Similar to the calculation of the number of effective tests by Gao et al.,40 we estimate M by applying an eigen decomposition to the Jaccard matrix. Suppose that λ1 ≥ λ2 ≥ … ≥ λp are the eigenvalues arranged in descending order. We estimate M by the smallest value such that . It should be noted that a smaller C will result in more conservative p values because the number of “effective tissues” is smaller, e.g., M = 124 when C = 99.5% and M = 96 when C = 95%. A threshold C that is too large or too small could cause M to be either overly liberal or overly conservative. The p values were calculated on the basis of C = 99.5%.
Availability of Code and Functional Scores
We have implemented the LDA algorithm into an R package, FUNLDA. The package and FUN-LDA scores for all genome-wide positions across all 127 tissues and cell types are available online (see Web Resources).
Results
Overview of the FUN-LDA Approach
We propose here an integrative functional score for predicting the functional effects of genetic variants at a tissue- and cell-type-specific level. The model is based on the LDA model, a generative probabilistic model commonly used in the topic modeling literature. The variant scores in each tissue are modeled as a mixture over latent functional classes. In the mixture distribution, we assume that the mixture components are shared across all tissues, whereas the mixture proportions for the different functional classes can vary from tissue to tissue (more details on the model and inference algorithm are given in the Material and Methods). Because our primary goal is to provide a functional score (as opposed to a functional element annotation), we focus on integrating four activating histone modifications (i.e., H3K4me1, H3K4me3, H3K9ac, and H3K27ac) and DNase. For the data on the four activating histone modifications, we compute “valley” scores (Material and Methods) on the basis of previous work showing that within regions of high histone acetylation, local minima (or valleys) are strongly associated with transcription factor binding sites.33 We fit the LDA model with multiple functional classes to these data and compute for each position its posterior probability of belonging to a functional class in a specific tissue. We define the functional score at a position as the sum of posterior probabilities for the designated active functional classes (see next sub-section).
FUN-LDA Model with Nine Classes
Here, we use data on four activating histone modifications, namely H3K4me1, H3K4me3, H3K9ac, and H3K27ac, and DNase for 127 different cell types and tissues represented in the Roadmap datasets (see Tables S3 and S4). Not all of the histone marks were profiled for each of the 127 different cell types and tissues. However, using the relationships between different marks within and across tissues, previous studies have predicted signal tracks for each of these marks across all tissues.10, 20 We make use of these predicted signal tracks to compute integrated functional scores for every possible position in the human genome for 127 cell types and tissues. Specifically, using the perplexity-based criterion (see Material and Methods), visual inspection of the resulting classes, and prior knowledge of the relationship between histone modifications and chromatin states, we investigate models with varying numbers of classes and chose as our final model a model with nine classes (as shown in Figure S2, the perplexity measure begins to plateau with models with nine classes). We fit the LDA model with nine classes to the valley scores for the data on active histone modification and quantitative DNase and compute posterior probabilities at each position for the different functional classes in the different tissues and cell types. The active functional classes correspond to active promoters and active enhancers (Figure S1). As in genome segmentation approaches such as ChromHMM (25-state model), Segway, and IDEAS, we also make a similar partition (see Material and Methods and Table S1). For each position, both our method and ChromHMM use the sum of the posterior probabilities for the classes in the functional group to score the position. Segway and IDEAS provide only a functional-class assignment for each position for each cell type and tissue in Roadmap, and we use these assignments to identify the functional variants. The proportions of positions in the functional groups across different tissues and cell types for each method are shown in Figure S3. FUN-LDA, ChromHMM, and DNase-narrow (DNase narrow peaks) estimate that an average of 2% of the genome is functional in a cell type or tissue in Roadmap, and the remaining methods produce higher estimates for the size of the functional component.
Sharing Predicted Functional Variants across Tissues and Cell Types
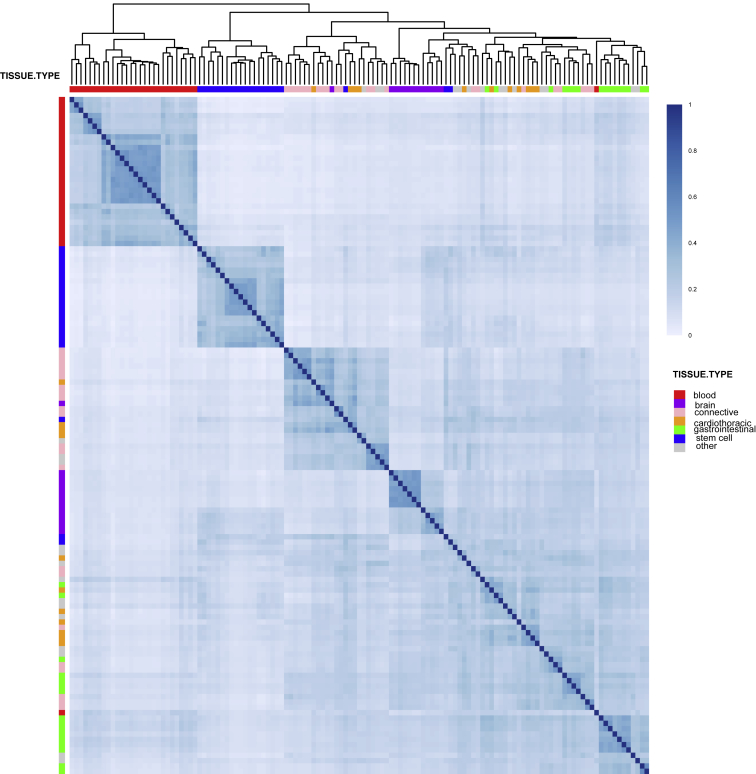
For each variant in the 1000 Genomes Project, we separately compute the probability that each Roadmap tissue is in the functional class. In Figure 1, we provide a global picture of the sharing of predicted functional variants across tissues in Roadmap by using the generalized Jaccard similarity index, a measure of overlap between predicted functional variants in two tissues (see Material and Methods). General tissue groupings are indicated in different colors. As expected, tissues that are functionally related tend to cluster together. There are roughly three major groups: blood cells (indicated in red), including various primary immune cell subtypes, stem cells (indicated in blue), and a third group corresponding to various solid organs (this grouping is also apparent in the multi-dimensional scaling visualization of the correlations between the functional scores in Figure S4; see also Kundaje et al.,10 Ernst and Kellis,19 and Figure S5 for related results using single histone marks and DNase).
Figure 1.
Jaccard Index of Overlap among Predicted Functional Variants in Different Cell Types and Tissues in Roadmap Epigenomics
Hierarchical clustering is used for clustering the different cell types and tissues.
Overall, the median Jaccard index across all pairs of tissues is 0.24. As a comparison, we also compute the Jaccard overlap indices by using predicted functional variants that fall in promoters and separately in enhancers (Material and Methods; see also Figures S6 and S7, which show the Jaccard indices for all pairs of tissues by using predicted functional variants that fall in promoters and enhancers, respectively). The median Jaccard index for variants falling in promoters is 0.33, and that for variants falling in enhancers is 0.16, concordant with existing literature showing that there tends to be more tissue specificity (less sharing) for predicted functional variants in enhancers than for those in promoters.41
Enrichment Analyses Using eQTLs from the GTEx, Geuvadis, and TwinsUK Data
eQTLs from the GTEx Project
The GTEx project is designed to establish a comprehensive data resource on genetic variation, gene expression, and other molecular phenotypes across multiple human tissues.37 We focus here on the cis-eQTL results from the GTEx v.6 release, which comprises RNA sequencing data on 7,051 samples in 44 tissues, each with at least 70 samples (Table S2). For each GTEx tissue, we are interested in identifying the Roadmap tissue that has a more significant enrichment of eQTLs from that GTEx tissue than from other Roadmap tissues (see Material and Methods). We exclude from analysis the sex-specific GTEx tissues (ovary, vagina, uterus, testis, prostate, and breast), most of which have no relevant counterpart in Roadmap. In Table 1, we show the top Roadmap tissue for each remaining GTEx tissue, along with the p value and the enrichment ratio from the enrichment test. In most cases, eQTLs from a GTEx tissue show the most significant enrichment in the functional component of a relevant Roadmap tissue. For example, for liver tissue in GTEx, liver is the Roadmap tissue with the most significant enrichment; for pancreas tissue in GTEx, pancreas is the Roadmap tissue with the most significant enrichment; for skeletal muscle tissue in GTEx, skeletal muscle is the corresponding Roadmap tissue. However, there are also a few cases where the top tissue is not necessarily the most intuitive one, such as for lung and several brain tissues. Generally, the tissues with unexpected combinations tend to either have small sample sizes for eQTL discovery in GTEx (such as brain tissues) or inadequate representation in Roadmap (e.g., thyroid, pituitary gland, tibial artery, coronary artery, gastroesophageal junction of the esophagus, etc.). Controlling the false-discovery rate (FDR) at the 0.05 level, we find that most of the mismatches are not significant at this level (Table 1). We note that enrichment ratios tend to be small, especially for those tissues above the FDR threshold. This might be due to considerable tissue sharing of cis-eQTL effects reported in the GTEx study.42
eQTLs from the Geuvadis and TwinsUK Data
We sought to perform similar analyses by using eQTLs identified in other studies, particularly those in lymphoblastoid cell lines (LCLs) from the Geuvadis project and four tissues (fat, LCLs, skin, and whole blood) in individuals from the TwinsUK cohort. We have focused here on the lead eQTLs, i.e., those variants most associated with gene expression levels.38 As shown in Table 1, in each case, the most significantly enriched Roadmap tissue corresponds very well to the tissue of origin used in the eQTL discovery, providing an independent validation of the findings using the eQTLs from GTEx.
Prediction of Causal Tissues for 21 Complex Traits
As an application of our scores to the genetics of complex traits, we use the recently developed stratified LD-score regression framework39 to identify the most relevant cell types and tissues for 21 complex traits for which moderate to large GWASs have been performed (Table 2).43, 44, 45, 46, 47, 48, 49, 50, 51, 52, 53, 54, 55, 56, 57, 58, 59, 60, 61, 62, 63 The stratified LD-score regression approach uses information from all SNPs and explicitly models LD to estimate the contribution to heritability of different functional classes of variants. We modify this method to weigh SNPs by their tissue-specific functional score (e.g., FUN-LDA), and in this way we assess the contribution to heritability of predicted functional SNPs in a particular Roadmap cell type or tissue (see Material and Methods for more details).
Table 2.
Top Cell Types and Tissues in Roadmap for 21 GWAS Traits according to FUN-LDA Posterior Probabilities
| Trait | Roadmap Epigenomics Name | −log10(p) | nGWAS |
|---|---|---|---|
| Schizophrenia | fetal brain female | 14.69 | 82,315 |
| Height | mesenchymal-stem-cell-derived chondrocyte cultured cells | 12.27 | 133,653 |
| Rheumatoid arthritis | GM12878 lymphoblastoid cells | 6.92 | 58,284 |
| Crohn disease | primary B cells from cord blood | 6.24 | 20,883 |
| Age at menarche | H9-derived neuronal progenitor cultured cells | 6.14 | 132,989 |
| Educational attainment | fetal brain female | 5.83 | 101,069 |
| BMI | brain germinal matrix | 4.79 | 123,865 |
| HDL | liver | 4.72 | 99,900 |
| Coronary artery disease | liver | 4.60 | 86,995 |
| Ulcerative colitis | primary T helper 17 cells PMA-I stimulated | 4.44 | 27,432 |
| Type 2 diabetes | pancreatic islets | 4.20 | 69,033 |
| Epilepsy | brain anterior caudate | 4.11 | 34,853 |
| Triglycerides | liver | 4.10 | 96,598 |
| LDL | liver | 4.08 | 95,454 |
| Alopecia areata | primary T cells from cord blood | 3.90 | 7,776 |
| Alzheimer | primary hematopoietic stem cells G-CSF-mobilized male | 3.78 | 54,162 |
| IGAN | primary natural killer cells from peripheral blood | 3.28 | 11,946 |
| Bipolar disorder | fetal brain female | 3.19 | 16,731 |
| Ever smoked | brain inferior temporal lobe | 2.67 | 74,035 |
| Autism | primary monocytes from peripheral blood | 2.40 | 10,263 |
| Fasting glucose | pancreatic islets | 1.44 | 58,074 |
The p value from the stratified LD-score regression and the GWAS sample size are reported for each trait.
In Table 2, we show the top Roadmap cell type or tissue (the one with the smallest p value from testing whether predicted functional variants in a tissue contribute significantly to SNP heritability) for each of the 21 complex traits from using FUN-LDA to predict functional variants in specific cell types and tissues. For most disorders, the top tissue has previously been implicated in their pathogenesis. For example, the top tissues for body mass index (BMI) are brain tissues, consistent with recent findings indicating that BMI-associated loci are enriched with expression in the brain and central nervous system.64 Similarly, the brain represents the top tissue for most neuropsychiatric disorders, education levels, and smoking. Blood-derived and immune cells represent the top tissue for virtually all of the autoimmune conditions available for analysis. For example, GWAS findings for ulcerative colitis map specifically to the regulatory elements in T helper 17 cells, whereas LCLs represent the top cell type for rheumatoid arthritis. Another interesting finding involves primary hematopoietic stem cells for Alzheimer disease, consistent with emerging data on the involvement of bone-marrow-derived immune cells in the pathogenesis of neurodegeneration.65 For the top cell type or tissue for each trait, we further estimate the contribution to heritability of variants in different functional classes (in Figure S1) and show that, overall, the variants in active promoters and active enhancers for the top tissue are the most enriched in heritability, whereas many of the non-functional classes show no enrichment or disenrichment.
Although in Table 2 we report only the top-ranked tissue for each trait, clearly other top-ranked tissues might be relevant as well. Indeed, as a result of high correlation among particular tissue and cell types in Roadmap (e.g., many different blood cell types, including various primary immune cell subtypes), it is difficult to distinguish among the top tissues. As shown in Figure S8, for autoimmune and inflammatory conditions (including Crohn disease, alopecia areata, rheumatoid arthritis, and immunoglobulin A [IgA] nephropathy), because of the large number of immune cells in Roadmap, several top cell types show similar patterns of enrichment. On the other hand, for traits such as low-density lipoprotein (LDL), high-density lipoprotein (HDL), and triglycerides, the top tissue (liver) is substantially more enriched than the next few tissues.
Results for other methods are shown in Tables S5–S7. Estimates of enrichment (defined as the proportion of SNP heritability in the category divided by the proportion of SNPs in that category) for the functional component in the top tissues in Tables S5–S7 are shown in Figure S9. On average across traits, the functional component for the top tissue as defined by FUN-LDA shows a higher enrichment than other methods. FUN-LDA is followed closely by DNase-narrow and ChromHMM. Methods such as DNase-gapped, GenoSkyline, Segway, and IDEAS show substantially lower enrichments. However, in terms of actual heritability explained, methods such as DNase-gapped, Segway, and IDEAS lead to more explained heritability than methods such as FUN-LDA (Figure S9). For example, for FUN-LDA, 2% of the SNPs (functional in the top tissue) explain an estimated 32% of SNP heritability, whereas for IDEAS, 7.1% SNPs explain an estimated 52% of heritability. Essentially, FUN-LDA tends to have higher specificity at the expense of lower sensitivity, whereas methods such as IDEAS and Segway tend to have higher sensitivity and lower specificity. In the next sub-section, we compare these different methods on the basis of the AUROC (area under a receiver operating characteristic curve), which rigorously accounts for sensitivity-specificity tradeoffs, for different test datasets.
In terms of the top tissues identified by each method, it is difficult to make an objective comparison given that the underlying tissues and cell types are not known for many complex traits. However, looking at the results in Tables S5–S7, one can point out several likely mismatches, such as “lung” identified for coronary artery disease by both GenoSkyline and DNase-narrow or “Dnd41 T cell leukemia cell line” and “fetal thymus” identified for epilepsy by DNase and DNase-narrow, respectively. Notably, for type 2 diabetes, FUN-LDA, Segway, and DNase-gapped are the only methods to point to pancreatic tissue, well-known to be relevant to type 2 diabetes.
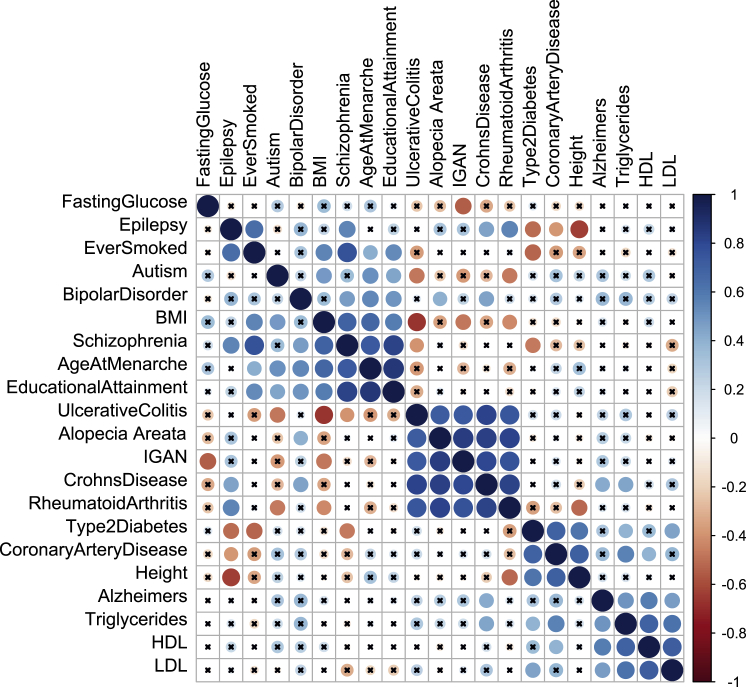
In Figure 2, we show the correlation matrix for the 21 traits on the basis of the Z scores from the LD-score regressions (see Material and Methods for more details on how these pairwise correlations were estimated). This correlation matrix reflects the extent to which traits share the same causal tissues rather than the genetic correlation.66 Three large phenotypic clusters are clearly evident. The most tightly correlated cluster contains autoimmune and inflammatory conditions, including Crohn disease, alopecia areata, rheumatoid arthritis, and IgA nephropathy. As expected, these conditions share the highest functional scores in blood-derived immune cells. The second most strongly inter-correlated cluster is driven by scores in neuronal tissues and consists of BMI, age at menarche, educational attainment, schizophrenia, and smoking history, as well as somewhat weaker correlations with autism, epilepsy, and bipolar disorder. Lastly, there is a clear co-clustering of cardiometabolic traits that map to the tissues of liver, pancreas, and small intestine. Also, as shown, Alzheimer disease clusters with LDL, HDL, and triglycerides, concordant with recent reports on a link between cardiovascular disease and Alzheimer disease.67
Figure 2.
Tissue Correlations for 21 Common Traits
Hierarchical clustering (average linkage method) is used for clustering diseases. The x symbol indicates that those correlations are not significant at the 0.0001 level.
Validations of Our Model’s Predictions and Comparisons with Existing Methods for Functional Annotation
To further assess the accuracy of our predictions and compare with existing approaches for functional prediction, we use variants that have been shown in the literature to have some evidence of a regulatory function. We focus on several main lists of variants with tissue- and/or cell-type-specific functional evidence: (1) eight variants that have been implicated in Mendelian and complex diseases and have additional experimental validation of their functional effects;68, 69, 70, 71, 72, 73, 74, 75 (2) confirmed regulatory variants from a multiplexed reporter assay in LCLs;76 (3) regulatory motifs in 2,000 predicted human enhancers from a MPRA in two human cell lines: liver carcinoma (HepG2) and erythrocytic leukemia (K562);77 (4) a collection of dsQTLs (DNase I sensitivity quantitative trait loci) in LCLs;78 and (5) validated enhancers in 167 ultra-conserved sequence elements.79 We also employed several non-tissue-specific datasets: (6) manually curated, experimentally validated regulatory SNPs;80 (7) allelic-imbalanced SNPs in chromatin accessibility from a large number of DNase sequencing (DNase-seq) assays;81 (8) refined causal SNPs in noncoding regions from different sources (including the Human Gene Mutation Database [HGMD], ClinVar, and OregAnno) and variants from fine-mapping candidate causal SNPs for 39 immune and non-immune diseases in a recent fine-mapping study;80 and (9) eQTLs from 11 uniformly processed fine-mapping studies.38
Tissue- and Cell-Type-Specific Functional Predictions
Noncoding Variants Implicated in Mendelian and Complex Traits with Experimentally Predicted Regulatory Function
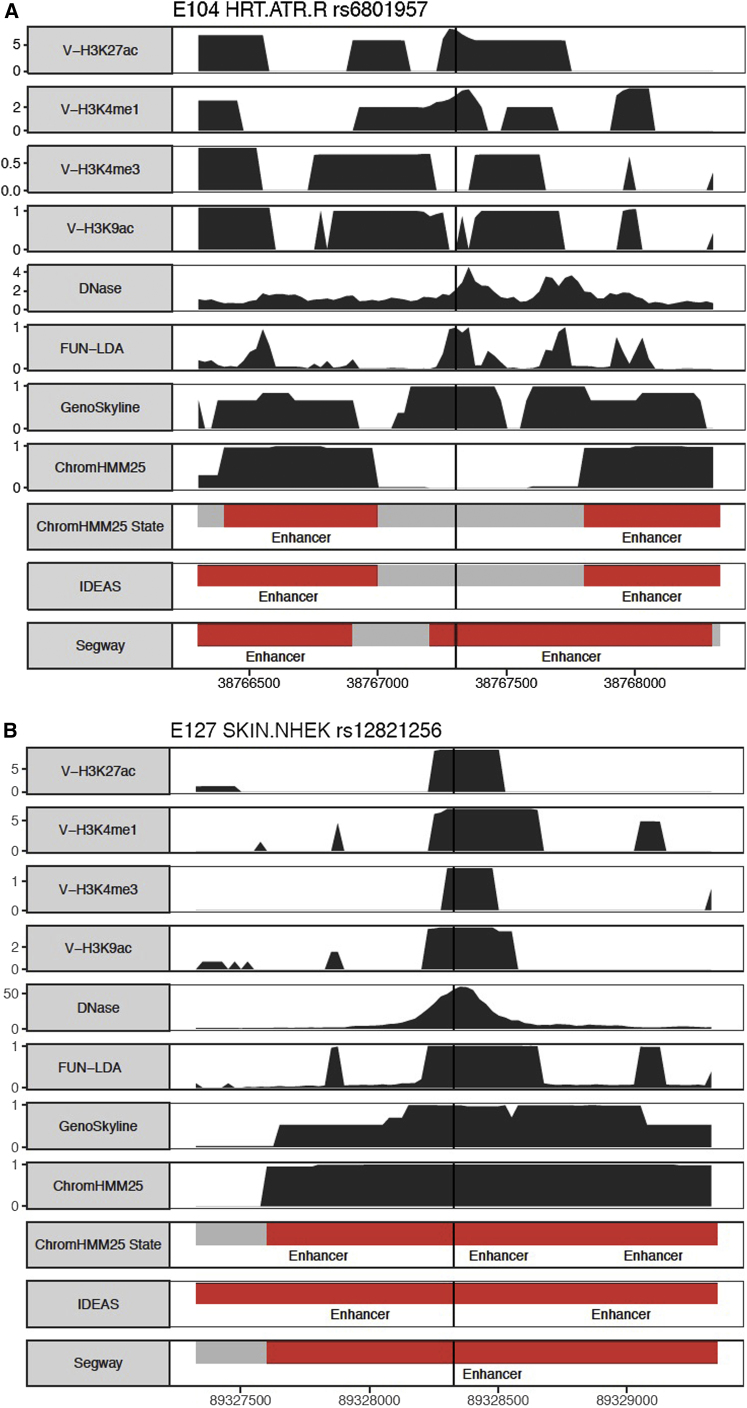
We selected the following eight SNPs that have been shown experimentally to have a regulatory function in particular tissues: rs6801957,68 rs12821256,69 rs12350739,70 rs12740374,71 rs356168,72 rs2473307,73 rs227727,74 and rs144361550.75 In Figure 3 and Figures S10–S15, we show the predictions in ∼2 kb windows centered at these SNPs from the different approaches: FUN-LDA, GenoSkyline, ChromHMM (25-state model), Segway, and IDEAS. For each of these SNPs, we selected the Roadmap tissue that we believe is closest to the tissue used in the original functional studies68, 69, 70, 71, 72, 73, 74, 75 (Table S8). We summarize below the results for two of the SNPs (rs6801957 and rs12821256) that showed more tissue specificity than the other SNPs in the set (i.e., were predicted to be functional in a small number of Roadmap tissues). For the remaining six SNPs, the results are summarized in the Supplemental Material and Methods and Figures S10–S15.
Figure 3.
Functional Predictions from Different Methods
Valley scores for four activating histone marks and DNase, posterior probabilities from FUN-LDA, GenoSkyline, and ChromHMM, and segmentations from ChromHMM, IDEAS, and Segway are shown in 2 kb windows centered around the lead SNPs. For clarity, we highlight in the segmentations only the type of states we consider functional (enhancer states in red and promoter states in blue) for the different segmentation approaches.
In their report, van den Boogaard et al.68 showed that SNP rs6801957, found to be associated with electrocardiography measures in GWASs, is associated with lower SCN5A (MIM: 600163) expression in heart tissue in humans and mice. In Figure 3, we show the predictions for Roadmap tissue E104 (right atrium).
SNP rs12821256, associated with blond hair color in Iceland and the Netherlands, is located in an enhancer and influences expression of KITLG (MIM: 184745) in cultured human keratinocytes.69 In Figure 3, we show the predictions for Roadmap tissue E127 (primary normal human epidermal keratinocytes).
For both SNPs, FUN-LDA assigns a posterior probability of 1 that they are functional in the corresponding tissues. Compared with that of existing integrative methods (last five rows in Figure 3), the region predicted to be functional by FUN-LDA tends to be substantially smaller, and therefore FUN-LDA has a better ability to predict the causal variant in a region of interest than commonly used approaches.
Confirmed Regulatory Variants (emVars) from a Multiplexed Reporter Assay
Tewhey et al.76 applied a new version of the MPRA to identify variants with effects on gene expression. In particular, they applied it to 32,373 variants from 3,642 cis-eQTLs and control regions in LCLs and identified 842 variants showing differential expression between alleles, or emVars (expression-modulating variants). We used this set of 842 emVars as positive control variants. We paired each positive control with four variants tested by using the MPRA where neither allele showed expression different from that of the control and applying a threshold of 0.1 for the Bonferroni-corrected p value. After removing from the list of positive and negative control variants those variants that we could not map to a genomic location by using Ensembl (see Web Resources), we were left with 693 positive control variants and 2,772 negative control variants.
We computed the AUROC and AUPR (area under the precision-recall curve) values for several methods, including FUN-LDA, GenoSkyline, ChromHMM (250state model), Segway, IDEAS, and cepip (both cepip_cell and cepip_combined). For ChromHMM, we partitioned the 25 states into two groups, functional and non-functional; the functional group consisted of TssA (active transcription start site [TSS]), PromU (promoter upstream TSS), PromD1 (promoter downstream TSS 1), PromD2 (promoter downstream TSS 2), EnhA1 (active enhancer 1), EnhA2 (active enhancer 2), and EnhAF (active enhancer flank). For each variant, we used the sum of ChromHMM posterior probabilities for the classes in the functional group above to score the variant. For FUN-LDA, we similarly grouped the designated active promoter and active enhancer classes to form the functional class (see the Material and Methods and Table S1). Segway and IDEAS provided only a functional-class assignment for each position, and we used these assignments to identify the functional variants. Results are shown in Table 3. As shown, FUN-LDA had a higher AUROC (0.707) than the existing methods: ChromHMM (0.669), GenoSkyline (0.673), IDEAS (0.645), Segway (0.622), cepip_cell (0.653), and cepip_combined (0.642). Compared with DNase, FUN-LDA performed significantly better than the two binarized versions—DNase-narrow (0.666) and DNase-gapped (0.659), the two versions commonly used in practice—but it did not outperform the quantitative DNase (0.718). We report the results in terms of both AUROC and AUPR, which exhibited similar patterns.
Table 3.
Tissue- and Cell-Type-Specific Functional Predictions
| Method | AUROC | AUPR |
|---|---|---|
| emVars in Tewhey et al.,76E116 | ||
| FUN-LDA | 0.707 | 0.468 |
| GenoSkyline | 0.673 | 0.394 |
| ChromHMM | 0.669 | 0.420 |
| Segway | 0.622 | 0.356 |
| IDEAS | 0.645 | 0.321 |
| DNase | 0.718 | 0.540 |
| DNase-narrow | 0.666 | 0.406 |
| DNase-gapped | 0.659 | 0.335 |
| cepip_cell | 0.653 | 0.321 |
| cepip_combined | 0.642 | 0.373 |
| Regulatory Motifs in Kheradpour et al.,77E118 and HepG2 | ||
| FUN-LDA | 0.691 | 0.445 |
| GenoSkyline | 0.629 | 0.331 |
| ChromHMM | 0.606 | 0.344 |
| Segway | 0.618 | 0.334 |
| IDEAS | 0.546 | 0.290 |
| DNase | 0.719 | 0.506 |
| DNase-narrow | 0.561 | 0.312 |
| DNase-gapped | 0.550 | 0.291 |
| cepip_cell | 0.592 | 0.300 |
| cepip_combined | 0.641 | 0.364 |
| Regulatory Motifs in Kheradpour et al.,77E123 and K562 | ||
| FUN-LDA | 0.645 | 0.287 |
| GenoSkyline | 0.620 | 0.256 |
| ChromHMM | 0.634 | 0.263 |
| Segway | 0.585 | 0.241 |
| IDEAS | 0.615 | 0.231 |
| DNase | 0.656 | 0.337 |
| DNase-narrow | 0.524 | 0.191 |
| DNase-gapped | 0.565 | 0.205 |
| cepip_cell | 0.606 | 0.217 |
| cepip_combined | 0.625 | 0.247 |
| dsQTLs in Degner et al.,78E116 | ||
| FUN-LDA | 0.750 | 0.374 |
| GenoSkyline | 0.740 | 0.368 |
| ChromHMM | 0.639 | 0.303 |
| Segway | 0.580 | 0.277 |
| IDEAS | 0.677 | 0.330 |
| DNase | 0.823 | 0.474 |
| DNase-narrow | 0.665 | 0.345 |
| DNase-gapped | 0.662 | 0.313 |
| cepip_cell | 0.741 | 0.379 |
| cepip_combined | 0.760 | 0.398 |
| deltaSVM | 0.751 | 0.589 |
| dsQTLs and eQTLs in Degner et al.,78E116 | ||
| FUN-LDA | 0.793 | 0.476 |
| GenoSkyline | 0.756 | 0.372 |
| ChromHMM | 0.721 | 0.403 |
| Segway | 0.648 | 0.340 |
| IDEAS | 0.700 | 0.334 |
| DNase | 0.832 | 0.529 |
| DNase-narrow | 0.713 | 0.376 |
| DNase-gapped | 0.701 | 0.327 |
| cepip_cell | 0.753 | 0.381 |
| cepip_combined | 0.769 | 0.473 |
| deltaSVM | 0.708 | 0.509 |
AUROC and AUPR values for discriminating between variants likely to be functional and control variants are shown. Results are shown for several datasets (three different cell lines) with experimental validation (MPRA) of potential regulatory variants and one dsQTL dataset (dsQTLs and eQTLs contains a subset of dsQTLs that are also eQTLs). Methods include FUN-LDA, GenoSkyline, ChromHMM (25-state model), Segway, IDEAS, DNase (quantitative, DNase-narrow, and DNase-gapped), cepip, and deltaSVM (note that deltaSVM predictions are available only for the dsQTL dataset).
Regulatory Motifs in 2,000 Predicted Human Enhancers from a MPRA
Kheradpour et al.77 used a MPRA to measure the transcriptional levels produced by targeted motif disruptions in 2,104 candidate enhancers in two human cell lines, liver carcinoma (HepG2) and erythrocytic leukemia (K562) cell lines, providing one of the largest resources of experimentally validated enhancer manipulations in human cells. We used as positive control variants those variants where the p value comparing expression values for the sequence including the motif with expression values for the sequences including scrambled versions of the motif was less than 0.05. We used as negative control variants those variants where this p value was greater than 0.1. After removing those variants whose genomic coordinates we could not resolve, we were left with 525 positive and 1,451 negative control variants for HepG2 and 342 positive and 1,578 negative control variants for K562. For all methods, we calculated the scores for these motifs by averaging across all bases in the motifs. As shown in Table 3, FUN-LDA had better accuracy than GenoSkyline, ChromHMM, IDEAS, Segway, and cepip, and for HepG2, the improvement was substantial (e.g., AUROC = 0.691 for FUN-LDA, 0.606 for ChromHMM, 0.629 for GenoSkyline, 0.592 for cepip_cell, and 0.641 for cepip_combined).
We attempted to form the functional group in an objective manner on the basis of prior knowledge of which functional classes from the different segmentation approaches (ChromHMM, Segway, and IDEAS) should be considered active functional elements. We performed an additional analysis where we computed the AUROC for all combinations of states (individual AUROC ≥ 0.5) for each segmentation method and selected the state combination with the highest AUROC for the three datasets above. The results from these analyses are shown in Table S10. Even with this optimized state combination, the AUROCs for the various methods were mostly lower than for our (unbiasedly selected) state combination for FUN-LDA. Furthermore, the state combination with the maximum AUROC often contained states such as poised and bivalent promoters, which would not be considered functional a priori.
dsQTLs in LCLs
We also utilized a collection of dsQTLs in human LCLs, originally identified with the use of DNase I sequencing data from human LCLs.78 Lee et al.29 further processed this list of dsQTLs and generated 579 dsQTLs (with p value < 1 × 10−5) and randomly selected as controls a larger set of common SNPs (minor allele frequency > 5%) only from the top 5% of DHSs that had been used for identifying dsQTLs in the original study.78 After removing variants with missing functional predictions, we were left with 560 dsQTLs in the positive control set. We paired each of these dsQTLs with four randomly selected controls (2,236 negative controls). In addition, Degner et al.78 observed that a substantial fraction (16%) of dsQTLs are also associated with variation in the expression levels of nearby genes (i.e., these loci are also eQTLs). Therefore, we also separately considered 102 dsQTLs that are also eQTLs and paired them with 408 randomly selected (from the set above) negative controls. We present the results in Table 3. It should be noted that the vast majority of dsQTLs reside close to the target DHS, and hence methods such as DNase and deltaSVM are expected to perform well for these datasets. Despite this, FUN-LDA attained an AUROC similar to that of deltaSVM for the dsQTL dataset and a substantially higher value for the dsQTL and eQTL dataset (0.793 for FUN-LDA and 0.708 for deltaSVM). In terms of AUPR, FUN-LDA performance was third after DNase and deltaSVM.
In Figure S16, we use hierarchical clustering to show how the different tissue-specific methods are grouped together. Methods such as DNase and FUN-LDA are closest together in terms of AUROC and tend to perform best across the various tissue-specific datasets, whereas segmentation methods such as ChromHMM, Segway, and IDEAS are grouped together along with the binary DNase scores (DNase-gapped and DNase-narrow).
Ultra-conserved Sequence Elements
Pennacchio et al.79 used extreme evolutionary sequence conservation as a filter to identify putative gene regulatory sequences. Using this approach, they identified 167 ultra-conserved sequence elements and then used a transgenic mouse enhancer assay that linked each of these candidate elements to a mouse promoter fused to a lacZ reporter gene. In total, 75 of 167 candidate sequences functioned reproducibly as tissue-specific enhancers of gene expression by the readout of lacZ expression at mouse embryonic day 11.5 (E11.5). Out of 75 positive fragments, 50 mapped to a single anatomical structure in the E11.5 embryonic tissue, whereas the remaining 25 enhancers directed expression to two or more anatomical structures. Here, we compare the functional scores for the variants falling into these 75 positive enhancers with scores of variants in the remaining 92 elements. In Table S11, we show the top Roadmap tissue for each method and the corresponding AUROC values. Notably, most methods, including FUN-LDA, selected embryonic tissue as the top tissue, consistent with the conducted experiment. Importantly, FUN-LDA outperformed all other methods except for GenoSkyline in predicting functional elements on the basis of these enhancer assays. Results based on AUPR values show similar patterns. It is important to bear in mind that the negative controls we used in this analysis (and more generally in our other analyses) could in fact be functional in different experimental environments, and the performance of the prediction methods we considered was affected by such misclassifications.
Widths of Predicted Functional Regions for Each Method
In Figure S17, we show the distribution of the widths of predicted functional regions that include validated functional variants from the three lists above for the tissue- and cell-type-specific methods. We determined the width of the functional region around a variant by finding the width of the window around the variant for which the value of the score (the probability) was greater than 0.5. Widths were truncated at 20,000 bp (so all widths greater than 20,000 bp were represented as 20,000 bp). Regions predicted by FUN-LDA were predicted to be substantially narrower than those predicted by the other methods (median width was 600 bp for FUN-LDA, 2,100 bp for ChromHMM, 1,800 bp for GenoSkyline, 1,100 bp for DNase-narrow, 4,400 bp for DNase-gapped, 4,100 bp for IDEAS, 1,300 bp for Segway, and 2,300 bp for cepip_cell); hence, compared with existing methods, FUN-LDA has the ability to more precisely localize the functional variants in a region of interest.
Comparison with Organism-Level Functional Prediction Methods on Tissue- and Cell-Type-Specific Datasets
Whereas above we focused on comparing FUN-LDA with other tissue- and cell-type-specific functional prediction methods on datasets with available tissue- and cell-type-specific predictions for variants, we also compared them on the same datasets with some of the more popular organism-level prediction methods, including phyloP,36 CADD, DANN, Eigen, DeepSea, and LINSIGHT. The results are in Table S12. In most cases, FUN-LDA had a higher AUROC and AUPR than the organism-level methods, illustrating that prediction at the level of the tissue or cell type is more informative than it should be given that the underlying functional effects of variants are tissue or cell-type specific and can vary from tissue to tissue. Note, however, that for the dsQTLs, DeepSea performed better than FUN-LDA most likely because DeepSea was trained to predict DHSs, and the vast majority of dsQTLs reside close to the target DHSs. This is similar to what we observed with DNase as well, namely that DNase substantially outperformed other methods, including FUN-LDA, on those datasets.
Organism-Level Functional Predictions
In addition to considering tissue- and cell-type-specific functional predictions, we also considered applications to datasets where the functional evidence is not restricted to a particular tissue. In such cases, for the tissue and cell-type functional prediction methods discussed above, we defined the functional score for a variant as the maximum of the functional scores across the 127 tissues in ENCODE and Roadmap (this is the most severe functional score and is similar to common practice, e.g., when a variant matches multiple functional categories in the Ensembl Variant Effect Predictor). We were unable to include cepip and deltaSVM in these comparisons because these two methods’ scores are not yet available across all 127 tissues and cell types in Roadmap. We compared our proposed method with several popular organism-level functional prediction methods, including phyloP (primate), CADD, Eigen, DANN, DeepSea, and LINSIGHT.
Validated Regulatory SNPs
We used a set of 76 manually curated experimentally validated regulatory SNPs80 and the same set of 156 frequency-matched background SNPs within 10 kb of the curated causal variants as in Li et al.80 The results are shown in Table 4. FUN-LDA achieved an excellent AUROC of 0.878, substantially outperforming the organism-level functional prediction methods phyloP (0.575), CADD (0.718), Eigen (0.806), DANN (0.711), DeepSea (0.774), and LINSIGHT (0.818).
Table 4.
Organism-Level Functional Prediction
| Method | AUROC | AUPR |
|---|---|---|
| Validated Regulatory SNPs | ||
| FUN-LDA-max | 0.878 | 0.764 |
| GenoSkyline-max | 0.846 | 0.647 |
| ChromHMM-max | 0.865 | 0.796 |
| Segway-max | 0.711 | 0.461 |
| IDEAS-max | 0.694 | 0.451 |
| DNase-max | 0.885 | 0.818 |
| DNase-narrow-max | 0.828 | 0.707 |
| DNase-gapped-max | 0.807 | 0.590 |
| Eigen | 0.806 | 0.679 |
| CADD | 0.718 | 0.492 |
| DANN | 0.711 | 0.531 |
| LINSIGHT | 0.818 | 0.615 |
| PhyloP | 0.575 | 0.417 |
| DeepSea | 0.774 | 0.680 |
| Allelic-Imbalanced SNPs in Chromatin Accessibility | ||
| FUN-LDA-max | 0.935 | 0.899 |
| GenoSkyline-max | 0.906 | 0.849 |
| ChromHMM-max | 0.863 | 0.846 |
| Segway-max | 0.793 | 0.688 |
| IDEAS-max | 0.794 | 0.694 |
| DNase-max | 0.968 | 0.952 |
| DNase-narrow-max | 0.869 | 0.859 |
| DNase-gapped-max | 0.849 | 0.778 |
| Eigen | 0.753 | 0.732 |
| CADD | 0.692 | 0.638 |
| DANN | 0.619 | 0.557 |
| LINSIGHT | 0.880 | 0.815 |
| PhyloP | 0.581 | 0.584 |
| DeepSea | 0.865 | 0.839 |
| Refined Causal SNPs | ||
| FUN-LDA-max | 0.803 | 0.534 |
| GenoSkyline-max | 0.811 | 0.529 |
| ChromHMM-max | 0.748 | 0.504 |
| Segway-max | 0.714 | 0.332 |
| IDEAS-max | 0.720 | 0.342 |
| DNase-max | 0.807 | 0.510 |
| DNase-narrow-max | 0.680 | 0.411 |
| DNase-gapped-max | 0.756 | 0.418 |
| Eigen | 0.655 | 0.206 |
| CADD | 0.591 | 0.122 |
| DANN | 0.587 | 0.109 |
| LINSIGHT | 0.775 | 0.435 |
| PhyloP | 0.560 | 0.257 |
| DeepSea | 0.686 | 0.379 |
| Fine-Mapped eQTLs | ||
| FUN-LDA-max | 0.775 | 0.727 |
| GenoSkyline-max | 0.785 | 0.725 |
| ChromHMM-max | 0.680 | 0.671 |
| Segway-max | 0.687 | 0.603 |
| IDEAS-max | 0.686 | 0.605 |
| DNase-max | 0.778 | 0.721 |
| DNase-narrow-max | 0.615 | 0.611 |
| DNase-gapped-max | 0.707 | 0.655 |
| Eigen | 0.653 | 0.616 |
| CADD | 0.621 | 0.562 |
| DANN | 0.573 | 0.506 |
| LINSIGHT | 0.777 | 0.685 |
| PhyloP | 0.548 | 0.519 |
| DeepSea | 0.684 | 0.629 |
AUROC and AUPR values for discriminating between variants likely to be functional and control variants for are shown for four non-tissue specific datasets. Methods include FUN-LDA-max (maximum across 127 different tissues), GenoSkyline-max, ChromHMM-max (25-state model), Segway-max, IDEAS-max, and DNase-max (quantitative, DNase-narrow, and DNase-gapped). In addition, results for several organism-level functional prediction methods, including phyloP (primate), Eigen, CADD, DANN, DeepSea, and LINSIGHT, are reported.
Allelic-Imbalanced SNPs in Chromatin Accessibility
We also considered a dataset of allelic-imbalanced SNPs in chromatin accessibility (9,456 positive controls and 9,678 negative controls) identified by a large number of DNase-seq assays.81 The negative controls were frequency-matched background SNPs around the nearest TSS of randomly selected genes. After removing variants with missing functional predictions, we were left with 8,592 dsQTLs and 9,610 controls. It should be noted that the allelic-imbalanced SNPs were identified by DNase-seq assays, and hence DNase was expected to perform well for this dataset. As shown in Table 4, FUN-LDA performed very well with an AUROC of 0.935, higher than that of tissue-specific functional prediction methods GenoSkyline (0.906), ChromHMM (0.863), Segway (0.793), and IDEAS (0.794) and substantially better than that of organism-level functional prediction methods phyloP (0.581), CADD (0.692), Eigen (0.753), DANN (0.619), DeepSea (0.865), and LINSIGHT (0.880).
Refined Causal SNPs
We used a collection of 5,229 refined “causal” SNPs in noncoding regions from different sources, including the HGMD, ClinVar, and ORegAnno, and variants from fine-mapping candidate causal SNPs for 39 immune and non-immune diseases in a recent fine-mapping study.80 The controls consisted of 20,916 randomly selected frequency-matched noncoding SNPs. FUN-LDA performed very well with an AUROC of 0.803, outperforming almost all the other functional prediction methods, especially the organism-level prediction methods (Table 4): phyloP (0.560), CADD (0.591), Eigen (0.655), DANN (0.587), DeepSea (0.686), and LINSIGHT (0.775).
Fine-Mapped eQTLs
Finally, we used a collection of eQTLs (31,118 positive controls and 36,540 negative controls) from the uniformly processed eQTL fine-mapping data in Brown et al.38 The eQTLs were originally identified by multi-trait Bayesian linear regression models from 11 studies on 7 tissues and cell lines and then pre-processed by Li et al.80 for the generation of a dataset of 31,118 most likely functional eQTLs (our positive controls) and 36,540 frequency-matched background SNPs around the nearest TSS of randomly selected genes (our negative controls). FUN-LDA performed very well with an AUROC of 0.775, which was the same as that of LINSIGHT but substantially better than that of phyloP (0.548), CADD (0.621), Eigen (0.653), DANN (0.573), and DeepSea (0.684). With AUROCs of 0.785 and 0.778, respectively, GenoSkyline and DNase performed slightly better than FUN-LDA for this dataset.
In Figure S18, we use hierarchical clustering to show how the different organism-level methods are grouped together. As with the tissue-specific datasets, methods such as DNase and FUN-LDA are closest together in terms of AUROC and tend to perform best across the various datasets, whereas segmentation methods such as ChromHMM, Segway, and IDEAS are grouped together along with the binary DNase scores (DNase-gapped and DNase-narrow).
Discussion
Here, we have introduced FUN-LDA, an unsupervised approach that uses histone modification and DNase data from the ENCODE and Roadmap Epigenomics projects for the functional prediction of genetic variation in specific cell types and tissues, and have provided comparisons with commonly used functional annotation methods both at the tissue- and cell-type-specific level and at the organism level. FUN-LDA is based on a mixture model that focuses on identifying the narrow genomic regions whose disruption is most likely to interfere with function in a particular cell type or tissue. Such context-specific functional prediction of genetic variation is essential for understanding the function of noncoding variation across cell types and tissues and for interpreting genetic variants uncovered in GWASs and sequencing studies. Although existing segmentation approaches can be used to derive a numeric functional score as well, we have shown that they tend to be less accurate at predicting functional effects and tend to predict wider functional regions than the proposed approach. Relative to other recently developed functional scores, such as GenoSkyline, FUN-LDA can have substantially better prediction accuracy, can use annotation data on the original scale (e.g., quantitative or binary), and furthermore makes it explicit which classes are considered functionally active, namely active promoters and active enhancers, providing an attractive tool for functional scoring of variants.
In terms of prediction accuracy, we have shown that, overall, FUN-LDA outperforms existing methods over a variety of test datasets, sometimes substantially. In particular, FUN-LDA has substantially better accuracy than popular organism-level functional scores, such as phyloP, CADD, Eigen, DANN, DeepSea, and LINSIGHT. We have also shown that quantitative DNase can have a higher predictive power than FUN-LDA and other tissue- and cell-type-specific functional prediction methods, although the difference between FUN-LDA and DNase is minor in most comparisons and is smaller than the difference between FUN-LDA and other integrative methods (except for the DNase-based datasets, such as dsQTLs and allelic-imbalanced SNPs in chromatin accessibility, where DNase has an inherent advantage). This observation is concordant with a recent study showing that within open chromatin regions, transcription factor binding is strongly correlated with the quantitative level of chromatin accessibility (as measured by DNase-seq).82 Therefore, the proposed FUN-LDA method, by being able to integrate annotation data with arbitrary distributions, has clear advantages over other mixture-based methods such as GenoSkyline and ChromHMM, which make use of binary peak calls. However, not being a probabilistic score is a significant deficiency of DNase (e.g., it is more difficult to implement and interpret enrichment analyses shown here for eQTLs and LD-score regression analyses), and in practice, in the vast majority of cases, researchers use binary DNase peak calls (DNase-narrow and DNase-gapped) rather than quantitative DNase scores; as we have shown, FUN-LDA significantly outperforms DNase peaks on the metrics we considered. One approach of interest would be to develop a probabilistic DNase score. Using FUN-LDA or other non-parametric mixture models with only DNase is not straightforward, given that in general, non-parametric mixture models are not identifiable with only one dimension, so one needs to either add additional annotations as we have done here or make parametric assumptions on the distribution of DNase (which is not straightforward). We leave this extension to future work.
These cell-type- and tissue-specific functional scores have numerous applications. We have shown here that eQTLs from several large studies, such as GTEx, Geuvadis, and the TwinUK cohort, are most enriched in the functional components from relevant Roadmap tissues. As previously shown,39 and as illustrated here as well, they can be used for inferring the most relevant cell types and tissues for a trait of interest and can help focus the search for causal variants in complex traits by restricting the set of candidate variants to only those that are predicted to be functional in tissues relevant for the trait under consideration. Beyond the applications shown here, such functional predictions have numerous other applications. They can naturally be used in gene-discovery studies to potentially improve power in sequence-based association tests such as SKAT and burden83, 84 and in fine-mapping studies.85, 86 They can also be used in identifying regulatory regions that are depleted in functional variation in a specific tissue, similar to recent efforts to identify coding regions that are depleted in functional variation (e.g., missense, nonsense, and splice acceptor or donor variants).9 Other applications include improving power of trans-eQTL studies by using cell-type- and tissue-specific functional predictions as prior information. Similarly, studies on gene-gene and gene-environment interactions can benefit from an analysis focused on variants predicted to be functional in a cell type or tissue relevant to the trait under study.
Choosing the number of functional classes in the LDA model is not an easy task, partly because the number of functional classes is not well defined. Here, we focused on a model that includes nine functional classes and is based on combining an objective measure (such as the perplexity of the model), visual inspection of the resulting states, and biological knowledge. In our investigations, the results were not very sensitive to the number of classes, but models with fewer classes (i.e., 3–7) were not able to distinguish among different functional classes (such as enhancers and promoters). There is some subjectivity in any method that seeks to partition the genome into functional classes both in terms of the number of such classes and in terms of their interpretation. Further experiments that produce catalogs of specific types of elements with validated tissue-specific functions would aid in determining the number of states that a genomic annotation model should have and interpreting those states, leading to potential improvements in the accuracy of such functional predictors. Such tissue-specific experimental data would also allow the use of supervised methods that could lead to improved tissue-specific functional scores.
Unlike our method, most of the existing segmentation methods smooth the genomic signal spatially. Although they thereby use information from neighboring regions in making predictions for a particular variant, they can be less able to predict functionality of narrow regions with different histone-modification profiles from neighboring regions. Another difference between our method and those that use binary peak calls is that ours can incorporate the quantitative level of the functional annotations, which can be important; for example, in the case of DNase, it has been recently shown that the quantitative level of chromatin accessibility is strongly correlated with transcription factor binding.82 Furthermore, the use of the valley score allows our method to predict narrower functional regions than existing methods.
Overall, we propose a general framework for integrating various features in order to predict the functional effects of variants in noncoding regions of the genome. Although the epigenetic features we integrate are mostly helpful for predicting the effects of variants in cis-regulatory elements, such as promoters, enhancers, silencers, and insulators, the integration of additional features can lead to the discovery of other types of functional variants, such as those with effects on post-transcriptional regulation by alteration of RNA secondary structure or RNA-protein interactions. Similar to segmentation approaches (such as ChromHMM and Segway), our LDA framework could also be used for segmenting the genome into detailed functional classes.
We have computed FUN-LDA posterior probabilities for every position in the human genome for 127 tissue and cell types available in Roadmap. These scores are available on our website and can be imported into the UCSC Genome Browser. Note also that it is easy to make predictions in a new tissue once the model has been fit to the tissues in Roadmap. Furthermore, as with some other existing methods,19 it is possible to make predictions in a new tissue even if not all the epigenetic features we included are available, as long as one can impute the missing features by taking advantage of the correlations of epigenetic signals across both marks and samples as in ChromImpute.20
Acknowledgments
We gratefully acknowledge support by National Institutes of Health grants MH106910 and MH095797 (D.B., Z.H., and I.I.-L.), AR065963 (L.P. and A.C.), DK105124 (K.K.), and MH100233 (J.D.B.), the Herbert Irving Scholars Award (K.K.), the Seaver Foundation (J.D.B.), and the ATIP-Avenir program (V.B.). We thank Bin Xu and Badri Vardarajan for helpful discussions. We thank Andrew Brown for making the data on lead eQTLs in the Geuvadis and TwinsUK cohort available to us.
Published: May 3, 2018
Footnotes
Supplemental Data include Supplemental Material and Methods, 18 figures, and 13 tables and can be found with this article online at https://doi.org/10.1016/j.ajhg.2018.03.026.
Web Resources
1000 Genomes, http://www.1000genomes.org/
cepip, http://jjwanglab.org/cepip/
ChromHMM, http://compbio.mit.edu/ChromHMM/
deltaSVM, http://www.beerlab.org/deltasvm/
ENCODE, https://www.encodeproject.org/
Ensembl, http://grch37.ensembl.org/index.html
FUNLDA: Genomic Latent Dirichlet Allocation, https://cran.r-project.org/web/packages/FUNLDA
FUN-LDA, http://www.funlda.com/
GenoSkyline, http://genocanyon.med.yale.edu/GenoSkyline
GIANT consortium data files (BMI, height), http://www.broadinstitute.org/collaboration/giant/index.php/GIANT_consortium_data_files
GTEx Portal, http://www.gtexportal.org/home/
GTEx analysis methods, https://www.gtexportal.org/home/documentationPage#staticTextAnalysisMethods
Reg2Map, https://personal.broadinstitute.org/meuleman/reg2map/HoneyBadger2-intersect_release/
Roadmap Epigenomics, http://www.roadmapepigenomics.org/
UCSC Genome Browser, https://genome.ucsc.edu/
GWAS Summary Statistics:
Age at Menarche, http://www.reprogen.org/Menarche_Nature2014_GWASMetaResults_17122014.zip
Alopecia areata (link no longer available), http://www.broadinstitute.org/∼sripke/sharelinks/sRSxpynHPaYRJ1SnYXD17eo3qK8IE6daner_ALO4_1011b_mdsex/
Alzheimer disease, http://web.pasteur-lille.fr/en/recherche/u744/igap/igap_download.php
Autism, http://www.med.unc.edu/pgc/shared-methods/data-access-portal
Bipolar disorder, http://www.med.unc.edu/pgc/files/resultfiles/pgc.bip.2012-04.zip
Crohn disease, ftp://ftp.sanger.ac.uk/pub/consortia/ibdgenetics/cd-meta.txt.gz
Coronary artery disease, ftp://ftp.sanger.ac.uk/pub/cardiogramplusc4d/cardiogram_gwas_results.zip
Educational attainment, http://ssgac.org/documents/SSGAC_Rietveld2013.zip
Epilepsy, http://www.epigad.org/gwas_ilae2014/ILAE_All_Epi_11.8.14.txt.gz
Ever smoked, http://www.med.unc.edu/pgc/files/resultfiles/tag.evrsmk.tbl.gz
Fasting glucose, ftp://ftp.sanger.ac.uk/pub/magic/MAGIC_Manning_et_al_FastingGlucose_MainEffect.txt.gz
HDL, http://www.broadinstitute.org/mpg/pubs/lipids2010/HDL_ONE_Eur.tbl.sorted.gz
IgA nephropathy, https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000431.v2.p1
LDL, http://www.broadinstitute.org/mpg/pubs/lipids2010/LDL_ONE_Eur.tbl.sorted.gz
Rheumatoid arthritis, http://plaza.umin.ac.jp/yokada/datasource/files/GWASMetaResults/RA_GWASmeta_European_v2.txt.gz
Schizophrenia, http://www.med.unc.edu/pgc/files/resultfiles/scz2.snp.results.txt.gz
Triglycerides, http://www.broadinstitute.org/mpg/pubs/lipids2010/TG_ONE_Eur.tbl.sorted.gz
Type 2 diabetes, http://www.diagram-consortium.org/downloads.html
Ulcerative colitis, ftp://ftp.sanger.ac.uk/pub/consortia/ibdgenetics/ucmeta-sumstats.txt.gz
Supplemental Data
References
- 1.Lindblad-Toh K., Garber M., Zuk O., Lin M.F., Parker B.J., Washietl S., Kheradpour P., Ernst J., Jordan G., Mauceli E., Broad Institute Sequencing Platform and Whole Genome Assembly Team. Baylor College of Medicine Human Genome Sequencing Center Sequencing Team. Genome Institute at Washington University A high-resolution map of human evolutionary constraint using 29 mammals. Nature. 2011;478:476–482. doi: 10.1038/nature10530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Khurana E., Fu Y., Colonna V., Mu X.J., Kang H.M., Lappalainen T., Sboner A., Lochovsky L., Chen J., Harmanci A., 1000 Genomes Project Consortium Integrative annotation of variants from 1092 humans: application to cancer genomics. Science. 2013;342:1235587. doi: 10.1126/science.1235587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.ENCODE Project Consortium An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Altshuler D., Daly M.J., Lander E.S. Genetic mapping in human disease. Science. 2008;322:881–888. doi: 10.1126/science.1156409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Khurana E., Fu Y., Chakravarty D., Demichelis F., Rubin M.A., Gerstein M. Role of non-coding sequence variants in cancer. Nat. Rev. Genet. 2016;17:93–108. doi: 10.1038/nrg.2015.17. [DOI] [PubMed] [Google Scholar]
- 6.Kellis M., Wold B., Snyder M.P., Bernstein B.E., Kundaje A., Marinov G.K., Ward L.D., Birney E., Crawford G.E., Dekker J. Defining functional DNA elements in the human genome. Proc. Natl. Acad. Sci. USA. 2014;111:6131–6138. doi: 10.1073/pnas.1318948111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Melnikov A., Murugan A., Zhang X., Tesileanu T., Wang L., Rogov P., Feizi S., Gnirke A., Callan C.G., Jr., Kinney J.B. Systematic dissection and optimization of inducible enhancers in human cells using a massively parallel reporter assay. Nat. Biotechnol. 2012;30:271–277. doi: 10.1038/nbt.2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cong L., Ran F.A., Cox D., Lin S., Barretto R., Habib N., Hsu P.D., Wu X., Jiang W., Marraffini L.A., Zhang F. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Petrovski S., Wang Q., Heinzen E.L., Allen A.S., Goldstein D.B. Genic intolerance to functional variation and the interpretation of personal genomes. PLoS Genet. 2013;9:e1003709. doi: 10.1371/journal.pgen.1003709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kundaje A., Meuleman W., Ernst J., Bilenky M., Yen A., Heravi-Moussavi A., Kheradpour P., Zhang Z., Wang J., Ziller M.J., Roadmap Epigenomics Consortium Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kircher M., Witten D.M., Jain P., O’Roak B.J., Cooper G.M., Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 2014;46:310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fu Y., Liu Z., Lou S., Bedford J., Mu X.J., Yip K.Y., Khurana E., Gerstein M. FunSeq2: a framework for prioritizing noncoding regulatory variants in cancer. Genome Biol. 2014;15:480. doi: 10.1186/s13059-014-0480-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ionita-Laza I., McCallum K., Xu B., Buxbaum J.D. A spectral approach integrating functional genomic annotations for coding and noncoding variants. Nat. Genet. 2016;48:214–220. doi: 10.1038/ng.3477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Quang D., Chen Y., Xie X. DANN: a deep learning approach for annotating the pathogenicity of genetic variants. Bioinformatics. 2015;31:761–763. doi: 10.1093/bioinformatics/btu703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou J., Troyanskaya O.G. Predicting effects of noncoding variants with deep learning-based sequence model. Nat. Methods. 2015;12:931–934. doi: 10.1038/nmeth.3547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang Y.F., Gulko B., Siepel A. Fast, scalable prediction of deleterious noncoding variants from functional and population genomic data. Nat. Genet. 2017;49:618–624. doi: 10.1038/ng.3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bannister A.J., Kouzarides T. Regulation of chromatin by histone modifications. Cell Res. 2011;21:381–395. doi: 10.1038/cr.2011.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Friedman N., Rando O.J. Epigenomics and the structure of the living genome. Genome Res. 2015;25:1482–1490. doi: 10.1101/gr.190165.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ernst J., Kellis M. ChromHMM: automating chromatin-state discovery and characterization. Nat. Methods. 2012;9:215–216. doi: 10.1038/nmeth.1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ernst J., Kellis M. Large-scale imputation of epigenomic datasets for systematic annotation of diverse human tissues. Nat. Biotechnol. 2015;33:364–376. doi: 10.1038/nbt.3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hoffman M.M., Buske O.J., Wang J., Weng Z., Bilmes J.A., Noble W.S. Unsupervised pattern discovery in human chromatin structure through genomic segmentation. Nat. Methods. 2012;9:473–476. doi: 10.1038/nmeth.1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zacher B., Michel M., Schwalb B., Cramer P., Tresch A., Gagneur J. Accurate Promoter and Enhancer Identification in 127 ENCODE and Roadmap Epigenomics Cell Types and Tissues by GenoSTAN. PLoS ONE. 2017;12:e0169249. doi: 10.1371/journal.pone.0169249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mammana A., Chung H.R. Chromatin segmentation based on a probabilistic model for read counts explains a large portion of the epigenome. Genome Biol. 2015;16:151. doi: 10.1186/s13059-015-0708-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Biesinger J., Wang Y., Xie X. Discovering and mapping chromatin states using a tree hidden Markov model. BMC Bioinformatics. 2013;14(Suppl 5):S4. doi: 10.1186/1471-2105-14-S5-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang Y., An L., Yue F., Hardison R.C. Jointly characterizing epigenetic dynamics across multiple human cell types. Nucleic Acids Res. 2016;44:6721–6731. doi: 10.1093/nar/gkw278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang Y., Hardison R.C. Accurate and Reproducible Functional Maps in 127 Human Cell Types via 2D Genome Segmentation. BioRxiv. 2017 doi: 10.1101/118752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Song J., Chen K.C. Spectacle: fast chromatin state annotation using spectral learning. Genome Biol. 2015;16:33. doi: 10.1186/s13059-015-0598-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lu Q., Powles R.L., Wang Q., He B.J., Zhao H. Integrative Tissue-Specific Functional Annotations in the Human Genome Provide Novel Insights on Many Complex Traits and Improve Signal Prioritization in Genome Wide Association Studies. PLoS Genet. 2016;12:e1005947. doi: 10.1371/journal.pgen.1005947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee D., Gorkin D.U., Baker M., Strober B.J., Asoni A.L., McCallion A.S., Beer M.A. A method to predict the impact of regulatory variants from DNA sequence. Nat. Genet. 2015;47:955–961. doi: 10.1038/ng.3331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li M.J., Li M., Liu Z., Yan B., Pan Z., Huang D., Liang Q., Ying D., Xu F., Yao H. cepip: context-dependent epigenomic weighting for prioritization of regulatory variants and disease-associated genes. Genome Biol. 2017;18:52. doi: 10.1186/s13059-017-1177-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Blei D.M., Ng A.Y., Jordan M.I. Latent Dirichlet Allocation. J. Mach. Learn. Res. 2003;3:993–1022. [Google Scholar]
- 32.Silverman B.W. Chapman & Hall; 1986. Density Estimation for Statistics and Data Analysis. [Google Scholar]
- 33.Ramsey S.A., Knijnenburg T.A., Kennedy K.A., Zak D.E., Gilchrist M., Gold E.S., Johnson C.D., Lampano A.E., Litvak V., Navarro G. Genome-wide histone acetylation data improve prediction of mammalian transcription factor binding sites. Bioinformatics. 2010;26:2071–2075. doi: 10.1093/bioinformatics/btq405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hagai Attias (1999) Inferring parameters and structure of latent variable models by variational bayes. Proceedings of the Fifteenth conference on Uncertainty in artificial intelligence (Morgan Kaufmann Publishers), pp. 21–30.
- 35.Libbrecht M.W., Rodriguez O., Weng Z., Hoffman M., Bilmes J.A., Noble W.S. A unified encyclopedia of human functional DNA elements through fully automated annotation of 164 human cell types. BioRxiv. 2017 doi: 10.1101/086025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cooper G.M., Stone E.A., Asimenos G., Green E.D., Batzoglou S., Sidow A., NISC Comparative Sequencing Program Distribution and intensity of constraint in mammalian genomic sequence. Genome Res. 2005;15:901–913. doi: 10.1101/gr.3577405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.The GTEx Consortium Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348:648–660. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brown A.A., Viñuela A., Delaneau O., Spector T.D., Small K.S., Dermitzakis E.T. Predicting causal variants affecting expression by using whole-genome sequencing and RNA-seq from multiple human tissues. Nat. Genet. 2017;49:1747–1751. doi: 10.1038/ng.3979. [DOI] [PubMed] [Google Scholar]
- 39.Finucane H.K., Bulik-Sullivan B., Gusev A., Trynka G., Reshef Y., Loh P.R., Anttila V., Xu H., Zang C., Farh K., ReproGen Consortium. Schizophrenia Working Group of the Psychiatric Genomics Consortium. RACI Consortium Partitioning heritability by functional annotation using genome-wide association summary statistics. Nat. Genet. 2015;47:1228–1235. doi: 10.1038/ng.3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gao X., Starmer J., Martin E.R. A multiple testing correction method for genetic association studies using correlated single nucleotide polymorphisms. Genet. Epidemiol. 2008;32:361–369. doi: 10.1002/gepi.20310. [DOI] [PubMed] [Google Scholar]
- 41.Heintzman N.D., Hon G.C., Hawkins R.D., Kheradpour P., Stark A., Harp L.F., Ye Z., Lee L.K., Stuart R.K., Ching C.W. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459:108–112. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Battle A., Brown C.D., Engelhardt B.E., Montgomery S.B., GTEx Consortium. Laboratory, Data Analysis &Coordinating Center (LDACC)—Analysis Working Group. Statistical Methods groups—Analysis Working Group. Enhancing GTEx (eGTEx) groups. NIH Common Fund. NIH/NCI. NIH/NHGRI. NIH/NIMH. NIH/NIDA. Biospecimen Collection Source Site—NDRI. Biospecimen Collection Source Site—RPCI. Biospecimen Core Resource—VARI. Brain Bank Repository—University of Miami Brain Endowment Bank. Leidos Biomedical—Project Management. ELSI Study. Genome Browser Data Integration &Visualization—EBI. Genome Browser Data Integration &Visualization—UCSC Genomics Institute, University of California Santa Cruz. Lead analysts. Laboratory, Data Analysis &Coordinating Center (LDACC) NIH program management. Biospecimen collection. Pathology. eQTL manuscript working group Nature. 2017;550:204–213. [Google Scholar]
- 43.Perry J.R., Day F., Elks C.E., Sulem P., Thompson D.J., Ferreira T., He C., Chasman D.I., Esko T., Thorleifsson G., Australian Ovarian Cancer Study. GENICA Network. kConFab. LifeLines Cohort Study. InterAct Consortium. Early Growth Genetics (EGG) Consortium Parent-of-origin-specific allelic associations among 106 genomic loci for age at menarche. Nature. 2014;514:92–97. doi: 10.1038/nature13545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Betz R.C., Petukhova L., Ripke S., Huang H., Menelaou A., Redler S., Becker T., Heilmann S., Yamany T., Duvic M. Genome-wide meta-analysis in alopecia areata resolves HLA associations and reveals two new susceptibility loci. Nat. Commun. 2015;6:5966. doi: 10.1038/ncomms6966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lambert J.C., Ibrahim-Verbaas C.A., Harold D., Naj A.C., Sims R., Bellenguez C., DeStafano A.L., Bis J.C., Beecham G.W., Grenier-Boley B., European Alzheimer’s Disease Initiative (EADI) Genetic and Environmental Risk in Alzheimer’s Disease. Alzheimer’s Disease Genetic Consortium. Cohorts for Heart and Aging Research in Genomic Epidemiology Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat. Genet. 2013;45:1452–1458. doi: 10.1038/ng.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cross-Disorder Group of the Psychiatric Genomics Consortium Identification of risk loci with shared effects on five major psychiatric disorders: a genome-wide analysis. Lancet. 2013;381:1371–1379. doi: 10.1016/S0140-6736(12)62129-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Psychiatric GWAS Consortium Bipolar Disorder Working Group Large-scale genome-wide association analysis of bipolar disorder identifies a new susceptibility locus near ODZ4. Nat. Genet. 2011;43:977–983. doi: 10.1038/ng.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Speliotes E.K., Willer C.J., Berndt S.I., Monda K.L., Thorleifsson G., Jackson A.U., Lango Allen H., Lindgren C.M., Luan J., Mägi R., MAGIC. Procardis Consortium Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat. Genet. 2010;42:937–948. doi: 10.1038/ng.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schunkert H., König I.R., Kathiresan S., Reilly M.P., Assimes T.L., Holm H., Preuss M., Stewart A.F., Barbalic M., Gieger C., Cardiogenics. CARDIoGRAM Consortium Large-scale association analysis identifies 13 new susceptibility loci for coronary artery disease. Nat. Genet. 2011;43:333–338. doi: 10.1038/ng.784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jostins L., Ripke S., Weersma R.K., Duerr R.H., McGovern D.P., Hui K.Y., Lee J.C., Schumm L.P., Sharma Y., Anderson C.A., International IBD Genetics Consortium (IIBDGC) Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491:119–124. doi: 10.1038/nature11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Petukhova L., Christiano A.M. Functional Interpretation of Genome-Wide Association Study Evidence in Alopecia Areata. J. Invest. Dermatol. 2016;136:314–317. doi: 10.1038/JID.2015.402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xing L., Dai Z., Jabbari A., Cerise J.E., Higgins C.A., Gong W., de Jong A., Harel S., DeStefano G.M., Rothman L. Alopecia areata is driven by cytotoxic T lymphocytes and is reversed by JAK inhibition. Nat. Med. 2014;20:1043–1049. doi: 10.1038/nm.3645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yokoyama J.S., Wang Y., Schork A.J., Thompson W.K., Karch C.M., Cruchaga C., McEvoy L.K., Witoelar A., Chen C.H., Holland D., Alzheimer’s Disease Neuroimaging Initiative Association Between Genetic Traits for Immune-Mediated Diseases and Alzheimer Disease. JAMA Neurol. 2016;73:691–697. doi: 10.1001/jamaneurol.2016.0150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rietveld C.A., Medland S.E., Derringer J., Yang J., Esko T., Martin N.W., Westra H.J., Shakhbazov K., Abdellaoui A., Agrawal A., LifeLines Cohort Study GWAS of 126,559 individuals identifies genetic variants associated with educational attainment. Science. 2013;340:1467–1471. doi: 10.1126/science.1235488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.International League Against Epilepsy Consortium on Complex Epilepsies. Electronic address: Genetic determinants of common epilepsies: a meta-analysis of genome-wide association studies. Lancet Neurol. 2014;13:893–903. doi: 10.1016/S1474-4422(14)70171-1. epilepsy-austin@unimelb.edu.au [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tobacco and Genetics Consortium Genome-wide meta-analyses identify multiple loci associated with smoking behavior. Nat. Genet. 2010;42:441–447. doi: 10.1038/ng.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Manning A.K., Hivert M.F., Scott R.A., Grimsby J.L., Bouatia-Naji N., Chen H., Rybin D., Liu C.T., Bielak L.F., Prokopenko I., DIAbetes Genetics Replication And Meta-analysis (DIAGRAM) Consortium. Multiple Tissue Human Expression Resource (MUTHER) Consortium A genome-wide approach accounting for body mass index identifies genetic variants influencing fasting glycemic traits and insulin resistance. Nat. Genet. 2012;44:659–669. doi: 10.1038/ng.2274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Teslovich T.M., Musunuru K., Smith A.V., Edmondson A.C., Stylianou I.M., Koseki M., Pirruccello J.P., Ripatti S., Chasman D.I., Willer C.J. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010;466:707–713. doi: 10.1038/nature09270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kiryluk K., Li Y., Scolari F., Sanna-Cherchi S., Choi M., Verbitsky M., Fasel D., Lata S., Prakash S., Shapiro S. Discovery of new risk loci for IgA nephropathy implicates genes involved in immunity against intestinal pathogens. Nat. Genet. 2014;46:1187–1196. doi: 10.1038/ng.3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Okada Y., Wu D., Trynka G., Raj T., Terao C., Ikari K., Kochi Y., Ohmura K., Suzuki A., Yoshida S., RACI consortium. GARNET consortium Genetics of rheumatoid arthritis contributes to biology and drug discovery. Nature. 2014;506:376–381. doi: 10.1038/nature12873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schizophrenia Working Group of the Psychiatric Genomics Consortium Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–427. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Morris A.P., Voight B.F., Teslovich T.M., Ferreira T., Segrè A.V., Steinthorsdottir V., Strawbridge R.J., Khan H., Grallert H., Mahajan A., Wellcome Trust Case Control Consortium. Meta-Analyses of Glucose and Insulin-related traits Consortium (MAGIC) Investigators. Genetic Investigation of ANthropometric Traits (GIANT) Consortium. Asian Genetic Epidemiology Network–Type 2 Diabetes (AGEN-T2D) Consortium. South Asian Type 2 Diabetes (SAT2D) Consortium. DIAbetes Genetics Replication And Meta-analysis (DIAGRAM) Consortium Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nat. Genet. 2012;44:981–990. doi: 10.1038/ng.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lango Allen H., Estrada K., Lettre G., Berndt S.I., Weedon M.N., Rivadeneira F., Willer C.J., Jackson A.U., Vedantam S., Raychaudhuri S. Hundreds of variants clustered in genomic loci and biological pathways affect human height. Nature. 2010;467:832–838. doi: 10.1038/nature09410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Locke A.E., Kahali B., Berndt S.I., Justice A.E., Pers T.H., Day F.R., Powell C., Vedantam S., Buchkovich M.L., Yang J., LifeLines Cohort Study. ADIPOGen Consortium. AGEN-BMI Working Group. CARDIOGRAMplusC4D Consortium. CKDGen Consortium. GLGC. ICBP. MAGIC Investigators. MuTHER Consortium. MIGen Consortium. PAGE Consortium. ReproGen Consortium. GENIE Consortium. International Endogene Consortium Genetic studies of body mass index yield new insights for obesity biology. Nature. 2015;518:197–206. doi: 10.1038/nature14177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gjoneska E., Pfenning A.R., Mathys H., Quon G., Kundaje A., Tsai L.H., Kellis M. Conserved epigenomic signals in mice and humans reveal immune basis of Alzheimer’s disease. Nature. 2015;518:365–369. doi: 10.1038/nature14252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bulik-Sullivan B., Finucane H.K., Anttila V., Gusev A., Day F.R., Loh P.R., Duncan L., Perry J.R., Patterson N., Robinson E.B., ReproGen Consortium. Psychiatric Genomics Consortium. Genetic Consortium for Anorexia Nervosa of the Wellcome Trust Case Control Consortium 3 An atlas of genetic correlations across human diseases and traits. Nat. Genet. 2015;47:1236–1241. doi: 10.1038/ng.3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jefferson A.L., Beiser A.S., Himali J.J., Seshadri S., O’Donnell C.J., Manning W.J., Wolf P.A., Au R., Benjamin E.J. Low cardiac index is associated with incident dementia and Alzheimer disease: the Framingham Heart Study. Circulation. 2015;131:1333–1339. doi: 10.1161/CIRCULATIONAHA.114.012438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.van den Boogaard M., Smemo S., Burnicka-Turek O., Arnolds D.E., van de Werken H.J., Klous P., McKean D., Muehlschlegel J.D., Moosmann J., Toka O. A common genetic variant within SCN10A modulates cardiac SCN5A expression. J. Clin. Invest. 2014;124:1844–1852. doi: 10.1172/JCI73140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Guenther C.A., Tasic B., Luo L., Bedell M.A., Kingsley D.M. A molecular basis for classic blond hair color in Europeans. Nat. Genet. 2014;46:748–752. doi: 10.1038/ng.2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Visser M., Palstra R.J., Kayser M. Human skin color is influenced by an intergenic DNA polymorphism regulating transcription of the nearby BNC2 pigmentation gene. Hum. Mol. Genet. 2014;23:5750–5762. doi: 10.1093/hmg/ddu289. [DOI] [PubMed] [Google Scholar]
- 71.Musunuru K., Strong A., Frank-Kamenetsky M., Lee N.E., Ahfeldt T., Sachs K.V., Li X., Li H., Kuperwasser N., Ruda V.M. From noncoding variant to phenotype via SORT1 at the 1p13 cholesterol locus. Nature. 2010;466:714–719. doi: 10.1038/nature09266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Soldner F., Stelzer Y., Shivalila C.S., Abraham B.J., Latourelle J.C., Barrasa M.I., Goldmann J., Myers R.H., Young R.A., Jaenisch R. Parkinson-associated risk variant in distal enhancer of α-synuclein modulates target gene expression. Nature. 2016;533:95–99. doi: 10.1038/nature17939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gilks W.P., Hill M., Gill M., Donohoe G., Corvin A.P., Morris D.W. Functional investigation of a schizophrenia GWAS signal at the CDC42 gene. World J. Biol. Psychiatry. 2012;13:550–554. doi: 10.3109/15622975.2012.666359. [DOI] [PubMed] [Google Scholar]
- 74.Leslie E.J., Taub M.A., Liu H., Steinberg K.M., Koboldt D.C., Zhang Q., Carlson J.C., Hetmanski J.B., Wang H., Larson D.E. Identification of functional variants for cleft lip with or without cleft palate in or near PAX7, FGFR2, and NOG by targeted sequencing of GWAS loci. Am. J. Hum. Genet. 2015;96:397–411. doi: 10.1016/j.ajhg.2015.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Choi J., Xu M., Makowski M.M., Zhang T., Law M.H., Kovacs M.A., Granzhan A., Kim W.J., Parikh H., Gartside M. A common intronic variant of PARP1 confers melanoma risk and mediates melanocyte growth via regulation of MITF. Nat. Genet. 2017;49:1326–1335. doi: 10.1038/ng.3927. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 76.Tewhey R., Kotliar D., Park D.S., Liu B., Winnicki S., Reilly S.K., Andersen K.G., Mikkelsen T.S., Lander E.S., Schaffner S.F., Sabeti P.C. Direct identification of hundreds of expression-modulating variants using a multiplexed reporter assay. Cell. 2016;165:1519–1529. doi: 10.1016/j.cell.2016.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kheradpour P., Ernst J., Melnikov A., Rogov P., Wang L., Zhang X., Alston J., Mikkelsen T.S., Kellis M. Systematic dissection of regulatory motifs in 2000 predicted human enhancers using a massively parallel reporter assay. Genome Res. 2013;23:800–811. doi: 10.1101/gr.144899.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Degner J.F., Pai A.A., Pique-Regi R., Veyrieras J.B., Gaffney D.J., Pickrell J.K., De Leon S., Michelini K., Lewellen N., Crawford G.E. DNasecI sensitivity QTLs are a major determinant of human expression variation. Nature. 2012;482:390–394. doi: 10.1038/nature10808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pennacchio L.A., Ahituv N., Moses A.M., Prabhakar S., Nobrega M.A., Shoukry M., Minovitsky S., Dubchak I., Holt A., Lewis K.D. In vivo enhancer analysis of human conserved non-coding sequences. Nature. 2006;444:499–502. doi: 10.1038/nature05295. [DOI] [PubMed] [Google Scholar]
- 80.Li M.J., Pan Z., Liu Z., Wu J., Wang P., Zhu Y., Xu F., Xia Z., Sham P.C., Kocher J.P. Predicting regulatory variants with composite statistic. Bioinformatics. 2016;32:2729–2736. doi: 10.1093/bioinformatics/btw288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Maurano M.T., Haugen E., Sandstrom R., Vierstra J., Shafer A., Kaul R., Stamatoyannopoulos J.A. Large-scale identification of sequence variants influencing human transcription factor occupancy in vivo. Nat. Genet. 2015;47:1393–1401. doi: 10.1038/ng.3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Grossman S.R., Zhang X., Wang L., Engreitz J., Melnikov A., Rogov P., Tewhey R., Isakova A., Deplancke B., Bernstein B.E. Systematic dissection of genomic features determining transcription factor binding and enhancer function. Proc. Natl. Acad. Sci. USA. 2017;114:E1291–E1300. doi: 10.1073/pnas.1621150114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lee S., Wu M.C., Lin X. Optimal tests for rare variant effects in sequencing association studies. Biostatistics. 2012;13:762–775. doi: 10.1093/biostatistics/kxs014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.He Z., Xu B., Lee S., Ionita-Laza I. Unified Sequence-Based Association Tests Allowing for Multiple Functional Annotations and Meta-analysis of Noncoding Variation in Metabochip Data. Am. J. Hum. Genet. 2017;101:340–352. doi: 10.1016/j.ajhg.2017.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ionita-Laza I., Capanu M., De Rubeis S., McCallum K., Buxbaum J.D. Identification of rare causal variants in sequence-based studies: methods and applications to VPS13B, a gene involved in Cohen syndrome and autism. PLoS Genet. 2014;10:e1004729. doi: 10.1371/journal.pgen.1004729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kichaev G., Yang W.Y., Lindstrom S., Hormozdiari F., Eskin E., Price A.L., Kraft P., Pasaniuc B. Integrating functional data to prioritize causal variants in statistical fine-mapping studies. PLoS Genet. 2014;10:e1004722. doi: 10.1371/journal.pgen.1004722. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.