Abstract
Background:
Increasing evidence has supported the link of intestinal Fusobacterium nucleatum infection to colorectal cancer (CRC). However, the value of F. nucleatum as a biomarker in CRC detection has not been fully defined. In order to reduce the random error and bias of individual research, this meta-analysis aimed to evaluate the diagnostic performance of intestinal F. nucleatum in CRC patients and provide evidence-based data to clinical practice.
Methods:
An article search was performed from PubMed, Embase, Cochrane Library, and Web of Science databases up to December 2017, using the following key words: “Fusobacterium nucleatum”, ”Fusobacterium spp.”, ”Fn”, “colorectal cancer(s)”, “colorectal carcinoma(s)”, “colorectal neoplasm(s)”, and “colorectal tumor(s)”. Articles on relationships between F. nucleatum and CRC were selected according to the preestablished inclusion and exclusion criteria. This meta-analysis was performed using STATA 12.0 software, which included mapping of forest plots, heterogeneity tests, meta-regression, subgroup analysis, sensitivity analysis, and publication bias. The sensitivity, specificity, positive likelihood ratio (LR), negative LR, diagnostic odds ratio (DOR), and their corresponding 95% confidence interval (CI) of each eligible study were summarized.
Results:
Finally, data for 1198 participants (629 CRC and 569 healthy controls) in 10 controlled studies from seven articles were included. The summary receiver operator characteristic curve was mapped. The diagnostic performance of intestinal F. nucleatum infection on CRC was as follows: the area under the curve: 0.86 (95% CI: 0.83–0.89), the pooled sensitivity: 0.81 (95% CI: 0.64–0.91), specificity: 0.77 (95% CI: 0.59–0.89), and DOR: 14.00 (95% CI: 9.00–22.00).
Conclusion:
Intestinal F. nucleatum is a valuable marker for CRC diagnosis.
Keywords: Colorectal Neoplasms, Diagnosis, Fusobacterium nucleatum, Meta-Analysis
摘要
背景:
近年,肠道具核梭杆菌感染和结直肠癌发病的相关性的研究增多,但其作为生物标志物的临床诊断价值尚未完全明 确。为减少单个研究随机误差及偏倚,本研究采用荟萃(Meta)分析评估肠道具核梭杆菌检测对结直肠癌诊断的价值, 为临 床应用提供循证医学依据。
方法:
检索从建库起至2017年12月PubMed、Embase、 Cochrane Library、Web of Science,关键词为“具核梭杆菌”, “梭菌属”, “Fn”, “结直肠癌”, 和“结直肠肿瘤”。按照预先制定的纳入和排除标准筛选文献。本文使用Stata12.0软件进行统计学分析,包 括绘制森林图、异质性检验、Meta回归、亚组分析、敏感性分析、发表偏倚等。再汇总每项纳入研究的诊断的敏感性,特异 性,阳性似然比,阴性似然比,诊断比值比以及其相应的95%可信区间等参数。
结果:
最终本分析共纳入1198参与者,(结直肠癌629例,健康人群569例),资料来源于7篇文献,共10项研究。具核梭杆 菌诊断结直肠癌的综合参数如下:曲线下面积0.86(95% CI: 0.83–0.89)、敏感性0.81(95% CI: 0.64–0.89)、特异性0.77 (95% CI: 0.59–0.89)、诊断比值比14.00(95% CI: 9.00比0.89积)。
结论:
具核梭杆菌可作为诊断结直肠癌有价值的标志物。
INTRODUCTION
Colorectal cancer (CRC) ranks as the third-most common malignancy and the fourth leading cause of cancer-related deaths worldwide. CRC is more prevalent in Western than in developing countries. In recent years, the prevalence of CRC in Asian-Pacific areas increases rapidly.[1,2] The pathogenesis of CRC involves a multistep process including histological, morphological, and genetic changes. The transformation takes time from normal bowel epithelium to a precancerous state, i.e., adenomatous lesions (polyps), and finally to adenocarcinoma. Reduction of the incidence of CRC and its associated mortality primarily depend on the detection of premalignant and early-stage malignant colorectal neoplasia (polyps) and resection of the lesions before they become cancerous. As most CRCs grow slowly over time, development of diagnostic methods with good sensitivity and specificity for the early CRC is crucially important in clinical practice. Population-wide screening and prevention programs have been recommended by many consensus and guidelines, and various diagnostic strategies have been carried out in different countries according to their economic resources.[2]
Colonoscopy, which can provide convincing diagnosis with histology-based protocols, has been well established as the gold standard in CRC diagnosis. However, it is not an ideal tool in practice because not all patients are willing to accept its invasive nature. Advancing age, male gender, family history of CRC, smoking, and obesity are the risk factors for CRC. Several risk-stratified scoring systems, such as High Risk Factor Questionnaire and Quantitative Risk-Assessment Method, have been recommended to evaluate the degrees of CRC risk.[3,4] Colonoscopy is obligatory for patients at high risk and patients with positive signs, suggesting neoplastic lesions in order to discover and manage the possible lesions.[3,5,6,7]
Noninvasive methods have been applied to appeal to patients at average risk. Fecal occult blood tests (FOBTs) including guaiac-based FOBT (gFOBT) and fecal immunochemical test (FIT) have been commonly used clinically for a long time. FOBT is an inexpensive, simple, and widely available test, but its accuracy needs improvement.[8] As the development of CRC requires the accumulation of gene alterations, identification of aberrant gene alterations is theoretically an effective method for CRC detection. Fecal DNA tests or stool DNA (sDNA) testing has been developed, which has the advantages of being highly sensitive and convenient. The sDNA tests measure molecular biomarkers including a wide variety of genetic alterations such as mutant adenomatous polyposis coli, KRA, actin, FIT, tumor protein P53, retinoic acid receptor beta 2, p16, vimentin (VIM), tissue factor pathway inhibitor 2 (TFPI2) and aberrantly methylated VIM, TFPI2, N-Myc downstream-regulated gene 4 protein, bone morphogenetic protein 3, and secreted frizzled-related protein 2. Some of them are based on detecting DNA alteration (mutations and methylations) in a single gene, while others in a number of genes. The pooled sensitivities in multigene testing were higher than that in single gene, but the specificity remained similar to each other.[5,9,10,11] In 2014, a stool multitarget sDNA (Mt-sDNA) test consisting of multitarget mutant DNA, aberrantly methylated DNA across CRC-related genes, and FIT was commercially available. Mt-sDNA had high sensitivity, similar specificity, and high costs compared with those of FIT alone.[12] In 2016, the first blood-based test, i.e., mSEPT9 assay was evolved, as hypermethylated DNA of septin 9 gene might be shed into the bloodstream from CRC tissues and be detected. The mSEPT9 serves to increase the participants in CRC detection, although its sensitivity and specificity are similar to FIT.[8,13] Other noninvasive screening tests, including microRNAs and stool proteins, have been explored, but still are in their infancy and need further evaluation.[14]
Currently, the gut microbiome is increasingly recognized to play an important role in the pathogenesis of diseases. The gut dysbiosis exposes the colon to different metabolic and inflammatory stimuli, increases DNA mutations, and eventually develops CRC. Several bacterial species including Fusobacterium spp., Enterococcus faecalis, enterotoxigenic Bacteroides fragilis, enteropathogenic Escherichia coli, Parvimonas micra ATCC 33270, Streptococcus anginosus, and Proteobacteria were significantly increased in stools from patients with CRC compared to controls.[15,16] Among them, Fusobacterium nucleatum (F. nucleatum, Fn), a human's oral cavity colonizer, has been frequently reported. Besides periodontal disease, F. nucleatum is involved in a wide spectrum of disorders including gastrointestinal diseases, respiratory tract infections, cardiovascular diseases, rheumatoid arthritis, Lemierre's syndrome, and Alzheimer's disease.[17] Accumulating evidence has revealed that F. nucleatum overabundance was present in tumor tissues and stool samples of CRC patients compared with normal controls.[18,19,20] Two previous studies also demonstrated that the prevalence of F. nucleatum was significantly higher in CRC tissues than those in controls and was associated with CRC progress and metastasis.[16,21] Although mechanisms and causalities between F. nucleatum and CRC have been still uncovered, some studies have explored the diagnostic performance of the presence of F. nucleatum in feces or tumor tissues in CRC patients.[22] This meta-analysis summarized the published data in order to evaluate the value of this novel technique in CRC diagnosis.
METHODS
Data sources
A comprehensive literature search was performed from the following electronic databases: PubMed, Embase, Cochrane Library, and Web of Science. The search period was up to December 2017. The following keywords were used in the database literature search: “Fusobacterium nucleatum”, “Fusobacterium spp.”,”F. nucleatum”, “Fn”, “colorectal cancer(s)”, “colorectal carcinoma(s)”, “colorectal neoplasm(s)”, and “colorectal tumor(s)”. The study was performed according to the Meta-analysis ofObservational Studies in Epidemiology guidelines [Supplemental File 1].[23]
Supplemental File 1.
MOOSE checklist
| Item number | Recommendation | Reported on page number |
|---|---|---|
| Reporting of background should include | ||
| 1 | Problem definition | 2 |
| 2 | Hypothesis statement | 2 |
| 3 | Description of study outcome(s) | 2 |
| 4 | Type of exposure or intervention used | None |
| 5 | Type of study designs used | 2 |
| 6 | Study population | 2 |
| Reporting of search strategy should include | ||
| 7 | Qualifications of searchers (e.g., librarians and investigators) | 3 |
| 8 | Search strategy, including time period included in the synthesis and keywords | 3 |
| 9 | Effort to include groups available studies, including contact with authors | 3 |
| 10 | Databases and registries searched | 2 |
| 11 | Search software used, name and version, including special features used (e.g., explosion) | 2 |
| 12 | Use of hand searching (e.g., reference lists of obtained articles) | None |
| 13 | List of citations located and those excluded, including justification | 3,4 |
| 14 | Method of addressing articles published in languages other than English | None |
| 15 | Method of handling abstracts and unpublished studies | 2 |
| 16 | Description of any contact with authors | None |
| Reporting of methods should include | ||
| 17 | Description of relevance or appropriateness of studies assembled for assessing the hypothesis to be tested | 3 |
| 18 | Rationale for the selection and coding of data (e.g., sound clinical principles or convenience) | None |
| 19 | Documentation of how data were classified and coded (e.g., multiple raters, blinding, and interrater reliability) | None |
| 20 | Assessment of confounding (e.g., comparability of cases and controls in studies where appropriate) | 3 |
| 21 | Assessment of study quality, including blinding of quality assessors, stratification or regression on possible predictors of study results | 3 |
| 22 | Assessment of heterogeneity | 3 |
| 23 | Description of statistical methods (e.g., complete description of fixed or random effects models, justification of whether the chosen models account for predictors of study results, dose-response models, or cumulative meta-analysis) in sufficient detail to be replicated | 3 |
| 24 | Provision of appropriate tables and graphs | 3 |
| Reporting of results should include | ||
| 25 | Graphs summarizing individual study estimates and overgroups estimate | 3 |
| 26 | Table giving descriptive information for each study included | 4 |
| 27 | Results of sensitivity testing (e.g., subgroup analysis) | 4,5 |
| 28 | Indication of statistical uncertainty of findings | None |
| Reporting of discussion should include | ||
| 29 | Quantitative assessment of bias (e.g., publication bias) | 7 |
| 30 | Justification for exclusion (e.g., exclusion of non-English language citations) | None |
| 31 | Assessment of quality of included studies | 7 |
| Reporting of conclusions should include | ||
| 32 | Consideration of alternative explanations for observed results | 7 |
| 33 | Generalization of the conclusions (i.e., appropriate for the data presented and within the domain of the literature review) | 7 |
| 34 | Guidelines for future research | 7 |
| 35 | Disclosure of funding source | None |
MOOSE: Meta-Analysis of Observational Studies in Epidemiology.
Inclusion and exclusion criteria
The inclusion criteria were as follows: (1) a clinical study of CRC patients and healthy controls with intact data of F. nucleatum-positive and F. nucleatum-negative cases; (2) studies that reported necessary data for calculating true-positive (TP), false-positive (FP), true-negative (TN), and false-negative (FN) F. nucleatum in CRC detection; (3) the diagnosis of CRC should be based on histology; (4) the detection of F. nucleatum should be based on quantitative polymerase chain reaction analysis, fluorescence in situ hybridization, or 16S rRNA sequencing; (5) the samples (feces or tissues) should be stored at −20°C to −80°C soon after collection; and (6) the articles should be original articles.
The exclusion criteria were as follows: (1) the samples in control group were collected from adjacent nontumor tissues of CRC patients, but not from healthy controls; (2) the results of studies did not include TP, FP, FN, and TN F. nucleatum; (3) only the abstract forms were presented; and (4) articles that were not original articles but reviews or other compositions.
Data extraction and assessment
The data from each study were independently extracted by two independent reviewers and then reciprocally verified using the predefined selection criteria. Disagreements of two reviewers were resolved in consultation with a third investigator. The following information from each article was extracted: the first author's name, the year of publication, patient ethnicity, sample size, sample type, diagnostic techniques of CRC and F. nucleatum, andclinicopathological characteristics of CRC patients and healthy controls. The parameters in results were summarized by bivariate mixed-effects models. The pooled TPs, FPs, TNs, and FNs, positive likelihood ratio (PLR), negative likelihood ratio (NLR), diagnostic odds ratio (DOR), and their 95% confidence interval (CI) were extracted for mapping forest plot.
The quality of each study was assessed according to the Quality Assessment of Diagnostic Accuracy Studies (QUADAS) recommended by the Cochrane Collaboration. QUADAS tool includes 14 items covering patient spectrum, reference standard, disease progression bias, verification bias, review bias, clinical review bias, incorporation bias, test execution, study withdrawals, and indeterminate results. Each of the 14 items in the QUADAS checklist is scored as “yes”, “no”, or “unclear”. QUADAS tool allows for more transparent rating of bias and applicability of primary diagnostic accuracy studies. If the QUADAS score is <10 points, the study is classified as having low methodological quality.
Statistical analysis
All analyses were conducted by the STATA-12.0 software (Stata Corp., College Station, TX, USA). The numerical variables with normal distribution were expressed as mean ± standard deviation (SD), and qualitative variables were calculated as numbers and percentages. The 95% CI s were estimated for each predictive test. A two-tailed P < 0.05 was considered statistically significant. Bivariate mixed-effects models were used to process variable combination. The strength of association was represented as an overall odds ratio with corresponding 95% CI. The pooled sensitivity, specificity, and area under curve (AUC) of the summary receiver operator characteristic (SROC) with their 95% CI s were analyzed to determine the diagnostic accuracy of F. nucleatum for CRC detection. Between-study heterogeneity was assessed using Chi-square-based Q-statistic test and I2 test. If the Q-test showed P < 0.10 and I2 > 50%, which indicated significant heterogeneity, the random effects model (DerSimonian-Laird method) was conducted; otherwise if the Q-test showed P > 0.10 and I2 < 50%, the fixed effect model (Mantel-Haenszel method) was used. The sources of heterogeneity between studies were evaluated by meta-regression and subgroup analyses. Funnel plots were used to evaluate a potential publication bias, which was quantitatively estimated with Egger's linear regression test. If a potential publication bias existed, the meta-trim method was conducted.
RESULTS
Study characteristics
A total of 142 articles were identified using the search strategy described above. Totally, 135 articles were excluded after careful filtration. Among them, 32 articles were duplicates, 93 had inappropriate abstracts and titles, and 10 articles did not provide necessary data mentioned in inclusion criteria, i.e., TP, FP, TN, or FN. Among these 10 articles, data were unavailable in 9 articles and insufficient in 1 article. Finally, 7 articles with 10 studies were included in this meta-analysis, which involved 1198 participants (629 CRC patients and 569 healthy controls). The selection process is illustrated in Figure 1. Basic characteristics and F. nucleatum performance in CRC detection of these 10 studies are shown in Table 1.
Figure 1.
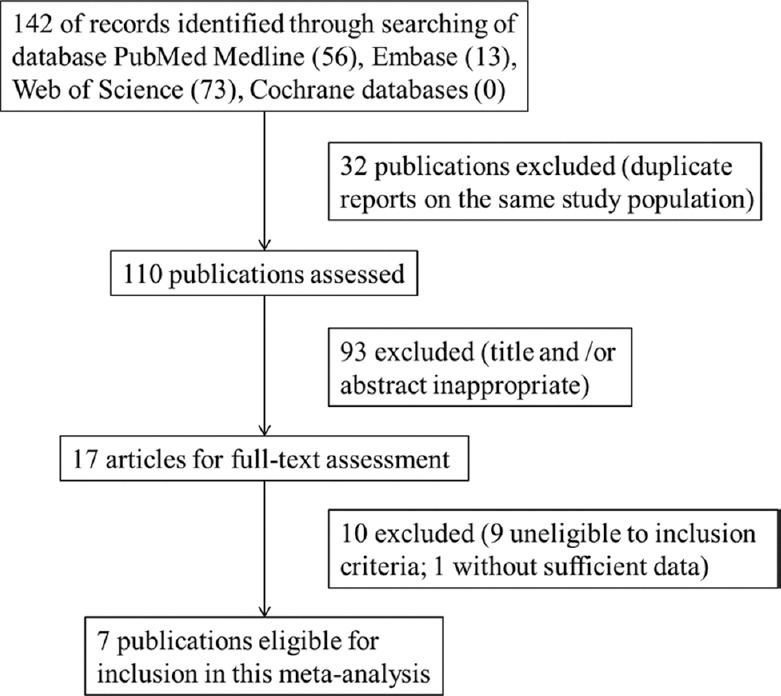
Flow diagram of the step-wise selection for this meta-analysis.
Table 1.
Basic characteristics and diagnostic performance of F. nucleatum in CRC
| Authors | Year | Country | Sample types | Detection methods | QUADAS scores | Sample collection | Sample size, n | TP, n | FN, n | FP, n | TN, n | Sensitivity (%) | Specificity (%) | PPV (%) | NPV (%) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Wong et al.[24] | 2017 | China | Feces | Real-time qPCR | 12 | Colonoscopy | 206 | 75 | 29 | 9 | 93 | 72.1 | 91.2 | 89.3 | 76.2 |
| Wong et al.[24] | 2017 | China | Feces | Real-time qPCR | 12 | Colonoscopy | 119 | 21 | 2 | 19 | 77 | 91.3 | 80.2 | 52.5 | 97.5 |
| Liang et al.[25] | 2017 | China | Feces | Probe-based duplex qPCR | 11 | Colonoscopy | 370 | 132 | 38 | 41 | 159 | 77.7 | 79.5 | 76.3 | 80.7 |
| Liang et al.[25] | 2017 | China | Feces | Probe-based duplex qPCR | 11 | Colonoscopy | 69 | 27 | 6 | 17 | 19 | 81.8 | 52.8 | 61.4 | 76.0 |
| Yu et al.[26] | 2016 | China | CRC tissues | FISH | 11 | Colonoscopy | 113 | 62 | 31 | 4 | 16 | 66.7 | 80.0 | 93.9 | 34.0 |
| Suehiro et al.[27] | 2017 | Japan | Feces | Droplet digital PCR | 11 | Colonoscopy or surgical operation | 218 | 85 | 73 | 6 | 54 | 53.8 | 90.0 | 93.4 | 42.5 |
| Kostic et al.[20] | 2013 | USA | Feces | qPCR | 10 | Surgical operation | 58 | 27 | 0 | 15 | 16 | 100.0 | 51.6 | 64.3 | 100.0 |
| Mira-Pascual et al.[19] | 2015 | Spain | Feces | Real-time qPCR | 10 | Colonoscopy | 16 | 6 | 1 | 2 | 7 | 85.7 | 77.8 | 75.0 | 87.5 |
| Mira-Pascual et al.[19] | 2015 | Spain | CRC tissues | Real-time qPCR | 10 | Colonoscopy | 12 | 2 | 5 | 0 | 5 | 28.6 | 100.0 | 100.0 | 50.0 |
| Fukugaiti et al.[28] | 2015 | Brazil | Feces | Real-time qPCR | 10 | Colonoscopy | 17 | 7 | 0 | 9 | 1 | 100.0 | 10.0 | 43.8 | 100.0 |
TP: True positive; FN: False negative; FP: False positive; TN: True negative; PPV: Positive predictive value; NPV: Negative predictive value; CRC: Colorectal cancer; F. nucleatum: Fusobacterium nucleatum; qPCR: Quantitative polymerase chain reaction; FISH: Fluorescence in situ hybridization; QUADAS: Quality Assessment of Diagnostic Accuracy Study.
Heterogeneity assessment
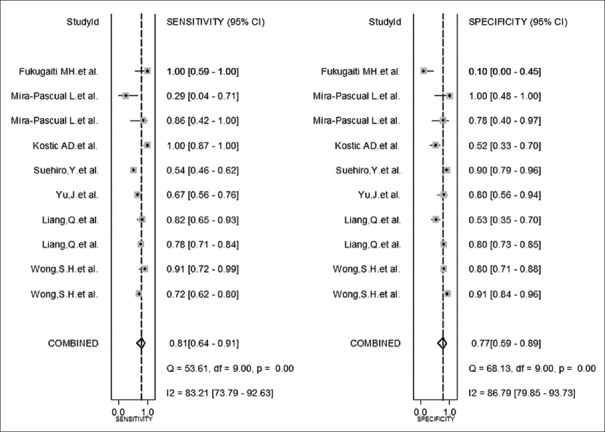
The performance of F. nucleatum for CRC diagnosis is shown in forest plot [Figure 2]: pooled sensitivity: 0.81 (95% CI: 0.64–0.91), specificity: 0.77 (95% CI: 0.59–0.89), PLR: 3.60 (95% CI: 2.10–6.00), NLR: 0.25 (95% CI: 0.15–0.42), and DOR: 14.00 (95% CI: 9.00–22.00).
Figure 2.
Forest plot of the pooled diagnostic accuracy of Fusobacterium nucleatum for colorectal cancer detection. CI: Confidence interval.
The sensitivity heterogeneity (I2 = 83.2%, 95% CI: 73.79–92.63, P < 0.01) and specificity heterogeneity (I2 = 86.8%, 95% CI: 79.85–93.73, P < 0.01) indicated significant heterogeneity between studies [Figure 2]. SROC distributed advisably [AUC: 0.86, 95% CI: 0.83–0.89; Figure 3], and Spearman's rank correlation coefficient was –0.98, calculated by an equation using logarithm of sensitivity and 1–specificity. Which indicated no statistical significance (P = 0.97). Both of them suggested that the pooled data in this meta-analysis were feasible since threshold effects did not exist.
Figure 3.
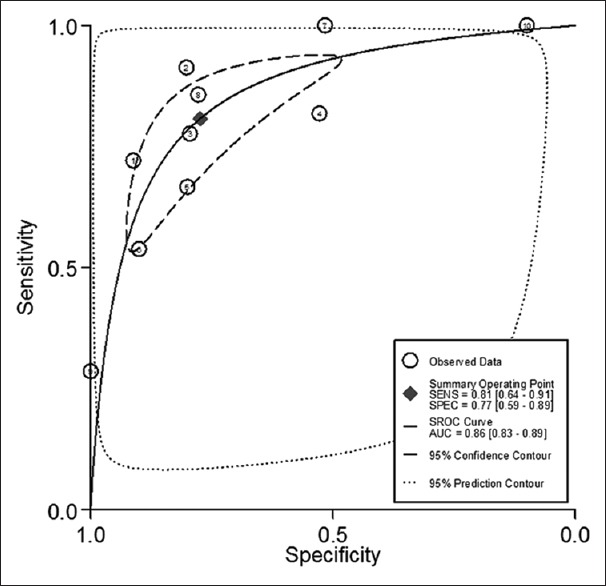
SROC assessment of diagnostic performance of Fusobacterium nucleatum for colorectal cancer. SROC: Summary receiver operator characteristic curve.
Meta-regression analysis
As forest plot revealed significant heterogeneity between studies, single-factor meta-regression analysis was applied to screen the potential variables impacting on the pooled data. The results showed that race, sample type, sample size, and QUADAS scores were not the source of heterogeneity [all P > 0.05; Table 2].
Table 2.
Single-factor meta-regression analysis for sources of heterogeneity between studies
| Variables | Coefficient | SE | t | P | 95% CI |
|---|---|---|---|---|---|
| Race | 0.02 | 0.85 | 0.02 | 0.98 | −1.94–1.98 |
| Sample type | −0.66 | 0.70 | −0.95 | 0.37 | −2.27–0.94 |
| Sample size | 0.00 | 0.00 | 0.54 | 0.60 | −0.00–0.01 |
| QUADAS scores | 0.71 | 0.35 | 2.06 | 0.07 | −0.09–1.52 |
SE: Standard error; QUADAS: Quality Assessment of Diagnostic Accuracy Study; CI: Confidence interval.
Subgroup analysis
As meta-regression analysis failed to identify any source of heterogeneity, subgroup analyses were performed. Since the data from eligible literature were limited, only three subgroup analyses, i.e., race, sample type, and sample size were carried out. The results showed significant heterogeneity in Asian and non-Asian subgroups. In six Asian subgroups, the pooled sensitivity was 0.74 (95% CI: 0.63–0.82, I2 = 84.4%, P < 0.01) and specificity was 0.82 (95% CI: 0.72–0.90, I2 = 82.4%, P < 0.01), and in four non-Asian subgroups, the pooled sensitivity was 0.97 (95% CI: 0.30–1.00, I2 = 88.5%, P < 0.01) and specificity was 0.65 [95% CI: 0.20–0.94, I2 = 78.9%, P < 0.01; Table 3]. Significant heterogeneity existed between the subgroups with different sample size. In five subgroups with sample size >100, the pooled sensitivity was 0.71 (95% CI: 0.61–0.80, I2 = 86.4%, P < 0.01) and specificity was 0.85 (95% CI: 0.79–0.90, I2 = 57%, P = 0.05), and in five subgroups, with sample size ≤100, the pooled sensitivity was 0.91 (95% CI: 0.47–0.99, I2 = 83.3%, P < 0.01) and specificity was 0.58 (95% CI: 0.27–0.84, I2 = 71.6%, P = 0.01). Significant heterogeneity was also found in sample type subgroups. In eight subgroups of stool studies, the pooled sensitivity was 0.86 (95% CI: 0.70–0.94, I2 = 85.5%, P < 0.01) and specificity was 0.72 (95% CI: 0.51–0.86, I2 = 89.5%, P < 0.01). Due to only two subgroups of CRC tissue studies, analysis data were unavailable [Table 3].
Table 3.
Subgroup analysis for sources of heterogeneity between studies
| Variables | AUC (95% CI) | Sensitivity (95% CI) | Specificity (95% CI) | PLR (95% CI) | NLR (95% CI) | DOR (95% CI) |
|---|---|---|---|---|---|---|
| Pooled data | 0.86 (0.83–0.89) | 0.81 (0.64–0.91) | 0.77 (0.59–0.89) | 3.60 (2.10–6.00) | 0.25 (0.15–0.42) | 14.00 (9.00–22.00) |
| Race | ||||||
| Asian | 0.84 (0.81–0.87) | 0.74 (0.63–0.82) | 0.82 (0.72–0.90) | 4.20 (2.70–6.40) | 0.32 (0.24–0.43) | 13.00 (8.00–21.00) |
| Non-Asian | 0.90 (0.87–0.92) | 0.97 (0.30–1.00) | 0.65 (0.20–0.94) | 2.80 (0.80–9.60) | 0.05 (0.00–1.82) | 57.00 (3.00–1163.00) |
| Sample type | ||||||
| Stool | 0.87 (0.83–0.89) | 0.86 (0.70–0.94) | 0.72 (0.51–0.86) | 3.00 (1.80–5.10) | 0.20 (0.11–0.37) | 15.00 (8.00–27.00) |
| CRC tissues | NA | NA | NA | NA | NA | NA |
| Sample size | ||||||
| >100 | 0.87 (0.84–0.90) | 0.71 (0.61–0.80) | 0.85 (0.79–0.90) | 4.90 (3.60–6.50) | 0.34 (0.25–0.45) | 14.00 (10.00–21.00) |
| ≤100 | 0.81 (0.77–0.84) | 0.91 (0.47–0.99) | 0.58 (0.27–0.84) | 2.20 (1.20–4.20) | 0.16 (0.03–0.96) | 14.00 (3.00–76.00) |
AUC: Area under the curve; PLR: Positive likelihood ratio; NLR: Negative likelihood ratio; DOR: Diagnostic odds ratio; CI: Confidence interval; NA: Not applicable; CRC: Colorectal cancer.
Sensitivity analysis
In order to obtain the stability of the pooled data and evaluate the influence of single study on whole studies, a sensitivity analysis was conducted. The alteration of efficient variables was not statistically significant (P > 0.05), which suggested that the polled results were not interfered by the data from single studies [Table 4].
Table 4.
Sensitivity analysis for stability of the pooled data and influence of single study on all studies
| Studies | AUC (95% CI) | Sensitivity (95% CI) | Specificity (95% CI) | PLR (95% CI) | NLR (95% CI) | DOR (95% CI) |
|---|---|---|---|---|---|---|
| Wong et al.[24] | NA | NA | NA | NA | NA | NA |
| Wong et al.[24] | 0.85 (0.82–0.88) | 0.80 (0.62–0.90) | 0.77 (0.56–0.90) | 3.50 (1.90–6.30) | 0.26 (0.16–0.44) | 13.00 (9.00–19.00) |
| Liang et al.[25] | 0.86 (0.83–0.89) | 0.82 (0.61–0.93) | 0.77 (0.56–0.90) | 3.50 (1.90–6.50) | 0.23 (0.11–0.47) | 15.00 (8.00–29.00) |
| Liang et al.[25] | 0.88 (0.84–0.90) | 0.83 (0.59–0.94) | 0.79 (0.61–0.90) | 4.00 (2.40–6.60) | 0.22 (0.09–0.51) | 18.00 (10.00–33.00) |
| Yu et al.[26] | 0.87 (0.83–0.89) | 0.83 (0.64–0.93) | 0.76 (0.55–0.89) | 3.50 (1.90–6.30) | 0.23 (0.12–0.43) | 15.00 (9.00–25.00) |
| Suehiro et al.[27] | 0.86 (0.83–0.89) | 0.82 (0.67–0.91) | 0.76 (0.54–0.89) | 3.40 (1.80–6.10) | 0.24 (0.15–0.37) | 14.00 (9.00–22.00) |
| Kostic et al.[20] | 0.85 (0.81–0.88) | 0.76 (0.61–0.87) | 0.80 (0.61–0.91) | 3.80 (2.10–7.20) | 0.30 (0.20–0.44) | 13.00 (8.00–22.00) |
| Mira-Pascual et al.[19] | 0.86 (0.83–0.89) | 0.81 (0.62–0.91) | 0.77 (0.57–0.90) | 3.60 (2.00–6.40) | 0.25 (0.14–0.45) | 14.00 (9.00–22.00) |
| Mira-Pascual et al.[19] | 0.86 (0.83–0.89) | 0.83 (0.69–0.92) | 0.74 (0.56–0.86) | 3.20 (2.00–5.10) | 0.23 (0.13–0.38) | 14.00 (8.00–23.00) |
| Fukugaiti et al.[28] | 0.86 (0.83–0.89) | 0.77 (0.63–0.86) | 0.82 (0.70–0.89) | 4.10 (2.80–6.20) | 0.29 (0.19–0.44) | 14.00 (10.00–22.00) |
| Pooled data | 0.86 (0.83–0.89) | 0.81 (0.64–0.91) | 0.77 (0.59–0.89) | 3.60 (2.10–6.00) | 0.25 (0.15–0.42) | 14.00 (9.00–22.00) |
AUC: Area under the curve; PLR: Positive likelihood ratio; NLR: Negative likelihood ratio; DOR: Diagnostic odds ratio; CI: Confidence interval; NA: Not available.
Fagan plot
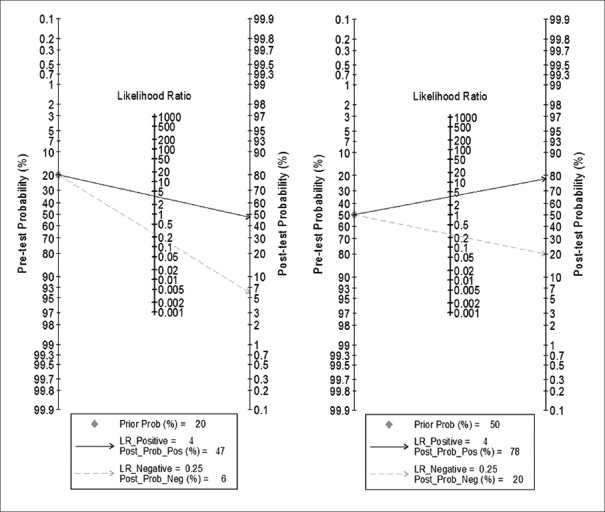
When prior probability was set at 0.20, the LRs of positive postprobability and negative postprobability were 0.47 and 0.06, respectively. The results implied that a positive result of intestinal F. nucleatum infection increased 47.0% possibility of CRC presence, whereas a negative result decreased 6.0% in comparison with pretest possibility. When prior probability was set at 0.50, the LRs of positive postprobability and negative postprobability were 0.78 and 0.20, respectively. The results implied that a positive result increased 78.0% possibility of CRC presence, whereas a negative result decreased 20.0% in comparison with pretest possibility [Figure 4].
Figure 4.
Fagan plot analysis to evaluate the intestinal Fusobacterium nucleatum infection for diagnosis of CRC. Prior Prob: Prior probability; Post Prob: Postprobability; LR: Likelihood ratio.
Bias of publication
Funnel plot and Egger's linear regression tests were conducted to evaluate potential publication bias. The spots were symmetrically distributed in Funnel plot and no significance was found in Egger's plot (t = –0.19, P = 0.86). Both suggested no publication bias in the analysis [Figures 5 and 6].
Figure 5.
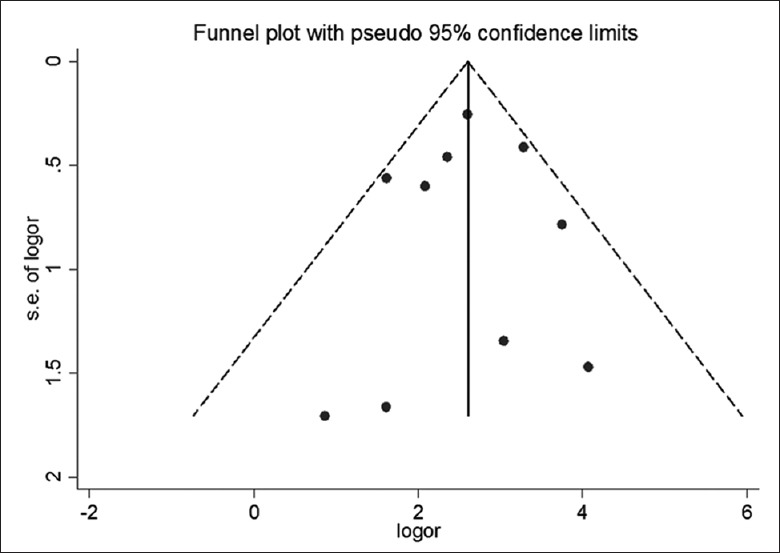
Funnel plot asymmetry test for publication bias. Each spot represents a separate study. logor: Log odds ratio; s.e. of logor: Standard error of log odds ratio.
Figure 6.
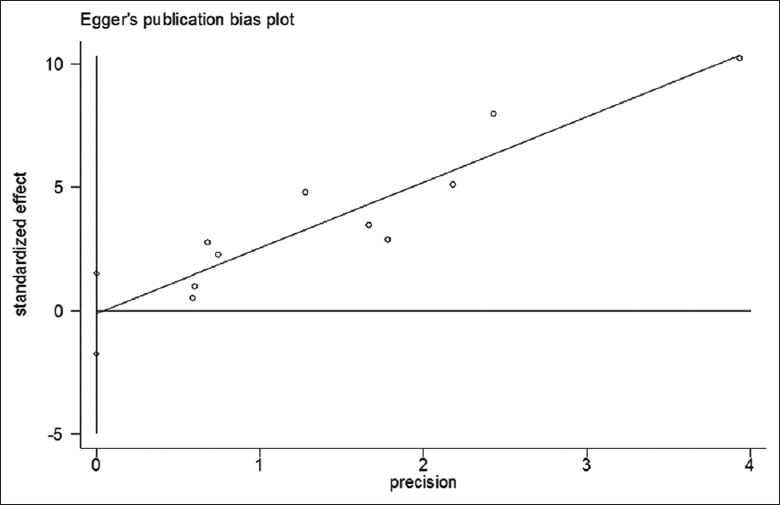
Egger's linear regression test for publication bias. Each spot represents a separate study.
DISCUSSION
The ideal strategies for CRC diagnosis should be efficient, safe, convenient, and cheap. So far, gFOBT/FIT followed by colonoscopy have been recommended as standard CRC program in most guidelines. Colonoscopy is obligatory for obtaining histological diagnosis. Great efforts have been made to develop cost-effective noninvasive methods. gFOBT and FIT have advantages such as being convenient, inexpensive (US 1 dollar and US 10 dollar), and quick (30 min), but their results are instable (sensitivity: gFOBT 6.2–83.3%, FIT 5.4–98.0% and specificity: gFOBT 80.0–98.4%, FIT 77.0– 99.0%). FIT tests an antibody to human hemoglobin in stools instead of testing heme by gFOBT. Therefore, FIT is comparatively steady because of no cross-reaction to dietary meat. Stool DNA tests have comparatively high sensibility, i.e., stool DNA tests (48.0–77.8%) and multi-target stool DNA test (52.0–92.3%), but similar specificity (92.7–97.0% and 86.6–94.4%) to FIT. Both stool DNA test and multi-target stool DNA test are expensive ($300 and $600) and time-consuming (1 day). Serum methylated SEPT9 has advantages of being convenient, compliant, and available at an acceptable price (US 170 dollar), but its accuracy (sensitivity 64.0–80.0% and specificity 75.0–88.0%) needs further confirmation.[5,7,8,9,10,11,12,13,16,29,30,31,32,33,34] This meta-analysis demonstrated that the performance of F. nucleatum test (sensitivity 64.0–91.0%, specificity 59.0–89.0%, and cost US 70 dollar) ranked in the dominated level among the noninvasive methods so far available. It had a reasonable diagnostic value compared with that of FIT and lower price compared with that of stool DNA and serum SEPT9 tests. A recent study showed that fecal F. nucleatum biomarker combined with FIT had significantly higher sensitivity than that of FIT alone in detecting CRC (92.3% vs 73.1%, P < 0.001) and advanced colorectal neoplasia (38.6% vs 15.5%, P < 0.001).[24] Intestinal F. nucleatum testing may be an additional option for detecting patients with CRC.
The association of F. nucleatum infection withclinical variables such as tumor node metastasis (TMN) stages and sites of CRC is an important issue for a new technique. In the eligible literatures, no study directly reported the data on early-stage CRC, but three articles documented that F. nucleatum was overabundance in malignant colorectal neoplasia (polyps, adenoma), compared with healthy controls.[19,20,27] Two articles revealed the sensitivity of F. nucleatum for CRC detection in different TMN stage,[24,25] one of which showed higher sensitivity in Stages 2, 3, and 4 than in Stage 1.[25] Two articles evaluated the link of F. nucleatum infection to the sites of CRC,[24,26] one of which concluded that F. nucleatum played more important roles in CRC development in proximal colon than in distal colon.[26] The above data were insufficient and not enough to perform further analysis.
In order to obtain an accurate result, this study selected healthy individuals as controls. The studies with CRC adjuvant tissues as controls were excluded because the abundance of F. nucleatum in CRC adjuvant mucosa was much higher than that in healthy controls.[16,29] Subgroup analyses were applied to identify the possible source of heterogeneity among studies. Significant heterogeneity between Asian and non-Asian groups in this study was consistent with the results in literatures. Gut microbial biomarkers had ethnical difference in patients with CRC.[35] Heterogeneity was also revealed between the subgroups of different sample sizes and sample types. In consistent with literatures, the amount of F. nucleatum in CRC tissues was significantly higher than that in stool.[36] Funnel plot and Egger's linear regression tests were applied to examine the heterogeneity, and no publication bias was found in this meta-analysis.
To our knowledge, this meta-analysis early evaluated the value of intestinal F. nucleatum test in CRC detection. This approach may take one step further toward a noninvasive, accurate, and affordable method for CRC detection. There were several limitations in this study. First, the number of eligible literatures and the sample size might not be large enough to support a powerful conclusion. In order to find more literatures, we searched Chinese articles and found 39 additional articles. However, the methodological quality of them was not good enough to meet the inclusion criteria. Second, there was heterogeneity between the subgroups, although no publication bias was found. Third, the techniques of intestinal F. nucleatum testing were not standard.
On the whole, this meta-analysis proved that intestinal F. nucleatum is a valuable marker for CRC diagnosis. However, it remains to be proven whether this method can improve diagnostic uptake compared with the established strategies.
Supplementary information is linked to the online version of the paper on the Chinese Medical Journal website.
Financial support and sponsorship
Nil.
Conflicts of interest
There are no conflicts of interest.
Footnotes
Edited by: Xin Chen
REFERENCES
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A, et al. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D, et al. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 3.Ye D, Huang Q, Li Q, Jiang X, Mamat M, Tang M, et al. Comparative evaluation of preliminary screening methods for colorectal cancer in a mass program. Dig Dis Sci. 2017;62:2532–41. doi: 10.1007/s10620-017-4648-1. doi: 10.1007/s10620-017-4648-1. [DOI] [PubMed] [Google Scholar]
- 4.Sung JJ, Ng SC, Chan FK, Chiu HM, Kim HS, Matsuda T, et al. An updated Asia Pacific consensus recommendations on colorectal cancer screening. Gut. 2015;64:121–32. doi: 10.1136/gutjnl-2013-306503. doi: 10.1136/gutjnl-2013-306503. [DOI] [PubMed] [Google Scholar]
- 5.Lee JK, Liles EG, Bent S, Levin TR, Corley DA. Accuracy of fecal immunochemical tests for colorectal cancer: Systematic review and meta-analysis. Ann Intern Med. 2014;160:171. doi: 10.7326/M13-1484. doi: 10.7326/M13-1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Katsoula A, Paschos P, Haidich AB, Tsapas A, Giouleme O. Diagnostic accuracy of fecal immunochemical test in patients at increased risk for colorectal cancer: A meta-analysis. JAMA Intern Med. 2017;177:1110–8. doi: 10.1001/jamainternmed.2017.2309. doi: 10.1001/jamainternmed.2017.2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhai RL, Xu F, Zhang P, Zhang WL, Wang H, Wang JL, et al. The diagnostic performance of stool DNA testing for colorectal cancer: A systematic review and meta-analysis. Medicine (Baltimore) 2016;95:e2129. doi: 10.1097/MD.0000000000002129. doi: 10.1097/MD.0000000000002129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Issa IA, Noureddine M. Colorectal cancer screening: An updated review of the available options. World J Gastroenterol. 2017;23:5086–96. doi: 10.3748/wjg.v23.i28.5086. doi: 10.3748/wjg.v23.i28.5086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kadiyska T, Nossikoff A. Stool DNA methylation assays in colorectal cancer screening. World J Gastroenterol. 2015;21:10057–61. doi: 10.3748/wjg.v21.i35.10057. doi: 10.3748/wjg.v21.i35.10057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Abola MV, Fennimore TF, Chen MM, Chen Z, Sheth AK, Cooper G, et al. Stool DNA-based versus colonoscopy-based colorectal cancer screening: Patient perceptions and preferences. Fam Med Community Health. 2015;3:2–8. doi: 10.15212/FMCH.2015.0125. [Google Scholar]
- 11.Imperiale TF, Ransohoff DF, Itzkowitz SH, Levin TR, Lavin P, Lidgard GP, et al. Multitarget stool DNA testing for colorectal-cancer screening. N Engl J Med. 2014;370:1287–97. doi: 10.1056/NEJMoa1311194. doi: 10.1056/NEJMoa1311194. [DOI] [PubMed] [Google Scholar]
- 12.Berger BM, Schroy PC, 3rd, Dinh TA. Screening for colorectal cancer using a multitarget stool DNA test: Modeling the effect of the intertest interval on clinical effectiveness. Clin Colorectal Cancer. 2016;15:e65–74. doi: 10.1016/j.clcc.2015.12.003. doi: 10.1016/j.clcc.2015.12.003. [DOI] [PubMed] [Google Scholar]
- 13.Johnson DA, Barclay RL, Mergener K, Weiss G, König T, Beck J, et al. Plasma septin9 versus fecal immunochemical testing for colorectal cancer screening: A prospective multicenter study. PLoS One. 2014;9:e98238. doi: 10.1371/journal.pone.0098238. doi: 10.1371/journal.pone.0098238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dong Y, Wu WK, Wu CW, Sung JJ, Yu J, Ng SS, et al. MicroRNA dysregulation in colorectal cancer: A clinical perspective. Br J Cancer. 2011;104:893–8. doi: 10.1038/bjc.2011.57. doi: 10.1038/bjc.2011.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shah MS, DeSantis TZ, Weinmaier T, McMurdie PJ, Cope JL, Altrichter A, et al. Leveraging sequence-based faecal microbial community survey data to identify a composite biomarker for colorectal cancer. Gut. 2017;(pii: gutjnl-2016-313189) doi: 10.1136/gutjnl-2016-313189. doi: 101136/gutjnl-2016-313189. [DOI] [PubMed] [Google Scholar]
- 16.Zhou Y, He H, Xu H, Li Y, Li Z, Du Y, et al. Association of oncogenic bacteria with colorectal cancer in South China. Oncotarget. 2016;7:80794–802. doi: 10.18632/oncotarget.13094. doi: 10.18632/oncotarget.13094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Han YW. Fusobacterium nucleatum: A commensal-turned pathogen. Curr Opin Microbiol. 2015;23:141–7. doi: 10.1016/j.mib.2014.11.013. doi: 10.1016/j.mib.2014.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Flanagan L, Schmid J, Ebert M, Soucek P, Kunicka T, Liska V, et al. Fusobacterium nucleatum associates with stages of colorectal neoplasia development, colorectal cancer and disease outcome. Eur J Clin Microbiol Infect Dis. 2014;33:1381–90. doi: 10.1007/s10096-014-2081-3. doi: 10.1007/s10096-014-2081-3. [DOI] [PubMed] [Google Scholar]
- 19.Mira-Pascual L, Cabrera-Rubio R, Ocon S, Costales P, Parra A, Suarez A, et al. Microbial mucosal colonic shifts associated with the development of colorectal cancer reveal the presence of different bacterial and archaeal biomarkers. J Gastroenterol. 2015;50:167–79. doi: 10.1007/s00535-014-0963-x. doi: 10.1007/s00535-014-0963-x. [DOI] [PubMed] [Google Scholar]
- 20.Kostic AD, Chun E, Robertson L, Glickman JN, Gallini CA, Michaud M, et al. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe. 2013;14:207–15. doi: 10.1016/j.chom.2013.07.007. doi: 10.1016/j.chom.2013.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li YY, Ge QX, Cao J, Zhou YJ, Du YL, Shen B, et al. Association of Fusobacterium nucleatum infection with colorectal cancer in Chinese patients. World J Gastroenterol. 2016;22:3227–33. doi: 10.3748/wjg.v22.i11.3227. doi: 10.3748/wjg.v22.i11.3227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Amitay EL, Werner S, Vital M, Pieper DH, Höfler D, Gierse IJ, et al. Fusobacterium and colorectal cancer: Causal factor or passenger? Results from a large colorectal cancer screening study. Carcinogenesis. 2017;38:781–8. doi: 10.1093/carcin/bgx053. doi: 10.1093/carcin/bgx053. [DOI] [PubMed] [Google Scholar]
- 23.Stroup DF, Berlin JA, Morton SC, Olkin I, Williamson GD, Rennie D, et al. Meta-analysis of observational studies in epidemiology: A proposal for reporting.Meta-analysis of observational studies in epidemiology (MOOSE) group. JAMA. 2000;283:2008–12. doi: 10.1001/jama.283.15.2008. doi: 10.1001/jama.283.15.2008. [DOI] [PubMed] [Google Scholar]
- 24.Wong SH, Kwong TNY, Chow TC, Luk AKC, Dai RZW, Nakatsu G, et al. Quantitation of faecal Fusobacterium improves faecal immunochemical test in detecting advanced colorectal neoplasia. Gut. 2017;66:1441–8. doi: 10.1136/gutjnl-2016-312766. doi: 10.1136/gutjnl-2016-312766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liang Q, Chiu J, Chen Y, Huang Y, Higashimori A, Fang J, et al. Fecal bacteria act as novel biomarkers for noninvasive diagnosis of colorectal cancer. Clin Cancer Res. 2017;23:2061–70. doi: 10.1158/1078-0432.CCR-16-1599. doi: 10.1158/1078-0432.CCR-16-1599. [DOI] [PubMed] [Google Scholar]
- 26.Yu J, Chen Y, Fu X, Zhou X, Peng Y, Shi L, et al. Invasive Fusobacterium nucleatum may play a role in the carcinogenesis of proximal colon cancer through the serrated neoplasia pathway. Int J Cancer. 2016;139:1318–26. doi: 10.1002/ijc.30168. doi: 10.1002/ijc.30168. [DOI] [PubMed] [Google Scholar]
- 27.Suehiro Y, Sakai K, Nishioka M, Hashimoto S, Takami T, Higaki S, et al. Highly sensitive stool DNA testing of Fusobacterium nucleatum as a marker for detection of colorectal tumours in a Japanese population. Ann Clin Biochem. 2017;54:86–91. doi: 10.1177/0004563216643970. doi: 10.1177/0004563216643970. [DOI] [PubMed] [Google Scholar]
- 28.Fukugaiti MH, Ignacio A, Fernandes MR, Ribeiro Júnior U, Nakano V, Avila-Campos MJ, et al. High occurrence of Fusobacterium nucleatum and Clostridium difficile in the intestinal microbiota of colorectal carcinoma patients. Braz J Microbiol. 2015;46:1135–40. doi: 10.1590/S1517-838246420140665. doi: 10.1590/S1517-838246420140665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tahara T, Yamamoto E, Suzuki H, Maruyama R, Chung W, Garriga J, et al. Fusobacterium in colonic flora and molecular features of colorectal carcinoma. Cancer Res. 2014;74:1311–8. doi: 10.1158/0008-5472.CAN-13-1865. doi: 10.1158/0008-5472.CAN-13-1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Song LL, Li YM. Current noninvasive tests for colorectal cancer screening: An overview of colorectal cancer screening tests. World J Gastrointest Oncol. 2016;8:793–800. doi: 10.4251/wjgo.v8.i11.793. doi: 10.4251/wjgo.v8.i11.793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Di Lena M, Travaglio E, Altomare DF. New strategies for colorectal cancer screening. World J Gastroenterol. 2013;19:1855–60. doi: 10.3748/wjg.v19.i12.1855. doi: 10.3748/wjg.v19.i12.1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ahlquist DA, Shuber AP. Stool screening for colorectal cancer: Evolution from occult blood to molecular markers. Clin Chim Acta. 2002;315:157–68. doi: 10.1016/s0009-8981(01)00712-4. doi: 10.1016/S0009-8981(01)00712-4. [DOI] [PubMed] [Google Scholar]
- 33.Haug U, Hundt S, Brenner H. Quantitative immunochemical fecal occult blood testing for colorectal adenoma detection: Evaluation in the target population of screening and comparison with qualitative tests. Am J Gastroenterol. 2010;105:682–90. doi: 10.1038/ajg.2009.668. doi: 10.1038/ajg.2009.668. [DOI] [PubMed] [Google Scholar]
- 34.Wu D, Zhou G, Jin P, Zhu J, Li S, Wu Q, et al. Detection of colorectal cancer using a simplified SEPT9 gene methylation assay is a reliable method for opportunistic screening. J Mol Diagn. 2016;18:535–45. doi: 10.1016/j.jmoldx.2016.02.005. doi: 10.1016/j.jmoldx.2016.02.005. [DOI] [PubMed] [Google Scholar]
- 35.Yu J, Feng Q, Wong SH, Zhang D, Liang QY, Qin Y, et al. Metagenomic analysis of faecal microbiome as a tool towards targeted non-invasive biomarkers for colorectal cancer. Gut. 2017;66:70–8. doi: 10.1136/gutjnl-2015-309800. doi: 10.1136/gutjnl-2015-309800. [DOI] [PubMed] [Google Scholar]
- 36.Chen W, Liu F, Ling Z, Tong X, Xiang C. Human intestinal lumen and mucosa-associated microbiota in patients with colorectal cancer. PLoS One. 2012;7:e39743. doi: 10.1371/journal.pone.0039743. doi: 10.1371/journal.pone.0039743. [DOI] [PMC free article] [PubMed] [Google Scholar]